@norsivaeb Tweets 
| 2022
| 2021
| 2020
| 2019
| 2018
|
Wed Nov 09 17:32:12 +0000 2022@UCDProteomics @M_plus_H @mrsalemi1 @HaoLab_GWU It is one of those intractable problems that appears as though it should only require a bit of effort, then a little more, a little more, a little more ... ad infinitum.
Wed Nov 09 17:14:47 +0000 2022@UCDProteomics @M_plus_H @mrsalemi1 @HaoLab_GWU It seems to be an evergreen topic, though.
Wed Nov 09 16:51:35 +0000 2022Also, if you are being hired as part of an institute or other type of special, non-departmental organization, be sure that your academic department head is 100% on board. If they are even a little wobbly about their support for the institution, don't go there.
Wed Nov 09 16:44:33 +0000 2022Also find out how long they are going to remain in that position: junior faculty hired by one head can be disfavored by an new head that wants to change gears within the department.
Wed Nov 09 16:41:36 +0000 2022If you happen to be interviewing for academic positions this season, be sure to ask any potential department head you meet how they assist junior faculty to succeed in their department & how they judge that success (with examples/case studies if possible).
Wed Nov 09 15:05:12 +0000 2022As someone who has sat through their share of such sessions on both sides of the table, the last question really should be first & take up most of the time in the interview. 🔗
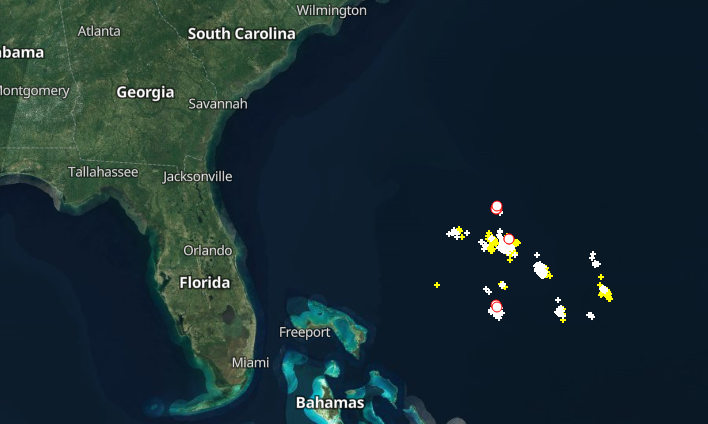
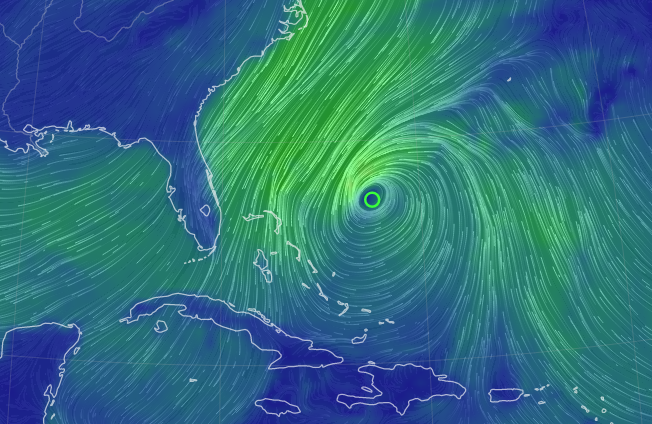
Wed Nov 09 13:31:05 +0000 2022Lightning strikes in Tropical Storm Nicole, as it heads towards Florida for a landfall tomorrow morning near West Palm Beach as a hurricane. It is expected to cross Florida & then turn north, running up from Tallahassee thru Charlotte to Washington DC. 🔗
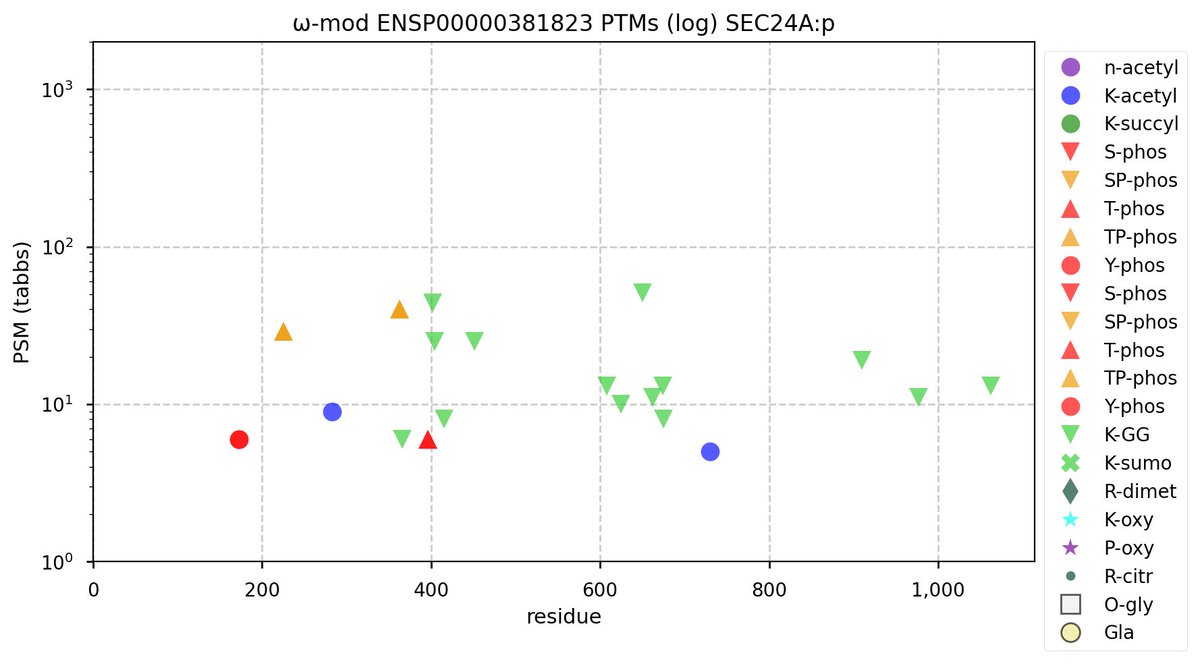
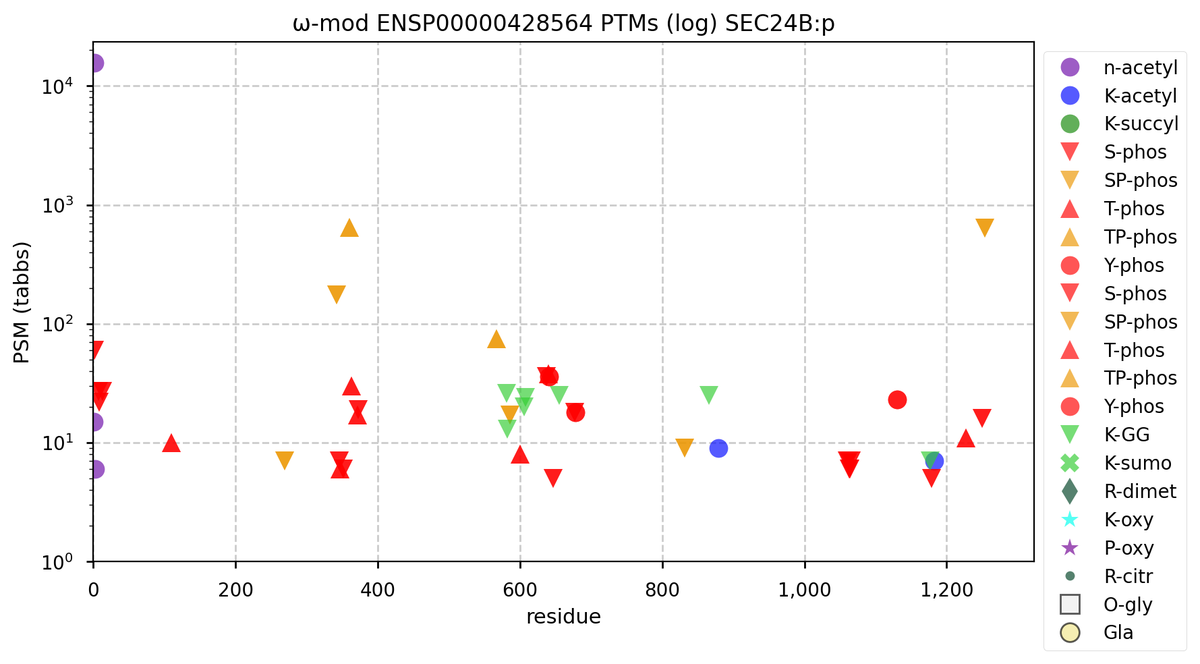
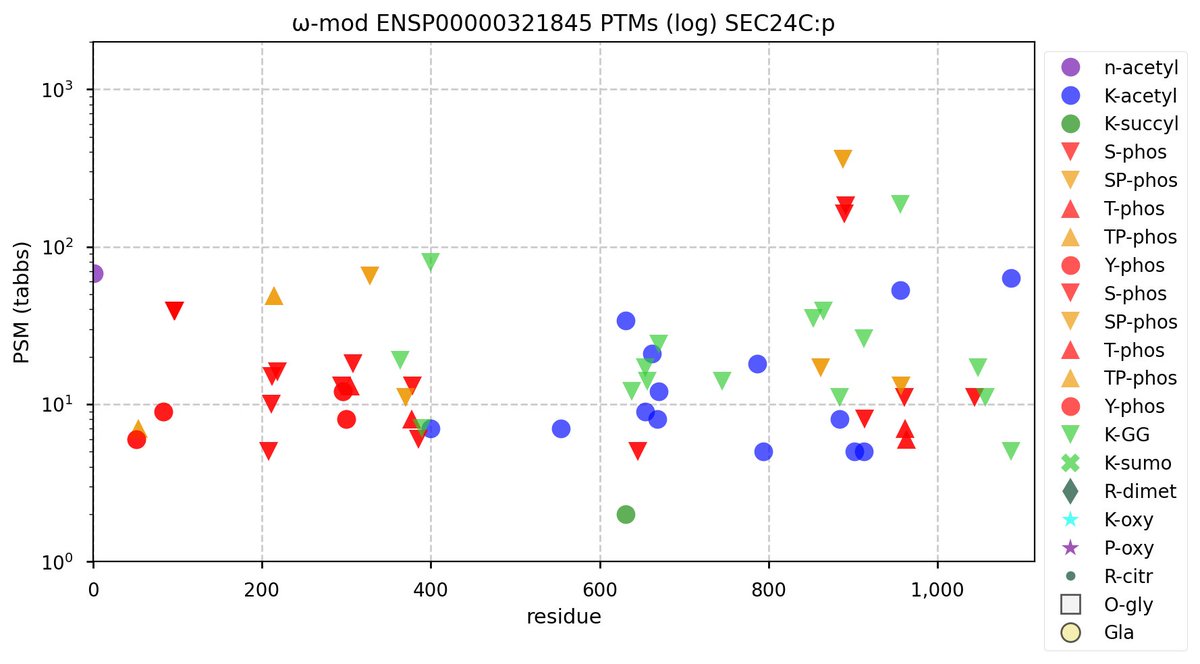
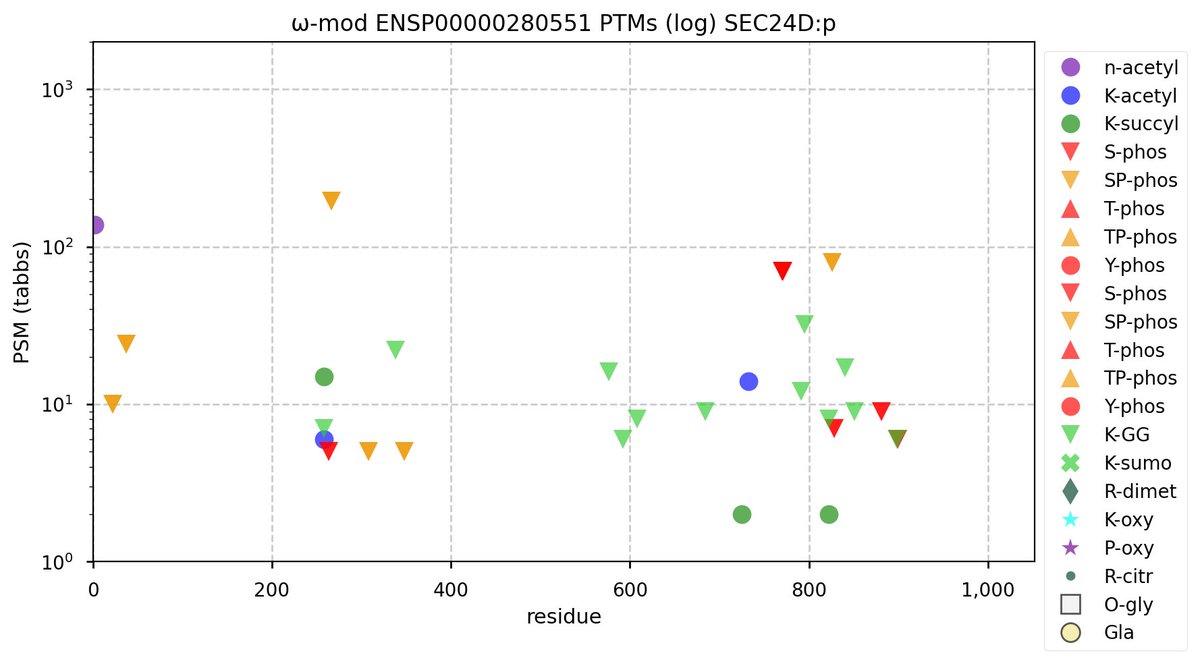
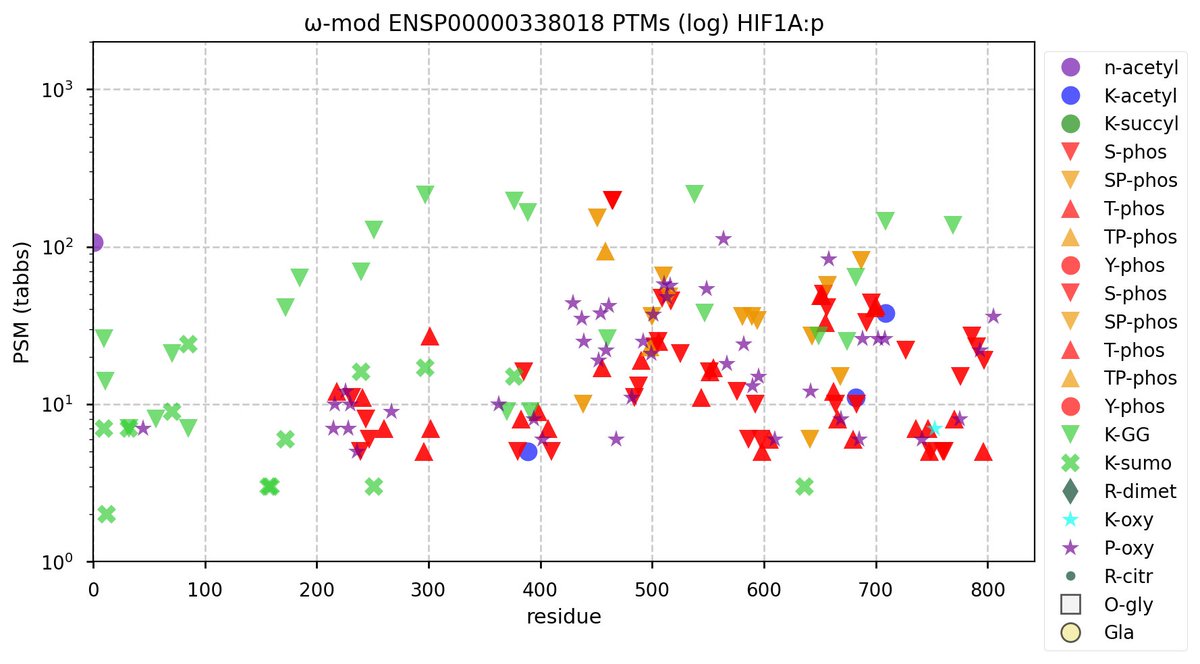
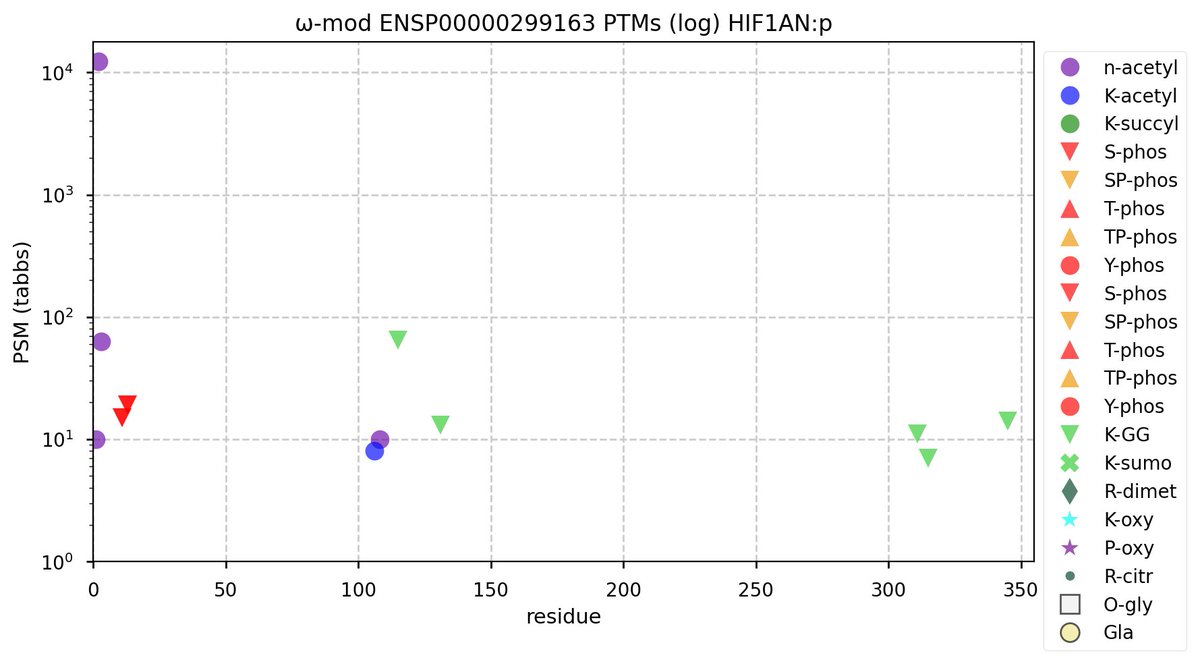
Wed Nov 09 12:26:35 +0000 2022Modification diagrams for SEC24A:p to SEC24D:p, 4 paralogous subunits of the COP II complex. While there are some shared notes and riffs, these patterns clearly diverged a long, long time ago. 🔗
Tue Nov 08 20:06:11 +0000 2022@riley_nm1 @Smith_Chem_Wisc Looking back through an old log book it turns out that I signed up for one in 2018 but never really got around to using it:
@RonBeavis@mastodon.social
Tue Nov 08 19:04:46 +0000 2022Even though I shouldn't be surprised, I still find it jarring when a paper says that "carbamidomethylation (C)" was used as a possible modification, even though IAA/CAA was not used in the sample workup.
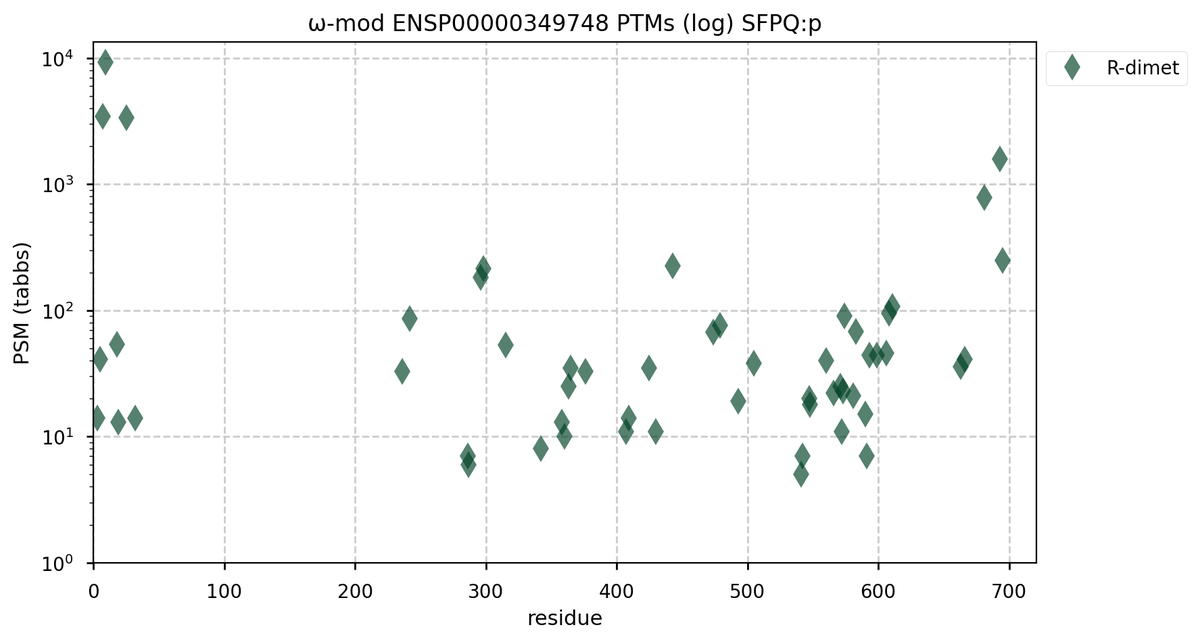
Tue Nov 08 16:05:11 +0000 2022SFPQ:p may hold the record for the most occupied R+dimethyl sites, with 54 of 60 arginines observed to be acceptors. 🔗
Tue Nov 08 15:41:43 +0000 2022@dawnnadon @stphnmaher Given what happened in Anaheim, "Deadpool Arena" is more likely.
Tue Nov 08 15:21:38 +0000 2022The CBC website still seems to be in denial about this one (of interest to Canadians only):
🔗
Tue Nov 08 15:06:17 +0000 2022There seems to have been a resurgence in phosphoproteomics data in PRIDE recently: it had gone quiet for a while but it seems to be back with a vengeance.
Tue Nov 08 13:45:04 +0000 2022@fcyucn If the software's development was privately funded, then closed source is OK. Otherwise, it isn't.
Tue Nov 08 12:45:21 +0000 2022While it may seem obvious to you, mention the model organism you are using in the Abstract & casually throughout the manuscript, not only in just one line of the Methods.
Tue Nov 08 12:40:36 +0000 2022Tropical storm Nicole is expected to make landfall near Palm Bay in Florida as a hurricane on Thursday morning. 🔗
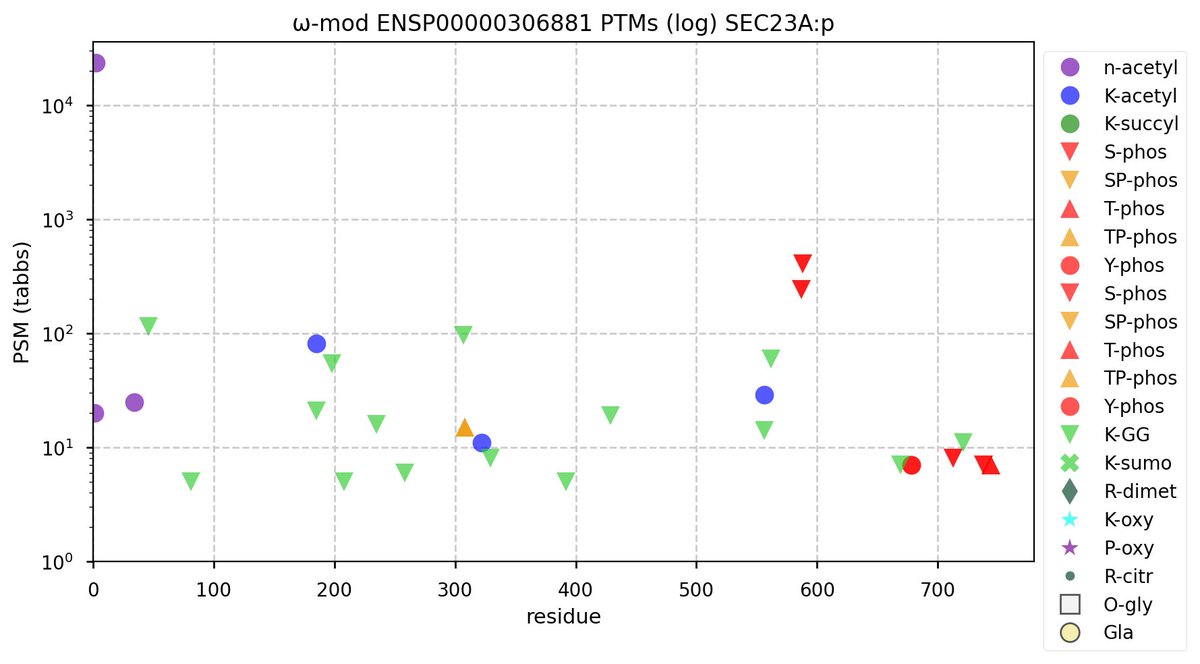
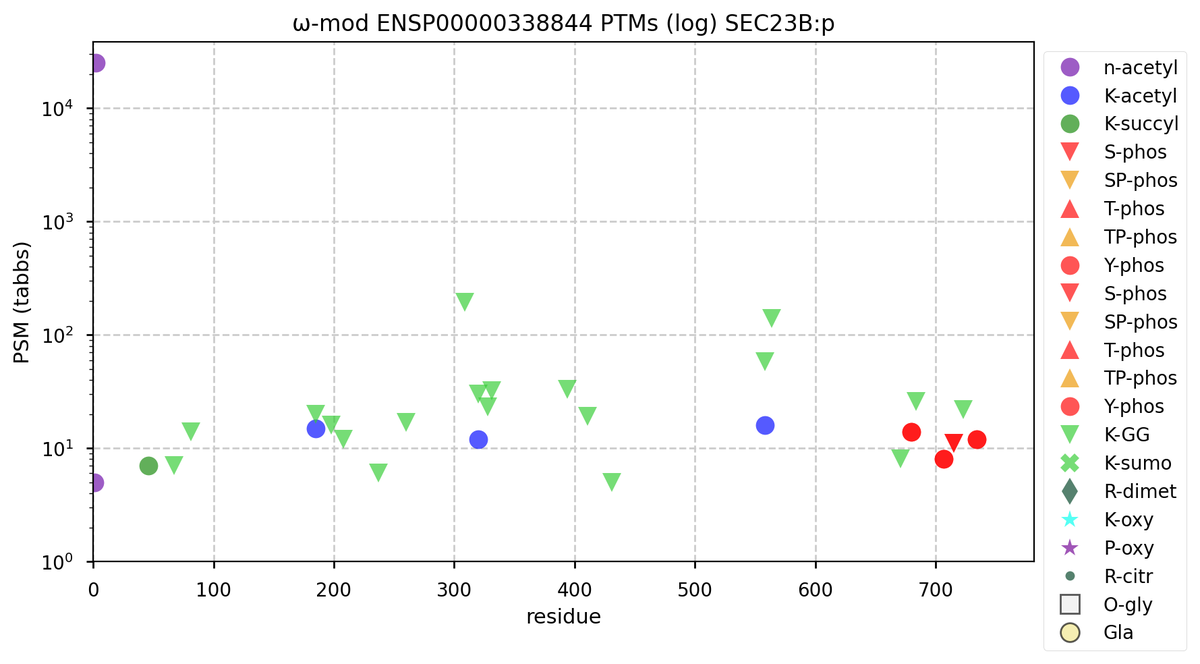
Tue Nov 08 12:25:36 +0000 2022Modification diagrams for SEC23A:p & SEC23B:p, paralogous subunits of the COP II complex. The PTM patterns are similar to each other & they have far fewer K+acetyl acceptors than most COP I subunits. 🔗
Mon Nov 07 18:25:11 +0000 2022I see quite a bit of comment wrt to Canadian university research funding. I have come around to the opinion that the problem isn't the level of funding. The problem is an entrenched academic system that can't adapt to changing funding environments.
Mon Nov 07 18:07:18 +0000 2022I'd be more interested in American politics if there was any real policy difference between the 2 parties. They are clearly different on a few edge cases (that make a lot of noise), but on the whole they are what would be called a "Conservative" party anywhere else.
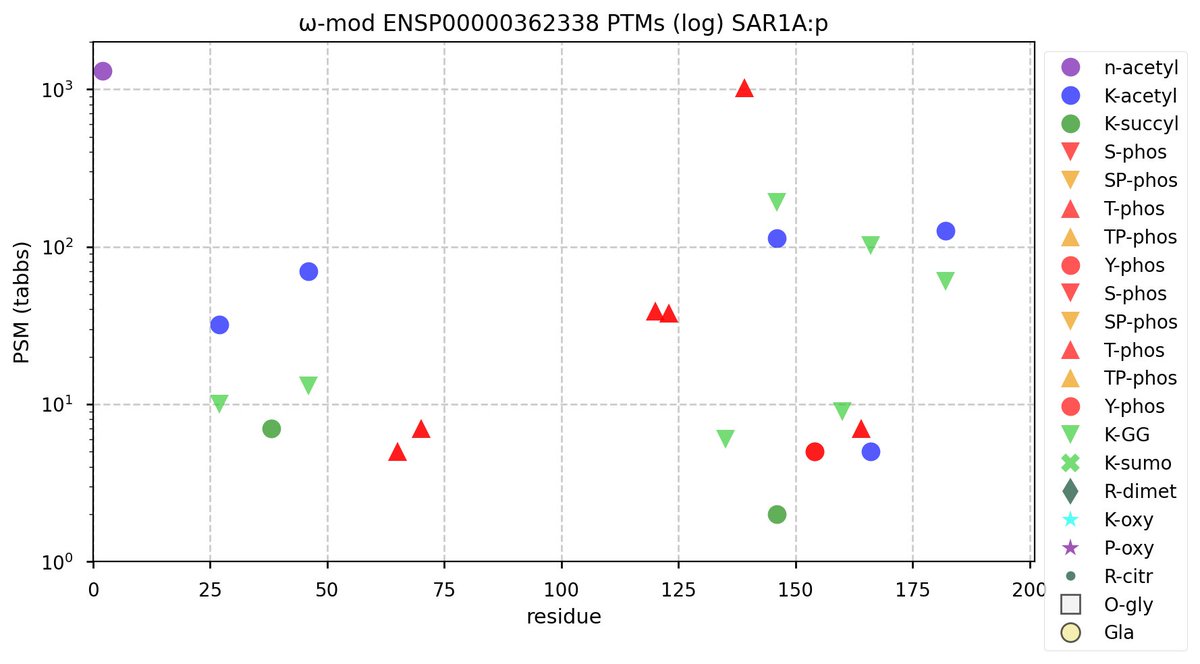
Mon Nov 07 14:01:29 +0000 2022COP II genes have more paralogues than COP I genes in vertebrates. They reflect rather old gene duplication events, so the genes are often on different chromosomes & the protein sequences do not have a high degree of sequence identity.
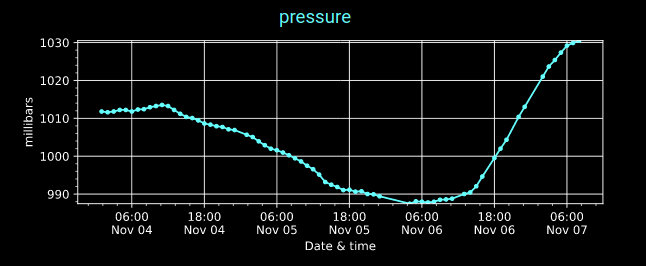
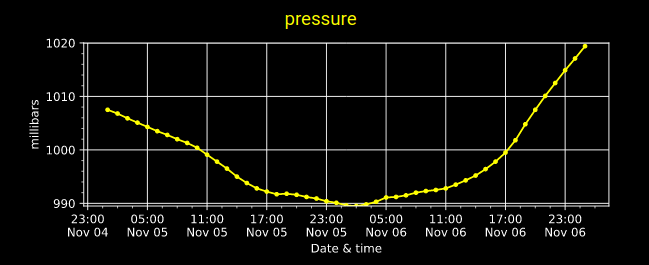
Mon Nov 07 13:39:12 +0000 2022The ground station recorded pressure measurements of the low pressure system predicted above. 🔗
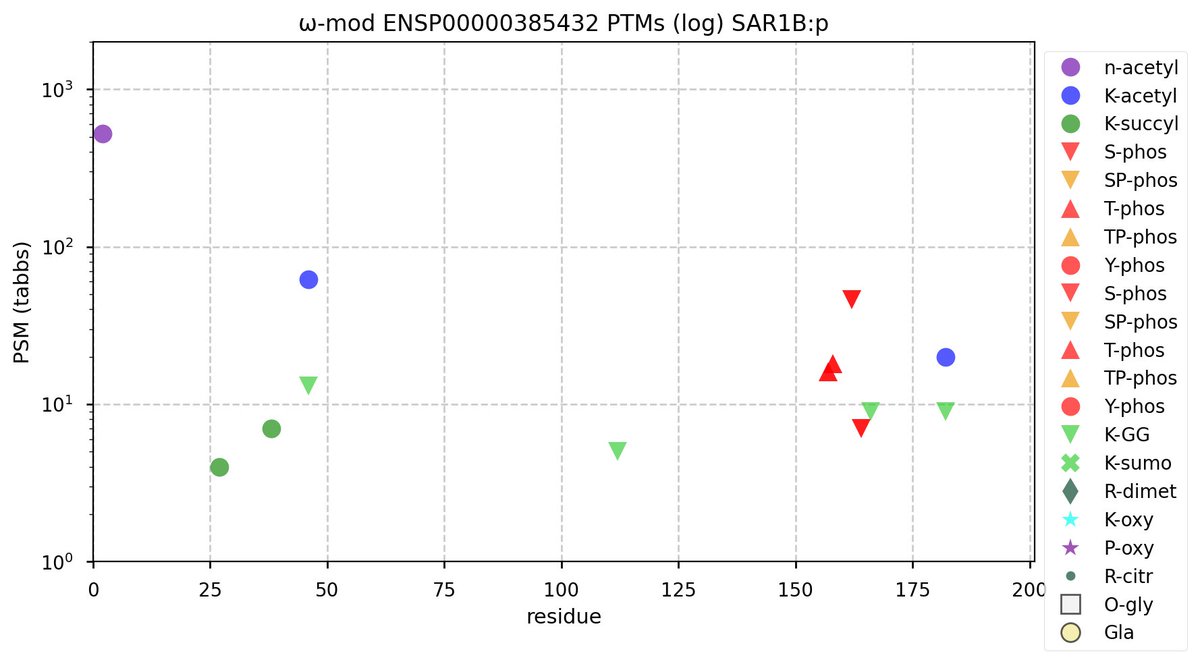
Mon Nov 07 12:50:04 +0000 2022Modification diagram for human SAR1A & B, paralog small GTP binding subunits in the Coat Protein Complex II (COP II). The 2 subunits have about 20% sequence identity, but very different PTM patterns. 🔗
Mon Nov 07 03:18:32 +0000 2022Not a big fan of daylight savings time, especially today. Time zones are a squirrelly kludge, particularly the non-integral ones left over from the British Empire.
Sun Nov 06 15:23:06 +0000 2022HUPO should provide some web design/technology workshops specifically designed for supervisors (no students allowed).
Sun Nov 06 15:20:09 +0000 2022Looking at another new proteomics data/information web site where clearly the students learned a lot, but the supervisor has a lot to learn.
Sun Nov 06 14:29:48 +0000 2022A very popular narrative structure for modern scientific papers is the Hero's Journey (monomyth):
🔗
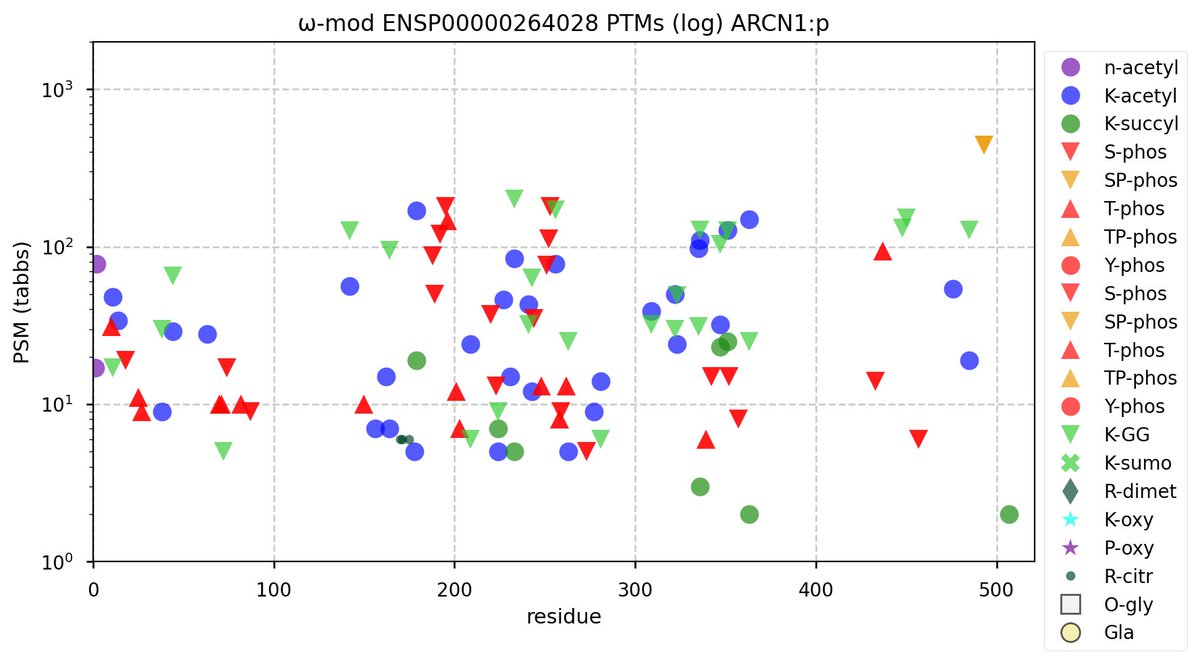
Sun Nov 06 12:41:29 +0000 2022Modification diagrams for human ARCN1:p (aka COPD, delta-COP), a traditionally-named subunit of the COP I complex. It has the K+acetyl/GGyl pattern common in the complex, with 34 of 38 lysines acting as acceptors. It also has multiple discrete, low occupancy phosphodomains. 🔗
Sat Nov 05 15:14:56 +0000 2022@ProteomicsNews @UCDProteomics @pwilmarth Everything I've ever written. I've never used Proteome Discoverer, but I doubt that it has anything to teach me about screwing things up.
Sat Nov 05 13:49:43 +0000 2022Listening to an "expert" talking on the radio (CBC) about the effects of tomorrow's time change on people: the pseudoscience came out quick & flowed deep.
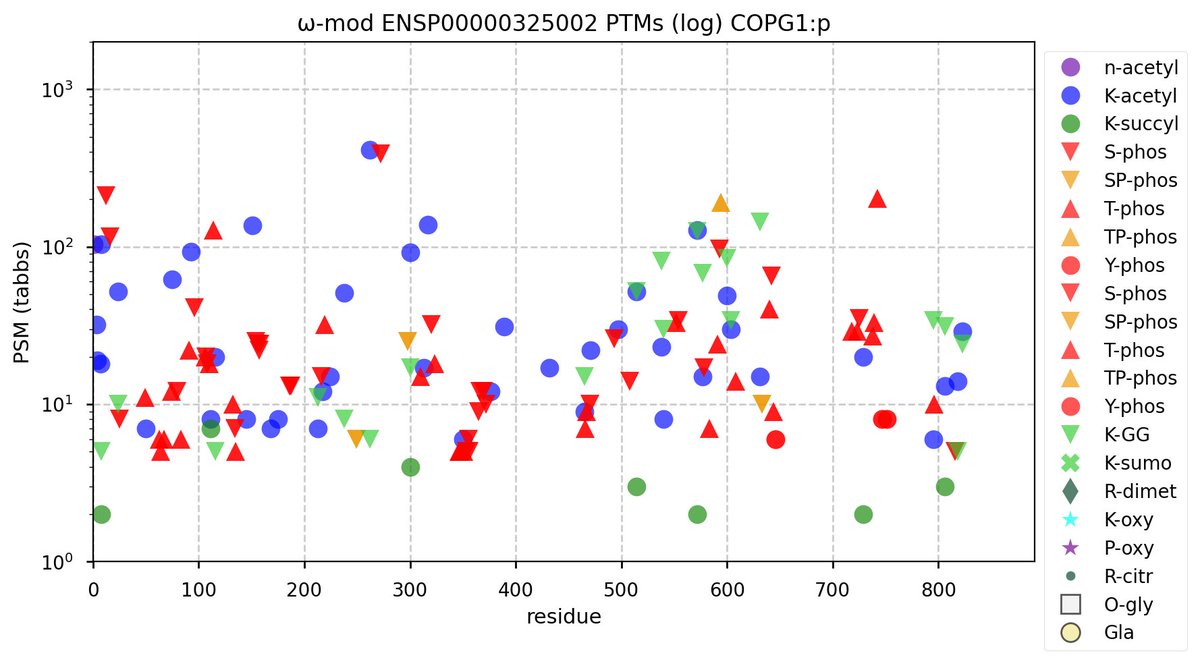
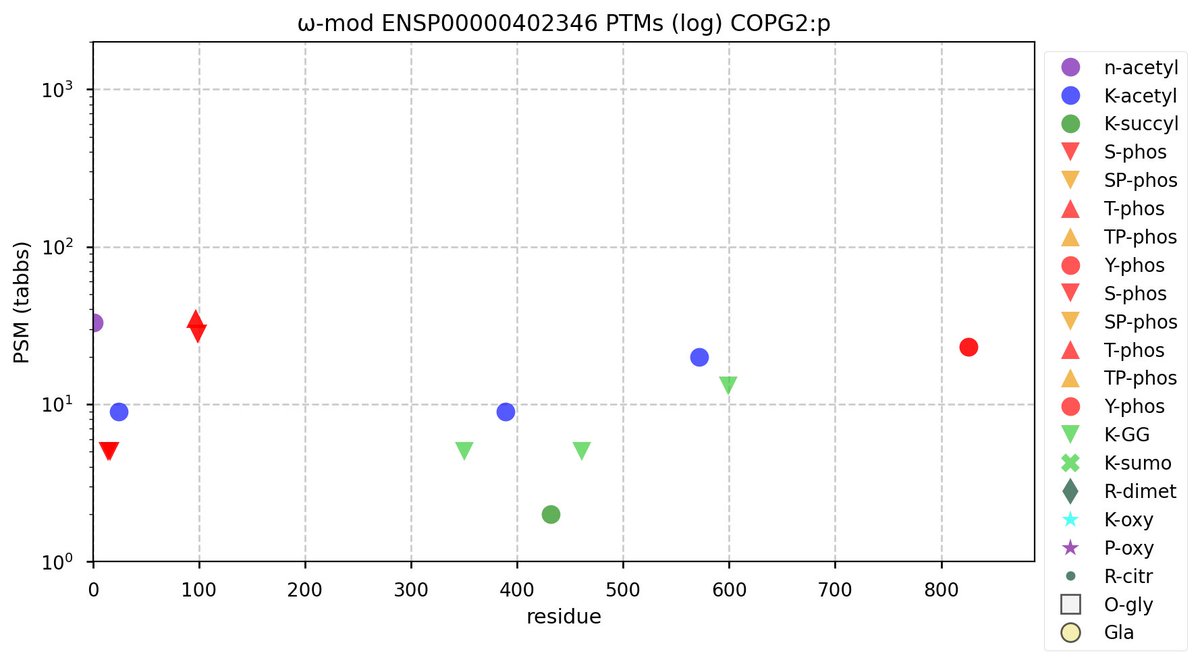
Sat Nov 05 13:28:57 +0000 2022For anyone who is interested, these diagrams are based on 71 k observations (853 kTa) of COPG1:p and 44 k observations (290 kTa) of COPG2:p.
Sat Nov 05 13:19:14 +0000 2022The local forecast for today's arrival & departure of an "Alberta Clipper", a common type of fast moving low pressure system that form in northern Alberta. 🔗
Sat Nov 05 13:04:38 +0000 2022@MagnusPalmblad @wfondrie I hesitate to move to Mastodon, because the fact that I could set up my own server pretty much guarantees the eventuality that I would: a classic battle between my DIY impulses & my indolence.
Sat Nov 05 11:52:55 +0000 2022The patterns are sufficiently different it might be worth while to determine if COPG2:p may have some additional activity/localization rather than relying on its relationship with its paralog.
Sat Nov 05 11:48:36 +0000 2022Even though these 2 sequences are homologous, they only share 5 observable tryptic peptides.
Sat Nov 05 11:36:26 +0000 2022Modification diagrams for human COPG1:p & COPG2:p, paralogous subunits of the COP I complex. The 2 sequences have gone very different ways wrt their PTM decoration, with COPG1:p being in line with the other COP I subunits, while COPG2:p takes a minimalist approach. 🔗
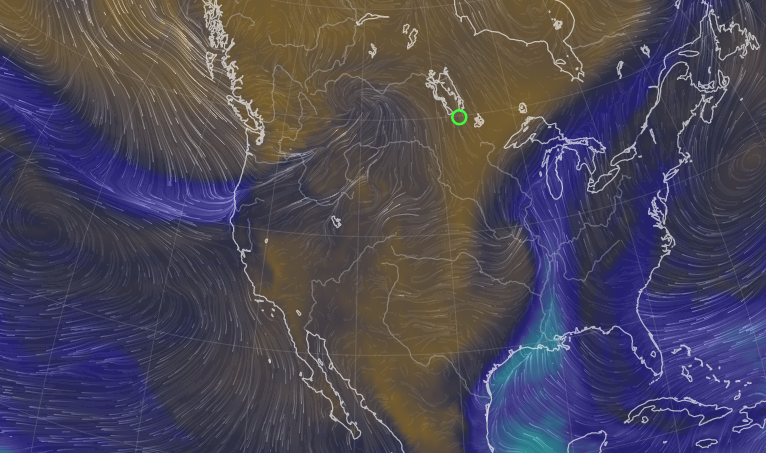
Sat Nov 05 11:05:20 +0000 2022The "atmospheric river" has dropped down to the Oregon-California border, while another one has popped up running wet air from the Gulf of Mexico to the Great Lakes. 🔗
Fri Nov 04 15:42:18 +0000 2022@mjmaccoss @MattWFoster @neely615 @LaurelOld I agree. Just to keep perspective, 0.5-1% of PSMs can be easily assigned to SAVs associated with SNVs with population frequencies > 1%. So, if you have assigned 100,000 PSMs, you are going to have about 500-1,000 that identify an SAV.
Fri Nov 04 14:10:09 +0000 2022@neely615 @mjmaccoss @MattWFoster @LaurelOld There is no question that it is possible to find a sufficiently large set of SAVs in any comprehensive proteomics data set to theoretically identify an individual. The field has simply decided that it isn't an issue, because there isn't any practical way to go about doing so.
Fri Nov 04 13:59:39 +0000 2022@mjmaccoss @neely615 @MattWFoster That is certainly my understanding of the current idea regulating the distribution of human raw proteomics data. The CPTAC group have advocated that position & made their proteomics data files available on that basis.
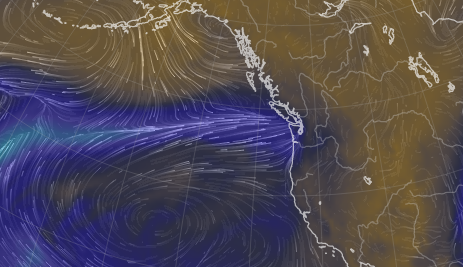
Fri Nov 04 12:24:57 +0000 2022Looks like there will be a few wet days in the Vancouver/Seattle area, with an "atmospheric river" pointed right at them (surface winds + total precipitable water) 🔗
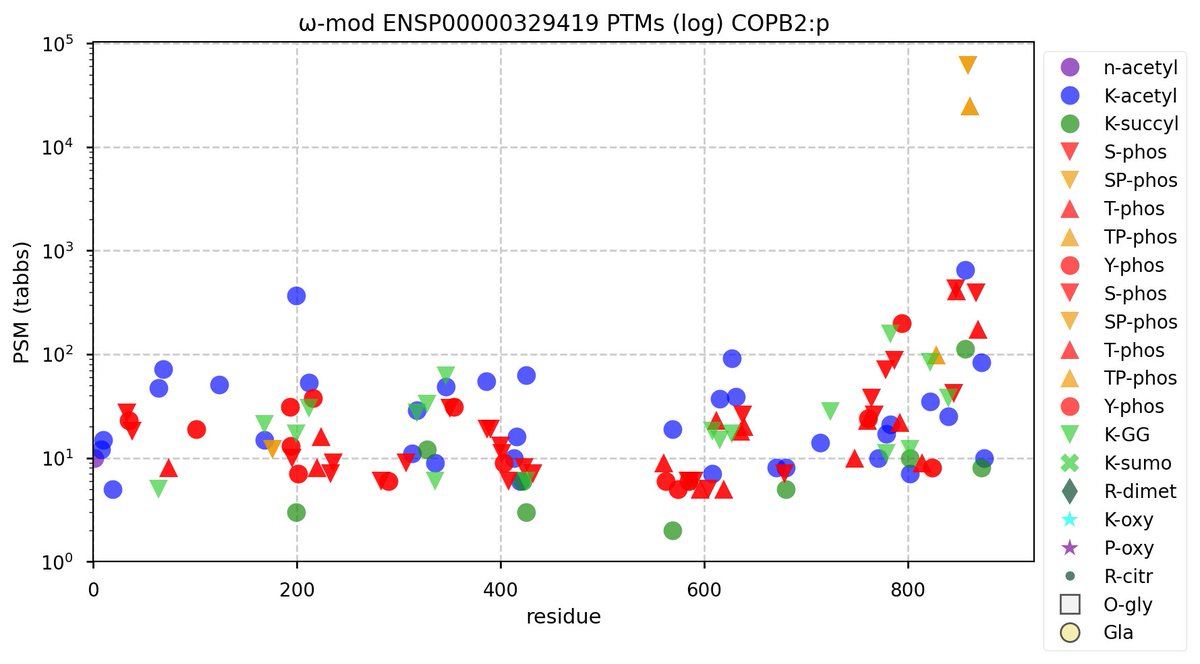
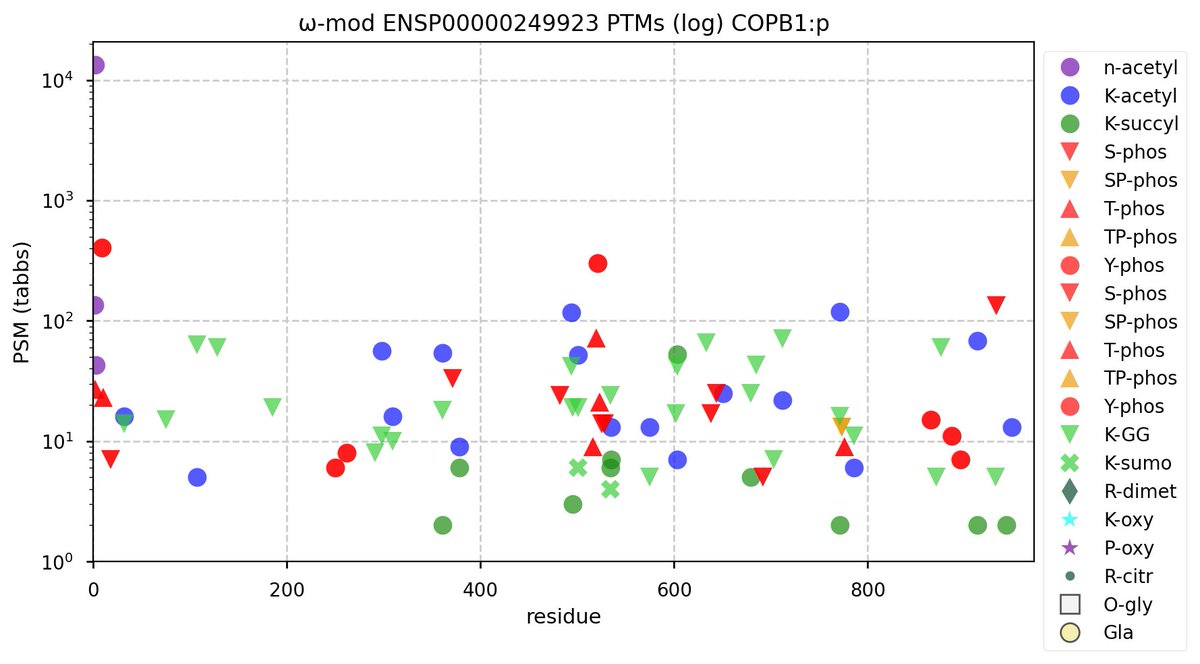
Fri Nov 04 12:06:56 +0000 2022COPB1 & COPB2 have been observed with similar frequency in publicly available data (73,118 & 75,873 LC/MS/MS runs respectively), but they are not homologous and do not share any tryptic peptides.
Fri Nov 04 11:54:41 +0000 2022The modification gap (433-568) is not caused by an absence of observable tryptic peptides.
Fri Nov 04 11:36:44 +0000 2022It also has a smattering of low occupancy phosphoryl acceptors, with 98% of phosphorylated PSMs assigned to 2 adjacent ΦP sites near the C-terminus.
Fri Nov 04 11:31:51 +0000 2022Modification diagram for human COPB2:p, a subunit of the COP I complex. The K modification pattern is similar to COPA & B1:
2× K+GGyl
16× K+acetyl/GGyl
19× K+acetyl
with 36 observed K+acetyl acceptors, out of 55 K residues. 🔗
Thu Nov 03 14:40:23 +0000 2022If anyone knows of a more recent review or any study that addresses the functional role of K+GGyl/acetyl alternating modifications in COP I subunits, please feel free to add it to this thread.
Thu Nov 03 11:43:33 +0000 2022The COP I complex has been studied in considerable depth, as it is part of the easily told story of Golgi retrograde transport. For anyone interested in the subject, Luo & Boyce (2019) provides a good overview
🔗
Thu Nov 03 11:38:23 +0000 2022Modification diagram for human COPB1:p, a subunit of the COP I complex. Like COPA:p, this sequence has a busy K modification acceptor pattern:
1× K+acetyl/GGyl/SUMOyl,
1× K+GGyl/SUMOyl,
5× K+acetyl,
11× K+acetyl/GGyl &
13× K+GGyl 🔗
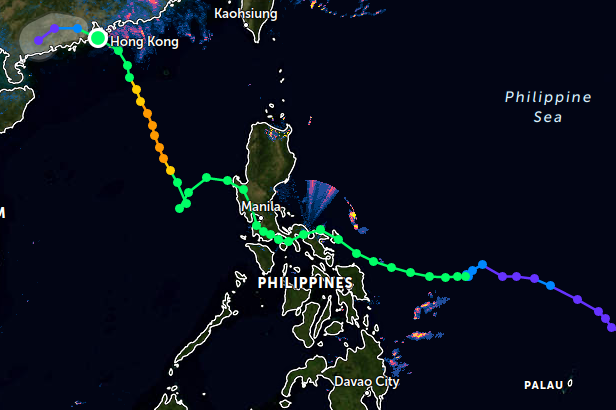
Thu Nov 03 11:27:40 +0000 2022Tropical Storm Nalgae shifted its course to the north & is making landfall at Hong Kong today. 🔗
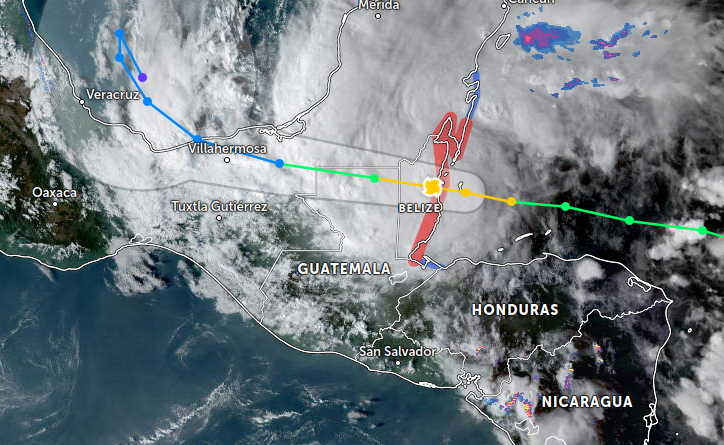
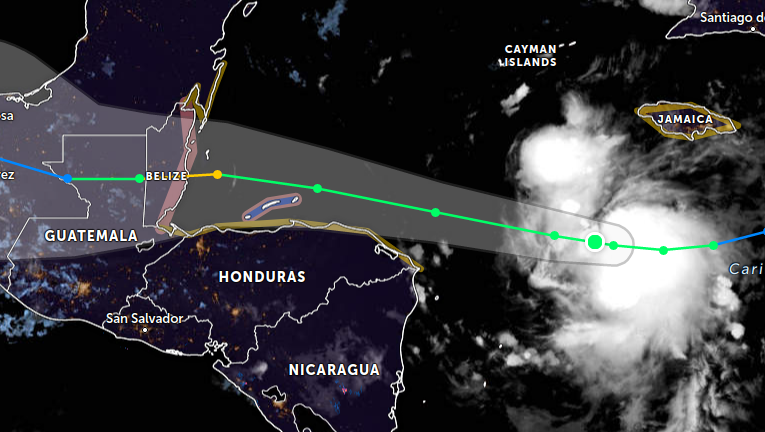
Wed Nov 02 22:47:58 +0000 2022Hurricane Lisa has come ashore in Belize, right where & when it was predicted to arrive. 🔗
Wed Nov 02 20:48:37 +0000 2022@byu_sam We are further north, but you are a kilometer higher. You win the snow lottery this year!
Wed Nov 02 20:42:45 +0000 2022My various flavours of Ubuntu seem to have all patched the OpenSSH issues.
Wed Nov 02 19:26:59 +0000 2022Very much in the "learn from learners not from teachers" tradition I have always found helpful.
Wed Nov 02 19:05:36 +0000 2022Machine learning should be taught by machines.
Wed Nov 02 15:41:51 +0000 2022If nothing else, you have to admire their persistence. I'm pretty sure that the idea of “A Function for Each Protein” doesn't make any sense, but neither did "Fortune Favors the Bold" ...
🔗
Wed Nov 02 15:35:22 +0000 2022The COP I complex coats the surface of vesicles which move proteins that should be retained by the ER back from the Golgi. Coating a vesicle with COP I initiates this reverse transport, but the coat is removed prior to their re-integration with the ER.
Wed Nov 02 15:13:07 +0000 2022We are so desperate for attention some times that we are willing take credit for the things that we really should keep quiet about. Oh Canada!🇨🇦
🔗
Wed Nov 02 14:34:07 +0000 2022Of the 4 mechanisms, alternative translation is probably the one that make biomedical types the most nervous when it it brought up. "Are you sure that is the only splice variant involved?" is a question that is never welcome in a review or following a talk.
Wed Nov 02 14:28:55 +0000 2022Non-tryptic cleavage (other than experimental artifacts) is involved in the co- & post-translational maturation of most proteins. It is particularly prominent in extracellular proteins. Necessary for both normal cell function & organism wide processes.
Wed Nov 02 14:25:38 +0000 2022Amino acid variants are caused by the normal variability of alleles in a population. Not necessary for normal cell function, but affects overall fitness of a population.
Wed Nov 02 14:23:47 +0000 2022Alternative splicing is when more than one combination of exons from a gene result in a viable protein. Necessary for normal cell function.
Wed Nov 02 14:22:13 +0000 2022For those who would like to know:
Alternative initiation is when the annotated start of translation (an M) is passed over by a ribosome & the 2nd (or rarely the 3rd) M in line is where the translation starts. Necessary for normal cell function.
Wed Nov 02 14:15:42 +0000 2022Thanks to everyone who shared an opinion. Alternative splicing won out in a squeaker, although given the nature of the question, the loser (alternative initiation) should probably be declared the winner.
Wed Nov 02 13:26:24 +0000 2022@VATVSLPR Bypassing the ballast is easy when the fixture is sitting on a table in front of you: it is trickier when you are on top of a ladder reaching around a corner without a clear view of what you are doing.
Wed Nov 02 13:20:29 +0000 2022@VATVSLPR I agree, but I pretty much have to take a wall apart to get at the fixture. I have done bypasses on the other fluorescent fixtures in the place.
Wed Nov 02 13:07:16 +0000 2022For any one interested, "GE Universal Cool White T8 32W and T12 40W Replacement LED G13 Base 48-in T8 Linear Tube" (# 93128862) worked properly with the magnetic ballast in my fixtures.

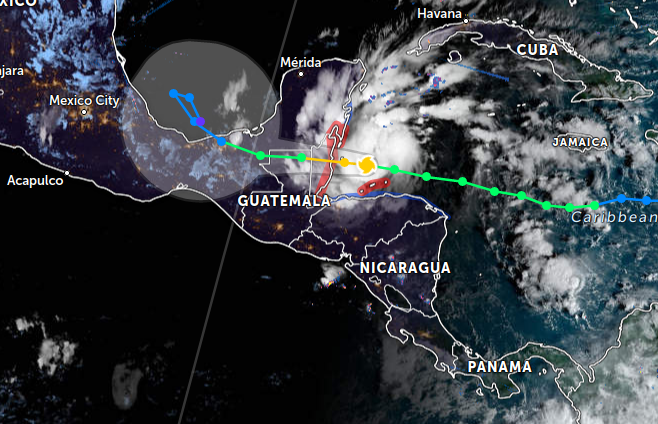
Wed Nov 02 12:42:55 +0000 2022Hurricane Lisa is still on track to strike the central Belize coast today, pass thru Belize into northern Guatemala, continuing on into southern Mexico, then heading north into the Gulf of Mexico. 🔗
Wed Nov 02 12:29:07 +0000 2022My experience with start ups is that when they succeed it is usually in spite of their CEO, rather than because of them.
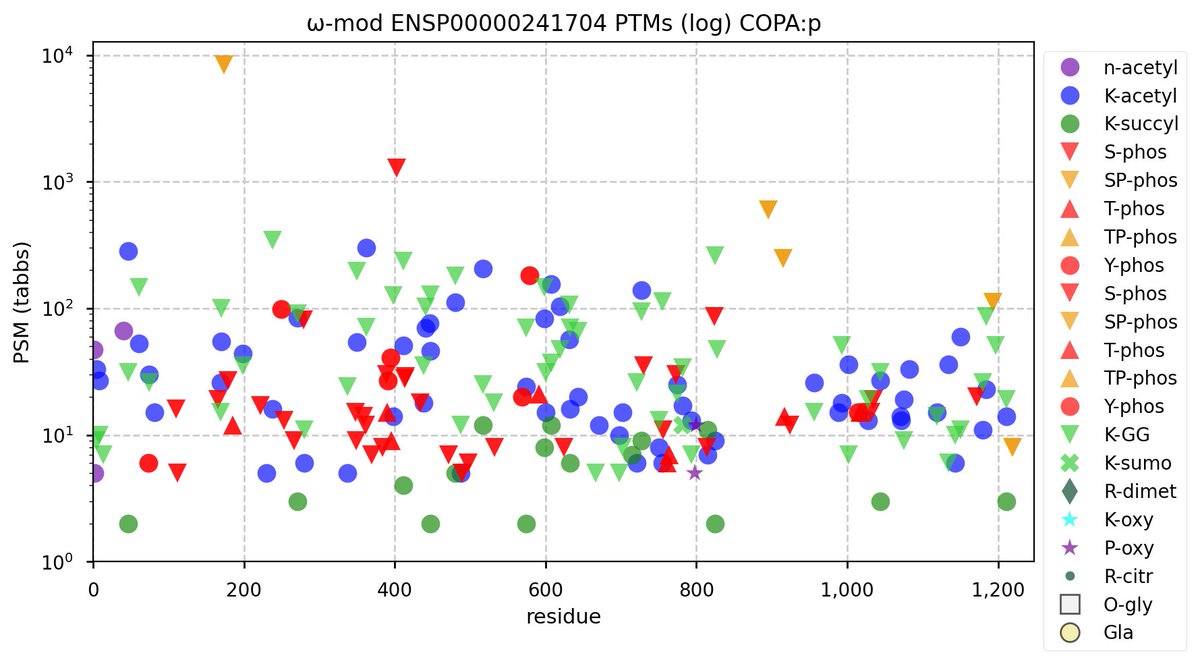
Wed Nov 02 12:19:24 +0000 2022There are 2 commonly observed splice variants associated with COPA:g, CCDS1202 (1,224 aa) & CCDS41424 (1,233 aa).
Wed Nov 02 12:11:33 +0000 2022Modification diagram for human COPA:p, a subunit of the coatomer protein complex I (COPI). Unlike COG subunits that have no use for K+acetyl, ~75% of the lysines in COPA:p may serve as acetyl acceptors. Of those 62 lysines, 51 have been shown to also serve as GGyl acceptors. 🔗
Tue Nov 01 17:42:36 +0000 2022@sciencehacker FacebookProt.
Tue Nov 01 17:00:52 +0000 2022@UCDProteomics Hallelujah, brother, hallelujah! Say it again!
Tue Nov 01 15:16:21 +0000 2022It must be Nov. 1: yet another pointless set of time zone re-definitions have arrived … reboot now!
Tue Nov 01 14:10:29 +0000 2022@Smith_Chem_Wisc Our good friends in genetics have constructed what is a much better funded & higher profile field than proteomics is currently doing nothing but looking for variants.
Tue Nov 01 13:37:45 +0000 2022Which of these would you nominate to be the Rodney Dangerfield of proteomics?
Tue Nov 01 12:15:50 +0000 2022The lack of an n-dash (‒), an m-dash (―) or an ellipsis (…) on a standard keyboard is annoying.
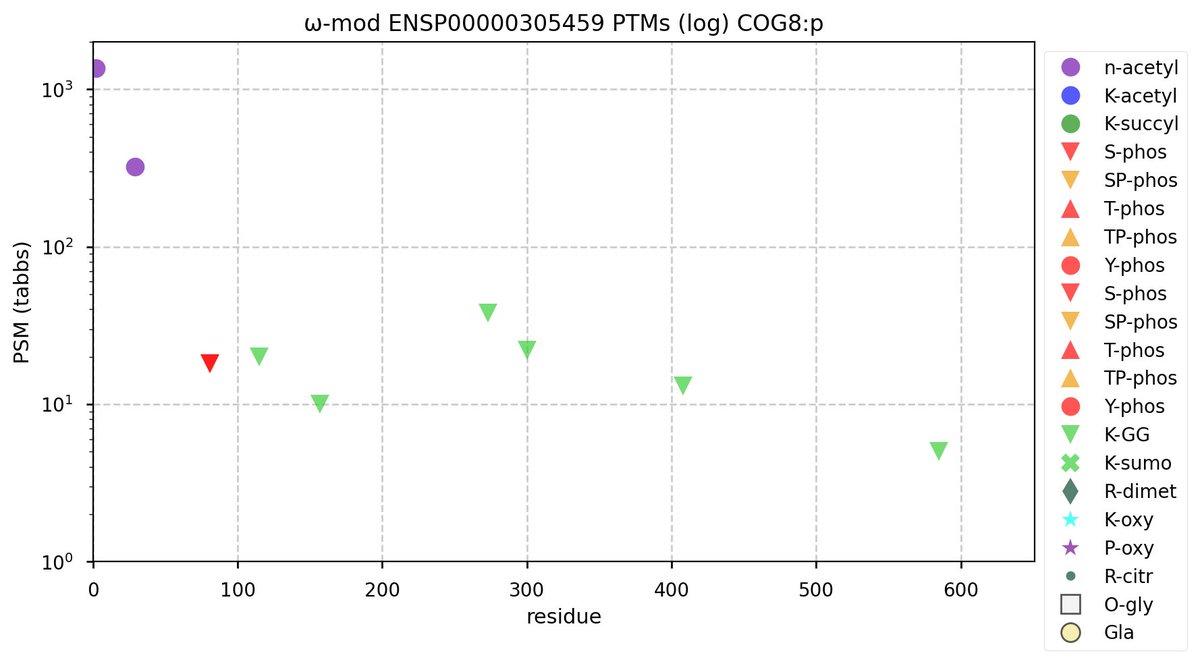
Tue Nov 01 12:00:24 +0000 2022Modification diagram for human COG8:p, 8/8 subunit of the oligomeric golgi complex. About 20% of observed sequence starts at A29+acetyl, because of alternate initiation at M28. Otherwise, it is a pretty typical COG subunit: a scattering of K+GGyl acceptors across the sequence. 🔗
Tue Nov 01 02:24:03 +0000 2022Tropical Storm Lisa is on track to make landfall in Belize as a Category 1 hurricane on Wednesday afternoon. 🔗
Mon Oct 31 18:03:15 +0000 2022Sounds like the guys at Tesla & SpaceX will finally be able to get some stuff done.
Mon Oct 31 17:58:36 +0000 2022TPR (Translocated Promoter Region) protein? Someone on an HGNC panel really phoned that one in.
Mon Oct 31 17:45:03 +0000 2022@UCDProteomics @pwilmarth My hypothesis is that it is somewhere in the Metaverse.
Mon Oct 31 16:16:46 +0000 2022PXD033168: there are the many that talk about clinical applications & then there are the few actively pursuing them.
Mon Oct 31 15:30:26 +0000 2022It can be tricky to use protein co-ordinates properly in a paper. Just checking GoogleProt often doesn't work out as well as you might hoped. A canonical sequence is not necessarily the most abundant one: it doesn't have to exist at all!
Mon Oct 31 14:50:44 +0000 2022PXD032110: most of the samples have good signals from pretty much all of the proteins characteristic of azurophilic granules, like BPI, ELANE & MPO.
Mon Oct 31 14:32:11 +0000 2022@pwilmarth @astacus @J_my_sci Pretty sure most 'omics is done on the mode, rather than the mean, so "proteomics à la mode" would probably be a more accurate formulation. 🇫🇷
Mon Oct 31 11:51:50 +0000 2022COGs tend to be pretty stingy wrt PTMs: it is their interaction partners, particularly golgins, that really know how to put on a show. 🔗
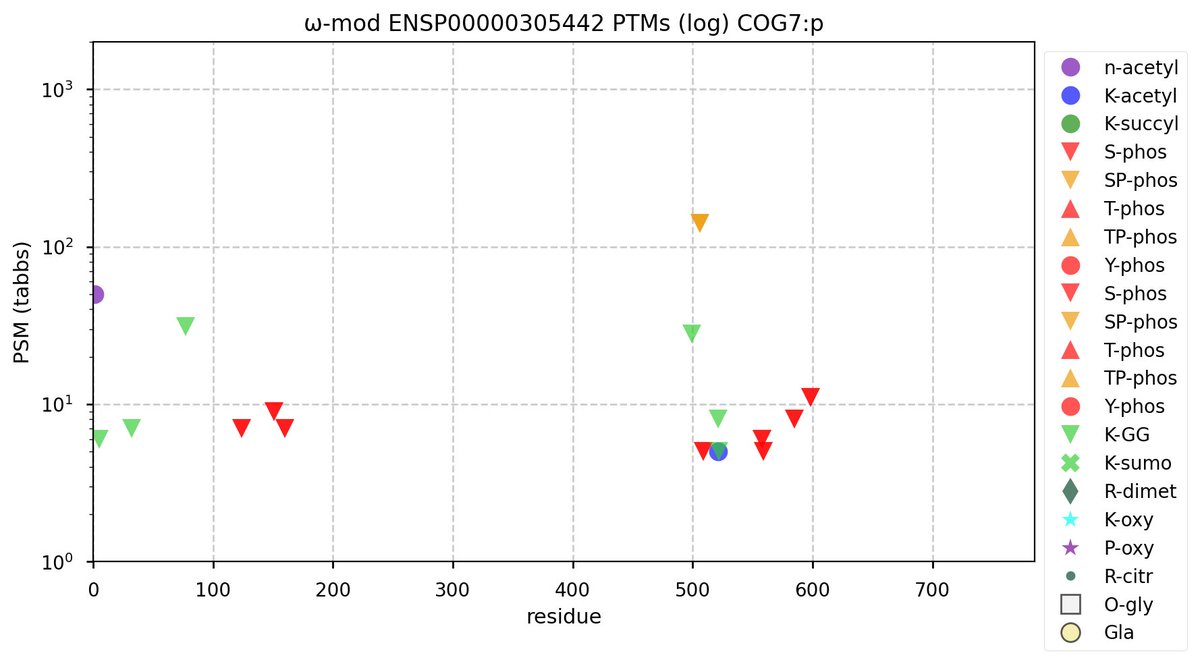
Mon Oct 31 11:46:46 +0000 2022Modification diagram for human COG7:p, 7/8 subunit of the oligomeric golgi complex. 6× K+GGyl acceptors in 2 domains & one high occupancy, ΦP-type S+phosphoryl acceptor on an IDR connecting helical domains. 🔗
Sun Oct 30 19:11:40 +0000 2022@AlexUsherHESA Sound like a job for a Royal Commission!
Sun Oct 30 17:43:12 +0000 2022@J_my_sci Yes. It is an uncontrolled variable (or more properly a big bag of variables).
Sun Oct 30 17:19:30 +0000 2022@theoneamit The technical capability is there, but the theoretical understanding is probably a generation away.
Sun Oct 30 15:59:28 +0000 2022A significant problem with many proteomics experiments is they can't be used to look for cell-cycle-associated changes in proteome composition & modification.
Sun Oct 30 13:43:11 +0000 2022For me, the worst parts of scientific conferences were always the plenary lectures. Sometime entertaining, but usually without much content. And they got much worse once TED talks came in to vogue.
Sun Oct 30 12:29:22 +0000 2022Tropical storm Nalgae (Paeng) has moved into the South China Sea, about to turn north. It is predicted to increase intensity to typhoon strength and then turn towards Hainan. 🔗
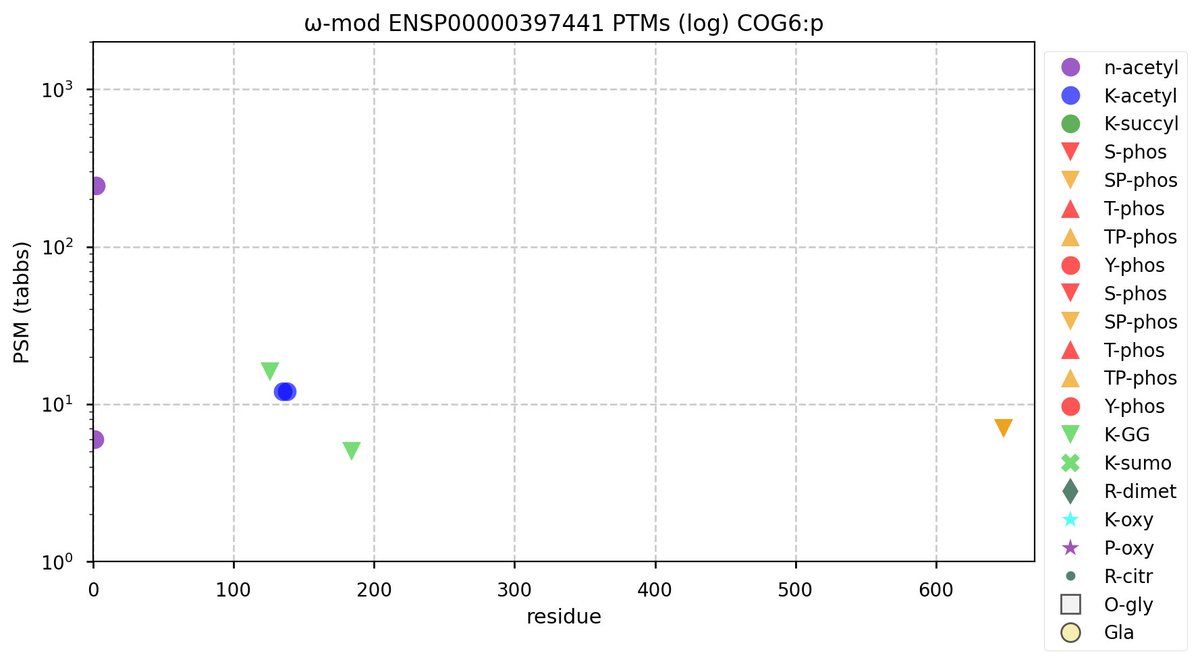
Sun Oct 30 11:23:13 +0000 2022Modification diagram for human COG6:p, 6/8 subunit of the oligomeric golgi complex. Nothing much going on: only a few low occupancy acceptors. 🔗
Sat Oct 29 17:29:41 +0000 2022Academic tenure is a rather silly thing that simply shouldn't exist.
Sat Oct 29 16:18:13 +0000 2022Are PTM pattern changes drivers or consequences of speciation?
Sat Oct 29 15:39:48 +0000 2022@BenCollinsLab The "green" (hard to see) tryptic peptides are assigned by 4 simple rules:
1. peptide length <6;
2. peptide length >35;
3. >2/3 of residues are [LIVMFWY]; or
4. has >6 residues in a row that are [LIVMFWY].
Sat Oct 29 14:31:22 +0000 2022Listening to someone on the radio lamenting that Canada is losing its global influence, even though I am unaware of any point in the last 60 years when Canada had any global influence.
Sat Oct 29 14:08:42 +0000 2022But maybe it is just my eternal optimism that leads me to be "surprised".
Sat Oct 29 14:07:53 +0000 2022It is surprising how often I look at a new published project's web site & conclude the supervisor shouldn't be doing this sort of work. The students have done as good a job as they could, but the boss clearly doesn't know what they are doing.
Sat Oct 29 14:03:26 +0000 2022@Oblivious_ @Smith_Chem_Wisc GoogleProt
Sat Oct 29 13:31:50 +0000 2022My secret recipe for Dawn Powerwash refill fluid:
50 ml isopropanol
15 ml Dawn Ultra
2 ml essential oil (eucalyptis/rosemary/lemon)
make up to 750 ml with water & stir.
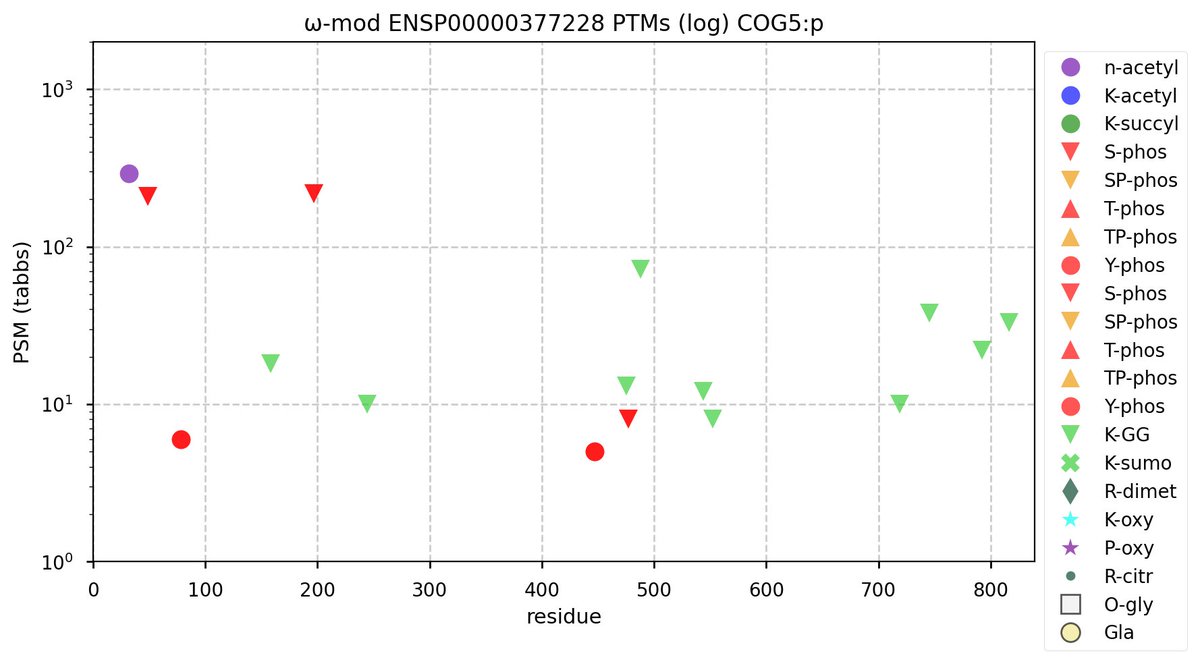
Sat Oct 29 11:54:23 +0000 2022Modification diagram for human COG5:p, 5/8 subunit of the oligomeric golgi complex. A simple pattern: 2 high occupancy, GΦ motif S+phosphoryl acceptors & 10× K+GGyl acceptors scattered across its many helical domains. 🔗
Fri Oct 28 22:24:44 +0000 2022@cajun_science In ENSEMBL, it is exon 19 (ENSMUSE00001340298), chr 9:49,418,953-49,418,153.
🔗
Fri Oct 28 22:22:44 +0000 2022@cajun_science It is the rather long one, second from the end (show in blue). It is where the additional S/T+phosphoryl acceptors are in both the mouse & rat proteome. 🔗

Fri Oct 28 16:25:20 +0000 2022There seems to be a big difference in S/T+phosphoryl acceptor pattern between the two that is located on the "missing" exon. 🔗
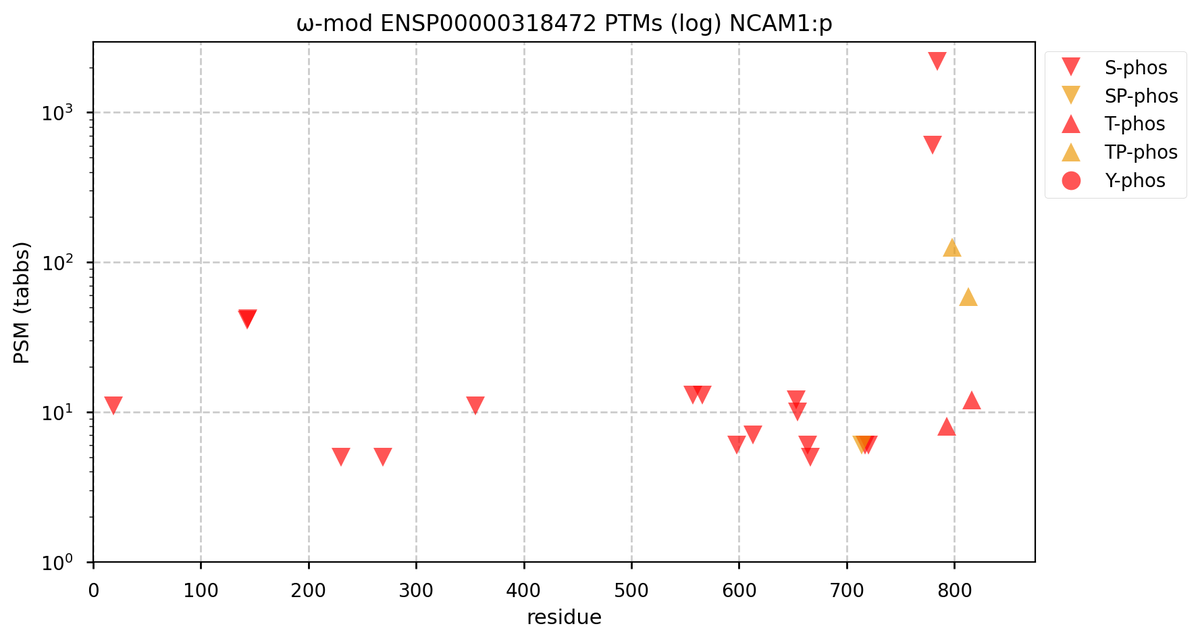
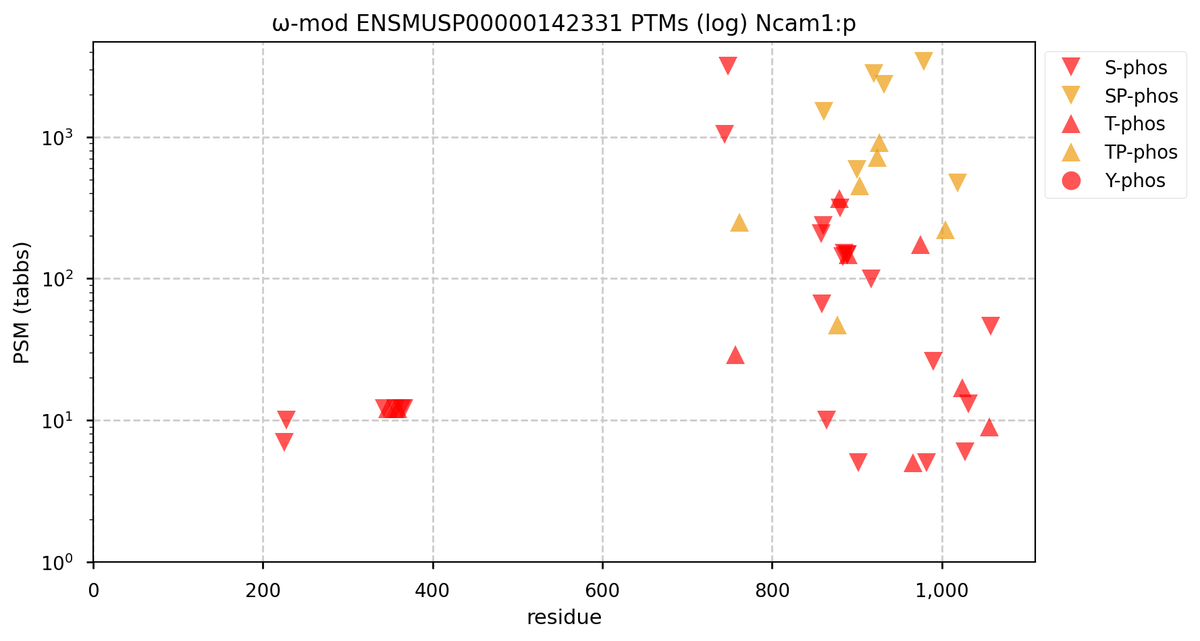
Fri Oct 28 16:15:50 +0000 2022Does anybody know why human NCAM1:p seems to be missing an exon compared to mouse or rat?
Fri Oct 28 12:12:44 +0000 2022@VassB "comfortable competition" is a great description of how Canada works.
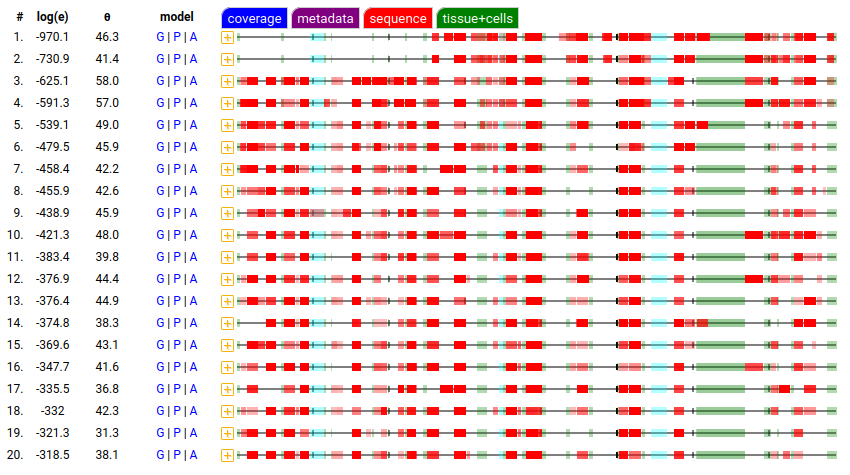
Fri Oct 28 11:55:11 +0000 2022Two of these things are not like the others, but they are all from the same dataset. 🔗
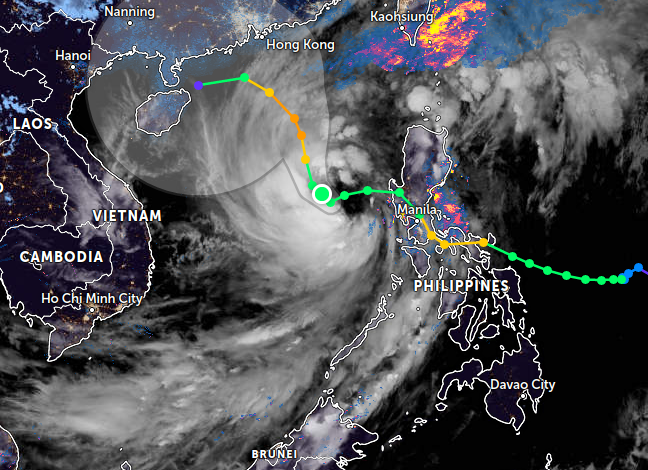
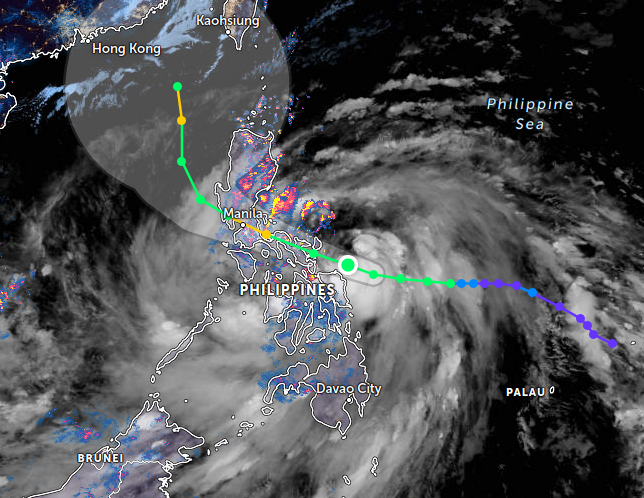
Fri Oct 28 11:43:35 +0000 2022Tropical storm Nalgae (Paeng) is going to spend the next few days traversing the north central Philippines, including Manila. 🔗
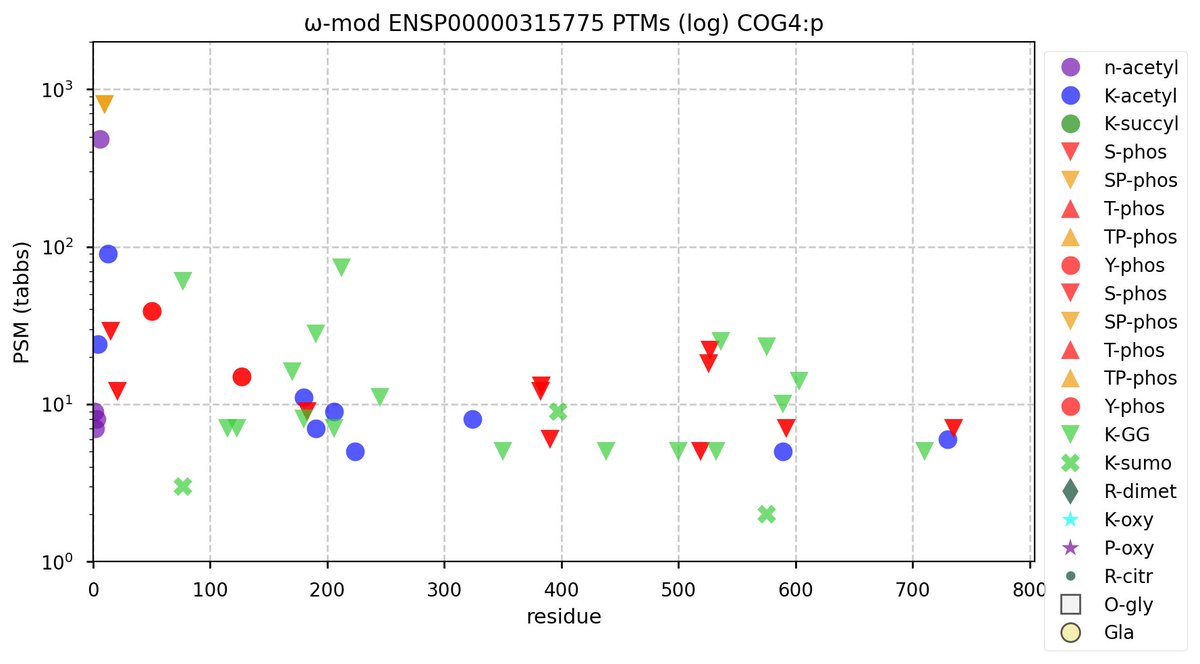
Fri Oct 28 11:33:08 +0000 2022Modification diagram for human COG4:p, the 4/8 subunit of the oligomeric golgi complex. Unlike COG3:p, it has plenty of K-mods:
1× K+SUMOyl (9 Ta, 1.8%)
2× K+GGyl/SUMOyl (88 Ta, 17.8%)
4× K+acetyl/GGyl (85 Ta, 17.2%)
5× K+acetyl (133 Ta, 27.0%)
12× K+GGyl (178 Ta, 36.1%) 🔗
Thu Oct 27 19:18:46 +0000 2022@scalzi Believe it or not, Haligonian is the correct term.
Thu Oct 27 16:30:30 +0000 2022I realize this is the best you can do, but IMHO this particular vision of GOLGA3 doesn't look quite as "optimized" as I would like it.
🔗 🔗

Thu Oct 27 15:17:21 +0000 2022Word to the wise: if it takes 12 slides to explain how to use a feature in your user interface, no one (other than the developer) is ever going to use it.
Thu Oct 27 14:43:15 +0000 2022Regardless of how you fund universities to do research, they are going to produce, within error, the same result. Expecting anything else requires more optimism than I can muster.
🔗
Thu Oct 27 14:30:57 +0000 2022Is there a good term for describing the extent of tryptic peptide identity between 2 proteins? For example, BACE1:p & BACE2:p are considered to be very homologous, but they do not share any observable tryptic peptides.
Thu Oct 27 14:15:19 +0000 2022@tombrodbeck That is why run-offs are used in many jurisdictions. In a case like this, with only 37% of eligible voters participating and a squeaker of a result, the winner is stuck in a lame duck position until the next election.
Thu Oct 27 13:51:36 +0000 2022If the PTM patterns are any guide (& I don't know that they really are) then COG3:p has the main control switches for the complex & it is long lived compared to the other COG complex component subunits.
Thu Oct 27 13:47:19 +0000 2022For example, if you want to know what is in your "Tide Pods", simply search for "tide pods safety data sheet"
🔗
and click on one of the PDFs that pop up, like
🔗
Thu Oct 27 13:11:12 +0000 2022If you want to know the composition of most commercial cleaners, the magic words are
"SAFETY DATA SHEET"
Thu Oct 27 12:48:38 +0000 2022@tombrodbeck That is true, but no one was given a mandate to execute a collection of policies. They were all rejected.
Thu Oct 27 12:43:37 +0000 2022@tombrodbeck But we have ended up with a mayor whose candidacy was rejected by more than 70% of the voters.
Thu Oct 27 12:09:40 +0000 2022@slavov_n Many people are willing to accept the proposition that if they aren't burning the stuff personally, they have done all they can and "the problem" now belongs to somebody else.
Thu Oct 27 11:59:52 +0000 2022COG3:p is also the only member of the complex with multiple isolated high occupancy S/T phosphoryl acceptor domains with several main motifs:
1× SxxΦL (1226 Ta, 17.3%)
2× ΦP (1171 Ta, 16.5%)
3× RxxΦ (1600 Ta, 22.5%)
8× ΦL/I/V (716 Ta, 10.1%)
Thu Oct 27 11:48:09 +0000 2022Modification diagram for human COG3:p, 3/8 subunit of the Conserved Oligomeric Golgi complex. It is the only COG subunit that has no evidence of K+GGyl, although it does generate MHC class I peptides.
🔗
Wed Oct 26 20:34:18 +0000 2022Golgin B1 is just weird.
Wed Oct 26 16:27:04 +0000 2022@Smith_Chem_Wisc It depends on how old it is. The statute of limitations period for citations seems to be 3-5 years now, depending on the sub-field.
Wed Oct 26 14:48:57 +0000 2022@scalzi 🔗
Wed Oct 26 14:34:35 +0000 2022@TrostLab @uniprot If you look at the source for the new site, displaying information depends on Javascript-based calls from your browser rather than straight-up HTML retreival, with the information routed thru to 🔗 Probably no easy way to figure out where slow downs occur.
Wed Oct 26 14:25:38 +0000 2022@MagnusPalmblad @EvelynRampler @an_chem @DorferV @fwf In Canadian academic language, this is often referred to as "punching above our weight".🧐
Wed Oct 26 14:09:35 +0000 2022@EvelynRampler @MagnusPalmblad @an_chem @DorferV @fwf As a Canadian, I prefer the full figure, showing just where we fit into the discussion.
🔗
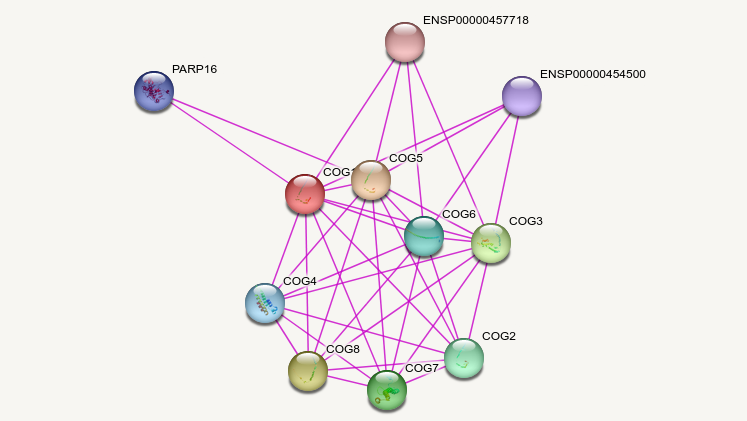
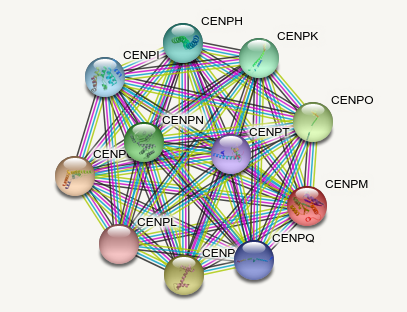
Wed Oct 26 13:30:09 +0000 2022The STRING protein-protein interaction diagram for COG1:p (experimental data only). 🔗
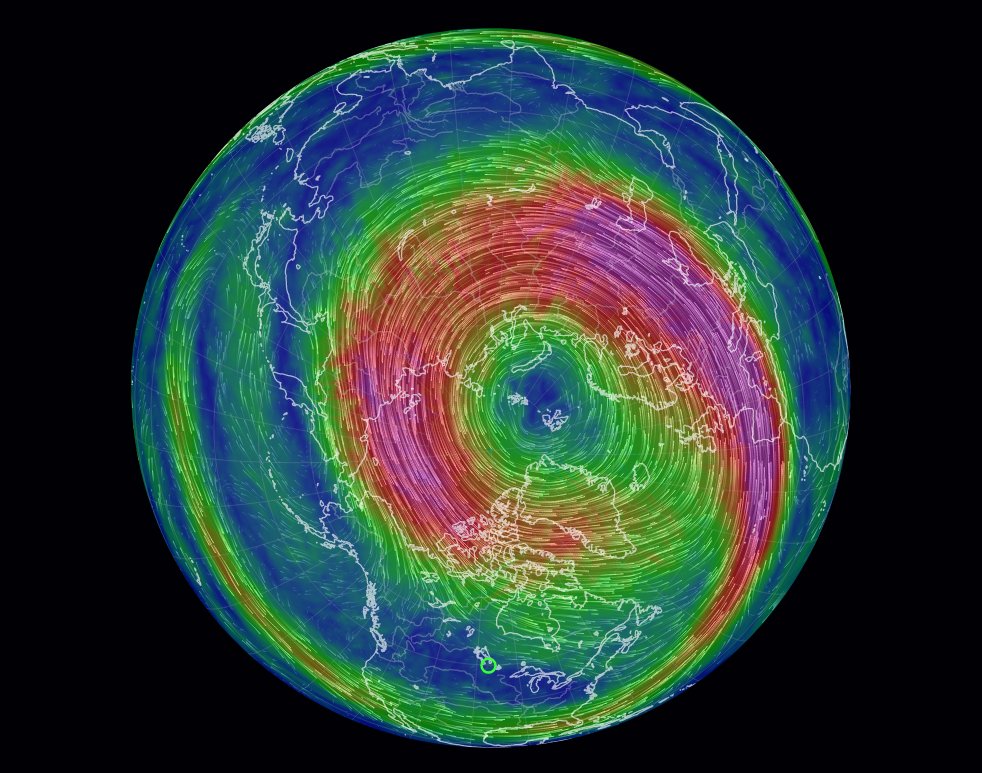
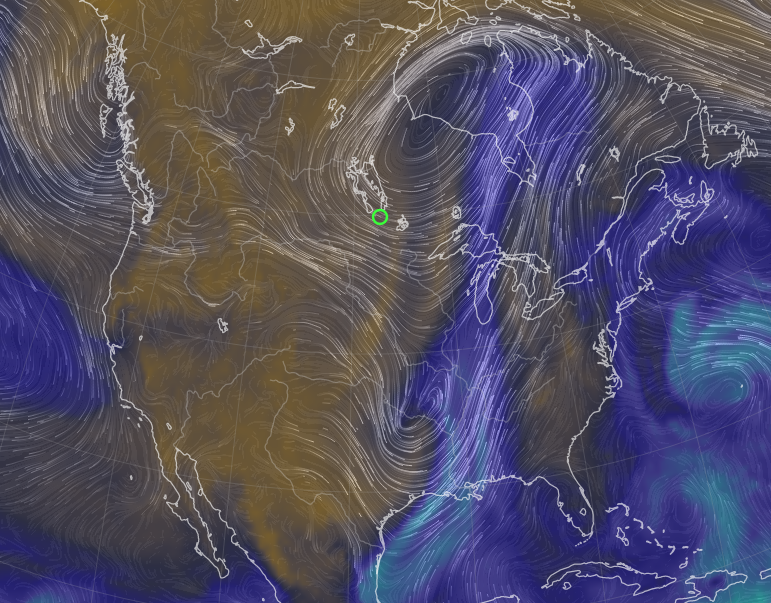
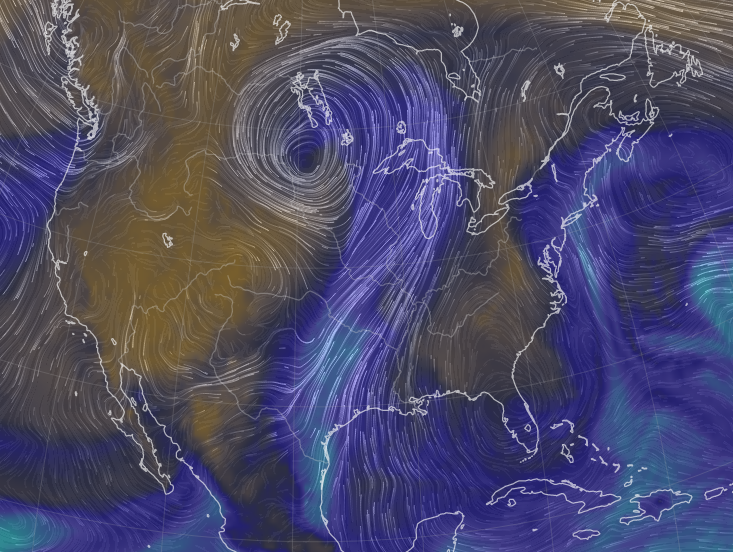
Wed Oct 26 12:28:08 +0000 2022The polar vortex (winds at 10 hPa). 🔗
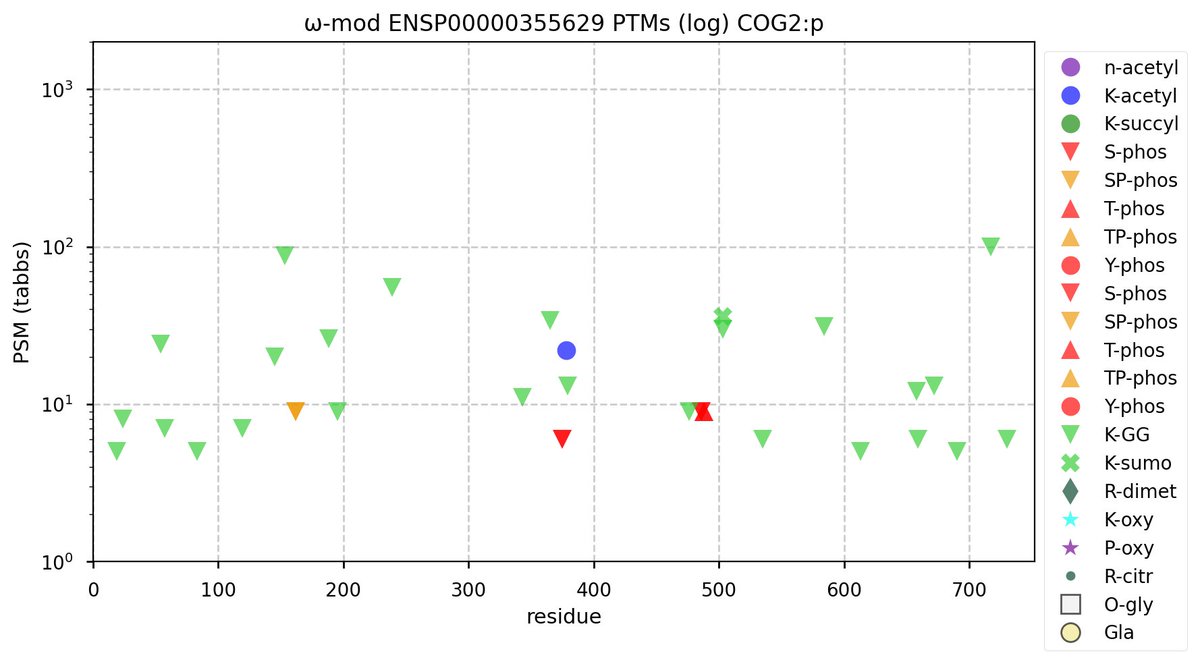
Wed Oct 26 11:55:19 +0000 2022Modification diagram for human COG2:p, 2/8 subunits of the Conserved Oligomeric Golgi (COG) complex. 25 out of 53 K's have been observed to serve as GGyl acceptors but only 1 of those is GGyl/acetyl. 🔗
Tue Oct 25 17:43:04 +0000 2022@tomlau From my own odd point of view, it is any set of things that make it a waste of time to download and process data from a particular lab.
Tue Oct 25 17:34:36 +0000 2022@tomlau Consistently bad sample handling/separations is the usual problem, although there are a few groups that mix up their sample labeling so often that it doesn't seem to be accidental.
Tue Oct 25 15:23:43 +0000 2022Looks like everyone is staying put for another 6 months.
Tue Oct 25 15:09:33 +0000 2022@bioinformagic Ingenuity is much less valued than even the most puerile novelty.
Tue Oct 25 12:56:36 +0000 2022I'm in the middle of reviewing labs on "the black list". Hopefully some will have fixed their issues, but it doesn't happen very often.
Tue Oct 25 12:31:32 +0000 2022The Colorado Low has moved northeast to near the Manitoba-Ontario border on Hudson Bay. The moist Gulf of Mexico air is still being pumped north by the remnants of Hurricane Roslyn that is now in Texas. 🔗
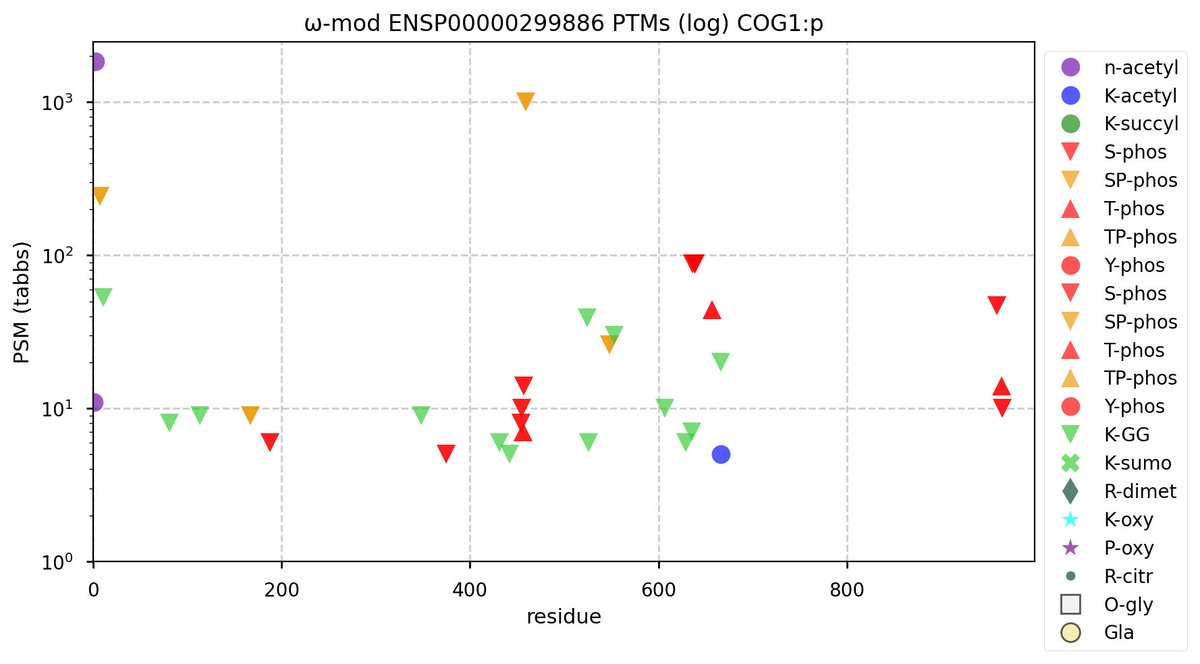
Tue Oct 25 12:07:06 +0000 2022The modification diagram for human COG1:p, 1/8 subunits of the Conserved Oligomeric Golgi (COG) complex. The 3D structure has plenty of gaps where the S/T+phosphoryl acceptors (red) perform their mysteries. The K+GGyl acceptors are scattered through the helices. 🔗
Mon Oct 24 19:13:30 +0000 2022It is much easier to build things than to keep them running.
Mon Oct 24 18:01:47 +0000 2022PXD029424: another good example of how analyzing gel bands is still a viable sub-genre in the field.
Mon Oct 24 14:21:21 +0000 2022@stateofthecity Are you sure it isn't ennui? They have very similar aromas.
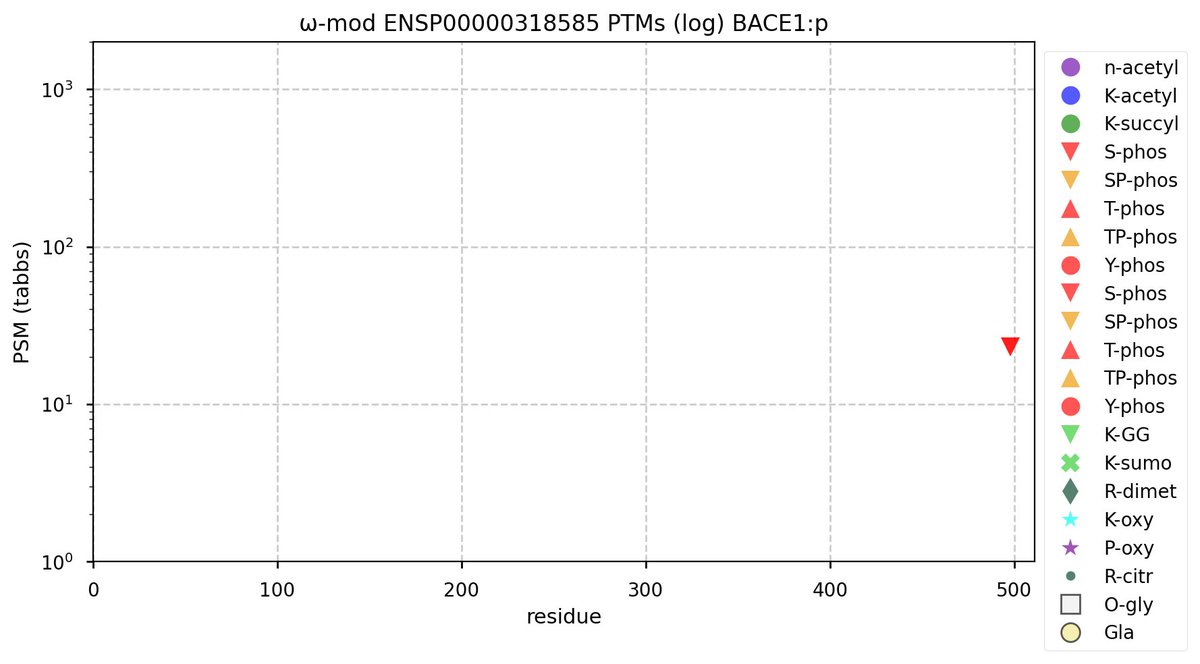
Mon Oct 24 13:59:28 +0000 2022BACE1:p isn't frequently seen in proteomics datasets: I only have IDs in 512 LC/MS/MS . It is most commonly found in CNS-related tissue samples. 🔗

Mon Oct 24 12:00:29 +0000 2022Modification diagram for human BACE1:p, a proteolytic enzyme normally associated with the trans-Golgi network. The sole S+phosphoryl acceptor has the ΦL/I/V motif. 🔗
Mon Oct 24 11:45:04 +0000 2022A classic Colorado Low (winds at 850 kPa), with moisture (blue) pumped up from the Gulf of Mexico by a high pressure system centred in northern Florida. 🔗
Mon Oct 24 02:52:09 +0000 2022@neely615 I don't think it would hurt: the books fill in a lot of details, although the how & what of the "Jackpot" is still pretty vague.
Sun Oct 23 18:16:24 +0000 2022A bedazzled transcription "master" regulator & its more abundant, but understated, inhibitor. 🔗
Sun Oct 23 18:02:15 +0000 2022@MagnusPalmblad It seems as though the defense posture is similar to that taken in the "Baltimore incident" from many years ago.
Sun Oct 23 15:15:49 +0000 2022I'm currently using Roboto & Roboto Mono, but there is something about the number glyphs that I'm not crazy about.
Sun Oct 23 15:10:07 +0000 2022What is your favourite font for a information-dense web site with lots of numbers?
Sun Oct 23 14:21:33 +0000 2022The only reason I change an interface is that I forget how much of a PITA changing an interface can be.
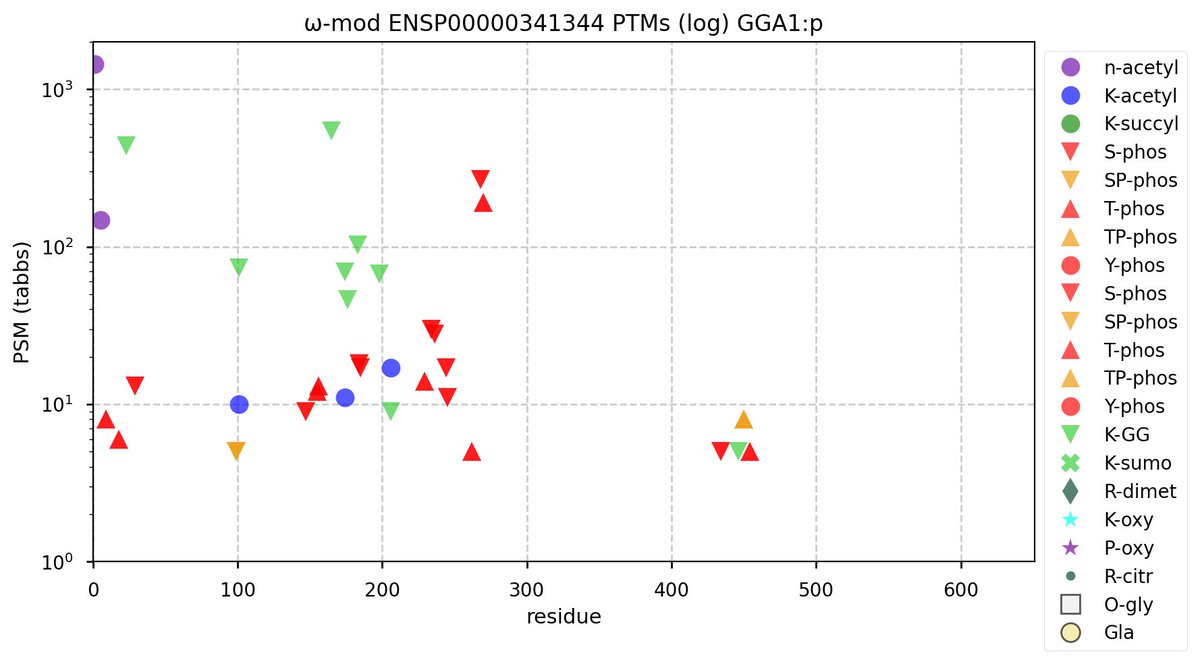
Sun Oct 23 12:50:49 +0000 2022Modification diagram for human GGA1:p, a subunit involved in trans-Golgi vesicular transport. Most of the PTM acceptors occur in the so-called "GGAs and TOM1" (GAT) domain (171-314). All 3 of the K+acetyl acceptors are also K+GGyl acceptors. 🔗
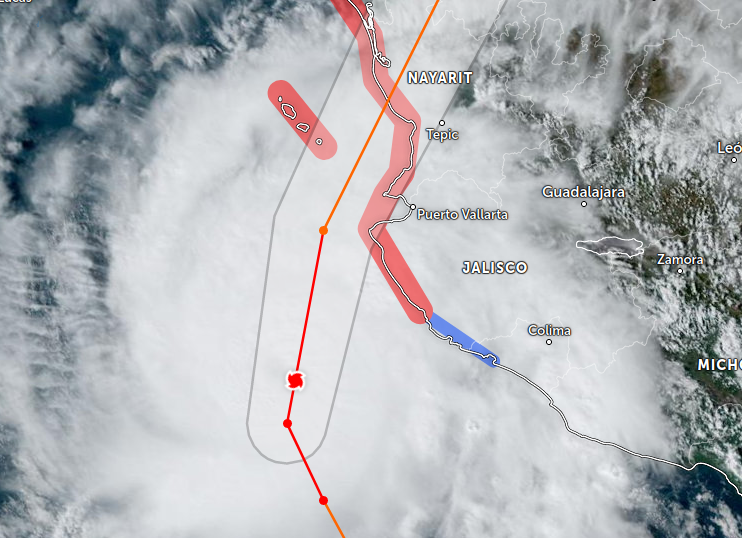
Sat Oct 22 21:12:50 +0000 2022Major Hurricane Roslyn is expected to make landfall between Puerto Vallarta and Mazatlan Mexico in the morning of Oct 23rd. 🔗
Sat Oct 22 19:27:27 +0000 2022@TrostLab I should add that I have read the books. I don't know how well someone who hasn't would be able to pick up on the plot.
Sat Oct 22 18:26:23 +0000 2022@Hayes_McDonald I'm not saying it is unusually difficult for an MD to be a scientist, but the media seem to assume that any random MD they talk to is a "scientist" & able to comment on the validity of "scientific evidence".
Sat Oct 22 15:33:53 +0000 2022Was there a particular point in time where MDs became scientists by default? The MDs I know are much closer to being sociologists than scientists.
Sat Oct 22 14:54:18 +0000 20221 font in 2 sizes & underline.
Sat Oct 22 14:42:16 +0000 2022On a bit of a "simplify" kick at the moment.
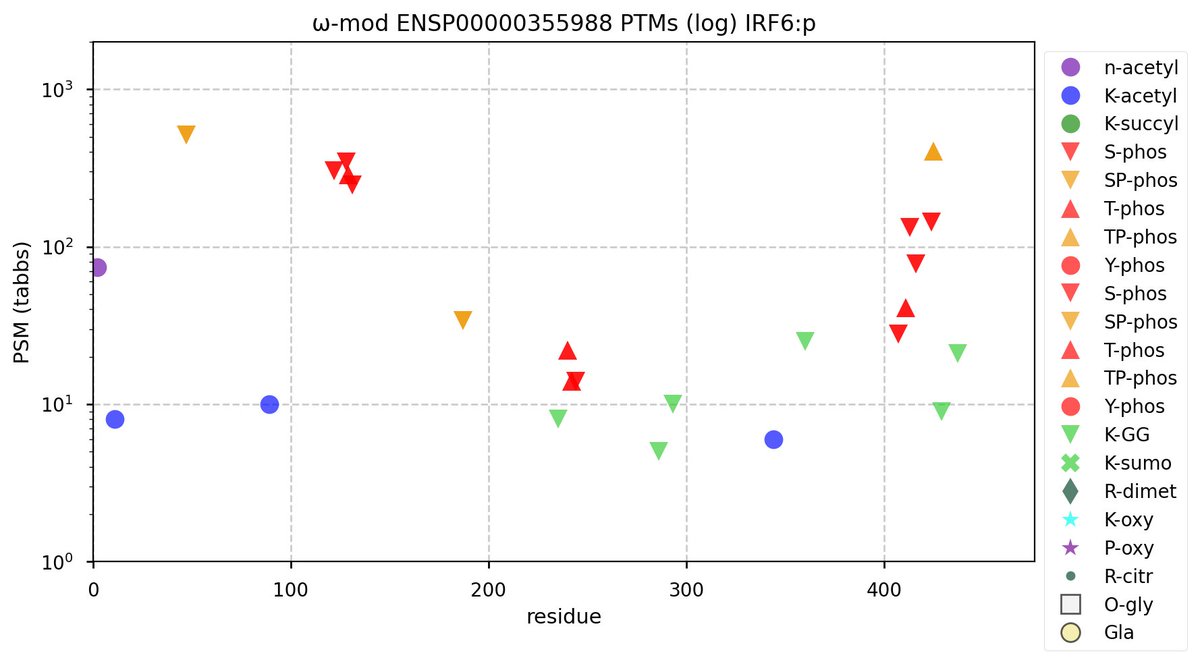
Sat Oct 22 12:03:25 +0000 2022Modification diagram for IRF6:p, a transcription factor. It has more S/T+phosphoryl acceptors than is typical of the IRF family. It is most commonly observed in keratinocytes & hepatocytes: it is rare in leukocytes or lymphocytes. 🔗
Fri Oct 21 23:49:36 +0000 2022@TrostLab It was a good adaptation of the story. I will be looking forward to the release of next episodes.
Fri Oct 21 18:07:10 +0000 2022First 2 episodes of the Peripheral are up: everything else must wait for about 2 hours ...
Fri Oct 21 12:41:42 +0000 2022The sequence has several P-rich domains & a poly-E no-complexity domain (142-149). 🔗
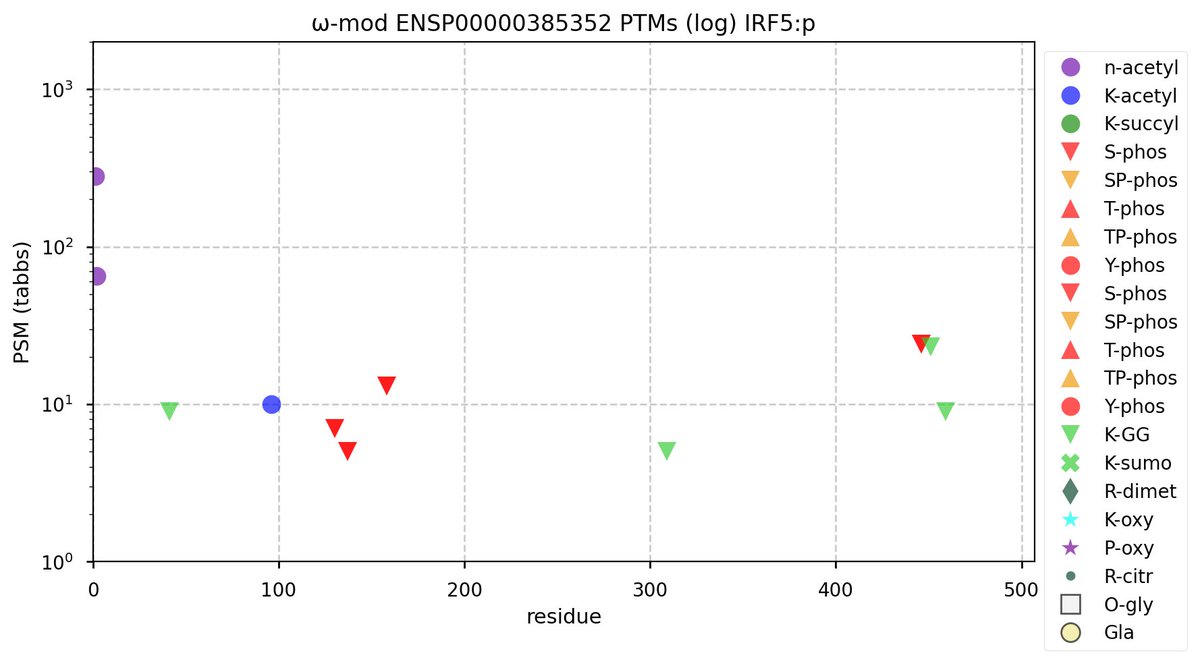
Fri Oct 21 12:06:40 +0000 2022Modification diagram for human IRF5:p, a transcription factor most abundant in monocyte-lineage leukocytes. Very modest, low abundance PTM acceptors, reminiscent of the IRF4:p pattern. 🔗
Thu Oct 20 17:28:15 +0000 2022Thanks to the participants. Clearly, the people have spoken.
Thu Oct 20 17:00:20 +0000 2022@UCDProteomics @neely615 @ProteomicsNews Hopefully v 1.1 will also support sparklines.
Thu Oct 20 16:01:08 +0000 2022@neely615 I await the Nature Biotech paper, with its inevitable sequel: a Nature Methods crossover thriller involving top-down data.
Thu Oct 20 15:29:26 +0000 2022@byu_sam @paulbfrandsen @sciencehacker I am biased because I have switched over to using it pretty much exclusively, except for a few web development boxes where I use Firefox because its has some nice interfaces to detect & correct HTML/CSS/Javascript problems.
Thu Oct 20 15:25:50 +0000 2022@byu_sam @paulbfrandsen @sciencehacker Brave is also pretty good at blocking trackers.
🔗
Thu Oct 20 14:41:35 +0000 2022@sciencehacker @byu_sam Switching to Brave works pretty well.
Thu Oct 20 13:32:42 +0000 2022@slavov_n @JohnRYatesIII Hopefully he gives some insight into how the associated patent shaped public and private sector research in the field for almost 2 decades.
Thu Oct 20 12:13:50 +0000 2022Normally only found in cell lines derived from lymphocytes. Abundant in lymphocyte MHC class I peptide experiments. Alternate initiation at M14 has been observed in lymphoblastoid cell lines. No issue with KR-poor regions with this one.
🔗
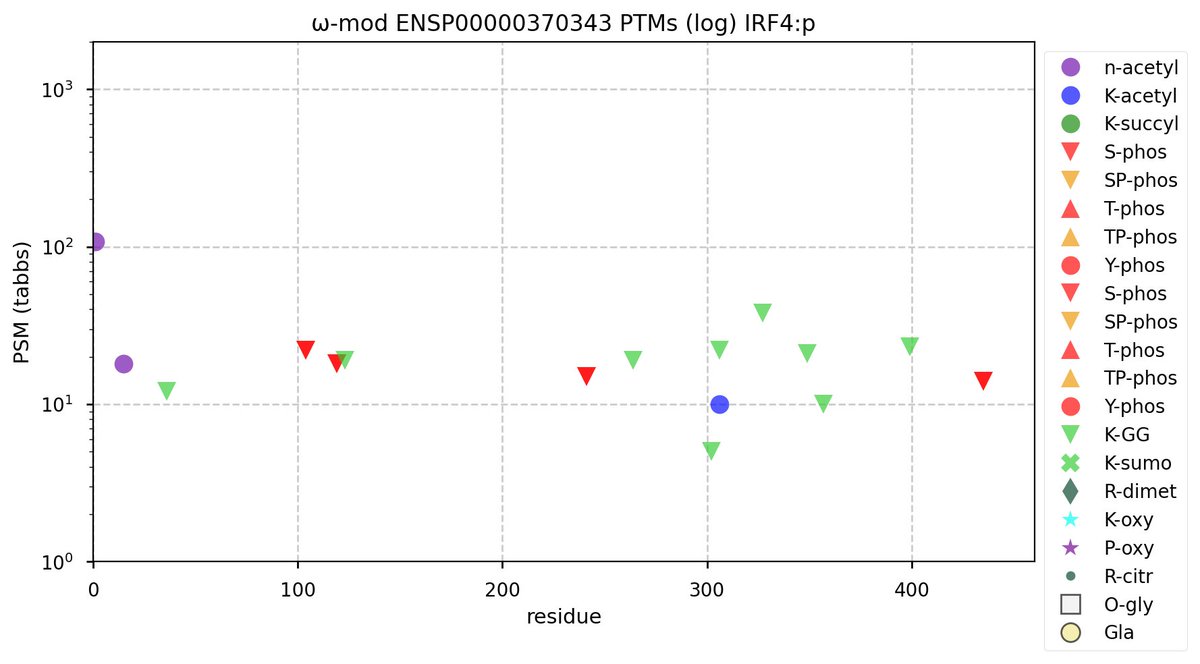
Thu Oct 20 12:12:24 +0000 2022Modification diagram for human IRF4:p, a transcription factor involved in lymphocyte maturation. Moderation rules the day, with 4 isolated, low occupancy S+phosphoryl acceptors in IDRs & 9×K+GGyl acceptors in intrinsically ordered regions. 🔗
Wed Oct 19 17:30:38 +0000 2022@RickBagshaw For an example of its use:
🔗
Wed Oct 19 17:24:09 +0000 2022I like to make up new terms as much as the next guy, but does "proxisome" really need to exist?
Wed Oct 19 17:13:39 +0000 2022Passwords would be better if more people took advantage of non-ASCII UTF-8 characters.
"∫🎄😃⊗😎😲😱⅝✋☀☁❄⚡ᶜ" would take quite a few clock cycles to guess.
Wed Oct 19 15:38:44 +0000 2022@cstross Hugo Weaving would be a good choice. He seems to like that sort of role.
Wed Oct 19 15:30:19 +0000 2022@PeterBOConnor It would change the way that people write grants.
Wed Oct 19 15:04:45 +0000 2022"Instead of funding grants that get the highest average score from reviewers, a funder should use the variance (or kurtosis or some similar measurement of disagreement)" 🔗
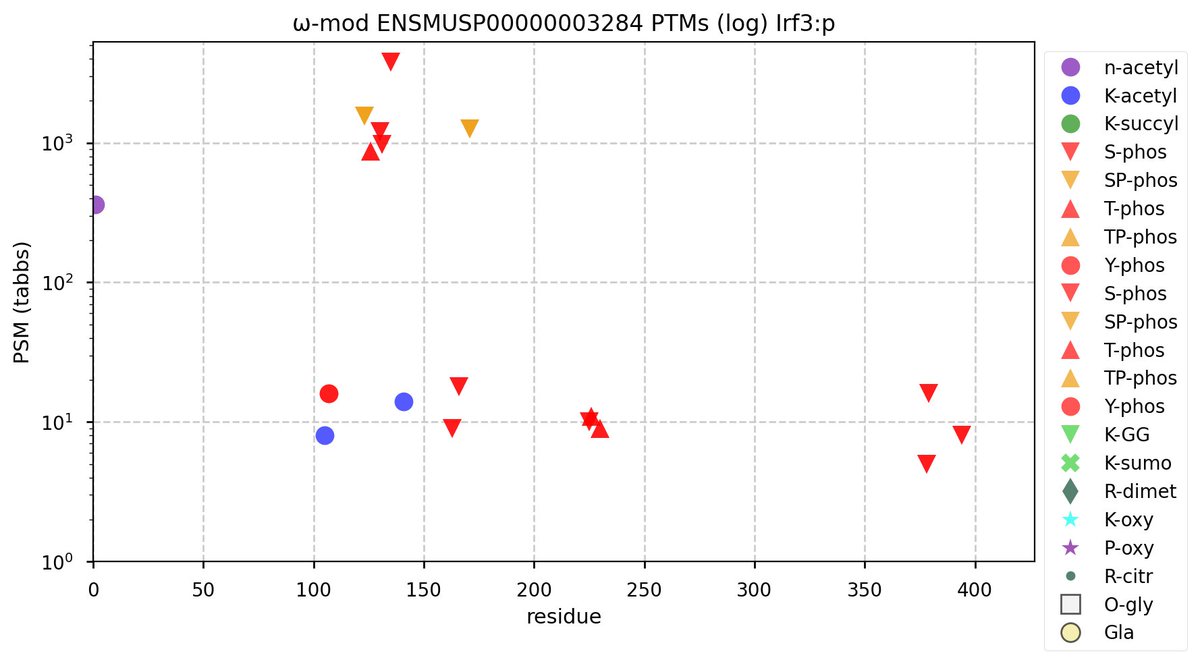
Wed Oct 19 14:04:06 +0000 2022It is also worth noting that the human IRF3:p sequence has a unusually variable C-terminal residue: it has an SAV―S427T―with a 50% maf. Many cell lines are either homozygous (TT) or heterozygous (ST) for this variant.
Wed Oct 19 12:28:13 +0000 2022@stphnmaher The Ottawa police have been at the forefront of the movement to make municipal police forces an independent branch of government.
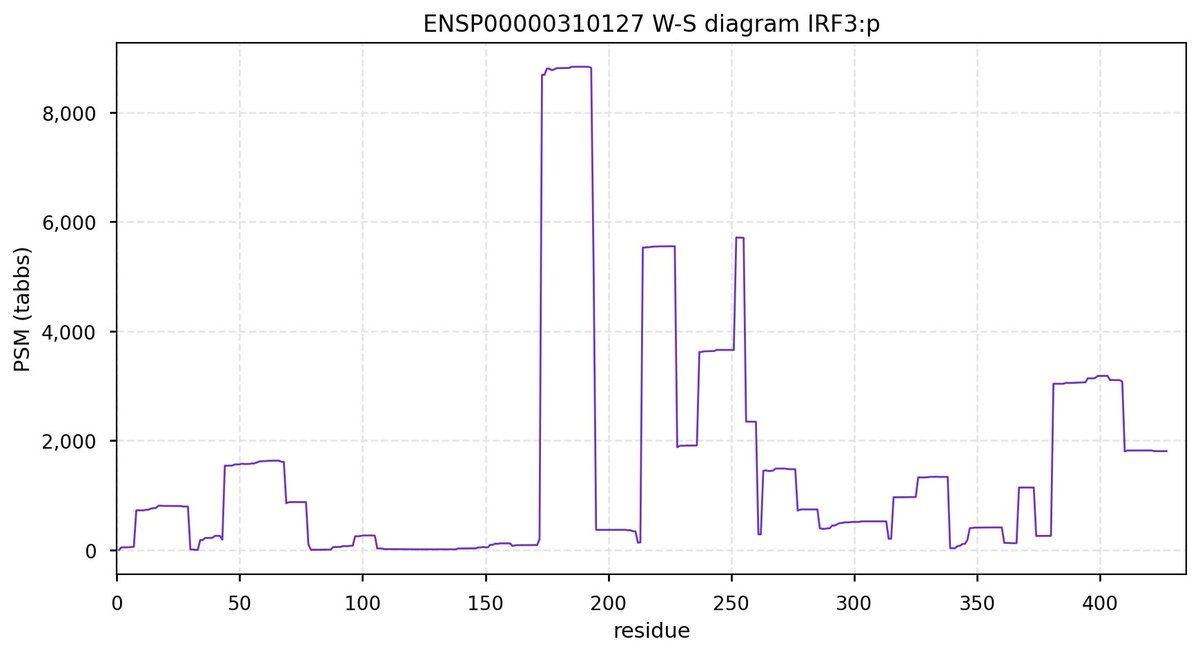
Wed Oct 19 12:12:20 +0000 2022IRF3:p is the interferon regulatory factor most commonly observed in proteomics data & like other IRF family members it is frequently found in lymphocyte MHC class I peptide experiments.
Wed Oct 19 12:00:40 +0000 2022The mouse sequence was used because a KR-poor region (106-171) in the human sequence blocks observation of the highest occupancy iHOP domain (123-135), as illustrated by their Westmore-Standing diagrams 🔗
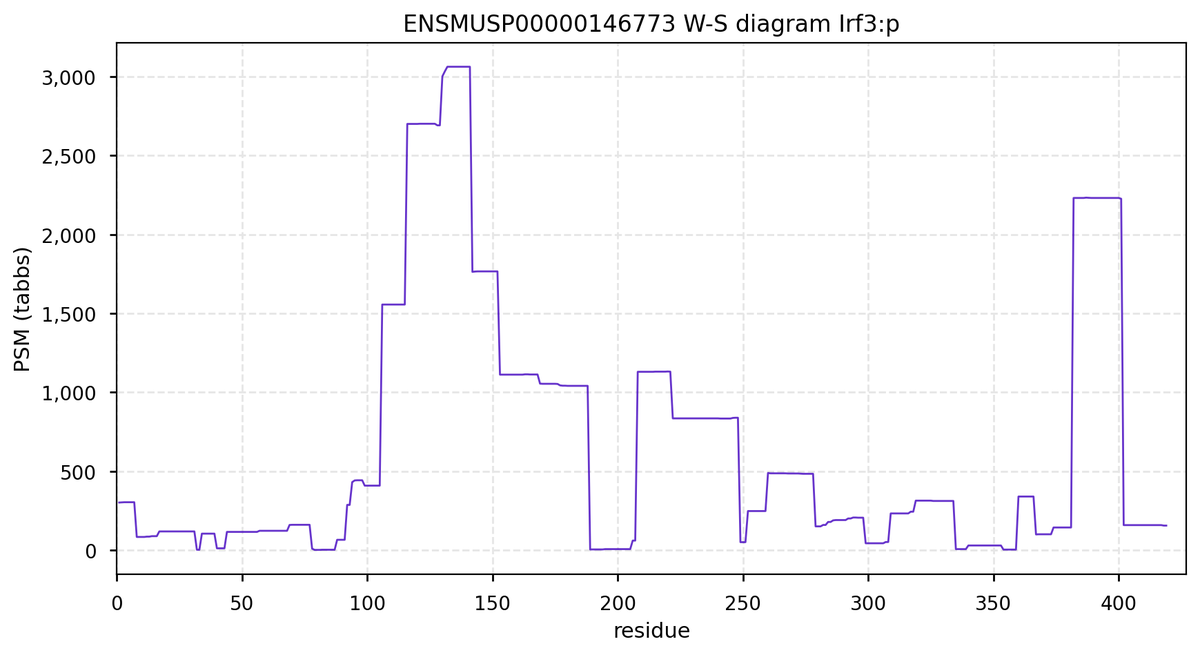
Wed Oct 19 11:56:28 +0000 2022Modification diagram for mouse IRF3:p, a subunit of the transcriptional regulator of type I interferon system. S+phosphoryl acceptors dominate:
1× ΦxExL/I/V (1687 Ta, 57.6%)
1× ΦQ (17 Ta, 0.6%)
1× ΦxxD/E (10 Ta, 0.3%)
2× ΦP (965 Ta, 32.9%)
4× ΦL/I/V (251 Ta, 8.6%) 🔗
Tue Oct 18 20:15:58 +0000 2022It also notes that "only one out of three new hires in 2021" stay with the company for 90 or more days. 🔗
Tue Oct 18 15:29:04 +0000 2022I am retweeting in the hopes that someone knows. I have tried local lighting/electrical stores, but they are all pushing the idea of light fixtures with built in, non-servicable LED boards, rather than replaceable assemblies like an LED tube. 🔗
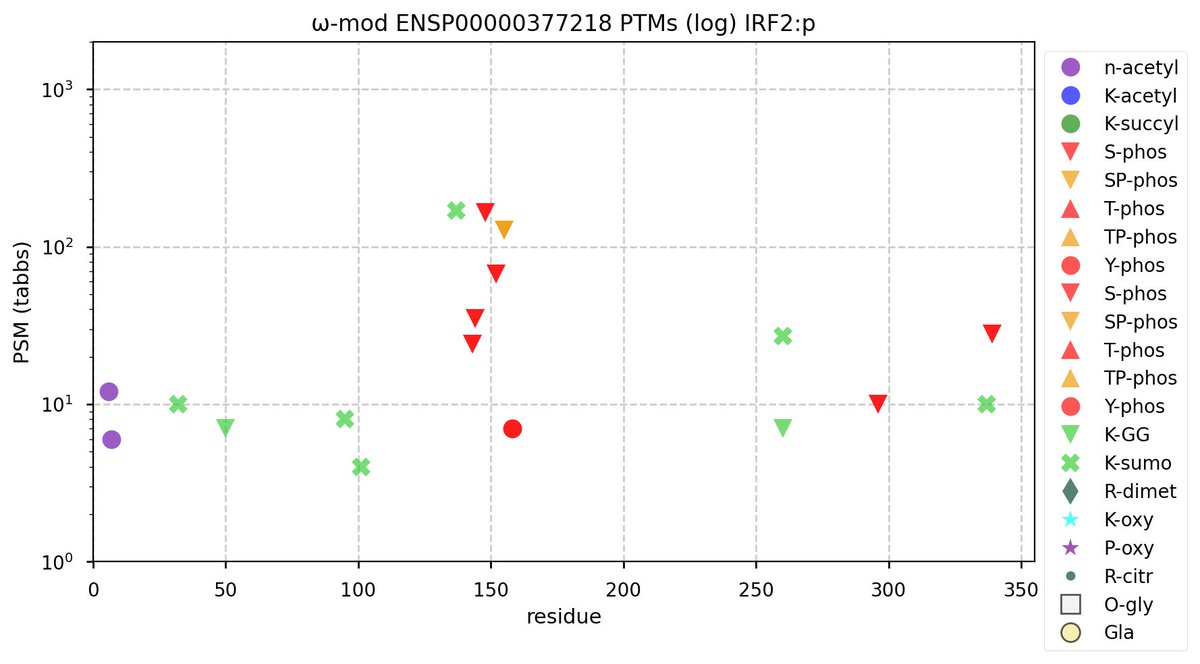
Tue Oct 18 14:17:23 +0000 2022IRF2:p has an extended KR-poor domain (167-252), similar to the KR-poor domain in IRF1:p (138-231).
Tue Oct 18 14:12:42 +0000 2022Modification diagram for human IRF2:p, a transcription factor most abundant in leukocytes. It has a busier PTM pattern than IRF1:p, especially the S+phosphoryl iHOP acceptor domain (143-155) that has no analog in IRF1. 🔗
Tue Oct 18 14:04:59 +0000 2022@neely615 XY-poor is a pretty good suggestion!
Tue Oct 18 13:57:22 +0000 2022What do you call a domain in which several amino acids are underrepresented or simply absent?
Tue Oct 18 13:16:50 +0000 2022@kyle_minogue Carbon dioxide is surprisingly soluble in many types of organic solvents, including (as you demonstrate) plasticizers.
Tue Oct 18 12:27:21 +0000 2022@uniprot @sciencehacker It would have been much better to have created a new entry type so that these made up names could be easily identified by a user. Replacing the usually accurate "Uncharacterized" with an authoritative sounding name of unknowable provenance isn't helpful (at least for me).
Tue Oct 18 11:53:10 +0000 2022@uniprot @sciencehacker You do understand that it can't be both from the gene "Sptbn4" & called a "Col1A1-like" protein. "uncharacterized" would be correct in this case.
Tue Oct 18 00:48:36 +0000 2022@Smith_Chem_Wisc PS. The biggest volume of publicly available "brain" data is from mice: for some reason or other humans just don't seem to like giving up those tissue samples.
Mon Oct 17 21:48:09 +0000 2022@J_my_sci Just add it to your wiki page. No one will check.🤠
Mon Oct 17 15:51:34 +0000 2022@dtabb73 Both. But not looking for examples: just whether or not a specific algorithm takes the possibility into consideration.
Mon Oct 17 15:49:41 +0000 2022@Smith_Chem_Wisc PXD005629 or PXD002516 for all criteria
PXD010138 doesn't quite fit the criteria, but may be worth a look.
PXD014606 if you want to compare with low res fragment ion data set.
Mon Oct 17 15:03:13 +0000 2022🔗
Mon Oct 17 15:02:05 +0000 2022I check my email just about as often as I check my fax machine.
Mon Oct 17 14:58:47 +0000 2022Does anybody know which of the existing PSM ID algorithms & spectrum library curation schemes incorporate z=2 fragment ions?
Mon Oct 17 14:07:24 +0000 2022The only consistent eccentricity associated with "^H.+[^KR]$" or "^K.+[^KR]$" is an over fondness for b-ions with z=2.
Mon Oct 17 13:57:48 +0000 2022My current city (Winnipeg) makes a fetish out of trying to look like a spiffy version of 1890's Chicago. I really wish someone from the city planning department would spend some time in 2020's Chicago to get some new ideas.
Mon Oct 17 12:56:23 +0000 2022"Irrational optimism" is the tone of most CBC local morning shows across the country. As I remember it, the show in Vancouver was about 3 hours of real estate sales promos.
Mon Oct 17 12:53:12 +0000 2022Listening to a hilariously positive person describe a downtown Winnipeg that doesn't really exist on CBC's morning show.
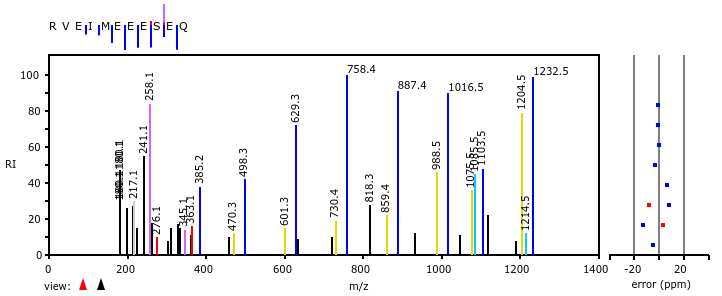
Mon Oct 17 12:46:49 +0000 2022Thanks to the bold few who participated in this poll. The correct answer is "^R.+[^KR]$". The attached spectrum of RVEIMEEESEQ (z=2, C-terminal tryptic peptide of USP14:p) has a-ions in yellow & b-ions in blue. 🔗
Mon Oct 17 12:13:25 +0000 2022Fortune favours the bold, but survival favours the timid.
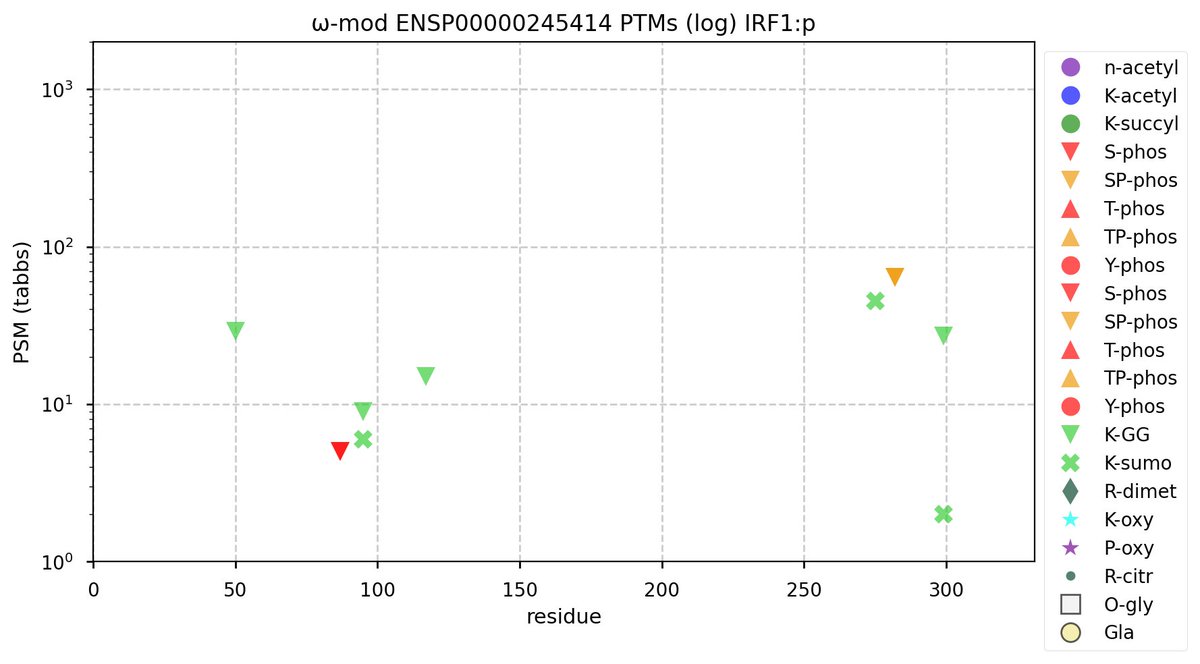
Mon Oct 17 12:04:34 +0000 2022IRF1:p-derived peptides are significantly enriched in T-cell MHC class 1 peptide experiments.
Mon Oct 17 12:03:07 +0000 2022Modification diagram for human IRF1:p, a transcription factor most abundant in leukocytes. The K-mods are quite conventional: 1×K+SUMOyl, 2×K+GGyl, 2×K+GGyl/SUMOyl. as are the low occupancy S+phosphoryl: 1×ΦxxD/E, 1×ΦP. K+acetyl is conspicuous by its absence. 🔗
Sun Oct 16 22:41:29 +0000 2022PXD035020: mouse milk proteomics is pretty rare.
Sun Oct 16 14:23:03 +0000 2022@AlexUsherHESA A quick survey of Canada's senior political leaders should answer that question. It is not a group that values skills: they value opportunity.
Sun Oct 16 13:10:05 +0000 2022For the non-regex folks out there:
^X means "starts with X"
.+ means "a string of any letters"
[^KR]$ means "ends with any letter except K or R"
Sun Oct 16 13:00:01 +0000 2022Typhoon Nesat (Neneng) has skirted north of Luzon and is headed into the South China Sea. It is expected to pass south of Hainan (海南) and make landfall as a tropical storm in northern Vietnam sometime on Friday. 🔗
Sun Oct 16 12:46:44 +0000 2022Which of these sequence motifs is most likely to generate an extensive pattern a-ions in its MS/MS spectrum:
Sun Oct 16 12:26:33 +0000 2022Even though it is only 81 aa long, CENPX:p still manages to squeeze in 5 exons. 🔗
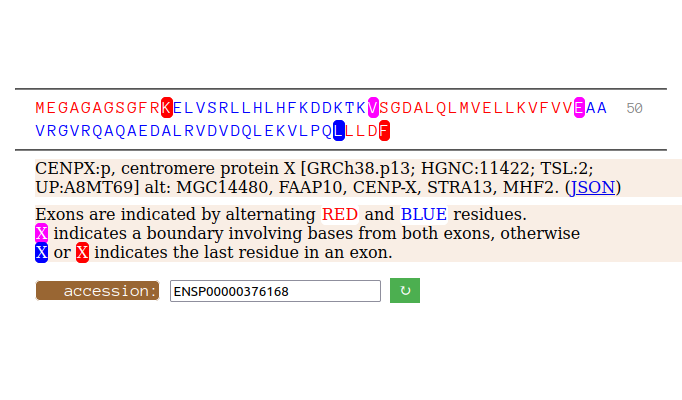
Sun Oct 16 11:51:39 +0000 2022Modification pattern for human CENPX:p, a centromere subunit. Eschews bling. 🔗
Sat Oct 15 15:15:48 +0000 2022@DavidMorgenst17 That is certainly the prevailing opinion. It is the dominant, tried-and-true logical premise used in the development funding documents and publications.
Sat Oct 15 14:51:16 +0000 2022@bkives Big box developments like that are more industrial than commercial, but I take your point. Any case studies of pure-play industrial developments recouping municipal investment via property taxes would be informative.
Sat Oct 15 14:19:50 +0000 2022@bkives I am sure that the details matter. But, are there any case studies available that attempt to quantify the case for public investment in "shovel-ready" land? Cases that worked (e.g., IKEA) vs those that didn't (e.g., Target).
Sat Oct 15 14:10:54 +0000 2022Related Unpopular Opinion℠―There are few, if any, real world situations that can adequately expressed using a string of directed acyclic graphs.
Sat Oct 15 14:05:47 +0000 2022Unpopular Opinion℠― "Metadata" is a common red herring tossed out while discussing data repositories. It is almost always the opening foray in someone's plan to develop THE ontology to rule them all.
Sat Oct 15 13:33:24 +0000 2022@bkives How long does it take to recoup the costs of making land "shovel-ready" through property taxes?
Sat Oct 15 13:31:10 +0000 2022Does anybody know an LED replacement tube that uses a T12 compatible magnetic ballast? I've tried a few that are supposed to work, but they don't. I don't particularly want to go with a type B replacement, because the lamp is in a very inconvenient place.
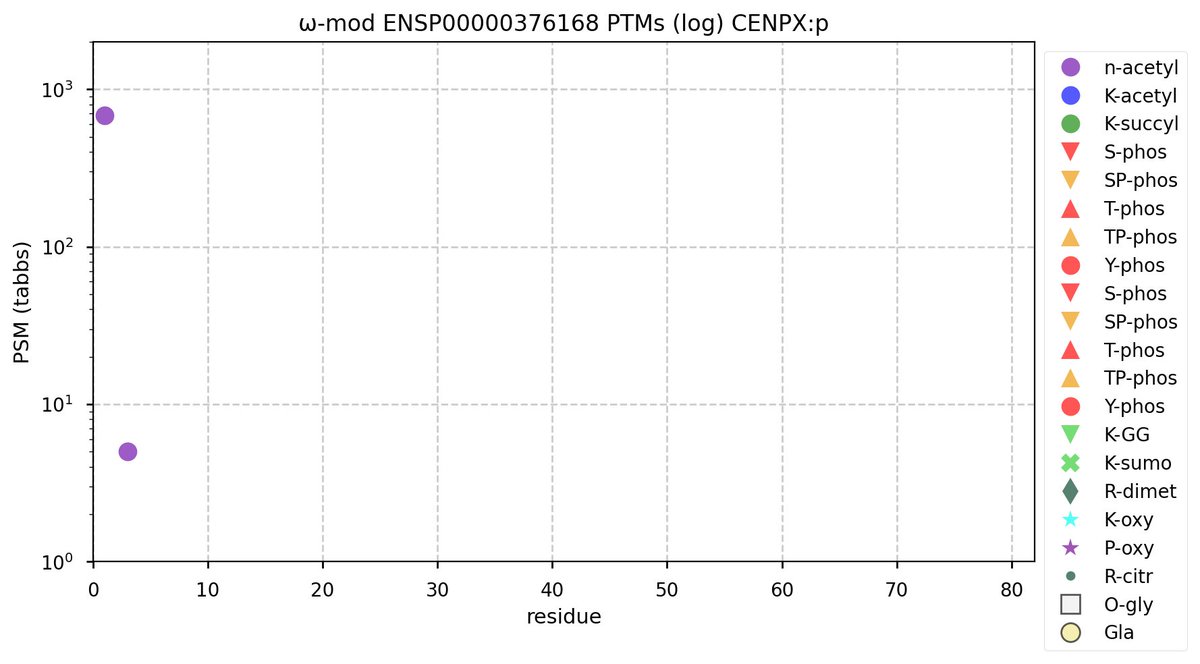
Sat Oct 15 12:07:23 +0000 2022Modification diagram for human CENPW:p, a centromere subunit. This small sequence (88 aa) is the least frequently observed of the CENP genes. 🔗
Fri Oct 14 18:13:10 +0000 2022The main thing I have achieved from trying a lot of things & failing at many of them is being better able to realize when something genuinely isn't worth pursuing.
Fri Oct 14 15:45:27 +0000 2022@pwilmarth @neely615 @sciencehacker Who knows, maybe their new Google-branded names will catch on.
Fri Oct 14 15:39:46 +0000 2022Downloaded some data from a repository I don't use very much & was reminded of why I don't use that repository very much.
Fri Oct 14 15:29:17 +0000 2022UnProtein has not yet renamed this sequence.
Fri Oct 14 13:30:19 +0000 2022Whenever you become too optimistic about the impact of modern science on medicine, just review the CDC's equivalent of “Memento mori”:
🔗
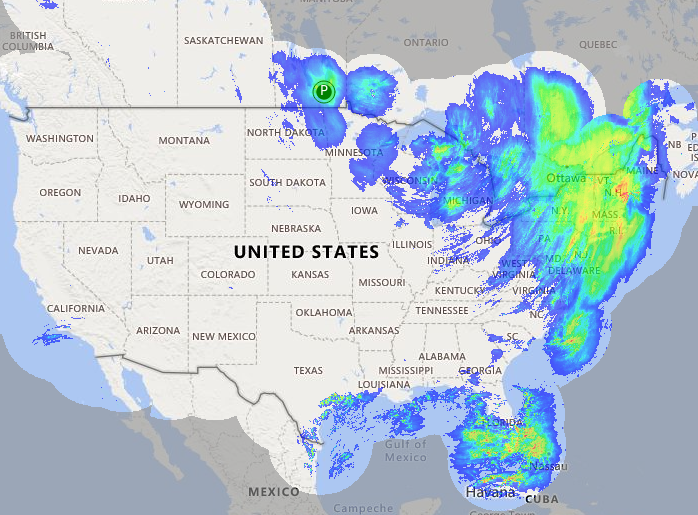
Fri Oct 14 13:21:10 +0000 2022& unfortunately it means it is still snowing here (radar-based estimate of precipitation in the last 24 hours). 🔗
Fri Oct 14 13:14:28 +0000 2022@astacus @LMcLean51 They share some of the aspects associated with AI-generated visualizations: they look great & are clearly showing some sort of differences, but it is impossible to understand what they mean without extensive followup investigations.
Fri Oct 14 12:30:47 +0000 2022Unpopular Opinion℠: PTM acceptor site localization usually isn't that important. Rejecting a site because you can't prove exactly which residue it was on biases your results.
Fri Oct 14 12:13:48 +0000 2022Yesterday's low near Hudson Bay has split in 2, with a secondary circulation centred in Northern Ontario. But it is still pumping cold air as far south as Kansas, pulling it back up through Chicago & Detroit. 🔗
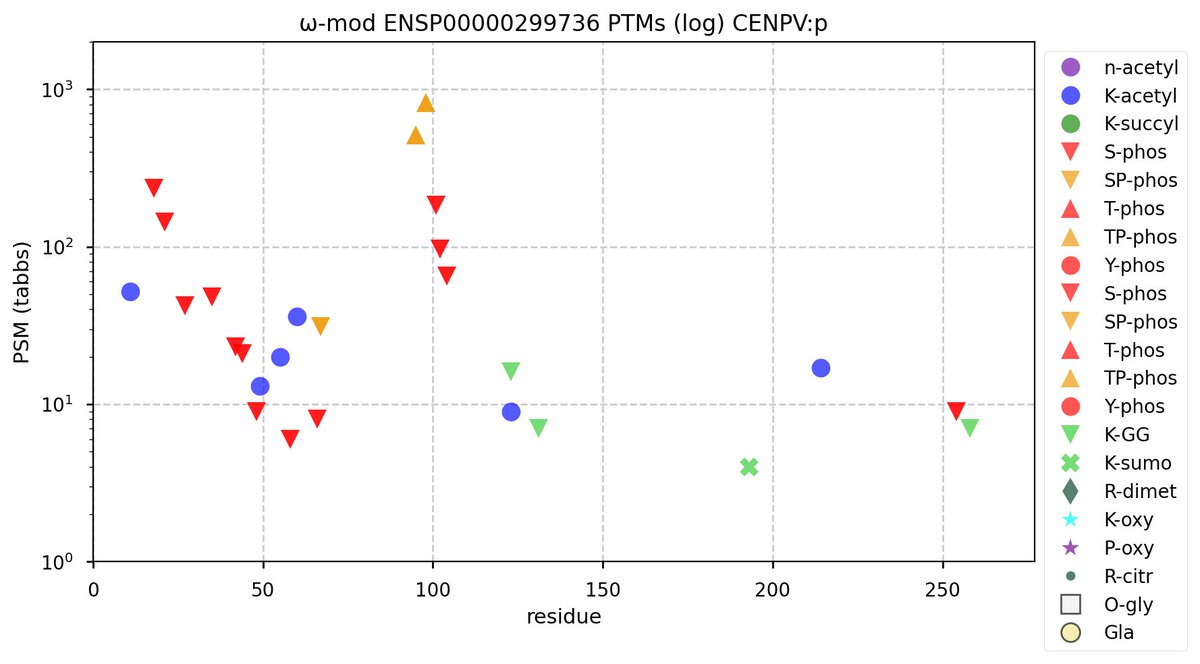
Fri Oct 14 12:04:41 +0000 2022Little CENPV:p is the #2 most frequently observed human centromeric protein in GPMDB (272 aa, 17,047×). #1 is the much larger CENPF:p (3114 aa, 21,000×). Most other centromere subunit are in the range 1,000-2,500×.
Fri Oct 14 11:49:14 +0000 2022Modification diagram for human CENPV:p, a centromere-associated subunit. It has several swarms of S/T+phosphoryl across an N-terminal region (1-104) in multiple low complexity domains. The presence of multiple K+acetyl acceptors is unusual for a centromeric protein. 🔗
Thu Oct 13 22:58:38 +0000 2022@sciencehacker This one's new name is a classic piece of AI goofery
🔗
Thu Oct 13 21:42:32 +0000 2022@pwilmarth @sciencehacker There probably will be a brisk black market in UP FASTAs that have been re-annotated to use the standard protein names biologists expect.
Thu Oct 13 21:06:41 +0000 2022@pwilmarth @sciencehacker Unfortunately, mice no longer have "arginine vasopressin", so that name isn't available. It has been mad libed to "Vasopressin-neurophysin 2-copeptin"
🔗
Thu Oct 13 20:00:49 +0000 2022@sciencehacker Looking further down the list, there have been a lot of unhelpful changes made. Hopefully one of the grownups at EBI will realize this didn't work & revert back to the original annotation.
Thu Oct 13 19:44:14 +0000 2022@sciencehacker Lots of fun to be had by the whole proteomics community dealing with this development.
Thu Oct 13 19:43:13 +0000 2022@sciencehacker For example, what used to be called
"Sptbn4, spectrin beta, non-erythrocytic 4"
has been renamed by Google to be
"Collagen alpha-1(I) chain-like"
🔗
Thu Oct 13 19:39:51 +0000 2022@sciencehacker It turns out you can see at least some subset of the annotations affected using
🔗)
Thu Oct 13 19:19:01 +0000 2022@sciencehacker Hmm, indeed. I hope this new "information" is clearly marked as to its origins.
Thu Oct 13 17:21:44 +0000 2022@lkpino @byu_sam I'd settle for "stable".
Thu Oct 13 15:18:37 +0000 2022The same system is causing lake effect snow here (Winnipeg) & rain 2,600 km away in NYC.
Thu Oct 13 13:33:19 +0000 2022Hmm, there appears to be crystallized water falling from the sky this morning.
Thu Oct 13 13:30:42 +0000 2022@docdez @AlexUsherHESA And it wouldn't be immediate: I'd say having the election 12 months after the leadership change would give the public a good chance to get to know the new leader.
Thu Oct 13 13:22:44 +0000 2022@docdez @AlexUsherHESA In my experience, once a politician obtains a leadership position they are loath to give it up. And any opposition that did that even once would no longer be the opposition following the election.
Thu Oct 13 13:11:03 +0000 2022@AlexUsherHESA I am leaning towards the notion that a change in the leader of either the governing or the official opposition parties should trigger an election.
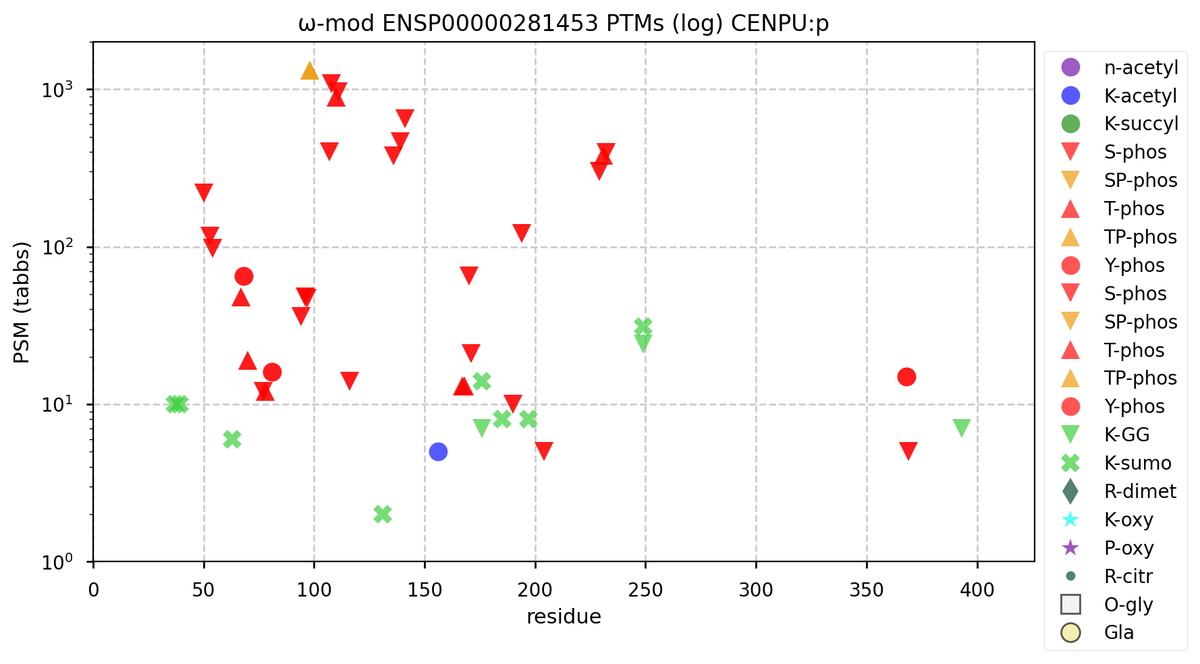
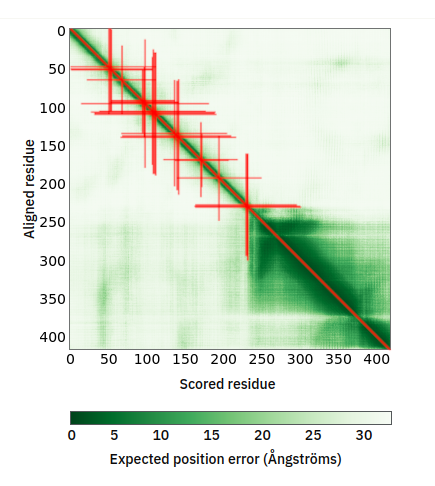
Thu Oct 13 12:09:29 +0000 2022Modification diagram for human CENPU:p, a centromere complex subunit. There are several swarms of S/T+phosphoryl in an N-terminal region (1-246) containing multiple low complexity domains. K-mod acceptors are dominated by SUMO2: 1×K+GGyl, 2×K+GGyl/SUMO2yl, 6×K+SUMO2yl. 🔗
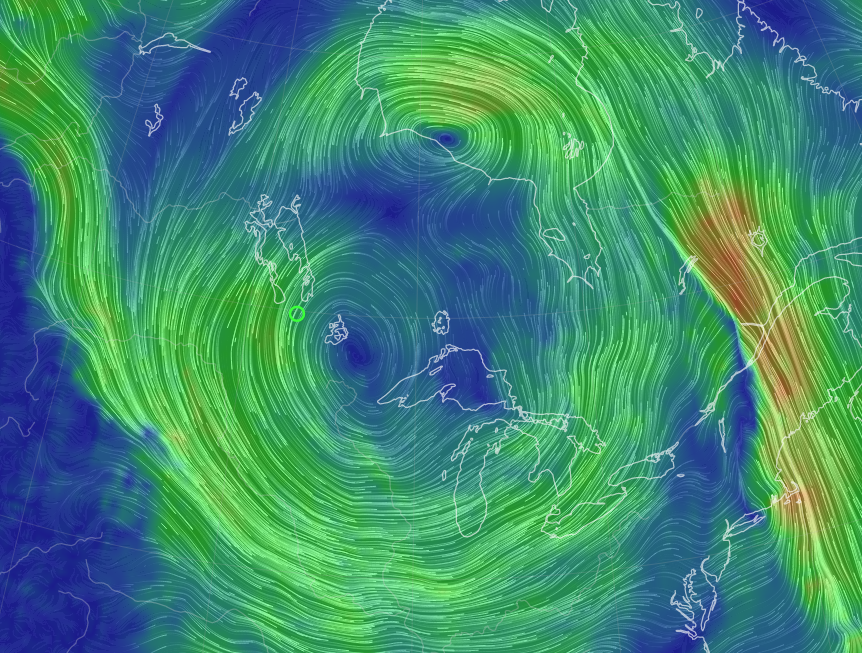
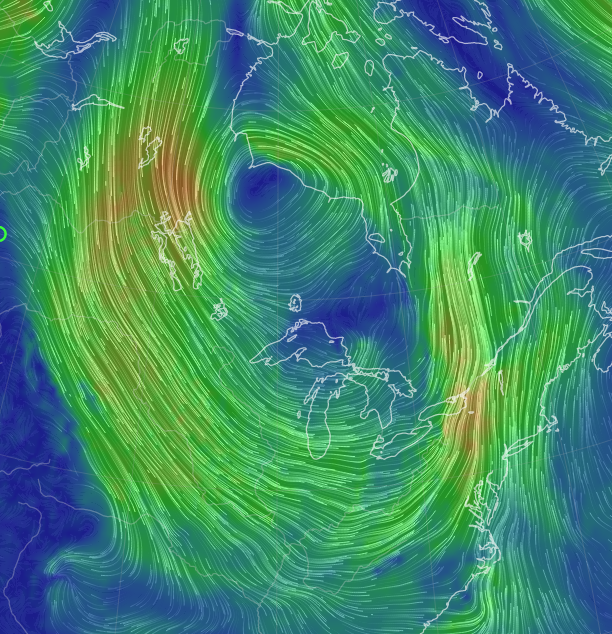
Thu Oct 13 12:04:38 +0000 2022A low pressure system centred just southeast of Churchill Manitoba is causing a cold front that is moving east towards the US Atlantic coast (winds at 850 hPa). 🔗
Thu Oct 13 00:02:06 +0000 2022There are 2 types of people: those who put tools back where they got them from & those who put tools in a place they think they will remember next time the tools are needed. These 2 types of people should not collaborate on anything that involves tools.
Wed Oct 12 18:45:37 +0000 2022POGZ:p (Pogo transposable element with ZNF domain). Don't know what it is, but it plays by the rules. 🔗
Wed Oct 12 16:23:51 +0000 2022@KentsisResearch @willwbooker @CedricFeschotte How can it be that proteome & transcriptome measurements agree?
Wed Oct 12 15:38:28 +0000 2022journals editors + peer-review = ? 🔗
Wed Oct 12 14:58:34 +0000 2022@macintoshmaggie Hopefully they can at least figure out how to not collect & then refund school tax.
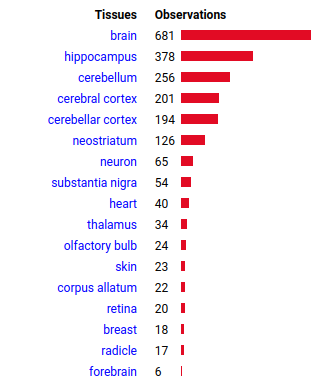
Wed Oct 12 14:39:20 +0000 2022@KentsisResearch @willwbooker @CedricFeschotte It's observed tissue distribution definitely shows a preference. I do wish that people would be a bit more specific than "brain" when they state the origin of a tissue sample, but beggars can't be choosers ... 🔗
Wed Oct 12 14:23:18 +0000 2022@slashdot Makes me feel a lot better about my $650 Lenovo 16-core AMD bruiser.
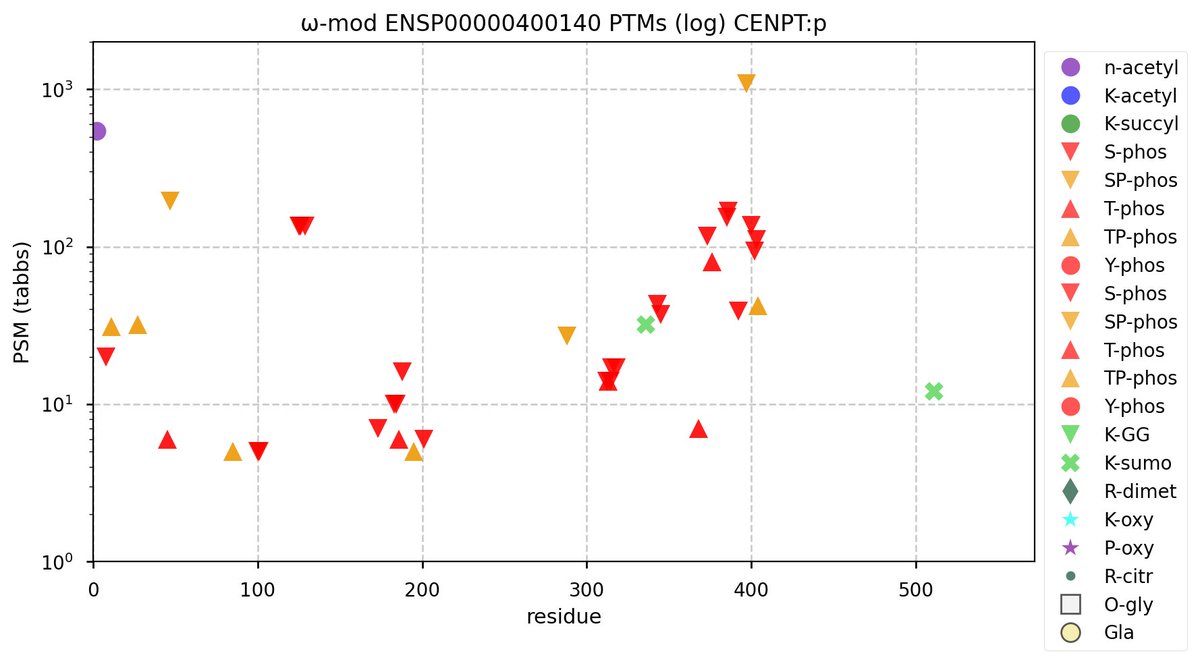
Wed Oct 12 12:27:42 +0000 2022For the non-SUMO-obsessed, KxE for SUMO2yl is pretty much equivalent to ΦP for S/T+phosphoryl.
Wed Oct 12 12:24:31 +0000 2022@AlexUsherHESA The fourth season of "Community" pretty much did in the whole sector.
Wed Oct 12 12:19:43 +0000 2022The two K+SUMO2yl acceptors have the common KxE motif. The S/T+phosphoryl acceptors are split between ΦP, ΦL/I/V, and ΦxxD/E motifs, with the usual occupancy pattern.
Wed Oct 12 11:58:16 +0000 2022Its lack of K+GGyl is unusual for a centromere subunit, but the absence of K+acetyl is par for the course. Unlike RNA-interacting proteins, the centromere subunits all lack R+dimethyl acceptors.
Wed Oct 12 11:55:40 +0000 2022Modification diagram for human CENPT:p, a centromere complex subunit. The pattern is dominated by S/T+phosphoryl acceptors scattered across an IDR with multiple low complexity regions (1-400). The domain with multiple α-helices (400-561) is left undecorated. 🔗
Tue Oct 11 17:45:13 +0000 2022@Ribrob The engineer who proposed it probably did so as a joke, but the MBAs up the chain just went with it.
Tue Oct 11 16:45:53 +0000 2022@astacus Very much on the nose! 😎
Tue Oct 11 15:03:38 +0000 2022In the unlikely case that anyone wants to look at the patterns of observed PTMs for an entire species, an up-to-date list is made available monthly at 🔗 in the "ω-mod" folder for H. sapiens, M. musculus, R. norvegicus & S. cerevisiae.
Tue Oct 11 14:43:28 +0000 2022There really has to be some sort of moratorium on silly acronyms associated every methods paper that comes out.
Tue Oct 11 13:59:38 +0000 2022Cheap switches have been the bane of my existence. 🔗
Tue Oct 11 11:56:28 +0000 2022You could probably construct a pretty good "Protein Micrometer" from a selected basket of sparsely modified centromere subunits.
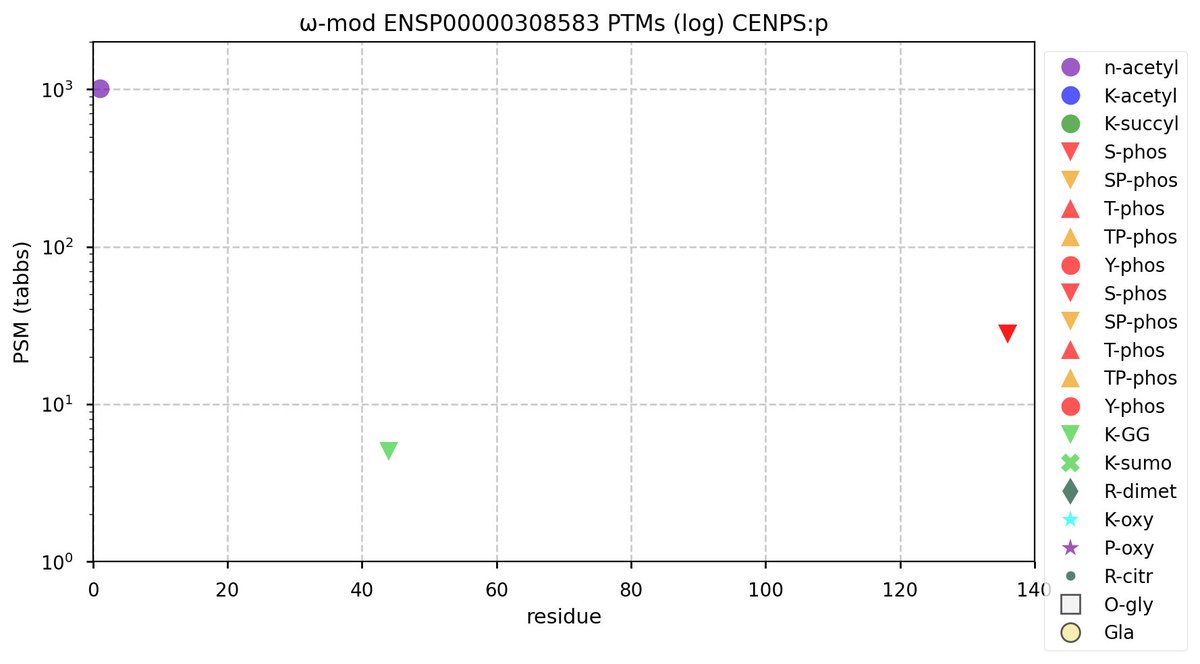
Tue Oct 11 11:47:32 +0000 2022Modification diagram for human CENPS:p, a subunit of the centromere complex. A single S+phosphoryl decorates the C-terminal IDR, while a single K+GGyl, with a ζ-K-ζ motif, is located at the N-terminal end of the 2nd α-helical domain. 🔗
Mon Oct 10 13:31:48 +0000 2022State of the art in vehicle automation
🔗
Mon Oct 10 13:11:06 +0000 2022@AJ_Brenes @labs_mann Is mandatory retirement at 65 still the rule in Germany?
Mon Oct 10 12:46:56 +0000 2022The sequence shown is from the splice variant (CCDS30736, 177 aa) that appears to be the most abundant. There is also an observed longer variant (CCDS55603, 216 aa), with 2 exons replacing the 2nd translated exon in the short version. 🔗
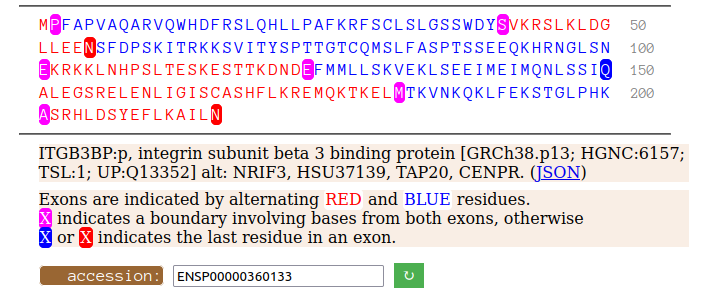
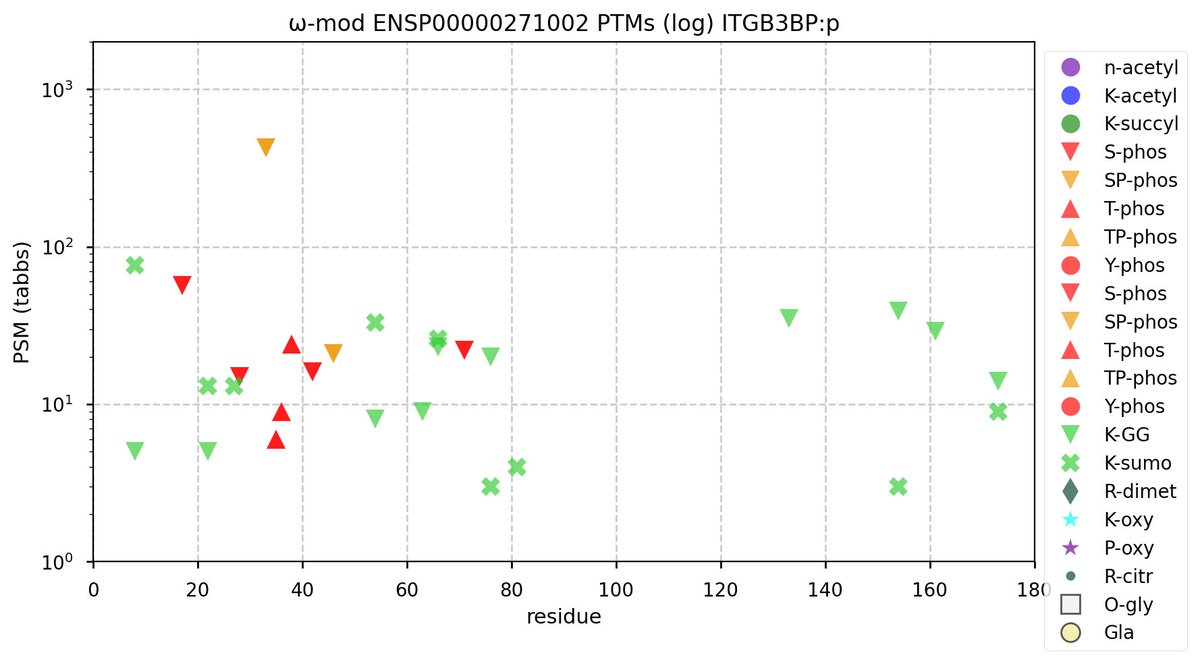
Mon Oct 10 12:32:38 +0000 2022Modification diagram for ITGB3BP:p, aka CENPR:p, a subunit of the centromere (& other things). The K-mods are a mix, with 2×K+SUMOyl, 3×K+GGyl, & 7× K+GGyl/SUMOyl. As with other centromere subunits, K+acetyl is absent. It has a RIP domain in its N-terminal IDR. 🔗
Mon Oct 10 12:17:49 +0000 2022@ItsBiniR @MattWFoster @Smith_Chem_Wisc All of the proteins in a good saliva sample:
🔗
Just the bacterial proteins for the same sample:
🔗
From Murr, et al. 🔗
Sun Oct 09 22:38:23 +0000 2022@MattWFoster @Smith_Chem_Wisc Saliva contains a lot of immunoglobulins: properly accounting for all immunoglobulin peptides is not normally done, so you will have quite a few rather abundant peptides that aren't assigned but are available to confuse & amaze you.
Sun Oct 09 22:36:18 +0000 2022@MattWFoster @Smith_Chem_Wisc But remember, the golden rule of proteomics applies: "You are most likely to see the most abundant proteins."
If you aren't seeing the DNA transcription apparatus, ribosomes, chaperonins & most abundant metabolic enzymes, don't trust some off-brand single protein id.
Sun Oct 09 22:34:41 +0000 2022@MattWFoster @Smith_Chem_Wisc Neisseria subflava
Prevotella melaninogenica
...
Sun Oct 09 22:32:14 +0000 2022@MattWFoster @Smith_Chem_Wisc Streptococcus sanguinis
Streptococcus suis
Streptococcus thermophilus
Streptomyces avermitilis
Tannerella forsythia
Thiomicrospira crunogena
Veillonella parvula
Weeksella virosa
Alloprevotella rava
Fusobacterium periodonticum
Klebsiella sp OBRC7
Leptotrichia goodfellowii
...
Sun Oct 09 22:31:55 +0000 2022@MattWFoster @Smith_Chem_Wisc Streptococcus agalactiae
Streptococcus dysgalactiae
Streptococcus equi
Streptococcus gordonii
Streptococcus mitis
Streptococcus oralis
Streptococcus parasanguinis
Streptococcus pneumoniae
Streptococcus pseudopneumoniae
Streptococcus pyogenes
Streptococcus salivarius
...
Sun Oct 09 22:31:35 +0000 2022@MattWFoster @Smith_Chem_Wisc Myxococcus fulvus
Neisseria gonorrhoeae
Neisseria lactamica
Nostoc punctiforme
Pasteurella multocida
Prevotella denticola
Prevotella melaninogenica
Prevotella ruminicola
Riemerella anatipestifer
Rothia dentocariosa
Rothia mucilaginosa
Staphylococcus aureus
...
Sun Oct 09 22:31:18 +0000 2022@MattWFoster @Smith_Chem_Wisc Fusobacterium nucleatum
Haemophilus influenzae
Haemophilus parainfluenzae
Lactobacillus plantarum
Lactococcus garvieae
Leifsonia xyli
Leptotrichia buccalis
Magnetospirillum magneticum
Megasphaera elsdenii
Methanosphaera stadtmanae
Methylomicrobium alcaliphilum
...
Sun Oct 09 22:30:59 +0000 2022@MattWFoster @Smith_Chem_Wisc Chitinophaga pinensis
Chloroflexus aggregans
Chromohalobacter salexigens
Citrobacter koseri
Corynebacterium aurimucosum
Croceibacter atlanticus
Enterococcus faecalis
Erysipelothrix rhusiopathiae
Eubacterium rectale
Fibrobacter succinogenes
Flavobacteriaceae bacterium
...
Sun Oct 09 22:30:39 +0000 2022@MattWFoster @Smith_Chem_Wisc Anaeromyxobacter Fw109 5
Arcanobacterium haemolyticum
Archaeoglobus fulgidus
Atopobium parvulum
Bacillus clausii
Bacteroides fragilis
Bacteroides helcogenes
Bacteroides thetaiotaomicron
Bifidobacterium adolescentis
Brachybacterium faecium
Butyrivibrio proteoclasticus
...
Sun Oct 09 22:30:20 +0000 2022@MattWFoster @Smith_Chem_Wisc My better safe than sorry list is
Acetobacter pasteurianus
Achromobacter xylosoxidans
Acidimicrobium ferrooxidans
Acidovorax avenae
Actinobacillus pleuropneumoniae
Actinobacillus succinogenes
Aggregatibacter actinomycetemcomitans
Agrobacterium vitis
Anabaena variabilis
...
Sun Oct 09 12:22:58 +0000 2022@byu_sam CODECs. They are as varied & oddly documented as bioinformatics CSV formats.
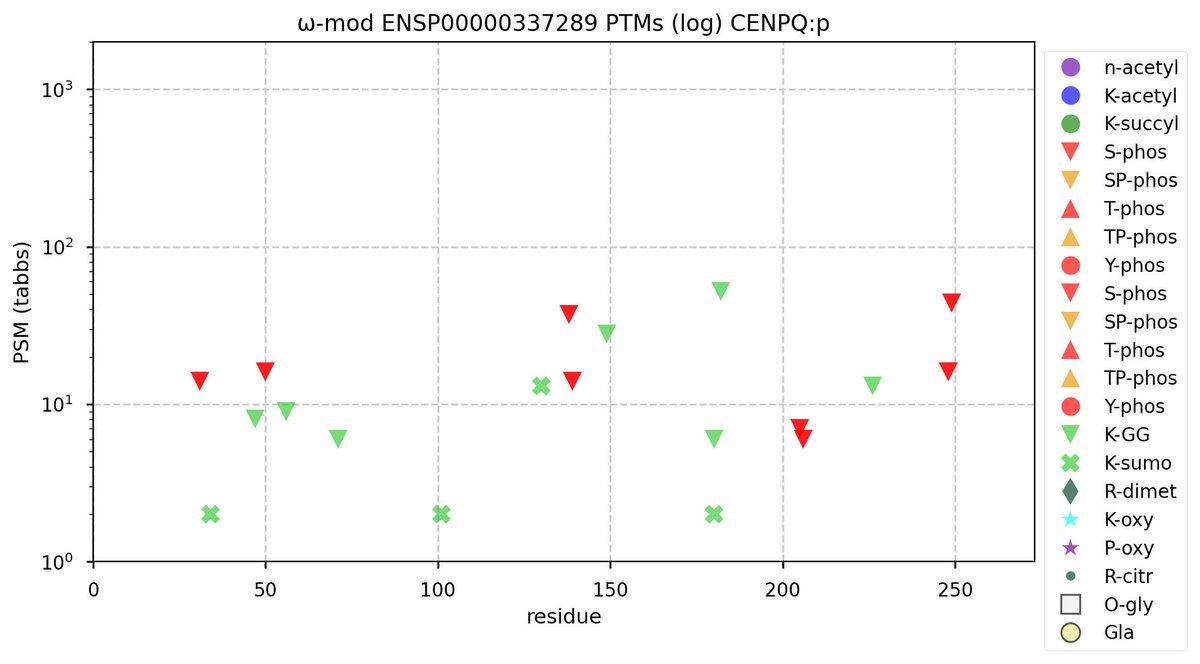
Sun Oct 09 11:37:47 +0000 2022Modification diagram for human CENPQ:p, a subunit of the centromere complex. Mainly individual K-mods:
1× K+GGyl/SUMO2yl,
3× K+SUMO2yl, &
6× K+GGyl.
As with most centromere subunits, K+acetyl is absent. The 8×S+phosphoryl acceptors are grouped into 5 domains, with 3 NxΦ motifs. 🔗
Sat Oct 08 15:29:07 +0000 2022@LewisGeer @pwilmarth @VATVSLPR In keeping with the field's primary maxim:
"Bioinformatics abhors a standard."
Sat Oct 08 13:55:53 +0000 2022@pwilmarth @VATVSLPR However, FASTA has resisted standardization since it originally came on the scene. There have been lots of efforts to rein it in, but none have managed to tame it.
Sat Oct 08 13:09:05 +0000 2022@pwilmarth I suspect it did, but it may have been a DECwriter artifact.
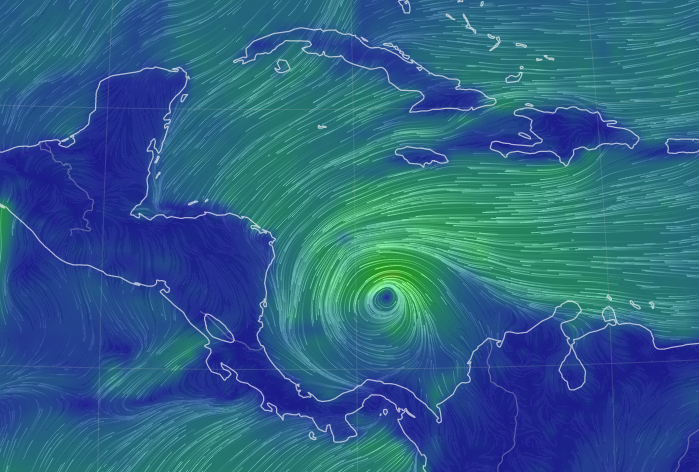
Sat Oct 08 12:27:08 +0000 2022Tropical Storm Julia is expected to make landfall on Nicaragua's Mosquito Coast as a hurricane on Sunday morning. 🔗
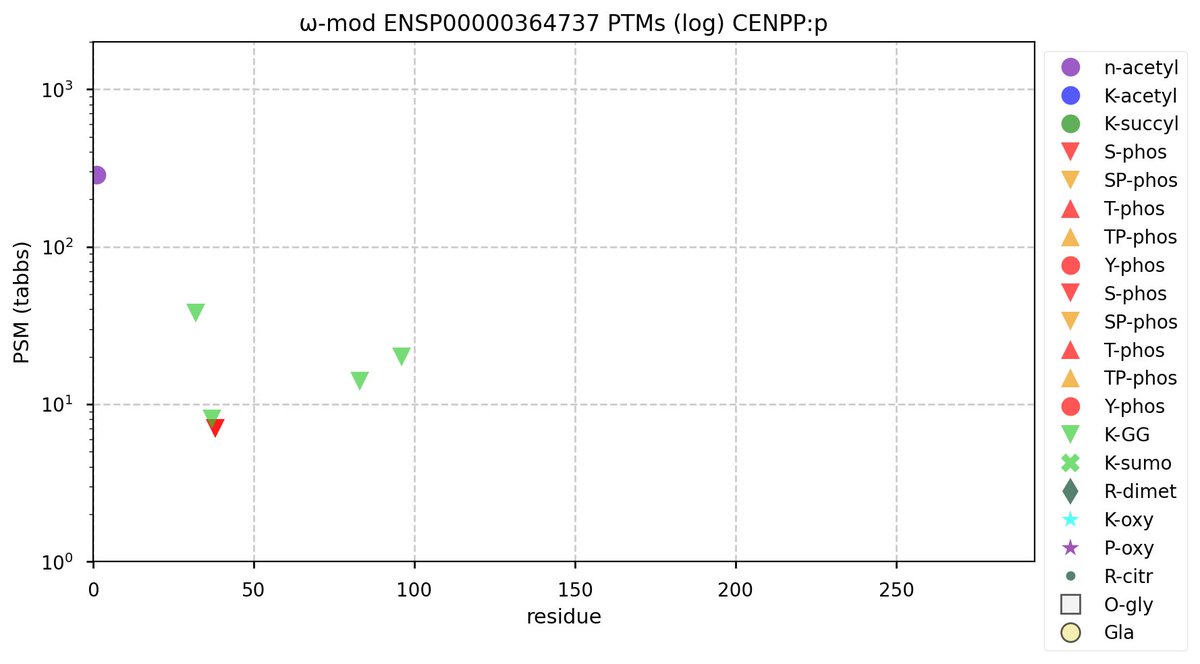
Sat Oct 08 12:11:46 +0000 2022Modification diagram for CENPP;p, a subunit of the centromere complex. The four K+GGyl acceptors share a ζ-K-ζ motif (ζ = hydrophilic residue). 🔗
Fri Oct 07 17:56:41 +0000 2022@neely615 @wfondrie @thabangh I'm pretty sure that the DECwriter (which replaced cards) didn't have lower case characters either.
Fri Oct 07 17:42:11 +0000 2022@neely615 @wfondrie @thabangh It was originally on punch cards, which didn't have lower case characters.
Fri Oct 07 14:51:05 +0000 2022@byu_sam Thanks, Sam. I was just wondering if Un!pr*t had taken any steps towards linking their sequence info with the underlying genome coordinates: I guess the answer is "no".
Fri Oct 07 11:56:09 +0000 2022@ItsBiniR @ProteomicsNews @sabbatiello0310 Comparing a new method to a previous method is a predominant lietmotif in the proteomics literature. New recipes are necessary if you want to publish in JPR, MCP, Nature Biotech, Nature Methods, etc.
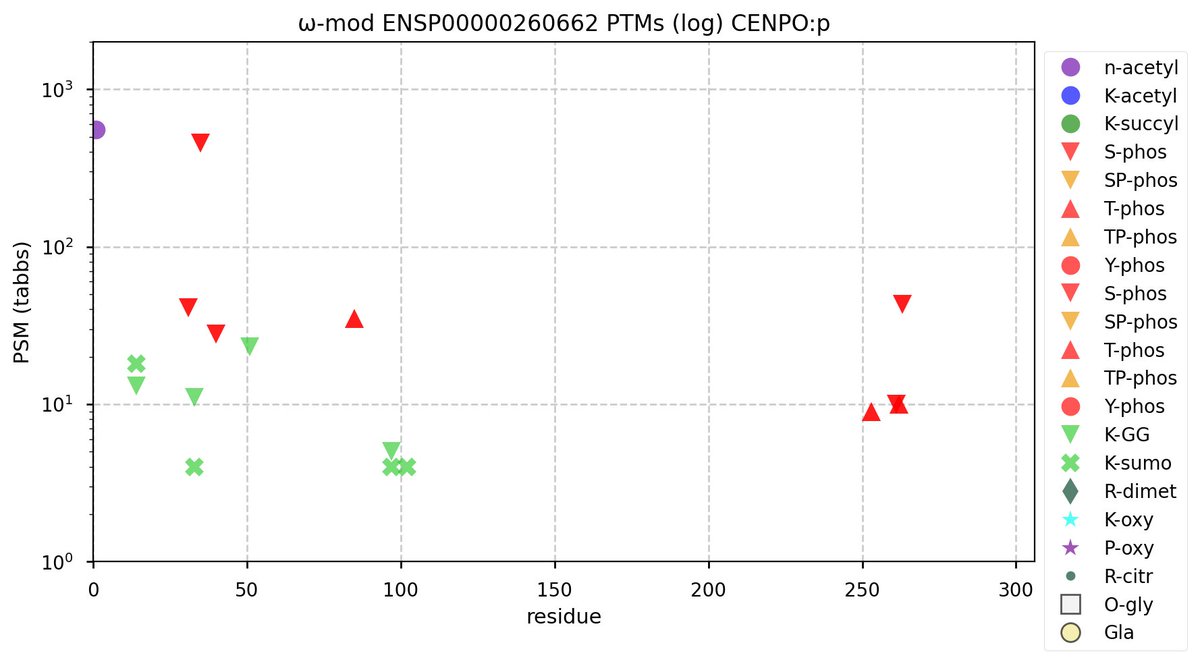
Fri Oct 07 11:52:01 +0000 2022Modification diagram for CENPO:p, a subunit of the centromere complex. It has 3 iHOP domains with major acceptor types RxxΦ, NxΦ & ΦxxD/E. Lysine acceptors are a GGyl/SUMO2yl mix: 1×K+GGyl, 1×K+SUMO2yl & 3×K+GGyl/SUMO2yl. 🔗
Fri Oct 07 11:41:25 +0000 2022@ProteomicsNews @ItsBiniR @sabbatiello0310 Most proteomics PIs value improvisation over just playing the notes.
Fri Oct 07 11:28:21 +0000 2022@KVittingSeerup CPTAC Lung Adenocarcinoma (LUAD) Discovery Study
Thu Oct 06 22:50:10 +0000 2022For Uniprot users: is there some way to display where exon boundaries are in a sequence?
Thu Oct 06 17:33:35 +0000 2022@nesvilab @oleg8r @YMerbl D-to-E or N-to-Q SAAVs fill the bill for D/N, but once again they would be too rare to show up here.
Thu Oct 06 17:29:16 +0000 2022@nesvilab @oleg8r @YMerbl That being said, I don't know of any biological mechanism to generate D/E/N/Q+methyl either.
Thu Oct 06 17:05:36 +0000 2022@nesvilab @oleg8r @YMerbl Maybe I am just a bit dense about this stuff, but I don't know what you are trying to say. There is no biological/artifact mechanism that I know of that can generate L+methyl. The nearest thing to it would be an L to Q SAAV, but that would be too rare to show up here.
Thu Oct 06 16:18:32 +0000 2022@nesvilab @YMerbl How does leucine get methylated?
Thu Oct 06 15:51:10 +0000 2022@Smith_Chem_Wisc No.
Thu Oct 06 15:02:05 +0000 2022It may be a cancer-thing, though, as many human MHC studies are done on cancer tissue or cancer-derived cell lines.
Thu Oct 06 14:59:56 +0000 2022Worthy of note―but apropos of nothing―most centromere subunits show up more prominently than one would expect in MHC class I peptide experiments.
Thu Oct 06 14:31:47 +0000 2022PXD032003: another good example of the pervasive infestation of XMRV in lab mice. At this point, it is close to being an endogenous retrovirus in the little critters.
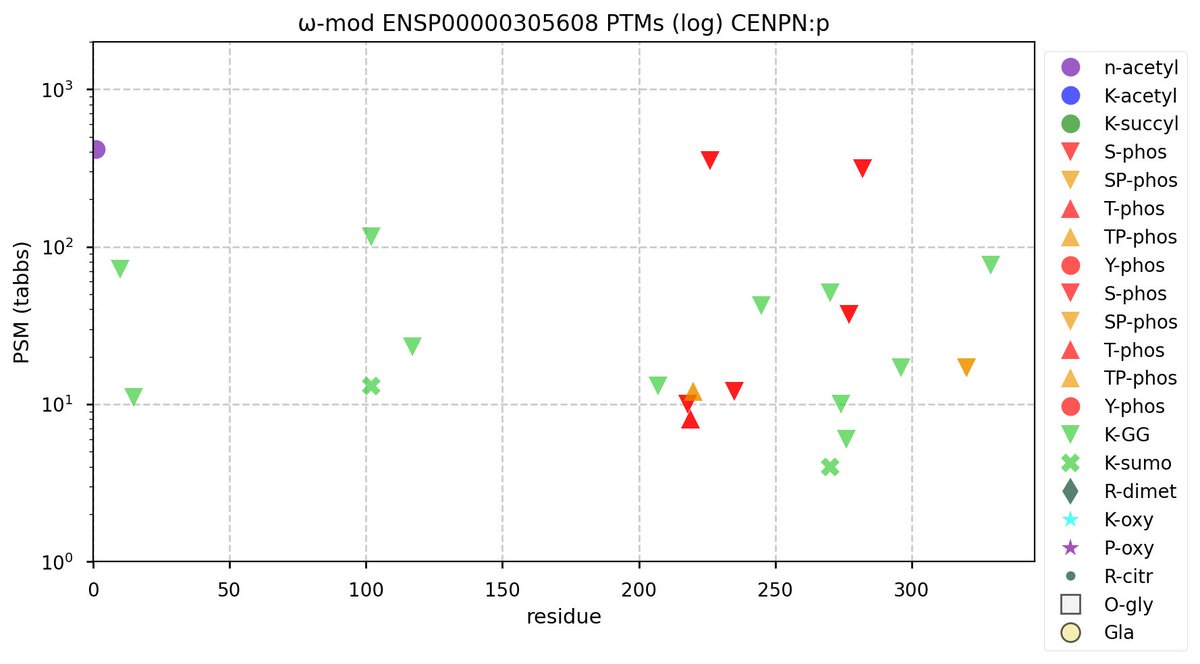
Thu Oct 06 12:34:41 +0000 2022Modification diagram for human CENPN:p, a subunit of the centromere complex. It has 2 high occupancy S+phosphoryl acceptors with different motifs: ΦL/I/V and NxΦ. It also has 2×K+GGyl/SUMOyl &
9×K+GGyl acceptors. 🔗
Wed Oct 05 20:16:05 +0000 2022@pwilmarth I wouldn't rule out Harvard, Oxford & Dartmouth.
Wed Oct 05 16:36:43 +0000 2022PXD034897 has an unusually prominent pattern of peptide isoelectric points in each of the LC/MS runs, because of the way the high pH fractions were mixed.
Wed Oct 05 15:35:34 +0000 2022@byu_sam They are pretty common but episodic: you need good coverage of any sequence to see them. When you have abundant proteins, like CES1, CPS1 or PDIA4 in liver tissue, you see them all the time. If you are getting an protein id'd with 2 or 3 peptides, though, you have to be lucky.
Wed Oct 05 14:01:23 +0000 2022Email is the worst thing to ever happen to academics. 2nd worst is any web site the administration describes as a "portal". 🔗
Wed Oct 05 13:58:43 +0000 2022@chrashwood @VATVSLPR @Smith_Chem_Wisc I don't doubt that some good stuff has resulted, but only being able to point out 5 or 6 counter-examples from the hundreds (or thousands) of projects announced in the literature over the last 20 years kind of supports for my original assertion.
Wed Oct 05 13:10:22 +0000 2022@byu_sam @KVittingSeerup @MichaelTress @thabangh That is the dominant zeitgeist in the field. Everyone seems nervous that if they look around a bit more thoroughly they may find something they don't like.
Wed Oct 05 12:45:22 +0000 2022This is a really bad idea. 🔗
Wed Oct 05 12:14:16 +0000 2022Although it gets a bit more interesting if you exclude everything other than experimental evidence 🔗
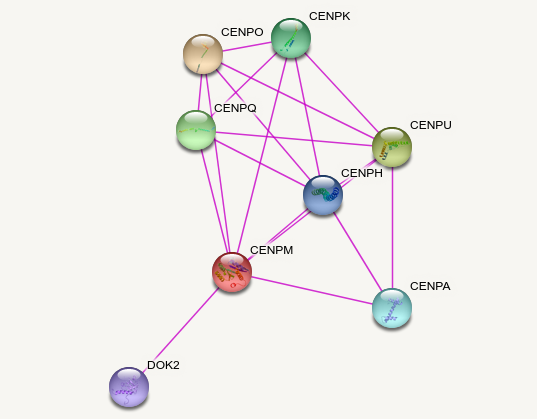
Wed Oct 05 12:12:23 +0000 2022Kind of predictable protein-protein interaction diagram (from STRING) 🔗
Wed Oct 05 11:48:36 +0000 2022Modification diagram for human CENPM:p, a subunit involved in the centromere complex.
🔗
Wed Oct 05 00:11:52 +0000 2022@dexivoje Those guys were hilarious back when I was faculty there.
Tue Oct 04 20:47:43 +0000 2022@gangulyteena Never had it, never wanted it. You can try selling it back to France, though.
Tue Oct 04 19:44:33 +0000 2022@dtabb73 @amazon I spent an hour yesterday walking to a specialty battery store to avoid ordering from those guys.
Tue Oct 04 16:49:14 +0000 2022This virus is a bit of an odd duck: no one has ever deliberately studied it, but it volunteers in samples so often that there is quite a bit known about its translated gene products, especially their S/T/Y+phosphoryl acceptor patterns.
Tue Oct 04 16:07:18 +0000 2022@dtabb73 @chrashwood @Smith_Chem_Wisc I was surprised when they managed to get several manufacturers to allow them to distribute proprietary file access APIs with their package, something that hadn't been allowed previously.
Tue Oct 04 15:08:30 +0000 2022PXD029659 has some very nice examples of protein data from the always unexpected Parainfluenza virus 5.
Tue Oct 04 14:56:09 +0000 2022Equally bad in both English et française
Tue Oct 04 14:50:24 +0000 2022The worst example of signing I see regularly. To open the door, you have to put your hand on the small hand in the center. The rest of the yellow area is inert. Almost nobody gets this right, other than by accident when they are frantically trying to get the door open. 🔗

Tue Oct 04 14:14:05 +0000 2022The same idea has been the bedrock of academic software projects for many years. 🔗
Tue Oct 04 13:48:54 +0000 2022Only an hour left to weigh in. Not a very pro-AF response so far! 🔗
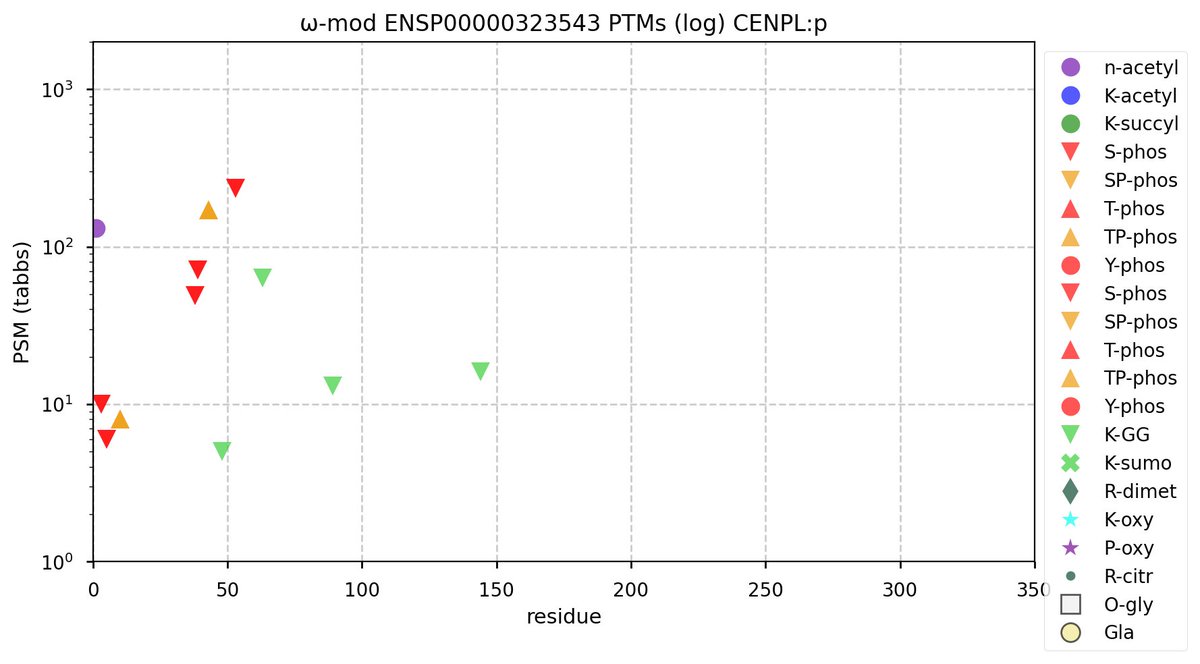
Tue Oct 04 12:03:54 +0000 2022Modification diagram for human CENPL:p, part of the centromere complex. The pattern rhymes with CENPI:p & CENPK:p, with an N-terminal phosphorylation arpeggio followed by a GGylation stanza. Acetyl & SUMOyl are conspicuous by their absence. 🔗
Tue Oct 04 11:56:47 +0000 2022I am very much in favour of the European idea of age limits for professors. The North American idea of letting profs hang around until they have to be forced out is cruel.
Mon Oct 03 15:19:32 +0000 2022Across the street I am seeing an oft-observed syllogism play out:
craft table + teamsters = movie shoot
Mon Oct 03 14:53:19 +0000 2022Does anyone think this structure is correct?
Mon Oct 03 14:29:31 +0000 2022Lysine modification domains are more like a bunch of these strung together. 🔗

Mon Oct 03 14:21:39 +0000 2022My phone's software update is promising me that this time, they got it right (unlike all of the previous times).
Mon Oct 03 12:02:57 +0000 2022I really don't like science prizes: we are in the Nobel Prize promotion season, which is particularly annoying.
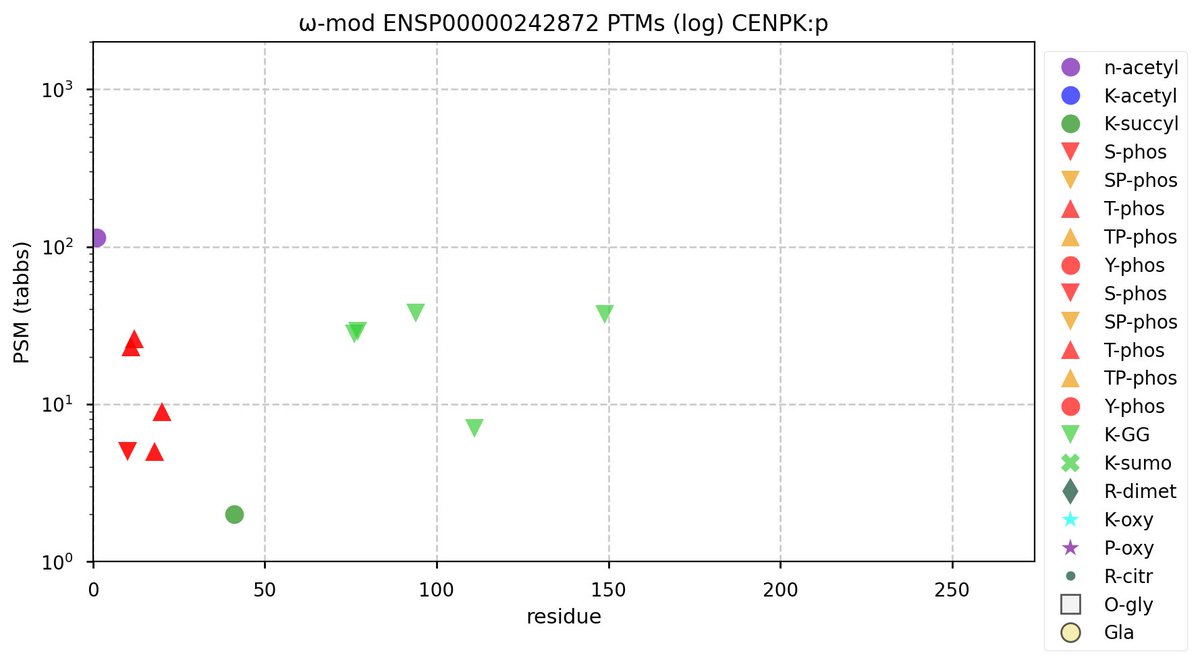
Mon Oct 03 11:58:20 +0000 2022Modification diagram for human CENPK:p, a centromere complex subunit. N-terminal phosphodomain is similar to the one in CENPI:p, but with T acting as the major acceptor type. 🔗
Sun Oct 02 13:38:55 +0000 2022When I think about phosphodomains, this is the electrical analogue that comes to mind. 🔗
Sun Oct 02 12:15:39 +0000 2022@LewisGeer Things have gone surprisingly well: the box has been working great on all 16 cores & no problems with any of the Bluetooth peripherals. Now I need some thick black tape to cover up the annoying logos on the case.
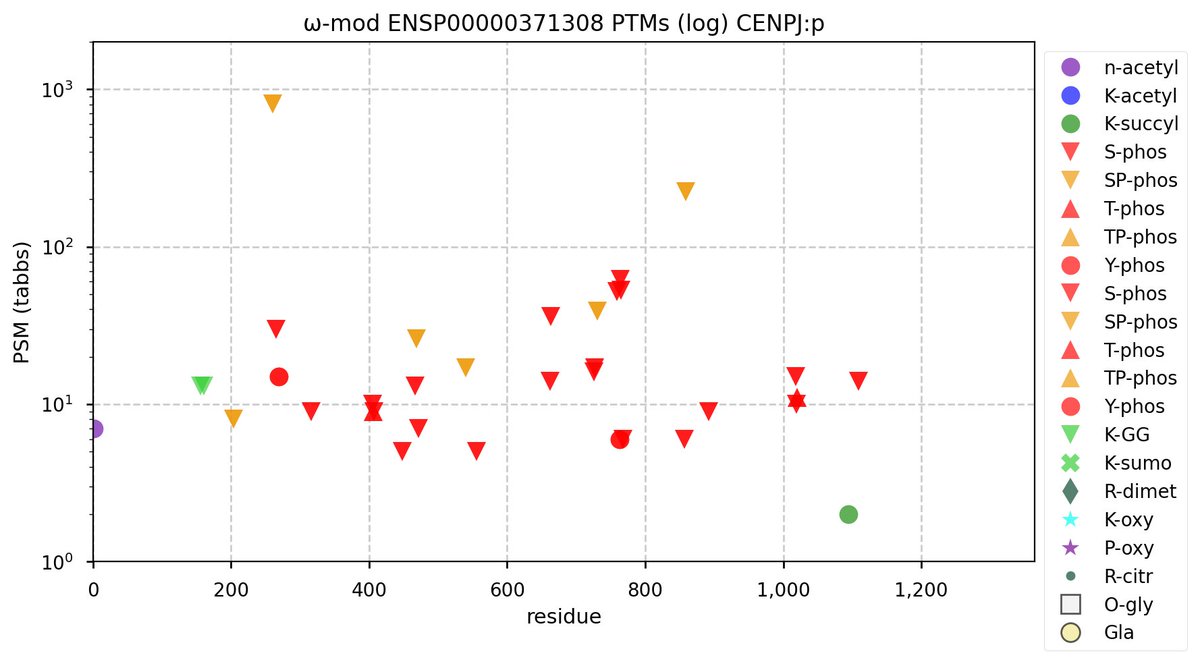
Sun Oct 02 12:08:05 +0000 2022Modification diagram for CENPJ:p, a transitory part of the centromere complex, as well as the centrosome & centriole complexes. WARNING: the pattern 27×S, 2×T+phosphoryl may cause some to quickly look away, muttering pejoratives, e.g., "junk", "noise" or "background". 🔗
Sat Oct 01 20:32:51 +0000 2022@statesdj @Noahpinion Everything was going well for them in Afghanistan, too, until they left. Same for the British (19th century) & US (21st century).
Sat Oct 01 20:20:23 +0000 2022Every time I look at some data, I feel I should thank Wade Hines for showing me SVG, back when everybody else was telling me that I was crazy for not using Flash.
Sat Oct 01 17:05:07 +0000 2022@LewisGeer Thanks for the advice. I've gone to the store, bought the device, went through the Windows install, updated the BIOS & installed Ubuntu 22. So far, so good.
Sat Oct 01 14:21:19 +0000 2022I'm planning to get a ThinkPad E15 Gen 3 to use as an Ubuntu laptop, replacing my last functional Win 10 box. Any warnings?
Sat Oct 01 14:00:14 +0000 2022I have never trusted a "scientist" who can't cook. To me, the two things are intimately related.
Sat Oct 01 12:11:43 +0000 2022K+acetyl, K+SUMOyl, K+succinyl, O-glycosyl, R+dimethyl, R-deimidation: you are missed. 😞
Sat Oct 01 11:46:27 +0000 2022Modification diagram for CENPI:p, part of the centromere complex. Nearly textbook pattern with 5×K+GGyl & 8×S, 3×T,0×Y+phosphoryl in 3 phosphodomains. The 2 high occupancy ΦP motif acceptors are flanked by low occupancy ΦL/I/V motif acceptors.
🔗
Fri Sep 30 14:31:28 +0000 2022@Smith_Chem_Wisc They also have trouble with litotes, antiphrasis, hyperbole, satire, sarcasm & pretty much any other literary device that makes language worth reading.
Fri Sep 30 14:23:09 +0000 2022@Smith_Chem_Wisc Irony, like negation, are difficult concepts for keyword pattern matching to deal with effectively.
Fri Sep 30 14:15:32 +0000 2022Dear Twitter, once again I feel that I should mention to you that your "Based on your Retweets" recommendations are annoying & contain nothing I would ever be interested in seeing. Please stop making them.
Fri Sep 30 14:01:24 +0000 2022@kyle_minogue No better at the councilor level, at least for me.
Fri Sep 30 13:00:54 +0000 2022@harrisonspecht @aleks_petelski @_AndrewLeduc Inline documentation is the only thing that I find helpful, as pseudocode usually doesn't state where a particular statement is expressed in the code. The other thing I look at immediately is the use of try-catch blocks: a lot of prototype code relies rather heavily on them.
Fri Sep 30 12:40:00 +0000 2022@harrisonspecht @aleks_petelski @_AndrewLeduc Pseudocode has a tendency to describe "what we were hoping to do" rather than "what was done".
Fri Sep 30 12:29:24 +0000 2022@harrisonspecht @aleks_petelski @_AndrewLeduc I find UML to be more helpful than arbitrarily constructed pseudocode.
Fri Sep 30 12:10:48 +0000 2022Quid pretii users?
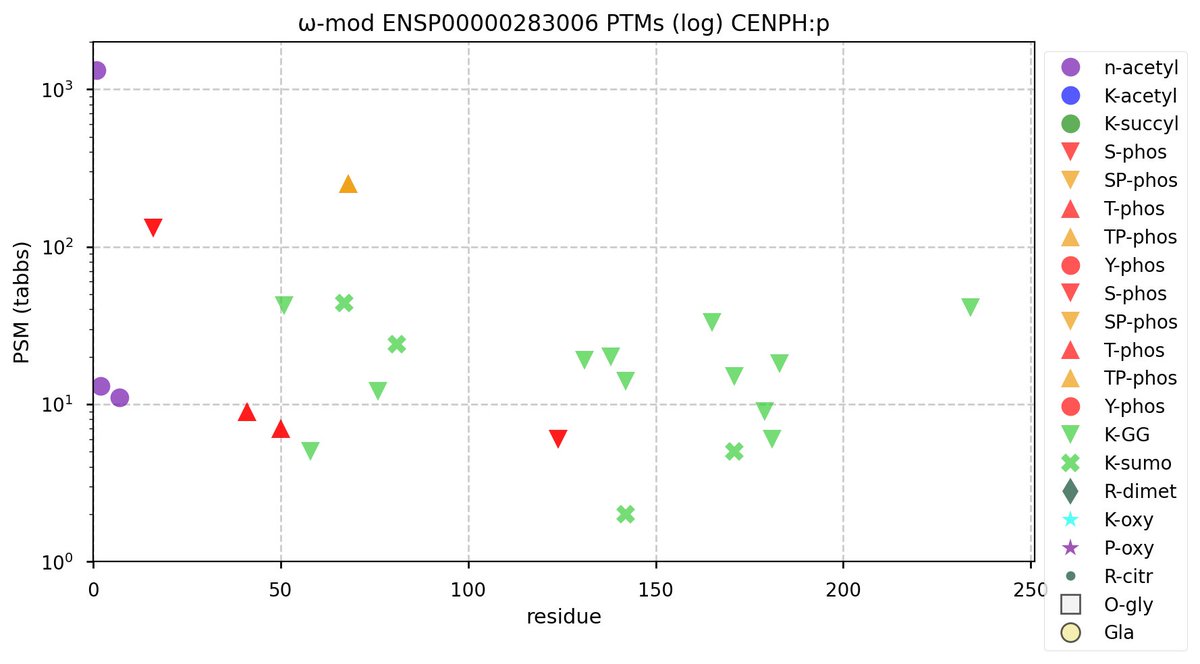
Fri Sep 30 12:02:45 +0000 2022K+acetyl acceptors are conspicuous by their absence.
Fri Sep 30 11:53:54 +0000 2022The K-mod pattern is more abundant than it should be (½ of available K's are observed acceptors), but people don't seem very emotionally invested in small protein tags:
2× K+SUMOyl (68 Ta, 22.0%)
2× K+GGyl/SUMOyl (36 Ta, 11.7%)
10× K+GGyl (205 Ta, 66.3%)
Fri Sep 30 11:49:43 +0000 2022Modification diagram for human CENPH:p, part of the centromere complex. The S/T+phosphoryl pattern could have been in Lehninger:
1× ΦxxD/E (131 Ta, 32.4%)
1× ΦL/I/V (9 Ta, 2.2%)
1× RxxΦ (7 Ta, 1.7%)
1× ΦP (251 Ta, 62.1%)
1× RxxΦL/I/V (6 Ta, 1.5%) 🔗
Thu Sep 29 21:23:08 +0000 2022@camtflower @pwilmarth I actually prefer the original to the homage:
🔗
Thu Sep 29 16:18:40 +0000 2022Are there lab mice without an active XMRV infection? It has been a while since I've seen mouse tissue sample proteomics data without XMRV proteins.
Thu Sep 29 15:42:37 +0000 2022@JohnLeesonTO @AlexUsherHESA It looks like the racoon problem is more serious than I thought.
Thu Sep 29 15:17:18 +0000 2022Just sitting here wondering why E. histolytica prefers to use S > T > A for its proteins' acetyl-blocked N-terminal residue, while mammals prefer A > S > T.
Thu Sep 29 14:13:12 +0000 2022@KristinaHakans2 I knew Per Håkansson as a colleague for more than a decade when I was involved in desorption research. Careful scientist & one of the most optimistic people I have ever met.
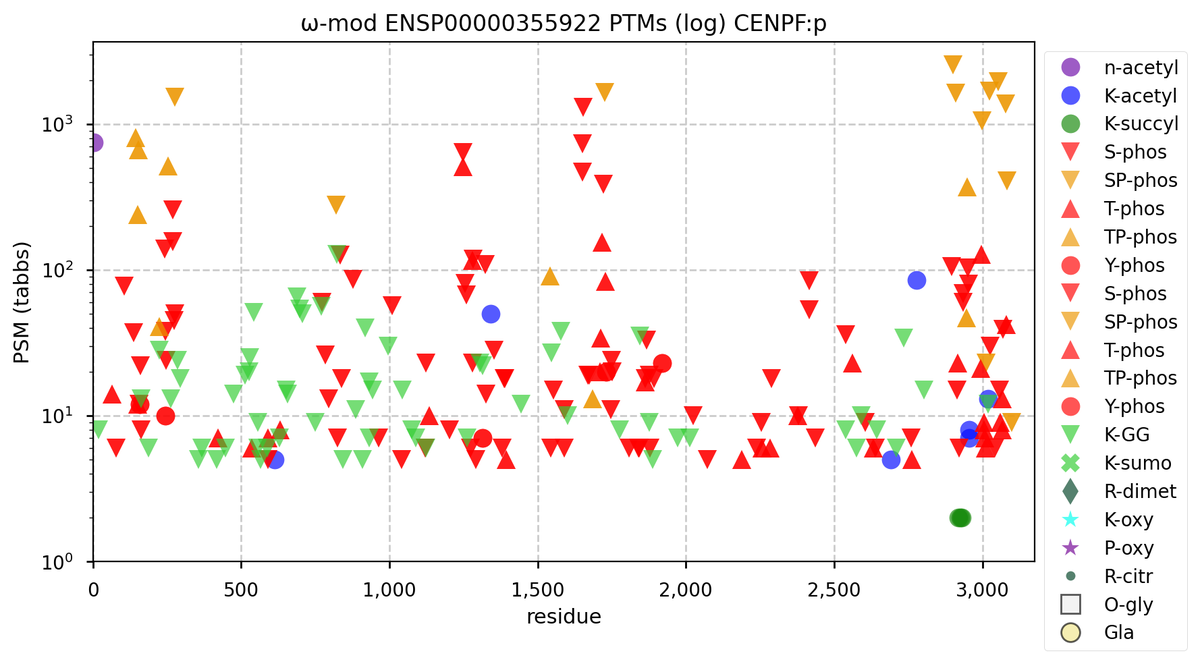
Thu Sep 29 11:56:12 +0000 2022The big noise on this one is S/T+phosphoryl acceptors, clustered into multiple domains. 82% of observed occupied acceptors are associated with ΦP motifs, including 1× ΦPxK, 2× RxxΦP, 2× PxΦP, 2× GxxΦP & 14× ΦP. Of the lower occupation acceptors, 11 are ΦL/I/V & 19 are ΦxxD/E.
Thu Sep 29 11:46:54 +0000 2022The sequence is broadly decorated by K+GGyl acceptors, but few K+acetyl and no K+SUMOyl:
1× K+acetyl/GGyl (1.9%)
6× K+acetyl (12.0%)
61× K+GGyl (86.1%)
Thu Sep 29 11:43:12 +0000 2022Modification diagram for human CENPF1:p, a cell-cycle dependent subunit of the centromere complex. WARNING: this sequence is an example of non-canonical PTMs that some biomedical researchers may find disturbing. 🔗
Wed Sep 28 22:59:18 +0000 2022@cajun_science @byu_sam And don't forget alternate initiation: lots of data & plenty of publications, too.
Wed Sep 28 18:33:43 +0000 2022Hurricane Ian comes to Punta Gorda Airport (FL) in 2 graphs. 🔗
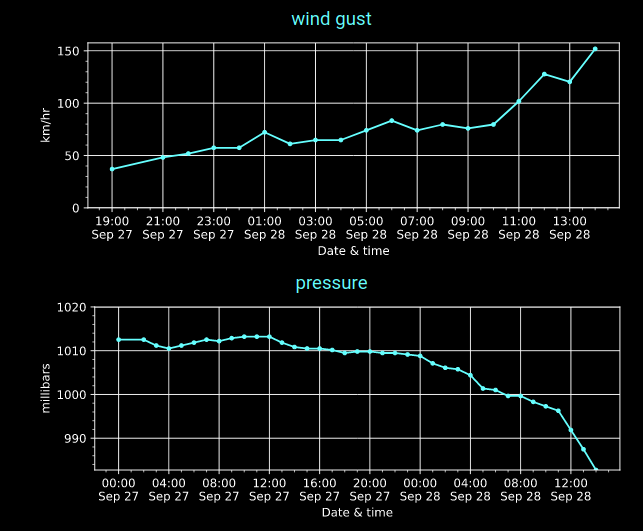
Wed Sep 28 15:36:41 +0000 2022Quite a few weather stations in the Fort Myers/Cape Coral area have already recorded over 130 mm of rain (~5 inches) in the last 24 hours.
Wed Sep 28 14:21:48 +0000 2022@MaartenvSmeden Try to use "Manichean" as often as possible during these discussions, as it lends a hint of superior erudition & linkage to dark arts of the past.
Wed Sep 28 14:17:47 +0000 2022@stphnmaher Public policy fits nicely into an envelope provided by the insurance & bond markets: everything else is terra incognita.
Wed Sep 28 14:14:13 +0000 2022@stphnmaher When insurance companies make it so.
Wed Sep 28 14:09:51 +0000 2022@astacus @winstanley_c Our weather bunch here in the colonies―Environment & Climate Change Canada―provide next to nothing to help people with solar installations.
Wed Sep 28 14:07:56 +0000 2022@astacus @winstanley_c Does the UK Met Office offer any help to people who want solar panel installations. They have a lot of data that would be of use in operating this type of system, particularly in forecasting usable insolation.
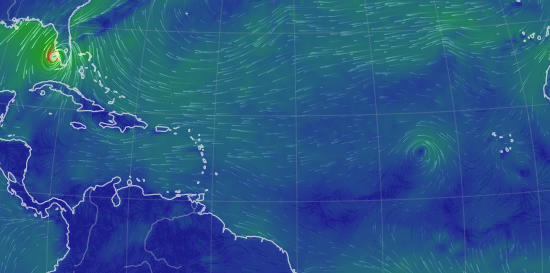
Wed Sep 28 13:58:19 +0000 2022And even as Ian is about to come ashore, there is another spinning low pressure system due east of Cape Verde (known for its islands & hurricanes). 🔗
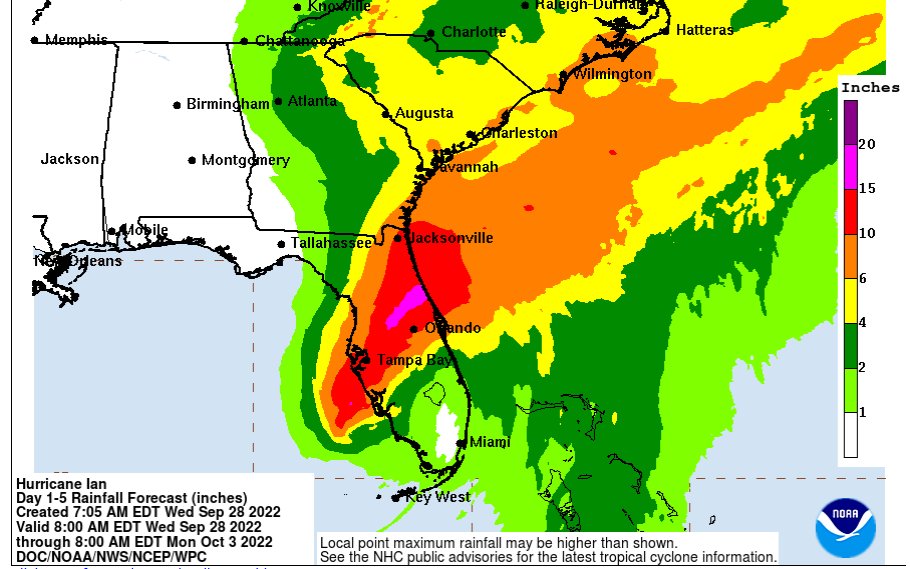
Wed Sep 28 13:52:57 +0000 202210-20 inches of rain (250-500 mm) in a few days is going to cause a lot of distress throughout central Florida. 🔗
Wed Sep 28 13:04:47 +0000 2022@MagnusPalmblad @byu_sam This wikipedia article does a pretty good job of lining out the tortured process by which the territory of the US evolved. Ohio was part of the "Northwest Territory" at one point.
🔗
Wed Sep 28 12:51:54 +0000 2022@JProteomeRes The beginning of the end for OS search algorithms.
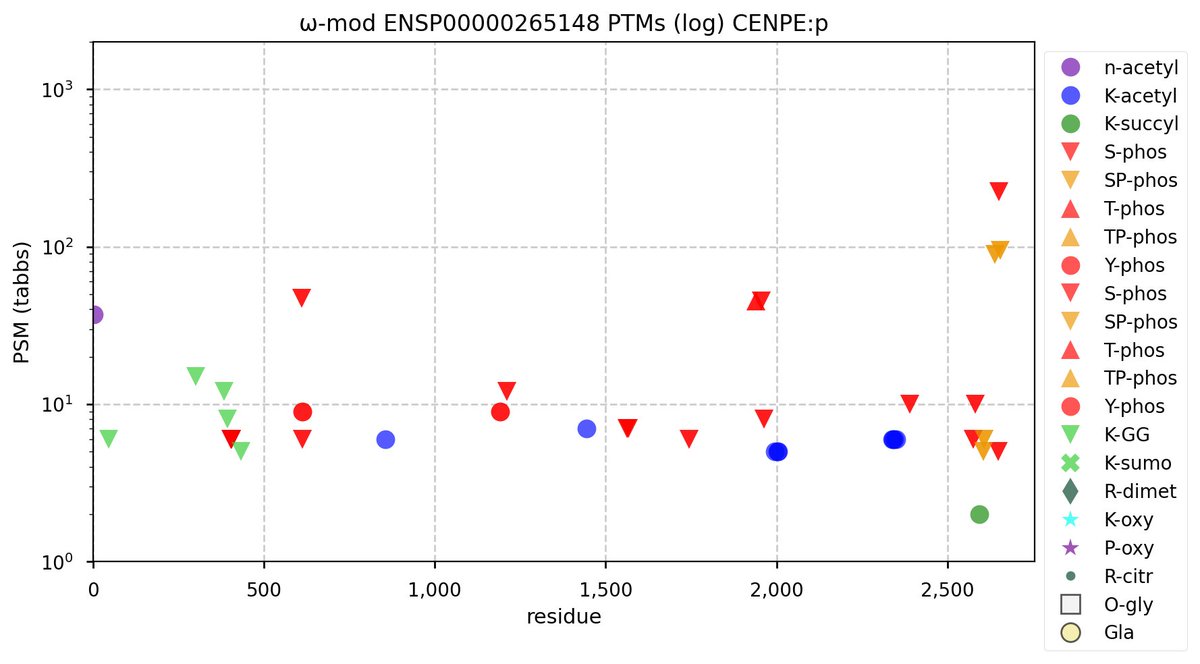
Wed Sep 28 12:13:57 +0000 2022PTM diagram for human CENPE:p, a large, cell-cycle dependent subunit of the centromere complex. It has a smattering of low occupancy acceptors, with 3 nearly canonical "phosphorylation sites". 🔗
Wed Sep 28 11:59:45 +0000 2022@neely615 @ron_alfa It is the same human failing known as "testing into compliance" in the pharma business. One of the legal concepts they bang into your head vigorously.
🔗
Wed Sep 28 02:27:29 +0000 2022Hurricane Ian via weather radar (NEXRAD). 🔗

Tue Sep 27 20:27:39 +0000 2022@astacus @winstanley_c It would appear to be a bit little less in the winter months, but predictable.
Tue Sep 27 13:56:16 +0000 2022@astacus @winstanley_c In tab-separated value format:
🔗
The times are in GMT, without correction for daylight-savings time.
Tue Sep 27 13:32:42 +0000 2022@astacus @winstanley_c There is a NOAA MADIS weather station that is probably pretty close to you. I have 5 years of solar radiation data (24 obs/day in W/m^2) for that station, if it would be of any interest to you. 🔗
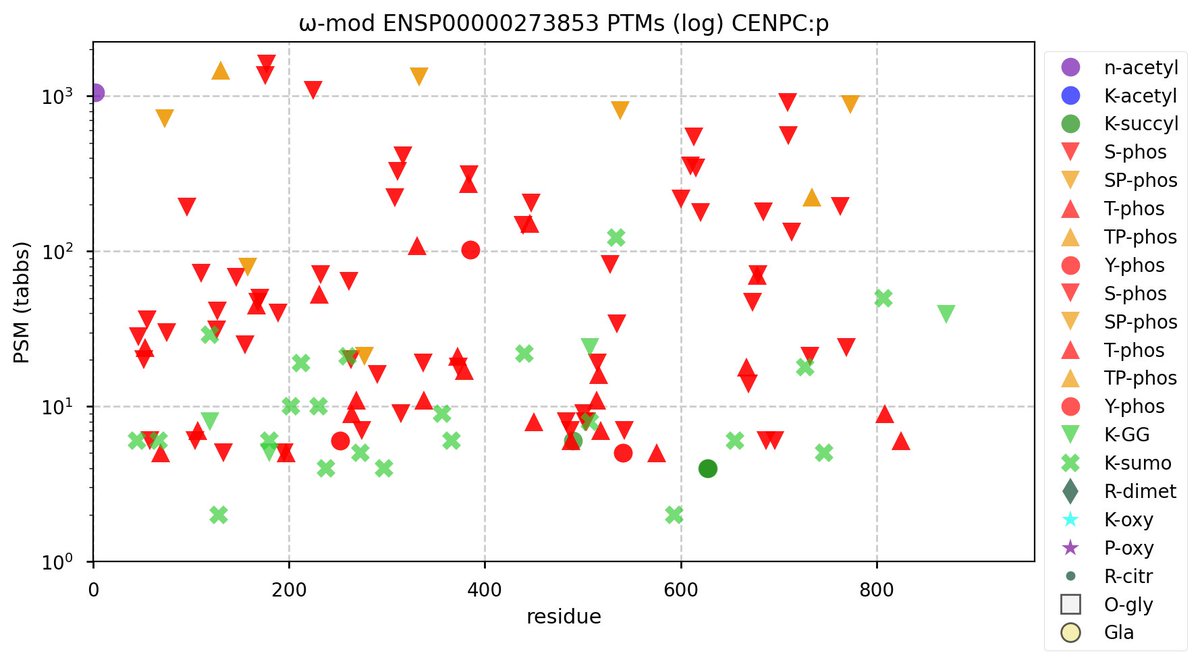
Tue Sep 27 12:01:53 +0000 2022The big show here is S/T+phosphoryl, scattered across a broad N-terminal Really Interesting Phosphodomain (RIP). Within the RIP, 3 acceptor site patterns dominate the observed occupancy: ΦP, ΦL/I/V & ΦxxD/E.
Tue Sep 27 11:58:06 +0000 2022PTM diagram for human CENPC:p, a subunit of the centromere complex. The lysine modification acceptor distribution points to a role for SUMO2 in CENPC:p function:
1× K+GGyl (39 Ta)
3× K+GGyl/SUMO2yl (80 Ta)
19× K+SUMO2yl (327 Ta) 🔗
Tue Sep 27 11:36:38 +0000 2022@astacus @winstanley_c How many square meters of solar panelling do you have?
Mon Sep 26 19:11:50 +0000 2022PXD019869: Welcome to Collagen City! 🐭
Mon Sep 26 17:48:27 +0000 2022PXD031451: sounds kind of interesting. I don't remember anyone using TMT labeling with MHC class I peptides before.
Mon Sep 26 17:34:41 +0000 2022PXD034970: if you really want to see how well you can do using not-the-exactly-right-strain (or species) genomes to interpret data, this one looking at S. aureus antibiotic resistant strains might be a good one to use for testing ideas.
Mon Sep 26 15:13:45 +0000 2022@neely615 @pwilmarth @uniprot Always amazed by how many bytes in those files are taken up by the repeated "Copyright" & "License" declarations.
Mon Sep 26 14:52:41 +0000 2022CENPD:p still shows up in many database entries describing centromere proteins referencing MIM 117142. If you go to 🔗, it has been corrected to point to 🔗, the CENPB:p entry.
Mon Sep 26 14:33:19 +0000 2022CENPB:g was mistakenly assigned the name CENPD:g as well. Once it was realized that they were the same gene, the name CENPD:g was retired (it is no longer used).
Mon Sep 26 14:21:48 +0000 2022I have come to the considered opinion that the structure of the federal & provincial governments of Canada renders them unable to create or execute any effective policy that encourages scientific research or technological innovation in either the public or private sectors.
Mon Sep 26 13:54:15 +0000 2022Typhoon Noru (Karding) has made its way across the Philippines, passing just north of Manila. It is now in the South China Sea and it will make landfall in Vietnam near Da Nang late tomorrow. 🔗
Mon Sep 26 12:45:22 +0000 2022The C-terminal portion of the sequence has multiple low-to-no complexity domains, e.g.
406 EEEEEEEEEEEEEE 419
515 EEEDDEEEDDEDEDDDDDEED 535
Mon Sep 26 12:02:02 +0000 2022Modification diagram for human CENPB:p, a subunit in the centromere complex. There are several discrete phosphodomains & some low occupancy K mods:
1× K+acetyl (5 Ta)
1× K+GGyl/SUMOyl (162 Ta)
1× K+acetyl/GGyl (14 Ta)
2× K+SUMOyl (16 Ta)
2× K+GGyl (38 Ta) 🔗
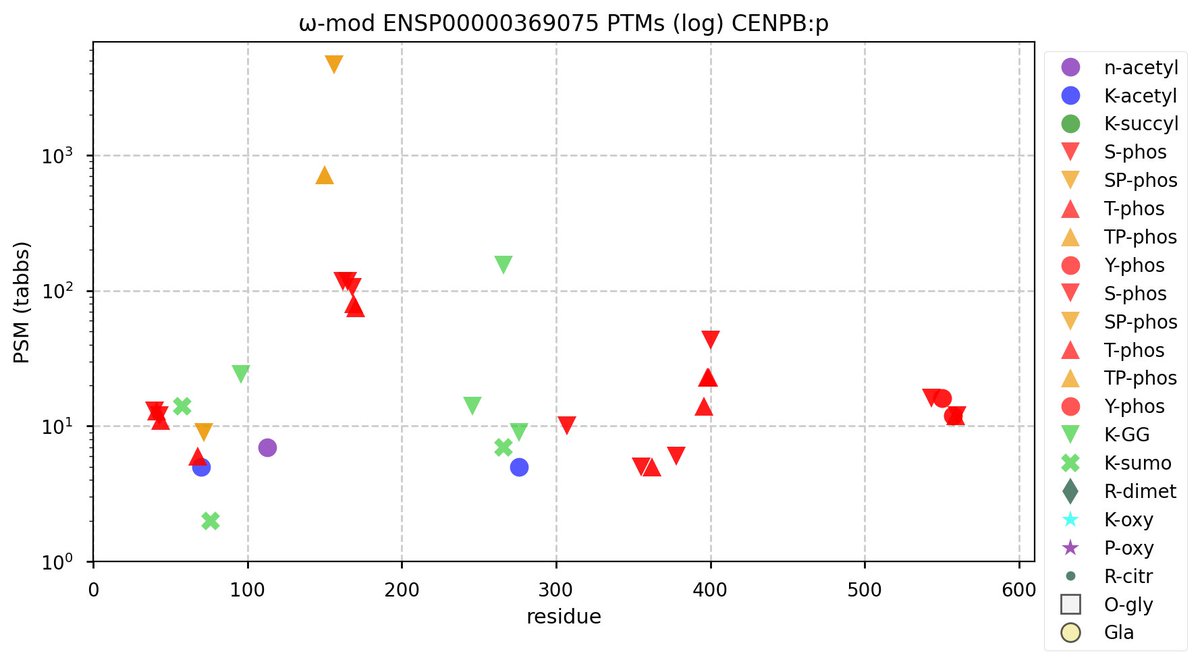
Sun Sep 25 19:33:47 +0000 2022I am still surprised by how little overall abundance has to do with how large a signal you get from a protein in MHC peptide experiments. Relatively rare sequences like PATJ:p or MARCH6:p are right up there with VIM:p or CAD:p.
Sun Sep 25 16:07:15 +0000 2022@YishaiLevin It really appears as if they are trying to discourage me from retweeting anything, via negative reinforcement.
Sun Sep 25 16:00:40 +0000 2022I need an automobile for about for about 1 hour every 2 weeks. There doesn't seem to be any viable economic model for providing automobiles that effectively meets my needs.
Sun Sep 25 15:34:03 +0000 2022@jwoodgett They are trying to make it clear than only truly dedicated career bureaucrats need apply.
Sun Sep 25 15:27:46 +0000 2022@chrashwood I would actually equate these entries with feedback on a PA system: an annoying squealing sound that makes it hard to listen.
Sun Sep 25 15:21:50 +0000 2022@chrashwood So do I, but these new entries―entitled something like "Based on your Retweets"―just started to show up a few days ago (or at least that is when I noticed them).
Sun Sep 25 14:41:15 +0000 2022The centromere complex is a visible feature of eukaryote chromosomes essential for proper mitosis. It is composed of some combination of ~26 subunits (CENPA-Z), changing with the cell cycle. It is often described as specialized chromatin, although it is a poor analogy.
Sun Sep 25 14:03:45 +0000 2022Canadian creativity in action
🔗
Sun Sep 25 13:56:30 +0000 2022@YishaiLevin Machine learning can't know anything.🥴
Sun Sep 25 13:25:27 +0000 2022@AlexUsherHESA @CADInnovators A glimpse into the Balsillie-brain.
Sun Sep 25 13:00:04 +0000 2022I suppose they are hoping I will actively assist them in training an algorithm to do something I don't want them to do in the first place. Nope.
Sun Sep 25 12:57:44 +0000 2022Twitter has started to show me things it thinks I will like. I don't like either seeing these things or any of the things I am seeing. Please stop.
Sun Sep 25 12:43:21 +0000 2022PXD024417: better than expected examples of class I and II MHC peptide data. Really shows off how the class II peptides reflect the extracellular environment (in this case FBS) and an eccentric collection of intracellular proteins.
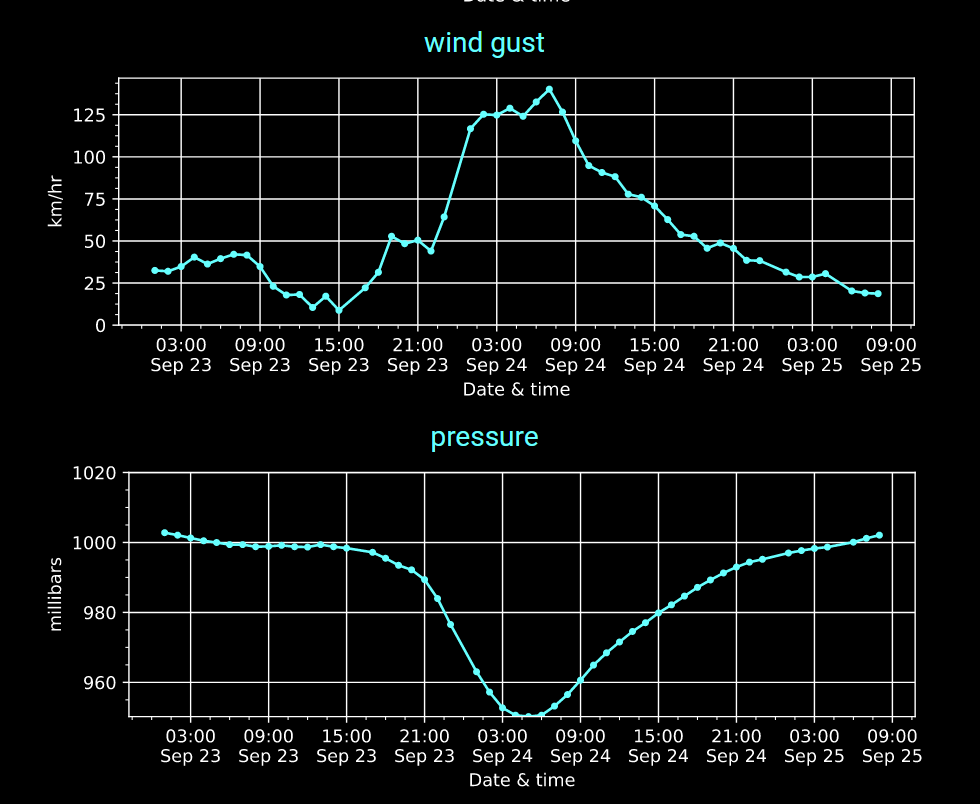
Sun Sep 25 12:17:55 +0000 2022Fiona comes ashore & departs Louisbourg, NS, in 2 graphs 🔗
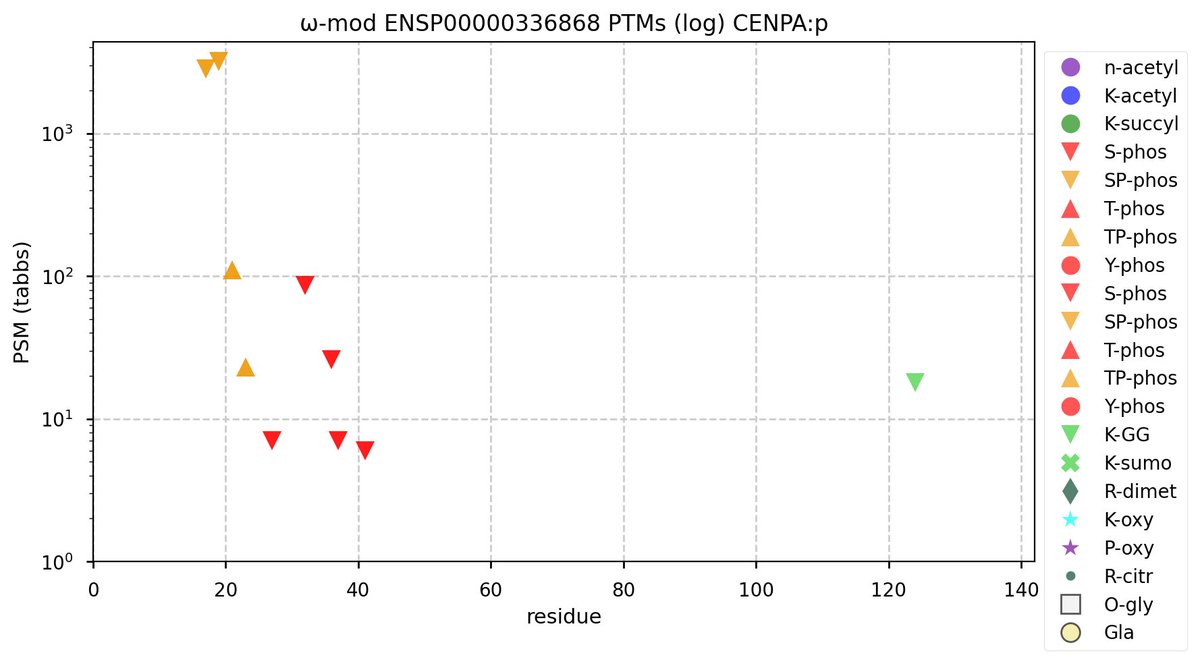
Sun Sep 25 12:08:56 +0000 2022Modification diagram for human CENPA:p, part of the centromere complex. S/T+phosphoryl acceptors on the N-terminal IDR and a singleton K+GGyl on the C-terminal H3-like domain. No observed acetylation or SUMOylation (or anything else for that matter). 🔗
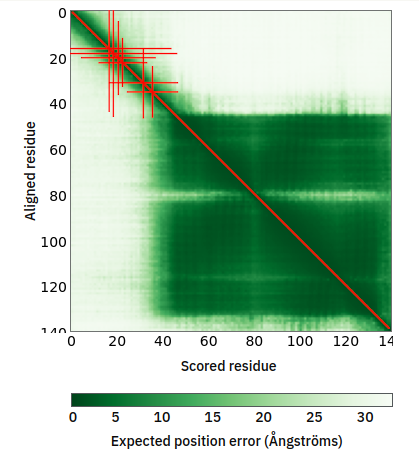
Sat Sep 24 16:44:35 +0000 2022@AlexUsherHESA @stephenjwild I wouldn't be quite so generous about the process. "One of us" is much higher on the list of priorities than "intellectual rigor" or "research excellence" when it comes to faculty hiring.
Sat Sep 24 14:48:26 +0000 2022I have left several faculty positions, but I've never had an exit interview. Once you say you are leaving, the whole place develops a certain type of protective amnesia about your ever having been there.
Sat Sep 24 14:35:30 +0000 2022Just to finish up my little stint on SUMO2ylated proteins: there is no known reason for multiple SUMO2 acceptors on particular proteins & no theoretical framework even to discuss SUMO2ylation sites that also serve as acetyl or ubiquitinyl acceptors.
Sat Sep 24 14:15:53 +0000 2022The CNN report on Fiona's landfall was better, with a good interview & much (much) better maps. CBC, in particular, was scrambling to find anyone to provide on the ground reports in a region they normally ignore.
Sat Sep 24 14:06:37 +0000 2022Canadian news channels are all showing some poor correspondent trying to stand still outside in a hurricane telling people how dangerous it is to be outside in a hurricane.
Sat Sep 24 13:31:45 +0000 2022Dear Windows 10/11, I have never used Cortana & I never will. Stop pestering me. No means No.
Sat Sep 24 13:22:29 +0000 2022Tropical Storm Ian is now expected to make landfall near Tampa (Florida) as a major hurricane some time on Sept. 28th. 🔗
Sat Sep 24 12:52:49 +0000 2022@AlexUsherHESA For the last 20 years, every advertised STEM faculty position has generated 100s of applications. The first step in dealing with all of this paperwork is a triage, in which "letterhead" is one of the factors, biasing the outcome towards a select set of institutions.
Sat Sep 24 12:27:20 +0000 2022This is ground station data from the Sydney Airport AWOS station, as reported by ECCC. 🔗
Sat Sep 24 12:23:31 +0000 2022It looks like Fiona is just passed the half-way point in Sydney, Nova Scotia 🔗
Sat Sep 24 11:58:57 +0000 2022Multiple S+phosphoryl isolated, high-occupancy phophorylation (iHOP) domains are nicely slotted in to the spaces between the 10 C2H2 zinc-finger domains.
Sat Sep 24 11:53:21 +0000 2022Modification diagram for human MECOM:p, a nuclear subunit. It is near the top of any protein list generated from SUMO2-enriched data, with
1× K+GGyl (6 Ta),
1× K+acetyl (11 Ta), and
44× K+SUMOyl (2714 Ta) acceptors. 🔗
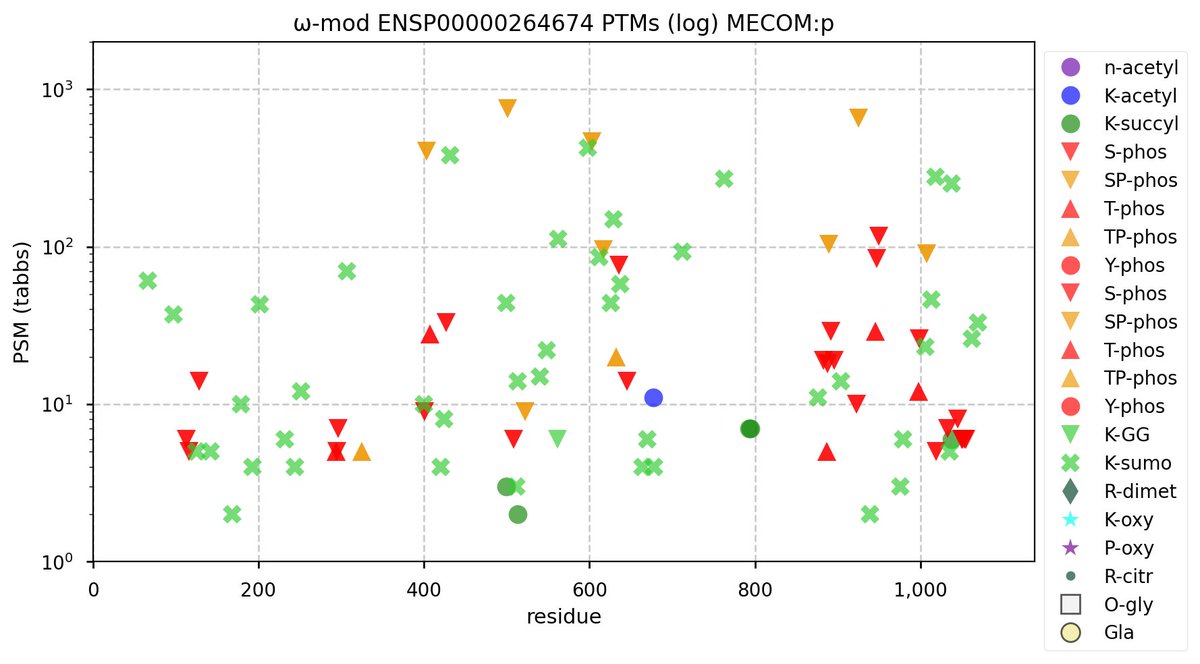
Fri Sep 23 18:21:39 +0000 2022Is there some technical reason that pulldown studies like PXD030720 do not reduce & block cysteines as part of the sample workup?
🔗
Fri Sep 23 15:01:55 +0000 2022The NOAA model is showing ~125 mm (5") of rain from now through 6 PM Saturday. 🔗
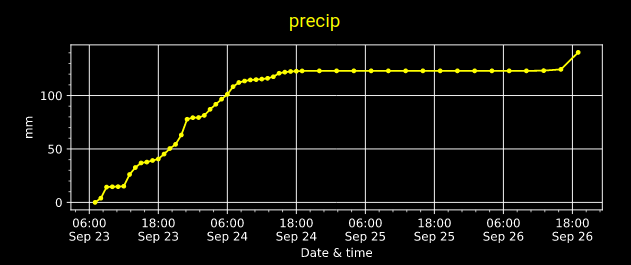
Fri Sep 23 14:36:20 +0000 2022@rabbleca Promoting the hilarious idea that Canadian provinces are really in competition with each other for "the best" people. Talent tends to pursue opportunity, not housing prices or access to Broadway touring companies.
Fri Sep 23 14:25:56 +0000 2022No-complexity domains are scattered throughout the proteome, like little black obelisks waiting for the apes to notice them & wonder.
Fri Sep 23 14:08:55 +0000 2022Predicted sustained wind speeds for Louisbourg, Nova Scotia. It should be >80 km/hr for ~ 12 hours. 🔗
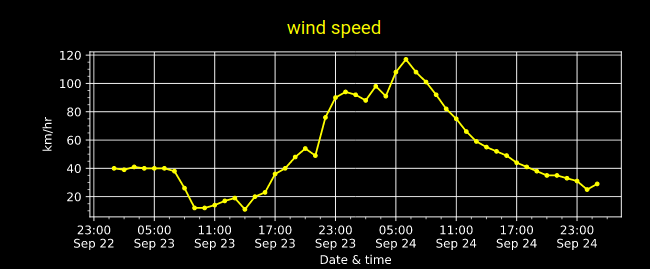
Fri Sep 23 13:23:59 +0000 2022@pwilmarth There is a lot known about the frequency of protein observation. It is complicated and requires attention to the details of protein translation, transport, modification & recycling. These projects are great subjects for a "white paper", but are soon forgotten.
Fri Sep 23 13:01:25 +0000 2022@pwilmarth I have lost count of the number of "projects" taking aim at straw men constructed out of the idea of "missing" proteins. I consider it to be an intellectually lazy, impractical approach.
Fri Sep 23 12:54:36 +0000 2022@pwilmarth After looking at the web site & pondering the organizing committee, I expect nothing to come of this project.
🔗
Fri Sep 23 12:30:25 +0000 2022Fiona is now expected to make landfall somewhere between Ecum Secum & Canso (NS) tonight around midnight as a category 3 hurricane. It should then head north & weaken, passing near Happy Valley/Goose Bay (NL) around noon on Sunday with 85 km/hr winds.
Fri Sep 23 12:19:50 +0000 2022@BioDataGanache Nope, just common sense. Good for you.
Fri Sep 23 12:03:43 +0000 2022The acceptor pattern changes dramatically following a no-complexity domain (389-400)
...HHHHHHHHHHHH...
Fri Sep 23 12:01:39 +0000 2022Modification diagram for CBX4:p, a small subunit the acts as an E3-ligase. Lots of K-mod acceptors with alternate PTMs:
2× K+acetyl/GGyl/SUMOyl (354 Ta)
3× K+GGyl/SUMOyl (319 Ta)
9× K+acetyl (86 Ta)
10× K+acetyl/SUMOyl (913 Ta)
11× K+SUMOyl (267 Ta) 🔗
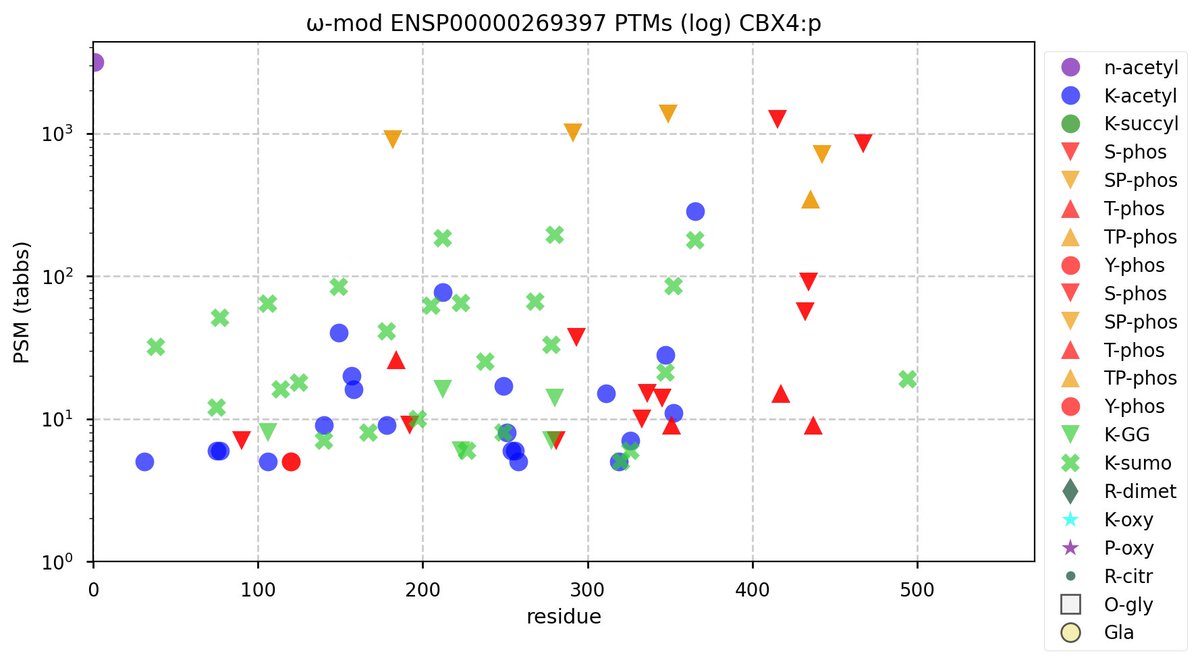
Thu Sep 22 16:43:38 +0000 2022@michaelhoffman 0: Fortran
1: C
2: C++
3: SQL
4: Java
5: Perl
...
Thu Sep 22 16:07:56 +0000 2022@byu_sam Anything that demonstrates the design & construction of a workable taxonomy would fill the bill.
Thu Sep 22 16:05:50 +0000 2022@AlexUsherHESA I was a STEM prof at 3 different Canadian universities & I cannot recall a single conversation (or open discussion) about federal/provincial politics between faculty members. I am sure my colleagues all had opinions, but it is considered extremely poor form to talk about them.
Thu Sep 22 15:59:08 +0000 2022@mjmaccoss So, in spite all of the hubbub about Orbis, LTQs rule.
Thu Sep 22 15:41:37 +0000 2022Fiona is coming to town: the predicted barometric pressure for Sydney, Nova Scotia for the next few days. 🔗
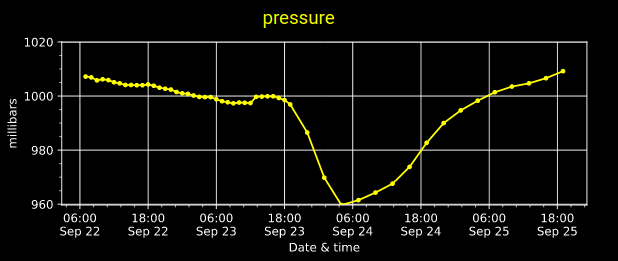
Thu Sep 22 15:29:08 +0000 2022@byu_sam Taxonomy.
Thu Sep 22 14:20:35 +0000 2022A story only possible at a Canadian university
🔗
Thu Sep 22 14:11:04 +0000 2022Slow week at GPMDB: only 20 megatabbs of new information added.
Thu Sep 22 14:03:42 +0000 2022@neely615 @bioinformer @Nature @Smith_Chem_Wisc @nesvilab It is common place in Canada for large, state run degree mills to consider themselves "elite", but the US has always been more discerning about these things. Since it was in Nature, though, it may be a bit of British sneering at colonial pretensions.
Thu Sep 22 13:25:42 +0000 2022@neely615 @bioinformer @Nature When did big state unis like Michigan, Wisconsin & UC Berkeley become "elite"?
Thu Sep 22 13:12:28 +0000 2022@CharlieSaid3 Ion trap-type CID produces more sequence specific ions than beam-type CID. So even though the masses are more characteristic, there are fewer of them―it ends up being a bit of a wash.
Thu Sep 22 12:24:25 +0000 2022The S/T+phosphoryl acceptor types are more varied:
1× RxxΦL/I/V;
1× ΦQ;
1× PxΦP;
1× RxxΦP;
2× GxxΦP;
2× RxxΦ;
2× ΦF;
2× GΦ;
3× ΦPxK;
4× ΦL/I/V;
5× NxΦ;
9× ΦP; and
13× ΦxxD/E.
Thu Sep 22 12:20:24 +0000 2022Modification diagram for MIS18BP1:p, aka M18BP1, FLJ11186, KIAA1903, KNL2 & C14orf106. Lot of K-mods, except acetyl:
1× K+acetyl/GGyl;
10× K+GGyl;
22× K+SUMOyl; and
31× K+GGyl/SUMOyl acceptors. 🔗
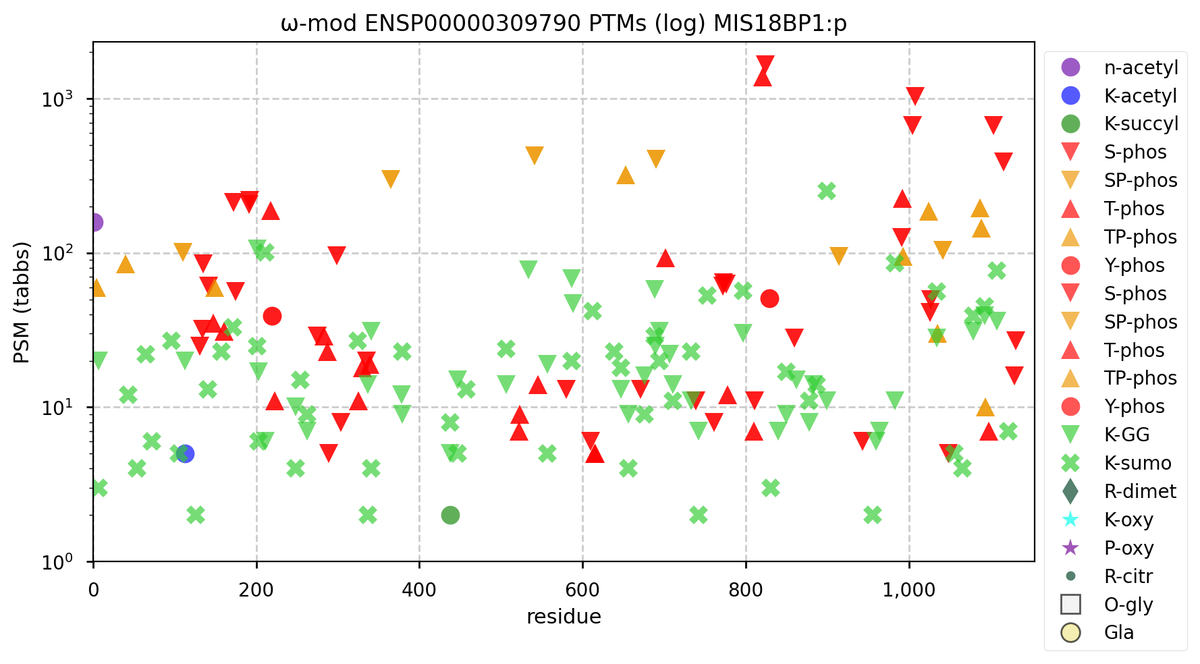
Thu Sep 22 01:18:37 +0000 2022@CharlieSaid3 It has a similar effect for peptide spectra: many algorithm-lineages were originally designed for LTQ data & don't take advantage of higher mass accuracy.
Wed Sep 21 17:46:22 +0000 2022@chrashwood As in 0.5 - 1 Da.
Wed Sep 21 17:00:19 +0000 2022Does anybody know why people seem to like to run the Eclipse instruments using high resolution parent but low resolution fragment ion masses? Can they get more cycles per second or is it something else?
Wed Sep 21 16:18:01 +0000 2022ADNP:p activity dependent neuroprotector homeobox phosphoryl acceptor occupation overlaid on its AF predicted aligned error. Given its name, I would expect this to be a relatively low concentration protein, but it is frequently observed. 🔗
Wed Sep 21 14:49:38 +0000 2022@StanKutcher @jwoodgett @Abe_McGill @cafreeland @AndyFillmoreHFX @FP_Champagne @jyduclos @CIHR_IRSC @SSHRC_CRSH @NSERC_CRSNG I support the sentiment, but I don't think anyone in government is really listening.
Wed Sep 21 14:31:31 +0000 2022🔗
Wed Sep 21 14:27:00 +0000 2022@statesdj Hurricane Juan was the last big storm to hit Nova Scotia, which was also at this time of the year (it made landfall on Sept. 28, 2003).
🔗
Wed Sep 21 14:10:33 +0000 2022@emily_m_arndt @MHendr1cks If it is a fake, they have done a great job of providing a fake provenance, e.g.
🔗
🔗
Wed Sep 21 14:07:05 +0000 2022An Environment Canada map showing the same thing, but with the agency's characteristic 19th century hand-drawn appearance. 🔗
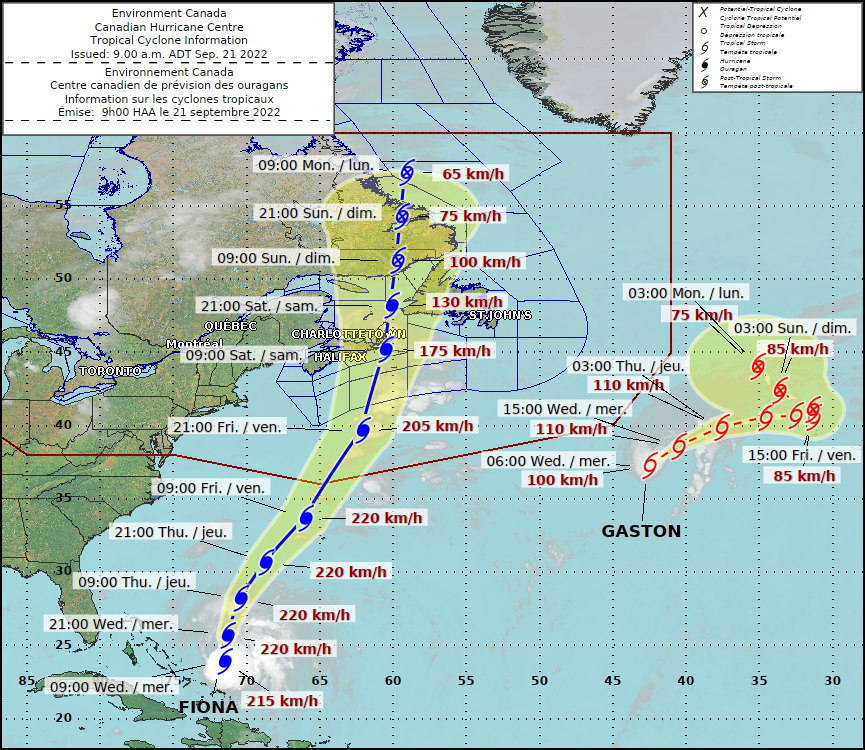
Wed Sep 21 13:58:47 +0000 2022From NOAA, regarding Fiona's arrival in Bermuda on Thursday and Nova Scotia/Newfoundland on Saturday. 🔗
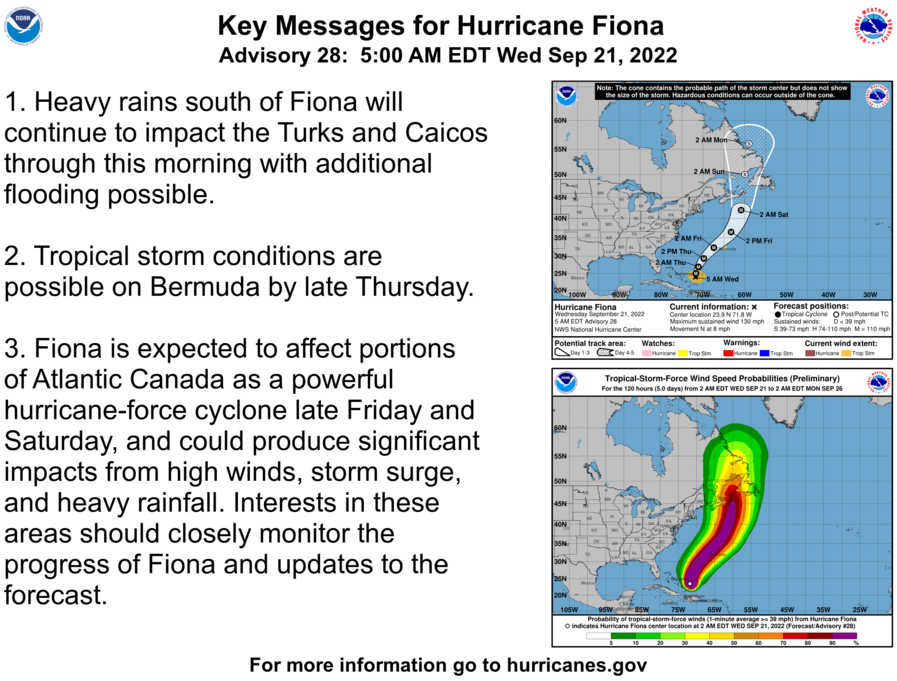
Wed Sep 21 12:37:18 +0000 2022Modification diagram for human ZNF107:p, a subunit that may do things. It has 25 C2H2 zinc finger domains and a full slate of K-mods:
1× K+GGyl/SUMOyl
1× K+SUMOyl
1× K+acetyl/SUMOyl
2× K+acetyl/GGyl
3× K+acetyl
5× K+acetyl/GGyl/SUMOyl
9× K+GGyl
🔗
Tue Sep 20 21:03:19 +0000 2022PXD031455: an étude of just how deep into the carbamylated amine background an Eclipse can take you.
Tue Sep 20 19:18:17 +0000 2022A contemplation of protein low complexity domains from an "RNA is what really matters" point of view:
🔗
Tue Sep 20 18:39:08 +0000 2022Major Hurricane Fiona is centered just north of the Turks & Caicos Islands. It looks like it will miss the US East Coast, but pass right through the Cabot Strait as a Category 1 storm Saturday afternoon. If you are taking the ferry to Port aux Basques or Argentia, don't delay! 🔗
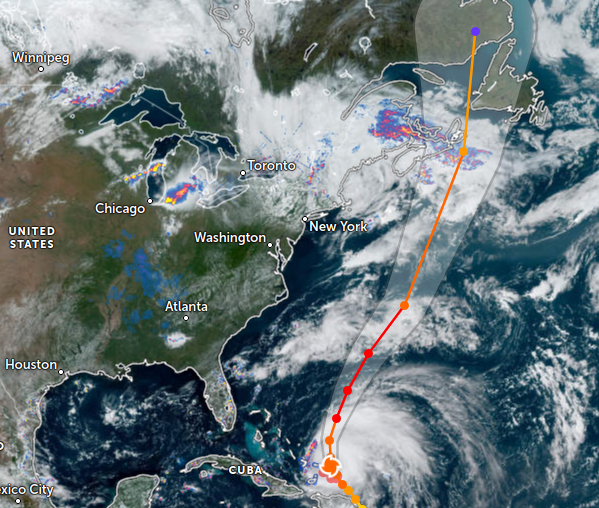
Tue Sep 20 18:27:55 +0000 2022The "H key got stuck" domain of CBX4:p (378-400):
…HPPSHHPHPHPHHHHHHHHHHHH…
Tue Sep 20 16:53:23 +0000 2022@VATVSLPR @neely615 @JohnRYatesIII @stshuken @GarciaLabMS @MagnusPalmblad Actually, in the US, the DMCA provides rather draconian penalties for anyone caught doing that sort of thing. It certainly would stop me from trying.
🔗
Tue Sep 20 16:35:25 +0000 2022Just including the words "cell line" after "acute myeloid leukemia" in the title makes it very clear to all what you are doing.
Tue Sep 20 16:33:06 +0000 2022IMO this is the right way to deal with using a cell line for measurement, but focusing on its precursor properties
🔗
Tue Sep 20 16:07:14 +0000 2022The US of A can always be counted on to produce the most seriously toned, unintentional self-parody. 🔗
Tue Sep 20 15:38:59 +0000 2022PXD028509: an interesting slice of outer membrane proteins from the always entertaining H. pylori.
Tue Sep 20 15:09:20 +0000 2022@AJ_Brenes Immunopeptide ―MHC-presented class I & II― purification schemes have a similar upside (good source for low concentration proteins) and downside (highly variable methodology).
Tue Sep 20 14:52:55 +0000 2022@AJ_Brenes In their defense, though, pull down experiments tend to be difficult to analyze because of the wide variety of methods used by investigators. It is an area where many publications come with their own clever take on bait-prey methodology (& its associated forest of acronyms).
Tue Sep 20 14:39:49 +0000 2022@AJ_Brenes I suspect that the problem is that the way those sites select their underlying datasets underweight pull down/interaction experiments, which tend to be the best source of data for many low concentration proteins.
🔗
Tue Sep 20 14:15:48 +0000 2022@stateofthecity @AlexUsherHESA Tactical, rather than strategic, approaches seem to be necessary in any policy arena where a longer-term strategic plan is unlikely to survive changes in party leader.
Tue Sep 20 14:02:01 +0000 2022@AlexUsherHESA @stateofthecity Canada's traditional response has been through immigration or importing transient laborers rather than training.
Tue Sep 20 13:41:51 +0000 2022@pwilmarth @dtabb73 An easy tee-up for this one
🔗
Tue Sep 20 13:36:23 +0000 2022@neely615 @MagnusPalmblad @ProteomicsNews Enough that I stopped using the text-based representations of LC/MS/MS runs available on repository sites. They were simply unreliable.
Tue Sep 20 13:09:04 +0000 2022@MagnusPalmblad @ProteomicsNews @neely615 IMHO, mzML/MGF re-imaginings of raw data tend to suffer more from the removal of inconvenient spectra than from the insertion of fabricated ones.
Tue Sep 20 13:08:56 +0000 2022@MagnusPalmblad @ProteomicsNews @neely615 It isn't a recent development. It has been going on for a long time (at least since I was a grad student).
Tue Sep 20 13:00:31 +0000 2022@dtabb73 The lack of an established, rational nomenclature for PTMs on proteins is a big confounder: most of the practical problems that crop up are associated with having to create one from scratch.
Tue Sep 20 12:31:32 +0000 2022Protecting elastin is an important job: elastin molecules have an estimated half life of ~70 years in humans.
🔗
Tue Sep 20 12:27:17 +0000 2022I would guess that none of these protein tags are present in the exported form. Instead they probably function in intracellular processes such as targeting for vesicular transport & folding QC.
Tue Sep 20 12:14:13 +0000 2022Is a wiki a good way to document PTM sites? I tried it years ago, but shut it down for lack of useful contributions, the general idea that all wiki sites are somehow Wikipedia & it is kind of an awkward way to document this sort of thing. Thoughts?
Tue Sep 20 11:58:54 +0000 2022Modification diagrams for human FBN2:p, a large ECM subunit believed to be involved in protecting elastin. It has 7× N+glycosyl, 4× K+GGyl, 17× K+GGyl/SUMOyl & 19× K+SUMOyl acceptors. N-glycosylation is pretty common in ECM proteins, but SUMO2yl/GGylation is not. 🔗
Mon Sep 19 19:13:29 +0000 2022@NAT_machinery @rosiebell94 @VendruscoloLab @jrkumita @JMolBiol @proteintermini Nit-picking: the gene name "SCNA" in the Abstract is incorrect. It should be "SNCA", as it is in the body of the article.
Mon Sep 19 16:18:44 +0000 2022@JoePlatelet I'm pretty sure that nobody has an answer for that type of question. About all I know is that sequences with tandem zinc-finger repeats are over-represented in SUMO2 pull-down experiments (~30% of proteins id'd) & tend to have many acceptors across the repeats.
Mon Sep 19 13:42:23 +0000 2022Unfortunately, these modifications have no obvious links to a disease (or NIH Institute), so it probably won't be investigated in my life time.
Mon Sep 19 12:10:38 +0000 2022It would be interesting to know which of the SUMO2-specific E3-ligases were responsible for this type of selective modification, which happens in so many sequences with tandem zinc finger domains.
Mon Sep 19 12:01:58 +0000 2022The sequence has 16 C2H2-type zinc finger domains. The SUMO2yl acceptors are spread across these domains. The 5 solo phosphoryl acceptors are placed in gaps between them. S78 is a ΦP-type acceptor, while the other 4 are base-directed RxxΦ-type acceptors.
Mon Sep 19 11:57:25 +0000 2022Modification diagram for RBAK:p, a subunit that may do things. It has multiple K ε-amino PTM acceptors:
36× K+SUMO2yl
1× K+GGyl/SUMO2yl
1× K+acetyl
1× K+acetyl/GGyl/SUMO2yl
1× K+acetyl/SUMO2yl
but relatively little involvement with acetylation or GGylation. 🔗
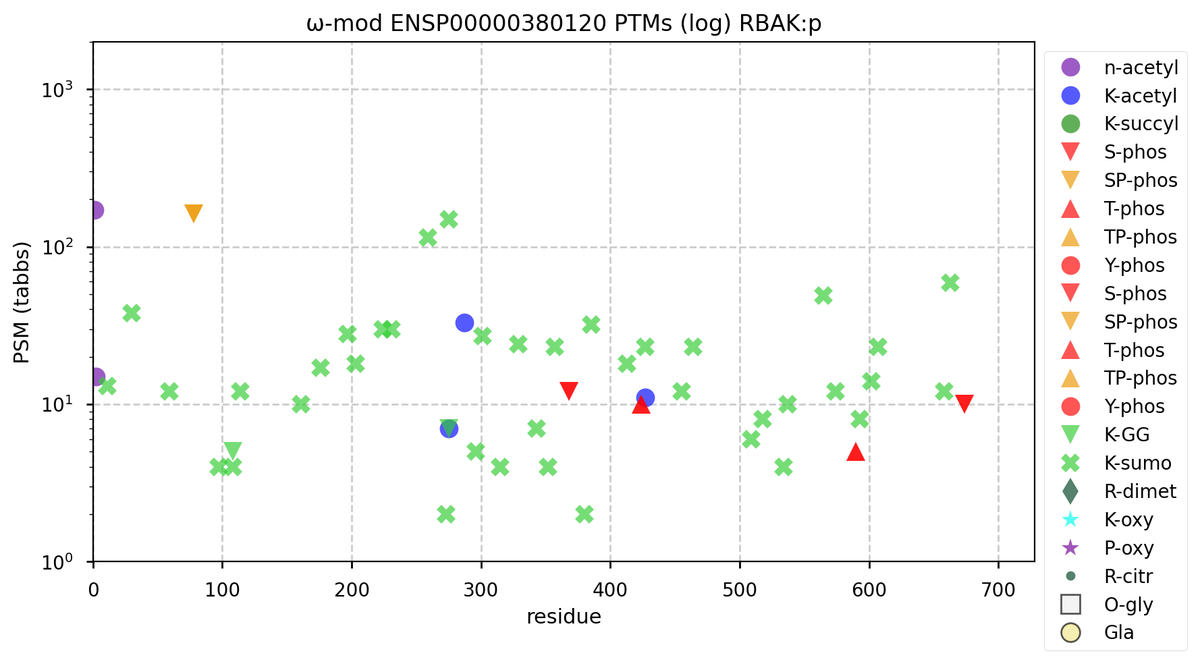
Sun Sep 18 14:51:02 +0000 2022Rather than trying to make itself a part of a "cradle-to-grave" value chain of measurements running all the way from DNA coding to HLA peptide presentation, proteomics has made an article of faith out of opposing the value of other technologies wrt "proteins".
Sun Sep 18 14:38:24 +0000 2022MS-based proteomics has spent decades trying to wall itself off from genomic & transcriptomic results. Is it any surprise that other protein-based platforms are trying to steal a march on it by making it easier to linking up with those other result sets?
Sun Sep 18 12:31:25 +0000 2022The protein modifier-based PTMs do not avoid the zinc finger domains: they seem "attracted" by them. This type of non-canonical protein modifier acceptor pattern has no known biological function: it is a weird one.
Sun Sep 18 12:06:25 +0000 2022It also has 4 isolated, high occupancy phosphorylation (iHOP) domains nicely placed in the gaps between the 10 zinc finger domains that span the sequence.
Sun Sep 18 12:03:36 +0000 2022Modification diagram for human ZNF451:p, a SUMO2 E3 ligase. It is also a substrate for SUMO2 & ubiquitin, with
1× K+acetyl/ubiquitinyl/SUMO2yl,
11× K+SUMO2yl,
20× K+ubiquitinyl, &
33× K+ubiquitinyl/SUMO2yl acceptors. 🔗
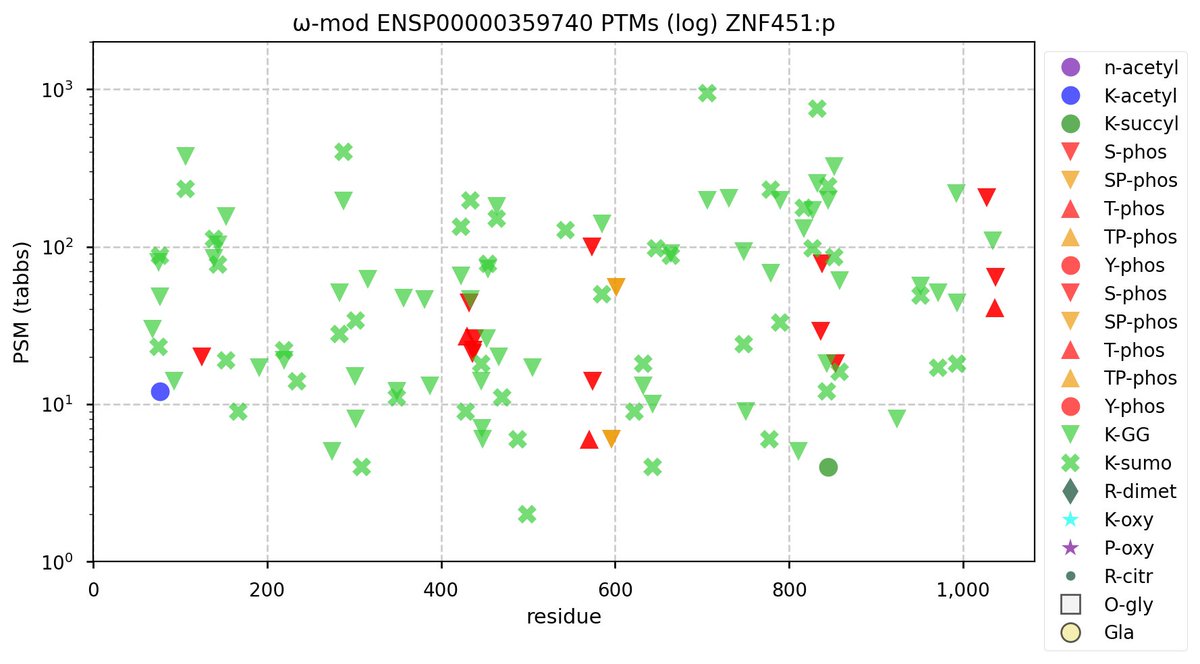
Sat Sep 17 16:26:02 +0000 2022I had seen Marvin around conferences in the 80's, but I really got to know him when his company Vestec was adapting Brian Chait's MALDI ion source design for a commercial instrument. I spent a few enjoyable days in Texas talking with Marvin & his lead technician, Randy Nelson.
Sat Sep 17 15:27:31 +0000 2022@AlexUsherHESA @TorontoStar This applies nicely to Winnipeg too, also in the midst of an election season.
Sat Sep 17 14:57:26 +0000 2022To me, Marvin was a figure like Edison or Tesla, an authentically American innovator who brought the future with him.
Sat Sep 17 14:45:53 +0000 2022He was loyal to his friends and enjoyed nothing more than a spirited discussion of theoretical & technological issues. He worked series expansion calculations for the joy of it, the way many people use crossword puzzles.
Sat Sep 17 13:49:08 +0000 2022Super Typhoon Nanmadol is expected to make landfall very near Kagoshima Japan on the 18th & then veer off to the northeast, cutting across Honshu until exiting near Sendai on the 21st. 🔗
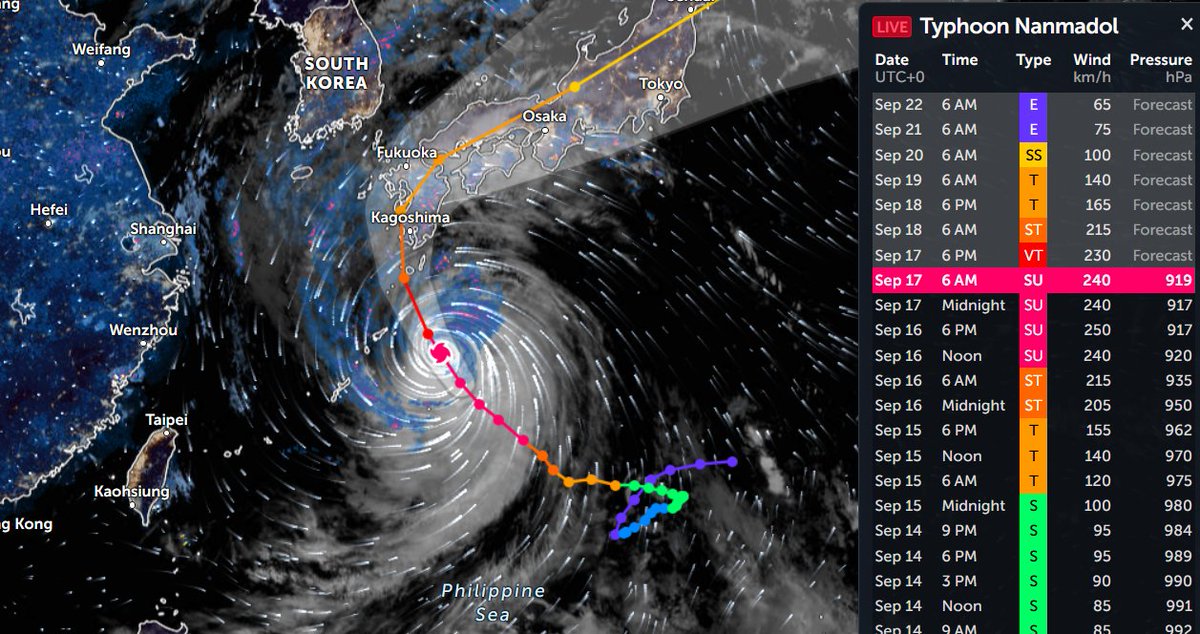
Sat Sep 17 12:55:42 +0000 2022And decomposing that pattern into S and T separately gives a bit of insight into the multiple kinases & phosphatases involved in generating the pattern. 🔗
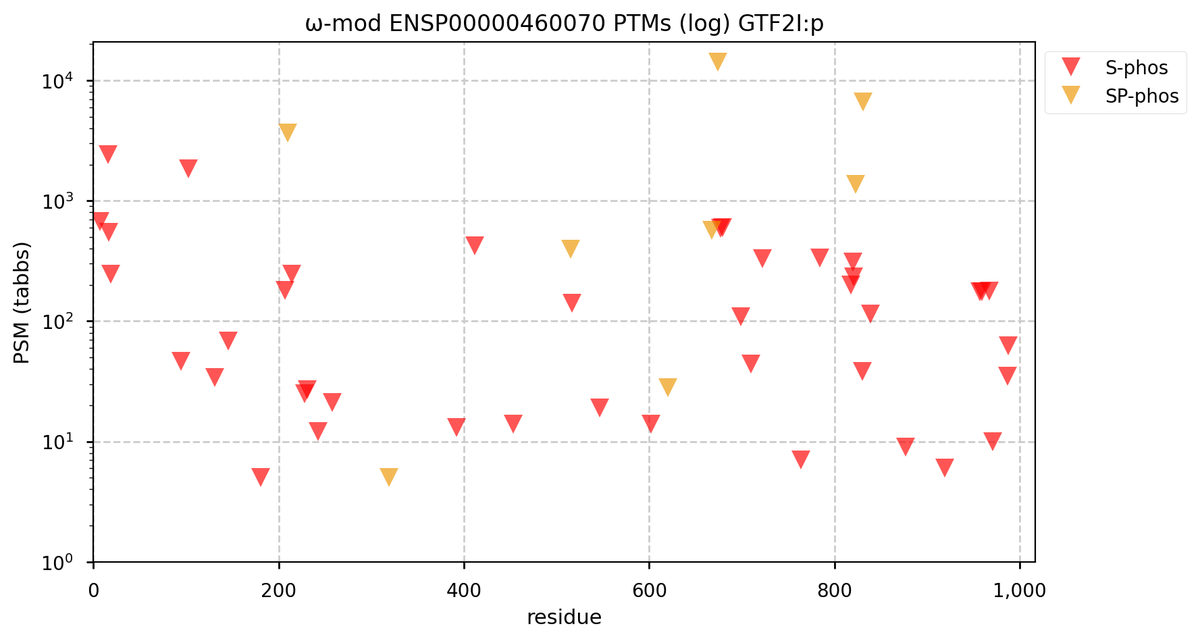
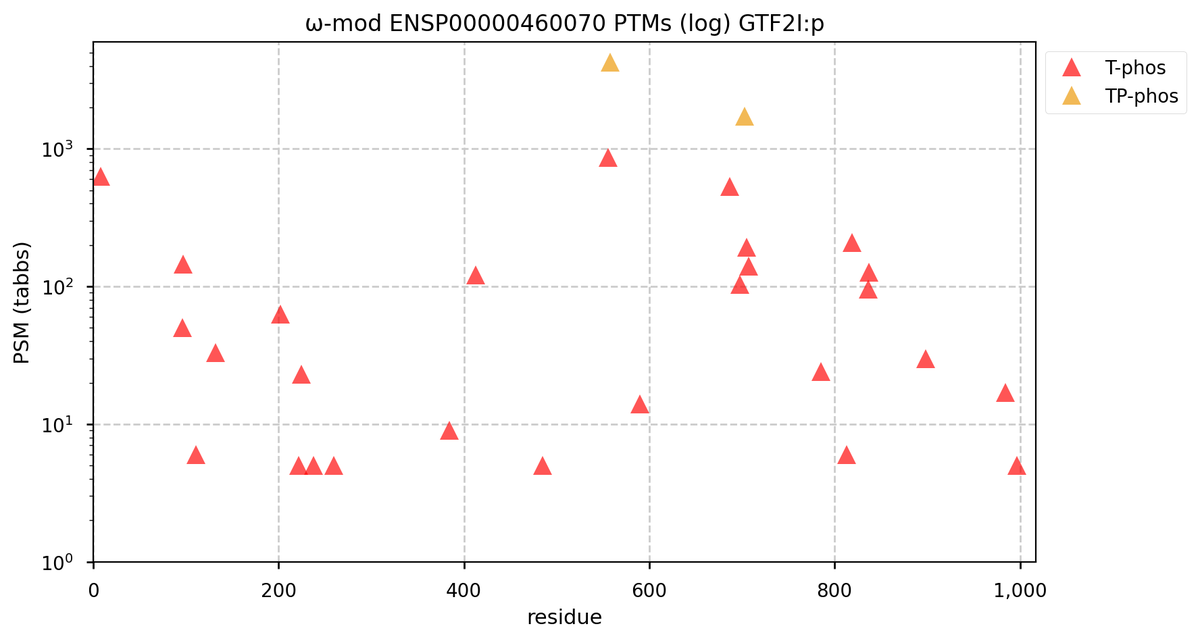
Sat Sep 17 12:50:10 +0000 2022GTF2I:p is more decorated than GTF2IRD1, with a lot more phospho-bling than its cousin. 🔗
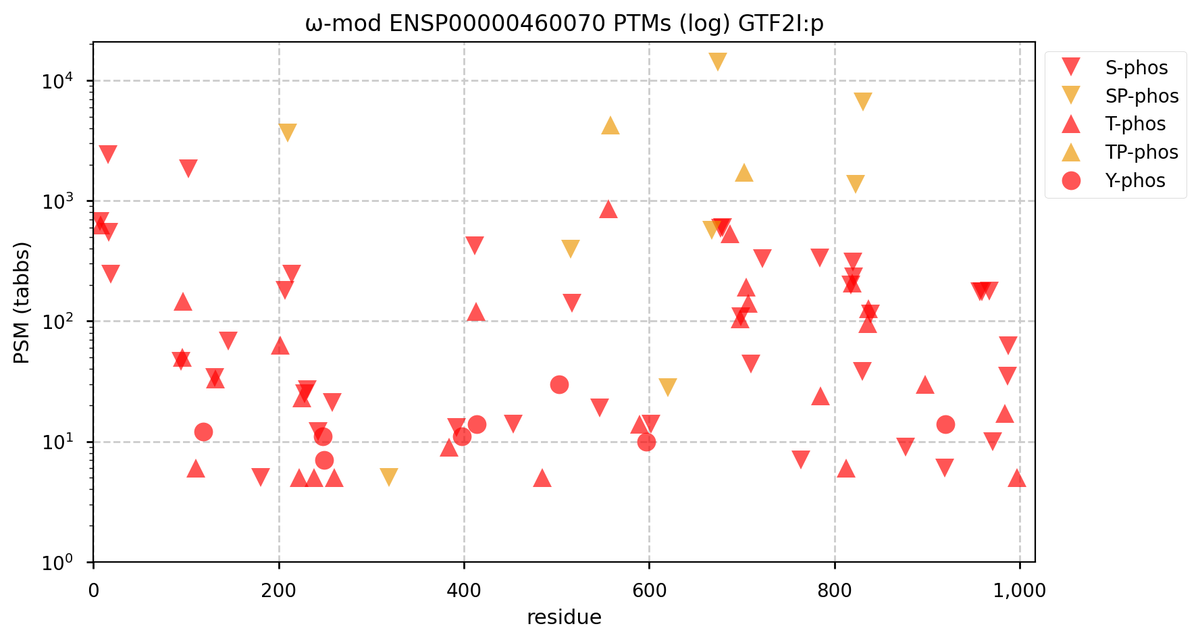
Sat Sep 17 12:12:19 +0000 2022GTF2I:p does not share any tryptic peptides with GTF2IRD1:p.
Sat Sep 17 12:04:56 +0000 2022This lack of acetylation is different from the subunit's namesake GTF2I:p, which has 23 of them, all but one of them shared as either K+acetyl/GGyl or K+acetyl/GGyl/SUMO2yl. 🔗
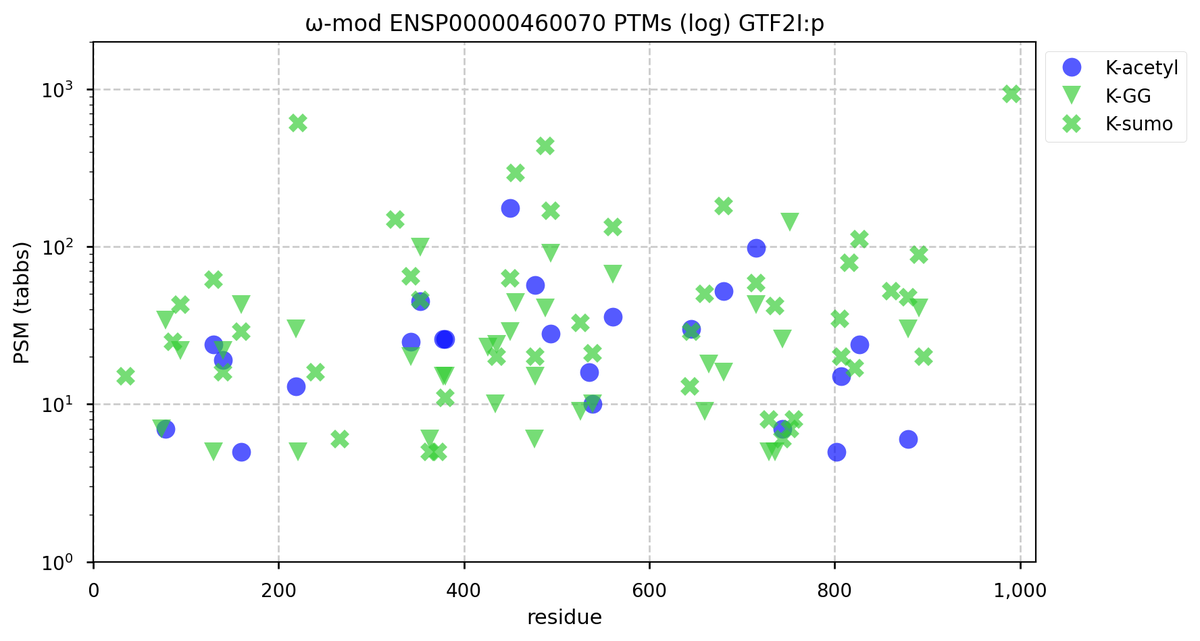
Sat Sep 17 12:04:34 +0000 2022The modification diagram for GTF2IRD1:p, a subunit that may be involved in a number of processes. It has 1 K+GGyl, 24 K+SUMO2yl and 8 K+SUMO2yl/GGyl acceptors. It has no observed use for K+acetyl. 🔗
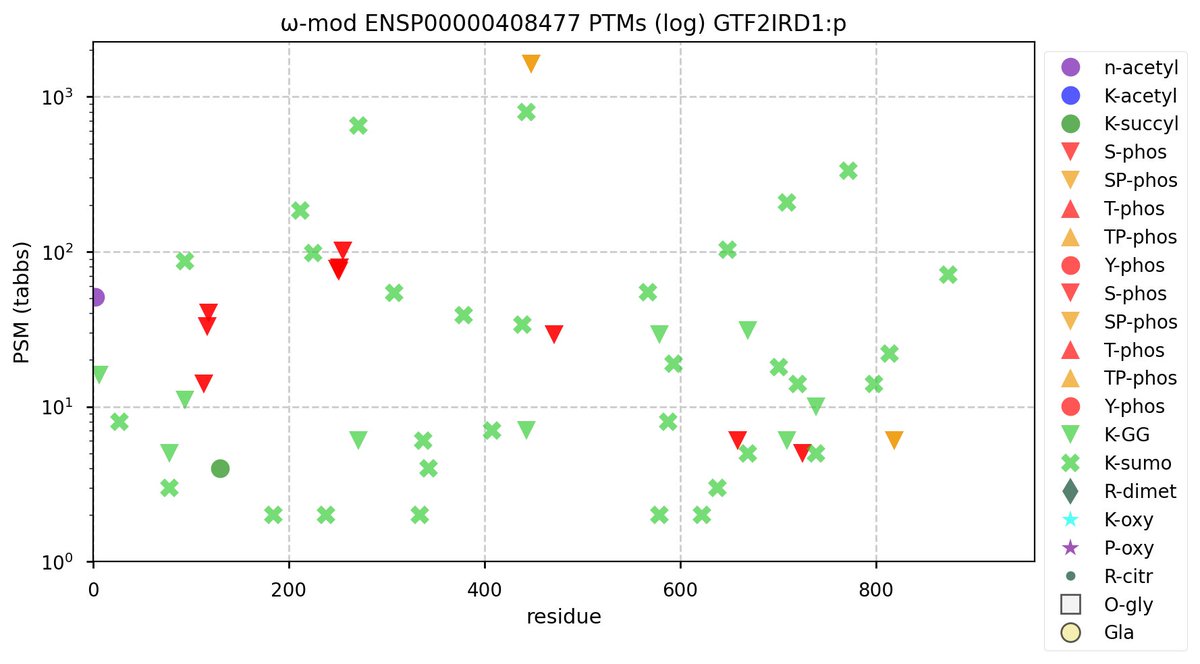
Fri Sep 16 19:00:25 +0000 2022While not all proteins with zinc finger domains have SUMOyl acceptors, many proteins with SUMOyl acceptors have zinc finger domains.
Fri Sep 16 15:02:54 +0000 2022Why do some people seem to think that long, disjointed email threads are a useful way to carry on a discussion?
Fri Sep 16 14:42:26 +0000 2022He was also a source of valuable advice: he had an unblinkered view of how the chaos that is science + academia + government + private business functions and was willing to share insights into this maelstrom with younger colleagues.
Fri Sep 16 14:32:02 +0000 2022@RobynUrback It is hard to forget it is also the path that eventually led the same party to believing the barbaric cultural practices snitch line was vote getter.
Fri Sep 16 14:20:21 +0000 2022@neely615 Our local ballet company (the Royal Winnipeg Ballet) is very successful at doing new things for a small market dance company. But, their long-term success is predicated on making sure that they perform "The Nutcracker" every year at Christmas.
Fri Sep 16 14:11:24 +0000 2022Thanks to the brave few willing to state an opinion. The majority felt it was important to group the K-based acceptors on their observed suite of potential PTMs, rather than characterizing the PTMs individually & hoping the others were completely independent of each other.
Fri Sep 16 14:03:20 +0000 2022Intellectually Marvin was a polymath, but practically he was the most valuable type of serial mono-tasker: once he decided on a course of action, he stuck with it until it succeeded/failed. He didn't make things because it was his job. He made things because he wanted to do it.
Fri Sep 16 13:22:44 +0000 2022"…RerunAfterDewarChange…" may be my new favorite descriptor concatenated into a data file name.
Fri Sep 16 13:21:02 +0000 2022As always, extra points to the student who can explain this pattern solely based on the Michaelis-Menten equation & Langmuir isotherms (some relativistic terms may be necessary).
Fri Sep 16 13:00:02 +0000 2022Protein S is such an odd ball. "Gla", "Gla", "Gla", always with the "Gla". 🔗
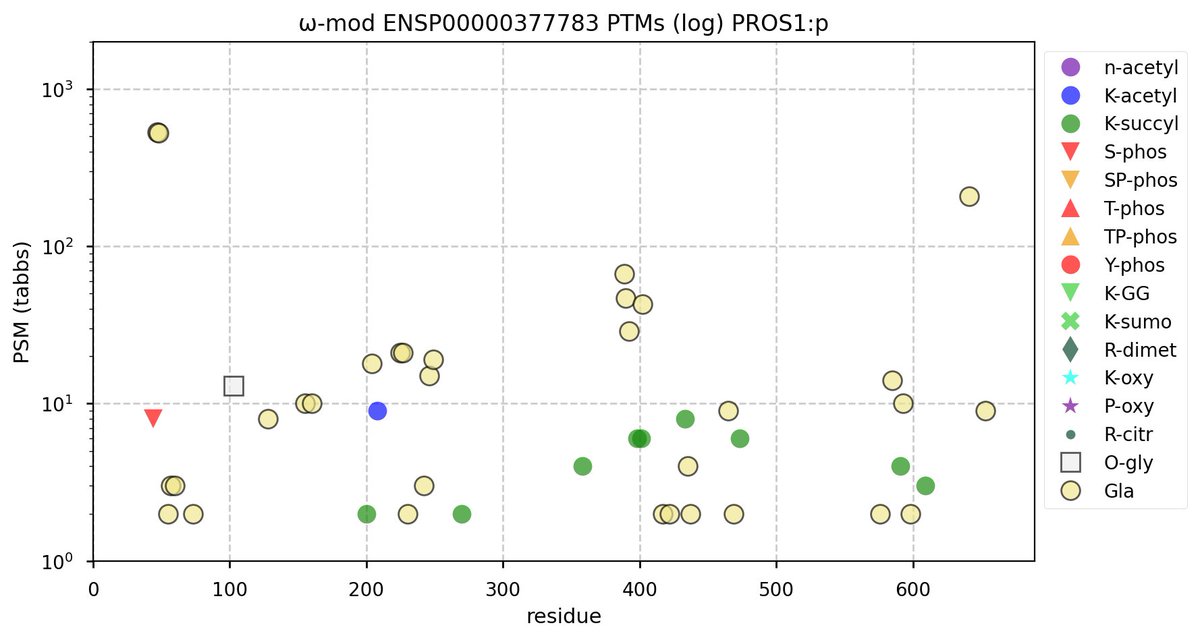
Fri Sep 16 12:41:36 +0000 2022He was an inventor: someone who could develop theory to the point that it could be translated into cut steel & new electronics.
Fri Sep 16 12:29:22 +0000 2022I was made aware yesterday of the passing of my friend, Marvin Vestal. He was instrumental in driving the creation of the modern mass spec business, particularly keeping TOF technology competitive with other analyzers. 🔗
Fri Sep 16 12:25:34 +0000 2022It also has one of "those" observable peptide coverage patterns. 🔗
Fri Sep 16 12:06:16 +0000 2022@leprevostfv 🔗
Fri Sep 16 11:55:38 +0000 2022It would appear that this is not a burning issue in the proteomics community. 🔗
Fri Sep 16 11:50:25 +0000 2022It also has a hopeful (but not very helpful) Un!Pr*t function assigned to it. 🔗

Fri Sep 16 11:48:37 +0000 2022Modification diagram for the subunit translated from the 776th gene on a list of human genes with a zinc finger, ZNF776:p. It has 2 K+GGyl, 7 K+SUMO2yl & 5 K+SUMO2yl/GGyl acceptors. It has no observed K+acetyl acceptors. 🔗
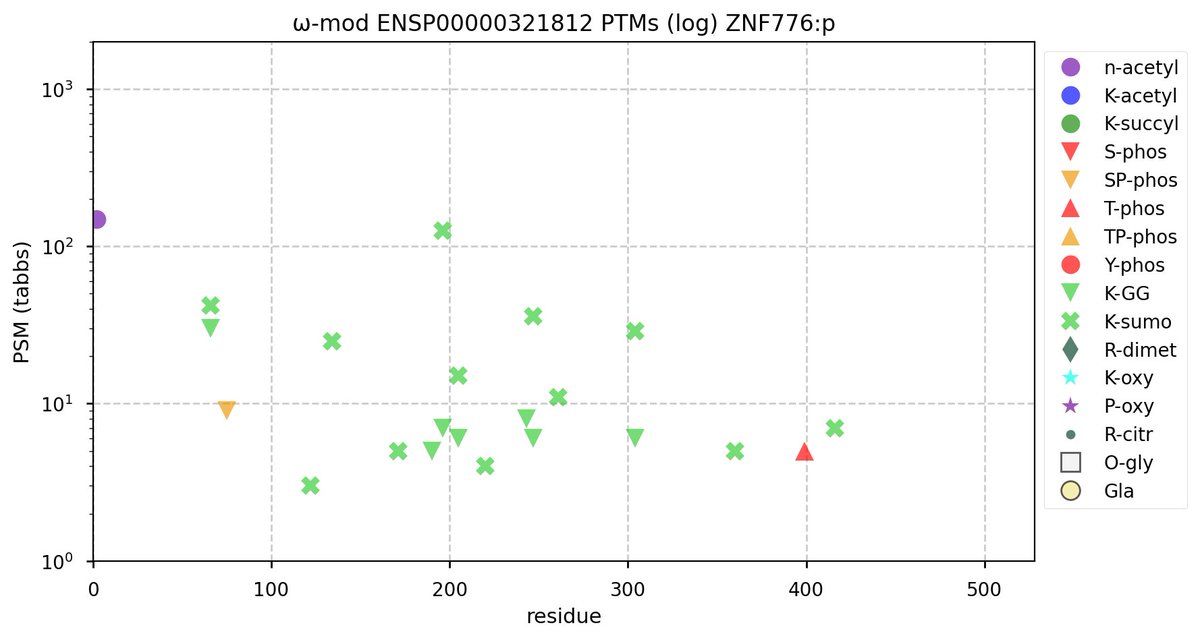
Fri Sep 16 01:51:13 +0000 2022PXD036085: if you've ever wondered how many different keratins you can detect in a single sample, this one is for you. Also has some nice examples of endogenous R+deimination (citrulline formation).
Thu Sep 15 15:29:17 +0000 2022@jwoodgett @MHendr1cks @ImogenRCoePhD @LiisaGalea @DimensionsTMU No doubt. But algorithm-based grant triage is coming & it will be implemented by the Common CV team.
Thu Sep 15 15:22:58 +0000 2022@jwoodgett @MHendr1cks @ImogenRCoePhD @LiisaGalea @DimensionsTMU Algorithms are currently used as the first step in the consideration of Scientific Research and Experimental Development (SR&ED) tax incentives by CRA. SRED is normally grouped in with TriCouncil grants as part of the Feds science expenditure envelope.
Thu Sep 15 14:15:43 +0000 2022@pwilmarth I'm pretty sure that is frowned-upon by the coding police.
Thu Sep 15 14:14:30 +0000 2022For the SUMO-fanciers: I have left out the paralogue complications because, well, tweets aren't that long. GGyl also hides 3 potential conjugate subunits. 🧐
Thu Sep 15 14:05:36 +0000 2022Is it reasonable to distinguish between K's observed to act as acceptors for combinations of PTMs:
K+acetyl,
K+acetyl/GGyl,
K+acetyl/SUMOyl,
K+GGyl,
K+SUMOyl,
K+GGyl/SUMOyl, or
K+acetyl/GGyl/SUMOyl?
Thu Sep 15 13:21:15 +0000 2022There is probably a phosphodomain (252-271), but a long tryptic peptide (246-336) makes it inaccessible to commonly used methods.
Thu Sep 15 12:19:20 +0000 2022Tropical Storm Muifa is approaching Qingdao, producing significant rainfall. Now Typhoon Nanmadol (bottom right) is now expected to turn as it passes over Fukuoka & proceed along the coast of the Sea of Japan. 🔗
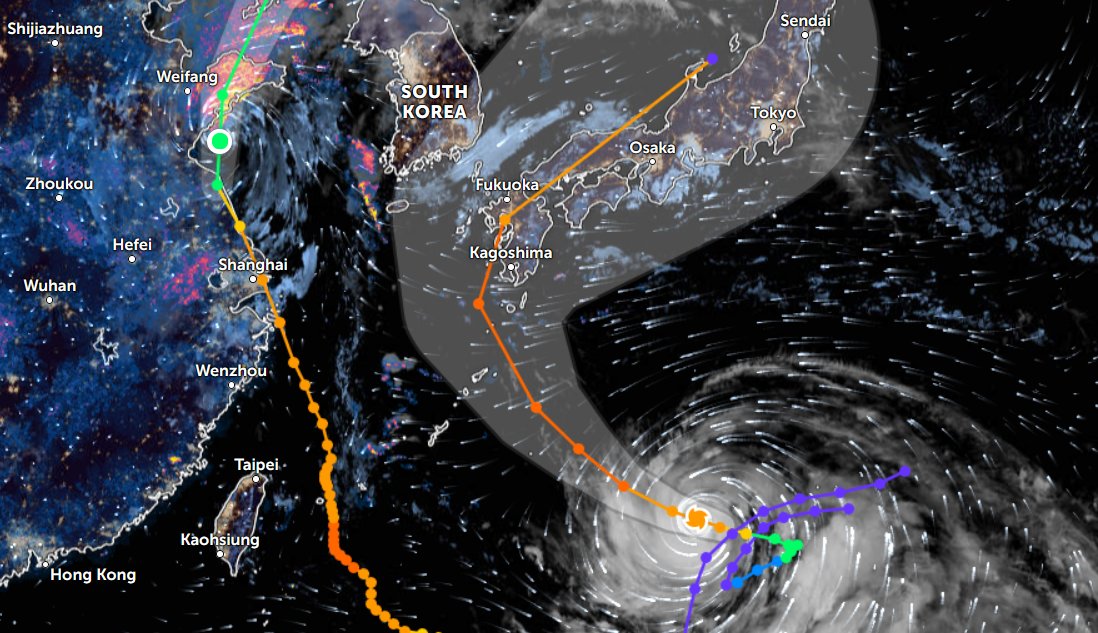
Thu Sep 15 12:08:43 +0000 2022exot() is my most frequently typed, but non-existent Python function. exot() and missing the final parenthesis on complicated print() expressions cause most of my Python debugging problems.
Thu Sep 15 11:53:30 +0000 2022Modification diagram for human RNF111:p, a large subunit with multiple low complexity domains. It has 12 observed K+SUMO2yl acceptors, 4 of which may be occupied by K+GGyl. All but one of these is in an N-terminal domain (19-218). 🔗
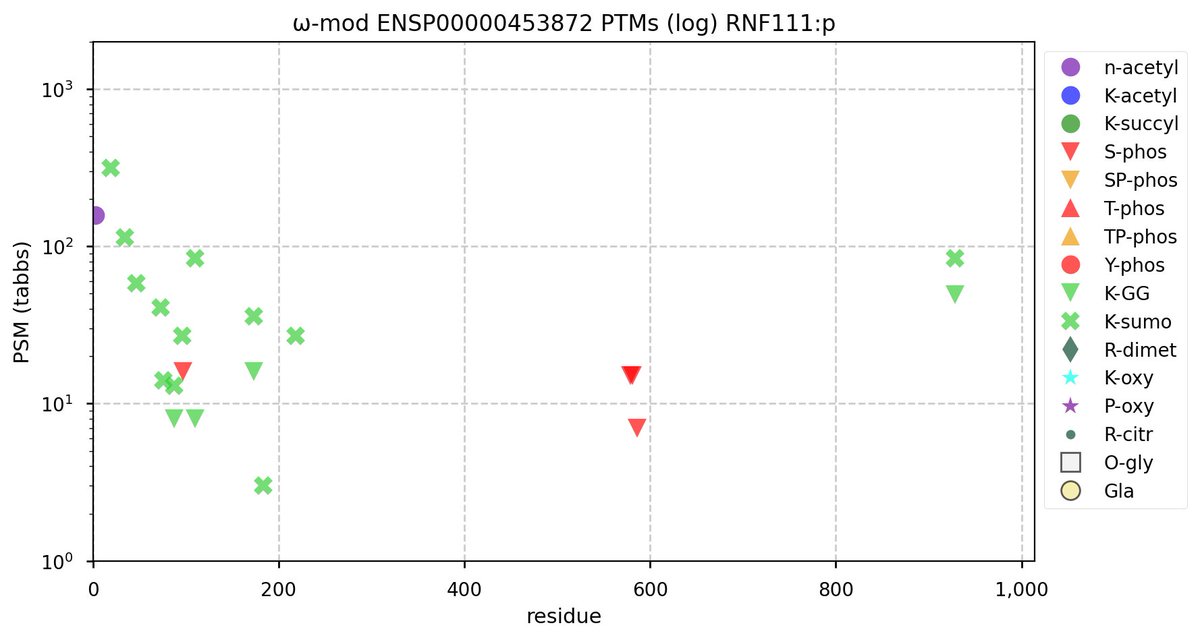
Wed Sep 14 16:40:09 +0000 2022The PRIDE FTP site has been acting up for the last few days.
Wed Sep 14 15:01:59 +0000 2022🔗
Wed Sep 14 14:54:25 +0000 2022Rain and winds from Typhoon Muifa's outer bands are being felt in Shanghai as it crosses Hangzhou Bay. 🔗
Wed Sep 14 14:37:47 +0000 2022Looking back on the many grants panels that I have sat on, I'm not really sure that "grantsmanship" is a real thing. I think it is an imagined intangible that was dreamed up trying to make logical sense of a predominantly stochastic process.
Wed Sep 14 13:33:12 +0000 2022@pwilmarth @ItsBiniR Unfortunately, the way the literature works at the moment, a straightforward route to publication (or at least a conference poster) is to come up with increasingly complex experimental schemes & claim incremental improvements.
Wed Sep 14 12:12:16 +0000 2022XPA:p was originally named "Xeroderma Pigmentosum, complementation group A", after a syndrome that can be caused by defective XPA:g.
Wed Sep 14 12:02:43 +0000 2022Modification diagram for XPA:p, a small subunit that volunteers at DNA damage sites. It has 6 K+SUMO2yl acceptors, one of which doubles as a K+acetyl acceptor. No phosphorylation or ubiquitinylation as far as the eye can see. 🔗
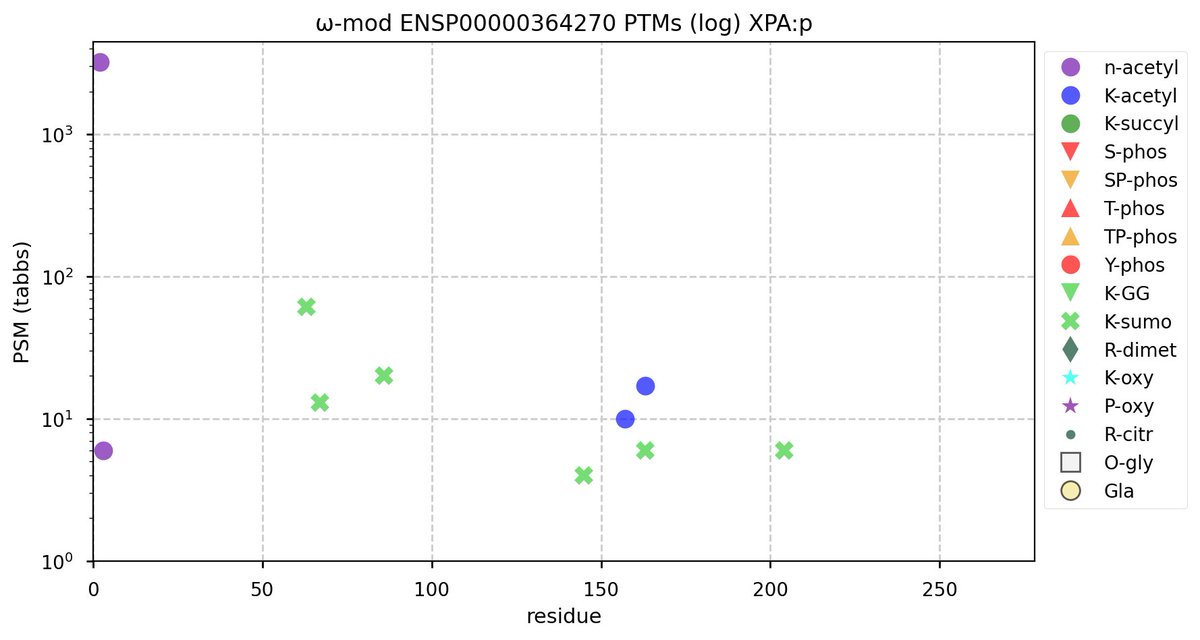
Tue Sep 13 23:52:49 +0000 2022Typhoon Muifa is beginning to affect the Chinese mainland & should pass near Shanghai within 24 hours. At the bottom right is a new storm (green dot), Tropical Storm Nanmadol, expected to pass near the coast of Fukuoka on its way into the Korean Strait as a Typhoon on the 18th. 🔗
Tue Sep 13 20:39:59 +0000 2022@byu_sam @dnusinow You mean Helacyton gartleri? 🔗
Tue Sep 13 16:04:32 +0000 2022@neely615 @MattWFoster @byu_sam @labs_mann But I pray thee forsooth, don't forget about the alphapapillomavirus 7 proteins, which are most assuredly the deepest of the "deep".
Tue Sep 13 13:11:15 +0000 2022As predicted Danielle has dipsy-doodled around the Atlantic & it is making landfall in northern Spain as a low pressure system. 🔗
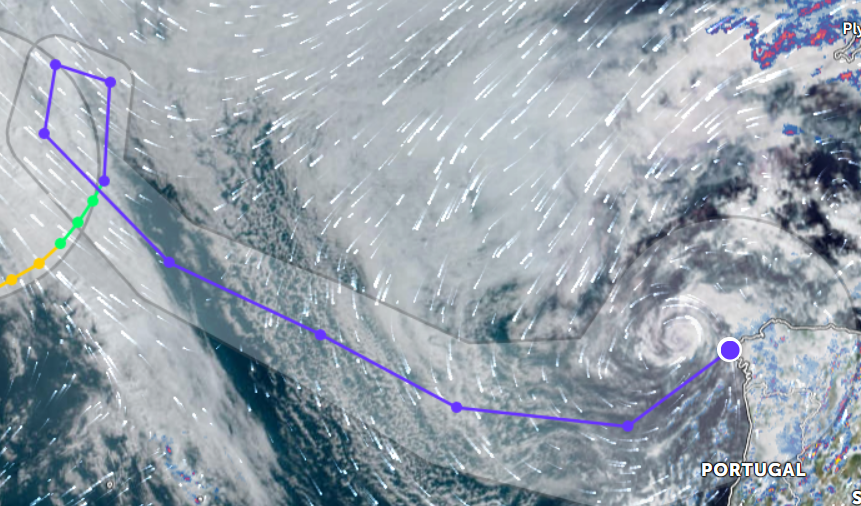
Tue Sep 13 12:26:56 +0000 2022@TrostLab @Rachel_Hx @marinfuenla Always heartened a bit when something I found a long time ago is still used by clever folks for discovering interesting things.
Tue Sep 13 12:21:28 +0000 2022If it wasn't for the pesky K+GGyl bling, this would be an example of canonical PTM decoration, as opposed to its over-the-top Harry Styles/Elton John/Liberacesque cousin PRKDC:p.
Tue Sep 13 12:12:55 +0000 2022Its use of phosphorylation is also broadly similar to ATM:p & ATR:p. It has 4 iHOP domains: 3× S+phosphoryl acceptor domains & 1× T+phosphoryl. It is also big: at 3829 residues it probably takes between 15-30 minutes to make a single copy of the nascent peptide.
Tue Sep 13 12:04:32 +0000 2022Modification diagram for TRRAP:p, a kinase subunit involved in DNA repair. It's pattern resembles kinase family members ATM:p & ATR:p: a frosting of 49 K+GGyl acceptors, 8 of which double as K+GGyl/acetyl acceptors.
🔗
Tue Sep 13 02:30:19 +0000 2022Typhoon Muifa has moved past Taiwan & is accelerating towards the Chinese coast. It should make landfall near Ningbo on the 14th and pass just south of Shanghai on its way inland. 🔗
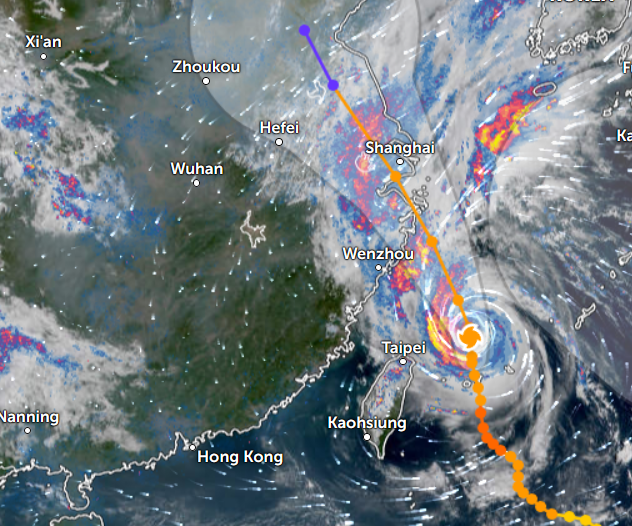
Mon Sep 12 23:42:41 +0000 2022PXD032089: some nice spermatocyte phosphorylation data. It is a rarely sampled cell type.
Mon Sep 12 22:03:19 +0000 2022@MattWFoster @byu_sam @labs_mann Any of the datasets that have good signals for LINE-1 ORF1 and the alphapapillomavirus 7 proteins (particularly E7) are "deep" in my book.
Mon Sep 12 21:51:31 +0000 2022@MattWFoster @byu_sam @labs_mann The HeLa data from 🔗 is also worth a look 🔗
Mon Sep 12 21:47:09 +0000 2022@MattWFoster @byu_sam @labs_mann I would also recommend that one, 🔗
Mon Sep 12 20:31:20 +0000 2022@byu_sam @labs_mann I don't think that one survived the TRANCHE apocalypse. I have the results, 🔗, but not the raw files. Their notion of what "deep" meant back in the day hasn't weathered very well, either.
Mon Sep 12 18:39:55 +0000 2022@camtflower I don't think I can give you a very satisfactory answer, as how AF evaluates the expected error isn't really meant to answer the question: Is this region an IDR? In many cases it is probably a good proxy, though.
Mon Sep 12 12:59:13 +0000 2022@cwvhogue It is an interesting subunit. Because of its length, it probably takes about 15 to 30 minutes on ribosome to make a single copy, but there is a lot of it in cells, with > 100,000 observations in publicly available data.
Mon Sep 12 12:14:38 +0000 2022However, like its family members, the origins and functional significance of its anomalous PTM acceptor patterns are an open question in DNA repair biology.
Mon Sep 12 12:05:16 +0000 2022Its phosphorylation pattern is also very different from ATM:p & ATR;p, with a frosting of low occupancy acceptors coating the length of the sequence with only a few gaps: ~50% of S & T residues participate as potential acceptors.
Mon Sep 12 12:02:55 +0000 20222 modification diagrams for human PRKDC:p, a subunit recruited to DNA repair assemblies. Unlike the other phosphatidylinositol 3-kinase-related kinase family members ATM:p & ATR:p, most of the K+GGyl acceptors are also K+acetyl acceptors. ~70% of K's participate in these PTMs. 🔗
Sun Sep 11 18:05:30 +0000 2022Earl's approach to St. John's, via the St. John's West ground station atmospheric pressure record 🔗
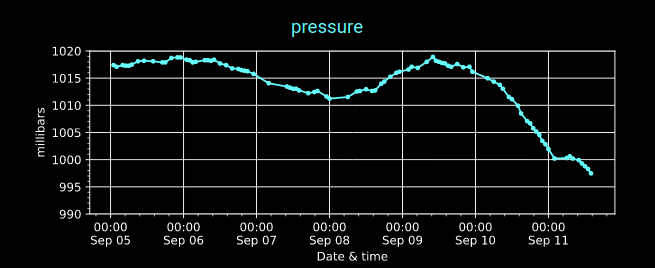
Sun Sep 11 15:16:42 +0000 2022@massspec4tw @Karl_Mechtler I'll let you guys get your story straight.
Sun Sep 11 12:30:10 +0000 2022@Karl_Mechtler @massspec4tw So Chimerys uses Kathy Resing's approach (🔗) and some added heuristics to increase the number of PSMs considered to be valid.
Sun Sep 11 12:04:43 +0000 2022Also like ATR:p, the functional implication of this very distinctive pattern of K+GGyl acceptors remains mysterious.
Sun Sep 11 11:59:48 +0000 2022Modification diagram for ATM:p, a kinase subunit recruited to DNA repair complexes. Like the somewhat shorter ATR:p, 30% of its 214 lysines are K+GGyl acceptors, only 2 of which are K+acetyl/GGyl acceptors. It has 3 very discrete iHOP phosphodomains. 🔗
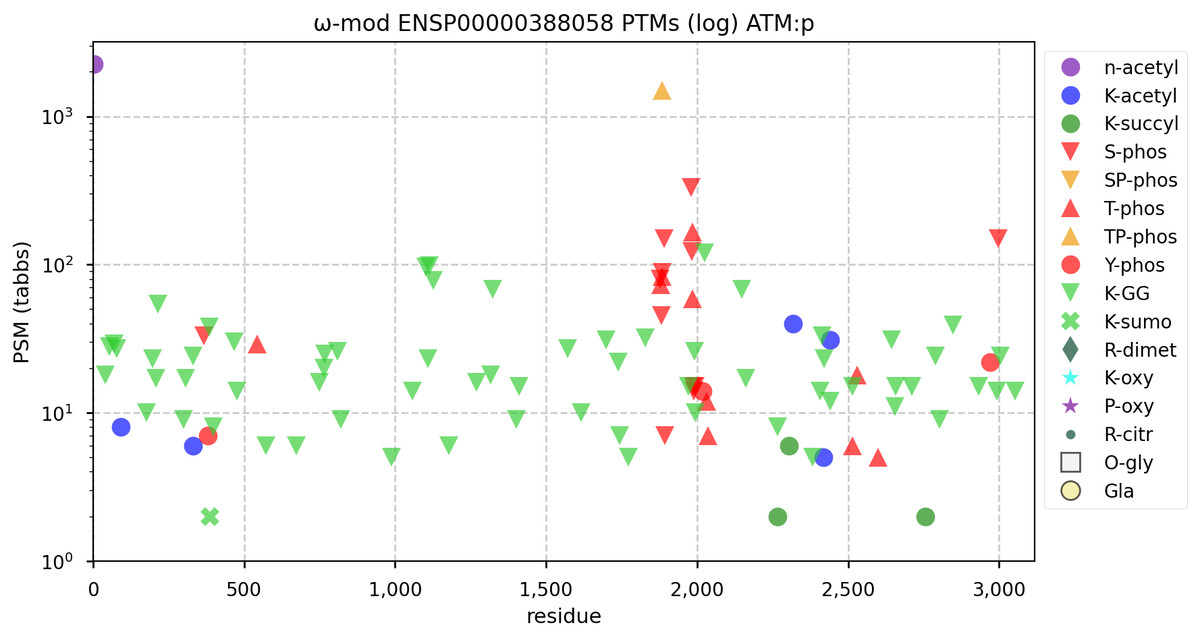
Sun Sep 11 11:30:09 +0000 2022Saw the Winter Hexagon when I got up this morning. It is coming.
Sun Sep 11 10:58:00 +0000 2022Super Typhoon Muifa (Inday, est 942 hPa) has begun its slow swing past Taiwan, with outer bands causing rain on the island. It is predicted to make landfall as a Typhoon on Sept. 15 near Shanghai. 🔗
Sun Sep 11 01:55:07 +0000 2022PM10 particulate hot spots & 2 low pressure systems. 🔗
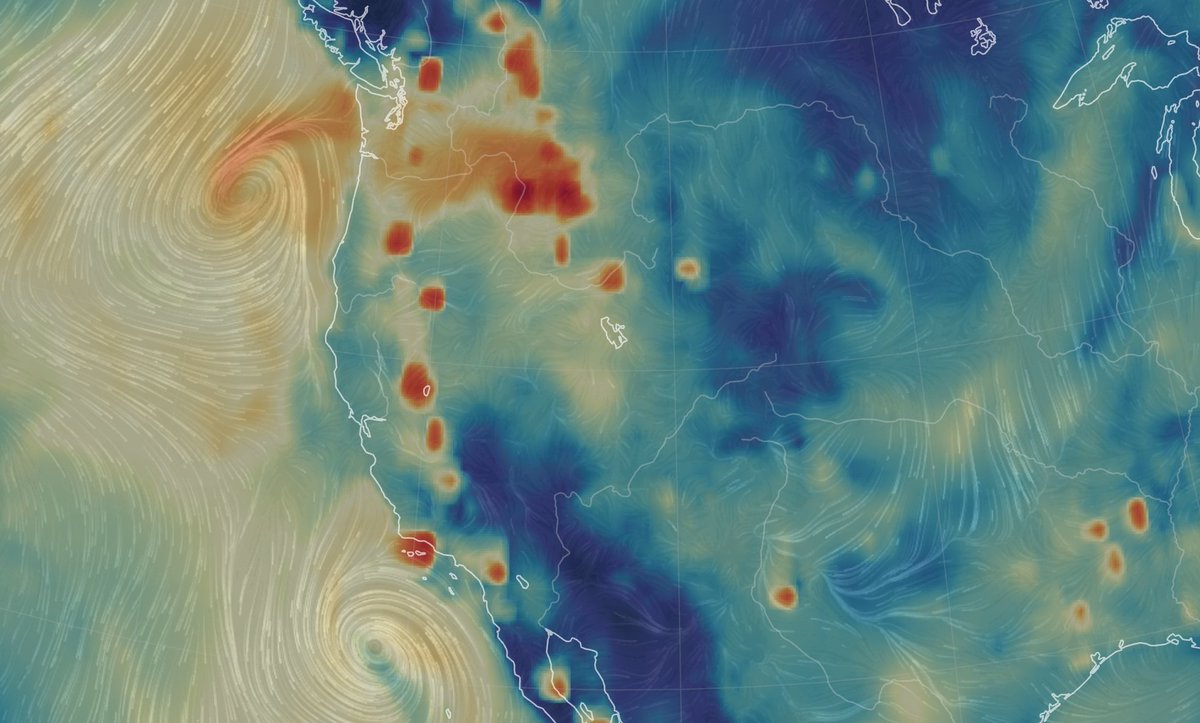
Sat Sep 10 19:42:28 +0000 2022@massspec4tw @Karl_Mechtler I was referring to the claim of associating 2× as many proteins with an IP experiment, compared to the lab's previous algorithm. Assigning more proteins is a matter of accepting assignments the previous algorithm rejected as too risky. How was the risk assessed in the 2 cases?
Sat Sep 10 16:38:14 +0000 2022It looks like Hurricane Earl is going to bring stormy weather to the Avalon Peninsula for the next few days as it executes a slow turn to an easterly heading. 🔗
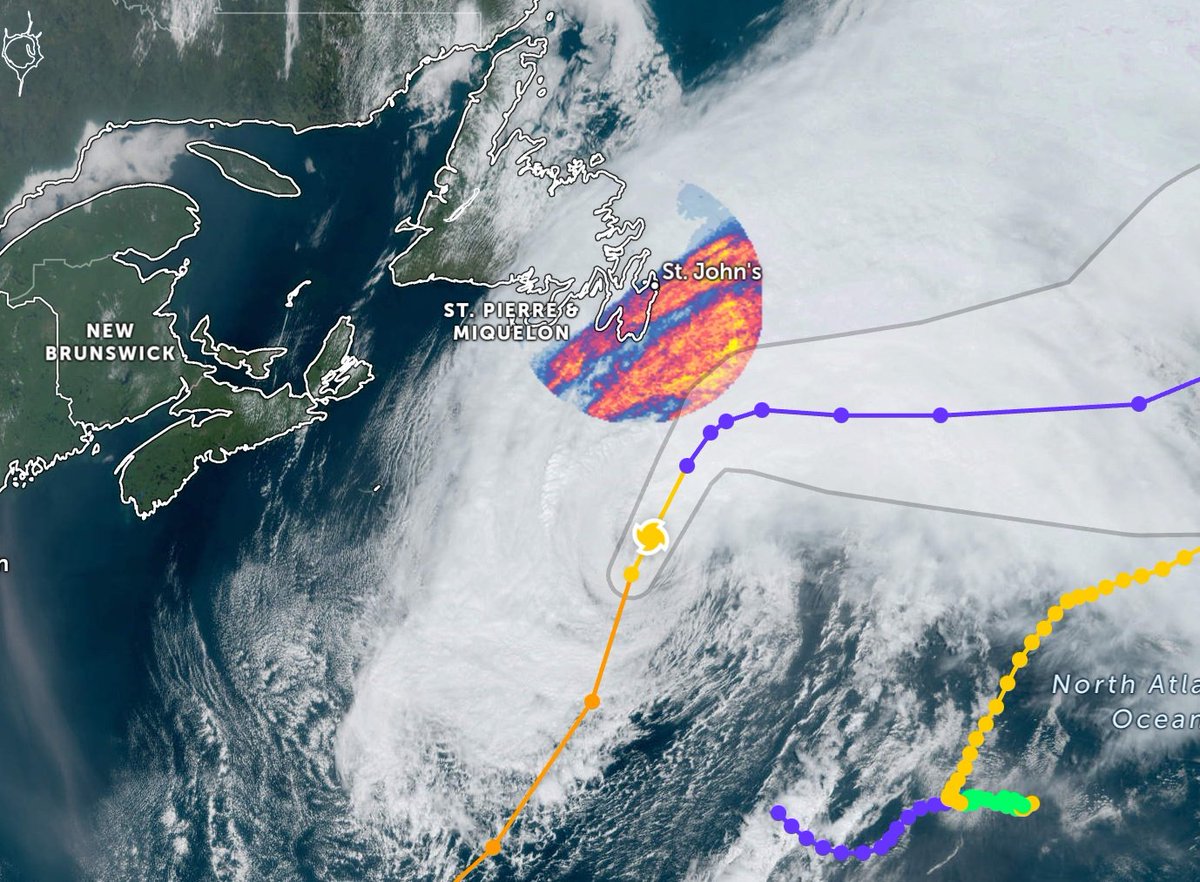
Sat Sep 10 14:47:11 +0000 2022They are both currently covering the signing of the paperwork that changes the Canadian Crown from being QE II to KC III, with the breathless intensity you would associate with developments in a matter that was in doubt.
Sat Sep 10 14:32:04 +0000 2022Current events have moved the Canadian TV news channels from boring irrelevance into full-blown Pythonesque absurdity.
Sat Sep 10 14:17:29 +0000 2022American Exceptionalism (& pursuit of old-fashioned modernity) on full display. 🔗
Sat Sep 10 14:08:16 +0000 2022@neely615 @chrashwood @mjmaccoss @Zeczycki_Lab @ProteomicsNews @JohnRYatesIII Not to mention phone calls from irate Congresspersons, who can get very involved in this sort of thing.
Sat Sep 10 14:03:31 +0000 2022@neely615 @chrashwood @mjmaccoss @Zeczycki_Lab @ProteomicsNews @JohnRYatesIII Federal agencies (US or otherwise) are always careful not to do anything that might throw shade on home country-based companies, unless they are specifically tasked with regulating them & even then only in the most extreme circumstances.
Sat Sep 10 13:39:36 +0000 2022@slavov_n I don't think there is a term analogous to "gene regulation" wrt proteins. Genes are either being transcribed or they aren't. There is no such set of Manichean alternatives for proteins.
Sat Sep 10 13:25:37 +0000 2022@slavov_n "Protein regulation" doesn't have any well-established meaning, so I would expect it to be a less useful descriptor in practice.
Sat Sep 10 13:18:28 +0000 2022@Karl_Mechtler It is a little unclear from the paper how ID reliability is assessed. Does it all hang off FDR's or is something more sophisticated being used?
Sat Sep 10 12:52:11 +0000 2022Typhoon Muifa (Inday, est 978 hPa) should make a close pass by Taipei as a Super Typhoon on the 13th & come very near Shanghai on the 15th. 🔗
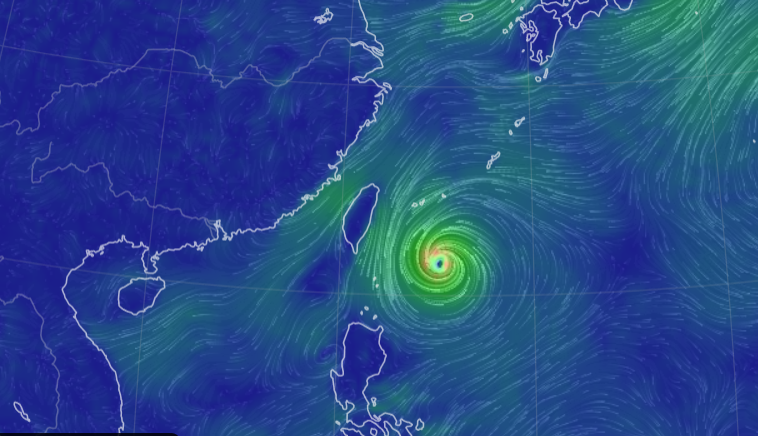
Sat Sep 10 12:25:15 +0000 2022Embarrassingly, the mechanism for generating this sort of GGylation pattern is unknown, as is its purpose & consequences.
Sat Sep 10 12:19:15 +0000 2022Modification diagram for ATR:p, a large kinase involved in DNA repair. It has 53 observed K+GGyl acceptors (30% of all K residues), 1 K+GGyl/SUMOyl acceptor & no K+acetyl acceptors. It also has 2× S+phosphoryl & 1× T+phosphoryl major iHOP domains.
🔗
Fri Sep 09 22:37:43 +0000 2022@VATVSLPR The current rain from Kay as it heads out into the Pacific, just grazing LA (NEXRAD) 🔗

Fri Sep 09 22:24:06 +0000 2022@astacus @byu_sam @nesvilab Possibly, but I would think the HLA presentation mechanism is a more likely source of selectivity. Each allele is pretty selective about with peptides they present and I don't think they are above being fussy about sidechains.
Fri Sep 09 21:32:16 +0000 2022@Smith_Chem_Wisc @byu_sam @nesvilab Just take a synthetic tryptic peptide & see if you can polymerize it with trypsin. It doesn't produce much product, but it is easy to get a feel for how much substrate you really need to force a protease into reverse.
Fri Sep 09 21:08:18 +0000 2022@byu_sam @nesvilab But PTMs aren't the issue: nearly all HLA peptides are unmodified, except for N-terminal processing, acetylation & pyroGlu when the peptide N-terminus is Q.
Fri Sep 09 21:05:53 +0000 2022@byu_sam @nesvilab I think it was just an honest mistake by the original group: many of the methods for QC in proteomics are not well adapted to sets of PSMs that are non-tryptic, all the same length (9±1 residues) & many heuristics like "2 peptides/protein" don't apply.
Fri Sep 09 20:00:09 +0000 2022@byu_sam If your really want to get into them, all of the public HLA class I expts can be found at
🔗
and class II are at
🔗
Fri Sep 09 19:58:58 +0000 2022@byu_sam This is a pretty typical, good HLA class II run:
🔗
and this a a pretty typical, good class I run:
🔗
Fri Sep 09 19:56:15 +0000 2022@byu_sam The way these samples are handled, the usual suspects (carbamyl, carbamidomethyl) aren't possible, but M/W+oxidation is common. Most C's will be cystines, unless you use derivatize them (not normally done).
Fri Sep 09 17:17:57 +0000 2022@VATVSLPR Rainfall caused by Kay in the last 48 hours, estimated from NEXRAD signals. Most of the heavy rains have been on the east side of the mountains, towards the Gulf of California. 🔗
Fri Sep 09 16:14:39 +0000 2022The data set has the added feature of a very well done TMT derivatization: nearly >99% of available amines derivatized with very few detectable side reactions. It makes the data suitable for SAAV detection, which is not always the case in such studies.
Fri Sep 09 15:53:27 +0000 2022Looking around the downtown skyline, it is surprising how few flags are at half-mast.
Fri Sep 09 15:41:54 +0000 2022Tropical storm Kay is in the process of veering off into the Pacific. It should be causing about as much trouble for southern Cali right now as it is going to, spreading cloudy weather as far away as Albuquerque NM. 🔗
Fri Sep 09 15:35:30 +0000 2022@DanZiemianowicz Try looking at this simple page explaining the basic properties Gaussian functions
🔗
Then go through the page & alter the equations to use base 2 rather than e. The added complexity should be apparent.
Fri Sep 09 15:18:54 +0000 2022@andy___jones @pwilmarth For your own sanity (& that of your students), delete that potential rat's nest of odd ball reviewer comments!
Fri Sep 09 14:25:17 +0000 2022@MHendr1cks I particularly like the use of "??" as delimiters for variables (??release_number??) that haven't been filled in, making reporting the error nearly impossible. Truly an example of contractor programming genius!
Fri Sep 09 14:05:49 +0000 2022@pwilmarth @andy___jones Put on your big-boy pants & delete them. Moving forward is much more important than your speculations about what you "should" do from 6 months ago. 🧐
Fri Sep 09 14:01:58 +0000 2022@DanZiemianowicz The only exception is if you are examining a process in which "doubling" is a fundamental process that should be clearly detectable in the results you will be plotting. Then base-2 makes sense.
Fri Sep 09 13:53:43 +0000 2022@DanZiemianowicz "e" is one of the fundamental constants that set how our universe works, like π, Plank's constant (h) or a whole bunch of other, less well known transcendental numbers. Substituting 2 for e is like substituting 3 for π: it is about right, but it leads to problems down the road.
Fri Sep 09 13:46:00 +0000 2022PXD036334: for anyone interested in using chromatographic separation effects to improve PSM id algorithms, this one has some very stark examples.
Fri Sep 09 12:36:12 +0000 2022You can also see that the N-terminal IDR is ripe for an RIP domain from the AF PAE diagram. 🔗
Fri Sep 09 12:27:13 +0000 2022That particular domain is a tough nut to crack: even chymotrypsin wouldn't help.
MSRESDVEAQ QSHGSSACSQ PHGSVTQSQG SSSQSQGISS SSTSTMPNSS QSSHSSSGTL SSLETVSTQE LYS …
Fri Sep 09 12:15:30 +0000 2022It has a 4 discrete major iHOP-type phosphodomains. The N-terminal S-rich low complexity domain (2-73) is almost certainly phosphorylated, but because the domain also lacks appropriately spaced K, R, E or D residues, it does not show up in MS-based proteomics experiments.
Fri Sep 09 12:08:33 +0000 2022Modification diagram for human CHEK2:p, a subunit recruited to DNA repair & replication assemblies. It has 6× K+GGyl acceptors, 2 of which are shared K+GGyl/acetyl acceptors. The 2× K+SUMOyl acceptors are not shared.
🔗
Fri Sep 09 00:53:37 +0000 2022@VATVSLPR @pwilmarth @UCDProteomics @DRAWheatcraft I guess I have had too much contact with young people who are Linux-native & are dismissive of Windows-related utilities.
Thu Sep 08 23:35:35 +0000 2022Tropical storm Muifa looks like it may affect Taipei as a Typhoon on the 13th. 🔗
Thu Sep 08 23:05:31 +0000 2022@pwilmarth @UCDProteomics @DRAWheatcraft If only there was some way for a professor to pass on knowledge to the young ... 🧐
Thu Sep 08 22:43:22 +0000 2022@UCDProteomics @DRAWheatcraft That & pigz for compression can archive things quite quickly.
Thu Sep 08 22:41:20 +0000 2022@UCDProteomics @DRAWheatcraft Command line "tar.exe" has been part of Windows since 10 was released, so the whipper-snappers should broaden their horizons.
Thu Sep 08 22:35:07 +0000 2022@UCDProteomics @DRAWheatcraft So why not use tar?
Thu Sep 08 21:18:38 +0000 2022Hurricane Kay is currently grazing the Pacific coast of the Baja Peninsula, with 130 km/hr winds. 🔗
Thu Sep 08 18:51:40 +0000 2022@astacus @neely615 @UCDProteomics Many of the investigators that do these experiments are very much aware of the kinetics of bait-prey interactions & have been putting some effort into understanding how it may influence their results for quite a long time, e.g. 🔗
Thu Sep 08 14:50:26 +0000 2022@astacus @neely615 @UCDProteomics More like:
observed in ≥ 8/10 replicates for baits A, B & C,
but ≤ 1/10 for other baits
Washing/binding stringency is the sine qua non in these experiments.
Thu Sep 08 14:31:28 +0000 2022@astacus @neely615 @UCDProteomics A commonly done experiment is bait-prey protein-protein interaction determination, which is of biological value but it relies on reproducibility rather than anything that could be legitimately called quantitation.
Thu Sep 08 14:23:28 +0000 2022@astacus @neely615 @UCDProteomics I hate/love to be the pedantic know-it-all, but it really depends on the hypothesis you are testing. Many don't require anything more than above or below LOD.
Thu Sep 08 14:04:52 +0000 2022People who use their email client as a document storage & retrieval system drive me nuts! 😱
Thu Sep 08 13:07:31 +0000 2022@Ben_Leo_Parker I would agree that trade shows like the ASMS still have a place in the sales cycle for large ticket equipment.
Thu Sep 08 12:42:36 +0000 2022Hurricane Kay (140 km/hr, est. 974 hPa) is moving up the Baja Peninsula, where ground stations are reporting up to 70 mm of rain. It is expected to veer off into the Pacific before reaching San Diego. 🔗
Thu Sep 08 12:28:23 +0000 2022S/T+phosphoryl acceptors steer clear of the protein kinase domain (9-265), mainly confining themselves to a high occupancy, S-rich RIP domain (279-357), as illustrated on the AF predicted aligned error diagram. 🔗
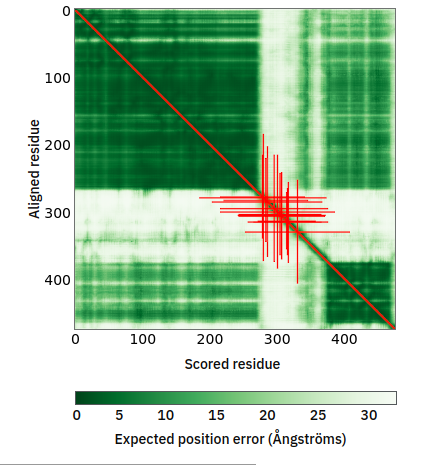
Thu Sep 08 12:22:42 +0000 2022Modification diagram for human CHEK1:p, a subunit recruited to DNA repair & reproduction assemblies. The 17× K+GGyl acceptors are spread across the sequence & include 4× GGyl/acetyl & 7× GGyl/SUMOyl shared acceptors. 🔗
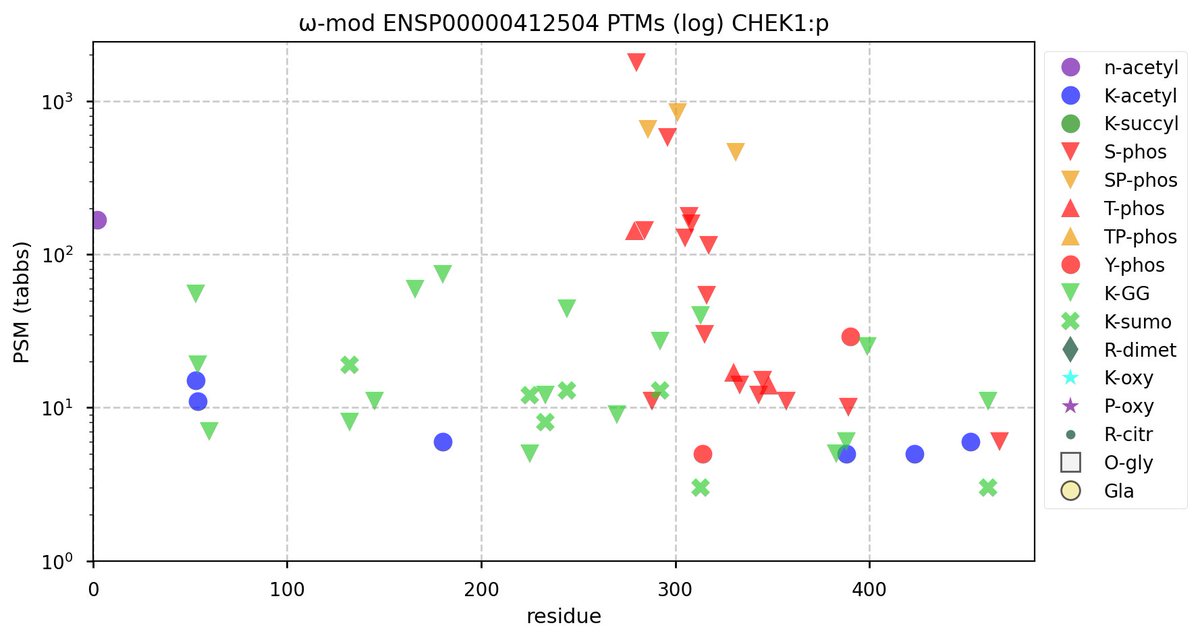
Thu Sep 08 11:25:44 +0000 2022@ronanlupton I don't know enough about that type of therapeutic to be able to comment. I am primarily interested in how proteins use post-translational modifications, not their role in disease.
Thu Sep 08 02:49:10 +0000 2022Huricane Danielle is still steaming northeast through the Atlantic. It is predicted to loop around & head for northern Spain, arriving on Monday evening as extratropical storm. 🔗
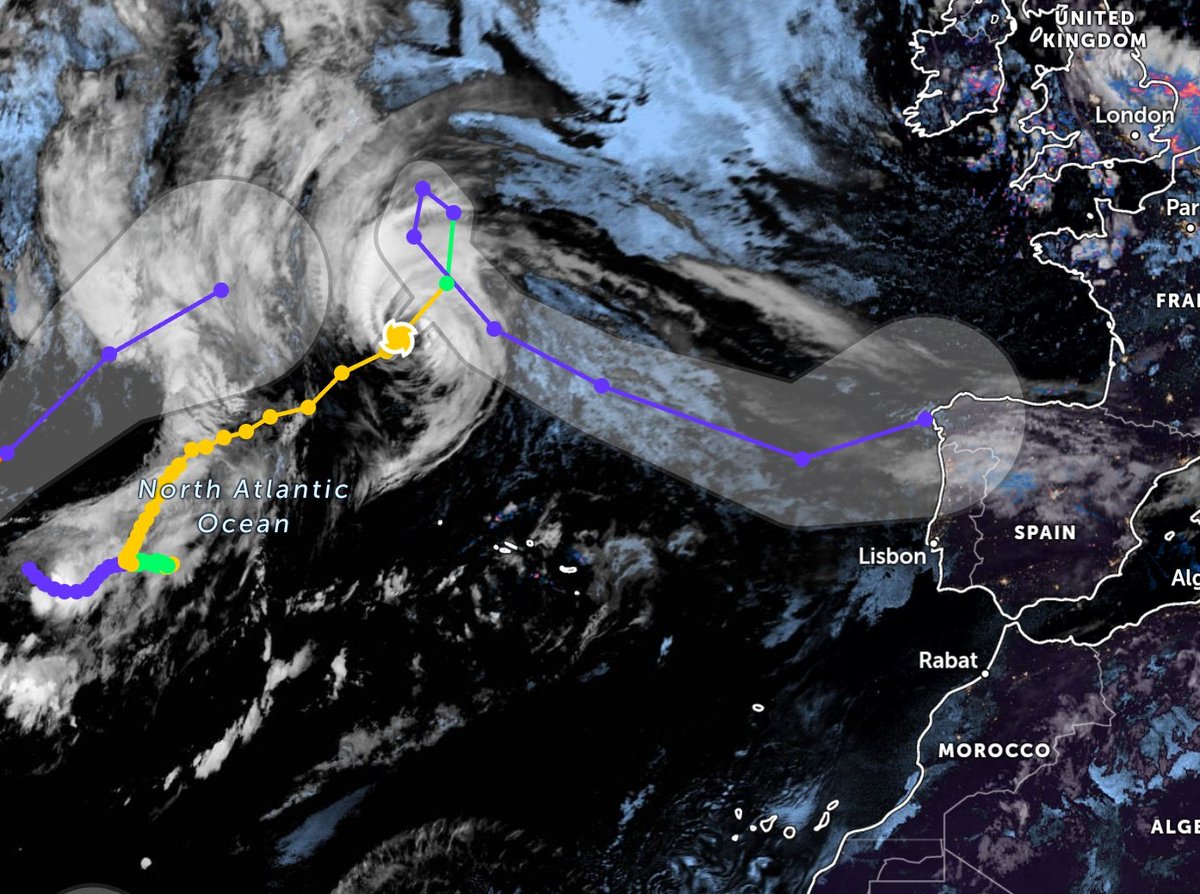
Wed Sep 07 18:02:01 +0000 2022@GarciaLabMS It may be that there is still some merit to the IRL/warm-handshake style of meeting, but I think most students now connect & network on-line long before they attend a conference.
Wed Sep 07 15:37:06 +0000 2022Someone has decided it would be a good idea to try to stuff a bunch of cats into a bag that already contains a number of wild squirrels. 🔗
Wed Sep 07 13:15:23 +0000 2022@pwilmarth They are more like high-pressure sales events (for condo time-shares & the like) held at conference hotels near the airport.
Wed Sep 07 13:02:50 +0000 2022Morning mirepoix 🔗

Wed Sep 07 12:40:12 +0000 2022When I see people make enthusiastic comments about conferences they have attended, all I can think of is: why does anyone still go to conferences? It all seems very 20th century to me.
Wed Sep 07 12:15:56 +0000 2022The AF structure isn't very helpful in this case. While amusing (& somewhat suggestive), I don't think it represents a realistic conformation of the molecule. It looks like the problem AF tends to have with coiled-coils. 🔗
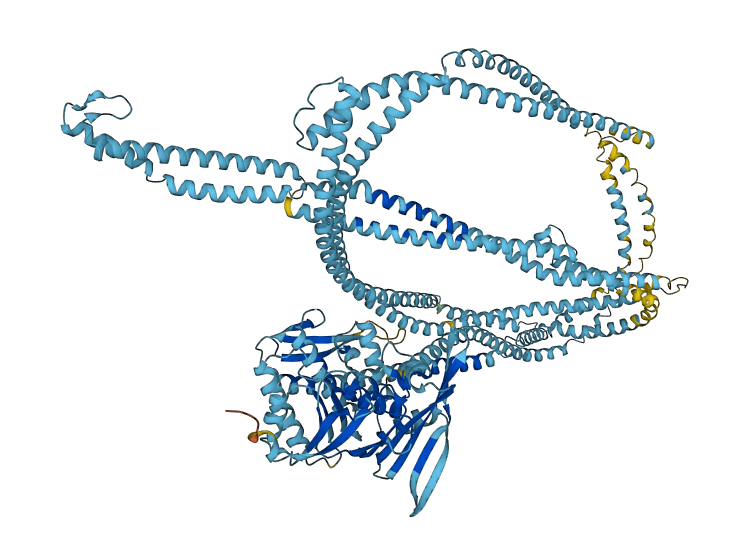
Wed Sep 07 12:11:47 +0000 2022Modification diagrams for RAD50:p, a subunit of DNA repair foci. Unlike RAD51:p, it is frosted with K+acetyl acceptors, 12 of which are K+acetyl/GGyl. It does not have any observed K+SUMOyl acceptors. It has a single major phosphodomain (623-690) & several minor RIP domains. 🔗
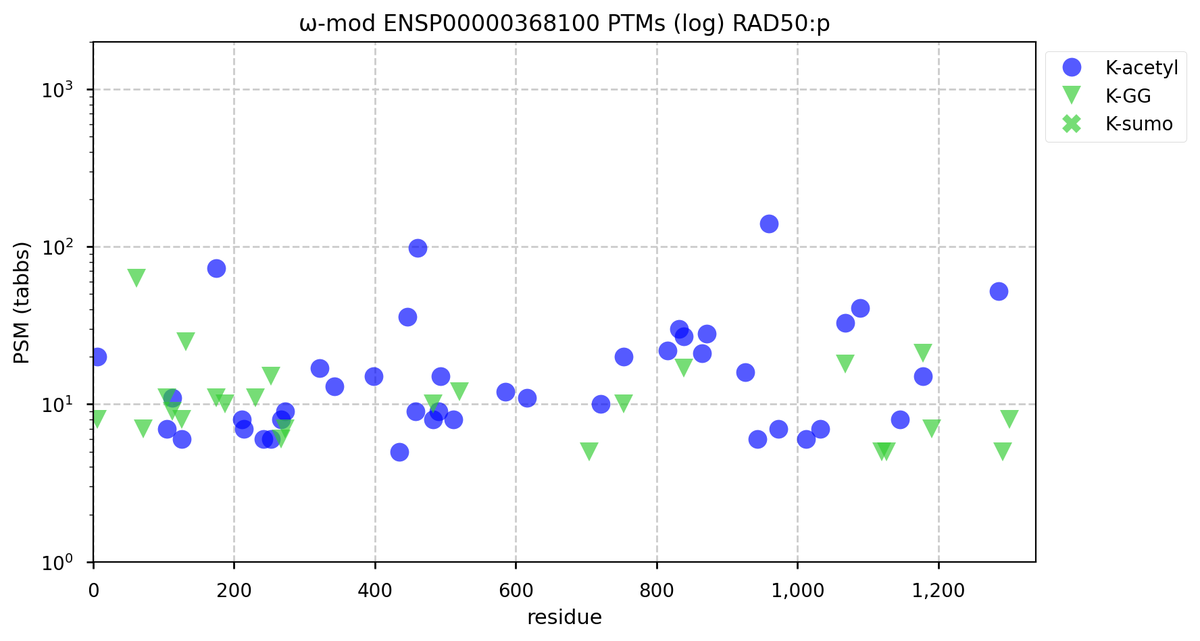
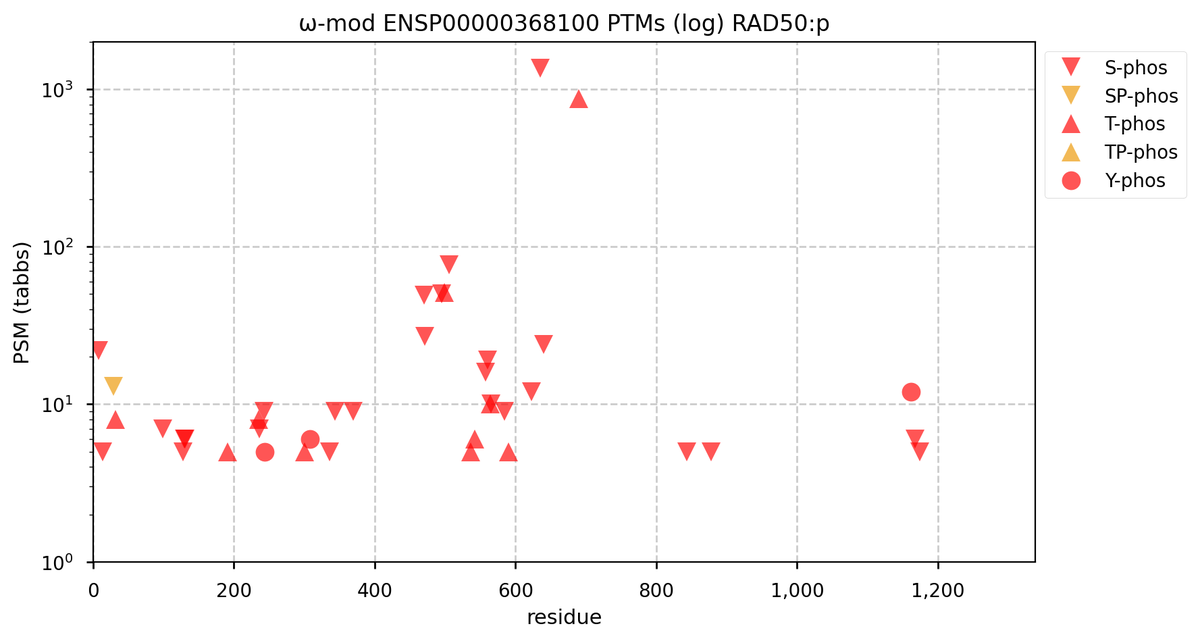
Wed Sep 07 11:58:13 +0000 2022Hurricane Kay (155 km/hr, est. 975 hPa) is heading north, up the Mexican coast. It is expected to graze the Baja Peninsula as a category 1 storm, starting Thursday evening. 🔗
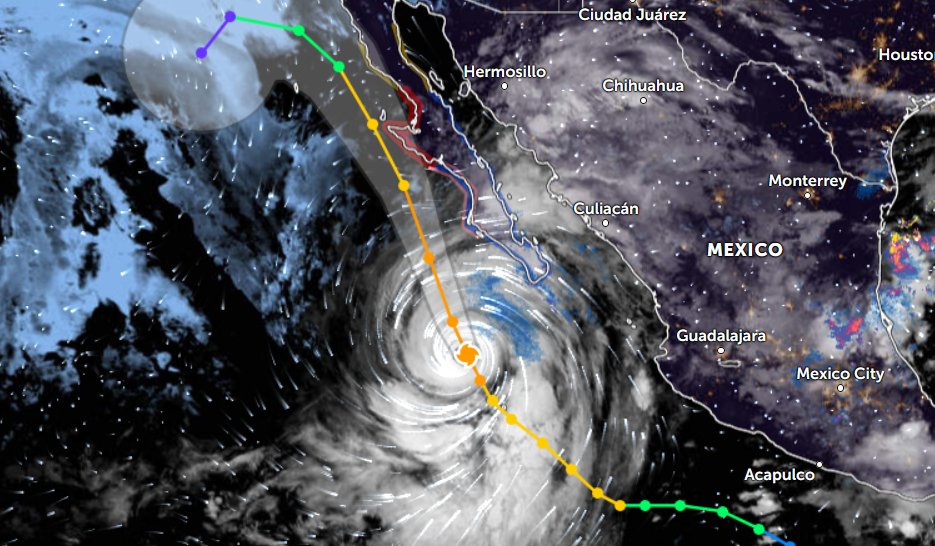
Wed Sep 07 00:57:00 +0000 2022@UCDProteomics @VATVSLPR @pwilmarth @neely615 @chrashwood I'll take you guys at your word, but I remain skeptical.
Tue Sep 06 18:49:36 +0000 2022@VATVSLPR @pwilmarth @neely615 @chrashwood Most people do not like that sort of algorithm-based parameter allocation. Hopeful the attitude will die out, but many practitioners see parameter-tinkering as one of the skills that makes them "experts".
Tue Sep 06 17:51:13 +0000 2022It is going to be a warm few days in Sacramento CA, but particularly today, topping out at 47° C 🔗
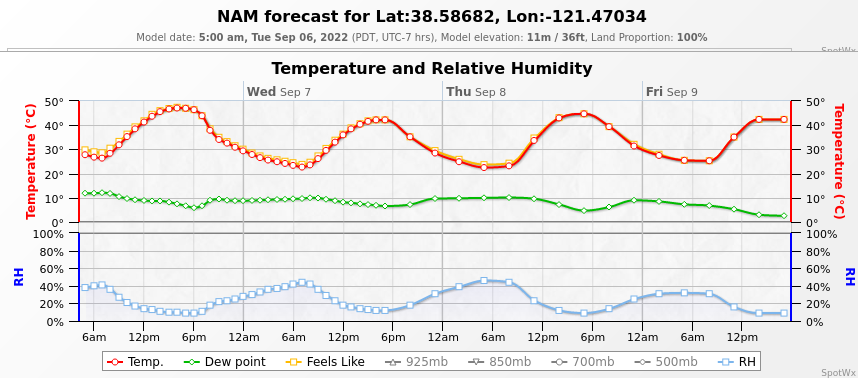
Tue Sep 06 14:55:51 +0000 2022Whenever possible, chose base e (~2.71828) logs rather than base 2 logs. Using base 2 makes your results look like they were done as a hobby project by a well-meaning country physician in olden-times.
Tue Sep 06 12:12:39 +0000 2022Typhoon Hinnamnor (140 km/hr winds, est. 967 hPa) is heading north through the Sea of Japan, about half way between Sapporo & Vladivostock. It should make landfall in Russia later on today. 🔗
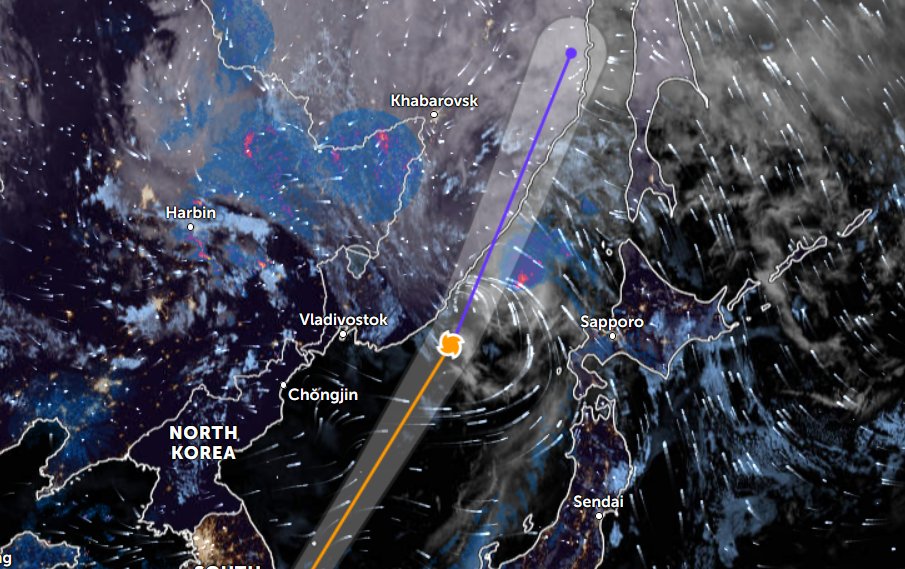
Tue Sep 06 12:01:20 +0000 2022Modification diagram for RAD51:p, a subunit of DNA repair foci. It eschews the PTM bling so popular with many DNA repair subunits, with only N-terminal acetyl, a low occupancy S/T+phosphoryl & 3 K+GGyl acceptors.
🔗
Tue Sep 06 00:17:15 +0000 2022PXD026371: nice data & just like PXD032355, pancreatic ductal adenocarcinoma tissue has lots of collagen
Mon Sep 05 23:30:28 +0000 2022🔗
Mon Sep 05 22:47:31 +0000 2022🔗
Mon Sep 05 22:45:45 +0000 2022Typhoon Hinnamnor (165 km/hr winds, est. 952 hPa) has passed over Busan KR & is headed out into the Sea of Japan. 🔗
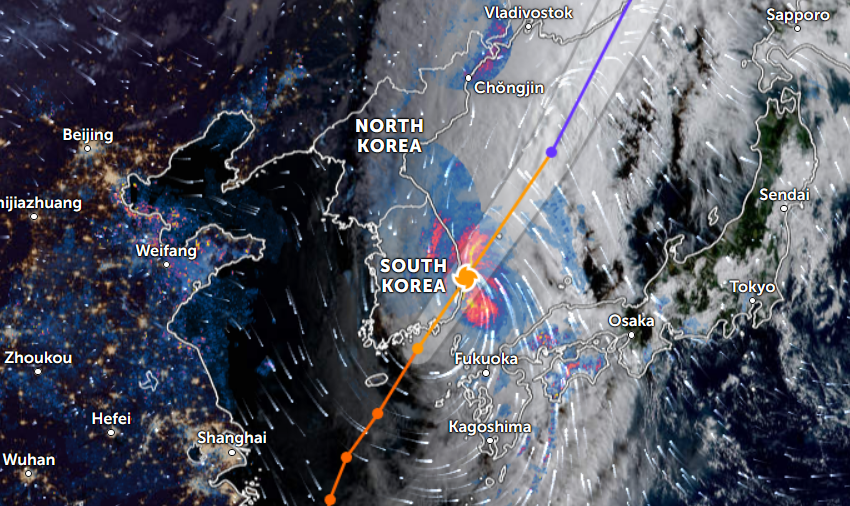
Mon Sep 05 15:45:09 +0000 2022🔗
Mon Sep 05 15:26:34 +0000 2022Very Strong Typhoon Hinnamnor is picking up speed & should make landfall near Busan KR later on today. It is bringing widespread rain & stormy conditions of South Korea at the moment and firmly blocking the southern end of the Korean Strait. 🔗
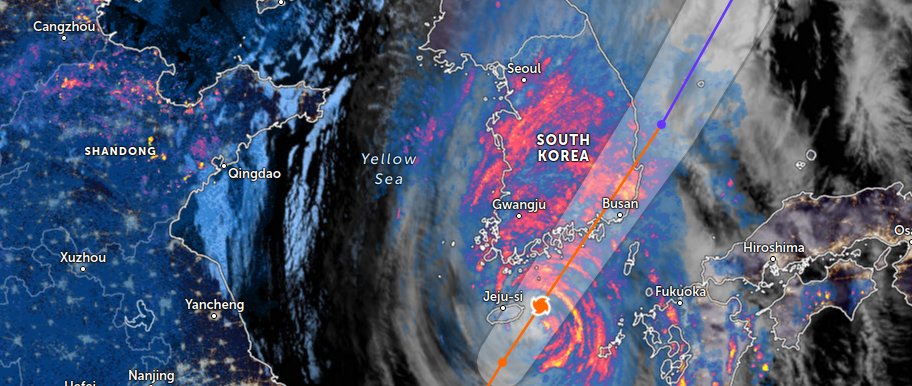
Mon Sep 05 14:42:31 +0000 2022Not a great pattern for harvest time. 🔗
Mon Sep 05 13:27:58 +0000 2022MDC1:p has lots of PTMs & 32 SAAVs with maf > 1%. The somewhat longer ATRX:p also has lots of PTMs but 0 SAAVs with maf > 1%. Proteomics is a bad field for anyone who feels the need to generalize about everything.
Mon Sep 05 13:10:24 +0000 2022Lysine modifications are dominated by K+acetyl in the front (10-1479), shifting to K+GGyl/SUMOyl in the back (1488-2490): the rarely seen ε-mullet domain pattern.
Mon Sep 05 12:28:26 +0000 2022Very Strong Typhoon Hinnamnor (aka Henry, est. 941 hPa) is still tracking north through the East China Sea, approaching South Korea. Its outer bands are affecting Fukuoka & it continues to disrupt this very busy shipping lane. 🔗
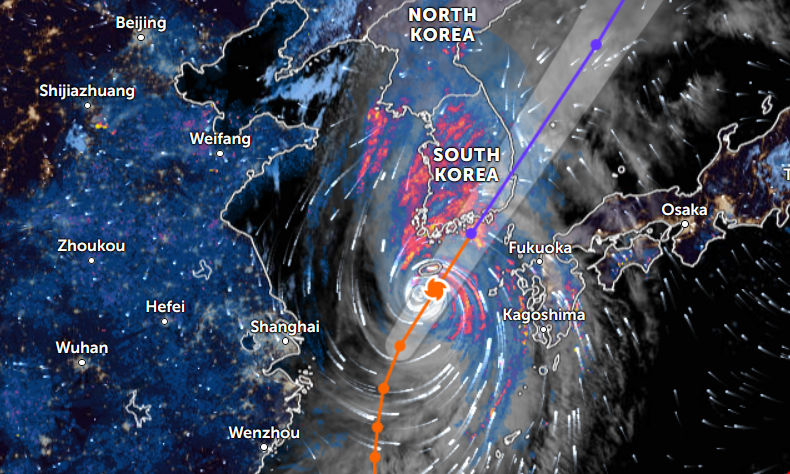
Mon Sep 05 12:13:47 +0000 2022The S/T+phosphoryl pattern follows the typical pattern of sticking to IDRs in a protein that only has "structure" in the C-terminal third of the sequence (phosphoryl, in red, superimposed on the AF predicted aligned error diagram). 🔗
Mon Sep 05 12:07:44 +0000 2022Modification diagrams for human ATRX:p, a subunit attracted to DNA damage. It has a hard to remember name (Alpha Thalassemia Retardation syndrome X-linked), but a lot of PTMs. 🔗
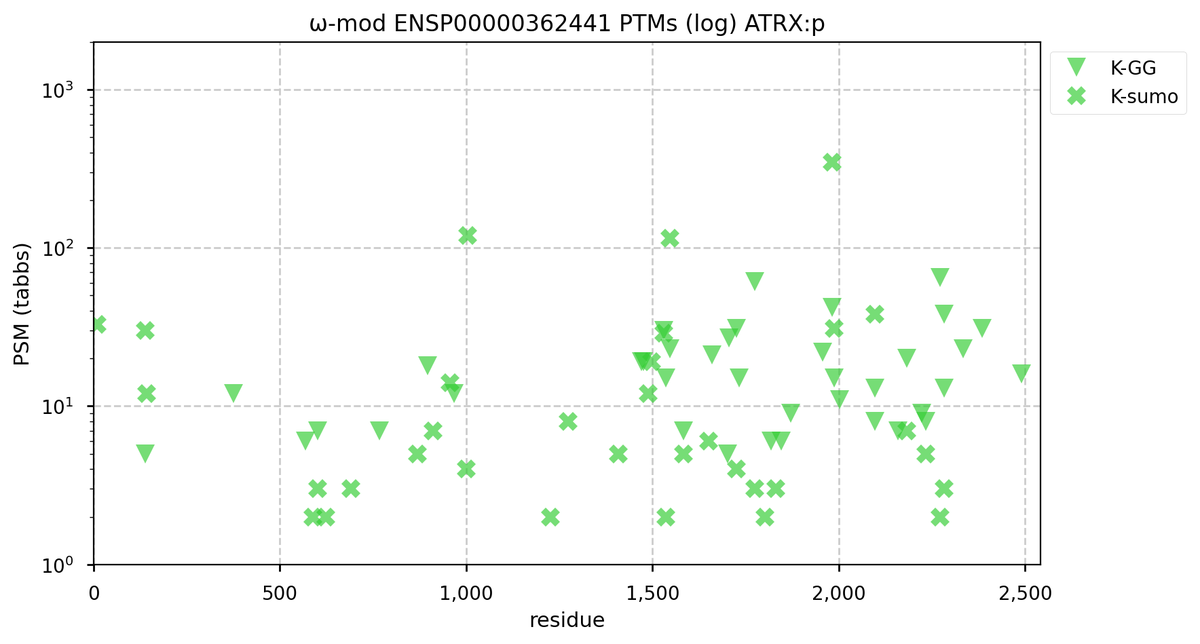
Mon Sep 05 01:51:33 +0000 2022PXD032042: provides an interesting look at L-A & L-BC virus levels in multiple strains of S. cerevisiae.
Mon Sep 05 01:15:51 +0000 2022Looks like there will be visible aurora here again tonight. 🔗
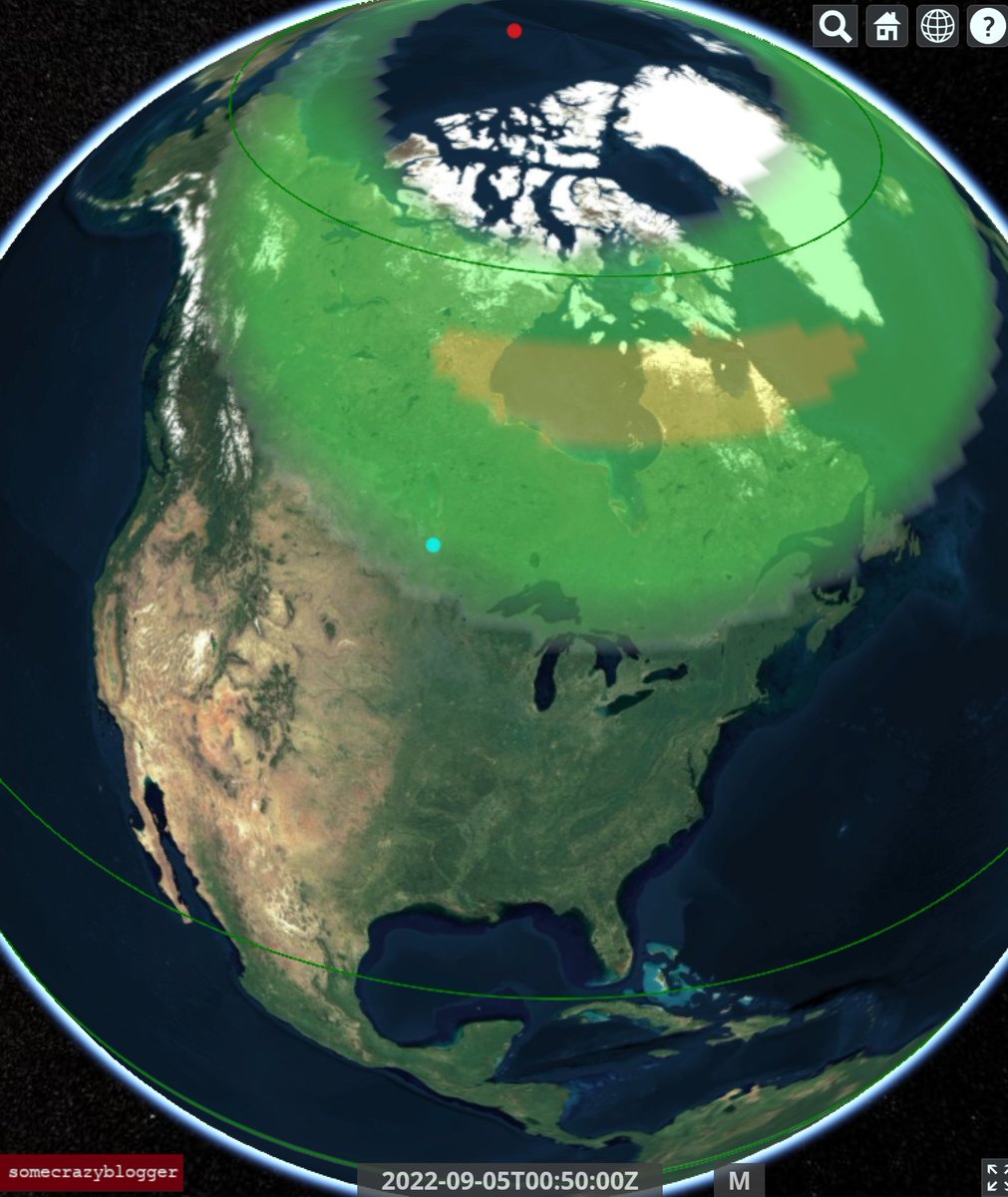
Sun Sep 04 16:09:54 +0000 2022PXD032355: nice data that really shows how big the collagen signals can be in cancer tissue samples (in this case ovarian clear cell carcinoma).
Sun Sep 04 15:52:53 +0000 2022Very Strong Typhoon Hinnamnor (aka Henry, est. 942 hPa) is now tracking north through the East China Sea. It's center is predicted to pass over Busan KR as an extratropical cyclone at about midnight Sept 6. 🔗
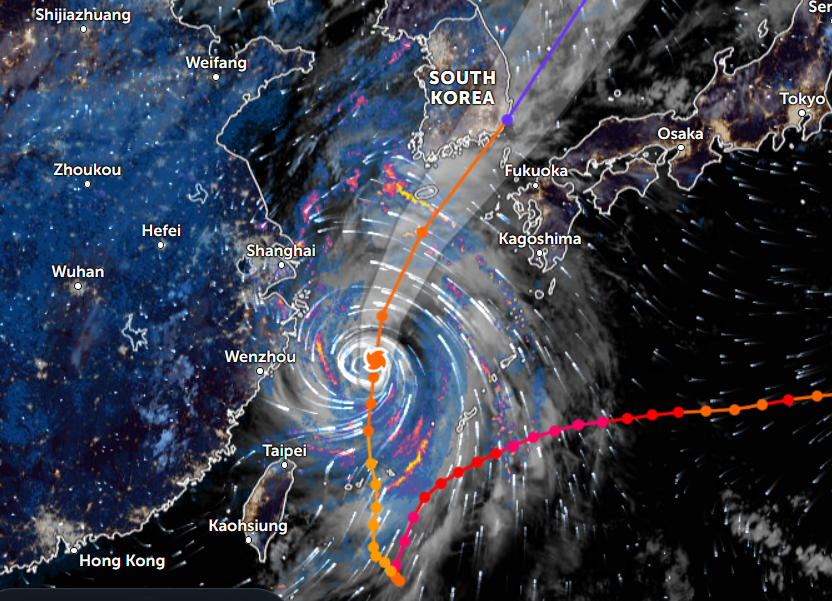
Sun Sep 04 15:34:24 +0000 2022Trigger warning: contains wide-spread evidence of non-canonical phosphorylation, which may upset some viewers.
Sun Sep 04 15:13:24 +0000 2022The potential for monsoon rainfall has dropped significantly in Pakistan & northern India (cyan means more total precipitable water). 🔗
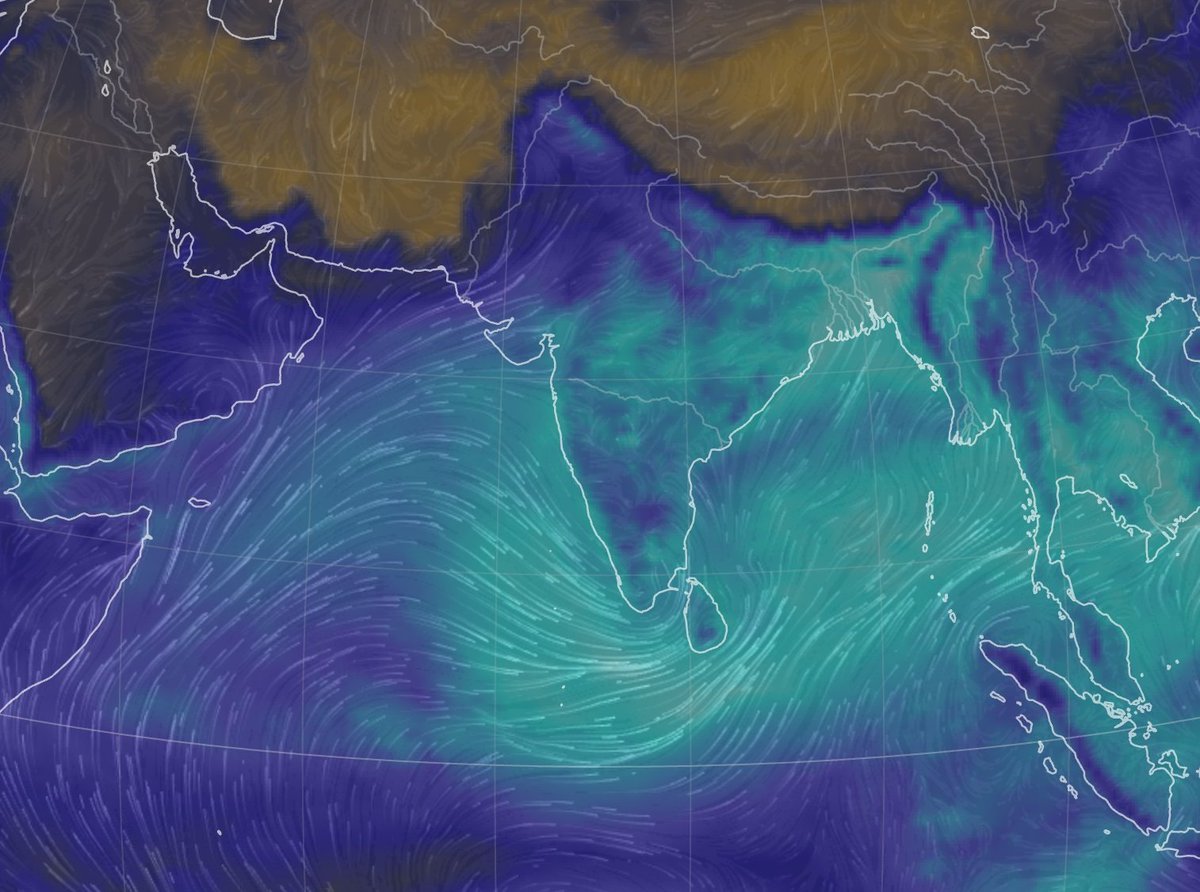
Sun Sep 04 12:50:44 +0000 2022MDC1:p also has 32 single amino acid variants with minor allele frequencies > 1% that have been observed in publicly available proteomics data, the most common of which is MDC1:p.T1623P (maf = 12%).
Sun Sep 04 12:43:10 +0000 202261×K+acetyl acceptors are present in 3 or 4 loose clusters, along with observation of
2×K+SUMOyl,
1×K+GGyl/SUMOyl,
2×K+acetyl/GGyl,
2×K+acetyl/SUMOyl, &
2×K+acetyl/GGyl/SUMOyl
acceptors.
Sun Sep 04 12:26:47 +0000 2022Modification diagrams for MDC1:p, a subunit of DNA damage foci. Dominated by IDRs: 65% of both S & T residues have been observed to act as phosphorylation acceptors. The high occupancy acceptors in (1-1086) are S+phosphoryl, but T+phosphoryl is the top dog in (1138-1820). 🔗
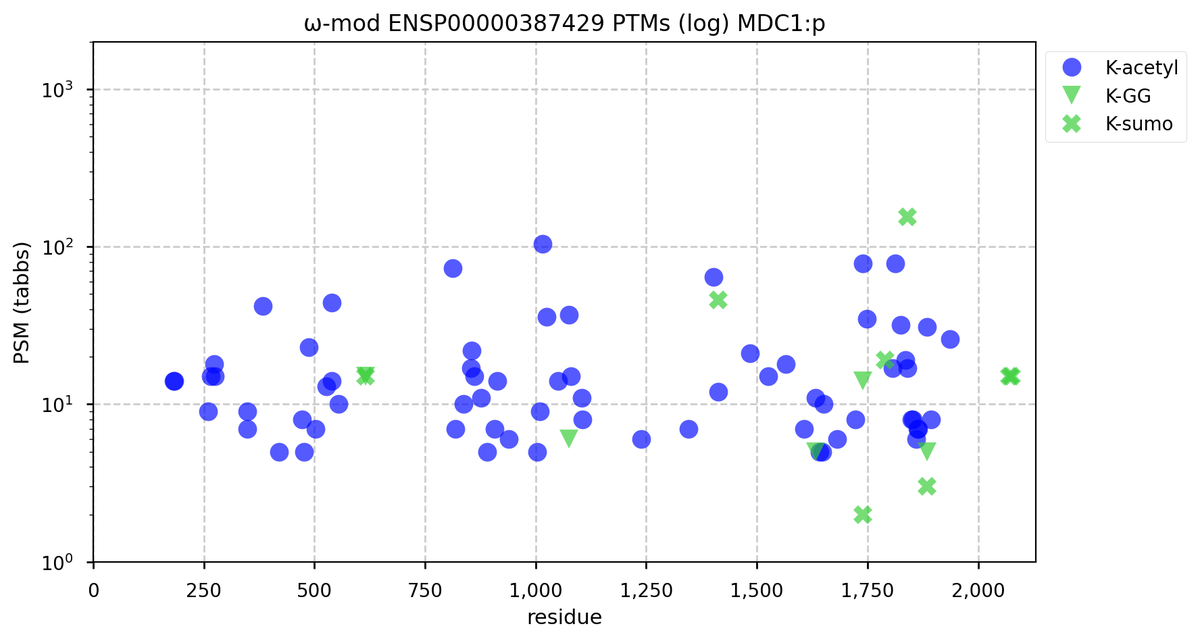
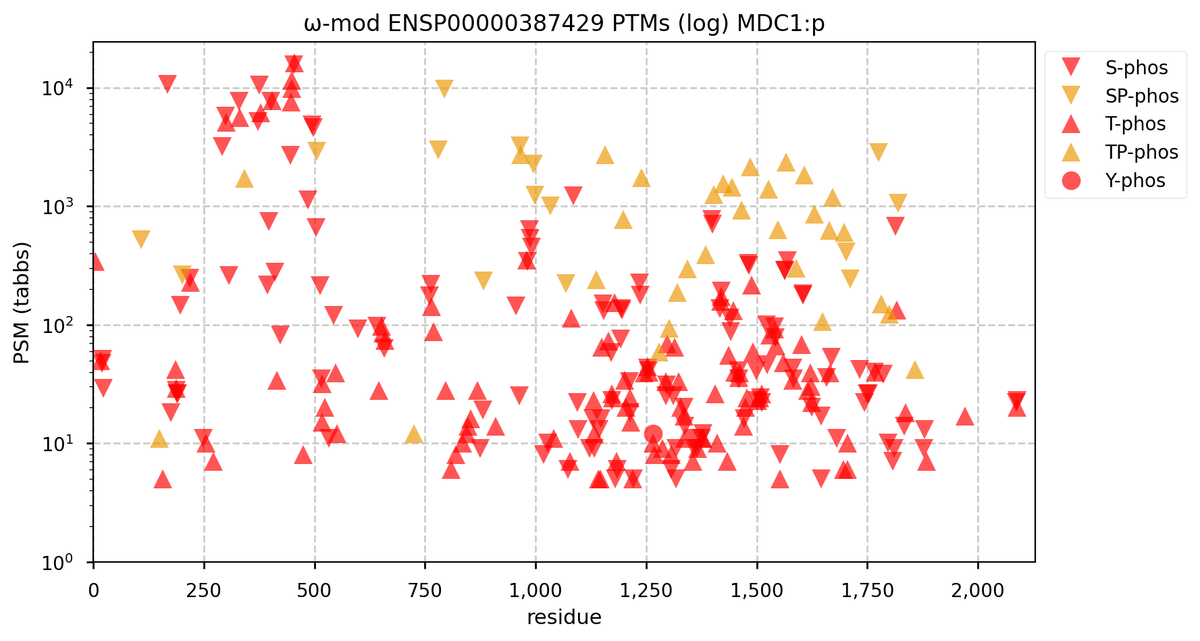
Sat Sep 03 19:37:45 +0000 2022@neely615 @VATVSLPR @pwilmarth Will do. Plants can be, well, challenging, particularly ones that have had humans tinkering around with their chromosomes for a few thousand years. And amphibians, too, although they seem to screw around with their own chromosomes without our interference.
Sat Sep 03 19:30:13 +0000 2022@neely615 @VATVSLPR @pwilmarth Was that for the octaploid or decaploid variety?
Sat Sep 03 19:09:49 +0000 2022@neely615 @pwilmarth As a practical example, the bovine proteome is pretty much useless for analyzing human plasma (or urine or CSF) data & vice versa.
Sat Sep 03 18:43:57 +0000 2022@VATVSLPR @pwilmarth I await the strawberry proteome.
Sat Sep 03 18:11:52 +0000 2022PXD025401 has some pretty top-drawer Trypanosoma brucei data. If you are into kinetoplastids (& who isn't), it may be just the amuse-bouche you needed to get interested in proteomics.
Sat Sep 03 17:25:44 +0000 2022PXD018214 is kind of interesting. It is unusually enriched in PSMs with multiple S/T+phosphoryl on the same peptide. Worth a look if that sort of thing floats your boat, but old-timey biochemists might want to give it a miss.
Sat Sep 03 17:00:10 +0000 2022<meta name="viewport" content="width=700,initial-scale=1" />
<script>
var W = 700;
var C = screen.width/W;
if(screen.availHeight > screen.availWidth){
document.querySelector('meta[name="viewport"]').setAttribute('content','width='+W+',initial-scale='+C+'');
}
</script>
Sat Sep 03 16:59:45 +0000 2022I have found that the following incantation smooths out a lot of the kinks:
Sat Sep 03 16:35:16 +0000 2022Getting web sites to look good on mobiles, tablets & laptops in both landscape & portrait modes is much easier than it used to be, but it still tests my soul every time I do it.
Sat Sep 03 15:03:04 +0000 2022@pwilmarth I have only really looked at this for mammals, but identifiable tryptic peptide identity from 1 species to another has an overall average of ~50%, ranging from a higher value (~70%) for mitochondrial proteins to <10% for many abundant circulating plasma proteins.
Sat Sep 03 14:36:05 +0000 2022Was anything done following this report? I see data that is supposed to be from a particular cell line that looks suspiciously like it is from another, but I always thought it was just a mismatch between what the boss wanted & what actually worked.
🔗
Sat Sep 03 13:23:54 +0000 2022@PingzhaoH @SchulichMedDent Our previous employer was a really tough place to do bioinformatics at any level. Hopefully the administration at Western has a better disposition towards the field.
Sat Sep 03 12:58:05 +0000 2022Both MMS22L:p & TONSL:p have about 1 K+GGyl acceptor per 100 residues. Looking at it another way, MMS22L:p has 16% of its K's serving as acceptors, while TONSL:p has 30% of its K's serving as acceptors. How many is too many?
Sat Sep 03 12:30:00 +0000 2022Typhoon Hinnamnor (aka Henry, est. 959 hPa) is still tracking north, on track to pass over Busan KR. Its outer bands are causing rain & stormy condition from Taiwan to Okinawa. 🔗
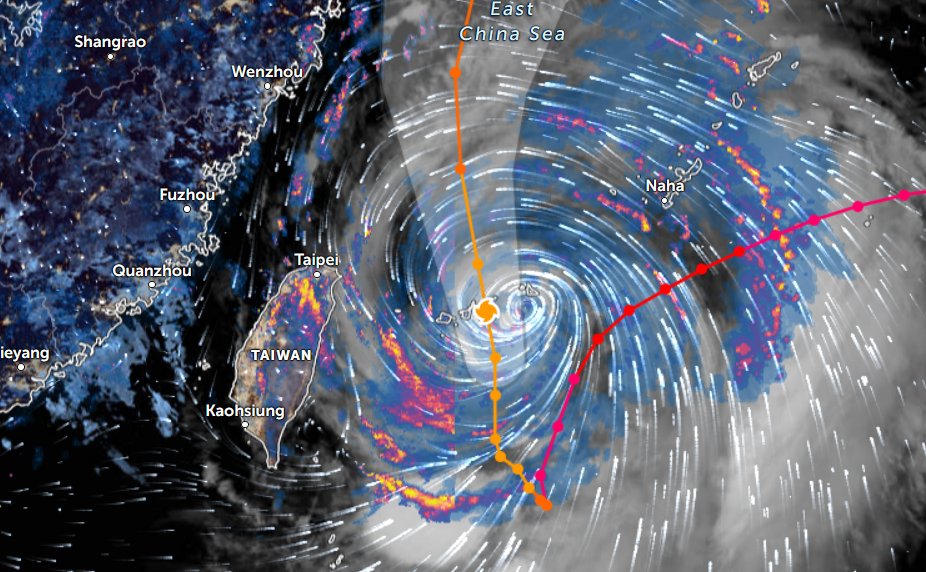
Sat Sep 03 12:20:35 +0000 2022How many experimentally confirmed ubiquitination acceptors does a protein need to have before it can be officially referred to as "short-lived"?
Sat Sep 03 12:09:23 +0000 2022TONSL:p has quite a bit of structural stuttering. (27-385) has 8× tetratricopeptide repeat domains, (528-626) has 3× ankyrin repeat domain & (1069-1354) has 7× leucine rich repeat domains.
Sat Sep 03 12:08:53 +0000 2022Modification diagrams for human MMS22L:p & TONSL:p, subunits of the aptly named MMS22L-TONSL complex. MMS22L:p has K+GGyl-only acceptors across the sequence, while TONSL:p mixes in a few K+SUMOyl acceptors. TONSL:p also features an RIP domain (669-908) in its main IDR. 🔗
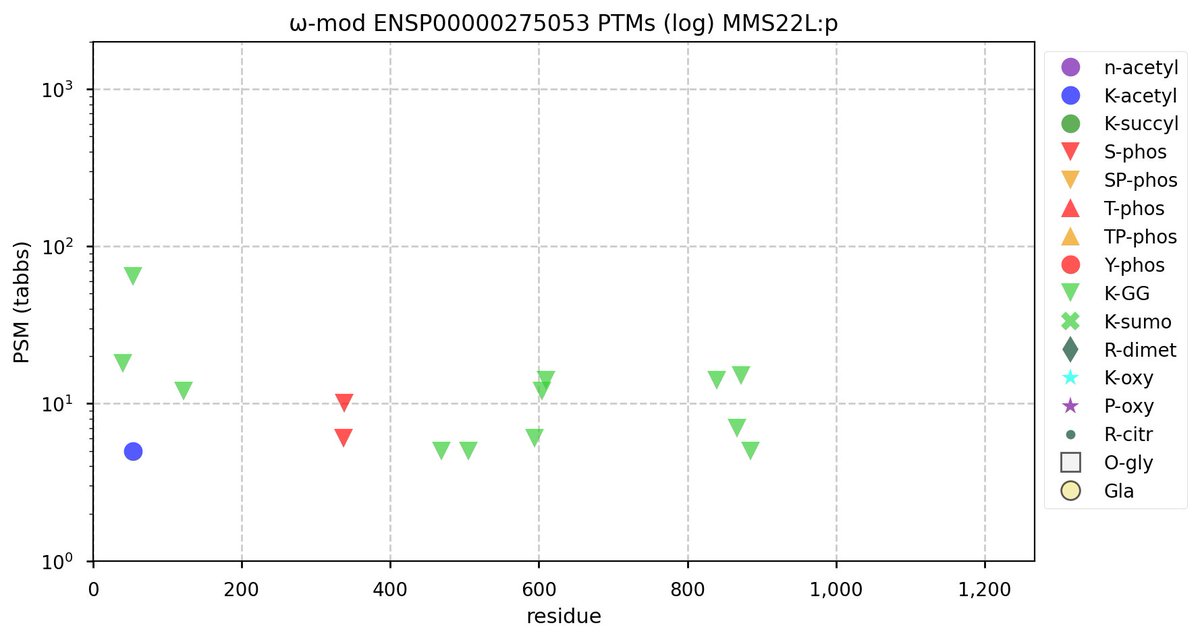
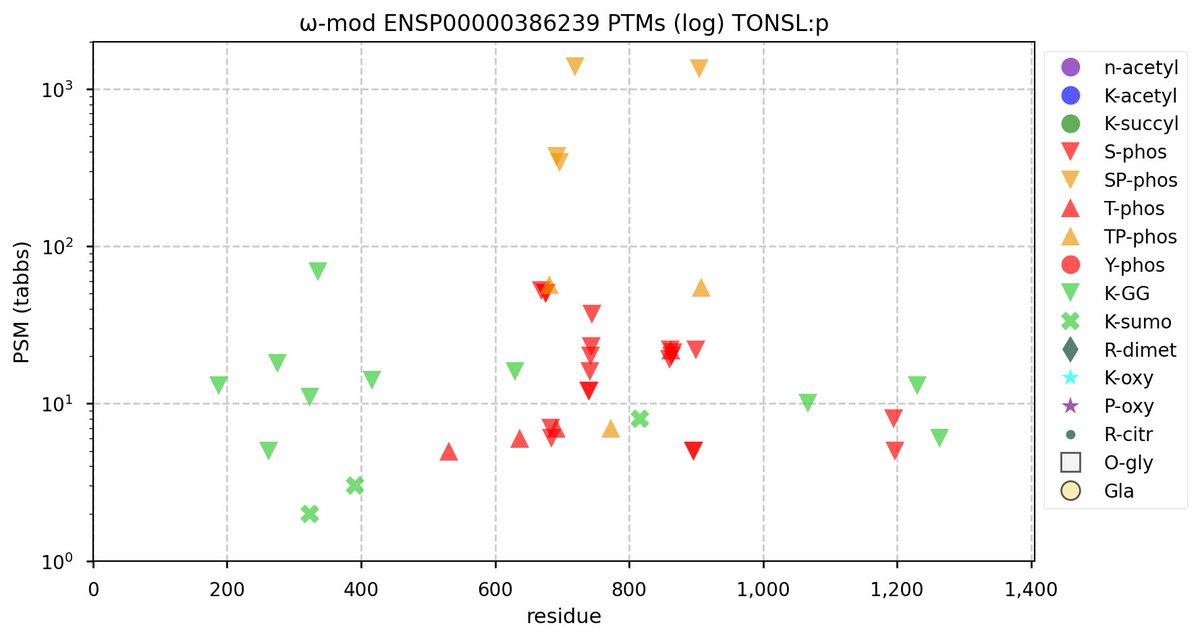
Sat Sep 03 02:07:06 +0000 2022I just opened up a console to start an ssh session & typed out a logged in to mysql instead. Maybe I should give it a rest for the evening ...
Sat Sep 03 01:18:02 +0000 2022@OverlordAnth Whoops.
Fri Sep 02 16:59:14 +0000 2022Typhoon Hinnamnor (aka Henry) has executed its predicted turn to the north & is now predicted to pass directly over the busy port city of Busan, KR sometime after noon on Sept. 5. as an extra-tropical cyclone (155 km/hr winds), 🔗
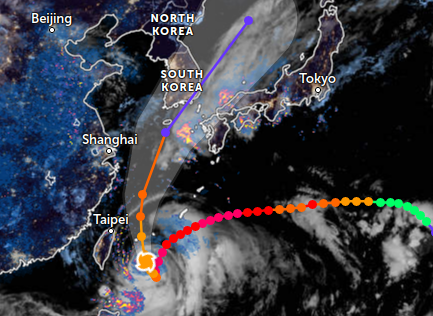
Fri Sep 02 12:14:40 +0000 2022K+GGyl/SUMOyl acceptors occur throughout the sequence, generalizing to K+GGyl/SUMOyl/acetyl acceptors in (298-510) and (609-682).
Fri Sep 02 12:14:18 +0000 2022Modification diagrams for human MRE11:p, a nuclease involved in DNA maintenance.The major S+phosphoryl RIP domains in (512-702) occupy the C-terminal IDR, with several minor isolated S+phosphoryl acceptors in (2-275). 🔗
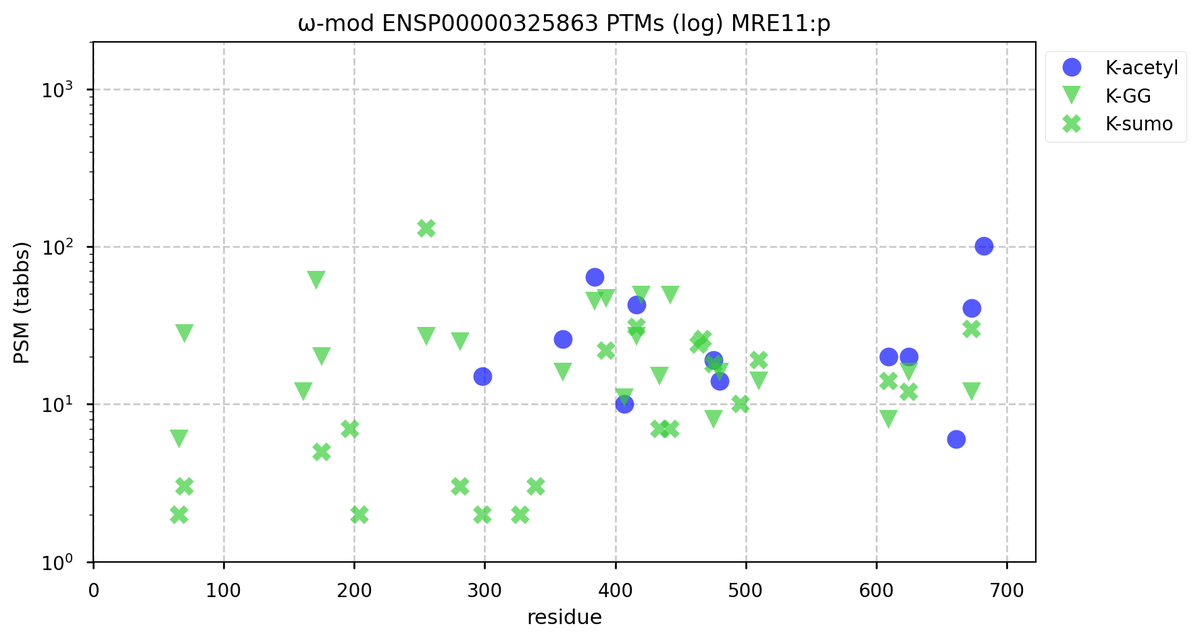
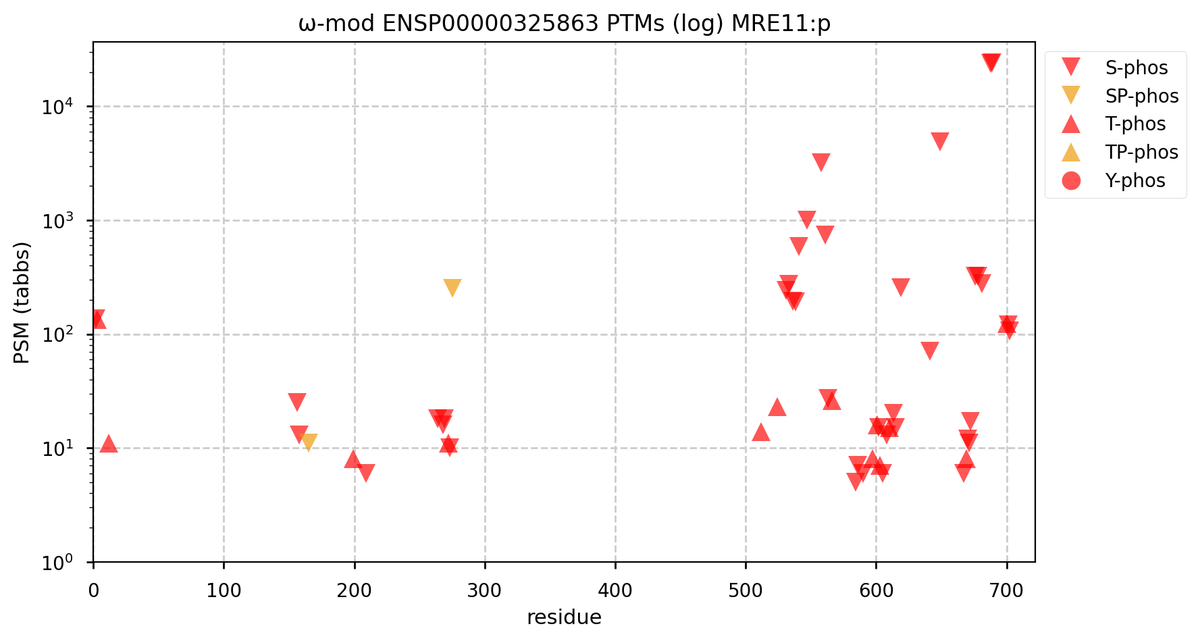
Fri Sep 02 03:14:55 +0000 2022@PingzhaoH @SchulichMedDent Congrats.
Fri Sep 02 02:57:22 +0000 2022@cwvhogue infovoriferous
Fri Sep 02 02:50:26 +0000 2022🔗
Fri Sep 02 02:17:45 +0000 2022@Zeczycki_Lab @neely615 @UCDProteomics It is an odd feature of "proteomics culture", but chromatography expertise is really the distinguishing feature of between labs: the mass specs are largely off-the-shelf components. The mass specs are the Dean Martin equivalent to the mad-cap escapades of the LC's Jerry Lewis
Fri Sep 02 01:28:19 +0000 2022Disturbance 91L east of the Leeward Islands this evening, moving northwest 🔗
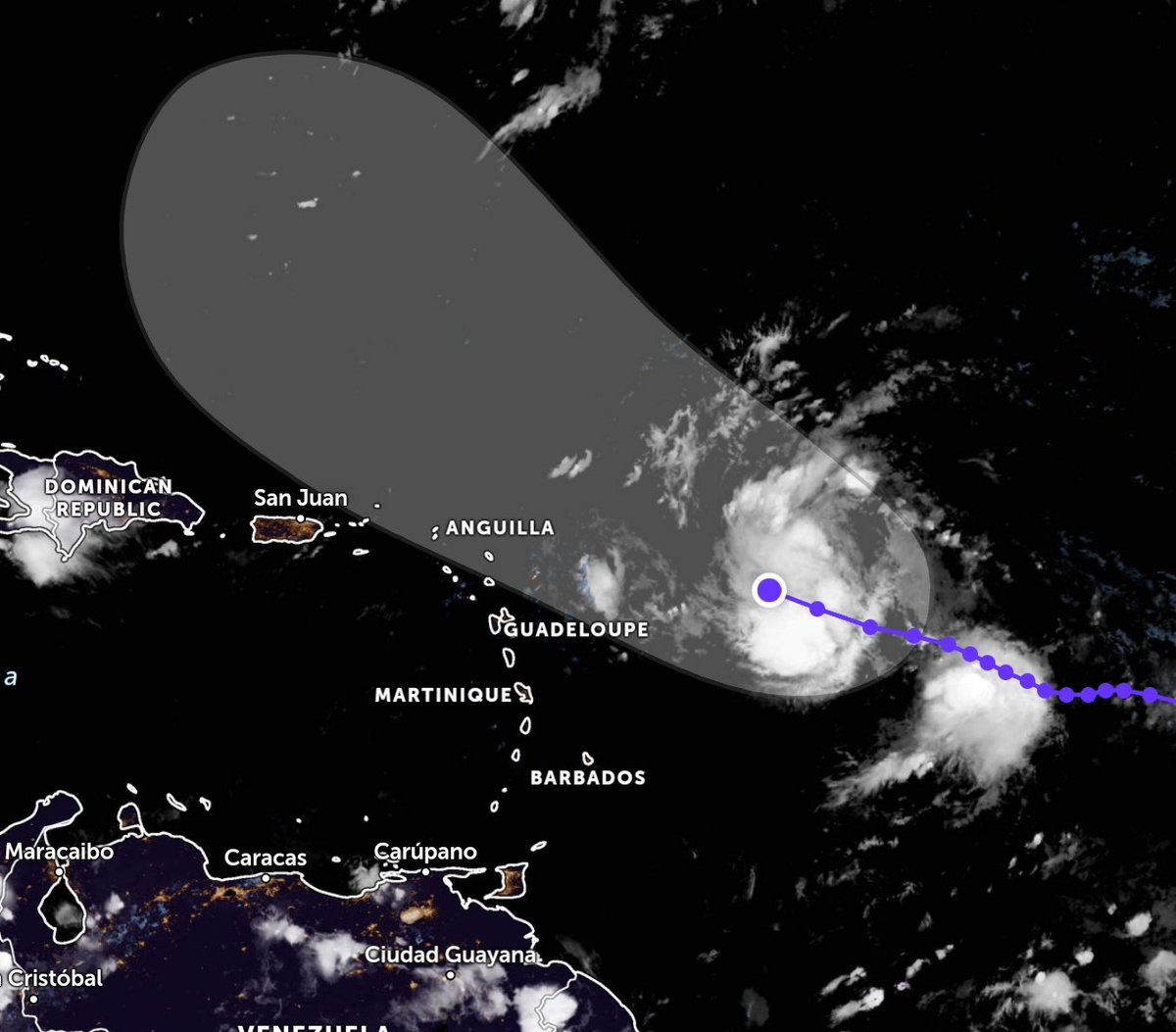
Fri Sep 02 00:49:15 +0000 2022Always wondered why some HEK-293T cell lines seem to translate SV40 small T antigen while others don't. They all make lots of the large T antigen, though, along with the usual E1B 55K & 19K from human adenovirus C.
Thu Sep 01 20:03:38 +0000 2022I'm not so sure that this provides the best landing page to recruit new people: 🔗
Thu Sep 01 14:38:36 +0000 2022Based on the frequency of observation, the subunits break out into 2 groups:
#1 [TERF2, TERF2IP]; &
#2 [TERF1, ACD, POT1, TINF2].
Group #2 would probably be the most fit-for-purpose as a set of signals to use as a reference level for low abundance proteins in nucleated cells.
Thu Sep 01 14:33:57 +0000 2022Now to figure out the basket of telosome proteins that would useful for a "proteome micrometer". Looking at the # of times each have been observed in public data:
TERF1: 3,879×
TERF2: 15,029×
TERF2IP: 25,603×
ACD: 3,103×
POT1: 2,777×
TINF2: 2,439×
Thu Sep 01 14:29:10 +0000 2022The absence of K+SUMOyl acceptors in the mouse data probably is caused by the very small number of studies looking at SUMOylation in anything other than human samples.
Thu Sep 01 14:27:14 +0000 2022Modification diagram for human & mouse TINF2:p, a subunit of the telosome/shelterin complex. Both show a cluster of S+phosphoryl acceptors in a phosphodomain (300-400), although the mouse phosphodomain is shifted a bit C-ward. 🔗
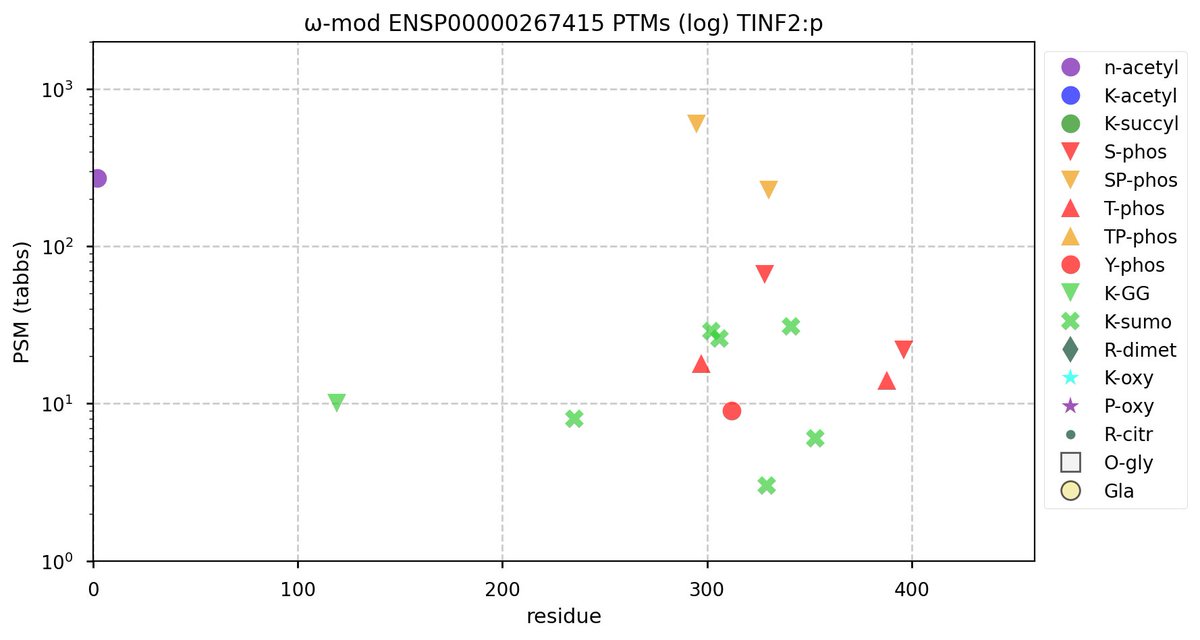
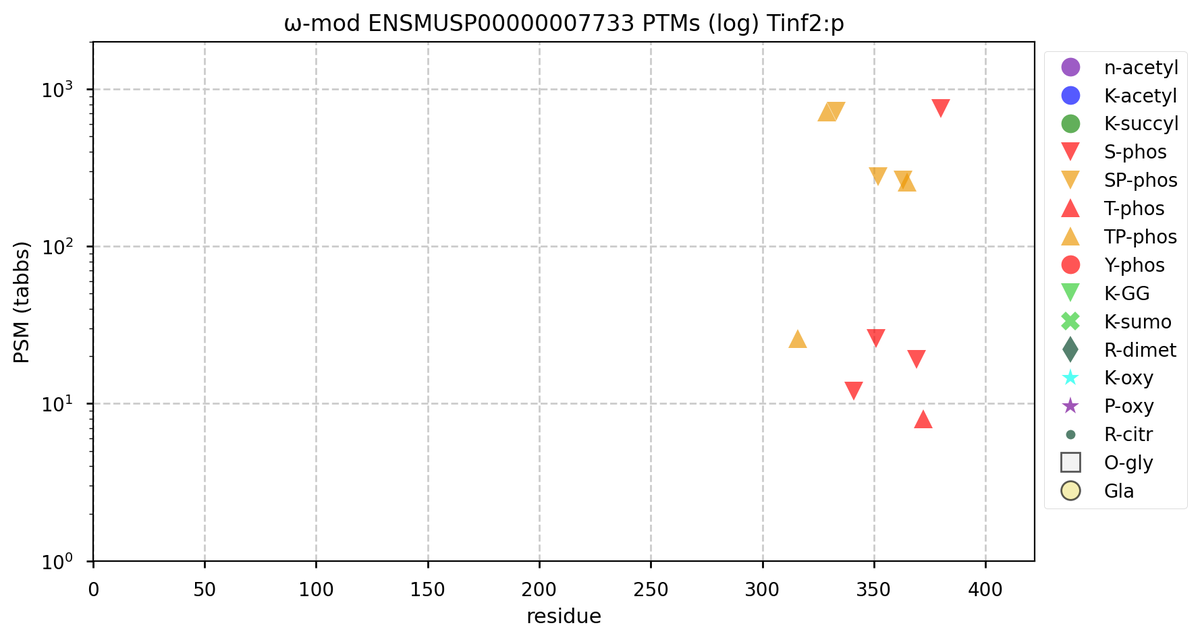
Thu Sep 01 13:24:12 +0000 2022I have been struck a number of times by just how varied people's understanding (& beliefs) about "the pandemic" really are in practice. It may be impossible to effectively communicate even a relatively simple scientific issue to the general public.
Thu Sep 01 13:15:27 +0000 2022A guy I see every day―but rarely talk to―spent some time yesterday explaining to me how a Chinese culinary preference was the cause of COVID-19.
Thu Sep 01 12:44:58 +0000 2022Super Typhoon Hinnamnor (the red icon) should be just about to change direction & head north. If this track holds, the storm should start to effect Kyushu on Monday, passing through the Korea Strait on Tuesday. It may also sideswipe Wenzhou & Shanghai earlier on Monday. 🔗
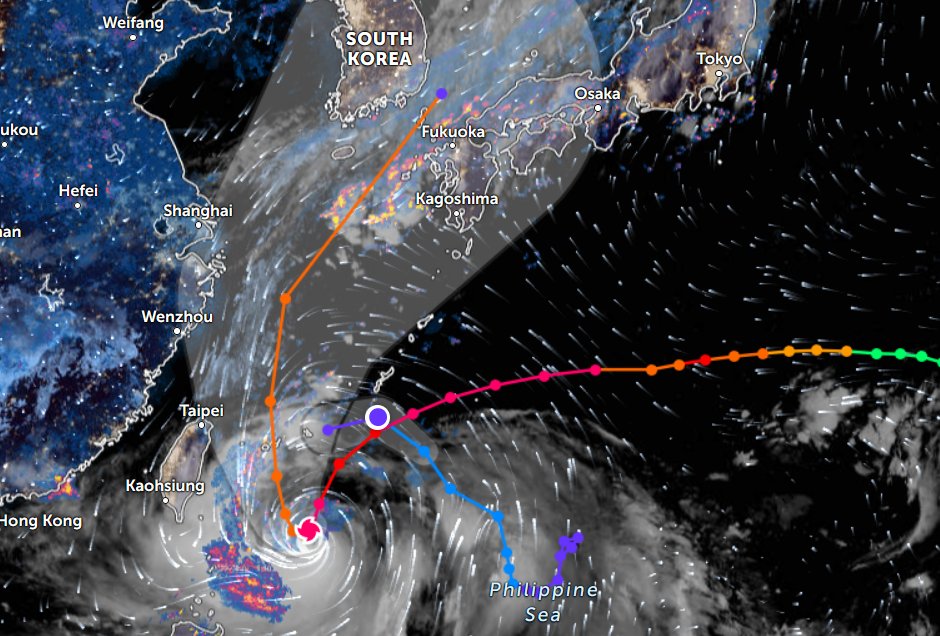
Wed Aug 31 20:11:49 +0000 2022If you are doing a proteomics study of mouse lymphocytes & it turns out that you detect significant amounts of XMRV viral proteins, is this anything to be concerned about wrt reproducibility?
Wed Aug 31 16:36:42 +0000 2022@bffo @LewisGeer @PLOS I agree: lining out the authors' contributions in the Acknowledgements should be done more often. But that doesn't always help, since many investigators trained in biomedical labs just read the first & last names. Biologists (non-medical) are much less prone to this failing.
Wed Aug 31 14:39:27 +0000 2022Violent Typhoon Hinnamnor has stayed on its predicted track & is now just south of Okinawa. It should make a hairpin turn on Friday & head straight up the East China Sea. 🔗
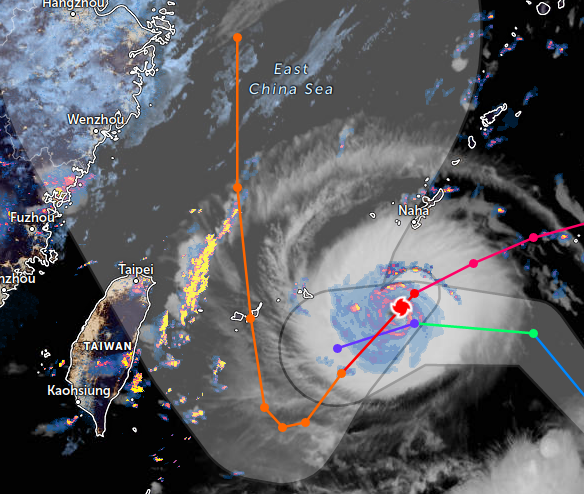
Wed Aug 31 12:18:38 +0000 2022POT1:p has managed to hold on to its original name, even if it is a bit on-the-nose wrt to its hoped for function: Protection of Telomeres 1. It has also been known as CMM10, HGLM9 & hPOT1.
Wed Aug 31 12:10:31 +0000 2022Modification diagram for human POT1:p, a subunit of the telosome/shelterin complex. Two K+acetyl/GGyl acceptors are part of a domain (289-469) with 9 K+GGyl or K+SUMOyl acceptors. There is also an N-terminal, low occupancy K+SUMOyl domain (33-121). 🔗
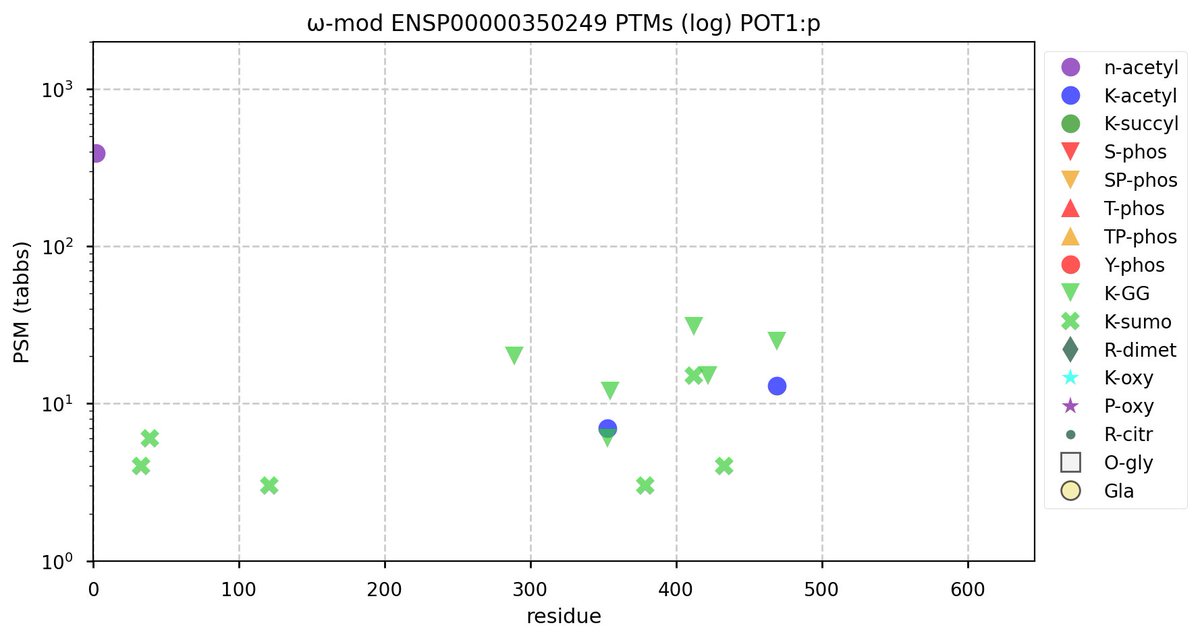
Wed Aug 31 00:05:36 +0000 2022Lot of lightning just west of New Orleans. 🔗
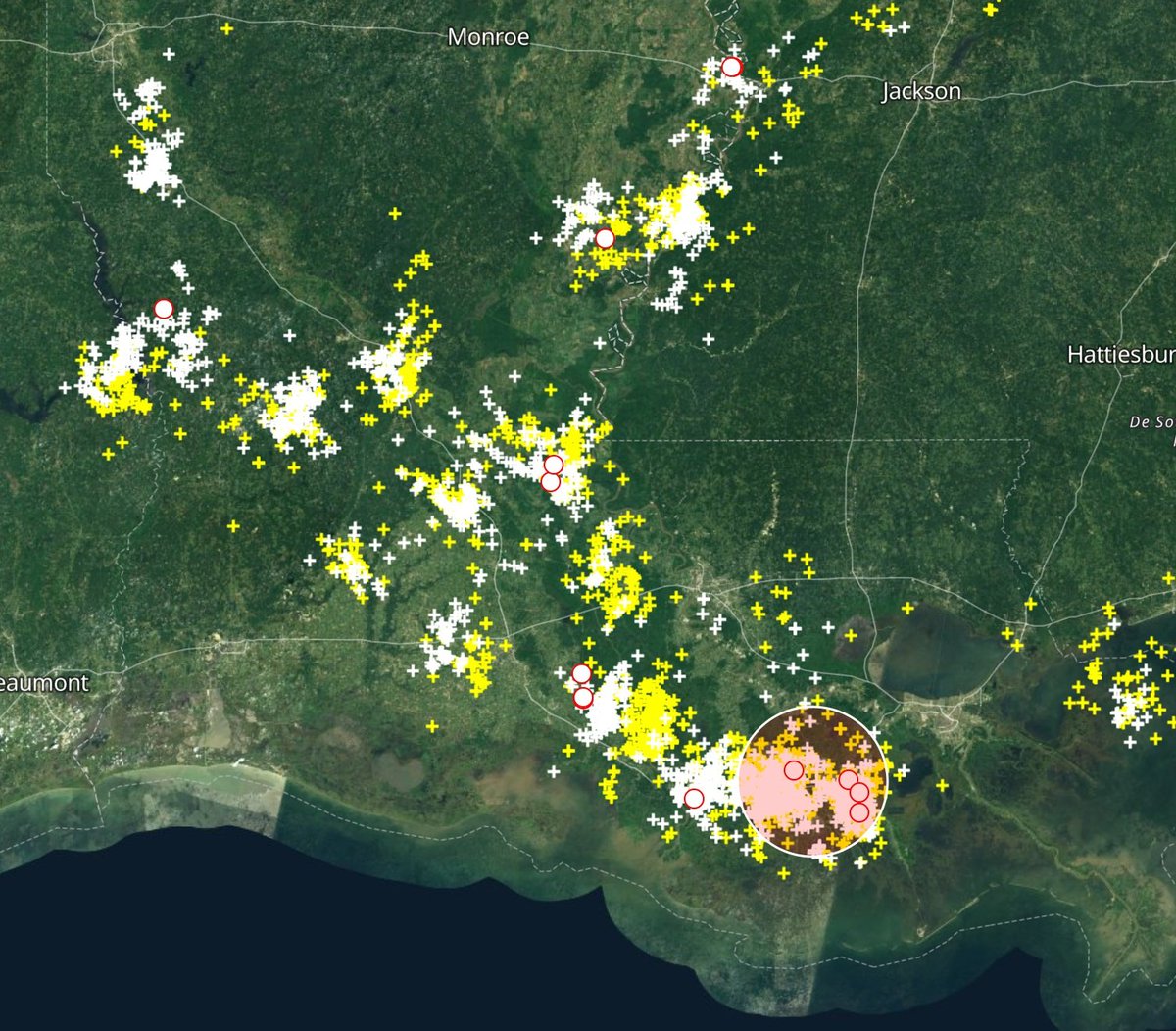
Tue Aug 30 23:46:36 +0000 2022I always examine prokaryote data for the species' preferences with respect to protein N-terminal acetylation. It is usually some combination of N-acetyl at either M1, S2 or T2, but there is quite a bit of species-to-species variability in their relative amounts.
Tue Aug 30 22:08:33 +0000 2022@bffo @PLOS I personally don't read anything at all into the order of authors and never have.
Tue Aug 30 19:48:44 +0000 2022Super Typhoon Hinnamnor is on an westerly heading, with winds up to 240 km/hr. The current prediction has the center passing south of Okinawa tomorrow, but it is a large storm & bad weather should be expected in Naha. 🔗
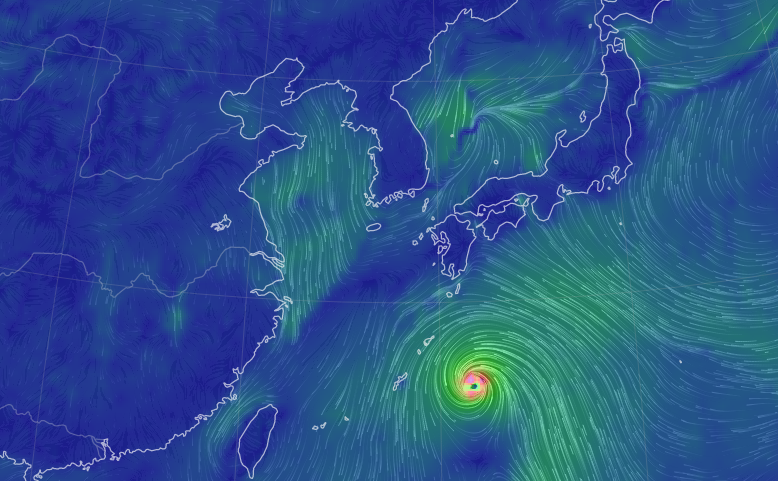
Tue Aug 30 13:15:13 +0000 2022It greatly reduces the complexity of storing non-indexed parameters, particularly those where using an object is a more straightforward solution than a group of linked tables of that are all flags, numbers & id strings.
Tue Aug 30 12:55:56 +0000 2022I've been using JSON object to store information (particularly things like permissions) in mysql databases recently and I'm never going back. Whoever pushed through 𝑗𝑠𝑜𝑛_𝑒𝑥𝑡𝑟𝑎𝑐𝑡() is my hero!
Tue Aug 30 12:11:18 +0000 2022Like TERF2IP:g, ACD:g was not the human gene's original name. It was originally called TPP1:g, but that name has been assigned to TriPeptidyl Peptidase 1. So, the gene name was revised to AdrenoCortical Dysplasia protein homolog, after the mouse ortholog.
Tue Aug 30 12:01:24 +0000 2022Modification diagram for human ACD:p, a subunit of the telosome/shelterin complex. It has 2 S+phophoryl acceptor domains & 3 K+GGyl acceptor domains. No K+acetyl has been observed.
🔗
Mon Aug 29 12:50:30 +0000 2022TERF2IP:g is the most recent symbolic name for this gene. It was originally called RAP1:g, but the name was changed to avoid confusion with Ras-related protein 1 (also originally RAP1:g), which has since changed to deal with its paralogs, now RAP1A:g & RAP1B:g.
Mon Aug 29 12:35:43 +0000 2022Tropical low 91L (green circle), currently east of the the Windward Islands, is predicted to have a 50% chance of developing into a cyclone in the next 48 hours (surface winds & total precipitable water). 🔗

Mon Aug 29 12:20:48 +0000 2022Modification diagram for human TERF2IP:p, a subunit of the telosome/shelterin complex. It has a bit more razzle-dazzle than the other telosome subunits, with multiple isolated S/T+phosphoryl, K+acetyl/SUMOyl & K+GGyl acceptors. 🔗
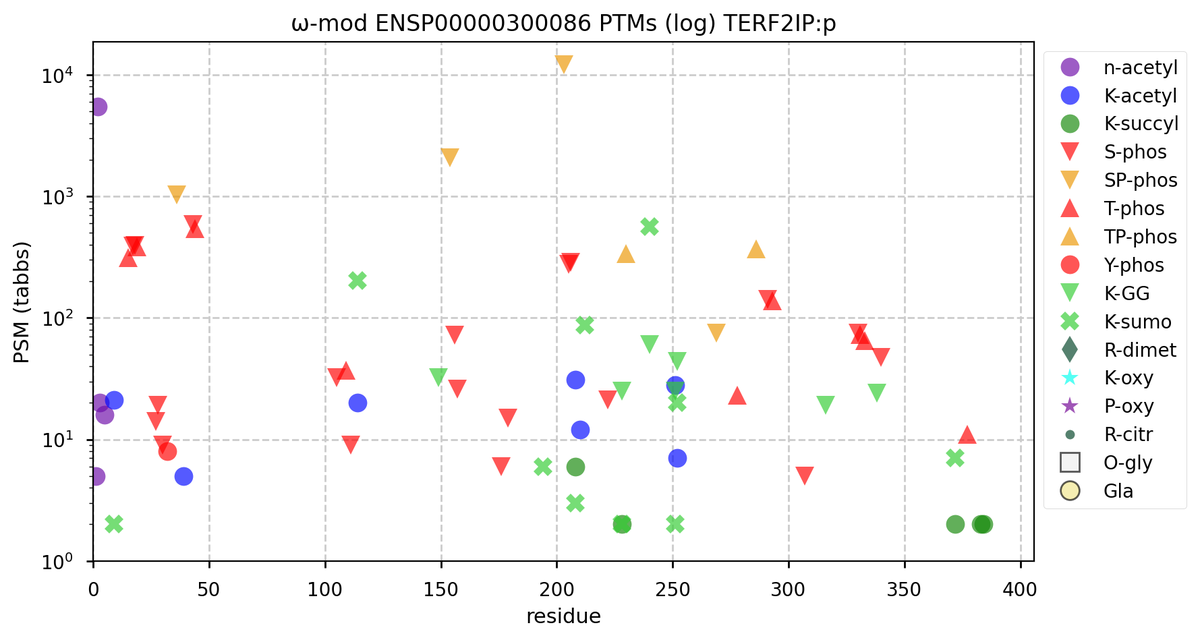
Sun Aug 28 15:30:17 +0000 2022As is the case for histones, this rather attractive idea doesn't work so well with non-diploid cells, 𝘦.𝘨., common cell lines like HeLa or HEK-293.
Sun Aug 28 14:30:35 +0000 2022Given this small, relatively rigid per-cell molecule count, signals from a basket of telosome subunits may make at least as good a yardstick for protein amount comparisons as the more abundant (& more variable) histone signals currently in use.
Sun Aug 28 13:14:47 +0000 2022@statesdj It is part of a more general trend, though.
🔗
Sun Aug 28 13:11:27 +0000 2022It has been a wet week along a band that includes Jackson, Mississippi (ground station precipitation 7 day totals, in mm) 🔗
Sun Aug 28 12:12:20 +0000 2022The telosome/shelterin complex has 6 subunits & protects telomeres from degradation. There are 2 of these complexes per chromosome, so for cells in G1 phase there should be a maximum of 92 per cell, 92-184 in S phase & 184 in G2 phase.
Sun Aug 28 11:57:58 +0000 2022Modification diagram of human TERF2:p, a subunit of the telosome/shelterin complex. It has several phosphodomains in IDRs and a prominent K+acetyl/GGyl/SUMOyl mixed acceptor domain (222-375) overlapping with its 2 protein-protein interaction domains.
🔗
Sat Aug 27 22:36:15 +0000 2022A narrow line of intense storms just south of Havana this afternoon. 🔗
Sat Aug 27 22:27:40 +0000 2022@neely615 @harrisonspecht Could you give an example of what you mean by interoperability?
Sat Aug 27 13:54:48 +0000 2022@astacus You will have to take that up with these guys:
🔗
Sat Aug 27 13:31:24 +0000 2022The "146" in the gene name is a not a molecular weight or some other physical property: it is a serial number. There are a lot of genes coding peptide sequences containing Really Interesting New Gene (RING) finger domains.
Sat Aug 27 12:06:02 +0000 2022@statesdj They are pretty tied up with the EBI mob, so I wouldn't want to be involved.
Sat Aug 27 11:44:22 +0000 2022Modification diagram for human RNF146:p, an E3 ligase responsible for ubiquitination of subunits PARylated by TNKS:p. It has 1 minor (87-90) & 1 major (272-300) phosphodomain. There are 2 K+GGyl acceptors and no evidence of K+acetyl or SUMOyl acceptors.
🔗
Sat Aug 27 00:39:44 +0000 2022I hate debugging projects that send SMS messages: it is bad enough that the software is screwing up, but beeping away on my phone is the proof.
Fri Aug 26 17:03:08 +0000 2022It is still quite dusty in the part of the Atlantic where Cape Verde hurricanes would normally develop this time of the year (shown: PM₁₀ particulates & surface winds) 🔗
Fri Aug 26 12:30:57 +0000 2022As advertised, the AF calculated 3D structure has the potential to revolutionize understanding of this subunit.😀 🔗
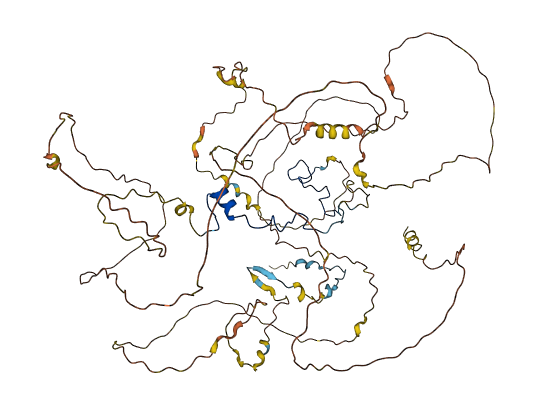
Fri Aug 26 12:21:12 +0000 2022Modification diagram for human CASC3:p, a substrate for PARylation by TNKS:p. It has a more complex phosphoryl acceptor pattern than TNKS' other substrates & it has a distinct grouping of K+GGyl acceptors (113-344) across its btz domain. 🔗
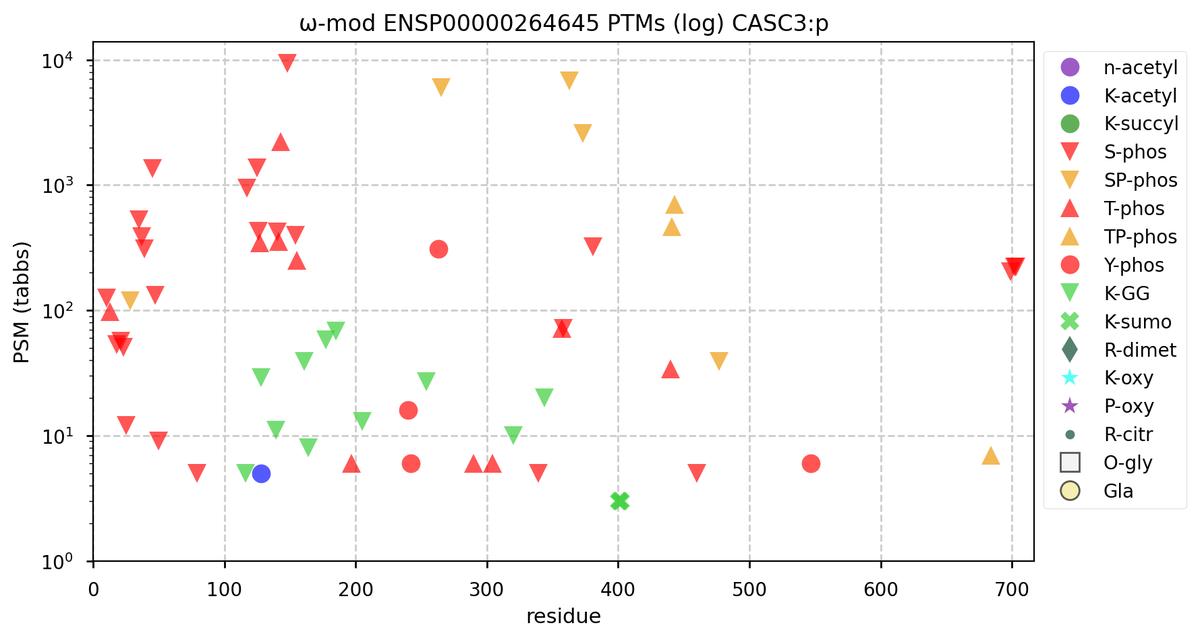
Thu Aug 25 17:08:56 +0000 2022The lack of observed K+GGyl for AXIN1:p is a little odd: the whole point of being a PARylation substrate of TNKS:p is to mark the subunit for ubiquitination, so it can be tossed into the cell's protein disposall (the proteasome).
Thu Aug 25 15:50:48 +0000 2022A creator after my own heart 🔗
Thu Aug 25 15:36:32 +0000 2022Florita/Ma-On is moving further inland as a tropical storm, with its center just east of the Chinese boarder. It is causing considerable rainfall along the northern coast of Vietnam. 🔗
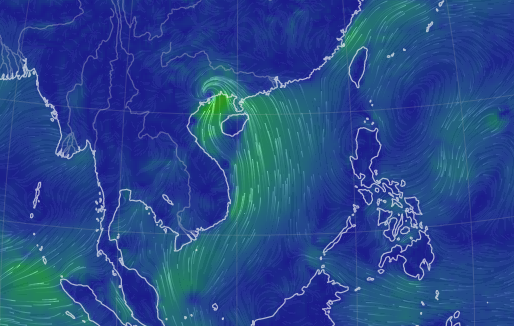
Thu Aug 25 15:15:04 +0000 2022@arneelof @lukas_k @LaineElodie What are you looking to find?
Thu Aug 25 12:06:59 +0000 2022Modification diagram for human AXIN1:p, a substrate for PARylation by TNKS:p. Unlike BLZF1:p, this subunit has a set of discrete high occupancy S/T+phosphoryl acceptors & no significant occupancy of K+GGyl, SUMOyl or acetyl acceptors. 🔗
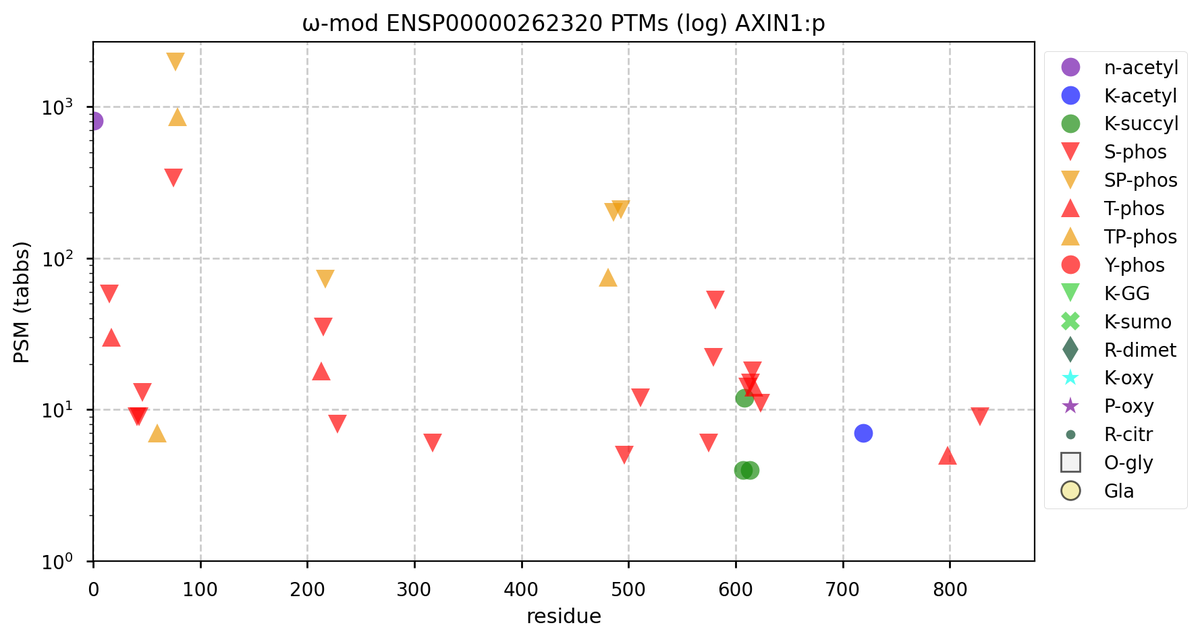
Wed Aug 24 21:41:04 +0000 2022@macro_momo Corrected recipe:
1 bottle =
6 large jalapeno peppers
6 cloves of garlic
8-10 small, thin Thai peppers
handful of cilantro
handful of mint
60 ml olive oil
30 ml vinegar
15 ml salt
2 ml cumin
15 ml lemon juice (optional)
Put chopped ingredients into blender & whirl until smooth.
Wed Aug 24 21:39:40 +0000 2022@macro_momo Yikes. I forgot the garlic. It needs 6 cloves of garlic! 😱
Wed Aug 24 20:23:13 +0000 2022@macro_momo Notes:
60 ml = ¼ cup,
15 ml = 1 tablespoon, &
2 ml = ½ teaspoon.
Wed Aug 24 20:20:17 +0000 2022@macro_momo 1 bottle =
6 large jalapeno peppers
8-10 small, thin Thai peppers
handful of cilantro
handful of mint
60 ml olive oil
30 ml vinegar
15 ml salt
2 ml cumin
Put chopped ingredients into a blender & whirl until smooth. Tada!
Wed Aug 24 18:41:48 +0000 2022Florita/Ma-On just about making landfall east of Hainan near Diancheng as a severe tropical storm (100 km/hr winds). 🔗
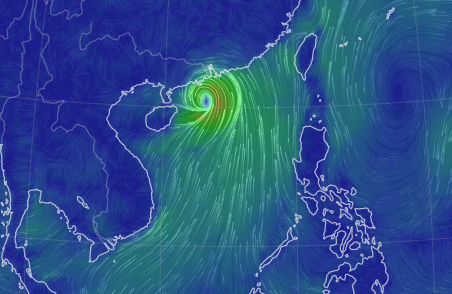
Wed Aug 24 15:04:38 +0000 2022"unfortunately results"
should read
"unfortunate results"
On-the-fly copy editing is not my strong suit.
Wed Aug 24 14:16:30 +0000 2022Another late summer heavy rain event in the southern US, this time ranging from Lake Charles to Tuskaloosa, with the heaviest rainfall near Union, MS, where a CoCoRaHS ground station reported 184 mm (7¼") yesterday. 🔗
Wed Aug 24 13:30:32 +0000 2022@stphnmaher 🔗
Wed Aug 24 13:14:24 +0000 2022If you have ended up using Occam's Razor to come to a conclusion, something about your argument went wrong near its beginning. It is the equivalent of using a brick wall to stop a careening car: it may work but it always has unfortunately results.
Wed Aug 24 13:01:22 +0000 2022Made a bottle of corriander-mint hot sauce over the weekend: perfection.
Wed Aug 24 12:30:53 +0000 2022While a brave effort (or at least is would be if bots had courage), the rather fanciful AF prediction for BLZF1:p isn't really very salutary, verging on the unsalutary. 🔗
Wed Aug 24 12:19:53 +0000 2022Modification diagram for human BLZF1:p, a substrate for PARylation by TNKS:p. It has a smattering of ΦP S+phosphoryl, but K+GGyl/SUMOyl acceptors are doing the lion's share of work for this subunit. K+acetyl is conspicuous by its absence. 🔗
Tue Aug 23 22:17:19 +0000 2022Rain estimated from radar for the last 48 hours, showing the extent of the rainfall event that clobbered Dallas. 🔗
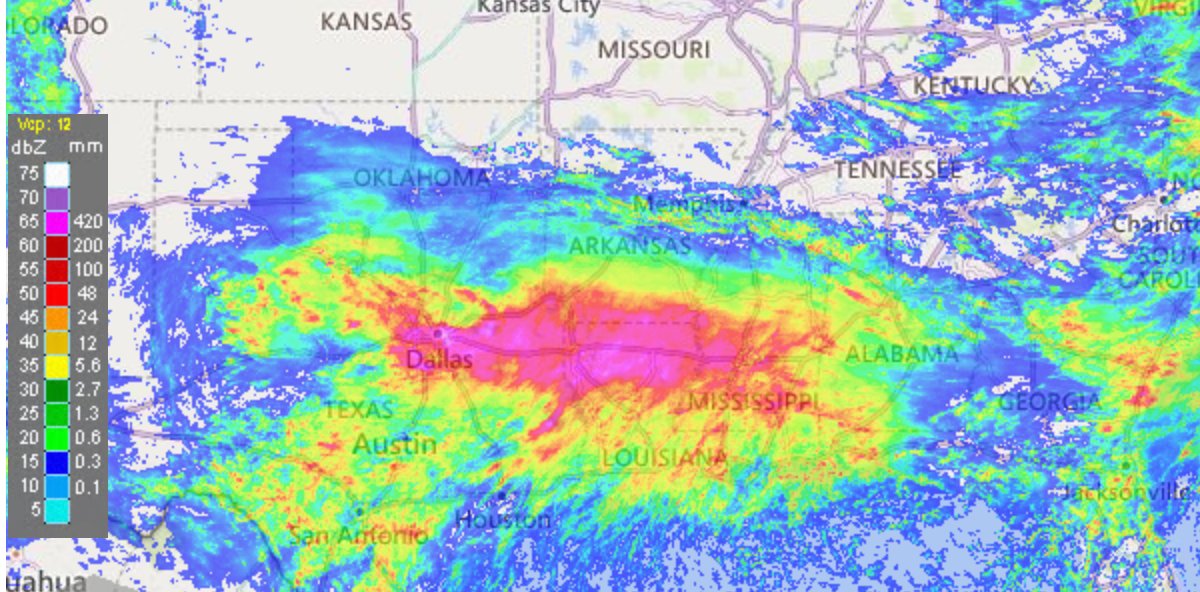
Tue Aug 23 20:43:37 +0000 2022Florita/Ma-On has passed over Luzon & it is heading through the South China Sea towards the mainland. It is now on track to make landfall near Yangjiang just after midnight on Thursday (UTC). 🔗
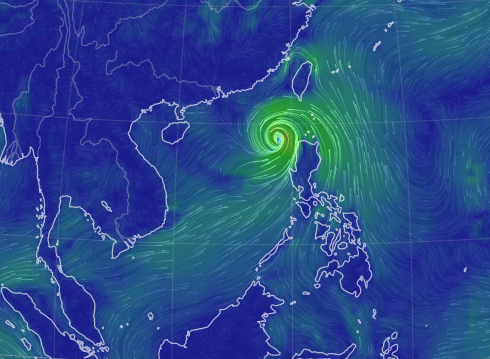
Tue Aug 23 20:33:24 +0000 2022It is about to get wet in Brandon, MB (radar). 🔗

Tue Aug 23 16:32:14 +0000 2022@ILikeCellBio @pwilmarth @slavov_n ∝ means "directly proportional to" 🔗
Tue Aug 23 11:42:51 +0000 2022@slavov_n Without regard for rational argument or counter examples, the axiom
[RNA] ∝ [protein]
is an article of faith for many biomedical researchers.
Tue Aug 23 11:12:17 +0000 2022Modification diagram for TERF1:p, the telosome/shelterin subunit bound by TNKS:p. It is dominated by a major ΦP phosphodomain (6-11) & a broader, low occupancy ΦxxD/E phosphodomain (219-296). There are a few scattered K+GGyl & SUMOyl acceptors.
🔗
Tue Aug 23 02:29:54 +0000 2022Florita/Ma-On making landfall on the northeastern coast of Luzon, not too far from Tuguegarao. 🔗
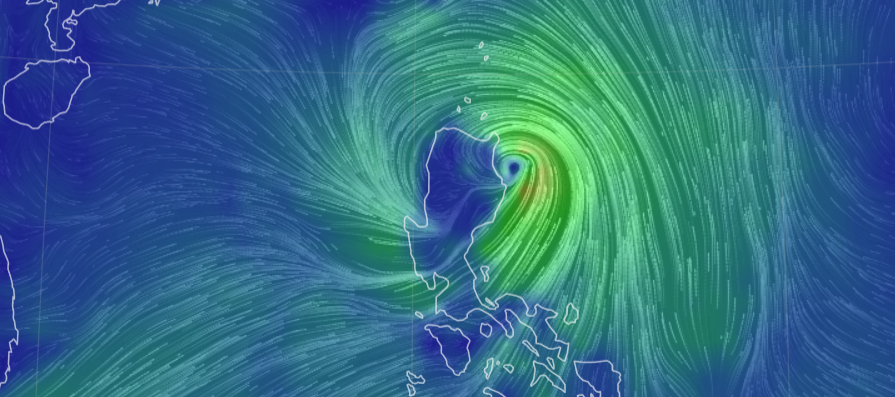
Mon Aug 22 15:28:11 +0000 2022I have only rummaged around in the literature wrt TNKS1BP1:p for an hour or two, but there does seem to be a general lack of appreciation for the potential consequences of the subunit's whole-hearted commitment to jacketing itself with phosphorylation.
Mon Aug 22 11:52:52 +0000 2022As you expect from this modification diagram, AF was unable to identify any significant amount of 3D structure in the sequence. 🔗
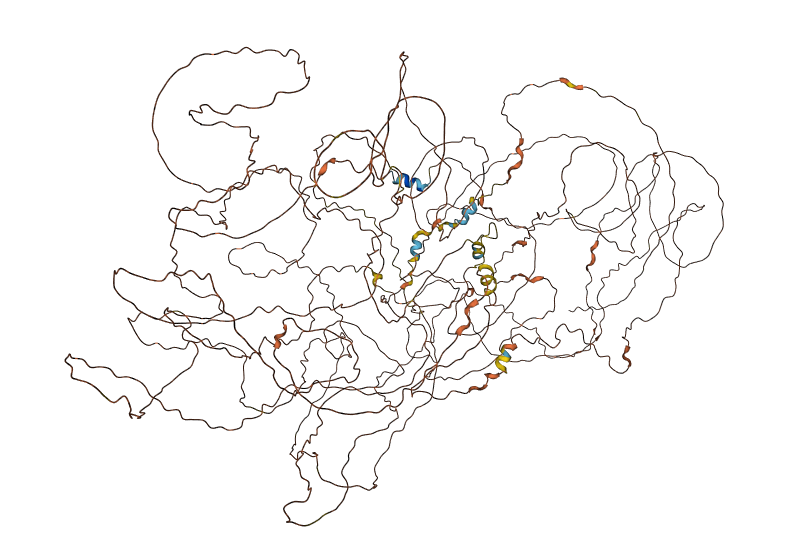
Mon Aug 22 11:49:23 +0000 2022Modification diagram for human TNKS1BP1:p, named for its ability to bind TNKS1:p. Unlikely the sparsely modified tankyrases, it is one big phosphodomain (dominated by S+phosphoryl acceptors) & it shuns K+GGyl. 🔗
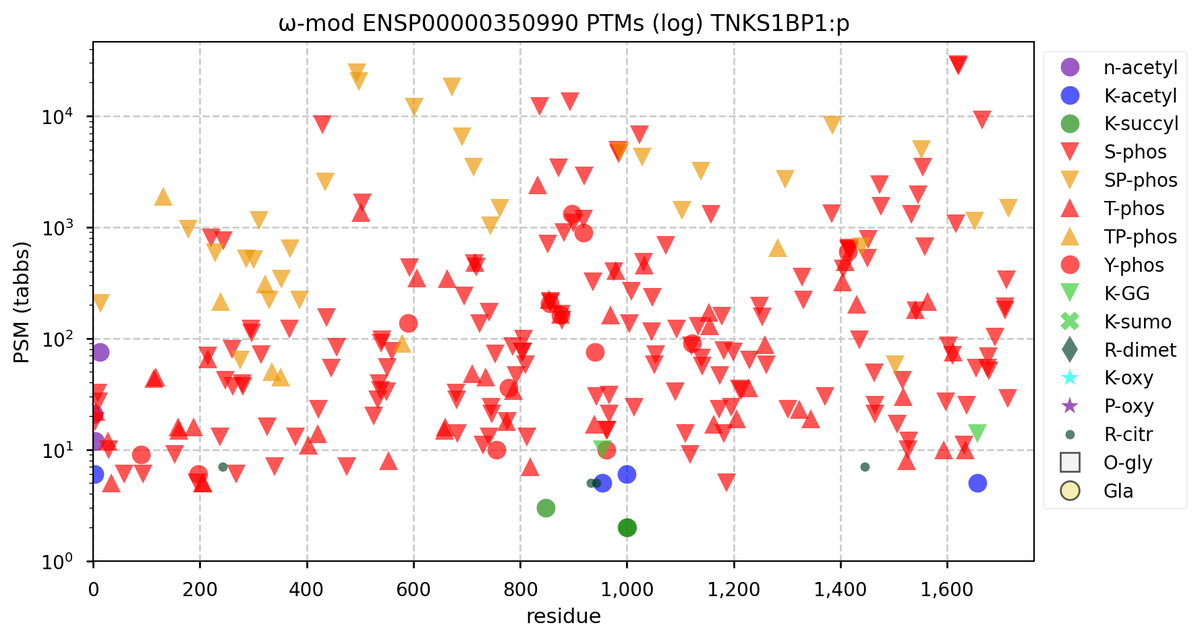
Mon Aug 22 11:28:38 +0000 2022The predicted track for Florita/Ma-On has changed. Now it is expect to cross northern Luzon & arrive just west of Hong Kong as a hurricane early Thursday morning. 🔗
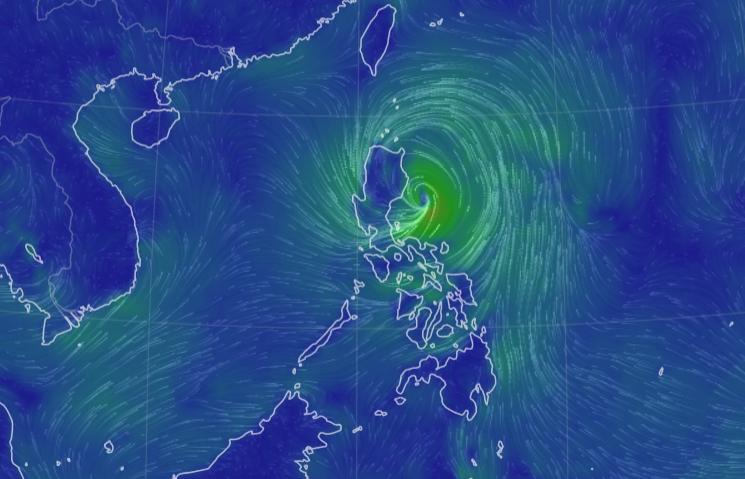
Mon Aug 22 02:30:50 +0000 2022Tropical depression Florita, just east of Luzon in the Philippines. It is currently predicted to pass north of the island & finally come ashore just east of Hong Kong on Wednesday with hurricane strength winds. 🔗
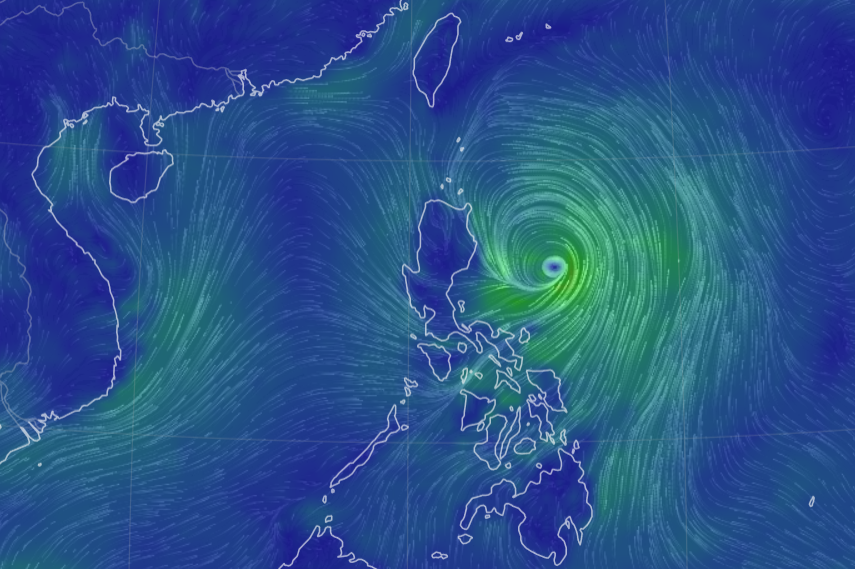
Sun Aug 21 18:36:38 +0000 2022@AlexUsherHESA @MemorialU I am misreading this or do the top 15 CIHR grant institutions constitute only a 58% share? There must be a lot of institutions in the rest of the list.
Sun Aug 21 15:53:52 +0000 2022Does anybody have anything bad to say about the JSON Lines format (🔗)? I use it all the time, but it hasn't seemed to have caught on in the larger biomedical/bioinformatics world.
Sun Aug 21 15:22:05 +0000 2022@neely615 @pwilmarth @MattWFoster "Kedi" is very good. It is a set of documentary stories about community cats in Istanbul, as told by kind of hard-case working class guys who consider the cats to be very helpful collaborators.
Sun Aug 21 14:29:50 +0000 2022Tankyrase is a portmanteau of the full gene name "telomeric repeat binding factor 1 (TRF1)-inter-acting ankyrin-related ADP-ribose polymerase", but it is also know as TIN1, TINF1, TNKS1, PARP-5a, PARP5A, pART5 or ARTD5.
Sun Aug 21 12:45:25 +0000 2022Does anybody know if there are any E3 ligases known to specialize in ankyrin-repeat domain ubiquitination?
Sun Aug 21 12:38:27 +0000 2022@neely615 If you want to follow the individual lightning strike activity, this site has a pretty good, real-time display.
🔗
Sun Aug 21 12:35:34 +0000 2022Modification diagrams for human TNKS1:p & TNKS2, 2 tankyrase paralogs. The K+GGyl acceptors are spread across their subunits' ankyrin-repeat & poly(ADP-ribose) polymerase domains. Phosphorylation & acetylation seem to have been relegated to bit parts. 🔗
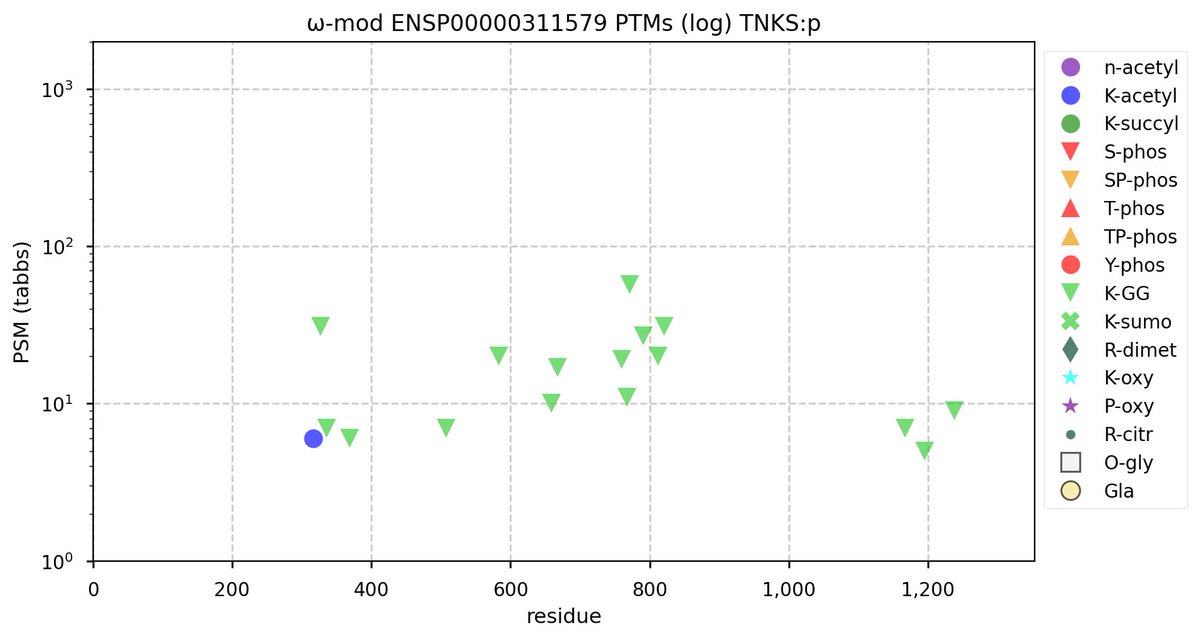
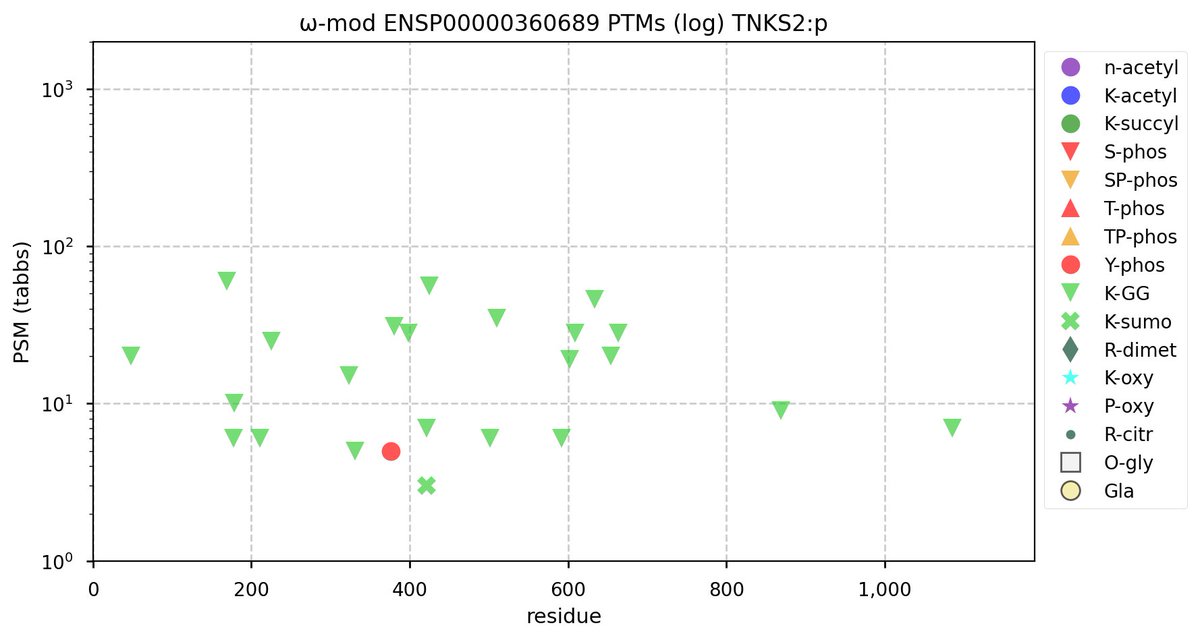
Sun Aug 21 01:23:46 +0000 2022And a line of weak storms in western Cuba, not far from Havana. 🔗
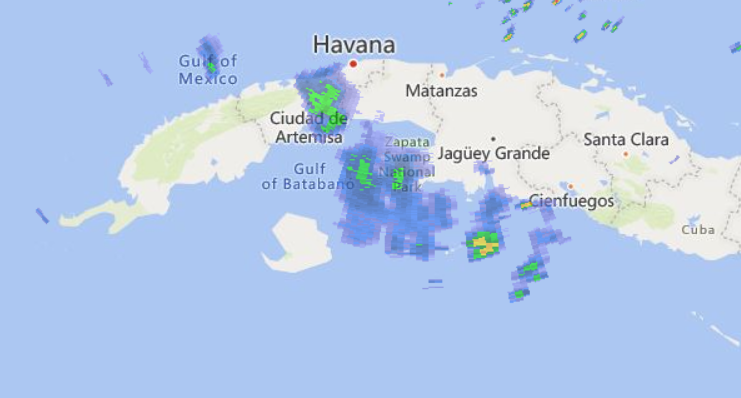
Sun Aug 21 01:19:37 +0000 2022A line of thunderstorms escaping Ohio, only to find themselves in Pennsylvania (NEXRAD radar) 🔗
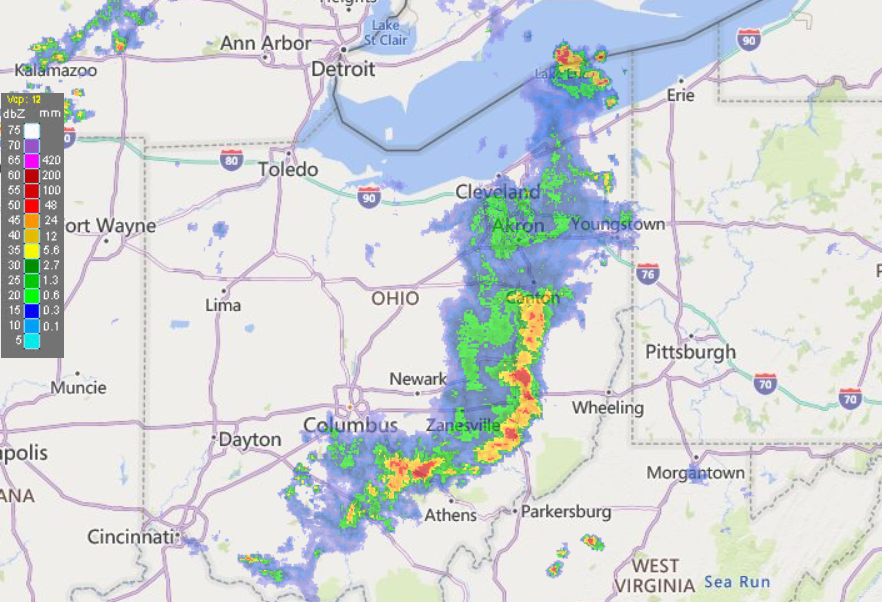
Sat Aug 20 22:27:45 +0000 2022Another unusually windy day across the Mediterranean & Adriatic Seas (surface wind vectors) 🔗
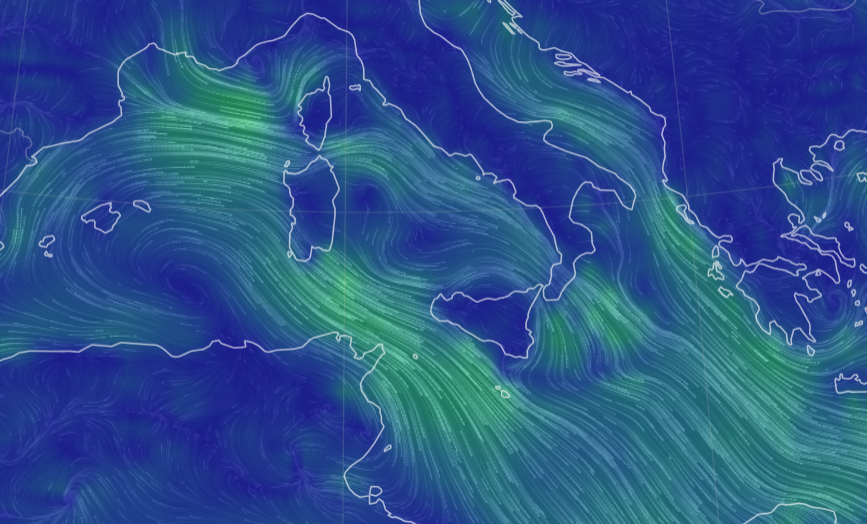
Sat Aug 20 18:06:28 +0000 2022It is a good example of using a private not-for-profit vendor to supply a public "good", where most of the private for-profit vendors failed pretty badly to supply the same thing.
Sat Aug 20 17:57:20 +0000 2022I have been using Let's Encrypt to handle HTTPS certificates for years, but using it still makes me happy. 😀
Sat Aug 20 16:26:59 +0000 2022A bunch of popcorn thunderstorms stretching from Beaumont to New Orleans this morning 🔗
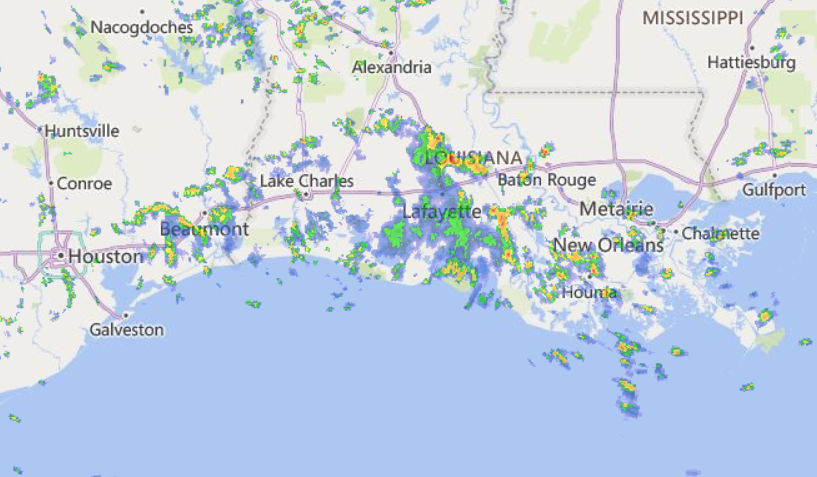
Sat Aug 20 15:46:52 +0000 2022@virtuous_sloth @AlexUsherHESA @StephanieCarvin Sounds very British. I suspect it would be a non-starter in North America.
Sat Aug 20 15:07:09 +0000 2022@rabbleca While I often find Canadian politics to be deliberately confusing low-stakes baffle-gab, I do find the endless, cynical ways politicians try to lever provincial "rights" as an issue perversely entertaining.
Sat Aug 20 14:30:05 +0000 2022@cstross A very interesting topic, but fraught with cultural & behavioral confounders.
Sat Aug 20 14:03:16 +0000 2022My experience with med students is that they excel if you give them specifics to remember (esp. if it involves polysyllabic words), but are easily stumped if you want them to expand outwards from a set of basic concepts.
Sat Aug 20 13:34:12 +0000 2022It doesn't surprise me when MDs struggle to explain scientific concepts, as I have always had trouble explaining scientific concepts to MDs.
Sat Aug 20 13:12:13 +0000 2022@pwilmarth Back in the olden times, Procter & Gamble had a big proteomics effort: those guys really knew their stuff wrt cleaning products and how to talk about their physical properties.
Sat Aug 20 13:00:19 +0000 2022I always feel like I should be wearing a wizard's hat when writing style sheets.
Sat Aug 20 12:57:09 +0000 2022Is CSS really anything other than some basic functionality + a bottomless bag of seemingly unrelated tricks thought up by some big group of warring committees?
Sat Aug 20 12:39:11 +0000 2022In cleaning solution of water with 5% isopropanol, would it be OK to refer to the alcohol as a "wetting agent"?
Sat Aug 20 11:54:26 +0000 2022Modification diagram for BRD9:p, the 9ᵗʰ (& final) subunit named for having at least 1 bromodomain. It has two major phosphodomains (14-139) & (478-588) and a suspiciously significant smattering of K+acetyl, K+GGyl & K+SUMOyl.
🔗
Sat Aug 20 11:39:51 +0000 2022@goodlettlab1 @Rogers @ATT Sounds about right. Always had issues with my Nay-dun cell phones once I stepped over the border, even in Mer-cah!
Sat Aug 20 02:33:08 +0000 2022Thunderstorms making their way towards Lake Michigan (marked are lightning strikes in the last 40 min) 🔗
Fri Aug 19 21:34:21 +0000 2022Haploid human cells tend to be little weirdos: they will often have splice variants, PTM patterns & translated proteins you rarely see in diploid cells.
Fri Aug 19 14:24:34 +0000 2022@astacus @TimothyKassis I had the same issue a while ago, so I made this:
🔗
Fri Aug 19 12:54:46 +0000 2022The difference between the splices is illustrated by the observed peptide diagram for the "top 20" observations of BRD8:p, plotted for the long (testes-specific) splice. One of these things is not like the others ... 🔗
Fri Aug 19 12:18:16 +0000 2022Lighting strikes this morning, along the northern coast of the Gulf of Mexico. 🔗

Fri Aug 19 12:03:51 +0000 2022Modification diagrams for 2 splice variants of human BRD8:p, the #8 subunit to be named for having at least 1 bromodomain. The shorter splice is the most commonly observed: the long form seems to be testes/spermatozoa-specific. 🔗
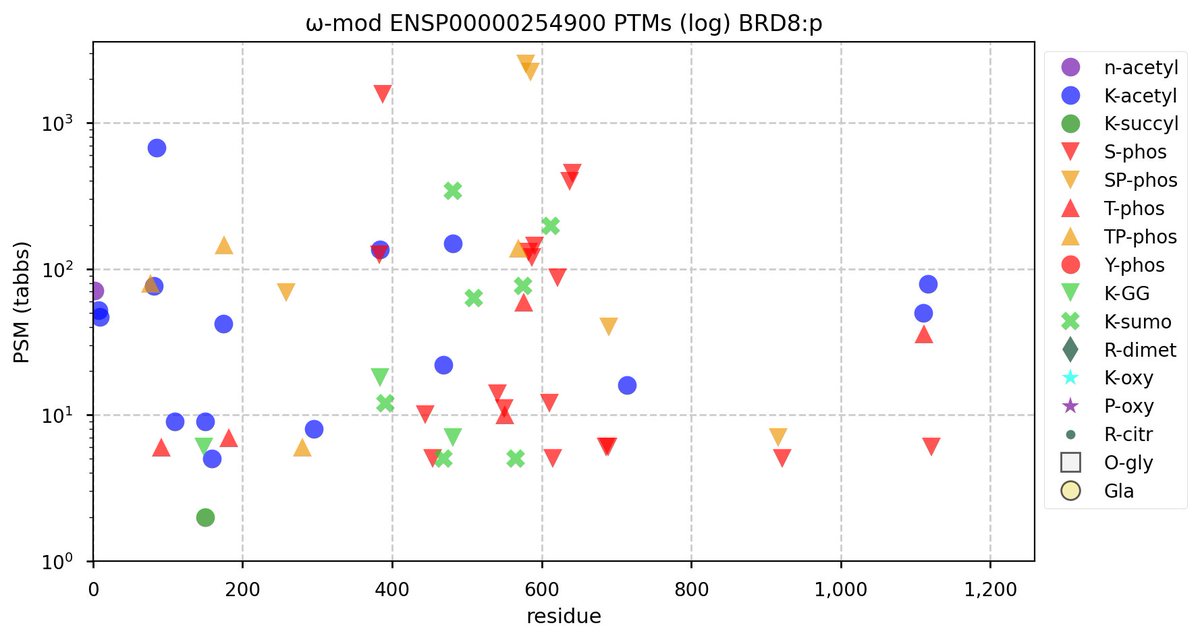
Thu Aug 18 13:01:42 +0000 2022A line of thunderstorm has just barreled through Fargo. 🔗
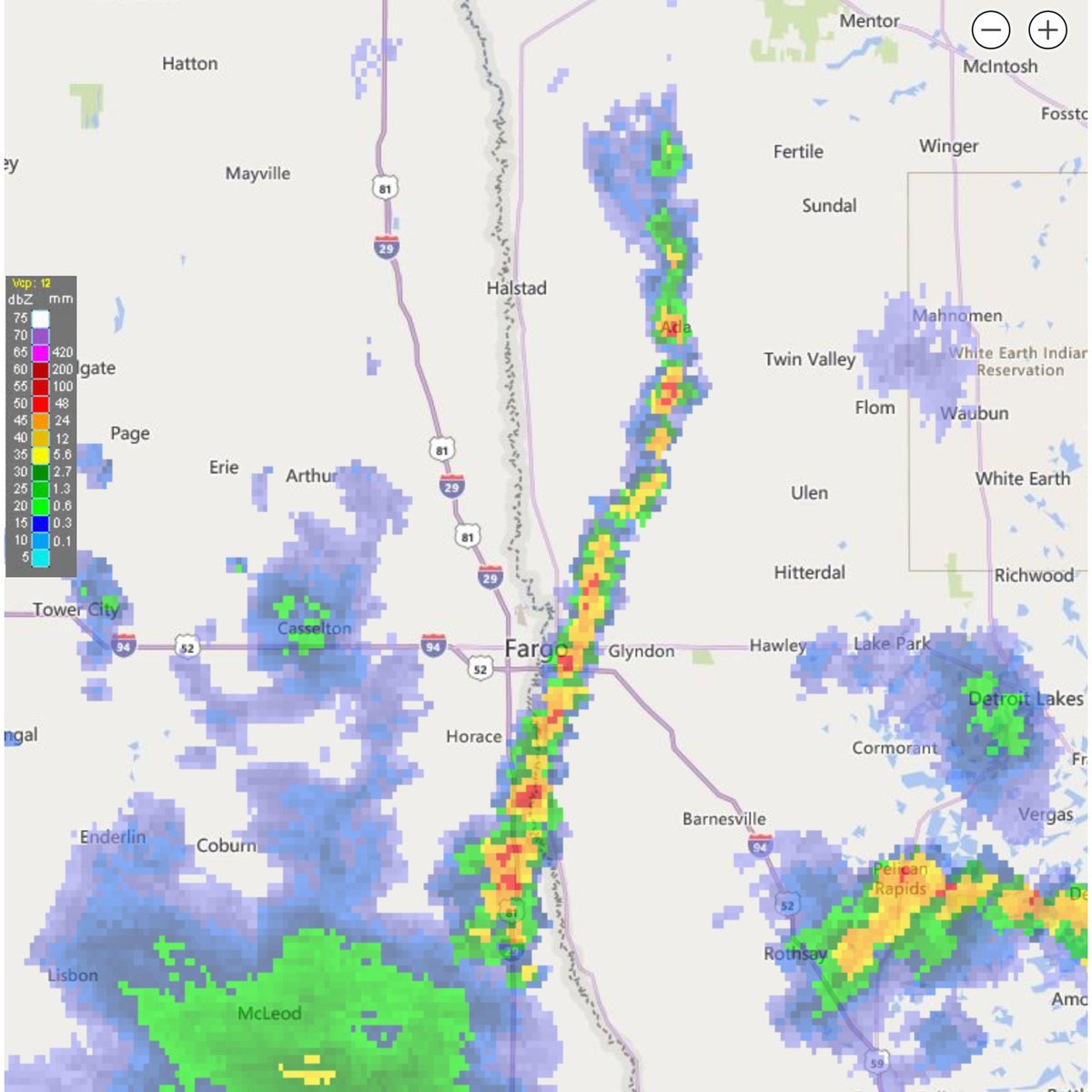
Thu Aug 18 12:49:57 +0000 2022Modification diagram for human BRD7:p, the 7ᵗʰ subunit to be named for having >1 bromodomain. It has 1 minor & 2 major phosphodomains, some pianissimo K+acetyl & a chorus of K+SUMOyl across the bromodomain (129-237)
🔗
Thu Aug 18 12:44:30 +0000 2022PXD032877: also has some nice phospho-peptide enrichment from tissue (liver).
Thu Aug 18 01:20:34 +0000 2022PXD027693: some nice phospho-peptide enrichment from lung tissue samples.
Wed Aug 17 22:21:09 +0000 2022@vapetyuk @AJ_Brenes Is this described in an open source? I'm not an academic so I don't have an institutional subscription.
Wed Aug 17 12:11:23 +0000 2022Modification diagram for human BRD4:p, the 4ᵗʰ subunit named for having a bromodomain (it has 2). Compared to the first 3, it has more K+acetyl & K+acetyl/SUMOyl acceptors in discrete domains. It also has 4 major S/T phosphodomains.
🔗
Tue Aug 16 12:59:57 +0000 2022I really don't like it when a weather story is reported as a climate story. It confuses public conversation on the issues.
🔗
Tue Aug 16 12:24:13 +0000 2022Modification diagrams for human & mouse BRD3:p, the 3ʳᵈ subunit named for having a bromodomain. They has 1 minor + 2 major phosphodomains & a few of K+acetyl. They have no observed K+GGyl acceptors, although the human form contributes at least 7 observed MHC class 1 peptides. 🔗
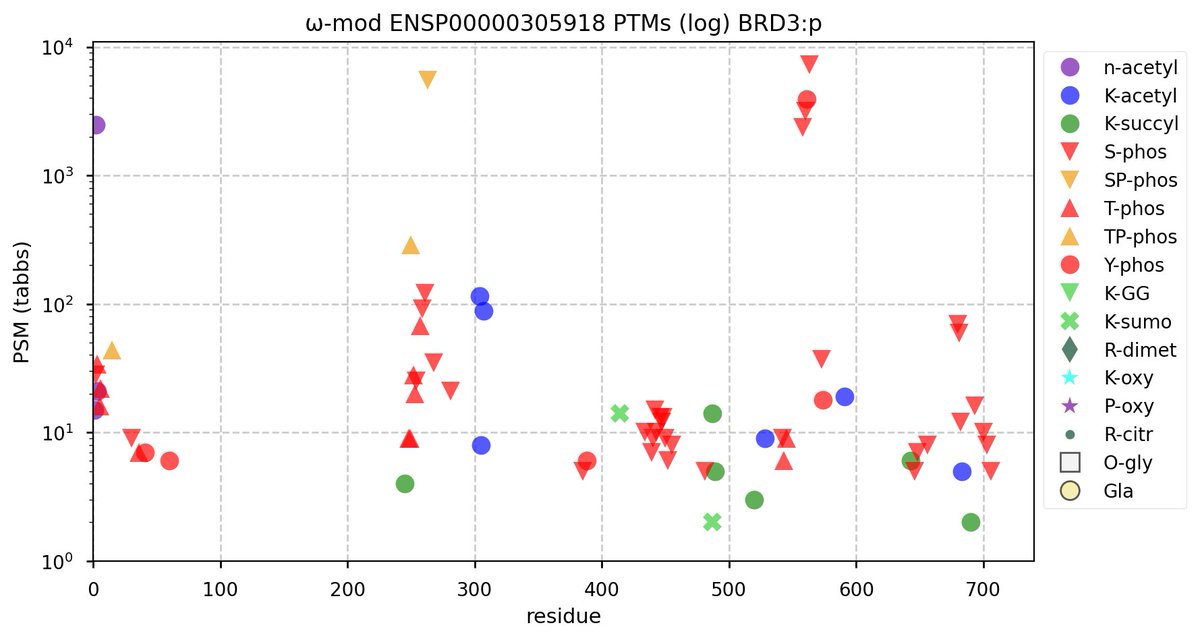
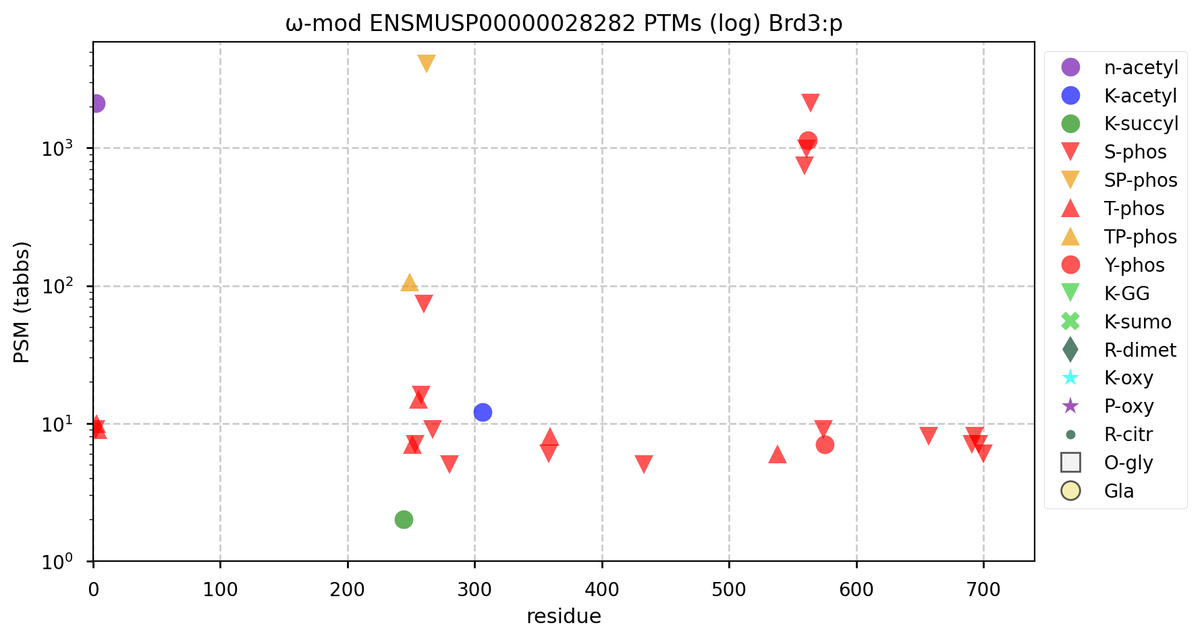
Tue Aug 16 11:47:00 +0000 2022Ended up with 86 mm of rain yesterday. Not a lot for some places, but a lot for here.
Tue Aug 16 03:57:51 +0000 2022An odd little blob of a storm that has dropped over 60 mm on my place in the last 4 hours. 🔗
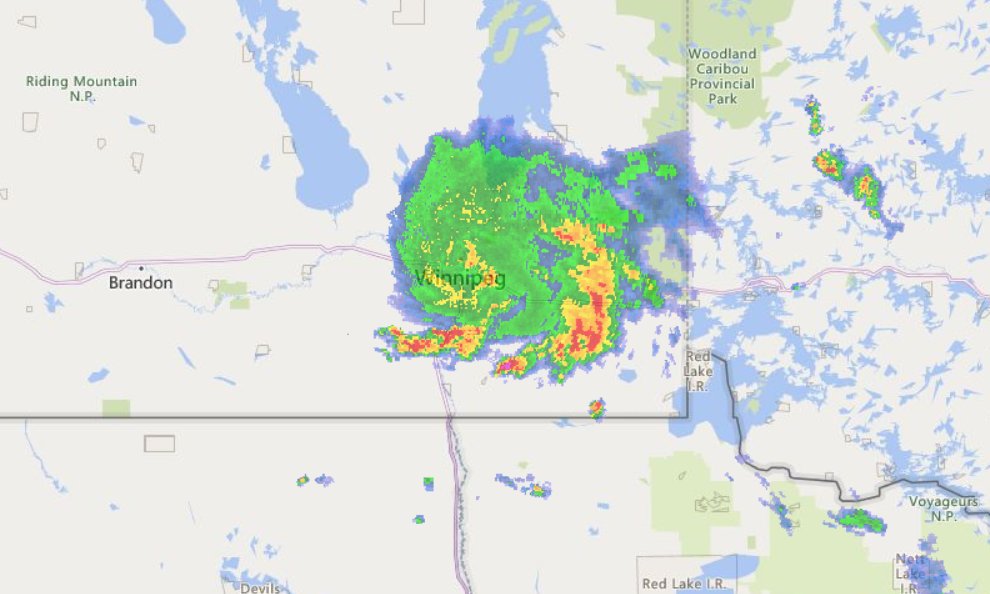
Tue Aug 16 01:33:47 +0000 2022@GeiszlerDaniel "Keratin" is pretty complex 🔗
Tue Aug 16 01:08:50 +0000 2022@GeiszlerDaniel It isn't in the paper, but it is a red-headed stepchild that just won't stay away throughout the data ...
Tue Aug 16 00:18:29 +0000 2022PXD034933: Keratin (proteomics' great Shaitan) you win again. Well played.
Mon Aug 15 23:46:01 +0000 2022I wonder how much academic research is not done because early on it became apparent that there would be no way to chose a single "first author" for the result.
Mon Aug 15 20:06:28 +0000 2022PXD022605: interesting.
Mon Aug 15 14:27:44 +0000 2022I immediately jump to the conclusion the PI isn't supervising properly & really shouldn't be involved in anything that requires reporting numbers. The editor & reviewers clearly didn't even bother to look at the tables, so why should I?
Mon Aug 15 14:04:10 +0000 2022I become irrationally dismissive when I see columns of numbers in which the authors have flamboyantly ignored the concept of significant figures, opting for "%f" instead.
Mon Aug 15 12:58:20 +0000 2022Even though BRD2:p has prominent displayed MHC class 1 peptides, it only has one rarely observed K+GGyl site, adjacent to a singular, similarly rarely observed K+SUMOyl acceptor.
Mon Aug 15 12:37:10 +0000 2022The protein-protein interaction map (from STRING) suggests direct engagement with many chromatin components 🔗
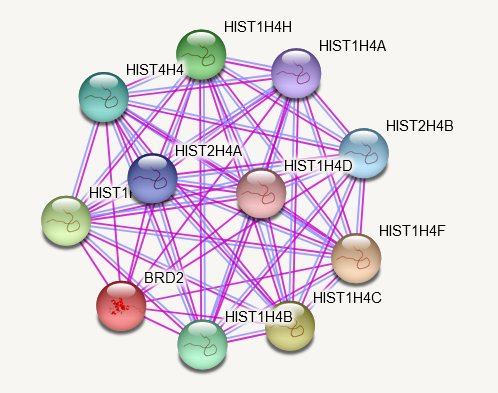
Mon Aug 15 12:06:52 +0000 2022Modification diagram for human BRD2:p, the 2ⁿᵈ subunit named for having >1 bromodomain. It has 2 of these domains―(91-163) & (364-436)―corresponding to gaps in the PMT pattern. It has fewer K+acetyl acceptors than BRD1:p.
🔗
Sun Aug 14 15:19:47 +0000 2022I once went to a biocuration conference: I realized within the first few hours that I had nothing at all in common with that particular community.
Sun Aug 14 15:07:00 +0000 2022A storm coming in off the Gulf of Mexico worrying Corpus Christi TX this morning 🔗

Sun Aug 14 14:45:15 +0000 2022It is tempting to include the AF predicted aligned error for this one, but the 3D structure has a very long, straight alpha helix (494-564) in the middle of it that I think is pretty iffy.
Sun Aug 14 13:18:24 +0000 2022A bromodomain has high affinity for K+acetyl in proteins. It was named (eccentrically) after the fruitfly gene "brahma" where it was originally described. The name has nothing to do with "bromine".
Sun Aug 14 13:17:50 +0000 2022Modification diagram for human BRD1:p, the 1ˢᵗ of the subunits named for containing a bromodomain (579-649). It has 3 prominent S+phosphoryl domains towards the terminii & 2 K+acetyl domains: the gap between them is the bromodomain.
🔗
Sun Aug 14 13:04:20 +0000 2022It is important to keep in mind that just because the same annotation occurs in multiple "databases" doesn't have any confirmatory implication: they are all just recirculating the same "information" amongst themselves.
Sun Aug 14 13:01:25 +0000 2022@stateofthecity When I was there (in the '00s) UBC was for all practical purposes a REIT with a sideline in housing profs & students.
Sun Aug 14 12:56:58 +0000 2022Although it does seem that the AF releases have lit a fire under most of them: they are all sporting an increase in structural "information", spurring a fiesta of cutting-&-pasting from each other.
Sun Aug 14 12:49:33 +0000 2022Spent an entertaining 30 minutes chasing a protein annotation around a dozen "databases" this morning. They all had the same annotation (& wording), but none of them seemed to have any idea where it came from.
Sun Aug 14 02:03:34 +0000 2022If you call a domain with lots of phosphorylation a "phosphodomain", what do you call one with lots of acetylation?
Sat Aug 13 21:00:29 +0000 2022@AJ_Brenes I stand corrected. 🔗
Sat Aug 13 18:54:48 +0000 2022Another warm, dry afternoon across Western Europe (ground station temperature). 🔗
Sat Aug 13 14:28:37 +0000 2022@ypriverol @byu_sam IMHO, the ones with the most sites aren't the most valuable. A lot of sites usually means it is from the sort of cell line/tumor expts that are dominated by cell cycle related phosphorylation processes. Single tissue expts are more interesting―fewer but rarer sites.
Sat Aug 13 14:00:11 +0000 2022@AlexUsherHESA Our federal government's not-so-long-ago enthusiastic promotion of the "Barbaric Cultural Practices Hotline" suggests that he is not the only one.
Sat Aug 13 13:14:43 +0000 2022@byu_sam @ypriverol & teleconferences. My only strong memory from being part of CPTAC 1 was the seemingly endless stream of teleconferences & complex email chains about what was said in the teleconferences.
Sat Aug 13 12:12:09 +0000 2022@byu_sam @ypriverol The one dataset to rule them all.
Sat Aug 13 11:57:54 +0000 2022CD247:p is only observed in T cells, T cell-derived cell lines & blood plasma extracellular vesicles (probably only those derived from T cells).
Sat Aug 13 11:55:40 +0000 2022Modification diagram for human CD247:p (aka CD3H, CD3Q, CD3Z), a type 1 membrane subunit of the T-cell receptor complex. In the intracellular domain (52-164), 7 of 7 Y residues are Y+phosphoryl acceptors & 8 of 9 K residues are K+GGyl acceptors.
🔗
Fri Aug 12 14:44:23 +0000 2022A high pressure system in North Africa cooperating with a low pressure system just west of Portugal, is pushing warm air up from Morocco as far north as England (winds at 700 mb). 🔗
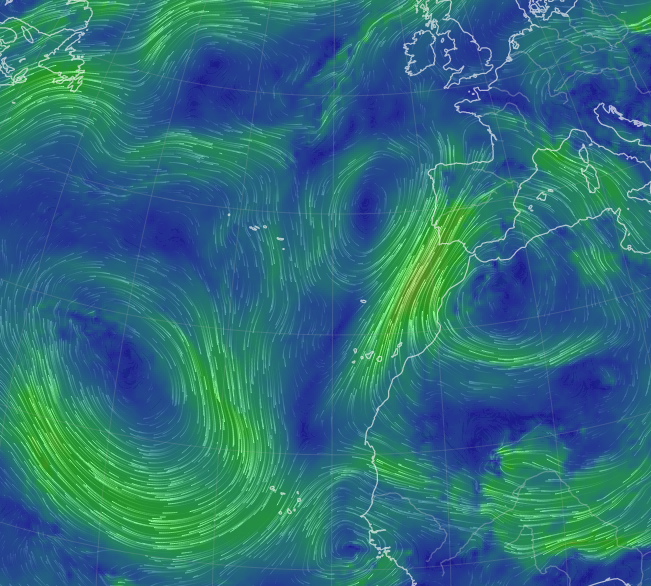
Fri Aug 12 14:22:17 +0000 2022A warm day across Western Europe (ground station temp). 🔗
Fri Aug 12 13:18:45 +0000 2022And an honorary degree for any who can identify all of the E3 ligases involved. 👨🏻🎓
Fri Aug 12 13:13:42 +0000 2022Extra points available if the student can identify the tyrosine kinase(s) involved.
Fri Aug 12 12:58:22 +0000 2022I love an article that echoes my own views!
🔗
Fri Aug 12 12:23:05 +0000 2022VAV1:p is not observable in many commonly used cell lines, such as HeLa, HEK-293, MCF-7 or MCF-10A. It can be found in leukocyte-derived cell lines, such as JURKAT or THP-1.
Fri Aug 12 12:19:39 +0000 2022Modification diagram for human VAV1:p, part of the guanine exchange factor family. It has 10 observed Y+phosphoryl acceptors across the sequence. It also has 27 scattered K+GGyl acceptors, with only 4 found with acetyl alternate modifications.
🔗
Fri Aug 12 02:52:51 +0000 2022Lots of little storms this evening, from Monterrey to Philadelphia 🔗
Thu Aug 11 14:31:01 +0000 2022@AlexUsherHESA I predict a race to the bottom (i.e., Ontario).
Thu Aug 11 14:25:26 +0000 2022@stateofthecity Are there any academics in Canada working on this particular policy issue? Software system procurement probably requires too much specialist knowledge & experience for most policy pros to work through without a good sherpa.
Thu Aug 11 13:28:03 +0000 2022@scalzi Science has the same problem: AI can gather together a lot of observations and generate interesting looking diagrams, but it still lacks anything like the ability to synthesize those observations into an actionable idea.
Thu Aug 11 13:16:54 +0000 2022@scalzi But first, someone will have to figure out how to make art out of odd-looking AI-generated images, which are not art any more than AI-generated text is an interesting story. AI fluff is not the expression of an idea, so it isn't inherently interesting (or art).
Thu Aug 11 12:53:58 +0000 2022@stateofthecity Is there a Canadian government at the municipal, provincial or federal level that has a good track record for developing or acquiring software-based systems?
Thu Aug 11 12:34:54 +0000 2022Its family member LYN:p has a rhyming pattern of Y+phosphoryl, but has a bit more jazz going on in the background.
🔗
Thu Aug 11 12:17:20 +0000 2022Modification diagram for human BLK:p, a SrcB family tyrosine kinase. The subunit itself has a very functional attitude towards PTM acceptors: an N-terminal myristoylation & a stanza of Y+phosphoryl acceptors.
🔗
Thu Aug 11 00:45:28 +0000 2022Some angry weather rumbling around Chesapeake Bay this evening & another bunch of squalls touring through South Carolina. 🔗
Wed Aug 10 20:49:52 +0000 2022@UCDProteomics @andreafossati9 And as I have mentioned before, I am no fan of the idea of re-calibrating the m/z data based purely on calculated results and then re-running the calculation on the new-and-improved data to get cosmetically prettier results.
Wed Aug 10 16:12:14 +0000 2022@UCDProteomics @andreafossati9 It can be dealt with, but by that point often the correction isn't very linear & there may be several peaks in the Δ mass distribution. Better to just do some routine QA on the instruments.
Wed Aug 10 16:08:56 +0000 2022IMHO everyone running an academic proteomics facility should have to spend some time in a GMP-regulated lab to get a good feel for how QA/QC works in practice.
Wed Aug 10 15:59:45 +0000 2022@UCDProteomics @andreafossati9 It is very common in the field. The highest I've seen in published data is ~40 ppm. 😱
Wed Aug 10 15:48:53 +0000 2022@rabbleca Senior Canadian university admins should have someone sit them down before they take the job and explain to them the fact that they are effectively provincial govt employees & they will be expected to execute govt policy in the same manner as an ADM.
Wed Aug 10 14:51:30 +0000 2022NEDD9 (Neural precursor cell Expressed Developmentally Down-regulated protein 9) is named for a differential expression experiment from the early 90s. Other proteins in the NEDDx clan are unrelated, other than via a PMID.
🔗
Wed Aug 10 14:24:51 +0000 2022Another, more abundant, CAS family member is BCAR1:p. It sports a rhyming PTM pattern, with a few more baroque flourishes. NEDD9:p & BCAR1:p are paralogs, but they only share 2 tryptic peptides. 🔗
Wed Aug 10 14:22:08 +0000 2022Modification diagram for human NEDD9:p, 1 of 4 members of the CAS (Crk-Associated Substrate) family of scaffolding proteins. "Hyperphosphorylation" is a term well suited to the main phosphodomain (92-395). 🔗
Tue Aug 09 18:50:03 +0000 2022PXD024491 is kind of a rarity: glam journal but interesting data.
Tue Aug 09 17:31:46 +0000 2022@neely615 @neilhimself It is the best thing I've seen on Netflix for a while.
Tue Aug 09 16:59:23 +0000 2022And they are compatible with other accessories for cat up-armouring
🔗
Tue Aug 09 16:03:45 +0000 2022The true apex of American exceptionalism & its never ending quest for technological defenses from lurking dangers.
🔗
Tue Aug 09 14:34:58 +0000 2022For added credit, the student should derive these three patterns using Michaelis–Menten kinetics & Langmuir adsorption isotherms.
Tue Aug 09 13:47:45 +0000 2022Also with the same domain structure, DOK2:p displays a quite different PTM pattern than its other family members. 🔗
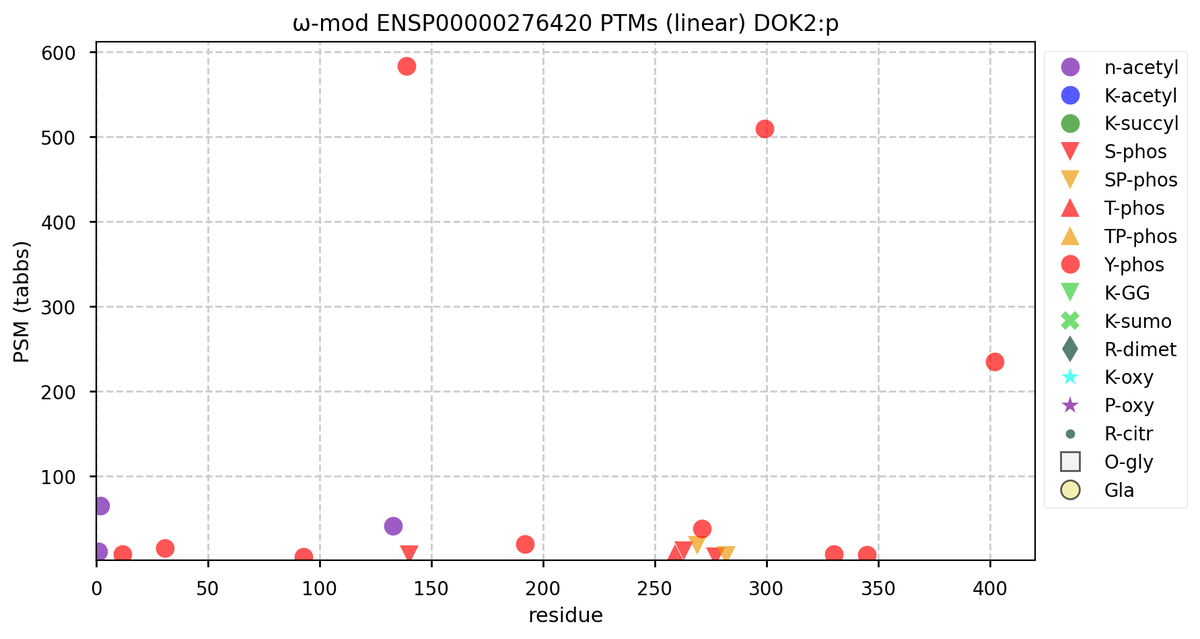
Tue Aug 09 13:35:43 +0000 2022Even though it has the same domain structure as DOK1:p, DOK3:p has a very different modification pattern in its C-terminal IDR phosphodomain
🔗
Tue Aug 09 13:24:24 +0000 2022For anyone interested in such things, the AF frankenstructure for DOK1:p can be found here 🔗 🔗
Tue Aug 09 12:48:10 +0000 2022Modification diagram (linear) for human DOK1:p, an adaptor protein. From the acetylated N-terminus: pleckstrin homology domain → insulin receptor substrate phosphotyrosine binding domain → phosphodomain (IDR). Lots of high occupancy Y+phosphoryl acceptors. 🔗
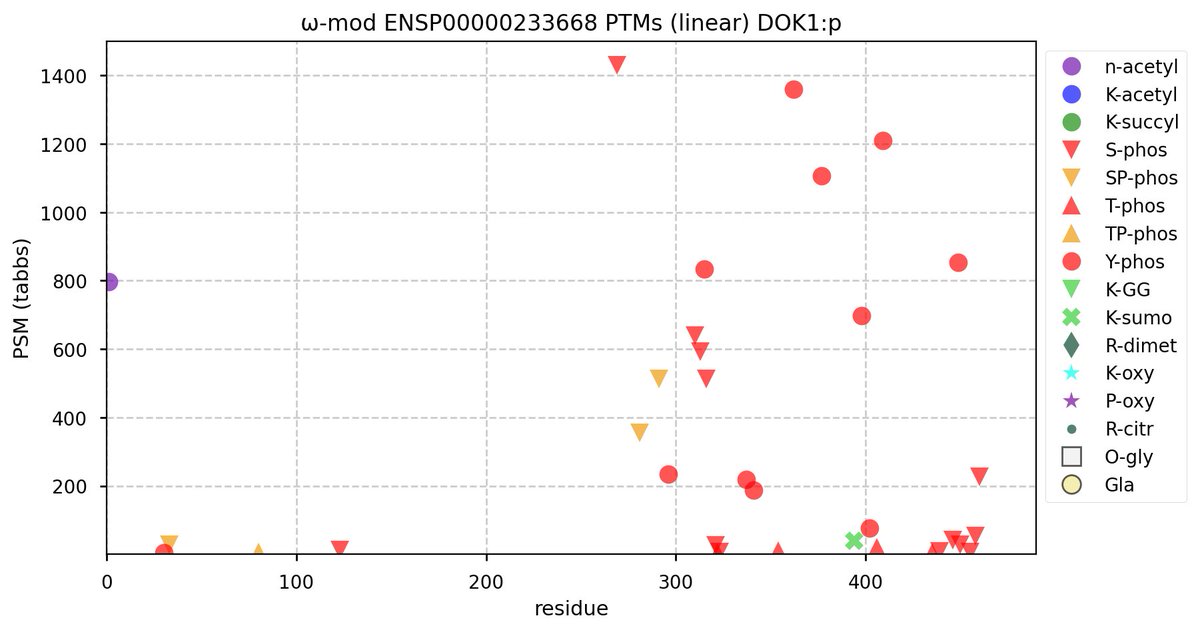
Mon Aug 08 19:53:46 +0000 2022@camtflower Something associated with their actual function, rather than what happens if you break them. Associating genes with cancer can be productive from the viewpoint of grantsmanship, but if you aren't interested in that particular subject, the description is rather pejorative.
Mon Aug 08 18:09:59 +0000 2022Status:Connecting to 193.62.193.138:21...
Status:Connection established, waiting for welcome message...
Error:Could not connect to server
Status:Disconnected from server
Mon Aug 08 16:55:17 +0000 2022It is like calling a car's steering wheel or brake pedal a "proto-crash device".
Mon Aug 08 15:51:20 +0000 2022The only term I hate more the "oncogene" is "proto-oncogene".
Mon Aug 08 15:26:59 +0000 2022@lgatt0 Be sure to get checked for an intermediate distance prescription (i.e., computer viewing distance). It gets rid of the need for large text & makes using a desktop/laptop computer much easier.
Mon Aug 08 15:05:27 +0000 2022The distribution of observations of MHC class 2 peptides has a greater contribution from the N-terminal extracellular domain, but maintains significant representation from the C-terminal, intracellular kinase domain. 🔗
Mon Aug 08 14:46:18 +0000 2022The absence of K+acetyl co-mods at K+GGyl acceptors is typical of type I membrane protein receptors. The presence of GGyl acceptors & the distribution of observations of MHC1 peptides (red bars) suggests that the cytoplasmic portion of AXL:p must end up in the proteasome. 🔗
Mon Aug 08 12:02:17 +0000 2022Modification & N-linked glycosylation diagrams for human AXL:p, a cell surface receptor Y-kinase with a single transmembrane domain (450-472). While everything is where it should be, there are a lot of Y+phosphoryl acceptors on the cytoplasmic kinase domain. 🔗
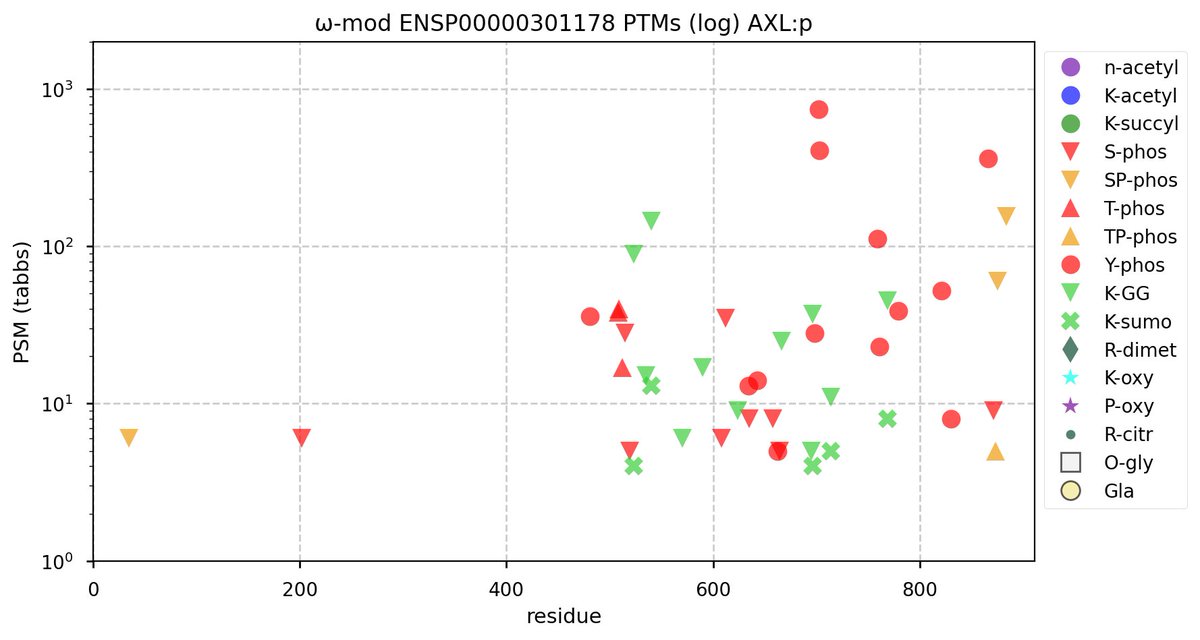
Sun Aug 07 17:02:17 +0000 2022@slavov_n @neely615 Perhaps, but it (& subsequent related experiences) informed the way I have viewed "authorship" for the last 35 years.
Sun Aug 07 15:58:04 +0000 2022@neely615 @slavov_n My favorite was some work I did as a postdoc with a colleague: we came up with the idea, did the experiments & wrote the paper. When the manuscript came back from the boss, we had acquired 8 co-authors.
Sun Aug 07 15:00:02 +0000 2022@slavov_n But, being too high minded about these things can end up causing profound disillusionment in trainees when they are first confronted with how the system actually works.
Sun Aug 07 14:32:46 +0000 2022@slavov_n If everyone who used monetary clout to become a co-author was to be investigated, there would be little else going on in academia (& government & industry).
Sun Aug 07 14:04:05 +0000 2022Do any of the chaperonin cognoscente out there have insights into the utility or damage caused by these lysine PTMs?
Sun Aug 07 13:24:08 +0000 2022While ST13:p (HIP) has a similarly baffling pattern of lysine PTMs, it does not share HOP's propensity for tyrosine phosphorylation 🔗
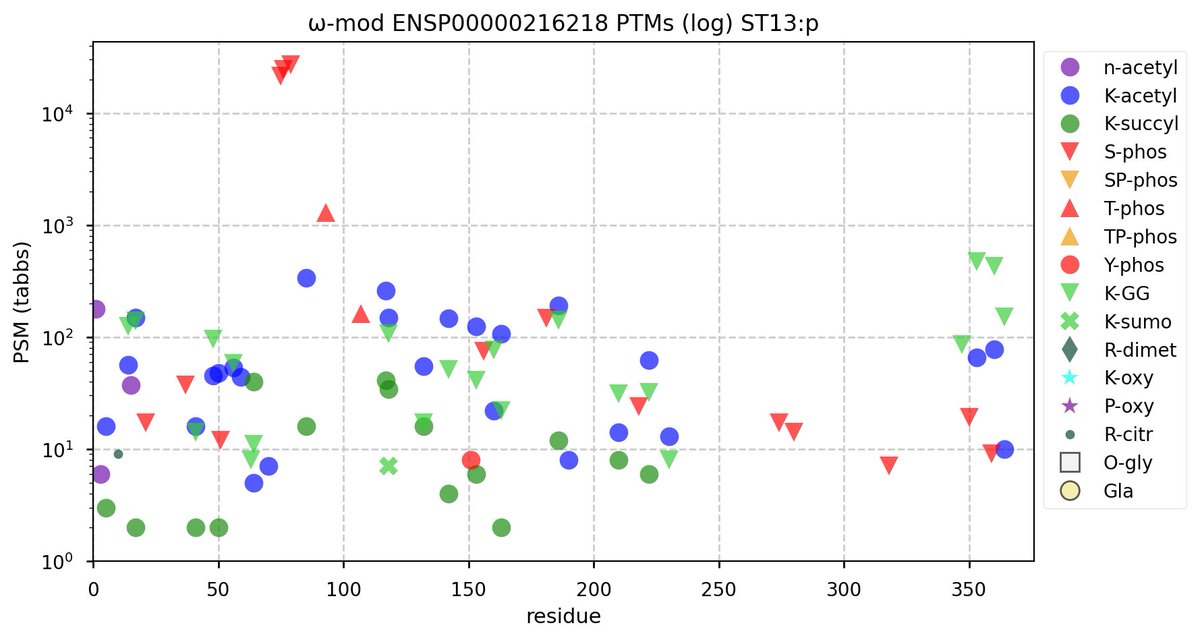
Sun Aug 07 12:35:57 +0000 2022HOP:p (Hsp70-Hsp90 Organizing Protein) has been proposed as an alternate name for STIP1:p, part of a clever name pairing with an alternate name HIP:p (Hsc70-Interacting Protein) for ST13:p, another co-chaperonin.
Sun Aug 07 12:19:39 +0000 2022Modification diagrams for human STIP1:p, a co-chaperonin in the HSP70-HSP90 system. The full diagram has the sort of complex lysine PTM pattern common in chaperonin subunits. The phosphoryl-only diagram highlights an unusual number of Y+phosphoryl acceptors (red circles). 🔗
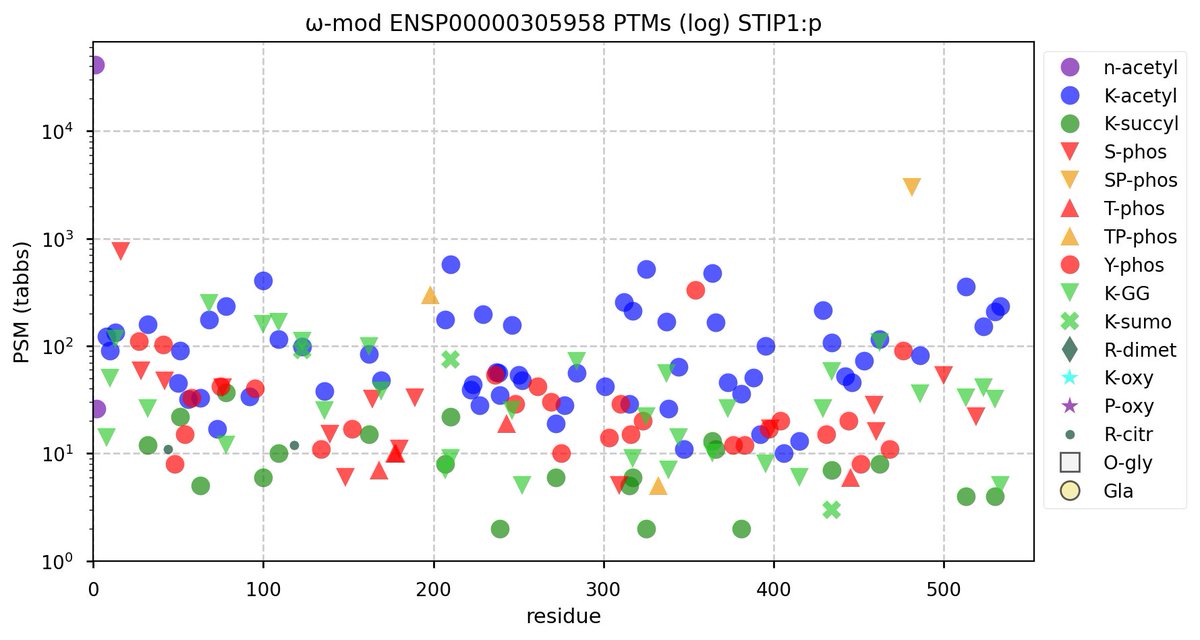
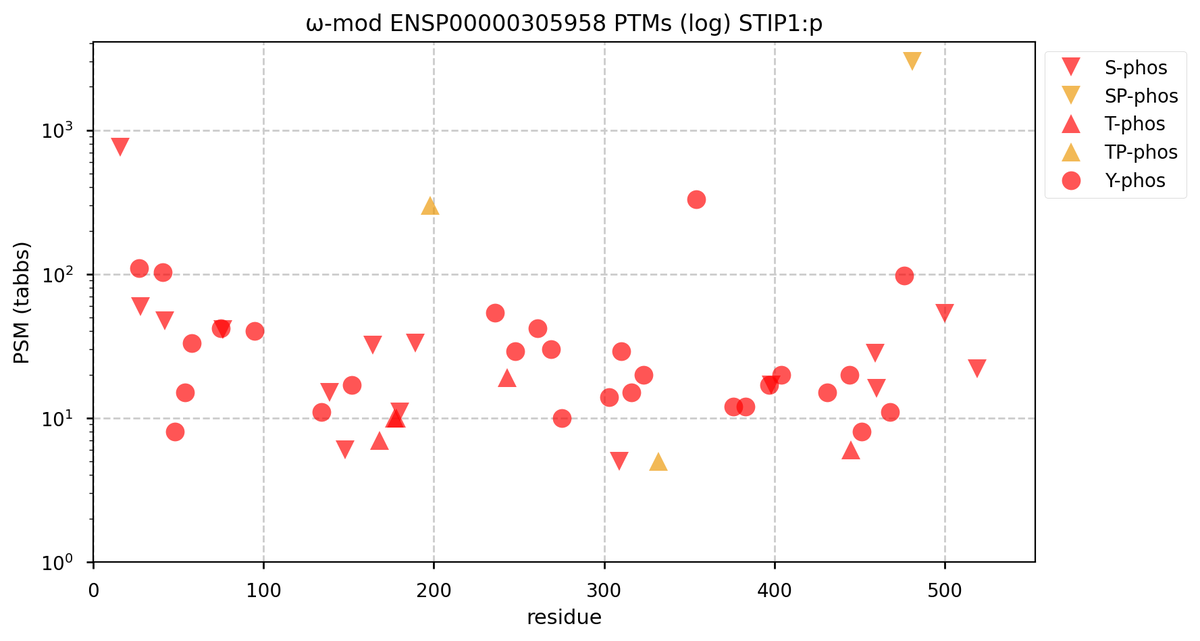
Sat Aug 06 23:08:21 +0000 2022🔗
Sat Aug 06 15:06:44 +0000 2022@astacus That is something I haven't put in yet: I'll get right on it! I use a Firefox extension (Consent-O-Matic) that handles GPDR consent forms automatically for me.
Sat Aug 06 14:21:29 +0000 2022@pwilmarth I keep "The Elements of Typographic Style" (Bringhurst) in my line of sight at all times to prevent just that sort of temptation from taking hold.
Sat Aug 06 13:40:18 +0000 2022To appear modern, I really should add some incredibly intrusive ads to my sites―especially those with little multiple floating video windows that skip around in the display window obscuring as much content as possible.
Sat Aug 06 12:45:09 +0000 2022Thanks to everyone who participated in the poll & added discussion. There doesn't seem to be a shared community QA level for non-tryptic cleavage in their samples: opinions span the range.
Sat Aug 06 12:08:52 +0000 20227 out of 14 K+GGyl acceptors are shared as K+SUMOyl acceptors. The lack of K+acetyl is a common feature of Kelch-like subunits.
Sat Aug 06 12:04:43 +0000 2022Modification diagram for human ENC1:p (aka PIG10, TP53I10, KLHL37), a Kelch-like family protein. It's domain structure: ID phosphodomain, BTB, BACK & 6 Kelch β-propeller. S406+phosphoryl is on a short disordered loop between Kelch domain #2 & #3.
🔗
Sat Aug 06 01:46:01 +0000 2022"Probably plays a crucial role ..."
Fri Aug 05 14:05:42 +0000 2022A notable exception is KBTBD6:p & KBTBD7:p, which overlap in 37% of their observable tryptic peptides.
Fri Aug 05 12:52:36 +0000 2022Even though there are many Kelch domain containing proteins, it is rare for any of them share even 1 observable tryptic peptide with any of the others.
Fri Aug 05 12:19:39 +0000 2022Modification diagram for human KLHL24:p, part of the Kelch-like protein family. This subunit has 1 low occupancy S+phosphoryl acceptor in an N-terminal intrinsic disorder domain & no K+acetyl acceptors.
🔗
Fri Aug 05 03:48:42 +0000 2022Maybe some Aurora tonight (I am the blue dot). 🔗
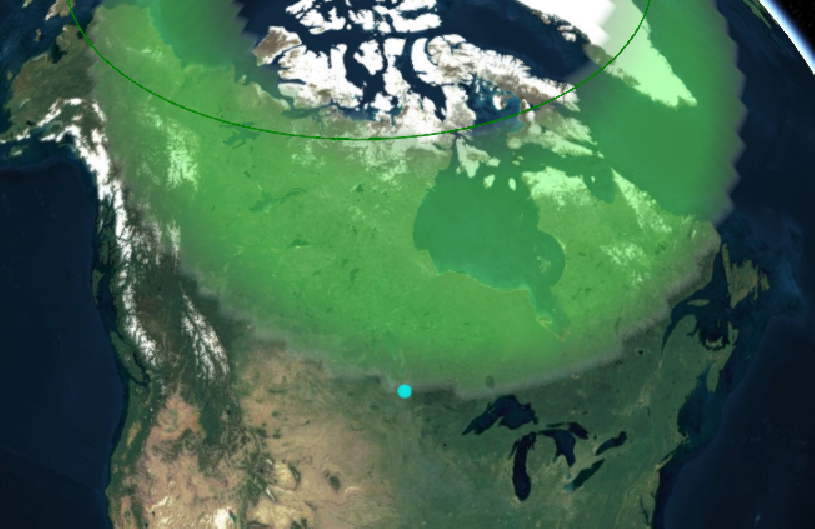
Thu Aug 04 23:40:55 +0000 2022@pitman_mark @ItsBiniR I would if I knew the answer. I would think it is more of a @pwilmarth question.
Thu Aug 04 18:16:54 +0000 2022From people's responses, it would seem that many have experience explaining the finer points of practical proteolytic digestion to the hoi polloi. 🔗
Thu Aug 04 17:13:09 +0000 2022@astacus Yes: I would normally consider K/R-P and D-P cleavage as "expected".
Thu Aug 04 17:02:20 +0000 2022@pwilmarth Let's say you are examining the results of a tryptic digest (Pierce sequencing-grade trypsin) carried out on a lyzed MCF-7 cell culture with a single, unfractionated LC/MS/MS run, excluding anything resulting from in-source fragmentation.
Thu Aug 04 16:40:34 +0000 2022@VATVSLPR Since it was looking at the results from a recent whole-cell lysate study that spurred the question, "unexpected non-tryptic cleavage in a whole-cell lysate" would be a better way to phrase it.
Thu Aug 04 16:23:30 +0000 2022Expected non-tryptic cleavage would be the result of normal protein maturation processes, e.g. N-terminal processing, pre- or pro-protein peptide removal or ER/mitochondrial signal peptide removal.
Thu Aug 04 16:21:43 +0000 2022How much unexpected non-tryptic cleavage in a proteomics sample prep is too much (as a % of identified PSMs)?
Thu Aug 04 15:06:14 +0000 2022@astacus So probably this one, aka "sticky willy":
🔗
Thu Aug 04 14:51:37 +0000 2022@astacus You are going to have to be more precise
🔗
Thu Aug 04 14:46:43 +0000 2022Many authors seem to shorten "breast cancer derived cell line" down to simply "breast cancer" when they are writing technical papers, as though the terms were really interchangable. It happens so often it seems to be editorial policy in some journals.
Thu Aug 04 14:37:29 +0000 2022This article from 2013 is still a pretty good survey of the properties of KLHL subunits
🔗
Thu Aug 04 14:25:36 +0000 2022@dtabb73 The version of Stirling's approximation in Eqn 27 here is my go to for big factorials 🔗
Thu Aug 04 14:15:47 +0000 2022@Smith_Chem_Wisc Stirling's approximation is another (& often better) way to get around the issue.
Thu Aug 04 11:59:24 +0000 2022It also has the PTM pattern typical of the family: few, if any, S/T+phosphoryl and scattered K+GGyl acceptors that do not accept K+acetyl as alternate modifications.
Thu Aug 04 11:56:05 +0000 2022Modification diagram for human KLHL28:p, part of the Kelch-like protein family. It has the domain structure characteristic of the family: N-terminal acetyl→BTB/POZ→BACK→6 Kelch domain β-propeller.
🔗
Wed Aug 03 20:53:10 +0000 2022The line of storms has become more organized & made its way through Chicago, on track to pass through Toronto, Detroit & Toledo next. Lots of lightning 🔗 🔗
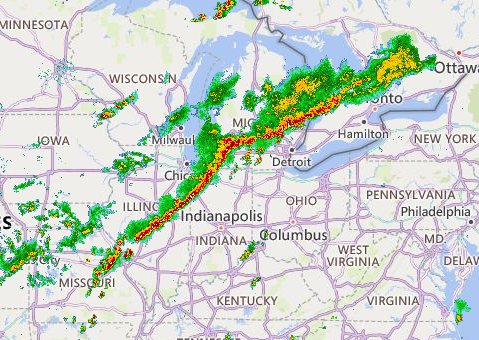
Wed Aug 03 18:06:53 +0000 2022But I like this one better
🔗
Wed Aug 03 18:06:14 +0000 2022Un!pr*t calls this splice variant "canonical"
🔗
Wed Aug 03 16:23:10 +0000 2022@byu_sam @neely615 @JohnRYatesIII Discuss
🔗
Wed Aug 03 16:14:07 +0000 2022Hard to imagine this one missing Chicago. Should be spicy weather through the Midwest this afternoon. 🔗

Wed Aug 03 16:00:06 +0000 2022The modification diagram for mouse HCFC1:p indicates a well-populated domain of O-linked glycosylation acceptors (450-880). 🔗
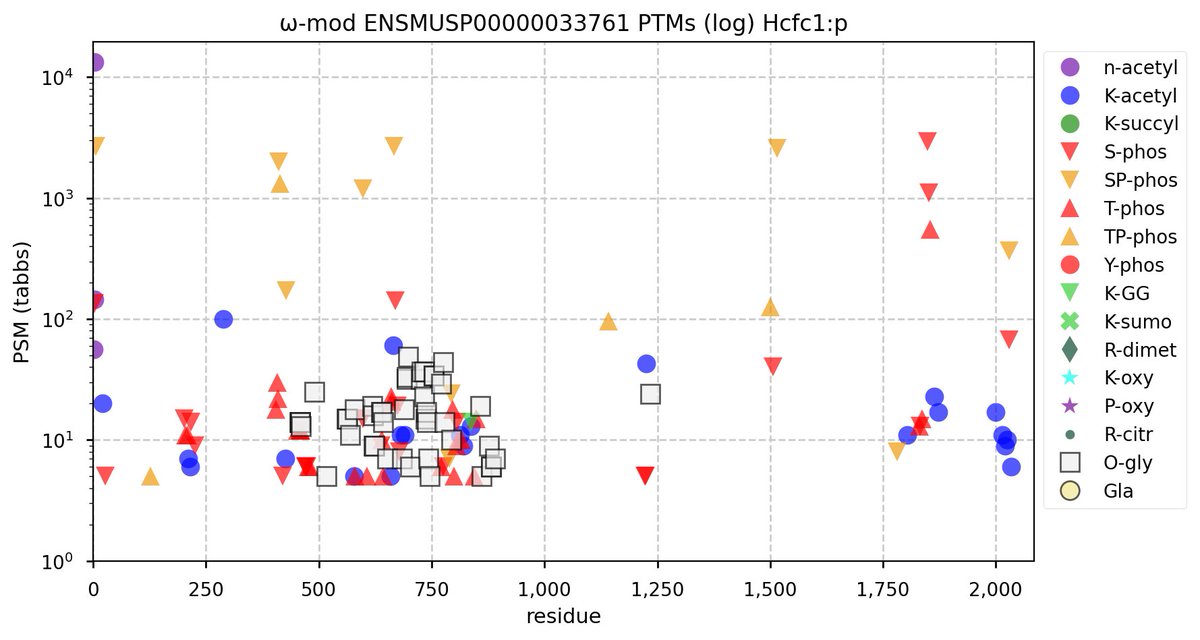
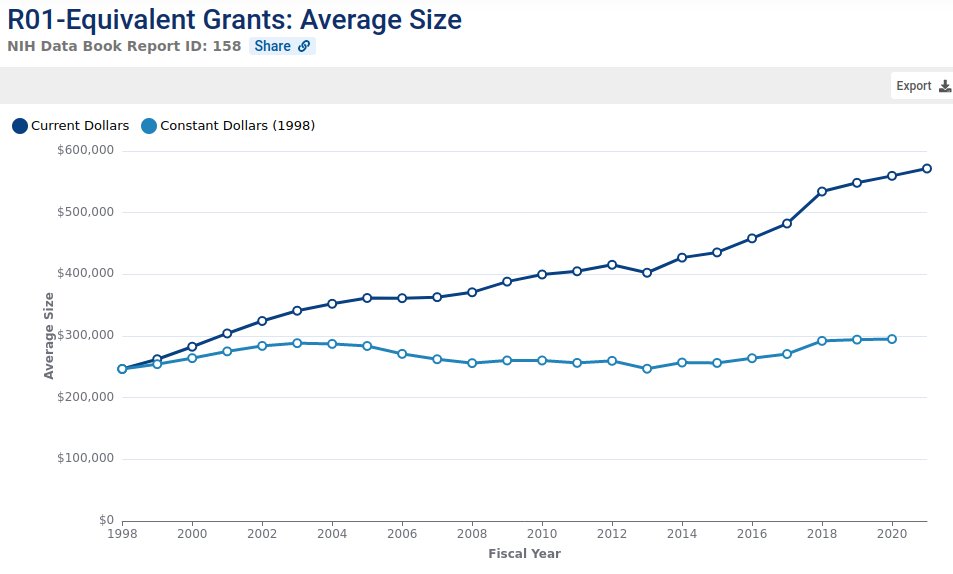
Wed Aug 03 13:57:07 +0000 2022Another feature of HCFC1:p is that the mature protein is created by proteolytic cleavage at 1 of 6 identical sites:
…VCSNPPCE⁓THETGTT…
The resulting peptide chains remain associated to form the protein. There is good evidence in public proteomics data for 5 of these cuts.
Wed Aug 03 12:13:14 +0000 2022The same diagram for human HCFC2:p, which also has a similarly placed patch of Kelch domains, has the sort of sparse PTM pattern more typical of Kelch-domain containing proteins
🔗
Wed Aug 03 12:08:34 +0000 2022Modification diagram for human HCFC1:p, aka HCF-1, HCF1, CFF, VCAF, MGC70925, PPP1R89, HFC1 & MRX3. It has a very unusual modification pattern for a sequence with a Kelch-type β-propeller (44-313), with multiple acceptors for many common PTMs.
🔗
Tue Aug 02 22:52:54 +0000 2022In differential proteomics experiments, if you know that some small number of proteins changed, the job isn't done until you know why the rest of the proteins didn't.
Tue Aug 02 19:35:44 +0000 2022It came back for a while last week, but otherwise the PRIDE FTP site has not been very cooperative for the last month.
Tue Aug 02 16:38:20 +0000 2022Many countries seem to be run by a small number of stubborn elderly people recklessly trying to prove something at each other's expense.
Tue Aug 02 14:37:35 +0000 2022Looks like it will pass west of Indianapolis, but Evansville & Bowling Green may not be so lucky 🔗

Tue Aug 02 12:53:11 +0000 2022Not a bad little survey article about the requirements (& plugs) for the 3 main categories EV chargers.
🔗
Tue Aug 02 12:33:55 +0000 2022One of the many examples of why "cyber-security" is a a chaotic mess.
🔗
Tue Aug 02 12:03:33 +0000 2022Modification diagram for KLHL12:p, a Kelch-like family subunit. Typical of this family, the domain structure is (from the N-terminus): BTB/POZ, BACK & 6×Kelch (β-propeller).
🔗
Mon Aug 01 17:05:25 +0000 2022@AlexUsherHESA Andoria.
Mon Aug 01 15:49:04 +0000 2022For some reason, MEGF8:p also has one of the most detailed Wikipedia entries of any protein without being associated with a fundable disease
🔗
Mon Aug 01 14:09:38 +0000 2022@goodlettlab1 It's Terry Fox Day here in Manitoba.
Mon Aug 01 12:46:05 +0000 2022MEGF8:p is commonly found in urine, blood plasma & cerebrospinal fluid. 🔗
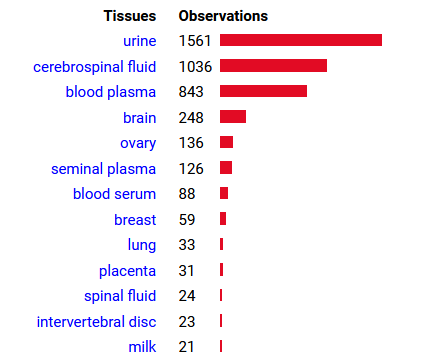
Mon Aug 01 12:26:46 +0000 2022Modification & N-glycosylation patterns for human MEGF8:p, a large type I membrane protein that has 12 Kelch domains arranged into what are probably 2 separate β-propellers. The N-terminal extracellular domain domain (28-2647) has all of the observed N+glycosyl acceptors. 🔗
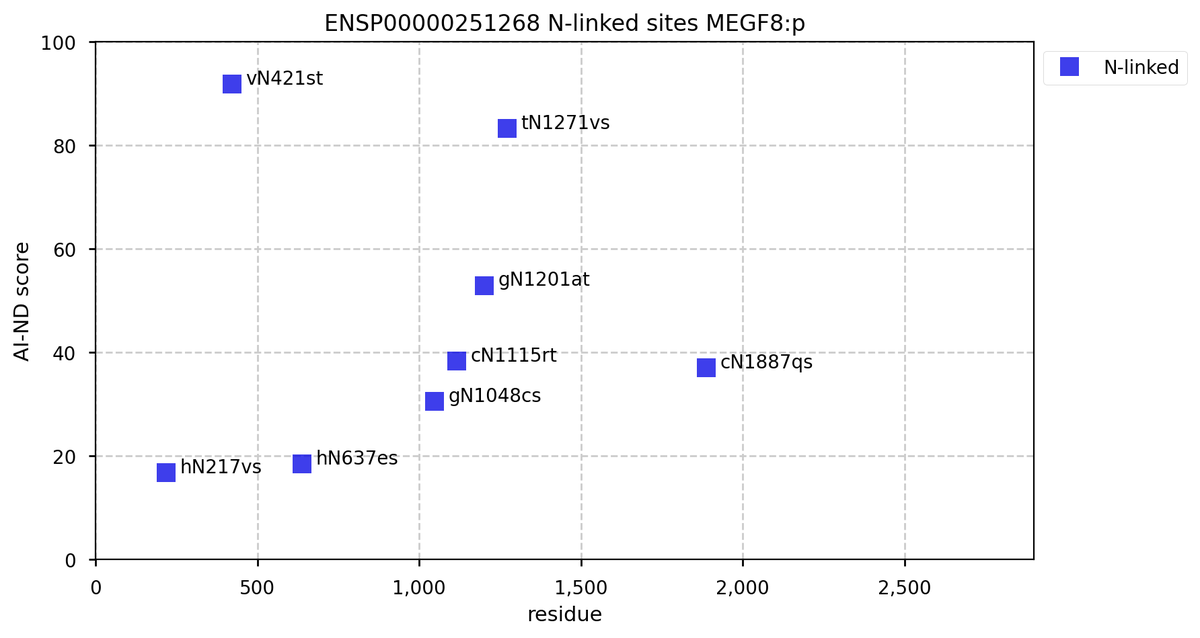
Mon Aug 01 01:47:34 +0000 2022Hurricane Frank (heading northwest) & Tropical Depression Georgette (heading north) in the Pacific west of Mexico (shown: TPW & surface winds). 🔗
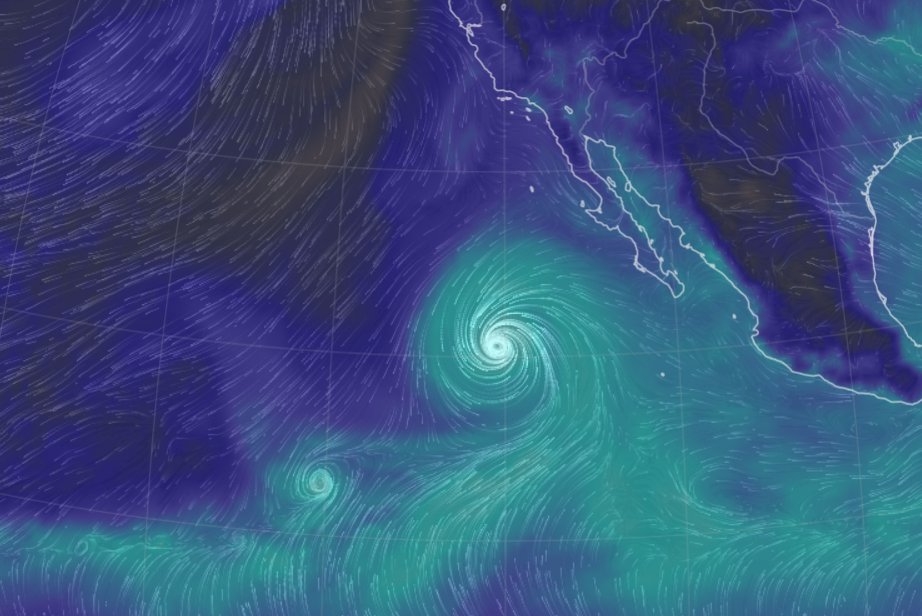
Mon Aug 01 01:19:15 +0000 2022Scattered storms this evening across the normally dry southwestern USA. 🔗
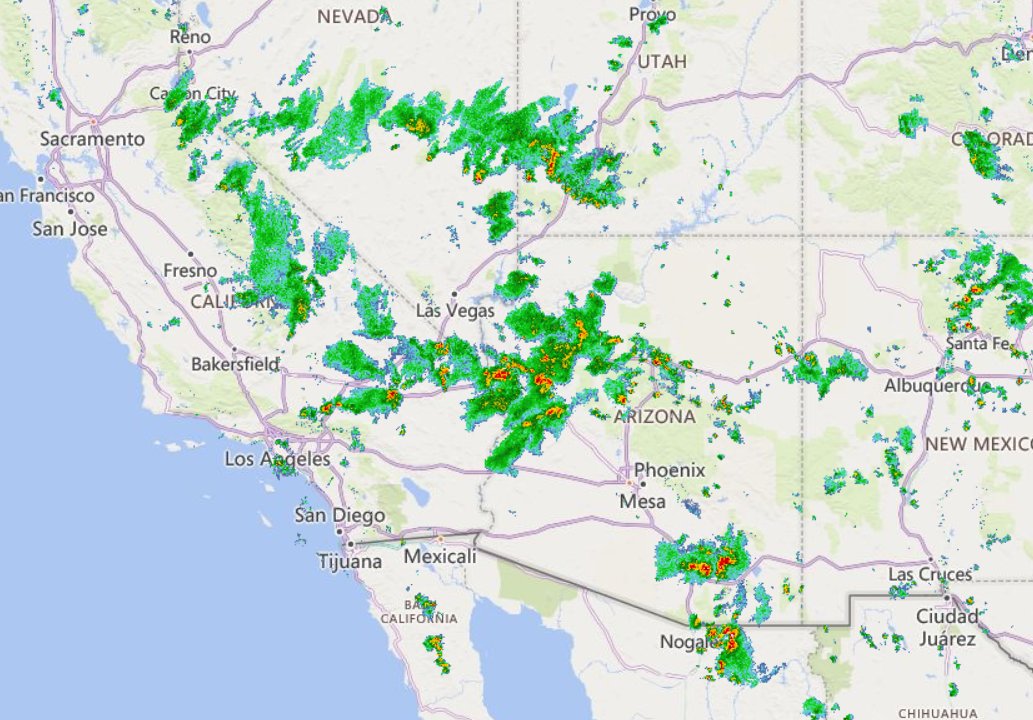
Sun Jul 31 17:01:55 +0000 2022@BiswapriyaMisra Yes. Many, many times. I don't know if "validation" has any precise meaning wrt to the AF list of predicted structures, though. It is just a bunch of best guesses of one possible set of structures for each sequence.
Sun Jul 31 13:16:54 +0000 2022Even though many proteins have Kelch-domain β-propellers, the function of these rather showy structures has been hard to pin down (not for lack of trying). Are they functional or are they more like the tail fins on a 1958 Plymouth Fury?
Sun Jul 31 12:38:13 +0000 2022Modification diagram for KLHDC4:p, a member of the Kelch-domain-containing family. It has 6 Kelch domains, with an 80 residue disordered phosphodomain between Kelch #5 & #6 (362-442). 🔗
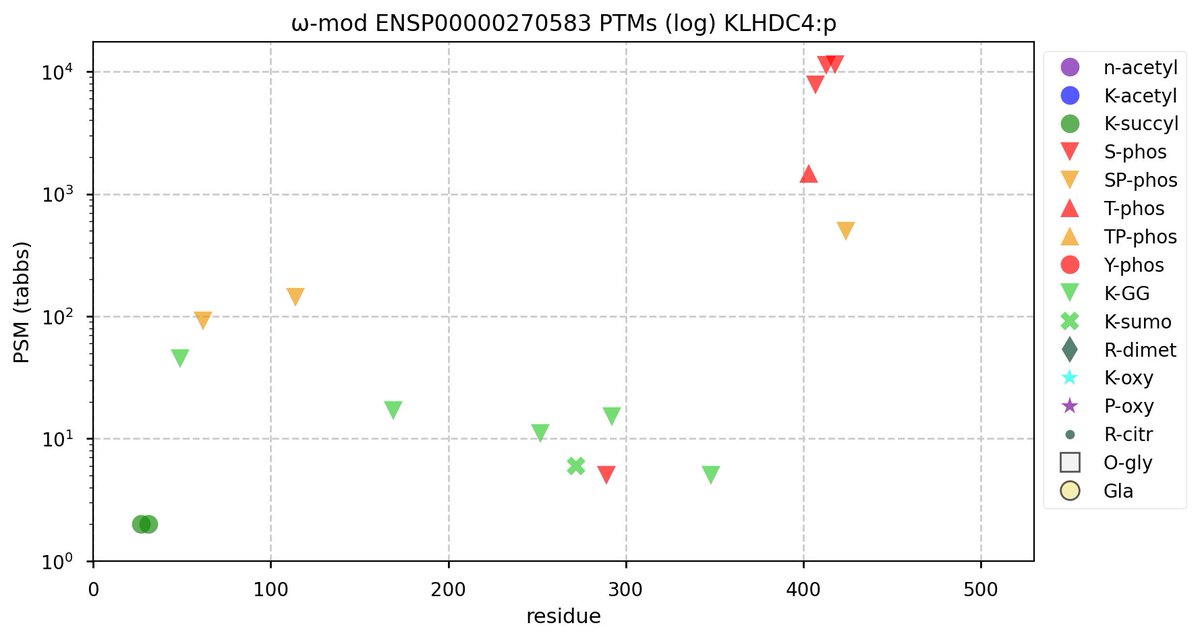
Sat Jul 30 19:12:35 +0000 2022While there are many modifications (both PTMs & artifacts) that can cause a peptide to become more acidic, there are only a few that can make one more basic.
Sat Jul 30 18:50:40 +0000 2022The region of moist air (cyan) that has been causing rain storms & squalls across the southeastern US for the last few days. 🔗
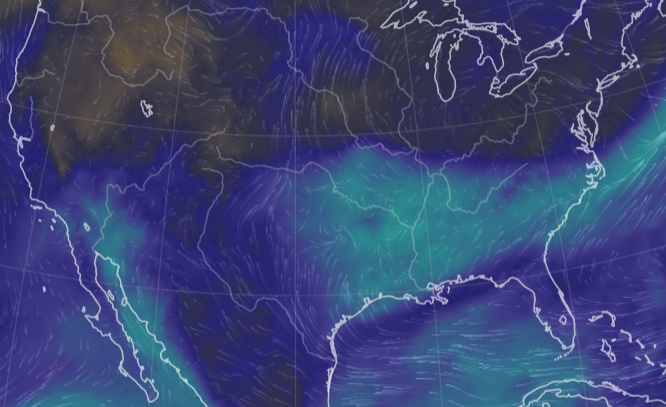
Sat Jul 30 17:15:26 +0000 2022An intensifying group of storms along the Gulf Coast from Houston to Mobile. 🔗
Sat Jul 30 16:22:27 +0000 2022The most surprising thing about AF is its PR budget.
Sat Jul 30 13:15:20 +0000 2022A frontal system with thunderstorms this morning, currently stretching from Amarillo to the Mississippi River, heading east towards Kentucky & Tennessee. 🔗

Sat Jul 30 12:40:05 +0000 2022As with most subunits containing a β-propeller made of Kelch domains, there are few PTMs other than a set of potential K+GGyl acceptors.
Sat Jul 30 12:09:11 +0000 2022Modification diagram for human KLHDC3:p, a member of the Kelch-domain-containing family of subunits. It has 5 Kelch domains that form a β-propeller spanning most of the sequence (25-301).
🔗
Fri Jul 29 23:45:27 +0000 2022@neely615 Weather types come from a geography tradition (academic meteorology is often part of Geography Depts), so they tend to describe things in a different way than hard science types & they have a thing for any esoterica associated with spherical geometry.
Fri Jul 29 22:09:03 +0000 2022@cajun_science @SciFiRobitaille @chrashwood The residual mass is 100.0160 Da, the same as 🔗
Fri Jul 29 20:26:57 +0000 2022@SciFiRobitaille @chrashwood It is rather episodic. It can range from undetectable to ~20% of PSMs, but it is consistent within a particular data set. I suspect it is a straightforward sample prep issue, e.g., not keeping the reaction as anhydrous as it should be.
Fri Jul 29 16:15:36 +0000 2022@chrashwood You should note that I am guessing wrt to the identity of the succinyl group. What I know for sure is that there is a substitution of peptide N-terminal TMTPro → X, where X is isobaric with succinyl.
Fri Jul 29 16:11:38 +0000 2022@chrashwood TMTPro → succinyl substitution also leads to an increase in retention time.
Fri Jul 29 15:54:40 +0000 2022While not a popular topic, thanks to the brave few who offered an opinion. The correct answer is the apparently counter-intuitive choice: "increase".
Fri Jul 29 15:10:09 +0000 2022If you take the time to understand the origins of a protein structural domain's name, you are usually none the wiser. Their tendency to be idiosyncratic historical footnotes rather than evocative of structure or function is a barrier for students & practitioners alike.
Fri Jul 29 14:52:52 +0000 2022@neely615 MJO + Australians = interesting graphical representation of a complicated motion.
🔗
Fri Jul 29 14:42:14 +0000 2022@neely615 It is also often referred to as the Bermuda-Azores High (or the North Atlantic Subtropical Anticyclone) because its center is somewhere between the 2 sites:
🔗
Fri Jul 29 14:18:23 +0000 2022Looks like there is going to be another day of thunderstorms running along a cold front at the southern edge of the polar jet stream, where it meets moist air being pumped up from the Gulf of Mexico by an Azores High. 🔗

Fri Jul 29 13:38:23 +0000 2022Kelch domain associated K+GGyl acceptors generally do not double as K+acetyl acceptors.
Fri Jul 29 11:46:39 +0000 2022The BACK domain is commonly found in this family, but it has not been assigned an independent function. It is named for its position between the other 2 ordered domains: "Btb And C-terminal Kelch" domain.
Fri Jul 29 11:41:34 +0000 2022Modification diagram for human KLHL13:p, a member of the Kelch-like family of subunits. N-to-C-terminal domains: intrinsic disorder (S/T+phosphoryl), BTB/POZ (homodimerization), BACK, 6×Kelch (β propeller, K+GGyl) & intrinsic disorder (S/T+phosphoryl).
🔗
Thu Jul 28 17:39:17 +0000 2022A big plume of dust in the Atlantic blown off North Africa is probably the reason that there have not been any Cape Verde type hurricanes yet this season (shown: surface winds & PM10 particulates) 🔗
Thu Jul 28 15:32:35 +0000 2022If you were to substitute the N-terminal TMT6 group on a derivatized peptide with a succinyl group, would you expect the peptide's RP-HPLC retention time to
Thu Jul 28 15:16:57 +0000 2022@kadzuis @Nature I'm afraid it is a ship that has already sailed. It will take years to unwind the impact of the current crop of dodgy predictions.
Thu Jul 28 15:02:28 +0000 2022And a related, but even odder, take in English
🔗
Thu Jul 28 14:59:50 +0000 2022Odd takes on public health issues is a problem not limited to US politicians: we have it up here, too, in both official languages
🔗
Thu Jul 28 12:01:44 +0000 2022Modification diagram for human MKLN1:p, an intracellular subunit (2-735) with an N-terminal acetylation. It has a C-terminal β-propeller made up of 6 kelch domains (285-650).
🔗
Thu Jul 28 11:54:57 +0000 2022@astacus I don't disagree, which was the reason for my early optimism. However, my reanalysis of the data they used to support their proposal of peptide-level splicing didn't lead me to the same conclusion.
Thu Jul 28 01:34:48 +0000 2022🔗
Wed Jul 27 16:28:17 +0000 2022@leprevostfv Me too. I was quite enthusiastic when I originally saw the claims, but after looking at the data I had to concede that they just were not there.
Wed Jul 27 15:30:59 +0000 2022PXD035524 has good examples of the products of the side reaction that generates N-succinylation, which can occur as an unintended result of TMT peptide derivatization (~14% of assignable PSMs).
Wed Jul 27 15:14:56 +0000 2022Answer = No.
🔗
Wed Jul 27 12:17:18 +0000 2022Modification diagram for human IPP:p, an intracellular subunit (2-582) with N-terminal acetylation, 1 BTB (homodimerization) & 6 kelch domains (β-propeller).
🔗
Tue Jul 26 16:37:24 +0000 2022Nestin has a very complex phosphotype. When present, it is frequently a source of phosphorylated PSMs even in experiments without phosphopeptide enrichment.
🔗
Tue Jul 26 14:02:41 +0000 2022A storm dumps 6-8 inches of rain in a narrow band north of the Missouri River, ending just east of St. Louis (ground station 24 hr precipitation in mm). 🔗
Tue Jul 26 13:14:54 +0000 2022The use of the alternate initiation site M32 is only observed in cell line data, particularly in HET-1A cells.
Tue Jul 26 12:49:03 +0000 2022Regardless of what you call it, this domain usually results in the subunit forming a homodimeric protein, as is the case for KEAP1.
Tue Jul 26 12:28:22 +0000 2022The alternate name "POZ" stands for "Pox virus and Zinc finger", echoes of proteins with this domain. To make the literature easier to read & comprehend, it is also sometimes referred to as a "BR-C/Ttk" or "ZiN" domain.😡
Tue Jul 26 12:27:12 +0000 2022Modification diagram for KEAP1:p, an intracellular subunit with 6 Kelch & 1 BTB/POZ domains. These 2 domain types are frequently found together. BTB is an acronym for "Broad-complex, Tramtrack, and Bric-à-brac", fruit fly proteins with this domain.
🔗
Mon Jul 25 20:29:28 +0000 2022A front of small, but intense, storms moving towards the Atlantic coast, stretching from New Orleans to the Bay of Fundy 🔗
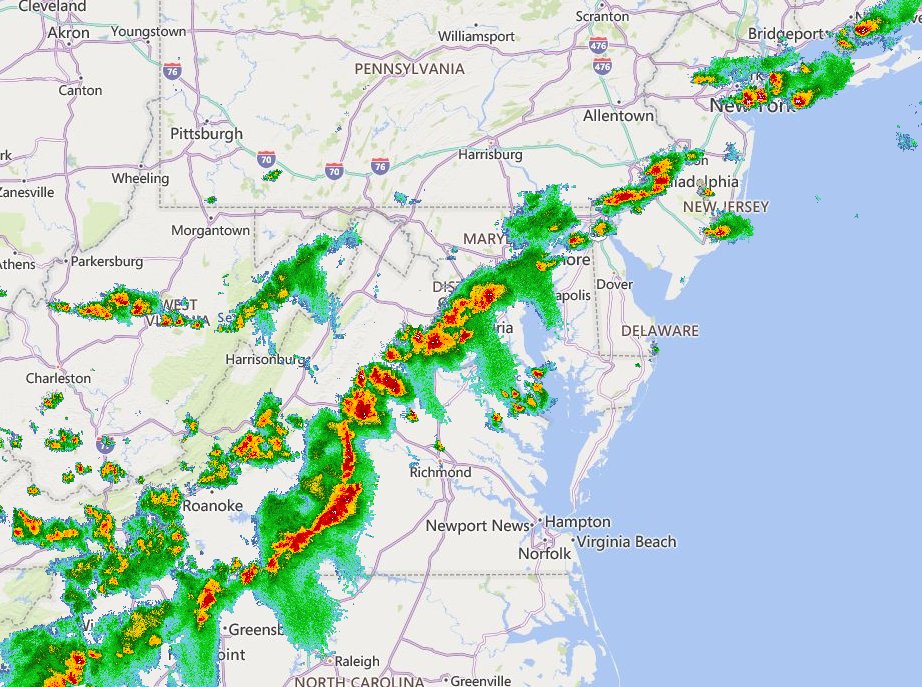
Mon Jul 25 19:53:27 +0000 2022Proteoglycan 4: a name well-earned
🔗
Mon Jul 25 19:20:58 +0000 2022@J_my_sci The problem is that the only sort of fusion reactor we know about requires you to back up about 150 million km before you turn it on.
Mon Jul 25 19:19:59 +0000 2022@J_my_sci Man made: not for quite a while unless someone has a great idea & we develop some materials we don't know how to make yet.
Natural: it is where almost all of our energy comes from, incl. fossil fuels, biomass & sunshine.
Mon Jul 25 17:51:41 +0000 2022@AlexUsherHESA Not a first for him, though, e.g.,
🔗
Mon Jul 25 17:35:09 +0000 2022"This helps explain why so much of waking life has been colonized by advertising" 🔗
Mon Jul 25 14:48:33 +0000 2022There should be some sort of international tribunal that calls the fruit-fly community to account for their irresponsible gene naming spree in the '80s & '90s.
Mon Jul 25 14:01:17 +0000 2022To Toronto-based media: everyone in Canada knows that Toronto has the largest population of any Canadian city. You do not have to preface every mention of it with this declaration. It make you sound desperate.
🔗
Mon Jul 25 11:59:14 +0000 2022Modification & N-linked glycosylation diagrams for human ATRN:p, a type I membrane protein with 6 kelch domains. Phosphorylation is uncommon in subunits with kelch domains, but this one has at least 13 N+glycosyl acceptors that may be occupied. TM domain (1279-1301). 🔗
Sun Jul 24 19:13:52 +0000 2022@statesdj Are they going to fold it in to Homeland Security?
Sun Jul 24 15:32:52 +0000 2022It does seem to be harder (or at least it takes longer) to get them back up & stable, though.
Sun Jul 24 14:36:15 +0000 2022One odd thing about cloud-based, high redundancy systems is that they seem to unavailable at about the same frequency as the old single-server systems they replaced.
Sun Jul 24 13:05:33 +0000 2022Discovering the Karl May phenomenon was probably the most surprising part of living in Germany, particularly having grown up in Winnipeg.
🔗 🔗
Sun Jul 24 12:28:20 +0000 2022SPP1:p is normally the main source of phosphorylated PSMs detectable in urine without affinity purification. It is also the most abundant source of phosphorylated PSMs in human milk―more than either alpha or beta casein.
Sun Jul 24 12:17:27 +0000 2022Modification diagrams for the 3 most commonly observed splice variants of human SPP1:p. These small phosphoproteins are abundant in some biological fluids (milk, urine, blood plasma, CSF, vitreous humor). 🔗
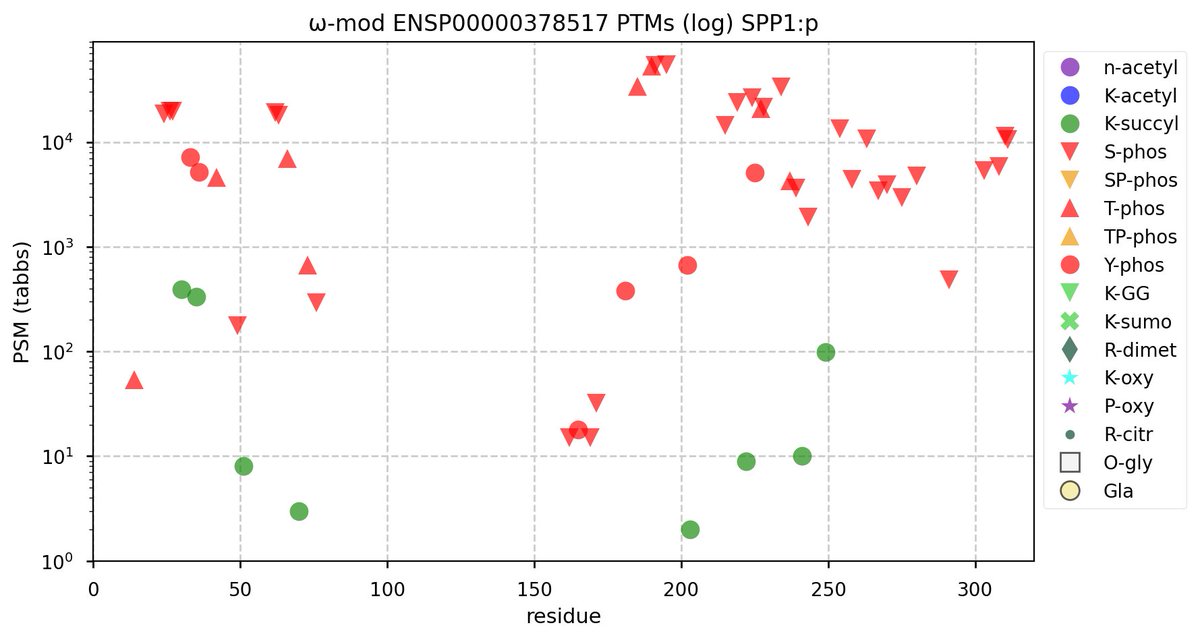
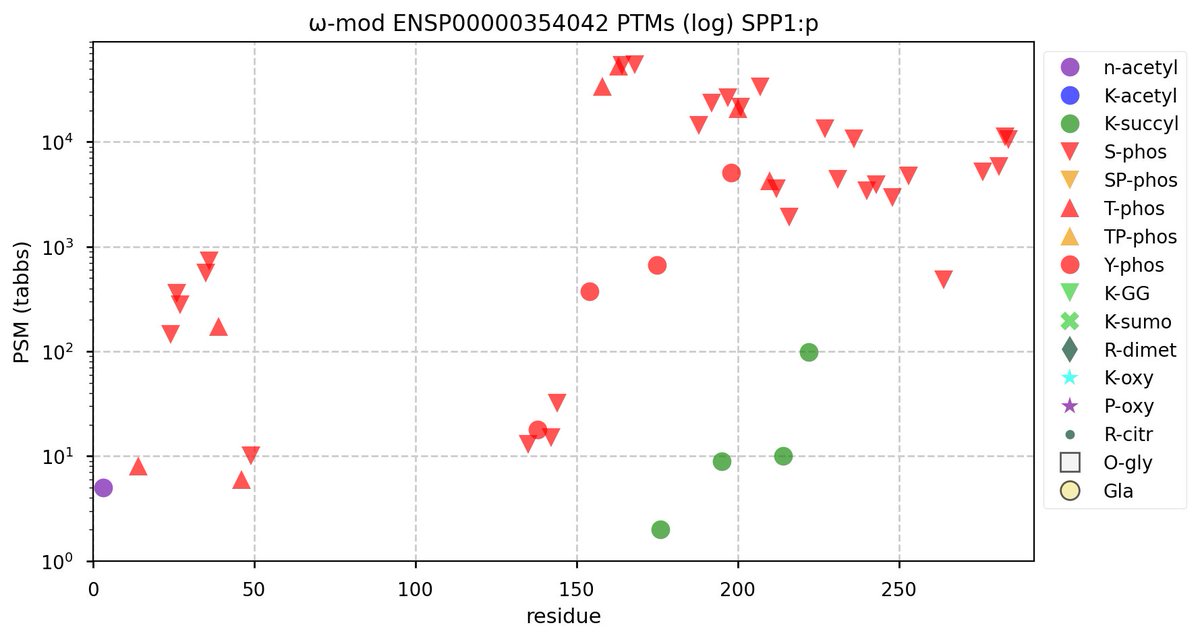
Sun Jul 24 11:34:02 +0000 2022Stormy weather traveling east across the lower Great Lakes region. 🔗
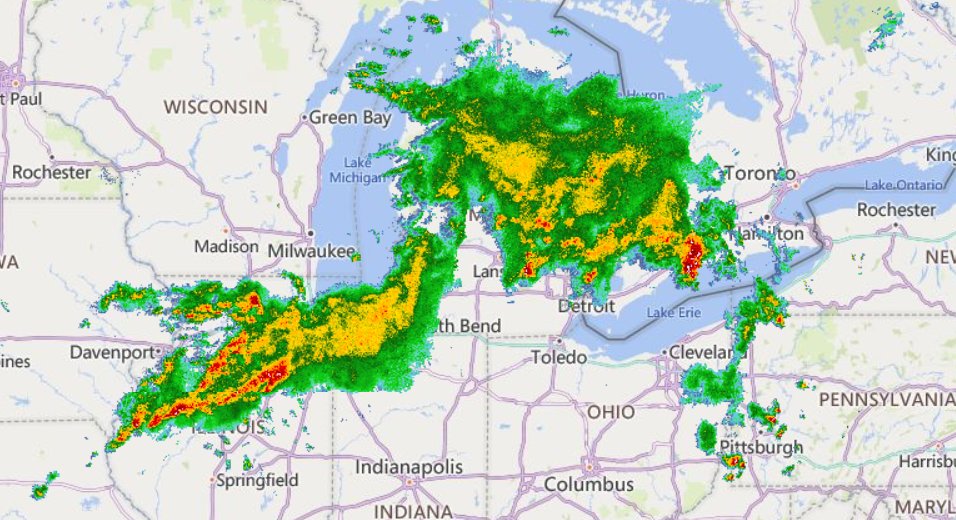
Sat Jul 23 17:21:40 +0000 2022@VATVSLPR @UCDProteomics @Zeczycki_Lab @leprevostfv Actually, it is my perception that informatics folks associated with proteomics MS lab groups that are the worst for getting lock-in bias towards small, non-representative data collections, usually from their own lab. Often the PI doesn't have the expertise to be of much help.
Sat Jul 23 16:35:21 +0000 2022The CBC news reader just referred to the lastest broadcast from the US Congress "January 6th" committee as its "season finale".
Sat Jul 23 16:10:02 +0000 2022@VATVSLPR @UCDProteomics @Zeczycki_Lab @leprevostfv You might be surprised how many informatics groups use a small, static selection of "representative" data sets for testing that is often left unchanged for years.
Sat Jul 23 15:06:01 +0000 2022A storm comes to Ohio. 🔗

Sat Jul 23 13:23:01 +0000 2022Like CSN1S1:p, the only real exception to the normal tissue distribution of CSN2:p is PXD020222. But that particular lab frequently generates exceptional tissue distributions, so it should be taken with a grain of salt (& maybe a shot of schnapps).
Sat Jul 23 12:03:10 +0000 2022Modification diagram for human CSN2:p, aka casein beta. Secreted into milk with a ragged N-terminus, resulting in three main mature forms (13,15,16-226). The human sequence does not share any tryptic peptides with the bovine sequence. Its biological function is as food. 🔗
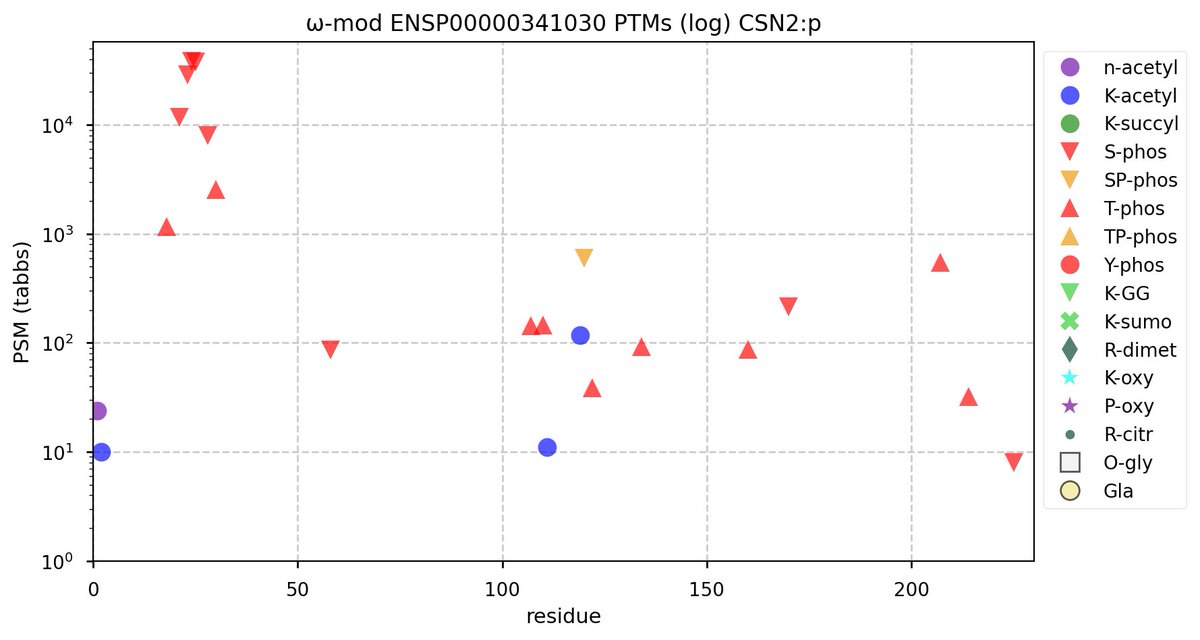
Fri Jul 22 17:54:45 +0000 2022@TanentzapfLab I think you mean to say "university professor" rather than "scientist".
Fri Jul 22 15:21:37 +0000 2022@sciencehacker That is correct.
Fri Jul 22 14:40:18 +0000 2022@ProtifiLlc PS. That particular data set also has strong signals from CSN2:p & LTF:p in it, so it is consistent with human milk being added, but again, don't know why.
Fri Jul 22 14:36:44 +0000 2022@VATVSLPR @UCDProteomics @Zeczycki_Lab @leprevostfv Amen, brother. Amen.
Fri Jul 22 14:23:01 +0000 2022@ProtifiLlc Human CSN1S1:p is only really found in milk, with the odd exception of PXD020222, where it is found in many affinity purification expts from A-549 cells. Don't know why.
Fri Jul 22 14:17:16 +0000 2022@ProtifiLlc Food biochemistry can be very complex (& interesting), but it is still meant to go in one end & out the other.
Fri Jul 22 14:12:37 +0000 2022@UCDProteomics @Zeczycki_Lab @leprevostfv I guess it is a consequence of the Highlander approach so commonly found in some schools of proteomics thought.
Fri Jul 22 13:07:10 +0000 2022@doctorow Ketchup. It is always some version of ketchup.
Fri Jul 22 11:56:55 +0000 2022@UCDProteomics @Zeczycki_Lab @leprevostfv It took a while for this to sink in with me. Did you mean that the software can get the hiccups if it finds identical sequences in the lists of proteins being used for identification?
Fri Jul 22 11:45:37 +0000 2022Modification diagram for human & bovine CSN1S1:p, aka casein alpha s1. Found in milk, although the bovine form can show up unexpectedly in the darndest places. The two sequences do not share any tryptic peptides. Its biological function is as food. 🔗
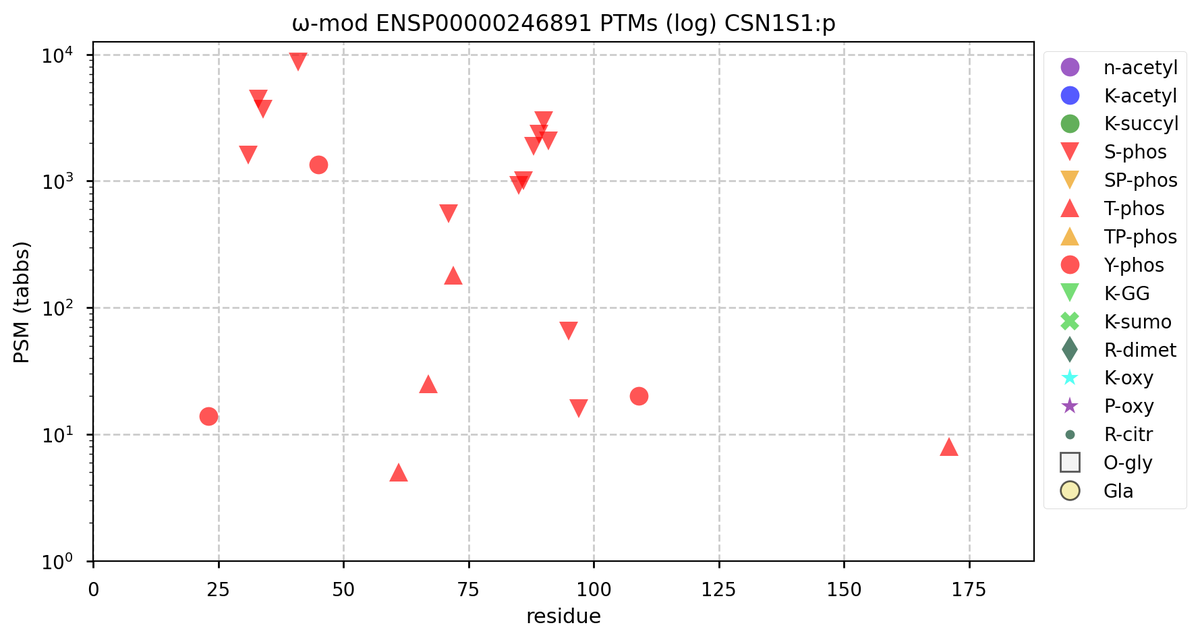
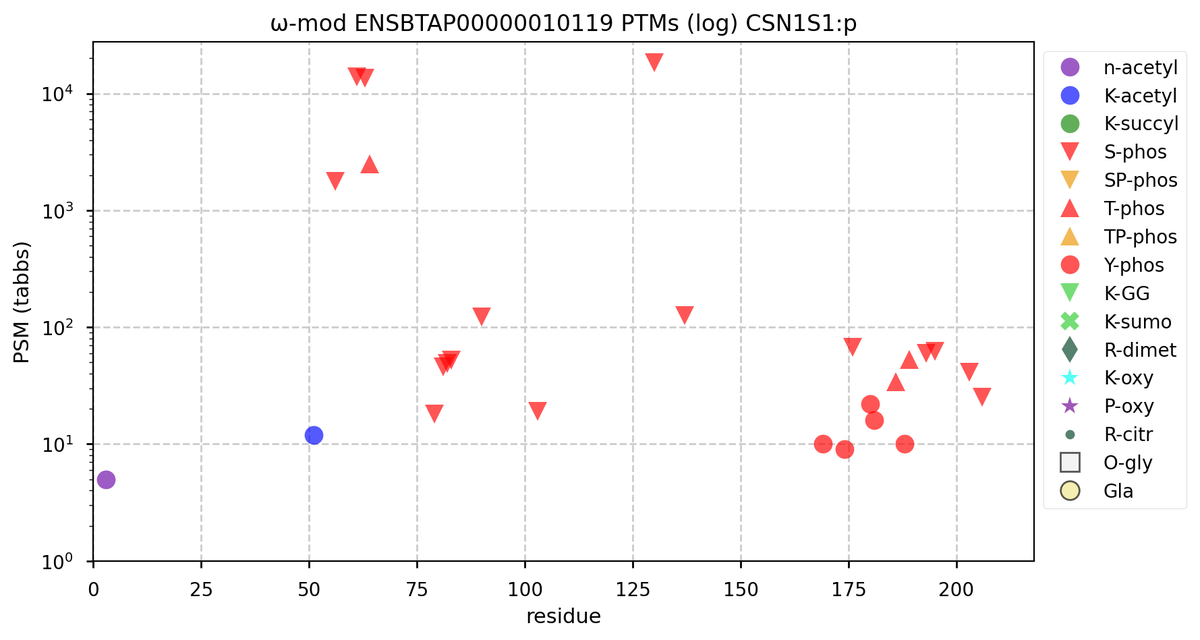
Fri Jul 22 02:34:09 +0000 2022And this one looks like its headed for Ann Arbor, Detroit & Toledo 🔗
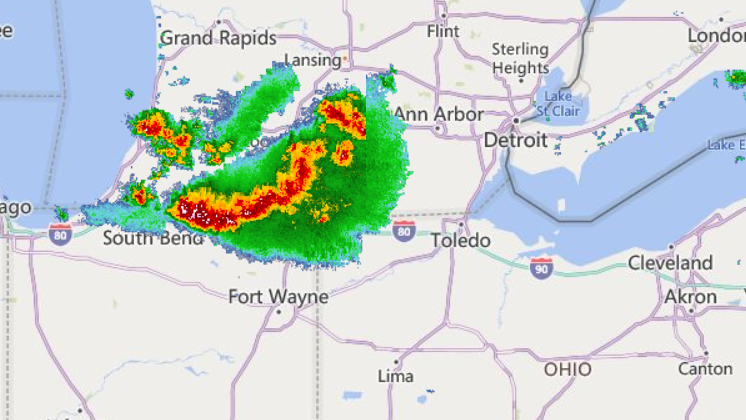
Fri Jul 22 02:29:21 +0000 2022Looks like this storm is headed for Mobile. 🔗
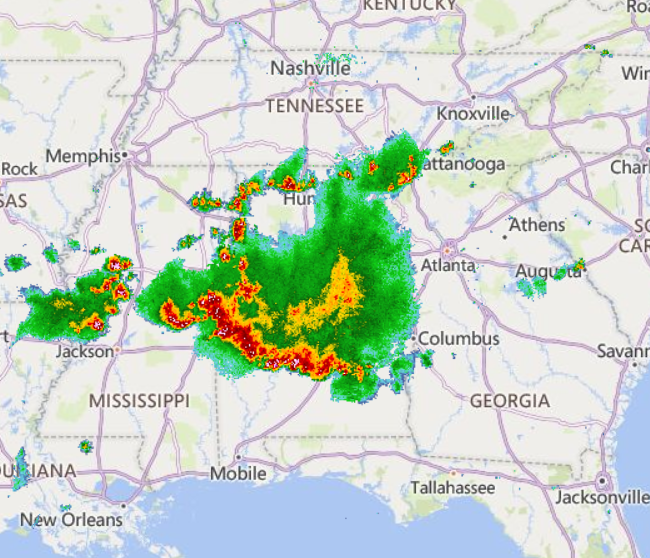
Fri Jul 22 02:17:20 +0000 2022The approved name for the human GART gene suggests there may have been some lack of consensus within the naming committee:
"phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase"
🔗
Thu Jul 21 23:28:53 +0000 2022@BestPixMN Do you really need to include Kansas?
Thu Jul 21 23:06:56 +0000 2022As is common in this sort of study, the class 1 expts show good evidence of small ORFs (such as 🔗), while the class 2 expts have none.
Thu Jul 21 19:35:00 +0000 2022It also contains quite a bit of information about IGHE:p, a rather elusive bit of sequence that is part of the final translated version of IgE.
Thu Jul 21 19:18:08 +0000 2022@savitski_lab @SinSri_1 @CuylenLab @wolfgangkhuber @pedrobeltrao @MBantscheff Nice work on a challenging problem.
Thu Jul 21 19:01:49 +0000 2022It also shows, yet again, that the MHC class 1 peptide presentation system seems to be obsessed with showing peptides from HERPUD1 & DERL1 (for some reason). Maybe ERAD is something the immune system likes to keep tabs on?
Thu Jul 21 18:21:18 +0000 2022PXD035324 has some excellent examples of how much fetal calf serum affects MHC class 2 peptide experiments, while it is undetectable in MHC type 1.
Thu Jul 21 14:57:32 +0000 2022They also tend to have more high frequency single amino acid variants than their much more conservative intracellular colleagues.
Thu Jul 21 14:54:41 +0000 2022Secreted transport proteins often have much greater species-to-species sequence variation than do intracellular proteins.
Thu Jul 21 14:33:24 +0000 2022I clearly need to rethink my Netflix subscription
🔗
Thu Jul 21 14:14:41 +0000 2022@pwilmarth @ItsBiniR @neely615 @UCDProteomics @MagnusPalmblad My Cassandra-status remains intact.
Thu Jul 21 14:09:58 +0000 2022Unremarkable summer weather across Western Europe, except maybe in southern Sweden (ground station temperatures) 🔗
Thu Jul 21 14:01:25 +0000 2022@stateofthecity Pretty much my take on Toronto ...
Thu Jul 21 13:03:13 +0000 2022@stateofthecity They are very pro-parkade.
Thu Jul 21 12:40:10 +0000 2022@peterfaull @leprevostfv Proteomics lab & informatics practitioners tend to be singularly disinterested in the details of how sequence lists are constructed.
Thu Jul 21 12:30:03 +0000 2022Sequence overlap diagrams for MGFE8:p comparing human on bovine (2 tryptic peptides) and mouse on bovine (1 tryptic peptide) & human on mouse (1 tryptic peptide). 🔗

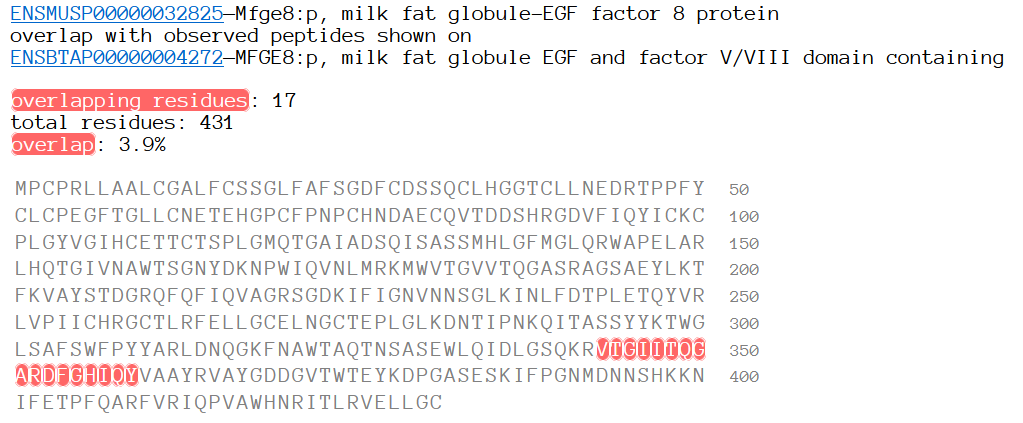
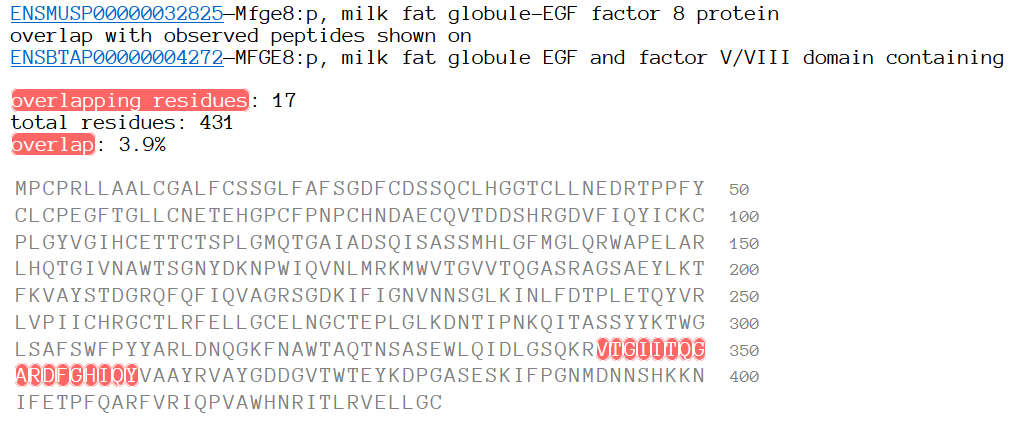
Thu Jul 21 12:12:23 +0000 2022Modification & N-linked glycosylation diagrams for human MGEF8:p, aka milk fat globule-EGF factor 8 protein. It is abundant in milk, but it is also abundant in heart tissue, urine, intervertebral discs & cell culture extracellular vesicles. 🔗
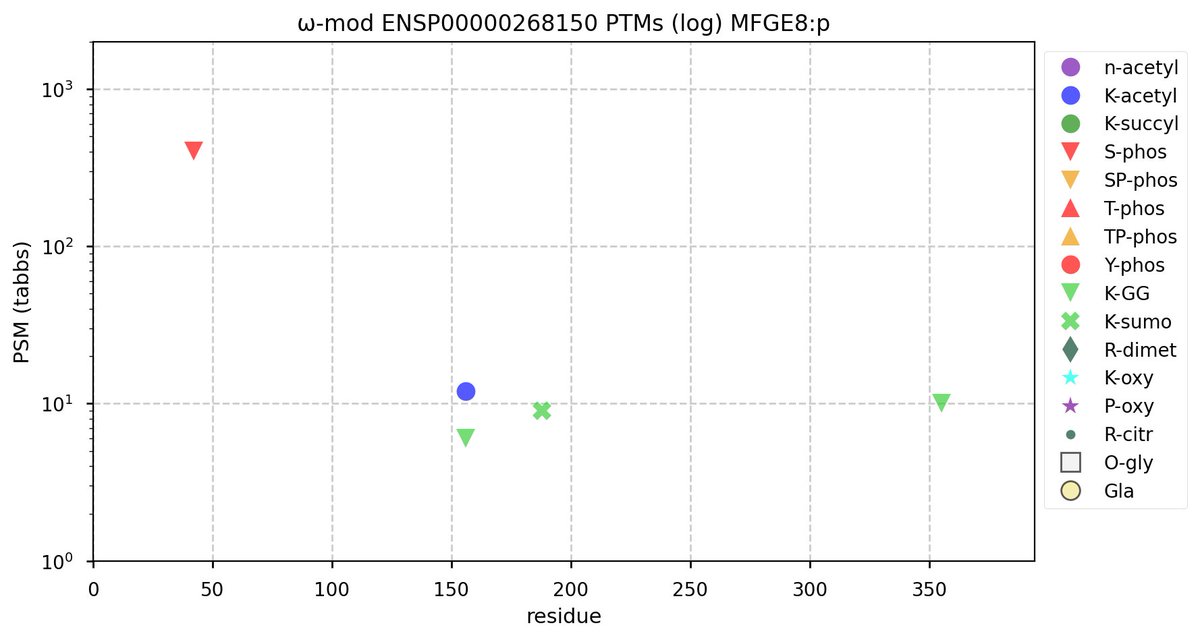
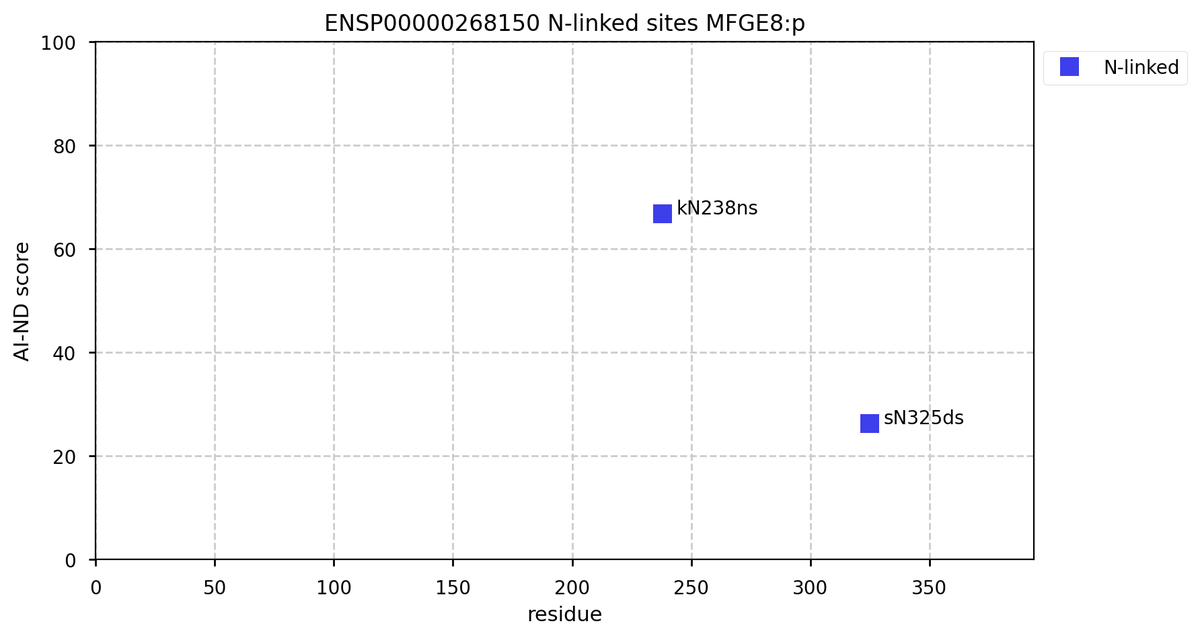
Thu Jul 21 00:05:55 +0000 2022@UCDProteomics @leprevostfv The name isn't one of my better ideas.
Wed Jul 20 21:13:49 +0000 2022@jke000 @leprevostfv If ftp is an issue, the info is available via HTTP using the same directory structure, 🔗
Wed Jul 20 21:07:56 +0000 2022@jke000 @leprevostfv ftp://ftp.thegpm.org/fasta/crap seems to still be there. What URL is causing a problem?
Wed Jul 20 15:32:41 +0000 2022@AlexUsherHESA @Wonkhe Amen, brother. Amen.
Wed Jul 20 15:25:59 +0000 2022Is there some way to disable the "More Tweets" section that trails the content you want when clicking to read a thread? For me, it is an annoying feature that leads to frequent internal-dialog-WTFs.
Wed Jul 20 14:37:25 +0000 2022One thing I've noticed about people who run mass spec facilities is that they become so invested in the repeated, strident arguments they must make to obtain funding for an instrument that they imbue the instruments they obtain with all sorts of almost mystical properties.
Wed Jul 20 14:02:51 +0000 2022The only way FAT1:p's pattern makes sense is if there is a significant amount of an intracellular form of the protein, if only for recycling. FAT2:p is common in CSF, while FAT1:p is rarely found in fluids.
Wed Jul 20 13:01:24 +0000 2022@neely615 @UCDProteomics @MagnusPalmblad Only a very few "old" proteins had tryptic peptides that spanned prokaryotes-to-vertebrates.
Wed Jul 20 13:01:11 +0000 2022@neely615 @UCDProteomics @MagnusPalmblad Back in olden times (the '00s, when I was enthusiastic about using spectral libraries for peptide ID) I did some detailed studies comparing how libraries from 1 species could be used to leverage others.
Wed Jul 20 12:46:02 +0000 2022@neely615 @UCDProteomics @MagnusPalmblad Human & yeast share almost no tryptic peptides. Human & mouse share 50% of their tryptic peptides.
Wed Jul 20 12:41:30 +0000 2022The same diagrams for human FAT1:p, a very large protein with a domain structure similar to FAT2:p including a single TM domain (4179-4201). While the PTMs for FAT2:p are easy to rationalize, only FAT1:p's N+glycosylation makes sense. 🔗
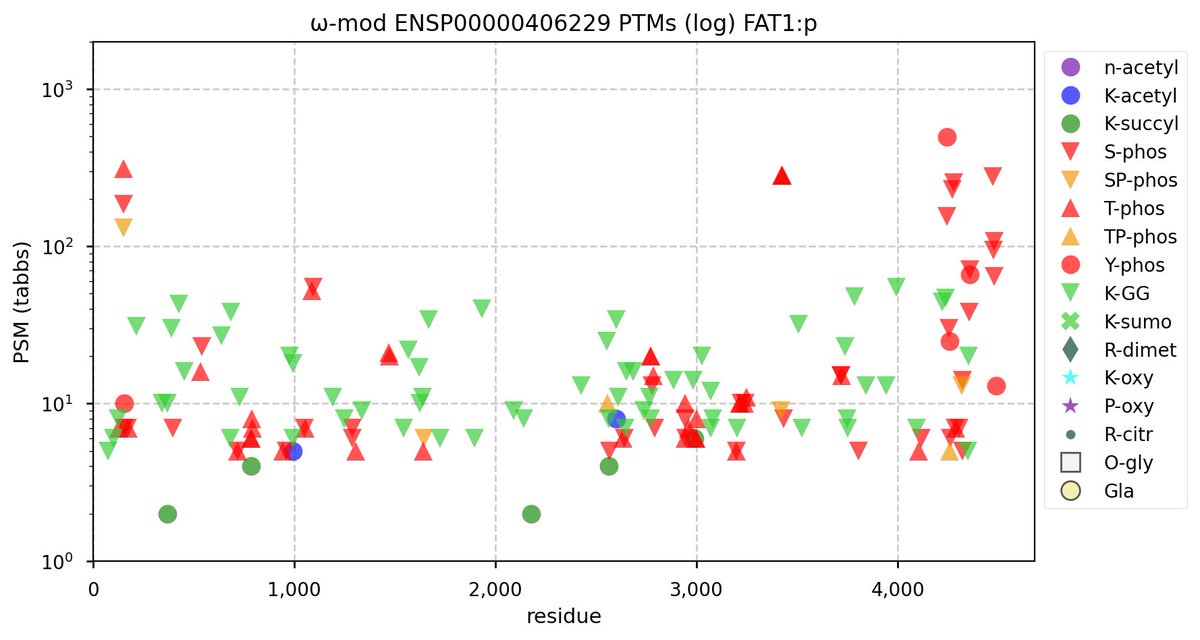
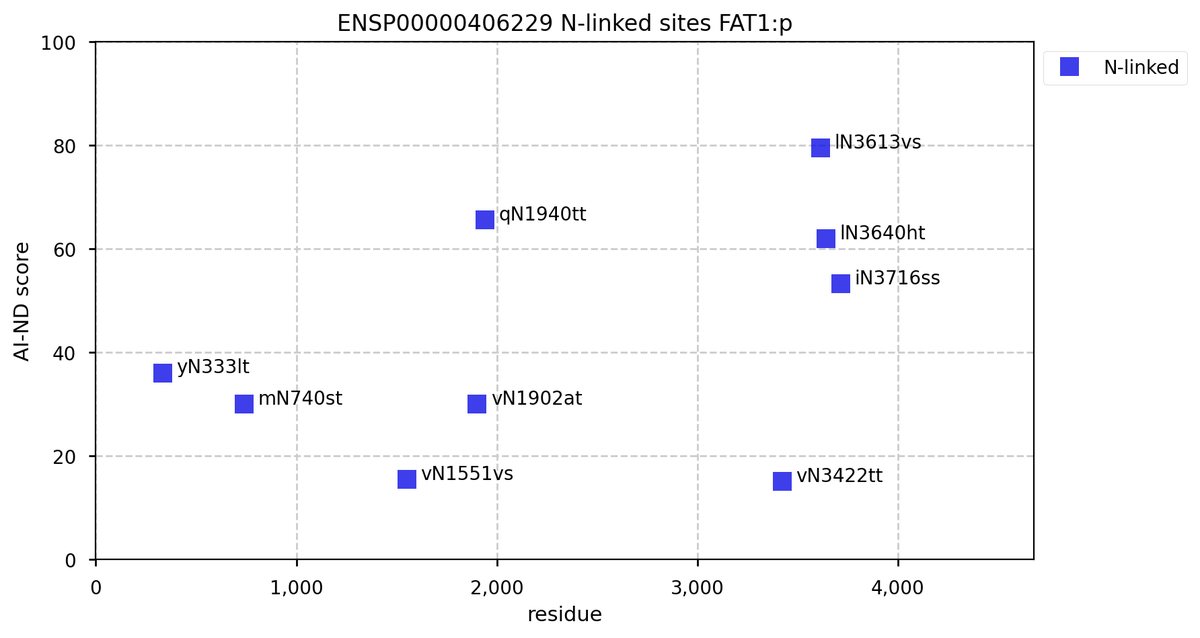
Wed Jul 20 12:14:10 +0000 2022Modification & N-linked glycosylation diagrams for human FAT2:p (aka MEGF1:p), a very large cell surface membrane protein with a multiple EGF & cadherin-like domains. There is a single transmembrane domain (4049-4071). The phosphodomain (4090-4343) is intracellular. 🔗
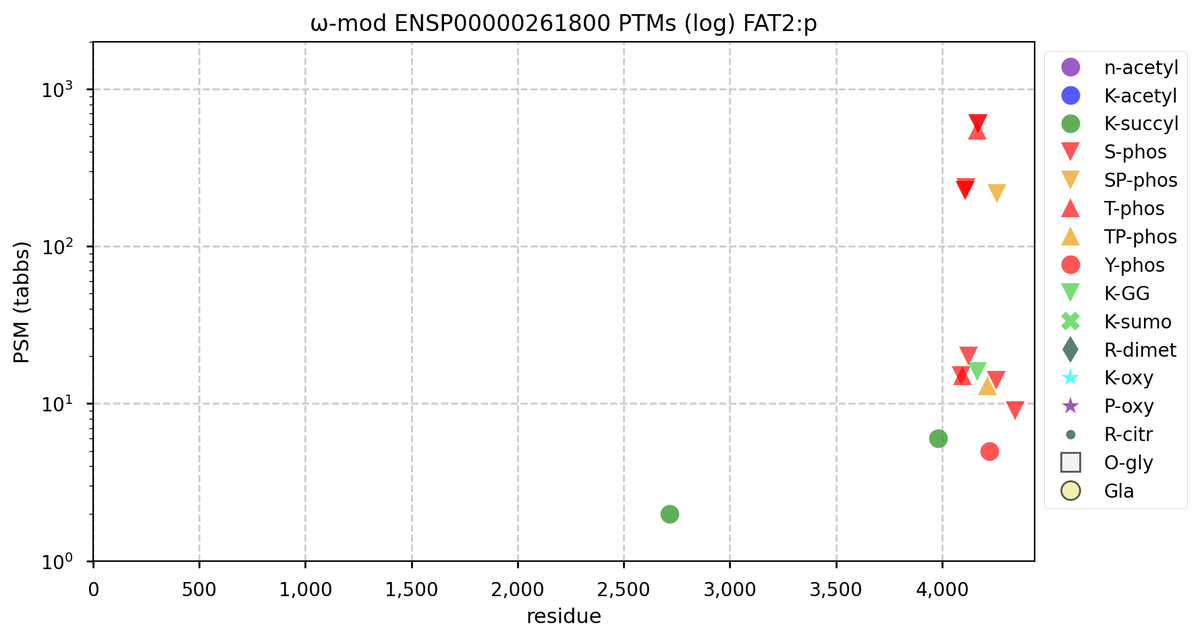
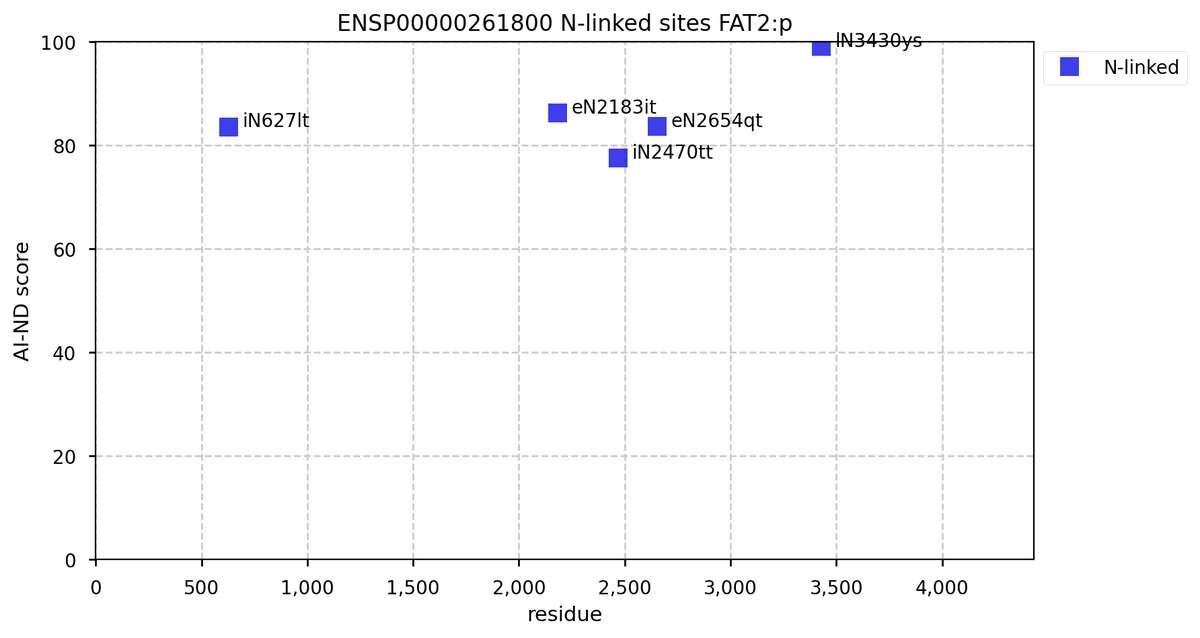
Wed Jul 20 11:54:01 +0000 2022The low pressure system that caused high temperatures in Western Europe has come ashore & is no longer bringing up hot air from North Africa (winds at 500 hPa). 🔗
Tue Jul 19 20:53:20 +0000 2022@nesvilab @UCDProteomics It is something I do all of the time. You would be surprised how often human (& bovine) proteins show up in samples you wouldn't expect to have them. Too many primates (with bags of cow's blood) around the lab!
Tue Jul 19 20:21:20 +0000 2022@camtflower If I could, I would. Nasty little poorly-designed protein in arguably the least robust pathways.
Tue Jul 19 16:20:17 +0000 2022@dtabb73 @JohnRYatesIII You should really judge software with the same standards as you would an ink jet printer: low expectations & suppressing the urge to apply a baseball bat.
Tue Jul 19 15:40:08 +0000 2022@RolandMBruderer 🔗
Tue Jul 19 13:58:20 +0000 2022Actually, any of the older phosphoprotein gene symbols that are "p" followed by a molecular weight in kiloDaltons (p42, p45, p58, p63, p105, etc) should be avoided.
Tue Jul 19 13:22:30 +0000 2022@chrashwood There is no corresponding reduction in the use of Orbitrap-based instruments, just a falloff in the proportion of studies that use isobaric quant reagents. Thermo is still king wrt the generation of public datasets.
Tue Jul 19 13:08:15 +0000 2022"p62" is a synonym for the following human proteins:
SQSTM1,
KHDRBS1,
NUP62,
GTF2H1,
DCTN4, &
IGF2BP2.
Try not to use "p62" in manuscripts: it may be traditional in your field, but it is very confusing for the unwashed masses.
Tue Jul 19 12:19:46 +0000 2022As this low moves closer to land, it has generated a line of rain storms across Cornwall, Wales and the west coast of Ireland. 🔗
Tue Jul 19 12:10:17 +0000 2022Modification & N-linked glycosylation diagrams for human MEGF8:p, a multiple EGF-domain containing protein. A largely extracellular glycoprotein, with a single transmembrane domain (2648-2670). Common in urine, CSF & blood plasma, particularly in extracellular vesicle fractions. 🔗
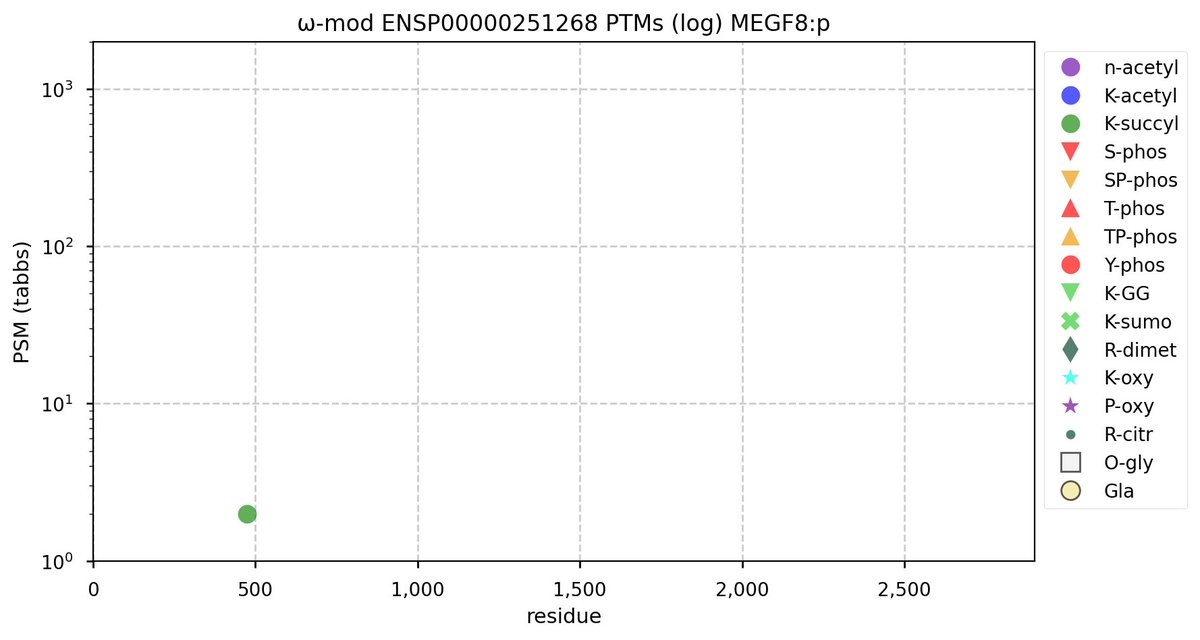
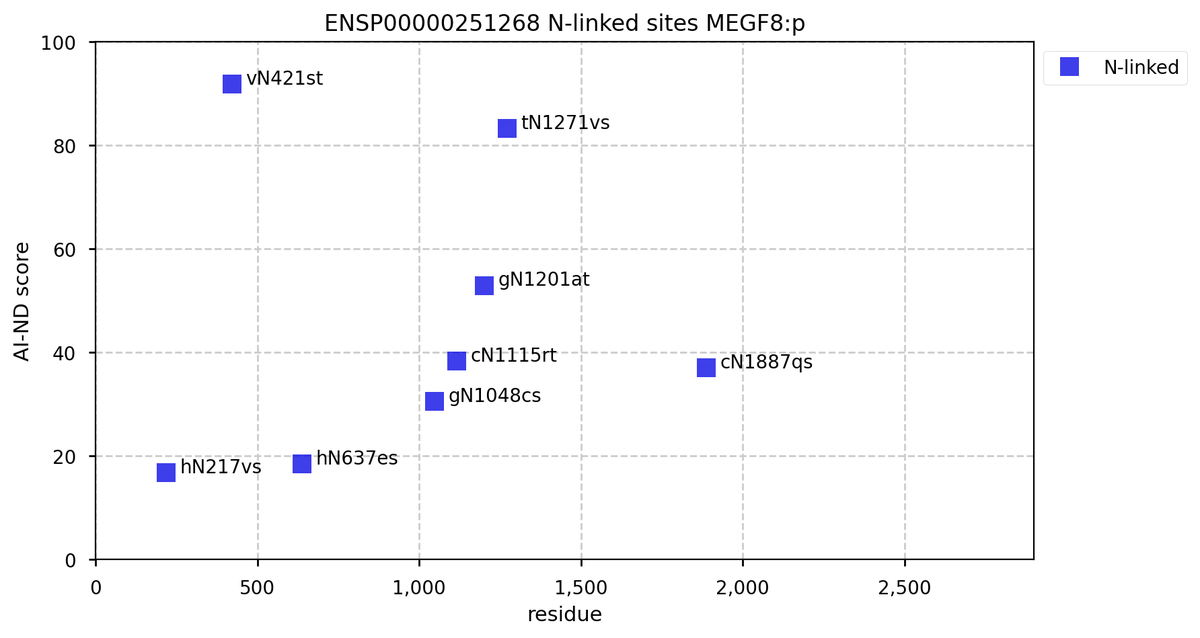
Tue Jul 19 11:17:02 +0000 2022The low pressure system causing high temperatures in Western Europe has moved northeast & is current centred in the Bay of Biscay (winds at 500 hPa). 🔗
Mon Jul 18 17:45:04 +0000 2022Someone from the RAF should have a word with their paving contractor about using more suitable materials.
Mon Jul 18 17:31:23 +0000 2022Same period of time, Denver International Airport 🔗
Mon Jul 18 17:24:17 +0000 2022🔗
Mon Jul 18 17:23:46 +0000 2022Temperature & dewpoint measurements for the Brize-Norton RAF airfield, just west of Oxford. 🔗
Mon Jul 18 14:17:56 +0000 2022ftp://ftp.pride.ebi.ac.uk has been down since yesterday afternoon.
Mon Jul 18 14:11:37 +0000 2022The sun is coming up on Hurricane Estelle, churning its way west-southwest through the Pacfic. 🔗
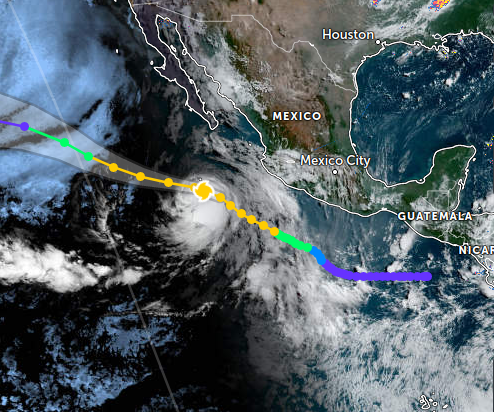
Mon Jul 18 12:13:42 +0000 2022Modification diagrams for human STRN:p, STRN3:p & STRN4:p (PPP2R6A/B/C:p), members of the protein phosphatase 2 regulatory subunit family 6. They have an unmodified C-terminal WD40-repeat domain (~300 residues) & phosphodomains in the intrinsically disordered N-terminal domain. 🔗
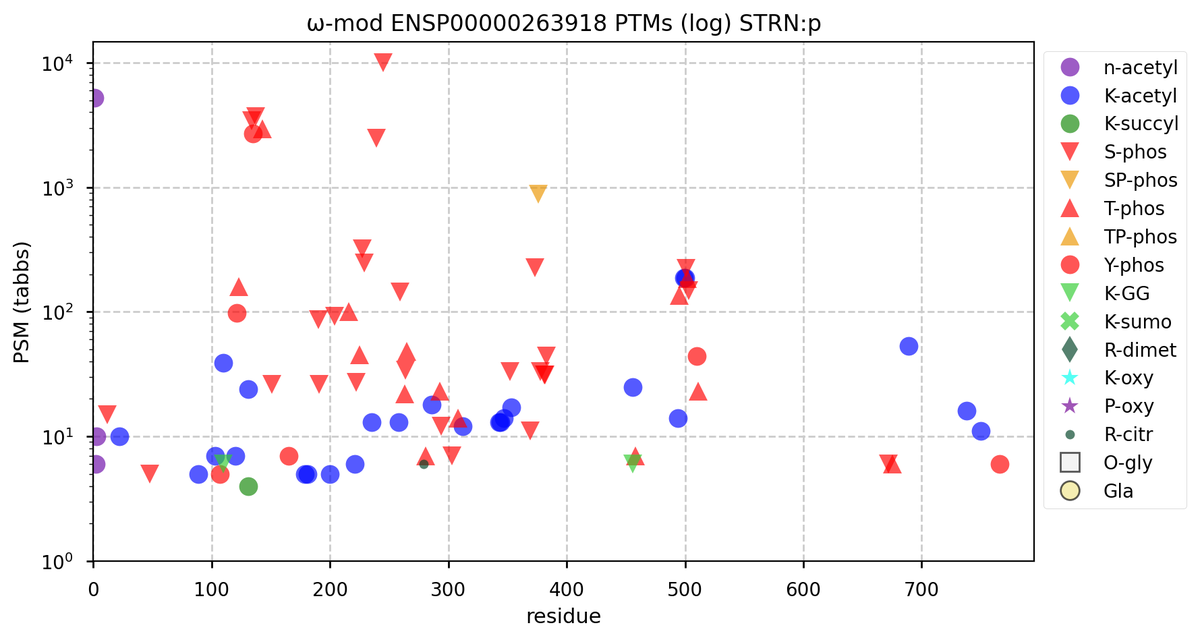
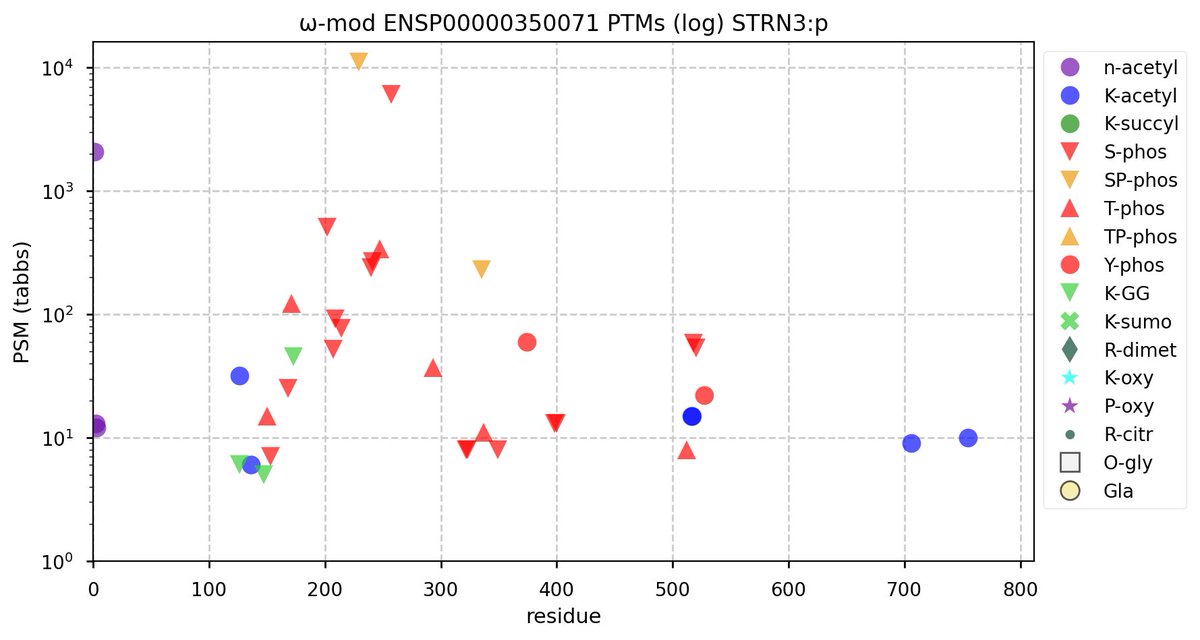
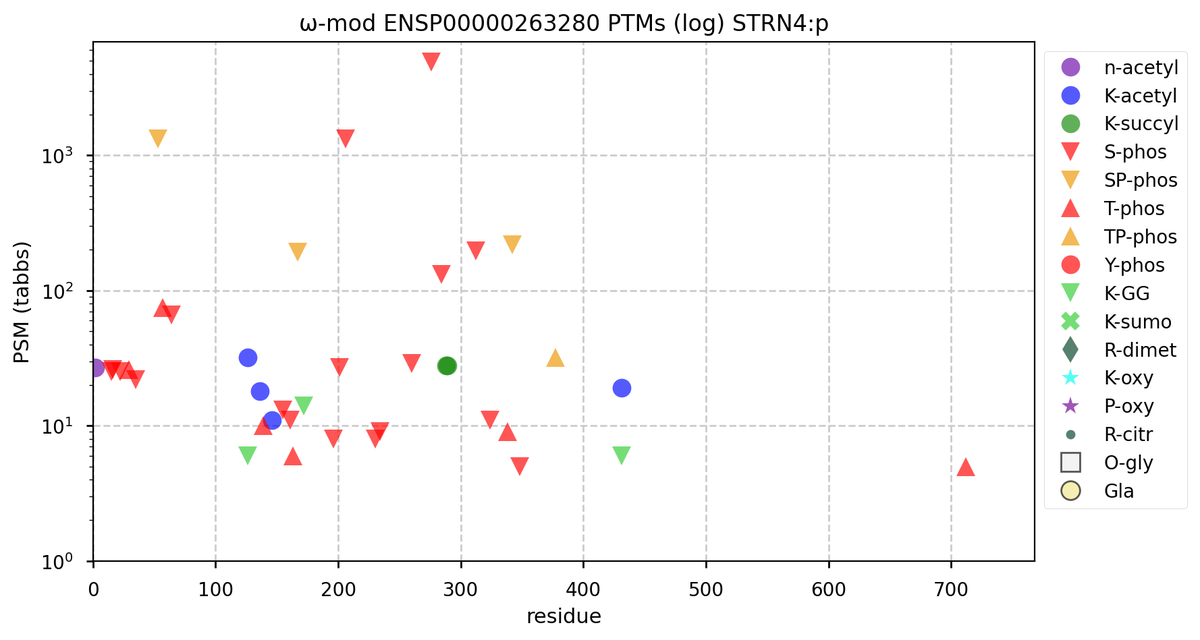
Mon Jul 18 11:48:36 +0000 2022The low pressure system causing unusually warm weather across Western Europe is slowly moving towards the Portuguese coast (winds at 500 hPa). 🔗
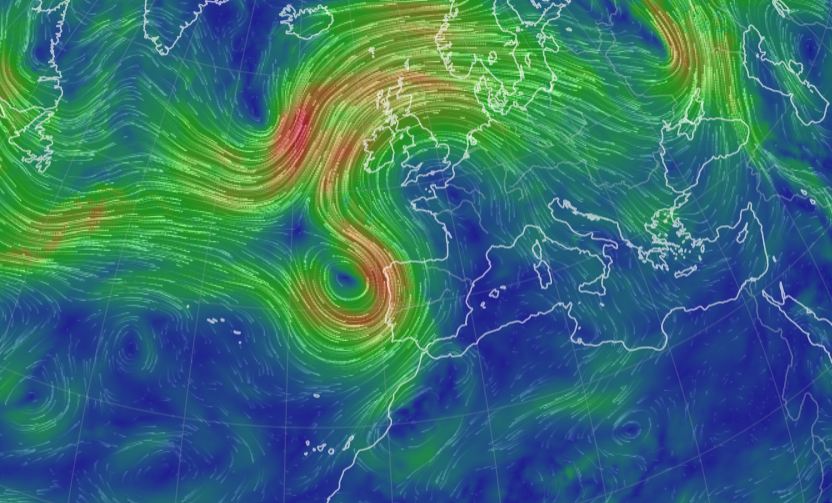
Sun Jul 17 22:38:47 +0000 2022Lots of good points wrt use of quant reagent-based methods. 🔗
Sun Jul 17 20:01:49 +0000 2022Unsettled weather this afternoon across the eastern US 🔗

Sun Jul 17 15:45:13 +0000 2022Any theories? I suspect that DIA may have made underivatized quant more respectable, but it may be something else entirely.
Sun Jul 17 15:22:41 +0000 2022PXD031078 has some pretty nice timsTOF Staphylococcus aureus data.
Sun Jul 17 15:19:42 +0000 2022Warming up the UK (ground station temperatures) 🔗
Sun Jul 17 15:07:38 +0000 2022Since their introduction, the trend in public data sets was a steady increase the the fraction of data sets that used peptide quantitation reagents (e.g., iTRAQ or TMT). For the last year, that trend seems to have reversed. I do not know why, though.
Sun Jul 17 12:37:48 +0000 2022Low pressure system stalled west of Portugal, isolated from high level steering winds, dragging air up from Africa into western Europe (winds at 250 hPa) 🔗
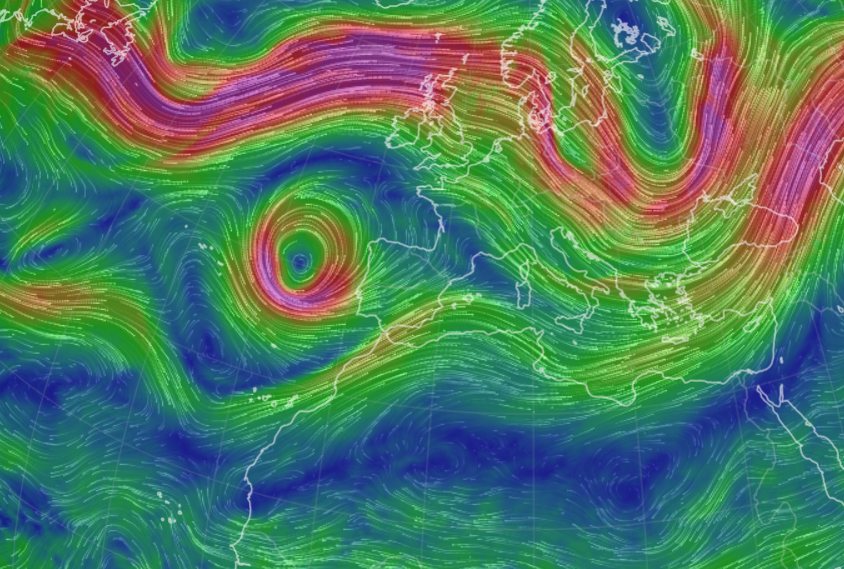
Sun Jul 17 12:05:31 +0000 2022PPP2R5E:p, for completeness. 🔗
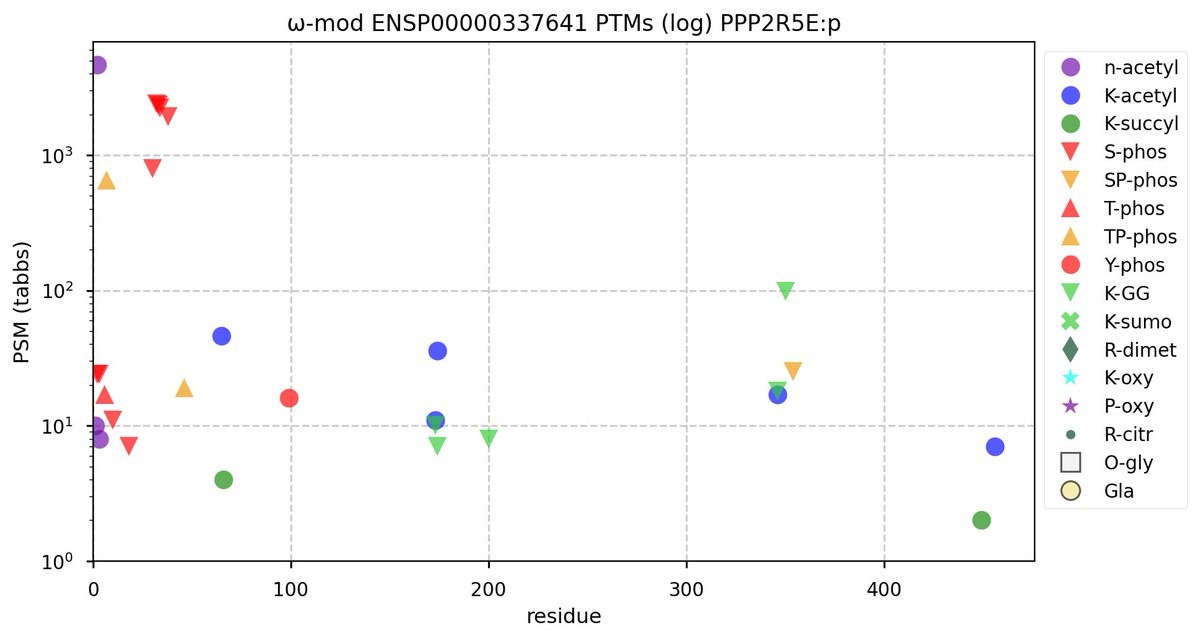
Sun Jul 17 12:04:40 +0000 2022Modification diagrams for PPP2R5A/B/C/D:p, four members of the protein phosphatase 2 subunit family 5. All members of the family have a central Armadillo-type domain (~400 residues) with a fringe of phosphodomains on either end. 🔗
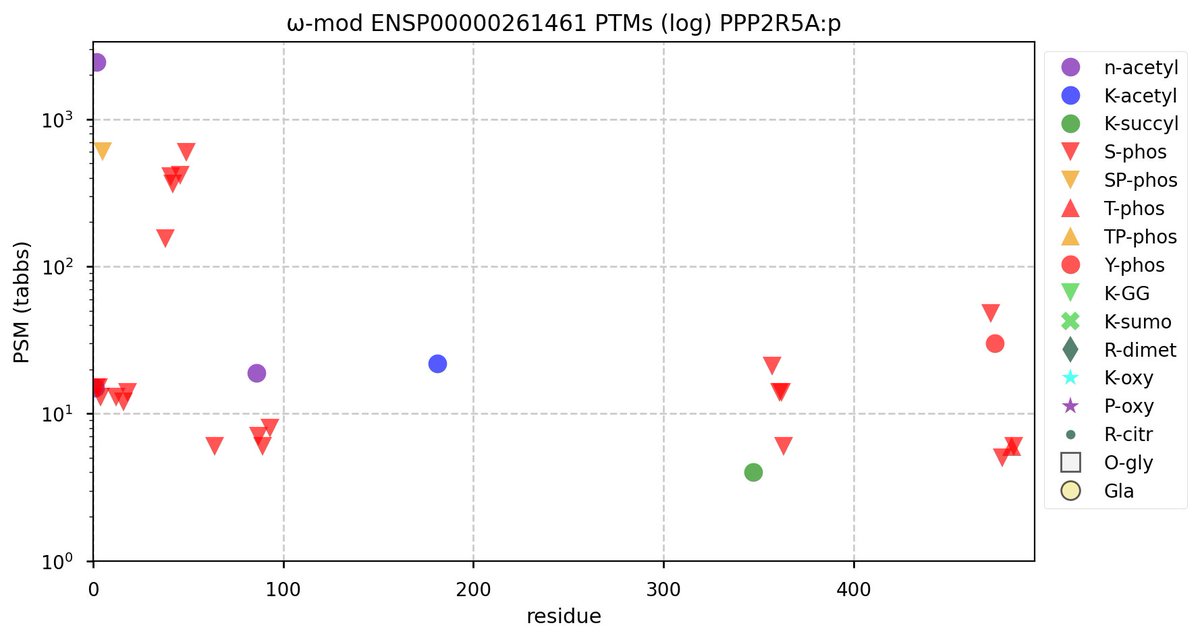
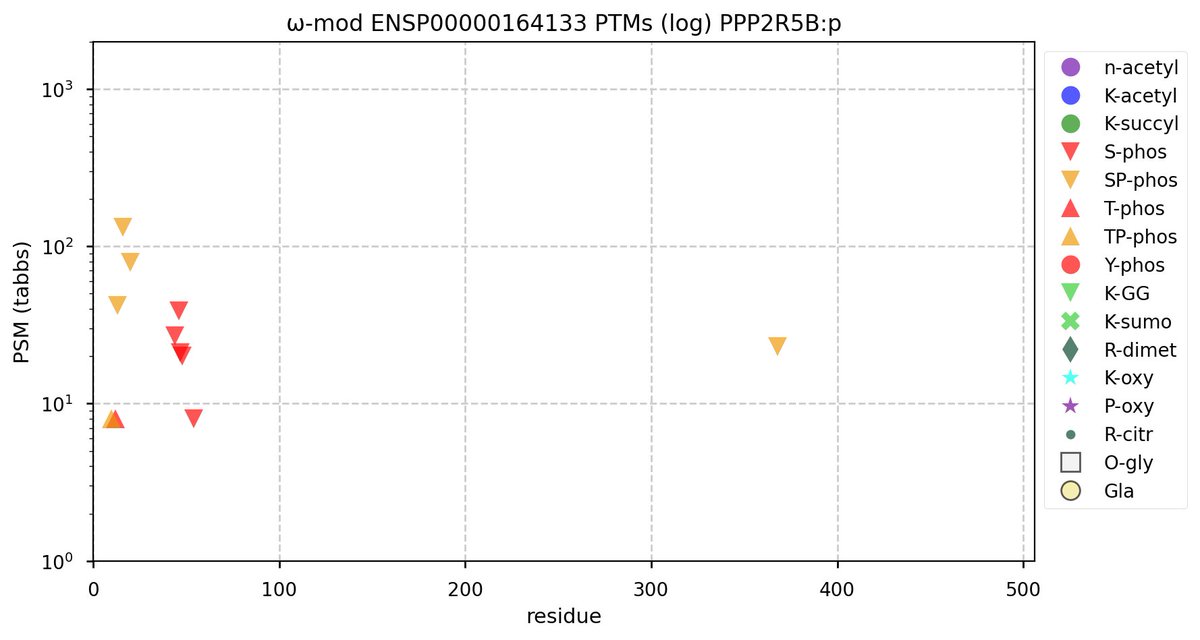
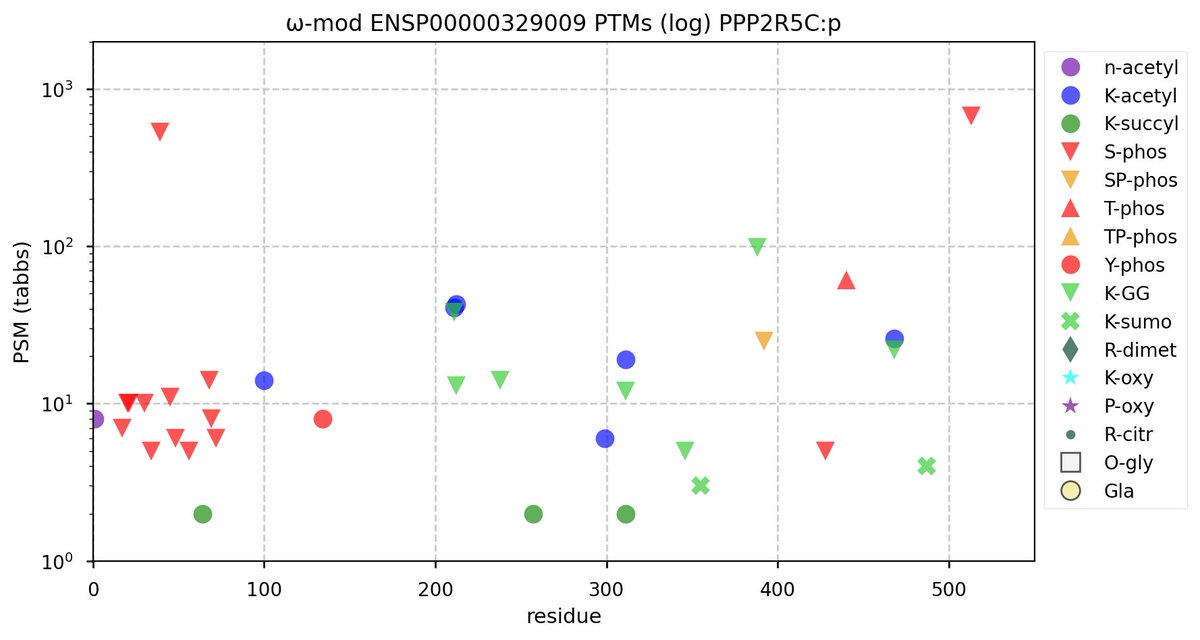
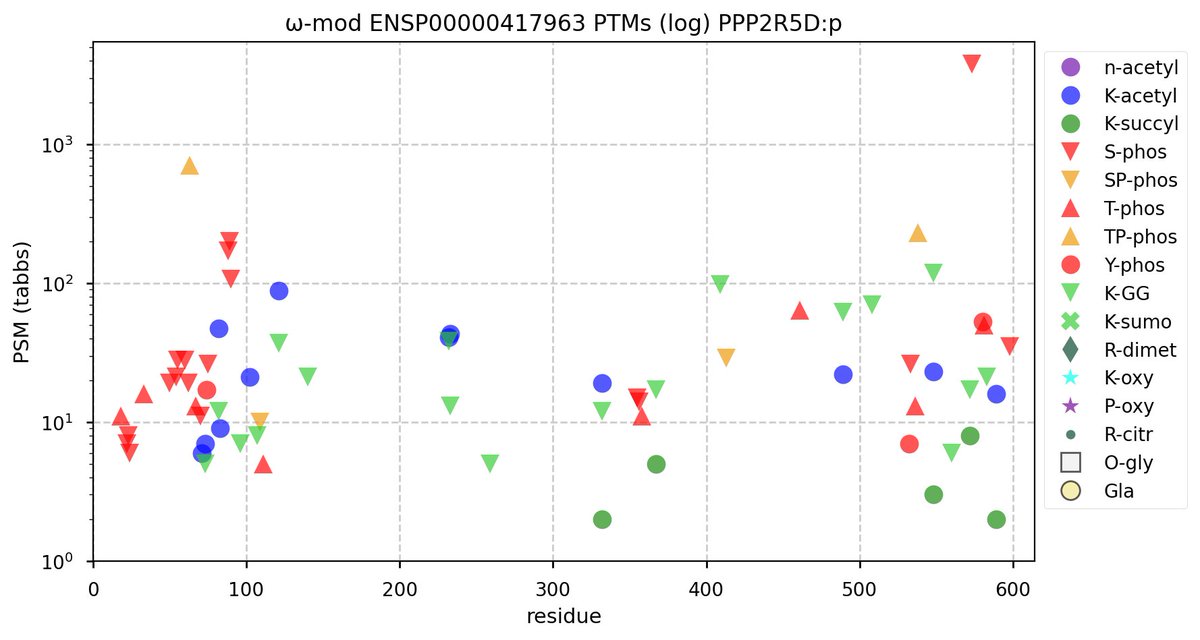
Sat Jul 16 21:48:10 +0000 2022A low pressure system pumping warm air from off North Africa up the Spanish coast towards the south of England (winds at 500 hPa) 🔗
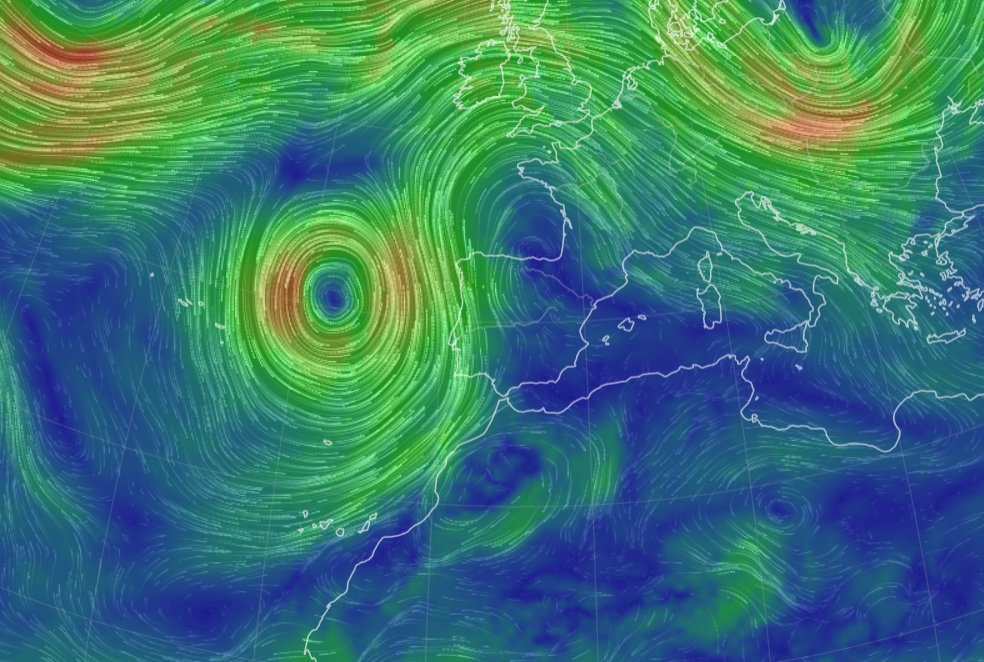
Sat Jul 16 19:46:02 +0000 2022Although sometimes it concerns me that I know people who would want to know what mouse plasma looks like ...
Sat Jul 16 19:40:15 +0000 2022If you want to look for interesting features in mouse blood plasma, PXD030303 is a good place to start.
Sat Jul 16 16:18:24 +0000 2022It would fix a lot of the problems with the current system.
Sat Jul 16 16:12:16 +0000 2022IMHO, all funded federal grant documents should be made public, 24 months after the start date of the grant.
Sat Jul 16 14:00:01 +0000 2022A warm few days in London, UK according to the GEM-GLB forecast model. 🔗
Sat Jul 16 13:05:43 +0000 2022I have seen some speculation that acetyl is used to block a K from ubiquitination, but the K+acetyl stoichiometry doesn't look right for that idea in most cases.
Sat Jul 16 13:02:22 +0000 2022Does observing, for example, a particular K+GGyl/acetyl mean something different than another K+GGyl only. Or observing a K+GGyl/SUMOyl mean something different than a K+SUMOyl only?
Sat Jul 16 12:58:27 +0000 2022I've probably asked this before, but has anyone ever tried to understand why some K residue can act as acceptors for either GGyl, acetyl or SUMOyl, while others are limited to 1 or 2 of those options?
Sat Jul 16 12:21:51 +0000 2022Modification diagram for human PTPA:p (aka PPP2R4:p), the only member of protein phosphatase 2 regulatory subunit family 4. Continues the low stress, calm altitude of most PPP2R families, with a selection of low occupancy K+GGyl/acetyl acceptors. 🔗
Sat Jul 16 01:07:55 +0000 2022Anything to do with printers tend to bring out "Office Space"-with-"Still"-playing-in-the-background energy in me.
Fri Jul 15 21:53:21 +0000 2022@sabbatiello0310 @Peptidome Chromatographers like to blame proline for everything from odd peak shapes to the Kennedy Assassination. While I'm sure it may have some role, 55-60% of all PSMs have at least one "P", so its presence alone cannot explain this relatively rare effect.
Fri Jul 15 15:14:05 +0000 2022PM10 particulates across Asia (dark red = highest concentration) 🔗
Fri Jul 15 14:52:34 +0000 2022@cstross The Sweeney!
Fri Jul 15 14:48:36 +0000 2022My new favorite example of a protein with N-linked glycosylation.
🔗
Fri Jul 15 14:24:10 +0000 2022The future
🔗
Fri Jul 15 12:36:03 +0000 2022Which of these Nature-branded journals do you consider the highest profile?
Fri Jul 15 12:31:18 +0000 2022Peak Thunder Bay.
🔗
Fri Jul 15 12:00:10 +0000 2022A storm moving out of Wisconsin & Iowa towards the east, into the other Upper Midwest states. 🔗
Fri Jul 15 11:53:00 +0000 2022Modification diagrams for PPP2R3A/B/C:p, members of the protein phosphatase 2 regulatory subunit family 3. One of these things is not like the others. 🔗
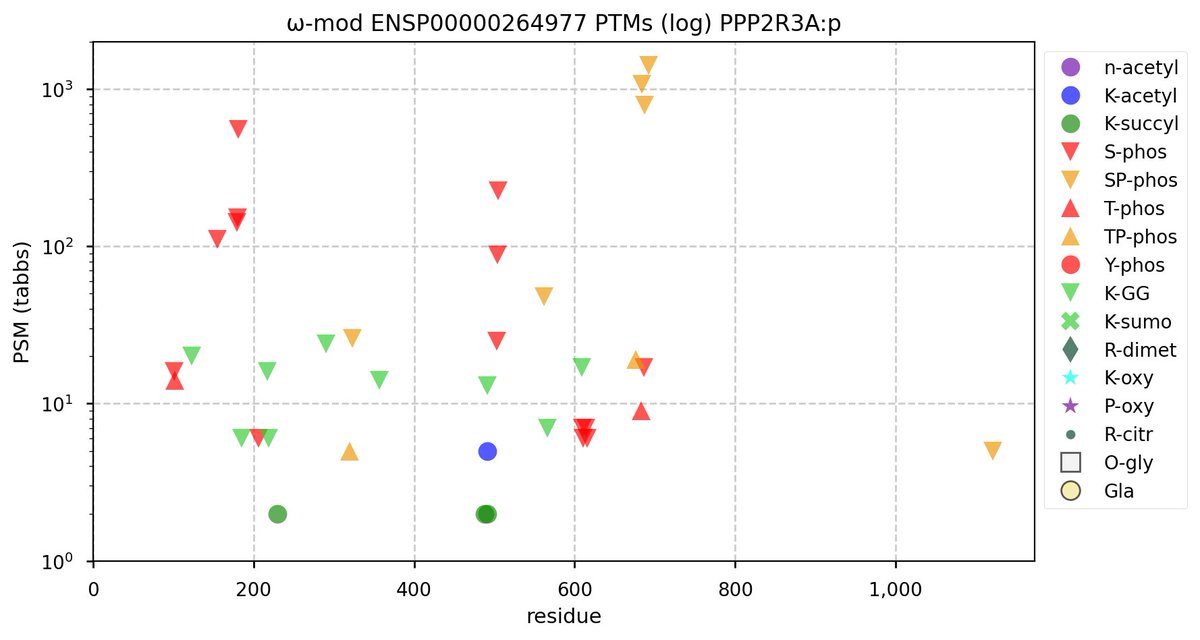
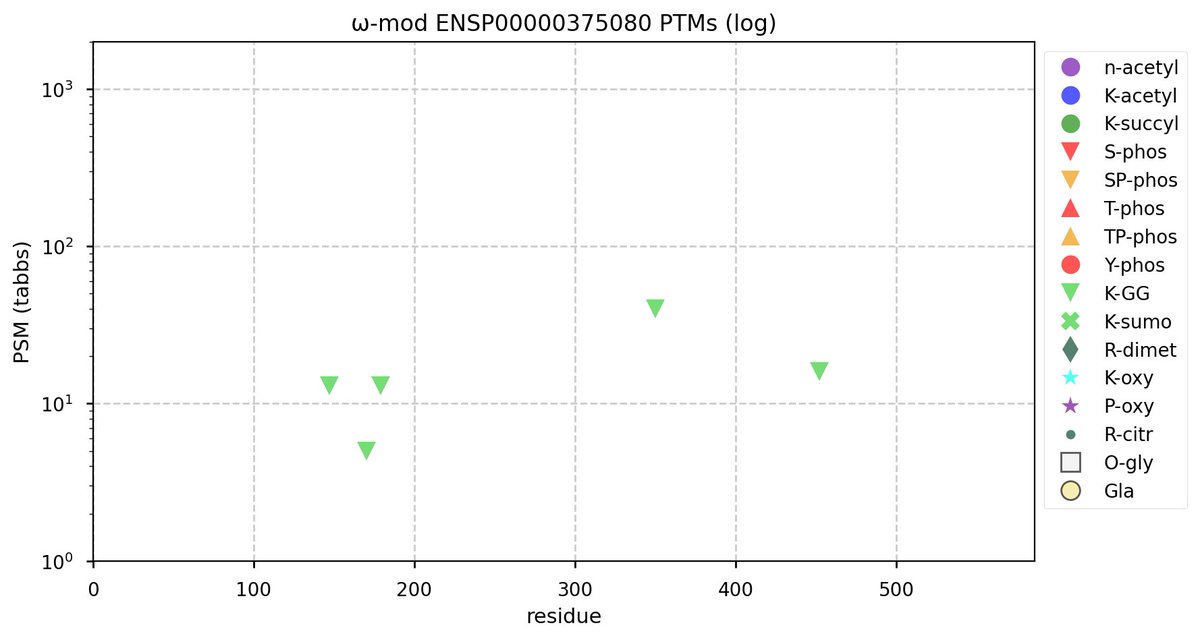
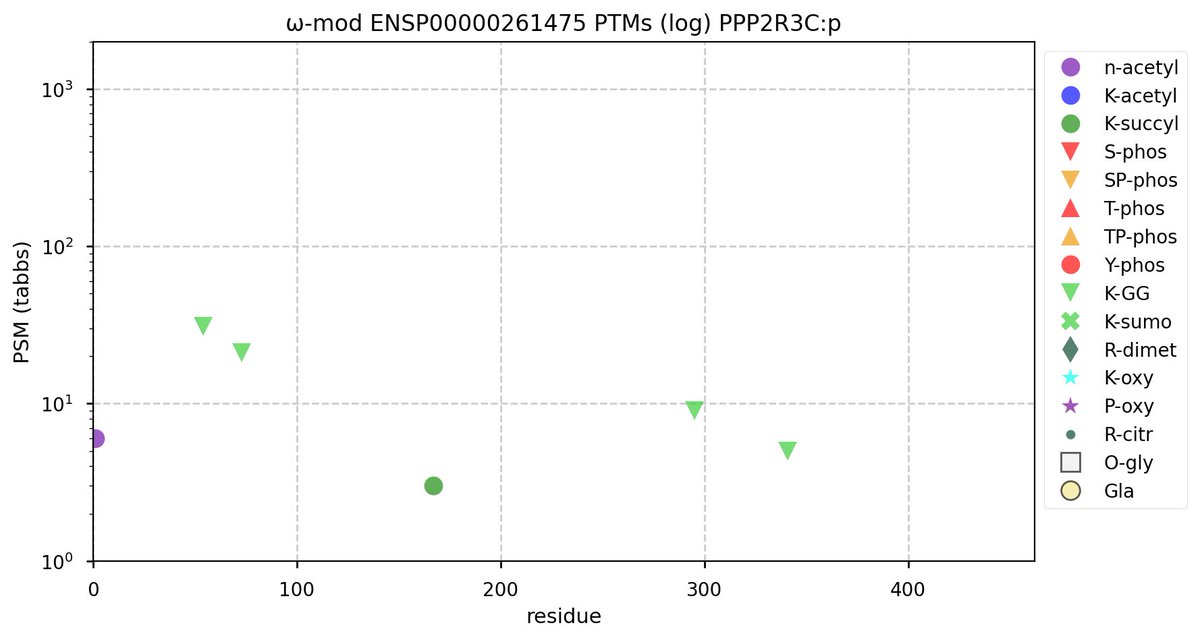
Fri Jul 15 01:12:57 +0000 2022PXD033413 has some nice high resolution data from mitochondria preparations obtained using human retinal pigment epithelium samples.
Thu Jul 14 23:01:25 +0000 2022@Peptidome Some SMEAR-effect peptides:
LLVVYPWTQR
LFETVEELSSPLTAHVTGR
YCGKPYTYAYGLGLNHFVPDR
LFETVEELSSPLTAHVTGR
IPLWLTGSLLR
FFSYFR
QAFEFPQINYQK
IWHHTFYNELR
Thu Jul 14 14:51:37 +0000 2022@LindorffLarsen To infinity and beyond!
Thu Jul 14 14:47:52 +0000 2022Is there a chromatographic term for the phenomenon that causes some peptides to elute in broad bands, while most are in sharp peaks?
Thu Jul 14 14:28:52 +0000 2022@RuudVeldhuizen @DBHardyLab @drcnatale @bryony_natale Intermittent reinforcement is a helluva thing. Just a bit of it & people will continue to push coins into the slot, no matter how bad the odds.
Thu Jul 14 14:26:20 +0000 2022I know the wheel is rediscovered about every 5 years, but sometimes the monotony of what-was-old becoming the shiny-new-thing can act as a brake on my enthusiasm.
Thu Jul 14 12:33:46 +0000 2022Most of the activity involves low-occupancy acceptors that can alternate between K+GGyl & K+acetyl.
Thu Jul 14 12:20:49 +0000 2022Modification diagrams for human PPP2R2A/B/C/D:p, the members of protein phosphatase 2 regulatory subunit family 2. Similar PTM-minimalist energy in this family, unlike the often frantic PPP1R subunits. 🔗
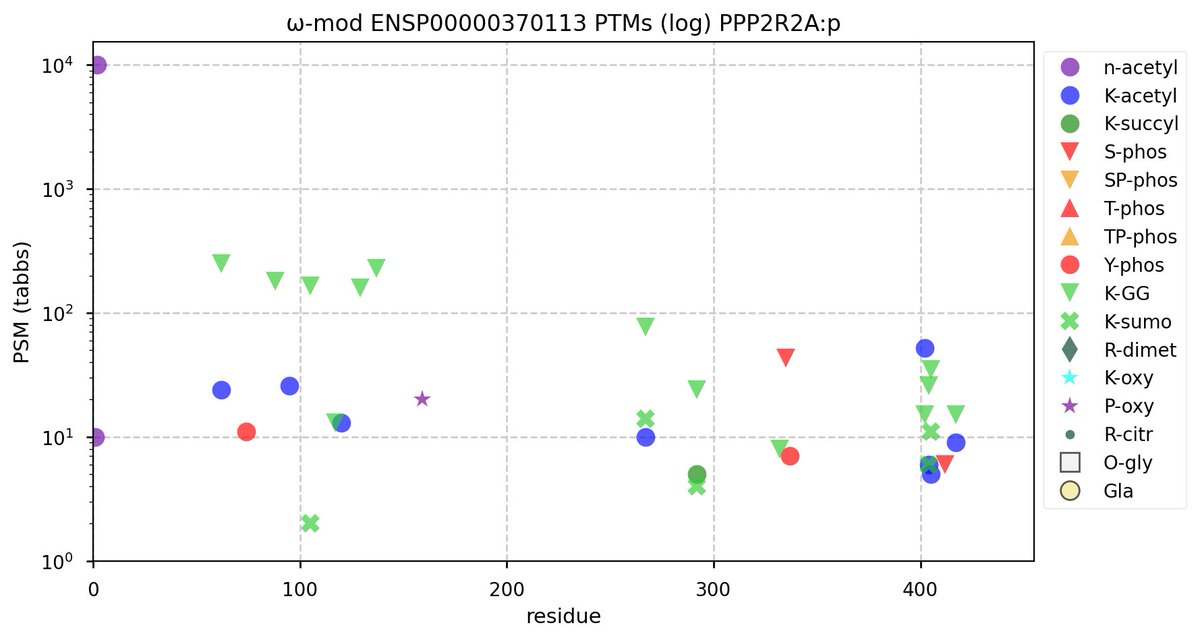
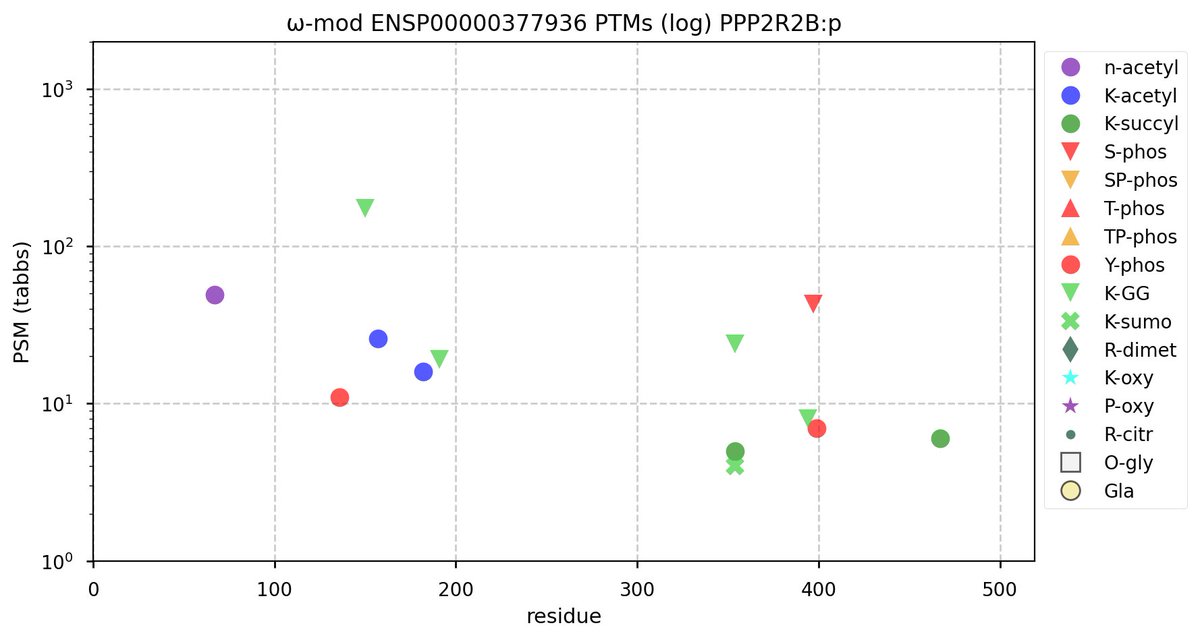
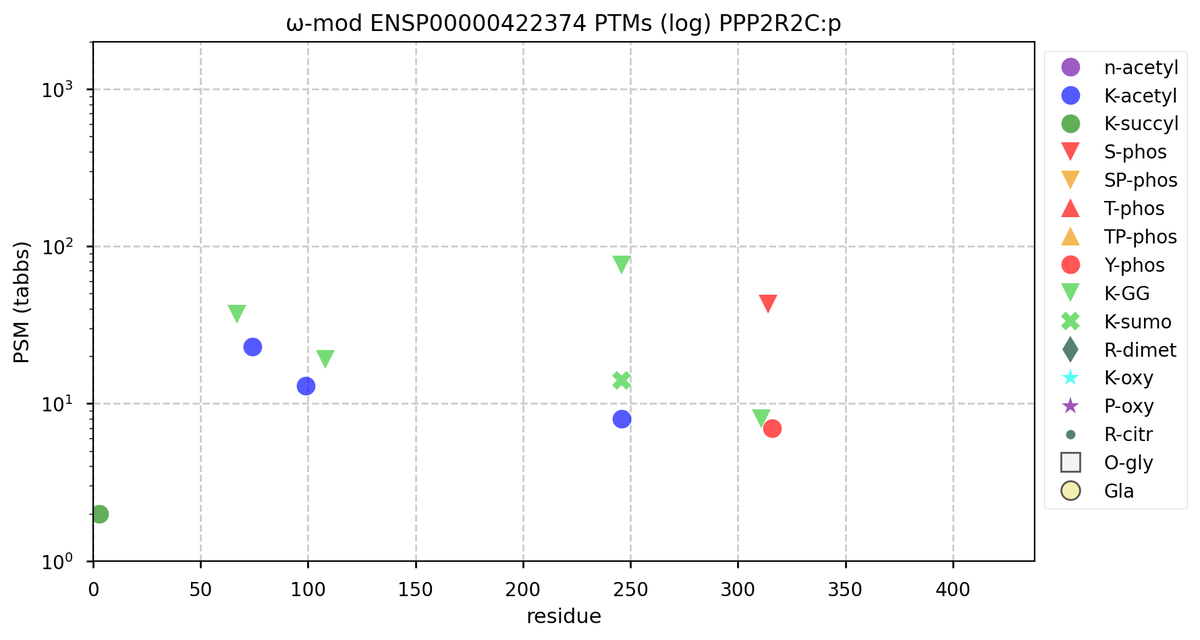
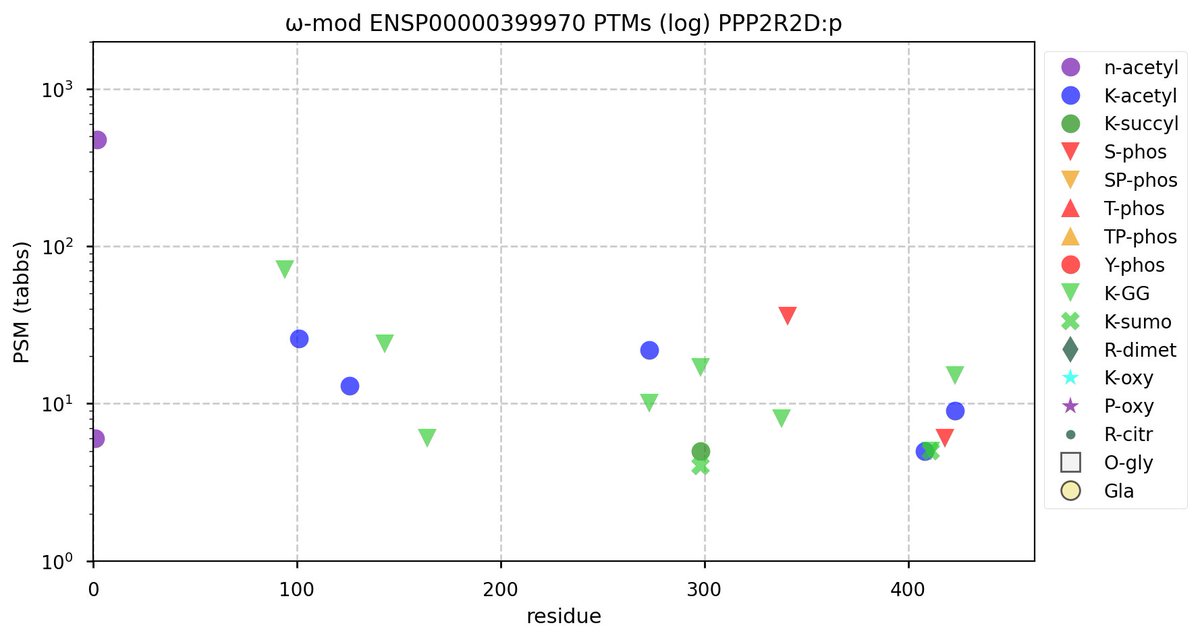
Wed Jul 13 15:35:06 +0000 2022A forecast of nice, warm weather in London, UK 🔗
Wed Jul 13 13:39:11 +0000 2022@TanentzapfLab You should probably mention that Stanley Park is a "day-time only" destination.
Wed Jul 13 12:06:11 +0000 2022PPP2R1A:p has 24 observed K+GGyl acceptors of the 32 available K residues; PPP2R1B:p has observed 26 K+GGyl acceptors of the 30 available K residues. "A" has 28 observed K+acetyl acceptors while "B" only has 6.
Wed Jul 13 11:51:59 +0000 2022In humans, there are 2 structural subunit genes, 12 regulatory subunit genes grouped into 4 families & 2 catalytic subunit genes.
Wed Jul 13 11:36:11 +0000 2022Modification diagrams for human PPP2R1A:p & PPP2R1B:p, members of the protein phosphatase 2 structural subunit family. Protein phosphatase 2 is a heterotrimer of 1 structural, 1 regulatory & 1 catalytic subunit. 🔗
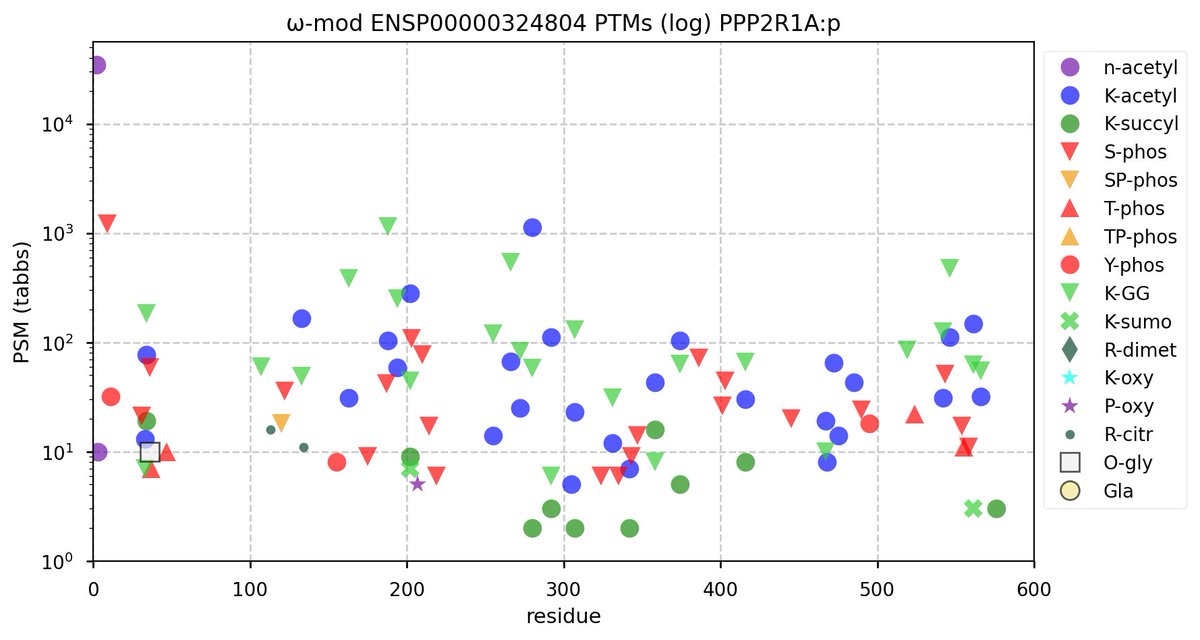
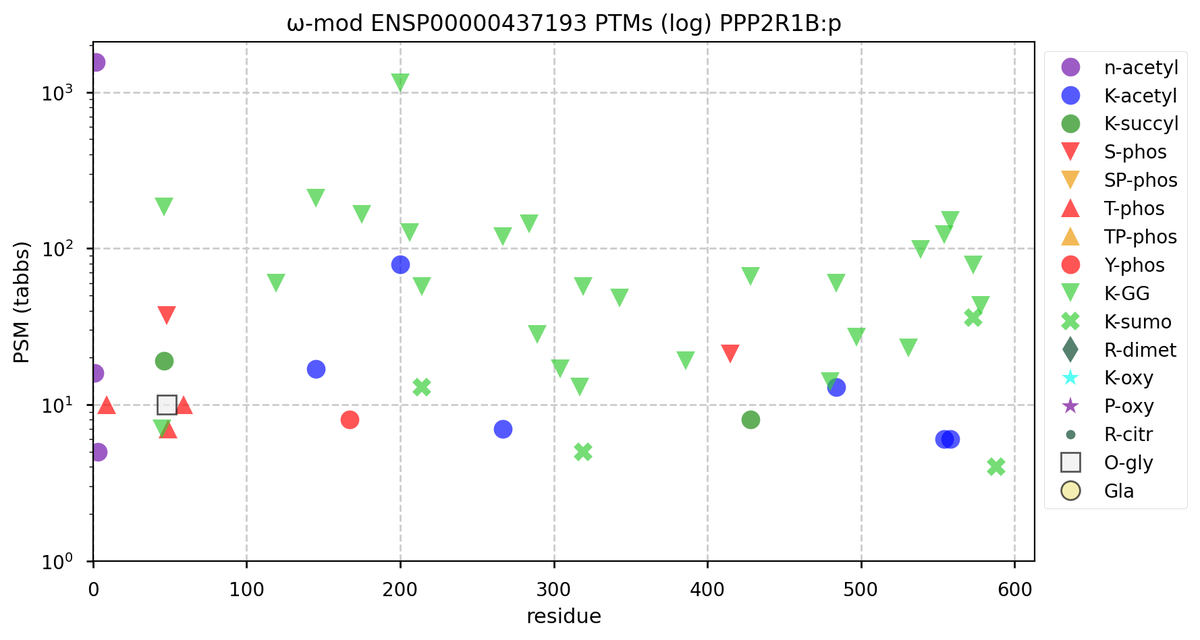
Tue Jul 12 20:46:56 +0000 2022I often wish I knew more about proteins.
Tue Jul 12 14:57:16 +0000 2022Joint appointments are nothing but trouble: avoid them at all costs.
Tue Jul 12 14:33:59 +0000 2022@neely615 @ypriverol If you have some time & want to see just how "modern" modern stuff really is, try watching the "Mother of All Demos" from December 9, 1968:
🔗
Tue Jul 12 14:22:09 +0000 2022@neely615 @ypriverol Probably counterproductive. Old versions that aren't actively used by developers/maintainers tend to fragment & decay fairly quickly.
Tue Jul 12 14:05:36 +0000 2022@neely615 @ypriverol Technically, it is an "alternate reality" rather than the "future".
Tue Jul 12 12:17:47 +0000 2022Modification diagram for human PPP1R15B:p, one of the protein phosphatase 1 regulatory subunit family 15. This less commonly observed family has a simpler pattern than many of the others & this particular member is quite involved with a K+GGyl mechanism.
🔗
Tue Jul 12 11:58:55 +0000 2022@neely615 You shouldn't feel too bad: those "olden times" were less than a month ago.
Mon Jul 11 14:10:22 +0000 2022@neely615 @JoePlatelet 🔗
Mon Jul 11 13:03:19 +0000 2022@neely615 @JoePlatelet It would only be 17 years to get through brewer's yeast!
Mon Jul 11 11:58:19 +0000 2022Modification diagrams for human & mouse PPP1R14A:p, members of the protein phosphatase 1 regulatory subunit family 14. Short sequences with S+phosphoryl acceptors on both ends. 🔗
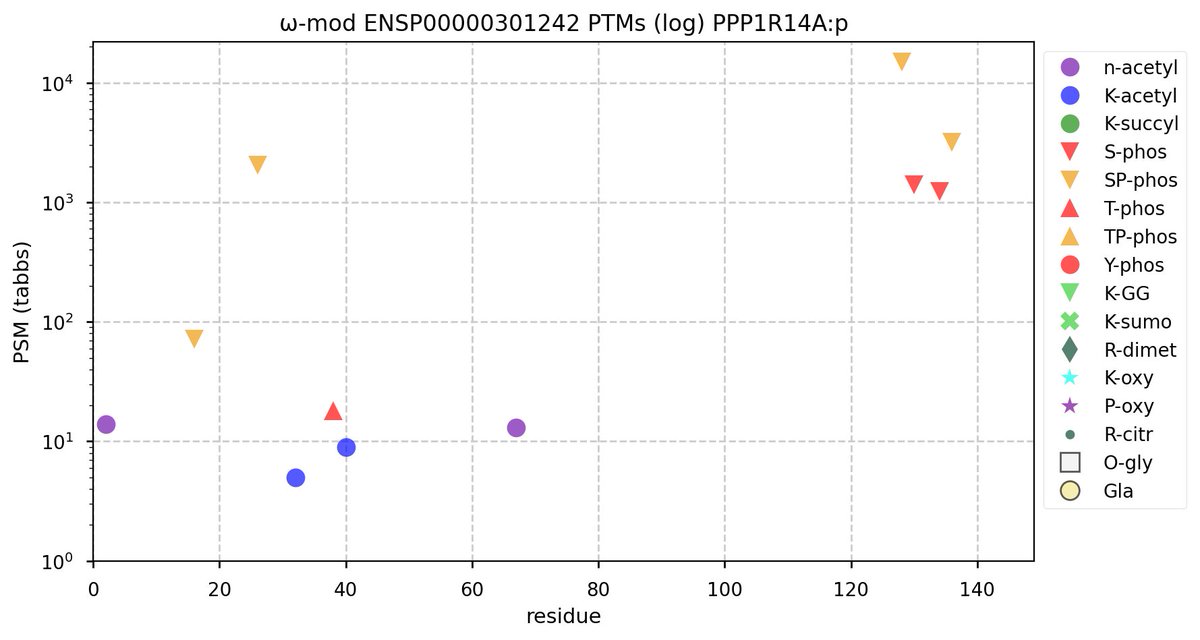
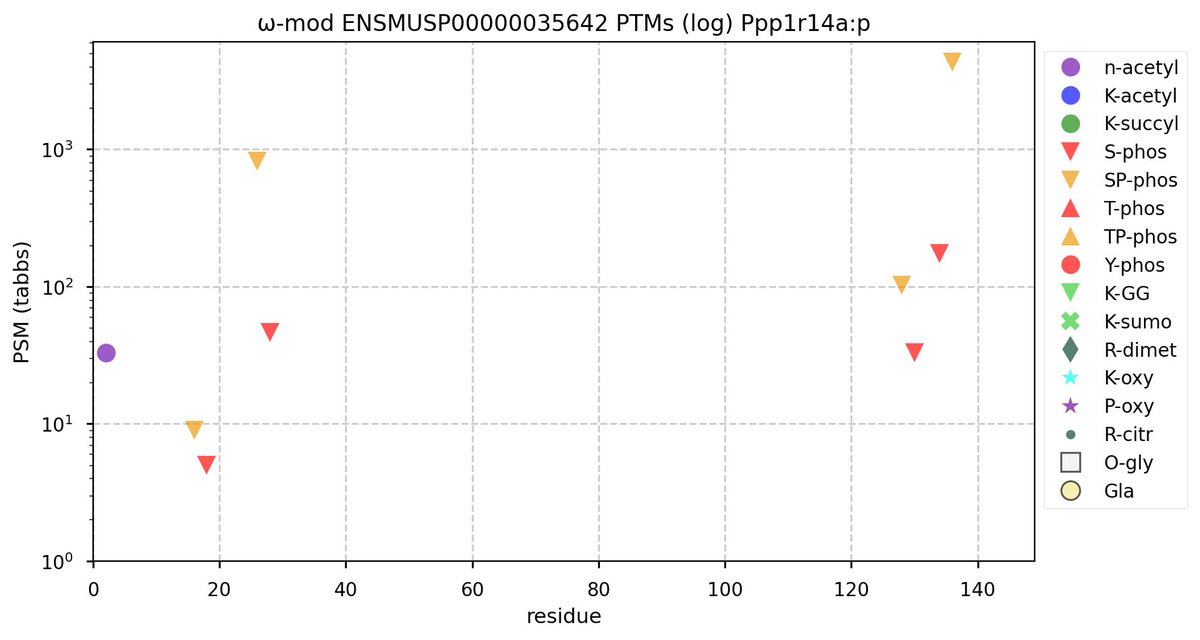
Mon Jul 11 11:23:00 +0000 2022@JoePlatelet Not yet: lots of proteins & only so many days in a year.
Sun Jul 10 20:50:56 +0000 2022@chrashwood And it does depend on where you look
🔗
Sun Jul 10 20:46:34 +0000 2022@chrashwood Phospho-enrichment studies are much more common than O-glycosylation enrichment studies, particularly with the NCI-funded crowd.
Sun Jul 10 15:59:07 +0000 2022When looking at a pattern like this, I wonder how many of the acceptor sites are occupied simultaneously: I suspect very few, but it would be nice to have some experimental evidence one way or the other. How about some top-down work on these more challenging molecules?
Sun Jul 10 14:44:35 +0000 2022The wacky looking TP53BP1:p isn't part of this family, although it does have a structured region right at the C-terminus (a BRCT domain), leaving a similar gap in phosphorylation.
🔗
Sun Jul 10 13:21:25 +0000 2022@pwilmarth @gingraslab1 @slavov_n It must be difficult a difficult point to make to students, when papers from experiments with either inadequate or non-existent controls are the norm, not the exception.
Sun Jul 10 12:58:38 +0000 2022Sometimes I wonder how much biomedical research is affected―hypothesis through to conclusions―by the ever evolving nomenclatural mess caused by the endless changing of human gene names & symbols.
Sun Jul 10 12:28:52 +0000 2022The N-terminal region is composed of multiple phospho-domains grouped in with short low complexity patches.
Sun Jul 10 12:24:59 +0000 2022Modification diagrams for human TP53BP2 (PPP1R13A) & PPP1R13B, subunits in the protein phosphatase 1 regulatory family 13. Their geometry is the reverse of family 12, with an ankyrin repeat-containing domain at the C-terminus (the gap in phosphorylation). 🔗
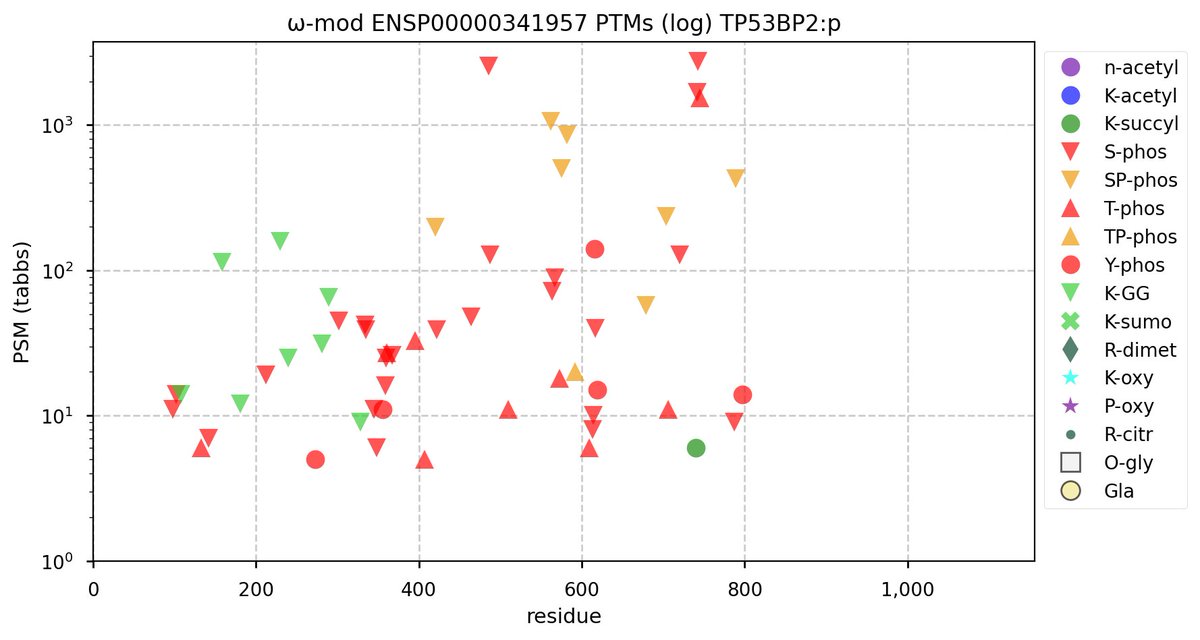
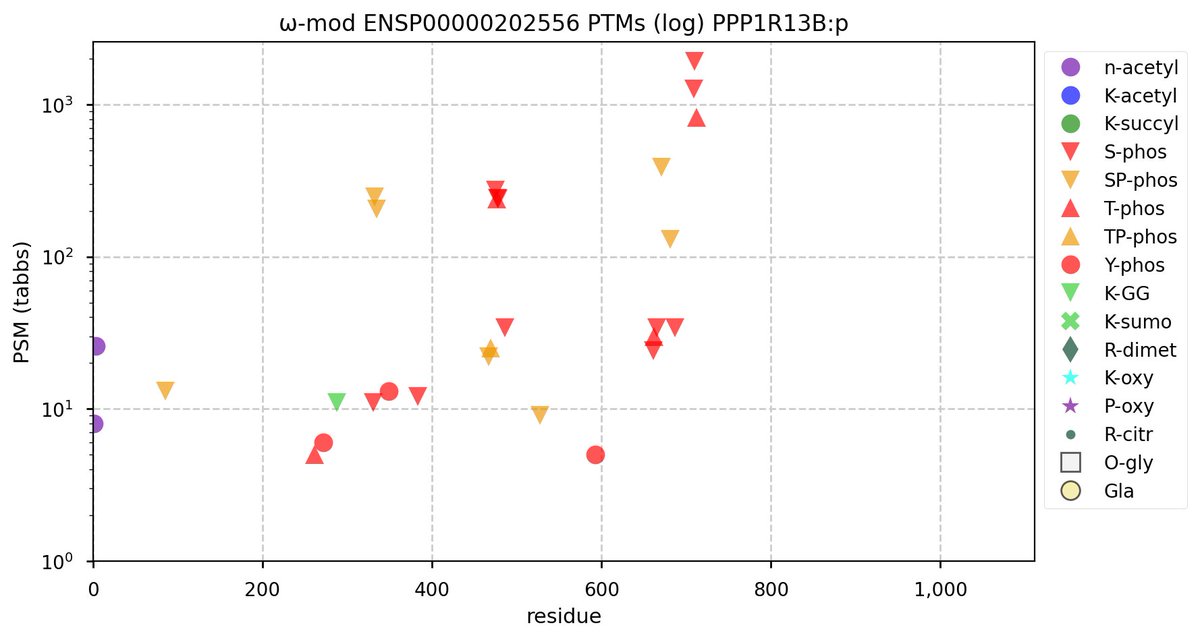
Sat Jul 09 19:09:08 +0000 2022🔗
Sat Jul 09 15:29:24 +0000 2022@ProtifiLlc So from this response, I would assume that you are a "sequence maximalist" wrt to these lists: you would like to see items included that are known to present, even though they are unlikely to be observed. How would you feel about including bovine INS?
Sat Jul 09 13:20:11 +0000 2022Even though these subunits are in the same family & show rhyming patterns of modification, when compared pair-wise, they share < 5% of their sequences.
Sat Jul 09 13:00:12 +0000 2022@SLIM_technology Reminds me of a colleague who did GC-MS of wine, who managed to convince growers that they needed to send him a case to do the analysis.
Sat Jul 09 12:15:43 +0000 2022Modification diagrams for human PPP1R12A/B/C:p, the family 12 protein phosphatase 1 regulatory subunits. All 3 have an N-terminal ankyrin repeat-containing domain (gap in phosphorylation), followed by a longish region of intrinsic disorder with multiple low complexity patches. 🔗
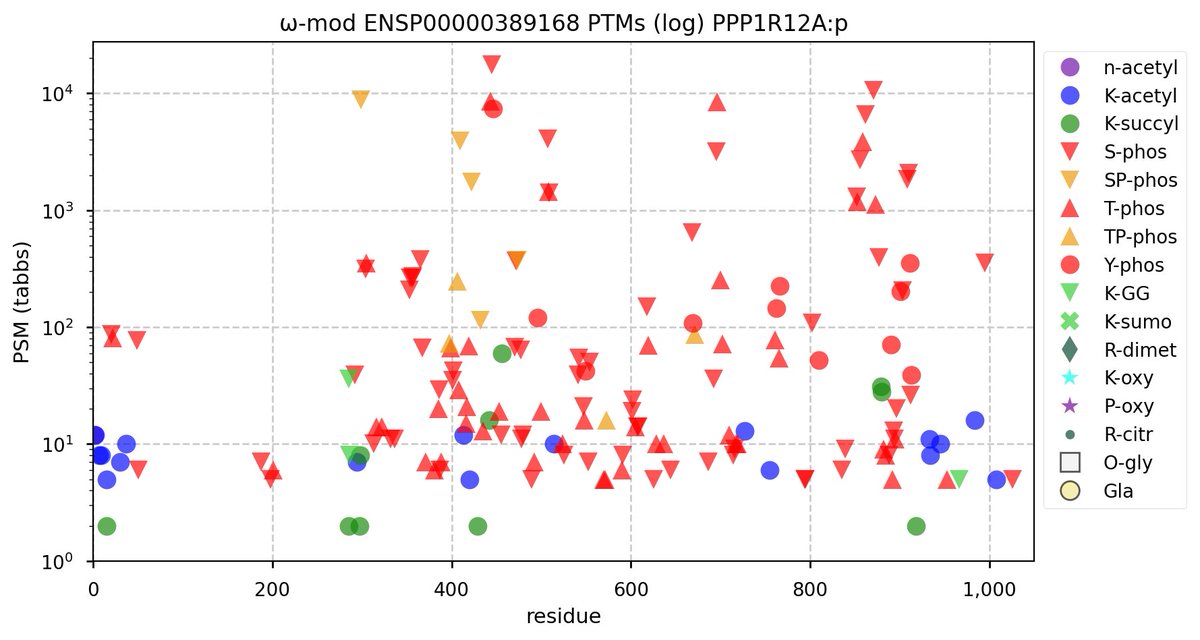
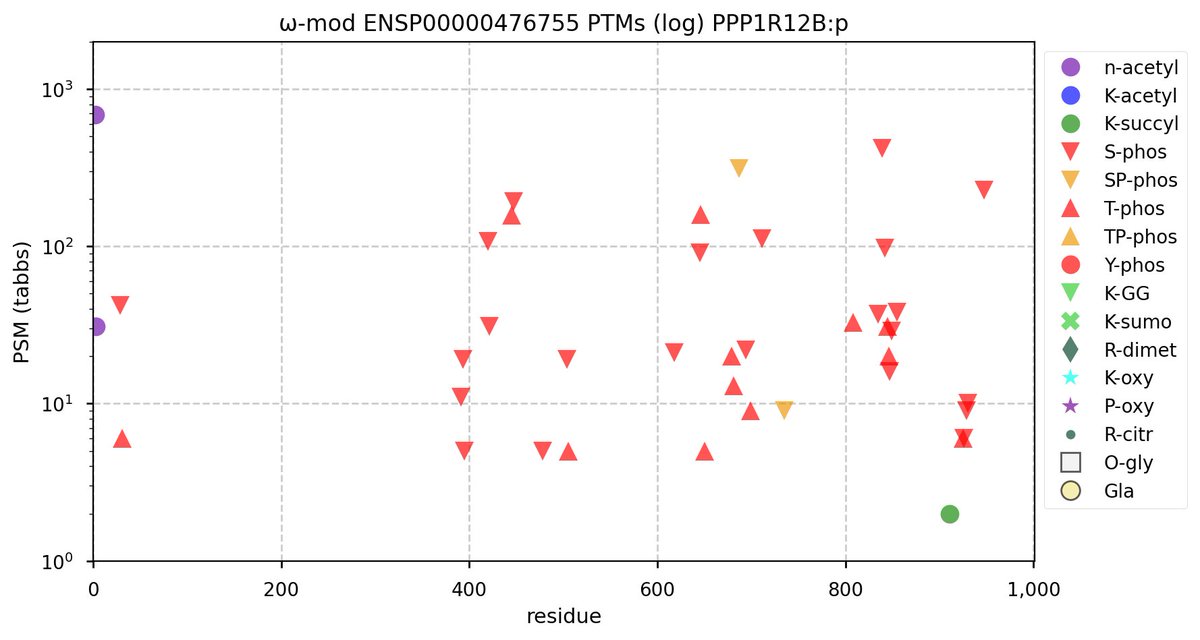
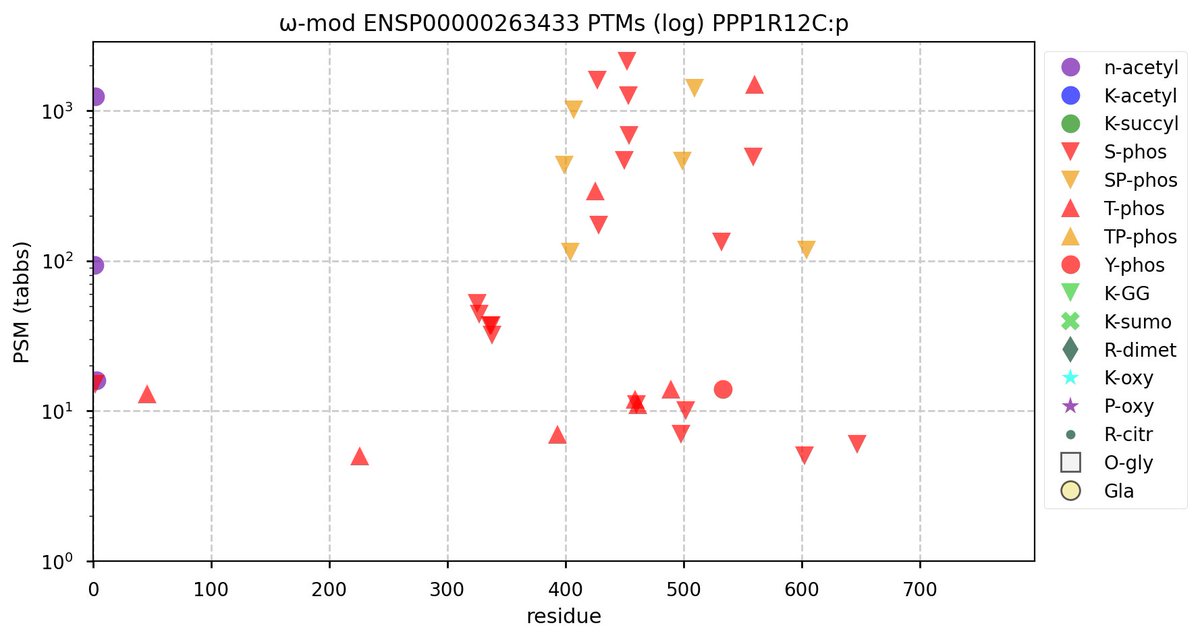
Sat Jul 09 11:44:36 +0000 2022Vertical stability numbers from the forecast, as displayed at SpotWx: 🔗
Sat Jul 09 11:36:05 +0000 2022Rumours of storms to come: NAM forecast for Winnipeg for the next couple of days. 🔗
Sat Jul 09 11:32:42 +0000 2022A summer storm just west of Bismark steaming its way across the prairie towards Grand Forks and Fargo this morning. 🔗
Sat Jul 09 11:23:20 +0000 2022@neely615 I've never had much luck getting answers back via email: I gave up on that approach a decade ago. I am, however, interested in the rationale for creating lists like this, particularly how a group balances their collective ideas about what "should" be there against what "is".
Fri Jul 08 23:11:28 +0000 2022I was reading an article about creating a standard list of contaminant sequences, but it didn't really go into the reasons for choosing particular proteins. Looking through the FASTA, the presence of B. taurus thyroglobulin stood out: anybody know why TG would be a contaminant?
Fri Jul 08 19:44:02 +0000 2022A complex of storms making a run across South Carolina this afternoon. 🔗

Fri Jul 08 16:02:24 +0000 2022The proteins you are most likely to find from Wolbachia in a fruit fly sample are (not very surprisingly) those abundant in many prokaryotes:
DnaK, ATP synthase F0F1, GroEL, TUF & rpoBC
As well, Wolbachia-only proteins like Wsp, WD0745 & WD0722 may also be prominent.
Fri Jul 08 15:22:18 +0000 2022The Southern Ocean's storms are looking particularly symmetric today. 🔗
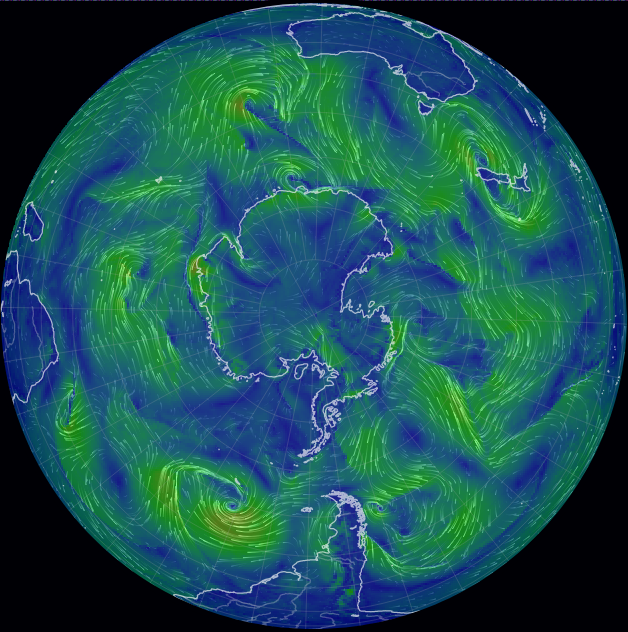
Fri Jul 08 15:12:20 +0000 2022Wolbachia endosymbiont of Drosophila melanogaster, the "other" proteome in many fruit fly samples:
🔗
Fri Jul 08 12:56:46 +0000 2022🔗
Fri Jul 08 12:51:26 +0000 2022The AF proposed structure isn't very helpful wrt to understanding this pattern, other than it suggests a wide-open playing field for PTM related enzymes. 🔗
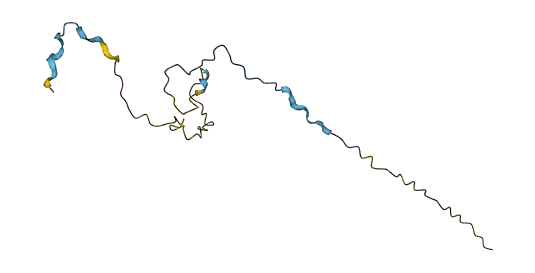
Fri Jul 08 12:50:01 +0000 2022Modification diagram for human PPP1R11:p, the only member of protein phosphatase 1 regulatory subunit family 11. Unlike family 10, this subunit has no significant involvement with K+acetyl or R+dimethyl mechanisms. 🔗
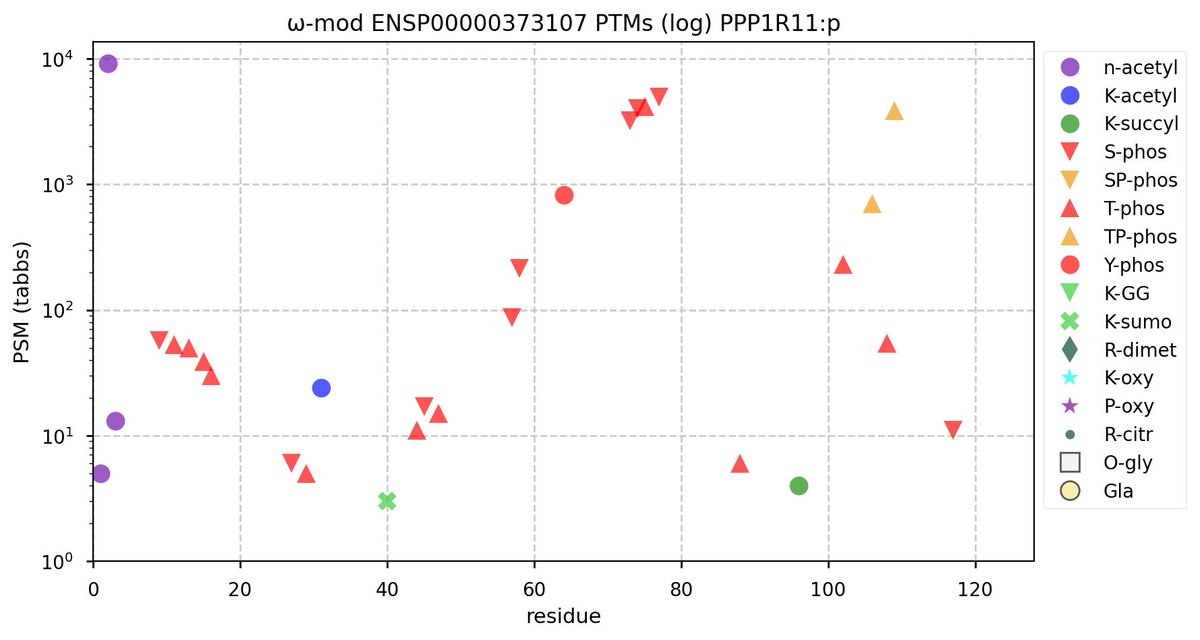
Fri Jul 08 02:56:19 +0000 2022To the SCOPE-MS cognoscenti: would you characterize 🔗 as genuine single cell work or put it in some type of single-cell-adjacent category.
Thu Jul 07 23:43:08 +0000 2022@Peptidome If the current conditions hold, it should be visible from Chicago, Cleveland & Boston once it gets dark tonight.
Thu Jul 07 21:25:34 +0000 2022There is a geomagnetic storm going on at the moment, which has generated a fairly broad area of aurora, extended to lower than normal latitudes. The image is the current aurora extent (4:30 CDT). 🔗
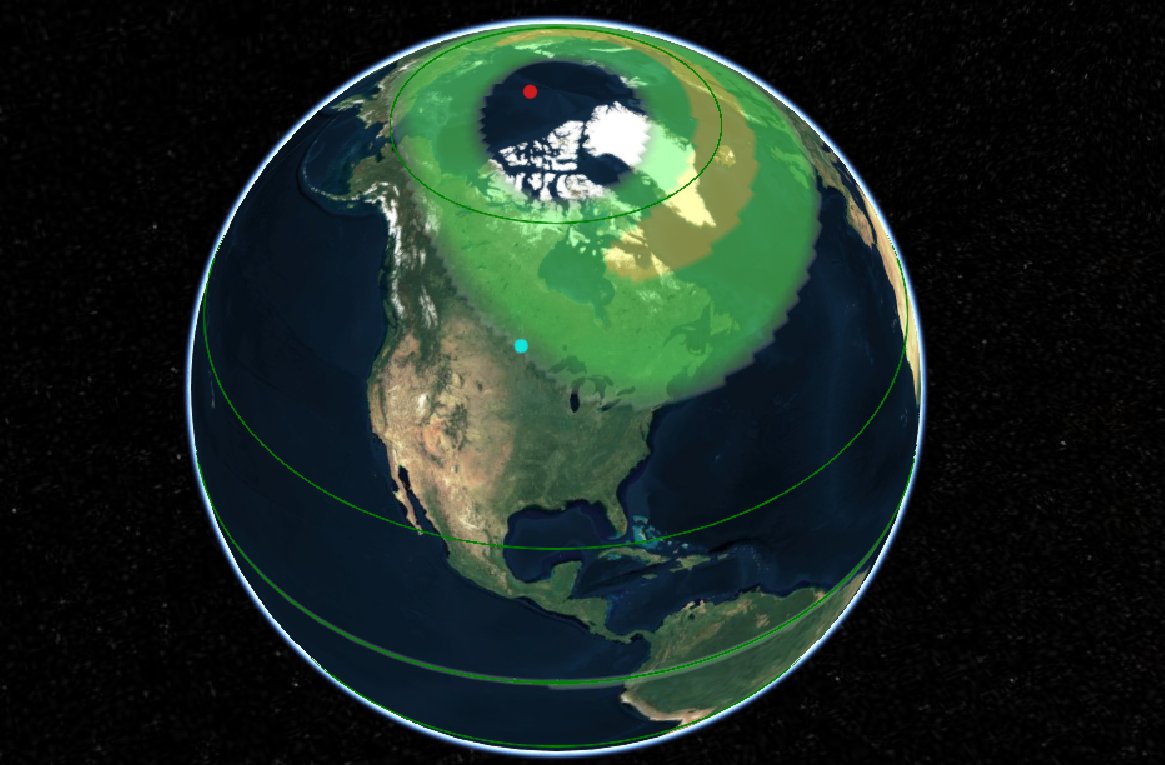
Thu Jul 07 14:32:45 +0000 2022It is almost endearing that the Canadian media, with all of world events to chose from, still think this odd, inside-baseball story about small time Ontario politics is worthy of national coverage
🔗
Thu Jul 07 14:16:45 +0000 2022@astacus Not admirable, but consistent. He seems to be able to convince enough people to think it isn't possible that he could be doing "that" again, while he proceeds to do just "that" again.
Thu Jul 07 13:03:32 +0000 2022May be I'm not understanding events, but it would appear that the UK PM has survived an effort by his Cabinet who resigned to get him to leave, by saying that he may leave at some time in the future & in the mean time he is busily appointing a new Cabinet.
Thu Jul 07 12:34:18 +0000 2022Particularly if you were planning to camp out at the Folk Festival this weekend.
Thu Jul 07 12:30:53 +0000 2022Well, this is unexpected & worth keeping track of is you happen to live in or around Winnipeg (from the North American Mesoscale Forecast System for 49.877,-97.157) 🔗
Thu Jul 07 12:20:41 +0000 2022I am no great fan of common law, at least once in a while the Canadian Supremes do something that isn't goofy: 🔗
Thu Jul 07 12:03:50 +0000 2022Modification diagram for human PPP1R10:p, the only member of protein phosphatase 1 regulatory subunit family 10. The pattern of occupied K+acetyl & R+dimethyl sets this subunit apart from the other PPP1R families: it is decorated more like an hnRNP than a phosphatase. 🔗
Thu Jul 07 11:20:55 +0000 2022@neely615 Once you get out of it, it is mildly embarrassing to have been in & contributed to the literature cult for so long.
Wed Jul 06 15:03:11 +0000 2022@seejbrown @fireinlab Looks like Indianapolis has changed since I was there. All anyone wanted to do back then was have an "ice cream social".
Wed Jul 06 14:32:56 +0000 2022The entire UK seems transfixed, at the moment, waiting for a shameless man to display shame. It is like someone who sees a tiger coming towards them and stops to ponder whether this particular tiger might be a vegetarian.
Wed Jul 06 13:06:39 +0000 2022Looks like Columbus is about to get pummeled again this morning. 🔗
Wed Jul 06 12:26:22 +0000 2022Has a fairly active literature subgenre describing speculative functions under the noms de guerre "neurabin 2" & "spinophilin".
Wed Jul 06 12:23:56 +0000 2022Modification diagram for human PPP1R9B:p, a regulatory subunit in the protein phosphatase 1 family. Very little involvement in K+GGyl/acetyl mechanisms.
🔗
Wed Jul 06 00:15:11 +0000 2022An oddly shaped storm appears to be having its way with the Chicagoland region. 🔗
Tue Jul 05 21:24:23 +0000 2022And a major known-unknown:
Why are the ratios of PSMs with Y/T/S acceptors about 1:10:80?
Tue Jul 05 19:14:32 +0000 2022Some corrections, apologies & compute time later: human CKB:p 🔗
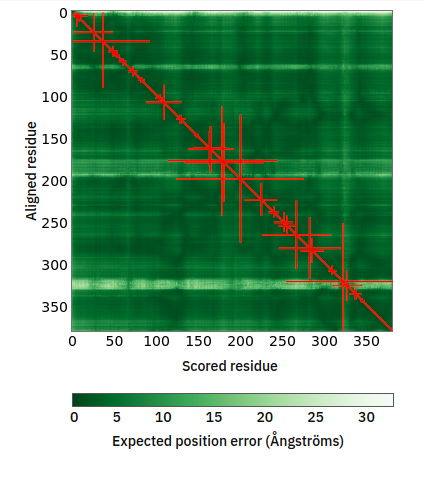
Tue Jul 05 18:29:06 +0000 2022The two most obvious cons:
- over representation of sites associated with the cell cycle; &
- over representation of sites in domains enriched with acidic residues.
Tue Jul 05 18:06:53 +0000 2022PXD034264 exemplifies the pros & cons of using cell lines & metal oxide affinity purification methods with LC-MS/MS to identify phosphorylation acceptor sites.
Tue Jul 05 17:28:31 +0000 2022Found it: 🔗
Tue Jul 05 17:17:22 +0000 2022Interesting. PTM eXchange? 🔗
Tue Jul 05 15:58:47 +0000 2022Look, you stupid AI, I told you I wanted "an example of an exception", not an "an exceptional example"! 🔗
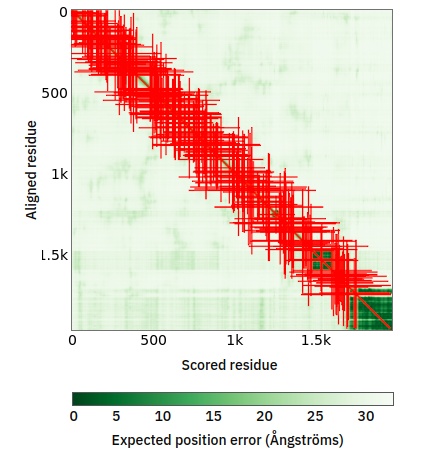
Tue Jul 05 14:27:34 +0000 2022A pretty angry-looking line of squalls heading for the Columbus, OH area, including a satellite storm just to the south that may tag Dayton, once it get done with Indiana. 🔗

Tue Jul 05 14:21:25 +0000 2022@AlexUsherHESA Ontario College of Trades and Apprenticeship Act, 2009?
Tue Jul 05 12:24:42 +0000 2022Modification diagram for human PPP1R3D:p, 1 of 7 subunits in protein phosphatase 1 regulatory chain family 3. Also shown is the unsuprising AF predicted aligned error plot (no overlay required). 🔗
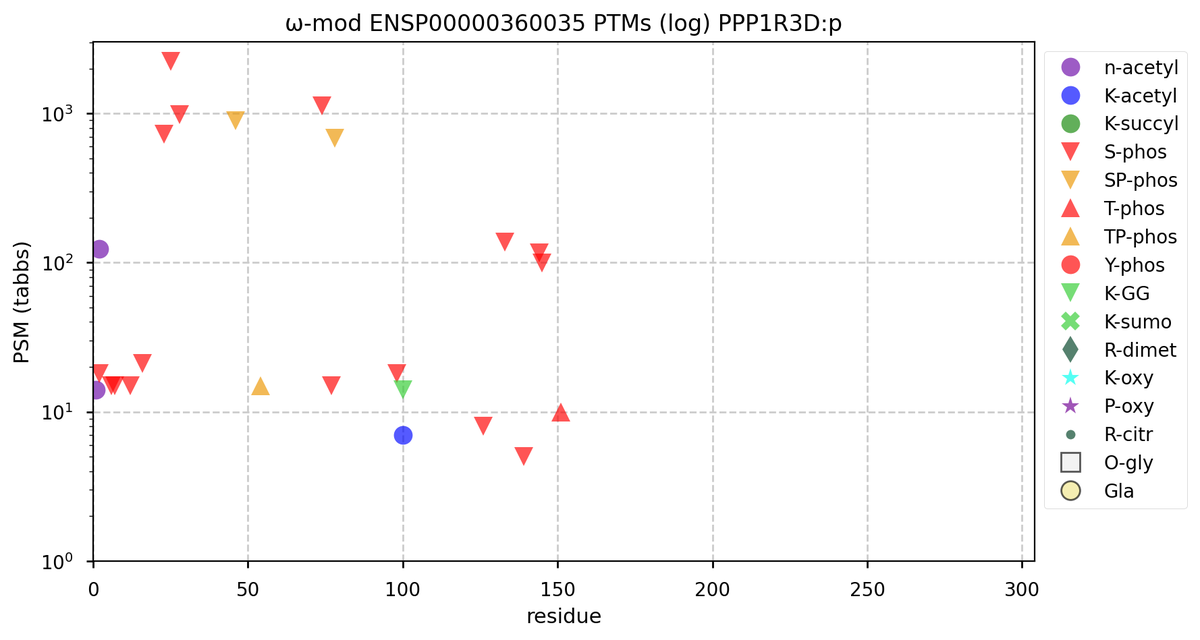
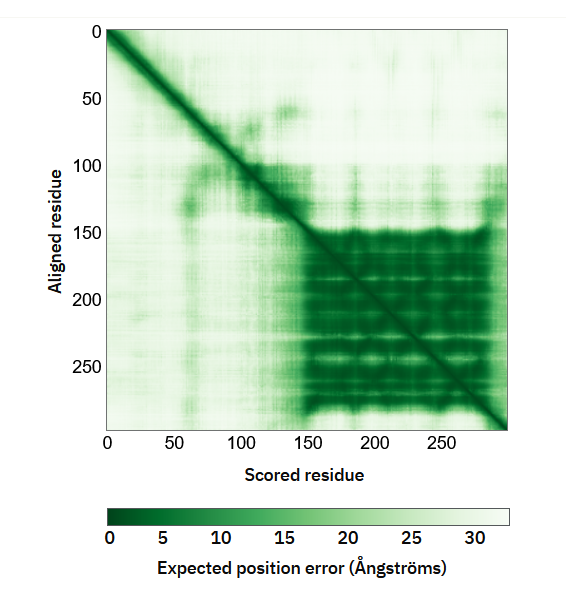
Tue Jul 05 01:47:43 +0000 2022An odd line of small heavy rain squalls heading due east through Milwaukee WI. 🔗
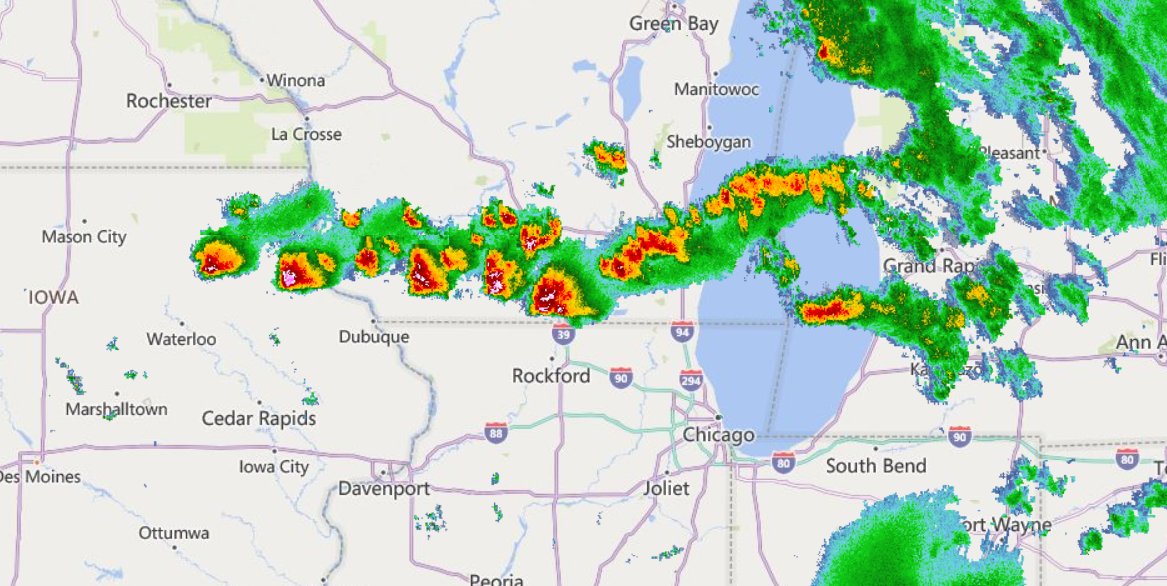
Mon Jul 04 21:44:56 +0000 2022@EdHuttlin @HMSBioPlex It is just an impression on my part, but the large T antigen seems to show up more often in affinity preps than I would naïvely expect.
Mon Jul 04 18:05:26 +0000 2022Control protein E1A [Human mastadenovirus C] also shows up pretty prominently in HEK-293T phosphopeptide work.
Mon Jul 04 17:42:32 +0000 2022Go, Canada! 🔗
Mon Jul 04 16:03:47 +0000 2022@michaelhoffman @mmgrabow Its hard to predict how long it will take an inexperienced person to do something. When someone Julienne's a carrot with a knife the first time, it tends to take a long time (& maybe a short hospital excursion).
Mon Jul 04 15:47:08 +0000 2022@michaelhoffman They are asymptotic values, only approached after many repetitions.
Mon Jul 04 15:09:25 +0000 2022The 3 viral proteins you should always find in HEK-293T samples (all others are bonus):
- large T antigen [Simian virus 40]
- control protein E1B 55K [Human mastadenovirus C]
- control protein E1B 19K [Human mastadenovirus C]
Mon Jul 04 13:11:20 +0000 2022@ucdmrt Nuclear binding energy determination was a central theme in mass spectrometry: there were sessions devoted to it at the ASMS. When I was a grad student, half the floor below our lab was devoted to testing binding energy computations via high accuracy nuclear mass determination.
Mon Jul 04 12:44:26 +0000 2022Is SARS-COV2 "over" at grants panels to the same extent as it is "over" in civil society?
Mon Jul 04 11:46:41 +0000 2022Modification diagrams for human & mouse PPP1R2:p, a family 2 regulatory subunit of protein phosphatase 1. 🔗
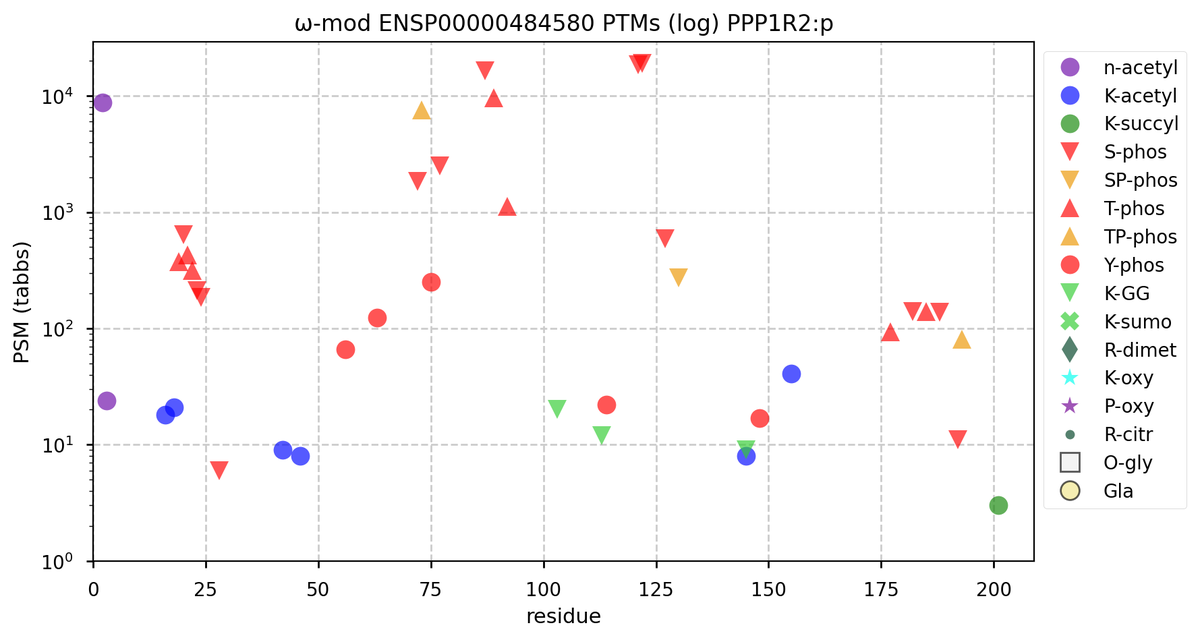
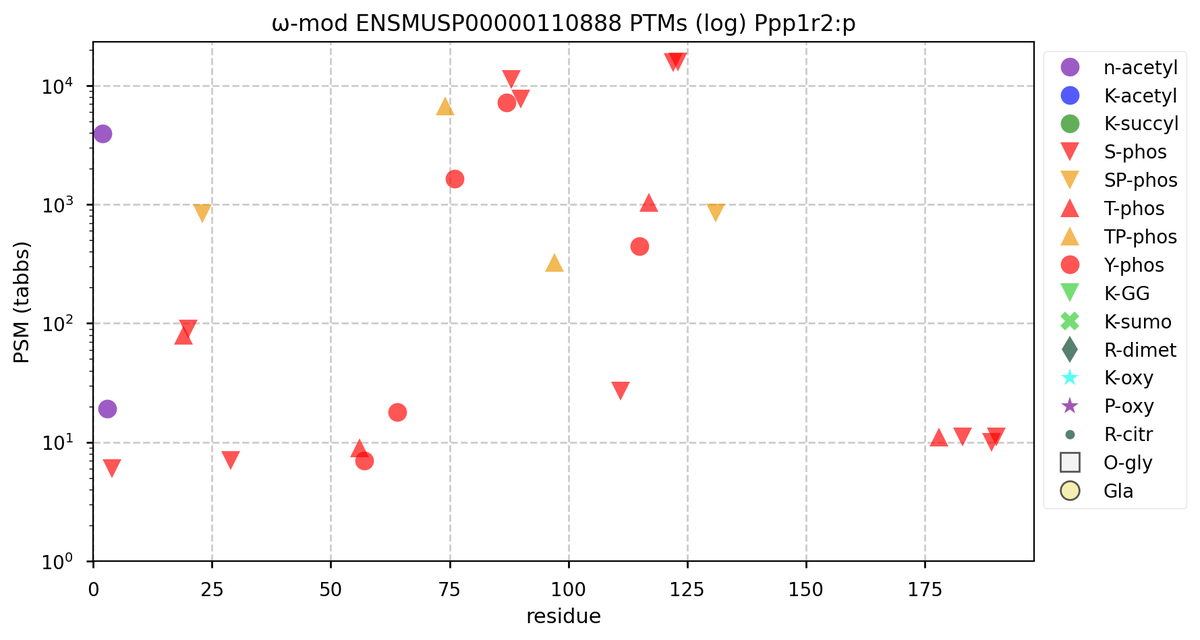
Sun Jul 03 18:03:12 +0000 2022@neely615 Our weird version of metric means a stick of butter here is the same as the US: 1/4 of a 454 gram package (with or without its associated 4 ml of salt).🇨🇦
Sun Jul 03 17:09:59 +0000 2022@neely615 In Canuckistan―we are sort-of metric―that type of packaging has to include the # of milliliters corresponding to the tablespoon count, so in most cases the packaging does not include those measures any more. But grams are always better (don't get me started on fluid ounces).
Sun Jul 03 16:21:05 +0000 2022Added note: 10 years ago, I was a charter member of the "junk phosphorylation" club, but it is increasingly difficult to square with the evidence.
Sun Jul 03 15:21:00 +0000 2022It is hard for me to come up with a reasonable proposal explaining why there is so much agreement between these composites of thousands of LC/MS/MS analyses of both species if the underlying mechanism is largely stochastic, i.e., the "junk phosphorylation" hypothesis.
Sun Jul 03 14:23:49 +0000 2022@pwilmarth @Peptidome Its discussion is reminiscent of the Clampett v Drysdale feud. I would be surprised if you could ever find 6 reviewers to agree on the value any particular narrative.
Sun Jul 03 14:11:35 +0000 2022And for the brave few who are interested in such things, I've attached the diagram for mouse PPP1R1B, which rhymes rather nicely with the human subunit (right down to the T34,75+phosphoryl backbeat) 🔗
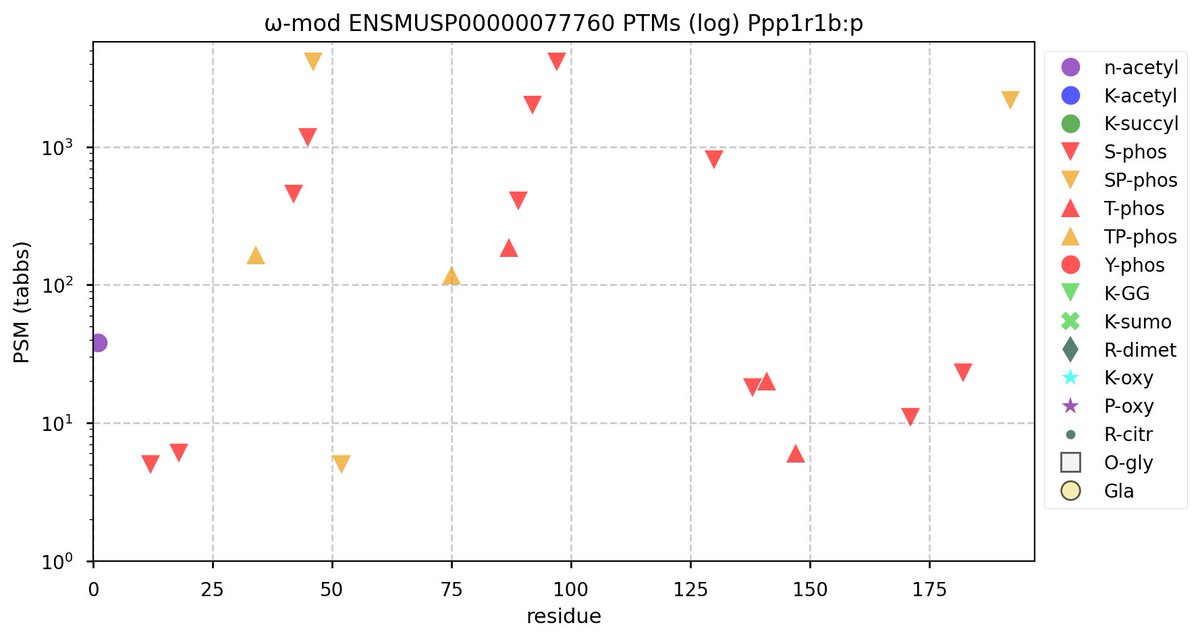
Sun Jul 03 13:39:15 +0000 2022Not at all profound, but annoying me at the moment: the most confounding thing about North American recipes is their insistence on the use of cups & tablespoons to measure butter instead of weight.
Sun Jul 03 12:32:41 +0000 2022These primary structure constraints mean that PTMs found for a Catalytic subunit are composites: patterns of that subunit when paired with multiple Regulatory subunits, as well as PTMs on identical tryptic peptides from the other 2 paralogs.
Sun Jul 03 12:24:39 +0000 2022The 3 Catalytic paralog subunits share ~70% of their sequence: a highly conserved central domain with unique N- & C-terminal domains. Regulatory subunits are much more variable, even within family groupings.
Sun Jul 03 12:20:34 +0000 2022Modification diagram for human PPP1CB:p & PPP1R1B:p, subunits of protein phosphatase 1. Protein phosphatase 1 is created by forming a heterodimer (trimer) of 1 of 3 Catalytic subunits (PPP1CA/B/C) & 1 (or 2) of 29 Regulatory subunits that have been organized into 16 families. 🔗
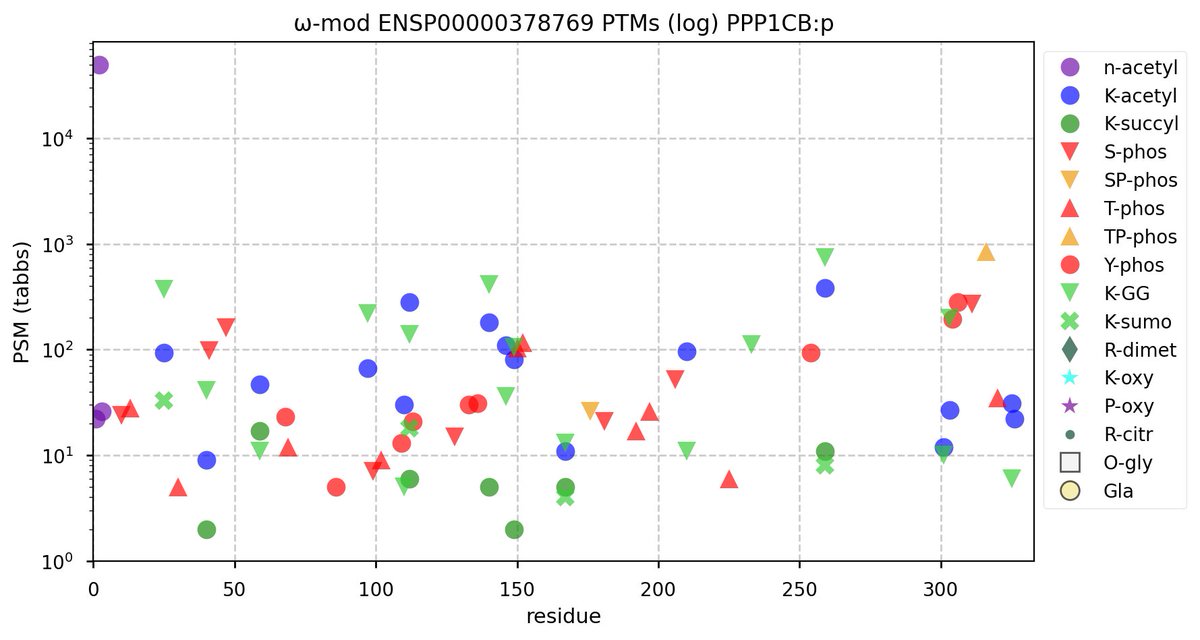
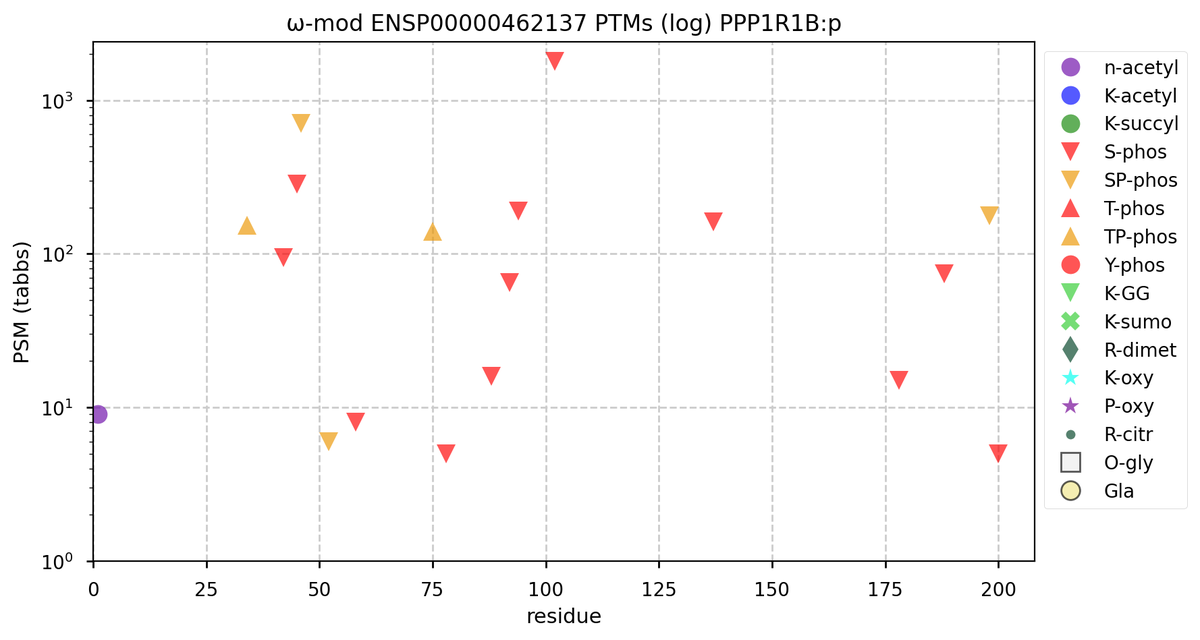
Sat Jul 02 17:31:29 +0000 2022PXD029730 has really grown on me. Nice work.🧐
Sat Jul 02 13:44:52 +0000 2022@astacus I particularly love the real weirdos, e.g. POLR2A:p.
Sat Jul 02 13:34:21 +0000 2022@astacus A lot of proteins are pretty dull wrt PTMs. I tend to show the ones that are all glitter & sequins.
Sat Jul 02 13:25:58 +0000 2022An arc of storms developing along a cold front running from Nebraska to Prince Edward Island 🔗
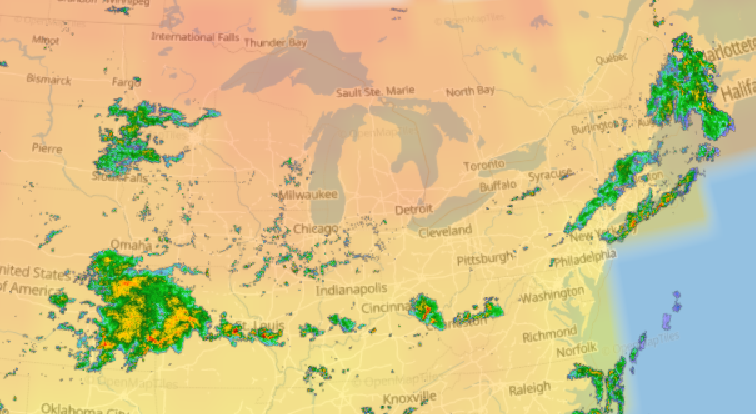
Sat Jul 02 13:19:29 +0000 2022One of many examples where common law (with its obsession with logic) & science (that champions rationality) go in different directions 🔗
Sat Jul 02 12:39:26 +0000 2022Tomorrow I will move on from the cool, spartan MMP family into the elegant, rococo decoration of the phosphatome's subunits.
Sat Jul 02 12:07:55 +0000 2022Modification diagram for human MMP10:p, a monomeric ECM protease. The proprotein (18-476) is generated by the co-translational removal of the signal peptide (1-17); the active form (99-476) is made upon secretion. Fibroblasts produce a lot, but it is absent from many cell lines. 🔗
Sat Jul 02 01:42:24 +0000 2022@UCDProteomics @DRockstar420 @Fridgwick I'd take a look at the SMART information for the hard drives. Whenever I've had trouble that resulted in the need for multiple reboot attempts, it was always a wobbly drive.
Sat Jul 02 01:26:10 +0000 2022Tropical Storm Bonnie is making landfall, headed directly west towards Lake Nicaragua. 🔗
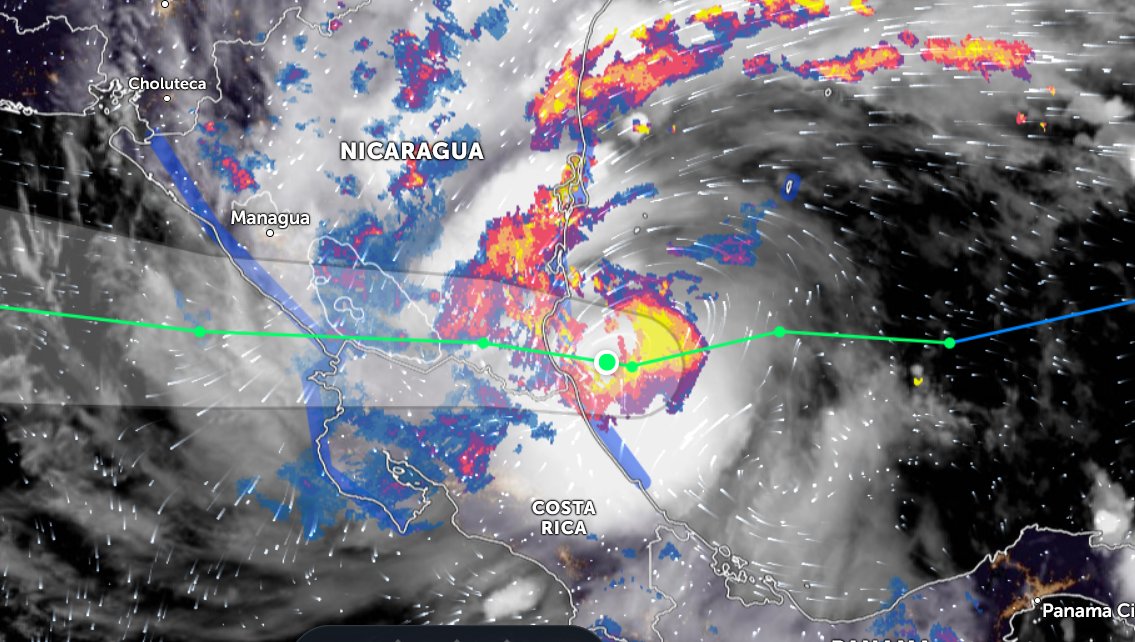
Sat Jul 02 01:04:22 +0000 2022Typhoon Chaba has moved closer to shore & its winds have increased. It seems to be capable of producing a lot of rain as it moves inland. 🔗
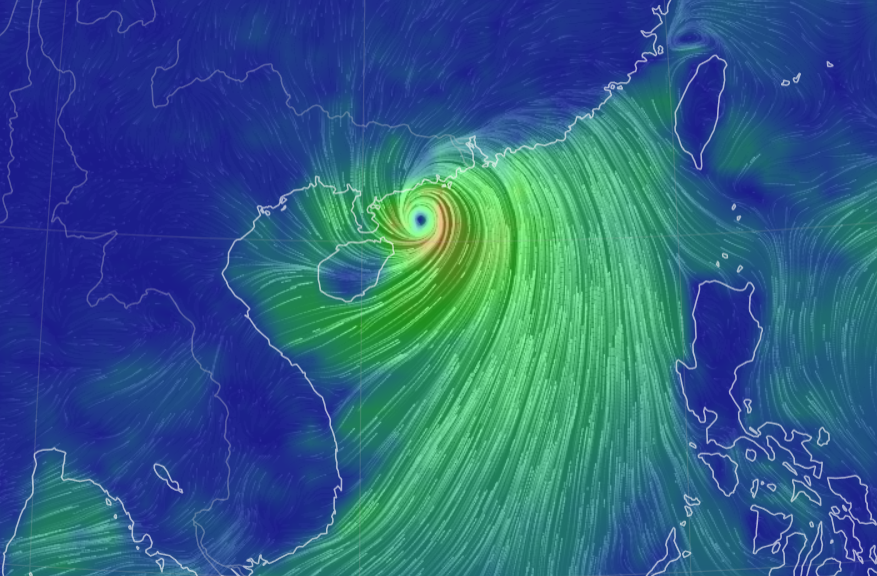
Fri Jul 01 19:50:21 +0000 2022@theoneamit Sounds like a Star Wars villain, but it seems Wikipedia has made it the official term:
🔗
Fri Jul 01 18:47:26 +0000 2022If the accepted word for the collection of kinases in an organism is "kineome", what would you call the collection of protein phosphatases? The "anti-kineome"?
Fri Jul 01 18:38:19 +0000 2022Something, something, hubris, something, something ...
🔗
Fri Jul 01 17:32:14 +0000 2022@UCDProteomics Not necessary. You just have to be able to snap out of it when they start whirring again.
Fri Jul 01 17:26:52 +0000 2022@dexivoje Back in ancient times (when I first published), there would normally be 1, or at most 2, reviewers along with a detailed critique from the Editor. Now editors seem to believe that their only role is to organize a poll from a focus group.
Fri Jul 01 17:13:26 +0000 2022@dexivoje Who really wants to be reviewer #6?
Fri Jul 01 15:46:12 +0000 2022The British never really got the value proposition of the Americas, other than as somewhere to send troublesome groups.
Fri Jul 01 15:11:33 +0000 2022It is Dominion Day in Canada. Sort of like July 4th, but we just asked whether it would be OK if we were self-governing & the Brits said "fine, sure, go ahead".
Fri Jul 01 13:57:51 +0000 2022Is there a good review of MS/MS-based phosphopeptide detection with an emphasis on the biological processes involved? No sequence logos, please!
Fri Jul 01 13:08:52 +0000 2022Personally, I would say it is the other way around, but I must admit to well documented bias. 🔗
Fri Jul 01 12:34:21 +0000 2022If you like looking at weather patterns, particularly storms in progress, this site is worth a look 🔗
Fri Jul 01 12:31:37 +0000 2022Weak upper atmosphere steering winds have left Typhoon Chaba drifting just off shore of Guangdong, now on track to make landfall somewhere between Hong Kong & Hainan. 🔗
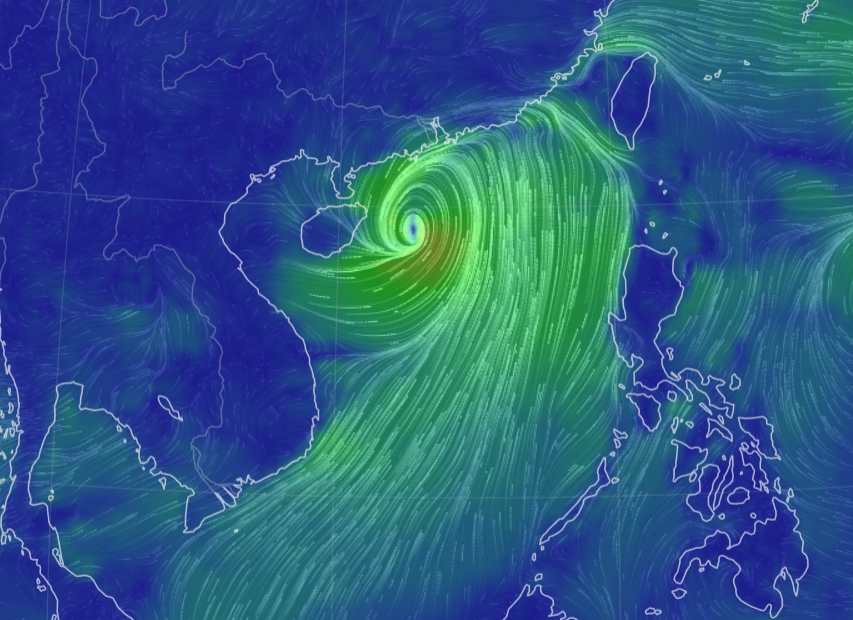
Fri Jul 01 12:11:02 +0000 2022Modification diagram for human MMP8:p, a monomeric collagenase. The proprotein (21-467) is generated by the co-translational removal of the N-terminal signal peptide (1-20). It is found in saliva & urine, but absent from many common cell lines. 🔗
Fri Jul 01 02:13:22 +0000 2022Now Typhoon Chaba is on track to make landfall very near Hong Kong & Dongguan. 🔗
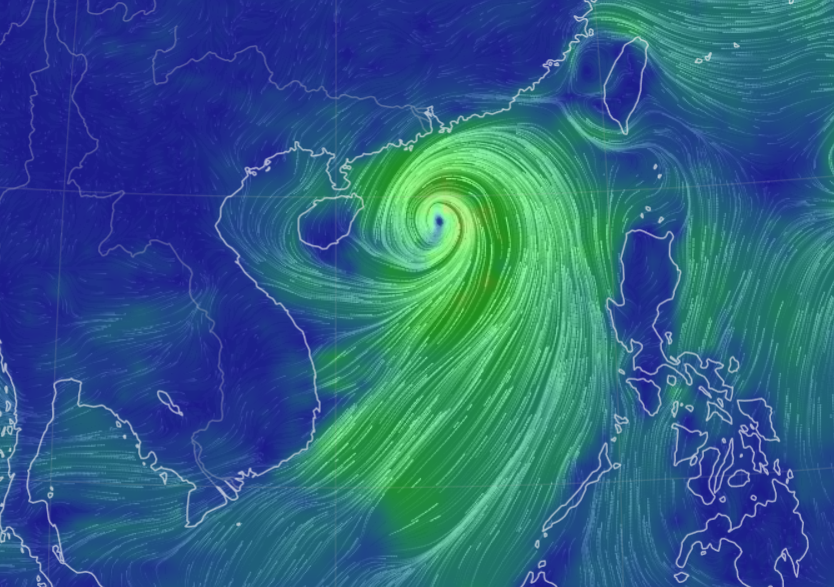
Thu Jun 30 14:59:52 +0000 2022Stormy weather this morning from Houston to Mobile. 🔗
Thu Jun 30 14:54:26 +0000 2022Papers that fit into the "I found a gene/transcript/protein" genre of stories often lose me when they pick an incongruous (but convenient) "function" for the associated gene product & using that "function" go to town trying to interpret its role in their system.
Thu Jun 30 14:18:49 +0000 2022@idpemery They will eventually agree & then demand the money back.
Thu Jun 30 14:15:05 +0000 2022@slashdot No. It cannot.
Thu Jun 30 12:19:50 +0000 2022I am running through some of the genes in the MMP family as a bit of a palate cleanser from my usual fare that tend to be garishly decorated with PTMs. Keep in mind that that most likely # of S/T/Y+phosphoryl acceptors in any gene product is 0 (followed by 1, 2, 3, 4 ...).
Thu Jun 30 12:14:12 +0000 2022Modification diagram for human MMP3:p, a monomeric collagenase. The proprotein (20-477) is generated by the co-translational removal of the N-terminal signal peptide (1-19). The proprotein is most abundant in fibroblasts & tissues that contain them. 🔗
Thu Jun 30 10:59:24 +0000 2022Precipitable water vapour & surface winds, showing the extent of the Indian monsoon & Tropical Storm Chaba churning away in the South China Sea. 🔗
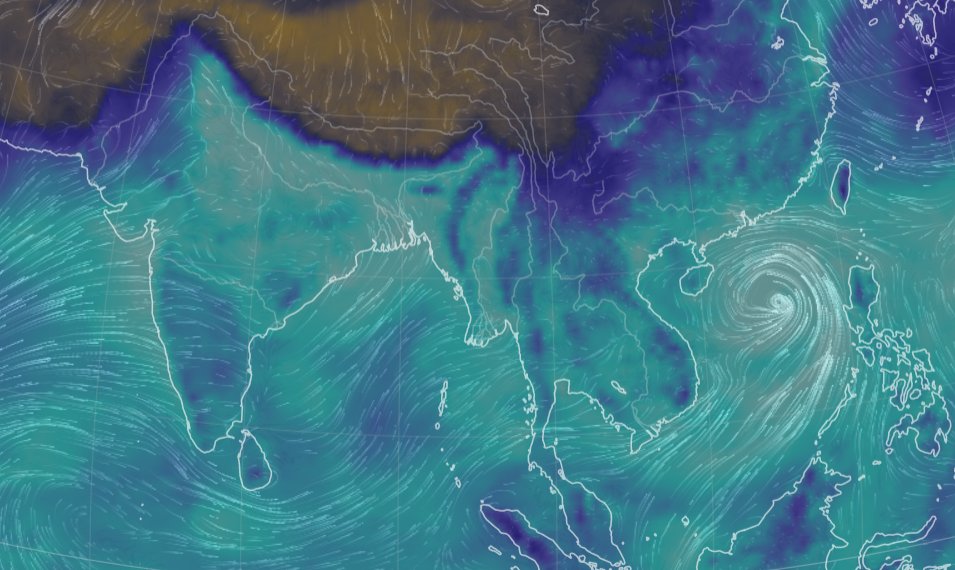
Wed Jun 29 19:48:09 +0000 2022I don't think anyone has ever deliberately studied XMRV proteins via MS/MS proteomics, but it doesn't seem to stop them from showing up as unwelcome guests.
Wed Jun 29 15:58:36 +0000 2022@ypriverol I'll look around.
Could you elaborate a bit on what you mean by "more phospho coverage"?
S/T+phosphoryl & Y+phosphoryl are also experimentally different sample preps, so are you more interested in S/T or Y?
Wed Jun 29 15:49:55 +0000 2022If you want a high-res data set from a human cell line (MHCC97H) to look for SAAVs, PTMs detectable without purification (K+GGyl, S/T/Y+phospho, R+dimethyl), viruses or which FBS proteins you might expect to find, PXD032201 would be a good choice.
Wed Jun 29 15:49:29 +0000 2022Normally I find data sets constructed specifically for bioinformatics purposes to be not so great, but PXD032201 is the exception that proves the rule.
Wed Jun 29 14:10:04 +0000 2022IMHO, this is not a great idea.
🔗
Wed Jun 29 12:17:57 +0000 2022The proprotein is found in fibroblast samples & a surprising (to me) amount of it in platelet samples. It is also frequently detected in extracellular exosomes in skin cancer/melanoma tissues.
Wed Jun 29 12:09:38 +0000 2022Modification diagram for human MMP1:p, a monomeric collagenase. The proprotein (20-469) is generated by the co-translational removal of the N-terminal signal peptide (1-19). The mature form (100-469) is produced upon secretion into the ECM.
🔗
Wed Jun 29 01:01:31 +0000 2022@neely615 It isn't me: I'm a big HGNC fan! I just noticed that with the new page format, the Un!Pr*t Recommended Name was much more prominent than the HGNC standard name, for instance 🔗
Wed Jun 29 00:48:01 +0000 2022PXD029368 has some pretty good examples of iodo-tyrosine PSMs from R. rattus tissue samples.
Tue Jun 28 17:17:26 +0000 2022@NCI_HRodriguez @theNCI @broadinstitute @JHUPress @PNNLab @IcahnMountSinai @MayoClinic @OHSUKnight @WUSTL @UPMC @bcmhouston @fredhutch Exited?
Tue Jun 28 15:56:42 +0000 2022@pwilmarth @chrashwood @MattWFoster Isn't CHIMERYS just a spinout of something generated by the Kuster mob at TUM at Garching (MSAID)?
Tue Jun 28 14:21:58 +0000 2022Un!Pr*t has finally moved over to their long promised new page layout. They also seem to be emphasizing their own naming taxonomy for proteins, as opposed to the standard HGNC gene names that many of us use.
Tue Jun 28 12:48:44 +0000 2022@ypriverol @ProteomeXchange Congrats to PRIDE for maintaining & expanding their offerings over more than a decade of service.
Tue Jun 28 12:46:26 +0000 2022@ypriverol @ProteomeXchange Whenever I have been at a meeting where people have talked about data repositories, I've noticed most people do not appreciate how challenging it can be to move around large numbers of large files, particularly with a collection that rapidly grows with time.
Tue Jun 28 12:29:20 +0000 2022This gene product uses K+GGyl/acetyl acceptors more than the other FAM83 proteins. The S/T+phosphoryl acceptors avoid the ordered FAM83 domain (16-283). 🔗
Tue Jun 28 12:22:37 +0000 2022Modification diagram for human FAM83H:p, a subunit of unknown function. It's C-terminal phosphodomain has a scheme similar to that in FAM83B/G: an extended region of short low complexity sequences & heterogenous S+phosphoryl acceptors.
🔗
Mon Jun 27 20:44:07 +0000 2022@Simon_Hammann @astacus Trypical, it will be. And it has me hankering for some hypertrypical peptides, too.
Mon Jun 27 16:38:00 +0000 2022I'm pretty sure no one should expound on Schrödinger’s Cat unless they know that "ganz burleske Fälle" does not mean "thought experiment"
🔗
Mon Jun 27 14:26:34 +0000 2022In light of the last few entries, I should repeat the trigger warning: I'm going through the FAM83A-H genes, whose mature, translated forms may contain examples of non-canonical PTM patterns. If this sort of thing upsets you, please look away.
Mon Jun 27 13:49:29 +0000 2022Some may see a mess, where others see opportunity … 🔗

Mon Jun 27 13:22:57 +0000 2022If you want to hear a minister responsible for post-secondary education say ridiculous things about post-secondary education, tune in to 🔗
Mon Jun 27 12:14:58 +0000 2022The N-terminal S+phosphoryl acceptor is conserved in all FAM83 members, where it can be observed. The S+phosphoryl acceptors in the disordered "gap" in the FAM83 domain is also found in FAM83E/F:p 🔗
Mon Jun 27 12:08:34 +0000 2022Modification diagram for human FAM83G:p, a subunit of unknown function. A region with multiple S+phosphoryl acceptors starts C-terminal to the FAM83 domain (15-308), which also contains multiple, short low complexity sequences.
🔗
Mon Jun 27 00:06:02 +0000 2022@goodlettlab1 🔗
Sun Jun 26 22:55:31 +0000 2022@gurugav69 @mjmaccoss @UCDProteomics @briansearle @LewisGeer @chrashwood @riley_nm1 @Smith_Chem_Wisc @J_my_sci @HaynesLabMQ Yup, that is definitely more like it.
Sun Jun 26 19:18:59 +0000 2022PXD029747 has some good examples of the human herpesvirus 4 (Epstein-Barr virus) proteins you can expect to find in samples derived from lymphoblastoid cell lines (LCLs)
Sun Jun 26 14:13:12 +0000 2022@gurugav69 @mjmaccoss @UCDProteomics @briansearle @LewisGeer @chrashwood @riley_nm1 @Smith_Chem_Wisc @J_my_sci @HaynesLabMQ Thanks for the list: they were interesting. I was hoping for something hypothesizing what the transition state might look like. I can imagine how it might look for something like S-X-S, but I'm having trouble with other sequence scenarios, e.g., S-S or S-X-X-S.
Sun Jun 26 13:13:49 +0000 2022Tropical Storm Celia has been stalled just off the Pacific coast of Mexico, sitting almost stationary for the last 10 days. 🔗
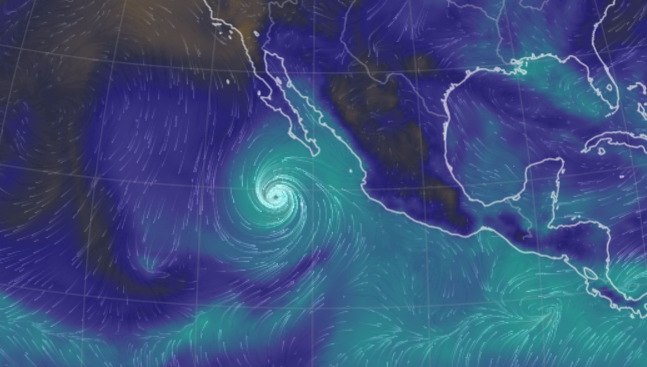
Sun Jun 26 12:31:35 +0000 2022Modification diagram for FAM83F:p, a protein of unknown function that is absent from many cell lines. Its S+phosphoryl follows a similar pattern to FAM83E:p, but FAM83F:p has some K+GGyl, which is spotty in FAM83.
🔗
Sat Jun 25 22:33:47 +0000 2022@gurugav69 @mjmaccoss @UCDProteomics @briansearle @LewisGeer @chrashwood @riley_nm1 @Smith_Chem_Wisc @J_my_sci @HaynesLabMQ Which ones would you recommend reading?
Sat Jun 25 20:48:21 +0000 2022@mjmaccoss @UCDProteomics @briansearle @LewisGeer @gurugav69 @chrashwood @riley_nm1 @Smith_Chem_Wisc @J_my_sci @HaynesLabMQ Has anyone ever figured out the chemistry associated with the rearrangement?
Sat Jun 25 17:19:48 +0000 2022@astacus @LewisGeer I realize that many people in the proteomics biz have a positive feeling about the deamidation work from the Robinsons, you really should take a good look through their web site (🔗) if you want to assess their credibility, in general.
Sat Jun 25 16:26:07 +0000 2022I really wish you Americans would knock this stuff off. Canadians tend to be insufferably smug at the best of times & this is just throwing gasoline on that particular dumpster fire.
Sat Jun 25 16:23:20 +0000 2022Some spicy weather right now, in and around Chicagoland along a cold front (NEXRAD). 🔗
Sat Jun 25 16:21:01 +0000 2022I'm no expert in monsoons, but this one looks like a bit of a beast. Dark to light blue indicates total precipitable water & vectors are surface winds. 🔗
Sat Jun 25 16:00:47 +0000 2022@astacus @LewisGeer I ran a peptide synthesis outfit at NYU Med School & a peptide/protein characterization lab at Eli Lilly, which gave me some practical grounding in the weird world of peptides in solution.
Sat Jun 25 15:54:31 +0000 2022@astacus @LewisGeer Sure do. It is interesting in general, but because of the way peptides get treated during sample prep (including charge-based separations) it can't give you anything more quantitative than trends. They also missed out on what happens when the N or Q is at a peptide terminus.
Sat Jun 25 15:26:55 +0000 2022@chrashwood @byu_sam @LewisGeer @mjmaccoss @kusterlab I have tried it, but it isn't a particularly useful approach. It is far better to grab an unfractionationed blood plasma data set & focus on ALB or TF peptide assignments.
Sat Jun 25 15:13:12 +0000 2022@astacus @LewisGeer They are all non-stochastic ratios set at observation time, with some contribution from computational errors. Their specific values are unknown, but their trends are pretty easy to suss out of the data.
Sat Jun 25 15:08:22 +0000 2022@astacus @LewisGeer N:Q deamidation isn't the only ratio I pay attention to for QA purposes. M:W oxidation, M:M & W:W oxidation:dioxidation and K:K carbamyl:acetyl (when available) can be very useful too.
Sat Jun 25 13:50:56 +0000 2022@ProteomicsNews Try blocking some keywords. It works wonders.
Sat Jun 25 13:46:29 +0000 2022@astacus @LewisGeer To be responsive, for a randomly selected LC/MS/MS run from the phosphoryl-enriched data, the deamidation ratio of NG:NV is 530:0, while for NG:NA is 530:21.
Sat Jun 25 13:25:00 +0000 2022@astacus @LewisGeer N-X-S/T is also a standout in deamidated PSMs for any organism that makes use of canonical N-linked glycosylation.
Sat Jun 25 13:10:51 +0000 2022@astacus @LewisGeer You can see a similar effect in individual fractions of a study that uses ion-exchange chromatography, where the acidic fractions will have many more deamidated PTMs than the basic fractions.
Sat Jun 25 13:09:08 +0000 2022@astacus @LewisGeer Yep, that is the way things work. -NG- dominates, followed by N or Q at the N-terminus of a peptide. Phosphopeptide affinity methods (e.g. TiO₂) usually favour acidic residues, so there is some enhancement of deamidated products.
Sat Jun 25 12:48:40 +0000 2022@LewisGeer @astacus There is no way to know, without site specific reaction rates for all of the conditions the sample was held at so you could do a proper integral across time. But it is a fixed ratio for a data set, frozen at the time of observation. The bulk ratio of unmodified N:Q ≈ 1:1.4
Sat Jun 25 12:31:43 +0000 2022Unfortunately, the (82-105) domain doesn't violate the general rule that S/T phosphorylation doesn't occur in regions with 2°+3° structure: the AF structure contains a disordered domain smack in the middle of the FAM83 domain 🔗
Sat Jun 25 12:23:48 +0000 2022Modification diagram for human FAM83E:p, a protein of unknown function. The S/T+phosphoryl pattern rhymes with the other family members, although the phosphodomain (82-105) appears to violate the boundaries of the N-terminal FAM83 domain (19-290)
🔗
Sat Jun 25 12:18:28 +0000 2022@astacus @LewisGeer N:Q is also a handy measure of the reliability of a subset of assignments, e.g. K+acetyl or SAAVs. Other ratios, like M:W oxidation, can also be used for the same purpose.
Sat Jun 25 12:12:04 +0000 2022@astacus @LewisGeer Not a great way to put it. I use deamidation as an indicator of how well a set of PSM assignment parameters is working by monitoring the ratio N:Q deamidation. For PXD022435, that ratio is about 6:1 for the phosphopeptide data & 4:1 for the proteome data.
Sat Jun 25 00:14:53 +0000 2022PXD034877: blast from the past or just some closet cleaning?
Fri Jun 24 20:53:16 +0000 2022@neely615 @ProtifiLlc Of the 70,516 LC/MS/MS runs I have with EIF3C in them, only 74 are from blood plasma. Of those 74 assignments, they are either very weak or data from a single paper (🔗) that probably is mislabeled as plasma.
Fri Jun 24 20:44:29 +0000 2022@neely615 @ProtifiLlc I'm in more of a Hotel California situation at the moment. Do you know the parent charge state for that MS/MS spectrum? I can't find any exemplar spectra for that peptide that look like that one.
Fri Jun 24 20:04:10 +0000 2022@byu_sam @LewisGeer @mjmaccoss @kusterlab It isn't suitable for testing algorithms. The data is quite messy & there was some confusion about which peptides were made.
Fri Jun 24 18:26:49 +0000 2022@LewisGeer The "proteome" data has a similar feature set, but with low res MS/MS.
Fri Jun 24 18:25:46 +0000 2022@LewisGeer If you really want a challenge, I'd recommend the phosphopeptide data from PXD022435. It has it all: superSILAC, low level carbamylation, easy to distinguish deamidation, modest amounts of M & W oxidation and good strong signals from iRT chromatography calibrant peptides.
Fri Jun 24 15:47:13 +0000 2022Correction: the YBX1:p peptide
132EDVFVHQTAIKKNNPR 147
also has GGylation simultaneously on both K142 & K143, but it is much more rarely observed.
Fri Jun 24 15:13:43 +0000 2022The RPS10:p peptide
130 SAVPPGADKKAEAGAGSATEFQFR 153
is the only case I know where two adjacent GGylation sites (K138 & K139) are frequently found to be both occupied simultaneously in a PSM.
Fri Jun 24 14:56:16 +0000 2022@astacus I can't "like" this, but I can't disagree with the sentiment.
Fri Jun 24 14:34:24 +0000 2022@astacus A bit of envy would be much better than the alternate universes approach we have now.
Fri Jun 24 14:01:19 +0000 2022One of the worst miscalculations made by funders in the last 20 years was to set up genomics & proteomics as competitors.
Fri Jun 24 12:47:15 +0000 2022@astacus @FrankSobott PS: Canadian passenger rail service (run by an odd type of entity called a "Crown Corporation") is so bad people in most of the country have forgotten that it exists.
Fri Jun 24 12:41:37 +0000 2022@astacus @FrankSobott I guess that is true: many of those stories had plot points that required the trains to run on time.
Fri Jun 24 12:33:36 +0000 2022@astacus @FrankSobott But it doesn't have the charming, "Sherlock-Holmes-being-pursued-by-Professor-Moriarity" vibe I always got on British trains.
Fri Jun 24 12:11:24 +0000 2022So who had "the discovery of a centrimetre-long bacteria" in the end-of-times sweepstakes?
Fri Jun 24 12:07:25 +0000 2022Modification diagram for human FAM83D:p, member of a family of proteins of unknown function. Nothing out of the ordinary here: a scattering of K+GGyl acceptors (but no K+acetyl) & S/T+phosphoryl acceptors avoid the structured FAM83 domain (20-296).
🔗
Fri Jun 24 02:17:24 +0000 2022Missed out on my chance to have a data set associated with trousers!
Fri Jun 24 01:54:13 +0000 2022rather than the correct reference 🔗
Fri Jun 24 01:53:46 +0000 2022Amusing (at least to me). In PXD033584, the PubMed ID is 1 digit short, pointing to 🔗
Fri Jun 24 00:37:36 +0000 2022🔗
Thu Jun 23 21:28:07 +0000 2022@neely615 @Smith_Chem_Wisc @riley_nm1 @J_my_sci @chrashwood @AlexHgO @wfondrie I would not have guessed "dodgy tech consulting on a sci fi TV space opera".
Thu Jun 23 20:34:42 +0000 2022🔗
Thu Jun 23 19:41:54 +0000 2022@neely615 The only explanation I could give would require an unfortunate event during sample prep or a wonky antibody.
Thu Jun 23 17:47:00 +0000 2022Sustainable animals? 🔗
Thu Jun 23 17:07:47 +0000 2022I just noticed that the Biomedical Research Centre at UBC has joined the list of institutes where I have worked but subsequently vanished, leaving a building to be administered by others.
Thu Jun 23 15:42:08 +0000 2022@Smith_Chem_Wisc @J_my_sci Keep everything at voltage as clean & smooth as possible. Anything that comes to a point (including microscopic dust, burrs, scratches) has the potential to emit electrons that generate discharges.
Thu Jun 23 15:12:14 +0000 2022@theoneamit @astacus @GarciaLabMS @dantyrr Two is one too many.
Thu Jun 23 14:55:56 +0000 2022@chrashwood That is correct: they leave that type of training to academia & expect PhD hires to act as internal experts of those subject.
Thu Jun 23 13:40:01 +0000 2022@macro_momo Not to mention writing: thousands of papers are published every week demonstrating that PIs could use access to basic & advanced courses in scientific writing & editing.
Thu Jun 23 13:15:20 +0000 2022@macro_momo I was thinking about things that would be useful, but are out-of-scope for particular expertise. E.g.: useful HR practices, copyright law in the context of academic publishing, current thinking on post-secondary teaching or best practices in student advising (undergrad v grad).
Thu Jun 23 12:53:56 +0000 2022In my experience, a major difference between working in pharma compared to academia is that pharma provides a lot of valuable training for its employees. Academia does not, even though it employs legions of potential trainers whose daily tasks are mainly, at best, busy work.
Thu Jun 23 12:00:28 +0000 2022Modification diagram for human FAM83C:p, a member of a protein family of unknown function. The main phosphodomain (318-572) is just C-terminal of the FAM83 domain (30-306). Like many FAM83 proteins, it is most commonly observed in ectodermal tissue/cell lines. 🔗
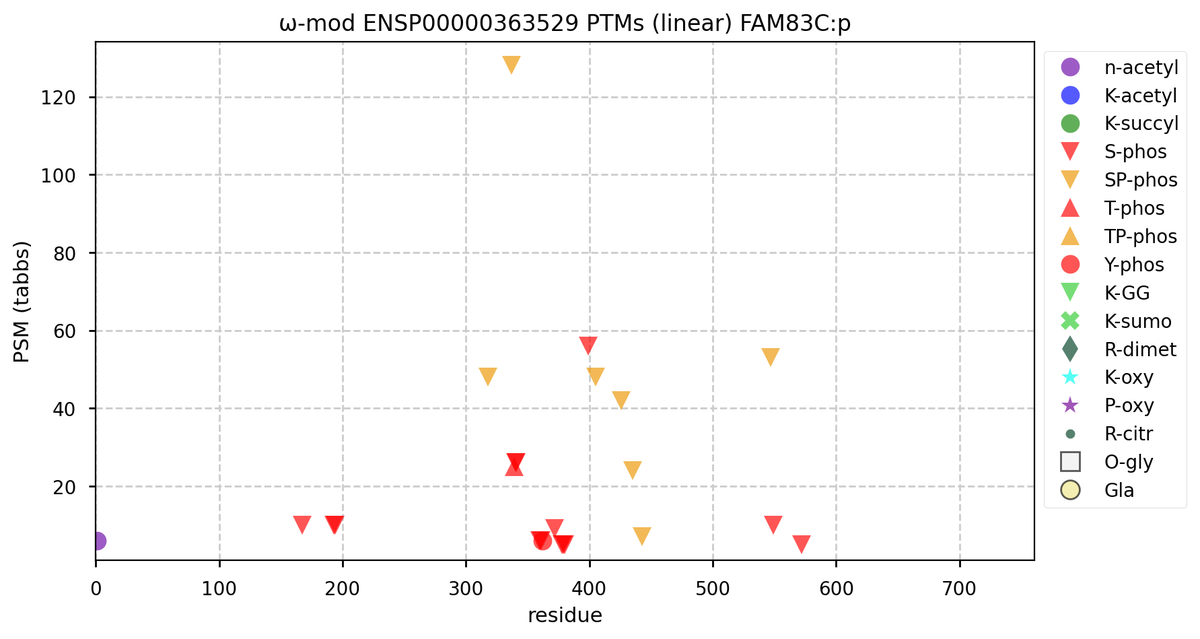
Wed Jun 22 14:33:31 +0000 2022@stateofthecity "The illusion of change is a good thing. Gives the people a jolt of hope. Makes people believe in the possibility of things... What we need is the illusion of change on the surface, continuity underneath." Tom Kane, from "Boss"
Wed Jun 22 14:09:22 +0000 2022The lineup on the street at the Passport Office across the street is only about 100 people right now.
Wed Jun 22 14:02:43 +0000 2022If my experiences are to be believed, it would appear that the unstated goal of internet search engine development has been to produce an abridged version of the ads in the Yellow Pages with just 1 page. Dynamite, innovative business model.
Wed Jun 22 12:12:32 +0000 2022Modification diagram for human FAM83B:p, a member of a protein family of unknown function. The FAM83 domain (15-281) has a lone K+GGyl acceptor, but the rest of the sequence is bullish on phosphorylation.
🔗
Wed Jun 22 01:52:18 +0000 2022@GeiszlerDaniel @UCDProteomics I've been all in with UniMod for at least a decade. But I have a fondness for clutter ...
Tue Jun 21 21:56:59 +0000 2022@chrashwood Negative mode, counterintuitively, is necessary for positively charged domains, not negatively charged ones. It is like the way tryptic peptides are predominantly acidic, even though you normally have a guaranteed basic group at the C-terminus.
Tue Jun 21 16:55:10 +0000 2022@chrashwood There are some protein sequence domains that are currently hard to see where it would be nice to have an alternate source of info, like sequences that are low complexity in K, R or H. But I don't know if that is a good enough argument to justify the expense.
Tue Jun 21 15:37:46 +0000 2022@doctorow I would add "librarians" to the list of early adopters of web tech, but their motivations may be different than the groups you focused on in this thread.
Tue Jun 21 15:14:18 +0000 2022@chrashwood Negative electrospray sparks like a SOB unless you are very careful and peptides with too few protons don't fragment as nicely as peptides with too many protons.
Tue Jun 21 15:08:48 +0000 2022@GoldbergTrader @davidmackau @internetofshit If you are interested, the findings linked to in this news release as well as the remarks of the privacy commissioner do a good job of explaining the details
🔗
Tue Jun 21 14:58:42 +0000 2022@CNPN_org Reading this made me a bit nostalgic, but I realized that I have forgotten who was the first president of CNPN, even though I was one of the founding members. John Bergeron, maybe?
Tue Jun 21 14:51:22 +0000 2022I am squarely on the 👻 side wrt eukaryote H+phosphoryl, although I find the formulation of the issue as a dichotomy a bit rigid given our tenuous understanding of most PTMs.
🔗
Tue Jun 21 14:40:46 +0000 2022S+phosphoryl decoration on a low complexity domain near residue 100 (in this case, 98-113) is found in most, but not all, FAM83 family mature subunits.
Tue Jun 21 14:10:18 +0000 2022This paper does a good job of explaining what is known about FAM83. It puts some effort into underlining the fact that they are not phospholipases (or pseudo-phospholipases), which I'm guessing is a bone of contention amongst the cognoscenti..
🔗
Tue Jun 21 13:56:16 +0000 2022Trigger warning: I'm going through the FAMily with sequence similarity 83 (FAM83) genes this week, whose mature, translated forms may contain examples of non-canonical PTM patterns. If this sort of thing upsets you, please look away.
Tue Jun 21 13:19:14 +0000 2022@dexivoje @J_my_sci @TimothyKassis Even a bad RESTful style JSON API would be a much better vehicle for disseminating the information & a far more instructive excercise for the students involved. A clumsy, never-to-get-passed-beta-version web site is not.
Tue Jun 21 12:55:39 +0000 2022@dexivoje @J_my_sci @TimothyKassis While these sites could be training opportunities for students, unless the PIs know enough to really supervise the work, they tend to simply make the same basic mistakes that anyone would when confronted with the problem of storing & displaying this type of information.
Tue Jun 21 12:52:51 +0000 2022@dexivoje @J_my_sci @TimothyKassis May be it is just me, but I think that the elaborate web sites that experimental groups try to generate themselves in an effort to disseminate the information tend to be wasted effort.
Tue Jun 21 12:39:31 +0000 2022Modification diagram for human FAM83A, a subunit that is part of a family of proteins without known function. It has 4 main phosphodomains (2 on each end) in disordered regions & a central K+GGyl/SUMOyl domain in a structured region.
🔗
Mon Jun 20 22:21:18 +0000 2022@dexivoje @J_my_sci @TimothyKassis Most large data generation projects in proteomics have produced pretty dodgy (& often mislabeled) data, the main exceptions being a subset of the CPTAC projects & BioPlex.
Mon Jun 20 17:01:02 +0000 2022RETREG3:p (human) ― works better than it should, even on this VERY modest quality AF structure 🔗
Mon Jun 20 15:53:44 +0000 2022@AlexUsherHESA A dance-off judged by the Governor General & 2 Lt. Governors from neutral provinces.
Mon Jun 20 14:05:43 +0000 2022Monsoon! Shown are surface winds & total precipitable water (lighter blue mean more water in the air) 🔗
Mon Jun 20 13:06:44 +0000 2022WASHC5:p is the last of the normally included WASH complex subunits. WASH4P:g & WASH6P:g are officially classified as pseudogenes (& therefore not included), but there is very good evidence that both of these genes can produce observable concentrations of proteins.
Mon Jun 20 12:23:42 +0000 2022Proteomics publications often make authoritative sounding claims based on homebrew informatics methods that make me nervous.
🔗
Mon Jun 20 12:04:50 +0000 2022Modification diagram for human WASHC5:p, a subunit of the endosomal WASH complex. This sequence has a pattern of K+GGyl similar to WASHC4:p, but without the associated K+acetyl alternative modifications (with a single exception). 🔗
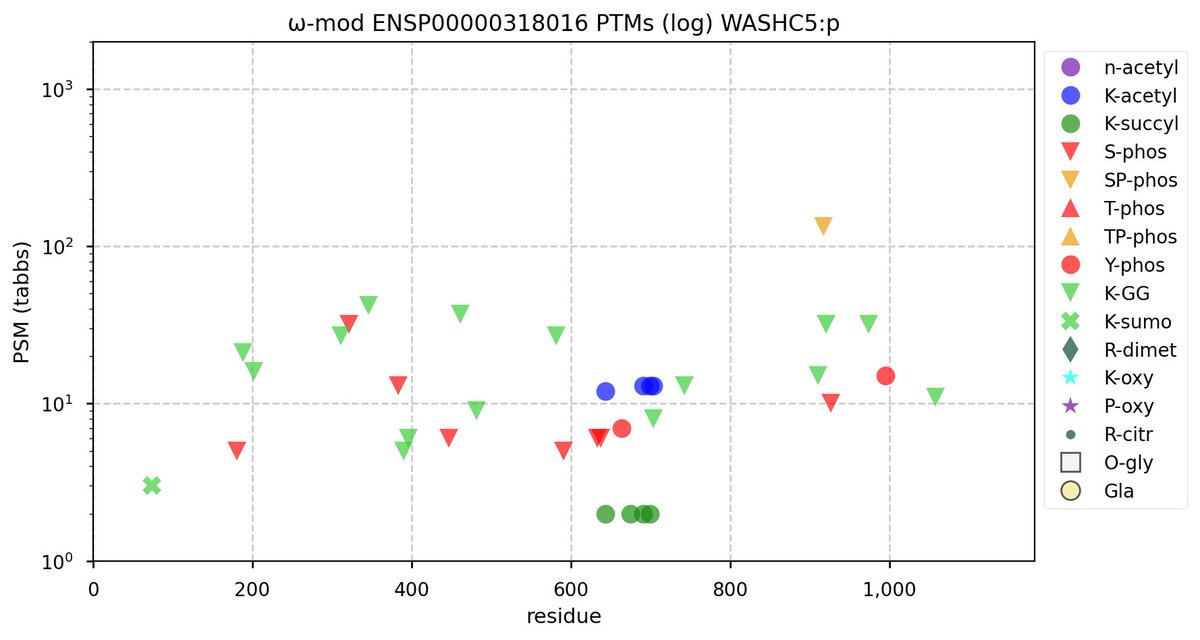
Mon Jun 20 11:43:00 +0000 2022Good edition of Canadaland today, about the weird world of policing academic conduct at Canadian universities 🔗
Mon Jun 20 01:06:11 +0000 2022The temperature gradient Winnipeg-to-Houston, summer-style. 🔗
Sun Jun 19 19:45:46 +0000 2022A low pressure system that has been slowly moving north up the Spanish coast (it is centered in the Bay of Biscay at the moment) continues to pump hot air up into Spain, France & Germany (winds @ 500 hPa) 🔗
Sun Jun 19 19:05:16 +0000 2022NIBAN1:p (aka FAM129A:p) ― more questions than answers. 🔗
Sun Jun 19 19:00:11 +0000 2022Reasonable Agreement by @doctorow 🔗
Sun Jun 19 18:09:26 +0000 2022I have to agree with this post 🔗 Even when people use the most fancy-pants-of-all-time super-Trypsin, they dump in some expensive but often not very good Lys-C, negating any possible advantage the super-Trypsin may have provided.
Sun Jun 19 17:27:39 +0000 2022After checking through some sites that classify proteins by function, it would appear that they get around the problem by simply making the substitution
"may be an XXX" ⟶ "is an XXX".
Sun Jun 19 15:52:17 +0000 2022One thing you realize if you read a lot of protein descriptions is that the modal verb "may" is wildly over-used. It must drive AI to all sorts of odd conclusions.
Sun Jun 19 14:34:33 +0000 2022Today I'm back to the service I'm paying for: 82 Mbits down, 8 Mbits up.
Sun Jun 19 13:50:26 +0000 2022Even though the Southern Ocean is often stormy, it seems to be particularly roiled today (shown: surface winds) 🔗
Sun Jun 19 12:31:52 +0000 2022Modification diagram for human WASHC4:p, a subunit of the WASH complex. This subunit has much more involvement with K+GGyl/acetyl than the other components of this complex. The C-terminal S/T+phosphoryl acceptors are in a low complexity domain. 🔗
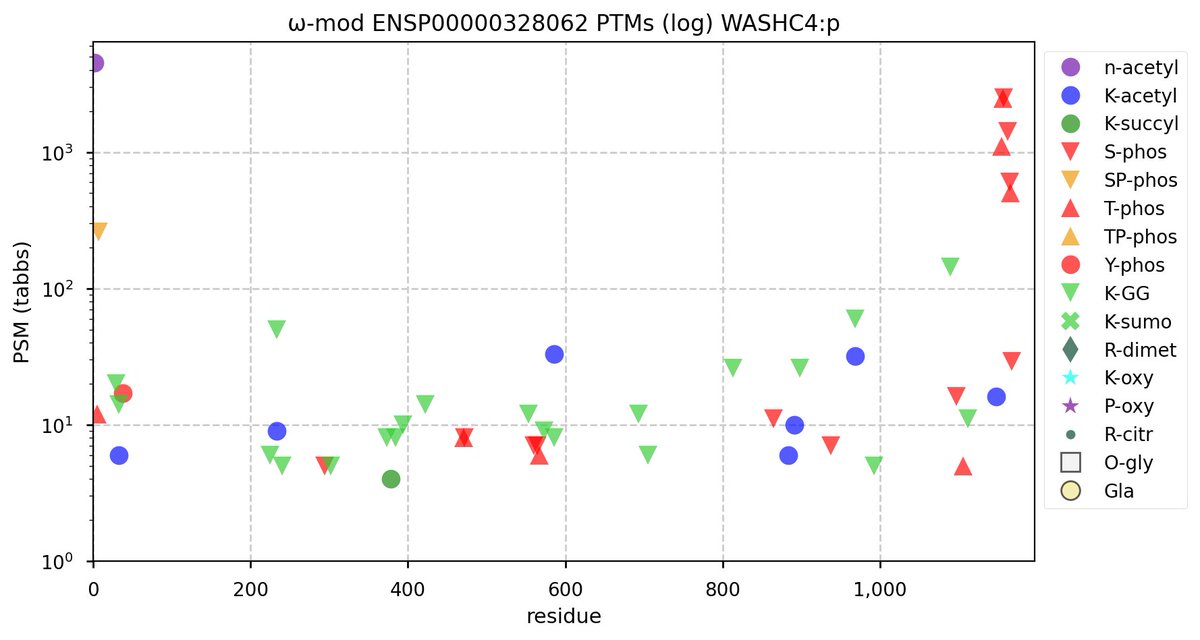
Sat Jun 18 15:38:27 +0000 2022@GarciaLabMS @BiswapriyaMisra @MetabolomicsSoc At a minimum they should supply some type of "all in" carbon foot print accounting for their shindig so that attendees/members would have a better idea of their impact.
Sat Jun 18 14:49:04 +0000 2022@GarciaLabMS @BiswapriyaMisra @MetabolomicsSoc It wouldn't hurt if conferences had to do some type of carbon offsetting, too. Many scientists pay lip service to GHG concentrations, but they somehow don't see themselves as agents of umweltverschmutzung.
Sat Jun 18 14:38:49 +0000 2022Hot up the middle & chill on the East coast today (winds @ 850 hPa) 🔗
Sat Jun 18 14:03:21 +0000 2022@MagnusPalmblad ... not that there is anything wrong with that!
Sat Jun 18 13:49:49 +0000 2022@MagnusPalmblad If you haven't built an MS yourself, it is hard to rise above simply being a knob-twiddler.
Sat Jun 18 13:48:12 +0000 2022I've learned more about parallel arm door closers in the last week than I imagined possible.
Sat Jun 18 12:44:03 +0000 2022Day #4: 1.1 Mbits down, 0.8 Mbits up.
Sat Jun 18 12:35:00 +0000 2022Heterogeneous S/T+phosphorylation acceptor sites serving as modulators within low complexity protein-protein interaction domains is a pretty common theme in eukaryotes.
Sat Jun 18 12:08:46 +0000 2022Modification diagrams for human & mouse WASHC3:p, a subunit of the endosomal WASH complex. The most obvious feature is a nice little patch of S+phosphoryl acceptors on the C-terminal protein-protein interaction domain. 🔗
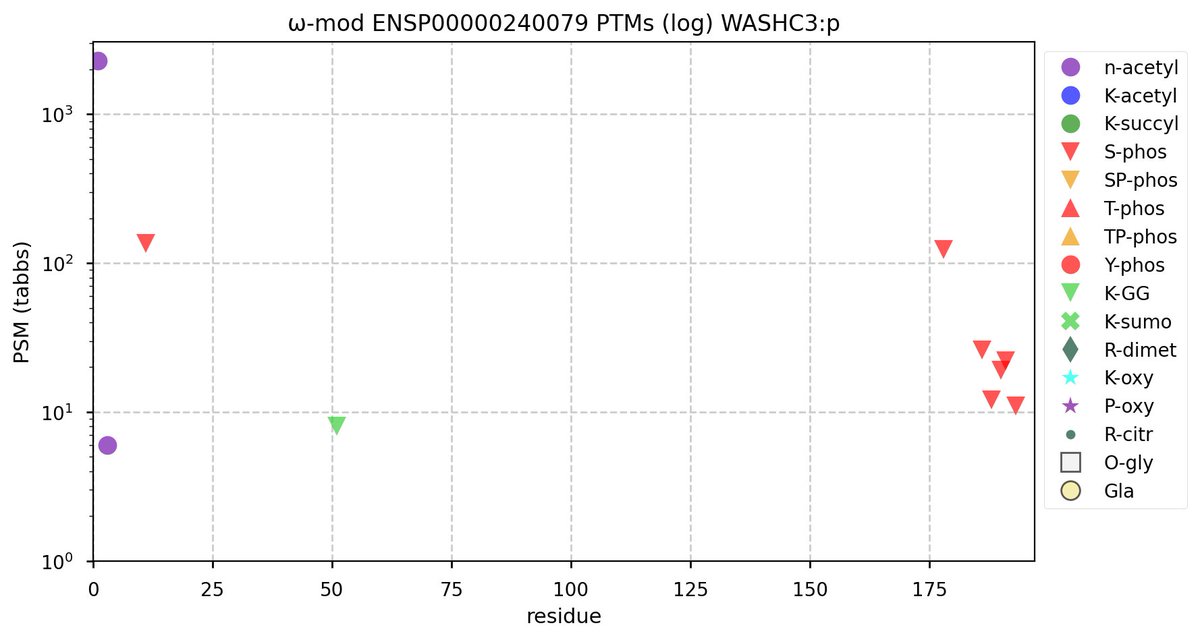
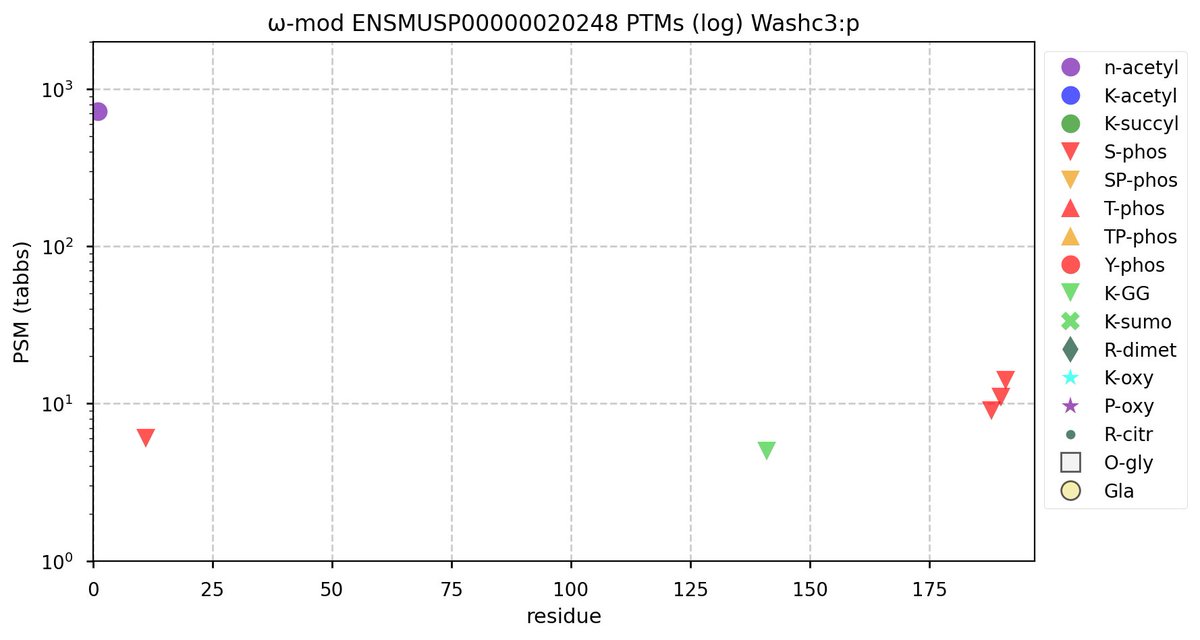
Fri Jun 17 15:39:43 +0000 2022Thanks to everyone who participated in the poll. It looks like a solid Canadian-style majority are still good with the phrase "proteotypic peptide", even though precisely what it means is a matter of some debate. 🔗
Fri Jun 17 14:50:48 +0000 2022🔗
Fri Jun 17 14:16:59 +0000 2022@slashdot Common charger legislation would be a welcome addition here in Canada.
Fri Jun 17 14:04:29 +0000 2022I suspect some sort of uniqueness constraint is what has led to the oddly curtailed list of phosphorylation sites in Un!pr*t (although you can never tell with those guys) 🔗
Fri Jun 17 14:00:41 +0000 2022For the third day this week, my ISP has slowed my home connection down to what I used to get out of a telephone modem.
Fri Jun 17 13:00:14 +0000 2022This particular pair can serve as a useful example when considering how to applying notions about peptide sequence uniqueness to phospho-proteomics data analysis.
Fri Jun 17 12:43:18 +0000 2022Most of the references in the literature to these 2 proteins use their older gene names, FAM21A & FAM21C.
Fri Jun 17 12:40:55 +0000 2022These paralogues are only present in primates very closely related to humans, e.g., gorillas. Every other species has a single gene, i.e., WASHC2:g. There is no "WASHC2B": while there was speculation of a third copy, it was demonstrated that "A" & "B" were the same gene.
Fri Jun 17 12:36:28 +0000 2022Modification diagrams for human WASHC2A:p & WASHC2C:p, paralogous subunits of the endosomal WASH complex. S+phosphoryl acceptors are the big story here. 83% of the tryptic peptides in these 2 paralogues are identical. 🔗
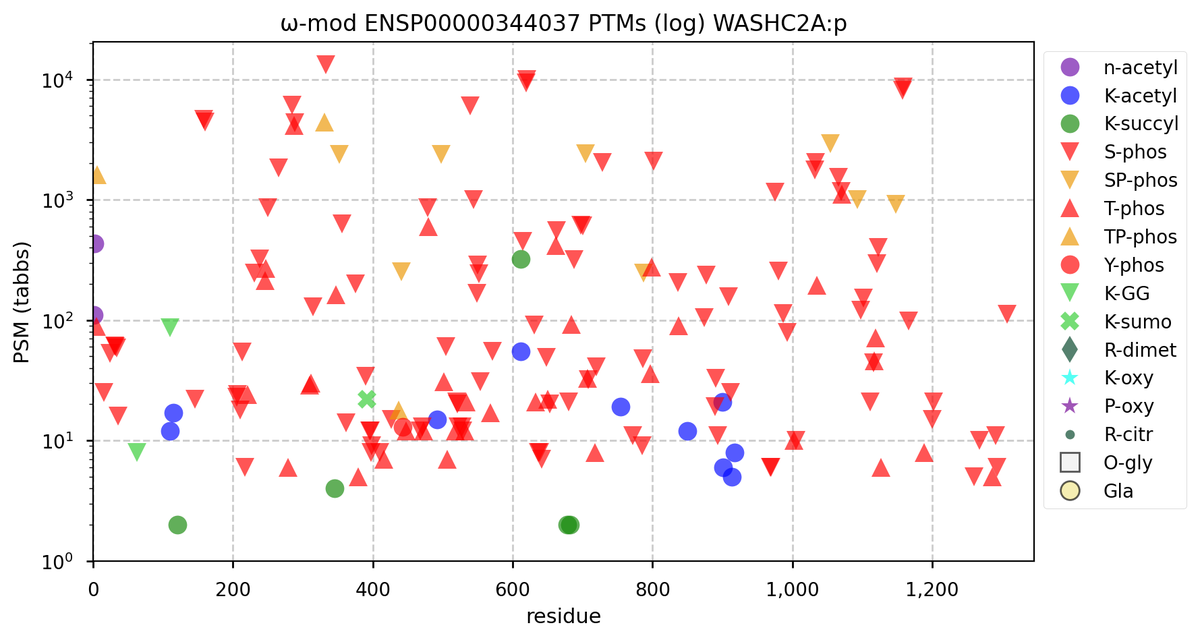
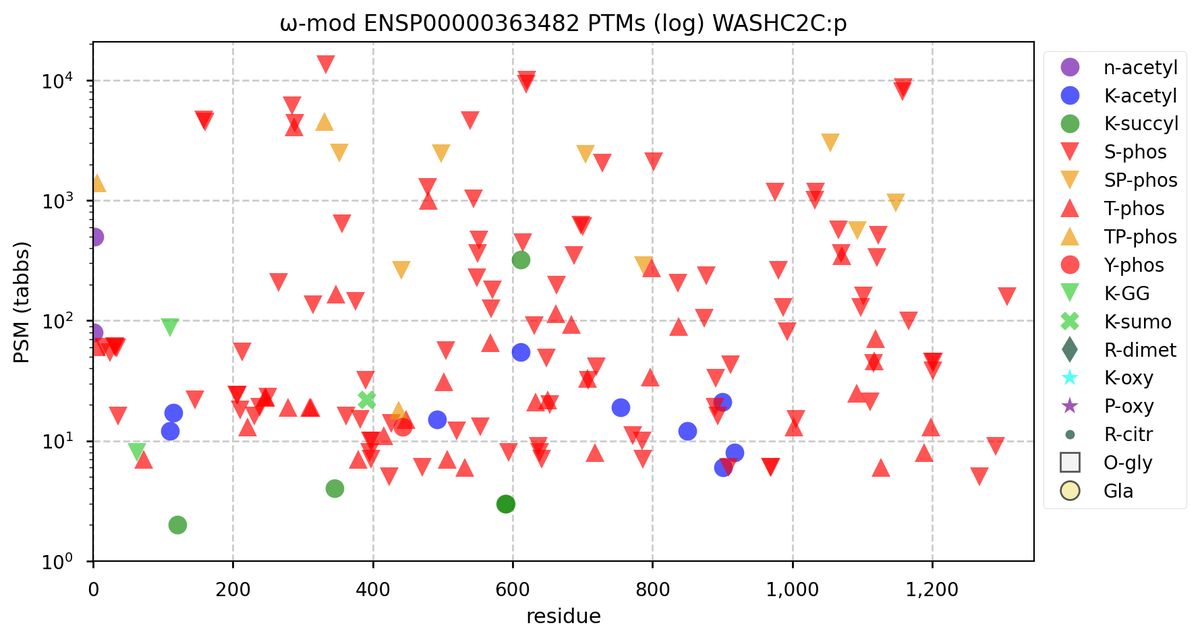
Fri Jun 17 12:21:07 +0000 2022Some intense weather driving south east through Indiana towards Kentucky after spending some time in northern Missouri & southern Illinois. 🔗
Thu Jun 16 20:53:42 +0000 20222 lines of T-storms roughing up the East, running from Columbus to Montreal & Athens to Harrisburg. 🔗
Thu Jun 16 19:28:04 +0000 2022@ypriverol @JohnRYatesIII Although I guess they are probably feeling quite a bit of heat from DeepMind moving into their backyard. I personally suspect they will stage a take over of UniProt's annotation functionality at some time in the near future.
Thu Jun 16 19:07:17 +0000 2022@ypriverol @JohnRYatesIII The only cases I know of are cases were the protein has a pharmaceutical use & it is necessary to fully characterize its properties for regulatory purposes.
Why would anyone at PDB care about this sort of thing?
Thu Jun 16 19:00:48 +0000 2022@dtabb73 Its definition has been bent, folded & mutilated by many, although I can't really put my finger on where it came from originally (probably sometime in the early aughts).
Thu Jun 16 17:59:19 +0000 2022@btaplatt It will require a graceful landing into a pool of Western Canada Select.
Thu Jun 16 16:39:32 +0000 2022Time to batten down the hatches, Ottawa. From the EC HRRR forecast via SpotWx. 🔗
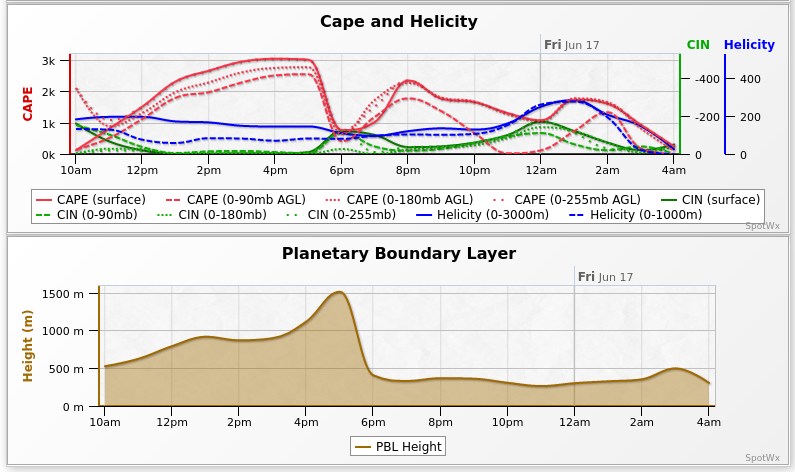
Thu Jun 16 15:30:21 +0000 2022Do people still take the phrase/concept "proteotypic peptide" seriously?
And if your answer is "Yes I do!" what do you believe it means?
Thu Jun 16 15:25:56 +0000 2022@AlexUsherHESA While I don't disagree, local Spanish politics makes any such comparison a potentially combustible subject.
Thu Jun 16 13:57:33 +0000 2022The endosomal WASH complex (a whimsical acronym for Wiskott Aldrich Syndrome protein and scar Homologue), connects the vesicles with the actin cytoskeleton. WASH complex subunits have had many names, making it hard to associate the literature with current gene nomenclature.
Thu Jun 16 13:15:29 +0000 2022Monsoon! Blue to cyan indicates increasing TPW (total precipitable water) in the atmosphere 🔗
Thu Jun 16 12:33:27 +0000 2022Modification diagram for human WASHC1:p, a subunit of the WASH complex. The modifications are sparse & nicely isolated. Their functions could be investigated using methods like alanine scanning or differential proteomics.
🔗
Wed Jun 15 17:07:12 +0000 2022Now show me the same scenes with Charles Barkley in place of the rather petite model.
🔗
Wed Jun 15 16:41:07 +0000 2022@AJ_Brenes Give up? I have never found a way to winkle info about data sets out of authors.
Wed Jun 15 13:50:55 +0000 2022Two storms in the Pacific west of Central America, both of which are headed further out to sea 🔗
Wed Jun 15 12:54:44 +0000 2022DOCK10:p phosphorylation has a prominent disordered domain (1228-1440) that has order-disorder transition sites at both ends & an RIP in the middle. 🔗
Wed Jun 15 12:50:59 +0000 2022Modification diagrams for human & mouse DOCK10:p, a guanine nucleotide exchange factor. While DOCK10:p has more in common with DOCK6-9 than DOCK1-5, it really seems to be doing its own thing, with more involvement in K+GGyl/acetyl associated mechanisms. 🔗
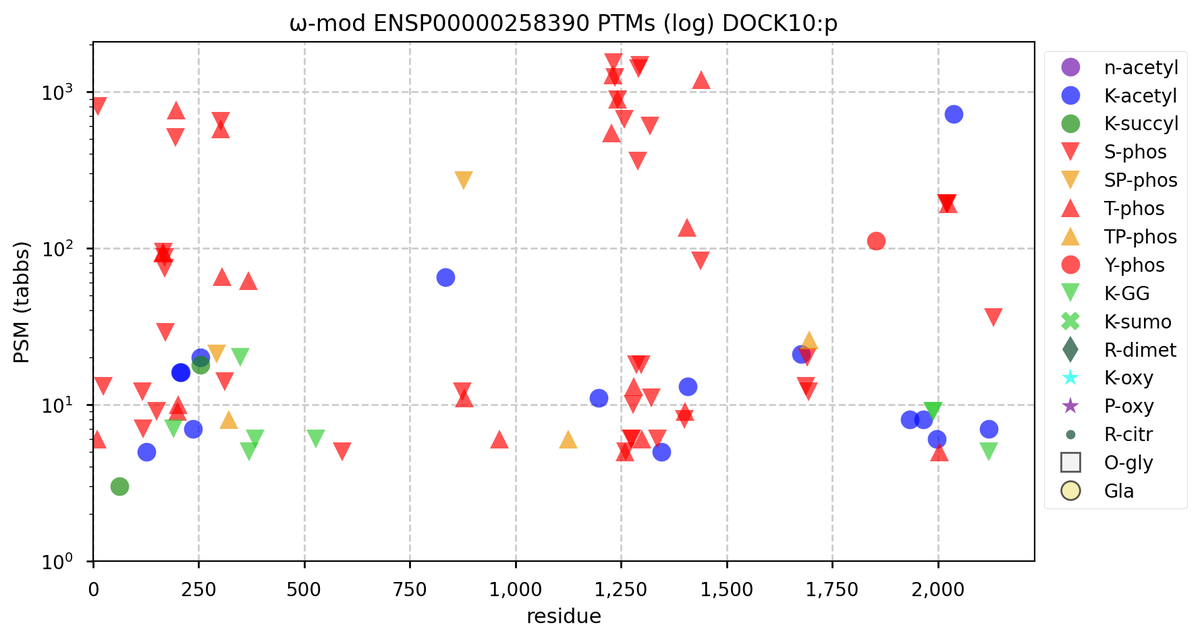
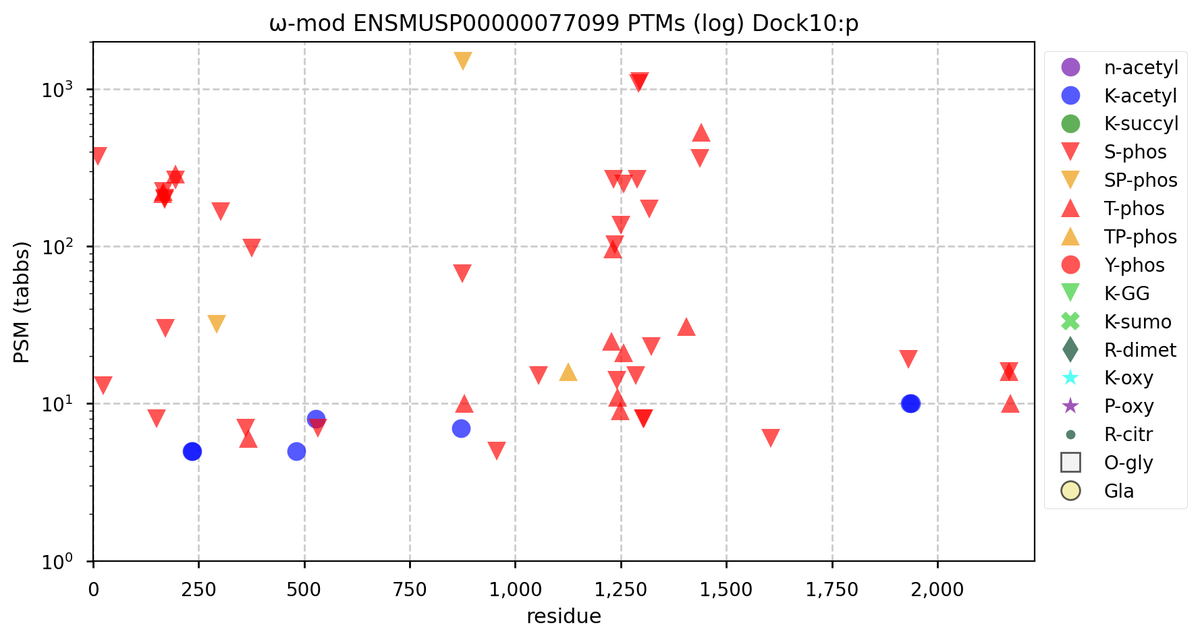
Wed Jun 15 12:03:07 +0000 2022@stateofthecity Heritage status for what were very ordinary commercial buildings when they were built is always suspect (& usually bad for the city).
Tue Jun 14 23:31:46 +0000 2022The 2007 monkeypox data (actually quite a bit of it) was from PNNL, part of a data set they called "Orthopox".
Tue Jun 14 23:30:12 +0000 2022But, will the monkeypox virus fill the void left by SARS-CoV2 overload?
Tue Jun 14 19:29:22 +0000 2022First Monkeypox data set I've seen since 2007―PXD034494 from the Jean Armengaud mob.
Tue Jun 14 19:22:37 +0000 2022@mjmaccoss @chrashwood Wouldn't argue. It was an odd study with lots of politics. I didn't see the manuscript until it was published.
Tue Jun 14 18:36:05 +0000 2022@chrashwood I'd forgotten all about that one.
Tue Jun 14 13:12:35 +0000 2022Frick'n storms. 🔗

Tue Jun 14 13:02:02 +0000 2022@GeiszlerDaniel You will have to check with the AlphaFold guys: their text descriptions are a little vague. But the notion that phosphorylation avoids regions of "green" seems to be fairly general & it applies to many rather broad domains. On the flip side, GGylation loves the green. 🔗
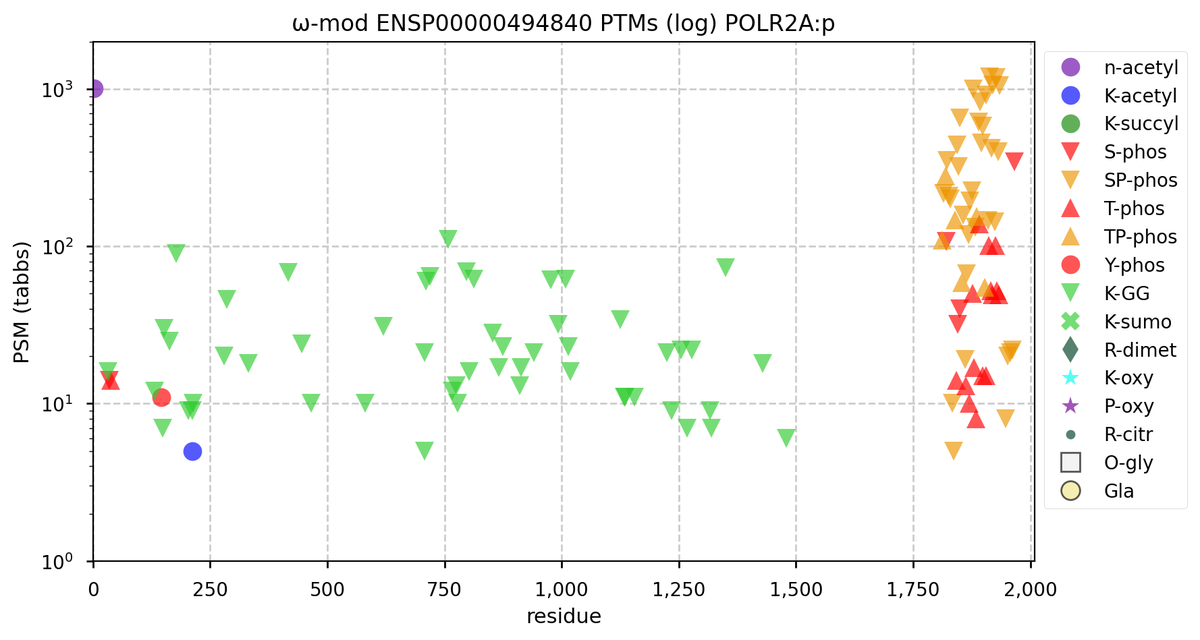
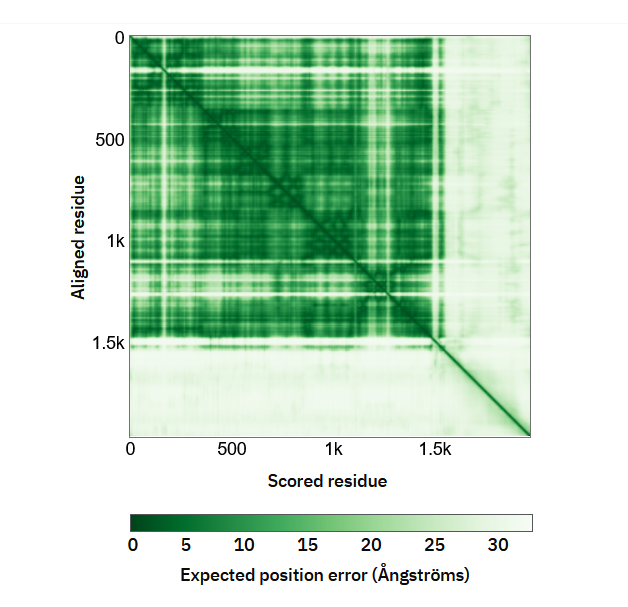
Tue Jun 14 12:19:07 +0000 2022Modification diagram for human DOCK9:p, a guanine nucleotide exchange factor. The pattern rhymes with the DOCK8:p PTM distribution, mainly composed of phosphorylation with very little participation in K+acetyl/GGyl/SUMOyl mechanisms. 🔗
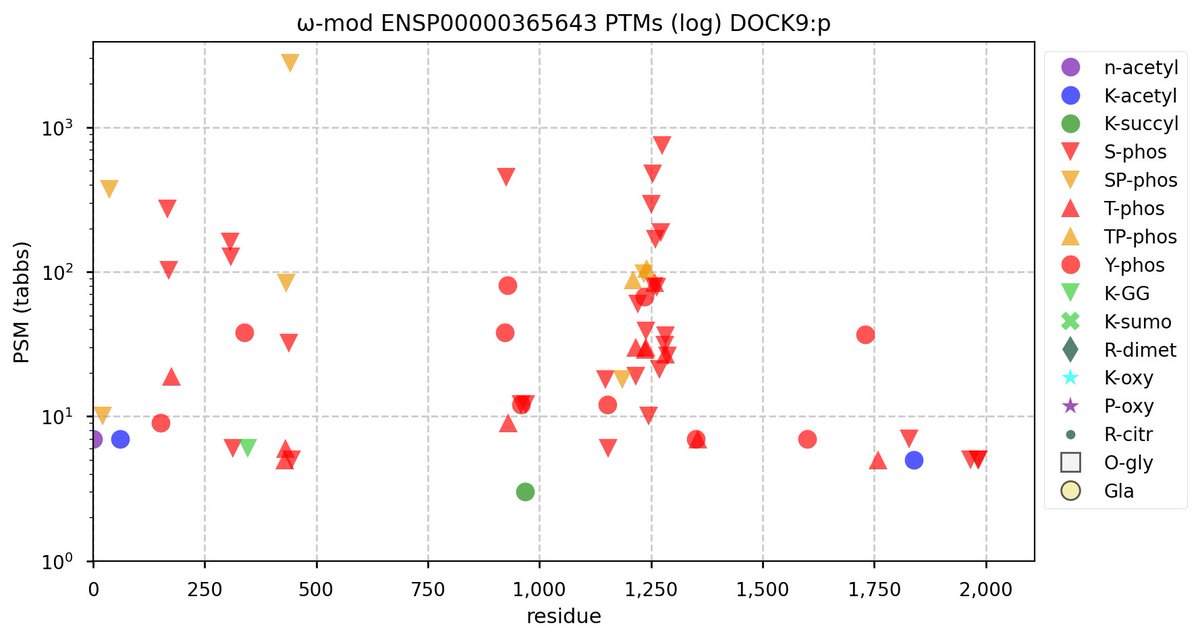
Mon Jun 13 18:32:22 +0000 2022@GeiszlerDaniel In this particular case, they are relatively narrow, although the one centered at 900 is about 100 residues across. They can be quite a bit more extensive, for example human SRRM1:p 🔗
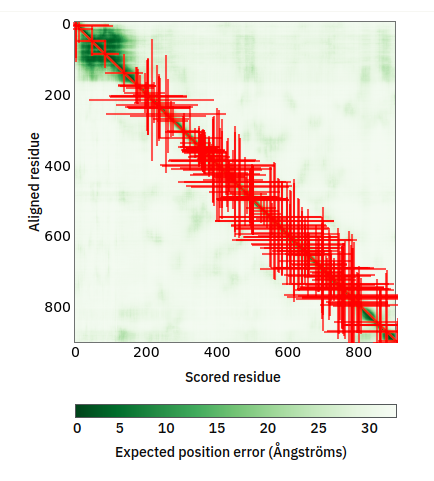
Mon Jun 13 17:49:21 +0000 2022@GeiszlerDaniel Ordered on one side & disordered on the other.
Mon Jun 13 17:01:55 +0000 2022@MattWFoster @neely615 @andy___jones @J_my_sci @r_l_moritz @JohnRYatesIII @GarciaLabMS @LewisGeer @byu_sam @asmsnews @MagnusPalmblad Hospitality suites were probably more active hot zones. Any idea about masking/physical distancing in those rooms? I wasn't there so I don't know how these were managed.
Mon Jun 13 16:56:45 +0000 2022To be useful in the PTM context, I can think of 4 main types of transition domains. Using the domain symbols
α = alpha-helical
β = beta-strand
ω = disordered
and going from N-to-C-terminal, then the transition types would be
α-ω
β-ω
ω-α
ω-β
Mon Jun 13 16:06:05 +0000 2022Two hours later, the line is longer & quite a few folding chairs are in use.
Mon Jun 13 16:02:53 +0000 2022Is there any term/nomenclature commonly used in protein structural biology to describe what I am calling order-disorder transition domains?
Mon Jun 13 15:00:40 +0000 2022@CBCSamSamson @EDWinnipeg Who ever chooses these slogans really likes a good non sequitur.
Mon Jun 13 14:23:15 +0000 2022There are at least 100 people lined up on the sidewalk across the street this morning, waiting to get into the Passport Office. Haven't seen this type of lineup since the Harper administration.
Mon Jun 13 13:04:02 +0000 2022For those who may be interested, the AF predicted aligned error (green) overlaid with observed phosphorylation frequency (red), showing many acceptors to be at order-disorder transitions. 🔗
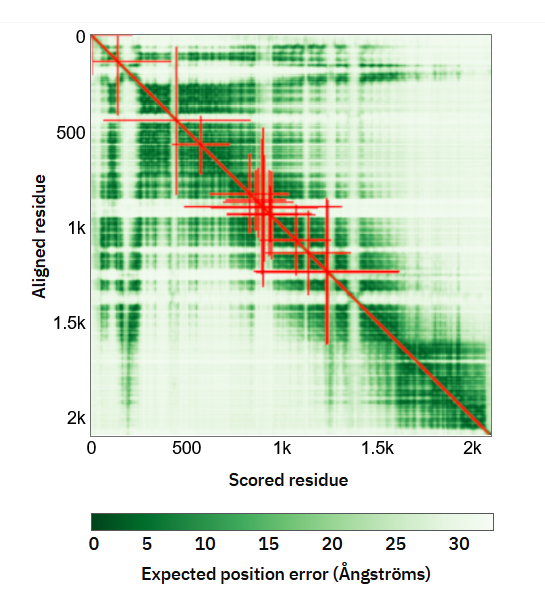
Mon Jun 13 11:53:47 +0000 2022Both the human & mouse patterns show a significant amount of alternate initiation (splicing?) that starts at T70+acetyl (initiation at M69) in both species.
Mon Jun 13 11:49:04 +0000 2022Modification diagrams for human & mouse DOCK8:p, a guanine nucleotide exchange factor. It has a PTM scheme different from that in the DOCK1/2/5, DOCK3/4 or DOCK6/7 groups, with significantly fewer S/T+phosphoryl acceptors in 4 iHOP domains. 🔗
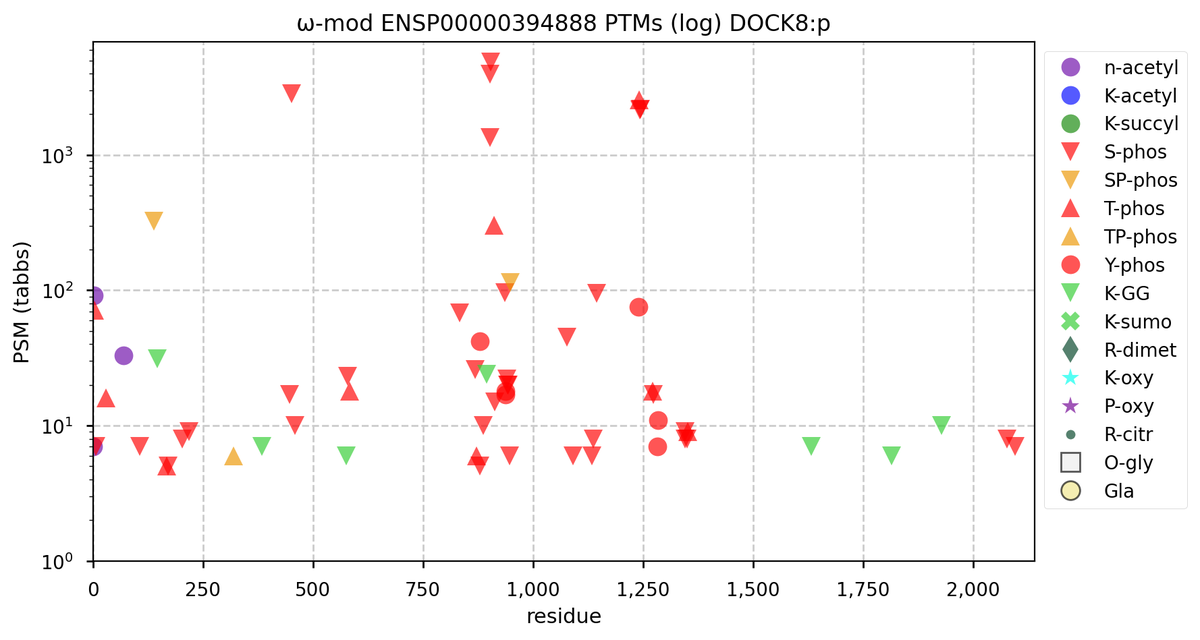
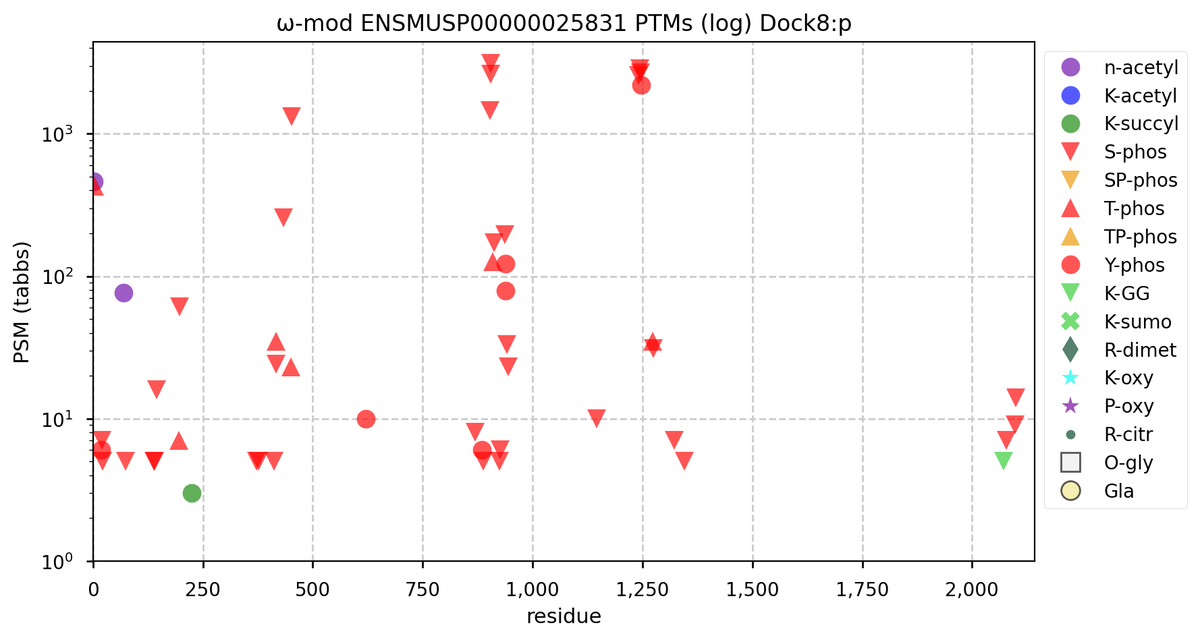
Sun Jun 12 21:46:16 +0000 2022@JoePlatelet I guess so, but it any but trivial cases it would take more manual checking than I have the resources to carry out.
Sun Jun 12 16:33:25 +0000 2022@J_my_sci I'm grabbing the AF predicted aligned error diagrams from EBI's resource via the UP accession, e.g.
🔗
Sun Jun 12 15:31:40 +0000 2022@J_my_sci I haven't found any good counter examples when there is a decent AF structure with enough secondary structure prediction to be probative.
Sun Jun 12 13:12:44 +0000 2022Events from the rarely discussed, but coldly disputed Canada-Denmark border
🔗
Sun Jun 12 13:02:38 +0000 2022@neely615 @SCwxFrankStrait Still (!) going as a weaker, but still churning, low pressure system in the Norwegian Sea. 🔗
Sun Jun 12 12:32:56 +0000 2022Shining a "phosphorescent" light on the portions of the proteome previously referred to as dark.
Sun Jun 12 12:29:13 +0000 2022The AF predicted aligned error plot (in green), with observed phosphoryl frequency (in red). The phosphorylation domains are on the disorder side of order-disorder boundaries. The exception to this is (863-970), which also has an RIP domain. 🔗
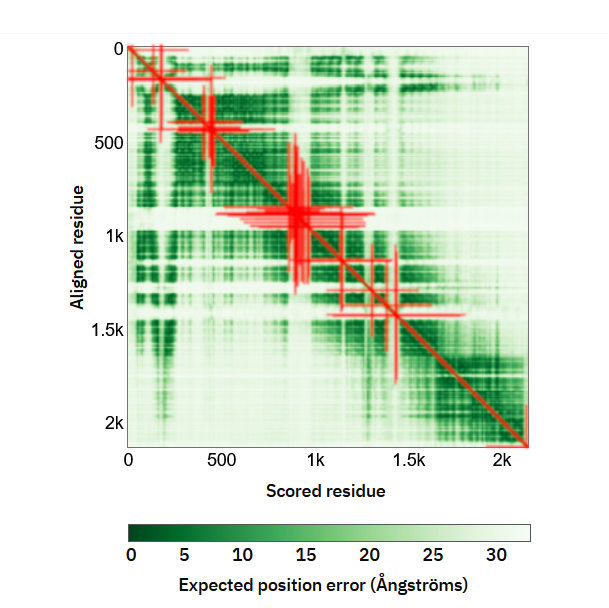
Sun Jun 12 12:21:23 +0000 2022Modification diagrams for human & mouse DOCK7:p, a guanine nucleotide exchange factor. The patterns rhyme quite nicely, they are consistent with the DOCK6:p pattern, & they are unlike DOCK1-5. 🔗
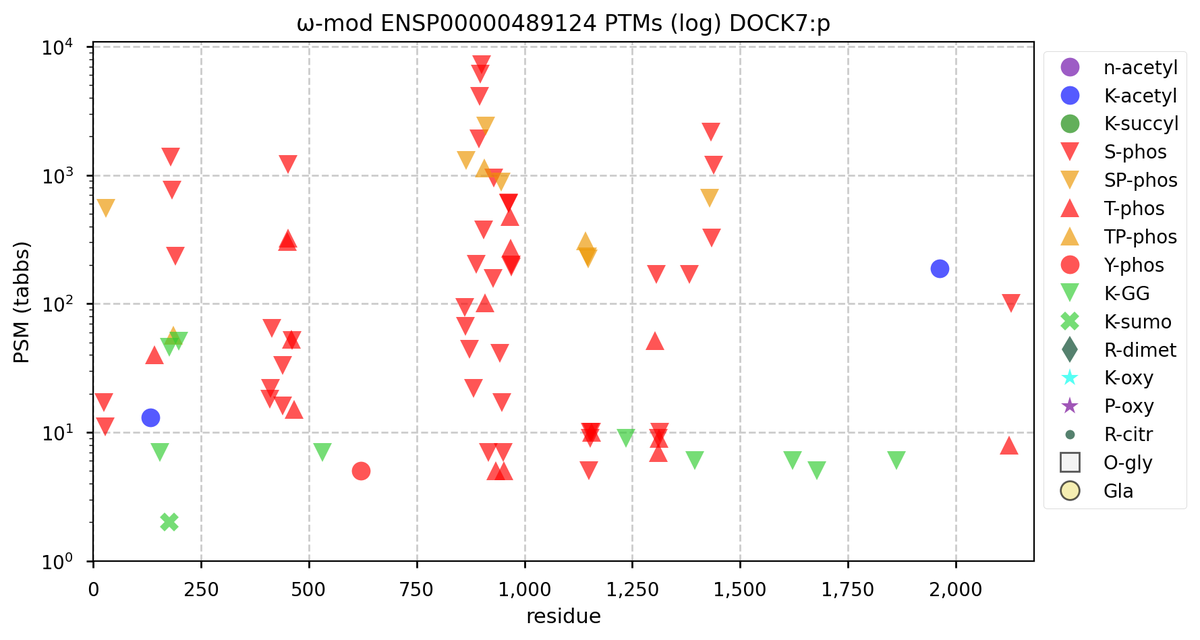
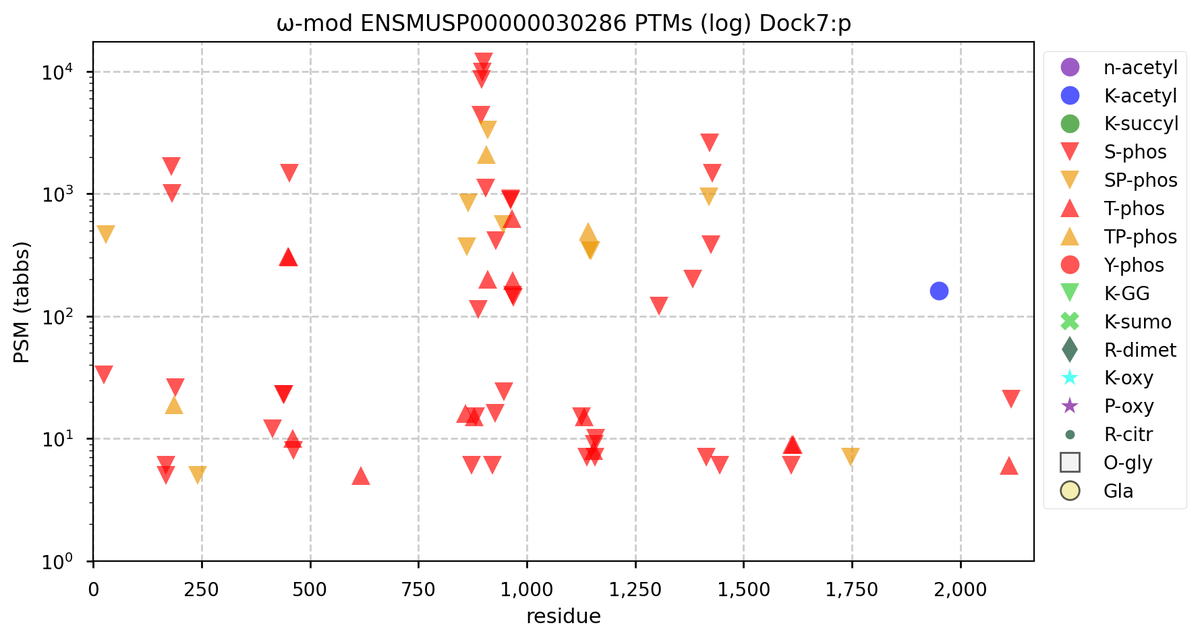
Sat Jun 11 22:27:53 +0000 2022@astacus But, if modifying a domain is purely just an artifact without consequence, it most likely wouldn't be evolutionarily conserved.
Sat Jun 11 22:02:30 +0000 2022@astacus Perfectly reasonable alternate hypothesis. As are general mechanisms like CDK phosphorylation/dephosphorylation or integrative mechanisms that require more than one site modified to have full effect.
Sat Jun 11 20:24:30 +0000 2022@ucdmrt @pwilmarth This Wikipedia entry sums up the normal usage quite well:
🔗
Sat Jun 11 18:25:48 +0000 2022@MattWFoster @UCDProteomics @AmandaLSmythers @NUProteomics @PMF_UNLBiotech @chemical_laura @BrukerMassSpec Mislabeled data files are something that I still find annoying (& often disquieting), but it is something you just have to learn to live with when doing large scale information generation.
Sat Jun 11 17:50:02 +0000 2022@UCDProteomics @MattWFoster @AmandaLSmythers @NUProteomics @PMF_UNLBiotech @chemical_laura @BrukerMassSpec I don't think it is even theoretically possible to produce a formal LIMS system that would be useful to enough proteomics groups to make it a viable product.
Sat Jun 11 17:37:03 +0000 20222. broad regions in disordered domains, often associated with low complexity sequences
hypothetical function: modulate protein/complex quaternary structures
3/3
Sat Jun 11 17:36:21 +0000 2022These phosphorylated regions can be divided into two main types:
1. narrow regions on the disordered side of order-disorder transition domains
hypothetical function: modulate changes in subunit tertiary structure
2/3
Sat Jun 11 17:35:58 +0000 2022The simplified theory of protein subunit phosphorylation:
Phosphorylated regions of a subunit are mainly in domains with no conventional secondary structure, e.g., no alpha helices or beta sheets
1/3
Sat Jun 11 14:18:22 +0000 2022I'm always flummoxed when I read a paper that used FDR as a measure to "tune" an algorithm or method.
𝘜𝘯𝘱𝘰𝘱𝘶𝘭𝘢𝘳 𝘰𝘱𝘪𝘯𝘪𝘰𝘯𝘴℠
Sat Jun 11 12:40:52 +0000 2022The AF predicted error plot with observed phosphorylation overlaid (in red) shows 10 phosphodomains, almost all in order-disorder transition domains on the disordered side. 🔗
Sat Jun 11 12:37:03 +0000 2022Modification diagram for human & mouse DOCK6:p, a guanine nucleotide exchange factor. This distribution does not resemble those of DOCK1, 2, 3, 4 or 5, with many iHOP domains & no C-terminal RIP domain. 🔗
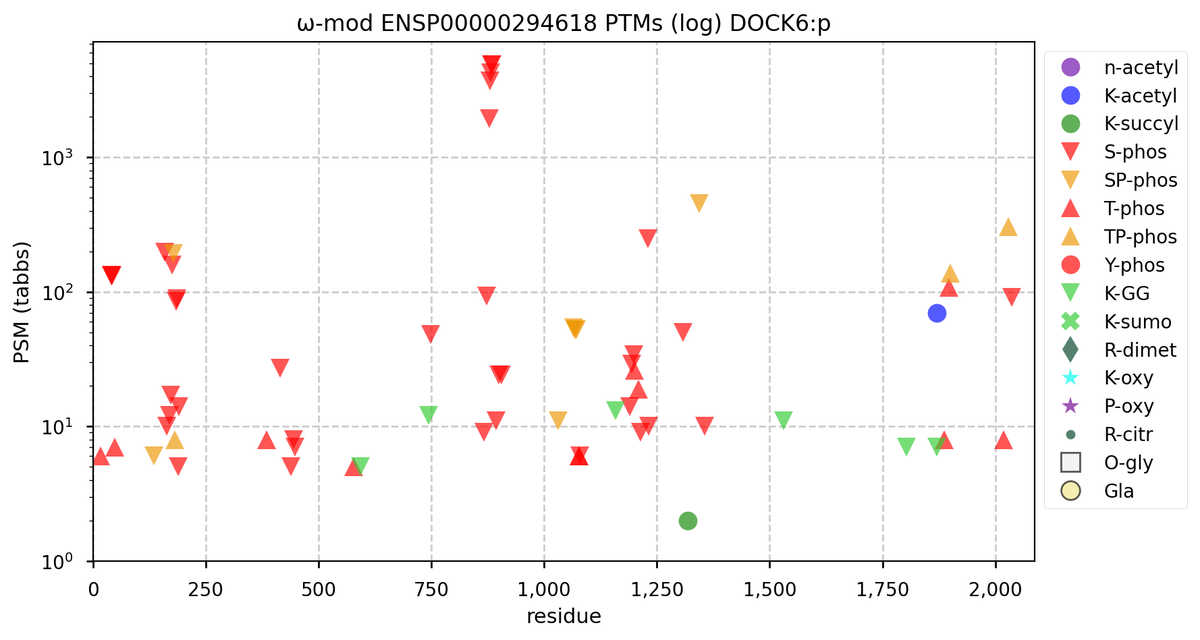
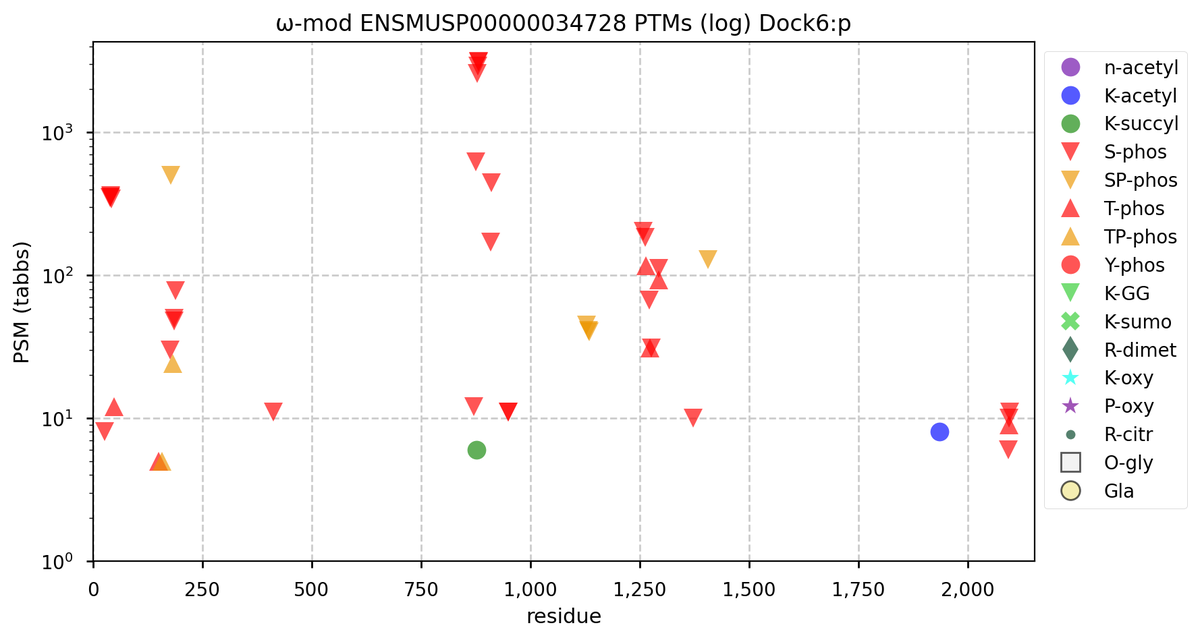
Fri Jun 10 16:41:26 +0000 2022@neely615 @SCwxFrankStrait Still going & about to pass from the North Atlantic into the Norwegian Sea. 🔗
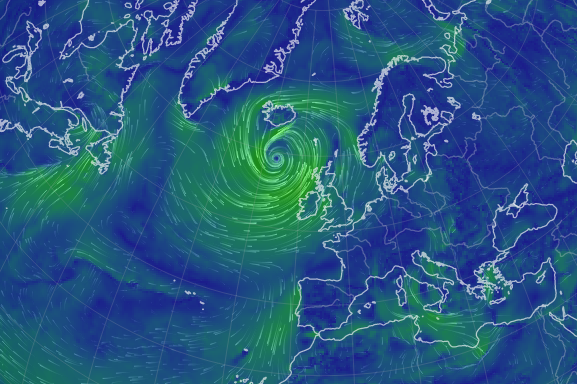
Fri Jun 10 16:35:36 +0000 2022Oh No!
🔗
Fri Jun 10 14:09:58 +0000 2022The AF predicted aligned error plot for human DOCK4:p with the observation frequency of S/T+phosphoryl (in red), showing that the iHOP domain (1608-1640) is just C-terminal to an order-disorder transition, while the RIP domain is disordered all the way. 🔗
Fri Jun 10 12:21:40 +0000 2022Modification diagrams for human, rat & mouse DOCK4:p, a guanine nucleotide exchange factor. All 3 show rhyming patterns, with an S+phosphoryl iHOP domain followed by a C-terminal RIP domain. 🔗
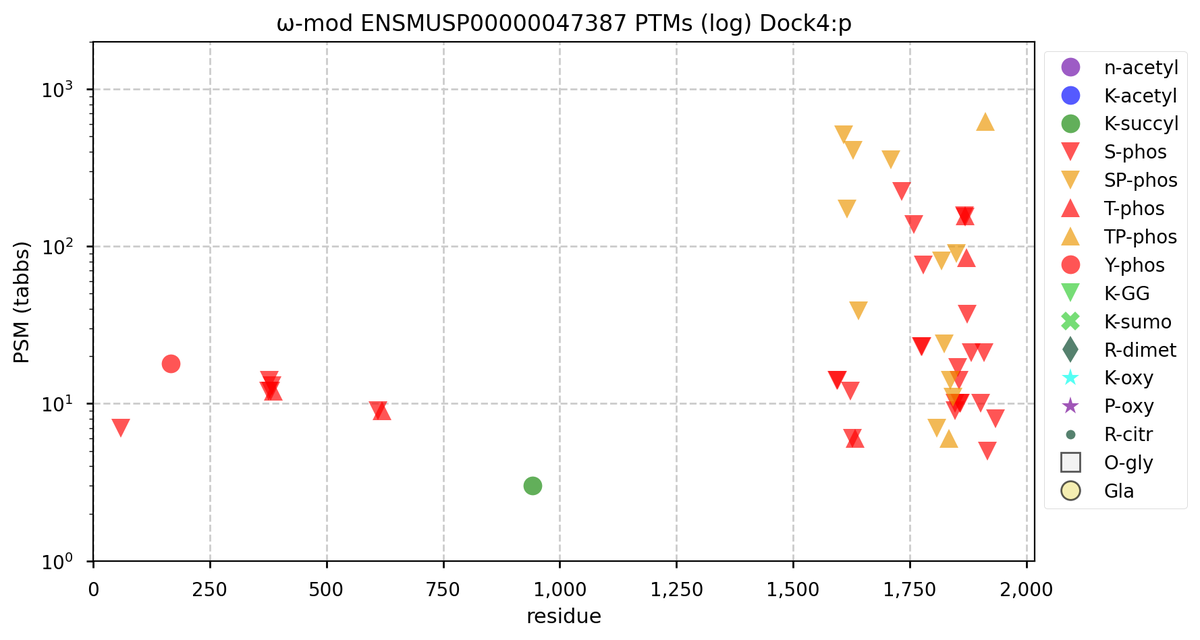
Fri Jun 10 01:22:51 +0000 2022@stateofthecity Americans are like what Canadians would be like if they had an unrelenting instinct for aggressively marketing all aspects of everything.
Thu Jun 09 23:08:35 +0000 2022And all of the things that vanished were govt. lab sponsored resources that swore on a stack of bibles that they were really permanent, reliable, never-going-to-go-away services.
Thu Jun 09 23:02:23 +0000 2022I was looking through some system code this afternoon & I was surprised by how many things were no longer used because information resources had come & gone during the last 15 years. Really underlined the value of inline documentation for me.
Thu Jun 09 18:08:15 +0000 2022@GeiszlerDaniel @OliverMBernhar1 @msaid_de Why would you write a search engine, other than for fun?
Thu Jun 09 18:01:38 +0000 2022I must say that I agree with this assessment
🔗
Thu Jun 09 15:36:50 +0000 2022@JoePlatelet Nope. I'll finish the DOCK-Bs tomorrow, start the Zirs on Saturday, to be followed by the Zizimins on Tuesday.
Thu Jun 09 14:36:33 +0000 2022@ucdmrt @TheCoxLab No. They don't work like that at all.
Note: this is one of my "𝘜𝘯𝘱𝘰𝘱𝘶𝘭𝘢𝘳 𝘖𝘱𝘪𝘯𝘪𝘰𝘯𝘴℠".
Thu Jun 09 12:26:43 +0000 2022@ucdmrt @TheCoxLab It still surprises (& annoys) me how accepted spectral libraries are in DIA analysis, while they are still treated with intense skepticism wrt DDA analysis.
Thu Jun 09 12:15:35 +0000 2022Modification diagram for human & mouse DOCK3:p, a brain tissue specific guanine nucleotide exchange factor. Like DOCK1/2/5:p, this subunit has a C-terminal S+phosphoryl acceptor RIP hotspot, although by comparison the DOCK3:p domain is broader. 🔗
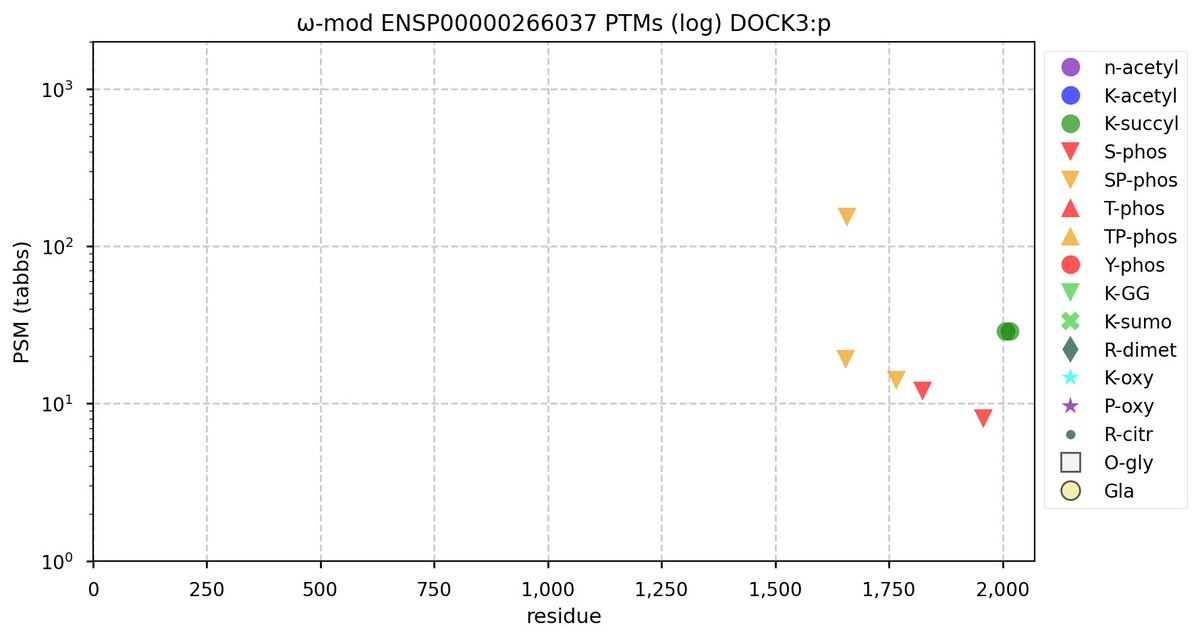
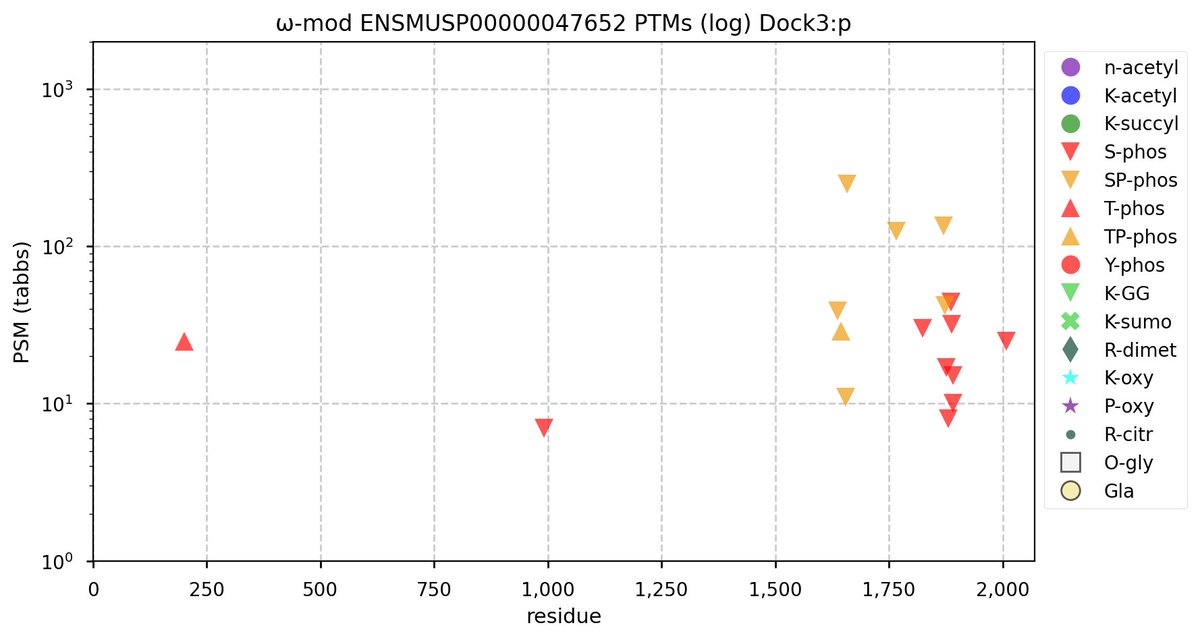
Wed Jun 08 15:06:26 +0000 2022I really don't like making on-line purchases of small items. Shipping is ridiculously expensive, purchase portals are squirrely & many payment completion services seem to be just trying to pump you for information.
Wed Jun 08 14:14:30 +0000 2022@neely615 @SCwxFrankStrait Agatha (reborn as Alex) is still churning away in the North Atlantic as a low pressure system, with its center about halfway between Newfoundland and Ireland. 🔗
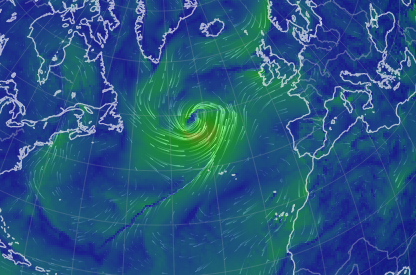
Wed Jun 08 14:08:24 +0000 2022Some spicy weather this morning across Oklahoma, Kansas, Arkansas, Alabama & Illinois 🔗
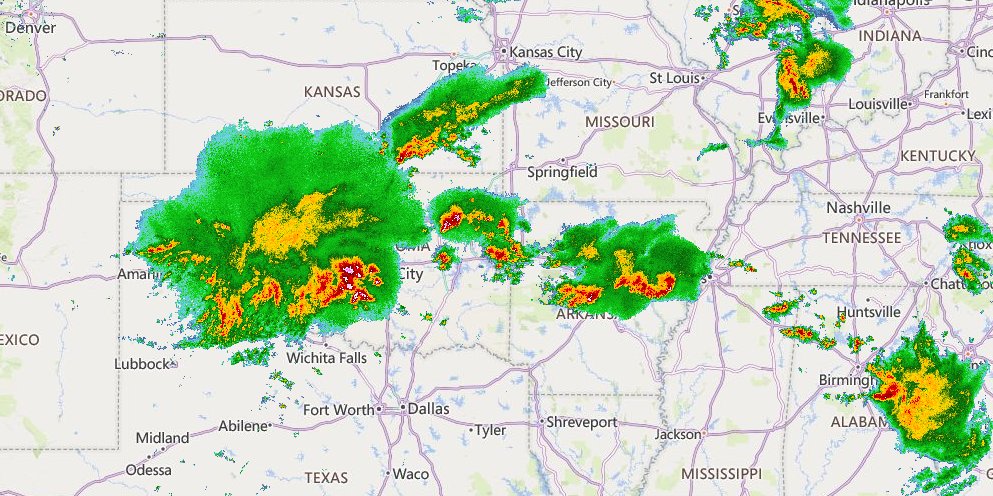
Wed Jun 08 13:58:05 +0000 2022As with DOCK2:p, the AF structure isn't very good, but the predicted aligned error plot with obs. phosphorylation overlay (red) shows the C-terminal RIP domain nicely avoids the dark green regions, while the iHOP acceptors occur in order-disorder transition domains. 🔗
Wed Jun 08 11:50:29 +0000 2022Modification diagram for human DOCK5:p, a guanine nucleotide exchange factor. Like DOCK1 & 2, it has a C-terminal RIP domain & scattered GGyl acceptor sites. Unlike DOCK1 & 2, it has observable K+acetyl
acceptors.
🔗
Tue Jun 07 14:38:40 +0000 2022The AF predicted aligned error plot for DOCK2:p with phosphoryl acceptor frequency of observations (log) overlaid in red. The AF prediction isn't very good, but most of the red still avoids the darker green areas, with iHOP domains nicely placed at order-disorder boundaries. 🔗
Tue Jun 07 14:33:47 +0000 2022Some of the iHOP acceptors would be suitable for alanine scanning, but the RIP hotspot probably wouldn't be detectable. I should note that MS/MS based differential phosphoproteomics expts don't work very well for RIP hotspots either.
Tue Jun 07 14:19:56 +0000 2022The usual crowd of about 75 lined up on the sidewalk in front of Passport Canada (across the street from my office). A daily reminder of why I don't travel any more.
Tue Jun 07 13:08:57 +0000 2022"Tera-scale" is a new term for me, although analyzing data sets with > 1 TB of data is rather old hat.
🔗
Tue Jun 07 12:29:22 +0000 2022Modification diagram for human DOCK2:p, a subunit that functions as a guanine nucleotide exchange factor. Like DOCK1:p, it has a scattering of isolated GGyl acceptors & a panic button RIP hotspot domain. Unlike DOCK2:p, it has 5 iHOP domains. 🔗
Tue Jun 07 02:01:59 +0000 2022@Glen4Climate Not a real success story, though. Within a month of the city completing that work, a Hydro crew came by & cut down the trees that had been painstakingly preserved during the installation.
Mon Jun 06 17:25:42 +0000 2022@Pdorrestein1 It pre-dates "modern" proteomics & probably would need Josh Lederberg + Klaus Biemann on the stage (but posthumous awards aren't allowed).
Mon Jun 06 14:55:56 +0000 2022I've never really understood why scientists who make their software freely available would want to have a lot of "users". I've had some good interactions with enthusiastic early adopters, but most "users" are time-consuming & generally "useless".
Mon Jun 06 12:47:02 +0000 2022This type of "hotspot" RIP phosphodomain would probably go undetected using an alanine-scan approach.
Mon Jun 06 12:05:28 +0000 2022Modification diagram for DOCK1:p, a human guanine nucleotide exchange factor. It has modest engagement with GGylation across its length & a prominent "panic button" style S/T+phosphoryl domain at the C-terminus, like the POLR2A C-terminus.
🔗
Sun Jun 05 21:50:01 +0000 2022@koblaatsu @dtabb73 Finally!
Sun Jun 05 13:28:20 +0000 2022Note: the "canonical" form of TXNRD1:p isn't observed very often. Two of the shorter splice variants dominate in proteomics data.
Sun Jun 05 13:01:12 +0000 2022Phosphorylation diagrams for human TXNRD1:p & TXNRD2:p, the cytoplasmic and mitochondrial forms of thioredoxin reductase. The role of Y+phosphoryl in the cytoplasmic form is unknown. Both have the C-terminal sequence -GCUG. 🔗
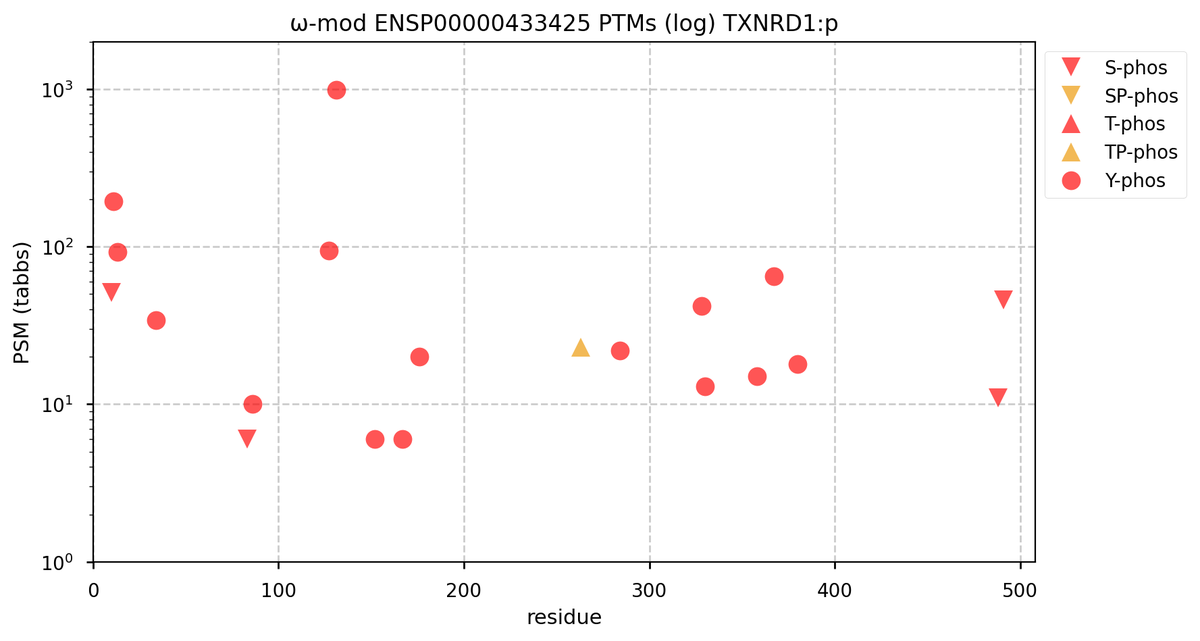
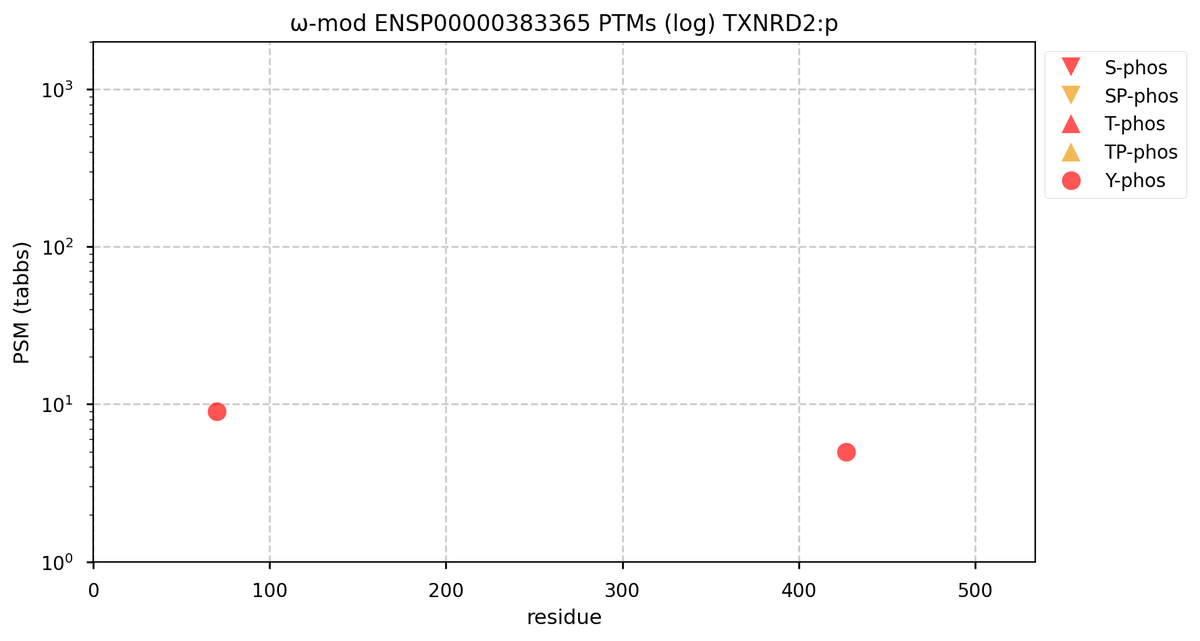
Sun Jun 05 12:49:29 +0000 2022@neely615 @SCwxFrankStrait After a brief trip through the mouth of the Gulf of Mexico as a tropical storm, Agatha has been reborn as Hurricane Alex, heading off-shore into the Atlantic. 🔗
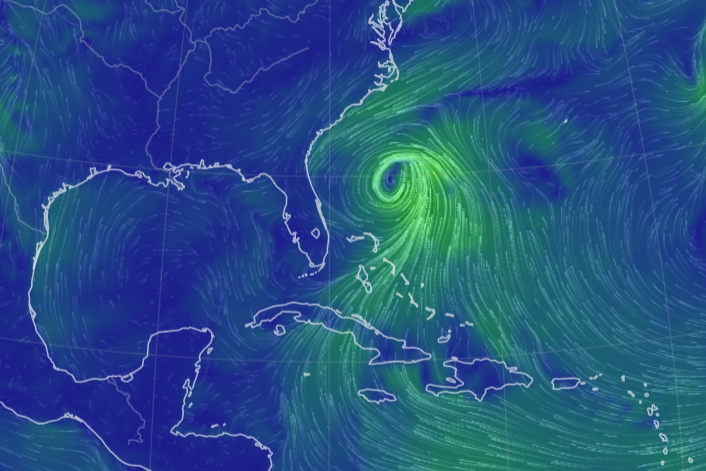
Sat Jun 04 22:44:54 +0000 2022Seems like the general opinion is "nothing"! 🔗
Sat Jun 04 16:45:59 +0000 2022@astacus @mjmaccoss @neely615 With or without ID. 2D gels don't have any inherent "ID" capability, but they are one of the many proteomics technologies used in the hopes that an invited-trip-to-Stockholm-worthy pattern change will reveal itself.
Sat Jun 04 15:42:07 +0000 2022@mjmaccoss @astacus @neely615 The idea of "skipping the hard part" via naïve pattern detection is still very attractive to many in the biomedical research business.
Sat Jun 04 14:10:06 +0000 2022What discovery involving proteomics should result in a Nobel Prize?
Sat Jun 04 13:16:37 +0000 2022Modification diagrams for human CUBN:p & PRR12:p, two large subunits. These two sequences both have 51 occurrences of the CDK phosphorylation motif -X-(S/T)-P-X- 🔗
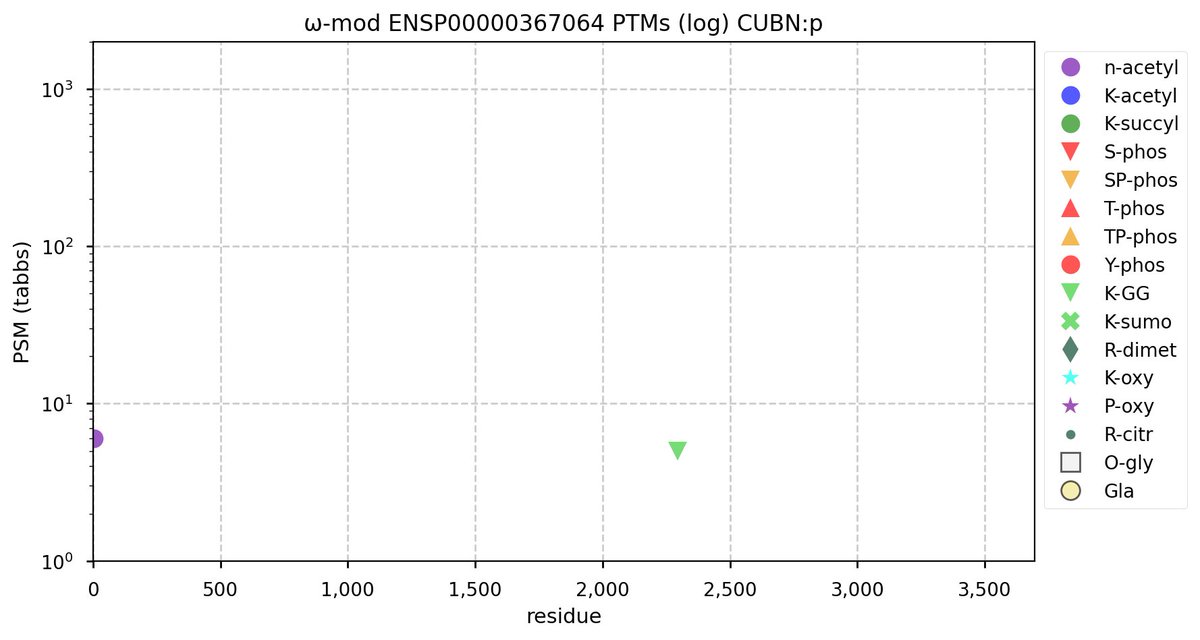
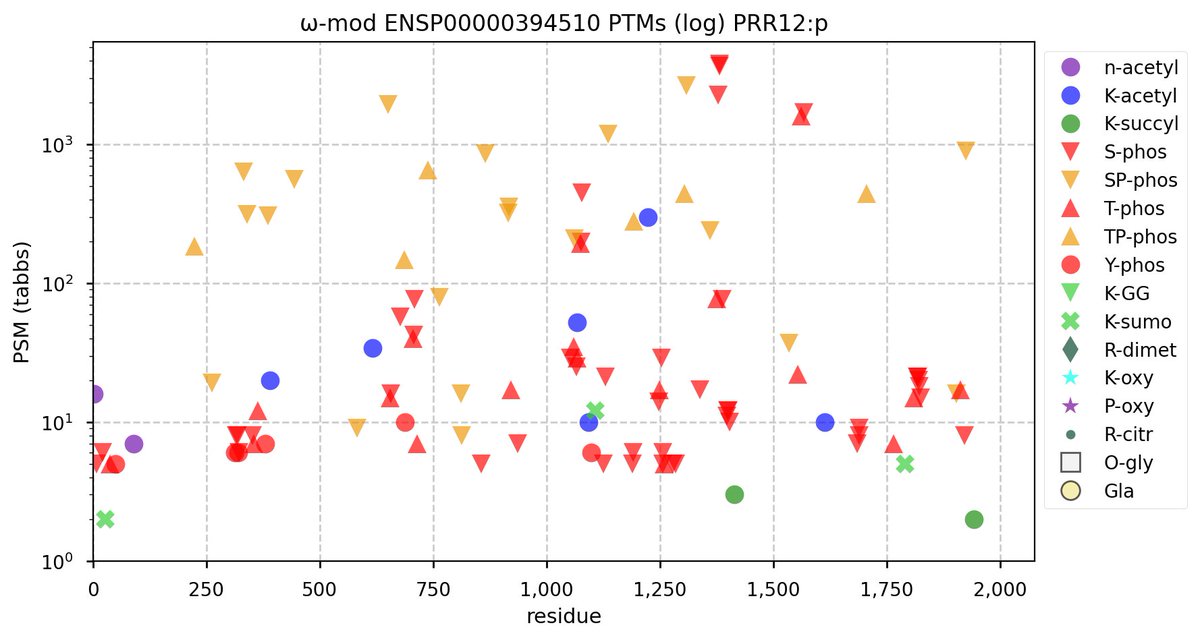
Fri Jun 03 14:40:05 +0000 2022🔗
Fri Jun 03 13:29:47 +0000 2022Details some excellent examples of the proteomics "walled garden" approach to information creation & storage.
🔗
Fri Jun 03 13:16:47 +0000 2022This aligns the provincial PC party with the provincial branch of the federal Liberals, who have had a similar makeup in Ontario since the dissolution of the federal PCs & the formation of the current CPC.
Fri Jun 03 12:56:20 +0000 2022Ontario is following the same pattern as Manitoba, with a PC v NDP provincial scene. It is different, however, in that in Manitoba the Liberals mainly went over to team orange, while in Ontario they have gone to team blue.
Fri Jun 03 12:41:39 +0000 2022@neely615 @SCwxFrankStrait The remnants of Hurricane Agatha (which started in the Pacific) are starting to show up on weather radar over western Cuba & southern Florida 🔗
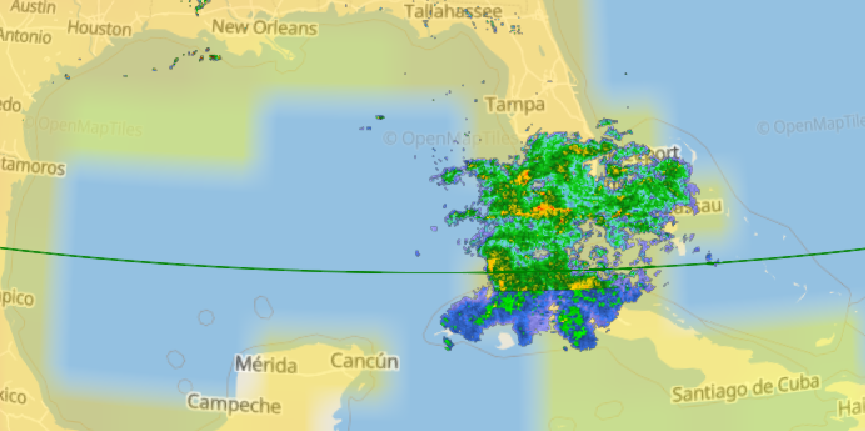
Fri Jun 03 12:30:23 +0000 2022ZNF398:p would probably be a good candidate for acceptor site function determination via alanine scan, while applying the same method to ZNF106:p would probably lead to the conclusion that it was not modified in a biologically significant manner.
Fri Jun 03 12:17:37 +0000 2022Modification diagrams for ZNF398:p & ZNF106:p, human subunits that may be involved in many things. The designation "ZNF" indicates the presence of a predicted zinc finger domain in the nascent sequence, which makes involvement in nucleic acid binding a go-to speculation. 🔗
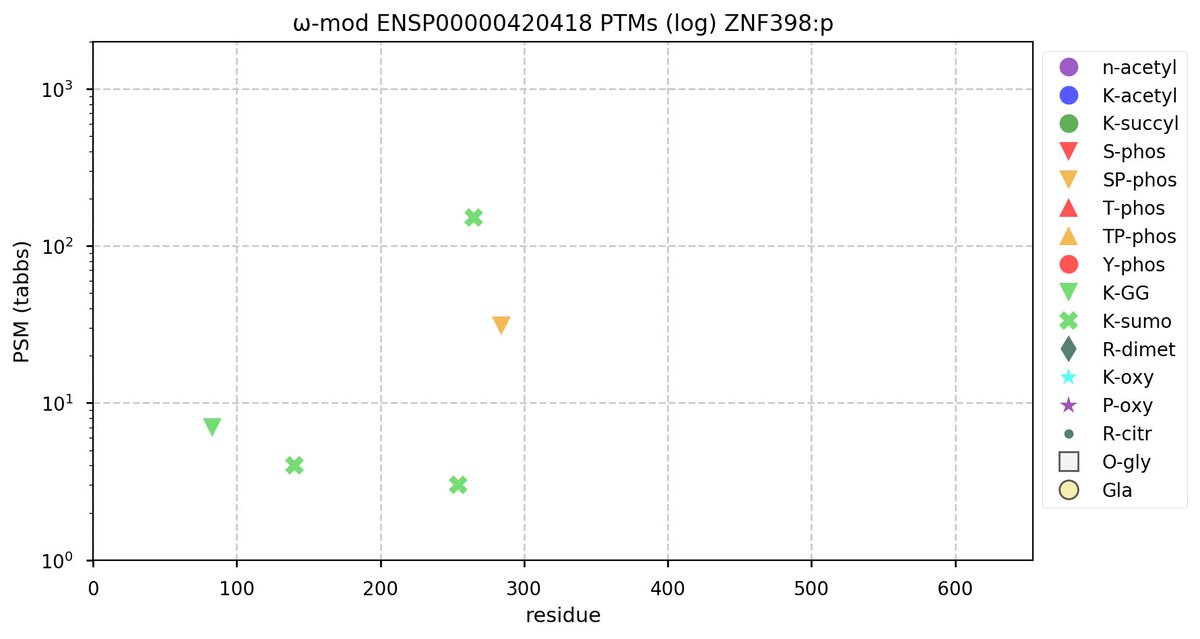
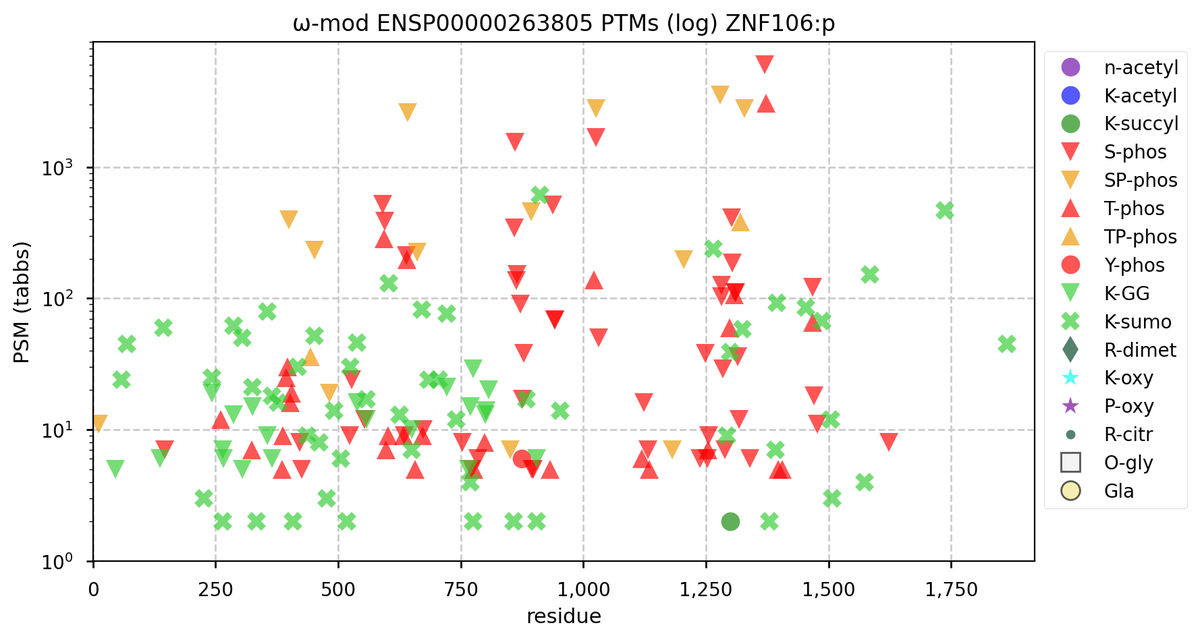
Thu Jun 02 13:27:17 +0000 2022I suspect that an alanine scan would be much more likely to detect functional phosphorylation in PRUNE1:p than in PRUNE2:p.
Thu Jun 02 12:30:52 +0000 2022Watch out, Shreveport! 🔗

Thu Jun 02 12:18:39 +0000 2022Modification diagrams for human PRUNE1:p & PRUNE2:p, named for a phosphodiesterase domain believed to be homologous to the Drosophila PRUNE gene. While that may be true, there may be some functional differences between these 2 not captured by this naming decision. 🔗
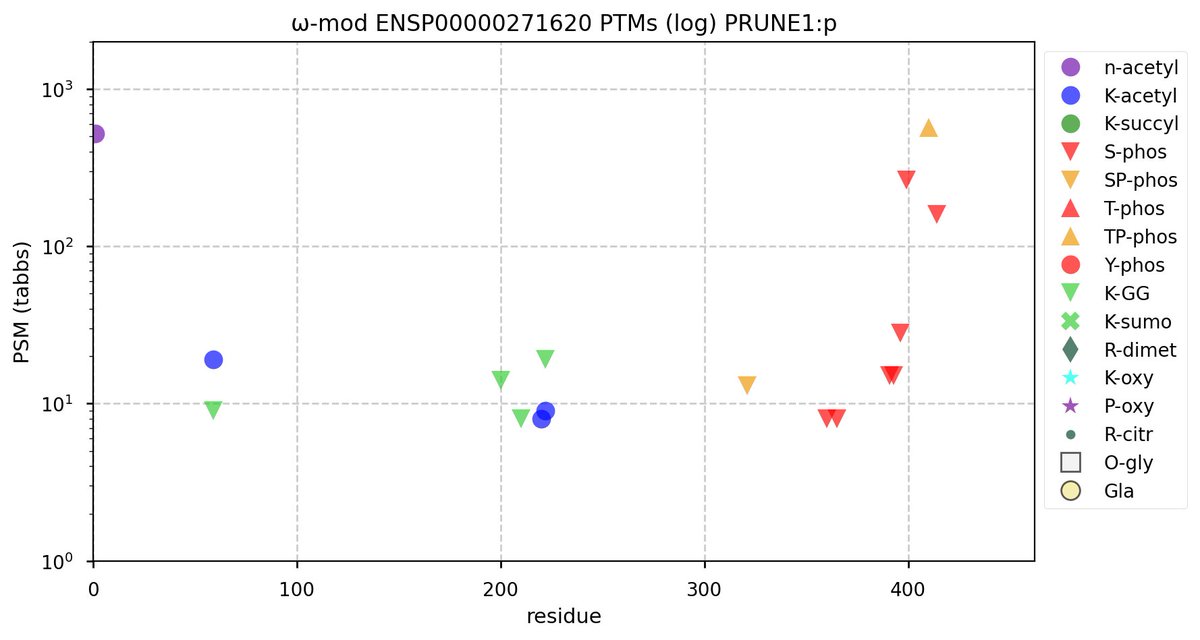
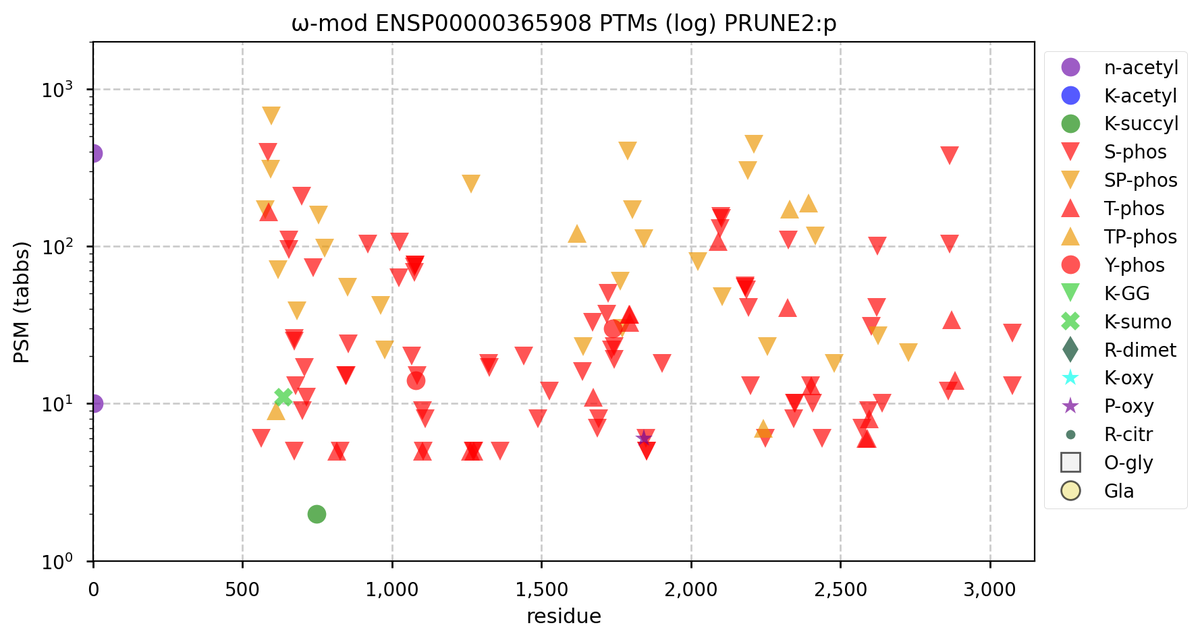
Thu Jun 02 01:17:03 +0000 2022@JoePlatelet Not that I know of.
Wed Jun 01 14:38:57 +0000 2022@ucdmrt @AlexHgO It depends on bilateral tax treaties. When I was there, I had the option of either paying German taxes or Canadian taxes (& I stupidly chose Canadian).
Wed Jun 01 13:11:19 +0000 2022Needless to say, but I'm going to diss alanine scans for the next couple of weeks.
Wed Jun 01 12:53:46 +0000 2022This sort of PTM scheme would be unsuitable for function determination using alanine scans.
Wed Jun 01 12:43:58 +0000 2022Modification diagram for human WIZ:p, a commonly observed protein that may do many things. Its PTM scheme is dominated by clusters of high occupancy S+phosphoryl & K+SUMOyl acceptor sites.
🔗
Wed Jun 01 12:24:30 +0000 2022@Smith_Chem_Wisc Why would they care about activity?
Tue May 31 16:01:14 +0000 2022@michaelhoffman Just a habit. One of the ghost behaviors you acquire over time.
Tue May 31 15:30:44 +0000 2022@michaelhoffman They may, but the party leader is free to reject that choice and substitute their own candidate.
Tue May 31 15:26:47 +0000 2022@michaelhoffman As someone who has been a member for ~40 years, I wouldn't recommend it, other than as an exercise. Canadian political parties at the riding level now have very little influence on anything except during the brief period of a leadership contest.
Tue May 31 14:41:49 +0000 2022I suspect that many people think about a scheme like this when they think about PTMs, even though it is only one of many possibilities. It is one of the few patterns for which site-directed mutagenesis (alanine scanning) would be a useful method for exploring function.
Tue May 31 13:16:17 +0000 2022Modification diagram for TSEN2:p, a subunit of the human tRNA splicing endonuclease. The PTM scheme is in the '80s textbook classic style: a single, isolated high occupancy S+phosphoryl & very limited low occupancy K+acetyl, GGyl or SUMOyl.
🔗
Mon May 30 14:58:43 +0000 2022@stateofthecity I guess I was hoping that there was some centralized problem that could be fixed. I live downtown & I have to walk around failed/stalled public & private projects all of the time.
Mon May 30 14:44:38 +0000 2022While the function of this sort of domain may be a bit mysterious (but probably just an on-off switch), at least the AF predicted aligned error plot shows it is in the "right place": in a dark (& mysterious) part of the sequence. 🔗
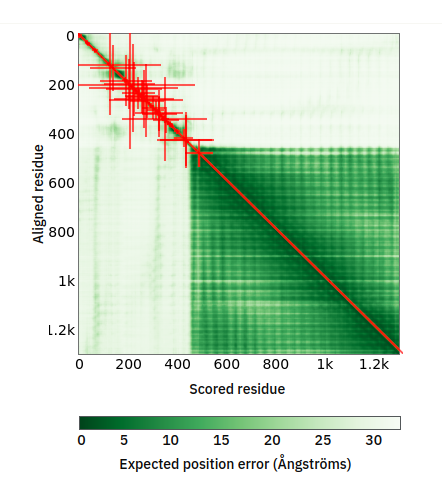
Mon May 30 14:42:13 +0000 2022The high occupancy T+phosphoryl ΦP acceptor domain is clearer with a linear y-axis: linear plots for both the human & mouse subunits are given below. This domain has the same sort of "panic button" feel as the C-terminal S+phosphoryl ΦP acceptor domain in POLR2A. 🔗
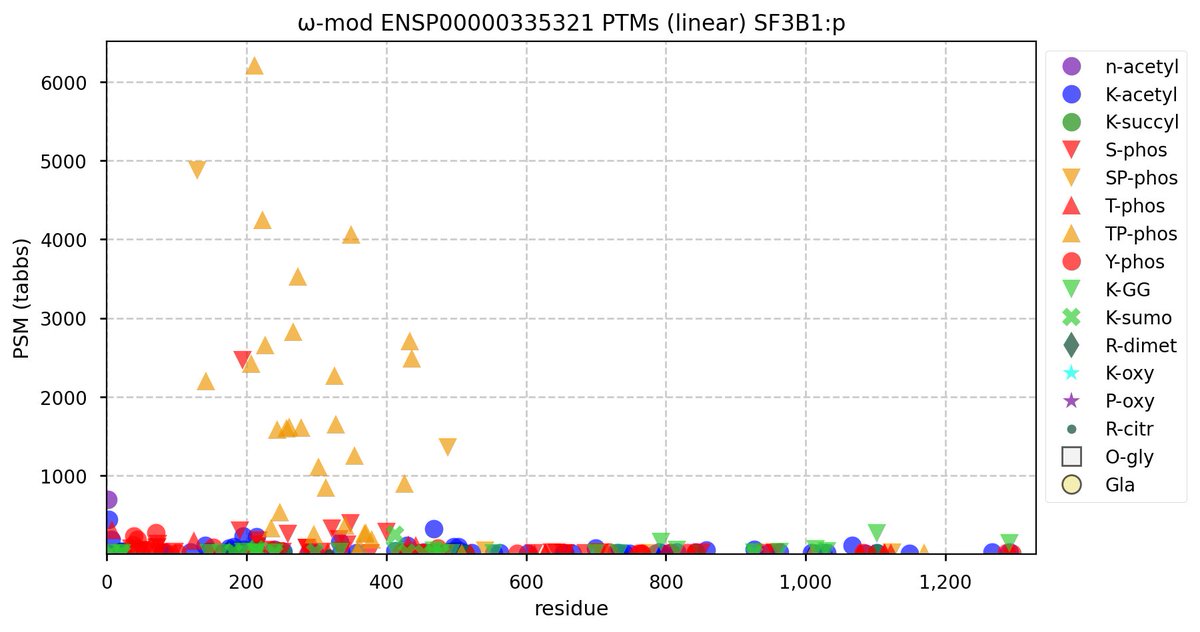
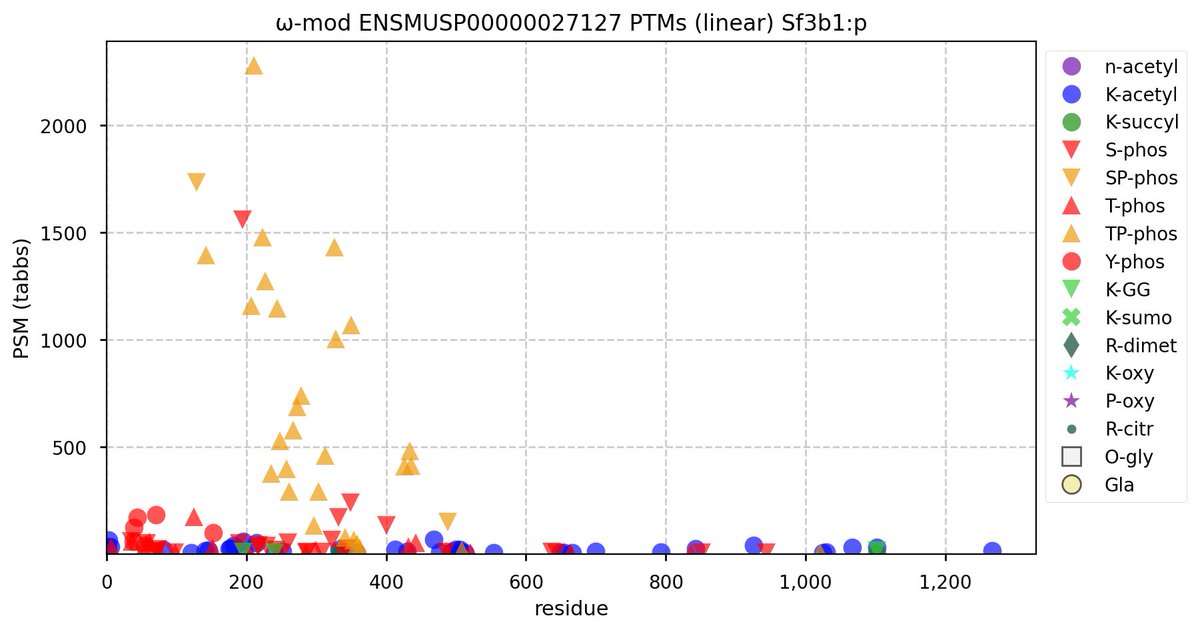
Mon May 30 14:25:11 +0000 2022Modification diagram for SF3B1:p, a subunit of the human SF3B complex, which is in turn part of the U2 snRNP. There are low occupancy K+GGyl/acetyl/SUMOyl acceptors spread across the sequence. 🔗
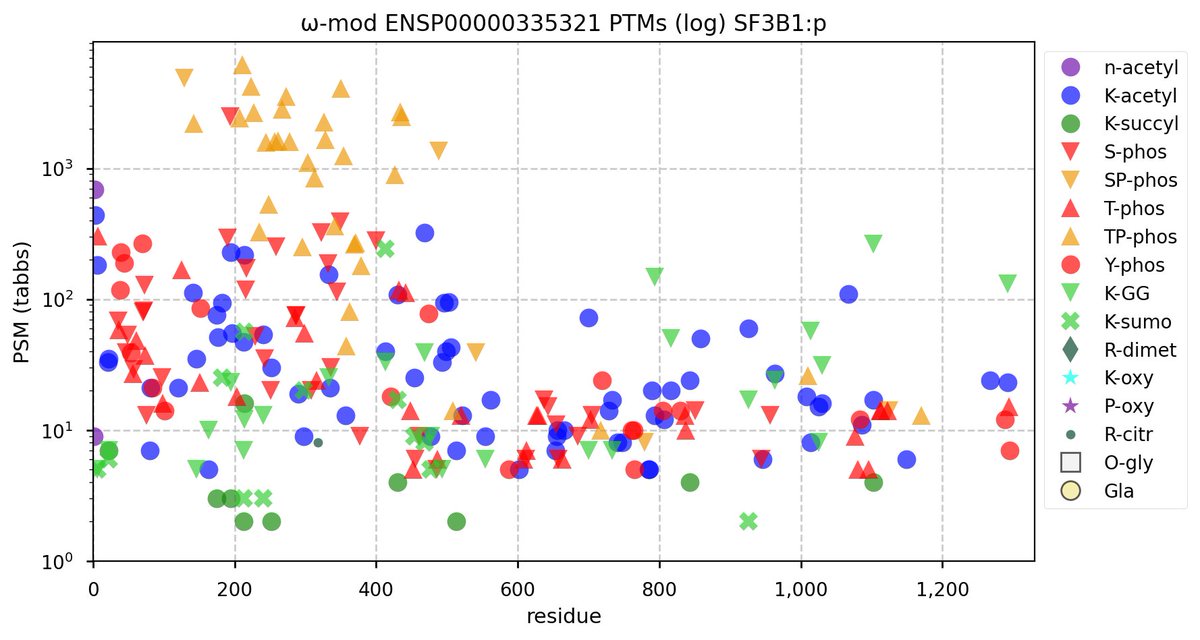
Mon May 30 13:12:52 +0000 2022@stateofthecity Are the winning designs for this type of project selected directly by Council as a whole or is there an expert committee that makes recommendations?
Mon May 30 12:24:57 +0000 2022Hurricane Agatha just west of the Gulf of Tehuantepec, heading for shore this morning. 🔗
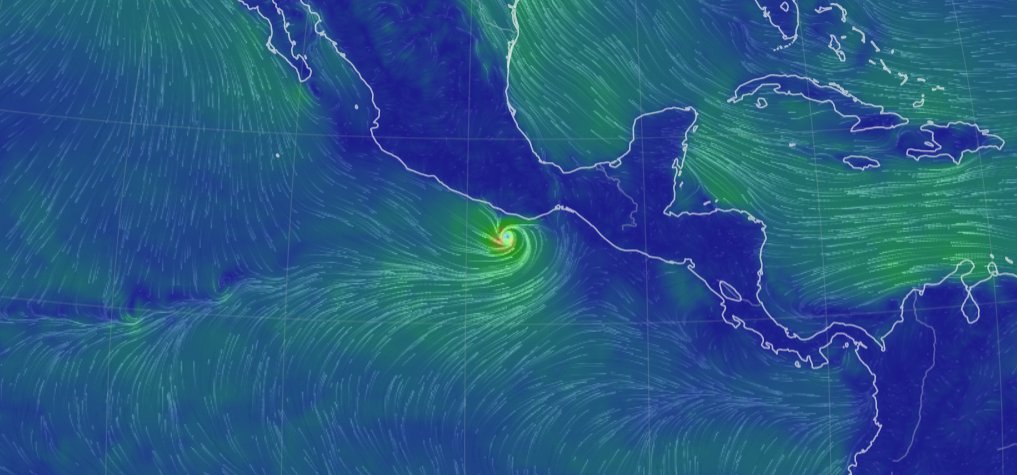
Sun May 29 22:56:54 +0000 2022@BMarshChem @taghioskoui @the_ion_doctor @neely615 @mjmaccoss @UCDProteomics @slavov_n @chrashwood @GarciaLabMS It might be useful, but many institutions don't have the mechanical & electronics shops needed to develop new instruments.
Sun May 29 19:35:28 +0000 2022Oh no!
🔗
Sun May 29 13:17:20 +0000 2022@chrashwood @neely615 @mjmaccoss @UCDProteomics @slavov_n @GarciaLabMS People involved in proteomics seem to forget just how crucial PCR is to almost everything that can be done with nucleic acid sequencing. Very few of the methods made available by genomics core facilities would work at all if you took away their thermal cyclers.
Sun May 29 13:05:45 +0000 2022@chrashwood @neely615 @mjmaccoss @UCDProteomics @slavov_n @GarciaLabMS A peptide PCR would even the playing field quite a bit.
Sun May 29 12:56:09 +0000 2022Note: there is no human NatG (NAA70:p). That transferase is found in plant chloroplasts only. There is no known α-amino N-acetyl transferase activity in mitochondria.
Sun May 29 12:48:20 +0000 2022NAA80:p acetylates actin subunits that begin with MXXX, where X = D, E. The ribosome produces acetyl-MXXX into the cytoplasm, where the N-terminal acetyl-Met is removed by an unknown aminopeptidase activity & then NAA80:p adds the acetyl to form the mature actin acetyl-XXX.
Sun May 29 12:42:11 +0000 2022Modification diagram for NAA80:p, the sole subunit of human NatH. Unlike the NatA/B/C/D/E/F, It is a cytoplasmic enzyme, not associated with ribosomes. It has more involvement in ΦP phosphorylation (7 sites) than the ribosomal transferases. 🔗
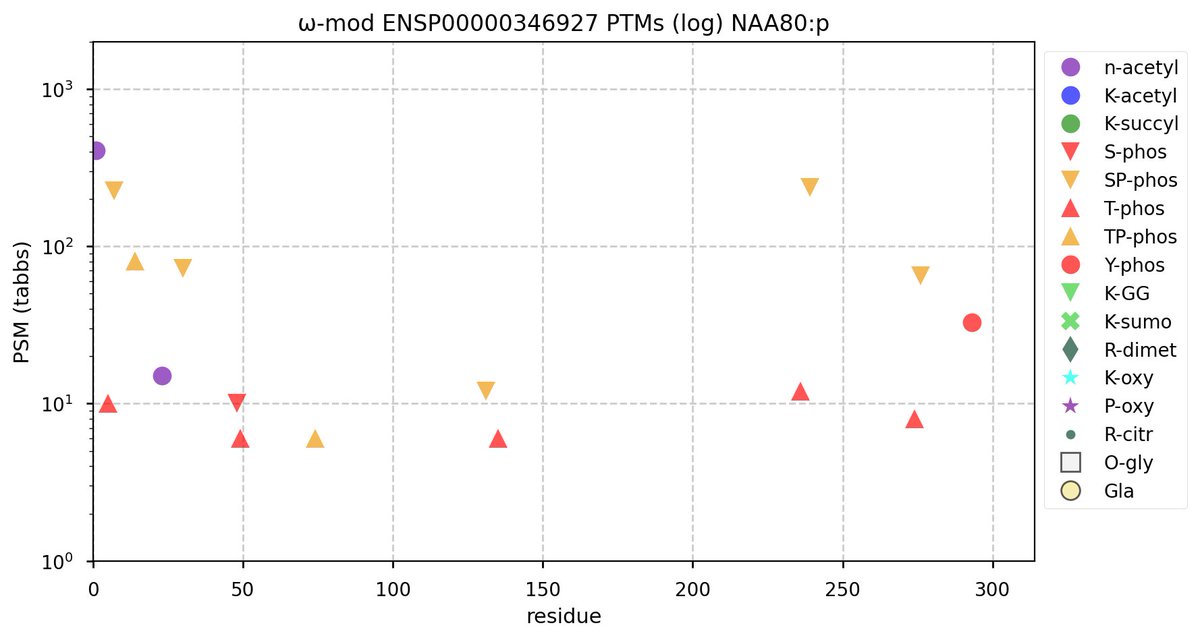
Sat May 28 17:11:42 +0000 2022@slavov_n @chrashwood @neely615 @GarciaLabMS Unpopular opinion #1: popularity is all that matters in the end.
Sat May 28 17:01:21 +0000 2022@chrashwood @neely615 @GarciaLabMS @slavov_n I've got nothing but unpopular opinions on the subject that are best kept to myself.
Sat May 28 15:03:51 +0000 2022@chrashwood @neely615 @GarciaLabMS @slavov_n It has never been necessary to know the mass of the parent ion: it isn't actively used in the PSM assignment process, other than to make the calculation somewhat more tractable.
Sat May 28 14:50:50 +0000 2022@neely615 @chrashwood @GarciaLabMS @slavov_n As it has in the past, if anything is going to kill off a proteomics technology, it will be overconfidence in the results.
Sat May 28 12:18:29 +0000 2022Modification diagram for NAA60:p, the sole subunit of human NatF. Adds an acetyl to the N-terminus of nascent peptides beginning with MK-, ML-, MI-, MW- or MF-. T2+acetyl (by NatA) is the only observed mod. #natases 🔗
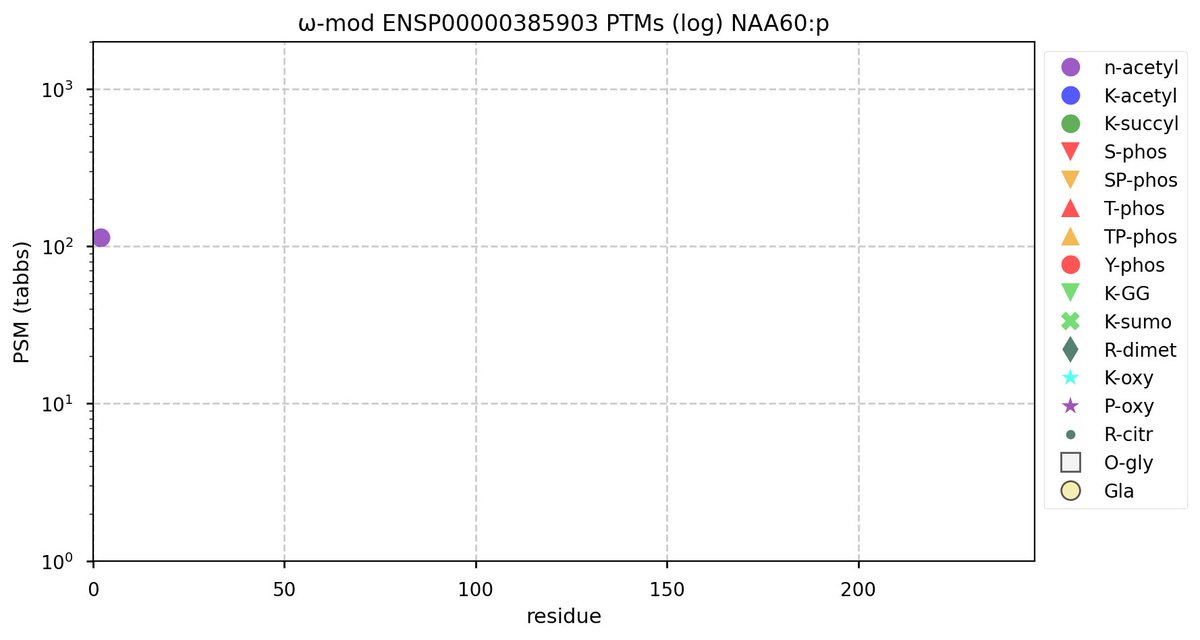
Fri May 27 18:24:45 +0000 2022@J_my_sci Just the last one about bad annotation of N-terminal processing by a well known "knowledgebase".
Fri May 27 17:18:24 +0000 2022As much as I enjoy tilting at windmills, I think this will be the last of these.
Fri May 27 17:17:28 +0000 2022CDKAL1-202, CDK5 regulatory subunit associated protein 1 like 1
🔗
1 – 579 → 2 – 579 (N-acetyl-Pro, ο = 3%, Nat?)
#realNt
Fri May 27 13:07:40 +0000 2022@neely615 @byu_sam In Canada, this sort of thing only happens in Newfoundland & Cape Breton.
Fri May 27 12:58:01 +0000 2022Modification diagram for NAA50:p, a small subunit of the enzyme NatE. The other 2 subunits of this enzyme (NAA10:p & NAA15:p) are shared with NatA. Transfers acetyl to N-termini starting with ML-, MA-, MK- & MM-. #natases 🔗
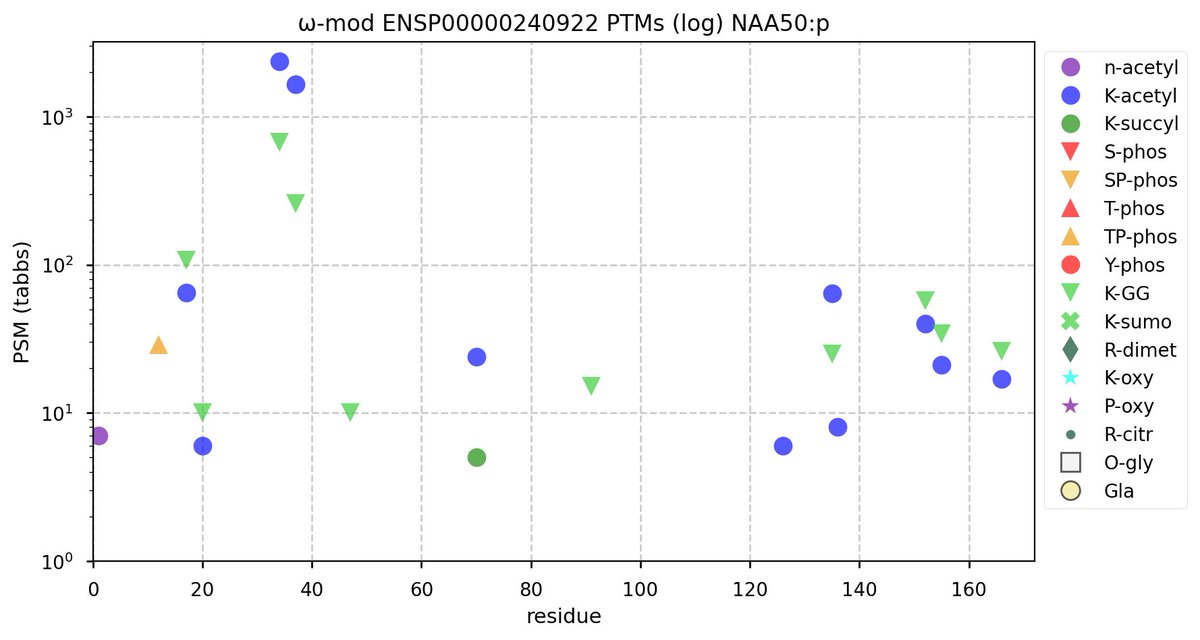
Fri May 27 12:50:55 +0000 2022UBE3C-201, ubiquitin protein ligase E3C
🔗
1 – 1083 → 1 – 1083 (N-acetyl-Met, ο = 100%, NatC)
Fri May 27 01:44:29 +0000 2022@jwoodgett Have the same, but got it locally (no flying). Not COVID-19, but there are lots of other respiratory viruses kicking around.
Thu May 26 15:15:03 +0000 2022ABF1, ARS-Binding Factor 1
🔗
1 – 731 → 1 – 731 (N-acetyl-Met, ο = 100%, NatB)
Thu May 26 15:06:28 +0000 2022DARS1-201, aspartyl-tRNA synthetase 1
🔗
1 – 501
↓
2 – 501 (N-acetyl-Pro, ο = <1%)
18 – 501 (N-acetyl-Met, ο = 98%)
#realNT
Thu May 26 13:36:33 +0000 2022The alpha-amino N-acetyltransferases are believed to be very old enzymes: current thinking suggests that they are little changed from the Last Eukaryote Common Ancestor. #natases
Thu May 26 13:21:41 +0000 2022Modification diagram for NAA40:p, the only subunit of human NatD. This enzyme catalyzes N-terminal acetylation in histone 2A and histone 4 (& apparently nothing else). It eschews phosphorylation & has a limited number of GGyl/acetyl/SUMOyl acceptor in the domain (25-93). 🔗
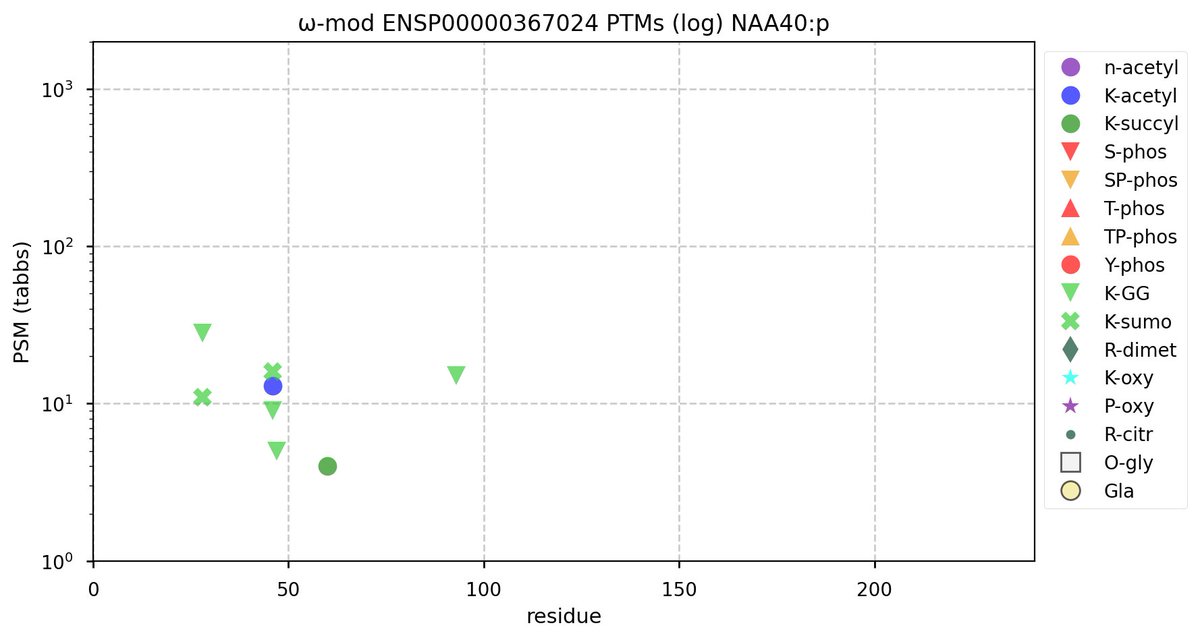
Wed May 25 17:15:07 +0000 2022TUT1-205, terminal uridylyl transferase 1, U6 snRNA-specific
🔗
1 – 874 → 2 – 874 (N-acetyl-Ala, ο = 97%, NatA)
Wed May 25 17:10:04 +0000 2022DES-201, desmin
🔗
2 – 470 → 2 – 470 (N-acetyl-Ser, ο = >99%, NatA)
Wed May 25 14:45:31 +0000 2022An RIP (Really Interesting Phosphorylation) domain is a stretch of intrinsically disordered sequence least 100 residues long with >5% of the residues serving at S/T+phosphoryl acceptor sites. The NAA30:p example is 200 residues long & 10% of the residues are acceptors.
Wed May 25 13:49:36 +0000 2022@ntheberge12 @justinemercier So Canadian Crown Copyright is perpetual, unlike copyright for the rest of us that expires?
Wed May 25 13:28:42 +0000 2022@justinemercier I was wondering about this: does the Crown have some sort of perpetual copyright in Canada? Normally a copyright declaration has a year explicitly stated.
Wed May 25 13:26:16 +0000 2022@justinemercier Aussi, ROC 🔗
Wed May 25 13:04:25 +0000 2022NatC comes at the PTM decoration problem with a different approach than either NatA or NatB. NAA35:p & NAA38:p have some spotty involvement with GGylation. NAA30:p has no GGyl acceptors, but a fairly aggressive S+phosphoryl strategy across its N-terminal RIP domain. 🔗
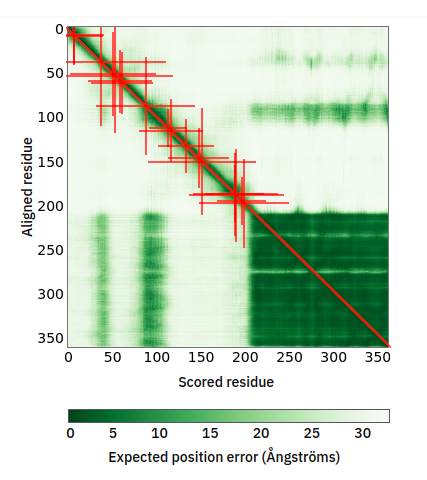
Wed May 25 12:52:26 +0000 2022Modification diagram for human NAA30:p, NAA35:p & NAA38:p, subunits of the enzyme NatC. It is responsible for N-terminal acetylation of sequences that begin with MI-, ML-, MW- & MF-. 🔗
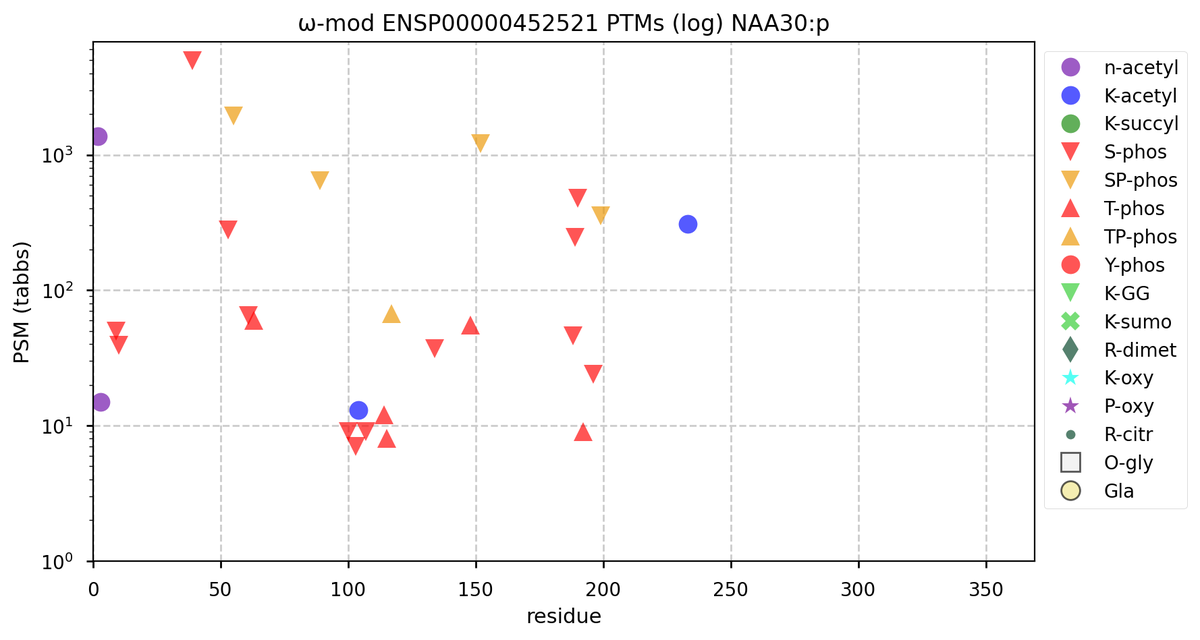
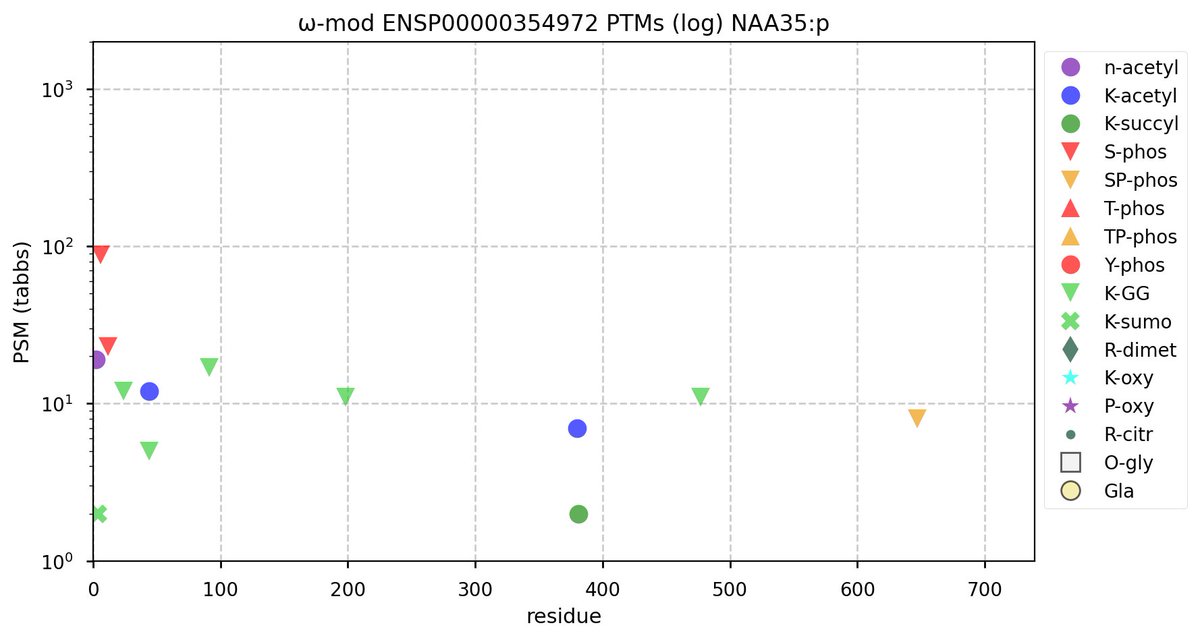
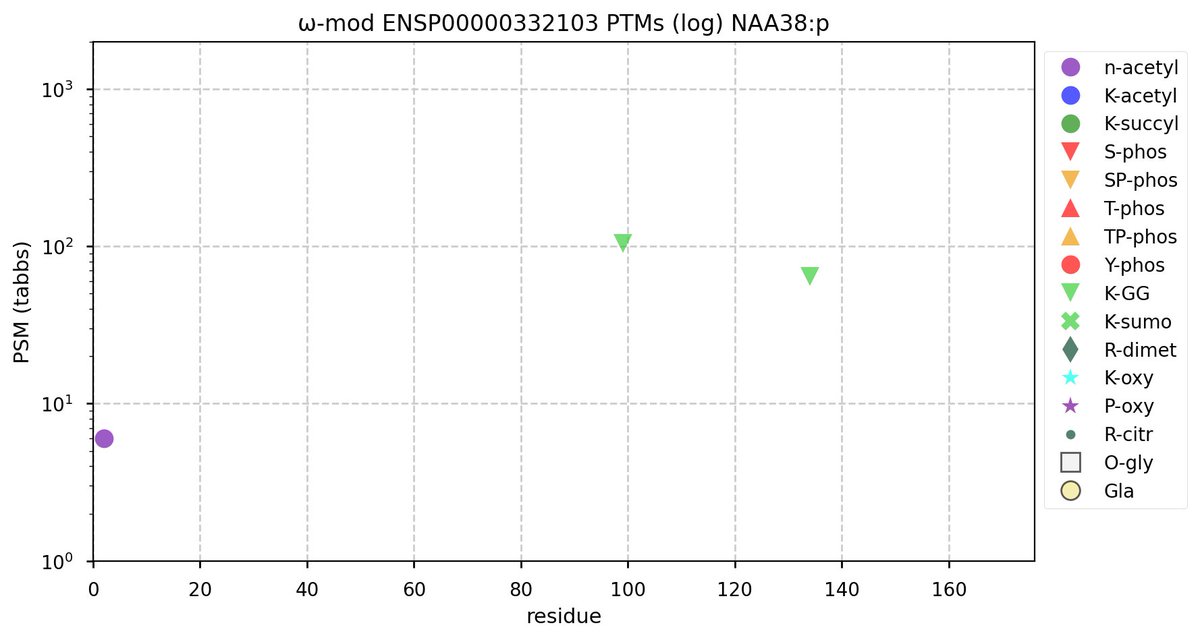
Tue May 24 18:44:48 +0000 2022About 2% of observations are 1-639 with no N-acetyl, but this proteoform is only found in cell lines.
Tue May 24 18:39:30 +0000 2022CD2AP-201, CD2 associated protein
🔗
1 – 639 → 2 – 639 (N-acetyl-Val, ο = 99%, NatA)
Tue May 24 16:11:21 +0000 2022C5orf24-201, chromosome 5 open reading frame 24
🔗
1 – 188
↓
1 – 188 (N-acetyl-Met, ο = 100%)
2 – 188 (N-acetyl-Met, ο = 100%)
#realNT
Tue May 24 14:35:53 +0000 2022If you have any interest in α-amino-NATs, this article has a pretty good description of the subject from an evolutionary point of view, although rather predictably subunit PTMs are left out of the discussion.
🔗
Tue May 24 14:25:17 +0000 2022Both NAA20:p & NAA25:p are N-acetylated by NatA.
Tue May 24 13:08:48 +0000 2022This big difference in PTM patterns between these 2 enzymes is not what I would have expected, but proteins are often full of surprises.
Tue May 24 12:50:12 +0000 2022For comparison, the diagrams for the 2 subunits of NatA (NAA10:p & NAA15:p), an enzyme that does the same job with different N-terminal sequence specificity. 🔗
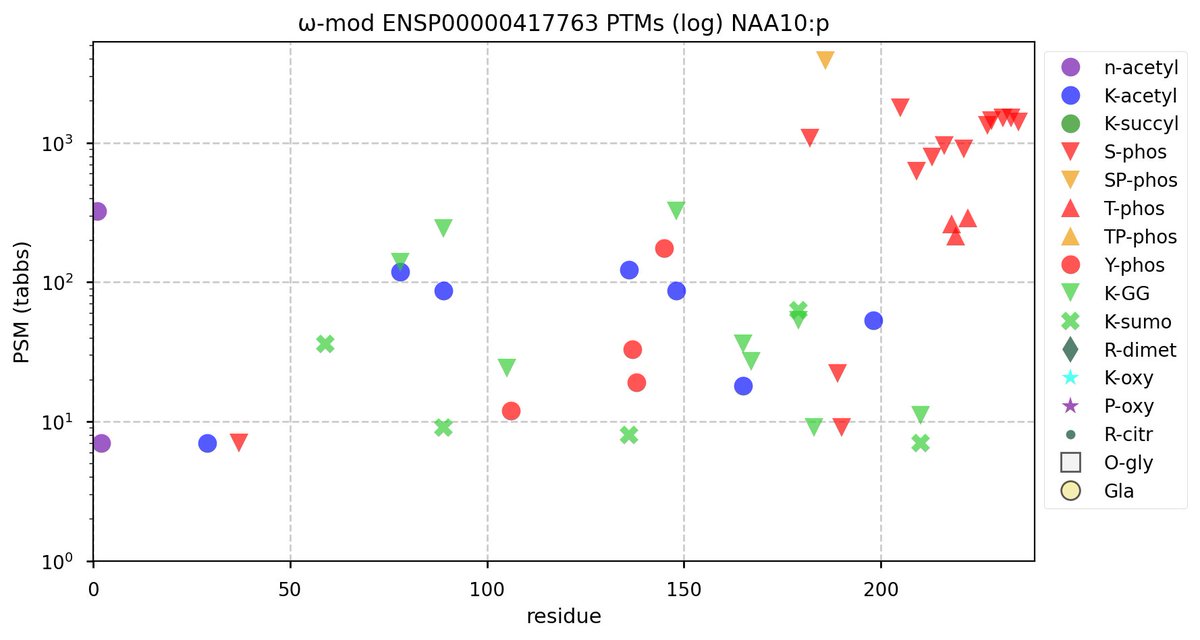
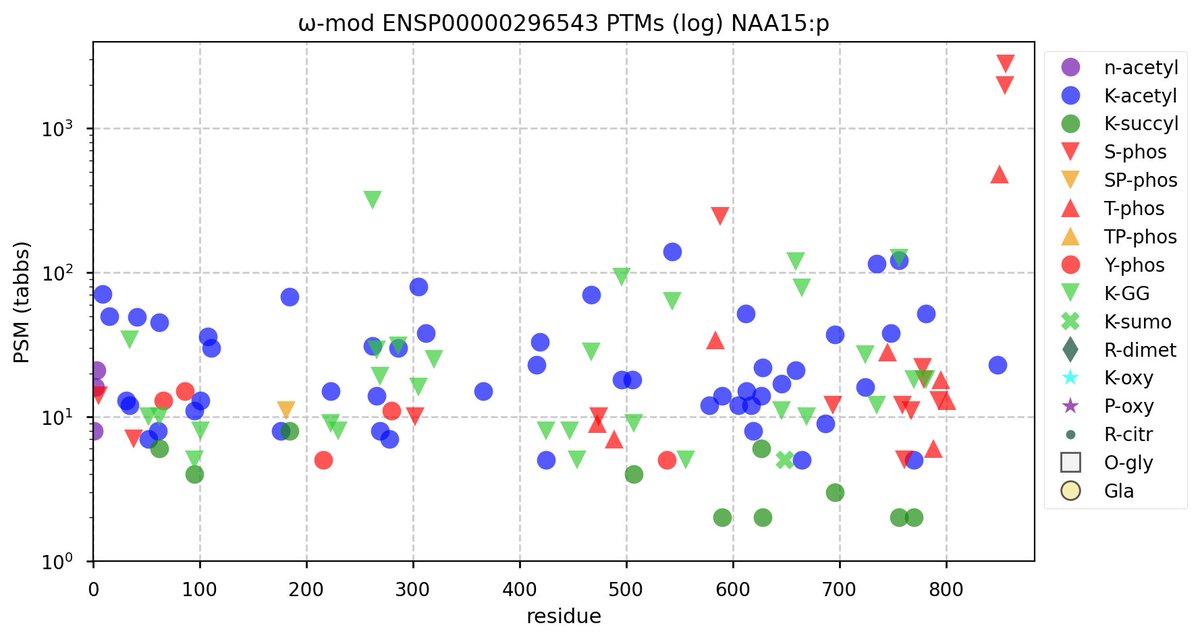
Tue May 24 12:42:36 +0000 2022Modification diagrams for human NAA20:p & NAA25:p, the 2 subunits of NatB, an enzyme that blocks the initiator Met in nascent peptides starting with MD-, ME-, MQ- or MN-. This enzyme has a scattering of GGyl with a bit of alternating GGyl/acetyl on the larger subunit. #natases 🔗
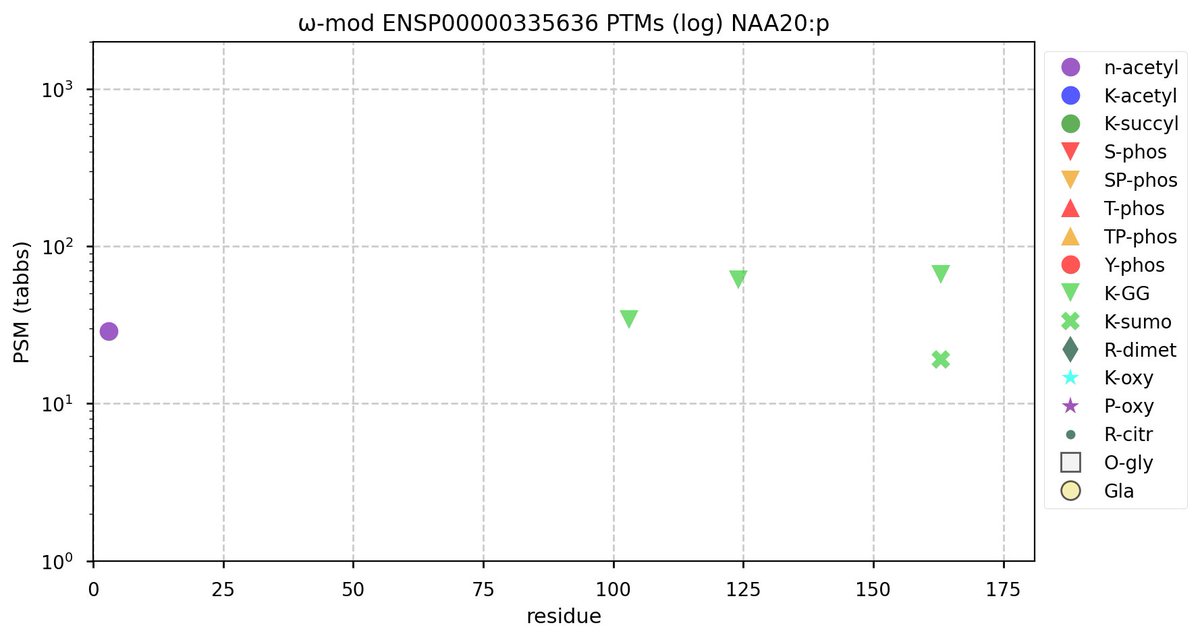
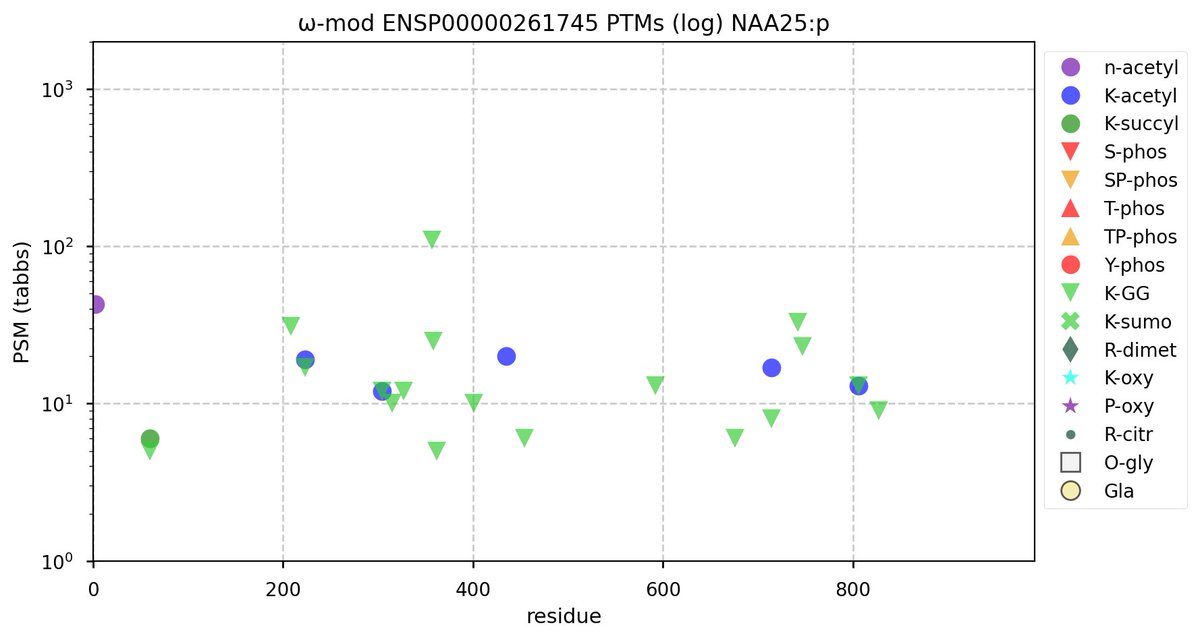
Mon May 23 20:54:16 +0000 2022I was reminded of this advice while trying to find a tweet about a recent paper discussing what the authors call "Deep Visual Proteomics" via its initials. I DO NOT RECOMMEND DOING THIS YOURSELF.
Mon May 23 20:50:23 +0000 2022Note to students: if you have what you believe to be a novel acronym for your latest, buzz-worthy technology, you should always (always) try searching that acronym on all major platforms (e.g. Twitter, Instagram) prior to publication.
Mon May 23 20:01:09 +0000 2022@J_my_sci @OllyMCrook @slavov_n I won't read much into "unbiased". It is just one of the performative adjectives frequently used to inject excitement into an otherwise workman-like method, e.g., "robust", "ultra", "high", "seamless", "intuitive", or any use of "rich".
Mon May 23 19:43:39 +0000 2022A political commercial came on TV last night & I couldn't figure out why Mark Proksch was telling me about Ontario political issues using his "Colin Robinson" character, until, at the end, the ad said it was from the leader of the Ontario Liberal party.
Mon May 23 19:21:00 +0000 2022CYSRT1-202, cysteine rich tail 1
🔗
1 – 144 ⟶ 1 – 144 (N-acetyl-Met, ο = 100%, NatB)
#realNT
Mon May 23 19:14:01 +0000 2022SULT1E1-201, sulfotransferase family 1E member 1
🔗
1 – 294 ⟶ 1 – 294 (N-acetyl-Met, ο = 100%, NatB)
#realNT
Mon May 23 16:57:21 +0000 2022KATNA1-202, katanin catalytic subunit A1
🔗
1 – 491 ⟶ 2 – 491 (N-acetyl-Ser, ο = 100%, NatA)
#realNT
Mon May 23 16:24:47 +0000 2022ZNF385A-201, zinc finger protein 385A
🔗
1 – 386 ⟶ 1 – 386 (N-acetyl-Met, ο = 100%, NatC)
#realNT
Mon May 23 16:17:21 +0000 2022PSRC1-202, proline and serine rich coiled-coil 1
🔗
1 – 363 ⟶ 1 – 363 (N-acetyl-Met, ο = 100%, NatB)
#realNT
Mon May 23 15:44:56 +0000 2022In about 10% of observations, the N-terminus is
1 – 432 (N-acetyl-Met, ο = 99%)
Mon May 23 15:42:51 +0000 2022GRAMD2B-201, GRAM domain containing 2B
🔗
1 – 432 ⟶ 2 – 432 (N-acetyl-Thr, ο = 93%)
#realNT
Mon May 23 14:20:10 +0000 2022@Smith_Chem_Wisc @theoneamit I'm almost surprised that JPR hasn't implemented a checklist for cover letters, requiring a additional supplemental information attachment.
Mon May 23 13:29:24 +0000 2022NAA15 does not have N-terminal acetylation, but the initiator M residue is removed, resulting in the mature sequence:
2 PAVSLPPK …
N-terminal prolines are rarely acetylated via the co-translation NAT modification mechanism.
Mon May 23 13:28:55 +0000 2022Modification diagrams for mouse & human NAA15:p, a midsized subunit of the NatA enzyme. The patterns rhyme nicely, with 4 broad K+GGyl/acetyl domains, several low occupancy phosphorylation domains & a C-terminal high occupancy S+phosphoryl domain. #natases 🔗
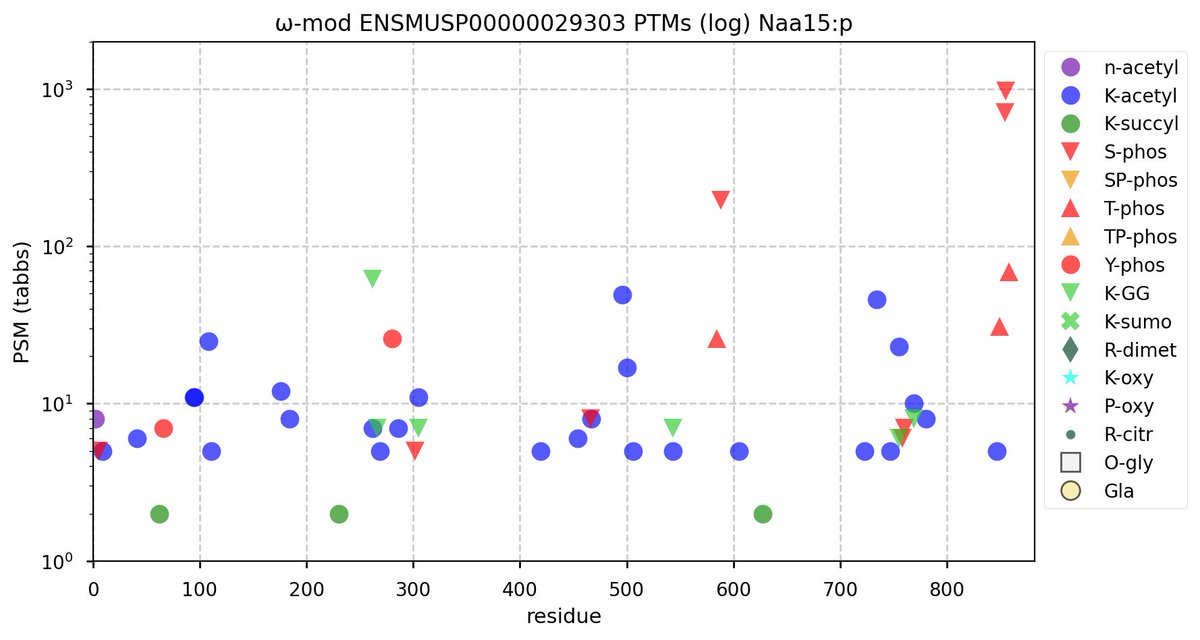
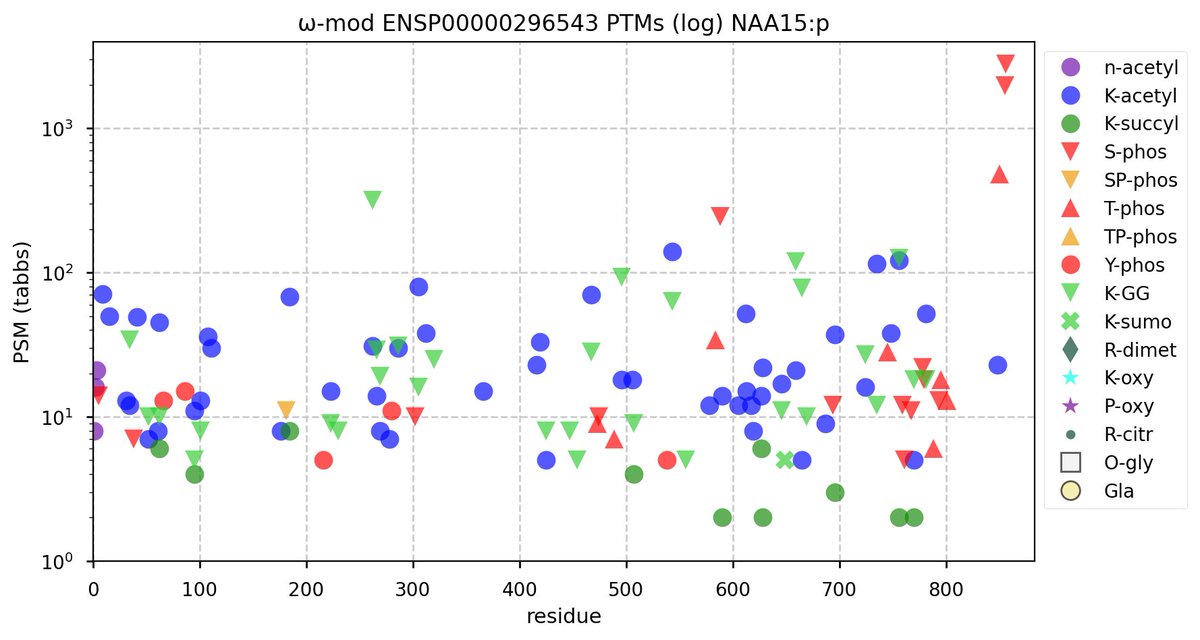
Mon May 23 01:54:47 +0000 2022POC1A-201, POC1 centriolar protein A
🔗
1 – 407 ⟶ 2 – 407 (N-acetyl-Ala, ο = 100%)
#realNT
Mon May 23 01:30:13 +0000 2022FLNB-201, filamin B
🔗
1 – 2602 ⟶ 2 – 2602
#realNT
Mon May 23 01:25:33 +0000 2022ZBTB7B-202, zinc finger and BTB domain containing 7B
🔗
1 – 539 ⟶ 2 – 539 (N-acetyl-Gly, ο = 97%)
#realNT
Mon May 23 01:16:26 +0000 2022DAPK3-202, death associated protein kinase 3
🔗
1 – 454 ⟶ 2 – 454 (N-acetyl-Ser, ο = 100%)
#realNT
Mon May 23 00:36:48 +0000 2022KPTN-201, kaptin, actin binding protein
🔗
1 – 436 ⟶ 3 – 436 (N-acetyl-Gly, ο = 100%)
#realNT
Sun May 22 17:38:18 +0000 2022Both human and mouse NAA10 are co-tranlationally N-terminal acetylated at M1 by their respective NatB enzymes.
Sun May 22 13:04:05 +0000 2022The high occupancy human S186+phosphoryl acceptor (SP-type, CDK?) is not present in the mouse sequence.
Sun May 22 12:58:03 +0000 2022NatA transfers acetyl to exposed S, C, G, T, V or A N-terminii formed by the removal of the translation initiator M residue.
Sun May 22 12:52:04 +0000 2022Normal protein N-acetyltransferase (NAT) activity in mammals requires 7 different NAT enzymes, composed of at least 14 different gene products. Six of the enzymes are co-translational, integrally associated with ribosomes. One of the enzymes is post-translational.
Sun May 22 12:47:15 +0000 2022Modification diagrams for mouse & human NAA10:p, a subunit of the enzyme NATA. The patterns show a consistent C-terminal S+phosphoryl domain & a Y+phosphoryl domain (100-150). GGyl/SUMOyl/acetyl acceptors are scattered through (50-200).
#natases 🔗
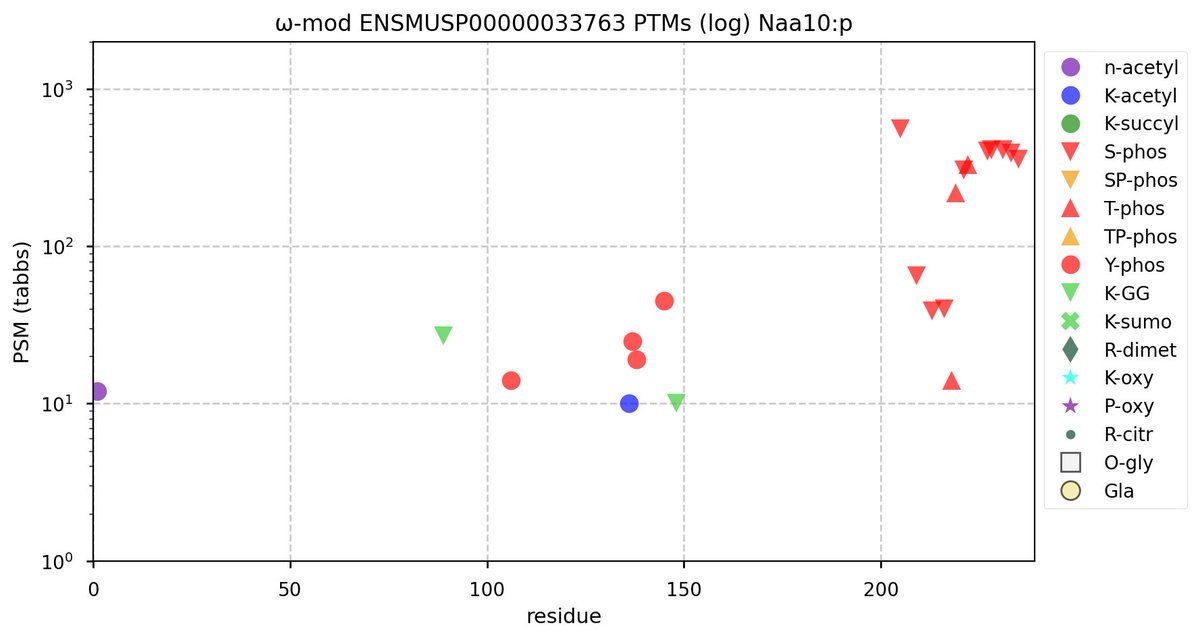
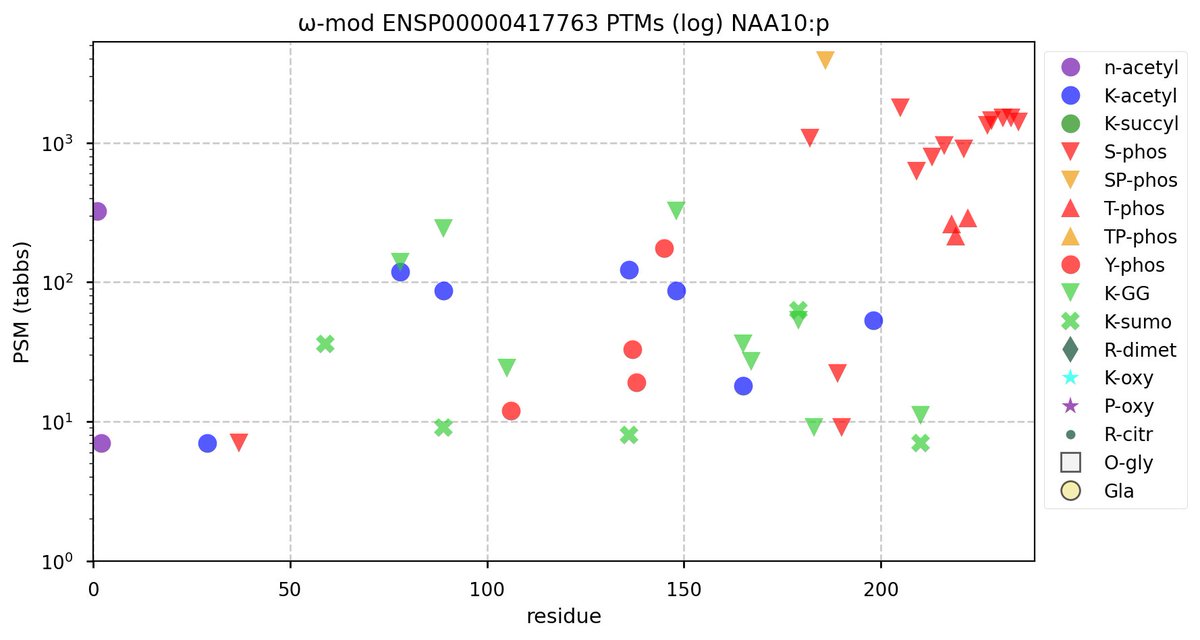
Sat May 21 22:20:53 +0000 2022Lots of storm activity along the cold front & further east. 🔗
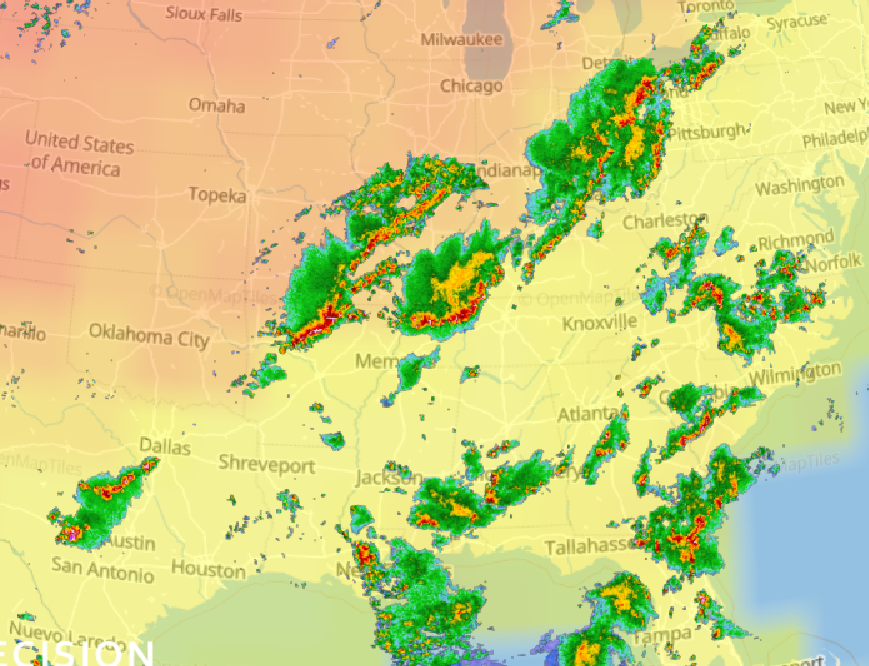
Sat May 21 19:00:46 +0000 2022Storms, as a cold front passes through the Midwest, right now 🔗
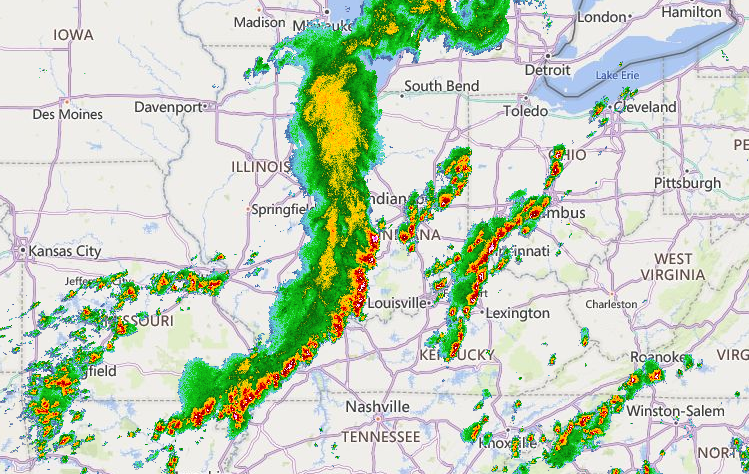
Sat May 21 18:15:19 +0000 2022Anyone who thinks that some individual is going to "cure cancer" hasn't been keeping up with the literature.
Sat May 21 17:53:45 +0000 2022🔗
Sat May 21 15:04:00 +0000 2022Cause & effect: the jet stream (250 hPa) winds across the same region 🔗
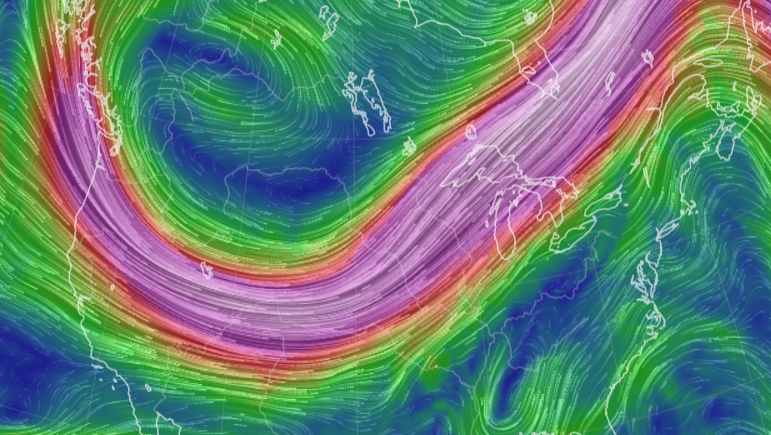
Sat May 21 15:01:21 +0000 2022The opening salvo in what promises to be an active day along a cold front running from Toronto to Dallas. 🔗
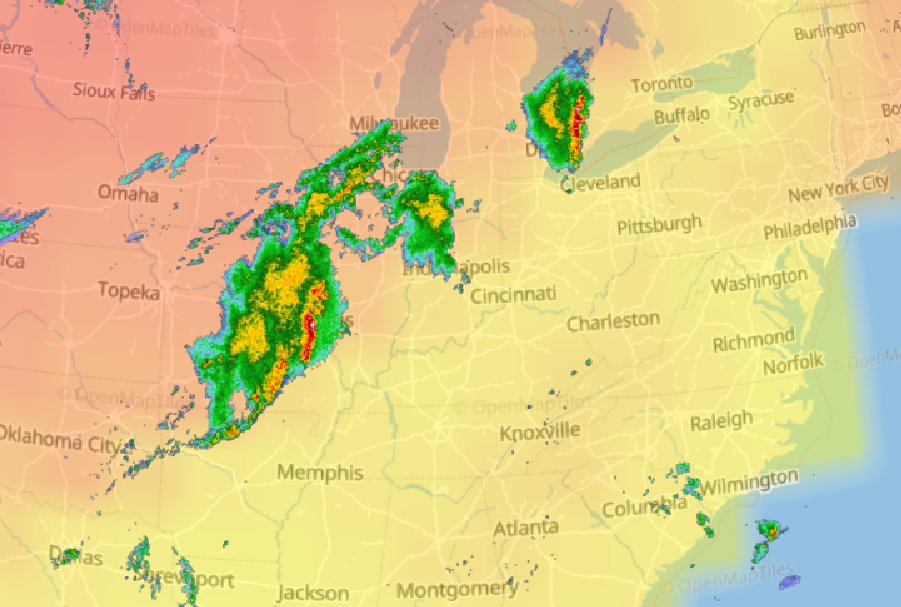
Sat May 21 13:58:20 +0000 2022The GRINL1A CTU phenomenon has been explored in some depth at the mRNA level, but how many of the observed mRNAs result in physiologically relevant proteins is unknown. The non-Central-Dogma nature of this effect leads to notation problems for biocurators.
Sat May 21 13:57:10 +0000 2022The absence of GGyl/SUMOyl acceptors is odd for a nuclear protein. POLR2M:g is part of an unusual locus: the GRINL1A complex translation unit (CTU) that includes the upstream MYZAP:g. This CTU produces multiple mRNAs composed of exons from both genes.
Sat May 21 13:36:54 +0000 2022Modification diagram for POLR2M:p, a subunit associated with human RNA polymerase II. This pattern is discordant with the other POLR2 complex subunits, resembling the pattern of POLR2 regulator proteins (e.g., RPRD1A:p) instead.
🔗
Fri May 20 20:43:56 +0000 2022HYPK-201, huntingtin interacting protein K
🔗
1 – 121 ⟶ 2 – 121 (N-acetyl-Ala, ο = 100%)
#realNT
Fri May 20 17:00:01 +0000 2022Guess the year of the flood. 🔗
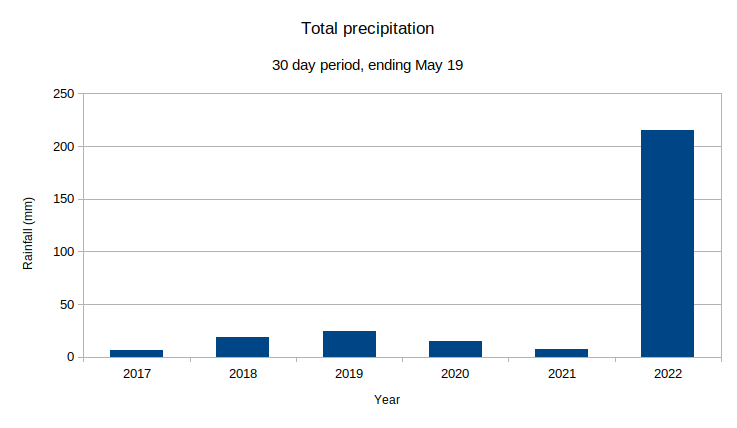
Fri May 20 15:51:17 +0000 2022Gina seems to be headed for New Caledonia next. 🔗
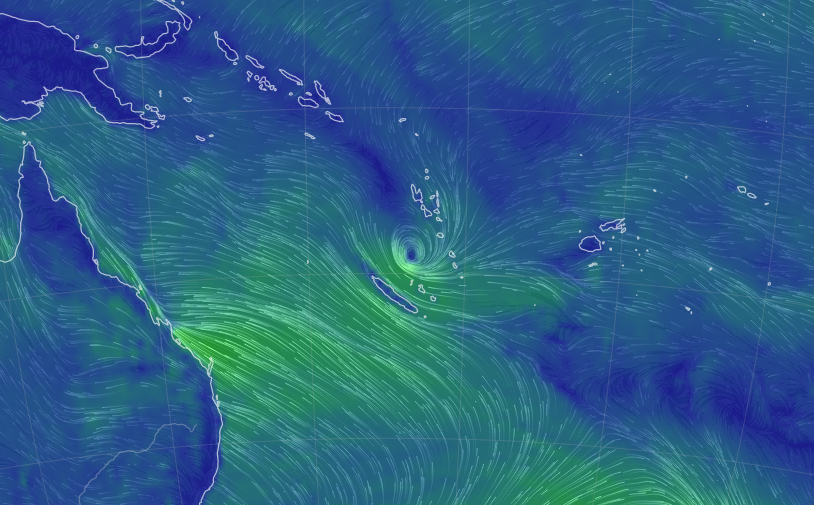
Fri May 20 14:54:33 +0000 2022@astacus I was thinking of Duns Scotus-style hats, but sure, a fascinator will do.
Fri May 20 14:45:32 +0000 2022OSBPL1A-204, oxysterol binding protein like 1A
🔗
1 – 437 ⟶ 2 – 437 (N-acetyl-Ser, ο = 100%)
#realNT
Fri May 20 14:42:03 +0000 2022OSBPL1A-201, oxysterol binding protein like 1A
🔗
1 – 950 ⟶ 1 – 950 (N-acetyl-Met, ο = 100%)
#realNT
Fri May 20 14:21:19 +0000 2022The only unusual feature is the lack of acetylation on the N-terminal Met residue. The mature protein begins with the rather hydrophobic sequence
1 MIIPV…
which seems to prevent the co-translational NATs from doing their work
Fri May 20 13:45:00 +0000 2022Modification diagram for POLR2L:p, a small subunit of human RNA polymerase I, II & III. The pattern rhymes nicely with the other subunits that are common to all 3 nuclear RNA polymerase complexes. #POLR2
🔗
Fri May 20 13:13:53 +0000 2022To be accurate, the one that set me off today was originally isolated in 1964, so it has actually been used for 58 years.
Fri May 20 13:07:59 +0000 2022One thing that is guaranteed to get me to stop reading a paper is when the text starts discussing results obtained from a cell line that has been commonly used for 40 years as though it was really an examination of cancer tissue.
Thu May 19 20:22:49 +0000 2022OLFML1-204, olfactomedin like 1
🔗
1 – 28 (transit peptide)
29 – 402 (mature sequence)
↓
1 – 26 (transit peptide)
27 – 402 (mature sequence, Q27+ammonia-loss, 100%)
#realNT
Thu May 19 17:46:26 +0000 2022@VATVSLPR Splice variant.
Thu May 19 17:35:02 +0000 2022@cajun_science It is often used to indicate that sequences are difficult to distinguish using a particular method. GSK3α & GSK3β are frequently referred to as isoforms because they can be hard to resolve via immunochemistry-based methods, even though they are quite different paralogs.
Thu May 19 16:47:01 +0000 2022Isoform is probably the most frequently misused term to describe a relationship between protein sequences. It should be officially retired as a descriptor & anyone using it should be made to wear a funny hat <:‑|
Thu May 19 16:17:12 +0000 2022@neely615 @JohnRYatesIII The best way to understand how these outfits work is to understand the tax laws (& loopholes) that make them possible.
Thu May 19 15:41:57 +0000 2022Tropical cyclone Gina making a direct hit on Vanuata 🔗
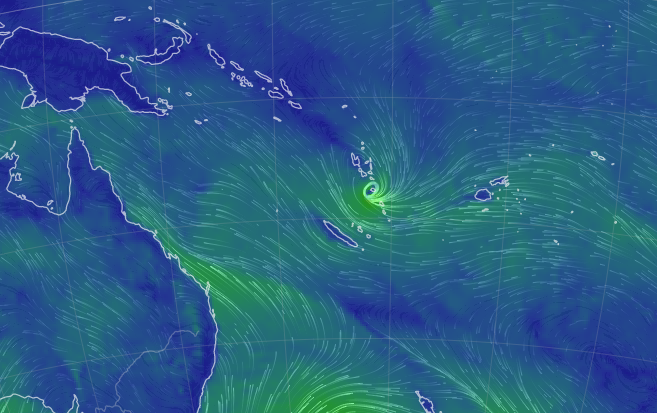
Thu May 19 15:07:58 +0000 2022POLR2M-201, RNA polymerase II subunit M
🔗
1 – 368 ⟶ 2 – 368 (N-acetyl-Cys, ο = 100%)
#realNT
Thu May 19 12:35:06 +0000 2022Like many other 𝑚𝑖𝑛𝑖 (𝑚𝑖𝑐𝑟𝑜. 𝑛𝑎𝑛𝑜?) proteins, this subunit is an integral part of a large complex. It has a yeast ortholog (🔗), so it doesn't seem to be easily replaceable, although the nomenclature for these little guys has remained fluid.
Thu May 19 12:26:34 +0000 2022Modification diagram for POLR2K:p, a very small subunit of human RNA polymerase I, II & III. The sequence is a bit short to declare a "trend", but it rhymes best with the N-terminus of POLR2A:p. #POLR2
🔗
Thu May 19 00:45:49 +0000 2022TTYH3-201, tweety family member 3
🔗
1 – 523 ⟶ 2 – 523 (N-acetyl-Ala, ο = 97%)
Wed May 18 16:54:30 +0000 2022ZWINT-203, ZW10 interacting kinetochore protein
🔗
1 – 277 ⟶ 1 – 277 (N-acetyl-Met, ο = 95%)
Wed May 18 15:02:08 +0000 2022CENPC-201, centromere protein C
🔗
1 – 2342 ⟶ 1 – 2342 (N-acetyl-Met, ο = 100%)
Wed May 18 12:56:38 +0000 2022Modification diagram for POLR2J:p, a small subunit of human the RNA polymerase II complex. Its pattern rhymes with the N-terminal domain of POLR2A:p―no phosphorylation & K acceptors specific for GGylation. #POLR2 🔗
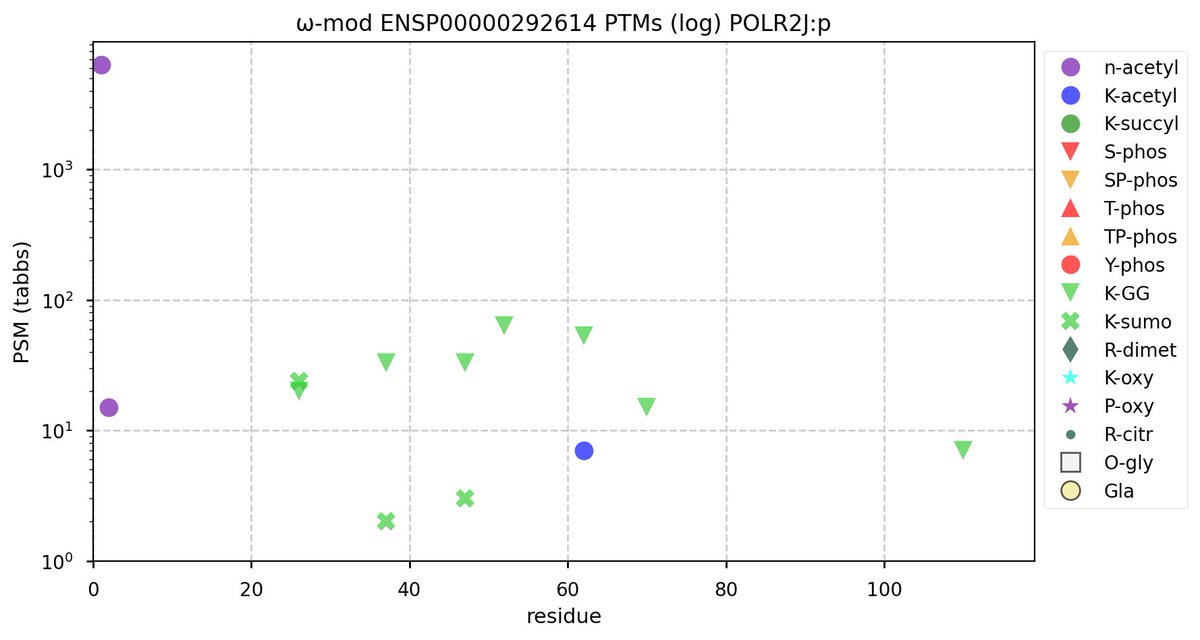
Tue May 17 17:15:04 +0000 2022RPS6KA4-202, ribosomal protein S6 kinase A4
🔗
1 – 772
↓
1 – 772 (N-acetyl-Met, ο = 13%)
2 – 772 (N-acetyl-Gly, ο = 40%)
Tue May 17 16:45:44 +0000 2022GFM2-201, GTP dependent ribosome recycling factor mitochondrial 2
🔗
1 – 779
↓
1 – 45 (transit peptide)
46 – 779 (mature sequence)
Tue May 17 16:31:40 +0000 2022TIPRL-202, TOR signaling pathway regulator
🔗
1 – 272
↓
1 – 272 (N-acetyl-Met, ο = 100%)
2 – 272 (N-acetyl-Met, ο = 100%)
Tue May 17 13:35:53 +0000 2022@AlexUsherHESA A trend toward treating Masters & PhD students like employees.
Tue May 17 13:28:52 +0000 2022The 4 main PTM acceptor types for #POLR2 (& subunit archetypes):
1. S+phosphoryl ΦP madness ("A" C-term domain);
2. K+GGyl ("A" N-term domain);
3. K+GGyl/SUMOyl/acetyl ("B"); &
4. Y+phosphoryl IHOPs ("I").
Tue May 17 12:54:12 +0000 2022Modification diagram for POLR2I:p, a small subunit of human RNA polymerase II. The subunit does not have detectable participation in K-based PTMs, but is the archetype for Y+phosphoryl involvement in this complex. #POLR2
🔗
Mon May 16 18:56:00 +0000 2022@LindorffLarsen Someone has figured out how to make posters an even worse experience.
Mon May 16 16:46:32 +0000 2022Still looking around for the kinase(s) involved: the first few dozen papers on kinase specificity haven't shed any light on the issue.
Mon May 16 12:48:13 +0000 2022EPPK1-202, epiplakin 1
🔗
1 – 5088 ⟶ 2 – 5088 (N-acetyl-Ser, ο = 100%)
Mon May 16 12:40:36 +0000 2022Modification diagram for POLR2H:p, a small subunit of human RNA polymerase I, II & III. The 3 Y+phosphoryl acceptors suggest this subunit may have some regulatory function. #POLR2
🔗
Sun May 15 21:09:19 +0000 2022I prefer Structural Transition Domain, but its acronym is problematic.
Sun May 15 19:59:07 +0000 2022🔗
Sun May 15 19:35:02 +0000 2022S100A16-202, S100 calcium binding protein A16
🔗
1 – 103 ⟶ 2 – 103 (N-acetyl-Ser, ο = 98%)
Sun May 15 18:03:29 +0000 2022Lightning beat Leafs in a forgone conclusion.
Sun May 15 17:59:55 +0000 2022POLR2K-201, RNA polymerase II, I and III subunit K
🔗
1 – 58 ⟶ 1 – 58 (N-acetyl-Met, ο = 100%)
Sun May 15 17:27:52 +0000 2022Trying to come up with a term that describes the phosphorylation that occurs at an acceptor in an intrinsically disordered domain immediately adjacent to an ordered domain: Transition Zone Phosphorylation (TZP) is my favorite at the moment.
Sun May 15 13:09:56 +0000 2022Modification diagram for POLR2G:p, a small subunit of human RNA polymerase II. The pattern rhymes with the N-terminal domain of POLR2A:p & is discordant with POLR2B:p. #POLR2
🔗
Sun May 15 00:46:45 +0000 2022Extra point for anyone who knows which kinase phosphorylates N-acetyl-Serine (I have no clue).
Sat May 14 13:27:18 +0000 2022An acetylated N-terminal serine serving as an Isolated High Occupancy Phosphorylation (IHOP) acceptor is not uncommon, although its significance is unknown.
Sat May 14 13:20:50 +0000 2022Modification diagram for POLR2F:p, a small subunit of human RNA polymerase I, II & III. It has few mods, but it does have a K+acetyl/GGyl/succinyl acceptor & a high occupancy N-terminal S+phosphoryl.
🔗
Sat May 14 12:19:57 +0000 2022Just because somebody has enough money to fund research doesn't turn them into Vannevar Bush. Most research doesn't generate very much new science: it is very easy to tip that balance towards no new science at all.
Fri May 13 19:55:04 +0000 2022🔗
Fri May 13 17:38:37 +0000 2022If you want to test a general purpose PTM-discovery PSM assignment algorithm & need a big volume of very challenging data, PXD022435 is pretty much tailor-made for the purpose.
Fri May 13 17:22:32 +0000 2022RRM2B-201, ribonucleotide reductase regulatory TP53 inducible subunit M2B
🔗
1 – 351 ⟶ 2 – 351 (N-acetyl-Gly, ο = 100%)
Fri May 13 15:02:25 +0000 2022TAMM41-205, TAM41 mitochondrial translocator assembly and maintenance homolog
🔗
1 – 452 ⟶ 2 – 452 (N-acetyl-Ala, ο = 100%)
#realNT
Fri May 13 14:55:16 +0000 2022Does anyone know when Uniprot is going to roll out their new web site design as their default interface? I thought it was supposed to happen last Monday (May 9).
Fri May 13 13:48:37 +0000 2022🔗
Fri May 13 13:13:20 +0000 2022Modification diagram for POLR2E:p, a subunit of human RNA polymerase I, II & III (yes, it is part of all 3 complexes). Like POLR2A:p, it has no K+acetyl site sharing with its complement of K+GGyl acceptors.
🔗
Fri May 13 02:33:03 +0000 2022Does anybody know if 🔗 has a publication associated with it?
Thu May 12 19:51:27 +0000 2022S100A2-202, S100 calcium binding protein A2
🔗
1 – 98
↓
1 – 98 (N-acetyl-Met, ο = 51%)
3 – 98 (N-acetyl-Cys, ο = 97%)
#realNT
Thu May 12 16:10:38 +0000 2022To whom it may concern: if you are looking at tumor lesion tissue from exterior-facing layers (skin, stomach, bladder), adding in sequences from well-known, site specific opportunistic pathogens (S. aureus, H. pylori, P. aeruginosa) will often produce interesting results.
Thu May 12 15:59:26 +0000 2022RHOD-201, ras homolog family member D
🔗
1 – 207 ⟶ 2 – 207 (N-acetyl-Thr, ο = 89%)
#realNT
Thu May 12 12:31:09 +0000 2022The modification diagram for POLR2D:p, one of the small subunits of human RNA polymerase II: the mature form weighs in at 141 aa. It is not involved in PTM high jinks in the manner of most of its fellow complex members. #POLR2
🔗
Wed May 11 15:19:30 +0000 2022If you are interested in the pathogen Pseudomonas aeruginosa (commonly observed in urine samples), PXD025827 has some excellent examples of what you should be seeing in clinical isolates.
Wed May 11 14:24:10 +0000 2022BTW, in humans the RNA polymerase II complex has 13 subunits, while the analogous complex in prokaryotes has 5. Mitochondrial RNA polymerase only has 1 subunit, as do those in chloroplasts & many bacteriophages.
Wed May 11 14:14:56 +0000 2022POLR2D-201, RNA polymerase II subunit D
🔗
1 – 142 ⟶ 2 – 142 (N-acetyl-Ala, ο = 100%)
#realNT
Wed May 11 13:18:38 +0000 2022@edward_marcotte @JohnRYatesIII @jurirappsilber @NatureBiotech @GeorgKus @gingraslab1 @lilley_ks @guomics @Prof_Lundberg @hhermjakob @TreyIdeker @RalserLab @tpc_tom @RappsilberLab They are the most frequently observed, but I doubt whether most investigators could give you a few coherent extemporaneous paragraphs on keratin structure & function or albumin's journey from liver to lysosome.
Wed May 11 12:08:17 +0000 2022Modification diagram for human POLR2C:p, a subunit of the RNA polymerase II complex. It has no K+acetyl alternates at K+GGyl acceptors, but has significant K+SUMOyl alternates instead. Two prominent Y+phosphoryl acceptors. #POLR2
🔗
Wed May 11 11:52:41 +0000 2022Cyclone Asani making landfall this morning. 🔗
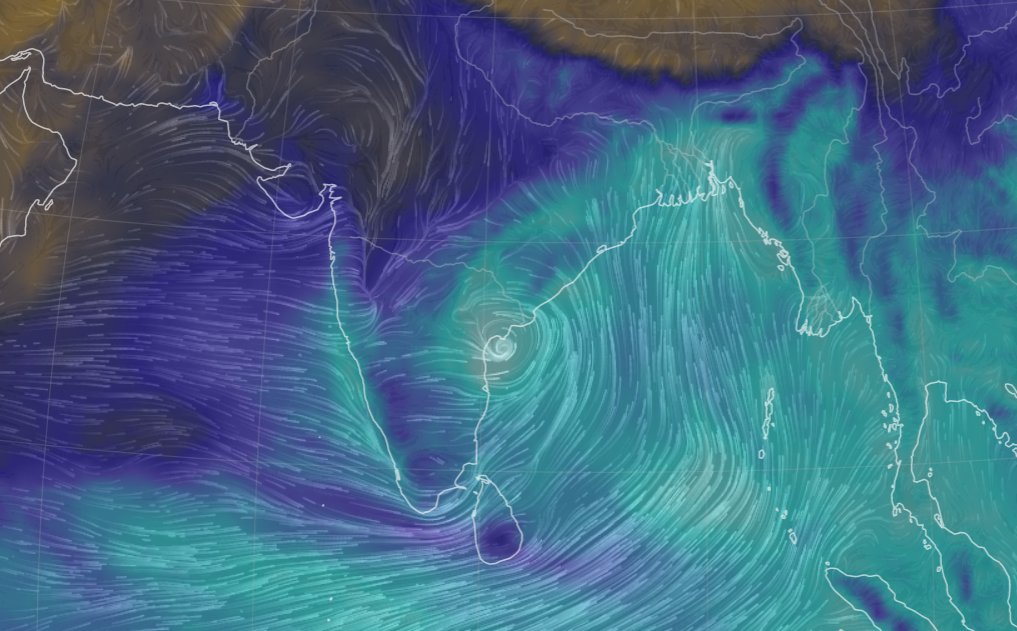
Tue May 10 18:24:06 +0000 2022@J_my_sci jacarandas
Tue May 10 16:53:14 +0000 2022I wish you folks the best of luck
🔗
Tue May 10 15:53:52 +0000 2022@dtabb73 @Smith_Chem_Wisc Do "GIF" next!
Tue May 10 15:26:45 +0000 2022If you are interested in the human pathogen Proteus mirabilis & want to know what you can see using proteomics methods, examining PXD031222 would be a good place to start.
Tue May 10 13:25:04 +0000 2022Further unsolicited advice: never use the same statistical methods for QC that were used for QA on a dataset.
Tue May 10 13:03:15 +0000 2022The differences between the A & B subunits illustrate how ill-advised it is to apply naïve statistical methods to PTM experimental results, even though these 2 subunits are physically close to each other in the same complex.
Tue May 10 12:55:18 +0000 2022Modification diagram for human POLR2B:p, part of the RNA polymerase II complex. #POLR2
Compared to POLR2A:p
Like:
- lots of K+GGyl
Unlike:
- no extended phosphodomains
- many GGyl alternate with acetyl
- multiple Y+phosphoryl acceptors
🔗
Mon May 09 17:44:21 +0000 2022ODF1-201, outer dense fiber of sperm tails 1
🔗
1 – 250 ⟶ 2 – 250 (N-acetyl-Ala, ο = 100%)
#realNT
Mon May 09 16:17:16 +0000 2022Cyclone Asani in the Bay of Bengal (surface winds) 🔗
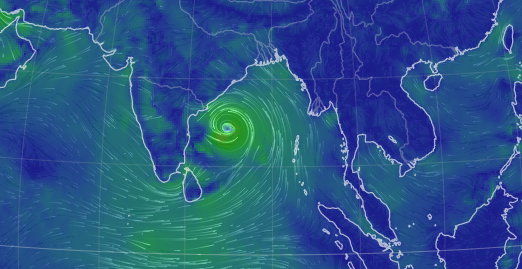
Mon May 09 14:24:11 +0000 2022POLRMT-202, RNA polymerase mitochondrial
🔗
1 – 41 (transit peptide)
42 – 1230 (mature sequence)
↓
1 – 40 (transit peptide)
41 – 1230 (mature sequence)
#realNT
Mon May 09 14:15:01 +0000 2022And, for emphasis, a rhyming pattern many M. musculus perished to produce
🔗
Mon May 09 13:15:57 +0000 2022It should be noted that there are probably a lot more C-terminal phosphorylation acceptors, but 2 big tryptic peptides (1482-1603) & (1604-1810) are too large to be observed by commonly used LC/MS/MS methods.
Mon May 09 12:57:46 +0000 2022Modification diagram for human POLR2A:p, a subunit of the RNA Polymerase II complex. The rather garish C-terminal phosphodomain grabs the eye, but the lack of acetylation alternating at all of those GGylation acceptors is a bit of a surprise. #POLR2
🔗
Mon May 09 12:49:59 +0000 2022It would be nice if Texas stopped pumping so much wet air up to the northern Great Plains. 3rd big storm in 3 weeks. 🔗
Mon May 09 12:00:41 +0000 2022#resMods: Tyr/Y
N-amino: none
Sidechain: reactive phenol
«bio―hydroxyl»
+phosphoryl; +sulfonyl
«bio―aromatic¹»
+iodo; +diiodo; +triiodo
C-carboxyl: none
Note:
1. On thyroglobulin in thyroid tissue only.
Sun May 08 19:52:51 +0000 2022γ-carboxyglutamic acid (Gla) is a PTM that only occurs on a few blood plasma proteins, but when it is present, it isn't subtle.
🔗
Sun May 08 18:30:47 +0000 2022I wonder if anyone at Nature honestly believes that I'm going to pay US$199 to read a 50 year old paper.
Sun May 08 13:22:39 +0000 2022The entire mechanism associated with the mitochondrial ribosome has been maintained to translate the 13 protein-coding genes encoded on the mitochondrial chromosome.
Sun May 08 13:14:30 +0000 2022Modification diagram for RPL21:p & MRPL21, subunits of the cytosolic & mitochondrial ribosomes, respectively. The cytosolic assembly has a lot more bells & whistles on it than the PTM-minimalist prokaryotic-style mitoribosome.
#rpsubs 🔗
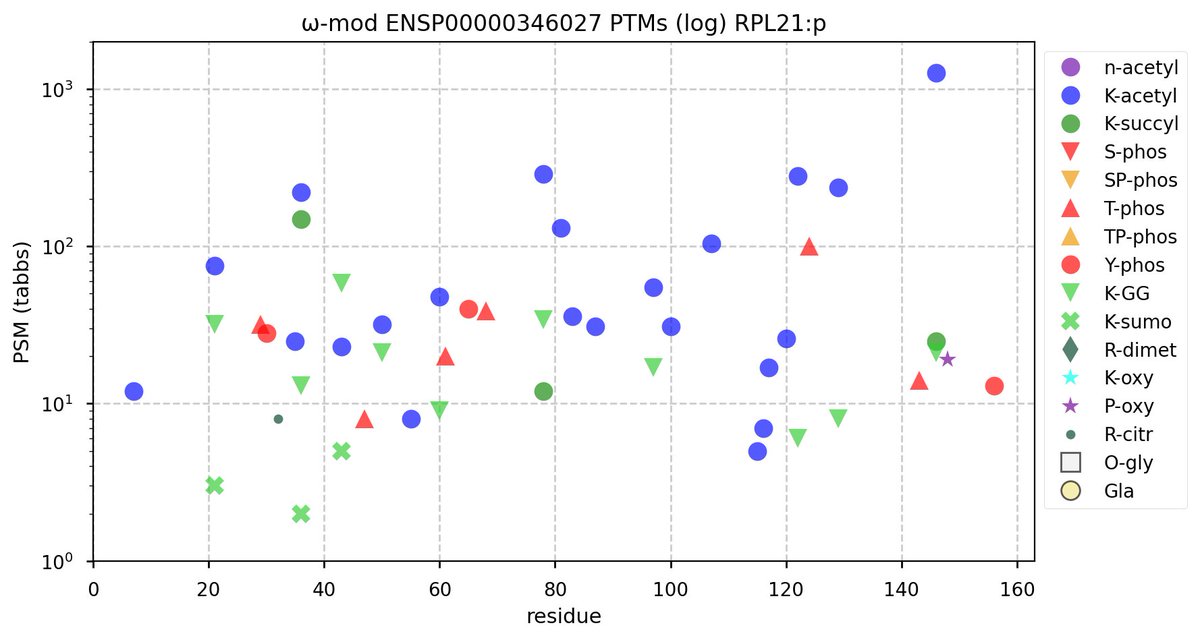
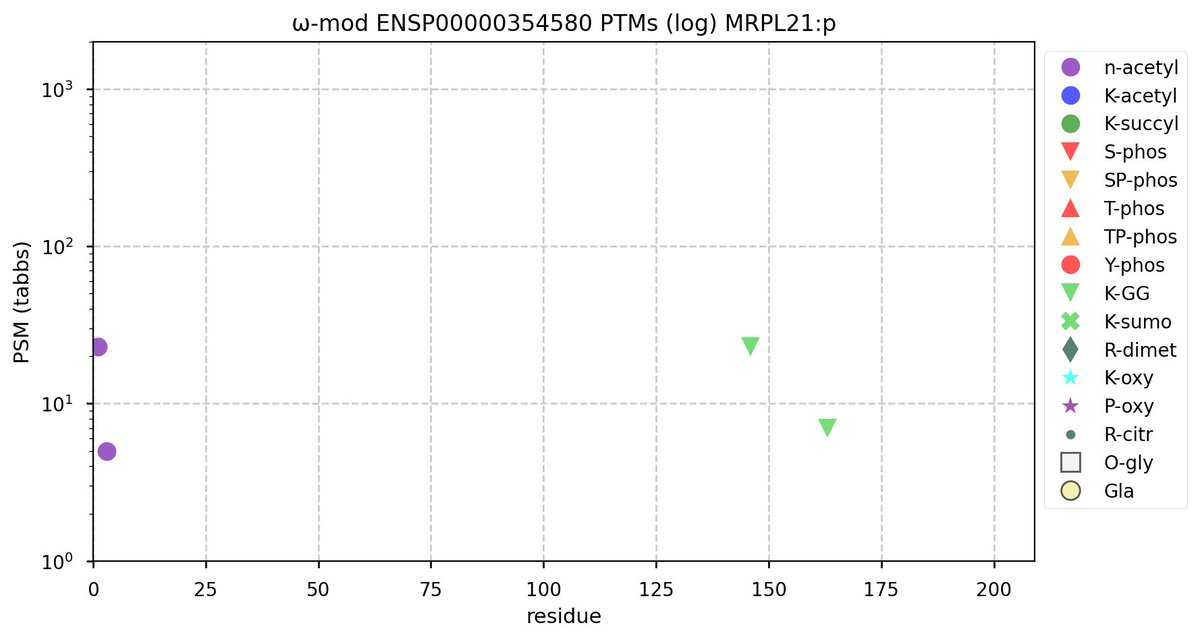
Sun May 08 13:02:14 +0000 2022@canto_general @HeerJeet CBC considers it to be BC regional news
🔗
Sun May 08 12:24:33 +0000 2022#resMods: Trp/W
N-amino: none
Sidechain: reactive indole
«artifact―redox»
+O; +2O
C-carboxyl: none
Sat May 07 15:32:30 +0000 2022Mitochondrial subunits are kind of boring, in that they aren't going to show up on any list of differentially expressed proteins, but they are the components of little devices that are, for my money, the most amazing machines ever to come in to existence.
Sat May 07 13:37:01 +0000 2022MRPS21-202, mitochondrial ribosomal protein S21
🔗
1 – 87 → 2 – 87 (N-acetyl-Ala, ο = 100%)
#realNT
Sat May 07 13:16:03 +0000 2022Modification diagrams for RPS21:p & MRPS21:p, subunits of the cytosolic & mitchondrial ribosomes, respectively. The data suggests that MRPS21:p gets into the mitochondrial matrix as the intact, N-acetylated (2-87), just the way it rolls off the "other" ribosome. #rpsubs 🔗
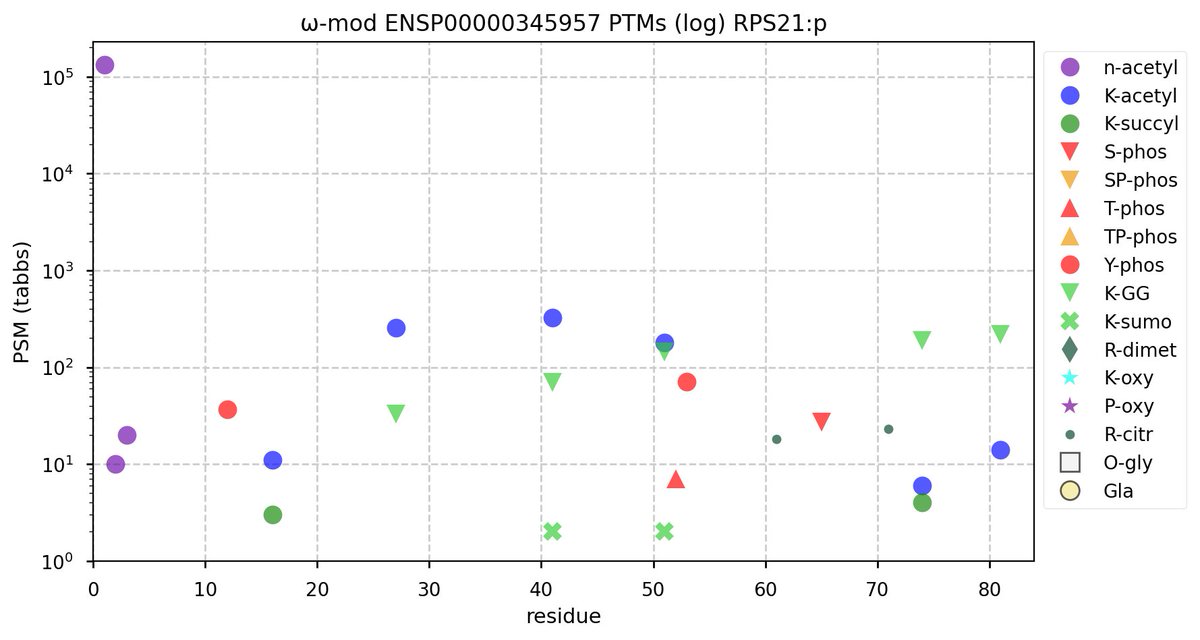
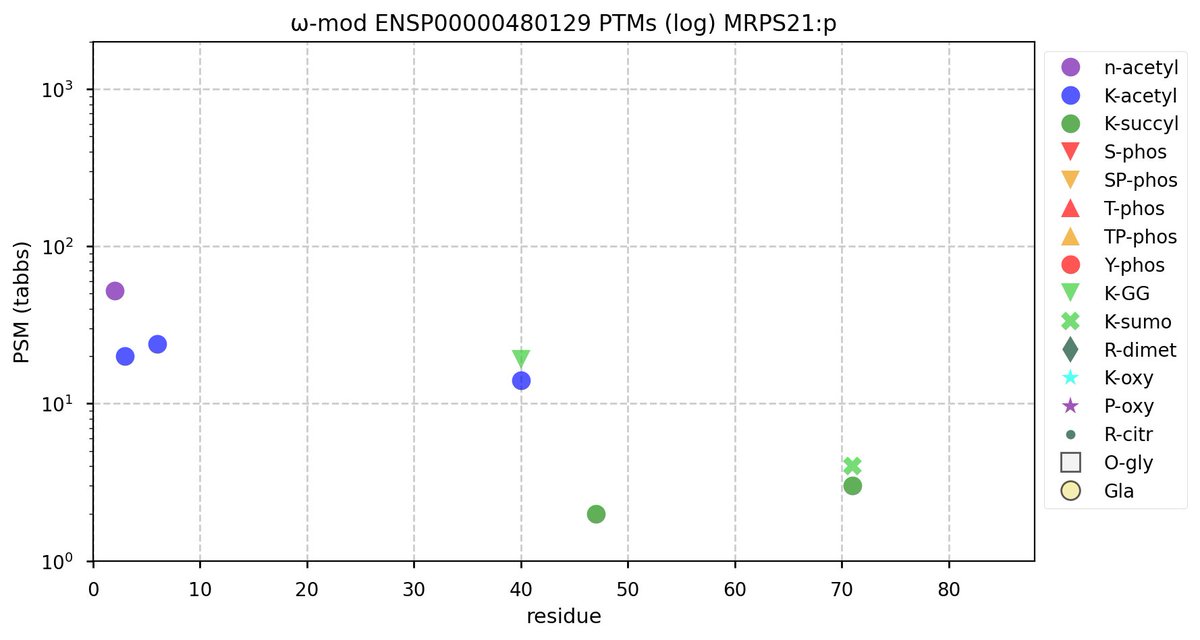
Sat May 07 12:09:56 +0000 2022#resMods: Val/V
N-amino: +acetyl¹
Sidechain: not reactive
C-carboxyl: none
Notes:
1. Protein N-terminus.
Sat May 07 11:39:21 +0000 2022@YishaiLevin That there is a significant difference in the patterns of modification between the the cytosolic & mitochondrial ribosomes, most noticeably in the alternating lysine modifications acetyl/succinyl/GGyl/SUMOyl.
Fri May 06 20:04:54 +0000 2022Got this automated message this morning:
"GPMDB build 6666 complete."
Sounds ominous (& a little biblical). My own bots creep me out sometimes.
Fri May 06 15:47:40 +0000 2022A shorter version of RPL10:p―missing (1-51)―is commonly found in some cell lines, e.g. HEK-293T & MDA-MB-231. It is unclear whether this is caused by alternate splicing, alternate translation initiation or a specific proteolytic cleavage (the most likely suspect).
Fri May 06 15:46:38 +0000 2022Modification diagrams for human RPL10:p & MRPL10:p, subunits of the cytosolic & mitochondrial ribosomes, respectively. "1812 Overture" (with cannons) vs "Ambient 1: Music for Airports"
#rpsubs 🔗
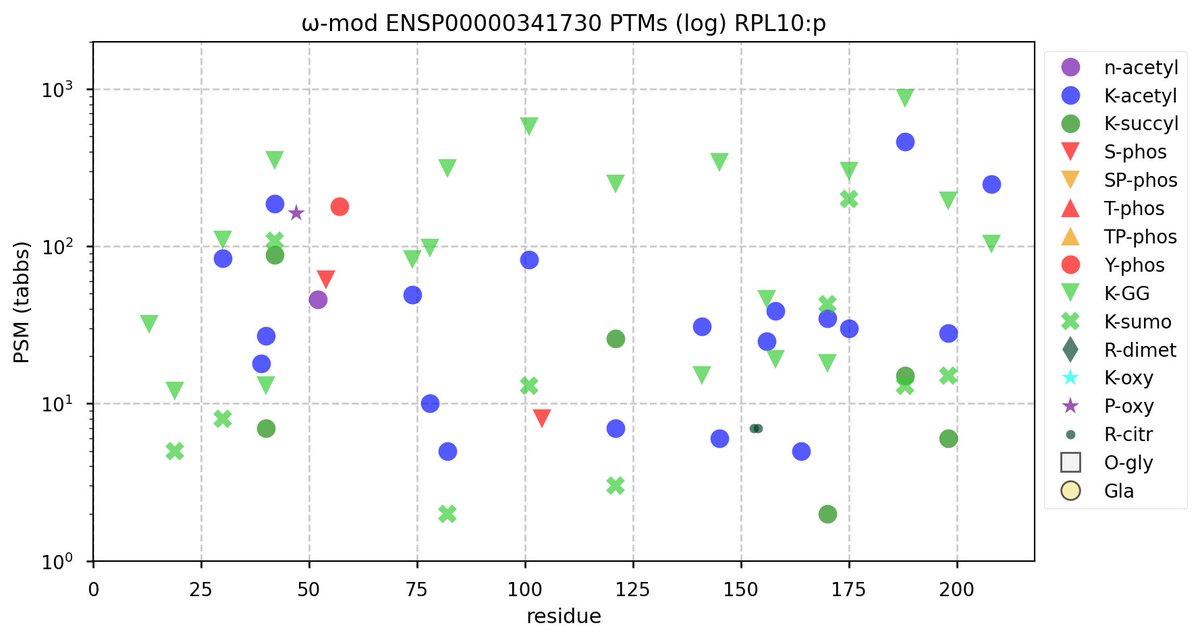
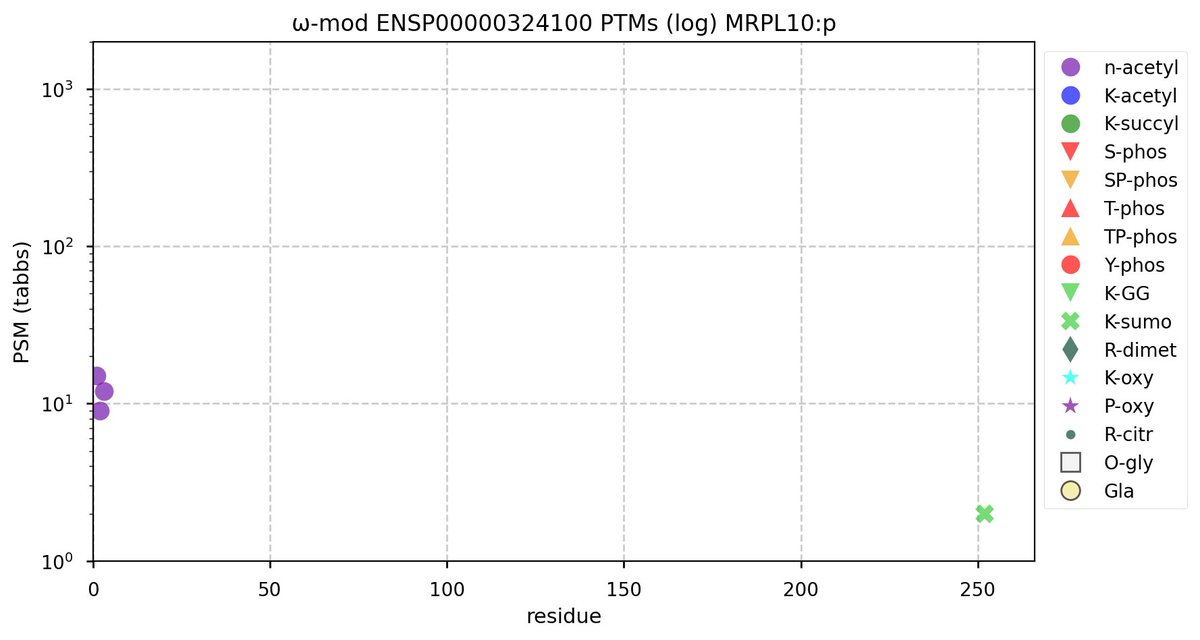
Fri May 06 11:58:04 +0000 2022#resMods: Sec/U ¹
N-amino: none
Sidechain: reative 1° selenol
«artifact»
+carboxamidomethyl ²
C-carboxyl: none
Notes:
1. A sparingly-used translation of the UGA codon.
2. Selenol is derivatized by most sulfhydryl blocking reagents.
Fri May 06 10:45:22 +0000 2022A line of intense storms heading towards Tallahassie. 🔗

Fri May 06 01:26:44 +0000 2022Some heavy weather running along a cold front stretching from San Antonio to Nashville. 🔗

Thu May 05 15:56:51 +0000 2022& Genome Canada & Compute Canada & Innovation Canada & on & on ...
Thu May 05 15:53:02 +0000 2022Sometimes I forget that Canada still has Superclusters!
Thu May 05 14:46:16 +0000 2022@VATVSLPR Thanks to those who contributed, but I think just how this happened is still in the "unsolved mystery" column. I do find this phenomenon occurring from time-to-time: hopefully what ever is going wrong will eventually die out with improved training.
Thu May 05 14:34:17 +0000 2022Emphasis on the "Mini"
🔗
Thu May 05 14:22:37 +0000 2022Modification diagrams for human RPL3:p & MRPL3:p, uL3 subunits of the cytosolic & mitochondrial ribosomes. While lysine mods are not absent from mitoribosomal subunits, they are timorous pianissimo compared to their cytosolic counterparts raucous fortissimo.
#rpsubs 🔗
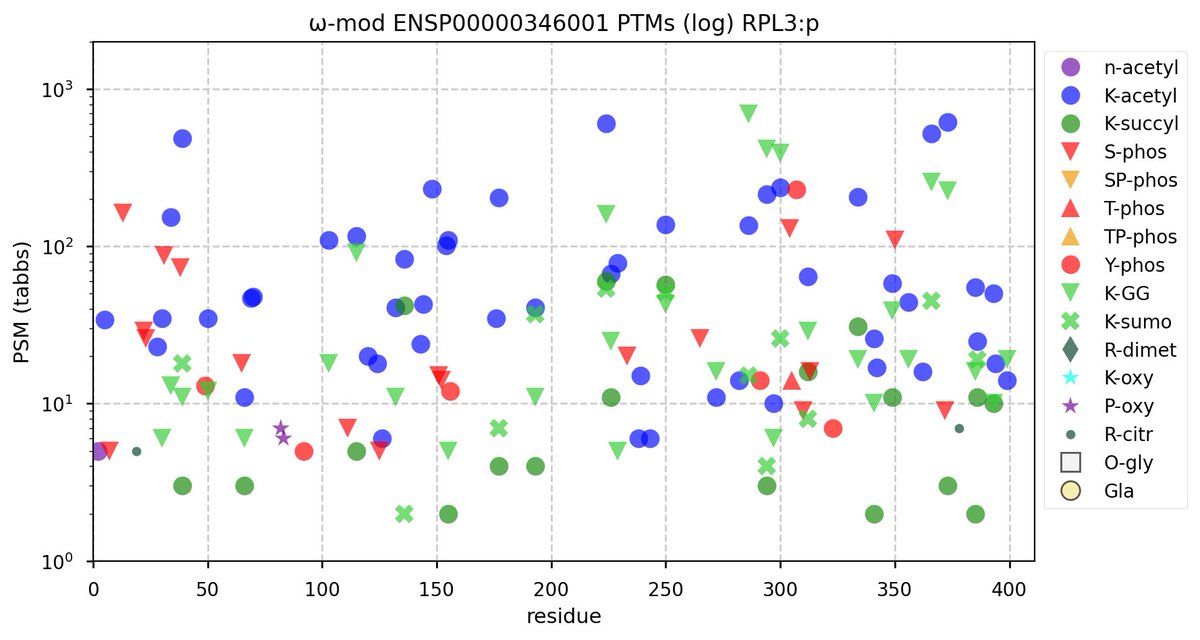
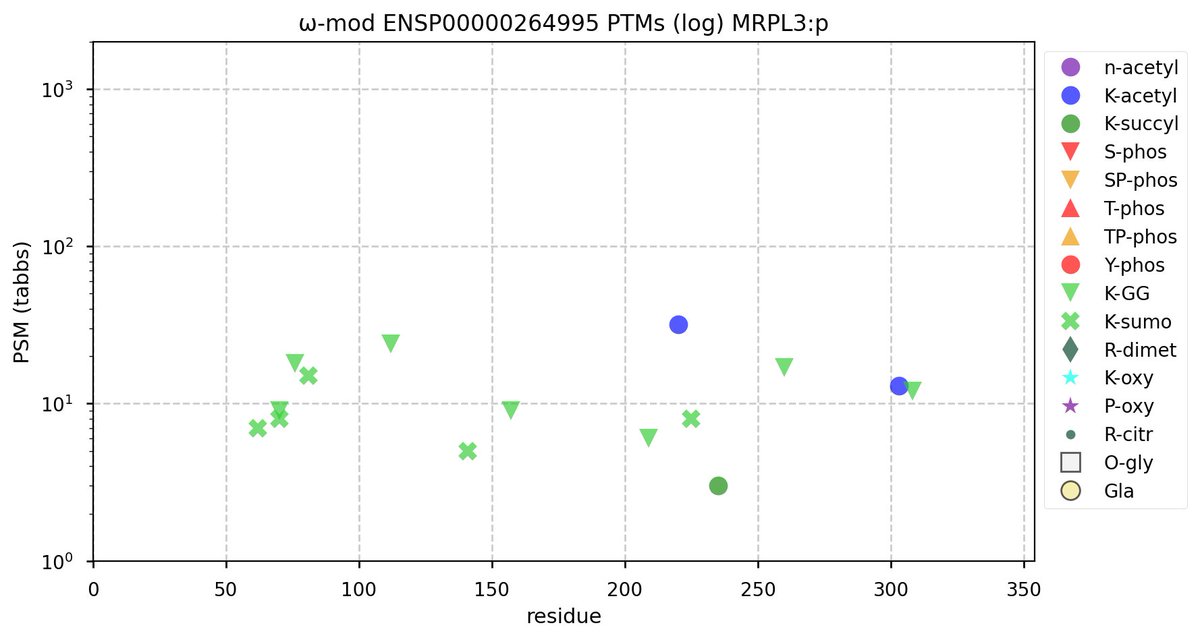
Thu May 05 13:10:18 +0000 2022@neely615 @astacus @ProteomicsNews Once you have processed your grief, I'll break the bad news about Carrie-Anne Moss & Ryan Reynolds.
Thu May 05 12:55:16 +0000 2022A line of rather spicy-looking weather heading east, just north of the Mississippi Delta (NEXRAD). 🔗

Thu May 05 12:26:18 +0000 2022@astacus @neely615 @ProteomicsNews Many find Canadians off-putting.
Thu May 05 12:23:29 +0000 2022#resMods: Thr/T
N-amino: +acetyl¹
Sidechain: reactive 2° alcohol
«bio»
+phosphoryl; +glycosyl²;
«artifact»
-H₂O ³
C-carboxyl: none
Notes:
1. Protein N-terminus.
2. O-linked glycosylation: many glycoforms are possible.
3. β-elimination.
Thu May 05 12:04:41 +0000 2022MRPS12-201, mitochondrial ribosomal protein S12
🔗
1 – 29 (transit peptide)
30 – 138 (mature sequence)
↓
1 – 30 (transit peptide)
31 – 138 (mature sequence)
#realNT
Thu May 05 00:40:21 +0000 2022@VATVSLPR They didn't mention the glutathionyl derivative: I found it while reanalyzing their data. With the IAA settings they used they probably didn't find any C-containing peptides at all.
Wed May 04 22:35:26 +0000 2022If anyone can explain to me how the method described here 🔗 & repeated here 🔗 generates C+glutathionyl & no C+carbamidomethyl, I will declare you "Proteomics Master™".
Wed May 04 15:07:43 +0000 2022MRPS6:p is one of a small number of proteins that manage to scoot from the cytosol, through 2 mitochondrial membranes & end up in the mitochondrial matrix without losing a transit peptide (mature form 2-125).
Wed May 04 15:07:19 +0000 2022Modification diagrams for human RPS6:p & MRPS6:p, subunits of the cytosolic & mitochrondrial ribosomes, respectively. Even though the names suggest homology, these proteins are part of different families (PF01092 vs PF01250). #rpsubs 🔗
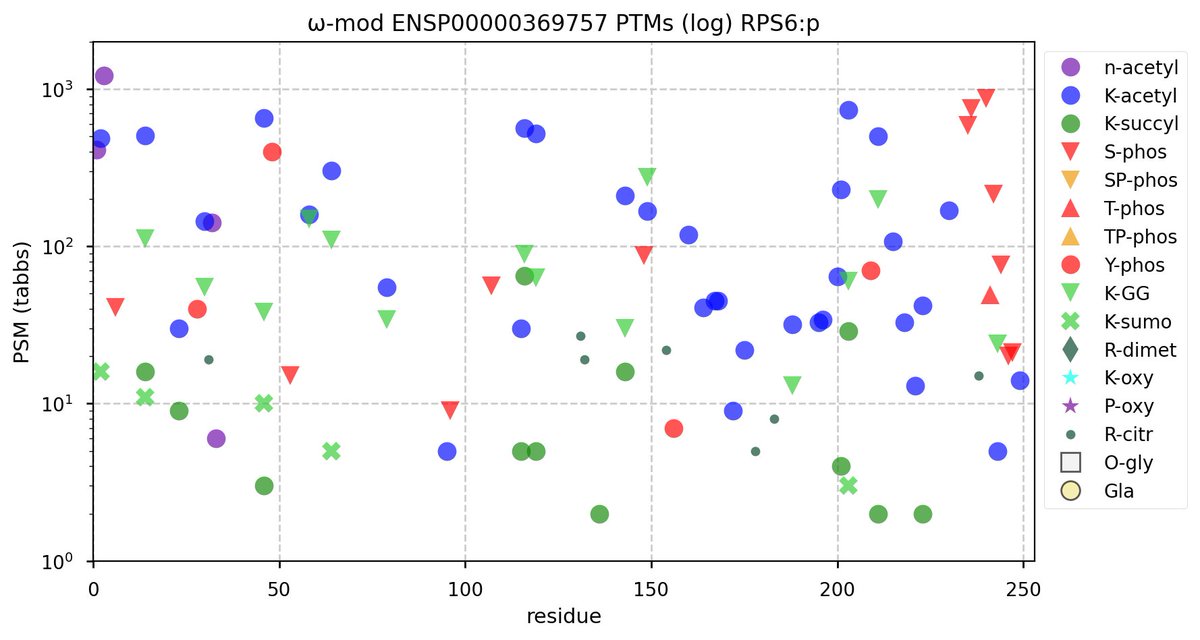
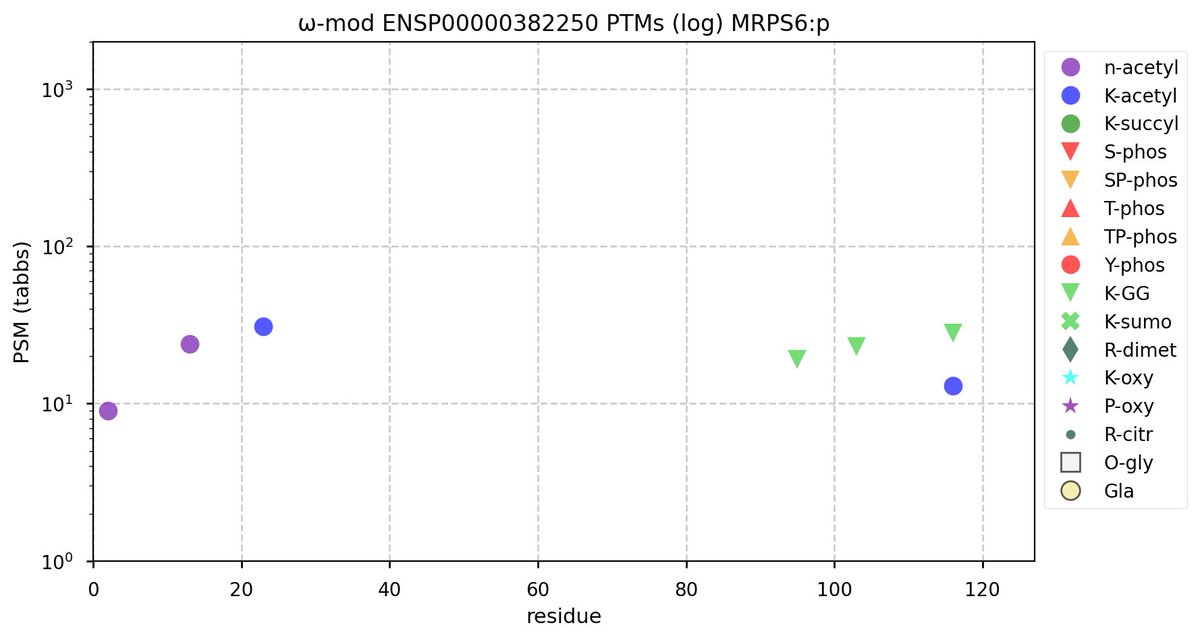
Wed May 04 14:27:00 +0000 2022MRPS6-201, mitochondrial ribosomal protein S6
🔗
1 – 125 ⟶ 2 – 125 (N-acetyl-Pro, ο = 0.3%)
#realNT
Wed May 04 12:38:42 +0000 2022#resMods: Ser/S
N-amino: +acetyl¹
Sidechain: reactive 1° alcohol
«bio»
+phosphoryl; +glycosyl²;
«artifact»
-H₂O ³
C-carboxyl: none
Notes:
1. Protein N-terminus.
2. O-linked glycosylation: many glycoforms are possible.
3. β-elimination.
Tue May 03 16:09:59 +0000 2022To anyone interested in protein subunit nomenclature, Ban, et al. (2014) does a pretty good job of explaining how to think through the problem for all types of ribosomes.
🔗
Tue May 03 16:09:44 +0000 2022The same pair of diagrams for mouse RPS2:p & MRPS2:p. 🔗
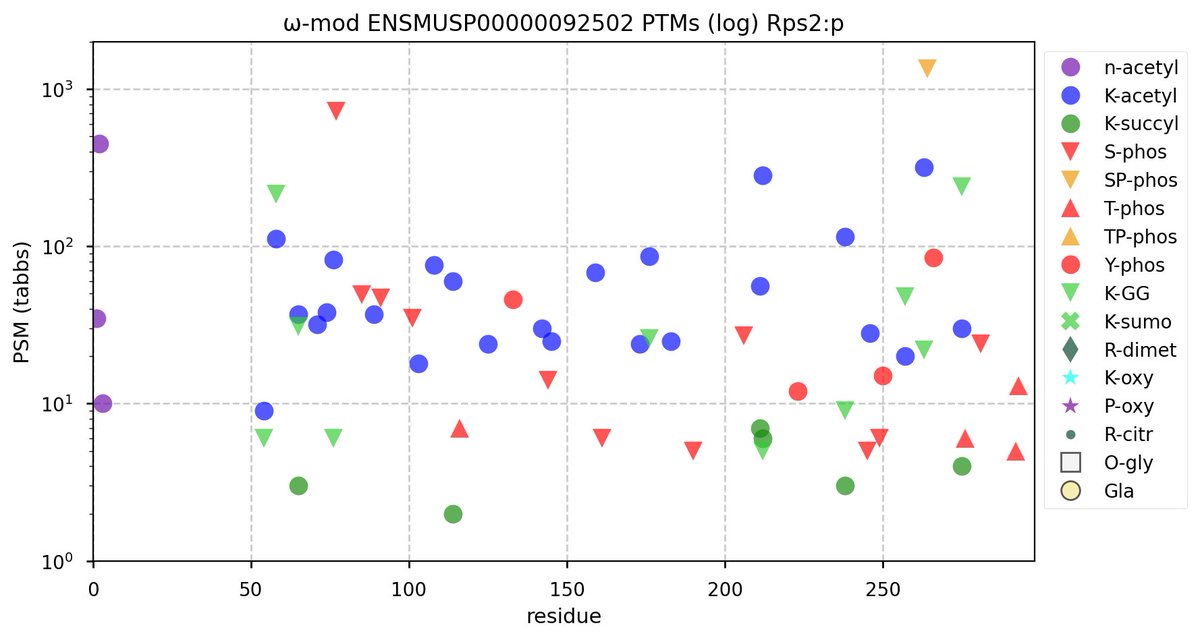
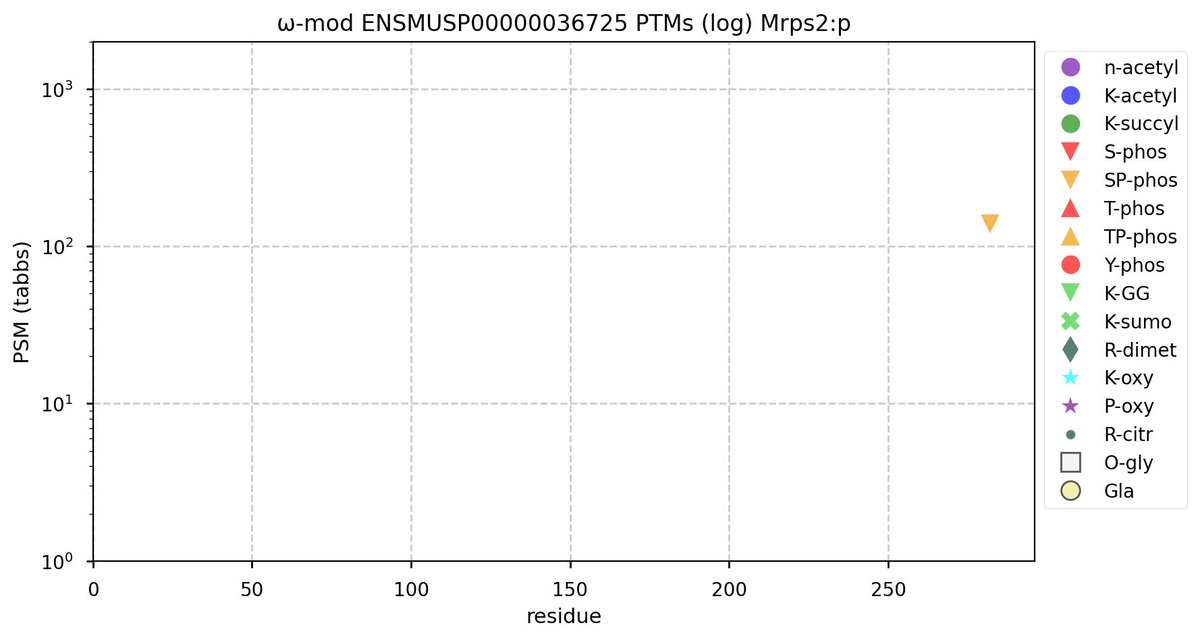
Tue May 03 16:09:19 +0000 2022Modification diagrams for human RPS2:p & MRPS2:p, the homologous uS5 subunits of the cytosolic and mitochondrial ribosomes, respectively. #rpsubs 🔗
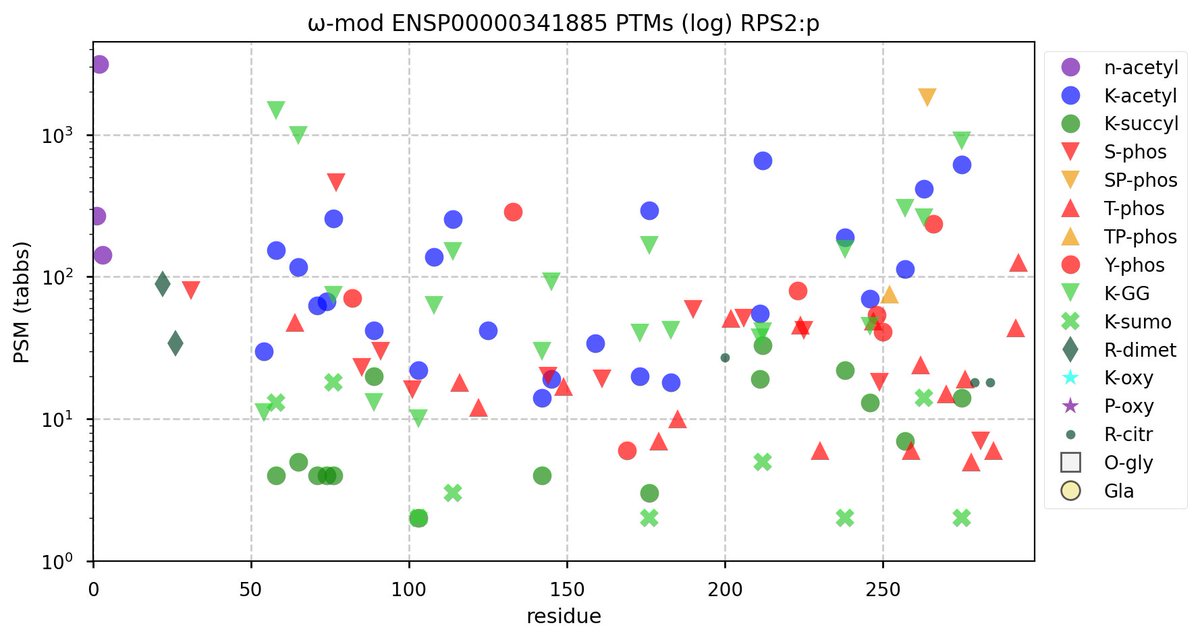
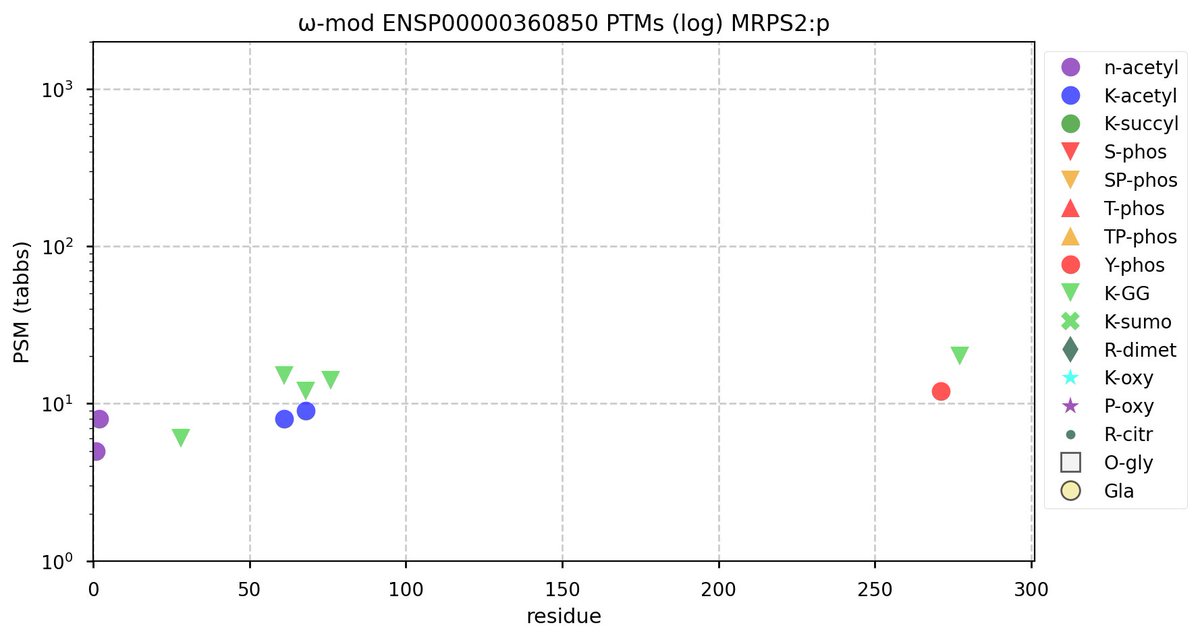
Tue May 03 14:31:24 +0000 2022Students should note that although deamidation (N,Q) & deimidation (R) both result in the same change in atomic composition (NH → O, Δm = 0.984016 u), they are very different reactions with unrelated biological consequences.
Tue May 03 14:14:07 +0000 2022#resMods: Arg/R
N-amino: none
Sidechain: reactive guanadinyl
«bio»
+deimidation¹; +methyl; +dimethyl; +phosphoryl²
C-carboxyl: none
Notes:
1. Forms citrulline.
2. Absent from chordates.
Tue May 03 13:52:36 +0000 2022MRPS2-202, mitochondrial ribosomal protein S2
🔗
1 – 296
↓
1 – 41,42 (transit peptide)
42,43 – 296 (mature sequence)
#realNT
Mon May 02 17:52:50 +0000 2022#resMods: Gln/Q
N-amino: none
Sidechain: reactive -CONH₂
«bio»
+cyclization¹; deamidation²
C-carboxyl: none
Notes:
1. Protein (or peptide) N-terminus: forms pyroglutamic acid.
2. Results in Glu: rate depends on adjacent residues.
Mon May 02 14:18:48 +0000 2022Note to newspaper sites: if I am not a paying subscriber, reminding me of this fact in ever escalating banner pages or vague teaser paragraphs is not going to win me over.
Mon May 02 12:46:01 +0000 2022Modification diagrams for yeast & human IST1:p, orthologs of an "accessory" ESCRT-III subunit. Seems pretty different to me. 🔗
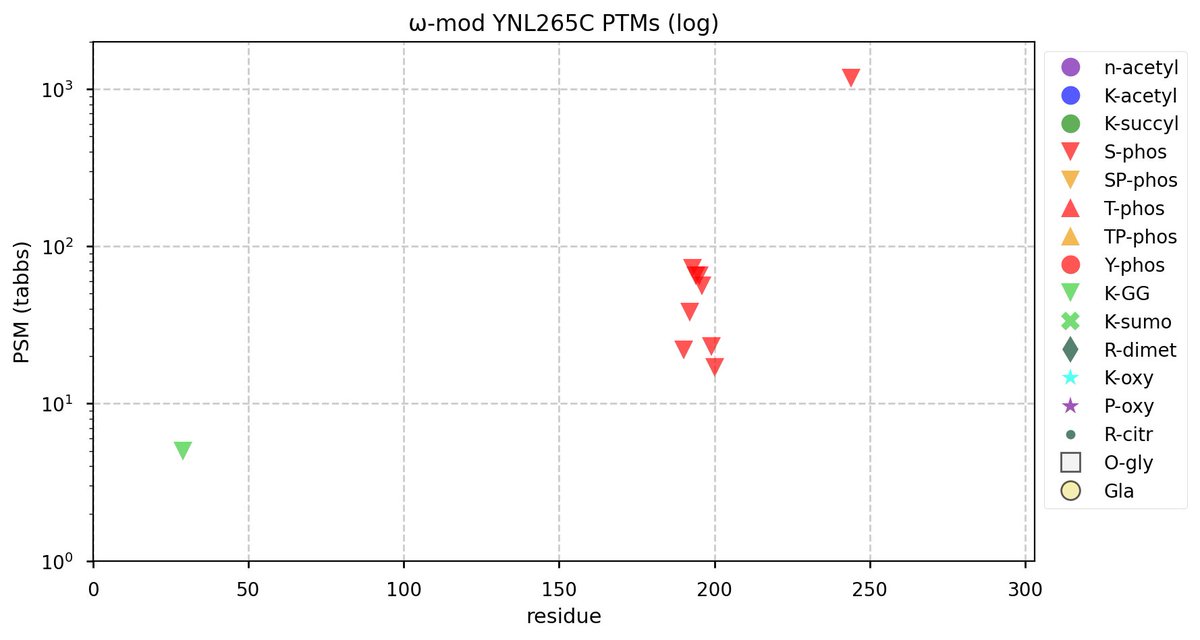
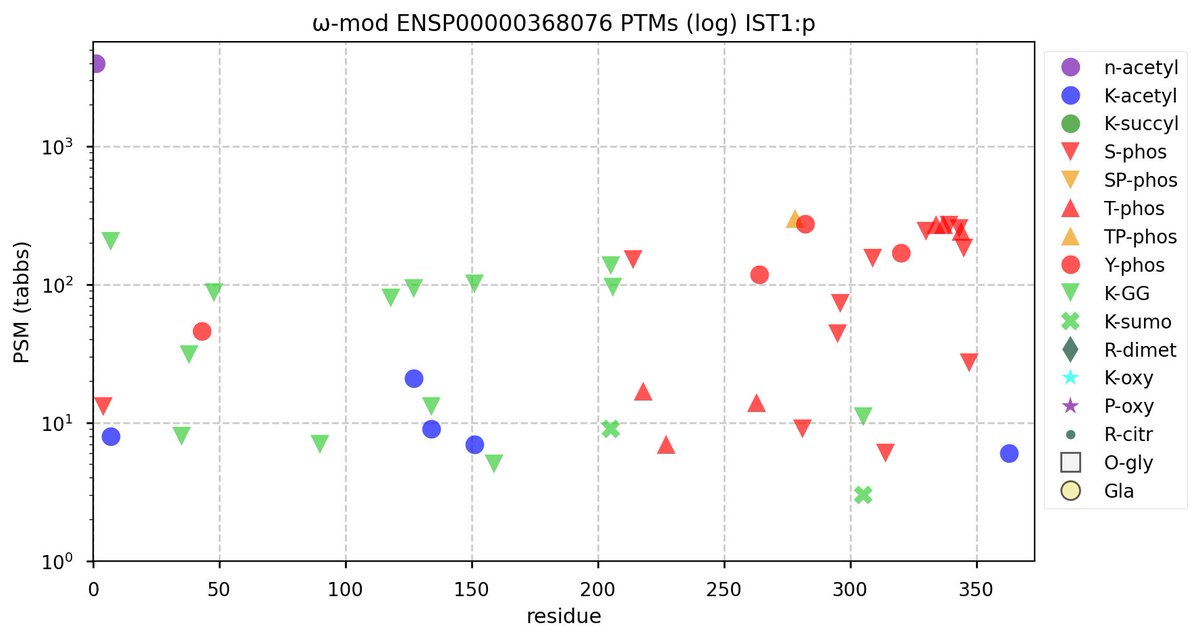
Sun May 01 15:13:21 +0000 2022Modification diagrams for yeast DID2:p & human orthologs CHMP1A:p & CHMP1B:p, an ESCRT-III complex subunit. The human paralogs share a rhyming pattern of GGylation in their N-terminal domain, even though they do not share any tryptic peptides. 🔗
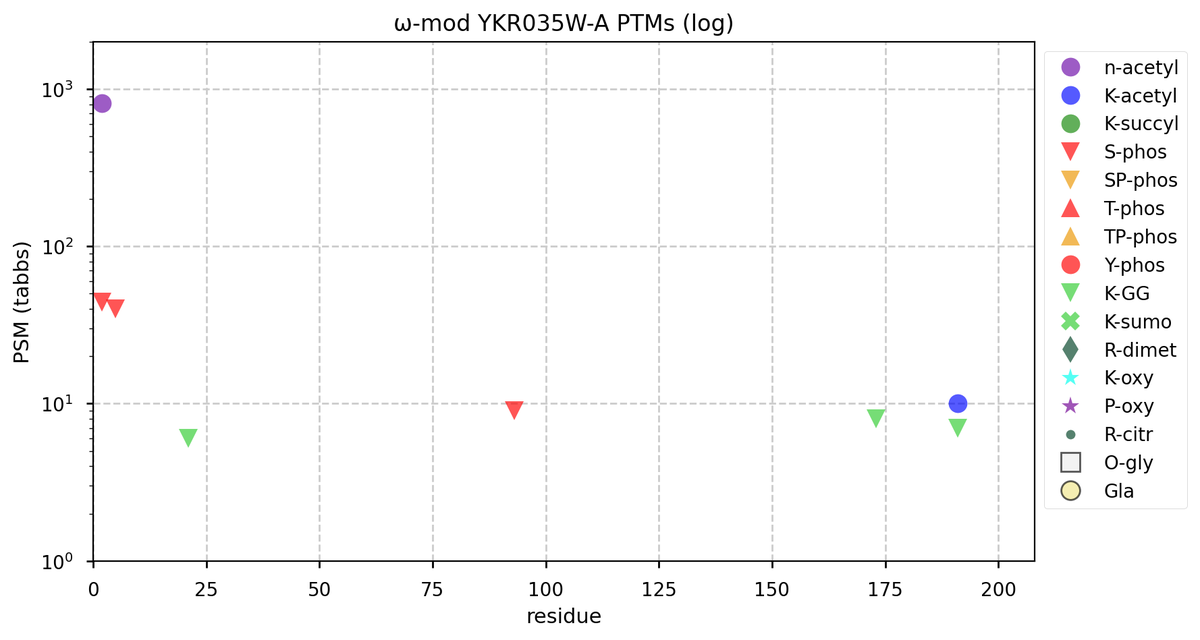
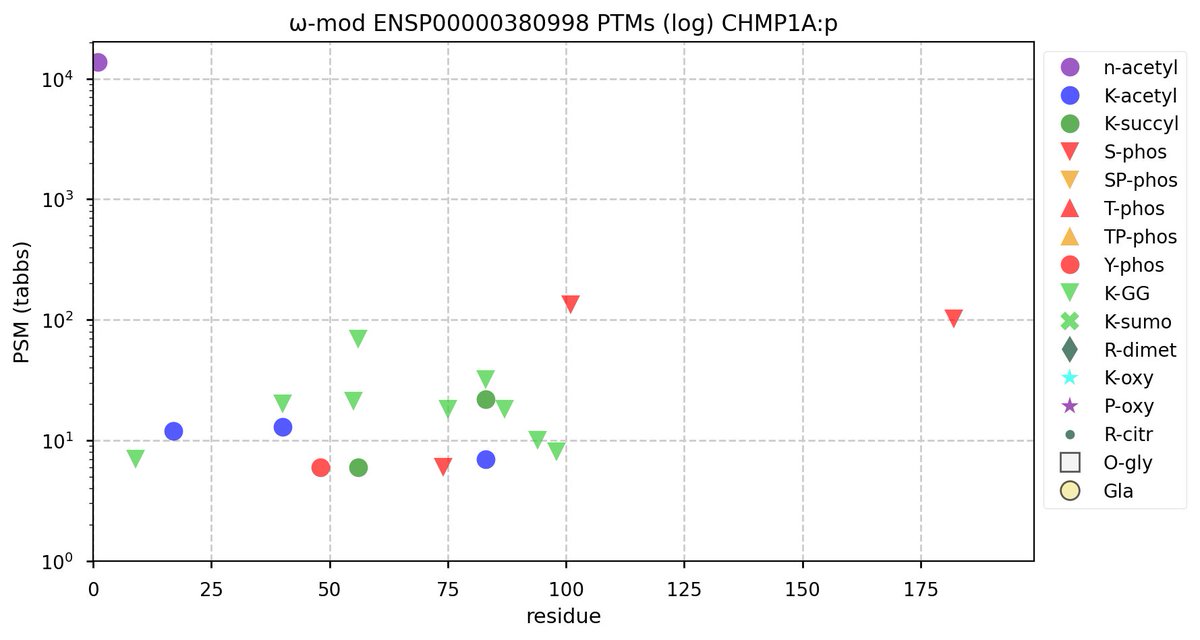
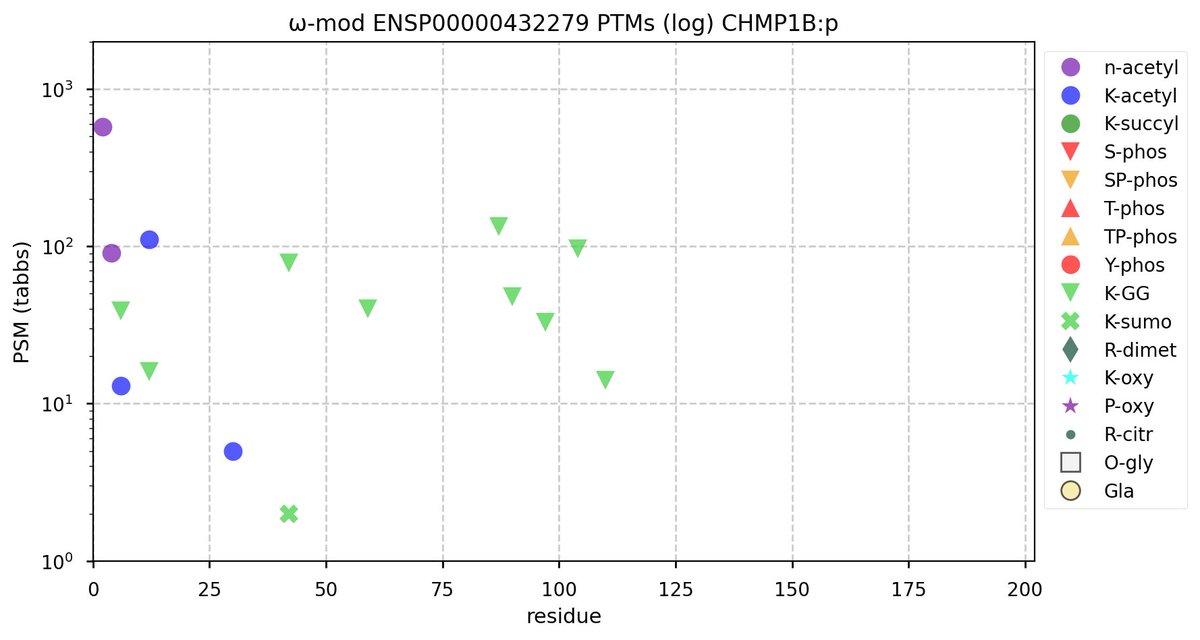
Sun May 01 13:56:42 +0000 2022#resMods: Pro/P
N-amino: +acetyl¹
Sidechain: reactive pyrrolidine
«bio»
+hydroxy² ³
C-carboxyl: none
Notes:
1. Protein N-terminus
2. 4-Hydroxyproline (Hyp), found in collagens & plant cell wall glycoproteins.
3. Modified residue used as a site for intermolecular cross-linking.
Sun May 01 13:25:26 +0000 2022Even when something undeniable happens (PI goes to jail), there is no attempt at a postmortem analysis of how it happened or was dealt with: instead the most common approach is never to speak their name again.
Sun May 01 13:13:07 +0000 2022Highly moral people I know & like will develop a weird sort of realpolitik attitude (a sort of ethical tunnel vision) when discussing candidates (or colleagues) if they can make it rain money.
Sun May 01 12:54:37 +0000 2022The current issue around hiring at NYU Med School is really a symptom of the biggest problem in academia at the moment: almost everyone is willing to overlook/downplay bad behavior if a PI has a good angle on getting federal or private money.
Sun May 01 01:17:09 +0000 2022The overlaid data is from the Iowa Environmental Mesonet (Iowa State's great weather data resource).
Sun May 01 01:14:55 +0000 2022You can see today's Colorado Low in the NEXRAD rainfall total estimates for the last 48 hrs. 🔗
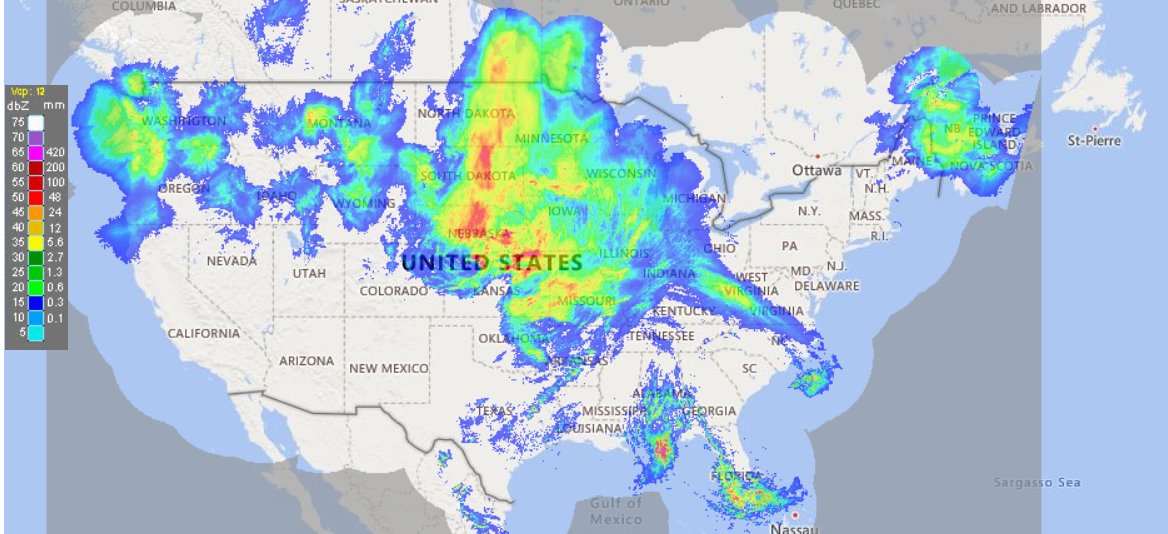
Sat Apr 30 20:15:17 +0000 2022@scalzi 🔗
Sat Apr 30 14:15:42 +0000 2022Modification diagrams for yeast VPS60:p & human CHMP5:p, orthologs of a "non-essential" ESCRT-III complex subunit. The human version shows considerably more engagement with PTM signalling mechanisms than its yeast counterpart. 🔗
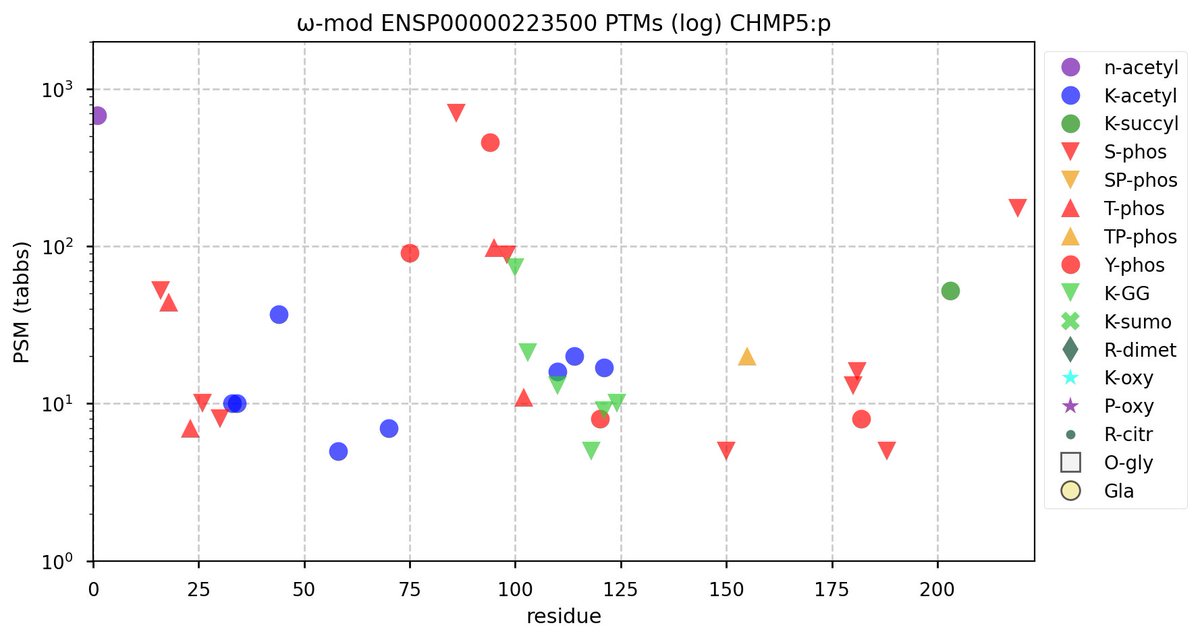
Sat Apr 30 13:06:26 +0000 2022@TanentzapfLab The whole operation seems to be focused on funding bad (but simple) ideas.
Sat Apr 30 12:30:30 +0000 2022#resMods: Pyl/O¹
N-amino: none
Sidechain: reactive pyrroline
«bio»
none
C-carboxyl: none
Notes:
1. Prokaryotes only: pyrolysine is encoded as a special translation of the mRNA codon UAG.
Sat Apr 30 12:28:52 +0000 2022@neely615 @slavov_n @EastCoast55 Sounds like keeping students away may be the point.
Sat Apr 30 12:01:01 +0000 2022For the 3rd weekend in a row, a classic Colorado Low centered just north of the Missouri River in South Dakota with a big patch of warm, damp Gulf air to tap into. 🔗
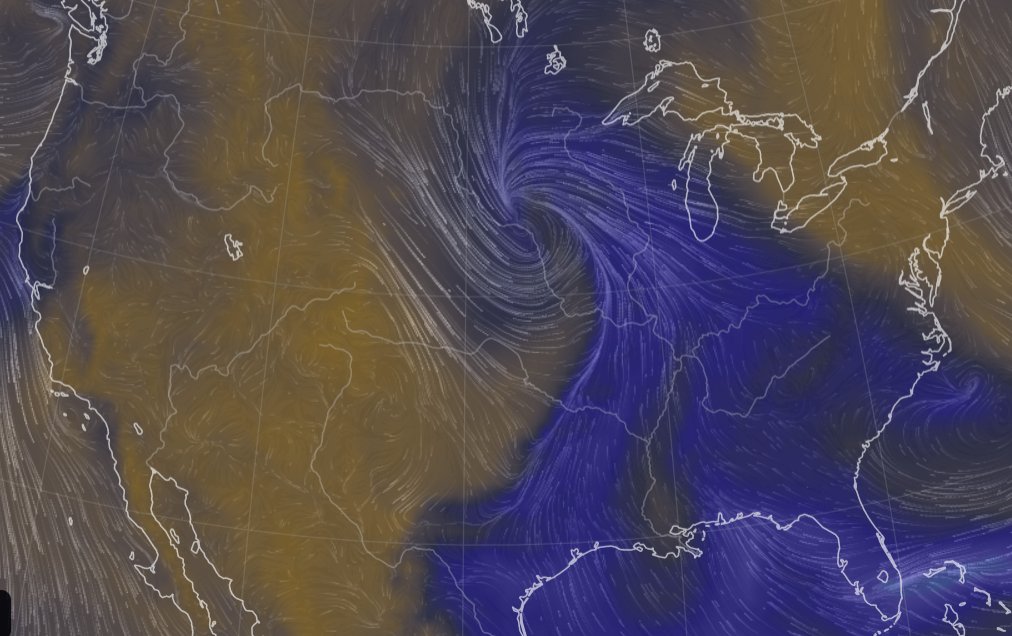
Fri Apr 29 15:03:02 +0000 2022Human CHMP4B:p seems to be playing a somewhat different game than its paralogs, with many more acetyl/succinyl/GGyl modifications than the other sequences.
Fri Apr 29 14:58:55 +0000 2022Modification diagrams for yeast SNF7:p and its human orthologs CHMP4A:p, CHMP4B:p & CHMP4C:p, subunits of the ESCRIT-III complex. Unlike most ESCRT subunits, the yeast sequence has more phosphorylation acceptor sites than the 3 human paralogs. 🔗
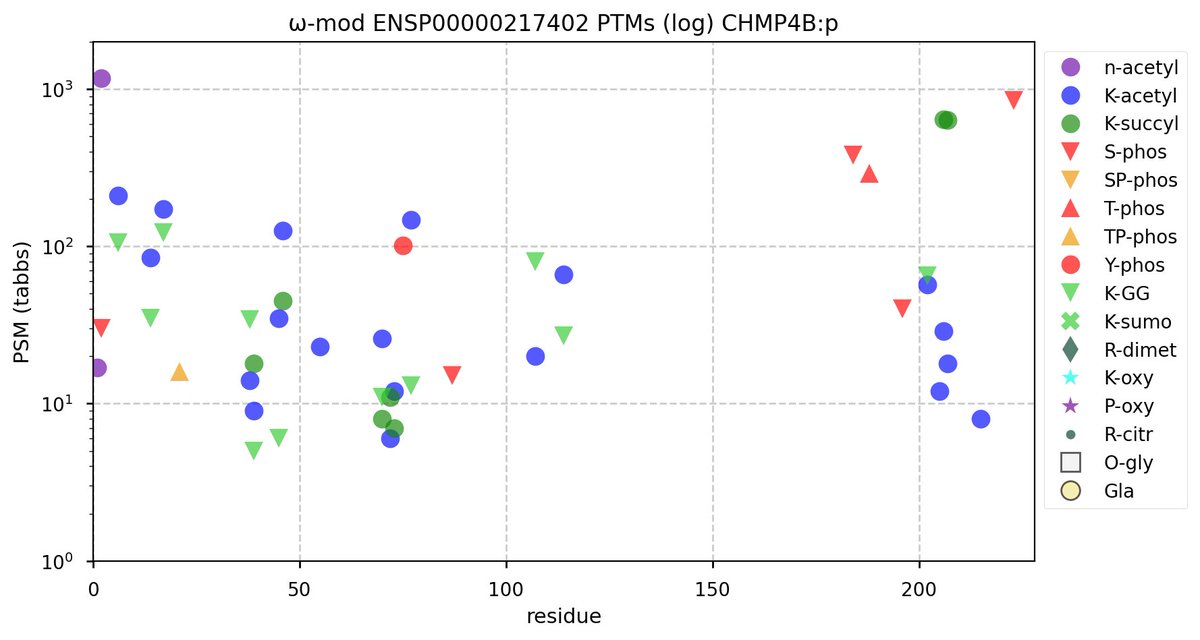
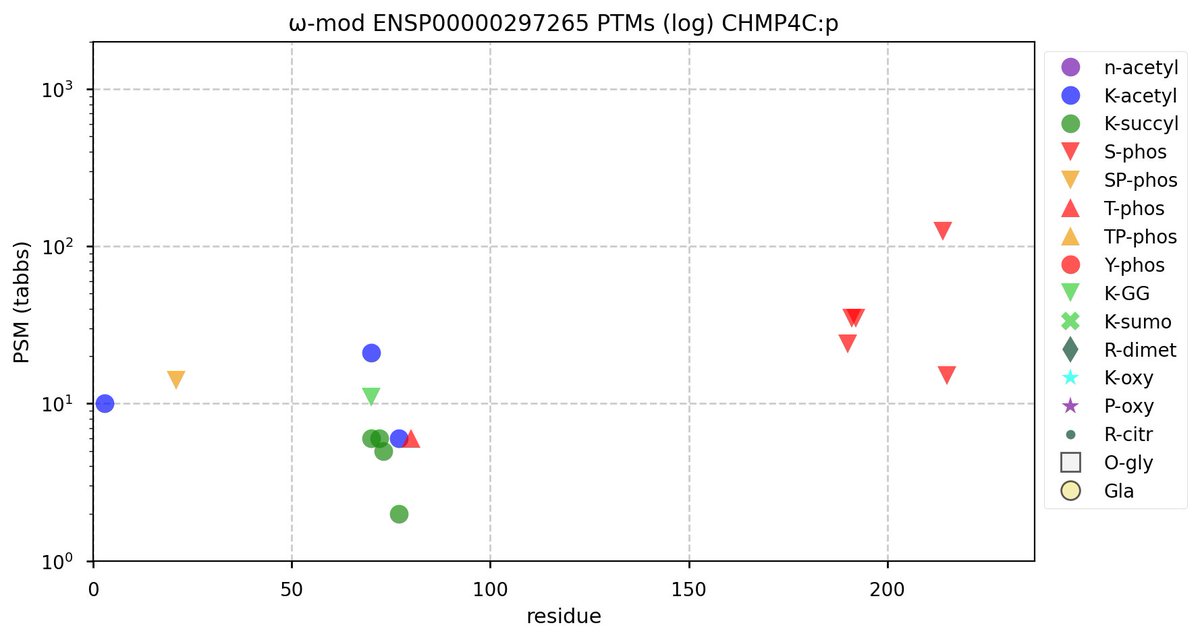
Fri Apr 29 13:42:21 +0000 2022#resMods: Asn/N
N-amino: none
Sidechain: reactive -CONH₂
«bio»
+glycosyl¹; deamidation²
C-carboxyl: none
Notes:
1. "N-linked" glycosylation: many glycoforms possible.
2. Results in either Asp or isoAsp: rate depends on adjacent residues.
Fri Apr 29 13:27:52 +0000 2022@slashdot I'll stick with the VPN that has been in Opera for quite a while: I can't bring myself to use Edge.
Fri Apr 29 12:37:02 +0000 2022FBP2-201, fructose-bisphosphatase 2
🔗
1 – 339 ⟶ 2 – 339 (N-acetyl-Thr, ο = 92%)
#realNT
Thu Apr 28 23:31:42 +0000 2022Another Colorado Low is shaping up to generate the 3rd such storm soaking the Dakotas in 3 weeks. 🔗
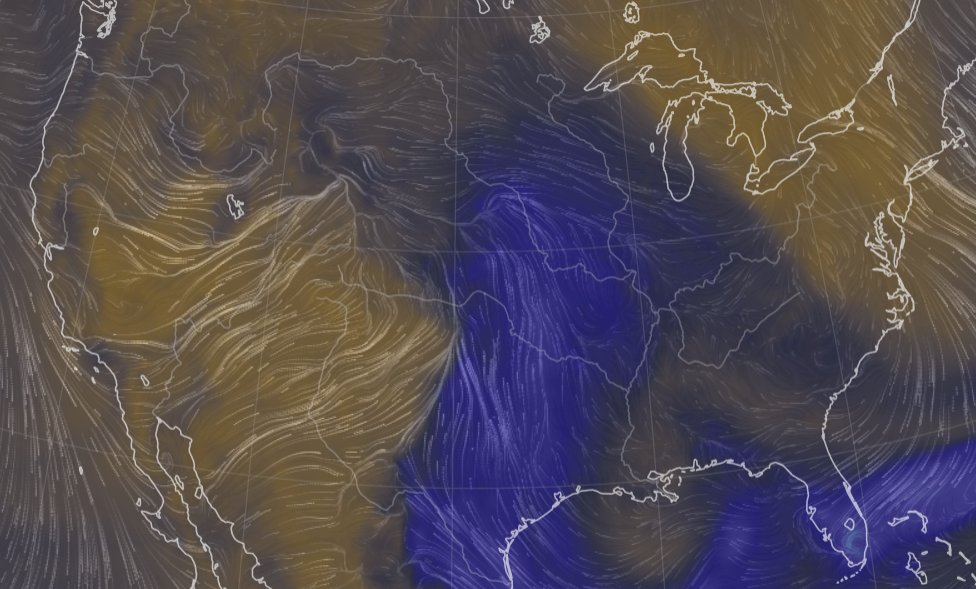
Thu Apr 28 21:16:34 +0000 2022I often see threads from students & junior faculty about the PhD process that describe expectations, processes & events in a way that I don't recognize from my own experience (admittedly I went through it back in the '80s).
Thu Apr 28 20:59:50 +0000 2022JPH2-202, junctophilin 2
🔗
1 – 696 ⟶ 2 – 696 (N-acetyl-Ser, ο = 100%)
#realNT
Thu Apr 28 20:57:10 +0000 2022FBP2-201, fructose-bisphosphatase 2
🔗
1 – 339 ⟶ 2 – 339 (N-acetyl-Thr, ο = 92%)
#realNT
Thu Apr 28 18:56:42 +0000 2022"Benevolent Ponzi Scheme" method of research funding 😀 🔗
Thu Apr 28 16:46:25 +0000 2022@byu_sam It actually produces a pretty nice z=3 spectrum, although just where the charges are located may cause some debate among the cognoscenti
Thu Apr 28 16:25:38 +0000 2022#resMods: Met/M
N-amino: +acetyl¹
Sidechain: reactive -S-CH₃
«artifact―redox»
+O, +2O
«artifact―tRNA loading²»
-Se-CH₃
C-carboxyl: none
Notes:
1. Protein N-terminus.
2. Selenomethioine (SeMet): may substitute for Met―at high concentrations.
Thu Apr 28 15:46:42 +0000 2022It has been hot in New Dehli, even by Indian standards. 🔗
Thu Apr 28 14:46:07 +0000 2022#resMods: Met/M
N-amino: +acetyl¹
Sidechain: -S-CH₃
«artifact―redox»
+O, +2O
«artifact―tRNA loading²»
-Se-CH₃
C-carboxyl: none
Notes:
1. Protein N-terminus.
2. Selenomethioine (SeMet): may substitute for Met (at high concentrations).
Thu Apr 28 14:33:14 +0000 2022@theoneamit @AcademicDilemma If there is an in-person meeting of the reviewers involved, say "yes". Otherwise, "no".
Thu Apr 28 14:31:25 +0000 2022@CBCSamSamson @CBCManitoba You've got to admire the chutzpah of bringing up a 70 year plan as a defense for dumping raw sewage last week.
Thu Apr 28 14:24:08 +0000 2022@AlexUsherHESA Not so much common sense as it is tangential to common sense.
Thu Apr 28 14:19:54 +0000 2022Modification diagrams for yeast VPS24:p & human CHMP3:p, orthologs of an ESCRT-III subunit. The human subunit seems more attuned to input from kinases than its yeast counterpart, an evolutionary trend common to most ESCRT subunits. 🔗
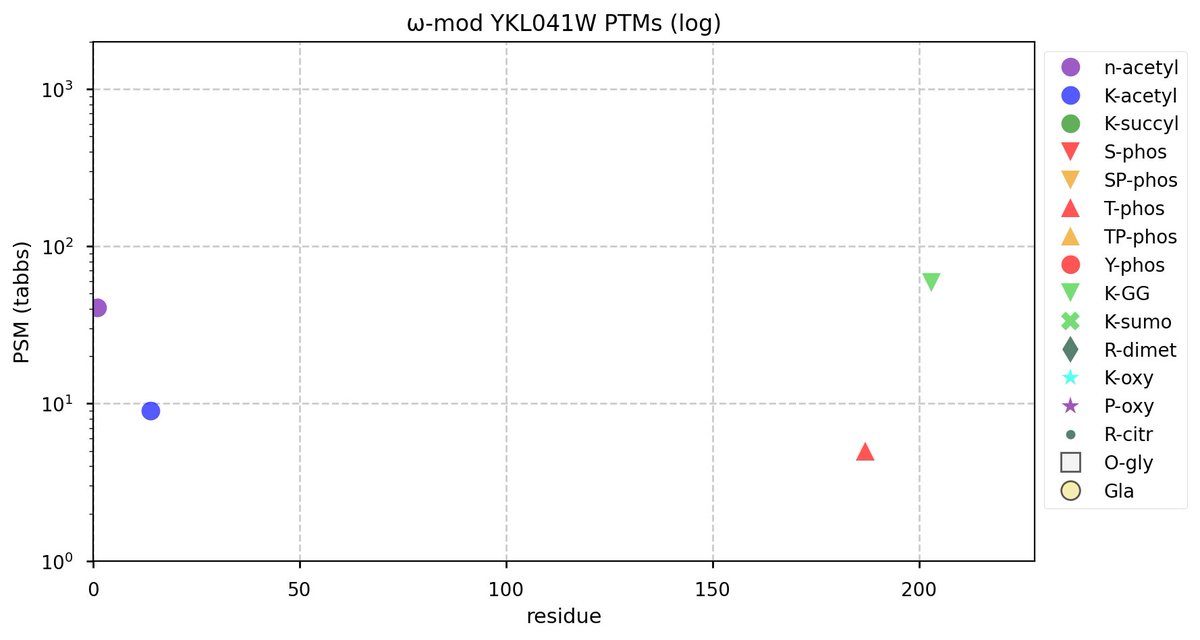
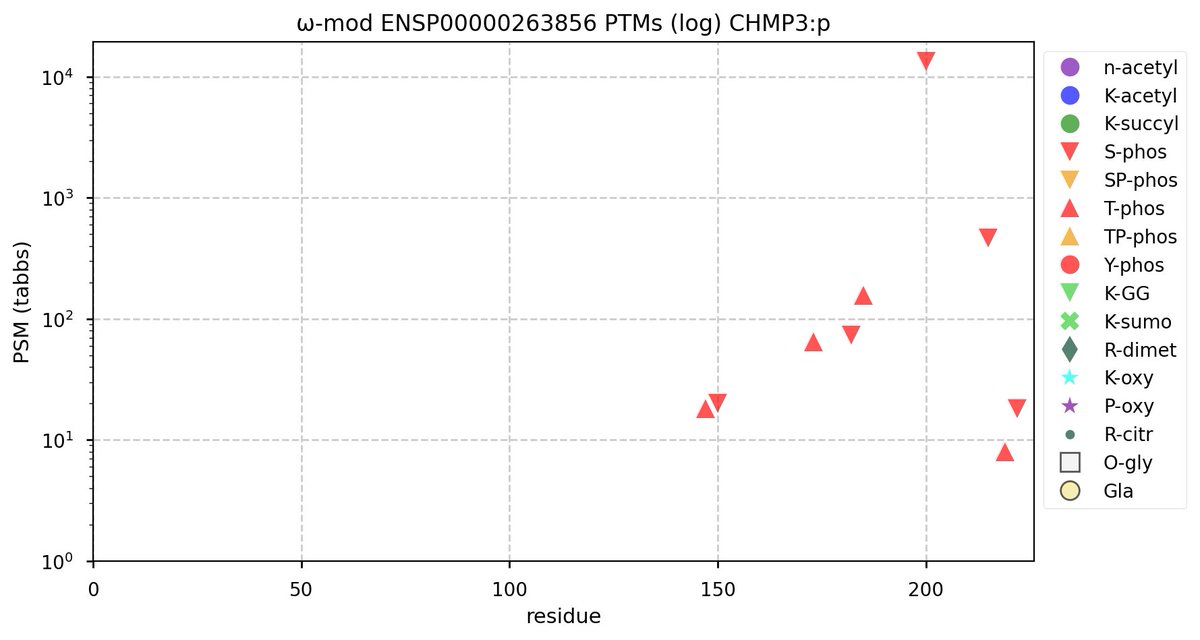
Thu Apr 28 13:00:38 +0000 2022@neely615 @JoelisSteele @eliaslab1 Saliva (at the start of the process) has the same potential for food-based protein contributors, along with the bacterial usual suspects.
Thu Apr 28 12:01:14 +0000 2022@neely615 @JoelisSteele I pretty much automagically slot things into a food web without thinking about it
🔗
Thu Apr 28 11:53:26 +0000 2022@neely615 @JoelisSteele My like/unlike are my zoologist brain talking: the FBS & E. coli systems are supplying nutrients to the organism of interest, while Wolbachia & H. pylorii are taking nutrients.
Thu Apr 28 11:33:53 +0000 2022@neely615 @JoelisSteele Yes, they seem to make a pretty good concentrate of their E. coli food in their gut, which can generate significant signals. It is like FBS in cell culture, not like the Wolbachia endosymbionts you get in some fruitfly tissue or H. pylori infections in human gastric cancer.
Wed Apr 27 17:30:18 +0000 2022HNNHNNQQNNHHHHHQR
My choice for the ideal tryptic peptide to test the general applicability of your favorite MS/MS spectrum prediction software.
Wed Apr 27 15:48:57 +0000 2022Modification diagrams for yeast VP20:p & human CHMP6:p, orthologs of a subunit in the ESCRT-III complex. ESCRT-III differs from -I & -II in that it isn't an assembly, rather it is a transient association of subunits that forms filaments. 🔗
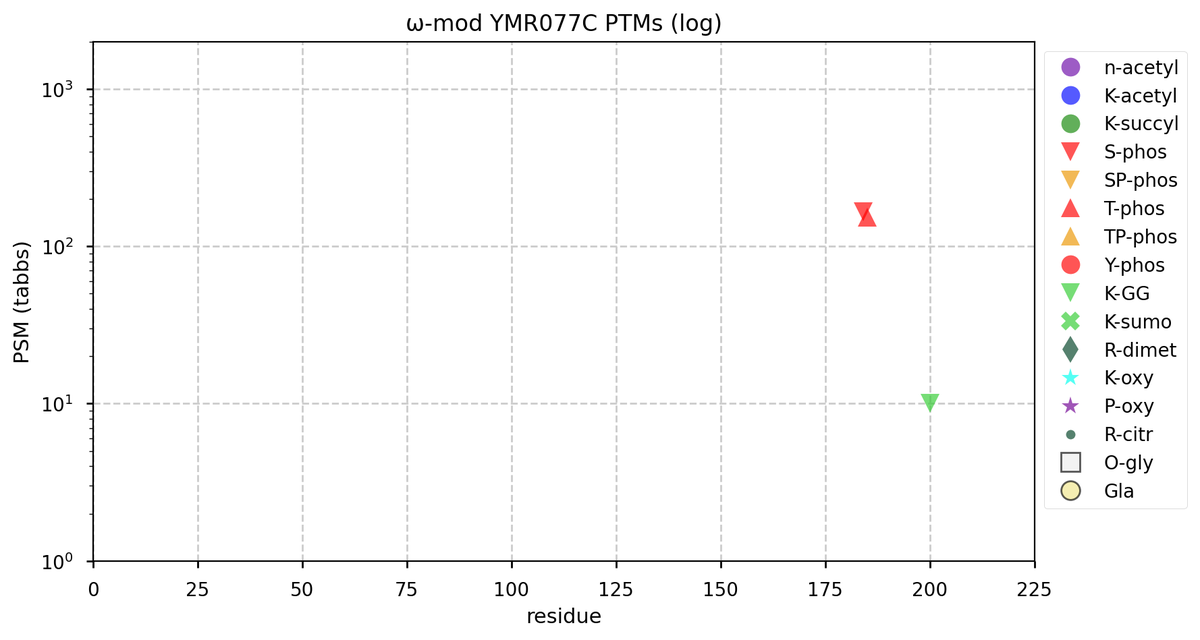
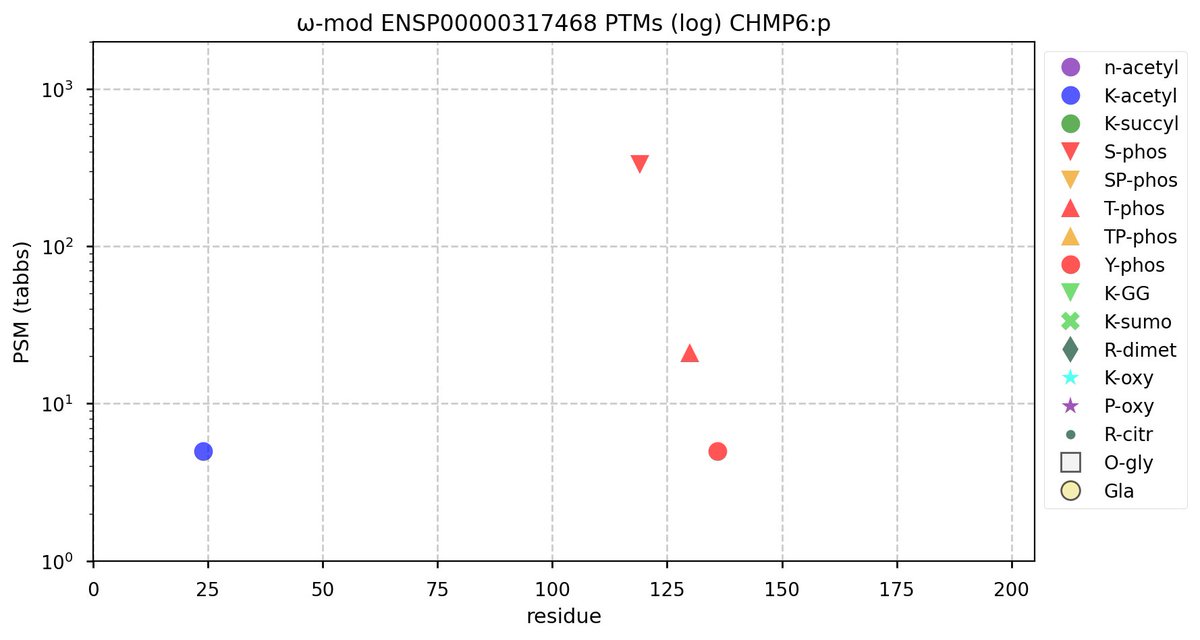
Wed Apr 27 14:32:06 +0000 2022#resMods: Leu/L
N-amino: +acetyl¹
Sidechain: not reactive
C-carboxyl: none
Notes:
1. Only possible if the Leu RNA codon CUG has been read as a translation initiation site.
Wed Apr 27 14:01:18 +0000 2022@byu_sam The degree to which university professors are have little formal training wrt teaching―they are mainly autodidacts―can be painful to watch once you are in the club.
Wed Apr 27 13:46:12 +0000 2022@byu_sam I found it impossible to take courses once I had spent some time teaching them.
Wed Apr 27 13:03:23 +0000 2022@stateofthecity To me, the interesting part of the last "crisis" was all of the politicians on TV talking about the police as though they were an independent branch of government.
Wed Apr 27 12:47:01 +0000 2022@MattWFoster @neely615 @nesvilab And the way US law works, obvious text formats are just as protected as highly encrypted binary formats.
Wed Apr 27 12:42:56 +0000 2022@MattWFoster @neely615 @nesvilab If you want to find out how many lawyers work at your university, ask the grants office if it is OK to reverse engineer a file format with Fed funding. They will all be lined up in front of your office the next morning.
Wed Apr 27 12:42:09 +0000 2022@MattWFoster @neely615 @nesvilab The MaxQuant license has some language specifically forbidding reverse engineering, making any sort of format hacking extremely risky.
Wed Apr 27 12:16:22 +0000 2022@neely615 @MattWFoster @nesvilab The MQ people would have to publish a specification & make it clear they were OK with others using the format.
Wed Apr 27 12:14:28 +0000 2022But remember that for any C. elegans data analysis it is always best to include a E. coli B strain proteome.
Wed Apr 27 12:10:40 +0000 2022If you need some protein phosphorylation data to work with & you have grown bored of mammalian samples, PXD032260 is the best C. elegans TiO₂ experiment I've seen in quite a while.
Tue Apr 26 21:10:55 +0000 2022Notes:
1. Other carboxylic acid-containing molecules can be added enzymatically
2. Any sugar with a reducing end can add non-enzymatically
3. 5-hydrolysine (Hyl)―in collagen
4. Ubiquitin (-like) protein crosslink
5. Hypusine, single residue in EIF5A homologs―K50 in human.
2/2
Tue Apr 26 21:08:33 +0000 2022#resMods: Lys/K
N-amino: none
Sidechain: reactive ε-amine
«bio―acyl¹»
+acetyl; +succinyl; +malonyl; +myristoyl
«bio―glyco²»
+glycation
«bio―methyl»
+CH₃; +(CH₃)₂; +(CH₃)₃
«bio―oxidation³»
+O
«bio―amino»
protein C-terminal ligation⁴; +hydroxyaminobutyl⁵
C-carboxyl: none
1/2
Tue Apr 26 15:41:56 +0000 2022For want of better words, I like to refer to similarities in PTM patterns using terms related to "rhyming".
Tue Apr 26 15:29:58 +0000 2022🔗
Tue Apr 26 15:10:22 +0000 2022Modification diagrams for yeast DID4:p and its 2 human orthologs CHMP2A:p & CHMP2B:p, subunits of the ESCRT-III complex. Their decorations are consistent with other ESCRT subunits. The 2 human paralogs have significantly different patterns, but there is some rhyming going on. 🔗
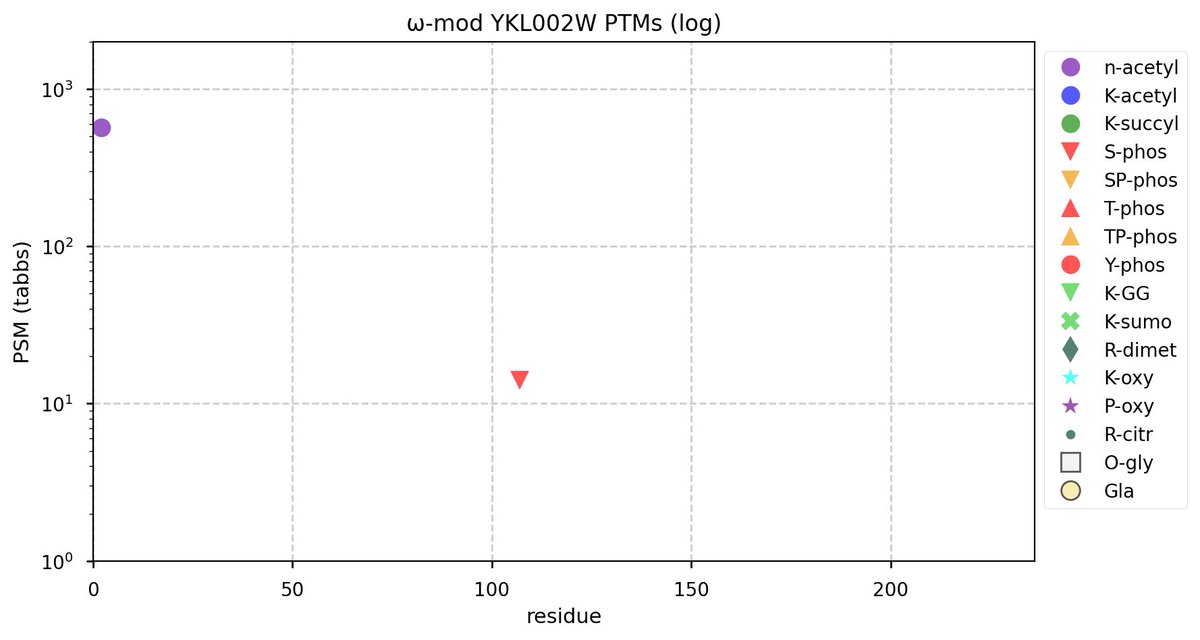
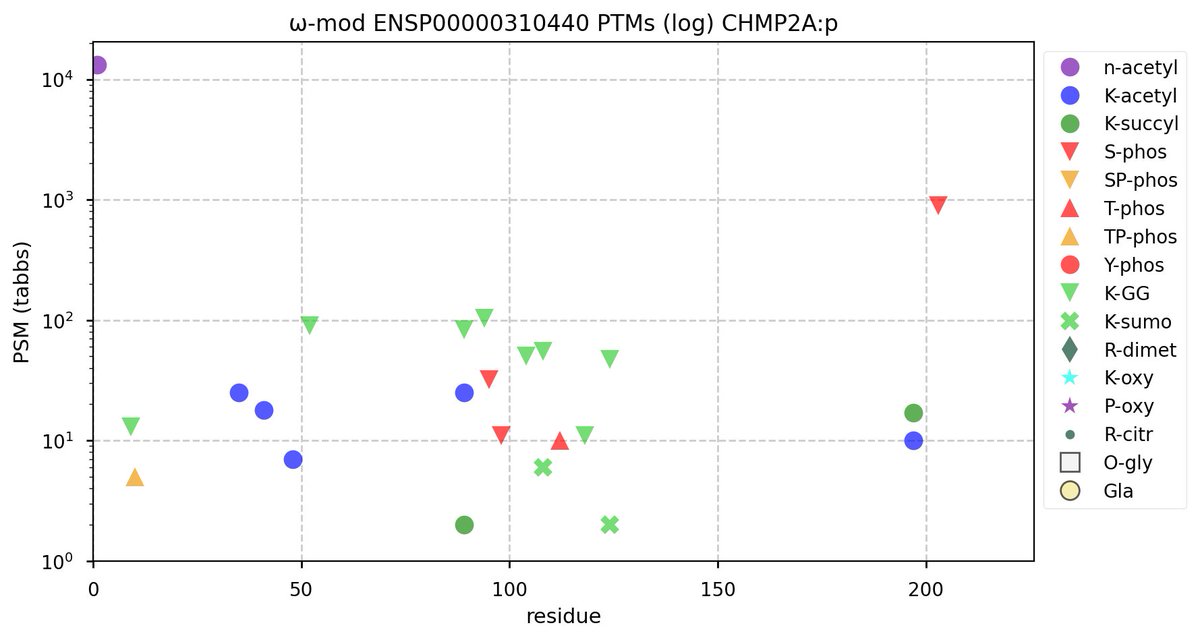
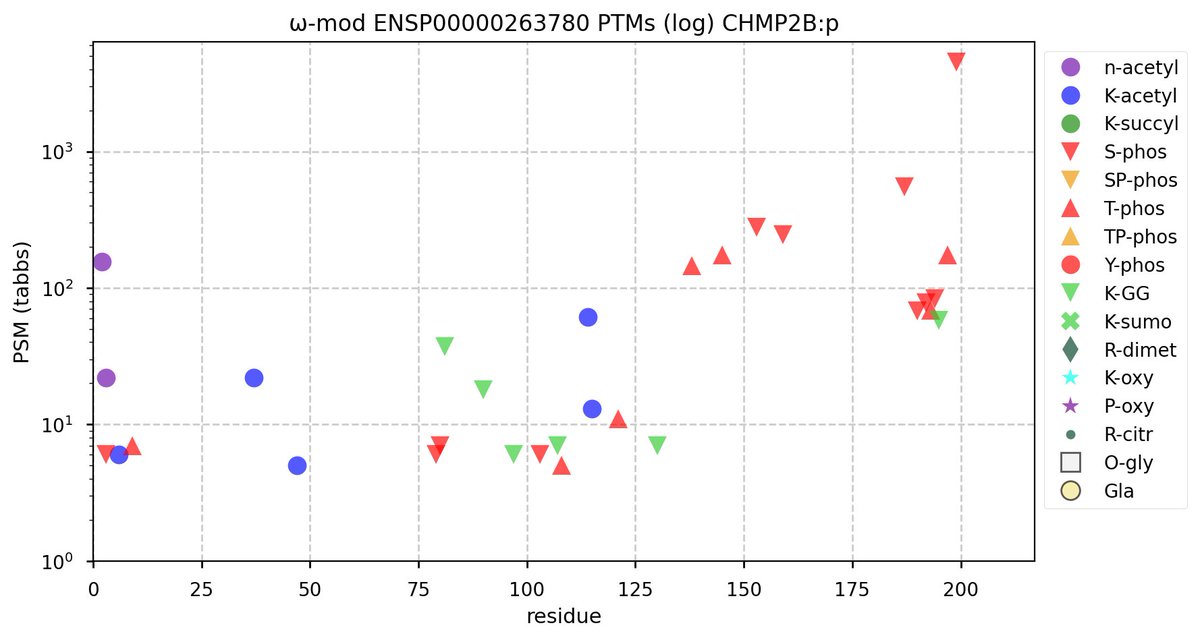
Tue Apr 26 15:01:46 +0000 2022@scalzi Editorial note: "Sorry." is not a very cat-like thing to say. "Macht schnell." is more plausible.
Tue Apr 26 13:24:47 +0000 2022Even though it has been well known for 20 years, it is one of those clear violations of Watson's "Central Dogma" that seems to cause cognitive dissonance/conversion disorder/hope-it-will-just-go-away among many with a molecular biology background.
Tue Apr 26 13:14:02 +0000 2022For those of you unfamiliar with the causes & effects of alternate initiation on the resulting translated sequence, this note tries to make it "fun". I'd guesstimate 3-5% of human mRNA show this pattern of translation (i.e. 6-10% of human proteins).
🔗
Tue Apr 26 12:58:34 +0000 2022MAVS-202, mitochondrial antiviral signaling protein
🔗
1 – 540
↓
2 – 540 (N-acetyl-Pro, ο = 2%)
143 – 540 (N-acetyl-Pro, ο = 1%, alt initiation M142)
#realNT
Tue Apr 26 12:50:41 +0000 2022A cold front line of storms running from New Orleans LA to Burlington VT, the last remnant of the mechanics that gave rise to the weekend's very successful Colorado Low 🔗
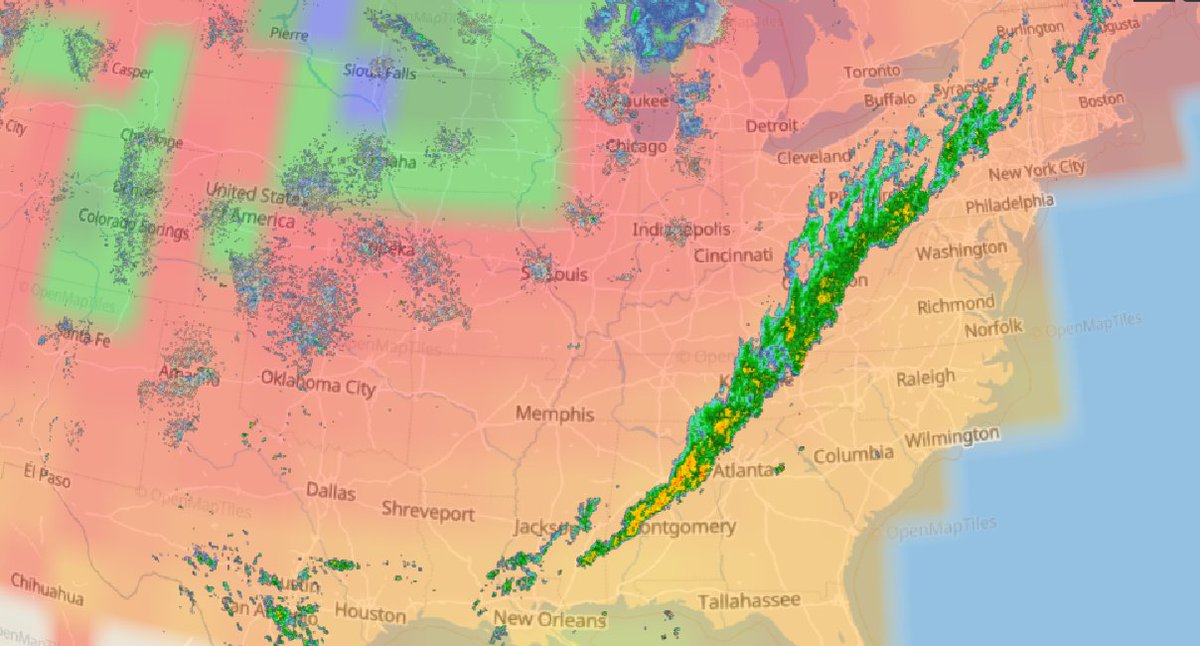
Tue Apr 26 01:24:34 +0000 2022@aschuller17 @NewsfromScience His dad was a big wheel at NYU's Med School when I was there.
Tue Apr 26 00:37:05 +0000 2022PXD010700: once again the Armengaud group does not disappoint.
Mon Apr 25 15:17:15 +0000 2022Sounds like the guys at Tesla are in for a bit of a reprieve.
Mon Apr 25 14:58:02 +0000 2022Modification diagrams for yeast & human VPS36:p orthologs, a component of the ESCRT-II complex. Both patterns are consistent with other ESCRT component subunits. 🔗
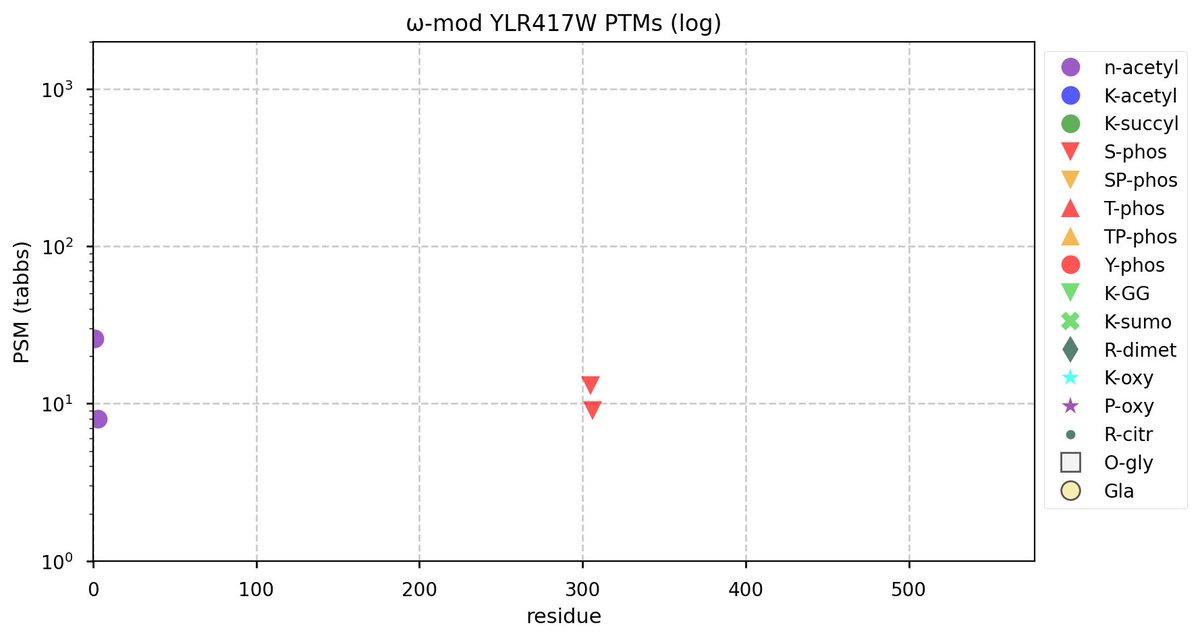
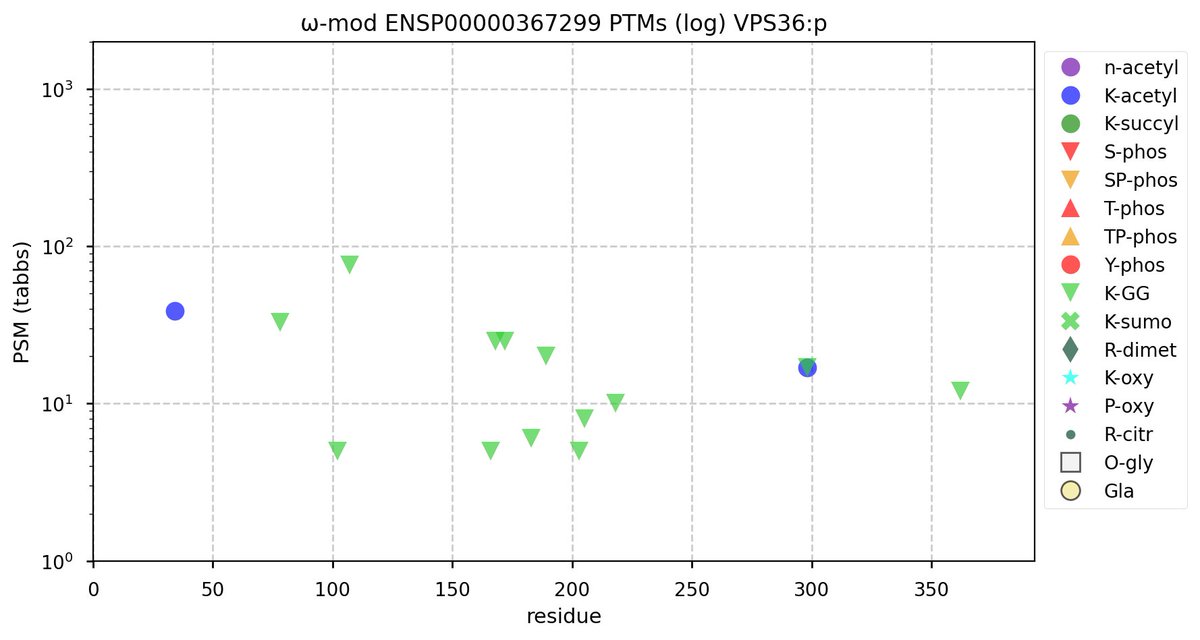
Mon Apr 25 14:55:01 +0000 2022At the moment, it looks like Tropical Cyclone Jasmine is going to stay north of the hard-hit east coast of South Africa. 🔗
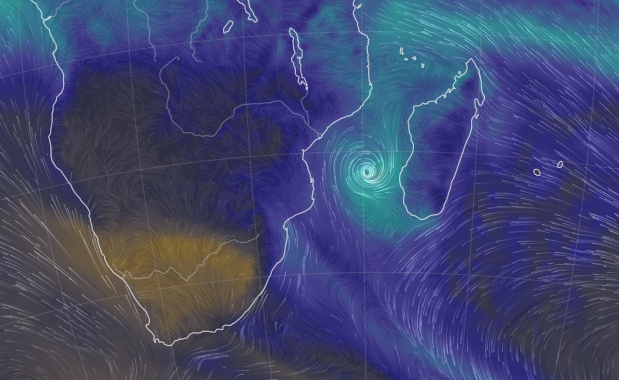
Mon Apr 25 12:51:43 +0000 2022#resMods: Ile/I
N-amino: none
Sidechain: not reactive
C-carboxyl: none
Mon Apr 25 12:45:18 +0000 2022UBR7-201, ubiquitin protein ligase E3 component n-recognin 7
🔗
1 – 425 ⟶ 2 – 425 (N-acetyl-Ala, ο = 100%)
#realNT
Mon Apr 25 12:28:07 +0000 2022UBR3-201, ubiquitin protein ligase E3 component n-recognin 3
🔗
1 – 1888 ⟶ 2 – 1888 (N-acetyl-Ala, ο = 99%)
#realNT
Mon Apr 25 11:56:30 +0000 2022UBR4-204, ubiquitin protein ligase E3 component n-recognin 4
🔗
1 – 5183 ⟶ 2 – 5183 (N-acetyl-Ala, ο = 100%)
#realNT
Mon Apr 25 01:40:00 +0000 2022@dccc_phd @michaelhoffman @GordonConf Hypocrisy is the lifeblood of the academy.
Sun Apr 24 20:07:38 +0000 2022ACTN3-203 (actinin alpha 3) has an N-terminus that looks like a key got stuck, but ribosomes use all four possible translation initiation sites (same for the mouse ortholog):
1 MMMVMQPEGLGAGEGR ...
Sun Apr 24 19:08:19 +0000 2022A new data set from one of my favorite groups!
Sun Apr 24 16:39:02 +0000 2022Quite a productive Colorado Low here in Winnipeg: 72 mm of rain in the last 2 days. Not much for a rainy place, but Winnipeg is not a rainy place.
Sun Apr 24 16:16:36 +0000 2022Modification patterns for human & yeast orthologs of VPS25, a subunit of the ESCRT-II complex. Both patterns are consistent with the subunits of ESCRT complexes for their respective species. 🔗
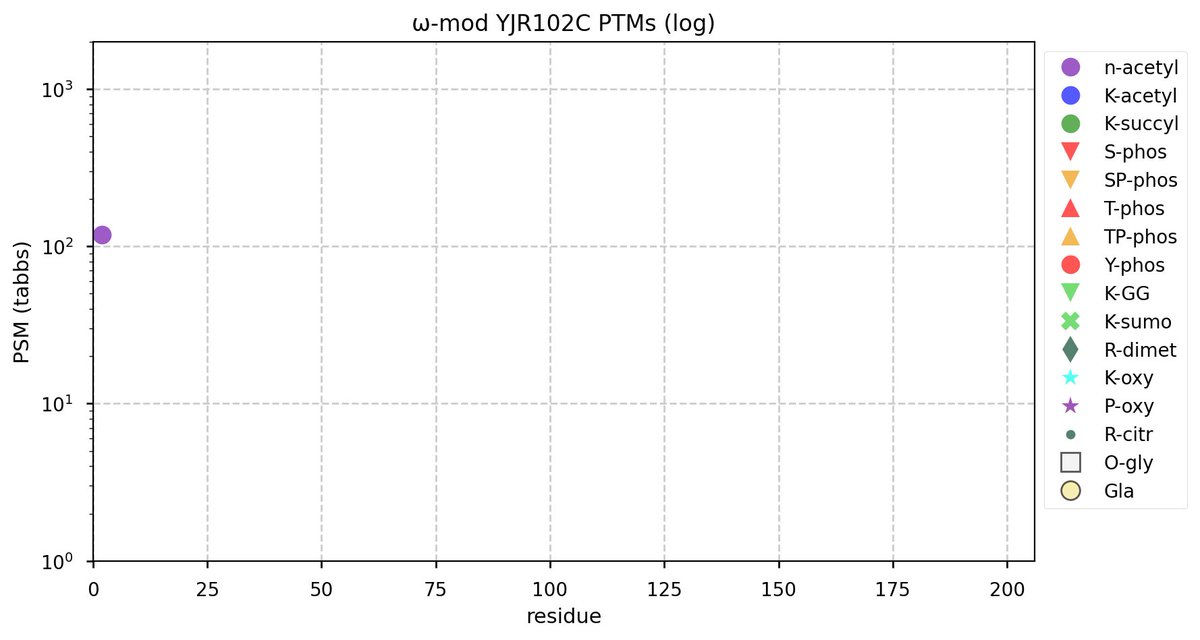
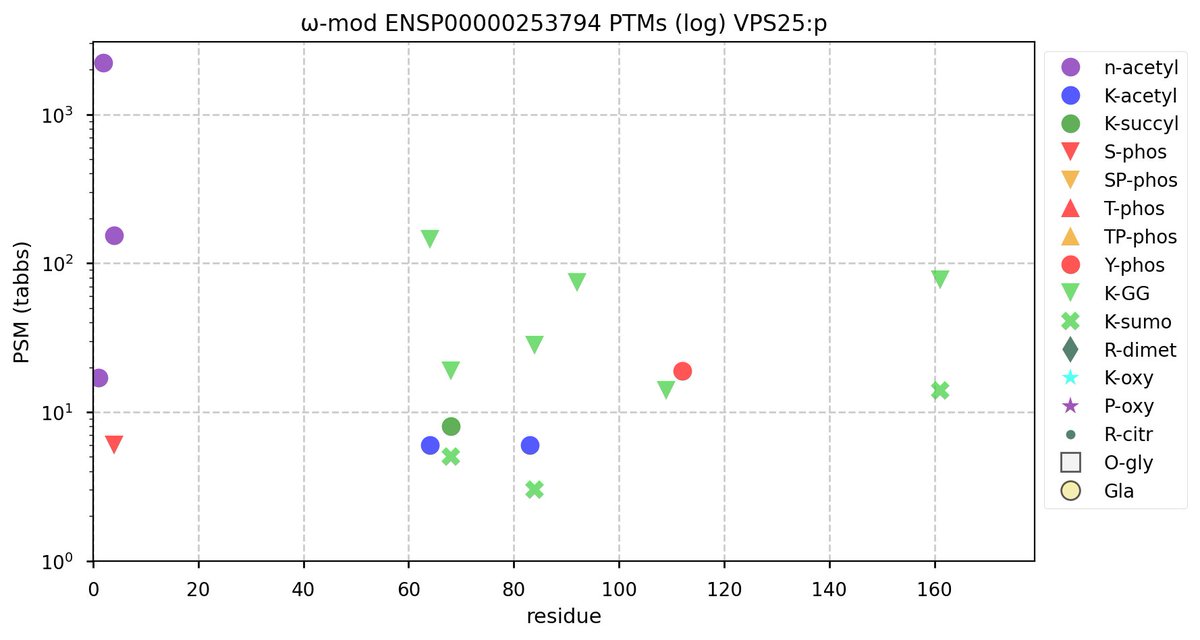
Sun Apr 24 14:02:08 +0000 2022@ItaiYanai I should also say that these highly successful academics all believe (to this day) that they are really good in the lab.
Sun Apr 24 13:52:49 +0000 2022I find PTMs like diphthamide interesting/disturbing. It only occurs at a single residue but takes several enzymes to synthesize & install. Is it a remnant of a once common PTM that died out or the result of an ongoing residue optimization process we don't understand?
Sun Apr 24 13:45:36 +0000 2022#resMods: His/H
N-amino: none
Sidechain: reactive imidazole
«bio»
+phosphoryl¹; +diphthamide²
«artifact―redox»
+O
C-carboxyl: none
Notes:
1. In prokaryotes.
2. Single site in proteome: eukaryote/archaeal EEF2 homologs, e.g., H715 in human.
Sun Apr 24 13:34:53 +0000 2022MYH2-201, myosin heavy chain 2
🔗
1 – 1941 ⟶ 2 – 1941 (N-acetyl-Ser, ο = 94%)
#realNT
Sun Apr 24 13:20:27 +0000 2022@ItaiYanai Not to put a damper on this notion, but the most successful academics I know were (& are) terrible in the lab.
Sun Apr 24 13:13:00 +0000 2022NUTF2-201, nuclear transport factor 2
🔗
1 – 127 ⟶ 2 – 127 (N-acetyl-Gly, ο = 100%)
#realNT
Sun Apr 24 13:10:03 +0000 2022MYH7-201, myosin heavy chain 7
🔗
1 – 1935 ⟶ 2 – 1935 (N-acetyl-Gly, ο = 95%)
#realNT
Sun Apr 24 00:39:37 +0000 2022@astacus @DRAWheatcraft @pwilmarth Unknown, but sometimes having a zoology degree comes in handy.
Sun Apr 24 00:28:57 +0000 2022@DRAWheatcraft @pwilmarth @astacus Looks like a coati.
Sat Apr 23 16:42:42 +0000 2022#resMods: Gly/G
N-amino: +acetyl¹; +myristoyl¹
Sidechain: non-reactive
C-carboxyl: lysine sidechain²
Notes:
1. Protein N-terminus.
2. Ubiquitin(-like) protein crosslink.
Sat Apr 23 16:27:48 +0000 2022The current Colorado Low is firmly entrenched in the Dakotas at the moment, with lots of access to moisture laden air being circulated up from the Gulf of Mexico by a high pressure system centered on the Mid-Atlantic coast 🔗
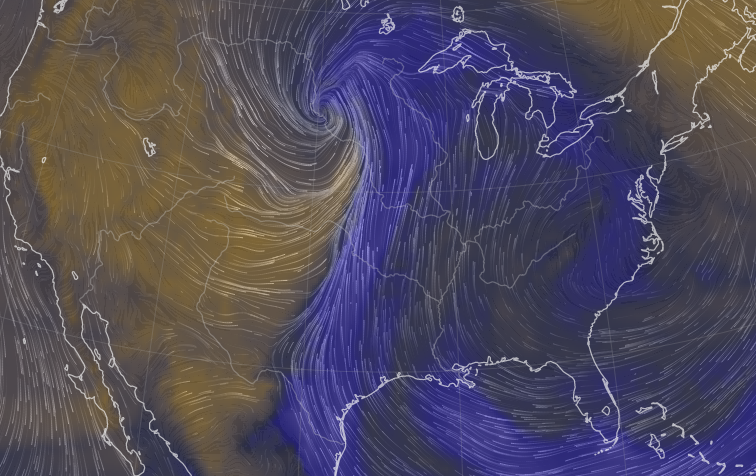
Sat Apr 23 16:19:03 +0000 2022@VATVSLPR @theoneamit @DRAWheatcraft Another difference is that writing user documentation has no value to the author, particularly if they have documented the code as they wrote it. Most people write code because they need it: other users are―at best―an afterthought.
Sat Apr 23 14:55:21 +0000 2022@bkives Have they announced the architects for the project?
Sat Apr 23 14:24:41 +0000 2022@Niigaanwewidam @WinnipegNews From the announcements, it sounds like it will be a very challenging rebuild, given the limitations of the existing structure. Has there been any indication of the architects who will be involved?
Sat Apr 23 13:46:24 +0000 2022Even though many of the ESCRT subunits have occupied tyrosine phosphorylation acceptors, the role of these sites in ESCRT function remains unexamined.
Sat Apr 23 13:30:42 +0000 2022@Peptidome @JesseBrown The attacks all seem to be automated: I'm pretty sure they just run through ranges of IP addresses wacking-away, trying to find a vulnerability.
Sat Apr 23 13:16:12 +0000 2022Modification diagrams for yeast & human SNF8:p, orthologs of one of the 3 components of the ESCRT-II heterotetramer. The yeast subunit does not have any observable PTMs, while human has GGyl/SUMOyl/acetyl co-acceptors & a single Y+phosphoryl acceptor. 🔗
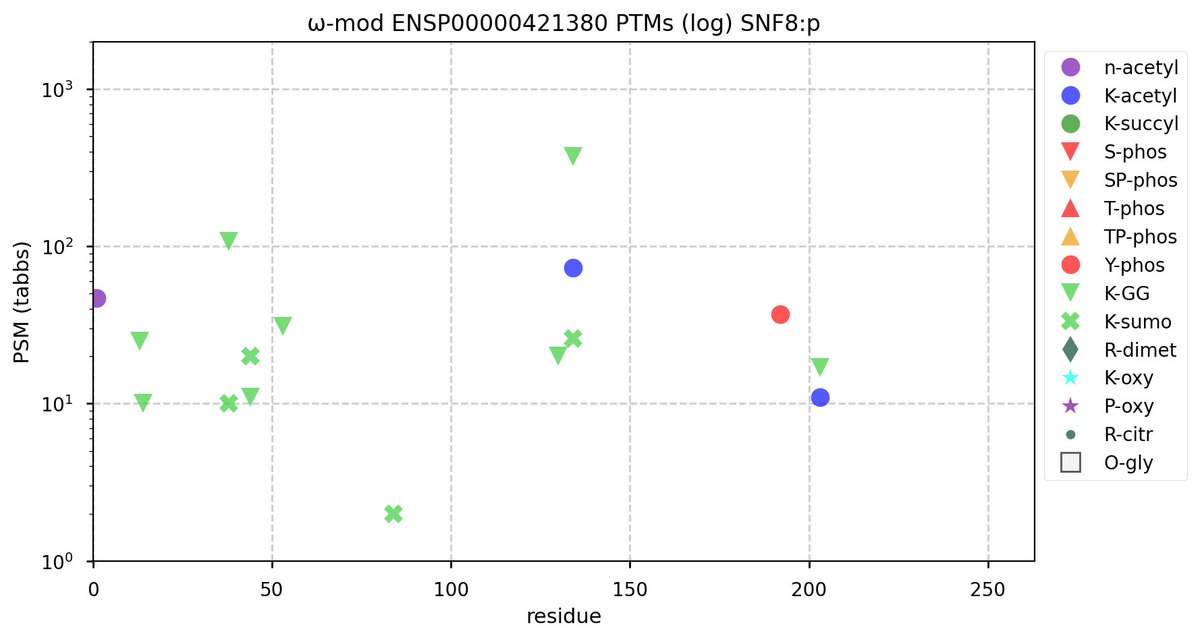
Sat Apr 23 12:56:17 +0000 2022CEP43-202, centrosomal protein 43
🔗
1 – 399 ⟶ 2 – 399 (N-acetyl-Ala, ο = 98%)
#realNT
Sat Apr 23 12:52:02 +0000 2022ABCB1-211, ATP binding cassette subfamily B member 1
🔗
1 – 1280 ⟶ 1 – 1280 (N-acetyl-Met, ο = 100%)
#realNT
Fri Apr 22 23:19:56 +0000 2022@neely615 @makingions @leprevostfv @Smith_Chem_Wisc I'm not aware of any. I made the first version available in 2004, so it would turning 18.
Fri Apr 22 22:24:05 +0000 2022@makingions @neely615 @leprevostfv @Smith_Chem_Wisc That may be the only publication on the topic.
Fri Apr 22 20:20:53 +0000 2022@neely615 @Smith_Chem_Wisc @leprevostfv These list are of practical use, but because their creation & maintenance will not result in a publication (or product), it is hard to justify putting a lot of effort into them.
Fri Apr 22 18:57:44 +0000 2022@VATVSLPR @DRAWheatcraft A tutorial is much (much) easier to generate than coherent documentation. It is also more likely to produce positive feedback for the creator.
Fri Apr 22 16:50:12 +0000 2022@dtabb73 I forgot to mention that the "N-formyl-Met initiation residue that is then removed" paradigm often mentioned wrt prokaryote translation isn't very consistent with existing data. For example, the mature form of E. coli's rpoA, rrpoB, rpoB' & rpoD all begin at an unmodified M1.
Fri Apr 22 15:23:46 +0000 2022MRPS9-201, mitochondrial ribosomal protein S9
🔗
1 – ? ⟶ 1 – 49 (transit peptide)
? – 396 ⟶ 50 – 396 (mature sequence)
#realNT
Fri Apr 22 15:15:30 +0000 2022OSTC-201, oligosaccharyltransferase complex non-catalytic subunit
🔗
1 – 149 ⟶ 1 – 149 (N-acetyl-Met, ο = 90%)
#realNT
Fri Apr 22 15:11:56 +0000 2022UQCRFS1-201, ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
🔗
1 – 78 ⟶ 1 – 78 (N-acetyl-Met, ο = 100%)
#realNT
Fri Apr 22 14:46:07 +0000 2022@ItaiYanai Even though the modern, rigid, corporate, rote scientific paper format has done its best to get rid of any hint of mischievousness.
Fri Apr 22 14:42:10 +0000 2022The human subunits show the more nuanced, redundant, multiple kinase/phosphatase pattern common in multicellular organisms. The 2 human paralogs do not share any observable tryptic peptides.
Fri Apr 22 14:40:08 +0000 2022Modification diagrams for yeast MVB12 & its human orthologs MVB12A & MVB12B, a subunit of the ESCRT-I complex. The yeast subunit could have been an illustration for a 1980's biochem text (e.g. Lehninger) explanation of phosphorylation. 🔗
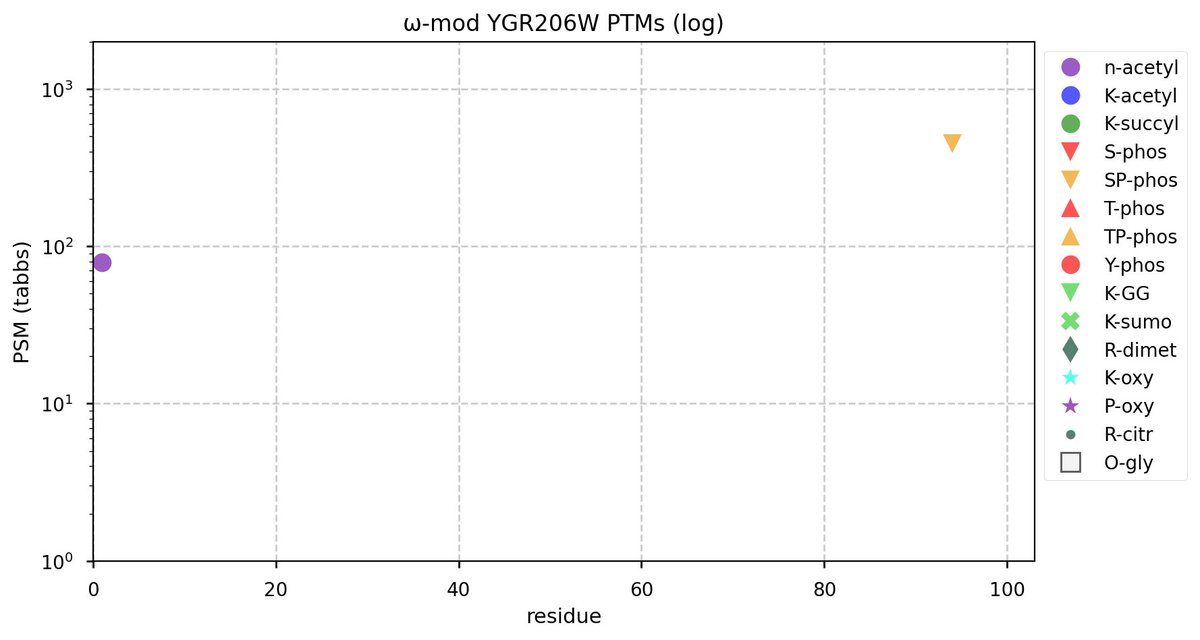
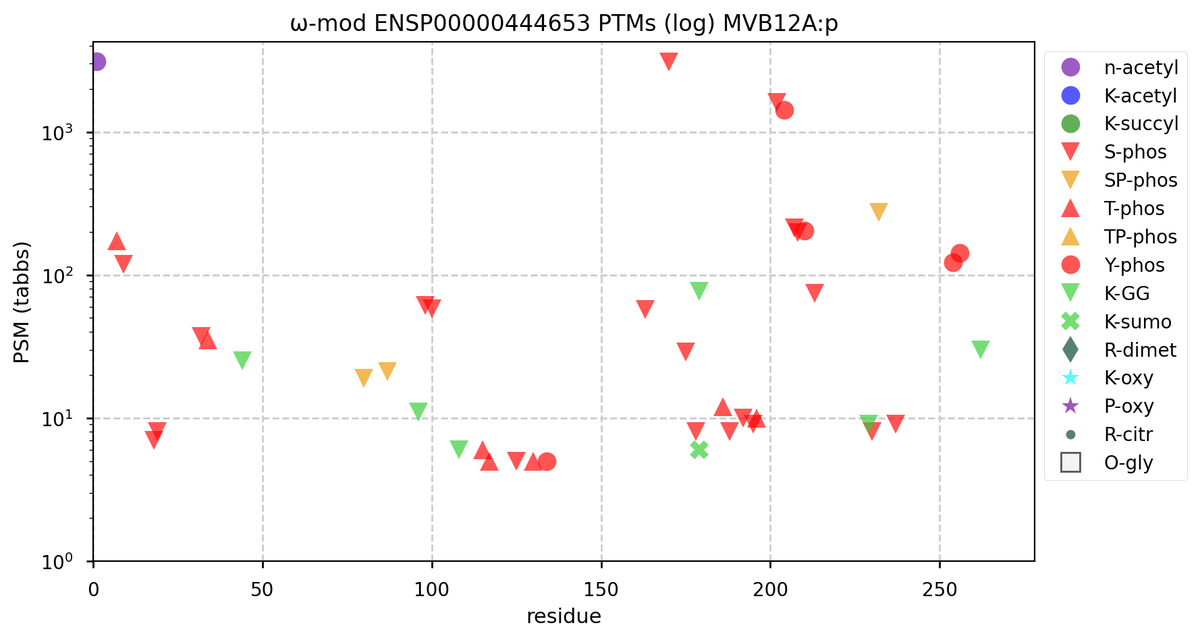
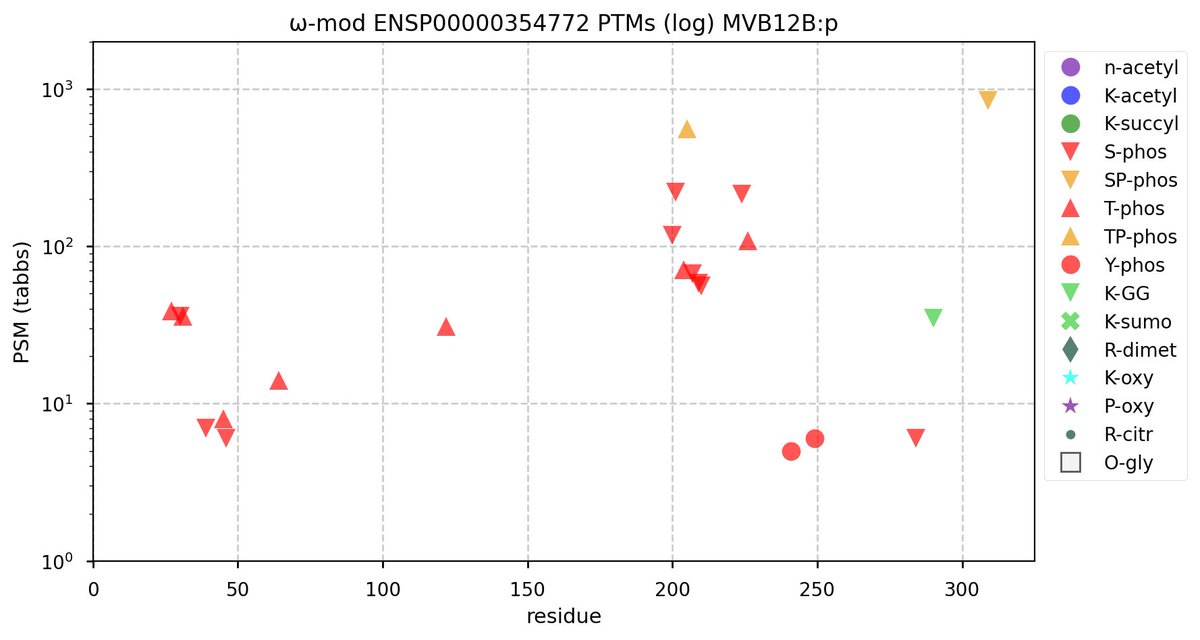
Fri Apr 22 14:33:30 +0000 2022@AlexUsherHESA At the time, Montreal was one of the few teams with an effective defense.
Fri Apr 22 13:39:46 +0000 2022Interesting thread if you are interested in the details of user tracking as applied to PDFs in the modern "scholarly" publication biz. 🔗
Fri Apr 22 12:42:25 +0000 2022@JesseBrown & just now my feed has filled up with ads from "cybersecurity" companies.
Fri Apr 22 12:39:02 +0000 2022It looks like it has quite a bit of wet air to draw on today (precipitable water & surface wind vectors) 🔗
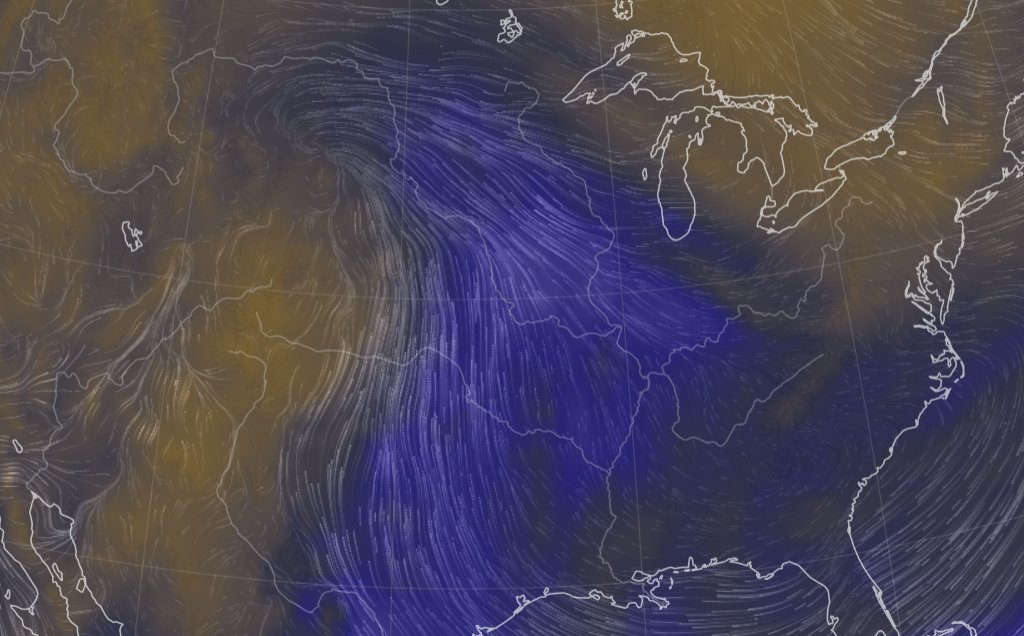
Fri Apr 22 12:36:41 +0000 2022Rain & snow (NEXRAD) along the cold fringes of today's Colorado Low. 🔗
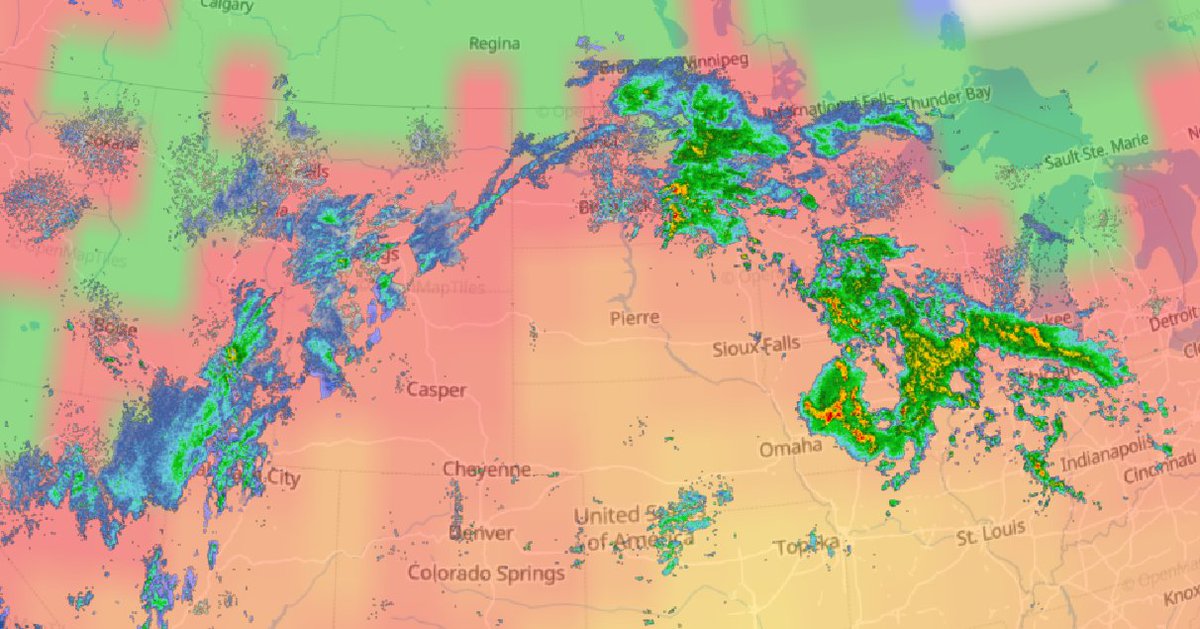
Fri Apr 22 12:24:15 +0000 2022@JesseBrown I run a set of web sites only interesting to a small set of academic scientists, but they get DDOS (& other) attacks pretty regularly.
Fri Apr 22 12:19:07 +0000 2022#resMods: Phe/F
N-amino: none
Sidechain: non-reactive
C-carboxyl: none
Thu Apr 21 20:28:08 +0000 2022@cajun_science PS: when present, Gla modifications can be a bit more rambunctious than you might expect, e.g., coagulation factor II (thrombin) 🔗
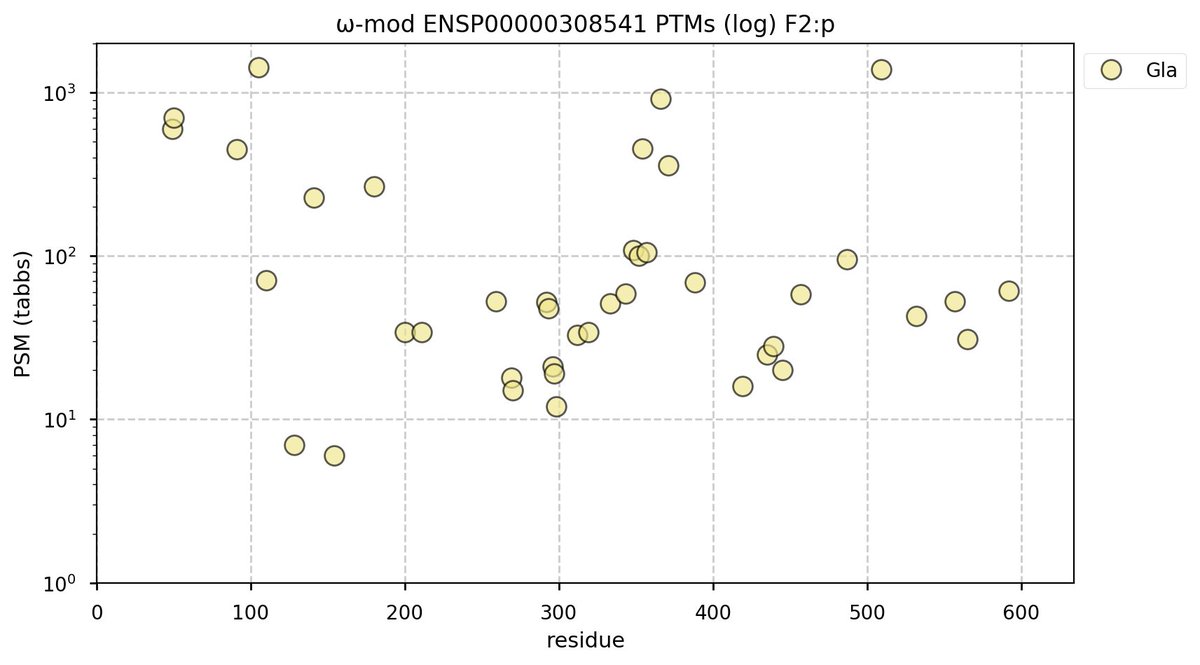
Thu Apr 21 17:28:18 +0000 2022@Paul_M_Thomas Because isoaspartate is normally produced from Asn as 1 of the 2 products of its deamidation, I was going to wait for the "Asn/N" entry to mention it.
Thu Apr 21 15:56:05 +0000 2022@cajun_science 4. Common in select blood plasma proteins, e.g., F2:p (extracellular enzymatic reaction)
5. Pyroglutamate at peptide N-terminus (extracellular enzymatic reaction).
6. Pyroglutamate at peptide N-terminus (in source reaction).
Thu Apr 21 15:55:44 +0000 2022@cajun_science So, final version:
Glu/E
N-amino: +acetyl¹
Sidechain: reactive -COOH
«bio»
+ADP-ribosylation²; γ-glutamyl³ ⁴; water-loss⁵
«artifact»
water-loss⁶
C-carboxyl: none
Notes:
1. Protein N-terminus.
2. Rare without enrichment.
3. Modified residue symbol 'Gla'.
Thu Apr 21 15:35:45 +0000 2022Modification diagrams for yeast SRN2:p & its human homologs VPS37A:p, VPS37B:p & VPS37C:p, subunits of ESCRT-I. They have sparse, unrelated modification patterns. SRN2 & VPS37C have observed alternate initiation sites. The 3 human paralogs share no observable tryptic peptides. 🔗
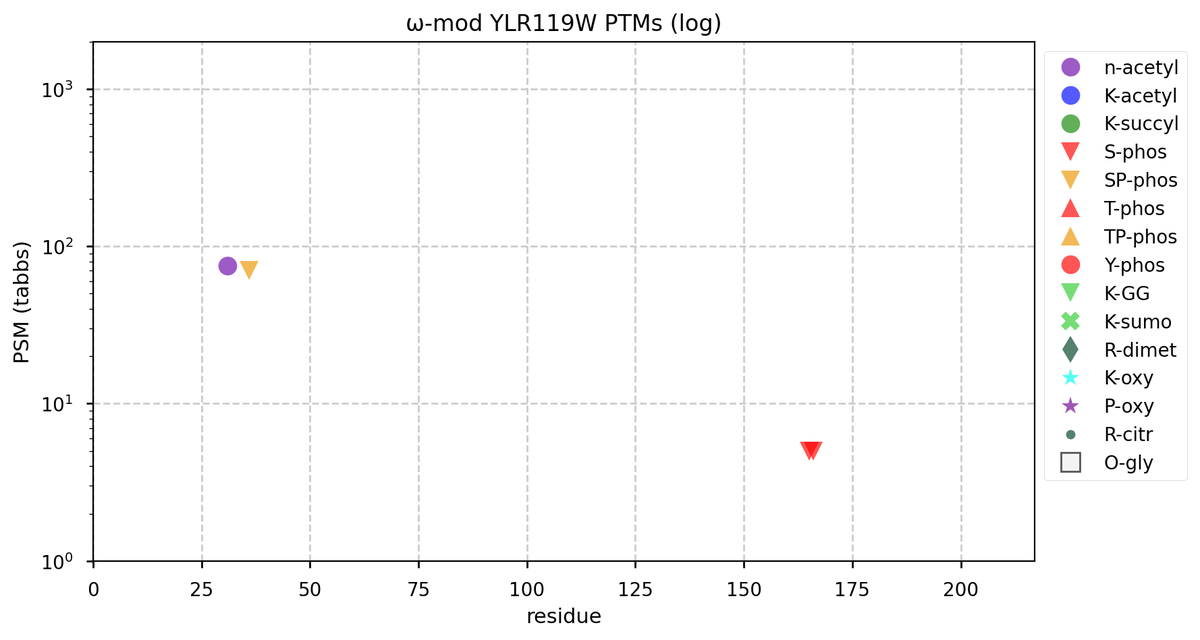
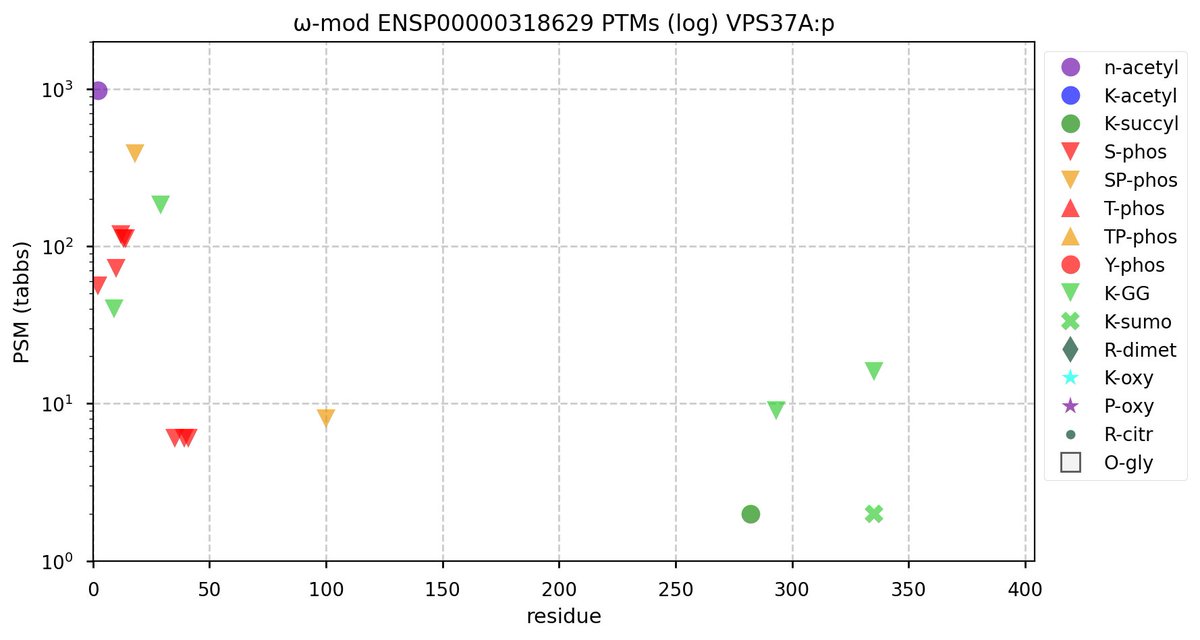
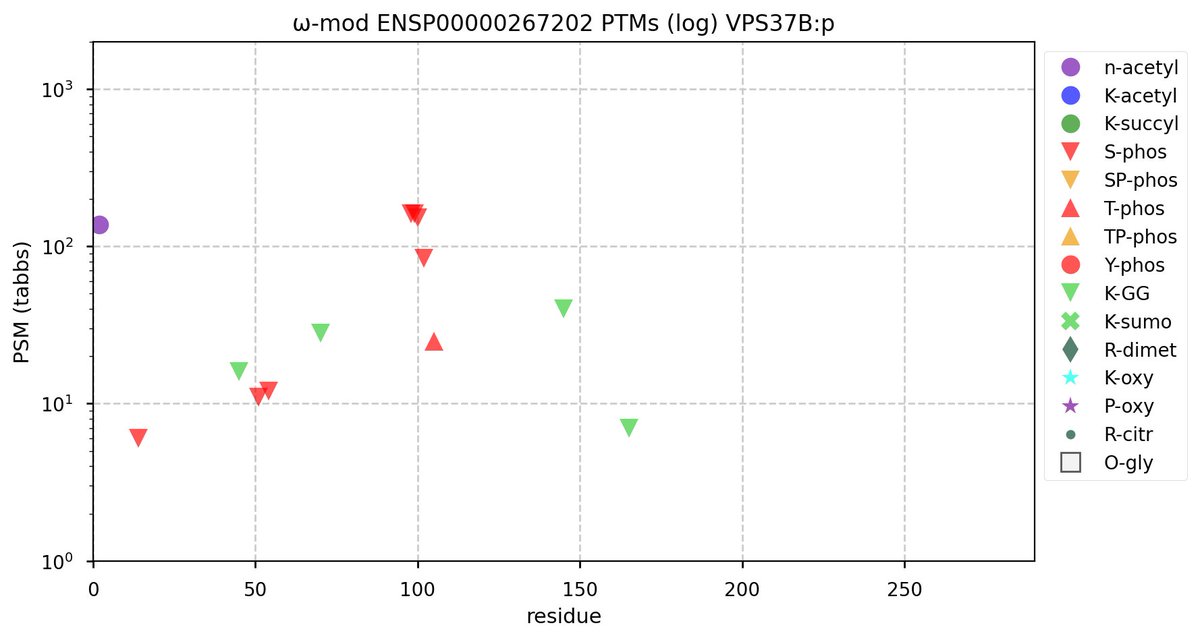
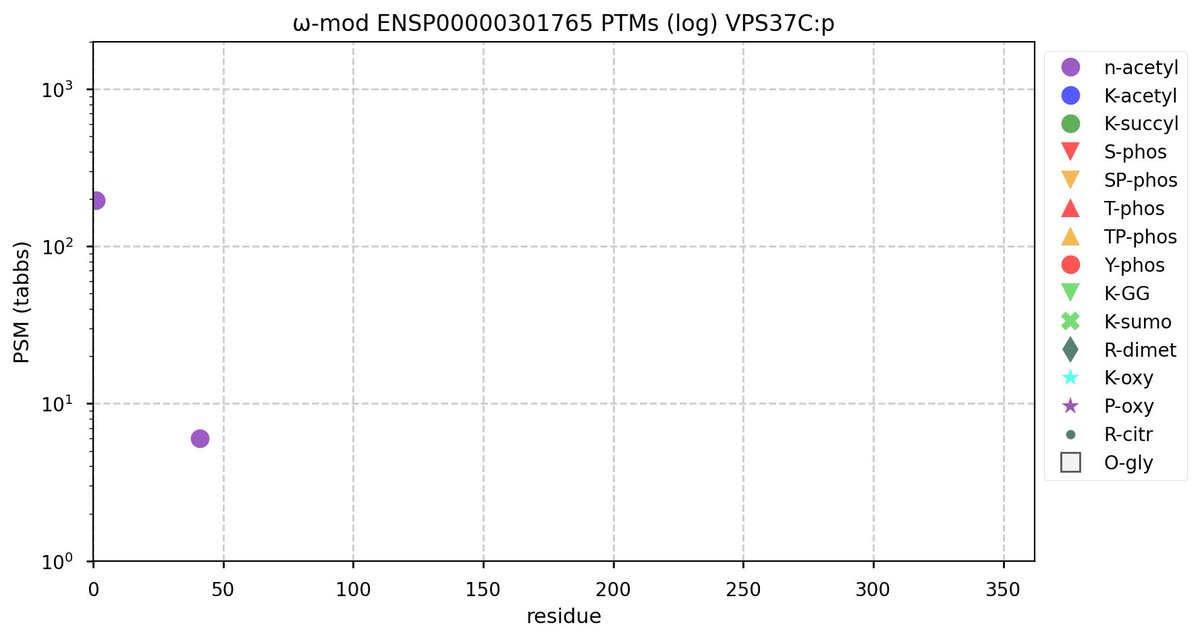
Thu Apr 21 15:26:32 +0000 2022@cajun_science How does QPCT sneak back into the cell? Or does the amyloid peptide make its way into the ER?
Thu Apr 21 14:53:55 +0000 2022@cajun_science Has there been any work to determine what enzyme performs this reaction? Water-loss into an aqueous solution usually requires some sort of catalytic push.
Thu Apr 21 14:44:31 +0000 2022@cajun_science 4. Common in select blood plasma proteins, e.g., F2:p.
5. Cyclization with N-amino to form pyro-glutamic acid (in source reaction).
Thu Apr 21 14:43:19 +0000 2022@cajun_science #resMods: Glu/E
N-amino: +acetyl¹
Sidechain: reactive -COOH
«bio»
+ADP-ribosylation²; γ-glutamyl³ ⁴
«artifact»
water-loss⁵
C-carboxyl: none
Notes:
1. Protein N-terminus.
2. Rare without enrichment.
3. Modified residue symbol "Gla".
Thu Apr 21 14:42:04 +0000 2022@cajun_science I have never been convinced that it is an in vivo or in vitro reaction. I would place it as an in vacuum (source) reaction. But maybe it should be here, so
Thu Apr 21 14:20:05 +0000 2022#resMods: Glu/E
N-amino: +acetyl¹
Sidechain: reactive -COOH
«bio»
+ADP-ribosylation²; γ-glutamyl³ ⁴
C-carboxyl: none
Notes:
1. Protein N-terminus.
2. Rare without enrichment.
3. Modified residue symbol "Gla".
4. Common in select blood plasma proteins, e.g., F2:p.
Thu Apr 21 13:57:11 +0000 2022@statesdj I'm OK, but the bike required quite a bit of work. The SUV rolled through a stop sign (& then took off).
Thu Apr 21 13:03:10 +0000 2022@statesdj As someone who was T-boned by an SUV last week at an intersection while riding in a protected bike lane, I am skeptical ...
Thu Apr 21 13:00:58 +0000 2022If you are interested in using machine learning, this is an interesting examination of an unusual aspect of its sensitivity to the details during the learning process
🔗
Thu Apr 21 11:55:37 +0000 2022Forgot sidechain +S-nitrosylation.
Wed Apr 20 19:39:13 +0000 2022Say what you will about 280 characters, trying to shoehorn information into its restraints while attempting to retaining clarity can be a useful exercise.
Wed Apr 20 19:01:29 +0000 2022Things are shaping up for another Colorado Low rain/snow storm. 🔗
Wed Apr 20 18:06:14 +0000 2022@DRAWheatcraft @neely615 @UCDProteomics That is true, but only once you realize that professors are, by definition, amateur scientists.
Wed Apr 20 17:43:12 +0000 2022#resMods: Asp/D
N-amino: +acetyl¹
Sidechain: reactive -COOH
«bio»
+ADP-ribosylation²; +phosphoryl² ³
C-carboxyl: none
Notes:
1. Protein N-terminus.
2. Rare without enrichment.
3. Found in prokaryotes.
Wed Apr 20 17:40:38 +0000 2022@neely615 @DRAWheatcraft @UCDProteomics I go full Sherlock Holmes: "consulting scientist".
Wed Apr 20 15:15:51 +0000 2022Modification diagrams for yeast & human VPS28:p, homologs of a subunit of the ESCRT-I heterotetramer. The human sequence has more PTM acceptor activity, including Y+phosphoryl & SUMOylation sites. 🔗
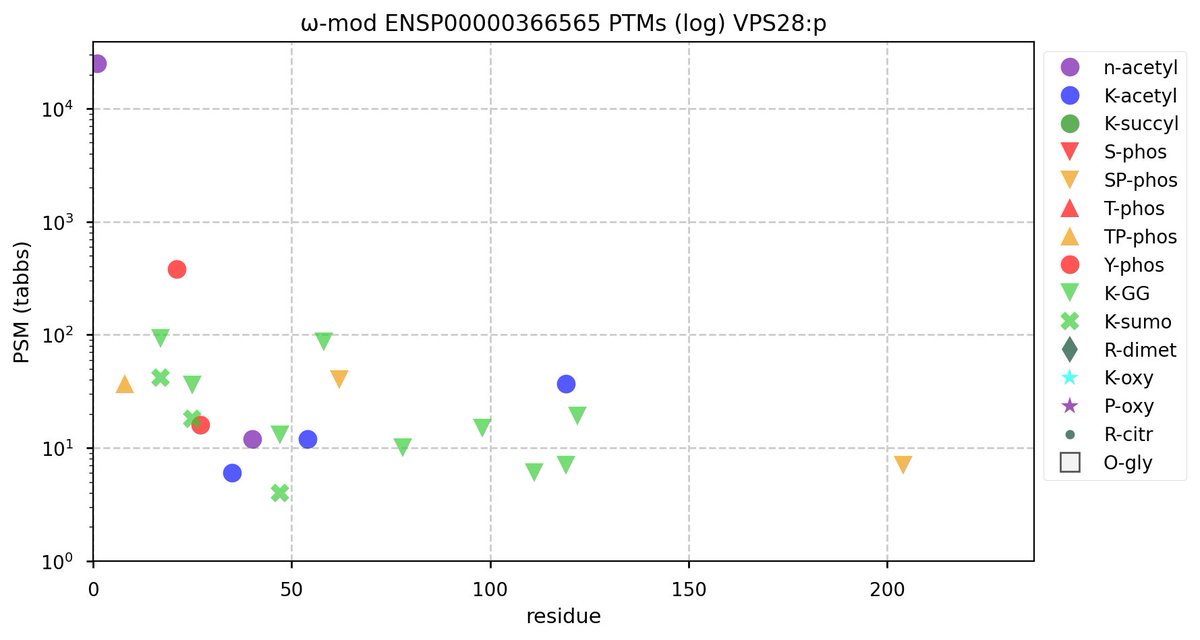
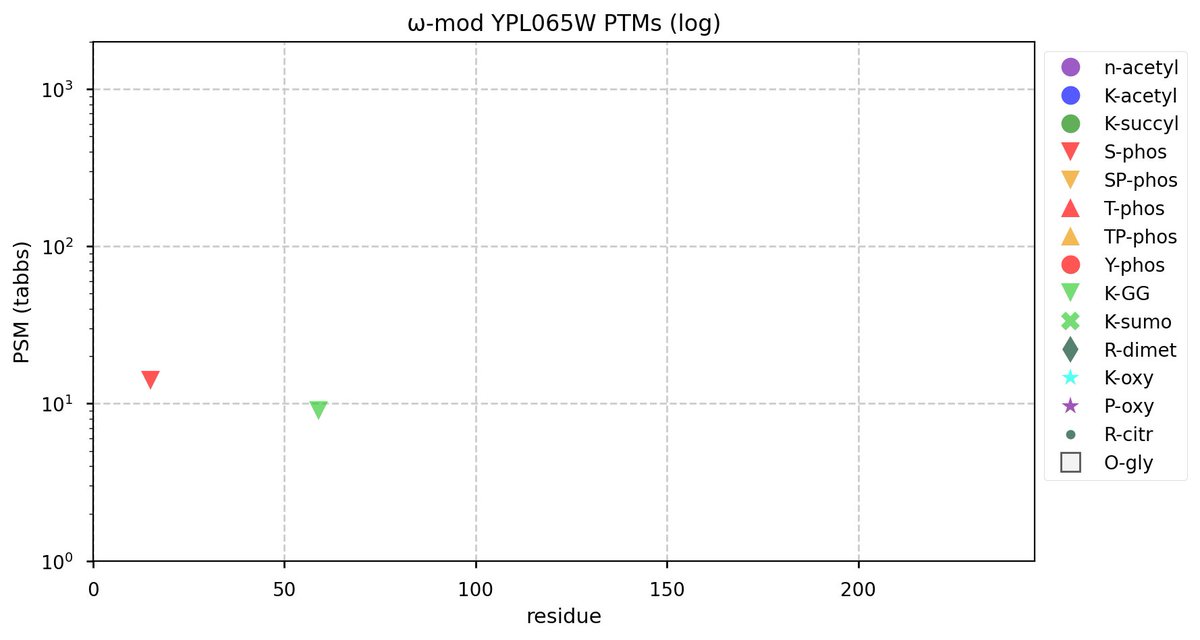
Wed Apr 20 13:11:24 +0000 2022@dtabb73 As I have pointed out with my #realNT running screed, Un!Pr*t's protein N-terminal annotation is really quirky.
Wed Apr 20 13:01:24 +0000 2022@dtabb73 I would suggest caution before betting the farm on that interpretation: I'm pretty sure either derivatization would block the formation of the other.⚠️
Wed Apr 20 12:52:12 +0000 2022@dtabb73 So the amino group has both formyl & acetyl attached?
Wed Apr 20 12:45:15 +0000 2022@dtabb73 I thought the initiation N-formyl-Met was always removed co-translationally in E. coli.
Tue Apr 19 16:49:32 +0000 2022@VATVSLPR @pwilmarth I have seen it, but rarely & coupled with the lack of any shift in retention time led me to conclude that it was in the ion source (like peptide N-terminal Glu dehydration).
Tue Apr 19 16:41:09 +0000 2022@VATVSLPR @pwilmarth #resMods: Cys/C+carbamidomethyl
N-amino: +acetyl (protein N-term);
Sidechain: ammonia-loss (cyclization with N-amino)
C-carboxyl: none
Note:
1. This is not a native amino acid, but many proteomics protocols generate this derivative on purpose.
Tue Apr 19 14:47:21 +0000 2022Modification diagrams for yeast STP22:p & human TSG101:p, homologous subunits of the #ESCRT-I heterotetramer. STP22:p doesn't participate in these PTMs, while TSG101:p does, including an unusual C-terminal Y+phosphoryl acceptor. 🔗
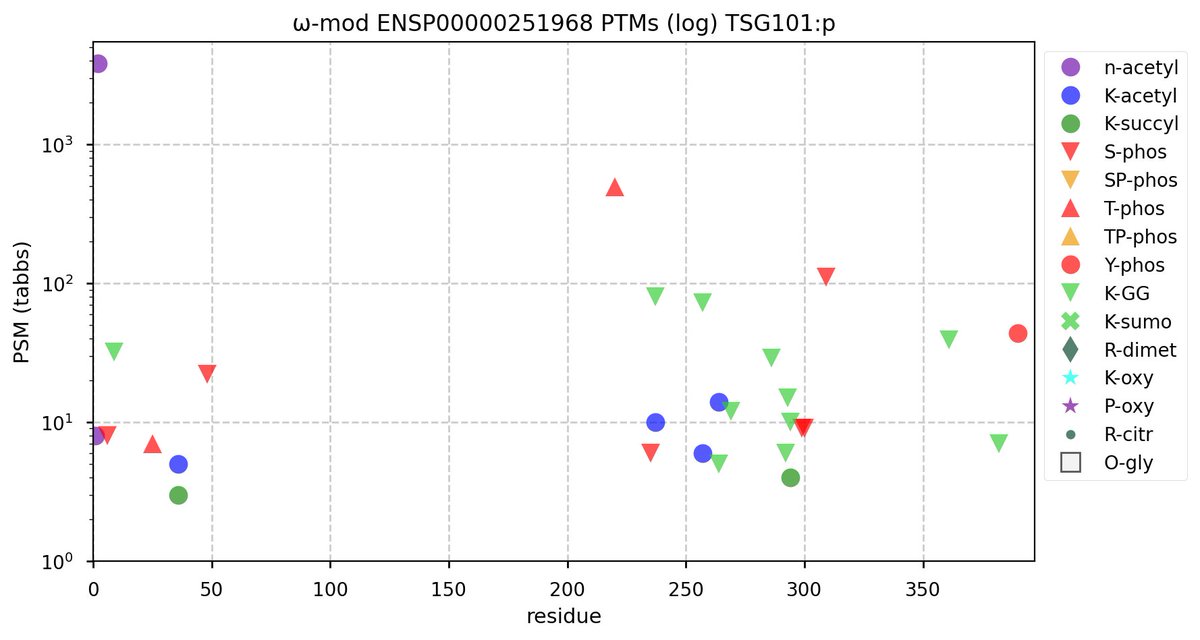
Tue Apr 19 14:31:54 +0000 2022@VATVSLPR @pwilmarth That is the intent, but given how often it happens (most expts use IAA or CAA), mentioning the cyclization reaction here is a reasonable addition. It also has a significant effect on chromatographic retention, shifting the peptide to later elution.
Tue Apr 19 14:11:42 +0000 2022@pwilmarth @cajun_science In the late 90's it was coined as an attempt to claim currency with the nucleic acid sequencing folks, but it sounds old-timey now. Enzymatic cleavage followed by peptide analysis was always the way protein sequencing was done, back to the days of AAA sequencing.
Tue Apr 19 13:06:43 +0000 2022🔗
Tue Apr 19 12:55:41 +0000 20224. C-carboxyl+methyl is present in a small number of C-prenylated proteins.
Tue Apr 19 12:55:03 +0000 2022NOTES:
1. The -SH is normally derivatized in proteomics experiments, abolishing redox mods.
2. Lipid mods are present in a limited number of proteins & affects only 1 C per protein.
3. Lipid modified peptides are too hydrophobic to be detected using typical LC/MS/MS experiments.
Tue Apr 19 12:54:08 +0000 2022#resMods: Cys/C
N-amino: +acetyl (protein N-term)
Sidechain: reactive -SH
«bio―redox»
+glutathionyl; +cysteinyl
«bio―lipid»
+palmitoyl; +farnesyl; +geranylgeranyl
«artifact―redox»
+3O; rarely +1 or +2
«artifact―PAGE»
+propionamide; +DeStreak
C-carboxyl: +methyl (protein C-term)
Mon Apr 18 15:25:14 +0000 2022@pwilmarth Tomorrow's residue (C) should be a good one to work out the kinks.
Mon Apr 18 15:24:25 +0000 2022@pwilmarth I agree these things are important, so I'll try to juggle being inclusive versus the "too-much-information-about-really-rare-stuff-no-one-but-me-has-ever-claimed" approach that has been taken in the past.
Mon Apr 18 14:52:27 +0000 2022I should also add that I know that protein C-terminal amidation has been described at some length in the literature, but I have never been able to find any (& I put some real effort into it) so I'm not going to mention it as a potential residue carboxyl-group mod.
Mon Apr 18 14:40:06 +0000 2022Any mods that are the result of deliberate modification during an experiment will not be mentioned, e.g. C+cam, K+TMT or peptide amino terminal+dimethyl.
Mon Apr 18 14:37:27 +0000 2022#resMods: alanine (A).
N-amino: protein N-terminus acetylation
Sidechain: none
C-carboxyl: none
Mon Apr 18 14:35:20 +0000 2022I will go through the residues in alphabetical order, posting an entry once a day with the hashtag #resMods. Anyone who has anything (informative) to add should feel free to chime in.
Mon Apr 18 14:32:03 +0000 2022Because of responses to a question regarding amino acid post-translational modification in proteins, it may be of some value to go through each residue type & list its commonly observed mods, attempting to distinguish between different sites & mechanisms of modification.
Mon Apr 18 12:50:05 +0000 2022The literature has assigned roles for the HSE:p & STAM:p proteins individually, but it is unclear whether those roles are real or simply misinterpretations of the subunits' ESCRT-0 function.
Mon Apr 18 12:46:33 +0000 2022Modification diagrams for yeast HSE1:p & human STAM:p, homologous subunits of the ESCRT-0 heterodimer. As with the VPS27/HGS homolog pair, the main change from yeast to human is the addition of Y+phosphoryl acceptors. 🔗
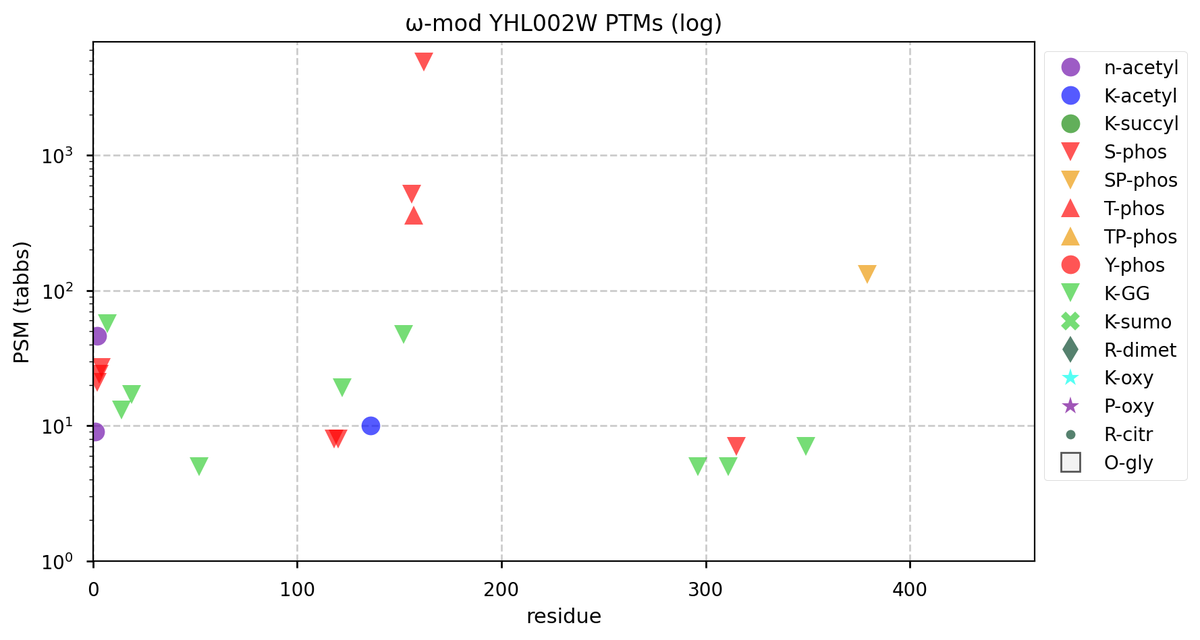
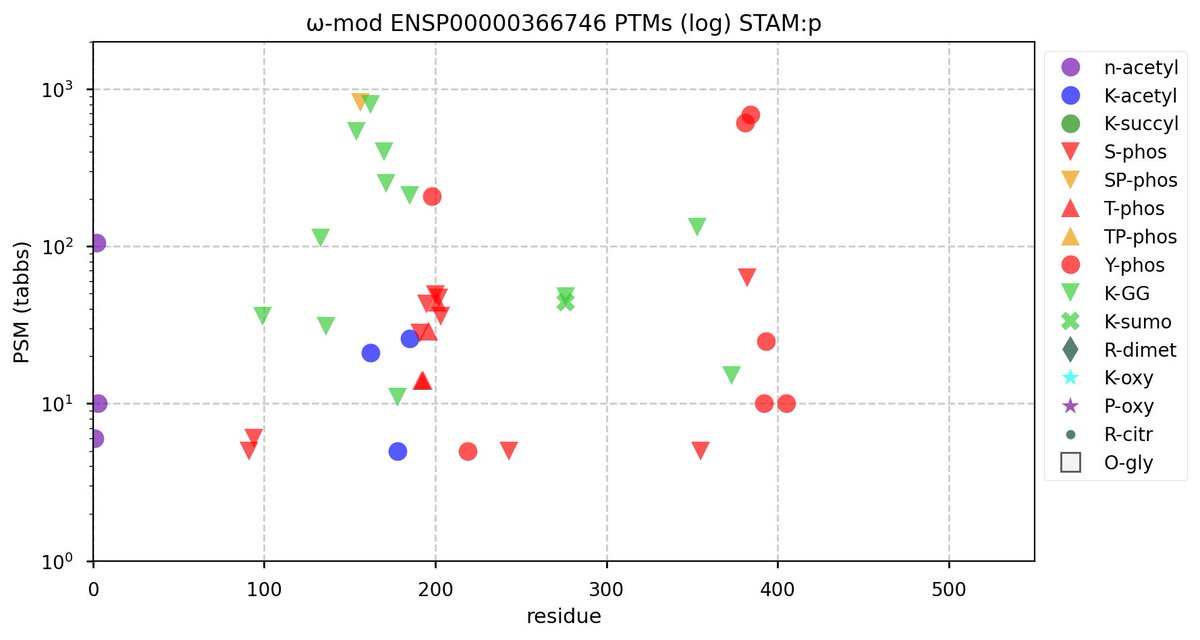
Mon Apr 18 12:40:47 +0000 2022It always annoys me when I go to a PI's web site & have to really look around to figure out where they are employed. It should be top line information.
Sun Apr 17 16:44:39 +0000 2022@cajun_science @JoelisSteele @TrumanLab I personally blame the "histone bros" for this: as a group they never met a modification too speculative to include in a paper.
Sun Apr 17 16:19:57 +0000 2022@cajun_science @JoelisSteele @TrumanLab Mods like N->citrulline or I+N-acetyl may be chemically possible, but they aren't what I would consider to be biological protein PTMs.
Sun Apr 17 16:05:16 +0000 2022@cajun_science @JoelisSteele @TrumanLab There has been a lot of speculation―based largely on the interpretation of MS/MS spectra―by groups that are motivated to find new (amazing) things. The field needs a "Koch's postulates" equivalent.
Sun Apr 17 15:02:21 +0000 2022I find cross-species PTM pattern comparisons interesting, although it isn't a popular past time within biomedical proteomics.
Sun Apr 17 14:01:35 +0000 2022@philbetts @ryanqnorth Non-Canadians often fail to recognize the pattern established by the fact that the country's 2 national sports (lacrosse & hockey) both involve beating each other with long wooden clubs: we really aren't very nice to each other.
Sun Apr 17 13:28:44 +0000 2022ESCRT (Endosomal Sorting Complexes Required for Transport) refers to heteromeric proteins (0, 1, 2, 3) involved in the formation & maintenance of endosomes/multivesicular bodies. ESCRT-0 recruits ubiquitinated cell membrane proteins into endosomes.
Sun Apr 17 13:16:35 +0000 2022Modification diagrams for yeast VPS27:p & human HGS:p, homologous subunits of the ESCRT-0 heterodimer. The palette of PTMs has changed during the last few 100s of million years. 🔗
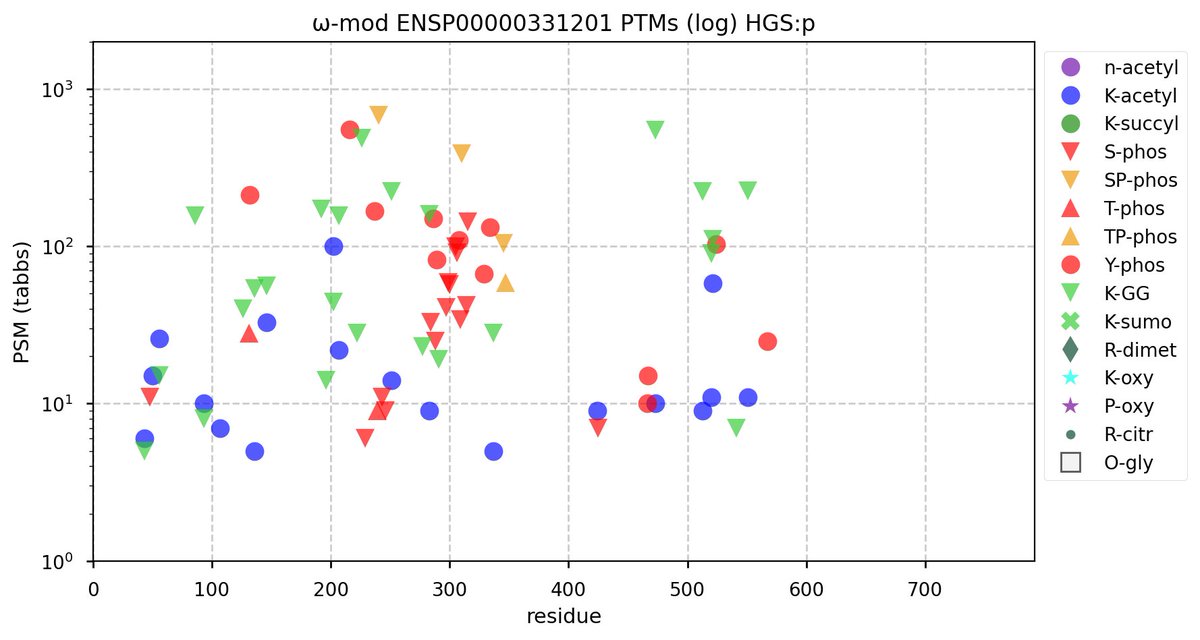
Sun Apr 17 02:28:53 +0000 2022S100a6-201, S100 calcium binding protein A6
🔗
1 – 89 ⟶ 2 – 89 (N-acetyl-Ala, ο = 96%)
#realNT
Sat Apr 16 22:32:40 +0000 2022@TrostLab @TrumanLab A & G side chains remain untouched. Same goes for V, I, F & L.
Sat Apr 16 21:27:47 +0000 2022@TrostLab @TrumanLab Gly can also be N-myristylated & it's C-terminus can be attached to a lysine side chain forming the protein-protein bridge in ubiquintin & ubi-like modifications, e.g., ubiqutinylation, nedylation or sumoylation.
Sat Apr 16 21:20:31 +0000 2022@TrostLab @TrumanLab They can be N-acetylated if they are the N-terminal residue of a protein.
Sat Apr 16 21:03:12 +0000 2022@UCDProteomics I am always interested in the rationale people use for the informal evaluation of the data-to-information transformation process.
Sat Apr 16 19:21:13 +0000 2022The greatest trick the Devil ever pulled was convincing the world that email was a good idea.
Sat Apr 16 16:38:26 +0000 2022@slashdot "Stick to your knitting" is always good advice.
Sat Apr 16 16:20:23 +0000 2022EIF4A2-201, eukaryotic translation initiation factor 4A2
🔗
1 – 407 ⟶ 2 – 407 (N-acetyl-Ser, ο = 100%)
#realNT
Sat Apr 16 15:42:06 +0000 2022@UCDProteomics How do you determine when you've got the right combination?
Sat Apr 16 15:25:02 +0000 2022CNRIP1-202, cannabinoid receptor interacting protein 1
🔗
1 – 164 ⟶ 2 – 164 (N-acetyl-Gly, ο = 99%)
#realNT
Sat Apr 16 15:14:14 +0000 2022PDE1A-205, phosphodiesterase 1A
🔗
1 – 535 ⟶ 2 – 535 (N-acetyl-Gly, ο = 99%)
#realNT
Sat Apr 16 14:06:32 +0000 2022CTNNA2-209, catenin alpha 2
🔗
1 – 953 ⟶ 2 – 953 (N-acetyl-Thr, ο = 98%)
#realNT
Sat Apr 16 13:38:43 +0000 2022BBS5-201, Bardet-Biedl syndrome 5
🔗
1 – 341 ⟶ 2 – 341 (N-acetyl-Ser, ο = 100%)
#realNT
Sat Apr 16 12:19:46 +0000 2022Modification diagram for #DENND10:p, a subunit containing a tripartite DENN domain. The rather compact DENN domain (2-357) comprises the entire sequence. Sometimes a cigar is just a cigar. 🔗
Sat Apr 16 01:00:29 +0000 2022I found "Everything Everywhere All at Once" to be well worth the price of admission.
Fri Apr 15 16:50:11 +0000 2022@NathHawkins Fortunately few people are taking advantage of full blown UTF-8 file names:
😡✋☀☁☔❄⚡✔❌.raw
Fri Apr 15 16:29:36 +0000 2022VPS37A-201, VPS37A subunit of ESCRT-I
🔗
1 – 397 ⟶ 2 – 397 (N-acetyl-Ser, ο = 100%)
#realNT
Fri Apr 15 16:14:33 +0000 2022@eliaslab1 Then there are many that go with kind of a Mad Libs style combining dates, instrument names, student's initials, fractions & experiment shorthand, e.g.
20110906_Velos2_JrHe_SA_RevCTF108CAF00_Phospho_Fr03.raw
Fri Apr 15 16:09:39 +0000 2022@eliaslab1 Depends on the group. Some are quite rigid about using a serial naming convention using some combination of letters & numbers, while others just jot down notes about the sample.
Fri Apr 15 15:55:55 +0000 2022SERPINB2-201, serpin family B member 2
🔗
Chain 1 – 415
Signal peptide 1 – ?
↓
1 – 415 (N-acetyl-Met, ο = 95%)
no signal peptide (not even close)
#realNT
Fri Apr 15 15:11:06 +0000 2022Credit: PXD028931, made available supporting 🔗
Fri Apr 15 13:24:05 +0000 2022In common with most DENN domain containing proteins, they have limited engagement with ubiquitination & no K+acetyl/succinyl acceptor sites.
Fri Apr 15 13:20:21 +0000 2022Modification diagrams for #DENND5a:p and #DENND5B:p, subunits with tripartite DENN domains. While genes are paralogs & the PTM patterns rhyme, they only share 12 of 151 observable 1° tryptic peptides, none of which include the phosphodomains. 🔗
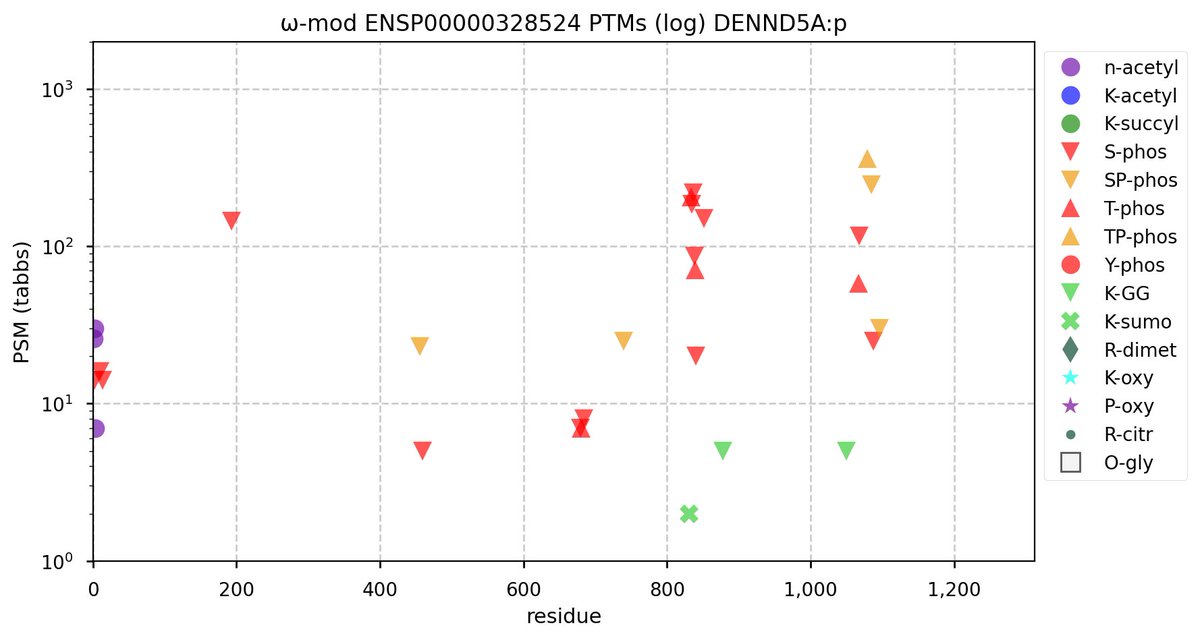
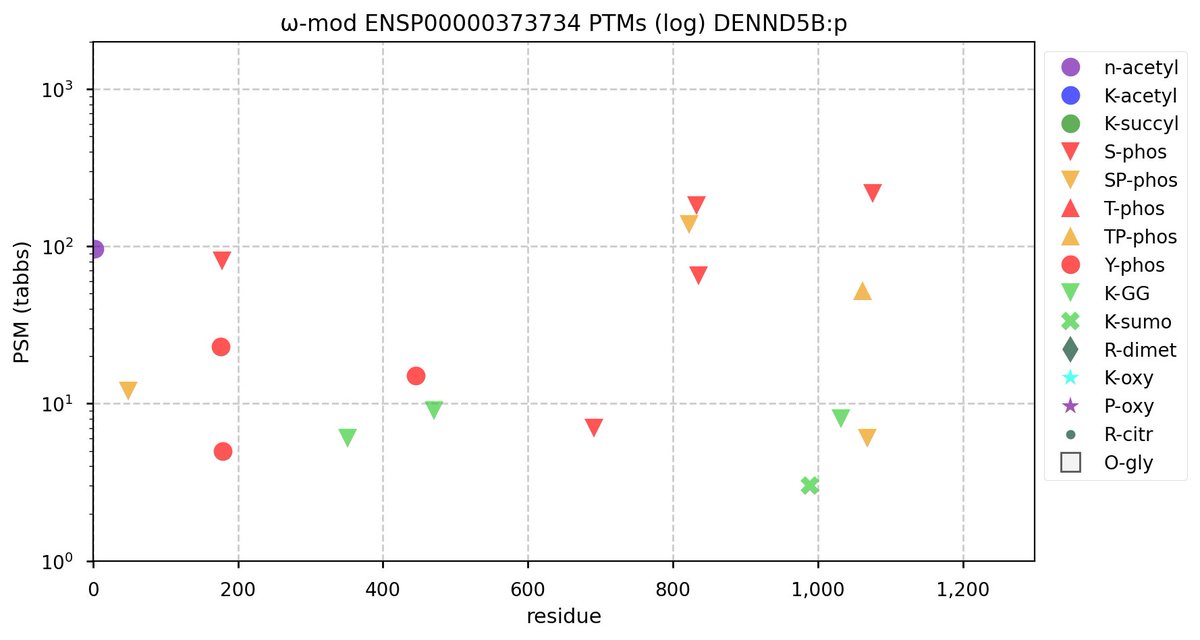
Fri Apr 15 11:59:45 +0000 2022The combination of a low pressure system over Vancouver Island & the Colorado low currently centered in northern Ontario is causing an odd pattern in the winds near the tropopause (250 hPa), aka "jet stream". 🔗
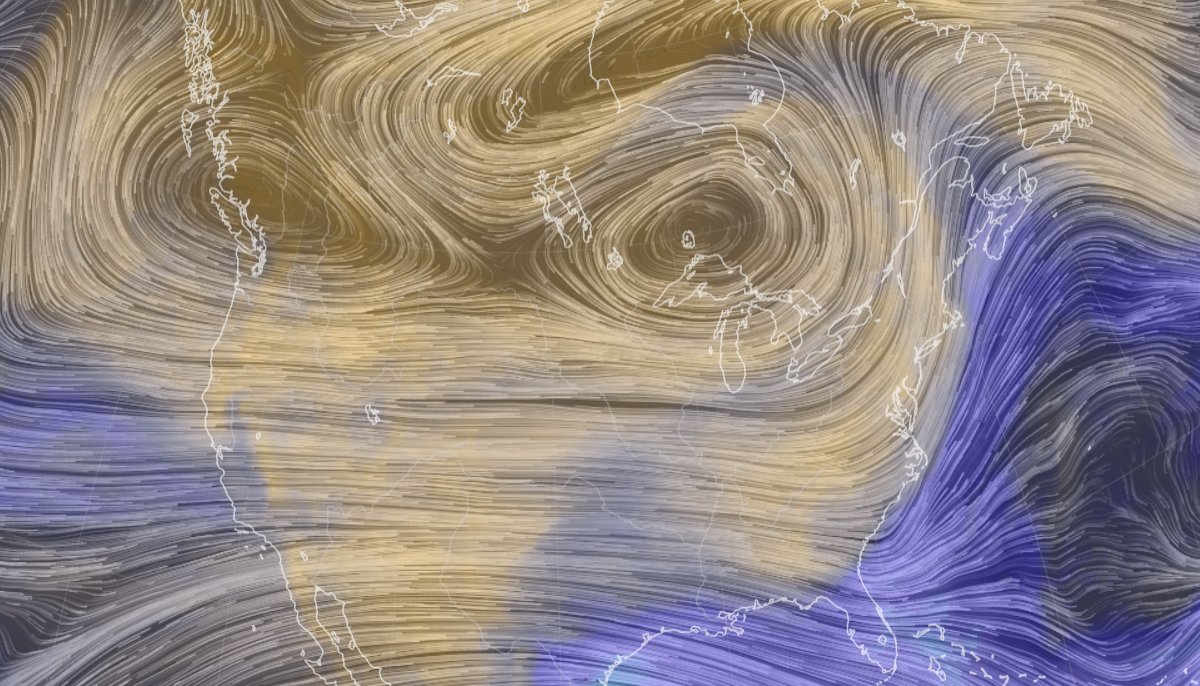
Fri Apr 15 11:35:15 +0000 2022My new favorite RAW file name in a public data set:
31_Lost some spray in the middle_UCSFslide10_2B4 with 350fmol pepmix_120min_3.5ul injection - Copy.raw
Thu Apr 14 12:49:58 +0000 2022(14-395) was the correct answer. Compact tripartite DENN domains do not contain S/T/Y+phosphoryl acceptors.
Thu Apr 14 12:40:04 +0000 2022Although the media made a big deal about what turned out to be a rather modest storm here in Winnipeg, it did produce a lot of rain locally across a band running from New Orleans to Chicago, particularly around the Mississippi Delta (in mm) 🔗
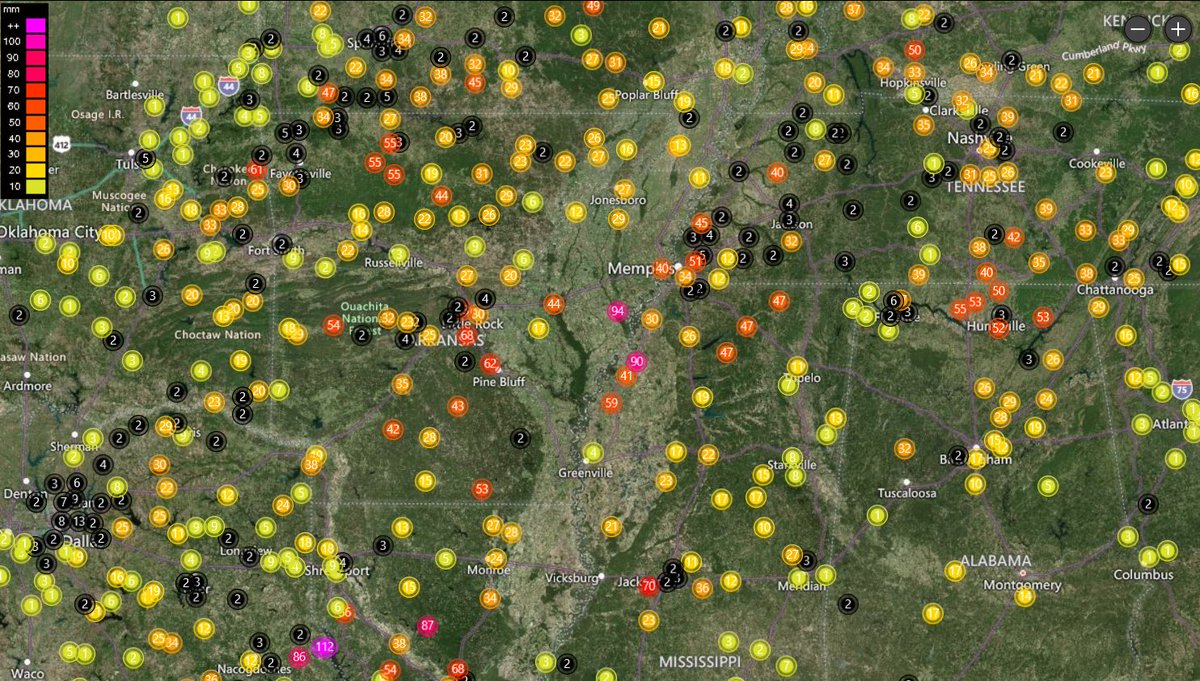
Thu Apr 14 12:25:22 +0000 2022Thanks to everyone who voted (all correctly). I will go back to spoon-feeding the information.
Thu Apr 14 12:23:29 +0000 2022These spaces result in longer domains, (48-910) & (86-558) that take up most of the subunit. This addition room between the non-IDRs means that it is possible for phosphodomains to sneak into the structure either as IHOPs adjacent to domain boundaries (FLCN) or an RIP (SMCR8). 🔗
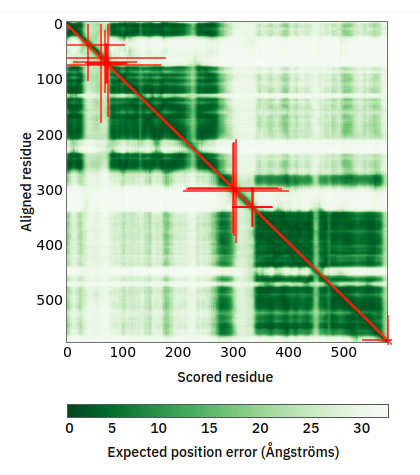
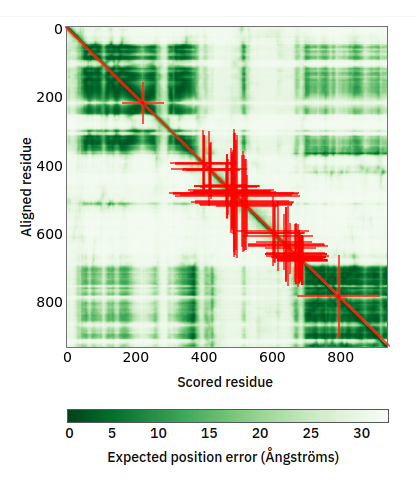
Thu Apr 14 12:16:26 +0000 2022Modification diagrams for #SMCR8:p & #FLCN:p, members of the DENN domain containing family of proteins (aka DENND8A & DENND8B). Unlike the DENN proteins mentioned earlier this week, in these proteins the 3 parts of the tripartite DENN domain have spaces between them. 🔗
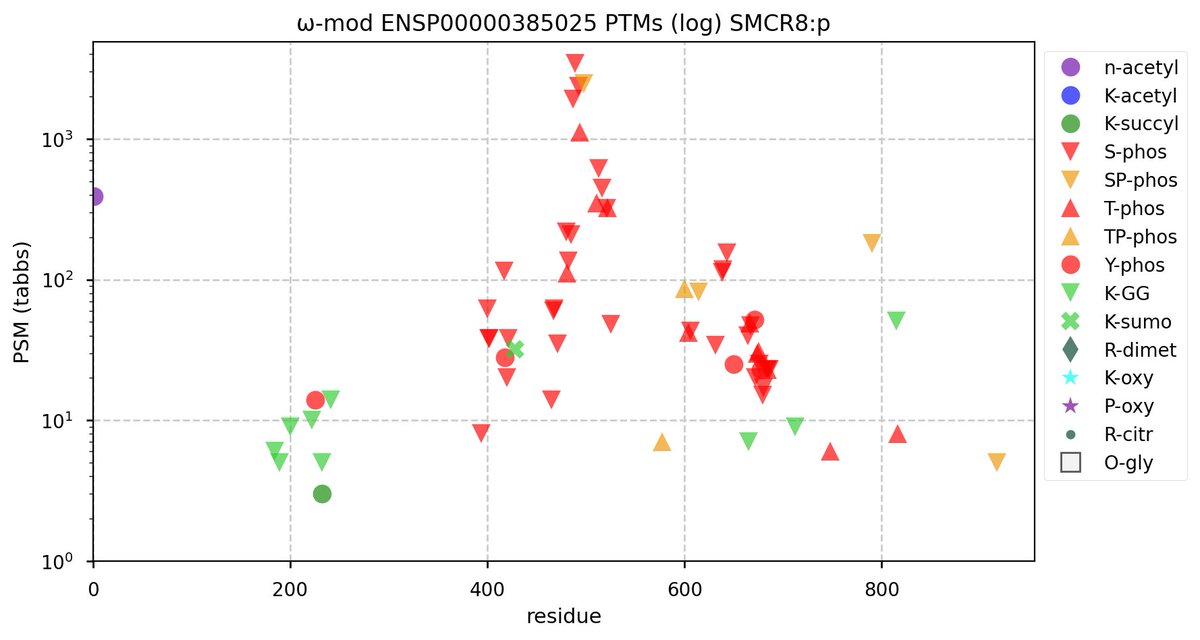
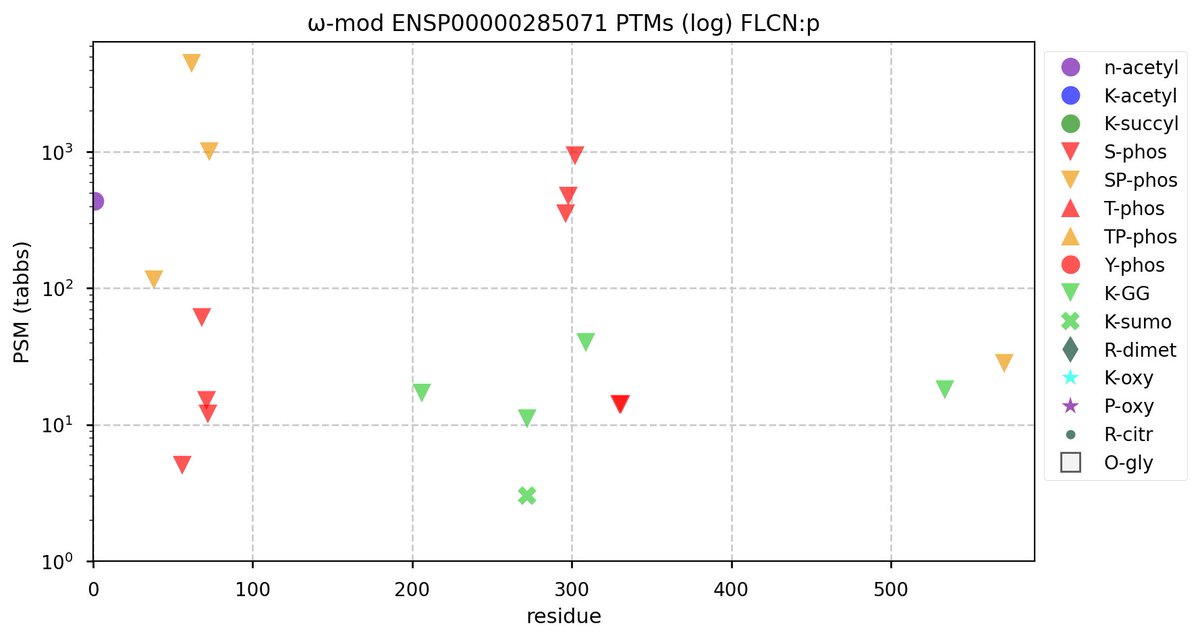
Thu Apr 14 12:08:39 +0000 2022🔗
Thu Apr 14 00:06:24 +0000 2022Things are going to be spicy along this front separating the cold outflow from the Colorado low in North Dakota and the flow up from the Gulf caused by a Bermuda high. Batten down the hatches. 🔗
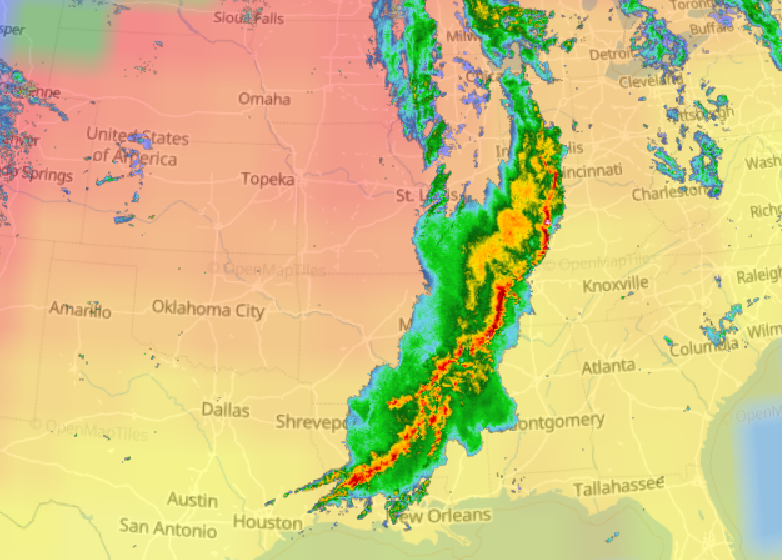
Wed Apr 13 17:08:05 +0000 2022@UCDProteomics @MattWFoster @sabbatiello0310 The objections were moral/religious rather than monetary: Las Vegas has good deals wrt conventions.
Wed Apr 13 16:32:20 +0000 2022Great quote: "which is why houses in Vancouver aspires to command prices on par with places like New York and Singapore despite being a second-rate deadzone of a city" 🔗
Wed Apr 13 15:31:23 +0000 2022I find it remarkable that so many of these basic annotations are wrong after 25 years of large scale proteomics, but I also have a notoriously skewed view of the subject.
Wed Apr 13 15:29:05 +0000 2022DENND10-201, DENN domain containing 10
🔗
1 – 357 → 2 – 357 (N-acetyl-Ala, ο = 100%)
#realNT
Wed Apr 13 14:49:03 +0000 2022@lenjf Which department offers the degrees?
Wed Apr 13 14:34:08 +0000 2022Coda: my office building door yesterday & today. 🔗


Wed Apr 13 14:28:09 +0000 2022At least in biomedical research. When the 1st set of genome sequences were made available, a long form report―similar to the now rare monograph―should have been introduced to deal with the new scale of investigations. The lack of such a format has tied the literature into knots.
Wed Apr 13 13:19:48 +0000 2022IMHO the answer has been "yes" for about 20 years.
🔗
Wed Apr 13 13:14:10 +0000 2022Does anybody know when & where the term "data scientist" emerged? Are there any dedicated PhD programs/departments in "data science" or is it an informal specialty associated with an applied math?
Wed Apr 13 13:02:10 +0000 2022It will continue to pump cold air down & across, running into warm air being pumped up from the Gulf of Mexico by a high off the mid-Atlantic coast generating a nearly static front & lots of intense stormy weather from Wisconsin down to Oklahoma.
Wed Apr 13 12:59:04 +0000 2022The low pressure circulation over the Dakotas & southern Manitoba is expected to stay put for a few days, dropping ~30 cm of snow were I am. 🔗
Wed Apr 13 12:44:41 +0000 2022Based on the information in this image, which of the following is the correct set of coordinates for the DENN domain: (2/2)
Wed Apr 13 12:42:29 +0000 2022Modification diagram for DENND1B:p, one of the tripartate DENN domain containing family of proteins.
🔗
1/2 (poll attached)
Wed Apr 13 12:34:00 +0000 2022Thanks to the brave few that took a shot at this one. All 3 were correct: a tripartite DENN domain, like most named domains, does not normally participate in phosphorylation.
Wed Apr 13 02:31:19 +0000 2022The frontal storm in Nebraska is along a line where the cold air from northern Saskatchewan meets the warm moist air from Texas, swirling around an axis stretching from the origin of the low up to a new secondary low pressure circulation now in South Dakota. 🔗
Wed Apr 13 01:36:54 +0000 2022While the northern portion of the storm has remained relatively static, there is now a very intense front running from Kansas, through Nebraska & headed for Iowa. There is also a storm in Texas just east of Dallas & a more diffuse front now in Louisiana & Mississippi. 🔗
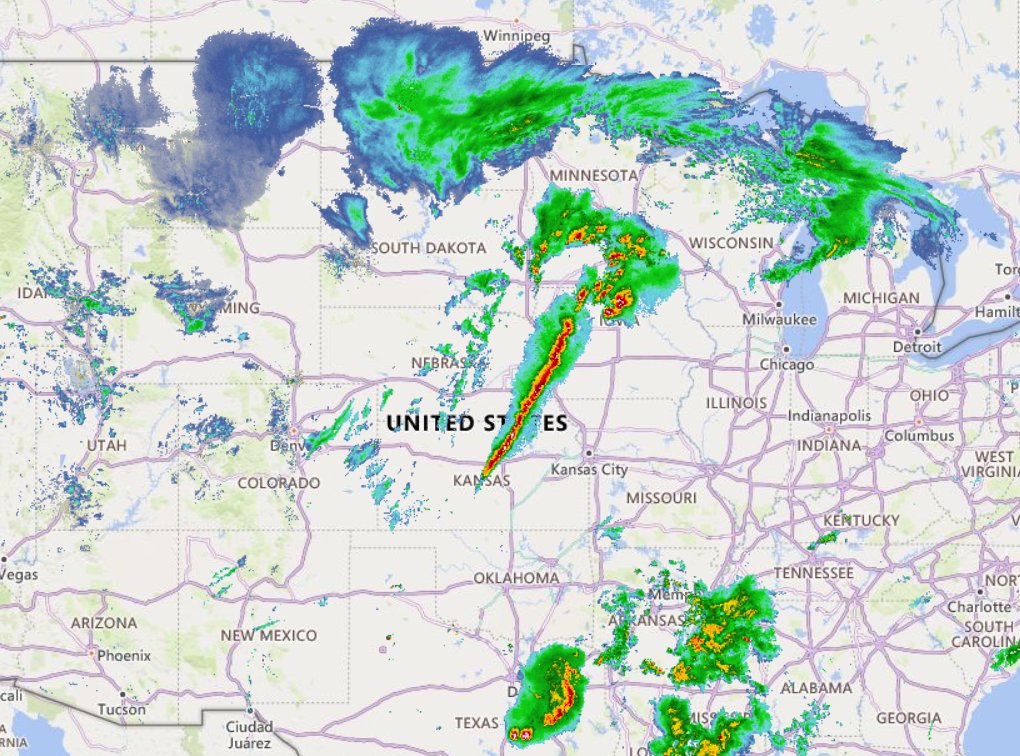
Wed Apr 13 00:40:16 +0000 2022@MattWFoster @sabbatiello0310 Probably stems from the same impulses that have always kept the ASMS (& HUPO) out of Las Vegas.
Tue Apr 12 22:43:16 +0000 2022From a traffic camera near Bismark, ND (now) 🔗

Tue Apr 12 22:40:18 +0000 2022The northern edge of the low has started to kick up snow fall across North Dakota, Minnesota & Wisconson, encroaching on southern Manitoba & Ontario (NEXRAD radar image) 🔗
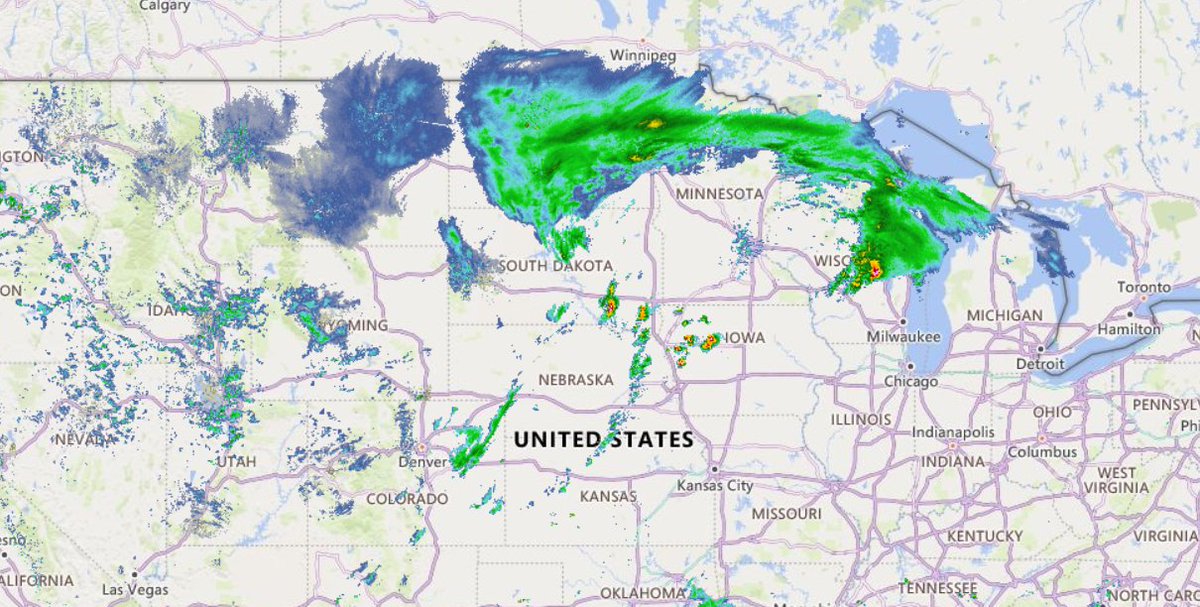
Tue Apr 12 21:03:35 +0000 2022There doesn't seem to be much interest in the thread-attached skill-testing question. 🔗
Tue Apr 12 20:01:25 +0000 2022MRPS30-202, mitochondrial ribosomal protein S30
🔗
1 – 439 → 18,26 – 439
#realNT
Tue Apr 12 19:08:03 +0000 2022RPL13-201, ribosomal protein L13
🔗
1 – 211 → 2 – 211 (N-acetyl-Ala, ο = 2%)
#realNT
Tue Apr 12 18:51:43 +0000 2022@dtabb73 I lost track of the number of times some jackass did this to me. You might want to include conference abstracts in this category.
Tue Apr 12 17:52:25 +0000 2022PRRX1-201, paired related homeobox 1
🔗
1 – 245 → 2 – 245 (N-acetyl-Ala, ο = 100%)
#realNT
Tue Apr 12 17:42:11 +0000 2022The low pressure system has become increasingly organized, with a good inflow of warm moist air from the south east and cold air straight down from northern Saskatchewan. A bad mix (unless you like a vigorous snow storm). 🔗
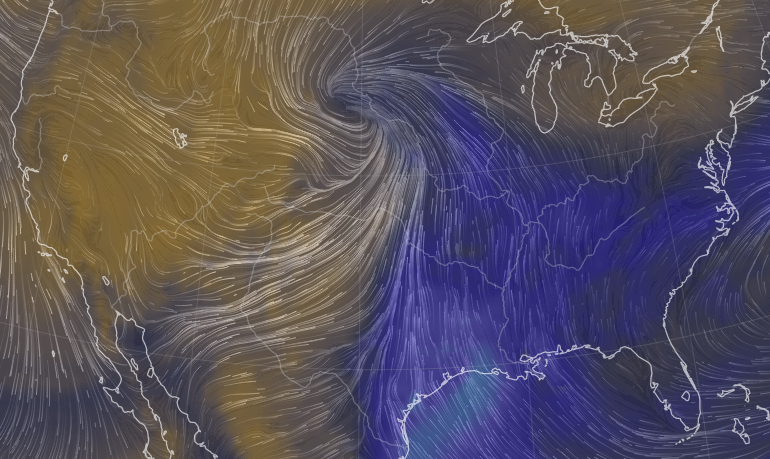
Tue Apr 12 15:20:40 +0000 2022@pwilmarth "Semantic semtex" is right up there with "noise mining" 😀
Tue Apr 12 15:15:54 +0000 2022A nasty-looking little storm cruising along the Kentucky-Tennessee border this morning 🔗

Tue Apr 12 14:49:16 +0000 2022@idpemery kismet
Tue Apr 12 14:41:17 +0000 2022Which knob on a Lumos (real or virtual) do you have to twiddle to get rid of this sort of fragmentation (the grey lines are y-CH₄SO ions from an oxidized M residue)? 🔗
Tue Apr 12 14:32:18 +0000 2022C11orf68-201, chromosome 11 open reading frame 68
🔗
1 – 292 → 2 – 292 (N-acetyl-Ala, ο > 99%)
#realNT
Tue Apr 12 12:15:06 +0000 2022Using this diagram only, which of the following domains is the correct location for the DENN domain structure:
Tue Apr 12 12:13:58 +0000 2022Modification diagram for a human DENND family protein that contains an ~400 residue tripartite DENN domain. 🔗
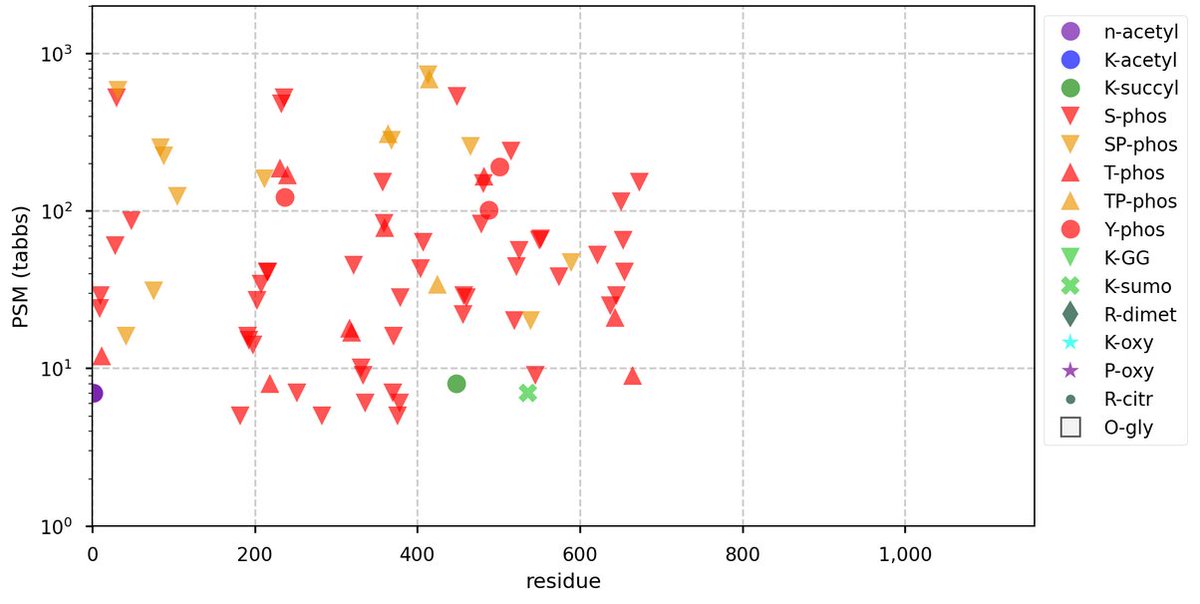
Tue Apr 12 11:50:54 +0000 2022For those not in the northern Plains, the potential disruptions associated with this particular storm has not gone unnoticed by the locals
🔗
Tue Apr 12 11:32:35 +0000 2022The intensifying low pressure system, now centered near the Colorado-Nebraska border, has started to draw moist air from the earlier high up into itself. The amount of moist air pulled in will determine how much rain/snow will be caused by this low. 🔗
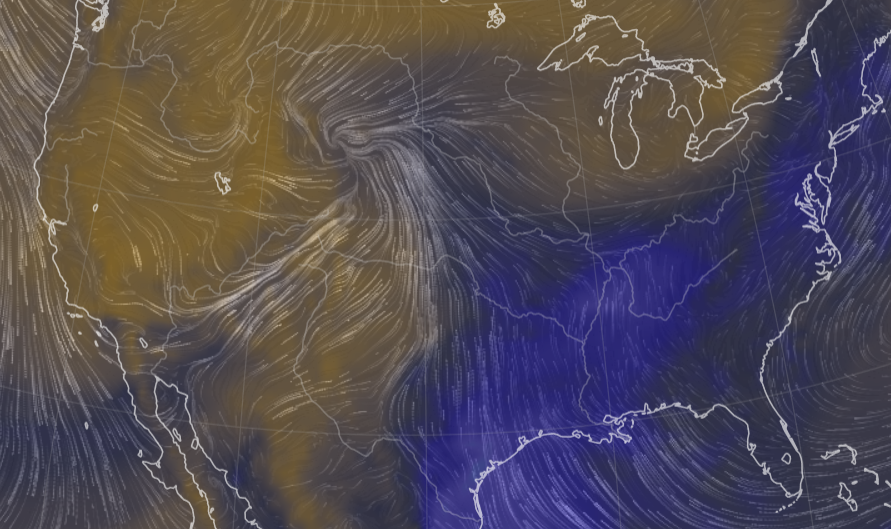
Tue Apr 12 00:07:33 +0000 2022The moist warmer air has now established itself in a band from the Gulf of Mexico up to the Great Lakes. Strong winds across the Rockies show the atmosphere destabilizing across the south-central Great Plains. 🔗
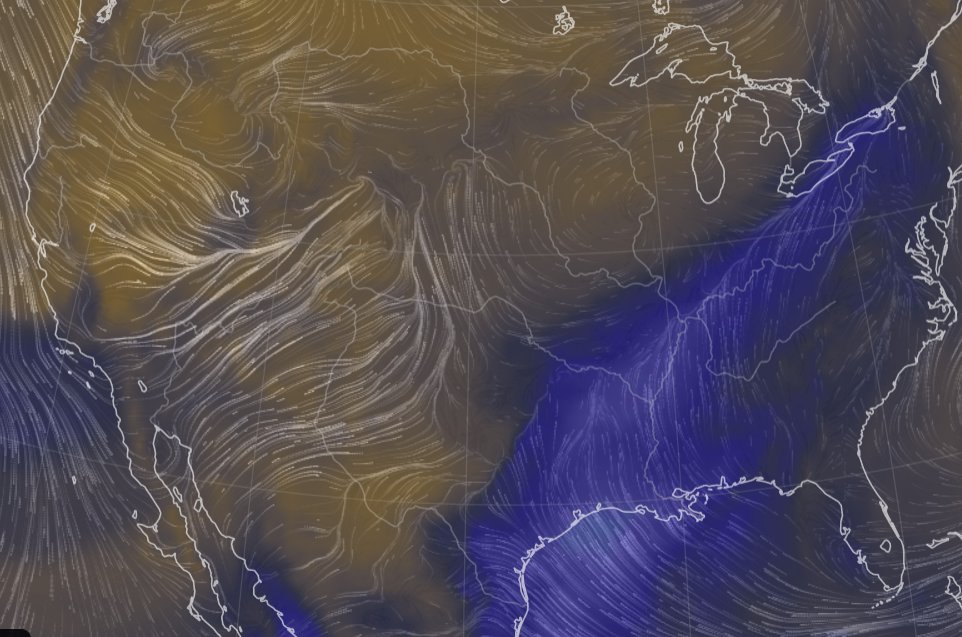
Mon Apr 11 15:11:54 +0000 2022Is the similarity adjacent to the N-terminus sufficient to keep DENND4B in the same naming category as A/C, when rest of the sequence has such a different phosphodomain pattern?
Mon Apr 11 14:16:12 +0000 2022Forgot the hash tags yesterday: DENN4A:p & DENN4C:p are featured here 🔗
Mon Apr 11 14:12:25 +0000 2022The only similarity between the rest of B and the A/C pattern is that all 3 have an IHOP domain just N-terminal to a tetratricopeptide-like helical domain located at (772-868) in B: the first phospho-domain on this plot. 🔗
Mon Apr 11 14:09:59 +0000 2022Modification diagram for DENND4B:p, a large #DENN domain containingsubunit. A PTM-free N-terminal tripartite DENN domain (195-644) is located similarly to #DENND4A or #DENND4C. Unlike A or C, B has no ER signal peptide & has an N-terminal A2+acetyl.
🔗
Mon Apr 11 12:31:24 +0000 2022@graemedmoffat @AlexUsherHESA 🔗
Mon Apr 11 12:24:53 +0000 2022PPP1R18-202, protein phosphatase 1 regulatory subunit 18
🔗
1 – 613 → 2 – 613 (N-acetyl-Ala, ο = 97%)
#realNT
Mon Apr 11 12:17:37 +0000 2022DENND4B-201, DENN domain containing 4B
🔗
1 – 1496 → 2 – 1496 (N-acetyl-Ala ο = 100%)
#realNT
Mon Apr 11 12:04:43 +0000 2022The "Bermuda High" that is driving this effect is currently centered in northern Florida, as seen by the winds at 850 hPa 🔗
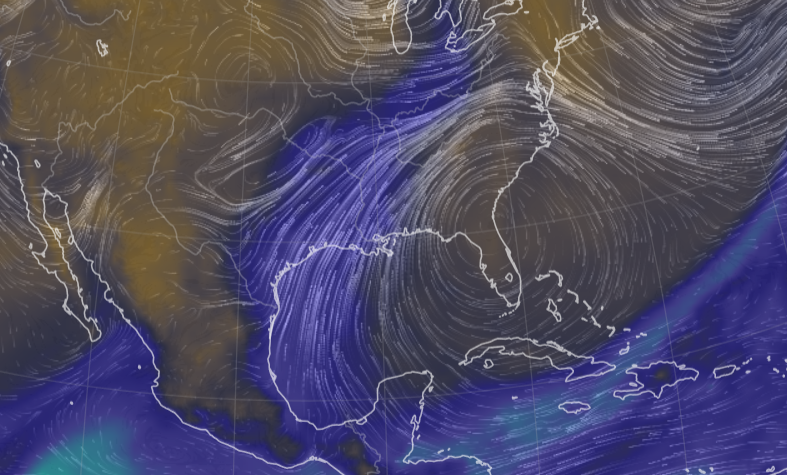
Mon Apr 11 11:59:37 +0000 2022The area covered by moist air has broadened & its northern tip has been pushed east by the high level wind pattern (winds at 250 hPa overlaid on total precipitable water). 🔗
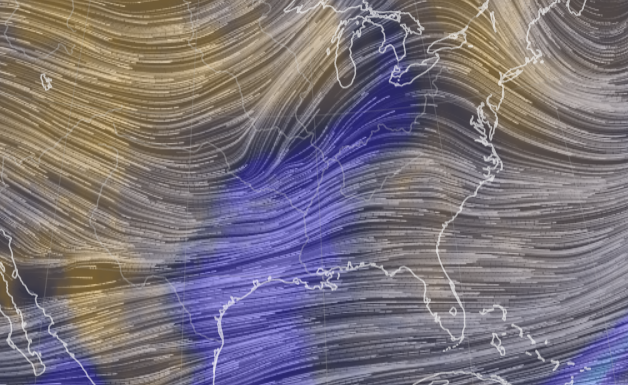
Mon Apr 11 01:10:37 +0000 2022The moist air from the Gulf of Mexico has reached well up into the northern US plains & some low pressure circulation has started to form. 🔗
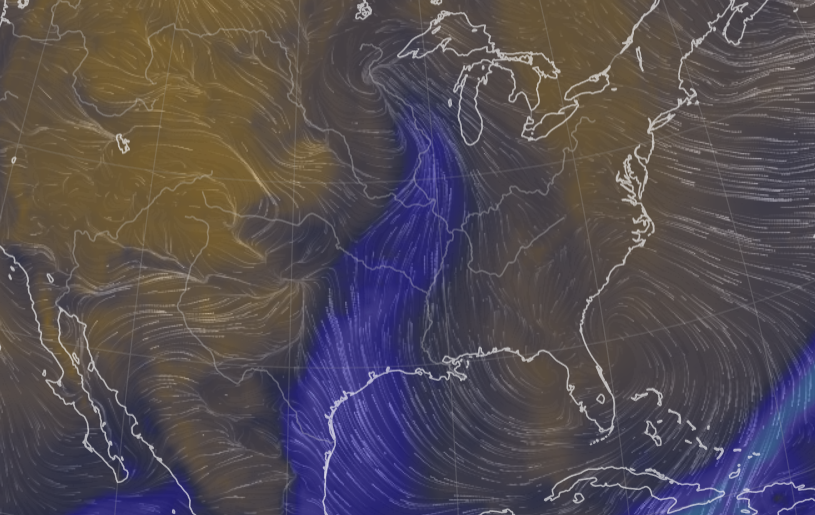
Sun Apr 10 23:06:33 +0000 2022TUBGCP6-201, tubulin gamma complex associated protein 6
🔗
1 – 1819 → 2 – 1819 (N-acetyl-Ala ο = 99%)
#realNT
Sun Apr 10 16:52:27 +0000 2022Not very surprisingly, the IHOP and RIP domains (red) avoid―but often cozy up to―AF-predicted non-IDR domains (green), as illustrated for DENND4C ... 🔗
Sun Apr 10 16:10:51 +0000 2022The beginnings of a Colorado Low (surface winds on total precipitable water, bluer mean more water). 🔗
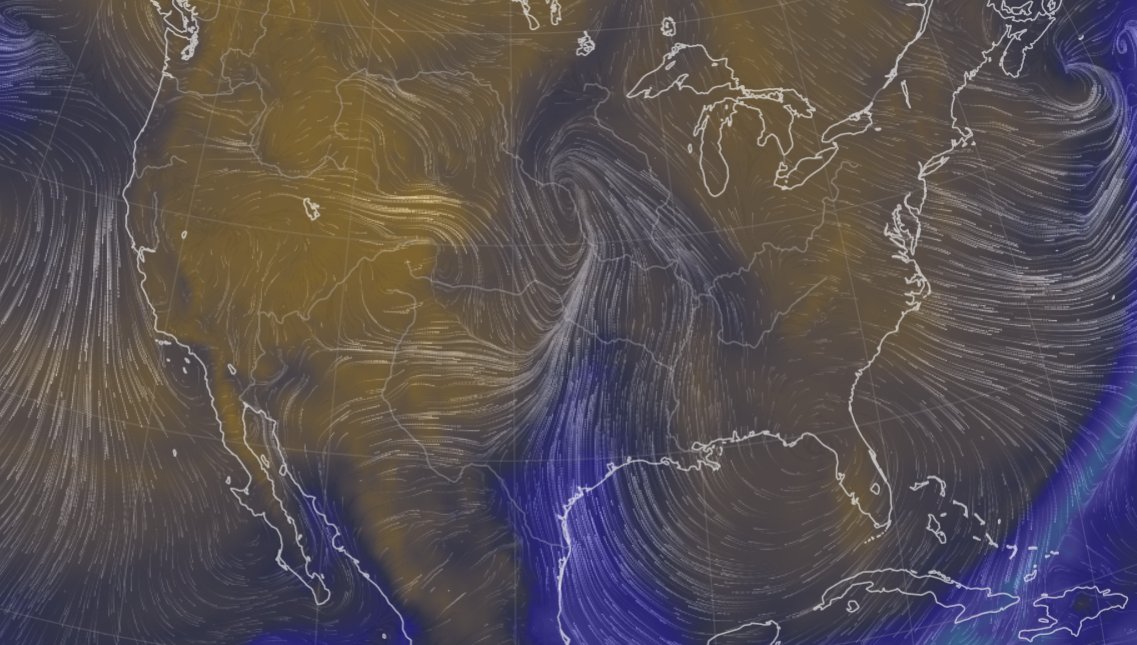
Sun Apr 10 15:21:45 +0000 2022As a student, I realized that a big part of the dissonance between how physicist view things & how biologists do is how they think of "time". To physicists, time is a dimension, every bit as physical as height or width. To biologists, it is the x-axis.
Sun Apr 10 13:53:40 +0000 2022Tripartite DENN domains are an aggregate of 3 adjacent domains: uDENN+cDENN+dDENN (upstream+core+downstream). DENN is a study-section/wishful-thinking acronym, derived from some version of the phrase "Differentially Expressed in Neoplastic versus Normal cells".
Sun Apr 10 13:41:13 +0000 2022While these sequences are paralogs, they only share 2 observable tryptic peptides.
Sun Apr 10 13:36:14 +0000 2022Modification diagrams for DENND4A:p & DENND4C:p, #DENN domain family paralogs. The N-terminal tripartite DENN domains―(192-641) & (191-641) respectively―have few PTMs, followed by rhyming mix of IHOP & RIP domains. There is little, if any, use of lysine PTMs. 🔗
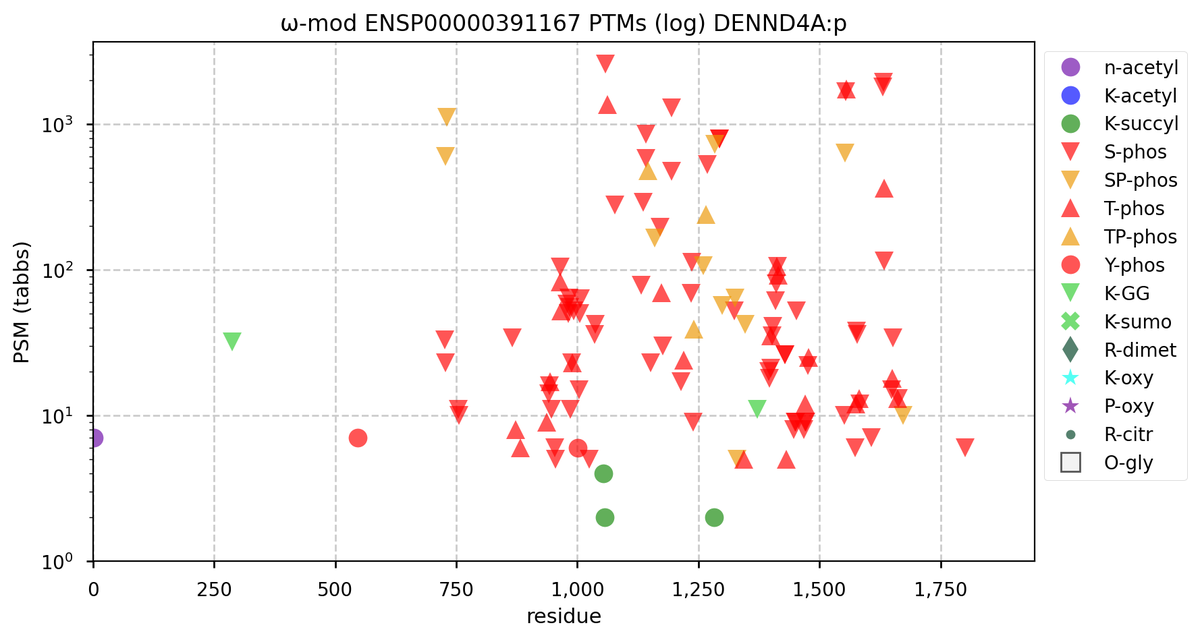
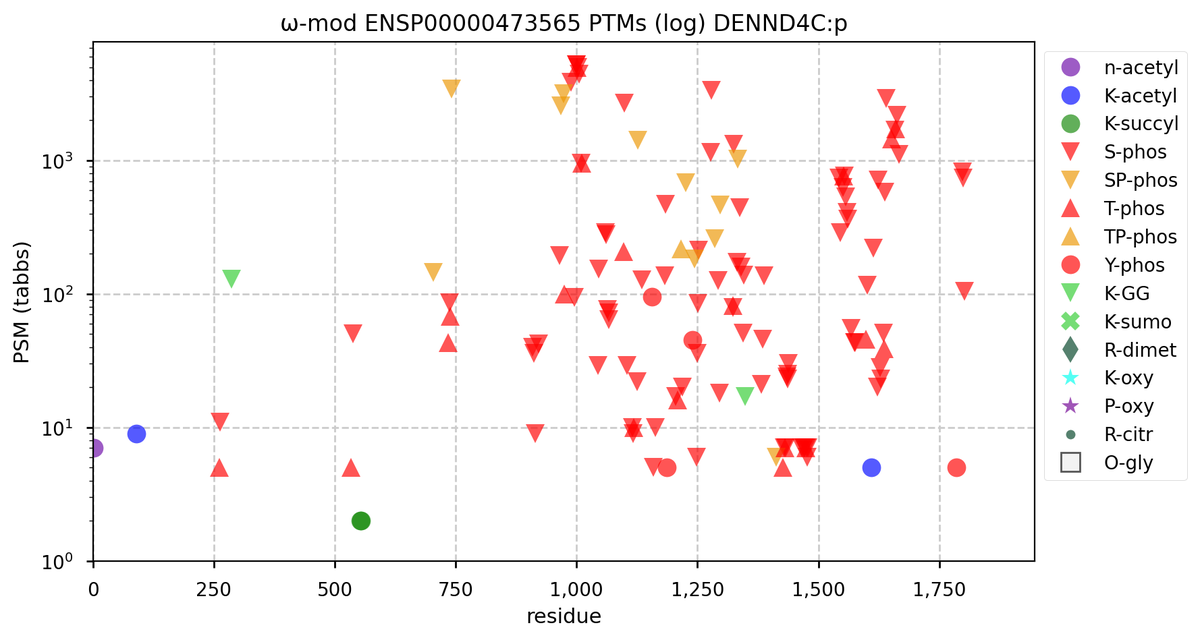
Sat Apr 09 14:11:27 +0000 2022@StatModeling Noise-mining is such a great term! Is there another source or was it coined here?
Sat Apr 09 14:00:37 +0000 2022P4HB:p has a number of short sequence overlaps with other proteins involved in redox chemistry, which may lead to some difficulty when using immuno-reagents to determine its presence. Its PTM pattern may also cause some confusion wrt immunostaining.
Sat Apr 09 13:12:45 +0000 2022The unusual PTM pattern may be part of the explanation for the multi-role functionality of this odd ball protein.
Sat Apr 09 13:09:48 +0000 2022This subunit has also been known as PDIA1, PROHB, DSI, GIT, PDI, PO4HB, P4Hbeta, PO4DB or ERBA2L. The literature suggests (rather confusingly) that it is used in multiple roles, particularly as a stand-alone disulphide isomerase.
Sat Apr 09 13:06:35 +0000 2022Modification diagram for #P4HB:p, the other subunit of P4H. The mature form (18-508) has an extraordinary pattern of high occupancy K+acetyl/succinyl/GGyl acceptors, as well as S/T/Y+phosphoryl. 🔗
Sat Apr 09 13:01:23 +0000 2022Modification diagrams for #P4HA1:p, a subunit of P4H (a tetramer of 2 A & 2 B chains). The mature form (18-534) is an ER glycoprotein with a not very unusual pattern of low occupancy K+acetyl/GGyl/SUMOyl co-acceptors. 🔗
Sat Apr 09 12:26:21 +0000 2022KRT1-201, keratin 1
🔗
2 – 644 → 2 – 644 (N-acetyl-Ser ο = 100%)
#realNT
Sat Apr 09 00:33:42 +0000 2022KRT36-201, keratin 36
🔗
1 – 467 → 2 – 467 (N-acetyl-Ala ο = 100%)
#realNT
Sat Apr 09 00:25:22 +0000 2022KRT8-201, keratin 8
🔗
1 – 483 → 2 – 483 (N-acetyl-Ser, ο = 100%)
#realNT
Fri Apr 08 18:09:40 +0000 2022XDH-201, xanthine dehydrogenase
🔗
1 – 1333
↓
1 – 1333 (N-acetyl-Met, ο = 100%) &
2 – 1333 (N-acetyl-Thr, ο = 30%)
#realNT
Fri Apr 08 17:56:15 +0000 2022One of these days I really should try to get interested―or at least learn how to feign interest―in a disease.
Fri Apr 08 15:56:00 +0000 2022KIAA1191-201, KIAA1191
🔗
1 – 305 → 2 – 305 (N-acetyl-Ala, ο = 100%)
#realNT
Fri Apr 08 15:18:47 +0000 2022KRT17-201, keratin 17
🔗
1 – 432 → 2 – 432 (N-acetyl-Thr, ο = 100%)
#realNT
Fri Apr 08 14:46:16 +0000 2022Modification diagrams for #EP300:p & #CREBBP:p, multifunctional proteins that have been accused of being #E4_ligases (among other things). They having rhyming patterns of PTM acceptor sites, suggesting a similar functional domain structure. 🔗
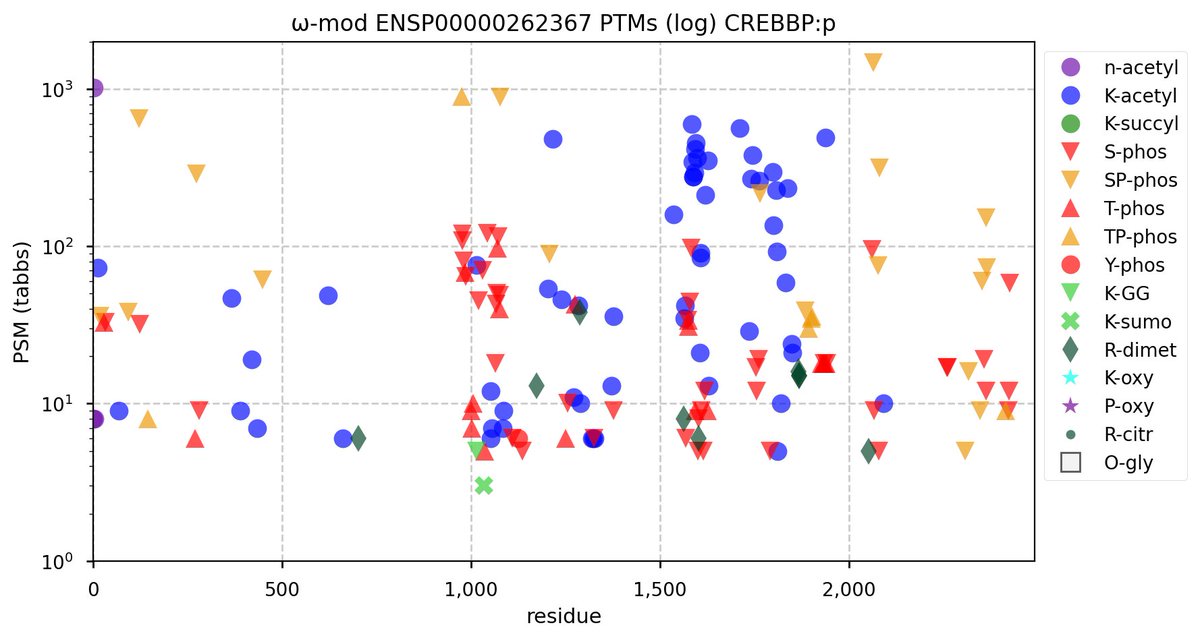
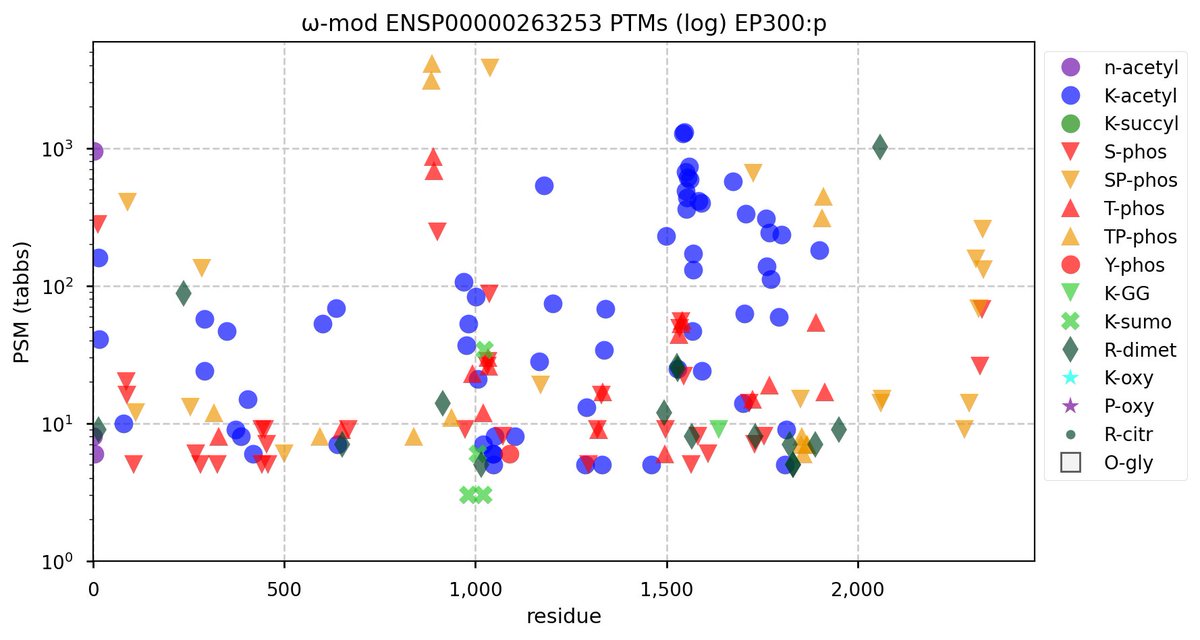
Fri Apr 08 14:17:46 +0000 2022KRT14-201, keratin 14
🔗
1 – 472 → 2 – 472 (N-acetyl-Thr, ο = 100%)
#realNT
Fri Apr 08 13:20:52 +0000 2022TRAF3IP2-208, TRAF3 interacting protein 2
🔗
1 – 574 → 1 – 574 (N-acetyl-Met, ο = 100%)
#realNT
Thu Apr 07 15:51:34 +0000 2022Modification diagram for #UBOX5:p, a potential #E3_ligase or #E4_ligase, with an internal UBOX domain (258-338) & a C-terminal RING Zn finger domain (480-541). Does not seem to actively participate in any of the PTM modalities listed.
🔗
Thu Apr 07 15:15:02 +0000 2022@neely615 @pwilmarth Exactly the purpose for which it was created. They are the proteins you can't wash off. Like the bad guy in a horror movie, they are already inside of the house!
Thu Apr 07 14:17:07 +0000 2022Would be totally out of line to suggest that the assembly/adhesion properties TNS1 are the consequence of its S/T+phosphoryl pattern? The Heterogenous, Programmable Adhesive Domain (H-PAD) hypothesis.
Wed Apr 06 20:46:56 +0000 2022I'm pretty sure an alanine scan wouldn't be very revealing.
Wed Apr 06 20:28:41 +0000 2022Maybe not the best example of the subtle, elegant signalling provided by site-specific phosphorylation.
🔗
Wed Apr 06 15:26:59 +0000 2022@neely615 @pwilmarth One thing that can surprise people is how different bovine plasma proteins are when compared to human. Bovine TF or APOB only share ~6% of tryptic sequence coverage with their human orthologs.
Wed Apr 06 15:07:01 +0000 2022Modification diagram for #STUB1:p, an #E3_ligase with a centrally located UBOX (226-300). It is unlike most E3's in that it has 3 high occupancy ΦP S+phosphoryl acceptors & 2 domains with K+GGyl/acetyl/SUMOyl alternate modification sites.
🔗
Wed Apr 06 14:49:36 +0000 2022@neely615 @pwilmarth 🔗
It is the list of bovine proteins commonly detected in data from HeLa cell lines that used FBS media.
Wed Apr 06 14:28:37 +0000 2022Does anyone know of a study that attempts to unwind the effects caused by of alternate initiation in large scale proteomics data interpretation?
Wed Apr 06 14:15:37 +0000 2022& also, the more frequently observed alternate-initiation-at-M39 version
40 – 189 (N-acetyl-Ala, ο = 100%)
#realNT
Wed Apr 06 14:12:35 +0000 2022FUNDC2-201, FUN14 domain containing 2
🔗
1 – 189 → 1 – 189 (N-acetyl-Met, ο = 90%)
#realNT
Wed Apr 06 12:07:39 +0000 2022MPRIP-202, myosin phosphatase Rho interacting protein
🔗
2 – 1025 → 2 – 1025 (N-acetyl-Ser, ο = 100%)
#realNT
Wed Apr 06 11:35:06 +0000 2022This morning, another Spring frontal storm, this time spinning off a low pressure system currently centred in Minnesota. It is describing an arc down from Thunder Bay, over to Chicago & all the way to Memphis. 🔗
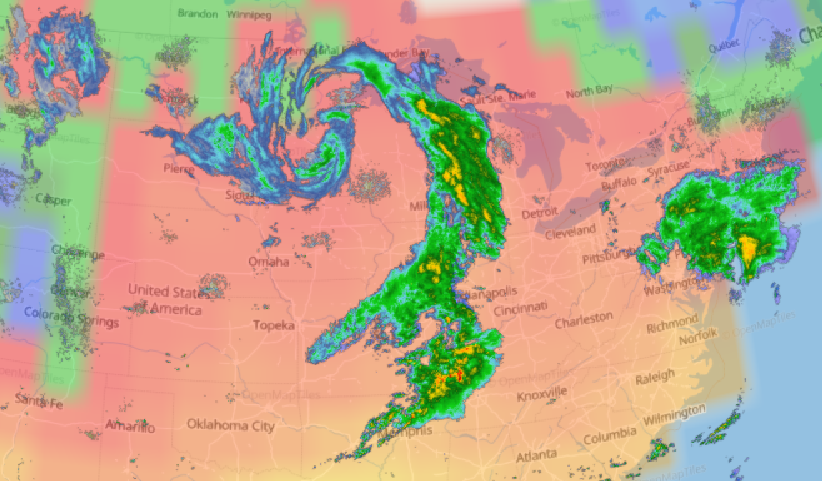
Wed Apr 06 01:26:05 +0000 2022The most intense line of rain has moved off-shore in the south, having passed through Charleston. It is still quite active in the north, just east of Rayleigh NC. 🔗

Tue Apr 05 20:43:09 +0000 2022LACTB2-201, lactamase beta 2
🔗
1 – 288 → 2 – 288 (N-acetyl-Ala, ο = 100%)
#realNT
Tue Apr 05 19:35:00 +0000 2022The storm is moving through eastern Georgia & the Florida Panhandle on its way to the Carolinas. 🔗

Tue Apr 05 16:39:28 +0000 2022SLC25A18-201, solute carrier family 25 member 18
🔗
1 – 315 → 2 – 315 (N-acetyl-Thr, ο = 93%)
#realNT
Tue Apr 05 15:43:08 +0000 2022And if you are at all interested in how mammalian UBOX proteins came to be, this guy has an informed take on explaining their origins
🔗
Tue Apr 05 15:38:36 +0000 2022For the interested few, RIPD is a "Really Interesting Phospho-Domain". Feel free to use it (no applicable © ® ™ ℠).
Tue Apr 05 14:59:19 +0000 2022Even though UBE3A & B are paralogs, they do not share any observable tryptic peptides. They are very similar to each other in the number of times they have been observed and the total number of assigned PSMs.
Tue Apr 05 14:57:06 +0000 2022Modification diagram for #UBE4B:p, an #E4_ligase with a C-terminal UBOX domain (1227-1300). It's PTM pattern is generally like its paralog #UBE4A:p, with the addition of an N-terminal RIPD (1-240).
🔗
Tue Apr 05 13:56:45 +0000 2022Thanks to everyone who participated. Apparently, consensus is that it is the collective fault of the students that they haven't been properly supervised.🧐
Tue Apr 05 13:06:19 +0000 2022The annotated version isn't wrong, but it is the rarest of the 4 possible states.
Tue Apr 05 13:04:23 +0000 2022SFXN3-201, sideroflexin 3
🔗
1 – 321 (N-acetyl-Met)
↓
1 – 321 (N-acetyl-Met, ο = 1%) &
2 – 321 (N-acetyl-Gly, ο = 75%)
M1:G2 = 4:1
#realNT
Tue Apr 05 12:16:57 +0000 2022A patch of angry-looking weather heading east from Mississippi, that looks bound for North & South Carolina. It will be making things a bit spicy through Alabama, Georgia & Tennessee for the next few hours. 🔗

Tue Apr 05 11:56:24 +0000 2022ACO1-201, aconitase 1
🔗
1 – 889 → 2 – 889 (N-acetyl-Ser, ο = 100%)
#realNT
Mon Apr 04 19:01:31 +0000 2022@J_my_sci If you using a standing desk, wearing the right shoes make a big difference.
Mon Apr 04 16:14:19 +0000 2022CDC34-201, cell division cycle 34
🔗
1 – 236 → 2 – 236 (N-acetyl-Ala, ο = 100%)
#realNT
Mon Apr 04 14:49:24 +0000 2022Modification diagram for #UBE4A:p, an #E4_ligase with a C-terminal UBOX domain (995-1066). This protein is 1 of 2 human orthologs of yeast #UFD2:p, with a similar pattern of PTMs except for a rather diffuse N-terminal S+phosphoryl domain (2-102).
🔗
Mon Apr 04 13:24:22 +0000 2022What is the likely cause of this "feature":
Mon Apr 04 13:16:13 +0000 2022I look at quite a bit of academic R code associated with data from papers. One oddity is that very little effort seems to be put in to error/exception checking: most of it seems to be full-steam-ahead-damn-the-torpedoes/get-to-the-graphics-quick.
Mon Apr 04 11:53:21 +0000 2022PTBP1-202, polypyrimidine tract binding protein 1
🔗
none → 2 – 527 (N-acetyl-Ser, ο = 85%)
#realNT
Mon Apr 04 00:51:43 +0000 2022🔗
1 – 511 → 2 – 511 (N-acetyl-Ser, ο = 100%)
#realNT
Mon Apr 04 00:19:42 +0000 2022🔗
1 – 120 → 2 – 120 (N-acetyl-Ser, ο = 99%)
#realNT
Sun Apr 03 22:15:16 +0000 2022🔗
1 – 1066 → 2 – 1066 (N-acetyl-Thr, ο = 95%)
#realNT
Sun Apr 03 21:44:00 +0000 2022🔗
1 – 385 → 2 – 385 (N-acetyl-Ser, ο = 75%)
#realNT
Sun Apr 03 19:31:01 +0000 2022🔗
1 – 375 → 2 – 375 (N-acetyl-Ser, ο = 100%)
#realNT
Sun Apr 03 15:23:14 +0000 2022I am regularly reminded that elegantly written, high profile papers are often built on top of a large mass of really messy, 2nd (or 3rd) tier experimental data.
Sun Apr 03 13:18:51 +0000 2022The UBOX domain is generally considered to be a variant of the RING domain motif (believe it or not "RING" stands for "Really Interesting New Gene"). RING domains are found in hundreds of E3 ligases.
Sun Apr 03 13:16:16 +0000 2022It is involved in creating the branched ubiquitination that improves the efficiency of protein degradation. The UBOX domain is named after the "U" in UFD2 (at the time it was trendy to call a sequence motif a "box")
Sun Apr 03 13:11:25 +0000 2022Modification diagram for S. cerevisiae #UFD2, the first #E4_ligase described. It has a C-terminal UBOX domain (881-953). It has a scattering of K+GGyl acceptors & 2 low occupancy K+acetyl acceptors.
🔗
Sat Apr 02 21:57:16 +0000 2022Looking at some data from a new R-mono/di-methyl enrichment expt is a nice reminder just how reproducible proteomics has become. ~1000 acceptor sites & none of them new!
Sat Apr 02 16:01:38 +0000 2022🔗
1 – 159 → 2 – 159 (N-acetyl-Ser, ο = 95%)
#realNT
Sat Apr 02 15:11:25 +0000 2022I think at least one way to think about the whole phosphorylation-in-IDRs issue is that few, if any, kinases can phosphorylate residues in highly organized secondary structures, e.g., alpha helices or beta sheets.
Sat Apr 02 14:34:54 +0000 2022PTM diagram for #WWP1, an #E3_ligase with a C-terminal HECT domain (586-922). The protein's other domains are a C2 lipid binding domain (19-113), WW protein-protein interaction domains (342-528) & S+phosphoryl acceptors at inter-domain boundaries. 🔗
Sat Apr 02 13:26:28 +0000 2022The correct order is:
1. JSON;
2. XML;
3. Any other format; &
4. CSV.
Sat Apr 02 13:15:06 +0000 2022Thanks to everyone who voted or commented. CSV was the clear preference (although JSON was the correct answer).
Sat Apr 02 12:20:12 +0000 2022I don't know how they are calling N-termini (the site suggests manual interpretation for this one), but it doesn't seem to work very well for A. thaliana chloroplast proteins.
Sat Apr 02 12:16:34 +0000 2022🔗
1 – 43 → 1 – 45 (chloroplast signal peptide)
44 – 257 → 46 – 257 (mature protein)
#realNT
Sat Apr 02 00:13:52 +0000 2022If it is of any help, unlike ubiquitin, ISG15 has low levels of GGylation itself so it probably isn't forming ubiquitin-like branching structures.
Fri Apr 01 21:08:54 +0000 2022It would also be great if someone on the lab side came up with a way to easily distinguish ISG15 modified proteins from ubiquitin modified proteins. There are a lot of GGylation sites on viral (& other) proteins that may be due to ISGylation, but there is no way to tell.
Fri Apr 01 20:48:51 +0000 2022@michaelhoffman @CSRpeerreview Leading to the #1 problem with grant reviews:
1. Lazy, vague general statements that cannot be addressed without considerable guesswork.
Fri Apr 01 15:10:54 +0000 2022I would expect at some point this family will be broken up into at least 2 separate groups.
Fri Apr 01 15:09:27 +0000 2022HERC3-6 are much smaller (1000 aa), with few modifications other than K+GGyl. 🔗
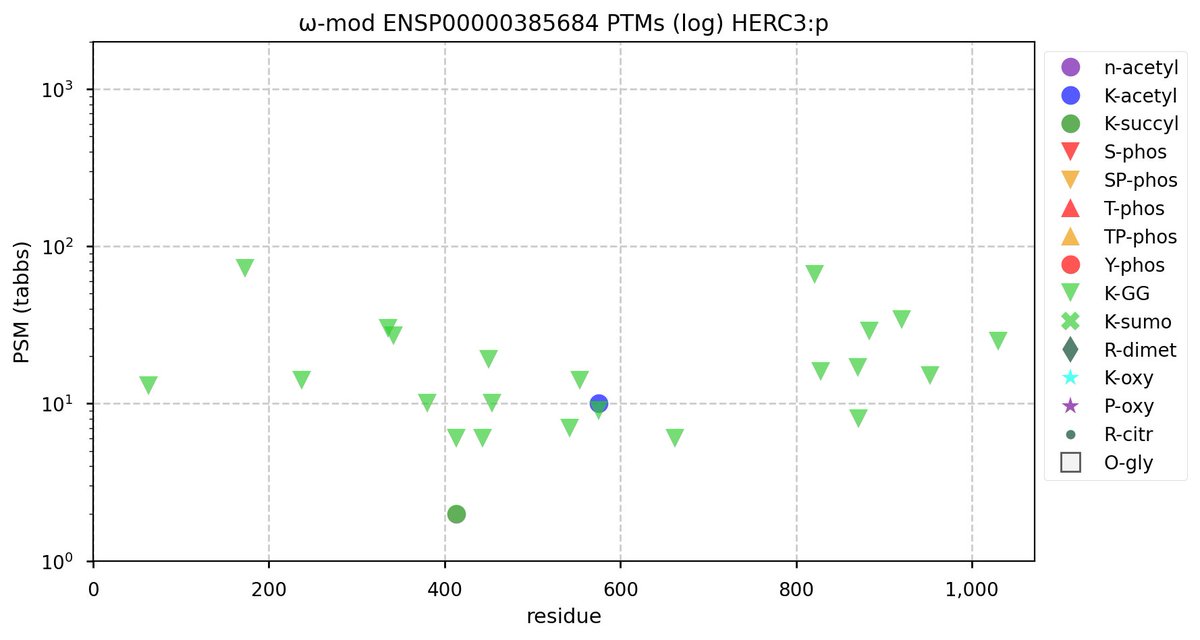
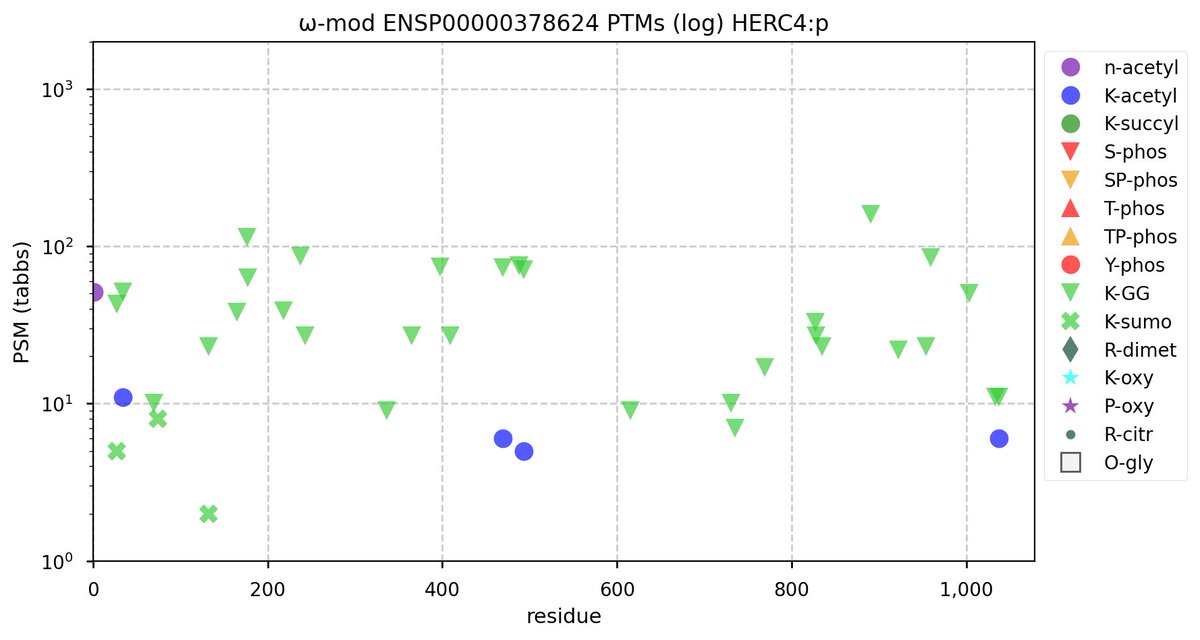
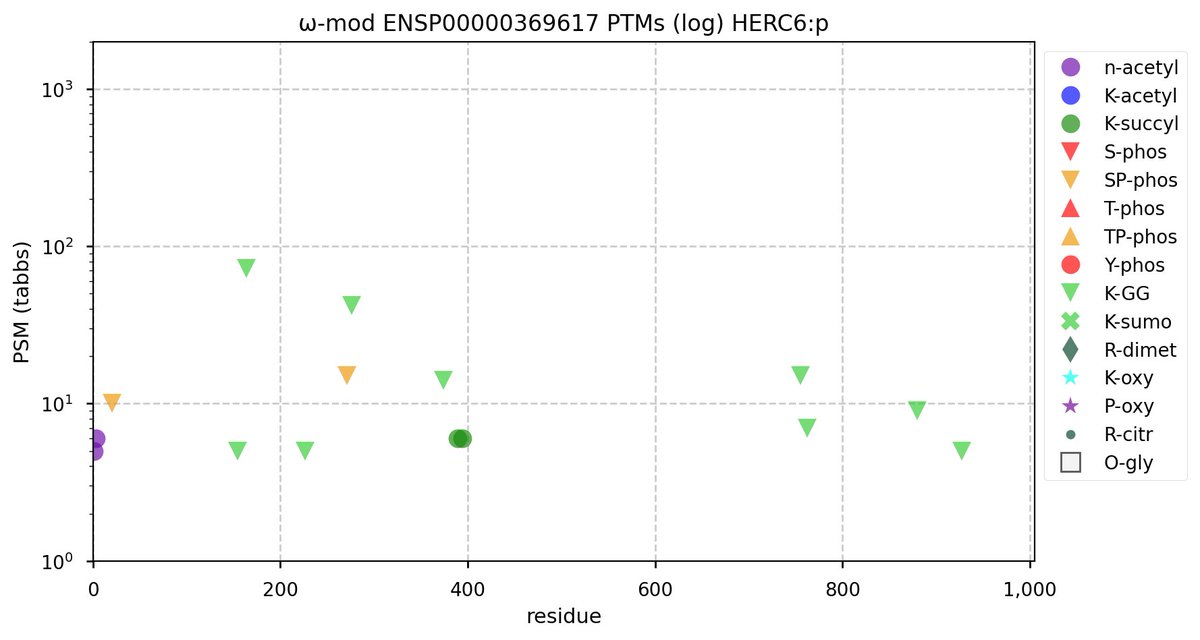
Fri Apr 01 15:07:02 +0000 2022The HERC family of #E3_ligase protein fall into 2 categories. #HERC1:p and #HERC2:p are large (4800 aa) & highly modified. 🔗
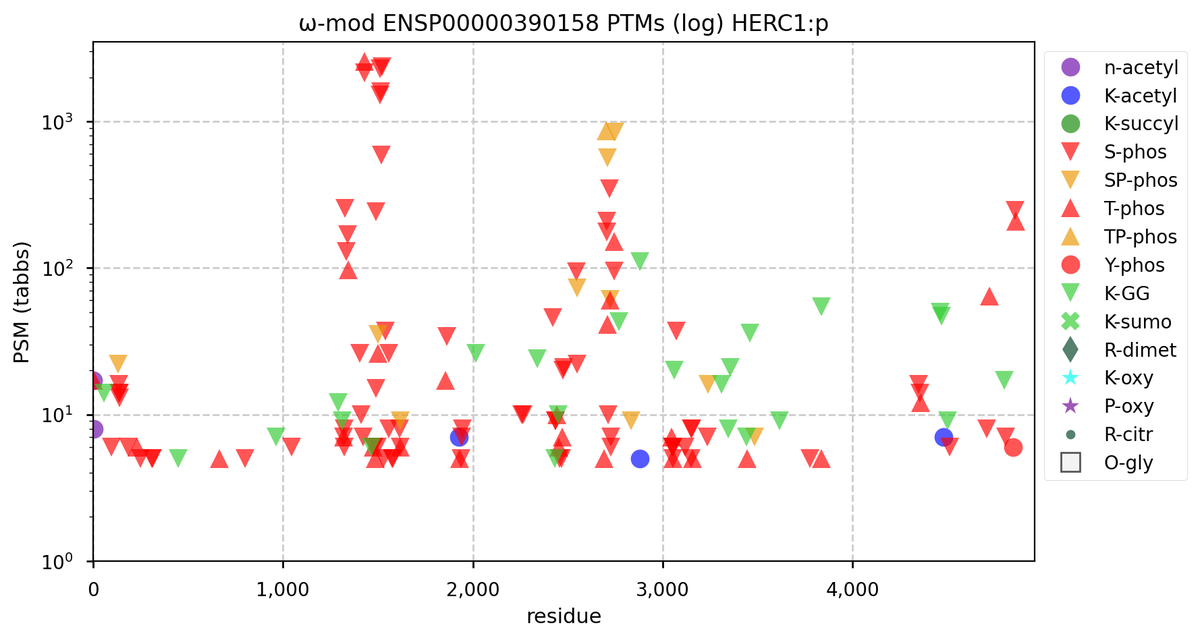
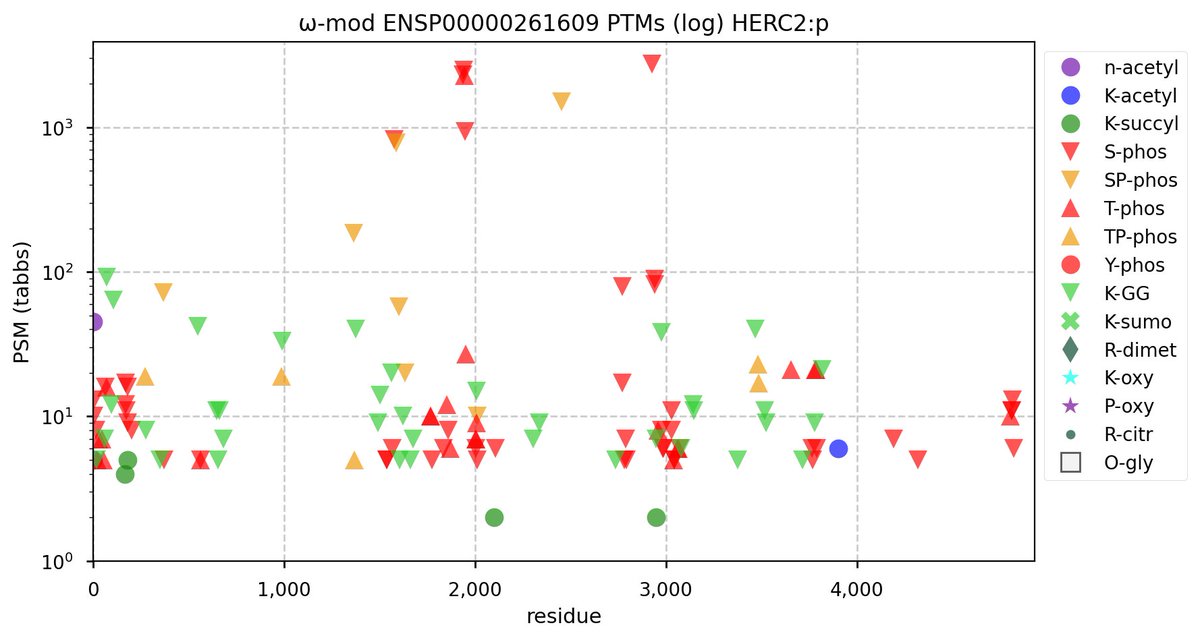
Fri Apr 01 15:03:44 +0000 2022Modification diagram for #HERC5, an #E3_ligase with a C-terminal HECT domain (700-1024) & multiple RCC1 domains (70-398). It has 13 observed K+GGyl acceptors. HERC5 does not facilitate the transfer of ubiquitin: instead it transfers the ubiquitin-like protein #ISG15:p. 🔗
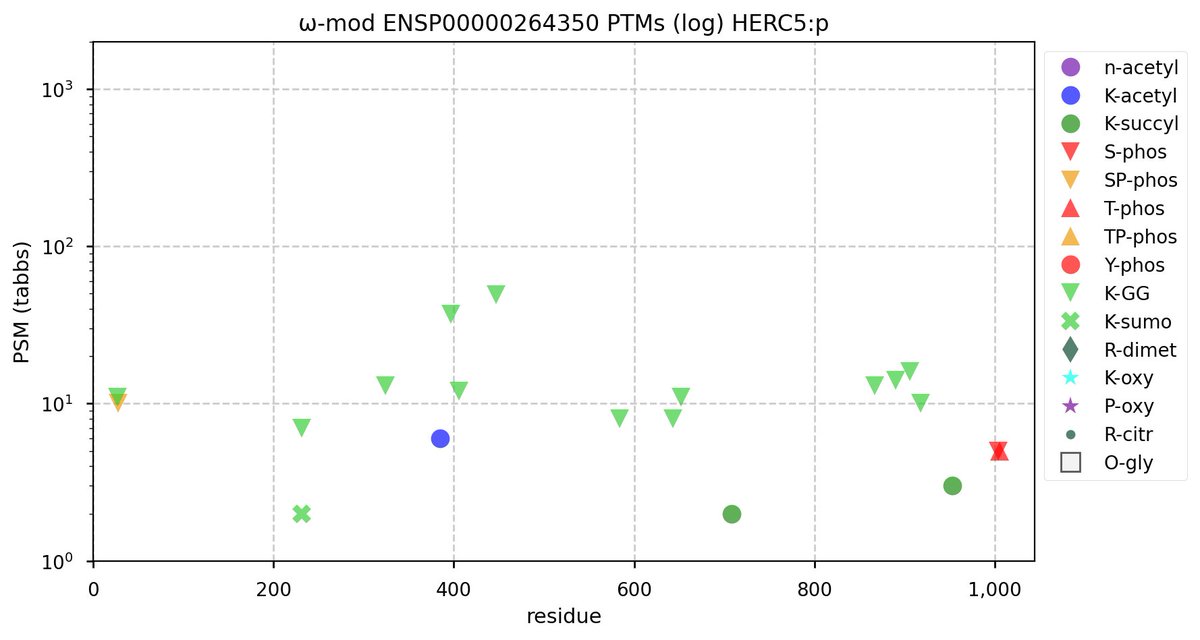
Fri Apr 01 14:27:41 +0000 2022@neely615 I forgot to include "bitmap graphics + OCR" as an option.
Fri Apr 01 13:11:09 +0000 2022A proteomics information resource offers a lot of information in multiple electronic formats. Which format would you prefer to use for non-tabular information:
Fri Apr 01 12:48:37 +0000 2022Thanks to everyone who participated in this poll. REST seems to be the preferred option, with GraphQL aspiring to take over.
Thu Mar 31 20:47:00 +0000 2022Based on this poll so far, the once highly-hyped SOAP appears destined to join XSLT in Bioinformatics Valhalla. 🔗
Thu Mar 31 18:33:59 +0000 2022@leprevostfv Realpolitik
Thu Mar 31 17:01:25 +0000 2022🔗
1 – 33 → 1 – 57 (chloroplast signal peptide)
34 – 387 → 58 – 387 (mature protein)
#realNT
Thu Mar 31 16:02:04 +0000 2022When someone provides an "Advanced Search" option―decked out with lots of drop-downs, text boxes and BOOLEAN operations―I always take a look at it, but I can't think of any that I use on a regular basis.
Thu Mar 31 15:13:57 +0000 2022Modification diagram for #SMURF1:p, an #E3_ligase with a C-terminal HECT domain (449-756). It has a domain structure similar to #ITCH:p, but it lacks the prominent K+GGyl acceptor activity in the parts of its sequence corresponding to ITCH's C2, WW & HECT domains. 🔗
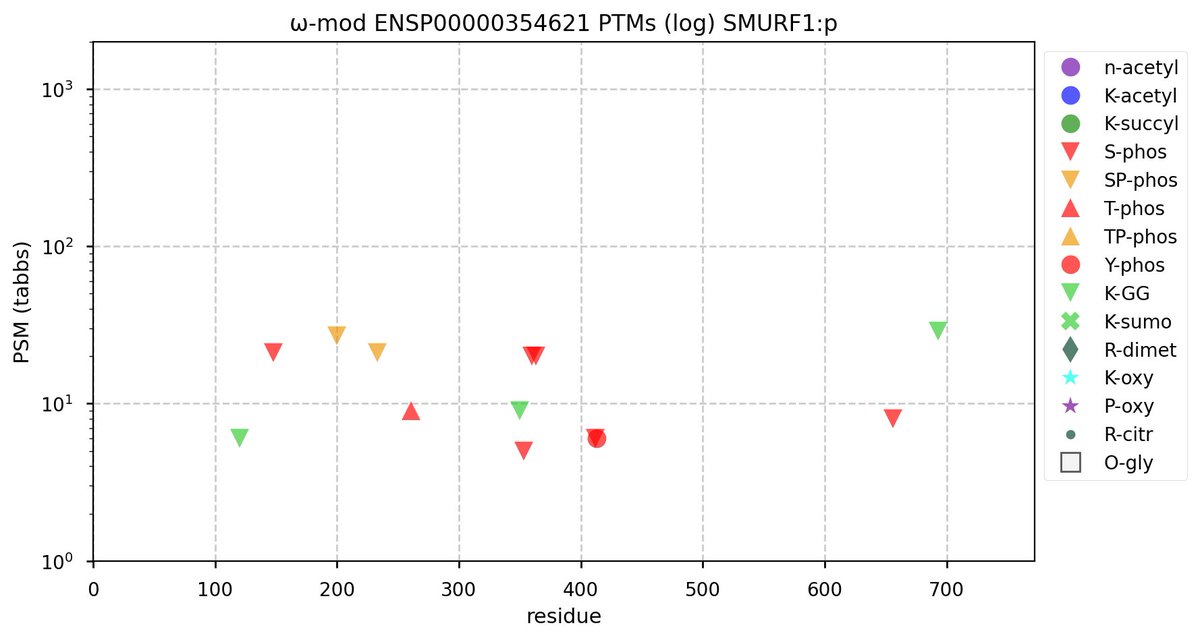
Thu Mar 31 13:34:39 +0000 2022It has joined other, once hot, up-and-coming ideas, like CORBA & "Objects in Biology", in the Valhalla of Bioinformatics (which I believe is located in the basement of the EMBL-EBI South Building in Cambridge).
Thu Mar 31 12:45:06 +0000 2022If you were creating an interface to provide information from a new proteomics resource, which of these standard styles of interface would you prefer to use
Thu Mar 31 12:36:11 +0000 2022Thanks to the brave few that answered. It would appear that XSLT has gone on to its reward―in this case being fully implemented in all web browsers but unused.
Wed Mar 30 20:35:08 +0000 2022XSLT would appear to have fallen from grace (unless someone steps up to bolster its stock). 🔗
Wed Mar 30 19:07:32 +0000 2022A recurring problem I have when reading articles is that it often requires considerable effort to translate the gene/protein names they discuss into those currently used to annotate the genome/proteome. And even then it can be difficult to be sure the translation was correct.
Wed Mar 30 16:16:27 +0000 2022🔗
1 – ? → 1 – 36 (chloroplast signal peptide)
? – 394 → 37 – 394 (mature protein)
#realNT
Wed Mar 30 14:48:03 +0000 2022The WW domain repeats (325-510) have spaced, low occupancy Y+phosphoryl acceptors. The C2 domain (19-114), the WW repeats and the HECT domain all have K+GGyl acceptors, some of which alternate as K+acetyl sites.
Wed Mar 30 14:47:02 +0000 2022Modification diagram for #ITCH, an #E3_ligase with a C-terminal HECT domain (599-901). It is a modular protein with 5 distinct domains, 2 of which are serine phosphodomains, (2-10) & (202-242). 🔗
Wed Mar 30 14:31:06 +0000 2022🔗
1 – 50 → 1 – 51 (chloroplast signal peptide)
51 – 253 → 52 – 253 (mature protein)
#realNT
Wed Mar 30 12:30:12 +0000 2022Back when XML was the newest, bestest thing, a mechanism for converting XML into HTML called XSLT was widely advertised as the future of the web. Does anybody still use XSLT for developing anything new?
Wed Mar 30 02:25:49 +0000 2022🔗
1 – 36 → 1 – 37 (chloroplast signal peptide)
37 – 409 → 38 – 409 (mature protein)
#realNT
Tue Mar 29 23:01:30 +0000 2022The I-29 gradient is only 23°C today: they will be in shorts today in Fargo at +6°C! 🔗
Tue Mar 29 21:06:21 +0000 2022@PastelBio @ProteomicsNews Good one!
Tue Mar 29 17:42:57 +0000 2022Just to rub it in a bit, the AF error diagram with phosphorylation overlay for #AREL1:p 🔗
Tue Mar 29 16:28:42 +0000 2022🔗
1 – 33 → 1 – 57 (chloroplast signal peptide)
34 – 387 → 58 – 387 (mature protein)
#realNT
Tue Mar 29 15:54:40 +0000 2022@camtflower @chrashwood Also, while certainly a quibble about wording, this sort of thing isn't a "promise": it is a "current deliverable".
Tue Mar 29 15:10:48 +0000 2022@chrashwood @camtflower For me, it is a nice way to think about why phosphorylation acceptor distributions often look more expressionist than pointillist.
Tue Mar 29 14:56:42 +0000 2022@chrashwood @camtflower S+phosphoryl only in this case (LRRK2:p).
Tue Mar 29 14:22:12 +0000 2022It also does not participate in the common K-acceptor modification modalities: acetyl, succinyl, GGyl (maybe a bit) or SUMOyl
Tue Mar 29 14:20:04 +0000 2022Modification diagram of #AREL1:p, an #E3_ligase with a C-terminal HECT domain. Like #HACE1:p, it has a phosphodomain (327-368) about 150 residues prior to the HECT domain (513-822).
🔗
Tue Mar 29 14:00:30 +0000 2022Phospho-Proteomics Shines a Bright Light on the Dark Proteome: How Proteomics Predicts Where 3D Structural Techniques Will Fail
😱 🔗
Tue Mar 29 00:33:50 +0000 2022@UCDProteomics @pwilmarth Old data always looks quaint.
Mon Mar 28 19:02:25 +0000 2022🔗
1 – 563 → 2 – 563 (N-acetyl-Ala, ο = 7%)
#realNT
Mon Mar 28 15:08:24 +0000 2022@J_my_sci Congrats on the new job. I have always thought that the ease of mobility is one of the best parts of the US academic system.
Mon Mar 28 14:19:09 +0000 2022It is up to the current generation of researchers to agree on some ideas that involve how to think about the information generated by proteomics collectively, rather than the one-off findings of single studies that has obsessed the previous generation.
Mon Mar 28 14:07:13 +0000 2022🔗
1 – 620 → 2 – 620 (N-acetyl-Val, ο = 1%)
#realNT
Mon Mar 28 13:38:36 +0000 2022The I-29 gradient: hang in there Fargo, tomorrow you should be above zero! 🔗
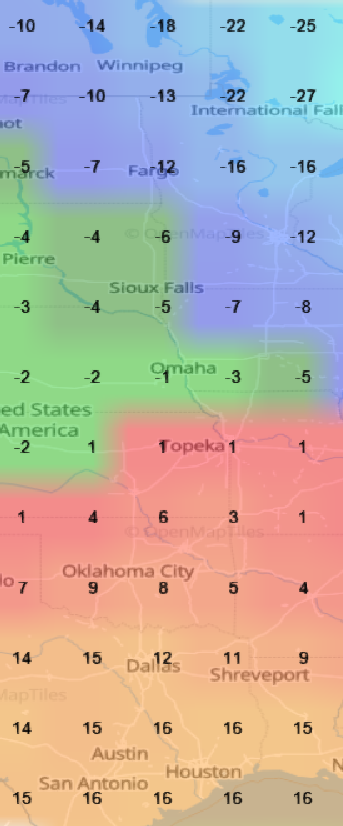
Mon Mar 28 13:03:34 +0000 2022Modification diagram for #HACE1:p, an #E3_ligase with a C-terminal HECT domain. Unlike UBE3A:p, it does not have K+GGyl acceptors. It has an N-terminal ankyrin repeat (28-260) domain, a phospho-domain (337-416) & the HECT (574-909) domain.
🔗
Mon Mar 28 11:37:59 +0000 2022🔗
1 – 731 → 1 – 731 (N-acetyl-Met, ο = 100%)
#realNT
Mon Mar 28 02:41:58 +0000 2022🔗
1 – 498 → 2 – 498 (N-acetyl-Arg, ο = 82%)
#realNT
Mon Mar 28 01:35:40 +0000 2022🔗
none → 2 – 265 (N-acetyl-Ala, ο = 100%)
#realNT
Sun Mar 27 18:24:06 +0000 2022Except, of course, for an odd fixation on the use of capital letters in the site names.
Sun Mar 27 18:10:43 +0000 2022I spent some time this morning browsing around entries on other proteomics data sites (MaxQB, ProteomicsDB, MassIVE). As a field, we really don't seem to be evolving towards a common representation of information (or anything else for that matter).
Sun Mar 27 17:49:53 +0000 2022UBE3A is rather aggressively targeted by other ubiquitin ligases, with at least 35 experimentally observed K+GGyl acceptors.
Sun Mar 27 17:17:27 +0000 2022@LindorffLarsen No strong opinions, but pretty much every journal will add the hyphen for you in proof.
Sun Mar 27 17:00:01 +0000 2022🔗
1 – 351 → 1 – 351 (N-acetyl-Met, ο = 100%)
#realNT
Sun Mar 27 16:50:49 +0000 2022🔗
1 – 313 → 2 – 313 (N-acetyl-Thr, ο = 100%)
#realNT
Sun Mar 27 14:34:13 +0000 2022🔗
Sun Mar 27 14:00:09 +0000 2022HECT stands for "Homologous to the E6-ap Carboxyl Terminus", in which E6-AP stands for "human papillomavirus E6-Associated Protein". E6-AP is the (very) old name for #UBE3A that has been immortalized in this particularly nostalgic form of domain nomenclature.
Sun Mar 27 13:56:36 +0000 2022Modification diagram for #UBE3A, an #E3_ligase with a C-terminal HECT domain (524-852). It has three S+phosphoryl acceptor domains & one low occupancy Y+phosphoryl acceptor (Y636) in the middle of the HECT domain.
🔗
Sun Mar 27 12:33:57 +0000 2022If you are at the southern tip of New Zealand or Tasmania, you might be able to see the aurora tonight. 🔗
Sun Mar 27 12:27:36 +0000 2022Winter makes a comeback at the top of I-29. -16°C in Fargo this morning. 🔗
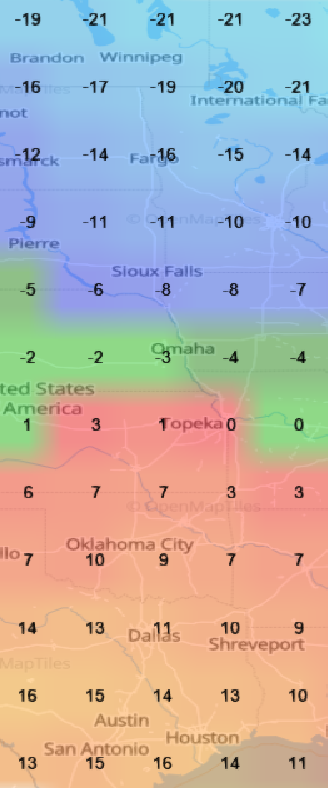
Sun Mar 27 02:03:45 +0000 2022I'm only a few experiments in, but PXD032710 is looking like pretty high quality data from an interesting set of tissue samples.
Sat Mar 26 21:22:10 +0000 2022@neely615 @J_my_sci @drstarbird @jbiolchem There are inexpensive bot options for pretty much everything associated with social media. Followers, votes, likes: you name it & you can pay someone a few dollars to get lots of it for you.
Sat Mar 26 20:44:09 +0000 2022🔗
1 – 76 → 2 – 76 (N-acetyl-Ala, ο = 100%)
#realNT
Sat Mar 26 20:11:39 +0000 2022🔗
1 – 351 → 2 – 351 (N-acetyl-Ala, ο = 100%)
#realNT
Sat Mar 26 18:24:57 +0000 2022@TanentzapfLab Most biologists in positions of academic influence are loath to have things (like ergodicity) explained to them by physicists.
Sat Mar 26 16:23:28 +0000 2022🔗
1 – 576 → 2 – 576 (N-acetyl-Ser, ο = 100%)
#realNT
Sat Mar 26 16:15:57 +0000 2022🔗
1 – 284 → 2 – 284 (N-acetyl-Thr, ο = 100%)
#realNT
Sat Mar 26 16:11:15 +0000 2022🔗
1 – 519 → 2 – 519 (N-acetyl-Thr, ο > 99%)
#realNT
Sat Mar 26 15:21:59 +0000 2022@slavov_n The "post hoc ergo propter hoc" method of QA/QC.
Sat Mar 26 14:30:40 +0000 2022I'm going to start using the lower case (minuscule) form of the Greek letter omicron "ο" to represent PTM acceptor occupancy.
Sat Mar 26 14:23:48 +0000 2022🔗
1 – 329 → 2 – 329 (N-acetyl-Thr, ο = 92%)
#realNT
Sat Mar 26 14:05:47 +0000 2022Modification diagram for #RFFL:p, an #E3_ligase with a C-terminal RING Zn finger domain (316-350). It has 2 distinct phosphodomains, 2 low occupancy K+GGyl acceptors & no K+acetyl acceptors. The evidence for the existence of an N-terminal FYVE/PhD Zn finger domain is modest.
Sat Mar 26 13:20:21 +0000 2022The whole messy business of gene/protein naming can only be understood when thought of as a consequence of a crippling reliance on nostalgia as a scientific principle.
Sat Mar 26 13:14:35 +0000 2022Grafton Tanner came up with the term "babbling corpse" to describe the destructive effects of nostalgia, but it is so evocative of my understanding of the scientific literature that I can't keep from calling it to mind when reading papers.
Fri Mar 25 15:19:50 +0000 2022PS - I understand that I am probably the only person who thinks that characterizing groups of subunits by their PTM acceptor patterns is good idea, but it "is my theory, it is mine, and belongs to me and I own it, and what it is too" - Anne Elk
Fri Mar 25 15:08:42 +0000 2022Is there an accepted symbol for PTM acceptor occupancy? "Occupancy" is rather long to write down all the time & "occ." seems sort of weak.
Fri Mar 25 15:06:33 +0000 2022🔗
1 – 468 ☞ 2 – 468 (N-acetyl-Ala, occ. 100%)
#realNT
Fri Mar 25 15:03:50 +0000 2022Modification diagram for #TRIM11, an #E3_ligase with a RING Zn finger domain (16-57). It belongs to the clade of E3 ligases characterized by a PTM acceptor pattern featuring heterogeneous K+GGyl, absence of K+acetyl & prominent ΦP S+phosphoryl.
🔗
Thu Mar 24 14:57:03 +0000 2022Like most of the E3 ligases, the subunit doesn't have any K+acetyl receptors. It also doesn't seem to partake of ubiquitination itself, with only 1 (just barely above threshold) K+GGyl acceptor.
Thu Mar 24 14:53:42 +0000 2022Modification diagram from #UNK, a RING Zn finger #E3_ligase, with 5 Zn fingers in a row (34-320). The mature subunit (1,2-810, M1,S2+acetyl) has several narrow phosphodomains, with some prominent ΦP sites. 🔗
Thu Mar 24 13:59:25 +0000 2022It passes the initial QA round, so into production it goes! 🧐
Thu Mar 24 13:46:15 +0000 2022Downloaded a TB dataset overnight: now to check whether it was worth the bandwidth.😎
Thu Mar 24 12:17:37 +0000 2022🔗
1 – 810 ☞ 2 – 810 (N-acetyl-Ala, occ. 83/83)
#realNT
Wed Mar 23 16:31:54 +0000 2022As with everything else in the GPM/GPMDB system, the displayed information & graphics have no copyright associated with them, they can be used without permission (although attribution is always nice).
Wed Mar 23 16:25:27 +0000 2022The display-creating software will be made available through 🔗
Wed Mar 23 15:22:34 +0000 2022After looking at this for a bit, I may want to revise upwards my guesstimate at the number of phosphodomains to ">5".
Wed Mar 23 15:15:27 +0000 2022As is typical of E3 ligases, there is no co-modification of GGyl acceptors with acetylation. The phosphorylation acceptors follow the usual pattern of sticking to the dark proteome, with the exceptions of S40, S261 & S406 that are in the BIR1, BIR3 & UBA domains. 🔗
Wed Mar 23 15:06:31 +0000 2022Modification diagram for #XIAP, an #E3_ligase that is the human proteome's most effective inhibitor of apoptosis. The mature sequence (2-497, T2+acetyl) has a RING Zn finger domain (450-484); 5 distinct phosphodomains & 16 K+GGyl acceptors (4 double as SUMOyl acceptors). 🔗
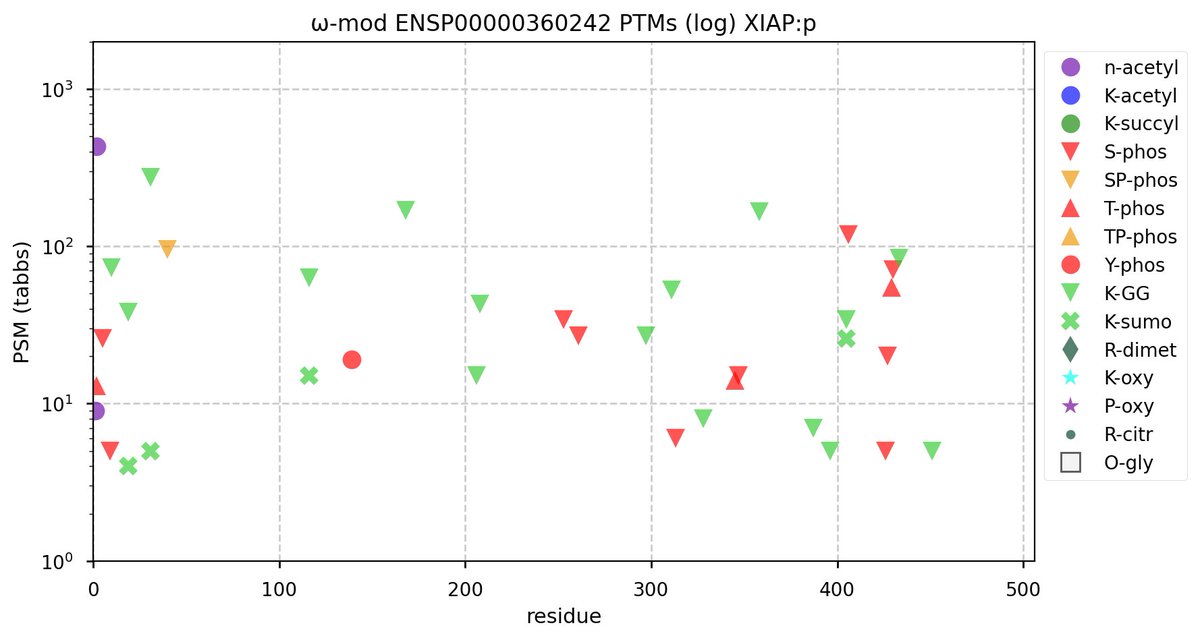
Wed Mar 23 13:35:20 +0000 2022Most of the more computationally intensive displays have been moved from the main GPMDB servers (AMD processors) to a new system based on energy efficient ARM 64-bit processor SoCs (Broadcom BCM2711) under the banner Intrinsic Disorder.
Wed Mar 23 12:38:17 +0000 2022This system dropped a lot of rain (100 mm, 4 in) across areas adjacent to the Mississippi Delta in Arkansas, Louisiana & Mississippi.
Wed Mar 23 12:21:37 +0000 2022The frontal system that caused a tornado in New Orleans overnight has moved through Mississippi & Alabama. It is currently affecting the Florida Panhandle and Georgia (NEXRAD & ground station temps). 🔗
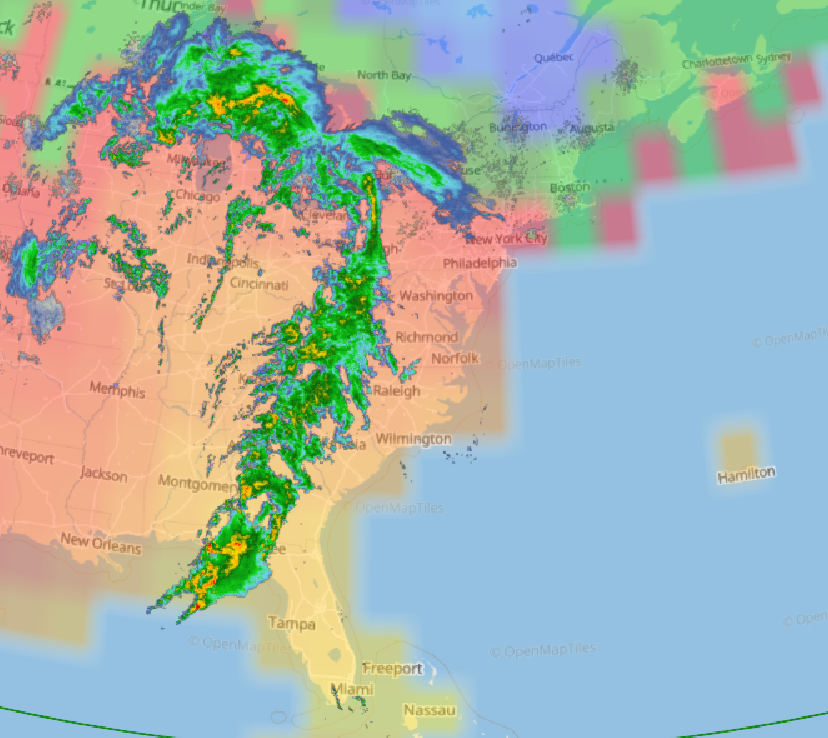
Wed Mar 23 12:13:46 +0000 2022The polar vortex (a high atmosphere low pressure system) has been displaced towards Central Asia by a high pressure vortex currently over Alaska (winds at 10 hPa shown) 🔗
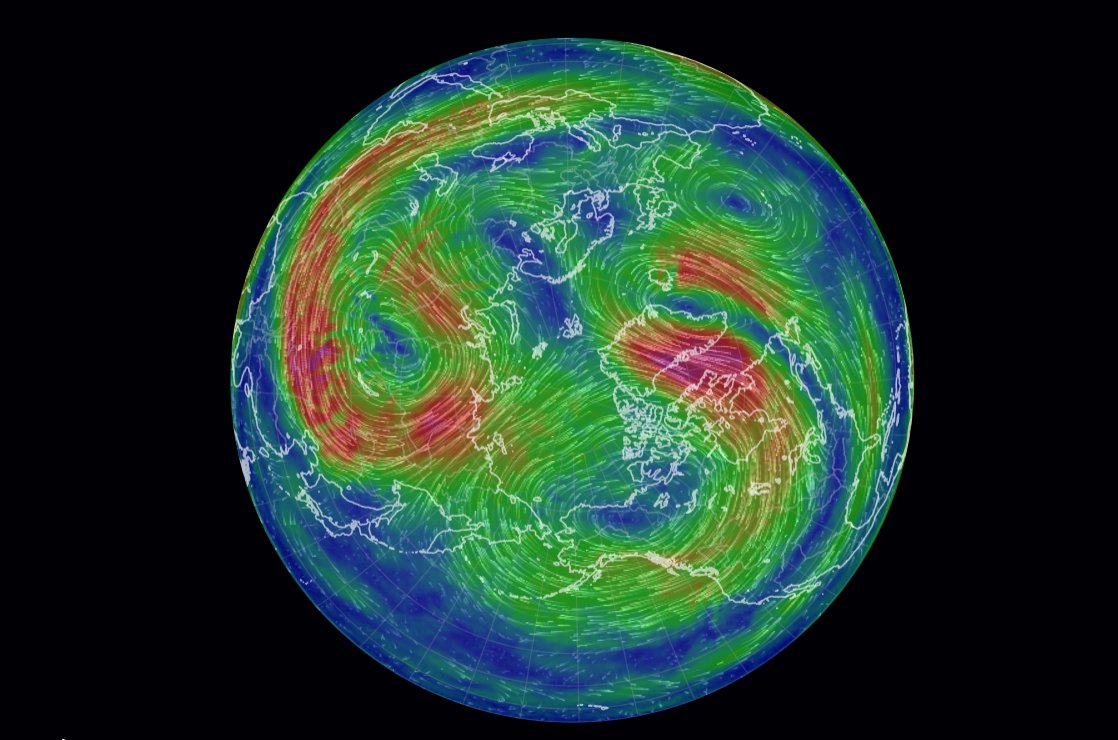
Tue Mar 22 22:17:09 +0000 2022@JoePlatelet I don't know of any proteomics MS/MS experiments that have tried to measure site occupancy for glutathione acceptors.
Tue Mar 22 15:47:25 +0000 2022They are also both rather shy about being photographed, as demonstrated by the AF error diagram for BRCA1. While it is true that the phosphorylation acceptors for this one are not in the structured (dark green) regions, that isn't really saying much in this case. 🔗
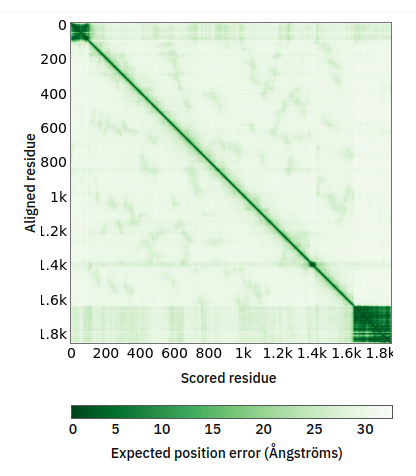
Tue Mar 22 15:00:51 +0000 2022PTM diagrams for #BARD1 & #BRCA1, subunits of an unnamed heterodimer #E3_ligase. Both have N-terminal RING zinc fingers―(50-86) & (24-65) respectively―and share a fondness for K+SUMOyl. BRCA1 has a weakness for S+phosphoryl bling (1000-1750). Both are averse to K+acetyl. 🔗
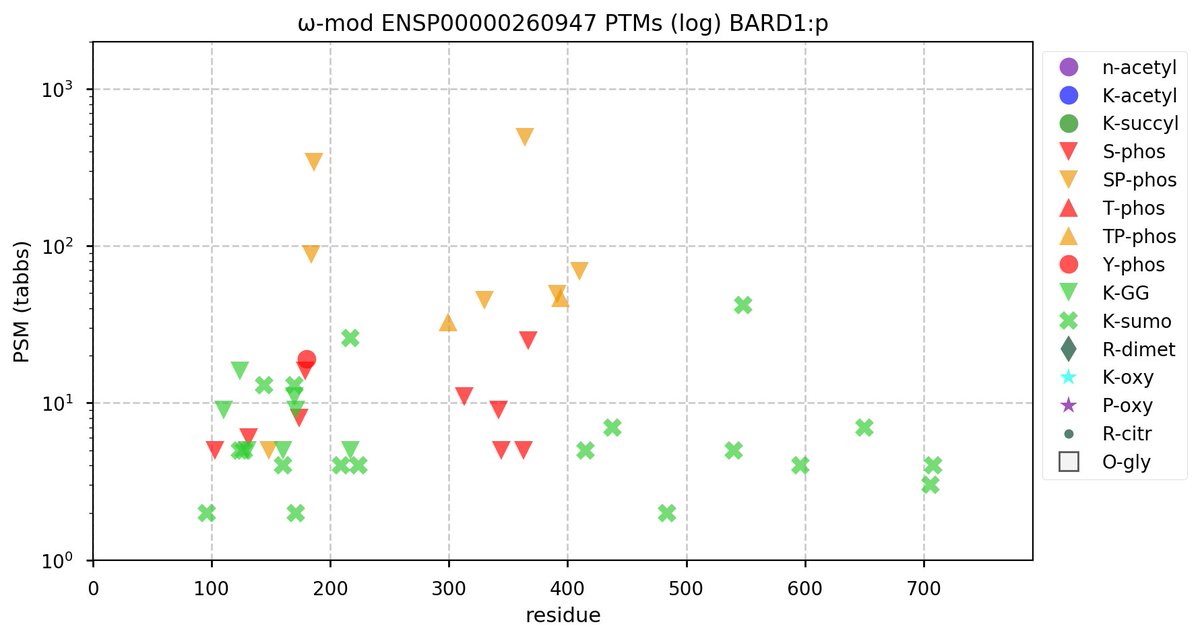
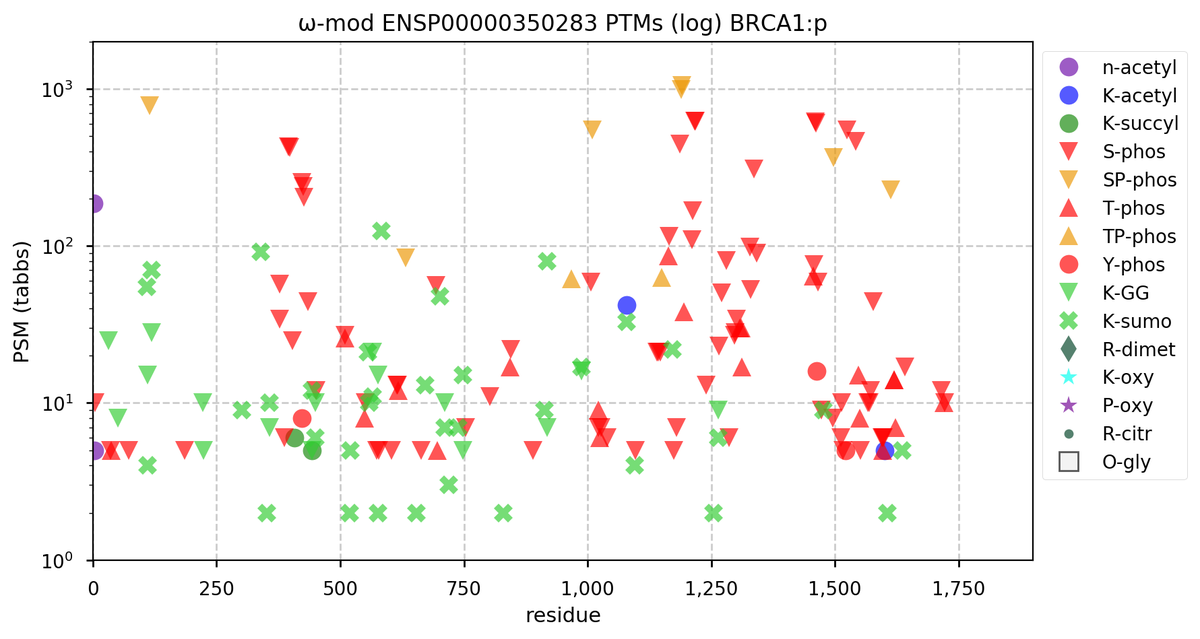
Tue Mar 22 14:20:29 +0000 2022I hadn't noticed before just how popular a target the spike protein was for E3 ligases. 🔗
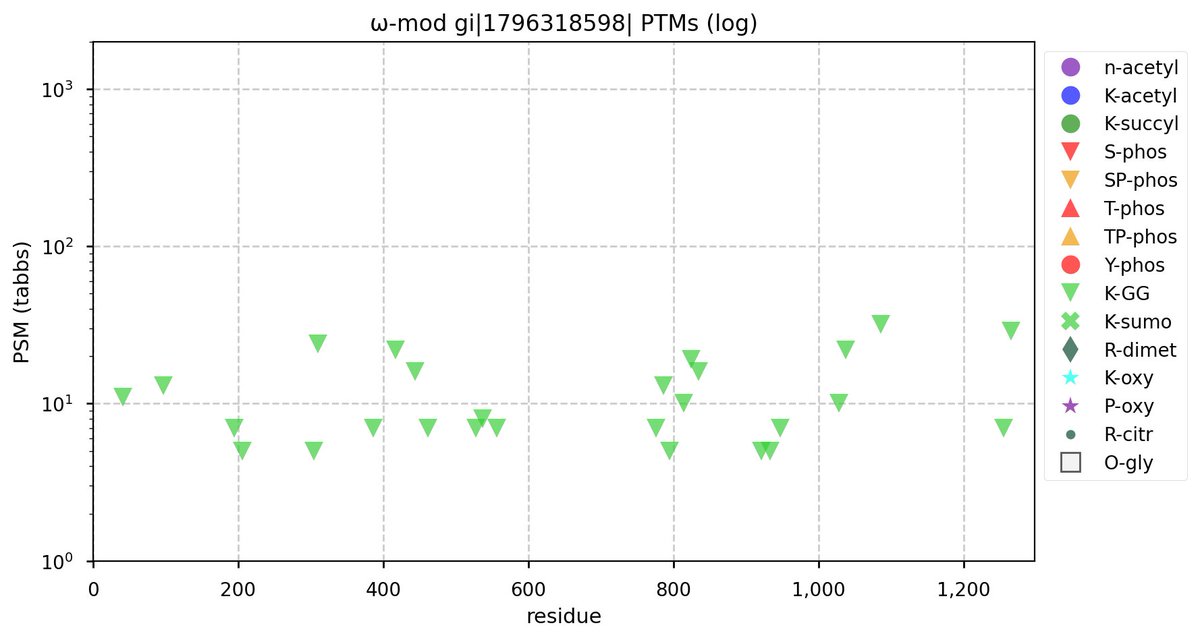
Mon Mar 21 19:39:16 +0000 2022It has a nice mix of Hyp-rich collagens (COL1A1, COL1A2, COL3A1 & COL3A2) with Hyp-lite collagens (COL6A1, COL6A2 & COL6A3).
Mon Mar 21 19:34:18 +0000 2022If you are interested in finding some hydroxyproline-containing peptides in cell culture data where the collagen signals aren't too overwhelming, PXD031053 looks like good data to try out. It has some issues (IAA & urea artifacts), but nothing out of the ordinary.
Mon Mar 21 19:16:20 +0000 2022I am a little surprised that 🔗 was still available. Most of the URLs that I think up have been snapped up long ago.
Mon Mar 21 16:21:55 +0000 2022@UCDProteomics @pwilmarth And, as official Devil's Advocate, the LC is a lot more useful than the MS without an LC.
Mon Mar 21 14:55:35 +0000 2022Ubiquitinylated ubiquitin is sufficiently abundant that it can easily be detected in proteomics data from cellular samples that were prepared without GG-enrichment.
/fin
Mon Mar 21 14:53:37 +0000 2022The most abundantly ubiquitinated protein is ubiquitin: any of its 7 lysines may be so modified to form large, branched poly-ubiquitins attached to target proteins.
/4
Mon Mar 21 14:52:58 +0000 2022There are multiple genes associated with each of these activities in the human genome:
9 E1 genes;
32 E2 genes; &
>500 E3 genes.
/3
Mon Mar 21 14:52:37 +0000 20222. Ubiquitin-conjugating enzyme (E2) transfers ubiquitin from E1 to one of its cysteines;
3. Ubiquitin ligase (E3) recruits an E2 with ubiquitin & facilitates the transfer of ubiquitin to the lysine of a target protein.
/2
Mon Mar 21 14:52:20 +0000 2022Ubiquitin is added to proteins via a process that requires three discrete subunit activites.
1. Ubiquitin-activating enzyme (E1) forms a covalent bond between ubiquitin & one of its cysteines;
/1
Mon Mar 21 14:51:42 +0000 2022The phosphorylation acceptor sites (red) follow the common pattern of being positioned in dark proteome domains (white) in the AF alignment error diagram.
The protein's RING zinc finger is (571-606) just C-terminal to the last of the phospho-acceptors. 🔗
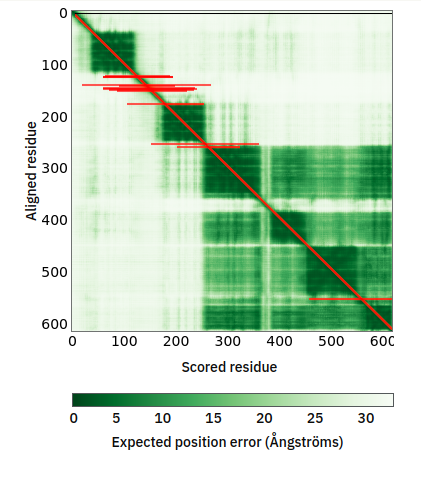
Mon Mar 21 14:45:44 +0000 2022PTM diagram for #BIRC2, an #E3_ligase. Unlike RNF11, the protein has many potential GGyl acceptors, but no evidence of acetyl co-modification at those lysines. The sequence created by alternate initiation at M20 is frequently observed. 🔗
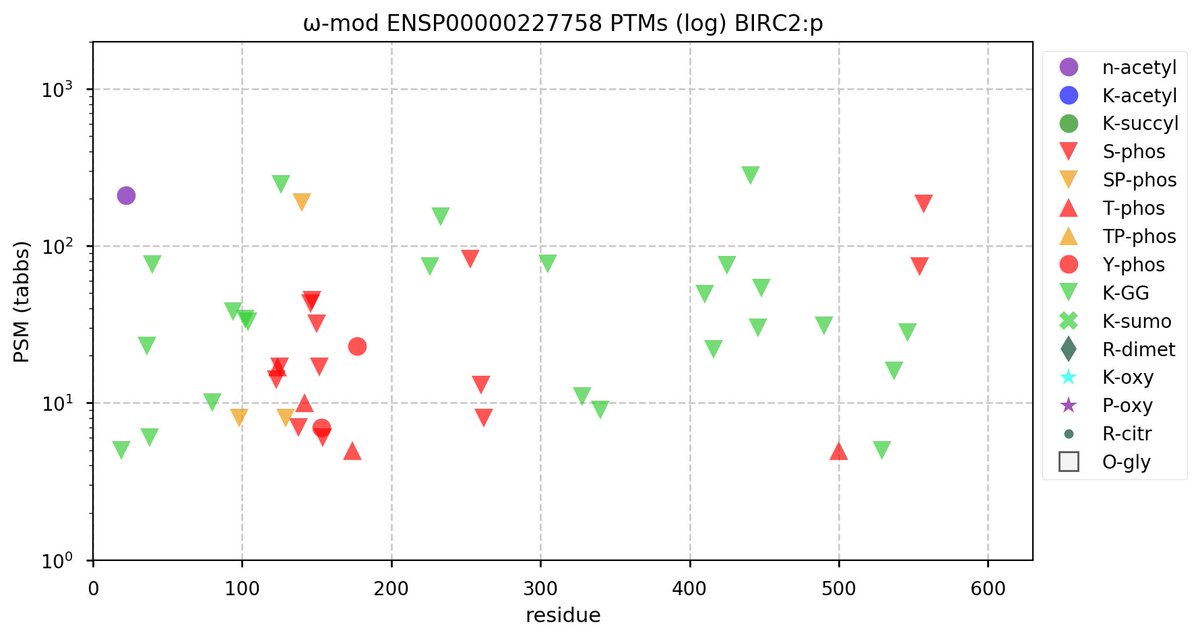
Mon Mar 21 12:43:17 +0000 2022While things are quiet here in North America, the party is still going on in far off lands 🔗
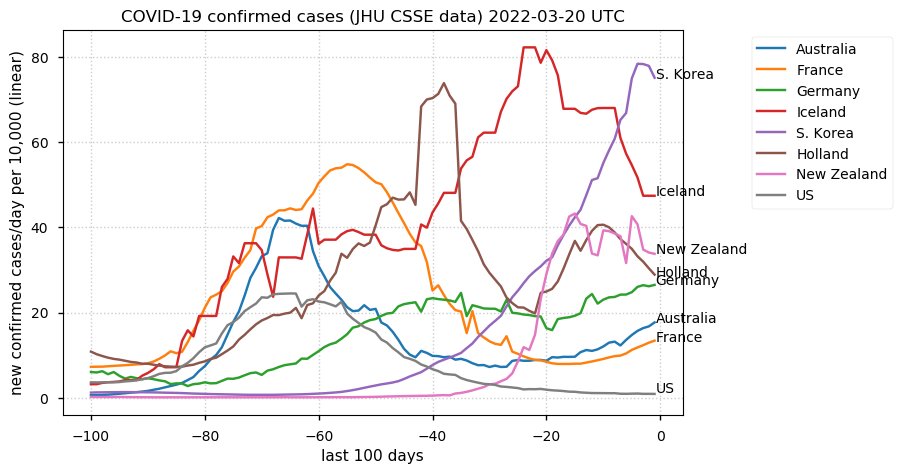
Mon Mar 21 12:17:03 +0000 2022The I-29 corridor is finally above zero from Winnipeg to Houston, with a spicy +5°C at Fargo ND! 🔗
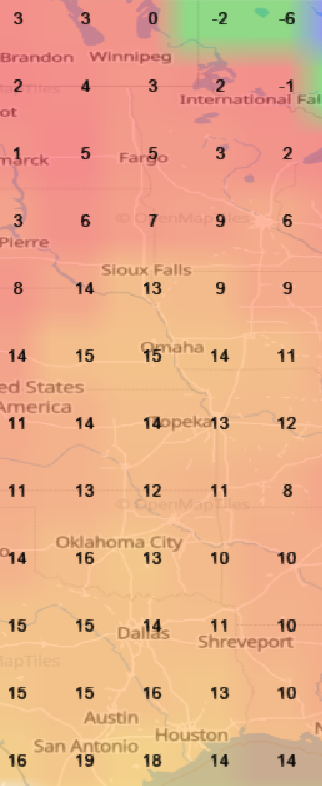
Mon Mar 21 02:01:25 +0000 2022Having tried to watch the current Batman twice, I haven't been able to make it through. Just standing around outside was much more entertaining.
Sun Mar 20 18:48:13 +0000 2022The device "sees the things I'm trying to photograph as a problem to solve," 🔗
Sun Mar 20 17:57:50 +0000 2022@pwilmarth Proteomics turns even the most mass spec of mass spec guys (e.g., Dick Smith) into at least closet chromatographers if they want to get anything done.
Sun Mar 20 17:13:10 +0000 2022@pwilmarth I don't know: proteomics has a foot in both camps. I have always insisted that it is good LC that distinguishes good from not-so-good proteomics data sets.
Sun Mar 20 14:50:22 +0000 2022@bkives I understand that, but there is a general impression amongst the citizenry that the city's administration is still in need of attention (& daylight). Keep up the good work.
Sun Mar 20 14:36:21 +0000 2022@LogueSus It works for me now.
Sun Mar 20 14:35:13 +0000 2022@bkives Hopefully, the candidate for Mayor in this election cycle will place emphasis on their plans to clean up the city's administration. Otherwise, we end up as Canada's version of Gotham City.
Sun Mar 20 14:11:12 +0000 2022@LogueSus There seems to be a problem with the link at the moment.
Sun Mar 20 13:40:45 +0000 2022Today's surface wind field indicates a large low pressure system steaming across the North Atlantic & an unusually windy day on Cape Breton, NS. 🔗
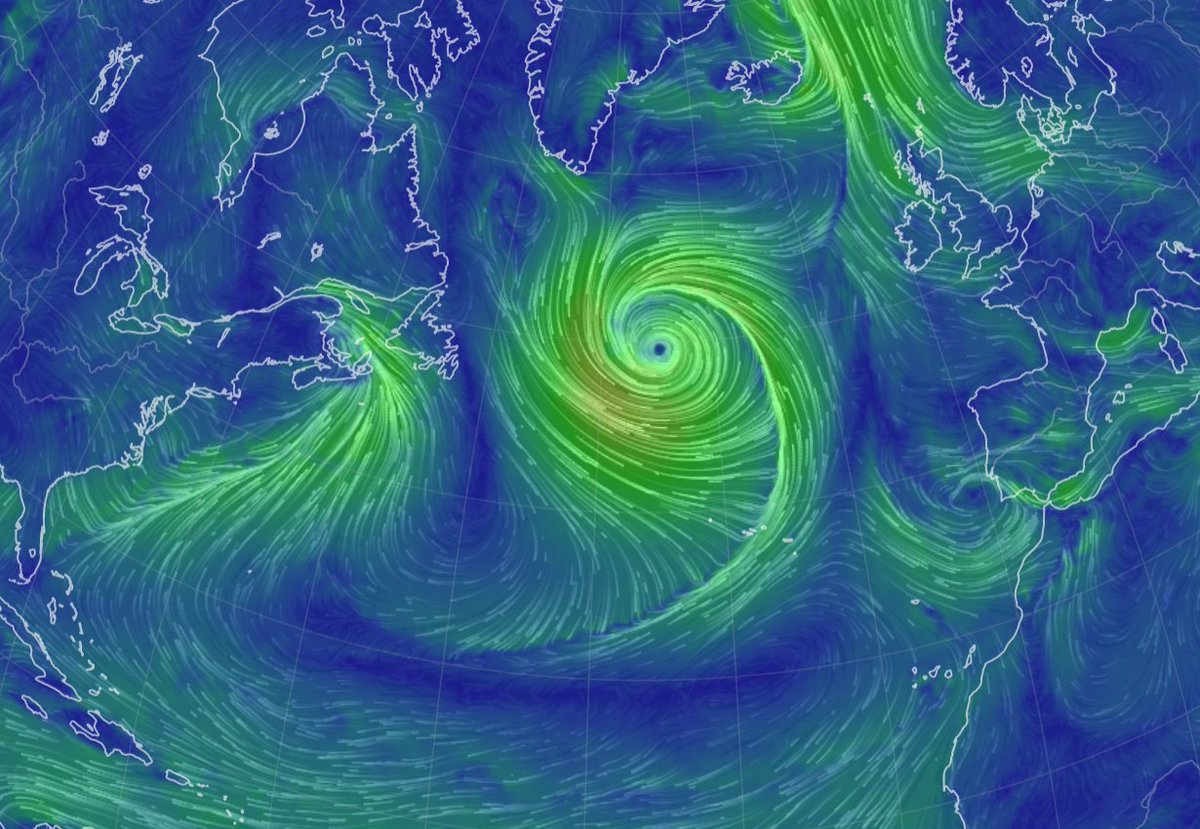
Sun Mar 20 13:27:49 +0000 2022The protein does not seem to partake of ubiquitination itself: it is only observable in MHC class 2 experiments.
Sun Mar 20 13:24:50 +0000 2022PTM diagram for #RNF11, an E3 ubiquitin ligase downstream participant in the #EGFR signaling mechanism. The mature membrane protein (2-154, G2+myristoyl) has a messy N-terminal phosphodomain; the unmodified C-terminal segment is a zinc finger RING domain. 🔗
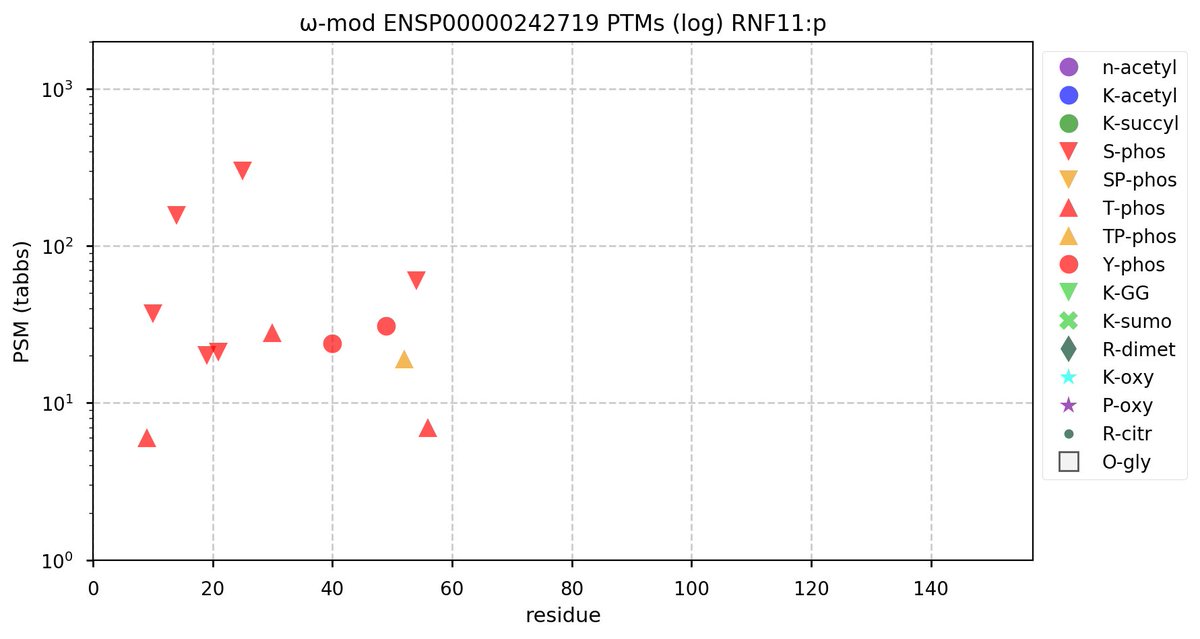
Sat Mar 19 22:52:11 +0000 2022@marcoyannic @mjmaccoss @byu_sam @Angrist_Fusion I'm always impressed by any system that works. But I am a protein snob 🧐
Sat Mar 19 17:30:44 +0000 2022@marcoyannic @mjmaccoss @byu_sam @Angrist_Fusion Someone I respect once told me that you usually only get one chance to impress people with something new, so be careful choosing examples.
Sat Mar 19 17:13:56 +0000 2022@Niigaanwewidam @HStefansonMB The current MB govt has shown a real willingness to dig deeper―rather than to step out of―any hole they find themselves in.
Sat Mar 19 16:57:10 +0000 2022🔗
none ☞ 2–375 (N-acetyl-Ser, occ. 32/32)
#realNT
Sat Mar 19 15:36:33 +0000 2022@mjmaccoss @byu_sam @marcoyannic @Angrist_Fusion And that interaction should be undetectable post hoc: those same proteins will not interact with TOMM70:p once they have been successfully imported.
Sat Mar 19 15:31:46 +0000 2022@mjmaccoss @byu_sam @marcoyannic @Angrist_Fusion TOMM70:p is also a bit of an odd duck in that its function is to perform a highly choreographed protein-protein interaction with almost every protein that is present in the mitochondrion
Sat Mar 19 15:02:08 +0000 2022@mjmaccoss @byu_sam @marcoyannic @Angrist_Fusion A mitochondrial membrane protein like TOMM70 isn't really the best example to use wrt the idea of cellular "concentration". A soluble protein like SARS1 (or many others like it) would be a better exemplar.
Sat Mar 19 14:18:50 +0000 2022Even though NFKB1 & NFKB2 are paralogs, they do not share any observable tryptic peptides. NFKB2 has significantly more observed K+GGyl & SUMOyl sites (30) in its N-terminal mature domain than NFKB1 (11).
Sat Mar 19 14:11:53 +0000 2022PTM diagrams for #NFKB2 & #RELB―that form 1 of the dimers generically called NF-κB―a downstream component of the #EGFR pathway. Like NFKB1, full length NFKB2 (1–899, M1+acetyl) can't pair with RELB (1–579, M1+acetyl): its C-terminal phosphodomain (455–899) must be removed. 🔗
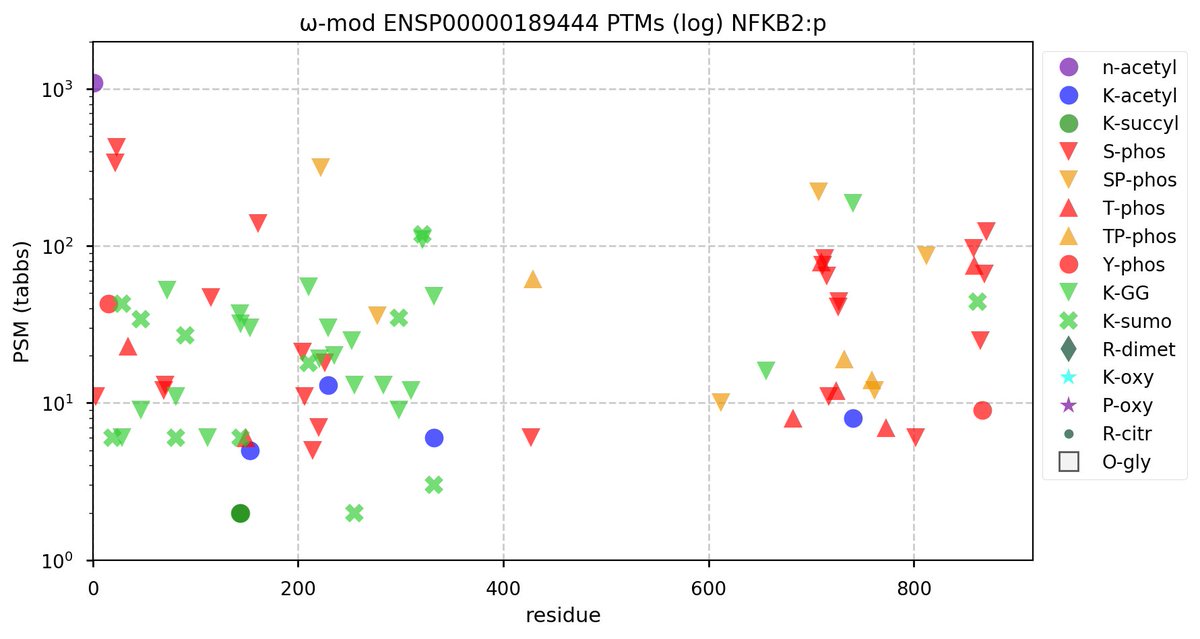
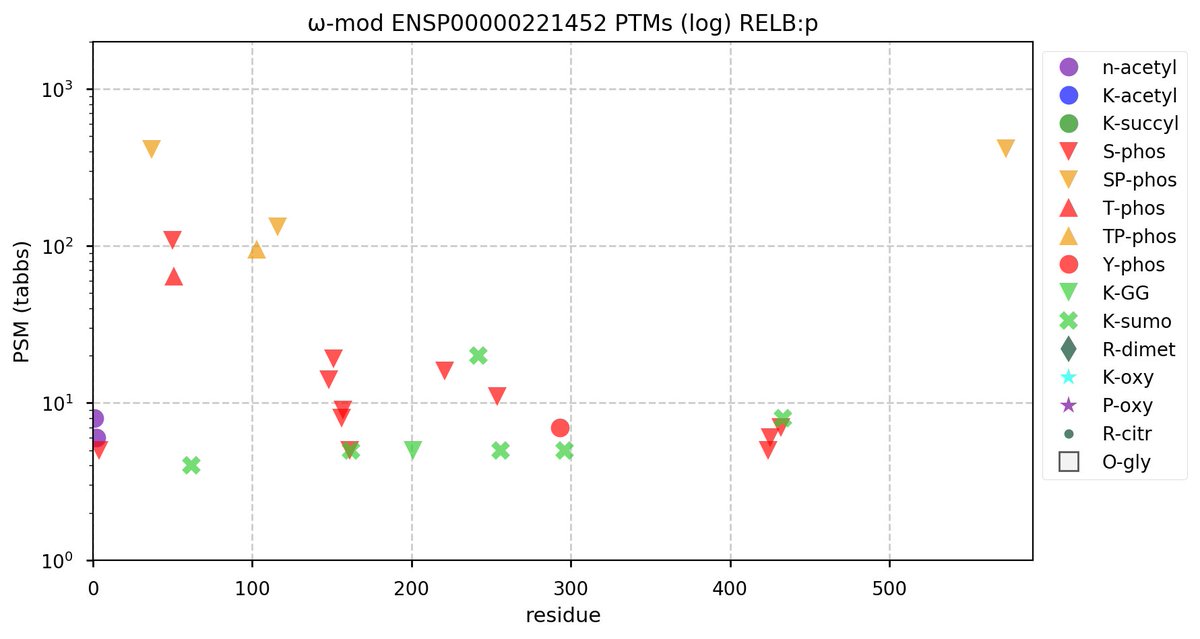
Sat Mar 19 13:26:23 +0000 2022@JDHL18 @NabaLabUIC Fibroblasts do all sorts of odd things when they encounter the unexpected: it is like having a swarm of angry, hyperactive beavers swimming around looking for any excuse to build something.
Sat Mar 19 12:57:40 +0000 2022Pleasant spring temperatures across Europe today 🔗

Sat Mar 19 12:21:58 +0000 2022@JDHL18 @NabaLabUIC The cancer cells themselves don't produce the collagen: it is supplied by annoyed fibroblasts that don't like the look of the cancer in tissue.
Sat Mar 19 12:20:33 +0000 2022@JDHL18 @NabaLabUIC It can also be surprisingly abundant in urine, blood plasma & CSF.
Sat Mar 19 12:16:18 +0000 2022@JDHL18 @NabaLabUIC It is rare in cell culture, unless there are fibroblast lineages involved. It is abundant in many types of tissue-derived samples, particularly from many cancers.
Fri Mar 18 15:43:44 +0000 2022Modification diagrams for #NFKB1 & #RELA―that form 1 of the heterodimers referred to as NF-κB―a downstream component of the #EGFR pathway. Full length NFKB1 (2–968, A2+acetyl) can't pair with RELA (1–548, M1+acetyl): its C-terminal phosphodomain (434–968) must be removed first. 🔗
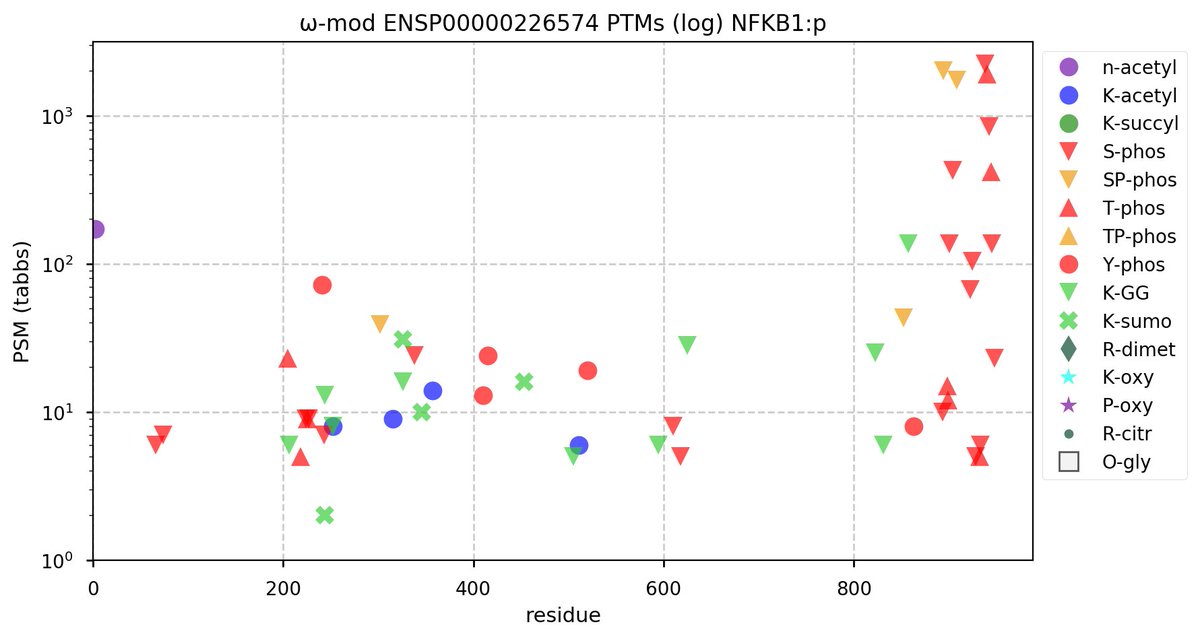
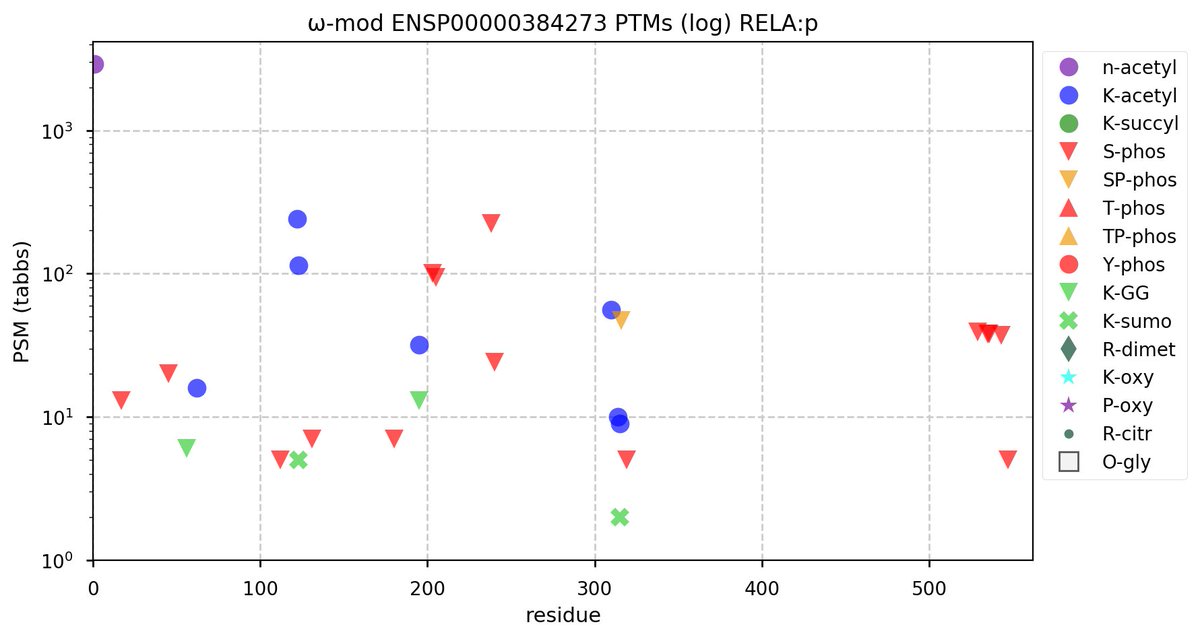
Fri Mar 18 14:23:10 +0000 2022@pwilmarth @ProteomicsNews @UCDProteomics @neely615 Intrinsic Disorder (dot) com (© 2022 ® ™ ℠)
Fri Mar 18 12:59:36 +0000 2022@aledmedwards Not to worry. There will be a new institute at the U of T (possibly taking over some space from one of the AI outfits), an NRC build-out in Montreal & a chair or 2 at UBC. Then back to slow decline ...
Fri Mar 18 12:21:38 +0000 2022Thanks to everybody who participated in this poll. It would appear that "don't bother me kid: look it up on the Internet" is the general approach to training in academic proteomics. The "field" needs a Stack Overflow.
Fri Mar 18 11:37:53 +0000 2022The storms predicted yesterday seem to be sparking up this morning 🔗
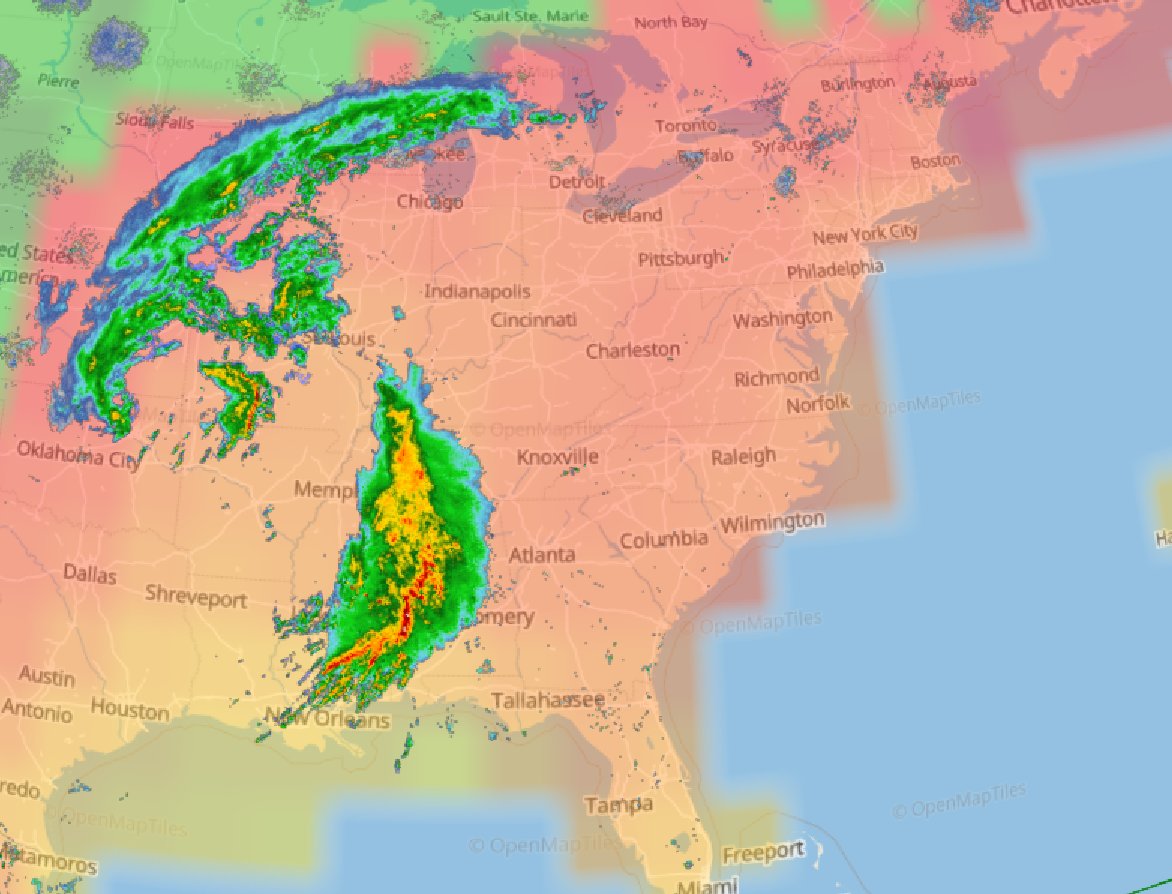
Thu Mar 17 21:44:24 +0000 2022The only "good" thing about the poll so far is that because so few votes have been registered, I can pretend that the results are a statistical anomaly. 🥺 🔗
Thu Mar 17 17:05:59 +0000 2022@UCDProteomics @Smith_Chem_Wisc @neely615 @Clubhouse It sounds like you may have been telling some tall-tales of life in the northern wilderness to your more gullible Californian colleagues.
Thu Mar 17 16:37:05 +0000 2022@jwoodgett There are languages (e.g., Chinese or Japanese) in which character counts make the most sense.
Thu Mar 17 16:15:43 +0000 2022@slavov_n About once in a career, it turns out to be like Lennon-McCartney. Usually it trends to be more like Lenin-Stalin.
Thu Mar 17 15:50:15 +0000 2022Sorting out the PTM patterns for these 3 sequences would seem to be an ideal opportunity for top-down proteomics.
Thu Mar 17 15:48:58 +0000 2022Modification diagrams for the 3 human paralogs #HRAS, #KRAS & #NRAS that may be part of the #EGFR signalling mechanism. The N-terminal domains of these 3 sequences are identical, so the PTMs may be composites of results from 1 or more of these subunits. 🔗
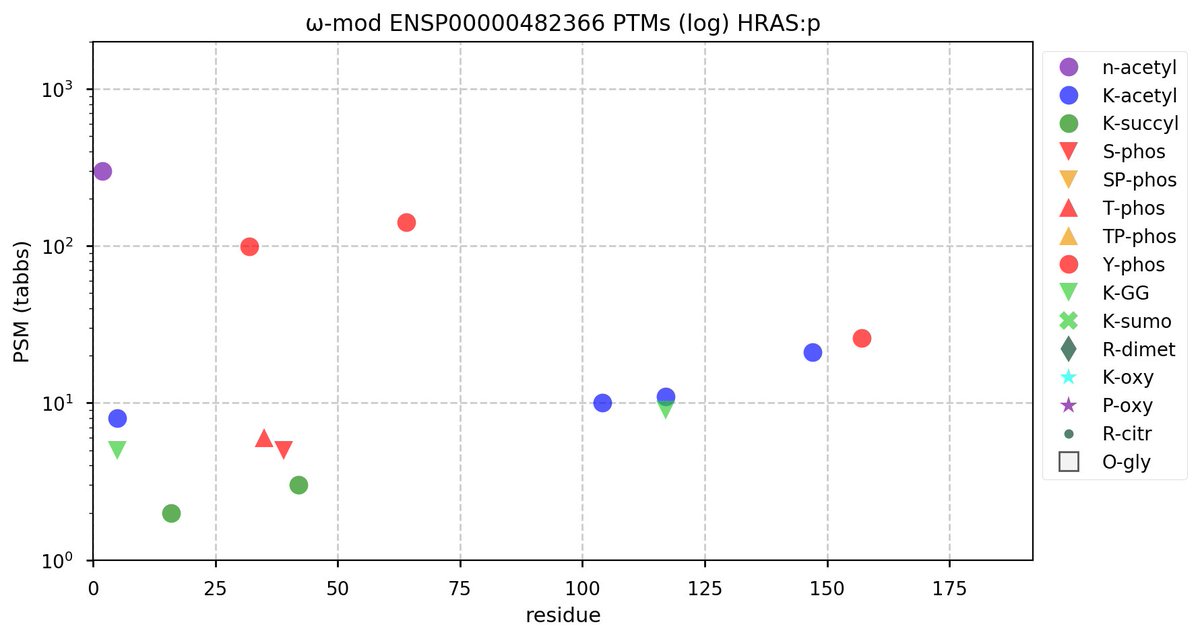
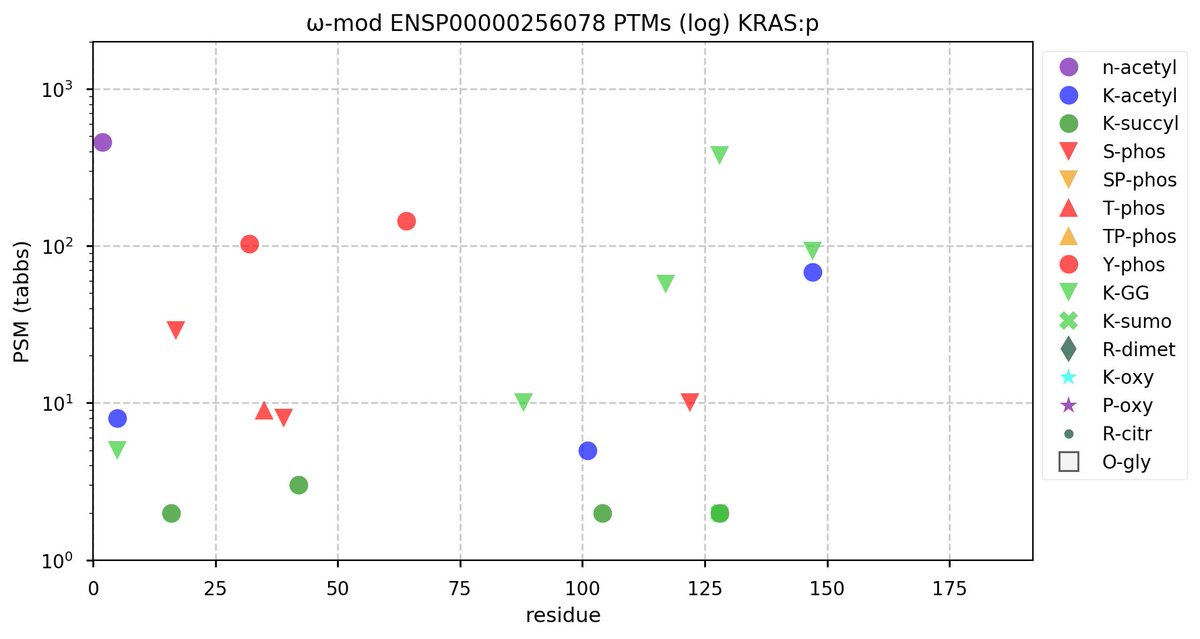
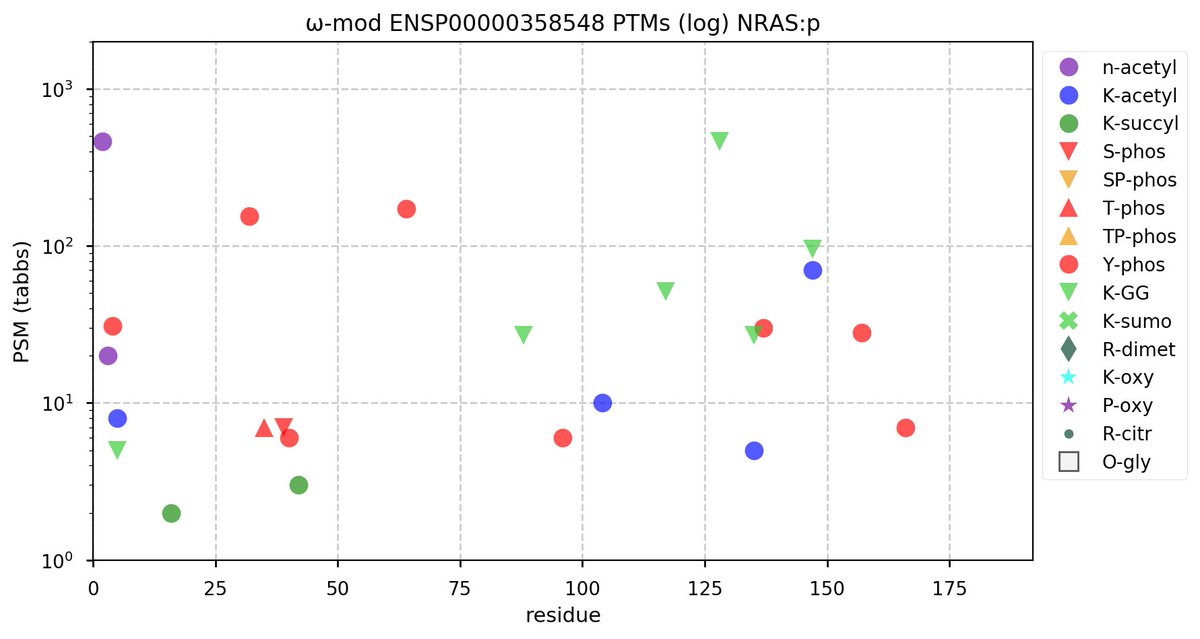
Thu Mar 17 15:30:04 +0000 2022🔗
none ☞ 1 – 1049 (N-acetyl-Met, occ. 183/183)
#realNT
Thu Mar 17 15:24:21 +0000 2022🔗
none ☞ 28 – 793 (mitochondrial signal 2 – 29)
#realNT
Thu Mar 17 15:12:56 +0000 2022🔗
none ☞ 32 – 555 (mitochondrial signal 1-31)
#realNT
Thu Mar 17 14:32:15 +0000 2022It is still early, but my money would be on a field-wide average of <1 course.
Thu Mar 17 13:35:36 +0000 2022@UCDProteomics @Smith_Chem_Wisc @neely615 @Clubhouse No pining for the glory days at MSU?
Thu Mar 17 12:49:16 +0000 2022The winter pattern along I-29 has started to break down. Hang in the Fargo! 🔗
Thu Mar 17 12:14:05 +0000 2022Although, on thinking about it, I am assuming something not in evidence.
How many graduate level courses that primarily cover topics specifically about proteins (biochemistry, synthesis, signalling, etc.) does a student need to take to obtain a Ph.D. in "proteomics"?
Wed Mar 16 21:55:24 +0000 2022Maybe someone can explain this to me. Why is it that it is still uncommon for people to check for hydroxyproline (& lysine) in ECM samples, even though every intro proteins text mentions its role of in collagen's biological function & that collagen is the most abundant protein?
Wed Mar 16 18:00:18 +0000 2022🔗
1 – 1242 ☞ 2 – 1242 (N-acetyl-Ala, occ. 2983/2988)
#realNT
Wed Mar 16 16:17:31 +0000 2022@herrtschmidt @RalfGabriels The PTM specification, e.g. ...A[Acetyl]K..., doesn't state which residue is modified in protein coordinates. While it isn't a big deal to count the intervening residues, it makes the whole thing too fragile for automated use & adds a lot of unnecessary alignment work.
Wed Mar 16 15:30:42 +0000 2022@herrtschmidt @RalfGabriels I can't think of use for it, though. The lack of coordinates makes it too cumbersome for doing anything at scale.
Wed Mar 16 15:30:01 +0000 2022@herrtschmidt @RalfGabriels I was looking at it yesterday. It seems like a standard way to come up with your own dialect for representing protein sequences.
Wed Mar 16 15:17:50 +0000 2022🔗
1 – 251 ☞ 2 – 251 (N-acetyl-Thr, occupancy 3011/3016)
#realNT
Wed Mar 16 14:55:56 +0000 2022@RuneLinding An M 7.3 at 14:36:30 (UTC) 37.700°N 141.700°E 🔗
Wed Mar 16 14:41:50 +0000 2022This highly phosphorylated domain (red bars) corresponds to an IDR in the AF proposed structure 🔗
Wed Mar 16 14:38:31 +0000 2022Modification diagram for #SOS1:p, part of the #EGFR pathway. It has several PTM domains, most prominently a C-terminal domain of high occupancy S+phosphoryl ΦP acceptors (1070-1333). 🔗
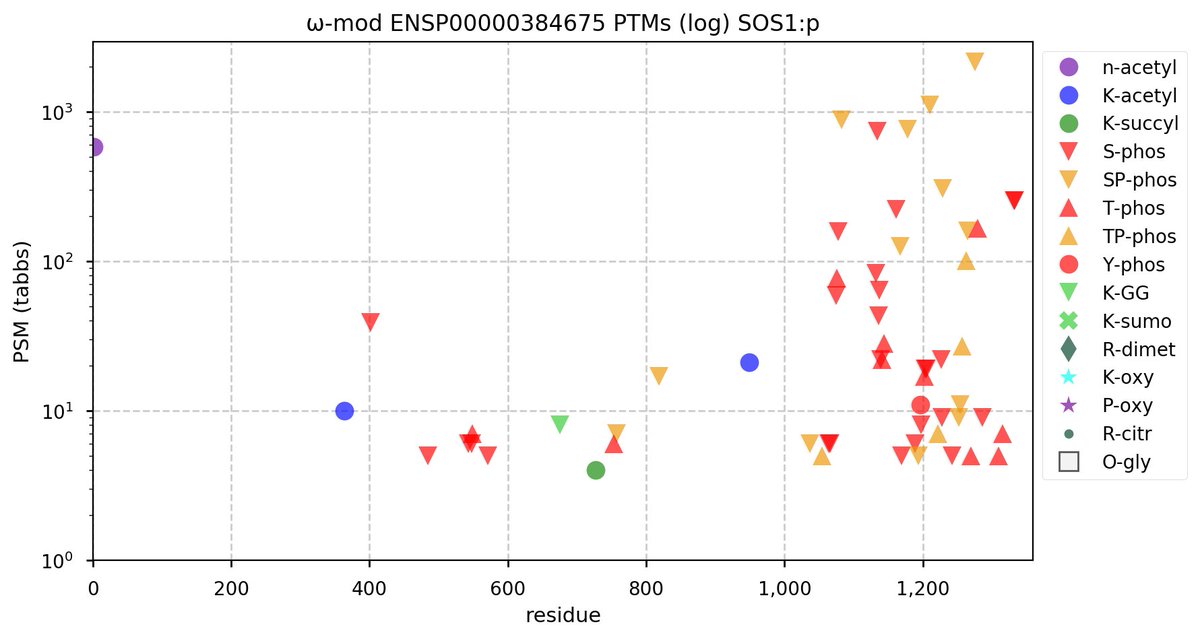
Wed Mar 16 14:31:08 +0000 2022🔗
none ☞ 2-1463 (N-acetyl-Ser, occupancy 1574/1597)
#realNT
Wed Mar 16 14:00:34 +0000 2022@AlexUsherHESA It is Canada's University of Michigan.
Wed Mar 16 13:14:00 +0000 2022🔗
1 – 397 ☞ 2 – 397 (N-acetyl-Ala, occupancy 472/476)
#realNT
Tue Mar 15 20:59:17 +0000 2022@chrashwood Maybe next year ...
Tue Mar 15 20:53:14 +0000 2022@chrashwood & play "Still" at disturbingly high volume throughout the workshop
Tue Mar 15 20:46:10 +0000 2022@chrashwood Only if you throw in "Ballistic Methods in Computer Repair"
Tue Mar 15 20:44:12 +0000 2022🔗
1 – 504 ☞ 1 – 504 (N-acetyl-Met, occupancy 100%)
#realNT
Tue Mar 15 16:14:57 +0000 2022🔗
1 – 559 ☞ 2 – 559 (N-acetyl-Ala, occupancy >99%)
#realNT
Tue Mar 15 15:47:42 +0000 2022@cajun_science It is a PTM that only occurs at a single residue in one protein, but it requires some orchestration to produce 🔗 & (as always) "The biological function of diphthamide is still not well understood. "
Tue Mar 15 14:44:08 +0000 2022🔗
1 – 326 ☞ 1 – 326 (N-acetyl-Met, occupancy 100%)
#realNT
Tue Mar 15 14:38:07 +0000 2022Just for clarity, here are the two acceptor classes broken out, using a linear scale for PSM observation frequency 🔗
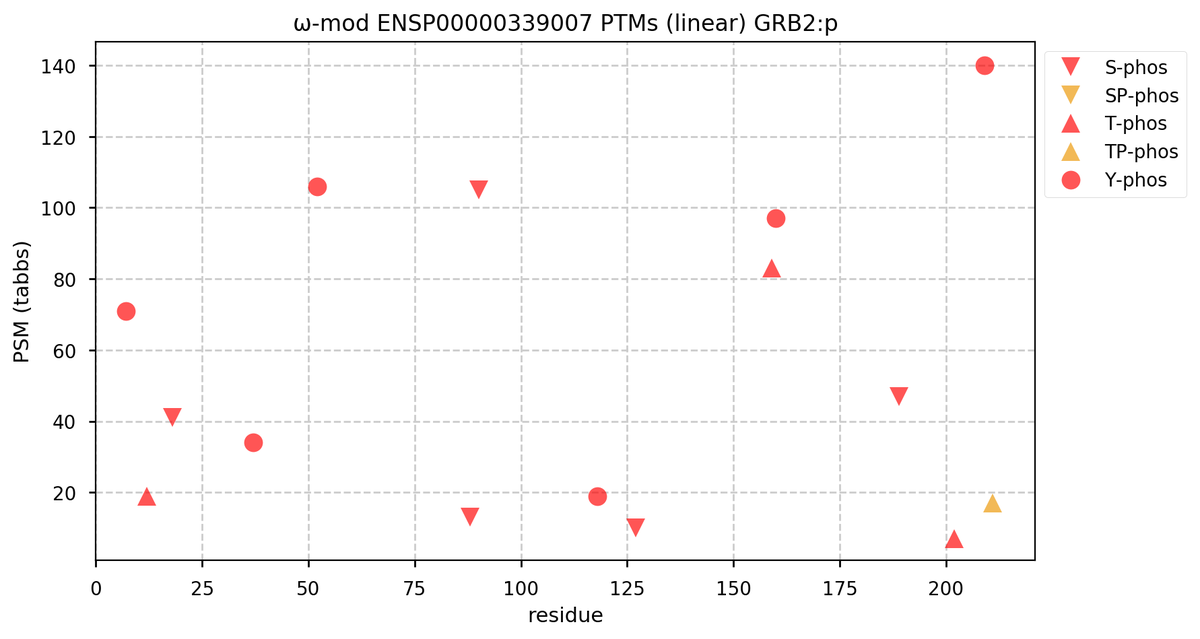
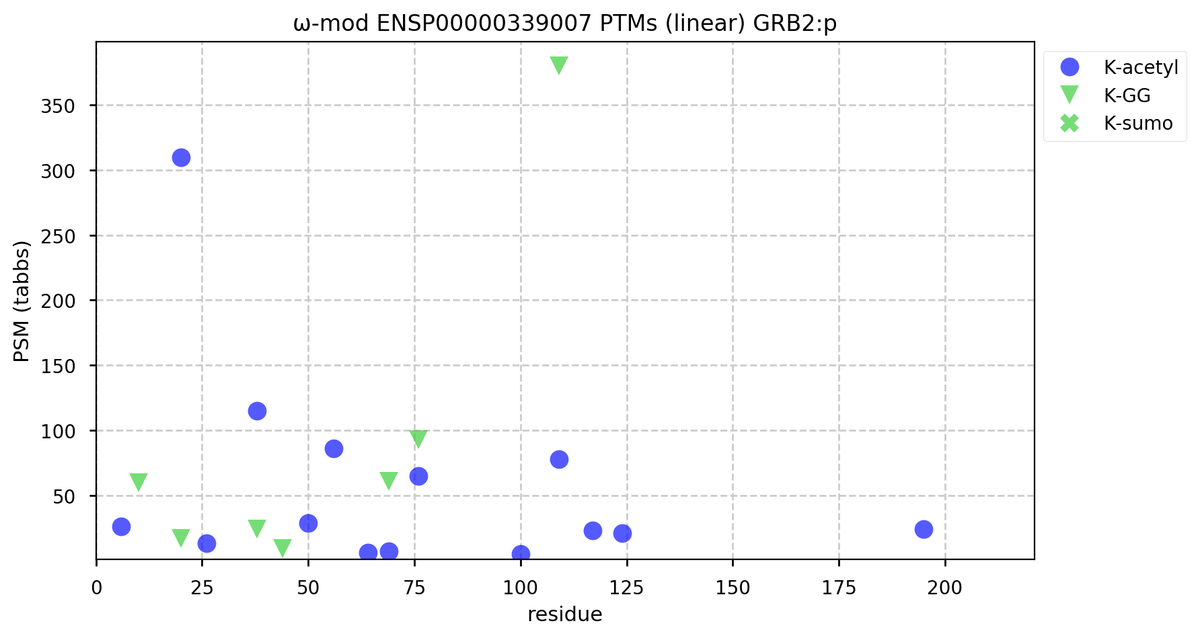
Tue Mar 15 14:27:44 +0000 2022Modification diagram for #GRB2:p, an adaptor that interacts directly with #EGFR:p. It has an N-terminal domain (1-125) with multiple K+acetyl/GGyl co-acceptors. Y+phosphoryl acceptors are concentrated in the two SH3 domains (1-58) & (156-215) 🔗
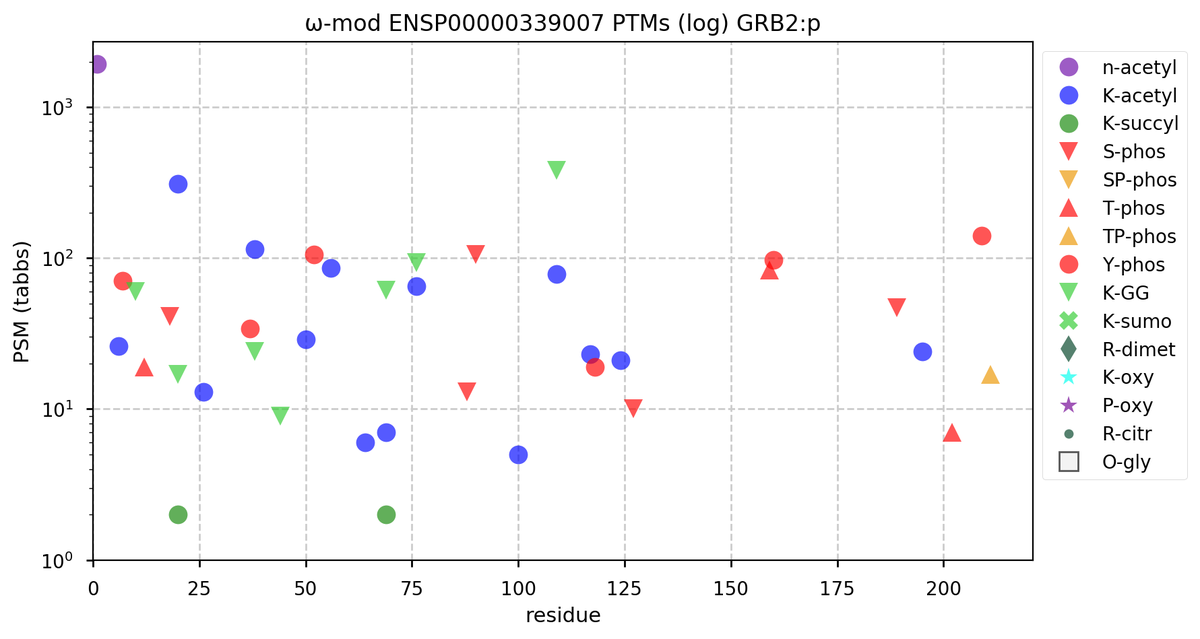
Tue Mar 15 13:03:31 +0000 2022@MHendr1cks @CIHR_IG @NSERC_CRSNG @SSHRC_CRSH And at least 2 additional sections in the CCV (length & format to vary by agency applied to).
Tue Mar 15 12:12:41 +0000 2022The same spectrum interpreted without trimethylamine loss & with trimethylamine loss at diphthamide (a modified H). 🔗
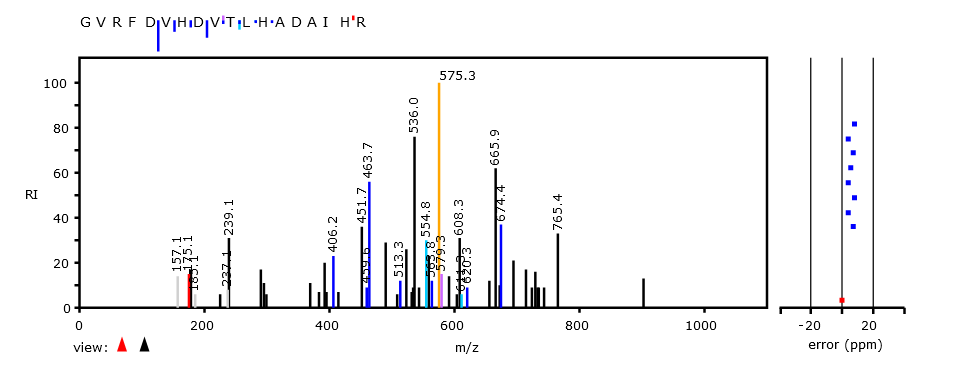
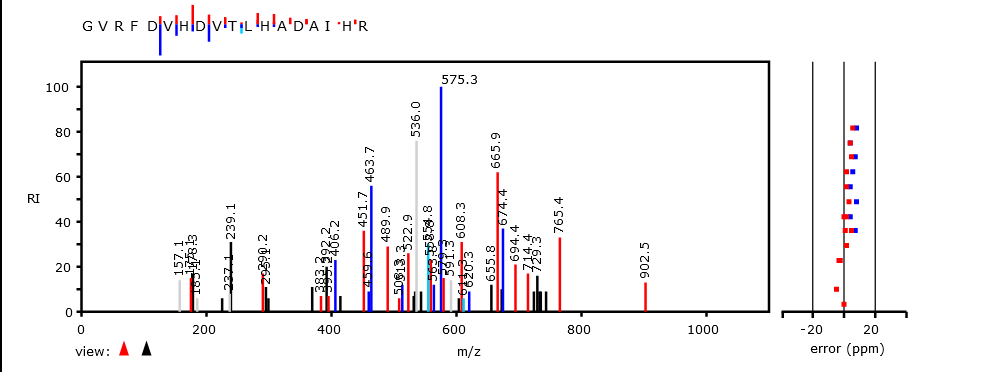
Tue Mar 15 02:03:01 +0000 2022@pwilmarth @Smith_Chem_Wisc Yup. >LOD but <LOQ
Mon Mar 14 22:41:05 +0000 2022I knew that the exopeptidase read the residue in the +1 position, but I did not know the NAT did too.
Mon Mar 14 22:28:23 +0000 2022I did not know that in a nascent peptide that starts with
MAP...
once the M is removed, the presence of P3 seems to block the co-translational formation of N-acetyl-Ala.
Mon Mar 14 21:16:42 +0000 2022🔗
1 – 592 ☞ 2 – 592 (N-acetyl-Ala, occupancy 1%)
#realNT
Mon Mar 14 20:35:08 +0000 2022Trimethylamine seems to be a particularly good neutral leaving group from diphthamide-containing peptides.
Mon Mar 14 17:12:23 +0000 2022An evergreen story from Vancouver
🔗
Mon Mar 14 15:56:36 +0000 2022Is there some practical reason that Methods sections are often shy about revealing whether cells were from male or female sources?
Mon Mar 14 14:16:33 +0000 2022And for the shortest splice variant, aka p60 🔗
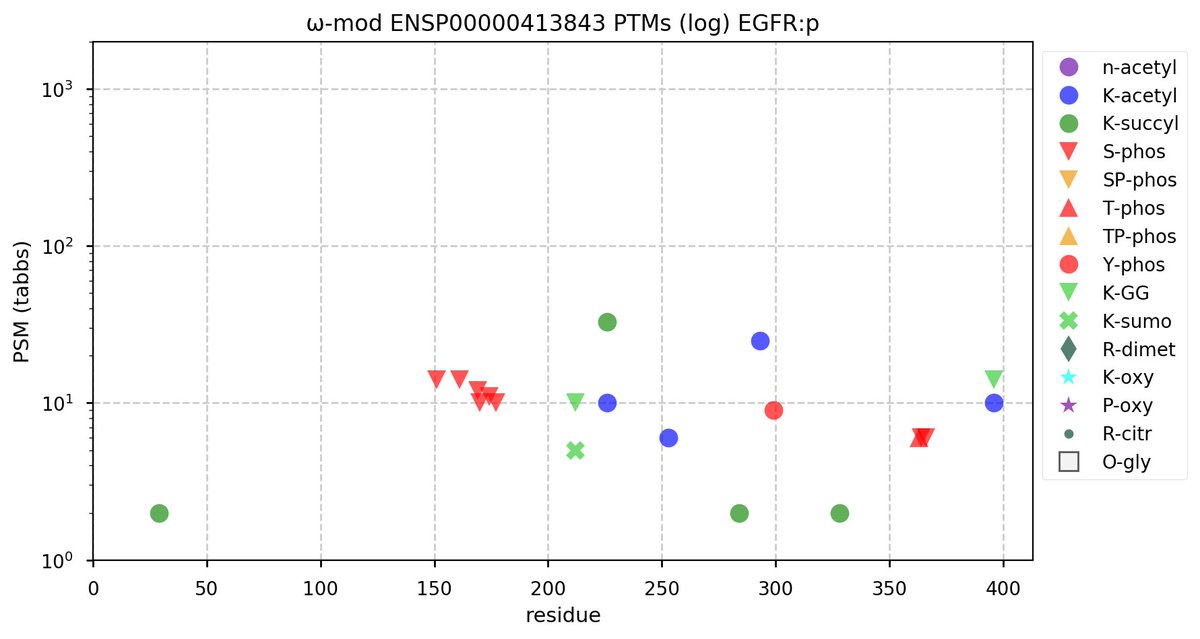
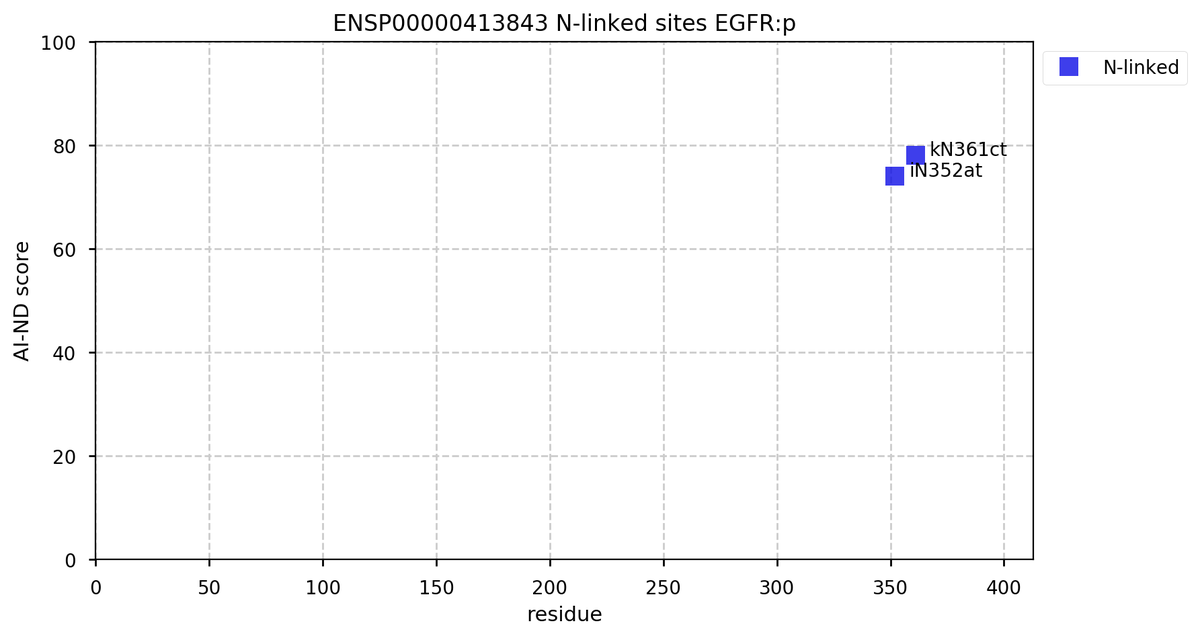
Mon Mar 14 14:15:51 +0000 2022Diagrams for the shorter splice variant, aka p110 🔗
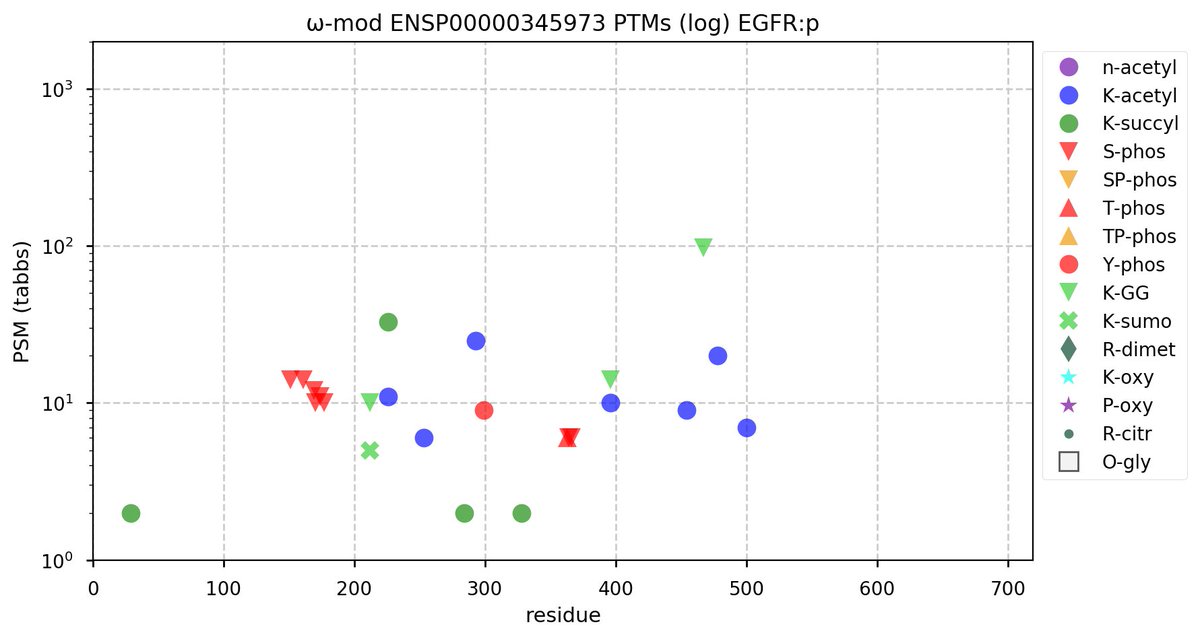
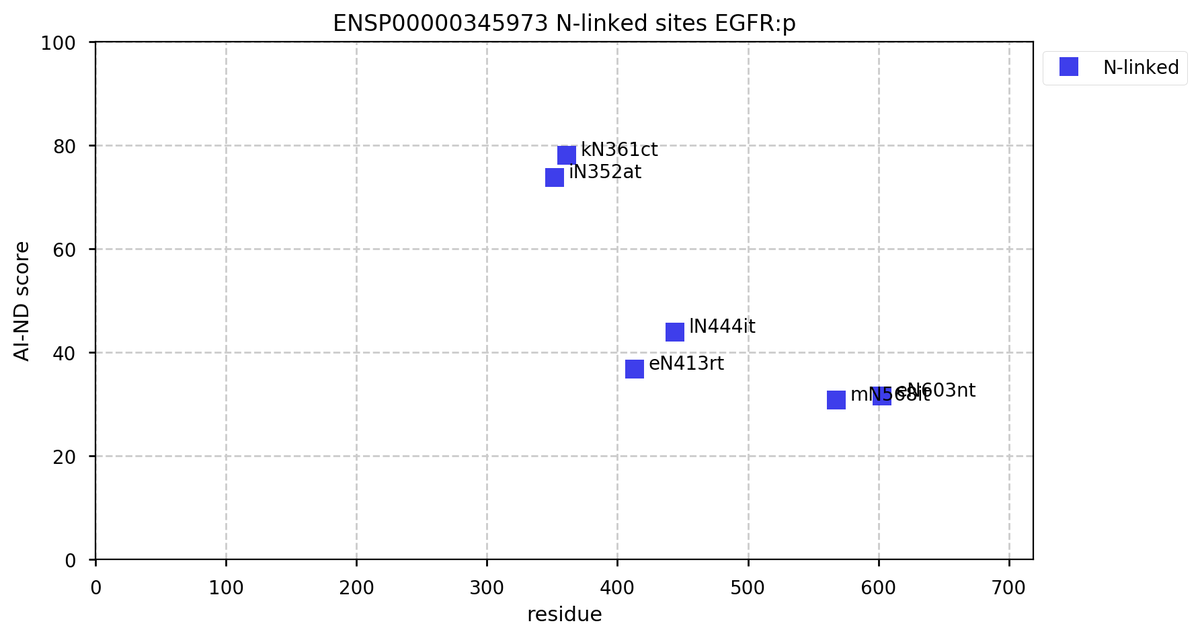
Mon Mar 14 14:12:24 +0000 2022Modification diagrams for the longest splice variant of #EGRF (aka p170). Unlike #EGF, this protein is highly decorated with PTMs, particularly the C-terminal intracellular domain (669-1210). N-linked glycosylation is exclusive to the extracellular domain (25-645). 🔗
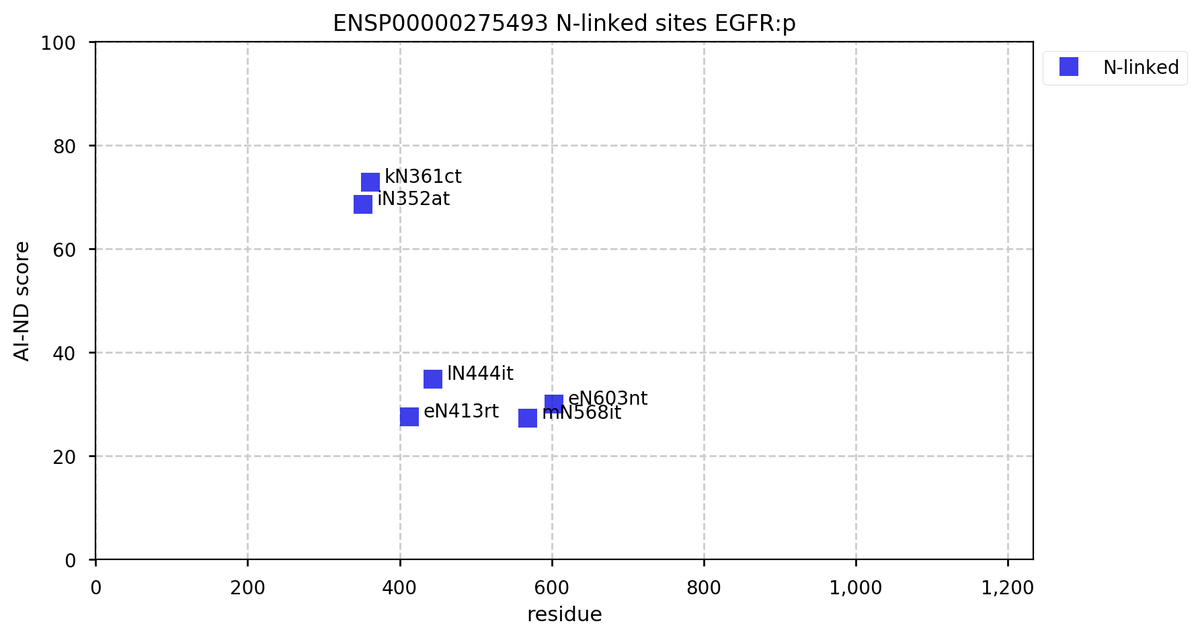
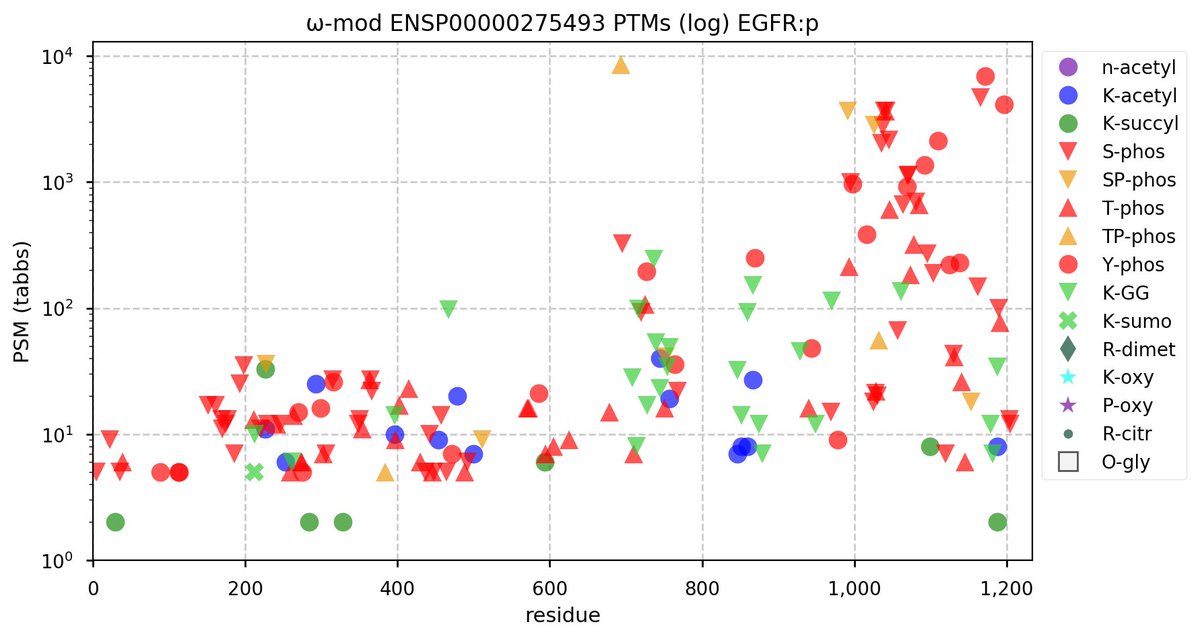
Mon Mar 14 00:58:35 +0000 2022🔗
1 – 2644 ☞ 2 – 2644 (N-acetyl-Gly, occupancy 100%)
#realNT
Mon Mar 14 00:55:24 +0000 2022Surrendering to my baser instincts:
🔗
1 – 1096 ☞ 2 – 1096 (N-acetyl-Gly, occupancy 90%)
#realNT
Sun Mar 13 14:40:45 +0000 2022Modification diagram from #EGF:p, the eponymous first mover in the #EGFR pathway. These PTMs are clearly not part of this subunit's life cycle. Most proteomics observations of EGF are from urine studies. 🔗
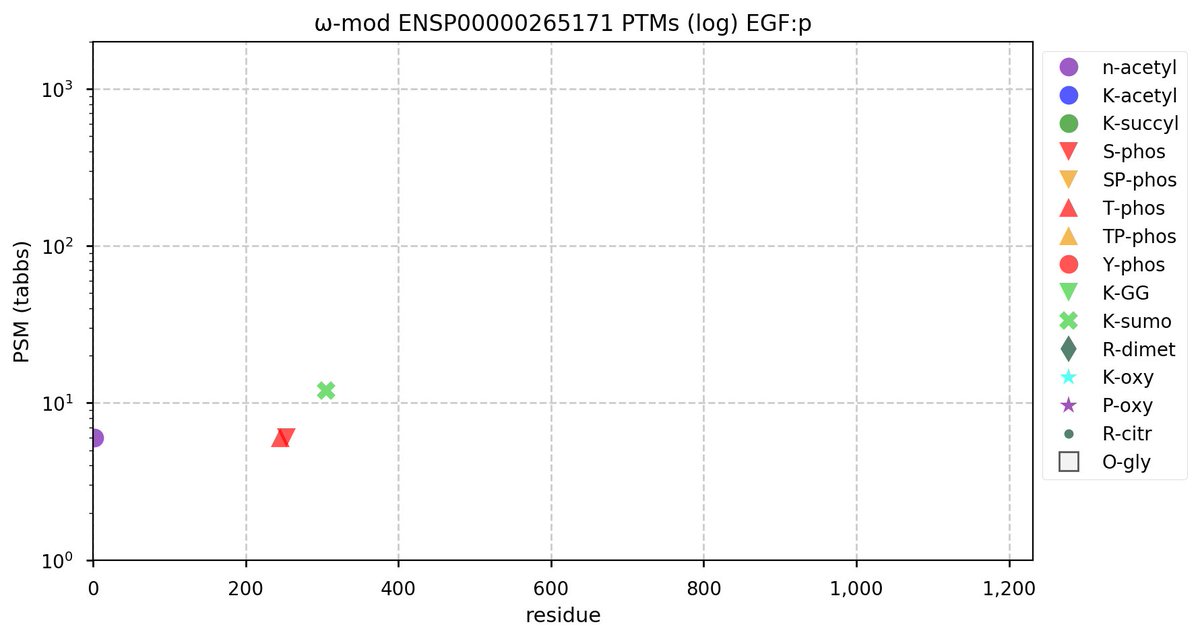
Sat Mar 12 20:05:12 +0000 2022I have to stop doing this. Just shooting fish in a barrel at this point (but it is fun anyway).
Sat Mar 12 16:54:16 +0000 2022Modification diagram for #NTNG1, which is sort of related to #NETRIN. The listed PTMs aren't part of this subunit's lifecycle. A signal peptide (1-28) is removed co-translationally. 🔗
Sat Mar 12 16:01:50 +0000 2022🔗
1 – 250 ☞ 2 – 250 (N-acetyl-Ser, occupancy 100%)
#realNT
Sat Mar 12 15:37:47 +0000 2022@Smith_Chem_Wisc @pwilmarth "PTM quantum signature" would be a wonderful combination of more accurate & more batshit at the same time ...
Sat Mar 12 14:00:40 +0000 2022A front of unstable weather stretching north from Disney World to the Irving Oil Refinery. 🔗
Sat Mar 12 13:39:59 +0000 2022It is still winter, top to bottom, on I-29 ❄️ 🔗
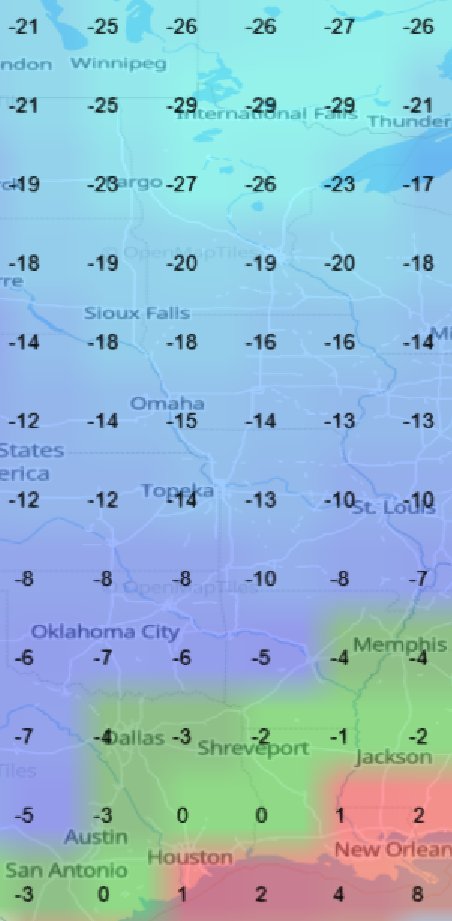
Sat Mar 12 13:36:16 +0000 2022🔗
1 – 461 ☛ 1 – 461 (N-acetyl-Met, occupancy 90%)
#realNT
Sat Mar 12 13:35:27 +0000 2022🔗
1 – 319 ☛ 1 – 319 (N-acetyl-Met, occupancy 74%)
#realNT
Sat Mar 12 00:59:30 +0000 2022🔗
1 – 253 ☛
1 – 253 (N-acetyl-Met 100%)
or
2 – 253 (N-acetyl-Met 100%)
or
3 – 253 (N-acetyl-Met, 100)
ratio of isoforms: M1/M2/M3 1.0/0.33/0.14
#realNT
Fri Mar 11 18:37:17 +0000 2022@byu_sam It is a feature, not a bug. Time zones are all about local/regional politics.
Fri Mar 11 16:50:26 +0000 2022"PTM crosstalk" is my new favorite example of the misapplication of an electrical engineering term in a biological context.
Fri Mar 11 16:27:50 +0000 2022Modification diagrams for NCK1:p & NCK2:p, adaptor proteins that have been associated with the #NETRIN mechanism. They have also associated with other pathways that involve tyrosine phosphorylation. 🔗
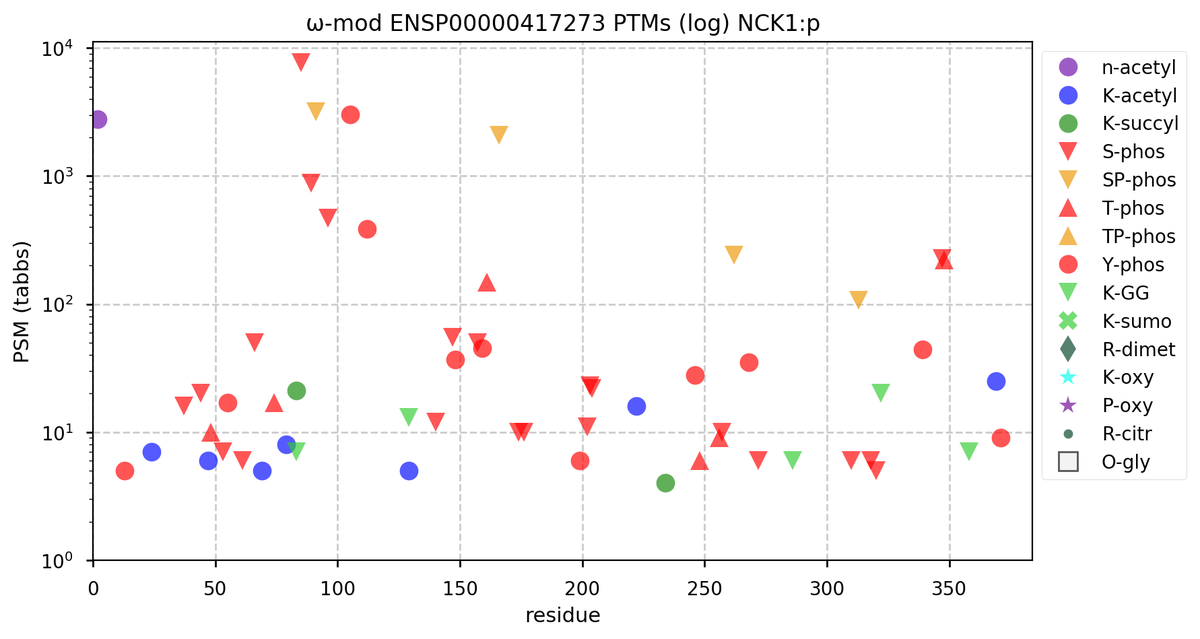
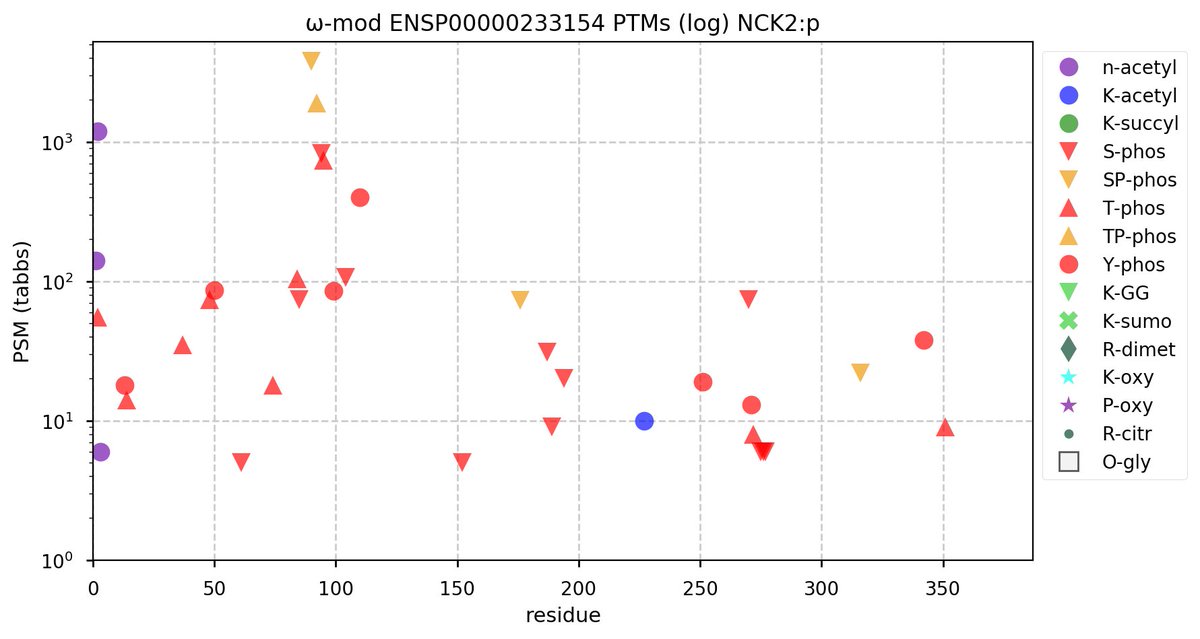
Fri Mar 11 14:39:02 +0000 2022🔗
1 – 302 ☛ 2 – 302 (N-acetyl-Ala, occupancy >99%)
#realNT
Fri Mar 11 14:34:40 +0000 2022🔗
1 – 399 ☛ 2 – 399 (N-acetyl-Ala, occupancy >99%)
#realNT
Fri Mar 11 13:11:01 +0000 2022🔗
1 – 300 ☛ 1 – 300 (N-acetyl-Met, occupancy 100%)
#realNT
Thu Mar 10 19:59:34 +0000 2022🔗
1 – 341 ☛ 2 – 341 (N-acetyl-Ser, 100% occupancy)
#realNT
Thu Mar 10 15:06:20 +0000 2022Modification diagram from PTK2:p, part of the intracellular #NETRIN mechanism. It is also widely referred to as focal adhesion kinase (FAK). The very modest number of GGyl acceptors for such a large, relatively abundant subunit is striking. 🔗
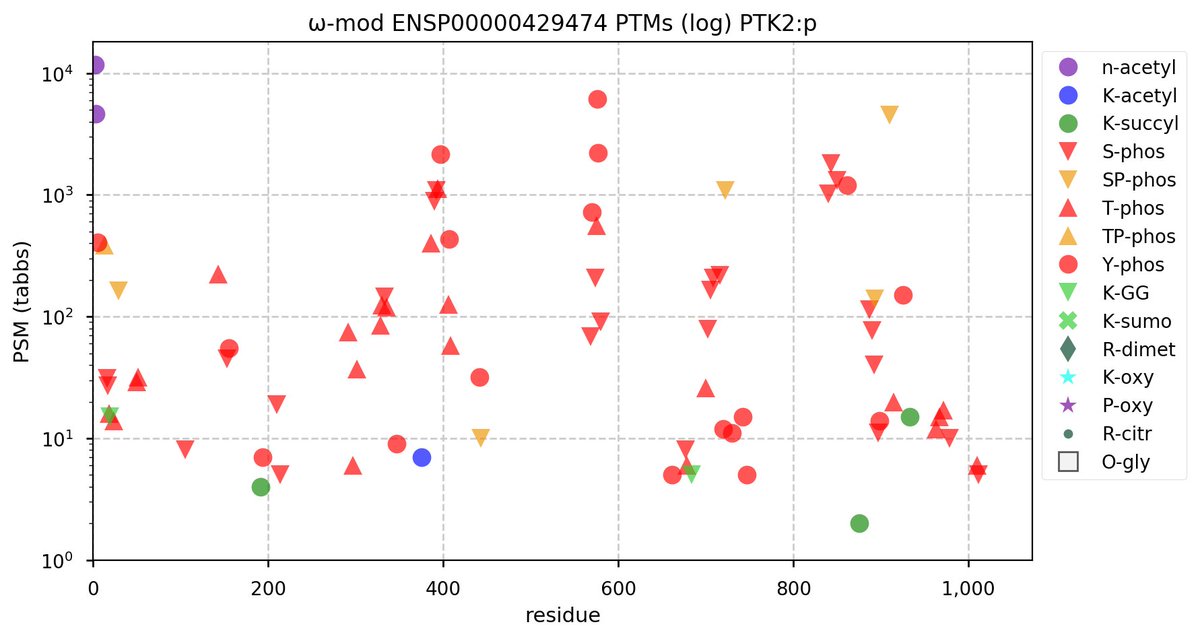
Thu Mar 10 14:58:30 +0000 2022One thing everyone in high-throughput fields should keep in mind all of the time is that machine-learning AI is completely dependent on accurate experimentally derived information. It repeats what it is told, trumpeting bad information with the same zeal as good.
Thu Mar 10 14:53:25 +0000 2022🔗
1 – 3703 ☛ 1 – 3703 (N-acetyl-Met, 80% occupancy)
#realNT
Thu Mar 10 13:36:25 +0000 2022🔗
1 – 1119 ☛ 3 – 1119 (N-acetyl-Thr, 100% occupancy)
#realNT
Thu Mar 10 13:14:34 +0000 2022The I-29 temperature gradient is still very much in a winter pattern (although the 1st Canada goose has been seen in Winterpeg) 🔗
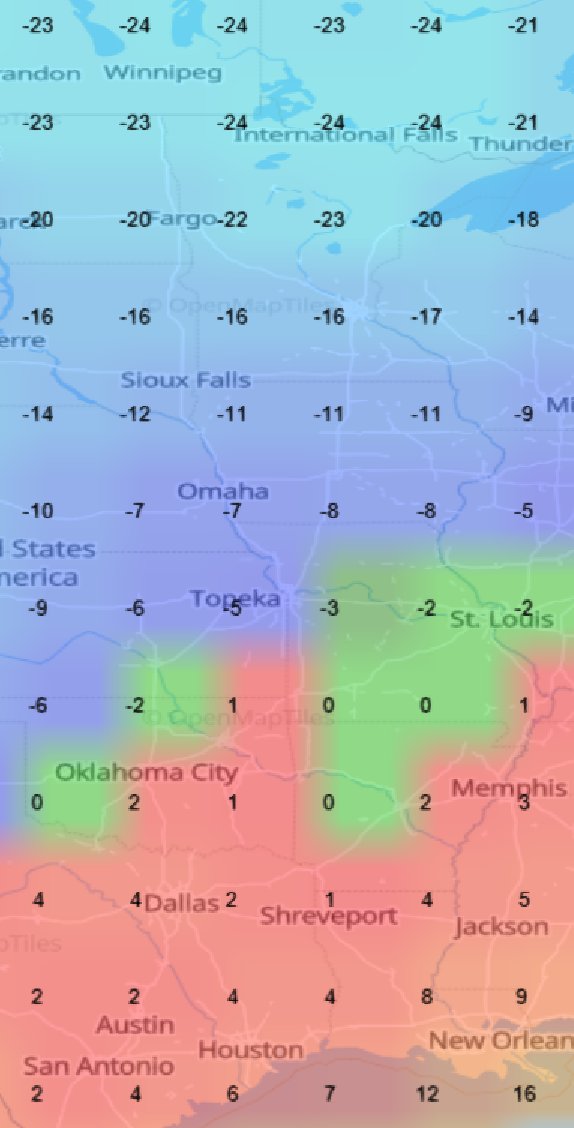
Thu Mar 10 13:05:48 +0000 2022🔗
1 – 4967 ☛ 2 – 4967 (N-acetyl-Ala, 100% occupancy)
#realNT
Wed Mar 09 17:20:46 +0000 2022Human PTK2:p phosphorylation site observation frequency (red) plotted on top of AF predicted aligned error (green) 🔗
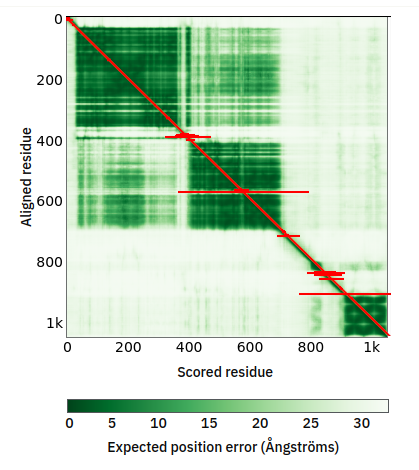
Wed Mar 09 16:05:31 +0000 2022Modification diagrams for #UNC5B, a receptor in the #NETRIN signaling mechanism. It is a type I membrane protein (TM 376-398). Observed phosphoryl acceptors are intracellular; the N-glycosyl acceptor is extracellular. It has a co-translationally removed signal peptide (1-26) . 🔗
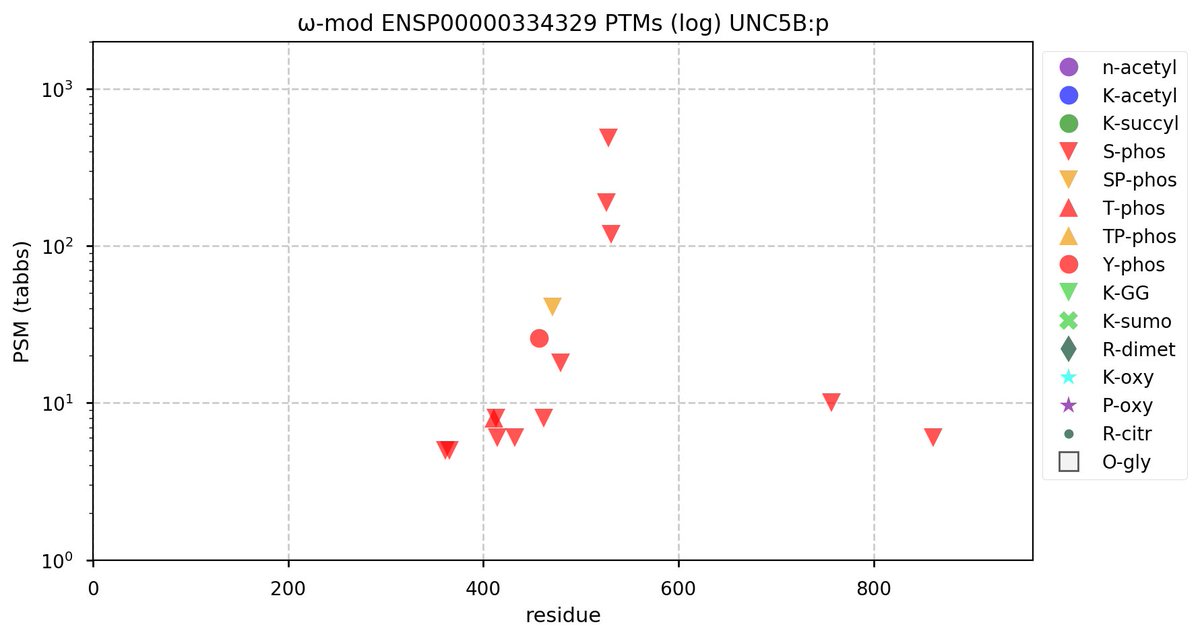
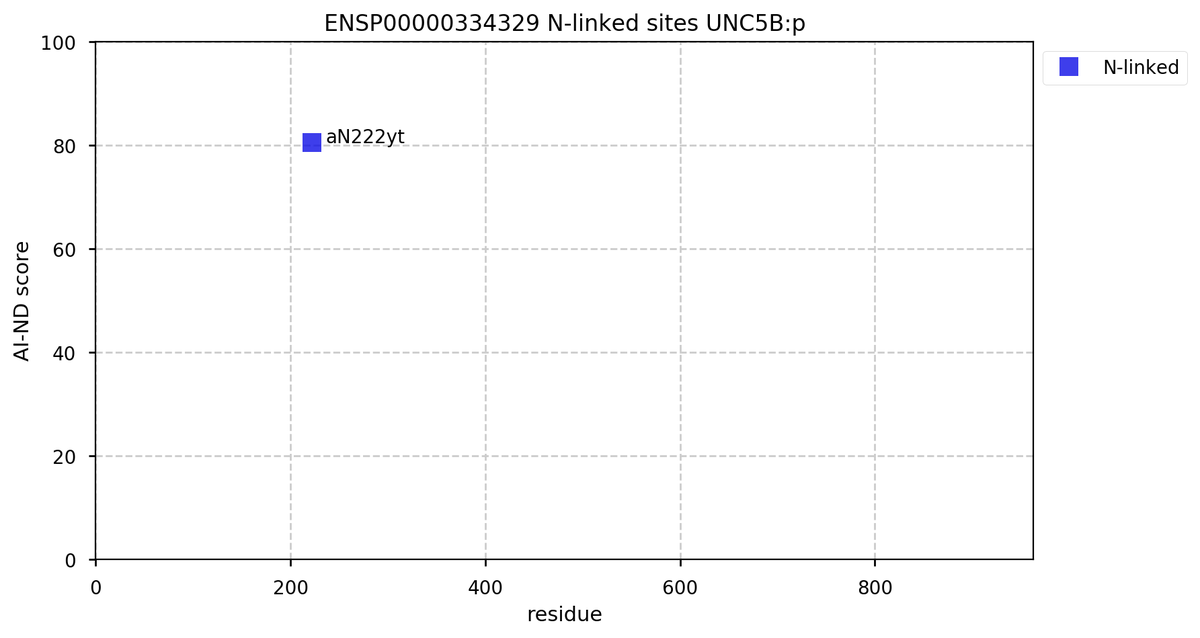
Wed Mar 09 15:00:45 +0000 2022🔗
1 – 323 ☛ 1 – 323 (N-acetyl-Met, occupancy 100%)
#realNT
Wed Mar 09 13:40:55 +0000 2022🔗
1 – 320 ☛ 2 – 320 (N-acetylvaline, occupancy 0.6%)
#realNT
Wed Mar 09 02:29:47 +0000 2022🔗
1 – 892 ☛ 2 – 892 (N-acetylthreonine, >99% occupancy)
#realNT
Tue Mar 08 20:54:16 +0000 2022@UCDProteomics I am the proverbial "choir" for this one.
Tue Mar 08 19:17:18 +0000 2022🔗
1 – 1002 ☛ 2 – 1002 (N-acetylalanine, 100% occupancy)
#realNT
Tue Mar 08 18:09:46 +0000 2022Unlike the other 2 #NETRIN receptors (#UNC5B & #DCC), the extracellular domain of #NEO1 is frequently observed in urine, blood plasma & cerebrospinal fluid.
Tue Mar 08 15:26:28 +0000 2022🔗
Tue Mar 08 14:54:19 +0000 2022@fmeierX @dfg_public Seite nicht gefunden (404)
Tue Mar 08 14:33:56 +0000 2022Modification diagrams for #NEO1, a receptor in the #NETRIN signaling mechanism. It is a type I membrane protein (TM 1105-1127). Observed phosphoryl acceptors are intracellular; N-glycosyl acceptors are extracellular. It has a co-translationally removed signal peptide (1-33) . 🔗
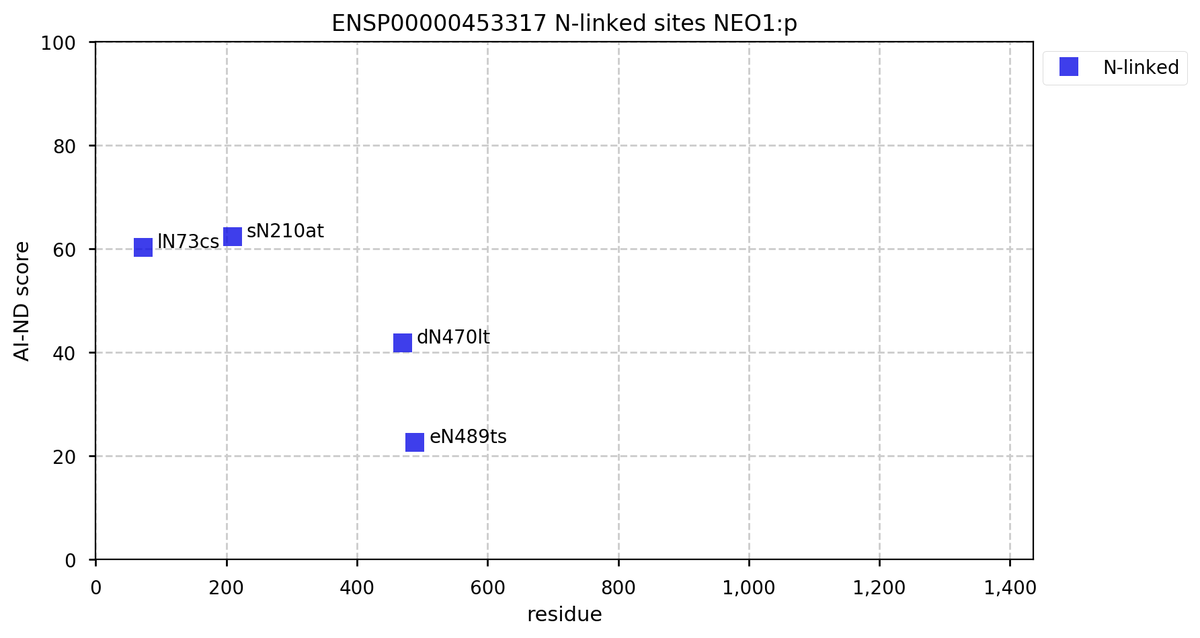
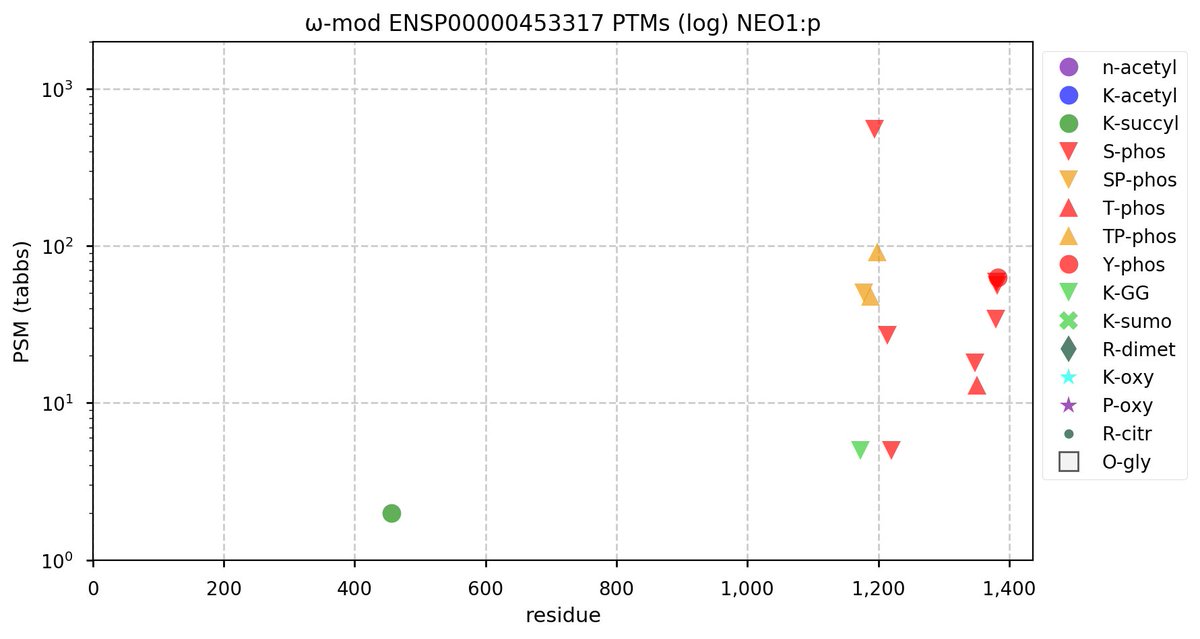
Tue Mar 08 13:40:46 +0000 2022🔗
1 – 403 ☛ 2 – 403 (N-acetylalanine, >99% occupancy)
#realNT
Mon Mar 07 16:15:35 +0000 2022@attilacsordas I'm seeing a lot of large projects where the hypothesis seems to be "maybe machine learning will find something".
Mon Mar 07 14:01:35 +0000 2022Modification diagram for #DCC, a membrane receptor & part of the #NETRIN signaling mechanism. It is a type I membrane protein (TM 1099-1121): observed S+phosphoryl acceptors are intracellular. It has a co-translationally removed signal peptide (either 1-25 or 1-31) . 🔗
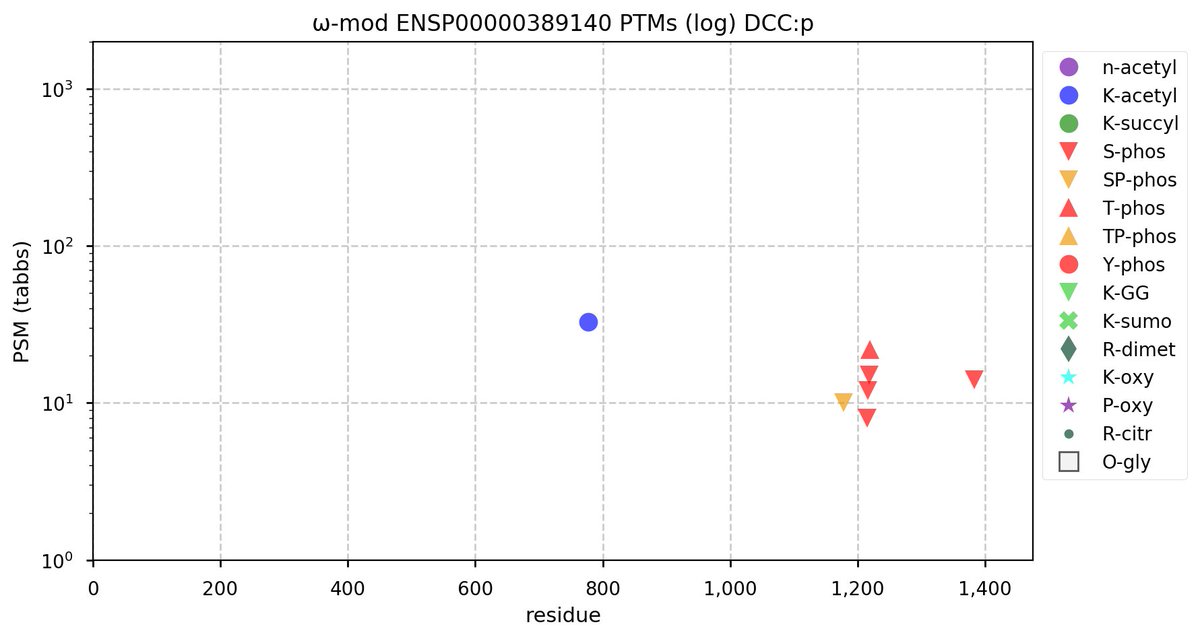
Sun Mar 06 19:42:38 +0000 2022@neely615 @nesvilab If you are going to reanalyze the data, note that they didn't derivatize the cysteine sidechain, so if you want to recover any Cys-containing peptides you should set Cys+cysteinyl (cystine) as a fixed modification.
Sun Mar 06 19:20:59 +0000 2022@neely615 @nesvilab It appears to be an error in the link. The FTP link in PRIDE is to
/pride/data/archive/2022/01/PXD023175
but the data is at
/pride/data/archive/2022/02/PXD023175
Sun Mar 06 17:03:08 +0000 2022@nesvilab The PRIDE FTP archive isn't publicly available yet.
Sun Mar 06 15:37:30 +0000 2022To be clear, I believe that clinically oriented proteomics research is a very good thing. The problem, for me, is the inappropriate linkage of clinical research to selling MS/MS instruments for routine proteomics-oriented clinical lab tests.
Sun Mar 06 15:09:01 +0000 2022Guys my age that I used to have some respect for bang away on this notion as though it made sense & would be "good for the field".
Sun Mar 06 13:44:33 +0000 2022Modification diagram from #NTN1, the most commonly observed soluble initiator of the #NETRIN pathways. The subunit has signal peptide (1-23) removed cotranslationally. 🔗
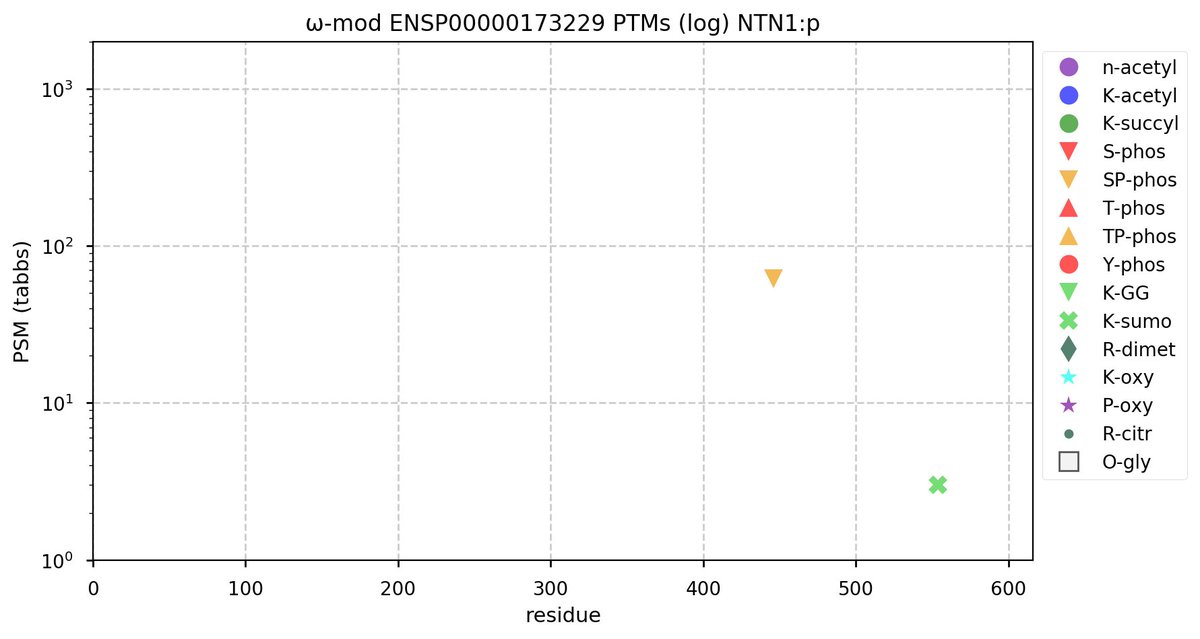
Sat Mar 05 19:25:28 +0000 2022Every time I read an article that yearns for MS/MS proteomics "in the clinic" I can't help but be reminded how the mass spec companies have completely captured the discussion.
Sat Mar 05 15:23:02 +0000 2022As someone who took years of circuit theory courses, the justification for the "wiring diagram" analogy used in cell biology has always evaded me
Sat Mar 05 13:55:57 +0000 2022Most species have a single NOMO gene. Human NOMO1 has two paralogs (NOMO2 & NOMO3) with >95% protein sequence overlap, making it impossible to determine which of the 3 possible proteins is associated with these unusual modifications.
Sat Mar 05 13:55:27 +0000 2022The mouse PTM pattern conserves the intracellular S+phosphoryl acceptors, but not the human GGyl & SUMOyl tagging or extracellular domain phosphorylation. 🔗
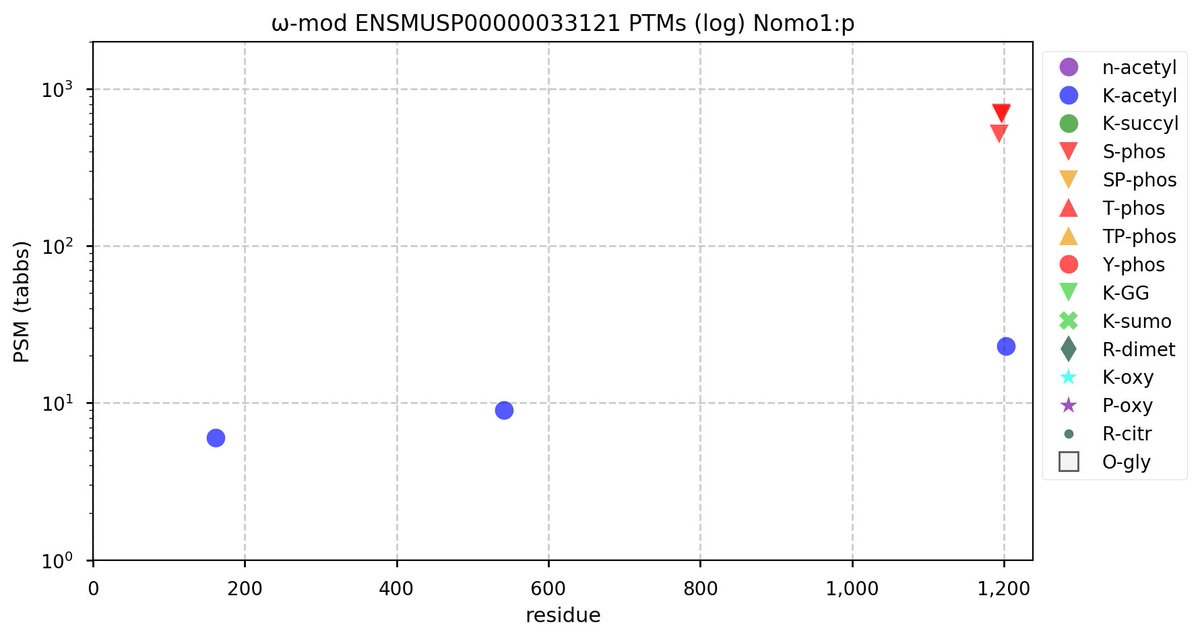
Sat Mar 05 13:54:59 +0000 2022None of the rest of the modifications make any sense at all. They are not consistent with the proposed role of the protein or conserved in other species.
Sat Mar 05 13:54:20 +0000 2022Modification diagram for human #NOMO1, an abbreviation for #NODAL modulator 1. It is a type 1 membrane protein, with a signal sequence domain (1-23) & a TM domain (1156–1176). The intracellular domain (1177-1222) has two S+phosphoryl acceptors, S1202 & S1205. 🔗
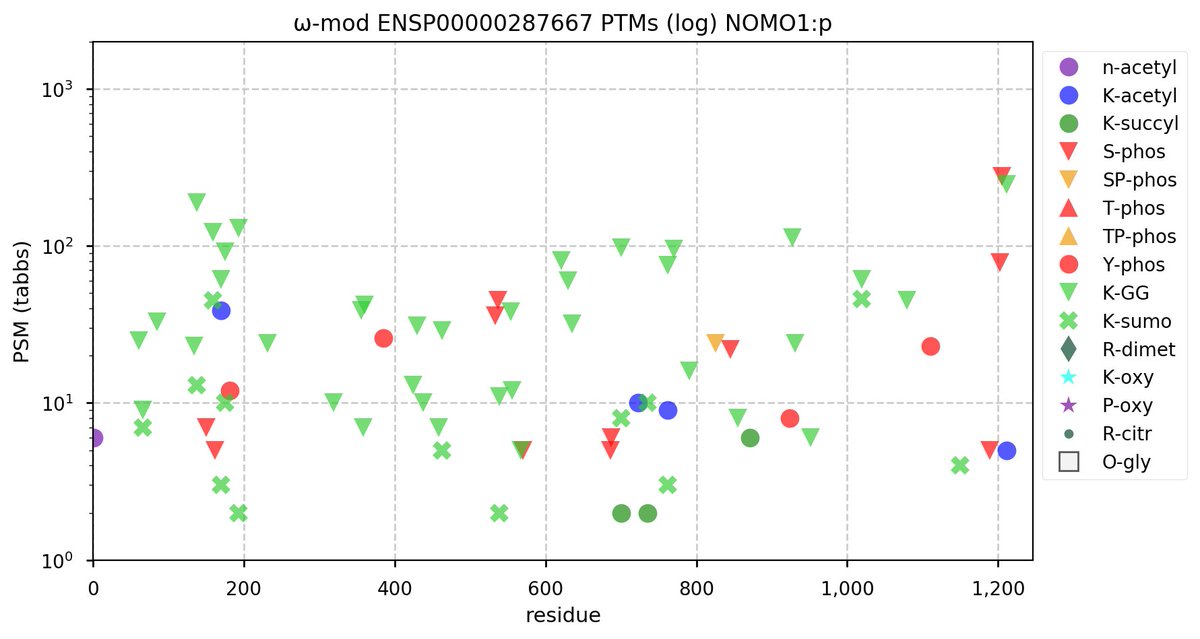
Fri Mar 04 17:19:09 +0000 2022@neely615 @pwilmarth I wouldn't be very concerned about "knowledgebases". Most only select a few papers to use for annotations. The majority of publications are not used.
Fri Mar 04 16:25:32 +0000 2022@pwilmarth Biological information is rarely cut & dried: stating that the phosphorylation is within a domain is more useful than twisting the data into knots to try to put it at one site or another.
Fri Mar 04 16:24:46 +0000 2022@pwilmarth I understand trying to do the best job you can do with the data, but if the data isn't good enough to distinguish between a few options, my attitude has always been to report all of them.
Fri Mar 04 15:35:32 +0000 2022Modification diagram for #TRIM33, a competitive inhibitor of #SMAD2 or #SMAD3 that can modulate the #NODAL mechanism. The protein is much more elaborately decorated than most of the other subunits in this pathway (K+SUMOyl is particularly prominent). 🔗
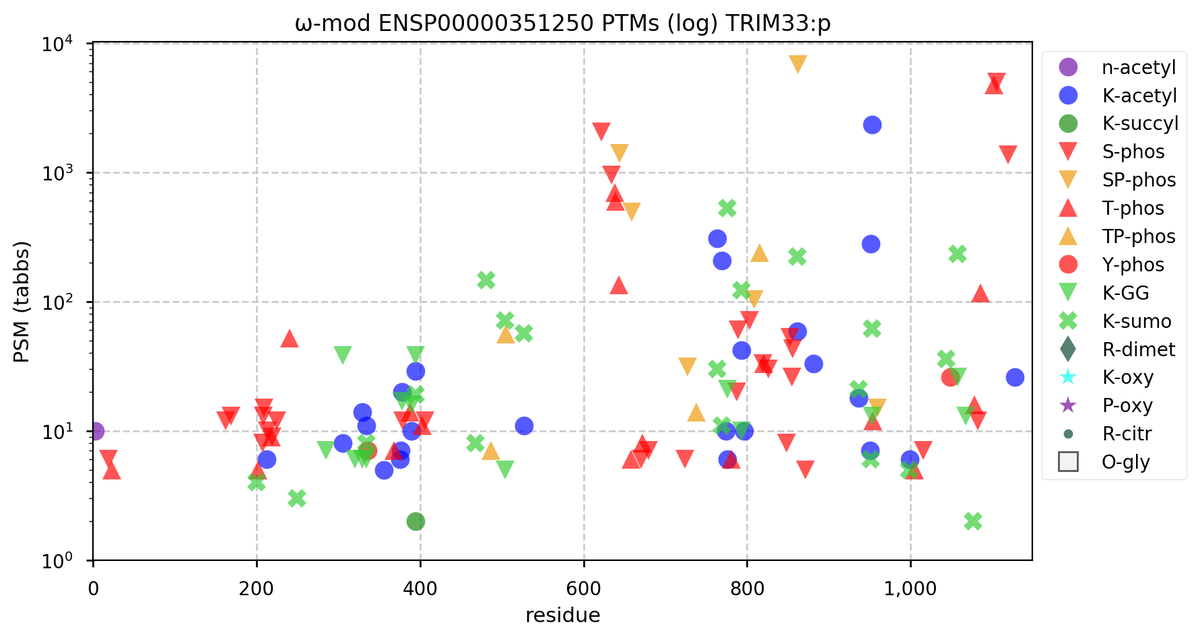
Fri Mar 04 14:05:25 +0000 2022I will never understand the logic of job postings that don't include a salary range.
Fri Mar 04 13:22:42 +0000 2022The I-29 corridor this morning. Fargo is up 12° from yesterday this time & Houston is nearly at the same air temperature as nearby regions of the Gulf. 🔗
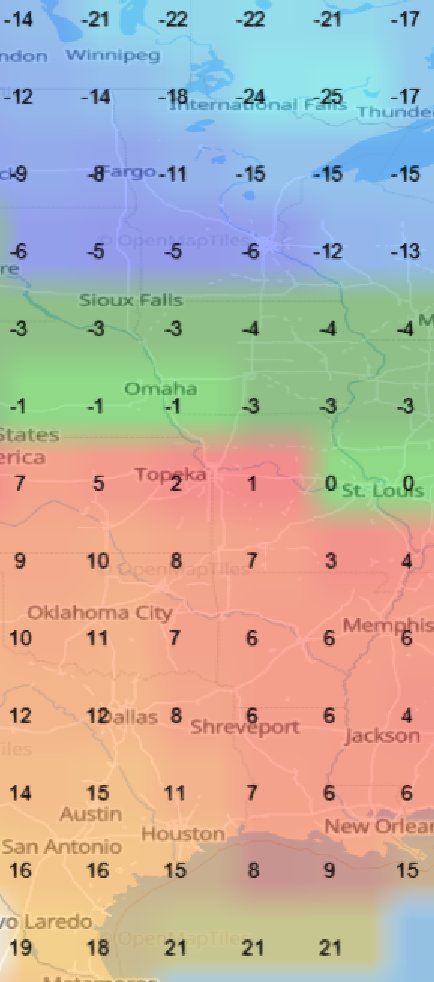
Fri Mar 04 13:03:55 +0000 2022🔗
17 – 757 ☛ 21 – 757
#realNT
Thu Mar 03 18:10:34 +0000 2022@scalzi Maybe it is just Canada, but many library systems here have a longer list, e.g. 🔗 | 🔗
Thu Mar 03 15:26:22 +0000 2022And, just cause I'm still interested in this, a comparison of phosphorylation acceptor site occupancy & the AF structure (you can also see the Ig-like domains in the EC domain) 🔗
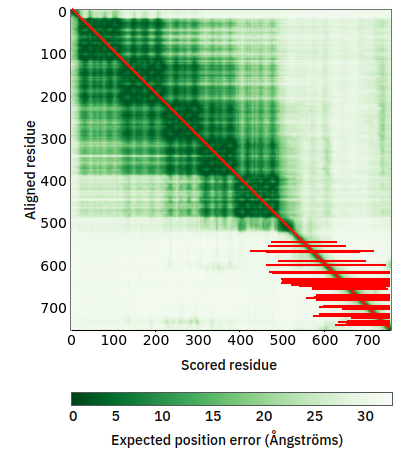
Thu Mar 03 15:18:50 +0000 2022The N-terminal extracellular domain has at least 3 occupied N+glycosyl acceptor sites. It is composed of 5 Ig-like C2-set domains that provide the mechanism for interacting with environment factors. 🔗
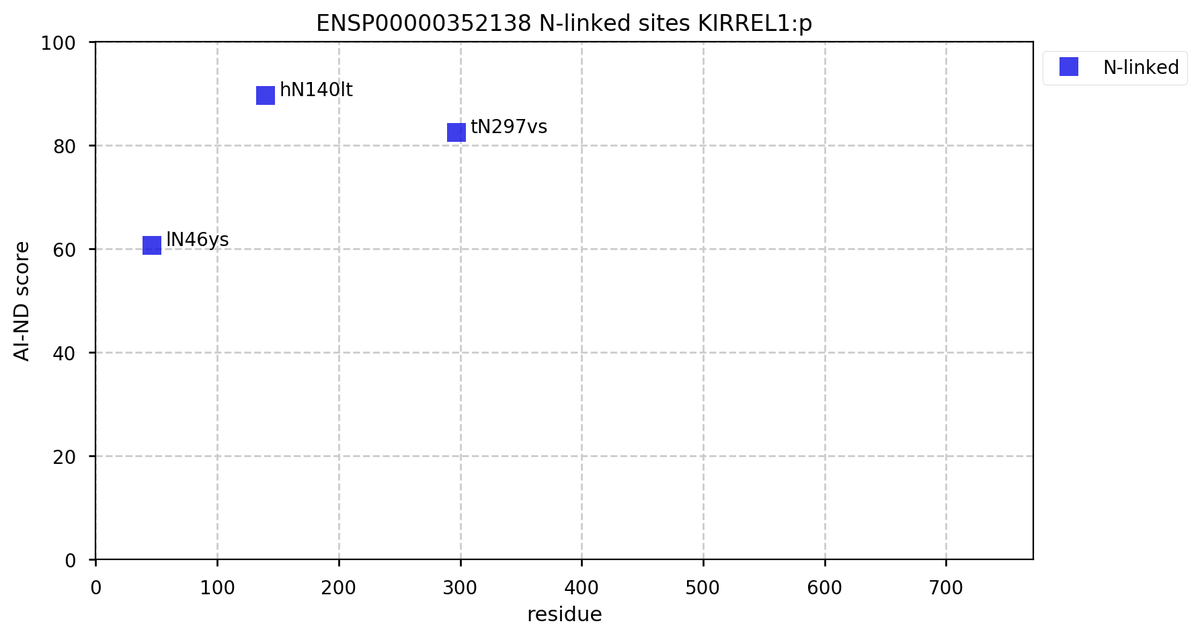
Thu Mar 03 15:15:45 +0000 2022Modification diagram for #KIRREL1, a type I membrane protein involved in signal transduction, but not currently positively associated with a pathway. C-terminal to the TM domain (497-519) is an intracellular domain with a number of high occupancy Y+phosphoryl acceptors 🔗
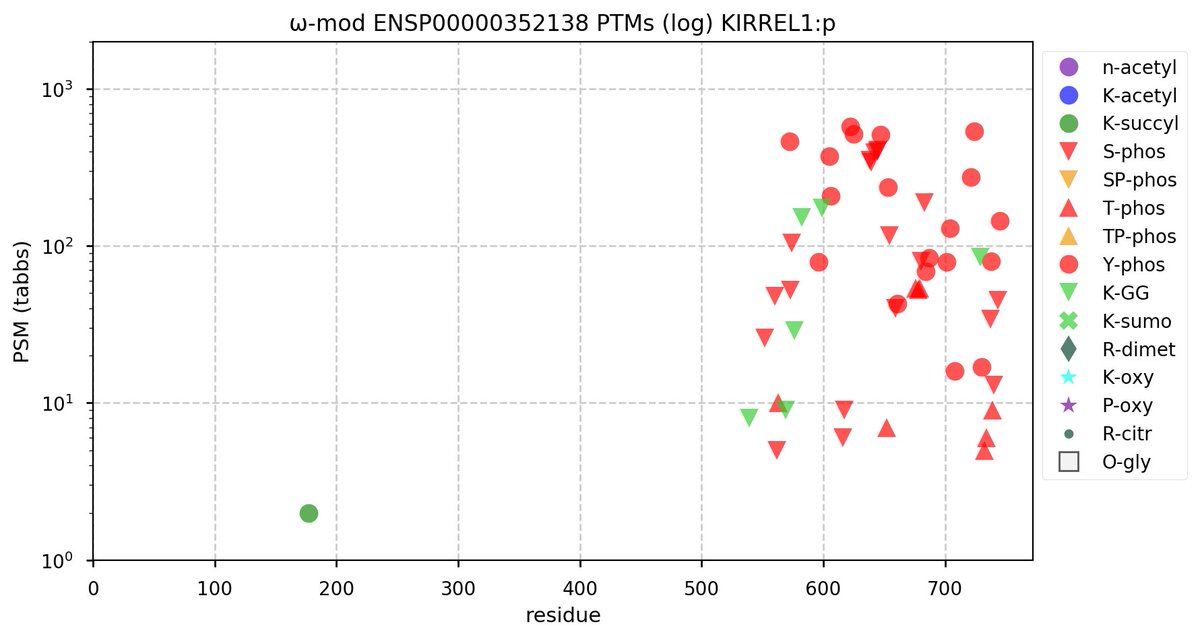
Thu Mar 03 14:15:31 +0000 2022Along the I-29 corridor, the line between winter & spring is moving north, but it is still a sharp line. Hang in there, Fargo! 🔗
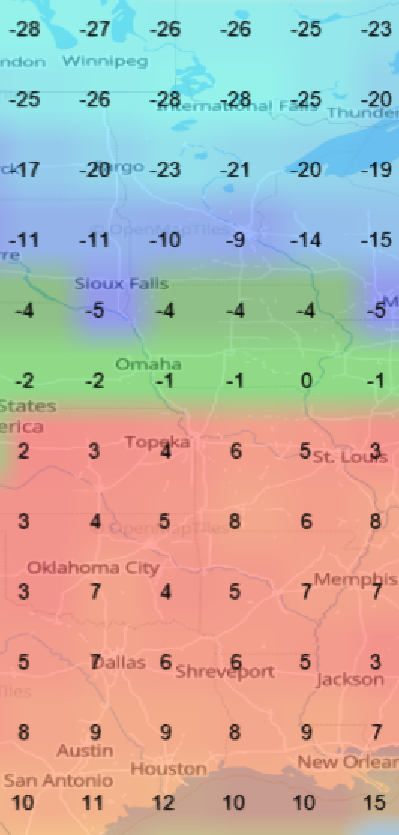
Wed Mar 02 21:34:45 +0000 2022And a final one, for the fences, TP53:p 🔗
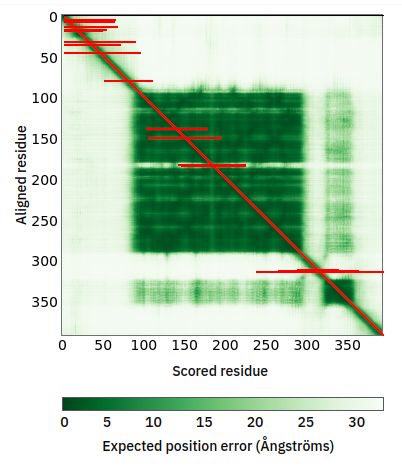
Wed Mar 02 19:19:00 +0000 2022@JoePlatelet @ozgunbabur It actually does have to # of observations built in: the length of the bar is proportional to the log(# of obs).
Wed Mar 02 17:31:37 +0000 2022Seems to hold up reasonably well in more complicated cases where screen resolution is a bit of an issue, like DYSF:p 🔗
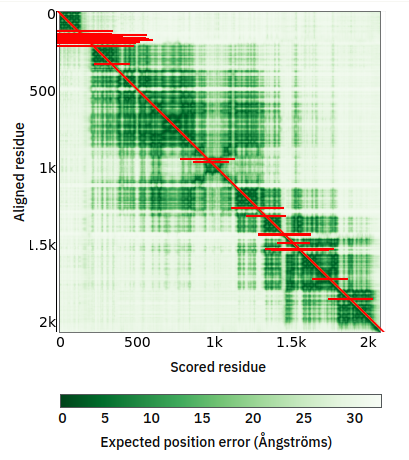
Wed Mar 02 15:43:13 +0000 2022The two sequences have limited observable peptide overlap. 🔗
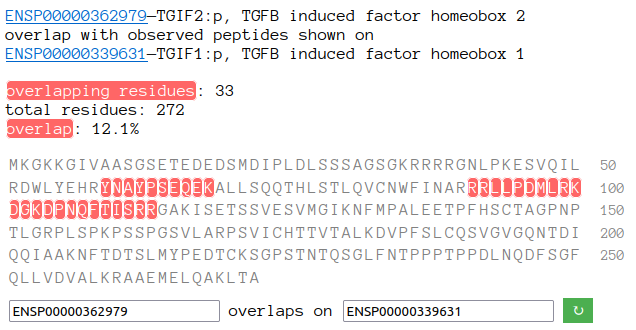
Wed Mar 02 15:41:56 +0000 2022Modification diagram for #TGIF1 & #TGIF2, competitive inhibitors of #SMAD2 that can modulate the #NODAL mechanism. When they bind #SMAD2, it is effectively n longer available for use in the pathway. 🔗
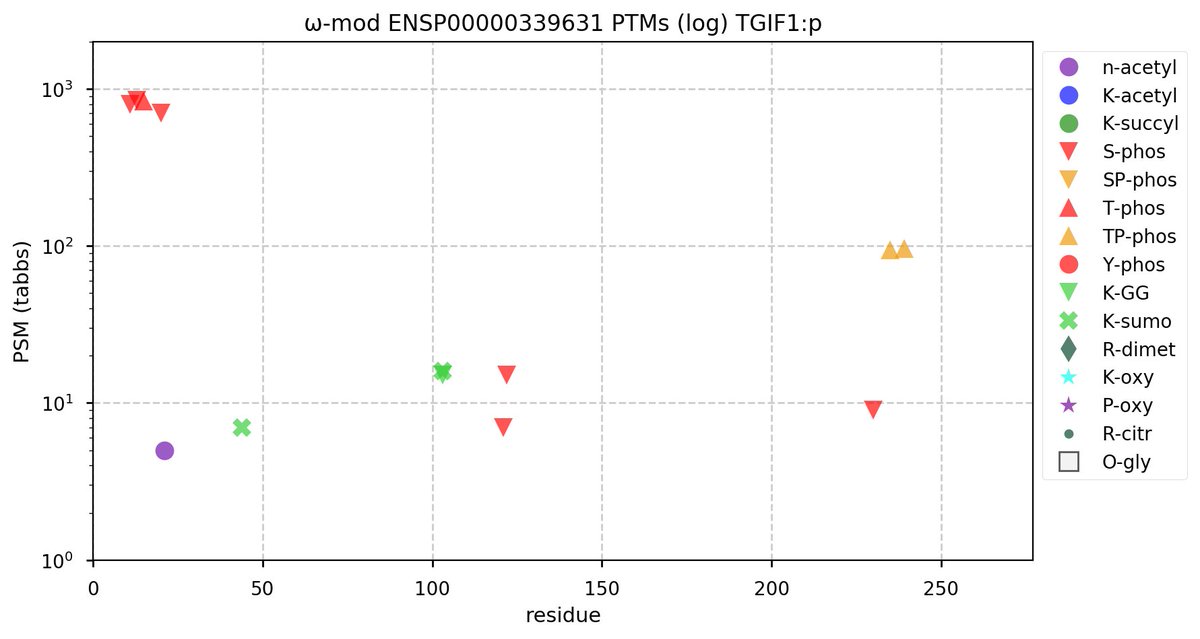
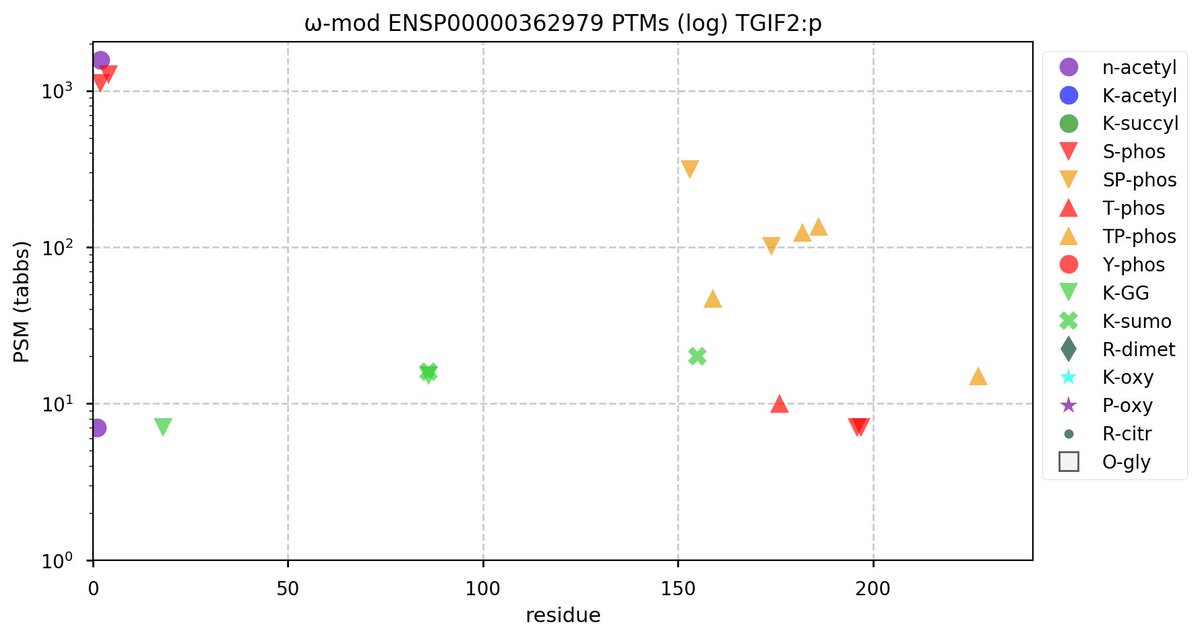
Wed Mar 02 15:23:20 +0000 2022🔗
1 – 540 ⮕ 2 – 540 (N-acetylproline, 2% occupancy)
#realNT
Wed Mar 02 13:16:02 +0000 2022🔗
1 – 475 ⮕ 2 – 475 (N-acetylalanine, 100% occupancy)
#realNT
Wed Mar 02 13:08:48 +0000 2022The cold has retreated to its traditional home at the north end of I-29. 🔗
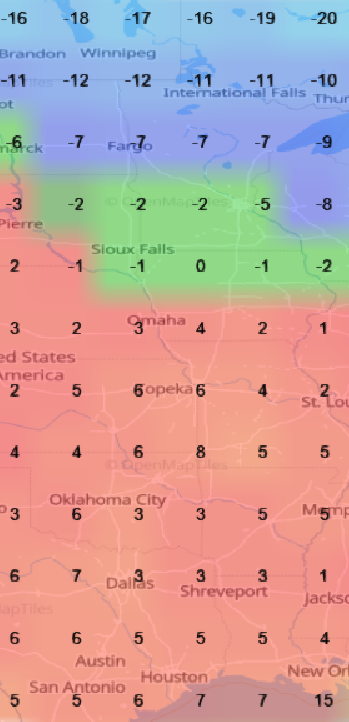
Wed Mar 02 12:20:18 +0000 2022@AchimTreumann @JoePlatelet Thanks.
Wed Mar 02 12:19:52 +0000 2022@dwendscheck Script to make the overlay & then fitting the two together by hand. When I'm happy with it, I'll automate the process.
Tue Mar 01 23:10:07 +0000 2022@FourMohrYears I'm aware of that particular work. I'm just trying to come up with a simple diagram that combines quantitative information about the structure with quantitative information about site-specific PTM observation frequency/occupancy.
Tue Mar 01 20:52:21 +0000 2022Getting there: phosphorylation occupancy overlaid on AF aligned error diagram for WWTR1 🔗
Tue Mar 01 16:14:26 +0000 2022SMAD is an awkward portmanteau of the C. elegans gene name ("Sma") & the flippant fruitfly gene name ("Mothers Against Decapentaplegic").
Tue Mar 01 16:08:59 +0000 2022Modification diagram for #SMAD2 & #SMAD4, intracellular signal transducers in the #NODAL mechanism. When SMAD2 is phosphorylated, it complexes with SMAD4 & enters the nucleus, where it affects transcription factors. 🔗
Tue Mar 01 13:54:36 +0000 2022There is no MS/MS evidence for a protein product generated by translation initiation at M37 (but lots for M1).
Tue Mar 01 13:05:40 +0000 2022🔗
1 – 518 ⮕ 2 – 518 (N-acetylalanine, 100% occupancy)
#realNT
Mon Feb 28 21:17:39 +0000 2022@JoePlatelet And just to be pedantic, the predicted aligned error plot for the AF structure (🔗) pretty nicely tracks with the phosphorylation pattern, with the high occupancy phosphodomains nicely avoiding the high confidence (dark green) parts of the structure. 🔗
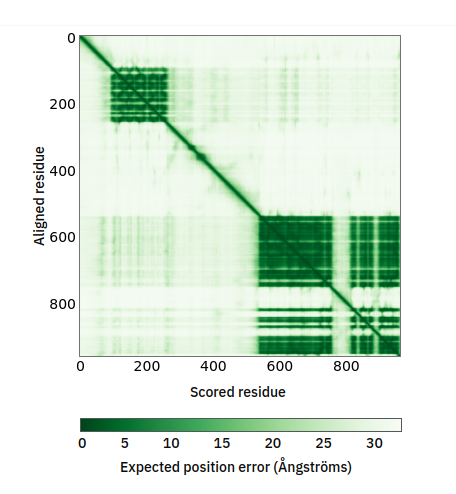
Mon Feb 28 20:23:23 +0000 2022PXD020343 has good examples of what you could expect to detect in human platelet preparations using single, long HPLC runs.
Mon Feb 28 17:35:38 +0000 2022@JoePlatelet And mouse 🔗
Mon Feb 28 17:35:08 +0000 2022@JoePlatelet How about PACS1 for 2 species?
Human 🔗
Mon Feb 28 15:53:48 +0000 2022The name #FURIN has nothing to do with the protein itself. Its gene was originally described as being immediately 5' to the already known feline sarcoma oncogene (FES), so the gene was called Fes Upstream Region (FUR) and the protein was dubbed FURIN.
Mon Feb 28 15:48:31 +0000 2022#FURIN itself is first produced as an ER proprotein, with the signal sequence (1-26) removed cotranslationally & the conversion of Q27 to pyroglutamate. Its propeptide (27-107) must be removed by autolysis for the protease to gain its desired biological activity.
Mon Feb 28 15:41:21 +0000 2022Modification diagram for #FURIN, a type I membrane protein that activates #NODAL by the removal of a propeptide. The few PTMs are in the intracellular domain (739-794) C-terminal to TM domain (716-738). 🔗
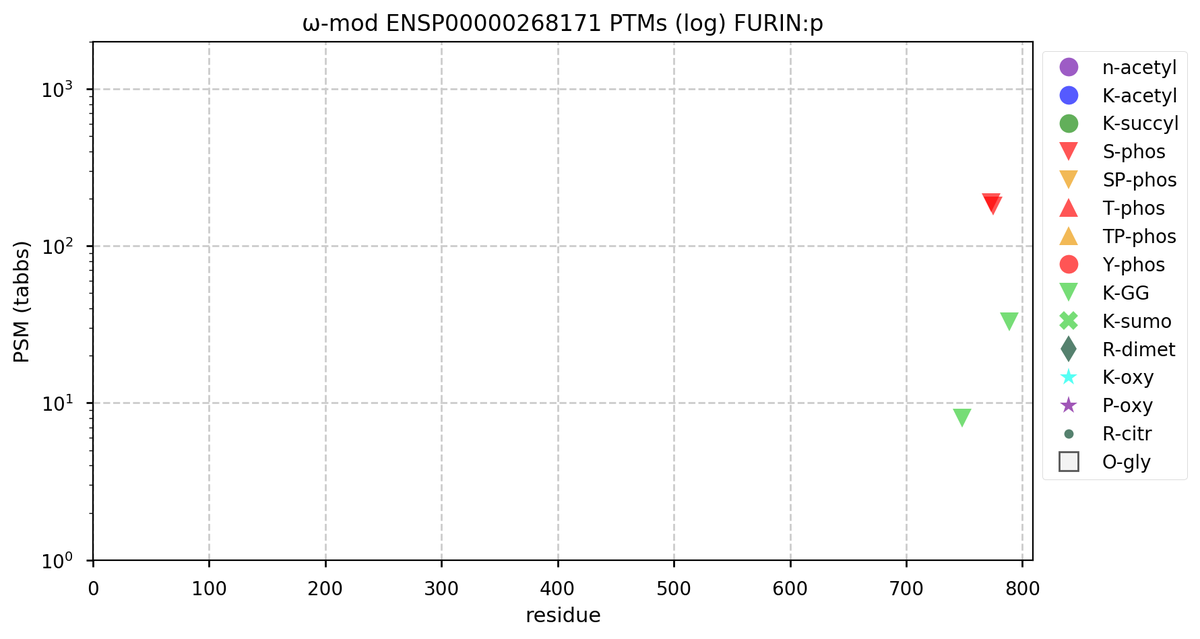
Mon Feb 28 13:04:00 +0000 2022🔗
1 – 544 ⮕ 2 – 544 (N-acetylserine, 100% occupancy)
#realNT
Mon Feb 28 02:30:58 +0000 2022🔗
1 – 518 ⮕ 2 – 518 (N-acetylalanine, 100% occupancy)
#realNT
Sun Feb 27 19:25:22 +0000 2022The GPM has added some temporary access restrictions. 🔗
Sun Feb 27 16:08:50 +0000 2022@AlexUsherHESA I lived on Roosevelt Island when the main connection to Manhattan was a tram. It was nifty, but didn't easily connect into the rest of the public transportation system. In a place like Toronto with no geographical barriers isolating parts of the city, it makes little sense.
Sun Feb 27 13:53:06 +0000 2022Modification diagram for #LEFTY1, an extracellular inhibitor of the #Nodal signaling mechanism. Other than a few GGylation tags, PTMs aren't a major part of this subunit's life cycle. The N-terminal ER-targeting domain―either (1-18) or (1-21)―is removed during translation. 🔗
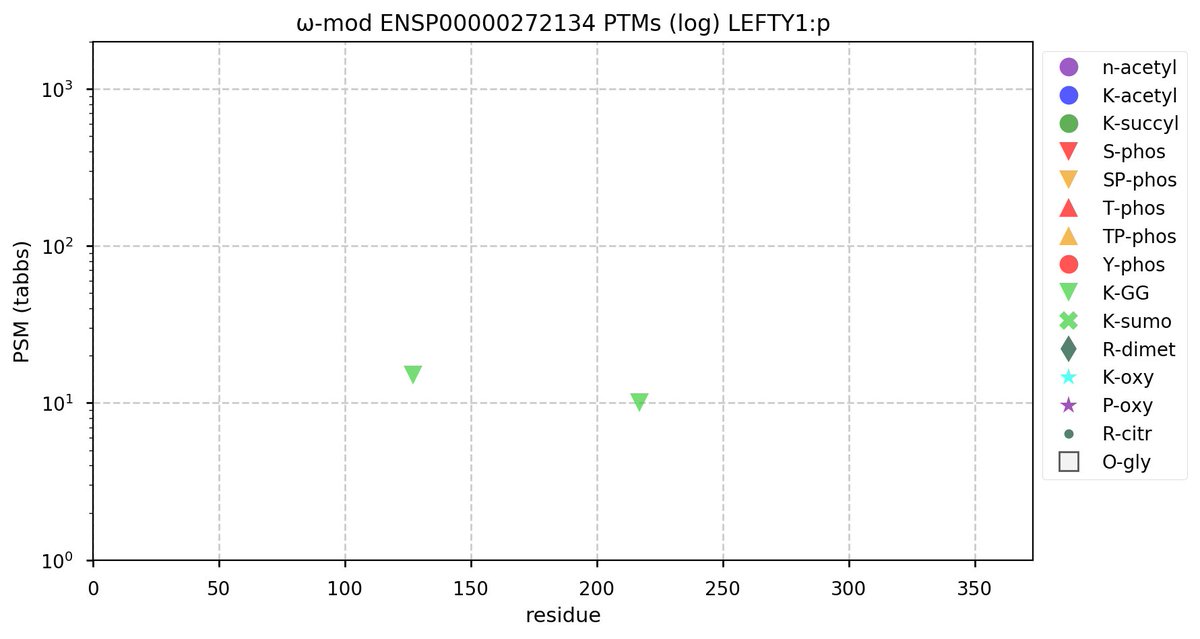
Sat Feb 26 21:11:08 +0000 2022@JoePlatelet What would you consider to be the best method to identify those sites? Some specific sequence motif?
Sat Feb 26 17:30:34 +0000 2022The only real exception to the pattern for 14-3-3 proteins is #SFN, which adds in a significant decoration with R+deimination (citrullination) on the N-terminal domain (1-150) 🔗
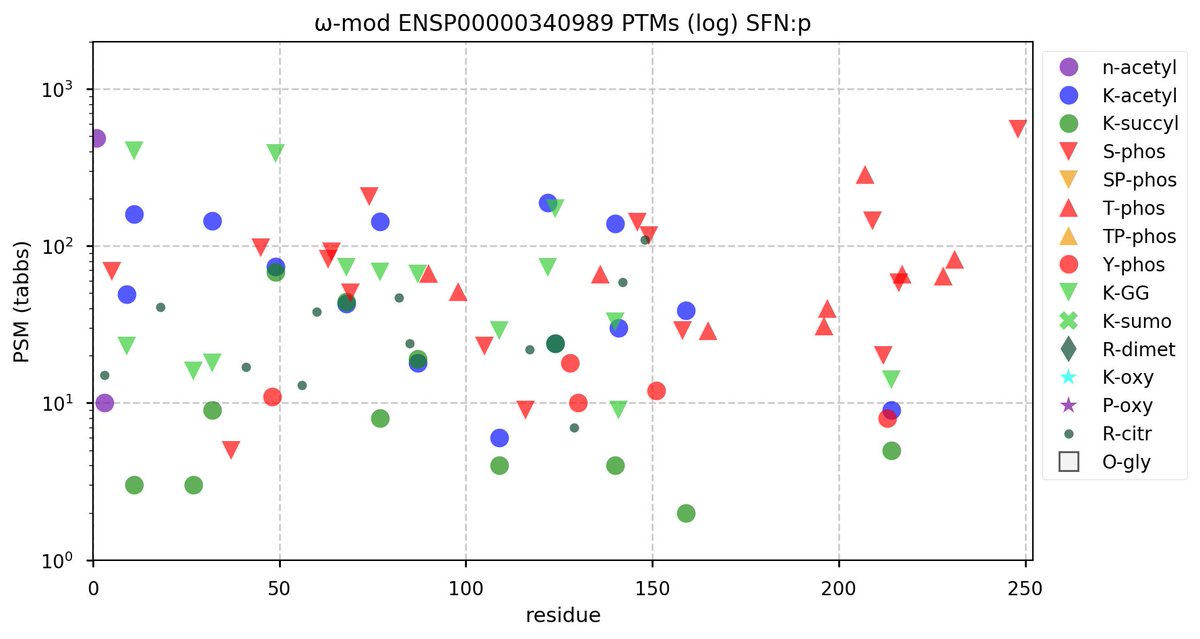
Sat Feb 26 17:20:06 +0000 2022The N-terminus of the protein is ragged: N-acetyl M1, T2 or M3 may be the first residue, in the ratio 1/2/5 (if the MS data is to be believed). None of the other 14-3-3 proteins show this pattern. The pattern is similar in mouse, but the ratio is different: 1/4/0.5.
Sat Feb 26 17:10:47 +0000 2022Modification diagram for #YWHAB, part of the signalling system involving the #mTORC2 structure. Like the other 14-3-3 proteins, a mix of phosphorylation & alternating acetylation/GGylation/succinylation patterns give this small protein lots of scope. 🔗
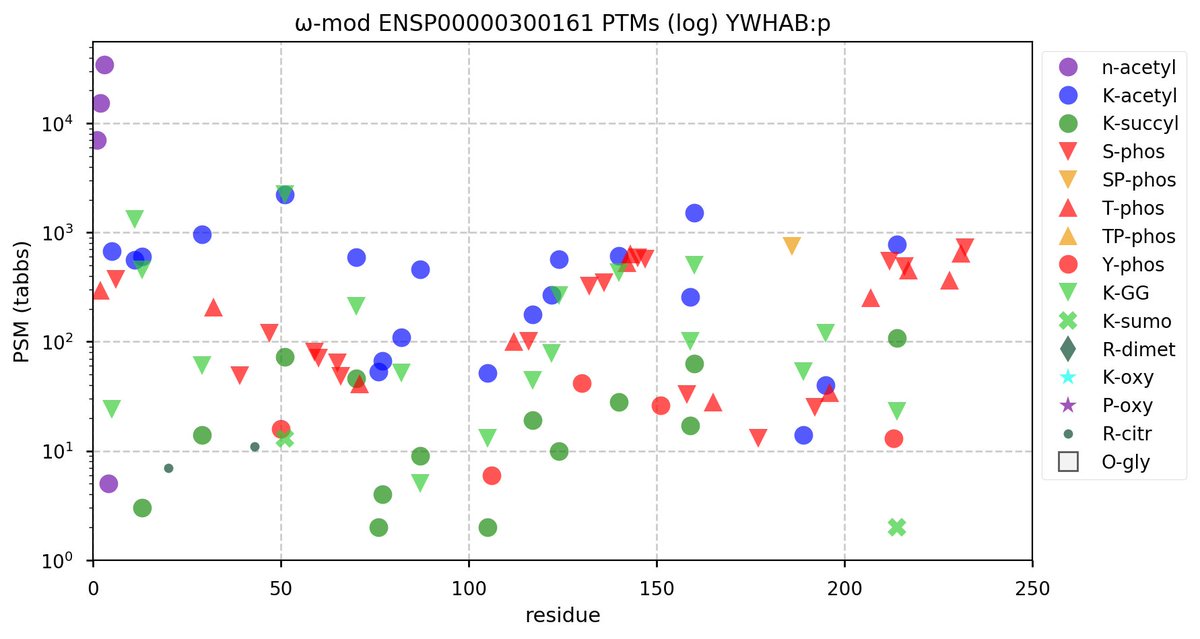
Sat Feb 26 13:56:26 +0000 2022🔗
1 – 328 ⮕
1 – 328 (N-acetylmethionine, 100% occupancy)
or
3 – 328 (N-acetylserine, 100% occupancy)
#real_NT
Fri Feb 25 17:32:55 +0000 2022This S+phosphoryl rich regulatory domain corresponds to the loopy part on the left in the AF "structure" below 🔗
Fri Feb 25 17:25:25 +0000 2022Modification diagram for #DEPTOR, a subunit in the #mTORC2 structure. The prominent grouping of S+phosphoryl acceptors in the center of the sequence (234-299) is nicely snuggled in between the 2nd DEP domain (145-219) the subsequent PDZ domain (330-407). 🔗
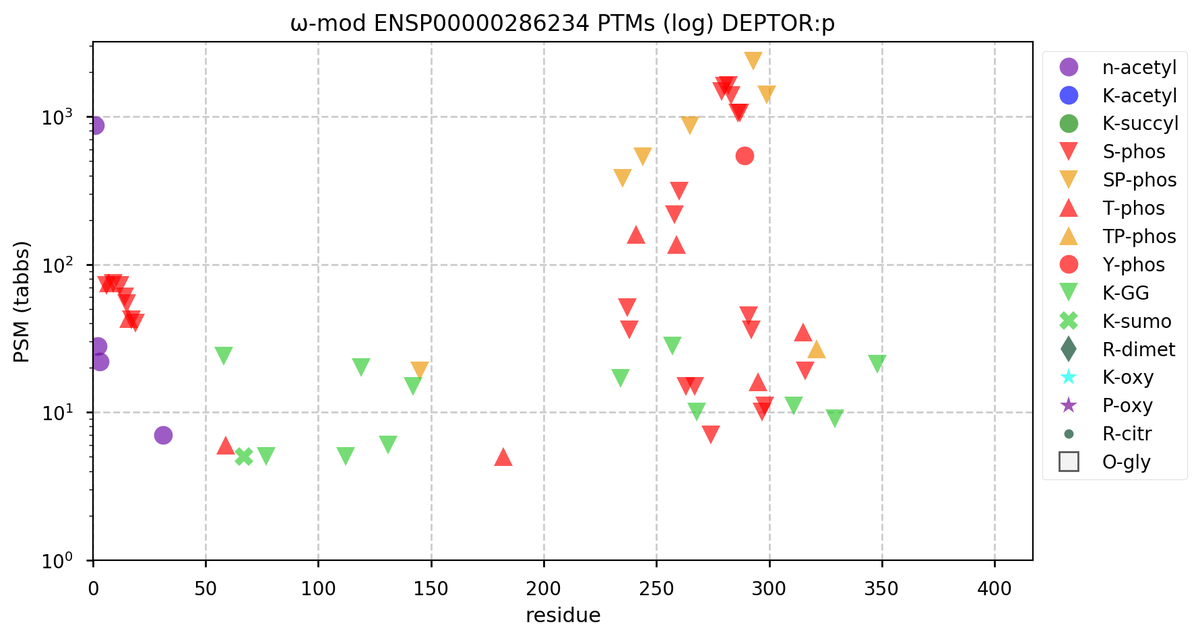
Fri Feb 25 16:02:39 +0000 2022@macro_momo To me this is a geeky form of "stolen valor": "stolen authority"
Fri Feb 25 16:01:10 +0000 2022@macro_momo The problem with that approach is that there may be (& should be) hundreds of data sets supporting a particular PTM. That form of citation would allow one to simply select the highest profile paper in the list & use it as your citation for the PTM.
Fri Feb 25 15:11:56 +0000 2022Is it fair to cite the manuscript associated with a data set as the source of a PTM assignment if that assignment was not explicitly made in that manuscript? And if so, how?
Fri Feb 25 15:09:52 +0000 2022🔗
1 – 187 ⮕ 2 – 187 (N-acetylalanine, 100% occupancy)
#real_NT
Fri Feb 25 13:44:38 +0000 2022🔗
1 – 691 ⮕ 1 – 691 (N-acetylmethionine, >99% occupancy)
#real_NT
Fri Feb 25 13:26:05 +0000 2022🔗
1 – 418 ⮕ 1 – 418 (N-acetylmethionine, 64% occupancy)
#real_NT
Fri Feb 25 01:46:04 +0000 2022Seems to be holding, although I'm not sure how well the list of IP ranges really corresponds to the target geography.
Thu Feb 24 21:26:10 +0000 2022It is a bit of a pain to block a country using Apache HTTPD, but I think I've got it working.
Thu Feb 24 17:56:55 +0000 2022🔗
1 – 81 ⮕ 1 – 81 (N-acetylmethionine, 19% occupancy)
#real_NT
Thu Feb 24 17:50:27 +0000 2022🔗
1 – 262 ⮕ 1 – 262 (N-acetylmethionine, >99% occupancy)
#real_NT
Thu Feb 24 17:46:12 +0000 2022🔗
1 – 701 ⮕ 1 – 701 (N-acetylmethionine, >99% occupancy)
#real_NT
Thu Feb 24 17:39:00 +0000 2022@labs_mann @BludauIsabell I agree with these results. Many PTMs, particularly phosphorylation, have an uncanny knack of avoiding regions with solved 3D structures, preferring to inhabit the much more interesting "dark proteome".
Thu Feb 24 17:18:36 +0000 2022🔗
1 – 149 ⮕ 1 – 149 (N-acetylmethionine, 95% occupancy)
#real_NT
Thu Feb 24 17:07:00 +0000 2022🔗
1 – 1045 ⮕ 2 – 1045 (N-acetylglycine, 99% occupancy)
#real_NT
Thu Feb 24 16:40:58 +0000 2022Modification diagram for #mSIN1 (#MAPKAP1), a subunit in the #mTORC2 structure. Relatively few PTMs, with a short riff of K+acetyl & S+phosphoryl acceptors just N-terminal of the middle CRIM domain (139-267). 🔗
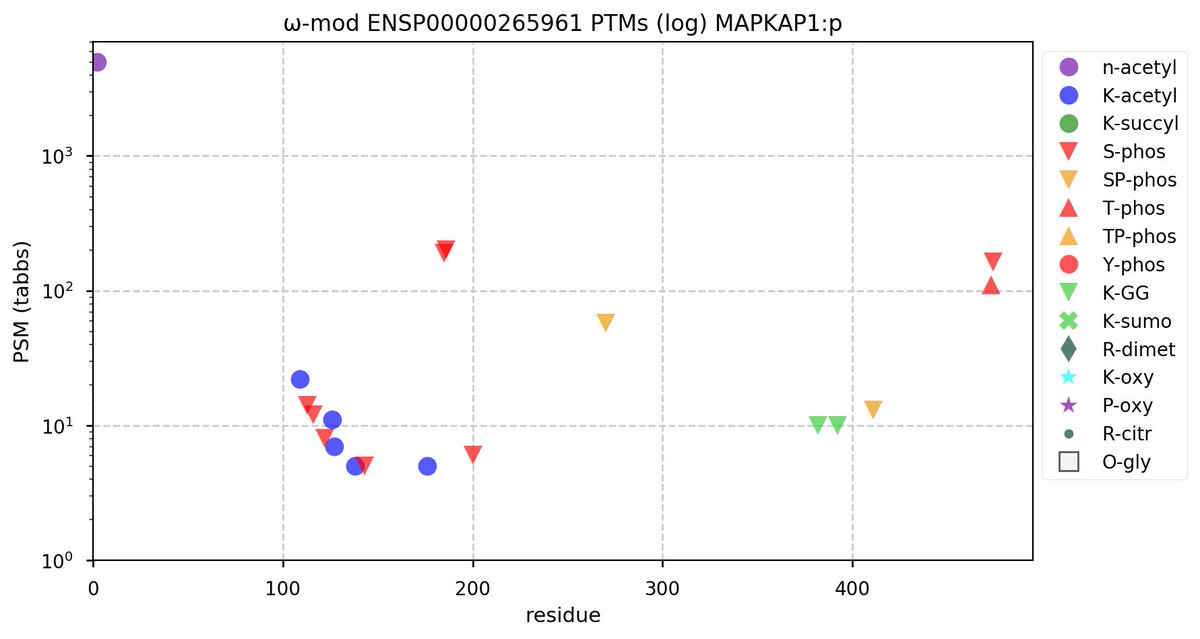
Thu Feb 24 15:11:37 +0000 2022🔗
1 – 305 ⮕ 1 – 305 (N-acetylmethionine, 98% occupancy)
#real_NT
Thu Feb 24 13:08:48 +0000 2022Another brisk morning along the I-29. Only the Gulf of Mexico is holding the line on warmer temps. 🔗
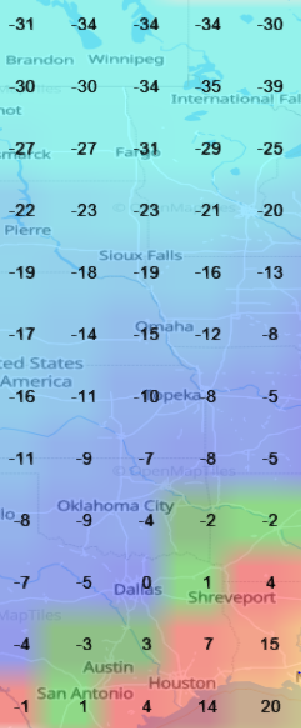
Wed Feb 23 19:30:46 +0000 2022@neely615 @byu_sam @pwilmarth @chrashwood 🔗 is an example of this approach: the GF in MS-GF.
Wed Feb 23 16:26:37 +0000 2022@neely615 I think it is "Deep" if it is DDA & "Ultra" if it is DIA, but there is quite a bit of flexibility in the literature's use of the terms.
Wed Feb 23 16:02:07 +0000 2022Even though their naming suggests significant similarity, the two subunits have no overlap in their observed tryptic peptides 🔗
Wed Feb 23 15:59:59 +0000 2022Madagascar getting hit yesterday by yet another tropical storm (Cyclone Emnati) 🔗
Wed Feb 23 15:51:56 +0000 2022Modification diagram for #PROCTOR1 (#PRR5) & #PROCTOR2 (#PRR5L), alternative small subunits in the #mTORC2 structure. There are significant differences in their S+phosphoryl acceptor patterns. 🔗
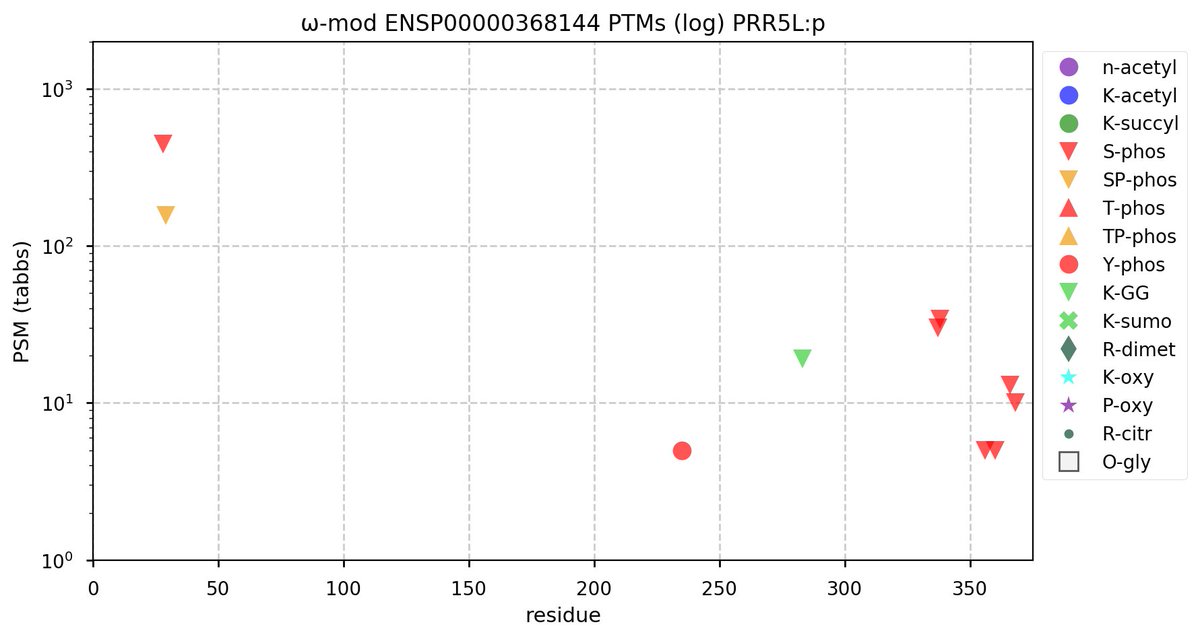
Wed Feb 23 15:42:19 +0000 2022SCP finally moves into the "Ultra" era: PXD024043.
Wed Feb 23 13:45:29 +0000 2022@byu_sam @pwilmarth @chrashwood The target-decoy simulation is an under powered heuristic: it will always have significant, implementation-specific variability. It is better than the system it replaced, but it doesn't give rock solid results.
Wed Feb 23 13:28:39 +0000 2022🔗
1 – 702 ⮕ 1 – 702 (N-acetylmethionine)
#real_NT
Tue Feb 22 16:02:00 +0000 2022Modification diagram for #RICTOR, a large subunit of the #mTORC2 structure. The N-terminal domain is dominated by Armadillo tandem repeats, while the C-terminal phospho-domain looks to be the fun part. 🔗
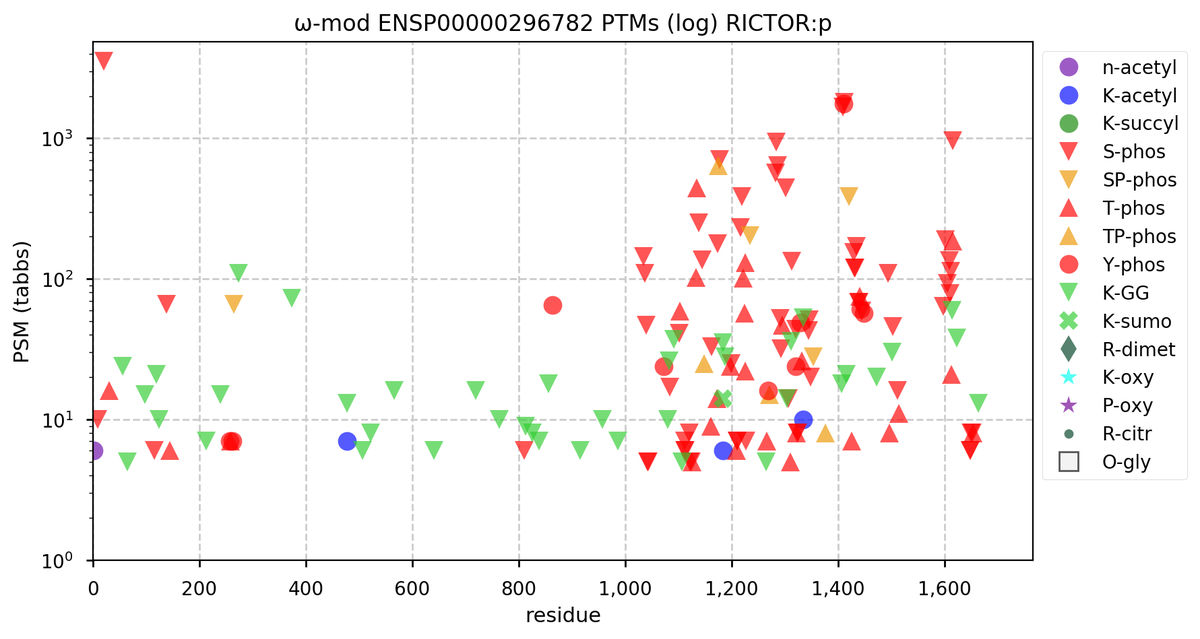
Tue Feb 22 14:08:56 +0000 2022🔗
1 – 342 ⮕ 2 – 342 (N-acetylalanine)
#real_NT
Tue Feb 22 03:31:22 +0000 2022PXD024807 isn't so bad, either. Different, but not bad.
Tue Feb 22 01:27:00 +0000 2022Gave up watching the sad spectacle of the Canadian House of Commons trying to vote for to ratify emergency powers via a mishmash of crap apps, POTS & yelling. GoC has problems dealing with any sort of tech (except for spending money on it, which it does generously).
Tue Feb 22 01:08:43 +0000 2022As phosphorylation data goes, PXD031405 is pretty good.
Mon Feb 21 16:48:38 +0000 2022While these two proteins share some MHC class 1 peptides, they only share one observable tryptic peptide. 🔗
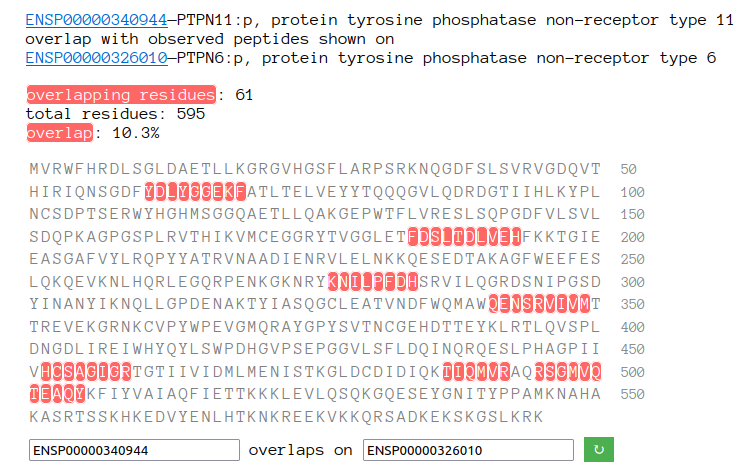
Mon Feb 21 16:46:43 +0000 2022Modification diagrams for #PTPN6 (#SHP1) & #PTPN11 (#SHP2), tyrosine phosphatases involved in regulating #JAK_STAT mechanisms. Both show significant numbers of Y+phosphoryl acceptors themselves. /1 🔗
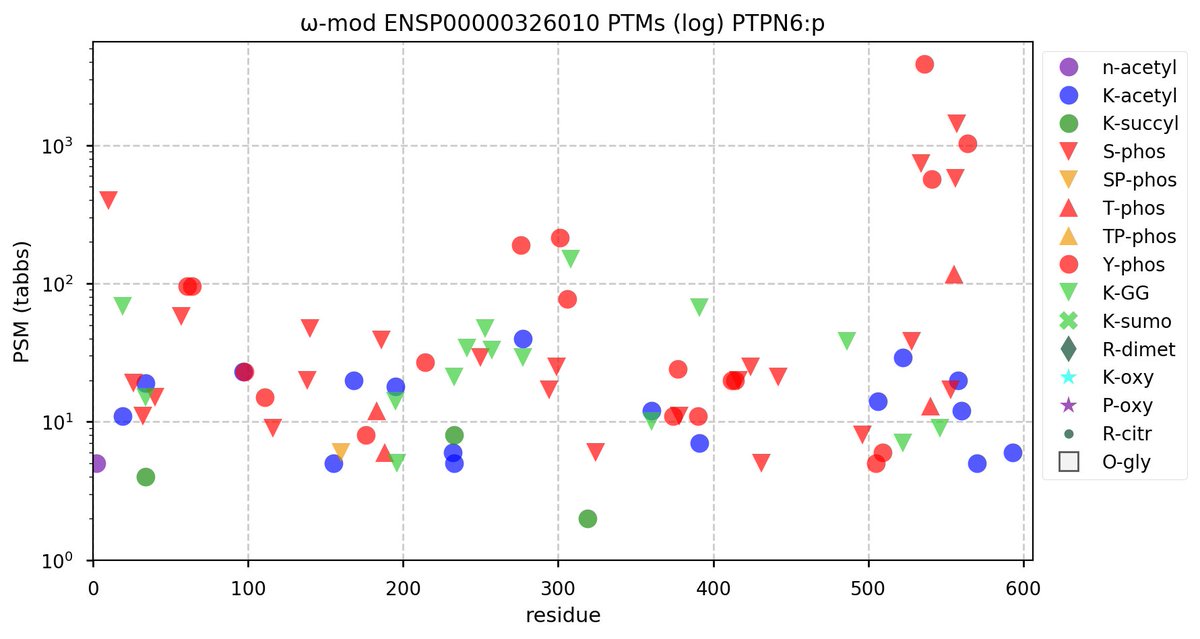
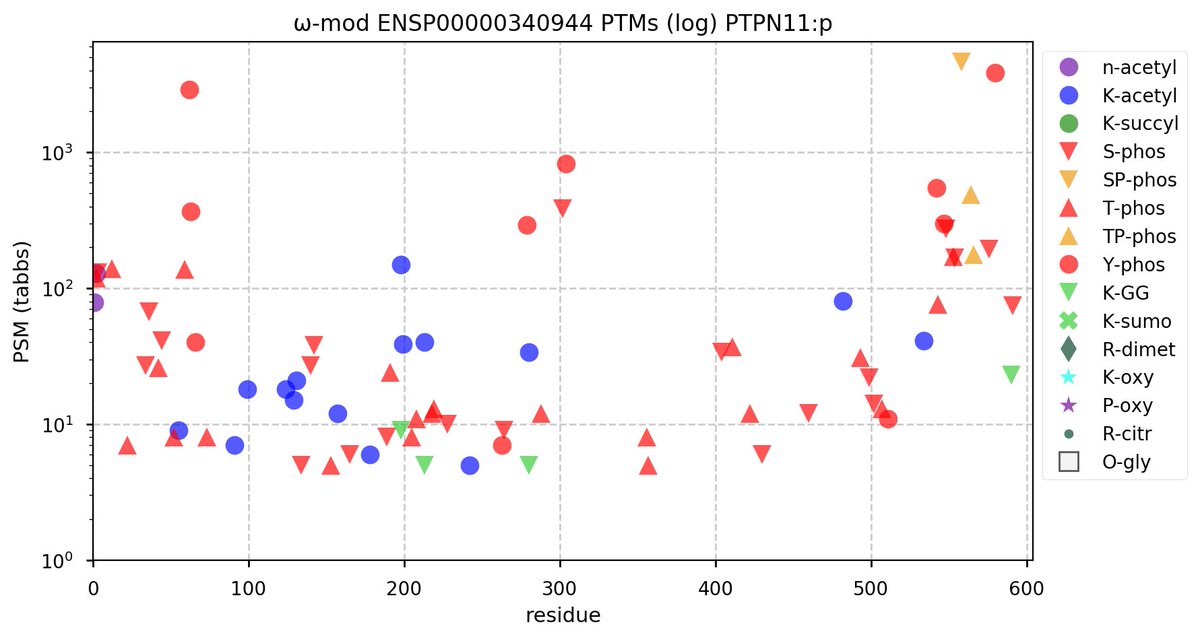
Mon Feb 21 15:09:42 +0000 2022@kadzuis @dexivoje Go ahead and be excited (& don't get me started about fuddy-duddy govt. agency projects).
Mon Feb 21 14:50:03 +0000 2022The I-29 temperature gradient this morning: Houston is looking good today ... 🔗
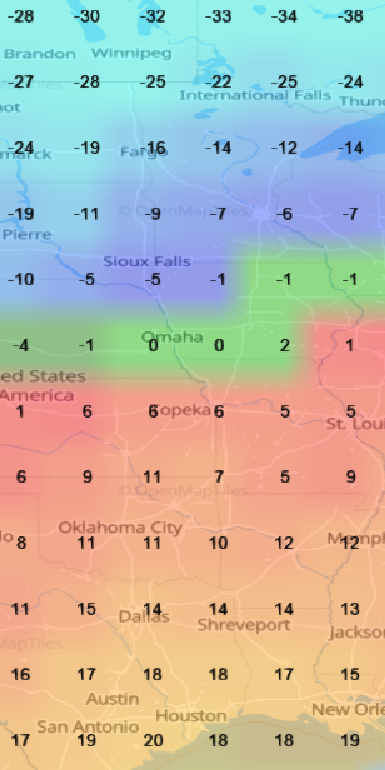
Mon Feb 21 14:35:24 +0000 2022@kadzuis @dexivoje As a few traveler, do what you have to do. Most users assume that on-line scientific resources are publicly funded, even though that is often not the case.
Mon Feb 21 14:11:25 +0000 2022🔗
1 – 759 ⮕ 2 – 759 (N-acetylated)
#real_NT
Sun Feb 20 18:18:16 +0000 2022@lacour_c @LindorffLarsen Norway & Sweden may be less enthusiastic about their inclusion in the consortium, given that Denmark's contribution was rather limited ...
Sun Feb 20 15:24:37 +0000 2022A framework for understanding the phenomenon of alternating PTMs at specific acceptor sites is one of the many missing pieces of understanding with respect to the practical functioning of proteins in vivo. /4
Sun Feb 20 15:11:51 +0000 2022None of the PIAS's show evidence of acetylation as an alternate modification at SUMOylation/ubiquitination acceptor sites. /3
Sun Feb 20 15:07:55 +0000 2022Like the other PIAS's, these subunits have very limited overlap of observable peptide sequences (in this case a single MHC class 1 peptide) /2 🔗
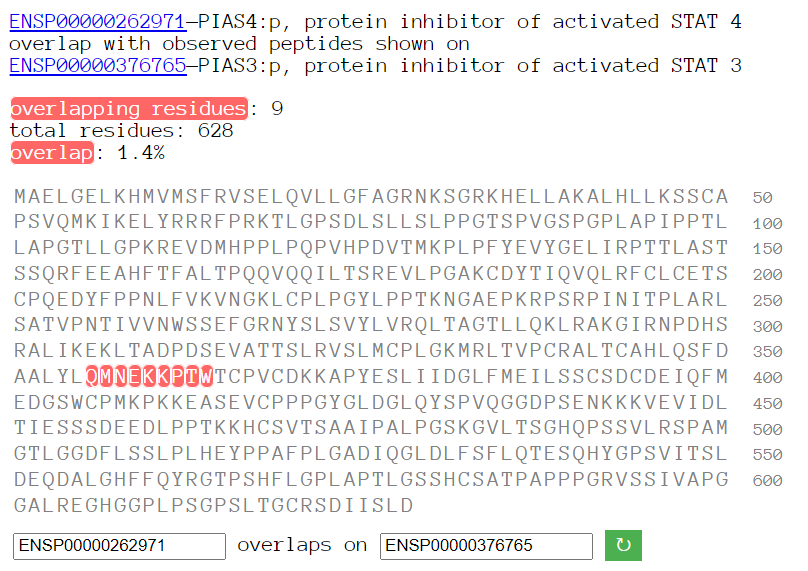
Sun Feb 20 15:05:53 +0000 2022Modification diagrams for #PIAS3 & #PIAS4, involved in #JAK_STAT mechanisms. They are Protein Inhibitors of Activated Stat, that act via STAT SUMOylation. Like #PIAS1 & #PIAS2, they have an unusual number of SUMOylation/ubiquitin acceptor sites themselves. /1 🔗
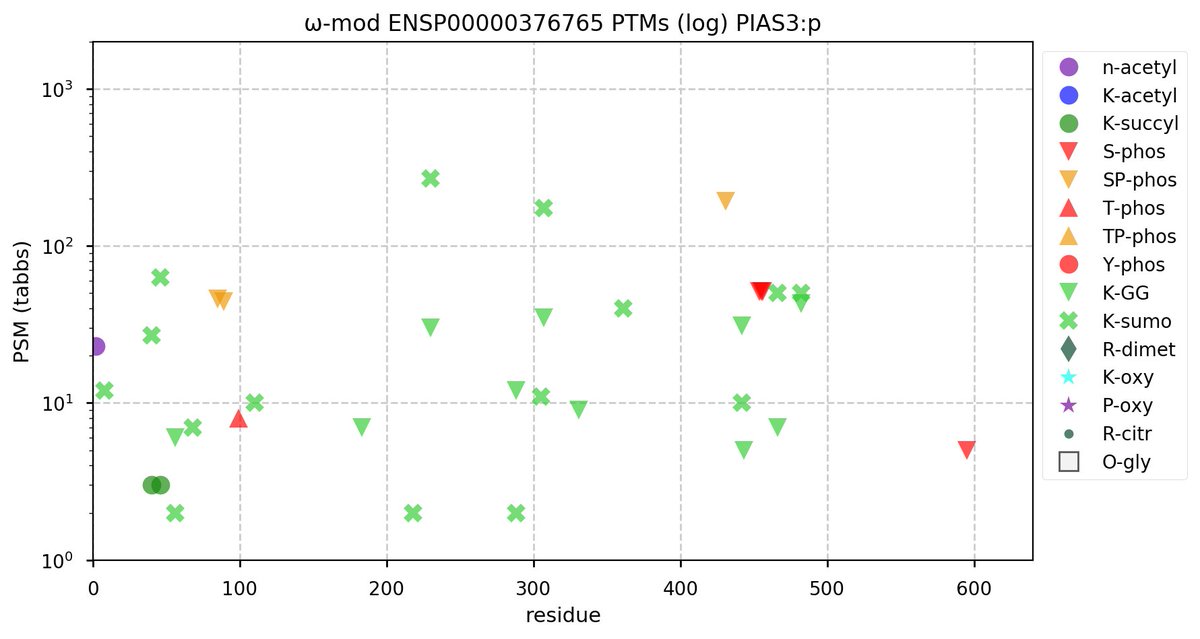
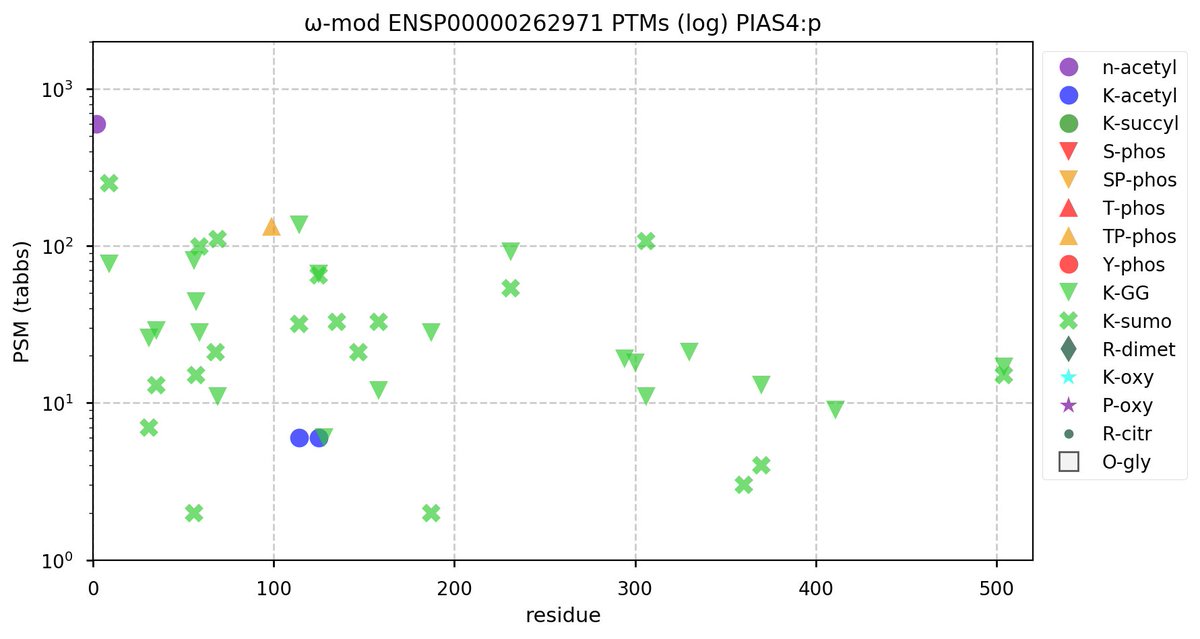
Sun Feb 20 14:04:29 +0000 2022🔗
1 – 628 ⮕ 2 – 628 (N-acetylated)
#real_NT
Sat Feb 19 15:49:58 +0000 2022These subunits have about a 10% overlap of observable peptide sequences. /2 🔗
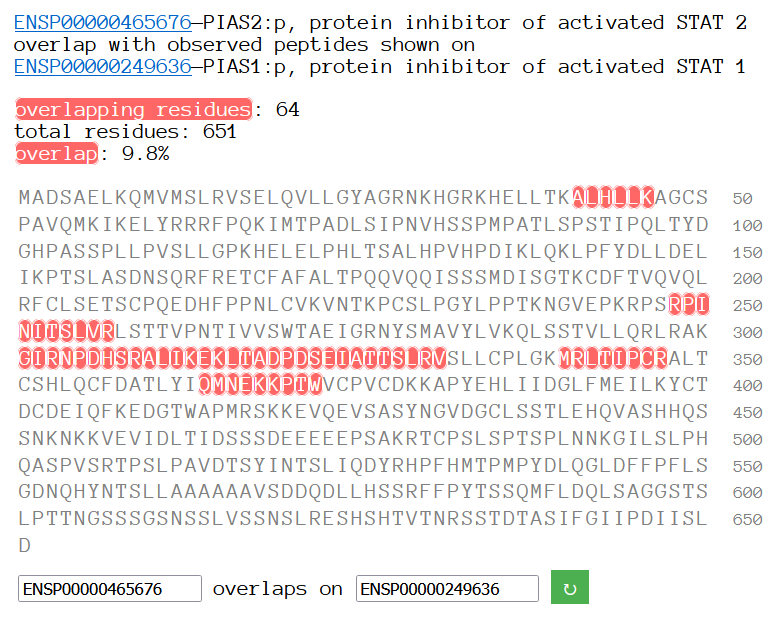
Sat Feb 19 15:47:41 +0000 2022Modification diagrams for #PIAS1 & #PIAS2, involved in #JAK_STAT mechanisms. They are 2 of 4 Protein Inhibitors of Activated Stat, that act via STAT SUMOylation. They also have an unusual number of SUMOylation/ubiquitin acceptor sites themselves. /1 🔗
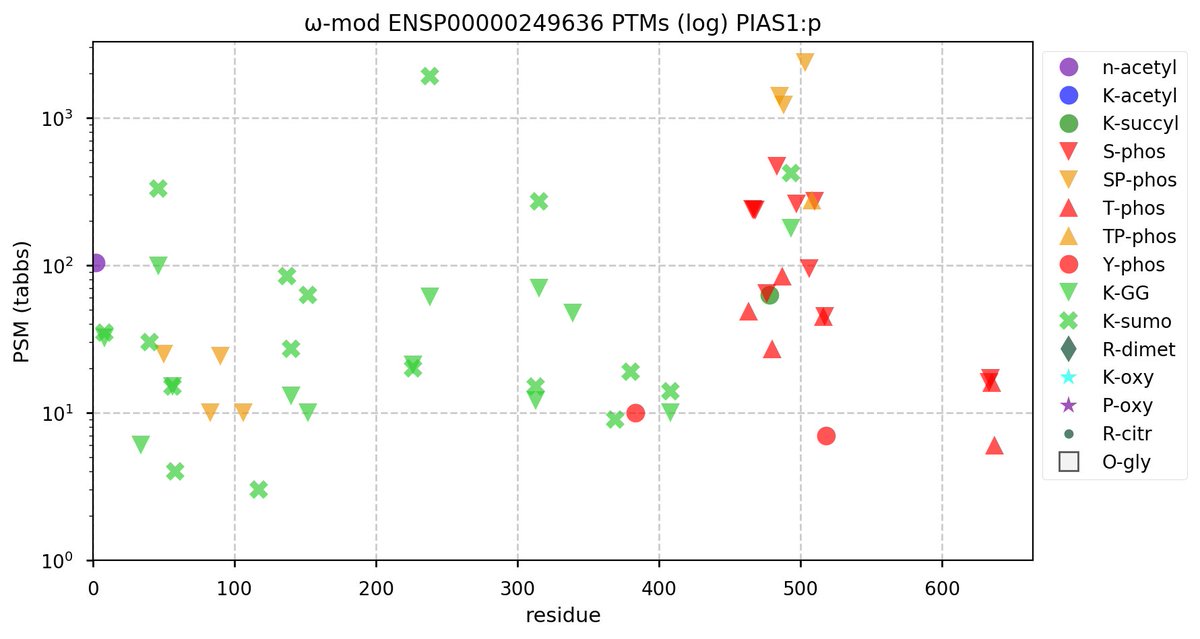
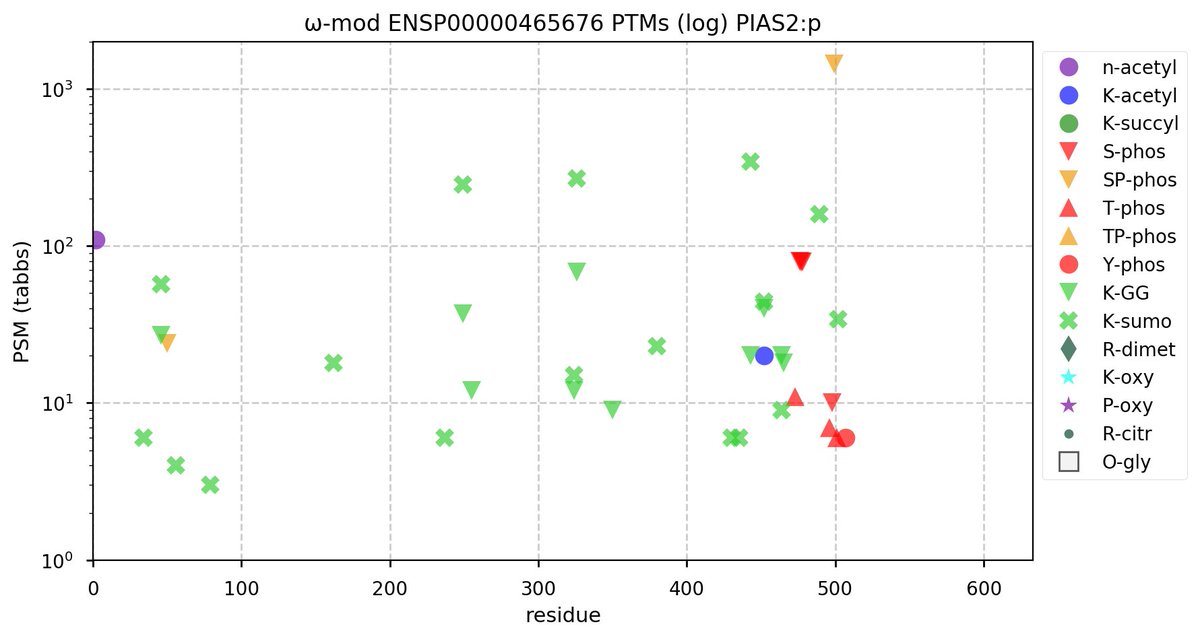
Sat Feb 19 14:57:17 +0000 2022These 2 molecules are among the main authors of "The Histone Code": they appear to have a rather involved PTM code all of their own.
Sat Feb 19 13:55:21 +0000 2022For me, the worst thing about Z**m meetings is the tendency for people to want to insert silly slide shows to illustrate a point. I would be OK with puppets, but not slides 😡😡😡😡
Sat Feb 19 13:38:16 +0000 2022🔗
1 – 621 ⮕ 2 – 621 (N-acetylated)
#real_NT
Fri Feb 18 17:11:27 +0000 2022Modification diagrams for #EP300 & #CREBBP, two acetyltransferases involved in #JAK_STAT mechanisms. /1 🔗
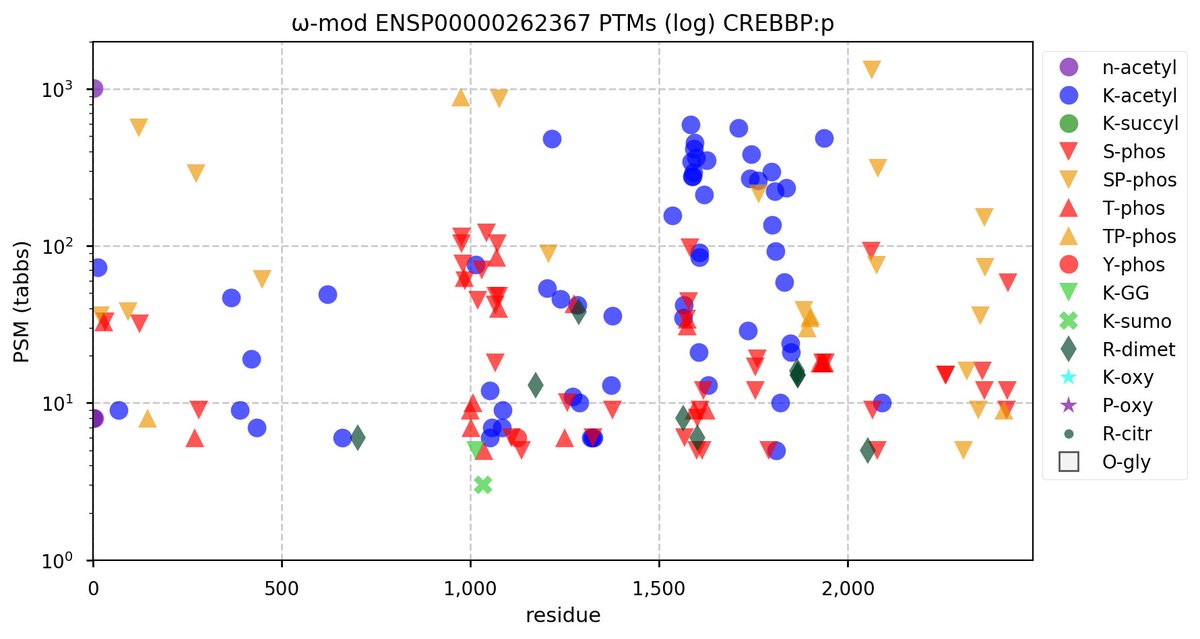
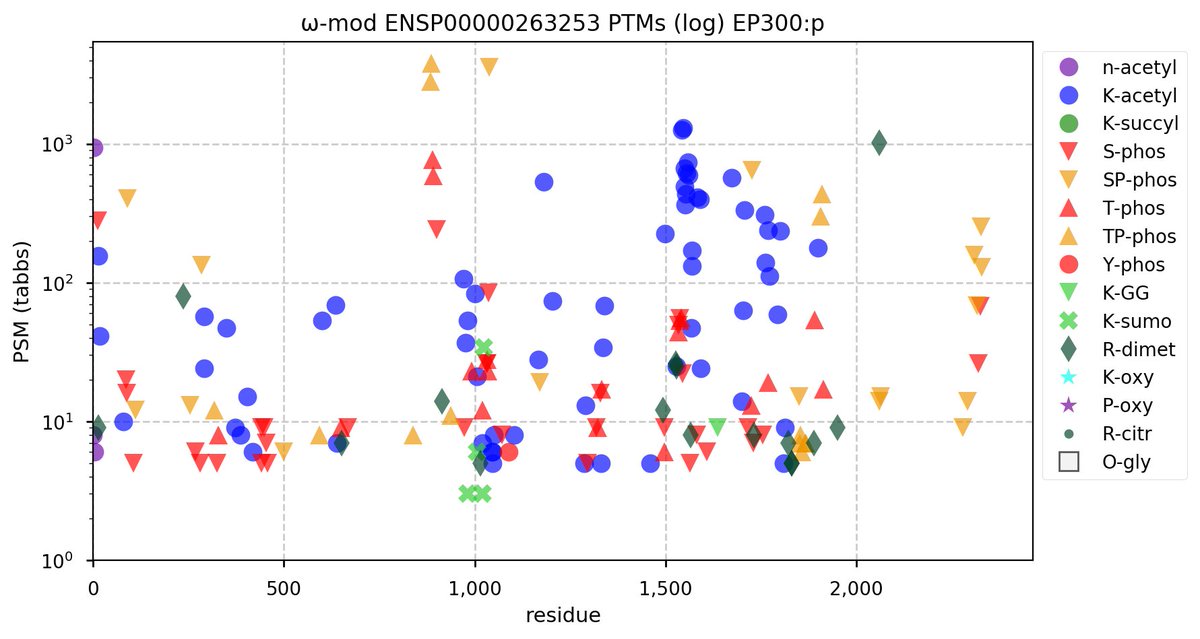
Fri Feb 18 15:55:22 +0000 2022Thanks to the brave few with an opinion. Experimental data from mammalian cells usually has this ratio at about 0.025, so the winning answer was correct. This means that you should detect about 40 acetylated N-terminal PSMs starting with 'A' for every 1 starting with 'G'.
Fri Feb 18 13:50:55 +0000 2022Strong surface winds from Nova Scotia to the UK caused by a large high pressure system affecting much of the northern Atlantic & assisted by a low off the southern tip of Greenland 🔗
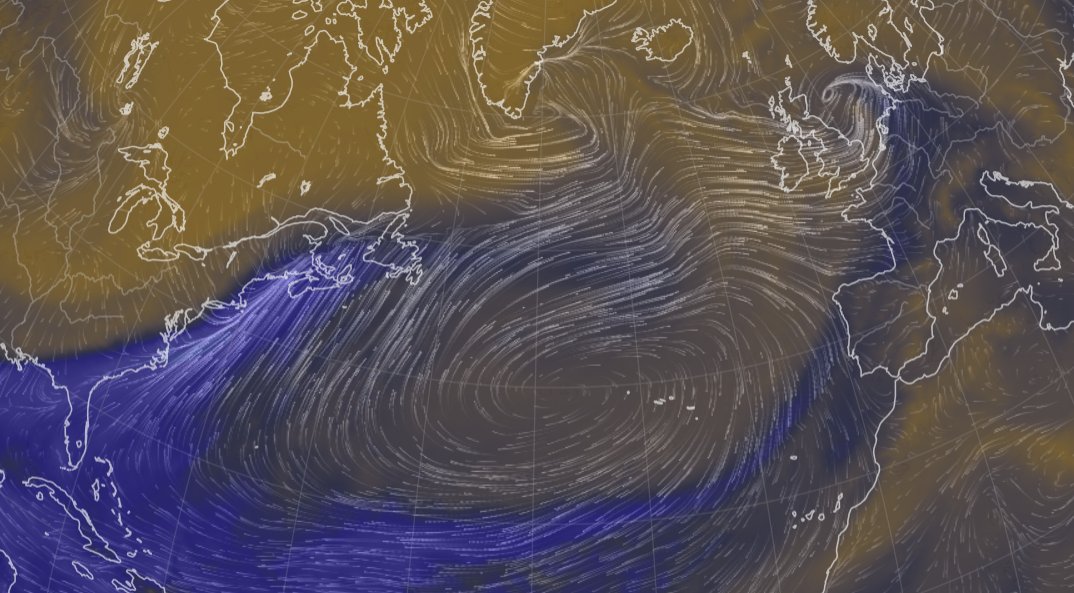
Fri Feb 18 13:36:20 +0000 2022🔗
1 – 674 ⮕ 2 – 674 (N-acetylated)
#real_NT
Thu Feb 17 20:02:49 +0000 2022@ChampionNd I was thinking eukaryote. For the sake of this example, let's say the results from analyzing a mammalian cell line.
Thu Feb 17 19:18:29 +0000 2022@DRAWheatcraft The usual cure is to put the boss on a flight out of town.
Thu Feb 17 15:34:10 +0000 2022In proteins the ratio of G:A residues is a bit less than 1 (≈0.9). Which ratio is the closest to that observed for PSMs assigned to N-acetylated G vs A protein N-terminii.
Thu Feb 17 15:29:06 +0000 2022#real_NT Many intracellular proteins are co-translationally modified by removing the nascent N-terminal Met & the acetylation of the new N-terminal residue. /1
Thu Feb 17 13:59:42 +0000 2022@VATVSLPR Temperature in K is always best, but it lacks popular appeal.
Thu Feb 17 13:27:39 +0000 2022🔗
1 – 787 ⮕ 2 – 787 (N-acetylated)
#real_NT
Thu Feb 17 13:05:14 +0000 2022That is a 90°F gradient for my American friends.
Thu Feb 17 12:58:38 +0000 2022Back to a 50°C north-south gradient along I-29. 🔗
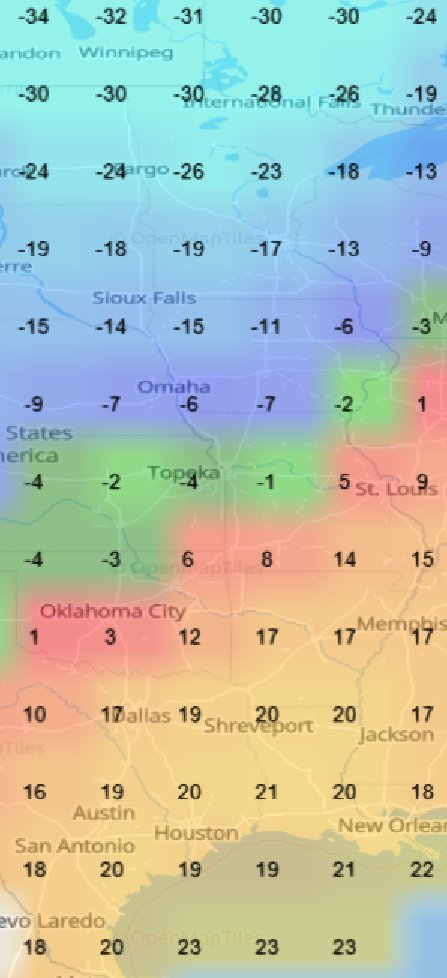
Wed Feb 16 16:53:20 +0000 2022For people not intimately familiar with #JAK_STAT, STATs are often blamed for pretty much everything that can happen in cells 🔗
Wed Feb 16 16:01:38 +0000 2022The #STAT3 pattern has considerable similarity to the #STAT1 pattern, including the C-terminal phosphodomain. /2
Wed Feb 16 16:01:18 +0000 2022Modification diagrams for #STAT3 & #STAT4, two of the possible transducer/transcription activators in the #JAK_STAT mechanism. The only similarity is the phosphorylation pattern in the C-terminal domain. /1 🔗
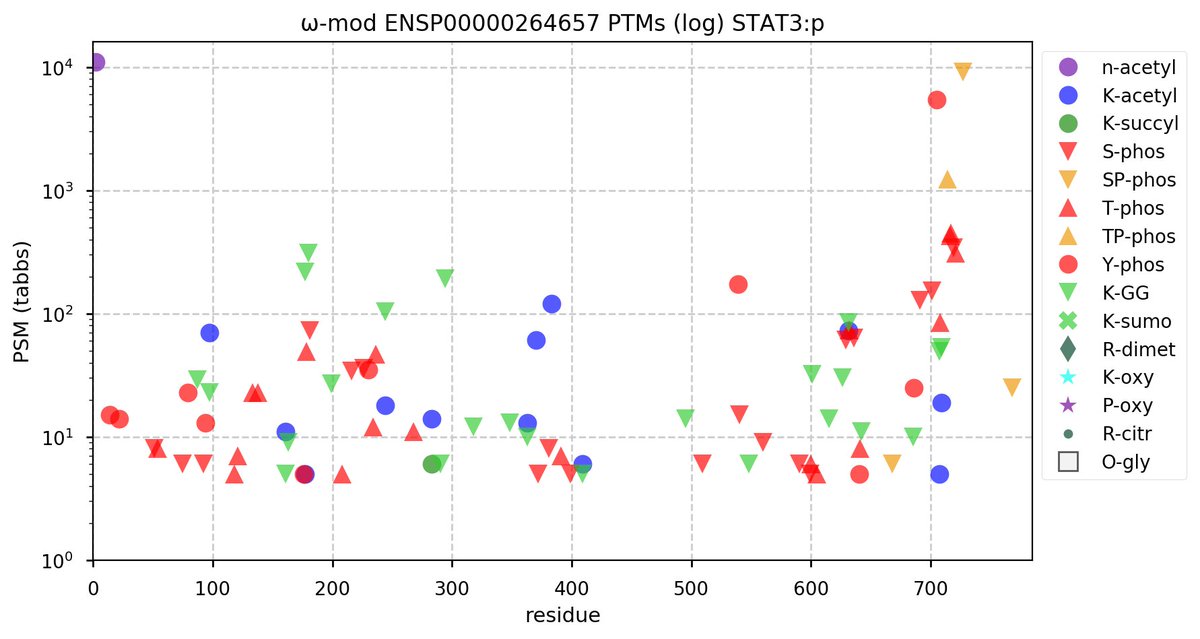
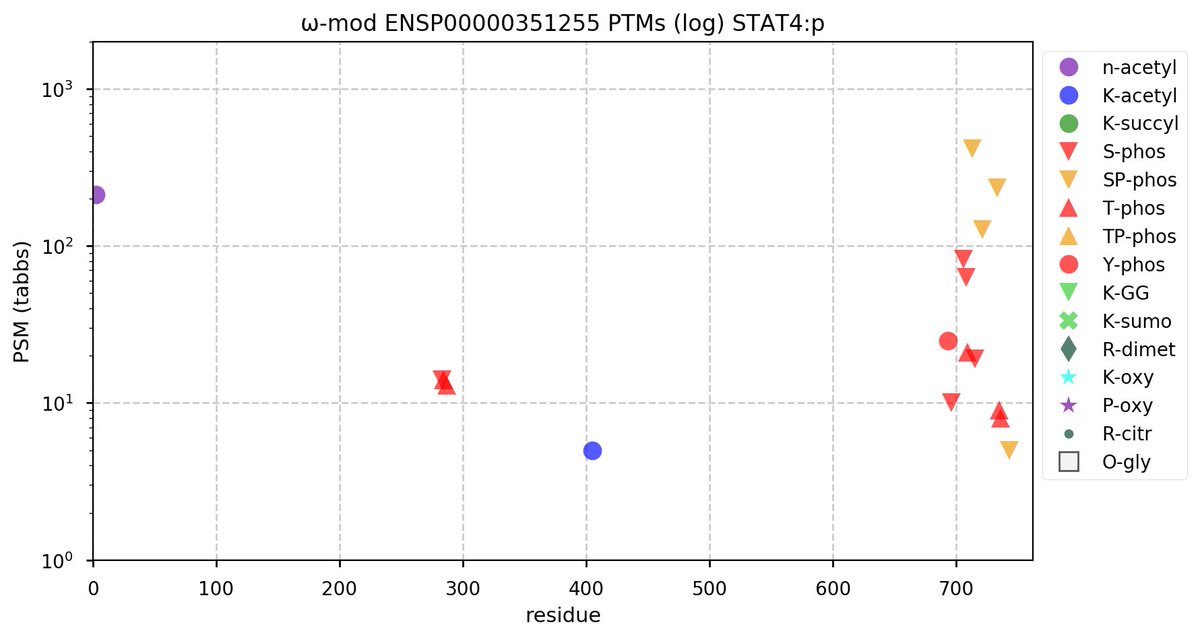
Wed Feb 16 14:08:44 +0000 2022🔗
1 – 794 ⮕ 2 – 794 (N-acetylated)
#real_NT
Tue Feb 15 18:33:32 +0000 2022@OliverMBernhar1 @UCDProteomics Spoken like someone with 10 years of experience. 🧓
Tue Feb 15 17:51:37 +0000 2022PXD030591 is a great example data set demonstrating how highly represented blood plasma proteins can be in HLA-DP presented class 2 peptides, even when the those proteins are from bovine FBS.
Tue Feb 15 15:21:06 +0000 2022#STAT1 & #STAT2 have no overlapping tryptic peptides but share a single MHC class 1 peptide: 🔗
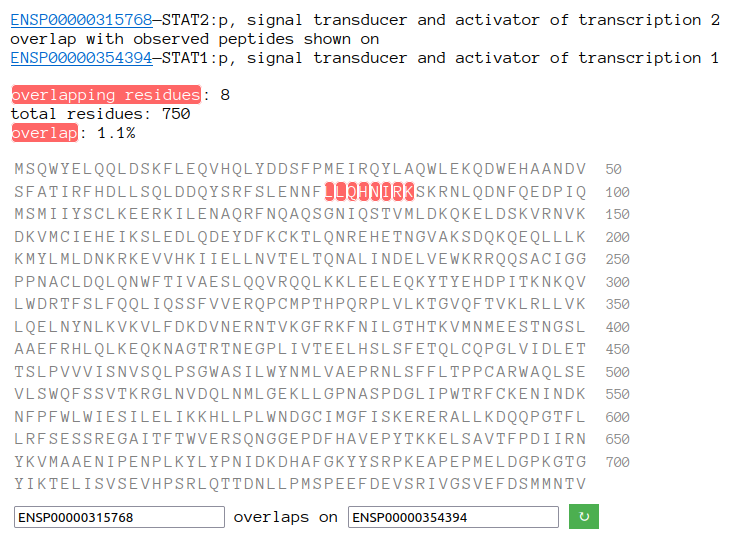
Tue Feb 15 15:19:01 +0000 2022Modification diagrams for #STAT1 & #STAT2, two of the possible transducer/transcription activators in the #JAK_STAT mechanism. Other than a rather generalized affinity for GGylation, these patterns show little overall similarity. /1 🔗
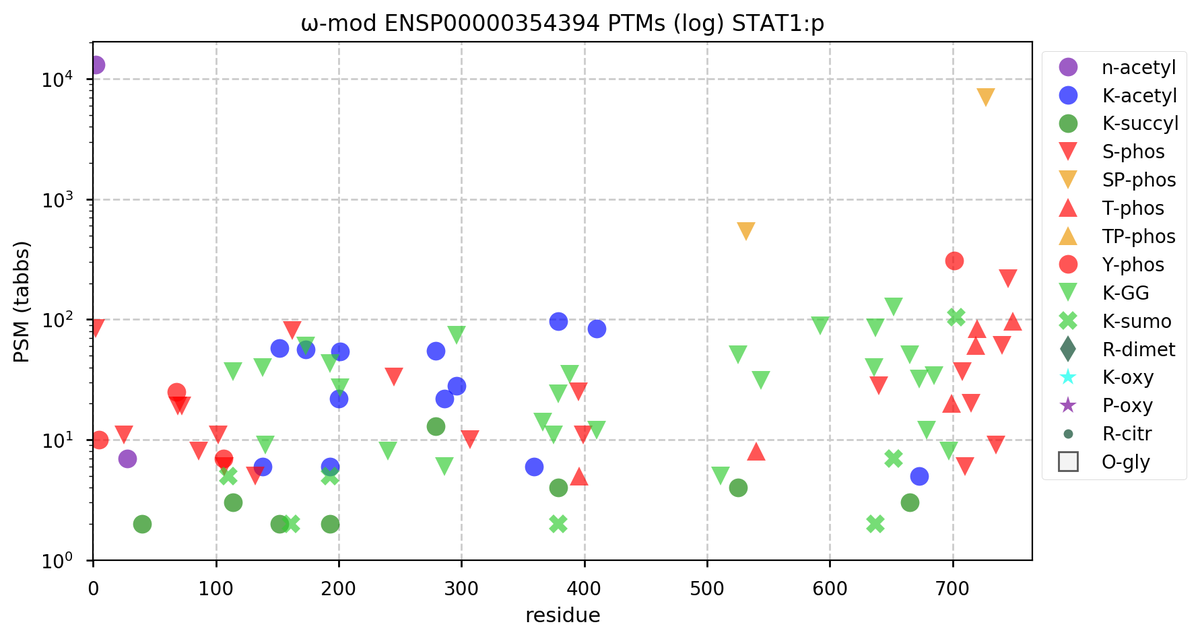
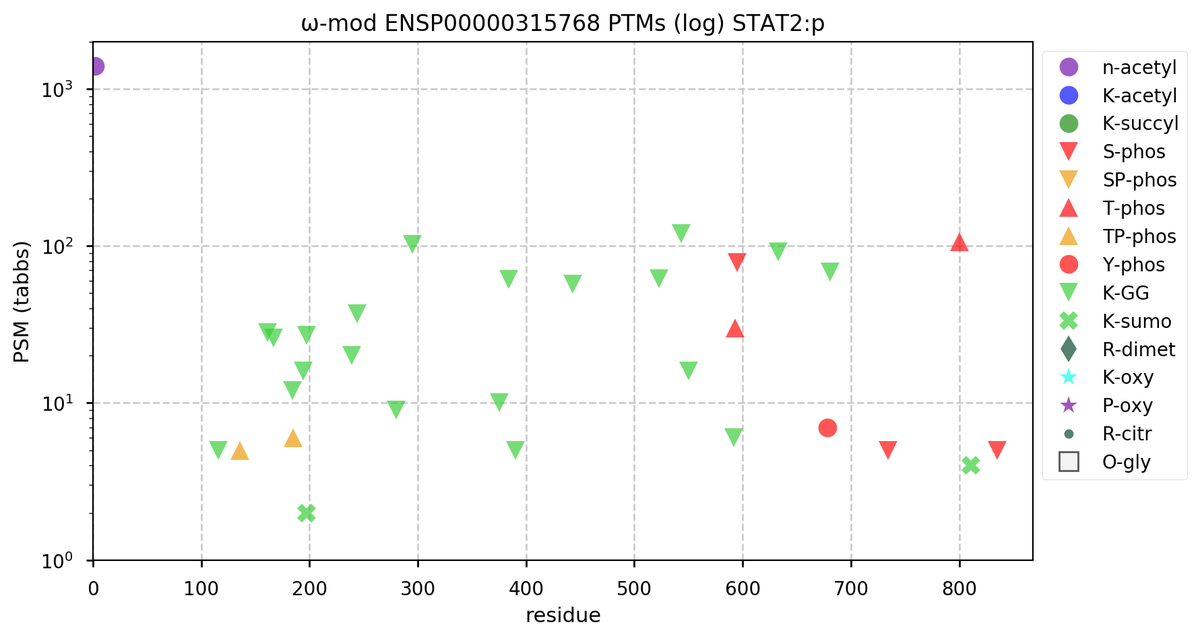
Tue Feb 15 13:36:04 +0000 2022🔗
1 – 748 ⮕ 2 – 748 (N-acetylated)
#real_NT
Mon Feb 14 20:16:06 +0000 2022@bffo @PLOSCompBiol That one is good & automating responses to failures is often overlooked. The first paragraph of Rule 1 is strong, too: "processing data at large scale is a substantial undertaking".
Mon Feb 14 18:41:39 +0000 2022Every time I think I understand mitochondrial protein importing, I run across some exception like MRPL23:p & have to go back to square 1.
Mon Feb 14 15:48:47 +0000 2022The two sequences have limited observed peptide sequence overlap, much of which is in MHC peptides: 🔗

Mon Feb 14 15:45:54 +0000 2022Modification diagrams for #JAK1 & #TYK2, two of the initiator tyrosine kinases in #JAK_STAT pathways. The patterns show a small number of phosphodomains & more general K+GGyl engagement without K+acetyl alternate modifications. /1 🔗
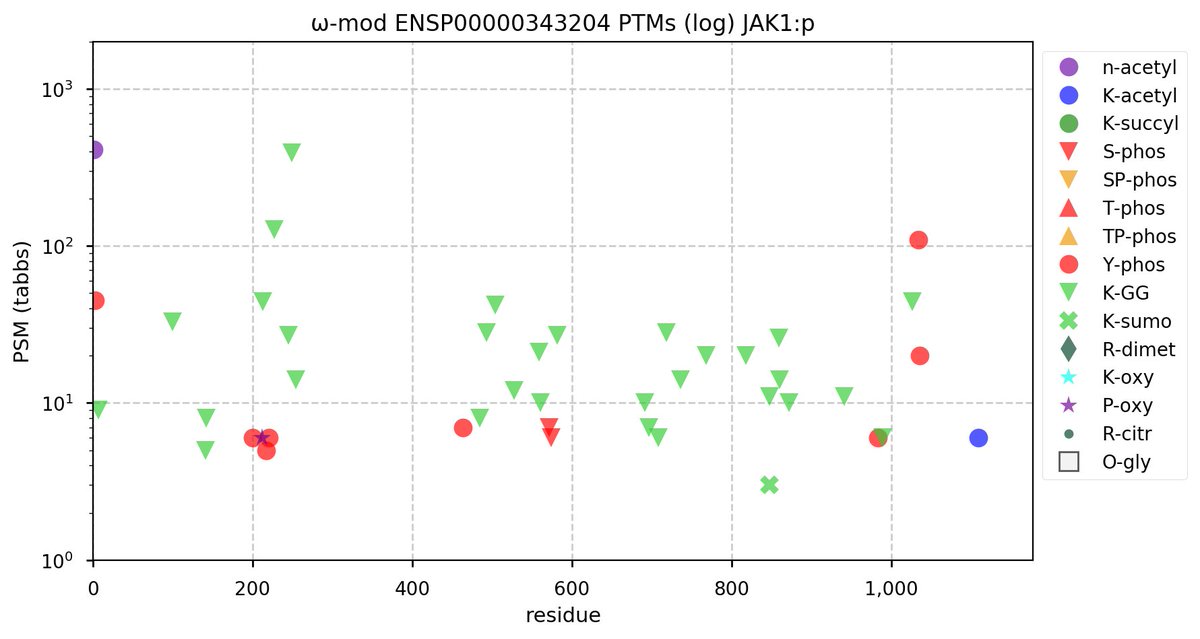
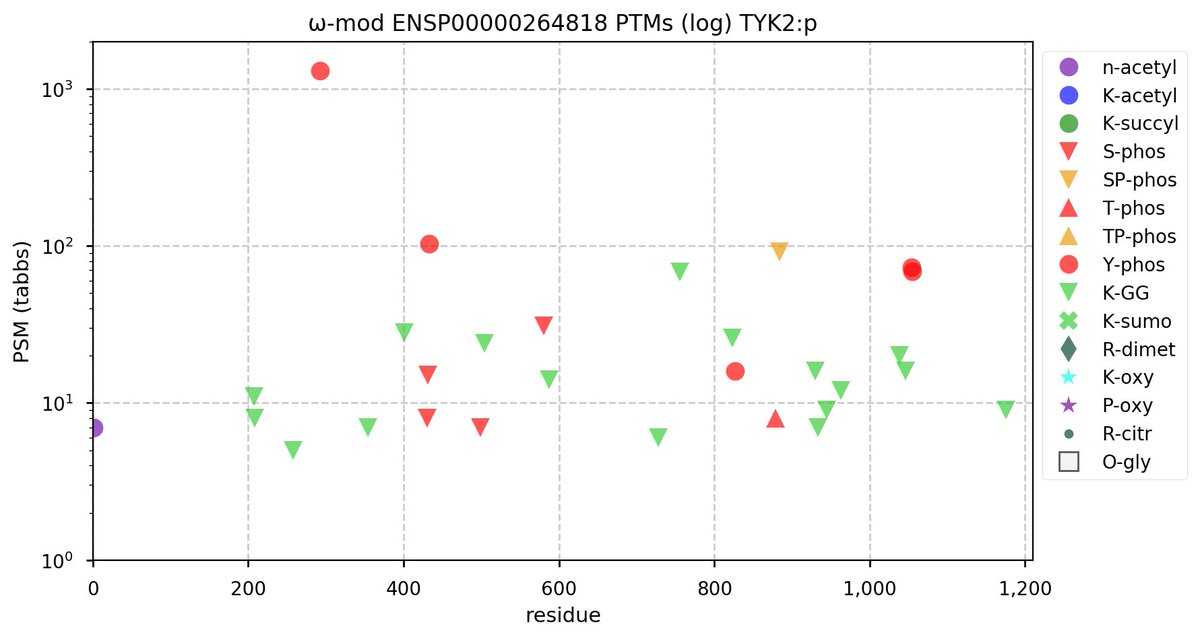
Mon Feb 14 15:16:02 +0000 2022I'm not sure why this particular sort of screw up bugs me, but it does.🧐
Mon Feb 14 13:56:40 +0000 2022🔗
1 – 851 ⮕ 2 – 851 (N-acetylated)
#real_NT
Mon Feb 14 00:15:05 +0000 2022CNPY3:p is an ER protein: it is via the imported signal peptide mechanism that relies on a hydrophobic N-terminal peptide to translocate the protein across the ER membrane. The peptide is removed co-translationally, prior to folding. /3
Sun Feb 13 19:54:44 +0000 2022@EdHuttlin @GermanRosano @gingraslab1 @HMSBioPlex Thanks for the discussion, Ed. It is a little beside, but adjacent to, the point I was originally trying to make, which was that when trying to determine a protein's normal N-terminus, steer clear of data sets in which that protein was being explicitly manipulated.
Sun Feb 13 19:33:25 +0000 2022@EdHuttlin @GermanRosano @gingraslab1 @HMSBioPlex You are right Ed: it is marked up as an ER protein so the removal of the N-terminal signal sequence would be co-translational, rather than subsequent to import. But that is actually a bit more problematic, since there shouldn't be any protein with the signal at all.
Sun Feb 13 17:55:32 +0000 2022The heterogeneous phosphodomain (in green) nicely avoids the WW domains (in blue) and the SARAH domain (in red) that are involved in protein-protein interactions. 🔗

Sun Feb 13 17:53:07 +0000 2022Modification diagram for #SAV1―homolog of fruitfly #Salvador― that is considered to be a scaffold protein involved in the #HIPPO pathway. The N-terminal half of the sequence has several phosphodomains, including a rather busy Y+phosphoryl domain. /1 🔗
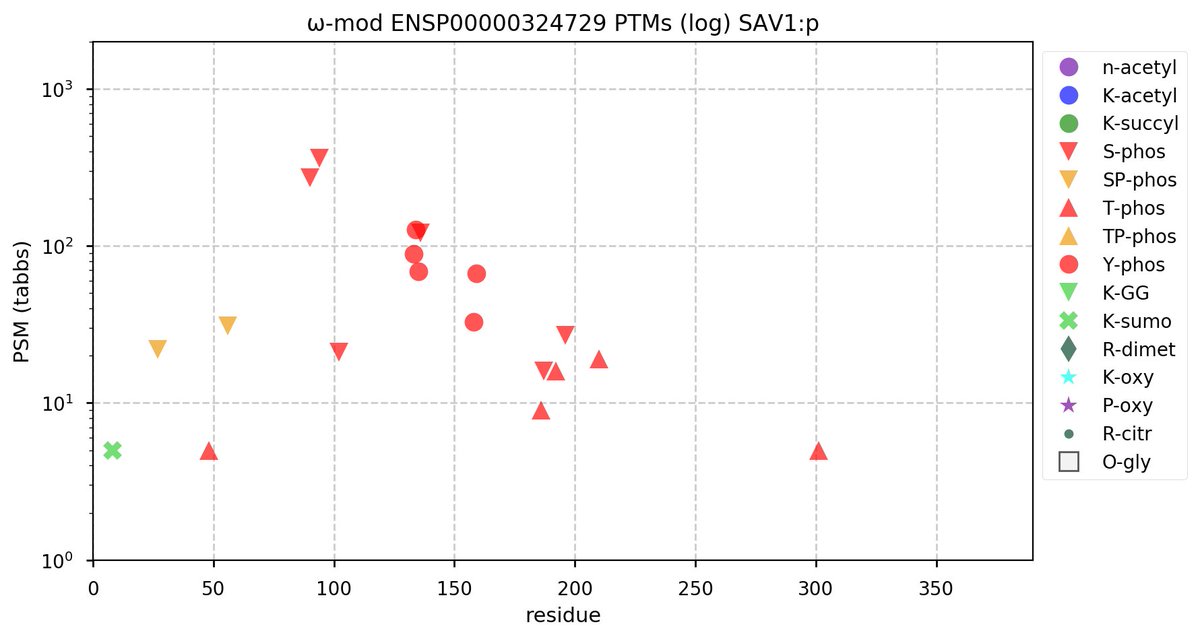
Sun Feb 13 17:00:06 +0000 2022@GermanRosano I'm not sure how this type of thing is accounted for in the affinity pull-down world: maybe one of the doyens @EdHuttlin or @gingraslab1 could elaborate?
Sun Feb 13 15:06:40 +0000 2022In this case, the top 4 observations are from affinity purification expts that used recombinant CNPY3:p as the bait, so it was never processed via TOMM/TIMM, resulting in the nascent sequence N-terminus (M1) rather than the mature N-terminus (E38). /fin
Sun Feb 13 15:06:03 +0000 2022The diagram shows the peptides observed (red bars) for the 20 best identifications of CNPY3:p. The 4 observations on top indicate that the protein begins at M1. In the other 16 observations, the most N-terminal peptide is a semi-tryptic peptide starting at E38. /2
Sun Feb 13 15:05:37 +0000 2022If you are ever interested in determining the real N-terminus of a protein using existing proteomics data (#real_NT), the coverage diagram for CNPY3:p (278 aa) below illustrates something you should watch for when looking at the information. /1 🔗
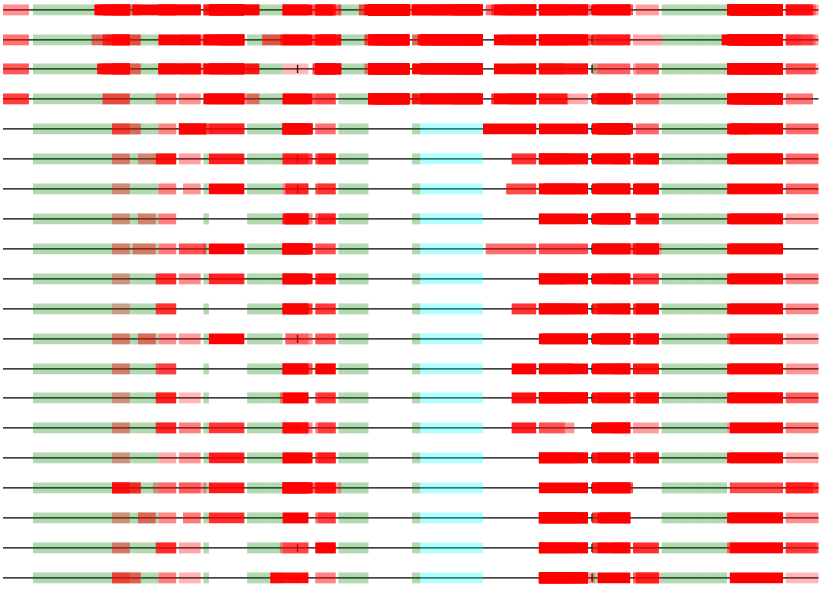
Sun Feb 13 14:20:28 +0000 2022🔗
1 – 41 ⮕ 1 – 42
42 – 615 ⮕ 43 – 615
#real_NT
Sun Feb 13 02:34:03 +0000 2022@stateofthecity @jm_mcgrath Navigator is on the job!
Sun Feb 13 01:40:46 +0000 2022Weird commercial from the Govt of Canada 🔗 about "gun violence". After living in New York and Chicago, downtown Winnipeg is not really very scary.
Sat Feb 12 22:53:55 +0000 2022@Duszynski116 @howardtayler @JamesSACorey One Froggy Evening
Sat Feb 12 18:24:44 +0000 2022But it serves me right: I should know better than to highlight a pathway where the proteins were "named" by high-on-banana-fumes Fruitfly Bros.
Sat Feb 12 18:21:02 +0000 2022In the last week I have been surprised by just how many #hippo tagged photographs of hippos end up being tweeted every day.
Sat Feb 12 16:52:23 +0000 2022The sequences do not share any tryptic peptides: the only shared observed peptide is an MHC1 class 1 sequence. YAP1:p peptides projected onto WWTR1:p are shown below: 🔗
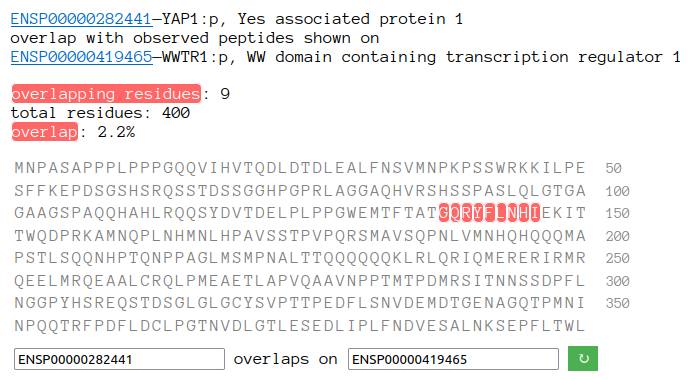
Sat Feb 12 16:50:10 +0000 2022Modification diagrams for #YAP1 and #WWTR1 (TAZ)―homologs of fruitfly #Yorkie― that are downstream targets of phosphorylation in the #HIPPO pathway. The N & C terminal phosphodomains have similar patterns, but the central domains are quite different. /1 🔗
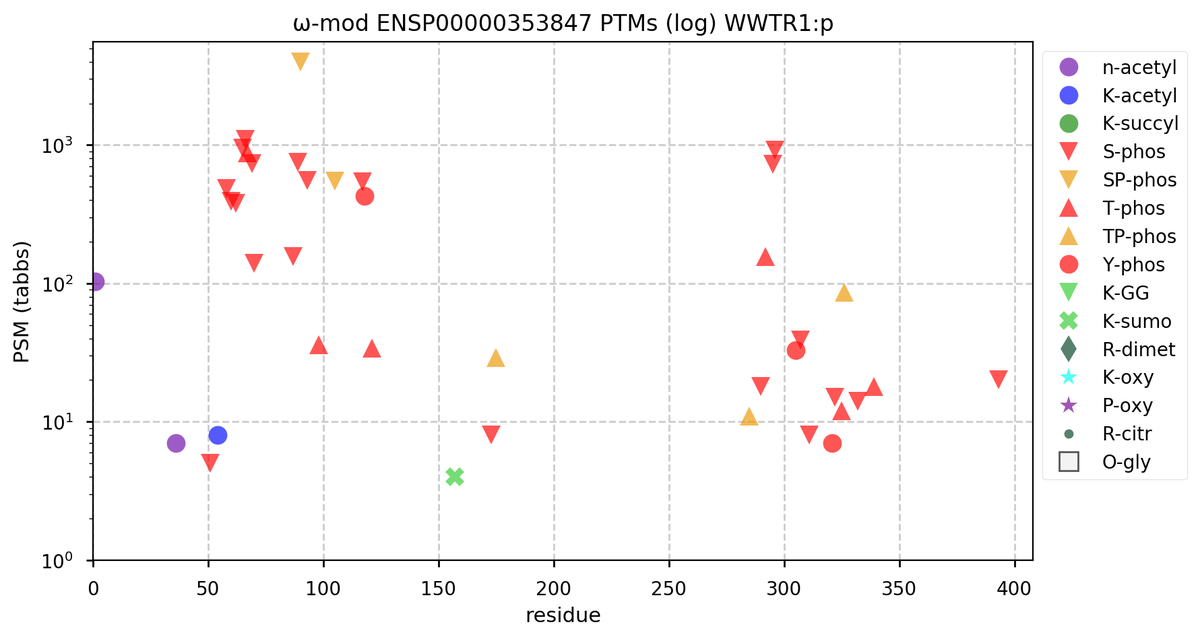
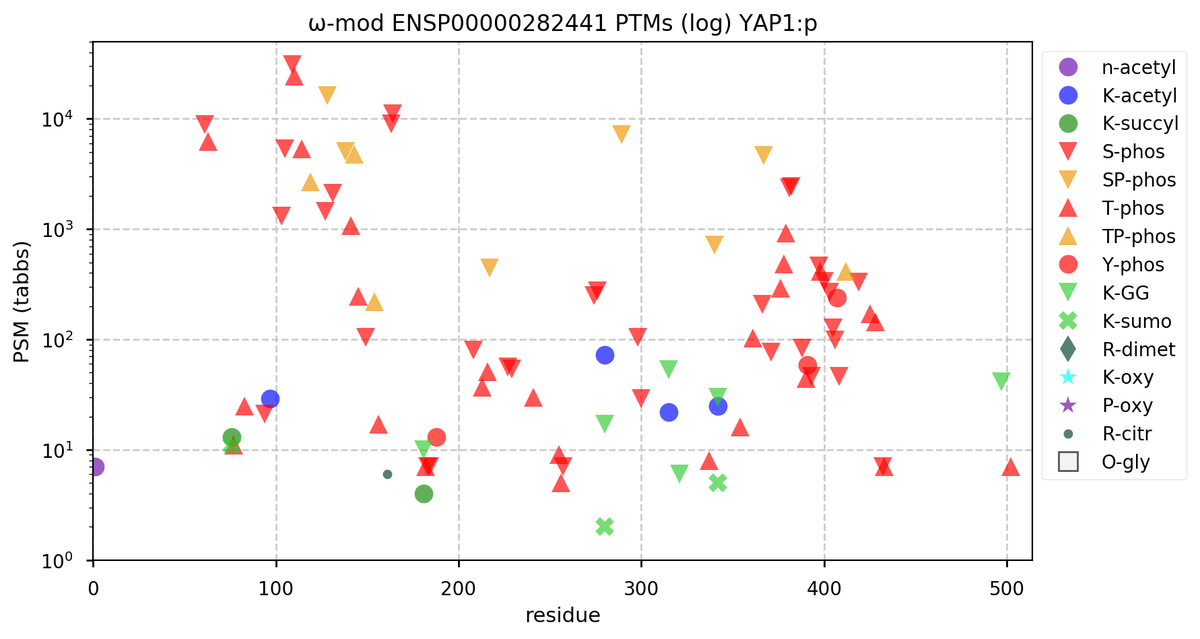
Sat Feb 12 14:17:02 +0000 2022@andy___jones @astacus I would suggest the ENSEMBL page for the gene, e.g.
🔗
It gives you a summary of the current list protein sequences, their status & direct access to each one's protein, mRNA and DNA sequences
Sat Feb 12 14:03:00 +0000 2022🔗
1 – 802 ⮕ 2 – 802
#real_NT
Fri Feb 11 18:15:43 +0000 2022I'm never quite sure how to deal with legit users who end up over taxing system resources by making a bunch of requests in parallel.
Fri Feb 11 16:52:42 +0000 2022@statesdj @Dereklowe However, when I lived in Newfoundland, I did have some flipper pie.
Fri Feb 11 16:44:39 +0000 2022@statesdj @Dereklowe None, as far as I can remember, but I will check. The only one I've seen in ProteomeXchange is PXD020112 (Phoca largha), but when I looked it didn't seem to have an annotated proteome available.
Fri Feb 11 16:37:46 +0000 2022It is very Canadian to declare a state of emergency to deal with blow back from the messy execution of the last few declared states of emergency.
Fri Feb 11 16:32:37 +0000 2022The subunits have some observable sequence overlap. Shown is the overlap of #LATS2 projected onto #LATS1. Note the various low complexity domains in (1-600). 🔗

Fri Feb 11 16:29:04 +0000 2022Modification diagram from #LATS1 & #LATS2 (fruitfly homolog Warts), which are phosphorylated by #STK3 or #STK4 in the #HIPPO pathway activating their own kinase activity. They have quite different patterns of observed phosphoryl acceptors. /1 🔗
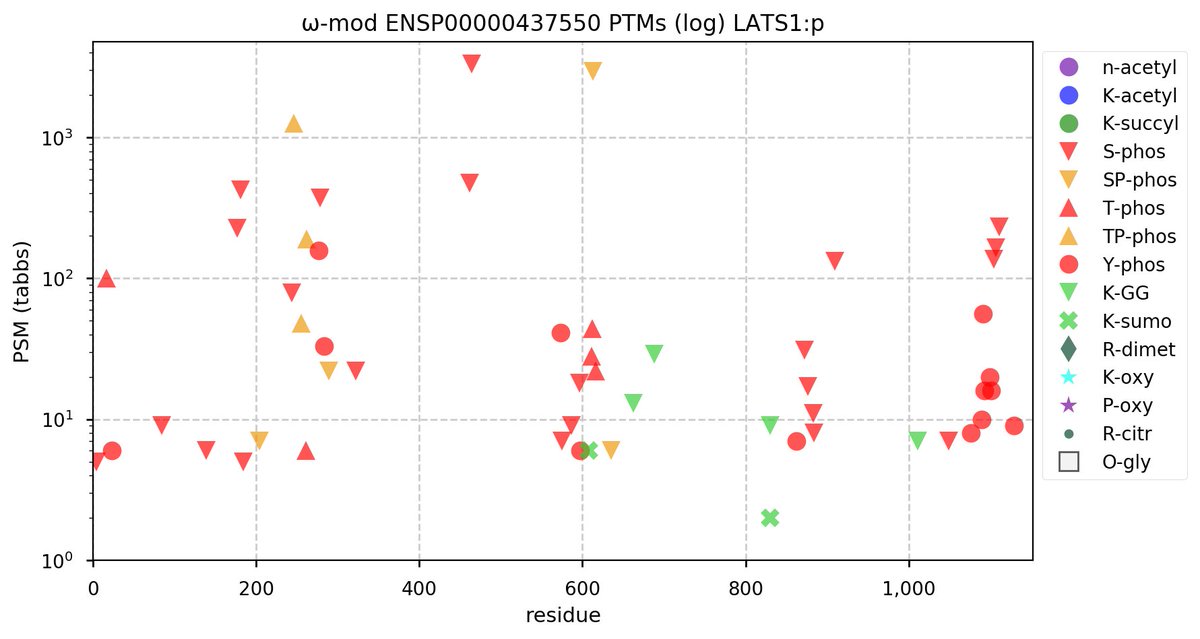
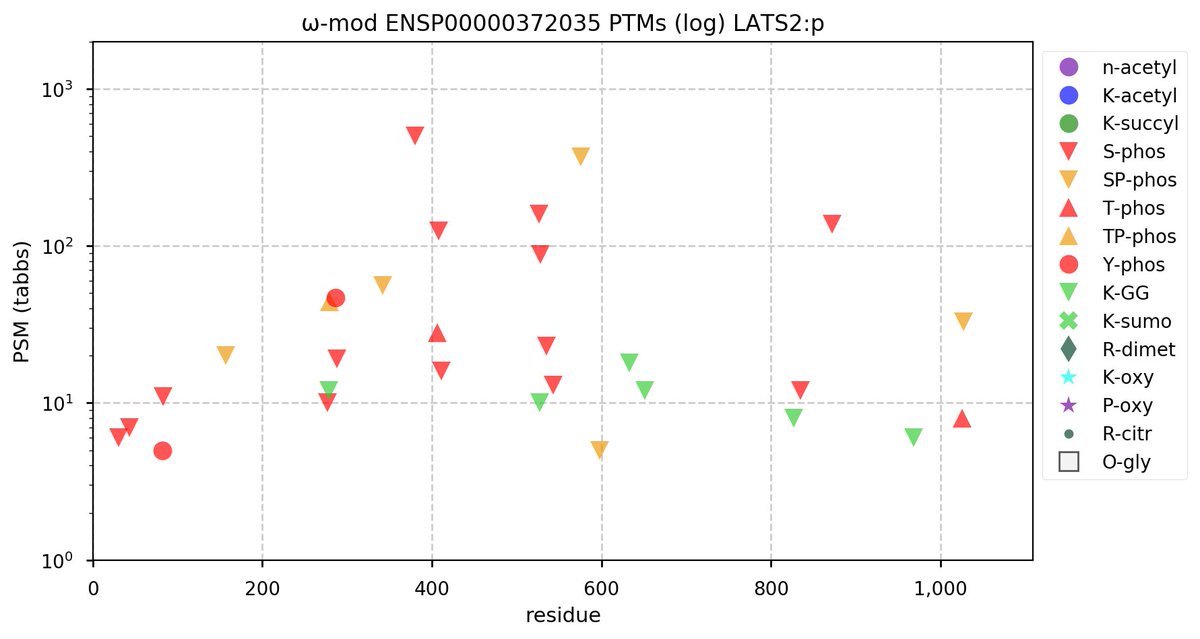
Fri Feb 11 13:51:24 +0000 2022🔗
1 – 50 ⮕ 1 – 66
51 – 351 ⮕ 67 – 351
#real_NT
Fri Feb 11 03:00:20 +0000 2022@GuiomarPedro @HAntonicka @VamsiMootha @mito_gene It has some similarity to way that selenocysteine is made using the appropriate tRNA charged with serine 🔗
Thu Feb 10 23:42:56 +0000 2022@GuiomarPedro @HAntonicka @VamsiMootha @mito_gene It is one of the many cases in protein biochemistry where "that doesn't make sense" doesn't matter because that is the way that it is.
Thu Feb 10 22:01:47 +0000 2022@bittremieux @slavov_n @thabangh @wfondrie An unusual charge state of a not very frequently observed, low complexity peptide from UNC5C seemed like it must have some meaning, but random it is!
Thu Feb 10 18:47:47 +0000 2022@MattWFoster It is a little different with some of the kits. They really look more like ion exchange: the first fraction is all acidic peptides & the last is all basic peptides. They don't act very much like RP separations.
Thu Feb 10 17:29:23 +0000 2022Also, unlike many of the kit-based high pH MDC methods, the one used in this study doesn't fractionate the peptides by pI.
Thu Feb 10 17:23:21 +0000 2022If you want a TMTpro dataset to fool around with, PXD024513 (🔗) is worth a look. Nearly complete derivatization, very few detectable side reactions & >95% recovery of Cys-containing peptide. The only downside is that 10-12% of the PSMs are semi-tryptic.
Thu Feb 10 16:42:35 +0000 2022From back when I used to think reviewing things was a useful activity, my best experience was 2 terms on an NIH study section. My worst was 2 terms on a CIHR peer review committee.
Thu Feb 10 15:46:23 +0000 2022The two proteins have significant peptide sequence overlap in their N-terminal domains. Shown in the overlap of STK3 on STK4. /2 🔗

Thu Feb 10 15:44:50 +0000 2022Modification diagram from #STK3 & #STK4 (fruitfly homolog Hippo), the central actors in #HIPPO core kinases. aka MST2 & MST1 respectively. The C-terminal phosphodomains show some significant differences. /1 🔗
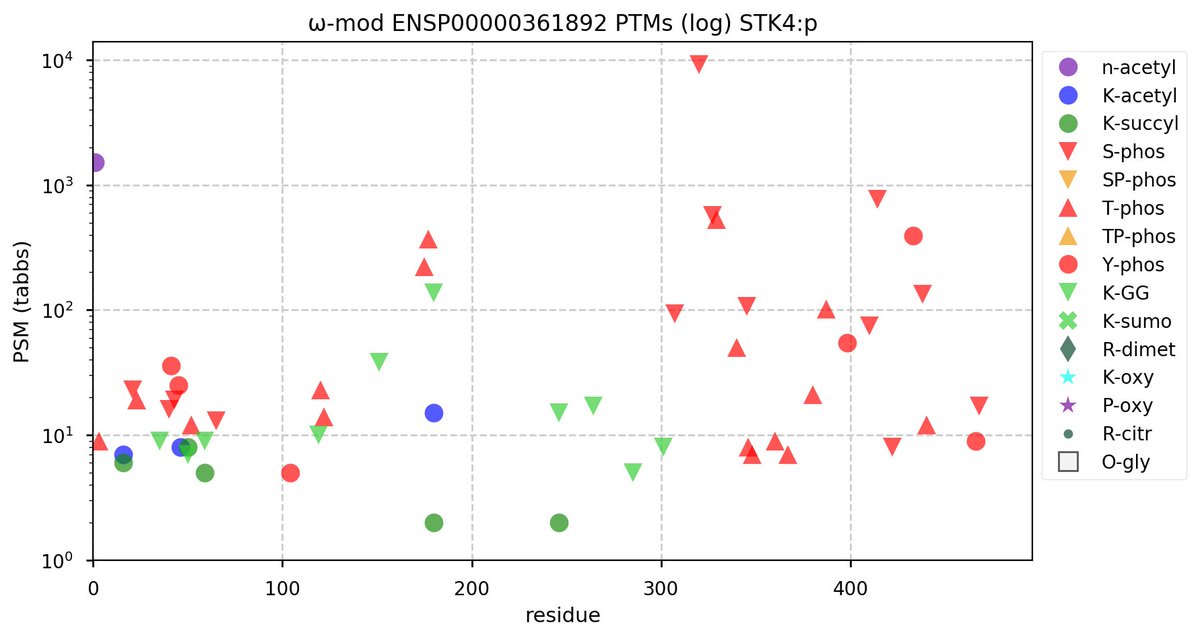
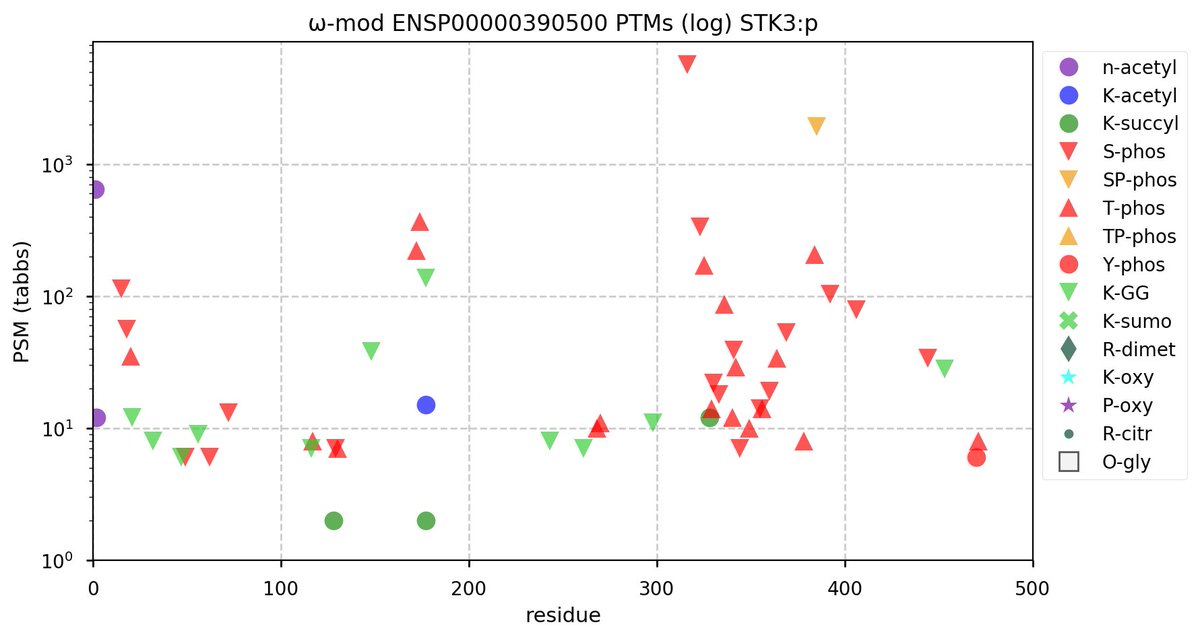
Thu Feb 10 13:11:39 +0000 2022🔗
1 – 921 ⮕ 2 – 921
#real_NT
Wed Feb 09 21:54:08 +0000 2022@slavov_n @thabangh @wfondrie When I see an example like the annotated spectrum in this paper, I am always curious why a particular spectrum/sequence were singled out as an exemplar.
Wed Feb 09 19:40:06 +0000 2022@marcoyannic Makes a nice spectrum, although it is pretty sensitive to charge state: 🔗
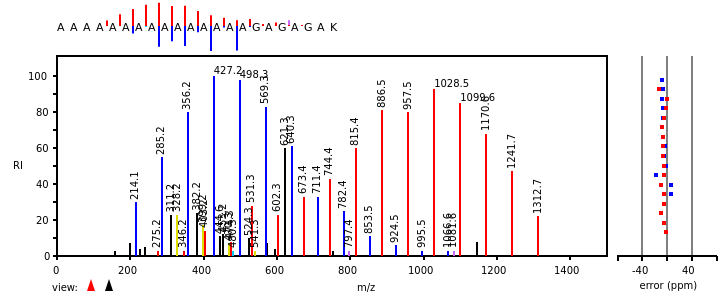
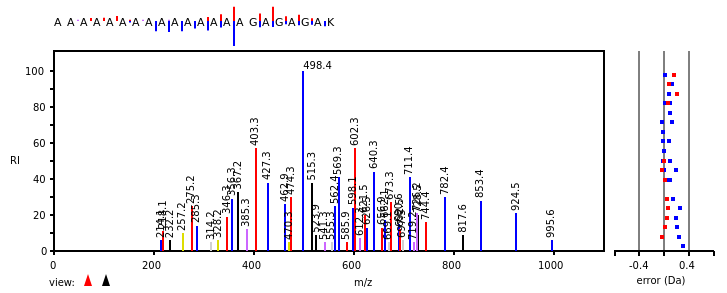
Wed Feb 09 17:16:22 +0000 2022🔗
1 – 595 ⮕ 2 – 595
#real_NT
Wed Feb 09 15:44:26 +0000 2022@michaelhoffman There is also the embarrassment of having to admit that they have looked the other way more than once when they have seen this type of behavior in colleagues.
Wed Feb 09 15:40:24 +0000 2022Modification diagram from #NF2 (fruitfly homolog Merlin), an upstream regulator of the #HIPPO core kinase. Isolated S/T+phosphoryl in 4 domains, but considerable K+GGyl. 🔗
Wed Feb 09 13:21:38 +0000 2022@MattWFoster @ProteomicsNews @chrashwood The literature isn't clear about how to classify omics research wrt clinical: I still see pubs where the title & abstract indicate research into specific cancers, but in the method section you discover they used a common cell line derived from a similar cancer 50 years ago.
Tue Feb 08 19:41:29 +0000 2022@J_my_sci @UCDProteomics It has been tried, 🔗, but people don't like to use detectability as a criterion. As far as I can tell, most are quite confident that their particular methods are so much better/different than everybody else's that their needs are exceptional.
Tue Feb 08 18:37:28 +0000 2022@sha_zhiqiang Disappointing lack of raw data.
Tue Feb 08 16:27:46 +0000 2022Does everyone have problems downloading data from jPOST? I can never seem to get download speeds any faster than 1 MB/s, which is too slow for large, multi-file data sets.
Tue Feb 08 15:55:26 +0000 2022As is so often the case, the unnamed portion of the protein is where the fun is happening: phosphodomain (338-578). /2
Tue Feb 08 15:53:11 +0000 2022Modification diagram for #FRMD6 (homolog of fruitfly Expanded), an upstream regulator of the #HIPPO core kinase. The named domains are pretty dull: (16-110) Ubiquitin-like, (115-226) FERM central domain & (233-324) FERM C-terminal PH-like domain. /1 🔗
Tue Feb 08 15:34:44 +0000 2022A Bonspiel Thaw in progress 🔗
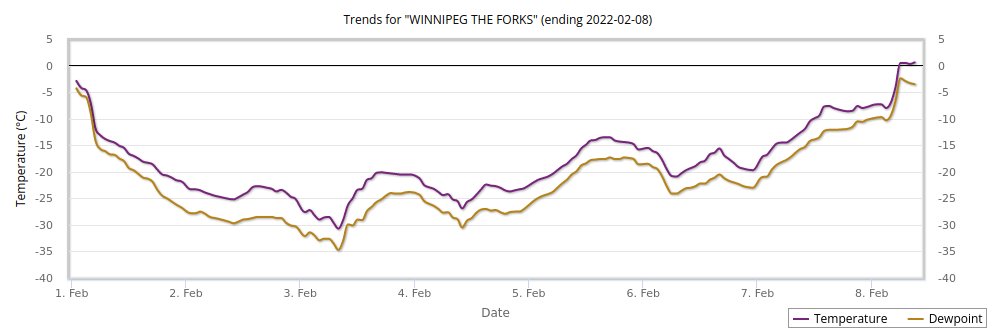
Tue Feb 08 13:54:52 +0000 2022@neely615 @Smith_Chem_Wisc @pwilmarth @nesvilab @OliverMBernhar1 IMHO, using unknown analytes for calibration is bad practice. Maintaining those analytes in the final results is very bad practice.
Tue Feb 08 13:38:29 +0000 2022🔗
1 – 30 ⮕ 1 – 37
31 – 278 ⮕ 38 – 278
#real_NT
Tue Feb 08 13:25:53 +0000 2022Bismarck ND & Austin TX (which is about 2000 km south of Bismarck) should not be at the same temperature this time of year. 🔗
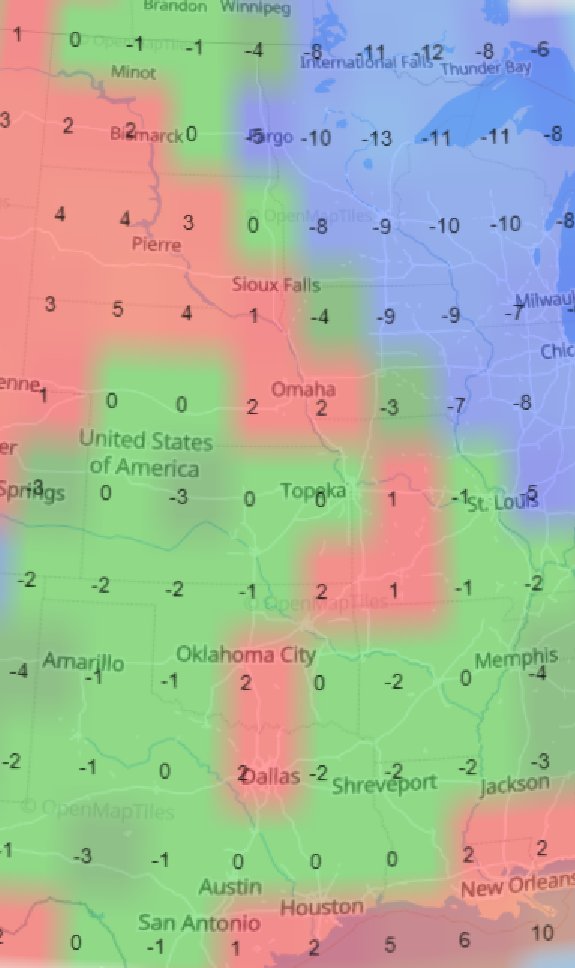
Mon Feb 07 16:49:56 +0000 2022🔗
1 – 39 ⮕ 1 – 40
40 – 403 ⮕ 41 – 403
#real_NT
Mon Feb 07 13:48:54 +0000 2022These are large subunits (~ 4500 residues) but with very limited (<1%) tryptic peptide overlap.
Mon Feb 07 13:33:36 +0000 2022Modification diagrams for the 4 human homologs of fruit fly FAT gene: #FAT1, #FAT2, #FAT3, #FAT4. Involved in/affect the #HIPPO pathway. FAT1 seems to be playing a more involved game than 2, 3 & 4, which are simple membrane transducers with cytoplasmic S/T+phosphoryl acceptors. 🔗
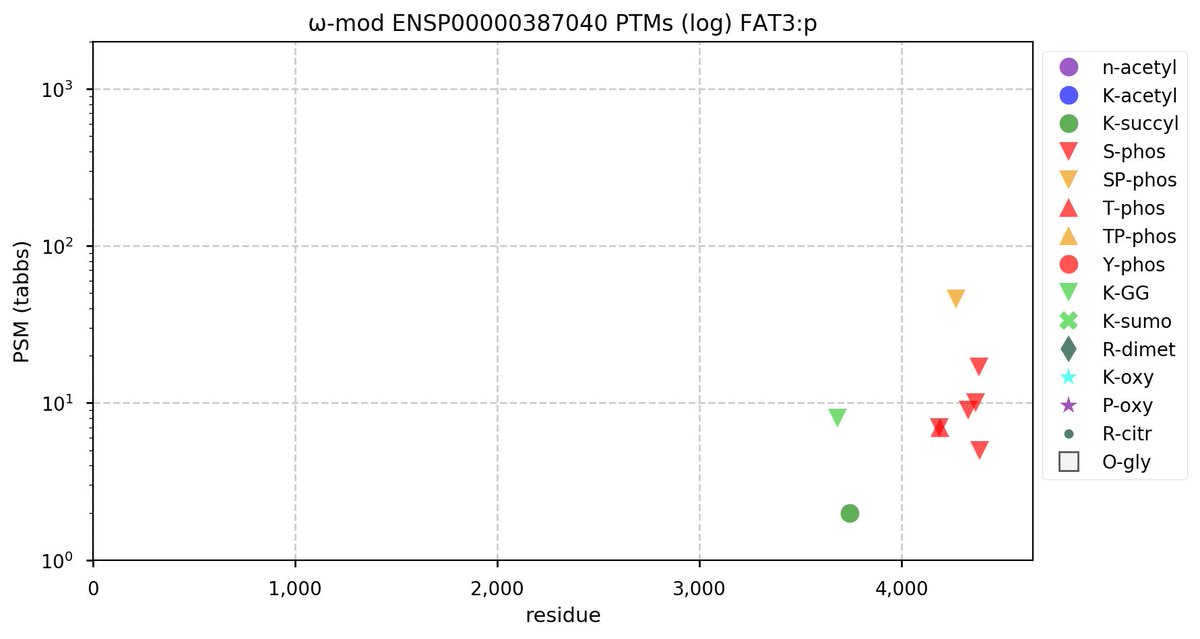
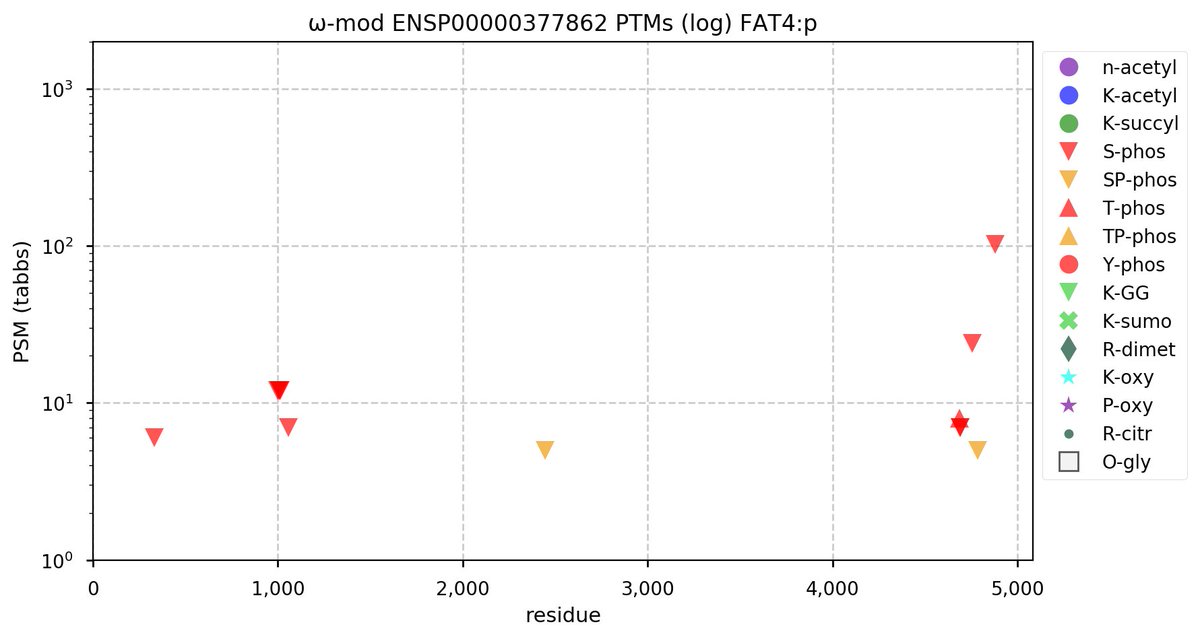
Sun Feb 06 18:06:06 +0000 2022🔗
1 – 292 ⮕ 2 – 292 & 4 – 292 (both acetylated)
#real_NT
Sun Feb 06 16:14:03 +0000 2022Modification diagram for #CTNNB1, the effector molecule in canonical #WNT pathways. It is a frequently observed phosphoprotein with 17 K+GGyl acceptors. Some of these sites are involved of the pathway's "destruction complex" CTNNB1 concentration regulation scheme. 🔗
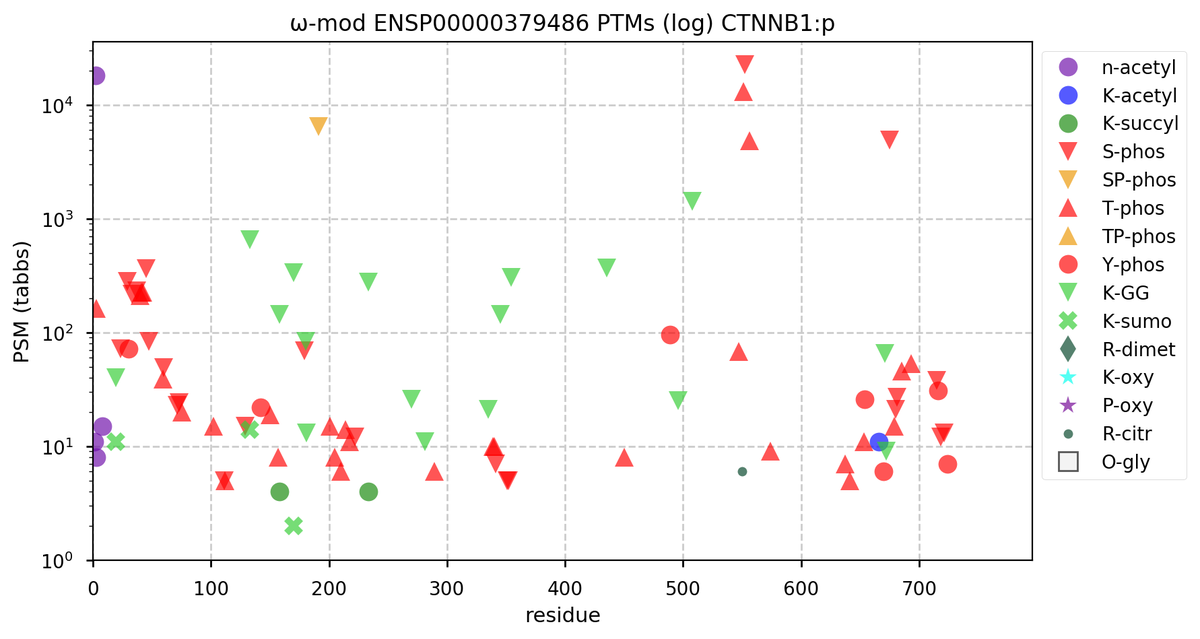
Sun Feb 06 14:26:16 +0000 2022@neely615 @ucdmrt @lgatt0 @molcellprot For comparison, EBI sites (ENSEMBL, UniProt) have 1 & NCBI sites have 2 or 3.
Sun Feb 06 14:17:08 +0000 2022@neely615 @ucdmrt @lgatt0 @molcellprot Brave is showing 18 trackers on the MCP home page.
Sun Feb 06 14:16:02 +0000 2022@neely615 @ucdmrt @lgatt0 @molcellprot I just tried a sampling of El$evier journals: they all have 10 trackers (according to Brave's tracker detector).
Sat Feb 05 19:54:10 +0000 2022🔗
1 – 881 ⮕ 2 – 881
#EXP_NT
Sat Feb 05 15:38:18 +0000 2022@stphnmaher EM may not realize that this particular group of protesters are very (very) unlikely to be in what might be called the "pro electric vehicle" camp. Hopefully an enterprising reporter may ask for their opinions on the subject.
Sat Feb 05 15:16:24 +0000 2022There is significant sequence overlap in the central domain of the subunits, but the N & C terminii are unique. Overlap of observed residues of GSK3B on GSK3A is shown below. 🔗

Sat Feb 05 15:11:42 +0000 2022Modification diagram for #GSK3A & #GSK3B, part of the #WNT pathways. Either may be part of the "Destruction complex" that adds ubiquitin to β-catenins. Phosphorylation of the N-terminal domain is distinctive. /1 🔗
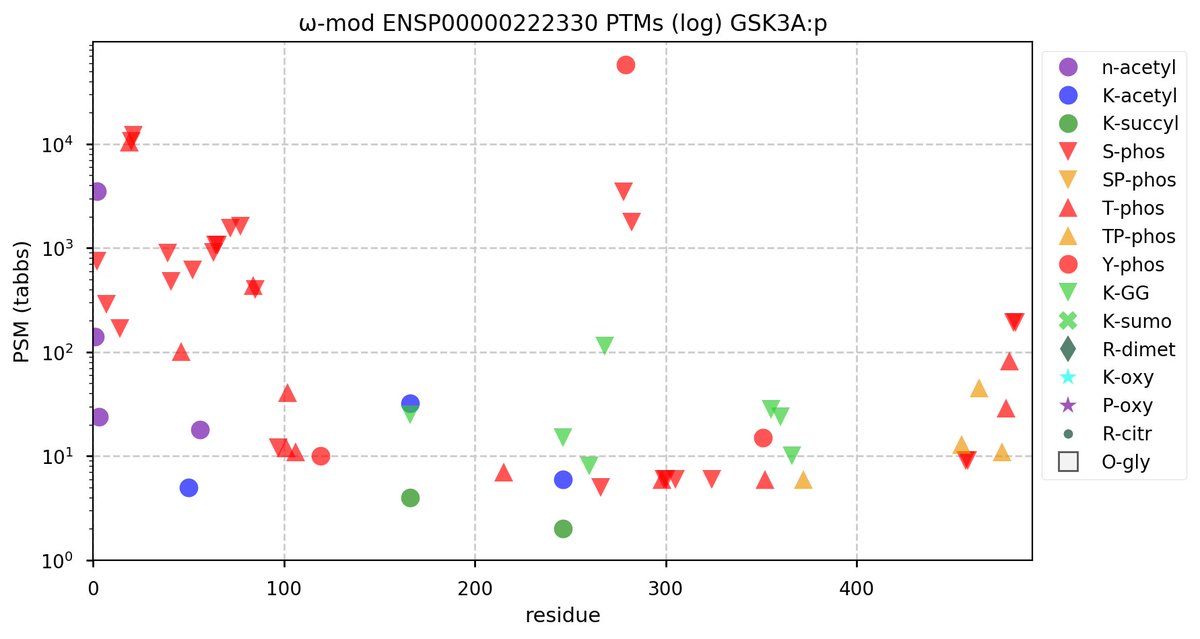
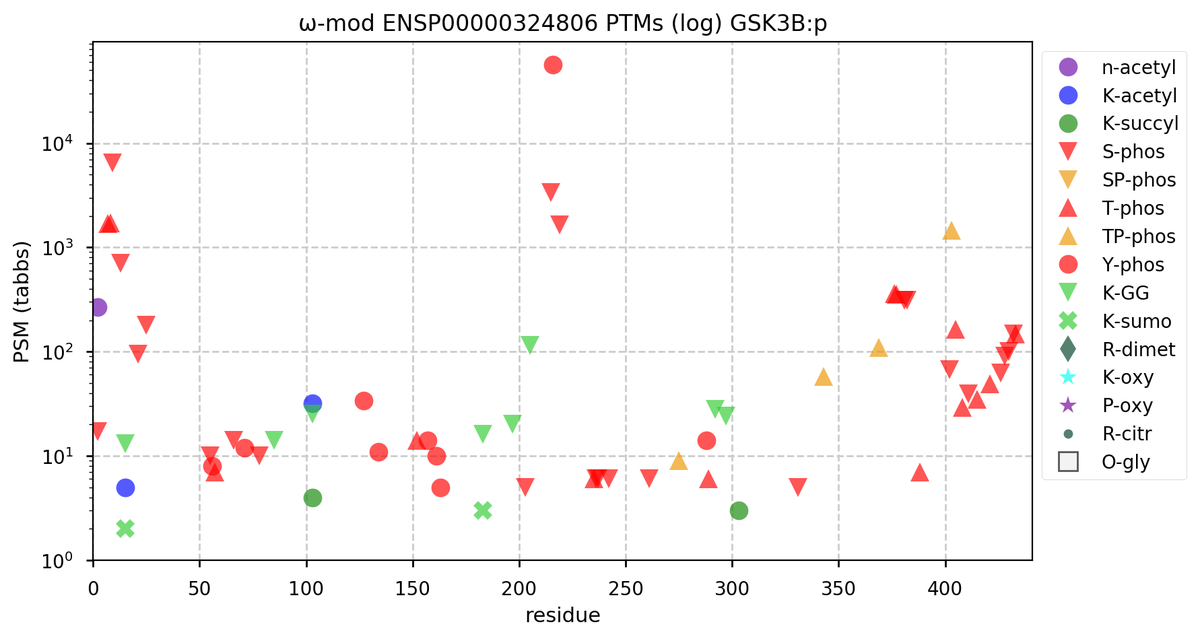
Sat Feb 05 14:51:32 +0000 2022The continental cold blob is is being encroached upon by a finger of warmer air from the Pacific, while it has almost pushed its way through to the south eastern Atlantic coast 🔗
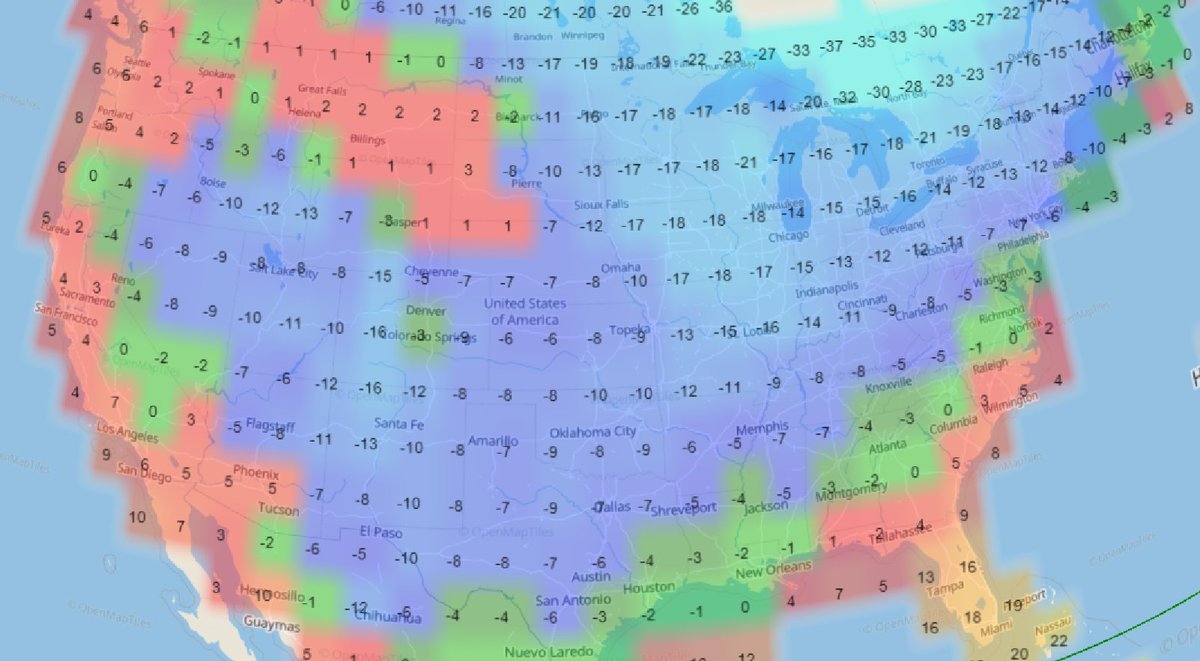
Sat Feb 05 13:32:55 +0000 2022@UCDProteomics @JohnRYatesIII @neely615 @pwilmarth @MattWFoster @DRAWheatcraft @seifert_steph This particular type of analysis would also be confounded by how much non-human keratin is present in labs. Based on the data, I would have to conclude that most European proteomics labs are populated with part-time shepherds.
Fri Feb 04 18:50:57 +0000 2022@neely615 @pwilmarth @MattWFoster @UCDProteomics @DRAWheatcraft @seifert_steph It is written as more of a "Vic Mackey" anti-hero role.
Fri Feb 04 17:52:05 +0000 2022When a site tells me I need to register to continue, I stop & go elsewhere. Every time.
Fri Feb 04 17:32:35 +0000 2022@pwilmarth @MattWFoster @UCDProteomics @DRAWheatcraft @neely615 @seifert_steph I was thinking more of a young, Vin Diesel type.
Fri Feb 04 16:31:44 +0000 2022@MattWFoster @UCDProteomics @pwilmarth @DRAWheatcraft @neely615 @seifert_steph "The Keratin Code" my latest thriller, is soon to be a major motion picture ... 🔗
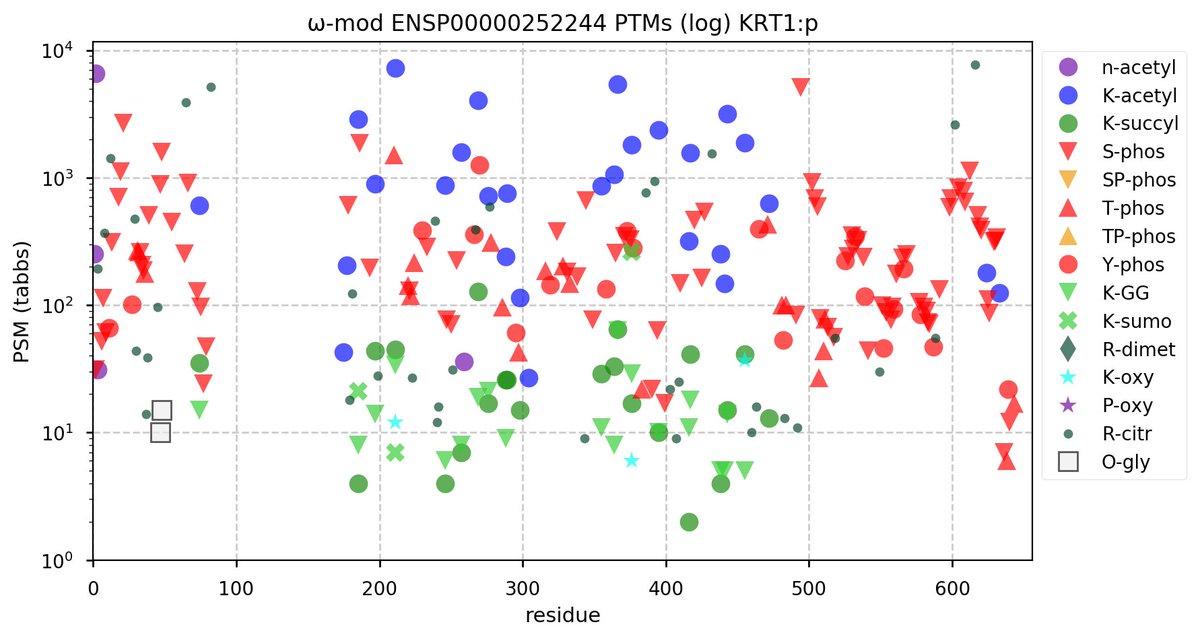
Fri Feb 04 15:52:01 +0000 2022The central deep freeze continues, extending well beyond the Great Plains. 🔗
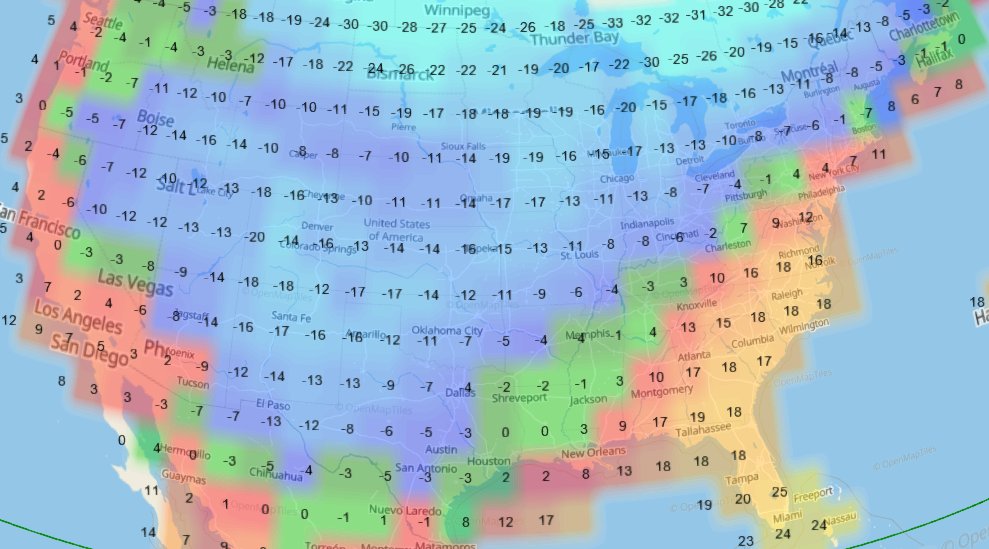
Fri Feb 04 15:28:06 +0000 2022🔗
1 – 296 ⮕ 41 – 296
#EXP_NT
Fri Feb 04 14:58:40 +0000 2022#APC is named after a disease syndrome caused by its malfunction, the also arch-sounding "Adenomatous polyposis coli". It is often abundant in nucleated diploid cells.
Fri Feb 04 14:58:17 +0000 2022Modification diagram for #APC, part of the #WNT pathways. Normally functions as part of the archly named "Destruction complex" that adds ubiquitin to β-catenins, limiting their concentration. APC is prolifically phosphorylated, but has almost no detectable ubiquitination. /1 🔗
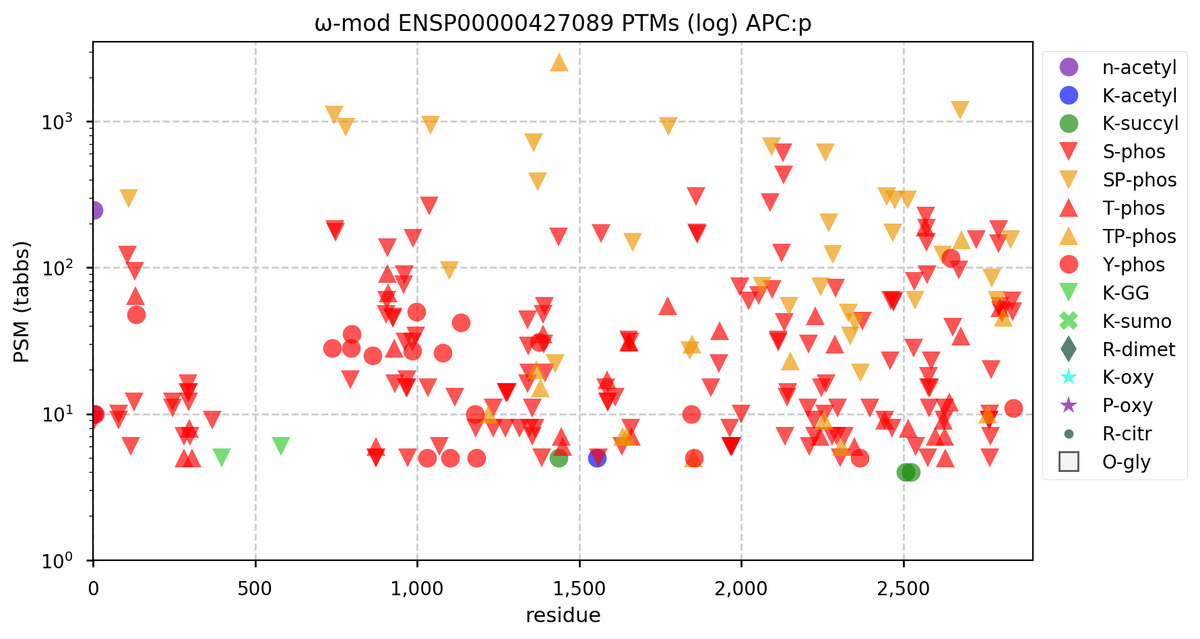
Thu Feb 03 21:37:19 +0000 2022Unusually cold in the south central US. For comparison, at this date & time last year, it was +21 °C in Austin TX. The current winter storm is along the cold front running from San Antonio TX to St John NB 🔗
Thu Feb 03 19:19:30 +0000 2022@AlexUsherHESA The latter.
Thu Feb 03 19:01:04 +0000 2022PXD020972 has some nice high resolution MS/MS runs (from 🔗).
Thu Feb 03 16:26:22 +0000 2022🔗
1 – 382 ≟ 20 – 382
#EXP_NT
Thu Feb 03 16:02:46 +0000 2022Neither subunit seems to be a target of ubiquitin signalling, which I find kind of odd.
Thu Feb 03 15:31:50 +0000 2022@andy___jones @dtabb73 On C, it serves as an alternate Cys blocking reagent generating -57+71 = +14 if you are using fixed C+carbamidomethyl in the id software.
Thu Feb 03 15:22:03 +0000 2022@andy___jones @dtabb73 Propionamide, +71.037114 Da. Depending on sample prep, it can be quite common on peptide C, K and N-terminii
Thu Feb 03 15:09:11 +0000 2022Modification patterns for human #AXIN1 & #AXIN2 subunits, which are used in #WNT pathways, interacting with the membrane transducer LRP5/6. Phosphorylation is concentrated into discrete domains in both. The 2 sequences have no tryptic peptide overlap. 🔗
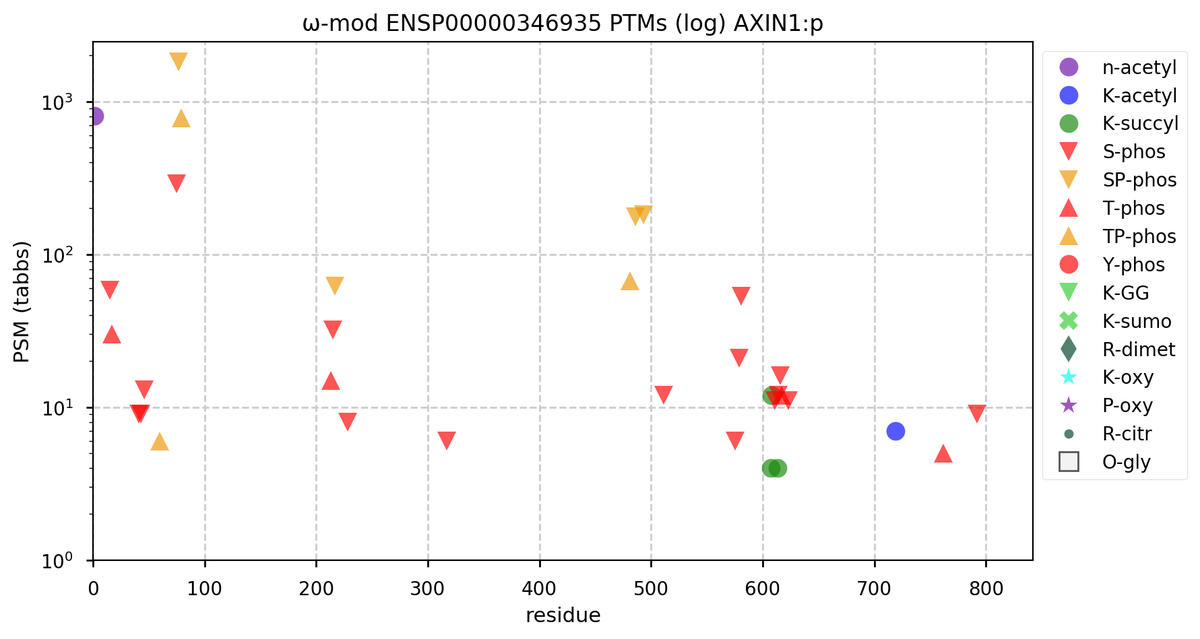
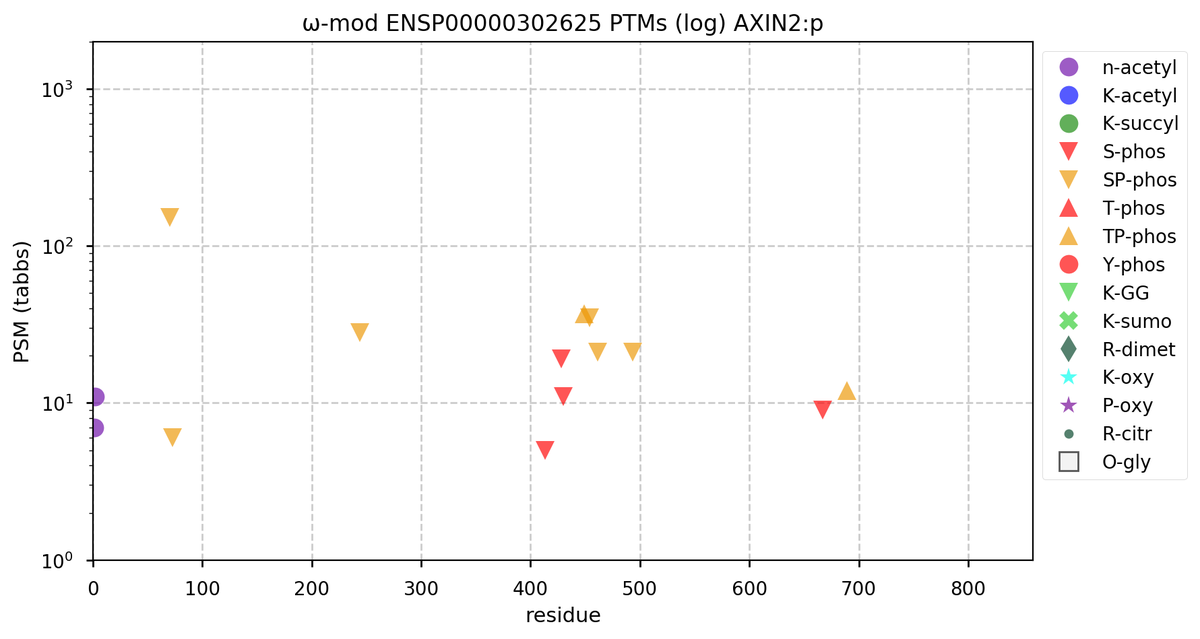
Thu Feb 03 13:09:44 +0000 2022@MattWFoster Is that number based on the LOD or LOQ?
Wed Feb 02 21:33:56 +0000 2022Thanks to everyone who participated. A majority of respondents found nothing wrong with the practice. I am happy to be the minority.
Wed Feb 02 17:38:31 +0000 2022@nesvilab It came through here yesterday. But it looks like MI will be getting more snow (~25 cm) than we got (only ~5 cm) 🔗
Wed Feb 02 17:12:54 +0000 2022'And wikipedia is about a zillion times better than the scientific literature ... in correcting mistakes when they are clearly labeled.' 🔗
Wed Feb 02 16:48:16 +0000 2022Is there a unicode math/logic/editorial symbol to replace 'should be' in:
X should be Y.
I'm using:
X ≟ Y
but the ? makes it feel more like:
should X be Y?
Wed Feb 02 16:11:23 +0000 2022Modification patterns for the LRP5 & LRP6 subunits (LRP5/6), which are used alternately in human #WNT pathways. The phosphorylation state of the C-terminal cytoplasmic domain signals extracellular WNTx binding to Frizzled #LRP5 #LRP6 🔗
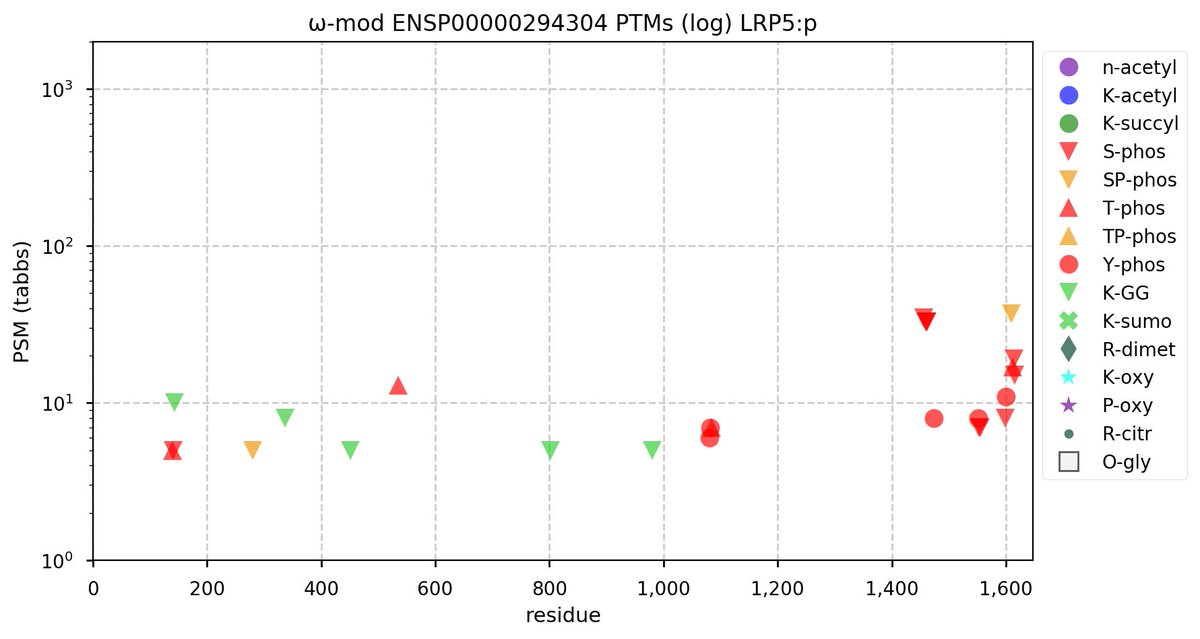
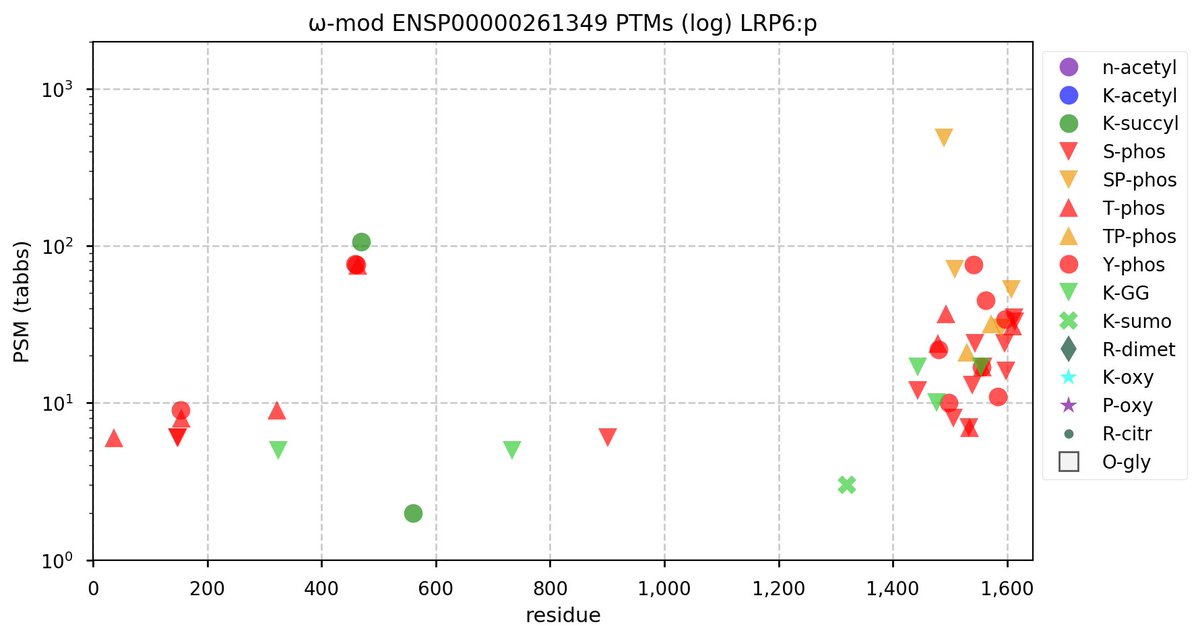
Wed Feb 02 13:42:38 +0000 2022🔗
1 – 21 ≟ 1 – 30
22 – 398 ≟ 31 – 398
Tue Feb 01 21:55:34 +0000 2022I have to admit spending the afternoon cycling through every available remix of "Being Boiled" hasn't had the calming effect I had imagined that it would.
Tue Feb 01 21:25:03 +0000 2022Is it OK to formulate the title of a talk or paper as a question you have no intention of answering?
Tue Feb 01 20:49:38 +0000 2022@scalzi "Industry insider"
Tue Feb 01 20:27:10 +0000 2022@miclugo @michaelhoffman Probably from someone who is going to name their next kid "66a9f304b2bd6f5b4f1ca3c44f32222b10e43265ce795541977852bad1e817a6".
Tue Feb 01 19:55:13 +0000 2022🔗
1-? = 1-37
?-457 = 38-457
Tue Feb 01 16:20:41 +0000 2022All three subunits show alternate initiation of the messenger RNA, generating novel N-termini at the second M (start) codon in the respective sequences (indicated by the purple dots to the left of residues 1 or 2).
Tue Feb 01 16:04:23 +0000 2022Modification patterns for the 3 Dishevelled subunits in human. Phosphorylation of the individual subunits is the primary step in cytoplasmic signaling from the Frizzled membrane transducer in the #WNT pathway. #DVL1 #DVL2 #DVL3 🔗
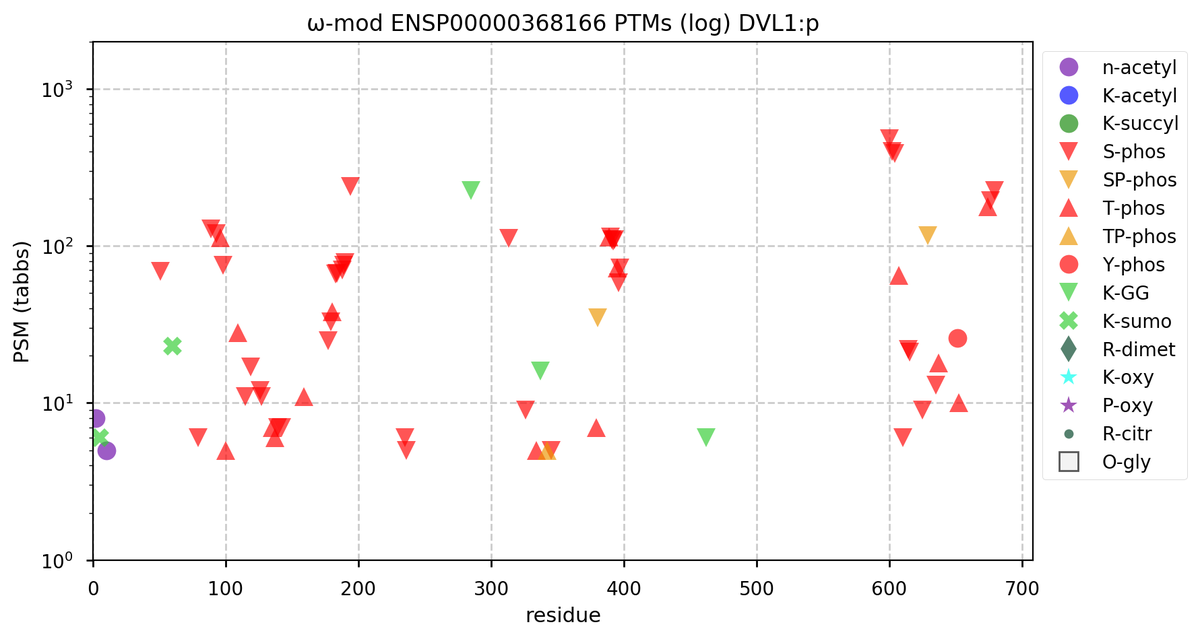
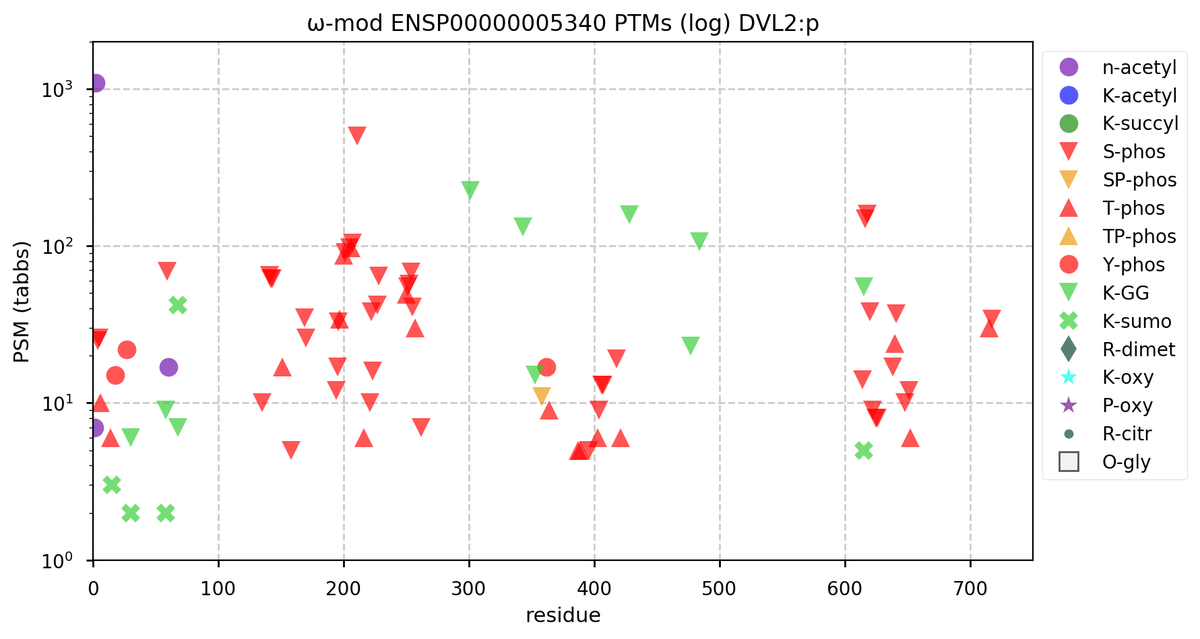
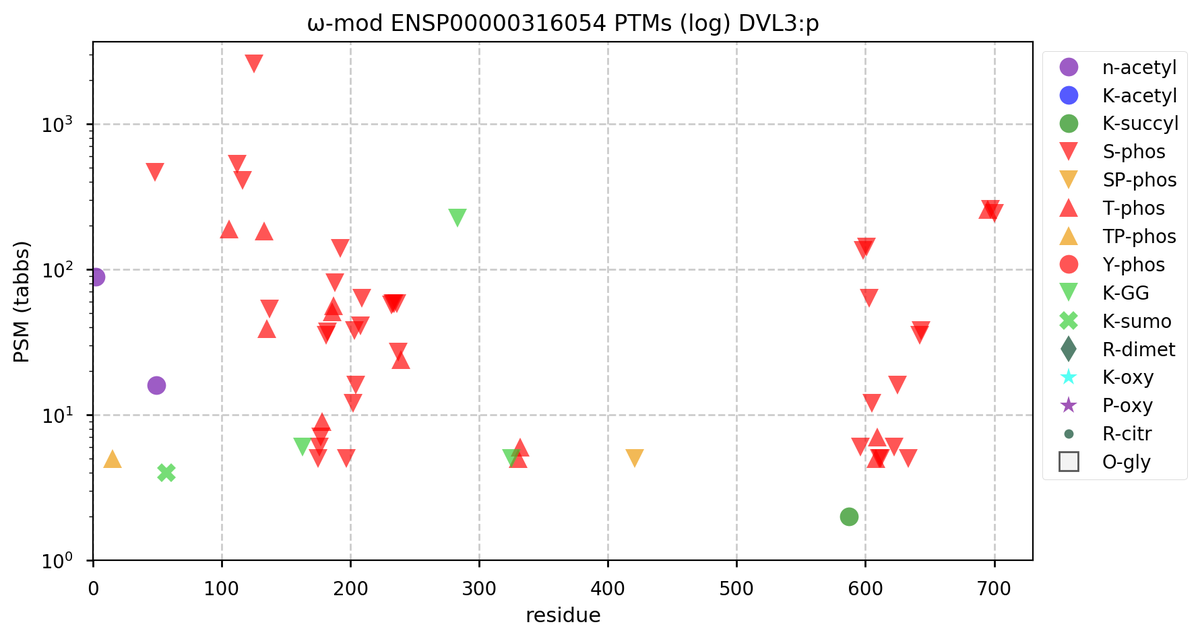
Mon Jan 31 16:13:10 +0000 2022@neely615 @astacus I tried it, but unless I'm doing something wrong, this seems to be an index to point you to the various existing silos, rather than an attempt to integrate anything.
Mon Jan 31 15:39:39 +0000 2022@neely615 @astacus I frankly don't know an example of "the opposite". I suspect anything exploring the idea of broader integration would be unfundable.
Mon Jan 31 15:38:15 +0000 2022@neely615 @astacus It is the tendency of bioinformatics resources to silo single types of molecule (DNA/RNA/proteins/sugars/lipids) & annotate them as though the others didn't exist (or at least weren't very important).
Mon Jan 31 14:55:15 +0000 2022PXD024716 (from 🔗) has some great examples of side-reaction peptide N-terminal succinylation in TMT10 preps, particularly how that modification segregates in high pH RP chromatography.
Sun Jan 30 19:02:27 +0000 2022Odd flex in the jet stream (winds at 250 hPa) resulting in Storm Corrie/Malik across the northern British Isles. 🔗
Sun Jan 30 16:26:26 +0000 2022@NLKProteomics @KentsisResearch For example, 🔗 assigns the N-terminal acetylation to index "0", rather than the more compatible "2", which is the common practice for locating modifications using protein coordinates.
Sun Jan 30 16:22:50 +0000 2022@NLKProteomics @KentsisResearch Thanks for fixing that problem: the new certificate works. Another question: why restart numbering of modifications using a 0-based index on the proteoforms.
Sun Jan 30 15:51:20 +0000 2022IMHO, molecular siloing is the worst (& most common) tendency among protein bioinformatics resources.
Sat Jan 29 15:40:07 +0000 2022Not-smartness (pakledism) is always my go-to explanation. 🔗
Sat Jan 29 14:55:57 +0000 2022I deal with weird/screwed-up data every day, some of which looks very fishy. Have the authors of 🔗 come up with something that would be useful to detect fraud as opposed to some mix of poor-training/bad-record-keeping/not-smartness?
Sat Jan 29 13:18:22 +0000 2022C1orf194:p, name recently changed to CFAP276. Homologs present across the metazoan lineage. Referred to as 1700013F07Rik:p in mouse. 4/ 🔗
Sat Jan 29 13:18:22 +0000 2022C1orf194:p.S145F 1:g.109106540G>A, rs62623711 (all tissue S:F 0.943:0.057) vaf=3%, Δm=60.0364, VAF by population group: african <1%, american 3%, east asian <1%, european 5%, south asian 2%. 3/
Sat Jan 29 13:18:21 +0000 2022C1orf194:p, θ(max) = 69. aka CFAP276. Not observed in MHC peptide experiments. Found in spermatozoa & lung epithelium: observed in some ovarian cancer tissue. 2/ 🔗

Sat Jan 29 13:18:21 +0000 2022>C1orf194:p, chromosome 1 open reading frame 194 (Homo sapiens) Small subunit; CTMs: P2+acetyl; PTMs: 0×K+acetyl; 0×K+GGyl; 0×K+SUMOyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: S145F (3%); mature form: (2-169) [139×, 1.1 kTa]. #ᗕᕱᗒ #俪㳅쭉 1/ 🔗
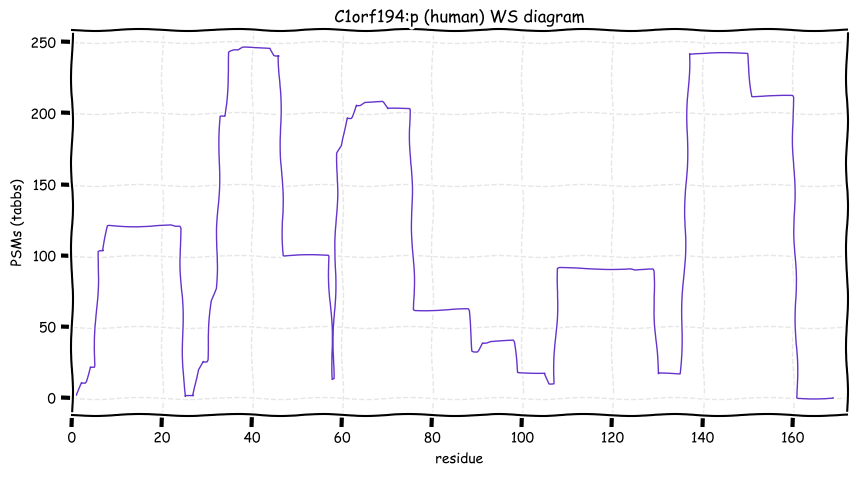
Fri Jan 28 19:52:11 +0000 2022There really aren't that many things that commonly screw up proteomics data, but there are lots of possible combinations of those things 😱
Fri Jan 28 17:54:33 +0000 2022@neely615 @UCDProteomics It is 4 °F here right now and I would describe the weather (without irony) as "pleasant".
Fri Jan 28 17:46:33 +0000 2022@NCI_HRodriguez @byu_sam I like the approach. I currently have to rely on a PI blacklist.
Fri Jan 28 16:02:20 +0000 2022@pmxpapers The code is very well documented. Good job.
Fri Jan 28 15:46:22 +0000 2022Thanks to everyone who participated: the tongue-in-cheek answer won out. While it is very speculative―in Nature―this review discusses some of the general possibilities for viral ubiquitination 🔗
Fri Jan 28 15:11:18 +0000 2022@KentsisResearch @NLKProteomics The HTTPS certificate also appears to be incorrectly configured.
Fri Jan 28 13:09:23 +0000 2022CFAP107:p, name recently changed from C1orf158. Homologs present across the metazoan lineage. Referred to as 1700012P22Rik:p in mouse. Pic from OrthoDB. 4/ 🔗

Fri Jan 28 13:09:23 +0000 2022CFAP107:p.M174I 1:g.12760888G>A, rs200686257 (all tissue M:I 0.955:0.045) vaf=<1%, Δm=-17.9564, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Fri Jan 28 13:09:22 +0000 2022CFAP107:p, θ(max) = 80. aka MGC35194, C1orf158. Not observed in MHC peptide experiments. Abundant in spermatozoa & lung epithelium: found in ciliated cells only. The pic shows the coverage diagram for observed peptides in spermatozoa samples. 2/ 🔗
Fri Jan 28 13:09:22 +0000 2022>CFAP107:p, cilia and flagella associated protein 107 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 0×K+acetyl; 0×K+GGyl; 0×K+SUMOyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: T45I (2%), M174I (<1); mature form: (1-194) [108×, 0.9 kTa]. #ᗕᕱᗒ #靌쯾槴 1/ 🔗
Thu Jan 27 18:27:17 +0000 2022@chrashwood Note: A-549 is just what is says in the paper (🔗 ). The data shows strong signals from large T antigen [Simian virus 40], control protein E1B 19K & E1B 55K [Human mastadenovirus C], which is consistent with the infected cells being HEK-293T instead.
Thu Jan 27 17:21:45 +0000 2022@chrashwood That is, lack of PTMs other than N-linked glycosylation.
Thu Jan 27 17:11:22 +0000 2022@chrashwood The GGylation was found in a ubi enrichment study done in A-549 cells (the only one I have). But the lack of all other PTMs is across A-549, HEK-293T and Vero cells.
Thu Jan 27 17:07:40 +0000 2022@dtabb73 @ReneZahedi "tabb" is my official unit for the number of PSMs assigned.
Thu Jan 27 15:51:56 +0000 2022In order to understand this information in a way that allows it to be used practically, what would be required?🧐
Thu Jan 27 15:32:03 +0000 2022I regret to admit that although this must mean something, I have no idea what that might be. 🔗
Thu Jan 27 13:01:15 +0000 2022C1orf159:p, has homologs across the vertebrate lineage. Refered to as 9430015G10Rik:p in mice and RGD1311517:p in rats 3/ 🔗
Thu Jan 27 13:01:14 +0000 2022C1orf159:p, θ(max) = 18. aka FLJ20584. Very rarely observed in MHC class 1 peptide experiments. Found in urine & HEK-293T cells. Domains: (19-110) extracellular, (111-133) TM, (134-344) cytoplasmic. 2/ 🔗

Thu Jan 27 13:01:13 +0000 2022>C1orf159:p, chromosome 1 open reading frame 159 (Homo sapiens) Small subunit; CTMs: none; PTMs: N92+glycosyl; SAAVs: none; mature form: (19-344) [691×, 1.0 kTa]. #ᗕᕱᗒ #檦䢻䀒 1/ 🔗
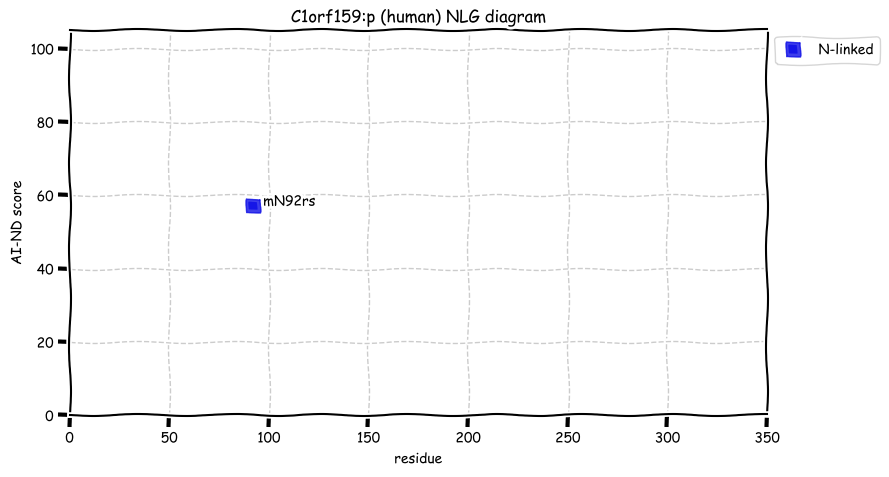
Wed Jan 26 15:08:57 +0000 2022@MagnusPalmblad @astacus @neely615 Those places are full of "friendly strangers".
Wed Jan 26 14:07:10 +0000 2022@astacus @neely615 Instruments are for later: hospitality suites are the gateways.
Wed Jan 26 13:36:53 +0000 2022@astacus @neely615 A criticism that lines up with my view that the field is currently 3-mass-spec-companies-in-a-trench-coat. 😉
Wed Jan 26 13:03:00 +0000 2022C1orf56:p, has homologs in several mammalian lineages. Referred to as Gm128:p in rodents. 4/ 🔗
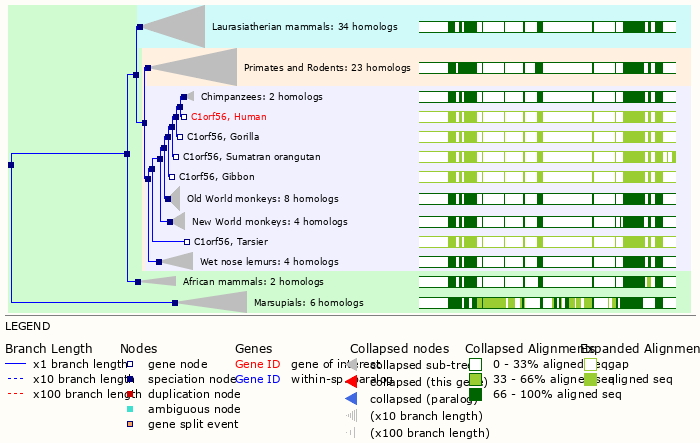
Wed Jan 26 13:02:59 +0000 2022C1orf56:p.S267N 1:g.151048647G>A, rs142217636 (all tissue S:N 0.955:0.045) vaf=<1%, Δm=27.0109, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Wed Jan 26 13:02:59 +0000 2022C1orf56:p, θ(max) = 70. aka FLJ20519, MENT. Very rarely observed in MHC class 1 peptide experiments. Found in spermatozoa, testis, urine, blood plasma & CSF: very rare in cell lines. 2/ 🔗
Wed Jan 26 13:02:58 +0000 2022>C1orf56:p, chromosome 1 open reading frame 56 (Homo sapiens) Small subunit; CTMs: none; PTMs: 0×K+acetyl; 0×K+GGyl; 0×K+SUMOyl; 5×S, 1×T, 0×Y+phosphoryl; 3×S, 1×T+glycosyl; SAAVs: S267N (<1%); mature form: (28-341) [1,197×, 7.9 kTa]. #ᗕᕱᗒ #翶絸없 1/ 🔗
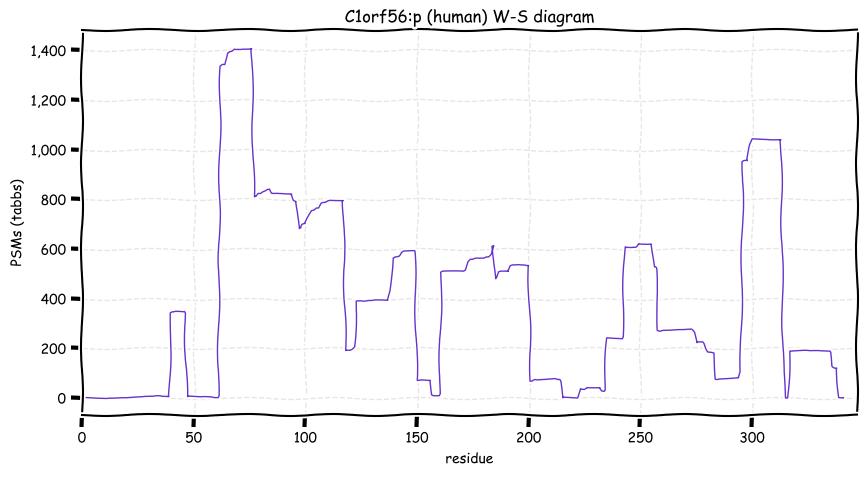
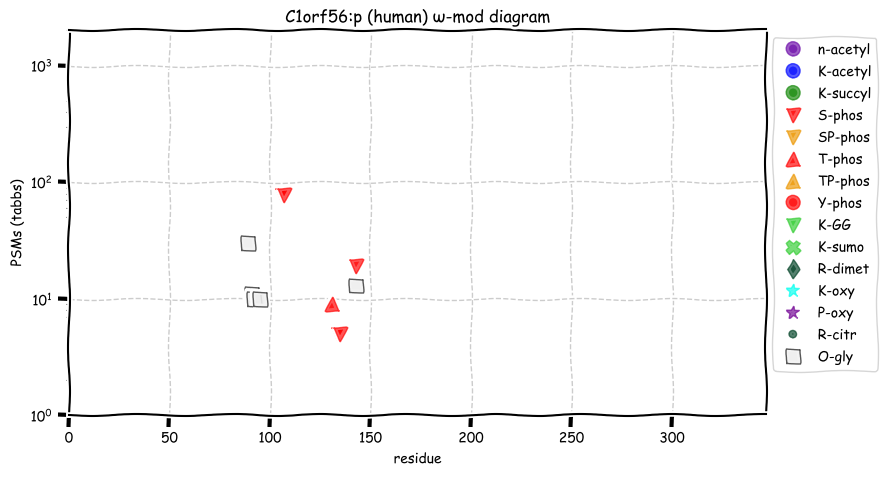
Tue Jan 25 18:00:42 +0000 2022@leprevostfv @byu_sam @ypriverol The most recent data set using iTRAQ in PRIDE is from 🔗
Tue Jan 25 17:57:09 +0000 2022@byu_sam @ypriverol TMT does have higher maximum multiplexing: I think iTRAQ tops out at 8 channels (but I could be wrong). Groups do tend to stick with their own "tried-and-true" + there are plenty of expts in which 8 channels is plenty. I would expect iTRAQ to continue to be used.
Tue Jan 25 16:58:41 +0000 2022ERVK3-1:p, observation of this protein is strongly correlated with the observation of translated LINE-1 ORF1. /4
Tue Jan 25 16:31:13 +0000 2022@ypriverol I would say "significant" rather than "huge": the various TMT reagents are more commonly used. iTRAQ does have its fans, though, like Steve Carr's mob.
Tue Jan 25 15:43:52 +0000 2022@ensembl @pwilmarth The move to align ENSEMBL annotation with gnomAD 3.1 is good news for me.
Tue Jan 25 15:19:34 +0000 2022... When attempting to analyze or reproduce results, keep in mind the likelihood that key parts of the experimental methods may have been recorded incorrectly in the associated metadata or manuscript."
Tue Jan 25 15:18:47 +0000 2022"Many datasets/papers contain serious errors in their metadata/methods sections. ... it is important to be skeptical of any experimental parameter (cell line, tissue type, modification reagents, quantitation methods, etc.) that may impact on your use of the data. /2
Tue Jan 25 15:17:44 +0000 2022IMHO the main barrier to useful implementation the FAIR principles are summed up in my warning to users of my own information repository: /1
Tue Jan 25 14:01:00 +0000 2022@AlexUsherHESA Canada has an additional issue: if someone has ambitions of participating in this type of endeavor, they are a few hours drive from somewhere that does these things really well. Fighting unis, govts & banks to do it locally isn't a very attractive alternative.
Tue Jan 25 13:04:21 +0000 2022ERVK3-1:p, homologues found in a small number of primates. This endogenous viral sequence normally has its translation supressed & it has no known function. It does not share any tryptic peptides with other human proteins. 3/ 🔗
Tue Jan 25 13:04:20 +0000 2022ERVK3-1:p, θ(max) = 86. aka none. Observed in MHC class 1 peptide experiments involving tumours. Found mainly in cancer tissue & cell lines, but may be present in extracellular vesicles in some conditions. 2/ 🔗
Tue Jan 25 13:04:19 +0000 2022>ERVK3-1:p, endogenous retrovirus group K3 member 1 (Homo sapiens) Small protein; CTMs: M1+acetyl; PTMs: 2×K+succinyl; 2×K+GGyl; 4×K+SUMOyl; 1×S, 1×T, 1×Y+phosphoryl; SAAVs: none; mature form: (1-109) [3,246×, 8.3 kTa].#ᗕᕱᗒ #饋앫ꦕ 1/ 🔗
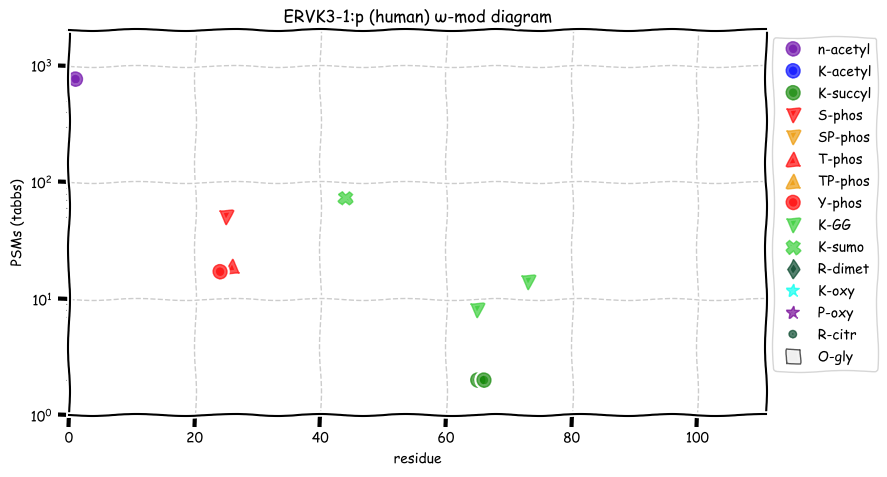
Mon Jan 24 12:57:40 +0000 2022C2or16:p, the most commonly observed translation (C2orf16-201) was recently removed from the GRC reference genome. The details of the gene's translation have changed at least 3 times: hopefully an improved translation will be made available in a future revision. 4/
Mon Jan 24 12:57:40 +0000 2022C2orf16:p.A660E 2:g.27578551C>A, rs1919126 (all tissue A:E 0.289:0.711) vaf=56%, Δm=58.0055, VAF by population group: african 74%, american 69%, east asian 85%, european 50%, south asian 51%. 3/
Mon Jan 24 12:57:39 +0000 2022C2orf16:p, θ(max) = 37. aka DKFZp434G118, P-S-E-R-S-H-H-S repeats containing. Rarely observed in MHC class 1 peptide experiments. Found in male & female reproductive tissue: very rare in cell lines. 2/ 🔗

Mon Jan 24 12:57:39 +0000 2022>C2orf16:p, chromosome 2 open reading frame 16 (Homo sapiens) Large subunit; CTMs: none; PTMs: K36+SUMOyl; S152, Y132+phosphoryl; SAAVs: A660E (56%), V685A (31%), I774V (27%); mature form: (1?-1984) [454×, 2.3 kTa]. #ᗕᕱᗒ #宝槕覬 1/ 🔗
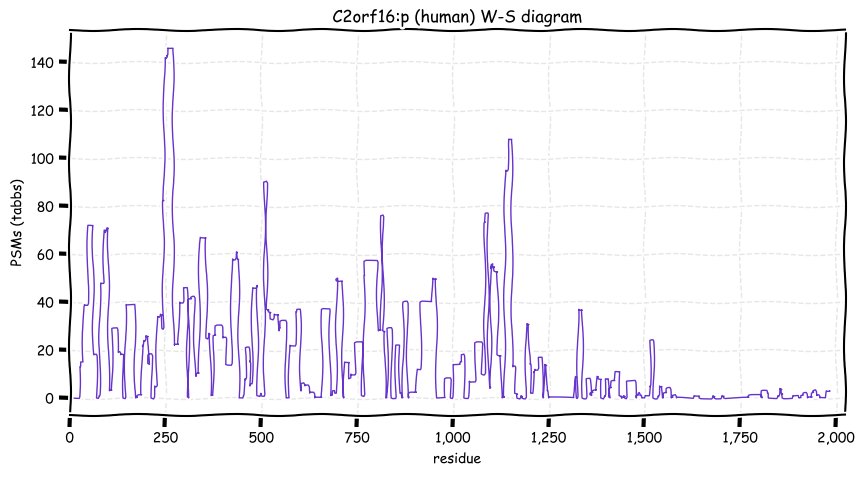
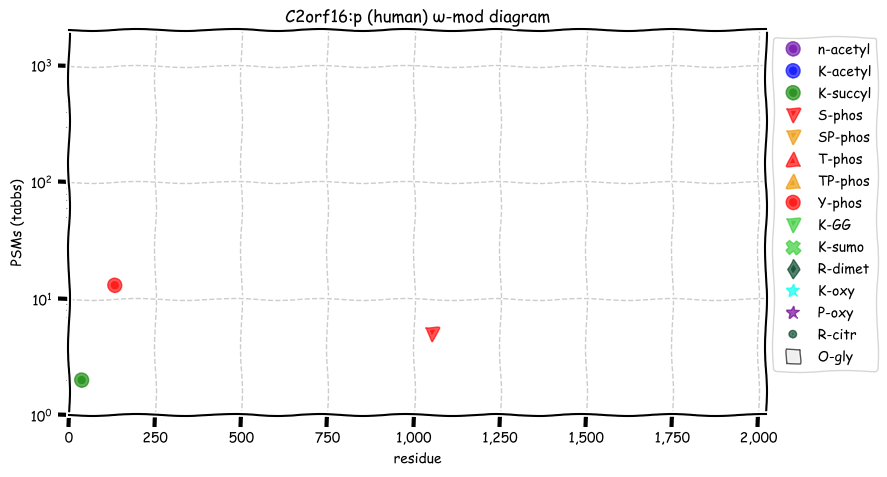
Sun Jan 23 22:30:05 +0000 2022@alistair604 @J_my_sci That looks like the explanation. I would not have guessed the postal regulations. Thanks!
Sun Jan 23 14:17:06 +0000 2022Does anybody know why Cell (for as long as I can remember) labels papers with the caveat "Advertisement"?
Sun Jan 23 14:08:36 +0000 2022@astacus @Putuma It reminds me of my stint running a GMP-regulated lab where we had to determine the amount of each protein and all contaminants in a vial (in grams). Scary stuff.😱
Sun Jan 23 13:37:54 +0000 2022C1orf100:p, this open reading frame is 1 of many uncharacterized proteins only translated in spermatozoa. They are abundant in sperm & probably involved in the unusual structure/function of sperm cells, but not necessary in other cell types. 3/
Sun Jan 23 13:37:54 +0000 2022C1orf100:p, θ(max) = 53. aka none. Not observed in MHC peptide experiments. Abundant in spermatozoa only. 2/ 🔗

Sun Jan 23 13:37:54 +0000 2022>C1orf100:p, chromosome 1 open reading frame 100 (Homo sapiens) Small subunit; CTMs: none; PTMs: 0×K+acetyl; 0×K+GGyl; 0×K+SUMOyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: none; mature form: (2?-147) [63×, 0.3 kTa]. #ᗕᕱᗒ #砚컒媈 1/ 🔗
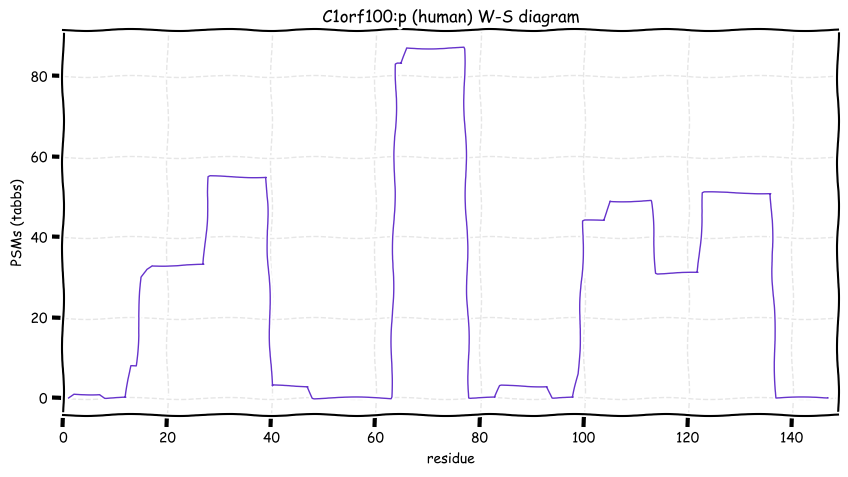
Sat Jan 22 21:48:07 +0000 2022@astacus @Putuma Imputation also helps the reproducibility quite a bit.
Sat Jan 22 21:44:34 +0000 2022@astacus @Putuma Also, it is best viewed on a log-log plot, where its "linearity" may manifest.
Sat Jan 22 21:42:26 +0000 2022@astacus @Putuma Linear? What sort of fancy-pants magic is that? Proteomics quant may be reproducible, but beyond that it is more of the "horseshoes & hand grenades" variety.
Sat Jan 22 20:38:12 +0000 2022@Putuma @astacus I'm not sure how you would go about identifying a protein without generating some type of "quantity" associated with it. Even the very limited quant you get from most types of PAGE gel staining has been put to good use.
Sat Jan 22 13:03:19 +0000 2022C7orf61:p, has homologs across the mammalian lineage. Absent in mice but present in voles and hamsters. 4/ 🔗
Sat Jan 22 13:03:19 +0000 2022C7orf31:p.A158T 7:g.25155134C>T, rs12535348 (all tissue A:T 0.941:0.059)vaf=42%, Δm=30.0106, VAF by population group: african 5%, american 39%, east asian 16%, european 49%, south asian 43%. 3/
Sat Jan 22 13:03:19 +0000 2022C7orf61:p, θ(max) = 56. aka IMAGE:4839025. Observed in MHC class 1 peptide experiments. Found in spermatozoa & samples likely to contain spermatozoa. 2/ 🔗
Sat Jan 22 13:03:18 +0000 2022>C7orf61:p, chromosome 7 open reading frame 61 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 0×K+acetyl; 0×K+GGyl; 0×K+SUMOyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: A158T (<1%); mature form: (1-206) [263×, 2.4 kTa]. #ᗕᕱᗒ #畣띝嵑 1/ 🔗
Fri Jan 21 14:29:10 +0000 2022@slavov_n If none of the originating groups have the resources (& the idea has merit), often a 3rd group that does have the necessary resources/reputation will take over the idea and run with it.
Fri Jan 21 13:36:14 +0000 2022@slavov_n It is mainly determined by resources, i.e., which group has the ability to follow up, producing a stream of related papers & being invited to promote the work at conferences.
Fri Jan 21 12:45:52 +0000 2022C7orf31:p, has homologs across the vertebrate lineage. Referred to as 4921507P07Rik:p in rodents. 4/ 🔗
Fri Jan 21 12:45:52 +0000 2022C7orf31:p.A158T 7:g.25155134C>T, rs12535348 (all tissue A:T 0.941:0.059)vaf=42%, Δm=30.0106, VAF by population group: african 5%, american 39%, east asian 16%, european 49%, south asian 43%. 3/
Fri Jan 21 12:45:51 +0000 2022C7orf31:p, θ(max) = 65. aka none. Rarely bserved in MHC class 1 peptide experiments. Found in spermatozoa & samples likely to contain spermatozoa. Cell line observations involve SUMOylation enrichment expts. 2/ 🔗

Fri Jan 21 12:45:51 +0000 2022>C7orf31:p, chromosome 7 open reading frame 31 (Homo sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 0×K+acetyl; 1×K+GGyl; 4×K+SUMOyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: A158T (42%); mature form: (1-590) [252×, 2.3 kTa]. #ᗕᕱᗒ #㥍䦝㣛 1/ 🔗
Fri Jan 21 01:36:02 +0000 2022PXD029418 has some examples of good recovery of cysteine-containing-peptides with the cysteines converted to the corresponding sulfonic acid derivative.
Thu Jan 20 16:49:53 +0000 2022@pwilmarth @andy___jones @uniprot @pride_ebi @ypriverol @juan_vizcaino By vague, I mean their description is not precise enough for me to reproduce their method. I can't go to UniProt and retrieve the same FASTA file they used, based on their description.
Thu Jan 20 15:19:10 +0000 2022@andy___jones @uniprot @pride_ebi @ypriverol @juan_vizcaino People are often rather vague when describing the sequences they used, e.g., "DDA data was searched ... against the Swissprot reviewed subset of the human UniProt database". I am often not sure exactly what they are referring to in their methods descriptions.
Thu Jan 20 12:56:32 +0000 2022C10orf82:p, high accuracy, high resolution AF2 3-dimensional structure available 🔗, but of limited utility. 5/ 🔗
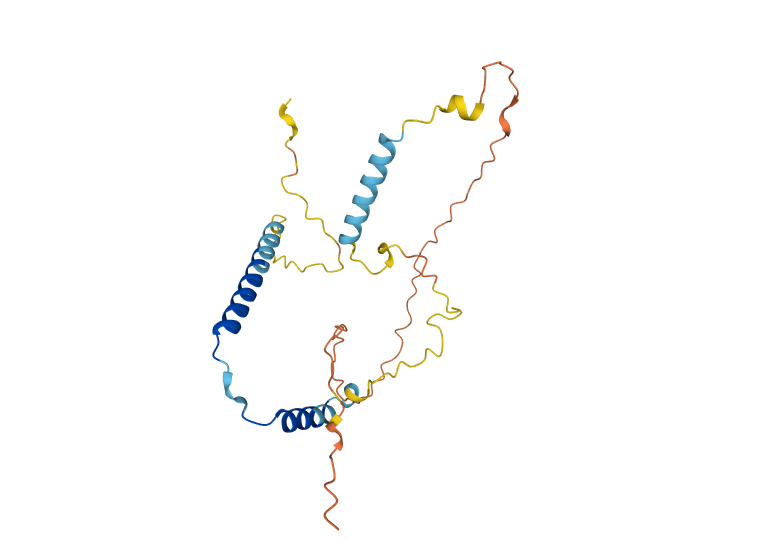
Thu Jan 20 12:56:31 +0000 2022C10orf82:p, has homologs across the vertebrate lineage. Referred to as 1700019N19Rik:p in rodents. 4/ 🔗
Thu Jan 20 12:56:30 +0000 2022C10orf82:p.L100F 10:g.116664924G>A, rs145415319 (all tissue L:F 0.750:0.250) vaf=<1%, Δm=33.9843, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Thu Jan 20 12:56:30 +0000 2022C10orf82:p, θ(max) = 87. aka MGC33547, Em:AC016825.4. Not observed in MHC peptide experiments. Found in spermatozoa & samples likely to contain spermatozoa. Observations in HEK-293T are the result of artificial (non-endogenous) translation. 2/ 🔗
Thu Jan 20 12:56:30 +0000 2022>C10orf82:p, chromosome 10 open reading frame 82 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 0×K+acetyl; 0×K+GGyl; 0×K+SUMOyl; 0×S, 0×T, 0×Y+phosphoryl; SAAVs: L100F (<1%); mature form: (1-234) [142×, 1.7 kTa]. #ᗕᕱᗒ #ꢌ饤捄 1/ 🔗
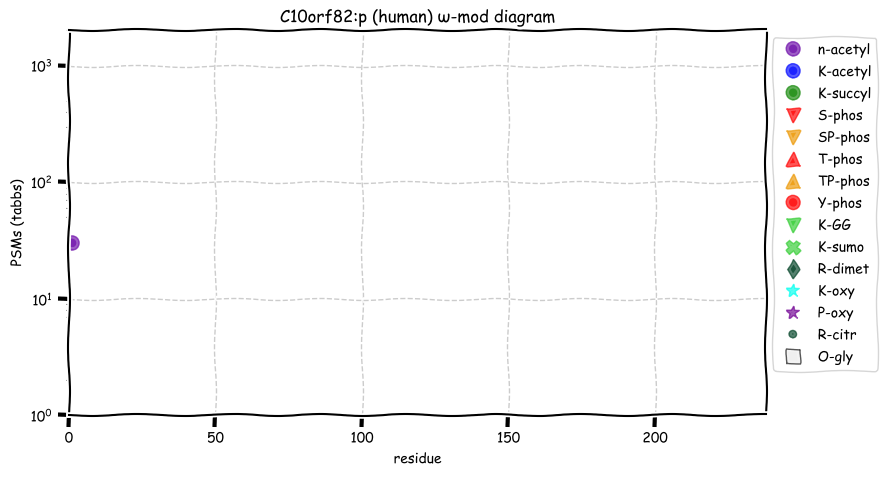
Wed Jan 19 20:50:52 +0000 2022@pwilmarth @slavov_n The problem (IMHO) is a lack of theory to use when interpreting the data. This particular sort of impasse has occurred repeatedly in science, when a measurement technology surges ahead of the understanding necessary to use it.
Wed Jan 19 17:06:28 +0000 2022Working through the data would be a good way to introduce students (professors?) to the difference between the concepts of species (+ ssp) and clinical strains/isolates in medical microbiology.
Wed Jan 19 16:56:24 +0000 2022PXD010504: if you are interested in pathological bacteria, this is a nice set of data from 🔗
Wed Jan 19 12:57:47 +0000 2022C19orf47:p, high accuracy, high resolution AF2 3-dimensional structure available 🔗, but of limited utility. 5/ 🔗
Wed Jan 19 12:57:46 +0000 2022C19orf47:p, has homologs across the chordate lineage. Referred to as 2310022A10Rik:p in rodents. 4/ 🔗
Wed Jan 19 12:57:46 +0000 2022C19orf47:p.P327L 19:g.40322171G>A, rs370321945 (all tissue P:L 0.999:0.001) vaf=<1%, Δm=16.0313, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Wed Jan 19 12:57:46 +0000 2022C19orf47:p, θ(max) = 46. aka FLJ36888. Observed in MHC class 1 peptide experiments. Found in many tissues & cell lines: absent in fluids. 2/ 🔗
Wed Jan 19 12:57:45 +0000 2022>C19orf47:p, chromosome 19 open reading frame 47 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 1×K+acetyl; 1×K+GGyl; 11×K+SUMOyl; 16×S, 5×T, 0×Y+phosphoryl; SAAVs: P327L (<1%); mature form: (1-422) [4,684×, 12 kTa]. #ᗕᕱᗒ #ꠔ䪒뱾 1/ 🔗
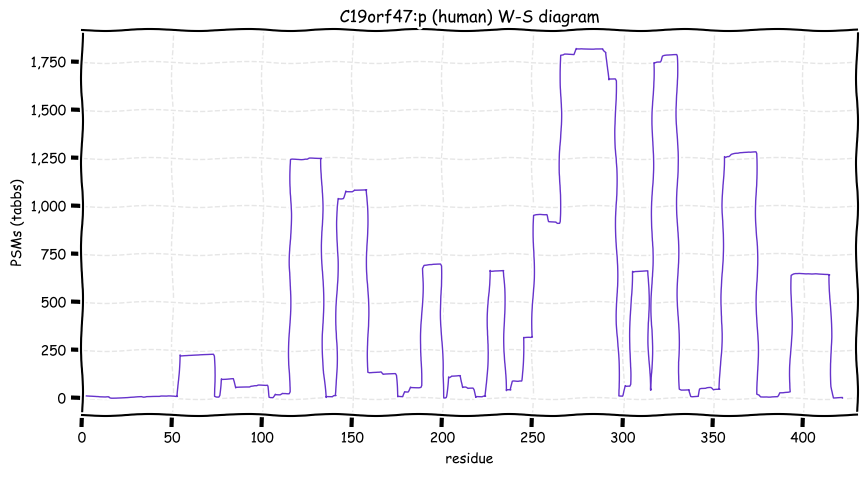
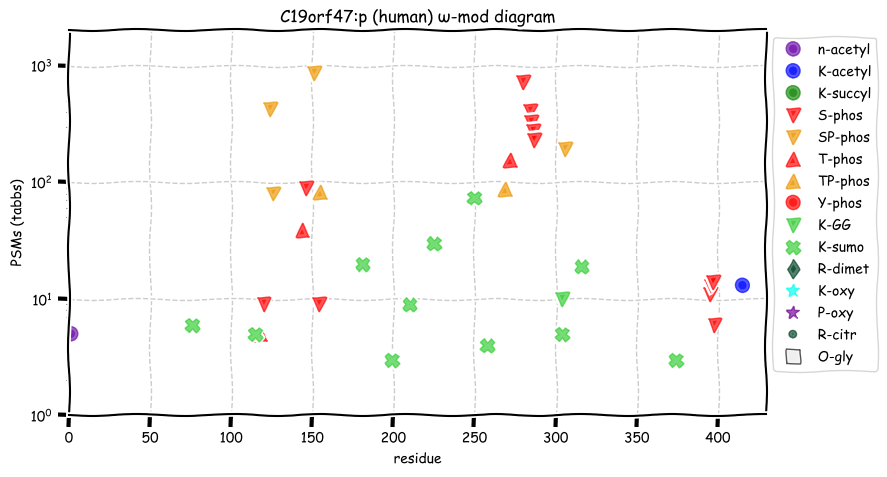
Tue Jan 18 18:27:20 +0000 2022HEK-293 is getting on my nerves a bit, too.
Tue Jan 18 18:22:28 +0000 2022I'm suffering from severe HeLa fatigue ☹️
Tue Jan 18 16:46:09 +0000 2022@ChrisRWiebe I started at UBC in 2006, so it has been a part of CRCs for quite a while. National standards would be good, but it would take a complete overhaul of the program as it has always been very "hands-off", leaving implementation up to the institutions.
Tue Jan 18 16:38:06 +0000 2022@ChrisRWiebe @CRC_CRC It sounds like U of W is using its CRCs better than most. I had a Tier 1 at UBC where it was a title only and having it was actually a significant disadvantage.
Tue Jan 18 12:50:40 +0000 2022C18orf25:p, high accuracy, high resolution AF2 3-dimensional structure available 🔗, but of limited utility. 5/ 🔗
Tue Jan 18 12:50:39 +0000 2022C18orf25:p, has homologs across the vertebrate lineage. Referred to as 8030462N17Rik:p in rodents. 4/ 🔗
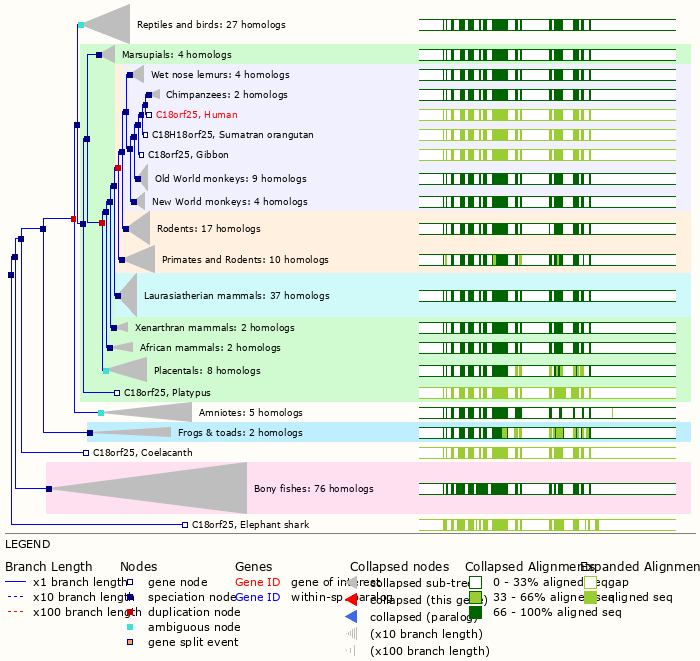
Tue Jan 18 12:50:38 +0000 2022C18orf25:p.D242E 18:g.46253707T>A, rs201644841 (all tissue G:D 0.999:0.001) vaf=<1%, Δm=14.0157, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Tue Jan 18 12:50:38 +0000 2022C18orf25:p, θ(max) = 47. aka MGC12909, ARKL1, RNF111L1, Ark2N. Observed in MHC class 1 peptide experiments. Found in many tissues & cell lines: rare in fluids. Alternative translation initiation at M3 observed in human, rat & mouse. 2/ 🔗
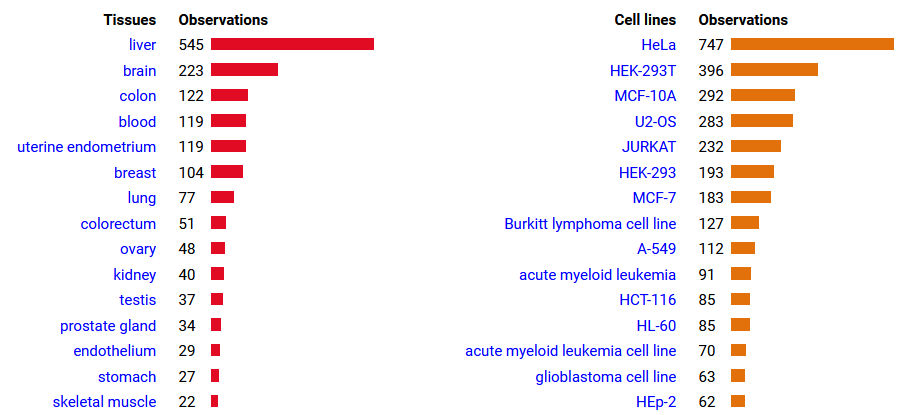
Tue Jan 18 12:50:38 +0000 2022>C18orf25:p, chromosome 18 open reading frame 25 (Homo sapiens) Small subunit; CTMs: M3+acetyl; PTMs: 2×K+acetyl; 1×K+GGyl; 6×K+SUMOyl; 19×S, 5×T,1×Y+phosphoryl; SAAVs: D242E (<1%); mature form: (3-343) [8,992×, 35 kTa]. #ᗕᕱᗒ #슭芣謆 1/ 🔗
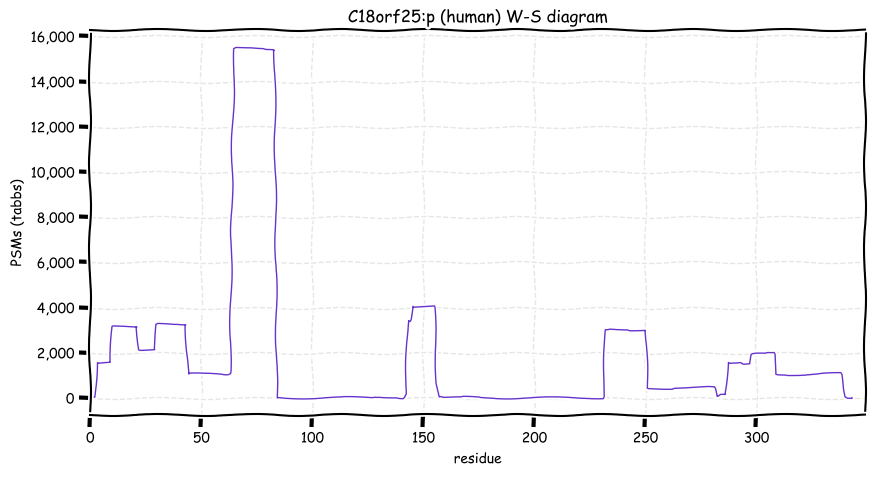
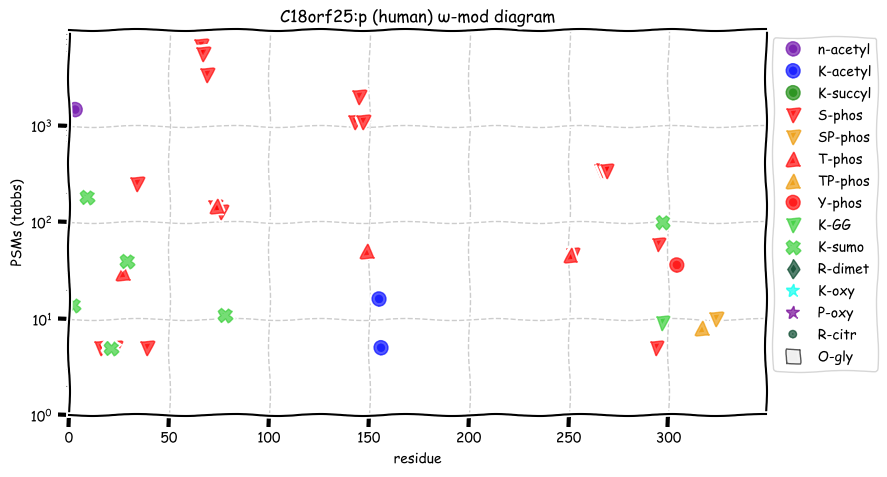
Mon Jan 17 17:48:22 +0000 2022Candiano_F_1017_ESO_urine_MSK_35_073.raw is a beauty 🔗
Mon Jan 17 15:24:51 +0000 2022C1orf52:p, high accuracy, high resolution AF2 3-dimensional structure available 🔗 /5 🔗
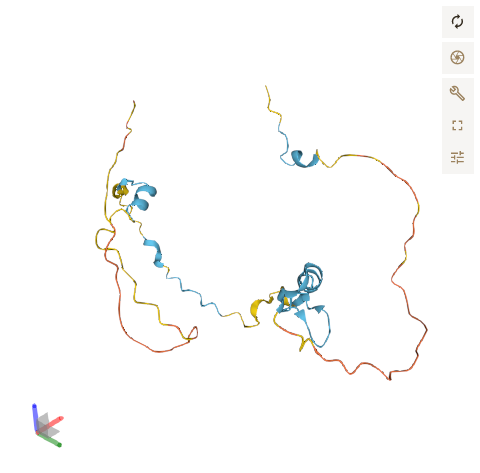
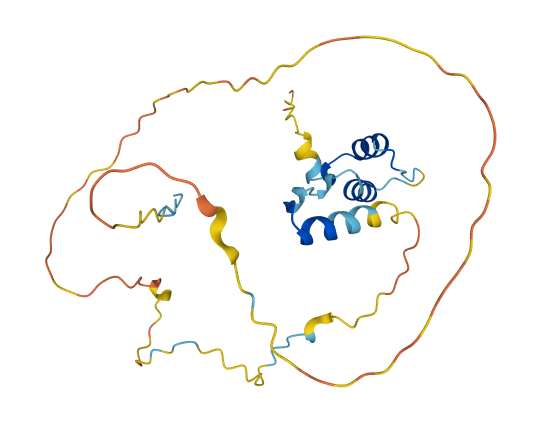
Mon Jan 17 13:32:37 +0000 2022Only a 12°C gradient up I-29 between Houston and Winnipeg. 🔗
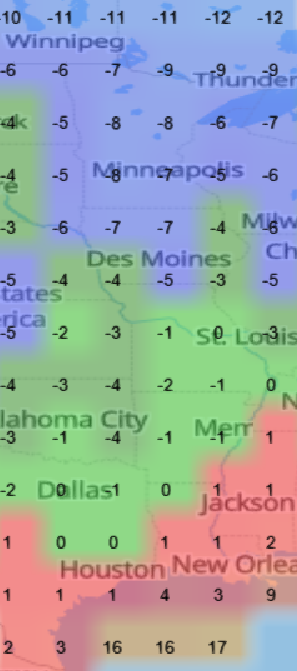
Mon Jan 17 12:53:50 +0000 2022C2orf49:p, has homologs across the vertebrate lineage. The PTM pattern suggests that this protein (aka AI597479:p in rodents) is all business in the front & party in the back. 4/ 🔗
Mon Jan 17 12:53:49 +0000 2022C2orf49:p.G185D 2:g.105343135G>A, rs28930676 (all tissue G:D 0.947:0.053) vaf=41%, Δm=58.0055, VAF by population group: african 8%, american 4%, east asian 2%, european 4%, south asian 6%. 3/
Mon Jan 17 12:53:49 +0000 2022C2orf49:p, θ(max) = 68. aka MGC5509, asw, Ashwin. Observed in MHC class 1 peptide experiments. Found in many tissues, cell lines: absent from fluids. 2/ 🔗
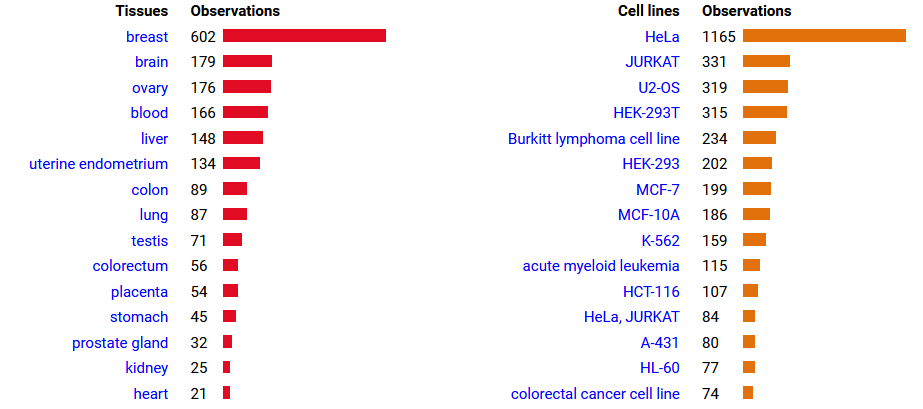
Mon Jan 17 12:53:48 +0000 2022>C2orf49:p, chromosome 2 open reading frame 49 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 3×K+acetyl; 1×K+GGyl; 1×K+succinyl; 14×S, 5×T,1×Y+phosphoryl; SAAVs: G185D (4%), P220L (1%); mature form: (2-232) [10,243×, 34 kTa]. #ᗕᕱᗒ #㟻톆蝊 1/ 🔗
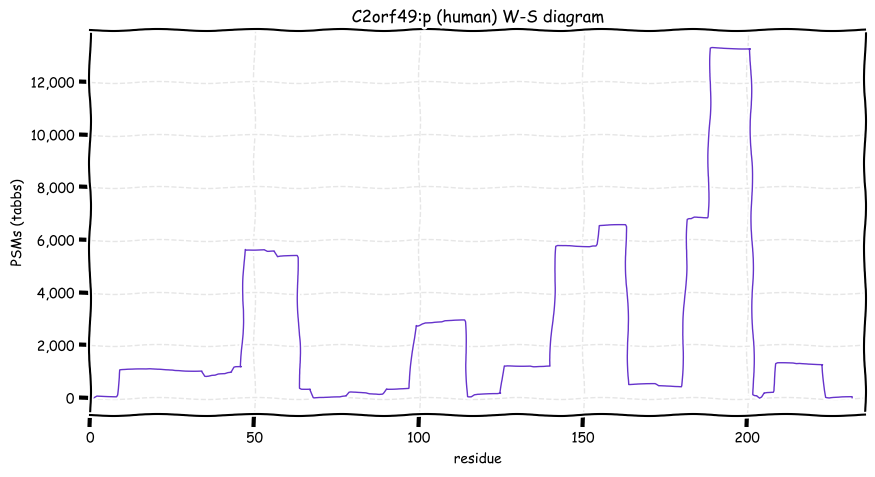
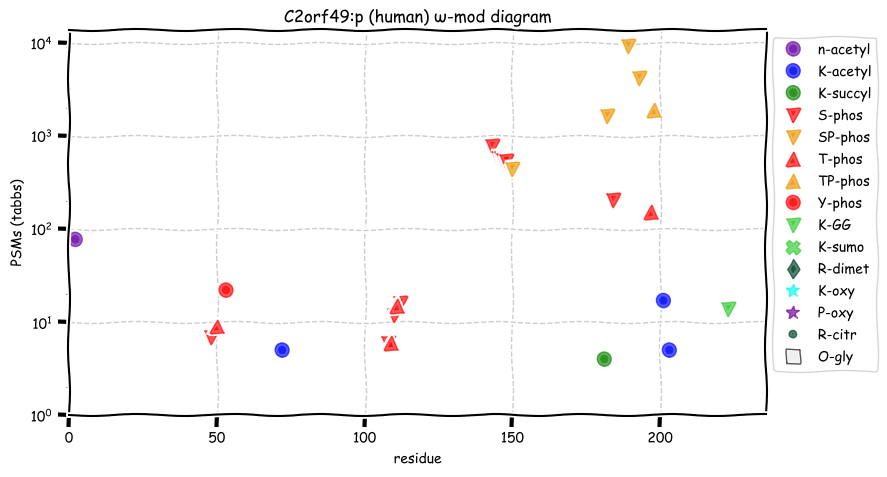
Sun Jan 16 17:00:16 +0000 2022HLA class I results are always interesting, particularly the juxtaposition of commonly & rarely observed proteins at similar intensities.
Sun Jan 16 15:12:09 +0000 2022PXD022884 from 🔗 has some spiffy high resolution HLA class I peptide data
Sun Jan 16 13:19:47 +0000 2022C1orf52:p, has homologs across the vertebrate lineage. 4/ 🔗
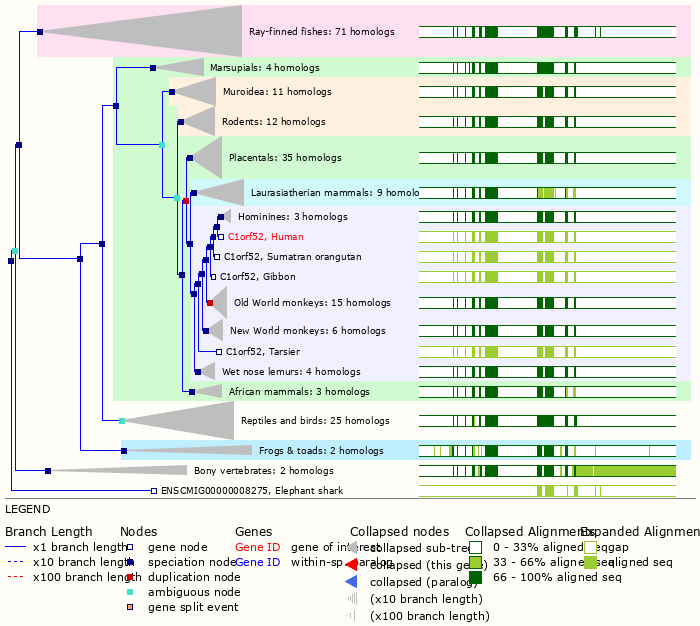
Sun Jan 16 13:19:46 +0000 2022C1orf52:p.I131V 1:g.85258608T>C, rs199632342 (all tissue I:V 0.998:0.002) vaf=<1%, Δm=-14.0156, VAF by population group: african <1%%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Sun Jan 16 13:19:46 +0000 2022C1orf52:p, θ(max) = 68. aka gm117, FLJ44982. Observed in MHC class 1 peptide experiments. Found in many tissues, cell lines: absent from fluids. 2/ 🔗
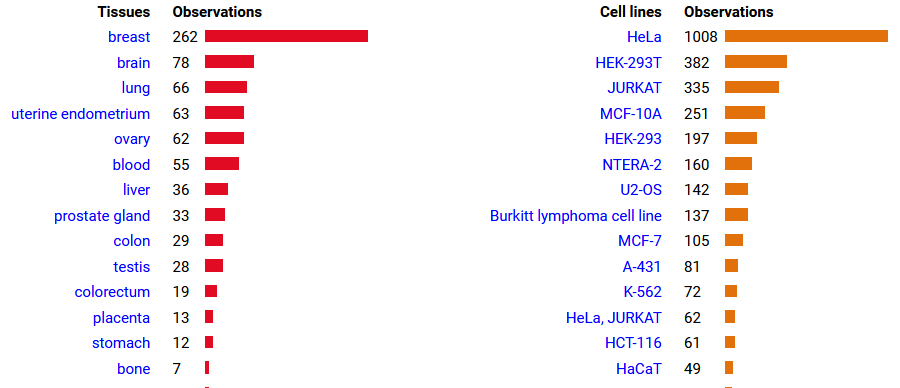
Sun Jan 16 13:19:45 +0000 2022>C1orf52:p, chromosome 1 open reading frame 52 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 5×K+acetyl; 4×K+GGyl; 2×K+succinyl; 12×S, 6×T,6×Y+phosphoryl; SAAVs: I131V (<1%); mature form: (2-182) [6,841×, 24 kTa]. #ᗕᕱᗒ #鎕횂輍 1/ 🔗
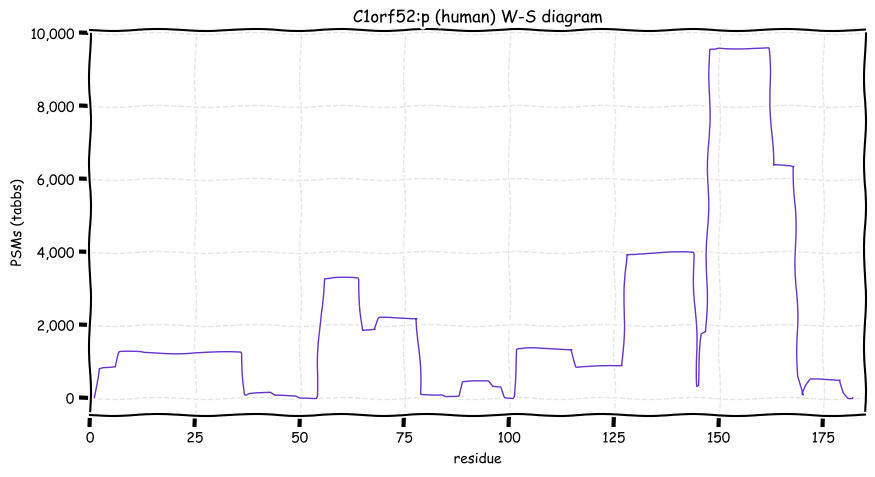
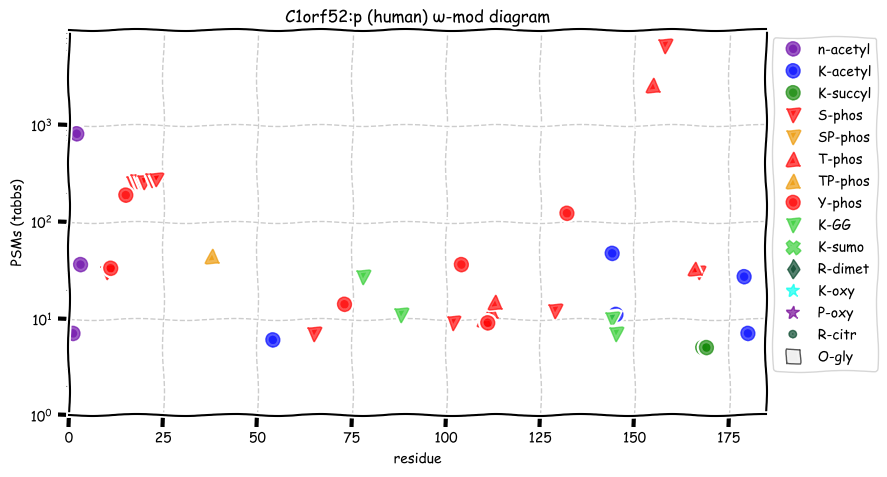
Sat Jan 15 19:47:54 +0000 2022A good exercise for students would be to take the data from a couple of the more flamboyantly infected urine samples and try to determine the species (& strains) of the bacteria present, as well as a best guess at their relative abundance.
Sat Jan 15 19:19:35 +0000 2022Bio-typing is also the Rodney Dangerfield of proteomics.
Sat Jan 15 19:08:08 +0000 2022And, just a reminder, bio-typing bacterial infections is still the premier clinical application of MS-based proteomics.
Sat Jan 15 18:52:12 +0000 2022What many people really believe about reviewer #3: 🔗
Sat Jan 15 18:43:26 +0000 2022PXD025547 (M. Bruschi, et al., 🔗) has some great examples of bacterial proteins from patients with undiagnosed urinary tract infections.
Sat Jan 15 16:49:27 +0000 2022@statesdj It is in stark contrast to the loopy tactic being used in another Canadian province of stopping people from entering govt liquor stores if they don't have a vaccine passport card.
Sat Jan 15 15:21:16 +0000 2022@statesdj I should also note that this is one of the few things that the govt here has done wrt to the pandemic that isn't panicky or silly.
Sat Jan 15 14:41:50 +0000 2022@statesdj I also walked out with a can of Crumb Queen Munich Dunkel, which was surprisingly good.
Sat Jan 15 14:38:21 +0000 2022@macro_momo @slavov_n Unless you are primarily involved in a bureaucratic process, in which case the appearance & documented evidence of the relentlessly logical pursuit of a 5 year plan (regardless of how imaginary) is all that matters.
Sat Jan 15 14:28:41 +0000 2022@statesdj When I walked in to the govt liquor store yesterday, the guard at the door handed me a free box of 20 KN-95s
Sat Jan 15 13:59:10 +0000 2022@neely615 It is being complemented by a high pressure system to its south east that is driving a 'Pineapple Express" type flow of moist air originating from Hawaii. 🔗
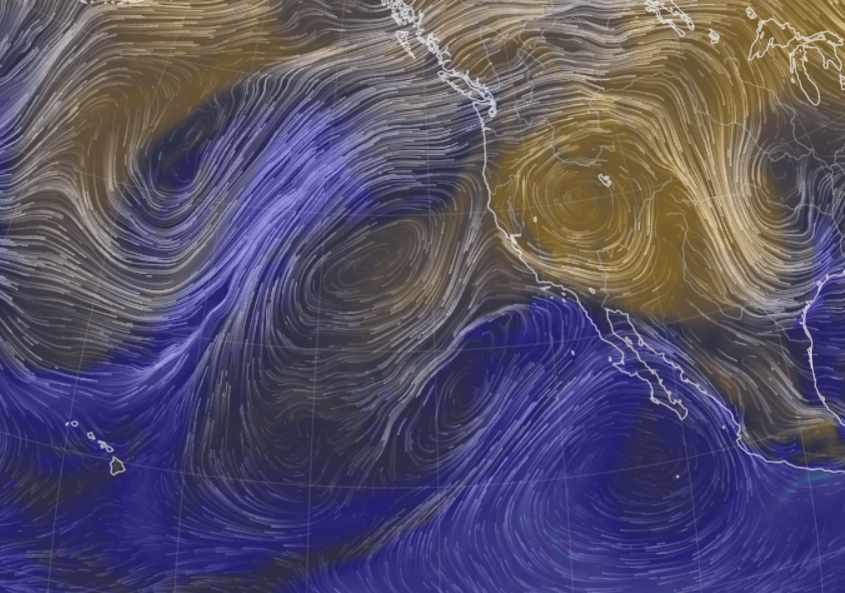
Sat Jan 15 13:15:25 +0000 2022C12orf57:p, has homologs across the vertebrate lineage. 4/ 🔗
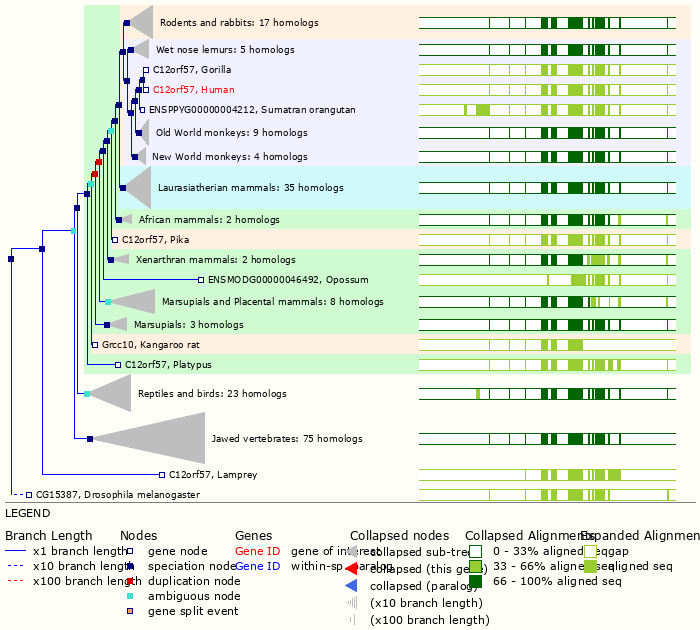
Sat Jan 15 13:15:24 +0000 2022C12orf57:p.A21T 12:g.6944484G>A, rs142743155 (all tissue A:T 0.998:0.002) vaf=<1%, Δm=30.0106, VAF by population group: african <1%%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Sat Jan 15 13:15:24 +0000 2022C12orf57:p, θ(max) = 44. aka GRCC10, C10. Observed in MHC class 1 peptide experiments. Found in many tissues, cell lines & urinary extracellular vesicles. 2/ 🔗
Sat Jan 15 13:15:23 +0000 2022>C12orf57:p, chromosome 12 open reading frame 57 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 3×K+acetyl; 4×K+GGyl; 0×K+SUMOyl; 1×S, 1×T,0×Y+phosphoryl; SAAVs: A21T (<1%); mature form: (2-126) [13,370×, 44 kTa]. #ᗕᕱᗒ #펣묃骲 1/ 🔗
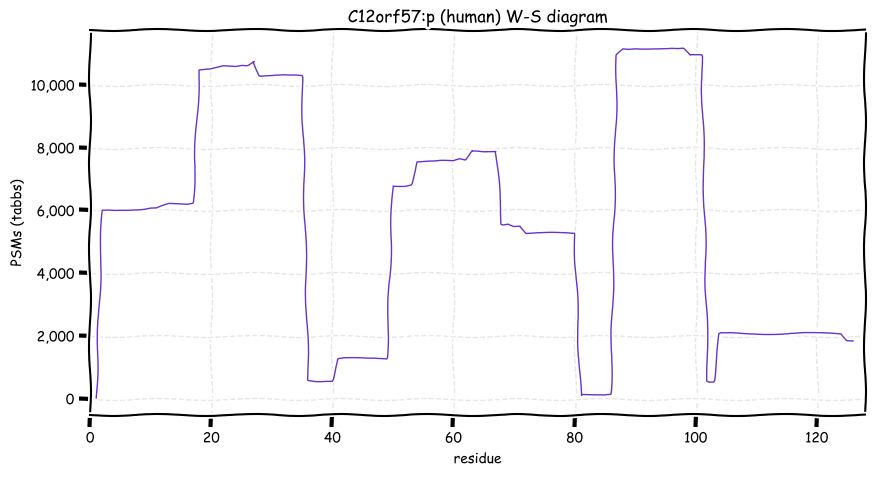
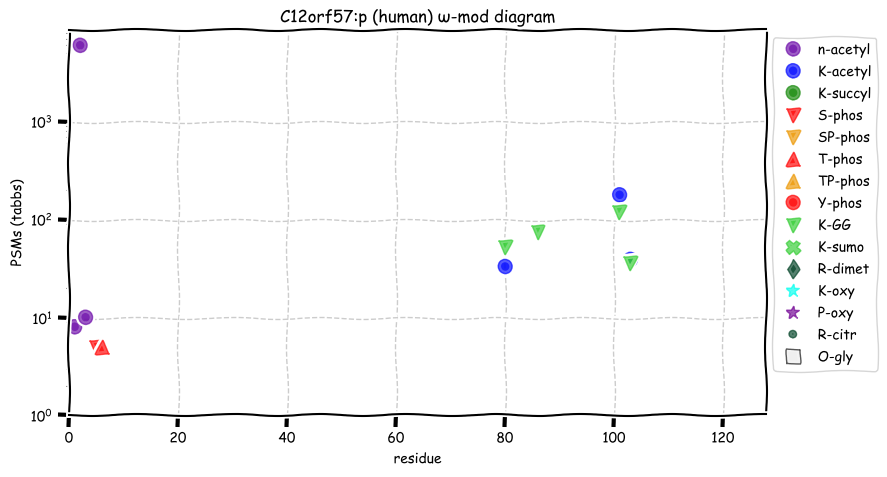
Fri Jan 14 19:47:56 +0000 2022@chrashwood I realized quite a while ago that what I think doesn't matter.
Fri Jan 14 19:02:40 +0000 2022After 3600 s, it appears the future will be pretty much the same as the current.
Fri Jan 14 18:00:50 +0000 2022Tropical storm Cody is drifting down towards the North Island. 🔗
Fri Jan 14 17:56:36 +0000 2022300 s until The Future of Proteomics Panel
Fri Jan 14 13:51:57 +0000 2022At least this trend seems to be holding 🔗
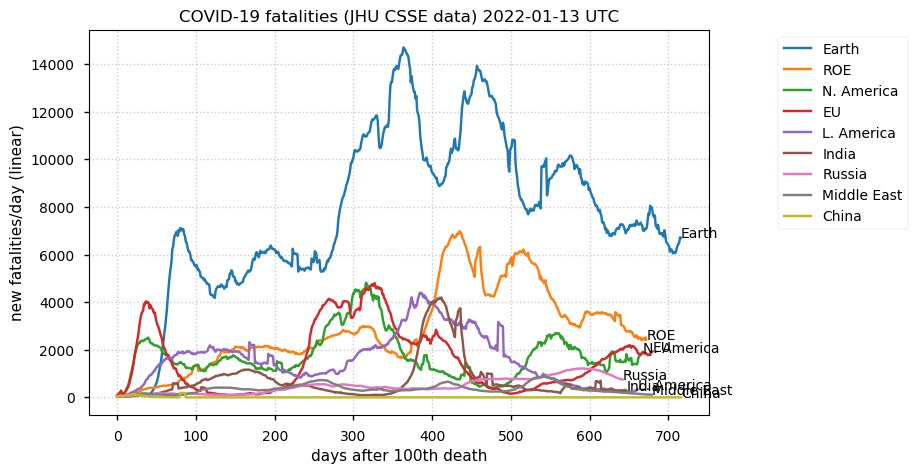
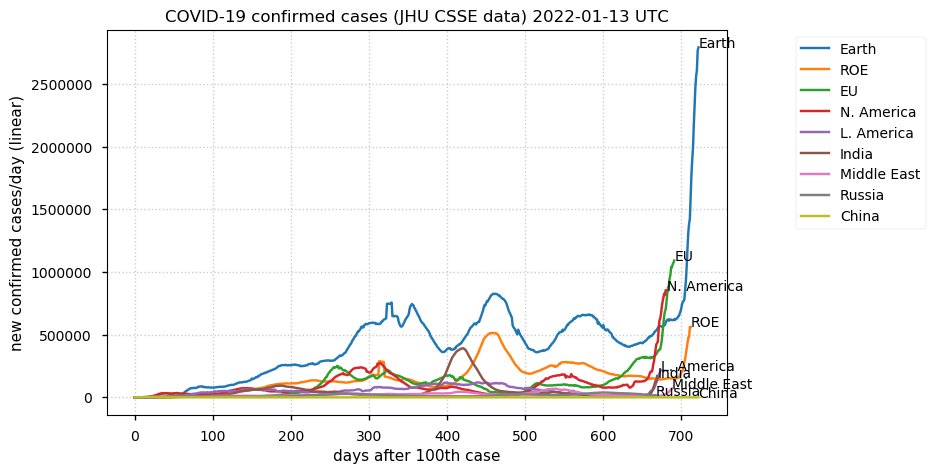
Fri Jan 14 12:57:07 +0000 2022C16orf58:p, has homologs across the metazoan lineage. The gene symbol RUSF1 was recently associated with this gene (but I don't expect it to last very long). 4/ 🔗
Fri Jan 14 12:57:06 +0000 2022C16orf58:p.T309I 16:g.31493635G>A, rs114108939 (all tissue T:I 0.999:0.001) vaf=<1%, Δm=12.0364, VAF by population group: african 3%, american af=<1%, east asian af=<1%, european af=<1%, south asian af=<1%. 3/
Fri Jan 14 12:57:06 +0000 2022C16orf58:p, θ(max) = 44. aka RUSF1, FLJ13868. Observed in MHC class 1 peptide experiments. Observed in many tissues & cell lines: absent from fluids. Domain (248-270) is predicted to be TM, although the membrane involved or the orientation is unclear. 2/ 🔗
Fri Jan 14 12:57:05 +0000 2022>C16orf58:p, RUS family member 1 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 1×K+acetyl; 11×K+GGyl; 0×K+SUMOyl; 2×S, 0×T,0×Y+phosphoryl; SAAVs: T309I (<1%); mature form: (2-468) [6,948×, 37 kTa]. #ᗕᕱᗒ #뛐酗쨊 1/ 🔗
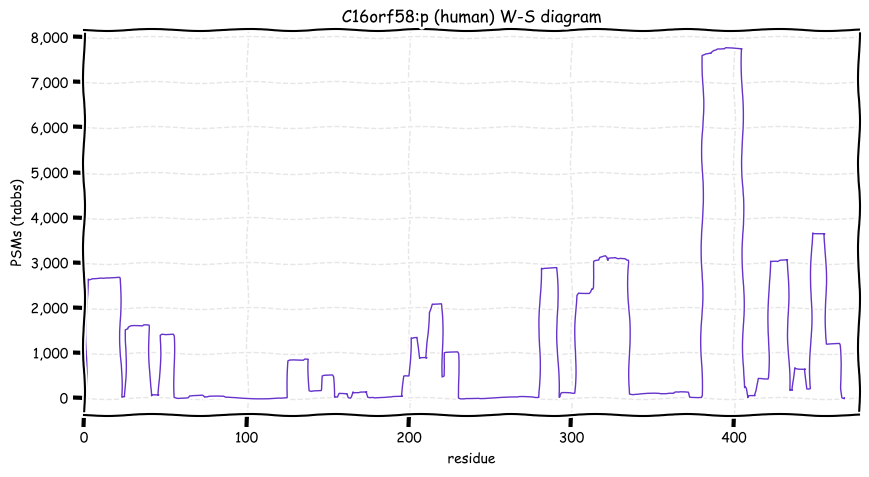
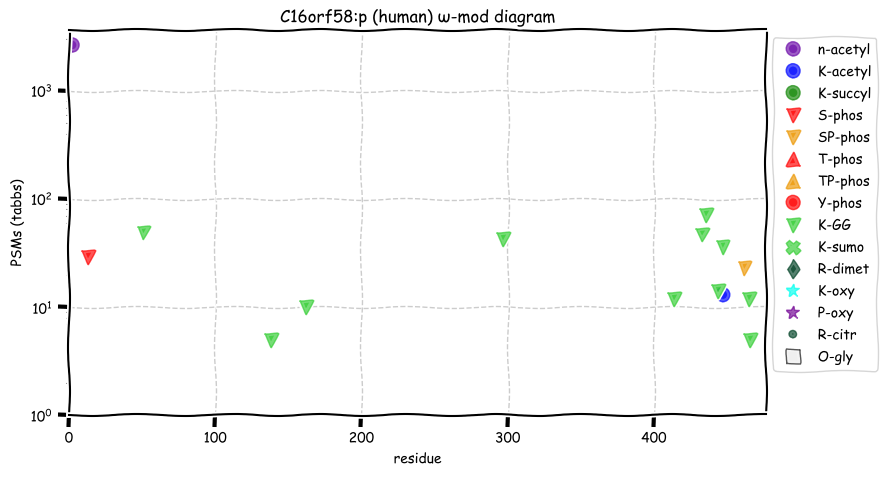
Thu Jan 13 22:53:18 +0000 2022@cajun_science @byu_sam It isn't the source of all problems, but it is a major contributing factor.
Thu Jan 13 16:33:25 +0000 2022@joshroby @lindseywiebe Try asking a local for directions to "Portage la Prairie", as pronounced correctly in Parisian French.
Thu Jan 13 16:30:59 +0000 2022The FLAG purified samples in PXD026306 have the interesting feature that you can see the phosphorylation state of the bait (MAPT) in the data
Thu Jan 13 16:04:12 +0000 2022@neely615 The 2nd low pressure system has gone north & is just off the south coast of Alaska now: the northern Pacific is very busy with low pressure systems at the moment. 🔗
Thu Jan 13 13:02:17 +0000 2022C5orf22:p, has homologs across the metazoan lineage. Referred to as MESR6:p in D. melanogaster and 6030458C11Rik:p in rodents. 4/ 🔗
Thu Jan 13 13:02:16 +0000 2022C5orf22:p.T235P chr 5:g.31538585>>C, rs17410000 (all tissue T:P 0.811:0.189) vaf=28%, Δm=-3.9949, VAF by population group: african 7%, american 18%, east asian 13%, european 31%, south asian 32%. TP in JURKAT cells. 3/
Thu Jan 13 13:02:16 +0000 2022C5orf22:p, θ(max) = 57. aka FLJ11193. Observed in MHC class 1 peptide experiments. Observed in many tissues & cell lines: absent from fluids. 2/ 🔗
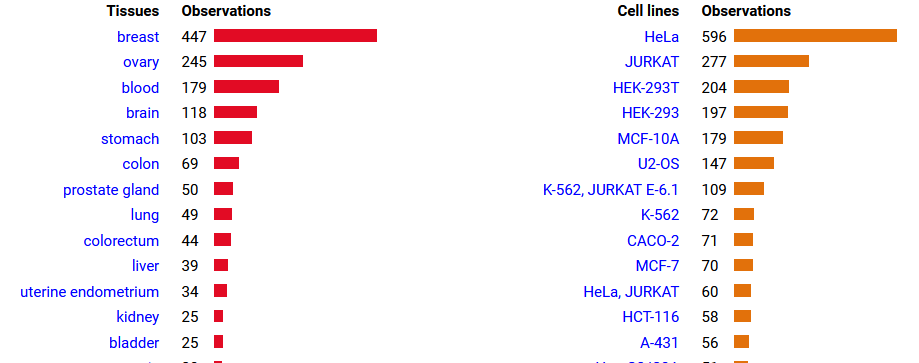
Thu Jan 13 13:02:15 +0000 2022>C5orf22:p, chromosome 5 open reading frame 22 (Homo sapiens) Small subunit; CTMs: S2+acetyl; PTMs: 0×K+acetyl; 7×K+GGyl; 9×K+SUMOyl; 8×S, 2×T,0×Y+phosphoryl; SAAVs: T235P (28%), D405E (5%); mature form: (2-442) [6,948×, 20 kTa]. #ᗕᕱᗒ #筟饥춬 1/ 🔗
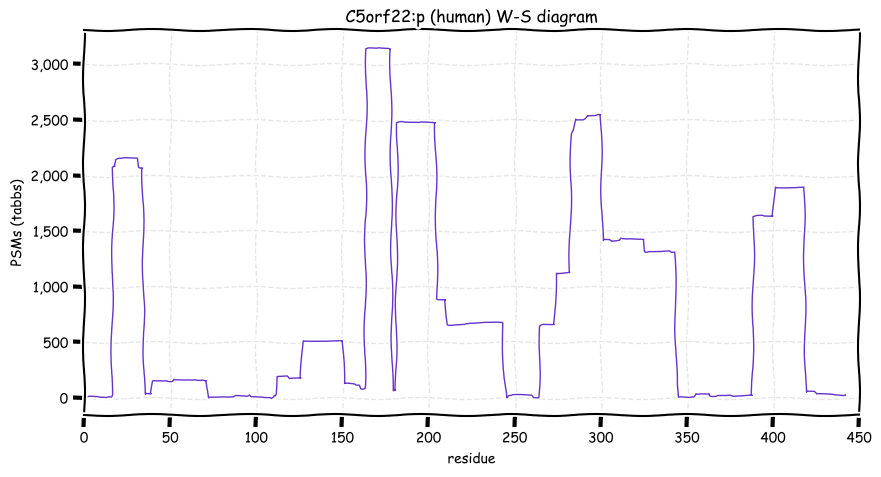
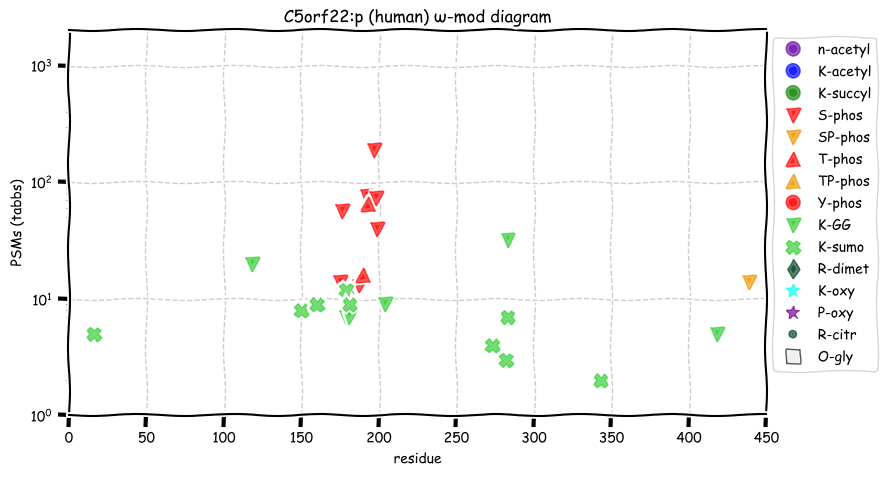
Wed Jan 12 23:49:30 +0000 2022@neely615 The distortion of the jet stream caused by the two low pressure systems seems to be what is pulling the wet air up from its normal place nearer to the equator. 🔗
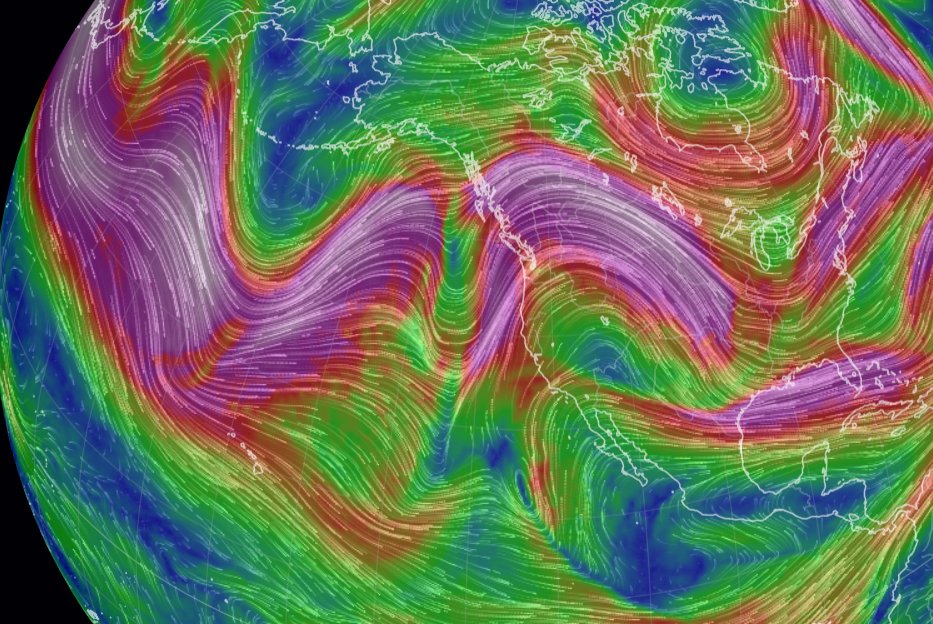
Wed Jan 12 23:19:59 +0000 2022@neely615 Two atmospheric "rivers" (blue mean lots of water in the air) & swirly bits (low pressure systems) bearing down on the Vancouver/Victoria/Seattle area 🔗
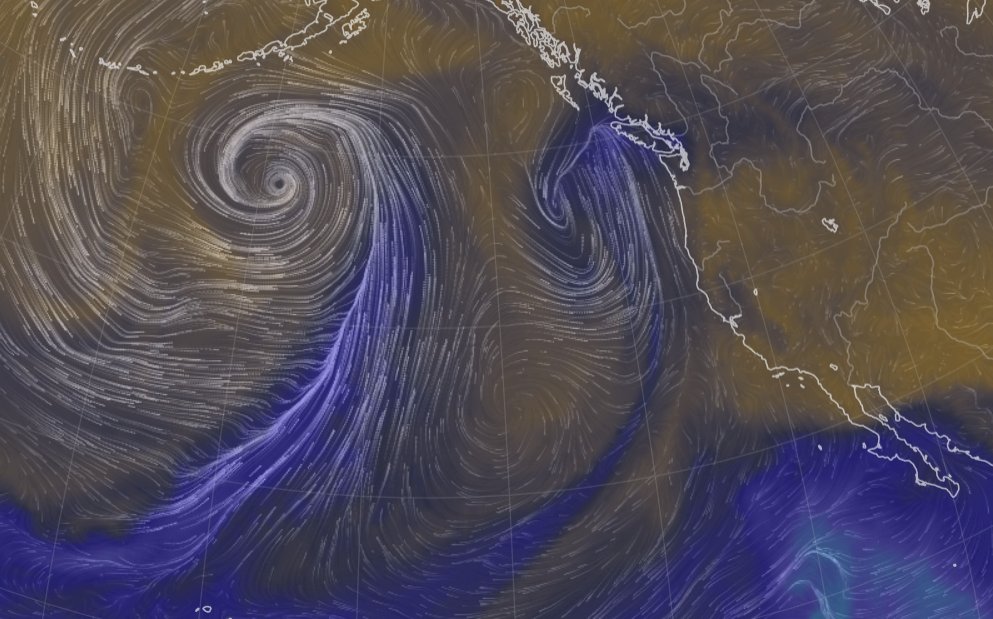
Wed Jan 12 17:45:03 +0000 2022@slavov_n @Coon_Labs @TheCoxLab @ATGTAC Splice variants are the third rail of proteomics.
Wed Jan 12 14:58:19 +0000 2022Only 23°C between Winnipeg & Houston this morning. 🔗
Wed Jan 12 14:21:02 +0000 2022If you want a high quality dataset with relatively large, consistent single shot HPLC runs (95k per) from human samples with very few experimental artifacts, PXD025174 is worth a look.
Wed Jan 12 13:06:38 +0000 2022C8orf82:p, has homologs across the metazoan & protist lineage, discussed in unusual detail in its Wikipedia entry 🔗 4/
Wed Jan 12 13:06:38 +0000 2022C8orf82:p.R110Q chr 8:g.144527664C>T, rs550232031 (all tissue R:Q 0.999:0.001) vaf=<1%, Δm=-28.0425, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Wed Jan 12 13:06:38 +0000 2022C8orf82:p, θ(max) = 77. aka MGC70857. Observed rarely in MHC class 1 peptide experiments. Observed in many tissues & cell lines: absent from fluids. Has an N-terminal mitochondrial targeting sequence that is removed in the mature sequence. 2/ 🔗
Wed Jan 12 13:06:37 +0000 2022>C8orf82:p, chromosome 8 open reading frame 82 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 0×K+acetyl; 0×K+GGyl; 2×S, 0×T,0×Y+phosphoryl; SAAVs: T10I (7%), R110Q (<%1); mature form: (21,22,23-216) [12,965×, 47 kTa]. #ᗕᕱᗒ #捂쎁䓇 1/ 🔗
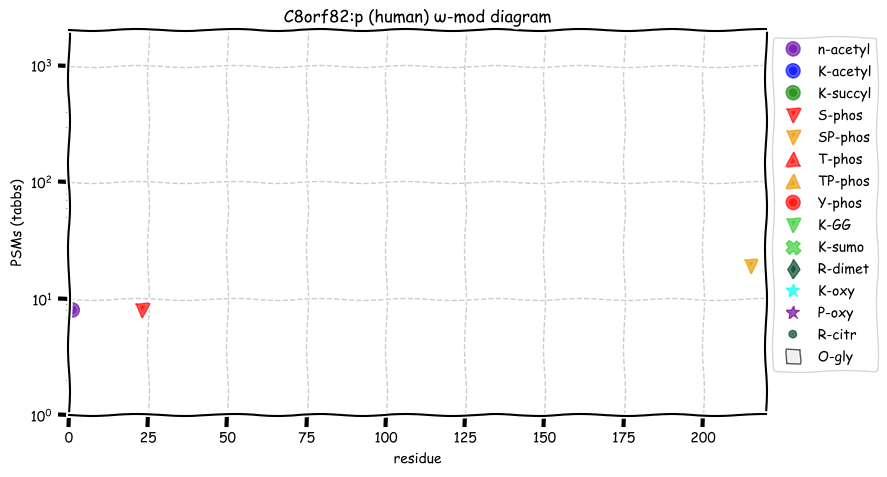
Tue Jan 11 15:23:11 +0000 2022@VATVSLPR @pwilmarth There is a reason scientists are not allowed to name the family pets.
Tue Jan 11 15:06:52 +0000 2022@seer_bio @sciex @DiscLS @SCIEXnews If I was to stop now, it would take them until 2035 to catch up.
Tue Jan 11 15:01:38 +0000 2022@seer_bio @sciex @DiscLS @SCIEXnews Only a billion? Rather modest goal IMHO.
Tue Jan 11 14:02:46 +0000 2022@ProteomicsNews Proteomics at all levels is inextricably linked to MS vendors & purchasing decisions, including the actions taken by editors and reviewers.
Tue Jan 11 13:13:18 +0000 2022C20orf27:p, has homologs across the vertebrate lineage. Referred to as 1700037H04Rik:p in rodents and C4C20orf27 in Zebra finch. 4/ 🔗
Tue Jan 11 13:13:17 +0000 2022C20orf27:p.P86L chr 20:g.3755513G>A, rs140915844 (all tissue P:L 0.999:0.001) vaf=<1%, Δm=16.0313, VAF by population group: african 1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Tue Jan 11 13:13:17 +0000 2022C20orf27:p, θ(max) = 92. aka FLJ20550. Observed in MHC class 1 & 2 peptide experiments. Observed in many tissues & cell lines: rare in fluids. 2/ 🔗
Tue Jan 11 13:13:16 +0000 2022>C20orf27:p, chromosome 20 open reading frame 27 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 1×K+acetyl; 1×K+GGyl; 3×S, 0×T,2×Y+phosphoryl; SAAVs: P86L (<1%); mature form: (2-174) [14,117×, 40 kTa]. #ᗕᕱᗒ #秬ꈺ닠 1/ 🔗
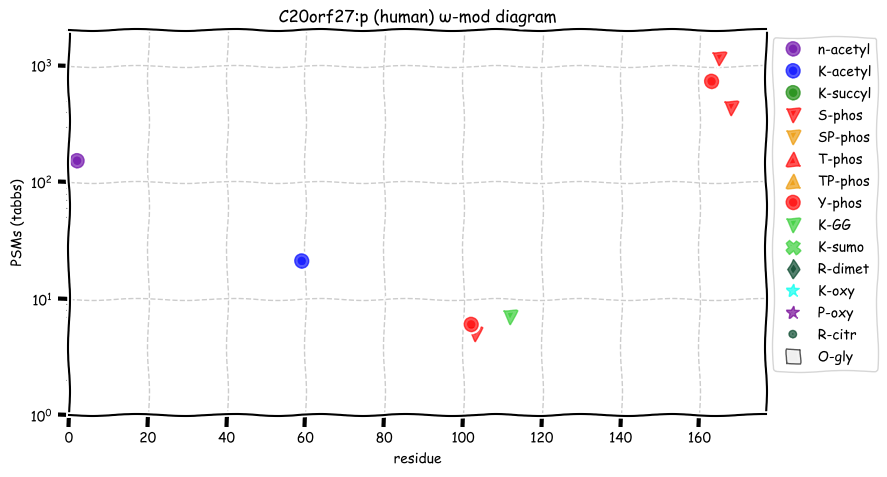
Mon Jan 10 12:54:05 +0000 2022C9orf64:p, has homologs across the metazoan lineage. Referred to as 2210016F16Rik:p in rodents, C09752 in D. melanogaster & C11D2.4:p in C. elegans. 4/ 🔗
Mon Jan 10 12:54:04 +0000 2022C9orf64:p.S120W chr 9:g.83955619G>A, rs7848149 (all tissue S:W 0.979:0.021) vaf=1%, Δm=99.0473, VAF by population group: african 3%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Mon Jan 10 12:54:04 +0000 2022C9orf64:p, θ(max) = 71. aka MGC10999. Observed in MHC class 1 peptide experiments (& rarely in class 2). Observed in many tissues, cell lines, & urine extracellular vesicles. 2/ 🔗
Mon Jan 10 12:54:03 +0000 2022>C9orf64:p, chromosome 9 open reading frame 64 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 4×K+acetyl; 9×K+GGyl; 0×S, 0×T,0×Y+phosphoryl; SAAVs: S120W (1%), S136Y (1%), K201R (1%), V219A (1%); mature form: (1-341) [14,117×, 58 kTa].#ᗕᕱᗒ #닯䱴溚 1/ 🔗
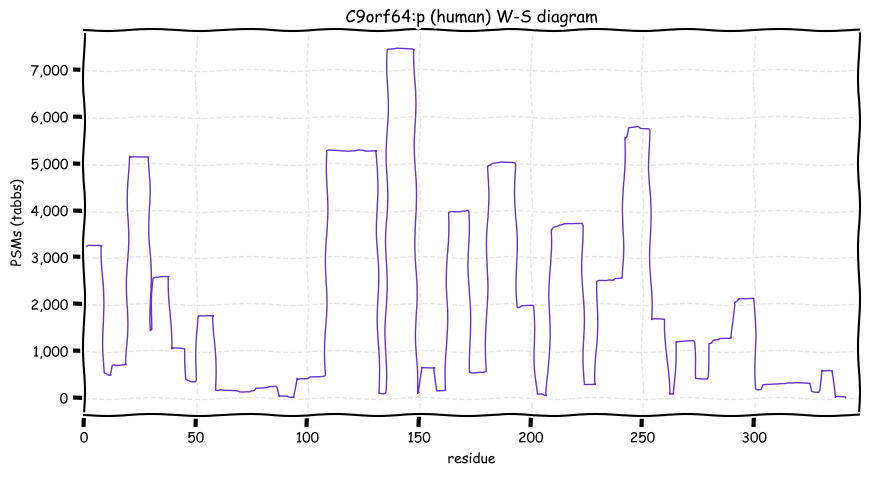
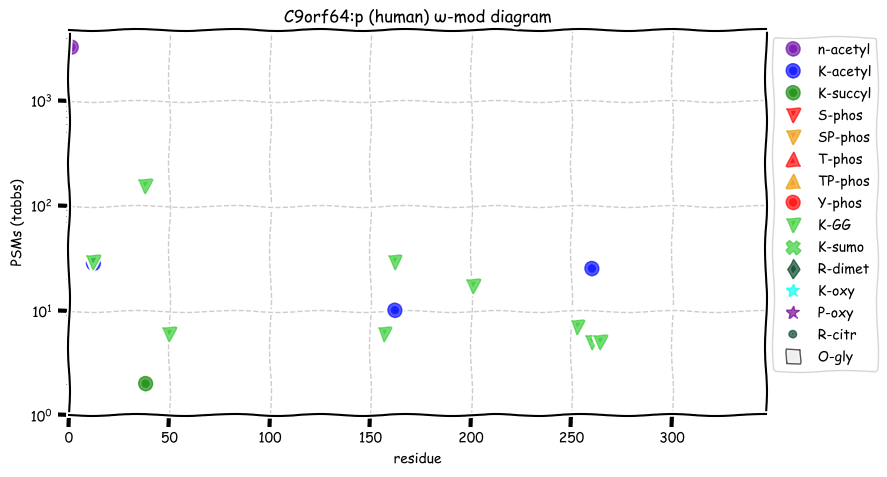
Mon Jan 10 00:34:36 +0000 2022Same front, 10 hours later 🔗
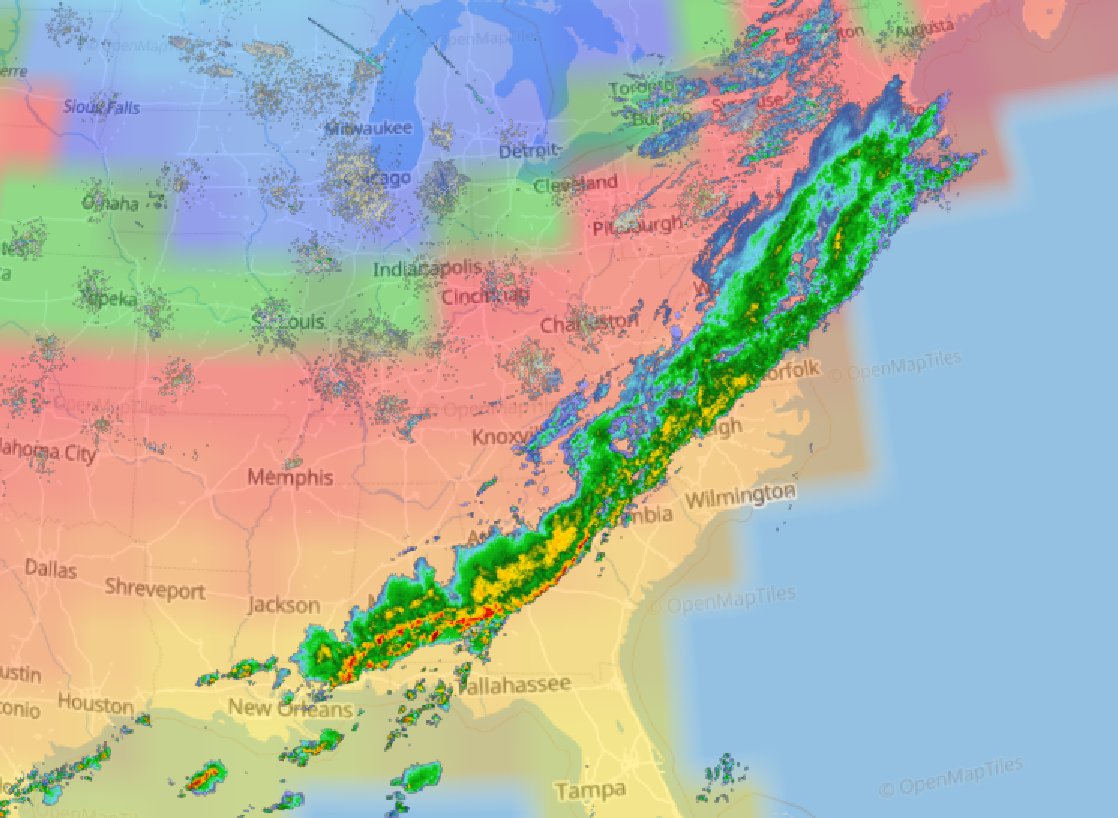
Sun Jan 09 19:57:24 +0000 2022@cwvhogue Well, they are going to have to rethink it & come up with a contest that can't be won by a DL-like approach.
Sun Jan 09 17:29:51 +0000 2022Conversely, if you are setting up a scientific competition & there is any way to use DL to win it, then it is a poorly designed competition.
Sun Jan 09 17:11:16 +0000 2022Silly stuff like AF2 (& most other DL) is the natural consequence of gamification in science.
Sun Jan 09 13:51:56 +0000 2022A colorful storm front running from Shreveport LA to Saint John NB. 🔗
Sun Jan 09 13:48:03 +0000 2022Today's I-29 gradient (2022-01-09 13:00 UTC) 🔗
Sun Jan 09 13:36:59 +0000 2022C19orf33:p, has homologs in mammals only. Referred to as 2200002D01Rik:p in rodents & C17HC19orf33:p in sperm whales. 4/ 🔗
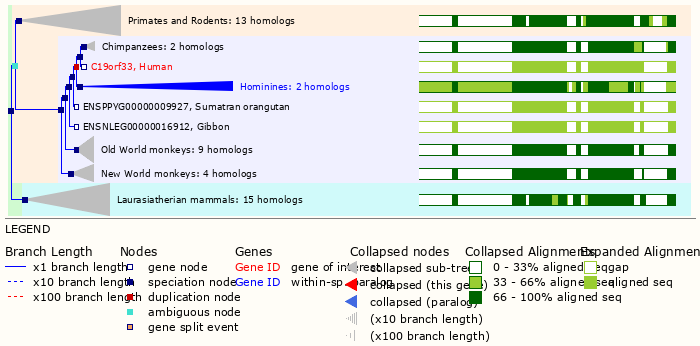
Sun Jan 09 13:36:58 +0000 2022C19orf33:p.D25H chr 19:g.38304425G>C, rs547355038 (all tissue D:H 1.000:0.000) vaf=<1%, Δm=22.032, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Sun Jan 09 13:36:58 +0000 2022C19orf33:p, θ(max) = 75. aka IMUP-1, IMUP-2, H2RSP, IMUP. Usually absent from MHC peptide experiments. Observed in many tissues, cell lines, saliva & blood plasma. 2/ 🔗
Sun Jan 09 13:36:57 +0000 2022>C19orf33:p, chromosome 19 open reading frame 33 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 1×K+acetyl; 0×K+succinyl; 0×K+GGyl; 16×S, 2×T,0×Y+phosphoryl; R36+deimidation; SAAVs: D25H (<1%), L515P (45%); mature form: (1-106) [13,048×, 51 kTa]. #ᗕᕱᗒ #忩䟖乻 1/ 🔗
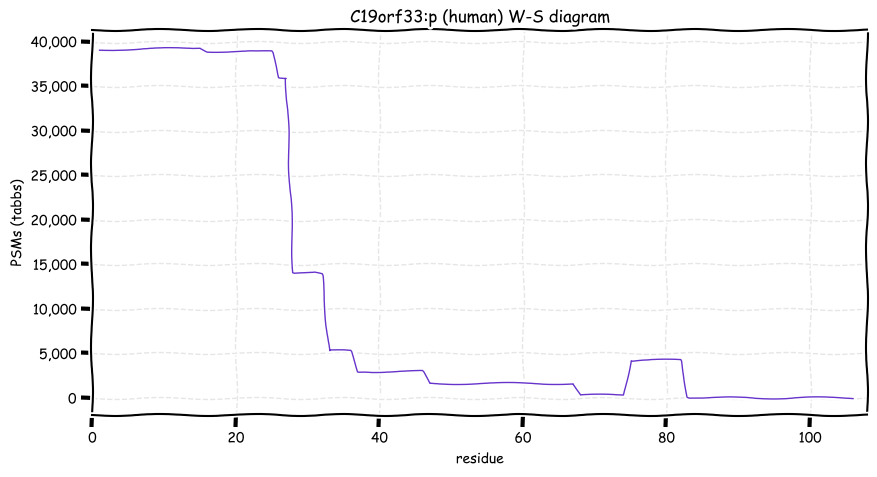
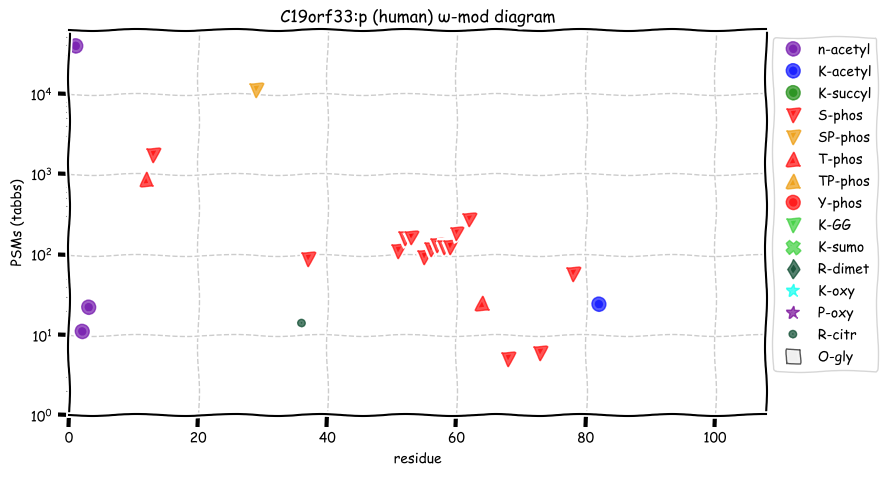
Sat Jan 08 16:05:29 +0000 2022I am finding unnamed human ORFs to be much more interesting than I thought they would be.
Sat Jan 08 12:58:53 +0000 2022C6orf132:p, the literature/databases are equivocal whether to characterize this gene as protein coding or as long non-coding RNA. The proteomics data is pretty clear―C6orf132:r is translated into a protein with multiple high occupancy phosphorylation sites. 4/
Sat Jan 08 12:58:53 +0000 2022C6orf132:p.L515P chr 6:g.42106368A>G, rs9394866 (all tissue L:P 0.718:0.282) vvaf=45%, Δm=-16.0313, VAF by population group: african 87%, american 41%, east asian 31%, european 47%, south asian 47%. 3/
Sat Jan 08 12:58:53 +0000 2022C6orf132:p, θ(max) = 38. aka bA7K24.2. Found in MHC class 1 experiments. Observed in some tissues, cell lines & extracellular vesicles in urine and saliva. 2/ 🔗

Sat Jan 08 12:58:52 +0000 2022>C6orf132:p, chromosome 6 open reading frame 132 (Homo sapiens) Large subunit; CTMs: none; PTMs: 0×K+acetyl; 2×K+succinyl; 1×K+GGyl; 41×S, 24×T, 12×Y+phosphoryl; 2×R+deimidation; SAAVs: K4N (15%), L515P (45%); mature form: (1-1188) [5,783×, 24 kTa]. #ᗕᕱᗒ #ꚣ茿㸎 1/ 🔗
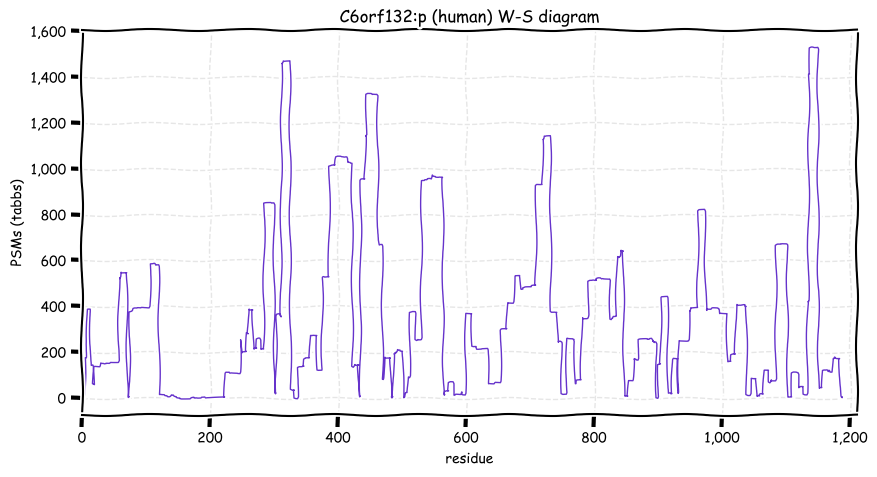
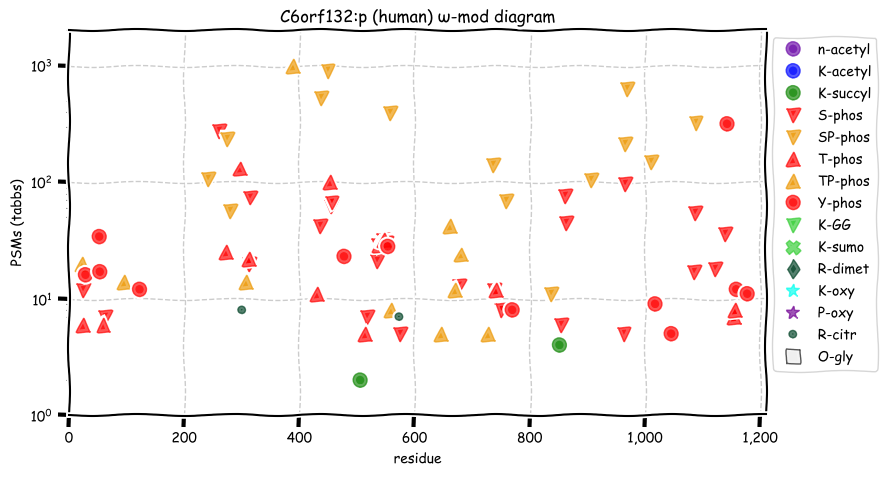
Fri Jan 07 18:25:18 +0000 2022The world hits 6000 for the first time in > 400 days 🔗
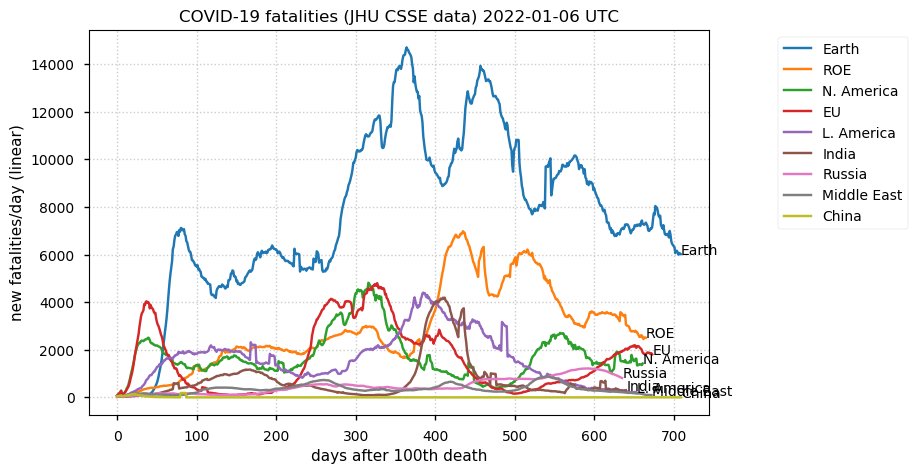
Fri Jan 07 16:10:13 +0000 2022Another bonnie (but seasonal) day down I-29 🔗
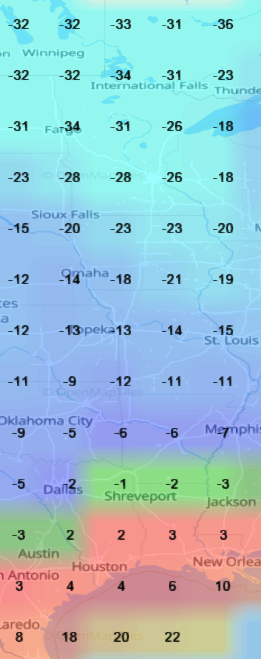
Fri Jan 07 13:18:00 +0000 2022C1orf116:p, has homologs across the vertebrate lineage. Referred to as AA986860:p in mice & zgc:158258:p in Red-bellied piranha. 4/ 🔗
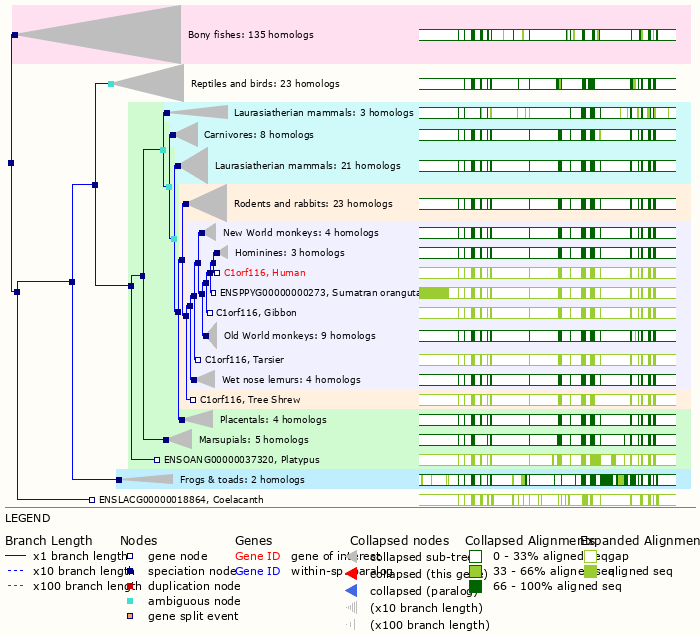
Fri Jan 07 13:18:00 +0000 2022C1orf116:p.N157T chr 1:g.207023294T>G, rs34660159 (all tissue N:T 0.991:0.009) vaf=3%, Δm=-12.9952, VAF by population group: african 36%, american 4%, east asian <1%, european 1%, south asian <1%. 3/
Fri Jan 07 13:17:59 +0000 2022C1orf116:p, θ(max) = 81. aka SARG, FLJ36507, MGC2742, MGC4309. Found in MHC class 1 experiments. Observed in some tissues, cell lines & extracellular vesicles in urine and saliva. 2/
Fri Jan 07 13:17:59 +0000 2022>C1orf116:p, chr 1 open reading frame 116 (H sapiens) Midsized subunit; CTMs: M1+acetyl; PTMs: 2×K+succinyl; 1×K+GGyl; 57×S,23×T,4×Y+phosphoryl; 4×R+deimidyl; SAAVs: N157T(3%), R258G(90%), F514S(2%), N586S(3%); mature form: (1,2-601) [10,645×, 69 kTa]. #ᗕᕱᗒ #贿꼌牚 1/ 🔗
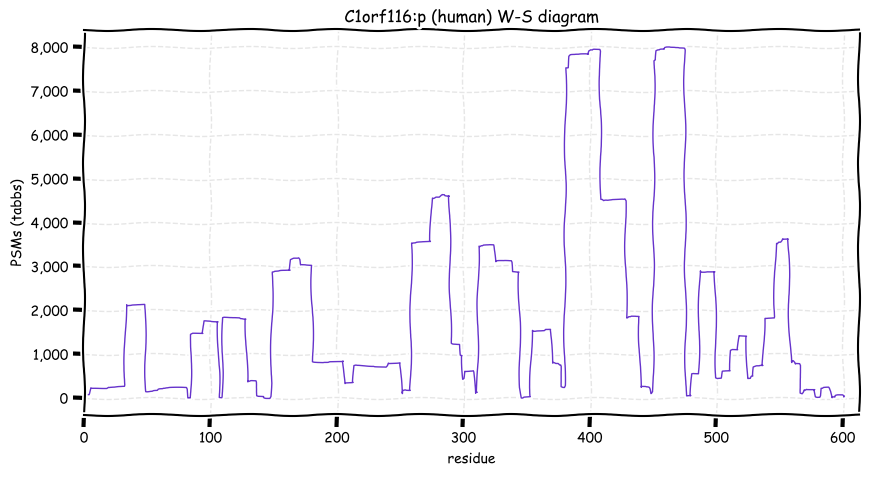
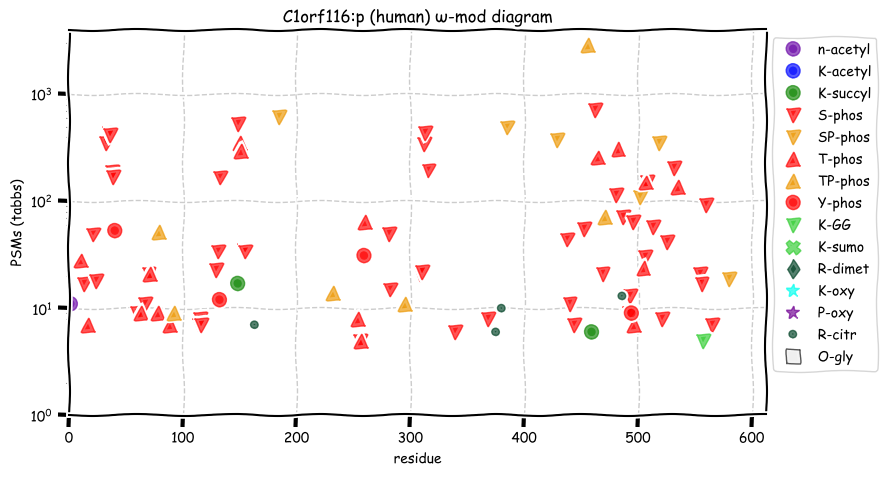
Thu Jan 06 17:53:37 +0000 2022@GarciaLabMS @neely615 @pwilmarth @oleg8r Does your institution have any written guidelines for how these sessions are conducted or is it up to the Department to determine the way this is done?
Thu Jan 06 15:56:41 +0000 2022Please.
Thu Jan 06 15:55:47 +0000 2022To the proteomics community in general:
cSNP is NOT the abbreviation for a SNP that results in a change in amino acid coding.
cSNP is the abbreviation for a SNP as represented in the associated cDNA sequence.
Use nsSNP or nsSNV instead.
Thu Jan 06 14:57:34 +0000 2022C11orf68:p, the experimental 3D structures for this protein are for the shorter variant, e.g., 🔗 /7
Thu Jan 06 14:53:31 +0000 2022C11orf68:p, this shorter variant also does not have the 2 R+dimethyl acceptor sites, a modification usually associated with protein-RNA/DNA interactions. /6
Thu Jan 06 14:49:12 +0000 2022C11orf68:p, the splice variant/alternative translation product C11orf68-203 (251 aa) has been observed in some data sets. This variant, in blue, does not have the N-terminal low complexity domain (in red) 5/ 🔗
Thu Jan 06 12:54:37 +0000 2022C11orf68:p, has homologs across the vertebrate lineage. Referred to as AI837181:p in mice & CUNH11orf68:p in Beluga whale. 4/ 🔗
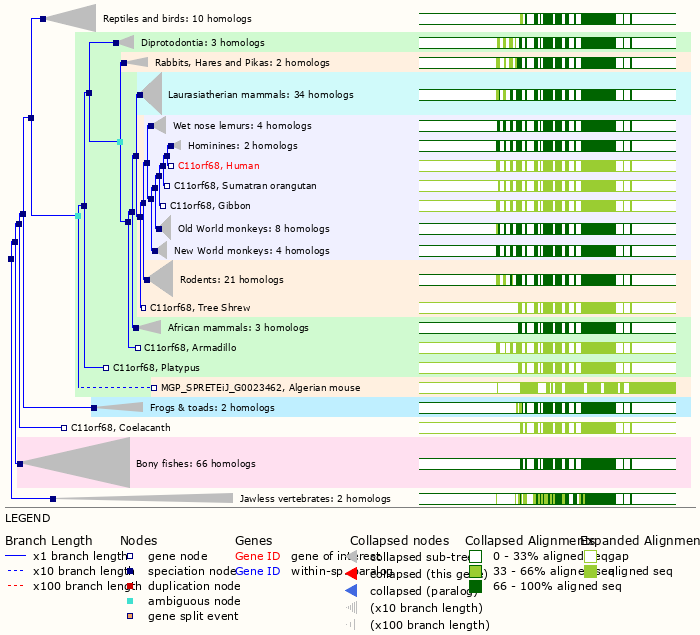
Thu Jan 06 12:54:37 +0000 2022C11orf68:p.Q195R chr 11:g.65917754T>C, rs7947504 (all tissue Q:R 0.975:0.025) vaf=97%, Δm=28.0425, VAF by population group: african 66%, american 95%, east asian 100%, european 98%, south asian 99%. Results in a new tryptic site generating 2 small peptides. 3/
Thu Jan 06 12:54:36 +0000 2022C11orf68:p, θ(max) = 54. aka P5326, BLES03. Found in MHC class 1 experiments. Observed in many tissues & cell lines: absent from fluids. 2/
Thu Jan 06 12:54:36 +0000 2022>C11orf68:p, chromosome 11 open reading frame 68 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 2×K+acetyl; 1×K+succinyl; 1×K+GGyl; R14, R29+dimethyl; 4×S, 0×T, 0×Y+phosphoryl; SAAVs: M172I (1%), Q195R (97%); mature form: (2-292) [11,024×, 68 kTa]. #ᗕᕱᗒ #㴊䘧礁 1/ 🔗
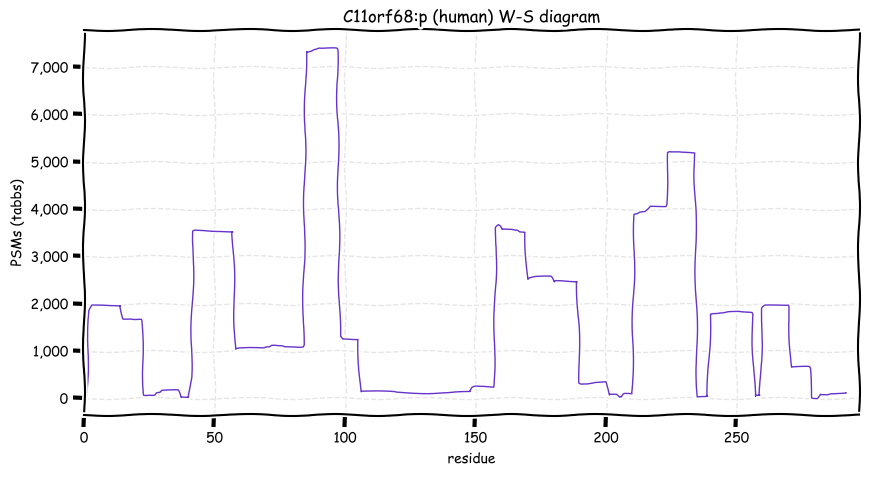
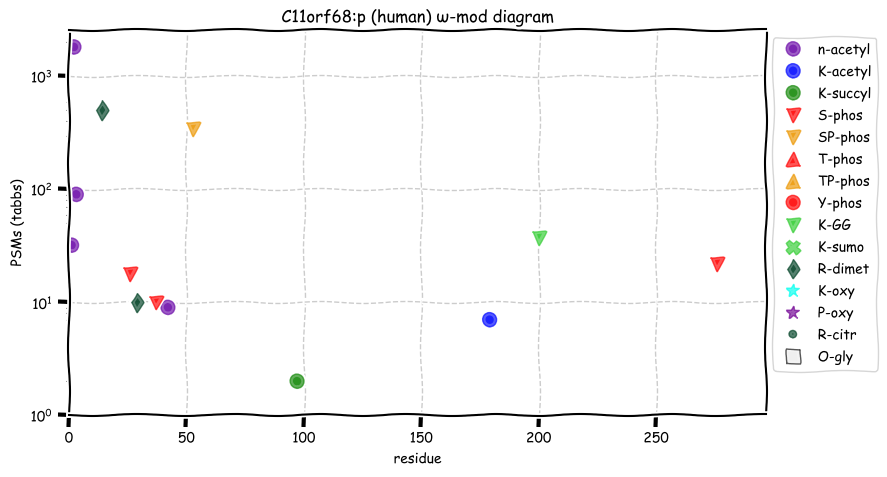
Wed Jan 05 23:38:30 +0000 2022@oleg8r @GarciaLabMS I'm sure that there is value to be had from these presentations. I was more interested in when they became a standard part of the interview process & whether they are used widely or only at specific institutions.
Wed Jan 05 20:31:51 +0000 2022When did "chalk talks" become entrenched in faculty hiring processes? I've had 4 faculty jobs & I've never had to participate in one.
Wed Jan 05 15:34:29 +0000 2022@KentsisResearch @ionicwoman They all have good signals from your protein of interest.
Wed Jan 05 15:33:18 +0000 2022@KentsisResearch @ionicwoman If you are interested in phosphorylation, try 🔗 from 🔗
Wed Jan 05 15:23:17 +0000 2022@KentsisResearch @ionicwoman I am unaware of multi-protease studies on human brain. My pick of the litter would be 🔗 from 🔗
Wed Jan 05 14:43:22 +0000 2022Seasonal weather down I-29 today―only 37°C between Winnipeg (-27°C) and Houston (+10°C ) 🔗
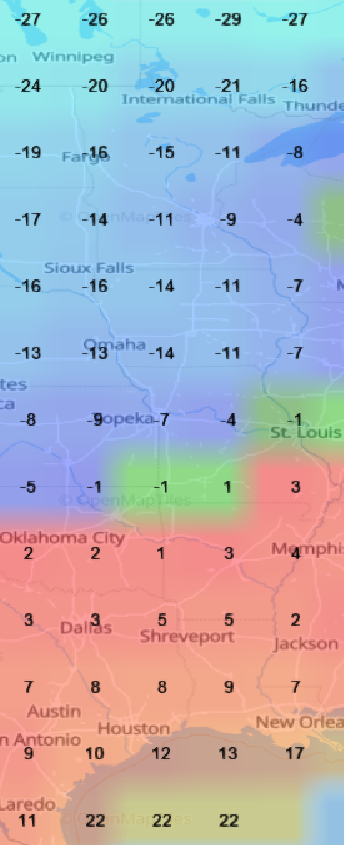
Wed Jan 05 14:21:12 +0000 2022Fortunately, these two rates seem to have decoupled in the last few weeks 🔗
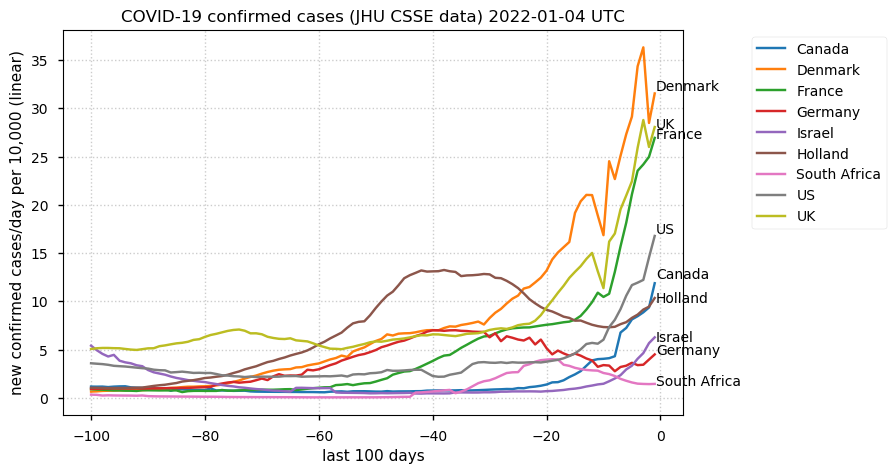
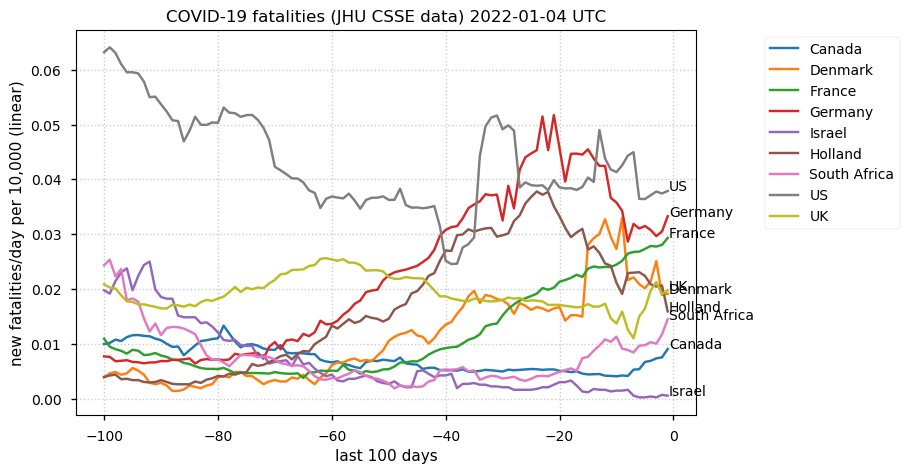
Wed Jan 05 13:39:52 +0000 2022C9orf78:p, has homologs across the metazoan lineage. Referred to as BC005654:p in mice, CG7974:p in D. melanogaster & T23G11.4 in C. elegans. 4/ 🔗
Wed Jan 05 13:39:51 +0000 2022C9orf78:p.R11C chr 9:g.129835191G>A, rs761344335 (all tissue R:C 0.999:0.001) vaf=<1%, Δm=-53.0919, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%. 3/
Wed Jan 05 13:39:51 +0000 2022C9orf78:p, θ(max) = 76. aka HSPC220, HCA59, CSU2. Found in MHC class 1 experiments. Observed in many tissues & cell lines: generally absent from fluids. 2/
Wed Jan 05 13:39:50 +0000 2022>C9orf78:p, chromosome 9 open reading frame 78 (Homo sapiens) Small subunit; CTMs: none; PTMs: 6×K+acetyl; 0×K+succinyl; 4×K+GGyl; 3×K+SUMOyl; 11×S, 6×T, 6×Y+phosphoryl; SAAVs: R11C (<1%); mature form: (2-289) [16,098×, 61 kTa]. #ᗕᕱᗒ #ό˃ɼ 1/ 🔗
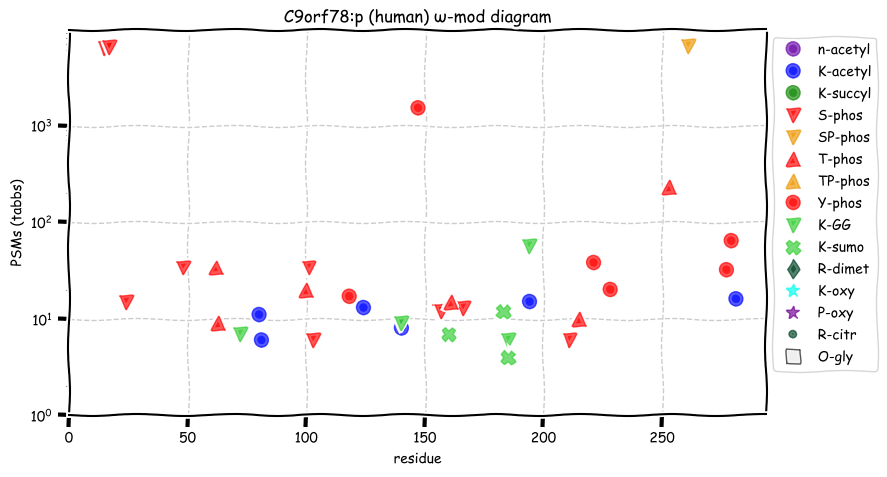
Tue Jan 04 20:03:10 +0000 2022@theoneamit @VATVSLPR @UCDProteomics @pwilmarth @nesvilab @dtabb73 @MagnusPalmblad @neely615 The discussion got a little side-tracked. In my experience, open search is best at spotting artifacts & experimentally enriched PTMs. It is not so good at SAAVs, rare PTMs or PTMs that tend to be clustered together.
Tue Jan 04 13:21:42 +0000 2022@VATVSLPR @UCDProteomics @pwilmarth @nesvilab @dtabb73 @MagnusPalmblad @neely615 OK, I'll bite. Which new PTMs have been discovered by open search, so far?
Tue Jan 04 13:19:35 +0000 2022C11orf54:p also has a good, full length 3D structure (🔗) and a reasonable looking protein-protein interaction map (🔗). But still, no name or accepted biological function. /5
Tue Jan 04 13:03:21 +0000 2022C11orf54:p, has homologs across the metazoan lineage. In mice, it is referred to as 4931406C07Rik:p, LOC411385:p in Apis melifera & c11orf54.L:p in Xenopus laevis. /4 🔗
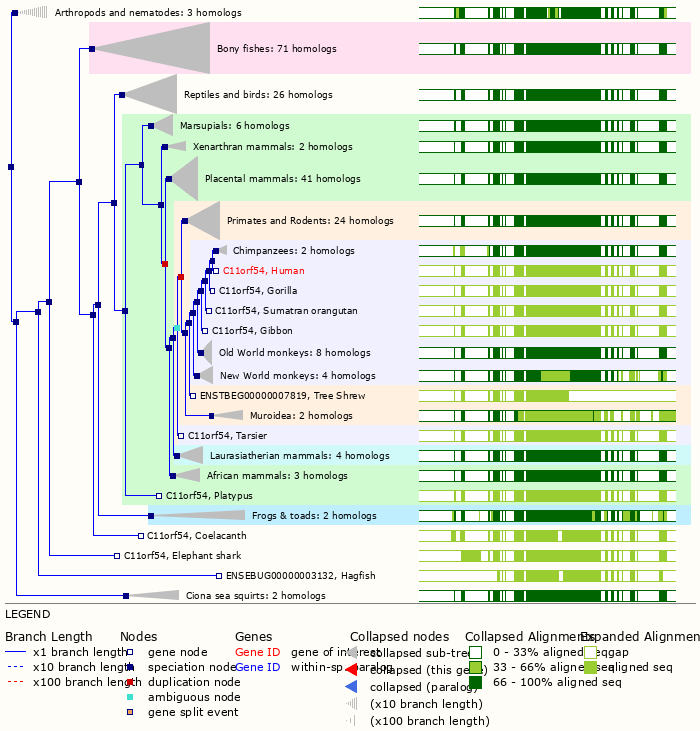
Tue Jan 04 13:03:20 +0000 2022C11orf54:p.I312T chr 11:g.93761675T>C, rs114772094 (all tissue I:T 0.995:0.005) vaf=0.5%, Δm=-12.0364, VAF by population group: african 7%, american <1%, east asian <1%, european <1%, south asian <1%. /3
Tue Jan 04 13:03:20 +0000 2022C11orf54:p, θ(max) = 76. aka PTD012. Found in tissue MHC class 1 & 2 experiments. Observed in many tissues & cell lines: most abundant in urine extracellular vesicles. /2
Tue Jan 04 13:03:20 +0000 2022>C11orf54:p, chromosome 11 open reading frame 54 (Homo sapiens) Small subunit; CTMs: A2+acetyl; PTMs: 4×K+acetyl; 0×K+succinyl; 4×K+GGyl; 2×K+SUMOyl; 1×S, 0×T, 0×Y+phosphoryl; SAAVs: I312T (0.5%); mature form: (2-315) [21,729×, 85 kTa]. #ᗕᕱᗒ /1 🔗
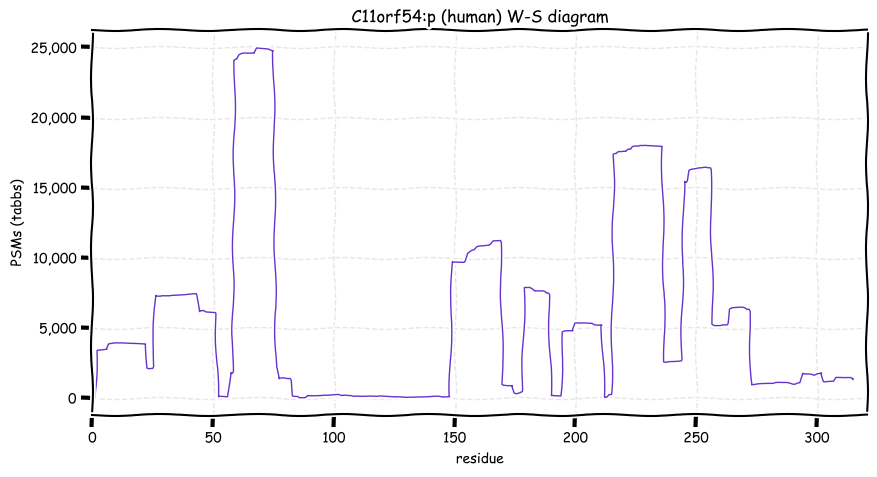
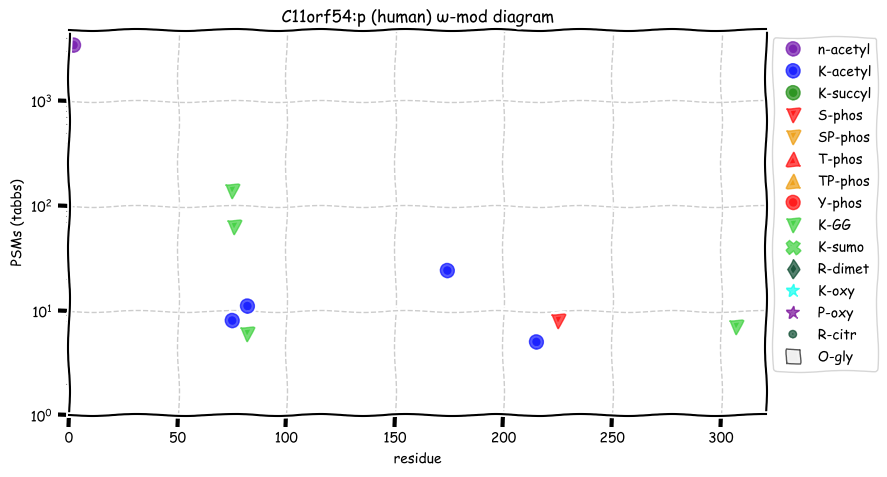
Mon Jan 03 21:46:08 +0000 2022@VATVSLPR @pwilmarth @UCDProteomics @nesvilab @dtabb73 @MagnusPalmblad @neely615 The cognoscenti have tended to oppose the idea of using prior knowledge to assist in proteomics data analysis.
Mon Jan 03 16:34:38 +0000 2022"adjunct" is a weird position/title.
Mon Jan 03 12:55:55 +0000 2022C7orf50:p, has homologs across the chordate lineage. In mice, it is referred to as 3110082I17Rik:p. 🔗
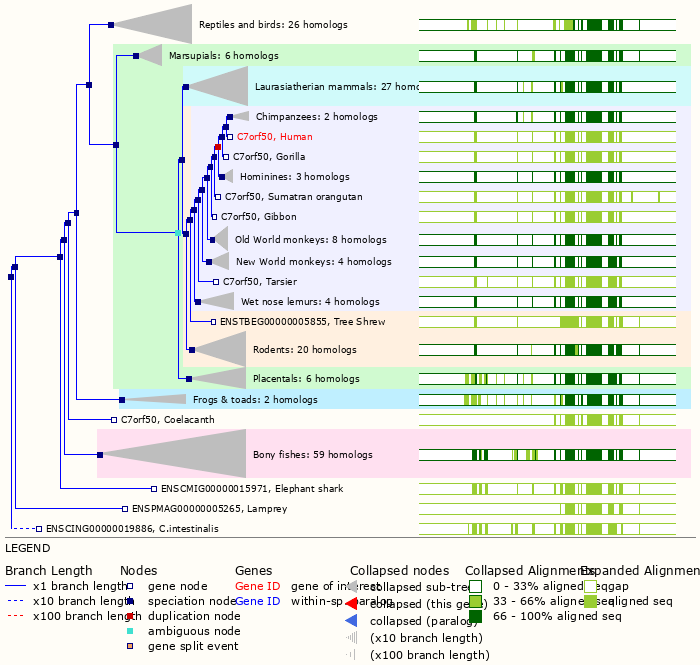
Mon Jan 03 12:55:55 +0000 2022C7orf50:p.G51R chr 7:g.1010122C>T rs61746340 (all tissue G:R 0.968:0.032) vaf=3%, Δm=99.0796, VAF by population group: african 10%, american 1%, east asian <1%, european <1%, south asian <1%.
Mon Jan 03 12:55:54 +0000 2022C7orf50:p, θ(max) = 77. aka MGC11257, YCR016W. Found, rarely, in MHC class 1 experiments. Found in many tissues & cell lines: rare in fluids.
Mon Jan 03 12:55:54 +0000 2022>C7orf50:p, chromosome 7 open reading frame 50 (Homo sapiens) Small subunit; CTMs: none; PTMs: 2×K+acetyl; 1×K+succinyl; 0×K+GGyl; 2×K+SUMOyl; 7×S, 0×T, 1×Y+phosphoryl; SAAVs: G51R (3%); mature form: (2-194) [23,203×, 85 kTa].#ᗕᕱᗒ 🔗
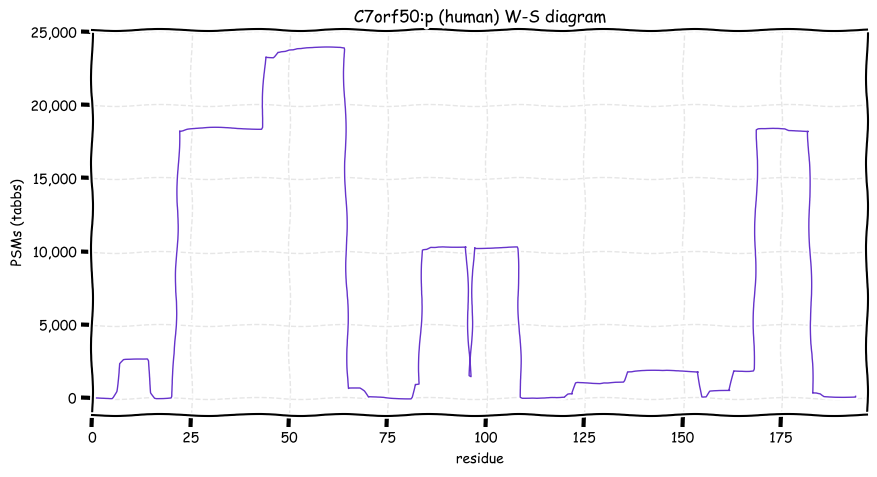
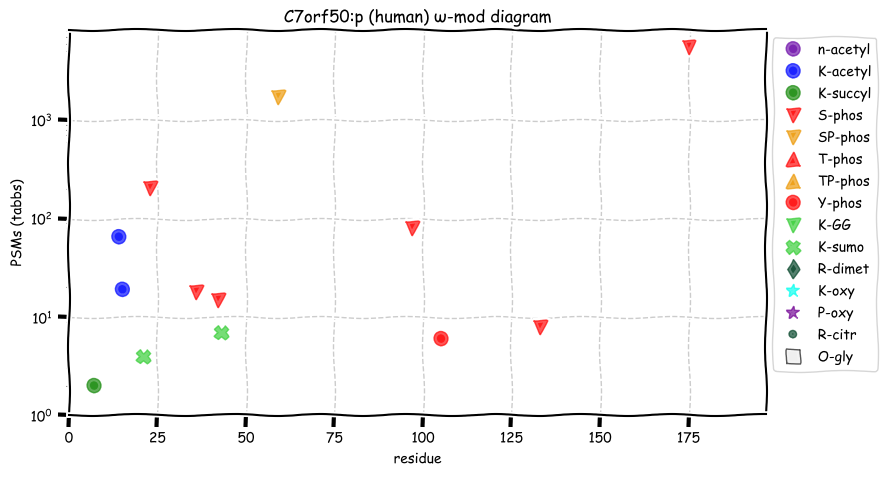
Mon Jan 03 12:53:24 +0000 2022@neely615 The blotch of weather northwest of the front that affected you is now a winter storm centered on Maryland & Delaware: your front is now a narrow ribbon out over the Atlantic. It just got chilly in the Carolinas, though. 🔗

Mon Jan 03 03:07:14 +0000 2022Same storm front at about 20:00 EST 🔗

Sun Jan 02 17:55:44 +0000 2022The big thing I learnt about proteins last year was the prevalence (& importance) of alternative translation initiation in mammalian cells.
Sun Jan 02 17:48:26 +0000 2022@Nick_E_Scott @ypriverol @dtabb73 I've been doing it for a quite a while: strongly recommend using this tactic 👍
Sun Jan 02 14:42:27 +0000 2022A front of locally intense weather crossing Alabama, Georgia & the Carolinas 🔗

Sun Jan 02 13:38:12 +0000 2022C1orf35:p, has a low complexity domain (182-263) enriched in basic residues. This feature reduces the number of observable peptides in the domain. 🔗

Sun Jan 02 13:38:12 +0000 2022C1orf35:p, has homologs across the vertebrate lineage. In mice, it is referred to as 2310033P09Rik:p. 🔗
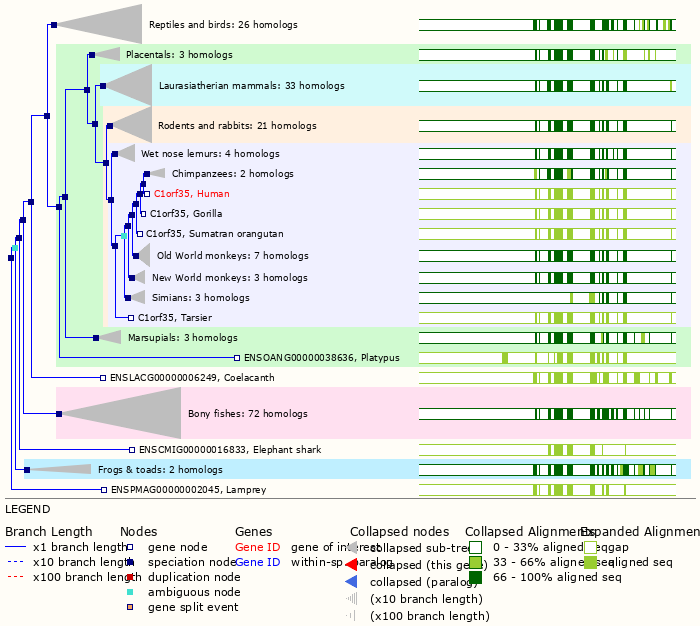
Sun Jan 02 13:38:11 +0000 2022C1orf35:p.F99L chr 1:g.228102555G>C rs371201234 (all tissue F:L 0.999:0.001) Δm=-33.9843, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sun Jan 02 13:38:11 +0000 2022C1orf35:p, θ(max) = 60. aka MGC4174, MMTAG2. Found in MHC class 1 experiments. Found in many tissues, lymphocytes & cell lines: absent from fluids.
Sun Jan 02 13:38:11 +0000 2022>C1orf35:p, chromosome 1 open reading frame 35 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 8×K+acetyl; 0×K+succinyl; 1×K+GGyl; 4×K+SUMOyl; 17×S, 6×T, 0×Y+phosphoryl; SAAVs: F99L (<1%), K142R (1%), R243Q (1%); mature form: (1-263) [23,980×, 189 kTa]. #ᗕᕱᗒ 🔗
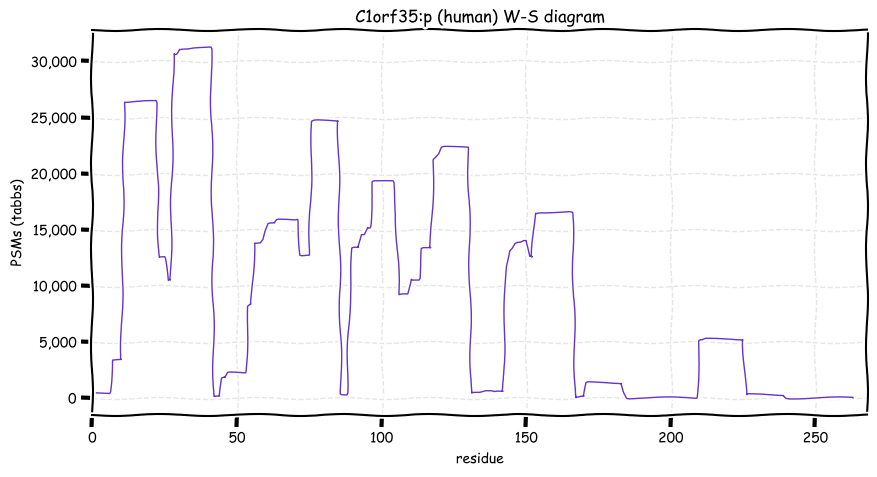
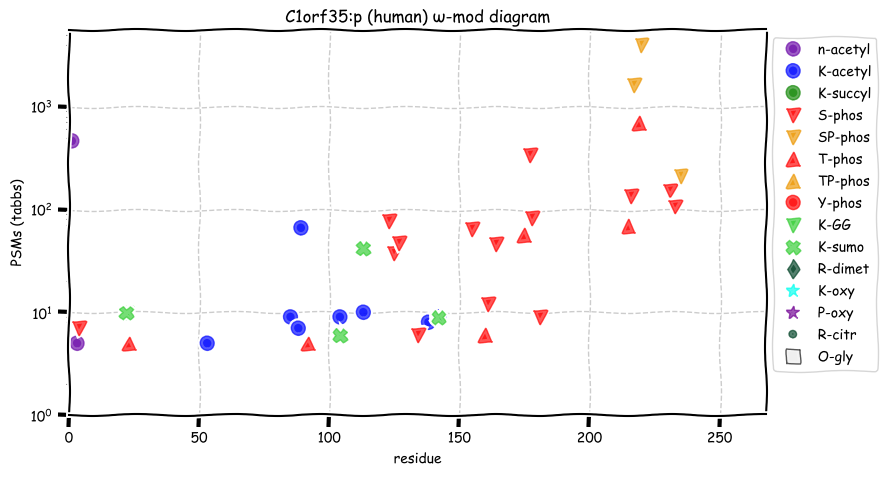
Sat Jan 01 20:30:31 +0000 2022Nasty looking New Year's Day storm running up the US Midwest this afternoon from Arkansas to upstate New York. 🔗

Sat Jan 01 18:43:25 +0000 2022Went to just over 13 billion PSMs overnight. How much is enough?
Sat Jan 01 14:51:11 +0000 202260°C (-35°C to 25°C) temperature gradient along I-29 this morning 🔗
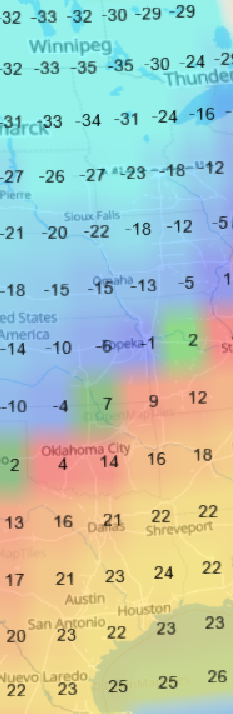
Sat Jan 01 13:22:18 +0000 2022C12orf10:p, the splice variant recommended in databases (MYG1-201) does not seem to be translated. MYG1-201 is only observed in protein-protein interaction expts when it is the bait. The observed splice (MYG1-204) does not have a mitochondrial targeting sequence.
Sat Jan 01 13:22:18 +0000 2022C12orf10:p, has homologs reaching across to fungi. 🔗
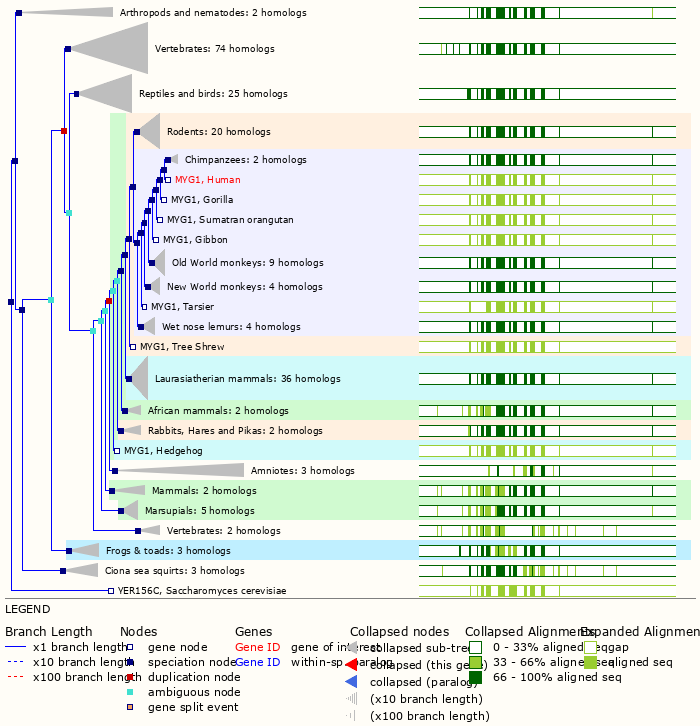
Sat Jan 01 13:22:17 +0000 2022C12orf10:p.V7I chr 12:g.53299828G>T rs376769284 (all tissue V:I 0.992:0.008) vaf=<1%, Δm=14.0156, VAF by population group: african <1%, american <1%, east asian <1%, european <1%, south asian <1%.
Sat Jan 01 13:22:17 +0000 2022C12orf10:p, θ(max) = 70. aka MYG, MYG1, Gamm1. Found in MHC class 1 & 2 experiments. Found in many tissues & cell lines: more rarely in fluids. This also initiates at M18.
Sat Jan 01 13:22:17 +0000 2022>C12orf10:p, chromosome 12 open reading frame 10 (Homo sapiens) Small subunit; CTMs: M1+acetyl; PTMs: 3×K+acetyl; 0×K+succinyl; 2×K+GGyl; 0×K+SUMOyl; 4×S, 0×T, 0×Y+phosphoryl; SAAVs: V7I (<1%), T274I (97%); mature form: (1-301) [27,581×, 115 kTa]. #ᗕᕱᗒ 🔗
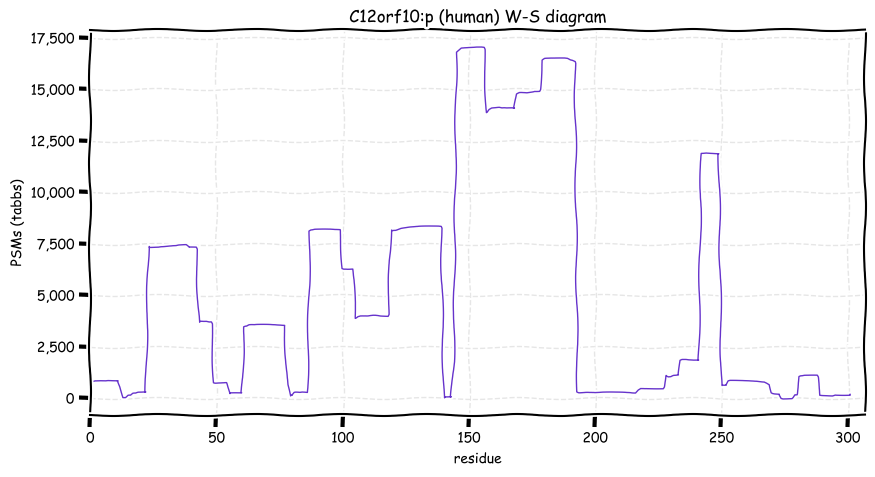
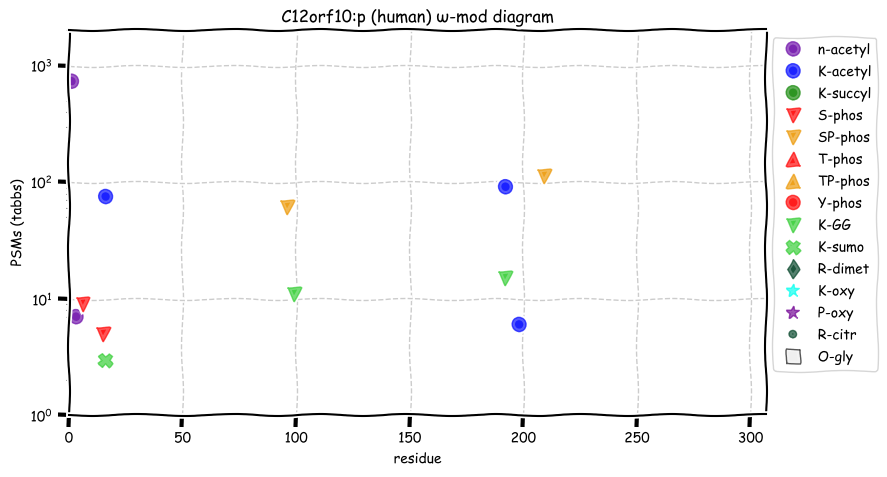
tweets = 3793