@norsivaeb Tweets 
| 2022
| 2021
| 2020
| 2019
| 2018
|
Fri Dec 31 13:50:39 +0000 2021@MattWFoster @mjmaccoss @chrashwood @ucdmrt I wish them luck, as it is will allow these sequential remove-and-read ideas to work.
Fri Dec 31 13:04:24 +0000 2021C10orf88:p, belongs to an broadly held lineage of chordate genes 🔗
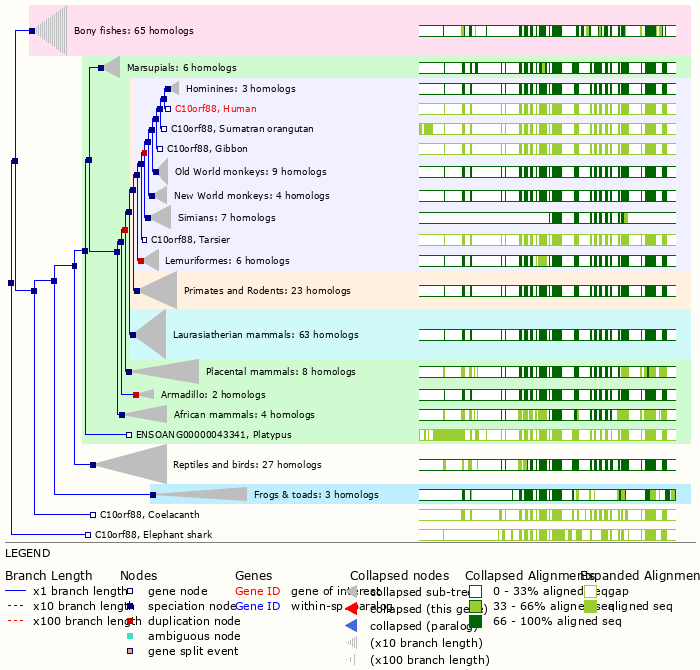
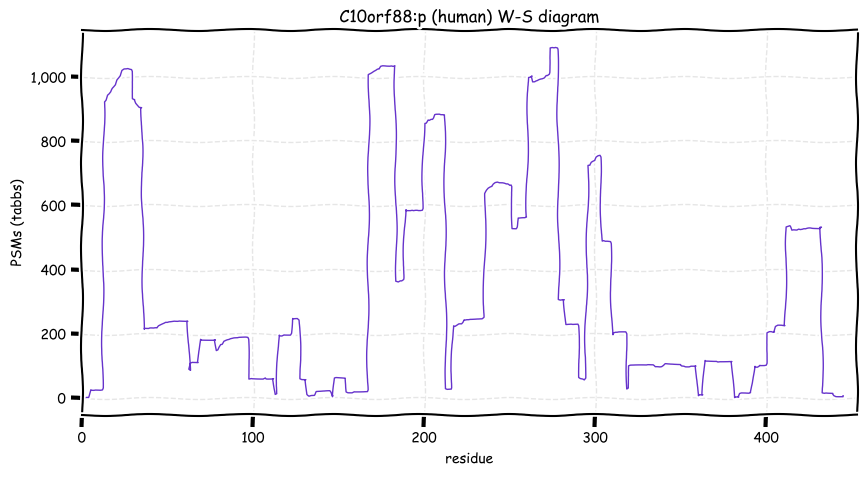
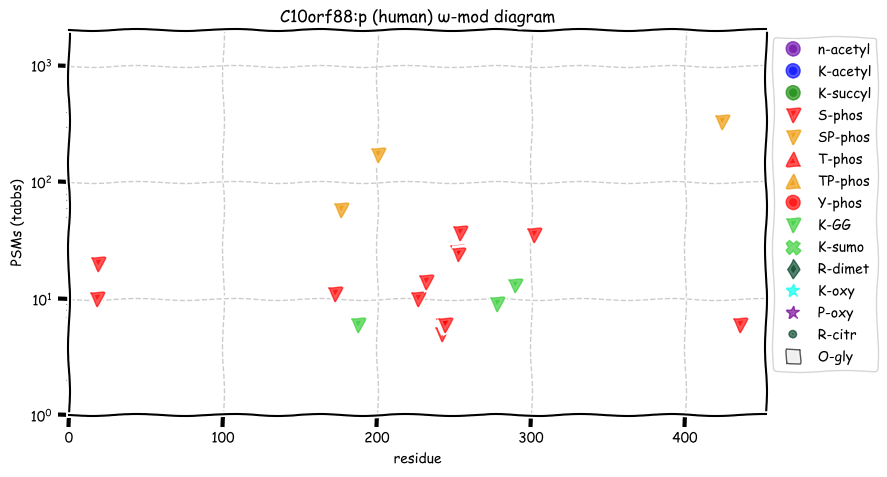
Fri Dec 31 13:04:23 +0000 2021C10orf88:p.A26S chr 10:g.122954103C>A rs751489489 (all tissue A:S 0.973:0.027) vaf=<1%, Δm=15.9949, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. AS in HEK-293T cells.
Fri Dec 31 13:04:23 +0000 2021C10orf88:p, θ(max) = 74. aka FLJ13490, Em:AC073585.5, PAAT. Found in MHC class 1 experiments. Found in a limited number of tissues tissues & cell lines: absent from most fluids (rarely in urine).
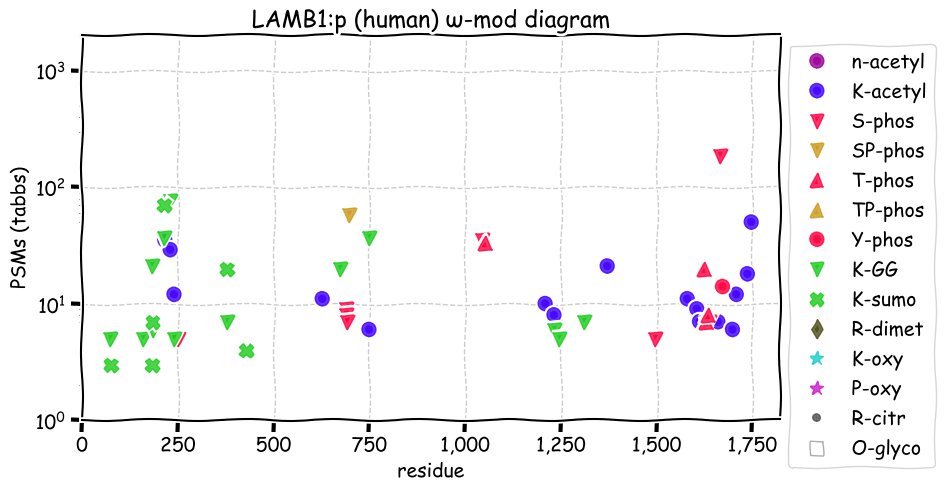
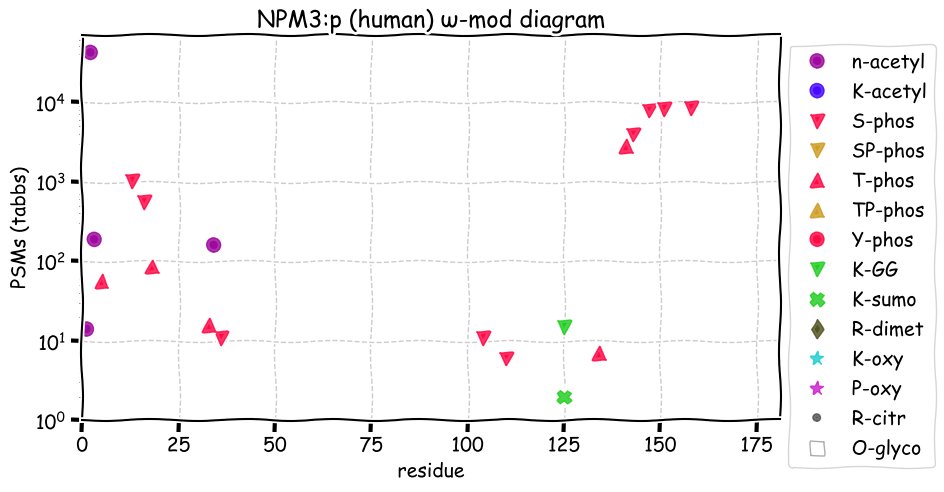
Fri Dec 31 13:04:23 +0000 2021>C10orf88:p, chromosome 10 open reading frame 88 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 0×K+acetyl; 0×K+succinyl; 3×K+GGyl; 0×K+SUMOyl; 16×S, 0×T, 0×Y+phosphoryl; SAAVs: A26S (<1%); mature form: (1-445) [2,454×, 9 kTa].#ᗕᕱᗒ 🔗
Fri Dec 31 02:42:02 +0000 2021@neely615 But often they leave a pseudogene behind: 🔗
Thu Dec 30 19:28:58 +0000 2021@chrashwood @mjmaccoss @ucdmrt To me, getting that step to work is the real game-changer tech development, not how you would readout the result.
Thu Dec 30 19:19:17 +0000 2021@chrashwood @mjmaccoss @ucdmrt I'm always stumped by the step where they simply lop off the N-terminal residue. I don't know of any way to do that with any sort of reliable stoichiometry (& only someone who has never done Edman would suggest "Edman"at this point)
Thu Dec 30 16:56:43 +0000 2021@mjmaccoss @byu_sam @MehtaManal @ProteomicsNews @slavov_n @ardongre @DemichevLab @decodegenetics @kstefans @Google @JMRothberg @new299 The most interesting aspect for me is the paper from Google/Alphabet. I was unaware that they had an in-house proteomics analytical methods development group.
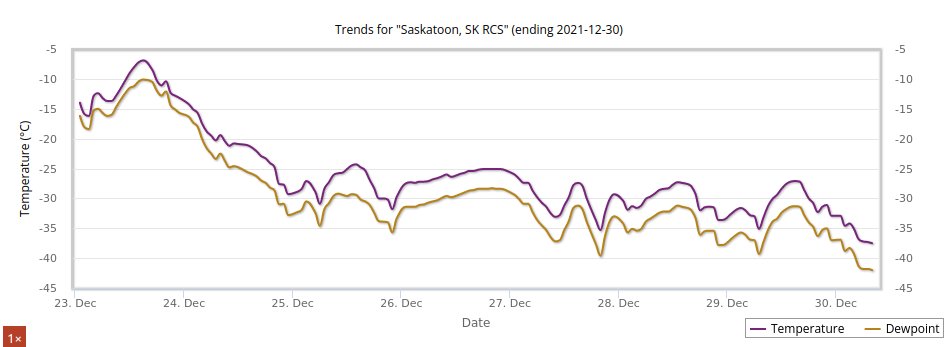
Thu Dec 30 15:02:53 +0000 2021Same thing, Saskatoon (where it is actually pretty cold) 🔗
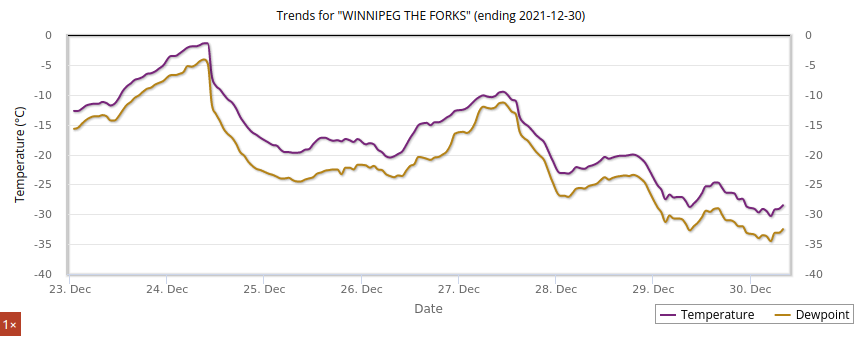
Thu Dec 30 14:55:08 +0000 2021Winter time in Winterpeg 🔗
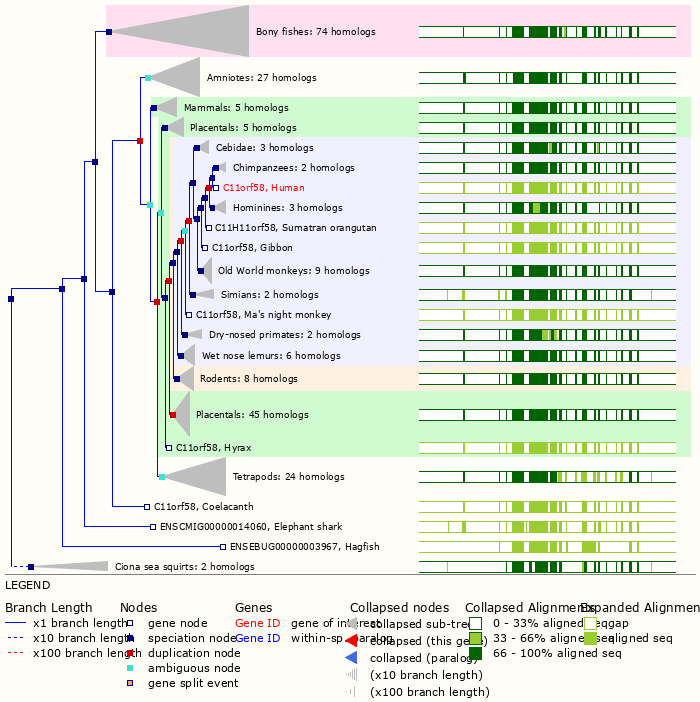
Thu Dec 30 13:06:41 +0000 2021C11orf58:p, belongs to an old lineage of chordate genes (including tunicates) 🔗
Thu Dec 30 13:06:40 +0000 2021C11orf58:p.E146K chr 11:g.16754988G>A rs56183939 (all tissue E:K 00.992:0.008) vaf=<1%, Δm=-0.9476, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Thu Dec 30 13:06:40 +0000 2021C11orf58:p, θ(max) = 69. aka SMAP. Found in MHC class 1 & 2 experiments. Found in many tissues & cell lines: absent from fluids. Domain (106-162) is highly enriched in acidic residues.
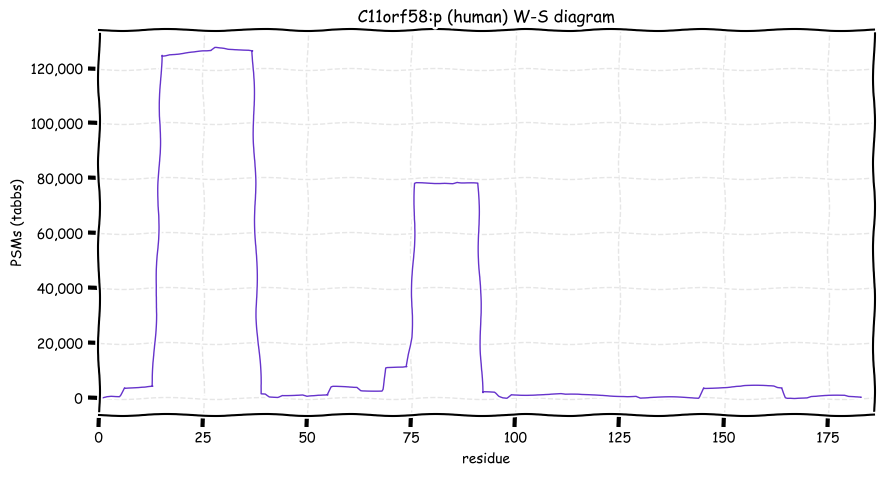
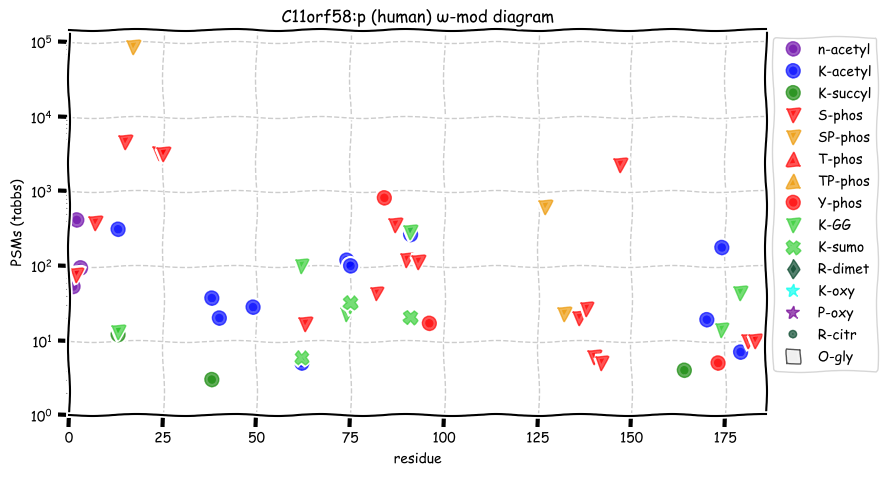
Thu Dec 30 13:06:40 +0000 2021>C11orf58:p, chromosome 11 open reading frame 58 (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 11×K+acetyl; 2×K+succinyl; 7×K+GGyl; 3×K+SUMOyl; 20×S, 0×T, 3×Y+phosphoryl; SAAVs: E146K (<1%); mature form: (2-183) [36,313×, 224 kTa]. #ᗕᕱᗒ 🔗
Wed Dec 29 18:53:37 +0000 2021Why do the Exploris results I look at all seem to have less parent ion mass accuracy than the Q-Exactive models I have grown so used to? Lots of spectra, but broader distributions.
Wed Dec 29 16:54:51 +0000 2021I spent about an hour this morning going through the proteomics repository site entries for particular protein: my main impression is that the field lacks any sort of shared vision as to what might be a useful way to represent the information it generates.
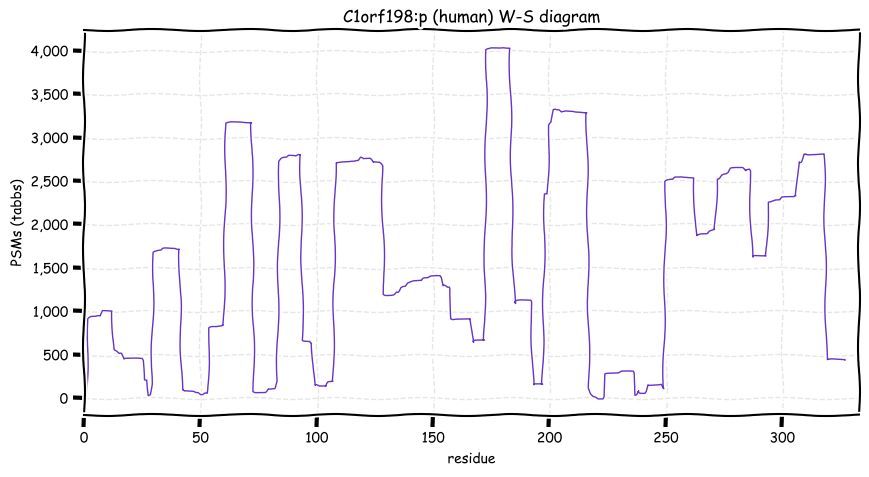
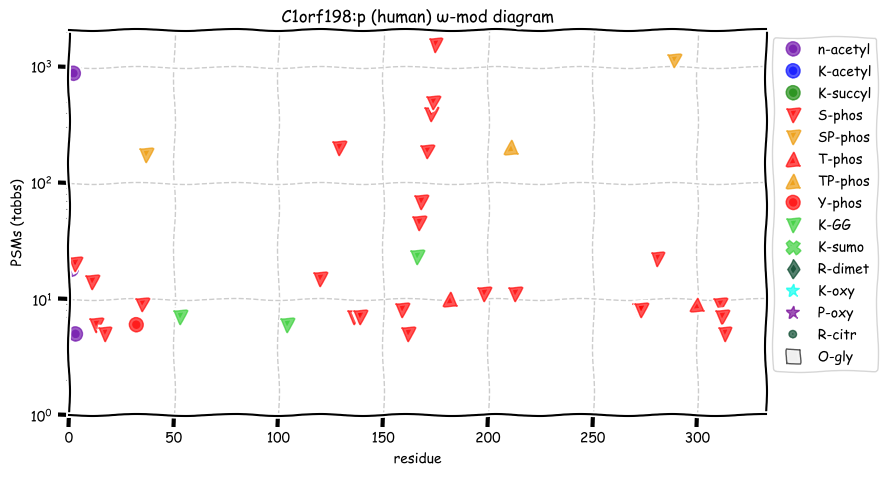
Wed Dec 29 14:08:33 +0000 2021C1orf198:p, I will admit I chuckled a bit when I noticed the coelacanth homolog 🔗
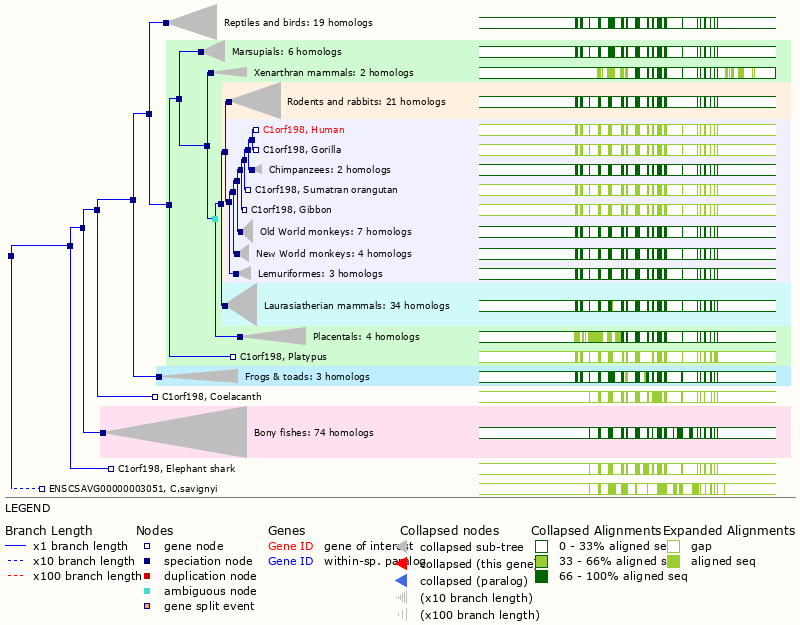
Wed Dec 29 13:50:40 +0000 2021C1orf198:p, I'm going to spend the next couple of weeks featuring frequently observed proteins that are still designated as "open reading frames".
Wed Dec 29 13:31:48 +0000 2021Today's polar vortex (winds at 10 hPa) 🔗
Wed Dec 29 12:57:49 +0000 2021C1orf198:p, in mice this gene is referred to a 2310022B05Rik:p.
Wed Dec 29 12:57:49 +0000 2021C1orf198:p.K306R chr 1:g.230843364T>>C, rs35115679 (all tissue K:R 0.995:0.005) vaf=0.3%, Δm=28.0061, VAF by population group: african 4%, american <1%, east asian <1%, european <1%, south asian <1%.
Wed Dec 29 12:57:48 +0000 2021C1orf198:p, θ(max) = 86. aka FLJ14525, MGC10710, FLJ16283, DKFZp667D152, FLJ38847. Found in MHC class 1 & 2 experiments. Found in many tissues & cell lines: absent from fluids.
Wed Dec 29 12:57:48 +0000 2021>C1orf198:p, chromosome 1 open reading frame 198 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 0×K+acetyl; 0×K+succinyl; 3×K+GGyl; 0×K+SUMOyl; 27×S, 3×T, 1×Y+phosphoryl; SAAVs: K306R (0.3%), A274S (9%); mature form: (2-327) [9,990×, 36 kTa]. #ᗕᕱᗒ 🔗
Tue Dec 28 16:25:26 +0000 2021There are no experimental structures for PLIN4 & the majority of the structure is colored to indicate that the features are in the lowest confidence range, but looking at it you almost feel as though you should try to explain things that are just attractive noise
Tue Dec 28 16:21:57 +0000 2021I was reminded of this yesterday by the AF2 model of perilipin 4 corresponding to Q96Q06 🔗
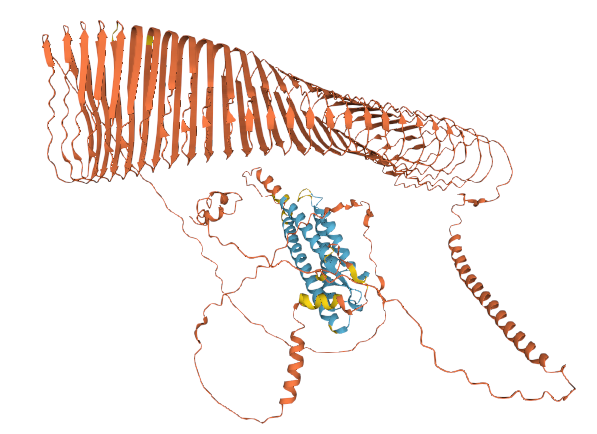
Tue Dec 28 15:45:00 +0000 2021The application of "Deep Learning" to science is interesting to me: it usually doesn't work very well but it often seems to create results that are much more attractive than those generated by experimental methods.
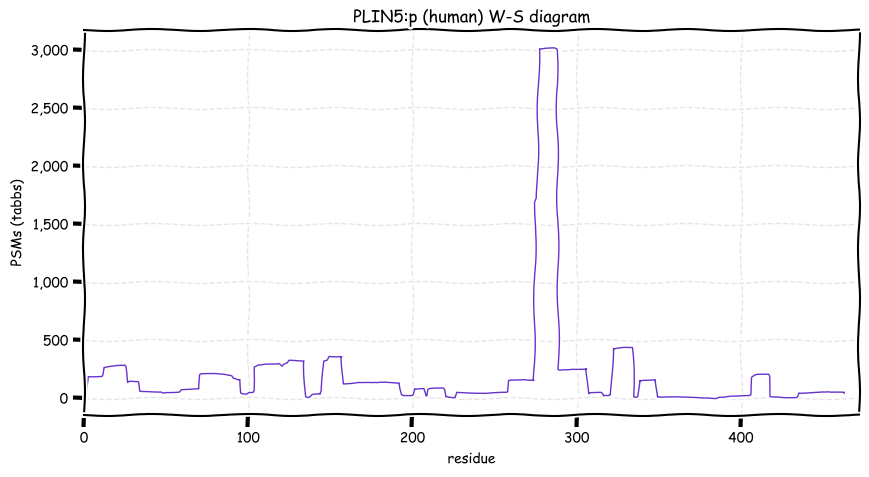
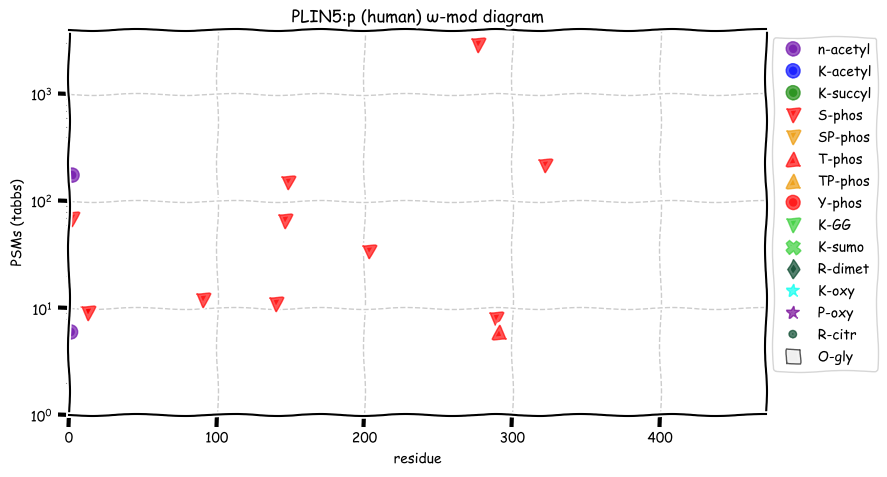
Tue Dec 28 13:16:30 +0000 2021PLIN5:p, 89% of phosphorylated PSMs observed are at RxxΦ acceptor sites.
Tue Dec 28 12:54:30 +0000 2021PLIN5:p.A6V. 19:g.4534058G>A, rs10407239 (all tissue A:V 0.972:0.028) vaf=3%, Δm=28.0313, VAF by population group: african 11%, american <1%, east asian <1%, european <1%, south asian <1%.
Tue Dec 28 12:54:29 +0000 2021PLIN5:p, θ(max) = 54. aka LSDP5, LSDA5, OXPAT, MLDP. Found in MHC class 1 ovary tissue experiments. Most prominent in breast, liver, muscle tissue, as well as MCF-7 cells, & more rarely in adipose tissue: absent from fluids.
Tue Dec 28 12:54:29 +0000 2021>PLIN5:p, perilipin 5 (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 0×K+acetyl; 0×K+succinyl; 0×K+GGyl; 0×K+SUMOyl; 11×S, 1×T, 0×Y+phosphoryl; SAAVs: A6V (3%); mature form: (2-463) [1,675×, 6.7 kTa]. #ᗕᕱᗒ 🔗
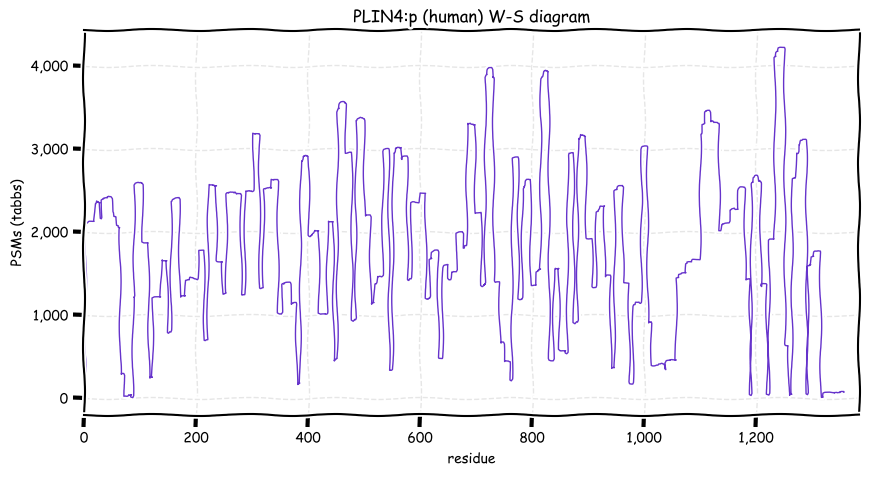
Mon Dec 27 13:07:43 +0000 2021PLIN4:p has an unusually large number of high frequency SAAVs, more like what would be expected of a pseudogene.
Mon Dec 27 13:03:41 +0000 2021PLIN4:p, the most recent version of this gene has a differnet N-terminal exon than the previous reference sequences, replacing
1 MQ... with
1 MSAPDEGRRDPPKPKGK...
which is more consistent with the data.
Mon Dec 27 13:03:41 +0000 2021PLIN4:p.V124A. 19:g.4513547A>G, rs4807597 (all tissue V:A 0.120:0.880) vaf=95%, Δm=-28.0313, VAF by population group: african 100%, american 97%, east asian 98%, european 96%, south asian 90%.
Mon Dec 27 13:03:41 +0000 2021PLIN4:p, θ(max) = 92. aka S3-12, KIAA1881. Found in MHC class 1 & 2 tissue experiments. Commonly observed in select tissues (particularly those containing adipocytes or hepatocytes) & cell lines: absent from fluids.
Mon Dec 27 13:03:40 +0000 2021PLIN4:p, SAAVs: V124A (95%), G329S (20%), M331V (89%), V333L (89%), T347I (12%), T421A (46%), M430V (11%), A596V (57%), S659G (88%), A668V (57%), M730T (93%), I734V (84%), K761N (85%), A784T (73%), C786L (27%), M802T (93%), A826E (89%), G852C (77%), A1124T (35%). 🔗
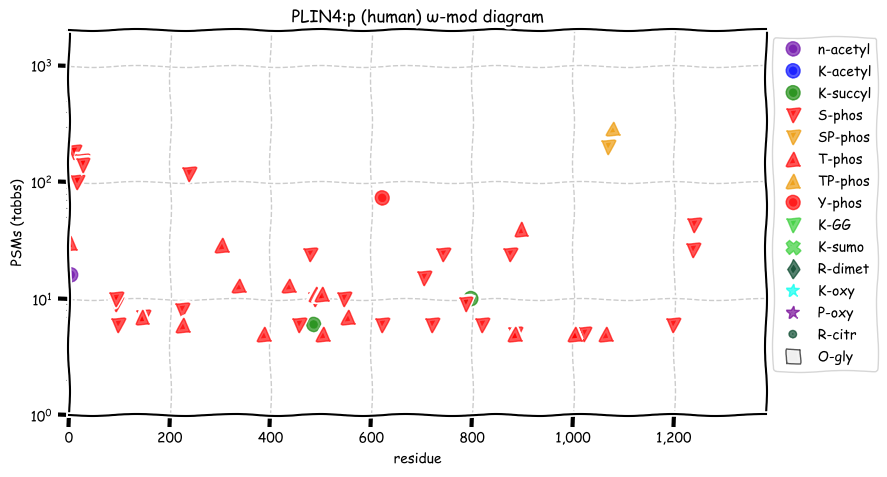
Mon Dec 27 13:03:38 +0000 2021>PLIN4:p, perilipin 4 (Homo sapiens) Large subunit; CTMs: T3+acetyl; PTMs: 0×K+acetyl; 2×K+succinyl; 0×K+GGyl; 0×K+SUMOyl; 29×S, 17×T, 1×Y+phosphoryl; mature form: (3-1157) [7,727×, 174 kTa]. #ᗕᕱᗒ
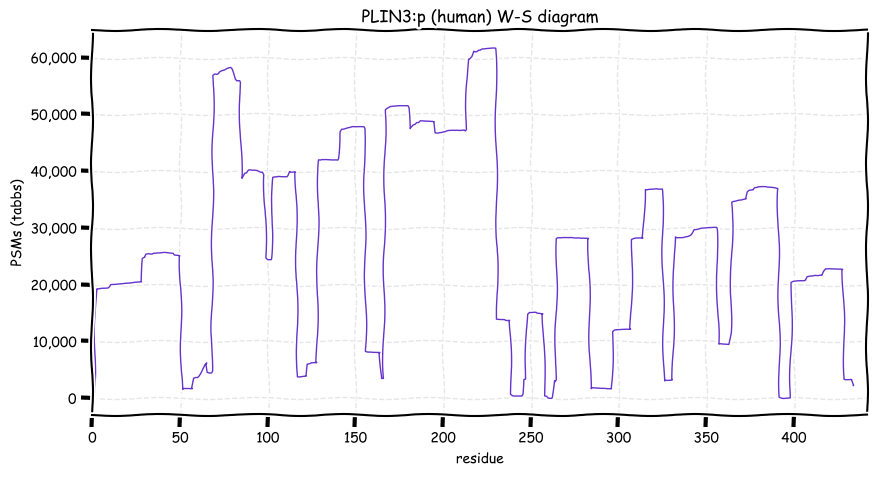
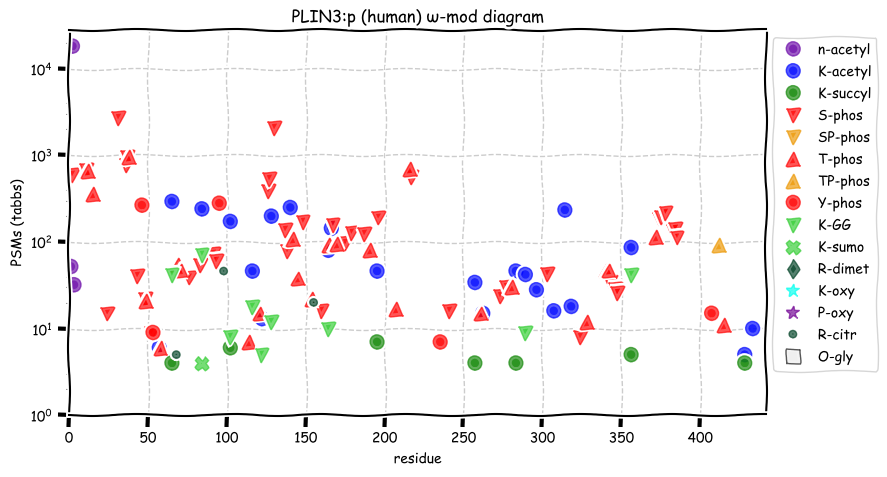
Sun Dec 26 13:15:41 +0000 2021PLIN3:p is the most frequently observed perilipin subunit:
PLIN1=5,075×,
PLIN2=12,913×,
PLIN3=62,528×,
PLIN4=7,744× &
PLIN5=1,675×
Sun Dec 26 13:15:40 +0000 2021PLIN3:p.V275A. 19:g.4847701A>G, rs9973235 (all tissue V:A 0.142:0.858) vaf=87%, Δm=-28.0313, VAF by population group: african 91%, american 93%, east asian 89%, european 88%, south asian 87%.
Sun Dec 26 13:15:40 +0000 2021PLIN3:p, θ(max) = 89. aka TIP47, PP17, M6PRBP1. Found in MHC class 1 & 2 experiments. Commonly observed in many tissues, cell lines & fluids.
Sun Dec 26 13:15:40 +0000 2021>PLIN3:p, perilipin 3 (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 22×K+acetyl; 7×K+succinyl; 9×K+GGyl; 1×K+SUMOyl; 40×S, 25×T, 5×Y+phosphoryl; SAAVs: F221L (87%), V275A (87%); mature form: (2-434) [62,508×, 690 kTa]. #ᗕᕱᗒ 🔗
Sun Dec 26 02:41:19 +0000 2021Not unusual, but still a substantial enough auroral zone tonight. 🔗
Sat Dec 25 15:42:49 +0000 2021PLIN2:p, perilipins are not lipid-associated membrane proteins: they are subunits that collectively form a membrane around lipid droplets. How that works in 3D is a matter of speculation at this point.
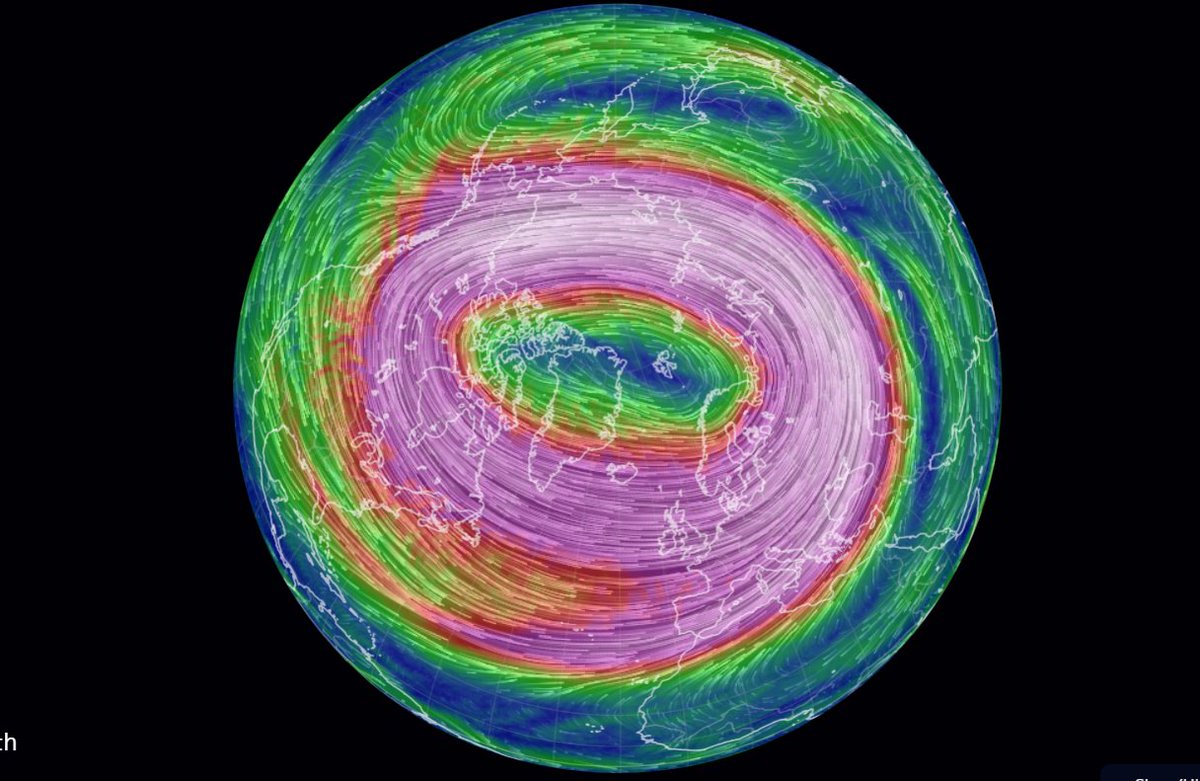
Sat Dec 25 14:29:34 +0000 2021The polar vortex (altitude 10 hPa) 🔗
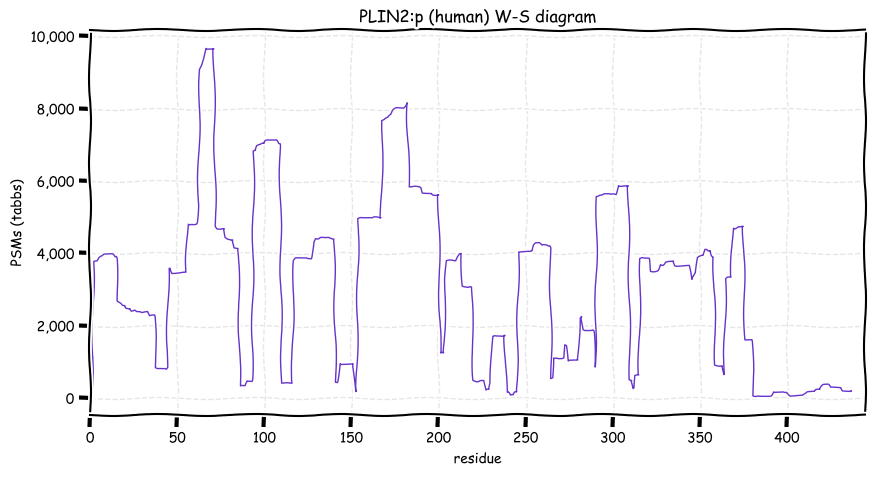
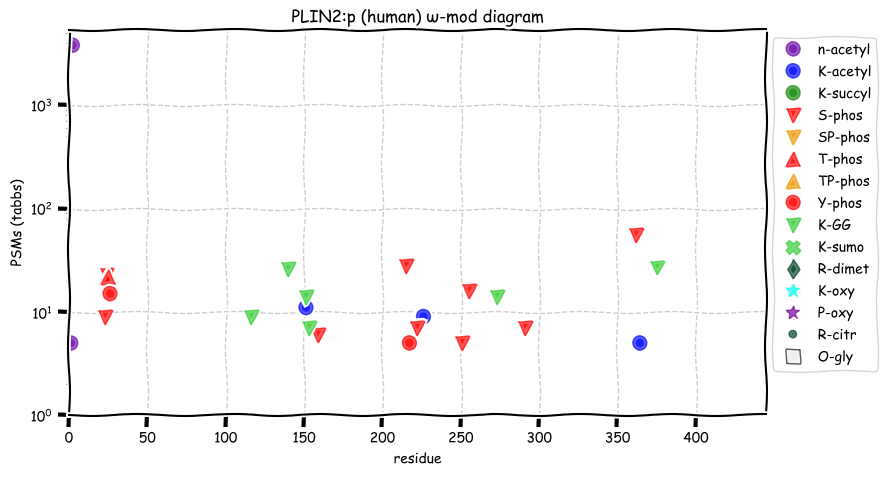
Sat Dec 25 13:38:28 +0000 2021PLIN2:p, rarely found in adipoctyes but common in hepatoctyes. The phosphorylation acceptor sites are much more rarely occupied than those in PLIN1:p and have different sequence motifs.
Sat Dec 25 13:38:28 +0000 2021PLIN2:p.N18S. 9:g.19126287T>C, rs202143072 (all tissue N:S 0.999:0.001) vaf=<1%, Δm=-27.0109, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sat Dec 25 13:38:28 +0000 2021PLIN2:p, θ(max) = 84. aka ADRP, ADFP. Found in MHC class 1 & 2 tissue experiments. Observed in many tissues & cell lines: abundant in milk but absent from other fluids.
Sat Dec 25 13:38:28 +0000 2021>PLIN2:p, perilipin 2 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 3×K+acetyl; 0×K+succinyl; 6×K+GGyl; 0×K+SUMOyl; 9×S, 1×T, 2×Y+phosphoryl; SAAVs: N18S (<%), S251P (2%); mature form: (2-437) [12,913×, 94 kTa]. #ᗕᕱᗒ 🔗
Fri Dec 24 13:28:22 +0000 2021How will talking to people who aren't involved in open source development help "improve" open source development? 🔗
Fri Dec 24 13:09:46 +0000 2021While it is supposed to be -20 °C for Christmas Day, but it is raining here right now.
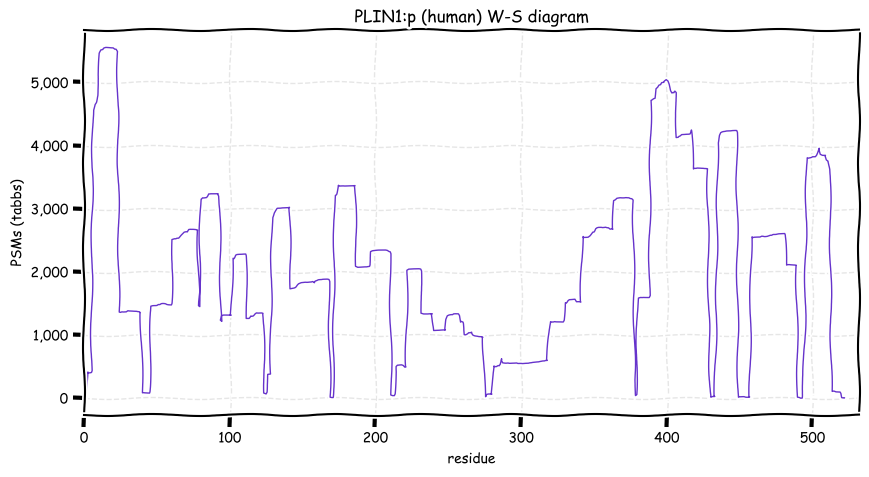
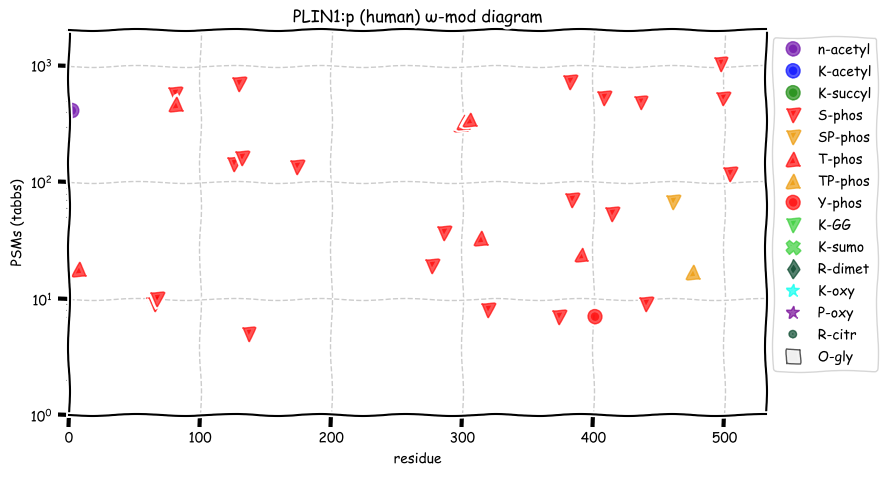
Fri Dec 24 12:53:48 +0000 2021PLIN1:p, 1 of 5 human perilipins that form a surface layer on intracellar lipid storage drops. Phosphorylation sites are dominsted by RxxS & TxxE motifs. Phophorylation is believed to increase the accessibility of the lipid droplet surface to metabolic enzymes.
Fri Dec 24 12:53:48 +0000 2021PLIN1:p.P194A. 15:g.89669998G>C, rs6496589 (all tissue P:A 0.067:0.933) vaf=93%, Δm=-26.0156, VAF by population group: african 98%, american 74%, east asian 86%, european 99%, south asian 87%.
Fri Dec 24 12:53:48 +0000 2021PLIN1:p, θ(max) = 93. aka PLIN. Found in MHC class 1 & 2 tissue experiments. Observed in adipose tissue (adipocytes) & liver (hepatocytes): absent from fluids & most cell lines.
Fri Dec 24 12:53:47 +0000 2021>PLIN1:p, perilipin 1 (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 0×K+acetyl; 0×K+succinyl; 0×K+GGyl; 0×K+SUMOyl; 23×S, 8×T, 1×Y+phosphoryl; SAAVs: P194A (93%), R231L (1%), S348L (1%), A380V (1%); mature form: (2-522) [5,075×, 70 kTa]. #ᗕᕱᗒ 🔗
Thu Dec 23 16:01:08 +0000 2021I say useless, mainly because the instructors repeatedly commented on how the methods being taught were too complicated to be used in practice, especially because if you skipped any step (or did one of them out of order) the whole process failed.
Thu Dec 23 15:43:21 +0000 2021They seemed to pass the tests I got from mask-training (part of a painful & largely useless 2 day course in infection control that I had to take when I was working at an institute attached to a hospital). They actually keep my glasses from fogging.
Thu Dec 23 15:38:48 +0000 2021Ha! I've been using the Dollarama KN-95's for over a year & everyone has told me they were probably just cheap knockoffs 🔗
Thu Dec 23 13:08:18 +0000 2021ATF7:p, family observation frequencies:
ATF1=6228×,
ATF2=8439×,
ATF3=830×,
ATF4=851×,
ATF5=63×,
ATF6=3935×, &
ATF7=8838×.
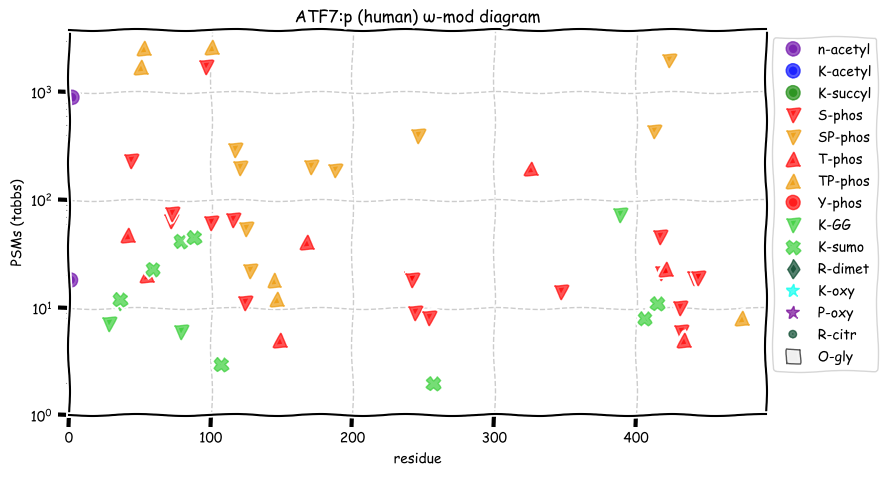
Thu Dec 23 13:06:53 +0000 2021ATF7:p.S97F. 12:g.53537527G>A, rs377338085 (all tissue S:F 1.000:0.000) vaf=<1%, Δm=60.0364, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Thu Dec 23 13:06:53 +0000 2021ATF7:p, θ(max) = 50. aka ATFA. Found in MHC class 1 experiments. Observed in many tissues and cell lines: absent from fluids. The gene symbol ATF7 may be confused with ATF5 in some older literature.
Thu Dec 23 13:06:53 +0000 2021>ATF7:p, activating transcription factor 7 (Homo sapiens) Small subunit; CTMs: G2, M11+acetyl; PTMs: 0×K+acetyl; 0×K+succinyl; 4×K+GGyl; 8×K+SUMOyl; 27×S, 13×T, 0×Y+phosphoryl; SAAVs: S97F (<1%); mature form: (2-483) [8,836×, 28 kTa]. #ᗕᕱᗒ 🔗
Wed Dec 22 17:26:23 +0000 2021@lukas_k ELITE-RSLC011436 to ELITE-RSLC011684 are trypsin
ELITE-RSLC012714 to ELITE-RSLC013143 are asp-N
ELITE-RSLC013149 to ELITE-RSLC012689 are chymotrypsin
ELITE-RSLC013152 to ELITE-RSLC013424 are glu-c
Wed Dec 22 17:05:04 +0000 2021@lukas_k Average is difficult to find in proteomics: almost every data set tries out some novel protocol. But, if you want:
MHC 1/2 peptides: PXD028633
Phosphopeptides: PXD027198
Lys-C (+8 SILAC): MSV000086426
Vanilla trypsin: PXD027258
Trypsin +TMT10: PXD023979
Wed Dec 22 16:55:52 +0000 2021@lukas_k If you don't mind low res fragment masses, PXD016924 has quite a bit going for it wrt testing. Good, consistent chromatography & MS/MS applied to multiple proteolytic enzymes.
Wed Dec 22 16:40:41 +0000 2021@lukas_k Any particular type of instrument, mass resolutions, sample prep and/or quant reagent required?
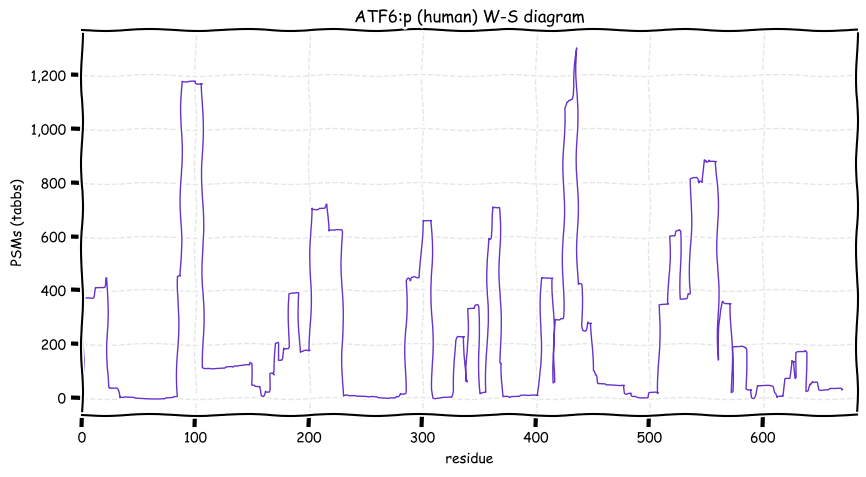
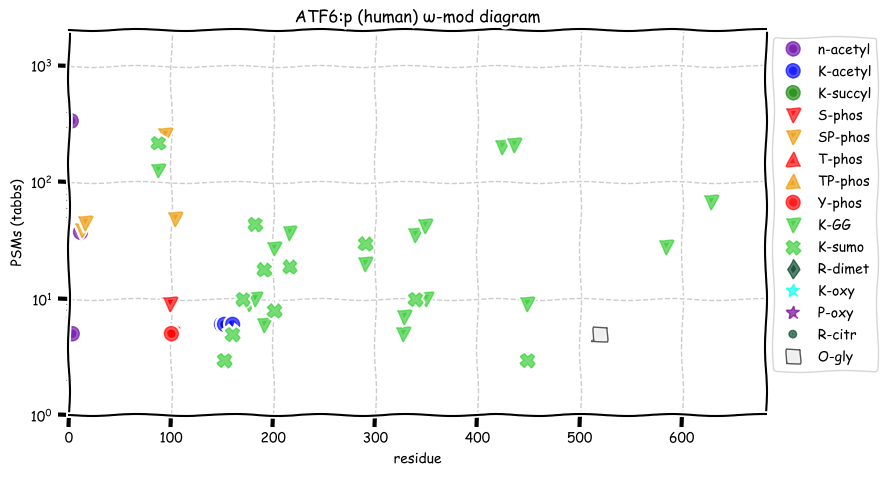
Wed Dec 22 13:49:29 +0000 2021ATF6:p, has a rather large number of ubiquitin (& ubiquitin-like) acceptor sites that are not acetylation/succinylation co-acceptors.
Wed Dec 22 13:19:36 +0000 2021ATF6:p, translation initiation at M11 is limited to human, among model species. It is unclear how an ER membrane protein can function as a transcription factor.
Wed Dec 22 13:13:34 +0000 2021ATF6:p, a transmembrane domain (378-398) is predicted by some algorithms with (2,11-377) cytoplasmic & (399-670) ER lumen. This is supported by the presence of N-linked glycosylation & the observed association of the protein with the ER membrane.
Wed Dec 22 13:13:34 +0000 2021ATF6:p.F20S. chr 1:g.161766419T>C, rs35284289 (all tissue F:S 0.994:0.006) vaf=1%, Δm=-60.0364, VAF by population group: african <1%, american <1%, east asian <1%, european 1%, south asian 1%.
Wed Dec 22 13:13:34 +0000 2021ATF6:p, θ(max) = 40. aka ATF6A. Found in MHC class 1 & 2 experiments. Most frequently observed in blood plasma, leukocytes & lymphocytes.
Wed Dec 22 13:13:34 +0000 2021>ATF6:p, activating transcription factor 6 (Homo sapiens) Midsized subunit; CTMs: G2, M11+acetyl; N472+glycosyl; PTMs: 3×K+acetyl; 0×K+succinyl; 17×K+GGyl; 11×K+SUMOyl; 8×S, 0×T, 1×Y+phosphoryl; SAAVs: F20S (1%); mature form: (2,11-670) [3,933×, 11 kTa]. #ᗕᕱᗒ 🔗
Wed Dec 22 02:30:00 +0000 2021@lenjf 🇨🇦 +1
Tue Dec 21 20:01:33 +0000 2021@DRAWheatcraft My recommendation would be Peter Lobel or someone from his group.
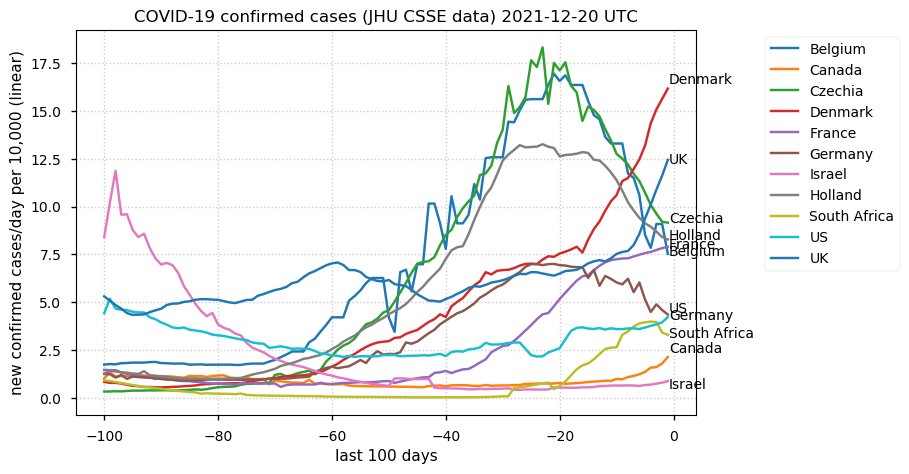
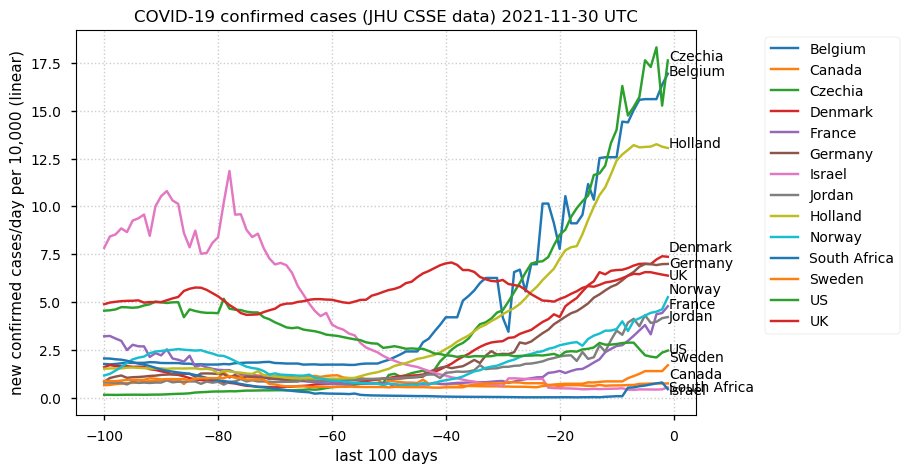
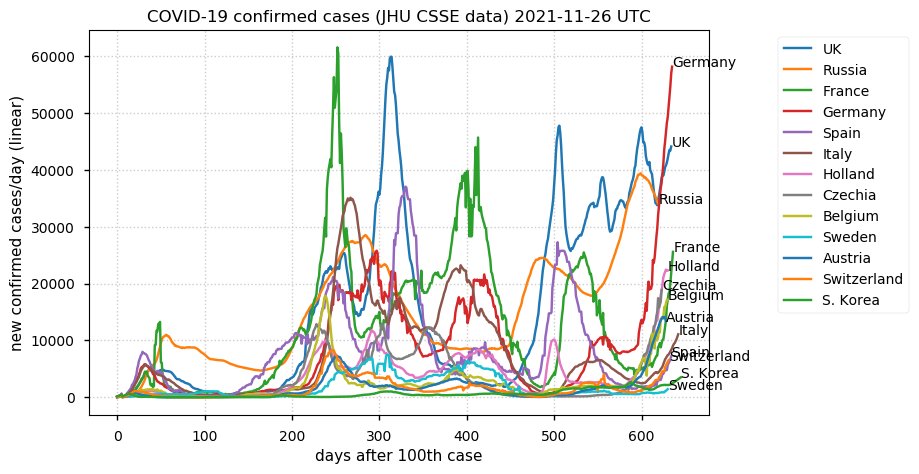
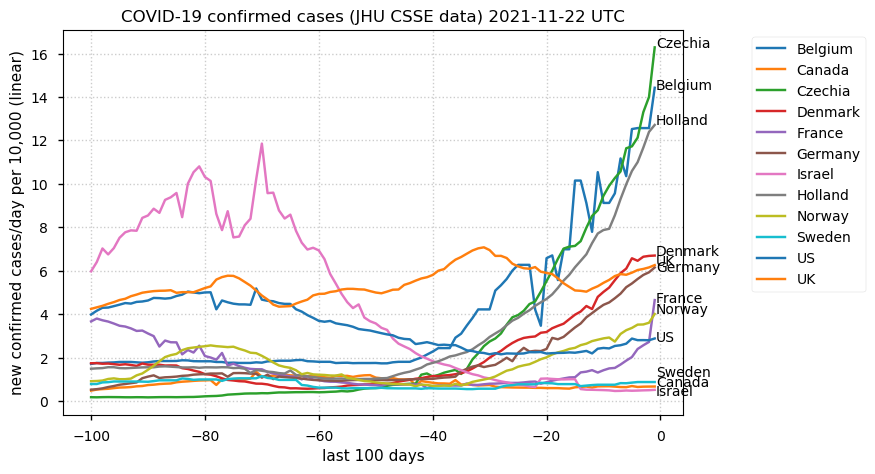
Tue Dec 21 16:09:36 +0000 2021Belgium, Holland, Germany, Czechia & South Africa are all coming down from their recent case number peaks. Canada is trending up but still modest compared to the recent northern European outbreak. 🔗
Tue Dec 21 15:26:25 +0000 2021@pwilmarth In Canada it tends to be a never ending stalemate between Faculty of Science (championing Comp Sci) & the Faculty of Medicine (championing Genetics) as to which would be the true home of bioinformatics.
Tue Dec 21 14:24:49 +0000 2021I see ads for candidates with Ph.D's in Bioinformatics. How many unis actually offer programmes that would satisfy this type of requirement? I don't think any Canadian U15 shop could credibly claim to support such a qualification.
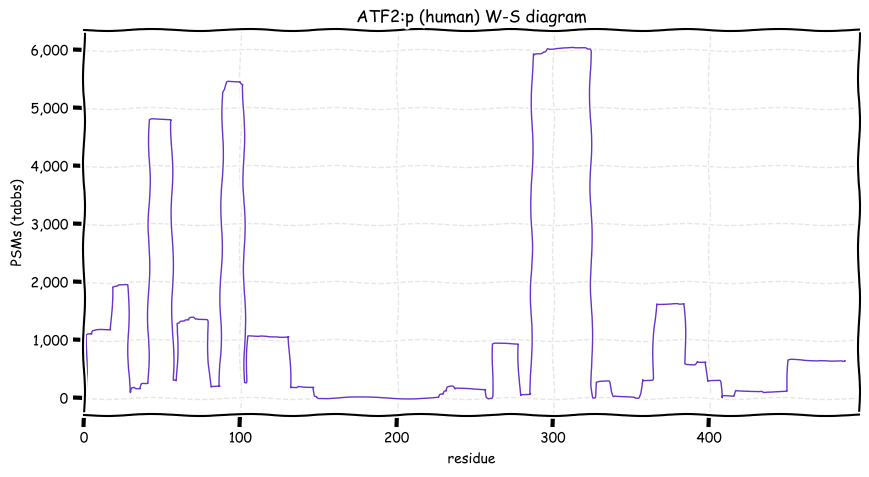
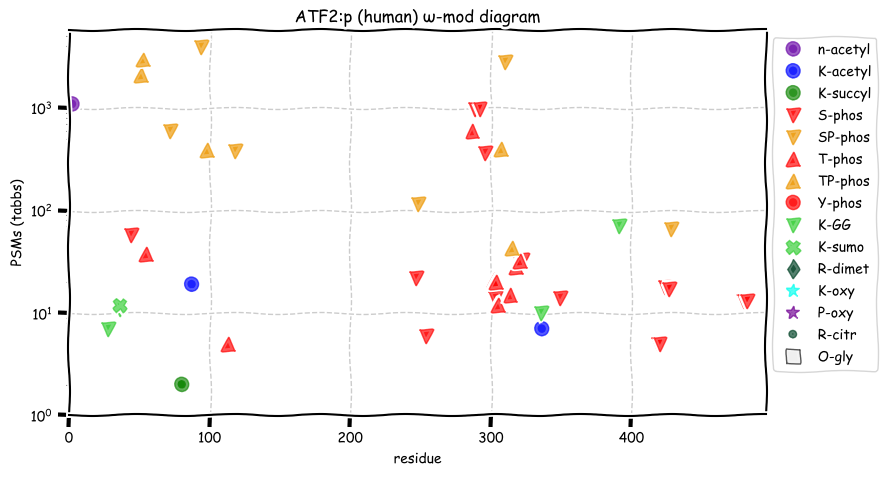
Tue Dec 21 13:33:29 +0000 2021ATF2:p is annotated as a leucine-zipper DNA-binding transcription factor, but it is also annotated as a histone specific deacetylase and as forming a heterodimer with JUN:p.
Tue Dec 21 13:28:11 +0000 2021ATF2:p, the "canonical" splice variant (ATF2-201, CCDS2262) is not consistent with most observations: the one shown here (ATF2-212, CCDS58738) is a much better fit to the data.
Tue Dec 21 13:05:41 +0000 2021ATF2:p.G323S. chr 2:175093225C>T, rs2230674 (all tissue G:S 0.999:0.001) vaf=<1%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Tue Dec 21 13:05:41 +0000 2021ATF2:p, θ(max) = 56. aka TREB7, CRE-BP1, HB16, CREB2. Found in MHC class 1 experiments. Observed in tissues & cell lines: absent from fluids. Has a very long 1° tryptic peptide (132-257) that leads to a gap in the information about this sequence.
Tue Dec 21 13:05:41 +0000 2021>ATF2:p, activating transcription factor 2 (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 2×K+acetyl; 2×K+succinyl; 4×K+GGyl; 1×K+SUMOyl; 23×S, 14×T, 0×Y+phosphoryl; SAAVs: G323S (<1%); mature form: (2-487) [8,614×, 28 kTa]. #ᗕᕱᗒ 🔗
Mon Dec 20 16:46:49 +0000 2021On reflection, you can annotate anything, too: making the annotation accurate & useful is the tricky part.
Mon Dec 20 15:24:21 +0000 2021Inspection of some data taken from Tuber melanosporum (black truffle) shows that while you may be able to sequence the genome of pretty much anything, annotating it is another thing entirely.
Mon Dec 20 14:41:23 +0000 2021@AlexUsherHESA Most Canadian unis require Asst. Profs to go up for tenure at 5 years. The increase over the period is probably an indication of how complex the post departmental approval process has become, even though it rarely disagrees with the Dept. Head's assessment.
Mon Dec 20 14:35:49 +0000 2021@MagnusPalmblad @pvanhouts @Imagine_Imaging @erwinboutsma Although it still bugs me once a year to remember how badly Franz and Michael got screwed by the "Prize".
Mon Dec 20 14:12:35 +0000 2021@MagnusPalmblad @pvanhouts @Imagine_Imaging @erwinboutsma It is a nice illustration. I still have some fondness for the method ...
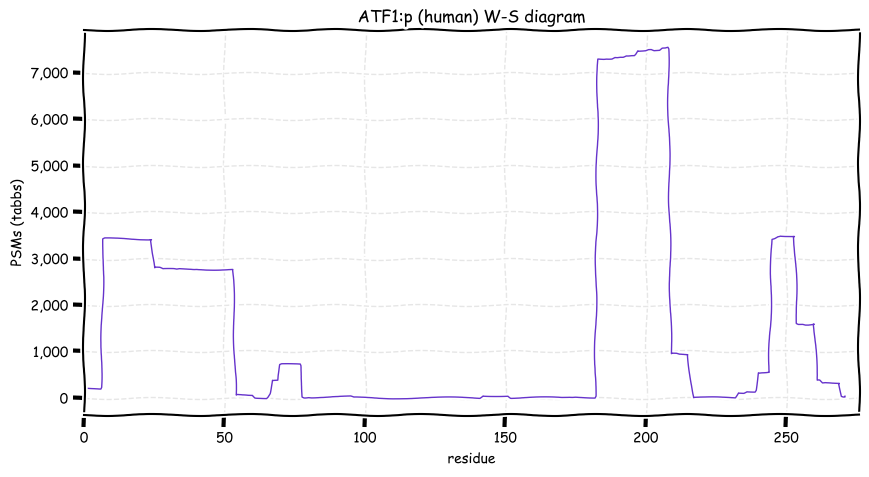
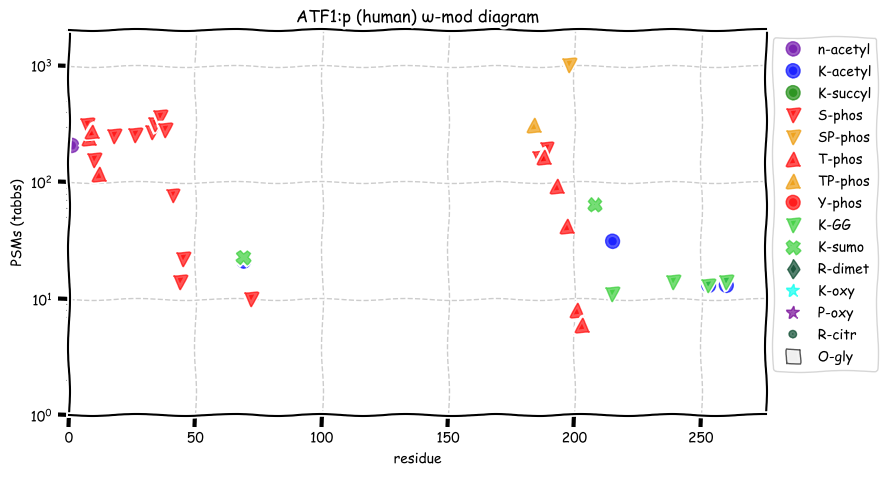
Mon Dec 20 13:37:44 +0000 2021ATF1:p, much of the variability in the observation of specific peptide domains is caused by an over-reliance on trypsin digestion and a narrow range of chromatographic conditions in proteomics studies, i.e., tradition.
Mon Dec 20 13:00:08 +0000 2021ATF1:pP191A. chr 12:g.50814339C>G, rs2230674 (all tissue P:A 0.995:0.005) vaf=0.2%, Δm=-83.0701, VAF by population group: african <1%, american 2%, east asian <1%, european 3%, south asian 1%.
Mon Dec 20 13:00:08 +0000 2021ATF1:p, θ(max) = 44. aka TREB36. Found in MHC class 1 experiments. Observed in tissues & cell lines: absent from fluids. Has a very long 1° tryptic peptide (80-182) that leads to a gap in the experimental information about this sequence.
Mon Dec 20 13:00:08 +0000 2021>ATF1:p, activating transcription factor 1 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 4×K+acetyl; 4×K+GGyl; 2×K+SUMOyl; 15×S, 9×T, 0×Y+phosphoryl; SAAVs: P191A (<1%); mature form: (1-271) [6,220×, 18 kTa].#ᗕᕱᗒ 🔗
Sun Dec 19 16:51:15 +0000 2021@slashdot IMHO, the worst conceivable choice, in every way.
Sun Dec 19 15:15:56 +0000 2021@macro_momo @oleg8r @jeffersle I have no problem with bureaucracy: it is just that reading a syllabus is more valuable to an acting assistant associate dean than it is to students taking the course.
Sun Dec 19 13:54:12 +0000 2021@oleg8r @jeffersle I have taken many university courses and written many syllabi, but I've never read one voluntarily. They are an odd ball admin document of no clear academic lineage or purpose beyond the bureaucratic.
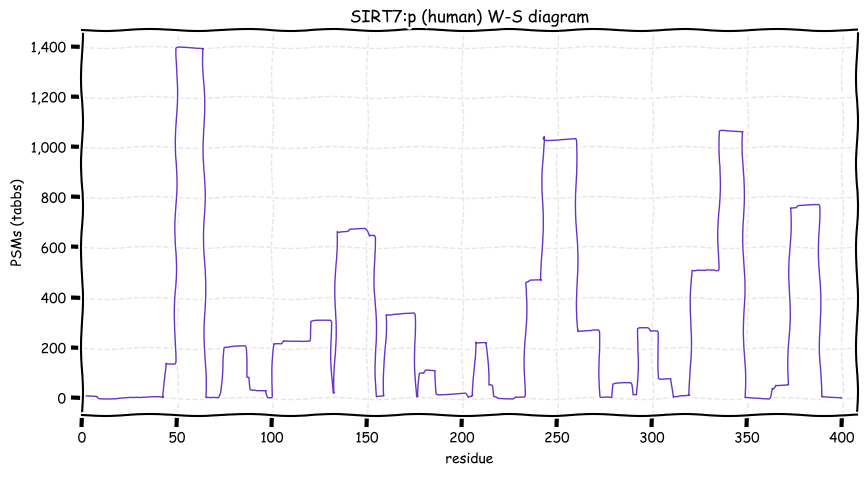
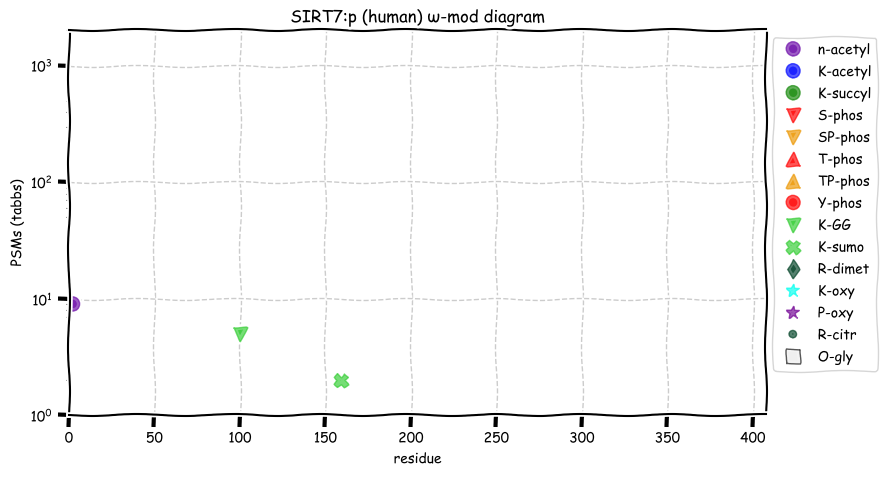
Sun Dec 19 13:09:52 +0000 2021SIRT7:p.W336C chr 17:g.81912611C>G, rs199923657 (all tissue W:C 0.992:0.008) vaf=<1%, Δm=-83.0701, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sun Dec 19 13:09:51 +0000 2021SIRT7:p, θ(max) = 61. aka SIR2L7. Found in MHC class 1 experiments. Observed in tissues & cell lines: absent from fluids.
Sun Dec 19 13:09:51 +0000 2021>SIRT7:p, sirtuin 7 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: K100+GGyl; K159+SUMOyl; SAAVs: W336C (<1%); mature form: (2-400) [1,560×, 8 kTa]. #ᗕᕱᗒ 🔗
Sat Dec 18 16:10:44 +0000 2021Is there some social norm that prevents people doing pull down experiments from checking to be sure the bait protein label matches with the protein observed in a data file?
Sat Dec 18 14:17:23 +0000 2021@dtabb73 @lgatt0 @byu_sam It is, however, the motto of IT organization everywhere.
Sat Dec 18 13:49:43 +0000 2021@dtabb73 @lgatt0 @byu_sam In the end, NCI has a tendency to try to generate a caBIG-like version of things, undeterred by a lack of previous successes.
Sat Dec 18 13:32:03 +0000 2021@lgatt0 @dtabb73 @byu_sam I find that site nearly impossible to navigate. The developers seem to have become so entranced by the elegance of their design that the site spends a lot of effort trying to teach dumb users how to conform to the designers' idea of what they should want.
Sat Dec 18 13:15:28 +0000 2021Seasonal weather (°C) across the Northern Great Plains 🔗
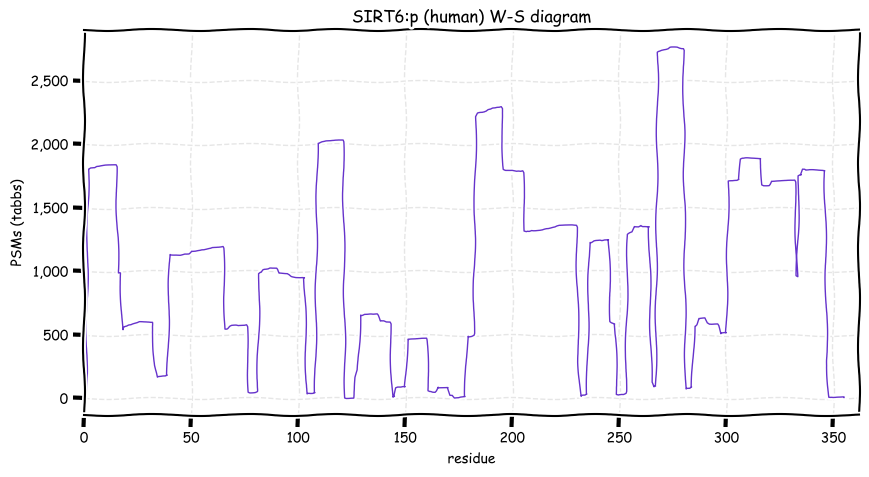
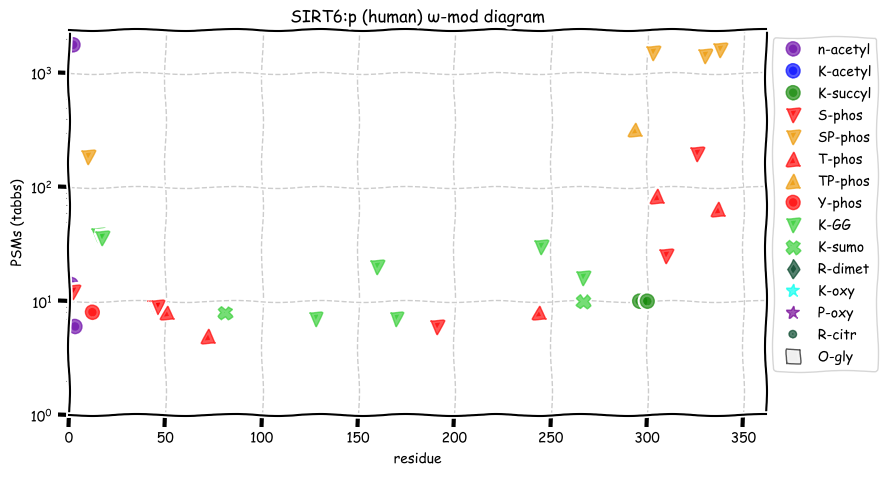
Sat Dec 18 12:46:57 +0000 2021SIRT6:p has a C-terminal phosphodomain (294-338) with several high occupancy ΦP-type sites. This domain that begins just C-terminal to the core catalytic domain (35-274).
Sat Dec 18 12:46:57 +0000 2021SIRT6:p.S46N chr 19:g.4180839C>T, rs352493 (all tissue S:N 0.894:0.106) vaf=88%, Δm=27.0109, VAF by population group: african 94%, american 78%, east asian 71%, european 93%, south asian 79%.
Sat Dec 18 12:46:57 +0000 2021SIRT6:p, θ(max) = 61. aka SIR2L6. Found in MHC class 1 experiments. Observed in tissues & cell lines: absent from fluids.
Sat Dec 18 12:46:56 +0000 2021>SIRT6:p, sirtuin 6 (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 0×K+acetyl; 7×K+GGyl; 2×K+SUMOyl; 2×K+succinyl; 11×S, 6×T, 1×Y+phosphoryl; SAAVs: S46N (88%); mature form: (2-355) [5,911×, 24 kTa]. #ᗕᕱᗒ 🔗
Fri Dec 17 13:13:24 +0000 2021@astacus @neely615 @UCDProteomics Editors used to police that sort of thing.
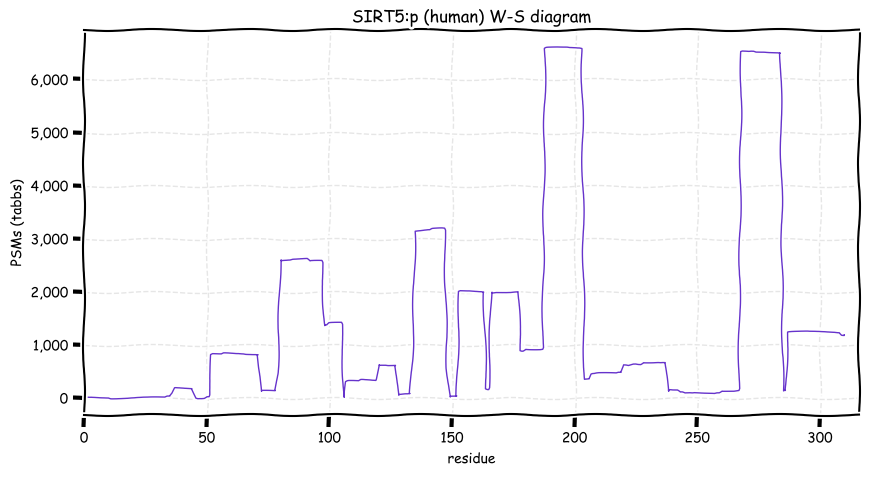
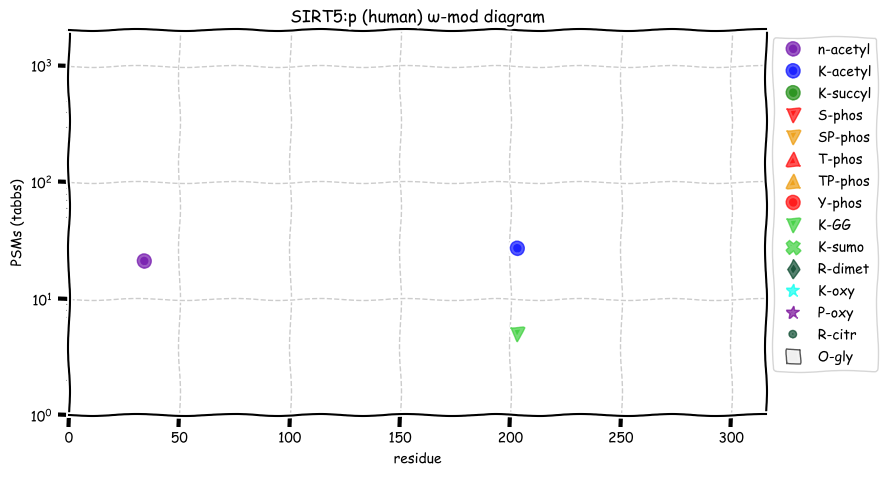
Fri Dec 17 13:03:12 +0000 2021SIRT5:p, for non-chemists:
HO₂C-R-CO₂H
R = CH₂ is malonic acid
R = (CH₂)₂ is succinic acid
R = (CH₂)₃ is glutaric acid
Fri Dec 17 12:46:03 +0000 2021SIRT5:p has a mitochondrial form―translation initiation M1, (1-36) removed mitochondrial transit peptide―and a cytoplasmic form―translation initiation M33, M33 removed and A34 N-acetylated. M. musculus & S. scrofa use the same 2-for-1 strategy.
Fri Dec 17 12:46:03 +0000 2021SIRT5:p.E305G chr 6:g.13611846A>G, rs3416262 (all tissue E:G 0.999:0.001) vaf=7%, Δm=-72.0211, VAF by population group: african 1%, american 19%, east asian <1%, european 5%, south asian 6%.
Fri Dec 17 12:46:03 +0000 2021SIRT5:p, θ(max) = 72. aka SIR2L5. Very rare in MHC class 1 or 2 experiments. Observed in tissues & cell lines: absent from fluids except urine extracellular vesicles. Responsible for the removal of malonyl, succinyl and glutaryl groups on lysine.
Fri Dec 17 12:46:03 +0000 2021>SIRT5:p, sirtuin 5 (Homo sapiens) Small subunit; CTMs: A34+acetyl; PTMs: K203+GGyl; K203+acetyl; SAAVs: E305G (7%); mature form: (34,37-310) [9,767×, 30 kTa]. #ᗕᕱᗒ 🔗
Fri Dec 17 12:27:19 +0000 2021@slavov_n Neither a good article or a persuasive argument.
Thu Dec 16 23:52:43 +0000 2021@ucdmrt I will leave that determination to wiser beings.
Thu Dec 16 22:21:57 +0000 2021The data does have some pretty extreme N/Q deamidation ratios though, clocking in at about 30/1.
Thu Dec 16 18:17:17 +0000 2021I realize this is only of interest to Canadians, but this mandate letter for Innovation, Science and Industry is so loopy it is hard to criticize: it is simply a bunch of buzz words & busy work
🔗
Thu Dec 16 17:25:24 +0000 2021Interesting thread: a premise taken seriously. 🔗
Thu Dec 16 17:11:35 +0000 2021Although I am not a fan of spiked-in SILAC-labelled mixtures for quant. For oocytes, it has a similar problem to "histone-ruler" type normalizations.
Thu Dec 16 16:48:45 +0000 2021The HGNC changed a bunch of gene symbols to stop spreadsheet problems associated with their original names. But, isn't changing histone names like "HIST1H1A" to "H1-1" just looking for spreadsheet-related trouble?
Thu Dec 16 16:01:49 +0000 2021PXD021331: while I always like oocyte data sets, the associated manuscript has a title any proteome-geek will love.
🔗
Thu Dec 16 15:39:40 +0000 2021Super Typhoon Odette (Rai) currently over the Philippines 🔗
Thu Dec 16 13:11:55 +0000 2021SIRT4:p has a different position in the frequency distribution for Mus musculus, where SIRT6:p is the least frequently observed:
SIRT2: 10,745×;
SIRT5: 4,947×;
SIRT1: 2,762×;
SIRT3: 2,470×;
SIRT4: 995×;
SIRT7: 732×; &
SIRT6: 556×.
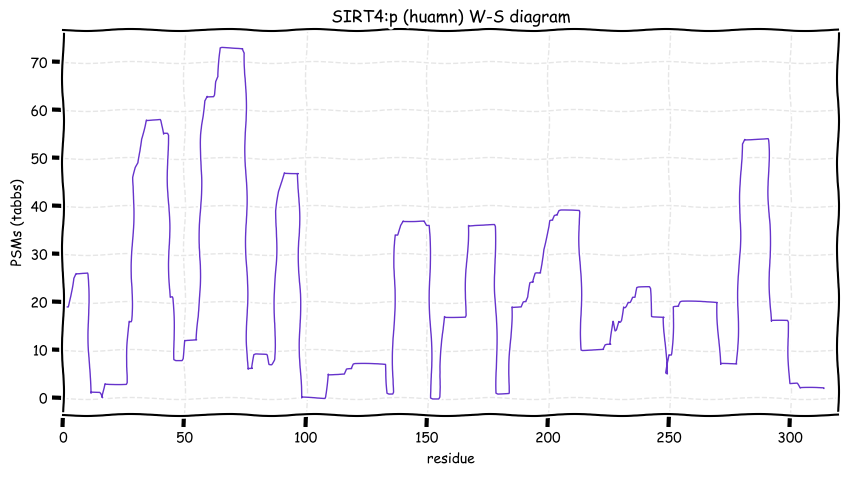
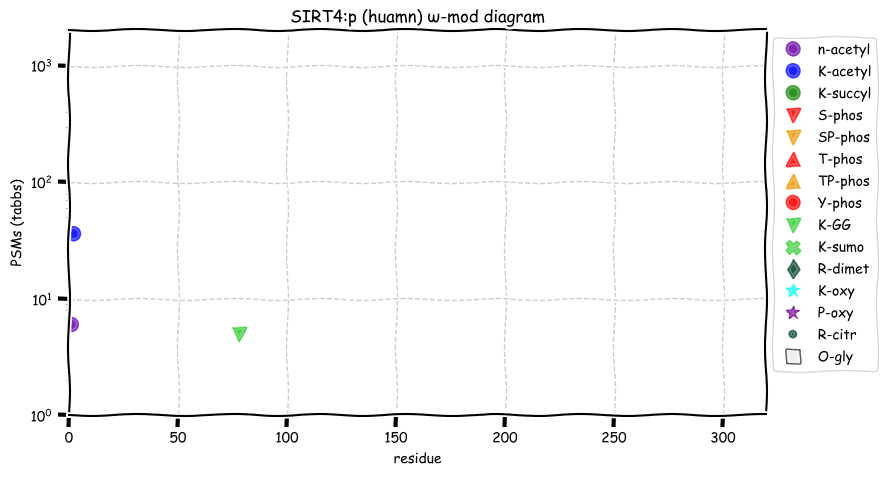
Thu Dec 16 13:10:07 +0000 2021SIRT4:p is the least frequently observed of the seven human sirtuins. The observation frequencies are as follows:
SIRT1: 13,425×;
SIRT2: 13,315×;
SIRT5: 11,556×;
SIRT3: 6,819×;
SIRT6: 5,912×;
SIRT7: 1,560×; &
SIRT4: 100×.
Thu Dec 16 13:10:07 +0000 2021SIRT4:p.I287V chr 12:g.120312950A>G, rs201288066 (all tissue I:V 0.967:0.033) vaf=<1%, Δm=-14.0156, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Thu Dec 16 13:10:07 +0000 2021SIRT4:p, θ(max) = 34. aka SIR2L4. Not observed in MHC class 1 or 2 experiments. Very rarely observed in tissues or cell lines.
Thu Dec 16 13:10:06 +0000 2021>SIRT4:p, sirtuin 4 (Homo sapiens) Small subunit; CTMs: none; PTMs: K78+GGyl; SAAVs: I287V (<1%); mature form: (29-314) [100×, 0.5 kTa]. #ᗕᕱᗒ 🔗
Wed Dec 15 17:41:44 +0000 2021@jwoodgett "Family MD" isn't really a thing for a lot of people in Canada: briefly talking to someone you've never seen before at a walk-in clinic is a more common experience.
Wed Dec 15 16:44:09 +0000 2021@neely615 @harrisonspecht I think the #CTPAC folks (like @NCI_HRodriguez) would be better people to ask. They have actually designed & tested methods by trying to reproduce them in multiple labs, esp. digestion & chromatographic methods.
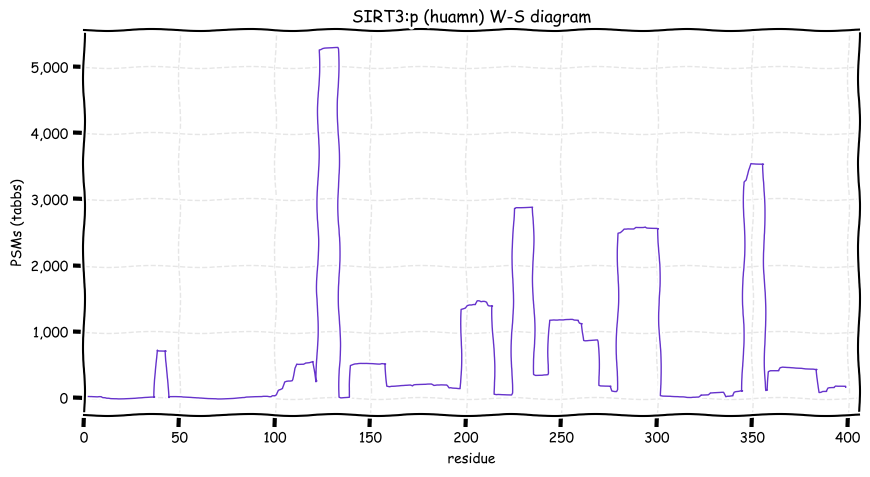
Wed Dec 15 13:00:54 +0000 2021SIRT3:p has an N-terminal mitochondrial targeting sequence that is removed on transit into a mitochondrion, but the cleavage site seems to be unusually imprecise.
Wed Dec 15 13:00:54 +0000 2021SIRT3:p.V208I chr 11:g.233067C>T, rs11246020 (all tissue V:I 0.973:0.027) vaf=17%, Δm=14.0156, VAF by population group: african 2%, american 15%, east asian 9%, european 20%, south asian 15%.
Wed Dec 15 13:00:54 +0000 2021SIRT3:p, θ(max) = 68. aka SIR2L3. Observed in rarely in MHC class 1 or 2 experiments. Found in tissues & cell lines: absent from fluids.
Wed Dec 15 13:00:54 +0000 2021>SIRT3:p, sirtuin 3 (Homo sapiens) Small subunit; CTMs: none; PTMs: none; SAAVs: A8V (2%), V208I (17%); mature form: (102,103,104,105-399) [6,814×, 21 kTa]. #ᗕᕱᗒ 🔗
Wed Dec 15 00:46:39 +0000 2021@UCDProteomics @NCBI It is also pretty much every grant request, without the jibber-jabber.
Tue Dec 14 22:32:25 +0000 2021@astacus Yes, it is. I was hoping to find out how you make a PTSD model mouse (& maybe be horrified about how you make a PTSD model mouse).
Tue Dec 14 20:15:12 +0000 2021@astacus 🔗 It is a crossover data set listing: the data is in iProX.
Tue Dec 14 19:38:02 +0000 2021@UCDProteomics @NCBI It's a consultancy staple. Pretty much the answer to any development project request.
Tue Dec 14 19:33:09 +0000 2021@UCDProteomics @NCBI Give me a research team of 10 & I'll let you know in 18-24 months.
Tue Dec 14 17:50:38 +0000 2021@jengerson Unspreadable butter.
Tue Dec 14 16:43:48 +0000 2021For those not familiar with the rather proficient pee pest Proteus mirabilis: 🔗
Tue Dec 14 16:29:16 +0000 2021It does have a bit of an issue with amine derivatization artifacts (both from iodoacetamide and urea), but not so much as to be a show stopper.
Tue Dec 14 16:26:26 +0000 2021If you are interested in mouse urine, PXD030347 is a good data set to look at. It has quality runs and several of the individuals show strong signals from UTIs: the strongest, HFX6_P6.raw, identifies >40 Proteus mirabilis proteins (out of 405 total).
Tue Dec 14 15:18:34 +0000 2021@neely615 Getting this cr*p on to and off of H1-1 isn't a subtle balletic dance that can be rendered into a clever illustration (unless you include some Mario Brothers)
🔗
Tue Dec 14 15:09:50 +0000 2021@astacus @BrukerMassSpec Does anyone use mass spec to detect the current "Portlandia"-style craze around the bovine A1/A2 alleles of beta casein (CSN2:p.P82H) in milk?
Tue Dec 14 15:03:03 +0000 2021@neely615 Protein acylation/deacylation is a fairly recent discovery & nobody seems to know exactly what to do with it. I suspect this is caused by the fact that it often isn't a delicate interplay of site-specific mods: it is more of a blue-collar "spackle on/chisel off" mechanism.
Tue Dec 14 14:58:05 +0000 2021@neely615 Sirtuins are named for the catalytic domain responsible for removing an acyl group from lysine's ε amino group: it would be like naming all serine proteases as "Serpro-X". The proteins & their biological activities diverged so long ago they really don't resemble each other.
Tue Dec 14 13:37:41 +0000 2021@PastelBio @ProteomicsNews Mass spec people love to complain about the size/complexity of their data files.
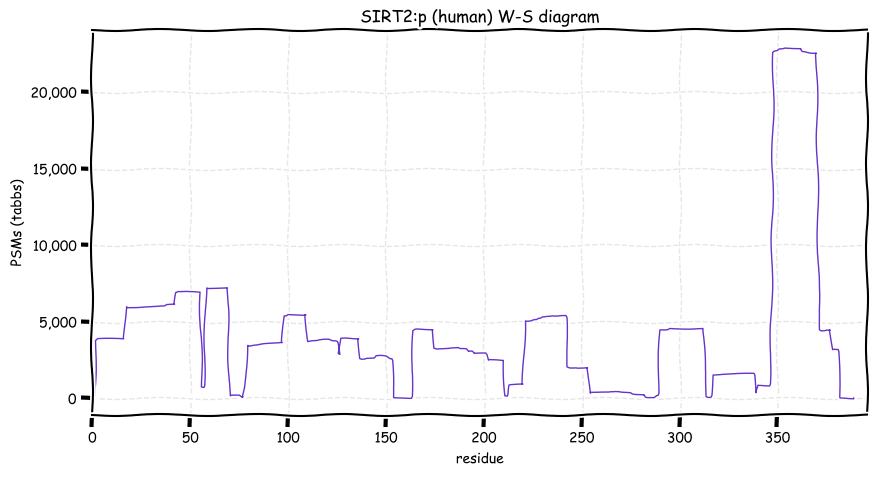
Tue Dec 14 12:56:58 +0000 2021SIRT2:p.L44F chr 19:g.38893508T>A, rs45535036 (all tissue L:F 0.999:0.001) vaf=<1%, Δm=33.9843, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Tue Dec 14 12:56:58 +0000 2021SIRT2:p, θ(max) = 83. aka SIR2L. Observed in MHC class 1 experiments. Found in tissues & cell lines: absent from fluids.
Tue Dec 14 12:56:58 +0000 2021>SIRT2:p, sirtuin 2 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 2×K+acetyl; 6×K+GGyl; 4×K+succinyl; 23×S, 7×T, 1×Y+phosphoryl; SAAVs: L44F (0.1%); mature form: (2-389) [13,310×, 58 kTa]. #ᗕᕱᗒ 🔗
Tue Dec 14 00:03:28 +0000 2021@astacus It would appear that stressed mice get UTI's too.
Mon Dec 13 22:21:14 +0000 2021It never occurred to me to check mouse urine for bacteria, but I guess that is just my perennial naivete showing ...
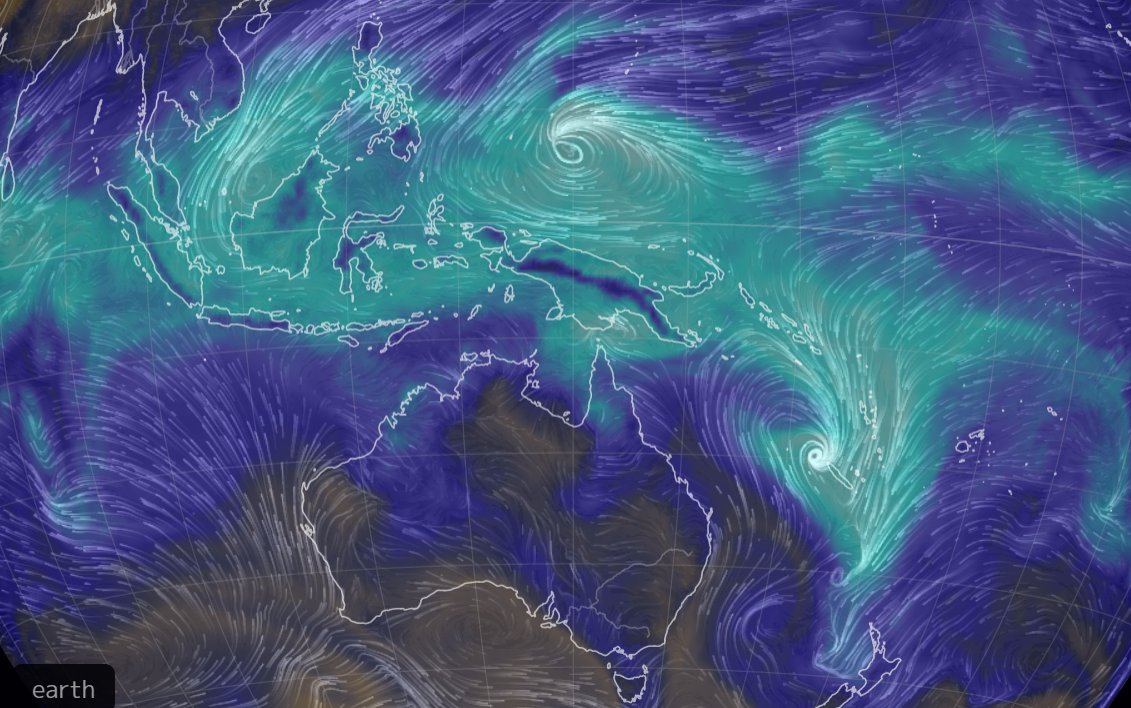
Mon Dec 13 13:56:35 +0000 2021Tropical Cyclone Ruby heading east, from just north of New Caledonia, while Tropical Storm Rai winds up for a run at the Philippines (light blue means lots of atmospheric water vapour) 🔗
Mon Dec 13 13:36:53 +0000 2021@J_my_sci Sounds like a nightmare for the authors.
Mon Dec 13 13:36:04 +0000 2021@J_my_sci Admirable, but I would be very surprised if this works in practice: "Readers of ERA publications will be able to inspect the code, modify it, and re-execute it directly in the browser".
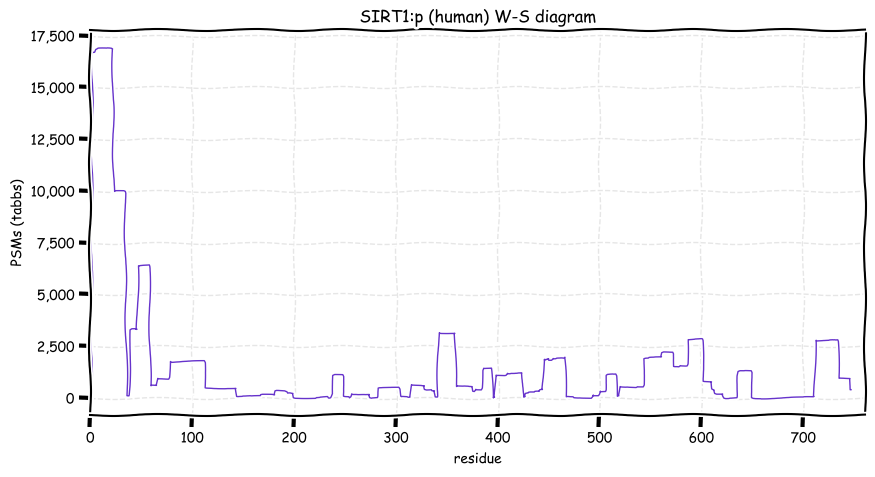
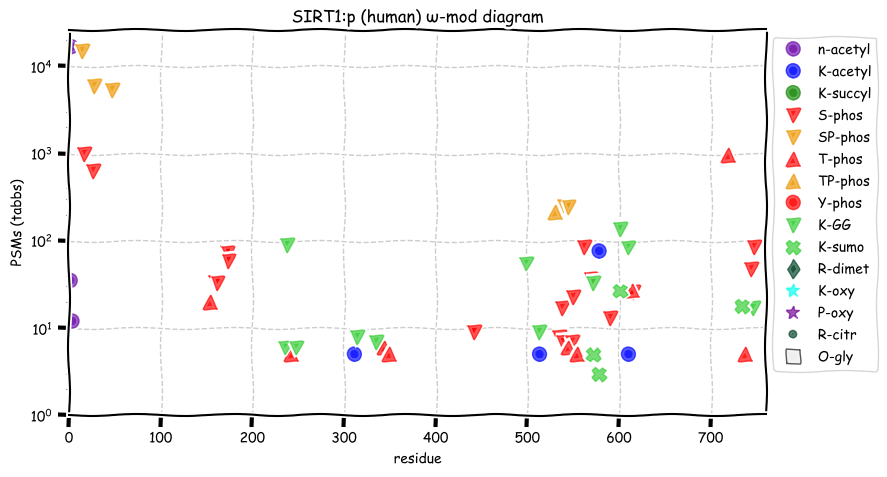
Mon Dec 13 12:54:00 +0000 2021SIRT1:p is one of seven human lysine deacylases referred to as sirtuins, named after an acronym for the yeast subunit "silent mating type information regulation 2" (Sir2).
Mon Dec 13 12:52:42 +0000 2021SIRT1:p has 4 distinct phosphodomains (14-147), (154-174), (530-615) & (714-747), all of which are outside of the core catalytic domain (244-498).
Mon Dec 13 12:52:42 +0000 2021SIRT1:p.M595T chr 10:g.67912900T>C, rs139635382 (all tissue M:T 0.999:0.001) vaf=0.1%, Δm=-29.9928, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Mon Dec 13 12:52:41 +0000 2021SIRT1:p, θ(max) = 60. aka SIR2L1, SIR2, hSIR2, SIR2alpha. Observed in MHC class 1 experiments. Found in tissues & cell lines: absent from fluids.
Mon Dec 13 12:52:41 +0000 2021>SIRT1:p, sirtuin 1 (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 4×K+acetyl; 11×K+GGyl; 4×K+SUMOyl; 25×S, 10×T, 0×Y+phosphoryl; SAAVs: M595T (0.1%); mature form: (2-747) [13,419×, 58 kTa]. #ᗕᕱᗒ 🔗
Sun Dec 12 13:58:24 +0000 2021@jovel_juan @BiswapriyaMisra Congrats for keeping this going. Funding this type of project is nearly impossible in Canada & yet Dave has managed it for 15 years!
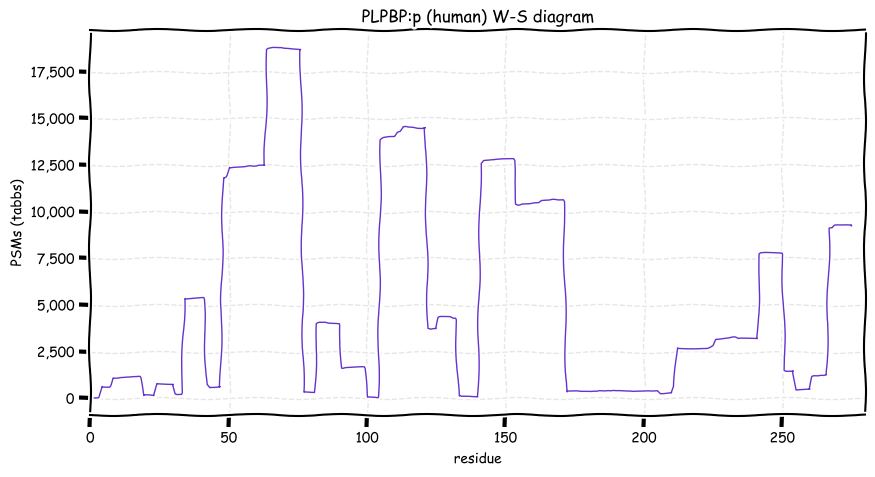
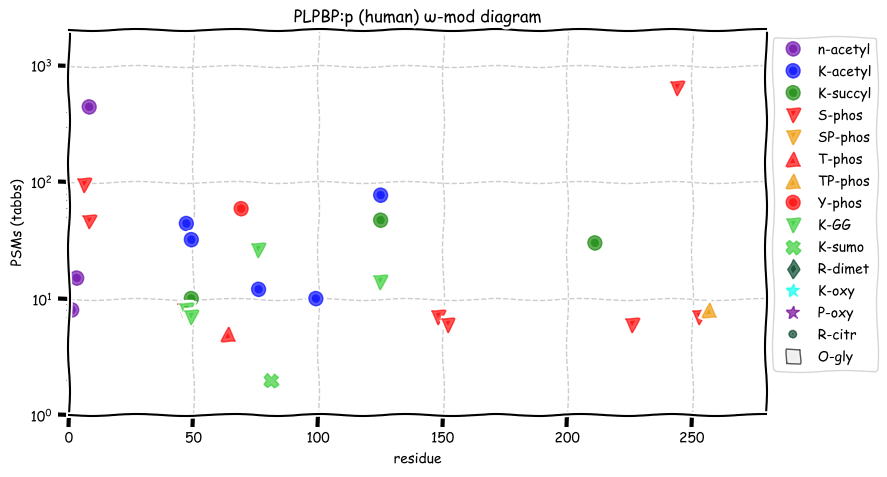
Sun Dec 12 12:53:56 +0000 2021PLPBP:p, sequence generated by the alternate initiation site M7 is obsserved about as often as the sequence using the reference inititation site M1.
Sun Dec 12 12:53:56 +0000 2021PLPBP:p.M53V chr 8:g.37765583A>G, rs79148472 (all tissue M:V 0.999:0.001) vaf=0.4%, Δm=26.0520, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian 1%.
Sun Dec 12 12:53:56 +0000 2021PLPBP:p, θ(max) = 74. aka PROSC. Observed in MHC class 1 experiments & very rarely in class 2. Found in many tissues, cell lines & urine extracellular vesicles. Has too few GO annotations.
Sun Dec 12 12:53:56 +0000 2021>PLPBP:p, pyridoxal phosphate binding protein (Homo sapiens) Small subunit; CTMs: M1, S8+acetyl; PTMs: 5×K+acetyl; 3×K+succinyl; 4×K+GGyl; 1×K+SUMOyl; 8×S, 2×T, 1×Y+phosphoryl; SAAVs: V24M (11%), M53V (1%); mature form: (1,8-275) [25,151×, 108 kTa]. #ᗕᕱᗒ 🔗
Sat Dec 11 16:18:09 +0000 2021@bffo @PLOSCompBiol Those two departments (+ Immunology) rely on computation rather heavily for many aspects of research, so a lack of teaching & research on the topic make them lower tier compared to departments that have it.
Sat Dec 11 16:02:43 +0000 2021@bffo @PLOSCompBiol Getting bioinformatics teaching/research capacity included as a requirement for the accreditation of Genetics & Biochemistry Departments is about the only way to make this happen at most places.
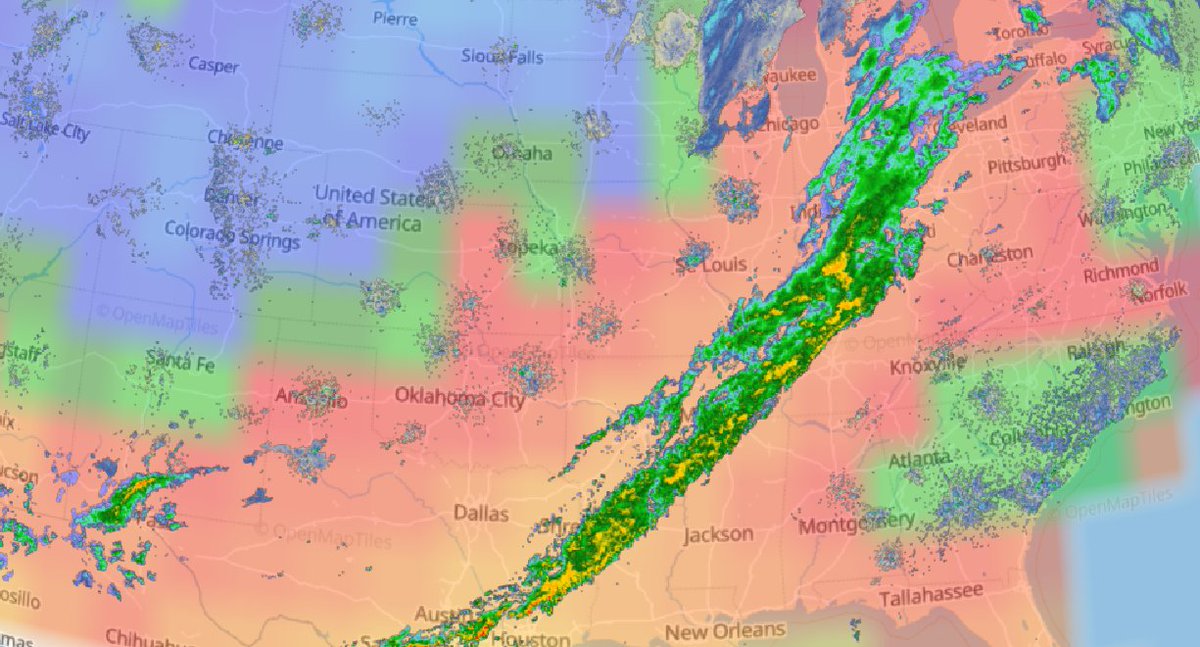
Sat Dec 11 15:32:54 +0000 2021After a stormy night, there is still a line of active storms stretching from Nuevo Laredo to Cleveland. 🔗
Sat Dec 11 15:08:16 +0000 2021MINDY1:p, endogenous cleavage resulting in a new N-terminus at S95 is quite common.
Sat Dec 11 14:53:11 +0000 2021Unexpected behavior: more the norm than the exception in my experience 🔗
Sat Dec 11 13:50:09 +0000 2021The data, PXD028633, shows that these methods can be used to prepare high quality class 1 & 2 samples that have nearly theoretical physical properties. If you want to either make samples or have some excellent exemplars for data analysis, this study is a good place to start.
Sat Dec 11 13:49:28 +0000 2021Sirois, et al, J Vis Exp. 2021 Oct 15;(176) tries to do something about this, providing methods that labs can use to get started in the field. 🔗
Sat Dec 11 13:48:34 +0000 2021MHC/HLA peptides are interesting on a number of levels. It is kind of tricky to prepare them as samples from MS-based proteomics, however. Many groups (even very well known ones) have produced data that demonstrates this difficulty.
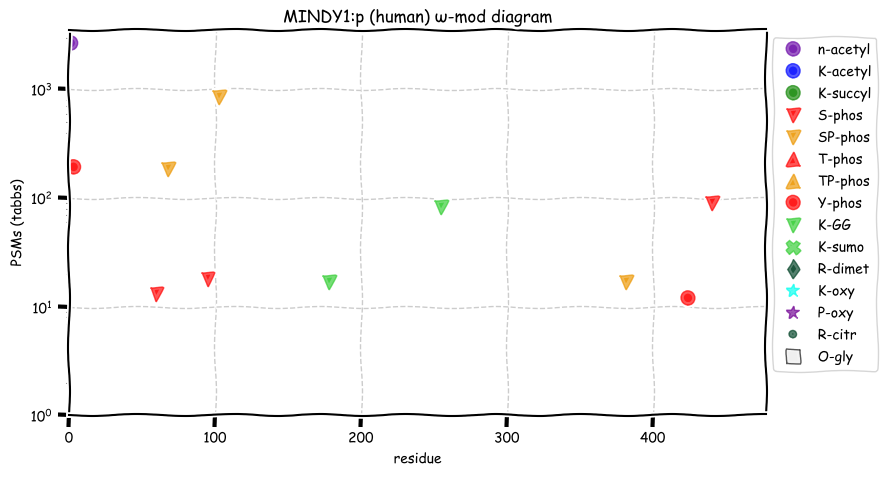
Sat Dec 11 13:06:28 +0000 2021MINDY1:p, the reference sequence has never been observed. Instead, all observations include T385K, which introduces a tryptic cleavage site into the sequence resulting in two peptides not in the reference sequence:
373 GPGAEGGSGSPE(T385K)
386 QLQVDQDYLIALSLQQQQPR
Sat Dec 11 12:51:07 +0000 2021MINDY1:p.T385K chr 1:g.150998101G>T, rs2925741 (all tissue T:K 0.000:1.000) vaf=99%, Δm=27.0473, VAF by population group: african <1%, american 79%, east asian 99%, european >99%, south asian >99%.
Sat Dec 11 12:51:06 +0000 2021MINDY1:p, θ(max) = 51. aka FLJ11280, MINDY-1, FAM63A. Observed in MHC class 1 experiments. Particularly abundant in blood platelets. Found in many tissues, cell lines & extracellular vesicles.
Sat Dec 11 12:51:06 +0000 2021>MINDY1:p, MINDY lysine 48 deubiquitinase 1 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 2×K+GGyl; 6×S, 0×T, 2×Y+phosphoryl; SAAVs: R41S (5%), T243K (99%); mature form: (1-469) [5,259×, 17 kTa]. #ᗕᕱᗒ 🔗
Fri Dec 10 20:52:24 +0000 2021@slashdot Hopefully Canada will maintain its lead in discussions about the possibilities of developing tech ...
Fri Dec 10 15:49:49 +0000 2021Any suggestions for a better shape or colour?
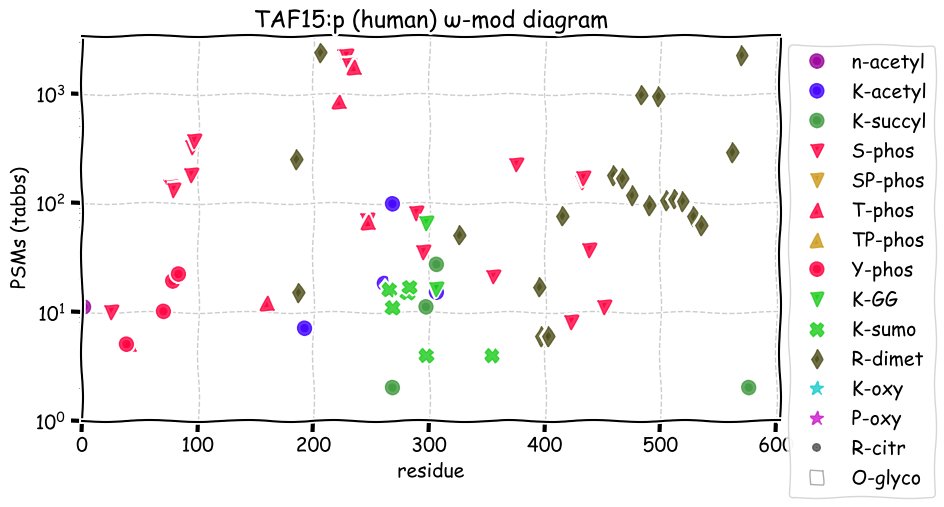
Fri Dec 10 15:48:09 +0000 2021In an my ongoing effort to make things more confusing, I have added succinylation (India green circles) to the ω-mod diagram of PTM acceptor site observed modification frequency 🔗
Fri Dec 10 15:06:53 +0000 2021Thanks to everyone who participated in this poll. It appears that the majority consider K-P to be a non-cleavable bond. I personally treat it the same as D-P (a spontaneously hydrolysed bond): test for its cleavage but don't include it in the count of "missed" cleavage sites.
Fri Dec 10 13:15:11 +0000 2021TAF15:p is a good example of a protein where you can get quite a bit of detailed functional info from its observed high-res PTM pattern & very little from its 3D structure.
Fri Dec 10 12:52:13 +0000 2021TAF15:p, most of the subunit does not produce good 3D structural information, as illustrated by the AlphaFold2 model: 🔗
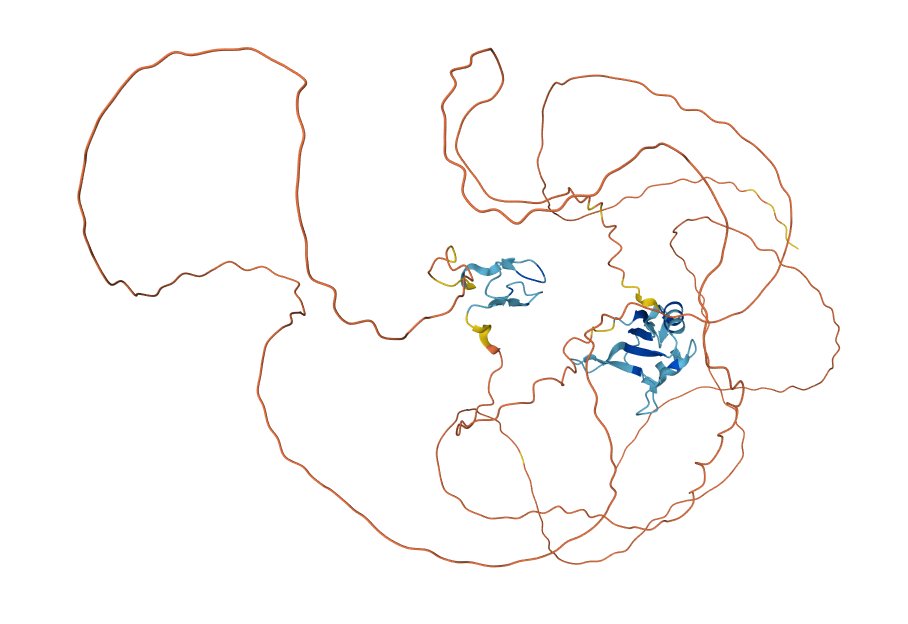
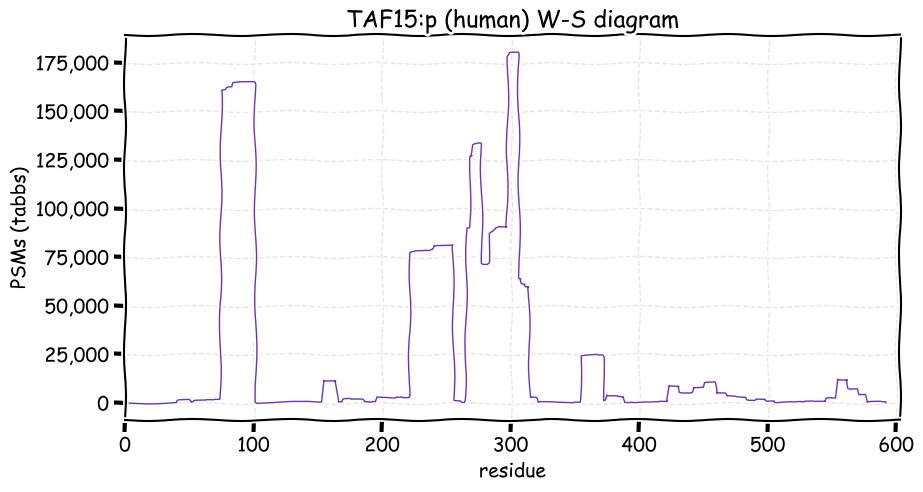
Fri Dec 10 12:41:48 +0000 2021TAF15:p along with FUS:p & EWS:p are the FET family of RNA-binding proteins. They all have unusually large numbers of R dimethylation acceptor sites, with TAF15 having the most, particularly (459-570). (261-306) has a high density of K acceptor PTMs.
Fri Dec 10 12:41:48 +0000 2021TAF15:p.S95L chr 17:g.35820431C>T, rs147936141 (all tissue S:L 0.999:0.001) vaf=<1%, Δm=26.0520, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Fri Dec 10 12:41:48 +0000 2021TAF15:p, θ(max) = 62. aka TAFII68, RBP56, Npl3, TAF2N. Observed in MHC class 1 & 2 experiments. Found in all cellular tissues & cell lines: rare in fluids. TAF15 has several repetitive, low complexity domains enriched in R, G or D residues
Fri Dec 10 12:41:48 +0000 2021>TAF15:p, TATA-box binding protein associated factor 15 (H sapiens) Midsized subunit; CTMs: M1, S2+acetyl; PTMs: 4×K+acetyl; 4×K+succinyl; 2×K+GGyl; 6×K+SUMOyl; 20×S, 5×T, 4×Y+phospho; 21×R+dimethyl SAAVs: S95L (<1%); mature form: (1,2-592) [74,009×, 772 kTa]. #ᗕᕱᗒ 🔗
Thu Dec 09 16:26:03 +0000 2021It is an odd juxtaposition 🔗
Thu Dec 09 14:55:09 +0000 2021So, in the current zeitgeist, if you have observed the tryptic peptide
MKQEPVKPEEGR
does it have 1 missed cleavage sites or 2?
Thu Dec 09 14:54:26 +0000 2021This was largely forgotten/ignored when the use of Lys-C as a pre-treatment came into vogue as it can cleave K-P bonds. However, in practice Lys-C also has a significantly reduced propensity to cleave K-P (just not as bad as trypsin).
Thu Dec 09 14:53:59 +0000 2021How do people feel about cleavage at K-P? There was a minor buzz in the literature back-in-the-day about whether or not K-P should be considered a legit tryptic scission site that be counted as a "missed" cleavage.
Thu Dec 09 13:52:37 +0000 2021PXD027198 is an interesting data set if you are interested in phosphorylation acceptor site analysis: made available as part of 🔗
Thu Dec 09 13:26:50 +0000 2021Anyone want to pitch for their favorite Lys-C vendor? 🔗
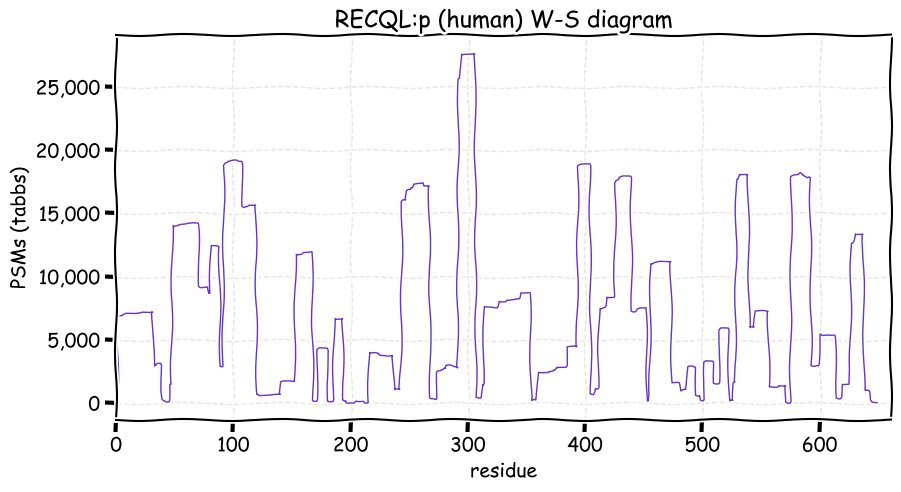
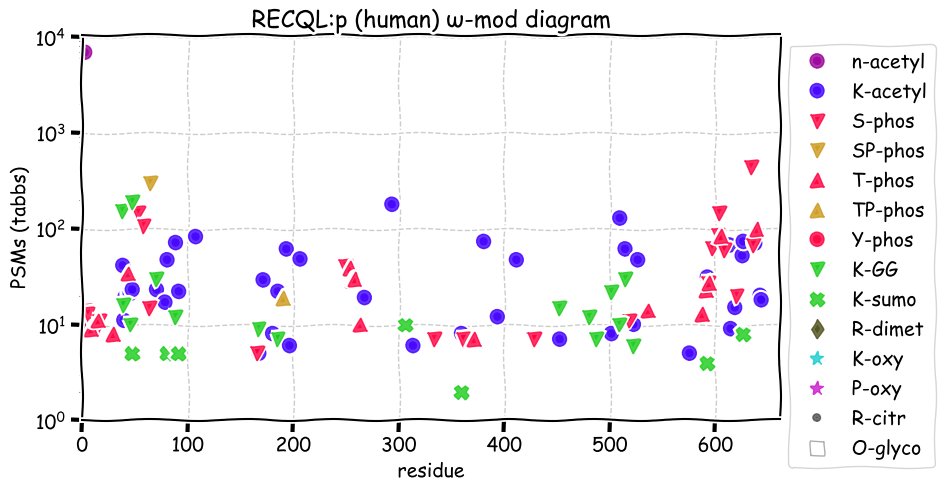
Thu Dec 09 12:45:28 +0000 2021RECQL:p is the most frequently observed of the RECQL helicases, even though its biological function is unclear. Compared to the longer REQC-like helicases WRN & BLM, RECQL:p has fewer K+SUMOylation acceptor sites, but many more K+acetylation acceptors.
Thu Dec 09 12:45:28 +0000 2021RECQL:p.V102I chr 12:g.21490289C>T, rs1065751 (all tissue V:I 0.986:0.014) vaf=1%, Δm=14.0156, VAF by population group: african 16%, american 1%, east asian <1%, european <1%, south asian <1%.
Thu Dec 09 12:45:28 +0000 2021RECQL:p, θ(max) = 76. aka RecQ1, RecQL1. Observed in MHC class 1 experiments and MHC2 rarely. Found in many tissues & cell lines: absent from fluids.
Thu Dec 09 12:45:28 +0000 2021>RECQL:p, RecQ like helicase (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 42×K+acetyl; 15×K+GGyl; 7×K+SUMOyl; 23×S, 16×T, 0×Y+phosphoryl; SAAVs: V102I (1%), K487T (2%); mature form: (2-649) [33,870×, 324 kTa]. ᗕᕱᗒ 🔗
Wed Dec 08 21:19:47 +0000 2021@ucdmrt @SeanJHumphrey I don't have a wet lab so I don't buy reagents, but I'm sure some one out there is willing to make recommendations about best-in-breed Lys-C preparations.
Wed Dec 08 17:00:50 +0000 2021@astacus @AJ_Brenes The tissues they identified as cartilage seem to have quite a few very abundant proteins more commonly found associated with skeletal muscle, e.g. TTN, NEB & most of the MYH's, but far less collagen than I was expecting.
Wed Dec 08 16:56:40 +0000 2021@astacus @AJ_Brenes So long as the student got some good training & thought about what it would be like to make an atlas, then good for them. I am kind of impressed how much they were able to get out of a disassembled mouse larynx, though.
Wed Dec 08 14:44:49 +0000 2021@SeanJHumphrey @ucdmrt Lys-C often has other proteolytic enzymes in a batch, that can sometimes generate an annoying amount of cleavage at other residues, particular A-X, E-X, L-X & D-X. This particular batch is generating >97 % of cleaves at K-X, with the usual hesitancy about K-P's.
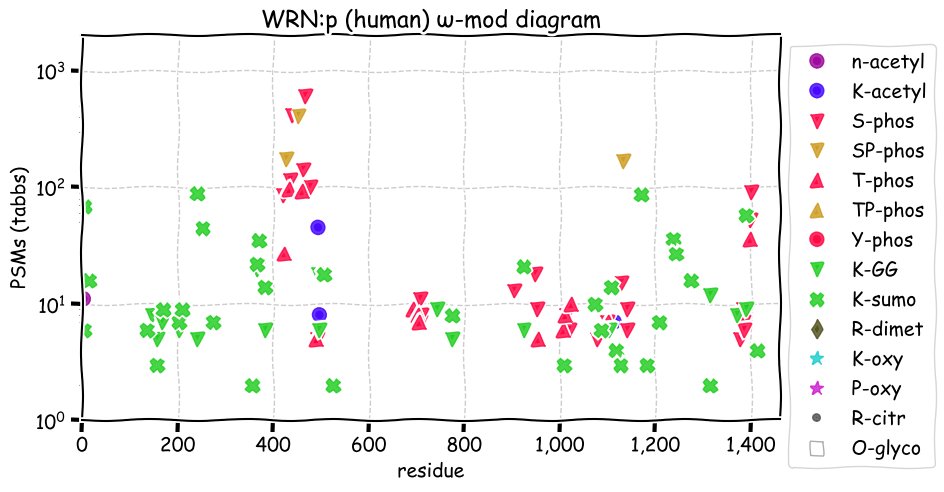
Wed Dec 08 12:44:47 +0000 2021WRN:p has an unusually large number of SUMOylation acceptor sites similar to BLM, another RecQ helicase.
Wed Dec 08 12:44:46 +0000 2021WRN:p.C1367R chr 8:g.31167138T>C, rs1346044 (all tissue C:R 0.769:0.231) vaf=24%, Δm=53.0919, VAF by population group: african 15%, american 17%, east asian 10%, european 27%, south asian 28%.
Wed Dec 08 12:44:46 +0000 2021WRN:p, θ(max) = 46. aka RECQL2, RECQ3. Observed in MHC class 1 experiments only. Found in many tissues & cell lines: absent from fluids.
Wed Dec 08 12:44:46 +0000 2021>WRN:p, Werner syndrome RecQ like helicase (Homo sapiens) Large subunit; CTMs: S2+acetyl; PTMs: 3×K+acetyl; 17×K+GGyl; 34×K+SUMOyl; 29×S, 13×T, 0×Y+phosphoryl; SAAVs: M387I (5%), L1074F (50%), S1079L (2%), C1367R (24%); mature form: (2-1432) [5,971×, 20 kTa]. ᗕᕱᗒ 🔗
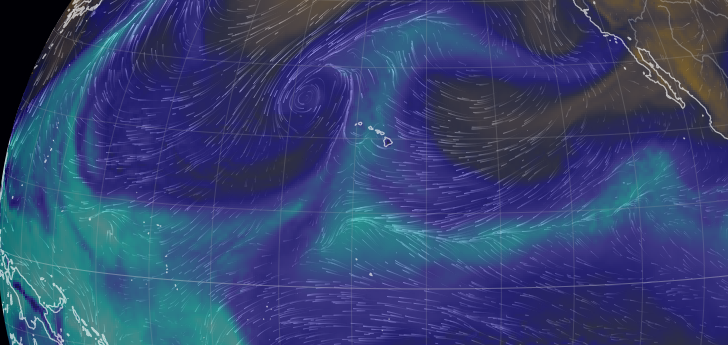
Tue Dec 07 18:37:51 +0000 2021Another "atmospheric river" (light blue means lots of water in the atmosphere): this one intersecting Hawaii 🔗
Tue Dec 07 17:31:01 +0000 2021Just something I find interesting
🔗
Tue Dec 07 16:33:23 +0000 2021I'm still working my way through the data, but it looks like these guys lucked out and got the good batch of Lys-C 🔗
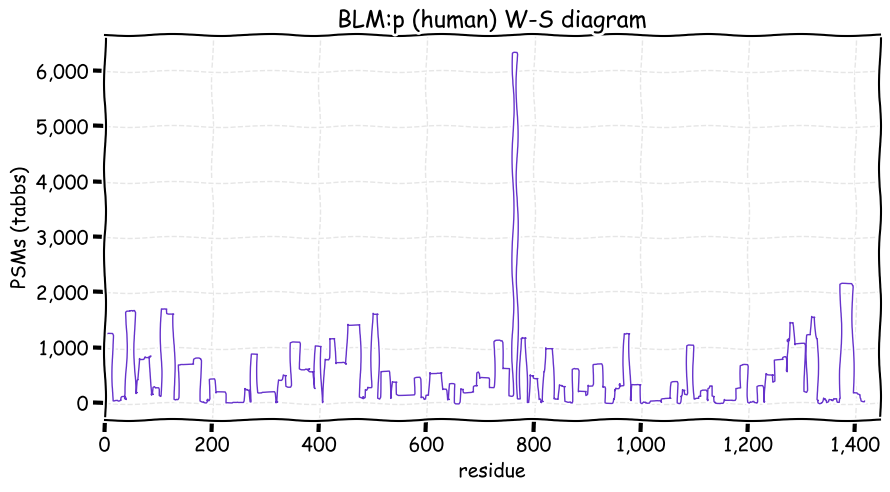
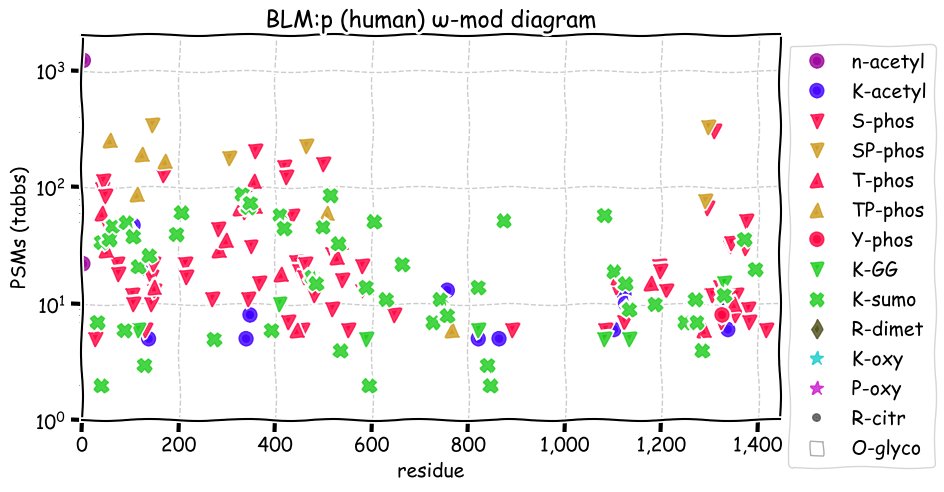
Tue Dec 07 12:55:28 +0000 2021BLM:p has an unusually large number of SUMOylation acceptor sites: why is an open question. Phosphorylation is absent in the central helicase domain of the protein.
Tue Dec 07 12:55:28 +0000 2021BLM:p.V1321I chr 15:g.90811291G>A, rs7167216 (all tissue V:I 0.926:0.074) vaf=7%, Δm=14.0156, VAF by population group: african 10%, american 5%, east asian 2%, european 8%, south asian 8%.
Tue Dec 07 12:55:28 +0000 2021BLM:p, θ(max) = 82. aka BS, RECQL3, RECQ2. Observed in MHC class 1 experiments only. Found in many tissues & cell lines: absent from fluids. Has too many GO annotations.
Tue Dec 07 12:55:27 +0000 2021>BLM:p, BLM RecQ like helicase (Homo sapiens) Large subunit; CTMs: A2+acetyl; PTMs: 17×K+acetyl; 7×K+GGyl; 51×K+SUMOyl; 79×S, 24×T, 1×Y+phosphoryl; SAAVs: T298M (2%), P868L (5%), V1321I (7%); mature form: (2-1417) [12,137×, 51 kTa]. ᗕᕱᗒ 🔗
Mon Dec 06 17:22:22 +0000 2021@oleg8r Do you think the problem is caused by distracted/stressed submitters or overly complex submission systems that may not be working properly?
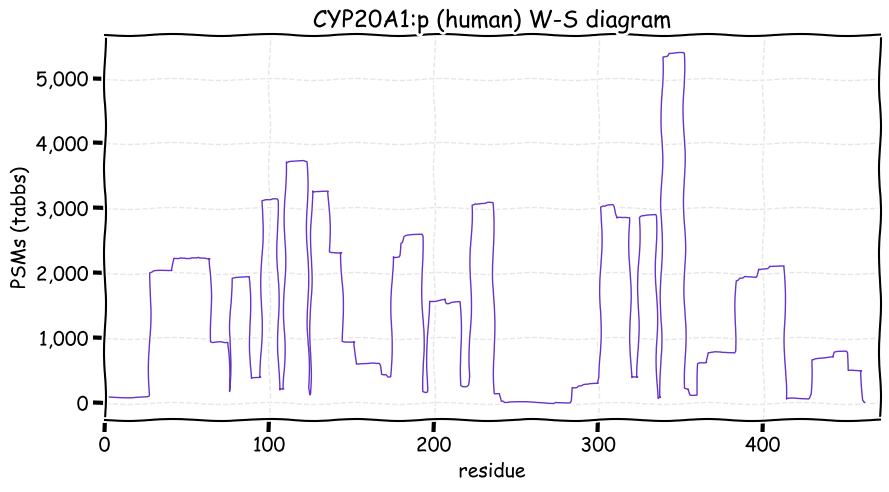
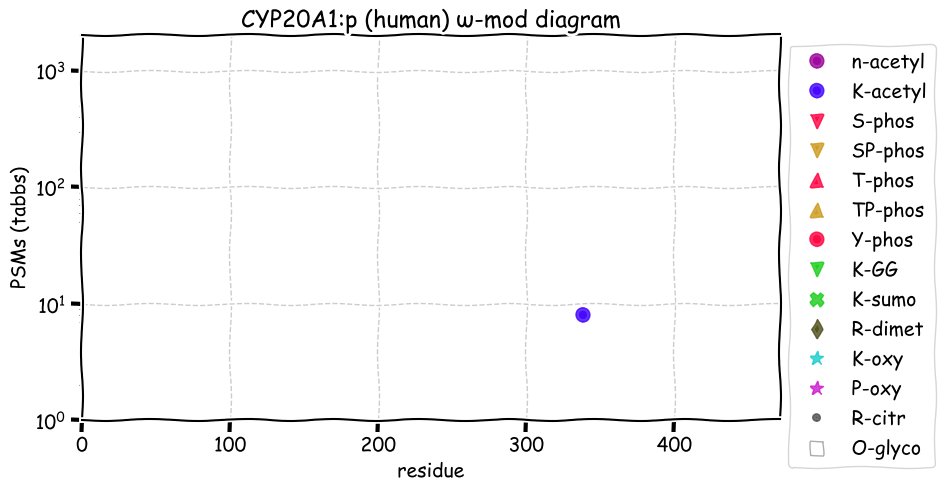
Mon Dec 06 12:51:48 +0000 2021CYP20A1:p is the most frequently observed cytochrome P450, but it is the only one without any known substrate specificity or biological function.
Mon Dec 06 12:51:48 +0000 2021CYP20A1:p.S97L chr 2:203251967C>T, rs2043449 (all tissue S:L 0.064:0.936) vaf=87%, Δm=26.052, VAF by population group: african 66%, american 94%, east asian 93%, european 96%, south asian 95%.
Mon Dec 06 12:51:48 +0000 2021CYP20A1:p, θ(max) = 66. aka CYP-M. Present in MHC class 1 experiments only. Found in many tissues, cell lines & extracellular vesicles: absent from fluids. Transmembrane domain: (5-24). There is no evidence that the N-terminal M is acetylated.
Mon Dec 06 12:51:47 +0000 2021>CYP20A1:p, cytochrome P450 family 20 subfamily A member 1 (Homo sapiens) Small subunit; CTMs: none; PTMs: K383+acetyl; 0×K+GGyl; 0×K+SUMOyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: S97L (87%), L346F (40%); mature form: (1-462) [10,826×, 49 kTa]. ᗕᕱᗒ 🔗
Sun Dec 05 17:28:35 +0000 2021@dancanplan @BCTransit The same thing was done here in Winnipeg, too: the online replacement was so awful I had to write my own.
Sun Dec 05 15:39:34 +0000 2021I find that reading code is a much better way to understand how someone thinks about a problem than reading the very formulaic manuscripts currently in vogue.
Sun Dec 05 15:29:49 +0000 2021My main interest in reading scientific source code is trying to figure out if the author is someone who should be taken seriously & if anyone (other than the author) actually read it prior to publication.
Sun Dec 05 13:30:42 +0000 2021LAMC3:p.H73Q chr 9:g.131009433T>G, rs3739512 (all tissue H:Q 0.669:0.331) vaf=53%, Δm=14.0156, VAF by population group: african 80%, american 55%, east asian 86%, european 45%, south asian 53%.
Sun Dec 05 13:30:42 +0000 2021LAMC3:p, θ(max) = 39. aka DKFZp434E202. Observed in tissue-based MHC class 1 & 2 peptide experiments. Found in many tissues, select cell lines & extracellular vesicles: absent from fluids.
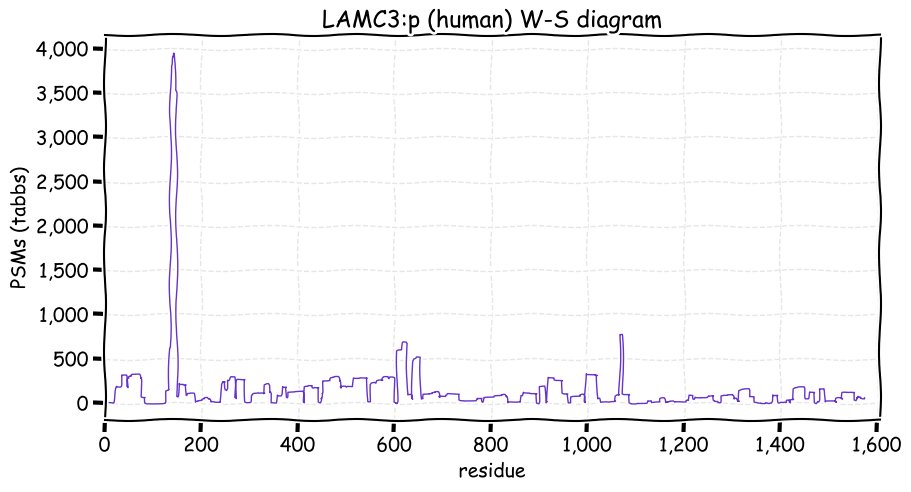
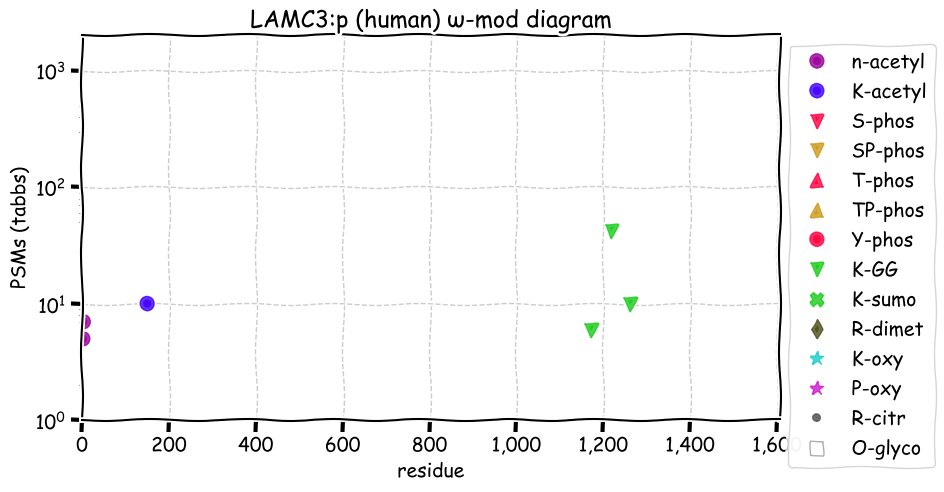
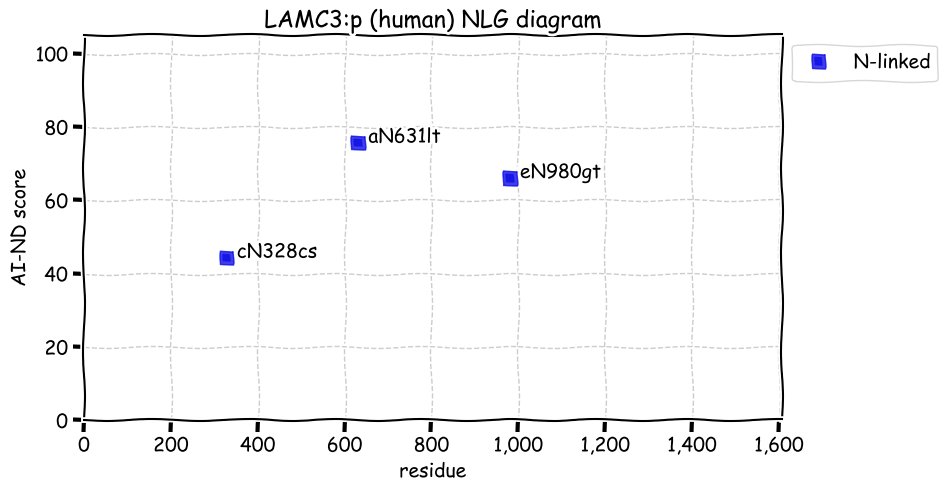
Sun Dec 05 13:30:41 +0000 2021>LAMC3:p, laminin subunit gamma 3 (Homo sapiens) Large subunit; CTMs: 3×N+glycosyl; PTMs: 1×K+acetyl; 3×K+GGyl; SAAVs: H73Q (52%), P522S (26%), E544G (47), R770G (21%), S1082G (30%), R1459Q (16%); mature form: (20,21-1575) [3,523×, 15 kTa]. #ᗕᕱᗒ 🔗
Sat Dec 04 19:44:26 +0000 2021Every once in a while, I see an old problem with new clarity. It is the intermittent positive reinforcement that keeps me bobbing along.
Sat Dec 04 18:55:46 +0000 2021@jjjvanderhooft @lgatt0 This can be quite intimidating for faculty (who in the end are simply employees) & can cause them to act very defensively wrt software.
Sat Dec 04 18:52:05 +0000 2021@jjjvanderhooft @lgatt0 Unlike bench work, which requires a conscious act (applying for a patent) to become IP, software is IP the moment it is written: copyright is affixed by definition. So who owns it or can license it becomes something University business layers feel should involve them.
Sat Dec 04 18:28:29 +0000 2021@jjjvanderhooft @lgatt0 Bayh-Dole is a controversial piece of US legislation that governs the ownership of IP developed using federal government money. It allows a not-for-profit institution receiving federal funding to retain ownership of IP developed using the that funding.
Sat Dec 04 17:29:08 +0000 2021@lgatt0 @jjjvanderhooft The most frequently advanced reason is institutional pressure using some legal pretext, such as the Bayh-Dole Act in the US.
Sat Dec 04 16:32:51 +0000 2021@lgatt0 Even in things I have released publicly, I always have a "pro" version that I don't release, either because it includes features that aren't ready for release (& may never be) or because a feature would simply be too controversial/hard-to-explain to the punters.
Sat Dec 04 15:55:00 +0000 2021@lgatt0 The main group that gets hurt by freebie closed-source projects is anyone who wants to commercialize software in the field. They don't get to benefit from publicly funded research.
Sat Dec 04 15:47:09 +0000 2021@lgatt0 Closed source projects may be very popular with users, but they tend to be academic dead ends. They either wall themselves off from developments in the field or they are viewed negatively by peers as taking advantage of ideas while not contributing any of their own.
Sat Dec 04 15:41:57 +0000 2021@lgatt0 I'm not very dogmatic about this point. In my view, if you don't want to make the code available & the journal agrees, then OK. But the paper probably won't be very influential among people doing research in the area—as opposed to users who may go gaga for the freebie.
Sat Dec 04 15:31:16 +0000 2021@hecklab @Karl_Mechtler @bmbSDU Do we have to call him Sir Ole now?
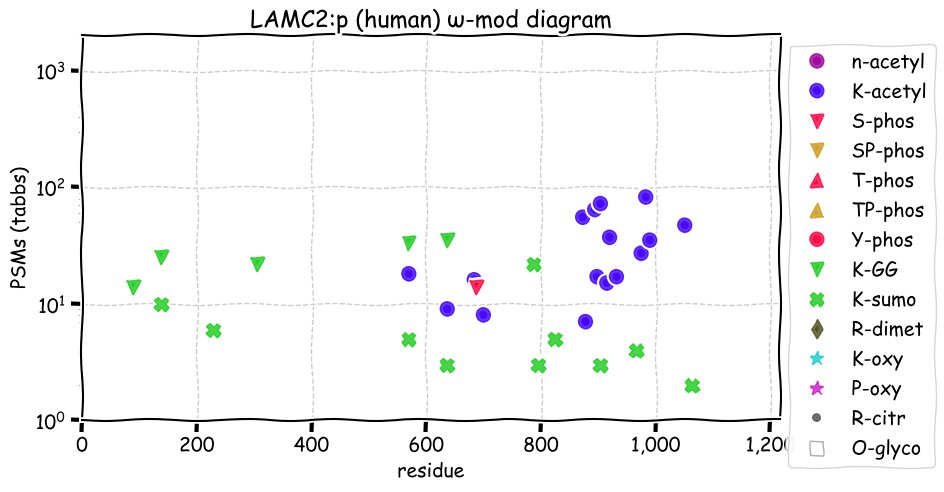
Sat Dec 04 13:51:41 +0000 2021LAMC2:p.S733T chr 1:g.183232835G>C, rs2296303 (all tissue S:T 0.828:0.172) vaf=14%, Δm=14.0156, VAF by population group: african 22%, american 16%, east asian 16%, european 10%, south asian 8%. TT in HaCat cells.
Sat Dec 04 13:51:41 +0000 2021LAMC2:p, θ(max) = 77. aka nicein-100kDa, kalinin-105kDa, BM600-100kDa, EBR2, LAMB2T, LAMNB2, EBR2A. Observed in tissue-based MHC class 1 & 2 peptide experiments. Found in many tissues, select cell lines & extracellular vesicles: absent from fluids.
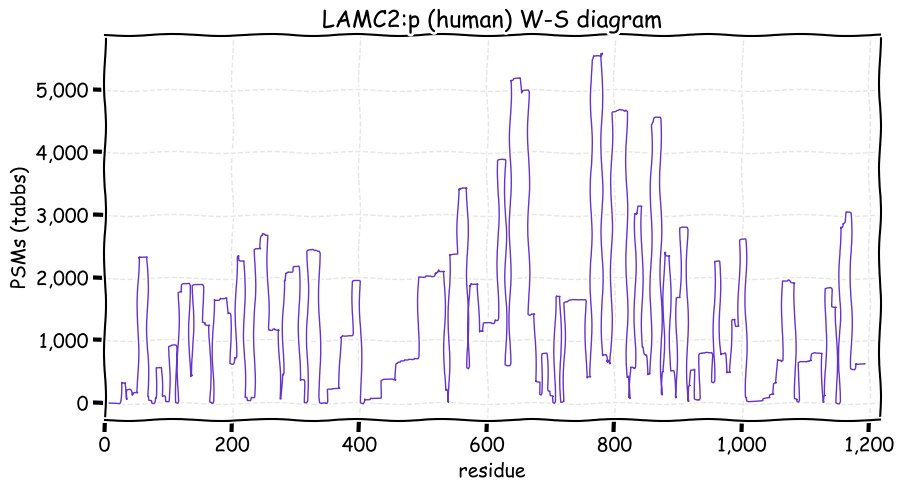
Sat Dec 04 13:51:41 +0000 2021>LAMC2:p, laminin subunit gamma 2 (Homo sapiens) Large subunit; CTMs: 2×N+glycosyl; PTMs: 17×K+acetyl; 5×K+GGyl; 10×K+SUMOyl; 1×S, 0×T, 0×Y+phosphoryl; SAAVs: A111P (3%), T124M (4%), D247E (6%), S733T (14%); mature form: (22?-1193) [11,066×, 107 kTa]. #ᗕᕱᗒ 🔗
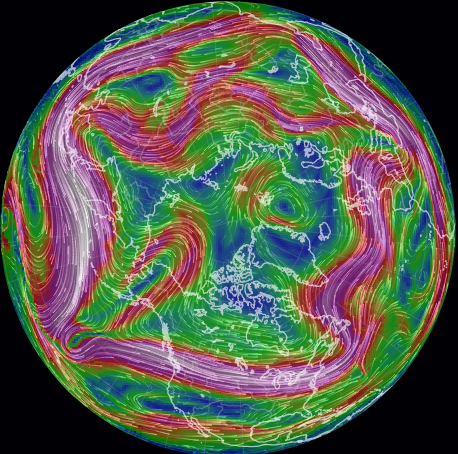
Fri Dec 03 18:14:10 +0000 2021The jet stream (250 hPa winds) are a straight-shot across North America today, but are in more of a ball-of-string-after-the-cat-played-with-it configuration for the rest of the hemisphere 🔗
Fri Dec 03 17:52:04 +0000 2021@TrostLab @BrewDog Less the crackers and crowns.
Fri Dec 03 13:56:44 +0000 2021@kabalak It is a clinical model & there is some proteomics data out there, but less I would expect. Are there any good pig cell lines?
Fri Dec 03 13:43:46 +0000 2021Given the quality of the genome sequence, Sus scrofa is probably an underutilized model species in proteomics. But I suspect the practical downsides outweigh its positives.
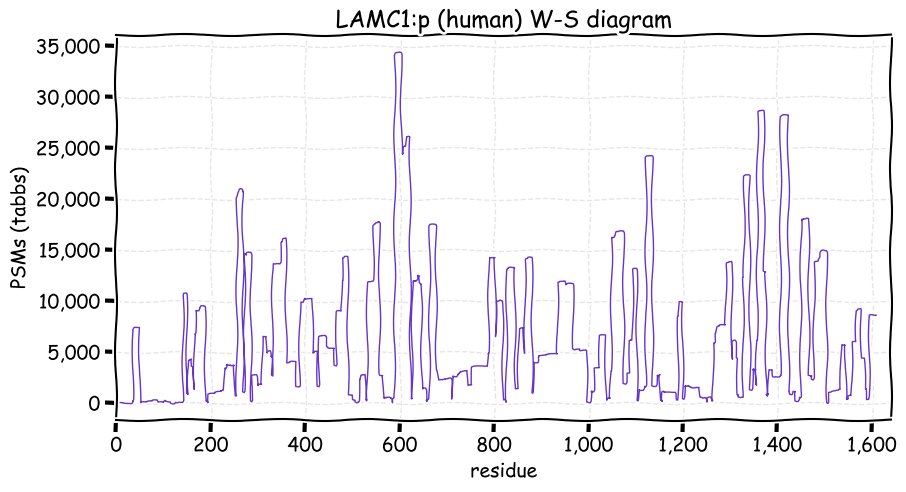
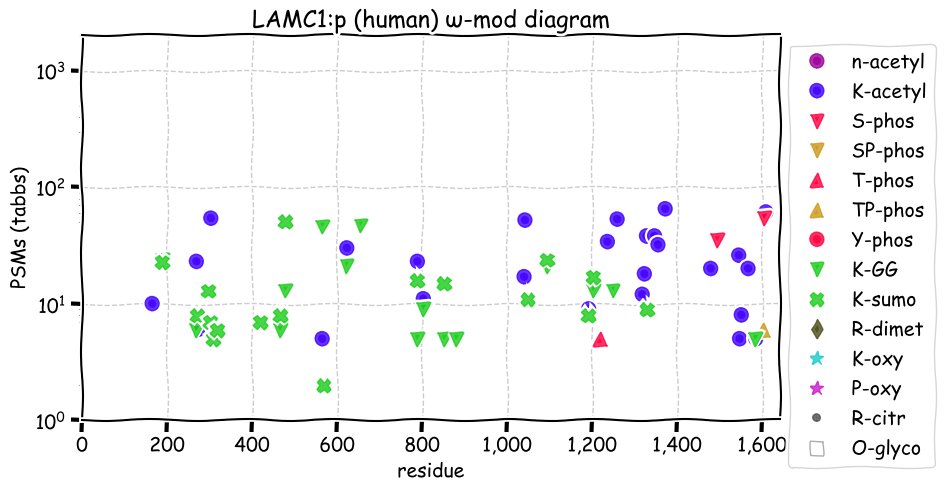
Fri Dec 03 13:11:14 +0000 2021LAMC1:p has the most occupied N-linked glycosylation acceptors of any of the laminin subunits. The role of SUMOylation acceptors in a primarily extracellular protein is unknown.
Fri Dec 03 13:11:14 +0000 2021LAMC1:p.I458V chr 1:g.183116620A>G, rs20563 (all tissue I:V 0.717:0.283) vaf=58%, Δm=-14.0156, VAF by population group: african 33%, american 63%, east asian 63%, european 56%, south asian 60%. VV in A-375 cells.
Fri Dec 03 13:11:14 +0000 2021LAMC1:p, θ(max) = 68. aka LAMB2. Observed in tissue-based MHC class 1 & 2 peptide experiments. Found in most tissues, cell lines & fluids. Laminin γ subunits have very little overlap in observable tryptic peptides (γ2:γ1 0% & γ3:γ1 1.6%).
Fri Dec 03 13:11:14 +0000 2021>LAMC1:p, laminin subunit gamma 1 (Homo sapiens) Large subunit; CTMs: 12×N+glycosyl; PTMs: 26×K+acetyl; 15×K+GGyl; 17×K+SUMOyl; 2×S, 2×T, 0×Y+phosphoryl; SAAVs: I458V (58%), N837K (53%), L888P (53%), N1205S (3%); mature form: (34,36-1609) [43,147×, 147 kTa]. #ᗕᕱᗒ 🔗
Thu Dec 02 19:31:23 +0000 2021@JDHL18 @KarlKadler Here are some patterns for selected collagens:
🔗
Thu Dec 02 16:09:57 +0000 2021I was just looking at the collagen 8 subunits & I am always surprised by how much hydroxy-proline/lysine there can be in a mature collagen 🔗
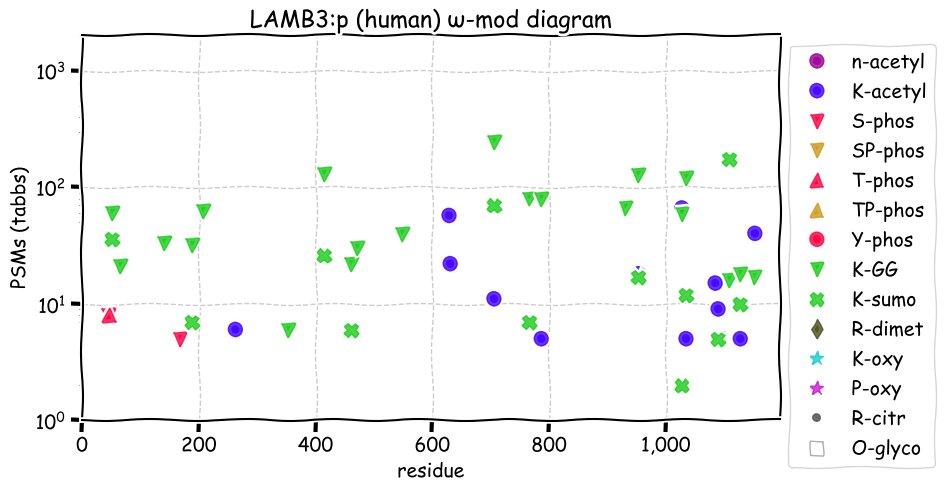
Thu Dec 02 12:46:25 +0000 2021LAMB3:p, has significantly different patterns of glycosylation, acetylation, phosphorylation, GGylation & SUMOylation compared to LAMB1 or LAMB2.
Thu Dec 02 12:46:25 +0000 2021LAMB3:p.N690S chr 1:g.209623908T>C, rs2229466 (all tissue N:S 0.959:0.041) vaf=6%, Δm=-27.0109, VAF by population group: african <1%, american 8%, east asian 26%, european 2%, south asian 10%. NS in HK-1 cells.
Thu Dec 02 12:46:25 +0000 2021LAMB3:p, θ(max) = 79. aka nicein-125kDa, kalinin-140kDa, BM600-125kDa, LAMNB1. Observed in tissue-based MHC class 1 & 2 peptide experiments. Found in most tissues & specific cell lines: absent from fluids.
Thu Dec 02 12:46:25 +0000 2021>LAMB3:p, laminin subunit beta 3 (H sapiens) >Large subunit; CTMs: 1×N+glycosyl; PTMs: 12×K+acetyl; 21×K+GGyl; 12×K+SUMOyl; 2×S, 1×T, 0×Y+phosphoryl; SAAVs: S438T (4%), V527M (26%), N690S (6%), M852L (10%), A926D (9%); mature: (18,21,22-1172) [13,793×, 147 kTa]. #ᗕᕱᗒ 🔗
Wed Dec 01 20:06:58 +0000 2021PXD027258: while I normally won't get out of bed for something that is anything less than "ultra", this mere "super" is kind of interesting.
Wed Dec 01 16:41:22 +0000 2021@astacus These guys go into some depth regarding how the specificity of chymotrypsin is achieved on a molecular level: 🔗
Wed Dec 01 15:51:00 +0000 2021@astacus V & I often get lumped in with L & M, but the β-branching really affects how they fit into tight spots.
Wed Dec 01 15:33:14 +0000 2021e.g., the % of PSMs by C-terminal AA for ELITE-RSLC012686.raw in PXD016924:
AA % of PSMs
A 0.5
C 0.5
D 0.5
E 0.07
F 25.7
G 0.2
H 2.0
I 0.2
K 1.6
L 34.0
M 2.7
N 1.1
P 0.2
Q 1.2
R 1.3
S 0.5
T 0.6
U 0
V 0.3
W 6.2
Y 20.8 🔗
Wed Dec 01 15:12:24 +0000 2021Even though the North American media is currently stressing the possible issues with a new variant, the old variants are still causing problems in Europe 🔗
Wed Dec 01 13:56:58 +0000 2021@VATVSLPR Amen, brother.
Wed Dec 01 13:13:25 +0000 2021Thanks to everyone who participated in this poll. The winner (FYWL-X) is a pretty good answer, although there is often some cleavage at M. Be sure to allow for several missed cleavage sites when using chymotrypsin (I use at least 3).
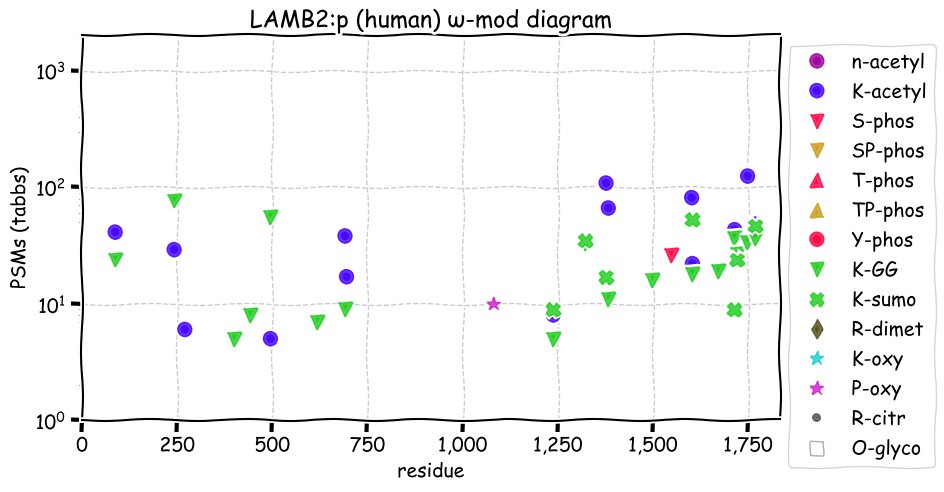
Wed Dec 01 12:59:45 +0000 2021LAMB2:p has a similar pattern of N-linked glycosylation when compared with LAMB1:p, but significantly different patterns of phosphorylation, GGylation and SUMOylation.
Wed Dec 01 12:59:44 +0000 2021LAMB2:p.A1765T chr 3:g.49121330C>T, rs74951356 (all tissue A:T 0.957:0.043) vaf=2%, Δm=30.0106, VAF by population group: african <1%, american 3%, east asian <1%, european 4%, south asian 1%.
Wed Dec 01 12:59:44 +0000 2021LAMB2:p, θ(max) = 71. aka NPHS5, LAMS. Observed in MHC class 1 & 2 peptide experiments. Observed in tissues, cell lines & fluids.
Wed Dec 01 12:59:44 +0000 2021>LAMB2:p, laminin subunit beta 2 (Homo sapiens) Large subunit; CTMs: 6×N+glycosyl; PTMs: 14×K+acetyl; 17×K+GGyl; 7×K+SUMOyl; 1×S, 0×T, 0×Y+phosphoryl; SAAVs: T398I (1%), G914R (3%), E987K (6%), A1765T (2%); mature form: (33-1798) [26,208×, 392 kTa]. #ᗕᕱᗒ 🔗
Tue Nov 30 21:43:24 +0000 2021When I want to see if a urine proteome data set is from a male subject, I usually check to see if SEMG1, SEMG2 and/or KLK3 is present. Is there a similar set of proteins that indicates female?
Tue Nov 30 19:14:04 +0000 2021nucleocapsid protein [Parainfluenza virus 5] is my least favorite protein (followed immediately by all other PIV5 proteins)
Tue Nov 30 17:49:16 +0000 2021Maybe it is just me, but it always strikes me as odd when a university professor states that their "job" is "being a scientist".
Tue Nov 30 14:59:32 +0000 2021@hecklab For example, pH = 8.0, 50 mM sodium phosphate buffer, T = 37° C?
Tue Nov 30 13:51:06 +0000 2021The pattern has shifted, so that there is another ribbon of high total precipitable water pointed at the Vancouver Island today 🔗
Tue Nov 30 13:18:51 +0000 2021@hecklab No doubt. Could you be specific?
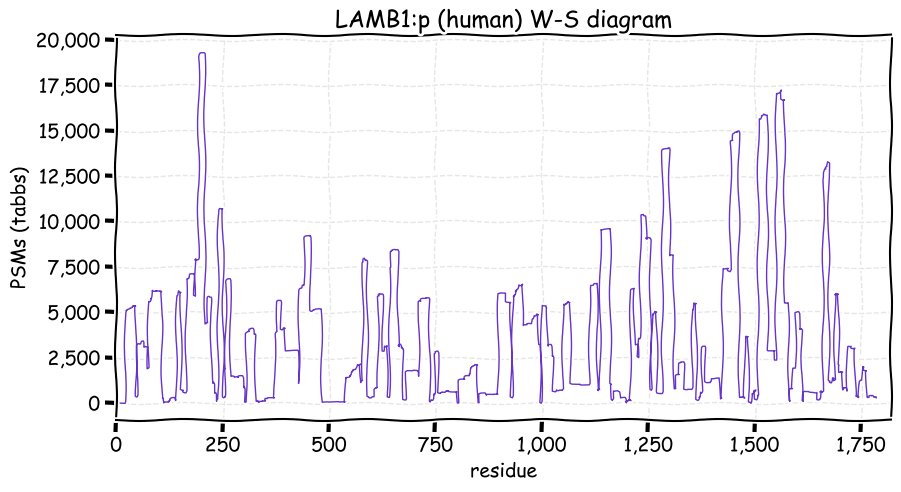
Tue Nov 30 12:59:45 +0000 2021LAMB1:p, like laminins α chains, its β chains have very little exact tryptic peptide sequence overlap: B2:B1 1.1%, B3:B1 0.6%, B4:B1 0.0%.
Tue Nov 30 12:59:45 +0000 2021LAMB1:p.Q1022R chr 7:g.107953544T>C, rs20556 (all tissue Q:R 0.762:0.238) vaf=65%, Δm=28.0425, VAF by population group: african 75%, american 66%, east asian 85%, european 59%, south asian 61%.
Tue Nov 30 12:59:45 +0000 2021LAMB1:p, θ(max) = 68. aka CLM. Observed in MHC class 1 & 2 peptide experiments. Observed in tissues, cell lines & fluids.
Tue Nov 30 12:59:44 +0000 2021>LAMB1:p, laminin subunit beta 1 (Homo sapiens) Large subunit; CTMs: 7×N+glycosyl; PTMs: 16×K+acetyl; 13×K+GGyl; 6×K+SUMOyl; 11×S, 4×T, 1×Y+phosphoryl; SAAVs: G860R (6%), Q1022R (65%), I1547T (2%); mature form: (22-1786) [32,006×, 400 kTa]. #ᗕᕱᗒ 🔗
Mon Nov 29 22:55:47 +0000 2021If you are analyzing a sample digested with chymotrypsin, which of the following cleavage pattern is the best choice?
Mon Nov 29 17:22:02 +0000 2021@graemedmoffat @antonhowes Some combination of the then recent Treaty of Paris (1763) that merged that region into Nova Scotia (finalizing the Acadian expulsion) & none-to-friendly behavior on the part of 13-Colonies-based privateers?
Mon Nov 29 15:49:54 +0000 2021@drmarcspooner @AlexUsherHESA I worked for a guy in Germany who would go postal if you didn't get prefix his name with "Prof Dr Dr" when speaking in German.
Mon Nov 29 15:41:00 +0000 2021PXD016924 combines some credible mitochondrial isolations with different protease treatments. Trypsin, Asp-N, Glu-C & chymotrypsin digests are done, in batches of 128 runs for each protease. May be particularly useful for the AI-prediction-of-RT crowd.
Mon Nov 29 15:16:43 +0000 2021It is pointed at Portland OR today 🔗
Mon Nov 29 13:46:33 +0000 2021@Raja_Nirujogi @Lange_Lab Those particular keratins aren't normally contaminants: KRT1, 2, 9 & 10 from shed human corneocytes are typical of environmental contamination.
Mon Nov 29 13:20:28 +0000 2021LAMA1:p also has a distinctive PTM pattern, when compared with LAMA3 or LAMA5: it has no lysine acetylation acceptor sites, but broadly distributed lysine GGylation acceptor sites.
Mon Nov 29 13:20:27 +0000 2021LAMA1:p.M1340V chr 18:g.7008592T>C, rs662471 (all tissue M:V 0.848:0.152) vaf=23%, Δm=-31.9721, VAF by population group: african 23%, american 19%, east asian 1%, european 30%, south asian 19%.
Mon Nov 29 13:20:27 +0000 2021LAMA1:p, θ(max) = 49. aka LAMA. Observed in MHC class 1 & 2 peptide experiments. Observed in tissues, some cell lines (HeLa & U2-OS) & extracellular vesicles.
Mon Nov 29 13:20:27 +0000 2021LAMA1:p, SAAVs: L349S (6%), N674T (15%), M1340V (23%), I1659V (11%), A1763V (14%), A1876T (14%), I2076T (27%) & T2611A (15%). 🔗
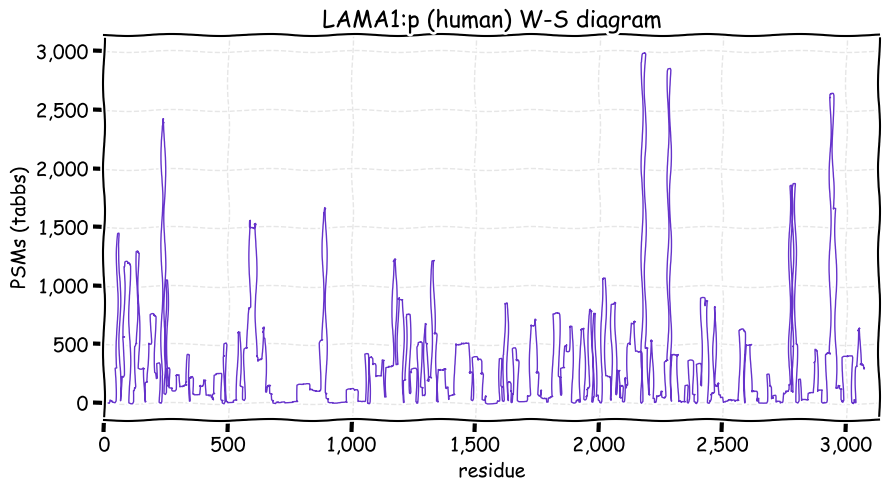
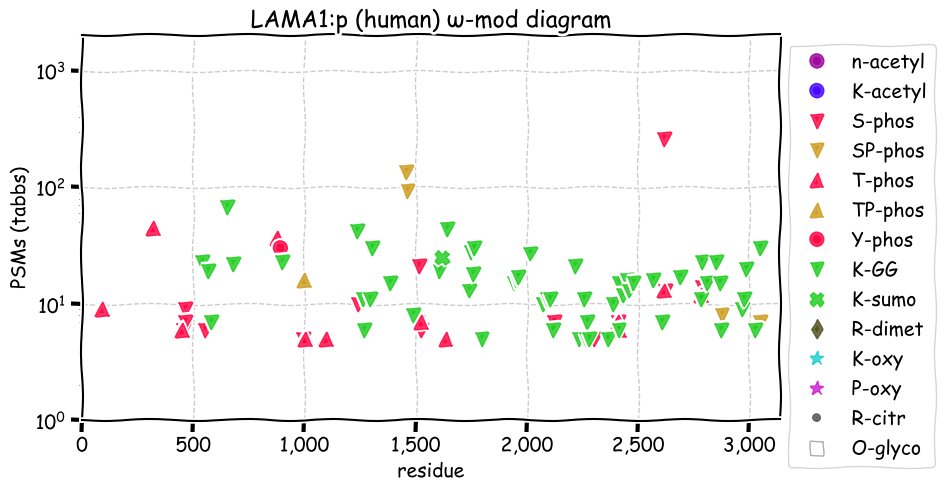
Mon Nov 29 13:20:23 +0000 2021>LAMA1:p, laminin subunit alpha 1 (Homo sapiens) Large subunit; CTMs: 5×N+glycosyl; PTMs: 0×K+acetyl; 56×K+GGyl; 21×S, 13×T, 1×Y+phosphoryl; mature form: (21?-3075) [6,127×, 70 kTa]. #ᗕᕱᗒ
Sun Nov 28 17:14:05 +0000 2021The "atmospheric river" aimed at the BC Lower Mainland right now (blue indicates elevated total precipitable water) 🔗
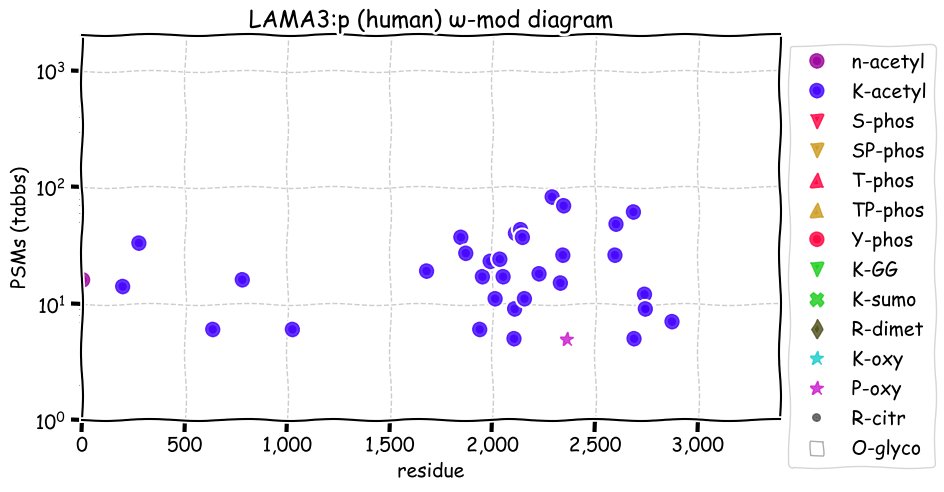
Sun Nov 28 13:10:42 +0000 2021LAMA3:p has a very different PTM pattern than LAMA5:p, with only 8 N-linked glycosylations & no GGylation or phosphorylation acceptor sites. It also has few high frequency amino acid variants by comparison.
Sun Nov 28 13:10:41 +0000 2021LAMA3:p, a shorter splice variant form, LAMA3-201 (1724 aa), is present in some cell lines―most notably MCF-10A cells.
Sun Nov 28 13:10:41 +0000 2021LAMA3:p.P1208T chr 18:g.23845027C>A, rs17202961 (all tissue P:T 0.985:0.015) vaf=94%, Δm=3.9949, VAF by population group: african 0%, american 4%, east asian 5%, european 7%, south asian 8%.
Sun Nov 28 13:10:41 +0000 2021LAMA3:p, θ(max) = 43. aka nicein-150kDa, kalinin-165kDa, BM600-150kDa, epiligrin, LAMNA. Observed in MHC class 1 & 2 peptide experiments. Observed in tissues, cell lines & blood plasma. Residue overlap with: LAMA1 0.2%, LAMA2 0.0%, LAMA4 0.2% & LAMA5 1.7%.
Sun Nov 28 13:10:41 +0000 2021>LAMA3:p, laminin subunit alpha 3 (Homo sapiens) Large subunit; CTMs: 8×N+glycosyl; PTMs: 33×K+acetyl; 0×K+GGyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: V1206A (8%), P1208T (5%), N2815K (16%); mature form: (34,35-3333) [12,819×, 135 kTa]. #ᗕᕱᗒ 🔗
Sun Nov 28 00:53:04 +0000 2021🔗
Sun Nov 28 00:51:54 +0000 2021Control protein E1B 55K [Human mastadenovirus C] is the most frequently observed viral protein, thanks to HEK-293. It will go over the 10,000 observations mark next week.
Sat Nov 27 19:59:10 +0000 2021I find it hard to disagree with this point of view 🔗
Sat Nov 27 15:58:36 +0000 2021@dtabb73 Sometimes I need an outlet to express how I feel about the farcical nature of received wisdom & ranting doesn't get me there.
Sat Nov 27 15:33:43 +0000 2021Canada bans travelers from South Africa, Namibia, Zimbabwe, Botswana, Lesotho, Eswatini, and Mozambique. 🔗
Sat Nov 27 14:44:27 +0000 2021Correction: θ(max) = 58. aka none.
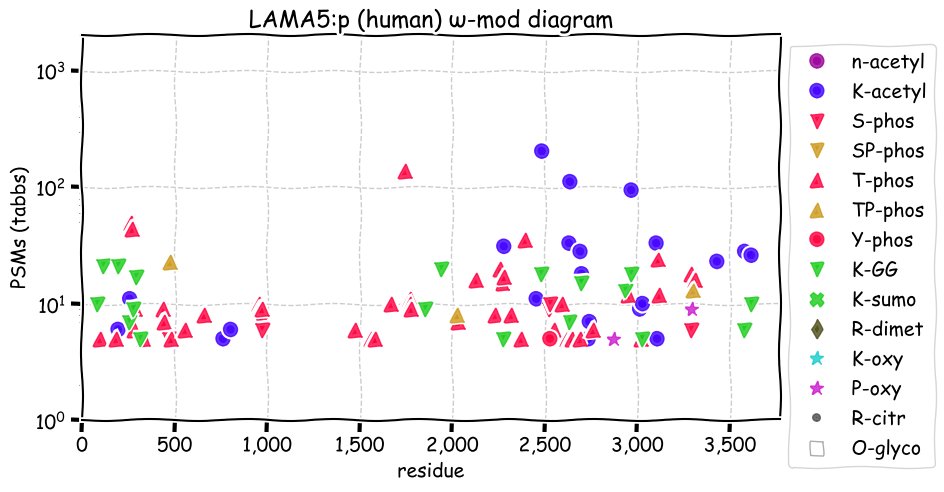
Sat Nov 27 13:57:41 +0000 2021LAMA5:p, laminins are heterotrimers of 3 subunit types: α (A), β (B) and γ (C), each of which are gene families of their own. They are named based on these gene families, so "laminin 511" is composed of LAMA5+LAMB1+LAMAC1 (α5β1γ1) 🔗
Sat Nov 27 13:47:38 +0000 2021@NAT_machinery @harrisonspecht @EMBOevents @proteintermini @DJ_Gibbs @NDissmeyer @Giglionelab @N_end_rules @mbarnalab @BukauLab @BeckmannLab @KrisGevaert_VIB @Fra_LicO2si Interesting topic.
Sat Nov 27 13:24:18 +0000 2021LAMA5:p, N-linked glycosylation is clearly doing some heavy lifting here, although its role in the function of the subunit has not been determined.
Sat Nov 27 13:18:23 +0000 2021LAMA5:p.T401A chr 20:g.62346587T>C, rs4925229 (all tissue T:A 0.019:0.981) vaf=94%, Δm=-30.0106, VAF by population group: african 61%, american 85%, east asian 96%, european 98%, south asian 97%.
Sat Nov 27 13:18:22 +0000 2021LAMA5:p, θ(max) = 99. aka B23, NPM. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in tissues, cell lines & extracellular vesicles. Has low levels of observable residue overlap with LAMA3 (1.6%) & LAMA1 (0.2%).
Sat Nov 27 13:18:22 +0000 2021LAMA:p, SAAVs (>10%): T401A (94%), V889M (11%), T956A (72%), M1258T (76%), F1807S (89%), V1900M (34%), A1908T (22%), H2036R (20%), R2226H (20%), R3079W (32%), R3085W (32%). It is very unlikely that the reference sequence will be present in any individual.
Sat Nov 27 13:18:22 +0000 2021>LAMA5:p, laminin subunit alpha 5 (Homo sapiens) Large subunit; CTMs: 18×N+glycosyl; PTMs: 21×K+acetyl; 18×K+GGyl; 6×S, 53×T, 1×Y+phosphoryl; mature form: (36-3695) [26,978×, 451 kTa]. #ᗕᕱᗒ 🔗
Fri Nov 26 16:59:20 +0000 2021@UCDProteomics Unfortunately, the PRIDE data hasn't been released yet.
Fri Nov 26 15:43:17 +0000 2021What are the best terms (& associated parameters) to describe the degree of overlap between sequences that may interfere with making a unique protein identification?
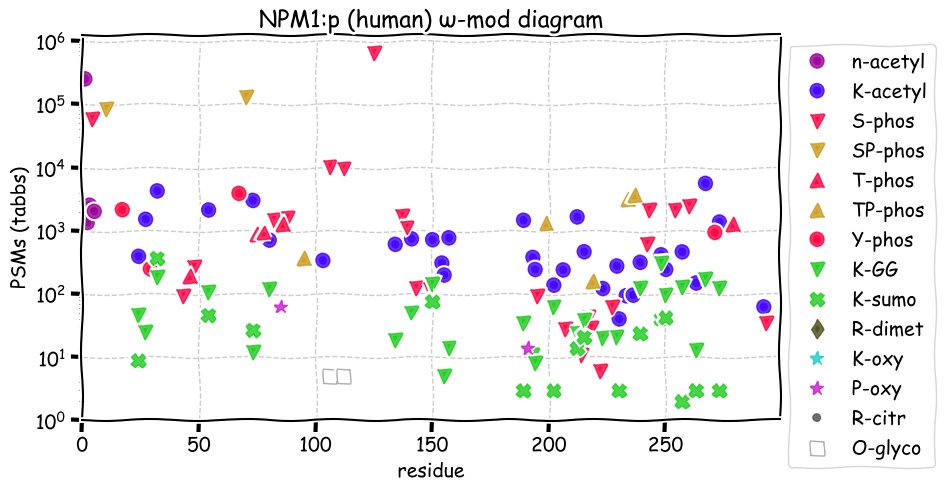
Fri Nov 26 15:26:48 +0000 2021NPM1:p, there is good evidence of translation initiation at either M1 or M5 in human and mouse: the M1 form predominates in both species.
Fri Nov 26 15:26:48 +0000 2021NPM1:p, is a good example of site-sharing alternate modification, sharing all 26 K+GGylation sites with K+acetylation & all 16 K+SUMOylation sites with K+GGylation and/or K+aceytlation. This pattern is completely different from NPM3:p.
Fri Nov 26 15:26:48 +0000 2021NPM1:p.A249V chr 5:g.171405378C>T, rs377348662 (all tissue A:V 0.999:0.001) vaf=<1%, Δm=28.0313, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Fri Nov 26 15:26:47 +0000 2021NPM1:p, θ(max) = 99. aka B23, NPM. Observed in MHC class 1 peptide experiments & class 2 in tissues. Commonly observed in tissues & cell lines: rare in fluids.
Fri Nov 26 15:26:47 +0000 2021>NPM1:p, nucleophosmin 1 (Homo sapiens) Small subunit; CTMs: M1, M5+acetyl; PTMs: 2×S+glycosyl; 33×K+acetyl; 26×K+GGyl; 16×K+SUMOyl; 26×S, 10×T, 4×Y+phosphoryl; SAAVs: A249V (<1%); mature form: (1,5-294) [120,726×, 3274 kTa]. #ᗕᕱᗒ 🔗
Fri Nov 26 14:51:04 +0000 2021White-knuckling my way through an unnecessary reorganization of my web server configuration files to make them more "elegant".
Thu Nov 25 16:01:47 +0000 2021@JeongKyowon Develop with yourself in mind: external users are fleeting & open source co-development is mainly wishful thinking.
Thu Nov 25 13:05:36 +0000 2021A Thanksgiving Day storm running ahead of a cold front from Houston to Cleveland 🔗
Thu Nov 25 12:50:08 +0000 2021SRA1:p is only one of the products produced from the gene SRA1:g, which can also produce biologically active lncRNA as illustrated here 🔗
Thu Nov 25 12:50:08 +0000 2021SRA1:p, there is good evidence of translation initiation at either M1 or M13: the M13 form predominates in human.
Thu Nov 25 12:50:08 +0000 2021SRA1:p.V110R chr 10:g.140552044_140552045insTCG, rs5871740 (all tissue V:R 0.427:0.573) vaf=66%, Δm=57.0327, VAF by population group: african 22%, american 51%, east asian 56%, european 48%, south asian 48%. The gene variant is a frameshifting insertion.
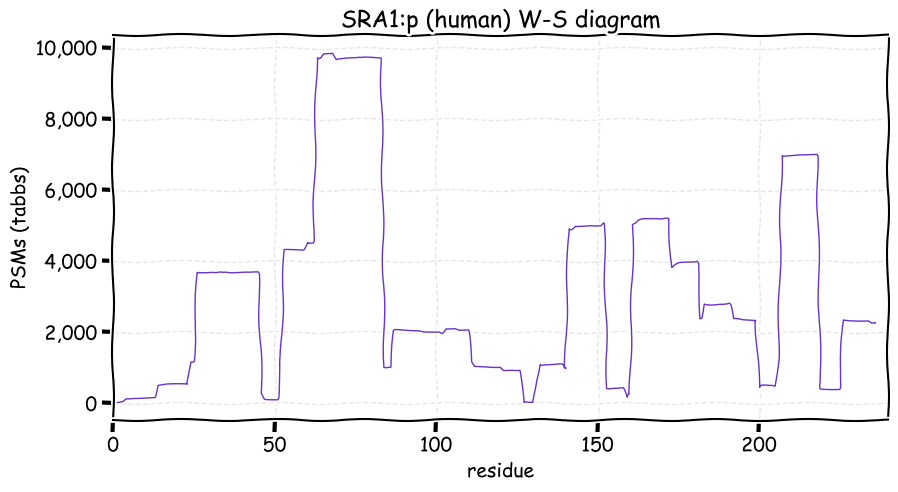
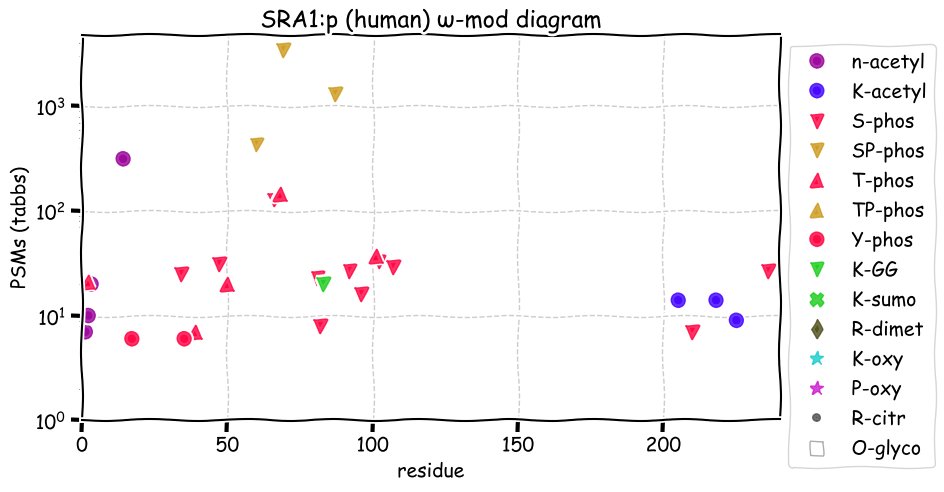
Thu Nov 25 12:50:08 +0000 2021SRA1:p, θ(max) = 71. aka SRA, STRAA1. Observed in MHC class 1 peptide experiments. Commonly observed in tissues & cell lines: rare in fluids. 99% of phosphorylated PSMs were observed in the N-terminal low complexity domain (14-107).
Thu Nov 25 12:50:08 +0000 2021>SRA1:p, steroid receptor RNA activator 1 (Homo sapiens) Small subunit; CTMs: A2, A14+acetyl; PTMs: 3×K+acetyl; 1×K+GGyl; 14×S, 5×T, 2×Y+phosphoryl; SAAVs: V110R (66%); mature form: (2,14-236) [12,715×, 48 kTa]. #ᗕᕱᗒ 🔗
Wed Nov 24 14:35:15 +0000 2021The 2 forms of the protein caused by alternate initiation may (at least in part) explain the ongoing nucleophosmin/nucleoplasmin feud.
Wed Nov 24 13:58:14 +0000 2021@mjmaccoss @lukas_k You would have to store more info about the decoy distributions & not simply the highest score: at least a histogram of the high scoring tail & the # of sequences tested. Then normalize that E distribution into a p distribution & re-scale it for use with other analyses.
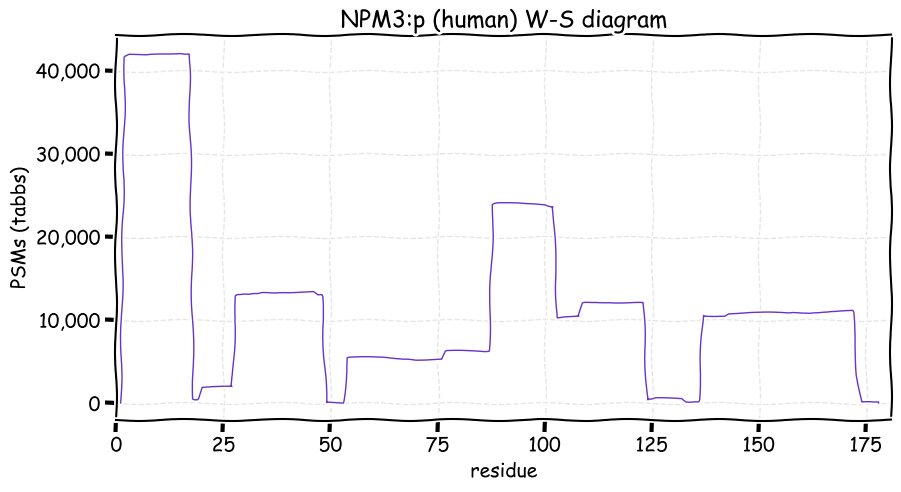
Wed Nov 24 12:28:34 +0000 2021NPM3:p, understanding of this protein is about where it was when it was discovered 🔗, even though it is frequently observed in protein-protein interaction studies.
Wed Nov 24 12:28:34 +0000 2021NPM3:p, There is good evidence of translation initiation at either M1 or M34 in human and mouse: the M1 form predominates in both species.
Wed Nov 24 12:28:34 +0000 2021NPM3:p.A3T chr 10:g.101783384C>T, rs146931276 (all tissue A:T 0.999:0.001) vaf=0.1%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Wed Nov 24 12:28:34 +0000 2021NPM3:p, θ(max) = 60. aka none. Very rarely observed in MHC peptide experiments. Commonly observed in tissues & cell lines: absent from fluids but may be present in some types of extracellular vesicle.
Wed Nov 24 12:28:33 +0000 2021>NPM3:p, nucleophosmin/nucleoplasmin 3 (Homo sapiens) Small subunit; CTMs: A2, M34+acetyl; PTMs: 0×K+acetyl; 1×K+GGyl; 1×K+SUMOyl; 9×S, 5×T, 0×Y+phosphoryl; SAAVs: A3T (<1%); mature form: (2,34-178) [34,033×, 115 kTa]. #ᗕᕱᗒ 🔗
Tue Nov 23 17:59:07 +0000 2021@mjmaccoss @lukas_k I was referring to the expectation-value distribution of decoy high-scoring values, rather than the p-value for individual spectra. If you want to re-use that distribution, you have to normalize it into a probability of decoy high-scoring values, which is somewhat more involved.
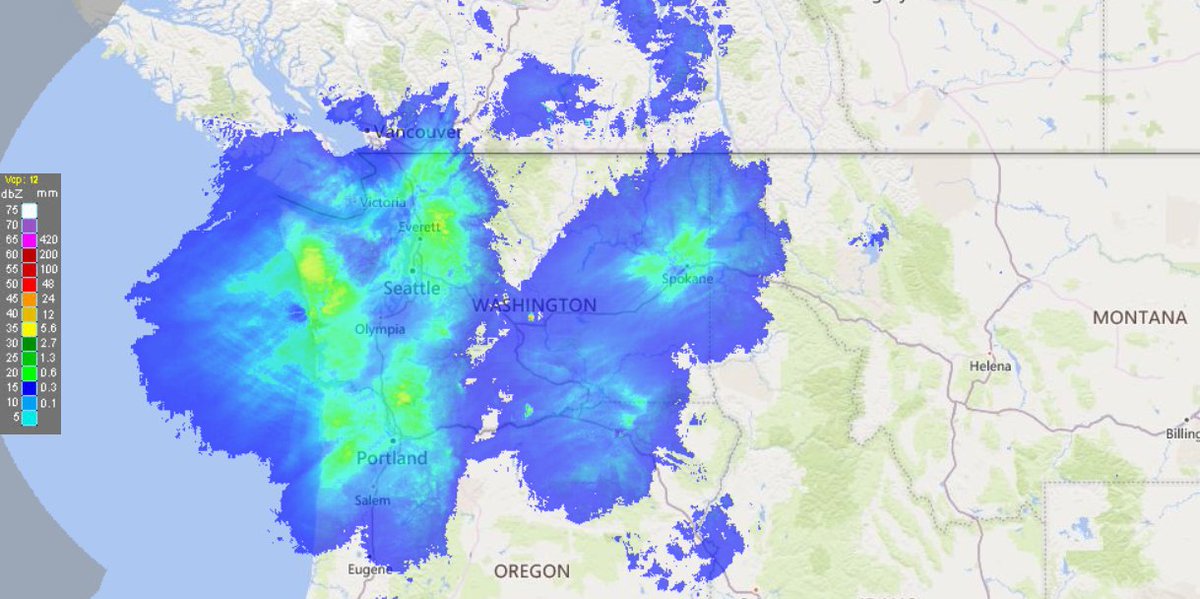
Tue Nov 23 16:38:52 +0000 2021A rainy day (last 24 hrs, estimated from radar density) in a region that doesn't need any rain at the moment 🔗
Tue Nov 23 15:41:36 +0000 2021Northern Europe seems to be well down the wrong road wrt infection rates 🔗
Tue Nov 23 15:31:47 +0000 2021Thanks to everyone who participated. Ironically, even though this method is in very wide use, the poll resulted in a statistical tie (std dev ≈ ± 15%). It would appear that the olds haven't done a great job of transferring how stuff works to the youngs.
Tue Nov 23 14:11:57 +0000 2021@lukas_k I suspect the problem's origin is that most stats courses emphasize the probability formulation of problems because the math is simpler, even though the E-values are much more important in scientific applications.
Tue Nov 23 14:09:41 +0000 2021@lukas_k There is a common misunderstanding of how to treat results that depend on expectation values (like TD FDR) that really depend on "how you got there", as opposed to those from a normalized probability calculation, which are much more portable.
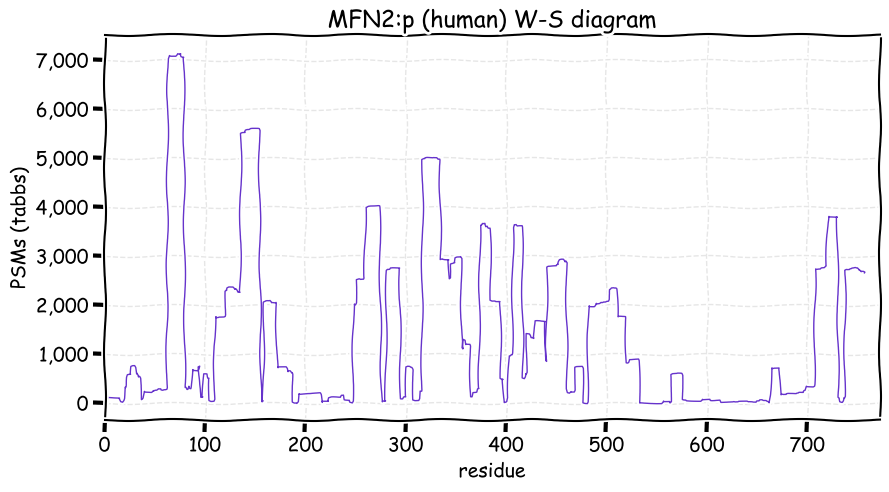
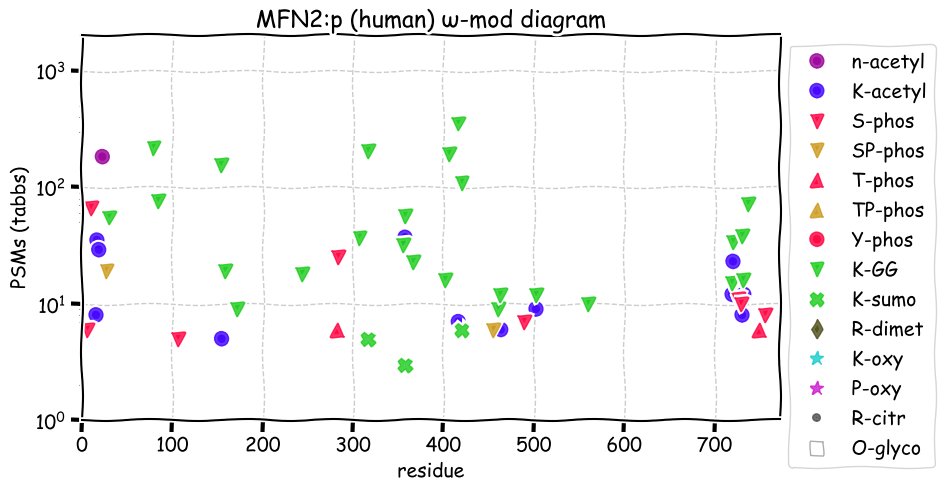
Tue Nov 23 12:56:41 +0000 2021MFN2:p, TM domains: (605–625) & (628-647). There is good evidence of translation initiation at either M1 or M21. The protein in a GTPase that participates in the dynamic structure of mitochondrial networks 🔗
Tue Nov 23 12:56:41 +0000 2021MFN2:p.A716T chr 1:g.12009668G>A, rs144860227 (all tissue A:T 0.997:0.003) vaf=0.1%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Tue Nov 23 12:56:41 +0000 2021MFN2:p, θ(max) = 59. aka CPRP1, KIAA0214, MARF, CMT2A2. Observed in class 1 MHC peptide expts. & rarely in class 2. Commonly observed in tissues & cell lines: very rare in fluids.
Tue Nov 23 12:56:40 +0000 2021>MFN2:p, mitofusin 2 (Homo sapiens) Midsized subunit; CTMs: S2, A22+acetyl; PTMs: 13×K+acetyl; 25×K+GGyl; 3×K+SUMOyl; 10×S, 2×T, 0×Y+phosphoryl; SAAVs: A716T (<1%); mature form: (2,22-757) [16,250×, 78 kTa]. #ᗕᕱᗒ 🔗
Tue Nov 23 03:38:27 +0000 2021A lot of NEXRAD ground clutter throughout the US mid-west this evening 🔗
Mon Nov 22 20:19:31 +0000 2021I'm looking at a Q Exactive HF data set that the authors say they analyzed with a fragment ion mass tolerance of 0.005 Da, which seems a bit too tight to me. Does anybody know of a benefit to having such a tolerance when using MaxQuant?
Mon Nov 22 19:03:14 +0000 2021@WinnipegNews The guns seized in Canadian drug raids often have a "Mad Max" quality to them.
Mon Nov 22 16:36:55 +0000 2021@mjmaccoss @thabangh @lukas_k It doesn't help that many of the commonly used implementations of the method are proprietary.
Mon Nov 22 15:12:14 +0000 2021Do you believe that proteomics use of the target-decoy FDR is the equivalent of the standard statistical method or it is something fundamentally different that simply shares the name:
Mon Nov 22 15:08:51 +0000 2021The alternate opinion was that there was no real connect, beyond the name and that ratio being taken. Since me getting things wrong is a literature sub-genre all its own, I'd like to hear from the community of users of this method.
Mon Nov 22 15:06:41 +0000 2021In a thread over the weekend, there was an airing of opinions about the underlying theory associated with the target-decoy FDR calculation. I asserted that it is fundamentally the same as the FDR described here 🔗
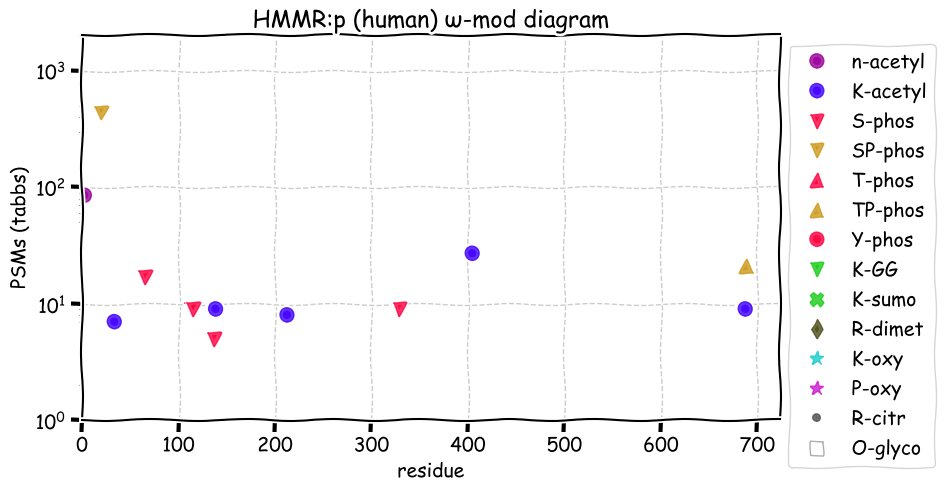
Mon Nov 22 12:57:33 +0000 2021HMMR:p, the idea that the protein is a soluble hyaluronan receptor is currently controversial 🔗
Mon Nov 22 12:52:09 +0000 2021HMMR:p.V353A chr 5:g.163475510T>C, rs299290 (all tissue V:A 0.672:0.328) vaf=28%, Δm=-28.0313, VAF by population group: african 29%, american 26%, east asian 48%, european 26%, south asian 25%. AA in HeLa cells.
Mon Nov 22 12:52:09 +0000 2021HMMR:p, θ(max) = 59. aka RHAMM, CD168. Observed in class 1 MHC peptide expts. Commonly observed in tissues & cell lines: rare in fluids.
Mon Nov 22 12:52:09 +0000 2021>HMMR:p, hyaluronan mediated motility receptor (Homo sapiens) Midsized subunit; CTMs: S2+acetyl; PTMs: 5×K+acetyl; 6×S, 1×T, 0×Y+phosphoryl; SAAVs: A240V (1%), R317H (2%), V353A (28%), A469V (78%); mature form: (2-709) [7,258×, 29 kTa]. #ᗕᕱᗒ 🔗
Sun Nov 21 23:14:06 +0000 2021@pwilmarth @fcyucn @GeiszlerDaniel I guess we'll have to agree to disagree about this one. To me, the stats of proteomics interpretation have been annoying & nuanced, but still just main stream stats. In the same way a std dev doesn't mean much with a distribution, a +ve/-ve formulation needs a hypothesis.
Sun Nov 21 17:28:10 +0000 2021@GeiszlerDaniel @fcyucn "Any H₀ negative PSM is a correct assignment" is a postulate (not a hypothesis) in this case because it can't be tested using the target-decoy approach
Sun Nov 21 17:24:56 +0000 2021@GeiszlerDaniel @fcyucn Yes. After making those tests, the H₀ negative results are re-framed using the postulate:
any H₀ negative PSM is a correct assignment.
Then you change TN→TP and FN→FP and you get the commonly used "FDR" language for describing the resulting set of PSMs.
Sun Nov 21 15:46:06 +0000 2021@GeiszlerDaniel In this common case, you make the simplifying assumption that any PSM with a score lower than the highest score indicates a positive H₀, so these the peptides for these PSMs are by definition random sequences.
Sun Nov 21 15:27:53 +0000 2021@GeiszlerDaniel It means that the test you perform on every PSM (10³ to 10⁵ for each spectrum) is "Is it consistent with H₀?". If it is consistent with H₀, it is discarded. If not, it is saved for further analysis. More than 99.9% of PSMs are consistent with H₀.
Sun Nov 21 14:20:21 +0000 2021UBAP2:p, there is evidence of translation initiation at either M1 or M2, although M2 seems to predominate.
Sun Nov 21 14:20:21 +0000 2021UBAP2:p.R14Q chr 9:g.34017108C>T, rs1785506 (all tissue R:Q 0.017:0.983) vaf=64%, Δm=-28.0425, VAF by population group: african 49%, american 73%, east asian 83%, european 62%, south asian 55%. QQ in JURKAT, HCT-116 & K-562 cells. RQ in HEK-293 cells.
Sun Nov 21 14:20:21 +0000 2021UBAP2:p, θ(max) = 51. aka KIAA1491, bA176F3.5, FLJ22435. Observed in class 1 MHC peptide expts & class 2 peptides from small intestine. Commonly observed in tissues & cell lines: absent from fluids.
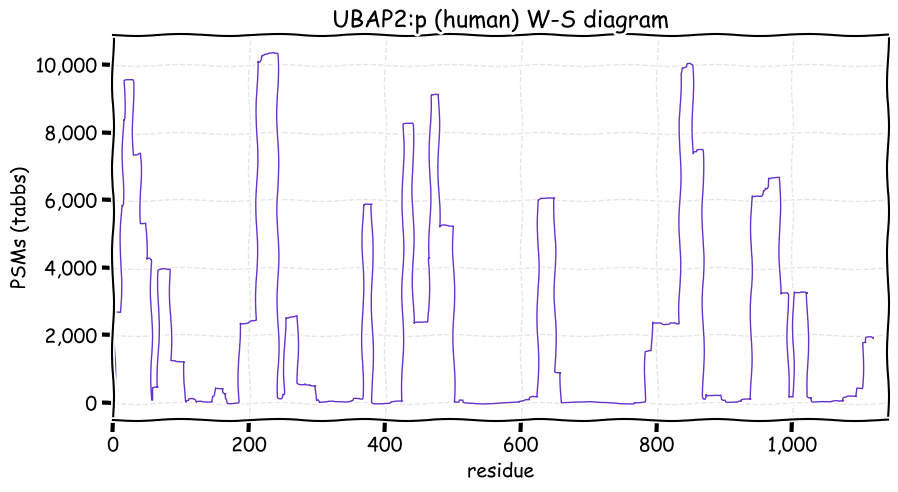
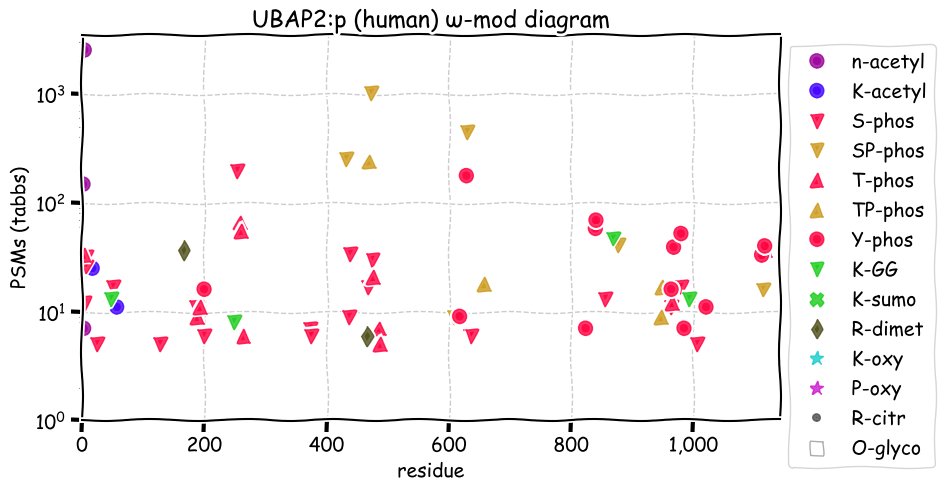
Sun Nov 21 14:20:21 +0000 2021>UBAP2:p, ubiquitin associated protein 2 (Homo sapiens) Large subunit; CTMs: M1, T3+acetyl; PTMs: 3×K+acetyl; 4×K+GGyl; 27×S, 15×T, 13×Y+phosphoryl; 2×R+dimethyl; SAAVs: R14Q (64%); mature form: (1,3-1119) [21,200×, 123 kTa]. #ᗕᕱᗒ 🔗
Sat Nov 20 20:23:07 +0000 2021Thanks to everyone for their comments. Based on the discussion it looks like this would be the null hypothesis being tested by the target-decoy simulation
H₀: peptide sequences are assigned at random 🔗
Sat Nov 20 16:07:01 +0000 2021@pwilmarth @fcyucn @wfondrie A little off the point: I am not looking for an argument that the target-decoy FDR method is a good (or bad) estimate. I was hoping to find a formulation of the problem that would be a suitable answer to the sort of question a student might be asked in a thesis oral exam.
Sat Nov 20 14:53:26 +0000 2021@fcyucn @wfondrie I was hoping that some one who teaches the target-decoy FDR idea in a course had created a clear formal statement of the problem, suitable for explaining it to people from outside of the cognoscenti
Sat Nov 20 14:45:41 +0000 2021@fcyucn @wfondrie That one comes closest so far, with the statement (although not really talking about the target-decoy FDR calculation):
"a p-value relative to the null hypothesis that the peptide is drawn randomly from all possible sequences."
Sat Nov 20 13:13:57 +0000 2021@fcyucn @wfondrie The paper from Bill Noble's group mentions several null hypotheses, but never explicitly defines them. The Levitsky paper doesn't mention it at all, simply classifying PSMs as either true or false.
Sat Nov 20 13:05:14 +0000 2021@wfondrie @GeiszlerDaniel @sangtaek That one isn't really part of the target-decoy FDR literature: it is concerned with assigning probabilities to each spectrum & it explicitly eschews the use of a null hypothesis.
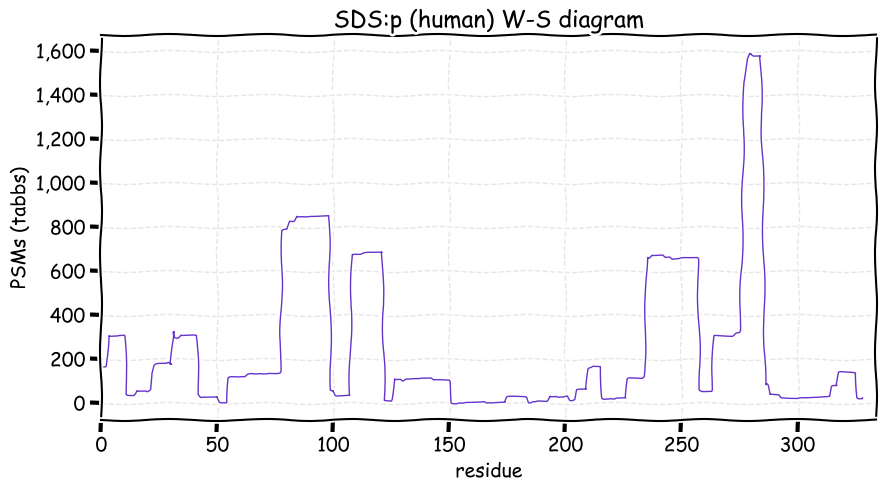
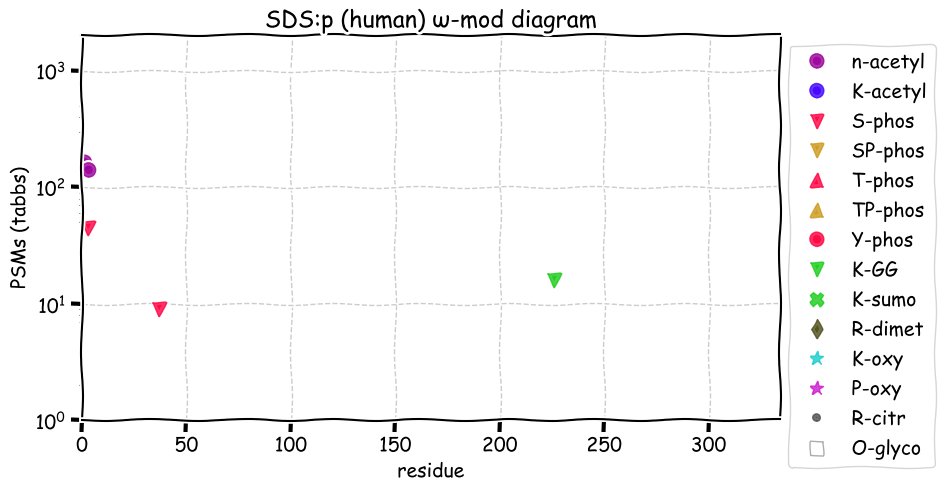
Sat Nov 20 12:59:52 +0000 2021SDS:p., there is evidence of translation initiation at either M1 or M2. Is SDS really a good name for a protein?
Sat Nov 20 12:59:52 +0000 2021SDS:p.A116T chr 12:g.113398594C>T, rs749693153 (all tissue A:T 0.997:0.003) vaf=<1%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. AT in HEK-293T cells.
Sat Nov 20 12:59:52 +0000 2021SDS:p, θ(max) = 61. aka SDH. Observed in class 1 MHC peptide expts & class 2 peptides from bone marrow. Most abundant in liver & liver-derived cell lines: absent from most fluids.
Sat Nov 20 12:59:51 +0000 2021>SDS:p, serine dehydratase (Homo sapiens) Small subunit; CTMs: M1, S3+acetyl; PTMs: 1×K+GGyl; 2×S, 0×T, 0×Y+phosphoryl; SAAVs: A116T (<1%); mature form: (1,3-328) [1,884×, 6 kTa]. #ᗕᕱᗒ 🔗
Fri Nov 19 20:03:55 +0000 2021@wfondrie It is an interesting approach, but it doesn't frame the problem around the usual 2x2 confusion matrix formulation, i.e., using a null hypothesis that is either confirmed or rejected.
Fri Nov 19 19:08:15 +0000 2021Is there a standard formulation of the null hypothesis being tested by the proteomics target-decoy simulation method? I've looked around a bit & I can't seem to find one.
Fri Nov 19 17:08:04 +0000 2021@BiswapriyaMisra That is hard to evaluate: most people think of mitochondria as little bean-shaped organelles, but mitochondria are normally extended networks that are difficult to count 🔗
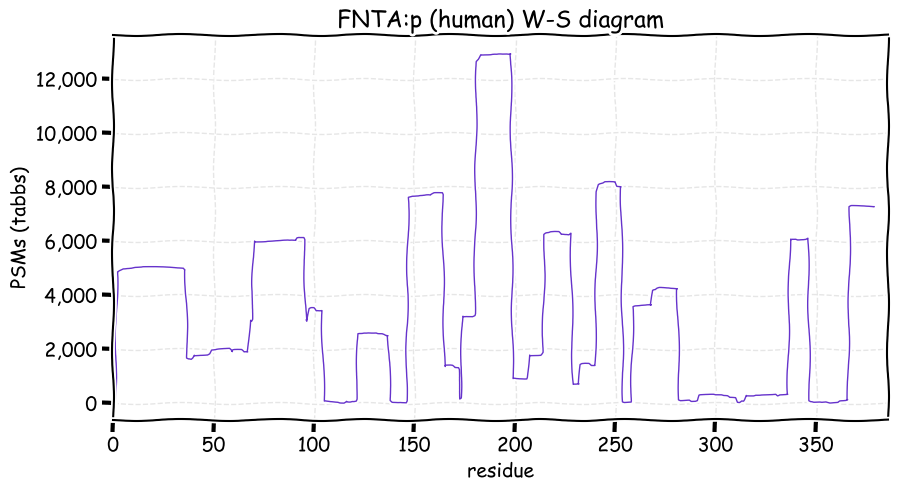
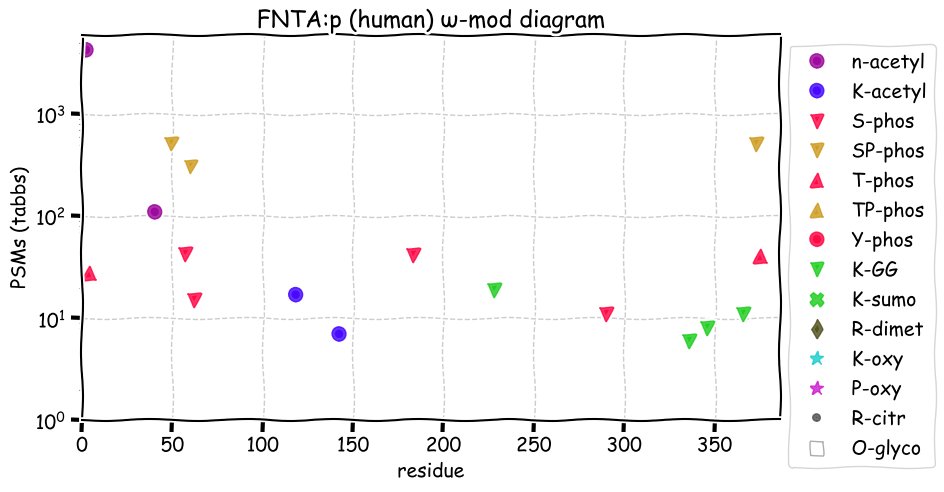
Fri Nov 19 16:09:29 +0000 2021FNTA:p, has no tryptic peptide overlap with the very similar sounding protein FNTB:p (farnesyltransferase, CAAX box, beta)
Fri Nov 19 13:56:08 +0000 2021FNTA:p, it is unknown if the M39 form of the enzyme has a different specificity than the M1 form, but the M39 form is missing a low complexity, proline-rich domain on the N-terminus that looks very much like a protein-protein interaction domain. 🔗

Fri Nov 19 13:11:25 +0000 2021FNTA:p.V64I chr 8:g.43056536G>A, rs143321089 (all tissue V:I 0.997:0.003) vaf=0.2%, Δm=14.0156, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Fri Nov 19 13:11:25 +0000 2021FNTA:p, θ(max) = 74. aka PTA, PGGT1A, PTAR2. Observed in class 1 MHC peptide expts. Observed in tissues & cell lines: absent from most fluids, except urine extracellular vesicles. There is good evidence for initiation at either M1 or M39.
Fri Nov 19 13:11:24 +0000 2021>FNTA:p, farnesyltransferase, CAAX box, alpha (Homo sapiens) Small subunit; CTMs: A2, A40+acetyl; PTMs: 2×K+acetyl; 4×K+GGyl; 7×S, 2×T, 0×Y+phosphoryl; SAAVs: V64I (<1%); mature form: (2,40-379) [20,527×, 85 kTa]. #ᗕᕱᗒ 🔗
Thu Nov 18 18:50:42 +0000 2021@r_l_moritz The files in PRIDE don't include a list connecting tissue information (e.g, disease state) with the raw data file names.
Thu Nov 18 18:30:09 +0000 2021@pwilmarth My main use of the approach is to determine the PSM FPRs in sub-populations, such as the error rate for a particular protein, hypothesis or PTM. It is a great way to determine overall FPRs, but that is more for algorithm development.
Thu Nov 18 16:47:19 +0000 2021@JooSeopPark Active biotin uptake in proximal tubules may be part of the effect 🔗
Thu Nov 18 16:25:40 +0000 2021In my hands, the histogram of observed-calculated parent ion masses is the most useful distribution for assessing PSM assignment error rates in proteomics results. It is always my go-to method.
Thu Nov 18 15:43:34 +0000 2021@lukas_k @r_l_moritz (or any of the co-authors) should be able to address that issue pretty easily. The lack of metadata tying data files to experimental conditions is so common I don't really even notice it any more.
Thu Nov 18 15:00:00 +0000 2021Just to be clear, I am talking about the usefulness & QA characteristics of the data set rather than any particular conclusions that may have been drawn from it.
Thu Nov 18 14:50:53 +0000 2021If you are interested in the problems & benefits of the systematic study of proteins obtained from clinical tissue samples, PXD024124 is a really good resource 🔗 (⭐️⭐️⭐️⭐️)
Thu Nov 18 12:50:04 +0000 2021Is the current outbreak in northwestern Europe caused by a new variant, behavior changes or something else? 🔗
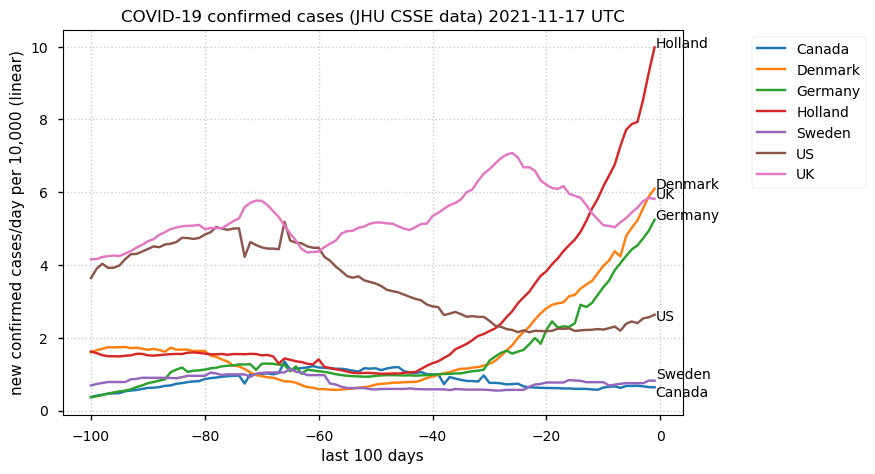
Thu Nov 18 12:46:13 +0000 2021SYK:p, good evidence for translation initiation at M1 and M6 in human: only M1 present in mouse or rat.
Thu Nov 18 12:46:13 +0000 2021SYK:p.A10T chr 9:g.90843926G>A, rs199606629 (all tissue A:T 0.997:0.003) vaf=<1%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Thu Nov 18 12:46:13 +0000 2021SYK:p, θ(max) = 84. aka none. Observed in class 1 & 2 MHC peptide expts. Observed in tissues, platelets, lymphocytes & leukocytes: common in urine & blood plasma. Has a lot of Y+phosphoryl acceptor sites.
Thu Nov 18 12:46:12 +0000 2021>SYK:p, spleen tyrosine kinase (Homo sapiens) Midsized subunit; CTMs: A2, A7+acetyl; PTMs: 1×K+acetyl; 7×K+GGyl; 14×S, 7×T, 18×Y+phosphoryl; SAAVs: A10T (<1%); mature form: (23-635) [16,256×, 221 kTa]. #ᗕᕱᗒ 🔗
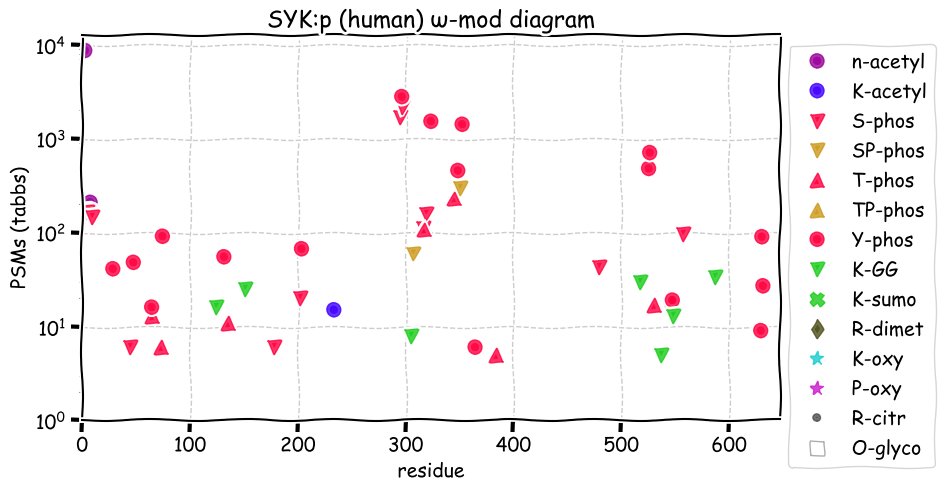
Wed Nov 17 17:27:39 +0000 2021@andreafossati9 The P's & Q's also seem to conspire to producing a nice crop of b-y type internal fragment ions in the spectra (in addition to summoning Nyarlathotep).
Wed Nov 17 16:47:15 +0000 2021(acetyl)AATEGVGEAAQGGEPGQPAQPPPQPHPPPPQQQHK: commonly observed tryptic peptide or Lovecraftian incantation that summons an eldritch horror?
Wed Nov 17 15:53:50 +0000 2021@slashdot Probably an interesting case study for copyright law students: is the cryptographic hash of a digital photograph of copyright-protected text also a protected, derived-work?
Wed Nov 17 15:47:19 +0000 2021The files with names containing "Ross_Birsoy_SPS-MS3_TMT" in PXD027673 give an unusually detailed peak inside of human mitochondria & use the TMTpro quantitation reagent.
Wed Nov 17 13:04:41 +0000 2021MIA3:p, TANGO1 (transport and Golgi organization protein 1) is probably a better name for this protein/gene given its subcellular localization and the current understanding of its function.
Wed Nov 17 12:22:04 +0000 2021MIA3:p.K605R chr 1:g.222629034A>G, rs2936052 (all tissue K:R 0.867:0.133) vaf=24%, Δm=28.0061, VAF by population group: african 32%, american 40%, east asian 31%, european 17%, south asian 30%. KR in Hep-G2 cell. RR in NTERA-2 cells.
Wed Nov 17 12:22:04 +0000 2021MIA3:p, θ(max) = 65. aka UNQ6077, FLJ39207, KIAA0268, TANGO, TANGO1. Observed in class 1 & 2 MHC peptide expts. Observed in tissues & cell lines: common in urine & extracellular vesicles. MIA3 is short for Melanoma Inhibitory Activity family, member 3.
Wed Nov 17 12:22:04 +0000 2021>MIA3:p, MIA SH3 domain ER export factor 3 (Homo sapiens) Large subunit; CTMs: Q23+ammonia-loss; N246, N250, N589+glycosyl PTMs: 13×K+acetyl; 6×K+GGyl; 76×S, 33×T, 1×Y+phosphoryl; SAAVs: K605R (24%), E881G (32%); mature form: (23-1907) [30,293×, 231 kTa]. #ᗕᕱᗒ 🔗
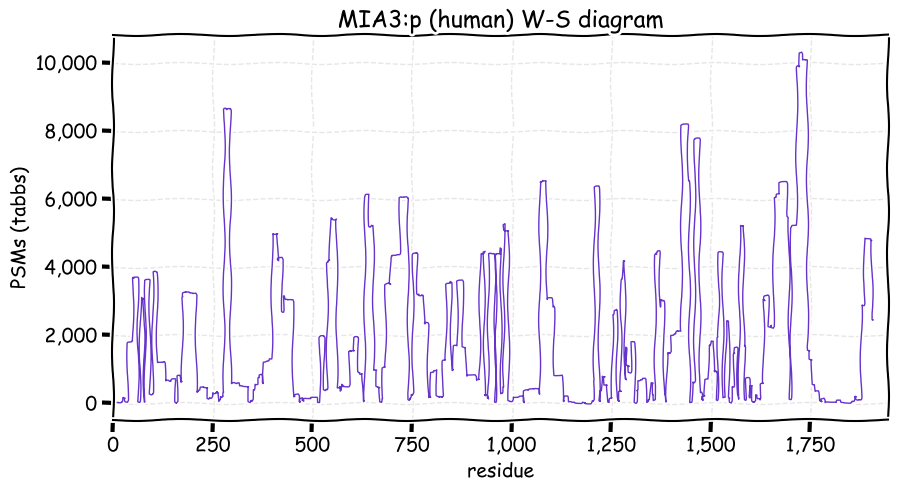
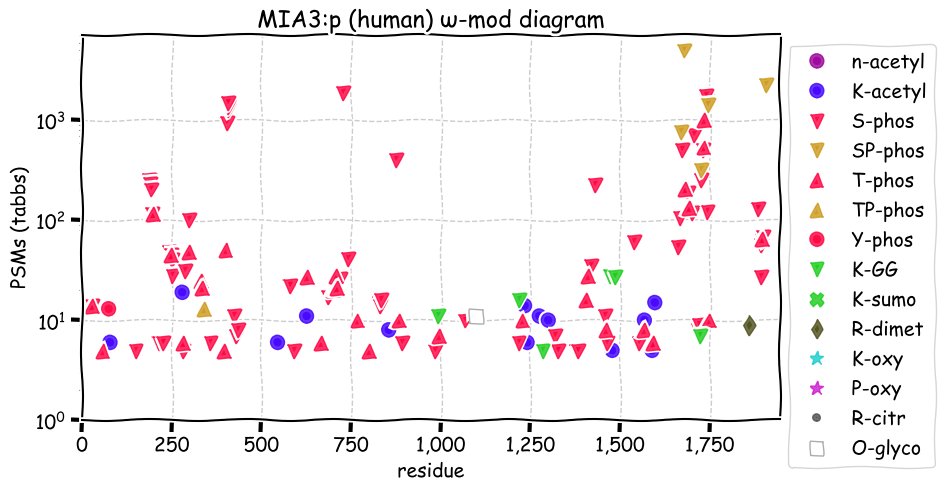
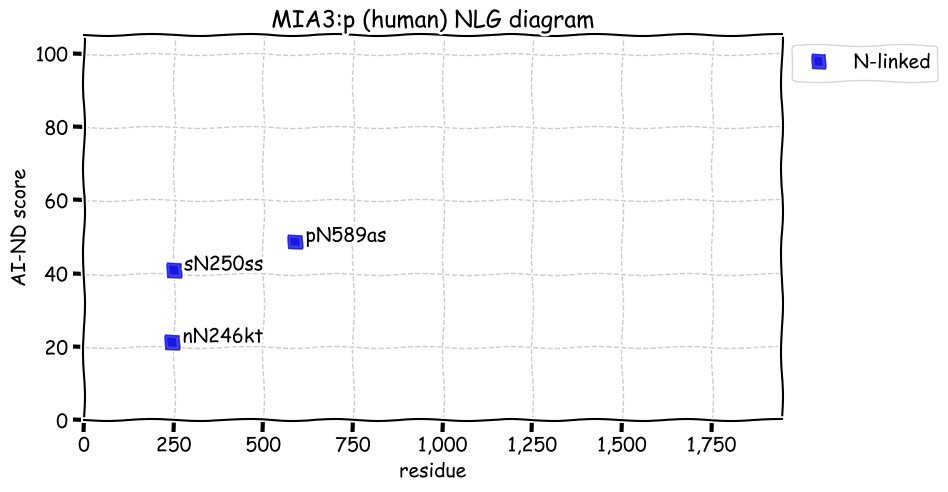
Tue Nov 16 20:28:21 +0000 2021@pwilmarth @nesvilab Steve Carr's mob is the only group I know of that consistently does IAA alkylation. Predictably, they are also the only ones that get good recovery of C-containing HLA peptides.
Tue Nov 16 15:20:36 +0000 2021There is an odd anti-correlation between named domains in proteins and PTMs: most named domains have very few PTMs, while adjacent unnamed domains are often bristling with them.
Tue Nov 16 12:50:14 +0000 2021GFER:p.F166L chr 16:g.1985906T>C, rs36041021 (all tissue F:L 0.937:0.063) vaf=7%, Δm=-33.9843, VAF by population group: african 1%, american 5%, east asian <1%, european 9%, south asian 2%. LL in DU-145 cells. FL in CACO-2 cells.
Tue Nov 16 12:50:14 +0000 2021GFER:p, θ(max) = 69. aka HSS, ERV1, ALR, HERV1, HPO1, HPO2. Observed in class 1 MHC peptide expts. Observed in tissues & cell lines: absent from fluids.
Tue Nov 16 12:50:14 +0000 2021>GFER:p, growth factor, augmenter of liver regeneration (Homo sapiens) Small subunit; CTMs: A2, M26+acetyl; PTMs: 3×K+acetyl; 7×K+GGyl; 5×S, 0×T, 0×Y+phosphoryl; SAAVs: F166L (7%); mature form: (2,26-205) [7,284×, 20 kTa]. #ᗕᕱᗒ 🔗
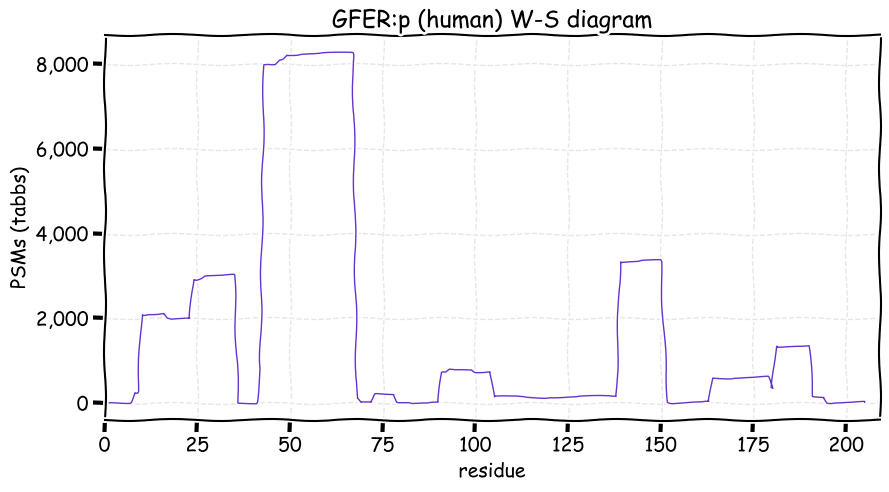
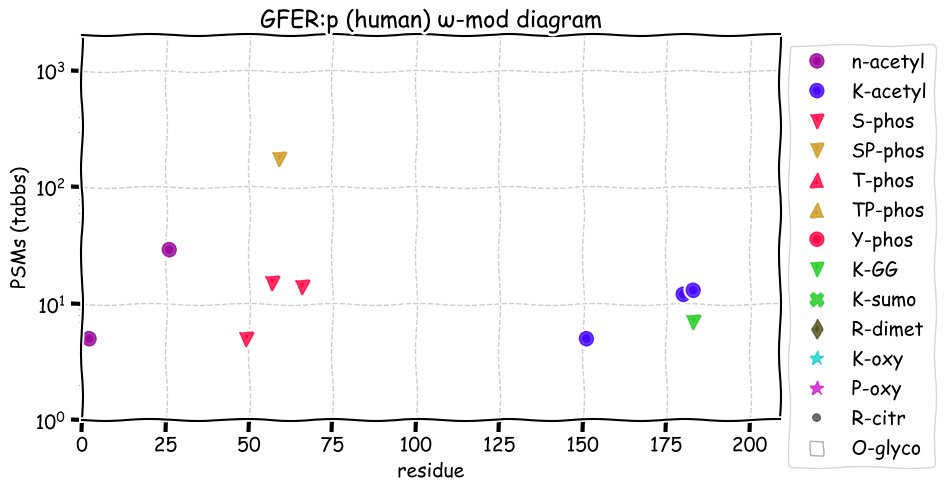
Mon Nov 15 20:13:01 +0000 2021🔗
Mon Nov 15 19:19:28 +0000 2021@WalterWChen1 The first 2, TNF & MAPT, also span the range of difficulty for this type of enumeration project: from trivially easy (TNF) to very hard (MAPT)
Mon Nov 15 13:13:39 +0000 2021NAXD:p, the mouse sequence (GRCm39 Naxd-202, J3QMM7) looks pretty good, though.
Mon Nov 15 12:47:49 +0000 2021NAXD:p, translation may initiate at M1 or M50 in humans (M1 or M30 in mice). Based on the proteomics data, it is uncertain that either of the 2 human splice variants in GRCh38.13 (NAXD-201, NAXD-202) or UniProt's Q8IW45 represent the protein accurately.
Mon Nov 15 12:47:49 +0000 2021NAXD::p.V149I chr 13:g.110627497G>A, rs3742192 (all tissue V:I 0.995:0.005) vaf=2%, Δm=14.0156, VAF by population group: african 11%, american 1%, east asian 7%, european <1%, south asian 9%.
Mon Nov 15 12:47:49 +0000 2021NAXD:p, θ(max) = 50. aka LP3298, FLJ10769, CARKD. Observed in class 1 & 2 MHC peptide expts. Observed in tissues & cell lines: rare in fluids but common in urine extracellular vesicles.
Mon Nov 15 12:47:48 +0000 2021>NAXD:p, NAD(P)HX dehydratase (Homo sapiens) Small subunit; CTMs: M1,M50+acetyl; PTMs: 1×K+acetyl; 2×K+GGyl; 3×K+SUMOyl; 1×S, 2×T, 2×Y+phosphoryl; SAAVs: V39I (2%); mature form: (1,50-390) [12,234×, 49 kTa]. #ᗕᕱᗒ 🔗
Mon Nov 15 01:19:53 +0000 2021@pwilmarth @UCDProteomics At least it isn't a Ciphergen ...
Sun Nov 14 18:04:39 +0000 2021@ypriverol @slavov_n @BiswapriyaMisra @MetabolomicsWB @MetaboLights I don't know about the DIA dataset (PXD002952), but the other 2 are really eccentric choices for popularity.
Sun Nov 14 15:28:15 +0000 2021@BiswapriyaMisra @slavov_n @ypriverol @MetabolomicsWB @MetaboLights Do you mean the depositing of data associated with a publication, or the use of existing data sets in other publications to come to new conclusions?
Sun Nov 14 14:52:21 +0000 2021Canadian politics at all level suffers from a real noblesse oblige problem.
Sun Nov 14 13:48:39 +0000 2021MTFR1L:p, translation initiates at M1 or M4 in humans. In mice, the only N-terminus corresponds to the human M4 sequence: the human M1 sequence does not exist.
Sun Nov 14 13:48:39 +0000 2021MTFR1L:p.P58S chr 1:g.25826344C>T, rs35448678 (all tissue P:S 0.993:0.007) vaf=0.6%, Δm=-10.0207, VAF by population group: african 8%, american <1%, east asian <1%, european <1%, south asian <1%.
Sun Nov 14 13:48:39 +0000 2021MTFR1L:p, θ(max) = 40. aka FAM54B. Observed in class 1 & 2 MHC peptide expts. Observed in tissues & cell lines: rare in fluids or extracellular vesicles.
Sun Nov 14 13:48:39 +0000 2021>MTFR1L:p, mitochondrial fission regulator 1 like (Homo sapiens) Small subunit; CTMs: A2,M4+acetyl; PTMs: 0×K+acetyl; 4×K+GGyl; 3×K+SUMOyl; 13×S, 5×T, 1×Y+phosphoryl; SAAVs: P58S (1%); mature form: (2,4-292) [12,231×, 679 kTa]. #ᗕᕱᗒ 🔗
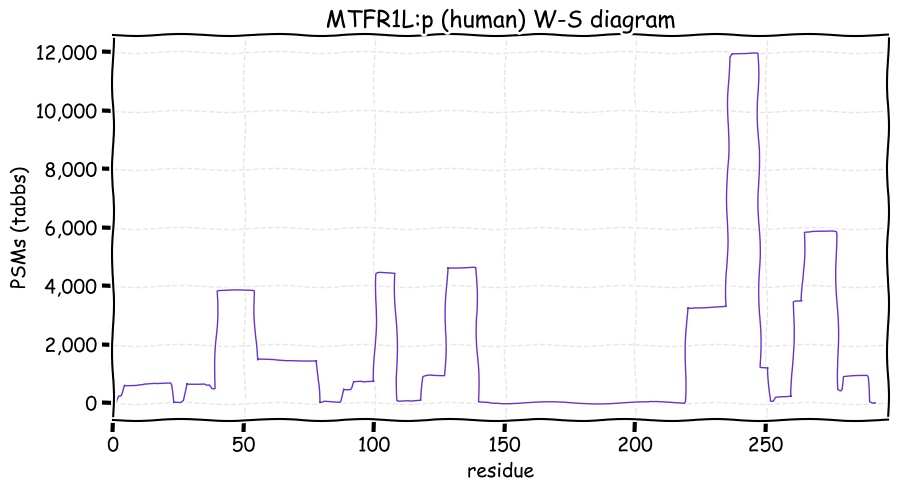
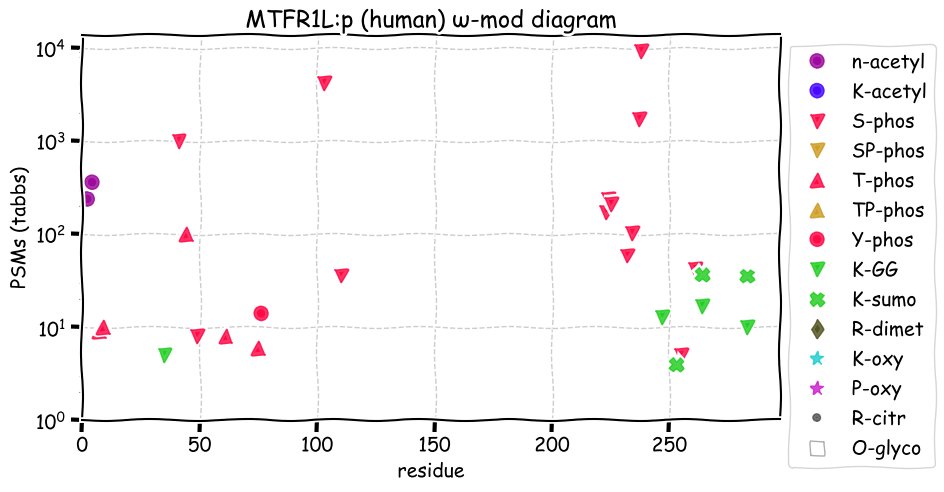
Sat Nov 13 18:52:37 +0000 2021@rajandelman @GreatDismal Canada = Robert Borden + William Shatner
Sat Nov 13 17:23:54 +0000 2021@ZacSpicer CRC is a pretty hands-off outfit. Their interaction with Chair Holders is limited to a year-end report. The money goes directly to the University and it is up to the University to decide how it is used and how appointments are handled.
Sat Nov 13 16:02:14 +0000 2021@pwilmarth @J_my_sci The DeepMind/AlphaFold play seems to be a little different: it implies that era of structure determination experiments is ending & calculation-only is the way forwards.
Sat Nov 13 15:54:42 +0000 2021@pwilmarth @J_my_sci This sort of thing is necessary from time to time: it is used by funding agencies to demonstrate to politicians that the money is being well-spent.
Sat Nov 13 15:21:34 +0000 2021@J_my_sci It is very reminiscent of the proteomics boom in the late '90s. Slightly updated language for the themes/goals/technology, but very similar.
Sat Nov 13 13:23:09 +0000 2021TALDO1:p, it may be worth noting that even though there are many observed GGyl acceptor sites, if any one of them is occupied, the protein is immediately recycled.
Sat Nov 13 13:00:33 +0000 2021TALDO1:p, translation initiates at M1 or M11 in humans & mice. 21 out of 21 K+GGyl sites are also K+acetyl acceptor sites: the protein is a nearly ideal example of this particular type of alternate modification.
Sat Nov 13 13:00:33 +0000 2021TALDO1:p.K321R chr 11:g.764414A>G, rs11302 (all tissue K:R 0.998:0.002) vaf=0.5%, Δm=28.0061, VAF by population group: african 4%, american <1%, east asian <1%, european <1%, south asian <1%.
Sat Nov 13 13:00:33 +0000 2021TALDO1:p, θ(max) = 89. aka none. Observed in class 1 & 2 MHC peptide expts. Observed in tissues & cell lines: common in urine, blood plasma & extracellular vesicles.
Sat Nov 13 13:00:33 +0000 2021>TALDO1:p, transaldolase 1 (Homo sapiens) Small subunit; CTMs: S2,M11+acetyl; PTMs: 27×K+acetyl; 21×K+GGyl; 1×K+SUMOyl; 20×S, 1×T, 1×Y+phosphoryl; SAAVs: K321R (1%); mature form: (2,11-337) [74,200×, 679 kTa]. #ᗕᕱᗒ 🔗
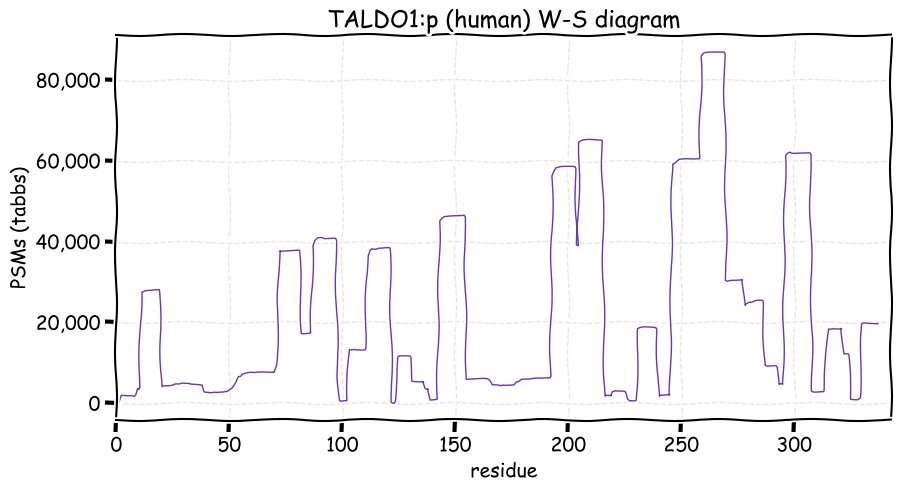
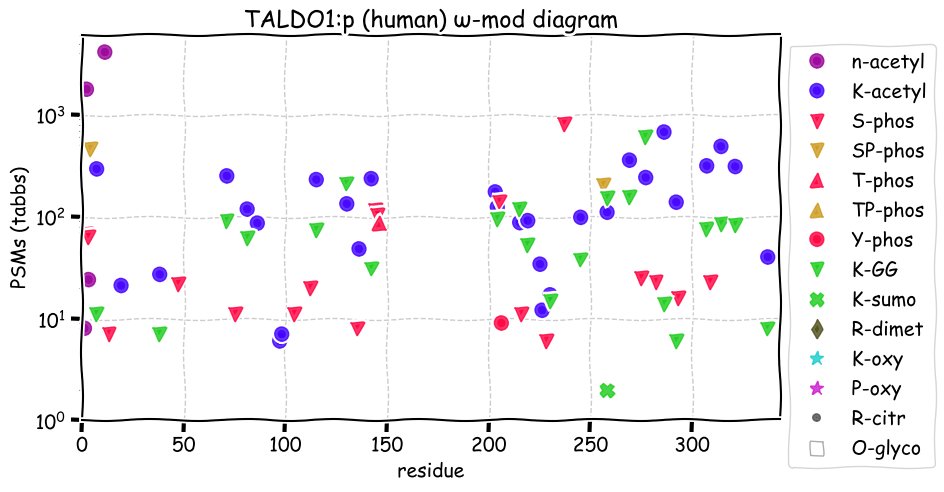
Fri Nov 12 21:21:48 +0000 2021@mjmaccoss @RyanRJulian I was at Rockefeller during the Baltimore affair (also played out in the NYT). Distressing for everyone, but no one was sure what was going on (to this day I'm not really sure what really happened).
Fri Nov 12 19:43:33 +0000 2021"Counterintuitive" is all I can say about this 🔗
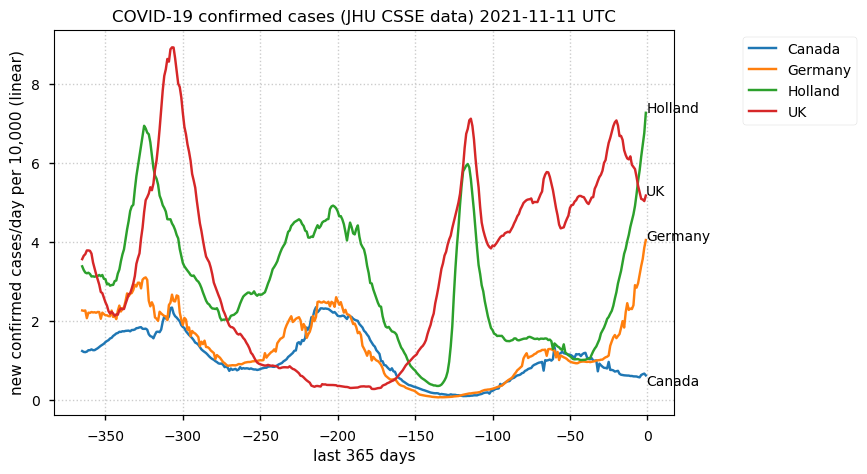
Fri Nov 12 18:34:00 +0000 2021🔗
Fri Nov 12 18:29:31 +0000 2021🔗
Fri Nov 12 17:38:22 +0000 2021This stuff doesn't go away because it was "called out". Most of the literature gets forgotten within 2-3 Ph.D. training cycles unless it proves to be useful, so time ends up removing the bad stuff, regardless of how big a splash it made at the time.
Fri Nov 12 15:05:26 +0000 2021Self-aggrandizing bad behavior & well-intentioned fraud have been part of academic research since I started (1981). I realize people tend to be surprised when they first stumble across it, but it isn't something new or something that MUST be addressed.
Fri Nov 12 14:53:41 +0000 202130 minutes of shoveling wet snow: Oh Canada!
Fri Nov 12 12:56:00 +0000 2021GSR:p, translation initiates at M1 (mitochondrial form) or M44 (cytosolic form) in humans & M1 (mito) or M27 (cyto) in mice.
Fri Nov 12 12:56:00 +0000 2021GSR:p.I357V chr 8:g.30684172T>C, rs75673983 (all tissue I:V 0.997:0.003) vaf=0.2%, Δm=-14.0156, VAF by population group: african 2%, american <1%, east asian <1%, european <1%, south asian <1%.
Fri Nov 12 12:55:59 +0000 2021GSR:p, θ(max) = 75. aka none. Observed in class 1 & 2 MHC peptide expts. Observed in tissues & cell lines: abundant in fluids & extracellular vesicles.
Fri Nov 12 12:55:59 +0000 2021>GSR:p, glutathione-disulfide reductase (Homo sapiens) Midsized subunit; CTMs: A45+acetyl; PTMs: 11×K+acetyl; 9×K+GGyl; 22×S, 19×T, 2×Y+phosphoryl; SAAVs: P14Q (1%), S15G (2%), R153C (4%), I357V (0.2%); mature form: (43,45-522) [51,697×, 405 kTa]. #ᗕᕱᗒ 🔗
Fri Nov 12 12:28:34 +0000 2021@GeiszlerDaniel It is an interesting study, in that it discusses mechanisms that may be involved in shaping chloroplast targeting sequences & presents some structural info about known sequences. I think it was hampered by the lack of sequences known at the time.
Thu Nov 11 20:04:19 +0000 2021But it concludes with "Despite the significance of the data presented, a large proportion of the variation among these sequences remains unexplained"
Thu Nov 11 20:02:17 +0000 2021This one is in the ball park
🔗
Thu Nov 11 18:24:32 +0000 2021While this isn't right on point, it is adjacent & interesting 🔗
Thu Nov 11 18:17:25 +0000 2021@astacus If my experience is any guide: no.
Thu Nov 11 16:21:34 +0000 2021@DRAWheatcraft Depends on the expt. I always check for under (& over alkylation) and adjust the final analysis accordingly. Detecting underivatized C's is rare, but there may be Cys+3O, Cys, β-met, proprionamide, glutathione, etc. Over alkylation is a more common situation.
Thu Nov 11 15:57:04 +0000 2021@mjmaccoss @astacus Nomenclature aside, I was simply venting a bit of frustration after reading yet another method section claiming to analyze data with a static C+57 mod when the description of the expt (& the data) show no signs of IAA treatment. Just yelling at the TV ...
Thu Nov 11 14:32:54 +0000 2021Seems like rather a lot of unintentional mistakes that must have taken quite a bit of work to get wrong. The group's leadership attribution (the 1st author) seems odd for a study of this type. 🔗
Thu Nov 11 14:13:47 +0000 2021Does anybody know of literature trying to understand the evolutionary diversity of mitochondrial targeting sequences?
Thu Nov 11 14:05:57 +0000 2021It's a winter wonderland this morning (25 cm of snow in the last 24 hrs) and more to come: 🔗
Thu Nov 11 13:24:52 +0000 2021YIF1B:p, there is good evidence for translation initiation at M1 or M32 in humans; M1 or M31 in mice. Domains: (1,33-156) cytosolic (from the PTM pattern); TMs: (157-176), E-I(191-213), (220-242), (246-264) & (293-314).
Thu Nov 11 13:24:52 +0000 2021YIF1B:p.P56S chr 19:g.38309536G>A, rs11556992 (all tissue P:S 0.986:0.014) vaf=8%, Δm=-10.0207, VAF by population group: african 3%, american 14%, east asian 30%, european 6%, south asian 15%. SS in IOSE-397 cells & PS in HEK-293 cells.
Thu Nov 11 13:24:52 +0000 2021YIF1B:p, θ(max) = 43. aka FinGER8. Observed in class 1 & 2 MHC peptide expts. Observed in many tissues & cell lines: abundant in blood but rare in blood plasma.
Thu Nov 11 13:24:52 +0000 2021>YIF1B:p, Yip1 interacting factor homolog B, membrane trafficking protein (Homo sapiens) Small subunit; CTMs: M1, A33+acetyl; PTMs: 5×K+GGyl; 8×S, 2×T, 1×Y+phosphoryl; SAAVs: P56S (8%); mature form: (1, 33-314) [12,257×, 37 kTa]. #ᗕᕱᗒ 🔗
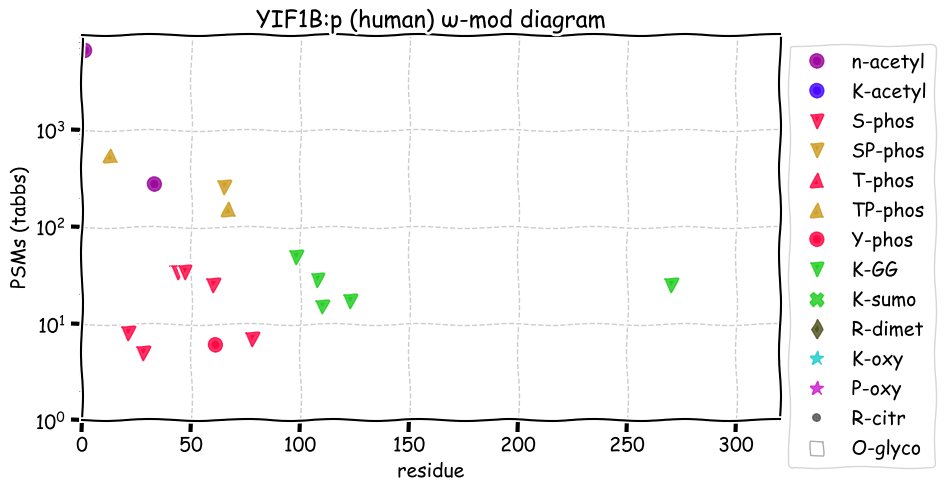
Wed Nov 10 19:43:54 +0000 2021@astacus The kids now refer to the amide version as IAA pretty consistently. I don't remember the last time I saw someone using the acid version in a data set.
Wed Nov 10 18:14:37 +0000 2021@TonyBurnetti @slavov_n It kind of defeats the purpose of a thesis: demonstrating that you can write a coherent monograph describing your research.
Wed Nov 10 17:40:29 +0000 2021Pro tip: if you didn't use IAA/CAA to derivatize your peptides, you don't have to set static C+carbamidomethyl as a search parameter
Wed Nov 10 17:36:45 +0000 2021@dtabb73 I thought it was a physical meeting: after checking the web site I see I was wrong. Good luck with the presentation.
Wed Nov 10 17:04:38 +0000 2021@dtabb73 What is the venue like? (pictures!)
Wed Nov 10 14:46:15 +0000 2021@LewisGeer I don't know of one.
Wed Nov 10 13:02:27 +0000 2021CLOCK:p, the central phosphodomain in human (401-463) is very similar to the corresponding phosphodomain in mouse (403-461). Both S836+phosphoryl (human) & S845+phosphoryl (mouse) acceptor sites share the "RxxΦL/I/V" motif. 🔗
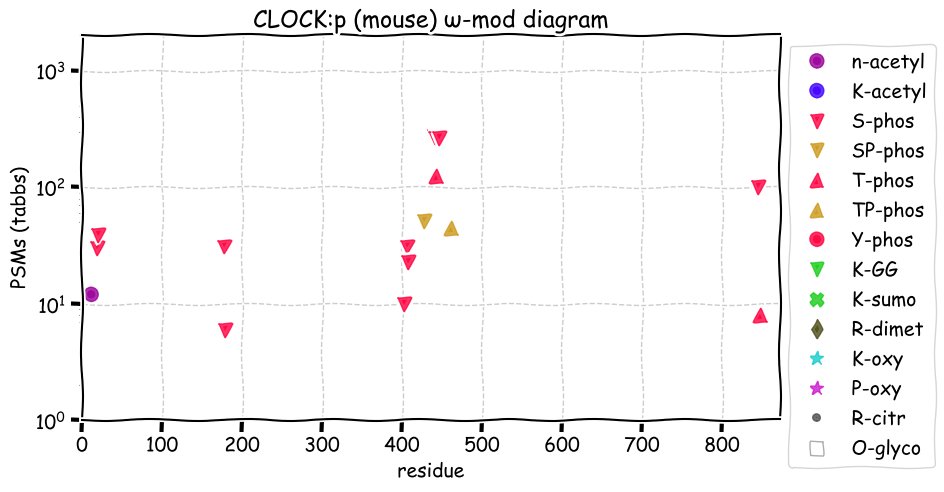
Wed Nov 10 13:02:25 +0000 2021CLOCK:p, there is good evidence for translation initiation at M10 in humans and mice, but no evidence of M1 initiation.
Wed Nov 10 13:02:25 +0000 2021CLOCK:p.H542L chr 4:g.55444700T>A, rs3762836 (all tissue H:L 0.988:0.013) vaf=<1%, Δm=-23.9748, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Wed Nov 10 13:02:25 +0000 2021CLOCK:p, θ(max) = 32. aka KIAA0334, KAT13D, bHLHe8. Observed in class 1 MHC peptide expts. Observed in many tissues & cell lines: absent from fluids.
Wed Nov 10 13:02:25 +0000 2021>CLOCK:p, clock circadian regulator (Homo sapiens) Midsized subunit; CTMs: S11+acetyl; PTMs: 1×K+GGyl; 1×SUMOyl; 9×S, 5×T, 1×Y+phosphoryl; SAAVs: H542L (<1%); mature form: (11-846) [1,812×, 4 kTa].#ᗕᕱᗒ 🔗
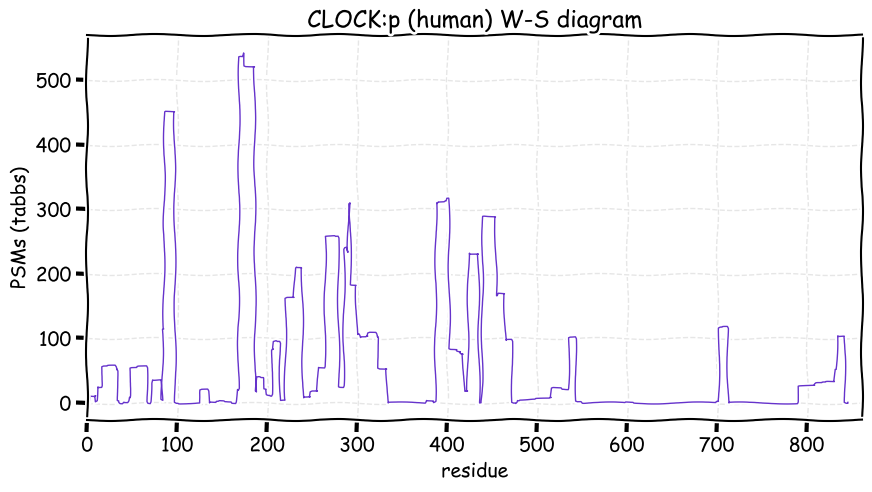
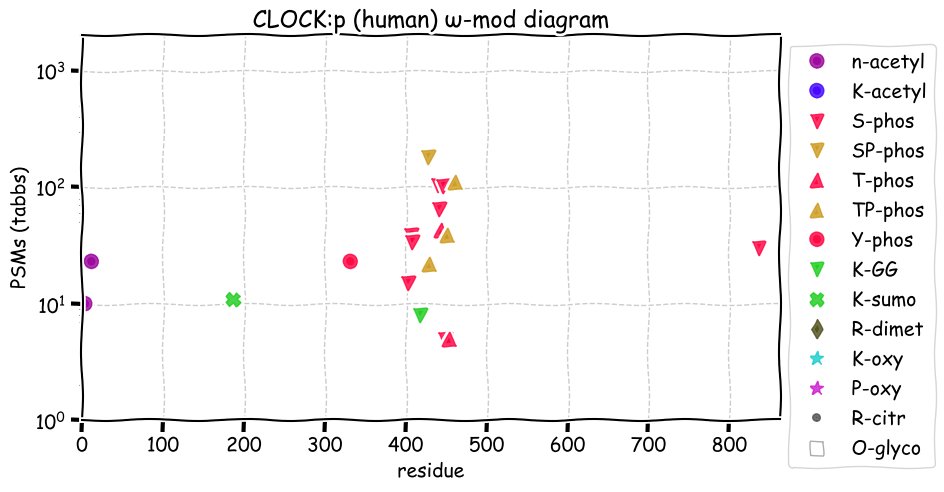
Tue Nov 09 17:37:04 +0000 2021I don't know what about their protocol makes it so, but PXD023979 did a great job of TMT10 derivatization.
Tue Nov 09 16:26:22 +0000 2021@mjmaccoss @dtabb73 @lkpino @hupo_org @NLKProteomics He's going to have to get a proper microphone & a mixing board.
Tue Nov 09 15:38:45 +0000 2021Every six months I rediscover that my cat does not auto-update for daylight savings time.
Tue Nov 09 14:31:02 +0000 2021Just a friendly reminder: you have to log back into PRIDE and release your data once your publication has become public 🔗
Tue Nov 09 12:59:59 +0000 2021TIMELESS:p.R831Q chr 12:g.56422138C>T, rs774047 (all tissue R:Q 0.065:0.935) vaf=50%, Δm=-28.0425, VAF by population group: african 23%, american 68%, east asian 74%, european 45%, south asian 54%. SAAV removes a tryptic cleavage site.
Tue Nov 09 12:59:59 +0000 2021TIMELESS:p, θ(max) = 39. aka hTIM, TIM, TIM1. Observed in class 1 MHC peptide expts. Observed in many tissues & cell lines: absent from fluids.
Tue Nov 09 12:59:58 +0000 2021>TIMELESS:p, timeless circadian clock (Homo sapiens) Large subunit; CTMs: M6+acetyl; PTMs: 2×K+acetyl; 5×K+GGyl; 8×S , 0×T, 0×Y+phosphoryl; SAAVs: R831Q (50%); mature form: (6-1208) [9,230×, 33 kTa]. #ᗕᕱᗒ 🔗
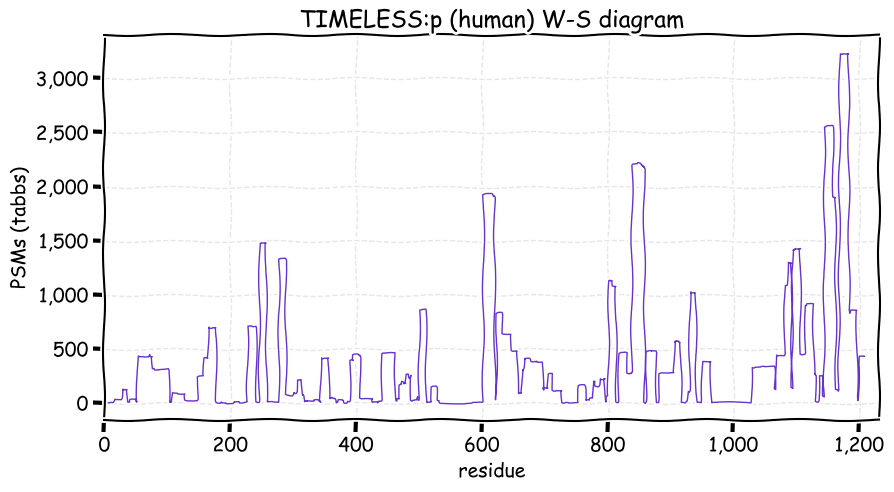
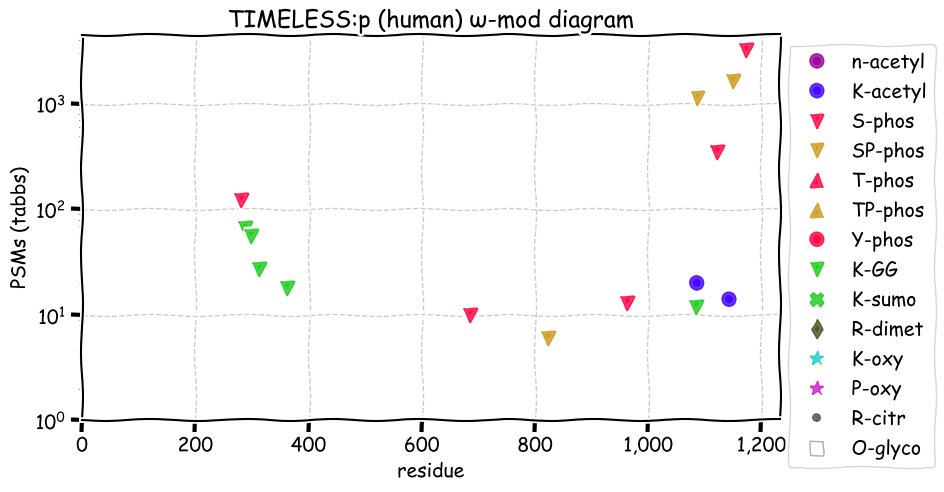
Mon Nov 08 15:45:48 +0000 2021@pwilmarth @chrashwood Or working for the government.
Mon Nov 08 12:54:15 +0000 2021HNRNPA0:p.S19T chr 5:g.137754011C>G, rs199553659 (all tissue S:T 0.998:0.002) vaf=<1%, Δm=14.0157, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Mon Nov 08 12:54:14 +0000 2021HNRNPA0:p, θ(max) = 71. aka HNRPA0. Observed in class 1 & (rarely) 2 MHC peptide expts. Observed in many tissues & cell lines: absent from fluids.
Mon Nov 08 12:54:14 +0000 2021>HNRNPA0:p, heterogeneous nuclear ribonucleoprotein A0 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 8×K+acetyl; 15×K+GGyl; 14×K+SUMOyl; 22×S , 2×T, 5×Y+phosphoryl; R209+dimethyl; SAAVs: S19T (0.02%); mature form: (1-305) [76,335×, 496 kTa]. #ᗕᕱᗒ 🔗
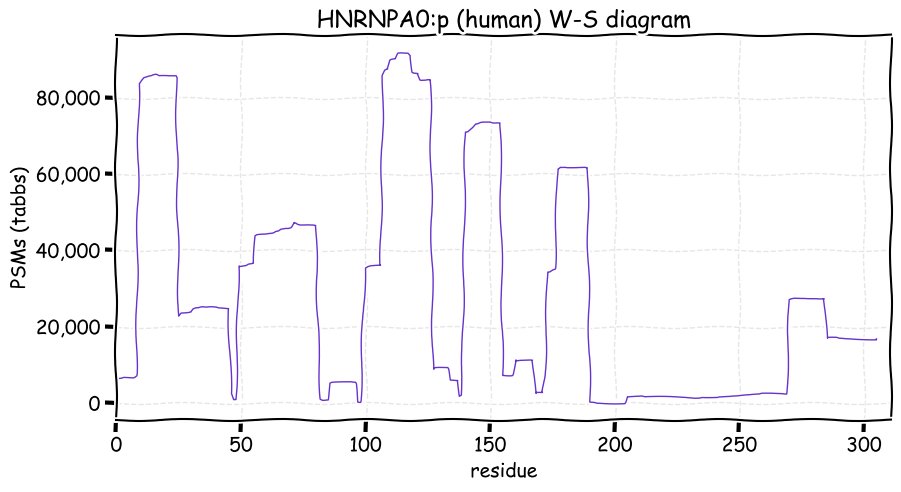
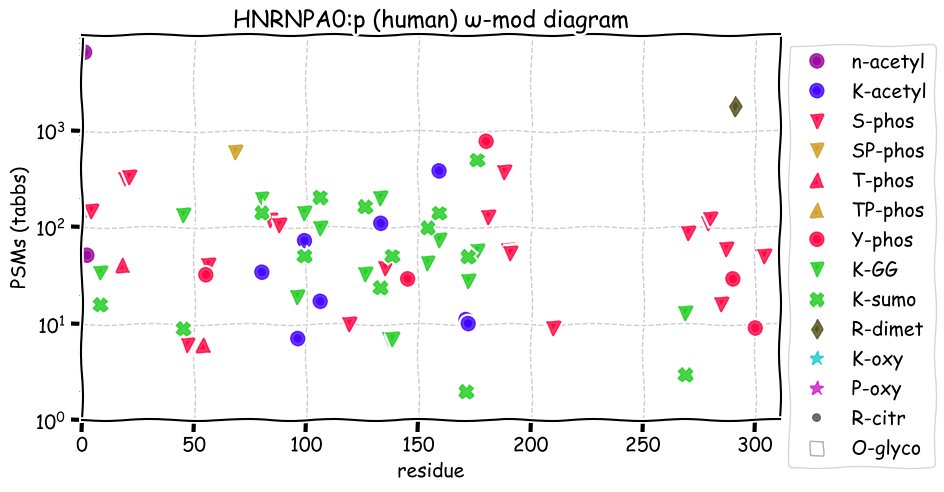
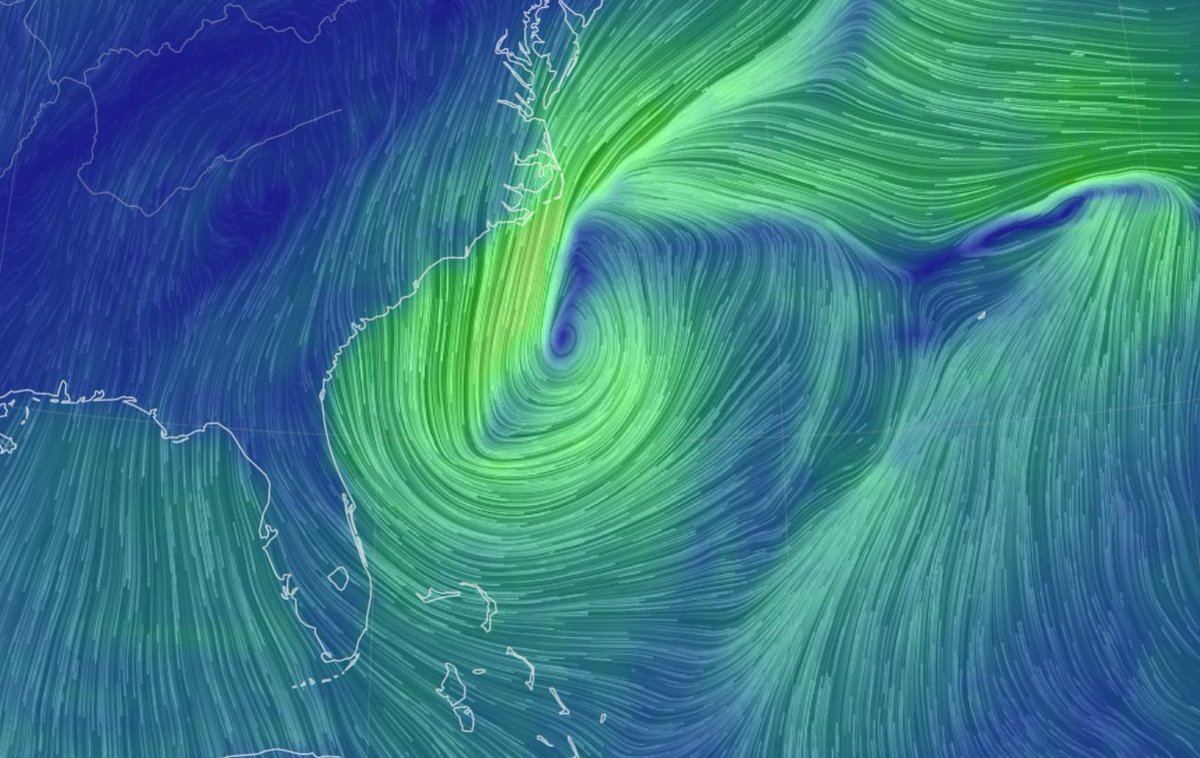
Sun Nov 07 14:22:24 +0000 2021An idea hasn't really made it into the scientific community until either:
1. no one remembers whose idea it was (or associates it with the wrong person), or
2. the idea has become so distorted with time that it is used in nearly the reverse of the original intention.
Sun Nov 07 13:45:58 +0000 2021@slashdot This sounds just awful. The main reason I stopped using Office was all the time I had to spend correcting its "corrections".
Sun Nov 07 12:51:36 +0000 2021MRPS12:p, translation initiation occurs at M1 only: the removal of the mitochondrial signal peptide (1-30) looks like it may be caused by an alternate translation at M30, but this is very unlikely because the mature N-terminal A30 is not N-acetylated.
Sun Nov 07 12:51:36 +0000 2021MRPS12:p.K122R chr 19:g.38932648A>G, rs752232760 (all tissue K:R 0.999:0.001) vaf=<1%, Δm=28.0061, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sun Nov 07 12:51:36 +0000 2021MRPS12:p, θ(max) = 63. aka RPS12, RPSM12, RPMS12. Observed in class 1 MHC peptide expts. Observed in most tissues & cell lines, not in fluids.
Sun Nov 07 12:51:36 +0000 2021>MRPS12:p, mitochondrial ribosomal protein S12 (Homo sapiens) Small subunit; CTMs: none; PTMs: 3×K+SUMOyl; SAAVs: K122R (0.0012%); mature form: (31-138) [6,024×, 17 kTa]. #ᗕᕱᗒ 🔗
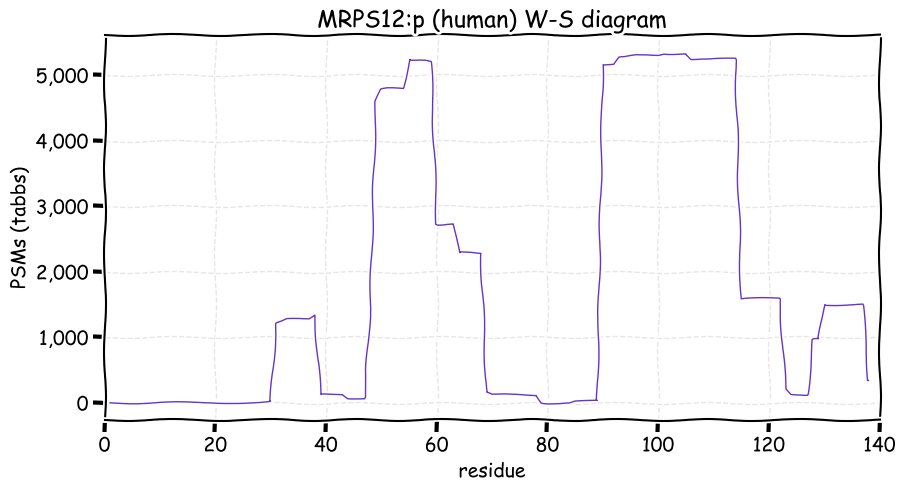
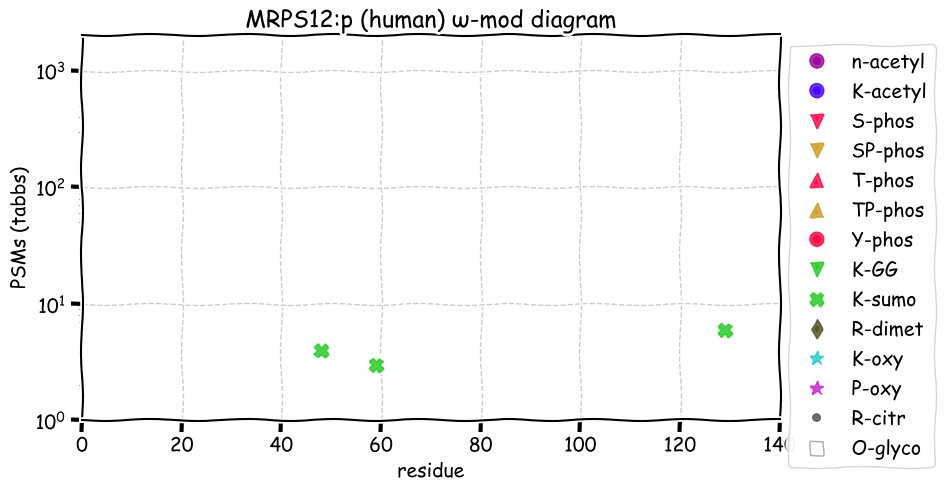
Sat Nov 06 18:13:57 +0000 2021Odd temperatures this year (except for Topeka).
Noon temps, 2021 (left, today) vs 2019 (right, more normal for this time of year) 🔗
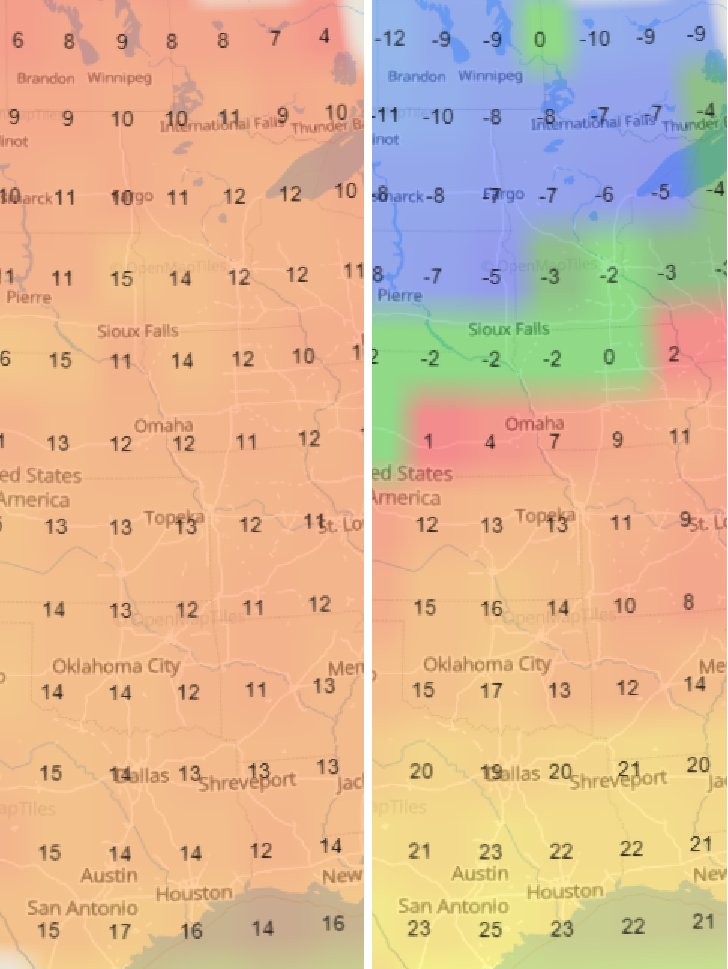
Sat Nov 06 14:01:24 +0000 2021Feel free to disagree, but to me "data science" belongs to "applied mathematics", not "science". It is kin to "statistics" (& the similarly misnamed "computer science"), rather than "physics" or "chemistry".
Sat Nov 06 11:58:22 +0000 2021STAU1:p.L84P chr 20:g.49151598A>G, rs61748376 (all tissue L:P 0.999:0.001) vaf=0.3%, Δm=-16.0313, VAF by population group: african 5%, american <1%, east asian <1%, european <1%, south asian <1%.
Sat Nov 06 11:58:22 +0000 2021STAU1:p, θ(max) = 85. aka PPP1R150, STAU. Observed in class 1 & rarely in class 2 MHC peptide expts. Observed in most tissues & cell lines: very rare in fluids or extracellular vesicles. Translation initiation occurs at either M1 or M8.
Sat Nov 06 11:58:21 +0000 2021>STAU1:p, staufen double-stranded RNA binding protein 1 (Homo sapiens) Small subunit; CTMs: M8+acetyl; PTMs: 3×K+acetyl; 7×K+GGyl; 16×S, 9×T, 2×Y+phosphoryl; 2×R+dimethyl; SAAVs: L84P (0.3%); mature form: (1,8-496) [33,063×, 209 kTa]. #ᗕᕱᗒ 🔗
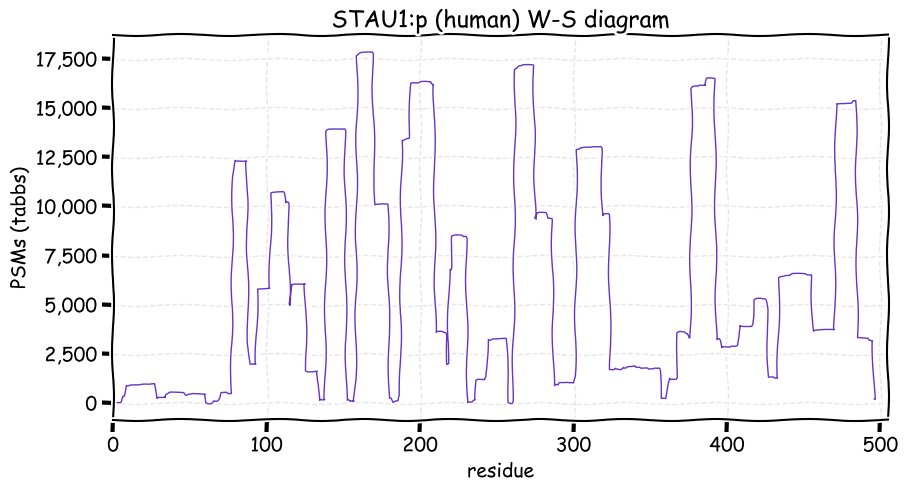
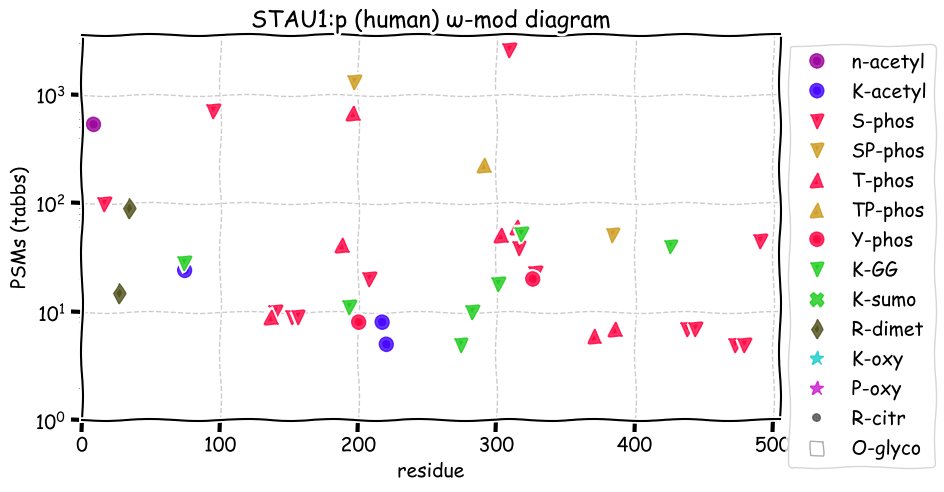
Fri Nov 05 20:41:54 +0000 2021"using the default settings in MaxQuant"
which are what exactly?
Fri Nov 05 11:51:36 +0000 2021ALDH9A1:p, uses a similar translation initiation mechanism yesterday's protein ALDH7A1:p (also an aldehyde dehydrogenase), but the two proteins do not share any observable peptides. 🔗

Fri Nov 05 11:51:36 +0000 2021ALDH9A1:p, translation initiation occurs at either M1 or M25. The M1 form has a mitochondrial targeting peptide (1-24) removed during passage through the mitochondrial inner membrane. The M25 cytosolic form has the N-terminal M removed & the S is N-acetylated.
Fri Nov 05 11:51:36 +0000 2021ALDH9A1:p.S221T chr 1:g.165680614C>G, rs1065756 (all tissue S:T 0.931:0.069) vaf=4%, Δm=14.0157, VAF by population group: african 4%, american 5%, east asian 1%, european <1%, south asian 8%.
Fri Nov 05 11:51:35 +0000 2021ALDH9A1:p, θ(max) = 73. aka E3, ALDH7, ALDH4, ALDH9. Observed in class 1 & 2 MHC peptide expts. Observed in most tissues & cell lines: common in urine extracellular vesicles & saliva. 16 K acceptor sites have also been observed to be GGyl acceptor sites.
Fri Nov 05 11:51:35 +0000 2021>ALDH9A1:p, aldehyde dehydrogenase 9 family member A1 (Homo sapiens) Midsized subunit; CTMs: S26+acetyl; PTMs: 22×K+acetyl; 16×K+GGyl; 1×K+SUMOyl; 17×S, 2×T, 2×Y+phosphoryl; SAAVs: S221T (4%); mature form: (29,30-518) [61,050×, 456 kTa]. #ᗕᕱᗒ 🔗
Thu Nov 04 19:31:20 +0000 2021If you are doing biomedical research, you should think about this one a bit (& maybe reconsider how you use the web). 🔗
Thu Nov 04 14:41:10 +0000 2021@statesdj I'm not sure if this is the case in the US, but FAX machines are still used in Canadian medical practice for legal reasons. Laws regulating things like prescriptions do not have provision for any electronic communication other than telephony (incl. FAXs).
Thu Nov 04 11:53:54 +0000 2021ALDH7A1:p, translation initiation at M1 or M29. The M1 form has a mitochondrial targeting peptide (1-28) that is removed in the mature mitochondrial protein. The M29 form has the N-terminal M removed & S30 is N-acetylated to form the mature cytosolic protein.
Thu Nov 04 11:53:54 +0000 2021ALDH7A1:p.T412A chr 5:g.126552104T>C, rs2306618 (all tissue T:A 0.992:0.008) vaf=0.7%, Δm=-30.0106, VAF by population group: african <1%, american <1%, east asian 8%, european <1%, south asian <1%.
Thu Nov 04 11:53:54 +0000 2021ALDH7A1:p, θ(max) = 89. aka EPD, PDE, ATQ1. Observed in class 1 & 2 MHC peptide expts. Found in most tissues & cell lines: common in urine & extracellular vesicles.
Thu Nov 04 11:53:54 +0000 2021>ALDH7A1:p, aldehyde dehydrogenase 7 family member A1 (Homo sapiens) Midsized subunit; CTMs: S30+acetyl; PTMs: 10×K+acetyl; 6×K+GGyl; 1×K+SUMOyl; 2×S, 0×T, 3×Y+phosphoryl; SAAVs: T412A (1%); mature form: (29,30-539) [47,589×, 512 kTa]. #ᗕᕱᗒ 🔗
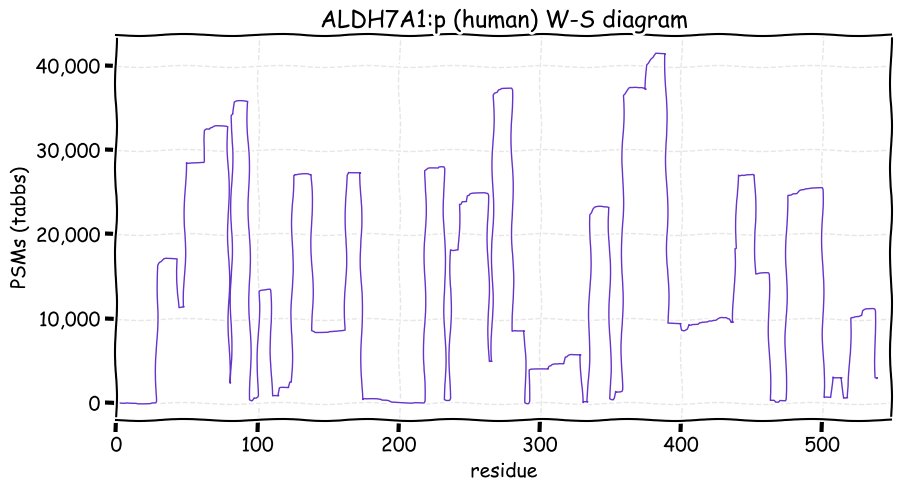
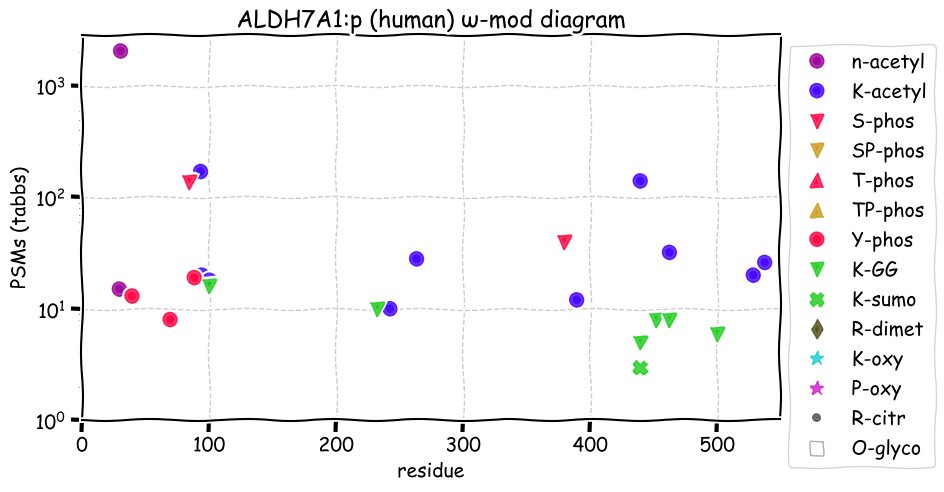
Wed Nov 03 19:03:28 +0000 2021@DRAWheatcraft @nesvilab For me the worst case scenario is one of the authors of a sub-project gets in a snit about something like work on HEK-293 cells & issues a take down notice.
Wed Nov 03 18:46:56 +0000 2021@nesvilab It may not be a big concern, but I have always struggled a bit with the "many licenses" issue when combining code from other open source projects. It may be more touchy in countries where "moral rights" are copyright protected when dealing with scientific applications.
Wed Nov 03 18:42:34 +0000 2021@RalfGabriels In practice, it is very similar to cysteine in both its chromatographic & MS/MS behavior. It is derivatived by common Cys-blocking reagents in the same way, too.
Wed Nov 03 15:42:28 +0000 2021Nobody likes to pay the bill for negative results.
Wed Nov 03 14:20:54 +0000 2021PXD004652 has some great data if you are interested in using isoelectric focusing as a method of peptide separation. Does a good job of producing reproducible analyses from multiple samples.
Wed Nov 03 14:15:12 +0000 2021@RalfGabriels Why does your implementation's call to Percolator reject peptides containing selenocysteine (U) by default?
Wed Nov 03 11:49:37 +0000 2021PLS3:p, translation initiation occurs at either M1 or M4, with good evidence for initiation both sites.
Wed Nov 03 11:49:37 +0000 2021PLS3:p.N385S chr X:g.115643479A>G, rs782600450 (all tissue N:S 0.998:0.002) vaf=<1%, Δm=-27.0109, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Wed Nov 03 11:49:37 +0000 2021PLS3:p, θ(max) = 89. aka T-plastin. Observed in class 1 & 2 MHC peptide expts. Observed in most tissues & cell lines: common in fluids & extracellular vesicles.
Wed Nov 03 11:49:37 +0000 2021>PLS3:p, plastin 3 (Homo sapiens) Midsized subunit; CTMs: M1, A5+acetyl; PTMs: 18×K+acetyl; 27×K+GGyl; 1×K+SUMOyl; 3×R+deimidation; 27×S, 17×T, 12×Y+phosphoryl; SAAVs: 385S (<1%); mature form: (1,5-630) [83,153×, 1062 kTa]. #ᗕᕱᗒ 🔗
Tue Nov 02 22:01:49 +0000 2021@Smith_Chem_Wisc I use RT predictions rather enthusiastically for all sorts of QA-related purposes. But, I'm skeptical about its use on an individual PSM-by-PSM basis.
Tue Nov 02 18:40:33 +0000 2021@VATVSLPR @chrashwood @JohnRYatesIII @MagnusPalmblad @goodlettlab1 @r_l_moritz @pwilmarth @neely615 @lkpino @OliverMBernhar1 @mjmaccoss It took a while to realize the ions produced by MALDI had substantial kinetic energy distributions, so most early attempts to trap them failed.
Tue Nov 02 15:30:58 +0000 2021I have seen many schemes for incorporating HPLC RT into PSM assignment methods that report at least some improvement. I am pretty skeptical as to whether this can be broadly applied, because of the way HPLC is done & non-ideal behavior of some common peptides. Comments?
Tue Nov 02 14:06:43 +0000 2021@nesvilab I was looking through DIA-NN & it is constructed from a variety of open source code projects, with 5 or 6 different licenses. From a practical point of view, how do you steer clear of licensing conflicts in this sort of case? Does the UofM provide legal advice?
Tue Nov 02 13:09:44 +0000 2021@pwilmarth The only significant price reduction I remember was the shift from sectors & triple quads to LCQs for MS/MS back in the 90's.
Tue Nov 02 12:38:30 +0000 2021TMUB1:p, initiation at M56 removes the first predicted TM domain, leaving the domains: (57-191) cytoplasmic, (192-214) TM, (215-220) non-cytoplasmic, (221-243) TM, (244-246) cytoplasmic.
Tue Nov 02 12:21:08 +0000 2021TMUB1:p, Translation initiation may occur at either M1 or M56, although there is very little evidence of M1 initiation. Detecting M1 initiation is complicated by the lack of a suitable tryptic peptide & the 3 predicted TM domains (in grey). 🔗

Tue Nov 02 12:21:08 +0000 2021TMUB1:p.P96L chr 7:g.151082277G>A, rs146736140 (all tissue P:L 0.983:0.017) vaf=<1%, Δm=16.0313, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Tue Nov 02 12:21:07 +0000 2021TMUB1:p, θ(max) = 37. aka SB144, HOPS, C7orf21. Observed in class 1 MHC peptide expts. Observed in many tissues & cell lines: absent from fluids.
Tue Nov 02 12:21:07 +0000 2021>TMUB1:p, transmembrane and ubiquitin-like domain containing 1 (Homo sapiens) Small subunit; CTMs: A57+acetyl; PTMs: K108+acetyl; K108, N119+GGyl; 5×S, 4×T, 0×Y+phosphoryl; SAAVs: P96L (<1%); mature form: (37,57-246) [8,865×, 21 kTa]. #ᗕᕱᗒ 🔗
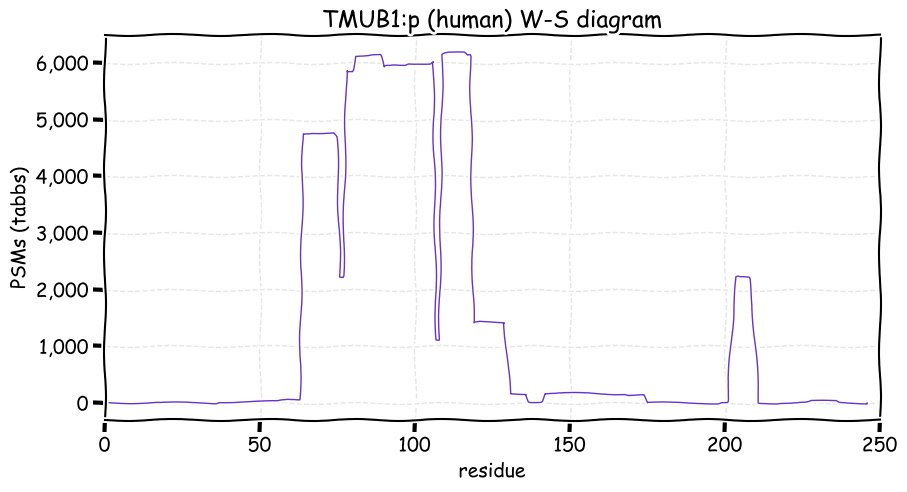
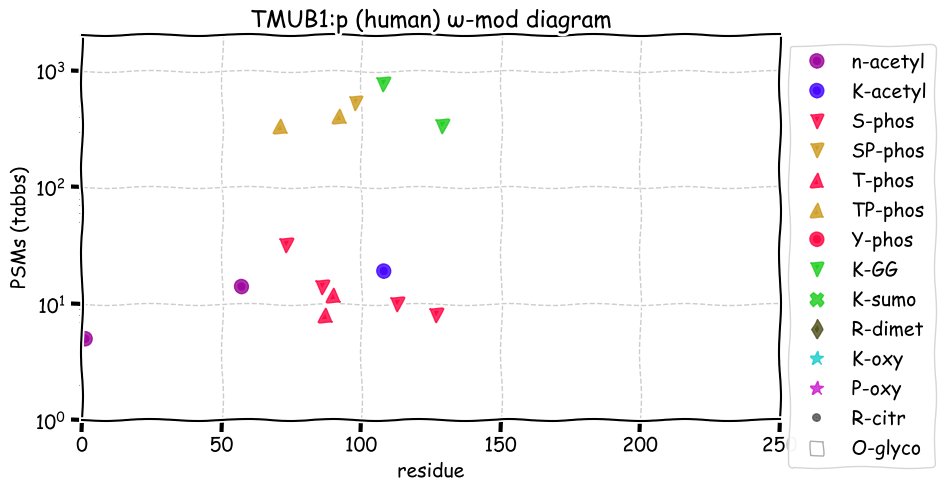
Mon Nov 01 11:43:19 +0000 2021PRAM1:p, translation initiation occurs at either M1 or M9 in humans. The M9 version N-terminal domain is homologous to the mouse or rat N-terminal domain.
Mon Nov 01 11:43:19 +0000 2021PRAM1:p.G135E chr 19:g.8499404C>T, rs58466313 (all tissue G:E 0.970:0.030) vaf=4%, Δm=72.0211, VAF by population group: african 58%, american 5%, east asian <1%, european <1%, south asian <1%.
Mon Nov 01 11:43:18 +0000 2021PRAM1:p, θ(max) = 67. aka PML-RAR. Observed in class 1 & 2 MHC peptide expts. Most abundant in leukocytes (e.g., neutrophils, eosinophils): absent from most cell lines, other than HL-60.
Mon Nov 01 11:43:18 +0000 2021>PRAM1:p, PML-RARA regulated adaptor molecule 1 (Homo sapiens) Midsized subunit; CTMs: A2, M9+acetyl; PTMs: 17×S, 6×T, 2×Y+phosphoryl; SAAVs: G135E (4%); mature form: (2,9-670) [3,421×, 28 kTa]. #ᗕᕱᗒ 🔗
Sun Oct 31 17:24:08 +0000 2021The 1st squalls of lake effect snow of the season: it must be Halloween!
Sun Oct 31 16:17:11 +0000 2021When interpreted in a larger context, the results show pretty clearly that if a K can be acetylated, there is a very good chance it can also be succinylated (& vice versa).
Sun Oct 31 15:54:49 +0000 2021PXD025621 does a great job of K+succinyl enrichment from murine brain tissue.
Sun Oct 31 13:40:06 +0000 2021@bkives The beginnings of a Manitoba Party?
Sun Oct 31 11:59:19 +0000 2021GNAQ:p, translation initiation occurs at either M1 or M7. Initiation at both sites has been observed in human, mouse & rat proteins. M7 version retains the G-protein alpha subunit, group Q domain (9-23, IPR000654).
Sun Oct 31 11:59:19 +0000 2021GNAQ:p.D319E chr 9:g.77721446G>C, rs183713546 (all tissue D:E 0.998:0.002) vaf=<1%, Δm=14.0157, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sun Oct 31 11:59:19 +0000 2021GNAQ:p, θ(max) = 78. aka G-ALPHA-q, GAQ. Observed in class 1 & 2 MHC peptide expts. Common in tissues, cell lines & urinary extracellular vesicles. Particularly abundant in blood platelets.
Sun Oct 31 11:59:19 +0000 2021>GNAQ:p, G protein subunit alpha q (Homo sapiens) Small subunit; CTMs: M1, A8+acetyl; PTMs: 11×K+acetyl; 11×K+GGyl; 13×S, 10×T, 1×Y+phosphoryl; SAAVs: D319E (<1%); mature form: (1,8-359) [25,130×, 140 kTa]. #ᗕᕱᗒ 🔗
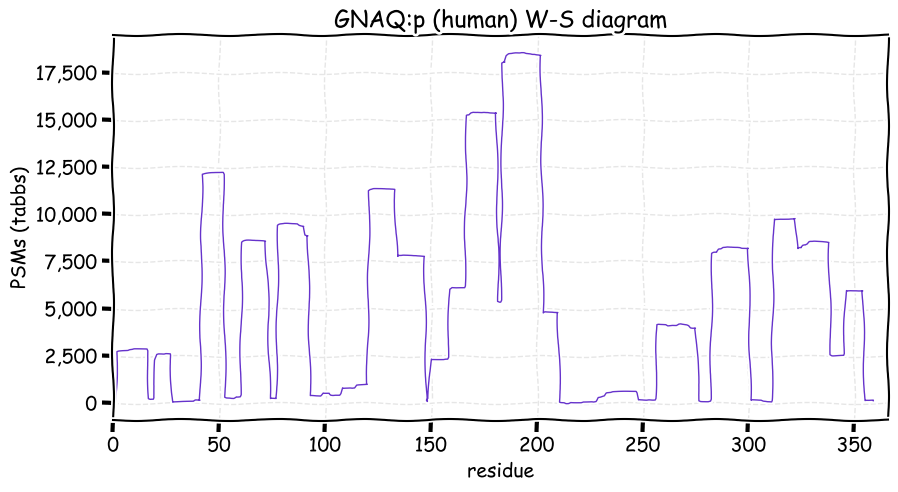
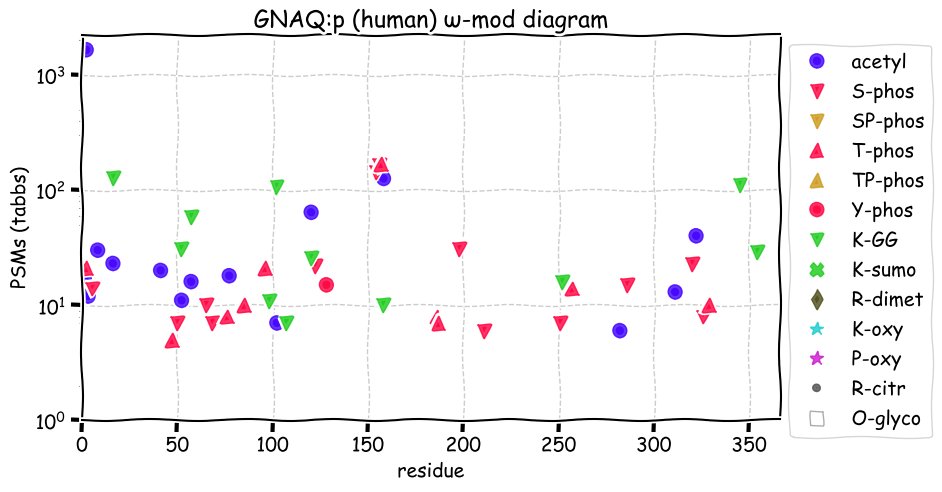
Sun Oct 31 04:51:36 +0000 2021It looked promising, but the aurora has largely dissipated (as of midnight CDT) 🔗
Sun Oct 31 00:40:15 +0000 2021The aurora associated with this storm is starting to spin up: hopefully more to come. 🔗
Sat Oct 30 14:55:40 +0000 2021@ProteomicsNews @DRAWheatcraft @JohnRYatesIII @RobinPa88093621 Looks like an HP case. The least they could have done is spring for one of these: 🔗
Sat Oct 30 13:22:50 +0000 2021If you want to use a TMT-based dataset, but need one where the derivatization was done really well (very few artifacts), PXD021038 is a pretty good exemplar.
Sat Oct 30 13:06:49 +0000 2021I am irrationally annoyed when I see lower case characters being used to differentiate between human & non-human names for the "same" gene, e.g. using SACS (human) vs Sacs (mouse). All caps, all the time.
Sat Oct 30 12:39:27 +0000 2021YARS:p, translation initiation occurs at either M1 or M37. Initiation at M37 produces a protein sequence with an N-terminal sequence similar to most YARS proteins. The N-terminal, positively-charged, low complexity domain (1-36) is present in mouse & rat mRNA.
Sat Oct 30 12:39:26 +0000 2021YARS:p.A36D chr 4:129083807C>A, rs27493376 (all tissue A:D 0.989:0.011) vaf=5%, Δm=43.9898.
Sat Oct 30 12:39:26 +0000 2021YARS:p, θ(max) = 83. aka none. Common in tissues & cell lines: absent from fluids, but may be present in extracellular vesicles.
Sat Oct 30 12:39:26 +0000 2021>YARS:p, tyrosyl-tRNA synthetase (Mus musculus) Midsized subunit; CTMs: M1, G38+acetyl; PTMs: 10×K+acetyl; 3×K+GGyl; 3×S, 0×T, 2×Y+phosphoryl; SAAVs: A36D (5%); mature form: (1,38-564) [13,703×, 187 kTa]. #ᗕᕱᗒ 🔗
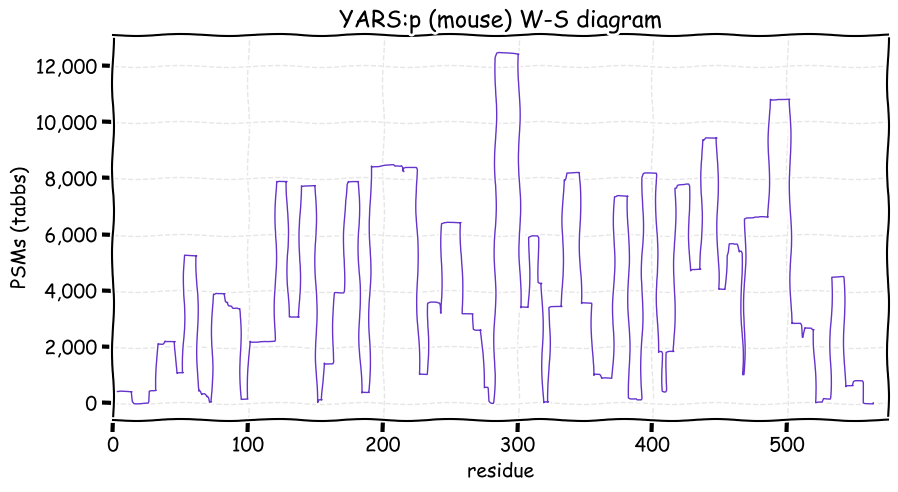
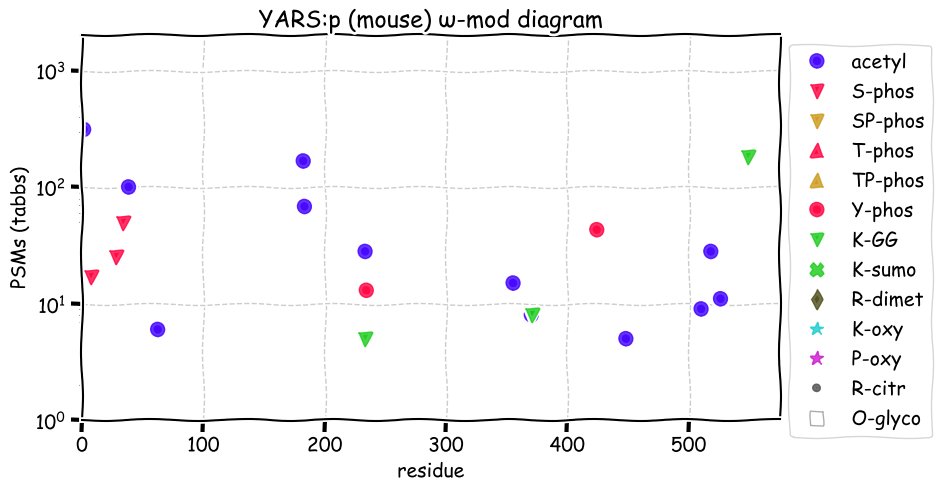
Fri Oct 29 23:46:50 +0000 2021There is supposed to be a strong (G3) geomagnetic storm tomorrow, as a coronal mass ejection produced by a solar flare yesterday passes the earth. The aurora may be visible as far south as Portland and Chicago.
Fri Oct 29 17:39:42 +0000 2021Seattle has a (not very) surprisingly large number of web-available weather stations that record rainfall data (this is yesterday's 24 h totals). Black circles indicate clusters of stations too close together to be viewed at this zoom level. 🔗
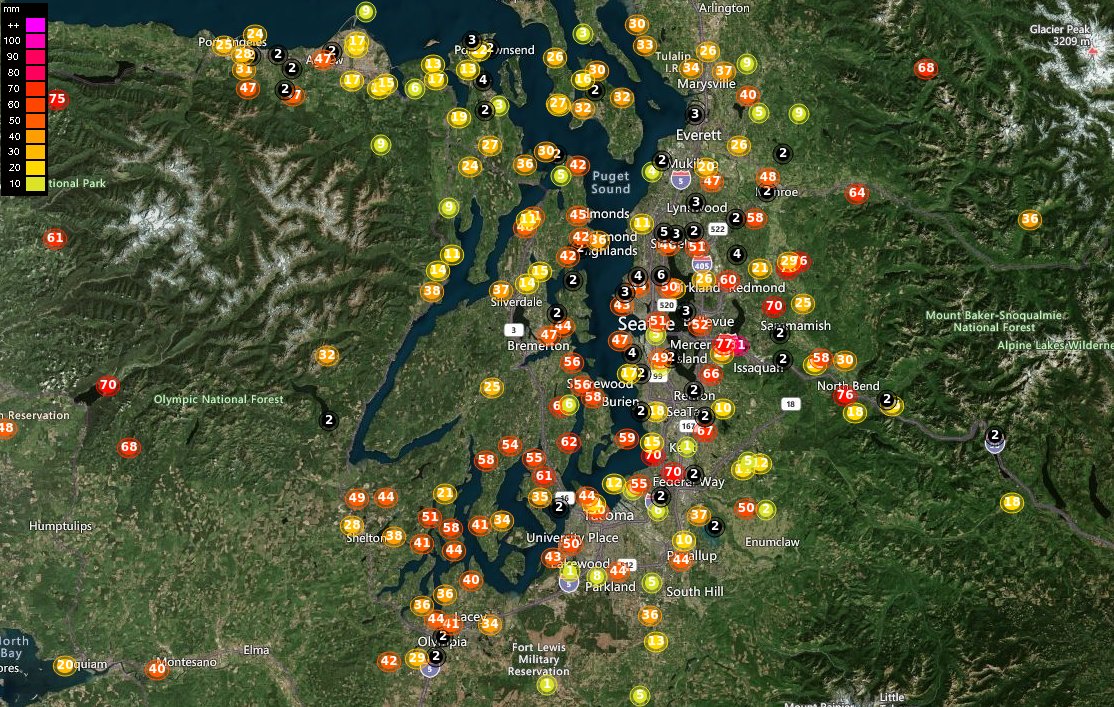
Fri Oct 29 14:24:49 +0000 2021@neely615 @danjbecker @ProteomicsNews Having "mass spectrometrist" as part of your credentials is much more important than "protein biochemist".
Fri Oct 29 14:23:39 +0000 2021@neely615 @danjbecker @ProteomicsNews I think one of the most limiting consequences has been that "proteomics" meetings have fallen into a structure & pattern of presentations similar to the ASMS, rather than (for example) the ASBMB.
Fri Oct 29 12:35:48 +0000 2021As a "field", proteomics has always suffered from being dominated by those interested in taking the data, rather than those interested in its use.
Fri Oct 29 11:56:32 +0000 2021I haven't mentioned it for a while, but the annotation style that I use in these daily blurbs is based on the HGVS annotation recommendations for substitutions (🔗) as extended for PTMs (🔗)
Fri Oct 29 11:52:22 +0000 2021CTBP1:p, translation initiation occurs at M1 or M9. Because of sequence similarity it is not possible to determine whether this alternate initiation occurs on CTBP1:r, CTBP2:r or both mRNAs, although there is unique evidence for M1 initiation in both cases.
Fri Oct 29 11:52:21 +0000 2021CTBP1:p.V358I chr 4:g.1212947C>T, rs138269585 (all tissue V:I 0.998:0.002)vaf=<1%, Δm=14.0156, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Fri Oct 29 11:52:21 +0000 2021CTBP1:p, θ(max) = 65. aka none. Observed in tissue class 1 MHC peptide expts. Common in most tissues & cell lines: absent from fluids, but has been found in cell line extracellular vesicles.
Fri Oct 29 11:52:21 +0000 2021>CTBP1:p, C-terminal binding protein 1 (Homo sapiens) Small subunit; CTMs: S2, M9+acetyl; PTMs: 3×K+acetyl; 5×K+GGyl; 1×S, 1×T, 0×Y+phosphoryl; R5+dimethyl; SAAVs: V358I (<1%); mature form: (2,9-429) [29,901×, 152 kTa]. #ᗕᕱᗒ 🔗
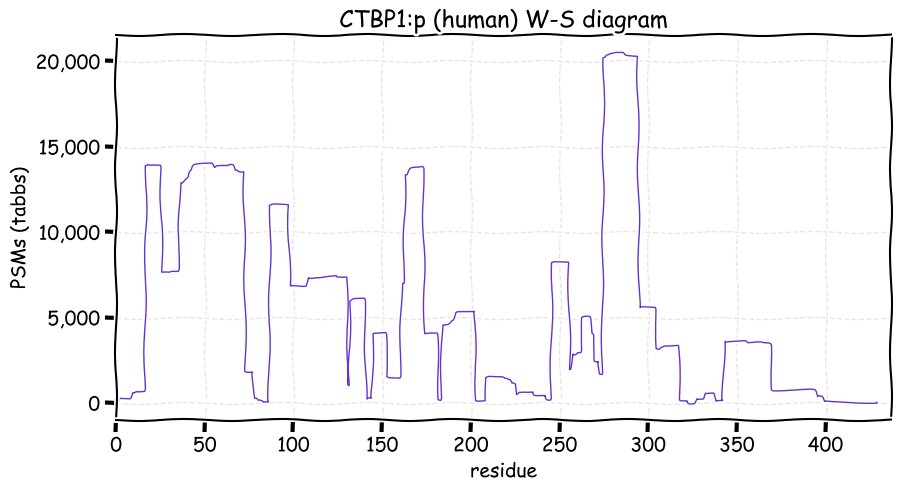
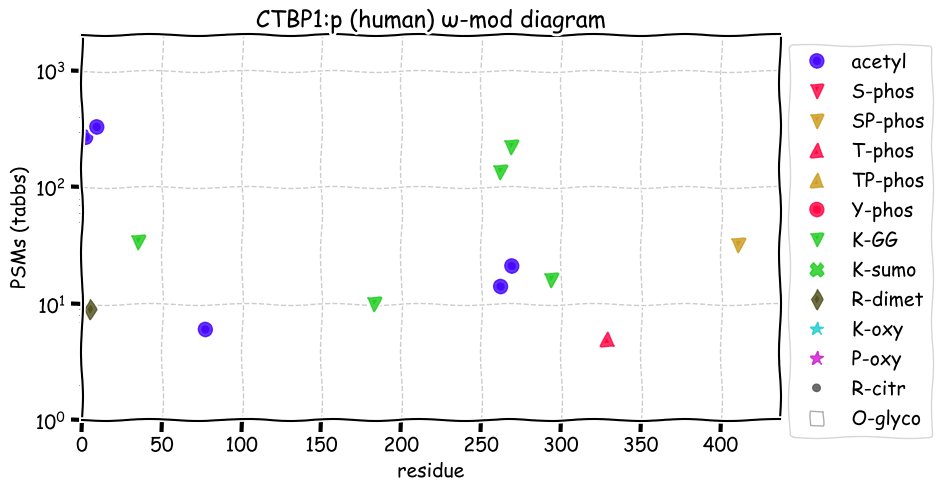
Thu Oct 28 16:31:06 +0000 2021It's foggy here this morning 🔗
Thu Oct 28 15:56:49 +0000 2021@astacus Seems a little "retro", but if that is the UK equivalent ...
Thu Oct 28 15:00:44 +0000 2021Are there any practical alternatives to cars like Teslas if you want to run an automobile on coal?
Thu Oct 28 14:16:42 +0000 2021@jwoodgett We're showing fealty to our liege by making them look good.
Thu Oct 28 12:36:22 +0000 2021Is there an abbreviation for protein variant sequences caused by alternate translation initiation events? If not, I'm calling dibs on "ATIV".
Thu Oct 28 11:58:57 +0000 2021CUL4B:p, translation initiation occurs at M1 or M116. Initiation at M116 removes the highly modified N-terminal low complexity domain, leaving the functional cullin domain. The resulting protein is similar to CUL4A in size & PTM pattern.
Thu Oct 28 11:58:56 +0000 2021CUL4B:p.R464K chr X:g.120541654C>T, rs188531002 (all tissue R:K 0.999:0.001) vaf=<1%, Δm=-28.0061, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Thu Oct 28 11:58:56 +0000 2021CUL4B:p, θ(max) = 75. aka none. Observed in tissue class 1 MHC peptide expts. Common in most tissues & cell lines: may be present in urinary extracellular vesicles.
Thu Oct 28 11:58:56 +0000 2021>CUL4B:p, cullin 4B (Homo sapiens) Midsized subunit; CTMs: M1, A117+acetyl; PTMs: 5×K+acetyl; 39×K+GGyl; 1×K+SUMOyl; 53×S, 18×T, 4×Y+phosphoryl; SAAVs: R464K (<1%); mature form: (1,117-895) [39,654×, 271 kTa].#ᗕᕱᗒ 🔗
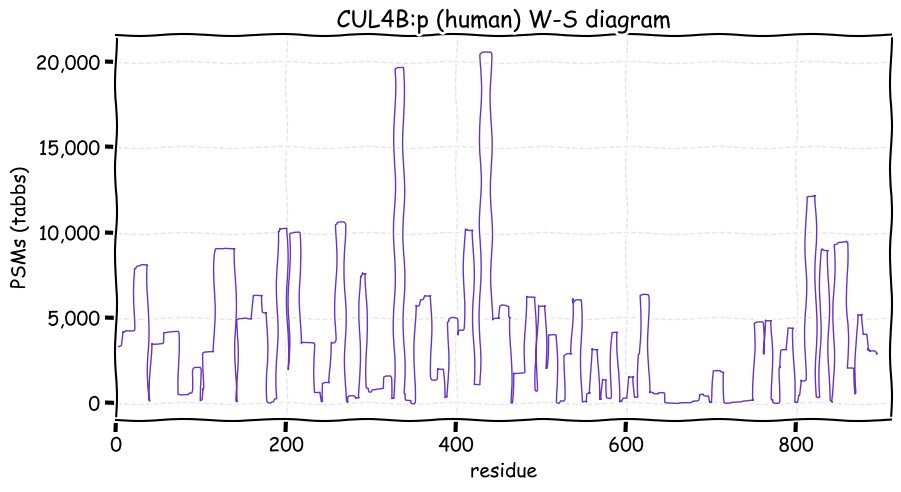
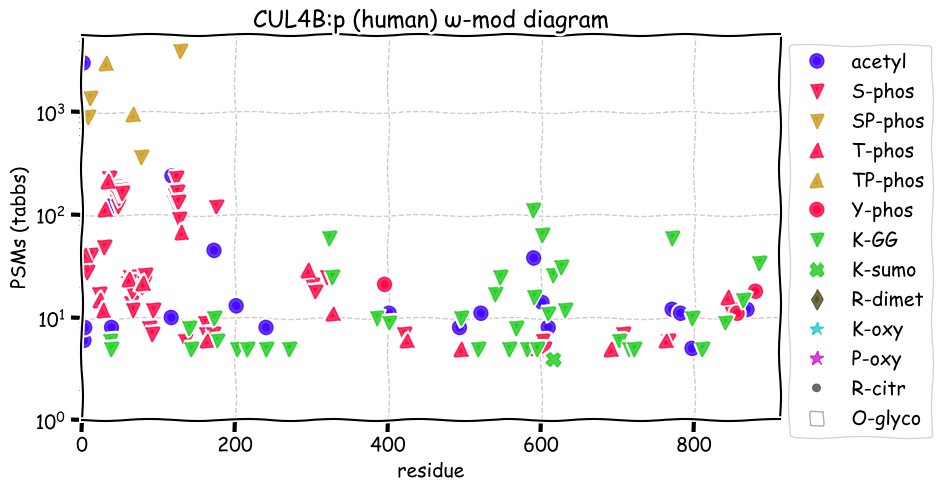
Wed Oct 27 20:17:09 +0000 2021@astacus @Peptidome @neely615 @MagnusPalmblad @IDsignals To be the devil's advocate: insulin C-peptide in humans has a similar relationship between translation & urine concentration, but insulin does not.
Wed Oct 27 13:00:35 +0000 2021KRR1:p, 98% of phosphorylated PSMs are associated with the proline-directed acceptor site S3+phosphoryl.
Wed Oct 27 12:14:21 +0000 2021KRR1:p.R134Q chr 12:g.75506602C>T, rs11540407 (all tissue R:Q 0.033:0.967) vaf=25%, Δm=-28.0425, VAF by population group: african 19%, american 36%, east asian 12%, european 23%, south asian 31%. QR in HeLa cells. SAAV abolishes a tryptic cleavage site.
Wed Oct 27 12:14:20 +0000 2021KRR1:p, θ(max) = 59. aka RIP-1, HRB2. Observed in tissue class 1 MHC peptide expts. Common in most tissues & cell lines: absent from fluids.
Wed Oct 27 12:14:20 +0000 2021>KRR1:p, KRR1 small subunit processome component homolog (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 5×K+acetyl; 6×K+GGyl; 8×K+SUMOyl; 5×S, 0×T, 3×Y+phosphoryl; SAAVs: R134Q (25%); mature form: (2-381) [18,585×, 78 kTa]. #ᗕᕱᗒ 🔗
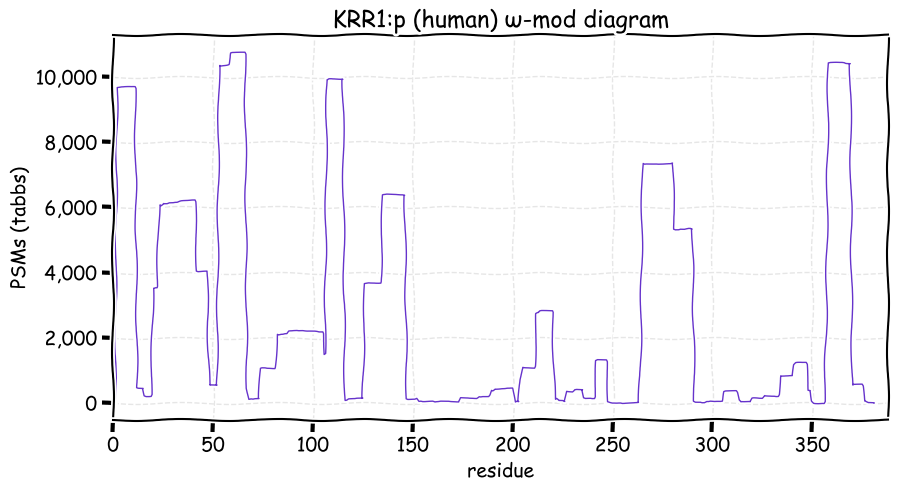
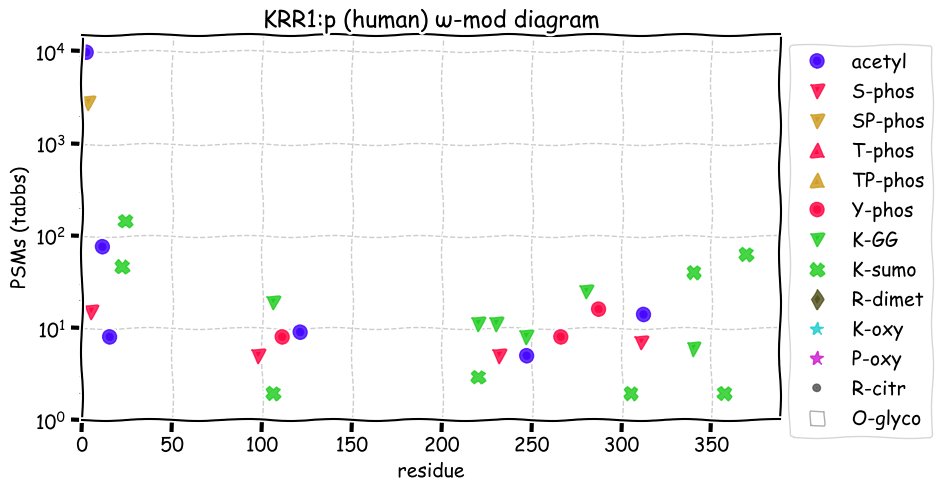
Tue Oct 26 15:46:12 +0000 2021@gingraslab1 @pedrobeltrao Does anybody know of an analogous paper for ubiquitination site motifs?
Tue Oct 26 13:51:55 +0000 2021@gingraslab1 @pedrobeltrao PS: I haven't given this order any more than the most preliminary of considerations.
Tue Oct 26 13:43:45 +0000 2021@gingraslab1 @pedrobeltrao The motif list is as follows:
ΦP,
RxxSxxΦ,
LxRxxΦ,
SxxΦL,
RxxΦ,
ΦxxD/E,
RxxΦL/I/V,
RRxΦ,
NxΦ,
R/KRxxΦ,
GΦ,
ΦD/ExD/E,
RSxΦ,
ΦxExL/I/V,
ΦF,
NxΦL/I/V,
ΦQ,
ΦL/I/V
The algo goes down the list & reports the first one that matches.
Tue Oct 26 12:46:11 +0000 2021@gingraslab1 I am using the motifs suggested by these folks 🔗
Tue Oct 26 12:38:48 +0000 2021Altered my PTM diagram a bit (as of today) to break out proline directed phosphorylation in a different color (gold) and including a column for known phosphorylation site motifs 🔗
Tue Oct 26 11:50:39 +0000 2021KBTBD11:p, translation initiation occurs at M1 or M197 in human (M1 or M203 in mouse). Initiation at M197 excludes the (140-196) BTB/POZ dimerization domain leaving the Kelch repeat structure intact.
Tue Oct 26 11:50:38 +0000 2021KBTBD11:p.R477C chr 8:g.2002621C>T, rs144532869 (all tissue R:C 0.999:0.001) vaf=0.1%, Δm=-53.0919, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. SAAV removes a tryptic cleavage site.
Tue Oct 26 11:50:38 +0000 2021KBTBD11:p, θ(max) = 65. aka KIAA0711, KLHDC7C. May be observed in tissue class 1 MHC peptide expts. Most abundant in brain & liver tissue: may be present in urinary extracellular vesicles.
Tue Oct 26 11:50:38 +0000 2021>KBTBD11:p, kelch repeat and BTB domain containing 11 (Homo sapiens) Midsized subunit; CTMs: M1, A197+acetyl; PTMs: 2×K+GGyl; 23×S, 1×T, 2×Y+phosphoryl; SAAVs: R477C (<1%); mature form: (1,197-623) [6,953×, 43 kTa]. #ᗕᕱᗒ 🔗
Mon Oct 25 16:17:40 +0000 2021How many S/T-kinases, other than CDKs, require an (S/T)P motif at their acceptor sites?
Mon Oct 25 15:01:49 +0000 2021🔗
Mon Oct 25 12:16:31 +0000 2021CDC42EP1:p.R76H chr 22:g.37566576G>A, rs61739954 (all tissue R:H 0.990:0.010) vaf=1%, Δm=-19.0422, VAF by population group: african 5%, american <1%, east asian <1%, european <1%, south asian <1%. SAAV removes a tryptic cleavage site.
Mon Oct 25 11:59:04 +0000 2021CDC42EP1:p, θ(max) = 61. aka MSE55, CEP1, Borg5. Occasionally observed in class 1 MHC peptide expts. Found in many tissues & cell lines: absent from fluids. Translation initiation occurs at both M1 and M13 in human & mouse sequences.
Mon Oct 25 11:59:04 +0000 2021>CDC42EP1:p, CDC42 effector protein 1 (Homo sapiens) Midsized subunit; CTMs: M1, P2, S14+acetyl; PTMs: 1×K+GGyl; 2×R+dimethyl; 36×S, 9×T, 3×Y+phosphoryl; SAAVs: R76H (1%), R97Q (1%); mature form: (1,14-391) [23,136×, 171 kTa]. #ᗕᕱᗒ 🔗
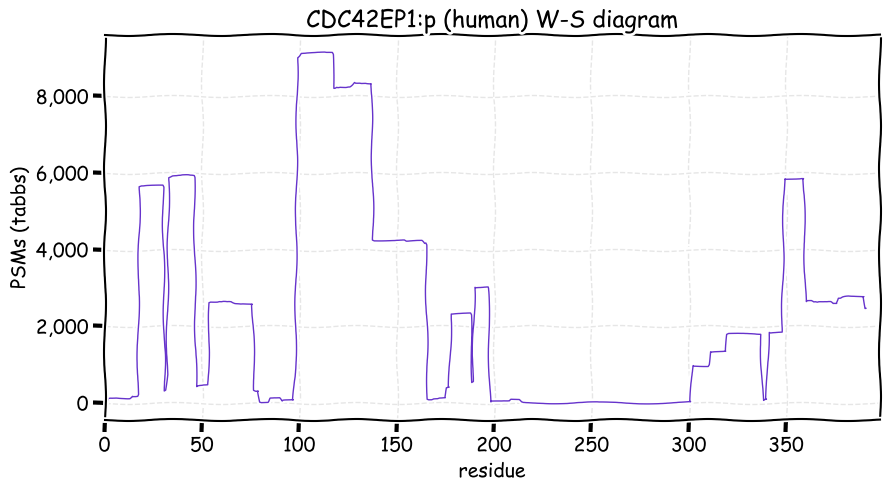
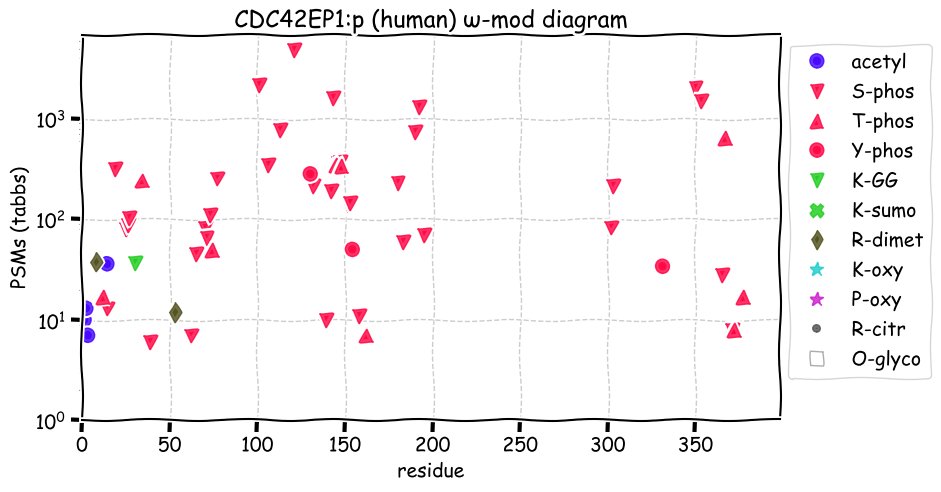
Sun Oct 24 23:26:20 +0000 2021Looks like one of those nasty fall storms everyone forgets to mention before you move to Vancouver. 🔗
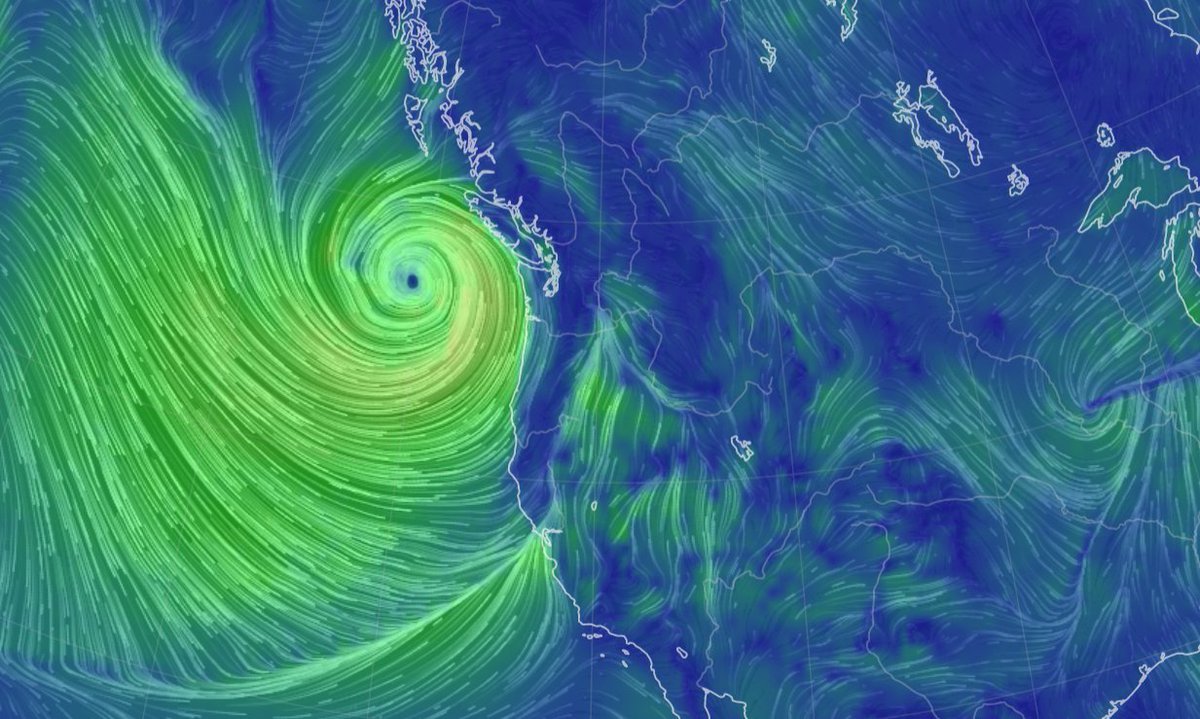
Sun Oct 24 12:32:27 +0000 2021CD3EAP:p.K259T chr 19:g.45408744A>C, rs735482 (all tissue K:T 0.224:0.776) vaf=21%, Δm=-27.0473, VAF by population group: african 31%, american 33%, east asian 46%, european 13%, south asian 28%. KT in SK-MEL-28 cells. TT in HEp-2 & HeLa cells.
Sun Oct 24 12:32:27 +0000 2021CD3EAP:p, θ(max) = 88. aka ASE-1, CAST, PAF49, RPA34, A34.5, CD3EAP. Occasionally observed in class 1 MHC peptide expts. Found in many tissues, cell lines & in extracellular vesicles.
Sun Oct 24 12:32:27 +0000 2021>CD3EAP:p, CD3e molecule associated protein (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 2×K+acetyl; 2×K+GGyl; 5×K+SUMOyl; 18×S, 23×T, 0×Y+phosphoryl; SAAVs: K259T (21%), T282A (2%); mature form: (1-510) [23,136×, 171 kTa]. #ᗕᕱᗒ 🔗
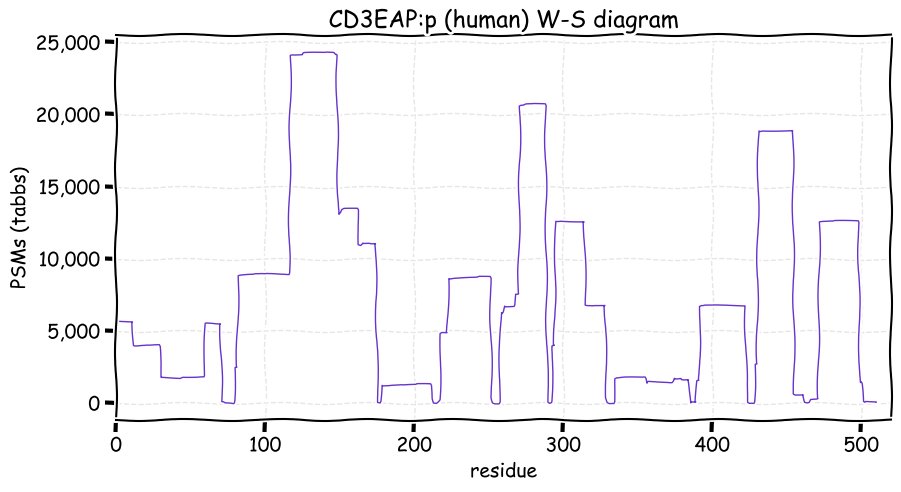
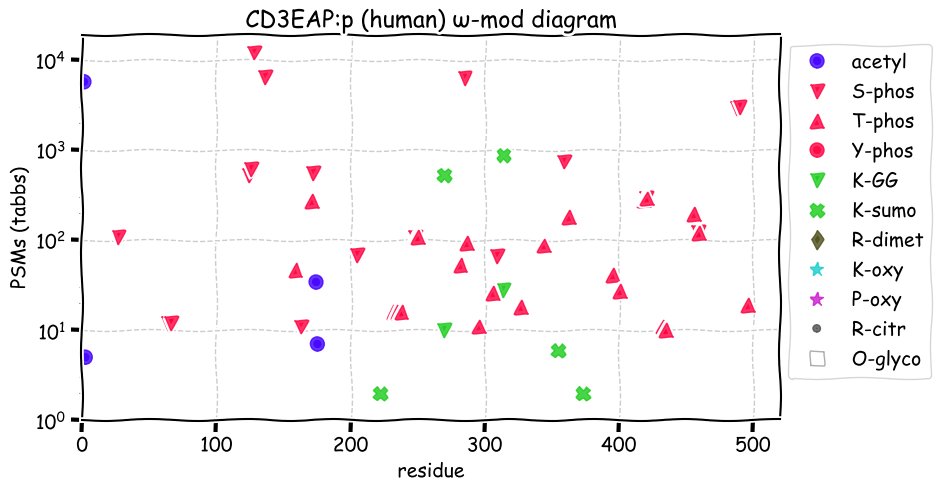
Sat Oct 23 16:31:48 +0000 2021@jwoodgett Good work summarizing the curiously convoluted conundrum that is Canadian biomedical research funding. ⭐️⭐️⭐️⭐️
Sat Oct 23 14:13:55 +0000 2021@leprevostfv It is a tricky problem: showing a lot of disparate information over > 4 orders of magnitude of dynamic range, where the low occupancy sites may be more important physiologically than the high occupancy ones.
Sat Oct 23 13:58:29 +0000 2021@leprevostfv If you want to see how the table/plot was generated using the GPMDB's REST interface, the code is at:
🔗
Sat Oct 23 13:57:24 +0000 2021@leprevostfv If you just want a tabular display of the information, you can use
🔗
Sat Oct 23 13:21:10 +0000 2021After struggling with the disease for some time, Cuba seems to be near the end of this wave of fatalities. The US & Russia (!) are still in the midst, however. 🔗
Sat Oct 23 13:04:42 +0000 2021@leprevostfv It is all public, but there is quite a bit of it (~860,000 PSMs from 64,632 LC-MS/MS runs). Did you want a list of the PXD/Massive/TRANCHE accessions?
Sat Oct 23 12:10:07 +0000 2021OTUD7B:p, alternate translation initiation observed at M5: this translation also occurs in rodents.
Sat Oct 23 12:10:06 +0000 2021OTUD7B:p.Y631H chr 1:g.149944498A>G, rs28365868 (all tissue Y:H 0.999:0.001) vaf=<1%, Δm=-26.0044, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sat Oct 23 12:10:06 +0000 2021OTUD7B:p, θ(max) = 51. aka CEZANNE, ZA20D1. Observed in class 1 MHC peptide expts. Found in many tissues & cell lines: may be present in urine extracellular vesicles.
Sat Oct 23 12:10:06 +0000 2021>OTUD7B:p, OTU deubiquitinase 7B (Homo sapiens) Midsized subunit; CTMs: T2, M5+acetyl; PTMs: 2×K+acetyl; 7×K+GGyl; 34×S, 6×T, 3×Y+phosphoryl; SAAVs: V11L (0.4%); mature form: (2,3-843) [8,468×, 31 kTa]. #ᗕᕱᗒ 🔗
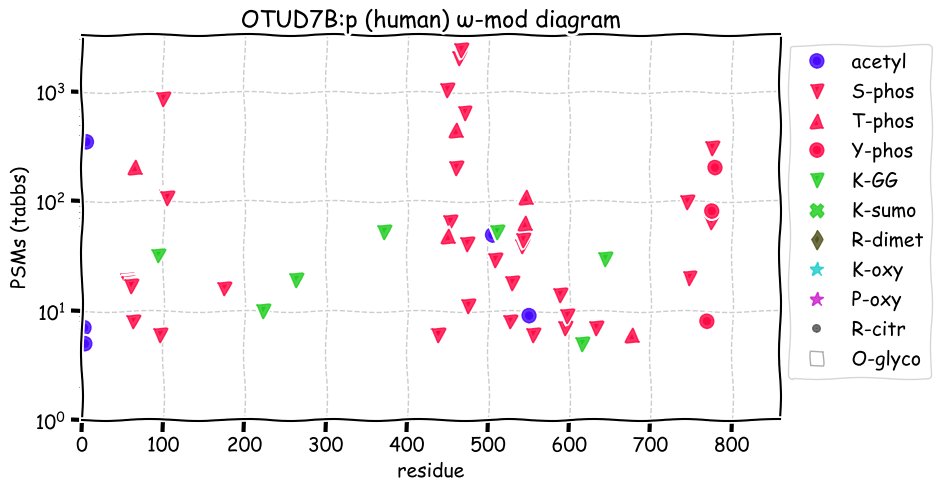
Fri Oct 22 17:24:20 +0000 2021@jjc2718 A problem for non-developer students who want to use software is that they don't know how to talk about it on a technical level. Having them exposed to occasional discussions of how it works helps them & nudges the local coders to explain things to a more general audience.
Fri Oct 22 16:39:01 +0000 2021@jjc2718 And it should be part of the normal lab meeting schedule.
Fri Oct 22 14:56:06 +0000 2021Just finished configuring a DNS forwarder for an internal split DNS implementation. Made me re-learn the details of how BIND services work: designing and implementing one would probably make a decent DNS tutorial.
Fri Oct 22 12:18:47 +0000 2021@matthewhirschey It was a job title. Any large company had computers in accounting, but most of the work was adding up columns of numbers for bookkeeping. They used a variety of mechanical calculators, but a most of the work was done with a pencil & paper.
Fri Oct 22 12:01:35 +0000 2021NASP:p, alternate translation initiation observed at M3: this particular translation appears to be unique to humans. Has one of the least charismatic names of any human protein.
Fri Oct 22 12:01:34 +0000 2021NASP:p.V11L chr 1:g.45584177G>C, rs41265176 (all tissue V:L 0.989:0.011) vaf=0.4%, Δm=14.0156, VAF by population group: african <1%, american <1%, east asian 3%, european <1%, south asian <1%. VL in 143B cells.
Fri Oct 22 12:01:34 +0000 2021NASP:p, θ(max) = 81. aka FLB7527, FLJ31599, FLJ35510, MGC19722, MGC20372, MGC2297, DKFZp547F162, PRO1999. Observed in class 1 MHC peptide expts & in some tissue class 2 expts. Found in many tissues & cell lines: very rare in fluids..
Fri Oct 22 12:01:34 +0000 2021>NASP:p, nuclear autoantigenic sperm protein (Homo sapiens) Midsized subunit; CTMs: A2, M3+acetyl; PTMs: 53×K+acetyl; 26×K+GGyl; 3×K+SUMOyl; 56×S, 32×T, 2×Y+phosphoryl; SAAVs: V11L (0.4%); mature form: (2,3-788) [64,632×, 859 kTa]. #ᗕᕱᗒ 🔗
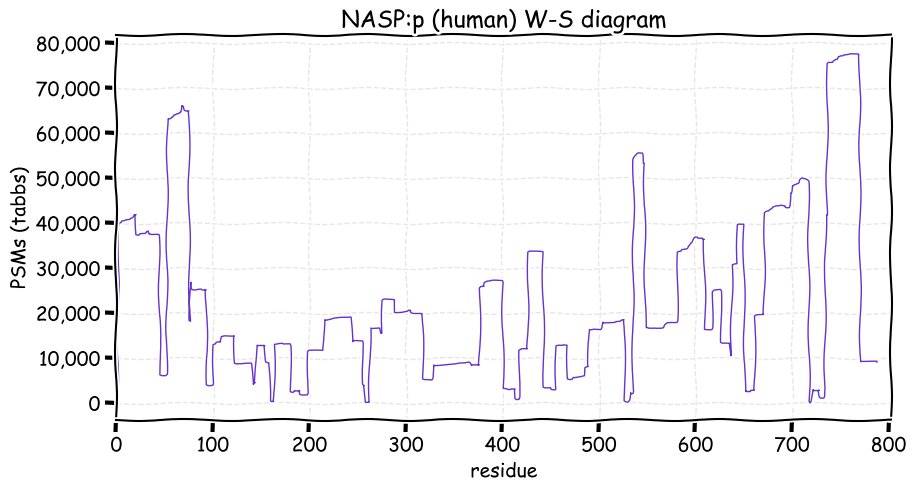
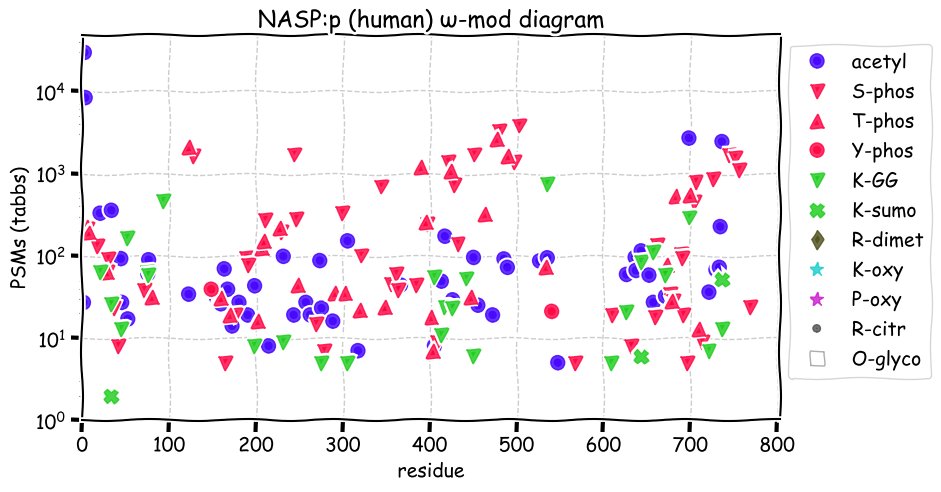
Thu Oct 21 17:46:43 +0000 2021Until yesterday, these 2 attractive (& very reflective) coasters were platters in one of the OS hard drives in the GPMDB assembly. 🔗

Thu Oct 21 17:16:59 +0000 2021If you are contemplating whether or not to start the latest Win10 update process, the statement "This may take a while" is a bit of an understatement.
Thu Oct 21 14:49:43 +0000 2021These kits really should say this on the label.
Thu Oct 21 14:47:06 +0000 2021To whom it may concern: using a commercial high pH RP 8 fraction kit is not a good choice for phosphopeptide studies.
Thu Oct 21 12:05:45 +0000 2021YIF1A:p, Predicted TM domains: (139-158), (170-192), (212-234) & (271-293); (2-138) intracellular phosphodomian. 🔗

Thu Oct 21 12:05:45 +0000 2021YIF1A:p.G475D chr 7:g.86838938G>A, rs17161026 (all tissue G:D 0.966:0.034) vaf=1%, Δm=58.0055, VAF by population group: african 1%, american <1%, east asian <1%, european 2%, south asian <1%.
Thu Oct 21 12:05:45 +0000 2021YIF1A:p, θ(max) = 37. aka YIF1P, 54TM, FinGER7, YIF1. Observed in class 1 MHC peptide experiments. Found in many tissues & cell lines: very rare in fluids.
Thu Oct 21 12:05:44 +0000 2021>YIF1A:p, Yip1 interacting factor homolog A, membrane trafficking protein (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 4×S+phosphoryl; SAAVs: G475D (1%); mature form: (2-293) [11,905×, 29 kTa]. #ᗕᕱᗒ 🔗
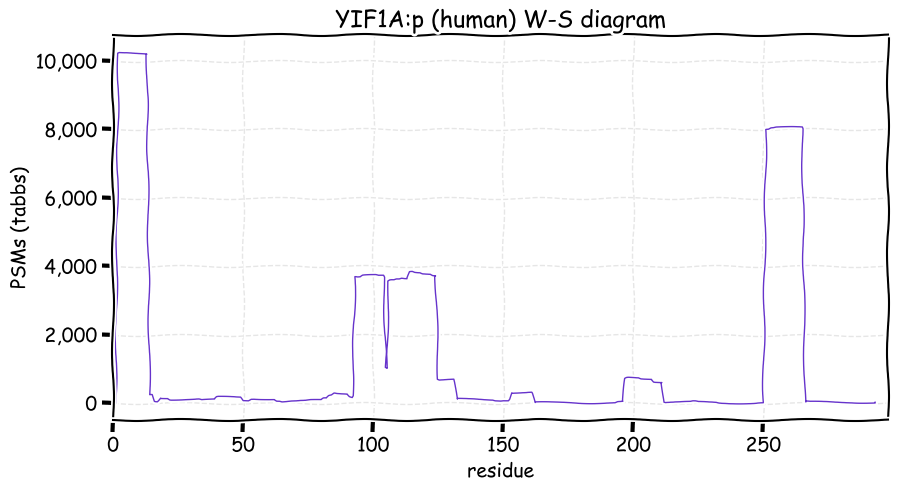
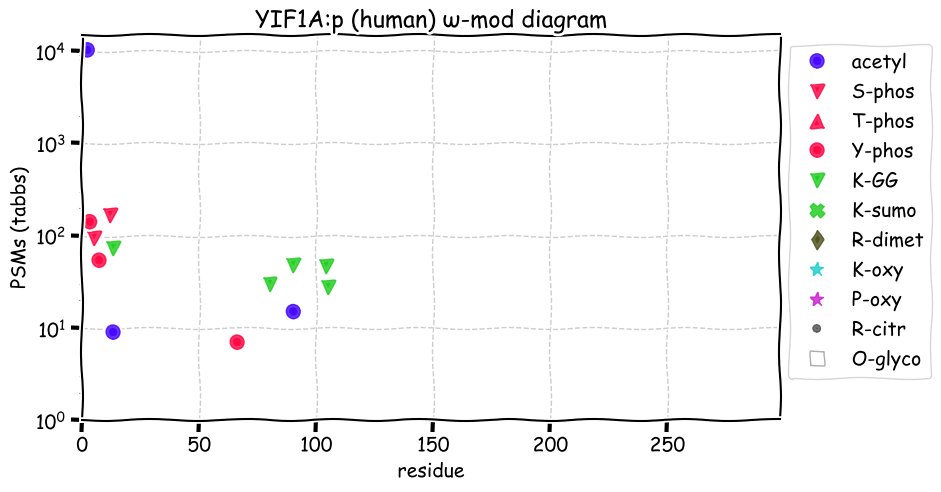
Wed Oct 20 20:35:54 +0000 2021@ypriverol @leprevostfv I got the files from both, but both threw the following error once:
"Could not read from socket: ECONNRESET - Connection reset by peer"
It didn't stop the download, but it appears in the download log.
Wed Oct 20 19:30:20 +0000 2021@leprevostfv @ypriverol Same here.
Wed Oct 20 19:13:58 +0000 2021@ypriverol @leprevostfv Sure.
Wed Oct 20 18:04:27 +0000 2021GRM3:p, predicted TM domains: (577-599), (612-634), (687-709), (737-756), (771-793) & (806-828); (25-576) extracellular, (794-879) intracellular phosphodomain. 🔗

Wed Oct 20 18:04:26 +0000 2021GRM3:p.G475D chr 7:g.86838938G>A, rs17161026 (all tissue G:D 0.966:0.034) vaf=1%, Δm=58.0055, VAF by population group: african 1%, american <1%, east asian <1%, european 2%, south asian <1%.
Wed Oct 20 18:04:26 +0000 2021GRM3:p, θ(max) = 31. aka GPRC1C, mGlu3, MGLUR3. Observed in class 1 MHC peptide experiments. Found in brain tissue: rare in cell lines.
Wed Oct 20 18:04:25 +0000 2021>GRM3:p, glutamate receptor, metabotropic 3 (Homo sapiens) Midsized subunit; CTMs: none; PTMs: 4×S+phosphoryl; SAAVs: G475D (1%); mature form: (25-879) [1,010×, 5 kTa]. #ᗕᕱᗒ 🔗
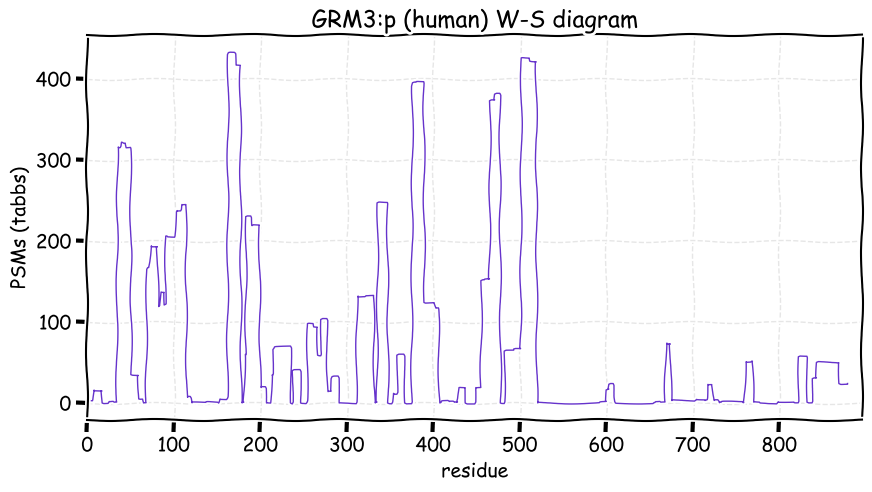
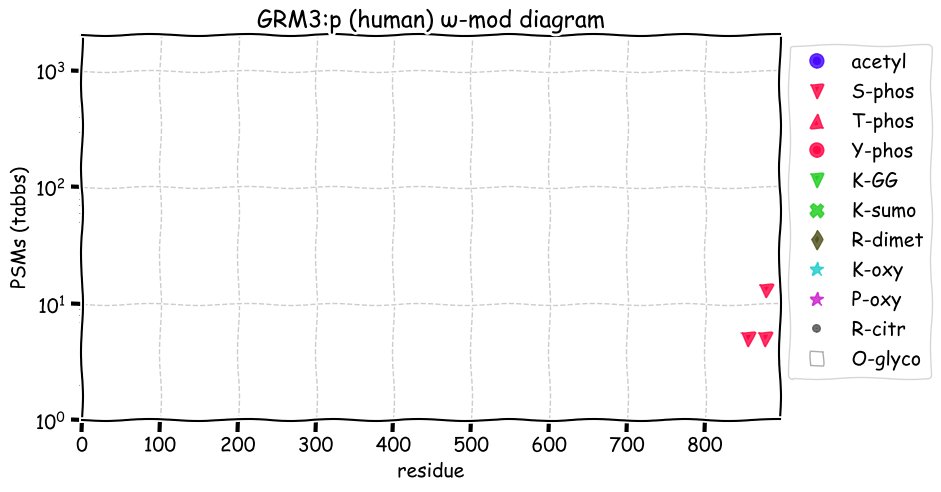
Tue Oct 19 16:22:17 +0000 2021Is it just me or are T*mo RAW files more fragile to copying errors than a lot of other binary file formats.
Tue Oct 19 15:16:26 +0000 2021Someone should write a paper (really, it is very low hanging fruit). While it may not be as obviously compelling as FH or GARS1, it shouldn't be too hard to turn it into a story someone might be interested in printing.
Tue Oct 19 15:00:48 +0000 2021LYPLA1:p (lysophospholipase 1) initiates translation at M6: M6 is then removed and S7 N-acetylated in human, mouse & rat sequences. M1 is rarely, if ever, the site of translation initiation.
Tue Oct 19 12:15:57 +0000 2021GCGR:p, predicted TM domains (plasma membrane): (145-167), (176-198), (227-249), (262-284), (304-326), (347-369) & (382-404); (27-144) is extracellular & (405-477) is in the cytoplasm & has all observed phosphorylation sites. 🔗

Tue Oct 19 12:15:56 +0000 2021GCGR:p.S456T chr 17:g.81813622G>C, rs766891858 (all tissue S:T 0.996:0.004) vaf=<1%, Δm=14.0157, VAF by population group: african <1%, american <1%, east asian<1%, european <1%, south asian <1%. ST in HEK-293 cells.
Tue Oct 19 12:15:56 +0000 2021GCGR:p, θ(max) = 12. aka CGI-119, S1R, ZPRO, LFG4, GAAP. Observed in class 1 MHC peptide experiments. May be found in liver & specialized cell lines.
Tue Oct 19 12:15:56 +0000 2021>GCGR:p, glucagon receptor (Homo sapiens) Small subunit; CTMs: none; PTMs: 7×S, 2×T, 0×Y+phosphoryl; SAAVs: S456T (<1%); mature form: (27-477) [809×, 2.7 kTa]. #ᗕᕱᗒ 🔗
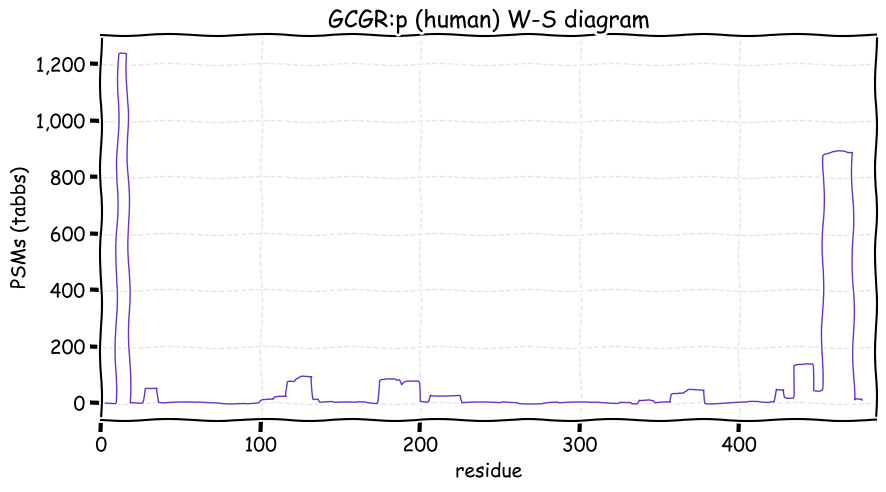
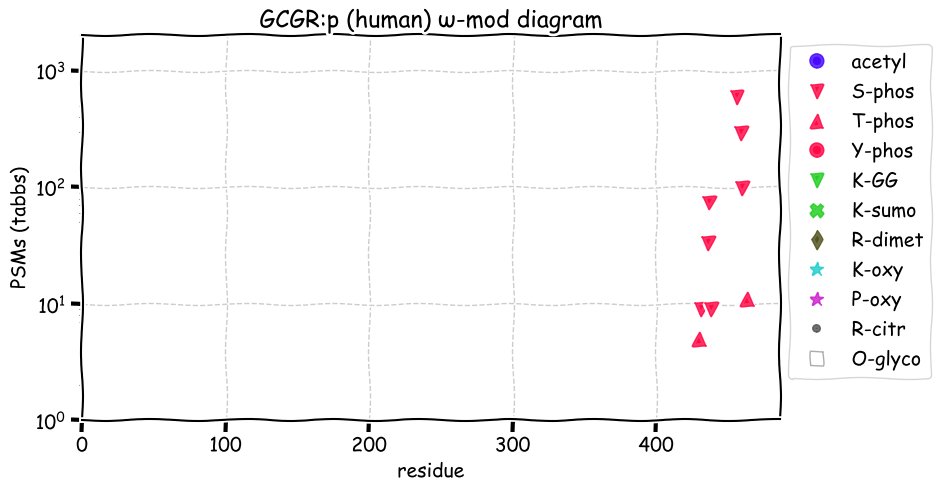
Mon Oct 18 19:42:06 +0000 2021@emblebi @embl 75% sounds a little high ...
Mon Oct 18 18:47:53 +0000 2021"human microvascular endothelial cells derived from adipose tissue" is one of those sample descriptions that doesn't really suit itself to representation using standard ontologies.
Mon Oct 18 15:39:39 +0000 2021@AaronWherry Pearson, 1963 (🔗)
Mon Oct 18 15:26:49 +0000 2021@DRAWheatcraft @neely615 @faunabio Sometimes my zoology degree comes in handy, at least for surprising people with odd animals ...
Mon Oct 18 15:15:55 +0000 2021@neely615 @faunabio Fat-tailed dwarf lemur, hands down 🔗
Mon Oct 18 12:06:46 +0000 2021TMBIM4:p, predicted TM domains (Golgi membrane): (38-60), (70-89), (96-118), (123-145), (152-174), (178-197) & (210-232); (2-37) is cytoplasmic & (233-238) is in the Golgi lumen. 🔗

Mon Oct 18 12:06:45 +0000 2021TMBIM4:p.A2V chr 12:66169947G>A, rs11176070 (all tissue A:V 0.667:0.333) vaf=<1%, Δm=-28.0313, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. AV in HeLa cells.
Mon Oct 18 12:06:45 +0000 2021TMBIM4:p, θ(max) = 31. aka CGI-119, S1R, ZPRO, LFG4, GAAP. Rarely observed in studies other than class 1 MHC peptide experiments. Rare, but found in tissues or cell lines.
Mon Oct 18 12:06:45 +0000 2021>TMBIM4:p, transmembrane BAX inhibitor motif containing 4 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: Y19+phosphoryl; SAAVs: A2V (<1%); mature form: (2-238) [505×, 0.8 kTa]. #ᗕᕱᗒ 🔗
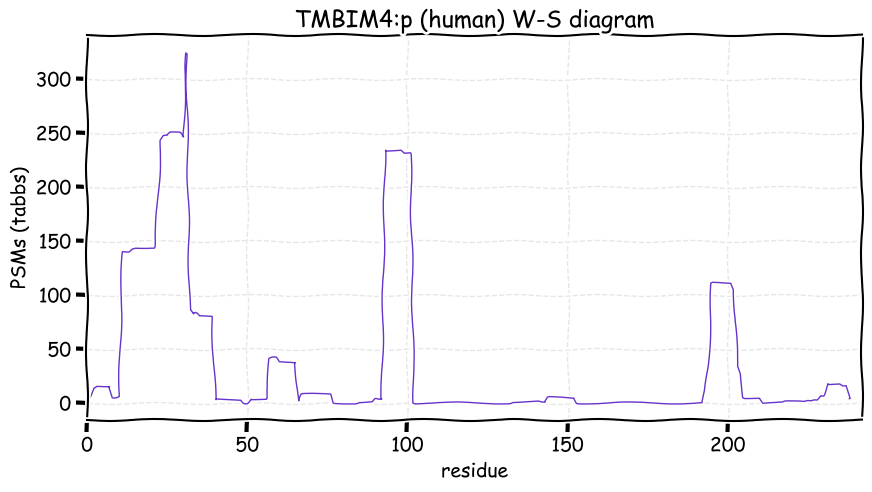
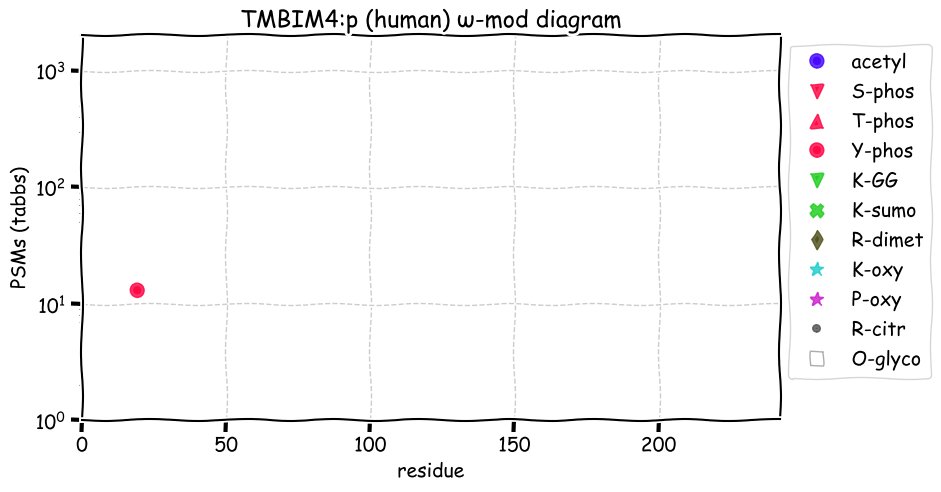
Mon Oct 18 01:12:07 +0000 2021My least favorite server problems have to be the ones caused by room temperature.
Sun Oct 17 12:31:20 +0000 2021TSPO:p, predicted TM domains in grey (mitochondrial external membrane): (4-26), (46-68), (78-100), (107-129) & (133-155); (1-3) is in the mitochondrial intermembrane space & (156-169) is cytoplasmic. 🔗

Sun Oct 17 12:31:19 +0000 2021TSPO:p.R162H chr 22:g.43162966G>A, rs6972 (all tissue R:H 0.709:0.291) vaf=23%, Δm=-19.0422, VAF by population group: african 6%, american 16%, east asian 24%, european 26%, south asian 34%.
Sun Oct 17 12:31:19 +0000 2021TSPO:p, θ(max) = 87. aka PBR, MBR, PKBS, mDRC, DBI, IBP, pk18, BZRP. Observed in class 1 & 2 MHC peptide experiments. Found in tissues & cell lines; also observed in blood plasma extracellular vesicles & urine from patients with UTIs.
Sun Oct 17 12:31:19 +0000 2021>TSPO:p, translocator protein (Homo sapiens) Small subunit; CTMs: M1, A2, P3+acetyl; PTMs: none; SAAVs: T147A (83%), R162H (23%); mature form: (1,2-169) [13,764×, 51 kTa]. #ᗕᕱᗒ 🔗
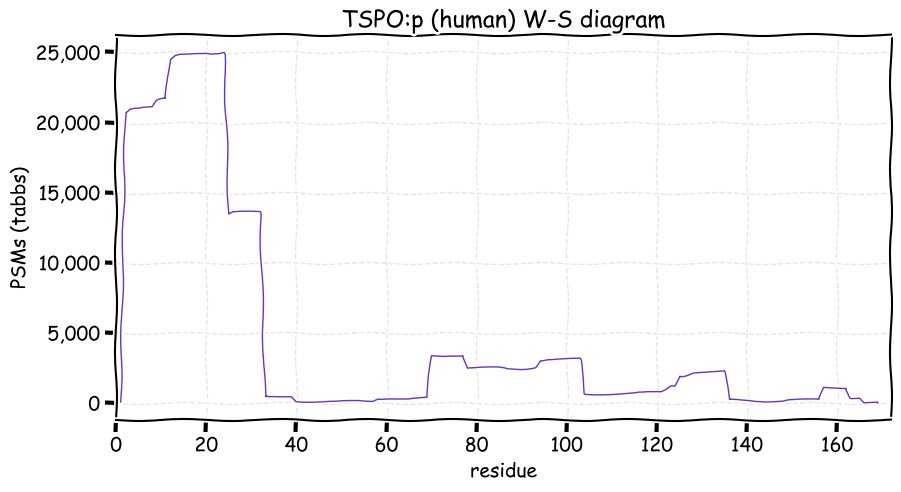
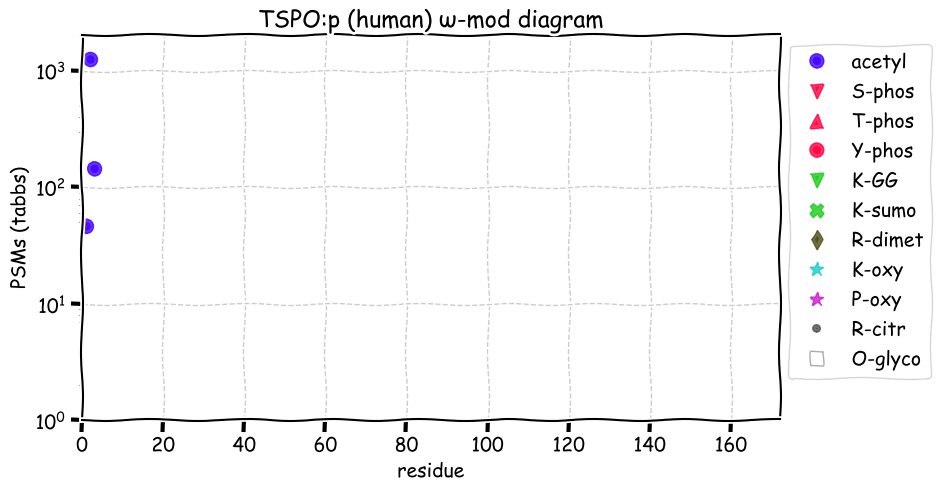
Sat Oct 16 14:37:49 +0000 2021@dtabb73 I spent an afternoon in Eastern Tennessee & didn't understand a single word that was said to me.
Sat Oct 16 13:05:42 +0000 2021FUS:p, it is very unusual to have such a high density of observed MHC class II peptides in a single domain.
Sat Oct 16 12:55:29 +0000 2021FUS:p, the domain (2-211) has very few charged residues (0×K, 0×R, 0×E & 4×D), so many proteolytic cleavage methods do not generate observable peptides from this region. Fortunately, >40 unique MHC class II peptides from this domain have been observed.
Sat Oct 16 12:39:32 +0000 2021FUS:p.P431L chr 16:31190398C>T, rs186547381 (all tissue P:L 0.994:0.006) vaf=<1%, Δm=16.0313, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sat Oct 16 12:39:31 +0000 2021FUS:p, θ(max) = 64. aka TLS, FUS1, hnRNP-P2, HNRNPP2, ALS6. Observed in class 1 & 2 MHC peptide experiments, with high coverage. Found in tissues & cell lines: absent from fluids.
Sat Oct 16 12:39:31 +0000 2021>FUS:p, FUS RNA binding protein (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 9×K+acetyl; 8×K+GGyl; 5×K+SUMOyl; 20×S, 10×T, 8×Y+phosphoryl; 35×R+dimethyl; SAAVs: P431L (<1%); mature form: (2-526) 92,049×, 1374 kTa]. #ᗕᕱᗒ 🔗
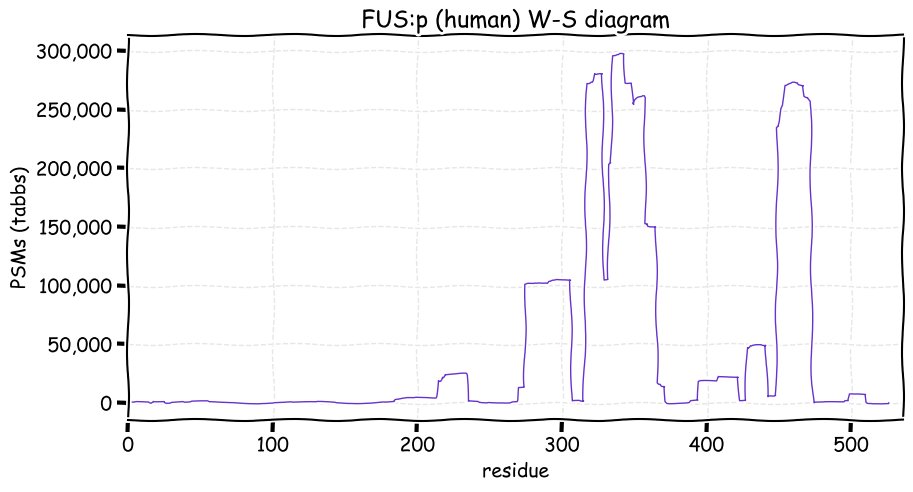
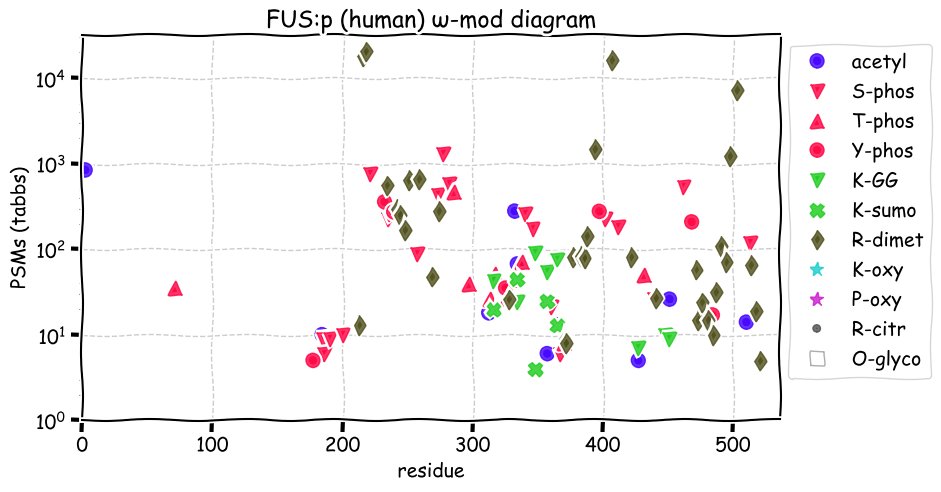
Fri Oct 15 14:44:07 +0000 2021@JennyRohn I saw something similar yesterday in an email reply from someone at Rockefeller U in New York.
Fri Oct 15 13:00:53 +0000 2021Something in their infrastructure seems to have gone down at 🔗
Fri Oct 15 12:24:58 +0000 2021SHCBP1:p.M21T chr 16:g.46621298A>G, rs6598679 (all tissue M:T 0.010:0.990) vaf=>99%, Δm=-29.9928, VAF by population group: african 94%, american >99%, east asian >99%, european >99%, south asian >99%.
Fri Oct 15 12:21:26 +0000 2021SHCBP1:p, θ(max) = 71. aka FLJ22009, PAL. Observed in class 1 MHC peptide experiments. Found in tissues & cell lines: very rare in fluids.
Fri Oct 15 12:21:26 +0000 2021>SHCBP1:p, SHC binding and spindle associated 1 (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 16×K+GGyl; 18×S, 3×T, 2×Y+phosphoryl; SAAVs: M21T (99%), M60R (1%); mature form: (2-672) [7,764×, 31 kTa]. #ᗕᕱᗒ 🔗
Thu Oct 14 17:13:24 +0000 2021Canadian innovation policy makers at work
🔗
Thu Oct 14 14:20:55 +0000 2021@AlexUsherHESA To be clear, I have cats & I am not recommending swinging them in anything other than a metaphorical sense.
Thu Oct 14 14:19:02 +0000 2021@AlexUsherHESA A primary indicator of which governments are successful wrt innovation vs all others is to count the number of STEMM PhD's in their senior decision making organizations. I suspect the PMO has about 0, while you can't swing a cat in the White House without hitting one.
Thu Oct 14 12:17:42 +0000 2021COPRS:p, the PTM pattern suggests two functional domains: (1-100) & (101-184). These domains have not been annotated or characterized biochemically.
Thu Oct 14 12:17:42 +0000 2021COPRS:p.S43G chr 17:g.31856838T>C, rs8068049 (all tissue S:G 0.256:0.744) vaf=88%, Δm=-30.0106, VAF by population group: african 100%, american 83%, east asian 87%, european 87%, south asian 90%. GG in HEK-293, MDA-MB-231 & MCF-7 cells.
Thu Oct 14 12:17:41 +0000 2021COPRS:p, θ(max) = 67. aka TTP1, HSA272196, COPR5, C17orf79. Observed in class 1 MHC peptide experiments only in thymus tissue. Found in tissues & cell lines: rarely in fluids.
Thu Oct 14 12:17:41 +0000 2021>COPRS:p, coordinator of PRMT5 and differentiation stimulator (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: R18+dimethyl; 10×S, 5×T, 0×Y+phosphoryl; SAAVs: S43G (88%); mature form: (1-184) [6,877×, 28 kTa]. #ᗕᕱᗒ 🔗
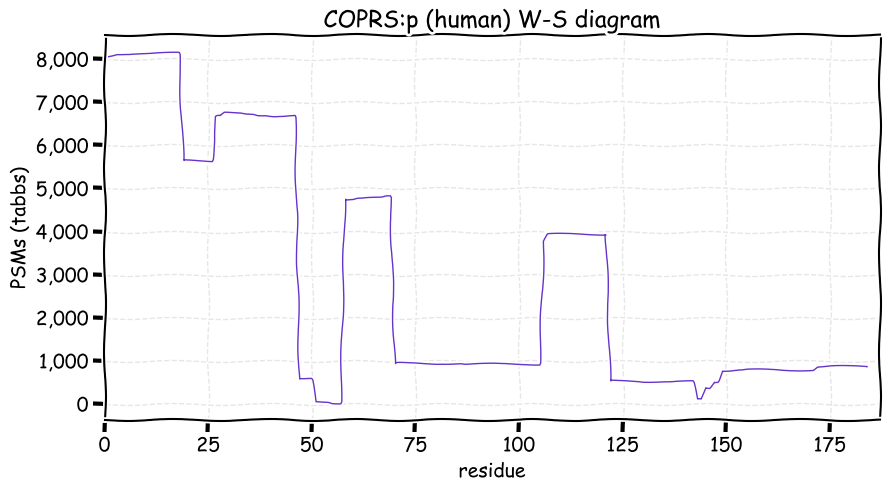
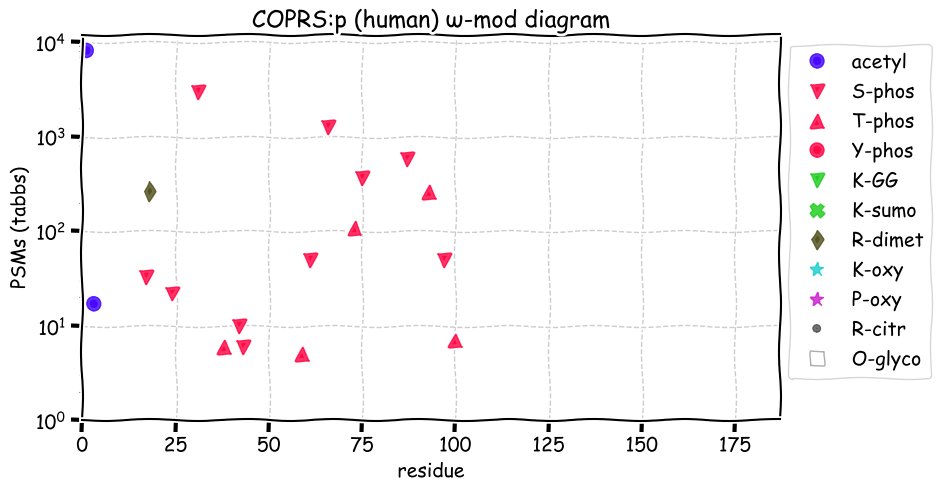
Wed Oct 13 14:41:32 +0000 2021@theoneamit It is an interesting take. I was surprised that it didn't mention department heads/chairs: they set the tone for attracting/repelling comp. bio. faculty. There isn't much the administration can do if the heads aren't on board.
Wed Oct 13 14:35:40 +0000 2021@goodlettlab1 @FRSClipper @HarbourAirLtd @southlakeunion Southern BC can be oddly isolated even at the best of times.
Wed Oct 13 12:16:23 +0000 2021TNKS:p, has too many GO annotations, but no detectable phosphorylation, SUMOylation or R-methylation & only 1 low occupancy acetylation acceptor site.
Wed Oct 13 12:00:22 +0000 2021TNKS:p.A342S chr 8:g.9679980G>A, rs201842298 (all tissue A:S 1.000:0.000) vaf=<1%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Wed Oct 13 12:00:22 +0000 2021TNKS:p, θ(max) = 40. aka TIN1, TINF1, TNKS1, PARP-5a, PARP5A, pART5, ARTD5. Observed in class 1 MHC peptide experiments. Found in tissues & cell lines: absent from fluids.
Wed Oct 13 12:00:21 +0000 2021>TNKS:p, tankyrase (Homo sapiens) Large subunit; CTMs: A2?+acetyl; PTMs: 1×K+acetyl; 16×K+GGyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: A342S (<1%); mature form: (2?-1327) [3,154×, 13 kTa]. #ᗕᕱᗒ 🔗
Tue Oct 12 20:02:23 +0000 2021@UCDProteomics N = 1 has been the standard method to cope with variability.
Tue Oct 12 19:36:01 +0000 2021@UCDProteomics @lkpino @tomlau @DemichevLab @ProteomicsNews @ProtifiLlc @pwilmarth You have to call them "workstations".
Tue Oct 12 17:48:46 +0000 2021@VATVSLPR It may also be that my expectations are a bit skewed, because for a while I ran a development lab at Eli Lilly creating release tests for recomb. insulin products, so I'm used to expecting pristine results from insulin digests.
Tue Oct 12 17:46:43 +0000 2021@VATVSLPR It is always possible that it is a sample prep artifact, but I look at a lot of this type of data & insulin's cleavage pattern feels more like endogenous cleavage/processing to me.
Tue Oct 12 17:22:30 +0000 2021Needless to say I am looking at a new data set of tissue isolated from pancreas right now.
Tue Oct 12 17:21:18 +0000 2021It always strikes me as odd how many non-tryptic cleaves are observed in insulin obtained from pancreatic samples, even once its known processing into the mature protein is taken into account. It is far more "chewed up" than other proteins found in the same sample.
Tue Oct 12 14:30:34 +0000 2021PRIDE is really moving today: 32 MBit downloads seem fast compared to the usual ~10 MBit speeds I've been seeing for the last while.
Tue Oct 12 14:20:51 +0000 2021TNKS1BP1:p, given its extensive phosphorylation pattern, is probably a very difficult protein to study using either genetic or biochemical approaches.
Tue Oct 12 12:23:38 +0000 2021TNKS1BP1, according to STRING, has been experimentally confirmed to physically interacts with many other proteins, although tankyrase 1 (TNKS) is not among them. 🔗
Tue Oct 12 12:23:36 +0000 2021TNKS1BP1:p.H1605R chr 11:g.57302094T>C, rs35272228 (all tissue H:R 0.928:0.072) vaf=4%, Δm=19.0422, VAF by population group: african 1%, american 3%, east asian <1%, european 4%, south asian 3%. HR in HCT-116 cells.
Tue Oct 12 12:23:36 +0000 2021TNKS1BP1:p, θ(max) = 82. aka TAB182, KIAA1741, FLJ45975. Observed in class 1 MHC peptide experiments. Common in tissues & cell lines: may be present in fluid extracellular vesicles.
Tue Oct 12 12:23:36 +0000 2021>TNKS1BP1:p, tankyrase 1 binding protein 1 (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 4×K+acetyl; 2×K+GGyl; 193×S, 55×T, 13×Y+phosphoryl; SAAVs: T322S (49%), P339R (4%), Q1097R (98%), H1605R (4%); mature form: (1-1729) [40,853×, 602 kTa]. #ᗕᕱᗒ 🔗
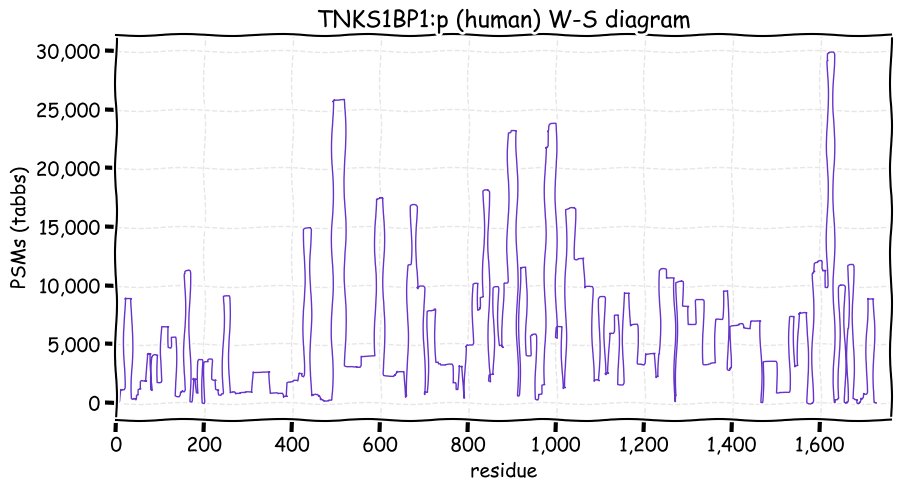
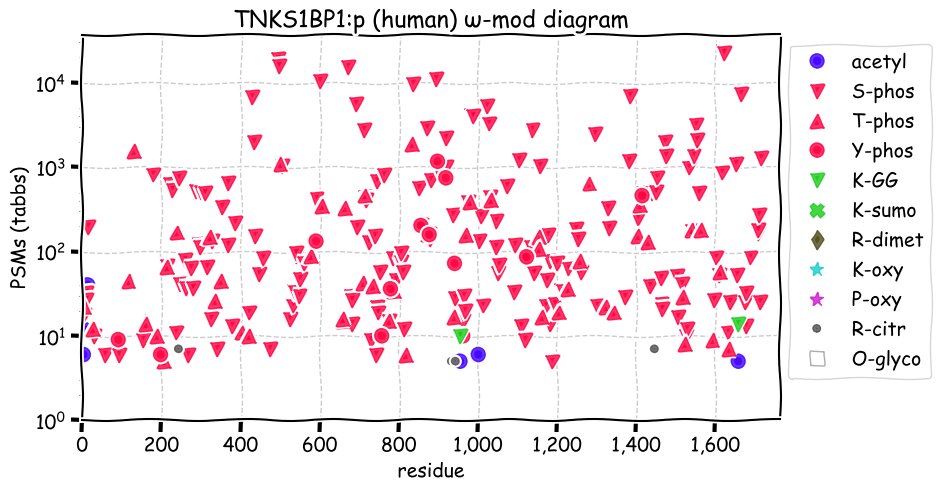
Mon Oct 11 20:57:42 +0000 2021@ProteomicsNews The whole CPTAC proteomics group resisted the idea of SAAVs for years: if they have come over to the dark side, good for them. David Fenyö may have finally nudged them over the edge ...
Mon Oct 11 20:33:20 +0000 2021Thanks to everyone who participated. The correct answer is probably "0".
Mon Oct 11 12:44:44 +0000 2021EP300:p.I997V chr 22:g.41152004A>G, rs20551 (all tissue I:V 0.828:0.172) vaf=31%, Δm=-14.0156, VAF by population group: african 2%, american 51%, east asian 5%, european 30%, south asian 38%. IV in A-431 & HEK-293 cells.
Mon Oct 11 12:44:43 +0000 2021EP300:p, θ(max) = 41. aka p300, KAT3B. Observed in class 1 MHC peptide experiments. Common in tissues & cell lines: absent from fluids.
Mon Oct 11 12:44:43 +0000 2021>EP300:p, E1A binding protein p300 (Homo sapiens) Large subunit; CTMs: A2+acetyl; PTMs: 63×K+acetyl; 1×K+GGyl; 4×K+SUMOyl; 51×S, 31×T, 1×Y+phosphoryl; 16×R+dimethyl; SAAVs: G211S (1%), I997V (31%), Q2223P (1%); mature form: (2-2414) [17,633×, 93 kTa]. #ᗕᕱᗒ 🔗
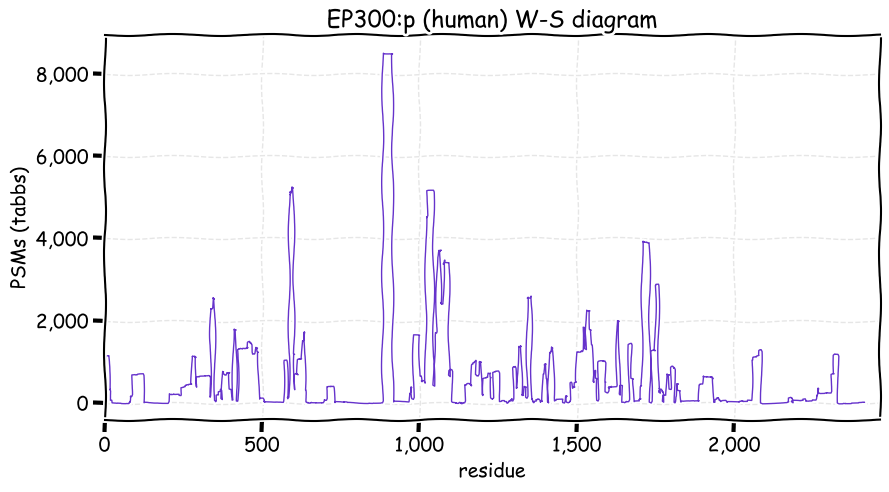
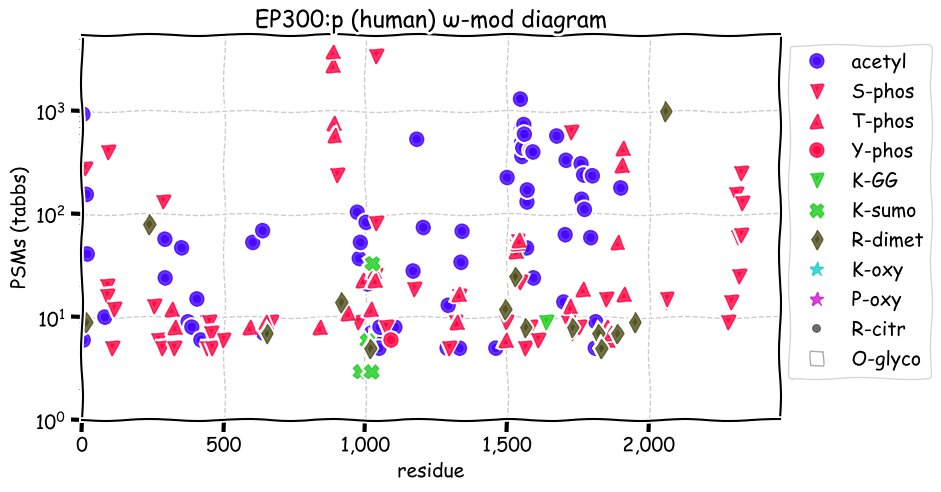
Sun Oct 10 14:57:16 +0000 2021@ypriverol @leprevostfv It is mainly a matter of the audience: if you have taught at a Med School, you would intuitively understand that "> 1" is far more convincing than a valid statistical argument (see also "< 0.05")
Sun Oct 10 14:39:54 +0000 2021@astacus @neely615 @pwilmarth I'd go with the more practical brass, Babbitt, solder & stainless steel.
Sun Oct 10 14:17:08 +0000 2021@neely615 @astacus @pwilmarth HPP went through endless hand wringing on which metallic finish to associate with protein identification quality. In the end, metals aren't good differentiators when shown on screen displays: it is easy to confuse bronze vs gold & platinum is indistinguishable from silver.
Sun Oct 10 13:09:33 +0000 2021There should be some Twitch channels where you watch famous mass spectrometrists wait for a run to end.
Sun Oct 10 12:00:43 +0000 2021LAPTM5:p, TM domains (in grey): (15-37), (63-85), (100-121), (142-164), (184-206). (207-262) is cytoplasmic & contains 100% of PTM-bearing PSMs. (259-262) is a Y-X-X-hydrophobic type lysosome targeting signal (in red). 🔗

Sun Oct 10 12:00:43 +0000 2021LAPTM5:p.R226K chr 1:g.30735195C>T, rs35351292 (all tissue R:K 0.857:0.143) vaf=20%, Δm=-28.0061, VAF by population group: african 4%, american 11%, east asian 8%, european 27%, south asian 10%. KR in JURKAT cells.
Sun Oct 10 12:00:42 +0000 2021IFNGR1:p, θ(max) = 23. aka none. Observed in class 1 & class 2 MHC peptide experiments. Common in leukocytes, lymphocytes & specific cell lines: rare in fluids.
Sun Oct 10 12:00:42 +0000 2021>LAPTM5:p, lysosomal protein transmembrane 5 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 5×K+GGyl; S238, Y239, Y259+phosphoryl; SAAVs: R226K (20%); mature form: (1-262) [3,135×, 16 kTa]. #ᗕᕱᗒ 🔗
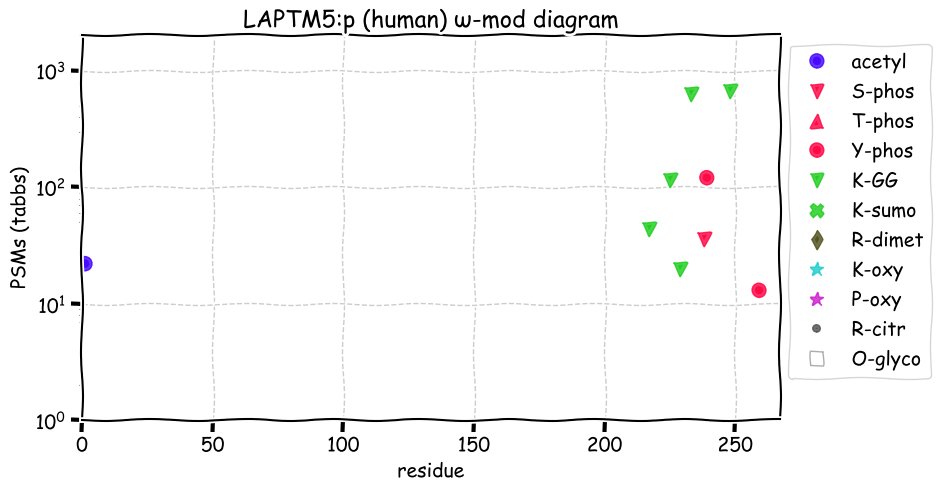
Sat Oct 09 23:58:30 +0000 2021@ProteomicsNews Otherwise, the reader has to try & find the reference and variant residues in the peptide sequences, which is kind of hard to scan out quickly (at least old-timers like me).
Sat Oct 09 23:56:55 +0000 2021@ProteomicsNews I took a look at your site & it looks good. My only suggestion would be to include the variant residue in the "variant site" column. For example, in the first row instead of just "T331" add the variant to make "T331A".
Sat Oct 09 18:00:19 +0000 2021@ProteomicsNews Does this mean that CPTAC is officially admitting the existence of single amino acid variants?
Sat Oct 09 12:31:07 +0000 2021IFNGR1:p, domains: extracellular (?-247), TM (248-270), intracellular (271-489). 98.4% of modified PSMs are associated with the intracellular domain.
Sat Oct 09 12:31:07 +0000 2021IFNGR1:p.L467P chr 6:g.137198101A>G, rs1887415 (all tissue L:P 0.811:0.189) vaf=8%, Δm=-16.0313, VAF by population group: african 2%, american 3%, east asian 6%, european 1%, south asian 31%. LP in JURKAT cells.
Sat Oct 09 12:31:07 +0000 2021IFNGR1:p, θ(max) = 22. aka none. Shares peptides in both MHC class 1 & 2 peptide experiments. Found in tissues & cell lines: absent from fluids.
Sat Oct 09 12:31:06 +0000 2021>IFNGR1:p, interferon gamma receptor 1 (Homo sapiens) Small subunit; CTMs: none; PTMs: 5×K+GGyl; 20×S, 6×T, 4×Y+phosphoryl; SAAVs: L467P (8%); mature form: (?-489) [4,907×, 18 kTa]. #ᗕᕱᗒ 🔗
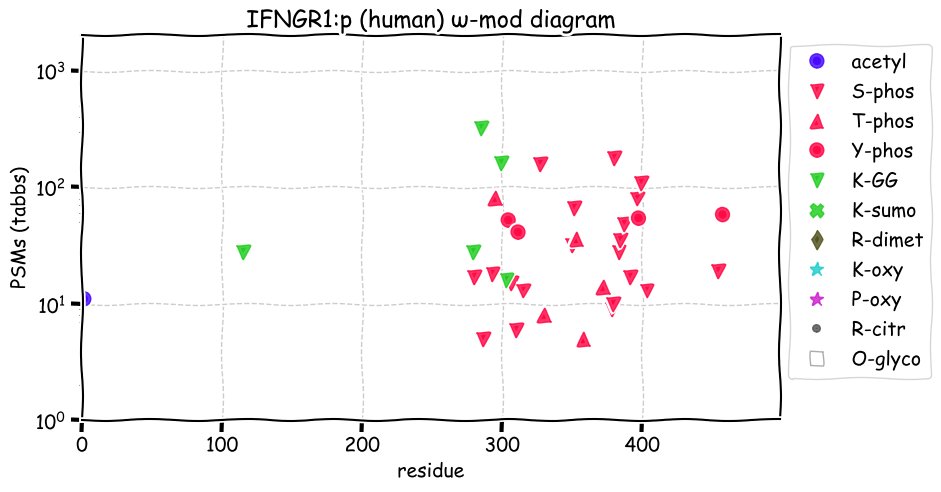
Fri Oct 08 17:15:50 +0000 2021@dlswaney @lenjf This particular type of carbamylation occurs during the proteolytic digestion process.
Fri Oct 08 15:20:22 +0000 2021Heading into Thanksgiving weekend (here in Canuckistan).
Fri Oct 08 14:27:10 +0000 2021@dtabb73 I've had to install that myself on a couple of boxes (not for this particular suite, but for other Python projects). If the project requires some compiled C/C++, usually the set up will try to compile them on your particular OS, rather than using binaries.
Fri Oct 08 12:32:04 +0000 2021@lenjf You can use this as a primer on how to get carbamylation going in your lab too! They manage to generate 10-15% of carbamylated PSMs in each pull down. 🔗
Fri Oct 08 12:27:04 +0000 2021FGA:p, O-linked glycosylation seems to be doing a lot of heavy lifting for this protein. May be a challenging subunit for top-down proteomics analysis.
Fri Oct 08 12:02:00 +0000 2021FGA:p.T331A chr 4:g.154586438T>C, rs6050 (all tissue T:A 0.926:0.074) vaf=29%, Δm=-30.0106, VAF by population group: african 43%, american 22%, east asian 43%, european 24%, south asian 25%.
Fri Oct 08 12:02:00 +0000 2021FGA:p, θ(max) = 71. aka none. Shares peptides in both MHC class 1 & 2 peptide experiments. Common in blood plasma, CSF & urine.
Fri Oct 08 12:01:59 +0000 2021>FGA:p, fibrinogen alpha chain (Homo sapiens) Midsized subunit; PTMs: 25×K+acetyl; 5×K+GGyl; 70×S, 43×T, 3×Y+phosphoryl; 45×S, 34×T+glysosyl; 24×K, 17×P+oxide; 11 R+deimination; SAAVs: I6V (2%), P302A (1%), T331A (29%); mature form: (20-644) [46,797×, 2447 kTa].#ᗕᕱᗒ 🔗
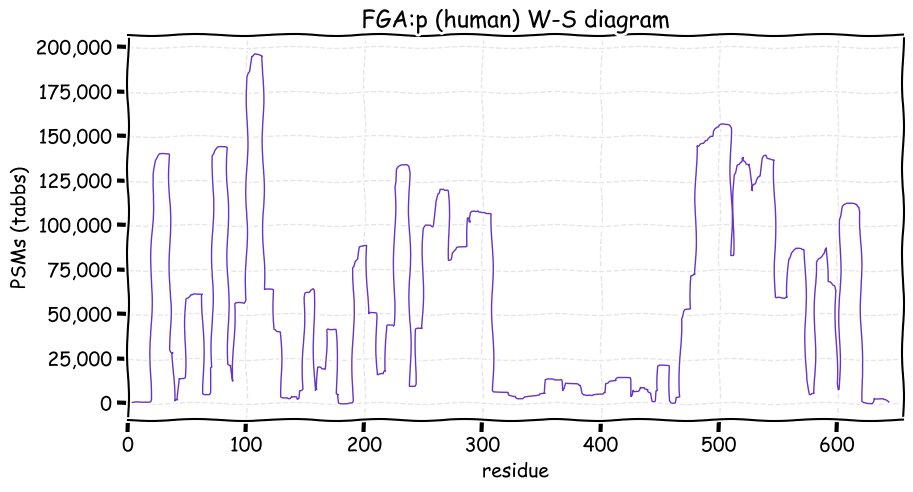
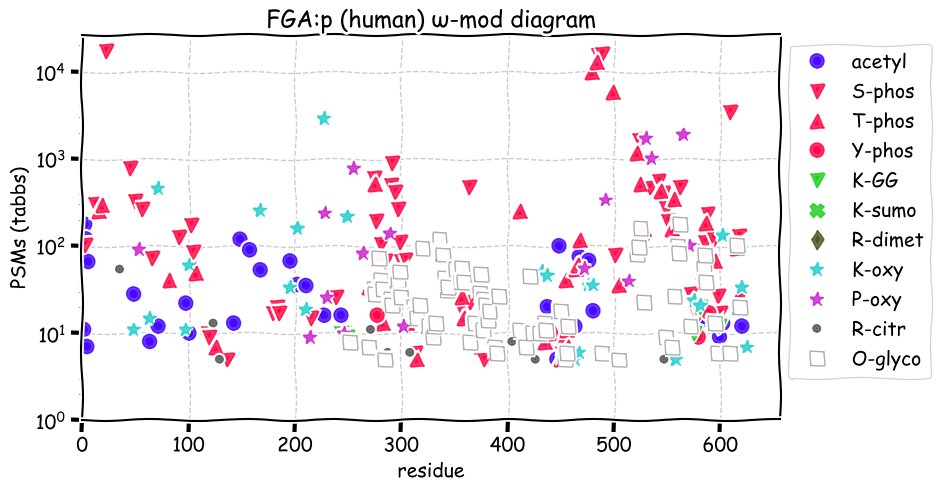
Fri Oct 08 00:40:55 +0000 2021🔗
Thu Oct 07 20:30:36 +0000 2021Just when I thought I had seen everything: some one has finally managed to generate significant amounts of peptide N-terminal carbamylation in a protein-protein pull down experiment. Normally this type of expt has very few artifacts, but I guess we in for some innovation...
Thu Oct 07 16:48:57 +0000 2021@fcyucn You have seen through my ruse! But teaching is a matter of repetition more than anything else & the results so far suggest that there is still some debate/confusion on the issue.
Thu Oct 07 15:00:01 +0000 2021Kind of interesting, if you have ever lived in London
🔗
Thu Oct 07 14:46:00 +0000 2021For those who care, if you do a trypsin digest on the sequence:
MGRRTHGRRFWSDEKK,
how many missed cleavage sites are present in the peptide:
RFWSDEKK
Thu Oct 07 12:05:04 +0000 2021LAD1:p, some of the references to this protein seem to be a bit tangled up with references to the disease syndrome "Leukocyte adhesion deficiency-1", also called "LAD1" caused by mutations in ITGB2:p.
Thu Oct 07 11:58:52 +0000 2021LAD1:p, the protein's pattern of observation in tissues and specific cell lines makes its commonly referenced role (filament in basement membrane ECM) difficult to support, particularly since it does not have an ER signal sequence.
Thu Oct 07 11:50:06 +0000 2021LAD1:p.T274A chr 1:g.201386541T>C, rs68021059 (all tissue T:A 0.960:0.040) vaf=7%, Δm=-30.0106, VAF by population group: african 14%, american 8%, east asian <1%, european 9%, south asian 4%.
Thu Oct 07 11:50:06 +0000 2021LAD1:p, θ(max) = 75. aka none. Present in MHC class 1 peptide experiments & rarely in class 2. Observed in tissues & cell lines: absent from fluids except saliva. Citrulline formation from R residues has been observed in saliva.
Thu Oct 07 11:50:05 +0000 2021>LAD1:p, ladinin 1 (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 4×K+acetyl; 3×K+GGyl; 53×S, 19×T, 0×Y+phosphoryl; 6×R+deimination; SAAVs: A56S (18%), L243P (12%), T274A (7%), V425I (1%), T503S (5%); mature form: (2-517) [16,481×, 125 kTa]. #ᗕᕱᗒ 🔗
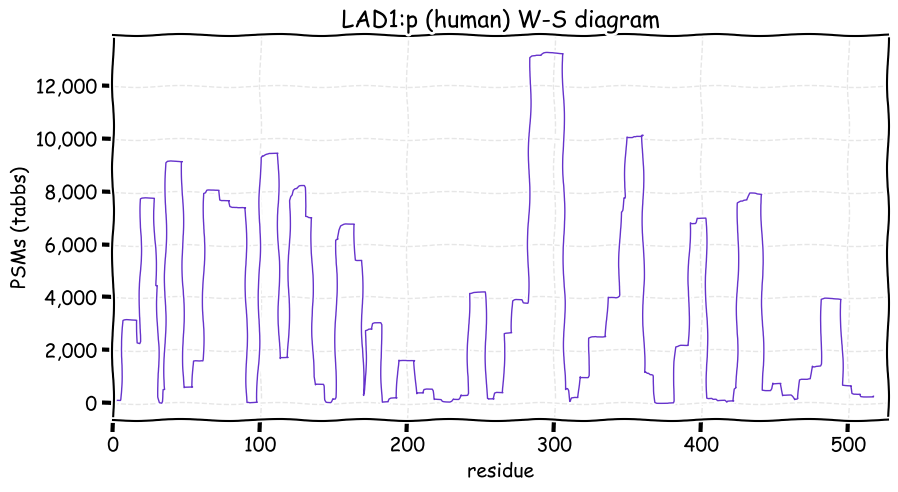
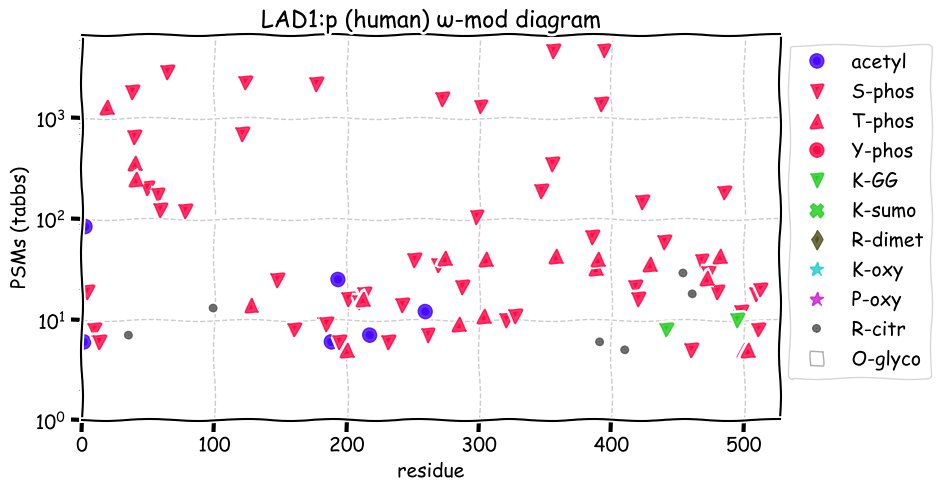
Wed Oct 06 15:54:32 +0000 2021IWS1:p, conforms to an odd pattern found in many proteins, in which the INTERPRO annotated sequence domains have relatively few PTMs, while the unnamed domains are highly modified.
Wed Oct 06 14:46:16 +0000 2021@rach_liv_uni @astacus Sites on university owned machines (& email addresses) are not very reliable if you want stability over time. Good move to put things under your control. :-)
Wed Oct 06 14:39:52 +0000 2021@neely615 If you are still interested, here are the obs. of Streptococcus salivarius enolase in human saliva samples 🔗 Click the metadata tab & you can scrape out the PXD numbers you find interesting. Enolase is A1 on the jukebox when Streptococcus sp. are present.
Wed Oct 06 11:51:42 +0000 2021IWS1:p, >99% of phosphorylated PSMs are found in the acidic low complexity domain (1-513). These phospho-PSMs are often found in data sets that haven't been affinity enriched for phosphopeptides, suggesting the presence of multiple high occupancy acceptor sites.
Wed Oct 06 11:51:41 +0000 2021IWS1:p.V425I chr 2:g.127503523Cgt;T, rs34785867 (all tissue V:I 0.995:0.005) vaf=0.5%, Δm=14.0156, VAF by population group: african 7%, american 1% east asian <1%, european <1%, south asian <1%.
Wed Oct 06 11:51:41 +0000 2021IWS1:p, θ(max) = 54. aka DKFZp761G0123, FLJ10006, FLJ14655, FLJ32319. Present in MHC class 1 peptide experiments. Observed in tissues & cell lines: absent from fluids.
Wed Oct 06 11:51:41 +0000 2021>IWS1:p, interacts with SUPT6H, CTD assembly factor 1 (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 3×K+acetyl; 1×K+GGyl; 1×K+SUMOyl; 91×S, 16×T, 1×Y+phosphoryl; SAAVs: A390V (1%), V425I (1%), T721A (1%); mature form: (1-819) [19,451×, 136 kTa].#ᗕᕱᗒ 🔗
Tue Oct 05 23:21:31 +0000 2021I usually find about 5% of PSMs in saliva to be from bacteria (S. salivarius, R. dentocariosa, P. denticola, V. parvula, et al.), although it isn't common for publications to mention them.
Tue Oct 05 12:19:30 +0000 2021SDHA:p.V657I chr 5:g.256394G>A, rs6962 (all tissue V:I 0.901:0.099) vaf=13%, Δm=14.0156, VAF by population group: african 37%, american 20% east asian 6%, european 11%, south asian 8%. VI in MCF-7 & HEK-293 cells.
Tue Oct 05 12:19:30 +0000 2021SDHA:p, θ(max) = 80. aka FP, SDHF, SDH2. Present in MHC class 1 & 2 peptide experiments. Frequently observed in tissues & cell lines. 1 of 4 subunits in mitochondrial respiratory complex II.
Tue Oct 05 12:19:30 +0000 2021>SDHA:p, succinate dehydrogenase complex flavoprotein subunit A (Homo sapiens) Midsized subunit; CTMs: none; PTMs: 27×K+acetyl; 19×K+GGyl; 2×K+SUMOyl; 2×S, 25×T, 6×Y+phosphoryl; SAAVs: D38V (2%), G184R (1%), V657I (13%); mature form: (33-664) [83,504×, 962 kTa]. #ᗕᕱᗒ 🔗
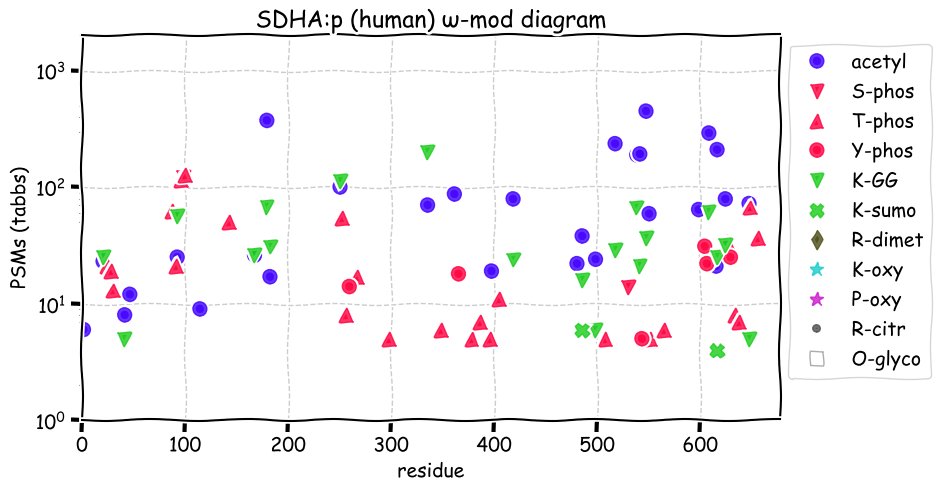
Tue Oct 05 00:45:11 +0000 2021@pwilmarth In terms of the # of peptides, it is a larger correction than the "except P" rule & any list of peptides that contains the single residues "K" or "R" is just not right.
Mon Oct 04 22:20:51 +0000 2021@pwilmarth It "works", but it doesn't properly deal with sequences like "..KK.." or "..RK.." or "..RR..".
Mon Oct 04 19:33:19 +0000 2021@slavov_n It has been standard practice for as long as I can remember, whenever an editor wants something to get through review quickly.
Mon Oct 04 18:36:47 +0000 2021🔗
Mon Oct 04 18:27:19 +0000 2021@juan_vizcaino @arxiv Is there any way, using this specification, to express the possibility of multiple different mods at the same site, e.g., a particular K observed to be modified with either ubiquitin or acetyl, but not both simultaneously.
Mon Oct 04 16:35:38 +0000 2021Just a reminder: regexes aren't great for generating lists of tryptic peptides, e.g.:
peps = re.findall(r'.(?:(?<).)*',proseq)
gives you a list of many, but not all, tryptic peptides & includes peptides that are not tryptic peptides.
Mon Oct 04 14:45:10 +0000 2021Looking at stuff like this is how I waste my Sunday afternoons.
Mon Oct 04 14:43:54 +0000 2021METAP2:p seem to have much more welcoming approach to PTMs
🔗
Mon Oct 04 14:41:50 +0000 2021Eukaryotes have 2 cytosolic exopeptidases that co-translationally remove the N-terminal Met residue from many nascent peptides. Both seem to do similar jobs & are present in similar amounts. I was struck by the differences in their PTM patterns. METAP1:p 🔗
Mon Oct 04 12:52:51 +0000 2021Have any of the CRC programs ever been used to recruit a Nobel laureate back to Canada (or to retain one at a Canadian university)?
Mon Oct 04 12:06:18 +0000 2021ANXA11:p, the N-terminal low complexity domain (2-200) only has 5 residues with charged side chains: 3 Asp & 2 Arg residues. This results in very few PSMs assigned to this domain, other than from MHC class 1 peptide experiments. 🔗

Mon Oct 04 12:06:17 +0000 2021ANXA11:p.I278V chr 10:g.80166110T>C, rs72807973 (all tissue I:V 0.993:0.007) vaf=0.3%, Δm=-14.0156, VAF by population group: african <1%, american <1% east asian <1%, european 1%, south asian <1%.
Mon Oct 04 12:06:17 +0000 2021ANXA11:p, θ(max) = 59. aka ANX11. Present in MHC class 1 & 2 peptide experiments. Common in tissues & cell lines: also observed in extracellular vesicles in urine & blood plasma.
Mon Oct 04 12:06:17 +0000 2021>ANXA11:p, annexin A11 (Homo sapiens) Midsized subunit; CTMs: S2?+acetyl; PTMs: 21×K+acetyl; 16×K+GGyl; 3×K+SUMOyl; 2×S, 1×T, 8×Y+phosphoryl; SAAVs: R230C (39%), I278V (0.3%); mature form: (2-505) [62,493×, 517 kTa]. #ᗕᕱᗒ 🔗
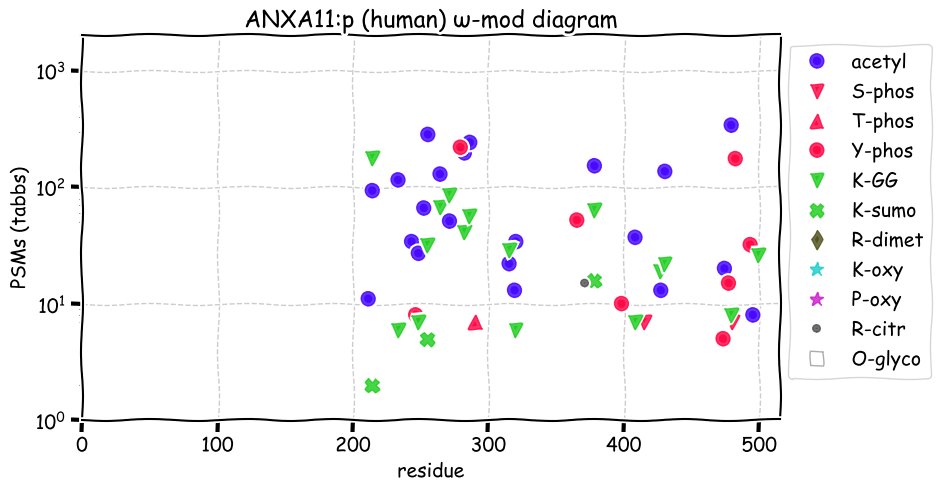
Sun Oct 03 18:49:55 +0000 2021@Peptidome The entire data set is also kind of interesting if you are at all curious about protein N-terminal acetylation in prokaryotes. Y. pestis seems to have some special quirks in its NAT(s?) sequence specificity.
Sun Oct 03 16:34:13 +0000 2021@Peptidome I'm not be critical of them: one thing that is very apparent if you look at data from many sources is how often simple errors/variations can occur in how methods are performed as well as in data archiving. It is "par for the course".
Sun Oct 03 16:05:23 +0000 2021@Peptidome The nice thing about the data is that the file labels are only approximately correct (like any set of real experimental data).
Sun Oct 03 14:18:28 +0000 2021I was looking through the list of Canadian Nobel prizes this morning: it does not paint research in Canadian academia or Canadian govt. funding in a very good light.
Sun Oct 03 14:07:30 +0000 2021If you an algorithm you think would be good for determining sequence specificity in proteases, the data in folder IPX0002310001 (part of PXD028891) would be pretty much ideal for testing it.
Sun Oct 03 14:00:57 +0000 2021I agree completely. But both funders and researchers have been actively ignoring anything to do with ethical issues in proteomics since it became an issue 15 years ago. Hopefully this group can influence the laggards by underlining the problem 🔗
Sun Oct 03 12:20:18 +0000 2021CKAP4:p, domains: cytoplasmic (1-108, blue), TM (109-131, grey), ER lumen (132-602, green). The cytoplasmic domain has several patches of very low complexity sequence. It is a type II membrane protein, so it does not have an N-terminal ER signal sequence. 🔗

Sun Oct 03 12:20:17 +0000 2021CKAP4:p.A348T chr 12:g.106239791C>T, rs3088113 (all tissue A:T 0.888:0.112) vaf=19%, Δm=26.0156, VAF by population group: african 7%, american 5%, east asian 55%, european 14%, south asian 22%.
Sun Oct 03 12:20:17 +0000 2021CKAP4:p, θ(max) = 85. aka P63, CLIMP-63, ERGIC-63. Present in MHC class 1 & 2 peptide experiments. Common in tissues & cell lines: also observed in fluids, e.g., urine & saliva.
Sun Oct 03 12:20:17 +0000 2021>CKAP4:p, cytoskeleton associated protein 4 (Homo sapiens) Midsized subunit; CTMs: M1, P2+acetyl; PTMs: 37×K+acetyl; 10×K+GGyl; 58×S, 19×T,1×Y+phosphoryl; SAAVs: A348T (19%), S396L (1%); mature form: (1,2-602) [55,106×, 855 kTa]. #ᗕᕱᗒ 🔗
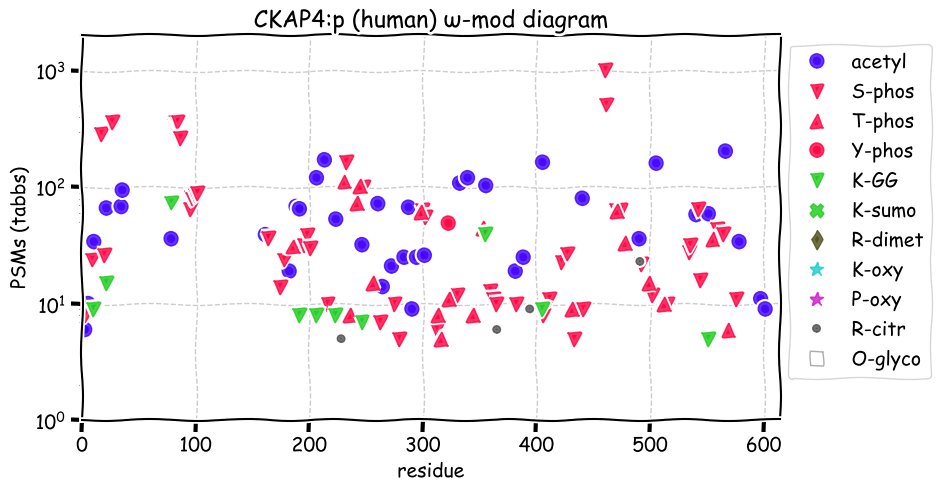
Sat Oct 02 14:08:15 +0000 2021@statesdj I think the old rate of "1 min⁻¹" is much more like "1 sec⁻¹" by now.
Sat Oct 02 13:20:49 +0000 2021As an exemplar of this particular mechanism, I prefer GARS1:p, but I am notoriously bad at choosing exemplars.
Sat Oct 02 12:15:40 +0000 2021It should be noted that translation starting at not-the-first-Met is not a new idea (or observation), it is just currently fashionable to ignore it 🔗
Sat Oct 02 11:50:42 +0000 2021ANAPC7:p, from observations of an N-terminal peptide (with N-acetylation), 98.5% of PSMs show evidence of the sequence beginning at the alternate initiation site M35 rather than the reference M1 site. The reference translation should be amended.
Sat Oct 02 11:50:42 +0000 2021ANAPC7:p.P74L chr 12:g.110396435G>A, rs370892553 (all tissue P:L 0.995:0.005) vaf=<1%, Δm=16.0313, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sat Oct 02 11:50:42 +0000 2021ANAPC7:p, θ(max) = 51. aka APC7. Present in MHC class 1 peptide experiments. Common in tissues & cell lines: very rare in fluids.
Sat Oct 02 11:50:41 +0000 2021>ANAPC7:p, anaphase promoting complex subunit 7 (Homo sapiens) Midsized subunit; CTMs: D2, M35+acetyl; PTMs: 7×K+acetyl; 25×K+GGyl; 4×K+SUMOyl; 12×S, 2×T,1×Y+phosphoryl; SAAVs: S33N (1%), P74L (<1%); mature form: (2,35-599) [18,739×, 80 kTa]. #ᗕᕱᗒ 🔗
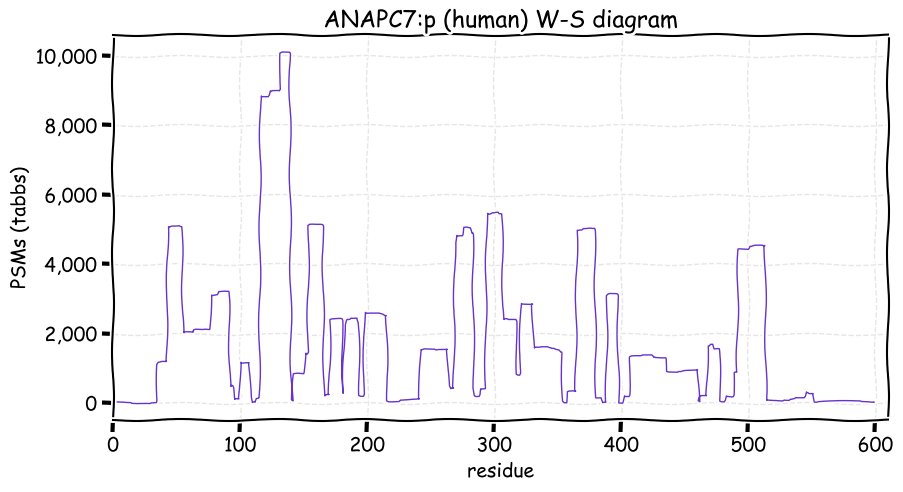
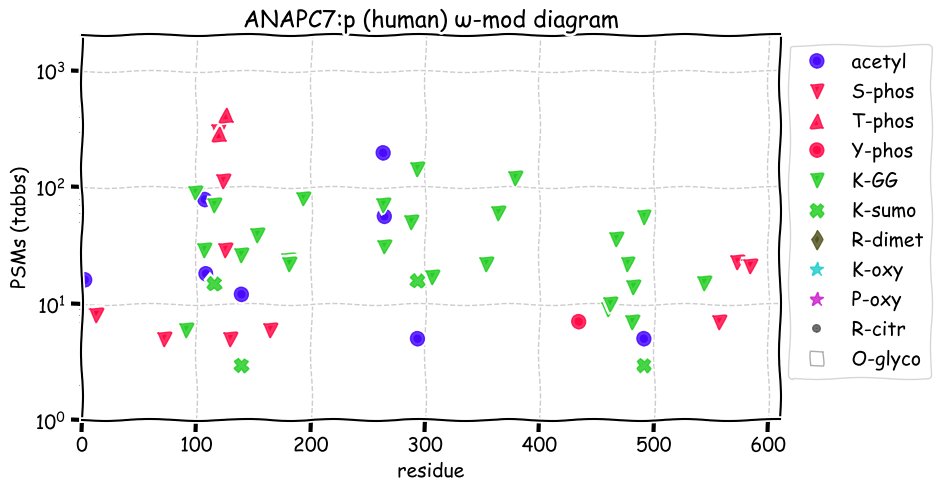
Fri Oct 01 15:34:57 +0000 2021The most zealously guarded resource in any university department is access to promising grad students who haven't committed to a supervisor yet.
Fri Oct 01 12:54:58 +0000 2021@AlexUsherHESA Maybe a "walk in the sand"?
Fri Oct 01 12:08:28 +0000 2021FURIN:p, the proprotein form is very rarely observed, other than in protein-protein interaction experiments in which FURIN:p is the bait.
Fri Oct 01 12:04:17 +0000 2021FURIN:p domains: extracellular (27-715), TM (716-738), intracellular (739-794). All PTMs observed on intracellular domain.
Fri Oct 01 11:55:25 +0000 2021FURIN:p.V109M chr 15:g.90876510G>A, rs116359616 (all tissue V:M 0.994:0.006) vaf=0.8%, Δm=31.9721, VAF by population group: african 3%, american <1% east asian <1%, european <1%, south asian <1%.
Fri Oct 01 11:55:25 +0000 2021FURIN:p, θ(max) = 59. aka SPC1, PCSK3, FUR, PACE. Present in MHC class 1 & 2 peptide experiments. Common in fluids (particularly saliva) & specific cell types.
Fri Oct 01 11:55:25 +0000 2021>FURIN:p, paired basic amino acid cleaving enzyme (Homo sapiens) Midsized subunit; CTMs: none; PTMs: 2×K+GGyl; 2×S, 0×T, 0×Y+phosphoryl; SAAVs: A43V(3%), V109M (1%); proprotein: (27-794), active form: (108-794) [3,405×, 31 kTa]. #ᗕᕱᗒ 🔗
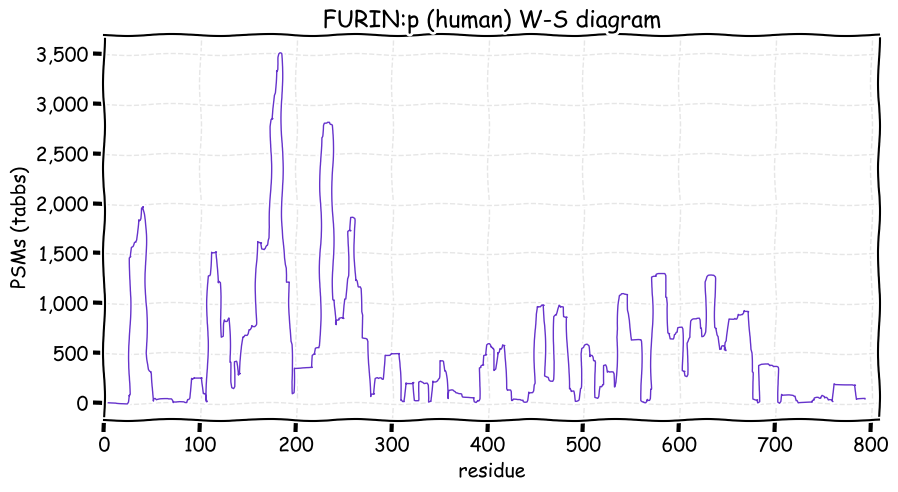
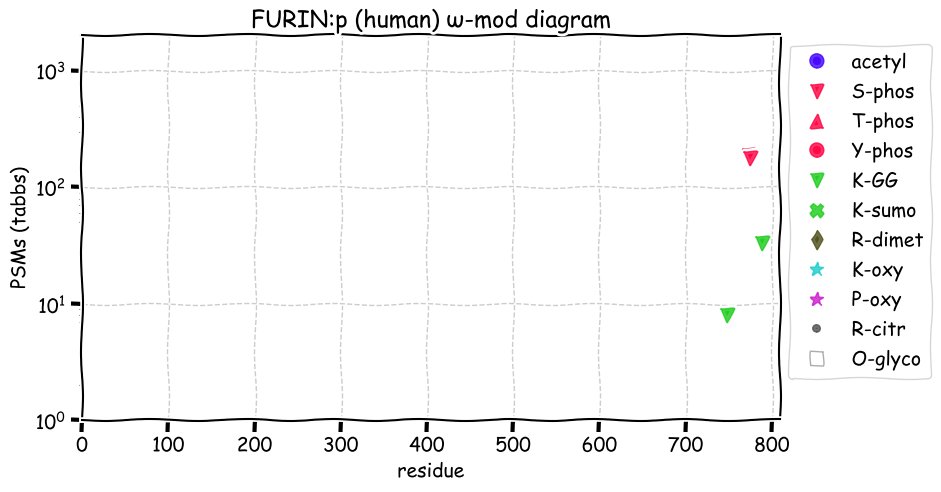
Thu Sep 30 16:14:10 +0000 2021@TopDownProteome @jchamotrooke @Ge_Lab_UW @NLKProteomics @JoeLoo85614818 @YTsybin @GenomeWeb @BBCScienceNews @NYTScience @GENbio @hupo_org I never know how to think about this type of project. The goals are admirable, but so general that I can't see exactly what success might look like. But, I joined anyway ...
Thu Sep 30 14:47:31 +0000 2021@jwoodgett @FPRoth I apologize for being unkind. My clumsy attempt at humour has been removed.
Thu Sep 30 14:21:19 +0000 2021@neely615 @eliaslab1 More broadly, though, I think any graphical display of GO categories (or any ontology) is the equivalent to a "knowledge graph": they describe higher order relationships between segments of a network 🔗
Thu Sep 30 13:10:40 +0000 2021@dexivoje Just made it up, but I may have read/heard it somewhere & don't remember the source.
Thu Sep 30 12:55:05 +0000 2021@dexivoje Especially when used together: "We have tentatively identified a potentially promising factor that may be a valuable clue to unlocking one of the puzzles associated with oncogenesis"
Thu Sep 30 12:13:00 +0000 2021LDLR:p, domains: extracellular (25-789), TM (789-810), intracellular (811-860). 99% of phospho-PSMs are from the intracellular domain & 100% of glyco-PSMs are from the extracellular domain.
Thu Sep 30 12:03:03 +0000 2021LDLR:p.A391T chr 19:g.11111624G>A, rs11669576 (all tissue A:T 0.871:0.129) vaf=4%, Δm=30.0106, VAF by population group: african 20%, american 4% east asian <1%, european 4%, south asian <1%.
Thu Sep 30 12:03:03 +0000 2021LDLR:p, θ(max) = 52. aka LDLCQ2. Present in MHC class 2 peptide experiments, mainly peptides from the domain (406-591). Commonly found in most tissues, cell lines and fluids.
Thu Sep 30 12:03:03 +0000 2021>LDLR:p, low density lipoprotein receptor (Homo sapiens) Midsized subunit; CTMs: N272, N657+glycosyl; PTMs: 2×K+acetyl; 9×K+GGyl; 5×S, 0×T, 3×Y+phosphoryl; 1×S, 3×T+glycosyl; SAAVs: A391T (4%); mature form: (25-860) [13,468×, 78 kTa]. #ᗕᕱᗒ 🔗
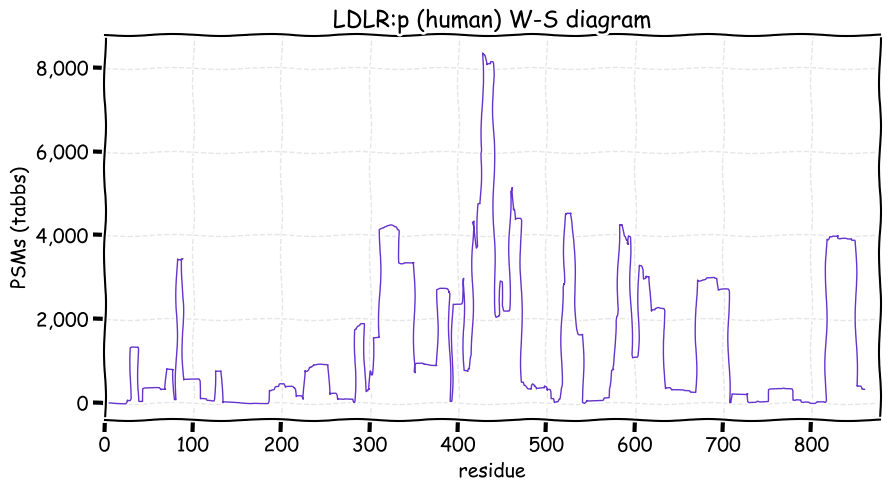
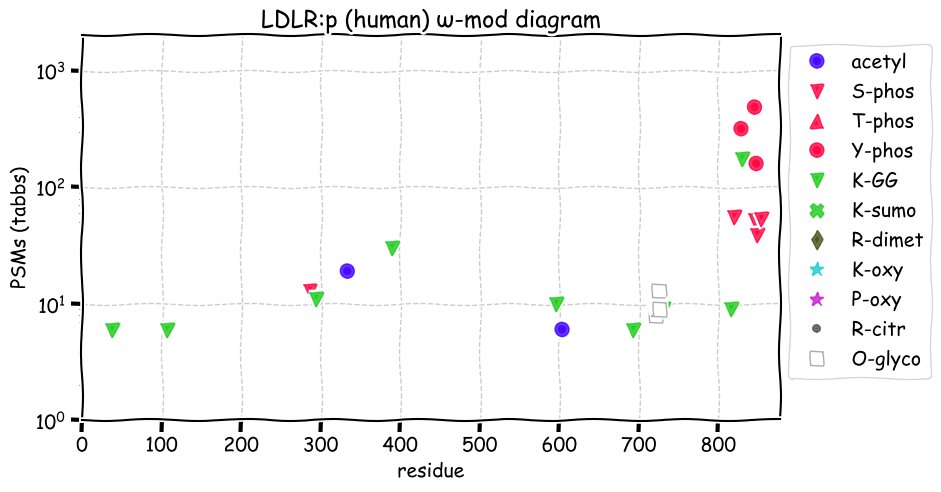
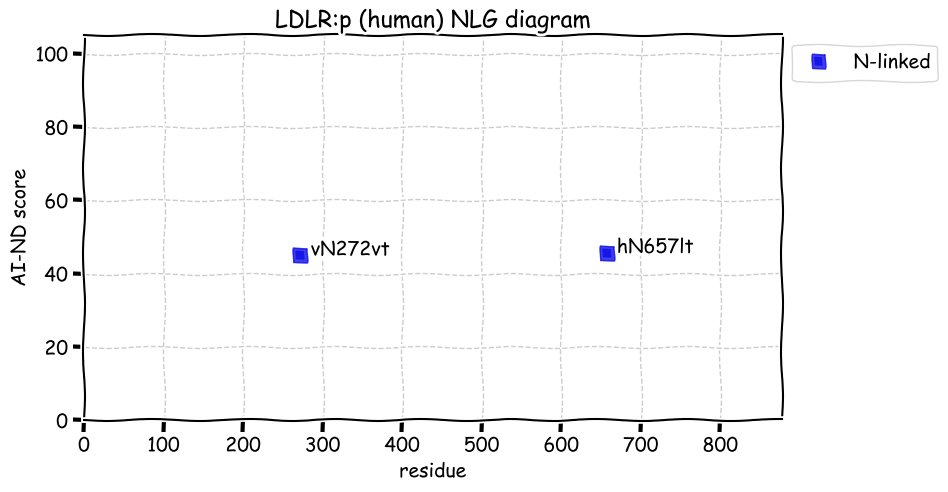
Thu Sep 30 01:35:06 +0000 2021@astacus Now the desired proteins are viewed through a filmy gauze of keratins.
Wed Sep 29 23:40:46 +0000 2021Whenever I look at "modern" proteomics data sets that use gels, it make me nostalgic for the old days when people used to run gels in flow hoods to keep out the dust.
Wed Sep 29 20:55:41 +0000 2021While there have been a lot of good ideas added to C++ in the last decade, the continued improvement in the implementation of "auto" has done the most for me (I use a lot of iterators).
Wed Sep 29 14:07:49 +0000 2021@neely615 Graph theory methods in id'ing spectra are most commonly used by de novo algorithms, including sequence tag approaches. In general, proteomics has never really been a "show the math" type of field, so there may be other applications that are buried in proprietary code.
Wed Sep 29 13:57:59 +0000 2021PCSK9:p has a role in LDL uptake in cells, inactivating LDLR:p via protein-protein interaction. Whether it functions as a proprotein convertase has not been thoroughly studied, but it is widely mentioned in the cholesterol homeostasis literature.
Wed Sep 29 12:25:56 +0000 2021PCSK9:p.A53V chr 1:g.55039995C>T, rs11583680 (all tissue A:V 0.953:0.047) vaf=11%, Δm=28.0313, VAF by population group: african 1%, american 7% east asian 10%, european 13%, south asian 16%. AV in A-431 & MDA-MB-231 cells.
Wed Sep 29 12:23:18 +0000 2021PCSK9:p, θ(max) = 66. aka NARC-1, FH3, HCHOLA3. Present in MHC class 1 & 2 peptide experiments. Most frequently observed in blood plasma & HeLa cells. The active form of the enzyme (153-692) is created by removal of a proprotein domain (29,31-152).
Wed Sep 29 12:23:18 +0000 2021>PCSK9:p, proprotein convertase subtilisin/kexin type 9 (Homo sapiens) Midsized subunit; CTMs: N533+glycosyl; PTMs: 1×K+SUMOyl; 5×K+GGyl; 4×S, 1×T, 0×Y+phosphoryl; SAAVs: A53V (11%), A443T (3%), V474I (13%), G670E (10%); secreted form: (29,31-692) [5,195×, 32 kTa]. #ᗕᕱᗒ 🔗
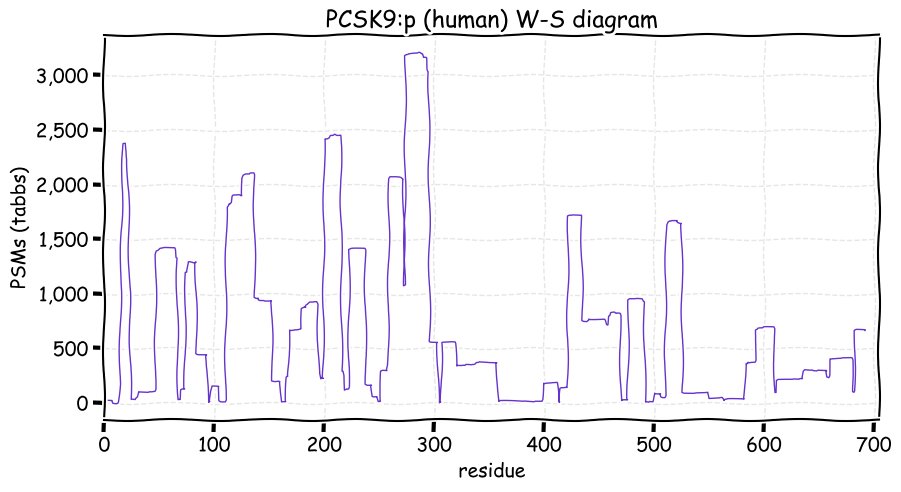
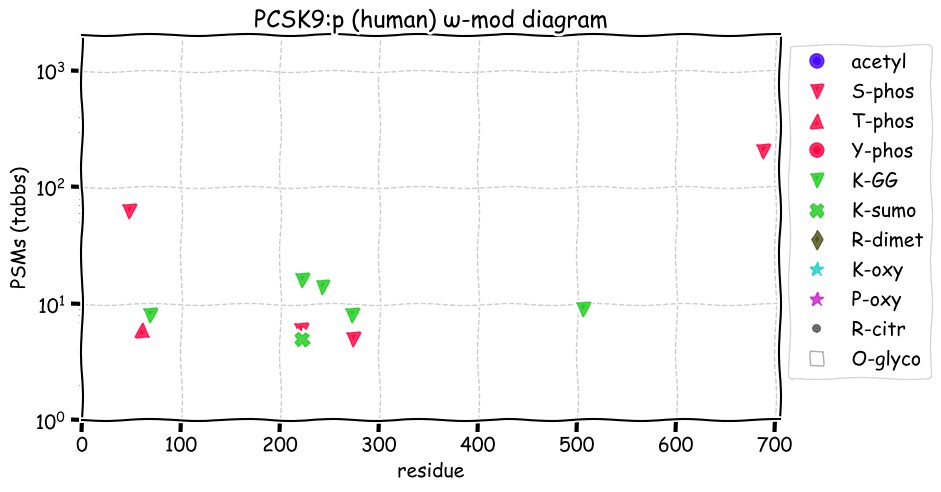
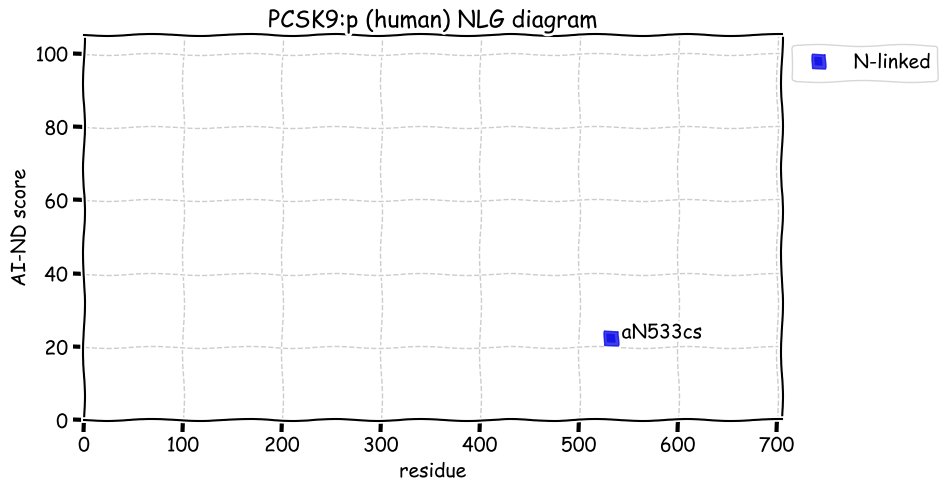
Tue Sep 28 14:58:08 +0000 2021@jwoodgett @MHendr1cks I had seen the recommendations when they came out, but they are no less valuable (or reasonable) now then when they were made. But given the way the CIHR Act works in practice, it has very little reason to change.
Tue Sep 28 14:49:41 +0000 2021@jwoodgett I guess a similar analysis from CIHR on the situation in Canada will be available soon? It would be great if we had a "Noni Byrnes" equivalent position here.
Tue Sep 28 11:38:14 +0000 2021PTPRF:p, its predicted transmembrane domain (1253-1275) is immediately followed by a short domain of positively charged residues (KRKR), a rule-of-thumb indicator that the C-terminal domain is cytoplasmic. This assertion is confirmed by the observed PTM pattern.
Tue Sep 28 11:38:14 +0000 2021PTPRF:p.Y450C chr 1:g.43591371A>G, rs3748796 (all tissue Y:C 0.997:0.003) vaf=0.3%, Δm=-60.0541, VAF by population group: african <1%, american 1% east asian 1%, european <1%, south asian <1%.
Tue Sep 28 11:38:14 +0000 2021PTPRF:p, θ(max) = 75. aka LAR. Present in MHC class 1 & 2 peptide experiments. Found in most tissues & cell lines. The extracellular domain (30-1245) can be found in urine, CSF, milk & blood plasma.
Tue Sep 28 11:38:13 +0000 2021>PTPRF:p, protein tyrosine phosphatase receptor type F (Homo sapiens) Large subunit; CTMs: N250, N721, N957+glycosyl; PTMs: 4×K+acetyl; 10×K+GGyl; 23×S, 3×T, 16×Y+phosphoryl; SAAVs: G1393R (26%); mature form: (30-1898) [22,709×, 160 kTa]. #ᗕᕱᗒ 🔗
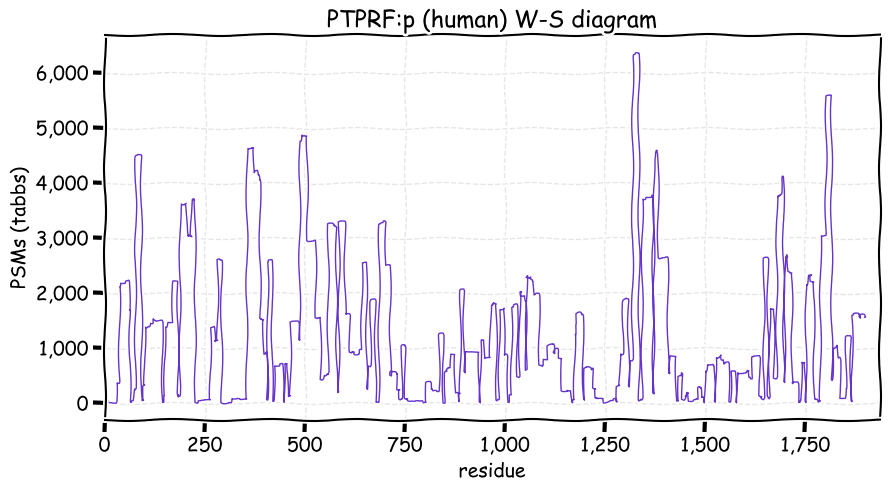
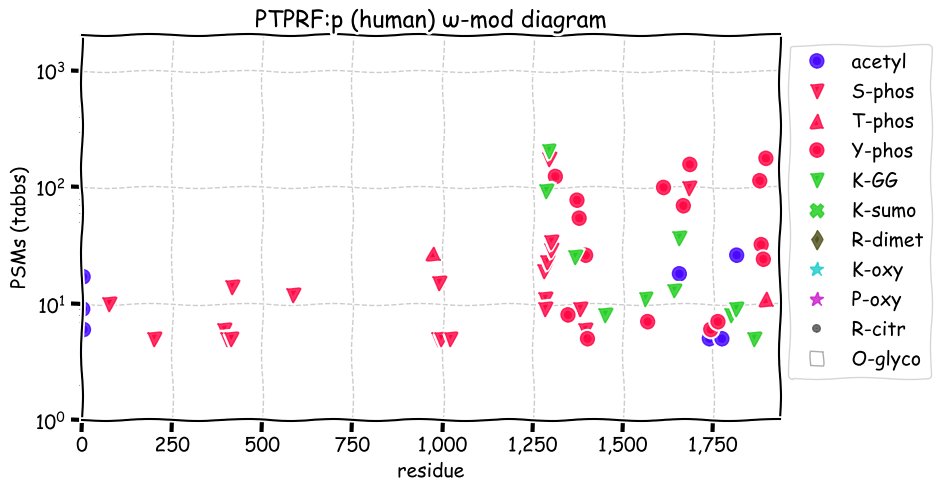
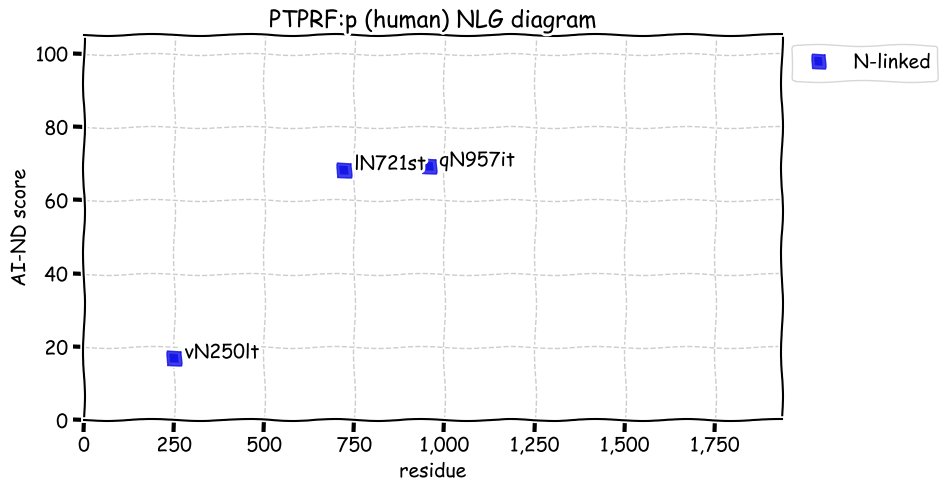
Mon Sep 27 12:22:21 +0000 2021PPFIA1:p, like its interactor PPFIBP1:p (yesterday's protein), has two N-terminal Met residues, resulting in translation initiation at either M1 or M2.
Mon Sep 27 12:00:09 +0000 2021PPFIA1:p.V71I chr 11:g.70272383G>A, rs546502 (all tissue V:I 0.788:0.212) vaf=>99%, vaf=22%, Δm=14.0156, VAF by population group: african 15%, american 25% east asian 11%, european 22%, south asian 27%.
Mon Sep 27 12:00:08 +0000 2021PPFIA1:p, θ(max) = 52. aka LIP.1, LIPRIN. Present in MHC class 1 peptide experiments. Found in most tissues & cell lines: very rare in fluids.
Mon Sep 27 12:00:08 +0000 2021>PPFIA1:p, PTPRF interacting protein alpha 1 (Homo sapiens) Large subunit; CTMs: M1, M2, C3+acetyl; PTMs: 7×K+acetyl; 5×K+GGyl; 73×S, 26×T, 1×Y+phosphoryl; SAAVs: V71I (22%), D540E (21%); mature form: (1,2-1202) [21,832×, 168 kTa]. #ᗕᕱᗒ 🔗
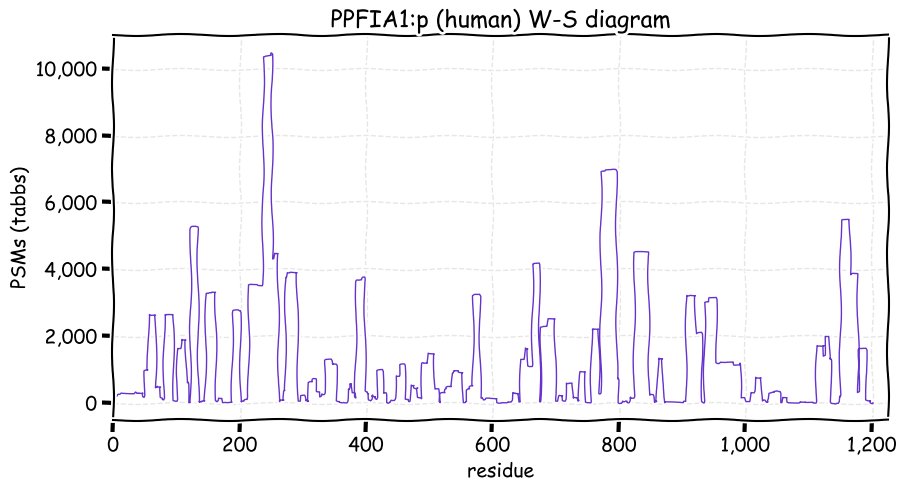
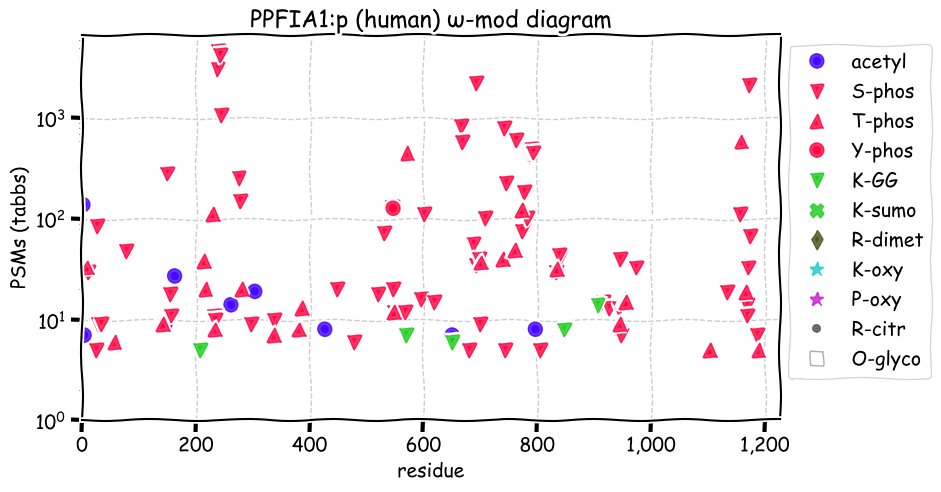
Sun Sep 26 13:52:13 +0000 2021@JodiesJumpsuit Maybe if it is part of a DPA?
Sun Sep 26 12:57:52 +0000 2021PPFIBP1:p, thanks for playing AlphaFold2, but I'm going to wait for an experimental structure 🔗 🔗
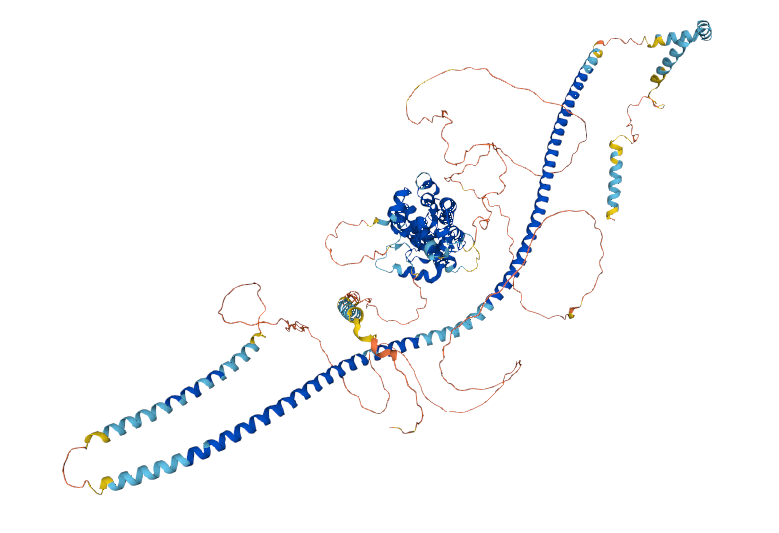
Sun Sep 26 12:24:15 +0000 2021PPFIBP1:p, its mRNA N-terminal translation begins with "MMS..." resulting in translation initiation at either M1 or M2, generating M1+acetyl or S3+acetyl as the mature N-terminus.
Sun Sep 26 12:16:12 +0000 2021PPFIBP1:p.V148L chr 12:g.27647813G>T, rs2194816 (all tissue V:L 0.016:0.984) vaf=>99%, Δm=14.0156, VAF by population group: african 98%, american >99% east asian >99%, european >99%, south asian >99%.
Sun Sep 26 12:16:12 +0000 2021PPFIBP1:p, θ(max) = 77. aka L2, hSGT2, hSgt2p, SGT2. Present in MHC class 1 peptide experiments. Found in most tissues & cell lines: absent from fluids.
Sun Sep 26 12:16:12 +0000 2021>PPFIBP1:p, PPFIA binding protein 1 (Homo sapiens) Large subunit; CTMs: M1, S3+acetyl; PTMs: 36×K+acetyl; 9×K+GGyl; 52×S, 25×T, 6×Y+phosphoryl; SAAVs: L95R (2%), V148L (99%), R949H (2%); mature form: (1,3-1011) [23,193×, 168 kTa]. #ᗕᕱᗒ 🔗
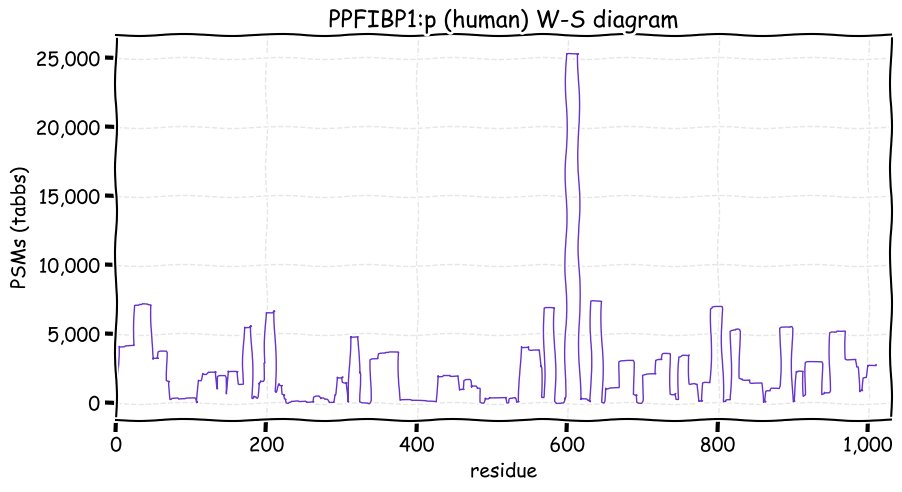
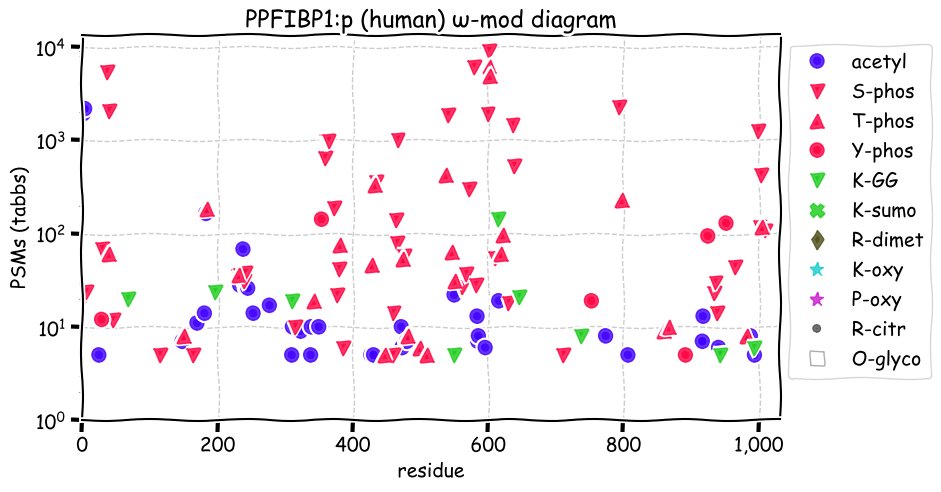
Sat Sep 25 17:41:01 +0000 2021@secretasianman @MHendr1cks The only approach that I've found that works with AC is to fly with any other airline or stay home.
Sat Sep 25 17:16:16 +0000 2021The DDA data in PXD022198 is interesting. I don't know how, but they were unusually effective at minimizing oxidation during their sample workup, while at the same time promoting N-deamidation.
Sat Sep 25 14:29:10 +0000 2021@statesdj I understand the implications in the US, but in the rest of world this particular issue has much lower (no?) impact.
Sat Sep 25 12:38:52 +0000 2021As is the fancy pants FANCD1 (which goes by the stage name BRCA2) 🔗
Sat Sep 25 12:35:34 +0000 2021FANCI:p is another good example of this GGylation pattern 🔗
Sat Sep 25 12:17:13 +0000 2021FANCA:p, Fanconi anemia complementation group proteins have many GGylation acceptor sites but there is little evidence of acetylation at those sites.
Sat Sep 25 12:13:14 +0000 2021FANCA:p.G501S chr 16:g.89783072C>T, rs2239359 (all tissue G:S 0.551:0.449) vaf=50%, Δm=30.0106, VAF by population group: african 70%, american 58% east asian 87%, european 41%, south asian 52%. GS in HEK-293 cells.
Sat Sep 25 12:13:13 +0000 2021FANCA:p, θ(max) = 29. aka FAA, FA-H, FAH, FACA, FANCH. Present in MHC class 1 peptide experiments. Found in most tissues & cell lines: rare in fluids.
Sat Sep 25 12:13:13 +0000 2021>FANCA:p, Fanconi anemia complementation group A (H sapiens) Large subunit; CTMs: S2+acetyl; PTMs: 24×K+GGyl; 13×S, 0×T, 1×Y+phosphoryl; SAAVs: V6D(4%), N8K(5%), T266A(70%), G501S(50%), G809D(67%), S1088F(2%); mature form: (1,2,3-1455) [2,921×, 8.2 kTa]. #ᗕᕱᗒ 🔗
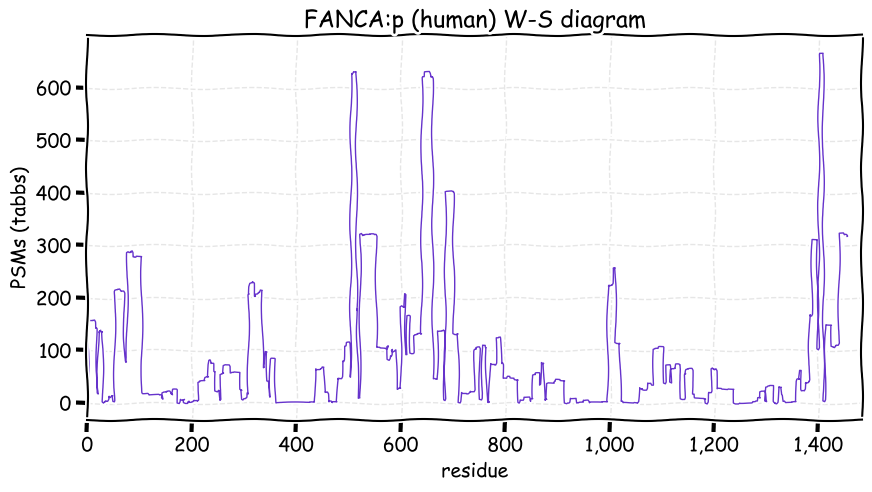
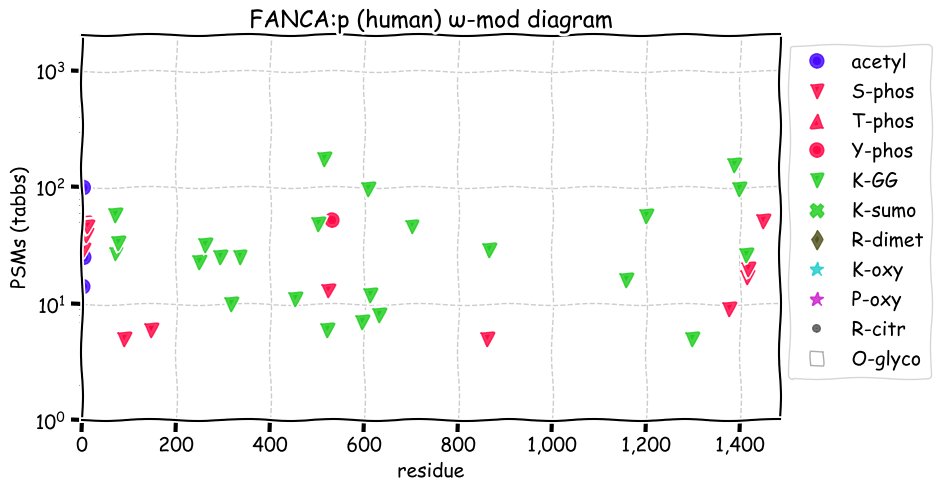
Fri Sep 24 14:13:22 +0000 2021I'm liking PXD016790 as an example of the effects you can expect to find in samples processed using the popular 8 fraction high pH MDC kit.
Fri Sep 24 12:18:03 +0000 2021FANCD2:p, whose ne'er do well cousin FANCD1 made it big by being awful at its job, changing its name to BRCA2 & moving to the bright lights of Bethesda.
Fri Sep 24 12:18:02 +0000 2021FANCD2:p.P714L chr 3:g.10064848C>T, rs3864017 (all tissue P:L 0.806:0.194) vaf=14%, Δm=16.0313, VAF by population group: african 28%, american 28% east asian 7%, european 16%, south asian 16%. PL in HeLa & HuH-7 cells. LL in NB-4 cells.
Fri Sep 24 12:18:02 +0000 2021FANCD2:p, θ(max) = 46. aka FAD, FA-D2, FACD, FANCD. Present in MHC class 1 peptide experiments. Found in most tissues & cell lines; absent from fluids.
Fri Sep 24 12:18:02 +0000 2021>FANCD2:p, FA complementation group D2 (Homo sapiens) Large subunit; CTMs: none; PTMs: 2×K+acetyl; 26×K+GGyl; 1×K+SUMOyl; 42×S, 16×T, 0×Y+phosphoryl; SAAVs: I172M (2%), P714L (14%); mature form: (1,2-1471) [14,634×, 73 kTa]. #ᗕᕱᗒ 🔗
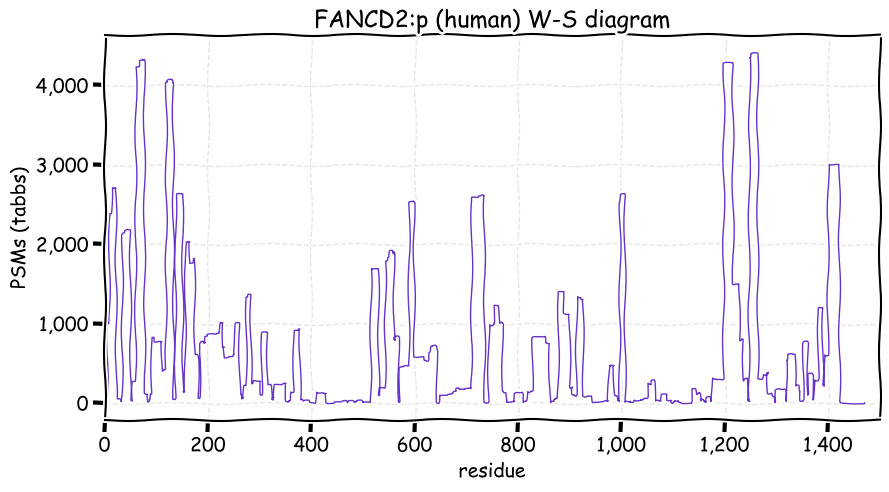
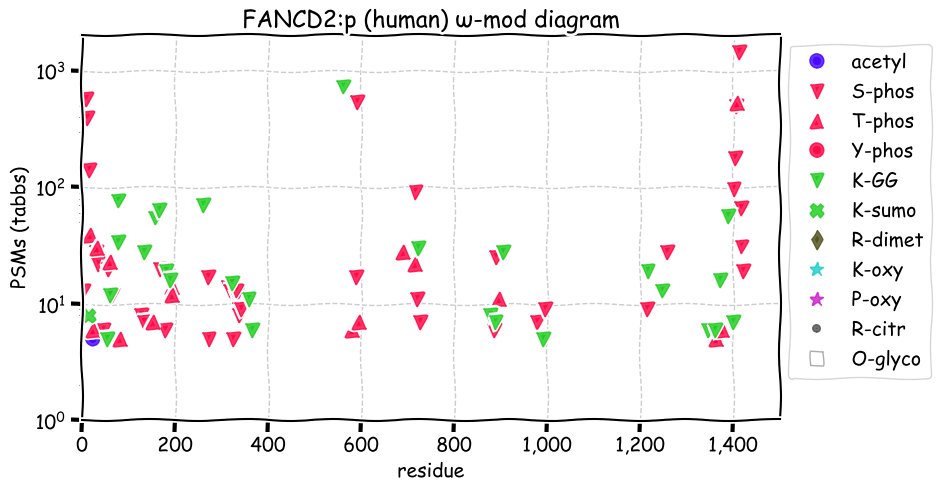
Thu Sep 23 19:38:13 +0000 2021@fcyucn @nesvilab @tomlau @ProteomicsNews @ProtifiLlc @pwilmarth @UCDProteomics @lkpino The only thing I would add is that if you are using a cloud-based solution, make sure you understand what you are paying for and how billing works. It is easy to run up a big bill if you aren't careful.
Thu Sep 23 15:23:45 +0000 2021This is the correct answer. 🧐 🔗
Thu Sep 23 12:32:21 +0000 2021EPHA2:p, domains: (25-535) extracellular, (536-558) transmembrane & (559-976) intracellular. The C-terminal intracellular domain has the lion's share of phosphorylation (99% of phospho-PSMs) 🔗

Thu Sep 23 12:32:21 +0000 2021EPHA2:p.G391R chr 1:g.16137994C>T, rs34192549 (all tissue G:R 0.974:0.026) vaf=3%, Δm=99.0796, VAF by population group: african <1%, american 9% east asian <1%, european 2%, south asian 1%.
Thu Sep 23 12:32:21 +0000 2021EPHA2:p, θ(max) = 66. aka ECK. Present in MHC class 1 & 2 peptide experiments. Found in most tissues, cell lines & extracellular vesicles in fluids.
Thu Sep 23 12:32:20 +0000 2021>EPHA2:p, EPH receptor A2 (Homo sapiens) Midsized subunit; CTMs: N407+glycosyl; PTMs: 3×K+acetyl; 14×K+GGyl; 43×S, 27×T, 18×Y+phosphoryl; S121+glycosyl; SAAVs: D232G (1%), G391R (3%), T511M (1%), V541M (1%), M631T (2%); mature form: (25-976) [22,767×, 296 kTa]. #ᗕᕱᗒ 🔗
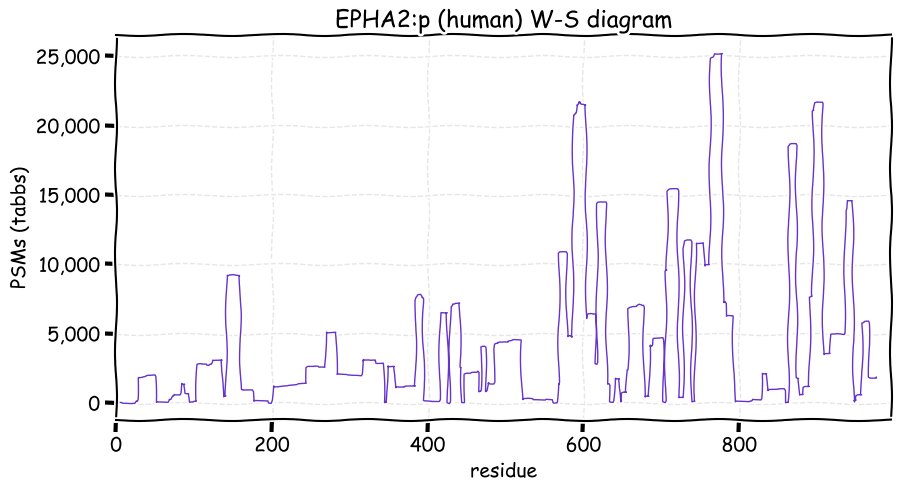
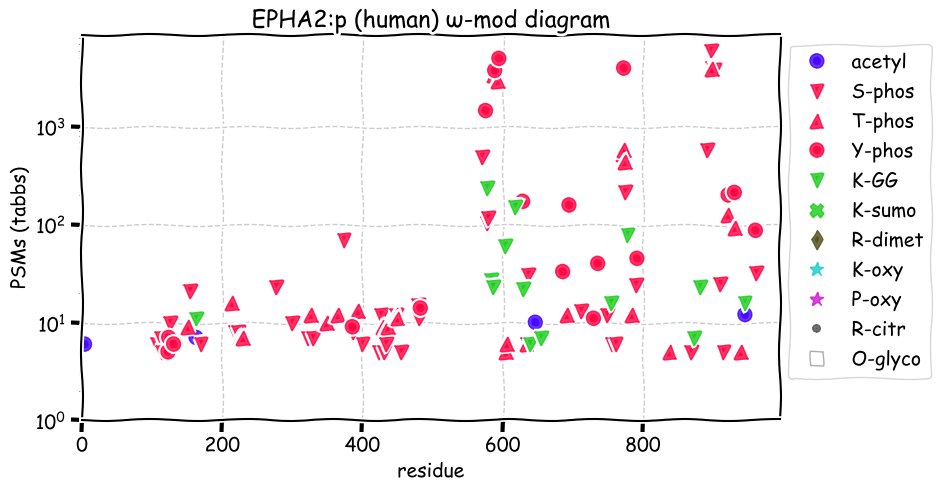
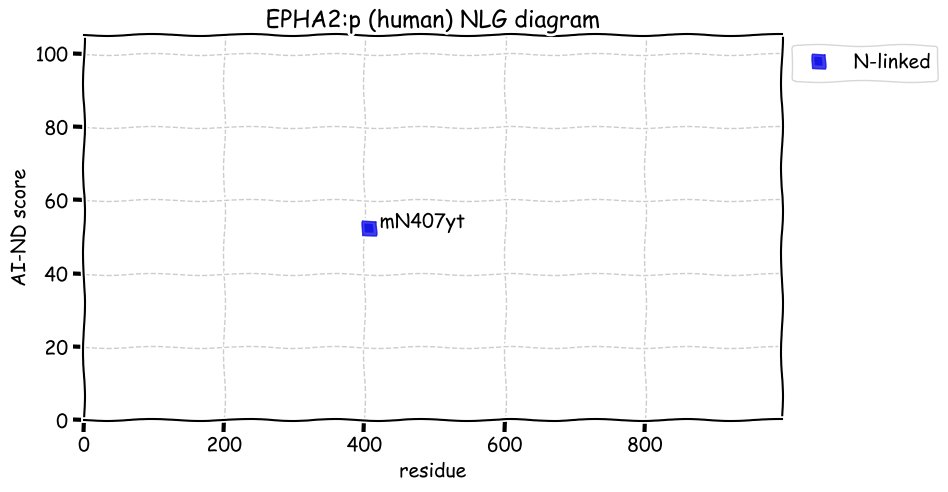
Wed Sep 22 19:06:55 +0000 2021@jengerson It also implies that for 2 elections in a row, electors did not understand the value proposition made by each of the parties and the only thing standing between the CPC and a majority was saying it better, louder or more often. Same for the LPC, NDP, GPC, BQ & PPC.
Wed Sep 22 16:19:59 +0000 2021While I try not to do it anyway, I hereby swear not to use any brand name on this platform.
Wed Sep 22 15:02:24 +0000 2021I'd treat common lab practices that misuse kits as a separate category of artifacts.
Wed Sep 22 14:37:33 +0000 2021I've often wondered how many published discoveries in biomedical research are batch-specific artifacts of commercial kits.
Wed Sep 22 12:03:48 +0000 2021LPCAT2:p, has 1 transmembrane domain (58-80), a pair of EF-hand domains (345-508) & an ER retention signal (541-544). Phosphorylation acceptor sites are in unnamed domains. 🔗
Wed Sep 22 12:03:47 +0000 2021LPCAT2:p.M163I chr 16:g.55528554G>A, rs837550 (all tissue M:I 0.273:0.727) vaf=62%, Δm=-17.9564, VAF by population group: african 34%, american 62% east asian 43%, european 65%, south asian 65%. II in THP-1 & Calu-3 cells.
Wed Sep 22 12:03:47 +0000 2021LPCAT2:p, θ(max) = 66. aka FLJ20481, AGPAT11, LysoPAFAT, AYTL1. Present in MHC class 1 peptide experiments. Found in most tissues, cell lines & urine extracellular vesicles.
Wed Sep 22 12:03:47 +0000 2021>LPCAT2:p, lysophosphatidylcholine acyltransferase 2 (Homo sapiens) Midsized subunit; CTMs: S2+acetyl; PTMs: 5×K+GGyl; 6×S, 2×S, 3×Y+phosphoryl; S121+glycosyl; SAAVs: Q1608H (27%); mature form: (2-544) [14,175×, 62 kTa]. #ᗕᕱᗒ 🔗
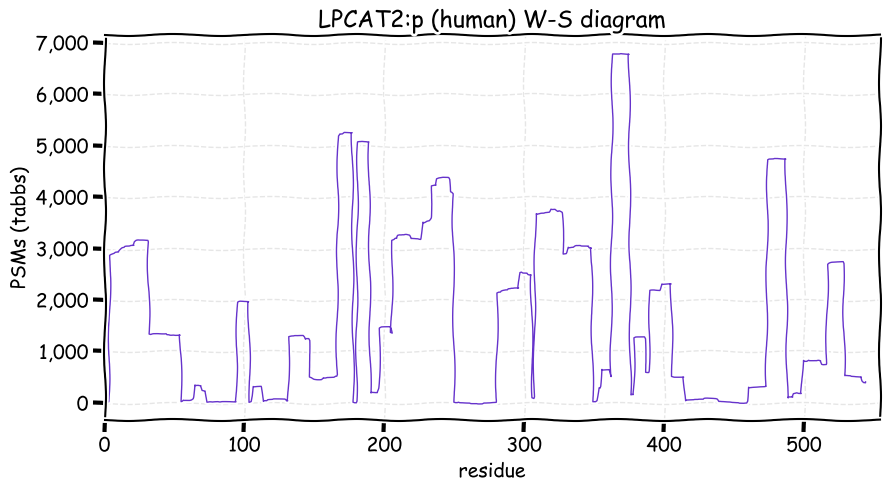
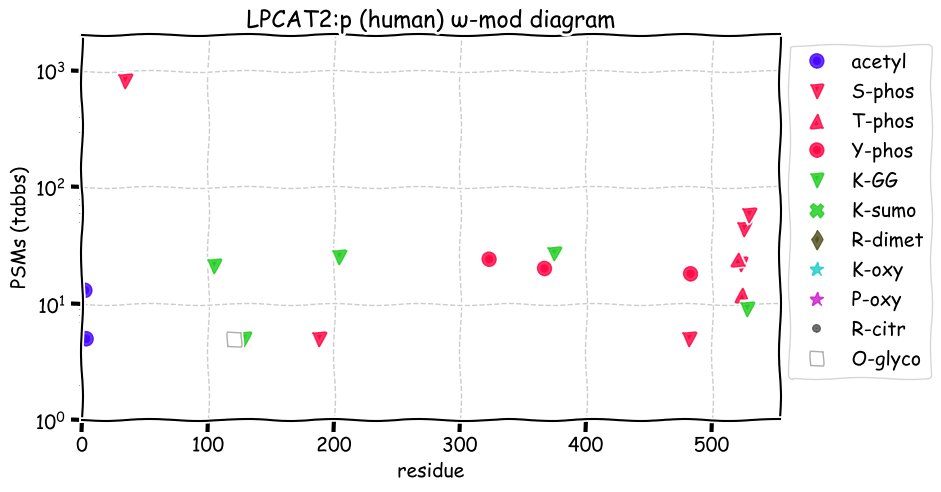
Tue Sep 21 15:59:42 +0000 2021A "career" is the story you tell people to rationalize your curriculum vitae, not a tangible (or valuable) thing.
Tue Sep 21 15:45:39 +0000 2021@pwilmarth Oddly enough, I feel exactly the same about the Canadian federal election yesterday: things are about the same as before but I find it marginally more annoying.
Tue Sep 21 14:10:18 +0000 2021@oleg8r There are some just-missing every day 🔗 (scroll to the heading "Near Earth Asteroids")
Tue Sep 21 12:57:08 +0000 2021@byu_sam Hopefully said in jest.
😃,😎,😲,😱,😡
11,-4.79,1.000987,Harry,+
157,-1.22279,57.00007,Jefferson,--
Tue Sep 21 11:57:33 +0000 2021KIDINS220:p, has 4 transmembrane domains: (495-517), (524-546), (658-680) & (687-706). The highly phosphorylated, unnamed C-terminal domain in humans (1250-1750) has the same decoration in mouse and rat. 🔗
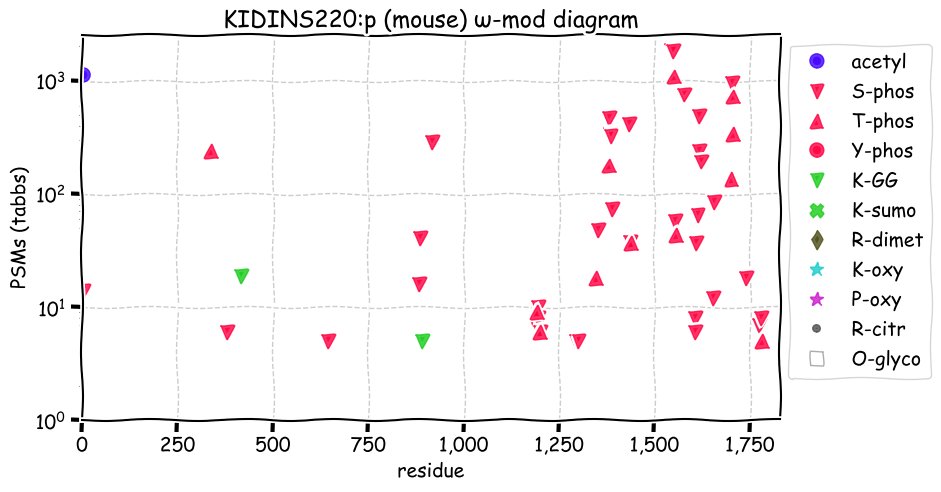
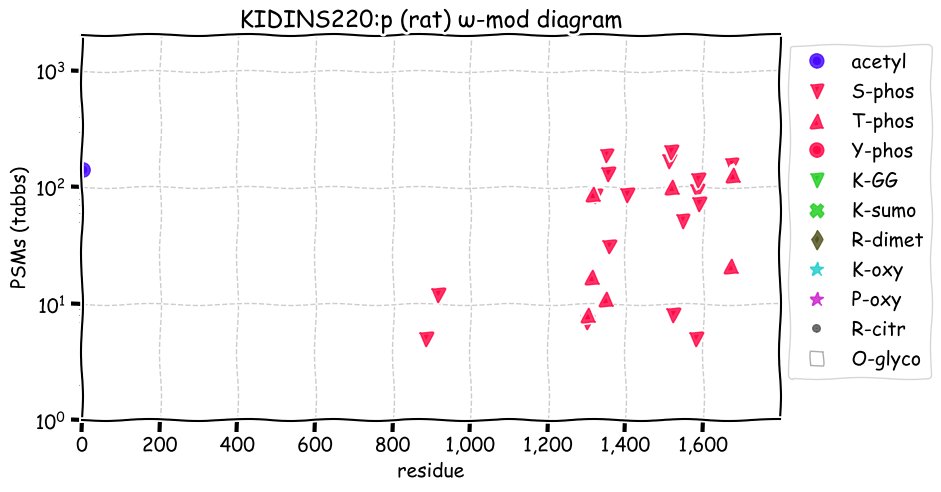
Tue Sep 21 11:57:32 +0000 2021KIDINS220:p.Q1608H chr 2:g.8731212C>A, rs1044280 (all tissue Q:H 0.917:0.083) vaf=27%, Δm=9.0003, VAF by population group: african 24%, american 14% east asian 16%, european 29%, south asian 24%. QH in HEP-2 & MCF-10A cells. HH in MDA-MB-231 cells.
Tue Sep 21 11:57:32 +0000 2021KIDINS220:p, θ(max) = 79. aka ARMS. Present in MHC class 1 & 2 peptide experiments. Found in most tissues & cell lines, but not common in fluids.
Tue Sep 21 11:57:31 +0000 2021>KIDINS220:p, kinase D interacting substrate 220 (Homo sapiens) Large subunit; CTMs: S2+acetyl; PTMs: 1×K+acetyl; 16×K+GGyl; 84×S, 24×S, 16×Y+phosphoryl; SAAVs: Q1608H (27%); mature form: (2-1771) [14,742×, 74 kTa]. #ᗕᕱᗒ 🔗
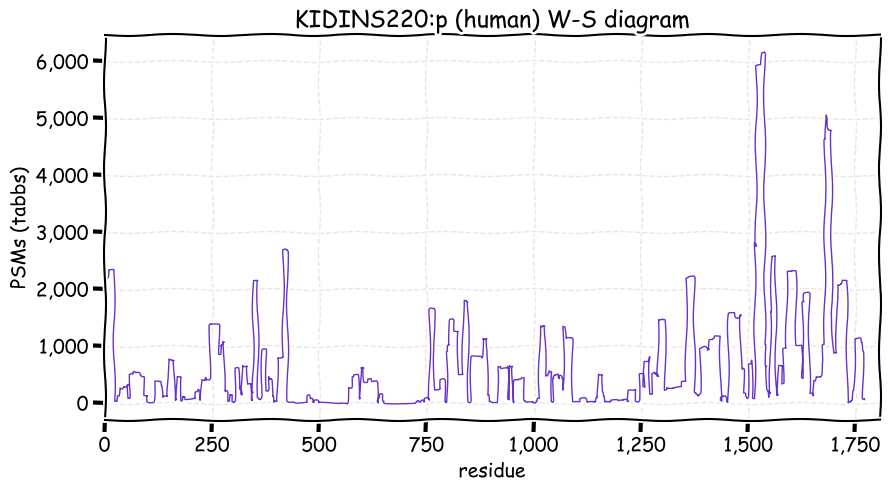
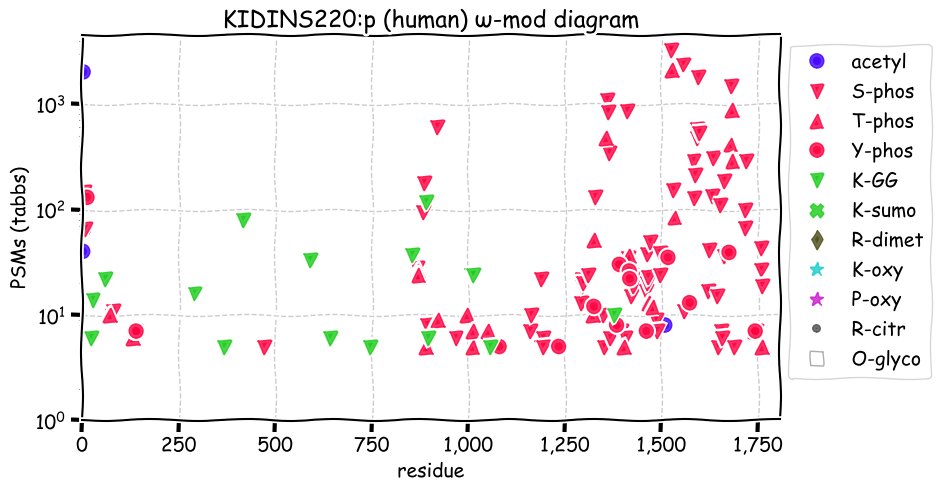
Mon Sep 20 15:39:16 +0000 2021I'm looking at you, InterProScan guys😠
Mon Sep 20 15:37:57 +0000 2021Pro tip: if you are using a simple table-based output for your software (e.g., TSV or CSV), it is never a waste of time to put good, descriptive headings on the columns.
Mon Sep 20 14:24:08 +0000 2021I recently was involved in trying to approve the minutes of a board meeting that was held via video conferencing for the first time. While trying to get approval (which is normally easy), it was apparent that each person experienced quite a different meeting.
Mon Sep 20 12:27:22 +0000 2021@aylwyn_scally @slavov_n None.
Mon Sep 20 12:20:35 +0000 2021NANS:p, there is no phosphorylation on the C-terminal SAF domain (292-351), but the pattern of acetyl/GGyl modifications sharing K-acceptor sites extends across the sequence. 🔗

Mon Sep 20 11:57:44 +0000 2021NANS:p.E68D chr 9:g.98060853G>C, rs1058446 (all tissue E:D 0.772:0.228) vaf=20%, Δm=-14.0157, VAF by population group: african 15%, american 19% east asian 7%, european 30%, south asian 15%. ED in SW-480 & HEK-293 cells. DD in Hs-578T cells.
Mon Sep 20 11:57:44 +0000 2021NANS:p, θ(max) = 79. aka SAS. Present in MHC class 1 & peptide experiments. Found in most tissues & cell lines, but not common in most fluids other than tears & milk.
Mon Sep 20 11:57:43 +0000 2021>NANS:p, N-acetylneuraminate synthase (Homo sapiens) Small subunit; CTMs: none; PTMs: 29×K+acetyl; 16×K+GGyl; 11×S, 3×S, 2×Y+phosphoryl; SAAVs: E68D (20%), P174S (1%); mature form: (2-359) [37,857×, 342 kTa]. #ᗕᕱᗒ 🔗
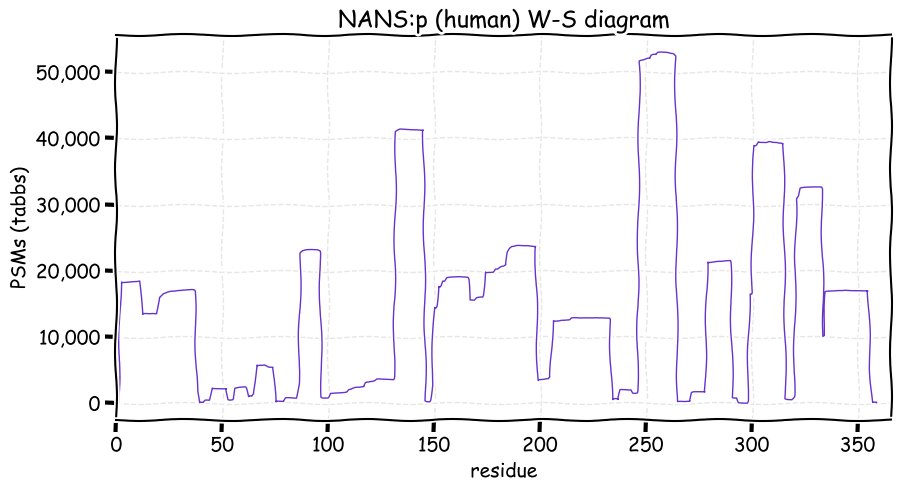
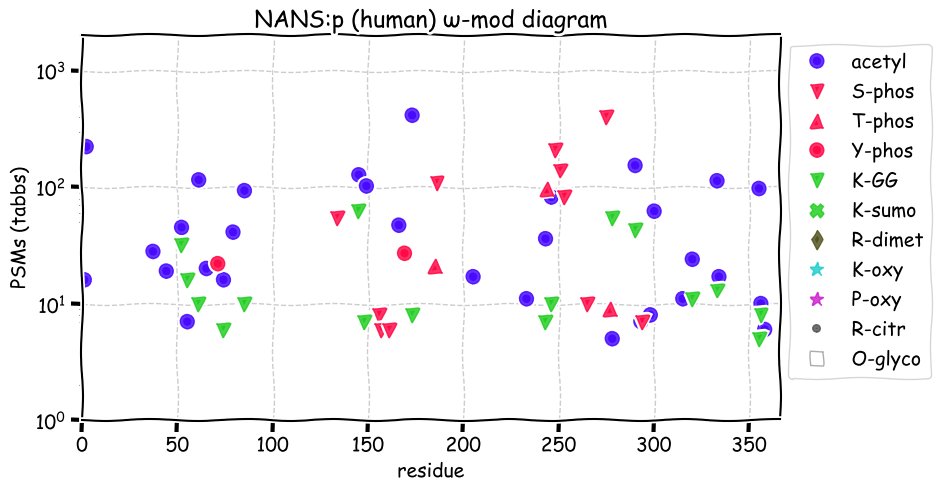
Sun Sep 19 12:59:54 +0000 2021IDE:p, is a peptidase that degrades a wide range of peptides, particularly peptide hormones: insulin B-chain was simply the first substrate discovered for this enzyme.
Sun Sep 19 12:28:52 +0000 2021IDE:p.V956I chr 10:g.92456389C>T, rs375559999 (all tissue V:I 0.981:0.019) vaf=<1%, Δm=14.0157, VAF by population group: african <1%, american <1% east asian <1%, european <1%, south asian <1%. II in JURKAT E-6.1 cells.
Sun Sep 19 12:28:52 +0000 2021IDE:p, θ(max) = 77. Present in MHC class 1 peptide experiments & more rarely in class 2. Detected in most tissues & cell lines; in fluids it is associated with extracellular vesicles.
Sun Sep 19 12:28:52 +0000 2021>IDE:p, insulin degrading enzyme (Homo sapiens) Large subunit; CTMs: none; PTMs: 17×K+acetyl; 19×K+GGyl; SAAVs: V956I (<1%); mature form: (1-1019) [31,957×, 243 kTa]. #ᗕᕱᗒ 🔗
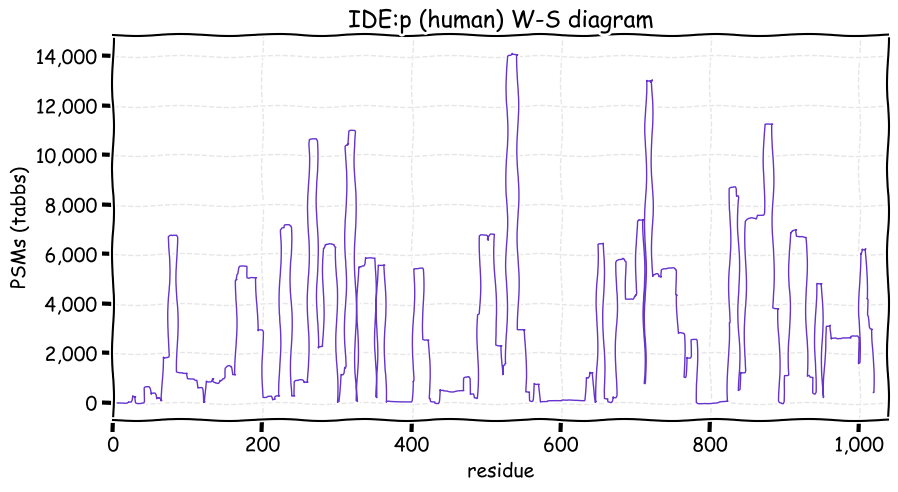
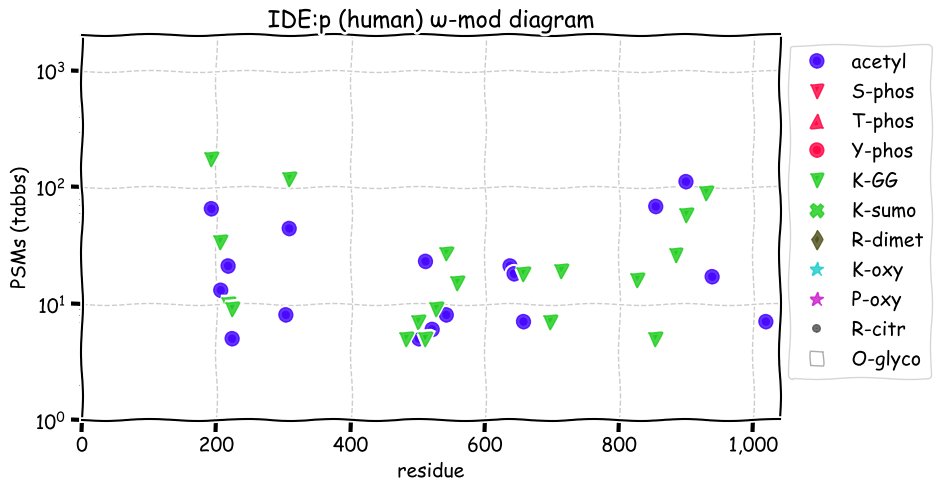
Sat Sep 18 18:05:44 +0000 2021It is entirely possible that I have never given, or received, useful advise.
Sat Sep 18 16:35:43 +0000 2021@pwilmarth It only makes you a top scientist if you think it does.
Sat Sep 18 15:55:33 +0000 2021That is not to say that I haven't met people who consider themselves to be "top scientists", but they have always been groomed (& promoted) by administrative organizations that can spot useful forms of hubris when it presents itself.
Sat Sep 18 15:52:11 +0000 2021I've never really understood the idea of a "top scientist". It has always seemed like politicians trying to impose their vision of the world onto a cadre of people who could care less.
Sat Sep 18 15:24:44 +0000 2021@dexivoje Attitude?
Sat Sep 18 15:04:56 +0000 2021I'm always pleasantly surprised when a new batch of sauerkraut works out well, even though I've never had one fail.
Sat Sep 18 12:40:50 +0000 2021@theoneamit @VATVSLPR @emwhitti Gels have retained some popularity as part of a protein-level separation MDC method, by cutting an entire gel into segments (5-10) and running each piece separately, e.g. PXD023266.
Sat Sep 18 12:36:08 +0000 2021IGFBP6:p, has 3 distinct domains: (28-107) C-rich, IGFBP domain; (108-150) O-glycosylated domain; (151-240) thyroglobulin type-1 domain with C-terminal phosphorylation 🔗

Sat Sep 18 11:54:21 +0000 2021IGFBP6:p.V187M chr 12:g.53101119G>A, rs145995968 (all tissue V:M 0.999:0.001) vaf=<1%, Δm=31.9721, VAF by population group: african <1%, american <1% east asian <1%, european <1%, south asian <1%.
Sat Sep 18 11:54:21 +0000 2021IGFBP6:p, θ(max) = 77. Present in MHC class 2 peptide experiments & more rarely in class 1. Detected in the CSF, blood plasma & urine, as well as the secretome of some cell lines via extracellular vesicles.
Sat Sep 18 11:54:21 +0000 2021>IGFBP6:p, insulin like growth factor binding protein 6 (Homo sapiens) Small subunit; CTMs: none; PTMs: 3×S+phosphoryl; T126, T146+glycosyl; SAAVs: V187M (<1%); mature form: (26,28-240) [6,087×, 42 kTa]. #ᗕᕱᗒ 🔗
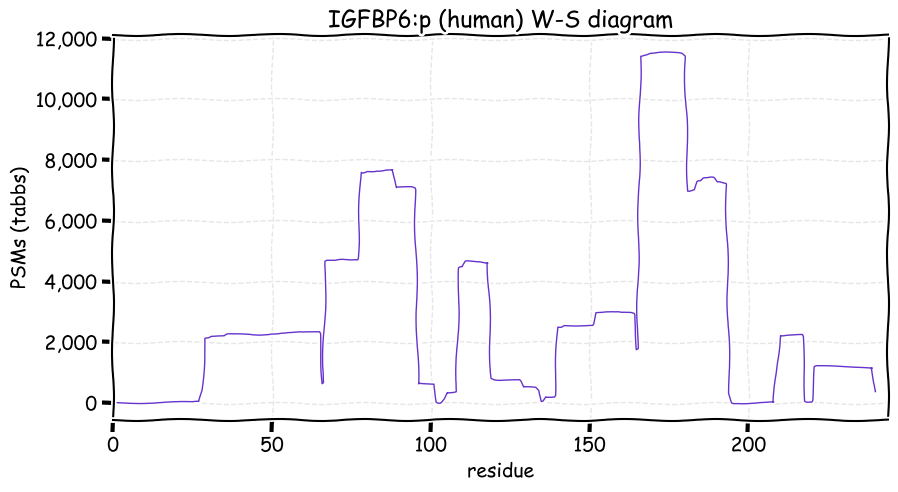
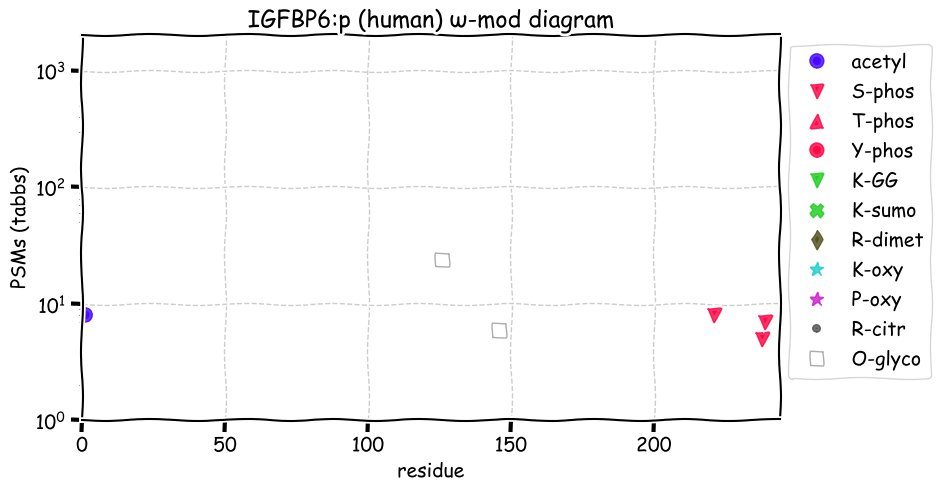
Fri Sep 17 17:36:16 +0000 2021@lenjf @VATVSLPR @emwhitti The largest problem I have had with gel-based results are that many people have an exaggerated sense of their ability to infer things from the relative position & staining of gel bands.
Fri Sep 17 14:43:11 +0000 2021I'm teeing this one up for you, @cwvhogue 🔗
Fri Sep 17 12:16:21 +0000 2021DUOX2:p has considerable tryptic peptide overlap with DUOX1:p & 6 TM domains (in grey): (594-616), (1044-1066), (1081-1103), (1149-1171), (1186-1208), (1221-1243) 🔗

Fri Sep 17 12:01:02 +0000 2021DUOX2:p.P138L chr 15:g.45111868G>A, rs2001616 (all tissue P:L 0.108:0.892) vaf=87%, Δm=16.0313, VAF by population group: african 36%, american 80% east asian 92%, european 91%, south asian 96%.
Fri Sep 17 12:01:01 +0000 2021DUOX2:p, θ(max) = 44. aka P138-TOX, P138(TOX), THOX2, LNOX2. Present in MHC class 1 peptide experiments & more rarely in class 2. Detectable mainly in the urine & the digestive tract.
Fri Sep 17 12:01:01 +0000 2021>DUOX2:p, dual oxidase 2 (Homo sapiens) Large subunit; CTMs: none; PTMs: T789+phosphoryl; SAAVs: P138L (87%), P982A (3%), S1067L (28%); mature form: (26-1548) [1,945×, 10.6 kTa]. #ᗕᕱᗒ 🔗
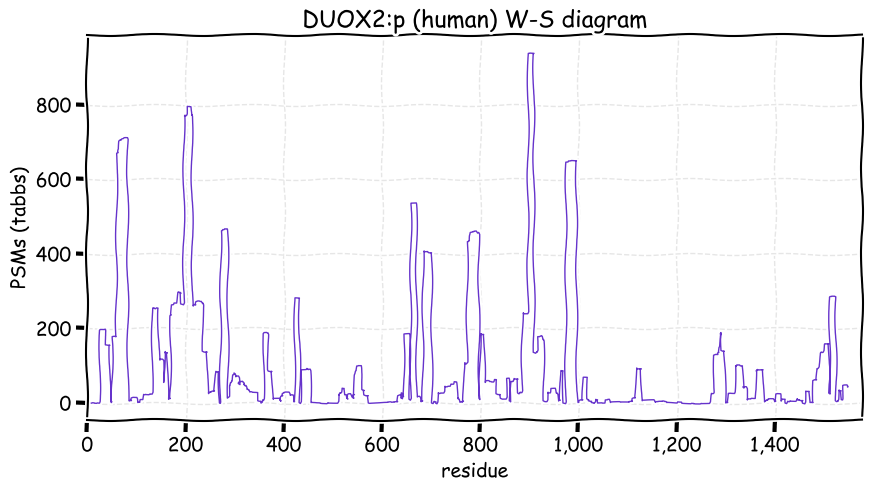
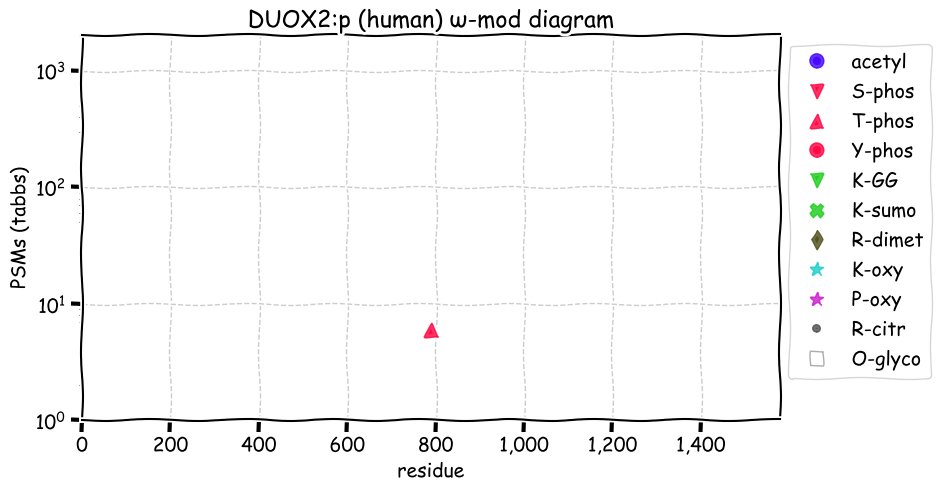
Thu Sep 16 18:12:46 +0000 2021I judge papers very harshly when they have titles that suggest they are studying tissue when they are really looking at a cell line. If you are willing to bait-and-switch, you aren't to be trusted.
Thu Sep 16 16:02:03 +0000 2021@slavov_n @embl @HHMIJanelia Underappreciated: having the Department Head/Chair solidly in your corner. If they are even a little wobbly about you & you don't have tenure, find somewhere else to work.
Thu Sep 16 15:53:16 +0000 2021Ha! And they said it couldn't be done ... (actually no one ever said this & I doubt if anyone other than me cares, but allow me to at least pretend to have small victories). 🔗
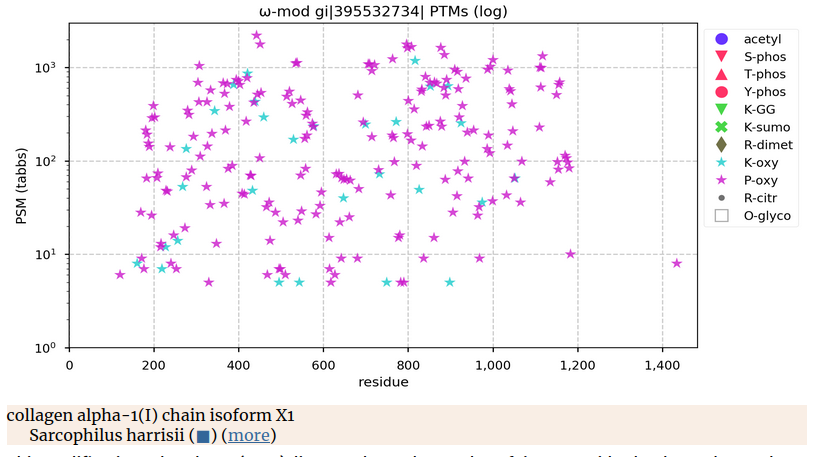
Thu Sep 16 15:09:44 +0000 2021@Smith_Chem_Wisc And I say this as someone who has been writing cross-platform software since the '90s (back when having both Windows & Mac versions with the same native GUI on both was a bit of a trick).
Thu Sep 16 14:55:59 +0000 2021@Smith_Chem_Wisc The only way referees should have this sort of authority is if they also supplied the funding, on the understanding that the result would be cross-platform.
Thu Sep 16 14:48:37 +0000 2021Initially confused by the observation of 🔗 in set of mouse breast cancer model data, then I figured out what the "PyMT" actually means ...
Thu Sep 16 12:48:15 +0000 2021TACC3:p, the acid rich domain (600-832) is dominated by 2 long, intertwined alpha helices that have very reduced PTM propensity compared to the N-terminal domain (2-600).
Thu Sep 16 12:16:23 +0000 2021TACC3:p.G514E chr 4:g.1731251G>A, rs17680881 (all tissue G:E 0.901:0.099) vaf=21%, Δm=72.0211, VAF by population group: african 13%, american 20% east asian 15%, european 26%, south asian 13%. EE in HEK-293 & U2-OS cells.
Thu Sep 16 12:16:23 +0000 2021TACC3:p, θ(max) = 95. aka ERIC1, ERIC-1, maskin, Tacc4. Present in MHC class 1 peptide experiments but not class 2. Detectable in many tissues & cell lines: may be found in extracellular vesicles.
Thu Sep 16 12:16:23 +0000 2021>TACC3:p, transforming acidic coiled-coil containing protein 3 (Homo sapiens) Midsized subunit; CTMs: S2+acetyl; PTMs: 2×K+acetyl; 6×K+GGyl; 1×K+SUMOyl; 33×S, 21×T, 5×Y+phosphoryl; SAAVs: G514E (21%), I57V (85%); mature form: (2-838) [17,973×, 97 kTa]. #ᗕᕱᗒ 🔗
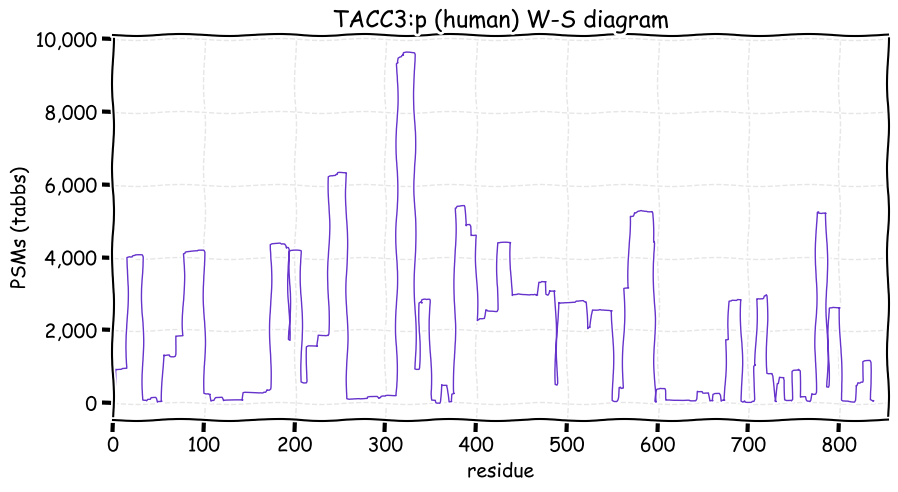
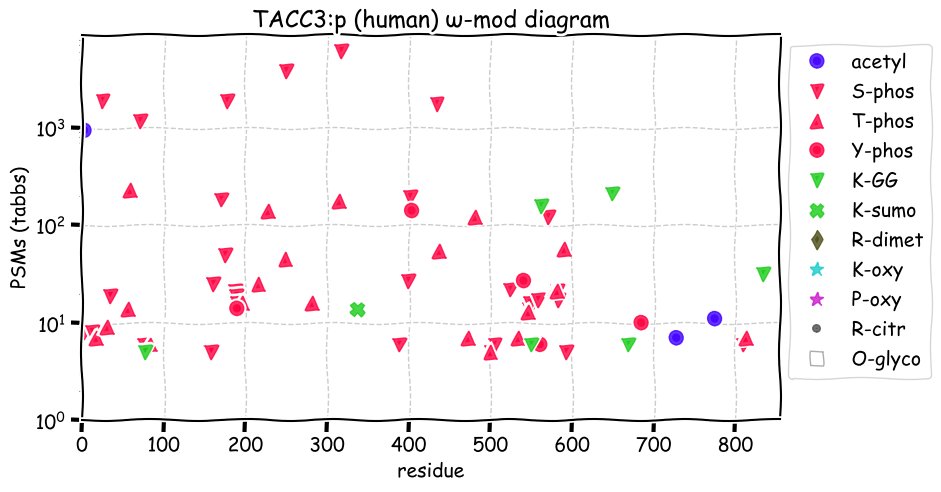
Wed Sep 15 16:10:59 +0000 20211 of the nice things about it is the data from peptides that were not phospho-enriched. This data has ~0% phosphorylation, so by analyzing it in the same way as the IMAC data, you can test how well you are able to discriminate against false positive phosphorylation assignments.
Wed Sep 15 15:53:04 +0000 2021@MagnusPalmblad It wasn't really forgotten, so much as bulldozed-over by some high profile papers in the 90's that did not properly refer to existing literature & methods.
Wed Sep 15 15:40:24 +0000 2021@AlexUsherHESA The primacy of program managers/officers in both DARPA and the NIH is completely antithetical to the Canadian way-of-doing-things.
Wed Sep 15 15:37:54 +0000 2021@AlexUsherHESA Amen.
Wed Sep 15 15:18:15 +0000 2021PXD020226 is very useful data if you are interested in S/T phosphorylation in prokaryotes 🔗
Wed Sep 15 12:51:34 +0000 2021@byu_sam @TrostLab I suspect the reviewers have been reading some piece of nostalgia-based commentary by a prominent old-timer, e.g. 🔗
Wed Sep 15 12:34:38 +0000 2021AURKA:p, has been assigned similar sounding names over time (STK6, STK7, STK15), making the literature references to the gene difficult to find & interpret reliably.
Wed Sep 15 11:50:35 +0000 2021AURKA:p.F31I chr 20:g.56386485A>T, rs2273535 (all tissue F:I 0.701:0.299) vaf=28%, Δm=-33.9843, VAF by population group: african 13%, american 29% east asian 69%, european 22%, south asian 30%. II in BT-474 cells.
Wed Sep 15 11:50:35 +0000 2021AURKA:p, θ(max) = 95. aka BTAK, AurA, STK7, ARK1, PPP1R47, AIK, STK15, STK6. May be present in MHC class 1 peptide experiments & more rarely in class 2. Detectable in many tissues & cell lines, not in fluids.
Wed Sep 15 11:50:34 +0000 2021>AURKA:p, aurora kinase A (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 2×K+acetyl; 7×K+GGyl; 3×K+SUMOyl; 0×S, 8×T, 1×Y+phosphoryl; SAAVs: F31I (28%), I57V (85%); mature form: (2-403) [13,531×, 147 kTa]. #ᗕᕱᗒ 🔗
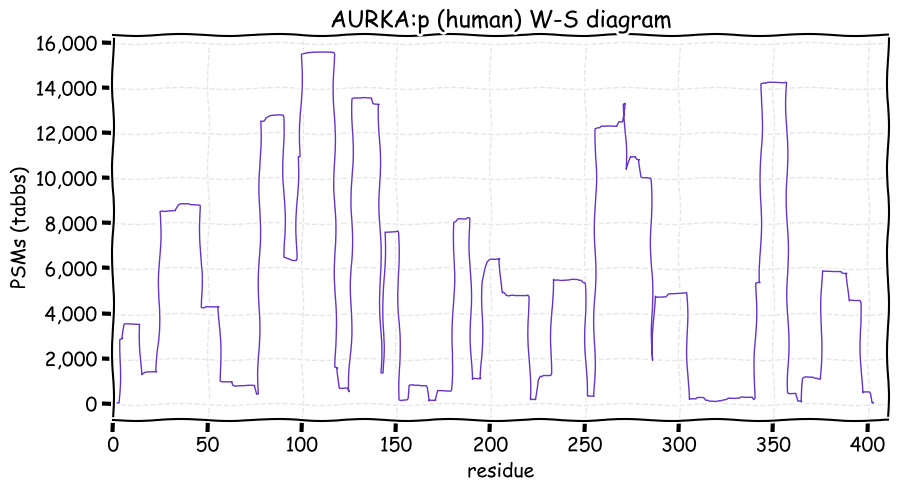
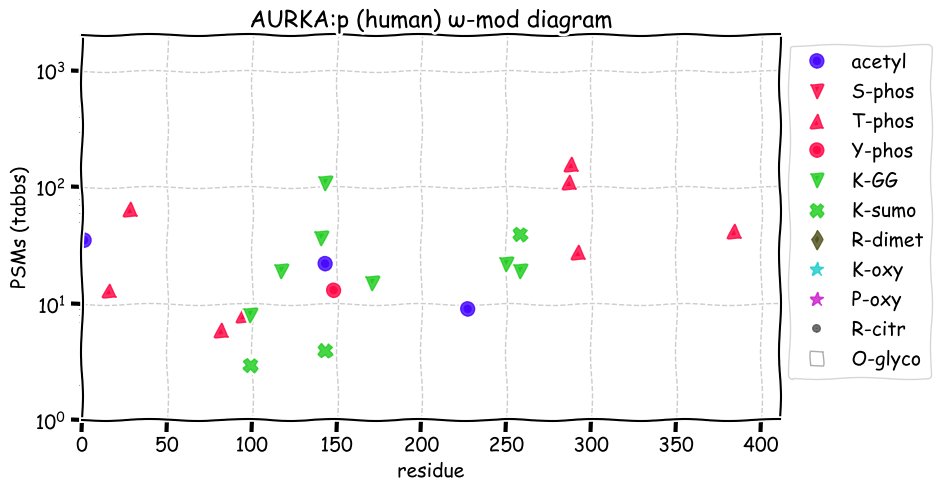
Wed Sep 15 00:13:53 +0000 2021An angry looking line of storms stretching from just north of Tulsa to just north of Montréal 🔗
Tue Sep 14 20:22:38 +0000 2021When MDC pooled fractions from the 1st D don't end up being quite as orthogonal as you were hoping in the 2nd D, even though the 2nd D is a linear gradient ... 🔗
Tue Sep 14 20:15:12 +0000 2021@Smith_Chem_Wisc I normally only write software for myself, so I'm not sure how to interpret the question in my case.
Tue Sep 14 19:44:41 +0000 2021@AlexUsherHESA Weltschmerzfußball?
Tue Sep 14 19:00:42 +0000 2021It is always pleasant to see a new data set from Jean Armengaud's lab. Something to look forward to analyzing.
Tue Sep 14 16:13:32 +0000 2021🔗
Tue Sep 14 16:09:14 +0000 2021When somebody tells you that "X is a team sport!", it usually means that they consider you to be equipment.
Tue Sep 14 15:18:41 +0000 2021@LindorffLarsen I would assert that it is biomedical research groups that have moved over to using increasingly larger data sets, mainly because biology is unable to provide much guidance about the questions they need to answer.
Tue Sep 14 14:58:58 +0000 2021@LindorffLarsen The article also seems to lump biology in with biomedical research. While the fields do share a lot of common ground, their goals are radically different, which leads to very different styles of research & data interpretation.
Tue Sep 14 14:50:36 +0000 2021@LindorffLarsen There is some merit to the position, although there is too much name-dropping in such a short article for my taste. However, it does sound a bit like an ode-to-yesteryear from someone who feels out of place with results based on modern methods.
Tue Sep 14 13:14:35 +0000 2021INCENP:p, if you are interested in the implications of long stable single alpha-helix domains, which are the antithesis of "globular" protein structures, this is a pretty good introduction 🔗
Tue Sep 14 12:06:59 +0000 2021INCENP:p has a long, low complexity domain highly enriched in charged residues (530-780), that forms a single alpha-helix with no PTMs (an SAH domain).
Tue Sep 14 12:06:59 +0000 2021INCENP:p.M506T chr 11:g.62140968T>C, rs2277283 (all tissue M:T 0.701:0.299) vaf=24%, Δm=-29.9928, VAF by population group: african 5%, american 19% east asian 7%, european 30%, south asian 15%. MT in U2-OS & MV4-11 cells.
Tue Sep 14 12:06:59 +0000 2021INCENP:p, θ(max) = 52. aka FLJ31633. May be present in MHC class 1 peptide experiments. Detectable in many tissues & cell lines, not in fluids.
Tue Sep 14 12:06:58 +0000 2021>INCENP:p, inner centromere protein (Homo sapiens) Midsized subunit; CTMs: G2+acetyl; PTMs: 7×K+acetyl; 3×K+GGyl; 4×K+SUMOyl; 54×S, 31×T, 6×Y+phosphoryl; SAAVs: R100H (3%), A137V (4%), M506T (24%), E644D (29%); mature form: (2-918) [13,984×, 76 kTa]. #ᗕᕱᗒ 🔗
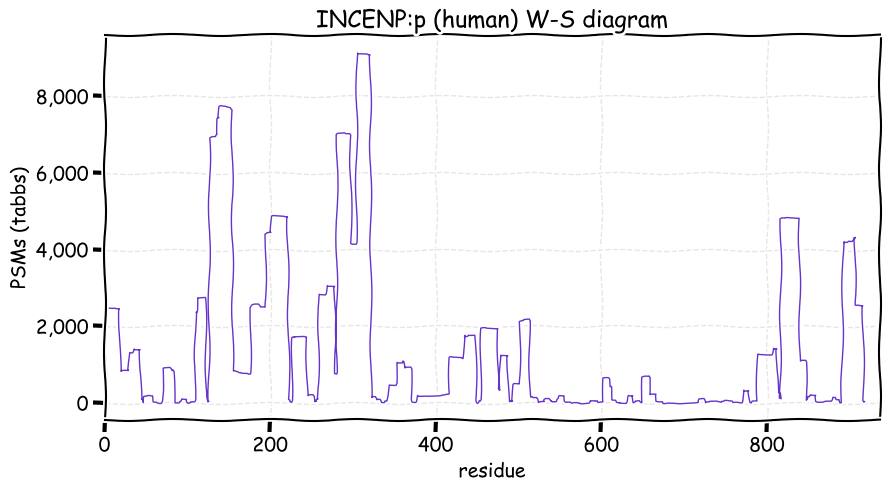
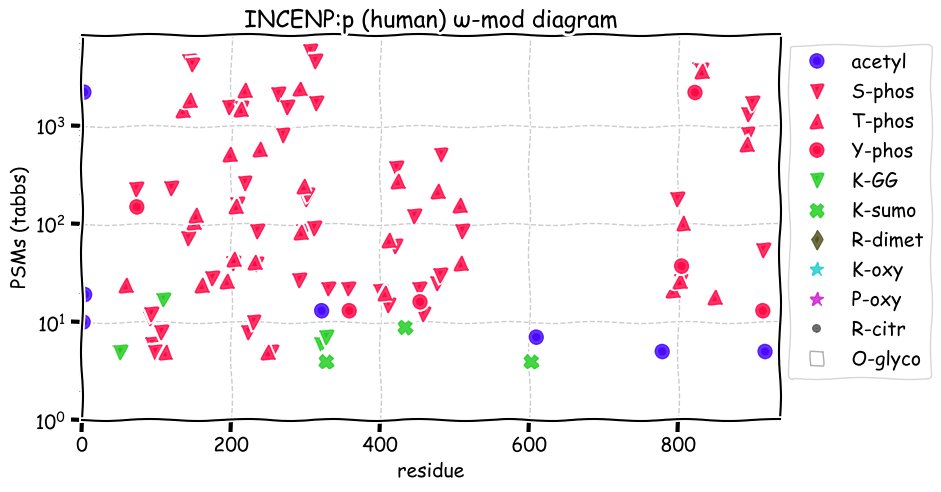
Mon Sep 13 16:41:06 +0000 2021The answer was zero, because there is no blood serum present in a human: it is a manufactured substance made from blood. The same thing can be said about plasma. There is simply no such thing. What is in it depends very much on how you make it.
Mon Sep 13 16:39:02 +0000 2021A colleague of mine used to like to stump unwary PhD students by asking them how many litres of serum were circulating in the average human.
Mon Sep 13 14:09:42 +0000 2021@ypriverol @byu_sam @stephenaramsey @bnlasse @arxiv LibreOffice Writer has an extension that allows you to export LaTeX documents (as well as a rather nifty equation editor).
Mon Sep 13 12:10:41 +0000 2021OCLN:p is commonly observed in some cell lines that are not thought to contain tight junctions, leading to speculation that the protein may be involved in other processes. 🔗
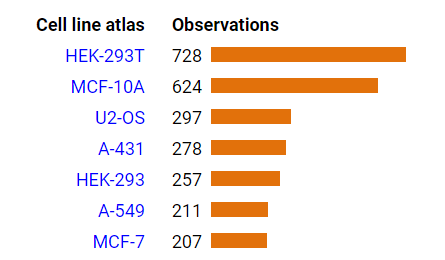
Mon Sep 13 12:08:46 +0000 2021OCLN:p has 4 transmembrane domains: (67-89), (139-161), (173-195) & (243-265). This is typical of tight junction forming proteins. The rather garish set of PTMs are on intracellular domains.
Mon Sep 13 12:08:46 +0000 2021OCLN:p.V308L chr 5:g.69534724G>C, rs369518478 (all tissue V:L 0.999:0.001) vaf=0.3%, Δm=14.0156, VAF by population group: african <1%, american <1% east asian <1%, european <1%, south asian <1%.
Mon Sep 13 12:08:46 +0000 2021OCLN:p, θ(max) = 37. aka PPP1R115. May be present in MHC class 1 peptide experiments. Detectable in many tissues & cell lines, as well as extracellular vesicles. Commonly associated with tight junctions.
Mon Sep 13 12:08:46 +0000 2021>OCLN:p, occludin (Homo sapiens) Midsized subunit; CTMs: S2+acetyl; PTMs: 7×K+GGyl; 24×S, 8×T, 14×Y+phosphoryl; SAAVs: V308L (<1%); mature form: (2-522) [10,875×, 48 kTa]. #ᗕᕱᗒ 🔗
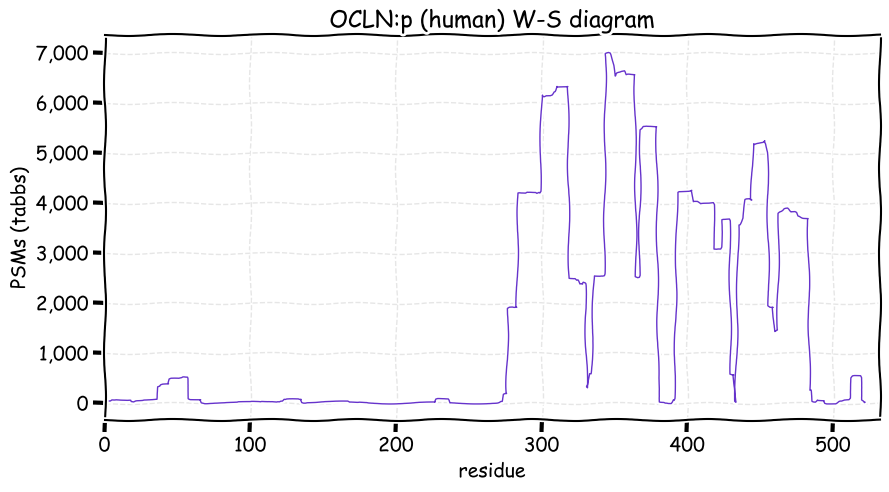
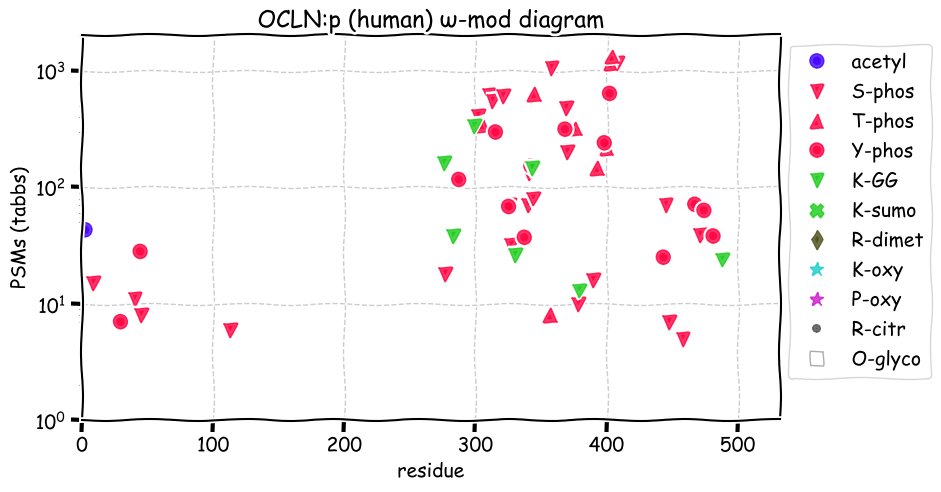
Sun Sep 12 18:36:38 +0000 2021@AlexUsherHESA So, next 🇨🇦 election Spring 2023?
Sun Sep 12 17:53:19 +0000 2021@schumacherbj Looks more like Oxbridge to me ...
Sun Sep 12 15:23:23 +0000 2021Especially if it allows for any transformation of postulate → hypothesis (no matter how poorly motivated).
Sun Sep 12 15:10:37 +0000 2021People are unlikely to see a data set as a noisy POC if it happens to be the best evidence for some postulated mechanism.
Sun Sep 12 12:16:53 +0000 2021GJB1:p has 4 transmembrane domains (shown in grey): (23-45), (76-95), (131-153) & (192-214). This is typical of gap junction forming proteins. The PTMs observed are all on intracellular domains. 🔗

Sun Sep 12 12:16:52 +0000 2021GJB1:p.V63I chr X:71223894G>A, rs116840818 (all tissue A:V 0.988:0.012) vaf=<1%, Δm=14.0156, VAF by population group: african <1%, american <1% east asian <1%, european <1%, south asian <1%.
Sun Sep 12 12:16:52 +0000 2021GJB1:p, θ(max) = 33. aka CX32, CMTX1, CMTX. May be present in MHC class 1 peptide experiments. Detectable in select tissues & cell lines, as well as extracellular vesicles in urine & cancer cell media.
Sun Sep 12 12:16:52 +0000 2021>GJB1:p, gap junction protein beta 1 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 1×K+GGyl; 4×S, 0×T, 0×Y+phosphoryl; SAAVs: V63I (<1%); mature form: (1-283) [1,139×, 2.8 kTa]. #ᗕᕱᗒ 🔗
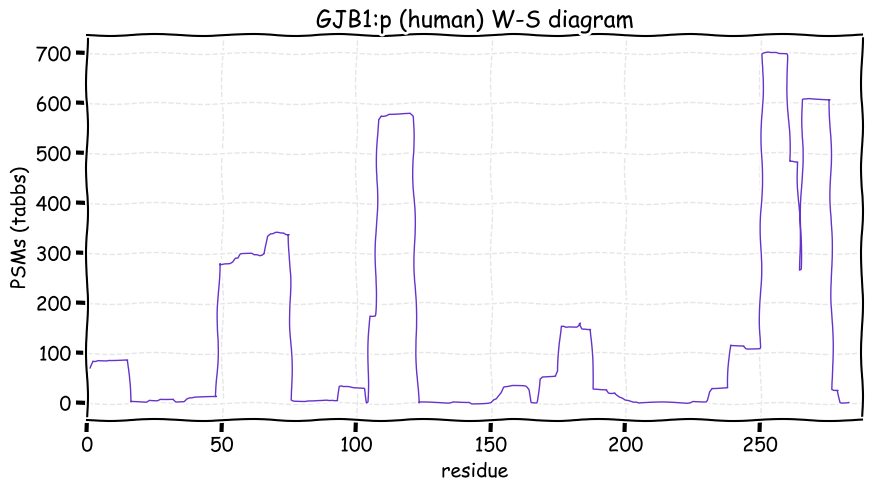
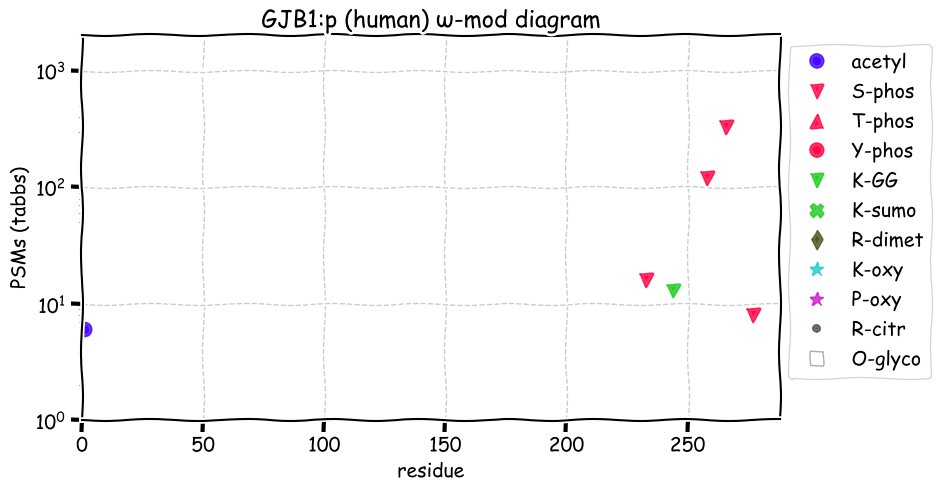
Sat Sep 11 15:28:26 +0000 2021But I do feel some sympathy for biocurators, who have no choice but to wade through the swamp of papers in "high profile" journals & try to find something of value.
Sat Sep 11 15:21:11 +0000 2021I do feel fortunate that I don't depend (at all) on the contents of papers to re-use their data for my own fiendish purposes.
Sat Sep 11 14:32:19 +0000 2021Although perhaps it was ever thus.
Sat Sep 11 14:31:03 +0000 2021When did it become OK to take a large volume of tricky data, naively analyze it with a dodgy algorithm at 5% FDR, focus on a very small number of weirdo PSMs as though they were ground truth & base a paper on speculative consequences of these "observations"?
Sat Sep 11 12:51:58 +0000 2021GJA1:p, has anyone ever looked into the consequences (if any) of extracellular vesicles having the necessary apparatus to form gap junctions?
Sat Sep 11 12:17:59 +0000 2021@RDUnwin @astacus Many labs simply have problems with the iTRAQ/TMT derivatization reactions. I suspect it depends on whether or not their is someone trained in organic chem in the lab.
Sat Sep 11 12:00:32 +0000 2021GJA1:p has 4 transmembrane domains (shown in grey): (24-46), (77-99), (155-177) & (209-231). This is typical of gap junction forming proteins. The PTMs observed are all on intracellular domains. 🔗

Sat Sep 11 12:00:31 +0000 2021GJA1:p.A253V chr 6:g.121447605C>T, rs17653265 (all tissue A:V 0.999:0.001) vaf=0.8%, Δm=28.0313, VAF by population group: african <1%, american 1% east asian <1%, european 1%, south asian <1%.
Sat Sep 11 12:00:31 +0000 2021GJA1:p,θ(max) = 55. aka CX43, ODD, ODOD, SDTY3, ODDD, GJAL. May be present in MHC class 1 or 2 peptide experiments. Detectable in many tissues, cell lines & urine extracellular vesicles.
Sat Sep 11 12:00:31 +0000 2021>GJA1:p, gap junction protein alpha 1 (Homo sapiens) Small subunit; CTMs: G2+acetyl; N294+glycosyl; PTMs: 13×K+GGyl; 24×S, 4×T, 8×Y+phosphoryl; SAAVs: A253V (0.8%); mature form: (2-382) [8,916×, 80 kTa]. #ᗕᕱᗒ 🔗
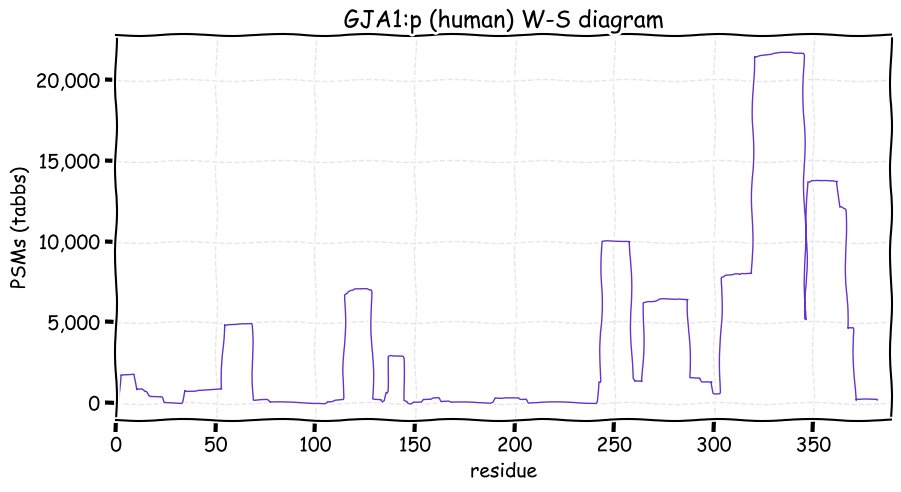
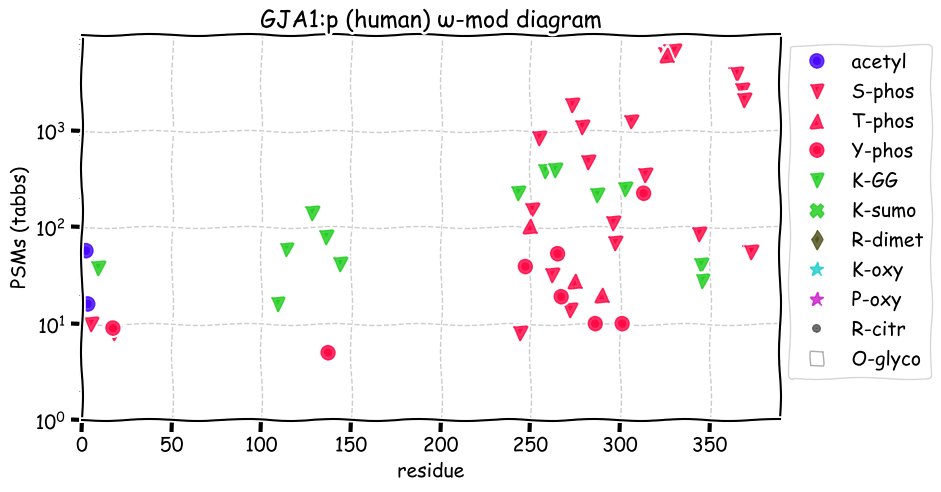
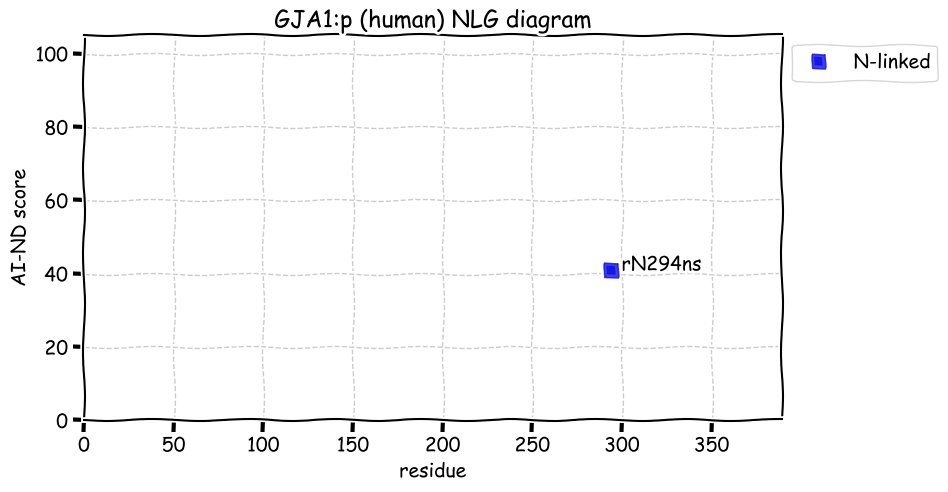
Fri Sep 10 19:34:39 +0000 2021@matthewhirschey Saccharomyces cerevisiae 🔗
Fri Sep 10 16:25:35 +0000 2021While the Canadian national media discusses how awful the debate was last night, Hurricane Larry is still on course to wallop St. John's tonight. 🔗

Fri Sep 10 12:12:41 +0000 2021PANX1:p has 4 transmembrane domains (38-60), (108-130), (213-235), (275-297), characteristic of the many subunits that can assemble into paired hexamer pores forming gap junctions 🔗 🔗

Fri Sep 10 12:01:55 +0000 2021PANX1:p.Q5H chr 11:g.94129327A>C, rs1138800 (all tissue Q:H 0.549:0.451) vaf=68%, Δm=9.0003, VAF by population group: african 86%, american 77% east asian 62%, european 62%, south asian 59%. QH in HEK-293 & HeLa cells.
Fri Sep 10 12:01:55 +0000 2021PANX1:p, θ(max) = 38. aka MRS1, UNQ2529, PX1. May be very abundant in MHC class 1 peptide experiments. May be detected in many tissues, cell lines & blood plasma, but very rarely in urine extracellular vesicles.
Fri Sep 10 12:01:54 +0000 2021>PANX1:p, pannexin 1 (Homo sapiens) Small subunit; CTMs: A2+acetyl; N255+glycosyl; PTMs: 5×K+GGyl; 2×S, 2×T, 0×Y+phosphoryl; SAAVs: Q5H (68%), T396M (1%); mature form: (2-426) [4,835×, 14 kTa].#ᗕᕱᗒ 🔗
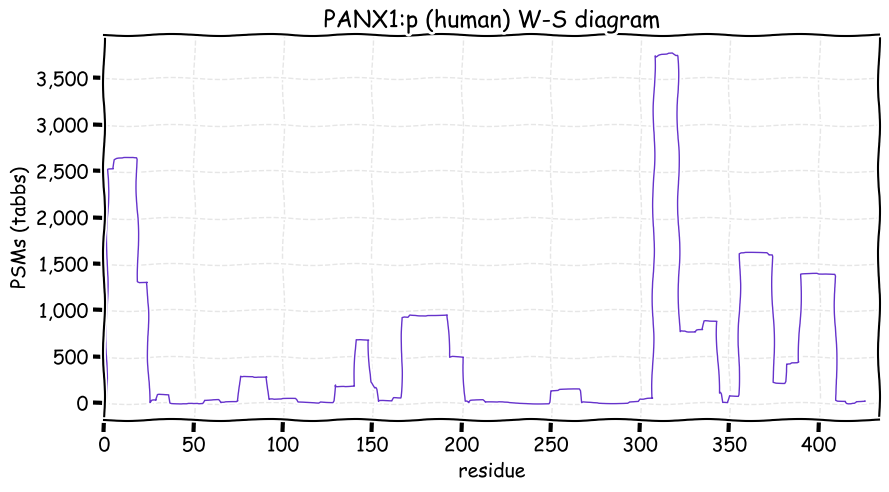
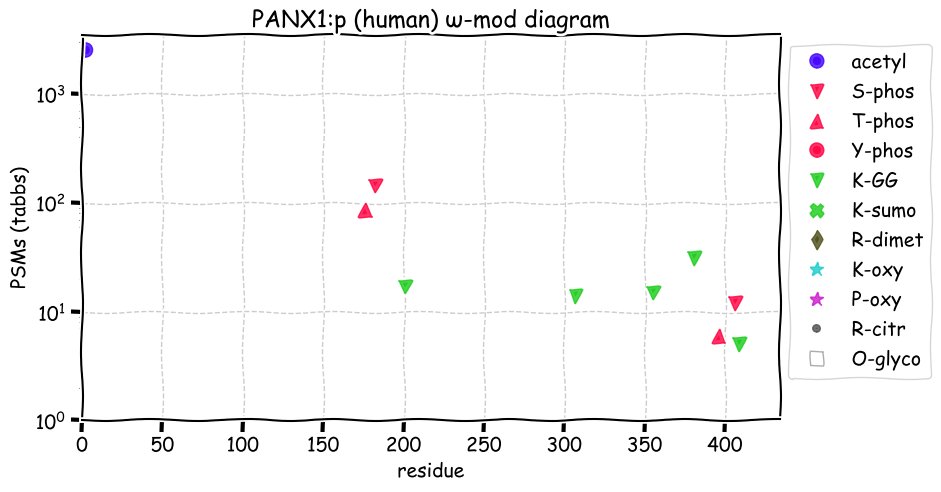
Thu Sep 09 15:45:09 +0000 2021I must admit to a childish glee when I see a new data set for Sarcophilus harrisii.
Thu Sep 09 15:42:25 +0000 2021@pwilmarth @ProteomicsNews I think he was actually referring to the 3rd statement in the tweet, regarding the inadequacy of all other methods (or maybe I was misinterpreting your reply).
Thu Sep 09 15:29:44 +0000 2021I suspect this is mainly a "US only" issue, but I'm not sure. 🔗
Thu Sep 09 15:17:24 +0000 2021Hurricane Larry is a fast moving son-of-a-gun expected to clobber the Avalon Peninsula tomorrow evening 🔗
Thu Sep 09 14:06:26 +0000 2021@Gagan90063345 To be clear: this doesn't involve measurements I took. It is data from 🔗 & it is true for all 175 of the LC/MS runs. The data is still useful for the purposes described, I was just interested in why this anomalous FWHM was happening (I hate mysteries).
Thu Sep 09 12:53:26 +0000 2021DMKN:p would be a great example of how important detecting splice variants & SAAVs was in proteomics, except no one really knows what DMKN:p does & whatever it is doing it hasn't been linked to a human disease.
Thu Sep 09 12:14:23 +0000 2021DMKN:p, most observations of this protein in cell lines are correlated with high levels of KRT1, KRT2, KRT9 & KRT10, i.e. contamination with castoff human skin.
Thu Sep 09 12:03:12 +0000 2021DMKN:p.V91A chr 19:g.35513204A>G, rs4806163 (all tissue V:A 0.337:0.663) vaf=80%, Δm=-28.0313, VAF by population group: african 99%, american 74%, east asian 87%, european 79%, south asian 83%.
Thu Sep 09 12:03:12 +0000 2021DMKN:p, θ(max) = 44. aka ZD52F10. May be very abundant in MHC class 1 or 2 peptide experiments. Common in urine, blood plasma & many types of epidermus. It has at least 6 splice variants distinguishable in proteomics data.
Thu Sep 09 12:03:12 +0000 2021>DMKN:p, dermokine (Homo sapiens) Small subunit; CTMs: none; PTMs: R76, R115+deiminyl; S26+phosphoryl; SAAVs: V91A (80%), N139S (11%), D427A (>99%); mature form: (22-476) [3,441×, 13 kTa]. #ᗕᕱᗒ 🔗
Wed Sep 08 19:54:21 +0000 2021Mass spec peeps: what would cause a Q Exactive HF-X to have a 20 ppm FWHM parent ion mass error distribution for PSMs, rather than the ~5 ppm value I normally see? Source needs cleaning, some subtle calibration problem, didn't turn on some widget, ...
Wed Sep 08 14:49:36 +0000 2021@CellsAndProts @michaelhoffman Bagged milk used to be common in Western Canada when I was a kid, but it is long gone now.
Wed Sep 08 14:36:33 +0000 2021Both Cuba and the UK seem to be having trouble shaking the 4th wave 🔗
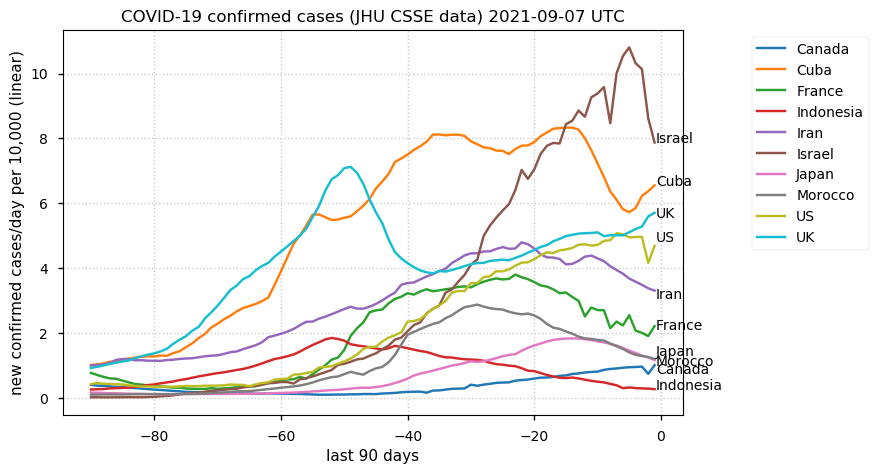
Wed Sep 08 12:08:14 +0000 2021ERBIN:p, the leucine-rich domains (47-413) & PDZ domain (1318-1419) have very different PTM patterns than the central (unnamed) domain (425-1310), which has 3 different patterns of phosphorylation (413-800), (801-1200) & (1201-1310).
Wed Sep 08 12:08:14 +0000 2021ERBIN:p.S1112L chr 5:g.66054653C>T, rs3805466 (all tissue S:L 0.868:0.132) vaf=10%, Δm=26.052, VAF by population group: african 1%, american 22%, east asian 13%, european 4%, south asian 7%.
Wed Sep 08 12:08:14 +0000 2021ERBIN:p, θ(max) = 73. aka LAP2, ERBB2IP. Abundant in MHC class 1 peptide experiments only. Common in tissues & cell lines: rare in fluids.
Wed Sep 08 12:08:13 +0000 2021>ERBIN:p, erbb2 interacting protein (Homo sapiens) Large subunit; CTMs: none; PTMs: 1×K+acetyl; 11×K+GGyl; 1×K+SUMOyl; 84×S, 27×T, 18×Y+phosphoryl; SAAVs: S1112L (10%); mature form: (2-1419) [22,689×, 149 kTa]. #ᗕᕱᗒ 🔗
Tue Sep 07 20:32:51 +0000 2021And it seems to be due to its unusually good chromatography & slightly flawed sample prep that just adds a little zest.
Tue Sep 07 20:28:50 +0000 2021The more I look at the analysis of PXD023938, the more interesting it looks as a potential model data set.
Tue Sep 07 20:19:36 +0000 2021@neely615 @PMSC34891253 The clinical use of MS-based proteomics (as seen on "The Expanse") seems like an odd fascination as they finish up careers that have been focused on discovery research.
Tue Sep 07 18:11:15 +0000 2021Nice to see that Sci-Hub has been restocked & is up-to-date.
Tue Sep 07 16:43:45 +0000 2021And now I can get files! 😀
Tue Sep 07 16:41:56 +0000 2021Note: CA$3B/year is how much is distributed; it does not include how much it costs CRA to run the program.
Tue Sep 07 16:17:47 +0000 2021Looks like Madison (& then Milwaukee) is in for some weather 🔗
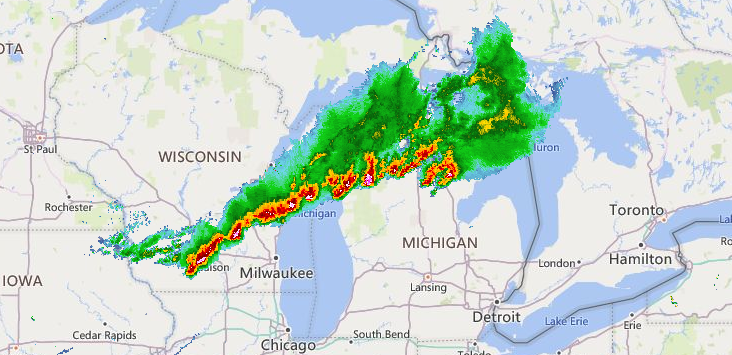
Tue Sep 07 16:04:22 +0000 2021I guess this makes it Canada's largest single scientific research/technical R&D program.
Tue Sep 07 16:02:44 +0000 2021Found it myself (figured out the appropriate Google keywords). More than CA$3B/year to 20k claimants 🔗 It isn't clear whether this is federal + provincial tax expenditures or federal alone, but either way it gives an order of magnitude.
Tue Sep 07 15:40:41 +0000 2021Does anybody know how much money is refunded annually through the federal portion of the Canadian Scientific Research and Experimental Development Tax Credit Program?
Tue Sep 07 15:29:37 +0000 2021The PRIDE FTP site is sort-of up: can list directories (😀) but can't download files (😪).
Tue Sep 07 15:01:19 +0000 2021@PatSchloss @pwilmarth There should be at least one in every Faculty of Medicine, as well as a Stats Club.
Tue Sep 07 14:57:06 +0000 2021@slashdot The number born/minute seems to have increased dramatically from the 19th century value of "1".
Tue Sep 07 14:54:08 +0000 2021@michaelhoffman @epdevilla Common law pronouncements often have unpredictable outcomes.
Tue Sep 07 14:40:01 +0000 2021Nothing loses my attention faster than coming across the word "histone" in a title.
Tue Sep 07 12:58:34 +0000 2021@MHendr1cks @mcgillu For those of us who are interested, but who are outside of la belle province, are the policies wrt COVID-19 at Université de Montréal or Université Laval different than McGill's?
Tue Sep 07 12:17:26 +0000 2021SDCBP:p, the N-terminal domain (2-107) has a significantly different pattern of modification than the two PDZ domains (108-206) & (207-270). Has too many GO annotations.
Tue Sep 07 12:07:36 +0000 2021SDCBP:p.T263A chr 8:g.58580553A>G, rs3739325 (all tissue T:A 0.996:0.004) vaf=0.5%, Δm=-30.0106, VAF by population group: african <1%, american <1%, east asian 2%, european <1%, south asian <1%.
Tue Sep 07 12:07:35 +0000 2021SDCBP:p, θ(max) = 91. aka SYCL, MDA-9. Abundant in MHC class 1 & 2 peptide experiments. Common in urine extracellular vesicles, but observable in many tissues and cell lines.
Tue Sep 07 12:07:35 +0000 2021>SDCBP:p, syndecan binding protein (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 2×K+acetyl; 7×K+GGyl; 19×S, 1×T, 5×Y+phosphoryl; SAAVs: T263A (0.5%); mature form: (2-298) [31,511×, 336 kTa]. #ᗕᕱᗒ 🔗
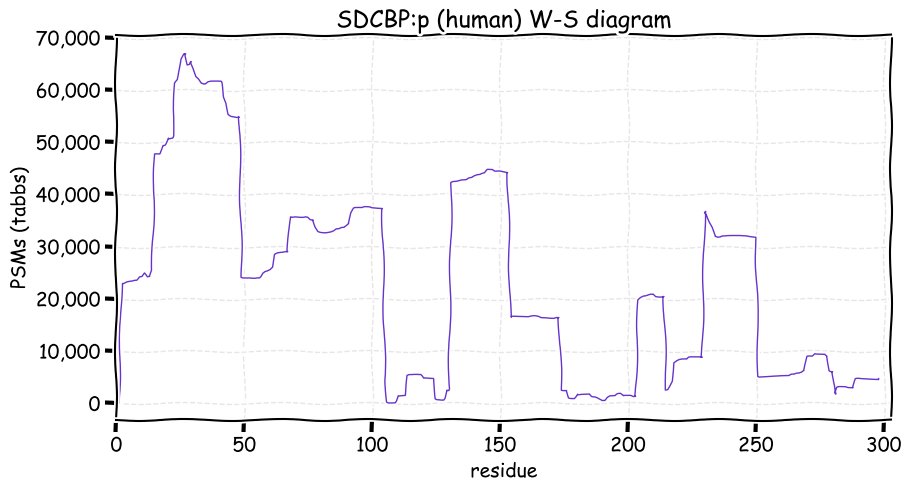
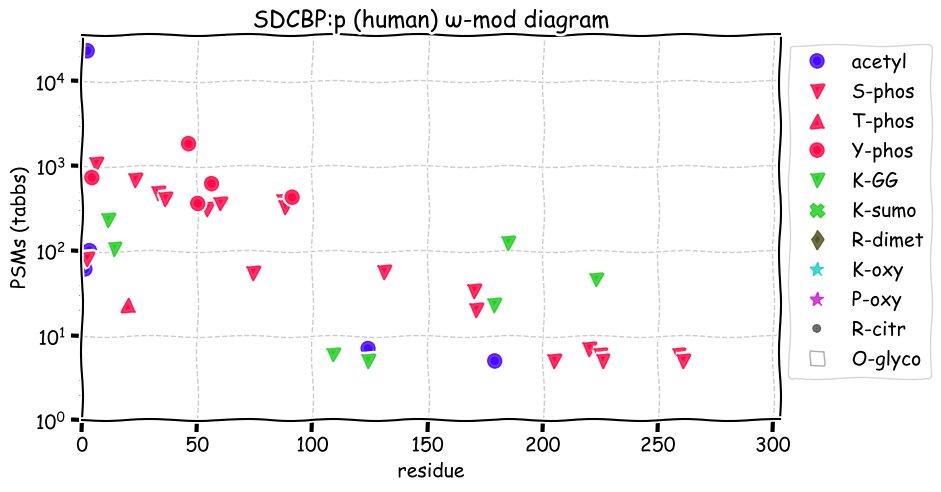
Mon Sep 06 19:13:36 +0000 2021@nesvilab @Smith_Chem_Wisc It would appear that my Cyrillic reading skills are worse than I had imagined. My Ukrainian grandfather would not be proud.
Mon Sep 06 18:36:47 +0000 2021@jwoodgett @BrownoftheGlobe @AlexUsherHESA What Canada does is spend a lot of money on Flavour-of-the-Month political consultants' pitches on R&D, none of which pan out but which end up as perennial budget line items anyway (e.g., Genome Canada, Compute Canada, Superclusters, etc.)
Mon Sep 06 12:38:21 +0000 2021NAV2:p's AlphaFold2 structure is characteristic of the algo when it doesn't have any existing structure to work from: a few basic 2° structure features wrapped up in a ball of string 🔗
Mon Sep 06 12:11:38 +0000 2021NAV2:p is observable in many types of cancer & it is not limited to CNS tissue as its name would suggest 🔗
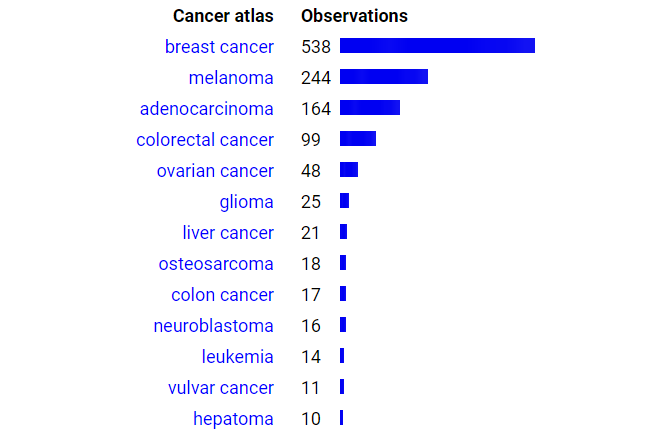
Mon Sep 06 11:57:51 +0000 2021NAV2:p.P990A chr 11:g.20044233C>G, rs3802800 (all tissue P:A 0.091:0.909) vaf=95%, Δm=-26.0156, VAF by population group: african 67%, american 89%, east asian 95%, european 99%, south asian 99%. Generates a novel tryptic cleavage site.
Mon Sep 06 11:57:51 +0000 2021NAV2:p, θ(max) = 28. aka FLJ10633, FLJ11030, HELAD1, KIAA1419, POMFIL2, RAINB1, FLJ23707. Common in MHC class 1 peptide experiments. Most abundant in tissues & a few cell lines: not found in fluids.
Mon Sep 06 11:57:51 +0000 2021>NAV2:p, neuron navigator 2 (Homo sapiens) Large subunit; CTMs: none; PTMs: 36×K+SUMOyl; 113×S, 19×T, 5×Y+phosphoryl; SAAVs: P990A (95%); mature form: (2-2365) [5,702×, 796 kTa]. #ᗕᕱᗒ 🔗
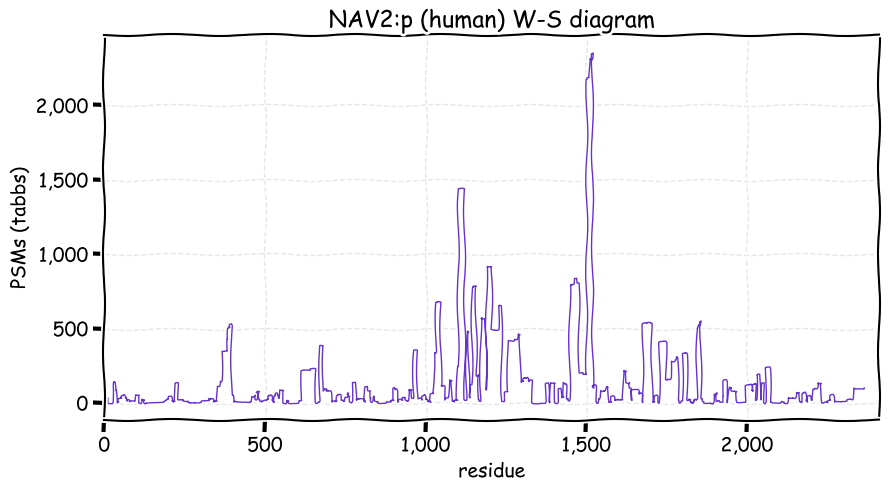
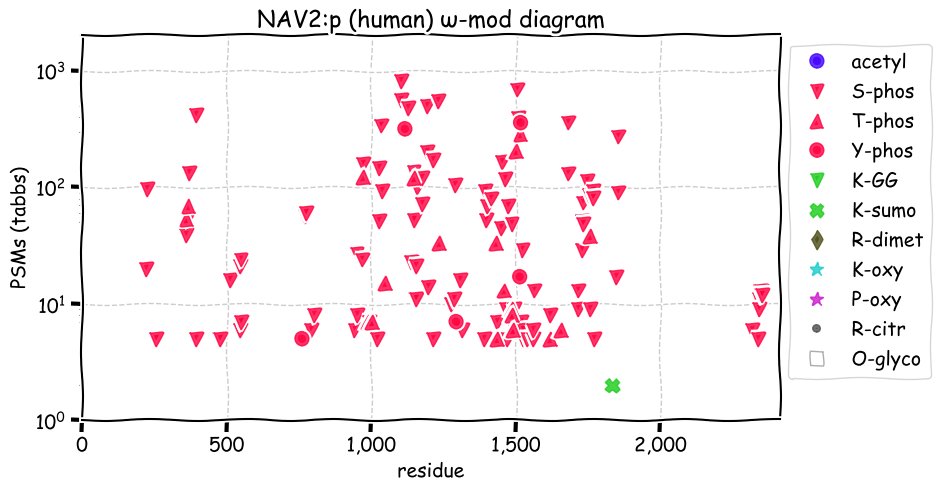
Sun Sep 05 15:50:41 +0000 2021@Smith_Chem_Wisc @nesvilab No Landau & Lifshitz books?
Sun Sep 05 14:21:40 +0000 2021RNAseq-based experiments that determine changes in RNA concentration have the same problem when it comes to analyzing the resulting information.
Sun Sep 05 13:56:09 +0000 2021AlphaFold2 represents these domains quite nicely, as they have been solved experimentally: 🔗 Pay no attention to the ball-of-string overall structure: the algo does that whenever it doesn't know what to do.
Sun Sep 05 13:29:47 +0000 2021The propagation of crappy annotation throughout publicly-available information retrieval systems is a significant problem in proteomics.
Sun Sep 05 12:56:46 +0000 2021HDLBP:p has 14 KH-type RNA-binding domains (in green) and its PTM pattern is consistent with a nuclear RNA-binding protein 🔗

Sun Sep 05 12:53:56 +0000 2021HDLBP:p.N418S chr 2:g.241253433T>C, rs7578199 (all tissue N:S 0.934:0.066) vaf=19%, Δm=-27.0109, VAF by population group: african 18%, american 19%, east asian 0%, european 27%, south asian 5%. NS in BT-474 & MDA-MB-453 cells.
Sun Sep 05 12:39:24 +0000 2021HDLBP:p, θ(max) = 75. aka HBP, VGL. Very common in MHC class 1 peptide experiments; class 2 peptides detected in specific tissues. Most abundant in tissue & cell lines: rare in fluids, but may be detectable in saliva.
Sun Sep 05 12:39:23 +0000 2021>HDLBP:p, high density lipoprotein binding protein (Homo sapiens) Large subunit; CTMs: S2+acetyl; PTMs: 20×K+acetyl; 36×K+GGyl; 51×S, 32×T, 10×Y+phosphoryl; SAAVs: N418S (19%), D1092E (12%); mature form: (2-1268) [54,559×, 796 kTa].. #ᗕᕱᗒ 🔗
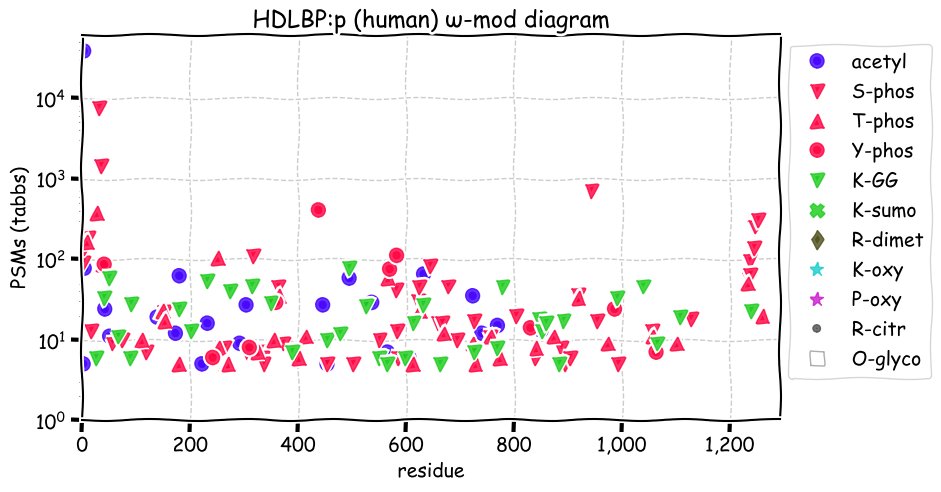
Sun Sep 05 00:25:01 +0000 2021@JoePlatelet @BiswapriyaMisra Laser microdissection setup?
Sat Sep 04 21:49:20 +0000 2021@leprevostfv "if what they claim" is where I get stuck. This is a pretty big data set (153 LC runs), analyzed using MaxQ with an FDR of 0.05 (!). They only included bacterial sequences that would support the hypothesis, so finding a few HLA peptides isn't very strong evidence.
Sat Sep 04 18:50:59 +0000 2021Based on the data, I'm skeptical.
Sat Sep 04 18:21:21 +0000 2021The data is at 🔗
Sat Sep 04 18:20:52 +0000 2021Opinions? 🔗
Sat Sep 04 16:47:52 +0000 2021Very often, when a paper states that a new method is generating "high quality" results, it is really trying to redefine what "high quality" means.
Sat Sep 04 12:19:55 +0000 2021SGPL1:p, the C-terminal phosphodomain looks like it may regulate function, but it's purpose has not been investigated (at least I couldn't find anything about it) 🔗

Sat Sep 04 12:03:51 +0000 2021SGPL1:p.V21L chr 10:g.70844506G>T, rs12770335 (all tissue V:L 0.911:0.089) vaf=9%, Δm=14.0156, VAF by population group: african 2%, american 7%, east asian 0%, european 14%, south asian 2%.
Sat Sep 04 12:03:51 +0000 2021SGPL1:p, θ(max) = 70. aka SPL. Common in MHC class 1 peptide experiments; class 2 peptides may be present in colon cancer tissue. Most abundant in tissue & cell lines: rare in fluids.
Sat Sep 04 12:03:51 +0000 2021>SGPL1:p, sphingosine-1-phosphate lyase 1 (Homo sapiens) Midsized subunit; CTMs: none; 21×K+acetyl; 10×K+GGyl; 10×K+SUMOyl; 18×S, 3×T, 0×Y+phosphoryl; SAAVs: V21L (9%); mature form: (2-568) [19,786×, 95 kTa]. #ᗕᕱᗒ 🔗
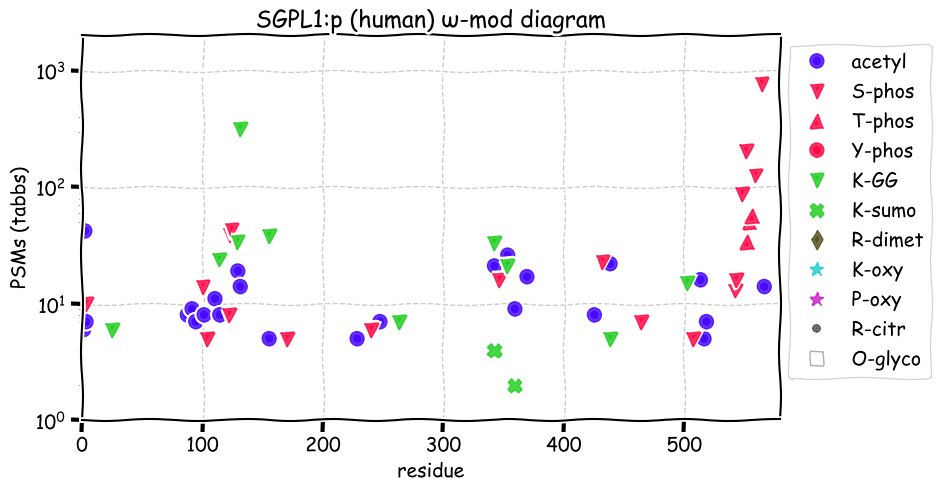
Fri Sep 03 15:47:29 +0000 2021Programming is a very perishable skill. Use it of lose it.
Fri Sep 03 12:43:26 +0000 2021Cuba is starting to come down from its recent high level, but Israel has a problem at the moment 🔗
Fri Sep 03 12:09:47 +0000 2021HRG:p, the C-terminal half of the sequence has repetitive patterns of of H, P & G 🔗

Fri Sep 03 11:56:35 +0000 2021HRG:p.R448C chr 3:g.186677647C>T, rs1042445 (all tissue R:C 0.407:0.593) vaf=27%, Δm=-53.0919, VAF by population group: african 24%, american 28%, east asian 28%, european 25%, south asian 33%.
Fri Sep 03 11:56:35 +0000 2021HRG:p, θ(max) = 71. aka HRGP, HPRG. Common in tissue-based MHC class 1 & 2 peptide experiments. Most abundant in blood plasma, urine & CSF. Has too many GO annotations.
Fri Sep 03 11:56:34 +0000 2021HRG:p, SAAVs: I180T (14%), P204S (51%), H340R (20%), R448C (27%), N493I (65%). It is unlikely that any individual will be homozygous for the reference sequence.
Fri Sep 03 11:56:34 +0000 2021>HRG:p, histidine rich glycoprotein (Homo sapiens) Midsized subunit; CTMs: N63, N125, N344, N345+glycosyl; PTMs: 6×K+acetyl; 13×K+GGyl; 7×S, 4×T, 0×Y+phosphoryl; 6×S, 4×T+glycosyl;mature form: (19-525) [29,223×, 472 kTa]. #ᗕᕱᗒ 🔗
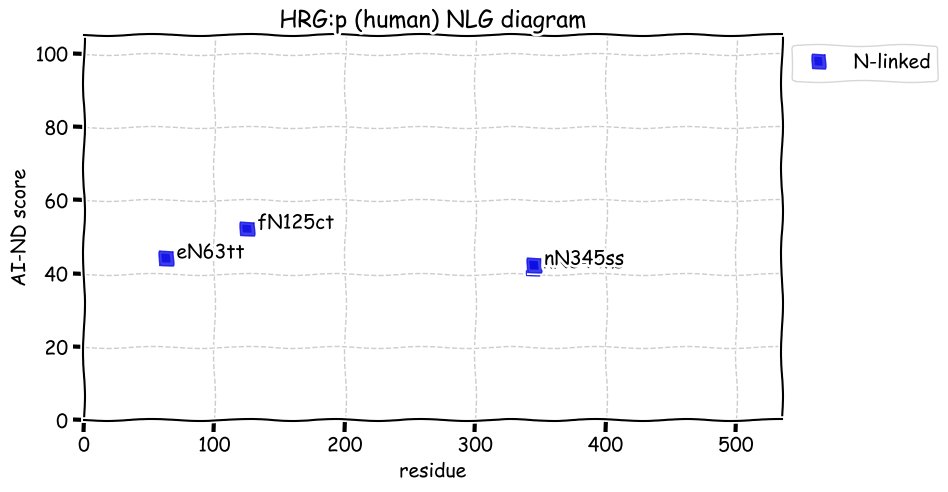
Thu Sep 02 11:57:06 +0000 2021CHORDC1:p does have a repeated C/H pattern, it isn't really "rich" in these 2 residues 🔗

Thu Sep 02 11:51:16 +0000 2021CHORDC1:p.A329D chr 11:g.90202418G>T, rs1045861 (all tissue A:D 0.458:0.542) vaf=70%, Δm=43.9898, VAF by population group: african 37%, american 65%, east asian 68%, european 70%, south asian 75%.
Thu Sep 02 11:51:16 +0000 2021CHORDC1:p, θ(max) = 83. aka CHP1, CHP-1. Common in MHC class 1 peptide experiments. Observed in tissues & cell lines: very rare in fluids.
Thu Sep 02 11:51:16 +0000 2021>CHORDC1:p, cysteine and histidine rich domain containing 1 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 9×K+acetyl; 13×K+GGyl; 12×S, 10×T, 2×Y+phosphoryl; SAAVs: A329D (70%); mature form: (2-332) [28,622×, 198 kTa]. #ᗕᕱᗒ 🔗
Wed Sep 01 14:54:18 +0000 2021@AlexUsherHESA @rasselin66 & superclusters were a tough act to beat ...
Wed Sep 01 14:19:17 +0000 2021@pwilmarth @byu_sam & Andromeda wasn't really a step forward, even at the time.
Wed Sep 01 13:22:27 +0000 2021Yup: great timing for a federal election 🇨🇦 🔗
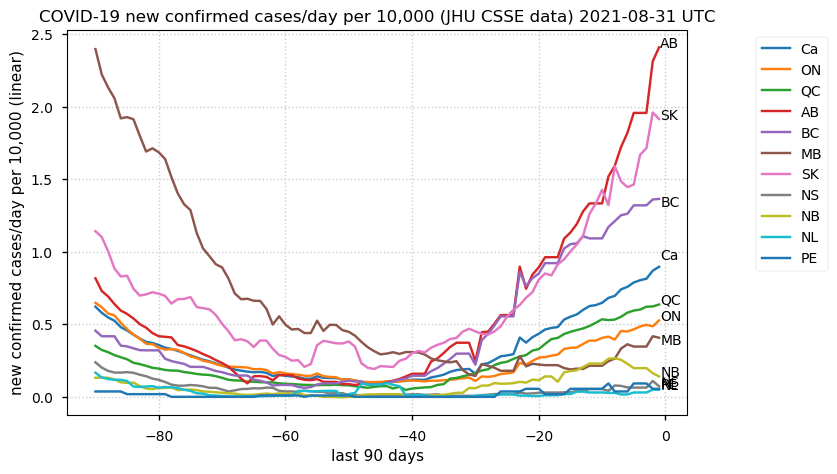
Wed Sep 01 12:19:18 +0000 2021HRC:p, is definitely H-rich, with basic domains like (288-300):
HHHRDPSHRHRSH
But for every H-rich domain, it also has E-rich acidic domains, e.g. (200-214):
EGEEEEEEEEEEEE
Wed Sep 01 12:07:41 +0000 2021HRC:p.S43N chr 19:g.49155110C>T, rs3745298 (all tissue S:N 0.602:0.398) vaf=46%, Δm=27.0109, VAF by population group: african 60%, american 51%, east asian 31%, european 43%, south asian 48%.
Wed Sep 01 12:07:41 +0000 2021HRC:p, θ(max) = 72. aka MGC133236. Found in heart MHC class 2 peptide experiments. Most common in urine & muscle fibres.
Wed Sep 01 12:07:40 +0000 2021>HRC:p, histidine rich calcium binding protein (Homo sapiens) Midsized subunit; CTMs: Q28-ammonia; N41+glycosyl PTMs: 35×S, 7×T, 2×Y+phosphoryl; SAAVs: S43N (%46), S96A (35%), S299G (2%), N305E (5%); mature form: (28-699) [2,479×, 23 kTa]. #ᗕᕱᗒ 🔗
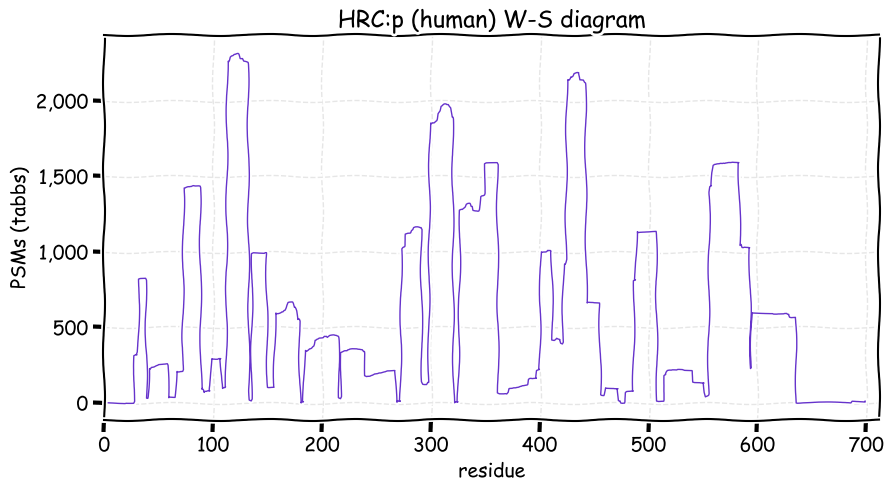
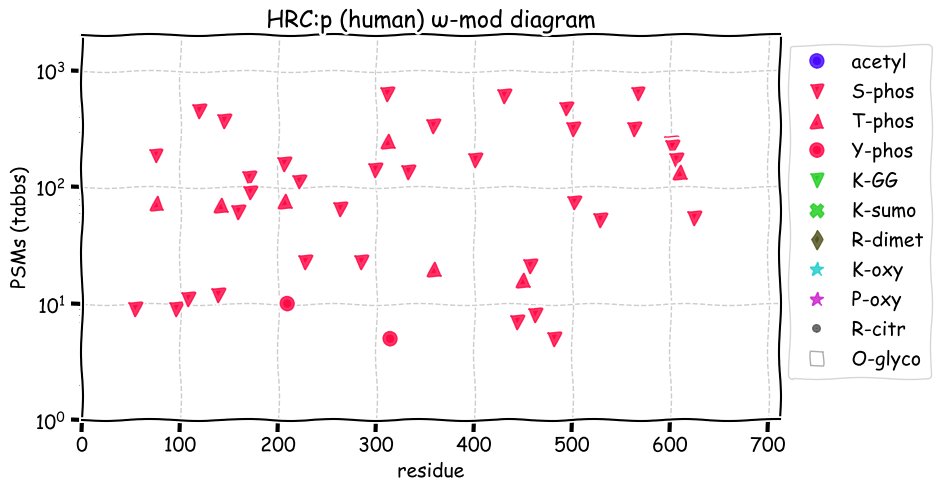
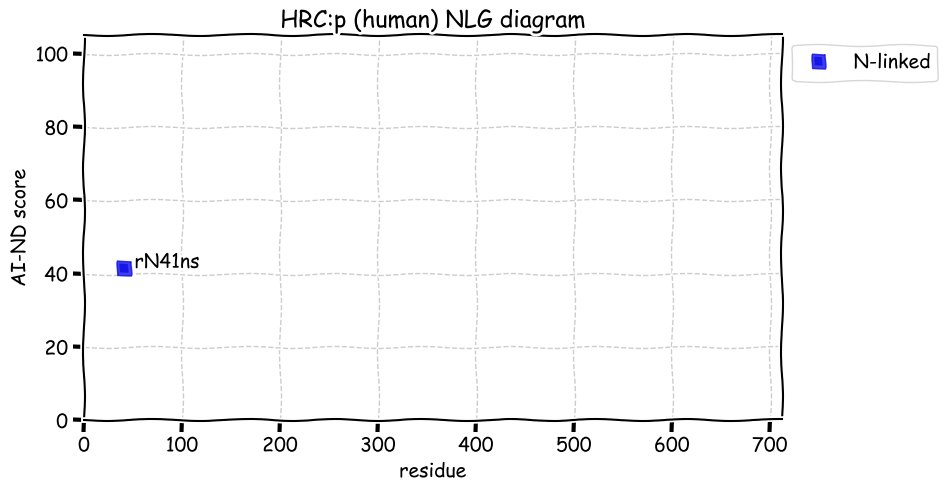
Tue Aug 31 22:04:27 +0000 2021@neely615 @bittremieux @ProteomeXchange @juan_vizcaino The largest project (in HPLC runs) I am aware of is currently outside of the PX system: BioPlex3 weighs in at 31,284 raw files.
Tue Aug 31 20:43:24 +0000 2021@neely615 @bittremieux @ProteomeXchange @juan_vizcaino What has increased is the # of MS/MS scans in a single file: modern instruments generate 10-20× as many scans from a chromatography run as those at the beginning of the PX era.
Tue Aug 31 16:04:22 +0000 2021🔗
Tue Aug 31 13:09:02 +0000 2021While UTIs are usually ignored in proteomics urine studies, they are easily detected & can be significant confounders
Tue Aug 31 12:49:15 +0000 2021@AlexUsherHESA It is the Canadian Way, though.
Tue Aug 31 12:38:51 +0000 2021If anyone is interested in biotyping pathogens in UTIs, PXD005219 has some nice examples.
Tue Aug 31 12:07:00 +0000 2021HAL:p.V439I chr 12:g.95980836C>T, rs7297245 (all tissue V:I 0.046:0.954) vaf=86%, Δm=14.0156, VAF by population group: african 99%, american 88%, east asian 95%, european 80%, south asian 90%.
Tue Aug 31 12:07:00 +0000 2021HAL:p, θ(max) = 73. aka MGA. Found in liver MHC class 1 peptide experiments. Most common in liver & keratinocytes.
Tue Aug 31 12:07:00 +0000 2021>HAL:p, histidine ammonia-lyase (Homo sapiens) Midsized subunit; CTMs: none; PTMs: 2×S, 1×T, 0×Y+phosphoryl; SAAVs: V439I (%86); mature form: (2-657) [3,565×, 22 kTa]. #ᗕᕱᗒ 🔗
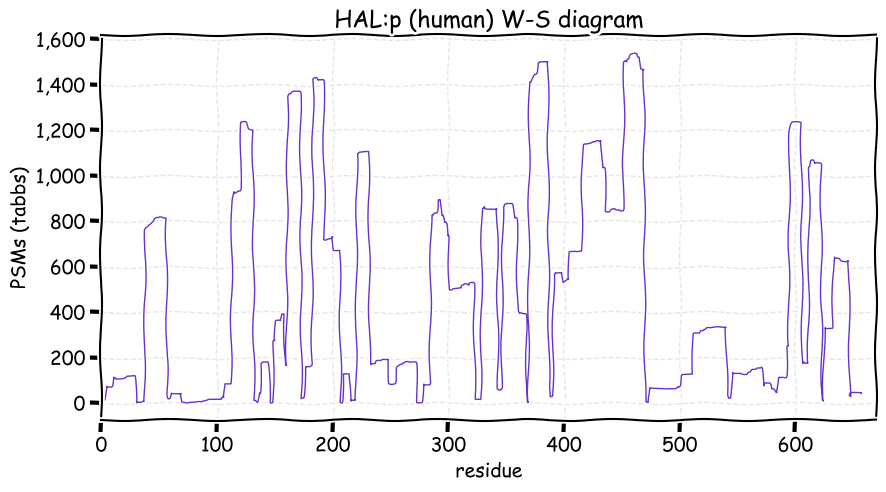
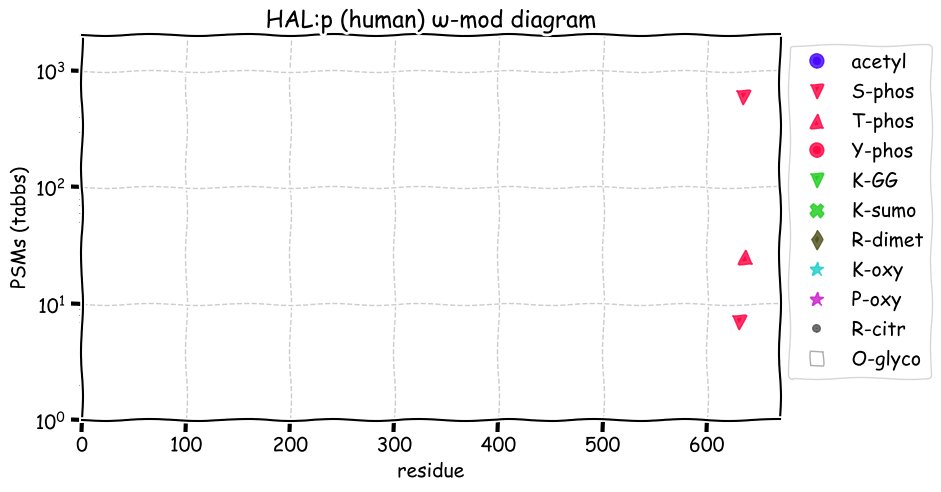
Mon Aug 30 14:20:30 +0000 2021@astacus It is also commonly found in neutrophil samples, which is hard to square with the amylase association.
Mon Aug 30 14:18:50 +0000 2021@astacus If you look at UniProt, the annotation for the full length sequence is 🔗, while a shorter splice gets a lot more enthusiasm 🔗 (proteomics supports the widespread existence of the E7ER45 form)
Mon Aug 30 13:50:23 +0000 2021@astacus It is marked up as a somewhat exotic starch digestive enzyme on the lumenal facing membrane of colon apical epithelium cells. If this is true, why is it abundant in urine & spermatozoa?
Mon Aug 30 12:36:36 +0000 2021MGAM:p, most of the annotation about this protein is odd.
Mon Aug 30 12:09:37 +0000 2021MGAM:p.P1424T chr 7:g.142063511C>T, rs185053832 (all tissue P:T 0.991:0.009) vaf=0.7%, Δm=3.9949, VAF by population group: african <1%, american <1%, east asian <1%, european 1%, south asian <1%.
Mon Aug 30 12:09:37 +0000 2021MGAM:p, domains: (2-13) cytoplasmic, (14-34) transmembrane, (35-2753) extracellular.
Mon Aug 30 12:09:37 +0000 2021MGAM:p, θ(max) = 56. aka MGA. Found rarely in tissue MHC class 1 & 2 peptide experiments in small intestine & kidney tissue. Most common in spermatozoa, urine, feces, colon, & stomach.
Mon Aug 30 12:09:36 +0000 2021>MGAM:p, maltase-glucoamylase (Homo sapiens) large subunit; CTMs: A2+acetyl; N1323, N1603+glycosyl; PTMs: none; SAAVs: Q404H (7%), P1424T (1%), M1572L (5%), Q1624H (3%), T2036M (%4); mature form: (2-2753) [5,936×, 195 kTa]. #ᗕᕱᗒ 🔗
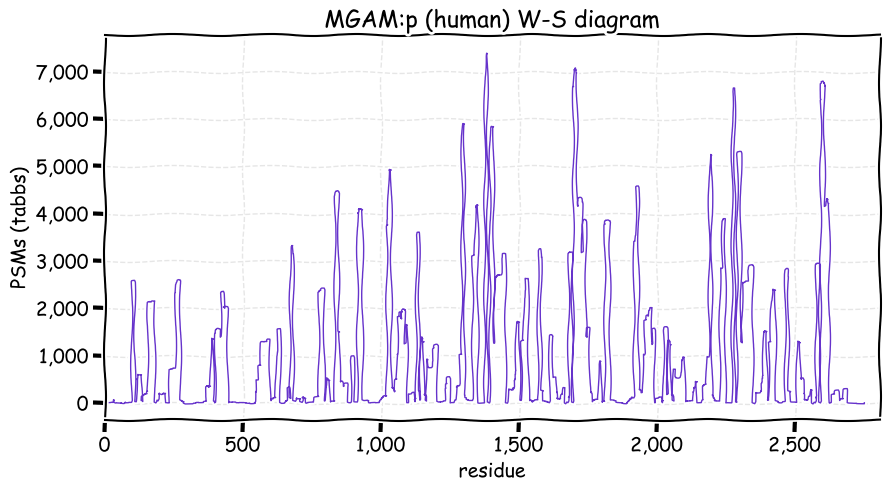
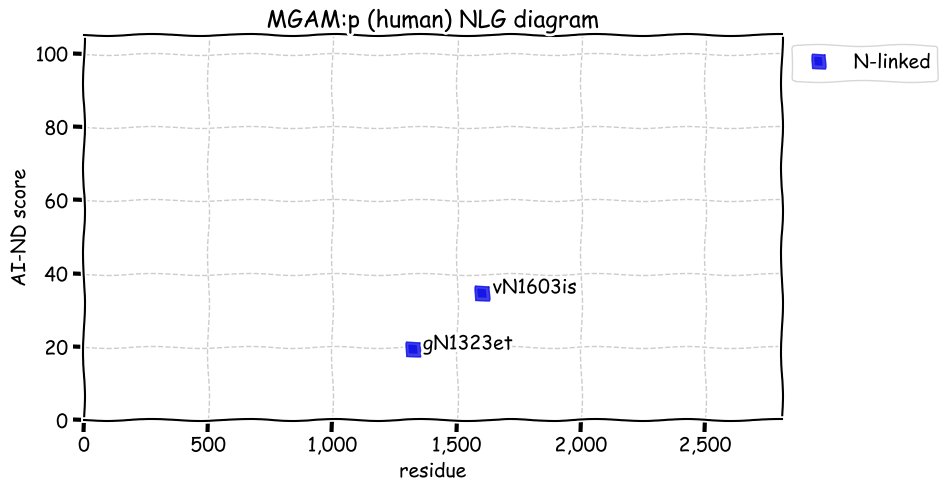
Sun Aug 29 16:55:29 +0000 2021@Peptidome @BiswapriyaMisra @CousinAmygdala I don't think diet is the issue. It is more of a "one-of-us" issue.
Sun Aug 29 16:47:45 +0000 2021This goes for any employer who throws shade at former employees 🔗
Sun Aug 29 15:19:59 +0000 2021@Peptidome @BiswapriyaMisra @CousinAmygdala I knew a department head who explicitly stated that a candidate's "lunch-ability" was an important consideration.
Sun Aug 29 13:27:59 +0000 2021Pressure & wind speed at the Mississippi Canyon (MDJ ASOS) oil production platform weather station in the Gulf of Mexico just south of New Orleans ~ 2 hours ago 🔗
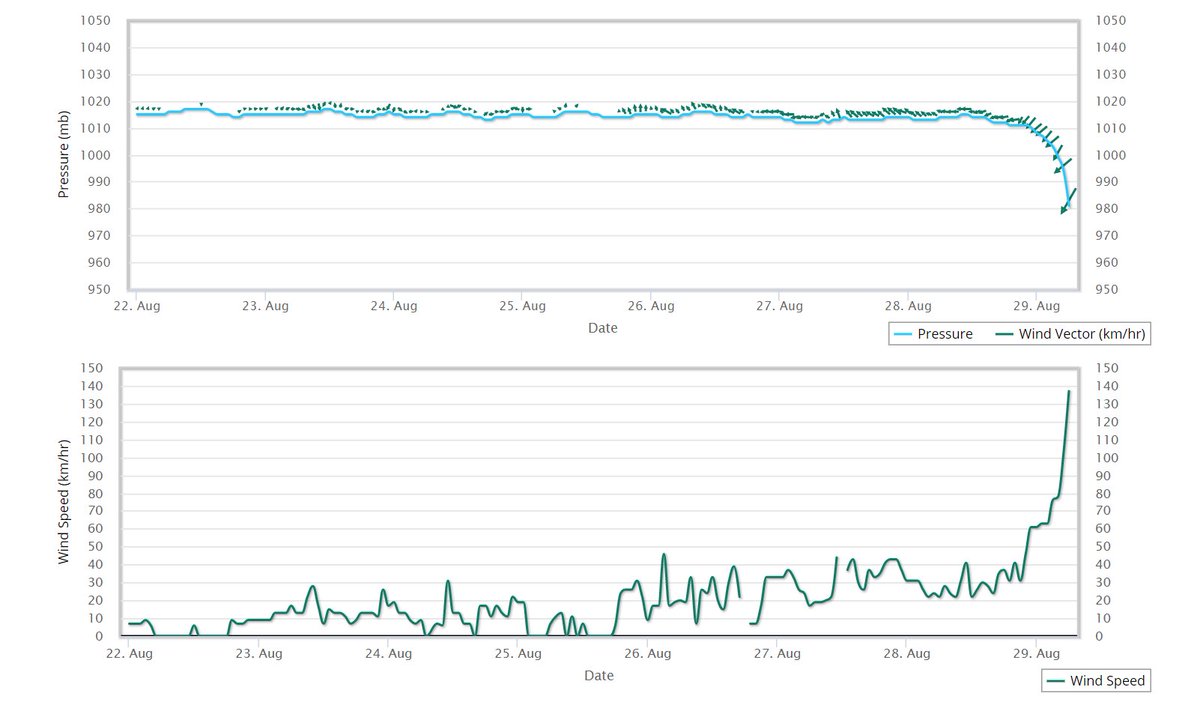
Sun Aug 29 12:18:19 +0000 2021BHMT:p, the exocytotic mechanisms that produce extracellular vesicles are the deus ex machina that allow ordinary cytoplasmic metabolic enzymes like this one to end up in blood & urine
Sun Aug 29 12:05:20 +0000 2021BHMT:p.R239Q chr 5:g.79126136G>A, rs3733890 (all tissue R:Q 0.268:0.732) vaf=30%, Δm=-28.0425, VAF by population group: african 20%, american 38%, east asian 32%, european 32%, south asian 29%. RR in HaCaT cells. Variant removes a tryptic cleavage site.
Sun Aug 29 12:05:20 +0000 2021BHMT:p, θ(max) = 84. aka BHMT1. Found rarely in tissue MHC class 1 & 2 peptide experiments. Most common in liver & kidney, as well as extracellular vesicles in urine & blood plasma.
Sun Aug 29 12:05:20 +0000 2021>BHMT:p, betaine-homocysteine S-methyltransferase (Homo sapiens) Small subunit; CTMs: none; PTMs: 2×S, 0×T, 0×Y+phosphoryl; SAAVs: R239Q (30%); mature form: (2-406) [6,994×, 115 kTa]. #ᗕᕱᗒ 🔗
Sat Aug 28 20:44:52 +0000 2021Nasty looking storm 🔗

Sat Aug 28 19:56:35 +0000 2021@slashdot By telling them to place it on Lucky Dan?
Sat Aug 28 14:01:37 +0000 2021Twitter threads can be quite effective for expressing the main argument of theoretical findings: much better than abstracts or editorial blurbs 🔗
Sat Aug 28 12:56:29 +0000 2021CASP7:p.D4E chr 10:g.113697505T>;G, rs11555408 (all tissue D:E 0.974:0.026) vaf=9%, Δm=14.0157, VAF by population group: african 15%, american 8%, east asian 9%, european 10%, south asian 9%. DE in MCF-10A cells.
Sat Aug 28 12:09:37 +0000 2021CASP7:p, θ(max) = 87. aka MCH3, CMH-1, ICE-LAP3. Found rarely in tissue MHC class 1 peptide experiments. Most common in tissue, cell lines & urine extracellular vesicles.
Sat Aug 28 12:09:37 +0000 2021>CASP7:p, caspase 7 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 4×K+acetyl; 9×K+GGyl; 7×S, 1×T, 0×Y+phosphoryl; SAAVs: D4E (9%), Q211R (27%); mature form: (2-303) [11,110×, 46 kTa]. #ᗕᕱᗒ 🔗
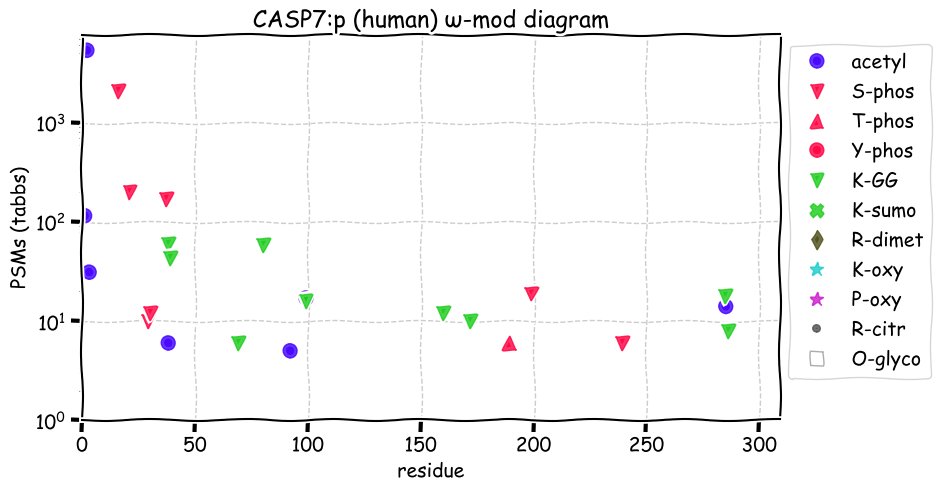
Sat Aug 28 01:09:50 +0000 2021Tonight's aurora will reach further south than usual 🔗
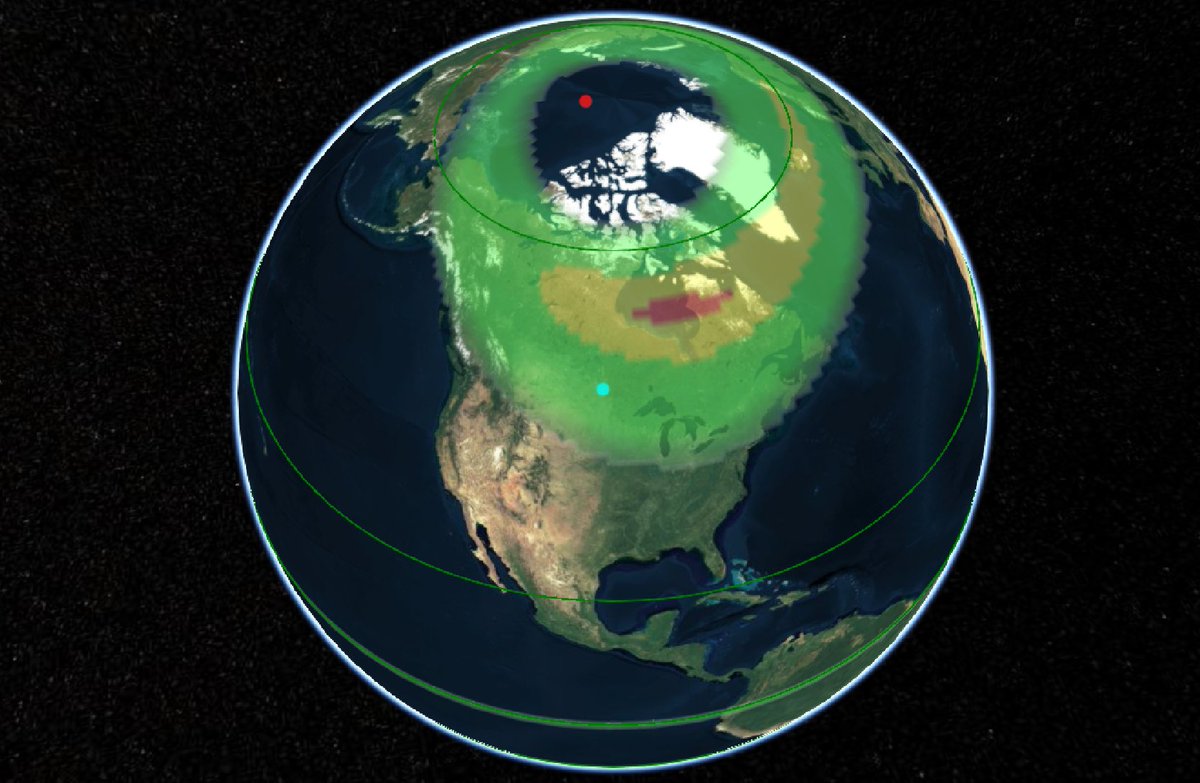
Fri Aug 27 14:49:12 +0000 2021@andy___jones More generally, DO NOT question the reliability of published results.
Fri Aug 27 14:34:49 +0000 2021@andy___jones But, of course, this is exactly the sort of thing that cannot be allowed into the literature (which has a lot of "Fight Club" aspects to it).
Fri Aug 27 14:15:16 +0000 2021@andy___jones Sometimes they clean up their act (usually after "discovering" open searching) so I still look at their data― at least in the initial screening stage―but once they are on the blacklist it takes a lot of proof to move them off.
Fri Aug 27 14:11:28 +0000 2021@andy___jones I found that the most effective QA method is to simply not include data from a small number of labs that generate most of the noise. There are a few labs that are really prolific noise generators.
Fri Aug 27 13:11:07 +0000 2021Alberta leads the way here in Canada 🔗
Fri Aug 27 12:51:13 +0000 2021@andy___jones The reviewers really didn't get what we were trying to do: it was a painful process. They kept on suggesting we run experiments ...
Fri Aug 27 12:33:36 +0000 2021@andy___jones Tried to published something like that a while ago. We had to de-emphasize most of the QC part and push mapping the sites to the genome to get it published 🔗
Fri Aug 27 12:27:13 +0000 2021CPNE1:p, most of the modifications are in the 2 N-terminal, membrane-targeting C2 domains (6-124) & (138-245). The C-terminal vWA_copine_like domain (251-509) has comparatively few.
Fri Aug 27 12:07:22 +0000 2021CPNE1:p.Q211R chr 20:g.35631574T>C, rs1987546 (all tissue Q:R 0.154:0.846) vaf=27%, Δm=28.0425, VAF by population group: african 42%, american 18%, east asian 14%, european 19%, south asian 35%.
Fri Aug 27 12:07:22 +0000 2021CPNE1:p, θ(max) = 59. aka CPN1. Found in MHC class 1 & 2 peptide experiments. Most common in tissue, cell lines & extracellular vesicles in urine & blood plasma.
Fri Aug 27 12:07:21 +0000 2021>CPNE1:p, copine 1 (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 18×K+acetyl; 16×K+GGyl; 2×K+SUMOyl; 16×S, 1×T, 1×Y+phosphoryl; SAAVs: V31I (8%), Q211R (27%); mature form: (2-537) [38,182×, 241 kTa]. #ᗕᕱᗒ 🔗
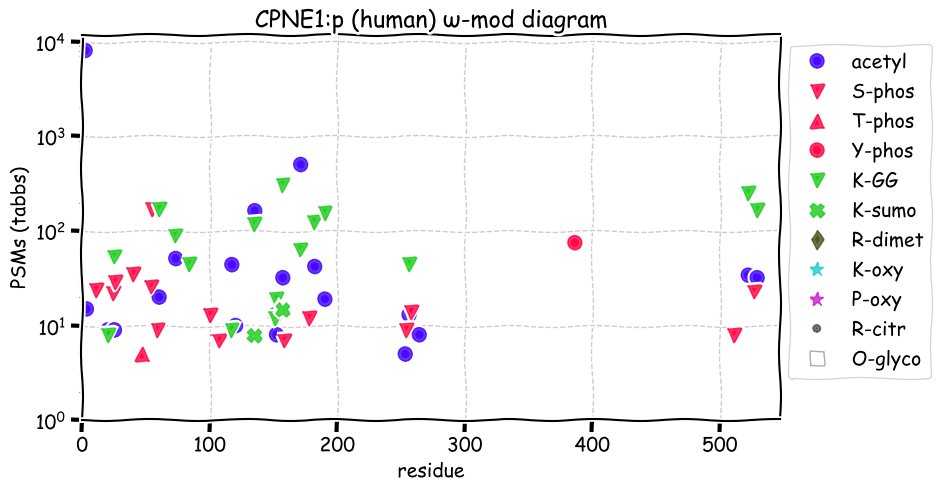
Thu Aug 26 16:50:05 +0000 2021@pwilmarth @byu_sam You aren't pulling your punches in this one.
Thu Aug 26 15:28:06 +0000 2021I often tell people that since all of the PSM assignment AI algos use the same physical chemistry, most of the difference between them is in the details. But your result show a big & consistent diff for only one of them. Is it true for any algorithm based on Mascot?
Thu Aug 26 15:23:51 +0000 2021@byu_sam, now that you folks have had some time to think about it, have you been able to rationalize the performance of Andromeda (MaxQuant) in your study 🔗?
Thu Aug 26 14:26:40 +0000 2021@dexivoje Specifically utopian science fiction.
Thu Aug 26 12:37:26 +0000 2021I always like to look at the data from extracellular vesicle expts: it is always surprising.
Thu Aug 26 11:55:11 +0000 2021HGFAC:p.F231L chr 4:g.3444405C>G, rs1987546 (all tissue F:L 0.373:0.627) vaf=71%, Δm=-33.9843, VAF by population group: african 66%, american 73%, east asian 92%, european 72%, south asian 66%.
Thu Aug 26 11:55:11 +0000 2021HGFAC:p, the mature protein is produced by scission of bonds between R372-V373 & R407-I408. The inactive proprotein (35-655) is the form most commonly observed.
Thu Aug 26 11:55:10 +0000 2021HGFAC:p, θ(max) = 59. aka none. Rare in MHC class 1 & 2 peptide experiments. Most common in blood plasma, CSF & urine in extracellular vesicles. Rare in cell lines.
Thu Aug 26 11:55:10 +0000 2021>HGFAC:p, HGF activator (Homo sapiens) Midsized subunit; CTMs: N290, N468+glycosyl; PTMs: 5×S, 1×T+glycosyl; SAAVs: F231L (71%), E245D (2%); mature form: (373-407, 408-655) [9,274×, 191 kTa]. #ᗕᕱᗒ 🔗
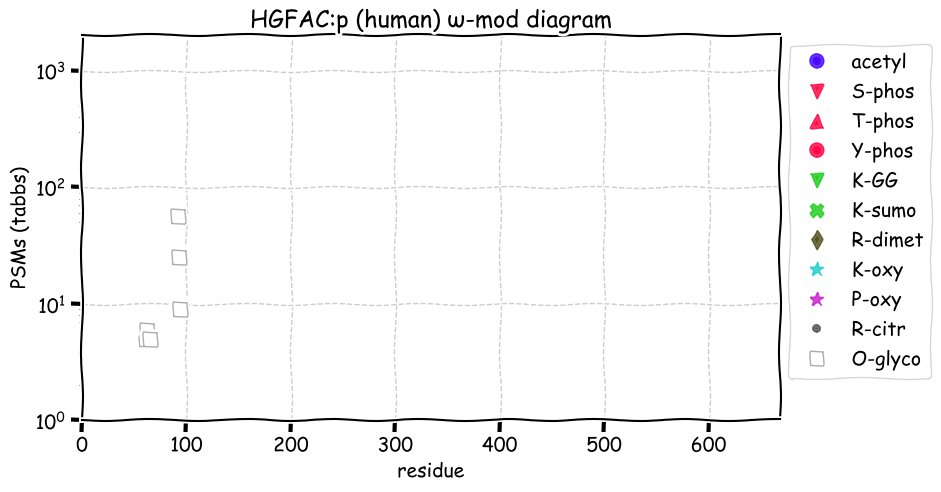
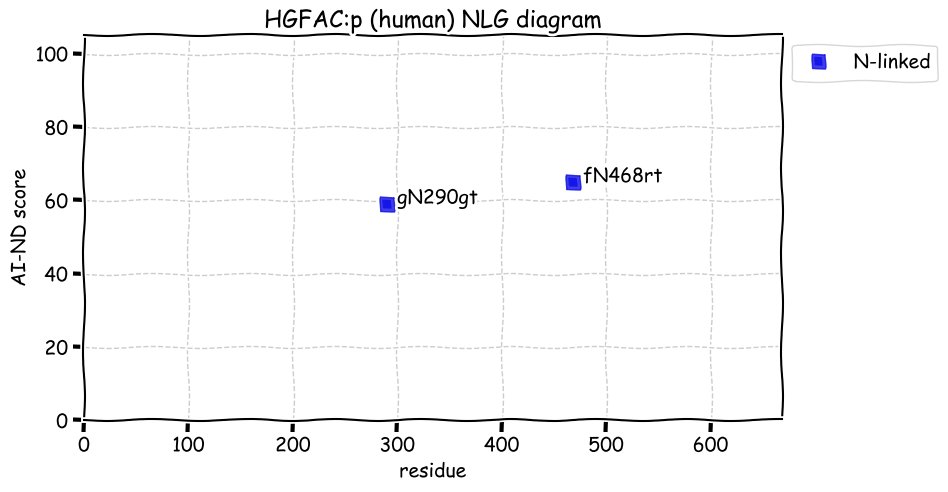
Wed Aug 25 17:24:02 +0000 2021@nesvilab @byu_sam @theNCI @NIHFunding In my experience, it is pretty rare that anyone on a panel will actually read code as part of a review.
Wed Aug 25 16:45:07 +0000 2021@MiguelCos I have a box connected direct-to-the-line specifically to test for for internal connectivity issues and got the same result. For me, at least, the problems have been on the EBI side.
Wed Aug 25 16:17:21 +0000 2021@lgatt0 I don't think anyone knows for sure if it is a sustainable (or cost effective) approach in the long term. It is all a bit of an experiment.
Wed Aug 25 16:11:38 +0000 2021@lgatt0 It is difficult to sort out problems with big cloud infrastructure: it is hard enough to do when you only have a few servers involved!
Wed Aug 25 15:42:14 +0000 2021The PRIDE FTP site has been a little unpredictable for the last week (or so). Is there some maintenance work going on?
Wed Aug 25 11:59:01 +0000 2021HIBCH:p.T46A chr 2:g.190296896T>C, rs1058180 (all tissue T:A 0.347:0.653) vaf=74%, Δm=-30.0106, VAF by population group: african 63%, american 75%, east asian 57%, european 76%, south asian 75%. AA in MCF-10A & HEK-293 cells. AT in MCF-7 cells.
Wed Aug 25 11:59:01 +0000 2021HIBCH:p, θ(max) = 75. aka none. Rare in MHC class 1 & 2 peptide experiments. Common in tissues & cell lines: episodically abundant in urine. Mitochondrial enzyme involved in the degradation of V, L & I.
Wed Aug 25 11:59:01 +0000 2021>HIBCH:p, 3-hydroxyisobutyryl-CoA hydrolase (Homo sapiens) Small subunit; CTMs: none; PTMs: 14×K+acetyl; 5×K+GGyl; 3×S, 0×T, 0×Y+phosphoryl; SAAVs: T46A (74%), E245D (2%); mature form: (30-386) [25,736×, 191 kTa]. #ᗕᕱᗒ 🔗
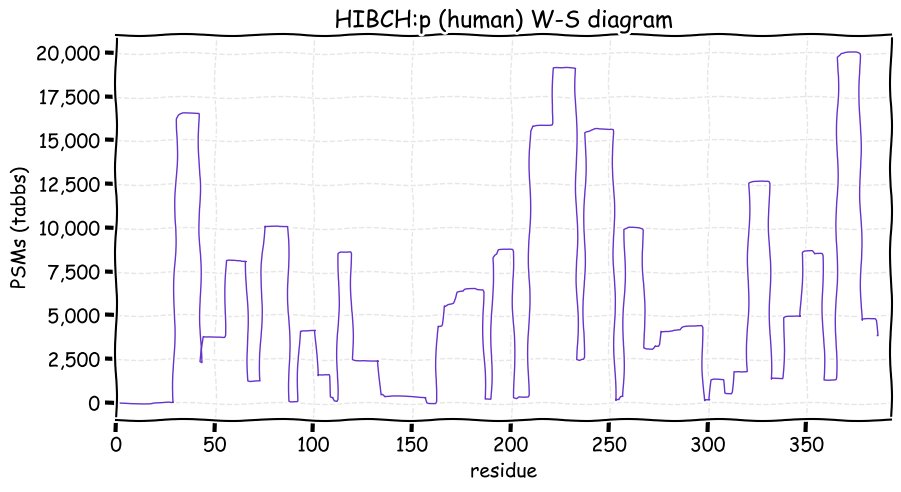
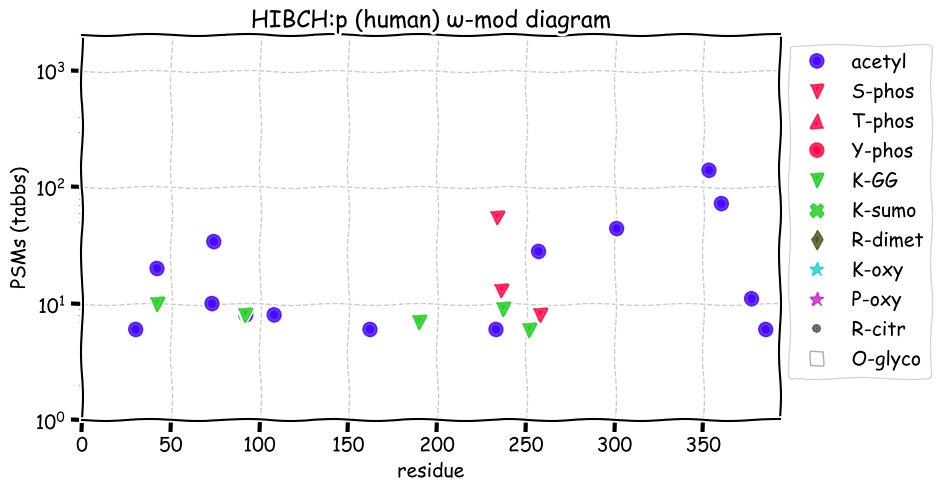
Tue Aug 24 19:34:52 +0000 2021Hint: do not run an overloaded tryptic digest immediately before you run an HLA peptide prep.
Tue Aug 24 19:33:38 +0000 2021I must admit people do find creative ways to screw up their experiments.
Tue Aug 24 12:01:02 +0000 2021PBK:p, the C-terminal phosphodomain (251-289) is part of a larger, D/E low complexity domain (250-322)
Tue Aug 24 11:53:36 +0000 2021PBK:p.E220D chr 8:g.27811070C>G, rs17057901 (all tissue E:D 0.943:0.057) vaf=0.5%, Δm=-14.0157, VAF by population group: african 7%, american 1%, east asian <1%, european <1%, south asian <1%. ED in HeLa & Hep-G2 cells.
Tue Aug 24 11:53:36 +0000 2021PBK:p, θ(max) = 98. aka TOPK, FLJ14385, Nori-3, SPK, CT84. Present in MHC class 1 peptide experiments. Most abundant in tissue & cell lines: rare in fluids.
Tue Aug 24 11:53:36 +0000 2021>PBK:p, PDZ binding kinase (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 7×K+acetyl; 8×K+GGyl; 8×K+SUMOyl; 16×S, 6×T, 3×Y+phosphoryl; SAAVs: N107S (22%), E220D (1%); mature form: (1-322) [15,085×, 1990 kTa]. #ᗕᕱᗒ 🔗
Tue Aug 24 00:53:45 +0000 2021It you want a few HLA class II runs to use as examples, you could do a lot worse than PXD023032.
Mon Aug 23 18:44:58 +0000 2021@AlexUsherHESA People always forget about the America/Atikokan time zone 🔗
Mon Aug 23 18:38:41 +0000 2021@AlexUsherHESA Kenora & Thunderbay-Rainy River?
Mon Aug 23 15:52:55 +0000 2021@astacus @MattWFoster But, probably apropos of nothing, the other major copper binding protein in plasma, A2M, also has an odd ball number of pY's
🔗
Mon Aug 23 15:51:05 +0000 2021@astacus @MattWFoster Occupancy is tricky, given the fact that most data is either:
1. not great at recovering phosphopeptides; or
2. phosphopeptide affinity pull downs that don't see anything else.
Mon Aug 23 15:12:11 +0000 2021These measurements are from a variety of ground stations, including those tracked via ASOS (airport weather stations), NOAA MADIS (automated private weather stations) & CoCoRaHS (manually read rain gauges).
Mon Aug 23 14:58:54 +0000 2021Lot of rain yesterday across New Jersey (in mm) 🔗
Mon Aug 23 14:52:01 +0000 2021@astacus It is a better suggestion than anything I've got. But the pY guys are usually pretty insistent that it is reserved for the most elegant of signalling pathways, rather than something as plebeian as metal ion coordination.
Mon Aug 23 12:19:43 +0000 2021CP:p, I am completely at a loss to explain (or understand) why there are so many Y+phosphoryl sites on this proteins.
Mon Aug 23 12:07:44 +0000 2021CP:p.E544D chr 3:g.149198448T>A, rs701753 (all tissue E:D 0.698:0.302) vaf=92%, Δm=-14.0157, VAF by population group: african 64%, american 96%, east asian 100%, european 94%, south asian 87%.
Mon Aug 23 12:07:44 +0000 2021CP:p, θ(max) = 92. aka none. Present in MHC class 1 & 2 peptide experiments. Most abundant in blood plasma, milk, CSF & urine. Because of its high copper content, purified CP:p is blue coloured.
Mon Aug 23 12:07:44 +0000 2021>CP:p, ceruloplasmin (Homo sapiens) Large subunit; CTMs: N138, N358, N397, N762+glycosyl; PTMs: 26×K+acetyl; 2×K+GGyl; 52×S, 8×T, 46×Y+phosphoryl; SAAVs: R367C (2%), E544D (92%), T551I (2%); mature form: (20-1065) [41,355×, 1990 kTa]. #ᗕᕱᗒ 🔗
Sun Aug 22 17:45:21 +0000 2021@byu_sam @neely615 @biorxivpreprint @pwilmarth @ryankellybyu I also like the abrupt onset of peptide containing spectra at ~5000 scans. There really is almost no carry over from run to run.
Sun Aug 22 12:01:52 +0000 2021The main circulation of Henri making landfall on Rhode Island 🔗
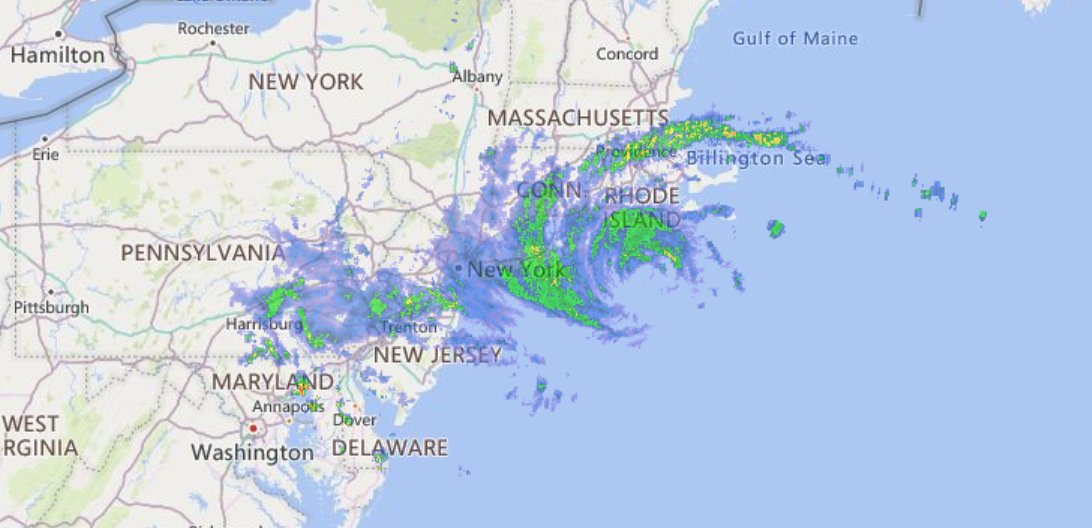
Sun Aug 22 11:53:26 +0000 2021APOA1:p.A61T chr 11:g.116837020C>T, rs12718465 (all tissue A:T 0.994:0.006) vaf=0.3%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian 4%, european <1%, south asian <1%.
Sun Aug 22 11:53:25 +0000 2021APOA1:p, θ(max) = 93. aka none. Present in MHC class 1 & 2 peptide experiments. Most abundant in blood plasma, milk, CSF & urine. Component of HDL particles.
Sun Aug 22 11:53:25 +0000 2021>APOA1:p, apolipoprotein A1 (Homo sapiens) Small subunit; CTMs: none; PTMs: 21×K+acetyl; 11×K+GGyl; 9×S, 3×T, 1×Y+phosphoryl; T221, S248+glycosyl; SAAVs: A61T (0.3%); mature form: (19-267) [50,634×, 2391 kTa]. #ᗕᕱᗒ 🔗
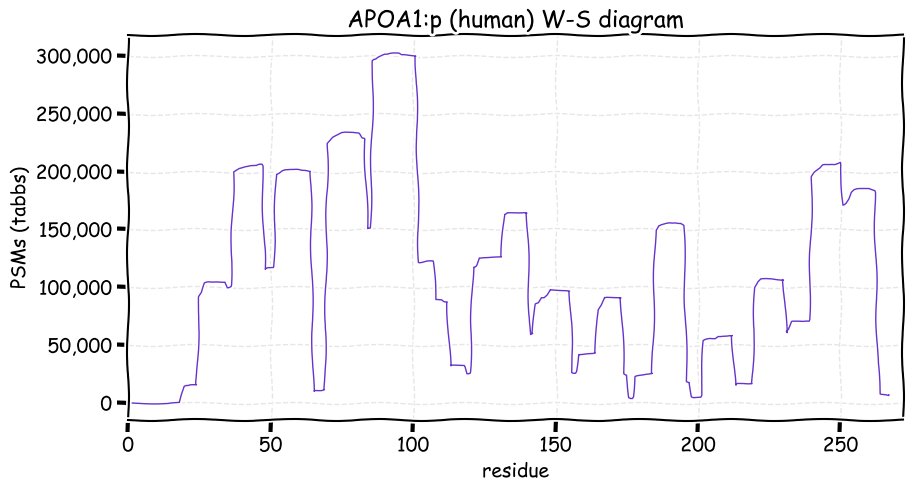
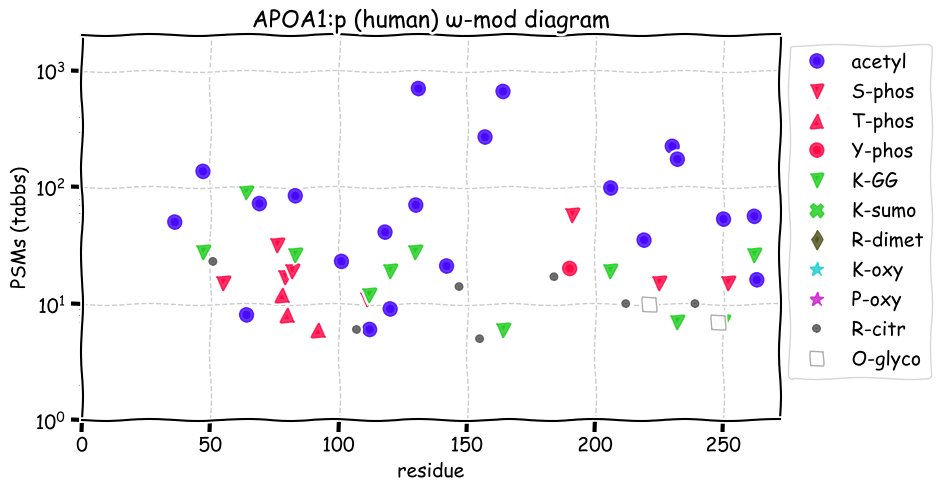
Sat Aug 21 23:54:05 +0000 2021A band of heavy rain over New York & Nassau County 🔗
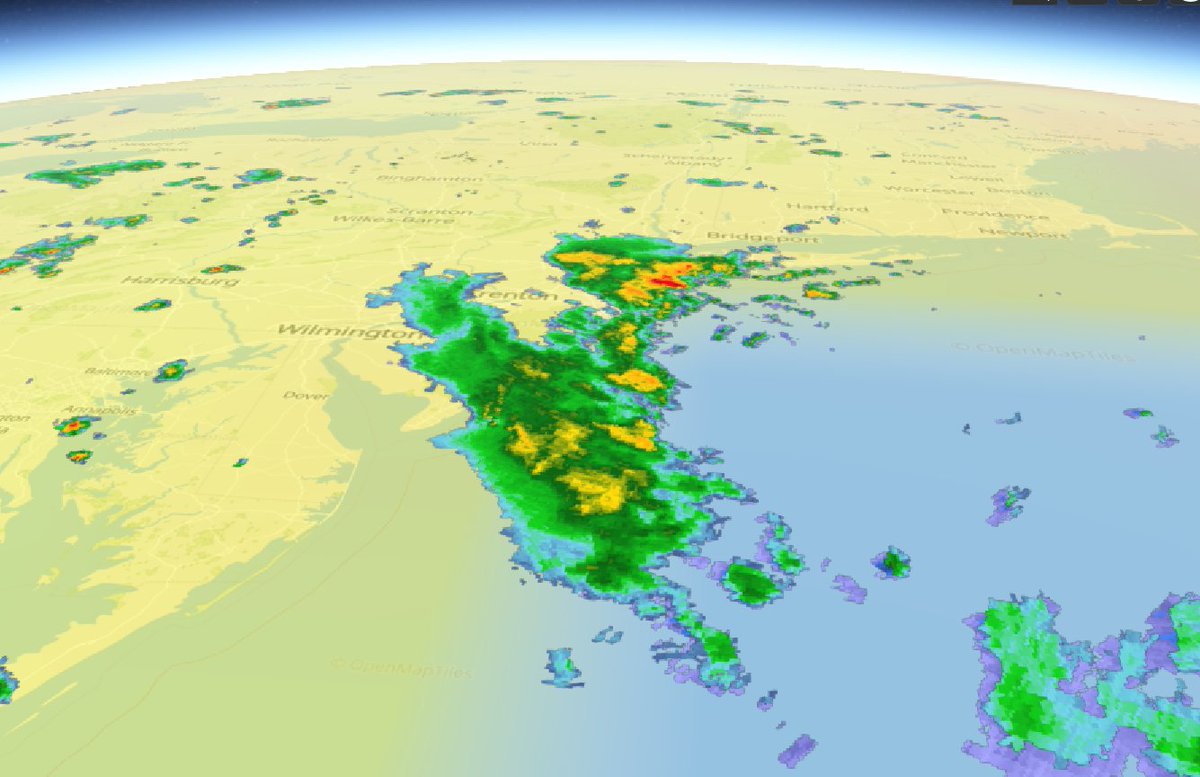
Sat Aug 21 23:18:07 +0000 2021@byu_sam @neely615 @biorxivpreprint @pwilmarth @ryankellybyu The MS/MS data is excellent. All of the things I would want to see in a test data set are there: local ID rates in the 40-60% range, very good residue-specific mod ratios, clean SAAV IDs, low # of missed & non-tryptic cleavages & nearly 100% recovery of C-containing peptides.
Sat Aug 21 18:02:01 +0000 2021@byu_sam @neely615 @biorxivpreprint @pwilmarth @ryankellybyu The modification pattern is typical of what happens during a digest if the urea isn't quite pristine: ~2-3% of PSMs with peptide N-terminal carbamylation but none with the mod on lysine ε-amino groups.
Sat Aug 21 17:59:27 +0000 2021Just about to make landfall on Long Island ... 🔗

Sat Aug 21 16:23:08 +0000 2021@neely615 @byu_sam @biorxivpreprint @pwilmarth Completely for my own information, do you think the carbamylation in the data was in the original sample from Pierce or was it an artifact of your sample workup?
Sat Aug 21 15:40:32 +0000 2021@SciBry Shaka, when the walls fell.
Sat Aug 21 14:49:00 +0000 2021I don't know if I ever mentioned this, but I spent a few years designing data systems for real-time and historic weather telemetry for agricultural applications, which is when I got interested in displaying weather info.
Sat Aug 21 14:38:24 +0000 2021NEXRAD radar showing the onshore areas currently affected 🔗

Sat Aug 21 14:05:24 +0000 2021Tropical Storm Henri (green circle) and the 500 hPa steering winds. Looks like it is headed for an opening on the Great White Way. 🔗
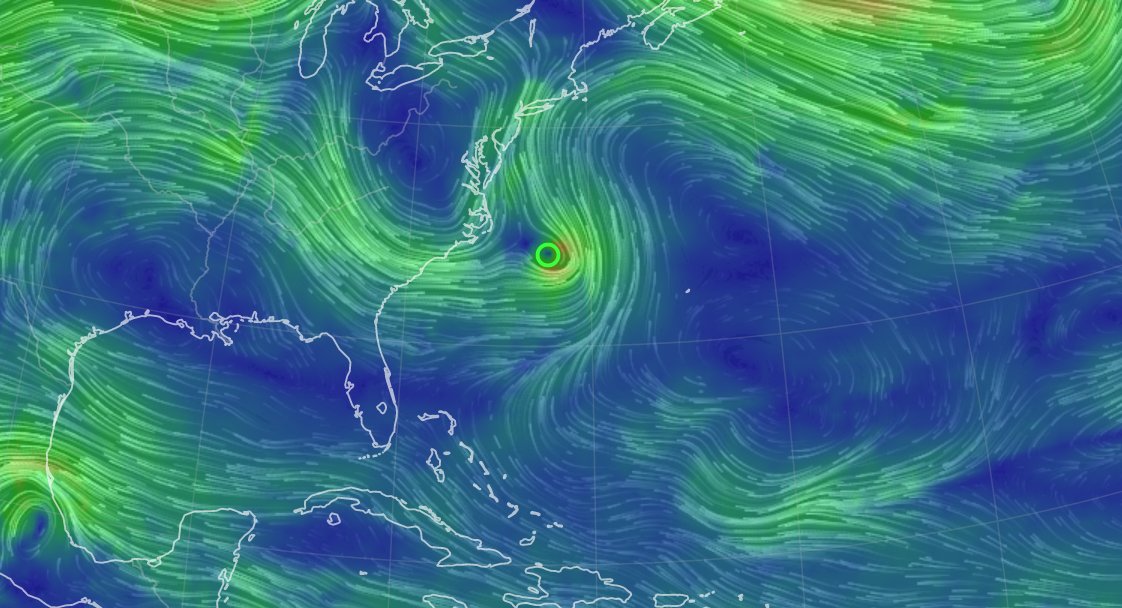
Sat Aug 21 13:46:27 +0000 2021Japan has moved past Brazil and is closing in on South Africa, Spain & Morocco. Cuba is still disturbing high. Canada will probably overtake Indonesia early next week. 🔗
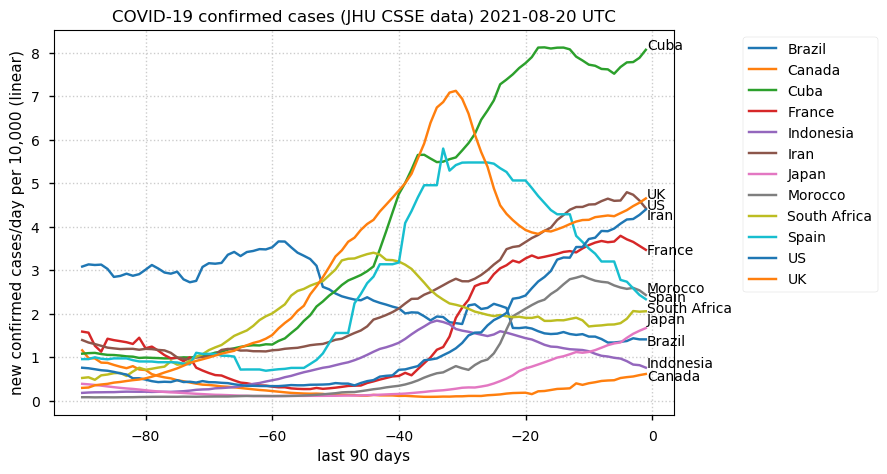
Sat Aug 21 13:30:49 +0000 2021A real evergreen 🔗 I have been occasionally looking back on this for years.
Sat Aug 21 12:59:52 +0000 2021PHIP:p.V663G chr 6:g.78998283A>C, rs7747479 (all tissue V:G 0.008:0.992) vaf=94%, Δm=-42.0470, VAF by population group: african 90%, american 89%, east asian 100%, european 96%, south asian 97%. GG in JURKAT, HEK-293 & HeLa cells.
Sat Aug 21 12:59:52 +0000 2021PHIP:p, θ(max) = 72. aka ndrp, FLJ20705, DCAF14, BRWD2, WDR11. Present in MHC class 1 peptide experiments; absent from class 2 expts. Found in tissues & cell lines; rare in fluids. WD40 repeat domain (181-551).
Sat Aug 21 12:59:52 +0000 2021>PHIP:p, pleckstrin homology domain interacting protein (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 41×K+acetyl; 18×K+GGyl; 11×K+SUMOyl; 66×S, 29×T, 7×Y+phosphoryl; SAAVs: V663G (94%), T874I (8%); mature form: (1-1821) [21,555×, 133 kTa]. #ᗕᕱᗒ 🔗
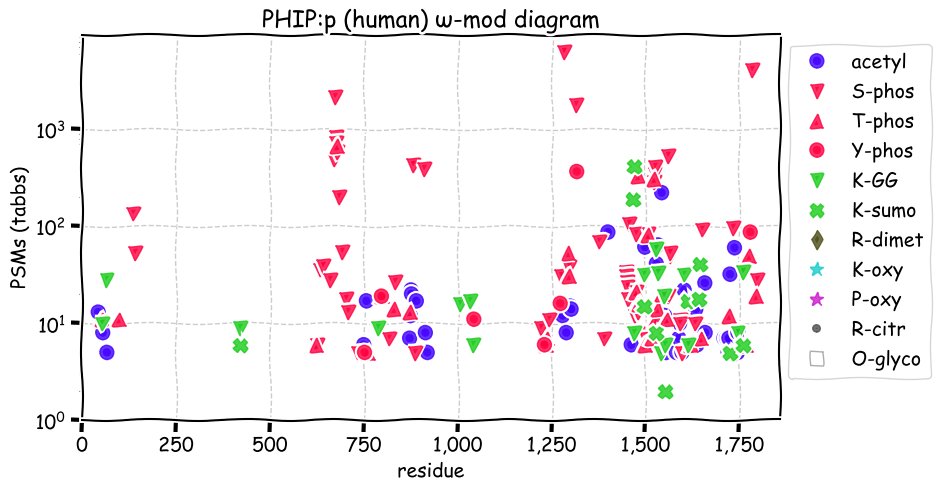
Fri Aug 20 20:03:49 +0000 2021Has anyone else looked at 🔗 ? I don't understand how the CTM would work practically: HuH-7 doesn't normally make Hyp-containing proteins & the enzymes aren't cytoplasmic even if are there.
Fri Aug 20 16:28:11 +0000 2021@byu_sam @biorxivpreprint The Massive directory for the data referred to in the manuscript, MSV000087689, doesn't seem to correspond to the description in the paper.
Fri Aug 20 13:58:51 +0000 2021PTPRC:p, in the initial post for this protein there was an accidental mix of annotations between 2 splice variants. The current version has the coordinate system corrected (corresponds to CCDS1398).
Fri Aug 20 13:58:51 +0000 2021PTPRC:p.N190S chr 1:g.198709705A>G, rs79141749 (all tissue N:S 0.998:0.002) vaf=0.2%, Δm=-27.0109, VAF by population group: african 5%, american <1%, east asian <1%, european <1%, south asian <1%.
Fri Aug 20 13:58:51 +0000 2021PTPRC:p, this enzyme is a tyrosine phosphatase that acts as a receptor to modulate its activity: it is not a receptor for tyrosine phosphatases. The role of its extensive N-linked glycosylation in its function is still not well understood.
Fri Aug 20 13:58:51 +0000 2021PTPRC:p, domains: (26-417) extracellular, (418-438) transmembrane, (439-1145) intracellular. All N-linked glycosylation is within the extracellular domain & the majority of other modification acceptor sites are part of the intracellular domain.
Fri Aug 20 13:58:51 +0000 2021PTPRC:p, θ(max) = 63. aka LCA, T200, GP180, CD45. Present in solid tissue MHC class 1 & 2 peptide experiments. Found in tissues, fluids & cell lines derived from lymphocytes. Has too many GO annotations.
Fri Aug 20 13:58:50 +0000 2021>PTPRC:p, protein tyrosine phosphatase receptor type C (H sapiens) Large subunit; CTMs: N73, N111, N117, N176, N260, N309, N329, N370+glycosyl; PTMs: 2×K+acetyl; 20×K+GGyl; 27×S,4×T,11×Y+phosphoryl; SAAVs: N190S (0.2%); mature form: (26-1145) [21,546×, 332 kTa]. #ᗕᕱᗒ 🔗
Fri Aug 20 12:48:43 +0000 2021Canada's 3 westernmost provinces are still trending up as a group. "Central" Canada is lagging, but appears to be heading in the same direction. 🔗
Fri Aug 20 01:01:32 +0000 2021@jjjotto If you mean the ion chromatograms, 🔗
Thu Aug 19 20:54:56 +0000 2021Time to take my weekly cigar break (today it is a Don Tomas maduro robusto )😀
Thu Aug 19 17:22:50 +0000 2021@FourMohrYears Thanks for putting in the effort. I really appreciate it.
Thu Aug 19 16:22:16 +0000 2021@FourMohrYears Apologies. 2 = B20171030-11_Huh7_PV3h_2.raw is correct.
Thu Aug 19 14:51:37 +0000 2021@VATVSLPR @FourMohrYears Sort of a "first pancake" situation?
Thu Aug 19 14:37:38 +0000 2021@FourMohrYears They are RAW files:
1 = B20171030-10_Huh7_PV3h_1.raw
2 = B20171030-10_Huh7_PV3h_2.raw
from PXD024546.
Thu Aug 19 14:21:53 +0000 2021It looks to me like the 1st rep is running a completely different chromatographic method than the 2nd rep.
Thu Aug 19 14:20:16 +0000 2021For MS instrument folks, the graphs below are for the 1st & 2nd tech reps of the same sample. Plotted is the calculated retention time of PSMs (blue) & the fraction of spectra resulting in PSMs (green) vs scan number. How can this happen? 🔗
Thu Aug 19 14:08:02 +0000 2021Briefly pondering "Why do HuH-7 cells produce so much LINE-1 ORF1?"
Thu Aug 19 13:02:59 +0000 2021But how to convincingly tell people that their cherished hypothesis is simply a great way to isolate high scoring false positives escapes me.
Thu Aug 19 12:25:18 +0000 2021It is the sort of analysis where Venn diagrams rule (& all judgement fled).
Thu Aug 19 12:23:52 +0000 2021Often the problem seems to be that people do a differential analysis & assume that the small number of left-overs has a similar confidence distribution to the results as a whole.
Thu Aug 19 12:06:49 +0000 2021It would be a good topic for an ASMS/HUPO short course.
Thu Aug 19 12:03:15 +0000 2021Particularly when they are seeing a pattern that doesn't exist, but it would be great for their hypothesis if it did.
Thu Aug 19 12:00:33 +0000 2021I have never figured out how to talk people down when they are over interpreting their data.
Thu Aug 19 11:54:17 +0000 2021HTATIP2:p.S197R chr 11:g.20383067T>G, rs3824886 (all tissue S:R 0.303:0.697) vaf=85%, Δm=69.0691, VAF by population group: african 59%, american 72%, east asian 47%, european 94%, south asian 89%. This variant adds a novel tryptic cleavage site.
Thu Aug 19 11:54:17 +0000 2021HTATIP2:p, θ(max) = 87. aka TIP30, CC3, FLJ26963, SDR44U1. Present in solid tissue MHC class 1 & leukocyte/lymphocyte class 2 peptide experiments. Found in tissues, cells lines & extracellular vesicles from multiple sources.
Thu Aug 19 11:54:16 +0000 2021>HTATIP2:p, HIV-1 Tat interactive protein 2 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 4×K+acetyl; 10×K+GGyl; 0×S, 0×T, 2×Y+phosphoryl; SAAVs: S197R (85%); mature form: (2-242) [15,904×, 74 kTa]. #ᗕᕱᗒ 🔗
Wed Aug 18 15:36:18 +0000 2021@MHendr1cks Is SNC-L in the mix?
Wed Aug 18 15:25:50 +0000 2021@byu_sam @AJ_Brenes I'd say it more like MDs are more stubbornly holding on to the idea. 10 years ago, many PhDs that are now OK with data sharing were dead set against it.
Wed Aug 18 15:00:37 +0000 2021@AJ_Brenes @byu_sam Neither do I. But it is a widely held opinion that people hold some type of IP in their data that should be respected, even though no such IP really exists.
Wed Aug 18 14:32:26 +0000 2021@byu_sam It seems odd to me, but based on the definition I am not a Research Parasite (even though I have aspired to be one). 🧐
Wed Aug 18 12:13:57 +0000 2021PALB2:p.P210L chr 16:g.23635917G>C, rs57605939 (all tissue P:L 0.921:0.079) vaf=2%, Δm=16.0313, VAF by population group: african 9%, american 1%, east asian <1%, european <1%, south asian <1%.
Wed Aug 18 12:13:57 +0000 2021PALB2:p, θ(max) = 76. aka FLJ21816, FANCN. Present in MHC class 1 peptide experiments. Found in tumour tissues & cell lines: absent from fluids. The N-terminal phosphodomain ends at the beginning of the C-terminal WD domain (837-1186).
Wed Aug 18 12:13:57 +0000 2021>PALB2:p, partner and localizer of BRCA2 (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 4×K+acetyl; 10×K+GGyl; 23×S, 4×T, 0×Y+phosphoryl; SAAVs: P210L (2%), G998E (1%); mature form: (1-1186) [2,260×, 7.5 kTa]. #ᗕᕱᗒ 🔗
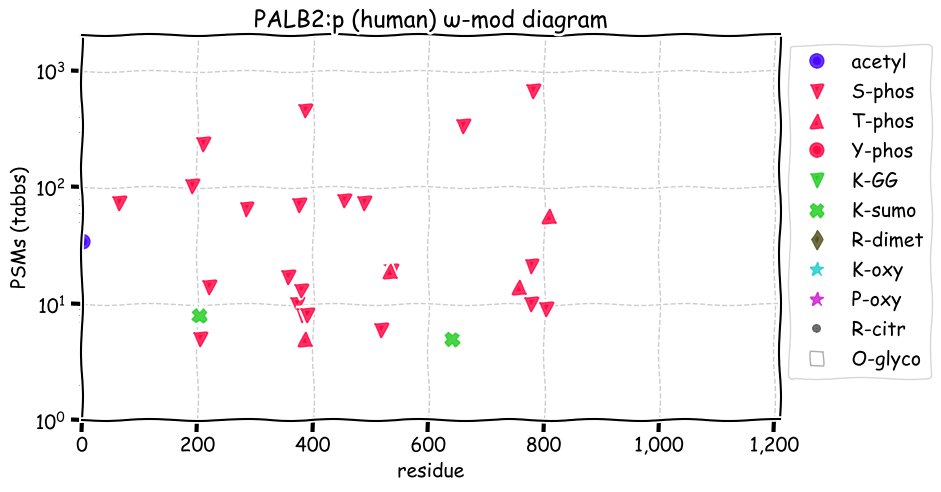
Tue Aug 17 19:23:28 +0000 2021I always feel like I'm white-knuckling it when I change old Perl scripts.
Tue Aug 17 15:07:45 +0000 2021Canada seems well on its way into a "4th wave", with the 3 westernmost provinces leading the way 🔗
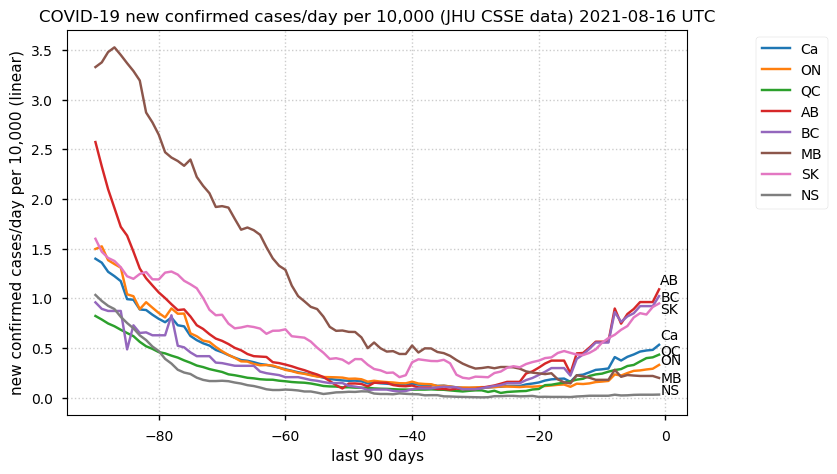
Tue Aug 17 12:33:45 +0000 2021MCAM:p is frequently found in extracellular vesicles in urine, blood plasma and shed by many cell lines in culture.
Tue Aug 17 12:12:12 +0000 2021MCAM:p.E89G chr 11:g.119314967T>C, rs34587557 (all tissue E:G 0.992:0.008) vaf=0.4%, Δm=-72.0211, VAF by population group: african 0%, american 5%, east asian 0%, european 5%, south asian 2%.
Tue Aug 17 12:12:12 +0000 2021MCAM:p, Domains: (24-559) extracellular, (560-583) transmembrane, (584-646) intracellular.
Tue Aug 17 12:12:11 +0000 2021MCAM:p, θ(max) = 67. aka MUC18, CD146, MelCAM, METCAM, HEMCAM, Gicerin. Present in MHC class 2 & tissue-based class 1 peptide experiments. Found in urine, CSF & blood plasma, as well as many tissues & cell lines.
Tue Aug 17 12:12:11 +0000 2021>MCAM:p, melanoma cell adhesion molecule (Homo sapiens) Midsized subunit; CTMs: N6, N418, N449+glycosyl; PTMs: 1×K+acetyl; 6×K+GGyl; 18×S, 0×T, 2×Y+phosphoryl; SAAVs: E89G (0.4%), L394M (2%); mature form: (23,24-646) [21,584×, 202 kTa]. #ᗕᕱᗒ 🔗
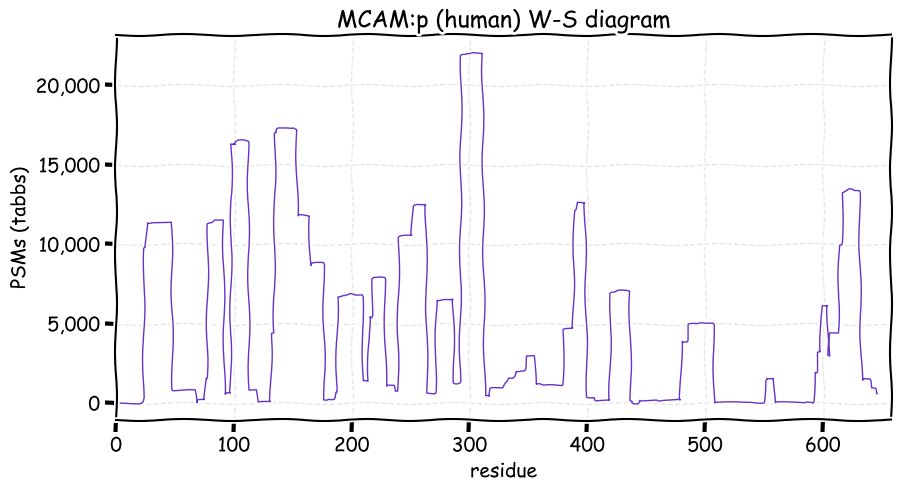
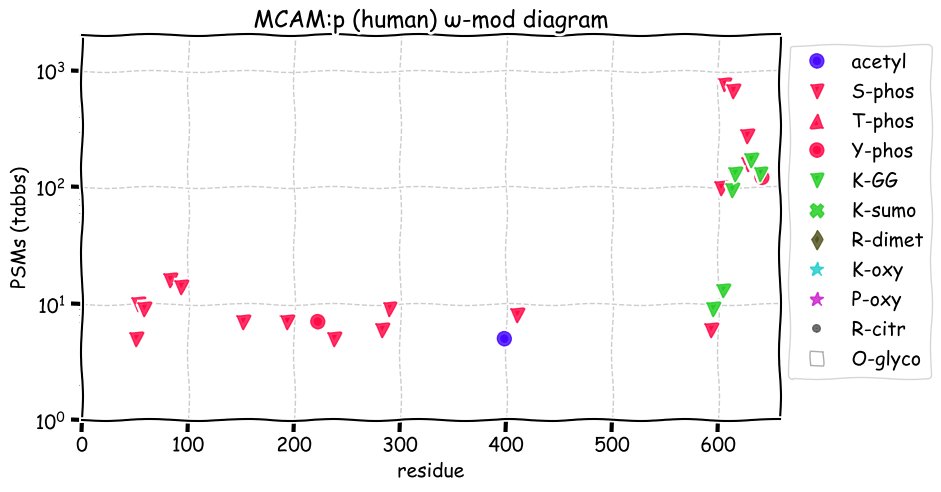
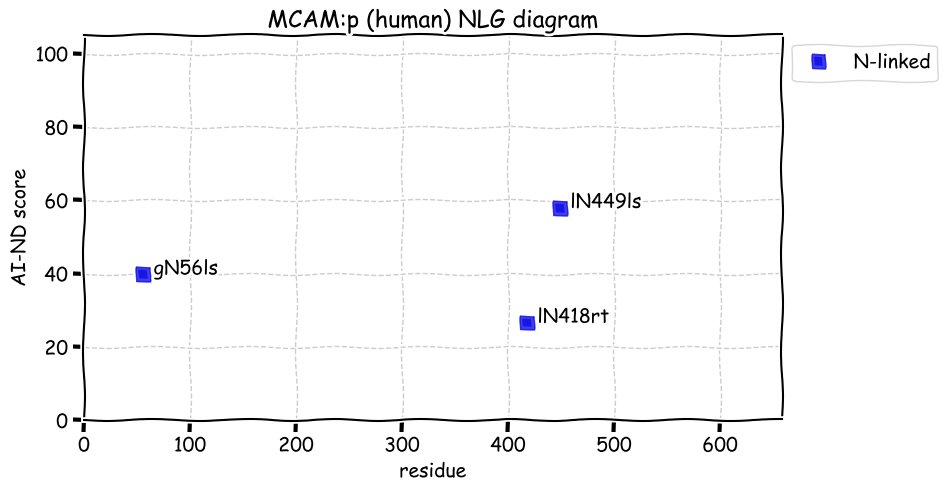
Mon Aug 16 16:06:55 +0000 202111. Try to remember that software apps are clockwork. Agreements are not subject to statistical variation, they are all deterministic.
Mon Aug 16 16:06:54 +0000 202110. If there is still a significant lack of agreement, try to understand why: parameter interpretations? low score cutoff artifacts? implementation differences?
Mon Aug 16 16:06:54 +0000 20219. If 8 suggests a problem with initial parameters, do over & iterate as often as necessary
Mon Aug 16 16:06:54 +0000 20218. Try to understand why the algorithms agree on most PSMs: view differences as a lack of agreement
Mon Aug 16 16:06:54 +0000 20217. Do the same thing, but for several different found parameters, scan number and (mass accuracy or score or modification)
Mon Aug 16 16:06:54 +0000 20216. Create a matrix of the observed PSMs for these proteins, based on their scan numbers & ID'd peptide
Mon Aug 16 16:06:54 +0000 20215. Do not look at proteins you are interested in: see 3 above
Mon Aug 16 16:06:53 +0000 20214. Do select 1‒3 proteins that are large and well represented in the data set (e.g., ALB, TRF, COL1A1, APOB or MYH9)
Mon Aug 16 16:06:53 +0000 20213. Interest is the mind killer: select test data you do not care about
Mon Aug 16 16:06:53 +0000 20212. Do not aggregate results or use overall statistics
Mon Aug 16 16:06:53 +0000 20211. Try to do the best job with each algorithm: do not try to level the playing field or "make things fair"
Mon Aug 16 16:06:53 +0000 2021Several people have mentioned the difficulty in comparing the results of protein id algorithms. Since this is something I have done a number of times, here is my 2¢ on the subject.
Mon Aug 16 12:39:53 +0000 2021FKBP15:p, FK506 (Tacrolimus) is an immunosuppressant and this protein is named for binding it. There is no mechanism connecting this activity to the immune system effects of the the drug.
Mon Aug 16 12:23:26 +0000 2021FKBP15:p.A106T chr 9:g.113206517C>T, rs1133618 (all tissue A:T 0.966:0.034) vaf=17%, Δm=30.0106, VAF by population group: african 33%, american 22%, east asian 2%, european 21%, south asian 14%. TT in MCF-10A cells.
Mon Aug 16 12:23:26 +0000 2021FKBP15:p, θ(max) = 71. aka PPP1R76, FKBP133, WAFL, KIAA0674. Present in MHC class 1 & 2 peptide experiments. Found in tissues & cell lines: rare in fluids.
Mon Aug 16 12:23:25 +0000 2021>FKBP15:p, FK506 binding protein 15 (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 40×K+acetyl; 3×K+GGyl; 67×S, 32×T, 1×Y+phosphoryl; SAAVs: A106T (19%), S303T (1%), R305H (2%), A847S (8%), A1069S (2%); mature form: (1-1219) [29,441×, 220 kTa]. #ᗕᕱᗒ 🔗
Sun Aug 15 14:18:30 +0000 2021The Beauveria bassiana proteome also has almost no overlap of tryptic peptide sequences with commonly studied species (inc. S. cerevisiae & S. pombe) making it good for testing machine learning algorithms trained on model species data.
Sun Aug 15 13:34:56 +0000 2021Cuba is still getting slammed & Japan is on the cusp of surpassing Brazil. 🔗
Sun Aug 15 13:17:41 +0000 2021If you are interested in looking at something other than biomedical model species, try the expts in PRIDE from the Beauveria bassiana, an entomopathogenic fungus. The data is very good quality, looks at a variety of common modifications & the proteome is well annotated.
Sun Aug 15 12:36:37 +0000 2021AP3B1:p.V585E chr 5:g.78129204A>T, rs6453373 (all tissue V:E 0.205:0.795) vaf=86%, Δm=-29.9742, VAF by population group: african 95%, american 96%, east asian 44%, european 93%, south asian 78%. EE in MCF-10A & HeLa cells.
Sun Aug 15 12:36:37 +0000 2021AP3B1:p, θ(max) = 55. aka ADTB3A, HPS2. Present in tissue-based MHC class 1 peptide experiments. Found in tissues & cell lines: rare in fluids.
Sun Aug 15 12:36:37 +0000 2021>AP3B1:p, adaptor related protein complex 3 subunit beta 1 (Homo sapiens) Large subunit; CTMs: S2+acetyl; PTMs: 46×K+acetyl; 13×K+GGyl; 53×S, 19×T, 8×Y+phosphoryl; SAAVs: V585E (86%); mature form: (2-1094) [34,816×, 229 kTa]. #ᗕᕱᗒ 🔗
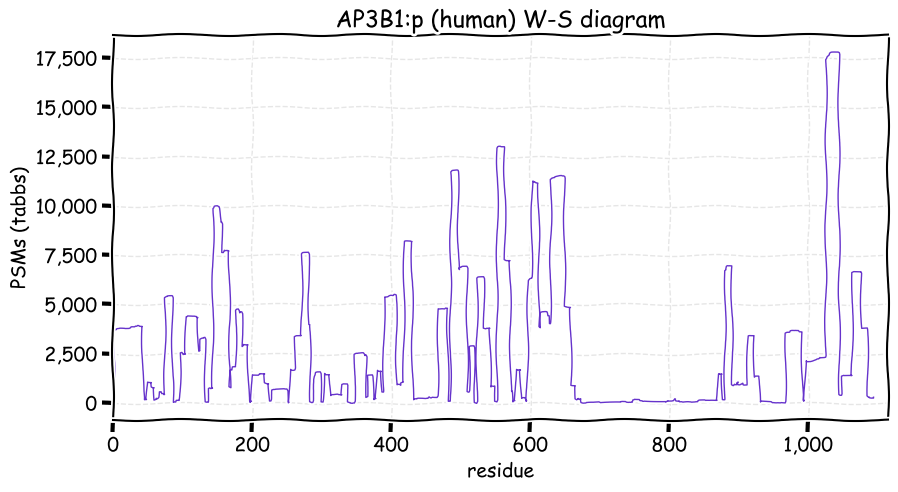
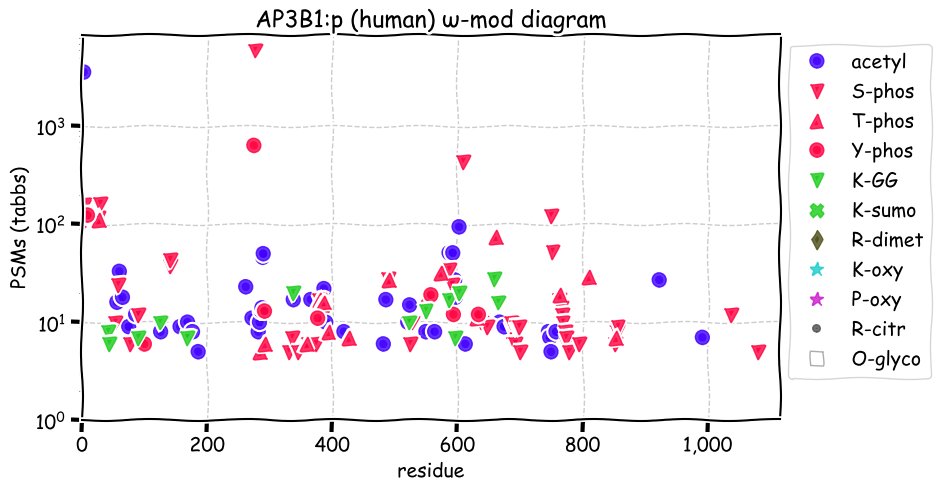
Sat Aug 14 16:16:51 +0000 2021The purpose of publishing academic software is to demonstrate the value of a new idea via an implementation. It should aim to move the field forward. It is not to provide some random group of "users" free "tools".
Sat Aug 14 14:27:20 +0000 2021@LewisGeer @pwilmarth @ypriverol @sackloo @olgavitek @JohnRYatesIII By parameters, I mean attempts to parameterize the results, not the input parameters. Unless there was an attempt to clone another piece of software, input parameters for one implementation often mean something slightly different in all others.
Sat Aug 14 13:45:32 +0000 2021@pwilmarth @ypriverol @sackloo @olgavitek @JohnRYatesIII Even the best "power-users" rarely understand what is going on well enough to compare algorithms in a useful manner. You end up with chili-cookoffs that focus on characterizing results by maximizing/minimizing a handful of "common sense" overall parameters.
Sat Aug 14 12:29:53 +0000 2021FCN2:p is one of the non-collagen proteins with hydroxy-P/K modified domains:
FCN1:p, FCN2:p, FCN3:p, FGA:p, C1QA:p, C1QB:p, C1QC:p, ADIPOQ:p, SFTPB:p, SFTPA2:p, SFTPA1:p, SFTPC:p, COLQ:p, MARCO:p, GLDN:p, EMID1:p, EMID2:p, EMILIN1:p, EMILIN2:p
Sat Aug 14 12:19:03 +0000 2021FCN2:p.T236M chr 9:g.134887180C>T, rs17549193 (all tissue T:M 0.814:0.186) vaf=26%, Δm=29.9928, VAF by population group: african 36%, american 25%, east asian 7%, european 28%, south asian 25%.
Sat Aug 14 12:19:02 +0000 2021FCN2:p, θ(max) = 63. aka P35, FCNL, EBP-37. Present in tissue-based MHC class 2 peptide experiments. Found in urine & blood plasma: rare in CSF. Absent from cell lines, except as a contaminant. Collagen-like domain (26-110).
Sat Aug 14 12:19:02 +0000 2021>FCN2:p, ficolin 2 (Homo sapiens) Small subunit; CTMs: N240+glycosyl; PTMs: 3×K+oxide; 15×P+oxide; SAAVs: T236M (26%), A258S (16%); mature form: (26-313) [4,389×, 19 kTa]. #ᗕᕱᗒ 🔗
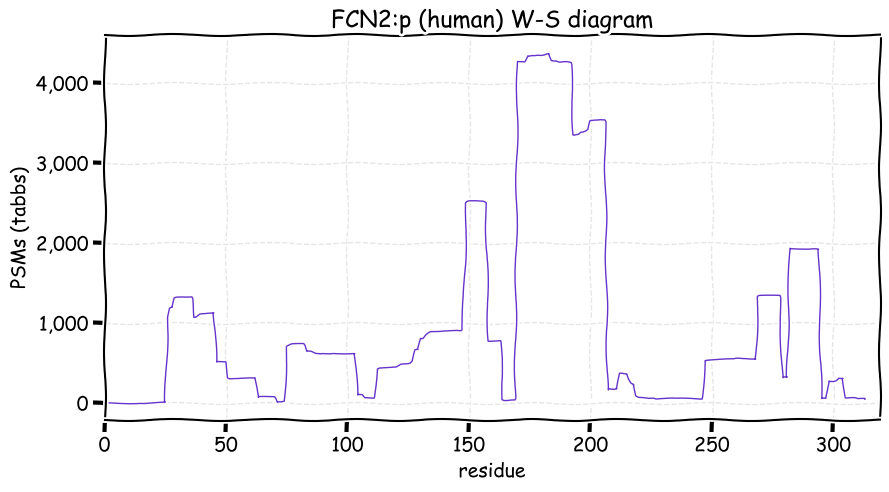
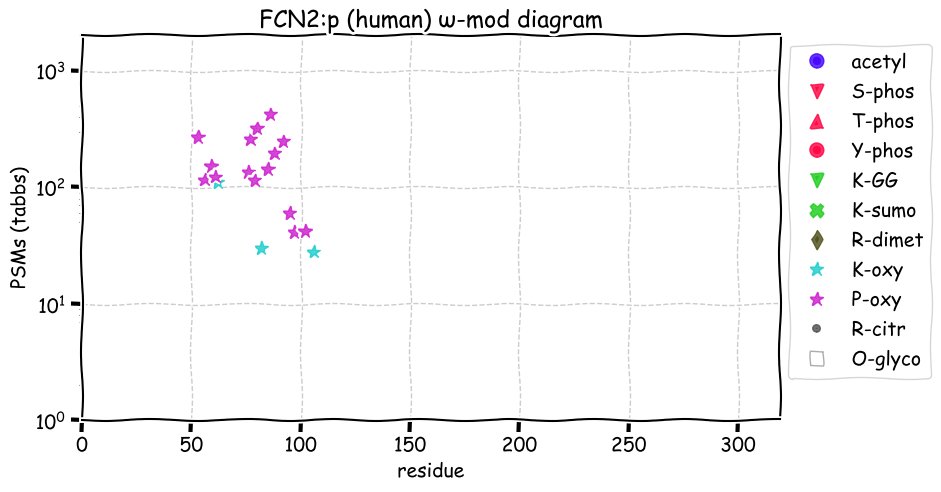
Fri Aug 13 12:34:05 +0000 2021FNC3:p, as is common with many proteins having collagen-like domains, the active protein is a trimer.
Fri Aug 13 12:19:45 +0000 2021FCN3:p.E166D chr 1:g.27370868C>G, rs56405086 (all tissue E:D 0.994:0.006) vaf=0.3%, Δm=-14.0157, VAF by population group: african <1%, american <1%, east asian 3%, european <1%, south asian <1%.
Fri Aug 13 12:19:45 +0000 2021FCN3:p, θ(max) = 87. aka FCNH, HAKA1. Rare in MHC peptide experiments. Found in blood plasma & CSF: rare in urine. Absent from cell lines, except as a contaminant. Collagen-like domain (24-118).
Fri Aug 13 12:19:45 +0000 2021>FCN3:p, ficolin 3 (Homo sapiens) Small subunit; CTMs: N189+glycosyl; PTMs: Q24+ammonia-loss; 3×K+oxide; 19×P+oxide; SAAVs: L117S (2%), E166D (0.3%); mature form: (24-299) [8,605×, 96 kTa]. #ᗕᕱᗒ 🔗
Thu Aug 12 15:32:50 +0000 2021@astacus I'm using a Python package called matplotlib to create the graphs. At the top of the python code you use:
import matplotlib.pyplot as plt
and then latter if you add
plt.xkcd()
it generates the plots a style evocative of illustrations on 🔗
Thu Aug 12 14:16:27 +0000 2021I find the looptail "g" in the new Twitter font distracting. Seems out of place with the other letterforms.
Thu Aug 12 12:31:20 +0000 2021@PastelBio @ProteomicsNews It is pretty good data that is quite reproducible, run to run and experiment to experiment. In spite of 20% of PSMs being from ALB, they do see good signals from ~1400 proteins.
Thu Aug 12 12:18:45 +0000 2021Search bots that ignore robots.txt still plague me from time to time (like this morning).
Thu Aug 12 12:07:07 +0000 2021MAD2L1:p forms a heterotetramer with MAD1L1:p to create an active protein that interacts with the mitotic spindle during metaphase. These two subunits have very different PTM patterns.
Thu Aug 12 11:58:44 +0000 2021MAD2L1:p.I190V chr 4:g.120060168>C, rs61752608 (all tissue I:V 0.997:0.003) vaf=0.4%, Δm=-14.0156, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Thu Aug 12 11:58:44 +0000 2021MAD2L1:p, θ(max) = 86. aka MAD2, HSMAD2. Present in MHC class 1 experiments. Found in most tissues and cell lines: rare in fluids.
Thu Aug 12 11:58:44 +0000 2021>MAD2L1:p, mitotic arrest deficient 2 like 1 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 2×K+acetyl; 8×K+GGyl; 2×K+SUMOyl; 3×S, 0×T, 1×Y+phosphoryl; SAAVs: I190V (0.4%); mature form: (2-205) [21,364×, 59 kTa]. #ᗕᕱᗒ 🔗
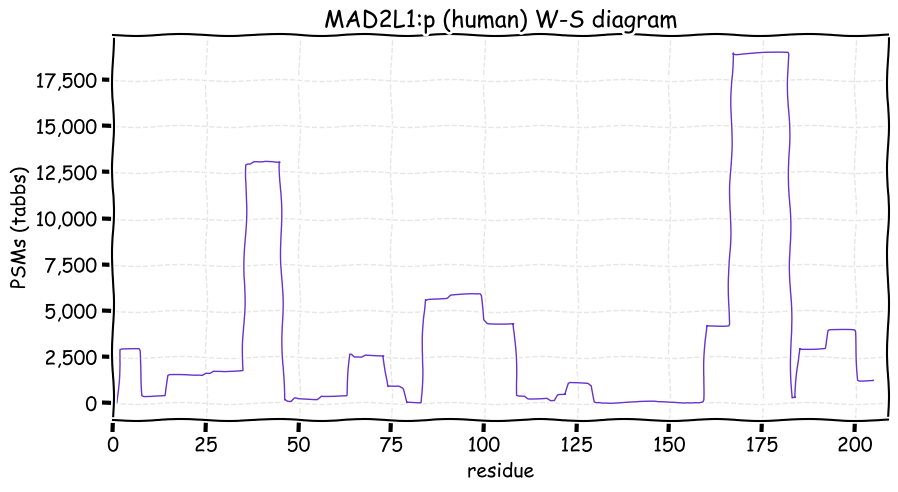
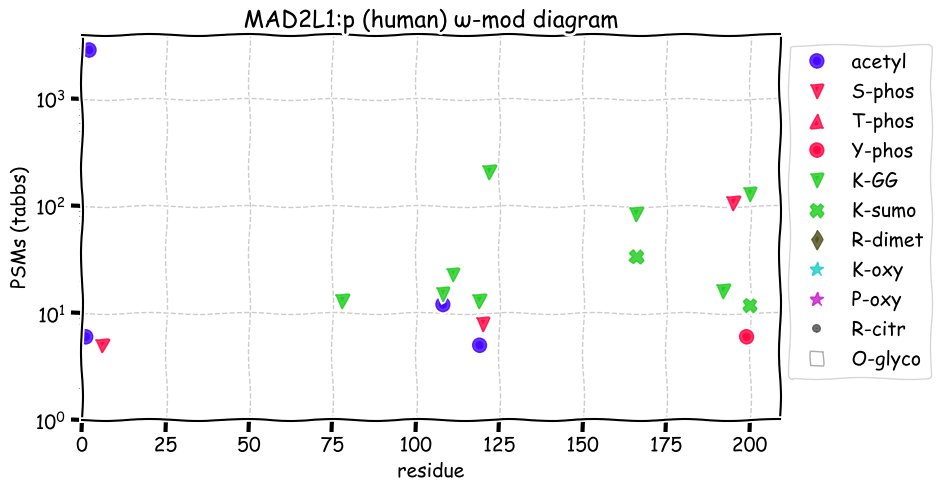
Wed Aug 11 19:06:14 +0000 2021A well-deserved shot at product management in software development
🔗
Wed Aug 11 12:12:31 +0000 2021MAD1L1:p.R558H chr 7:g.1936821C>T, rs1801368 (all tissue R:H 0.920:0.080) vaf=35%, Δm=-19.0422, VAF by population group: african 3%, american 44%, east asian 45%, european 39%, south asian 35%. RH in U2OS cells. SAAV removes a tryptic cleavage site.
Wed Aug 11 12:12:31 +0000 2021MAD1L1:p, θ(max) = 86. aka HsMAD1, TXBP181, MAD1, PIG9, TP53I9. Present in MHC class 1 experiments. Found in most tissues and cell lines: absent from fluids.
Wed Aug 11 12:12:31 +0000 2021>MAD1L1:p, MAD1 mitotic arrest deficient-like 1 (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 21×K+acetyl; 9×K+GGyl; 7×K+SUMOyl; 19×S, 10×T, 1×Y+phosphoryl; SAAVs: R558H (35%); mature form: (1-718) [17,552×, 82 kTa]. #ᗕᕱᗒ 🔗
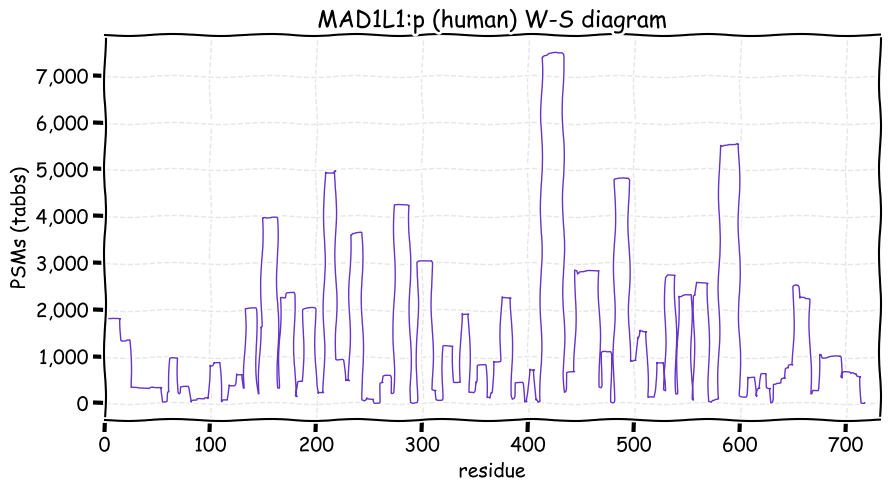
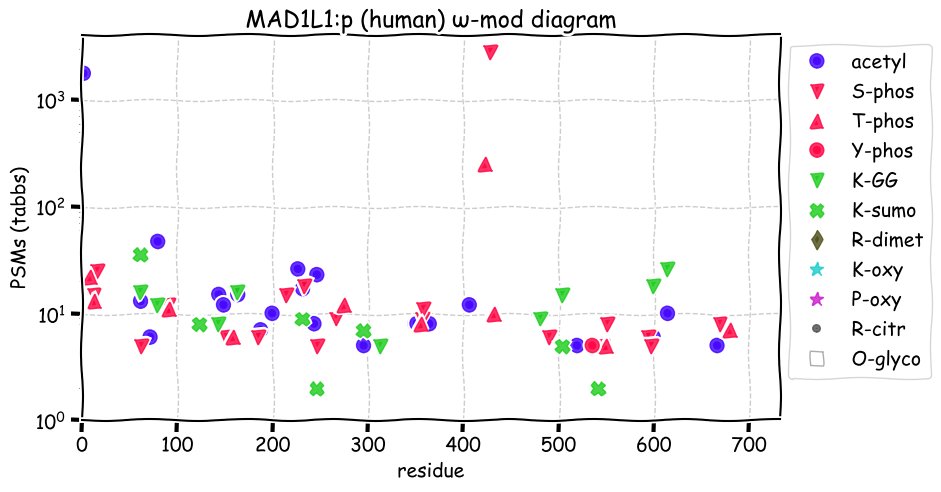
Tue Aug 10 20:47:13 +0000 2021@Smith_Chem_Wisc You should be able to see it in quite a few of the files in PXD025048, for example:
DT_01192021_20U_W_1.RAW.
There should also be a 1 residue longer form detectable, 57-88, as well as the intact insulin A chain (90-110).
Tue Aug 10 16:18:21 +0000 2021@DRAWheatcraft @pommie331 @ProtifiLlc @NBasisty @briansearle How many of the 21 MUPs do you see? They are a pretty good group of proteins to monitor in mouse plasma.
Tue Aug 10 15:09:09 +0000 2021@Smith_Chem_Wisc That is a reasonable approach. Have you identified the C-peptide in clinical samples?
Tue Aug 10 13:50:14 +0000 2021@mjmaccoss @qian_weijun @1jvaneyk It shows up intact in pancreatic tissue samples, along with mature insulin & produces quite good spectra (🔗).
Tue Aug 10 13:05:22 +0000 2021Assuming that you haven't FASP'd it into oblivion prior to analysis.
Tue Aug 10 12:59:30 +0000 2021How do people who do a lot of clinical plasma work cope with detecting insulin C-peptide in MS/MS runs? It tends to be ignored by search engines, unless you use semi-tryptic or non-specific cleavage. Do you just add it as a separate entry to the list of protein sequences file?
Tue Aug 10 12:18:03 +0000 2021SKA3:p.T254A chr 13:g.21161859T>C, rs17345690 (all tissue T:A 0.915:0.085) vaf=12%, Δm=-30.0106, VAF by population group: african 7%, american 7%, east asian 10%, european 14%, south asian 12%. TA in Hela cells.
Tue Aug 10 12:18:03 +0000 2021SKA3:p, θ(max) = 93. aka MGC4832; RAMA1; C13orf3. Present in MHC class 1 experiments. Found in most tissues and cell lines: absent from fluids.
Tue Aug 10 12:18:03 +0000 2021>SKA3:p, spindle and kinetochore associated complex subunit 3 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 2×K+acetyl; 10×K+GGyl; 5×K+SUMOyl; 23×S, 12×T, 5×Y+phosphoryl; SAAVs: V58I (1%), T254A (12%), D335E (3%); mature form: (1-412) [8,297×, 30 kTa]. #ᗕᕱᗒ 🔗
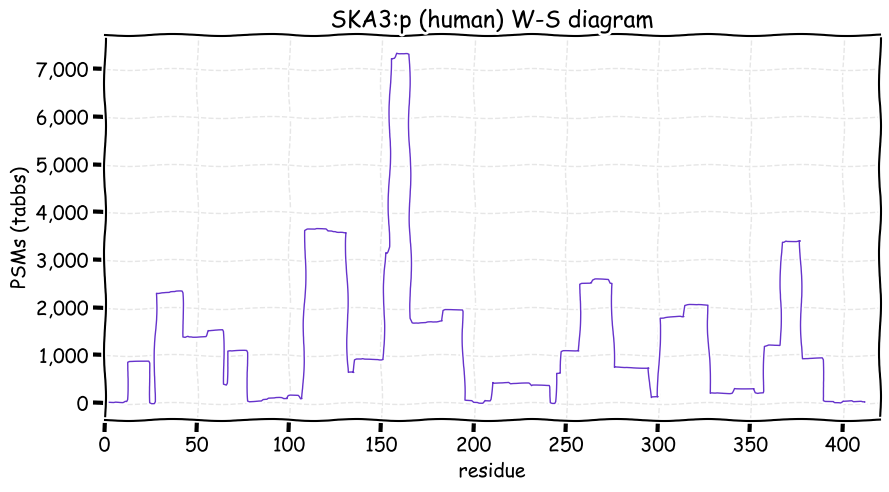
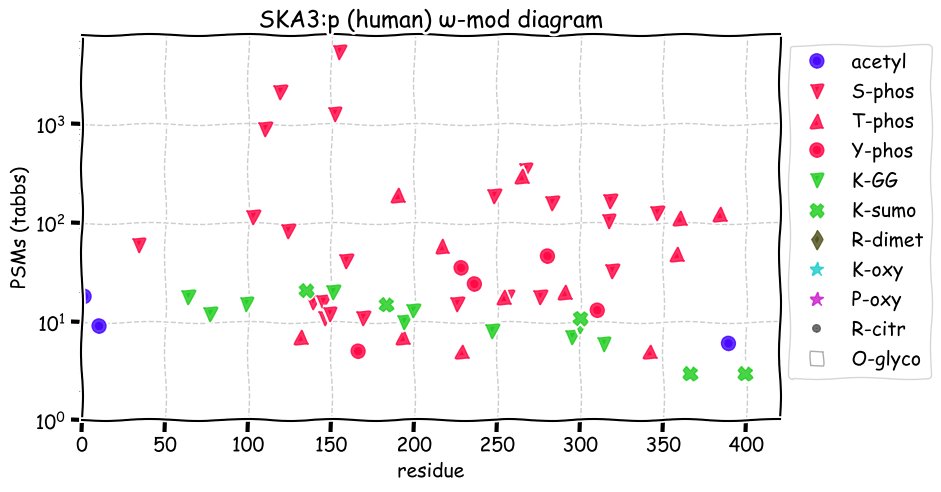
Mon Aug 09 16:11:19 +0000 2021Red Racer "Street Legal IPA" is the best low alcohol beer I have tried. It really tastes like a good American-style IPA, but with 0.5% alcohol.👍
Mon Aug 09 11:40:27 +0000 2021HRG:p, really has histidine rich domains, e.g. (351-410):
PHKHHSHEQHPHGHHPHAHH PHEHDTHRQHPHGHHPHGHH PHGHHPHGHHPHGHHPHCHD.
Mon Aug 09 11:40:26 +0000 2021HRG:p.I180T chr 3:g.186671770T>C, rs10770 (all tissue I:T 0.879:0.121) vaf=12%, Δm=-12.0364, VAF by population group: african 20%, american 11%, east asian 9%, european 12%, south asian 13%.
Mon Aug 09 11:40:26 +0000 2021HRG:p, θ(max) = 68. aka HRGP, HPRG. Present in MHC class 1 & class 2 experiments. Abundant in blood plasma, urine & CSF. Has too many GO annotations.
Mon Aug 09 11:40:26 +0000 2021>HRG:p, histidine rich glycoprotein (Homo sapiens) Midsized subunit; CTMs: N63, N125, N344, N345+glycosyl; PTMs: 5×K+acetyl; 6×S, 4×T+glycosyl; 6×S, 4×T, 0×Y+phosphoryl; SAAVs: I180T (12%), R448C (27%), N493I (55%); mature form: (19-525) [28,803×, 452 kTa].#ᗕᕱᗒ 🔗
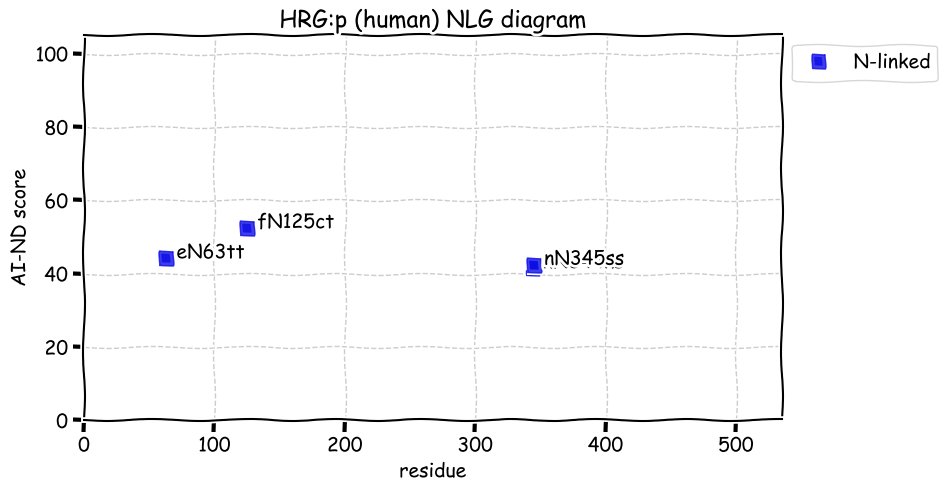
Sun Aug 08 19:20:34 +0000 2021@VATVSLPR Drying may well be the cause. It tends to be more prevalent in TMT/iTRAQ expts where the peptides spend a significant amount of time in non-aqueous solution.
Sun Aug 08 18:41:05 +0000 2021@nesvilab I thought once thought that too, but in many data sets the pyro-E peptides are detected at later LC elution times than the uncyclized peptides. Gas phase ion stability (particularly in any ion mobility expt) is definitely something that can cause similar effects, though.
Sun Aug 08 14:46:48 +0000 2021I have always been a bit puzzled by the formation of pyro-Glu on peptides with an N-terminal Glu, as opposed to the same reaction with an N-terminal Gln. Non-enzymatic loss of water in an aqueous solution just seems wrong ...
Sun Aug 08 12:18:47 +0000 2021ZFYVE16:p.I192T chr 5:g.80437260T>C, rs2544600 (all tissue I:T 0.172:0.828) vaf=87%, Δm=-12.0364, VAF by population group: african 44%, american 88%, east asian 92%, european 89%, south asian 82%. TT in MCF-7, Jurkat & Hep-G2 cells; II in Hela cells.
Sun Aug 08 11:55:11 +0000 2021ZFYVE16:p, θ(max) = 52. aka KIAA0305, PPP1R69. Present in MHC class 1 & class 2 experiments. Observable in most tissues & cell lines, but absent from fluids. The non-phophorylated C-terminal domain is classified as DUF3480 (pfam11979).
Sun Aug 08 11:55:11 +0000 2021>ZFYVE16:p, zinc finger FYVE-type containing 16 (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 1×K+acetyl; 24×K+GGyl; 60×S, 7×T, 10×Y+phosphoryl; SAAVs: I192T (87%), I598T (85%); mature form: (1-1539) [12,500×, 56 kTa].#ᗕᕱᗒ 🔗
Sat Aug 07 23:54:11 +0000 2021@pwilmarth @JoelisSteele @ProteomicsNews The company has been around since 2001. If anything, the naming issue is the other way around.
Sat Aug 07 18:17:39 +0000 2021@mjmaccoss @ucdmrt From the way it is described, I don't know how many residues they will be able to get from a peptide. I would guess they will run out of steam at about 20 residues, assuming that 5% of exopeptidase cycles remove 0 or >1 AAs.
Sat Aug 07 17:45:10 +0000 2021Pleasantly surprised by the equation editor in Libre Office. Nice way to deal with a difficult problem. 😎
Sat Aug 07 17:00:41 +0000 2021@VATVSLPR @mjmaccoss @ucdmrt Peptides in the 2-5 AA range are often not so useful because of they tend to be less unique than longer ones, but better access to peptides in the 6-12 AA range would be very useful in many proteins.
Sat Aug 07 16:35:32 +0000 2021@IgnatovaLab Very little needs to be added, even with 100 years of additional biomedical research.
Sat Aug 07 16:28:09 +0000 2021@mjmaccoss @ucdmrt In practice, the combination of the fact that they are short & the prevalence of incomplete cleavage means this doesn't impact protein coverage as much as one might naively imagine it would, but sometimes having them would be useful.
Sat Aug 07 16:26:03 +0000 2021@mjmaccoss @ucdmrt One thing they would have access to would be short peptides. The median length of a tryptic peptide is 6 AAs, which means ~50% of tryptic peptides are too short to be id'd.
Sat Aug 07 15:34:59 +0000 2021@pwilmarth It also doesn't help that "used BLAST a few times" seems to be enough for many people to justify adding "bioinformatics" to their CV's section on demonstrated expertise. It is the intellectual equivalent of adding "physics" expertise because they turned on & watched a TV.
Sat Aug 07 15:00:44 +0000 2021@pwilmarth There is a commonly held false equivalency in the minds of biomedical PI-types between anyone who claims to be involved in bioinformatics & "computer/stats/applied math wizard". It makes it nearly impossible to talk to them on the subject.
Sat Aug 07 11:45:54 +0000 2021FIZ1:p.T391A chr 19:g.55592770T>C, rs7247236 (all tissue T:A 0.220:0.780) vaf=79%, Δm=-30.0106, VAF by population group: african 90%, american 78%, east asian 56%, european 81%, south asian 84%. AA in Hela cells & TA in Jurkat cells.
Sat Aug 07 11:45:54 +0000 2021FIZ1:p, θ(max) = 32. aka FLJ14768, ZNF798. May be present in MHC class 1 experiments. Low levels in all tissues & cell lines, but absent from fluids.
Sat Aug 07 11:45:53 +0000 2021>FIZ1:p, FLT3 interacting zinc finger 1 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: K61, K226, K440+SUMOyl; SAAVs: V302L (1%), T391A (79%); mature form: (1-495) [1,405×, 3.7 kTa]. #ᗕᕱᗒ 🔗
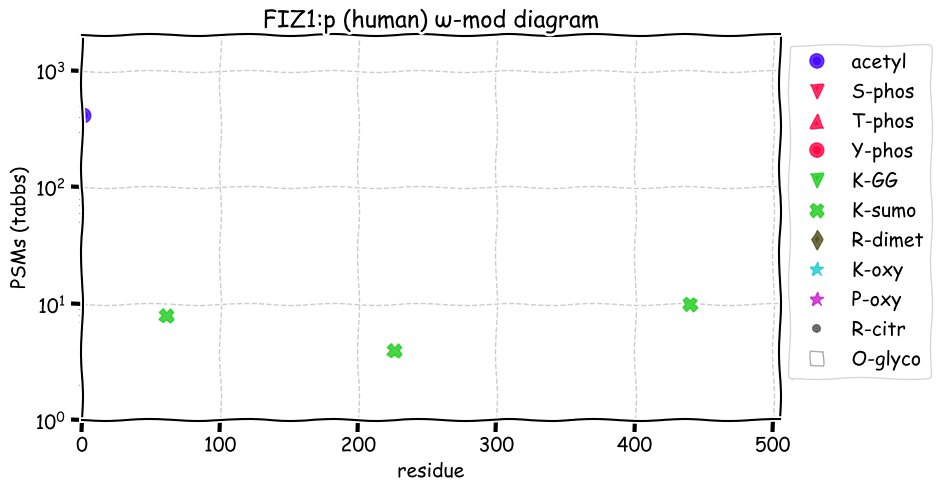
Fri Aug 06 14:15:34 +0000 2021The reason I ask is that I am writing something about the subject & I'd rather not re-invent-the-wheel wrt to notation. I checked a number of sources, but none of them were particularly rigorous & the notation wasn't particularly good.
Fri Aug 06 14:09:46 +0000 2021@ucdmrt If it works the way they describe, it actually would only be a relatively minor change for me. Just a different source of noisy peptide sequence data files.
Fri Aug 06 14:05:44 +0000 2021What is the best paper on the mathematical basis of MS/MS based peptide identification? Note: hand-waving arguments that involve mystery steps or "common sense" don't count.
Fri Aug 06 12:55:04 +0000 2021Rather anemic, patchy aurora australis today 🔗
Fri Aug 06 11:50:28 +0000 2021SRSF9:p.Y35F chr 12:g.120469506T>A, rs145350287 (all tissue Y:F 0.990:0.010) vaf=2%, Δm=-15.9949, VAF by population group: african 0%, american 2%, east asian 0%, european 3%, south asian 0%.
Fri Aug 06 11:50:28 +0000 2021SRSF9:p, θ(max) = 67. aka SRp30c, SFRS9. Common in MHC class 1 experiments; class 2 peptides observed in some tissues. Abundant in all tissues & cell lines, but not in fluids.
Fri Aug 06 11:50:27 +0000 2021>SRSF9:p, serine and arginine rich splicing factor 9 (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 5×K+acetyl; 8×K+GGyl; 1×K+SUMOyl; 1×R+dimethyl; 16×S, 2×T, 14×Y+phosphorylyl; SAAVs: Y35F (2%); mature form: (2-221) [45,220×, 328 kTa]. #ᗕᕱᗒ 🔗
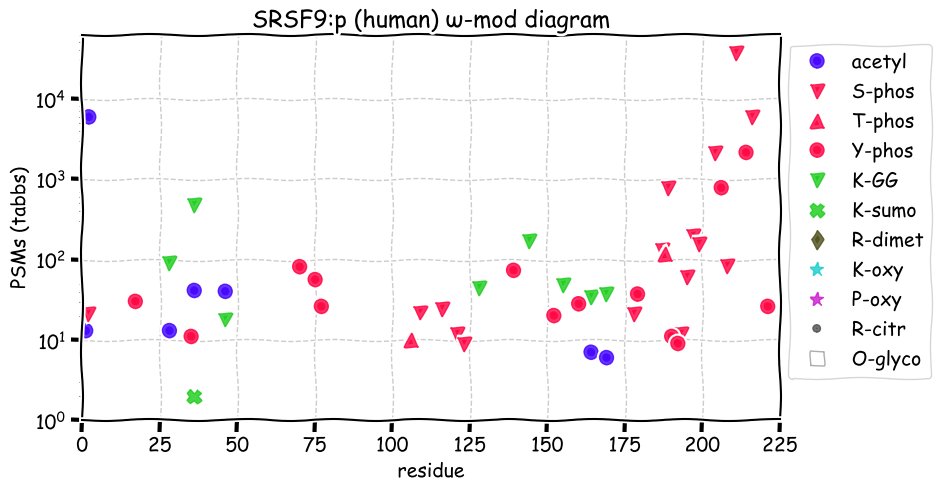
Thu Aug 05 14:58:30 +0000 2021If you want an interesting bacterial data set to look at, try PXD025119. The organism (T. halophilus) is rarely examined by proteomics, but it is of significant value in the food biz. It has a well annotated proteome & the data is good quality. From: 🔗
Thu Aug 05 14:42:53 +0000 2021@AlexUsherHESA I think that returns us quite nicely to your point regarding the avoidance of angering loud-mouthed jerks who are within your own party's apparatus.
Thu Aug 05 14:26:08 +0000 2021@AlexUsherHESA True, but the riding association can easily nominate some one else who does share their point of view.
Thu Aug 05 14:22:12 +0000 2021@AlexUsherHESA At least here in Manitoba, these concerns are also concentrated in specific ridings, amplifying their concerns to an existential level for the affected MP/MLA.
Thu Aug 05 12:13:26 +0000 2021ADAR:p.K384R chr 1:g.154601491T>C, rs2229857 (all tissue K:R 0.509:0.491) vaf=70%, Δm=28.0061, VAF by population group: african 37%, american 76%, east asian 67%, european 67%, south asian 77%.
Thu Aug 05 12:13:26 +0000 2021ADAR:p, θ(max) = 68. aka ADAR1, IFI4, G1P1. Found in MHC class 1 experiments; very rare in class 2. Abundant in all tissues & cell lines.
Thu Aug 05 12:13:26 +0000 2021>ADAR:p, adenosine deaminase RNA specific (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 23×K+acetyl; 33×K+GGyl; 12×K+SUMOyl; 1×R+dimethyl; 49×S, 39×T, 6×Y+phosphorylyl; SAAVs: K384R (70%); mature form: (1-1226) [51,554×, 568 kTa]. #ᗕᕱᗒ 🔗
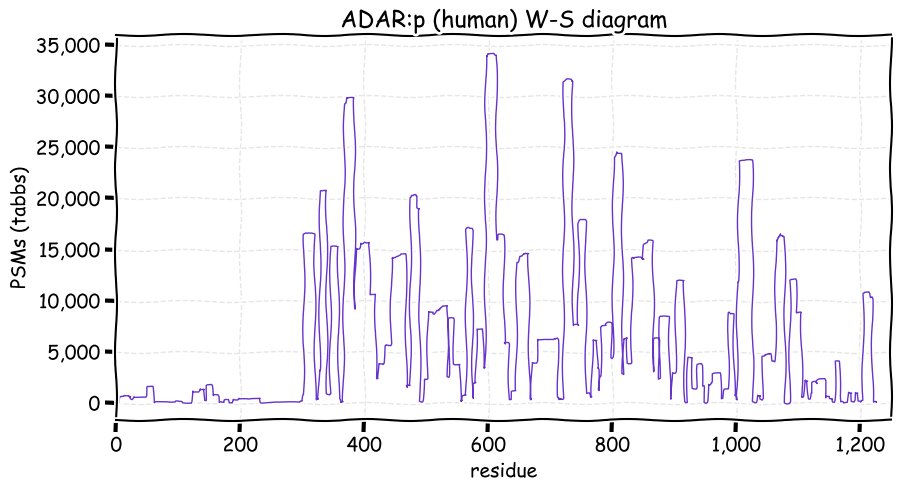
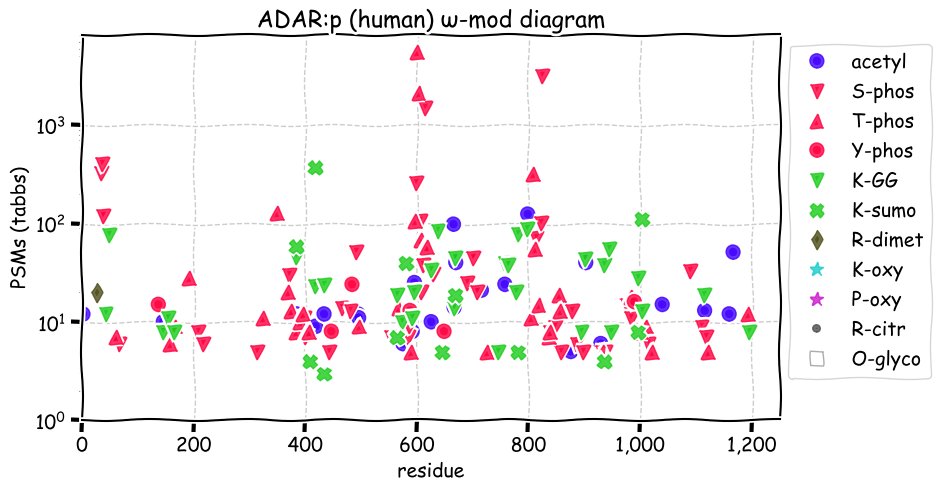
Wed Aug 04 18:44:40 +0000 2021@slashdot QA is hard, but QC is even harder.
Wed Aug 04 16:40:16 +0000 2021@chrashwood While this is a nifty feature for someone like me who is interested in the precision & accuracy of PTM assignments, it may be a significant disadvantage if you are using it in an expt looking for peptides that have been separately enriched for those mods.
Wed Aug 04 16:34:43 +0000 2021@chrashwood Note that for this particular kit, the 1st fraction is also enriched in mods that make a peptide more acidic. For the run above, the ratio of PSMs in f1/f8 with:
Q->pyro = 605/34;
protein N-term. acetyl = 1854/110;
N deamidation = 1144/164 &;
S,T,Y phosphoryl = 204/13.
Wed Aug 04 14:42:58 +0000 2021@chrashwood MDC with high pH first, followed by a good fraction mixing scheme & then low pH performs very well and ends up being pretty orthogonal.
Wed Aug 04 14:26:46 +0000 2021Calculated peptide pI vs calculated LC retention time (reversed phase low pH gradient) for the PSMs assigned to the first and last fraction from a Pierce high pH separation kit preparation 🔗
Wed Aug 04 12:25:34 +0000 2021KRT32:p.T395M chr 17:g.41462863G>A, rs2071563 (all tissue T:M 0.573:0.427) vaf=38%, Δm=29.9928, VAF by population group: african 43%, american 40%, east asian 34%, european 42%, south asian 24%.
Wed Aug 04 12:25:34 +0000 2021KRT32:p, θ(max) = 75. aka Ha-2, KRTHA2. Not found in MHC class 1 or class 2 experiments. Common in fluids, hair & epithelia. Has significant tryptic peptide overlap with other type I (acidic) keratins.
Wed Aug 04 12:25:34 +0000 2021>KRT32:p, keratin 32 (Homo sapiens) Small subunit; CTMs: T2+acetyl; PTMs: K213+acetyl; SAAVs: Q72R (67%), I171T (4%), S222Y (50%), R280H (18%), R369Q (12%), D371E (5%), T395M (38%), P427T (18%); mature form: (2-448) [34,144×, 705 kTa]. #ᗕᕱᗒ 🔗
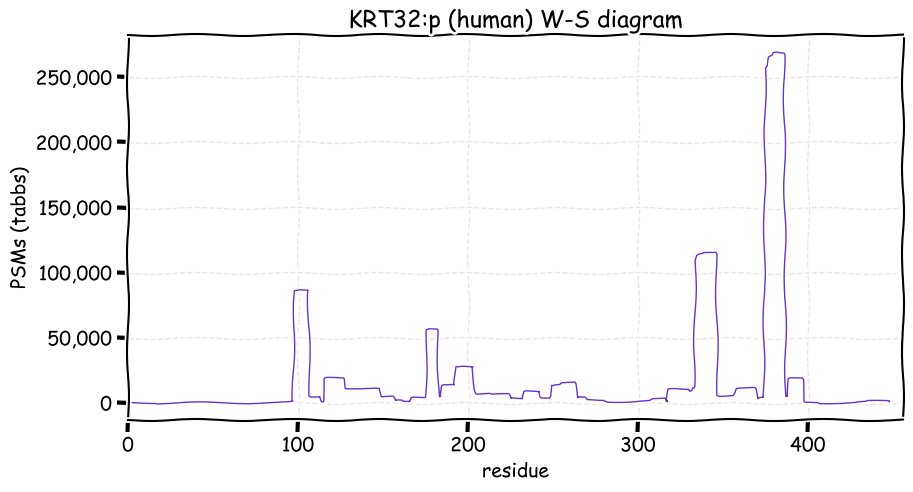
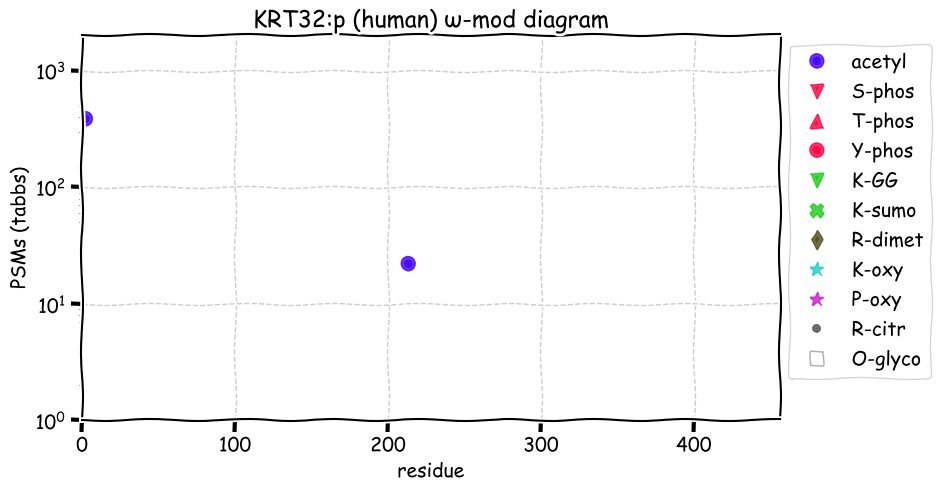
Tue Aug 03 20:41:02 +0000 2021What is the right ontology term to use when a sample says it is from a "cerebral organoid"?
Tue Aug 03 12:25:20 +0000 2021Nlrp5:p.L240P chr 7:23117044T>C, rs16790422 (all tissue L:P 0.573:0.427) vaf=56%, Δm=-16.0313.
Tue Aug 03 12:25:20 +0000 2021Nlrp5:p, θ(max) = 79. aka NALP5, MATER, PYPAF8, PAN11, CLR19.8. Abundant in oocytes & embryonic tissue; not found in cell lines except F9 cells
Tue Aug 03 12:25:19 +0000 2021>Nlrp5:p, NLR family, pyrin domain containing 5 (Mus musculus) Large subunit; CTMs: M1+acetyl; PTMs: none; SAAVs: R87K (40%), L240P (56%); mature form: (1-1095) [258×, 36 kTa]. #ᗕᕱᗒ 🔗
Mon Aug 02 20:05:33 +0000 2021@educhicano @AJ_Brenes I think of both of them as mainly "proteomics as imagined by a mass spectrometrist".
Mon Aug 02 18:08:44 +0000 2021This one just struck me as odd.
Mon Aug 02 18:06:58 +0000 2021YBR225W, Putative protein of unknown function; non-essential gene identified in a screen for mutants affected in mannosylphophorylation of cell wall components 🔗
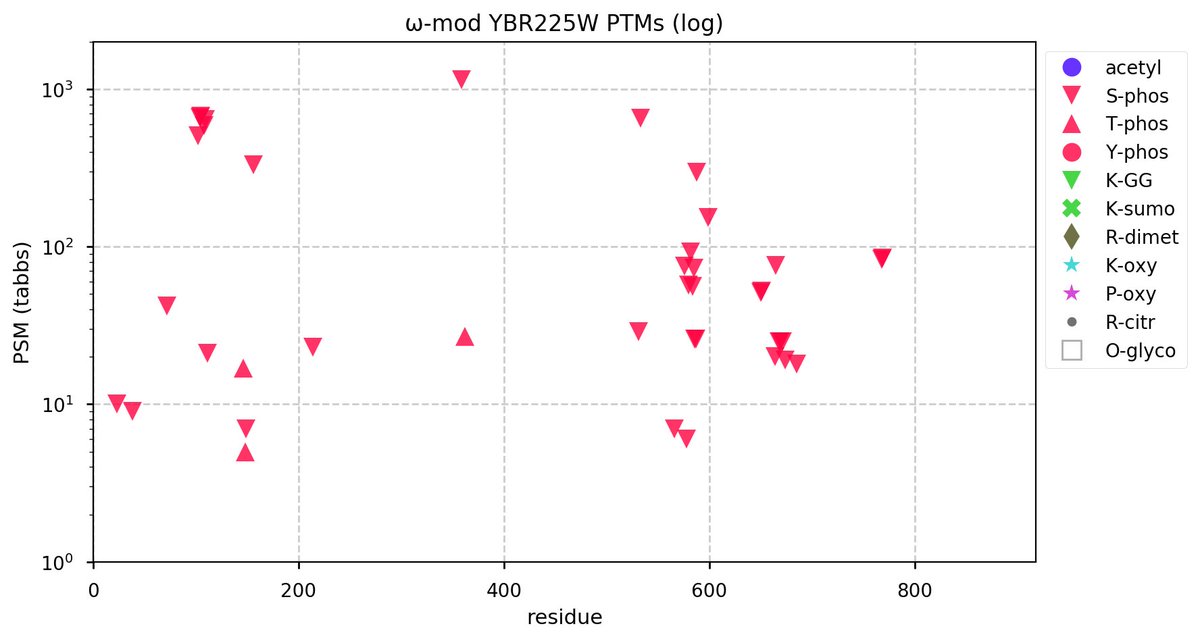
Mon Aug 02 12:25:24 +0000 2021Ebna1bp2:p.S51N chr 4:118478559G>A, rs31979678 (all tissue S:N 0.842:0.158) vaf=38%, vaf=22%, Δm=27.0109. N51 is the corresponding residue in the human proteome.
Mon Aug 02 12:25:24 +0000 2021Ebna1bp2:p, θ(max) = 56. aka 1810014B19Rik, B830003A16Rik, Ebp2, Nobp, p40. Most abundant in liver, brain & embryonic tissue.
Mon Aug 02 12:25:23 +0000 2021>Ebna1bp2:p, EBNA1 binding protein 2 (Mus musculus) >Small subunit; CTMs: M1+acetyl; PTMs: 2×K+GGyl; 6×S, 1×T, 0×Y+phosphoryl SAAVs: I42V (38%), T51I (33%), T248A (4%); mature form: (1-306) [4,230×, 75 kTa]. #ᗕᕱᗒ 🔗
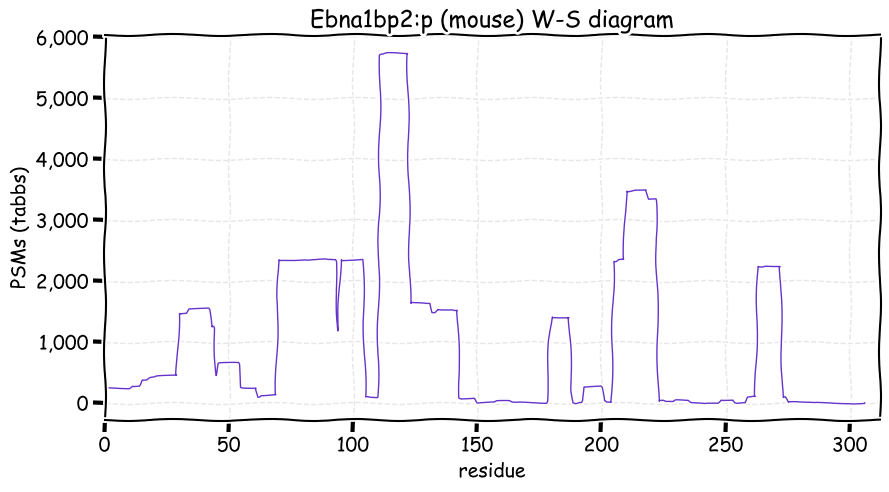
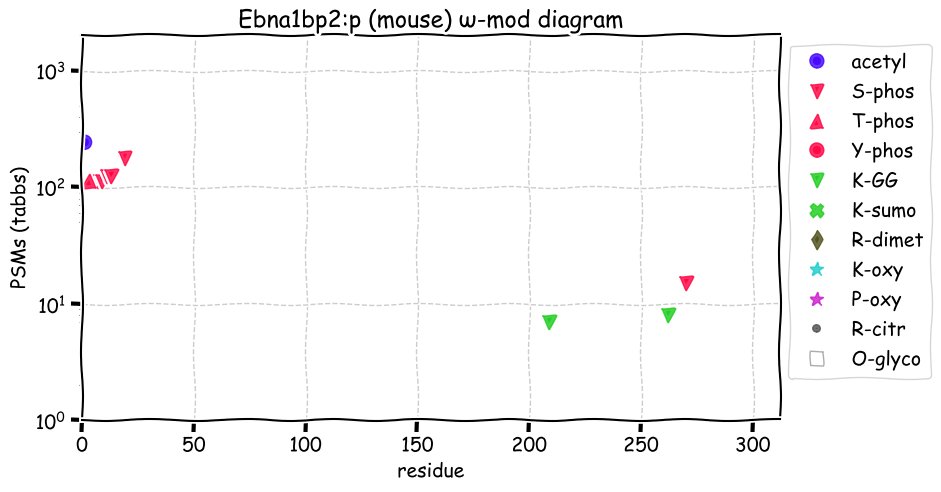
Sun Aug 01 15:01:35 +0000 2021Maybe "surprised" is the wrong word: "disappointed" is more like it.
Sun Aug 01 14:59:07 +0000 2021I am still surprised when people analyze C. elegans proteomics data without including an E. coli proteome in the mix.
Sun Aug 01 13:59:03 +0000 2021Cannot be repeated often enough ... 🔗
Sun Aug 01 12:28:37 +0000 2021Cuba is still on an upwards slope in its current wave of infections 🔗
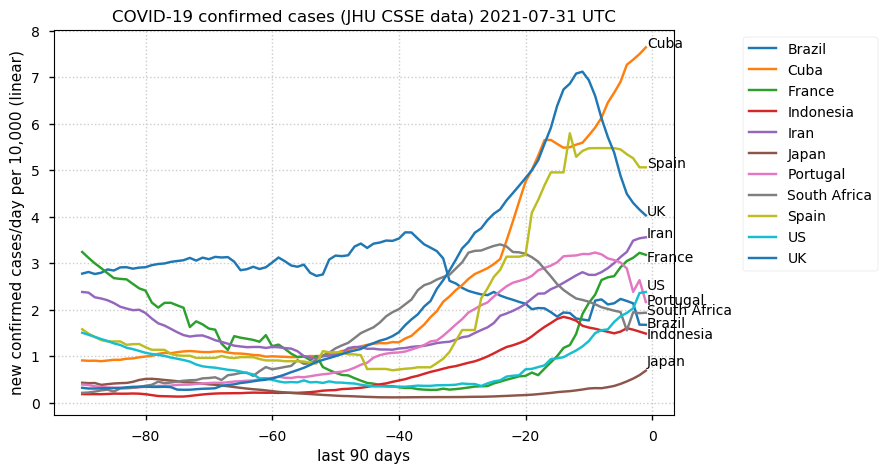
Sun Aug 01 12:03:39 +0000 2021SIRPB1:p.I42V chr 20:g.1611666T>C, rs72484087 (all tissue I:V 0.750:0.250) vaf=38%, Δm=-14.0156, VAF by population group: african 74%, american 52%, east asian 76%, european 51%, south asian 71%.
Sun Aug 01 12:03:39 +0000 2021SIRPB1:p, domains: extracellular (30-370), transmembrane (371-394) & intracellular (395-398).
Sun Aug 01 12:03:38 +0000 2021SIRPB1:p, θ(max) = 57. aka SIRP-BETA-1, CD172b. Observed rarely in MHC class 1 & 2 experiments. Most abundant in urine & CSF extracellular vesicles/exosomes.
Sun Aug 01 12:03:38 +0000 2021>SIRPB1:p, signal regulatory protein beta 1 (Homo sapiens) Small subunit; CTMs: N244, N269, N291, N318+glycosyl; PTMs: K97+GGyl; K342+acetyl; S116+phosphoryl SAAVs: I42V (38%), T51I (33%), T248A (4%); mature form: (30-398) [10,550×, 75 kTa]. #ᗕᕱᗒ 🔗
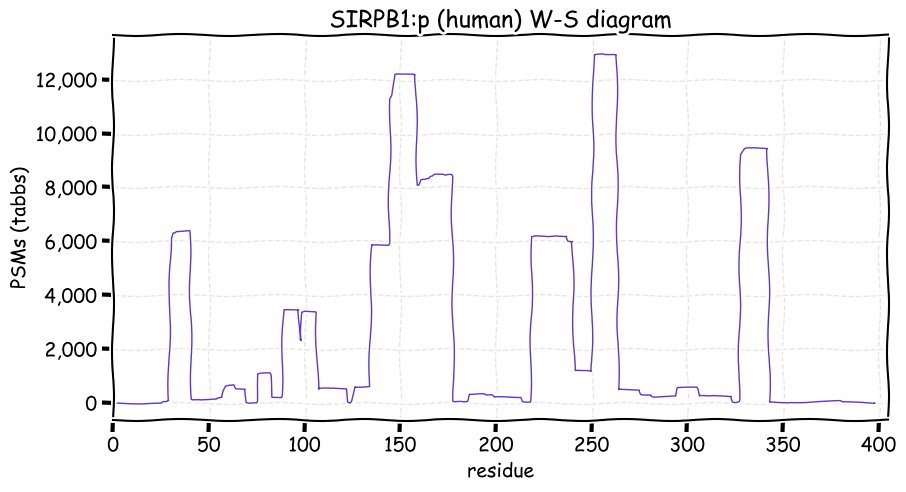
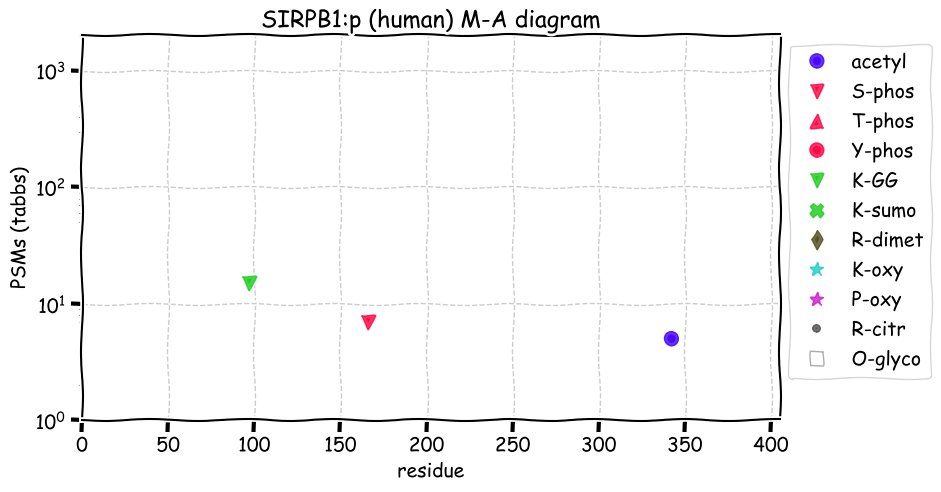
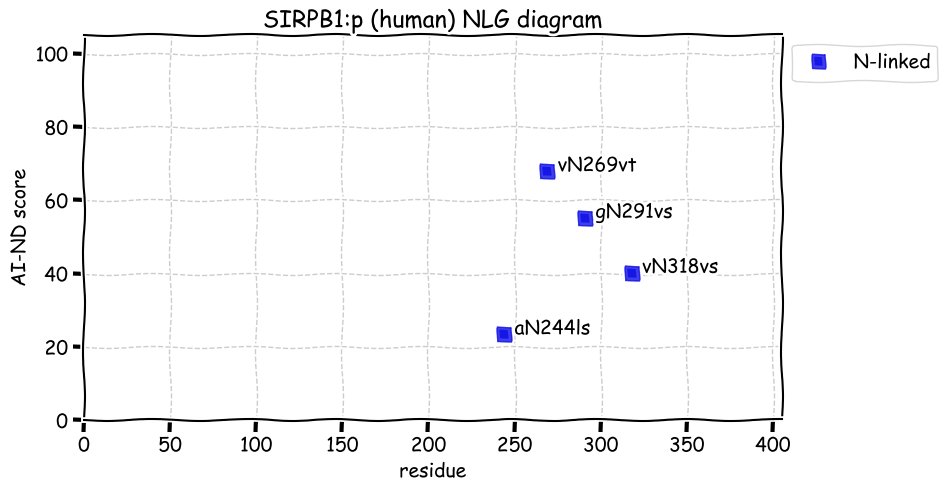
Sat Jul 31 14:23:34 +0000 2021Taken from my balcony this morning: the sun as seen through forest-fire smoke 🔗

Sat Jul 31 13:21:07 +0000 2021The release uses ENSEMBL v. 104 protein sequences. There is a (very) brief explanation of the file format included.
Sat Jul 31 13:19:04 +0000 2021ω-mod is a collection of JSON-lines format information about how often PTMs have been observed in the data sets stored in GPMDB. The initial release has human, mouse & rat info about acetyl (K), phosphoryl (STY), dimethyl (R) & GGyl (K) sites.
🔗
Sat Jul 31 12:37:42 +0000 2021ITGAM:p.M441T chr 16:g.31278075T>C, rs1143680 (all tissue M:T 0.938:0.062) vaf=11%, Δm=-29.9928, VAF by population group: african 19%, american 7%, east asian 0%, european 13%, south asian 4%.
Sat Jul 31 12:37:42 +0000 2021ITGAM:p, domains: extracellular (17-1105), transmembrane (1105-1128) & intracellular (1129-1152).
Sat Jul 31 12:37:41 +0000 2021ITGAM:p, θ(max) = 63. aka MAC-1, CD11b, CR3A, CD11B. Observed in MHC class 2 tissue experiments & to a lesser extent in class 1. Found in all tissues & cell lines; rare in fluids except saliva.
Sat Jul 31 12:37:41 +0000 2021>ITGAM:p, integrin subunit alpha M (Homo sapiens) Large subunit; CTMs: N240, N391+glycosyl; PTMs: none; SAAVs: M441T (11%), A859V (16%), P1147S (17%); mature form: (17-1152) [13,835×, 100 kTa]. #ᗕᕱᗒ 🔗
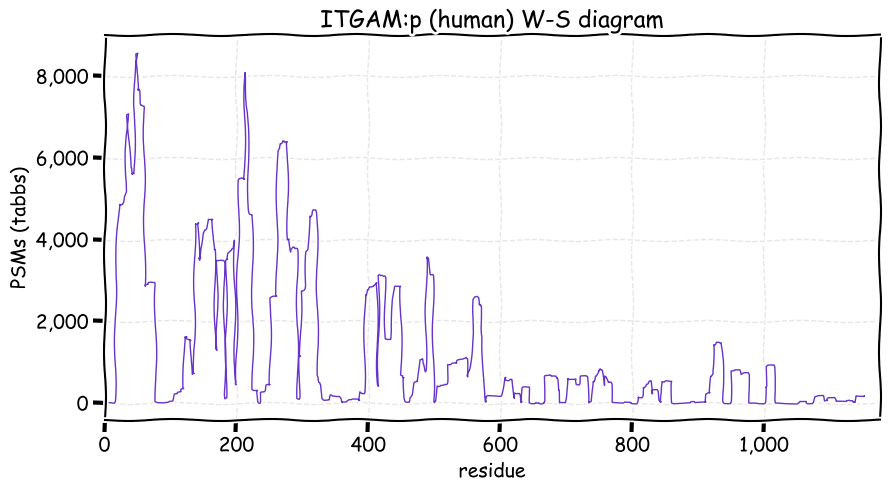
Fri Jul 30 14:42:54 +0000 2021@jamesdotcuff @michaelhoffman I once did an NIH site visit at a supercomputer center asking to build a large new system based on Alpha CPU architecture. When asked if that was a risk, they said absolutely not. Six months after funding ...
Fri Jul 30 12:00:56 +0000 2021SYNGR2:p.L21V chr 17:g.78168677C>G, rs11557900 (all tissue L:V 0.991:0.009) vaf=0.3%, Δm=-14.0156, VAF by population group: african <1%, american <1%, east asian 11%, european <1%, south asian <1%. LV in HeLa cells.
Fri Jul 30 12:00:55 +0000 2021SYNGR2:p, θ(max) = 64. aka none. Observed in MHC class 1 & 2 experiments. Found in all tissues & cell lines; rare in fluids except urine extracellular vesicles. Transmembrane domains: (26–46), (73-93), (105-125) & (47-167).
Fri Jul 30 12:00:55 +0000 2021>SYNGR2:p, synaptogyrin 2 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: K10+GGyl; S3, Y6, Y181+phosphoryl; SAAVs: L21V (<1%), A28S (5%); mature form: (1-224) [16,524×, 50 kTa]. #ᗕᕱᗒ 🔗
Thu Jul 29 16:08:44 +0000 2021@dtabb73 @ShevanWilkin And if you didn't derivatize the Cys residues, set Cys+O₃ as a fixed modification.
Thu Jul 29 15:57:58 +0000 2021@dtabb73 @ShevanWilkin Check for Q & N deamidation, M & W oxidation, as well as hydroxyproline + hydroxylysine if collagen is present.
Thu Jul 29 15:57:43 +0000 2021@dtabb73 @ShevanWilkin My experience with "ancient" proteins is that when they are really present, they aren't very degraded at all. If they weren't completely isolated from water & air, they would have long since disappeared, so looking for exotic products isn't helpful.
Thu Jul 29 13:54:33 +0000 2021Thanks to everyone who participated in this poll. While I can't say I am surprised by the result, I am disappointed.
Thu Jul 29 12:10:22 +0000 2021ASCC3:p.L146F chr 6:g.100848513G>A, rs9390698 (all tissue L:F 0.855:0.145) vaf=38%, Δm=33.9843, VAF by population group: african 6%, american 41%, east asian 23%, european 45%, south asian 17%. LF in HCT-116 & MCF-10A cells.
Thu Jul 29 12:00:14 +0000 2021ASCC3:p, θ(max) = 55. aka RNAH, ASC1p200, dJ121G13.4, dJ467N11.1, HELIC1. Observed in MHC class 1 experiments. Found in all tissues & cell lines; rare in fluids. An ATP-dependent DNA helicase involved in the removal of DNA alkylation.
Thu Jul 29 12:00:14 +0000 2021>ASCC3:p, activating signal cointegrator 1 complex subunit 3 (H sapiens) Large subunit; CTMs: A2+acetyl; PTMs: 59×K+GGyl; 6×K+SUMOyl; 5×K+acetyl; 20×S, 8×T, 2×Y+phosphoryl; SAAVs: L146F (38%), V1050I (1%), S1995C (65%), I2040V (1%); mature: (2-2202) [22,964×, 165 kTa]. #ᗕᕱᗒ 🔗
Wed Jul 28 19:02:25 +0000 2021While the UK and Holland seem to be passed the hump, Cuba is in tough at the moment after being relatively unscathed for most of the pandemic. 🔗
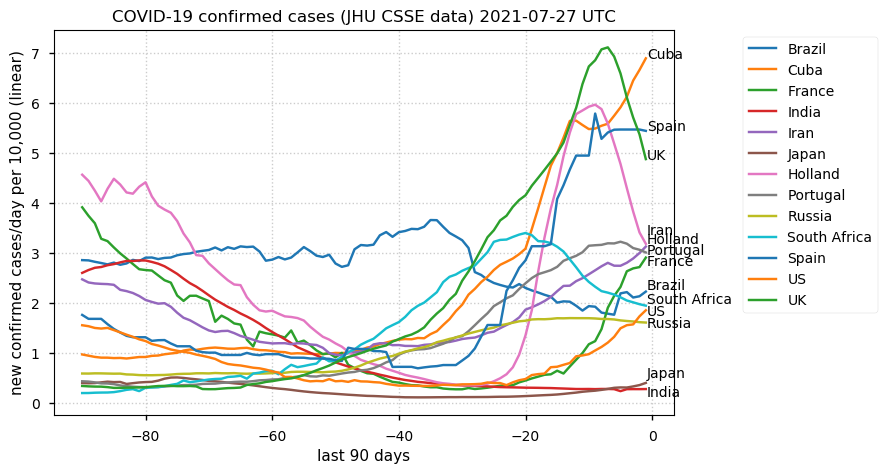
Wed Jul 28 16:39:11 +0000 2021@pwilmarth You have to pick it up and turn it over to get at the USB port. Not the best design ...
Wed Jul 28 13:21:00 +0000 2021Selenocysteine is a DNA-encoded amino acid represented by U in protein sequences. It reacts with C-blocking reagents (like IAA) resulting in the same modification as C. Do you normally set your search engine to test for this modified residue?
Wed Jul 28 13:10:59 +0000 2021@antonhowes Probably something similar to 🔗
Wed Jul 28 12:42:06 +0000 2021"NLM is expanding the size and scope of its IRP to address the growing demand for innovative data science and informatics approaches in biomedicine." 🔗
Wed Jul 28 11:57:56 +0000 2021HCFC1:p, I honestly don't see how having this #AlphaFold structure (🔗) available helps with anything.
Wed Jul 28 11:57:56 +0000 2021HCFC1:p has at least 6 internal peptide backbone cleavage sites that are thought to modulate function. The N-terminal and C-terminal chains remain associated following the cleavage.
Wed Jul 28 11:57:55 +0000 2021HCFC1:p.S1164P chr X:g.153954909A>G, rs1051152 (all tissue S:P 0.581:0.419) vaf=39%, Δm=10.0207, VAF by population group: african 74%, american 53%, east asian 78%, european 20%, south asian 62%. PP in MDA-MB-468 cells; SS in JURKAT cells.
Wed Jul 28 11:57:55 +0000 2021HCFC1:p, θ(max) = 58. aka HCF-1, HCF1, CFF, VCAF, MGC70925, PPP1R89, HFC1, MRX3. Observed in MHC class 1 experiments. Found in all tissues & cell lines; rare in fluids.
Wed Jul 28 11:57:55 +0000 2021HCFC1:p, host cell factor C1 (Homo sapiens) Large subunit; CTMs: A2+acetyl; PTMs: 27×K+GGyl; 8×K+SUMOyl; 38×K+acetyl; 63×S, 52×T, 3×Y+phosphoryl; SAAVs: S1164P (39%); mature form: (2-2035) [74,775×, 708 kTa]. #ᗕᕱᗒ 🔗
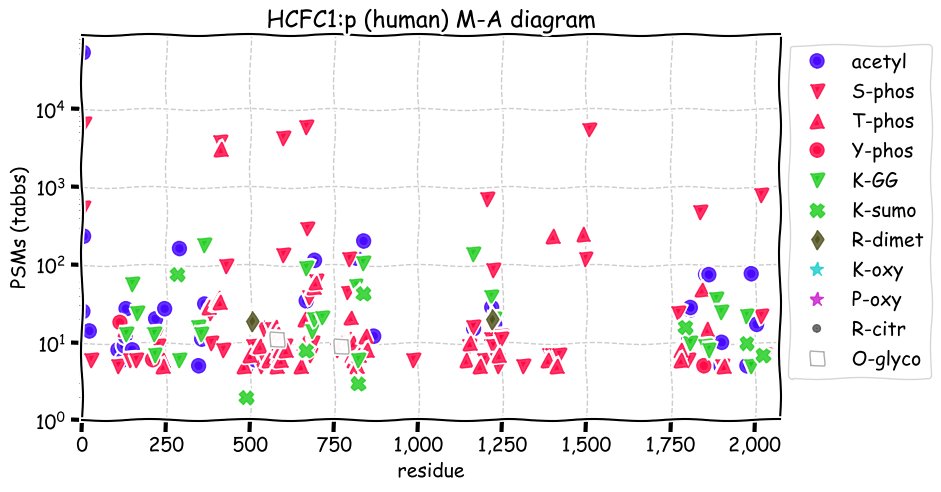
Tue Jul 27 16:18:58 +0000 2021I stopped being interested in the Olympics after the fiasco in Seoul in '88.
Tue Jul 27 15:31:05 +0000 2021@bittremieux @EdHuttlin I would strongly recommend the BioPlex3 data set to anyone who wants to try their algorithms out on a real, large scale experiment.
Tue Jul 27 15:08:43 +0000 2021@PastelBio Good review & a good issue on the not-very-well understood topic of extracellular vesicles (including a paper on MFGE8).
Tue Jul 27 14:55:50 +0000 2021This means shifting the service from a >200 W system over to a <5 W system. All of the electricity here is from hydroelectric dams, so no direct GHG implications, but it does make me feel virtuous (if just for one day).
Tue Jul 27 14:51:27 +0000 2021Just finished moving another service over to an ARM processor from an AMD Ryzen. Nearly done converting the whole shebang.
Tue Jul 27 12:55:57 +0000 2021MFGE8:p.L76M chr 15:g.88907356G>T, rs1878326 (all tissue L:M 0.373:0.627) vaf=62%, Δm=17.9564, VAF by population group: african 80%, american 62%, east asian 46%, european 64%, south asian 62%. MM in CACO-2 cells.
Tue Jul 27 12:55:37 +0000 2021MFGE8:p, θ(max) = 56. aka SED1, EDIL1, BA46, OAcGD3S, HsT19888, MFG-E8, hP47, SPAG10 . Observed in MHC class 1 & 2 experiments in tissue. Abundant in milk, but found in tissue, cell lines, urine & CSF.
Tue Jul 27 12:40:28 +0000 2021MFGE8:p, milk fat globule EGF and factor V/VIII domain containing (Homo sapiens) Small subunit; CTMs: N238+glycosyl; PTMs: 2×K+GGyl; 1×K+SUMOyl; 1×K+acetyl; 9×S, 0×T, 0×Y+phosphoryl; SAAVs: R3S (27%), L76M (62%); mature form: (24-335) [1,068×, 4 kTa]. #ᗕᕱᗒ 🔗
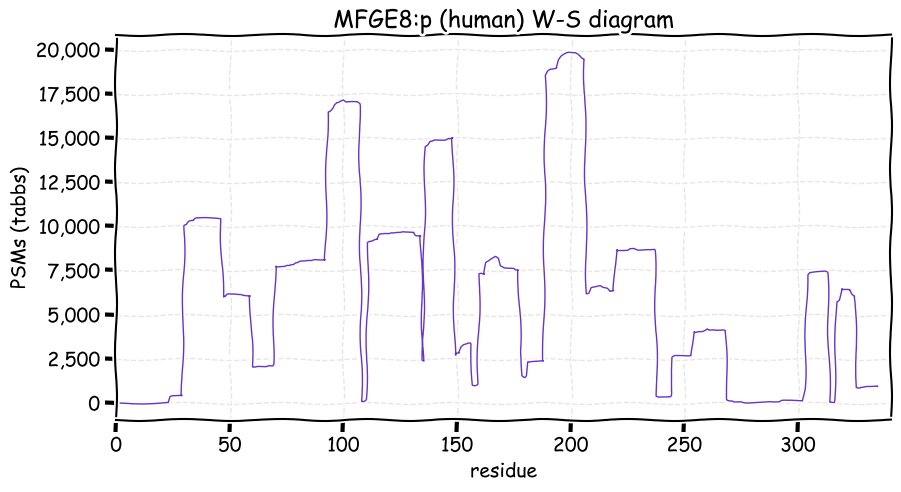
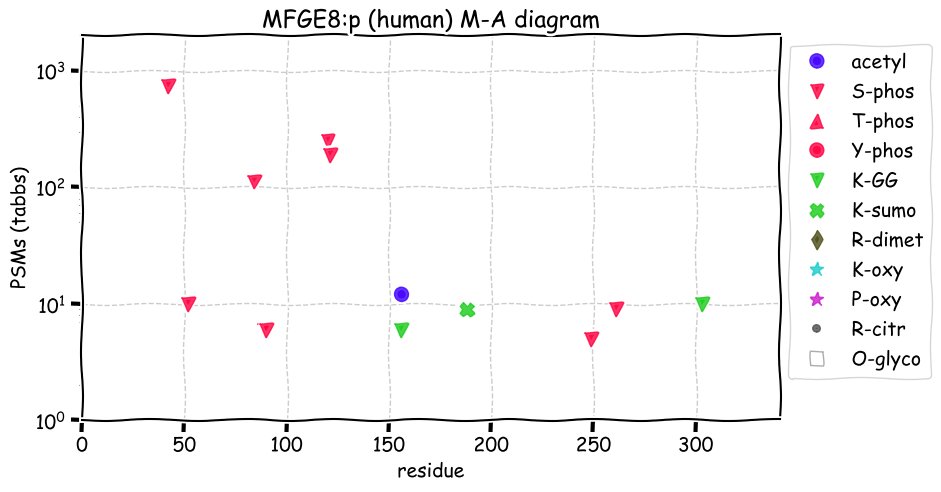
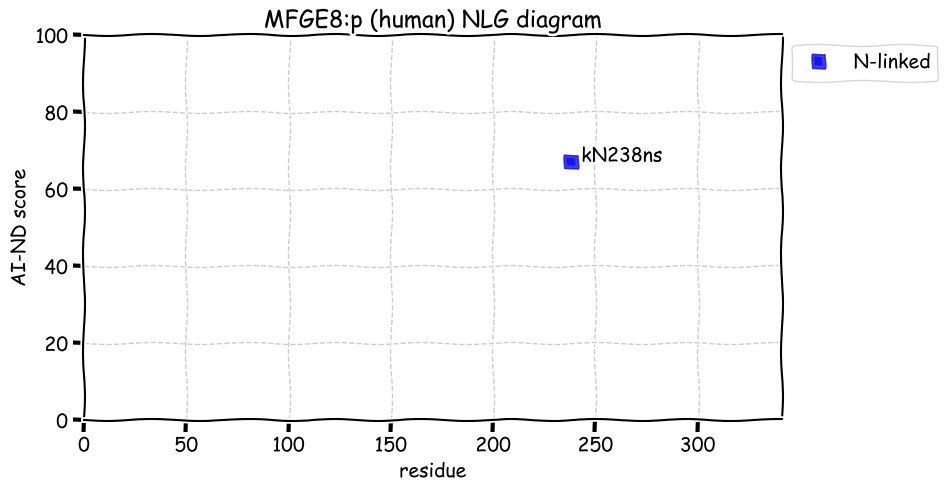
Mon Jul 26 15:09:28 +0000 2021@ProteomicsNews Welcome to the dark side ...
Mon Jul 26 11:57:34 +0000 2021SLAMF7:p, generates a particularly non-physical #AlphaFold cartoon: 🔗
Mon Jul 26 11:57:34 +0000 2021SLAMF7:p.G31S chr 1:g.160748229G>A, rs149217457 (all tissue G:S 0.996:0.004) vaf=<1%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Mon Jul 26 11:57:34 +0000 2021SLAMF72:p, domains: (23-226) extracellular, (227-247) transmembrane, (248-335) intracellular. Glycosylation is on the extracellular domain & phosphorylation on the intracellular domain.
Mon Jul 26 11:57:33 +0000 2021SLAMF7:p, θ(max) = 44. aka CRACC, 19A, CS1, CD319. Observed in MHC class 1 & 2 experiments in tissue. Observed in leukocyte and lymphocyte samples; not detectable in most commonly used cell lines.
Mon Jul 26 11:57:33 +0000 2021SLAMF7:p, SLAM family member 7 (Homo sapiens) Small subunit; CTMs: N172, N204+glycosyl; PTMs: 6×K+GGyl; 0×K+acetyl; 0×S, 0×T, 2×Y+phosphoryl; SAAVs: G31S (1%); mature form: (23-335) [1,068×, 4 kTa]. #ᗕᕱᗒ 🔗
Sun Jul 25 17:18:12 +0000 2021The UK & Netherlands both seem to be over the hump for the current wave, while the US and France are still on the up-slope. Spain & Portugal seem to be adrift at an apex. 🔗
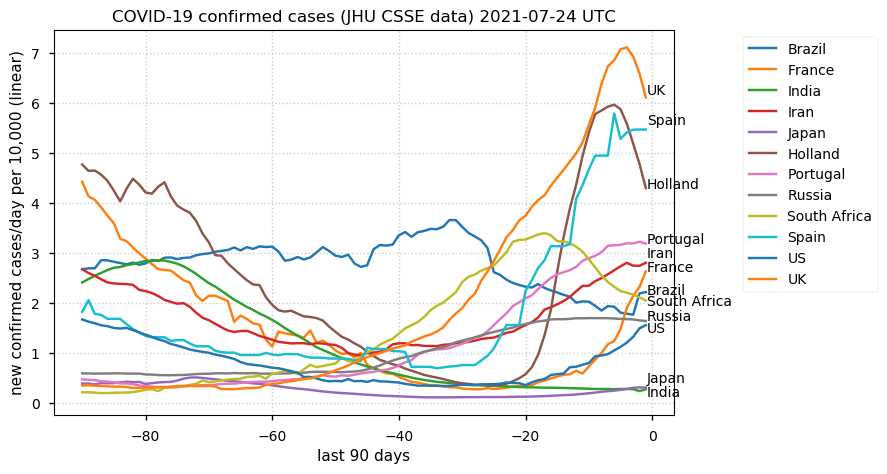
Sun Jul 25 15:14:33 +0000 2021@slavov_n The degree of hype is often inversely proportional to genuine utility in the AI game.
Sun Jul 25 14:46:18 +0000 2021Canadian "high-speed" internet advice:
"Restarting your Shaw modem is often the easiest way to improve the performance of your Internet connection. "
Sun Jul 25 13:23:56 +0000 2021@slavov_n I would say it is a step backwards for structural biology. Rather the good old days of carefully examined structures of suitable domains, pretending that you can generate models from gene translations & simply state that they are "accurate" is just being silly.
Sun Jul 25 12:27:15 +0000 2021ERBB2:p, 🔗 as with many of these #AlphaFold cartoons, creative but not even close.
Sun Jul 25 12:27:15 +0000 2021ERBB2:p.I655V chr 17:g.39723335A>G, rs1136201 (all tissue I:V 0.800:0.200) vaf=20%, Δm=-14.0156, VAF by population group: african 1%, american 14%, east asian 12%, european 25%, south asian 13%.
Sun Jul 25 12:27:14 +0000 2021ERBB2:p, domains: (22,23-652) extracellular, (653–675) transmembrane, (676-1255) intracellular. Glycosylation is on the extracellular domain & phosphorylation and GGylation on the intracellular domain.
Sun Jul 25 12:27:14 +0000 2021ERBB2:p, θ(max) = 67. aka NEU, HER-2, CD340, HER2, NGL. Observed in MHC class 1 & 2 experiments in tissue. Common in tissues & cell lines: rare in fluids, but often present in extracellular vesicles.
Sun Jul 25 12:27:14 +0000 2021>ERBB2:p, erb-b2 receptor tyrosine kinase 2 (Homo sapiens) Large subunit; CTMs: N530, N629+glycosyl; PTMs: 14×K+GGyl; 0×K+acetyl; 26×S, 17×T, 14×Y+phosphoryl; SAAVs: I655V (2%), I655V (20%); mature form: (22,23-1255) [17,234×, 116 kTa]. #ᗕᕱᗒ 🔗
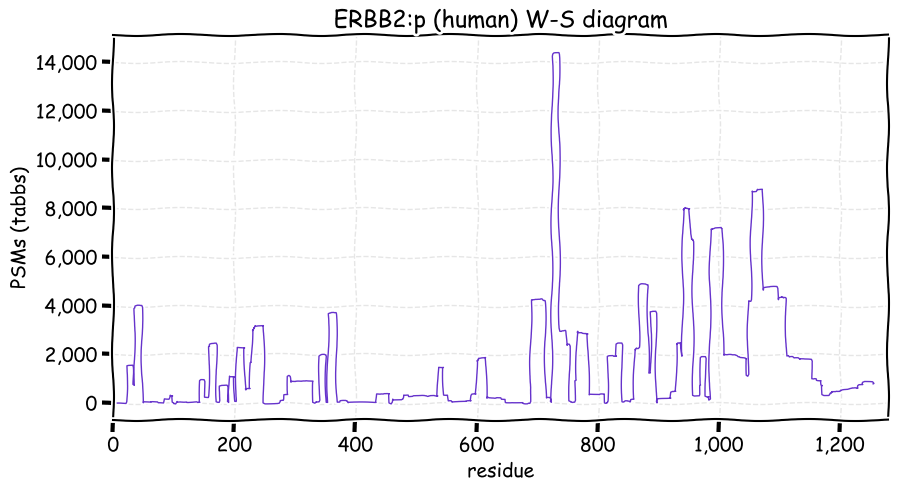
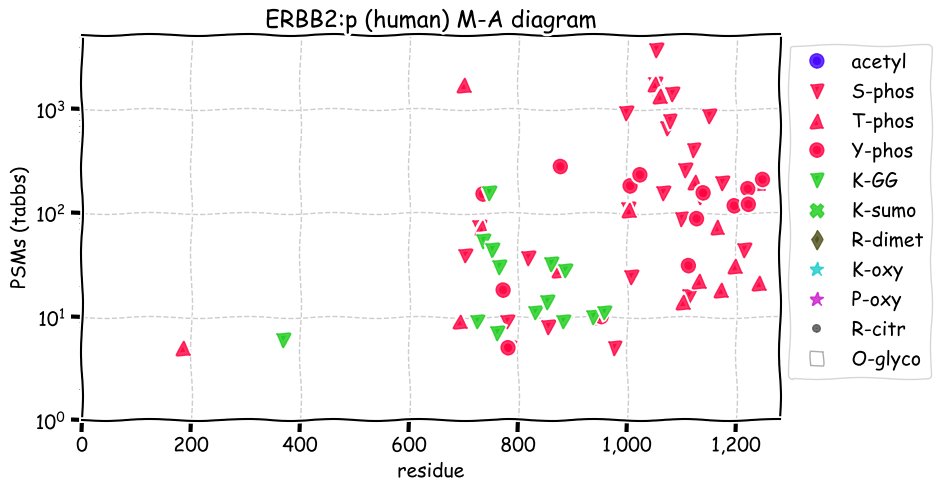
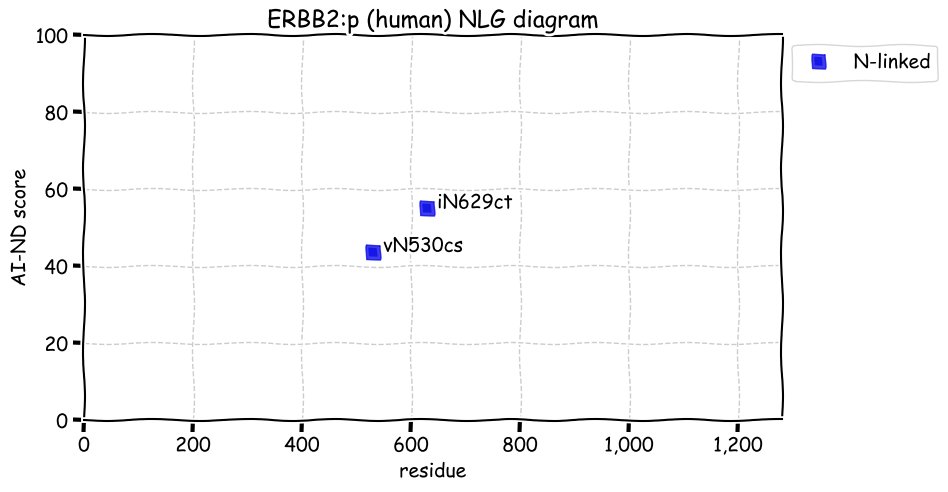
Sun Jul 25 01:12:11 +0000 2021Scripture written in the style of an Experimental Methods section ... 🔗
Sat Jul 24 14:26:42 +0000 2021@YishaiLevin Unfortunately, many of the structures are wrong, but because of the way they are rendered, non-experts are left with the impression that they may well be an "accurate", biologically relevant solution (as claimed by the adverts).
Sat Jul 24 12:25:07 +0000 2021PITRM1:p is believed to be the primary metallopeptidase for the degradation of the rather large peptides (can be >100 aa) left over after cleaving the targeting sequences from proteins imported into the mitochondrial matrix.
Sat Jul 24 12:08:38 +0000 2021PITRM1:p.Q1038R chr 10:g.3138035T>C, rs6901 (all tissue A:T 0.143:0.857) vaf=70%, Δm=28.0425, VAF by population group: african 69%, american 65%, east asian 74%, european 72%, south asian 72%.
Sat Jul 24 12:08:37 +0000 2021PITRM1:p, θ(max) = 69. aka MP1, KIAA1104, hMP1, PreP. Observed in MHC class 1 experiments. Common in tissues & cell lines: rarely in fluids.
Sat Jul 24 12:08:37 +0000 2021PITRM1:p, SAAVs: Q8R (5%), L145V (5%), I328V (62%), A397V (37%), Q516H (14%), F169S (13%), V964I (4%), Q1038R (72%): based on this distribution it is unlikely to find an individual homozygous for the reference sequence.
Sat Jul 24 12:08:37 +0000 2021>PITRM1:p, pitrilysin metallopeptidase 1 (Homo sapiens) Large subunit; CTMs: none; PTMs: 10×K+GGyl; 8×K+acetyl; 0×S, 0×T, 2×Y+phosphoryl; mature form: (42-1038) [7,818×, 292 kTa]. #ᗕᕱᗒ 🔗
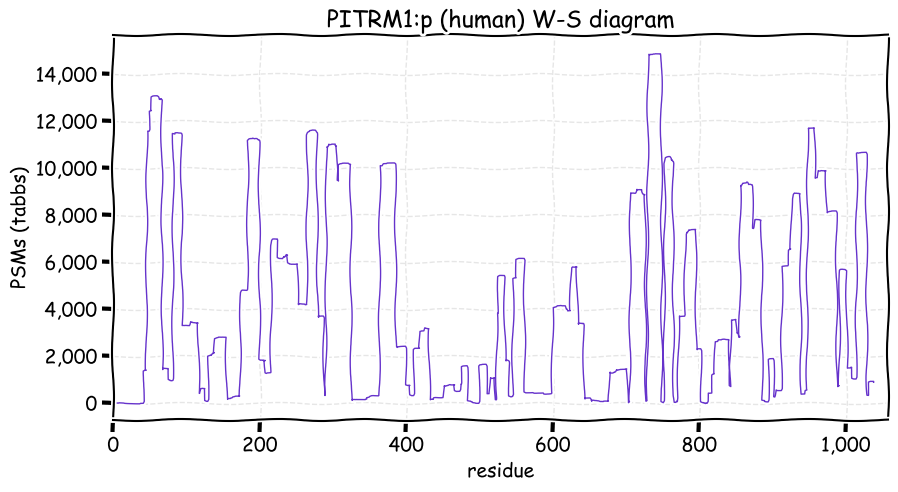
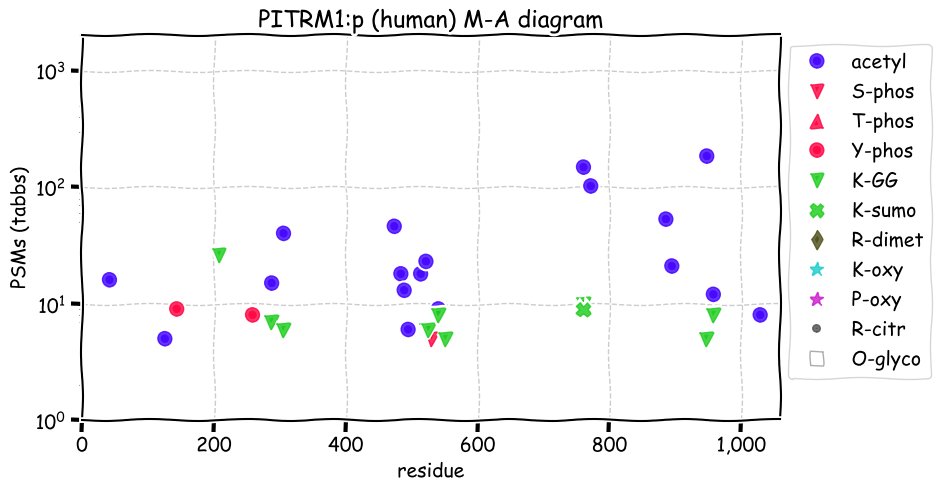
Sat Jul 24 11:54:51 +0000 2021@theoneamit PXD023446 is correct, but the data is stored in Massive at ftp://massive.ucsd.edu/MSV000086665
Fri Jul 23 17:06:34 +0000 2021It is also excellent for its designed purpose (detecting SAAVs), but hair really tests the limits of what is possible for protein inference.
Fri Jul 23 16:20:51 +0000 2021PXD023446 is a good data set to use as a torture-test any protein inference algorithm.
Fri Jul 23 15:05:40 +0000 2021@MosleyLab At least they still list the experimentally determined structures.
Fri Jul 23 14:23:42 +0000 2021@pwilmarth I can honestly say I have never learned anything about programming from lectures. It is more of a learn-while-you-are-doing-it sort of thing for me.
Fri Jul 23 13:57:43 +0000 2021Argh! SwissProt has replaced its PDB structure viewer with this AF nonsense! 😡
Fri Jul 23 12:23:00 +0000 2021ABCB1:p, 🔗 creative, but hilariously wrong.
Fri Jul 23 12:22:59 +0000 2021ABCB1:p.S893A chr 7:g.87531302A>C, rs2032582 (all tissue S:A 0.667:0.333) vaf=54%, Δm=-15.9949, VAF by population group: african 98%, american 57%, east asian 47%, european 57%, south asian 36%. #ᐯᐸᐱ
Fri Jul 23 12:22:59 +0000 2021ABCB1:p, 12 transmembrane domains: (45–67), (117–137), (187–208), (216–236), (293–316), (331–352), (712–732), (757–777), (833–853), (855–874), (935–957) & (974–995). Phosphodomain (644-675) is cytoplasmic.
Fri Jul 23 12:22:59 +0000 2021ABCB1:p, θ(max) = 51. aka P-gp, CD243, GP170, ABC20, PGY1, MDR1, CLCS. Commonly found in MHC class 1 & 2 peptide experiments. Common in urine extracellular vesicles, but detectable in many tissues & cell lines.
Fri Jul 23 12:22:59 +0000 2021>ABCB1:p, ATP binding cassette subfamily B member 1 (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 10×K+GGyl; 8×S, 0×T, 0×Y+phosphoryl; SAAVs: S893A (54%), S1141T (2%); mature form: (1-1280) [7,818×, 27 kTa]. #ᗕᕱᗒ 🔗
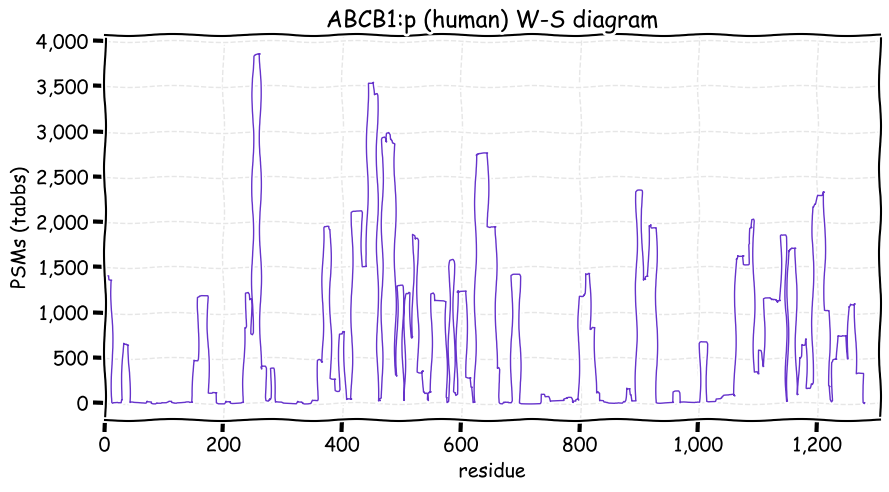
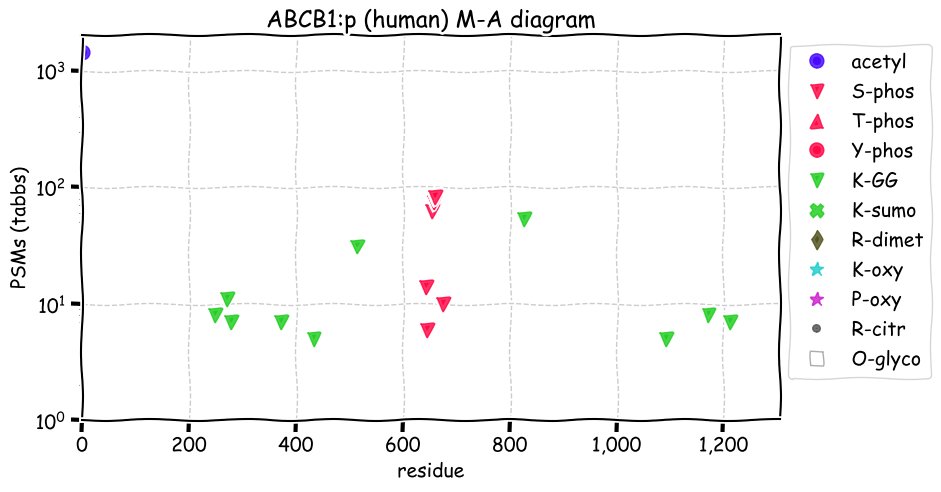
Thu Jul 22 21:21:38 +0000 2021@jwoodgett @MHendr1cks Either that or it wants to be shifted about 500 km SSE to the land of modern air conditioning and cream pies.
Thu Jul 22 21:16:28 +0000 2021It seems sharper than your run-of-the-mill ion exchange expt.
Thu Jul 22 21:15:28 +0000 2021@UCDProteomics, what type of chromatography did you use for the first dimension in PXD018133?
Thu Jul 22 18:58:02 +0000 2021@cwvhogue But, since I want to still be able to use my gmail account tomorrow, I will cease & desist all criticism.
Thu Jul 22 18:56:50 +0000 2021@cwvhogue I get that there are reasons for all the errors & that anyone with an encyclopedic knowledge of proteins should be able to easily spot and understand the mistakes, but the huge hype around the release is missing that type of nuanced consideration.
Thu Jul 22 18:47:00 +0000 2021@cwvhogue I am aware, but it isn't (the heading says "Insulin").
Thu Jul 22 18:34:43 +0000 2021@cwvhogue or insulin 🔗
Thu Jul 22 18:21:19 +0000 2021@cwvhogue & don't get me started on what it does with collagen subunits ...
Thu Jul 22 18:20:24 +0000 2021@cwvhogue It also doesn't seem to know what to do with the amphipathic helices in mitochondrial targeting sequences, e.g. 🔗
Thu Jul 22 18:01:38 +0000 2021@cwvhogue It seems to have issues with proteins that have transmembrane domains. Since it doesn't understand about membrane topology, it gets a bit too creative for my delicate sensibilities.
Thu Jul 22 17:25:22 +0000 2021🔗 not even close
Thu Jul 22 15:56:31 +0000 2021"Protein disulfate bonds were reduced by adding 160mM 2-chlor-acetamide (CAA) (Merck, C0267) and alkylated with 40mM tris (2-carboxyethyl)phosphin (TCEP) (Merck, C4706)."
I don't think I will pay too much attention to the method, as written.
Thu Jul 22 15:42:05 +0000 2021If you are looking for a good quality prokaryote data set using an interesting organism with a well annotated proteome that is NOT involved in disease, PXD019583 (Nitrospira moscoviensis, 🔗) would be a good choice.
Thu Jul 22 15:05:19 +0000 2021@byu_sam Come to a better understanding of what is meant by the term "predatory journals"?
Thu Jul 22 13:49:34 +0000 2021@Smith_Chem_Wisc @bittremieux @pride_ebi Just easily avoided urea & IAA artifacts, but the artifacts end up being 10-25% of the PSMs. It makes it difficult to find anything rare that might be there.
Thu Jul 22 13:30:38 +0000 2021@Smith_Chem_Wisc @bittremieux @pride_ebi I wouldn't put a lot of time examining that particular data set as a source of interesting ids as it has extremely high levels of experimental artifacts.
Thu Jul 22 12:22:25 +0000 2021STEAP4:p, the N-glycosylation of this protein must be post-translational, as is has no ER signal sequence. The protein must be translated in the cytoplasm and incorporated into the ER membrane during (or soon after) elongation.
Thu Jul 22 12:22:25 +0000 2021STEAP4:p.A122T chr 7:g.88283906C>T, rs34741656 (all tissue A:T 0.912:0.088) vaf=14%, Δm=30.0106, VAF by population group: african 5%, american 14%, east asian 0%, european 19%, south asian 6%. #ᐯᐸᐱ
Thu Jul 22 12:22:25 +0000 2021STEAP4:p, θ(max) = 68. aka FLJ23153, TIARP, STAMP2, SchLAH, TNFAIP9. Found in MHC class 2 peptide expts. Common in urine extracellular vesicles, but detectable in many tissues. Transmembrane domains: (202–224), (236–256), (293–313), (342–362), (381–401), (419–439).
Thu Jul 22 12:22:25 +0000 2021>STEAP4:p, STEAP4 metalloreductase (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 6×K+GGyl; N323+glycosyl; SAAVs: G75D (85%), A122T (14%); alternate translation initiation site M13; mature form: (1,14-459) [4,657×, 27 kTa]. #ᗕᕱᗒ 🔗
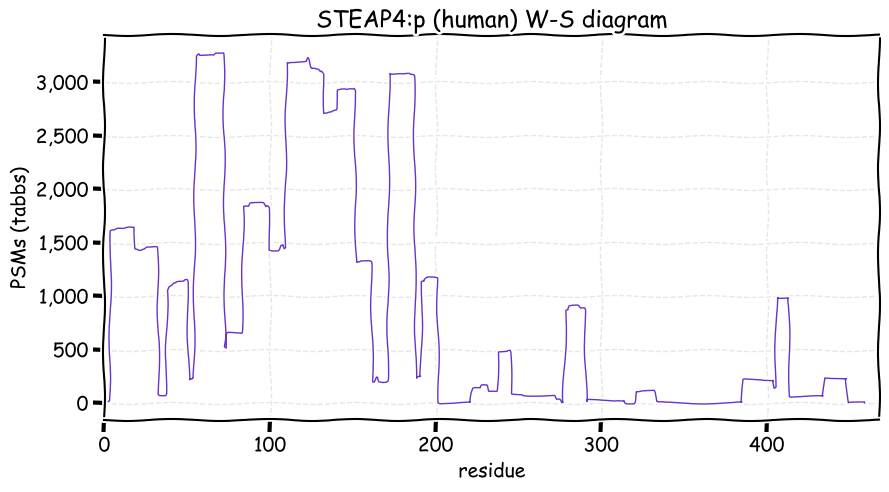
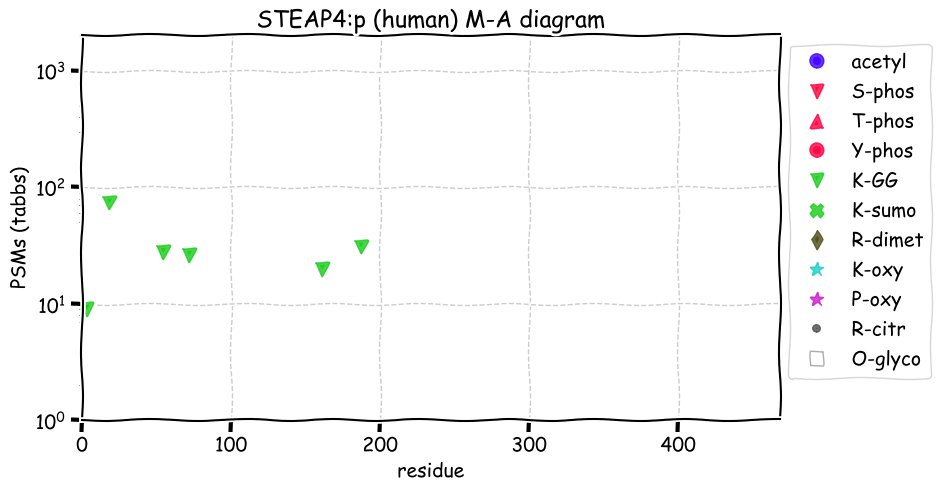
Wed Jul 21 22:13:58 +0000 2021@VATVSLPR I had seen them preparing to film, but I thought it was just one of the Hallmark TV movies they film in this area all the time. I wasn't expecting it in a "Taken/John Wick"-style revenge movie.
Wed Jul 21 20:31:26 +0000 2021I was watching the movie "Nobody" last night & I was surprised when I noticed that the gangster bar featured in the film was the ground floor of the building that also includes my office.
Wed Jul 21 15:28:35 +0000 2021🔗
Note: AIM is more commonly known as CD5L
Wed Jul 21 15:07:59 +0000 2021ITIH4:p, eight of the O-linked glycosylation sites are also phosphorylation sites
Wed Jul 21 13:26:24 +0000 2021@jwoodgett Does this mean you are moving to Vancouver or can you manage things from Toronto?
Wed Jul 21 12:04:42 +0000 2021ITIH4:p.I85N chr 3:g.52827195A>T, rs13072536 (all tissue I:N 0.775:0.225) vaf=25%, Δm=0.9589, VAF by population group: african 20%, american 38%, east asian 32%, european 26%, south asian 12%. #ᐯᐸᐱ
Wed Jul 21 12:04:42 +0000 2021ITIH4:p, θ(max) = 90. aka IHRP, H4P, ITIHL1. Commonly found in MHC class 2 peptide experiments & rarely in class 1. Common in urine, blood plasma & CSF, particularly in extracellular vesicles.
Wed Jul 21 12:04:41 +0000 2021>ITIH4:p, inter-alpha-trypsin inhibitor heavy chain 4 (H sapiens) CTMs: 3×N+glycosyl; PTMs: 4×K+acetyl; 5×S, 6×T+glycosyl; 9×S, 16×T, 0×Y+phosphoryl; SAAVs: I85N (25%), Q669L (18%), P698T (26%), M714I (5%), L791P (4%); mature form: (28,29-930) [36,081×, 1188 kTa] #ᗕᕱᗒ 🔗
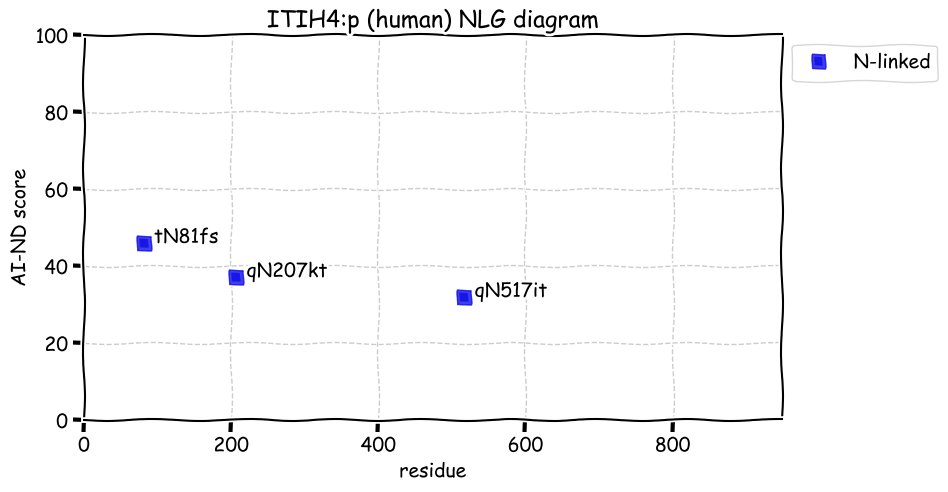
Wed Jul 21 00:49:58 +0000 2021Pretty active aurora borealis oval this evening 🔗
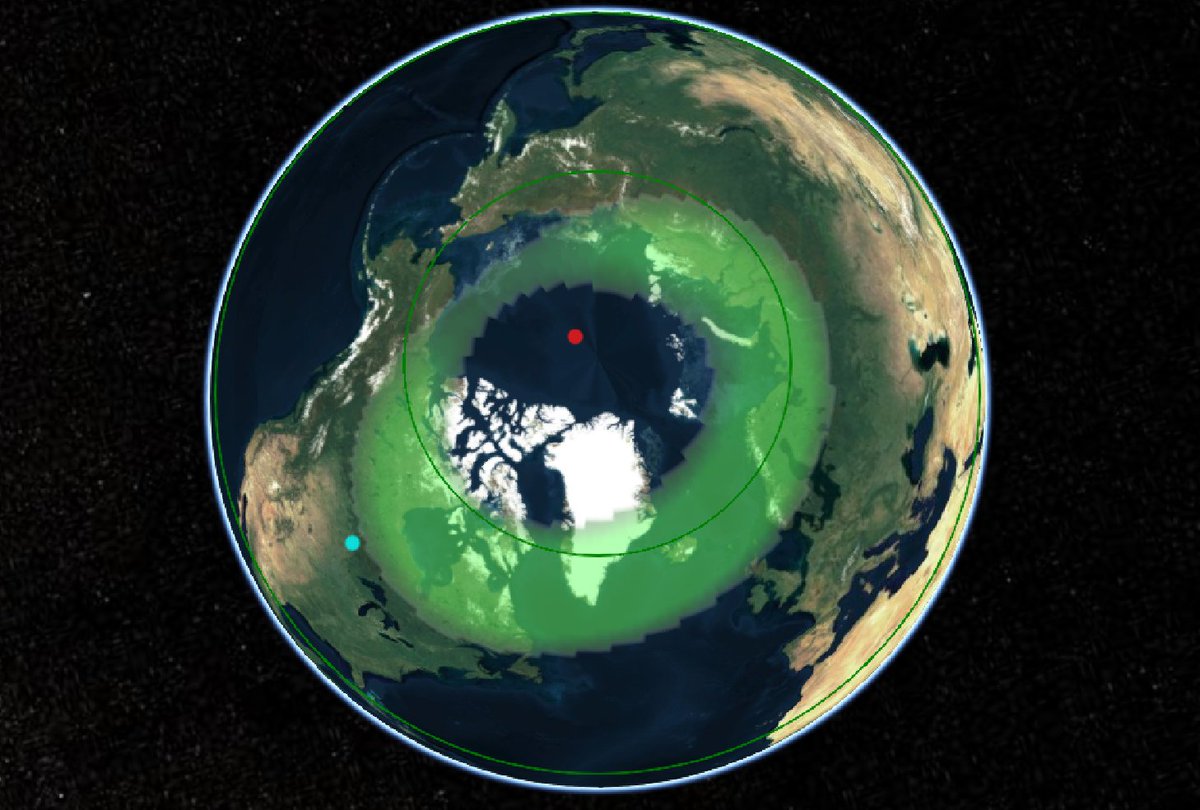
Tue Jul 20 23:42:45 +0000 2021For anyone bored with tryptic peptides, the protocol for protein Lys peracetylation followed by Arg-C type cleavage (🔗) generates a lot of PSMs & good coverage of protein domains frequently absent from conventional trypsin-based experiments.
Tue Jul 20 17:05:25 +0000 2021@pwilmarth At least it has given me the motivation to finally cancel my Prime membership.
Tue Jul 20 15:05:20 +0000 2021@astacus Sometimes the Guardian can be a bit alarmist ...
Tue Jul 20 14:51:33 +0000 2021These silly trips to the ionosphere remind me of summers in St. John's, when wealthy, but not very wise, individuals would try to sail to England in small, unusually designed boats that turned out to be better suited to conditions in the Gulf of Mexico than the North Atlantic.
Tue Jul 20 14:39:55 +0000 2021I guess I must have missed the memo describing when the ionosphere became "space".
Tue Jul 20 14:03:21 +0000 2021This has been a problem with standardized proteomics file formats for as long as I can remember. 🔗
Tue Jul 20 12:40:53 +0000 2021@Simon_Hammann @astacus 🔗
Tue Jul 20 11:59:56 +0000 2021GAK:p.K1265R chr 4:g.849932T>C, rs2306242 (all tissue K:R 0.971:0.029) vaf=4%, Δm=28.0061, VAF by population group: african 1%, american 3%, east asian 9%, european 4%, south asian 2%. #ᐯᐸᐱ
Tue Jul 20 11:59:56 +0000 2021GAK:p, θ(max) = 65. aka DNAJC26. Found in MHC class 1 peptide experiments but in rarely in class 2. common in tissues & cell lines: rarely in fluids. The domain (846-902) has no tryptic cleavage sites: it's residues has only been observed by immunopeptidomics.
Tue Jul 20 11:59:56 +0000 2021>GAK:p, cyclin G associated kinase (Homo sapiens) Large subunit; CTMs: S2+acetyl; PTMs: 7×K+acetyl; 17×K+GGyl; 1×K+SUMOyl; 55×S, 12×T, 12×Y+phosphoryl; SAAVs: P720S (1%), K1265R (4%); mature form: (2-1311) [29,161×, 403 kTa]. #ᗕᕱᗒ 🔗
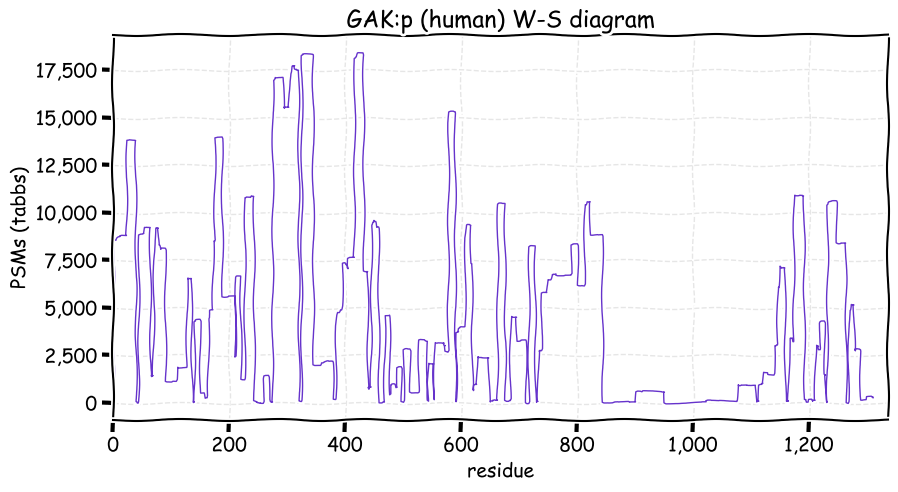
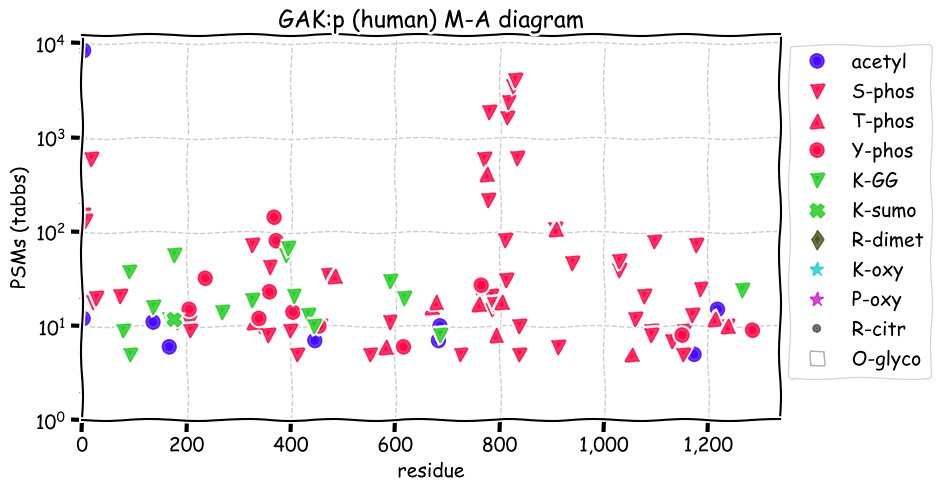
Mon Jul 19 16:10:01 +0000 2021People puzzle me. Why would you choose to use file names like:
191126_***_Mice_Brain_LE_R1_1.raw
for a data set generated exclusively from rat tissue?
Mon Jul 19 15:03:23 +0000 2021@bmss_official Took me a take-or-two to get the Farrokh Bulsara reference
Mon Jul 19 15:00:21 +0000 2021APOBR:p has been extensively studied in macrophages, but it is present in many other types of leukocyte & lymphocyte.
Mon Jul 19 14:29:53 +0000 2021@pwilmarth Many governments research funding programs are focused on the research-via-thesis mechanism, so I would have thought that someone (probably for a Ph.D. thesis in Commerce or Govt. Admin.) would have examined the utility of the underlying idea.
Mon Jul 19 14:25:49 +0000 2021@pwilmarth I was thinking more along the lines of someone examining various types of research problems & being able to show that Ph.D. theses were an effective method of solving those problems.
Mon Jul 19 14:16:06 +0000 2021🔗
Mon Jul 19 12:11:55 +0000 2021APOBR:p.P428A chr 16:g.28496323C>G, rs180743 (all tissue P:A 0.661:0.339) vaf=36%, Δm=-26.0156, VAF by population group: african 41%, american 43%, east asian 10%, european 35%, south asian 22%. PP in THP-1 cells. #ᐯᐸᐱ
Mon Jul 19 12:11:54 +0000 2021APOBR:p, θ(max) = 67. aka APOB48R, APOB100R. Found in MHC class 1 peptide experiments & in class 2 expts in brain, bone marrow + spleen. Found in tissue & cell lines: rarely in fluids.
Mon Jul 19 12:11:54 +0000 2021>APOBR:p, apolipoprotein B receptor (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 1×K+GGyl; 21×S, 19×T, 2×Y+phosphoryl; SAAVs: E61A (5%), R174G (5%), P428A (36%), Q684R (6%); mature form: (1-1097) [8,697×, 44 kTa]. #ᗕᕱᗒ 🔗
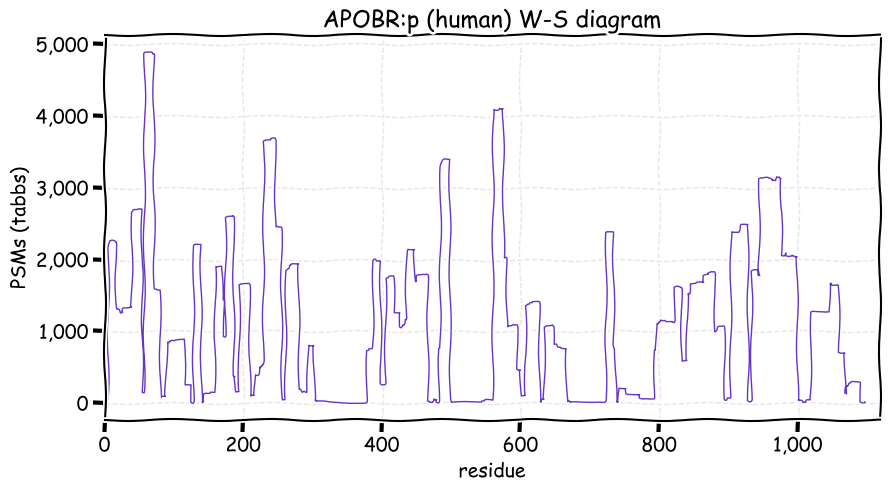
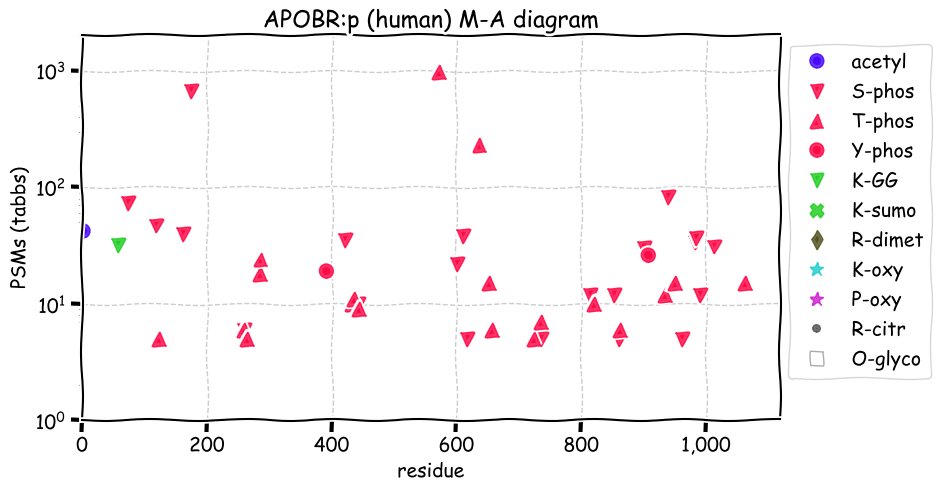
Mon Jul 19 00:56:42 +0000 2021You have to admire their willingness to maintain the use of strictly technical terms, when more vulgar language would be more evocative 🔗
Sun Jul 18 20:30:01 +0000 2021Storms scattered across a fairly uniformly warm US this afternoon 🔗
Sun Jul 18 13:36:13 +0000 2021Has anyone ever done any research into what types of scientific problems can best be solved solely through the generation of Ph.D. theses?
Sun Jul 18 12:59:23 +0000 2021@KentsisResearch No doubt. But since most studies focus on a single type of cancer, with tissue samples obtained in the same manner, the bias toward COL1 (10-20% of PSMs) can be hard to miss.
Sun Jul 18 12:31:46 +0000 2021After spending more time looking at proteomics results from many cancer tissues, it really does appear as though the pathologist is biased towards selecting tissue with significant collagen infiltration. You can usually tell cancer v. normal just by the number of COL1 PSMs.
Sun Jul 18 11:49:55 +0000 2021GFM2:p.N64S chr 5:g.74759384T>C, rs957680 (all tissue N:S 0.902:0.098) vaf=12%, Δm=-27.0109, VAF by population group: african 34%, american 12%, east asian 2%, european 14%, south asian 8%. NS in Hep-2 & HeLa cells. #ᐯᐸᐱ
Sun Jul 18 11:49:55 +0000 2021GFM2:p, θ(max) = 65. aka EFG2, FLJ21661, EF-G2mt. Found in MHC class 1 peptide experiments only. Found in tissue & cell lines: rarely in fluids. GFM1:p & GFM2:p have no tryptic peptide overlap and very different PTM distributions.
Sun Jul 18 11:49:55 +0000 2021>GFM2:p, GTP dependent ribosome recycling factor mitochondrial 2 (Homo sapiens) Midsized subunit; CTMs: none; PTMs: 1×K+acetyl; 11×K+GGyl; 1×K+SUMOyl; 0×S, 0×T, 1×Y+phosphoryl; SAAVs: N64S (12%), R774Q (12%); mature form: (46-779) [10,201×, 42 kTa]. #ᗕᕱᗒ 🔗
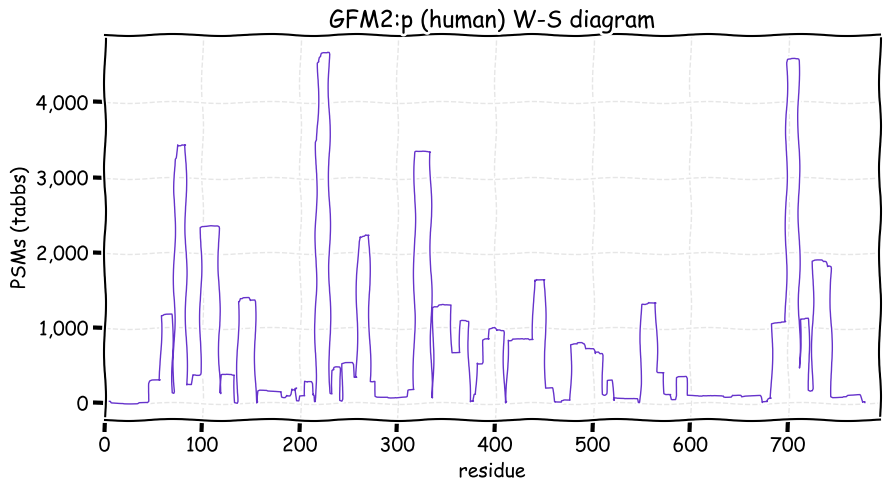
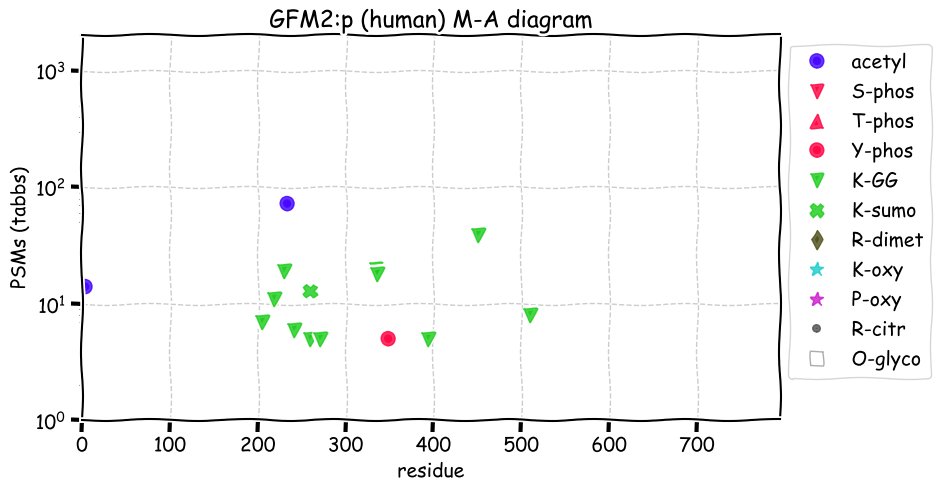
Sat Jul 17 20:23:13 +0000 2021@AlexUsherHESA I think I am being influenced by the fact that when I worked there, the administration (& most of the senior faculty) had a real Caddyshack vibe for me.
Sat Jul 17 19:45:27 +0000 2021@LewisGeer But there will probably be theses written about it for at least 50 years.
Sat Jul 17 19:42:56 +0000 2021@LewisGeer Probably won't really know until the epidemiology of this era gets settled: I'm guessing about a decade from now.
Sat Jul 17 19:37:37 +0000 2021Spent all day trying to figure out what is up with a data set: may be time to call it quits & discard as "non-archival"
Sat Jul 17 14:51:01 +0000 2021@LewisGeer A bit of an anomaly is that the current upturn in cases in north western Europe has not shown up in the fatality stats, unlike South Africa or Indonesia 🔗
Sat Jul 17 14:06:00 +0000 2021Extra points if the term has more than 8 syllables.
Sat Jul 17 13:58:49 +0000 2021Is there a genetics term for an SNV that is predominantly heterozygous in the population?
Sat Jul 17 12:46:33 +0000 2021PXD024126 is officially my current recommended data set for understanding experimental effects associated with using TMTPro, using a Th*rmo instrument. 🔗
Sat Jul 17 12:36:33 +0000 2021The UK seems to be staying just ahead of the Dutch, both at almost 3 times the Brazilian rate. 🔗
Sat Jul 17 12:22:22 +0000 2021@AlexUsherHESA Murray looks more like UBC to me.
Sat Jul 17 11:56:34 +0000 2021GFM1:p.V215I chr 3:g.158649111G>A, rs2303909 (all tissue V:I 0.0.573:0.427) vaf=54%, Δm=14.0156, VAF by population: african 77%, american 53%, east asian 38%, european 54%, south asian 61%. VI in Hep-G2, MCF-7, MCF-10A & SK-MEL-28 cells. II in K-562 cells. #ᐯᐸᐱ
Sat Jul 17 11:56:34 +0000 2021GFM1:p, θ(max) = 74. aka EFGM, GFM, EGF1, mtEF-G1. Found in MHC class 1 peptide experiments & very rarely in class 2. Found in tissues & cell lines: rarely in fluids.
Sat Jul 17 11:56:34 +0000 2021>GFM1:p, G elongation factor mitochondrial 1 (Homo sapiens) Midsized subunit; CTMs: none; PTMs: 22×K+acetyl; 1×K+SUMOyl; 5×K+GGyl; 13×S, 0×T, 1×Y+phosphoryl; SAAVs: K26R (1%), M190L (1%), V215I (54). V664I (1%); mature form: (36-751) [28,352×, 214 kTa]. #ᗕᕱᗒ 🔗
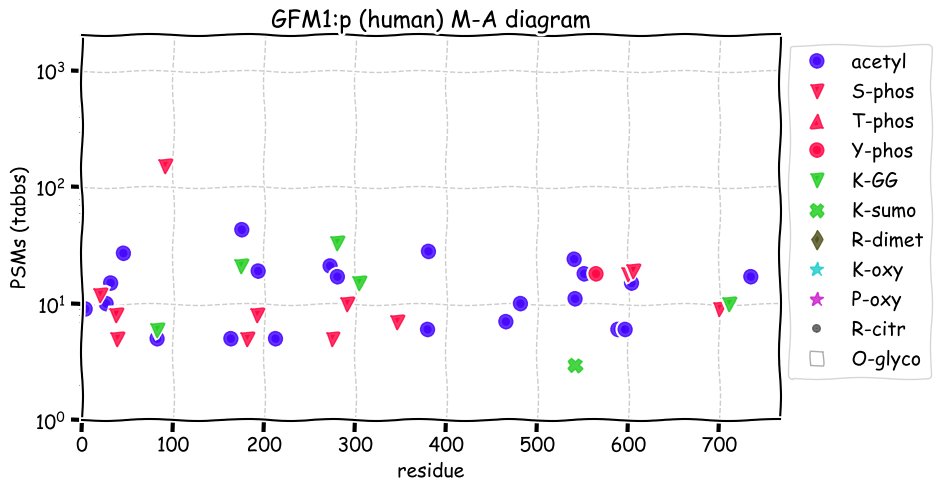
Fri Jul 16 15:26:24 +0000 2021The GPMDB Wiki is back on-line 🔗 It's downtime was just about exactly the length of time it took to get a replacement CPU fan mailed from Shanghai to Winnipeg.
Fri Jul 16 15:20:23 +0000 2021NCL:p (231–274, tryptic peptide, obs. 4243×)
NVAEDEDEEEDDEDEDDDDDEDDEDDDDEDDEEEEEEEEEEPVK
Sometimes it is easy to forget just how non-random real peptide sequences can be.
Fri Jul 16 12:22:13 +0000 2021KNL1:p is clearly being influenced by ubiquitin and/or ubiquitin-like proteins, but the signal is so loud and diffuse that investigating it is probably not possible given current theories & methods.
Fri Jul 16 11:55:42 +0000 2021KNL1:p.R43T chr 15:g.40606445G>C, rs7177192 (all tissue R:T 0.000:1.000) vaf=74%, Δm=-55.0534, VAF by population group: african 73%, american 61%, east asian 35%, european 86%, south asian 68%. SAAV removes a tryptic cleavage site. #ᐯᐸᐱ
Fri Jul 16 11:55:42 +0000 2021KNL1:p, θ(max) = 53. aka D40, AF15Q14, CT29, KIAA1570, hKNL-1, hSpc105, PPP1R55, Spc7, MCPH4, CASC5. Found in MHC class 1 peptide experiments. Found in tissue & cell lines, not fluids.
Fri Jul 16 11:55:42 +0000 2021>KNL1:p, kinetochore scaffold 1 (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 4×K+acetyl; 85×K+GGyl; 41×K+SUMOyl; 47×S, 21×T, 4×Y+phosphoryl; SAAVs: R43T (78%), A486S (65%), M598T (65%), R936G (71%); mature form: (1-2342) [8,196×, 44 kTa]. #ᗕᕱᗒ 🔗
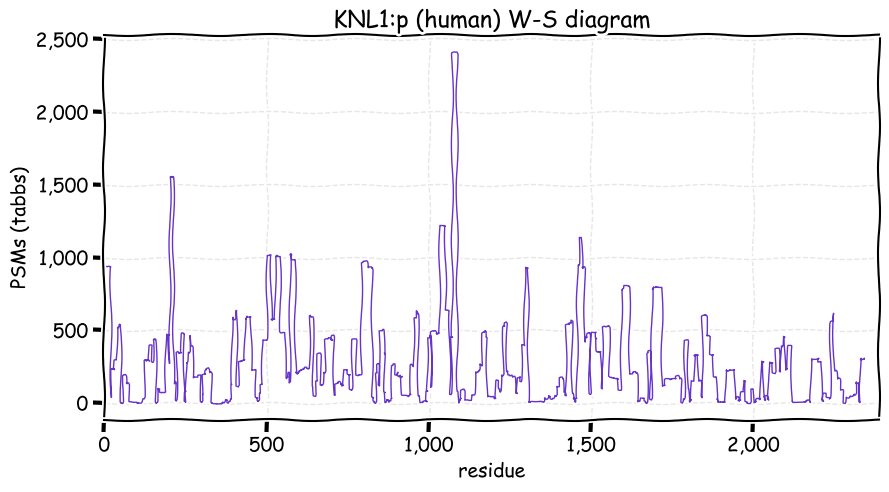
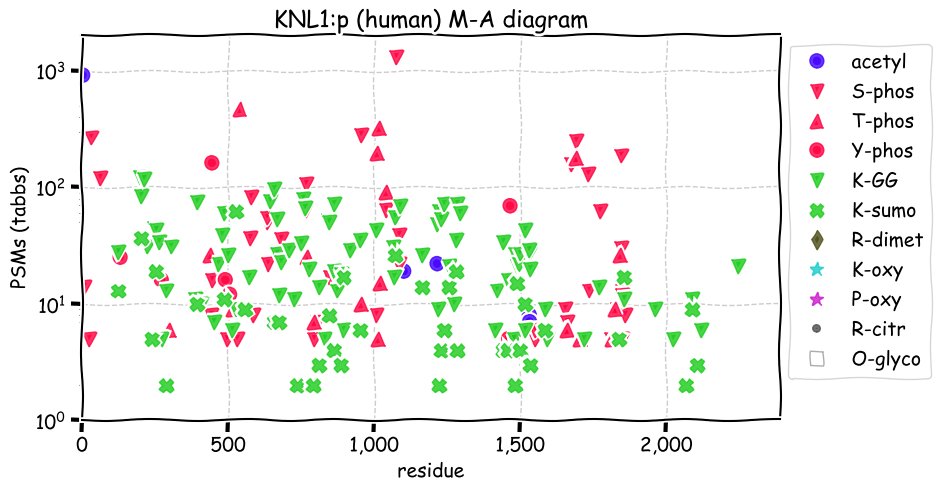
Thu Jul 15 18:49:25 +0000 2021PXD024126 is nice data for understanding experimental artifacts associated with TMTPro derivatization. High resolution MS/MS, good LC & enough of each common artifact to produce confirmatory distributions.
Thu Jul 15 18:10:29 +0000 2021C6:p and C7:p both have SAAVs with VAFs near 50%.
Thu Jul 15 18:10:29 +0000 2021C7:p.S389T chr 5:g.40955459G<C, rs1063499 (all tissue S:T 0.495:0.505) vaf=58%, Δm=14.0157, VAF by population group: african 30%, american 53%, east asian 52%, european 59%, south asian 53%. #ᐯᐸᐱ
Thu Jul 15 18:10:28 +0000 2021C7:p, θ(max) = 79. aka none. Found in MHC class 1 & 2 peptide experiments of most tissues. Commonly found in blood plasma, urine & CSF.
Thu Jul 15 18:10:28 +0000 2021>C7:p, complement component 7 (Homo sapiens) Midsized subunit; CTMs: N202, N754+glycosyl; PTMs: K132, K420, K744+acetyl; T696+glycosyl; SAAVs: S389T (58%), S428T (48%), T587P (19%), T598S (7%), T626P (19%); mature form: (19,23-843) [19,965×, 286 kTa]. #ᗕᕱᗒ 🔗
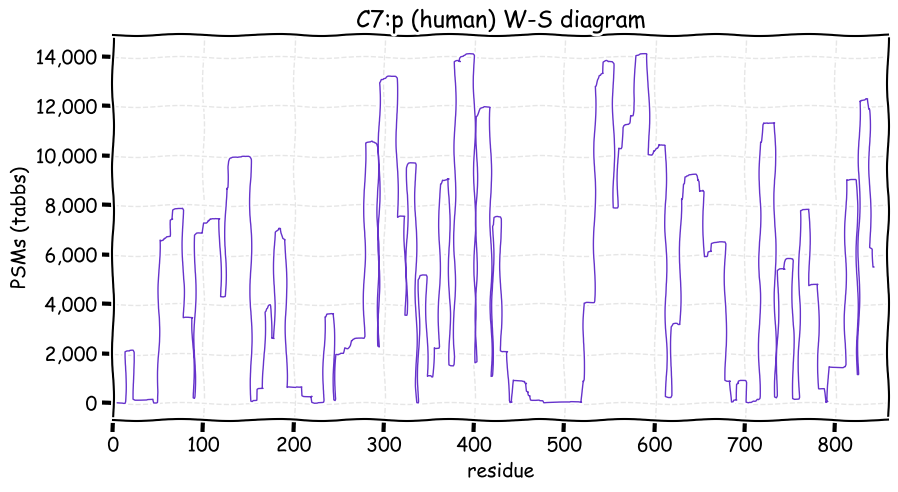
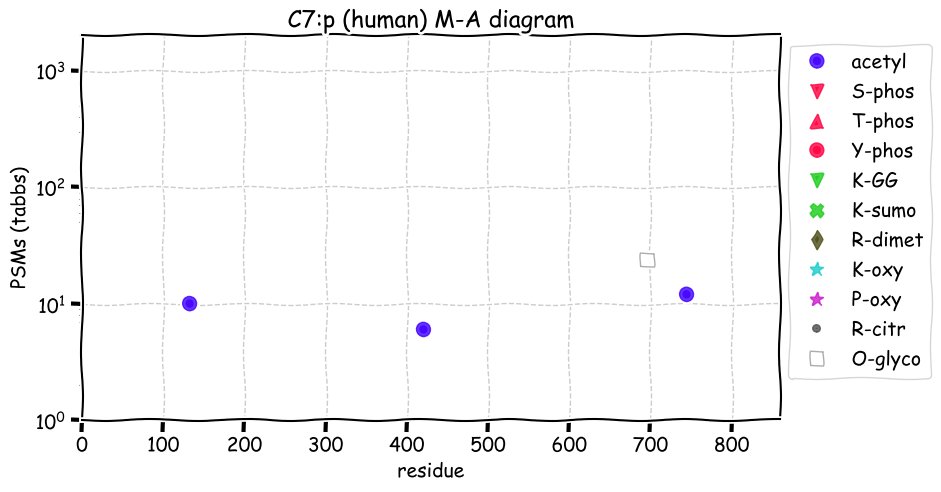
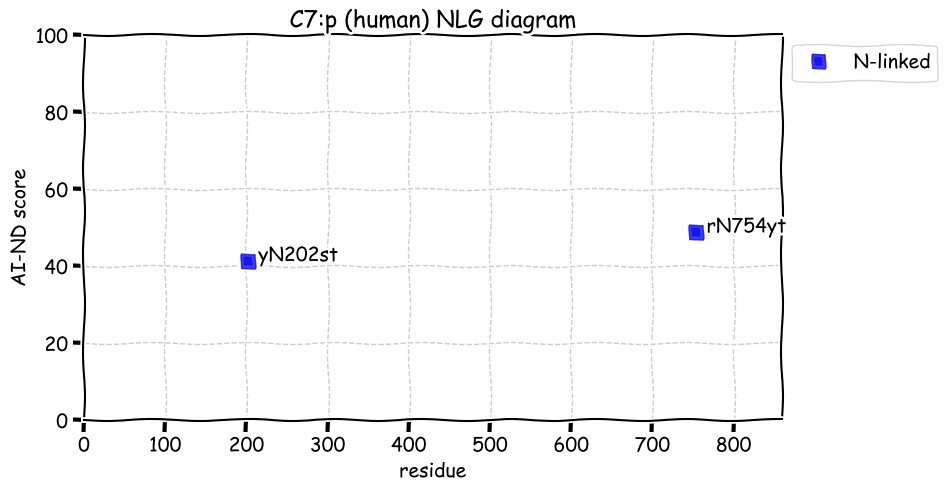
Thu Jul 15 16:57:33 +0000 2021Looking at it for a while, it looks like it is actually an RFC 4122 UUID, rather than an MD5.
Thu Jul 15 15:48:04 +0000 2021The "vaccine passport" QR code in Manitoba is an MD-5 hash of something. Anybody know what "it" is? I have tried the usual suspects.
Thu Jul 15 14:05:21 +0000 2021@TrumanLab My weirdest interviews (in order of descending weirdness):
1. McGill;
2. Indiana University;
3. Perkin Elmer.
Note: this is on a log scale.
Thu Jul 15 12:40:30 +0000 2021Looks like Holland is going to overtake the UK by the end of the week. 🔗
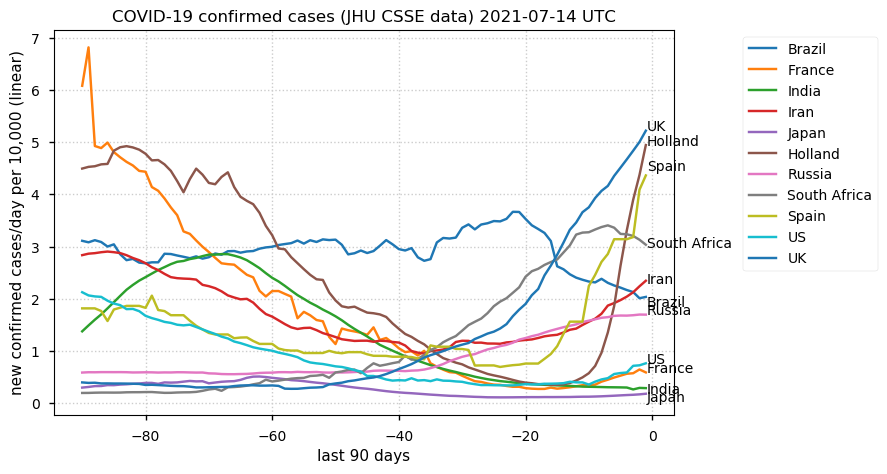
Thu Jul 15 12:20:51 +0000 2021One thing Canadians should keep in mind is that even though the Feds are paying for this research, they do not keep track of what is done & there are very few people in govt that would understand it even if they did.
Thu Jul 15 12:03:30 +0000 2021Listening to CanadaLand & they talked about the wacky "lets get CSIS involved in the grant review process" policy recently announced by the Feds.
Wed Jul 14 15:49:16 +0000 2021Madita Brauer, et al. PXD022830 is an excellent example of adventitious cysteine blocking by 2-mercaptoethanol
Wed Jul 14 14:39:31 +0000 2021Good work Caleb Walker, et al. PXD020854. Interesting data: a few artifacts, but Y. lipolytica has so many sequence differences from any model species that it can serve as a good test for algorithms trained on those models.
Wed Jul 14 12:37:37 +0000 2021@KentsisResearch @ProteomicsNews Is this a setup for one of those classic "Two search engines walk into a bar" jokes?
Wed Jul 14 12:22:28 +0000 2021Fixed a problem that ended up being a weirdo DNS issue, but I have no idea why the fix was a fix. DNS is miraculous when it works, but inexplicable when it is flaky.
Wed Jul 14 11:51:45 +0000 2021C6:p.A119E chr 5:g.41199857G>T, rs1801033 (all tissue A:E 0.465:0.535) vaf=53%, Δm=58.0055, VAF by population group: african 55%, american 55%, east asian 44%, european 65%, south asian 58%. #ᐯᐸᐱ
Wed Jul 14 11:51:45 +0000 2021C6:p, θ(max) = 83. aka none. Found in MHC class 2 peptide experiments & class 1 in liver tissue. Commonly found in blood plasma, urine & CSF.
Wed Jul 14 11:51:44 +0000 2021>C6:p, complement component 6 (Homo sapiens) Midsized subunit; CTMs: N324, N855+glycosyl; PTMs: 4×K+acetyl; 25×S+phosphoryl; SAAVs: A119E (53%), T226I (1%), F299L (3%), Q567H (1%), M616I (1%), K763R (1%); mature form: (22?-934) [18,822×, 276 kTa]. #ᗕᕱᗒ 🔗
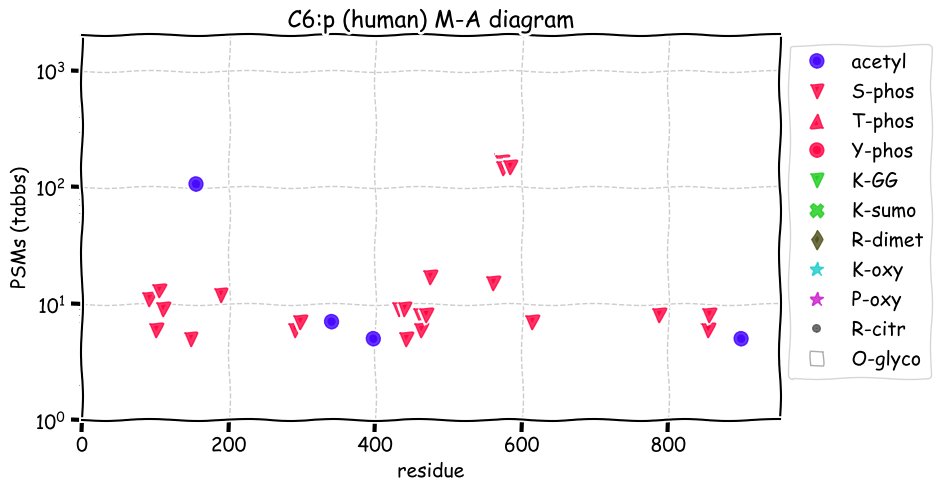
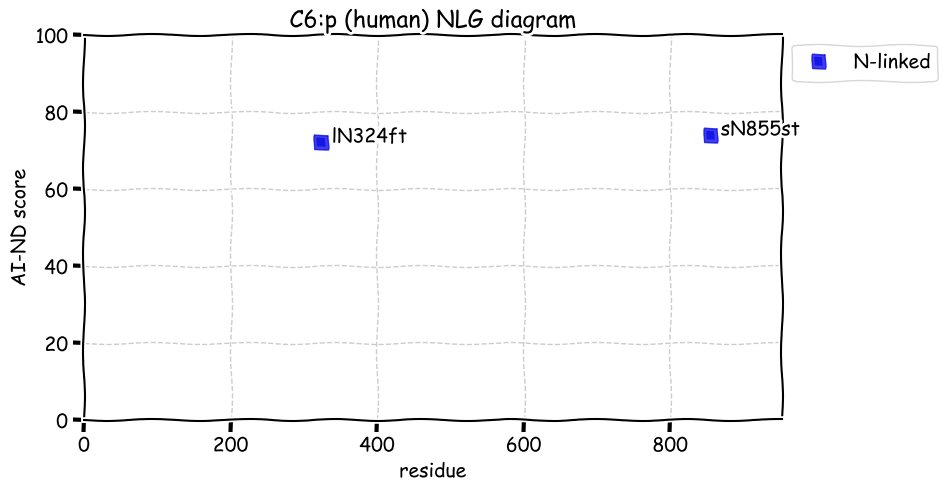
Wed Jul 14 01:07:43 +0000 2021Are there any mouse macrophage cell lines that produce Moloney murine leukemia virus proteins, other than RAW.264.7?
Tue Jul 13 21:26:15 +0000 2021Upper level (250 hPa) air currents over the northern hemisphere are looking a little chaotic at the moment 🔗
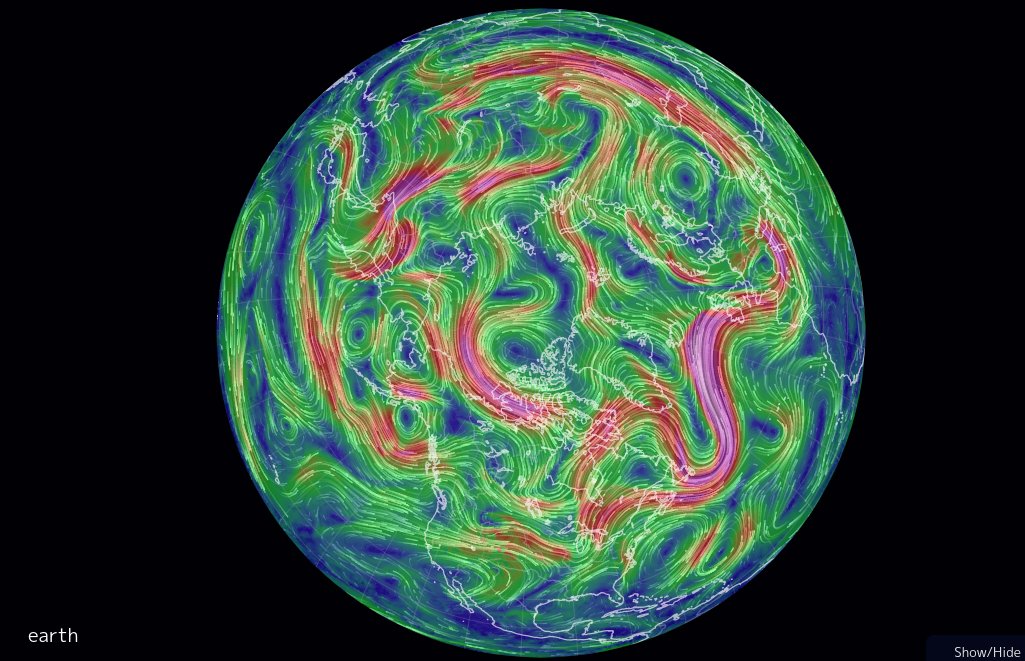
Tue Jul 13 14:34:21 +0000 2021Congrats to Yujiao Yang, et al. (PXD027205). Very nicely done. ⭐️⭐️⭐️⭐️
Tue Jul 13 14:07:32 +0000 2021I'm not sure why I am not hearing more push-back about the latest dopey idea from the Canadian government, effectively abolishing research into anything of potential commercial interest 🔗
Tue Jul 13 13:16:29 +0000 2021Looks like the Dutch will overtake the Limeys in short order. 🔗
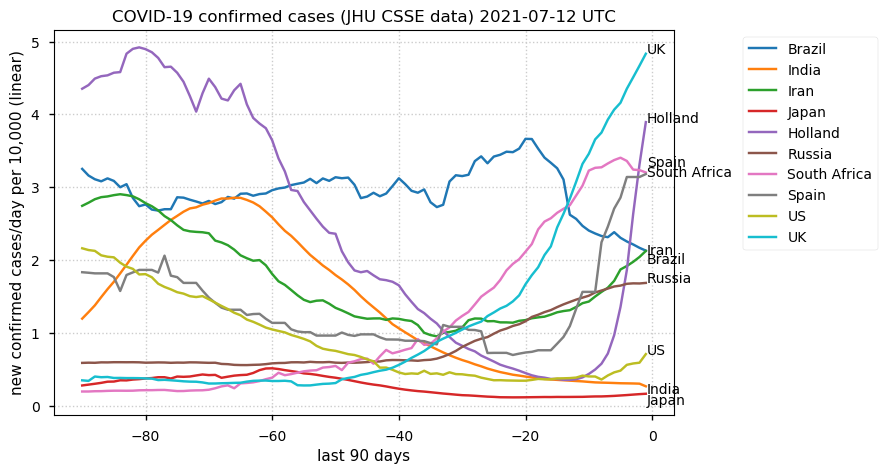
Tue Jul 13 11:53:12 +0000 2021KDM3B:p.A256T chr 5:g.138381576G>A, rs6865472 (all tissue A:T 0.041:0.959) vaf=>99%, Δm=30.0106, VAF by population group: african 92%, american 100%, east asian 100%, european 100%, south asian 100%. TT in all commonly used cell lines. #ᐯᐸᐱ
Tue Jul 13 11:53:12 +0000 2021KDM3B:p, θ(max) = 60. aka KIAA1082, NET22, C5orf7, JMJD1B. Found in MHC class 1 peptide experiments. Commonly found in tissues & cell lines: very rare in fluids. Has some tryptic peptide overlap with JMJD1C:p. Specifically demethylates histone H3:pm.K9+dimethyl.
Tue Jul 13 11:53:12 +0000 2021>KDM3B:p, lysine demethylase 3B (Homo sapiens) Large enzyme; CTMs: A2+acetyl; PTMs: 36×K+acetyl; 35×K+GGyl; 21×K+SUMOyl; 94×S, 33×T, 2×Y+phosphoryl; SAAVs: A5T (1%), A256T (>99%); mature form: (2-1761) [23,714×, 149 kTa]. #ᗕᕱᗒ 🔗
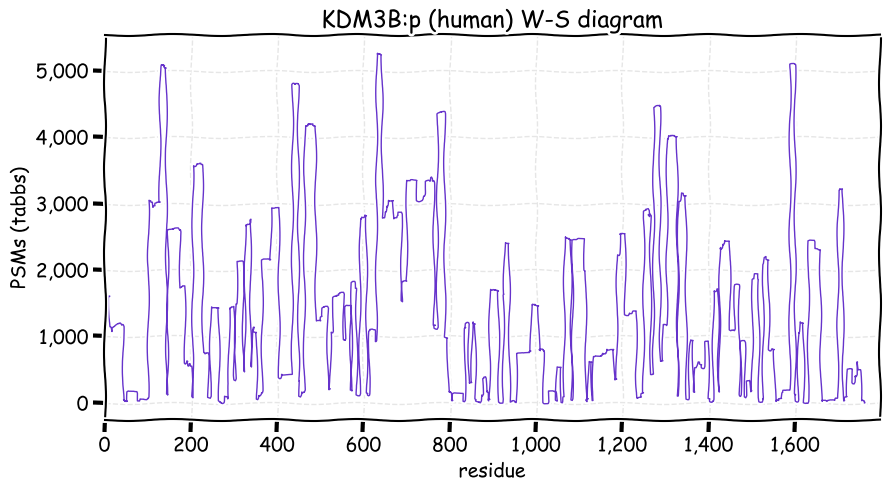
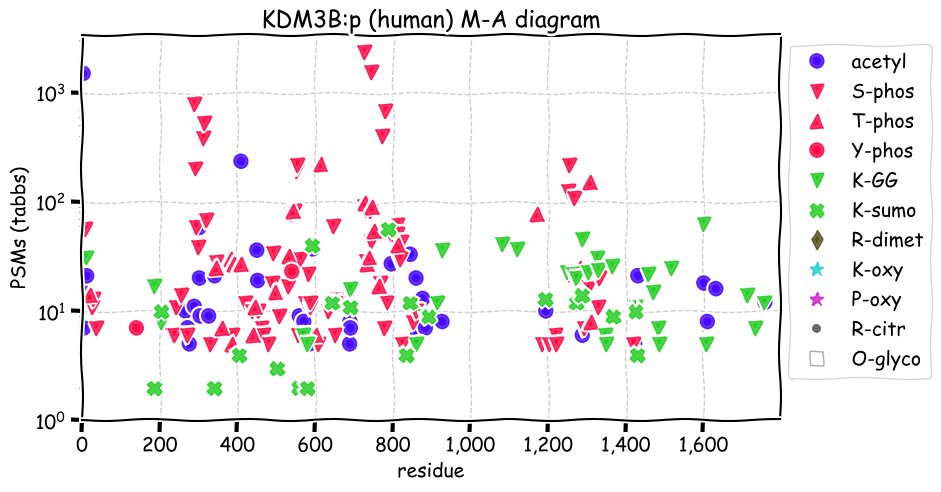
Mon Jul 12 21:05:46 +0000 2021@dk_munro @michaelhoffman @RachAelCMaxwell You are going to have to start including collaborators from the Faculty of Law ...
Mon Jul 12 20:50:36 +0000 2021Since when did Canadian universities start generating super-secret IP that anyone else wants to steal? 🔗
Mon Jul 12 12:02:36 +0000 2021GLB1:p.C521R chr 3:g.33014229A>G, rs4302331 (all tissue C:R 0.000:1.000)) vaf=98%, Δm=53.0919, VAF by population group: african 75%, american 97%, east asian 100%, european 100%, south asian 100%. Variant introduces a tryptic cleavage site. #ᐯᐸᐱ
Mon Jul 12 12:02:36 +0000 2021GLB1:p, θ(max) = 76. aka EBP, ELNR1. Found in MHC class 1 & 2 peptide experiments. Commonly found in urine, tissues & cell lines.
Mon Jul 12 12:02:35 +0000 2021>GLB1:p, galactosidase beta 1 (Homo sapiens) Midsized enzyme; CTMs: N247, N464, N498+glycosyl; PTMs: 1×K+acetyl; 4×K+GGyl; 1×S, 4×T, 2×Y+phosphoryl; SAAVs: P10L (43%), C521R (98%); mature form: (29-677) [36,960×, 114 kTa]. #ᗕᕱᗒ 🔗
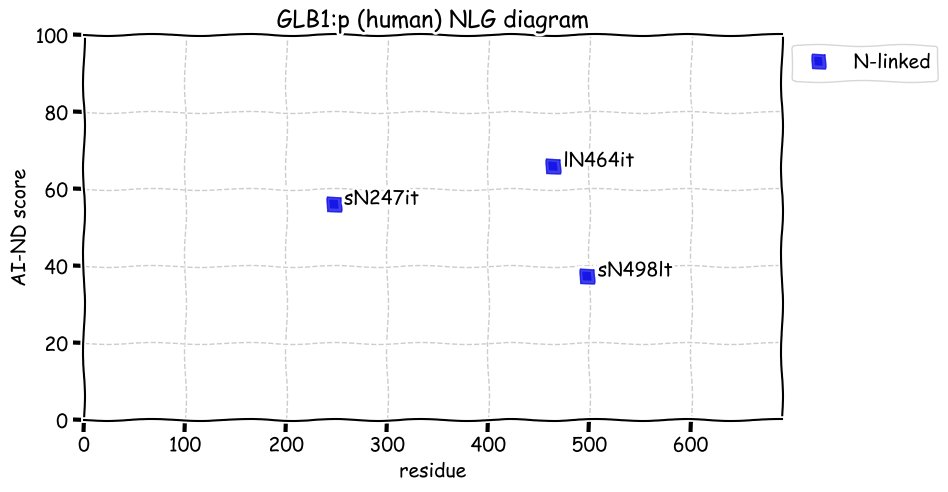
Sun Jul 11 16:44:59 +0000 2021HELZ:p, some sources suggest that there is a shorter splice variant protein (1-1183, from ENST00000579953.5) that has a different activity: there is no proteomics evidence to support the existence of this alternate sequence.
Sun Jul 11 12:51:25 +0000 2021HELZ:p enriched in RNA-protein interaction experiments. Based on the PTM distribution, (1-1100) & (1101-1883) are significantly different domains.
Sun Jul 11 12:40:07 +0000 2021HELZ:p.V74M chr 17:g.67215926C>T, rs8080100 (all tissue V:M 0.952:0.048) vaf=17%, Δm=31.9721, VAF by population group: african 25%, american 16%, east asian 20%, european 18%, south asian 16%. #ᐯᐸᐱ
Sun Jul 11 12:37:01 +0000 2021HELZ:p, θ(max) = 49. aka KIAA0054, HUMORF5, DHRC. Found in MHC class 1 peptide experiments. Commonly found in tissues & cell lines: rare in fluids.
Sun Jul 11 12:37:01 +0000 2021>HELZ:p, helicase with zinc finger (Homo sapiens) Large intracellular subunit; CTMs: M1+acetyl; PTMs: 2×K+acetyl; 22×K+GGyl; 45×S, 15×T, 2×Y+phosphoryl; SAAVs: V74M (17%), I1468V (1%); mature form: (1-1942) [14,764×, 114 kTa]. #ᗕᕱᗒ 🔗
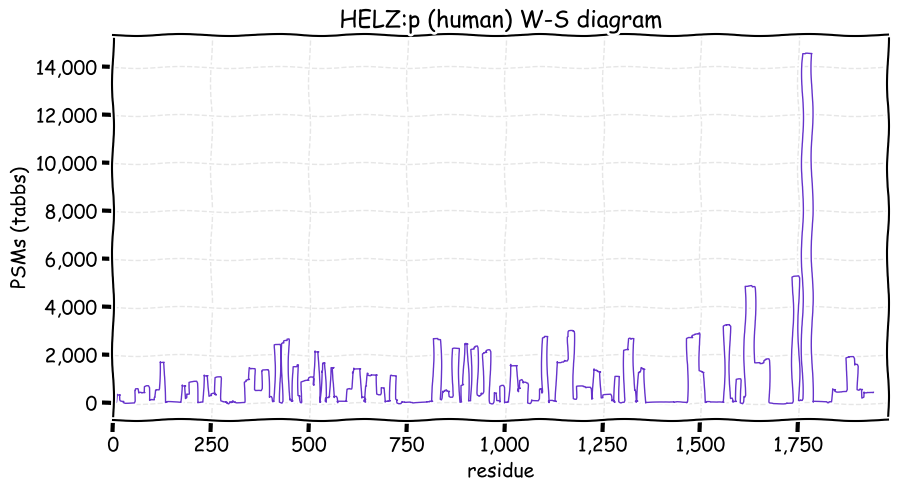
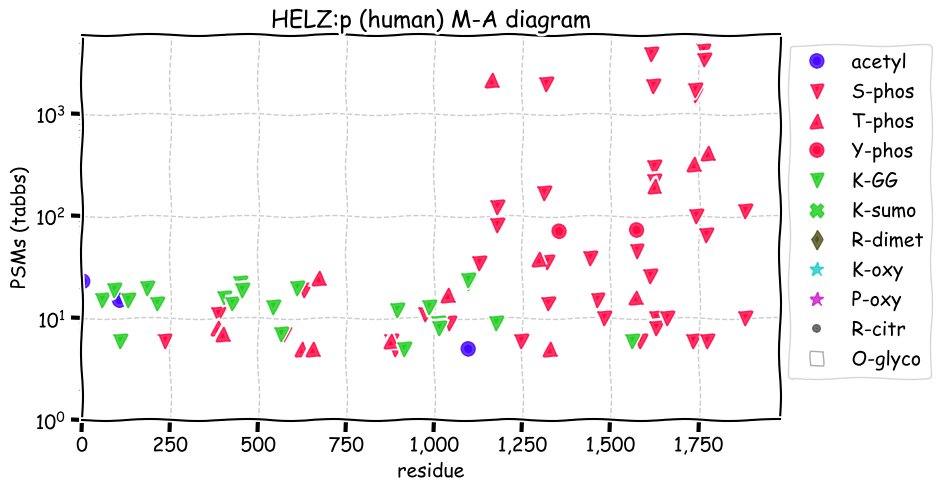
Sat Jul 10 16:14:05 +0000 2021@esnakayasu @NatureProtocols @cevansmo @paulpproteomics @brigittef @OmicsPNNL @sciencePNNL I was referring to UTIs as a cause of confounding effects in all proteomics-based urinary biomarker studies, rather than a consideration protein biomarkers for UTIs.
Sat Jul 10 14:39:30 +0000 2021@kadzuis @an_chem @scrippsresearch The layered security I've got in place seems to keep all but the most determined DDOS-type attack from affecting the main servers. Adding a DNS sinkhole can be quite useful, although that might not be practical for a cloud-based system.
Sat Jul 10 14:18:15 +0000 2021@kadzuis @an_chem @scrippsresearch GPMDB gets attacked regularly. I have no clue about motive, but I doubt it has anything to do with content.
Sat Jul 10 13:40:02 +0000 2021@esnakayasu @NatureProtocols @cevansmo @paulpproteomics @brigittef @OmicsPNNL @sciencePNNL While this is a useful discussion, I can't get fully behind a tutorial that includes a discussion of urinary biomarker discovery that doesn't at least mention the effects of urinary tract infections, particularly catheter associated UTIs.
Sat Jul 10 12:14:26 +0000 2021TAP1:p is part of the mechanism that transports antigen peptides from where they are made in the cytoplasm to the ER lumen, where they associate with MHC1 prior to the complex being moved to the plasma membrane.
Sat Jul 10 11:58:46 +0000 2021TAP1:p.D697G chr 6:g.32847198T>C, rs1135216 (all tissue D:G 0.952:0.048) vaf=17%, Δm=-58.0055, VAF by population group: african 27%, american 15%, east asian 13%, european 16%, south asian 20%. #ᐯᐸᐱ
Sat Jul 10 11:58:46 +0000 2021TAP1:p, θ(max) = 49. aka PSF1, RING4, D6S114E, ABCB2. Found in MHC class 1 peptide experiments. Commonly found in tissues & cell lines: rare in fluids.
Sat Jul 10 11:58:46 +0000 2021>TAP1:p, transporter 1, ATP binding cassette subfamily B member (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: K449+acetyl; 6×K+GGyl; 7×S, 1×T+phosphoryl; SAAVs: S346C (%), A430V (3%), G479C (2%), D697G (17%); mature form: (2-808) [19,977×, 114 kTa]. #ᗕᕱᗒ 🔗
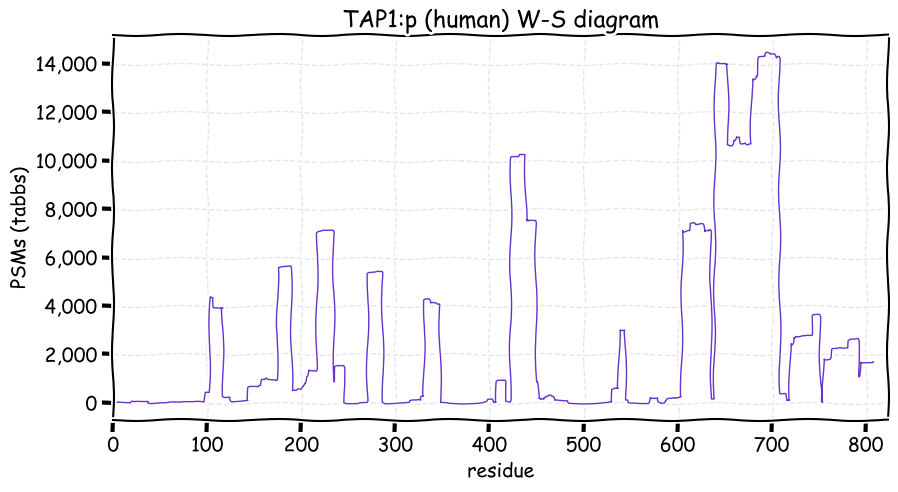
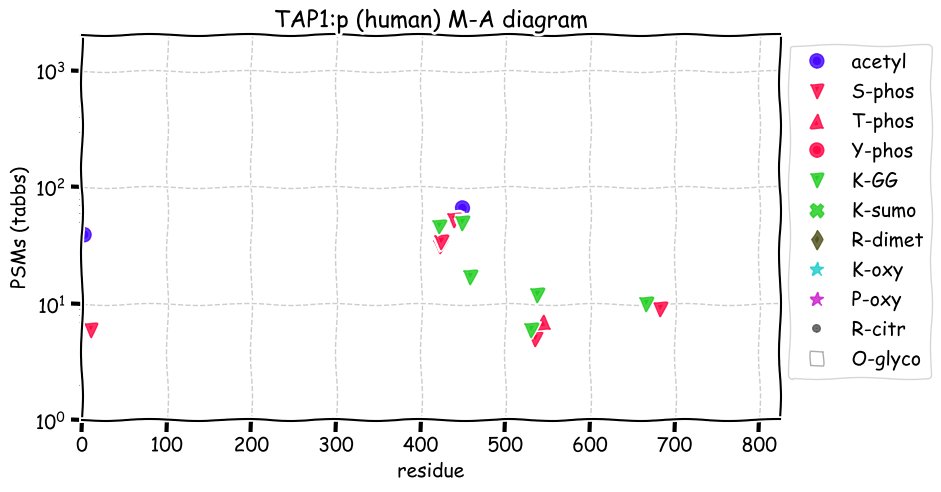
Fri Jul 09 15:26:54 +0000 2021While I know it sounds weird, but could anyone who uses a phrase like
"Cysteine carbamidomethylation was set as fixed modification"
in a paper or data annotation please check to make sure they actually used IAA/CAA in their experiment?
Fri Jul 09 15:01:19 +0000 2021Exosome: 🔗
Exosome: 🔗
Please change the name of one of these things, ASAP.
Fri Jul 09 12:47:30 +0000 2021SRGN:p is rarely observed in cancer tissue samples, except for leukemia. 🔗
Fri Jul 09 12:37:01 +0000 2021SRGN:p.R31Q chr 10:69097096G>A, rs2229498 (all tissue R:Q 0.000:1.000) vaf=41%, Δm=-28.0425, VAF by population group: african 35%, american 61%, east asian 50%, european 85%, south asian 73%. #ᐯᐸᐱ
Fri Jul 09 12:37:01 +0000 2021SRGN:p, θ(max) = 50. aka PPG, PRG, PRG1. Oddly promenient in MHC class 2 peptide experiments, otherwise mainly found in blood plasma, urine & CSF. O-glycosylated domain (94-110) not normally observed in experiments other than MHC peptide isolations.
Fri Jul 09 12:37:00 +0000 2021>SRGN:p, serglycin (Homo sapiens) Small subunit; CTMs: none; PTMs: 6×S+phosphoryl; SAAVs: R31Q (41%); mature form: (28-158) [5,153×, 38 kTa]. #ᗕᕱᗒ 🔗
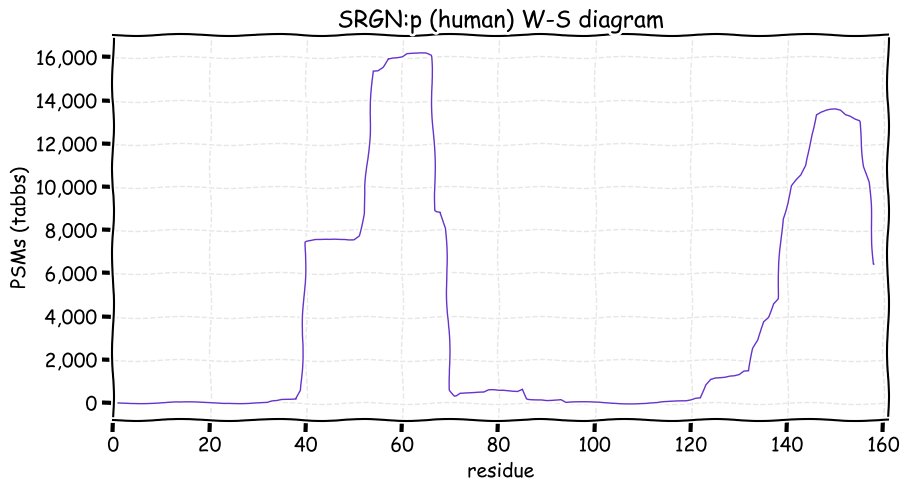
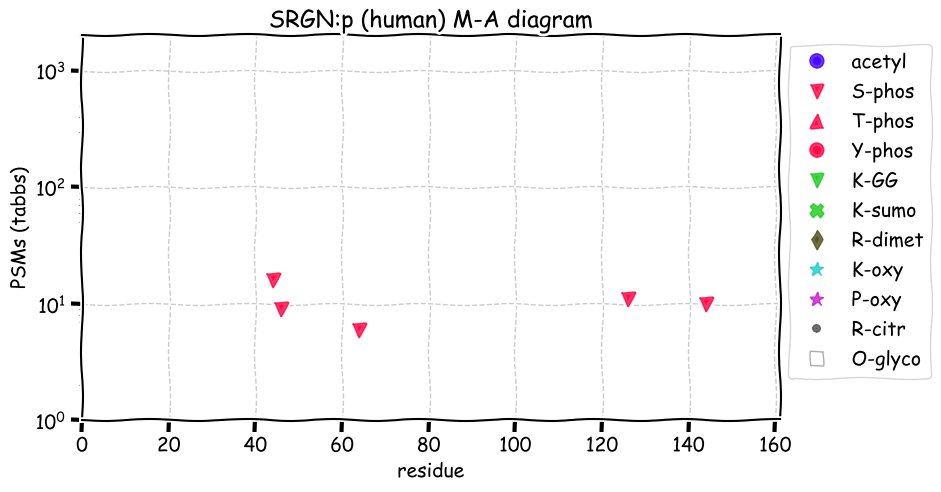
Thu Jul 08 16:07:47 +0000 2021Abundant MHC class II peptides are reproducibly from a list of proteins that seems to be kind of wacky, but it clearly means something.
Thu Jul 08 14:45:13 +0000 2021@VATVSLPR I've found those sources to be pretty unreliable wrt to N-linked glycosylation, as they tend to rely pretty heavily either on predictions or accounts given in single manuscripts.
Thu Jul 08 14:18:05 +0000 2021I'm guessing this isn't an issue that is top-of-mind for people involved in studying protein glycosylation. I was expecting someone to know (or at least tell me why this is a silly question).
Thu Jul 08 11:52:04 +0000 2021SORL1:p has many potential functions associated with it, but very little investigation I can find of why it has so many occupied N-linked glycosylation sites & how these modifications affect its use by a cell.
Thu Jul 08 11:46:00 +0000 2021SORL1:p.A528T chr 11:g.121522975G>A, rs2298813 (all tissue A:T 0.952:0.048) vaf=7%, Δm=30.0106, VAF by population group: african 12%, american 11%, east asian 12%, european 4%, south asian 13%. #ᐯᐸᐱ
Thu Jul 08 11:45:59 +0000 2021SORL1:p, θ(max) = 42. aka gp250, LR11, LRP9, SorLA, SorLA-1, C11orf32. Found in MHC class 1 & 2 peptide experiments. Commonly found in tissues, cell lines & fluids. Domains: 82-2137 (extracellular), 2138-2148 (transmembrane), 2149-2214 (intracellular).
Thu Jul 08 11:45:59 +0000 2021>SORL1:p, sortilin related receptor 1 (Homo sapiens) Large subunit; CTMs: 17×N+glycosyl; PTMs: 5×K+GGyl; 4×S+phosphoryl; SAAVs: A528T (7%), V1967I (98%); mature form: (82-2214) [13,741×, 104 kTa]. #ᗕᕱᗒ 🔗
Wed Jul 07 19:39:00 +0000 2021@dtabb73 I always reformat USB drives to use NTFS.
Wed Jul 07 18:49:37 +0000 2021@AlexUsherHESA Spain appears to be on a similar trajectory, but about a week behind the UK.
Wed Jul 07 16:05:27 +0000 2021Simply out of idle curiosity, does anybody know which human protein has the largest number of demonstrated N-linked glycosylation sites?
Wed Jul 07 15:53:34 +0000 2021@h_i_g_s_c_h Most mapping systems have a multi-finger gesture that allows for that type of navigation.
Wed Jul 07 14:29:56 +0000 2021Even though the UK and Spain are showing significant increases in the number of cases, neither are showing a rise in fatalities, unlike Russia and South Africa 🔗
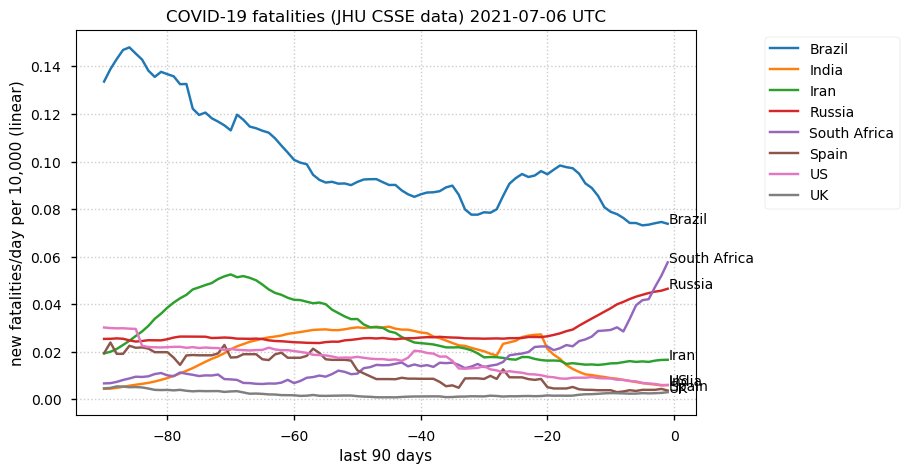
Wed Jul 07 12:02:12 +0000 2021CDKN2B:p.A19D chr 9:g.22008898G>T, rs184392037 (all tissue A:D 0.987:0.013) vaf=<1%, Δm=43.9898, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. #ᐯᐸᐱ
Wed Jul 07 12:02:12 +0000 2021CDKN2B:p, θ(max) = 74. aka P15, MTS2, INK4B, TP15, CDK4I, p15INK4b. Found in MHC class 1 peptide experiments. Commonly found in tissues & cell lines: rare in fluids. Tryptic peptide overlap with CDKN2A.
Wed Jul 07 12:02:12 +0000 2021>CDKN2B:p, cyclin dependent kinase inhibitor 2B (Homo sapiens) Small subunit; CTMs: none; PTMs: none; SAAVs: A19D (<1%); mature form: (1-138) [7,319×, 23 kTa]. #ᗕᕱᗒ 🔗
Tue Jul 06 12:33:01 +0000 2021@AlexUsherHESA @marypatcormier Being GG is a lot better gig than trying to win a Calgary seat as a Liberal.
Tue Jul 06 12:29:03 +0000 2021@marypatcormier @AlexUsherHESA Naheed Nenshi
Tue Jul 06 11:59:20 +0000 2021NDC80:p.A605P chr 18:g.2616458G>C, rs9051 (all tissue A:P 0.873:0.127) vaf=21%, Δm=26.0156, VAF by population group: african 12%, american 14%, east asian 12%, european 23%, south asian 31%. #ᐯᐸᐱ
Tue Jul 06 11:59:20 +0000 2021NDC80:p, θ(max) = 68. aka HEC, HEC1, hsNDC80, TID3, KNTC2. Found in MHC class 1 peptide experiments. Commonly found in tissues & cell lines: rare in fluids.
Tue Jul 06 11:59:20 +0000 2021>NDC80:p, NDC80 kinetochore complex component (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 26×K+GGyl; 2×K+acetyl; 7×K+SUMOyl; 14×S, 4×T, 2×Y+phosphoryl; SAAVs: E348D (9%), A605P (21%); mature form: (1-642) [8,568×, 36 kTa]. #ᗕᕱᗒ 🔗
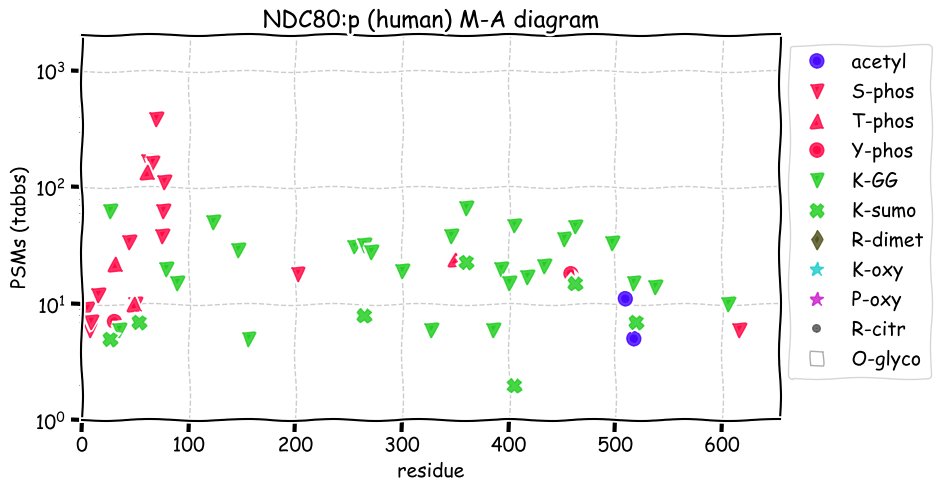
Tue Jul 06 00:36:25 +0000 2021If you are looking for a human data set to use for teaching, with reproducible reps, multiple cell lines & lots of interesting features in the results, PXD022002 could be what you are after.
Mon Jul 05 17:09:23 +0000 2021@J_my_sci I think they are more of a sign that you've already got a problem (& may need a friendly intervention).
Mon Jul 05 15:38:32 +0000 2021PM₁₀ atmospheric particulates, mainly from fires, from eastern Siberia to California (from 🔗) 🔗
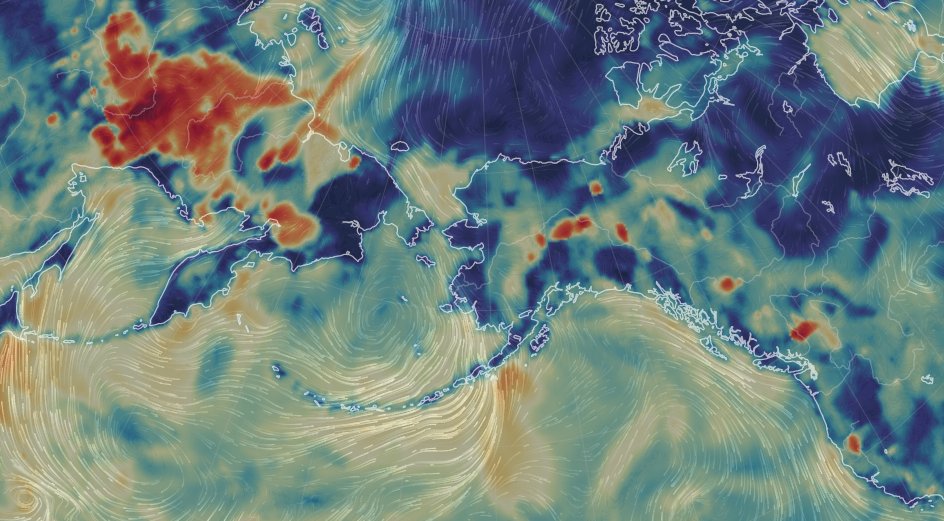
Mon Jul 05 13:15:44 +0000 2021@JuliaSkid11 @CommsBio @hecklab Got it this morning. Thanks for making it available.
Mon Jul 05 12:35:51 +0000 2021The UK seems to be running a little ahead of South Africa: a daily rate of 3 caused a huge furor in Manitoba just a month ago 🔗
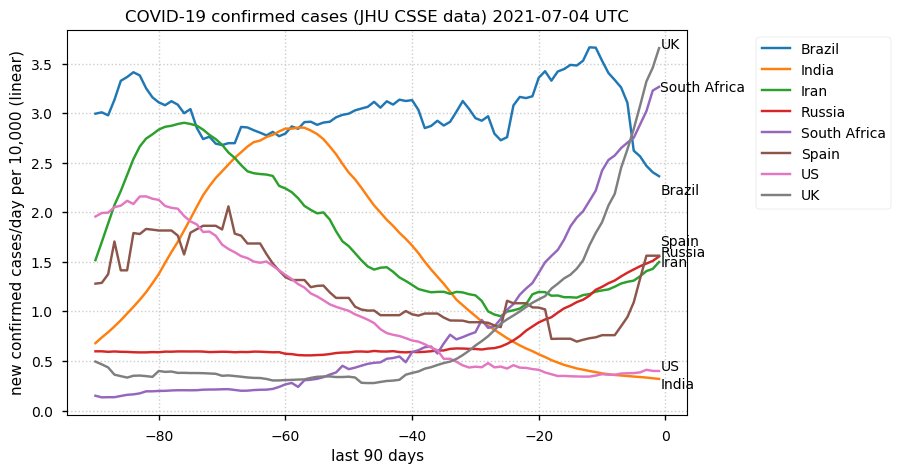
Mon Jul 05 12:01:16 +0000 2021ME1:p.V232I chr 6:g.83315320C>T, rs148600837 (all tissue V:I 0.995:0.005) vaf=0.4%, Δm=14.0156, VAF by population group: african 1%, american <1%, east asian <1%, european <1%, south asian <1%. #ᐯᐸᐱ
Mon Jul 05 12:01:16 +0000 2021ME1:p, θ(max) = 85. aka none. Found in MHC class 1 peptide experiments & by 1 peptide in class 2. Commonly found in tissues & cell lines: rare in fluids.
Mon Jul 05 12:01:15 +0000 2021>ME1:p, malic enzyme 1 (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 13×K+GGyl; 12×K+acetyl; 8×K+SUMOyl; 1×S, 1×T, 3×Y+phosphoryl; SAAVs: V232I (<1%), R254H (1%); mature form: (1-573) [23,220×, 174 kTa]. #ᗕᕱᗒ 🔗
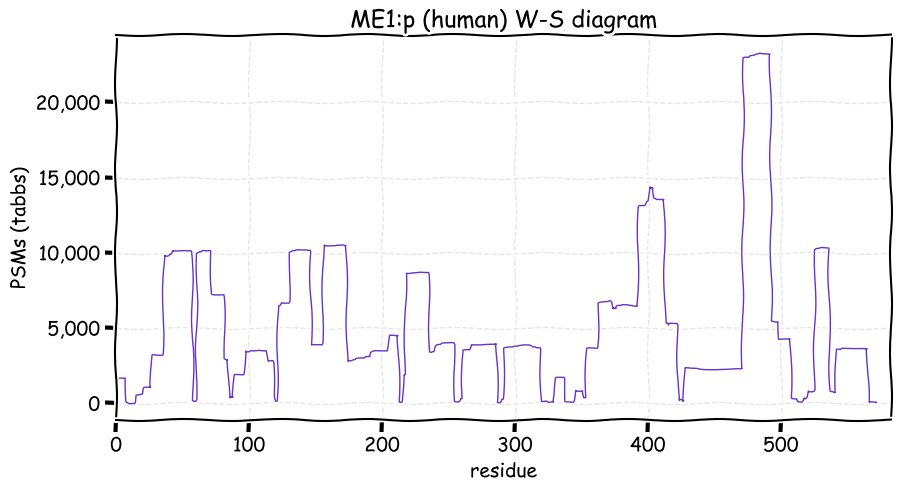
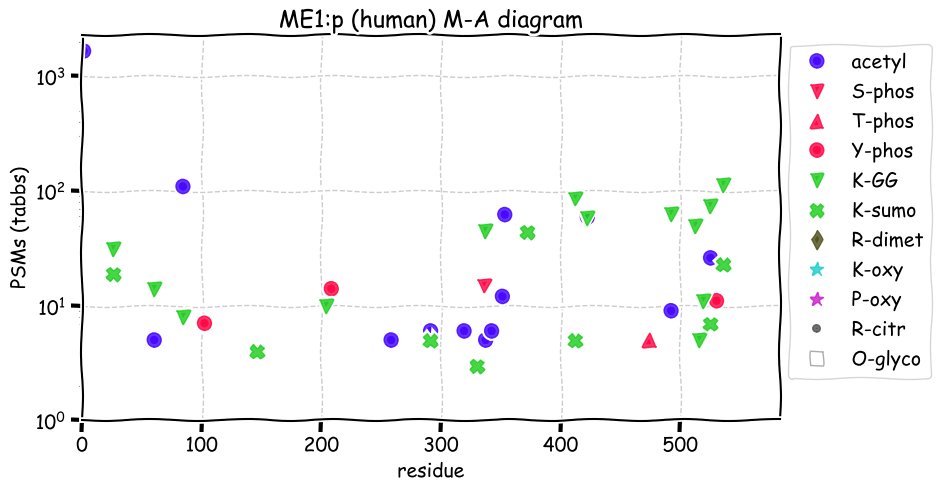
Mon Jul 05 01:51:54 +0000 2021"GAK is an association partner of cyclin G and CDK5.
Cyclin G-associated kinase received its name because it immunoprecipitated with cyclin G though it now appears to not be associated with it."
-Wikipedia
Sun Jul 04 12:03:41 +0000 2021SLK:p.C552Y chr 10:g.104002833G>A, rs805657 (all tissue C:Y 0.924:0.076) vaf=18%, Δm=60.0541, VAF by population group: african 36%, american 21%, east asian 10%, european 17%, south asian 16%. #ᐯᐸᐱ
Sun Jul 04 12:03:41 +0000 2021SLK:p, θ(max) = 65. aka STK2, se20-9, KIAA0204. Found in MHC class 1 peptide experiments & rarely in class 2. Commonly found in tissues, cell lines & urinary extracellular vesicles.
Sun Jul 04 12:03:41 +0000 2021>SLK:p, STE20 like kinase (Homo sapiens) Large subunit; CTMs: S2+acetyl; PTMs: 19×K+GGyl; 19×K+acetyl; 1×K+SUMOyl; 56×S, 41×T, 2×Y+phosphoryl; SAAVs: C552Y (18%), G666E (2%), T697I (21%), ; mature form: (2-1204) [36,578×, 464 kTa]. #ᗕᕱᗒ 🔗
Sat Jul 03 20:57:30 +0000 2021@pwilmarth @ucdmrt Be better to propose ML to isolate the signals from individual cells.
Sat Jul 03 17:52:40 +0000 2021@slashdot Sounds like a nightmare if you need to copyright the resulting code.
Sat Jul 03 17:42:26 +0000 2021@astacus Never seen it in mouse samples. XMRV shows up so often it is probably on the way to becoming an endogenous retrovirus in lab mouse strains.
Sat Jul 03 17:40:26 +0000 2021@pwilmarth @astacus In this notably odd-ball case, it would appear that the molecule is anxious to switch between an overall -ve charge (+phosphoryl, +acetyl) and a +ve charge (-phosphoryl, -acetyl), with a bunch of transport signals (+GGyl & +SUMOyl) we don't understand.
Sat Jul 03 16:21:26 +0000 2021@astacus Not me.
Sat Jul 03 14:44:10 +0000 2021@Hayes_McDonald @pwilmarth A gateway to super-hyperplexing (not to be confused with hyper-superplexing)
Sat Jul 03 14:22:33 +0000 2021Mouse proteomics data that doesn't have good evidence for XMRV proteins should have an asterisk beside it.
Sat Jul 03 13:07:00 +0000 2021fibronectin (for some reason spelling correction filters don't work so well with protein names).
Sat Jul 03 13:03:35 +0000 2021Do the visible effects of collagen infiltration bias tumor tissue sampling? I say this looking at the Nth tumor sample data set that is largely collagen & fibrinectin.
Sat Jul 03 12:18:13 +0000 2021While it is possible to consider the human proteome to be a collection of ~20,000 edge cases, MKI67:p is edgier than most.
Sat Jul 03 12:08:02 +0000 2021MKI67:p.N104S chr 10:g.128119296T>C, rs2071498 (all tissue N:S 0.583:0.417) vaf=65%, Δm=-27.0109, VAF by population group: african 58%, american 72%, east asian 64%, european 67%, south asian 61%. SS in U2-OS cells. #ᐯᐸᐱ
Sat Jul 03 12:08:02 +0000 2021MKI67:p, θ(max) = 73. aka MIB-1, PPP1R105. Found in MHC class 1 peptide experiments. Commonly found in tissues & cell lines: rare in fluids.
Sat Jul 03 12:08:02 +0000 2021MKI67:p, SAAVs: N104S (65%), W238R (75%), I631L (11%), L854V (77%), G1042S (19%), 1247I (10%), E1403V (14%), V1559M (10%), R1876Q (22%), I2101T (15%), R2649H (22%), D2760G (23%), R2786Q (18%), S2793N (23%), T2868S (18%), Q2904R (23%), T3150S (42%), K3217E (46%)
Sat Jul 03 12:08:02 +0000 2021>MKI67:p, marker of proliferation Ki-67 (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 104×K+GGyl; 313×K+acetyl; 68×K+SUMOyl; 240×S, 240×T, 11×Y+phosphoryl; mature form: (1-3256) [43,126×, 969 kTa]. #ᗕᕱᗒ 🔗
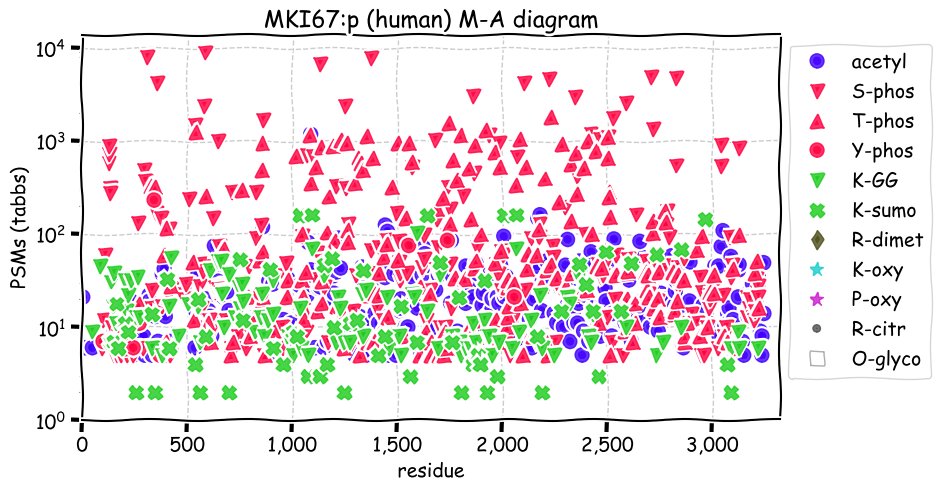
Fri Jul 02 15:08:52 +0000 2021And in local news 🔗
Fri Jul 02 14:21:37 +0000 2021@propris @_Yellow_Kid_ @doctorow @jbarro I spent a couple of weeks in Brazil & they seemed pretty convinced that chicken was a vegetable ...
Fri Jul 02 12:31:54 +0000 2021This morning, a string of storms running along a cold front tracing the path of upper level winds, running from northern Texas up along the Atlantic coast to southern Nova Scotia. 🔗
Fri Jul 02 11:53:00 +0000 2021EMC1:p.S345T chr 1:g.19238850C>G, rs709683 (all tissue S:T 0.656:0.344) vaf=44%, Δm=14.0157, VAF by population group: african 71%, american 53%, east asian 74%, european 35%, south asian 46%. TT in HeLa cells and ST in HEK-293 cells. #ᐯᐸᐱ
Fri Jul 02 11:53:00 +0000 2021EMC1:p, θ(max) = 83. aka KIAA0090. Found in MHC class 1 & 2 peptide experiments. Commonly found in tissues & cell lines: rare in fluids. Domains: ER lumen (22-960), membrane (961-981), cytosolic 982-993.
Fri Jul 02 11:53:00 +0000 2021>EMC1:p, ER membrane protein complex subunit 1 (Homo sapiens) Midsize subunit; CTMs: N370, N913+glycosyl; PTMs: 19×K+GGyl; 3×K+acetyl; 1×K+SUMOyl; 7×S, 0×T, 3×Y+phosphoryl; SAAVs: T309M (2%), S345T (44%), T544M (38%), R621C (1%); mature form: (22-993) [32,905×, 252 kTa]. #ᗕᕱᗒ 🔗
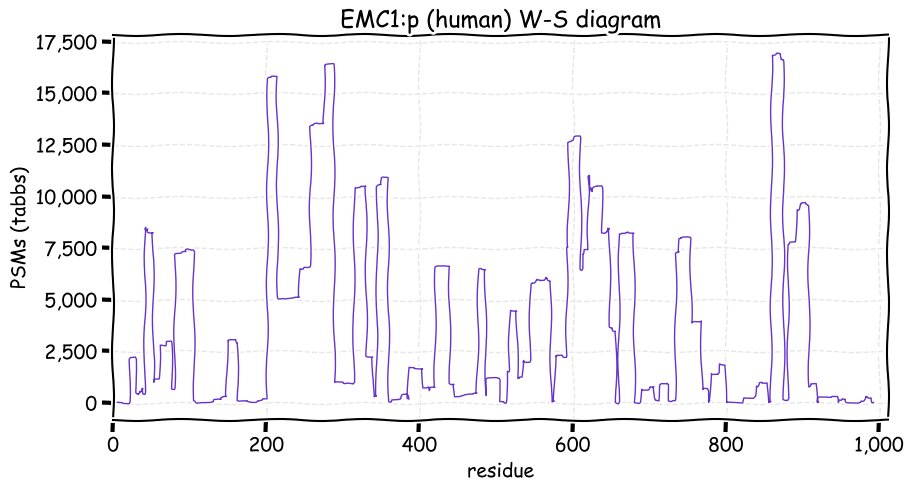
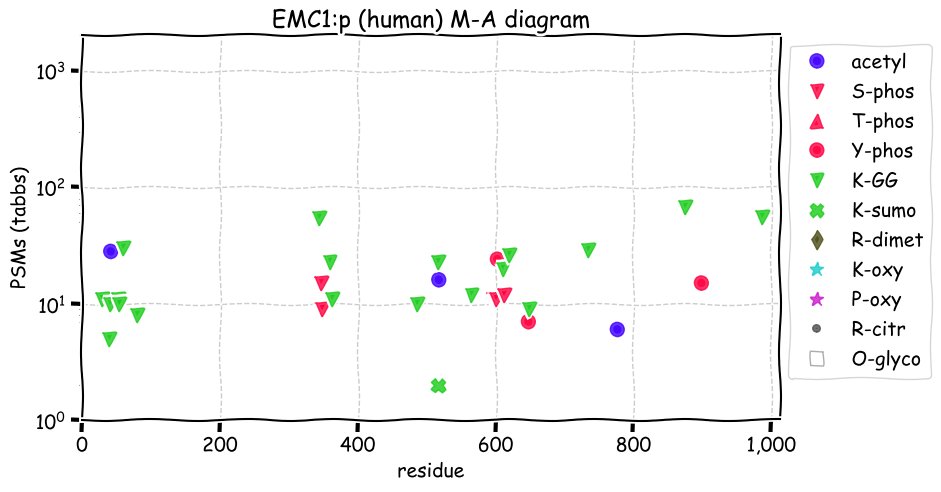
Fri Jul 02 10:51:47 +0000 2021@JuliaSkid11 @CommsBio @hecklab That isn't necessary. I will check their site periodically for the released data.
Fri Jul 02 00:22:33 +0000 2021Thanks to everyone who participated in the poll. Bovine apolipoprotein B produces the most PSMs of any bovine protein in this type of experiment: in fact it often produces more PSMs than any human protein. BSA usually produces no detectable PSMs in HLA II data.
Thu Jul 01 21:25:33 +0000 2021@JuliaSkid11 @CommsBio @hecklab When will the ProteomeXchange data be made public? I'm looking forward to exploring it.
Thu Jul 01 18:35:52 +0000 2021The UK and South Africa seem to be on similar trajectories for their next wave of infections: India's appears to have subsided. 🔗
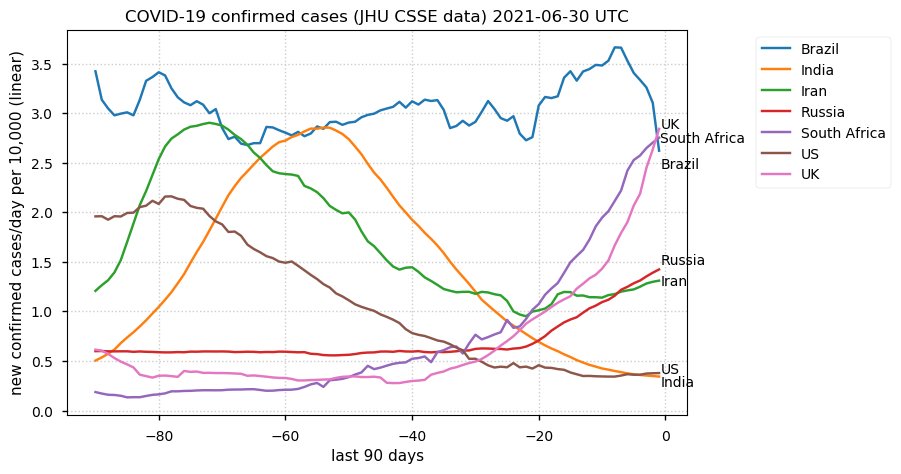
Thu Jul 01 12:39:03 +0000 2021FLVCR1:p, the full name of this protein is "Feline Leukemia Virus subgroup c Cellular Receptor 1 heme transporter 1", my pick for the worst protein name of the year so far.
Thu Jul 01 12:39:02 +0000 2021FLVCR1:p.A52P chr 1:g.212858606G>C, rs11120047 (all tissue A:P 0.524:0.476) vaf=48%, Δm=26.0156, VAF by population group: african 23%, american 31%, east asian 55%, european 56%, south asian 61%. PP in HCT-116 cells and AP in HEK-293 cells. #ᐯᐸᐱ
Thu Jul 01 12:39:02 +0000 2021FLVCR1:p, θ(max) = 41. aka FLVCR, MFSD7B, PCA, SLC49A1, AXPC1. Found in MHC class 1 peptide experiments (rarely). Commonly found in urine & platelets. Domains: extracellular (21-267), membrane (268-288), intracellular 289-339.
Thu Jul 01 12:39:02 +0000 2021>FLVCR1:p, FLVCR heme transporter 1(Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 8×K+GGyl; 3×S, 3×T, 1×Y+phosphoryl; SAAVs: A52P (48%), T544M (38%); mature form: (2-555) [2,055×, 16 kTa]. #ᗕᕱᗒ 🔗
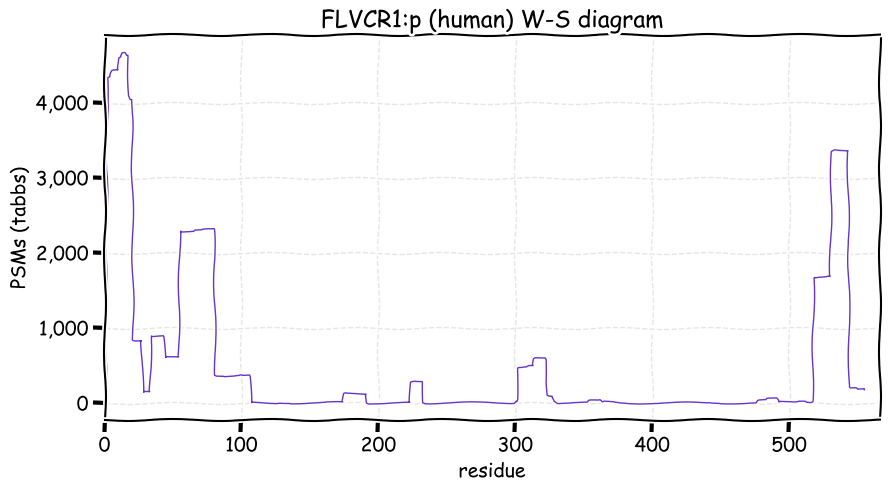
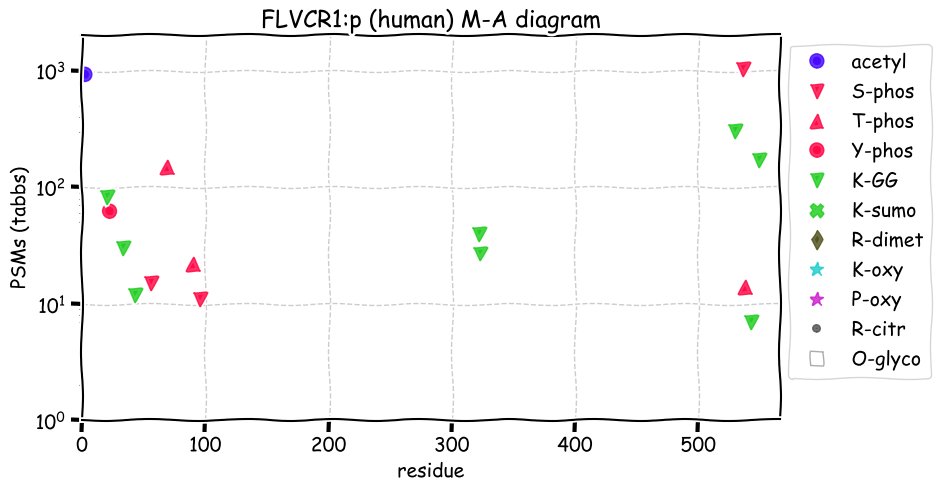
Thu Jul 01 00:12:48 +0000 2021In HLA class II peptide experiments in which the source cells have been in contact with bovine serum, which bovine protein produces the most class II peptide PSMs:
Wed Jun 30 16:59:22 +0000 2021@mekki Or Blackberry?
Wed Jun 30 15:54:34 +0000 2021@attilacsordas @nesvilab @ypriverol @labs_mann Perhaps, but if you really want to do absolute quant, just spiking in a known concentration of a mix of proteins can be used for any type of sample. It is easier to interpret & allows for unambiguous QA/QC testing of the results.
Wed Jun 30 15:13:53 +0000 2021@dtabb73 @ProteomicsNews Proteomics calculations don't actually scale that way. It is often a waste of processor cycles to go much beyond 16 cores.
Wed Jun 30 12:38:49 +0000 2021GP6:p, given the SAAV distributions, only a small fraction of the population will be homozygous for the reference sequence.
Wed Jun 30 12:38:49 +0000 2021GP6:p.L317Q chr 19:g.55014991A>T, rs1654413 (all tissue L:Q 0.260:0.740) vaf=80%, Δm=14.9745, VAF by population group: african 53%, american 83%, east asian 81%, european 84%, south asian 71%. #ᐯᐸᐱ
Wed Jun 30 12:38:49 +0000 2021GP6:p, θ(max) = 41. aka GPVI. Found in MHC class 1 peptide experiments (rare). Commonly found in urine & platelets. Domains: extracellular (21-267), membrane (268-288), intracellular 289-339.
Wed Jun 30 12:38:48 +0000 2021>GP6:p, glycoprotein VI platelet (Homo sapiens) Small subunit; CTMs: none; PTMs: none; SAAVs: L317Q (80%), N322H (85%); mature form: (21,23-339) [2,055×, 14 kTa]. #ᗕᕱᗒ 🔗
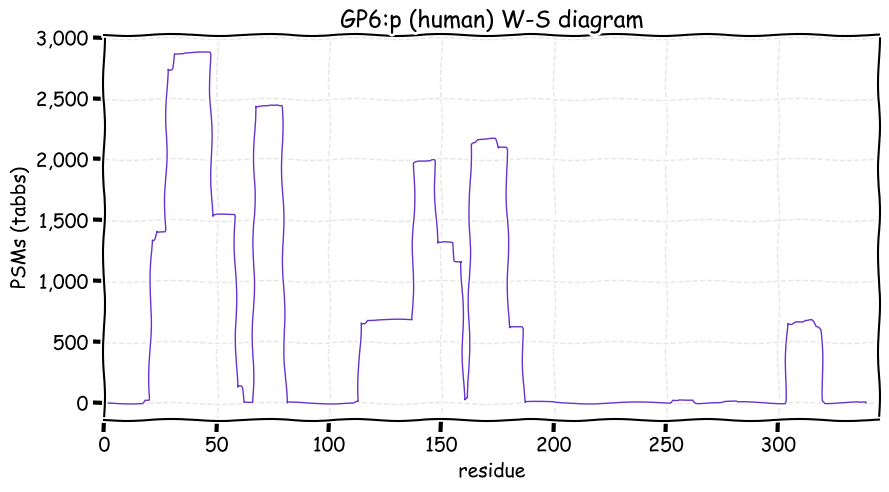
Wed Jun 30 12:00:50 +0000 2021@attilacsordas @nesvilab @ypriverol @labs_mann The lack of utility in tissue may be part of the problem with further study/adoption.
Wed Jun 30 11:37:39 +0000 2021@attilacsordas @nesvilab @ypriverol @labs_mann I'm not really sure about its current status. I have heard several people mention it on-line, but I can't remember the last time I read about it in a manuscript. Histones always seemed to me to be a problematic group of proteins to use as a reference.
Tue Jun 29 20:51:04 +0000 2021I guess the GPMDB version of a Universal Spectrum Identifier would be something like this?
mzspec:GPM06610084843:scan:56526:FLYDETQYGTETLWSK/2
Tue Jun 29 18:49:33 +0000 2021@UCDProteomics It has been pretty straightforward to do for a long time, but the marketing guys were dead set against it
Tue Jun 29 18:41:31 +0000 2021@UCDProteomics I have no idea why the platform companies wasted so much time not wanting to put this type of thing in place.
Tue Jun 29 16:12:45 +0000 2021@harrisonspecht Work in industry for a while before you go out on your own.
Tue Jun 29 12:21:37 +0000 2021As with most alternate initiation isoforms, the new N-terminal peptides (with CTMs) are the only PSMs that are characteristic of the isoform.
Tue Jun 29 12:18:07 +0000 2021SORBS3:p, the Bioplex project used a shorter form the gene (ENSP00000429764) that initiates at M343, resulting in the mature form 344-671 (CTM: A344+acetyl).
Tue Jun 29 12:04:24 +0000 2021SORBS3:p.I556T chr 8:g.22571145T>C, rs2449331 (all tissue I:T 0.115:0.885) vaf=95%, Δm=-12.0364, VAF by population group: african 73%, american 95%, east asian 100%, european 96%, south asian 97%. II in HCT-116 cells and TT in HeLa cells. #ᐯᐸᐱ
Tue Jun 29 12:04:24 +0000 2021SORBS3:p, θ(max) = 71. aka SCAM-1, SH3D4, vinexin. Found in MHC class 2 peptide experiments in tissue. Commonly found in tissues & cell lines: rarely in fluids.
Tue Jun 29 12:04:24 +0000 2021>SORBS3:p, sorbin and SH3 domain containing 3 (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 33×S, 14×T, 1×Y+phosphoryl; 2×acetyl; 2×GGyl; SAAVs: I556T (95%), T573A (13%); mature form: (1-671) [15,881×, 81 kTa]. #ᗕᕱᗒ 🔗
Tue Jun 29 11:58:01 +0000 2021@AlexUsherHESA This trend started some time ago. Of the Canadian millennials I know in tech-related fields, working remotely for a US company is often the only practical option.
Mon Jun 28 11:55:38 +0000 2021LUZP1:p, is commonly observed & highly modified but of unknown function, which has resulted in rather incoherent annotation.
Mon Jun 28 11:35:55 +0000 2021LUZP1:p.G458S chr 1:g.23092890C>T, rs477830 (all tissue G:S 0.776:0.224) vaf=75%, Δm=30.0106, VAF by population group: african 64%, american 59%, east asian 81%, european 83%, south asian 56%. SS in HeLa cells. #ᐯᐸᐱ
Mon Jun 28 11:35:55 +0000 2021LUZP1:p, θ(max) = 70. aka LUZP. Found in MHC class 1 peptide experiments. Commonly found in tissues & cell lines: rarely in fluids.
Mon Jun 28 11:35:55 +0000 2021>LUZP1:p, leucine zipper protein 1 (Homo sapiens) Large subunit; CTMs: A2+acetyl; PTMs: 77×S, 33×T, 3×Y+phosphoryl; 7×acetyl; 7×GGyl; SAAVs: S317A (5%), G458S (75%), T491I (2%), A896V (2%), R1061W (1%); mature form: (2-1076) [24,123×, 161 kTa]. #ᗕᕱᗒ 🔗
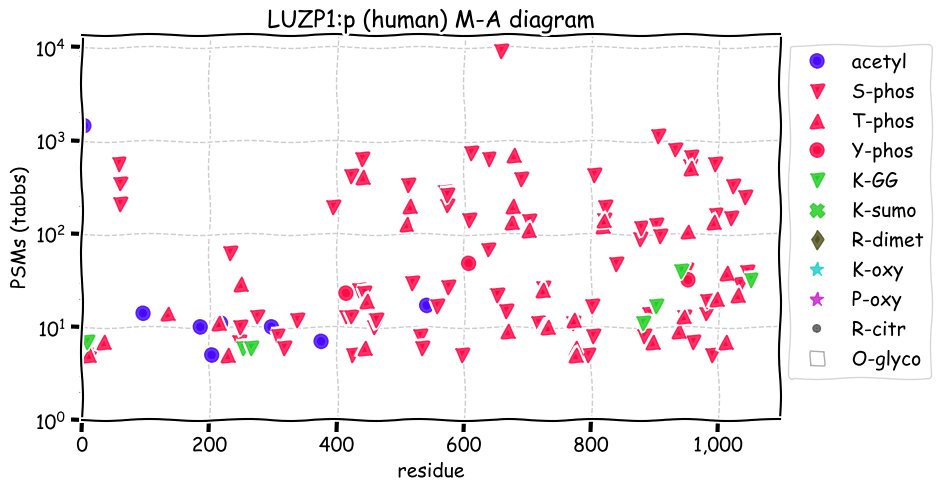
Sun Jun 27 19:38:13 +0000 2021Freakin' aphids ... 🔗

Sun Jun 27 12:35:53 +0000 2021As with other type I sulfatases, requires alpha-formylglycine (a PTM of S or C, 🔗) to provide a free aldehyde in its active site.
Sun Jun 27 11:58:10 +0000 2021SUMF2:p.D70E chr 7:g.56068567C<A, rs4245575 (all tissue D:E 0.026:0.974) vaf=>99%, Δm=14.0157, VAF by population group: african >99%, american >99%, east asian >99%, european >99%, south asian >99%. #ᐯᐸᐱ
Sun Jun 27 11:58:10 +0000 2021SUMF2:p, θ(max) = 69. aka DKFZp566I1024. Found in MHC class 1 peptide experiments: rarely in class 2. Commonly found in tissues & cell lines: rarely in fluids.
Sun Jun 27 11:58:10 +0000 2021>SUMF2:p, sulfatase modifying factor 2 (Homo sapiens) Small subunit; CTMs: none; PTMs: none; SAAVs: D51E (>99%), V290I (1%); mature form: (26-301) [31,127×, 40 kTa]. #ᗕᕱᗒ 🔗
Sat Jun 26 17:31:29 +0000 2021@MKupppusamy I have seen many studies in which either one works well, with minimal side reactions. However, if you don't check to be sure, either one can lead to unfortunate situations, particularly when comparing protocols or treatments.
Sat Jun 26 15:11:33 +0000 2021@pwilmarth @JoelisSteele @AchimTreumann They are like any 1st order reactions with peptides: the amounts range from undetectable to nearly complete, depending on time, temperature and pH.
Sat Jun 26 14:11:33 +0000 2021@pwilmarth @JoelisSteele @AchimTreumann And just to be the pedant that I am, C oxidation products are dominated by C+O3, with C+O1 & C+O2 being rare to undetectable.
Sat Jun 26 14:04:36 +0000 2021@pwilmarth @JoelisSteele @AchimTreumann M prefers to stick at M+O: M+O2 is always less abundant.
Sat Jun 26 14:03:47 +0000 2021@pwilmarth @JoelisSteele @AchimTreumann Actually, W tends to go straight to W+O2. W+O is always less abundant.
Sat Jun 26 13:47:29 +0000 2021Thanks to everyone who participated in this poll. The numbers show that the majority of respondents do not use a DB to store results (as expected). I am glad to see that there is a healthy minority (36%) that do.
Sat Jun 26 13:09:02 +0000 2021@JoelisSteele @AchimTreumann Examples of mods I normally check for and find 🔗
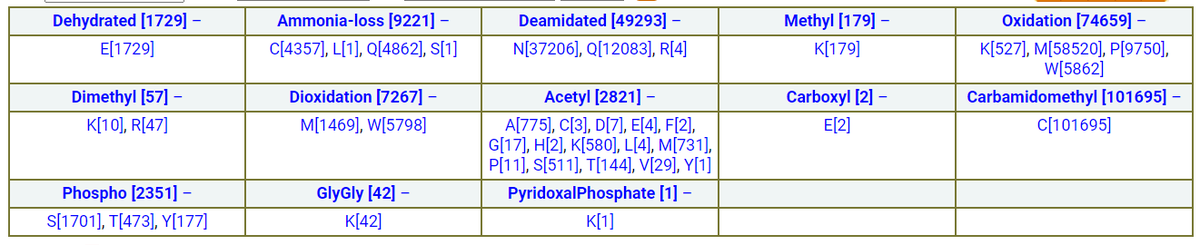
Sat Jun 26 12:51:15 +0000 2021@herrtschmidt @ypriverol I only use MongoDB for the reporting the results of a particular project within GPMDB & in that case using binary JSON. I use JSON all the time as an input/output format for analytical software.
Sat Jun 26 12:36:38 +0000 2021FTCD:p.V101M chr 21:g.46152973C>T, rs61735836 (all tissue V:M 0.937:0.063) vaf=6%, Δm=31.9721, VAF by population group: african 7%, american 2%, east asian 16%, european 5%, south asian 8%. #ᐯᐸᐱ
Sat Jun 26 12:36:38 +0000 2021FTCD:p, θ(max) = 84. aka none. Found in MHC class 1 &2 peptide experiments in liver & kidney. Commonly found in liver, kidney, urine extracellular vesicles as well as Hep-G2 & HEP-3B cells.
Sat Jun 26 12:36:38 +0000 2021>FTCD:p, formimidoyltransferase cyclodeaminase Midsized subunit; CTMs: S2+acetyl; PTMs: 7×S, 0×T, 0×Y+phosphoryl; SAAVs: V101M (6%), A438V (3%); mature form: (2-541) [4,272×, 85 kTa]. #ᗕᕱᗒ 🔗
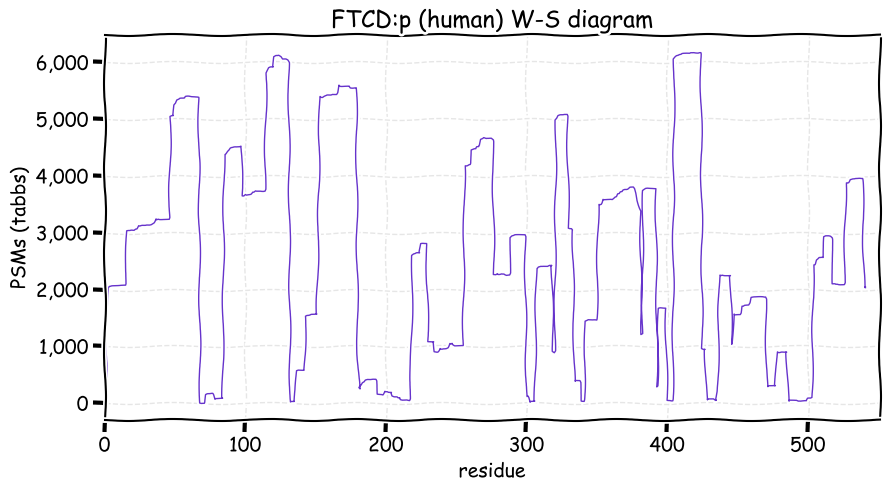
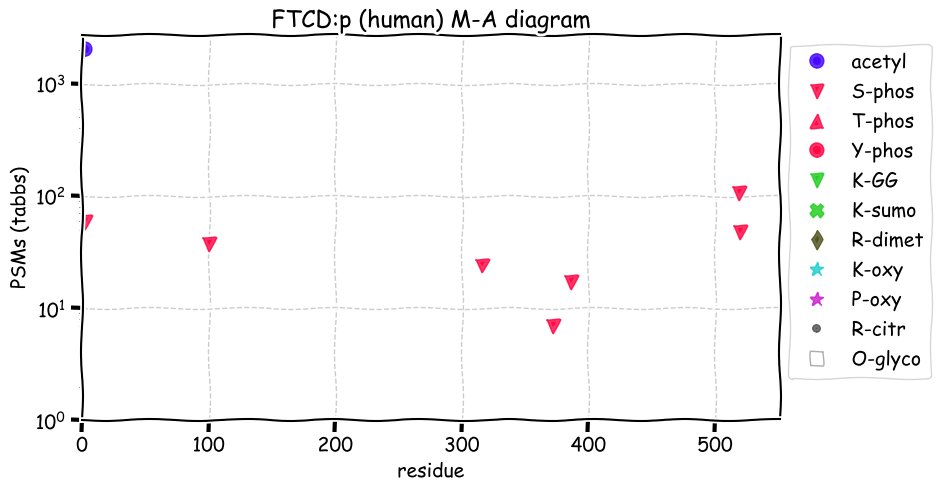
Fri Jun 25 18:44:22 +0000 2021@herrtschmidt @ypriverol I don't know enough about psql to have an opinion, but I have switched over to using JSON for all new projects: XML works but JSON is so easy to parse and validate that it is my format of choice at the moment.
Fri Jun 25 14:56:43 +0000 2021@herrtschmidt @ypriverol Some of the implementation has changed over the years (mainly practical accommodations to deal with size-related speed issues), but the way it works is still based on R. Craig, et al. J Proteome Res. Nov-Dec 2004;3(6):1234-42 (🔗)
Fri Jun 25 14:48:52 +0000 2021@ypriverol One of the things I've learned about using large tables in MySQL (> 10⁹ rows) is that you shouldn't try to be too fancy about the way you arrange things.
Fri Jun 25 14:46:42 +0000 2021@ypriverol No caches or precomputed views, other than MySQL's result caching. Most of the difference is just associated with the size of query: some peptides have been observed very frequently & marshaling the info takes time.
Fri Jun 25 14:37:20 +0000 2021But you have to appreciate the chutzpah of adding in the meta-sign underneath.
Fri Jun 25 14:36:27 +0000 2021Winnipeg's latest style of parking sign. Seems a bit over-the-top for such a prosaic purpose: it is reminiscent of many overly -detailed PowerPoint slides I've seen. 🇨🇦🤔 🔗

Fri Jun 25 14:31:53 +0000 2021@ypriverol A combination of MySql, MongoDB and indexed XML files.
Fri Jun 25 13:37:32 +0000 2021My prediction is that "we don't need no stinking databases" is going to be the winner.
Fri Jun 25 13:35:01 +0000 2021What type of database do you use to store your proteomics results?
Fri Jun 25 12:27:56 +0000 2021And South Africa is well into its 3rd wave 🔗
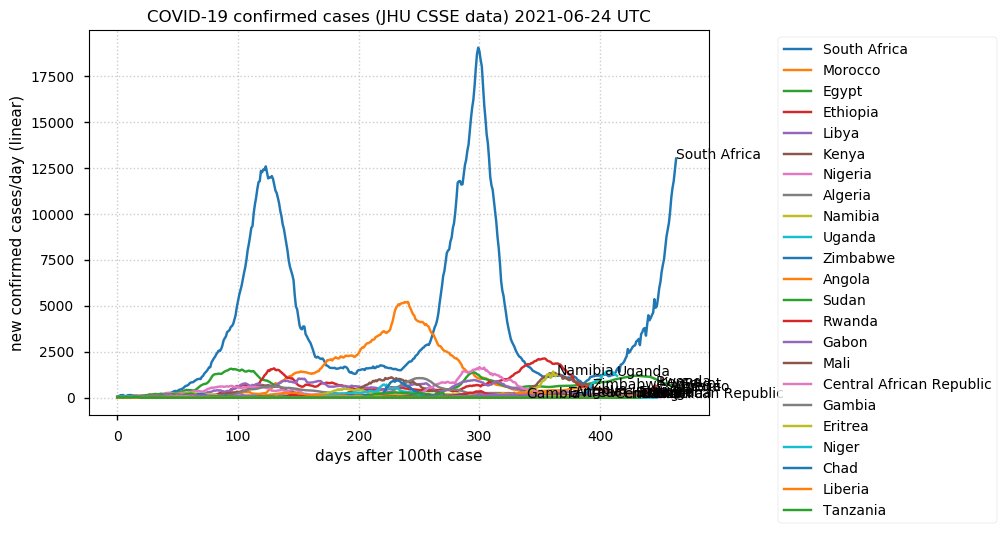
Fri Jun 25 12:26:24 +0000 2021Looks like Russia and the UK are trending up again 🔗
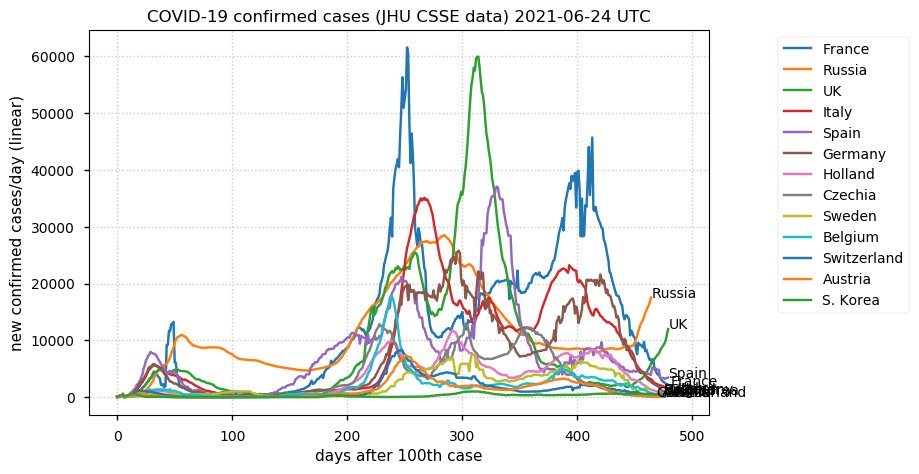
Fri Jun 25 12:18:48 +0000 2021BLVRA:p has a similar abundance & tissue distribution to BLVRB:p. The two proteins are similar size & general enzymatic activity, but have no tryptic peptide overlap & different PTM site patterns.
Fri Jun 25 11:55:22 +0000 2021BLVRA:p.A3T chr 7:g.43771165G>A, rs699512 (all tissue A:T 0.304:0.696) vaf=80%, Δm=30.0106, VAF by population group: african 96%, american 85%, east asian 68%, european 75%, south asian 84%. TT in MCF-7 cells & AT in HEK-293 cells. #ᐯᐸᐱ
Fri Jun 25 11:55:22 +0000 2021BLVRA:p, θ(max) = 83. aka BLVR. Found in MHC class 1 peptide experiments in many tissues; class 2 in thymus. Commonly found in most tissues & cell lines, as well as urine extracellular vesicles.
Fri Jun 25 11:55:21 +0000 2021>BLVRA:p, biliverdin reductase A Small subunit; CTMs: M1+acetyl; PTMs: 11×S, 0×T, 0×Y+phosphoryl; 18×K+acetyl; 7×K+GGyl; 18×K+SUMOyl; SAAVs: A3T (80%); mature form: (1-296) [31,857×, 18 kTa]. #ᗕᕱᗒ 🔗
Thu Jun 24 19:14:06 +0000 2021I had activated the SSH client and server on my Win 10 boxes some time ago, but I only recently realized that not only does Win 10 now have an SFTP command line client, but the boxes are also running an SFTP server as well.
Thu Jun 24 18:26:58 +0000 2021I've said it before & I'll say it again: if you are using CAA in place of IAA to derivatize cysteines, please check to make sure that it worked. 🧐
Thu Jun 24 17:51:19 +0000 2021SAG:p shows up rather strongly (& unexpectedly) in several of the tissues examined in the CPTAC Proteogenomic Analysis of Pediatric Brain Cancer Tumors Pilot Study.
Thu Jun 24 16:21:11 +0000 2021Thunderstorms currently along a mild cold front in the US upper Midwest 🔗
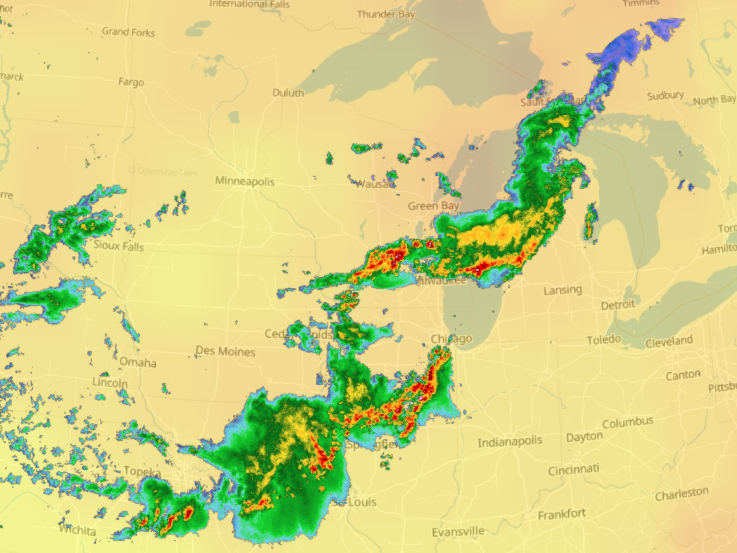
Thu Jun 24 12:04:54 +0000 2021SAG:p.I76V chr 2:g.233320674A>G, rs7565275 (all tissue I:V 0.950:0.050) vaf=6%, Δm=-14.0156, VAF by population group: african 11%, american 4%, east asian 3%, european 5%, south asian 11%. #ᐯᐸᐱ
Thu Jun 24 12:04:54 +0000 2021SAG:p, θ(max) = 98. aka ARRESTIN, RP47. Not found in MHC class 1 or 2 peptide experiments. Most abundant in retina. Tryptic peptide overlap with the other arrestins (e.g. ARR3, ARRB1 & ARRB2).
Thu Jun 24 12:04:54 +0000 2021>SAG:p, S-antigen visual arrestin Small subunit; CTMs: A2+acetyl; PTMs: 1×S, 2×T, 0×Y+phosphoryl; SAAVs: I76V (6%), A101T (1%), P364L (1%); mature form: (2-405) [2,314×, 18 kTa]. #ᗕᕱᗒ 🔗
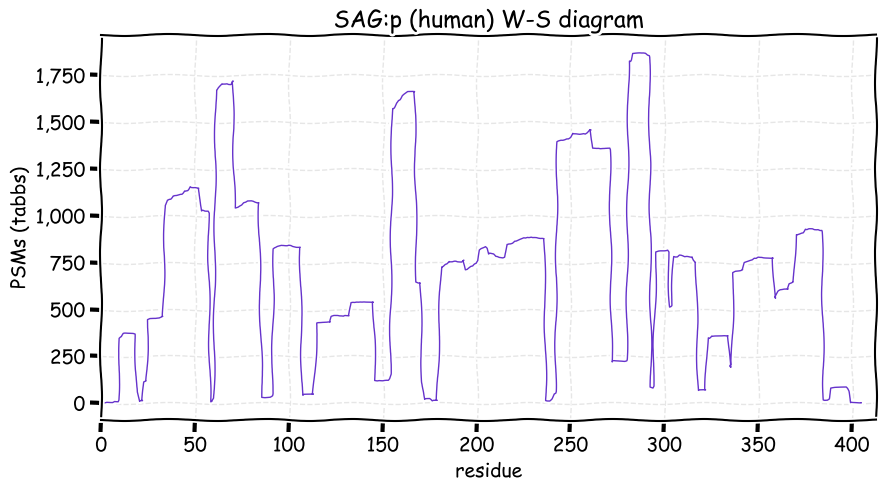
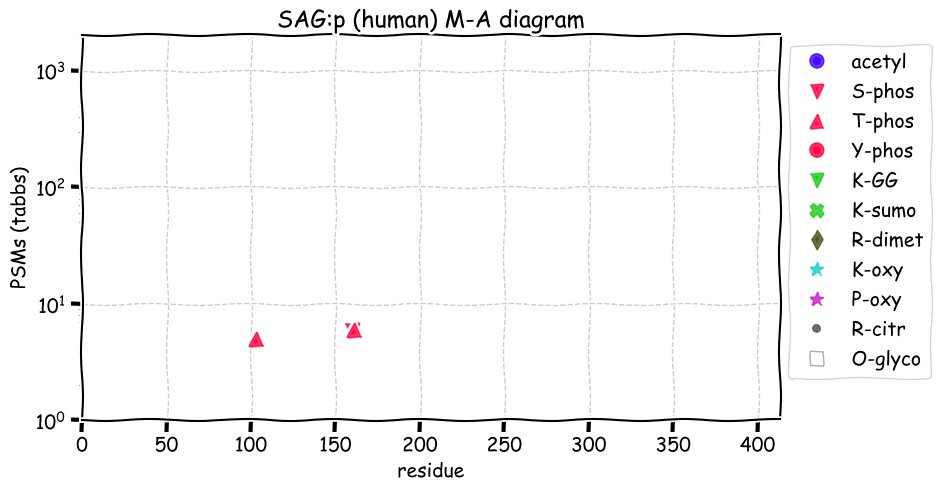
Wed Jun 23 15:11:28 +0000 2021@neely615 @UCDProteomics Another group I look at for thoroughness are the peripheral arm proteins of respiratory Complex I (🔗). There are 44 of them, they are present in a cell at similar stoichiometry, they are all detectable & their gene names all start with NDUF.
Wed Jun 23 11:54:52 +0000 2021GLRX3:p.Q21H chr 10:g.130136483G>C, rs13991 (all tissue Q:H 0.760:0.240) vaf=16%, Δm=9.0003, VAF by population group: african 12%, american 9%, east asian 11%, european 23%, south asian 25%. QH in HeLa cells. HeLa cells are also SS for GLRX3:p.P123S. #ᐯᐸᐱ
Wed Jun 23 11:54:52 +0000 2021GLRX3:p, θ(max) = 50. aka PICOT, bA500G10.4, GRX3, GLRX4, GRX4, TXNL2. Found in MHC class 1 & more rarely in class 2 peptide experiments. Observed in solid tissues, all cell lines & urine extracellular vesicles.
Wed Jun 23 11:54:52 +0000 2021>GLRX3:p, glutaredoxin 3 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 11×K+GGyl; 2×K+SUMOyl; 11×Kacetyl; 10×S, 0×T, 0×Y+phosphoryl; SAAVs: Q21H (16%), P123S (19%); mature form: (2-335) [35,707×, 276 kTa]. #ᗕᕱᗒ 🔗
Wed Jun 23 00:46:57 +0000 2021From 🔗
Wed Jun 23 00:45:25 +0000 2021Today's atmospheric sulfur dioxide concentration across Eurasia and Africa, the darker the purple, the higher the concentration 🔗
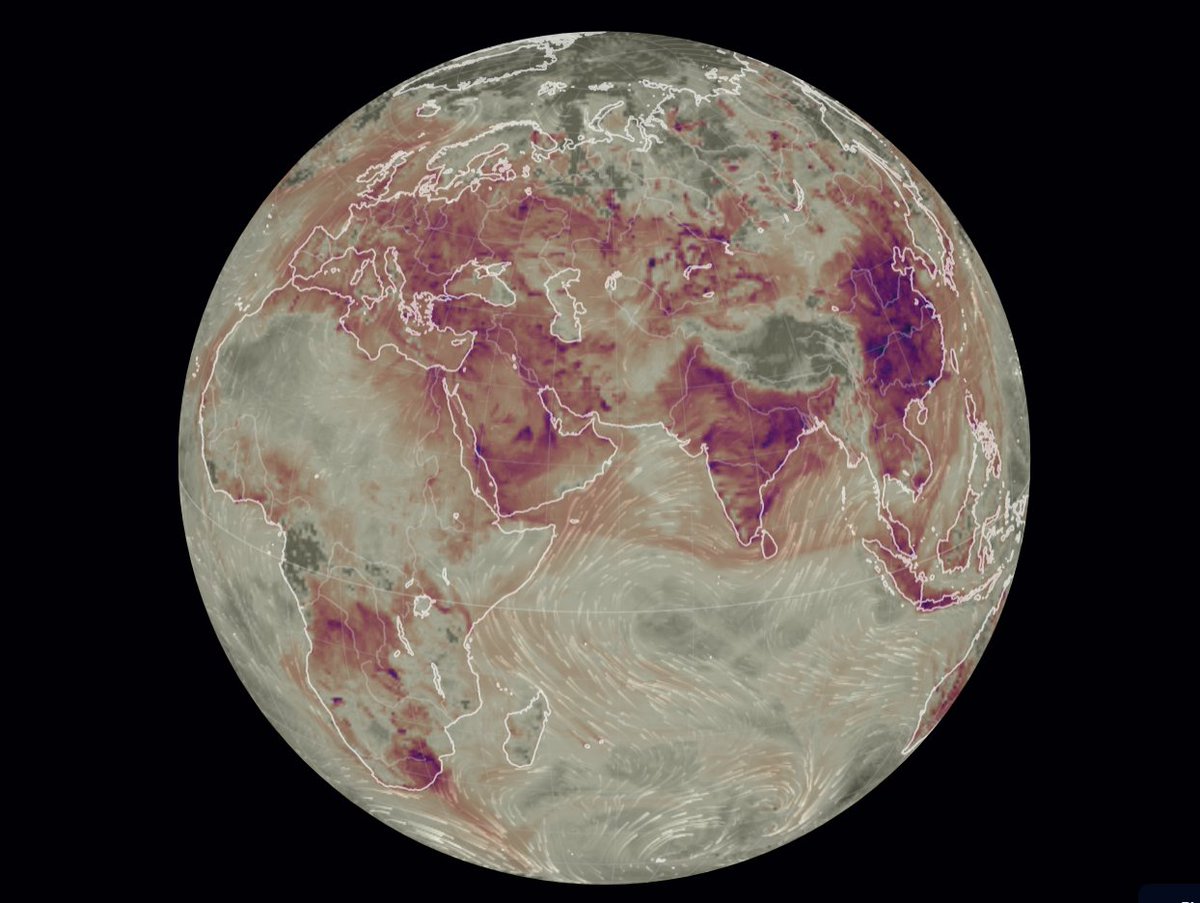
Tue Jun 22 22:52:15 +0000 2021@neely615 @UCDProteomics I just updated the article to use the new (& improved) convention for naming the cytosolic synthetases.
Tue Jun 22 22:44:31 +0000 2021@neely615 @UCDProteomics There are 17 mitochondrial aminoacyl tRNA synthetases, with the same gene names as the cytosolic ones, but with a "2". AARS (cyto) : AARS2 (mito). The article was written before the cytosolic gene names got a "1" to be "consistent".
Tue Jun 22 22:39:08 +0000 2021@neely615 @UCDProteomics 🔗 covers cytosolic aminoacyl tRNA synthetases & 🔗 covers proteins translated from the mitochondrial chromosome.
Tue Jun 22 22:34:02 +0000 2021Sometimes on-line troubleshooting advice on Linux configurations can be very helpful. But not today ...
Tue Jun 22 18:41:44 +0000 2021@VATVSLPR @RDUnwin @JohnRYatesIII @ypriverol @RyanRJulian True, but many protocols require the digested peptides to remain in solution for a relatively long time, e.g. peptide affinity purification (e.g., phosphorylation, GGylation), peptide deglycosylation, or multidimensional chromatography.
Tue Jun 22 18:30:07 +0000 2021@RDUnwin @JohnRYatesIII @ypriverol @RyanRJulian I agree: the extent of deamidation is very dependent on time course of conditions the peptides are kept in following protein cleavage. It is always possible to assign a miscalled A1 to a deamidation, but with high-res gear it is easy to correct this sort of misinterpretation.
Tue Jun 22 13:08:45 +0000 2021@statesdj @umichmedicine Worked on a sepsis drug when I was at Lilly: tough, tough problem.
Tue Jun 22 12:12:09 +0000 2021VASN:p, over 30 years after its discovery, its function is still a bit of a mystery 🔗
Tue Jun 22 12:01:16 +0000 2021VASN:p.T146M chr 16:g.4381314C>T, rs144719357 (all tissue T:M 0.995:0.005) vaf=0.3%, Δm=29.9928, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. #ᐯᐸᐱ
Tue Jun 22 12:01:16 +0000 2021VASN:p, θ(max) = 50. aka SLITL2. Found in MHC class 1 & 2 peptide experiments. Observed in urine, plasma & CSF. Found in some commonly used cell lines, e.g., HeLa. Domains: (23-575) extracellular, (576-596) transmembrane, (597-673) intracellular.
Tue Jun 22 12:01:16 +0000 2021>VASN:p, vasorin (Homo sapiens) Midsized subunit; CTMs: N117, N205, N528+glycosyl; PTMs: 2×K+GGyl; 14×S, 0×T, 3×Y+phosphoryl; SAAVs: T146M (<1%), R161Q (2%); mature form: (23-673) [14,240×, 147 kTa]. #ᗕᕱᗒ 🔗
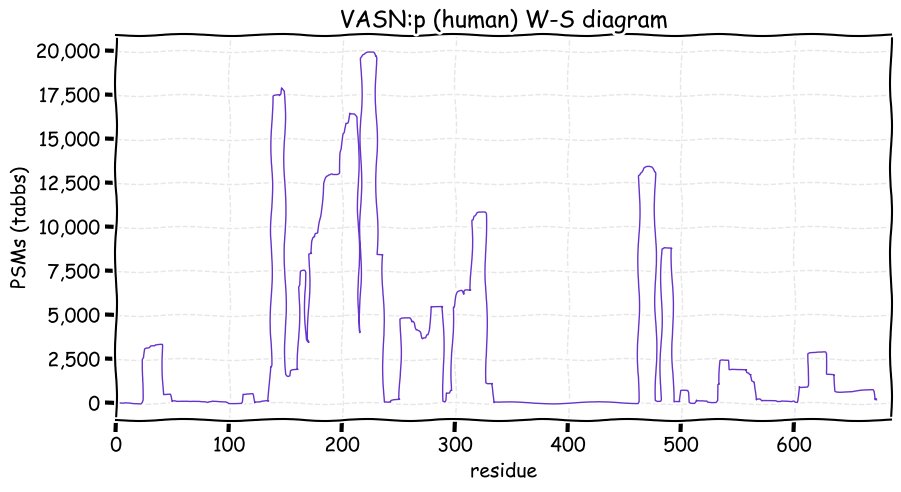
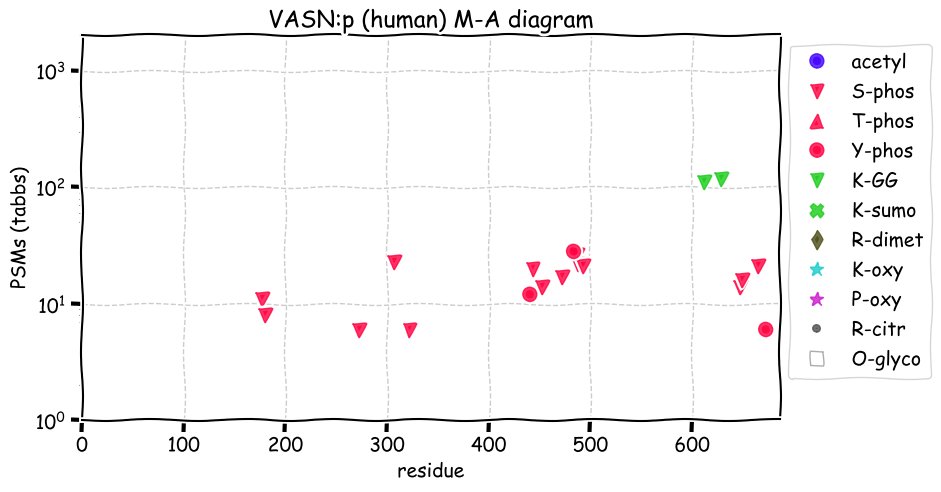
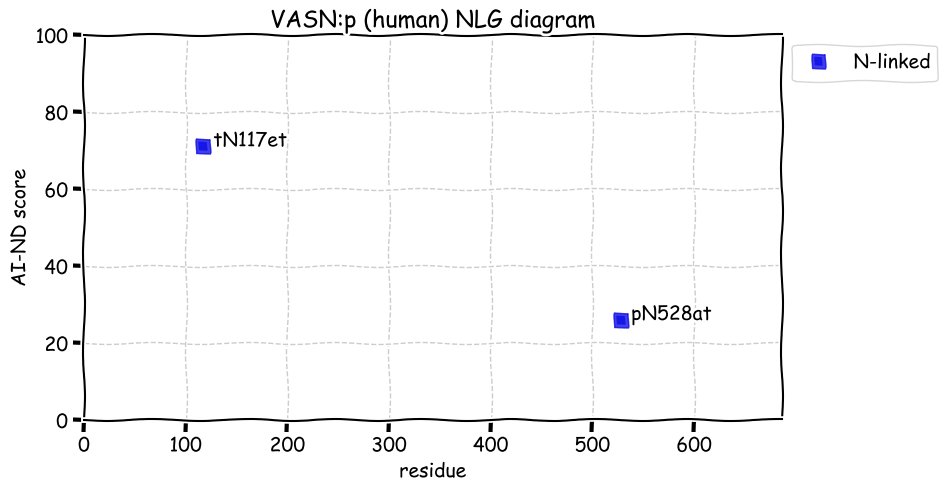
Tue Jun 22 11:59:26 +0000 2021@RobbinBouwmees Agree. Several of the IronWolf drives replaced WD Reds that had problems.
Mon Jun 21 17:42:08 +0000 2021@statesdj I predict it will also be a day of trying to fix up the index structure so that the queries don't take forever on a multi-billion row database.
Mon Jun 21 16:57:12 +0000 2021Controversial take: I have taken to using a KN95 mask. Works well, easily removed, durable and folds up nicely.
Mon Jun 21 15:01:04 +0000 2021@statesdj I agree, but translating what a customer wants into SQL can be challenging (at least for me).
Mon Jun 21 14:17:01 +0000 2021It is going to be one of those days dominated by writing complicated SQL queries & trying to figure out if what is being returned is what the customer really wanted ...
Mon Jun 21 14:13:38 +0000 2021@statesdj The problem seems to occur when the role of a protein doesn't fit in to framework used to build GO, i.e. directed acyclic graphs. Proteins are often involved in cyclic processes, which seems to lead to a sort of compensatory over-annotation.
Mon Jun 21 12:53:31 +0000 2021CD36:p has quite a bit of MS-based proteomics evidence for the removal of an N-terminal signal sequence 1-23, leaving G24 as the N-terminus. The UniProt annotation has the mature sequence starting at G2, with no signal sequence. Thoughts?
Mon Jun 21 12:12:50 +0000 2021CD36:p has too many GO annotations for them to be of much utility as classifiers for the protein/gene.
Mon Jun 21 12:08:15 +0000 2021CD36:p.Q388L chr 7:g.80672807A>T, rs20135571 (all tissue Q:L 00.999:0.001) vaf=<1%, Δm=-60.0542, VAF by population group: african 5%, american 0%, east asian 0%, european 0%, south asian 0%. #ᐯᐸᐱ
Mon Jun 21 12:08:15 +0000 2021CD36:p, θ(max) = 62. aka SCARB3, GPIV, FAT, GP4, GP3B. Found in MHC class 1 & 2 peptide experiments in most tissues. Observed in many tissues, platelets, urine & milk. Common in a limited number of cell lines, e.g., THP-1.
Mon Jun 21 12:08:15 +0000 2021>CD36:p, CD36 molecule (Homo sapiens) Small membrane subunit; CTMs: N79, N205, N220, N235, N321, N417+glycosyl; PTMs: P203+hydroxyl; SAAVs: Q388L (2%); mature form: (24-472) [9,199×, 70 kTa]. #ᗕᕱᗒ 🔗
Sun Jun 20 19:50:06 +0000 2021@KentsisResearch @ProteomicsNews Sure, but that type of system has been around for a long time: people just don't seem to like them. They would rather use a system where the two components come welded together.
Sun Jun 20 19:31:22 +0000 2021@KentsisResearch @ProteomicsNews Really any PSM assignment method that isolates pattern creation and pattern matching into separate activities should work OK. Coming up with testable patterns should be pretty straightforward.
Sun Jun 20 15:49:10 +0000 2021@ProteomicsNews @KentsisResearch But I'd be surprised if any existing algorithm for PTM detection would do a good job of finding that sort of thing.
Sun Jun 20 15:12:41 +0000 2021@ProteomicsNews @KentsisResearch I'd suggest focusing in on ubiquitinylation of ubiquitin initially. It is present at detectable levels in any most MDC experiments and with Lys-C the conjugated peptide will be "ESTLHLVLRLRGG", which should fit the bill wrt your stated parameters.
Sun Jun 20 14:53:16 +0000 2021@JoelisSteele @DavidGomezZep Novel GGylation sites in studies hit zero 2 or 3 years ago. If novel sites are the goal of collecting the data, that effort has largely succeeded. If understanding what the sites do is the goal, we are at the very beginning of the process.
Sun Jun 20 14:24:34 +0000 2021Has anyone else had good luck with IronWolf hard drives? I have 6 of them running now (added a new one yesterday) & they seem to be quite reliable.
Sun Jun 20 13:33:04 +0000 2021@JoelisSteele @DavidGomezZep I hadn't mentioned the source. When I've analyzed a large, PTM enriched data set I check to see how many of the significant results correspond to new PTM sites. The # of new sites has been asymptotically approaching 0 for some time, but this data set actually got there.
Sun Jun 20 12:26:55 +0000 2021@JoelisSteele @DavidGomezZep Here are links to everything about the experiments
🔗
🔗
Sun Jun 20 12:13:00 +0000 2021@DavidGomezZep @JoelisSteele The 50 runs were generated from 5 tissue samples, each generating 10 high pH fractions.
Sun Jun 20 12:06:07 +0000 2021@DavidGomezZep @JoelisSteele Yes. High pH HPLC fractionation first and then low pH HPLC/MS/MS.
Sun Jun 20 12:01:06 +0000 2021RPS6KB1:p.Y413C chr 17:g.20735746A>G, rs199581341 (all tissue Y:C 0.986:0.014) vaf=1.5%, Δm=-60.0542, VAF by population group: african 5%, american 0%, east asian 0%, european 0%, south asian 0%. #ᐯᐸᐱ
Sun Jun 20 12:01:06 +0000 2021RPS6KB1:p, θ(max) = 59. aka S6K1, p70(S6K)-alpha, PS6K, S6K, STK14A. Found in MHC class 1 peptide experiments only. Observed in solid tissue & cell lines: rare in fluids.
Sun Jun 20 12:01:06 +0000 2021>RPS6KB1:p, ribosomal protein S6 kinase B1 (Homo sapiens) Midsized enzyme; CTMs: P2+acetyl (rare); PTMs: 8×K+acetyl; 4×K+GGyl; 1×K+SUMOyl; 9×S, 9×T, 1×Y+phosphoryl; SAAVs: Y413C (1%); mature form: (1-525) [9,537×, 40 kTa]. #ᗕᕱᗒ 🔗
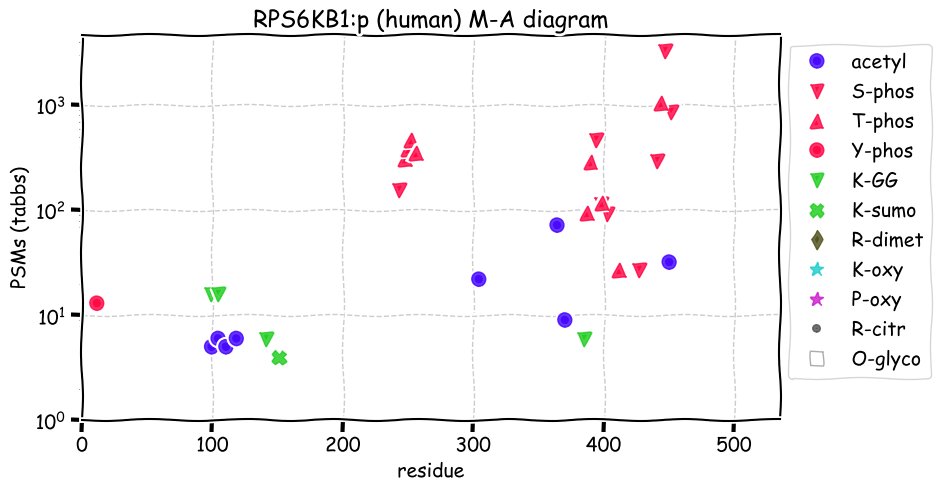
Sat Jun 19 23:10:27 +0000 2021Just finished analyzing a new data set made up of 50 LC/MS/MS runs of TMT-labelled, phosphopeptide enriched sample MDC from human tissue. It generated 250,000 phosphopeptide PSMs. All of the modification sites had been previously observed in other studies. 😎
Sat Jun 19 13:04:53 +0000 2021HMGN's show similar patterns & densities of modification as their more charismatic partners in the dynamics of nucleosome structure, the histones.
Sat Jun 19 12:01:19 +0000 2021HMGN1:p, 16% of the residues in this subunit are K+acetyl acceptors. All of the gene products named HMGNx have an unusually high density of high occupancy acetylation sites.
Sat Jun 19 11:51:52 +0000 2021HMGN1:p.A9T chr 21:g.39348568C>T, rs34613278 (all tissue A:T 0.969:0.031) vaf=6.6%, Δm=30.0106, VAF by population group: african 8%, american 7%, east asian 1%, european 6%, south asian 6%. #ᐯᐸᐱ
Sat Jun 19 11:51:52 +0000 2021HMGN1, θ(max) = 71. aka FLJ27265, FLJ31471, MGC104230, MGC117425, HMG14. Found in MHC class 1 & 2 peptide experiments. Observed in solid tissue & cell lines: rare in fluids.
Sat Jun 19 11:51:52 +0000 2021>HMGN1:p, high mobility group nucleosome binding domain 1 (Homo sapiens) Small subunit; CTMs: P2+acetyl (rare); PTMs: 16×K+acetyl; 7×K+GGyl; 1×K+SUMOyl; 9×S, 4×T+phosphoryl; SAAVs: A9T (7%); mature form: (2-100) [22,744×, 170 kTa]. #ᗕᕱᗒ 🔗
Fri Jun 18 14:51:18 +0000 2021First time I've included a print('\a') into a script in a long time. I'd forgotten how handy it can be ...
Fri Jun 18 14:48:15 +0000 2021To anyone who wants to produce a globally representative library of PSMs for AI & DIA research: try to use a cell line that has a Y chromosome in it.
Fri Jun 18 13:48:35 +0000 2021I have come to the conclusion I really don't like working with big files. They are slow to read and easily corrupted as well as costly to store & maintain. Someone out there should fix this problem ASAP.
Fri Jun 18 12:25:27 +0000 2021QPRT:p part of the large group of proteins that unknowingly provide a counter-argument to the "its mainly just random, junk phosphorylation" assertion.
Fri Jun 18 11:50:14 +0000 2021QPRT:p.A158V chr 16:g.29695123C>T, rs2303255 (all tissue A:V 0.912:0.088) vaf=16%, Δm=28.0313, VAF by population group: african 1%, american 10%, east asian 5%, european 17%, south asian 18%. AV in THP-1 & NK T cells. #ᐯᐸᐱ
Fri Jun 18 11:50:14 +0000 2021QPRT, θ(max) = 94. aka QPRTase. Found in MHC class 1 peptide experiments & rarely in class 2. Observed in solid tissue & cell lines: rare in fluids. Enzyme is a hexamer of 3 homodimers.
Fri Jun 18 11:50:14 +0000 2021>QPRT:p, quinolinate phosphoribosyltransferase (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 1×K+acetyl; 5×K+GGyl; 2×K+SUMOyl; SAAVs: A158V (16%), A185V (10%); mature form: (1-297) [14,421×, 81 kTa]. #ᗕᕱᗒ 🔗
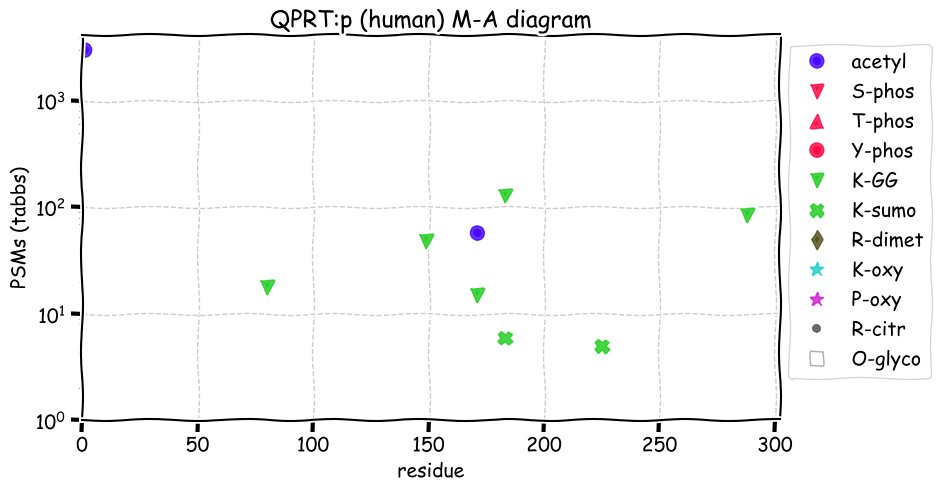
Thu Jun 17 18:04:12 +0000 2021I also think that adding "protein" (in any form) to a gene name isn't very useful. It is rather like insisting that every cat have the word 'cat' appended to its name.
Thu Jun 17 18:01:10 +0000 2021Thanks to everyone who participated in this poll. The majority of respondents who voiced a preference feel that "phosphoprotein" is no longer a useful term to include in a gene name.
Thu Jun 17 15:44:22 +0000 2021@YishaiLevin @pwilmarth It is a paradoxical TOF 🔗
Thu Jun 17 15:16:31 +0000 2021Thanks Yasset. Exactly what I wanted to know. 🔗
Thu Jun 17 14:22:03 +0000 2021@neely615 @pwilmarth I guess I should have included @uniprot in my original question, but can the UniProt folks confirm that the official title for their literature curator jobs is simply "curator"?
Thu Jun 17 13:39:26 +0000 2021@neely615 @pwilmarth That is RefSeq. Last time I heard, the SwissProt portion of UniProt still relies mainly on manual annotation provided by people who read papers and make additions to the entries. I was just wondering what their official job title was within the organization.
Thu Jun 17 12:54:45 +0000 2021Is there a job title for people who do manual annotation for UniProt?
Thu Jun 17 11:55:19 +0000 2021@camtflower Ahem, brother. I suspect many of the people on HGNC naming committees would not be allowed to name family pets when at home.
Thu Jun 17 11:51:35 +0000 2021CEBPZ:p.V102I chr 2:g.37228889C>T, rs2098386 (all tissue V:I 0.106:0.894) vaf=88%, Δm=14.0156, VAF by population group: african 99%, american 95%, east asian 100%, european 85%, south asian 89%. II in JURKAT & SW-480 cells. #ᐯᐸᐱ
Thu Jun 17 11:51:35 +0000 2021CEBPZ, all of the observed GGyl & SUMOyl sites are acetylation sites. Good evidence sequence generated by alternate translation initiation at M57.
Thu Jun 17 11:51:35 +0000 2021CEBPZ, θ(max) = 52. aka CBF2, CTF2, HSP-CBF, NOC1. Found in MHC class 1 lymphocyte-based peptide experiments. Observed in solid tissue & cell lines: rare in fluids.
Thu Jun 17 11:51:35 +0000 2021>CEBPZ:p, CCAAT enhancer binding protein zeta (Homo sapiens) Large subunit; CTMs: A2+acetyl; PTMs: 33×S, 18×T, 0×Y+phosphoryl; 58×K+acetyl; 8×K+GGyl; 4×K+SUMOyl; SAAVs: P15S (13%), V102I (88%), P201L (27%); mature form: (2-1054) [24,179×, 180 kTa]. #ᗕᕱᗒ 🔗
Thu Jun 17 11:38:59 +0000 2021NIH announces collaboration with itself
🔗
Wed Jun 16 15:52:54 +0000 2021Does adding the descriptor "phosphoprotein" to a gene's name really add value anymore?
Wed Jun 16 12:28:03 +0000 2021HDGF:p is the abbreviation for the old name for the gene (Hepatoma-Derived Growth Factor), which remained in place following the change to Heparin Binding Growth Factor. Since the protein is a normal constituent of all cell types, the new name is probably a better descriptive.
Wed Jun 16 12:05:44 +0000 2021Dust (PM10 particles) coming off North Africa crossing the Atlantic to the Americas today. 🔗
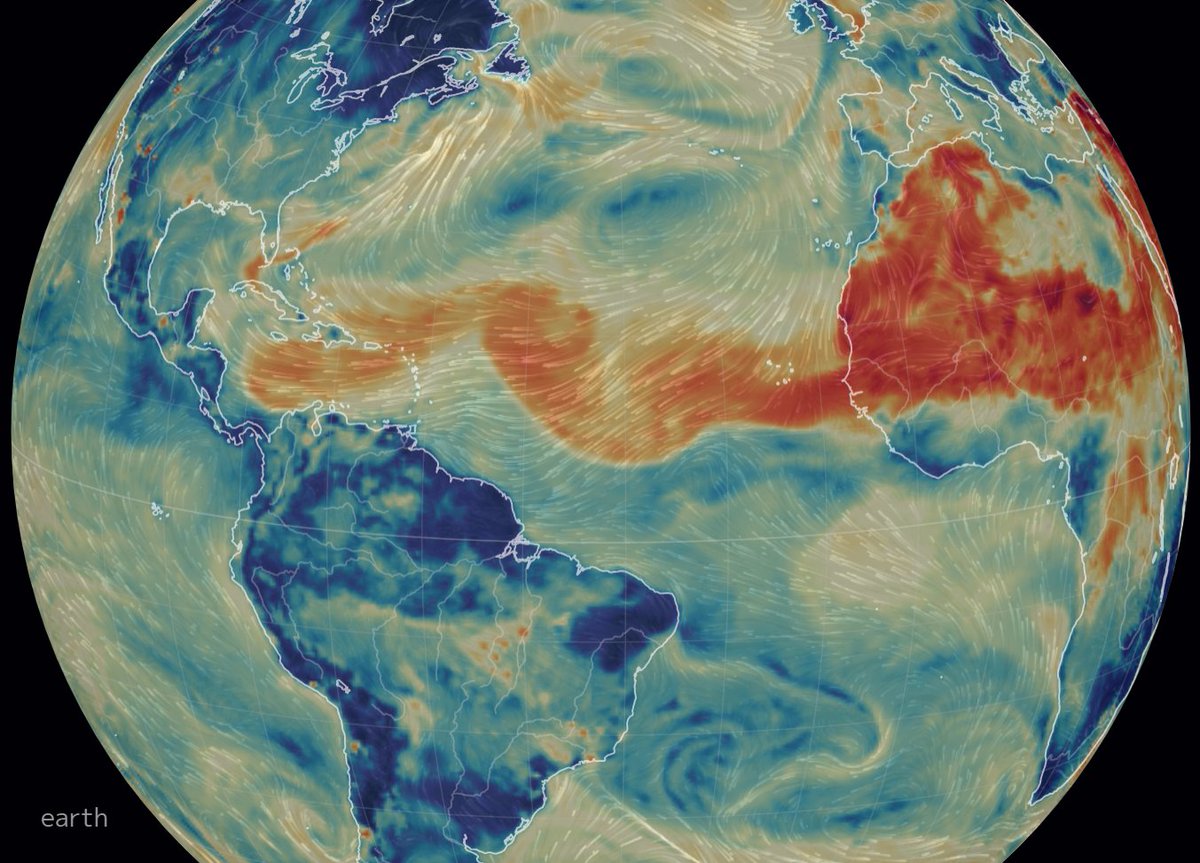
Wed Jun 16 11:59:35 +0000 2021HDGF:p.P201L chr 1:g.156743766G>A, rs4399146 (GPMDB all sample P:L 0.702:0.298) vaf=27%, Δm=16.0313, VAF by population group: african 7%, american 22%, east asian 12%, european 33%, south asian 26%. PL in A-431 & HaCaT cells. #ᐯᐸᐱ
Wed Jun 16 11:59:34 +0000 2021HDGF, θ(max) = 91. aka HMG1L2. Found in MHC class 1 peptide experiments in all tissues. Observed in solid tissue & cell lines: absent from fluids.
Wed Jun 16 11:59:34 +0000 2021>HDGF:p, heparin binding growth factor (Homo sapiens) Small subunit; CTMs: S2+acetyl); PTMs: 14×S, 6×T, 2×Y+phosphoryl; 26×K+acetyl; 14×K+GGyl; 2×K+SUMOyl; SAAVs: G177R (1%), P201L (27%); mature form: (2-240) [59,240×, 640 kTa]. #ᗕᕱᗒ 🔗
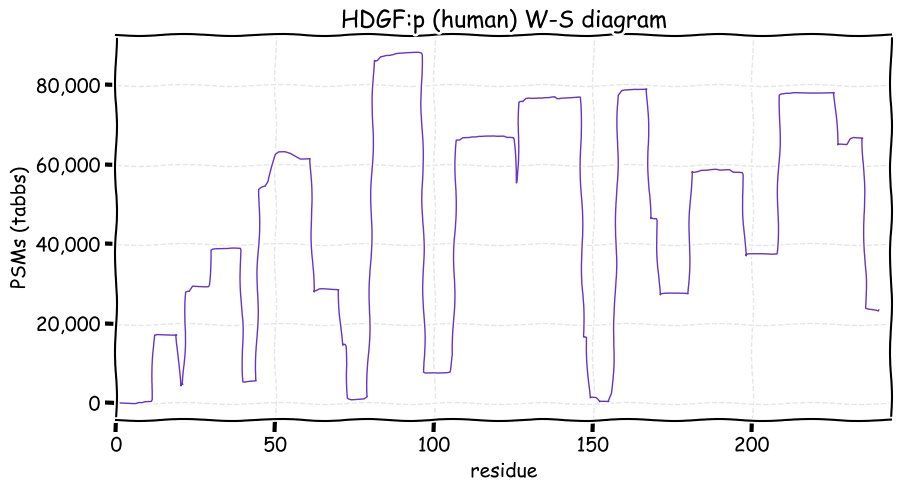
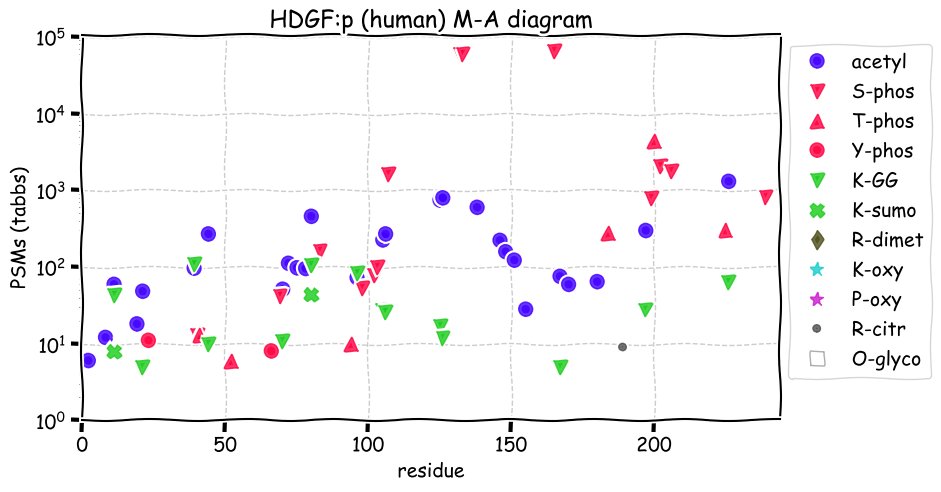
Wed Jun 16 00:32:32 +0000 2021@statesdj @StevenSalzberg1 Can't say: it is proprietary info at the moment.
Tue Jun 15 23:25:18 +0000 2021@statesdj @StevenSalzberg1 1. No we don't know; &
2. Yes, it is something I do regularly.
Tue Jun 15 20:31:51 +0000 2021@gingraslab1 @Lange_Lab @crc @CRC isn't pointing to where I think you think it does. I think you want @CRC_CRC.
Tue Jun 15 17:13:52 +0000 2021How should one interpret the results of a study that claims to provide "deep" proteome coverage of a cell lysate, but only one mitochondrial tRNA synthetase is detected? I'd say interpret it "with all due caution".
Tue Jun 15 15:19:40 +0000 2021miniMAVS:p would retain all of the high occupancy phosphorylation sites, but it does not have the caspase activation and recruitment domain (CARD, 3-93) believed to be a functional component of full length MAVS:p.
Tue Jun 15 15:03:10 +0000 2021"DARPA for Tractors" 🔗
Tue Jun 15 13:23:02 +0000 2021This is not a positive development 🔗
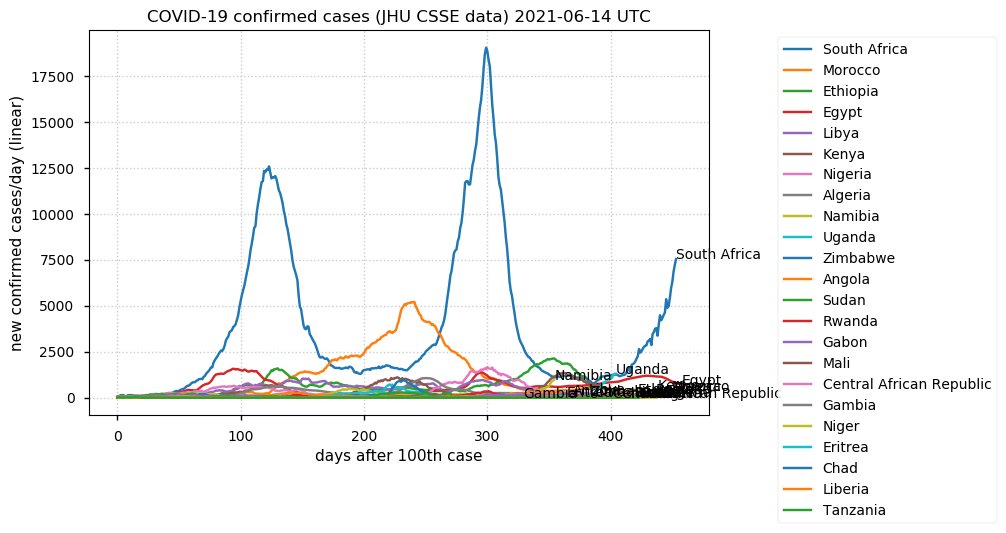
Tue Jun 15 12:59:21 +0000 2021@statesdj Thanks to everyone who participated in this poll. The majority opinion was the < 20% of human genes have a sufficiently well-defined function.
Tue Jun 15 12:48:35 +0000 2021MAVS:p, a shorter translation of the mRNA formed by alternate translation initiation at M142 (143-540), is referred to as miniMAVS.
Tue Jun 15 12:13:52 +0000 2021MAVS:p.S409F chr 20:g.3865750C>T, rs7269320 (all tissue S:F 0.942:0.058) vaf=16%, Δm=60.0364, VAF by population group: african 41%, american 13%, east asian 9%, european 6%, south asian 18%. SF in HEK-293T & MDA-MB-231 cells. #ᐯᐸᐱ
Tue Jun 15 12:13:52 +0000 2021MAVS:p, domains: (2-513) cytoplasmic , (514–534) transmembrane (outer mitochondrial membrane), (535-540) mitochondrial intermembrane space. All observed phosphorylation is within the cytoplasmic domain.
Tue Jun 15 12:13:52 +0000 2021MAVS:p, θ(max) = 83. aka VISA, KIAA1271, IPS-1, Cardif. Found in MHC class 1 peptide experiments in all tissues. Observed in solid tissue & cell lines: absent from fluids.
Tue Jun 15 12:13:52 +0000 2021>MAVS:p, mitochondrial antiviral signaling protein (Homo sapiens) Midsized mitochondrial membrane subunit; CTMs: P2+acetyl (trace); PTMs: 43×S, 17×T, 2×Y+phosphoryl; SAAVs: .Q93E (29%), R218C (5%), S409F (16%); mature form: (2-540) [25,575×, 141 kTa]. #ᗕᕱᗒ 🔗
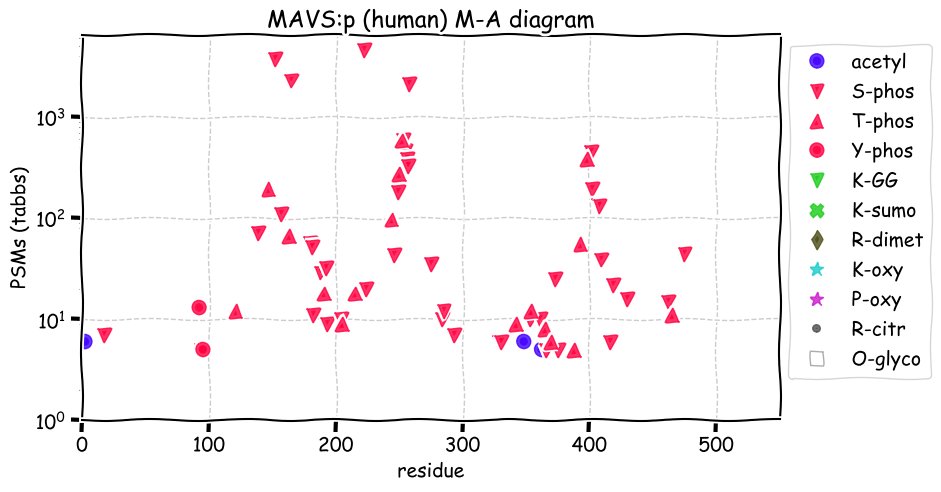
Mon Jun 14 13:29:49 +0000 2021@statesdj @neely615 That's why "do you feel" is in the question. Different people in different specialization will view proteins & their functions in a variety of alternate ways. Is being assigned to a "family" enough? Do you need to know an enzyme's substrate? For the poll, it is up to you.
Mon Jun 14 12:51:17 +0000 2021In response to a comment by @statesdj: What fraction of human protein coding genes do you feel have well-defined functions?
Mon Jun 14 12:22:41 +0000 2021I'm assuming this is a rhetorical flourish on the part of the authors: many of the ~19,500 currently annotated protein-coding genes have unknown functions.
Mon Jun 14 11:57:44 +0000 2021SBSN:p "Among the ~22,000 human genes, very few remain that have unknown functions. One such example is suprabasin (SBSN)" from "Suprabasin-A Review" 🔗
Mon Jun 14 11:57:44 +0000 2021SBSN:p.A419G chr 19:g.35527026G>C, rs10775583 (all tissue A:G 0.740:0.260) vaf=41%, Δm=-14.0157, VAF by population group: african 21%, american 47%, east asian 66%, european 36%, south asian 48%. AG in HEK-293 & HCT-116 cells. #ᐯᐸᐱ
Mon Jun 14 11:57:44 +0000 2021SBSN:p, θ(max) = 80. aka UNQ698, HLAR698. Found in MHC class 1 & 2 peptide experiments. Observed in blood plasma, saliva, urine & many commonly used cell lines, e.g. HEK-293, HeLa & HCT-116.
Mon Jun 14 11:57:44 +0000 2021>SBSN:p, suprabasin (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 5×S, 0×T, 0×Y+phosphoryl, 4×R+deimination; SAAVs: A250V (3%), T285A (6%) A419G (41%); mature form: (24-590) [12,299×, 130 kTa]. #ᗕᕱᗒ 🔗
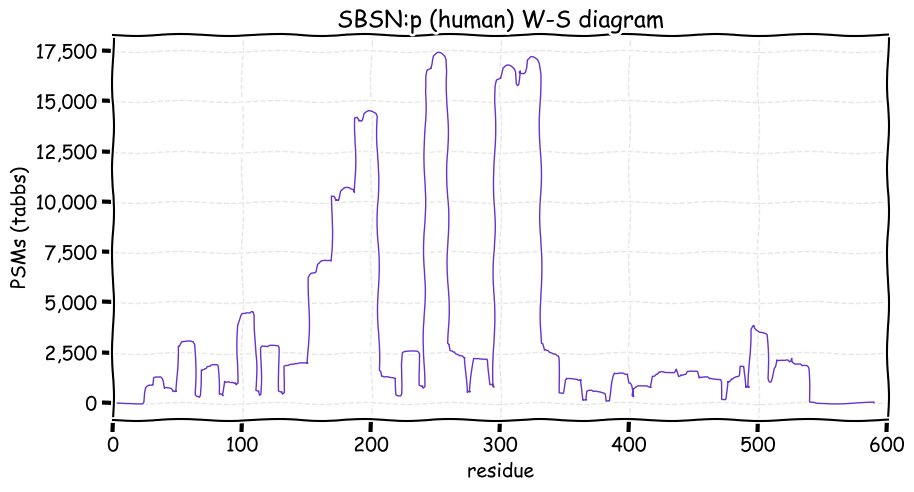
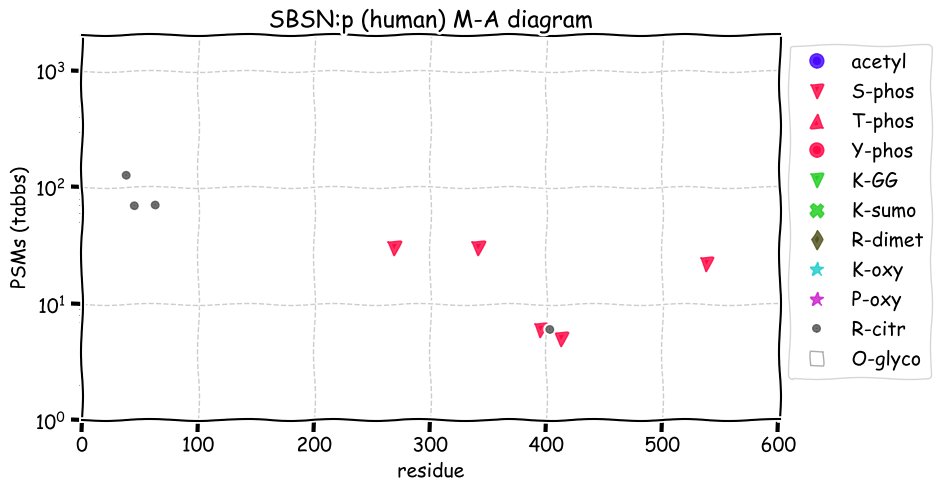
Sun Jun 13 17:57:40 +0000 2021@UCDProteomics @neely615 @lkpino Also, because of visa restrictions in the US, it is a good time for Canucks to apply for US private sector positions, because the relative ease of obtaining NAFTA TN-work permits.
Sun Jun 13 17:39:21 +0000 2021@UCDProteomics @neely615 @lkpino My reading of the current situation is that the US job market in bioscience is pretty strong in the private sector (outside of academia) & stagnant-to-receding in academia.
Sun Jun 13 17:08:53 +0000 2021The proteomics evidence regarding the observation of BCAS1:p in MCF-7 may be contrary to the statement made in RefSeq's entry for this gene wrt MCF-7: 🔗 🔗
Sun Jun 13 15:53:43 +0000 2021BCAS1:p is an interesting name for this protein/gene, however there is still no function associated with it, nor any causal relationship established between the protein & cancer or any other disease.
Sun Jun 13 12:22:52 +0000 2021BCAS1:p is in the group of 834 genes referred to as "MicroRNA protein coding host genes".
Sun Jun 13 12:19:46 +0000 2021BCAS1:p.Q472H chr 20:g.53957432T>G, rs35575210 (all tissue Q:H 0.920:0.080) vaf=95%, Δm=-9.0003, VAF by population group: african 70%, american 96%, east asian 89%, european 97%, south asian 96%. #ᐯᐸᐱ
Sun Jun 13 12:18:59 +0000 2021BCAS1:p, θ(max) = 65. aka NABC1, AIBC1, PMES-2. Found in MHC class 1 & 2 peptide experiments in spinal cord. Observed in many solid tissues & some cell lines, particularly MCF-7.
Sun Jun 13 12:05:30 +0000 2021>BCAS1:p, breast carcinoma amplified sequence 1 (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 32×S, 20×T, 0×Y+phosphoryl; SAAVs: T56M (1%), V163A (13%), G255E (14%), Q472H (13%), S583P (13%); mature form: (1-584) [6,143×, 60 kTa]. #ᗕᕱᗒ 🔗
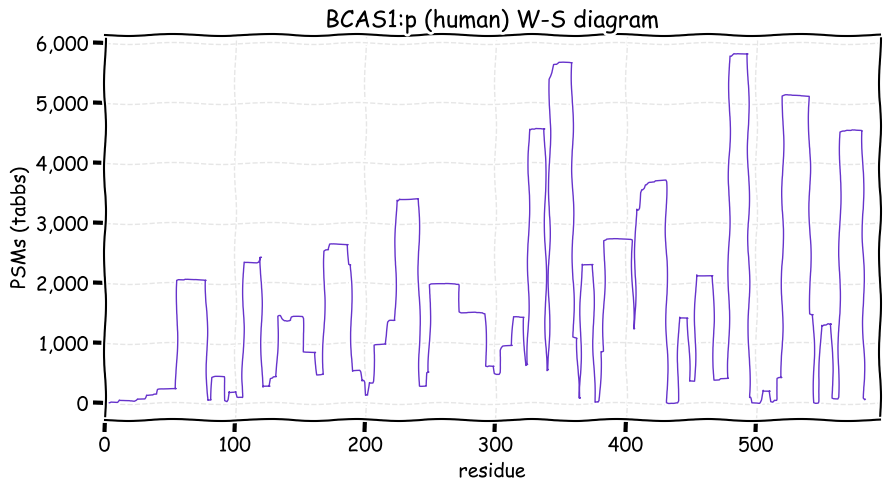
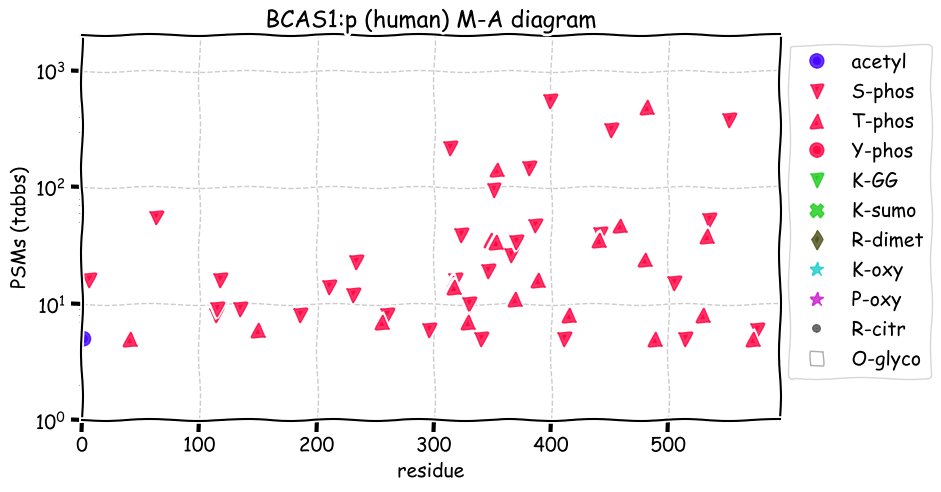
Sat Jun 12 12:26:59 +0000 2021@BiswapriyaMisra @kbseah Extensive analyte matrix adduct formation was one of the first things we noticed about the matrices used for peptide laser desorption.
Sat Jun 12 12:09:24 +0000 2021ZBF830:p needs the equivalent of a Kickstarter to get it a better name. Previously called "coiled-coil domain containing 16" & "orphan maintenance of genome 1".
Sat Jun 12 12:04:54 +0000 2021ZNF830:p.H99Q chr 17:g.34961863T>G, rs931196 (all tissue H:Q 0.114:0.886) vaf=95%, Δm=-9.0003, VAF by population group: african 70%, american 96%, east asian 89%, european 97%, south asian 96%. #ᐯᐸᐱ
Sat Jun 12 12:04:54 +0000 2021ZNF830:p, θ(max) = 65. Found in MHC class 1 peptide experiments. Commonly observed in solid tissues & many cell lines.
Sat Jun 12 12:04:54 +0000 2021>ZNF830:p, zinc finger protein 830 (Homo sapiens) Small nuclear subunit; CTMs: A2+acetyl; PTMs: 10×S, 4×T, 1×Y+phosphoryl; 2×K+acetyl; SAAVs: H99Q (95%), S154T (13%); mature form: (2-372) [8,595×, 28 kTa]. #ᗕᕱᗒ 🔗
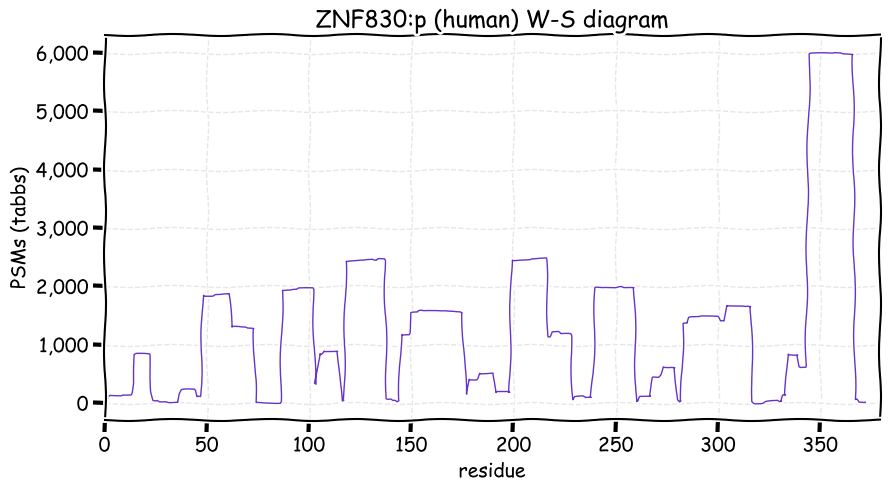
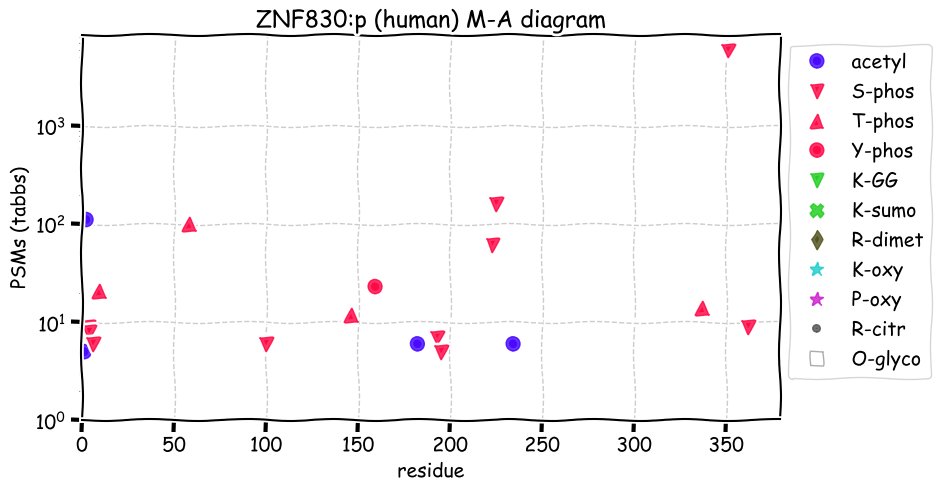
Fri Jun 11 14:31:13 +0000 2021@realBioMassSpec @JoelisSteele It is a rather nice study that shows what can be done using MS-based proteomics to examine exosomes. Any group involved in the subject should find the complete set of experimental data of interest. 🔗
Fri Jun 11 12:19:33 +0000 2021SAA1:p, SAA2:p & SAA3:p are secreted into the blood in response to an acute inflammation. SAA4:p is different in that it is secreted continuously by the liver.
Fri Jun 11 12:00:52 +0000 2021SAA4:p.C89Y chr 11:g.18231629C>T, rs2460827 (all tissue C:Y 0.202:0.798) vaf=95%, Δm=60.0541, VAF by population group: african 70%, american 96%, east asian 89%, european 97%, south asian 96%. #ᐯᐸᐱ
Fri Jun 11 12:00:52 +0000 2021SAA4:p, θ(max) = 74. Found in tissue MHC class 1 & 2 peptide experiments in many tissues. Most abundant in blood plasma & CSF; rare in cell lines & urine.
Fri Jun 11 12:00:52 +0000 2021>SAA4:p, serum amyloid A4 (Homo sapiens) Small subunit; CTMs: none; PTMs: K64+GGyl; K26, K74, K107+acetyl; SAAVs: C89Y (95%), R120C (1%); mature form: (23-130) [11,599×, 60 kTa]. #ᗕᕱᗒ 🔗
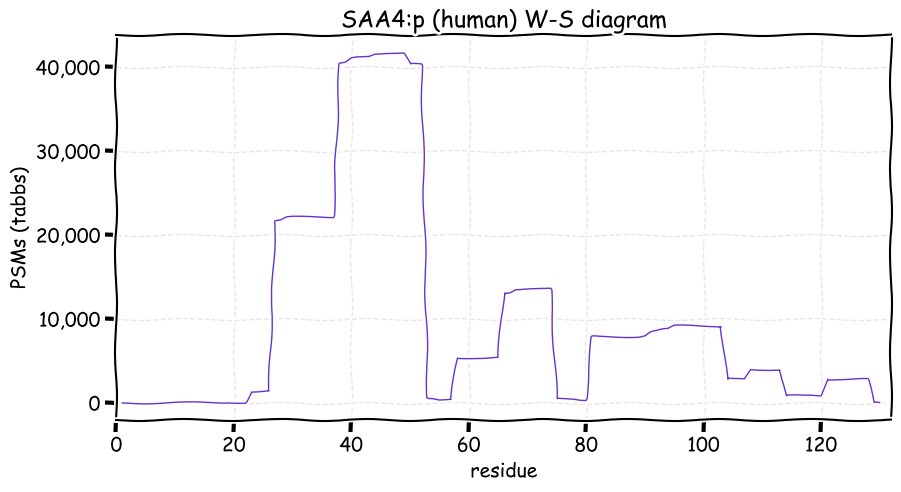
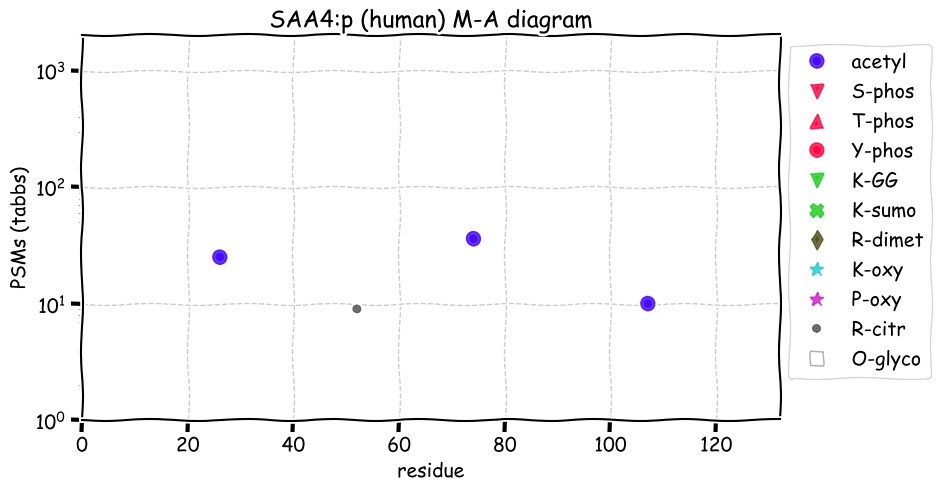
Thu Jun 10 19:08:45 +0000 2021@MHendr1cks Ended up taking an offer from UBC, where things were at least a bit more "modern".
Thu Jun 10 19:03:59 +0000 2021@MHendr1cks I can believe it. I interviewed for a position at McGill years ago & I was surprised by the poor condition of the buildings and physical plant even then.
Thu Jun 10 17:01:07 +0000 2021Blog post with a pretty good discussion of how science denial works in practice 🔗
Thu Jun 10 12:18:26 +0000 2021SAA2:p.R89H chr 11:g.18245480C>T, rs2468844 (all tissue R:H 0.202:0.798) vaf=84%, Δm=-19.0422, VAF by population group: african 61%, american 78%, east asian 93%, european 89%, south asian 70%. #ᐯᐸᐱ
Thu Jun 10 12:18:26 +0000 2021SAA2:p, θ(max) = 85. Found in tissue MHC class 1 & 2 peptide experiments in most tissues. Most abundant in blood plasma & CSF; rare in cell lines & urine.
Thu Jun 10 12:18:26 +0000 2021>SAA2:p, serum amyloid A2 (Homo sapiens) Small subunit; CTMs: none; PTMs: K64+GGyl; K121+acetyl; SAAVs: E74D (1%), R89H (84%); mature form: (19-122) [8,835×, 60 kTa]. #ᗕᕱᗒ 🔗
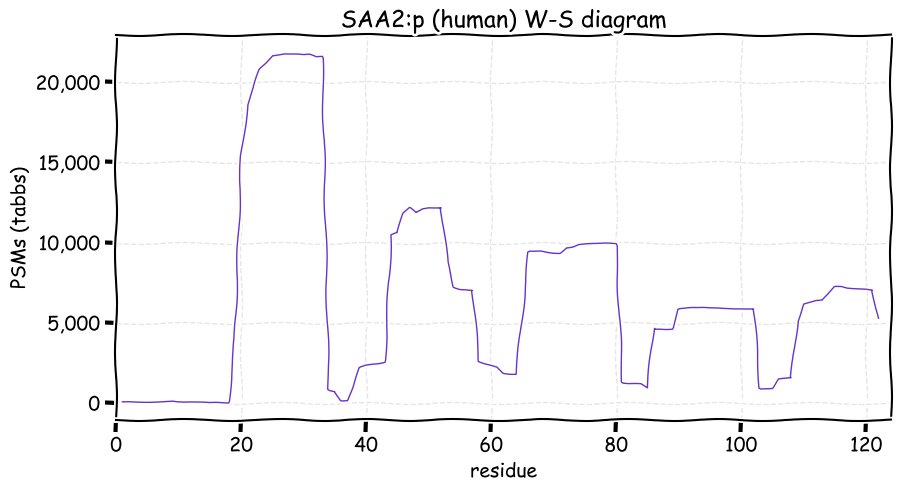
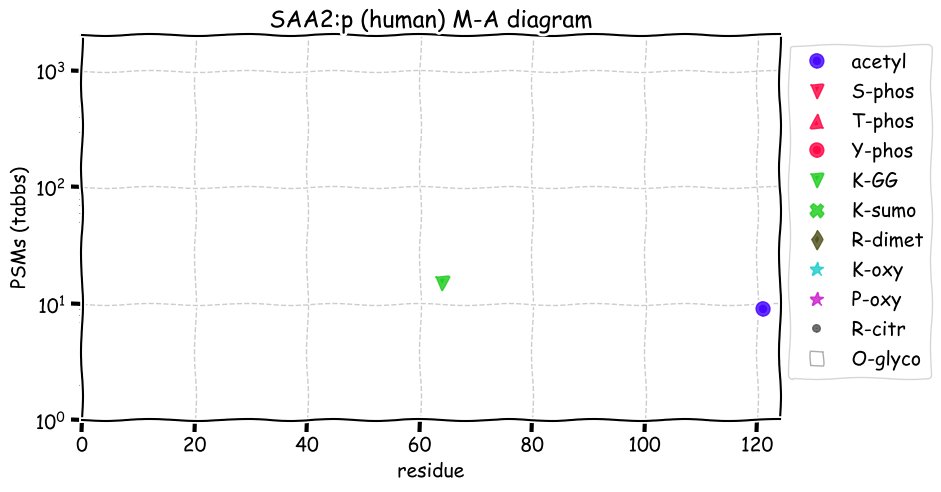
Wed Jun 09 15:50:57 +0000 2021@cwvhogue You are absolutely right.
Wed Jun 09 14:21:50 +0000 2021DDC:p The name DDC implies a specific target (DOPA), however it can decarboxylate many aromatic amino acids, e.g.
Tyr → tyramine,
His → histamine,
5-hydroxy-Tyr→ serotonin.
Why there is so much of it in urine is anybody's guess at this point.
Wed Jun 09 12:03:34 +0000 2021DDC:p.M17V chr 7:g.50544037T>C, rs6264 (all tissue M:V 0.042:0.958) vaf=>99%, Δm=-31.9721, VAF by population group: african 98%, american 100%, east asian 100%, european 100%, south asian 100%. VV in CACO-2 cells. #ᐯᐸᐱ
Wed Jun 09 12:03:33 +0000 2021DDC:p, θ(max) = 81. aka AADC. Found in tissue MHC class 1 & 2 peptide experiments in a limited number of tissues. Most frequently found in urine & some cell lines (e.g., CACO-2 cells). 🔗
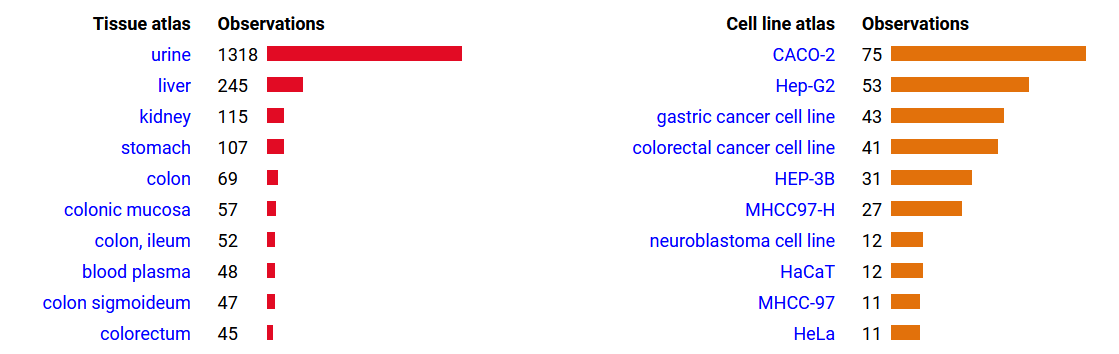
Wed Jun 09 12:03:33 +0000 2021>DDC:p, dopa decarboxylase (Homo sapiens) Small enzyme; CTMs: M1+acetyl; PTMs: S149, S220+phosphoryl; SAAVs: M17V (>99%), M217V (3%) R462Q (5%); mature form: (1-480) [4,359×, 28 kTa]. #ᗕᕱᗒ 🔗
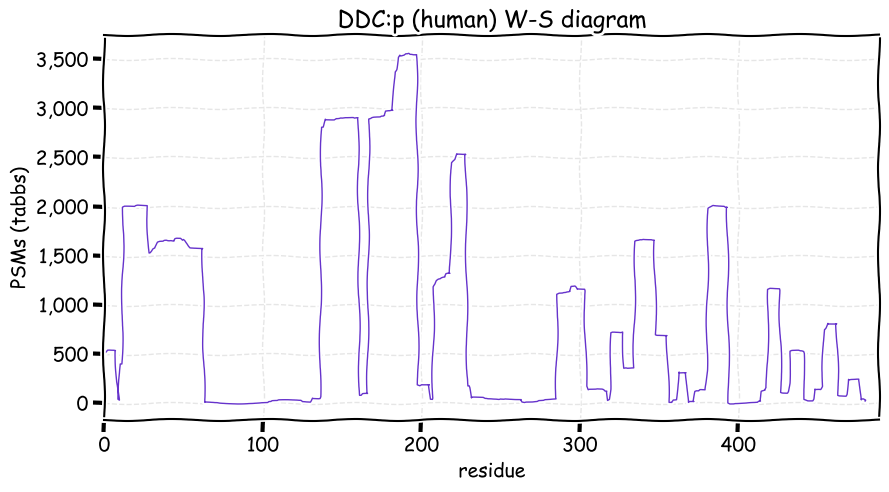
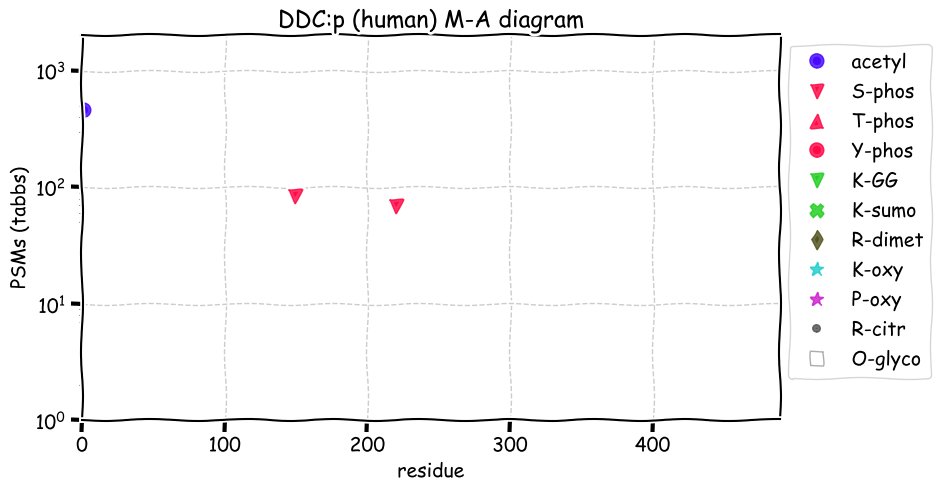
Wed Jun 09 01:45:13 +0000 2021While it wasn't the point of the study (PXD024967), the data files starting with "Urine_SARS_TP_TMT" represent the best example of urine from a Klebsiella pneumoniae UTI I've seen.
Wed Jun 09 01:02:06 +0000 2021@slavov_n There are patent applications available 🔗 but after reading a few of them I still don't understand how they are planning to sequence proteins.
Wed Jun 09 00:35:00 +0000 2021@slavov_n I'd have to know how their sequencing technology works. Nothing that I could find on their site mentions anything about how it works or an indication of demonstrated LOD/LOQ, either in vitro or in vivo.
Tue Jun 08 15:25:35 +0000 2021Because of this issue, I'm going to stop referring to K acceptor sites observed with GlyGly modification as being ubiquitinylated lysines & call them GGylated lysines instead. 4/4
Tue Jun 08 15:25:03 +0000 2021This method works & it can result in data with 80-90% of the PSMs matching GG-modified peptides. But there is a problem: 2 other proteins (NEDD8 & ISG15) conjugate with other proteins in the same way & also result in a GG-modified K sidechain. 3/4
Tue Jun 08 15:24:34 +0000 2021A common method of detecting this PTM is to digest a sample with trypsin, enrich for GG-modified K using an antibody & then perform LC/MS/MS, e.g. 🔗 2/4
Tue Jun 08 15:21:36 +0000 2021Ubiquitinylation (🔗) is a common PTM generated by conjugating the C-terminal of ubiquitin (…RGG-) to the sidechain of lysine. 1/4
Tue Jun 08 11:53:25 +0000 2021IFI16:p.S179T chr 1:g.159016687G>C, rs866484 (all tissue S:T 0.248:0.752) vaf=74%, Δm=14.0157, VAF by population group: african 41%, american 84%, east asian 59%, european 79%, south asian 68%. TT in JURKAT & HaCaT cells. ST in MCF-10A cells. #ᐯᐸᐱ
Tue Jun 08 11:53:25 +0000 2021IFI16:p, θ(max) = 81. aka IFNGIP1, PYHIN2. Found in tissue MHC class 1 peptide experiments from all tissues, but rarely in class 2. Found in most solid tissues & cell lines, but not in fluids.
Tue Jun 08 11:53:25 +0000 2021>IFI16:p, interferon gamma inducible protein 16 (H sapiens) Midsized subunit; CTMs: none; PTMs: 31×S, 29×T, 0×Y+phosphoryl; 21×K+acetyl; 44×K+ubiquitinyl; 18×K+SUMOyl; SAAVs: S179T(74%), R409S(63%), Y413N(63%), T723S(23%); mature form: (2-729) [31,059×, 318 kTa]. #ᗕᕱᗒ 🔗
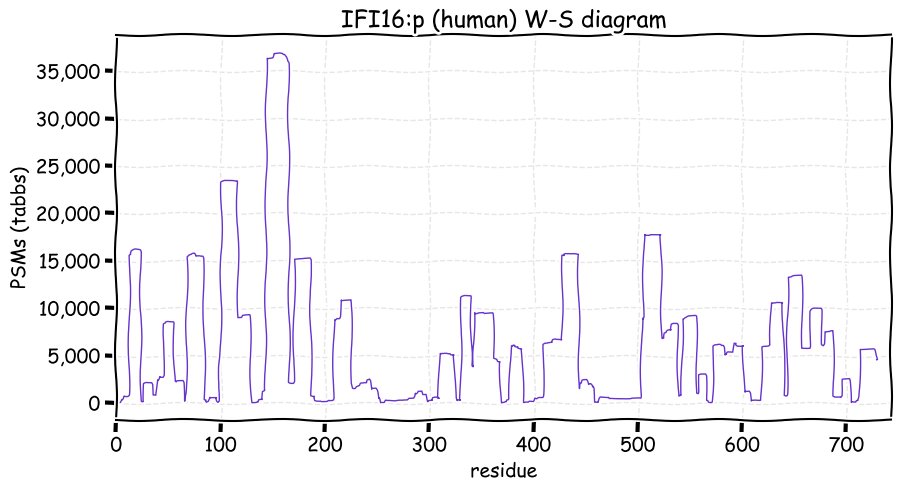
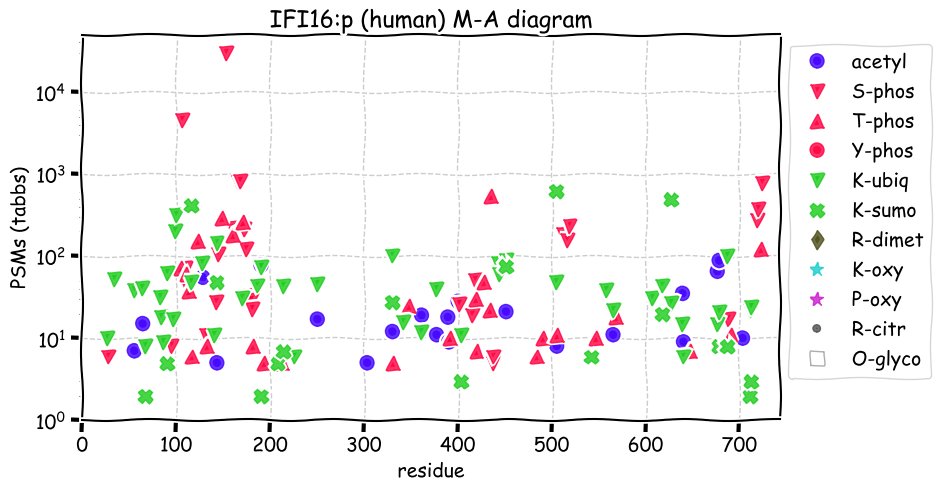
Mon Jun 07 12:38:59 +0000 2021Law of Conservation of Academic Positions:
The average number of students a tenured professor will train over the course of their career who will, in turn, become a tenured professor ≈ 1.
Mon Jun 07 11:52:29 +0000 2021CROCC:p.V1110M chr 1:g.16954740G>A, rs41272737 (all tissue V:M 0.964:0.036) vaf=4%, Δm=31.9721, VAF by population group: african 10%, american 2%, east asian 0%, european 5%, south asian 1%. #ᐯᐸᐱ
Mon Jun 07 11:52:29 +0000 2021CROCC:p, θ(max) = 87. aka rootletin, ROLT. Found in tissue MHC class 1 peptide experiments from ovary. Found in most solid tissues & urine. Has an observed alternate translation initiation site at M73, resulting in an A74+acetyl N-terminus.
Mon Jun 07 11:52:29 +0000 2021>CROCC:p, ciliary rootlet coiled-coil, rootletin (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 76×S, 7×T, 0×Y+phosphoryl; SAAVs: R372Q(9%), D463Y (2%), R903C (2%), V1110M (4%), T1361A (7%), G1471R (7%), S1744G (9%); mature form: (1-2017) [30,020×, 76 kTa]. #ᗕᕱᗒ 🔗
Sun Jun 06 19:05:56 +0000 2021Thanks to everyone who participated in this poll. On the basis of a majority (& the absence of a substantive counterargument), millidaltons it is!
Sun Jun 06 16:59:39 +0000 2021@pwilmarth I am willing to forgo the observation of such effects where v < 0.001c.
Sun Jun 06 15:54:23 +0000 2021@VATVSLPR @pwilmarth And automating the "Where's Waldo?" aspect of differential experiments does take a lot of the fun out of it.
Sun Jun 06 15:45:27 +0000 2021@VATVSLPR @pwilmarth Some times I like to look at my -metry mapped to -scopy 🔗
Sun Jun 06 12:27:18 +0000 2021LIPF:p.F234I chr 10:88673588T>A, rs6586145 (all tissue F:I 0.749:0.251) vaf=13%, Δm=-33.9844, VAF by population group: african 28%, american 8%, east asian 14%, european 6%, south asian 4%. #ᐯᐸᐱ
Sun Jun 06 12:27:18 +0000 2021LIPF:p, θ(max) = 87. aka HGL, HLAL. Found in tissue MHC class 1 & class 2 peptide experiments. Most abundant in GI tract, particular stomach tissue; absent from commonly used cell lines.
Sun Jun 06 12:27:17 +0000 2021>LIPF:p, lipase F, gastric type (Homo sapiens) Small extracellular digestive enzyme; CTMs: N44, N281, N377+glycosyl; PTMs: 3×P=hydroxyproline (very rare); SAAVs: T171A (15%), F234I (13%); mature form: (21-408) [802×, 8 kTa]. #ᗕᕱᗒ 🔗
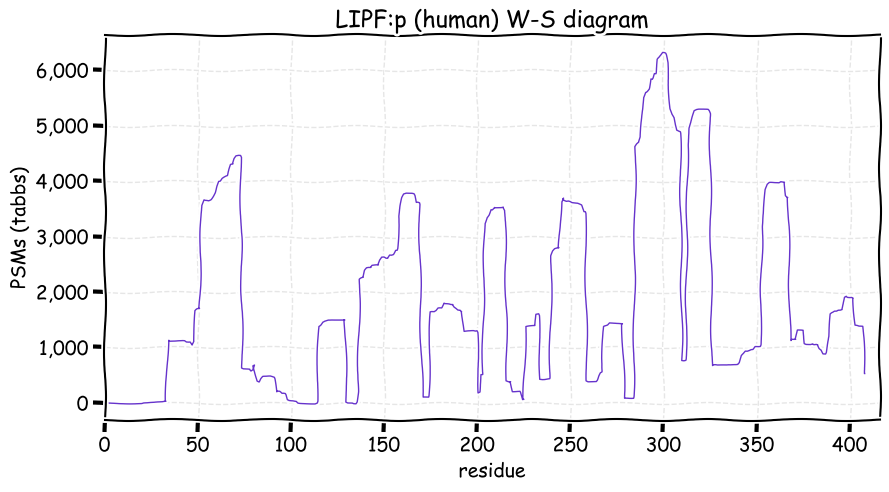
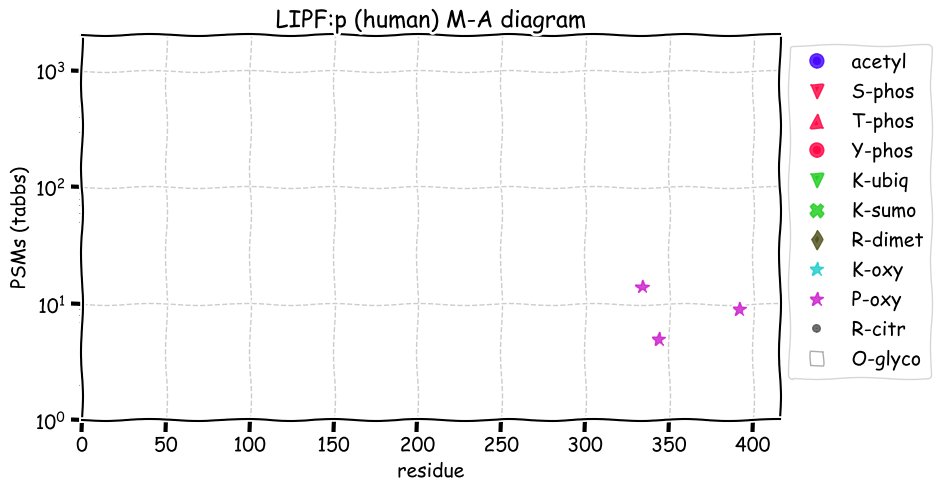
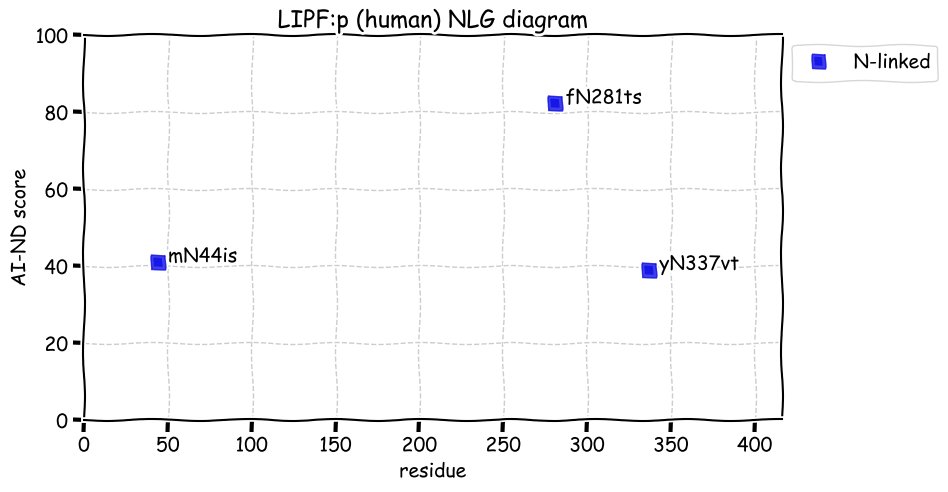
Sat Jun 05 19:50:07 +0000 2021I would personally say "No", but I am willing to be convinced otherwise.
Sat Jun 05 18:59:40 +0000 2021Is there any practical reason in proteomics data analysis to record the measured mass of a tryptic peptide to better than a millidalton?
Sat Jun 05 17:37:58 +0000 2021@olgavitek @pfizer I was a little amused by the very specific job skills qualification:
*Experience in analyzing “TMT-labeled” data sets
Sat Jun 05 12:04:14 +0000 2021RHBDF1:p.H58Y chr 16:g.64775G>A, rs147906988 (all tissue H:Y 0.988:0.012) vaf=1%, Δm=26.0044, VAF by population group: african 2%, american 1%, east asian <1%, european <1%, south asian <1%. #ᐯᐸᐱ
Sat Jun 05 12:04:13 +0000 2021RHBDF1:p, 7 transmembrane domain in the C-terminal region of the molecule, starting with (412–432). Most phosphorylations are within the N-terminal cytoplasmic domain (2-411).
Sat Jun 05 12:04:13 +0000 2021RHBDF1:p, θ(max) = 37. aka EGFR-RS, FLJ2235, Dist1, iRhom1, C16orf8. Found in immune cell MHC class 1 peptide experiments. Observed in solid tissue & cell lines, absent from fluids.
Sat Jun 05 12:04:13 +0000 2021>RHBDF1:p, rhomboid 5 homolog 1 (Homo sapiens) Midsized ER subunit; CTMs: A2+acetyl; PTMs: 24×S, 18×T, 0×Y+phosphoryl; 1×K+ubiquitinyl; 2×K+SUMOyl; SAAVs: H58Y (1%); mature form: (2-855) [3,284×, 8 kTa]. #ᗕᕱᗒ 🔗
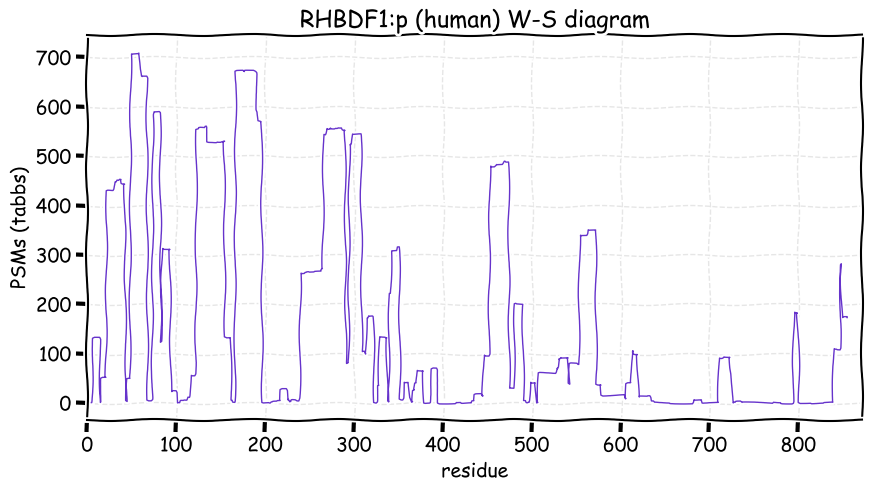
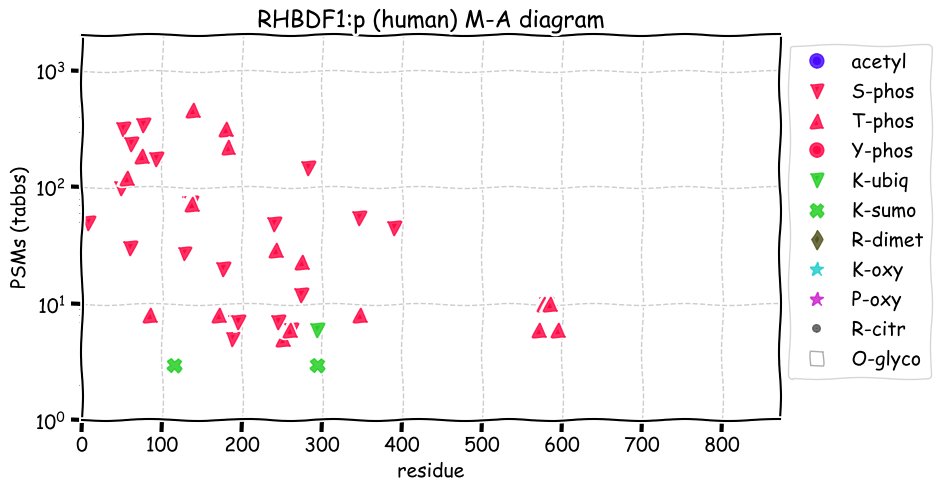
Fri Jun 04 14:30:59 +0000 2021Finally, a term that explains almost all interactions in faculty meetings 🔗
Fri Jun 04 14:01:41 +0000 2021Today's plasma PTM puzzler comes from complement C3, an abundant large protein involved in innate immunity. C3:p has 47 experimentally observed Y+phosphoryl sites (many with 100's of obs), but it's sequence does not have any of the well-known canonical Y+phosphoryl domains. 🔗
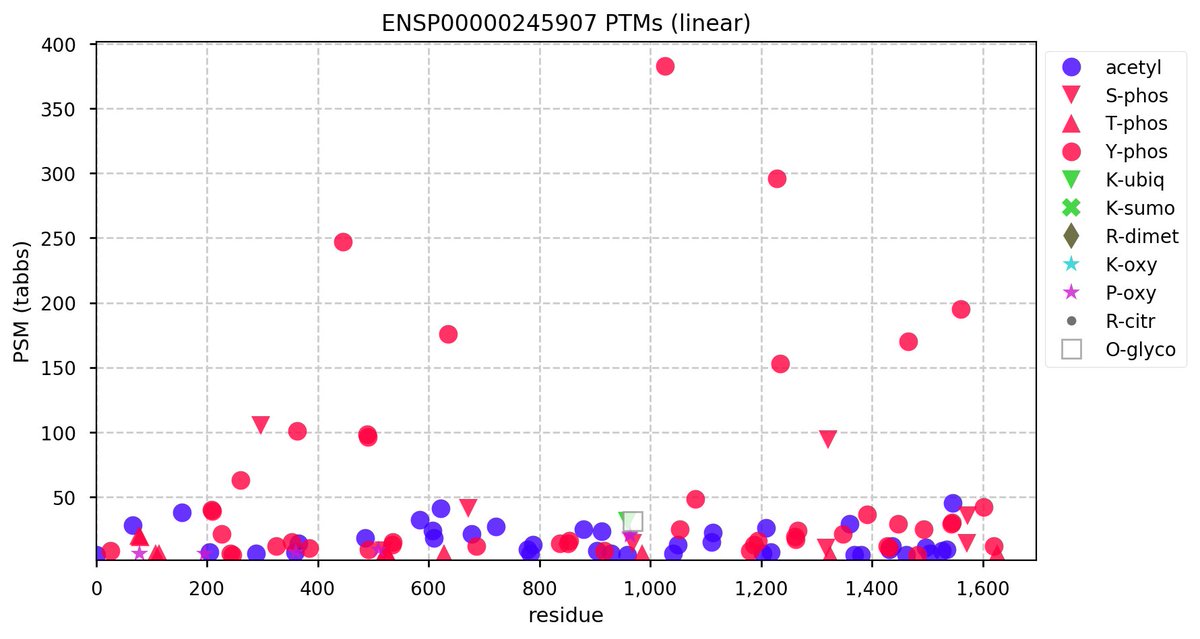
Fri Jun 04 11:58:48 +0000 2021ZC3HAV1:p.T851I chr 7:g.139047751G>A, rs3735007 (all tissue T:I 0.829:0.171) vaf=50%, Δm=12.0364, VAF by population group: african 71%, american 38%, east asian 26%, european 51%, south asian 59%. TI in HeLa cells. #ᐯᐸᐱ
Fri Jun 04 11:58:48 +0000 2021ZC3HAV1:p, θ(max) = 77. aka ZAP, FLB6421, FLJ13288, MGC48898, ZC3HDC2, ZC3H2, PARP13, ARTD13. Common in MHC class 1 peptide experiments. Observed in solid tissue & cell lines, rarely in fluids.
Fri Jun 04 11:58:48 +0000 2021>ZC3HAV1:p, zinc finger CCCH-type containing, antiviral 1 (Homo sapiens) Midsized intracellular subunit; CTMs: A2+acetyl; PTMs: 65×S, 29×T, 10×Y+phosphoryl; 25×K+ubiquitinyl; SAAVs: R485K (23%), H565Q (24%), T851I (50%); mature form: (2-902) [48,863×, 386 kTa]. #ᗕᕱᗒ 🔗
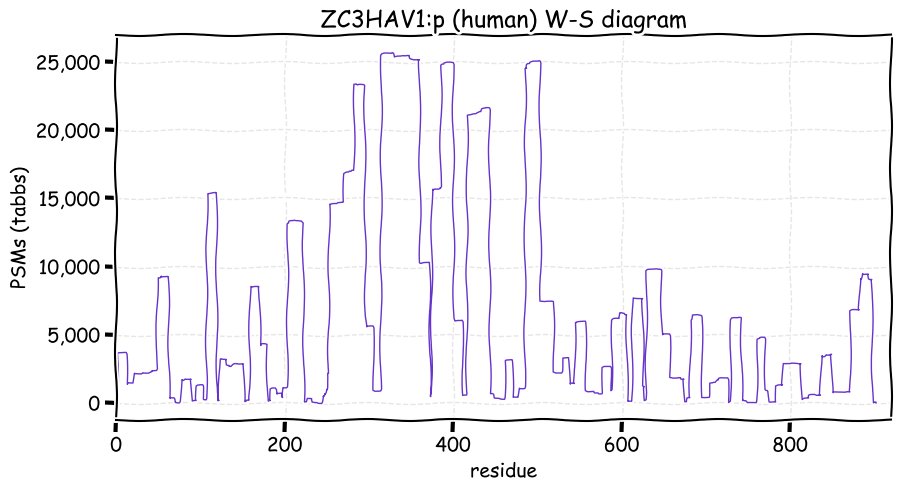
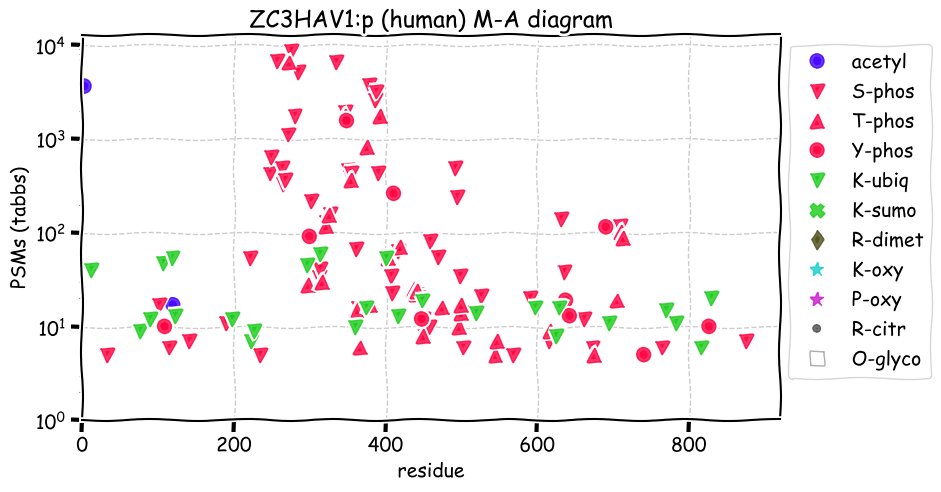
Thu Jun 03 20:41:58 +0000 2021@MHendr1cks From the wording, I suspect it was simply "happy". They seem to be taking that option off the board & replacing it with "not sick".
Thu Jun 03 16:34:34 +0000 2021@MattWFoster ALB:p tends to be a bit of a red-headed stepchild when it comes to discussing PTM observations: it is always there but better left unacknowledged.
Thu Jun 03 14:32:53 +0000 2021@dtabb73 A simple "Merde!" + a hangup is the usual idiom here in Canada.
Thu Jun 03 14:30:09 +0000 2021@MattWFoster Could be, but I've never seen any reference to that type of standard being used by them. Does anyone in CPTAC know if in vitro acetylated ALB:p is spiked into their KAc studies?
Thu Jun 03 14:05:18 +0000 2021@MattWFoster Just to add confusion, ALB:p is also one of the top contributors of K+ubiquitinyl PSMs in CPTAC ubiquitinylation studies.
Thu Jun 03 13:37:53 +0000 2021@MattWFoster I'm sure that type of reaction happens, but I don't think it would be enough to explain why ALB:p will frequently have the most acetylated PSMs in CPTAC KAc tissue studies.
Thu Jun 03 13:11:58 +0000 2021Does anybody know the mechanism that results in serum albumin K-acetylation?
Thu Jun 03 11:57:25 +0000 2021NSUN2:p.P760Q chr 5:g.6599951G>T, rs61744358 (all tissue P:Q 0.935:0.065) vaf=6%, Δm=31.0058, VAF by population group: african 9%, american 4%, east asian 0%, european 7%, south asian 6%. PQ in MCF-7 & RKO cells. #ᐯᐸᐱ
Thu Jun 03 11:57:25 +0000 2021NSUN2:p, θ(max) = 91. aka FLJ20303, TRM4, Misu, MRT5. Common in MHC class 1 peptide experiments. Observed in solid tissue & cell lines, rarely in fluids. 39 K acceptor sites are shared by at least two of acetyl, ubiquitinyl or SUMOyl & 18 share all three.
Thu Jun 03 11:57:24 +0000 2021>NSUN2:p, NOP2/Sun RNA methyltransferase 2 (Homo sapiens) Midsized subunit; CTMs: G2+acetyl; PTMs: 28×S, 30×T, 5×Y+phosphoryl; 46×K+acetyl; 38×K+ubiquitinyl; 28×K+SUMOyl; SAAVs: V627I (2%), D731E (83%), P760Q (6%); mature form: (2-767) [49,461×, 551 kTa]. #ᗕᕱᗒ 🔗
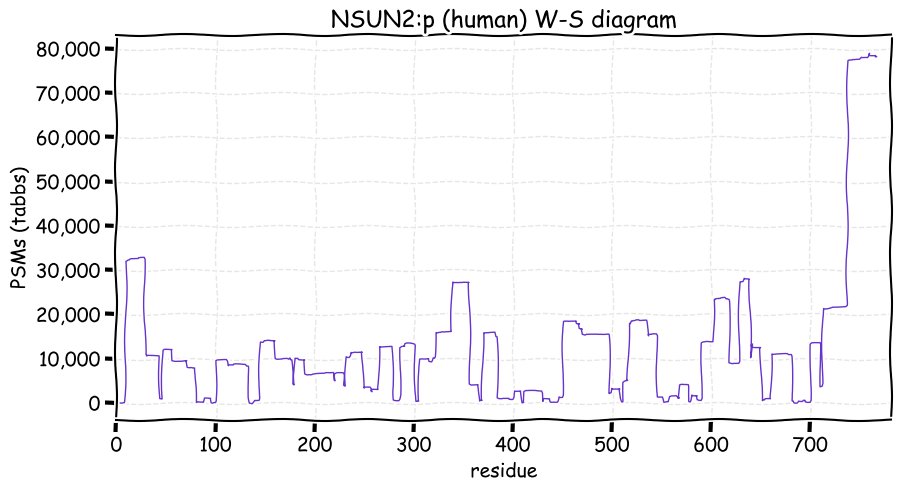
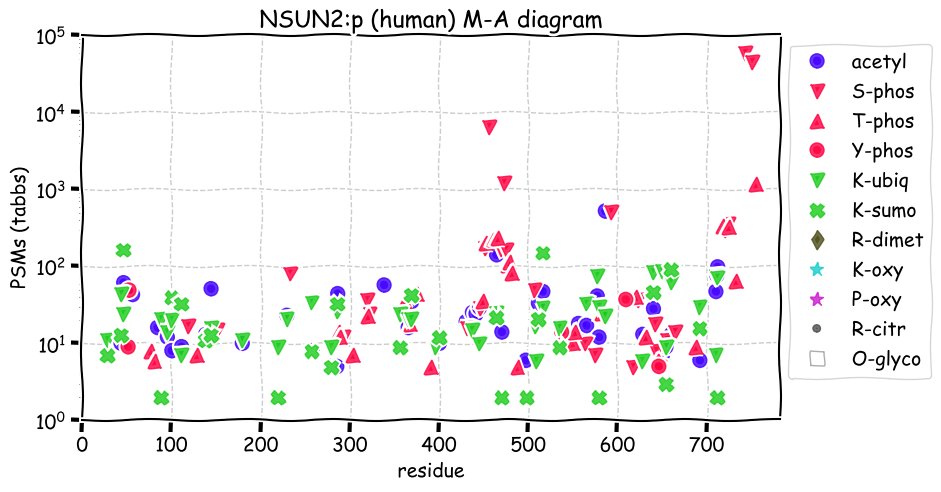
Wed Jun 02 16:07:16 +0000 2021@J_my_sci @pwilmarth @JohnRYatesIII @goodlettlab1 @r_l_moritz "Best weather in Canada" is a pretty low bar to hop over though. You quite literally only have to step over the US border to do better in most parts of the country.
Wed Jun 02 12:55:53 +0000 2021@ProtifiLlc For anyone who might be interested, the data set in question was the CPTAC LSCC proteome set of experiments.
Wed Jun 02 12:53:08 +0000 2021@ProtifiLlc With that taken into account, N-terminal acetylation completely blocked any TMT derivatization at that amino group.
Wed Jun 02 12:51:47 +0000 2021@ProtifiLlc Thanks a lot to everyone who commented. After some testing, the problem for the data set I was interested in was significant TMTylation of serine hydroxyls (with very little on T or Y). Once that was taken into account, it really cleaned up the results.
Wed Jun 02 11:51:21 +0000 2021NISCH:p.A1056V chr 3:g.52489389C>T, rs887515 (all tissue A:V 0.274:0.726) vaf=83%, Δm=28.0313, VAF by population group: african 31%, american 81%, east asian 98%, european 82%, south asian 88%. AV in JURKAT cells. VV in HCT-116 cells. #ᐯᐸᐱ
Wed Jun 02 11:51:21 +0000 2021NISCH:p, θ(max) = 35. aka KIAA0975, I-1, IRAS. Common in MHC class 1 peptide experiments. Observed in solid tissue & cell lines. Involved in processes that result in cell motility, via an unknown mechanism.
Wed Jun 02 11:51:21 +0000 2021>NISCH:p, nischarin (Homo sapiens) Large intracellular subunit; CTMs: A2+acetyl; PTMs: 8×S, 2×T, 3×Y+phosphoryl; 10×K+ubiquitinyl; SAAVs: V299I (1%), K460R (1%), A1028V (1%), A1056V (83%); mature form: (2-1504) [14,319×, 48 kTa]. #ᗕᕱᗒ 🔗
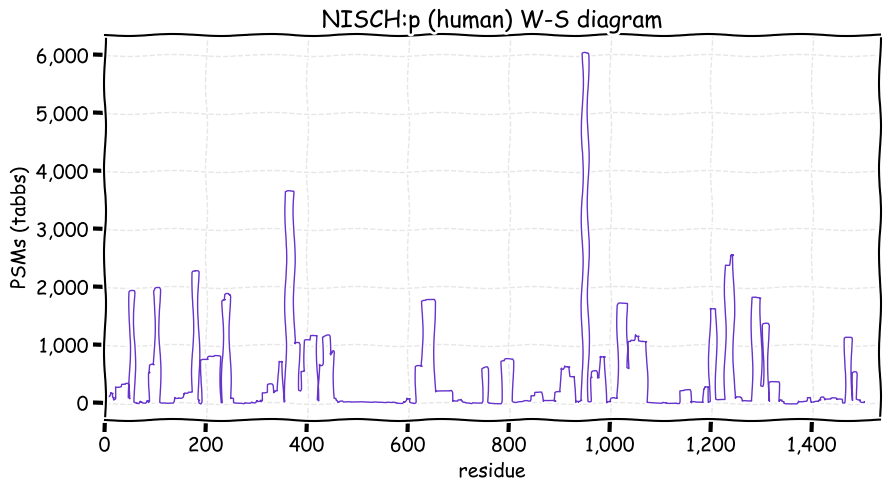
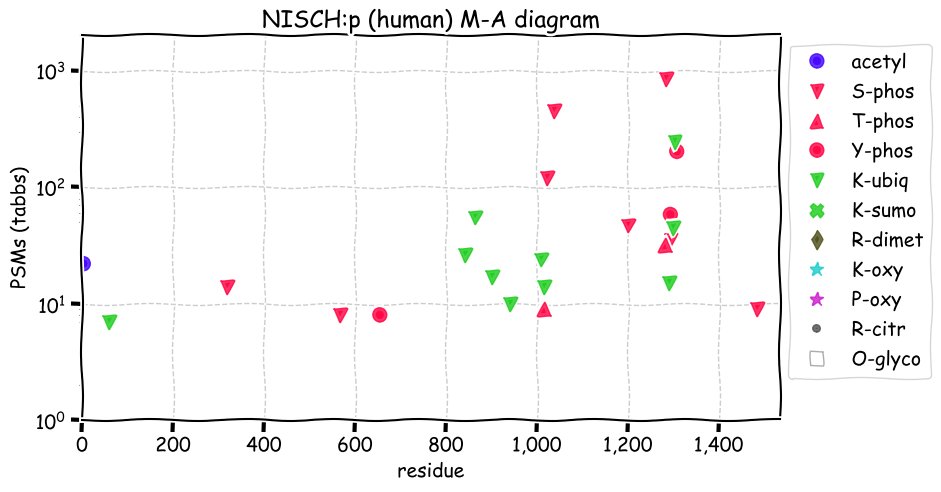
Tue Jun 01 17:10:33 +0000 2021@ProtifiLlc The most-difficult-to-explain peptide examples I can come up with have a serine at the 1, 2 or 3 position, so hydroxyl derivatization would certainly be an attractive explanation.
Tue Jun 01 16:50:50 +0000 2021🔗
Tue Jun 01 16:33:20 +0000 2021@kabalak The peptide in question would have both acetylation and TMTylation on the same amino group.
Tue Jun 01 15:49:17 +0000 2021Can a protein N-terminal tryptic peptide that has an endogenous N-terminal acetylation also be derivatized by a TMT-type reagent on the already acetylated amino group? (I thought I knew the answer but I'm not so sure anymore).
Tue Jun 01 15:15:35 +0000 2021@slavov_n Because it works. Serial exaggeration is a very effective strategy in academia, government and industry.
Tue Jun 01 13:49:36 +0000 2021Not a big fan of chili cook-off style comparisons of academic software. Completely misses the point as far as I am concerned.
Tue Jun 01 12:04:04 +0000 2021Part of an amusing spat back in 2001 about how to refer to homolog(ue)s 🔗
Tue Jun 01 11:46:43 +0000 2021FUCA1:p and FUCA2:p share a function & PTM pattern, but do not share any observable tryptic peptides.
Tue Jun 01 11:41:47 +0000 2021FUCA1:p.N96S chr 1:g.23868000T>C rs373805274 (all tissue N:S 0.997:0.003) vaf=<1%, Δm=-27.0109, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. #ᐯᐸᐱ
Tue Jun 01 11:41:46 +0000 2021FUCA1:p, θ(max) = 83. Common in MHC class 2 peptide experiments. Observed in solid tissue, fluids & cell lines.
Tue Jun 01 11:41:46 +0000 2021>FUCA1:p, alpha-L-fucosidase 1 (Homo sapiens) Small lysosomal enzyme; CTMs: N267, N382+glycosyl; PTMs: none; SAAVs: P10L (14%), Q286R (21%); mature form: (32-466) [17,691×, 83 kTa]. #ᗕᕱᗒ 🔗
Mon May 31 18:57:19 +0000 2021@jwoodgett Darn. I was just getting used to "A" & "B" ...
Mon May 31 17:58:59 +0000 2021TOM70 (ENSP00000284320) PTM observation frequency, linear vs log scale: same data, different energy 🔗
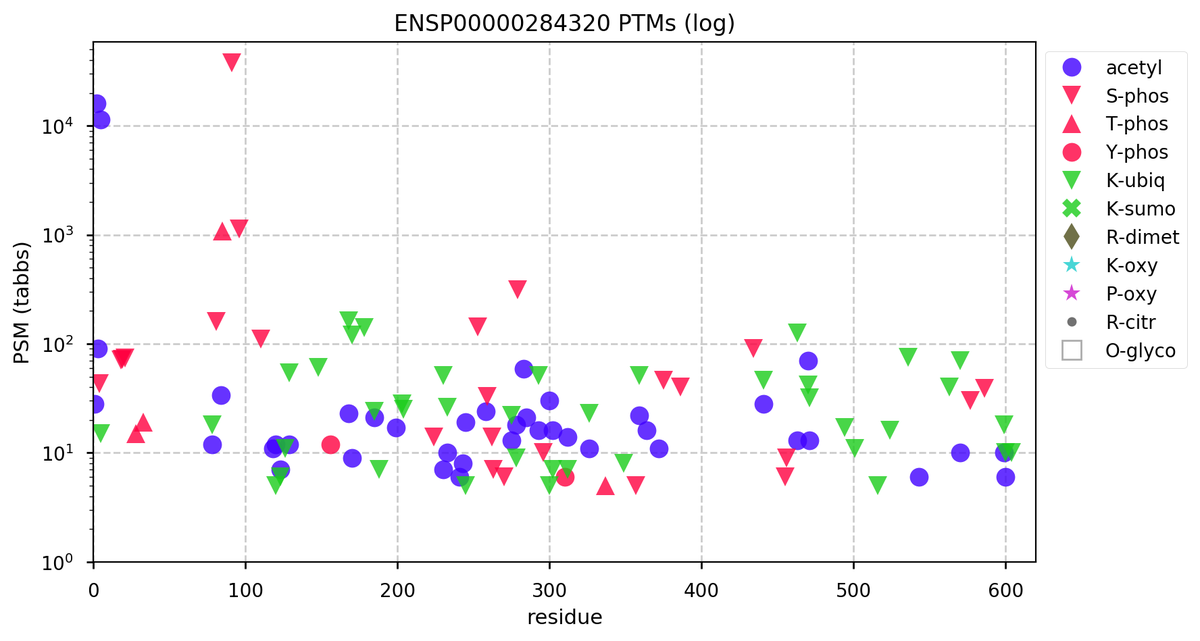
Mon May 31 12:42:35 +0000 2021@plantscientist9 @schumacherbj I was thinking about private sector research jobs. Academia has always been very difficult for immigrants in most countries (& no picnic for natives, either).
Mon May 31 12:38:42 +0000 2021What types of hypotheses are supported (or rejected) by the lack of FUCA2:p PTM observations?
Mon May 31 11:56:14 +0000 2021FUCA2:p.H371Y chr 6:g.143501975G>A, rs3762001 (all tissue H:Y 0.910:0.090) vaf=21%, Δm=26.0044, VAF by population group: african 12%, american 15%, east asian 17%, european 21%, south asian 19%. HY in RKO & SK-MEL-28 cells. #ᐯᐸᐱ
Mon May 31 11:56:14 +0000 2021FUCA2:p, θ(max) = 61. aka MGC1314, dJ20N2.5. Common in MHC class 1 & 2 peptide experiments. Observed in fluids & ovarian cancer. Removes fucose from glycoproteins.
Mon May 31 11:56:13 +0000 2021>FUCA2:p, fucosidase, alpha-L- 2, plasma (Homo sapiens) Small enzyme; CTMs: N377+glycosyl; PTMs: none; SAAVs: A233E (9%), H371Y (21%); mature form: (29-467) [16,147×, 56 kTa]. #ᗕᕱᗒ 🔗
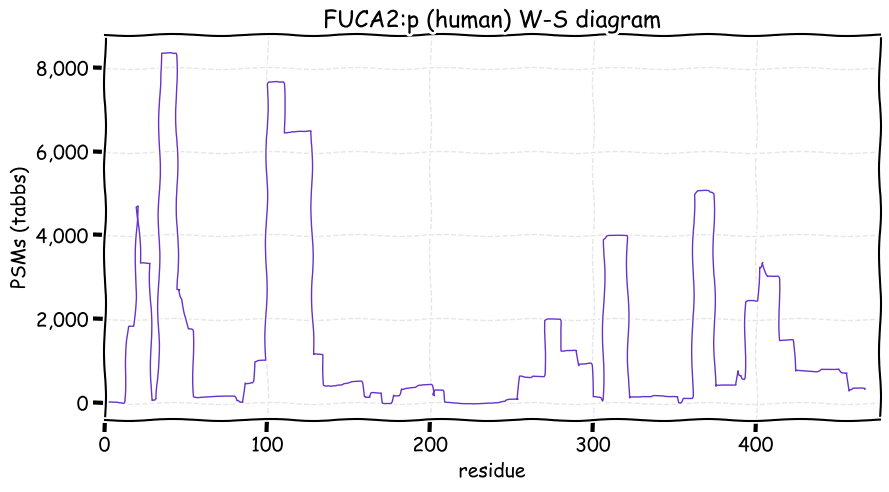
Sun May 30 15:54:39 +0000 2021Seems like the evolution of the "3rd wave" in Manitoba is going to be somewhere between that in Ontario & Alberta (which isn't really very surprising). At the moment, it looks like the declining leg of the curve is going to look more like AB's. 🔗
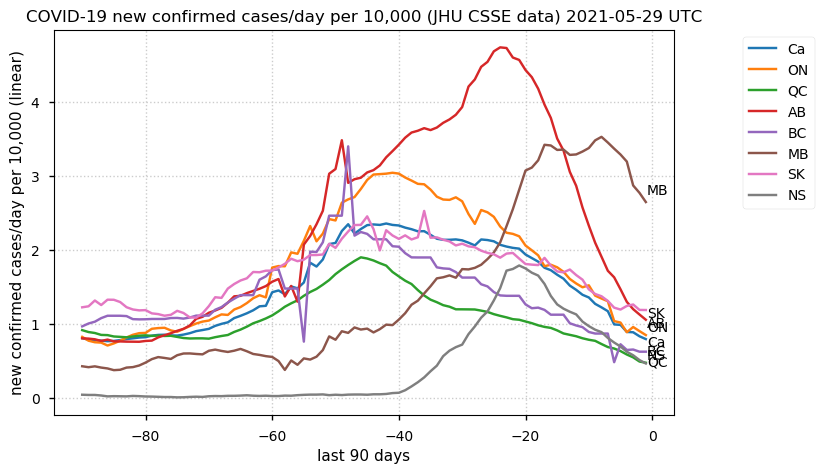
Sun May 30 13:56:59 +0000 2021@schumacherbj Although, we Canadians always seem to expect odd behavior wrt immigration by our southern neighbour to help with research activities here, but then our government's own anti-research tendencies kick in to balance things out.
Sun May 30 13:02:09 +0000 2021@schumacherbj I guess that now that GB is off the table as a research hub, DE should benefit as the destination for highly qualified personnel in Europe.
Sun May 30 12:10:15 +0000 2021PDLIM5:p has 2 domains (140-198) & (514-581) that are rarely observed because of an absence of tryptic cleavage sites. Shares a few tryptic peptides with ZNF318, but not the other Enigma family proteins.
Sun May 30 11:50:17 +0000 2021PDLIM5:p.S136F chr 4:g.94575731C>T, rs4546904 (all tissue S:F 0.797:0.203) vaf=27%, Δm=60.0364, VAF by population group: african 11%, american 24%, east asian 38%, european 31%, south asian 13%. SF in HeLa cells. SS in HCT-116 cells. #ᐯᐸᐱ
Sun May 30 11:50:16 +0000 2021PDLIM5:p, θ(max) = 44. aka LIM, Enh. Common in MHC class 1 peptide experiments. Observed in solid tissues, cell lines & urinary extracellular vesicles. May (or may not) be involved in many biological processes.
Sun May 30 11:50:16 +0000 2021>PDLIM5:p, PDZ and LIM domain 5 (Homo sapiens) Midsized subunit; CTMs: S2+acetyl; PTMs: 48×S, 25×T, 8×Y+phosphoryl; 7×K+ubiquitinyl; 1×K+SUMOyl 3×K+acetyl; SAAVs: S136F (27%), T381A (39%), S492N (2%); mature form: (2-596) [41,315×, 373 kTa]. #ᗕᕱᗒ 🔗
Sat May 29 14:03:46 +0000 2021@DonMartinCTV Unimportant tech note: the Quebec flag emoji is currently allowed 🔗 But, as with most of the other subdivision flag emojis, most font vendors aren't supporting them.
Sat May 29 13:19:05 +0000 2021PTGFRN:p is one of those proteins where much of the existing annotation looks kind of sketchy.
Sat May 29 12:45:19 +0000 2021PTGFRN:p is much more commonly observed than its proposed interaction partner PTGFR.
Sat May 29 12:38:39 +0000 2021PTGFRN:p, the low occupancy intracellular S875+phosphoryl is also observed in mice. Most observations of this modification in humans have been in CPTAC studies.
Sat May 29 12:08:00 +0000 2021PTGFRN:p.S277T chr 1:g.116945089T>A, rs4546904 (all tissue S:T 0.466:0.534) vaf=89%, Δm=14.0157, VAF by population group: african 75%, american 92%, east asian 85%, european 90%, south asian 93%. TT in HK-1 cells. ST in MCF-10A cells. #ᐯᐸᐱ
Sat May 29 12:08:00 +0000 2021PTGFRN:p, θ(max) = 63. aka FPRP, EWI-F, CD9P-1, FLJ11001, KIAA1436, SMAP-6, CD315. Common in MHC class 2 peptide experiments from solid tissue. Observed in solid tissues & cell lines. Extracellular (22-830), transmembrane (831-852), intracellular (853-879).
Sat May 29 12:08:00 +0000 2021>PTGFRN:p, prostaglandin F2 receptor inhibitor (Homo sapiens) Midsize membrane subunit; CTMs: N286, N300, N413, N525, N600, N618, N691+glycosyl; PTMs: S597, S875+phosphoryl; SAAVs: S277T (89%), R430Q (12%); mature form: (22-879) [16,260×, 118 kTa].#ᗕᕱᗒ 🔗
Fri May 28 16:36:31 +0000 2021@nesvilab Great minds think alike ... 🔗
Fri May 28 16:13:39 +0000 2021It has always seemed a little weird to me, given that all of these institutions loudly bang drums about how dedicated they are to the quality of education.
Fri May 28 16:08:41 +0000 2021As a prof, I have taught courses at 3 different universities. I have never received any type of instruction or policy guidance from any of these institutions about how I should assign grades to students.
Fri May 28 15:28:06 +0000 2021"The trouble is that top journals publish crap too." 🔗
Fri May 28 14:29:04 +0000 2021PFAS:p, T619+phosphoryl is believed to modulate the activity of the enzyme, with the phosphorylated form being much more active 🔗
Fri May 28 14:17:26 +0000 2021PFAS:p, the long name comes from the reaction it catalyzes:
ATP + N₂-formyl-N₁-(5-phospho-D-ribosyl)glycinamide + L-glutamine + H₂O ↔ ADP + phosphate + 2-(formamido)-N₁-(5-phospho-D-ribosyl)acetamidine + L-glutamate
Fri May 28 12:07:11 +0000 2021PFAS:p.L621P chr 17:g.8264282T>C, rs11078738 (all tissue L:P 0.466:0.534) vaf=64%, Δm=-16.0313, VAF by population group: african 52%, american 57%, east asian 27%, european 77%, south asian 52%. PP in HeLa cells. LP in HCT-116 cells. #ᐯᐸᐱ
Fri May 28 12:07:10 +0000 2021PFAS:p, θ(max) = 70. aka PURL, FGARAT, KIAA0361, GATD8. Common in MHC class 1 peptide experiments from solid tissue. Common in solid tissues, rarely in fluids.
Fri May 28 12:07:10 +0000 2021>PFAS:p, phosphoribosylformylglycinamidine synthase (Homo sapiens) Large enzyme; CTMs: A2+acetyl; PTMs: 32×S, 16×T, 1×Y+phosphoryl; 24×K+ubiquitinyl; 2×K+SUMOyl; 2×K+acetyl; SAAVs: P19S (63%), P367L (34%), L621P (64%); mature form: (2-1338) [42,426×, 416 kTa]. #ᗕᕱᗒ 🔗
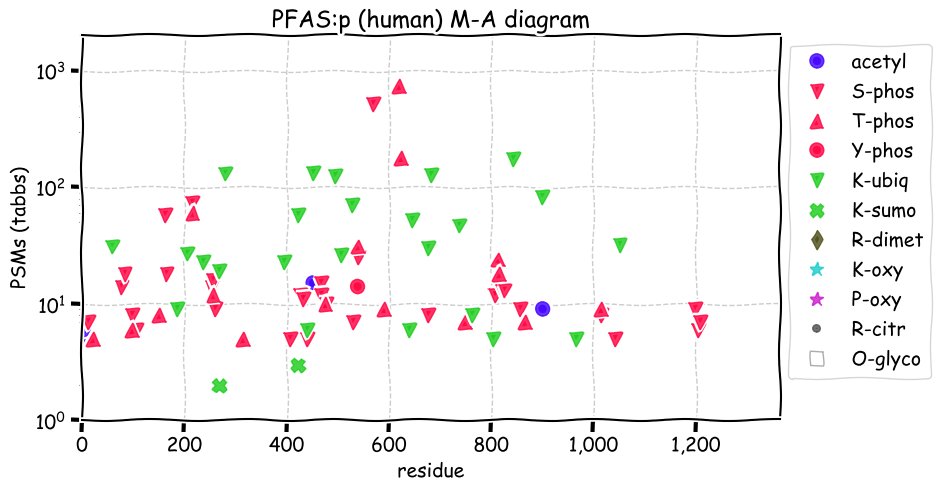
Fri May 28 01:21:22 +0000 2021@GeiszlerDaniel @nesvilab @pwilmarth In trypsin (& Lys-C) the specificity is achieved by having the basic sidechain form an ionic bond with a negatively charged Asp sidechain. The geometry of that ionic bond and the catalytic triad determine whether or not the peptide bond will be cleaved.
Thu May 27 20:28:31 +0000 2021And since it is C. elegans data, be sure to add an E. coli B strain proteome to the search parameters.
Thu May 27 19:11:35 +0000 2021@astacus @nesvilab Kids these days! 🧐
Thu May 27 18:55:00 +0000 2021@astacus @nesvilab I took my first biochem class in 1976.
Thu May 27 18:54:30 +0000 2021@astacus @nesvilab I agree: the Xtal structures make it clear that adding a GG to the sidechain should move the K-X far away from the catalytic triad. But I don't know if that particular structure is taught as thoroughly as it was back in the '70s, when it was still kind of exciting.
Thu May 27 18:46:04 +0000 2021If you want to try analyzing the data yourself (which I would strongly recommend), set the number of missed cleavage sites to 3.
Thu May 27 18:44:09 +0000 2021Since I tend not to believe myself, I checked a bunch of new GG-remnant data (PXD024338) for evidence of this type of cleavage. I am relieved to say I couldn't find any significant evidence of K+GG at the C-terminus of any of the tryptic peptides.
Thu May 27 18:41:41 +0000 2021Last week, I had a short tweet exchange with @nesvilab regarding the possibility of trypsin cleavage at lysines with ubiquitin-modified sidechains. I was adamant that it wasn't possible.
Thu May 27 17:09:45 +0000 2021I am sometimes struck by how often oncology researchers, who would normally rather be struck dead than attend a seminar on infectious disease, are now sagely giving their opinions on virology.
Thu May 27 14:19:48 +0000 2021HLA-A:p doesn't fit very well with the "reference sequence" idea commonly employed in proteomics, because of its large number of high VAF SAAVs. The immune system proteins encoded on chr 6 tend follow this pattern of high variability & linked AAV distributions.
Thu May 27 14:19:10 +0000 2021HLA-A:p.F33S chr 6:g.29942781T>A, rs2075684 (all tissue F:S 0.884:0.116) vaf=20%, Δm=-60.0364, VAF by population group: african 16%, american 16%, east asian 33%, european 15%, south asian 30%. #ᐯᐸᐱ
Thu May 27 14:16:40 +0000 2021HLA-A:p, θ(max) = 67. Common in MHC class 1 & class 2 peptide experiments. Common in solid tissues & fluids. Domains: (25-308) extracellular, (309-332) transmembrane & (333-365) cytoplasmic.
Thu May 27 11:57:19 +0000 2021HLA-A:p, SAAVs: E190D (21%), W191G (21%), D251H (40%), V255M (27%), E277Q (45%), L300P (31%), I306V (15%), L318F (46%), T323A (26%), K335N (15%), T345S (31%) 🔗
Thu May 27 11:57:17 +0000 2021HLA-A:p, SAAVs: G107R (20%), A114D (30%), I119F (22%), I121K (22%), I121K (31%), Y123C (15%), S129P (30%), G131R (24%), R138H (42%), Q139N (32%), D140H (42%), L150V (16%), N151K (46%), K168Q (34%), H175L (26%), E176A (19%), L180M (38%), T187K (21%)
Thu May 27 11:57:16 +0000 2021HLA-A:p, SAAVs: L10V (49%), F33S (20%), R41S (12%), R68K (9%), G80R (8%), Q86L (39%), E87D (18%), R89G (15%), R89W (15%), N90K (39%), V91L (9%), Q94H (32%), T97I (12), D98H (23%), V100A (22%), D101H (39%), G103R (19%), T104I (20%), L105P (20%), R106L (19%)
Thu May 27 11:57:16 +0000 2021>HLA-A:p, major histocompatibility complex, class I, A (Homo sapiens) Small subunit; CTMs: N110+glycosyl; PTMs: 24×S, 27×T, 17×Y+phosphoryl; 9×K+ubiquitinyl; 2×K+SUMOyl 1×K+acetyl; mature form: (19,25-365) [74,791×, 43 kTa]. #ᗕᕱᗒ
Wed May 26 15:25:24 +0000 2021@TrostLab @pwilmarth Do either one of them have an organic chemistry background? The derivatizations—both TMTpro and IAA—were done unusually well.
Wed May 26 14:07:42 +0000 2021The more I look at this one, the more I like it. If you want a TMTpro data set that was done very well from top-to-bottom, you need look no further than 🔗
Wed May 26 12:31:07 +0000 2021MATN2:p.T855M chr 8:g.98032300C>T, rs2255317 (all tissue T:M 0.952:0.048) vaf=12%, Δm=29.9928, VAF by population group: african 12%, american 12%, east asian 22%, european 9%, south asian 8%. TT in commonly used cell lines. #ᐯᐸᐱ
Wed May 26 12:31:07 +0000 2021MATN2:p, θ(max) = 52. Observed in MHC class 1 & class 2 peptide experiments. Common in many solid tissues & seminal plasma.
Wed May 26 12:31:07 +0000 2021>MATN2:p, matrilin 2 (Homo sapiens) Midsized extracellular subunit; CTMs: N221+glycosyl; PTMs: K102, K829+SUMOyl; SAAVs: T855M (12%), F867L (1%), S888N (1%); mature form: (24?-937) [6,593×, 43 kTa]. #ᗕᕱᗒ 🔗
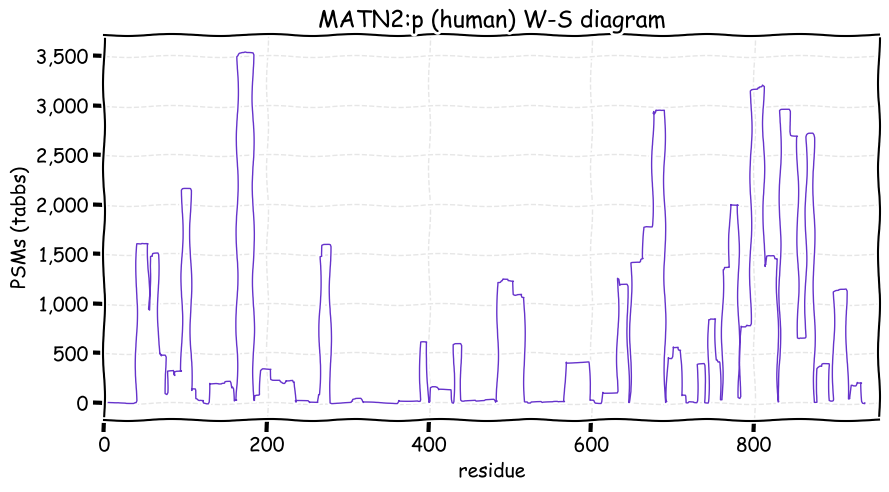
Tue May 25 20:38:45 +0000 2021Data from 🔗
Tue May 25 20:38:03 +0000 2021PXD022523: nicely done.
Tue May 25 16:42:27 +0000 2021@MiguelCos @astacus But ignoring UTIs seems to be SOP in proteomics "biomarker" papers.
Tue May 25 16:41:25 +0000 2021@MiguelCos @astacus PXD015289 does a pretty good job on extracellular vesicles in urine & PXD020722 on unfractionated urine. The write ups for both ignore effects associated with UTIs that are easily detected in the data (E. coli in the former & P. aeruginosa in the latter).
Tue May 25 14:55:15 +0000 2021@HellmutAugustin @HaoYin20 @Dev_Cell This is the paper associated with the well done phosphoproteomics data set I mentioned yesterday 🔗
Tue May 25 14:27:20 +0000 2021@dtabb73 @statesdj I am not surprised, but heartened. There has been comparatively little fundamental protein biochemistry/engineering work done since the "leptin fiasco" in the 1990's, even though there is so much more high quality primary sequence information available to work with.
Tue May 25 12:27:40 +0000 2021MELTF:p was named because the molecule was found on the plasma membrane of MELanoma cells & it had TransFerrin domains. It is also commonly observed in many other types of cancer. 🔗
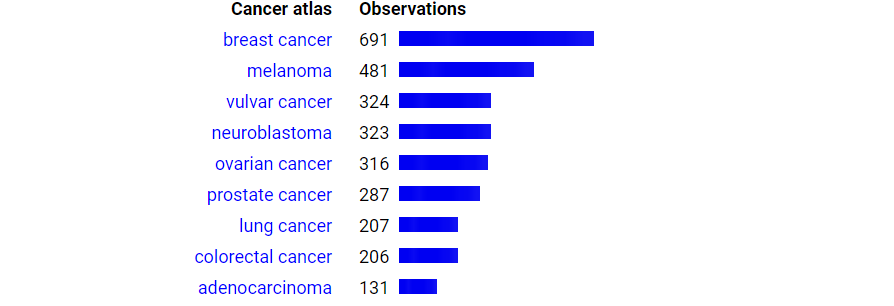
Tue May 25 11:56:22 +0000 2021MELTF:p.F480S chr 3:g.197009704A>G, rs41284047 (all tissue F:S 0.990:0.010) vaf=1.6%, Δm=-60.0364, VAF by population group: african 0%, american 2%, east asian 0%, european 3%, south asian 0%. FS in A-375 cells. #ᐯᐸᐱ
Tue May 25 11:56:22 +0000 2021MELTF:p, has two splice variants, 738 residues (above) & 302 residues. The short variant has the same 6 N-terminal exons as the longer one, but a different exon 7 incl. a stop codon. This exon 7 has 3 observable tryptic peptides not present in the longer form.
Tue May 25 11:56:22 +0000 2021MELTF:p, θ(max) = 66. aka CD228, FLJ38863, MAP97, MGC4856, MTF1, MTf, MFI2. Observed in MHC class 1 & rarely in class 2 peptide experiments. Most prominent in urine, many cancer tissues & cell lines.
Tue May 25 11:56:21 +0000 2021>MELTF:p, melanotransferrin (Homo sapiens) Midsized subunit; CTMs: N38, N515+glycosyl; PTMs: Y599+phosphoryl; K92, K392+SUMOyl; C709+GPI-anchor; SAAVs: V55I (1%), R387H (1%), V398M (1%), F480S (2%); mature form: (20?-709) [9,602×, 56 kTa]. #ᗕᕱᗒ 🔗
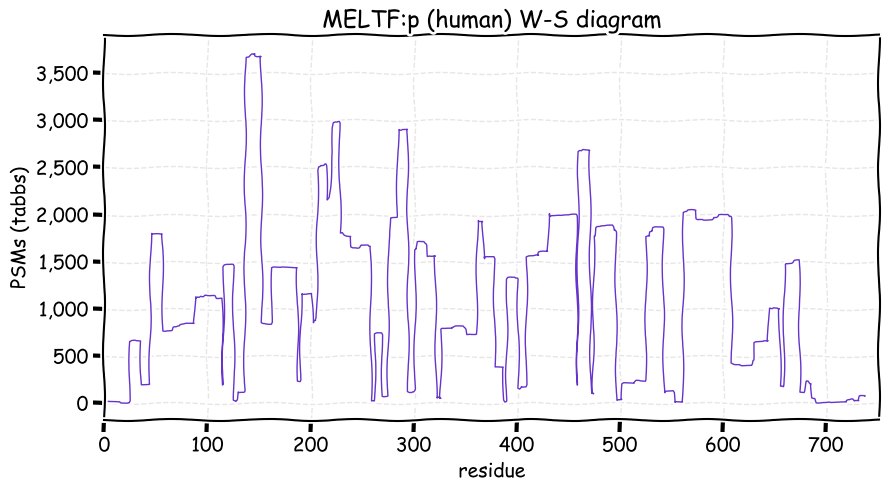
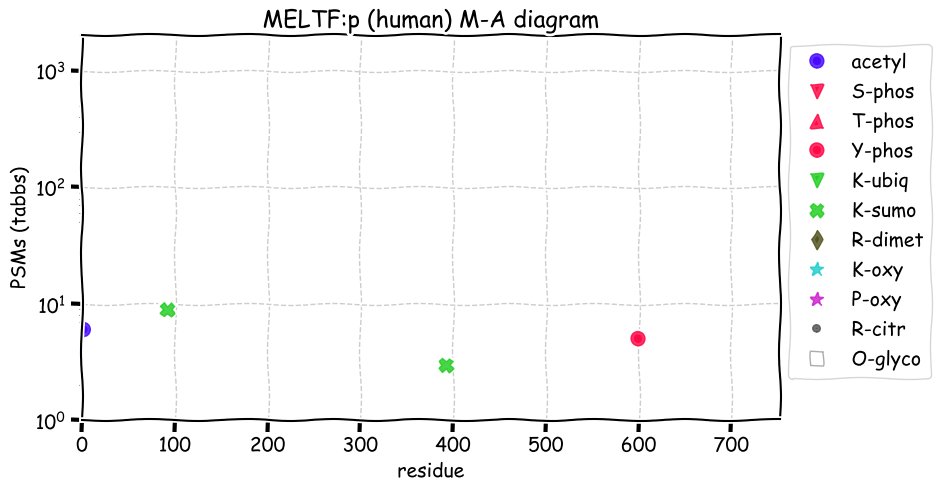
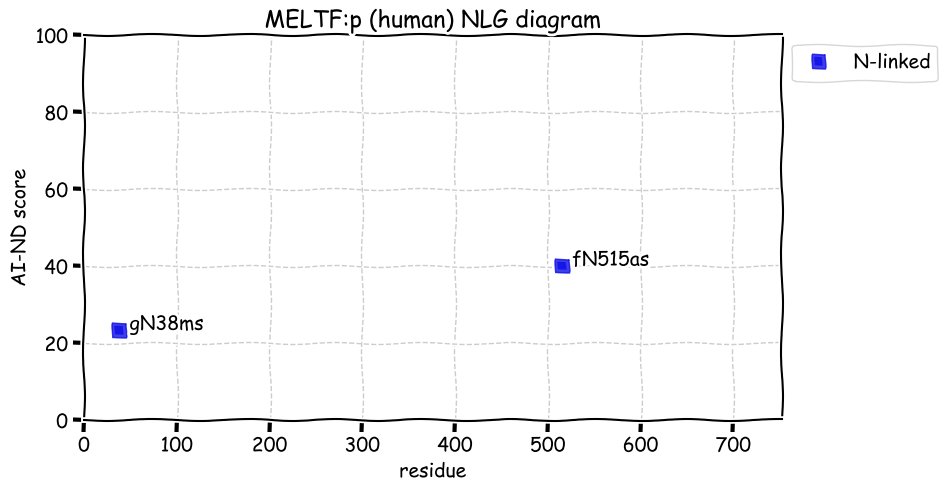
Mon May 24 17:01:36 +0000 2021If you would like a very good phosphopeptide data set that uses dimethyl isotopic labels for quantitation, PXD020805 is well worth a look. The protocol used achieved > 95% enrichment of phosphopeptides.
Mon May 24 15:46:38 +0000 2021@nesvilab @pwilmarth Anybody who has a better feeling for Xtal structures than I do (which is legion) should chime in if they think there is some way to spin-o-rama the extended sidechain back in to the catalytic pocket.
Mon May 24 15:46:08 +0000 2021@nesvilab @pwilmarth Lys-C and trypsin both are serine proteases with very similar His, Ser, Asp catalytic triads (& good Xtal structures). If you add GG to the substrate K side chain, there is no way +ve sidechain to attach & the K-X bond to slot itself into the catalytic pocket for scission.
Mon May 24 12:59:35 +0000 2021Weird Canadian practices: for reasons very few of us understand, today is a federally mandated holiday 🔗
Mon May 24 12:10:30 +0000 2021PCSK9:p, is secreted in an inactive form & activated by the removal of a proprotein domain (31-152) to form the active protease (153-692). 2 phosphorylation sites, 1 ubiquitinylation site & A53V are in the proprotein domain.
Mon May 24 12:01:35 +0000 2021PCSK9:p.G670E chr 1:g.55063514G>A, rs505151 (all tissue G:E 0.078:0.922) vaf=95%, Δm=72.0211, VAF by population group: african 71%, american 96%, east asian 96%, european 97%, south asian 96%. #ᐯᐸᐱ
Mon May 24 12:01:35 +0000 2021PCSK9:p, θ(max) = 66. aka NARC-1, FH3, HCHOLA3. Observed in MHC class 1 & 2 peptide experiments. Most prominent in blood plasma & CSF. Can be observed in HeLa & HCT-116 cell lines.
Mon May 24 12:01:34 +0000 2021>PCSK9:p, proprotein convertase subtilisin/kexin type 9 (Homo sapiens) Midsized enzyme; CTMs: N533+glycosyl; PTMs: 3×S, 1×T+phosphoryl; 5×K+ubiquitinyl; 1×K+SUMOyl; SAAVs: A53V (9%), A443T (3%), V474I (13%), G670E (95%); mature forms: (29,31-692) [4,932×, 30 kTa]. #ᗕᕱᗒ 🔗
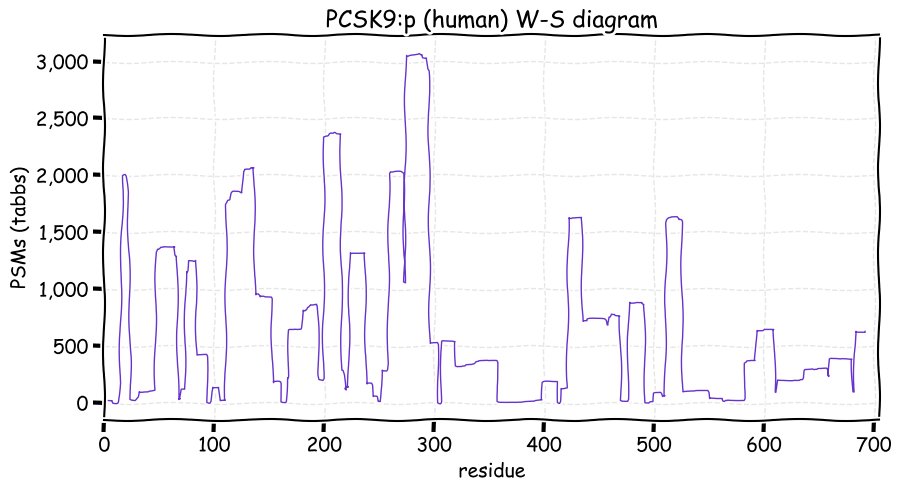
Sun May 23 15:10:49 +0000 2021If anyone ever rethinks the way GO ontologies are structured (particularly the biological processes segment), hopefully they will keep proteins like CCN2 in mind.
Sun May 23 12:31:43 +0000 2021Because the vaf = 100%, its corresponding MAF is usually listed as 0. Uniprot's protein sequence uses H83.
Sun May 23 12:27:55 +0000 2021CCN2:p.H83D chr 6:g.131950812G>C, rs7451102 (all tissue H:D 0.000:1.000) vaf=100%, Δm=-22.0320, VAF by population group: african 100%, american 100%, east asian 100%, european 100%, south asian 100%. #ᐯᐸᐱ
Sun May 23 12:27:55 +0000 2021CCN2:p, θ(max) = 82. aka IGFBP8, CTGF. Observed in MHC class 1 (& rarely class 2) peptide experiments. A prominent MHC class 1 peptide is from the signal sequence (2-10). Has too many GO annotations.
Sun May 23 12:27:54 +0000 2021>CCN2:p, cellular communication network factor 2 (Homo sapiens) Small subunit; CTMs: none; PTMs: 1×S+phosphoryl; 1×K+ubiquitinyl; 4×K+SUMOyl, T173, T193+glycosyl; SAAVs: H83D (>99%); mature forms: (27-349) [6,857×, 46 kTa]. #ᗕᕱᗒ 🔗
Sat May 22 16:08:07 +0000 2021Note: COL6A4 is a pseudogene in humans.
Sat May 22 16:06:44 +0000 2021For anyone interested in the PTM patterns associated with the "network" collagens IV & VI, here is a collection of M-A diagrams for the 11 subunits involved 🔗
Sat May 22 12:34:54 +0000 2021@pwilmarth @nesvilab If I'm not mistaken (& I often am), the Carr group came up with a protocol in which the GG did not get TMTylated, but that did not seem to catch on with others.
Sat May 22 12:21:38 +0000 2021@pwilmarth @nesvilab Yes, that is exactly what it is. I wasn't trying to saying anything about TMT addition, only cleavage.
Sat May 22 12:18:25 +0000 2021COL4A2:p.P718S chr 13:g.110469273C>T, rs9583500 (all tissue P:S 0.901:0.099) vaf=18%, Δm=-10.0207, VAF by population group: african 36%, american 22%, east asian 2%, european 22%, south asian 10%. #ᐯᐸᐱ
Sat May 22 12:18:25 +0000 2021COL4A2:p, θ(max) = 61. aka FLJ22259, DKFZp686I14213. Observed in MHC class 2 peptide experiments from tissue. Found in many tissues & urine. 1 of 6 subunits that can be used to make collagen IV.
Sat May 22 12:18:25 +0000 2021>COL4A2:p, collagen type IV α2 chain (Homo sapiens) Extracellular subunit; CTMs: none; PTMs: 3×S, 1×T, 0×Y+phosphoryl, 55×K, 233×P+oxidation; SAAVs: T552K (1%), K701R (2%), P718S (18%), V1399I (1%); mature forms: (18,184-1712) [22,745×, 205 kTa]. #ᗕᕱᗒ 🔗
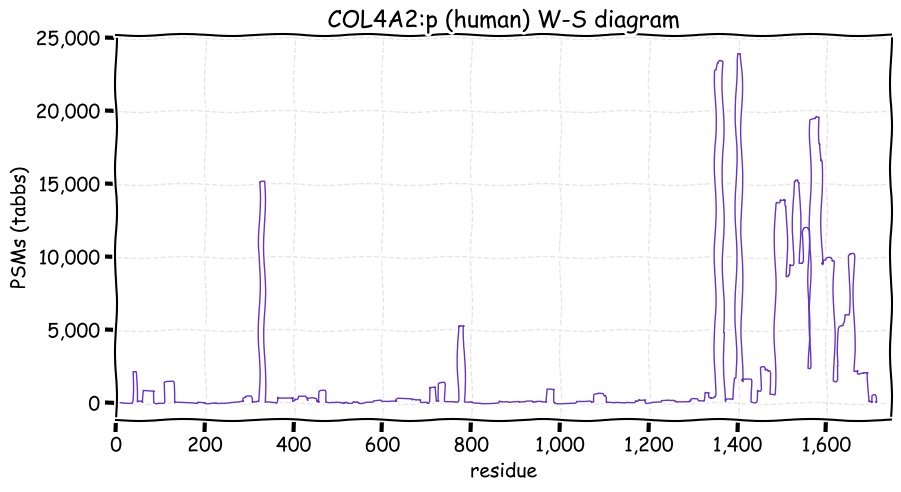
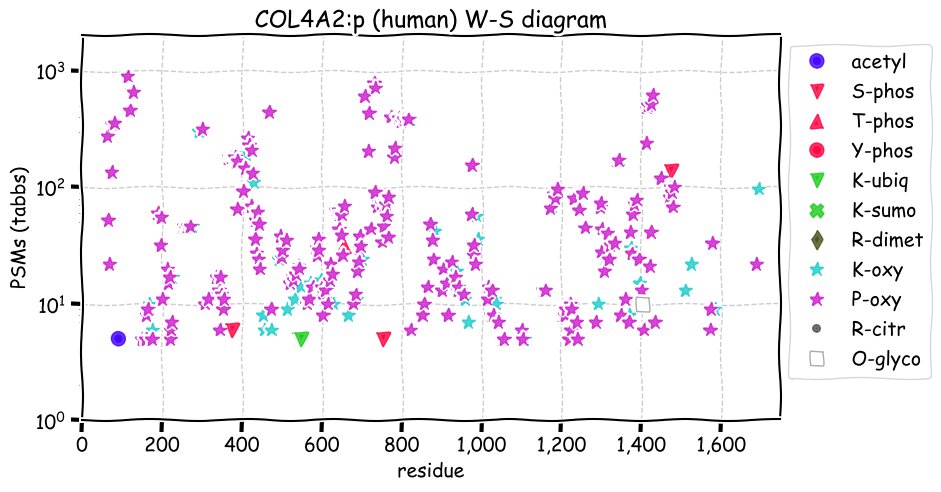
Fri May 21 18:09:48 +0000 2021@ProtifiLlc @nesvilab Formation of citrulline or methylation are the one's I normally run across in proteomics data.
Fri May 21 17:10:56 +0000 2021@nesvilab I should say that I exclude these solutions if & only if the R or K in question are the peptide's C-terminal residue.
Fri May 21 17:01:48 +0000 2021@nesvilab TMT samples can be problematic at the best of times. I explicitly reject any solution where the Lys sidechain amino group or Arg guanidino group are modified at the time of digestion.
Fri May 21 16:11:32 +0000 2021"when out on a leash, cats don't act like dogs"
🔗
Fri May 21 15:56:30 +0000 2021@cajun_science @michal_bassani I left out viral proteins as a source of peptides as it is very dependent on infection status. Except for EBV, which seems to be pretty much universal.
Fri May 21 14:31:33 +0000 2021Since the RPS27A transcript codes both for ubiquitin & the short ribosomal subunit RPS27A, this branching means that it will always be the most abundant ubiquitinated protein (equivalently true for the other 3 ubiquitin-containing transcripts). end/
Fri May 21 14:26:08 +0000 2021Thanks to the brave few that participated in this poll. RPS27A is the right answer. Once ubiquitin is attached to a protein, other ubiquitins often attach to the ubiquitin, forming a large branched structure. 1/
Fri May 21 12:52:20 +0000 2021@cajun_science @michal_bassani IMHO, HLA type I peptides from:
proteasome splicing: no
small ORF translation: yes
alternate splicing: yes
PTMs (except N-terminal acetylation): very rarely
Fri May 21 12:32:04 +0000 2021SVEP1:p has a long, fancy-sounding name but no known function (or Study Section). Maybe one of the cancer interest groups will adopt it & figure something out 🔗
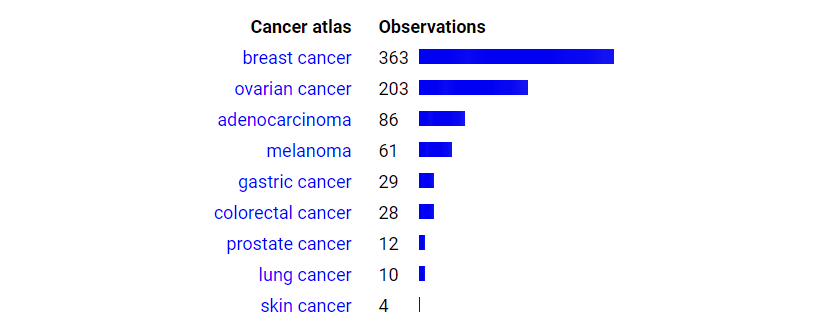
Fri May 21 12:15:35 +0000 2021SVEP1:p.A2727V chr 9:g.110407351G>A, rs7030192 (all tissue A:V 0.798:0.202) vaf=36%, Δm=28.0313, VAF by population group: african 57%, american 33%, east asian 44%, european 33%, south asian 39%. #ᐯᐸᐱ
Fri May 21 12:15:35 +0000 2021SVEP1:p,θ(max) = 52. aka bA427L11.3, POLYDOM, FLJ13529, C9orf13. Observed in MHC class 1 & class 2 peptide experiments from tissue. Observed in many tissues & blood plasma.
Fri May 21 12:15:35 +0000 2021SVEP1:p, SAAVs: G405S (4%), V484I (27%), K876R (80%), I1134V (18), A2727V (36%), F3138I (11%), T3536M (13%).
Fri May 21 12:15:34 +0000 2021>SVEP1:p, sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (Homo sapiens) Very large subunit; CTMs: N3562+glycosyl; PTMs: 7×S, 1×T, 0×Y+phosphoryl, 2×K+ubiquitinyl; mature form: (18,21-3571) [3,860×, 13 kTa]. #ᗕᕱᗒ 🔗
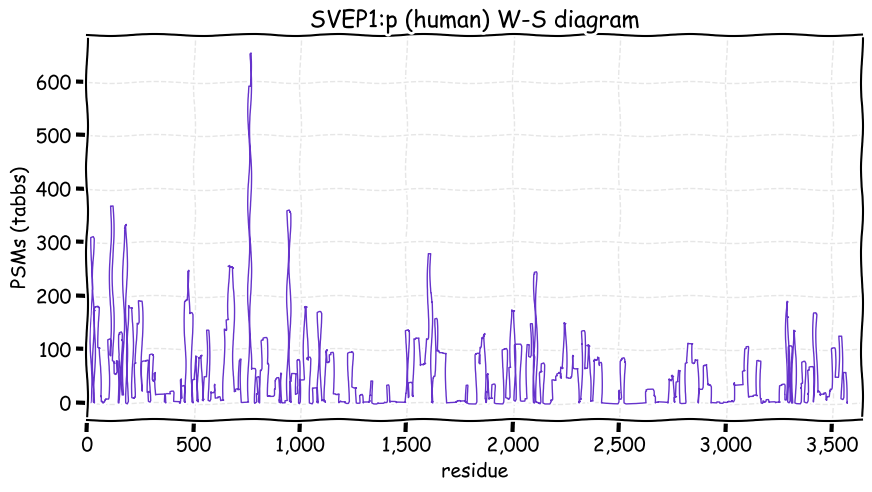
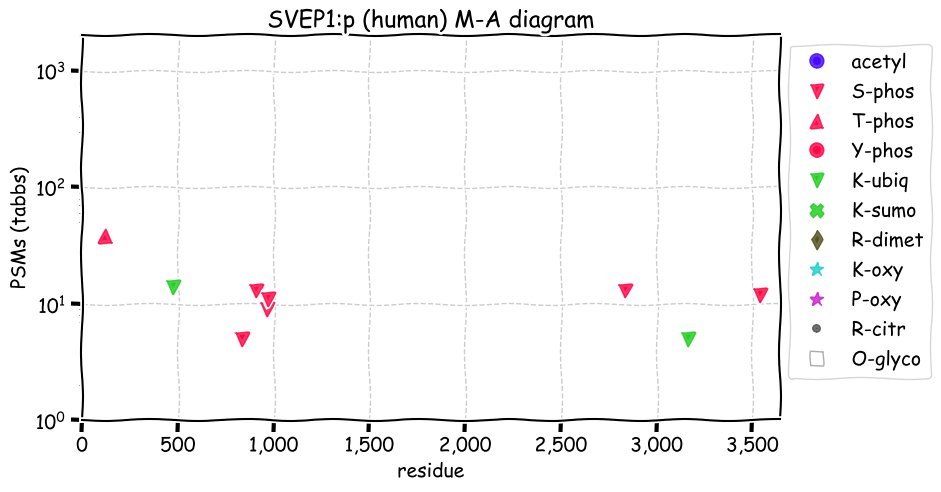
Thu May 20 16:10:52 +0000 2021Pet peeve: if you ran the control using CAD, don't switch to HCD for the interesting stuff! 🧐
Thu May 20 14:15:42 +0000 2021If you are looking for the Gly-Gly reminant of ubiquitinylation in a sample that has not been enriched for ubiquitinylation, which of the following proteins will produce the most GG-modified PSMs?
Thu May 20 12:14:37 +0000 2021S100A7:p.E28D chr 1:g.153458930C>G, rs3014837 (all tissue E:D 279:1994) vaf=96%, Δm=-14.0157, VAF by population group: african 99%, american 99%, east asian 100%, european 96%, south asian 99%. ED in HCT-116 cells. #ᐯᐸᐱ
Thu May 20 12:14:37 +0000 2021S100A7:p, θ(max) = 100. aka S100A7c, PSOR1. Observed in MHC class 1 & class 2 peptide experiments. Observed in many tissues, fluids and cell llines.
Thu May 20 12:14:37 +0000 2021>S100A7:p, S100 calcium binding protein A7 (Homo sapiens) Small subunit; CTMs: M1, S2+acetyl; PTMs: 4×S, 1×T, 0×Y+phosphoryl; SAAVs: E28D (96%); mature form: (1,2-101) [14,253×, 83 kTa]. #ᗕᕱᗒ 🔗
Wed May 19 20:33:23 +0000 2021I had some Sober Carpenter "Irish Red" non-alchoholic beer yesterday. It was the best na beer I'd ever had.
Wed May 19 13:01:23 +0000 2021I am irrationally pleased by the performance of my "AI" method for determining occupied N-linked glycosylation sites using deamidation patterns. Thanks to @pwilmarth for his skepticism wrt deamidation that made me look hard at the results.
Wed May 19 12:13:59 +0000 2021CHI3L1:p.R145G chr 1:g.203183673T>C, rs880633 (all tissue R:G 0.496:0.504) vaf=45%, Δm=-99.0796, VAF by population group: african 17%, american 33%, east asian 38%, european 54%, south asian 45%. #ᐯᐸᐱ
Wed May 19 12:13:59 +0000 2021CHI3L1:p, θ(max) = 93. aka GP39, YKL40, YK-40. Observed in MHC class 1 or class 2 peptide experiments. Commonly observed in CSF, blood plasma & urine as well as THP-1, U-87MG & MCF-10A cells. Rare in most other cell lines. Has no chitinase activity.
Wed May 19 12:13:59 +0000 2021>CHI3L1:p, chitinase 3 like 1 (Homo sapiens) Small subunit; CTMs: N60+glycosyl; PTMs: 6×S, 0×T, 0×Y+phosphoryl; SAAVs: P125L (1%), R145G (45%); mature form: (22-383) [7,883×, 96 kTa].#ᗕᕱᗒ 🔗
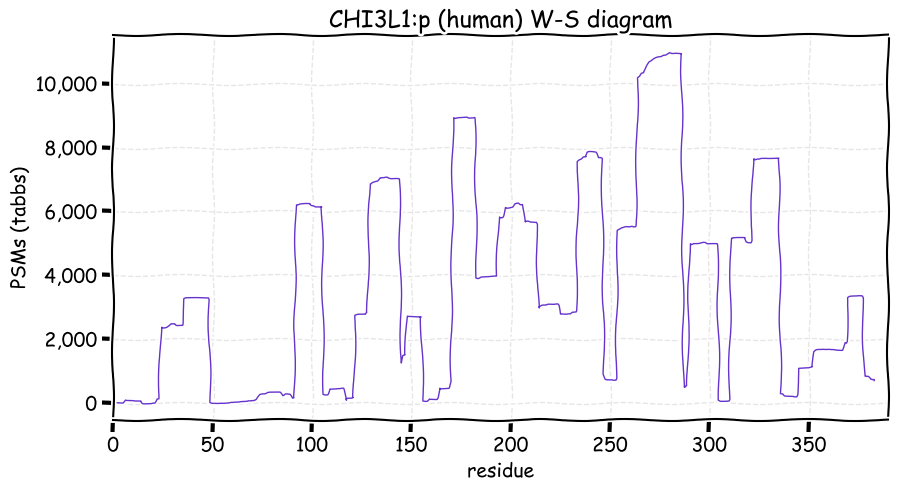
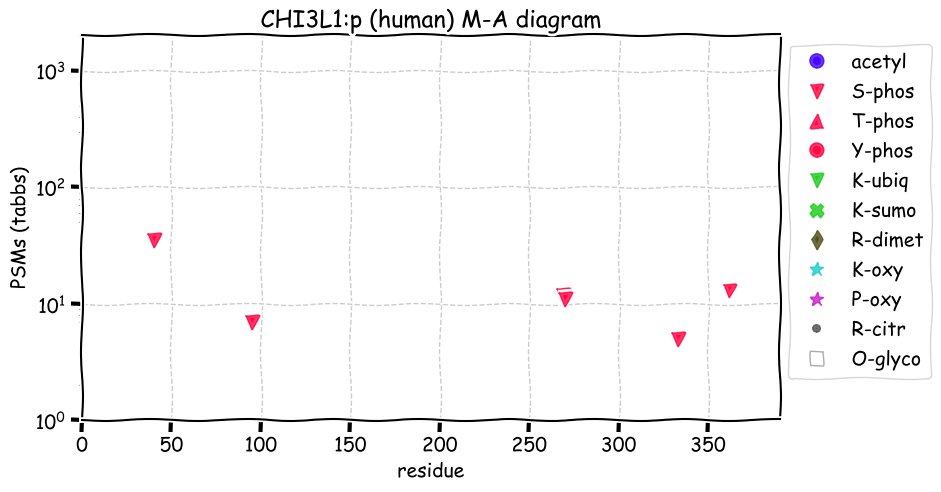
Tue May 18 18:47:09 +0000 2021@michaelhoffman Sounds like they are probably an IT group shared with the good folks at CommonCV 🇨🇦
Tue May 18 14:17:09 +0000 2021@TrumanLab A good example of why I don't even look at Nature Biotech any more.
Tue May 18 12:22:11 +0000 2021F12:p, MHC1 ligand peptides for this protein have only been observed in liver tissue (PXD019643), where the protein is synthesized.
Tue May 18 12:07:29 +0000 2021F12:p.A207P chr 5:g.177404825C>G, rs17876030 (all tissue A:P 0.076:0.924) vaf=95%, Δm=26.0156, VAF by population group: african 97%, american 88%, east asian 88%, european 99%, south asian 95%. #ᐯᐸᐱ
Tue May 18 12:07:29 +0000 2021F12:p, θ(max) = 71. Mature protein cleaves at R372-V373 to start contact-activated coagulation: further cleavage occurs during the coagulation cascade. Rarely observed in MHC class 1 or 2 peptide experiments. Commonly observed in blood plasma, urine & CSF.
Tue May 18 12:07:29 +0000 2021>F12:p, coagulation factor XII (Homo sapiens) Midsized protease; CTMs: N249, N433+glycosyl; PTMs: 3×S, 7×T+glycosyl; SAAVs: A207P (95%), R448C (1%); mature form: (20-615) [14,388×, 157 kTa]. #ᗕᕱᗒ 🔗
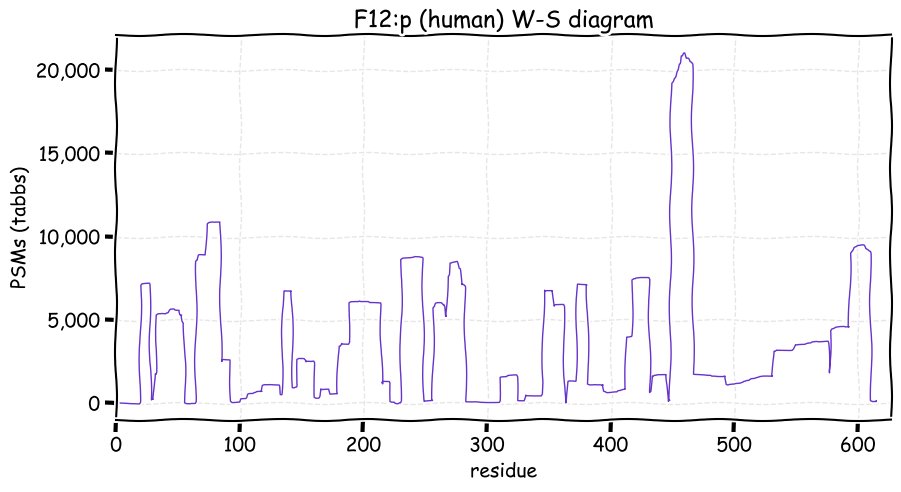
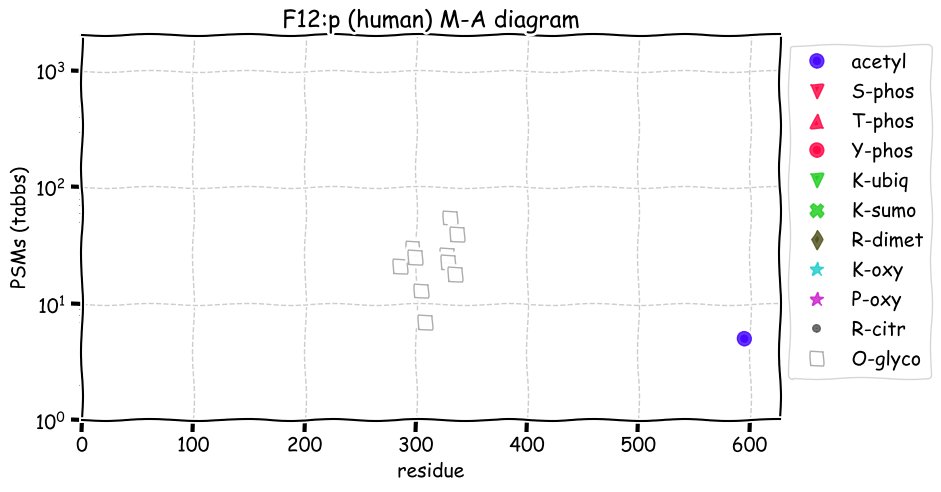
Mon May 17 16:26:03 +0000 2021Tropical Cyclone Tauktae makes landfall in Gujarat 🔗
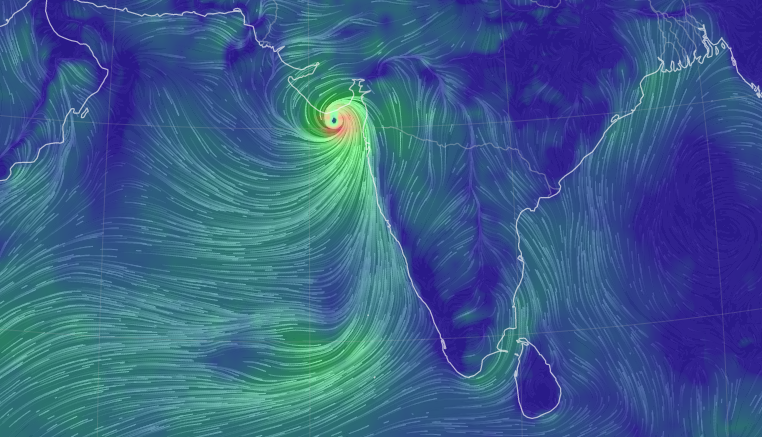
Mon May 17 14:18:22 +0000 2021Thanks to everyone who participated. It seems as though most respondents have a relatively small PV. Does anyone have any insight into why?
Mon May 17 12:37:02 +0000 2021🔗
Mon May 17 12:34:04 +0000 2021PIEZO1:p can serve as a useful counter-example when voicing skepticism about the generality of an overly simplified "new" method of analyzing protein sequences.
Mon May 17 12:11:59 +0000 2021PIEZO1:p.V394L chr 16:g.88737574C>G, rs6500493 (all tissue V:L 0.336:0.664) vaf=72%, Δm=14.0156, VAF by population group: african 74%, american 75%, east asian 89%, european 67%, south asian 72%. #ᐯᐸᐱ
Mon May 17 12:11:59 +0000 2021PIEZO1:p, θ(max) = 26. aka KIAA0233, FAM38A. Observed in MHC class 1 & some class 2 peptide experiments. Observable in tissues & cell lines, but not fluids. Has 36 transmembrane domains.
Mon May 17 12:11:58 +0000 2021PIEZO1:p, SAAVs: I83T (91%), P152L (18%), V394L (72%), P966L (18%), F1338L (87%), P1398L (18%), P1857S (17%), I2265V (20%). It is unlikely that any individual will be homozygous for the reference sequence. 🔗
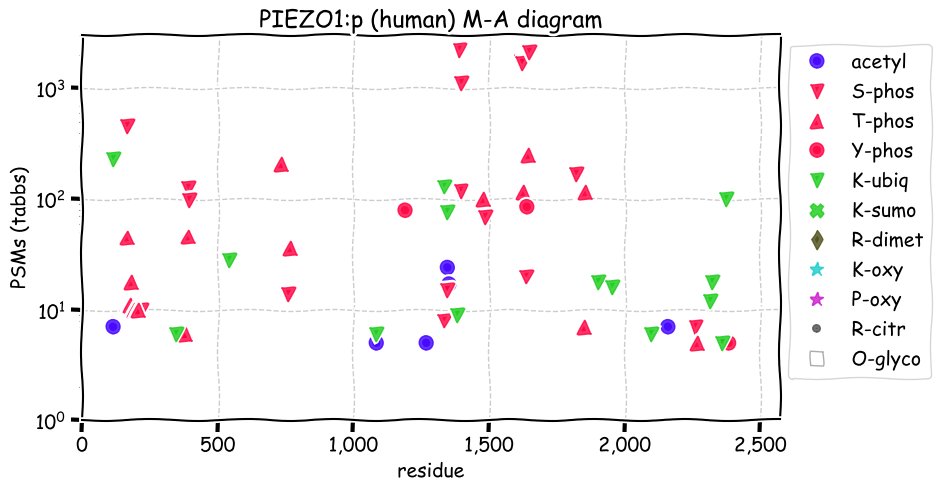
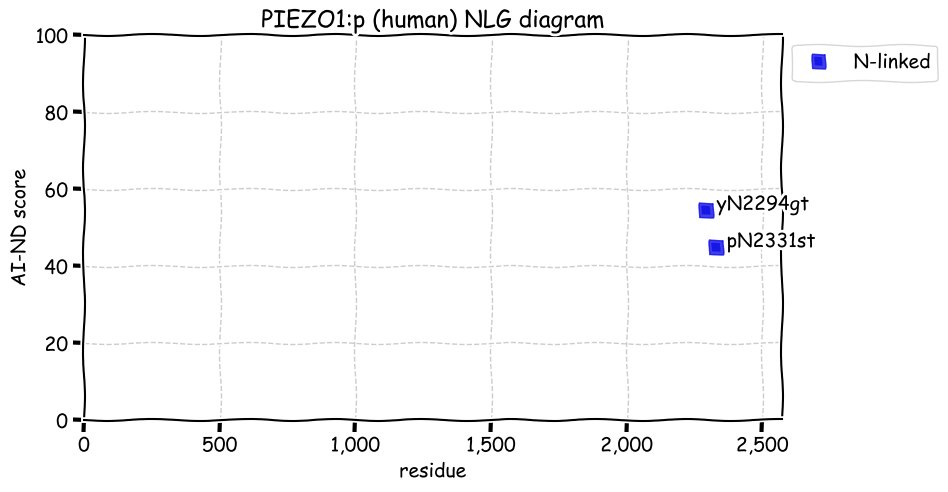
Mon May 17 12:11:56 +0000 2021>PIEZO1:p, piezo type mechanosensitive ion channel component 1 (Homo sapiens) Large membrane subunit; CTMs: N2294, N2331+glycosyl; PTMs: 18×S, 16×T, 3×Y+phosphoryl; 14×K+ubiquitinyl; 6×K+acetyl; ; mature form: (17-2521) [11,381×, 48 kTa]. #ᗕᕱᗒ
Sun May 16 15:19:41 +0000 2021Tropical Cyclone Tauktae off India 🔗
Sun May 16 14:11:44 +0000 2021How large is your protein vocabulary? Your PV is the set of proteins you are sufficiently familiar with so that you can associate them with useful, context-specific functional or structural information without looking at your phone.
Sun May 16 13:23:28 +0000 2021If you want to be involved in proteomics, it is very worth while to make a study of the SERPINs. The set of SERPINs observed in any data set often serve as a good basis for understanding it, both biochemically & analytically.
Sun May 16 13:06:28 +0000 2021Manitoba (MB) again living up to its reputation as a go-it-alone oddball province 🔗
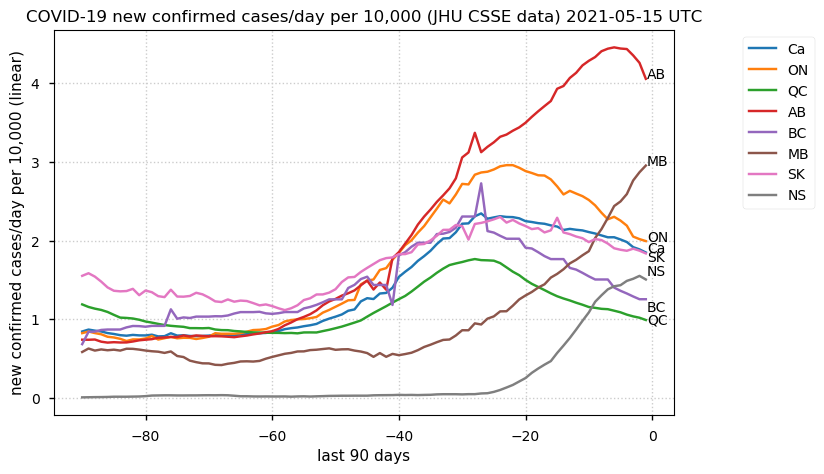
Sun May 16 12:39:39 +0000 2021SERPINB10:p.P140S chr 18:g.63919833C>T, rs9967382 (all tissue P:A 0.587:0.413) vaf=55%, Δm=-10.0207, VAF by population group: african 77%, american 40%, east asian 46%, european 22%, south asian 28%. #ᐯᐸᐱ
Sun May 16 12:39:39 +0000 2021SERPINB10:p, θ(max) = 80. aka bomapin, PI10. Observed in MHC class 1 & some class 2 peptide experiments. Observable in saliva & urine, but rarely in blood plasma. Rare in common cell lines, unless derived from lymphocyte/leukocyte progenitors.
Sun May 16 12:39:39 +0000 2021>SERPINB10:p, serpin family B member 10 (Homo sapiens) Small intracellular subunit; CTMs: M1+acetyl; PTMs: none; SAAVs: S3A (72%), I99T (25%), P140S (55%), R246C (47%); mature form: (1-397) [3,348×, 25 kTa]. #ᗕᕱᗒ 🔗
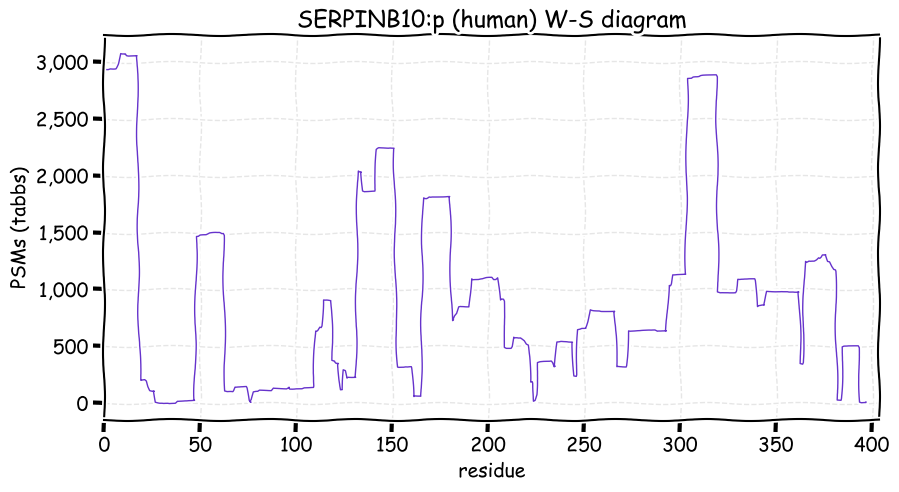
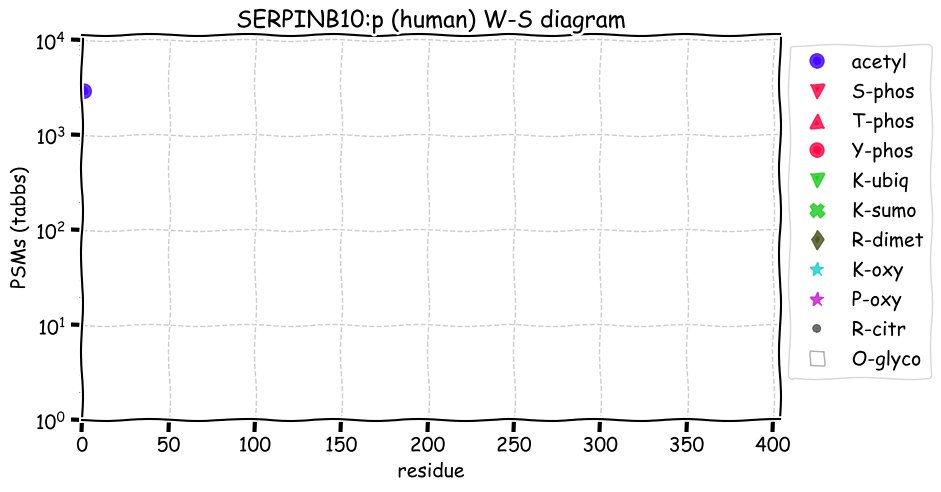
Sat May 15 14:11:50 +0000 2021Every video I have ever seen about clipping a cat's claws features a cat that behaves like a bean bag during the process. Mine, not so much ...
Sat May 15 12:55:12 +0000 2021Today's circle of storms surrounding Antarctica 🔗
Sat May 15 12:43:17 +0000 2021@J_my_sci The designer earned their pay with that one.
Sat May 15 11:56:07 +0000 2021ATP5MPL:p.I9V chr 14:g.103915165T>C, rs1053419 (all tissue I:V 0.905:0.095) vaf=11%, Δm=-14.0156, VAF by population group: african 43%, american 10%, east asian 5%, european 9%, south asian 15%. #ᐯᐸᐱ
Sat May 15 11:54:03 +0000 2021ATP5MPL:p,θ(max) = 93. aka MP68, MLQ, 6.8PL, C14orf2. Abundant in MHC class 1 & 2 peptide experiments. Observable in most cells and tissues, but absent from fluids.
Sat May 15 11:54:03 +0000 2021>ATP5MPL:p, ATP synthase membrane subunit 6.8PL (Homo sapiens) Very small intermembrane mitochondrial subunit; CTMs: M1+acetyl; PTMs: S51+phosphoryl; K49+ubiquitinyl; SAAVs: I9V (11%); mature form: (1-58) [3,640×, 11 kTa]. #ᗕᕱᗒ 🔗
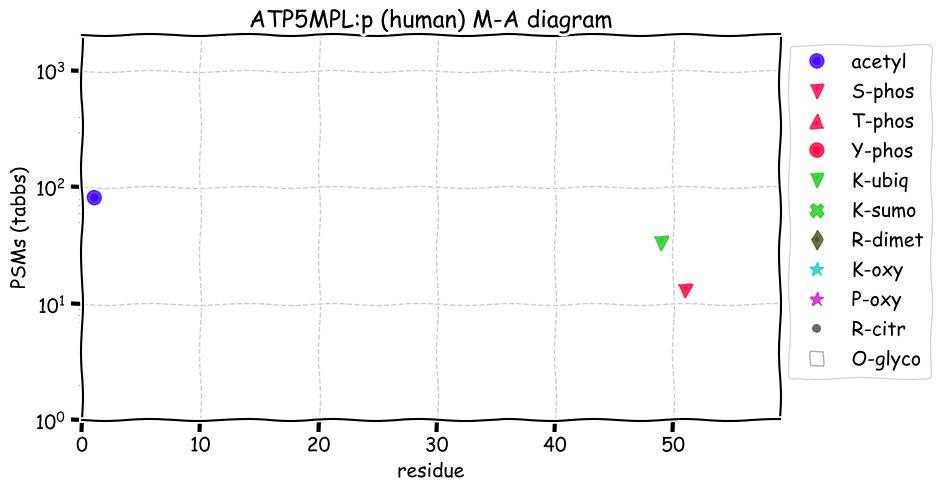
Fri May 14 17:10:31 +0000 2021Thanks to the people who contributed to this poll. It appears people get a range of download speed. I personally get either ~10 Mbits/sec or ~30 Mbits/sec, depending on the day.
Fri May 14 15:19:30 +0000 2021@jwoodgett @guilbourque @OICR_news @GenomeCanada Not a bad example. The GC group should really look in to the practical, behind-the-scenes steps taken by the GISAID folks to make that happen. GISAID has been around since 2008, so they must understand how to winkle data out of reluctant groups.
Fri May 14 14:57:00 +0000 2021@jwoodgett @guilbourque @OICR_news @GenomeCanada When people share do data, it is always reluctantly and in their own best interests.
Fri May 14 14:53:34 +0000 2021@jwoodgett @guilbourque @OICR_news @GenomeCanada It isn't an impossible hurdle to overcome, but it will take time.
Fri May 14 14:52:17 +0000 2021@jwoodgett @guilbourque @OICR_news @GenomeCanada Nobody ever wants to share their data. But, by rushing out the site with data largely from only 1 province while claiming that it is a "Canadian" resource will make the job of convincing anyone outside of that province more difficult.
Fri May 14 14:26:21 +0000 2021@jwoodgett @guilbourque @OICR_news @GenomeCanada They are going to have to continue to try hard, probably for several years. Submitters need confidence that a resource will be available for the long term. Association with GC & the site's initial restrictive geography are strikes against them that will have to be overcome.
Fri May 14 13:55:07 +0000 2021@jwoodgett @guilbourque @OICR_news @GenomeCanada Repository sites are like pipettors: regardless of how much you might wish them to, they will not fill themselves.
Fri May 14 13:46:25 +0000 2021@jwoodgett @guilbourque @OICR_news @GenomeCanada "If I build it, they will contribute to it" is a bit of a fantasy often used in grant documents. You have to go out & get content for a site. Relentlessly.
Fri May 14 12:22:59 +0000 2021TPO:p.A373S chr 2:g.1477383G>T, rs2280132 (all tissue A:S 0.341:0.659)) vaf=47%, Δm=15.9949, VAF by population group: african 32%, american 49%, east asian 52%, european 45%, south asian 32%. #ᐯᐸᐱ
Fri May 14 12:22:59 +0000 2021TPO:p, oxidizes iodide to iodine in thyrocyte vesicles. The iodine then non-specifically iodinates thyroglobulin Y residues. Domains: (19-846) in the vesicular lumen, (847-865) transmembrane & (866-933) in the cytoplasm.
Fri May 14 12:22:59 +0000 2021TPO:p, θ(max) = 74. aka TPX. Abundant in MHC class 1 & 2 peptide experiments of thyroid tissue. Common in thyroid samples only.
Fri May 14 12:22:59 +0000 2021>TPO:p, thyroid peroxidase (Homo sapiens) Midsized membrane-bound enzyme; CTMs: none; PTMs: 0×S, 0×T, 0×Y+phosphoryl; 2×K+ubiquitinyl; SAAVs: A373S (45%); mature form: (17,19-933) [337×, 5.8 kTa]. #ᗕᕱᗒ 🔗
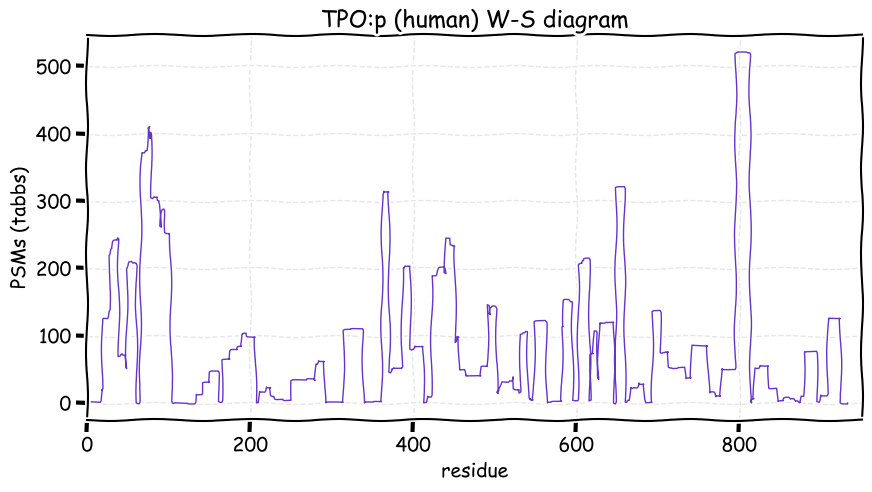
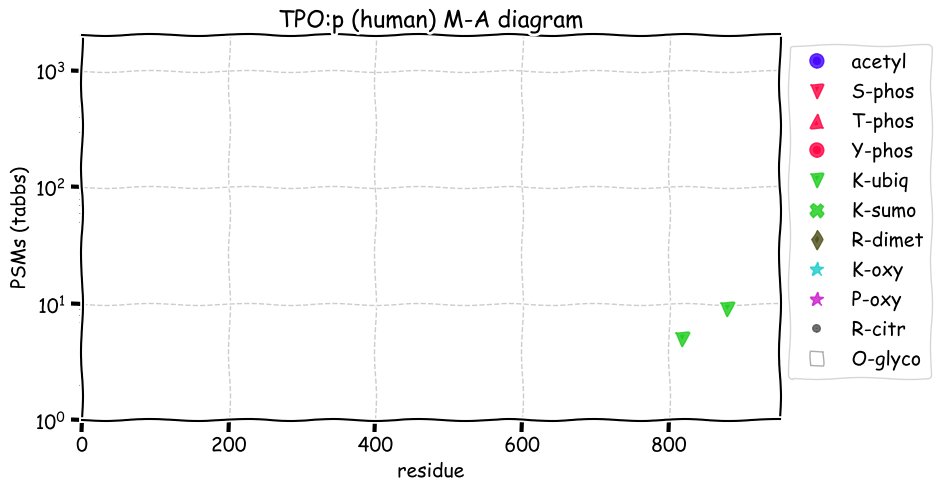
Thu May 13 20:12:32 +0000 2021Does anybody out there know is a protein C-terminal "WW" or "FF" is a targeting signal of some type?
Thu May 13 17:02:41 +0000 2021For people who use PRIDE for MS data, what answer best characterizes your best download speed from the repository:
Thu May 13 11:55:05 +0000 2021EPHX1:p.Y113H chr 1:g.225831932T>C, rs2791494 (all tissue Y:H 0.910:0.090) vaf=32%, Δm=-26.0044, VAF by population group: african 14%, american 32%, east asian 48%, european 30%, south asian 38%. YH in Hep-G2 & HaCaT cells. #ᐯᐸᐱ
Thu May 13 11:55:04 +0000 2021EPHX1:p, θ(max) = 93. aka EPHX. Abundant in MHC class 1 & 2 peptide experiments. Common in most solid tissues & cell lines. Rare in fluids, except saliva.
Thu May 13 11:55:04 +0000 2021>EPHX1:p, epoxide hydrolase 1 (H sapiens) Small enzyme; CTMs: M1+acetyl; PTMs: 5×S,2×T,3×Y+phosphoryl; 8×K+ubiquitinyl; 3×K+acetyl; SAAVs: R43T (1%), R49C (1%), R71H (1%), Y113H (32%), H139R (78%), S195P (1%); mature form: (1-455) [28,689×, 345 kTa] #ᗕᕱᗒ 🔗
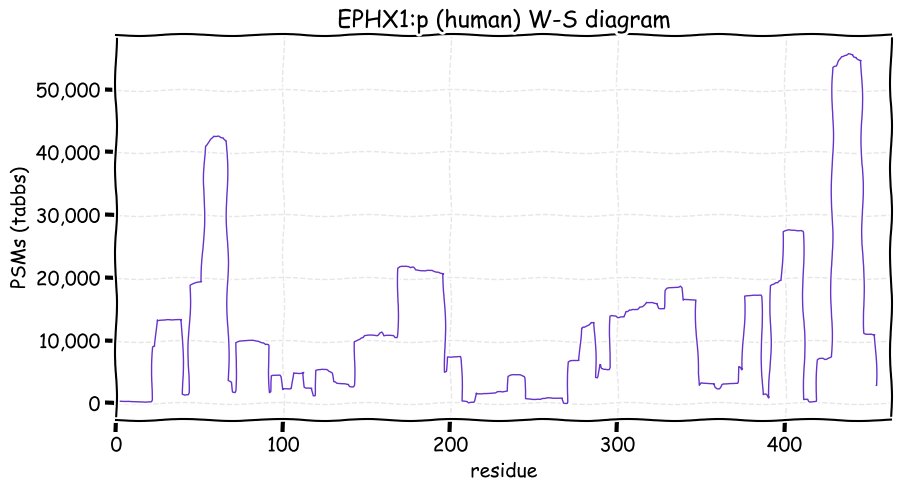
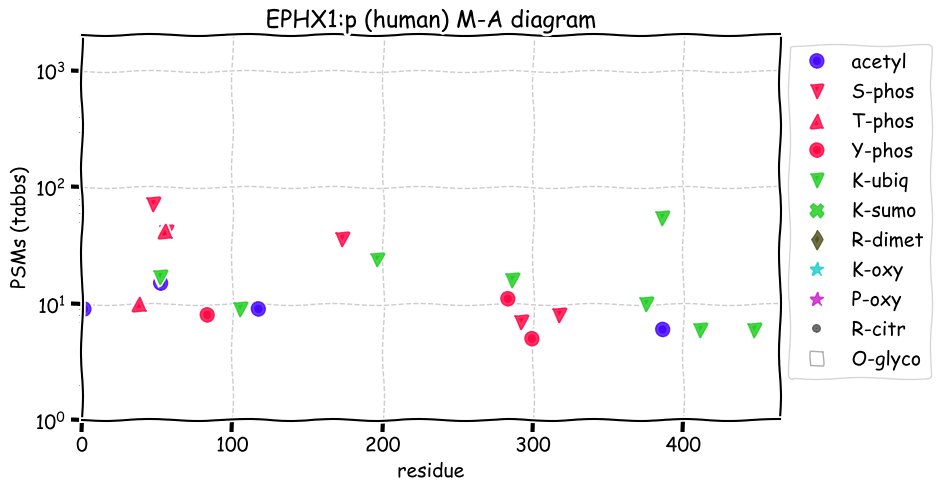
Wed May 12 15:36:00 +0000 2021The lysosome is a mechanism for cleaning up TG peptides remaining in thyrocyte vesicles after T3/T4 synthesis, resulting in the formation of these Y+iodo peptides. See the discussion of Fig 2-7 in 🔗 for details.
Wed May 12 15:28:21 +0000 2021Thanks to everyone who participated in the poll. According to the data provided along with 🔗, the answer is MHC class 2.
Wed May 12 15:13:51 +0000 2021This probably means that what ever was going on 2-3 weeks ago was the right thing to do. Unfortunately, all three totally changed their strategy & tactics over the weekend.
Wed May 12 13:04:45 +0000 2021It appears that the curves have nearly peaked in the current hotspot provinces: Alberta, Manitoba and Nova Scotia. 🔗
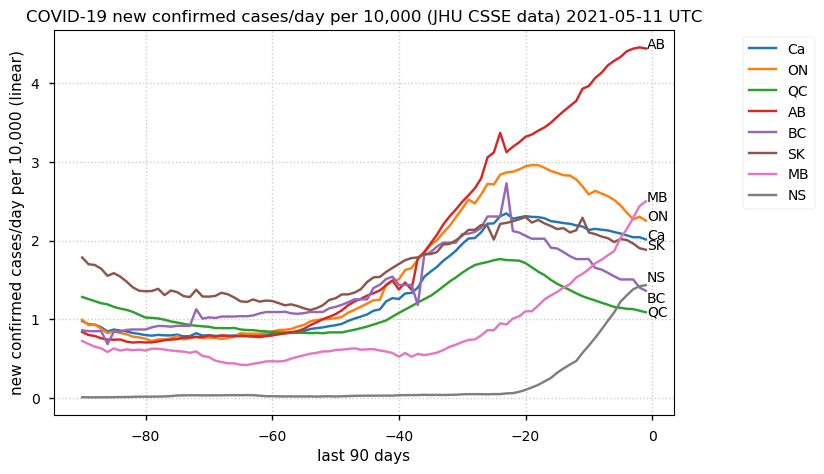
Wed May 12 12:11:04 +0000 2021CLCA1:p is often in the top 10 human proteins (by PSM count) in feces samples
Wed May 12 11:50:34 +0000 2021CLCA1:p.M524T chr 1:g.86493490T>C, rs2791494 (all tissue M:T 0.624:0.376) vaf=85%, Δm=-29.9928, VAF by population group: african 75%, american 88%, east asian 91%, european 82%, south asian 90%. 1 of 3 high VAF SAAVs in this protein. #ᐯᐸᐱ
Wed May 12 11:50:34 +0000 2021CLCA1:p, θ(max) = 83. aka CaCC, CLCRG1. Observed in MHC class 1 & 2 peptide experiments. Easily detected in the stomach, intestines & feces.
Wed May 12 11:50:34 +0000 2021>CLCA1:p, chloride channel accessory 1 (Homo sapiens) Midsized subunit; CTMs: N585, N770, N804, N810, N831+glycosyl; PTMs: T652+phosphoryl; SAAVs: L65F (88%), N357S (89%), M524T (85%); mature form: (22-914) [3,288×, 164 kTa]. #ᗕᕱᗒ 🔗
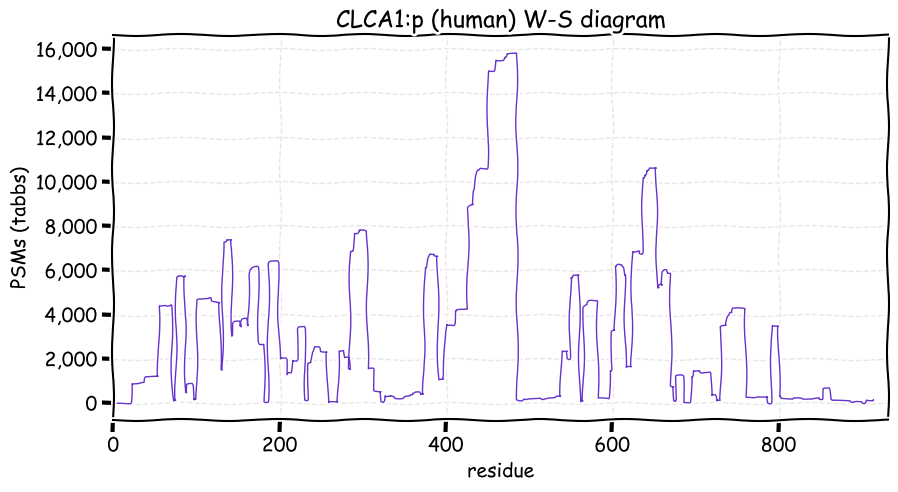
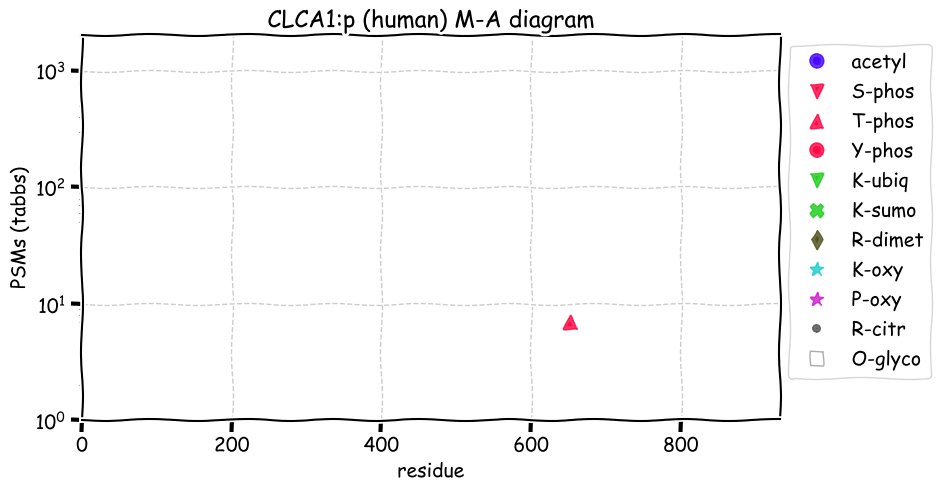
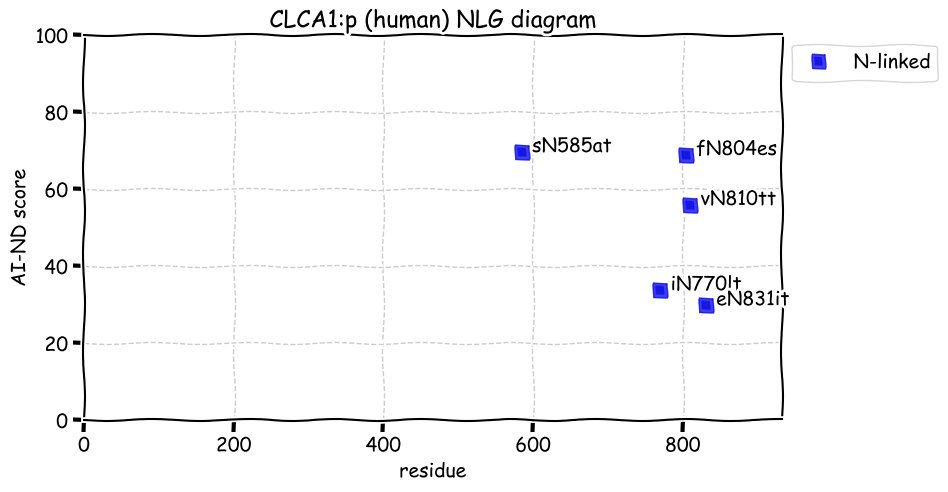
Tue May 11 18:08:48 +0000 2021@MHendr1cks Canadian programs tend to have an "all hat, no cattle" aspect to their written material.
Tue May 11 15:17:26 +0000 2021Which class of MHC peptides isolated from thyroid tissue samples should contain iodinated thyroglobulin (TG) peptides:
Tue May 11 12:24:41 +0000 2021Hint to on-line ppt presenters: noodling around with your mouse on the screen does not make your far-too-complex slides more easily understood.
Tue May 11 11:56:34 +0000 2021PRRC2C:p.A235T chr 1:g.171517773G>A, rs10913157 (all tissue A:T 0.935:0.065) vaf=14%, Δm=30.0106, VAF by population group: african 6%, american 11%, east asian 24%, european 13%, south asian 12%. #ᐯᐸᐱ
Tue May 11 11:56:33 +0000 2021PRRC2C:p is a large, highly modified, commonly observed intracellular protein that has no known function.
Tue May 11 11:56:33 +0000 2021PRRC2C:p, θ(max) = 48. aka KIAA1096, XTP2, BAT2D1, BAT2L2. Observed in MHC class 1 & more rarely in class 2 peptide experiments. Found in most solid tissues & cell lines.
Tue May 11 11:56:33 +0000 2021>PRRC2C:p, proline-rich coiled-coil 2C (H sapiens) Large subunit; CTMs: S2+acetyl; PTMs: 123×S,54×T,11×Y+phosphoryl; 26×K+acetyl; 5×K+ubiquitinyl; 5×R+dimethyl; SAAVs: A235T (14%), A906T (20%), A907T (20%), S1624C (20%); mature form: (2-2817) [43,504×, 477 kTa]. #ᗕᕱᗒ 🔗
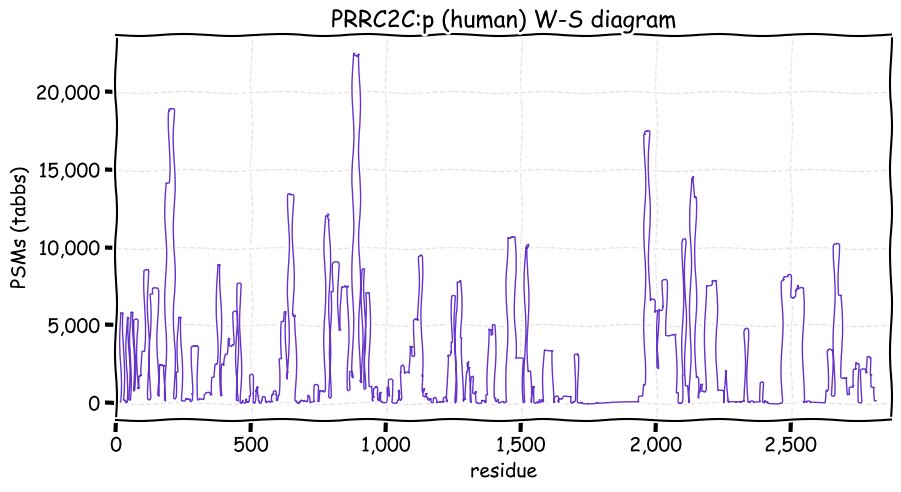
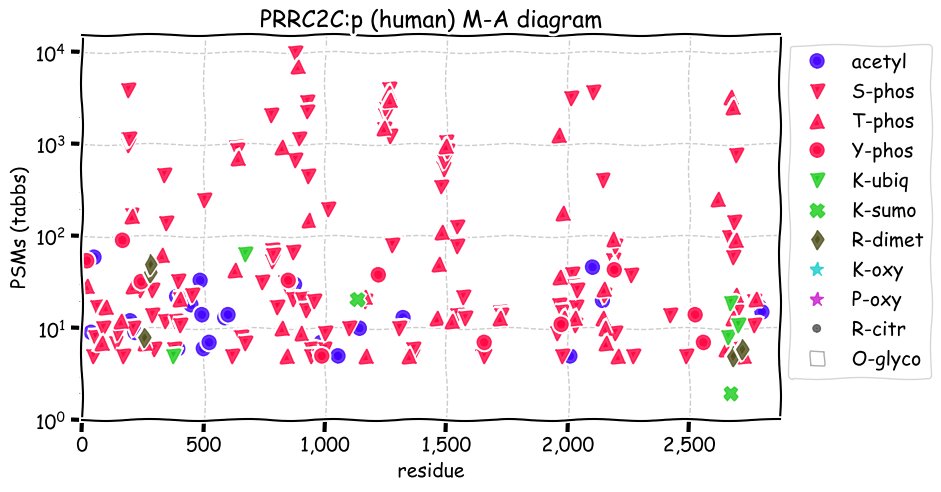
Mon May 10 16:02:45 +0000 2021I'm guessing this is caused by the trend towards the uncritical cutting & pasting of text into methods sections and the absence of effective peer review in most journals.
Mon May 10 15:45:19 +0000 20213rd data set in 4 days in which: the PX write up says they used IAA, the manuscript says they used IAA in their sample prep and that it was a fixed mod in their analysis. But no sign of it in the data.
Mon May 10 15:30:31 +0000 2021@AlexUsherHESA I'm sure if Réal Caouette was available & he had a chalk board ...
Mon May 10 15:26:50 +0000 2021@byu_sam Very meta. Maybe even meta-meta.
Mon May 10 13:01:19 +0000 2021@dtabb73 @harrisonspecht @fcyucn Low quality data is the #1 issue for many experiments.
Mon May 10 12:28:29 +0000 2021DIDO1:p is a large, highly modified, commonly observed intracellular protein that has no known function.
Mon May 10 12:00:35 +0000 2021DIDO1:p.T1568A chr 20:g.62881254T>C, rs910148 (all tissue T:A 0.011:0.989) vvaf=99%, Δm=-30.0106, VAF by population group: african >99%, american 99%, east asian >99%, european 99%, south asian >99%. Unlikely to find a TT individual. #ᐯᐸᐱ
Mon May 10 12:00:35 +0000 2021DIDO1:p, θ(max) = 59. aka DIO1, dJ885L7.8, FLJ11265, KIAA0333, DIO-1, BYE1, C20orf158, DATF1. Observed in MHC class 1 & 2 peptide experiments. Found in most solid tissues & cell lines.
Mon May 10 12:00:35 +0000 2021>DIDO1:p, death inducer-obliterator 1 (Homo sapiens) Large subunit; CTMs: M1+acetyl; PTMs: 114×S, 44×T, 5×Y+phosphoryl; 10×K+acetyl; 2×R+dimethyl; SAAVs: M544T (51%), A556T (28%), P1220Q (18%), T1568A (99%), V1743G (5%); mature form: (1-2240) [29,028×, 229 kTa]. #ᗕᕱᗒ 🔗
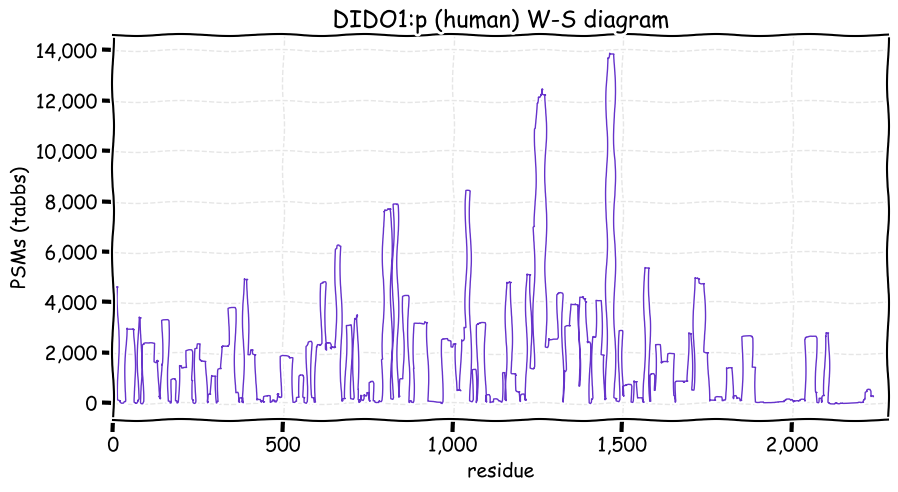
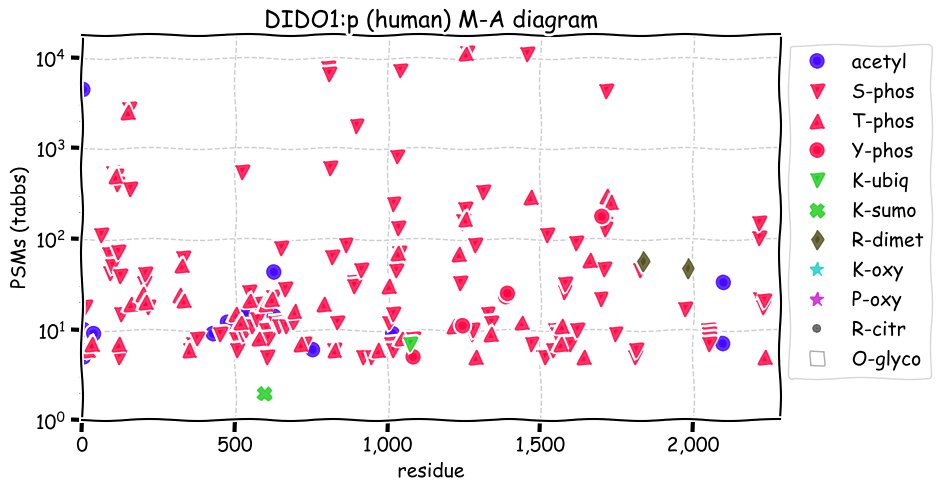
Sun May 09 23:35:10 +0000 2021A storm front, as of now, stretching from Dallas to Boston 🔗
Sun May 09 14:52:25 +0000 2021PXD025813 is one of those mysterious human cell culture experiments in which the data shows a surprisingly large number of PSMs for PIV5 proteins, even though there is no mention of PIV5 infection in the paper.
Sun May 09 13:33:11 +0000 2021Except for those who believe that many PTMs are caused by stochastic processes, for whom it is an example best forgotten.
Sun May 09 13:13:00 +0000 2021STING1:p seems to be one of those proteins on which investigators project the hopes and dreams of their respective study sections.
Sun May 09 12:27:31 +0000 2021STING1:p transmembrane domains: (18-34), (45-69), (92-106) & (117-134). The phosphodomain is cytoplasmic. Assigned to an unlikely number of biological functions and subcellular localizations.
Sun May 09 12:03:52 +0000 2021STING1:p.H232R chr 5:g.139478334T>C, rs1131769 (all tissue H:R 0.393:0.607) vaf=88%, Δm=19.0422, VAF by population group: african 89%, american 85%, east asian 86%, european 85%, south asian 90%. The variant introduces a novel tryptic cleavage site into the protein. #ᐯᐸᐱ
Sun May 09 12:03:52 +0000 2021STING1:p, θ(max) = 75. aka FLJ38577, NET23, ERIS, MPYS, STING, MITA, TMEM173. Observed in MHC class 1 & 2 peptide experiments. Found in most solid tissues & cell lines.
Sun May 09 12:03:51 +0000 2021>STING1:p, stimulator of interferon response cGAMP interactor 1 (Homo sapiens) Small subunit; CTMs: P2+acetyl; PTMs: 3×S, 2×T, 0×Y+phosphoryl; 2×K+ubiquitinyl; SAAVs: H232R (88%); mature form: (2-379) [9,896×, 47 kTa]. #ᗕᕱᗒ 🔗
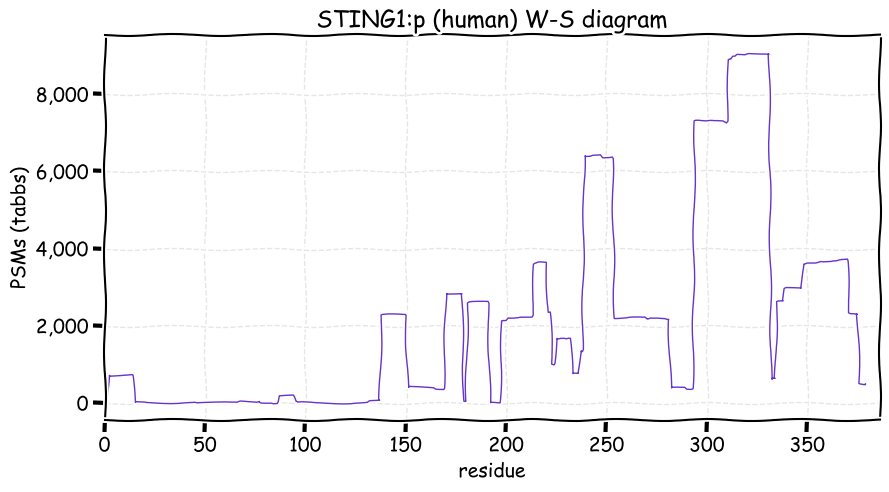
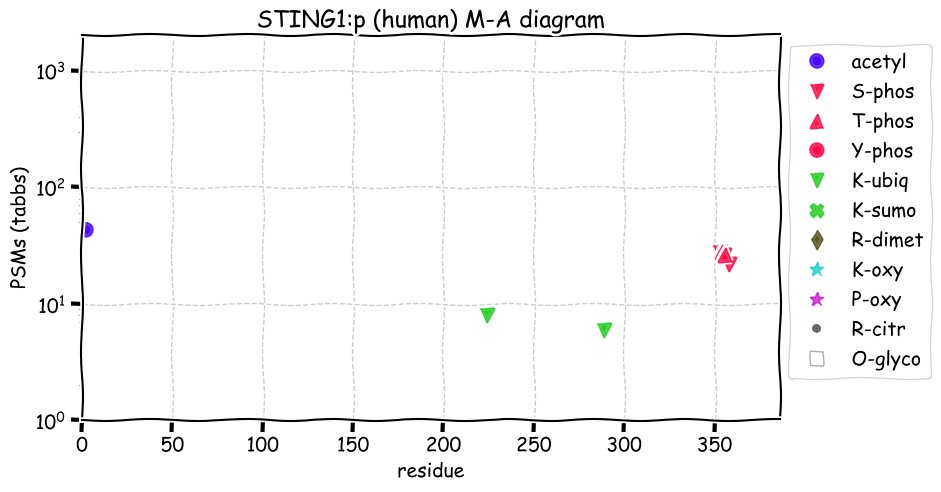
Sat May 08 19:54:35 +0000 2021"automating a flawed process creates flaws at scale"
C. Doctorow
Sat May 08 17:41:00 +0000 2021One of those samples from a human cell line where bovine A2M:p is the most abundant protein ...
Sat May 08 13:36:21 +0000 2021Possible ubiquitin/NEDD8 signaling?
Sat May 08 13:30:00 +0000 2021ALCAM:p, domains: extracellular (28-527), transmembrane (528-549) & intracellular (550-583). Y+phosphoryl & N+glycosyl sites are on the extracellular domain, while all K+ubiquitinyl sites are intracellular.
Sat May 08 12:34:42 +0000 2021@landlordsarebad @cstross It is possible he believes that Peaky Blinders was a documentary.
Sat May 08 11:59:30 +0000 2021ALCAM:p.T301M chr 3:g.105541676C>T, rs1044243 (all tissue T:M 0.947:0.053) vaf=9%, Δm=29.9928, VAF by population group: african 1%, american 5%, east asian 5%, european 11%, south asian 6%. Allele is TM in A-431 cell lines. #ᐯᐸᐱ
Sat May 08 11:59:30 +0000 2021ALCAM:p, θ(max) = 75. aka CD166, MEMD. Observed in MHC class 2 peptide experiments. Found in most solid tissues, fluids & cell lines.
Sat May 08 11:59:30 +0000 2021>ALCAM:p, activated leukocyte cell adhesion molecule (H sapiens) CTMs: N95, N167, N265, N306, N361, N457, N480, N499+glycosyl; PTMs: 1×T,5×Y+phosphoryl; 4×K+ubiquitinyl, 2×K+acetyl; SAAVs: N258S(8%), T301M(9%), M367I(2%); mature form: (28-583) [25,333×, 228 kTa]. #ᗕᕱᗒ 🔗
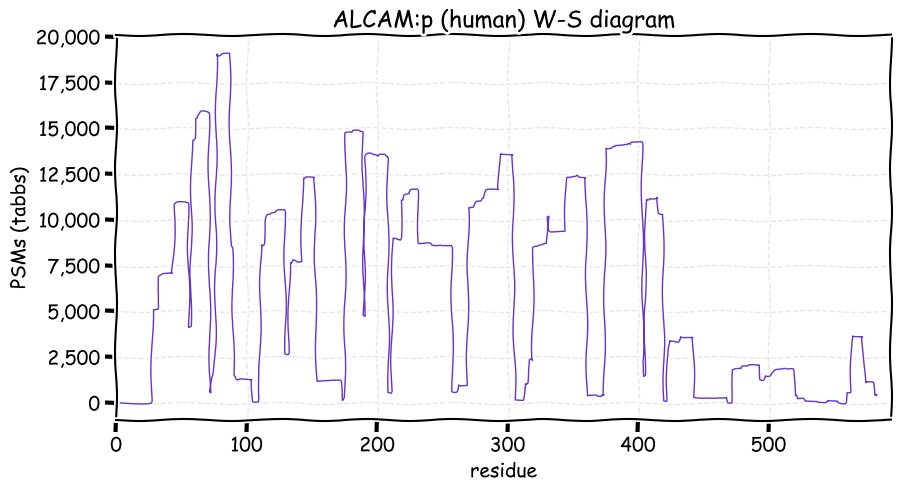
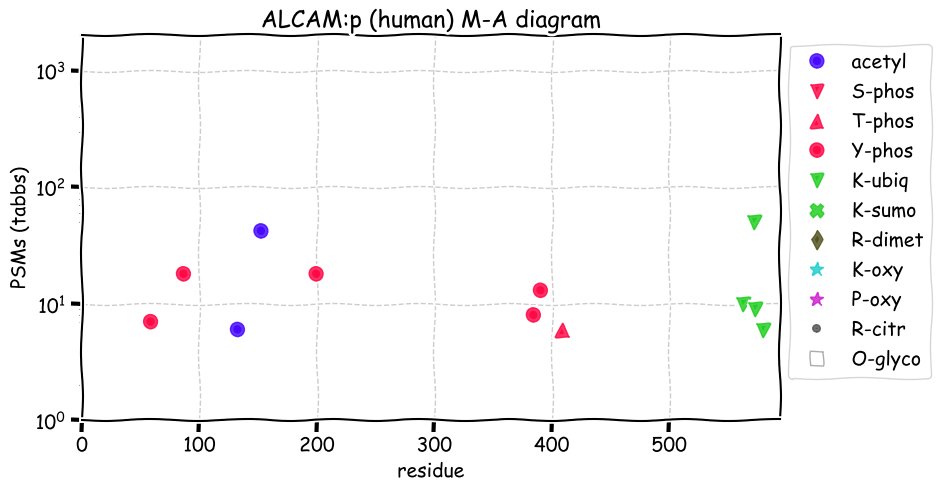
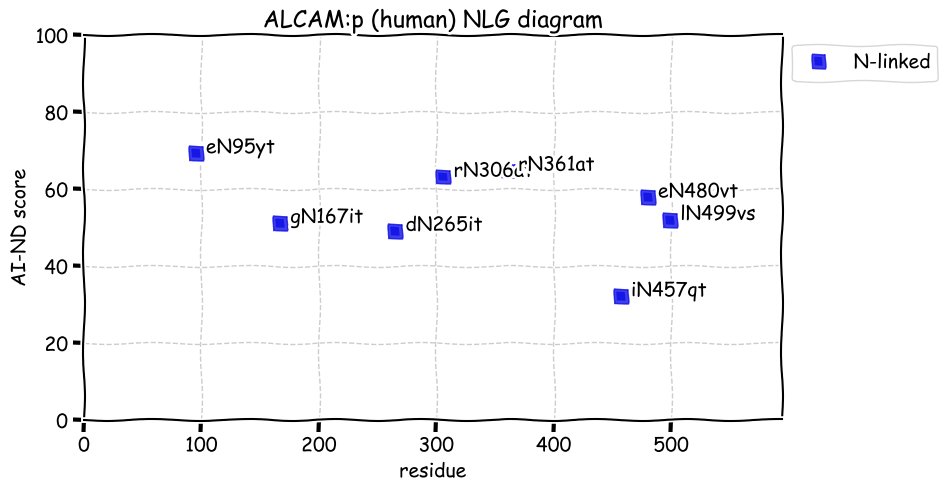
Fri May 07 15:01:01 +0000 2021@neely615 It would hopefully avoid situations like the NACA:g case, where much of the annotation for the short splice variant (🔗) really should be associated with the muscle splice variant (🔗).
Fri May 07 14:38:54 +0000 2021@CNPN_2021: are the times on the site for the meeting EDT (America/Toronto)?
Fri May 07 14:34:17 +0000 2021@neely615 Maybe using the word "exon" would overcome the natural resistance within the proteomics community to all things that involve alternate-splicing & its many consequences.
Fri May 07 12:52:01 +0000 2021@michaelhoffman "shockingly" 🔗
Fri May 07 12:45:32 +0000 2021Poster child for the development of a systematic exon-based functional annotation framework (XO?).
Fri May 07 12:01:16 +0000 2021ATR:p is not involved in the development of ataxia telangiectasia, which caused by mutations of a similar large kinase, ATM:p. ATM:p has a pattern of ubiquitinylation that is similar to ATR:p. 🔗
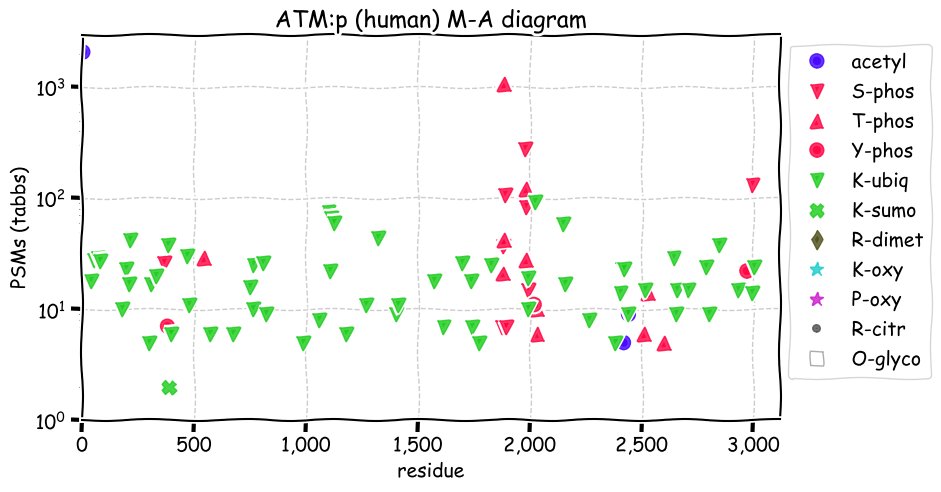
Fri May 07 12:01:14 +0000 2021ATR:p.F305S chr 3:g.142562488A>G, rs149129526 (all tissue F:S 0.992:0.008) vaf=0.003%, Δm=-60.0364, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. #ᐯᐸᐱ
Fri May 07 12:01:14 +0000 2021ATR:p, θ(max) = 39. aka FRP1, SCKL, SCKL1, MEC1. Observed in MHC class 1 peptide experiments. Found in most solid tissues and cell lines. Has 1 observed tryptic peptide that is shared with SMG1:p, another large kinase. ATR = Ataxia Telangiectasia & Rad3-related.
Fri May 07 12:01:14 +0000 2021>ATR:p, ATR serine/threonine kinase (Homo sapiens) Large enzyme; CTMs: A2+acetyl; PTMs: 17×S, 5×T, 0×Y+phosphoryl; 46×K+ubiquitinyl, 0×K+acetyl; SAAVs: F305S (<1%), R2425Q (10%); mature form: (2-2644) [9,577×, 43 kTa]. #ᗕᕱᗒ 🔗
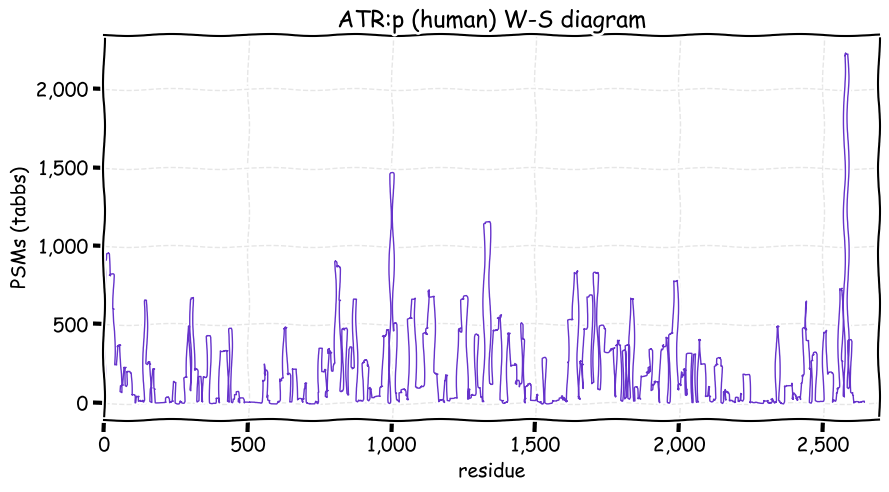
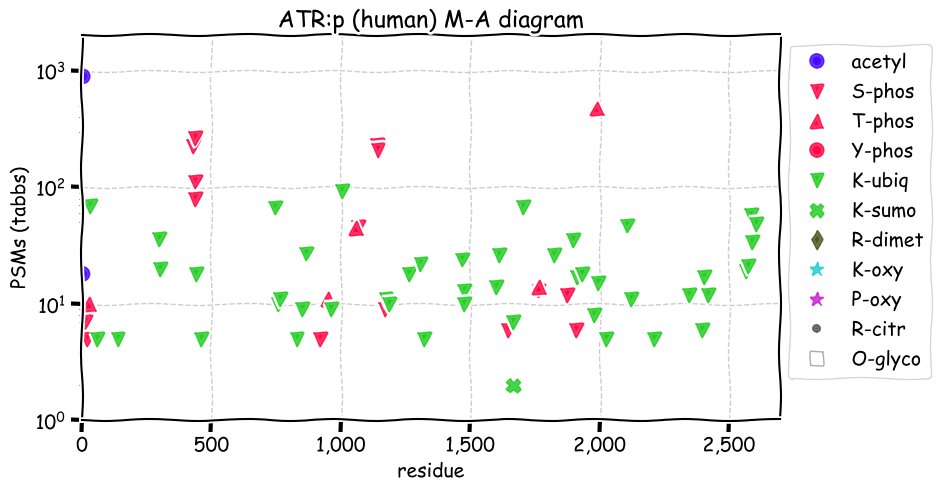
Thu May 06 20:06:37 +0000 2021Even with my new network connection, downloading 150 GB still takes more than a few minutes.
Thu May 06 15:15:56 +0000 2021@OliverMBernhar1 Anything but Musk! 😀
Thu May 06 15:04:22 +0000 2021@OdedRechavi @gangulyteena A part of the difference is that you are probably referring to "biomedical research" not "biology". Biomedical research traditions stem from its connections to medicine, an academic discipline with a longer history than science & a different way of valuing contributions.
Thu May 06 14:46:22 +0000 2021NACA1:g is represented by 2 different accessions in Uniprot: Q13765 (8 exon) & E9PAV3 (9 exon).
Thu May 06 14:42:15 +0000 2021NACA1:p, in muscle cells, a 9th exon (5,589 bp) is included, resulting in a very different 2,078 residue protein, with a long stretch of repetitive phosphodomains. 🔗
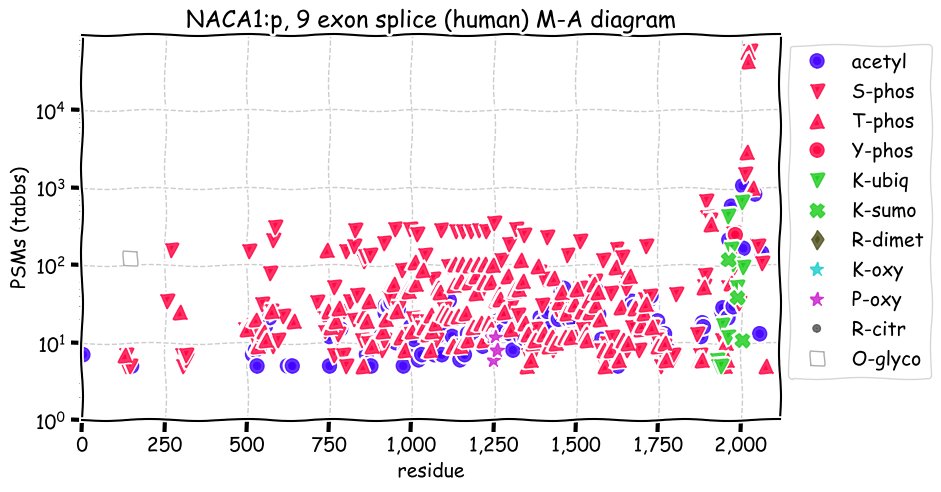
Thu May 06 14:42:14 +0000 2021NACA1:p is a nice example of tissue-specific alternate splicing. The gene has 8 exons in most tissues, resulting in a 215 residue protein that handles nascent peptides as they emerge from the ribosome exit tunnel. 🔗
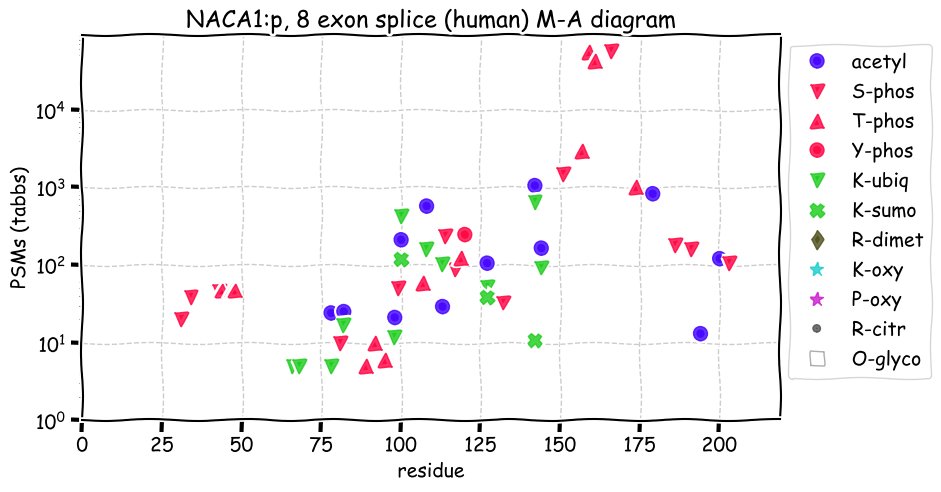
Thu May 06 12:03:58 +0000 2021ELAC2:p.S217L chr 17:g.13011692G>A, rs4792311 (all tissue S:L 0.711:0.289) vaf=27%, Δm=26.0520, VAF by population group: african 23%, american 24%, east asian 3%, european 32%, south asian 25%. #ᐯᐸᐱ
Thu May 06 12:03:58 +0000 2021ELAC2:p, θ(max) = 83. aka FLJ10530, HPC2. Observed in MHC class 1 peptide experiments. Found in most solid tissues and cell lines.
Thu May 06 12:03:57 +0000 2021>ELAC2:p, elaC ribonuclease Z 2 (Homo sapiens) Midsized protein; CTMs: M1+acetyl; PTMs: 24×S, 1×T, 0×Y+phosphoryl; 20×K+ubiquitinyl, 4×K+acetyl; 11×K+SUMOyl; SAAVs: S217L (27%), A541T (2%); mature form: (17?-826) [25,570×, 176 kTa]. #ᗕᕱᗒ 🔗
Wed May 05 12:06:54 +0000 2021UAP1L1:p.A319V chr 9:g.137079368C>T, rs7037849 (all tissue A:V 0.744:0.256) vaf=43%, Δm=28.0061, VAF by population group: african 48%, american 45%, east asian 65%, european 40%, south asian 45%. #ᐯᐸᐱ
Wed May 05 12:06:54 +0000 2021UAP1L1:p, θ(max) = 68. Observed in MHC class 1 peptide experiments. Observed in most solid tissues and cell lines. Has a very different PTM pattern than its namesake protein UAP1:p. 🔗
Wed May 05 12:06:53 +0000 2021>UAP1L1:p, UDP-N-acetylglucosamine pyrophosphorylase 1 like 1 (Homo sapiens) Midsized protein; CTMs: A2+acetyl; PTMs: 0×S, 0×T, 0×Y+phosphoryl; 2×K+ubiquitinyl; SAAVs: A319V (43%); mature form: (2-502) [13,800×, 51 kTa]. #ᗕᕱᗒ 🔗
Wed May 05 02:09:34 +0000 2021@ImmunoFever Yes. It was observed in the data (PXD008127) from 🔗
Tue May 04 15:12:12 +0000 2021@neely615 @ypriverol @MattWFoster Hopefully there is someone who can. I have been completely unsuccessful at getting anything published on the subject, other than a few oblique references in Methods sections.
Tue May 04 12:14:36 +0000 2021NCF2:p is part of the NADPH oxidase found in leukocytes, which generates reactive oxygen species, particularly superoxide, into cellular phagosomes.
Tue May 04 12:03:41 +0000 2021NCF2:p.K181R chr 1:g.183573252T>C, rs2274064 (all tissue K:R 0.565:0.435) vaf=50%, Δm=28.0061, VAF by population group: african 33%, american 60%, east asian 62%, european 47%, south asian 53%. #ᐯᐸᐱ
Tue May 04 12:03:41 +0000 2021NCF2:p, θ(max) = 94. aka NGN, p67phox, NOXA2. Observed in MHC class 1 & rarely in class 2 peptide experiments. Particularly abundant in samples from the CPTAC Lung Adenocarcinoma Discovery Study & leukocyte enriched samples.
Tue May 04 12:03:41 +0000 2021>NCF2:p, neutrophil cytosolic factor 2 (Homo sapiens) Midsized protein; CTMs: S2+acetyl; PTMs: 11×S, 1×T, 0×Y+phosphoryl; 3×K+ubiquitinyl; 4×K+SUMOyl; SAAVs: K181R (50%), H389Q (2%); mature form: (2-546) [7,795×, 48 kTa]. #ᗕᕱᗒ 🔗
Mon May 03 19:53:29 +0000 2021@ypriverol @MattWFoster But I would strongly advocate for as many people as possible exploring SAAVs & setting up as many resources and methods for finding & recording them as possible.
Mon May 03 19:52:29 +0000 2021@ypriverol @MattWFoster Actually, it is. You just have to look it up using the reference sequence (or the dbSNP accession rs373488172) rather than the variant sequence,
🔗
Mon May 03 16:21:18 +0000 2021Even though the number of cases in the 3rd wave is high in Canada, the fatality numbers suggest that something is different this time (more effective treatment?) 🔗
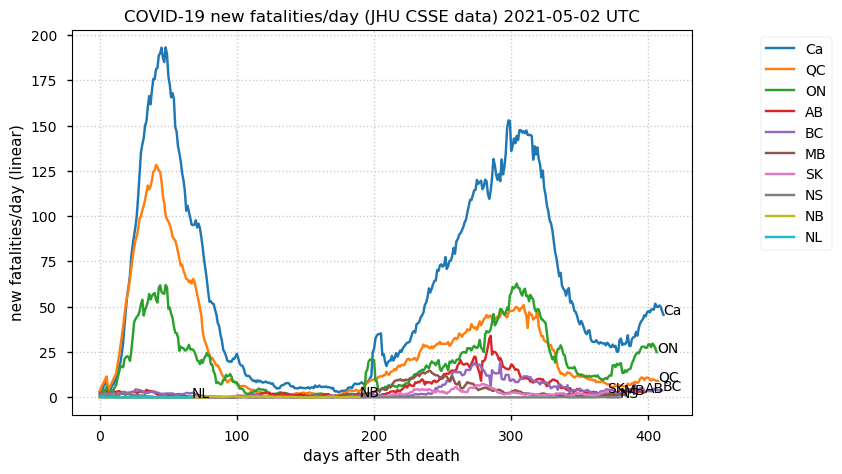
Mon May 03 14:04:23 +0000 2021@astacus Ha! More mouse urine (PXD018023). C57BL/6J mice this time.🐭
Mon May 03 11:44:35 +0000 2021NEO1:p.A288T chr 15:g.73126554G>A, rs192950211 (all tissue A:T 2145:1) vaf=0.002%, Δm=30.0106, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. #ᐯᐸᐱ
Mon May 03 11:44:35 +0000 2021NEO1:p, θ(max) = 50. aka NGN, HsT17534, IGDCC2, NTN1R2. Observed in MHC class 1 & 2 peptide experiments. Domains: extracellular glycodomain (36-1103), membrane spanning (1104 – 1126), intracellular phosphodomain (1127-1461).
Mon May 03 11:44:34 +0000 2021>NEO1:p, neogenin 1 (Homo sapiens) Large membrane protein; CTMs: N73, N210, N470, N489, N639+glycosyl; PTMs: 12×S, 4×T, 2×Y+phosphoryl; SAAVs: A288T (0.002%); mature form: (34,36,38-1461) [9,071×, 53 kTa]. #ᗕᕱᗒ 🔗
Sun May 02 15:14:25 +0000 2021For anyone interested, to see the complete list of the last 5 months worth of selected, easy-to-detect single amino acid variants, just click on the hash tag #ᐯᐸᐱ. If you want to look up a variant yourself, enter the rs number into SNAP and press "GO" 🔗
Sun May 02 13:56:00 +0000 2021It is a data set full of odd little facts about the relationships between the two classes of MHC peptides & how they are displayed in a tissue-specific manner.
Sun May 02 12:54:10 +0000 2021Ha! Just realized I hadn't updated the passive mode settings on the FTP site. Passive mode always gets me ...😡
Sun May 02 12:21:38 +0000 2021ASAH1:p.I93V chr 8:g.18069818T>C, rs1049874 (all tissue I:V 0.842:0.158) vaf=49%, Δm=-14.0156, VAF by population group: african 25%, american 57%, east asian 37%, european 48%, south asian 3%. #ᐯᐸᐱ
Sun May 02 12:21:38 +0000 2021ASAH1:p, the mature protein believed to be cleaved into alpha (22-242) & beta (243-395) subunits: there is no evidence supporting this idea in any proteomics data set.
Sun May 02 12:21:38 +0000 2021ASAH1:p, θ(max) = 80. aka AC, PHP32, FLJ21558, ACDase, ASAH. Observed in MHC class 1 & 2 peptide experiments. Commonly found in most solid tissues, urine & cell lines.
Sun May 02 12:21:37 +0000 2021>ASAH1:p, N-acylsphingosine amidohydrolase 1 (Homo sapiens) Small lysosomal enzyme; CTMs: N259, N286+glycosyl; PTMs: 1×S, 1×T, 3×Y+phosphoryl; 4×K+ubiqutinyl; 2×K+acetyl; SAAVs: I93V (49%), V246A (85%), V369I (1%); mature form: (22-395) [31,250×, 258 kTa]. #ᗕᕱᗒ 🔗
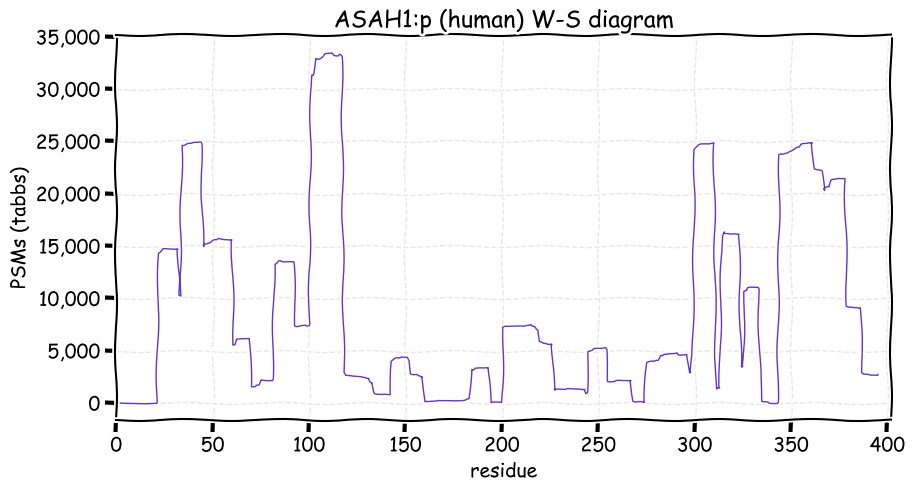
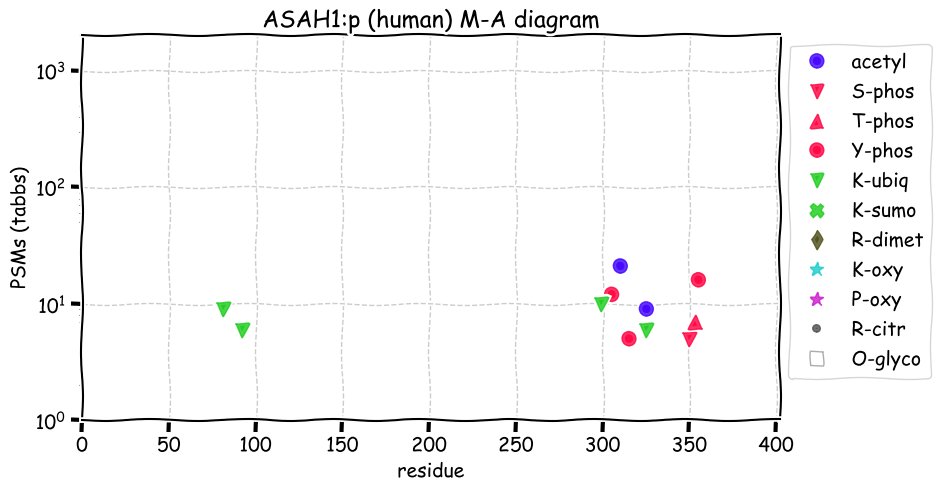
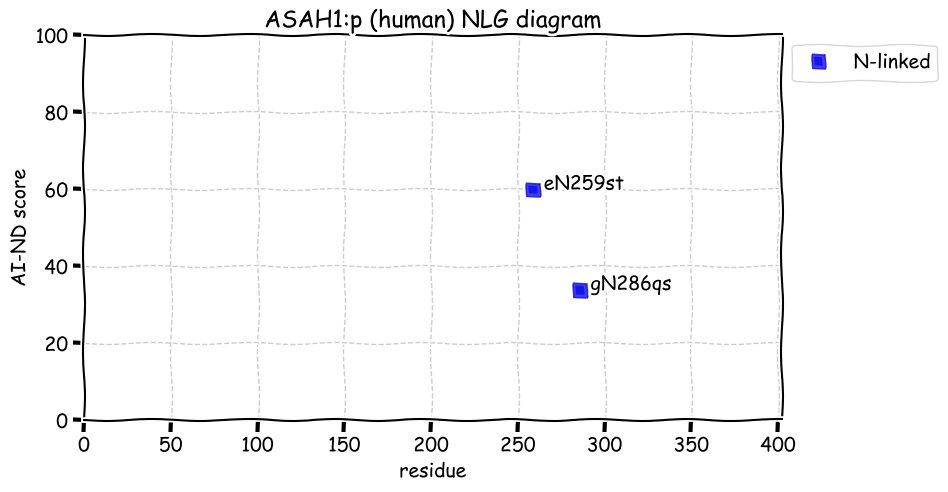
Sat May 01 21:29:09 +0000 2021If PXD019643 shows me anything, it would be that if you want lots of MHC class 1 or 2 peptides, skip every other tissue & go straight to for the thymus.
Sat May 01 16:24:12 +0000 2021@mjmaccoss @lkpino @MagnusPalmblad It is also easy to dismiss the lack of a PCR-equivalent in a sentence, but practically it is quite an thorny problem.
Sat May 01 16:20:38 +0000 2021At the moment: Ontario, Quebec, British Columbia & Saskatchewan have tentatively crested. Manitoba & especially Alberta both are still on the up-slope. Alberta currently has more reported cases/capita than Iran, Brazil or India (AB has 2.5× the current US rate). 🔗
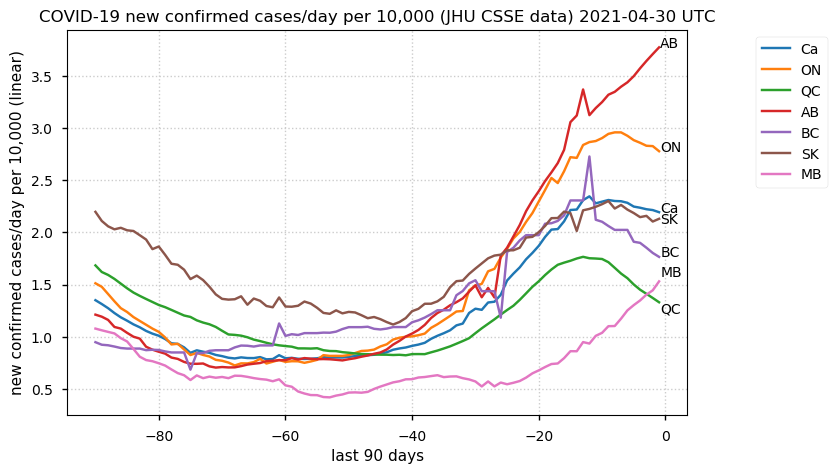
Sat May 01 16:00:08 +0000 2021Changed my external firewall, half of my external DNS settings & most of my internal IP addresses. Now I will wait & see to find out what I've screwed up ...
Sat May 01 12:26:23 +0000 2021@UCDProteomics It is a great data set. That group at U of Colorado has developed genuine expertise with human plasma methods (unlike most high profile groups that just phone it in).
Sat May 01 12:01:57 +0000 2021NCAN:p has a single, low occupancy phosphodomain (372-430), unlike the notionally related "lectican" family member versican (VCAN:p) which is boldly decorated with phosphorylation acceptor sites. 🔗
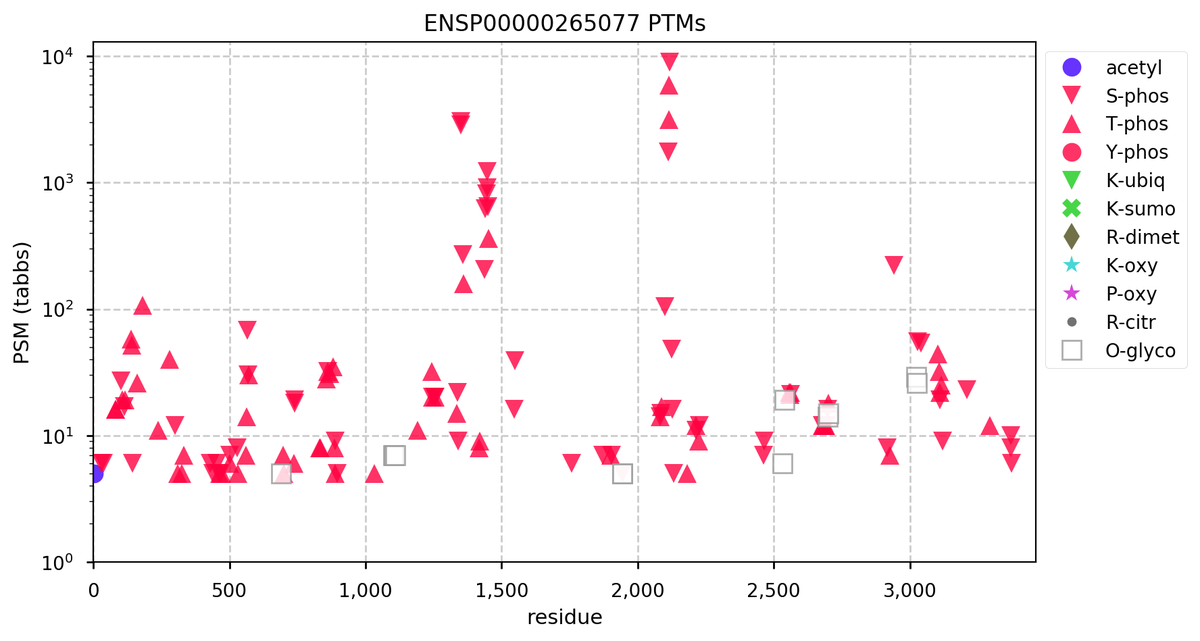
Sat May 01 11:52:49 +0000 2021NCAN:p.P92S chr 19:g.19219115C>A, rs2228603 (all tissue P:S 0.957:0.043) vaf=6%, Δm=-10.0207, VAF by population group: african 1%, american 2%, east asian 6%, european 7%, south asian 7%. #ᐯᐸᐱ
Sat May 01 11:52:49 +0000 2021NCAN:p, θ(max) = 45. aka CSPG3. Observed in MHC class 1 peptide experiments from brain tissue only. Commonly found in brain tissue & CSF. Has several difficult-to-observe domains because of a lack of tryptic cleavage sites. Shares tryptic peptides with BCAN:p & STAB1:p.
Sat May 01 11:52:48 +0000 2021>NCAN:p, neurocan (Homo sapiens) Large extracellular protein; CTMs: none; PTMs: 7×S, 3×T, 0×Y+phosphoryl; 3×T+glycosyl; SAAVs: P92S (4%); mature form: (22,23,24,25-1321) [3,619×, 54 kTa]. #ᗕᕱᗒ 🔗
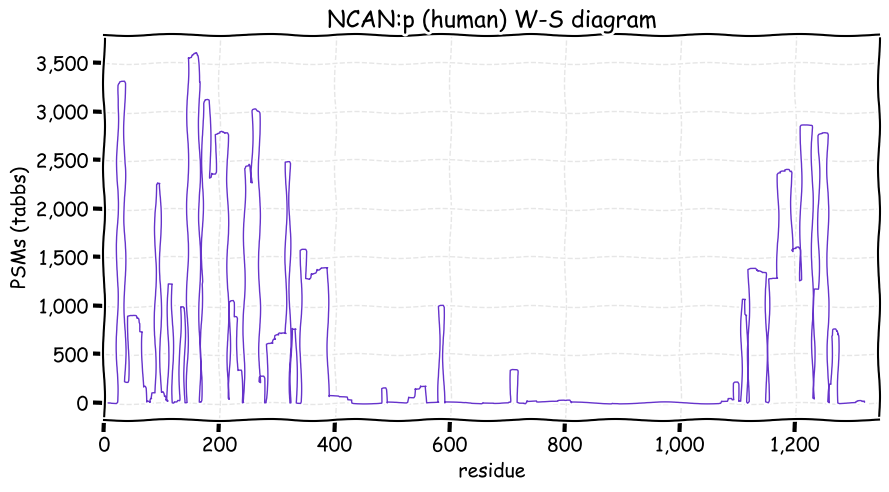
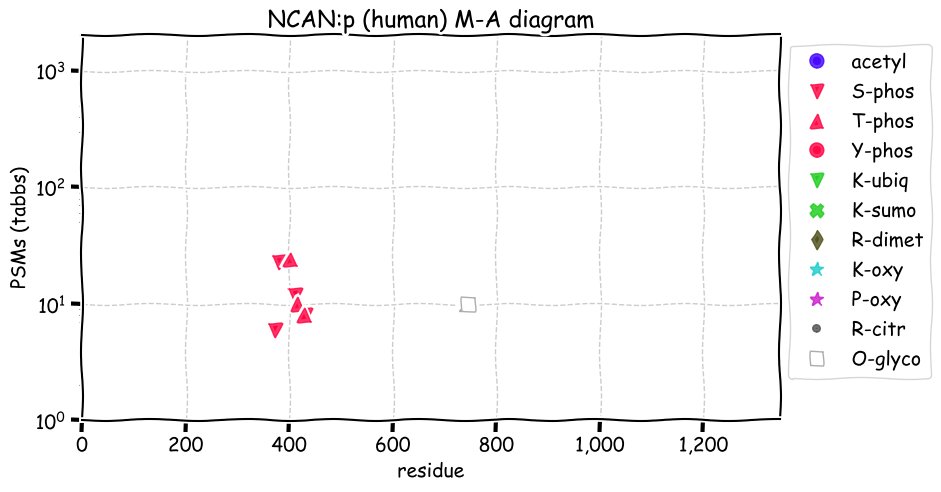
Fri Apr 30 14:56:55 +0000 2021@michaelhoffman @DrBrocktagon @NatureIndex @JohnRInglis @jessicapolka @HindleSamantha @dasaptaerwin Overthinking the problem seems to be the common theme of their suggestions.
Fri Apr 30 14:51:05 +0000 2021@JGMProteomics @CNPN_2021 It is an encouraging list of candidates.
Fri Apr 30 12:13:30 +0000 2021LYRM1, LYRM2 & LYRM4 all have similar PTM patterns to LYRM5: N-terminal acetylation, a few low occupancy K+acetyl acceptors and no S/T phosphorylation. NDUFB9 (which was LYRM3) is completely different.
Fri Apr 30 12:03:23 +0000 2021LYRM4:p.S11A chr 6:g.5260703A>C, rs2224391 (all tissue S:A 0.668:0.332) vaf=29%, Δm=-15.9949, VAF by population group: african 84%, american 33%, east asian 26%, european 25%, south asian 17%. #ᐯᐸᐱ
Fri Apr 30 12:03:23 +0000 2021LYRM4:p, θ(max) = 92. aka CGI-203, ISD11, C6orf149. Observed in MHC class 1 & 2 peptide experiments. Commonly found in solid tissue & many cell lines.
Fri Apr 30 12:03:23 +0000 2021>LYRM4:p, LYR motif containing 4 (Homo sapiens) Very small intracellular protein; CTMs: A2+acetyl; PTMs: K58+acetyl; SAAVs: S11A (29%); mature form: (2-91) [5,975×, 16 kTa]. #ᗕᕱᗒ 🔗
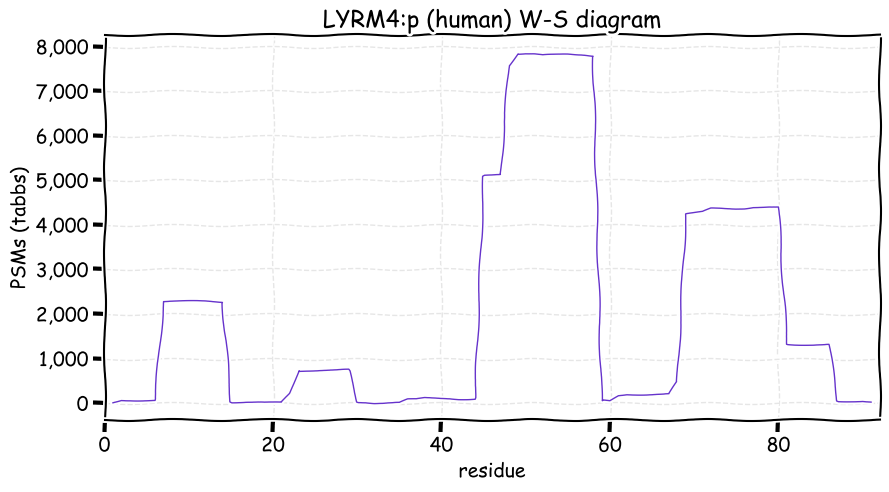
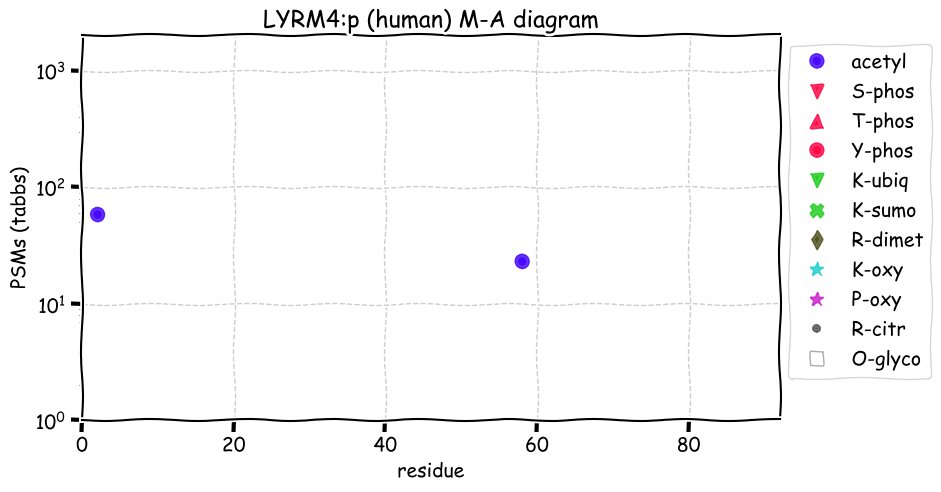
Thu Apr 29 19:32:56 +0000 2021Alberta seems to be taking the lead among the provinces here in Canada, while Manitoba & Quebec are just trading positions. Saskatchewan is hugging very close to the country-wide average right now, after leading the league for a months. 🔗
Thu Apr 29 12:28:11 +0000 2021Of the proteins named as "matrix remodeling associated X", only X = 5, 7 & 8 retain this nomenclature. None of these proteins were related by protein or genetic similarities: it is a functional designation.
Thu Apr 29 12:11:42 +0000 2021MXRA5:p has a similar pattern of posttranslational modification as versican (VCAN:p), another large ECM protein. Does anyone know which kinases are involved?
Thu Apr 29 12:04:19 +0000 2021MXRA5:p.G2000S chr X:g.3317683C>T, rs1635242 (all tissue G:S 0.590:0.410) vaf=65%, Δm=30.0106, VAF by population group: african 71%, american 67%, east asian 74%, european 65%, south asian 65%. #ᐯᐸᐱ
Thu Apr 29 12:04:19 +0000 2021MXRA5:p, θ(max) = 40. aka DKFZp564I1922. Observed in MHC class 1 & 2 peptide experiments. May be abundant in urine, but most commonly found in solid tissue. Rarely observed in commonly used cell lines
Thu Apr 29 12:04:18 +0000 2021>MXRA5:p, matrix remodeling associated 5 (H sapiens) ECM protein; CTMs: 11×N+glycosyl; PTMs: 39×S,31×T,0×Y+phosphoryl; SAAVs: T1094I (5%), A1128V (48%), A1351V (3%), G1394D (72%), G2000S (65%), L2531V (57%), E2700D (22%); mature form: (27-2828) [8,178×, 72 kTa]. #ᗕᕱᗒ 🔗
Wed Apr 28 22:16:09 +0000 2021@astacus @slavov_n If the acronym ends up in the dialogue of a movie, a sitcom or an X-Files-like TV series, sure.
Wed Apr 28 21:27:56 +0000 2021@astacus @slavov_n I'd say it has to have at least one passage that enters popular culture, like Schrödinger's paper with a joke involving a cat.
Wed Apr 28 21:07:41 +0000 2021@PastelBio The Homoiousian vs. Homoousian of our time ..
Wed Apr 28 12:40:31 +0000 2021It makes me nervous when someone describes their software interface with some phrase that sounds like "very flexible": almost always means "hellishly complicated".
Wed Apr 28 12:30:27 +0000 2021@NMechawar @MHendr1cks We are Canadian: "Network of Centres of Excellence"
Wed Apr 28 12:07:47 +0000 2021SAA1:p.T77S chr 11:g.18269333C>G, rs1671926 (all tissue T:S 0.017:0.983) vaf=99%, Δm=-14.01565, VAF by population group: african 98%, american 100%, east asian 100%, european 100%, south asian 100%. UniProt lists this variant as the reference allele. #ᐯᐸᐱ
Wed Apr 28 12:07:47 +0000 2021SAA1:p, SAA2:p & SAA4:p all appear to have missed the memo requiring them to place PTMs willy-nilly along their sequences.
Wed Apr 28 12:07:47 +0000 2021SAA1:p, θ(max) = 85. aka PIG4, TP53I4, SAA. Observed in MHC class 1 & 2 peptide experiments. Abundant in blood plasma & CSF but not in urine. Can be present in MCF-10A cell lines.
Wed Apr 28 12:07:47 +0000 2021>SAA1:p, serum amyloid A1 (Homo sapiens) Small extracellular protein; CTMs: none; PTMs: none; SAAVs: V51M (1%), T77S (99%), K108R (1%), K121R (4%); mature form: (19,20-122) [9,725×, 72 kTa]. #ᗕᕱᗒ 🔗
Tue Apr 27 17:56:27 +0000 2021@byu_sam @theNCI Does the interview really take 60 minutes?
Tue Apr 27 15:54:56 +0000 2021The new firewall settings seem to be doing a good job of blocking 🔗 crawlers.
Tue Apr 27 15:17:28 +0000 2021I get that India is having difficulty dealing with the rise in its current COVID-19 case load, but why do news media keep on adding references to Pakistan into the same story? 🔗
Tue Apr 27 15:04:42 +0000 2021While I do question the value of this type of project (🔗), if they can get people to stop talking about "proteosomal peptide splicing" as though it really exists, I may have to change my opinion.
Tue Apr 27 13:02:11 +0000 2021Web crawlers/spiders/every-dopey-goof-who-doesn't-want-to-do-anything-other-than-create-some-silly-list/index are the bane of my existence
Tue Apr 27 12:57:34 +0000 2021Why on earth is 🔗 ignoring a robots.txt file & bombing me with junk requests?
Tue Apr 27 12:49:55 +0000 2021SERPINF1:p.T72M chr 17:g.1769982C>T, rs1136287 (all tissue T:M 0.657:0.343) vaf=61%, Δm=29.9928, VAF by population group: african 88%, american 59%, east asian 49%, european 65%, south asian 43%. #ᐯᐸᐱ
Tue Apr 27 12:49:55 +0000 2021SERPINF1:p, θ(max) = 44. aka EPC-1, PIG35, PEDF. Observed in MHC class 1 & 2 peptide experiments. Abundant in most fluids: urine, blood plasma or CSF. Observable in some cell lines.
Tue Apr 27 12:49:55 +0000 2021>SERPINF1:p, serpin family F member 1 (Homo sapiens) Small protease inhibitor; CTMs: N285+glycosyl; PTMs: 31×S, 3×T, 0×Y+phosphoryl, 3×K+acetyl; SAAVs: V51M (1%), T72M (61%); mature form: (20-418) [29,150×, 407 kTa]. #ᗕᕱᗒ 🔗
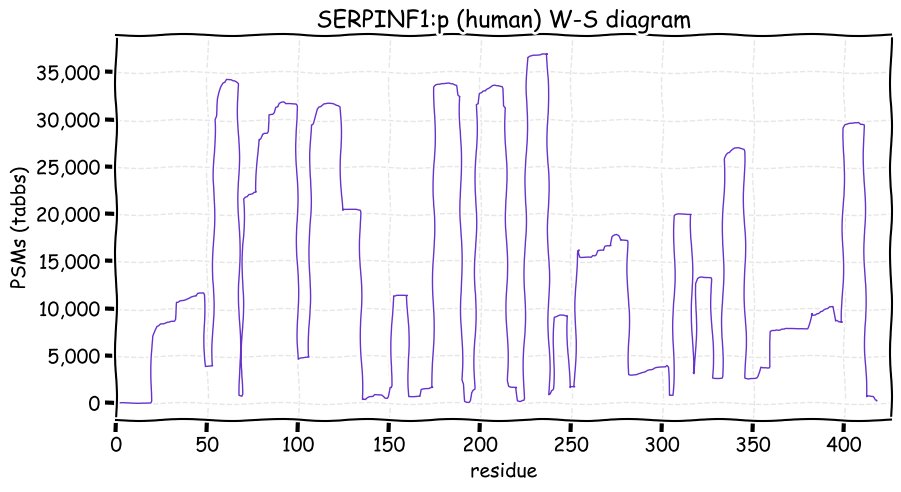
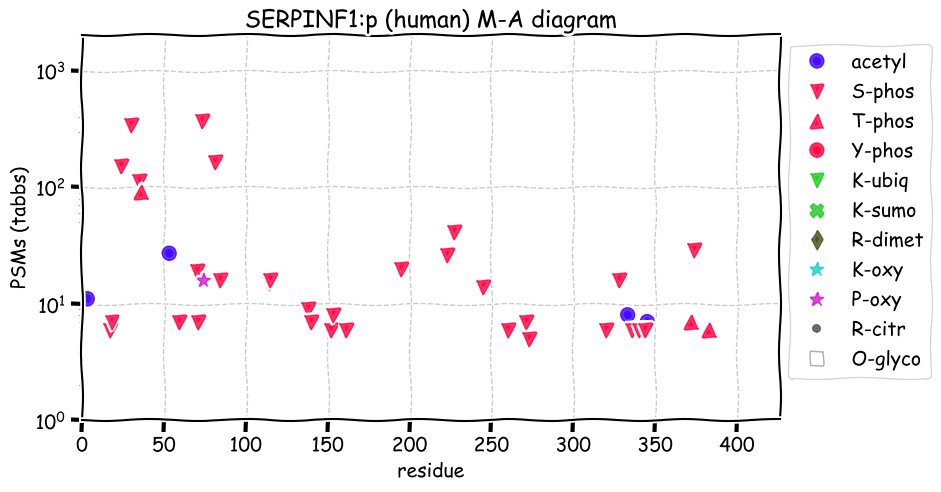
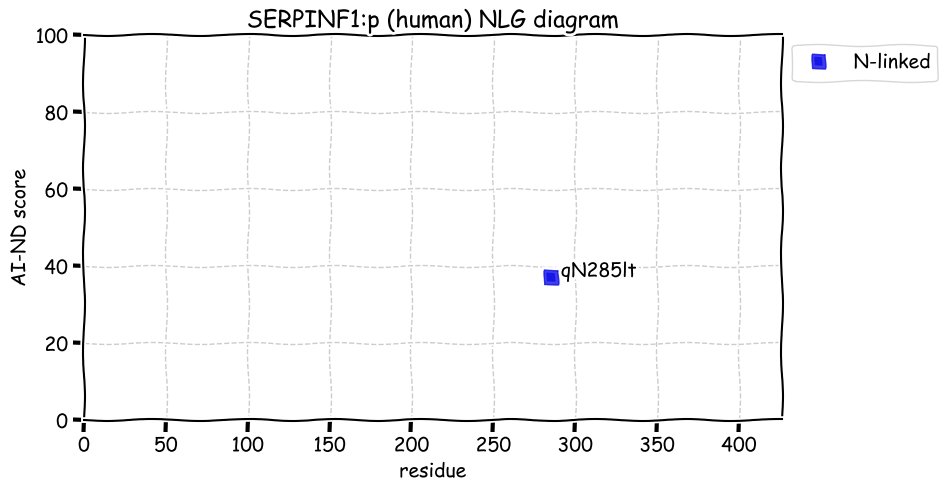
Mon Apr 26 17:11:53 +0000 2021VCAN also has a very different phosphorylation pattern than other members of the lectican family, e.g. NCAN
🔗
Mon Apr 26 16:52:43 +0000 2021VCAN:p, it is unusual for a protein to have about the same number of S+phosphorylation acceptor sites as T+phosphorylation sites
Mon Apr 26 16:46:11 +0000 2021Why do so many people believe e-mail is like instant messaging? E-mail gets there when it gets there (or not): it does not magically arrive at its destination immediately upon pressing "send".
Mon Apr 26 16:42:32 +0000 2021@andy___jones @pwilmarth @theoneamit So it sounds like a real problem with the algorithm/software implementation, rather than anything to do with the specifics of the additional modifications. Collagen must make it go nuts!
Mon Apr 26 14:23:56 +0000 2021@andy___jones @pwilmarth @theoneamit What sort of samples are you using where you suspect an A1 vs. deamidation problem?
Mon Apr 26 12:45:31 +0000 2021Viva Mexico! 🔗
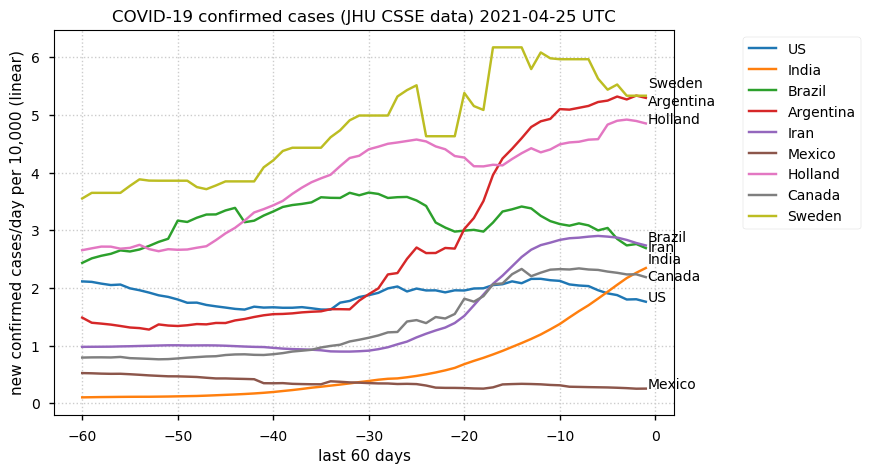
Mon Apr 26 12:11:55 +0000 2021VCAN:p.D2937Y chr 5:g.83541812G>T, rs160277 (all tissue D:Y 0.686:0.314) vaf=38%, Δm=48.0364, VAF by population group: african 36%, american 46%, east asian 32%, european 43%, south asian 27%. #ᐯᐸᐱ
Mon Apr 26 12:11:55 +0000 2021VCAN:p, θ(max) = 44. aka PG-M, CSPG2. Observed in MHC class 1 & 2 peptide experiments. Abundant in most solid tissues & fluids.
Mon Apr 26 12:11:55 +0000 2021>VCAN:p, versican (Homo sapiens) Large subunit; CTMs: N330, N1442, N1663, N1898, N2360, N3369+glycosyl; PTMs: 66×S, 63×T, 0×Y+phosphoryl, 5×S, 7×T+glycosyl; SAAVs: S300L (7%), G428D (21%), F2301Y (47%), D2937Y (38%); mature form: (21-3396) [21,405×, 362 kTa]. #ᗕᕱᗒ 🔗
Sun Apr 25 18:15:06 +0000 2021Does normalizing to population help, hurt or mean anything at all? 🔗
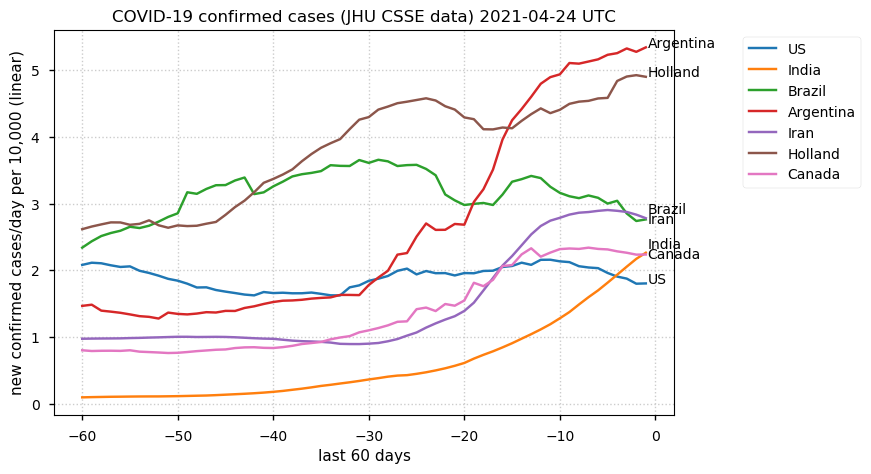
Sun Apr 25 15:14:53 +0000 2021But to listen to the network news in both countries, it sounds as though either:
US) things are looking up, or
CA) "catastrophe!!!"
Sun Apr 25 14:48:05 +0000 2021As of yesterday's numbers, the rate of new COVID-19 infections in Canada and the US were nearly equal, about 2 new cases per 10,000 people per day.
Sun Apr 25 12:52:26 +0000 2021UQCRFS1:p, it is unclear what happens to the domain (1-78) following the cleavage of 78-79, as tryptic peptides from the N-terminal domain are commonly observed.
Sun Apr 25 12:32:40 +0000 2021UQCRFS1:p is one of the few human proteins that still has the name of an individual ('Rieske') incorporated into the official HGNC descriptive gene name and symbol.
Sun Apr 25 12:18:23 +0000 2021UQCRFS1:p, further proteolytic processing occurs to form the observed complex III form of the protein (79-274), but it is not the conventional removal of a mitochondrial targeting peptide.
Sun Apr 25 12:06:44 +0000 2021UQCRFS1:p.S6A chr 19:g.29213103A>C, rs8100724 (all tissue S:A 280:2255) vaf=93%, Δm=-15.9949, VAF by population group: african 94%, american 93%, european 92%, east asian 100%, south asian 98%. #ᐯᐸᐱ
Sun Apr 25 12:06:44 +0000 2021UQCRFS1:p, θ(max) = 84. aka RIS1, RIP1, UQCR5, RISP. Observed in MHC class 1 & 2 peptide experiments. Abundant in most solid tissues & cell lines.
Sun Apr 25 12:06:43 +0000 2021>UQCRFS1:p, ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 4×S, 2×T, 1×Y+phosphoryl, 6×K+acetyl, 3×K+ubiquitinyl; SAAVs: S6A (93%); mature form: (1-274) [42,423×, 268 kTa]. #ᗕᕱᗒ 🔗
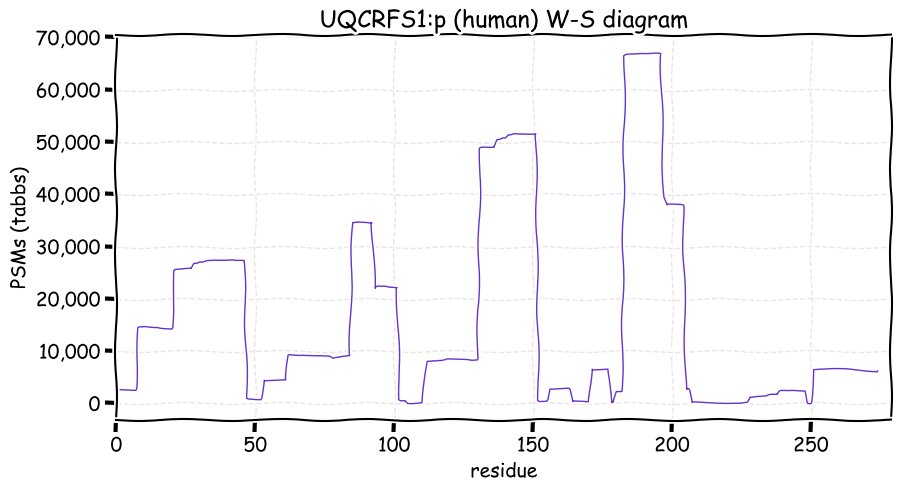
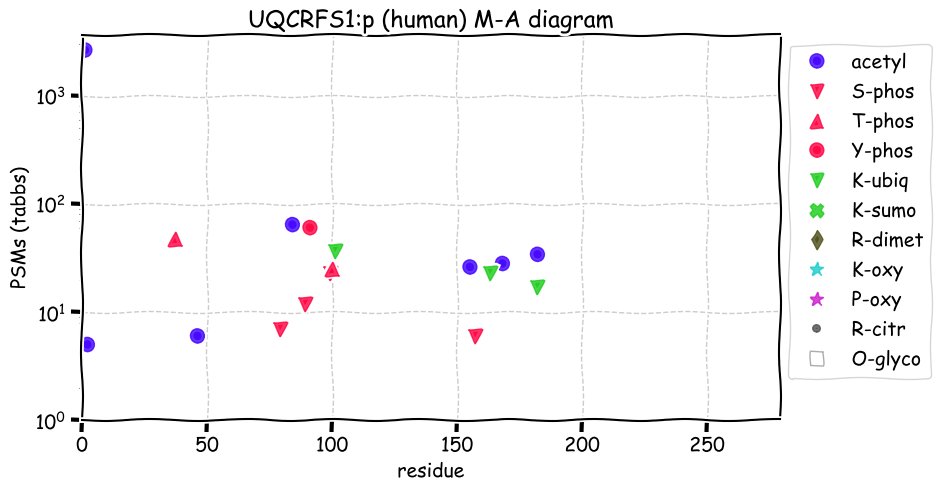
Sat Apr 24 17:42:32 +0000 2021ADH5:p is the only member of this family that show a significantly different pattern of modification
🔗
Sat Apr 24 15:18:59 +0000 2021India 🔗
Sat Apr 24 14:33:36 +0000 2021ADH4:p Note: this protein shares some tryptic peptides with ADH1A:p, ADH1B:p, ADH1C:p, ADH5:p & ADH6:p.
Sat Apr 24 11:56:35 +0000 2021ADH4:p.V374I chr 4:g.99124465C>T, rs1126673 (all tissue V:I 37:73) vaf=85%, Δm=14.01565, VAF by population group: african 86%, american 77%, european 70%, east asian 100%, south asian 88%. #ᐯᐸᐱ
Sat Apr 24 11:56:35 +0000 2021ADH4:p, θ(max) = 92. aka ADH-2, all-trans-retinol dehydrogenase [NAD(+)]. Observed in MHC class 1 & 2 peptide experiments. Most prominent in liver & Hep-G2 cell lines.
Sat Apr 24 11:56:34 +0000 2021>ADH4:p, alcohol dehydrogenase 4 (class II), pi polypeptide (Homo sapiens) Small subunit; CTMs: G2+acetyl; PTMs: 5×S, 2×T, 0×Y+phosphoryl; SAAVs: I309V (75%), V374I (85%); mature form: (2-380) [5,561×, 159 kTa]. #ᗕᕱᗒ 🔗
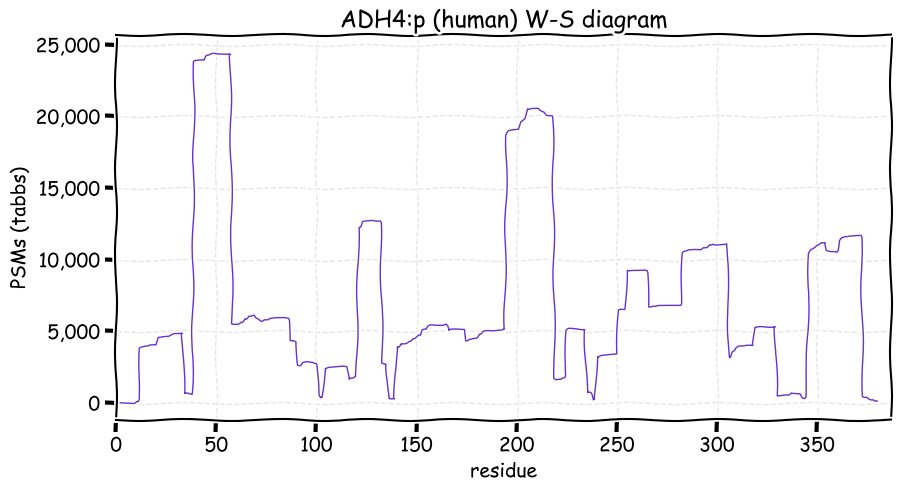
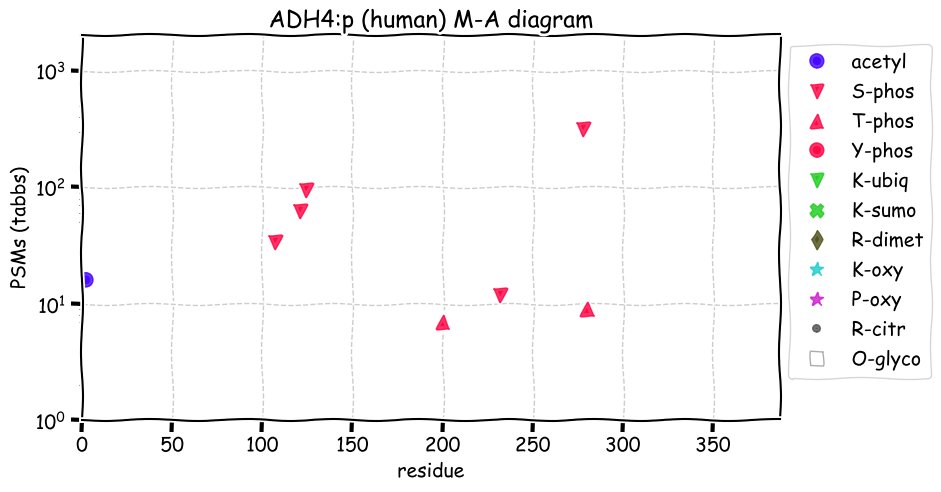
Fri Apr 23 15:34:41 +0000 2021@jwoodgett Yup, I'm sure that Mr. Ford is going to start making good public health decisions any day now. And a lot of teaching is done by adjuncts and TA's who, as a group, tend to be younger than LTRI faculty ...
Fri Apr 23 15:28:04 +0000 2021@AlexUsherHESA Don't know for sure about TO, but here in W'peg it is used to distinguish between office workers+condo dwellers and the poor (core) that live downtown.
Fri Apr 23 14:37:58 +0000 2021@dphannah @doctorow I particularly agree with ditching word processors. I code quite a bit so I use simple text editors all the time, but it took me an embarrassingly long time to realize that word processors are just a collection of bad marketing ideas wrapped up in annoying, scolding bloatware.
Fri Apr 23 14:23:12 +0000 2021@jwoodgett In Canuckistan, most students & many faculty are not old enough to get vaccinated. Hopefully that will change by the fall, but there is no guarantee.
Fri Apr 23 12:55:34 +0000 2021Although you would never know it from the SwissProt entry 🔗
Fri Apr 23 12:08:30 +0000 2021PDCD6IP:p, lysine acetylation seems to be doing something important wrt to this molecule's function, although just what is going on is a puzzler for me.
Fri Apr 23 12:04:51 +0000 2021PDCD6IP:p.S730L chr 3:g.33864074C>T, rs1127732 (all tissue S:L 0.802:0.198) vaf=23%, Δm=26.0520, VAF by population group: african 16%, american 31%, european 13%, east asian 39%, south asian 30%. #ᐯᐸᐱ
Fri Apr 23 12:04:51 +0000 2021PDCD6IP:p, θ(max) = 79. aka Alix, AIP1, Hp95. Observed in MHC class 1 & 2 peptide experiments. Prominent in urine as well as many tissues & cell lines. The K+ubiquitinyl sites are also K+acetyl sites. The P-rich domain (746-868) has no tryptic cleavage sites.
Fri Apr 23 12:04:50 +0000 2021>PDCD6IP:p, programmed cell death 6 interacting protein (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 57xK+acetyl; 27xK+ubiquitinyl; 35×S, 27×T, 15×Y+phosphoryl; SAAVs: V378I (29%), A309T (67%), S730L (23%); mature form: (2-868) [77,626×, 971 kTa]. #ᗕᕱᗒ 🔗
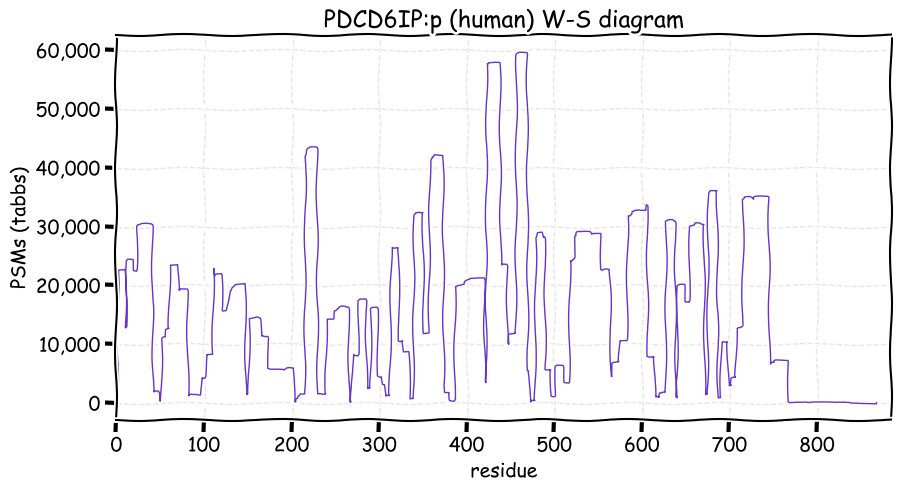
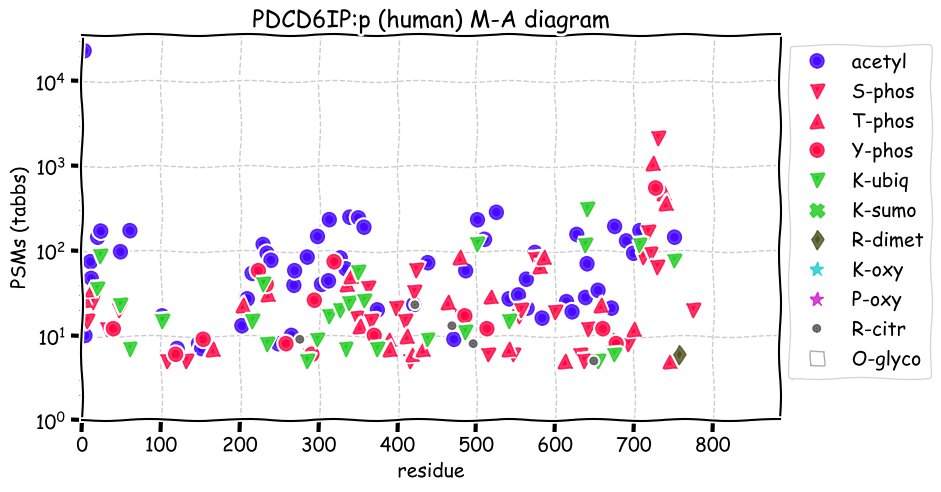
Thu Apr 22 17:36:40 +0000 2021@MattWFoster The data looks like unfractionated human plasma & the species listed is human, but that oddball description makes me more than a little nervous about using the data.
Thu Apr 22 17:21:56 +0000 2021@MattWFoster Is that commonly used to describe human embryo manipulations?
Thu Apr 22 16:53:57 +0000 2021Can anyone explain to me how the description in 🔗 makes any sense for what sounds to me like a human embryo secretome?
Thu Apr 22 14:12:38 +0000 2021@michaelhoffman @deevybee @dgmacarthur They are as close to a monograph as you can get at the moment.
Thu Apr 22 14:12:02 +0000 2021@michaelhoffman @deevybee @dgmacarthur You usually do it as a favor for a friend who agreed to edit the book. A lot of tenured faculty don't run active research labs but often have a better grasp of a field as a whole, which is perfect for book chapters.
Thu Apr 22 14:09:47 +0000 2021& you really should show something at least equivalent to this histogram if you want to prove that your sample prep worked properly for either class of peptide.
Thu Apr 22 14:04:49 +0000 2021@michaelhoffman *Unless you already have tenure.
Thu Apr 22 13:56:43 +0000 2021PXD019643 shows that even at scale, it is possible to do really good MHC class 1 or 2 sample preps. The peptide lengths observed look nearly theoretical. 🧐 These two distributions are from individual RAW files in their "adrenal gland" set of expts. 🔗
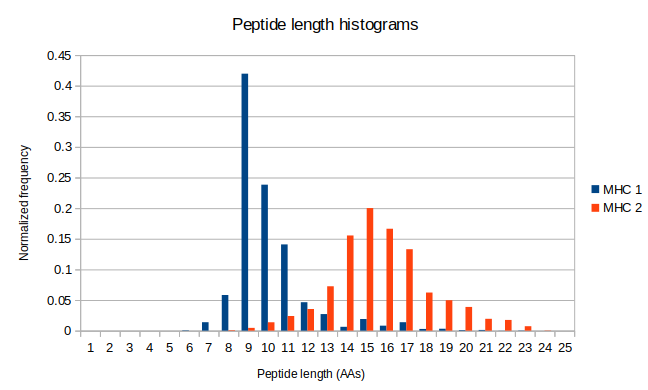
Thu Apr 22 11:49:59 +0000 2021SORCS3:p.G99S chr 10:g.104641622G>A, rs117144385 (all tissue G:S 0.979:0.021) vaf=8%, Δm=30.0106, VAF by population group: african 8%, american 15%, european 8%, east asian 1%, south asian 4%. #ᐯᐸᐱ
Thu Apr 22 11:49:59 +0000 2021SORCS3:p, θ(max) = 22. aka KIAA1059, SORCS. Observed in MHC class 1 peptide experiments. Most prominent in CSF & brain: very rarely observed in cell lines. Transmembrane domain (1126-1146).
Thu Apr 22 11:49:59 +0000 2021>SORCS3:p, sortilin related VPS10 domain containing receptor 3 (Homo sapiens) Large subunit; CTMs: none; PTMs: S1212+phosphoryl; SAAVs: G99S (8%); mature form: (34-1222) [990×, 3 kTa]. #ᗕᕱᗒ 🔗
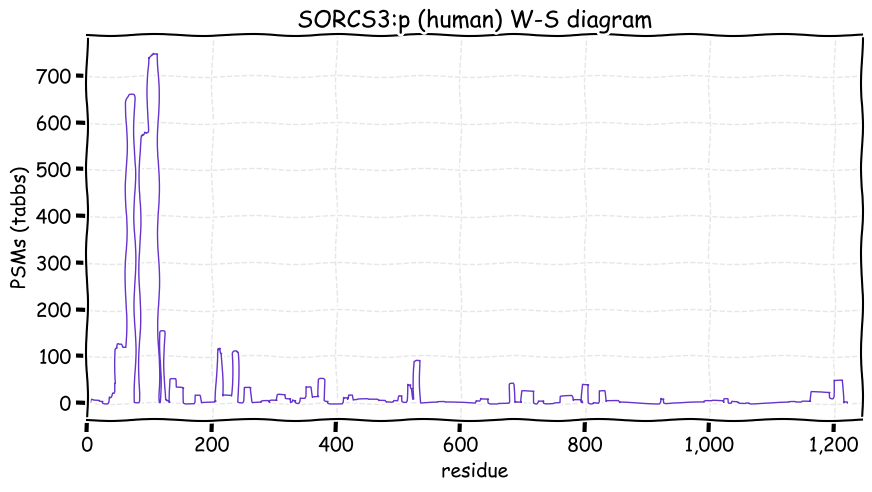
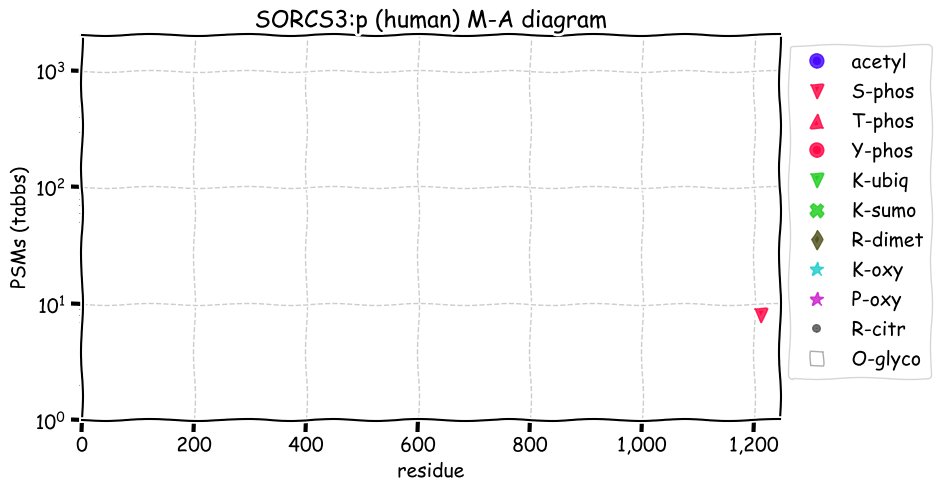
Wed Apr 21 11:45:49 +0000 2021PGLYRP2:p.R394Q chr 19:g.15472052C>T, rs34440547, (all tissue R:Q 0.767:0.233) vaf=22%, Δm=-28.0425, VAF by population group: african 16%, american 24%, european 22%, east asian 30%, south asian 23%. #ᐯᐸᐱ
Wed Apr 21 11:45:48 +0000 2021θ(max) = 65. aka PGRP-L, PGLYRPL, TAGL-like, tagL, tagL-alpha, tagl-beta, PGRPL. Observed in MHC class 2 peptide experiments. Most prominent in blood plasma, urine & CSF: rarely observed in cell lines.
Wed Apr 21 11:45:48 +0000 2021>PGLYRP2:p, peptidoglycan recognition protein 2 (H sapiens) CTMs: N77, N367, N485+glycosyl; PTMs: S177+phosphoryl; S177, T181+glycosyl; SAAVs: T46A (65%), R99Q (35%), G234V (1%), M270K (37%), R394Q (22%), H464Y (2%); mature form: (22,23-634) [19,239×, 262 kTa] #ᗕᕱᗒ 🔗
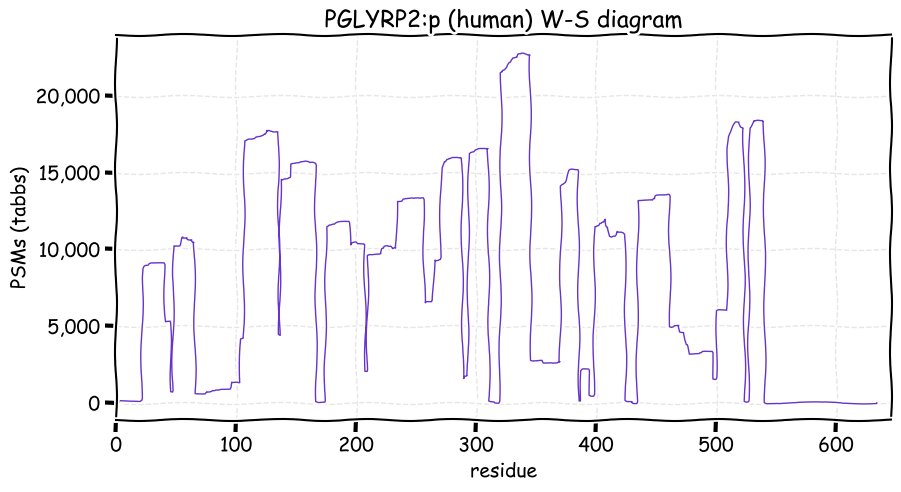
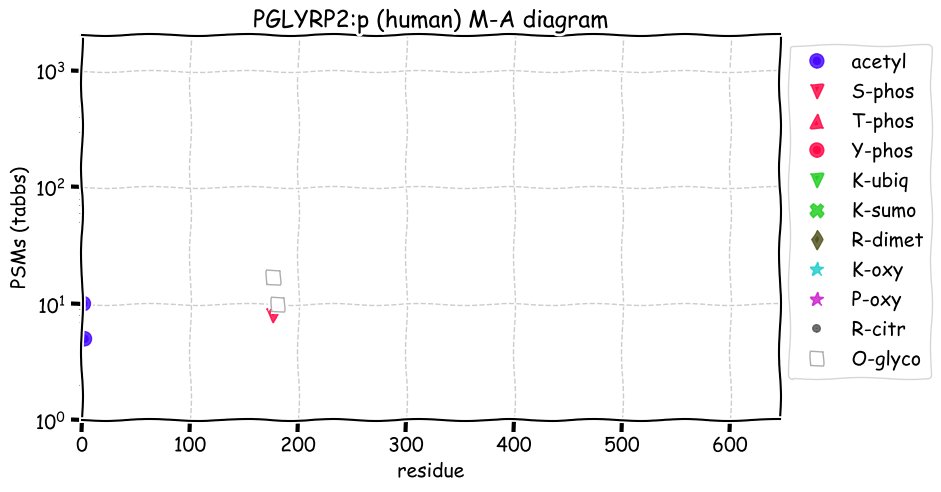
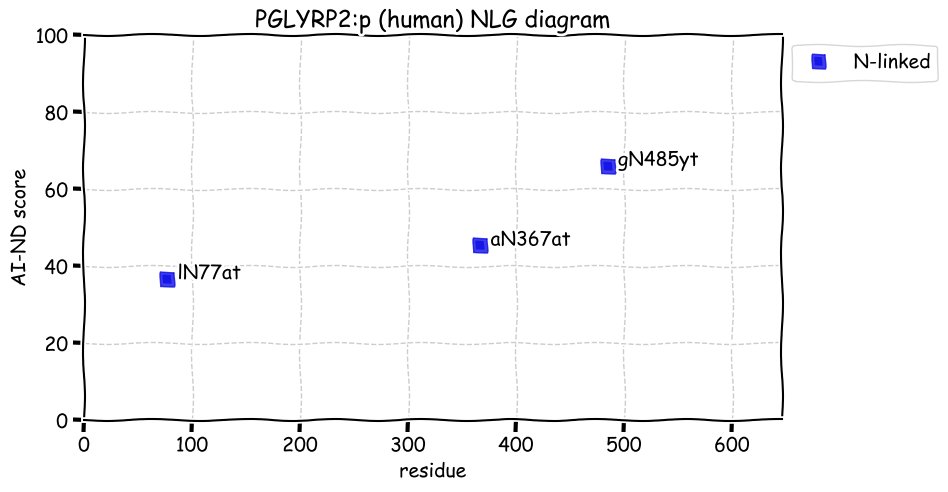
Tue Apr 20 19:41:17 +0000 2021In the spirit of #420, I give you CNR1's observed PTMs
🔗
Tue Apr 20 15:00:08 +0000 2021@jwoodgett @MHendr1cks @JimJohnsonSci I think at the moment, GC spends significant money in the riding of only 1 cabinet minister (Finance). How does that make any sense at all in terms of political sustainability?
Tue Apr 20 14:43:17 +0000 2021@jwoodgett @MHendr1cks @JimJohnsonSci The current Canadian science funding regime has shown no interest in funding outside of a very limited geographical footprint and until they do they can expect no significant increase in their base allotments.
Tue Apr 20 14:38:11 +0000 2021@jwoodgett @MHendr1cks @JimJohnsonSci Probably. Genome Canada sends almost all of its money to 3 or 4 postal codes at the moment, which is no way to win a vote in caucus.
Tue Apr 20 14:29:20 +0000 2021@MHendr1cks @jwoodgett @JimJohnsonSci Originally Genome Canada was supposed to do this sort of thing, but once it became focused on service platforms that sort of distributive function died out.
Tue Apr 20 14:27:58 +0000 2021@MHendr1cks @jwoodgett @JimJohnsonSci Yes. That initiative spends a lot of its money in ridings the Tricouncils do not supply with any funding, making it popular with a wide range of caucus members.
Tue Apr 20 14:20:52 +0000 2021@MHendr1cks @jwoodgett @JimJohnsonSci Bespoke programs do have the political advantage that they can be guaranteed to be placed in electoral districts that the Tricouncil approach does not (& never will) fund at any meaningful level.
Tue Apr 20 13:03:58 +0000 2021Note: the N-terminus of the mature protein corresponds to both the removal of an ER signal sequence and an alternate translation initiation site.
Tue Apr 20 12:56:09 +0000 2021FETUB:p.K360R chr 3:g.186652561A>G, rs7999, (blood plasma K:R = 2007:173 8%) vaf=5%, Δm=28.0061, VAF by population group: african 12%, american 6%, european 4%, east asian 8%, south asian 4%. #ᐯᐸᐱ
Tue Apr 20 12:56:09 +0000 2021FETUB:p, θ(max) = 68. Observed in MHC class 1 & 2 peptide experiments. Most prominent blood plasma & CSF: much more rarely observed in urine.
Tue Apr 20 12:56:09 +0000 2021>FETUB:p, fetuin B (Homo sapiens) Small subunit; CTMs: N37, N136+glycosyl; PTMs: 1×S, 0×T, 0×Y+phosphoryl; 2×S, 2×T+glycosyl; SAAVs: S33P (1%), R128G (2%), G202S (6%), K360R (5%); mature form: (19,20-382) [7,596×, 53 kTa]. #ᗕᕱᗒ 🔗
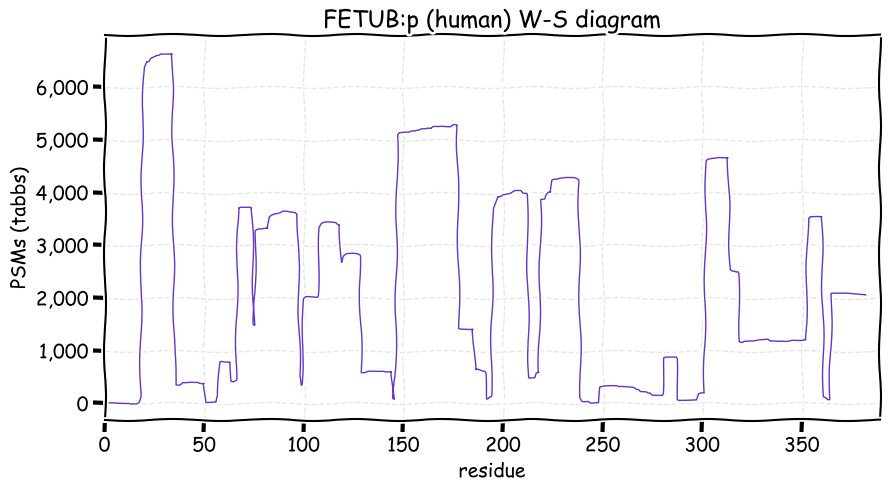
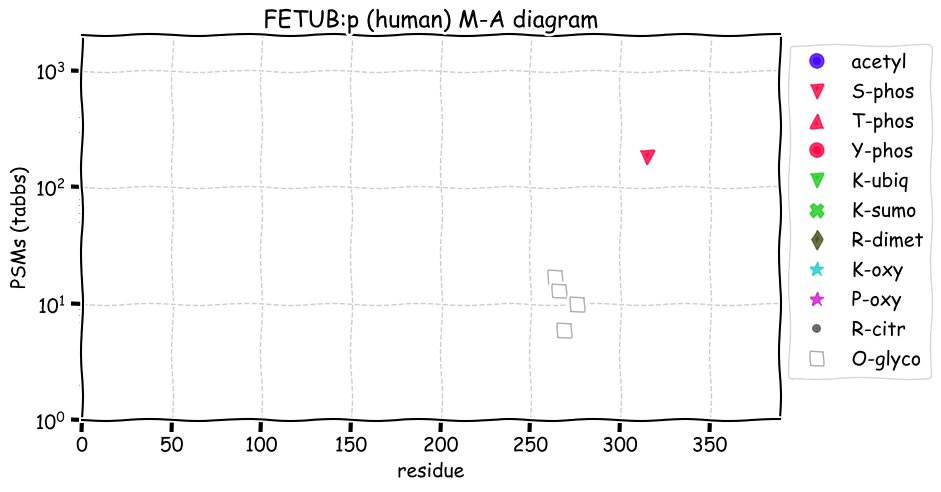
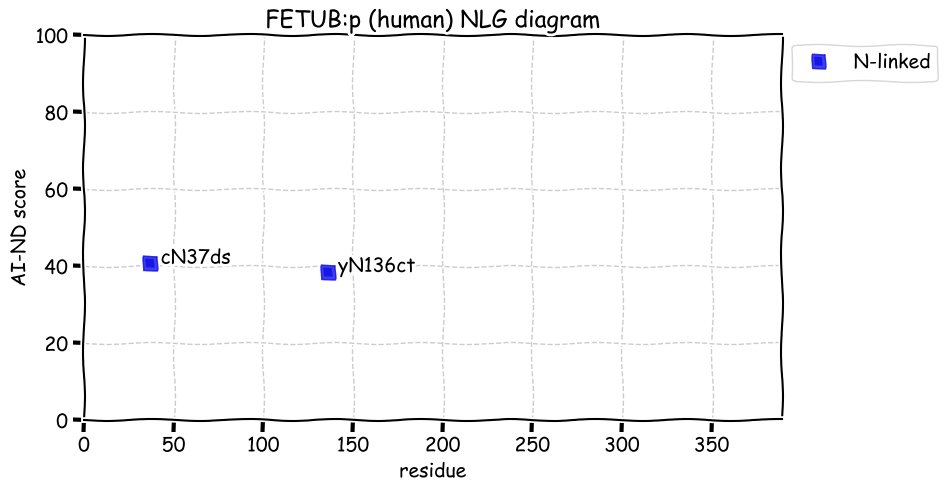
Mon Apr 19 11:52:43 +0000 2021IL6ST:p.L397V chr 5:g.55956103G>C, rs2228043, (cerebrospinal fluid L:V = 286:25 9%) vaf=12%, Δm=-14.0156, VAF by population group: african 38%, american 10%, european 11%, east asian 3%, south asian 8%. #ᐯᐸᐱ
Mon Apr 19 11:52:43 +0000 2021IL6ST:p, Transmembrane domain (620-641). The phosphodomain (659-905) is intracellular & the N-link glycosylation domain (131-390) is extracellular.
Mon Apr 19 11:52:42 +0000 2021IL6ST:p, θ(max) = 50. aka GP130, CD130, sGP130. Observed in MHC class 1 & 2 peptide experiments. Most prominent urine, CSF & blood plasma. Observed in many samples from cell lines, e.g., HeLa & MCF-10A.
Mon Apr 19 11:52:42 +0000 2021>IL6ST:p, interleukin 6 signal transducer (Homo sapiens) Midsized subunit; CTMs: N131, N157, N379, N383, N390+glycosyl; PTMs: 20×S, 7×T, 4×Y+phosphoryl; 3×K+ubiquitinyl; SAAVs: L397V (12%), V499I (1%); mature form: (23,34-918) [8,846×, 38 kTa]. #ᗕᕱᗒ 🔗
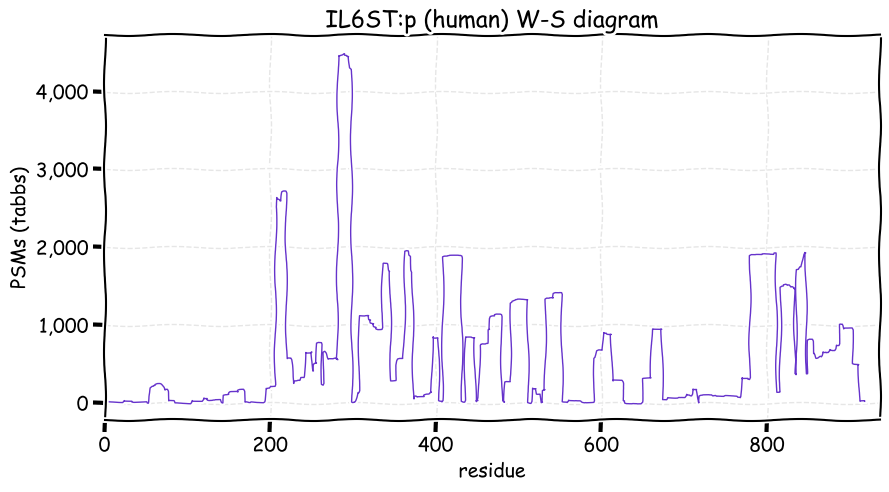
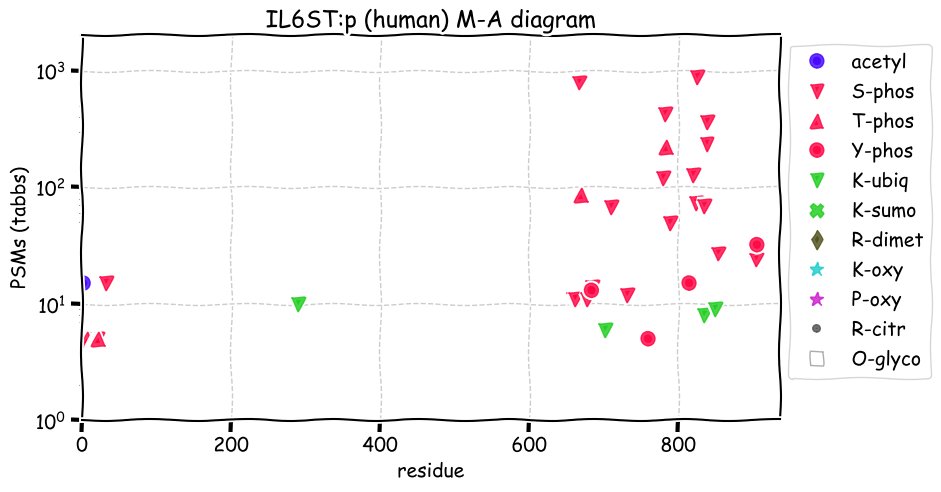
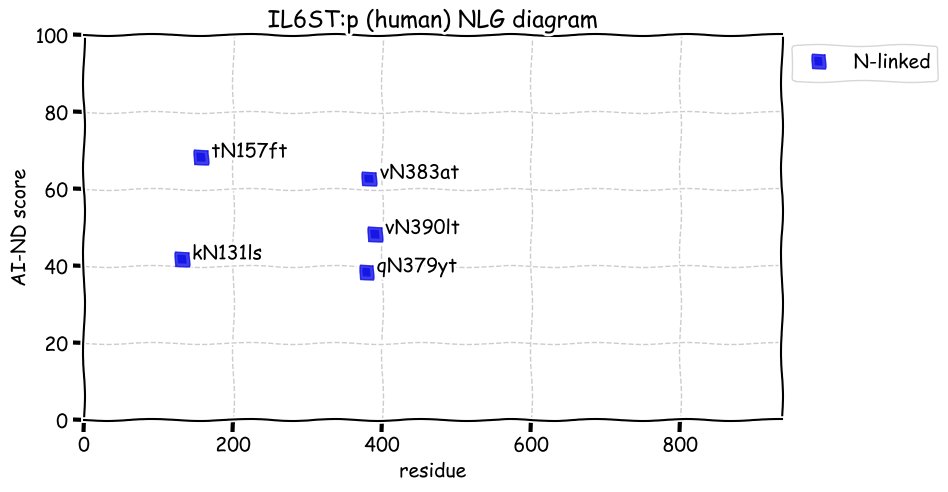
Sun Apr 18 15:44:05 +0000 2021Things don't seem to be slowing down in India 🔗
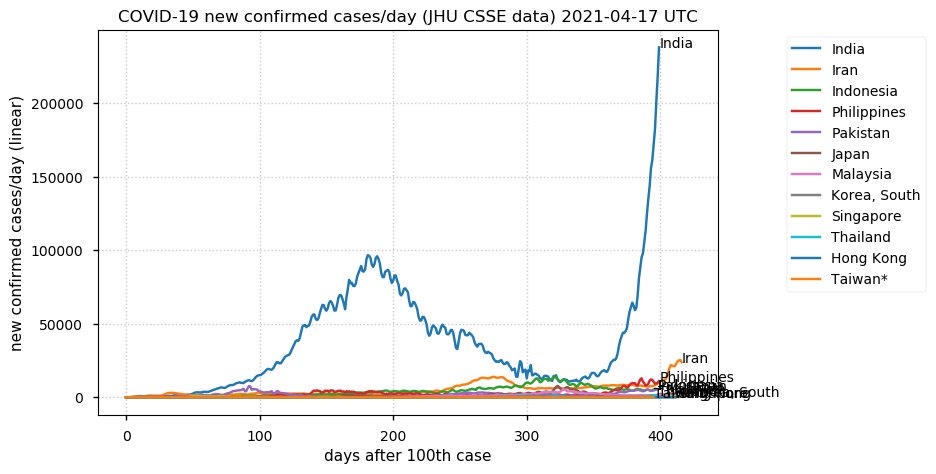
Sun Apr 18 14:48:05 +0000 2021& I never thought anything could knock "MudPIT" off of its top spot in the rankings.
Sun Apr 18 14:37:20 +0000 2021PXD018369 (CSF) is another state-of-the-art data set generated using timsTOF PASEF (which also wins the "worst acronym in proteomics" award).
Sun Apr 18 13:26:20 +0000 2021After analyzing about 100 of the LC/MS/MS runs, the data looks pretty good. For example, the PSMs found in kidney MHC class I peptide expts contain ~4% previously undetected peptides.
Sun Apr 18 12:38:23 +0000 2021GM2A:p.V153A chr 5:g.151267327T>C, rs61740602, (all tissues V:A = 18971:1305 7%) vaf=8%, Δm=-28.0313, VAF by population group: african 2%, american 5%, european 9%, east asian 2%, south asian 11%. #ᐯᐸᐱ
Sun Apr 18 12:38:23 +0000 2021GM2A:p, θ(max) = 83. aka SAP-3. Observed in MHC class 2 peptide experiments. Most prominent urine &l CSF: more rarely in blood plasma. Present in many samples from cell lines, most prominently HeLa, HEK-293 & THP-1.
Sun Apr 18 12:38:23 +0000 2021>GM2A:p, GM2 ganglioside activator (Homo sapiens) Small extracellular subunit; CTMs: none; PTMs: 12×K+ubiquitinyl; SAAVs: I59V (34%), M69V (86%), V153A (8%); mature form: (32,33,34-193) [14,373×, 85 kTa]. #ᗕᕱᗒ 🔗
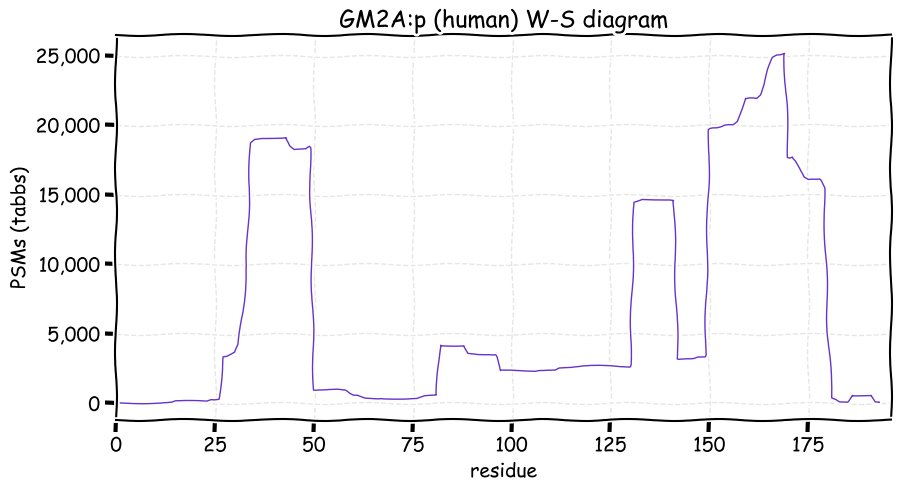
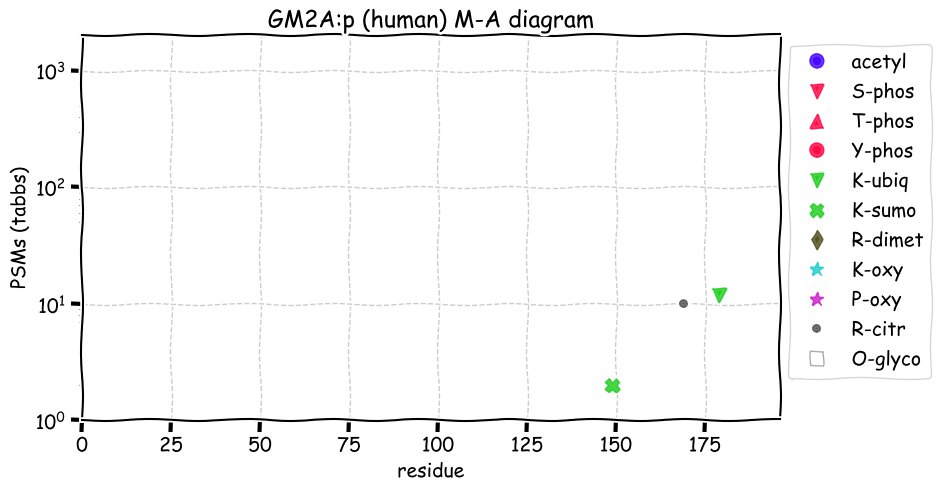
Sat Apr 17 18:17:38 +0000 2021@astacus Quite right. Phosphatase was what I was shooting for.
Sat Apr 17 15:34:38 +0000 2021@astacus These two alternatives also would imply that the observation of singleton phosphorylations should be rare: most real signaling/functional phosphorylation events should occur as phosphodomains rather than as individual sites.
Sat Apr 17 15:04:40 +0000 2021@astacus Both alternatives can exist happily together: in fact taken together they suggest that there should be lots of phosphorylation in proteins that utilize phosphorylation for either signaling or to alter their function (e.g., switch an enzyme on/off)
Sat Apr 17 14:57:15 +0000 2021@astacus Another alternative, championed by the late Tony Pawson, was that many phosphorylation events occur sequentially: a site starts the process, revealing another site, revealing another & so on until the final state is reached, with phosphorylases keeping things tidy along the way.
Sat Apr 17 14:52:04 +0000 2021@astacus My own notion on the issue of profligate PTM observation is that the very specific 1-site:1-function/signal notion commonly expressed in texts is a simplification. In real systems, there are several sites that may be involved: in an either-or manner or as a cumulative signal.
Sat Apr 17 12:45:27 +0000 2021NCAM2:p.L350P chr 21:g.21373867T>C, rs232518, (brain tissues L:P = 36:169 18%) vaf=35%, Δm=-16.0313, VAF by population group: african 38%, european 35%, east asian 58%, south asian 31%, american 29%. #ᐯᐸᐱ
Sat Apr 17 12:45:26 +0000 2021NCAM2:p, θ(max) = 62. aka NCAM21; MGC51008. Observed in MHC class 1 & 2 peptide experiments. Most prominent in most tissues brain, urine, CSF & blood plasma. Transmembrane domain (698-718), (20-697) is extracellular. Phosphodomain (765-824) is cytoplasmic.
Sat Apr 17 12:45:26 +0000 2021>NCAM2:p, neural cell adhesion molecule 2 (Homo sapiens) Midsized subunit; CTMs: N177, N419, N445, N474, N562+glycosyl; PTMs: 4×S, 4×T, 1×Y+phosphoryl; SAAVs: L350P (35%); mature form: (20-837) [5,372×, 55 kTa]. #ᗕᕱᗒ 🔗
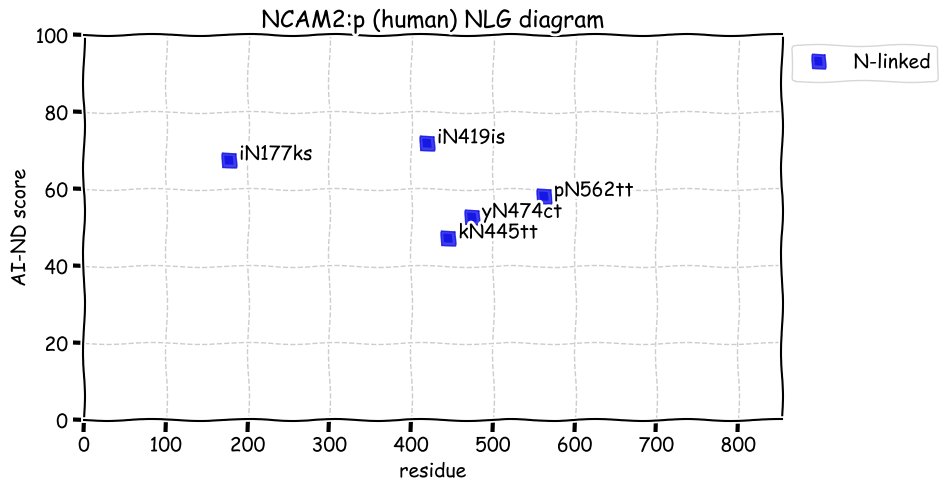
Fri Apr 16 20:17:05 +0000 2021@astacus Seems to rather like the phosphorylation portion of 🔗
Fri Apr 16 20:09:00 +0000 2021@astacus Like casein or SPP1 phosphorylation?
Fri Apr 16 19:12:34 +0000 2021It will depend on how the data works out, but PXD019643 certainly seems to be a rather ambitious MHC peptide project. The first few that I have analyzed look promising, though.
Fri Apr 16 19:06:46 +0000 2021@MHendr1cks It is much better than all of the kitschy Norman architecture they have in England.
Fri Apr 16 18:08:19 +0000 2021@pwilmarth Other scholarship suggests the "pro" may mean "too many" in a Sumerian dialect used by Akkadians to discuss mass spec related topics.
Fri Apr 16 17:55:19 +0000 2021@pwilmarth My theory is that "pro" is a mystical reference to "16" in some ancient Masonic texts.
Fri Apr 16 17:38:12 +0000 2021@astacus I was thinking about noise in the sense of incorrect PSM assignments because of the expanded solution space associated with PTM assignment. What is this "biological noise" of which you speak?
Fri Apr 16 16:25:03 +0000 2021What does the "pro" in "TMTpro" stand for? Amusing answers only ...
Fri Apr 16 14:38:07 +0000 2021This one is a puzzle for the never-to-be-dissuaded, tin-foil-hat club that are convinced the PTMs found by proteomics are all just noise.
Fri Apr 16 12:12:50 +0000 2021PLBD1:p.V265I chr 12:g.14535710C>T, rs7957558 (all tissues V:I = 35:789 96%) vaf=96%, Δm=14.0156 VAF by population group: african 63%, european 98%, east asian 99%, south asian 97%, american 96%. #ᐯᐸᐱ
Fri Apr 16 12:12:50 +0000 2021PLBD1:p, θ(max) = 59. aka MLC, KIAA0027, LVM, VL. Observed in MHC class 1 & 2 peptide experiments. Found in most tissues & urine, but rare in blood plasma or CSF.
Fri Apr 16 12:12:49 +0000 2021>PLBD1:p, phospholipase B domain containing 1 (Homo sapiens) Midsized subunit; CTMs: N71, N308, N411+glycosyl (high mannose); PTMs: none; SAAVs: V265I (96%), P534A (49%); mature form: (33-553) [9,264×, 56 kTa]. #ᗕᕱᗒ 🔗
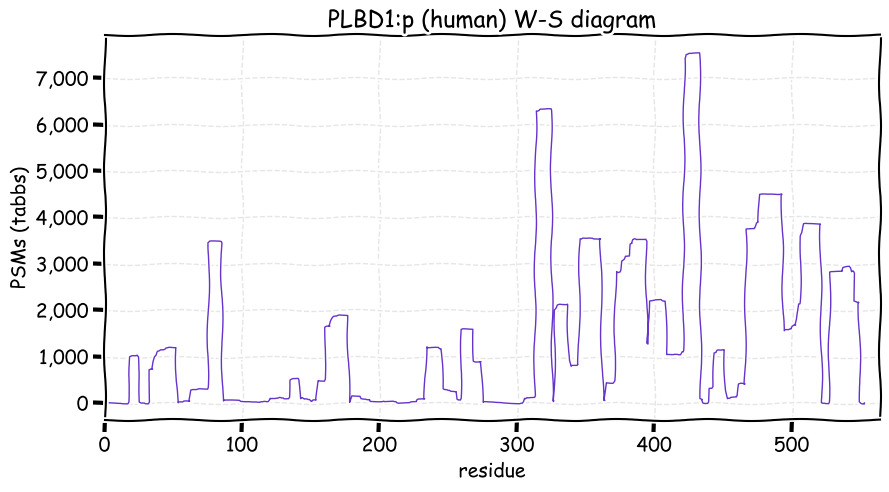
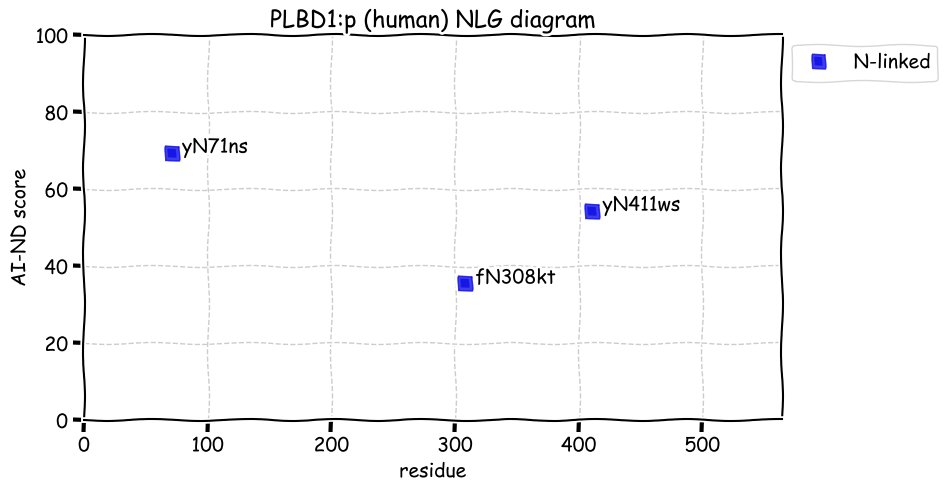
Thu Apr 15 15:13:29 +0000 2021I would love to see some top-down analysis done on it to see how many acceptor sites are simultaneously occupied.
Thu Apr 15 15:10:14 +0000 2021MAPT:p is one of those small proteins that seem to be PTM magnets for one reason or another.
🔗
Thu Apr 15 12:04:30 +0000 2021MLC1:p.N344S chr 22:g.50064062T>C, rs11568188, (all tissues N:S = 1502:131 8%) vaf=11%, Δm=-27.0109 VAF by population group: african 6%, european 13%, east asian 9%, south asian 24%, american 18%.. #ᐯᐸᐱ
Thu Apr 15 12:04:30 +0000 2021MLC1:p, the gene symbol comes from its association with megalencephalic leukoencephalopathy with subcortical cysts (MLC).
Thu Apr 15 12:04:29 +0000 2021MLC1:p, θ(max) = 35. aka MLC, KIAA0027, LVM, VL. Observed in MHC class 1 & 2 peptide experiments. Observed in brain tissue and cell lines derived from acute leukemia. Has 8 transmembrane domains.
Thu Apr 15 12:04:29 +0000 2021>MLC1:p, modulator of VRAC current 1 (Homo sapiens) Small intracellular membrane subunit; CTMs: M1, T2+acetyl; PTMs: 6×S, 1×T, 1×Y+phosphoryl; K33, K175+ubiquitinyl; SAAVs: C171F (12%), N344S (11%); mature form: (1,2-377) [1,730×, 25 kTa]. #ᗕᕱᗒ 🔗
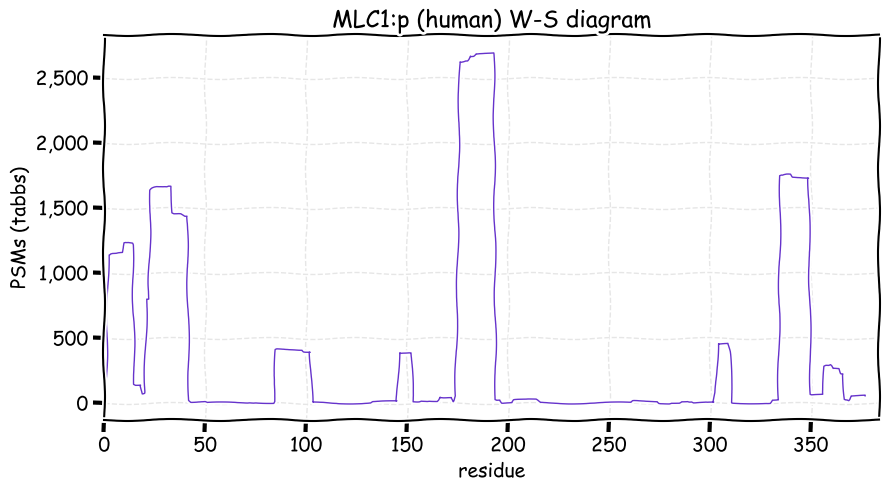
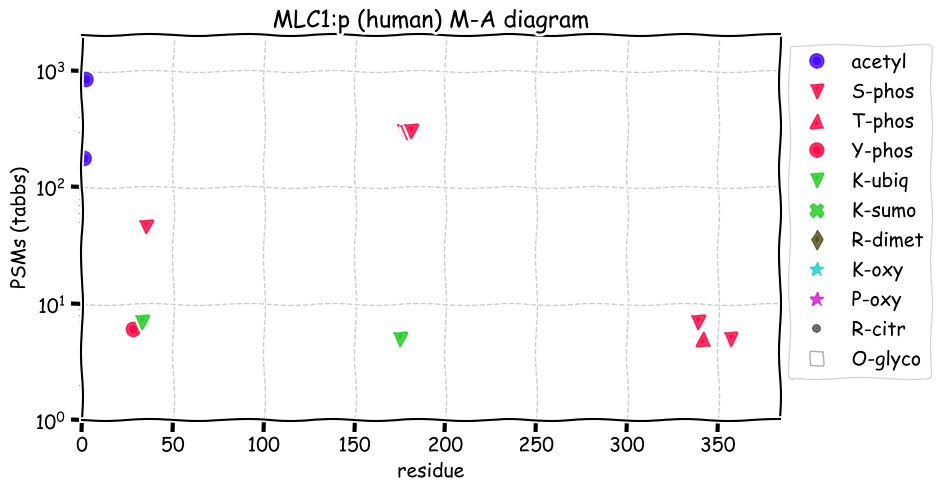
Wed Apr 14 13:00:31 +0000 2021@AlexUsherHESA Office staff + expenses, including any additional stipends/salary increases associated with the Dept. Head.
Wed Apr 14 12:58:42 +0000 2021@cajun_science A spreadsheet that evokes any other type of excited utterance (particularly those beginning with "F**k!" or "S**t!") I would probably judge as being not very well organized.
Wed Apr 14 11:41:00 +0000 2021CCN3:p.R42Q chr 8:g.119416784G>A, rs2279112, (urine R:Q = 55:24 30%) vaf=27%, Δm=-28.0425 VAF by population group: african 59%, european 21%, east asian 33%, south asian 31%, american 19%. #ᐯᐸᐱ
Wed Apr 14 11:41:00 +0000 2021CCN3:p, θ(max) = 51. aka IGFBP9, NOV, nephroblastoma overexpressed. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in urine, plasma & CSF.
Wed Apr 14 11:41:00 +0000 2021>CCN3:p, cellular communication network factor 3 (Homo sapiens) Small subunit; CTMs: none; PTMs: S316+phosphoryl, K272, K351+SUMOyl; SAAVs: R42Q (27%); mature form: (32-357) [3,711×, 25 kTa]. #ᗕᕱᗒ 🔗
Wed Apr 14 11:36:01 +0000 2021@cajun_science A well organized spreadsheet should be completely unremarkable: it should be obvious what it is & how to use it. A really well organized spreadsheet should evoke something like: "Wow, I wish I had thought of that!".
Wed Apr 14 10:52:47 +0000 2021@sjwill99 @astacus @NatureTractors Nature Pigeon Fancier
Tue Apr 13 23:06:18 +0000 2021After another day examining the same type of material, I'll revise that grade to an F+.
Tue Apr 13 16:26:06 +0000 2021@michaelhoffman @FEBS_Letters @SamanthaLWilson @gwaygenomics @bittremieux @valendraica @ontowonka "Eluding Excel's mangling of gene symbols can prove complicated."
Tue Apr 13 15:12:52 +0000 2021PXD000653 is worth a look as exemplar data for developing algorithms to detect deamidation deliberately caused by PNGase F (or endogenously by AGA). It uses older technology (low resolution fragment ions, LTQ Orbitrap Velos Pro), but the results are really pretty good.
Tue Apr 13 13:36:41 +0000 2021The North American news media I pay attention to do not seem to be tracking this COVID-19 story. I guess local concerns are saturating coverage to the detriment of world news. 🔗
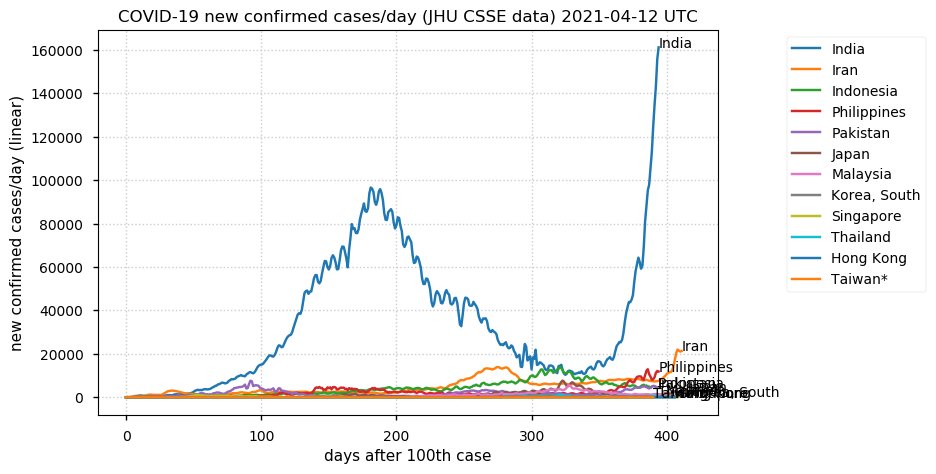
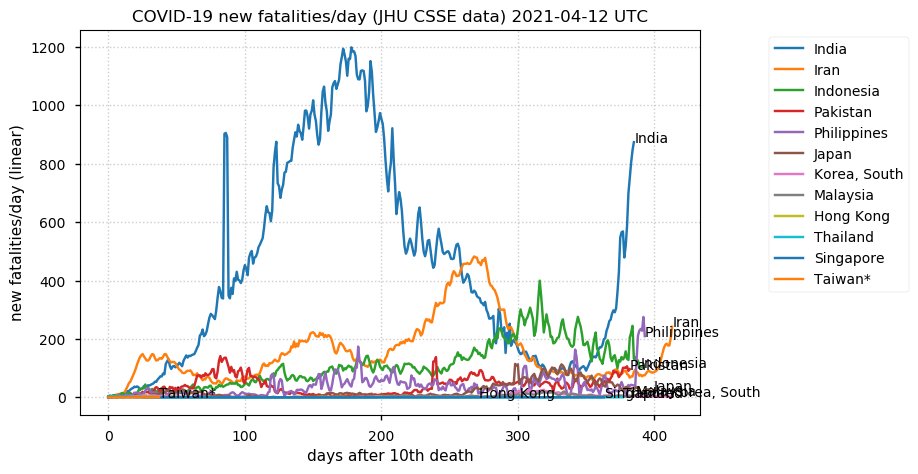
Tue Apr 13 12:50:52 +0000 2021I'd have to give the field a solid D- for spreadsheet organization and content.
Tue Apr 13 12:41:10 +0000 2021Spent the last few days going through the spreadsheets in manuscripts' Supplementary Data. Yikes! 😱
Tue Apr 13 11:52:21 +0000 2021GPC4:p.E391D chr X:g.133304844C>A, rs1129980, (urine E:D = 69:13 16% vaf=33%, Δm=-14.0157 VAF by population group: african 58%, european 30%, east asian 41%, south asian 49%, american 24%. #ᐯᐸᐱ
Tue Apr 13 11:52:21 +0000 2021GPC4:p, θ(max) = 67. aka K-glypican. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in urine & many tissues, but rare in plasma & CSF.
Tue Apr 13 11:52:20 +0000 2021>GPC4:p, glypican 4 (Homo sapiens) Midsized extracellular subunit; CTMs: none; PTMs: Y186+phosphoryl; SAAVs: E391D (33%), A442V (46%); mature form: (19?-529?) [8,707×, 51 kTa]. #ᗕᕱᗒ 🔗
Mon Apr 12 12:19:36 +0000 2021SERPINC1:p has one additional isoform (ENSP00000478688) that can be detected using MS-based proteomics via one tryptic peptide domain (183-197).
Mon Apr 12 12:19:35 +0000 2021SERPINC1:p.A296P chr 1:g.173909819C>G, rs372820797 (blood plasma H:R = 6469:86 1%) vaf=0.006%, Δm=26.0156 Has been associated with antithrombin II deficiency #ᐯᐸᐱ
Mon Apr 12 12:19:35 +0000 2021SERPINC1:p, θ(max) = 87. aka ATIII, MGC22579, AT3. Three domains observed in MHC class 2 peptide experiments. Commonly observed in blood plasma, urine & CSF.
Mon Apr 12 12:19:35 +0000 2021>SERPINC1:p, serpin family C member 1 (Homo sapiens) Small extracellular subunit; CTMS: N128, N187, N224+glycosyl; PTMs: 30×S, 6×T, 0×Y+phosphoryl; SAAVs: A296P (<1%); mature form: (33,35-464) [37,815×, 962 kTa] #ᗕᕱᗒ 🔗
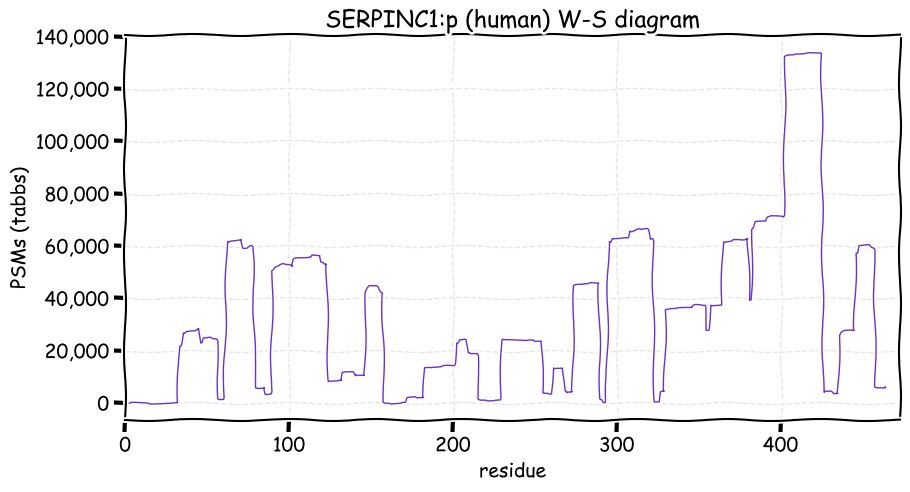
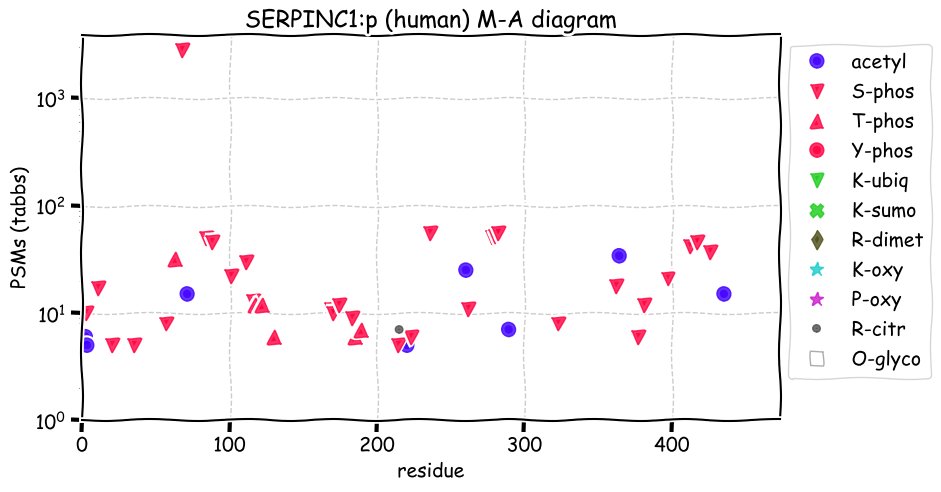
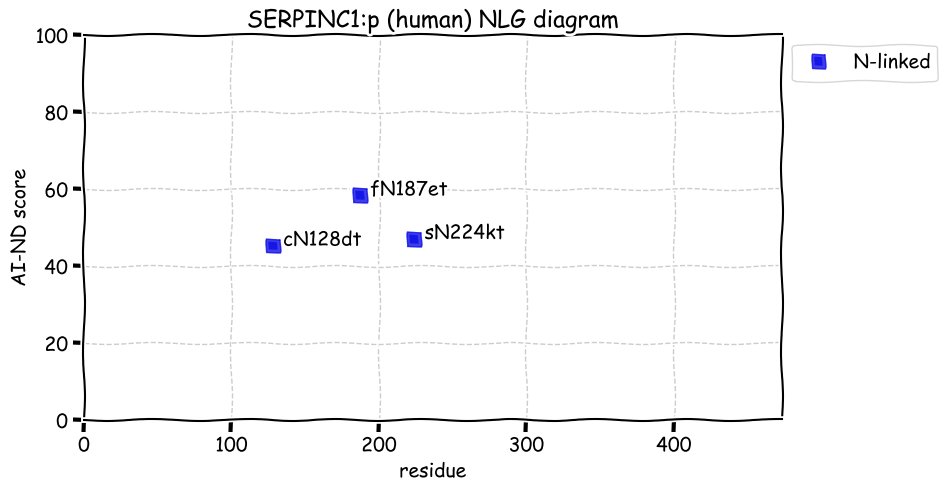
Sun Apr 11 15:27:28 +0000 2021The high enrichment of these enzymatically deamidated peptides (>70% of PSMs) makes it ideal for anyone wanting to test algorithms for discovering/localizing this rather challenging modification & distinguishing it from its analogous, non-enzymatic protein degradation product.
Sun Apr 11 15:23:50 +0000 2021An oldie but goodie: PXD004540. While probably not very useful for one of its original purposes because of poor coupling of TMT to peptides, it is a very good for the other: enriching peptides containing deglycosylated N-linked glycosylation. /1
Sun Apr 11 12:22:30 +0000 2021The sequence of the 13th protein, MT-ND4L:p, does not generate observable peptides in using typical enzymatic digestion methods. However, it is nicely observed in MHC class 1 peptide experiments. /fin
Sun Apr 11 12:19:05 +0000 2021Thanks to everyone who participated in the poll. According to Gencode, the mitochondrial chromosome has 13 protein-coding genes, that are all named with an "MT-" prefix. 12 of the proteins can be observed using standard proteomics methods. /1
Sun Apr 11 12:12:46 +0000 2021ADA2:p has an isoform caused by an alternate translation initiation site at M21. The peptide (22-34, S22+acetyl) is characteristic of this isoform.
Sun Apr 11 12:12:46 +0000 2021ADA2:p.H335R chr 22:g.17188416T>C, rs2231495, (JURKAT H:R = 0:21 homozygous) vaf=34%, Δm=19.0422 VAF by population group: african 36%, european 32%, east asian 23%, south asian 31%, american 32%. #ᐯᐸᐱ
Sun Apr 11 12:12:46 +0000 2021ADA2:p, θ(max) = 92. aka ADGF, IDGFL, CECR1. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in blood plasma & CSF, but not urine. Puzzling pattern of ubiquitinylation.
Sun Apr 11 12:12:46 +0000 2021>ADA2:p, adenosine deaminase 2 (Homo sapiens) Small extracellular subunit; CTMS: N174, N378+glycosyl; PTMs: 17×K+ubiquitinyl; SAAVs: H335R (34%); mature form: (30-511) [5,475×, 26 kTa] #ᗕᕱᗒ 🔗
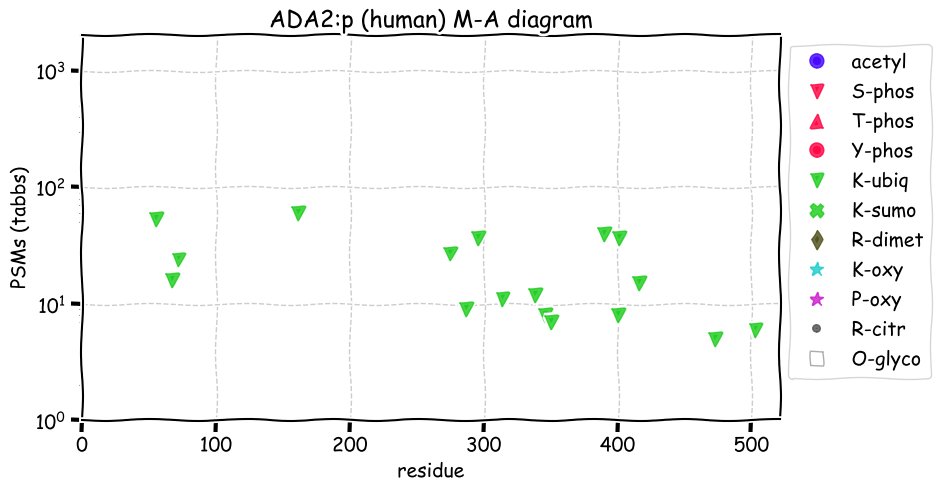
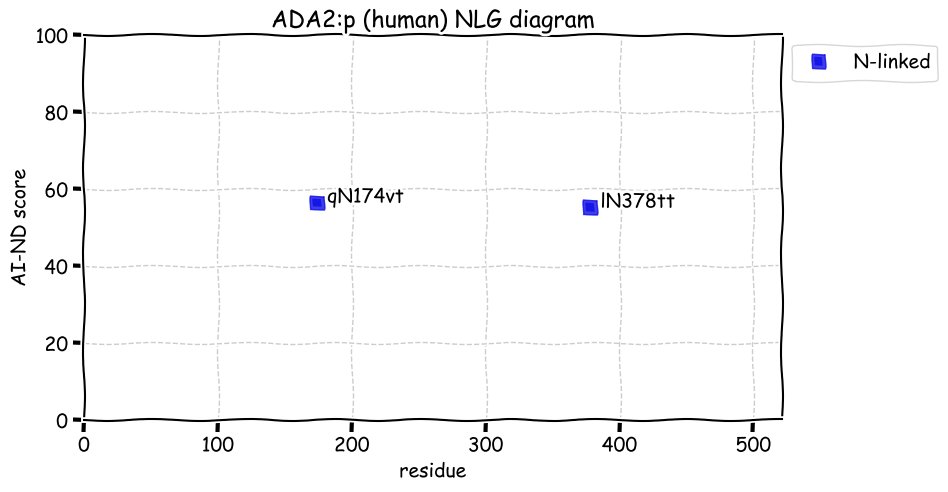
Sat Apr 10 13:29:07 +0000 2021@astacus Even here in Canada, where we have to put up with UK, US & French spellings and pronunciations, it is viewed askance. And for publishing, using them does screw up keyword searches.
Sat Apr 10 13:10:31 +0000 2021@astacus It went out of fashion when people started using typewriters. I personally still like to throw in the odd diaeresis & ligature myself―who doesn't prefer hæmoglobin to the rather pedestrian hemoglobin―but it is mildly eccentric these days.
Sat Apr 10 12:02:05 +0000 2021CHL1:p has three isoforms due to alternate splicing that can be readily distinguished using MS-based proteomics. DDX11:p has also been referred to as CLI1 in the literature.
Sat Apr 10 12:02:05 +0000 2021CHL1:p.I1050V chr 3:g.398280A>G, rs6442827, (cerebrospinal fluid I:V = 70:1473 95%) vaf=96%, Δm=-14.0156. VAF by population group: african 85%, european 100%, east asian 100%, south asian 100%, american 98%. #ᐯᐸᐱ
Sat Apr 10 12:02:05 +0000 2021CHL1:p, θ(max) = 60. aka CALL, L1CAM2, FLJ44930, MGC132578. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in biological fluids & brain tissue. Transmembrane domain (1098-1119), intracellular phosphodomain (1143-1224).
Sat Apr 10 12:02:04 +0000 2021>CHL1:p, cell adhesion molecule L1 like (H sapiens) Large subunit; CTMS: N87, N231, N492, N498, N578, N783, N1042, N1213+glycosyl; PTMs: 11×S,4×T,0×Y+phosphoryl; S260+glycosyl SAAVs: L17F (65%), T303A (15%), I1050V (96%); mature form: (25-1224) [7,461×, 96 kTa]. #ᗕᕱᗒ 🔗
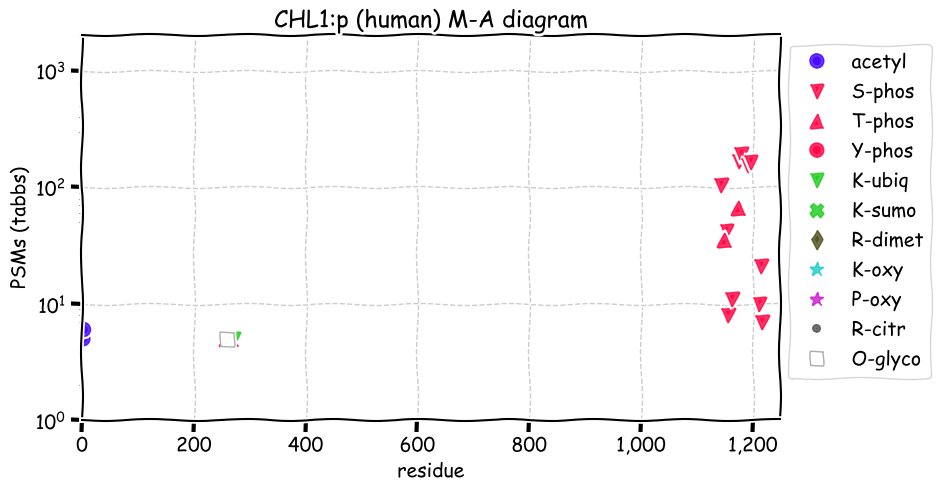
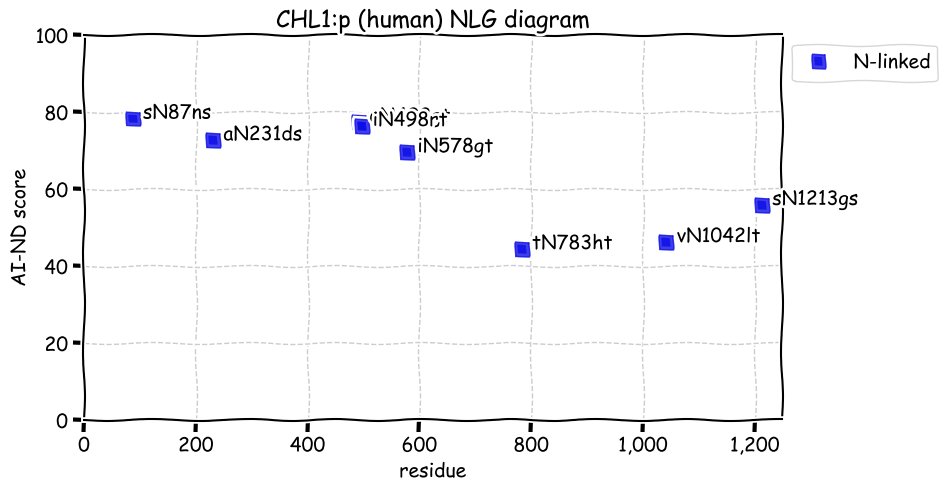
Sat Apr 10 11:57:37 +0000 2021In honor of the 40th anniversary of the human mitochondrial chromosome sequence, how many protein-coding genes are on that chromosome:
Fri Apr 09 16:11:44 +0000 2021I just noticed a manuscript title in Nat. Comm. that uses the now-archaic diaeresis form of naive (naïve). Is this some peculiar editorial practice or simply an attempt to confuse keyword searches? 🧐
Fri Apr 09 15:44:23 +0000 2021The manuscript says the sample was cerebrospinal fluid extracellular vesicles, but the data says it was corneocytes: which one should I believe?
Fri Apr 09 12:19:44 +0000 2021CHGA:p has two isoforms due to alternate splicing that can be readily distinguished using MS-based proteomics.
Fri Apr 09 12:10:08 +0000 2021CHGA:p.R399W, chr 14:g.92932756C>T, rs729940, (all tissue R:W = 1769:231 12%) vaf=15%, Δm=29.9782. VAF by population group: african 1%, european 16%, east asian 20%, south asian 10%, american 21%. #ᐯᐸᐱ
Fri Apr 09 12:10:08 +0000 2021CHGA:p, θ(max) = 96. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in biological fluids: most abundant in CSF. At least 18 domains of this molecule are formed by proteolytic cleavage to form bioactive peptides.
Fri Apr 09 12:10:08 +0000 2021>CHGA:p, chromogranin A (Homo sapiens) Small extracellular portein; CTMS: none; PTMs: 35×S,5×T,0×Y+phosphoryl; SAAVs: E264D (18%), R399W (15%), A274G (1%), G315S (1%), L332P (1%); mature form: (19-457) [6,878×, 150 kTa]. #ᗕᕱᗒ 🔗
Thu Apr 08 22:26:30 +0000 2021@bkives Just 1.1 mm at the Forks so far.
Thu Apr 08 22:15:38 +0000 2021@attilacsordas although there is no indication that they tested the rMSA for degradation caused by storage or artifacts associated with the recombinant expression & subsequent purification.
Thu Apr 08 22:13:35 +0000 2021@attilacsordas Reading the paper I'm not so sure about the "young" part, as the rMSA batch they are using is going to be older than the majority of the circulating MSA by the end of the study. But "undamaged" may be accurate /1
Thu Apr 08 16:18:13 +0000 2021@ucdmrt The α-amino group is also more likely to end up with the succinylation side reaction than the ε-amino group.
Thu Apr 08 15:53:12 +0000 2021@ucdmrt All of the TMT/iTRAQ reagents that use an amine-reactive NHS ester group show the same pattern.
Thu Apr 08 15:35:39 +0000 2021@WinnipegNews I can't help but look at 🔗 & be slightly cynical wrt this particular statement.
Thu Apr 08 15:29:41 +0000 2021@ucdmrt That is a pretty common pattern. The N-terminii are more reactive than K's to IAA and urea artifacts, but less than K's towards succinic anhydride derivatization.
Thu Apr 08 13:28:56 +0000 2021@astacus Did that exact study back in the 90's using synthetic peptides (I had a peptide synthesizer in the lab). Could not find a reviewer/editor that thought it was sufficiently interesting to publish.
Thu Apr 08 12:06:46 +0000 2021It is interesting to comparing the PTM pattern for SPARCL1 with the protein is is supposed to be "like" SPARC: it is clear that despite sequence similarities, the 2 proteins are being used quite differently 🔗
Thu Apr 08 12:02:03 +0000 2021SPARCL1:p.A49D, chr 4:g.87495036G>T, rs13051, (blood plasma A:D = 670:254 27%) vaf=45%, Δm=43.9898. VAF by population group: african 88%, european 38%, east asian 44%, south asian 56%, american 50%. #ᐯᐸᐱ
Thu Apr 08 12:02:02 +0000 2021SPARCL1:p,θ(max) = 79. aka MAST9. Observed in MHC class 2 peptide experiments. Commonly observed in biological fluids: most abundant in CSF.
Thu Apr 08 12:02:02 +0000 2021>SPARCL1:p, SPARC like 1 (Homo sapiens) Midsized matricellular subunit; CTMs: N169+glycosyl; PTMs: 33×S,10×T,1×Y+phosphoryl; 2×S,4×T+glycosyl; SAAVs: A49D (49%), H106D (58%), V253E (4%), T419A (22%), D575E (75%); mature form: (17-664) [9,230×, 119 kTa]. #ᗕᕱᗒ 🔗
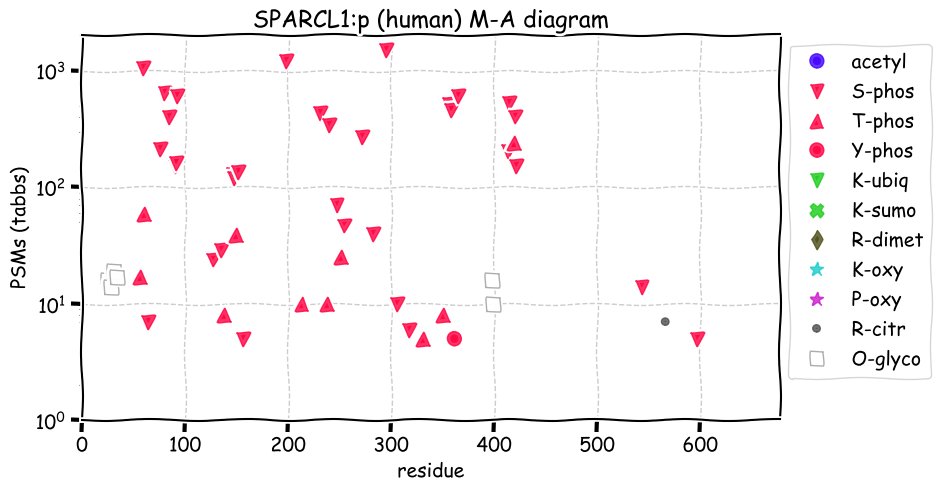
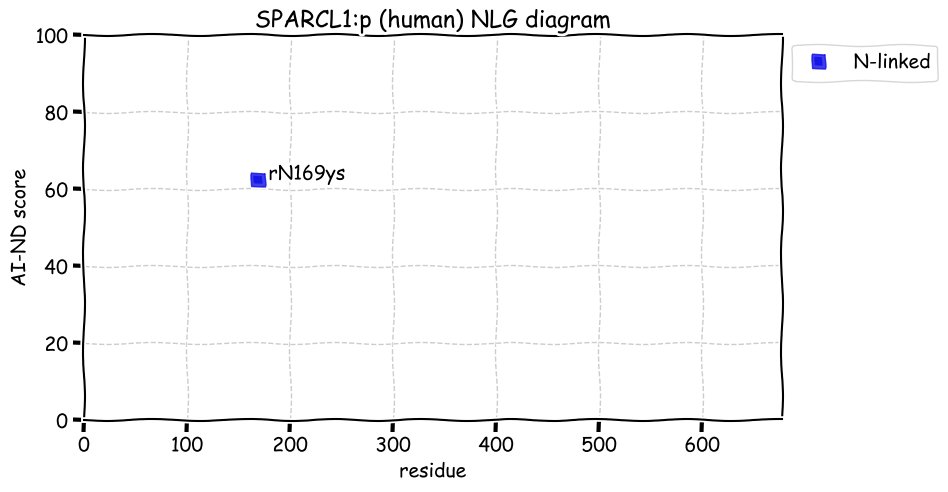
Wed Apr 07 23:04:19 +0000 2021uh-oh Canada 🔗
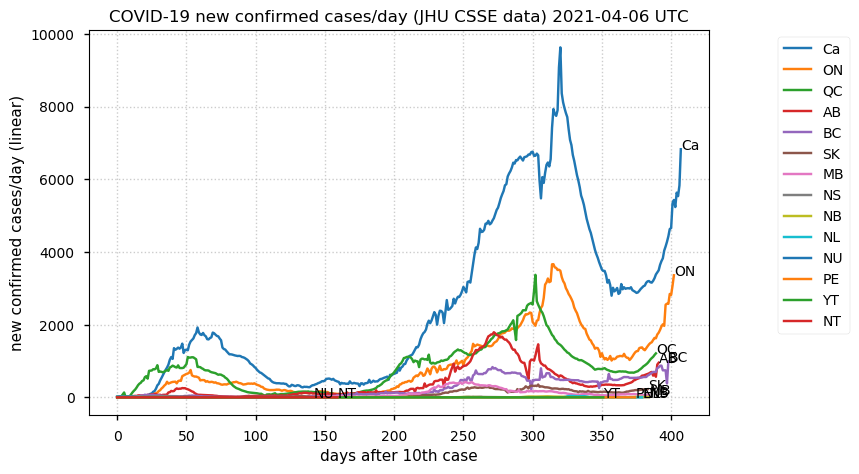
Wed Apr 07 20:38:05 +0000 2021When you are filling in a ProteomeXchange entry, don't be ashamed to admit that you are using a reagent like TMT or iTRAQ. Just declare it and move on. 🧐
Wed Apr 07 12:40:51 +0000 2021It seems like the answer is "no" …
Wed Apr 07 12:06:50 +0000 2021@ypriverol It seems to be up & nothing in the logs looks like there was a problem. Can you reach it now?
Wed Apr 07 12:03:23 +0000 2021C7:p.T587P, chr 5:g.40964750A>C, rs13157656, (blood plasma T:P = 4091:895 18%) vaf=24%, Δm=-3.9949. VAF by population group: african 3%, european 22%, east asian 21%, south asian 36%, american 21%. #ᐯᐸᐱ
Wed Apr 07 12:03:23 +0000 2021C7:p, θ(max) = 79. Observed in MHC class 2 peptide experiments. Commonly observed in biological fluids, but very rarely in common cell lines.
Wed Apr 07 12:03:22 +0000 2021>C7:p, complement C7 (Homo sapiens) Midsized subunit; CTMS: N202, N754+glycosyl; PTMs: 2×K+acetyl; T696+glycosyl; SAAVs: T587P (23%); mature form: (19,23-843) [19,243×, 271 kTa]. #ᗕᕱᗒ 🔗
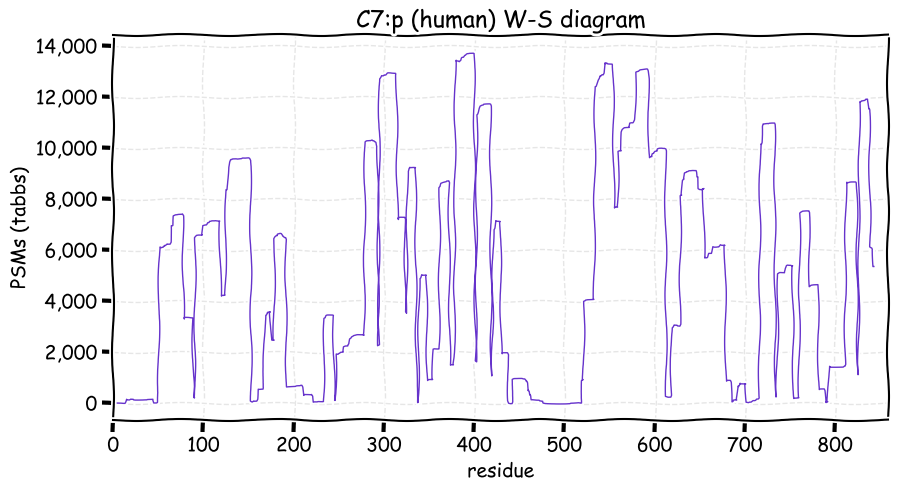
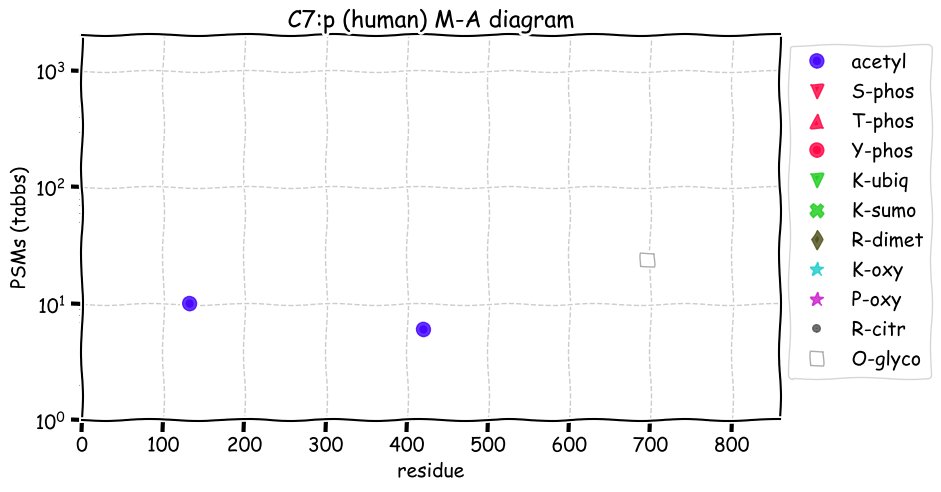
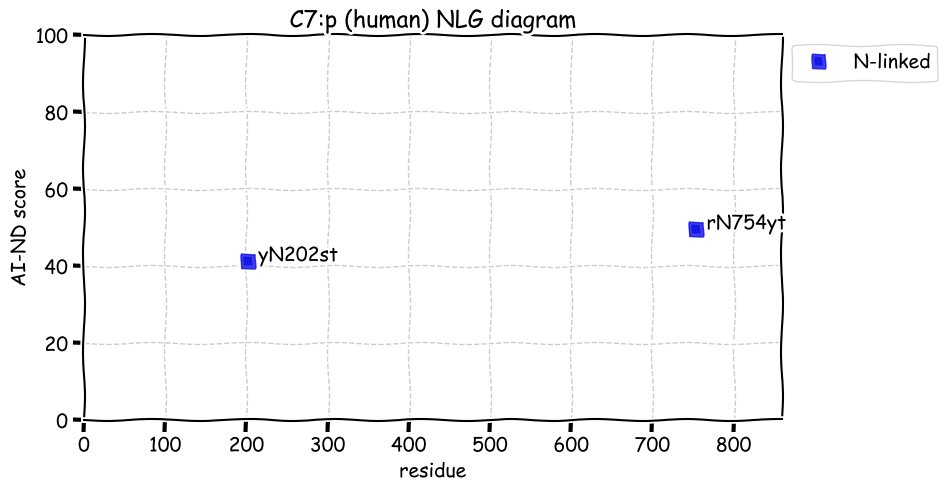
Tue Apr 06 16:21:30 +0000 2021Does anybody know of a good paper about the common alternate splice variants of ALB in humans?
Tue Apr 06 15:02:09 +0000 2021@attilacsordas I didn't realize I wasn't following you.
Tue Apr 06 14:48:14 +0000 2021@attilacsordas Next: molecular mortality ...
Tue Apr 06 14:11:35 +0000 2021@attilacsordas That is a normal use of "morbidity": illness at the level of the organism. A molecular morbidity would be the negative effect (illness) associated with the molecule itself, e.g., changes in 3D structure, function, activity or half-life.
Tue Apr 06 14:05:51 +0000 2021@attilacsordas No time like the present: a molecular morbidity affecting a chaperonin will probably result in molecular comorbidity involving many proteins.
Tue Apr 06 13:58:18 +0000 2021@attilacsordas Or, the molecular morbidity caused by sialylation variants of ENSP00000363827:pm.N89+glycosyl requires further investigation
Tue Apr 06 13:55:25 +0000 2021@attilacsordas The molecular morbidity associated with HSPG2:p.A1503V has not been determined.
Tue Apr 06 13:18:17 +0000 2021I have decided that "molecular morbidity" is a nifty term & I will start using it whenever possible.
Tue Apr 06 12:54:19 +0000 2021And, to be fair, large scale PTM comparative studies haven't produced much site-specific functional association information either.
Tue Apr 06 12:36:52 +0000 2021One comforting thing I have found examining human SAAVs is that not much more is known about their consequences than the effects associated with individual phosphorylation sites. GWAS has been a bit of a bust for this type of functional association.
Tue Apr 06 12:06:46 +0000 2021HSPG2:p.A1503V, chr 1:g.21864961G>A, rs897471, (all tissues A:V = 3975:5388 58%) vaf=73%, Δm=28.0313. VAF by population group: african 59%, european 73%, east asian 89%, south asian 53%, american 78%. #ᐯᐸᐱ
Tue Apr 06 12:06:45 +0000 2021HSPG2:p, θ(max) = 76. aka perlecan, PRCAN, SJS1. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in biological fluids, as well as many common cell lines.
Tue Apr 06 12:06:45 +0000 2021>HSPG2:p, heparan sulfate proteoglycan 2 (H sapiens) CTMS: N89,N554,N1755,N2121,N3279,N3780,N3836,N4068+glycosyl; PTMs: 22×S,13×T,0×Y+phosphoryl; 6×S,5×T+glycosyl; SAAVs: M638V(3%), N765S(7%), R1186Q(6%), A1503V(73%); mature form: (26-4391) [40,029 ×, 81 kTa] #ᗕᕱᗒ 🔗
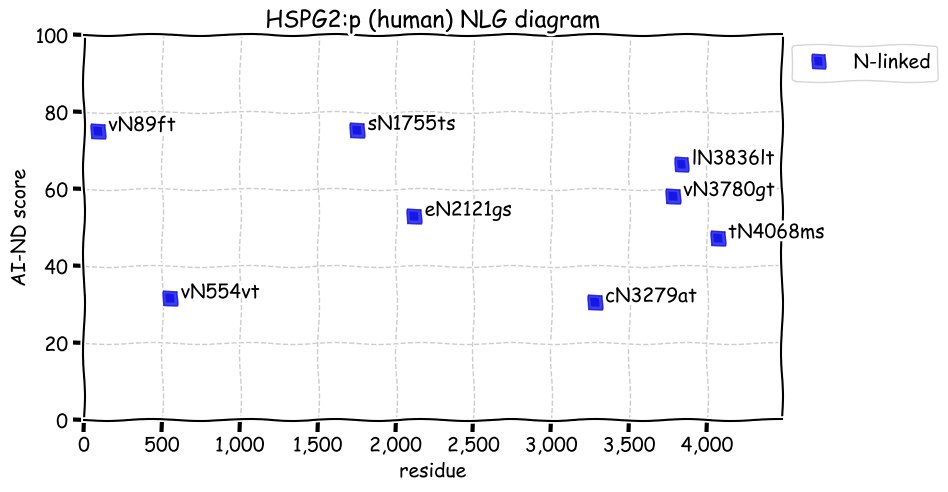
Mon Apr 05 16:05:38 +0000 2021@lgatt0 @TKockmann Many of the proteins are from keratinocytes and hair (38 keratins, SPRR5, HRNR, PKP1, LCE3D, SBSN, LCE1F, GSDMA, CDSN, LCD2B, ECM1, etc.), so it probably mainly represents lab environmental dust contamination.
Mon Apr 05 12:36:10 +0000 2021It appears that India has rather rapidly entered its second round of infections 🔗
Mon Apr 05 12:04:30 +0000 2021ENPP2:p.S545P, chr 8:g.119583783A>G, rs10283100, (cerebrospinal fluid S:P = 75:833 92%) vaf=96%, Δm=10.0207. VAF by population group: african 100%, european 95%, east asian 100%, south asian 99%, american 97%. #ᐯᐸᐱ
Mon Apr 05 12:04:30 +0000 2021ENPP2:p,θ(max) = 76. aka ATX, PD-IALPHA, PDNP2. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in cerebrospinal fluid, blood plasma & urine.
Mon Apr 05 12:04:29 +0000 2021>ENPP2:p, ectonucleotide pyrophosphatase/phosphodiesterase 2 (Homo sapiens) Midsized enzyme; CTMS: N54, N463+glycosyl; PTMs: none; SAAVs: S545P (96%); mature form: (36-915) [5,222×, 81 kTa]. #ᗕᕱᗒ 🔗
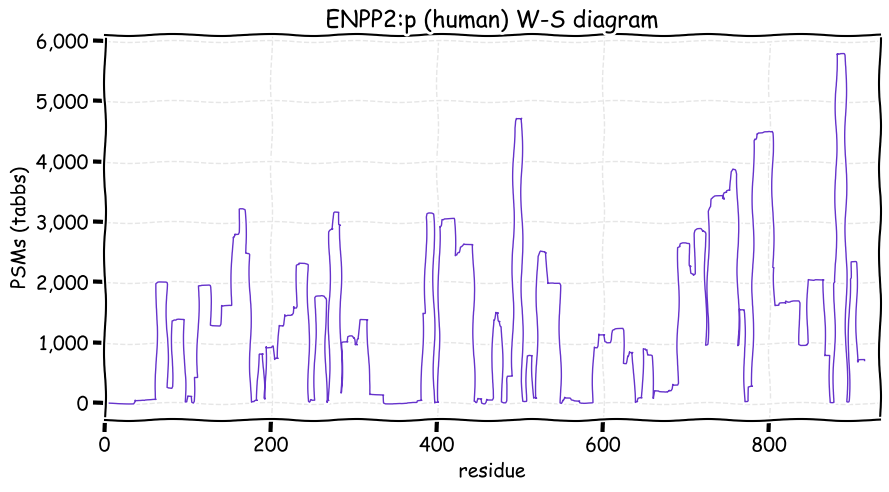
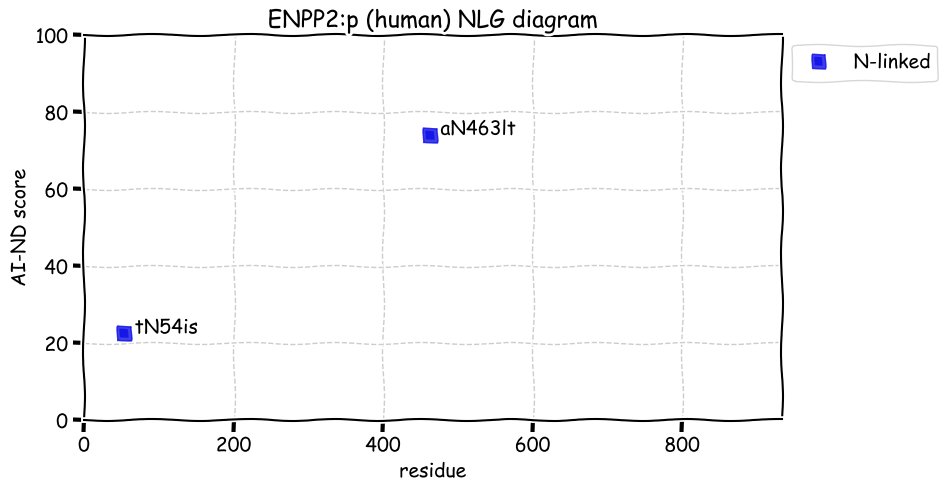
Sun Apr 04 16:47:52 +0000 2021The last hurrah of a supermarket whole roast chicken. 🔗

Sun Apr 04 14:53:42 +0000 2021To quote Robert Bringhurst on the subject: "They can also be used (as they often are) to shout at readers, putting them on edge and driving them away …"
Sun Apr 04 14:49:51 +0000 2021Thanks to everyone who participated. While people seemed reluctant to give their opinion, most thought text decoration was most often helpful. Personally, other than in headings & titles, I find the use of bold or underlining to be distracting & rarely added to the text.
Sun Apr 04 12:58:52 +0000 2021The server is 9×6×2.5 cm, 116 g, max. power consumption 6.4 W. The cat is of ill defined dimensions & power consumption, but weighs 3 kg.
Sun Apr 04 12:35:45 +0000 2021No one is really sure what the WASH complex is up to, but it does seem to associate with endosomes and actin components of the cytoskeleton.
Sun Apr 04 12:32:34 +0000 2021WASHC4:p is 1 of 6 subunits that participate in the Wiskott Aldrich Syndrome protein and scar homologue (WASH) complex. WASHC5:p is the only other subunit with a similar pattern of ubiquitinylation. WASHC2A & 2B are highly S/T phosphorylated (>140 acceptor sites).
Sun Apr 04 12:13:35 +0000 2021WASHC4:p.V901I, chr 12:g.105152394G>A, rs1663564, (breast tissues & cells V:I = 16:622 98%) vaf=95%, Δm=14.0156. VAF by population group: african 100%, european 92%, east asian 100%, south asian 99%, american 97%. #ᐯᐸᐱ
Sun Apr 04 12:13:35 +0000 2021WASHC4:p, θ(max) = 53. aka SWIP, KIAA1033. Observed in MHC class 1 & 2 peptide experiments. Commonly observed in most tissues & cell lines.
Sun Apr 04 12:13:34 +0000 2021>WASHC4:p, WASH complex subunit 4 (Homo sapiens) Large subunit; CTMS: A2+acetyl; PTMs: 21×K+acetyl; 12×S,7×T,1×Y+phosphoryl; 21×K+ubiquitinyl; SAAVs: Y38H (1%), V901I (95%); mature form: (2-1173) [21,109×, 110 kTa]. #ᗕᕱᗒ 🔗

Sat Apr 03 14:36:59 +0000 2021I frequently see people recommending the use of text decoration (bold, underline) in grant documents to indicate to reviewers which parts of the text are the most important. As a reviewer, do you find text decoration
Sat Apr 03 12:15:23 +0000 2021CNTN1:p.S775N, chr 12:g.41016821G>A, rs34326474, (all tissues S:N = 3074:33 1%) vaf=1%, Δm=27.0109. VAF by population group: african 5%, european <1%, east asian <1%, south asian <1%, american <1%. #ᐯᐸᐱ
Sat Apr 03 12:15:22 +0000 2021CNTN1:p, θ(max) = 75. aka F3, GP135. Rarely observed in MHC class 2 peptide experiments. Most commonly observed in urine, CSF & blood plasma. Also found in A-549 cell cultures.
Sat Apr 03 12:15:22 +0000 2021>CNTN1:p, contactin 1 (Homo sapiens) Midsized subunit; CTMS: N208, N258, N457, N494, N630, N933+glycosyl; PTMs: S993+GPI anchor; SAAVs: S775N (1%); mature form: (21-993) [8,578×, 195 kTa]. #ᗕᕱᗒ 🔗
Fri Apr 02 16:58:54 +0000 2021@oleg8r @slavov_n US govt. entities (e.g., NIST) can end up with several hundred % overhead, while a Canadian University is usually less than 10%. It is just a matter of influence and what can be negotiated.
Fri Apr 02 13:10:02 +0000 2021@neely615 @UCDProteomics The first of the new techs that is quantitative (g/L) wins.
Fri Apr 02 12:11:48 +0000 2021CHGB:p.S93T, chr 20:g.5922421T>A, rs6085324, (cerebrospinal fluid S:T = 371:208 36%) vaf=28%, Δm=14.0157. VAF by population group: african 7%, european 26%, east asian 38%, south asian 26%, american 7%. #ᐯᐸᐱ
Fri Apr 02 12:11:47 +0000 2021CHGB:p, θ(max) = 80. aka SgI, SCG1. Not observed in MHC class 1 or 2 peptide experiments. Most commonly observed in urine, CSF & blood plasma. Rarely observed in cell lines. Not unusual for an individual to have more than 1 SAAV.
Fri Apr 02 12:11:47 +0000 2021>CHGB:p, chromogranin B (H sapiens) Midsized enzyme; CTMS: none; PTMs: 21×K+acetyl, 42×S, 9×T, 5×Y+phosphoryl; SAAVs: S93T (28%), K117N (7%), R178Q (42%), N200H (16%), T243A (13%), A353G (48%), P413L (17%), R417H (23%); mature form: (21-677) [5,578×, 125 kTa]. #ᗕᕱᗒ 🔗
Thu Apr 01 15:26:30 +0000 2021@doctorow You didn't mention the Lastman story that has really stuck in the minds of Canadians outside of TO:
🔗
Thu Apr 01 15:14:40 +0000 2021@astacus Don't know what the issue was, but
🔗 is working. BALB/c female mice to be precise. The results are at 🔗
Lots of MUPs (& my favorite fluid phosphoprotein SPP1)
MUPs are also readily detectable in mouse plasma & esp. liver samples.
Thu Apr 01 12:36:26 +0000 2021CFI:p.T308A, chr 4:g.109757769T>C, rs11098044, (blood plasma T:A = 3:388 99%) vaf=99%, Δm=-30.0106. VAF by population group: african 88%, european 100%, east asian 100%, south asian 100%, american 99%. Reference sequence rarely observed in clinical samples. #ᐯᐸᐱ
Thu Apr 01 12:36:26 +0000 2021CFI:p,θ(max) = 70. aka FI, C3b-INA, KAF, IF. Rarely observed in MHC class 1 & 2 peptide experiments. Most commonly found in blood plasma, urine & CSF. May be secreted by some liver cell lines, e.g. HepG2.
Thu Apr 01 12:36:25 +0000 2021>CFI:p, complement factor I (Homo sapiens) Midsized extracellular subunit; CTMS: N70, N103, N177, N472, N502, N544+glycosyl; PTMs: S24, S90+phosphoryl; SAAVs: T308A (99%), R414H (4%); mature form: (19-591) [20,069×, 249 kTa]. #ᗕᕱᗒ 🔗
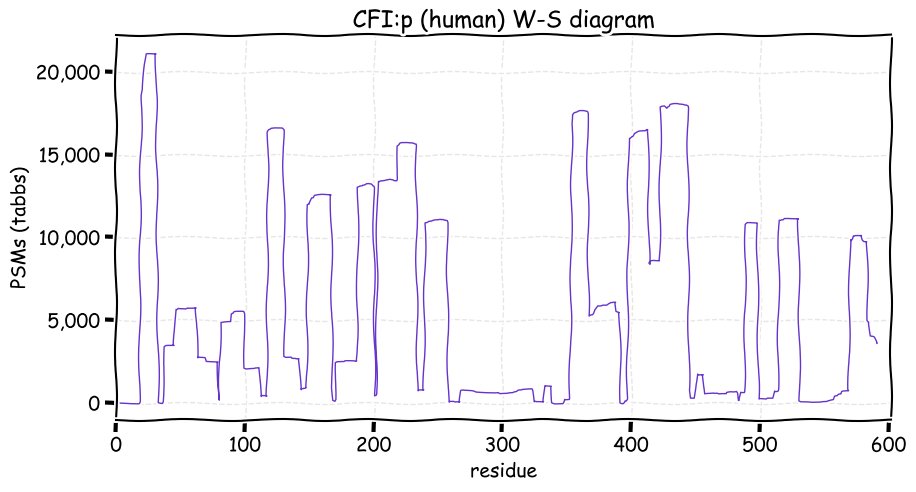
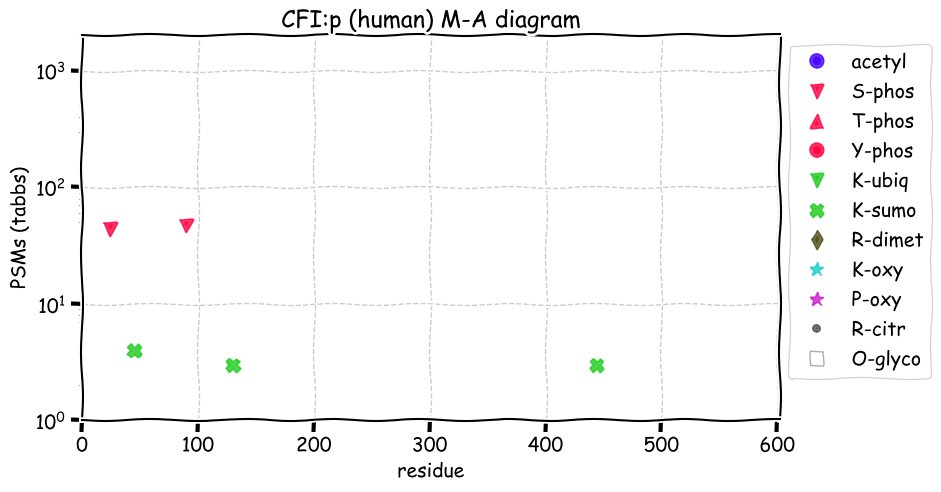
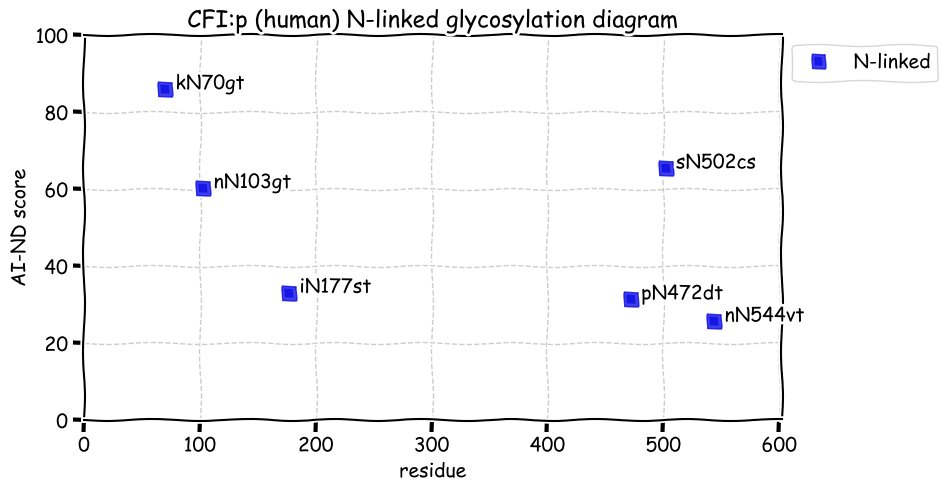
Wed Mar 31 22:51:17 +0000 2021I realized that my life is certainly at a strange place, when I got mildly excited by PXD022458, because it is the first data set I've seen taken from Mus musculus urine samples.
Wed Mar 31 17:28:20 +0000 2021@doctorow As used by the Northwest Mounted Police of Manitoba and Saskatchewan!
Wed Mar 31 12:48:31 +0000 2021@astacus Not all. Some of them are data types I'm not interested in (MRM/DIA), some of them are for critters with no good proteome sequence & some just don't look very promising. But I do look at quite a few of them.
Wed Mar 31 12:06:46 +0000 2021@astacus While I'm analyzing a public data set, I run a bunch of QA tests that both look for data quality and compare the results with previous results. If the new data is both top end in QA & it is better in some measures than previous ones, then I may give it a thumbs up.
Wed Mar 31 11:59:53 +0000 2021PRAP1:p.H101R, chr 10:g.133352286A>G, rs4369319, (all samples H:R = 0:240 100%) vaf=>99%, Δm=19.0422. VAF by population group: african 97%, european 100%, east asian 100%, south asian 100%. The reference sequence is rarely observed in clinical samples. #ᐯᐸᐱ
Wed Mar 31 11:59:53 +0000 2021PRAP1:p, θ(max) = 93. aka LRG. Observed in MHC class 1 & 2 peptide experiments. Most commonly observed in blood plasma, urine & CSF. May be secreted by some cell lines, e.g. HepG2.
Wed Mar 31 11:59:53 +0000 2021>PRAP1:p, proline rich acidic protein 1 (Homo sapiens) Small extracellular protein; CTMS: none; PTMs: none; SAAVs: P53L (1%), H101R (99%); mature form: (21-151) [1,547×, 5 kTa]. #ᗕᕱᗒ 🔗
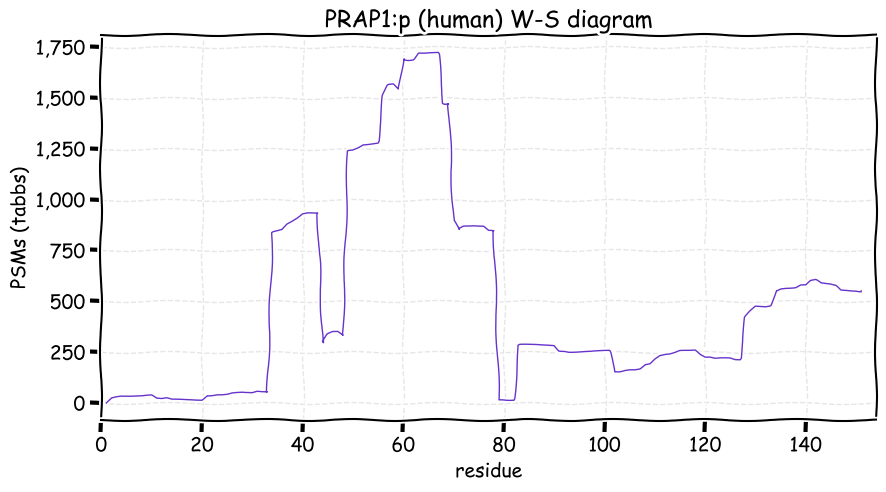
Tue Mar 30 23:56:30 +0000 2021A frequently disappointing type of manuscript involves trying to introduce & demonstrate the utility of a novel method on an "interesting" biological problem. Either the former or (more frequently) the latter suffer from the combination.
Tue Mar 30 15:21:49 +0000 2021@nesvilab I found the paper & the associated web site to be very disappointing. No data yet, so maybe that will be better. Hopefully the final manuscript will be improved by the review process.
Tue Mar 30 15:03:03 +0000 2021The study is also a nice illustration of the odd segregation phenomena you can see when using HILIC fractionation on phospho-peptide enriched samples
Tue Mar 30 14:46:24 +0000 2021@michaelhoffman @jwoodgett As someone who has the misfortune of having to regularly deal with pharmacists & RNs because of an illness in the family, they have no particular insight into what is going on wrt current situation. If there is a loop, they are not in it.
Tue Mar 30 14:38:30 +0000 2021@michaelhoffman @jwoodgett Many Canadians don't have a personal physician. They rely of walk-in clinics and ERs. Even if they do have one, getting them on the phone is nearly impossible unless there is some type of emergency.
Tue Mar 30 14:35:14 +0000 2021There are very (very) few proteomics studies of meiosis (even mitosis isn't frequently studied).
Tue Mar 30 14:32:19 +0000 2021If you are interested in the combination of mouse phospho-peptides and TMT 6-plex, you should definitely check out PXD023803/MSV000086764. Also good for meiosis fans! 🐭
Tue Mar 30 13:18:13 +0000 2021Maybe it is just me, but once the signal peptide is removed, this sequence doesn't seem to be particularly "leucine rich".
Tue Mar 30 12:49:50 +0000 2021LRG1:p.P133S, chr 19:g.4538587G>A, rs966384, (blood plasma P:S = 12572:2028 14%) vaf=37%, Δm=-10.0207. VAF by population group: african 9%, european 36%, east asian 53%, south asian 29%, american 45%. #ᐯᐸᐱ
Tue Mar 30 12:49:50 +0000 2021LRG1:p, θ(max) = 74. aka LRG. Observed in MHC class 2 peptide experiments. Most commonly observed in extracellular vesicle fraction of blood plasma, urine & CSF. May be secreted by some cell lines, e.g. HepG2.
Tue Mar 30 12:49:50 +0000 2021>LRG1:p, leucine rich alpha-2-glycoprotein 1 (Homo sapiens) Small extracellular protein; CTMS: N79, N186, N325+glycosyl; PTMs: none; SAAVs: P133S (37%); mature form: (36-347) [19,650×, 280 kTa]. #ᗕᕱᗒ 🔗
Mon Mar 29 21:16:53 +0000 2021From the crashing barometer readings, it seems like something wicked is on its way 🔗
Mon Mar 29 18:45:50 +0000 2021@CharlieSpec @AlexHgO The bottle of picric acid that has been in the cupboard under the fume hood for at least 40 years because nobody wants to touch it.
Mon Mar 29 14:36:02 +0000 2021@OgilvieTamra Only 2 chilly days & then back to nicer weather
🔗
Mon Mar 29 13:18:12 +0000 2021@neely615 @ProteomicsNews But is super-duper-ultra deep OK? "Ultra deep" is so 2014 ...
Mon Mar 29 12:15:07 +0000 2021CFH:p.E936D, chr 1:g.196740644G>T, rs1065489, (blood plasma E:D = 14120:7198 33%) vaf=20%, Δm=-14.0157. VAF by population group: african 6%, european 17%, east asian 50%, south asian 14%, Finnish 14%. #ᐯᐸᐱ
Mon Mar 29 11:53:48 +0000 2021CFH:p, θ(max) = 86. aka HUS, FHL1, ARMS1, ARMD4, HF, HF1, HF2. Observed in MHC class 2 peptide experiments. Most commonly observed in blood plasma, urine & CSF.
Mon Mar 29 11:53:48 +0000 2021>CFH:p, complement factor H (H sapiens) CTMS: N217, N529, N718, N882, N911, N1029+glycosyl; PTMs: 69×S, 49×T, 3×Y+phosphoryl, 11×K+SUMOyl, 11×K+acetyl; SAAVs: V62L/I (53%), H402Y (77%), S437A (16%), E936D (14%); mature form: (19-1231) [16,770×, 203 kTa]. #ᗕᕱᗒ 🔗
Sun Mar 28 14:38:41 +0000 2021@slashdot No.
Sun Mar 28 14:08:32 +0000 2021@PMSC34891253 @pwilmarth What I find the most interesting about this sort of discussion is that even after more than 2 decades, the field does not have any coherent framework for understanding the role of mass accuracy in PSM assignment accuracy.
Sun Mar 28 12:36:29 +0000 2021PON1:p.L55M, chr 7:g.95316772A>T, rs854560, (blood plasma L:M = 6468:2623 29%) vaf=29%, Δm=17.9564. VAF by population group: african 15%, european 36%, east asian 3%, south asian 23%, american 21%. The LM or MM alleles are associated with enhanced enzyme function. #ᐯᐸᐱ
Sun Mar 28 12:36:29 +0000 2021PON1:p, θ(max) = 91. Observed in MHC class 2 peptide experiments. Most commonly observed in blood plasma & CSF. Prevents oxidation in lipo-protein particles. Translated in the liver & secreted as part of lipoprotein particles.
Sun Mar 28 12:36:29 +0000 2021>PON1:p, paraoxonase 1 (Homo sapiens) Small enzyme; CTMS: A2+acetyl; PTMs: K3+acetyl; SAAVs: L55M (29%), R160G (2%), Q192R (64%); mature form: (2-355) [16,770×, 203 kTa]. #ᗕᕱᗒ 🔗
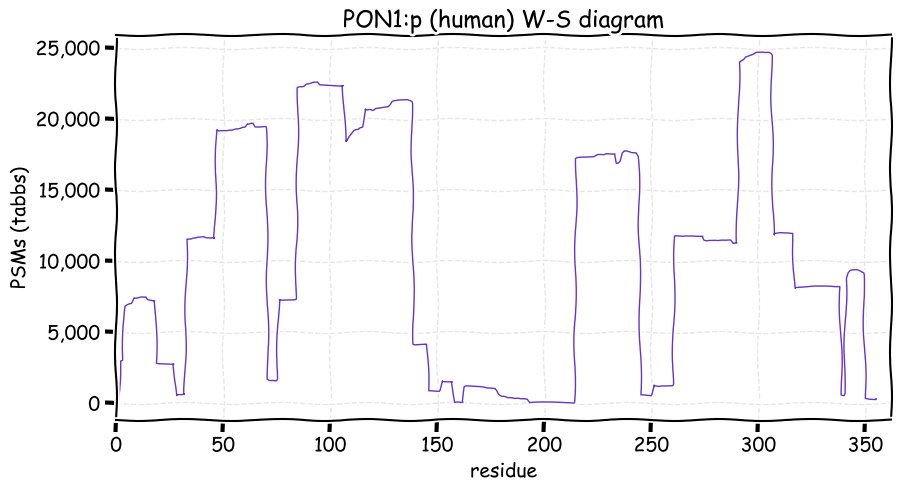
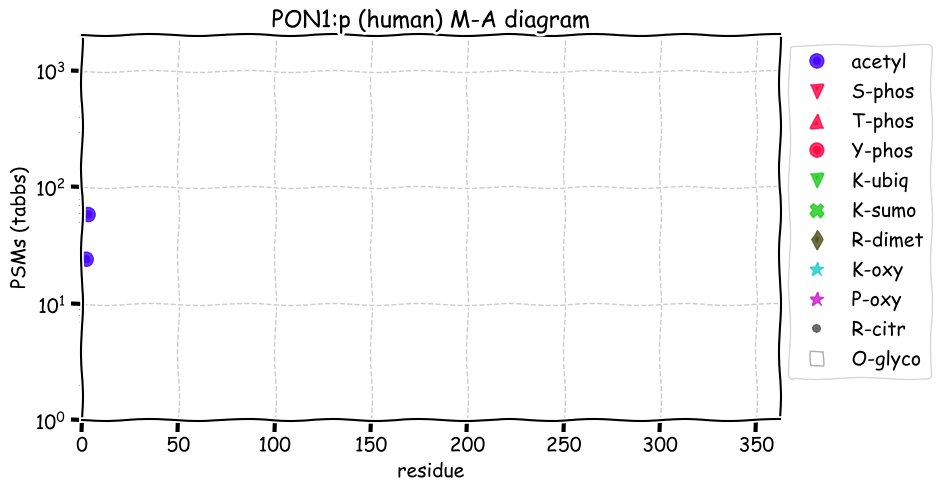
Sun Mar 28 02:32:40 +0000 2021Hope it stays clear tonight: it looks like the aurora is coming (I'm the blue dot) 🔗
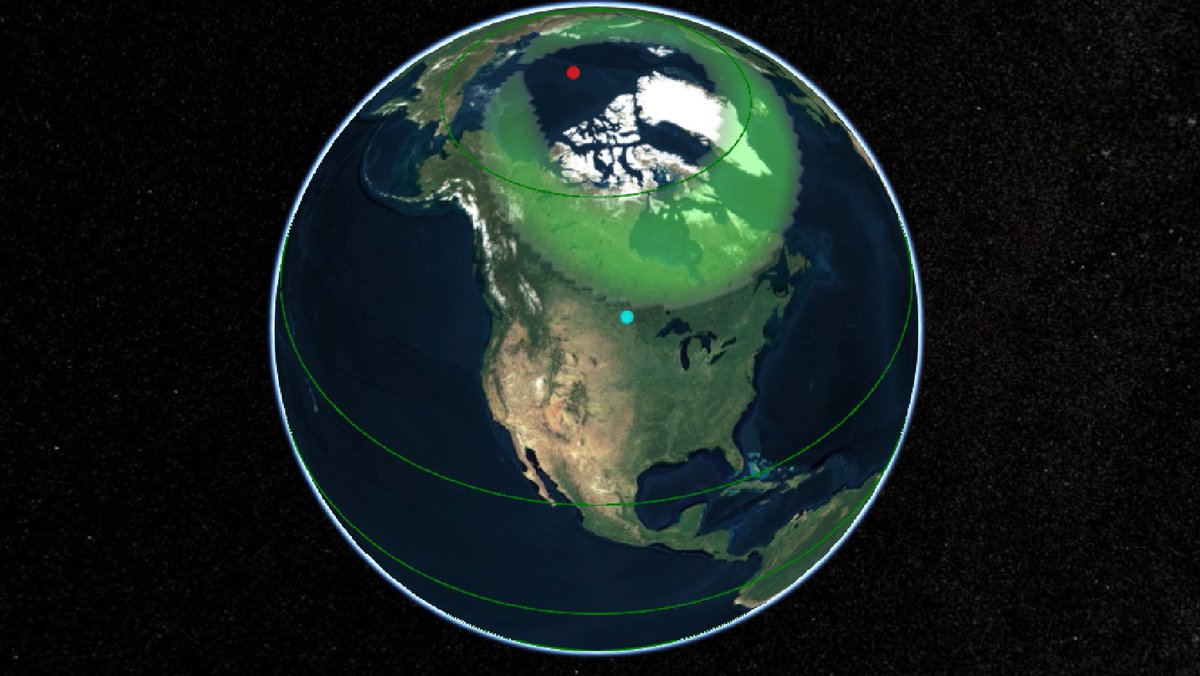
Sat Mar 27 15:16:00 +0000 2021@neely615 @pwilmarth I was right: the results are interesting!
Sat Mar 27 12:52:08 +0000 2021@NatureRevMicro @ShellyKalaora @samuels_yardena Has the data this manuscript used been made publicly available?
Sat Mar 27 12:46:38 +0000 2021Thanks to everyone who participated in this poll. It appears that there is still considerable skepticism regarding chat sessions as being a substitute for Ted-Talk style conference sales platforms.
Sat Mar 27 12:16:10 +0000 2021@MagnusPalmblad @neely615 @byu_sam 🔗 sounds authoritative.
Sat Mar 27 11:59:29 +0000 2021KLKB1:p.S381A, chr 4:g.186251858T>G, rs4253301, (blood plasma S:A = 3030:493 14%) vaf=12%, Δm=-15.9949. VAF by population group: african 5%, european 13%, east asian 7%, south asian 18%, american 11%. #ᐯᐸᐱ
Sat Mar 27 11:59:29 +0000 2021KLKB1:p, θ(max) = 85. Observed in MHC class 2 peptide experiments. Most commonly observed in blood plasma & CSF. Rarely observed in cell lines.
Sat Mar 27 11:59:29 +0000 2021>KLKB1:p, kallikrein B1 (Homo sapiens) Midsized extracellular enzyme; CTMS: N127, N308, N396, N453, N494+glycosyl; PTMs: T377, T384, T385+glycosyl; SAAVs: S143N (60%), T358A (1%), T361A (1%), S381A (12%), R560Q (23%); mature form: (20-638) [14,437×, 163 kTa]. #ᗕᕱᗒ 🔗
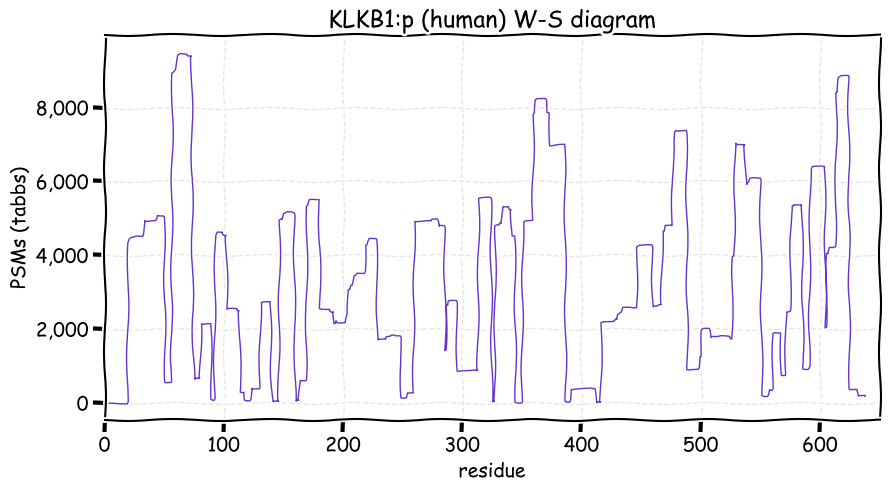
Fri Mar 26 19:35:17 +0000 2021@pwilmarth @YanYwang16 @neely615 A1+deamidation = 20%?
Fri Mar 26 15:08:36 +0000 2021@ProtifiLlc @JohnRYatesIII @pwilmarth Don't do it Phil. To quote a notable (but fictional) general "It's a trap!"
Fri Mar 26 14:40:50 +0000 2021@scottagerber @pwilmarth Using an LCQ Classic? Very few grad students or post-docs (or assistant profs) in the field will even know what that was, let alone how to apply the idea of "mass accuracy" to the results it would generate.
Fri Mar 26 13:42:18 +0000 2021@neely615 @pwilmarth I am interested to see what people think on this particular issue.
Fri Mar 26 13:04:41 +0000 2021@neely615 @byu_sam I agree: I think conferences have gone in a bad direction and they are due for replacement with something (anything) better.
Fri Mar 26 12:45:38 +0000 2021Given the frequency of SAAVs for this protein, it is unlikely to find an individual that is homozygous for the reference sequence.
Fri Mar 26 12:36:53 +0000 2021Will Clubhouse-style chat sessions replace (or at least supplement) conferences in science?
Fri Mar 26 12:04:26 +0000 2021SAA2:p.R89H, chr 11:g.18245480C>T, rs145801212, (all studies R:H = 949:3768) vaf=85%, Δm=-19.0422. VAF by population group: african 67%, european 83%, east asian 94%, south asian 71%, latin american 80%. Allele has been associated with cardiac disease. #ᐯᐸᐱ
Fri Mar 26 12:02:16 +0000 2021SAA2:p, θ(max) = 85. Observed in MHC class 1 & 2 peptide experiments. Most commonly observed in blood plasma & CSF as well as MCF-10A cell lines.
Fri Mar 26 12:02:16 +0000 2021>SAA2:p, serum amyloid A2 (Homo sapiens) Very small secreted protein; PTMs: none; SAAVs: S77T (99%), R89H (85%); mature form: (19-122) [8,097×, 5.5 kTa]. #ᗕᕱᗒ 🔗
Thu Mar 25 14:49:38 +0000 2021@Leili_Rohani More than one major scientific prize has been handed out to someone who claim-jumped an idea they heard at a seminar or read as a reviewer. Science just is (& always was) this way.
Thu Mar 25 14:47:10 +0000 2021@Leili_Rohani Can't be done: they just have to get used to the fact that once you've stated an idea out loud, it isn't yours any more.
Thu Mar 25 14:38:51 +0000 2021@kabalak @neely615 The coagulation (clotting) system is every bit as complex & involves even more proteolysis. The process of extracellular matrix building & re-modelling is no slouch either.
Thu Mar 25 14:19:59 +0000 2021@NLKProteomics @pwilmarth I guess spectrum library id algorithms would technically be classified as "deep learning" these days, but back when they were invented that terminology didn't exist.
Thu Mar 25 13:44:18 +0000 2021@NLKProteomics @pwilmarth It is often forgotten, but all PSM id algorithms are "expert systems", which was all the rage in AI research back in the day. It is pedestrian now, but without AI we'd still be figuring out peptide spectra by hand.
Thu Mar 25 13:40:14 +0000 2021@neely615 And for my next trick, if you are using Windows 10, try pressing ⊞+v (while holding down the Windows key press v)
Thu Mar 25 13:23:32 +0000 2021@neely615 If you are using Python & matplotlib.pyplot to draw graphs, then put matplotlib.pyplot.xkcd() in the code & it renders the graphs in a hand-drawn-appearing style, reminiscent of an xkcd cartoon.
Thu Mar 25 13:05:14 +0000 2021GC:p.D432E, chr 4:g.71752617A>C, rs7041, (PXD004242 D:E = 231:785) vaf=52%, Δm=14.0157. VAF by population group: african 9%, european 58%, east asian 30%, south asian 54%, american 54%. DD has been associated with reduced vitamin D levels in older adult smokers. #ᐯᐸᐱ
Thu Mar 25 12:12:23 +0000 2021@neely615 If you want to read some bang-your-head-against-a-wall type reviews, try proposing to do something about these anomalous sequence issues.
Thu Mar 25 12:09:02 +0000 2021GC:p, θ(max) = 95. aka DBP, VDBP, hDBP. Observed in MHC class 2 peptide experiments. Unusual proportion of T+phosphorylation acceptor sites. Given the SAAV distribution, it would be unlikely to find an individual that is homozygous for the reference sequence.
Thu Mar 25 12:09:02 +0000 2021>GC:p, GC vitamin D binding protein (Homo sapiens) Small protein; PTMs: 2×S, 25×T, 0×Y+phosphoryl, 11×Y+acetyl; SAAVs: D432E (52%), T436K (21%), H445R (99%); mature form: (17-474) [36,262×, 1060 kTa]. #ᗕᕱᗒ 🔗
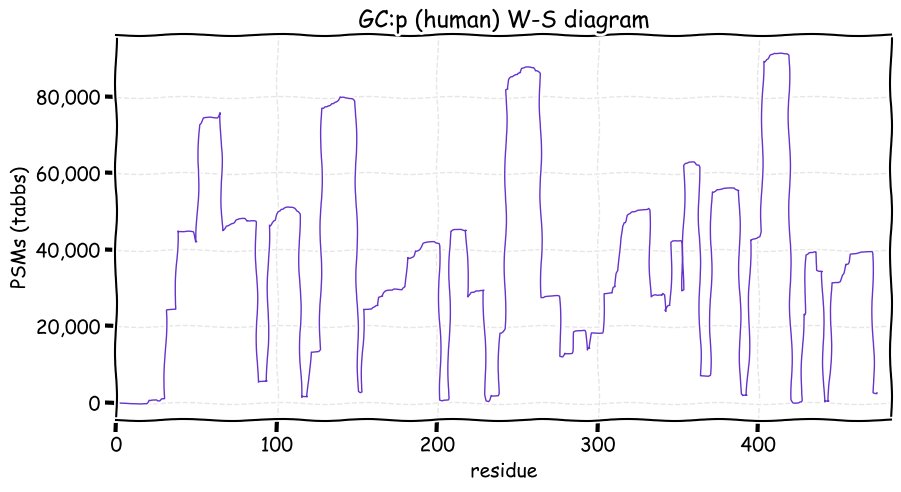
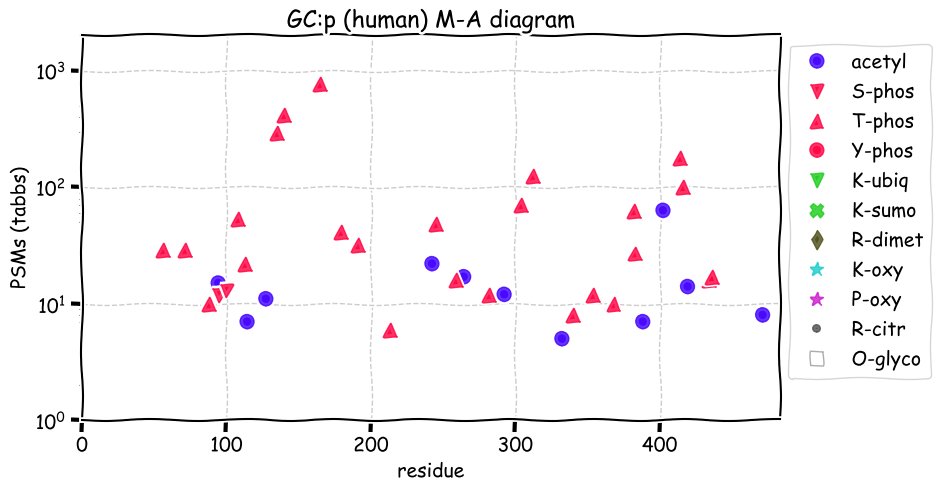
Thu Mar 25 12:03:40 +0000 2021@neely615 Sometimes the lack of interest in primary structure in proteomics still bugs me, but I have gotten used to it.
Thu Mar 25 12:02:42 +0000 2021@neely615 It is the opposite of immunoglobulins, that show up as dozens of genes, even though there are notionally only light and heavy subunits. Plasma proteins have quite a few features that don't fit in to the gene⇨protein⇨function
paradigm that works most of the time in cells.
Wed Mar 24 12:51:04 +0000 2021C3:p.P314L, chr 19:g.6713251G>A, rs1047286, (PXD004242 P:L = 1435:175) vaf=14%, Δm=16.0313. VAF by population group: african 1%, european 20%, east asian 0%, south asian 7%, american 9%. PL or LL carry increased risk of age-related macular degeneration. #ᐯᐸᐱ
Wed Mar 24 12:51:04 +0000 2021C3:p, θ(max) = 96. aka CPAMD1, ARMD9, C3a, C3b. Observed in MHC class 1 & 2 peptide experiments. Frequently observed scission at A667-R668. Biological significance of tyrosine phosphorylation as of yet unclear.
Wed Mar 24 12:51:04 +0000 2021>C3:p, complement C3 (Homo sapiens) Large protein; CTMS: N70, N107, N273+glycosyl; PTMs: 8×S, 10×T, 47×Y+phosphoryl, N85, S968+glycosyl; SAAVs: E469D (2%), P314L (14%), G1224D (1%); mature form: (23-1663) [59,888×, 4550 kTa]. #ᗕᕱᗒ 🔗
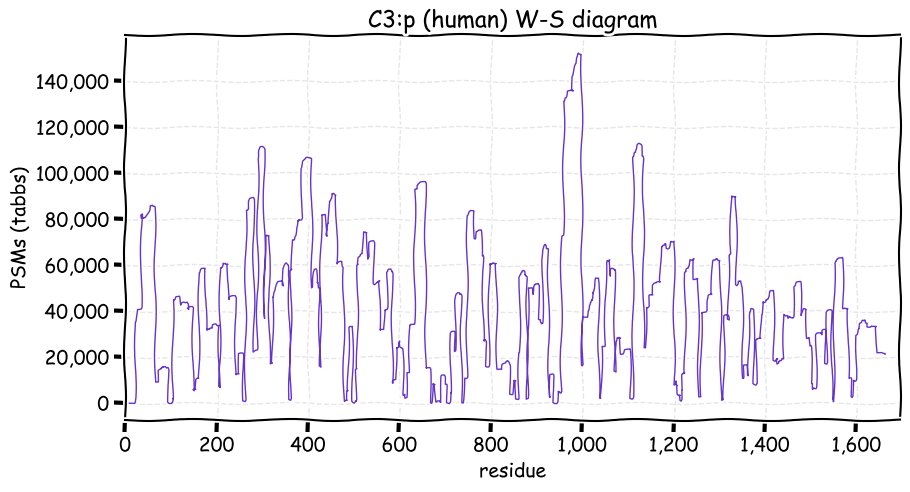
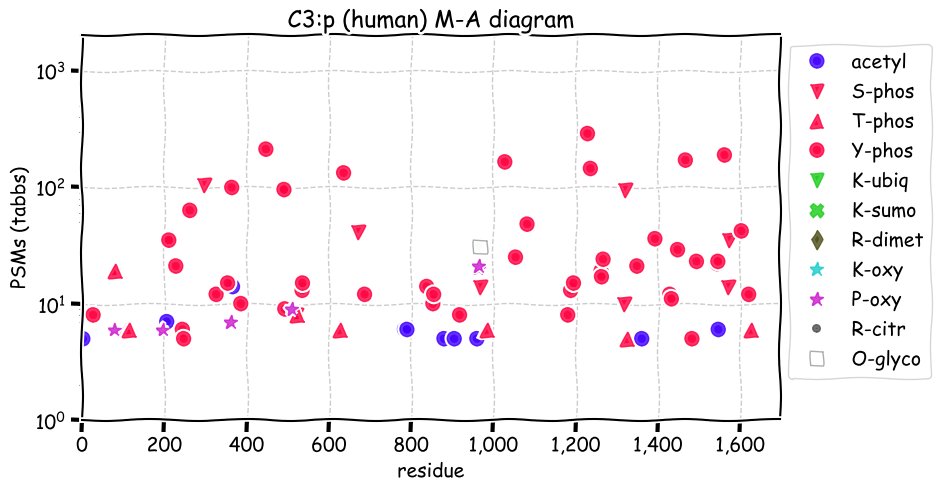
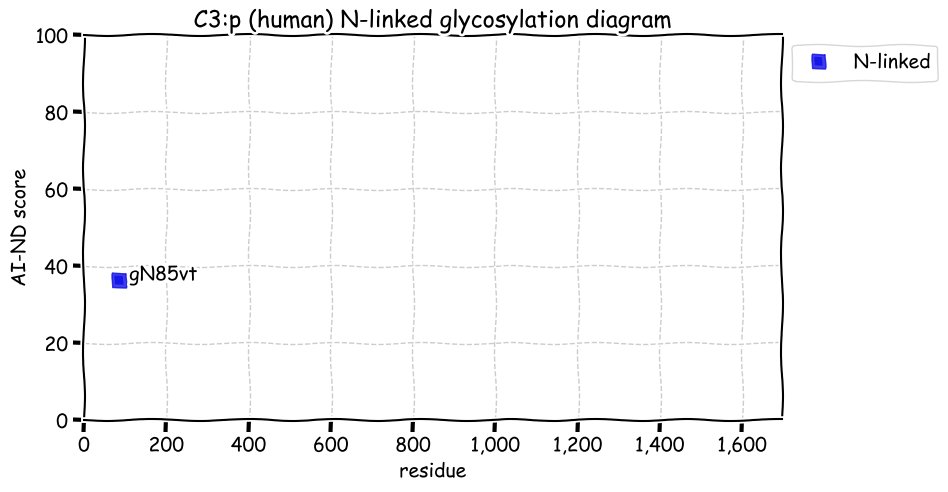
Tue Mar 23 14:11:28 +0000 2021@doctorow Canada is just as bad.
Tue Mar 23 14:07:26 +0000 2021I spent an enjoyable 5 minutes trying to get my phone to stop changing "palliative" to "Palpatine" in a text. I finally had to misspell is as "palitive" to get it to stop.
Tue Mar 23 12:57:49 +0000 2021@neely615 @pwilmarth @Smith_Chem_Wisc They can be used to create arrays, linked lists, objects, etc.: all of the usual CS fun. Search engines rarely use databases internally, but some do output directly into a database.
Tue Mar 23 12:46:37 +0000 2021@jvarga92 @neely615 @pwilmarth @Smith_Chem_Wisc Dr. Bairoch liked to edit SWISS-PROT with a text editor in which he could see all of the entries at once, rather than entry by entry. He also felt that databases were too slow for data retrieval (this is back in the 90's). He always referred to it as a databank.
Tue Mar 23 12:42:48 +0000 2021@neely615 @pwilmarth @Smith_Chem_Wisc database → FASTA → database is certainly possible for some (but not all) FASTA files, but the intermediate FASTA file isn't a database, in the same way that a MySQL dump file isn't a database (it is the instructions for making one).
Tue Mar 23 12:28:56 +0000 2021@neely615 @pwilmarth @Smith_Chem_Wisc The conversation (rather randomly) reminds me of the long tirades Amos Bairoch used to go into when someone accidently referred to SWISS-PROT as a "database", in which he would explain at length why it wasn't.
Tue Mar 23 12:11:58 +0000 2021SERPINA1:p.E400D, chr 14:g.94378506T>G, rs1303, (blood plasma E:D = 24822:6816, 21%) vaf=28%, Δm=-14.0157. VAF by population group: african 9%, european 25%, east asian 32%, south asian 48%, american 36%. #ᐯᐸᐱ
Tue Mar 23 12:11:58 +0000 2021SERPINA1:p, θ(max) = 88. aka AAT, A1A, PI1, alpha-1-antitrypsin, A1AT, alpha1AT, PI. Observed in MHC class 1 & 2 peptide experiments. Most abundant in clinical fluids, e.g., blood plasma & urine.
Tue Mar 23 12:11:58 +0000 2021>SERPINA1:p, serpin family A member 1 (Homo sapiens) Small protein; CTMS: N70, N107, N273+glycosyl; PTMs: 15×S, 12×T, 6×Y+phosphoryl; SAAVs: R125H (16%), V237A (75%), E288V (2%), E400D (28%),; mature form: (25-418) [46,601×, 2886 kTa]. #ᗕᕱᗒ 🔗
Mon Mar 22 23:46:00 +0000 2021@pwilmarth @Smith_Chem_Wisc The specification doesn't say anything about what has to be on the text description line: it doesn't have to contain any type of "key" or identifier. It is just a barely formatted text file. In no sense a database.
Mon Mar 22 23:42:30 +0000 2021@pwilmarth @Smith_Chem_Wisc The types of FASTA files you currently use come from databases so there is still the faint whiff of database about them. But the following is a perfectly well formed FASTA file:
>123
KIKIK
>123
KIQIK
>
TWITTER
Mon Mar 22 23:11:23 +0000 2021@pwilmarth @Smith_Chem_Wisc It is a database in the same sense that an ion source is a stapler.
Mon Mar 22 22:00:38 +0000 2021@Smith_Chem_Wisc Using "database" was a mistake made early on, incorrectly associating the FASTA file with the "database" it came from originally (& even that wasn't right for SWISS-PROT). It was fortunate though, as the use of the word in a key patent made open source dev possible.
Mon Mar 22 21:51:24 +0000 2021@Smith_Chem_Wisc I just call them sequence lists. Databanks is accurate but archaic now. Anything but "databases".
Mon Mar 22 16:33:49 +0000 2021@theoneamit Depends a lot on the experiment. Many expts. are just straight affinity enrichment (e.g. TiO2 for phosphopeptides), so comparing a sample run with & without enrichment would be a reasonable indicator.
Mon Mar 22 12:48:16 +0000 2021@rust_ruslan Such things used to exist: they were called monographs. They were usually longer than the oddly formulaic, short form manuscripts popular at the moment, but if they were any good they could enjoy a wide readership.
Mon Mar 22 12:41:49 +0000 2021I frequently see data in which the degree of enrichment is quite low, but it is enthusiastically (although imprecisely) touted in the associated manuscript.
Mon Mar 22 12:38:40 +0000 2021Reviewer request: if a manuscripts states that a sample has been enriched for some specific peptide feature (e.g. acetylated, phosphorylated or N-terminal peptides), please get them to provide some numerical measure of the degree of enrichment.
Mon Mar 22 12:09:58 +0000 2021APOH:p.W335S, chr 17:g.66212167C>G, rs1801690, (blood plasma W:S = 5890:731) vaf=5%, Δm=-99.0473. VAF by population group: african 0%, european 6%, east asian 5%, south asian 6%, american 3%. #ᐯᐸᐱ
Mon Mar 22 12:09:58 +0000 2021APOH:p, θ(max) = 76. aka BG, B2G1, Beta-2-glycoprotein 1. Observed in MHC class 2 peptide experiments. Most abundant in clinical fluids, e.g., blood plasma & urine.
Mon Mar 22 12:09:58 +0000 2021>APOH:p, apolipoprotein H (H sapiens) CTMS: N162, N183, N193, N253+glycosyl; PTMs: 17×S, 24×T, 0×Y+phosphoryl; 12×K+acetyl; SAAVs: N75S(1%), S107I(3%), I141T(7%), R154H(7%), A160D(1%), V266L(46%), C325G(2%), W335S (5%); mature form: (20,21-345) [30,575×, 638 kTa]. #ᗕᕱᗒ 🔗
Sun Mar 21 17:00:19 +0000 2021Who had the bright idea of calling the 1st sequenced version of SAVS CoV2 the "wild type"? I hope that all of the variants are also wild types, too.
Sun Mar 21 16:37:33 +0000 2021But I will freely admit that configuring BIND9 and Apache with a console text editor (nano in my case) is olden-times knowledge no longer commonly considered useful.
Sun Mar 21 15:55:12 +0000 2021Sometimes I still manage to impress myself with what you can with a few DNS and Apache configuration tricks
Sun Mar 21 13:51:36 +0000 2021The remnants of a G2 solar storm still lingering in the magnetosphere leading to a large area of aurora today 🔗
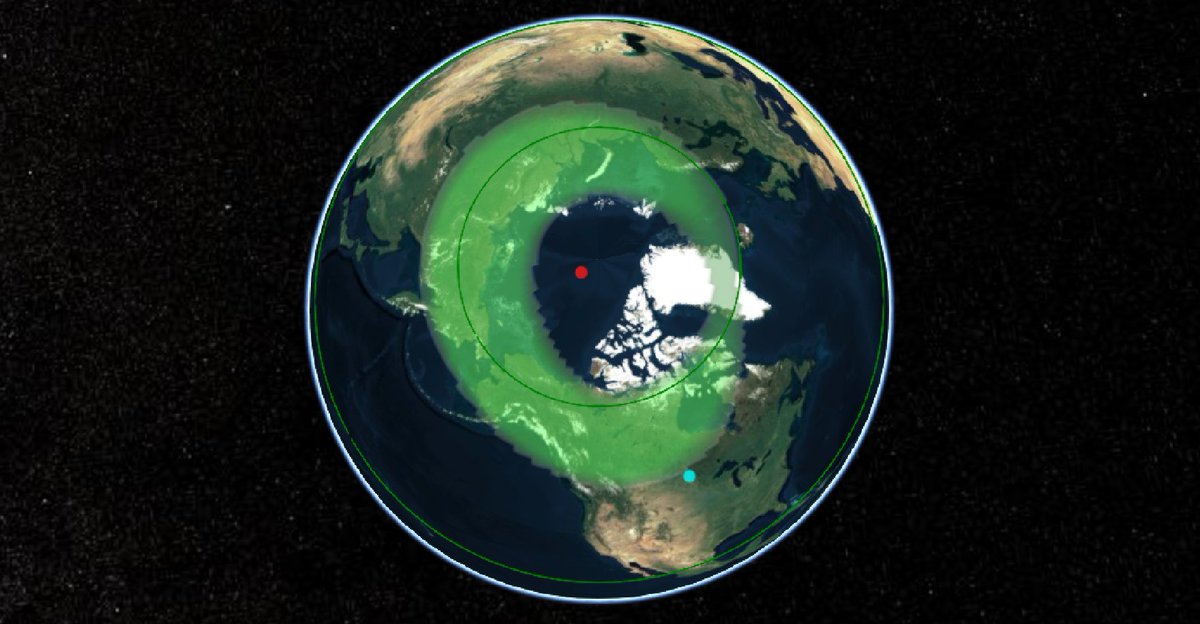
Sun Mar 21 13:35:49 +0000 2021@RohanMossBeach It is pretty nice looking code. The authors should get in there and do some source documentation while it is still fresh in their minds.
Sun Mar 21 13:17:09 +0000 2021The names for the allele states are APOE2 (C130+C176), APOE3 (C130+R176) & APOE4 (R130+R176). Homozygous APOE4 has a 91% risk of developing the disease. The status of this allele (& zygosity) is often available in plasma proteomics data.
Sun Mar 21 13:17:09 +0000 2021APOE:p.C130R, chr 19:g.44908684T>C, rs429358, (PXD004242 obs. 484×, PXD022817 obs. 32×) vaf=14%, Δm=53.0919. This SAAV & APOE:p.R176C form a polymorphic allele that can indicate risk of developing Alzheimer's disease. #ᐯᐸᐱ
Sun Mar 21 13:17:08 +0000 2021APOE:p, θ(max) = 92. Observed in MHC class 1 & 2 peptide experiments. Common in blood lipid particles & clinical fluids. Mainly translated in the liver, but it is not unusual to observe the protein in tissues & cells.
Sun Mar 21 13:17:08 +0000 2021>APOE:p, apolipoprotein E (Homo sapiens) Small subunit; PTMs: 2×S, 1×T, 0×Y+phosphoryl; K19+acetyl; T36, T212, S215, T307, S308, S314+glycosyl; SAAVs: C130R (14%), R163C (1%), R176C (8%); mature form: (19-317) [37,297×, 595 kTa]. #ᗕᕱᗒ 🔗
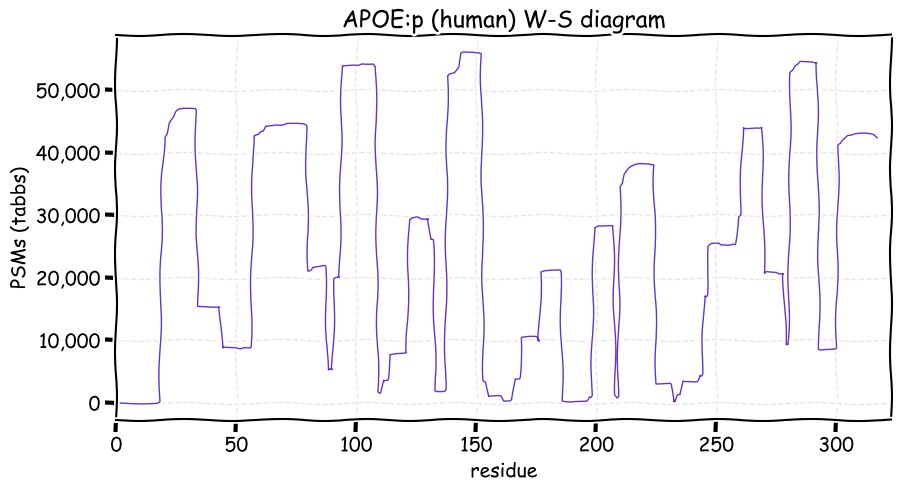
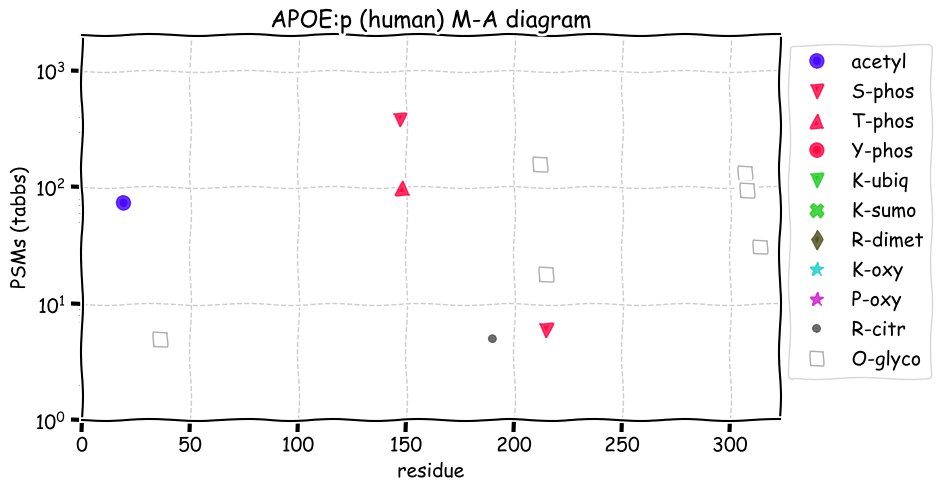
Sat Mar 20 13:50:25 +0000 2021If you ever what to explain what "disingenuous" means wrt academic discourse, refer the student to this article:
🔗
Sat Mar 20 12:01:42 +0000 2021APOA4:p.S147N, chr 11:g.116821618C>T, rs5104, (blood plasma A:V = 2361:5330) vaf=80%, Δm=27.0109. VAF by population group: african 89%, european 83%, east asian 64%, south asian 63%, american 76%. #ᐯᐸᐱ
Sat Mar 20 12:01:42 +0000 2021APOA4:p, θ(max) = 96. Observed in MHC class 1 & 2 peptide experiments. Common in blood lipid particles & clinical fluids. Translated in the intestine only, but it is not unusual to observe the protein in tissues & cells.
Sat Mar 20 12:01:42 +0000 2021>APOA4:p, apolipoprotein A4 (Homo sapiens) Small subunit; PTMs: 8×K+acetyl; SAAVs: S147N (80%), S353A (1%), T367S (10%), Q380H (2%); mature form: (19,20,21-398) [25,767×, 781 kTa]. #ᗕᕱᗒ 🔗
Fri Mar 19 19:48:49 +0000 2021@cwvhogue I've switched over to using these guys for fans:
🔗
Fri Mar 19 14:02:49 +0000 2021@neely615 @ideaproteomics To be a bit more clear, I do mean CROs that provide B2B services rather than govt-funded labs.
Fri Mar 19 13:17:31 +0000 2021Does anybody know if there are any CROs that could do this sort of thing anymore? There were quite a few at one point, but I think most of them have disappeared. All of the startup activity seems to be around developing methods rather than using them. 🔗
Fri Mar 19 12:10:38 +0000 2021HAUS6:p, is one of 8 subunits of the HAUS complex. These proteins have many K+ubiquitinyl sites, but (with the exception of HAUS4:p) there is no indication of alternating K+acetyl at the acceptor sites.
Fri Mar 19 11:52:16 +0000 2021HAUS6:p.A615V, chr 9:g.19058923G>A, rs62622380, (HCT-116 A:V = 9:9) vaf=12%, Δm=28.0313, is AV in HCT-116 cells. It is VV in Hep-2 cells. It is AA in JURKAT, U2-OS & HEK-293 cells. #ᐯᐸᐱ
Fri Mar 19 11:52:16 +0000 2021HAUS6:p, θ(max) = 62. aka FLJ20060, KIAA1574, dgt6, FAM29A. Observed in MHC class 1 peptide experiments. Common in cellular tissues and cell lines, rare in clinical fluids.
Fri Mar 19 11:52:16 +0000 2021>HAUS6:p, HAUS augmin like complex subunit 6 (Homo sapiens) Midsized subunit; CTMs: S2+acetyl; PTMs: 31×S, 8×T, 2×Y+phosphoryl; 29×K+ubiquitinyl; SAAVs: R322C (12%), S552T (2%), A615V (12%), H674Q (8%); mature form: (2-955) [12,824×, 52 kTa]. #ᗕᕱᗒ 🔗
Fri Mar 19 00:53:14 +0000 2021@MHendr1cks High risk innovation, yes. High risk accounting, no.
Thu Mar 18 20:49:06 +0000 2021@karthikskamath 🔗
Thu Mar 18 19:14:18 +0000 2021@ProtifiLlc The LC gradient takes 45 minutes.
Thu Mar 18 18:52:33 +0000 2021I'm looking at a data set of 1300 LC/MS/MS Q-Exactive DDA runs, 25,000 scans in each. Does anybody have any idea how much that would cost at normal facility rates & how long it would take to complete?
Thu Mar 18 13:08:15 +0000 2021It still amazes me how often failed experiments get written up as though they worked in the manner intended.
Thu Mar 18 12:52:08 +0000 2021TEX264:p.G292E, chr 3:g.51703949G>A, rs11553574, (HEK-293 G:A = 383:202) vaf=12%, Δm=72.0211, is GE in HEK-293, HeLa & RKO cells. It is GG in K-562, HAP-1 & A-549 cells. #ᐯᐸᐱ
Thu Mar 18 12:52:07 +0000 2021TEX264:p, θ(max) = 86. aka ZSIG11, FLJ13935. (17-31) transmembrane domain. Common in cellular tissues and cell lines, rare in clinical fluids.
Thu Mar 18 12:52:07 +0000 2021>TEX264:p, testis expressed 264 (Homo sapiens) Small subunit; PTMs: 20×S, 3×T, 1×Y+phosphoryl; 6×K+ubiquitinyl; SAAVs: G292E (12%), L302P (1%); mature form: (1-313) [12,411×, 42 kTa]. #ᗕᕱᗒ 🔗
Wed Mar 17 16:57:18 +0000 2021@IvisonJ For me, the issue is that the party leaders all rule by fiat at the moment, leaving the rest of the party to simply agree or be seen as "rebellious".
Wed Mar 17 15:40:58 +0000 2021@MattWFoster An optical quantum computer is like any other type of laser set up with discrete components: everything is screwed down onto a table covered with tapped holes.
Wed Mar 17 14:55:33 +0000 2021Thank you very much to everyone who participated in this poll. The results show that of people with an opinion, 75% believe checking for isoforms caused by alternate splicing is worth while. 25% do not share this belief.
Wed Mar 17 12:27:20 +0000 2021KRT7:p.G364A, chr 12:g.52245518G>C, rs2608009, (HeLa G:A = 0:293) vaf=84%, Δm=14.0157, is AA in HeLa, PC-3, Hep-2, MDA-MB-468 & BT-474 cells. It is GG in A-549 cells. #ᐯᐸᐱ
Wed Mar 17 12:27:20 +0000 2021KRT7:p, θ(max) = 92. aka K7, CK7, K2C7, SCL. 22 K+acetyl modification sites are shared ubiquitinyl or SUMOyl acceptors.
Wed Mar 17 12:27:19 +0000 2021>KRT7:p, keratin 7 (Homo sapiens) Small subunit; CTMS: S2+acetyl; PTMs: 44×S, 17×T, 9×Y+phosphoryl; 20×K+ubiquitinyl; 25×K+acetyl; 8×SUMOyl; SAAVs: F17L (1%), H186R (87%), A262T (2%), R313W (1%), G364A (84%), R464H (2%); mature form: (2-469) [125,713×, 1697 kTa]. #ᗕᕱᗒ 🔗
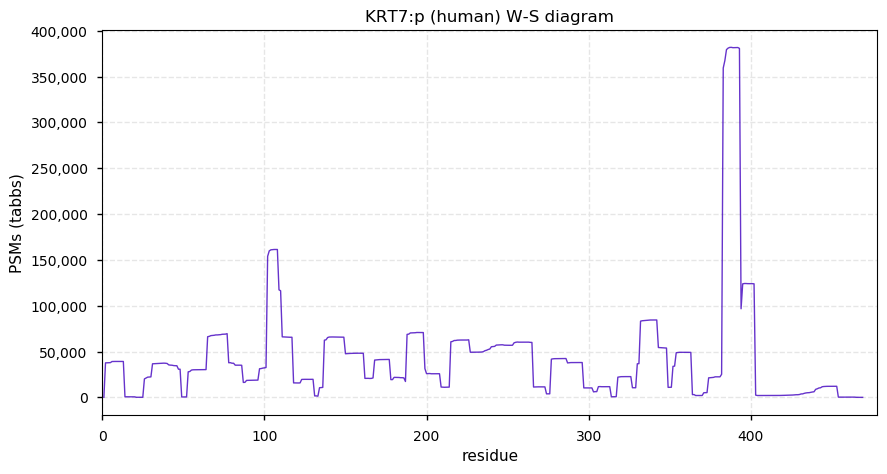
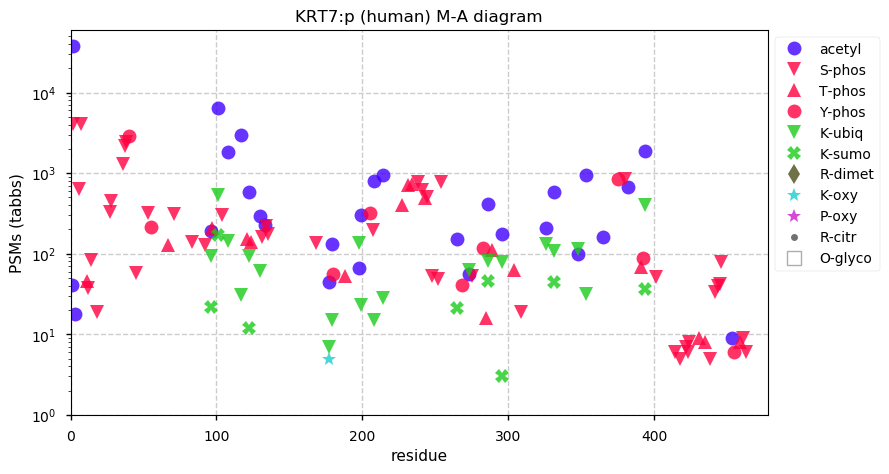
Tue Mar 16 17:30:01 +0000 2021@pwilmarth @dtabb73 So that's a "no"?
Tue Mar 16 16:21:08 +0000 2021@michaelhoffman It detracts a bit from the message when the domain 🔗 is unreachable because of a badly configured security certificate.
Tue Mar 16 15:20:47 +0000 2021@dtabb73 I have heard loud voices on both sides of this particular issue, but I have no idea how many people believe its utility outweighs its added complexity.
Tue Mar 16 14:47:16 +0000 2021Do you believe that there is any practical reason to routinely analyze MS-based proteomics data to ID isoform protein sequences caused by alternate RNA splicing?
Tue Mar 16 12:29:57 +0000 2021BIN1:p.T363M, chr 2:g.127050500G>A, rs112318500, (HeLa T:M = 765:1047) vaf=0.7%, Δm=29.9928, is TM in HeLa cells. It is TT in all other common cell lines. #ᐯᐸᐱ
Tue Mar 16 12:29:57 +0000 2021BIN1:p, θ(max) = 77. aka SH3P9, AMPH2, AMPHL. Found in HLA peptide class 1 (& rarely class 2) experiments. Common in cellular tissues, blood cells & common cell lines: rare in clinical fluids.
Tue Mar 16 12:29:57 +0000 2021>BIN1:p, bridging integrator 1 (Homo sapiens) Small subunit; CTMS: M1,A2+acetyl; PTMs: 13×S, 10×T, 0×Y+phosphoryl; 7×K+ubiquitinyl; 4×K+acetyl; SAAVs: T363M (0.7%); mature form: (2-424) [23,145×, 197 kTa]. #ᗕᕱᗒ 🔗
Mon Mar 15 15:16:22 +0000 2021@realsarahpolley It is a year in which "box office returns" were pretty much taken out of the equation.
Mon Mar 15 14:52:57 +0000 2021@pwilmarth @neely615 Excel is the spork of data analysis
Mon Mar 15 14:41:07 +0000 2021If anyone wants some piece of subcellular machinery to focus on to try to understand ubiquitin tagging, I nominate "The Condensin Complex"
🔗
Mon Mar 15 13:32:54 +0000 2021@lenjf Fortunately, none of this matters for we Canadians, as the possibility of the Canadian/provincial government getting involved in this type of thing is even lower than your estimated identification probability. The NIH is likely to change their policy, but CPTAC will fight it.
Mon Mar 15 12:19:06 +0000 2021LETM1:p.K587R, chr 4:g.1816898T>C, rs115233072, (HeLa K:R = 340:232) vaf=1%, Δm=28.0061, is KR in HeLa cells. It is RR in Hep-2 cells. It is KK in JURKAT, HEK-293, HCT-116 & most common cell lines. #ᐯᐸᐱ
Mon Mar 15 12:19:05 +0000 2021LETM1:p, θ(max) = 61. aka SLC55A1. Found in HLA peptide class 1 experiments only. (1-115) is a mitochondrial targeting domain that is removed upon entry to the organelle.
Mon Mar 15 12:19:05 +0000 2021>LETM1:p, leucine zipper and EF-hand containing transmembrane protein 1 (H sapiens) PTMs: 21×S, 0×T, 2×Y+phosphoryl; 6×K+ubiquitinyl; 45×K+acetyl; SAAVs: I82V(1%), N131K(1%), K587R(1%), R499H(1%), E665D(1%), K697R(1%); mature form: (116-739) [33,482×, 326 kTa]. #ᗕᕱᗒ 🔗
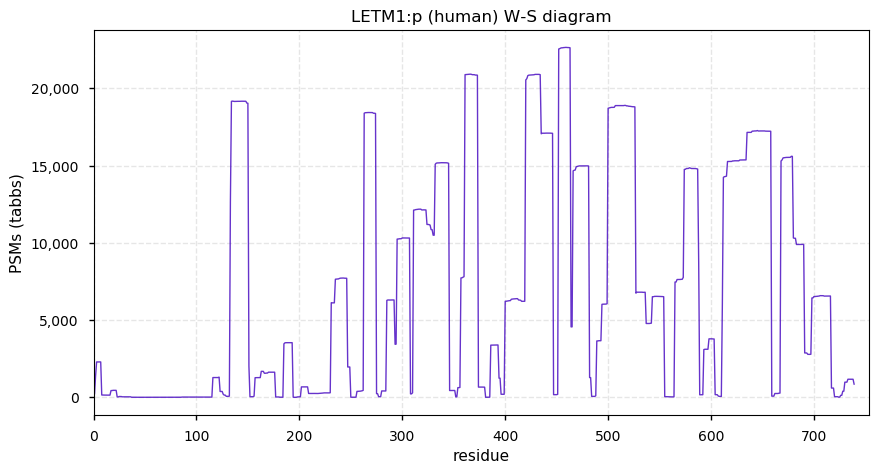
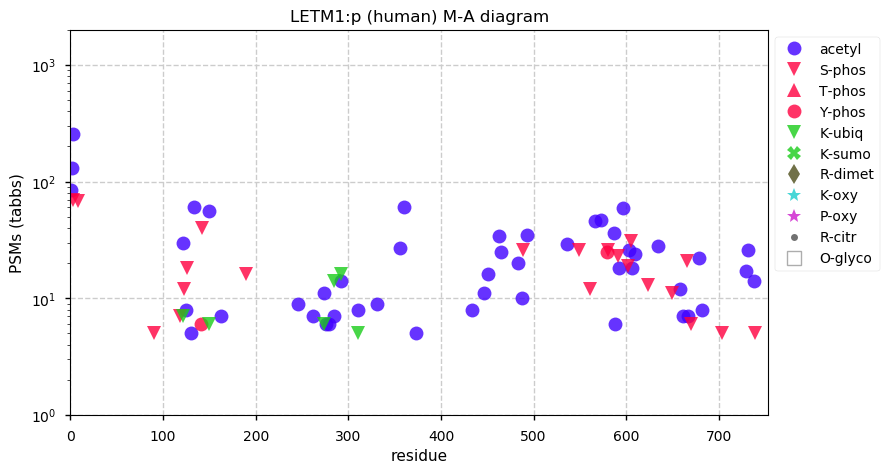
Sun Mar 14 21:38:51 +0000 2021@ypriverol But it hasn't "held back" genomics in any way, it simply leads to genomics repos being part of the larger discussion of how to use this new data in a beneficial manner. Proteomics repos aren't in that mix at the moment, even though they should be.
Sun Mar 14 21:27:50 +0000 2021@ypriverol Can't see how it could hold anything back. If anything, being ready for when the NIH (& other authorities) are forced to account for this reality should be a real advantage for informatics types, as they will need to provide funding for the additional repos.
Sun Mar 14 18:02:13 +0000 2021@neely615 @UCDProteomics That being said, IMO proteomics data from individual patients should not be stored in repositories that are not compliant with the regulations currently in use for genomic data.
Sun Mar 14 16:33:03 +0000 2021Thanks to everyone who participated in this poll. I am very pleased (& genuinely surprised) by the result: the majority of respondents believe that proteomics data can be used to identify an individual person.
Sun Mar 14 13:57:27 +0000 2021@mjmaccoss @MattWFoster Every blood/plasma/serum data set in GPMDB shows the status of the two SAAVs that determine the APOE status rs7412 (p.R176C) & rs429358 (p.C130R), if they were found in the data. These SAAVs are particularly easy to detect, as they add or subtract a tryptic cleavage site.
Sun Mar 14 13:15:25 +0000 2021DMXL2:p.S1288P, chr 15:g.51499362A>G, rs12102203, (HeLa S:P = 51:28) vaf=49%, Δm=10.0207, is SP in HeLa & MCF-10A cells. It is PP in JURKAT, HEK-293 & HCT-116 cells. It is SS in MCF-7 cells. #ᐯᐸᐱ
Sun Mar 14 13:15:25 +0000 2021DMXL2:p, θ(max) = 43. aka RC3, KIAA0856, DFNA71. Found in HLA peptide class 1 experiments only. Most commonly found in brain tissue. Too few GO annotations.
Sun Mar 14 13:15:25 +0000 2021>DMXL2:p, Dmx like 2 (Homo sapiens) Large subunit; PTMs: 67×S, 21×T, 2×Y+phosphoryl; 2×K+ubiquitinyl; 15×K+acetyl; SAAVs: T497M (13%), P956T (1%), S1287I (1%), S1288P (49%), D1481G (1%); mature form: (1-3036) [13,747×, 67 kTa]. #ᗕᕱᗒ 🔗
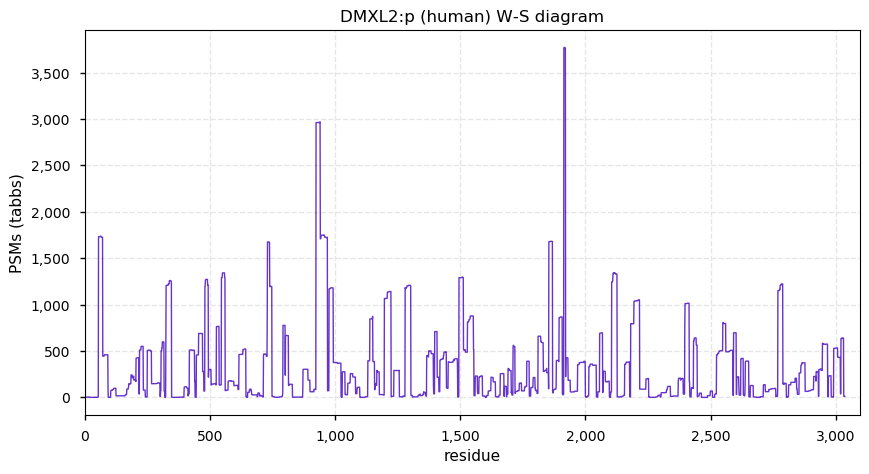
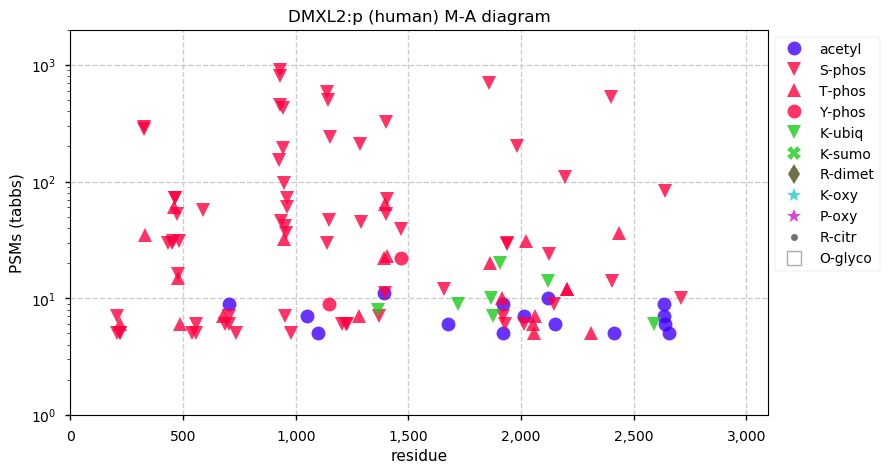
Sat Mar 13 16:30:28 +0000 2021Do you believe that MS-based human proteomics data can provide enough information to identify an individual?
Sat Mar 13 15:25:22 +0000 2021Are there any attendance figures for US HUPO available yet?
Sat Mar 13 14:49:10 +0000 2021@neely615 @UCDProteomics And because of the way it uses TMT10 encoding for individuals, CPTAC data wouldn't count as identifiable either (it is equivalent to a pooled sample)
Sat Mar 13 14:41:17 +0000 2021@neely615 @UCDProteomics It wouldn't change repositories that much, but it would take some effort. The majority of repository data is from cell lines, pooled samples & non-human sources that wouldn't be affected. There are really very few that use clinical samples from individuals.
Sat Mar 13 14:25:36 +0000 2021However one thing that was not mentioned is that if you are simply using the reference proteome for your PSM identifications, you are still detecting SAAVs, as many reference alleles are actually minor alleles in the human population.
Sat Mar 13 14:21:21 +0000 2021I think this is a useful starting point for the discussion 🔗
Sat Mar 13 14:00:02 +0000 2021XPC:p.A499V, chr 3:g.14158387G>A, rs2228000, (HeLa A:V = 121:131) vaf=24%, Δm=28.0313, is AV in HeLa, SH-SY5Y, JURKAT & MCF-7 cells. It is VV in CACO-2 & HEp-2 cells. It is AA in MCF-10A, A-431, A-549 & HCT-116 cells. #ᐯᐸᐱ
Sat Mar 13 13:50:07 +0000 2021θ(max) = 55. aka XPCC, RAD4. Found in HLA peptide class 1 experiments only. Commonly observed in cellular tissues and cell lines. Not detectable in clinical fluids.
Sat Mar 13 13:50:06 +0000 2021>XPC:p, XPC complex subunit, DNA damage recognition and repair factor (H sapiens) Midsized subunit; PTMs: 44×S, 11×T, 0×Y+phosphoryl; 17×K+ubiquitinyl; 4×K+acetyl; SAAVs: L48F (1%), P334R (1%), R492H (2%), A499V (24%); mature form: (2?-940) [13,747×, 76 kTa]. #ᗕᕱᗒ 🔗
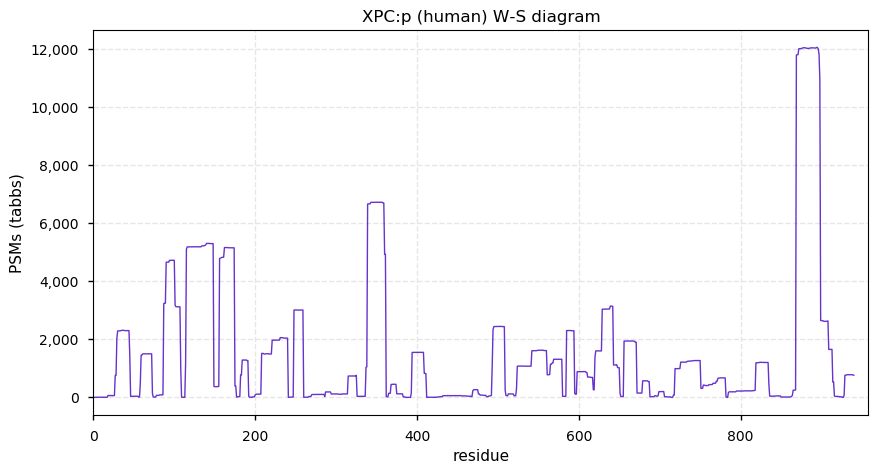
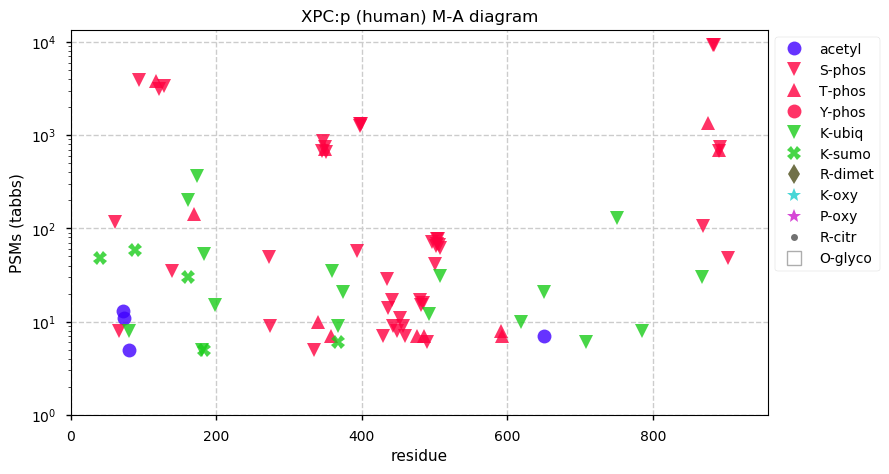
Fri Mar 12 18:45:54 +0000 2021@ianfroese & now we know why this change occurred
🔗
Fri Mar 12 18:39:10 +0000 2021@harrisonspecht Find some clinical chemists to talk to. Do not (& I cannot emphasize this enough) talk directly to docs.
Fri Mar 12 18:33:45 +0000 2021@stephen_taylor So this is the post-"buck-a-beer" phase of Ontario governance?
Fri Mar 12 18:17:52 +0000 2021@byu_sam @pride_ebi PXD006932 has some pretty good HeLa cell lysate QE-HF data & it is newish (2017). PXD011163 is newer (2020) , uses QE or Fusion. It is a bit more complicated (you should add in gi|130416| for the ids), but it is essentially a HeLa lysate.
Fri Mar 12 17:59:30 +0000 2021@byu_sam @pride_ebi What they are doing looks a lot more like QA to me ...
Fri Mar 12 16:26:07 +0000 2021@ianfroese So the release will be a "fait accompli".
Fri Mar 12 14:18:42 +0000 2021@jwoodgett Unfortunately, to be practical for America/Toronto to switch, you would have to get America/Montreal to go along, which is never going to happen so long as Toronto is proposing it.
Fri Mar 12 13:46:40 +0000 2021@jwoodgett You could always move to Atikokan: Ontario has 3 time zones & the America/Atikokan time zone does not observe daylight savings time. The other 2
(America/Toronto & America/Winnipeg) use DST.
Fri Mar 12 13:08:23 +0000 2021RIF1:p is an example of a protein in which its primary structure variability will make it challenging for most protein abundance measurement technologies.
Fri Mar 12 12:56:43 +0000 2021RIF1:p.V1362M, chr 2:g.151463604G>A, rs2123465, (HEK-293 V:M = 108:151) vaf=67%, Δm=31.9721, is VM in HEK-293 & MCF-7 cells. It is MM in HeLa, A-549 & MCF-10A cells. It is VV in JURKAT cells. #ᐯᐸᐱ
Fri Mar 12 12:56:43 +0000 2021RIF1:p, θ(max) = 50. aka FLJ12870, FLJ10599. Found in HLA peptide class 1 experiments only. Commonly observed in cellular tissues and cell lines. Not detectable in clinical fluids.
Fri Mar 12 12:56:43 +0000 2021>RIF1:p, replication timing regulatory factor 1 (H sapiens) CPTMs: M1, T2, A3+acetyl; PTMs: 136×S, 57×T, 11×Y+phosphoryl; 16×K+ubiquitinyl; 9×K+acetyl; SAAVs: G836S(64%), V1362M(67%), V1862I(7%), N2021Y(66%), L2418V(66%); mature: (1,2,3-2472) [28,814×, 272 kTa]. #ᗕᕱᗒ 🔗
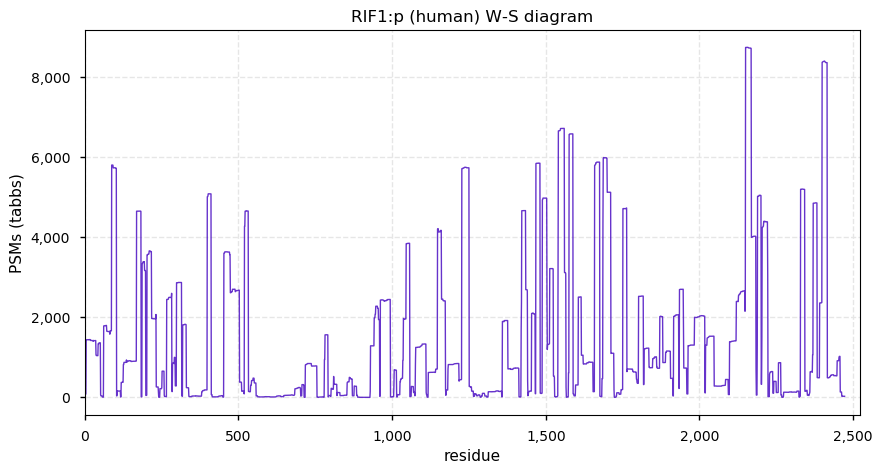
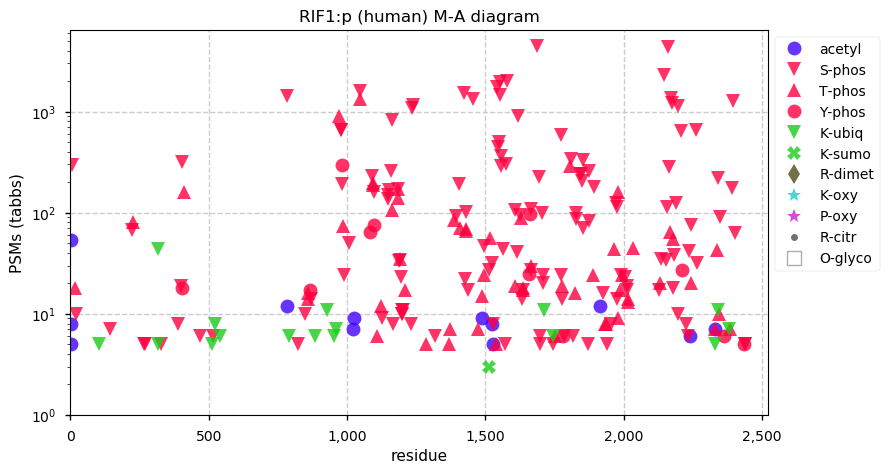
Thu Mar 11 21:37:47 +0000 2021@mjmaccoss Thanks, Mike. I read that one when it came out. It was just idle curiosity on my part.
Thu Mar 11 21:28:01 +0000 2021"CSF_Pool_Pool_Pool_Pool_Pool_Pool_01_Plate01.raw" may be my new favorite file name (from PXD009589).
Thu Mar 11 16:19:09 +0000 2021@dtabb73 The way I normally do it, I test for all SAAVs where the population vaf > 0.01%, so for any thorough study you usually end up with enough detected that are in the vaf<10% range to distinguish an individual.
Thu Mar 11 16:12:04 +0000 2021@dtabb73 Probably. You might need 50 or 100 (but there are lots to choose from).
Thu Mar 11 16:08:38 +0000 2021@dtabb73 Actually, all you have to do is select a basket of SAAVs from dbSNP that are present in abundant proteins & have a population frequency in the 5-10% range. A basket of 20 does a pretty good job of identifying an individual, particularly if any geographic info is attached.
Thu Mar 11 15:53:22 +0000 2021@MattWFoster I tried collaborating on an SAAV detection system some time ago, but my collaborators simply didn't get why encryption and good de-identification were necessary parts of the project.
Thu Mar 11 15:39:30 +0000 2021@MattWFoster You aren't wrong, but it is one of those issues that people would rather not address.
Thu Mar 11 15:30:37 +0000 2021I guess either nobody knows or it is a closely-held secret.
Thu Mar 11 14:45:54 +0000 2021@EUbOPEN @RockefellerUniv @scrippsresearch @CmdOxford Old ideas die hard.
Thu Mar 11 12:53:07 +0000 2021TSEN54:p.E4D, chr 17:g.75516572G>T, rs7216673, (HEK-293 E:D = 3:194) vaf=76%, Δm=-14.0157, is DD in HEK-293 & JURKAT cells. It is ED in HeLa, MCF-7 & A-549 cells. It is EE in HCT-116 cells. #ᐯᐸᐱ
Thu Mar 11 12:53:07 +0000 2021TSEN54:p, θ(max) = 58. aka SEN54, SEN54L. Found in HLA peptide class 1 experiments. Common in T-cells, breast cancer tissue & many cell lines.
Thu Mar 11 12:53:07 +0000 2021>TSEN54:p, tRNA splicing endonuclease subunit 54 (H sapiens) CPTMs: M1+acetyl; PTMs: 11×S, 0×T, 0×Y+phosphoryl; K489, K92+ubiquitinyl; K215, K340+SUMOyl; SAAVs: E4D (76%), H38Q (16%), Q389P (7%), A437V (44%), G525R (6%); mature form: (1-526) [6,342×, 16 kTa]. #ᗕᕱᗒ 🔗
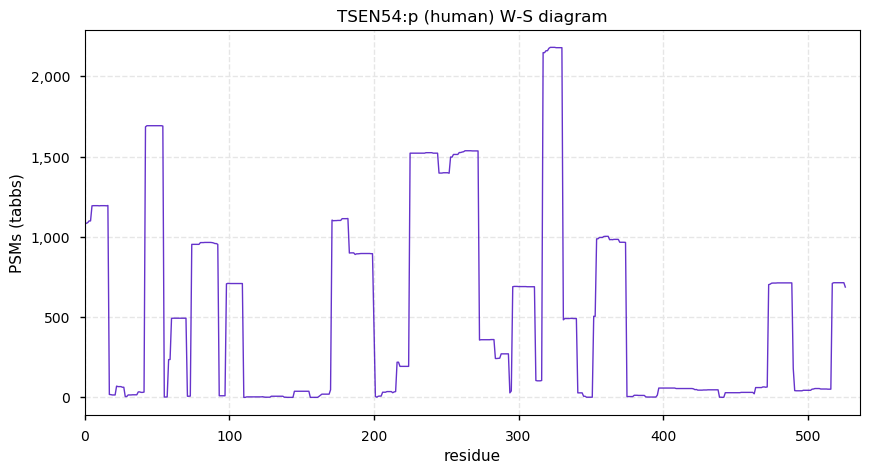
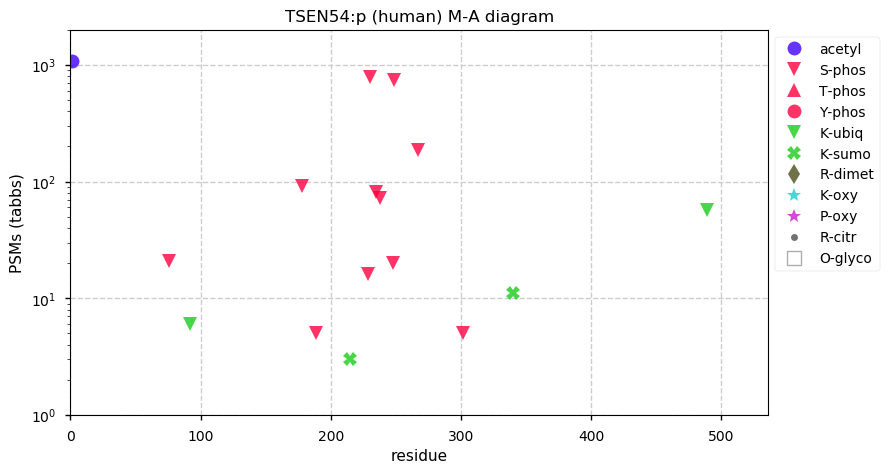
Wed Mar 10 17:49:44 +0000 2021@VDBiology @BiswapriyaMisra @EvosepBio @BrukerMassSpec @RalserLab @nesvilab But, it would be foolish to discount Thermo's formidable marketing operation: they have prospered in the face of competitors with technically superior equipment before.
Wed Mar 10 17:37:53 +0000 2021@VDBiology @BiswapriyaMisra @EvosepBio @BrukerMassSpec @RalserLab @nesvilab It would appear that Thermo (which has been engaged in a decade long, self-congratulatory victory dance wrt the orbitrap) has been out-hustled by Bruker. 😀
Wed Mar 10 15:51:17 +0000 2021This is for curiosity only: I am not looking to get into the DIA business. The data set I'm looking at is coming in at ~30%.
Wed Mar 10 15:40:30 +0000 2021What fraction of these sets of fragment ions have enough peaks to positively ID at least 1 peptide using a spectrum library ID algorithm alone?
Wed Mar 10 15:35:37 +0000 2021I'll try this again, hopefully wording it better.
Using a QExactive HF for DIA, you will obtain sets of fragment ions resulting from either
1. composites of multiple peptide parent ions,
2. one peptide parent &
3. no peptide parent. 1/2
Wed Mar 10 13:11:43 +0000 2021CBR4:p.L70M, chr 4:g.169007691G>T, rs2877380, (HeLa L:M = 0:118) vaf=77%, Δm=17.9564, is MM in HeLa, HCT-116 & HEK-293 cells. It is LM in JURKAT, MCF-7, LNCAP & MCF-10A cells. It is LL in A-549 cells. #ᐯᐸᐱ
Wed Mar 10 13:11:42 +0000 2021CBR4:p, θ(max) = 74. aka FLJ14431, SDR45C1. Found in HLA peptide class 1 experiments. Believed to be mitochondrial, but does not have an N-terminal mitochondrial targeting signal. Common in most tissues & cell lines, but rare in clinical fluids.
Wed Mar 10 13:11:42 +0000 2021>CBR4:p, carbonyl reductase 4 (Homo sapiens) Small subunit; CPTMs: M1+acetyl; PTMs: none; SAAVs: L70M (77%), T186M (1%); mature form: (1-237) [9,360×, 31 kTa]. #ᗕᕱᗒ 🔗
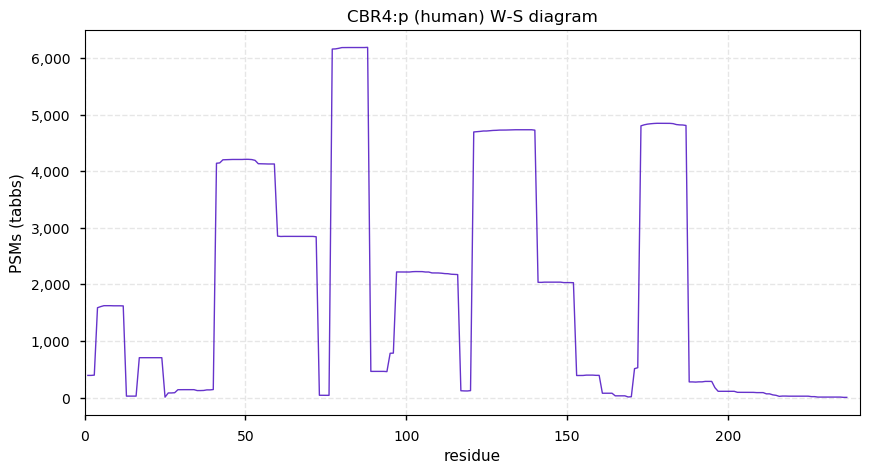
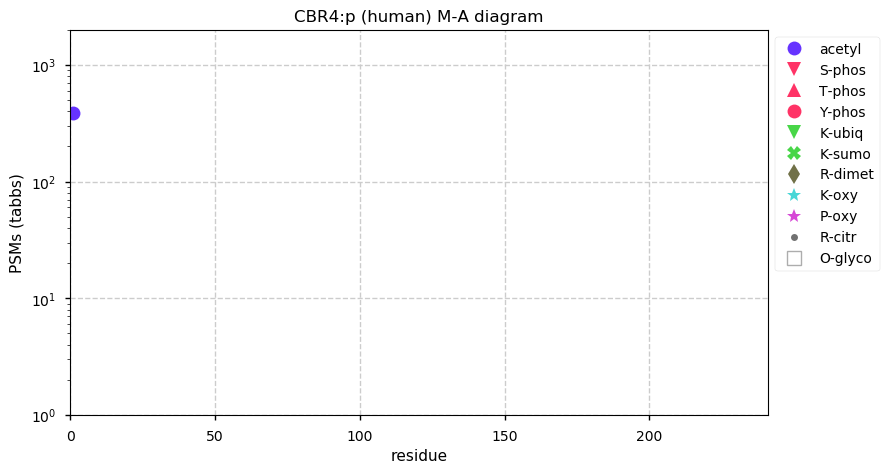
Wed Mar 10 00:50:27 +0000 2021@neely615 This is an example of an ID'd peptide match with the contents of one DIA bucket. Looks a spectrum to me (regardless of how many parent ion contributed to it) 🔗
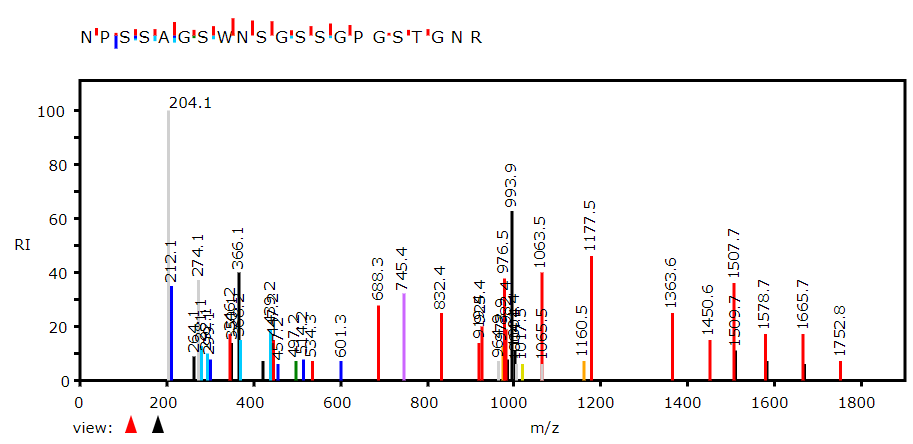
Wed Mar 10 00:35:07 +0000 2021@neely615 Some of them are 1. composites of multiple peptides, 2. some are the result of 1 peptide & 3. some have no ions from peptides. I was hoping someone knew off the top of their head what fraction were in 1+2.
Tue Mar 09 23:55:50 +0000 2021@J_my_sci Just a very conventional spectrum library id package I wrote some time ago.
Tue Mar 09 23:30:17 +0000 2021@J_my_sci I'm more interested in how many of the spectrum buckets contain significant matches to library spectra, rather than the aggregated info about proteins.
Tue Mar 09 19:39:36 +0000 2021& I know it is a vague questions & that "depends" is a good answer, but if 30% of the spectra are being ID'd would that be cause for celebration or simply another reason to start drinking early ...
Tue Mar 09 19:36:36 +0000 2021DIA peeps: when using a QExactive HF, what fraction of DIA spectra would you expect to be identifiable?
Tue Mar 09 17:00:04 +0000 2021@pwilmarth @UCDProteomics Since it was originally designed to serialize files onto magnetic tape (other uses were hacked into it later), maybe DTA would be a good acronym.
Tue Mar 09 13:06:44 +0000 2021MTDH:p.T317A, chr 8:g.97691089A>G, rs17854374 (JURKAT T:A = 28:17) vaf=5%, Δm=-30.0106, is TA in JURKAT & HEK-293 cells. It is AA in Hs-578T cells. It is TT in other commonly used cell lines. #ᐯᐸᐱ
Tue Mar 09 13:06:44 +0000 2021MTDH1:p, θ(max) = 80. aka LYRIC, 3D3, AEG-1. Found in HLA peptide class 1 experiments. (49-69) is a transmembrane domain. Common in most tissues & cell lines, but rare in clinical fluids.
Tue Mar 09 13:06:44 +0000 2021>MTDH:p, metadherin (Homo sapiens) Midsized subunit; CPTMs: A2+acetyl; PTMs: 59×S,33×T,1×Y+phosphoryl, 9×K+acetyl; 6×K+ubiquitinyl; SAAVs: L103V (1%), T317A (5%); mature form: (2-582) [45,939×, 373 kTa]. #ᗕᕱᗒ 🔗
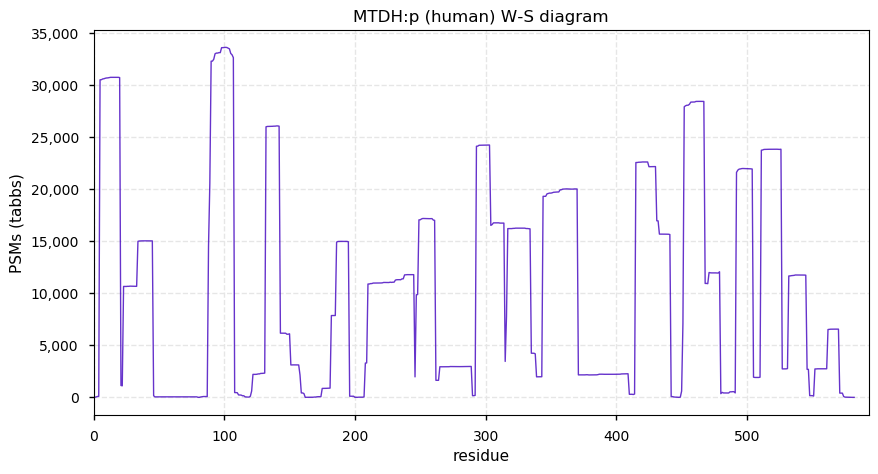
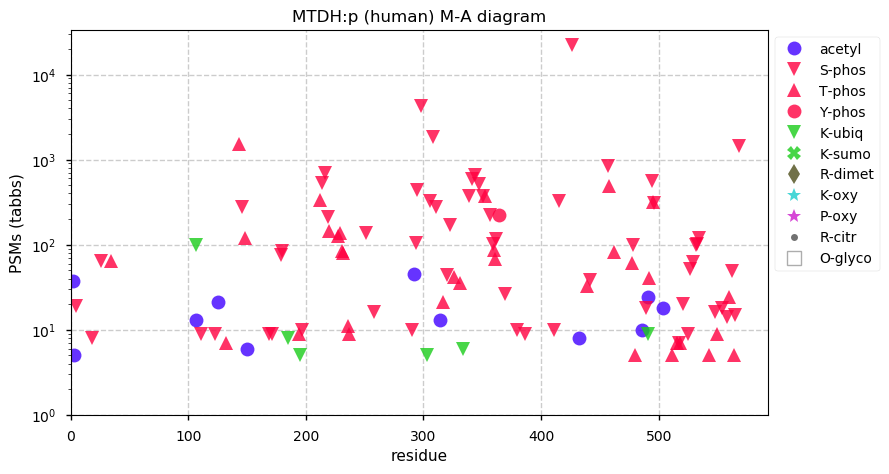
Tue Mar 09 02:36:17 +0000 2021@UCDProteomics Not really. It is the UNIX standard method of combining files together. It can just be less than intuitive if you are trying to use its features.
Tue Mar 09 02:00:19 +0000 2021@UCDProteomics I have said many things about "tar" (most of them not suitable for polite company), but "easy" has never been one of them.
Mon Mar 08 17:04:45 +0000 2021@chrashwood @JohnRYatesIII I do like MDC projects, though. The shifting patterns of PTM detection in the fractions really gets me thinking about what is going on (usually unintentionally) in the separations.
Mon Mar 08 16:10:59 +0000 2021@byu_sam @1jvaneyk Working on your first fragment of the Triforce of Wisdom?
Mon Mar 08 15:26:10 +0000 2021@chrashwood @JohnRYatesIII There has been a significant slow down in MDC projects submitted to PRIDE. Longer single runs have ramped up, I suspect because they are much cheaper & easier to reproduce. MDC is great for N=1, but technically challenging for N=100.
Mon Mar 08 12:50:54 +0000 2021HEATR1:p.E2017G, chr 1:g.236554626T>C, rs2275687 (MCF-10A E:G = 142:101) vaf=61%, Δm=-72.0211, is EG in MCF-10A, HEK-293, A-549, HCT-116 & JURKAT cells. It is GG in HeLa, Hep-G2 & MCF-7 cells. It is EE in WM-239, A-431 & LNCaP cells. #ᐯᐸᐱ
Mon Mar 08 12:50:54 +0000 2021HEATR1:p, θ(max) = 60. aka FLJ10359, BAP28, UTP10. Found in HLA peptide class 1 experiments. Given the SAAV distribution, it would be unlikely to find an individual with the reference sequence.
Mon Mar 08 12:50:54 +0000 2021>HEATR1:p, HEAT repeat containing 1 (H sapiens) CPTMs: T2+acetyl; PTMs: 35×S,1×T,1×Y+phosphoryl, 11×K+acetyl; 50×K+ubiquitinyl; SAAVs: H348R (66%), Y1433C (9%), S1559N (83%), N1694S (68%), V1854A (48%), E2017G (61%); mature form: (2-2144) [31,740×, 311 kTa]. #ᗕᕱᗒ 🔗
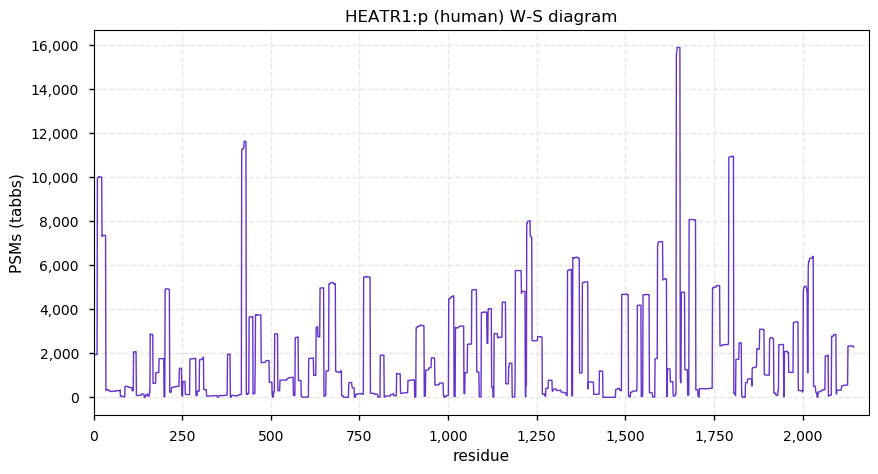
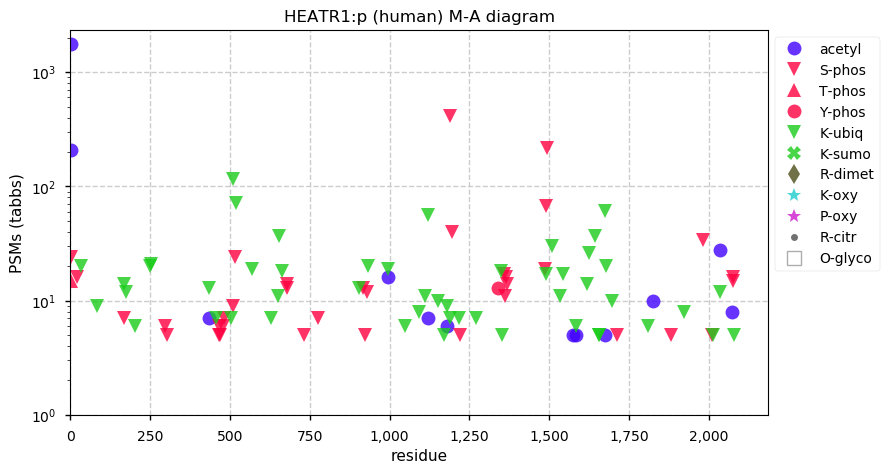
Sun Mar 07 23:58:55 +0000 2021Are ABRF & US-HUPO both having annual conferences over the same week?
Sun Mar 07 19:44:12 +0000 2021@neely615 @chrashwood @lkpino @ProteomicsNews The simplest way to look at it is to compare the calculated retention time for PSMs vs actual the scan number (or RT when available). A comparison of PSMs' calculated retention & isoelectric points can also be quite informative, particularly for MDC schemes.
Sun Mar 07 19:24:50 +0000 2021@chrashwood @lkpino @ProteomicsNews Proteomics has always had a passive-aggressive attitude towards HPLC. The quality of LC has always distinguished good from the no-so-good data, but I don't think any HUPO conference has every had an explicitly chromatography-themed session, workshop or training session.
Sun Mar 07 16:13:04 +0000 2021I never cease to be amazed at how easy it is to find detailed instructions on how to repair/fix just about anything, even as manufacturers make it increasingly difficult to do so.
Sun Mar 07 15:33:41 +0000 2021@dtabb73 Since I switched over to ARM-based platforms, I too have been using Joules used as my primary metric for software+hardware performance.
Sun Mar 07 14:45:07 +0000 2021A study after my own heart
🔗
Sun Mar 07 13:34:56 +0000 2021FABP1:p.V42M, chr 2:g.88126292C>T, rs1803563 (Hep-G2 V:M = 435:501) vaf=0.05%, Δm=31.9721, is VM in Hep-G2 cells. It is VV in all other commonly used cell lines. #ᐯᐸᐱ
Sun Mar 07 13:34:56 +0000 2021FABP1:p, θ(max) = 98. aka L-FABP. Most abundant in liver, urine, blood plasma and GI tissue.
Sun Mar 07 13:34:55 +0000 2021>FABP1:p, fatty acid binding protein 1 (Homo sapiens) Small subunit; CPTMs: S2+acetyl; PTMs: 3×S,1×T,0×Y+phosphoryl; SAAVs: V42M (0.05%), T94A (22%); mature form: (2-127) [7,519×, 267 kTa]. #ᗕᕱᗒ 🔗
Sat Mar 06 21:16:56 +0000 2021@AndrewWCraig @MHendr1cks @jwoodgett @JustinTrudeau @cdavidnaylor Did Naylor recommend ditching the CRC programs? They never really worked as a vehicle for attracting and retaining faculty, but they seem to still be going strong.
Sat Mar 06 14:58:20 +0000 2021@VDBiology @pride_ebi @ypriverol IMHO seminars like this should be a required part of biomedical grad student training.
Sat Mar 06 14:54:18 +0000 2021@VDBiology @pride_ebi @ypriverol Better yet, get them to come over to your department and give a seminar on how copyright law (& associated licensing) applies to research outputs.
Sat Mar 06 14:53:10 +0000 2021@VDBiology @pride_ebi @ypriverol If you aren't convinced, I would suggest you contact someone in the Faculty of Law who specializes in copyright law at your institution. Tell them some crazy guy on the internet said that MS files are not protected by copyright & therefore cannot be licensed & get their opinion
Sat Mar 06 13:53:20 +0000 2021@VDBiology @pride_ebi @ypriverol It isn't. It is actually very well established law. There is nothing creative about a mass spectrometer recording masses or a weather station recording the weather and therefore no copyright attached. Writing a line of code is creative & therefore protected by copyright.
Sat Mar 06 13:39:41 +0000 2021@ProteomicsNews @VDBiology @pride_ebi @ypriverol Fortunately, it is not. What you are doing is perfectly legal.
Sat Mar 06 13:35:28 +0000 2021@VDBiology @pride_ebi @ypriverol Yes it is. There is no copyright on the files (& there cannot be) so there is nothing to license and nothing requiring permission. If a machine generated it, you can only protect it by keeping it proprietary.
Sat Mar 06 13:30:04 +0000 2021@VDBiology @pride_ebi @ypriverol Same thing. Anything that is non-creative is not protected in any way, e.g. tables of numbers or data from instruments. Academic convention would require you to include a reference to the source material. Your R script can be protected, but the numbers are not.
Sat Mar 06 13:12:18 +0000 2021@VDBiology @pride_ebi @ypriverol They are like tables in a manuscript. You can use them any way you want.
Sat Mar 06 12:52:02 +0000 2021UQCC3:p.G89S, chr 11:g.62672097G>A, rs13941 (HEK-293T G:S = 0:512) vaf=74%, Δm=30.0106, is SS in HEK-293T, A-549 & HCT-116 cells. It is GS in HeLa & SK-MEL-28 cells. It is GG in MCF-10A & HAP-1 cells. #ᐯᐸᐱ
Sat Mar 06 12:52:02 +0000 2021UQCC3:p, θ(max) = 69. aka UNQ655, C11orf8. Commonly found in HLA class 1 experiments. Similar to cytochrome C, this protein has an N-terminal transit peptide that allows it to pass through the mitochondrial outer membrane, but it remains in the intermembrane space.
Sat Mar 06 12:52:02 +0000 2021>UQCC3:p, ubiquinol-cytochrome c reductase complex assembly factor 3 (Homo sapiens) Very small subunit; CPTMs: M1+acetyl; PTMs: none; SAAVs: W76G (1%), G89S (74%); mature form: (1-96) [4,120×, 11 kTa]. #ᗕᕱᗒ 🔗
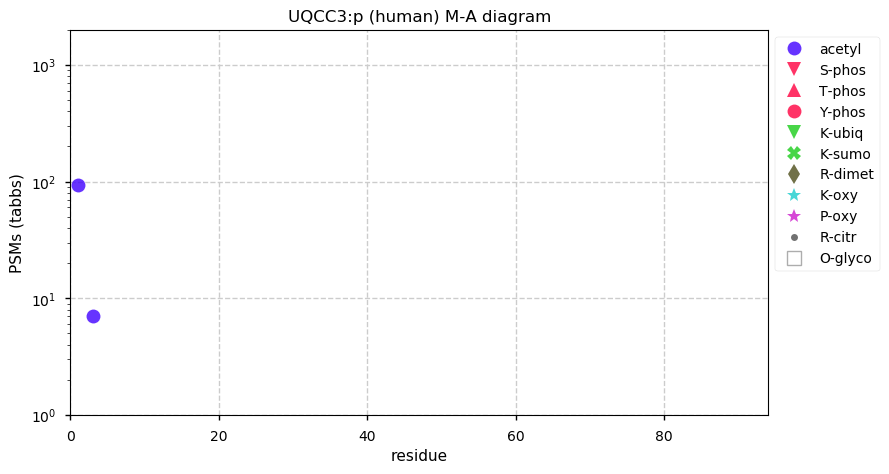
Fri Mar 05 21:18:16 +0000 2021@jwoodgett Reviews must now be written in the form of Hegelian dialectics including thesis, antithesis and synthesis.
Fri Mar 05 17:48:47 +0000 2021Today's polar vortex (10 hPa altitude), right where it should be 🔗
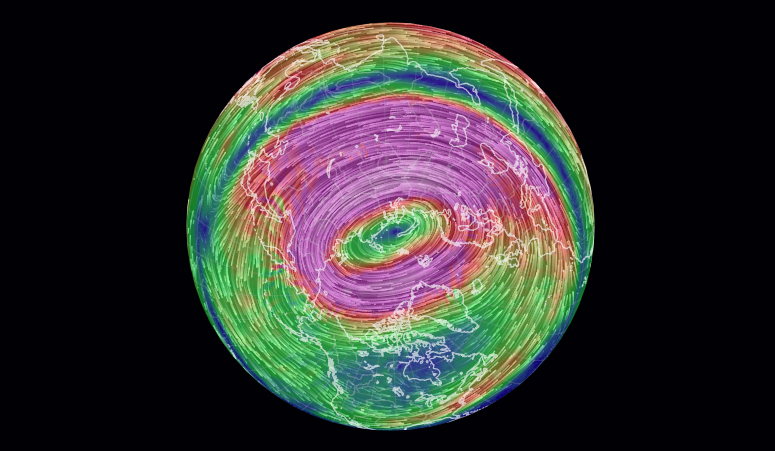
Fri Mar 05 17:29:33 +0000 2021@ucdmrt Deep learning from PRIDE submissions suggests "trypsin" is always the right answer! 😀
Fri Mar 05 14:43:13 +0000 2021@odd_opinions5 @Sci_j_my 4. demonstrated that within the limits of their experiments, COVID-19 produced no protein-level effects that were unusual for a viral infection.
If I was still on a regular study section & this was their preliminary data, I would be very positive about their chances.
Fri Mar 05 14:40:51 +0000 2021@odd_opinions5 @Sci_j_my OK, so now I have read the paper. To summarize:
1. developed (IMHO) the best LC/MS/MS method so far for clinical plasma samples;
2. applied the method to 74 COVID-19 positive & 37 COVID-19 negative patient samples;
3. analyzed the same samples with an orthogonal technique; &
Fri Mar 05 13:44:35 +0000 2021@odd_opinions5 @Sci_j_my Couldn't care less about the outcome of the study. I am only commenting on the quality of the data.
Fri Mar 05 12:58:38 +0000 2021TJP2:p.D482E, chr 9:g.69228107C>A, rs2309428 (Hep-G2 D:E = 1:247) vaf=81%, Δm=14.0157, is EE in Hep-G2, MCF-7, MCF-10A, U2-OS & HCT-116 cells. It is DE in HeLa cells. It is DD in HEP-3B cells. #ᐯᐸᐱ
Fri Mar 05 12:58:38 +0000 2021TJP2:p, θ(max) = 65. aka ZO-2, X104, ZO2, DFNA51. Commonly found in HLA class 1. Also found in class 2 experiments, but only peptides from (1011-1030). (1-20) is an R-rich low complexity domain that makes determining the mature N-terminal difficult.
Fri Mar 05 12:58:38 +0000 2021>TJP2:p, tight junction protein 2 (Homo sapiens) Large subunit; CPTMs: PTMs: 56×S,19×T,24×Y+phosphoryl; 2×K+acetyl; 3×K+ubiquitinyl; SAAVs: Q128K (13%), D482E (81%), R493K (1%), M668I (4%), S711P (3%), L937P (1%); mature form: (1?-1043) [32,117×, 282 kTa]. #ᗕᕱᗒ 🔗
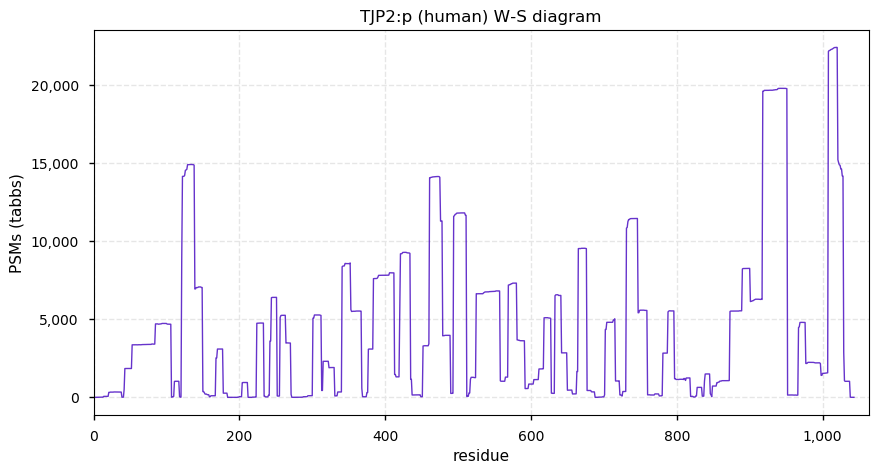
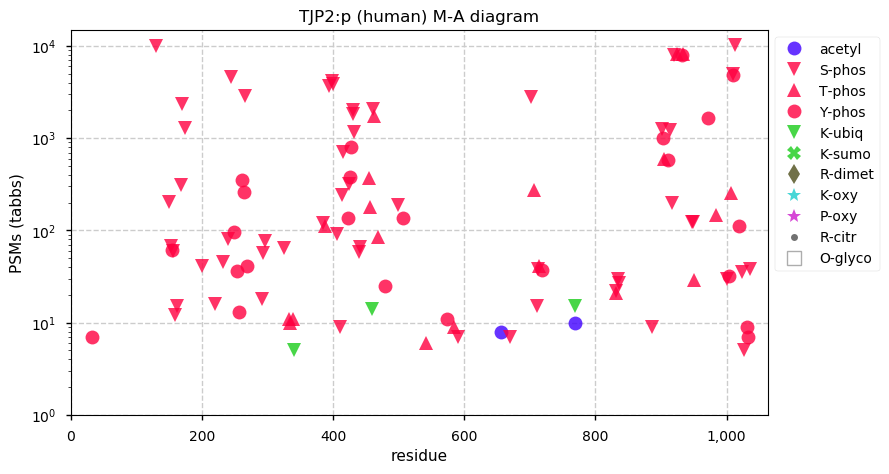
Fri Mar 05 12:46:19 +0000 2021PXD022817 is an excellent example of a state-of-the-art human plasma proteomics data set. Congrats guys: I'm going to take this one as the new standard 🔗
Thu Mar 04 22:27:34 +0000 2021You can always tell when a yeast study was done by a microbiology group or a technique-oriented group that considers it a simple model organism that is hard to screw up.
Thu Mar 04 17:46:45 +0000 2021@MattWFoster @UCDProteomics Being just a touch cynical, telling tumor from non-tumor tissue is really (really) easy in many types of tumor. Quant on a few peptides ends up being all you need.
Thu Mar 04 17:02:20 +0000 2021@UCDProteomics That is one way to think about it. But for me, the biggest issue is always dynamic range. It is great that you can see ALB & APOB in plasma, but I want to see INS too, in the same measurement.
Thu Mar 04 16:50:54 +0000 2021@UCDProteomics Smaller windows also drives up the LOD/LOQ dynamic range.
Thu Mar 04 16:50:16 +0000 2021@UCDProteomics At the moment, DIA is being run using similar separation methods as DDA, where you really only have a small number of parent ions being sampled at a time.
Thu Mar 04 16:49:35 +0000 2021@UCDProteomics But if you got the window size down, you could throw a lot more parent ions at the machine simultaneously, without producing overly complicated fragment signals in each window.
Thu Mar 04 16:13:59 +0000 2021@UCDProteomics My guess would be that the DIA acquisition windows shrink down to the point that there is no real distinction between DDA and DIA.
Thu Mar 04 15:28:12 +0000 2021👍😀 to the submitters of PXD022817. Including the MGF files really makes reusing the data easier for most non-Thermo raw file formats.
Thu Mar 04 15:21:06 +0000 2021@dk_munro @AlexUsherHESA You lost me at "Despite generous R&D tax credits ...". CRA is not the appropriate agency to deal with R&D: they try hard and operate in good faith, but their time scales and priorities just don't fit in with research-driven development.
Thu Mar 04 14:53:43 +0000 2021ESD:p is an interesting protein. It is a simple enzyme (formyl-GSH ⇨ formyl+GSH), but it has many PTM acceptors including a complex pattern of acetyl/ubiquitinyl/SUMOyl alternating modifications. It is found in extracellular vesicles, but it has no ER signal sequence.
Thu Mar 04 12:53:30 +0000 2021ESD:p.G257D, chr 13:g.46771495C>T, rs15303 (HCT-116 G:D = 543:227) vaf=0.6%, Δm=58.005, is GD in HCT-116. It is GG in HeLa, MCF-7, MCF-10A & most other common cell lines. #ᐯᐸᐱ
Thu Mar 04 12:53:30 +0000 2021ESD:p, θ(max) = 93. aka S-formylglutathione hydrolase. Commonly found in HLA class 1 & rarely in class 2 experiments. Observed in all tissues, cell lines & extracellular vesicles.
Thu Mar 04 12:53:30 +0000 2021>ESD:p, esterase D (Homo sapiens) Small enzyme; CPTMs: A2+acetyl; PTMs: 18×S, 2×T, 3×Y+phosphoryl; 19×K+acetyl; 14×K+ubiquitinyl, 4×K+SUMOyl; SAAVs: G190A (19%), G257D (1%); mature form: (2-282) [48,210×, 426 kTa]. #ᗕᕱᗒ 🔗
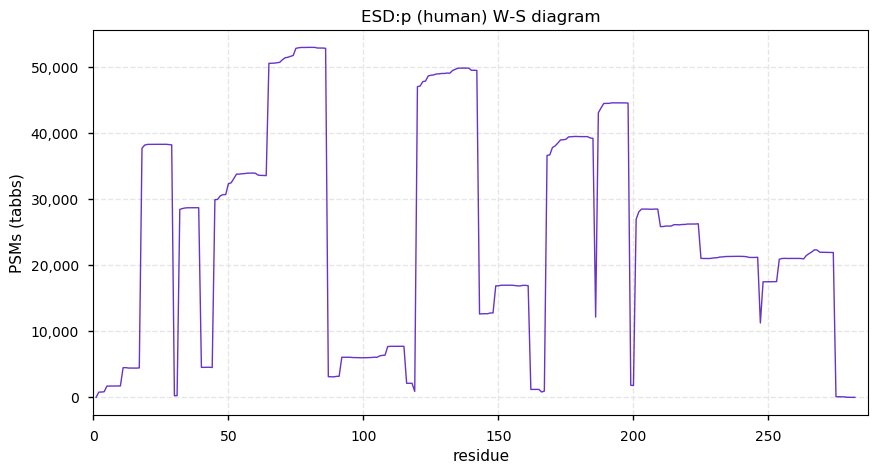
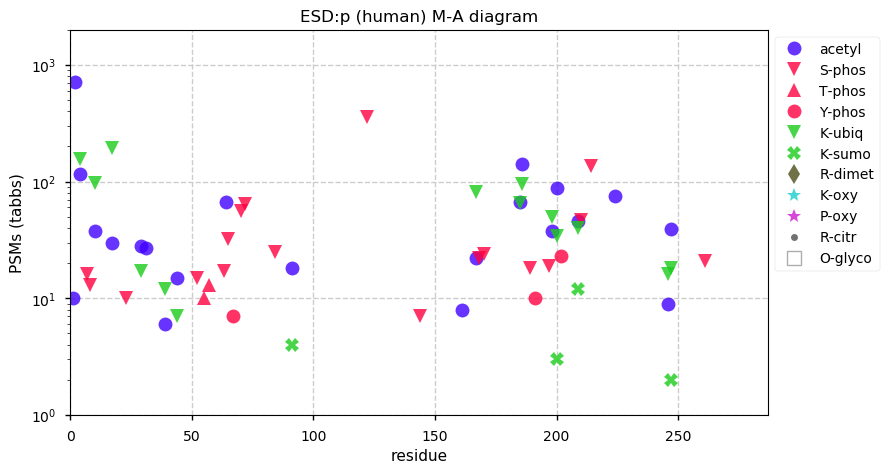
Thu Mar 04 00:41:41 +0000 2021PXD022621 is a really good example of yeast SILAC data using hi/low pH MDC separation. Excellent for use in tuning up algorithms and identification statistics for a midsized proteome. 🔗
Wed Mar 03 18:59:04 +0000 2021They (Flagship Pioneering FL69) have a number of proteomics-related job openings at the moment
Wed Mar 03 18:57:55 +0000 2021If you are in the Boston area (or would like to be)
🔗
Wed Mar 03 16:49:55 +0000 2021@AlexUsherHESA It is hard not to notice that the sectors the BCC wants funding for are almost exactly the same as those currently targeted by the Innovation Superclusters Initiative.
Wed Mar 03 16:24:45 +0000 2021Although it eventually lost out to spectrum libraries for interpreting organic molecule fragmentation patterns, Dendral involved the same methods as current peptide id algorithms use for interpreting peptide fragmentation patterns.
Wed Mar 03 15:52:21 +0000 2021A lot of what people do in proteomics data analysis had its origins in the Heuristic Dendral project, although I suspect very few current practitioners have ever heard of it. 🔗
Wed Mar 03 13:13:18 +0000 2021PDCD6IP:p.N550S, chr 3:g.33852495A>G, rs2041387 (CACO-2 N:S = 87:34) vaf=9%, Δm=-27.011, is NS in CACO-2, 16-HBE14o & MDA-MB-231 cells. It is SS in HCT-116 cells. It is NN in A-375, HeLa, JURKAT & most other common cell lines. #ᐯᐸᐱ
Wed Mar 03 13:13:18 +0000 2021PDCD6IP:p, θ(max) = 84. aka Alix, AIP1, Hp95. Commonly observed in HLA class 1 & 2 experiments. (717-860) is a low complexity, P-enriched domain. Peptides in the domain (768-868) are rarely observed.
Wed Mar 03 13:13:18 +0000 2021>PDCD6IP:p, programmed cell death 6 interacting protein (H sapiens) Midsized subunit; CPTMs: A2+acetyl; PTMs: 35×S,28×T,10×Y+phosphoryl; 27×K+ubiquitinyl, 57×K+acetyl; SAAVs: A309T (67%), V378I (29%), N550S (9%), S730L (25%); mature: (2-868) [77,186×, 964 kTa]. #ᗕᕱᗒ 🔗
Wed Mar 03 03:45:20 +0000 2021Looks like there will be some nice aurora borealis overhead tonight! 🔗
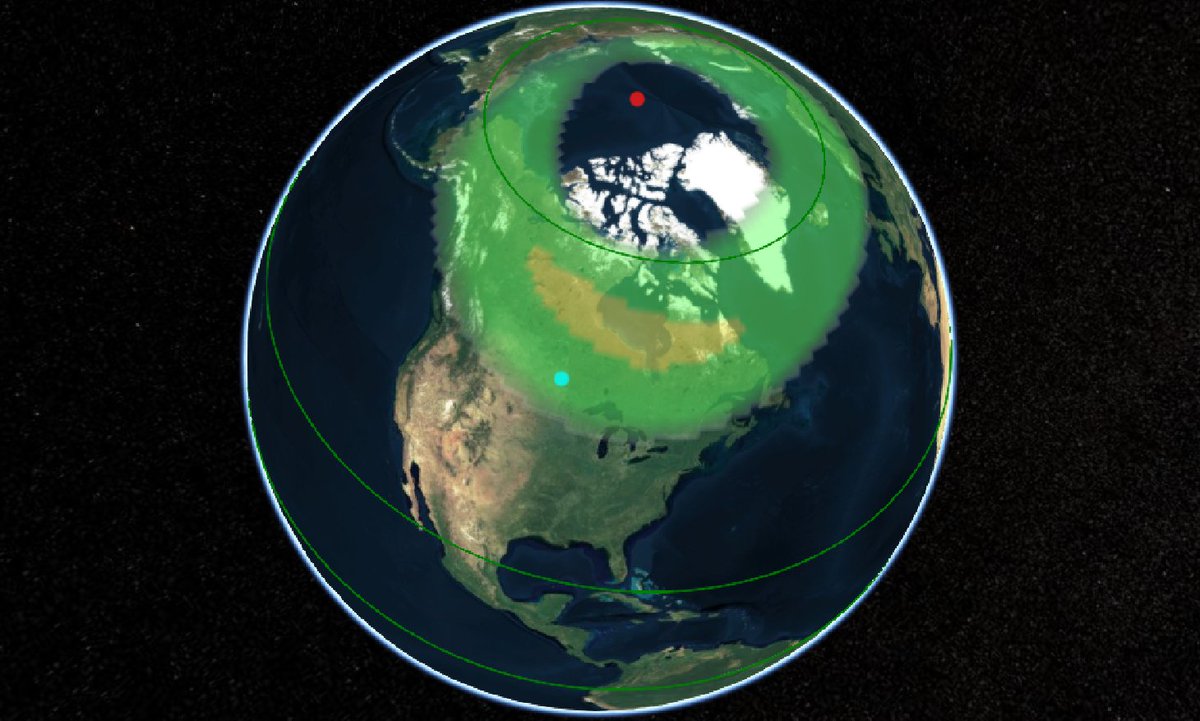
Wed Mar 03 00:24:52 +0000 2021It is also the home base for the GPM project site and wiki
🔗
Wed Mar 03 00:17:33 +0000 2021The oddball pile of bricks I live in. It was built the year after the Gunfight at the OK Corral. 🔗
Tue Mar 02 20:45:07 +0000 2021A warm wind is blowing north from Fargo. The temperature here is up 25 °C from yesterday.
Tue Mar 02 16:02:29 +0000 2021I must admit I find some of the old-school, simple KEGG pathway displays useful for getting a broader view of results 🔗
Tue Mar 02 15:25:36 +0000 2021@slashdot I would not like to be who ever has to go through that "transcript" and tries to use it to generate an accurate, coherent narrative.
Tue Mar 02 13:19:57 +0000 2021TAPBPL:p.A169V, chr 12:g.6453657C>T, rs2041387 (Hep-G2 A:V = 9:7) vaf=28%, Δm=28.031, is AV in Hep-G2 cells. It is AA in JURKAT & MHCC97-H cells. #ᐯᐸᐱ
Tue Mar 02 13:19:56 +0000 2021TAPBPL:p, θ(max) = 34. aka TAPBP-R, FLJ10143, TAPBPR. Commonly observed in HLA class 1 but not in class 2 experiments. Rare in cell lines but common in some lymphocytes. (407-416) is a membrane spanning domain.
Tue Mar 02 13:19:56 +0000 2021>TAPBPL:p, TAP binding protein like (Homo sapiens) Small membrane subunit; PTMs: K43, K166, K251+ubiquitinyl; SAAVs: C39W (23%), A42V (23%), T165A (69%), A169V (28%); mature form: (19-468) [3,691×, 9.2 kTa]. #ᗕᕱᗒ 🔗
Mon Mar 01 16:32:57 +0000 2021It is also a good example if you are interested in accurate modification site assignments from the results generated by open modification search algorithms.
Mon Mar 01 15:45:23 +0000 2021If you want to study the patterns of mouse hydroxylysine and hydroxyproline modifications in tissue, understanding the data in PXD019431 would be a good place to start.
Mon Mar 01 15:09:14 +0000 2021@AlexUsherHESA Note: both Federal and provincial (BC, Manitoba & Ontario) officials.
Mon Mar 01 15:08:17 +0000 2021@AlexUsherHESA Whenever I have spoken to Canadian govt. officials about anything related to research, they can barely get a sentence out without saying something so odd that I simply stop paying attention to them.
Mon Mar 01 12:56:43 +0000 2021NAT10:p.Y461H, chr 11:g.34131392T>C, rs2957516 (HEK-293 Y:H = 0:1375) vaf=100%, Δm=-26.004, is HH in HEK-293, HeLa, MCF-7, MCF-10A, HCT-116 & all commonly used cell lines. Normally listed as having a MAF = 0.00. #ᐯᐸᐱ
Mon Mar 01 12:56:42 +0000 2021NAT10:p, θ(max) = 73. aka hALP, FLJ10774, FLJ12179, NET43, KIAA1709, Kre33. Commonly observed in HLA class 1 but not in class 2 experiments. Commonly observed in most tissues & cell lines. The domain (900-1025) is more highly modified than the rest of the protein.
Mon Mar 01 12:56:42 +0000 2021>NAT10:p, N-acetyltransferase 10 (Homo sapiens) Large enzyme; PTMs: 20×S,20×T,7×Y+phosphoryl; 50×K+acetyl; 14×K+ubiquitinyl; 5×K+SUMOyl; SAAVs: P224L (1%), Y461H (100%), M638V (1%), A983T (1%); mature form: (1-1025) [42,342×, 413 kTa]. #ᗕᕱᗒ 🔗
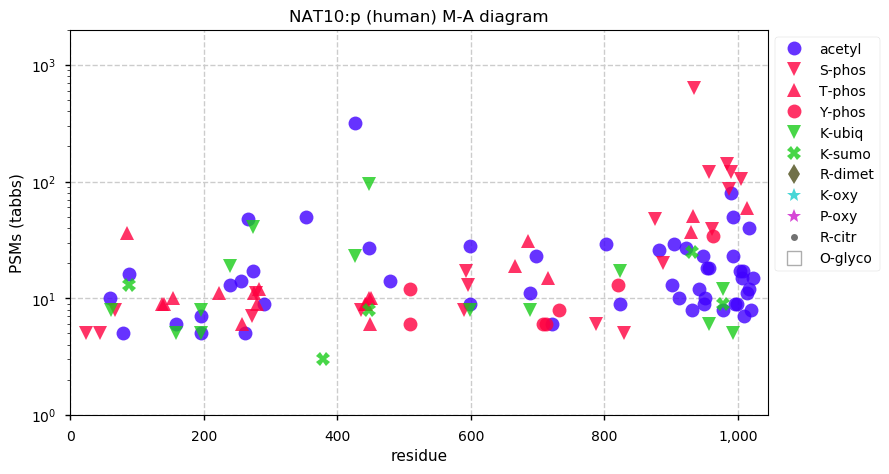
Sun Feb 28 20:09:49 +0000 2021@astacus The trypsin step is where you keep it nice & warm at a favorable pH for a long time. I wasn't suggesting an enzymatic mechanism: it is just isocyanate + heat + time. The peptide N-termini are more reactive in these conditions than the lysine ε-amino groups.
Sun Feb 28 15:40:02 +0000 2021A proteolytic cleavage appears to frequently create a new N-terminus at A55, although the purpose of this modification is unclear.
Sun Feb 28 14:45:22 +0000 2021@astacus Labs do seem to eventually figure it out, but it takes about a decade of operation. Some of the worst offenders in the 2000-2010 period now regularly have undetectable levels of +43.
Sun Feb 28 13:35:55 +0000 2021Cells seem to be very interested in sticking ubiquitin-type modifiers on this protein.
Sun Feb 28 13:31:58 +0000 2021NOP56:p.V576A, chr 20:g.2658236T>C, rs5856 (HeLa V:A = 204:2203) vaf=29%, Δm=-28.031, is AA in HeLa & RKO cells. It is VA in HCT-116 & MDA-MB-231 cells. It is VV in HEK-293, JURKAT, MCF-10A & MCF-7 cells. #ᐯᐸᐱ
Sun Feb 28 13:31:58 +0000 2021NOP56:p, θ(max) = 77. aka SCA36, NOL5A. Commonly observed in HLA class 1 & less frequently in class 2 experiments. Commonly observed in most tissues and cell lines but not in extracellular spaces or fluids.
Sun Feb 28 13:31:58 +0000 2021>NOP56:p, NOP56 ribonucleoprotein(Homo sapiens)Midsized subunit; CTMs: M1+acetyl; PTMs: 45×S,10×T,4×Y+phosphoryl; 13×K+acetyl; 18×K+ubiquitinyl; 13×K+SUMOyl; SAAVs: V576A (29%); mature form: (2-594) [61,471×, 749 kTa]. #ᗕᕱᗒ 🔗
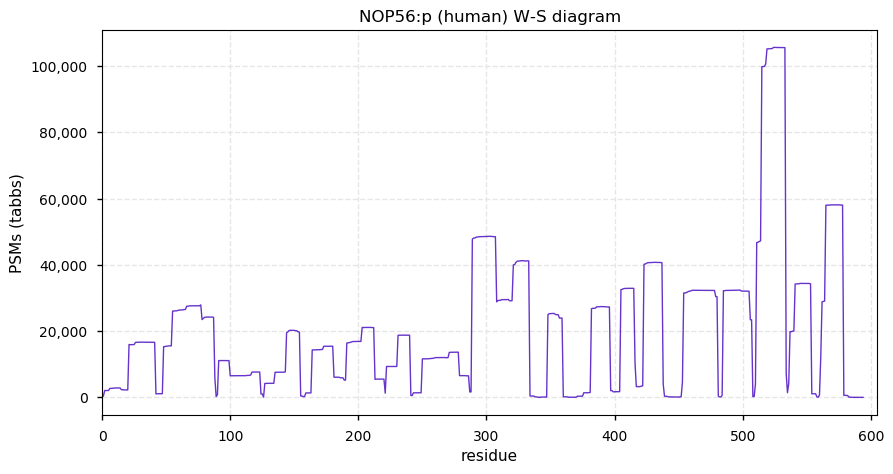
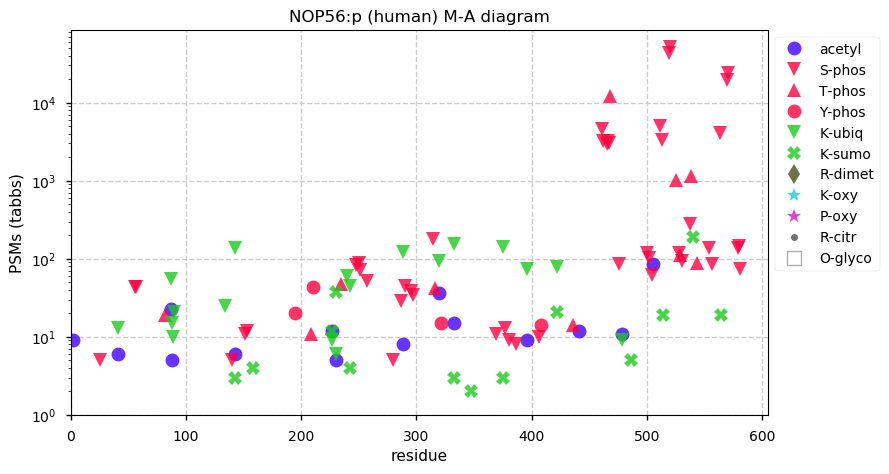
Sun Feb 28 13:01:10 +0000 2021@astacus It is real. Usually people mention using urea in protein isolation, but the derivatization occurs during trypsin digestion.
Sat Feb 27 17:47:46 +0000 2021Seeing a lot of carbamylation leads to irrational annoyance on my part. By now I should know people whose primary occupation is teaching are really, really bad at learning.
Sat Feb 27 13:37:27 +0000 2021LRRFIP1:p.H727D, chr 2:g.237764060C>G, rs3739038 (HeLa H:D = 266:178) vaf=29%, Δm=-22.032, is HD in HeLa, MDA-MB-231 & MCF-10A cells. It is DD in 143B cells. It is HH in JURKAT, MCF-7 & HCT-116 cells. #ᐯᐸᐱ
Sat Feb 27 13:37:26 +0000 2021LRRFIP1:p, θ(max) = 97. aka FLAP-1, FLIIAP1, TRIP, GCF-2, HUFI-1. Commonly observed in HLA class 1 & class 2 experiments . Commonly observed in most tissues and cell lines. Present in urine extracellular vesicles.
Sat Feb 27 13:37:26 +0000 2021>LRRFIP1:p, LRR binding FLII interacting protein 1 (H sapiens) Midsized subunit; CTMs: T2+acetyl; PTMs: 51×S,35×T,4×Y+phosphoryl; 12×K+acetyl; SAAVs: M338V (4%), N362S (85%), P589L (11%), H727D (49%); mature form: (2-752) [42,057×, 349 kTa]. #ᗕᕱᗒ 🔗
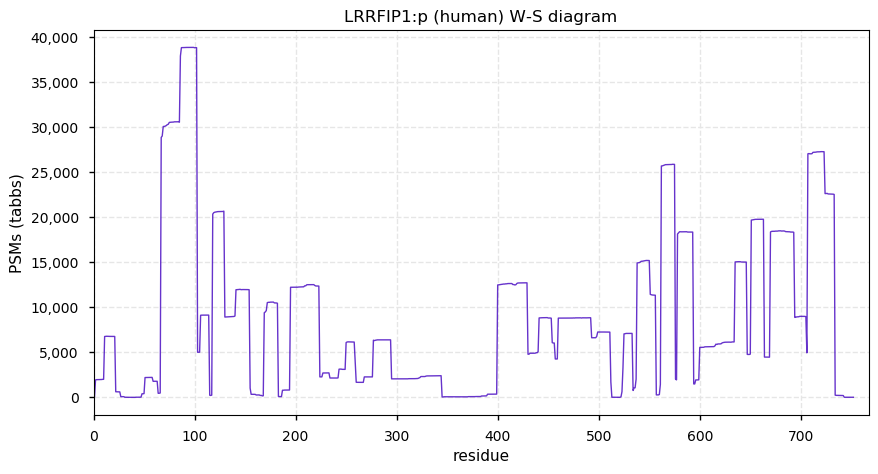
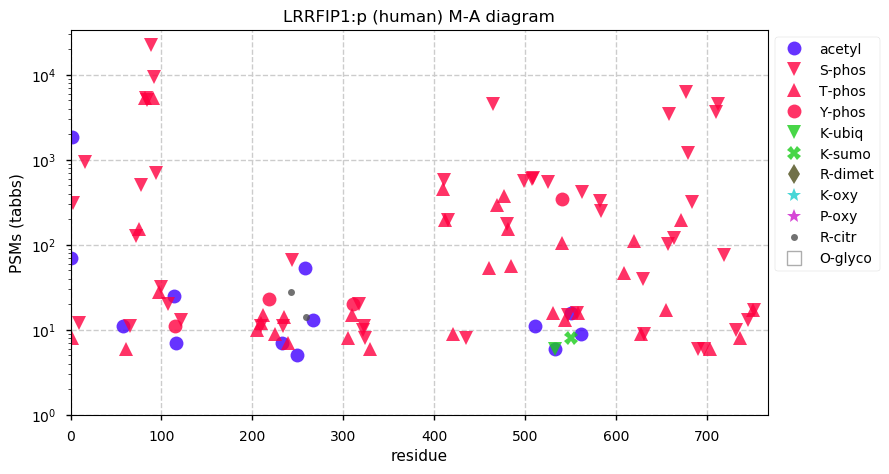
Fri Feb 26 18:44:05 +0000 2021@Sci_j_my If you are willing to pay for everything, I'll make you 3rd author!
Fri Feb 26 16:58:56 +0000 2021@WinnipegNews Possibly the least surprising set of conclusions ever reached by a political consultant.
Fri Feb 26 16:18:59 +0000 2021@astacus @oleg8r A possibly related pet-peeve of mine is lab web sites that do not make it clear what institution & department they are located within. PI labs are largely notional: anyone interested in working there is far more likely to be influenced by the institution than the PI.
Fri Feb 26 13:21:28 +0000 2021PYGB:p.A303S, chr 20:g.25278370G>T, rs2228976 (MHCC97-H A:S = 8:91) vaf=49%, Δm=31.972, is SS in MHCC97-H cells. It is AS in MCF-7, OVCA-429 & CACO-2 cells. It is AA in HCT-116, HeLa & HEK-293 cells. #ᐯᐸᐱ
Fri Feb 26 13:21:28 +0000 2021PYGB:p, θ(max) = 97. Commonly observed in HLA class 1 experiments & more rarely in class 2. Commonly observed in most tissues and cell lines. Present in urine extracellular vesicles.
Fri Feb 26 13:21:28 +0000 2021>PYGB:p, glycogen phosphorylase B (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 28×S,0×T,25×Y+phosphoryl; 13×K+acetyl; 36×K+ubiquitinyl; SAAVs: A303S (49%), D502N (16%), T514S (1%); mature form: (2-843) [15,822×, 945 kTa]. #ᗕᕱᗒ 🔗
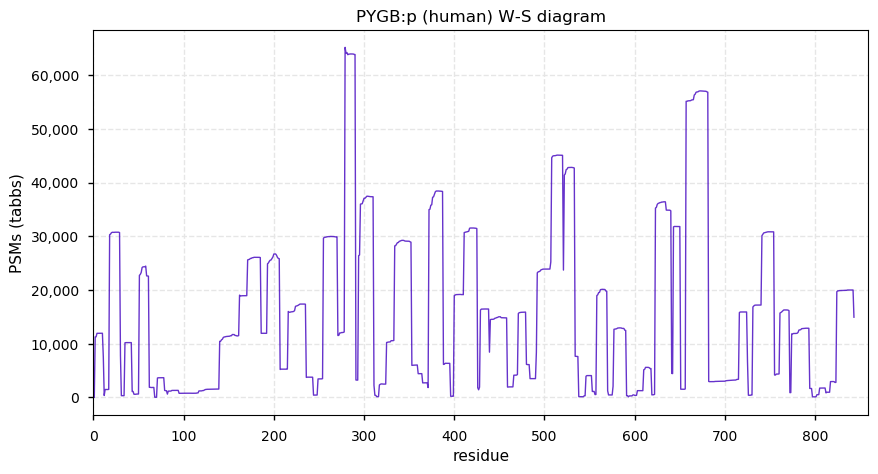
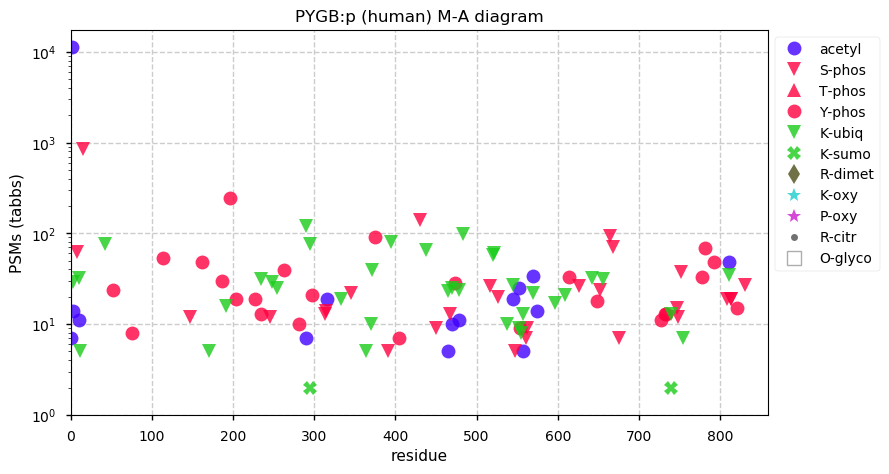
Thu Feb 25 20:54:06 +0000 2021@camtflower TMT in general. It depends on the way the experiment was done, so the amount can range from nearly undetectable to 20% of the PSMs.
Thu Feb 25 18:39:58 +0000 2021@camtflower Succinylation is an annoying side reaction of the derivatization process.
Thu Feb 25 18:24:12 +0000 2021Just about downloaded an interesting sounding succinylation data set, but then I noticed they used TMT-6-plex ... 😟
Thu Feb 25 17:50:51 +0000 2021Metabolomics in the news:
🔗
Thu Feb 25 15:52:46 +0000 2021@byu_sam @theoneamit @hupo_org @asmsnews Thanks for explaining: I've never seen one either. Sort of a mini satellite meeting-within-a-meeting.
Thu Feb 25 15:12:44 +0000 2021I would once again appeal to people's better natures: try very, very hard not to use any characters in the names of archived files other than A-Z, a-z, 0-9, "-", "." and "_". It may seem clever to use other symbols, but it is not.
Thu Feb 25 13:28:15 +0000 2021@Sci_j_my I think that is a reportable side effect.
Thu Feb 25 12:54:02 +0000 2021HS1BP3:p.V260M, chr 2:g.20624738C>T, rs2305458 (NTERA-2 V:M = 220:288) vaf=49%, Δm=31.972, is VM in NTERA-2, HCT-116, & U2-OS cell lines. It is MM in HeLa & HEK-293 cells. It is VV in MCF-7 cells. #ᐯᐸᐱ
Thu Feb 25 12:54:02 +0000 2021HS1BP3:p, θ(max) = 83. aka HS1-BP3, FLJ14249. Observed in HLA class 1 experiments: rarely in class 2. Commonly observed in most tissues and cell lines.
Thu Feb 25 12:54:02 +0000 2021>HS1BP3:p, HCLS1 binding protein 3 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 21×S,5×T,0×Y+phosphoryl; 11×K+acetyl; 0×K+ubiquitinyl; SAAVs: V260M (49%), G273R (2%), A388T (15%); mature form: (1-392) [15,822×, 63 kTa]. #ᗕᕱᗒ 🔗
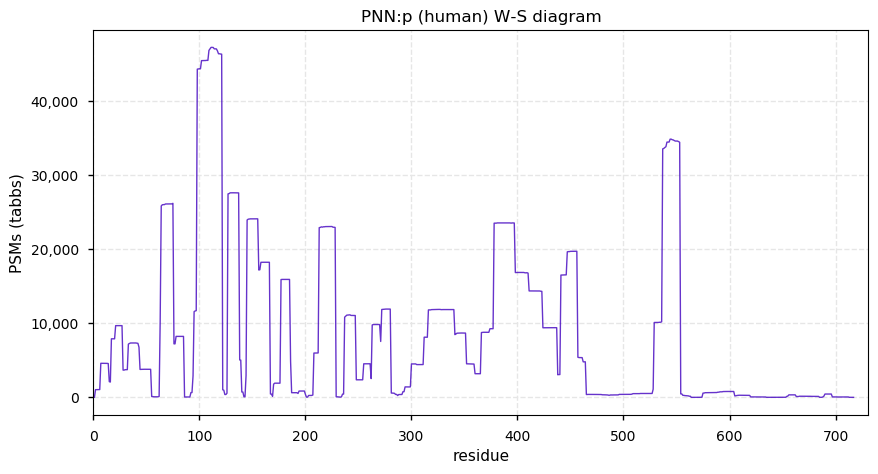
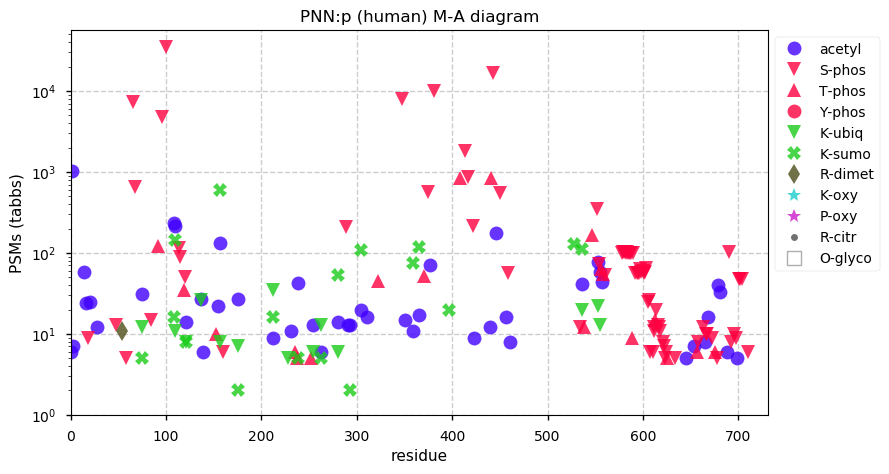
Wed Feb 24 17:53:29 +0000 2021@byu_sam Most implementations are pretty rough & ready hand-waving estimates that do not bear up under scrutiny very well.
Wed Feb 24 16:49:45 +0000 2021@MattWFoster The above & more. Immunoglobulins are patched together via a complicated rearrangement program from over 1000 genes: 1 gene ⇨ 1 peptide sequence doesn't happen. These genes also have a higher load of SAAVs than typical enzymes or structural proteins.
Wed Feb 24 16:00:07 +0000 2021For people unfamiliar with the process of creating immunoglobulin sequences (which is very different from normal gene expression), this on-line resource (🔗) does a pretty good job of explaining the mechanisms leading to complexity.
Wed Feb 24 15:39:02 +0000 2021Analysis of PXD024113 & PXD024123 has reminded me how little community effort has gone in to coping with variability in immunoglobulin sequences. Canonical sequences just don't do the job.
Wed Feb 24 15:23:15 +0000 2021@dtabb73 Living in Paris is so much better than simply a tourist visit.
Wed Feb 24 13:15:12 +0000 2021PNN:p.T441S, chr 14:g.39181030A>T, rs2180792 (HeLa T:S = 6:1427) vaf=91%, Δm=-14.016, is SS in HeLa, HCT-116, MCF-7, U2-OS, HEK-293, MCF-10A & most other cell lines. It is TT in 143B cells. #ᐯᐸᐱ
Wed Feb 24 13:15:12 +0000 2021PNN:p, θ(max) = 60. aka memA. Observed in HLA class 1 experiments. Usually absent from clinical fluids such as blood plasma and urine.
Wed Feb 24 13:15:12 +0000 2021>PNN:p, pinin, desmosome associated protein (Homo sapiens) Midsized subunit; CTMs: A2+acetyl; PTMs: 76×S,19×T,0×Y+phosphoryl; 44×K+acetyl; 14×K+ubiquitinyl; 17×K+SUMOyl; SAAVs: M290V (1%), T441S (91%), S671G (13%); mature form: (2-717) [39,464×, 416 kTa]. #ᗕᕱᗒ 🔗
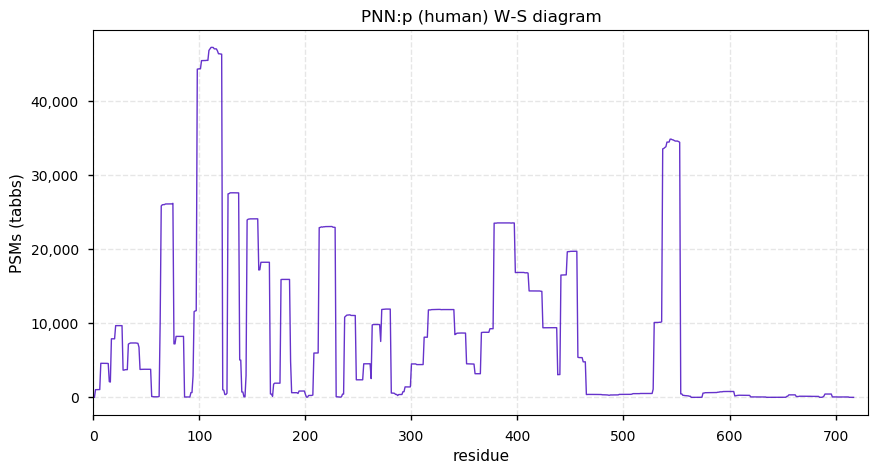
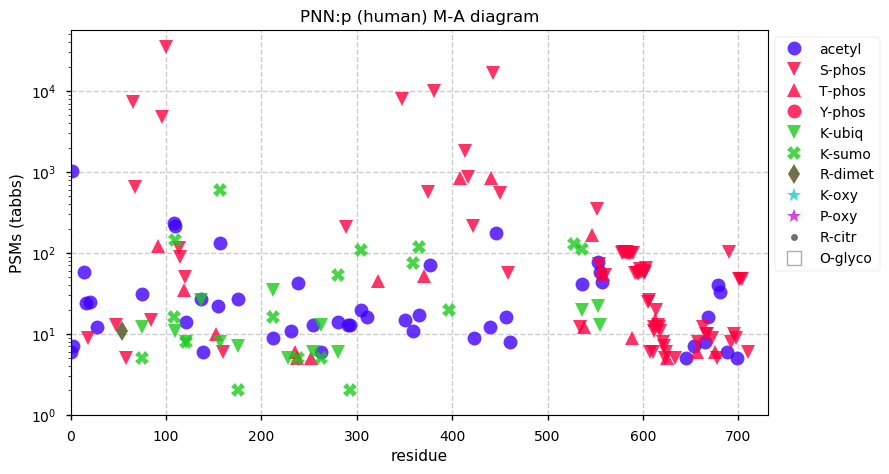
Tue Feb 23 22:51:03 +0000 2021@ypriverol Thanks for letting me know. I had changed a the fire wall setting yesterday, which I guess didn't work as well as I had hoped. It should be working for you now.
Tue Feb 23 22:45:49 +0000 2021@ypriverol It seems to be working. Is there a specific request that is failing?
Tue Feb 23 15:52:12 +0000 2021I don't think many people understand what the PHSCC does, how it is organized and how entwined it is with the US health research establishment. 🔗
Tue Feb 23 14:23:25 +0000 2021@AlexHgO I was make a sardonic jest: I had just read the nth (n >> 1) very positive blurb about the application of ML to yet another problem I don't care about, while staring at 100's of data files with names like "F001926" representing information from dozens of separate experiments.
Tue Feb 23 13:07:56 +0000 2021ACIN1:p.I311M, chr 14:g.23080576T>C, rs3811182 (HeLa I:M = 9:335) vaf=43%, Δm=17.956, is MM in HeLa cells. It is IM in TIG-3-20 cells. It is II in HCT-116, MCF-7, U2-OS, HEK-293 & MCF-10A cells. #ᐯᐸᐱ
Tue Feb 23 13:07:56 +0000 2021ACIN1:p, θ(max) = 59. KIAA0670, fSAP152, ACINUS. Observed in HLA class 1 experiments. Usually absent from clinical fluids & abundant in lymphocytes.
Tue Feb 23 13:07:55 +0000 2021>ACIN1:p, apoptotic chromatin condensation inducer 1 (H sapiens) CTMs: M1+acetyl; PTMs: 135×S,57×T,4×Y+phosphoryl; 78×K+acetyl (total); aPTMs:, 7×K+acetyl/ubiquitinyl; 13×K+acetyl/SUMOyl; SAAVs: I311M (43%), S467P (43%); mature form: (1-1341) [47,031×, 630 kTa]. #ᗕᕱᗒ 🔗
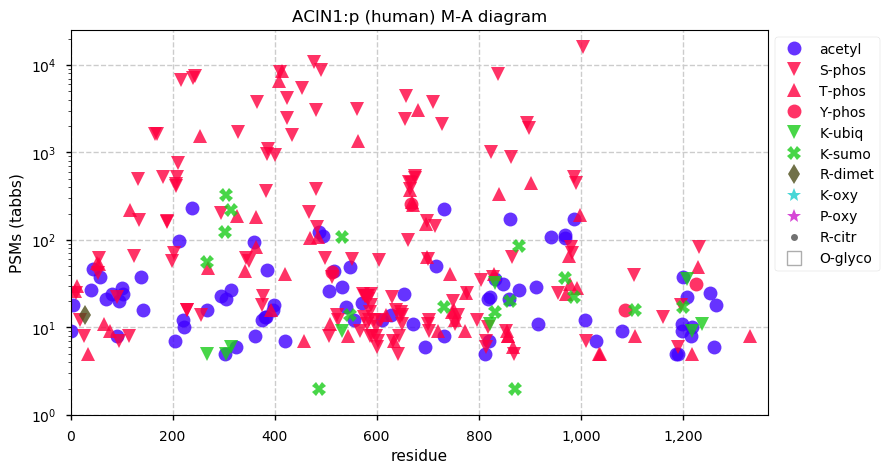
Tue Feb 23 02:59:48 +0000 2021@ProtifiLlc @dtabb73 Brett Phinney is the expert on that subject (e.g. PXD016169). If you see adventitious sheep keratins in a sample, it is almost always from a European group. The keratins from wool don't share tryptic peptides with human skin KRT1, KRT2, KRT9 & KRT10.
Mon Feb 22 19:27:09 +0000 2021@ProtifiLlc @dtabb73 Most European labs have such a thick coating of sheep keratin on all surfaces that a bit of extra cat keratin would be hard to detect.
Mon Feb 22 19:09:52 +0000 2021@dtabb73 Looks like somebody needs a lab cat.
Mon Feb 22 15:13:11 +0000 2021I should be working on a manuscript entitled:
"Using Machine Learning to Match Cryptic Data File Names with Experimental Descriptions in Manuscripts."
Mon Feb 22 13:45:31 +0000 2021UBR4:p is long enough that you can walk 3 km in the time it takes to translate a single molecule.
Mon Feb 22 13:02:14 +0000 2021UBR4:p.M4867L, chr 1:g.19086767T>A, rs12584 (JURKAT M:L = 2:142) vaf=59%, Δm=-17.956, is LL in JURKAT, HCT-116, U2-OS, HeLa & MCF-10A cells. It is ML in HEK-293 & SK-MEL-28 cells. It is MM in MCF-7 cells. #ᐯᐸᐱ
Mon Feb 22 13:02:14 +0000 2021UBR4:p, θ(max) = 59. aka KIAA1307, KIAA0462, RBAF600, ZUBR1. Observed in HLA class 1 & very rarely class 2 experiments. A component of the N-end rule pathway.
Mon Feb 22 13:02:13 +0000 2021>UBR4:p, ubiquitin protein ligase E3 component n-recognin 4 (Homo sapiens) Very large subunit; CTMs: A2+acetyl; PTMs: 170×S,98×T,21×Y+phosphoryl; 119×K+acetyl, 59×K+ubiquitinyl; SAAVs: T1107A (14%), M4867L (59%); mature form: (2-5183) [54,438×, 852 kTa]. #ᗕᕱᗒ 🔗
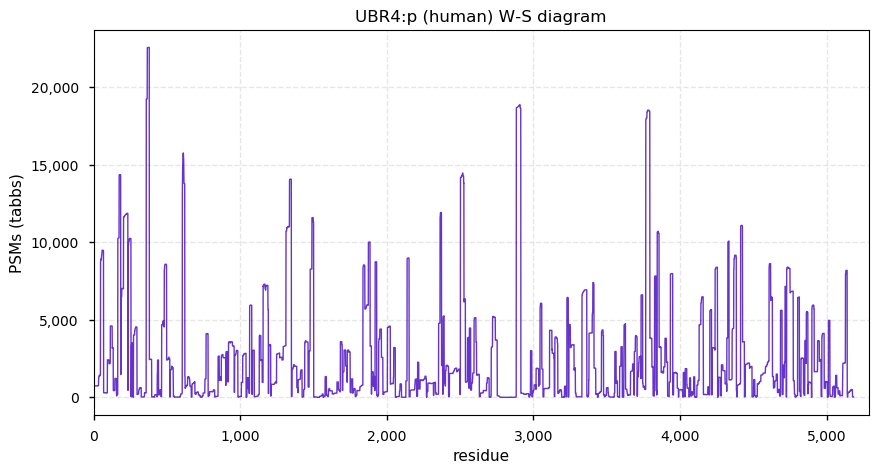
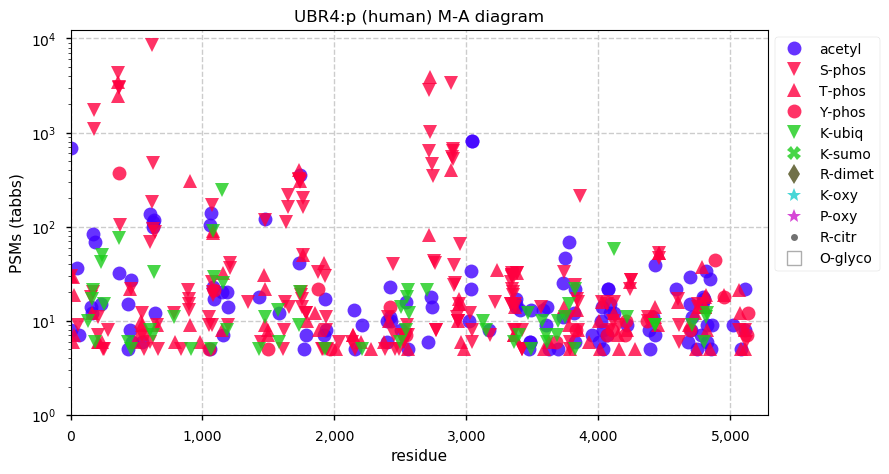
Sun Feb 21 18:45:40 +0000 2021@slashdot Nature jumps the shark ...
Sun Feb 21 16:39:48 +0000 2021It must be Sunday: a new crop of robots.txt ignoring bots have popped up.
Sun Feb 21 16:29:10 +0000 2021Thanks to everyone who took time to participate and comment. GO is one of the least examined (& tested) ideas in current bioinformatics: a critical examination of it would probably make a good thesis topic for anyone interested in the subject.
Sun Feb 21 13:38:26 +0000 2021HCLS1:p.T235A, chr 3:g.121634407T>C, rs2070179 (JURKAT T:A = 3:833) vaf=69%, Δm=-30.011, is AA in JURKAT, THP-1 & HEK-293 cells. It is TT in MCF-10A cells. #ᐯᐸᐱ
Sun Feb 21 13:38:26 +0000 2021HCLS1:p, θ(max) = 81. aka HS1, CTTNL. Observed in HLA class 1 & 2 experiments. Relatively abundant in lymphocytes and platelets. The domain (348-467) is difficult to observe. (428–486) is an SH3 domain.
Sun Feb 21 13:38:25 +0000 2021>HCLS1:p, hematopoietic cell-specific Lyn substrate 1 (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 21×S,6×T,11×Y+phosphoryl; 29×K+acetyl; aPTMs: 5×K+acetyl/ubiquitinyl; SAAVs: T235A (69%); mature form: (2-486) [18,876×, 361 kTa]. #ᗕᕱᗒ 🔗
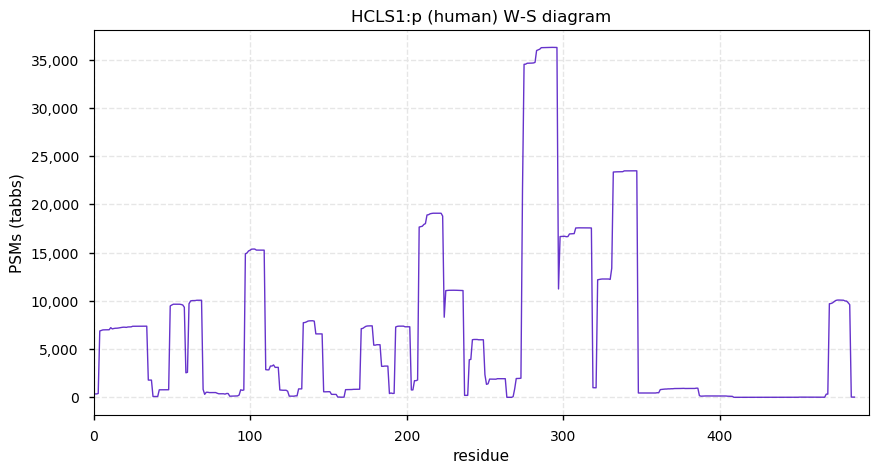
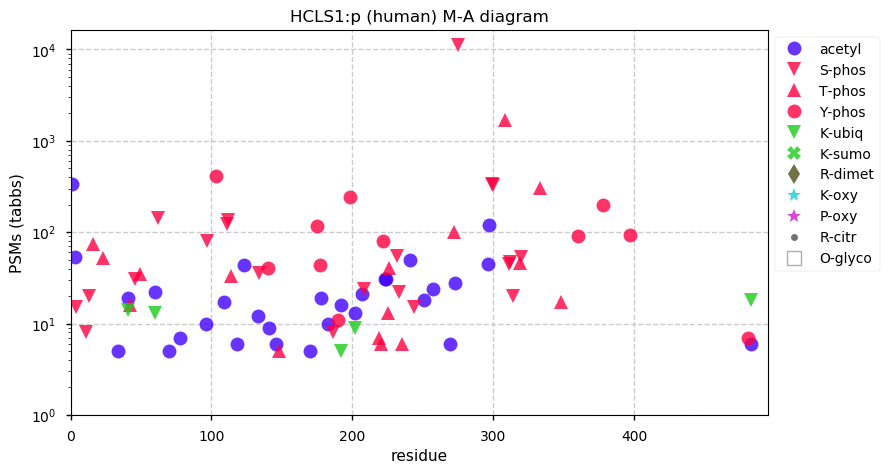
Sat Feb 20 16:24:58 +0000 2021@neely615 @pwilmarth I'm not a big fan of the broad use of directed acyclic graphs to describe living systems. They tend to produce unbalanced representations that skew towards the limited number of things that can be represented by DAGs in well-funded fields.
Sat Feb 20 15:26:59 +0000 2021An interesting take on a very (very) Canadian funding program. 🔗
Sat Feb 20 14:44:56 +0000 2021How many GO annotations associated with a single protein are too many to be useful?
Sat Feb 20 13:41:56 +0000 2021GLO1:p.E111A, chr 6:g.38682852T>G, rs4746 (MCF-10A E:A = 974:649) vaf=36%, Δm=-58.006, is EA in MCF-10A, HEK-293, HCT-116 & MCF-7 cells. It is AA in JURKAT cells. It is EE in 143B & HeLa. #ᐯᐸᐱ
Sat Feb 20 13:41:56 +0000 2021GLO1:p, θ(max) = 64. aka GLOD1. Observed in HLA class 1 & rarely in class 2 experiments. Commonly found in urine and saliva. Secreted via the extracellular vesicle mechanism.
Sat Feb 20 13:41:55 +0000 2021>GLO1:p, glyoxalase I (Homo sapiens) Small enzyme; CTMs: A2+acetyl; PTMs: 5×S,2×T,2×Y+phosphoryl; 8×K+ubiquitinyl, 8×K+acetyl; SAAVs: E111A (36%); mature form: (2-184) [57,436×, 361 kTa]. #ᗕᕱᗒ 🔗
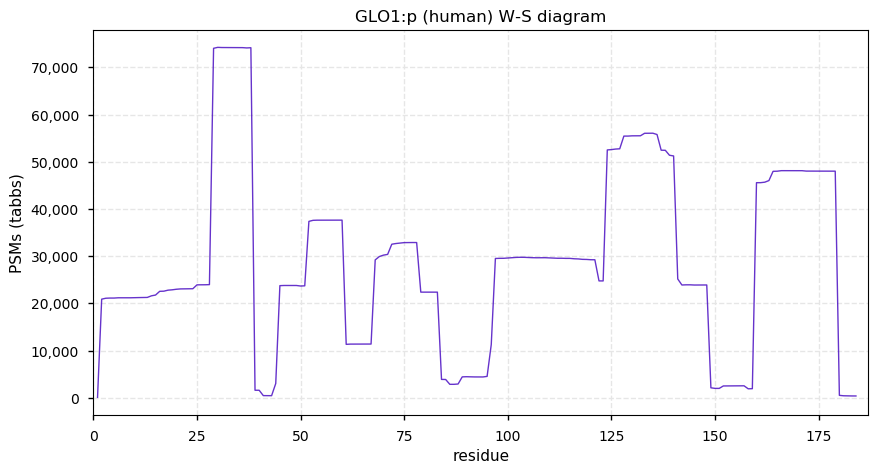
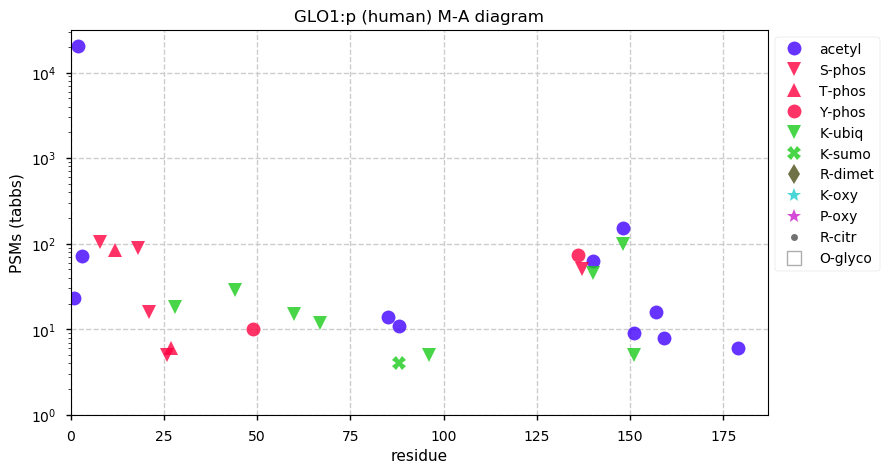
Fri Feb 19 17:11:46 +0000 2021@LewisGeer You might want to try PXD006110 (Lactobacillus delbrueckii ssp. bulgaricus). There are some IAA artifacts on amino groups, but the MS/MS is great, >50% PSM assignment & the LC is well done. Excellent for detecting N or Q deamidation, too.
Fri Feb 19 16:42:16 +0000 2021Thanks to everyone who participated. 86% of respondents indicated that they have changed their behavior wrt attending conferences. The comments were informative, suggesting a decrease in information exchange & satisfaction, even among those attending more events.
Fri Feb 19 16:00:45 +0000 2021@dtabb73 Turn that puppy off
🔗
Fri Feb 19 15:58:46 +0000 2021I happened across PXD004280 again: I had forgotten how good it was from a purely technical point of view. If you want to test an algorithm with excellent human cell line data where the % of identified spectra is high (>70%), you should try this one.
Fri Feb 19 15:38:32 +0000 2021Does anyone else have comments, before the poll closes? 🔗
Fri Feb 19 14:22:31 +0000 2021@bkives @joannehlev Good story. Keep digging.
Fri Feb 19 13:01:46 +0000 2021TACSTD2:p.D216E, chr 1:g.58576509G>T, rs14008 (MCF-10A D:E = 454:549) vaf=9%, Δm=14.016, is DE in MCF-10A, OVCA-429 & HK-1 cells. It is DD in MCF-7, HCT-116, HeLa & HEK-293 cells. #ᐯᐸᐱ
Fri Feb 19 13:01:46 +0000 2021TACSTD2:p, θ(max) = 64. aka TROP2, GA733-1, EGP-1, M1S1. Observed in HLA class 1 & rarely in class 2 experiments. Commonly found in urine and saliva. Secreted via the extracellular vesicle mechanism.
Fri Feb 19 13:01:46 +0000 2021>TACSTD2:p, tumor associated calcium signal transducer 2 (Homo sapiens) Small subunit; CTMs: N33,N168+glycosyl; PTMs: Y104,Y224,Y225,Y259,S322+phosphoryl; K312+ubiquitinyl; SAAVs: E147D (4%), D216E (9%), I297V (3%); mature form: (31-323) [11,192×, 68 kTa]. #ᗕᕱᗒ 🔗
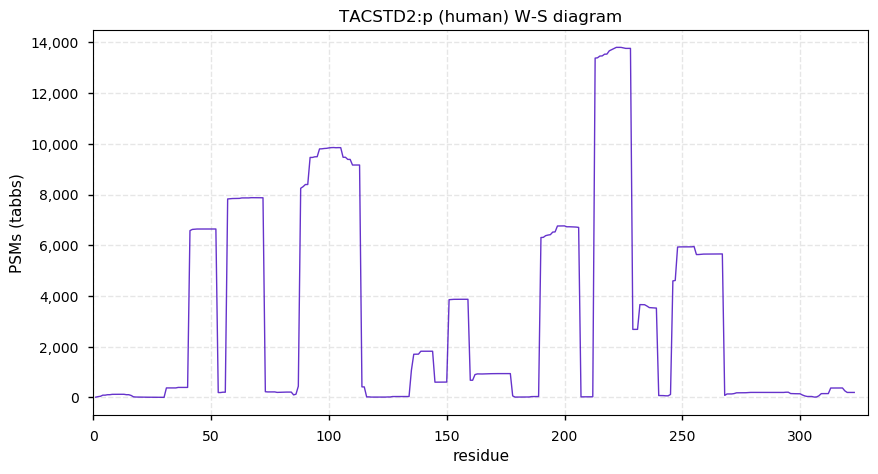
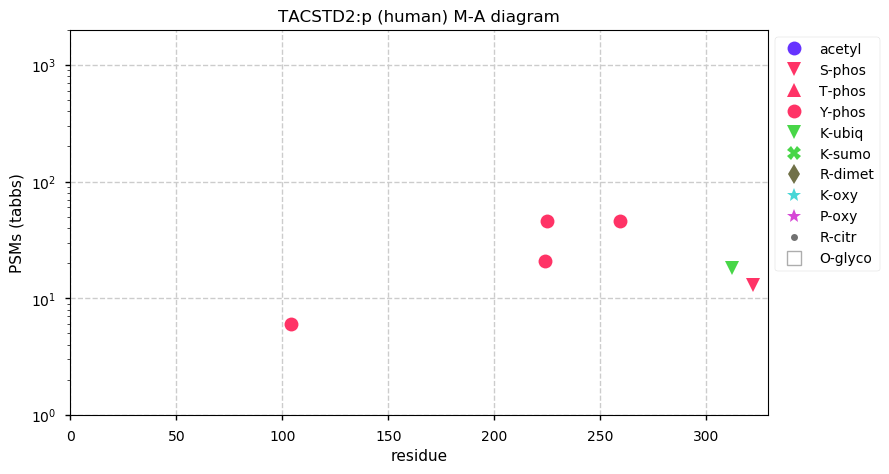
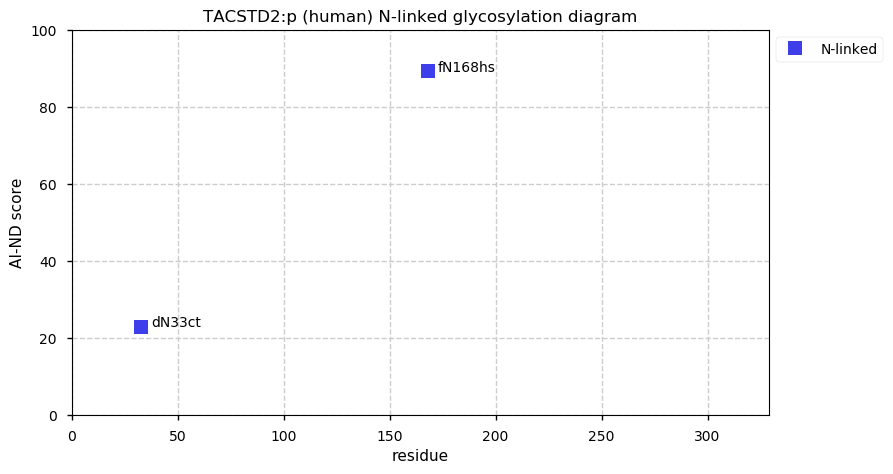
Thu Feb 18 20:24:12 +0000 2021Similarly, HLA-C*14:02 corresponds to searching for "MHC1 C1402" 🔗
Thu Feb 18 20:21:48 +0000 2021If you want to get the data for a specific allele, you can pull it out of GPMDB. For instance, if you want the data for the allele HLA-A*02:05, you can search for "MHC1 A0205" 🔗
Thu Feb 18 20:18:45 +0000 2021🔗 is a pretty interesting if you like HLA class I data sets. They isolate the epitope peptides from 95 specific HLA alleles, defining the sequence specificities for each of the alleles.
Thu Feb 18 19:34:21 +0000 2021At what point does optimistic data interpretation to conform with an idea become good old fashioned fabrication?
Thu Feb 18 16:33:07 +0000 2021Now that scientific conferences are virtual, many of the physical constraints on attending them are no longer relevant. Has this change led you to attend
Thu Feb 18 15:04:17 +0000 2021@AlexUsherHESA It will be a tough sell, given the attitude the Premier has consistently displayed wrt post-secondary education. No one I know here is willing to give the current government the benefit of doubt & accept any proposal as a real change of course.
Thu Feb 18 14:28:43 +0000 2021@dtabb73 I gave up on them a quite while ago. If you are a little clever about using M.2 NVMe SSDs, you can get very good performance without most of the problems.
Thu Feb 18 13:48:03 +0000 2021Still cold across the centre of North America, all the way down to the Gulf of Mexico 🔗
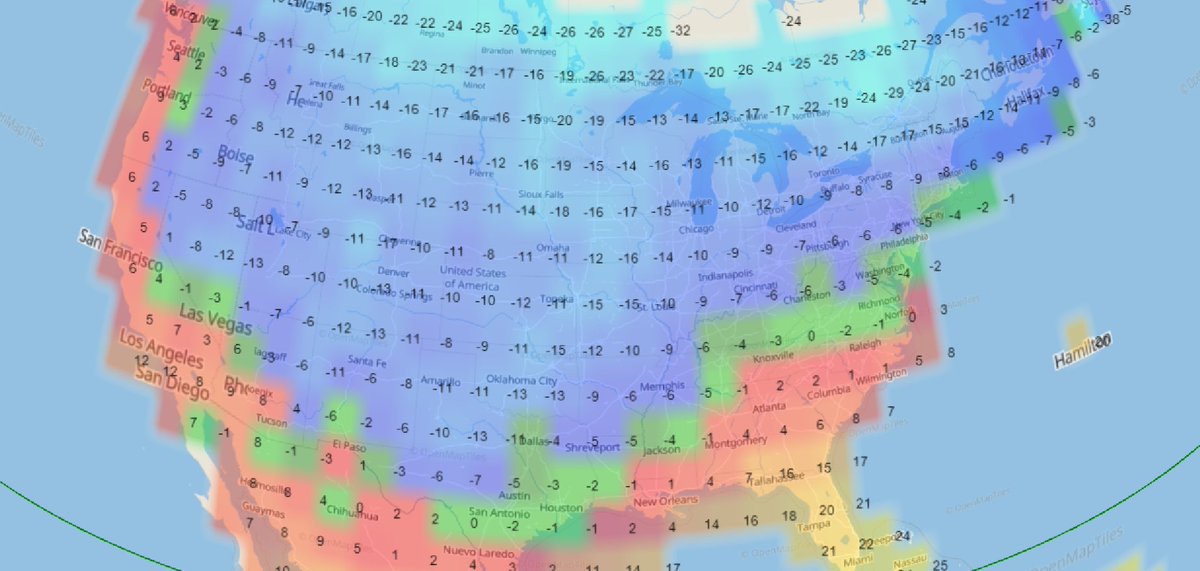
Thu Feb 18 13:00:51 +0000 2021DNAJC10:p.H646Q, chr 2:g.182757820T>G, rs288334 (MCF-10A H:Q = 97:210) vaf=63%, Δm=-9.000, is HQ in MCF-10A, Hep-G2 & HCT-116 cells. It is QQ in MCF-7, SK-MEL-28 & HeLa cells. It is HH in HEK-293 & HeLa cells. #ᐯᐸᐱ
Thu Feb 18 13:00:51 +0000 2021DNAJC10:p, θ(max) = 62. aka ERdj5, PDIA19. Observed in HLA class 1 & class 2 experiments. Reduces disulphide bonds present in misfolded proteins produced in the endoplasmic reticulum.
Thu Feb 18 13:00:51 +0000 2021>DNAJC10:p, DnaJ heat shock protein family (Hsp40) member C10 (Homo sapiens) Midsized ER subunit; PTMs: Y414+phosphoryl; 7×K+ubiquitinyl; SAAVs: H646Q (64%); mature form: (33-793) [52,705×, 434 kTa]. #ᗕᕱᗒ 🔗
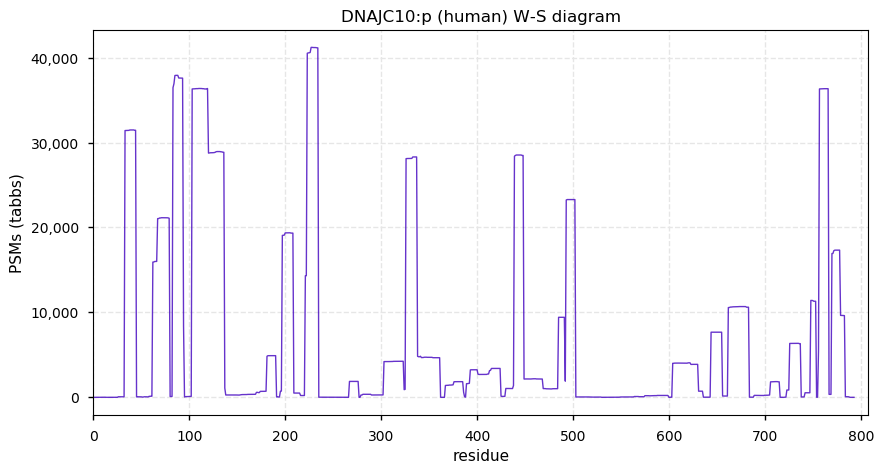
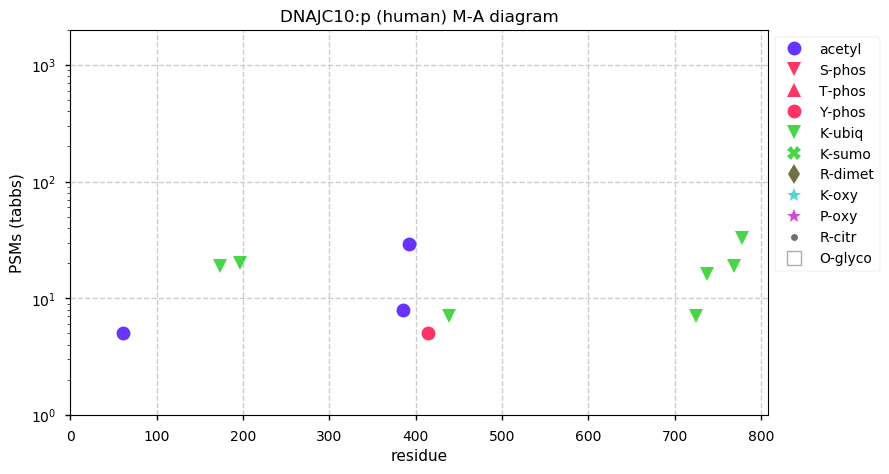
Wed Feb 17 21:20:30 +0000 2021@jwoodgett @MHendr1cks @DrDCourtney @AVoineskos @CIHR_IRSC 🔗
Wed Feb 17 17:53:02 +0000 2021@slavov_n @ass_deans Warning: while this works, every time you see that person again they will make a beeline to you & continue telling you about their experiences with CommonCV until you can escape. It has induced a PTSD-like psychosis in the affected Canadian academic community.
Wed Feb 17 15:19:11 +0000 2021@slavov_n @ass_deans If you ever have a Canadian academic in your office & nothing to talk about, mention this problem then tell them that you have heard of a great system Canada has for dealing with the issue: "CommonCV". You won't have to say anything else for the rest of your time with them.
Wed Feb 17 13:49:42 +0000 2021@slavov_n @ass_deans Really anything to do with a CV. It is the most asked for but least read document in academia.
Wed Feb 17 13:14:16 +0000 2021Congrats of Jiang Liu, et al. for the quality of PXD022673. The data is a single high/low pH multidimensional chromatography run with TMT, but is it a good one! ✅
Wed Feb 17 13:04:34 +0000 2021UQCRH:p.E22G, chr 1:g.46309111A>G, rs41292543 (MCF-10A E:G = 146:224) vaf=6%, Δm=-72.021, is EG in MCF-10A cells. It is GG in HAP-1 cells. It is EE in HEK-293, HeLa & MCF-7 cells. #ᐯᐸᐱ
Wed Feb 17 13:04:34 +0000 2021UQCRH::p,θ(max) = 91. aka QCR6, UQCR8. Observed in HLA class 1 experiments (but not class 2). Biomarker of inflamation when found in urine. Subunit of mitochondrial complex II. Evidence for alternate translation initiation at M9.
Wed Feb 17 13:04:33 +0000 2021>UQCRH:p, ubiquinol-cytochrome c reductase hinge protein (Homo sapiens) Very small subunit; PTMs: 4×S,5×T+phosphoryl; K8,K42,K85+acetyl/ubiquitinyl; SAAVs: E22G (6%); mature form: (14-91) [18,475×, 101 kTa]. #ᗕᕱᗒ 🔗
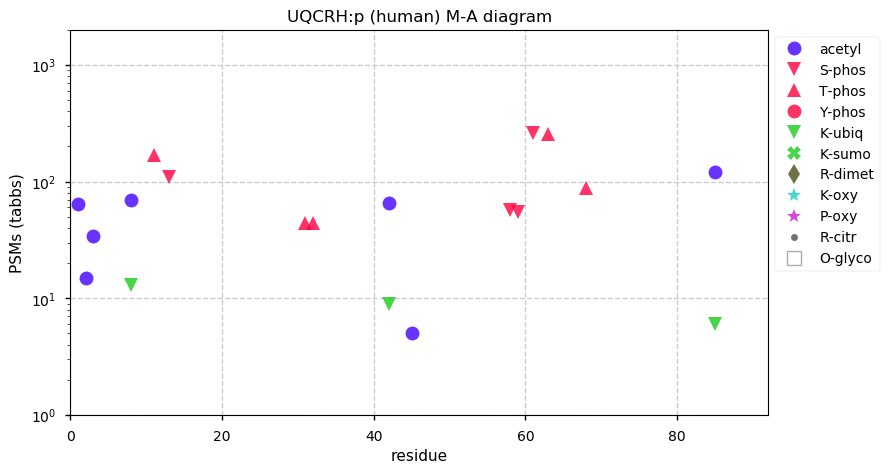
Tue Feb 16 22:18:26 +0000 2021@theoneamit Just out of idle interest, has anyone doing open searches found this particular modification?
Tue Feb 16 17:47:33 +0000 2021But that problem aside, the MS/MS and HPLC methods used for PXD021518 were particularly well done.
Tue Feb 16 17:04:11 +0000 2021Still chilly in Austin TX: 🔗
Tue Feb 16 16:44:52 +0000 2021I was just about to say how well executed the experiments leading to a data set were, until I noticed they had managed to lose 95% of the cysteine-containing peptides.
Tue Feb 16 13:47:08 +0000 2021@theoneamit Just to wrap it up, ufmylation leaves a VG (+170.105528) remnant on the modified lysine ε-amino group after trypsin cleavage.
Tue Feb 16 13:06:47 +0000 2021PCMT1:p.V178I, chr 6:g.149793609G>A, rs4816 (HEK-293 V:I = 16:288) vaf=46%, Δm=14.016, is II in HEK-293, A-431, HeLa & MCF-7. It is VV in CACO-2, A-549 & MCF-10A cells. #ᐯᐸᐱ
Tue Feb 16 13:06:47 +0000 2021PCMT1:p, θ(max) = 98. Observed in HLA class 1 experiments. PTMs all very near the LOD. Enzyme involved in the conversion of damaged protein residues (D-aspartyl & L-isoaspartyl) to L-aspartyl.
Tue Feb 16 13:06:46 +0000 2021>PCMT1:p, protein-L-isoaspartate (D-aspartate) O-methyltransferase (Homo sapiens) Small enzyme; CTMs: M1,A2+acetyl; PTMs: S11,S132,Y213+phosphoryl, K174+ubiquitinyl, SAAVs: V178I (46%); mature form: (1-228) [71,599×, 322 kTa]. #ᗕᕱᗒ 🔗
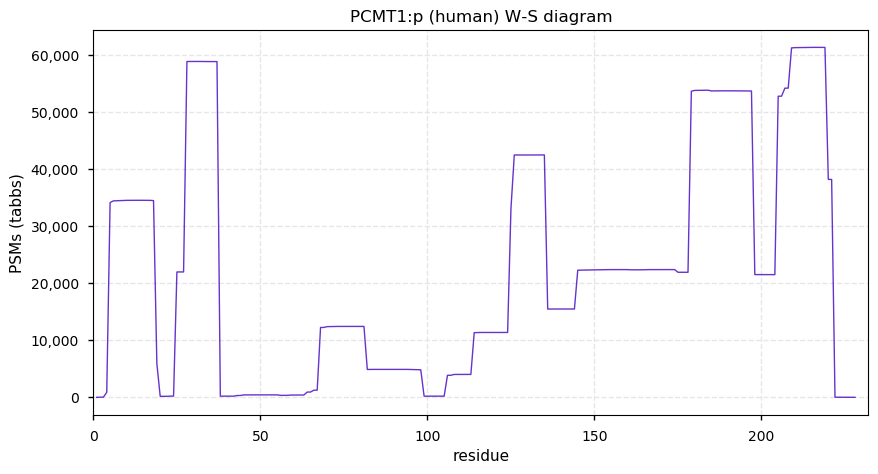
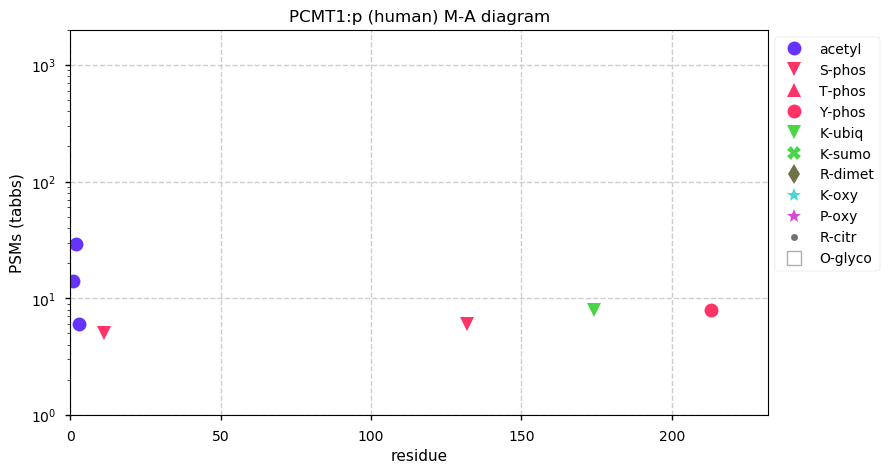
Mon Feb 15 21:34:13 +0000 2021Kind of makes me feel ancient, with Matthias' son (who I remember as being a little kid) being old enough to co-author papers 🔗
Mon Feb 15 20:00:27 +0000 2021@Smith_Chem_Wisc @nesvilab @UCDProteomics Wolverines live in colder climates - Wisconsin, Michigan (& St. Petersburg) are just too warm for them. 🔗
Mon Feb 15 19:36:25 +0000 2021Ufmylation? It sounds like you guys are just making things up now 🔗
Mon Feb 15 17:29:11 +0000 2021As of now, it is -12 °C in Austin TX and -10 °C in St. Petersburg RU.
Mon Feb 15 16:30:23 +0000 2021While it is seasonably cold up here, Austin TX (2400 km due south of Winnipeg) does not get down to -15 °C very often 🔗
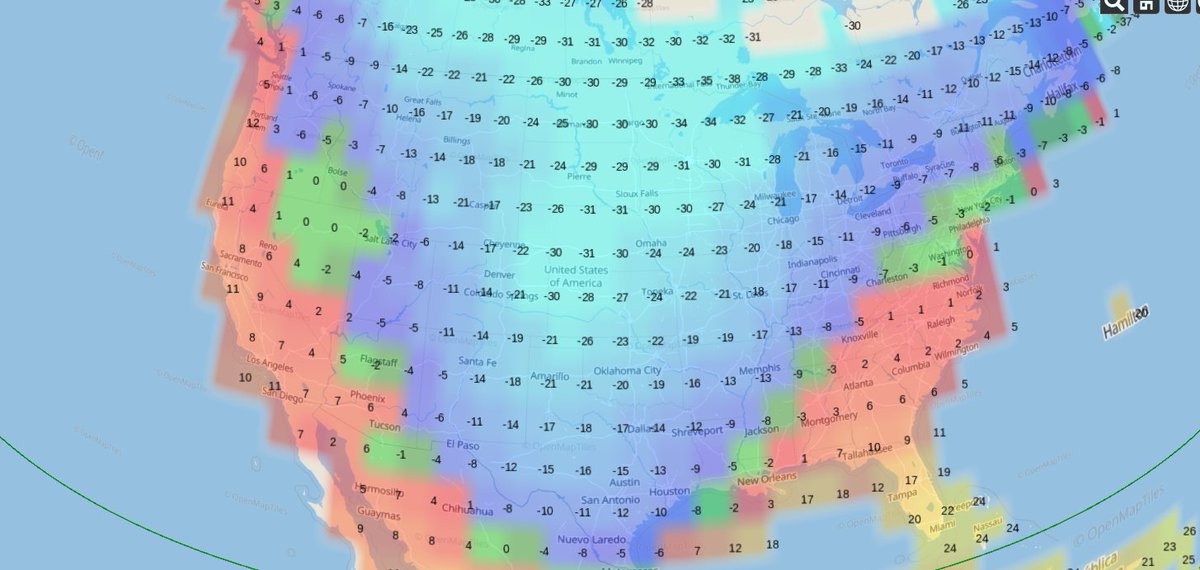
Mon Feb 15 14:53:22 +0000 2021@AlexUsherHESA @GlobalHigherEd NSERC is in a bit of hard place. It is mandated to partner with companies for tech development/transfer, but superclusters & other bespoke Fed funding projects have systematically strip-mined contributions from all of the eligible Canadian companies.
Mon Feb 15 13:26:23 +0000 2021MYH9:p.I1626V, chr 22:g.36288308T>C, rs2269529 (A-431 I:V = 3:458) vaf=26%, Δm=-14.016, is VV in A-431 & BT-474 cells. It is IV in SK-MEL-28 cells. It is II in HEK-293, MCF-7, MCF-10A & HeLa cells. #ᐯᐸᐱ
Mon Feb 15 13:26:23 +0000 2021MYH9:p, θ(max) = 92. aka NMMHCA, NMHC-II-A, MHA, FTNS, EPSTS, DFNA17. Observed in HLA class 1 & 2 experiments. A very commonly observed non-muscle class IIa myosin subunit found in most cells.
Mon Feb 15 13:26:22 +0000 2021>MYH9:p, myosin heavy chain 9 (Homo sapiens) Large protein; CTMs: A2+acetyl; PTMs: 77×S,74×T,26×Y+phosphoryl, 204×K+acetyl (total), 8×R+deimination; aPTMs: 80×K+acetyl/ubiquitinyl, SAAVs: I1626V (26%); mature form: (1-1960) [125,640×, 9584 kTa]. #ᗕᕱᗒ 🔗
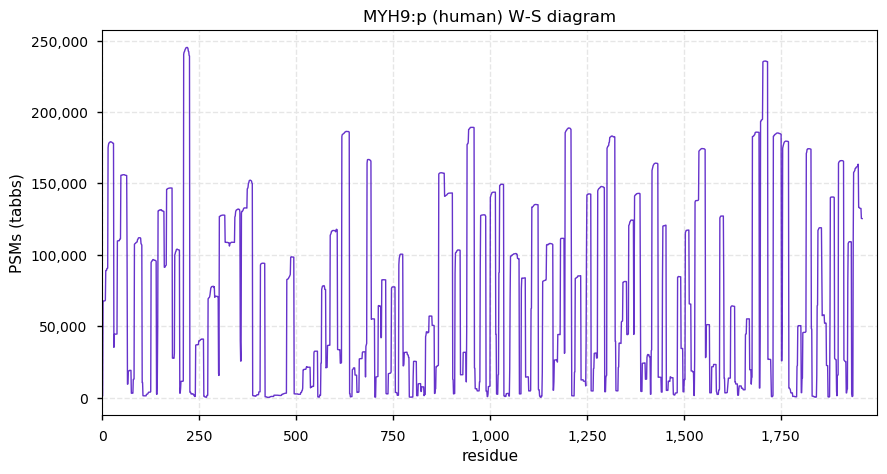
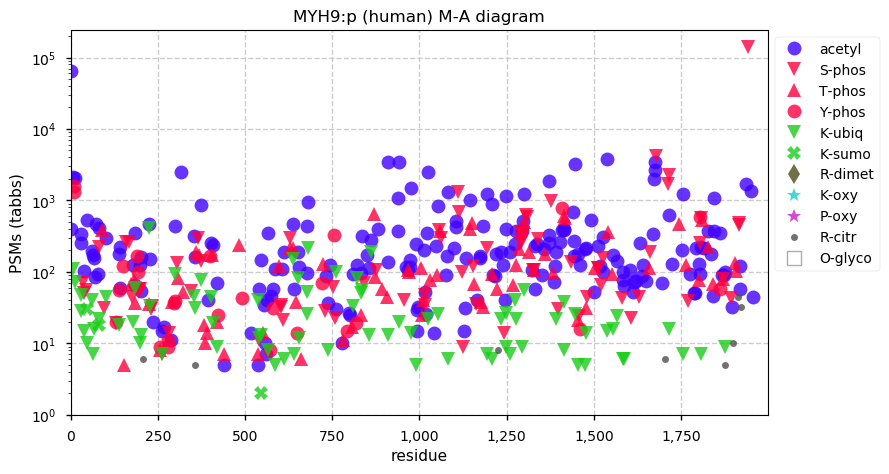
Sun Feb 14 20:14:56 +0000 2021@nesvilab @DanielLiebler1 Not that it is a contest ... 🔗
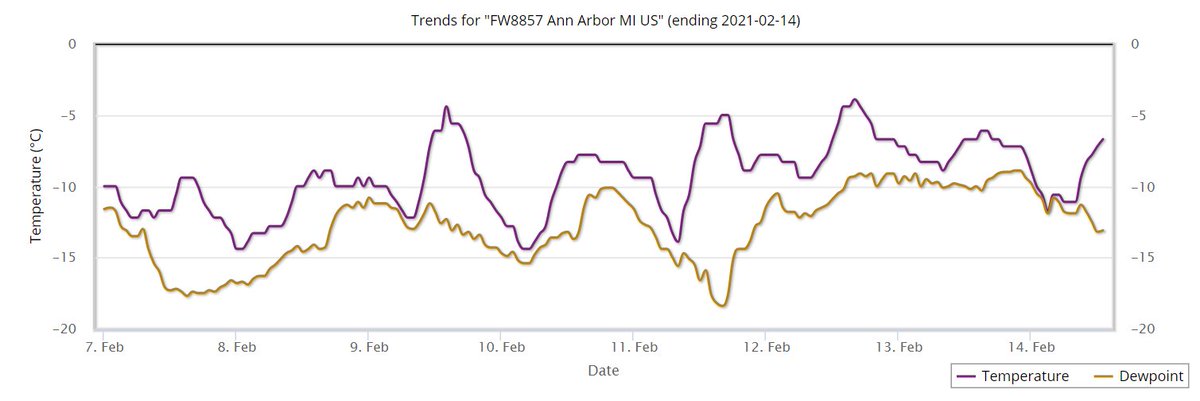
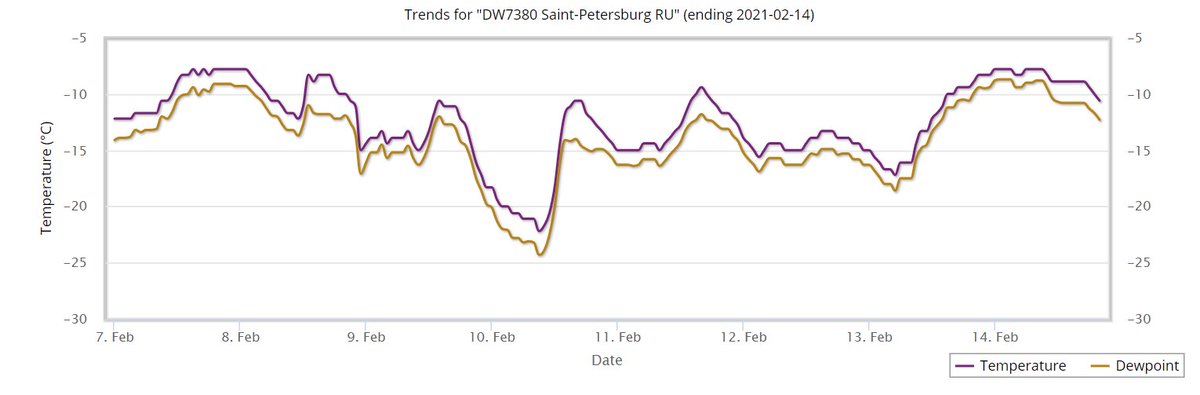
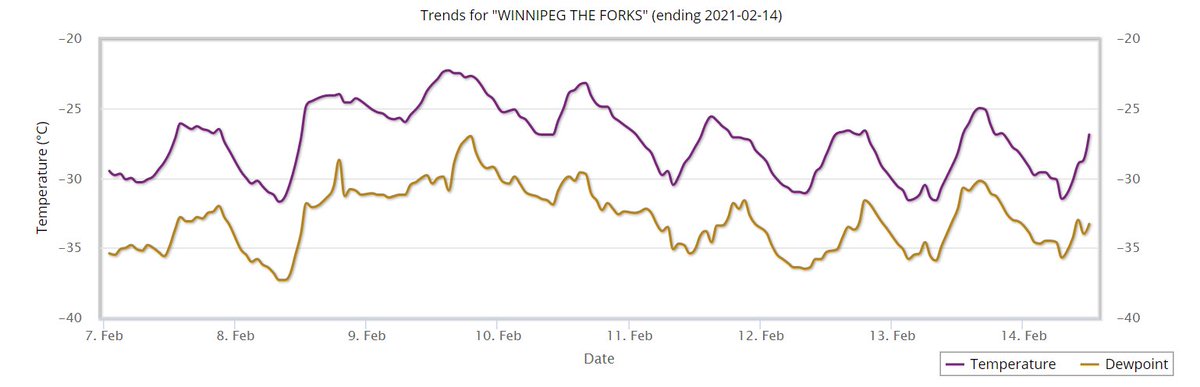
Sun Feb 14 15:51:47 +0000 2021Bracing weather across the Great Plains this morning 🔗
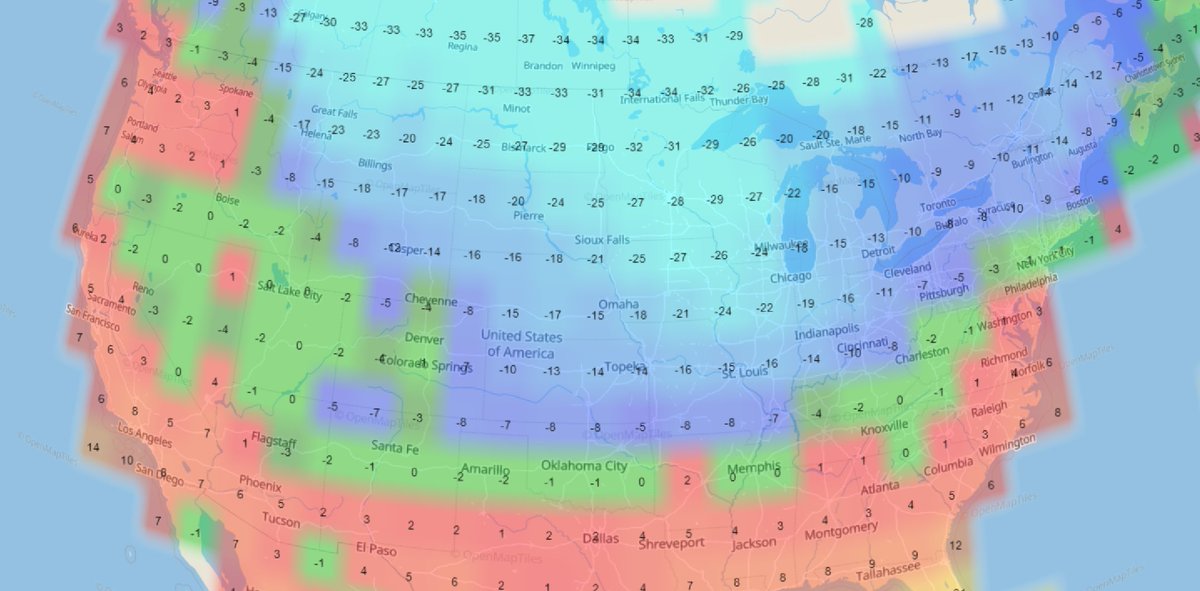
Sun Feb 14 14:56:46 +0000 2021Because IFITM1:p.P13A has a vaf=100%, it is usually listed as having a MAF=0. Uniprot lists the reference sequence P13 rather than the ancestral A13.
Sun Feb 14 14:53:14 +0000 2021IFITM1:p is part of a family of small proteins that inhibit infection by coronaviruses & other enveloped viruses by interfering with viral particle release into the cytoplasm. IFITM1, 2 & 3 have significant similarity, but they each produce unique detectable tryptic peptides.
Sun Feb 14 14:32:46 +0000 2021IFITM1:p.P13A, chr 11:g.314207C>G, rs9667990 (HeLa P:A = 0:904) vaf=100%, Δm=-26.016, is AA in HeLa & all commonly used cell lines. #ᐯᐸᐱ
Sun Feb 14 14:32:46 +0000 2021IFITM1:p, θ(max) = 44. aka 9-27, CD225, DSPA2a, IFI17. Observed in HLA class 1 & 2 experiments. Common in extracellular vesicles, particularly in urine. Membrane spanning domains (37-57) & (87-107).
Sun Feb 14 14:32:46 +0000 2021>IFITM1:p, interferon induced transmembrane protein 1 (Homo sapiens) Small protein; CTMs: M1+acetyl; PTMs: T17,T73,Y78+phosphoryl, K3,K67+ubiquitinyl; SAAVs: P13A (100%); mature form: (1-125) [14,796×, 56 kTa]. #ᗕᕱᗒ 🔗
Sat Feb 13 15:27:22 +0000 2021@slavov_n But at some point, in order to advance, you do have to take off the training wheels.
Sat Feb 13 14:57:56 +0000 2021I would dispute the idea that FOSS had anything to do with open source software in proteomics, 🔗 For me it was a matter of overreach by Thermo in pushing rights on a licensed patent.
Sat Feb 13 14:15:21 +0000 2021HSPG2:p is so long that it takes as much time for a ribosome to translate a single HSPG2:r mRNA as it does to bake a cake.
Sat Feb 13 14:06:21 +0000 2021HSPG2:p.S4331N, chr 1:g.21823627C>T, rs3736360 (HCT-116 S:N = 16:13) vaf=18%, Δm=27.011, is SN in HCT-116 cells. It is NN in A-431 & U2-OS cells. HSPG2:p is rarely observed in cell lines: in all sources (incl. tissue) S:N = 7376:2321. #ᐯᐸᐱ
Sat Feb 13 14:06:21 +0000 2021HSPG2:p, SAAVs: R1186Q (6%), G1230R (6%), A1503V (31%), S2995R (9%), A3168T (8%), H3256Y (15%), H3420Q (9%), R3632L (9%), D4104E(74%), S4331N (18%), V4332I (5%);
Sat Feb 13 14:06:21 +0000 2021HSPG2:p, θ(max) = 77. aka perlecan, PRCAN, SJS1. Observed in HLA class 1 & 2 experiments. Common in blood plasma, urine & CSF.
Sat Feb 13 14:06:20 +0000 2021>HSPG2:p, heparan sulfate proteoglycan 2 (Homo sapiens) Large protein; CTMs: N89, N554, N1755, N2121, N3279, N3780, N3836, N4068+glycosyl; PTMs: 22×S,13×T+phosphoryl, 6×S,5T+glycosyl; mature form: (26-4391) [38,517×, 1889 kTa]. #ᗕᕱᗒ 🔗
Fri Feb 12 20:21:41 +0000 2021@pwilmarth @dtabb73 PSM assignment inference only runs one way:
spectrum→peptides→proteins
Trying to loop backwards causes problems.
Fri Feb 12 18:45:06 +0000 2021PXD022851: nice data too👍 (no associated manuscript).
Fri Feb 12 17:28:23 +0000 2021PXD020908: nice data 👍🔗
Fri Feb 12 17:07:24 +0000 2021@AlexUsherHESA Canada produces talent: there never has been sustained interest in utilizing that talent domestically.
Fri Feb 12 15:40:14 +0000 2021@dtabb73 @theoneamit @mjmaccoss @thabangh @lukas_k @nesvilab Sounds like you are already in the thrall of those rascally French, who do some fabulous bacterial work with > 90% spectra resulting in PSMs!
Fri Feb 12 13:05:31 +0000 2021CPVL:p.A435V, chr 7:g.29030593G>A, rs7313 (HCT-116 A/V = 10/890) vaf=39%, Δm=28.03, is VV in HCT-116, MCF-10A, Hep-G2 & HEK-293 cells. It is AA in JURKAT, HCC-827 & MDA-MB-231 cells. #ᐯᐸᐱ
Fri Feb 12 13:05:31 +0000 2021CPVL:p, θ(max) = 85. Observed in HLA class 1 & 2 experiments. A combination of N-terminal processing and alternate translation initiation at M33 makes assigning an N-terminus to this protein ambiguous. Commonly observed in urine and CSF.
Fri Feb 12 13:05:31 +0000 2021>CPVL:p, carboxypeptidase vitellogenic like (H sapiens) Midsized protein; CTMs: N81,N132,N307,N346+glycosyl; PTMs: 3×S,2×T+phosphoryl, 6×K+ubiquitinyl; SAAVs: R25H (7%), M71V (1%), Y168H (1%), R398H (21%), A435V (39%); mature form: (30,33?-476) [34,263×, 204 kTa] #ᗕᕱᗒ 🔗
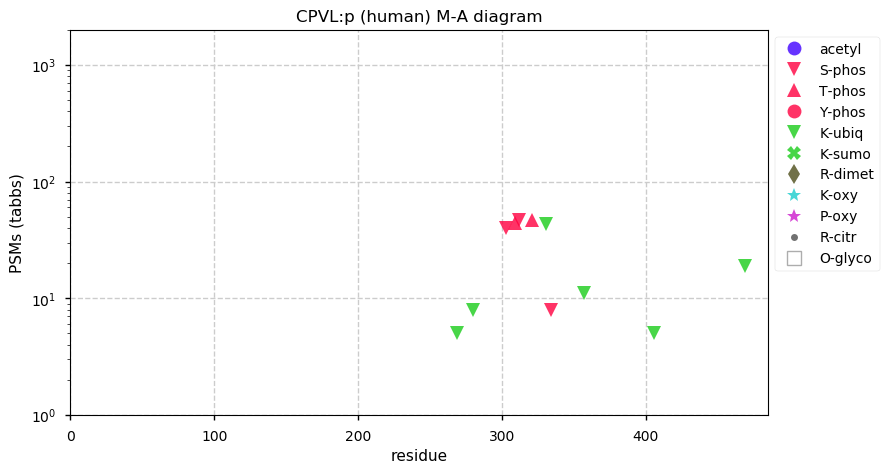
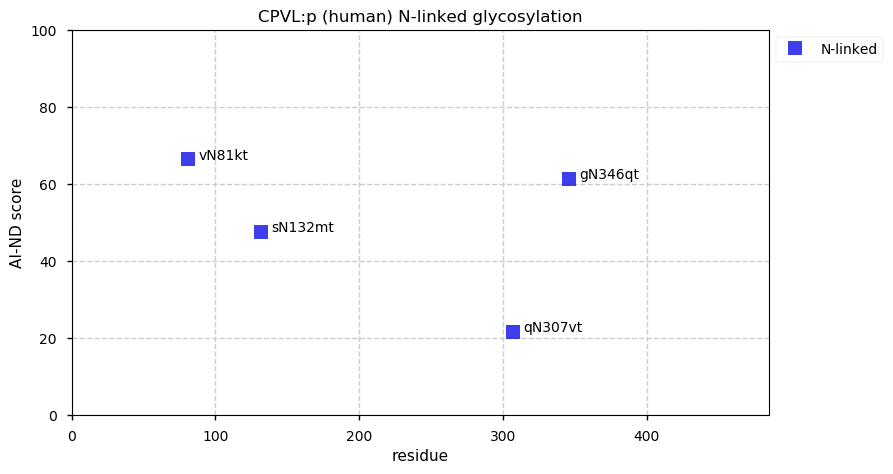
Thu Feb 11 20:19:25 +0000 2021@pwilmarth @molcellprot @byu_sam IMHO, making the raw data available obviates the need for reporting images and/or specialized viewers.
Thu Feb 11 18:55:43 +0000 2021@pwilmarth @byu_sam Does MCP still require bitmap images for every PTM? It always seemed an odd idea to me.
Thu Feb 11 16:04:53 +0000 2021Biggest space rock to just miss the earth today:
2021 CO, closest approach ~340,000 km, 31 meter diameter, orbit 🔗
Thu Feb 11 15:47:57 +0000 2021@TrumanLab @HombreRRo Given the results from this paper, I need to add NEDDylation to the list. It is reusing the same K-sites that SUMOyl, ubiquitinyl and acetyl share. 🔗
Thu Feb 11 15:38:35 +0000 2021@Sci_j_my @ProtifiLlc I'll be surprised if it can be reduced to practice, but I have been surprised before, so ... #thenewproteomics
Thu Feb 11 15:33:22 +0000 2021@Sci_j_my @ProtifiLlc Now I know! A panel of not-very specific antibodies/affinity reagents applied sequentially to build up a pattern that is hoped to be unique enough to provide an identification.
Thu Feb 11 14:19:31 +0000 2021@Sci_j_my This paper describes the options around public disclosure for US patents. 🔗
Thu Feb 11 13:59:30 +0000 2021@Sci_j_my Still an open question. I have looked fairly hard (patents, literature, web) & I have no clue as to what the technology might be. None of the principals are R&D types or inventors. Single molecule detection for proteins seems odd to me, but maybe there is an application?
Thu Feb 11 13:06:40 +0000 2021MPRIP:p.P327Q, chr 17:g.17142710C>A, rs3744137 (HEK-293 P/Q = 3/442) vaf=51%, Δm=31.006, is QQ in HEK-293 & HCT-116. It is PQ in MCF-7 & JURKAT cells. It is PP in MCF-7, A-549, A-431 & HeLa cells. #ᐯᐸᐱ
Thu Feb 11 13:06:40 +0000 2021MPRIP:p, θ(max) = 70. aka RHOIP3, M-RIP, p116Rip. Observed in HLA class 1 (but not class 2) experiments. Very rare in biological fluids.
Thu Feb 11 13:06:40 +0000 2021>MPRIP:p, myosin phosphatase Rho interacting protein (Homo sapiens) Large protein; CTMs: S2+acetyl; PTMs: 65×S,18×T,4×Y+phosphoryl, 3×K+ubiquitinyl; SAAVs: P327Q (51%), D1001E (1%); mature form: (2-1038) [25,279×, 210 kTa]. #ᗕᕱᗒ 🔗
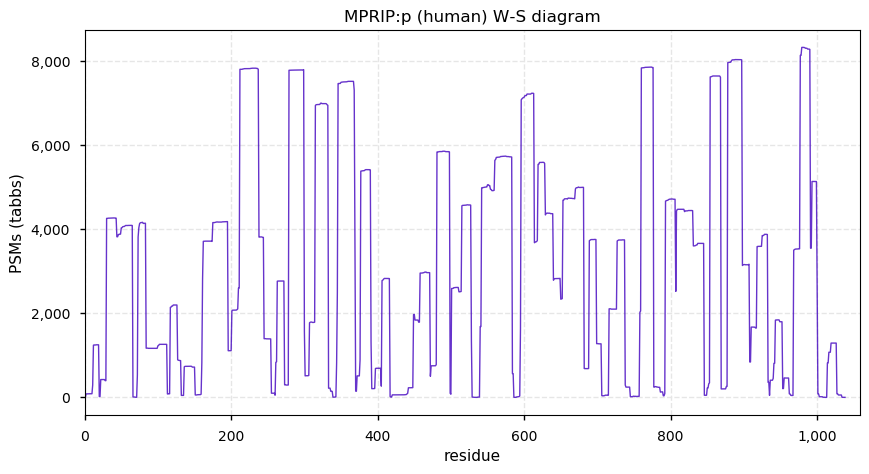
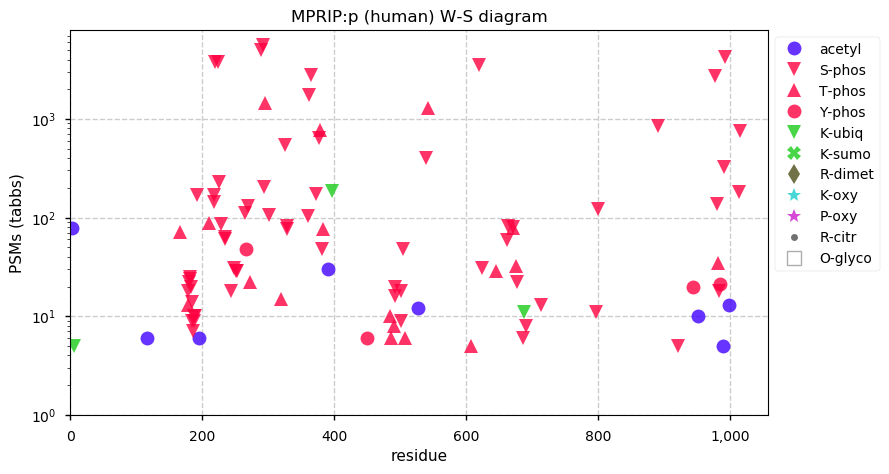
Thu Feb 11 00:15:30 +0000 2021Thanks to everyone who participated. Based on the results from PXD014917, the phosphorylated PSM ratios for unfractionated human milk are approximately
SPP1:CSN1S1:CSN2:CSN3
45 : 10 : 2 : 0
SPP1:p is also the dominant source of phosphopeptides in urine proteomics data.
Wed Feb 10 21:53:53 +0000 2021@MattWFoster There isn't a casein gamma gene: it was just my brain logically using Greek alphabetical order, even though protein naming is usually a logic-free zone. The gamma casein fraction is largely cleaved beta casein. CSN3, though, remains the same as it ever was.
Wed Feb 10 20:53:13 +0000 2021@pwilmarth @astacus Anything biomedical using log base 2 should be clearly marked as being for MDs only.
Wed Feb 10 19:47:37 +0000 2021@AlexUsherHESA I can't prove it, but I would be surprised if CBC had not linked the words "historic" & "Tim Horton's" at some point in time.
Wed Feb 10 17:47:59 +0000 2021@astacus logs with bases other than 10 or e should be banned, unless you write a 1000 word defense of using it in a particular application.
Wed Feb 10 17:04:28 +0000 2021@AJ_Brenes @pwilmarth STAT5A/B is a good example of one of the cases where general quant. solutions fail rather awkwardly. Looking at it for a while, I'm not sure that even a detailed, bespoke solution is going to be particularly accurate.
Wed Feb 10 15:15:30 +0000 2021@astacus At least they are keeping track of the batch number!
Wed Feb 10 14:16:36 +0000 2021IFI16:p is commonly observed in many tissues & cell lines. As per the literature, it is present in myeloid lineage cells. It is also present in many CPTAC tumor expts as well as common cell lines, such as HeLa, U2-OS & MCF10A. It is absent from HCT-116 cells.
Wed Feb 10 13:31:07 +0000 2021Sorry, CSN3 is casein kappa, not gamma.
Wed Feb 10 13:12:11 +0000 2021IFI16:p.S179T, chr 1:g.159016687G>C, rs866484 (JURKAT S/T = 3/442) vaf=74%, Δm=14.016, is TT in JURKAT, A-431 & HeLa cells. It is ST in HEK-293, TIG-3-20, SK-MEL-28 & MCF-10A cells. It is SS in RKO cells. #ᐯᐸᐱ
Wed Feb 10 13:12:10 +0000 2021IFI16:p, θ(max) = 81. aka IFNGIP1, PYHIN2. Observed in HLA class 1 experiments. Has 4 potential nuclear localization signal domains. Given the SAAV vafs, any observed protein will likely have at least 1 SAAV.
Wed Feb 10 13:12:10 +0000 2021>IFI16:p, interferon γ inducible protein 16 (H sapiens) PTMs: 31×S,28×T,0×Y+phosphoryl, 42×K+ubi; aPTMs: 12×K+acetyl/ubi, 16×K+acetyl/ubi/SUMOyl; SAAVs: S179T (74%), R409S (63%), Y413N (63%), S512F (2%), T723S (23%); mature form: (2-729) [29,608×, 307 kTa]. #ᗕᕱᗒ 🔗
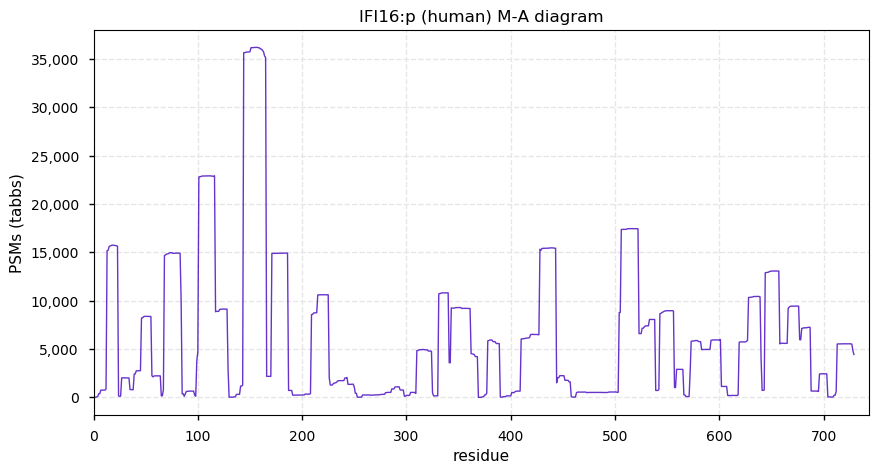
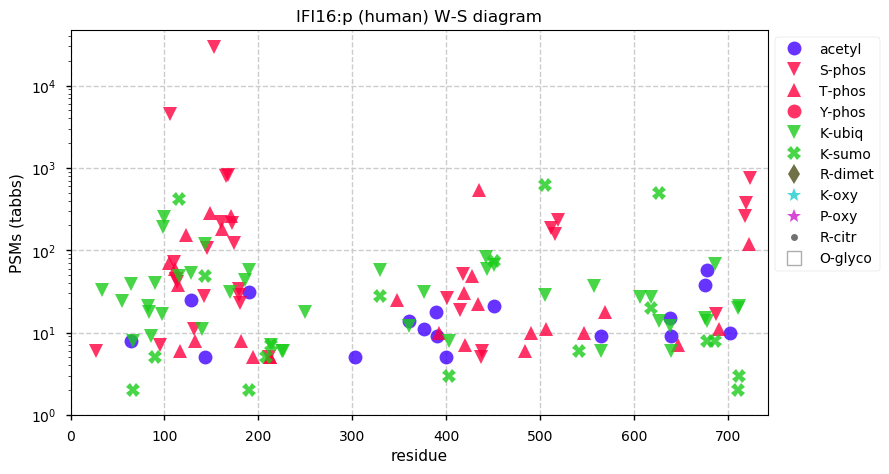
Wed Feb 10 00:01:37 +0000 2021In a human milk sample, which protein produces the largest number of phosphopeptide PSMs?
Tue Feb 09 22:39:56 +0000 2021Has anyone ever detected proteins from bacteria in human milk, e.g. L. fermentum?
Tue Feb 09 18:13:28 +0000 2021@UCDProteomics The conventional general compression methods are pretty slow & don't work that well for binary data, but work very well for XML files. I have custom utilities for mass-spec binary related stuff using differential compression.
Tue Feb 09 17:59:25 +0000 2021@UCDProteomics Amen, brother. Compress, compress, compress.
Tue Feb 09 16:54:34 +0000 2021@neely615 @astacus Don't know if this was cause or effect, though.
Tue Feb 09 16:54:16 +0000 2021@neely615 @astacus From my recollection of things, it happened during a period of growth in positions with titles like "Associate Dean of Research" or "Vice-President of Research", which didn't exist (or were less important) at most universities prior to the late 1980's.
Tue Feb 09 16:38:16 +0000 2021@neely615 @astacus I'm willing to bet it always involved a committee meeting.
Tue Feb 09 16:07:36 +0000 2021@byu_sam I have a odd way of thinking about this sort of thing, so I may not be getting it. Are you thinking about something like this, where we did an analysis of the data for each phosphorylation assignment & mapped significant ones to genomic coördinates: 🔗
Tue Feb 09 15:55:50 +0000 2021@byu_sam Not sure what "gene-level phosphorylation metric" means. Could you give a hypothetical example?
Tue Feb 09 15:50:58 +0000 2021@TrumanLab @HombreRRo IMHO the acetyl/ubiquitinyl/SUMOyl modification system should be a research priority for funders. It is clearly a major system in cellular protein function, but its complexity requires new ways of thinking about protein-based machinery.
Tue Feb 09 13:56:41 +0000 2021Also note that while the paper makes a point of discussing MHC class I peptides, the observations are an unseparated mix of class I & II peptides.
Tue Feb 09 13:43:13 +0000 2021About 35% of the observed Sarcophilus harrisii MHC peptides are identical to observed Homo sapiens MHC peptides.
Tue Feb 09 13:27:54 +0000 2021WDR1:p.I185V, chr 4:g.10097716T>C, rs13441 (HEK-293 I/V = 10/648) vaf=66%, Δm=-14.016, is VV in HEK-293, MCF-7, PANC-1, Hep-G2 & HCT-116. It is IV in MDA-MB-231, JURKAT & HeLa cells. It is II in MCF-10A cells. #ᐯᐸᐱ
Tue Feb 09 13:27:54 +0000 2021WDR1:p, θ(max) = 96. Observed in HLA class 1 & class 2 peptide experiments.
Tue Feb 09 13:27:53 +0000 2021>WDR1:p, WD repeat domain 1 (Homo sapiens) Midsized protein; CTMs: P2+acetyl; PTMs: 17×S,2×T,11×Y+phosphoryl, R17+deimination; aPTMs: 19 K+acetyl/ubiquitinyl; SAAVs: I185V (66%), H248R (1%); mature form: (2-606) [77,520×, 1217 kTa]. #ᗕᕱᗒ 🔗
Tue Feb 09 02:32:22 +0000 2021@goodlettlab1 The data comes from 🔗
Tue Feb 09 02:29:39 +0000 2021Reviewing results from some Tasmanian devil MHC peptide data, it is surprising how many of the sequences are identical to human MHC peptides.
Mon Feb 08 21:50:38 +0000 2021@KentsisResearch @dtabb73 @ProtifiLlc It is an interesting hobby, though.
Mon Feb 08 20:33:03 +0000 2021This is unfortunate news for Manitoba's post-secondary education sector. 🔗
Mon Feb 08 15:12:23 +0000 2021Surprises for this morning:
1. Tasmanian devil has a completed genome & proteome in RefSeq;
2. Someone has studied its MHC peptides (🔗); &
3. They made the data public (🔗)
Mon Feb 08 13:23:00 +0000 2021APEX1:p.D148E, chr 14:g.20456995T>G, rs1130409 (HCT-116 D/E = 2/422) vaf=56%, Δm=14.016, is EE in the HCT-116 cell line as well as MCF-7 & U2-OS cells. It is DE in JURKAT, MCF-10A & A-431 cells. It is DD-only in HeLa & HEK-293 cells. #ᐯᐸᐱ
Mon Feb 08 13:23:00 +0000 2021θ(max) = 96. aka APE, REF1, HAP1, APX, APEN, REF-1, APE-1, APEX. Observed in HLA class 1 peptide experiments (but not class 2). Too many GO annotations.
Mon Feb 08 13:23:00 +0000 2021>APEX1:p, apurinic/apyrimidinic endodeoxyribonuclease 1 (Homo sapiens) Small enzyme; CTMs: P2+acetyl; PTMs: 15×S,2×T,4×Y+phosphoryl; aPTMs: 10 K+acetyl/ubiquitinyl; SAAVs: Q51H (1%), I64V (1%), D148E (56%), G241R (1%); mature form: (2-318) [45,920×, 531 kTa]. #ᗕᕱᗒ 🔗
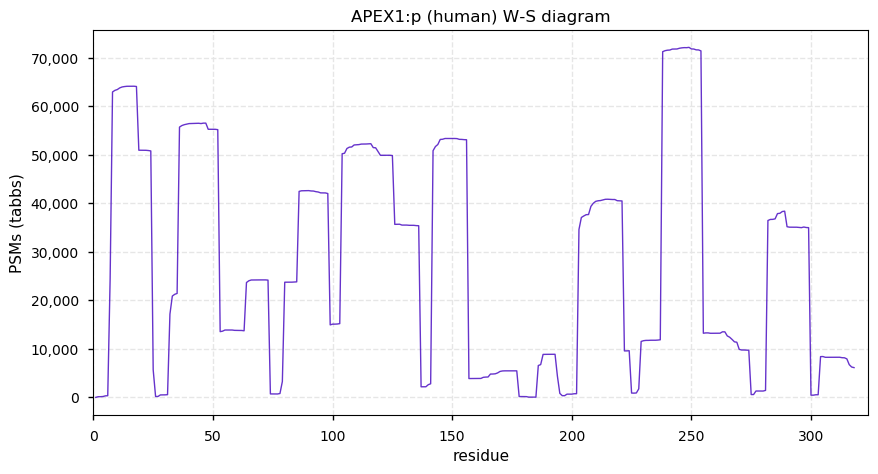
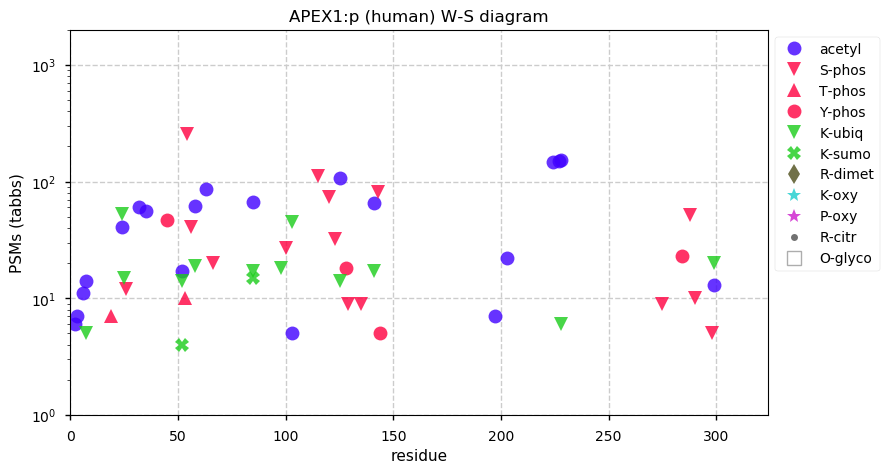
Sun Feb 07 19:05:40 +0000 2021@KentsisResearch @nesvilab @paolocifani @CompProteomics It isn't pessimism, it is just how the field works. The incentives for publication are strongly biased towards coming up with new methods generating data that is increasingly difficult to analyze.
Sun Feb 07 18:54:43 +0000 2021@KentsisResearch @nesvilab @paolocifani @CompProteomics Probably can't be done. There are simply too many weird & wonderful experimental methods used in proteomics.
Sun Feb 07 18:46:19 +0000 2021@jochwenk I really like the opening sentence:
"Genome-wide association studies (GWAS) have identified many risk loci for Alzheimer’s disease (AD) but how these loci confer AD risk is unclear."
Sun Feb 07 14:16:17 +0000 2021Seasonal cold weather across the Northern Plains this morning, making it chilly as far south as Chicago & Detroit. 🔗
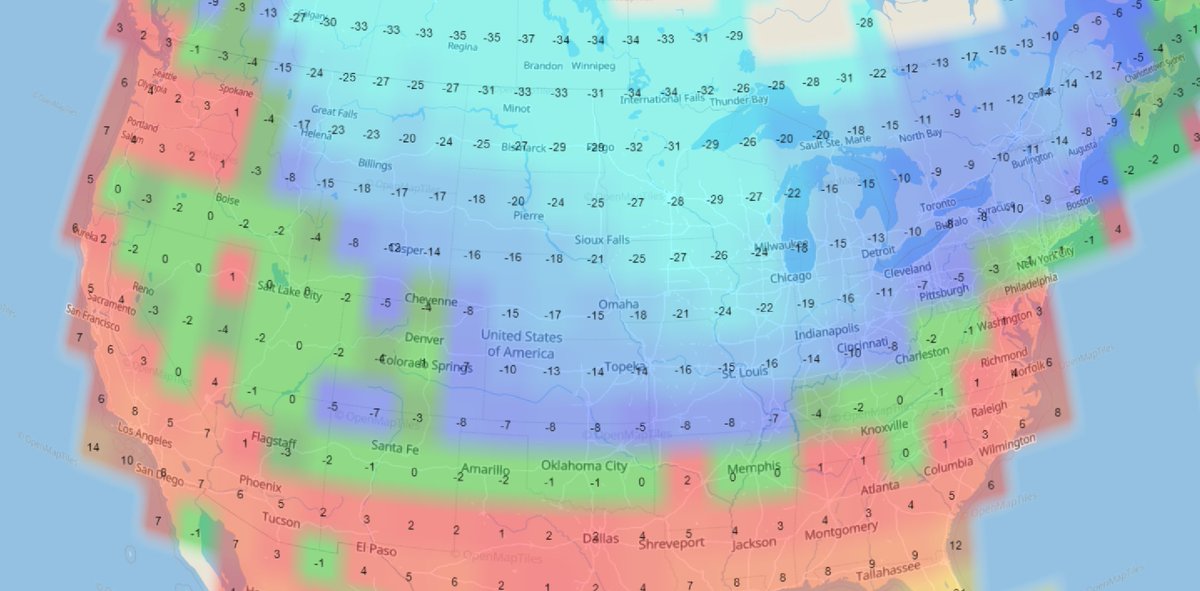
Sun Feb 07 14:10:00 +0000 2021-31 °C, precip 0.0 mm, 1024 mb↑, RH 62%, clear ☀, windchill 1800 W/m² (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Sun Feb 07 13:22:46 +0000 2021CEP55:p.H378L, chr 10:g.93519749A&T, rs2293277 (HEK-293 H/L=0/214) vaf=71%, Δm=-23.975, is LL in the HEK-293 & JURKAT cells. It is HL in HeLa & MCF-10A cells. It HH is MCF-7 cells. #ᐯᐸᐱ
Sun Feb 07 13:22:46 +0000 2021CEP55:p, θ(max) = 68. Observed in HLA class 1 &l class II peptide experiments. High occupancy phosphorylation in C-terminal domain (425-436).
Sun Feb 07 13:22:46 +0000 2021>CEP55:p, centrosomal protein 55 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 18×S,12×T,4×Y+phosphoryl, 25×ubiquitinyl; SAAVs: H57Q (27%), T99A (2%), G227D (3%), C236R (9%), A244P (9%), R348K (3%), H378L (71%); mature form: (2-464) [8,993×, 34 kTa]. #ᗕᕱᗒ 🔗
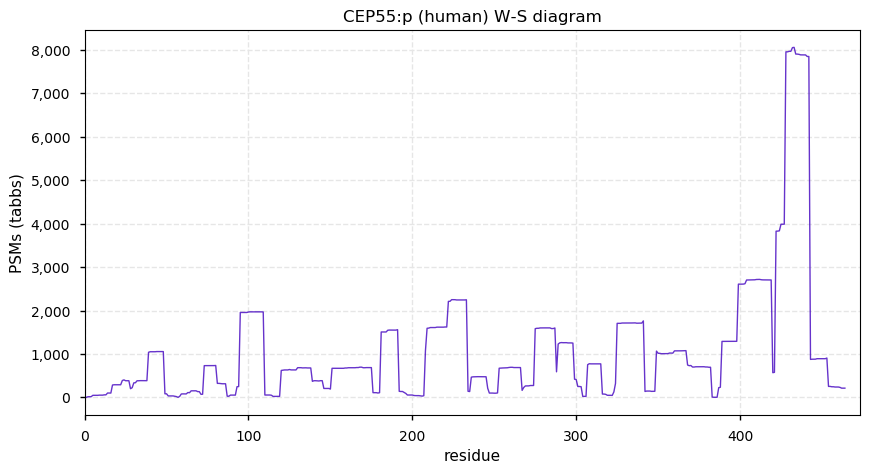
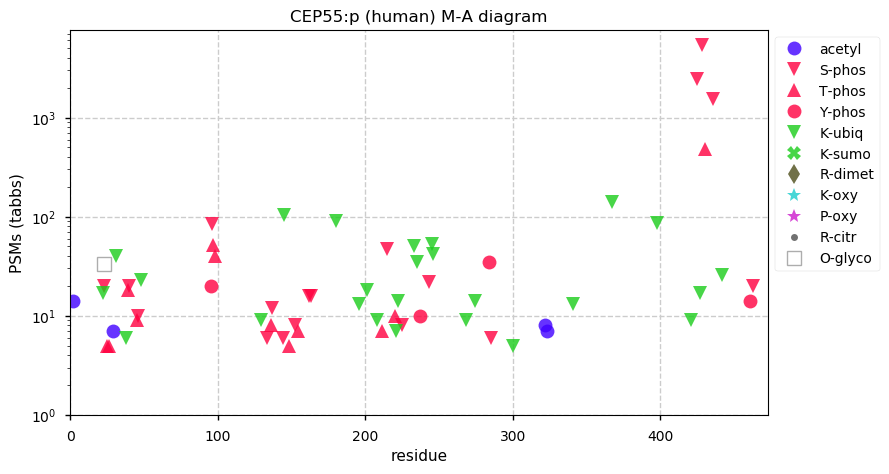
Sat Feb 06 13:52:48 +0000 2021@pwilmarth @neely615 @ypriverol @pride_ebi That is very true: proteomics expts. have a tendency towards the Rococo. However, I was talking about much less nuanced things, e.g., type of MS/MS instrument, Cys blocking reagent or LC types & fractionation.
Sat Feb 06 13:14:52 +0000 2021MTMR6:p.A599T, chr 13:g.25249303C>T, rs62619824 (HEK-293 A/T=45/32) vaf=8%, Δm=30.011, is AT in the HEK-293 & SK-MEL-28 cells. It is AA in HeLa, MCF-7 & MCF-10A cells. #ᐯᐸᐱ
Sat Feb 06 13:14:52 +0000 2021MTMR6:p,θ(max) = 81. Observed in HLA class 1 peptide experiments. High occupancy phosphorylation in a C-terminal domain (561-614). Surprising number of Y+phosphoryl acceptor sites.
Sat Feb 06 13:14:51 +0000 2021>MTMR6:p, myotubularin related protein 6 (Homo sapiens) Midsized enzyme; CTMs: M1+acetyl; PTMs: 11×S,1×T,6×Y+phosphoryl, 15×ubiquitinyl (total); aPTMs: 2×acetyl/ubiquitinyl; SAAVs: A131T (2%), I319V (49%), A599T (8%); mature form: (1-621) [10,735×, 42 kTa]. #ᗕᕱᗒ 🔗
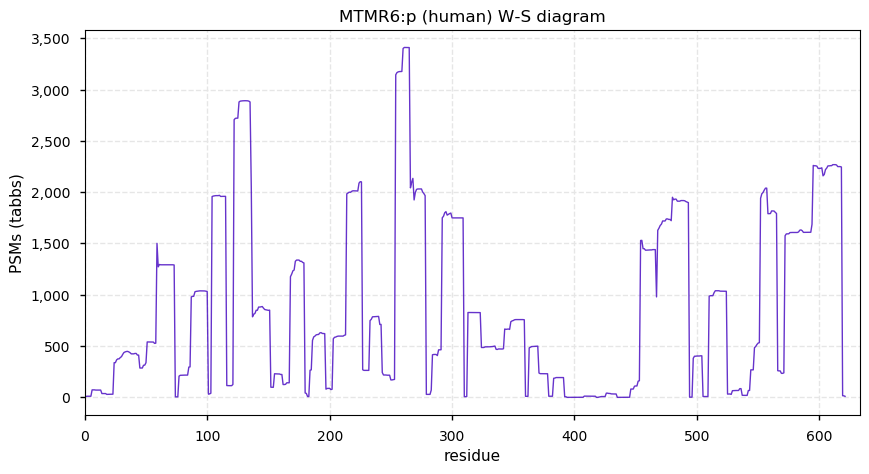
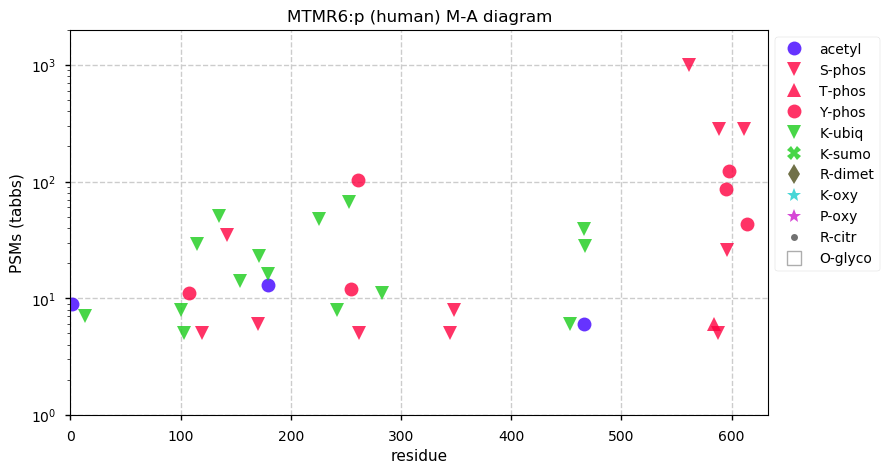
Fri Feb 05 18:03:10 +0000 2021@astacus @edemmott I'd recommend a 2018 Beaujolais.
Fri Feb 05 16:12:14 +0000 2021No matter how many times I've seen it, it still interests me when I see how many HLA class I & II peptides originate from FBS proteins in cell-line experiments. Bovine APOB & TF in particular are always very prominent.
Fri Feb 05 15:58:10 +0000 2021@astacus @dtabb73 GUI's are normally designed to help the guys in marketing.
Fri Feb 05 13:23:03 +0000 2021SLC1A5:p.V512L, chr 19:g.46775602C>G, rs3027961 (HCT-116 V/L=396/543) vaf=20%, Δm=14.016, is VL in the HCT-116 cell line as well as A-431, HT-29, JURKAT & A-549 cells. It is LL in Calu-4 cells. It is VV in HeLa cells. #ᐯᐸᐱ
Fri Feb 05 13:23:03 +0000 2021SLC1A5:p, θ(max) = 54. Observed in HLA class 1 & class 2 peptide experiments. 8 transmembrane domains: (52-81), (95-116), (131-153), (225-248), (258-285), (307-328), (374-400) & (461-482). (483-541) is cytoplasmic.
Fri Feb 05 13:23:02 +0000 2021>SLC1A5:p, solute carrier family 1 member 5 (Homo sapiens) Midsized subunit; CTMs: M1+acetyl, N212+glycosyl; PTMs: 14×S,8×T,2×Y+phosphoryl; aPTMs: 3×acetyl/ubiquitinyl; SAAVs: V512L (20%); mature form: (1-541) [48,127×, 291 kTa] #ᗕᕱᗒ 🔗
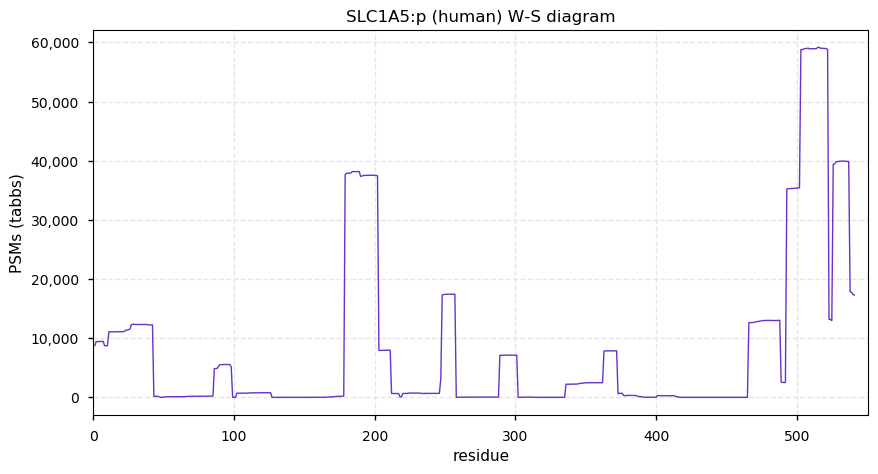
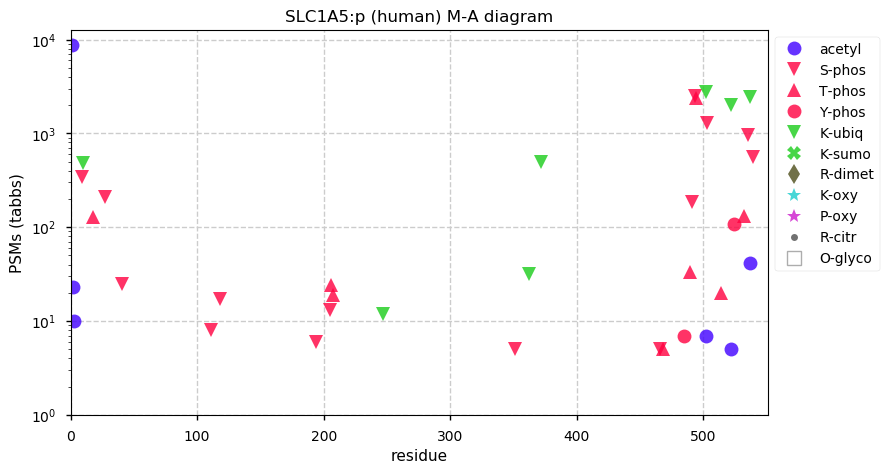
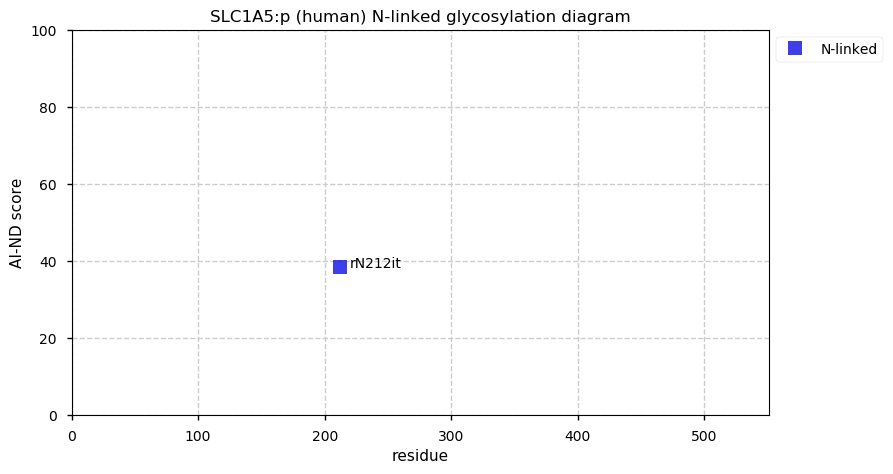
Fri Feb 05 12:45:38 +0000 2021@ypriverol @neely615 @pride_ebi @nextflowio @Twitter I would have to agree that accurate metadata is the most important problem in reanalysis at the moment. I have to spend a lot of effort to confirm what a dataset really represents & how expts were really performed.
Thu Feb 04 13:34:38 +0000 2021AK2:p.A209T, chr 1:g.33013276C>T, rs12116440 (HCT-116 A/T=1076/1279) vaf=0.7%, Δm=30.011, is AT in the HCT-116 cell line as well as HL-60 cells. It is AA in other common cell lines. #ᐯᐸᐱ
Thu Feb 04 13:34:37 +0000 2021AK2:p, θ(max) = 84. Observed in HLA class 1 peptide experiments. Characteristic of mitochondrial inter-membrane proteins, an N-terminal mitochondrial targeting sequence has not been removed in the mature enzyme.
Thu Feb 04 13:34:37 +0000 2021>AK2:p, adenylate kinase 2 (H sapiens) Small mitochondrial enzyme; CTMs: M1,A2+acetyl; PTMs: 13×S,12×T,4×Y+phosphoryl, 9×K+acetyl (total); aPTMs: 5×acetyl/ubiquitinyl, 3×acetyl/ubiquitinyl/SUMOyl; SAAVs: A209T (1%); mature form: (1,2,3,4-232) [70,972×, 658 kTa]. #ᗕᕱᗒ 🔗
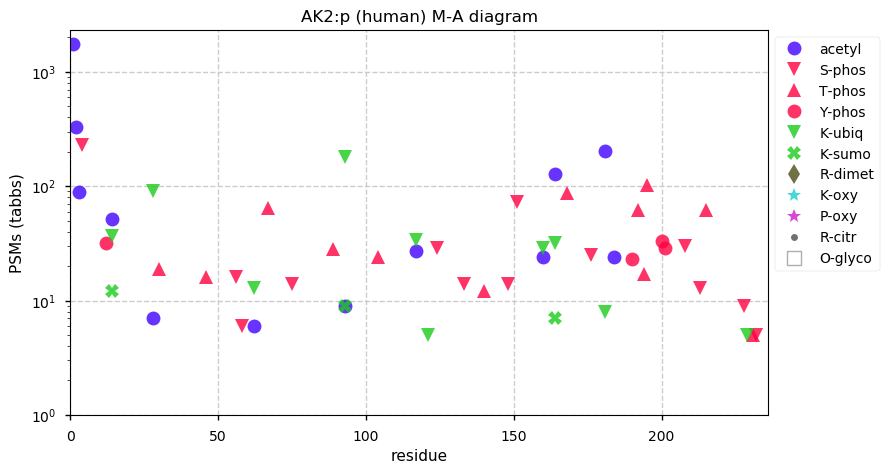
Wed Feb 03 12:55:54 +0000 2021GSTO1:p.A140D, chr 10:g.104263031C>A, rs4925 (HCT A/D=0/1257) vaf=25%, Δm=43.990, is DD in the HCT-116 cell line as well as GM00637 & Hep-G2 cells. It is AD in MHCC97-H & SW-480 cells. It is AA in JURKAT, MCF-7, MCF-10A & HeLa cells. #ᐯᐸᐱ
Wed Feb 03 12:55:54 +0000 2021GSTO1:p, θ(max) = 95. aka GSTTLp28, P28. Observed in HLA class 1 peptide experiments. Found in all tissues and cell lines.
Wed Feb 03 12:55:54 +0000 2021>GSTO1:p, glutathione S-transferase omega 1 (H sapiens) CTMs: S2+acetyl; PTMs: 2×S,1×Y+phosphoryl, 15×K+acetyl (total); aPTMs: 7×acetyl/ubiquitinyl, 1×acetyl/ubiquitinyl/SUMOyl; SAAVs: A140D (25%), E208K (4%); mature form: (2-241) [56,384×, 421 kTa]. #ᗕᕱᗒ 🔗
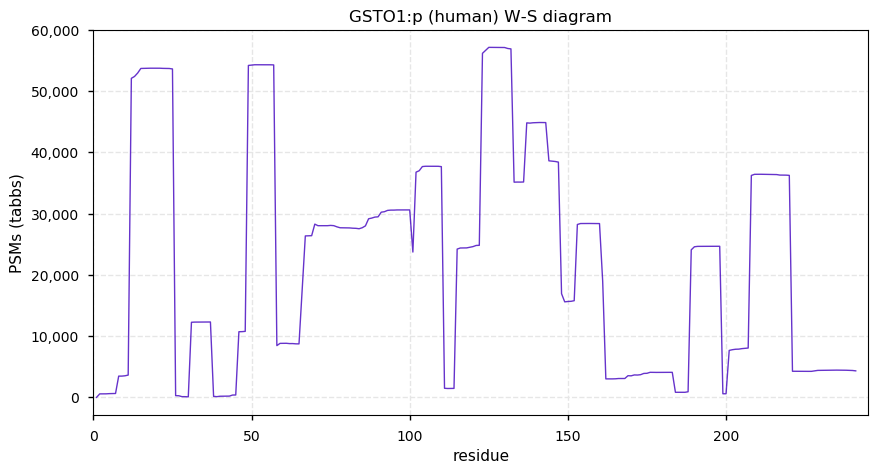
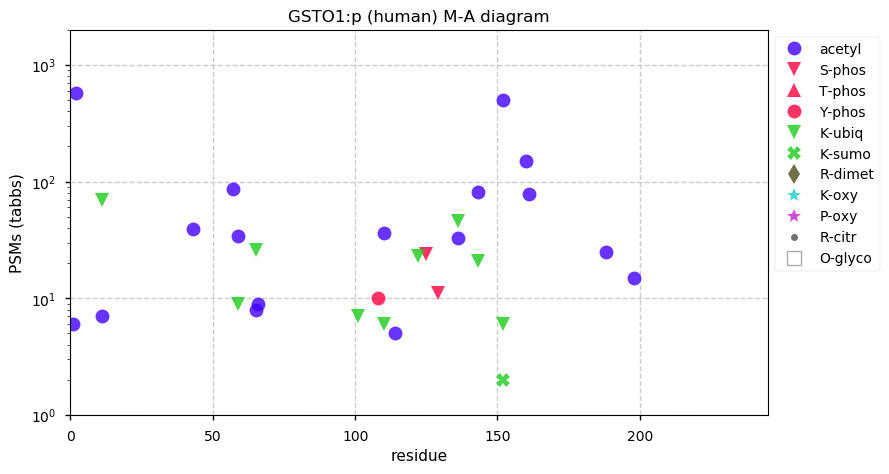
Tue Feb 02 20:34:50 +0000 2021@GenesnProts @seer_bio That helps. The section "10NPs and Plasma (Figure 4 and plasma in Figure 3C)" only lists part of the file name, but it is sufficient to figure it out.
Tue Feb 02 17:57:29 +0000 2021@Wybenga_Groot @lkpino @us_hupo And I should mention much better access to publishing in journals, which can have a significant effect on the range of jobs available for subsequent positions.
Tue Feb 02 17:28:06 +0000 2021@Wybenga_Groot @lkpino @us_hupo Libraries, seminars, access to journals, campuses, colleagues outside of a narrow range of interests, less pressure to perform: generally the benefits of being part of a large organization.
Tue Feb 02 17:01:35 +0000 2021@lkpino @us_hupo That sort of a split would probably do the trick. Just enough to provide some intellectual distance between the two types of presenters.
Tue Feb 02 16:55:05 +0000 2021@lkpino @us_hupo Has any thought been given to providing some separation between university-based pitches and start-up companies? Mixing the 2 together puts the companies at a significant disadvantage as universities have a lot of perks that companies can't provide.
Tue Feb 02 15:45:27 +0000 2021It isn't unusual to have difficulty correlating the experiments described in a paper with the raw data provided, but given the nature of the paper (demo of a tech platform) I was expecting it to be more straightforward.
Tue Feb 02 15:42:53 +0000 2021"London, for instance, has no shortage of think tanks but that hasn’t stopped the UK from becoming a quasi-failed state led by a cretinous troupe of simians stuffed into business suits ..." A. Usher (2021) 🔗
Tue Feb 02 12:57:07 +0000 2021RPL13:p.A112T, chr 16:g.89561665G>T, rs9930567 (HCT-116 A/T=9011/5729) vaf=16%, Δm=30.011, AT in the HCT-116 cell line as well as HaCaT & SK-MEL-28 cells. TT in U-373MG & A-549 cells. AA in JURKAT, U2-OS & HeLa cells. #ᐯᐸᐱ
Tue Feb 02 12:57:07 +0000 2021RPL13:p, θ(max) = 81. aka D16S444E, BBC1, L13. Common in HLA class 1 & 2 peptide experiments. Part of the 60S ribosomal subunit.
Tue Feb 02 12:57:07 +0000 2021>RPL13:p, ribosomal protein L13 (Homo sapiens) Small subunit; CTMs: M1,A2+acetyl; PTMs: 10×S,6×T,0×Y+phosphoryl; aPTMs: 9×acetyl/ubiquitinyl, 4×acetyl/ubiquitinyl/SUMOyl; SAAVs: A112T (16%); mature form: (1,2-211) [89,651×, 700 kTa]. #ᗕᕱᗒ 🔗
Mon Feb 01 18:01:24 +0000 2021@lkpino @seer_bio The experiments in the paper are based on that tech & some of the authors are listed as being from the company.
Mon Feb 01 16:40:51 +0000 2021Does anybody know exactly which experiments in this paper correspond to the DDA RAW files in PXD017052? After reading the paper a few times it isn't clear to me where the data fits in to what is being described. 🔗
Mon Feb 01 13:30:04 +0000 2021TFRC:p.G142S, rs3817672, chr 3:g.196073940C>T (LIM1863 G/S=0/46) vaf=45%, Δm=30.011, is SS in LIM1863 cells. It is GS in CACO-2 cells. It is GG in HCT-116, MCF-7 & MCF-10A cells. #ᐯᐸᐱ
Mon Feb 01 13:30:04 +0000 2021TFRC:p, θ(max) = 56. aka CD71, TFR1, p90. Common in HLA class 1 & 2 peptide experiments. Membrane spanning domain (68-88): (1-67) intracellular & (89-760) extracellular. A form of the protein (101-760) exists in plasma. M2 may be a translation initiation site.
Mon Feb 01 13:30:03 +0000 2021>TFRC:p, transferrin receptor (Homo sapiens) Midsized subunit; CTMs: M1,M2+acetyl, N251,N317,N727+glycosyl; PTMs: 32×S,17×T,17×Y+phosphoryl, 33×K+ubiquitinyl, 30×K+acetyl, T104+glycosyl; SAAVs: G142S (45%), G420S (1%); mature form: (1,2-760) [78,829×, 1004 kTa]. #ᗕᕱᗒ 🔗
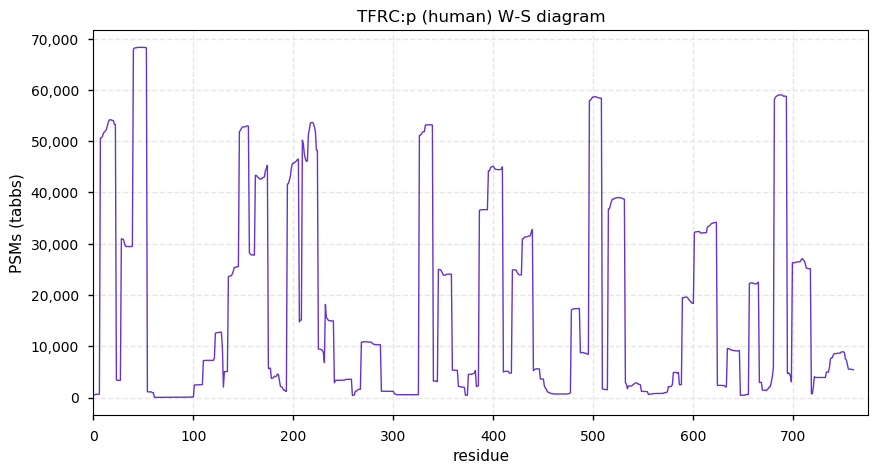
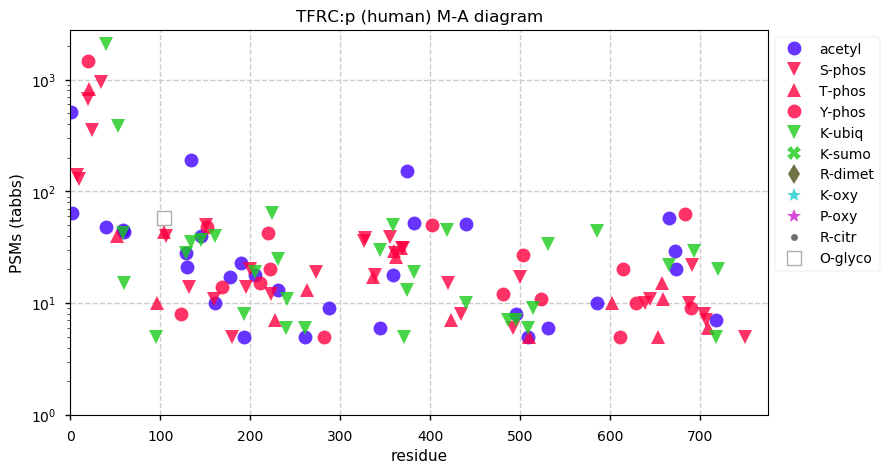
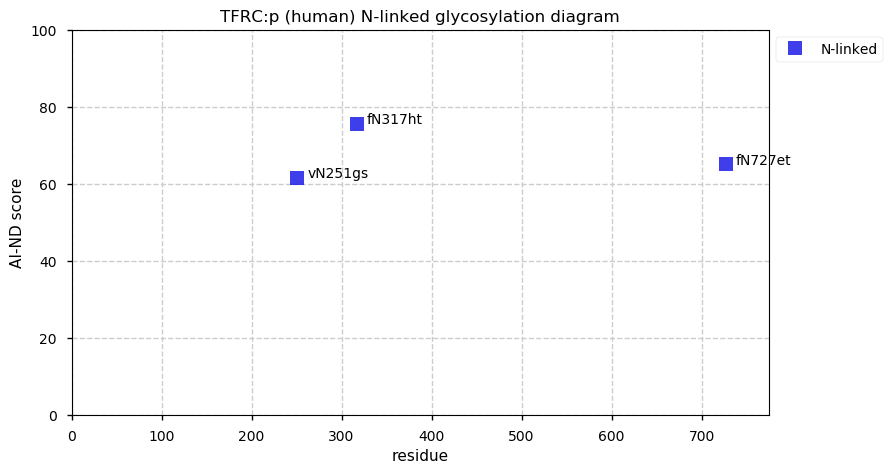
Sun Jan 31 17:35:28 +0000 2021@astacus Easier said than done in most common cases, especially when you really need to get all of the cleanup reactions to be near 100% completion.
Sun Jan 31 16:52:01 +0000 2021Protein domains where the cystines don't reduce or where there are extensive but variable O-linked glycosylation or many other types of PTMs or low complexity seqs don't affect the current tech much, but would stop any string-of-pearls reading tech in its tracks (or pores).
Sun Jan 31 16:42:39 +0000 2021One thing that proteomics-types should keep in mind when assessing new technologies for protein sequencing: the main strength of the current methods is that they can skip over the "hard" parts of a protein, even if they are the majority of a sequence.
Sun Jan 31 16:13:04 +0000 2021@michael15573267 @slavov_n Intelligence.
Sun Jan 31 15:20:15 +0000 2021I guess I'll ask this question again in 6 months. I have been asking people about parainfluenza 5's use in molecular biology since 2015, but I have yet to find anyone who knows (or is willing to say). It has been observed in >500 LC/MS/MS runs. 🔗
Sun Jan 31 13:09:03 +0000 2021PDPR:p.Y110H, chr 16:g.70127360T>C, rs2549532 (HCT-116 Y/H=5/383) vaf=46%, Δm=-26.004, is HH in HCT-116 cells. It is YH in HEK-293 & HeLa cells. It is YY in MCF-7 & MCF-10A cells. #ᐯᐸᐱ
Sun Jan 31 13:09:03 +0000 2021PDPR:p, θ(max) = 60. aka PDP3. Common in HLA class 1 (but not class 2) peptide experiments. All modifications appear to be very low occupancy. The SAAV Y640F occurs at the same site as the observed Y640+phosphoryl acceptor.
Sun Jan 31 13:09:03 +0000 2021>PDPR:p, pyruvate dehydrogenase phosphatase regulatory subunit (Homo sapiens) Midsized mitochondrial subunit; CTMs: none; PTMs: Y640+phosphoryl, 3×K+ubiquitinyl; SAAVs: Y110H (46%), Y640F (11%); mature form: (28-879) [17,096×, 81 kTa]. #ᗕᕱᗒ 🔗
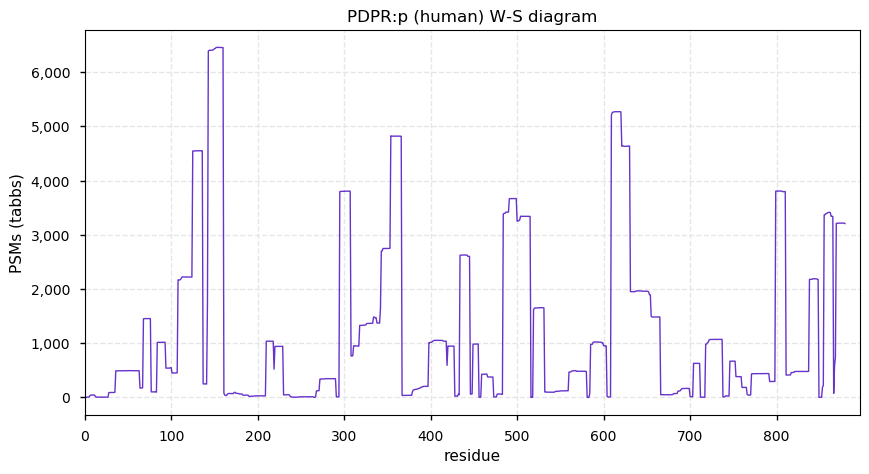
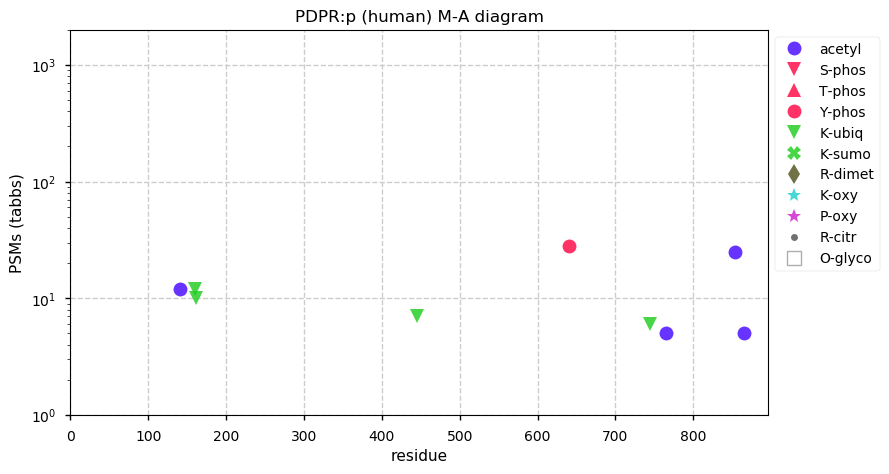
Sat Jan 30 18:45:52 +0000 2021For anyone interested in looking at some detailed ids from nanoparticle corona sample prep for plasma proteins (🔗), you can take a look at these results: 🔗
Sat Jan 30 17:08:58 +0000 2021@MattWFoster @byu_sam @ucdmrt @Sci_j_my I would personally go with #💩
Sat Jan 30 16:48:54 +0000 2021@Sci_j_my @byu_sam I am always surprised by how even very smart DNA/RNA guys try to oversimplify proteins to the point that they are like nucleic acids and then propose spherical-cow-style solutions as though they had any chance of working.
Sat Jan 30 14:33:38 +0000 2021I suspect we proteomics types don't appreciate just how much virus gets introduced during these studies.
Sat Jan 30 14:20:57 +0000 2021CRISPR stuff often has big signals from murine parainfluenza virus 1 (Sendai virus).
Sat Jan 30 14:18:56 +0000 2021I regularly see big signals from parainfluenza virus 5 proteins in data from samples where there RNA has been introduced to the cells being studied. Does anybody know which commercial systems use this virus?
Sat Jan 30 13:35:51 +0000 2021SLC1A5:p.V512L, chr 19:g.46775602C>G, rs3027961 (HCT-116 V/L=1252/1532) vaf=20%, Δm=14.016, is VL in JURKAT, HT-29, A-549 & A-431 cells. It is VV in U2-OS, CACO-2 & MCF-7 cells. #ᐯᐸᐱ
Sat Jan 30 13:35:51 +0000 2021SLC1A5:p does not have an N-terminal ER targeting signal peptide, suggesting that it is made in the cytoplasm and threaded into the ER membrane from the cytoplasmic side.
Sat Jan 30 13:35:51 +0000 2021SLC1A5:p, θ(max) = 54. aka AAAT, ASCT2, RDRC, M7V1. Common in HLA class 1 & 2 peptide experiments. 8 membrane spanning domains: the glycosylations are extracellular & the C-terminal high occupancy phosphodomain (485-539) is cytoplasmic.
Sat Jan 30 13:35:50 +0000 2021>SLC1A5:p, solute carrier family 1 member 5 (H sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 14×S,8×T,2×Y+phosphoryl, 4×K+ubiquitinyl, N163,N212+glycosyl; aPTMS: 3×K+acetyl/ubiquitinyl; SAAVs: P17A (20%), V512L (20%); mature form: (1-583) [47,787×, 290 kTa] #ᗕᕱᗒ 🔗
Fri Jan 29 22:39:55 +0000 2021After saying "What the f is that gi # doing there?", I realize that the new dataset I was testing is yet one more set of results with an abundance of parainfluenza virus 5 proteins for no apparent reason.
Fri Jan 29 22:13:35 +0000 2021@ProtifiLlc Usual RP column
Fri Jan 29 21:01:30 +0000 2021Inquiring minds want to know! 🔗
Fri Jan 29 20:37:09 +0000 2021gpmDB Jan 29 (build #6204)
proteins = 1.2 billion
distinct proteins = 3.5 million
peptides = 11.8 gTa
distinct peptides = 20.7 million
residues = 164 billion
Fri Jan 29 17:42:09 +0000 2021@andy___jones @astacus @ClaireEEyers itertools has saved me a lot of time solving combinatorial problems (& greatly reduced the amount of confused-head-banging-against-a-wall-frustration involved)
Fri Jan 29 17:36:01 +0000 2021@astacus @andy___jones @ClaireEEyers In Canadian coins, valued in ¢
from itertools import combinations_with_replacement
c = combinations_with_replacement([5, 10, 25,100,200], 5)
for i in list(c):
print(i,sum(i))
Fri Jan 29 16:37:13 +0000 2021Proteomics-phospho-chromo-peeps: what are the best ways to condition a column for phosphopeptide work?
Fri Jan 29 14:46:05 +0000 2021@dtabb73 C'est bon.
Fri Jan 29 12:57:42 +0000 2021NUDCD1:p.L252F, chr 8:g.109289818T>G, rs2980619 (MCF-10A L/F=0/90) vaf=58%, Δm=33.984, is FF in HCT-116 & MCF-10A cells. It is LF in HeLa cells. It is LL in HEK-293, U2-OS, MDA-MB-231,CACO-2 & MCF-7 cells. #ᐯᐸᐱ
Fri Jan 29 12:57:42 +0000 2021NUDCD1:p, θ(max) = 71. aka CML66, FLJ14991. Common in HLA class 1 peptide experiments, but not class 2.
Fri Jan 29 12:57:42 +0000 2021>NUDCD1:p, NudC domain containing 1 (Homo sapiens) Midsized subunit; CTMs: A2+acetyl, A3+acetyl; PTMs: 9×S, 5×T, 1×Y+phosphoryl, 6×K+ubiquitinyl; aPTMS: K222+acetyl/ubiquitinyl; SAAVs: L252F (58%), I269V (38%), N426S (8%); mature form: (1-583) [25,051×, 131 kTa]. #ᗕᕱᗒ 🔗
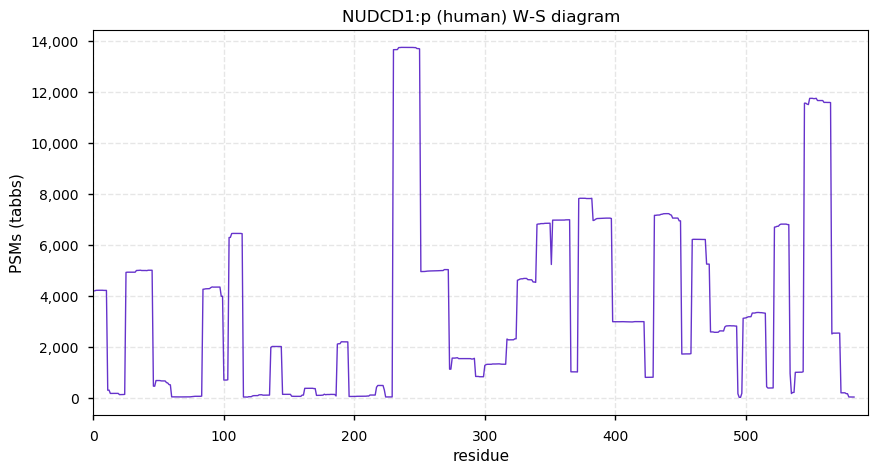
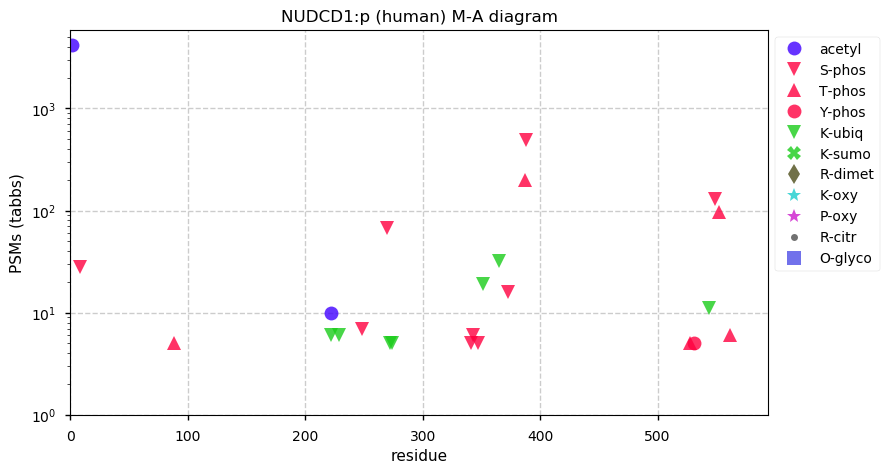
Thu Jan 28 23:14:44 +0000 2021@chrashwood Seemed like a good example to get the symbol I was using for O-links correct.
Thu Jan 28 23:14:19 +0000 2021@chrashwood I had just come across FGA as one of the major components of the "corona" in the data from 🔗 & I noticed how many O-linked glycosylation sites had been observed (most alternating with phosphorylation at the same site).
Thu Jan 28 19:10:55 +0000 2021@chrashwood So this image (for FGA:p mods) won't result in too many angry letters:
🔗
Thu Jan 28 18:27:18 +0000 2021@mjmaccoss @MattWFoster @pwilmarth @NLKProteomics Yes, even though I hate conferences.
Thu Jan 28 17:13:20 +0000 2021Glyco-types: what is the best symbol to use for representing a generic O-linked glycosylation? (colored square/circle/triangle/hexagon?)
Thu Jan 28 13:37:00 +0000 2021IRF2BP2:p.S78P, chr 1:g.234609263A>G, rs7545855 (MCF-7 S/P=0/819) vaf=61%, Δm=10.021, is PP in MCF-7, MCF-10A, U2-OS, CACO-2 & HEK-293 cells. It is SP in HL-60, A-431 & MDA-MB-231 cells. It is SS in MHCC-97 cells. #ᐯᐸᐱ
Thu Jan 28 13:37:00 +0000 2021IRF2BP2:p, θ(max) = 88. aka IRF-2BP2. Common in HLA class 1 peptide experiments, but not class 2. Many high occupancy serine phosphorylation sites.
Thu Jan 28 13:36:59 +0000 2021>IRF2BP2:p, interferon regulatory factor 2 binding protein 2 (H sapiens) Midsized subunit; CTMs: A2+acetyl, A3+acetyl; PTMs: 44×S,17×T+phosphoryl; aPTMS: K289,K303,K326,K327+ubiquitinyl/SUMOyl; SAAVs: S78P(61%), P118S(1%); mature form: (2-571) [28,077×, 200 kTa]. #ᗕᕱᗒ 🔗
Wed Jan 27 19:13:28 +0000 2021This bug is rather nasty, so if you are running Linux, you should apply the patch ASAP now that it is generally known in the community
🔗
Wed Jan 27 13:40:24 +0000 2021-27 °C, precip 0.0 mm, 1034 mb↑, RH 72%, clear ☀ , windchill 1700 W/m² (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Wed Jan 27 13:28:11 +0000 2021HNRNPH3:p.G284A, chr 10:g.68341660G>C, rs16925347 (HCT-116 G/A=3186/3130) vaf=8%, Δm=14.016, is heterozygous in the HCT-116 cell line as well as 143B, SK-BR-3 & MDA-MB-231 cells. It is reference only in HeLa cells. #ᐯᐸᐱ
Wed Jan 27 13:28:11 +0000 2021HNRNPH3:p, θ(max) = 85. aka 2H9, HNRPH3. Common in HLA class 1 peptide experiments, but not class 2. C-terminal low complexity domain (262-341) enriched in G & Y+phosphoryl. Unusual 7×Y+phosphoryation domain (143-181).
Wed Jan 27 13:28:10 +0000 2021>HNRNPH3:p, heterogeneous nuclear ribonucleoprotein H3 (H sapiens) CTMs: M1+acetyl; PTMs: R129,R323,R343+dimethyl, 18×S,7×T4,18×Y+phosphoryl; aPTMS: K6,K67+acetyl/ubiquitinyl/SUMOyl, K76+acetyl/ubiquitinyl; SAAVs: G284A (8%); mature: (1-346) [80,661×, 925 kTa]. #ᗕᕱᗒ 🔗
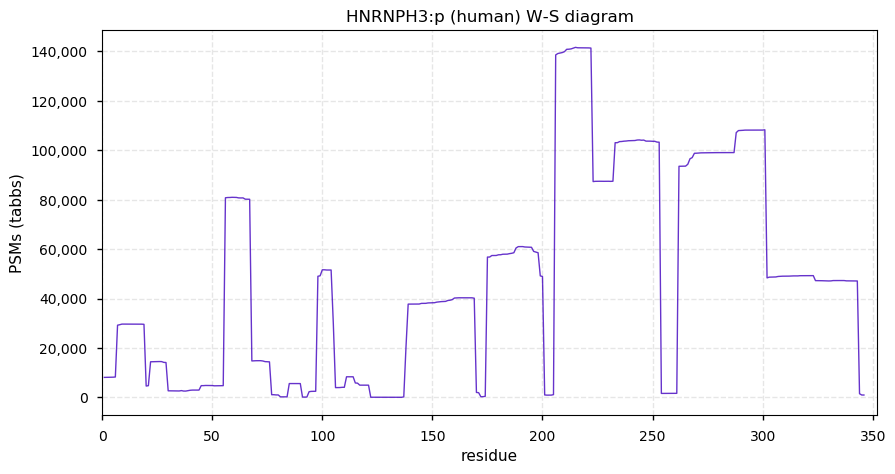
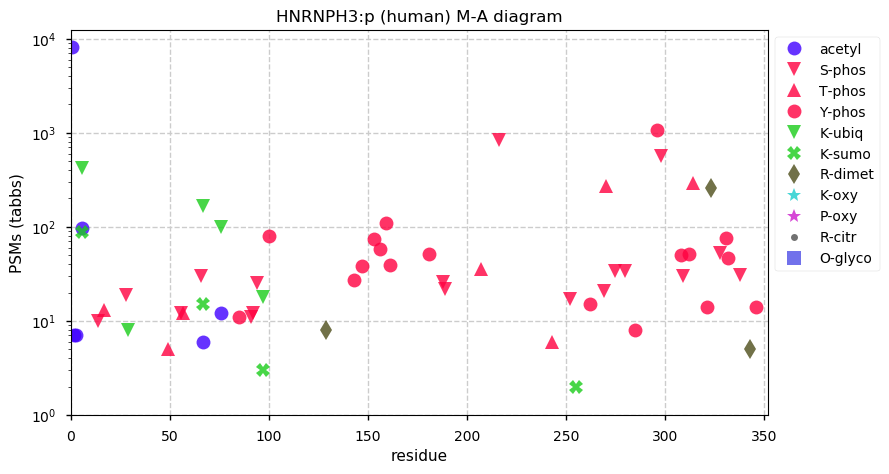
Tue Jan 26 18:25:51 +0000 2021@AaronWherry The term "vaccine" isn't testing well in focus groups.
Tue Jan 26 17:22:05 +0000 2021-27 °C, precip 0.0 mm, 1029 mb→, RH 72%, clear ☀, windchill: 1650 W/m² (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Tue Jan 26 15:54:10 +0000 2021@macro_momo @slavov_n It depends on the local academic tradition. I worked in a lab in Germany where a grad student "chatting" with Herr Professor Doctor simply never (ever) happened.
Tue Jan 26 13:13:03 +0000 2021DSG2:p.R773K, chr 18:g.31542836G>A, rs2278792 (HCT-116 R/K=0/1160) vaf=27%, Δm=-28.006, is KK in the HCT-116 cell line as well as HaCaT cells. It is RK in JURKAT cells & RR in HeLa cells. #ᐯᐸᐱ
Tue Jan 26 13:13:03 +0000 2021DSG2:p, θ(max) = 64. aka CDHF5. Frequently observed in HLA class I & II peptide experiments. Transmembrane domain (613-633). It has two distinct phosphodomains: extracellular low occupancy (24-560) & intracellular high occupancy (645-1118).
Tue Jan 26 13:13:02 +0000 2021>DSG2:p, desmoglein 2 (Homo sapiens) CTMs: N112, N182, N462, N1097+glycosyl; PTMs: 56×S,39×T4,11×Y+phosphoryl, 14× K+ubiquitinyl; aPTMS: 3×K+acetyl/ubiquitinyl; SAAVs: I293V(3%), R773K(27%), M863L(1%), S883P(1%), T903I(1%); mature: (24-1118) [43,496×, 354 kTa] #ᗕᕱᗒ 🔗
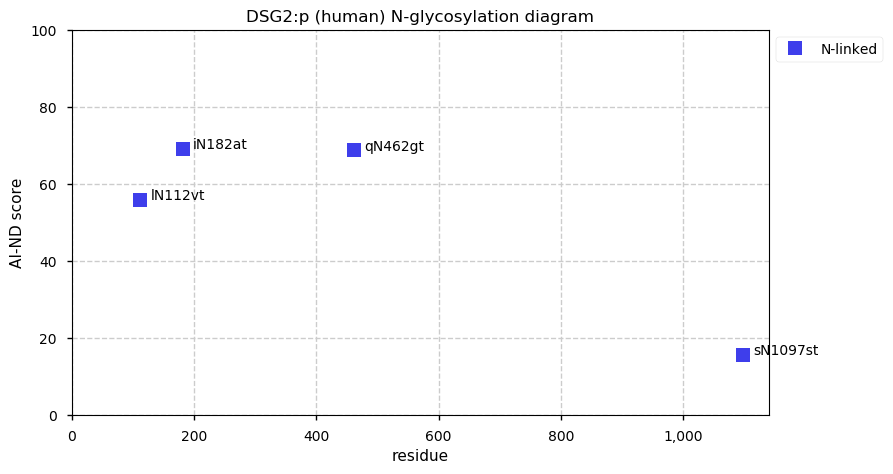
Mon Jan 25 22:09:11 +0000 2021Looks like there should be nice bright aurora tonight, down into the northern US Great Lakes region. 🔗
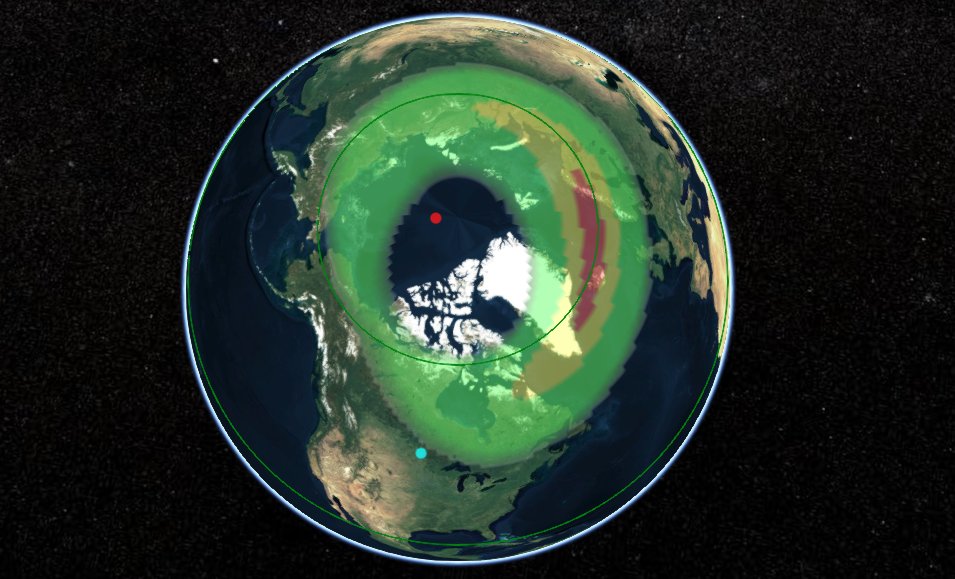
Mon Jan 25 13:12:00 +0000 2021ASNS:p.V210E, chr 7:g.97859257A>T, rs1049674 (HCT-116 V/E=16/297) vaf=79%, Δm=29.974, is EE in the HCT-116 cell line as well as LNCAP, U2-OS & HeLa cells. It is VE in JURKAT cells. VV only in MCF-7 & MCF-10A cells. #ᐯᐸᐱ
Mon Jan 25 13:12:00 +0000 2021ASNS:p, θ(max) = 95. Frequently observed in HLA class I & II peptide experiments. The involvement of acetyl|ubiquitinyl|SUMOyl mechanisms seems over-the-top for this metabolic enzyme.
Mon Jan 25 13:12:00 +0000 2021>ASNS:p, asparagine synthetase (glutamine-hydrolyzing) (Homo sapiens) Midsize enzyme; CTMs: none; PTMs: 2×S,0×T4,3×Y+phosphoryl; aPTMS: 4×K+acetyl/ubiquitinyl/SUMOyl, 8×K+acetyl/ubiquitinyl; SAAVs: V210E (79%), A277T (1%); mature form: (2-561) [30,148×, 267 kTa].#ᗕᕱᗒ 🔗
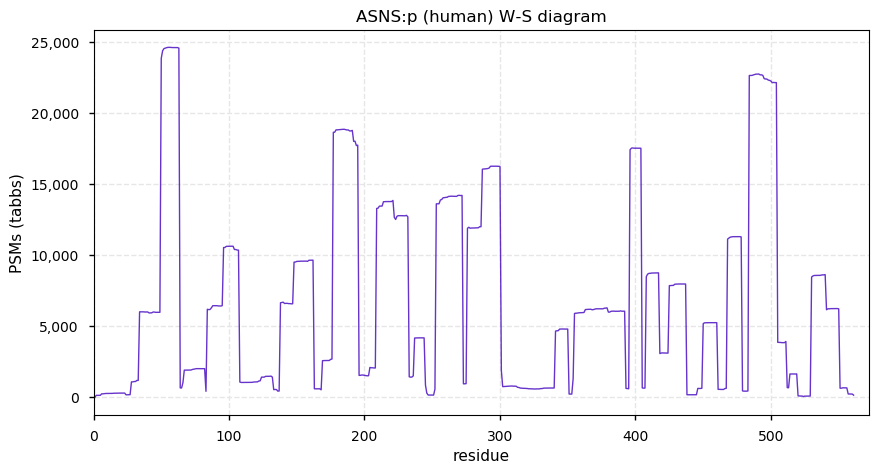
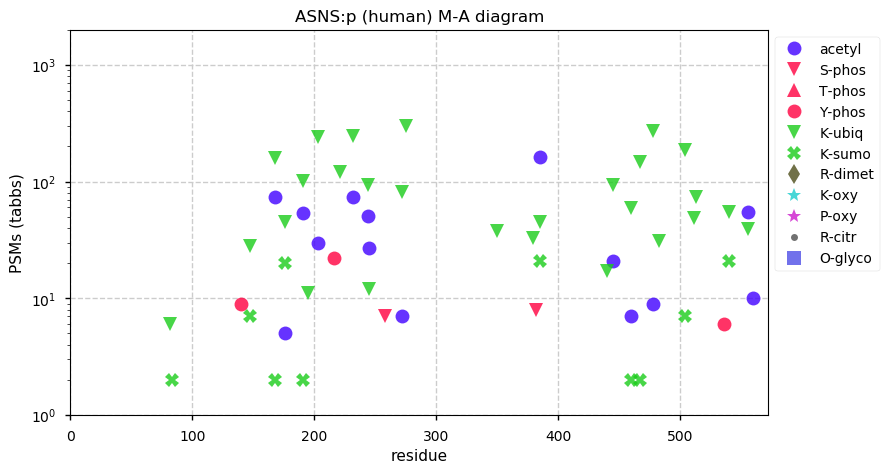
Sun Jan 24 17:48:59 +0000 2021@jwoodgett @Lange_Lab @SinaiHealth @laurpelletier I don't contest the value of viral phylogenic research: Canada should be able to do more of it, but can't because political "projects" like CanCOGen end up spending all of the allocated resources on tangentially related things.
Sun Jan 24 14:12:56 +0000 2021@jwoodgett @Lange_Lab @SinaiHealth @laurpelletier The Genome Canada project seems overly ambitious, given GC's track record & geographic footprint.
Sun Jan 24 14:01:10 +0000 2021RPA1:p.T351A removes an observed phosphorylation acceptor site.
Sun Jan 24 13:43:18 +0000 2021RPA1:p.T351A, rs5030755, chr 17:g.1879658A>G (HCT-116 T/A=204/478) vaf=8%, Δm=-30.011, is TA in the HCT-116 & JURKAT cell line. It is TT in HEK-293, MCF-7, MCF-10A & most common cells lines.
Sun Jan 24 13:43:18 +0000 2021RPA1:p, θ(max) = 88. aka REPA1, RPA70, HSSB, RF-A, RP-A. Frequently observed in HLA class I peptide experiments. Too many GO annotations.
Sun Jan 24 13:43:18 +0000 2021>RPA1:p, replication protein A1 (Homo sapiens] Midsize protein; CTMs: M1+acetyl; PTMs: 32×S,21×T4,3×Y+phosphoryl; aPTMS: 13×K+acetyl/ubiquitinyl/SUMOyl, 11×K+acetyl/ubiquitinyl; SAAVs: T351A (8%); mature form: (1,2-616) [57,433×, 441 kTa] #ᗕᕱᗒ 🔗
Sat Jan 23 15:46:16 +0000 2021This classic paper should be required reading for people involved in proteomics data analysis 🔗 (as it is for anyone who uses FDR to correct for multiple testing effects)
Sat Jan 23 14:08:20 +0000 2021@bffo Everyone knows mutations cause superpowers & make anyone with a PhD into a supervillain (with a few exceptions).
Sat Jan 23 13:28:04 +0000 2021ECHS1:p.T75I, rs1049951, chr 10:g.133370622G>A (HCT-116 T/I=1/501) vaf=92%, Δm=12.036, is II in the HCT-116 cell line as well as HEK-293, MRC-5, U2-OS, JURKAT, MCF-7, MCF-10A & most other common cell lines. HeLa is an exception: in HeLa T/I=3849/2742.
Sat Jan 23 13:28:04 +0000 2021ECHS1:p, θ(max) = 90. aka SCEH. Observed in HLA class I peptide experiments. The SAAV V11A is in the mitochondrial targeting domain (1-29) & is not part of the mature protein.
Sat Jan 23 13:28:04 +0000 2021>ECHS1:p, enoyl-CoA hydratase, short chain 1(Homo sapiens) Small mitochondrial subunit; CTMs: none; PTMs: 8×K+acetyl, 2×K+SUMOyl, S29, T46, S107, Y264+phosphoryl; SAAVs: V11A (32%), T75I (92%); mature form: (28,29,30-290) [26,489×, 606 kTa]. #ᗕᕱᗒ 🔗
Fri Jan 22 18:49:07 +0000 2021Well, at least someone is thinking outside-of-the-box
🔗
Fri Jan 22 18:02:52 +0000 2021@astacus @Sci_j_my Some scientists seem to be serial pareidoliacs (I know adding the -cs doesn't make a real word, but I have a facetious nature).
Fri Jan 22 17:45:55 +0000 2021@Sci_j_my For me, when a paper makes this type of over-the-top claim, I just stop being interested in the rest of the claims.
Fri Jan 22 17:37:42 +0000 2021@Sci_j_my It is pretty gutsy to claim that a log-log plot with r = 0.65 indicates high correlation, though.
Fri Jan 22 12:55:47 +0000 2021POLR2E:p.S44F, chr 19:g.1094005G>A, rs12459404 (HEK-293 S/F = 1/2482) vaf=>99%, Δm = +60.03, is FF in HEK-293, HeLa, JURKAT, HCT-116, U2-OS & all other common cell lines. Because the vaf is very nearly 100%, it is usually listed as having an maf = 0.0.
Fri Jan 22 12:55:46 +0000 2021POLR2E:p, θ(max) = 91. aka RPB5, RPABC1, XAP4, hRPB25, hsRPB5. Observed in HLA class I peptide experiments, but not in class II data.
Fri Jan 22 12:55:46 +0000 2021>POLR2E:p, RNA polymerase II, I and III subunit E (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 6×K+ubiquitinyl, 2×K+SUMOyl, T111, Y8+phosphoryl; SAAVs: S44F (100%); mature form: (1,2-210) [26,489×, 97 kTa]. #ᗕᕱᗒ 🔗
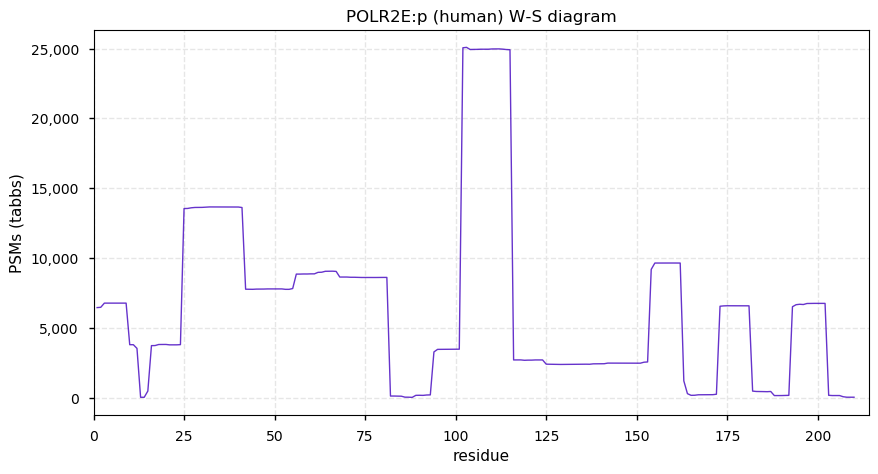
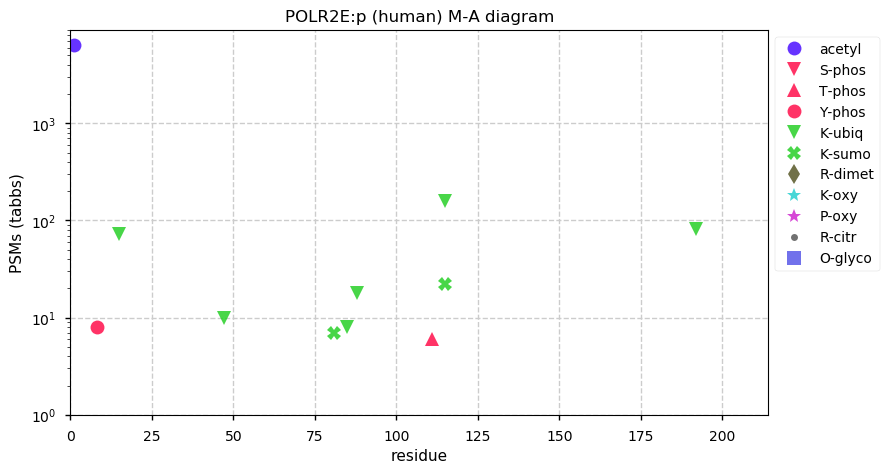
Thu Jan 21 16:10:35 +0000 2021@pwilmarth I've always attributed the "most-complicated-method-ever" tendency to the perverse incentives associated with the field's journals. There are a lot of venues to publish a complex method if you work in (or run) a facility lab, enough to have a hierarchy of IF's to choose from!
Thu Jan 21 14:59:35 +0000 2021@AlexUsherHESA It also makes any statement regarding a lack of candidates meaningless, because they may all be in low-skilled positions.
Thu Jan 21 14:55:42 +0000 2021@AlexUsherHESA Canada has an odd way of defining "tech" jobs. Everyone working for a "tech" company is considered to have a "tech" job, including admins, maintenance, financial, etc. It makes the numbers bigger than would be the case in many other countries for the same set of companies.
Thu Jan 21 13:46:37 +0000 2021@byu_sam @ypriverol @GygiLab Unfortunately TMT + multiplexing = no dice. Their Bioplex 3 data is great from SAAVs, though.
Thu Jan 21 13:29:02 +0000 2021@ypriverol From the specific purpose of SAAV's, yes CPTAC data isn't appropriate. The derivitization chemistry seems to introduce enough additional "noise" that finding SAAVs is difficult. Mixing multiple individuals also does't help, especially with heterozygous variants.
Thu Jan 21 13:05:45 +0000 2021@ypriverol I will check. The main criterion is the total protein coverage obtained: finding these things is a numbers game where the more coverage you get, the more likely you are to hit on a variant. You can also rule out any data sets that use quant. reagents, e.g. TMT or iTRAQ.
Thu Jan 21 12:57:20 +0000 2021@ypriverol Not sure what that means. Could you expand on "resolution" a bit?
Thu Jan 21 12:55:16 +0000 2021COMT:p.V158M, rs4680, chr 22:g.19963748G>A (HTC-116 V/M = 500/363 Ta) vaf=46%, Δm=31.972, is VM in the HCT-116 cell line as well as HEK-293, CACO-2 & THP-1 cells. It is MM in SK-MEL-28 cells. Reference allele VV in JURKAT, HeLa & MCF-10A cells.
Thu Jan 21 12:55:16 +0000 2021COMT:p, θ(max) = 82. Observed in HLA class I & class II peptide experiments. Observed in most cell lines & tissues―more broadly than its proposed function would require. Odd, low complexity membrane spanning domain (6-26).
Thu Jan 21 12:55:15 +0000 2021>COMT:p, catechol-O-methyltransferase (Homo sapiens) Small enzyme; PTMs: 7×K+acetyl, 4×S,2×T,3×Y+phosphoryl; aPTMs: K177, K259+acetyl/ubiquitinyl; SAAVs: A72S (1%), A72T (1%), V158M (46%); mature form: (1,2-271) [34,394×, 272 kTa] #ᗕᕱᗒ 🔗
Thu Jan 21 12:54:39 +0000 2021@ypriverol Sorry, I should comprehend the question before I answer. HEK-293 is not a cancer cell line: it is "only" transformed.
Thu Jan 21 12:26:05 +0000 2021@ypriverol Pretty much an even split between HEK-293, HCT-116, HeLa & MCF-7.
Thu Jan 21 03:19:19 +0000 2021@VATVSLPR I hadn't even considered Imposter Syndrome as possible reason …
Wed Jan 20 22:29:27 +0000 2021I was trying to come up with a poll about why proteomics scientists are always so nervous/jumpy/defensive about their results, but I couldn't get the number of possibilities down to four.
Wed Jan 20 16:22:19 +0000 2021GBE1:p.I334V, rs2172397, chr 3:g.81594016T>C (HCT-116 I/V = 0/190 Ta), vaf=98%, Δm=-14.016, is VV in the HeLa, HEK-293, MCF-7, SK-MEL-28, DLD-1 & pretty much all other commonly used cell lines.
Wed Jan 20 16:12:43 +0000 2021@TanentzapfLab Hopefully conferences die out. Their evolution into sales platforms in the last 20 years has made them more entertaining but increasingly divorced from scientific research.
Wed Jan 20 16:06:02 +0000 2021@ensembl @pwilmarth Good news.
Wed Jan 20 13:17:39 +0000 2021GBE1:p θ(max) = 83%. Observed in HLA class I & class II peptide experiments. Enzyme necessary for the proper formation of glycogen, which is present in all cells (incl. platelets and erythrocytes).
Wed Jan 20 13:17:39 +0000 2021>GBE1:p, 1,4-alpha-glucan branching enzyme 1 (Homo sapiens) Midsized enzyme; CTMs: A2+acetyl; PTMs: 7×K+acetyl, 13×S,4×T,9×Y+phosphoryl; aPTMs: 11×K+acetyl/ubiquitinyl; SAAVs: P40T (1%), R190G (32%), I334V (98%); mature form: (2-702). [32,896×, 240 kTa]. #ᗕᕱᗒ 🔗
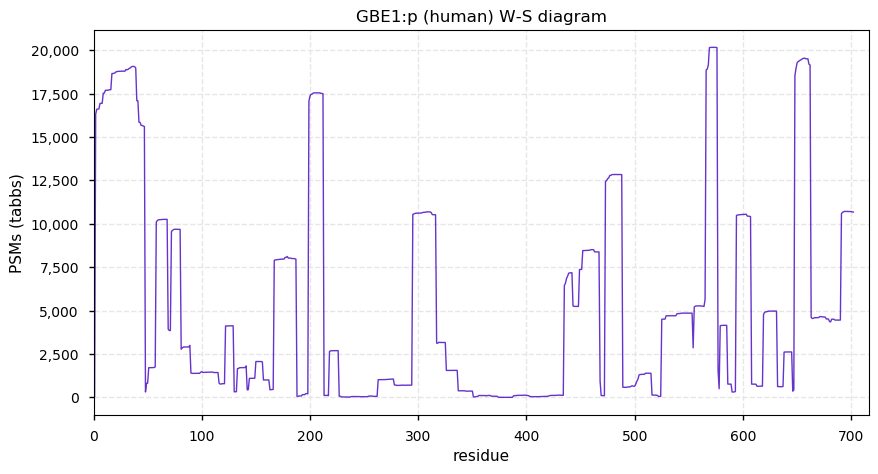
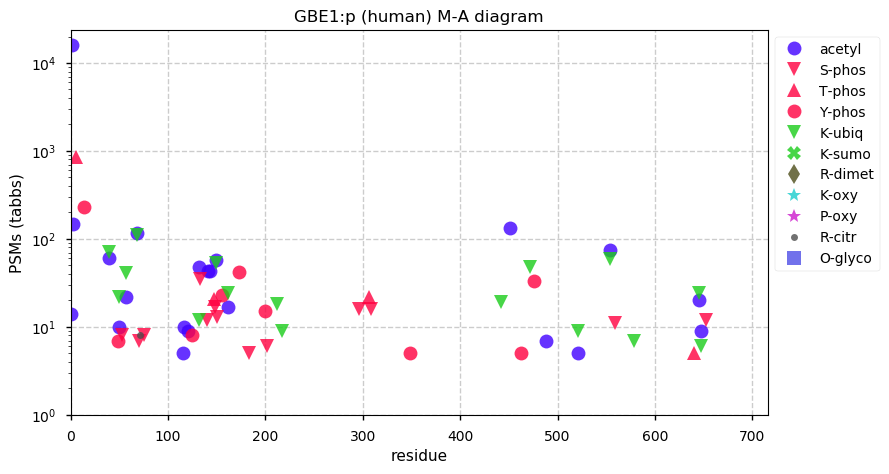
Tue Jan 19 15:39:25 +0000 2021@AlexUsherHESA Buck a beer?
Tue Jan 19 13:29:49 +0000 2021@theoneamit Presence of evidence is not evidence of translation! #RNASeq in one line.
Tue Jan 19 13:25:33 +0000 2021PLIN3:p θ(max) = 97%. aka TIP47, PP17, M6PRBP1. Observed in HLA class I & class II peptide experiments. It would be unusual to find a sample in which this protein is homozygous for the reference sequence.
Tue Jan 19 12:59:49 +0000 2021>PLIN3:p, perilipin 3 (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 14×K+acetyl, 40×S,25×T,5×Y+phosphoryl; aPTMs: 8×K+acetyl/ubiquitinyl; SAAVs: I56V (63%), F221L (87%),V275A (87%); mature form: (2-434) [51,327×, 598 kTa] #ᗕᕱᗒ 🔗
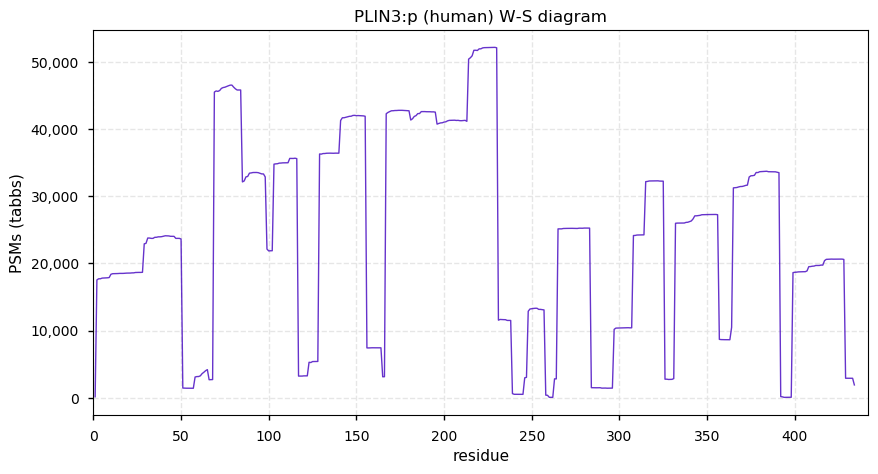
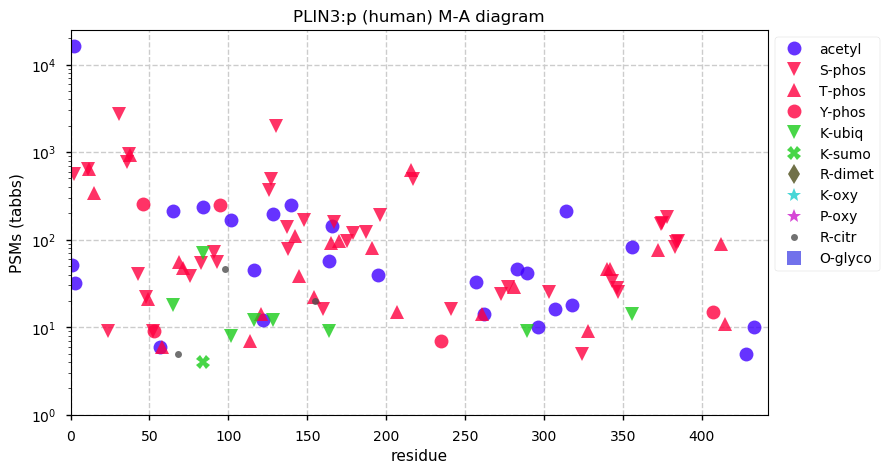
Mon Jan 18 16:08:48 +0000 2021If you are interested in MHC class I peptides, MSV000084172 is probably the most interesting data set currently available … 🔗
Mon Jan 18 15:00:50 +0000 2021@jwoodgett For the same reason that the temporal hijinx in the film made no sense.
Mon Jan 18 14:40:50 +0000 2021@jwoodgett It was not a good film, but there are no refunds.
Mon Jan 18 14:23:51 +0000 2021@AlexUsherHESA Ontario is becoming the new Grenada.
Mon Jan 18 13:46:10 +0000 2021CAP1:p θ(max) = 91. aka CAP. Observed in HLA class I & class II peptide experiments. Lysine acetylation appears to be doing some under-appreciated heavy lifting for this protein. Oddball proline rich domain (230-241).
Mon Jan 18 13:46:09 +0000 2021>CAP1:p, cyclase associated actin cytoskeleton regulatory protein 1 (Homo sapiens) CTMs: A2+acetyl; PTMs: 33× K+acetyl, 64× S,T,Y+phosphoryl; aPTMs: 8× acetyl/ubiquitinyl; SAAVs: S256A (100%); mature form: 2-475 [69,966×, 1347 kTa]#ᗕᕱᗒ 🔗
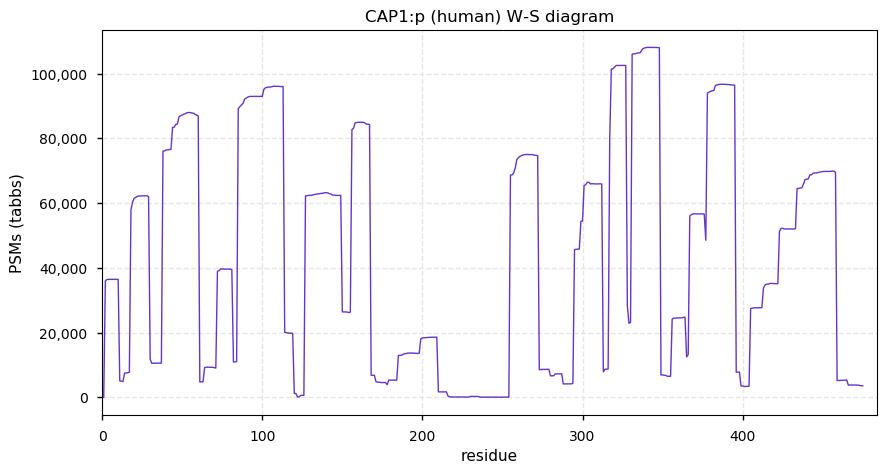
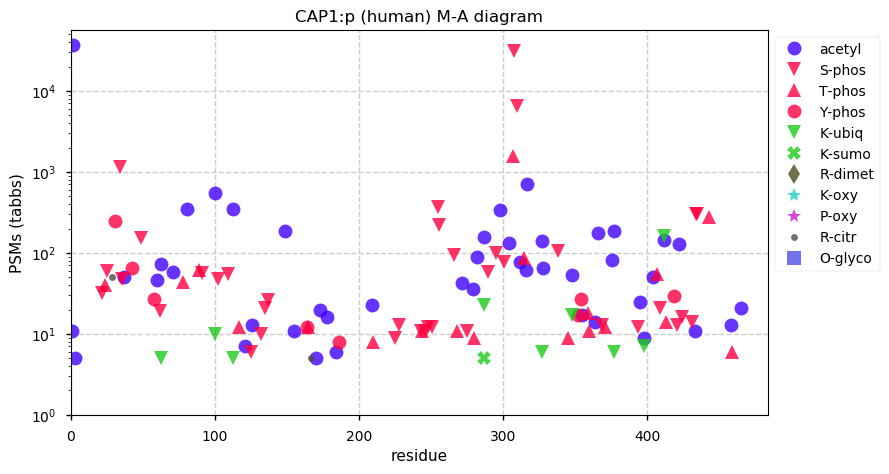
Sun Jan 17 17:11:12 +0000 2021Since I did mix up apples and oranges: 1st time for +ubi peptides amongst MHC I peptides, but not even close to the 1st time for latex antigenic peptides in this type of experiments.
Sun Jan 17 16:08:09 +0000 2021@jvarga92 They are from K+ubiquitinyl ubiquitin (as you'd expect) :
LIFAGKQLEDGR
FAGKQLEDGR
VDENGKISR
Sun Jan 17 15:38:22 +0000 2021There is a first time for everything. Just saw my first ubiquitinylated MHC class I peptides, thanks to the Broad bunch (🔗). Also has very good examples of MHC I peptides from HEVB3 (H. brasiliensis).
Sun Jan 17 14:42:56 +0000 2021It seems as though there are now quite a few bots that retrieve the contents of new links as they appear on twitter & ignore the robots.txt file. Very annoying. January is the worst month for new weirdness on the net.
Sun Jan 17 14:19:00 +0000 2021rs2230419, 1:g.225419442C>T, LBR:p.S154N, vaf=81%, Δm = 27.011, is homozygous in HEK-293 derived cell lines, as well as U2-OS, DU-145, JURKAT, K-562 & SH-SY5Y cells. Heterozygous in HCT-116, HeLa & THP-1 cells. #ᐯᐸᐱ
Sun Jan 17 13:34:38 +0000 2021LBR:p θ(max) = 38. aka DHCR14B, TDRD18. Observed in HLA class I (& rarely class II) peptide experiments. Has 8 membrane spanning domains enriched in residues with aromatic sidechains, but no ER signal sequence.
Sun Jan 17 13:34:37 +0000 2021>LBR:p, lamin B receptor (Homo sapiens) 🔗 Midsized membrane enzyme; PTMs: 34× S,T+phosphoryl; aPTMs: 12× acetyl/ubiquitinyl; SAAVs: S154N (81%), S537A (1%); mature form: 2-615 [34,262×, 219 kTa] [50,341, 545 kTa]#ᗕᕱᗒ 🔗
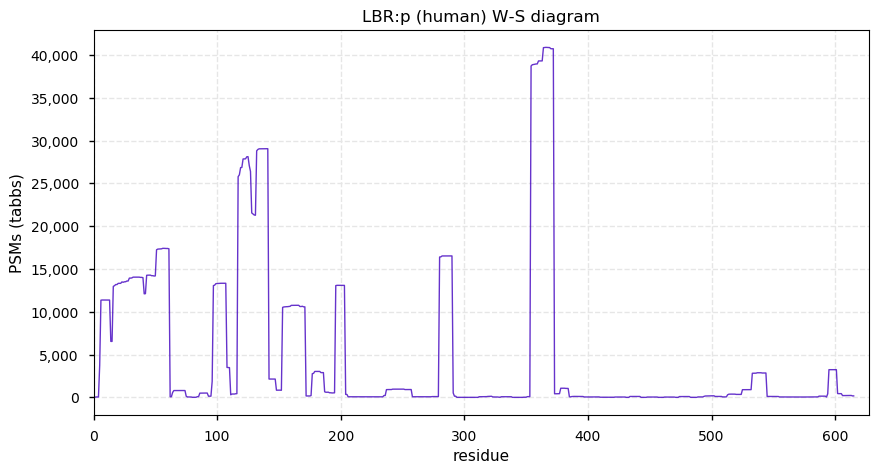
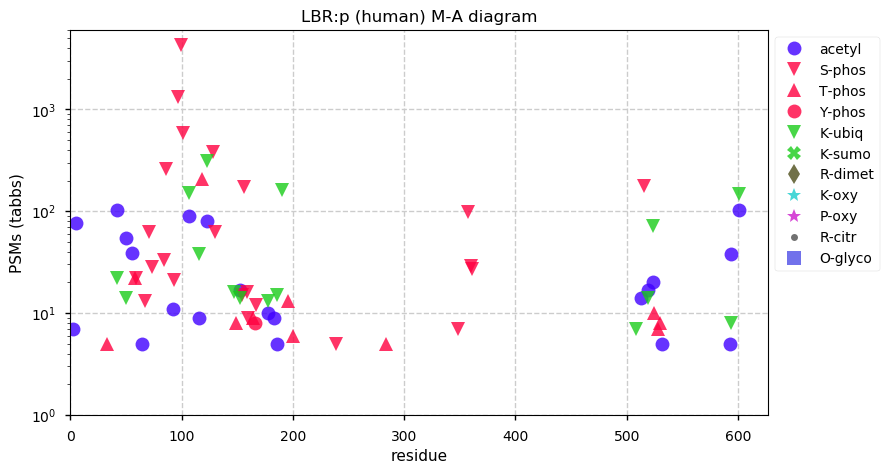
Sun Jan 17 13:25:19 +0000 2021-5 °C, precip 0.0 mm, 1011 mb↑, RH 88%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Sat Jan 16 14:38:27 +0000 2021NUP210:p, assuming variable occupation of the known sites of modification + SAAVs, there are 3.5e+41 possible molecular species (not including variability in glycosylation).
Sat Jan 16 13:51:39 +0000 2021rs2280084, removes a tryptic cleavage site, so to determine its specific expression you have to examine the peptides with & without cleavage at the site
Sat Jan 16 13:51:38 +0000 2021rs2280084, chr 3:g.13354079C>A NUP210:p.R786L, vaf=53%, Δm=-43.017, is homozygous in the HCT-116 cell line. It is heterozygous in HEK-293 and HeLa cells. #ᐯᐸᐱ
Sat Jan 16 13:30:34 +0000 2021NUP210:p, the high occupancy phosphodomain (1884-1886) is on the cytoplasmic domain. 24 K+ubiquitinyl sites and all of the high VAF SAAVs are in the nuclear domain.
Sat Jan 16 13:30:33 +0000 2021NUP210:p θ(max) = 52. aka GP210, POM210, FLJ22389, KIAA0906. Observed in HLA class I & class II peptide experiments. Transmembrane domain (1809-1828) separates the nuclear domain (26-1808) from the cytoplasmic domain (1829-1887).
Sat Jan 16 13:30:33 +0000 2021>NUP210:p, nucleoporin 210 (Homo sapiens) 🔗 CTMs: 9×N+glycosyl; PTMs: 98×S,T,Y+phosphoryl, 25×K+ubiquitinyl, 5×K+acetyl; SAAVs: A297T(48%), I608V(58%), A759V(18%), R786L(53%), R790L(58%), L1752S(89%); mature form: 26-1887 [50,341×, 545 kTa]#ᗕᕱᗒ 🔗
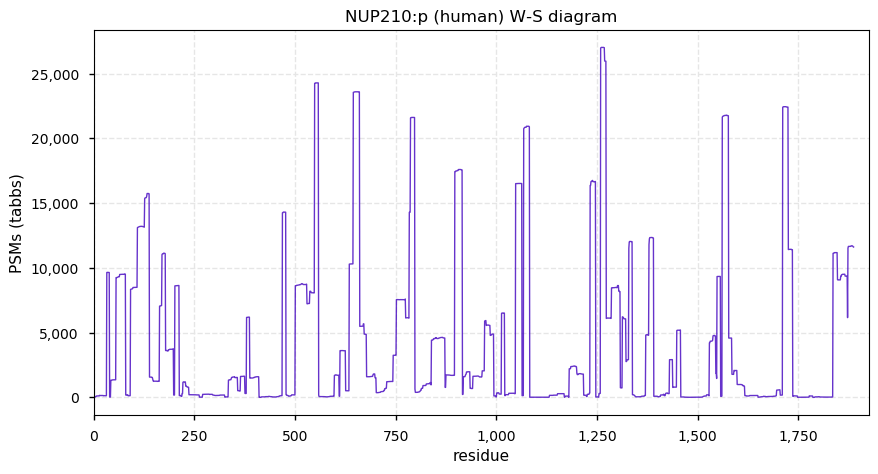
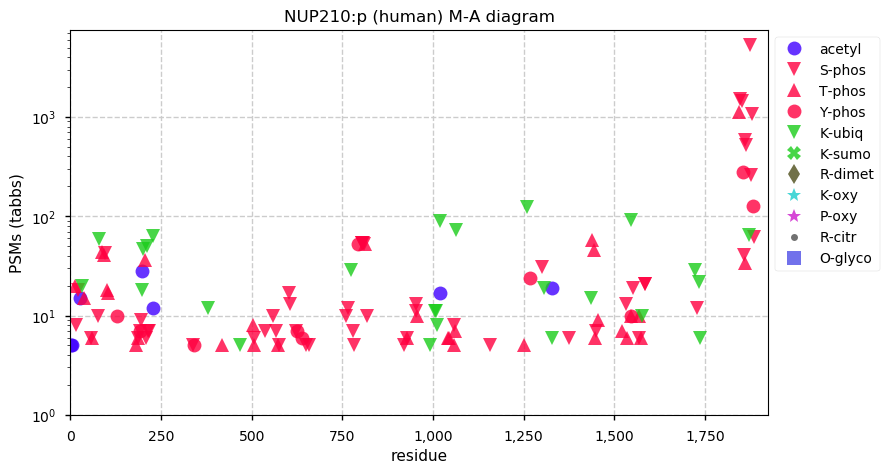
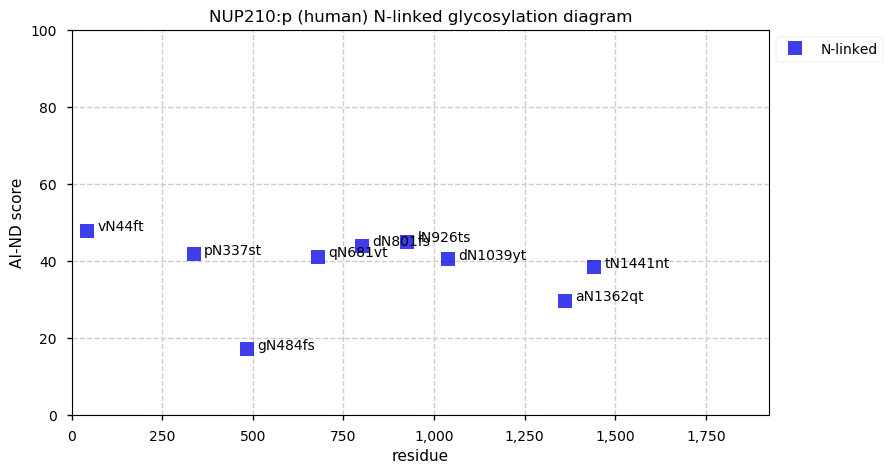
Sat Jan 16 13:29:16 +0000 2021-10 °C, precip 0.0 mm, 1013 mb↓, RH 86%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Fri Jan 15 22:32:26 +0000 2021A gust of solar wind should arrive at Earth on Sunday & remaining breezy thru Monday. The current aurora activity is modest. 🔗
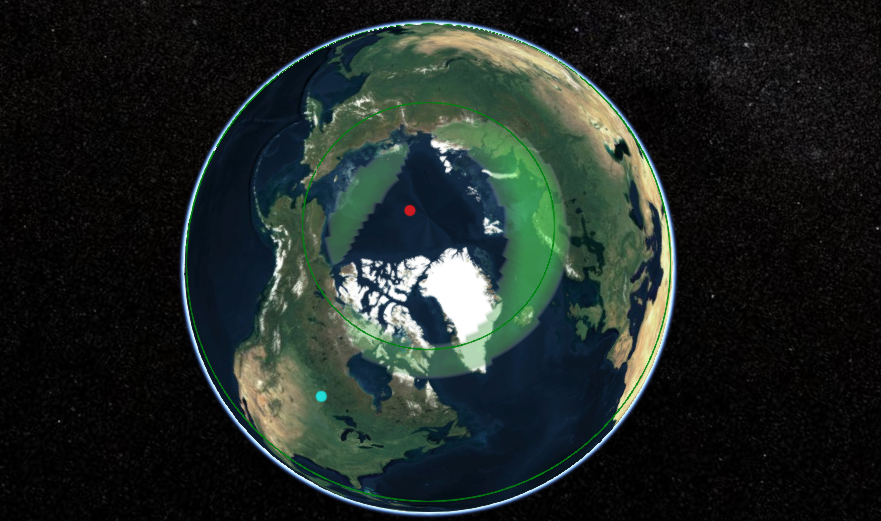
Fri Jan 15 21:01:37 +0000 2021Why do most transit web sites appear to have been designed by someone who has never waited for a bus?
Fri Jan 15 19:19:02 +0000 2021@AlexUsherHESA @FP_Champagne Your first sentence says it all: terse but accurate.
Fri Jan 15 15:24:42 +0000 2021A colorful low pressure system pushing a cold front across the US Midwest this morning (NEXRAD radar over) 🔗
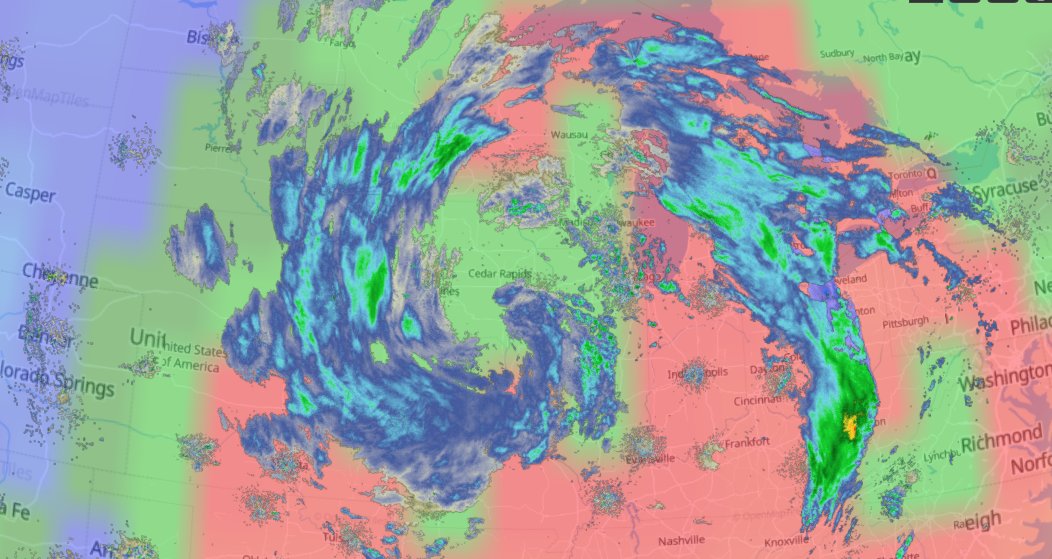
Fri Jan 15 13:29:31 +0000 2021rs1060622, 1:g.93154836G>A, TMED5:p.T175I, vaf=59%, Δm = +12.036, is heterozygous in HEK-293 derived cell lines as well as Hep-G2, JURKAT, MCF-7, CACO-2 & RKO cells. Reference allele only in HeLa, HCT-116 & THP-1 cells. #ᐯᐸᐱ
Fri Jan 15 13:05:18 +0000 2021TMED5:p θ(max) = 71. aka CGI-100, p24g2, p24gamma2. Observed in HLA class I & class II peptide experiments. Transmembrane domain (197-217) separates the lumenal domain (28-196) from the cytoplasmic domain (208-229). All PTMs in lumenal domain.
Fri Jan 15 13:05:17 +0000 2021>TMED5:p, transmembrane p24 trafficking protein 5 (Homo sapiens) 🔗 ER membrane subunit; PTMs: Y49+phosphoryl, K45, K164, K169+ubiquitinyl, aPTMs: K152+ubiquitinyl/SUMOyl; SAAVs: T175I (59%); mature form: 28-229 [13,482×, 56 kTa] #ᗕᕱᗒ 🔗
Fri Jan 15 12:58:40 +0000 2021-2 °C, precip 3.0 mm ❄, 1012 mb↑, RH 87%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Thu Jan 14 15:26:18 +0000 2021🔗
Thu Jan 14 14:57:57 +0000 2021No space rocks or geomagnetic storms today.
Thu Jan 14 13:14:03 +0000 2021rs12366, 2:g.74958729G>T, POLE4:p.G17V, vaf=34%, Δm = +42.047, is heterozygous in HEK-293 derived cell lines as well as HeLa, HEP-3B, & A-431 cells. Reference allele only in U2-OS, JURKAT, A-549, MCF-10A and MCF-7 cells.#ᐯᐸᐱ
Thu Jan 14 13:13:39 +0000 2021POLE4:p θ(max) = 86. aka p12. Observed in HLA class I (but not class II) peptide experiments. Not observed in biological fluids, e.g. urine, blood plasma or cerebrospinal fluid.
Thu Jan 14 13:13:39 +0000 2021>POLE4:p, DNA polymerase epsilon 4, accessory subunit (Homo sapiens) 🔗 Small subunit; CTMs: A2,A3+acetyl; PTMs: 5× S,T+phosphoryl, K90+acetyl, K51+SUMOyl; SAAVs: G17V (34%); mature form: 2,3-117 [8,492×, 21 kTa] #ᗕᕱᗒ 🔗
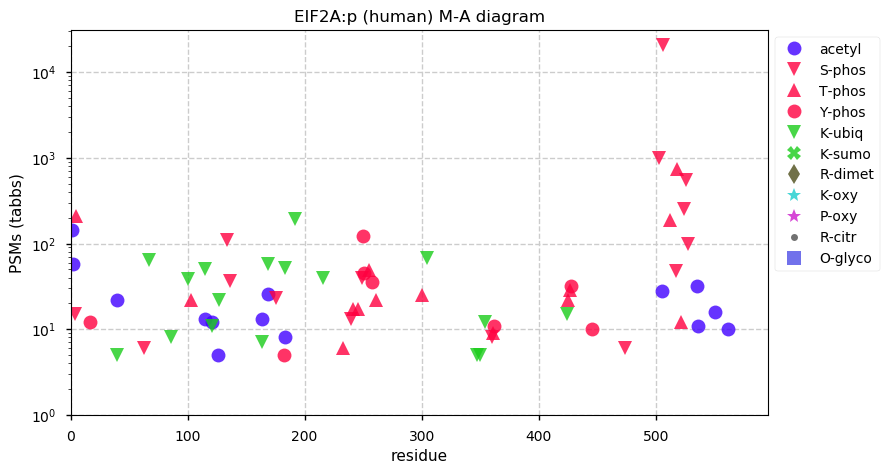
Thu Jan 14 13:10:05 +0000 2021+3 °C, precip 0.0 mm, 990 mb↓, RH 84%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Wed Jan 13 22:01:56 +0000 2021Only 8 lives left
🔗
Wed Jan 13 19:15:55 +0000 2021Thanks to everyone who participated in this poll. The clear favorite is "-f FILENAME" & nobody likes "-fFILENAME".
Wed Jan 13 16:34:01 +0000 2021@byu_sam Not because they were bad: it is just that implementation is so easy to criticize.
Wed Jan 13 16:29:27 +0000 2021@byu_sam I did 2 tours on an NIH informatics study section & pretty much any grant that spent significant effort describing implementation details was doomed.
Wed Jan 13 16:27:36 +0000 2021@byu_sam You should really stick to UML diagrams in a grant. Anything implementation specific (e.g., OS, DB platform or language) just invites tribal criticism.
Wed Jan 13 16:08:23 +0000 2021@byu_sam Rephrase the language specific parts of the grant & include a pilot study to determine best language for the tool.
Wed Jan 13 16:03:56 +0000 2021Still time to chime in on this one. 🔗
Wed Jan 13 15:30:33 +0000 2021Turned it back on with an amended robots.txt. Hopefully the googlebot will play nice from now on.
Wed Jan 13 15:18:48 +0000 2021rs1132979, chr 3:g.150562658C>G EIF2A:p.T79S, vaf=35%, Δm=-14.016, is heterozygous in the A-431 cell line as well as MCF-7 & U2-OS cells. It is homozygous in MDA-MB-231 & JURKAT cells. Reference allele only in A-549, HeLa, HEK-293 & MCF-10A cells #ᐯᐸᐱ
Wed Jan 13 13:34:35 +0000 2021Had to shutdown GPMDB's web access again. The problem today is a runaway googlebot.
Wed Jan 13 13:26:38 +0000 2021@ForrestMichelle @dr We'll see you when we see you ... 😀
Wed Jan 13 13:24:59 +0000 2021EIF2A:p θ(max) = 80. aka EIF-2A. Observed in HLA class I (& rarely class II) peptide experiments. The N-terminus is most commonly A2 (M1:A2 152:7580), but A2 is very infrequently acetylated (0.7%).
Wed Jan 13 13:24:58 +0000 2021>EIF2A:p, eukaryotic translation initiation factor 2A (Homo sapiens) 🔗 Midsized protein; CTMs: M1+A2+acetyl (rarel); PTMs: 37× S,T,Y+phosphoryl, 12× K+acetyl, 17× K+ubiquitinyl; SAAVs: T79S (35%); mature form: 2-585 [32,855×, 248 kTa] #ᗕᕱᗒ 🔗
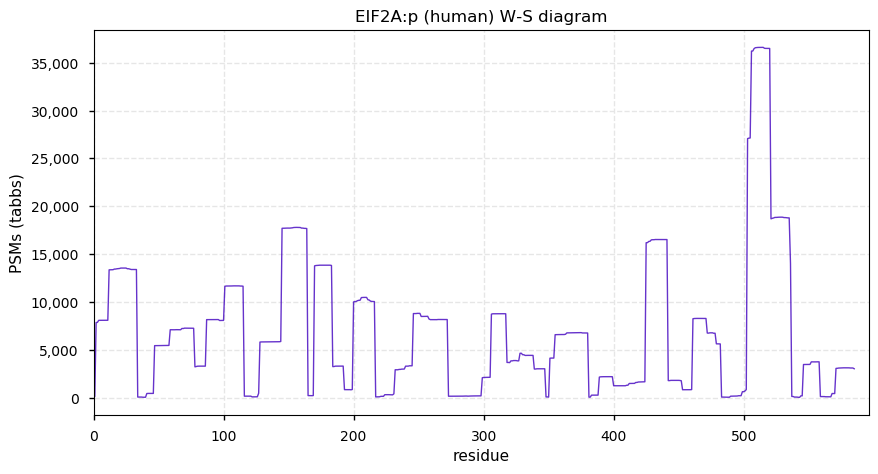
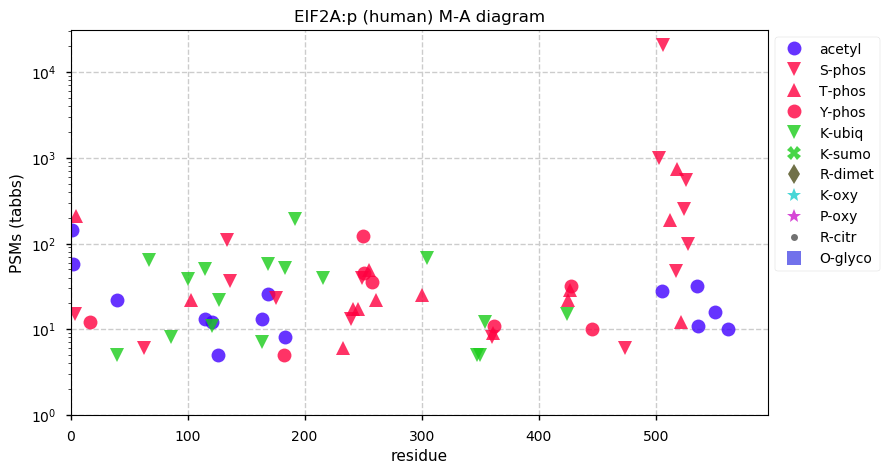
Wed Jan 13 13:24:14 +0000 2021-5 °C, precip 0.0 mm, 1000 mb↓, RH 79%, clear ☀ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Tue Jan 12 19:09:52 +0000 2021Which is the preferred form of passing a file name (FILENAME) in a command line interface?
Tue Jan 12 17:52:30 +0000 2021Space rock closest to hitting earth today:
2021 AX1, nearest approach 570,000 km, 11 meter diameter, orbit 🔗
Tue Jan 12 16:35:29 +0000 2021I don't know about anybody else, but I always seem to delay writing an appropriate CLI until the very last thing. Working on it is usually a sign I'm about done with a project.
Tue Jan 12 15:18:56 +0000 2021It seems the problem was enthusiastic overuse, rather than a malicious DOS. The web service has been back up for about 15 minutes and no problems so far.
Tue Jan 12 13:39:34 +0000 2021Had to turn off the GPMDB web service because of an apparent denial-of-service attack. Will turn it back on once the logs have been checked.
Tue Jan 12 13:27:36 +0000 2021rs35518193, chr 4:g.127820555T>C HSPA4L:p.I601T, vaf=4%, Δm=-12.036, is heterozygous in the HCT-116 cell line as well as Hep-G2 cells. It is homozygous in DLD-1 cells. Reference allele only in JURKAT, MCF-7, MCF-10A & HeLa cells #ᐯᐸᐱ
Tue Jan 12 13:27:35 +0000 2021rs1380154, chr 4:g.127801887T>C HSPA4L:p.L211S, vaf=70%, Δm=-26.052, is homozygous in the HEK-293 derived cell lines. It is heterozygous in HeLa cells. Reference allele only in CACO-2 & MCF-10A cells #ᐯᐸᐱ
Tue Jan 12 13:00:55 +0000 2021HSPA4L:p θ(max) = 81. aka APG-1, Osp94, HSPH3. Observed in HLA class I & class II peptide experiments. Significant tryptic peptide overlap with HSPA4.
Tue Jan 12 13:00:55 +0000 2021>HSPA4L:p, heat shock protein family A (Hsp70) member 4 like (Homo sapiens) 🔗 Large protein; CTMs: S2+acetyl; PTMs: 62× S,T,Y+phosphoryl, 21× K+acetyl, 23× K+ubiquitinyl; SAAVs: L211S (70%), I601T (4%); mature form: 2-839 [60,753×, 538 kTa] #ᗕᕱᗒ 🔗
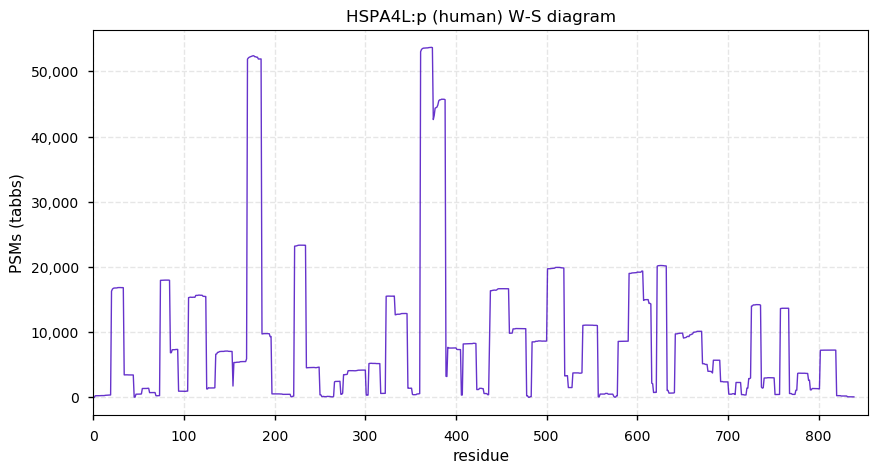
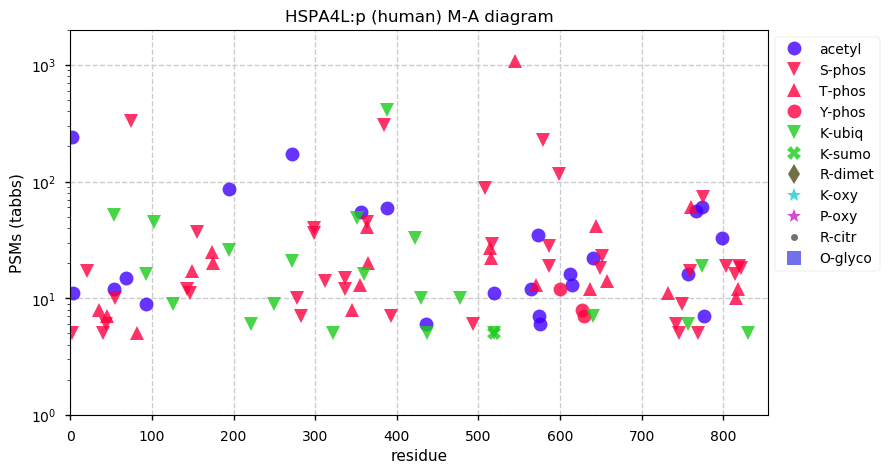
Tue Jan 12 12:57:12 +0000 2021-5 °C, precip 0.0 mm, 1006 mb↓, RH 86%, overcast️ ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Tue Jan 12 01:24:14 +0000 2021@RuneLinding That is 6 s for 20,000 spectra, or 300 μs per spectrum.
Mon Jan 11 22:46:07 +0000 2021As I am reminded, the SOA is ~6 seconds, not 30.
Mon Jan 11 13:30:24 +0000 2021rs1054486, chr 19:g.12663394G>C MAN2B1:p.L278V, vaf=25%, Δm = -14.016, is homozygous in HEK-293 derived cell lines as well as CACO-2 cells. Heterozygous in WM-239 cells. Reference only in HeLa, THP-1, A431 & JURKAT cells. #ᐯᐸᐱ
Mon Jan 11 13:30:23 +0000 2021rs1133330, chr 19:g.12661276C>T MAN2B1:p.R337Q, vaf=31%, Δm = -28.043, is homozygous in HEK-293 derived cell lines as well as MCF-7, WM-239 & CACO-2 cells. Heterozygous in SK-MEL-28 cells. The variant removes a tryptic cleavage site. #ᐯᐸᐱ
Mon Jan 11 13:24:33 +0000 2021MAN2B1:p θ(max) = 88. aka LAMAN, MANB. Observed in HLA class II (but not class I) peptide experiments. Has 6 SAAVs with vaf ≥ 1%.
Mon Jan 11 13:24:32 +0000 2021>MAN2B1:p, mannosidase alpha class 2B member 1 (Homo sapiens) 🔗 CTMs: N133, N310, N367, N497, N692, N766, N930+glycosyl PTMs: Y295+phosphoryl; SAAVs: L278V (25%), T312I (33%), R337Q (31%); mature form: 50,51-1011 [22,921×, 161 kTa] #ᗕᕱᗒ 🔗
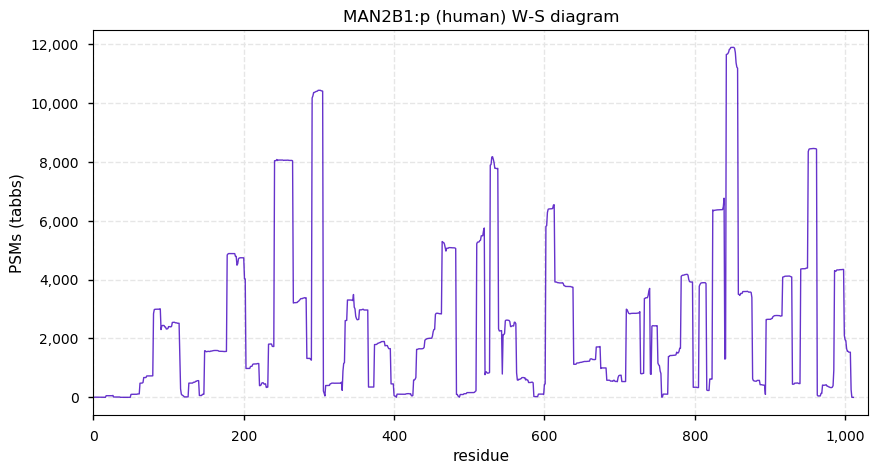
Mon Jan 11 13:00:27 +0000 2021-3 °C, precip 0.0 mm, 1010 mb↓, RH 92%, fog 🌫️ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Sun Jan 10 16:54:36 +0000 2021I thought it was surprising that people were OK with 30 minutes, even though I'm not sure what runs that slowly anymore. 30 seconds is about current state-of-the-art, so it was good that there was at least some support it.
Sun Jan 10 16:41:30 +0000 2021Thanks to everyone who participated in this pool. It was a tie between people who don't care about processing speed and those only willing to wait 5 minutes.
Sun Jan 10 13:35:40 +0000 2021rs910925, chr 17:g.746307G>C GEMIN4:p.A579G, vaf=63%, Δm = -14.016, is heterozygous in HEK-293 derived cell lines as well as HeLa cells. Homozygous in JURKAT cells. #ᐯᐸᐱ
Sun Jan 10 13:32:44 +0000 2021GEMIN4:p θ(max) = 56. aka HHRF-1, DKFZP434B131, p97, DKFZP434D174, HC56, HCAP1. Observed in HLA class I (but not class II) peptide experiments. Unusually large number of SAAVs (9) with vaf ≥ 1%.
Sun Jan 10 13:32:44 +0000 2021>GEMIN4:p, gem nuclear organelle associated protein 4 (Homo sapiens) 🔗 CTMS: M1+acetyl; PTMs: T84,S205+phosphoryl, 14× K+ubiquitinyl; SAAVs: A579G(63%), E593V(16%), I739T(7%), R1033C(29%); mature form: 1-1058 [16,823×, 85 kTa] #ᗕᕱᗒ 🔗
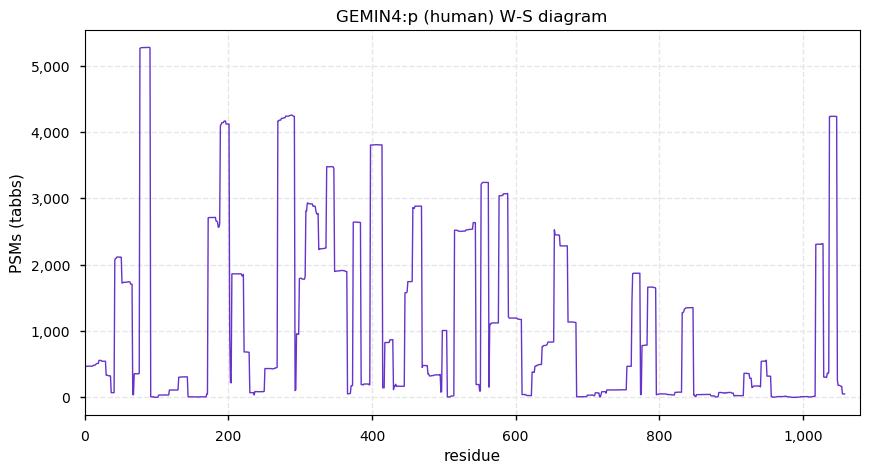
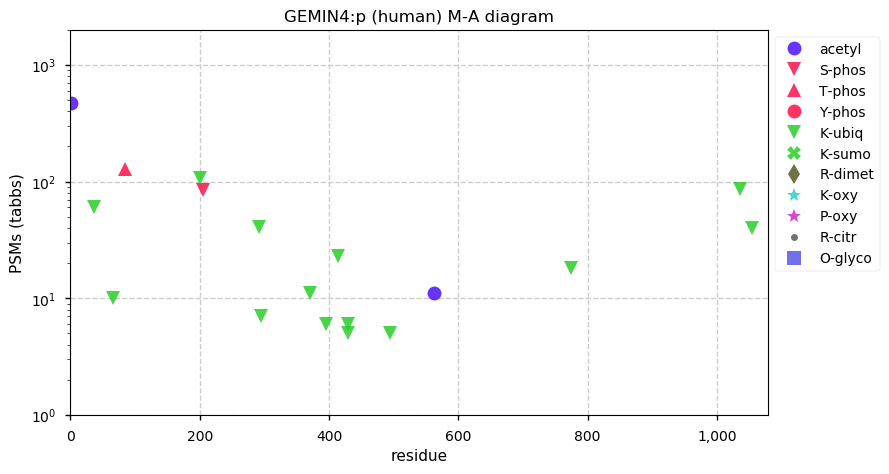
Sun Jan 10 13:30:16 +0000 2021-8 °C, precip 0.0 mm, 1020 mb↓, RH 89%, fog 🌫️ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Sat Jan 09 16:27:22 +0000 2021@MHendr1cks Although it may be simply formalizing the widespread practice of department heads expecting a co-PI kickback from junior faculty grants.
Sat Jan 09 16:22:15 +0000 2021@MHendr1cks It sounds like they are trying to establish a habilitation system.
Sat Jan 09 15:59:56 +0000 2021Time includes reading peak list files, FASTA files (+library and/or index files), writing output files and doing PSM-specific statistical analysis (assignment of p-values, E-values or their equivalent).
Sat Jan 09 15:54:08 +0000 2021What do you consider to be the maximum acceptable amount of time for PSM assignment (incl. PTM assignment and localization) with a data set of 20,000 high resolution (parent & fragment) spectra:
Sat Jan 09 15:05:39 +0000 2021Biggest space rock to just miss the earth today:
2015 NU13, closest approach 5.6 million km, 408 meter diameter, orbit 🔗
Sat Jan 09 14:04:47 +0000 2021rs9874077 note, testing for the SAAV is necessary to observe Y707+phosphoryl in homozygous cells.
Sat Jan 09 13:30:59 +0000 2021rs9874077, is located in the intracellular domain of CDCP1:p.
Sat Jan 09 13:30:59 +0000 2021rs9874077, chr 3:g.45086023T>C CDCP1:p.D709G, vaf=98%, Δm=-58.005, is homozygous in the HCT-116 cell line as well as HeLa, MCF-7, MCF-10A & most other cell lines. Reference allele only in 143B cells #ᐯᐸᐱ
Sat Jan 09 13:30:58 +0000 2021rs374919 is located in the extracellular domain of CDCP1:p. It also introduces a new tryptic cleavage site.
Sat Jan 09 13:30:58 +0000 2021rs374919, chr 3:g.45093330T>C CDCP1:p.Q525R, vaf=71%, Δm=28.043, is homozygous in the HeLa cell line as well as HK-1 & MDA-MB-231 cells. Reference allele only in HCT-116 & MCF-10A cells #ᐯᐸᐱ
Sat Jan 09 13:10:04 +0000 2021CDCP1:p θ(max) = 40. aka CD318, SIMA135. N-linked glycosylation on the extracellular domain (30-667) & 3× Y+phosphoryl on the intracellular domain (689-836).
Sat Jan 09 13:10:04 +0000 2021>CDCP1:p, CUB domain containing protein 1 (Homo sapiens) 🔗 CTMS: N39, N180, N270, N310, N339, N386, N512, N577+glycosyl; PTMs: Y283, Y707, Y734, Y806+phosphoryl; SAAVs: Q525R (71%), D709G (98%); mature form: 30-836 [5,862×, 34 kTa] #ᗕᕱᗒ 🔗
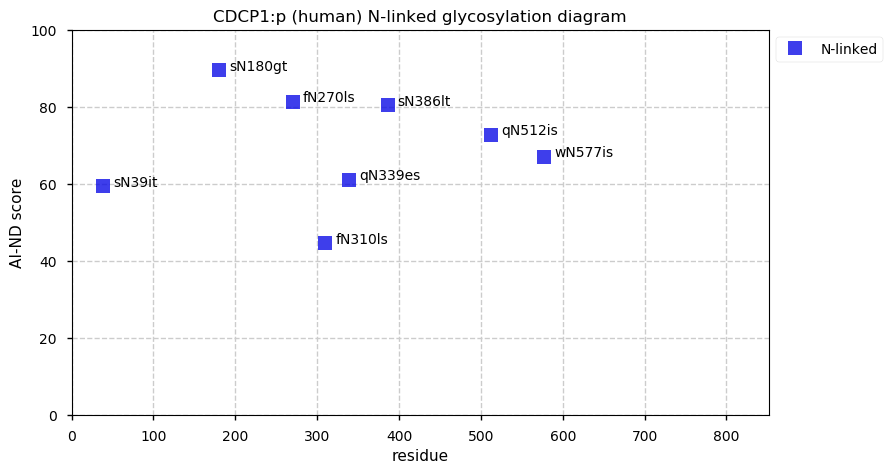
Sat Jan 09 12:53:49 +0000 2021-3 °C, precip 0.0 mm, 1026 mb→, RH 93%, fog 🌫️ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Fri Jan 08 16:13:54 +0000 2021Is there any way to get people to stop writing emails that sound as though they were a Nigerian prince? I have had to root around in my spam folder often in the last 6 months & it is always young people using spam as a style guide for their own correspondence. 🧐
Fri Jan 08 14:22:55 +0000 2021Part of the reason that the weather in Winnipeg is unusual this year (winds at 250 hPa, 🔗) 🔗
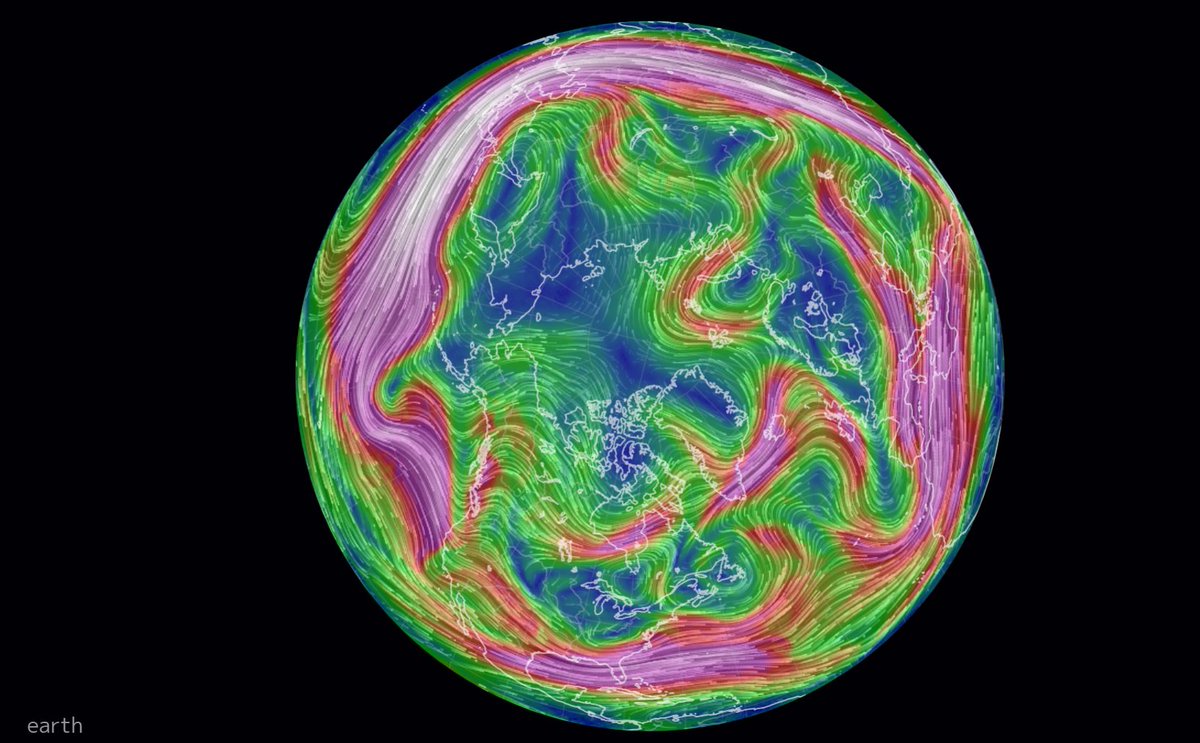
Fri Jan 08 13:27:43 +0000 2021rs381852 note, the UniProt canonical sequence for GPX8:p (Q8TED1-1) has the reference residue (K182) rather than the ancestral residue (R182).
Fri Jan 08 13:27:43 +0000 2021rs381852, chr 5:g.55164133A>G, GPX8:p.K182R, vaf=81%, Δm=28.006, is homozygous in HEK-293 derived cell lines as well as HaCaT, U2-OS, MDA-MB-231, A-549 and HeLa cells. Reference allele only in MCF-10A #ᐯᐸᐱ
Fri Jan 08 13:14:10 +0000 2021DDX39B:p θ(max) = 90. aka D6S81E, UAP56, BAT1. Unusually large number of acetyl/ubiquitinyl alternating modifications. Observed in HLA class I (but not class II) peptide experiments. Significant tryptic peptide sequence overlap with DDX39A.
Fri Jan 08 13:14:10 +0000 2021>DDX39B:p, DExD-box helicase 39B (Homo sapiens) 🔗 Small subunit; CTMS: M1, A2+acetyl; PTMs: 29× S,T,Y+phosphoryl; aPTMs: 18× K+acetyl/ubiquitinyl; SAAVs: none; mature form: 1,2-428 [76,284×, 873 kTa] #ᗕᕱᗒ 🔗
Fri Jan 08 12:46:59 +0000 2021-1 °C, precip 0.0 mm, 1026 mb↑, RH 92%, fog 🌫️ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Thu Jan 07 13:49:10 +0000 2021rs8418, chr 22:g.43983958A>G SAMM50:p.I345V, vaf=64%, Δm=-14.017, is homozygous in HEK-293 derived cell lines as well as SW-480, 3T3-L1, A-549 & CACO-2 cells. Heterozygous in MCF-7 cells. #ᐯᐸᐱ
Thu Jan 07 13:06:31 +0000 2021THOC7:p θ(max) = 88. aka NIF3L1BP1, FLJ23445, fSAP24. Found in most tissues and cell lines, except blood plasma derived fluids, e.g. CSF or urine. Observed in HLA class I & class II peptide experiments.
Thu Jan 07 13:06:31 +0000 2021>THOC7:p, THO complex 7 (Homo sapiens) 🔗 Small subunit; CTMS: G2+acetyl; PTMs: S197, T200+phosphoryl; aPTMs: K32, K36, K86, K153+acetyl/ubiquitinyl, K186+SUMOyl/ubiquitinyl; SAAVs: none; mature form: 2-204 [12,771×, 45 kTa] #ᗕᕱᗒ 🔗
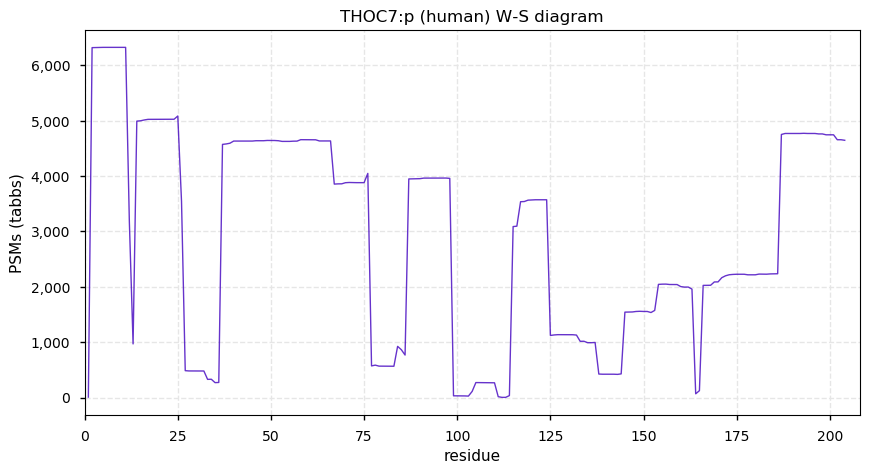
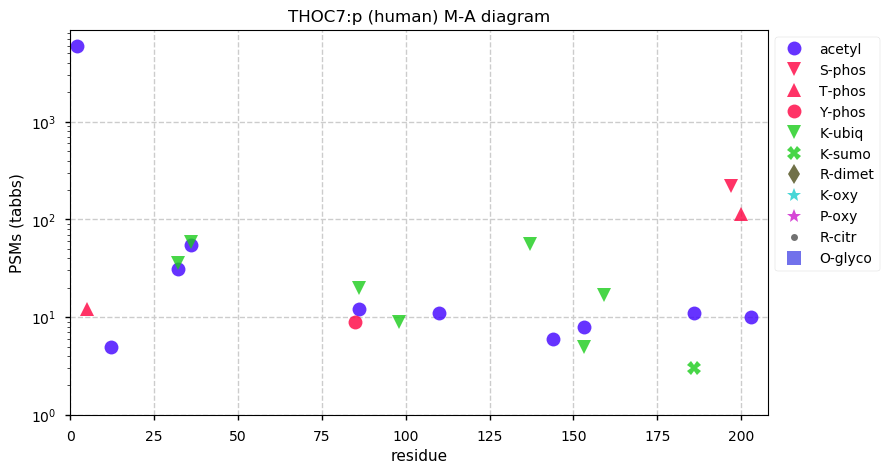
Thu Jan 07 12:57:43 +0000 2021-3 °C, precip 0.0 mm, 1022 mb↑, RH 84%, clear ☀ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Wed Jan 06 16:15:08 +0000 2021@Sci_j_my I believe that proteomics will always be red-headed-stepchild to DNA/RNA methods until good commercial platforms exist for doing routine analyses, e.g. blood and urine protein determination.
Wed Jan 06 16:08:03 +0000 2021@Sci_j_my There is plenty of room for the commercial development of platforms to fix the reproducibility problems that plague the home brew methods currently used in by practitioners.
Wed Jan 06 16:05:13 +0000 2021@Sci_j_my I do applaud these guys for putting together the company, though. Proteomics is ridiculously under-represented in the private sector: the 1st round of companies got hammered by the biotech bubble + 9/11 & they never came back.
Wed Jan 06 15:57:07 +0000 2021@Sci_j_my I took a look at the data. It looks like they are getting 700-800 protein ids per DDA run for some of the experiments (e.g., EXP18102_X1066_A_MSB37240A.raw) & 400-500 in others (e.g. EXP19002_X1818_A_MSB39028A.raw).
Wed Jan 06 15:27:50 +0000 2021Space rock just missing the earth today:
332446 is a 408 meter diameter object that is predicted to miss by 3.7 million kilometers (orbit: 🔗)
Wed Jan 06 15:07:51 +0000 2021@Sci_j_my The same holds true for urine: if you can get at the EV proteins, your method gains access to a lot more gene products.
Wed Jan 06 14:13:38 +0000 2021@Sci_j_my The results sound similar to 🔗 They obtained > 2,000 good quality protein ids per single injection. If you get good recovery of proteins from extracellular vesicles you get access to a wider diversity of sequences.
Wed Jan 06 13:37:56 +0000 2021rs4816 (chr 6:g.149793609G>A PCMT1:p.V178I, vaf=46%, Δm = +14.016) is homozygous in HEK-293 derived cell lines as well as A-431 cells. Heterozygous in U2-OS cells #ᐯᐸᐱ
Wed Jan 06 13:08:42 +0000 2021THOC6:p θ(max) = 88. aka MGC2655, fSAP35, WDR58. Found in most tissues and cell lines, except blood plasma derived fluids, e.g. CSF or urine. Observed in HLA class I but not in class II peptide experiments.
Wed Jan 06 13:08:42 +0000 2021>THOC6:p, THO complex 6 (Homo sapiens) 🔗 Midsized subunit; CTMS: M1+acetyl; PTMs: S180, S185, S249+phosphoryl; aPTMs: K67, K201, K349+SUMOyl/ubiquitinyl; SAAVs: none; mature form: 1-341 [24,583×, 98 kTa] #ᗕᕱᗒ 🔗
Wed Jan 06 13:04:54 +0000 2021-3 °C, precip 0.0 mm, 1019 mb↑, RH 83%, clear ☀ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Tue Jan 05 16:49:45 +0000 2021Note: there are only ~15,000 BioPlex 3 data sets available at the moment. I've been tied up with a lot of consulting work recently, so it may take a week or two to finish getting them all online.
Tue Jan 05 16:27:41 +0000 2021@jwoodgett @MHendr1cks @DGBassani They are still all listed on the CIHR site as being College Chairs 🔗
Tue Jan 05 15:49:19 +0000 2021Thanks to the cooperation of the group at Harvard, results from the data generated by the BioPlex 3 project are being added to GPMDB. If you want to get the results for any bait, e.g. THOC5, do a keyword search using a phrase like:
"bioplex3 THOC5"
🔗
Tue Jan 05 13:41:57 +0000 2021rs9294445 (chr 6:g.89692763G>A MDN1:p.H3423Y, vaf=54%, Δm = 26.004) is homozygous in HEK-293 derived cell lines as well as HEp-2, K-562 and Hep-G2 cells. Heterozygous in JURKAT cells. #ᐯᐸᐱ
Tue Jan 05 13:08:36 +0000 2021THOC5:p θ(max) = 76. aka PK1.3, KIAA0983, Fmip, fSAP79, C22orf19. Found in most tissues and cell lines, except blood plasma derived fluids, e.g. CSF or urine. Observed in HLA class I but not in class II peptide experiments.
Tue Jan 05 13:08:36 +0000 2021>THOC5:p, THO complex 5 (Homo sapiens) 🔗 Midsized subunit; CTMS: S2+acetyl; PTMs: 15× S/T+phosphoryl; aPTMs: K175+SUMOyl/ubiquitinyl; SAAVs: V525I (45%), V579I (22%); mature form: 2-683 [20,368×, 102 kTa] #ᗕᕱᗒ 🔗
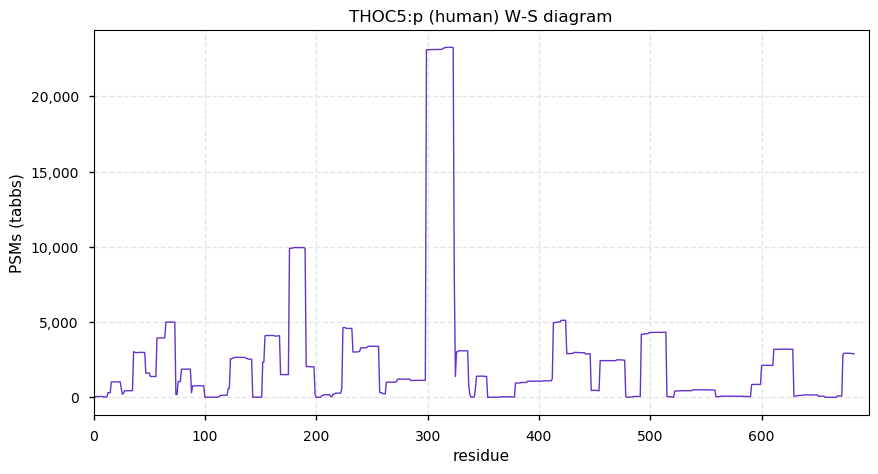
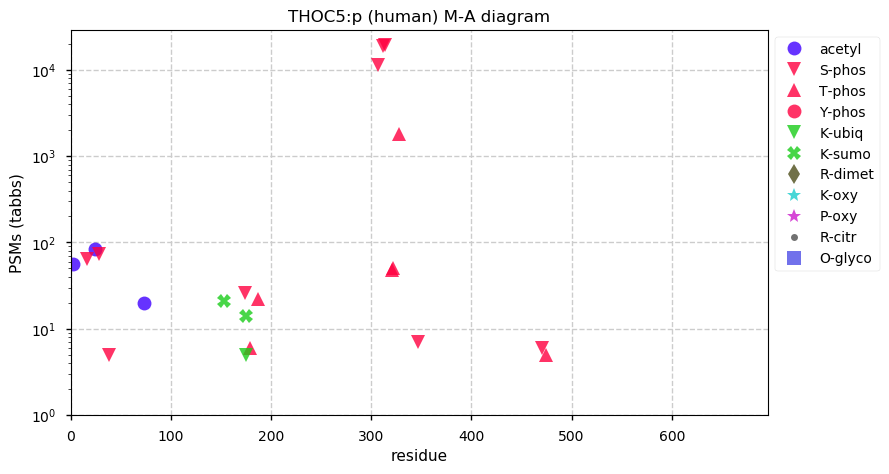
Tue Jan 05 13:04:10 +0000 2021-8 °C, precip 0.0 mm, 1013 mb↑, RH 84%, hazy ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Mon Jan 04 16:06:49 +0000 2021@pwilmarth I was thinking about Amino acid Residue Specific Expression ... 🙂
Mon Jan 04 15:41:02 +0000 2021In keeping with SAAV, maybe AASE for Amino Acid Specific Expression?
Mon Jan 04 15:03:36 +0000 2021The leading candidate for a new name would be something like the mRNA related concept of allele specific expression (ASE).
Mon Jan 04 15:02:01 +0000 2021Thank you to everyone who participated. There was an even split between respondents who felt that a new name was necessary & those that thought one of the older population genetics terms would be OK.
Mon Jan 04 13:57:06 +0000 2021Only 1 hour left to express your opinion🧐 🔗
Mon Jan 04 13:47:33 +0000 2021rs1049434, chr 1:g.112913924A>T SLC16A1:p.D490E, vaf=59%, Δm = 14.01565, is heterozygous in HEK-293 derived cell lines as well as MCF-10A, SW-480 & A-549 cells. Homozygous in HeLa cells. Reference allele only in JURKAT cells. #ᐯᐸᐱ
Mon Jan 04 13:32:11 +0000 2021Odd pattern in the stratosphere over the North Pole (winds at 10 hPa) 🔗
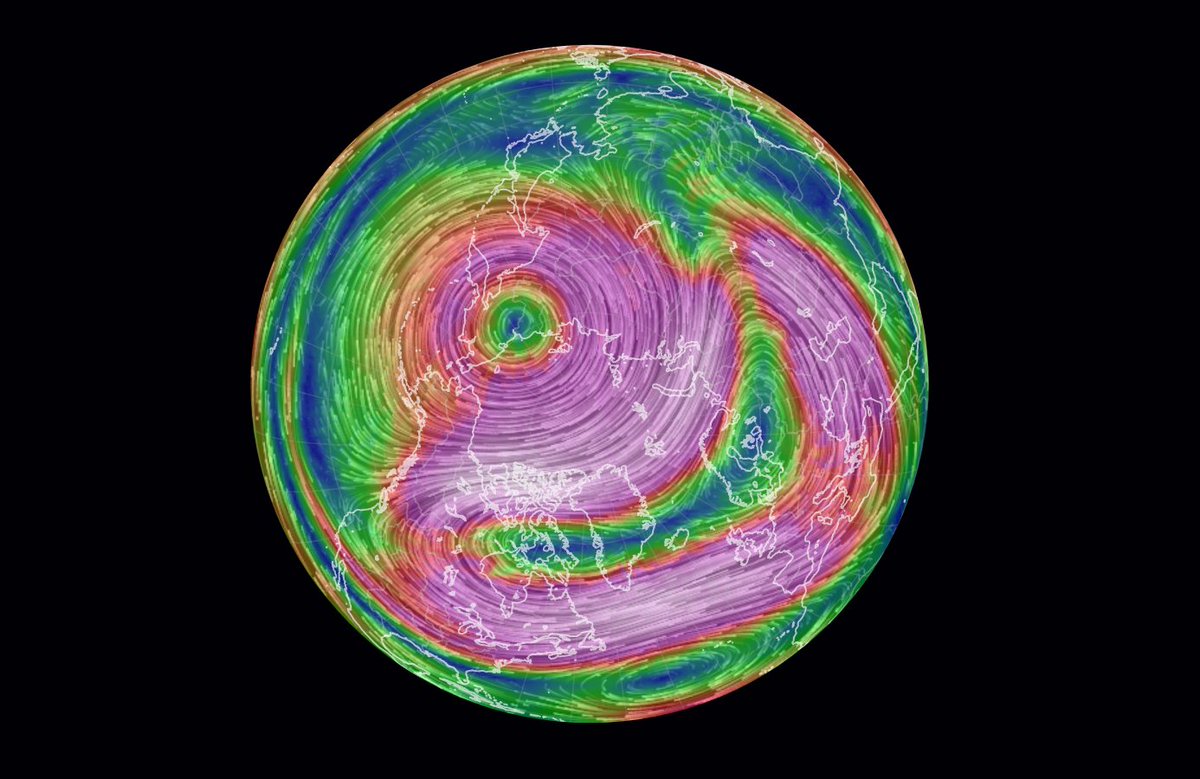
Mon Jan 04 13:15:41 +0000 2021ALYREF:p has too many GO annotations. It is the only THO complex protein that has R+dimethyl acceptor sites.
Mon Jan 04 13:15:41 +0000 2021ALYREF:p θ(max) = 74. aka ALY, BEF, ALY/REF, REF, THOC4. Found in most tissues and cell lines, except blood plasma derived fluids, e.g. CSF or urine. Observed in HLA class I and class II peptide experiments.
Mon Jan 04 13:15:40 +0000 2021>ALYREF:p, Aly/REF export factor (Homo sapiens) 🔗 Small subunit; PTMs: R45, R57, R70, R204, R211+dimethyl; aPTMs: K11, K21, K93, K168, K242+acetyl/ubiquitinyl; SAAVs: none; mature form: 2-351 [60,361×, 576 kTa] #ᗕᕱᗒ 🔗
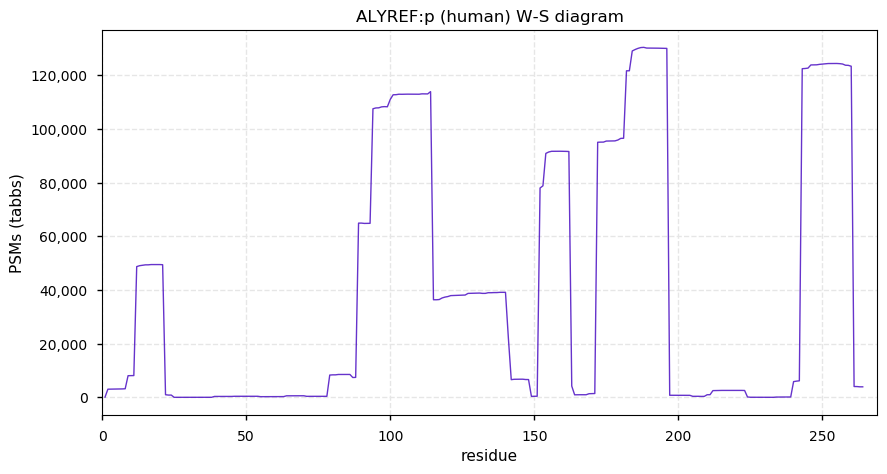
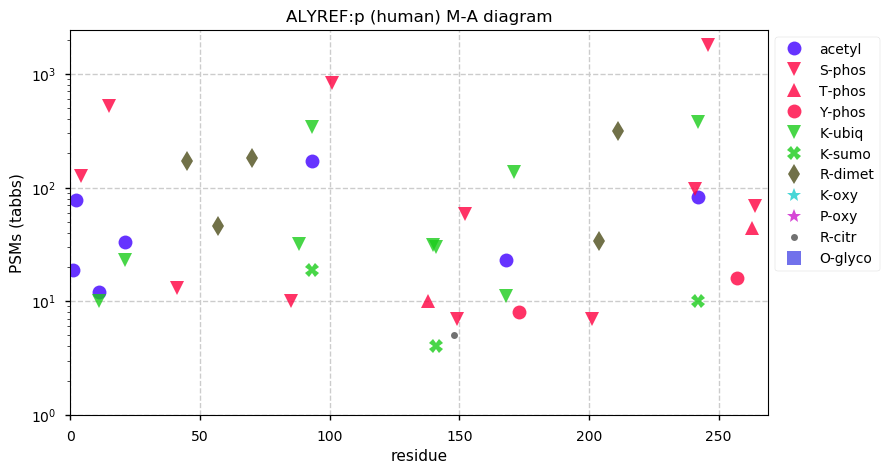
Mon Jan 04 13:10:13 +0000 2021+1 °C, precip 0.0 mm, 101 mb↑, RH 87%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Sun Jan 03 15:01:08 +0000 2021If you go with #3, feel free to propose a name.
Sun Jan 03 14:52:03 +0000 2021For a cell heterozygous for an SAAV, if the # of PSMs with the variant allele is "V" and the # of PSMs with the reference allele is "R", what would you call the ratio
? = V/(R+V)
Sun Jan 03 13:45:22 +0000 2021rs288334, chr 2:g.182757820T>G DNAJC10:p.H646Q, vaf=63%, Δm = -9.000, is heterozygous in HEK-293 derived cell lines. Homozygous in MCF-7, MCF-10A Hep-G2, MDA-MB-231, HEp-2 & HeLa cells. Reference allele only in U2-OS cells. #ᐯᐸᐱ
Sun Jan 03 13:23:52 +0000 2021-4 °C, precip 0.5 mm ❄, 998 mb→, RH 88%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Sun Jan 03 13:19:32 +0000 2021THOC3:p θ(max) = 73. aka TEX1, MGC5469. Found in most tissues and cell lines, except blood plasma derived fluids, e.g. CSF or urine. Observed in HLA class I peptide experiments, but not class II.
Sun Jan 03 13:19:32 +0000 2021>THOC3:p, THO complex 3 (Homo sapiens) 🔗 Small subunit; CTMs: A2+acetyl; aPTMs: K77, K91, K133, K171, K347+ubiquitinyl/SUMOyl, K86, K171, K329, K332+acetyl/ubiquitinyl; SAAVs: none; mature form: 2-351 [22,261×, 77 kTa] #ᗕᕱᗒ 🔗
Sat Jan 02 17:36:00 +0000 2021Just fixed a torn shirt sleeve with a Singer 401J. I had forgotten how well thought out and designed purely mechanical sewing machines were in their time.
Sat Jan 02 13:35:51 +0000 2021rs1466685 introduces a new trypsin cleavage site at the 184-185 bond. The UniProt sequence Q9Y5P6-1 has the reference sequence residue Q184 rather than the ancestral residue (R184). The reference mouse sequence has Q184 & no variant allele.
Sat Jan 02 13:35:51 +0000 2021rs1466685, the protein-level evidence: HEK-293 🔗 vs missing the variant Q184R cleavage 🔗
Sat Jan 02 13:35:50 +0000 2021rs1466685, chr 3:g.49722606T>C GMPPB:p.Q184R, vaf=>99%, Δm = 28.043, is homozygous in HEK-293 derived cell lines and all other commonly used cell lines. #ᐯᐸᐱ
Sat Jan 02 13:17:35 +0000 2021THOC2:p has a distinct C-terminal domain (1229-1593) that is enriched in K, E and S,T+phosphoryl acceptor sites.
Sat Jan 02 13:17:35 +0000 2021THOC2:p θ(max) = 56. aka THO2, dJ506G2.1, CXorf3, MRX12, MRX35. Found in most tissues and cell lines, except blood plasma derived fluids, e.g. CSF or urine. Observed in HLA class I peptide experiments, but not class II. Nuclear localization signal domain (923-930).
Sat Jan 02 13:17:34 +0000 2021>THOC2:p, THO complex 2 (Homo sapiens) 🔗 Large subunit; CTMs: A2,A3+acetyl; PTMs: 58x S,T+phosphoryl, 16x K+acetyl, 20x K+ubiquitinyl; SAAVs: none; mature form: 2,3-1593 [38,130 x, 259 kTa] [24,821×, 143 kTa] #ᗕᕱᗒ 🔗
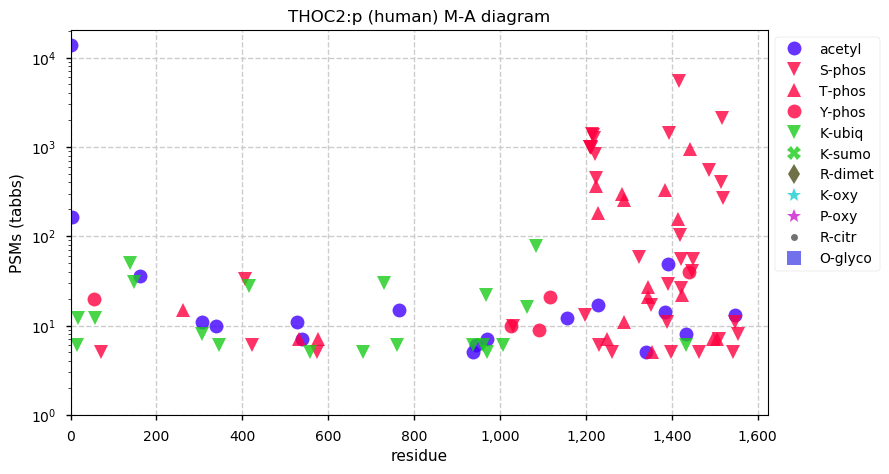
Sat Jan 02 13:15:11 +0000 2021-5 °C, precip 0.0 mm, 1006 mb↓, RH 93%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Fri Jan 01 16:43:28 +0000 2021@slashdot The very reason I set up my own OpenMapTiles server and switched to CesiumJS.
Fri Jan 01 16:08:28 +0000 2021Got my latest RPI4 yesterday (8 GB) yesterday & had it up and running data in about 15 minutes. First one I have set up using 64-bit Ubuntu server (20.04) as the operating system.
Fri Jan 01 15:14:12 +0000 2021And not to be out-done, the Chicago I remember:
After 3 years of progress, Chicago’s murder tally skyrockets in 2020 🔗 via @SunTimes
Fri Jan 01 15:11:30 +0000 2021This is the New York I remember:
NYC woman hit by stray bullet in possible first shooting of 2021 🔗 via @nypmetro
Fri Jan 01 14:16:20 +0000 2021rs747061326 note: the reference R135 is not a tryptic cleavage site because of P136. The results suggest low penetrance of the variant allele.
Fri Jan 01 14:16:20 +0000 2021rs747061326, chr 1:g.1455485G>A, ATAD3C:p.R135Q, vaf=0.0072%, Δm = -28.043, is heterozygous in HEK-293 derived cell lines and MCF-10A. #ᐯᐸᐱ
Fri Jan 01 13:52:03 +0000 2021THOC1:p θ(max) = 71. aka P84, HPR1. Found in most tissues and cell lines, except blood plasma derived fluids, e.g. CSF or urine. Observed in HLA class I peptide experiments, but not class II. Nuclear localization signal domain (415-430).
Fri Jan 01 13:52:03 +0000 2021>THOC1:p, THO complex 1 (Homo sapiens) 🔗 Midsized nuclear subunit; CTMs: M1,S2+acetyl; PTMs: 21× S,T+phosphoryl, 21 × K+acetyl, 7× K+ubiquitinyl, 3× K+SUMOyl; SAAVs: none; mature form: 1,2-657 [24,821×, 143 kTa] #ᗕᕱᗒ 🔗
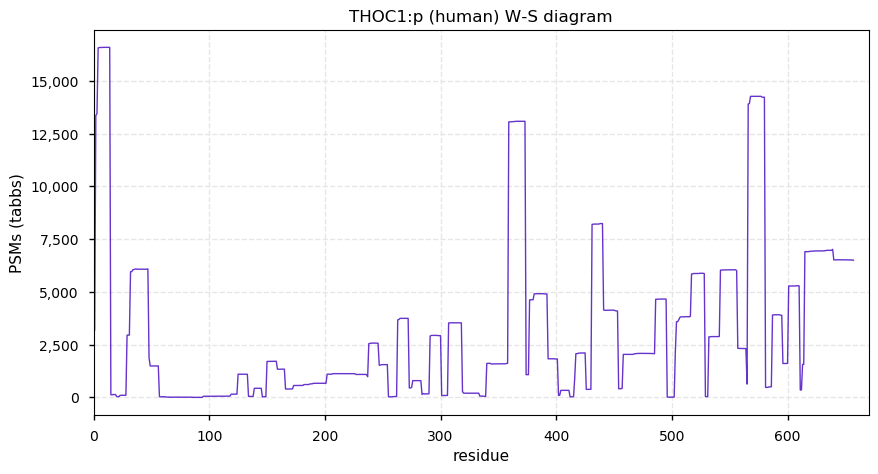
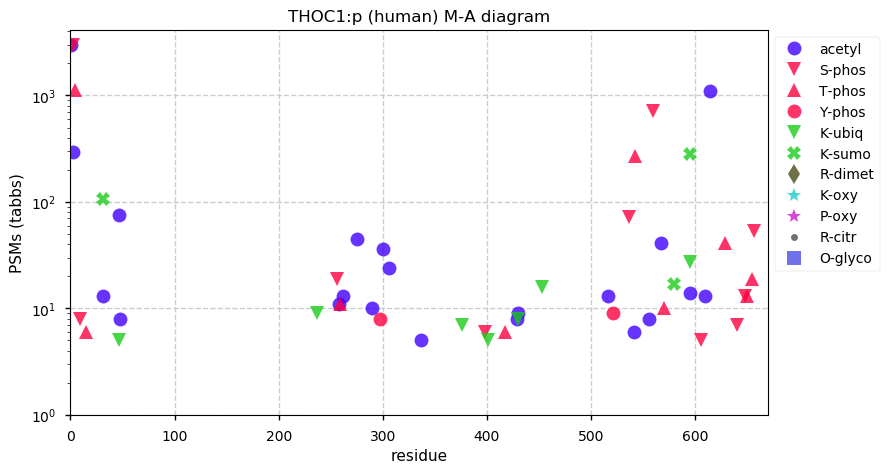
Fri Jan 01 13:37:41 +0000 2021Another rather artful looking radar image of a storm this morning, running along a cold front arcing from Oklahoma City, OK to Richmond, VA 🔗
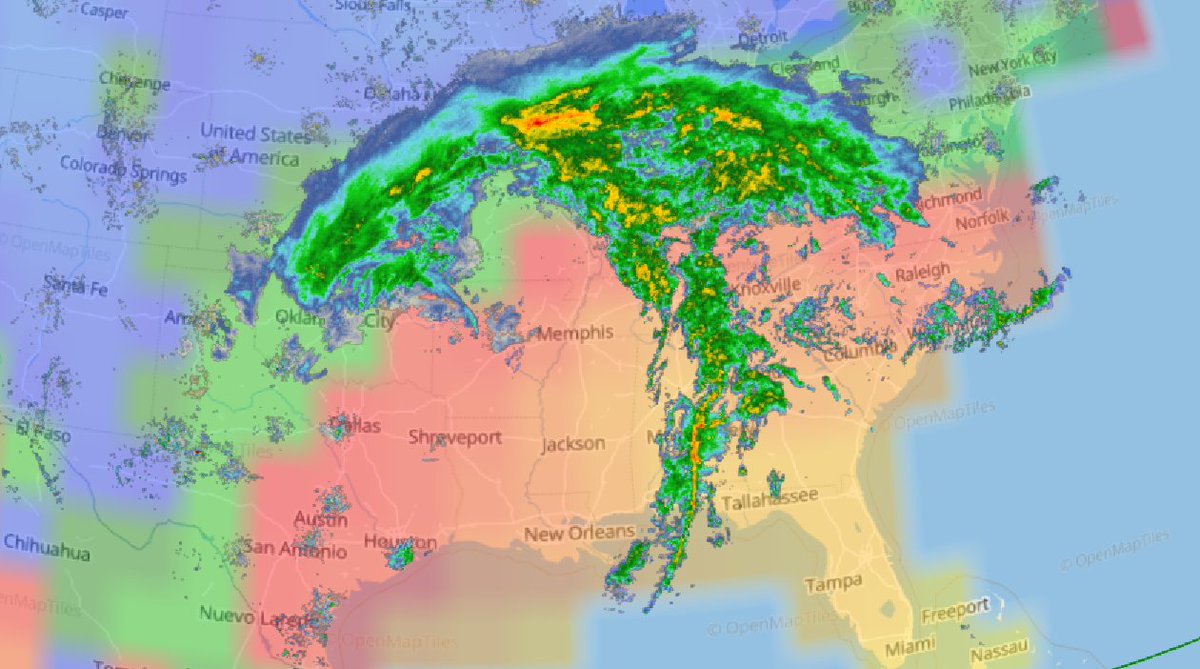
Fri Jan 01 13:25:33 +0000 2021-9 °C, precip 0.0 mm, 1016 mb→, RH 88%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
tweets = 4033