@norsivaeb Tweets 
| 2022
| 2021
| 2020
| 2019
| 2018
|
Thu Dec 31 16:39:05 +0000 2020Biggest space rock to just miss the earth today:
2020 YB4, 37 m in diameter, estimated miss ≈ 6.1 million kilometers, orbit: 🔗
Thu Dec 31 16:17:05 +0000 2020@FedExCanadaHelp After wasting yesterday trying to deal with this through your web site's various mechanisms, it is very unlikely that I will waste today trying to deal with it through your Twitter-based mechanisms.
Thu Dec 31 15:53:33 +0000 2020Just one of the many reasons I rarely buy anything on-line.
Thu Dec 31 15:27:34 +0000 2020It seems to be almost impossible to contact anyone to fix a delivery problem at FedEx in Canada.
Thu Dec 31 14:36:53 +0000 2020rs842381 note: UniProt canonical sequence (Q9Y2L9-1) has the reference allele (S234) rather than the ancestral (P234).
Thu Dec 31 14:36:53 +0000 2020rs842381, chr 13:g.46685919T>C LRCH1:p.S234P, vaf=>99%, Δm=10.021, is homozygous in HEK-293 derived cell lines and all other commonly used cell lines. #ᐯᐸᐱ
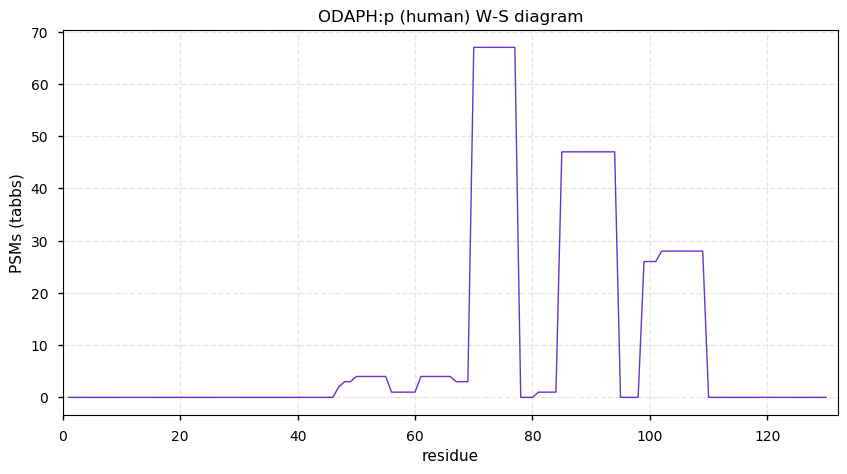
Thu Dec 31 13:41:59 +0000 2020ODAPH:p is the last entry in the "dark proteins and where to find them" series. The New Year will begin with an examination of the subunits of the THO complex.
Thu Dec 31 13:41:59 +0000 2020ODAPH:p θ(max) = 43. aka FLJ23657, AI2A4, C4orf26. Found in corneal endothelium, placenta & tooth enamel. Absent from cell lines and HLA experiments.
Thu Dec 31 13:41:58 +0000 2020>ODAPH:p, odontogenesis associated phosphoprotein (Homo sapiens) 🔗 Small protein; PTMs: none; SAAVs: none; mature form: 24?-130 [47×, 0.15 kTa] #ᗕᕱᗒ 🔗
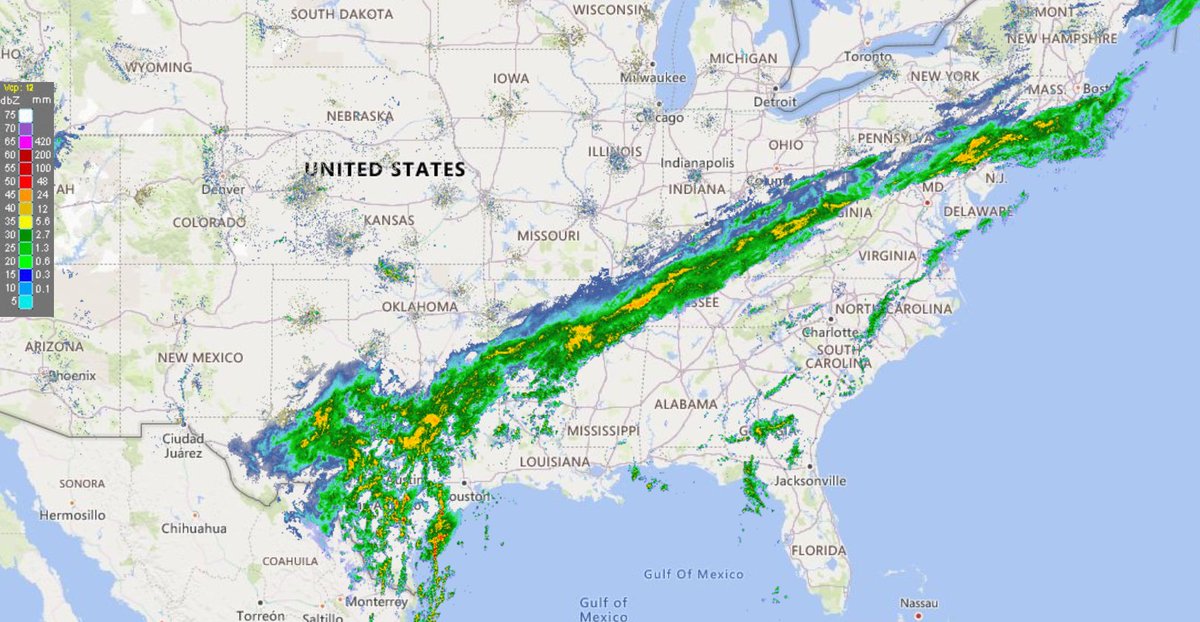
Thu Dec 31 13:02:23 +0000 2020It may be a nasty one on the ground, as the line of precipitation is the border between < °0 C in the west and > °0 C in the east (rain/freezing rain/snow), e.g. 🔗
Thu Dec 31 12:54:49 +0000 2020-10 °C, precip 0.0 mm, 1009 mb→, RH 87%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Thu Dec 31 12:49:19 +0000 2020A text-book quality cold front moving across the US this morning (NEXRAD radar, 12:45 UTC) 🔗
Wed Dec 30 21:50:16 +0000 2020@byu_sam It is one of the many things that seems to make people nervous when added to an id algorithm. I don't know why, but it just does.
Wed Dec 30 19:07:32 +0000 2020@jwoodgett @warren_weeks It has been a bad year for travel stories involving Finance Ministers who used to run Morneau-Shepell.
Wed Dec 30 17:34:07 +0000 2020@ASBMB @BiswapriyaMisra Does this merit some type of intervention or is it merely a curious but acceptable hobby?
Wed Dec 30 16:58:23 +0000 2020Biggest space rock to miss the earth today:
2012 UK171, 47 m in diameter, estimated miss ≈ 6 million kilometers, orbit: 🔗
Wed Dec 30 15:29:47 +0000 2020@Sci_j_my @byu_sam @GutsyBiotech @PhDVoice @OpenAcademics You might have to black out the references, too. Many PIs have a "distinctive" set of publications that they refer to in every article.
Wed Dec 30 15:18:10 +0000 2020@astacus Who manufactured it? Here in Canuckistan almost all of the slide rules were made by Hemmi, either via their own brand "Sun" or on contract to Hughes-Owen.
Wed Dec 30 15:10:42 +0000 2020@astacus Also flippantly: the British have long been the champions of "units-of-measurement-that-never-really-caught-on-even-in-the-pink-parts-of-the-map".
Wed Dec 30 14:37:50 +0000 2020Just heard the UK Health Secretary talking about vaccine distribution. He said the AZ vaccine produced immunity after a "fortnight". Is the use of Dickensian language mandatory for public school alumni?
Wed Dec 30 13:54:10 +0000 2020@astacus I use mine multiple times every day. I find it faster and it maintains your ability to do quick order-of-magnitude calculations.
Wed Dec 30 13:35:07 +0000 2020rs11576415 (chr 1:g.161212418C>G NDUFS2:p.P352A, vaf=8%, Δm = -26.016) is heterozygous in HEK-293 derived cell lines and HaCaT cells. Most other cell lines are homozygous for the reference allele, e.g., A-549. #ᐯᐸᐱ
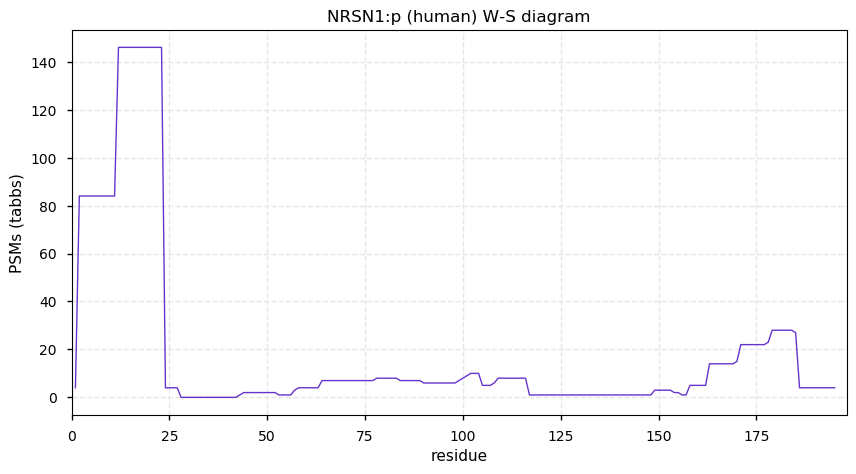
Wed Dec 30 13:13:31 +0000 2020NRSN1:p θ(max) = 61. aka p24, VMP. Found in brain tissue exclusively. Absent from cell lines and HLA experiments. Two membrane spanning domains: (64-86) & (117-140).
Wed Dec 30 13:13:31 +0000 2020>NRSN1:p, neurensin 1 (Homo sapiens) 🔗 Small membrane protein; CTMS: S2+acetyl; PTMs: none; SAAVs: none; mature form: 2-195 [199×, 0.31 kTa] #ᗕᕱᗒ 🔗
Wed Dec 30 13:04:54 +0000 2020-10 °C, precip 3.0 mm ❄, 1011 mb↑, RH 88%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Tue Dec 29 20:33:59 +0000 2020@VATVSLPR @pwilmarth @Sci_j_my @byu_sam Canadians like to think of it as either Western Northern Ontario or Eastern Saskatchewan.
Tue Dec 29 18:33:00 +0000 2020@Sci_j_my @byu_sam I shall restrain myself from mocking the so-called "winter" experienced by our friends in the US.
Tue Dec 29 17:10:32 +0000 2020I guess this might not be the right crowd to ask about this sort of thing 🧐
Tue Dec 29 14:04:46 +0000 2020And by "ClinVar", I mean this data system 🔗
Tue Dec 29 13:49:20 +0000 2020Does anybody use ClinVar? And if you do, what do you use it for?
Tue Dec 29 13:43:16 +0000 2020rs12584 (chr 1:g.19086767T>A UBR4:p.M4867L, vaf=59%, Δm = -17.956) is heterozygous in HEK-293 derived cell lines and SK-MEL-28. Homozygous in SW-480 & HeLa cells. #ᐯᐸᐱ
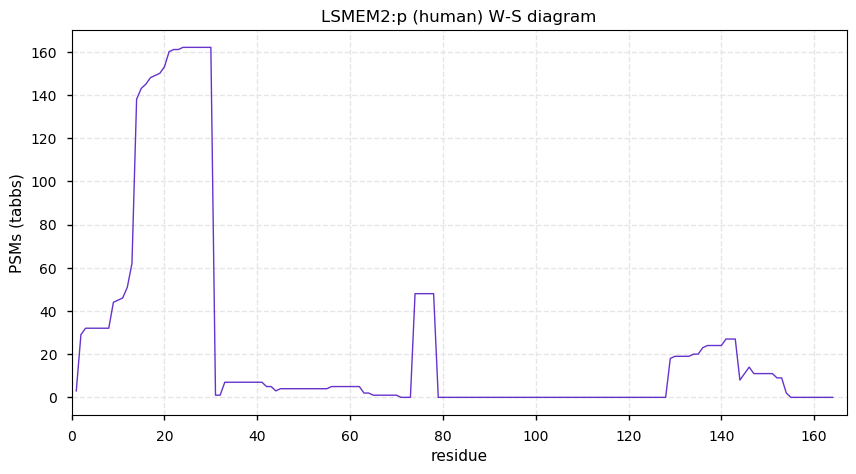
Tue Dec 29 13:06:58 +0000 2020LSMEM2:p θ(max) = 58. aka FLJ38608, C3orf45. Results consistent with alternate translation initiation site at M13 (mRNA context UACUGG[AUG]CCA … CUUGCC[AUG]CCU). As per the name, 1 membrane spanning domain (98-119).
Tue Dec 29 13:06:57 +0000 2020>LSMEM2:p, leucine rich single-pass membrane protein 2 (Homo sapiens) 🔗 Small protein; PTMs: none; SAAVs: none; mature form: 2,14-164 [45×, 0.27 kTa] #ᗕᕱᗒ 🔗
Tue Dec 29 13:03:28 +0000 2020-12 °C, precip 0.0 mm, 1017 mb↓, RH 79%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Mon Dec 28 16:28:13 +0000 2020rs1052637 (chr 2:g.117817639C>G, DDX18:p.T94S, vaf=37%, Δm = -14.016) is heterozygous in HEK-293 derived cell lines as well as MCF-10A and JURKAT cells. Homozygous in HeLa and WM-239 cells. Reference only in MCF-7 and Hep-G2 #ᐯᐸᐱ
Mon Dec 28 15:56:53 +0000 2020I have the same attitude about magazine articles: bad sig figs means no one with scientific training was involved in authoring or proofing the manuscript/web site.
Mon Dec 28 15:26:11 +0000 2020Thanks to everyone who participated in this poll. The USA won this one in a landslide.
Mon Dec 28 15:13:56 +0000 2020@slashdot A break-through in fuel production: the only thing that stands in its way is the 1st Law of Thermodynamics 🤔
Mon Dec 28 14:08:44 +0000 2020Only 1 hour left to express your opinion ... 🔗
Mon Dec 28 13:35:24 +0000 2020-20 °C, precip 0.6 mm, 1026 mb↑, RH 75%, clear ☀ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
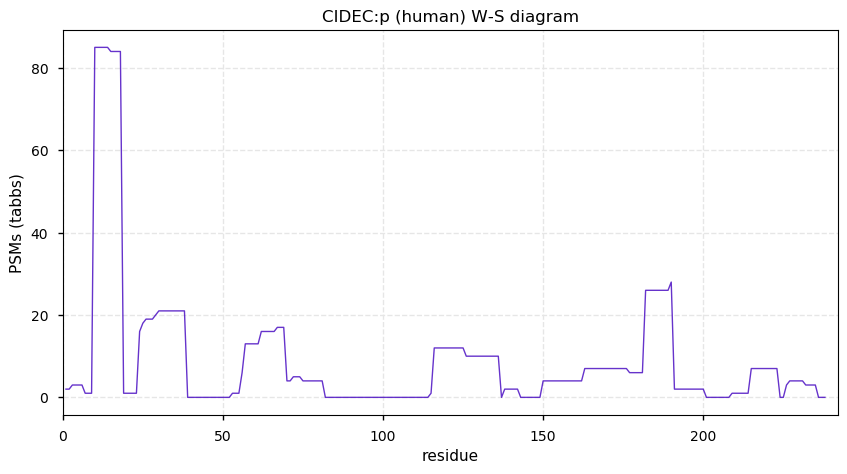
Mon Dec 28 13:32:26 +0000 2020CIDEC:p θ(max) = 58. aka CIDE-3, FLJ20871, Fsp27. Its best observation are from HLA class I experiments, but it has not been observed in class II experiments. It has not been detected in common cell lines.
Mon Dec 28 13:32:26 +0000 2020>CIDEC:p, cell death inducing DFFA like effector c (Homo sapiens) 🔗 Small protein; PTMs: none; SAAVs: A46P (1%); mature form: 1-238 [110×, 0.2 kTa] #ᗕᕱᗒ 🔗
Sun Dec 27 23:43:22 +0000 2020I can't take a data system seriously that doesn't use significant figures properly (I'm looking at you dbSNP).
Sun Dec 27 21:38:56 +0000 20203. d4 cxd4 🔗

Sun Dec 27 19:29:56 +0000 2020The results have been remarkably steady since the first few minutes.
Sun Dec 27 17:14:46 +0000 20203. d4 🔗

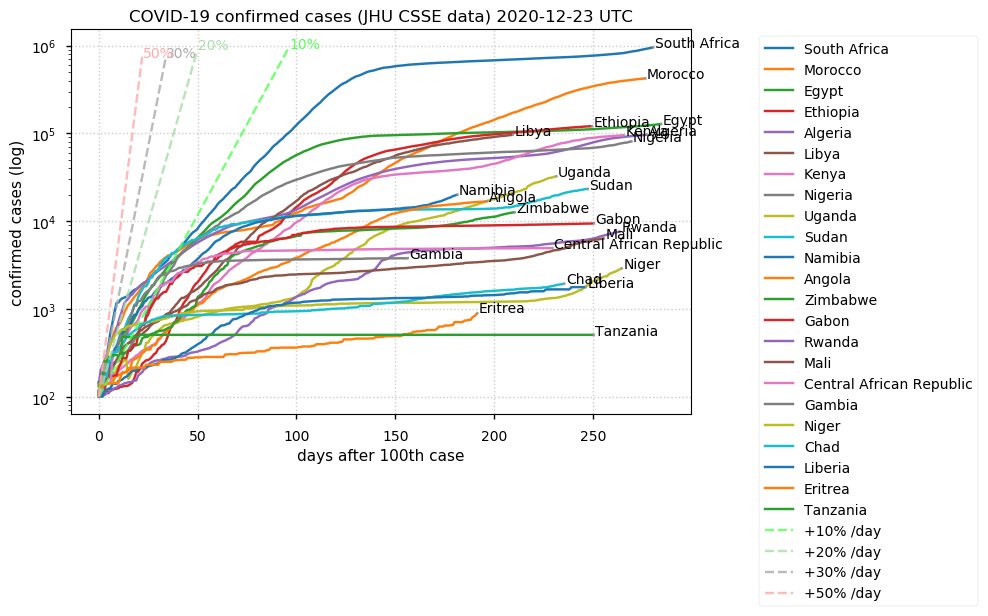
Sun Dec 27 15:24:06 +0000 2020Country whose reputation took the biggest hit from its handling of the COVID-19 epidemic:
Sun Dec 27 14:07:57 +0000 2020rs1801591 (chr 15:76286421G>A ETFA:p.T171I, vaf=7%, Δm = 12.036) is heterozygous in HEK-293 derived cell lines as well as HCT-116, A-549 & MHCC97-H cells. Reference only in HeLa, MCF-10A and MCF-7. #ᐯᐸᐱ
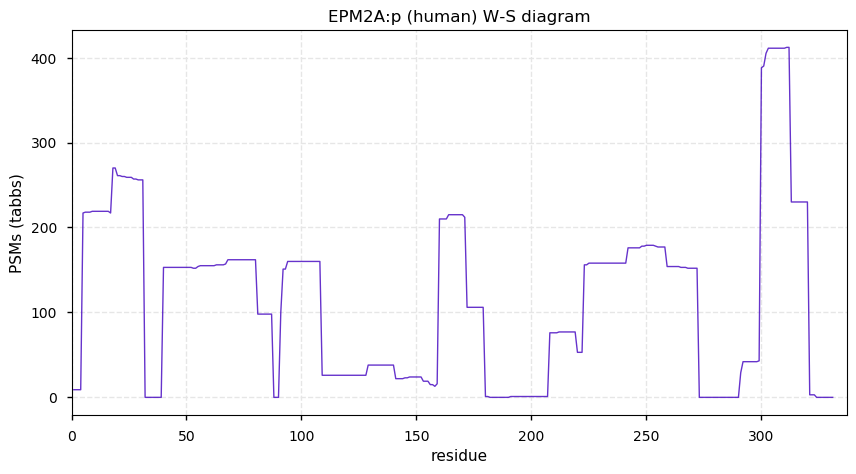
Sun Dec 27 13:46:42 +0000 2020EPM2A:p θ(max) = 64. aka LDE, LD. It has not been observed in HLA class I or II peptide experiments. Not commonly observed, but when present (e.g., cardiac, testis or brain tissue) produces good, unique PSMs. Associated with a form of hereditary epilepsy.
Sun Dec 27 13:46:42 +0000 2020>EPM2A:p, EPM2A glucan phosphatase, laforin (Homo sapiens) 🔗 Small protein; PTMs: none; SAAVs: A46P (1%); mature form: 1-331 [908× , 2.4 kTa] #ᗕᕱᗒ 🔗
Sun Dec 27 13:39:03 +0000 2020-6 °C, precip 0.0 mm, 1014 mb→, RH 80%, clear ☀ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Sat Dec 26 22:04:52 +0000 20202. Nf3 d6 🔗

Sat Dec 26 13:45:52 +0000 2020rs3182535 (chr 10:g.3158100G>A, PITRM1:p.A397V, vaf=66%, Δm = 28.031) is homozygous in HEK-293 derived cell lines as well as JURKAT & MCF-10A cells. Heterozygous in CACO-2 cells. Reference only in HeLa cells. #ᐯᐸᐱ
Sat Dec 26 13:27:44 +0000 20202. Nf3 🔗

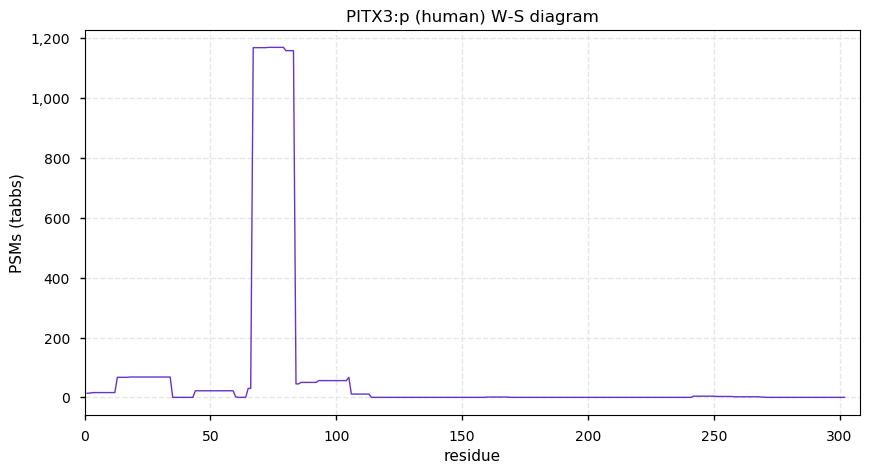
Sat Dec 26 13:13:13 +0000 2020PITX3:p θ(max) = 29. aka ASMD, paired like homeodomain 3. Significant tryptic peptide overlap with PITX1 & PITX2. The only HLA class I peptide observed is shared between PITX1, 2 & 3.
Sat Dec 26 13:13:13 +0000 2020>📦 PITX3:p, pituitary homeo-box 3 (Homo sapiens) 🔗 Small protein; CTMs: M1+acetyl; PTMs: S7, S13,S 17, S19, S52+phosphoryl; SAAVs: none; mature form: 1-302 [901 x, 1.4 kTa] #ᗕᕱᗒ 🔗
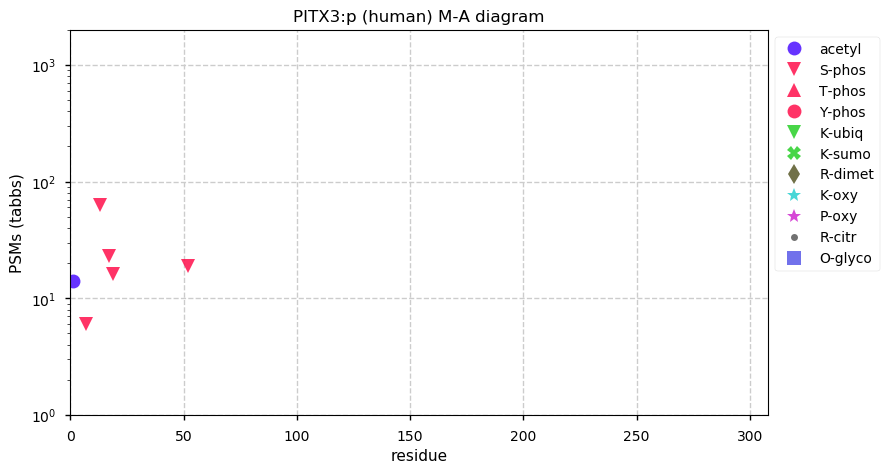
Sat Dec 26 12:59:24 +0000 2020-5 °C, precip 0.0 mm, 1012 mb↑, RH 83%, overcast ☁ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Fri Dec 25 22:25:02 +0000 20201. e4 c5 🔗

Fri Dec 25 16:53:14 +0000 20201. e4
#ᐁᐃ 🔗

Fri Dec 25 13:57:52 +0000 2020Because the existence of the variant removed the potential for cysteine sulphydryl blocking, the practical Δm will include the mass of the blocking reagent, e.g., if IAA was the blocking reagent, Δm = -(15.977+57.021) = -72.998
Fri Dec 25 13:57:52 +0000 2020And the protein-level evidence: HEK-293 🔗
Fri Dec 25 13:57:52 +0000 2020rs4329520 (chr 1:g.152760825T>A, KPRP:p.C413S, vaf=48%, Δm = -15.977) is heterozygous in HEK-293 derived cell lines as well as U2-OS & HeLa cells. #ᐯᐸᐱ
Fri Dec 25 13:24:59 +0000 2020@pwilmarth Not a bad suggestion. I had another one queued up, but if there is a box-ing day protein, why not?
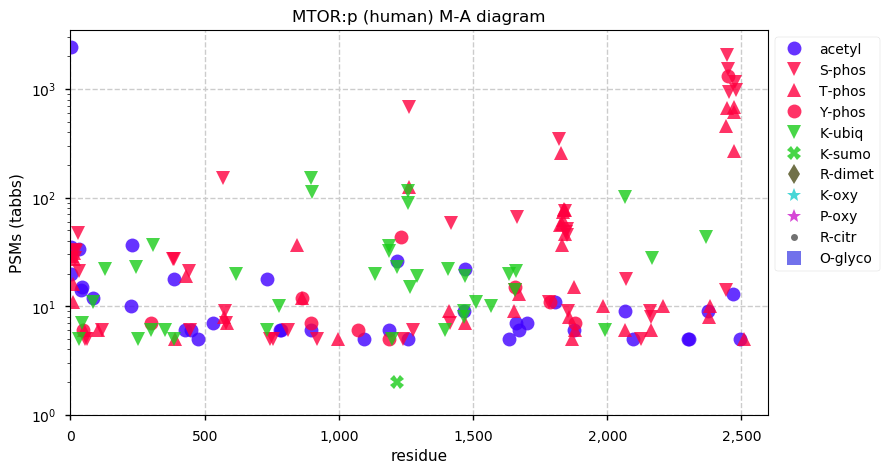
Fri Dec 25 13:02:40 +0000 2020MTOR:p is not part of the "dark proteins and where to find them" series. Back to that tomorrow until Dec. 31st.
Fri Dec 25 13:02:40 +0000 2020MTOR:p θ(max) = 66. aka RAFT1, RAPT1, FLJ44809, FRAP, FRAP2, FRAP1. It is commonly observed in HLA class I peptide experiments, but not class II experiments. Too many GO annotations. Chosen as today's protein because it is decorated like a Christmas tree.
Fri Dec 25 13:02:40 +0000 2020>🎄 MTOR:p, mechanistic target of rapamycin kinase (Homo sapiens) 🔗 Large protein; CTM: M1+acetyl; PTMs: lots of K+acetyl/ubiquitinyl & S,T+phosphoryl, no K+SUMOyl or +glycosyl SAAVs: none; mature form: 1-2549 [27,158× , 218 kTa] #ᗕᕱᗒ 🔗
Fri Dec 25 12:53:24 +0000 2020-14 °C, precip 0.0 mm, 1006 mb↓, RH 83%, clear ☀ (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Thu Dec 24 19:39:52 +0000 2020@astacus It is a serif, monospaced font in TrueType format (AMINONEW.TTF) that you can install on any computer:
🔗
Thu Dec 24 18:59:19 +0000 2020@astacus Back at you 🔗
Thu Dec 24 15:58:58 +0000 2020Sometimes I get the very clear impression that when people retweet paper announcements with positive statements ("at last", "great work"), they haven't actually read the paper yet …
Thu Dec 24 15:15:38 +0000 2020Probably good aurora australias on Heard Island at the moment 🔗
Thu Dec 24 13:26:59 +0000 2020Note: the UniProt cannonical sequence (P13646-1) has the reference T298 residue rather than the ancestral A289.
Thu Dec 24 13:26:59 +0000 2020Although KRT13:p has significant tryptic peptide overlap with other keratins, the peptide bearing this SAAV does not have any overlap with those other gene products.
Thu Dec 24 13:26:59 +0000 2020rs4796697 (KRT13:p.T298A, vaf=97%, Δm = -30.011) is homozygous in HEK-293 derived cell lines as well as HeLa, U2-OS & A-431 cells. The reference sequence is not observed in common cell lines. #ᐯᐸᐱ
Thu Dec 24 13:07:11 +0000 2020-22 °C, precip 0.0 mm, 1021 mb↑, RH 82%, cloudy (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
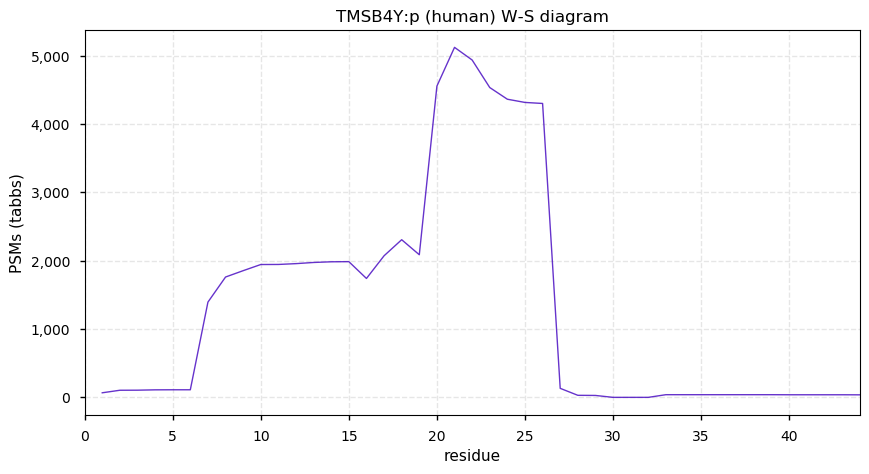
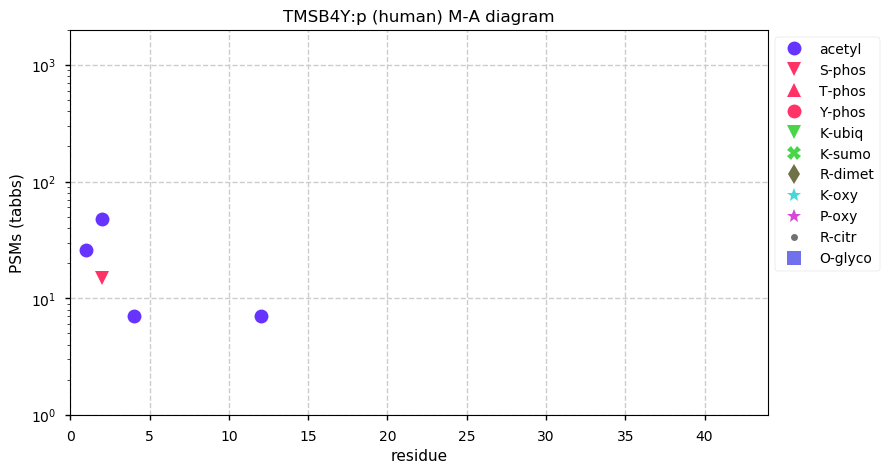
Thu Dec 24 12:44:45 +0000 2020TMSB4Y:p θ(max) = 55. aka TB4Y. Nearly complete peptide sequence overlap with the X-linked version of the protein, TMSB4X (they only differ at residues 6, 30 & 39). The best unique peptide observations are found in HLA class I experiments.
Thu Dec 24 12:44:45 +0000 2020>TMSB4Y:p, thymosin beta 4 Y-linked (Homo sapiens) 🔗 Very small protein; CTM: M1,S2+acetyl; PTMs: K4,K12+acetyl; SAAVs: none; mature form: 1,2-44 [1,806× , 6.3 kTa] #ᗕᕱᗒ 🔗
Thu Dec 24 12:13:45 +0000 2020It looks like SA is headed for another round of lockdowns. 🔗
Wed Dec 23 20:37:13 +0000 2020& realizing I'm not going to get anything done after that, Once More with Feeling.
Wed Dec 23 18:14:31 +0000 2020I will now pause for The Expanse, S05E04.
Wed Dec 23 17:24:46 +0000 2020@jwoodgett Some assembly required.
Wed Dec 23 16:20:37 +0000 2020@labs_mann @bruker @EvosepBio @fabian_theis Odd choice of a peptide to serve as an exemplar.
Wed Dec 23 15:44:20 +0000 2020@BrandonMurugan @dtabb73 Technical replicates? What type of sorcery is this?
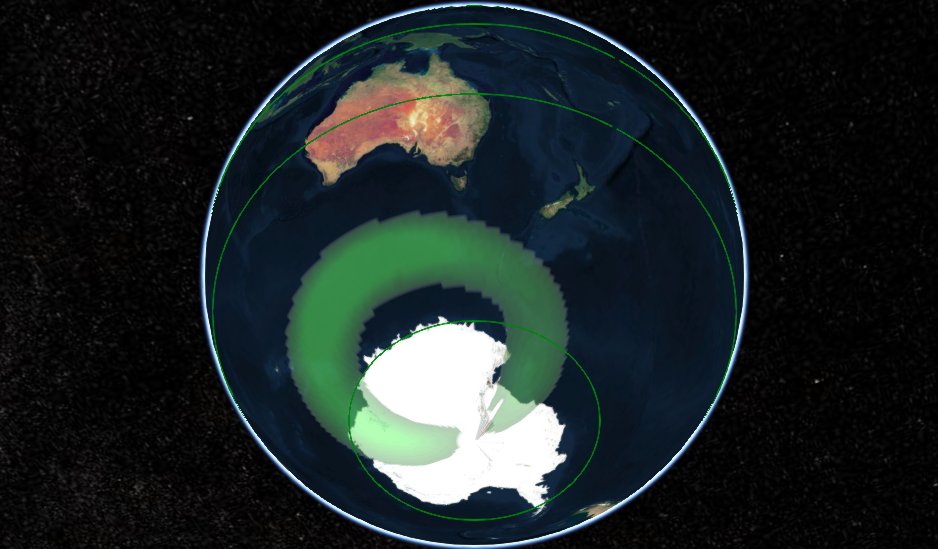
Wed Dec 23 15:27:25 +0000 2020Even though we are having some mild geomagnetic storms at the moment, it is still not enough to push aurora australis as far north as Tasmania or the South Island. 🔗
Wed Dec 23 14:13:06 +0000 2020In my experience, RTs are only useful for some PTMs. For example, peptide N-terminal acetylation can produce a big shift in RT, while S/T phosphorylation produces shifts small enough they can be difficult to reliably measure.
Wed Dec 23 13:31:35 +0000 2020rs2304497 (ACLY:p.E175D, vaf=10%, Δm = -14.016) is homozygous in HEK-293 derived cell lines and BT-474. Heterozygous in HEp-2, SKOV-3, A-431, HEp-2 & HeLa #ᐯᐸᐱ
Wed Dec 23 13:15:48 +0000 2020@Sci_j_my @BiswapriyaMisra @pwilmarth @Smith_Chem_Wisc Rather predictably, I check to see how many times the modification has been seen in previous studies.
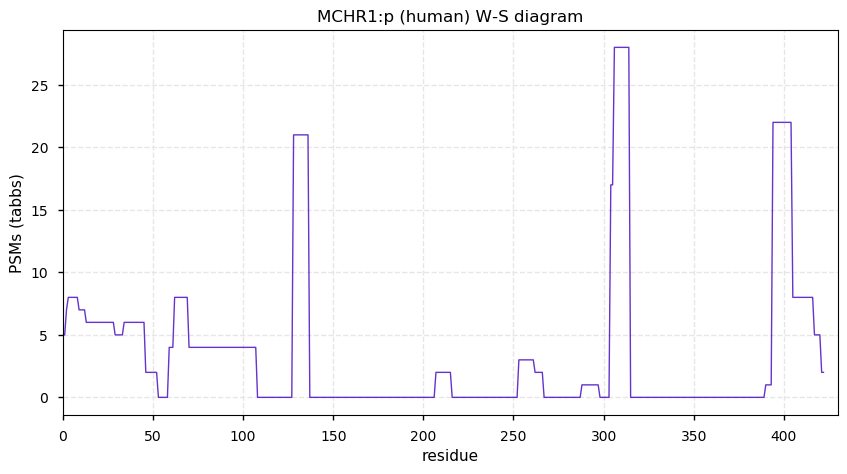
Wed Dec 23 13:09:26 +0000 2020MCHR1:p θ(max) = 23. aka SLC1, MCH1R, GPR24. The only convincing observations of the protein are from HLA type I experiments.
Wed Dec 23 13:09:26 +0000 2020>MCHR1:p, melanin concentrating hormone receptor 1 (Homo sapiens) 🔗 Small protein; PTMs: none; SAAVs: none; mature form: 2?-422 [51× , 0.10 kTa] #ᗕᕱᗒ 🔗
Wed Dec 23 13:03:38 +0000 2020-15 °C, precip 3.9 mm, 1013 mb↑, RH 85%, cloudy (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Tue Dec 22 20:22:00 +0000 2020My crypto-chirpty circles 🔗

Tue Dec 22 17:40:59 +0000 2020Just sitting here watching I've-lost-track-of-how-many job indicators creep towards 100%.
Tue Dec 22 16:38:35 +0000 2020@stephen_taylor 🔗

Tue Dec 22 13:32:53 +0000 2020Note: The UniProt canonical sequence for GPRIN1:p (Q7Z2K8-1) has the reference residue (M300) rather than the ancestral residue (V300).
Tue Dec 22 13:32:53 +0000 2020And the protein-level evidence for all tissues and cell lines: 🔗
Tue Dec 22 13:32:53 +0000 2020rs6556276 (GPRIN1:p.M300V, vaf=>99%, Δm = -31.97207) is homozygous in HEK-293 derived cell lines and all tissues and cell lines. #ᐯᐸᐱ
Tue Dec 22 13:06:14 +0000 2020-9 °C, precip 0 mm, 1008 mm↓, RH 90%, cloudy (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
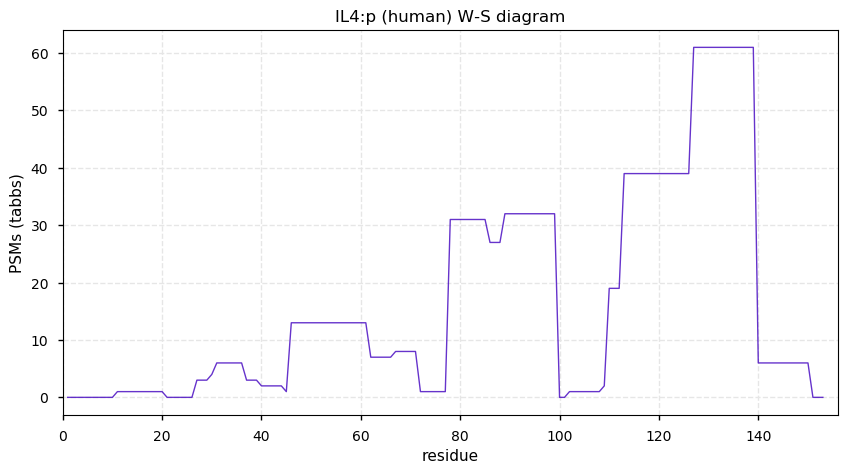
Tue Dec 22 13:03:06 +0000 2020IL4:p θ(max) = 35. aka BSF1, IL-4, BCGF1, BCGF-1, MGC79402. Other than a few PPI studies that have used IL4:p as a bait, the only convincing observations of the protein are from HLA type I experiments.
Tue Dec 22 13:03:06 +0000 2020>IL4:p, interleukin 4 (Homo sapiens) 🔗 SMall protein; PTMs: none; SAAVs: none; mature form: 25?-153 [36× , 0.16 kTa] #ᗕᕱᗒ 🔗
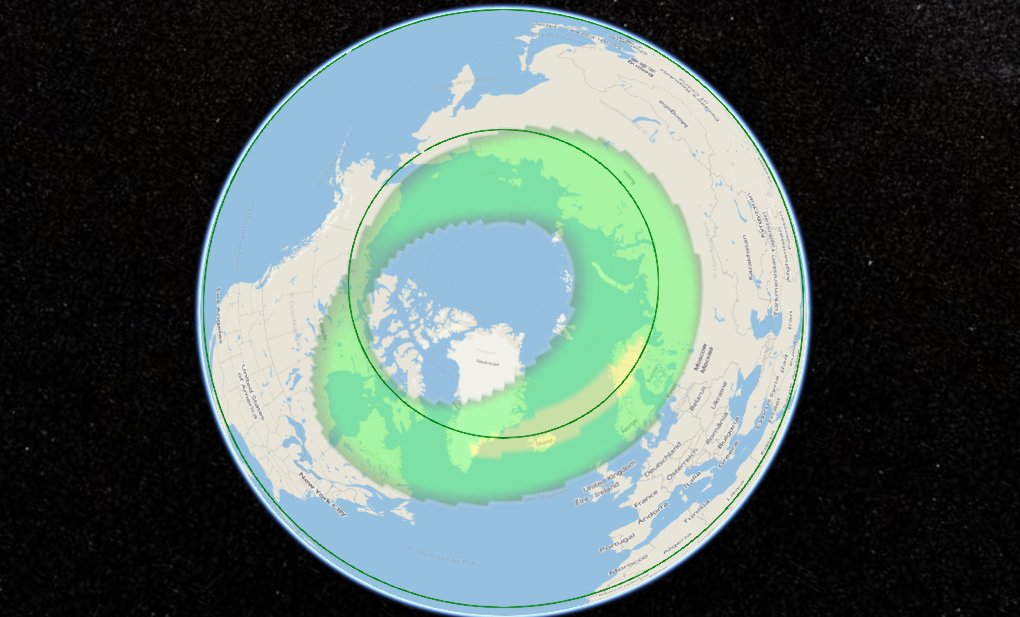
Mon Dec 21 21:59:35 +0000 2020Looks like the aurora should be visible from as far south as Inverness (SCT) & quite bright in Reykjavik (IS) right now. 🔗
Mon Dec 21 15:25:27 +0000 2020@oleg8r Cancer stuff probably inserted at the insistence of Reviewer #3.
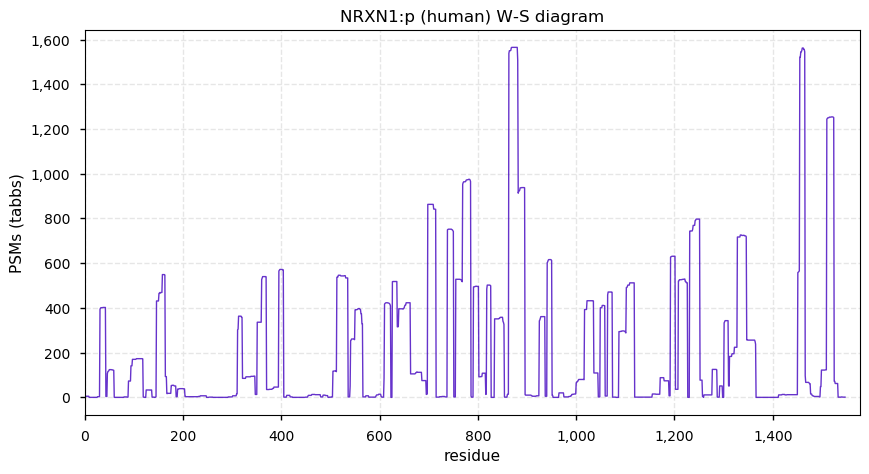
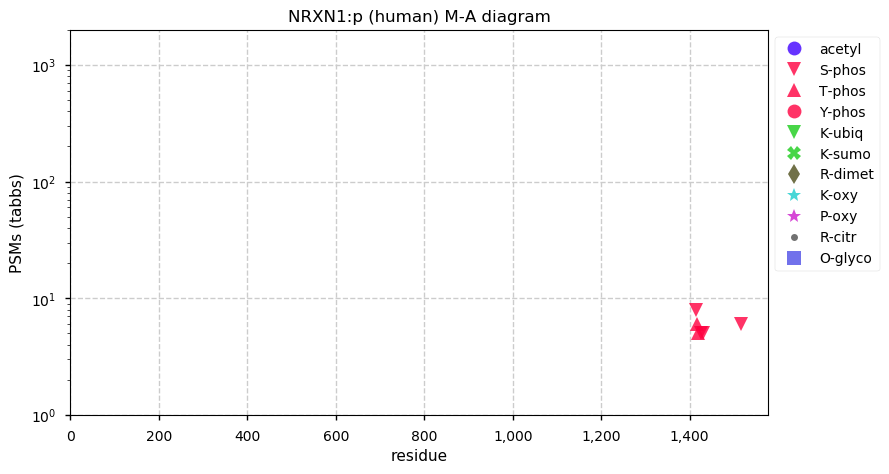
Mon Dec 21 13:21:51 +0000 2020NRXN1:p has considerable tryptic peptide overlap with DPYSL2, DPYSL3, DPYSL5, CRIMP1, NRXN2 & NRXN3, but good data (e.g. PXD004572 or PXD006109) contain unambiguous identifications.
Mon Dec 21 13:21:51 +0000 2020NRXN1:p θ(max) = 55. aka KIAA0578. High coverage observations in brain tissue and cerebrospinal fluid only. Type I membrane protein, with transmembrane domain (1472-1492): S,T+phosphoryl on cytoplasmic domain.
Mon Dec 21 13:21:51 +0000 2020>NRXN1:p, neurexin 1 (Homo sapiens) 🔗 Large plasma membrane protein; PTMs: 6× low occupancy S,T+phosphoryl; SAAVs: none; mature form: 31-1547 [2,554× , 24 kTa] #ᗕᕱᗒ 🔗
Mon Dec 21 13:18:19 +0000 2020rs1059476 (AURKB:p.M299T, vaf=83%, Δm = -29.993, chr17) is homozygous in HEK-293 derived cell lines as well as Jurkat, HCT-116 & HeLa cells. Reference only in A-431 and Hep-G2 #ᐯᐸᐱ
Mon Dec 21 12:55:41 +0000 2020@neely615 🔗
Mon Dec 21 12:47:43 +0000 2020-10 °C, precip 11.3 mm, 1004 mm↑, RH 85%, clear (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Mon Dec 21 01:15:04 +0000 2020SAAVs in the news
🔗
Sun Dec 20 17:39:52 +0000 2020While it is unfortunately disease oriented, this review is still a pretty good general introduction to nsSNVs in human tissue, without undo emphasis on the relatively minor contributions associated with somatic variants 🔗
Sun Dec 20 14:49:02 +0000 2020I would like to think that this sort of satellite deployment would require something more international than simple FCC approval, but it would seem to be enough. 🔗
Sun Dec 20 13:49:24 +0000 2020rs7535528 (PANK4:p.A547V, vaf=23%, Δm = 28.031) is heterozygous in HEK-293 derived cell lines as well as A-431 & HeLa cells. Homozygous in K-562 #ᐯᐸᐱ
Sun Dec 20 13:41:07 +0000 2020-8 °C, precip 0 mm, 1006 mb→, RH 70%, clear (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
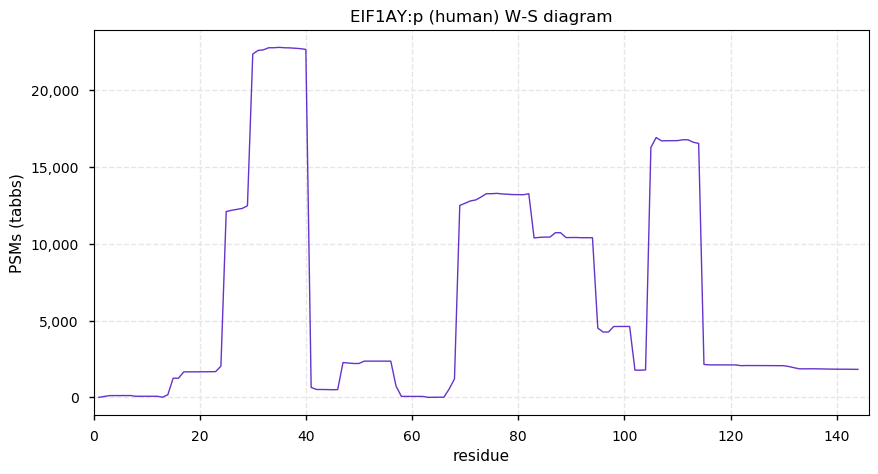
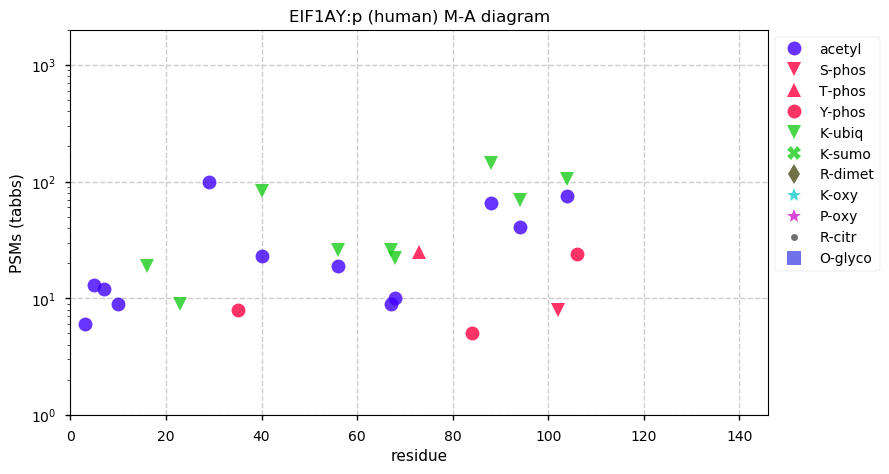
Sun Dec 20 13:34:08 +0000 2020EIF1AY:p θ(max) = 84. This is the Y-linked partner of EIF1AX:p, with significant tryptic peptide overlap. They can be distinguished by the corresponding peptide domains (47-56): LEALCFDGVK (EIF1AY:p, 1574 Ta) vs LEAMCFDGVK (EIF1AX:p, 9590 Ta).
Sun Dec 20 13:34:08 +0000 2020>EIF1AY:p, eukaryotic translation initiation factor 1A, Y-linked (Homo sapiens) 🔗 Small protein; PTMs: 5× K+acetyl, Y35, Y84, Y106+phosphoryl; aPTMs: 7× K+acetyl/ubiquitinyl; SAAVs: none; mature form: 2-144 [16,484× , 74 kTa] #ᗕᕱᗒ 🔗
Sat Dec 19 14:46:51 +0000 2020As the US military sinks into blistering self-satire
🔗
Sat Dec 19 14:42:48 +0000 2020Pretty much blows up the silly idea of "cybersecurity"
🔗
Sat Dec 19 14:22:54 +0000 2020Note: the peptide bearing this SAAV corresponds to Uniprot RBFOX2 isoform 6 or 8, O43251-6 or O43251-8. It does not exist in the "canonical" sequence O43251-1 or any of the other 7 isoforms.
Sat Dec 19 14:22:54 +0000 2020rs9607299 (RBFOX2:p.H8Q, vaf=>99%, Δm = -9.000) is homozygous in HEK-293 derived cell lines and all other common cell lines. #ᐯᐸᐱ
Sat Dec 19 13:24:16 +0000 2020-14 °C, precip 0 mm, 1012 mb↓, RH 70%, cloudy (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
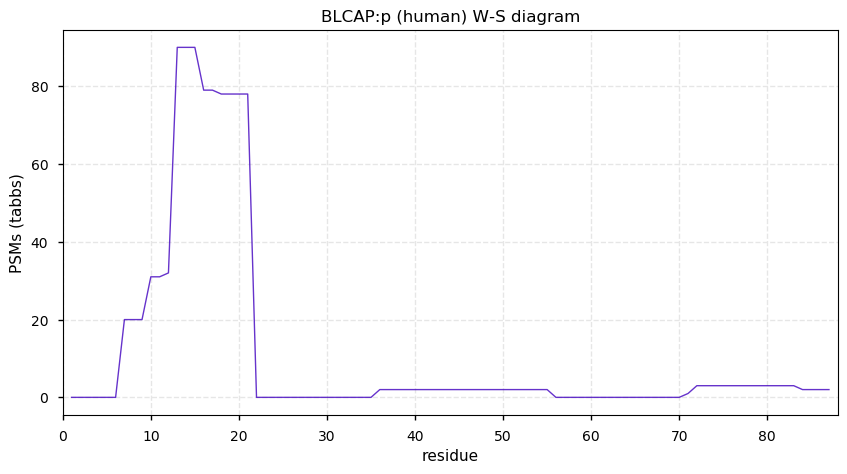
Sat Dec 19 13:15:35 +0000 2020BLCAP:p θ(max) = 17. aka BC10. It has 1 tryptic cleavage sites, but the 2 large tryptic peptides have not been observed. Each peptide has a membrane spanning domain: (19-39) or (43-63). The protein has been observed convincingly in HLA type I peptide experiments.
Sat Dec 19 13:15:35 +0000 2020>BLCAP:p, bladder cancer associated protein (Homo sapiens) 🔗 Very small protein; CTMs: none; PTMs: none; SAAVs: none; mature form: 1-87 [100× , 0.12 kTa] #ᗕᕱᗒ 🔗
Fri Dec 18 22:57:17 +0000 2020It is a good thing that
$ exot
isn't a Linux command.
Fri Dec 18 22:28:12 +0000 2020If it ins't cloudy, Trondheim should be seeing a pretty good aurora right now 🔗
Fri Dec 18 18:44:43 +0000 2020The release of season 5 of The Expanse is going to make working difficult for the next few days.
Fri Dec 18 16:09:38 +0000 2020So, just doing your normal analysis will always lead to at least some level of genotyping/identification without having to do any smarty-pants SAAV detection.
Fri Dec 18 15:47:35 +0000 2020Every time you detect the reference proteome allele in a peptide bearing an SAAV with a known MAF, by detecting that reference allele you are partially genotyping the individual. 🤔
Fri Dec 18 15:44:41 +0000 2020For those who don't like to think about SAAV detection: if you work with proteomics data from individuals (or cell lines) you have been doing it for years!
Fri Dec 18 15:03:25 +0000 2020🔗
Fri Dec 18 13:34:34 +0000 2020@Philipp_E_Geyer @labs_mann @MannPorsdam @PeterTreit Congrats on your paper & I hope that it helps bring broader recognition of this issue.
Fri Dec 18 13:28:58 +0000 2020rs2729835 (LACTB:p.R469K, vaf=71%, Δm = -14.017) is homozygous in HEK-293 derived cell lines. Heterozygous in MCF-10A & A-549 cells. #ᐯᐸᐱ
Fri Dec 18 13:28:36 +0000 2020@Philipp_E_Geyer @labs_mann @MannPorsdam @PeterTreit There is no question that SAAVs can be detected without any special effort using appropriate proteomics data and that it is possible to identify individuals. However, the NIH has very consistently pushed back against this fact.
Fri Dec 18 13:00:30 +0000 2020-1 °C, precip 0 mm, 1005 mb↓, RH 82%, cloudy (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Fri Dec 18 12:55:34 +0000 2020CCL4:p θ(max) = 70. aka MIP-1-beta, Act-2, AT744.1, LAG1, SCYA4. Shares significant sequence overlap with CCL4L1:p & CCL4L2:p. These 3 proteins can only be distinguished using data from HLA class II experiments.
Fri Dec 18 12:55:33 +0000 2020>CCL4:p, C-C motif chemokine ligand 4 (Homo sapiens) 🔗 Very small protein; CTMs: none; PTMs: none; SAAVs: S80T (18%); mature form: 24,26,27,28-92 [364×, 2.4 kTa] #ᗕᕱᗒ 🔗
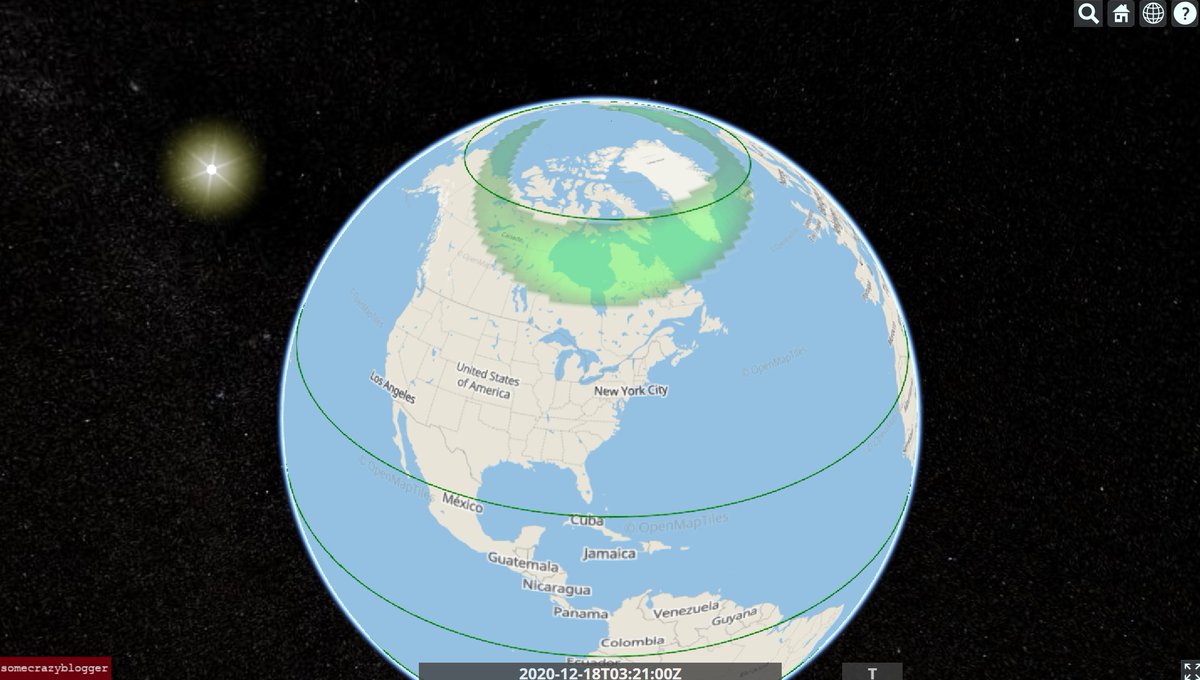
Fri Dec 18 03:50:51 +0000 2020Nothing much here, but the aurora should be nice in Churchill tonight 🔗
Thu Dec 17 18:36:18 +0000 2020@VATVSLPR @nesvilab @dtabb73 I use a special system (based on an fast search algorithm) that was designed for the purpose, which also detects which PTMs to include in a search. It save me a lot of time, esp. when dealing with heterogenous (& often dodgy) public data sets.
Thu Dec 17 18:20:56 +0000 2020@nesvilab @dtabb73 I personally use as much automation as possible to set parameters. But, there is a good argument to be made that having student analysts manually entering values is a useful training exercise, as it should motivate them to think a bit more deeply about what they are doing.
Thu Dec 17 16:54:17 +0000 2020rs2273526 (SEC23B:H489Q, vaf=12%, Δm = -9.000) is homozygous in HEK-293 derived cell lines as well as PANC-1 cells. Heterozygous in HEP-3B & SH-SY5Y cells. #ᐯᐸᐱ
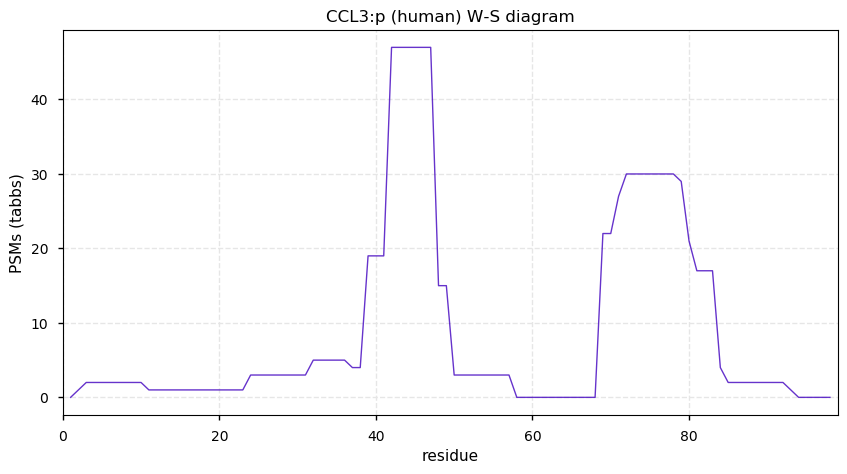
Thu Dec 17 13:16:32 +0000 2020The titles on the graphs say CCL3, but they are really CCL13.
Thu Dec 17 13:13:03 +0000 2020CCL13:p θ(max) = 35. aka MCP-4, NCC-1, SCYL1, CKb10, MGC17134, SCYA13. Protein observations are from HLA class II experiments and a set of experiments designed to enrich small proteins.
Thu Dec 17 13:13:03 +0000 2020>CCL13:p, C-C motif chemokine ligand 13 (Homo sapiens) 🔗 Very small protein; CTMs: none; PTMs: none; SAAVs: none; mature form: 17?-98 [56× , 0.088 kTa] #ᗕᕱᗒ 🔗
Thu Dec 17 13:03:20 +0000 2020-1 °C, precip 0 mm, 1012 mb↑, RH 89%, cloudy (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Thu Dec 17 12:38:44 +0000 2020@mjmaccoss Thanks Mike. I was hoping to find someone who had already done it for me 🙄. I guess the emphasis on disease causing variants has made neutral & +ve variants of lesser value, academically speaking, even though they are much more abundant.
Wed Dec 16 14:26:43 +0000 2020Correction: the reference SNP number is rs2289247 (missed the 'r' in my early morning copy-and-paste).
Wed Dec 16 14:03:59 +0000 2020Since the variant allele (M) is frequently oxidized, the practical Δm may be (31.972+15.995)=47.967.
Wed Dec 16 14:03:59 +0000 2020s2289247 (GNL3:p.V367M, vaf=40%, Δm = +31.972) is homozygous in HEK-293 derived cell lines as well as HeLa cells. Heterozygous in Hep-G2 & JURKAT cells. #ᐯᐸᐱ
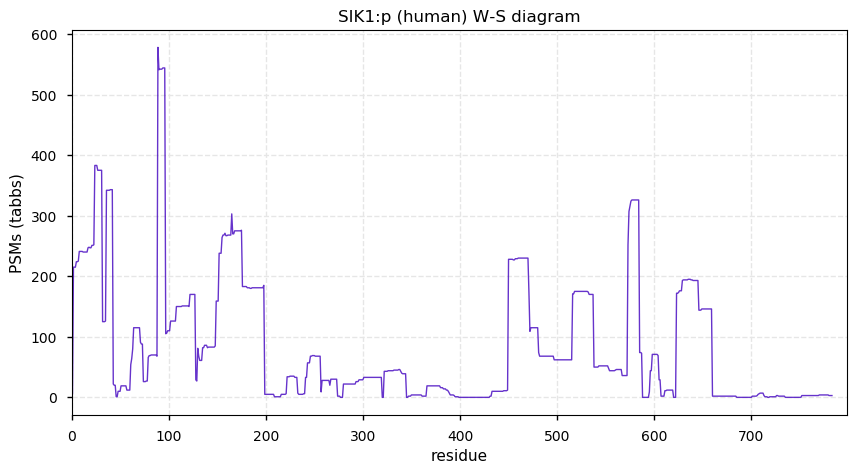
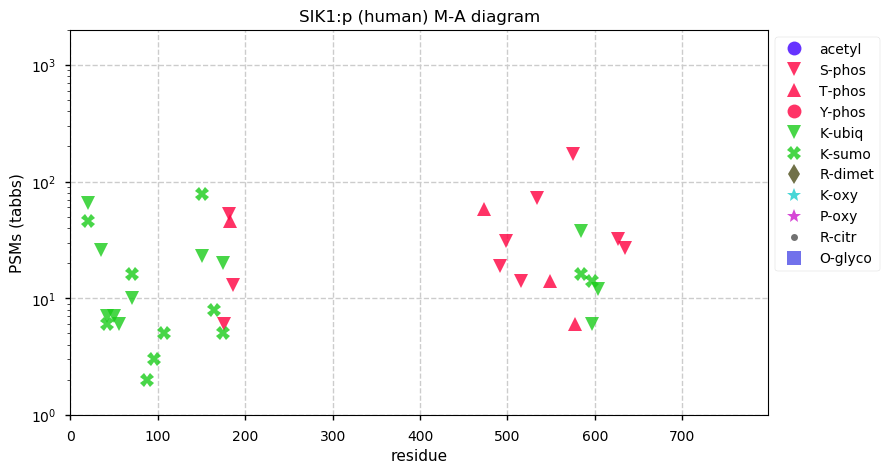
Wed Dec 16 13:44:44 +0000 2020SIK1:p the top 5 identifications are from protein-protein interactions studies, where SIK1:p was the bait protein.
Wed Dec 16 13:31:58 +0000 2020-6 °C, precip 0 mm, 1008 mb ↓, cloudy (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Wed Dec 16 13:22:50 +0000 2020@dtabb73 I was rather hoping for something in mammals, but I'd be interested in prokaryotes. Viruses, not so much.
Wed Dec 16 13:20:05 +0000 2020SIK1:p θ(max) = 70. aka msk, SNF1LK. Protein observations are mainly in studies enriching for SUMOyl, ubiquitinyl or phosphoryl modifications.
Wed Dec 16 13:20:05 +0000 2020>SIK1:p, salt inducible kinase 1 (Homo sapiens) 🔗 Midsized protein; CTMs: none; PTMs: 11× K+ubiquitinyl, 11× K+SUMOyl, 14× S,T+phosphoryl; SAAVs: G15S (17%), A615V (17%); mature form: 2-783 [1,766× , 4.5 kTa] #ᗕᕱᗒ 🔗
Wed Dec 16 01:21:27 +0000 2020It a bit further south than yesterday, but still no where near far enough to see here 🔗
Tue Dec 15 20:44:47 +0000 2020@bkives Except for the annual flu vaccine.
Tue Dec 15 20:36:02 +0000 2020Or at least improvement-of-function?
Tue Dec 15 20:34:48 +0000 2020Are there any good databases and/or reviews of gain-of-function nsSNVs?
Tue Dec 15 16:40:32 +0000 2020@jwoodgett GoogleBot is the only one of the allowed bots that doesn't have the directive
Disallow: */pdf*
in its list, so it is the only one allowed to rummage through the directories that starting with '/pdf'.
Tue Dec 15 16:17:41 +0000 2020@jwoodgett The 🔗 Science Direct robots directives are pretty clear that they only allow 10 search bots at the moment (3 of which are Google branded).
Tue Dec 15 15:38:08 +0000 2020@jwoodgett There is nothing that prevents a bot from ignoring these rules: if one does plow through anyway, you have to hunt down their IP addresses from the activity logs and block them manually.
Tue Dec 15 15:33:21 +0000 2020@jwoodgett You can get the information about blocking & allowing via the "robots.txt" file for any particular site. Here are the rules for the main NCBI site: 🔗
Tue Dec 15 15:32:17 +0000 2020@jwoodgett I could just let one or the other of the search engines have access. All of the scientific information providers (EBI, NCBI, etc) select which search bots to allow: this is not unusual or hidden. It is just not very widely known.
Tue Dec 15 15:17:32 +0000 2020@jwoodgett By default I block all search engine bots from anything I make available.
Tue Dec 15 15:15:38 +0000 2020@jwoodgett It is up to whoever runs the servers. Search engine bots can create very real problems for site admins, so they are fussy about which ones (if any) they allow to index their sites.
Tue Dec 15 13:40:37 +0000 2020There is protein-level evidence that the variant allele can serve as an alternate translation initiation site.
Tue Dec 15 13:40:37 +0000 2020rs10853751 (EXOSC5:p.T5M, vaf=61%, Δm = 29.993) is homozygous in HEK-293 derived cell lines as well as HeLa, HEp-2, HaCaT & SW-480 cells. Heterozygous in MDA-MB-468 & MCF-10A cells. #ᐯᐸᐱ
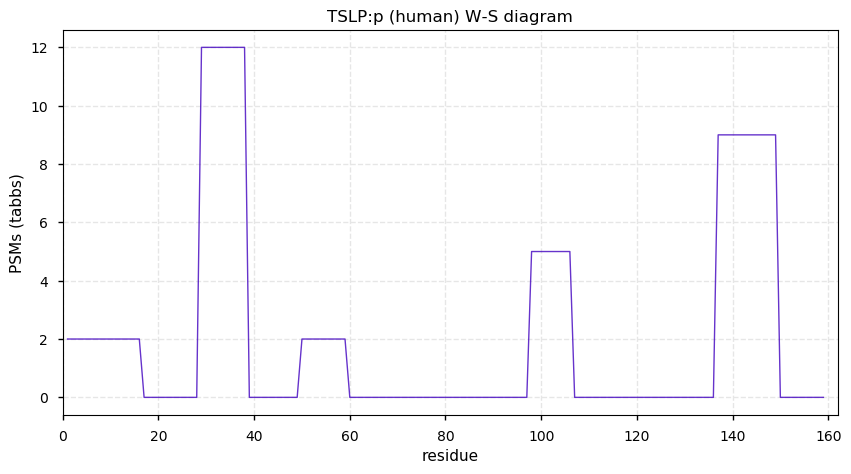
Tue Dec 15 13:04:56 +0000 2020TSLP:p θ(max) = 21. The only observations of this protein have been obtained from trachea tissue samples & HLA class I experiments.
Tue Dec 15 13:04:56 +0000 2020>TSLP:p, thymic stromal lymphopoietin (Homo sapiens) 🔗 Small protein; CTMs: none; PTMs: none; SAAVs: none; mature form: 29-159 [20× , 0.030 kTa] [24× , 0.035 kTa] #ᗕᕱᗒ 🔗
Tue Dec 15 12:57:28 +0000 2020-15 °C, 0 mm 24h precip, cloudy (#CoCoRaHS CAN-MB-361 49.8978°, -97.1358°)
Tue Dec 15 00:23:17 +0000 2020If you want to check for yourself anytime, 🔗 is a mashup of NOAA's data (🔗) and the Cesium 3D mapping platform.
Tue Dec 15 00:19:32 +0000 2020Looks like no aurora here tonight 🔗
Mon Dec 14 16:55:53 +0000 2020@pwilmarth @educhicano @ProteomicsNews I am a sucker for positive reinforcement.
Mon Dec 14 16:55:04 +0000 2020@pwilmarth @educhicano @ProteomicsNews Shifting from "contaminants" to "predictions" is just semantics, however, for me it changes my attitude towards these uninvited guests. Rather than being disappointed by observing cRAP, I am pleasantly surprised to see which of my predictions came true.
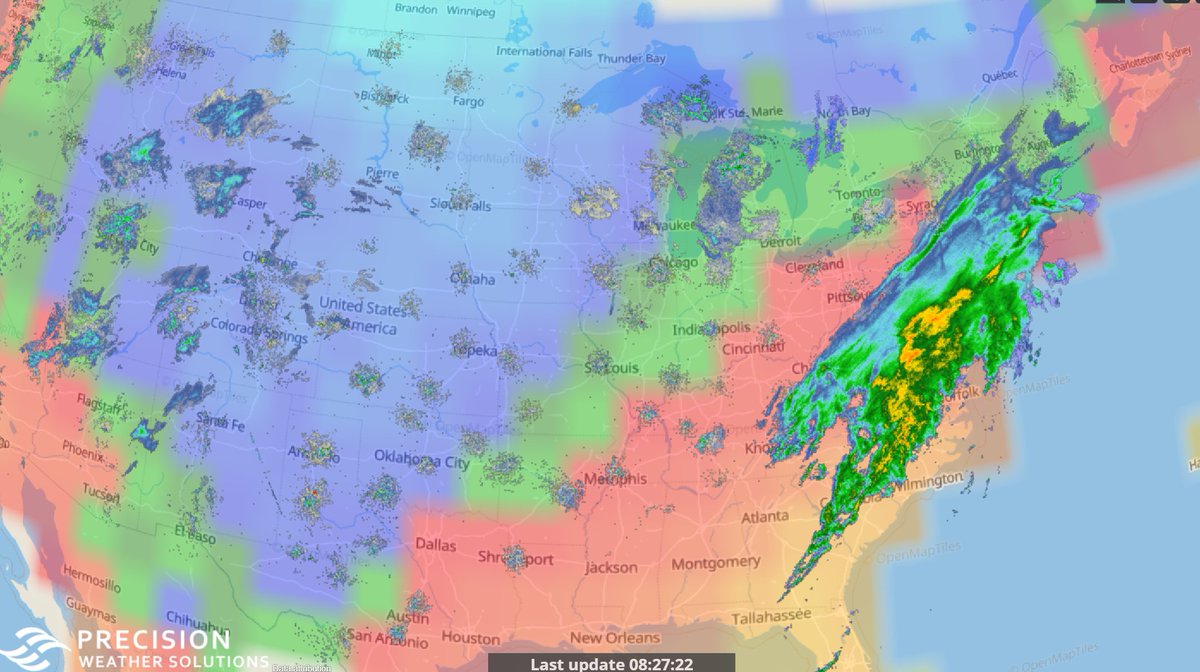
Mon Dec 14 14:35:27 +0000 2020Cold across the west (with lots of ground clutter) & stormy along the east coast this morning 🔗
Mon Dec 14 13:42:06 +0000 2020Note: Uniprot lists the variant allele as the reference.
Mon Dec 14 13:40:28 +0000 2020rs169547 (BRCA2:p.V2466A, vaf=98%, Δm = -28.031) is homozygous in HEK-293 derived cell lines as well as JURKAT & most other cell lines. #ᐯᐸᐱ
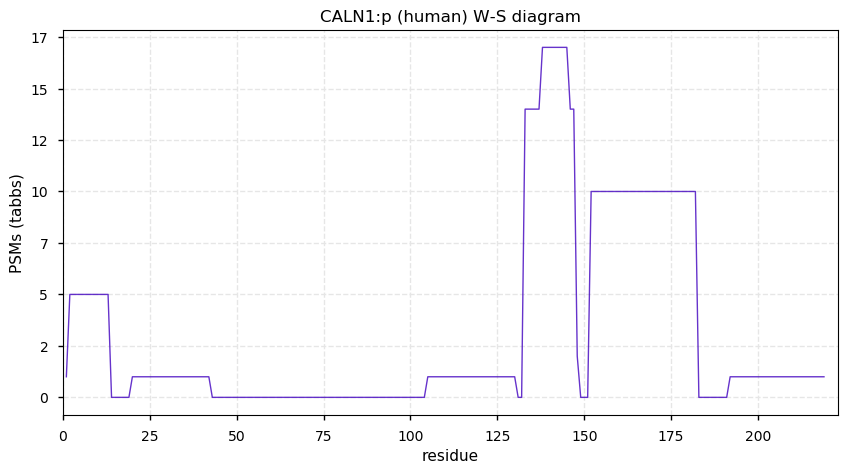
Mon Dec 14 13:30:35 +0000 2020CALN1:p θ(max) = 14. aka CABP8. The best observations of this protein have been obtained from HLA type II experiments.
Mon Dec 14 13:30:35 +0000 2020>CALN1:p, calneuron 1 (Homo sapiens) 🔗 Small protein; CTMs: none; PTMs: none; SAAVs: none; mature form: 2-219 [24× , 0.035 kTa] #ᗕᕱᗒ 🔗
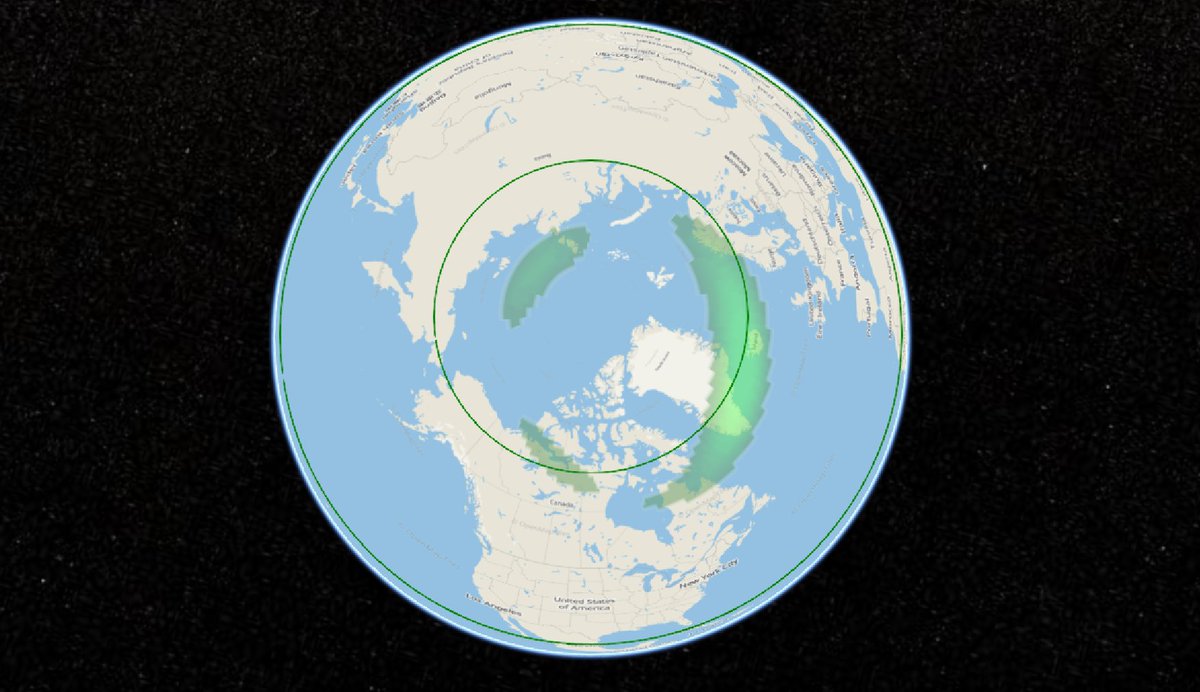
Mon Dec 14 02:09:33 +0000 2020Not much chance of seeing an aurora this evening ... 🔗
Sun Dec 13 18:58:17 +0000 2020@pwilmarth @educhicano @ProteomicsNews For most human-based samples, I add in ~ 2400 viral proteins & sets of bacterial proteins tailored to the source of the sample, e.g., bacteria commonly present in saliva, urine, BALF or stomach.
Sun Dec 13 18:23:38 +0000 2020@pwilmarth @educhicano @ProteomicsNews I also don't think of things as "contaminants" any more: it is more a matter of what may be in the sample. For example, I exclude Y-chromosome proteins from samples generated from female cells or tissues, e.g., HeLa, HEK-293 or ovarian tissue.
Sun Dec 13 14:17:19 +0000 2020I think I'll continue with the "dark proteins & where to find them" theme until New Years.
Sun Dec 13 13:55:41 +0000 2020🔗 is a way to contribute your local daily precipitation information for meteorological use. I (CAN-MB-361: 49.8978°,-97.1358°) have been doing it for years.
Sun Dec 13 13:22:21 +0000 2020rs2275689 (HEATR1:p.N1694S, vaf=68%, Δm = -27.011) is heterozygous in HEK-293 derived cell lines & JURKAT cells. It is homozygous in CACO-2 & HeLa cells. #ᐯᐸᐱ
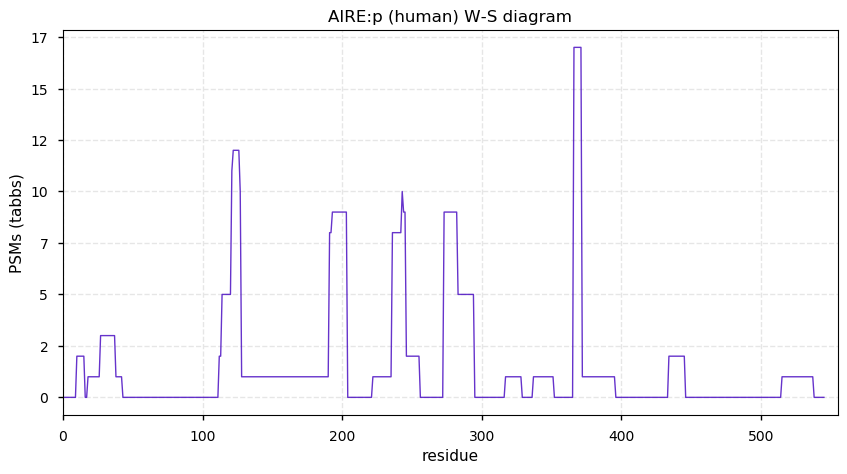
Sun Dec 13 13:03:46 +0000 2020AIRE:p θ(max) = 6.4. aka PGA1, APS1, APECED. The best observations of this protein have been obtained from HLA type I experiments. AIRE:p translation is largely restricted to the thymus, a tissue with no available public proteomics data.
Sun Dec 13 13:03:46 +0000 2020>AIRE:p, autoimmune regulator (Homo sapiens) 🔗 Midsized intracellular protein; CTMs: none; PTMs: none; SAAVs: none; mature form: 2?-545 [115× , 0.075 kTa] #ᗕᕱᗒ 🔗
Sat Dec 12 17:01:15 +0000 2020@pwilmarth @educhicano @ProteomicsNews No. The way I do it now would be very hard to implement using FASTA files.
Sat Dec 12 15:42:00 +0000 2020@educhicano @pwilmarth @ProteomicsNews I don't use that old list very often: it was a product of old-school proteomics methods. I use a set of sample/prep specific lists now.
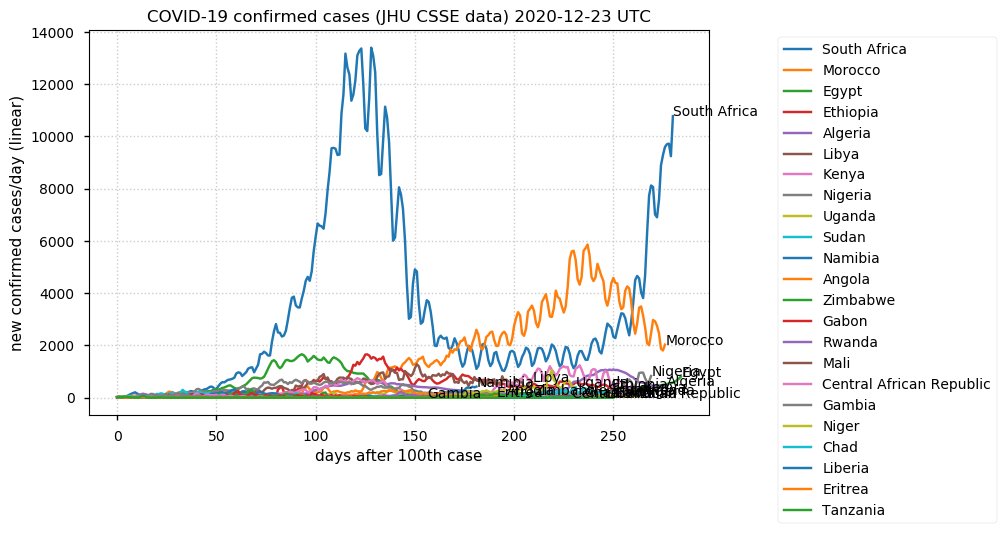
Sat Dec 12 14:01:22 +0000 2020Most of the Canadian provinces seem to be scrambling to catch up to Quebec. Numbers in the legend are provincial populations (in millions). 🔗
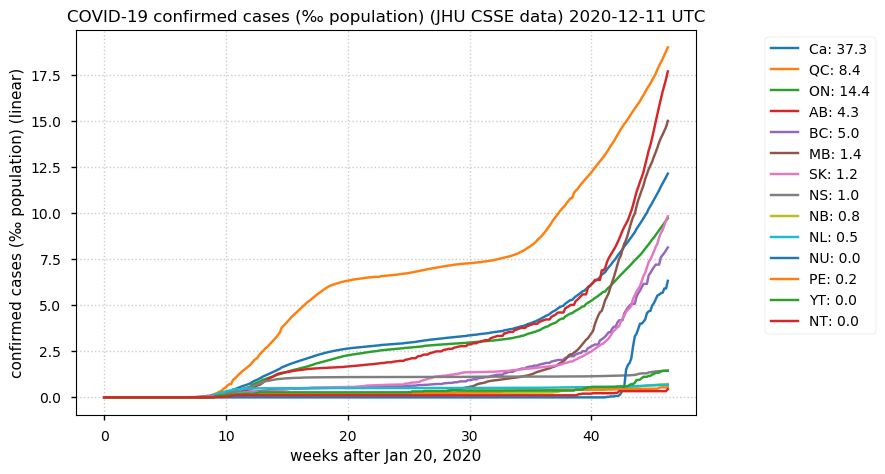
Sat Dec 12 13:44:11 +0000 2020rs7986131 (LMO7:p.M1113T, vaf=69%, Δm = -29.993) is homozygous in HEK-293 derived cell lines as well as RKO & HeLa cells. Heterozygous in CACO-2 cells. #ᐯᐸᐱ
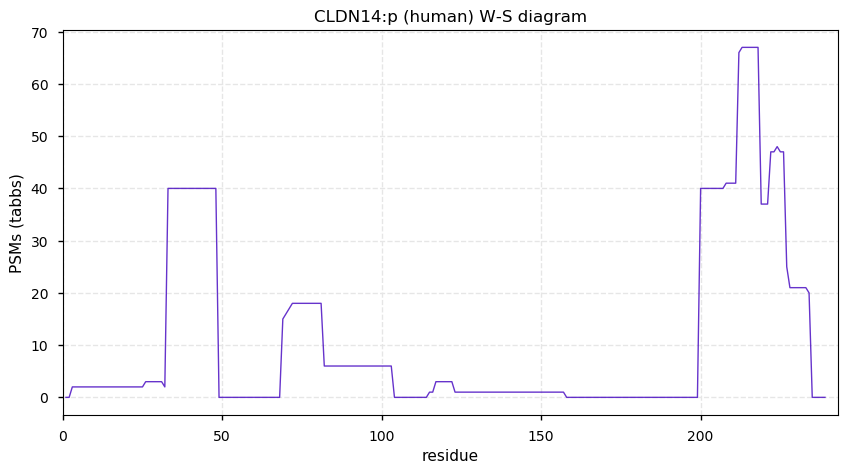
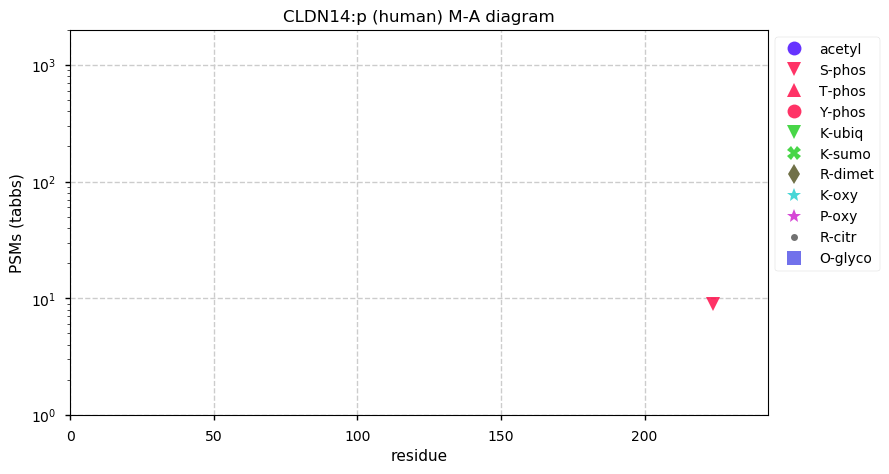
Sat Dec 12 13:21:42 +0000 2020CLDN14:p θ(max) = 25. aka DFNB29. The protein can be observed using conventional proteomics in urinary exosomes, but the best evidence has been obtained from HLA type II experiments. Four transmembrane domains: (8-28), (82-102), (117-138) & (163-183).
Sat Dec 12 13:21:41 +0000 2020>CLDN14:p, claudin 14 (Homo sapiens) 🔗 Small protein; CTMs: none; PTMs: S224+phosphoryl; SAAVs: none; mature form: 2?-239 [120× , 0.16 kTa] #ᗕᕱᗒ 🔗
Sat Dec 12 03:10:58 +0000 2020Looks like there will be a nice bright aurora tonight 😀 🔗
Fri Dec 11 17:14:20 +0000 2020@lgatt0 @theoneamit @slavov_n @dtabb73 @astacus I found it odd, because being Supplementary files I'm pretty sure the copyright was transferred to the Journal (Nat. Biotech.), which makes ownership of the code a bit of a mess.
Fri Dec 11 17:11:31 +0000 2020@lgatt0 @theoneamit @slavov_n @dtabb73 @astacus No. Just a copyright declaration giving rights to Max Planck (also struck me as odd at the time).
Fri Dec 11 16:31:45 +0000 2020@lgatt0 @theoneamit @slavov_n @dtabb73 @astacus Not a lot of error checking, either. It has been a while since I looked at it, but I remember at the time being surprised at the lack of exceptions.
Fri Dec 11 16:29:37 +0000 2020@lgatt0 @theoneamit @slavov_n @dtabb73 @astacus The code is in the Supplementary files. It has almost no code-level documentation, so it isn't very useful, other than to give you a sense of how the programmer thinks about the programming. I always thought it looked as though it would execute rather slowly.
Fri Dec 11 15:40:42 +0000 2020@TrostLab @TheCrick Recruiting people to the UK must be a bit more challenging these days.
Fri Dec 11 15:13:47 +0000 2020@KentsisResearch @lkpino @pierrepo @ypriverol Maybe a protein-oriented genomics course would be useful, too. 🦄
Fri Dec 11 15:07:39 +0000 2020And the protein-level evidence: HEK-293 🔗 v. HeLa 🔗 (red 🛑 highlighted residue means SAAV detected)
Fri Dec 11 14:04:04 +0000 2020rs25655 (CAPN2:p.D22E, vaf=99%, Δm = 14.016) is homozygous in HEK-293 derived cell lines as well as most other cell lines. Oddly, heterozygous in HeLa cells. #ᐯᐸᐱ
Fri Dec 11 13:54:10 +0000 2020@KentsisResearch @lkpino @pierrepo @ypriverol But this sort of training is hard to come by in while obtaining any standard, siloed university postgraduate degree.
Fri Dec 11 13:52:28 +0000 2020@KentsisResearch @lkpino @pierrepo @ypriverol It wouldn't hurt if they had designed and built some instrumentation, including computers.
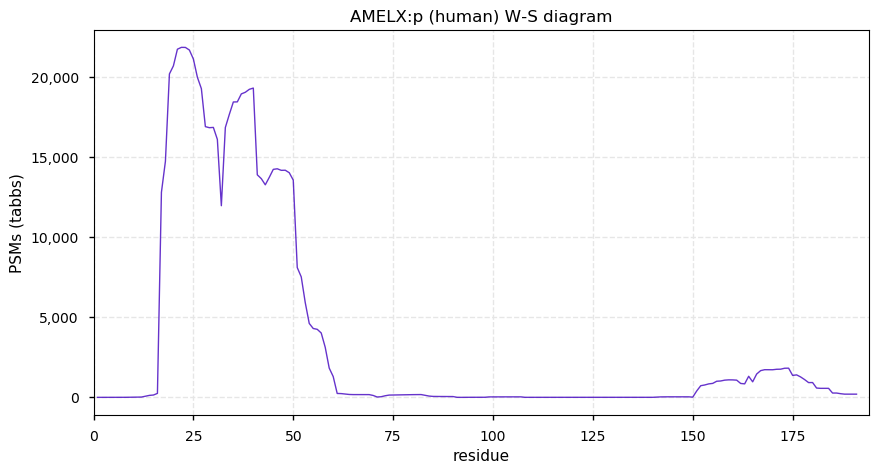
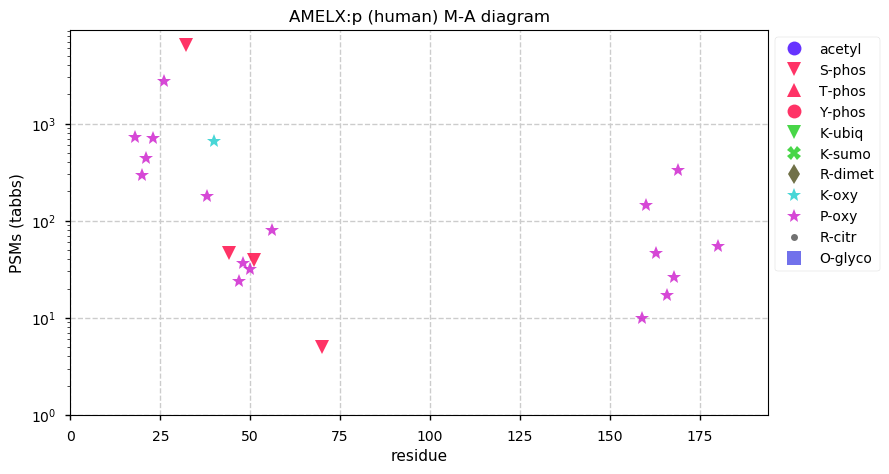
Fri Dec 11 13:33:12 +0000 2020AMELX:p θ(max) = 72. aka AMG, AIH1. The protein is easily observed using conventional proteomics, but it is only present in tooth enamel and prone to significant endogenous cleavage: it is not detectable in any other tissues or cell line.
Fri Dec 11 13:33:12 +0000 2020>AMELX:p, amelogenin X-linked (Homo sapiens) 🔗 Small extracellular protein; CTMs: none; PTMs: S32,S44,S51+phosphoryl, 18× P+oxidation (hydroxyproline); SAAVs: none; mature form: 17,18-191 [91× , 47 kTa] #ᗕᕱᗒ 🔗
Thu Dec 10 19:40:24 +0000 2020@CameronTFlower If you want an alternative, try PXD019909. Lots of challenges, but they are due to cell-specific protein biochemistry rather than experimental artifacts.
Thu Dec 10 18:50:47 +0000 2020@CameronTFlower On the N-terminal and K-ε amines of any of the tryptic peptides. Because of the way they did the expts, both reagents remained reactive during the trypsin cleavage.
Thu Dec 10 18:27:07 +0000 2020@CameronTFlower Its main failings are associated with amine derivatization artifacts caused by both IAA and urea, responsible for 10-30% of PSMs. Naïvely reanalyzing this data without compensating for this problem can lead students down a bad path.
Thu Dec 10 16:34:36 +0000 2020It has become the "bad penny" of proteomics data analysis: it just keeps on showing up.
Thu Dec 10 16:28:44 +0000 2020Why do people still use PXD000561 as an exemplar data set in studies? It has a lot of technical problems and unless you really want to show that you can identify and cope with those problems, it isn't a good choice for general purpose use.
Thu Dec 10 15:41:46 +0000 2020@ypriverol @pwilmarth It just seemed to me there was no obvious reason to allow shortened file names in the specification. The paper does list some examples with .mzXML in them, but many of them leave the extension out.
Thu Dec 10 15:35:46 +0000 2020@lkpino It is a recurring theme. The best way to get more PSMs out of a given set of MS/MS spectra is to come up with some rationale for accepting evidence based on fewer matched fragment ions. All of the schemes, so far, fall apart when you allow for a few PTMs or non-tryptic cleavage.
Thu Dec 10 14:30:19 +0000 2020@pwilmarth I thought that leaving out the file extension was an odd choice: why not distinguish between .raw, .mzml, & .mgf version of the file, which are often all present in the existing repositories.
Thu Dec 10 14:20:39 +0000 2020To make it a bit less confusing, I will use the abbreviation "vaf" to indicate that the frequency always corresponds to the presence of the variant in the population.
Thu Dec 10 14:17:09 +0000 2020rs45491898 (TCOF1:p.G1355A, vaf=1%, Δm = 14.016) is heterozygous in HEK-293 derived cell lines. #ᐯᐸᐱ
Thu Dec 10 14:13:07 +0000 2020@slashdot Note to writers: be sure to include this in the first chapter of your next dystopian novel.
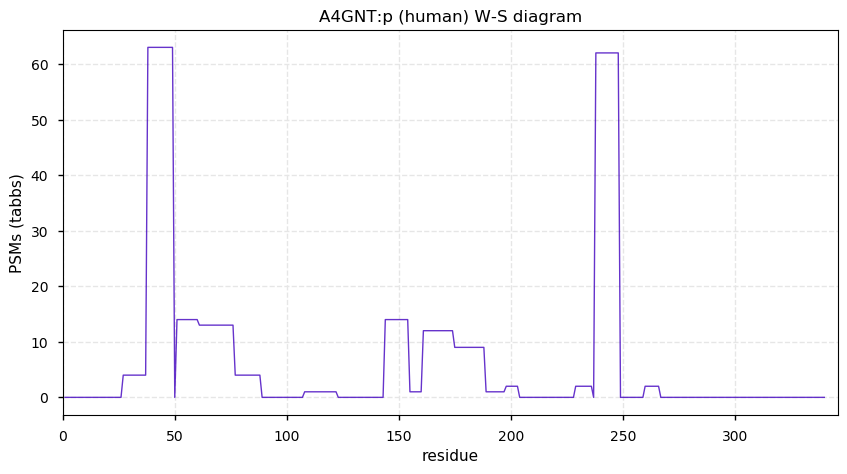
Thu Dec 10 13:47:22 +0000 2020A4GNT:p θ(max) = 54. aka alpha4GnT. The protein is easily observed using conventional proteomics, but it is only present in stomach tissue: it is not detectable in all other tissues or any cell line.
Thu Dec 10 13:47:21 +0000 2020>A4GNT:p, alpha-1,4-N-acetylglucosaminyltransferase (Homo sapiens) 🔗 Small enzyme; CTMs: none; PTMs: none; SAAVs: none; mature form: 1-340 [87×, 0.2 kTa] #ᗕᕱᗒ 🔗
Wed Dec 09 16:30:38 +0000 2020The article quoted does seem to make some good points, at least to a non-expert (i.e., me). 🔗
Wed Dec 09 13:27:06 +0000 2020I am using this alternate formulation because proteomics―with its Highlander approach to sequences―gives philosophical primacy to the reference sequence.
Wed Dec 09 13:21:42 +0000 2020Note: I am using the reference allele for computing the MAF, rather than the inferred ancestral allele. The more common approach in population genetics is to use the ancestral allele V, making the reference allele M the variant, making the MAF 6%.
Wed Dec 09 13:01:10 +0000 2020rs6659553 (POMGNT1:p.M623V, maf=94%, Δm = -31.972) is homozygous in HEK-293 derived cell lines, as well as HeLa, Hep-G2, MCF-10A, HCT-116, U-87MG, SW-480, NB-4 & A-549 cells. #ᐯᐸᐱ
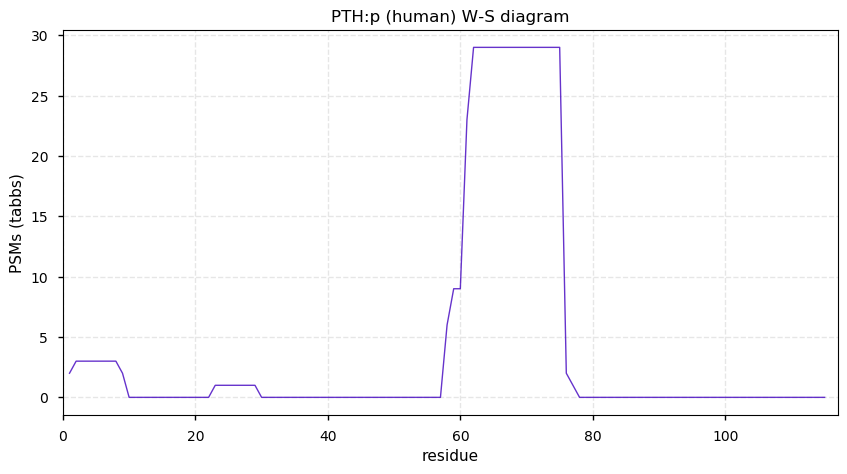
Wed Dec 09 12:55:47 +0000 2020PTH:p θ(max) = 26. aka PTH1. The protein has been convincingly observed using data produced by HLA class II experiments. Normal concentration range in blood is 8-50 pg/ml (ALB:p is 30-50 mg/ml).
Wed Dec 09 12:55:47 +0000 2020>PTH:p, parathyroid hormone (Homo sapiens) 🔗 Small peptide hormone; CTMs: none; PTMs: none; SAAVs: none; mature form: 32-115 [29×, 0.033 kTa] #ᗕᕱᗒ 🔗
Tue Dec 08 19:57:44 +0000 2020@Sci_j_my @pwilmarth @MagnusPalmblad I never left that blessed land.
Tue Dec 08 19:36:15 +0000 2020Downloading the BioPlex 3.0 raw data set sure takes a minute or two ...
Tue Dec 08 14:16:07 +0000 2020rs7313 (CPVL:p.A435V, maf=30%, Δm = +28.031) is heterozygous in HEK-293 derived cell lines. Homozygous in MCF-10A cells. #ᐯᐸᐱ
Tue Dec 08 13:05:08 +0000 2020My theme for the next few days is "dark proteins and where to find them".
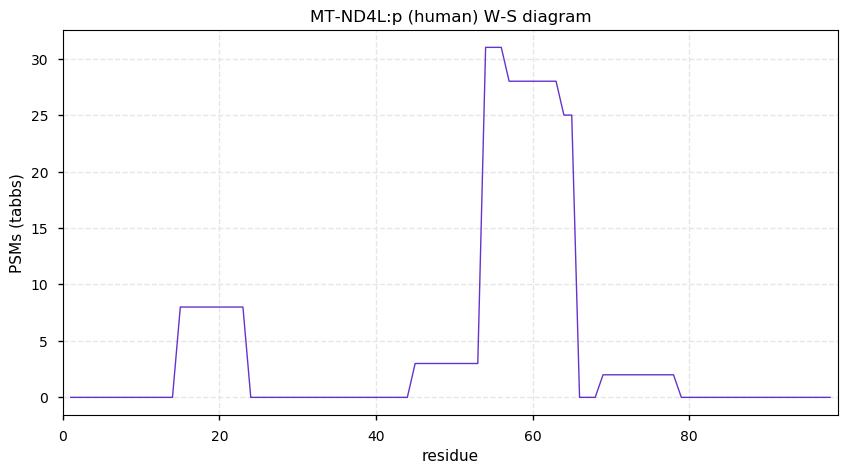
Tue Dec 08 12:50:23 +0000 2020MT-ND4L:p is 1 of the 13 proteins encoded on the mitochondrial chromosome and translated by MT-ribosomes. All other mitochondrial proteins are translated in the cytosol and imported into the organelle.
Tue Dec 08 12:50:22 +0000 2020MT-ND4L:p θ(max) = 7. Very hydrophobic protein with 3 membrane spanning domains: (1-21), (29-49) & (58-68). Sequence has no K or R residues & therefore no tryptic peptides. The protein can only observed using data produced by HLA class I experiments.
Tue Dec 08 12:50:22 +0000 2020>MT-ND4L:p, mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4L (Homo sapiens) 🔗 Very small subunit; CTMs: none; PTMs: none; SAAVs: none; mature form: 1-98 [35×, 0.041 kTa] #ᗕᕱᗒ 🔗
Mon Dec 07 22:28:17 +0000 2020@pwilmarth @Sci_j_my It is left as an exercise for the reader to get it down to one line in PERL.
Mon Dec 07 22:24:48 +0000 2020@pwilmarth @Sci_j_my h = [1, 2, 30, 44, 500, 40, 30, 2, 1]
L = len(h)
(m,i) = max((v,i) for i,v in enumerate(h))
M = (int)(0.5+m/100.0)
j = i
while j >= 0 and h[j] > M:
j -= 1
A = j
j = i
while j < L and h[j] > M:
j += 1
B = j
d = A+1+L-B
bgnd = sum(h[:A+1])/d+sum(h[B:])/d
Mon Dec 07 22:23:23 +0000 2020@pwilmarth @Sci_j_my For this type of histogram, I often use something like this for a quick estimate of the background PSMs per ppm:
Mon Dec 07 20:52:11 +0000 2020@UCDProteomics @Karl_Mechtler Giddy-up! Get that data flowing ...
Mon Dec 07 18:43:39 +0000 2020@mindymon @girlziplocked Teamsters
Mon Dec 07 17:21:07 +0000 2020@MagnusPalmblad Looking at beer proteins with mass spec is a recurring theme, even before "proteomics" existed.
Mon Dec 07 15:37:25 +0000 2020Does anybody know of a good site that I can use to provide links to genomic HEK-293 nsSNVs by rs number?
Mon Dec 07 15:32:25 +0000 2020I'm not in love with using the terms homozygous vs. heterozygous to refer to these SAAVs, but it is all I've got.
Mon Dec 07 13:25:12 +0000 2020Tweets with the #ᐯᐸᐱ hashtag will highlight SAAVs easily detectable in HEK-293 lineage cells using mass spec based proteomics.
Mon Dec 07 13:20:42 +0000 2020rs2280084 (NUP210:p.R786L, maf=43%) is heterozygous in HEK-293 derived cell lines. The SAAV removes a tryptic cleavage site #ᐯᐸᐱ
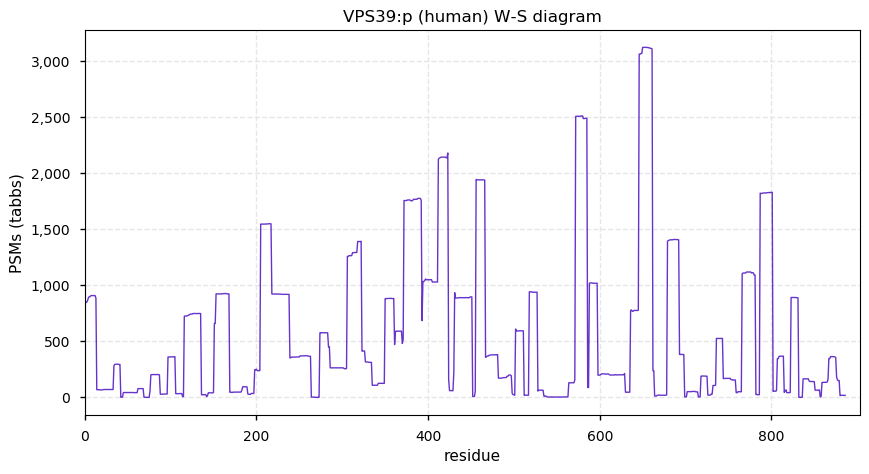
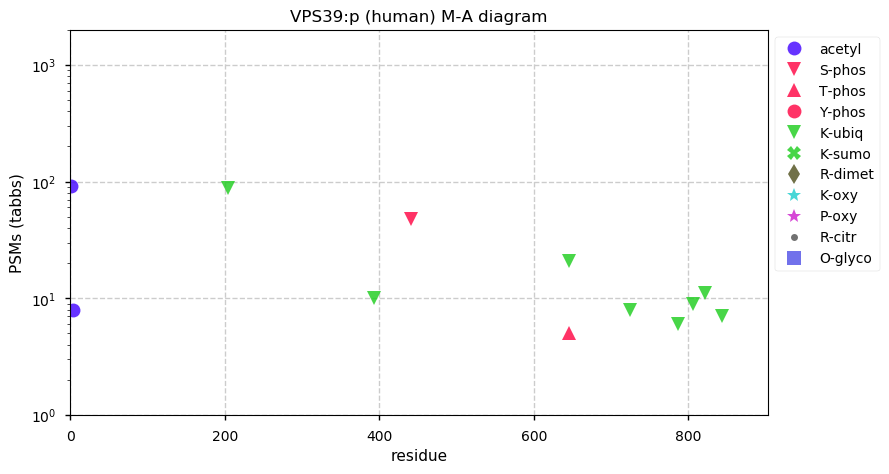
Mon Dec 07 13:03:19 +0000 2020VPS39:p θ(max) = 69. Found in HLA class I (but not class II) peptide experiments. Observed in cell lines & tissues, but rarely in fluids.
Mon Dec 07 13:03:19 +0000 2020>VPS39:p, HOPS complex subunit (Homo sapiens) 🔗 Midsized subunit; CTMs: M1+acetyl; PTMs: 8× K+ubiqutinyl, S441+phosphoryl; SAAVs: none; mature form: 1-886 [9,759×, 37 kTa] #ᗕᕱᗒ 🔗
Sun Dec 06 19:09:04 +0000 2020@cdsouthan I meant to say: "they have to take a serious look at MHC peptides". I didn't mean to imply that there were any plans on the part of the HPP to look at MHC peptides. I meant they need to start looking at MHC peptides of the reference proteome asap.
Sun Dec 06 16:28:48 +0000 2020@cdsouthan I have no question about the observation of smORF-derived peptides: they are reproducible true positives. I doubt whether smORFs produce proteins, though, in the sense that they are pieces of functional molecular devices. I suspect the little guys are doing something else.
Sun Dec 06 14:49:45 +0000 2020@cdsouthan Almost certainly some do not (I'd guess ~ 5%). For the rest, they are going to take a serious look at MHC peptides. I would have sworn that MT-ND4L:p would never be seen by MS-based proteomics, but it does show up quite nicely in class I experiments.
Sun Dec 06 14:24:20 +0000 2020I haven't been following this project for a few years, but they sound as if they are kind of stuck at the moment 🔗
Sun Dec 06 14:08:47 +0000 2020rs45491898 (TCOF1:p.G1355A, maf=1%) is heterozygous in HEK-293 derived cell lines. #ᐯᐸᐱ
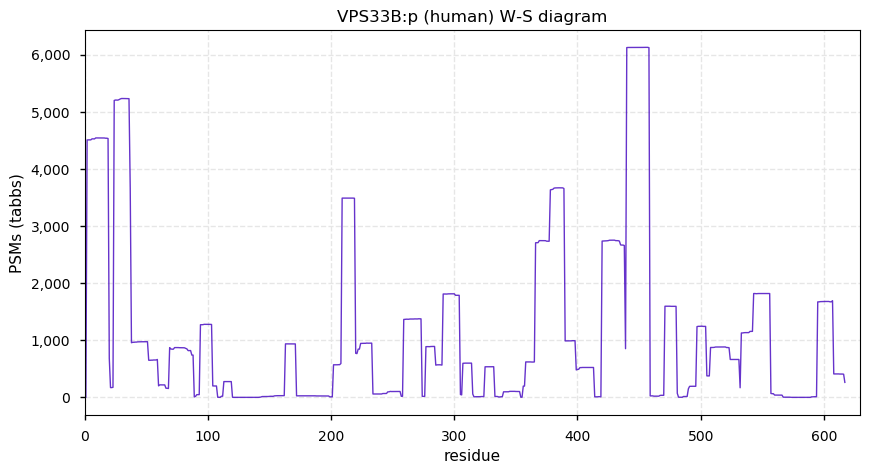
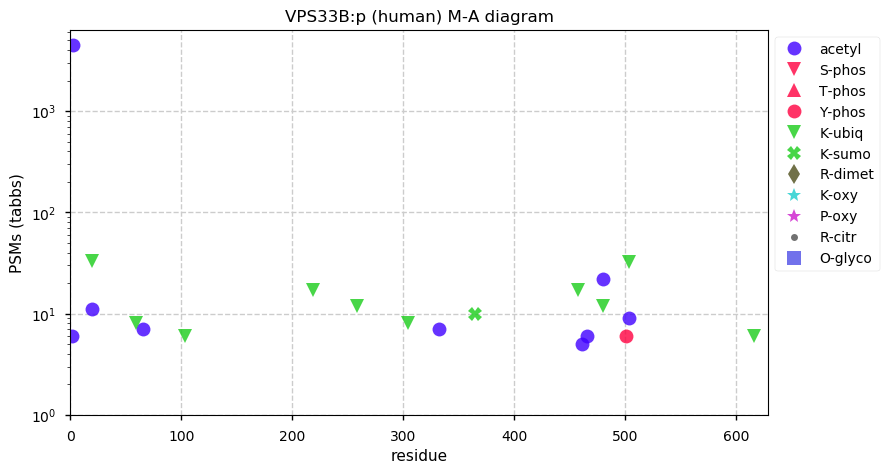
Sun Dec 06 13:36:52 +0000 2020VPS33B:p θ(max) = 56. Found rarely in HLA class I & class II peptide experiments. Abundant in cell lines & tissues, but not fluids. VPS33B PSMs do not overlap with VPS33A. VPS33A observed 13,185× & VSP33B observed 13,015×, but they don't physically interact.
Sun Dec 06 13:36:51 +0000 2020>VPS33B:p, late endosome & lysosome associated (Homo sapiens) 🔗 CTMs: A2+acetyl; PTMs: 7× K+ubiqutinyl, 4× K+acetyl; aPTMs: K19,K480,K504+acetyl/ubiquitinyl; SAAVs: G514S (23%); mature form: 2-617 [13,015×, 52 kTa] #ᗕᕱᗒ 🔗
Sat Dec 05 18:11:44 +0000 2020@Sci_j_my No. Unless the only other choice is Soylent Green.
Sat Dec 05 15:51:43 +0000 2020@bkives @CBCTheHouse Anyone who thought that Mr. Pallister was "courageous" for his statements regarding COVID-19 should listen to this report. He is a politician whose approach to the disease was to slyly minimize it, which has turned out to be a mistake. Now he is trying to rehabilitate his image.
Sat Dec 05 15:11:57 +0000 2020@MHendr1cks I would go so far as saying that universities are simply negligent when they expect profs to supervise employees with no institutional HR training.
Sat Dec 05 14:32:00 +0000 2020@MHendr1cks The HR and record keeping would be very useful for scientists in academia. Most profs are pretty awful at both (but are very defensive about it, as they all think they are great at both).
Sat Dec 05 14:01:09 +0000 2020@MHendr1cks I can only speak to the pharma business, but they train PhD's a lot. I had about 1 week a month of training: record keeping, communication, legal, business, HR, etc. The HR was particularly valuable.
Sat Dec 05 13:53:55 +0000 2020rs6960 (ENSP00000426514:p.Y548F, maf=18%) is homozygous in HEK-293 derived cell lines. #ᐯᐸᐱ
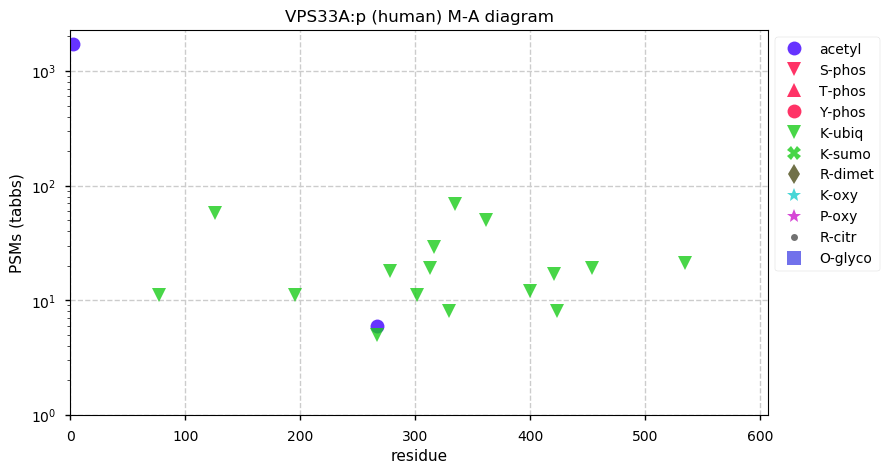
Sat Dec 05 13:07:34 +0000 2020VPS33A:p θ(max) = 65. Found in HLA class I & (more rarely) class II peptide experiments. Abundant in cell lines and tissues, but not fluids. VPS33A PSMs do not overlap with VPS33B (or any other protein).
Sat Dec 05 13:07:34 +0000 2020>VPS33A:p, CORVET/HOPS core subunit (Homo sapiens) 🔗 Midsized subunit; CTMs: A2+acetyl; PTMs: 15× K+ubiqutinyl; aPTMs: K267+acetyl/ubiquitinyl; SAAVs: none; mature form: 2-596 [13,185×, 54 kTa] #ᗕᕱᗒ 🔗
Fri Dec 04 22:42:29 +0000 2020Thanks to everyone who participated in this poll. It is pretty evenly split between the strict and laissez faire labelists. I have to come down on the side of strict: if you can get 5 labels wrong you (& your co-authors) aren't really on top of the material.
Fri Dec 04 15:14:13 +0000 2020For anyone who wants to look back at older "protein du jour" entries, click on the odd looking hashtag #ᗕᕱᗒ & you will get a list of them going back to July; click the "Latest" tab for a date ordered list. This hashtag is also on my profile page.
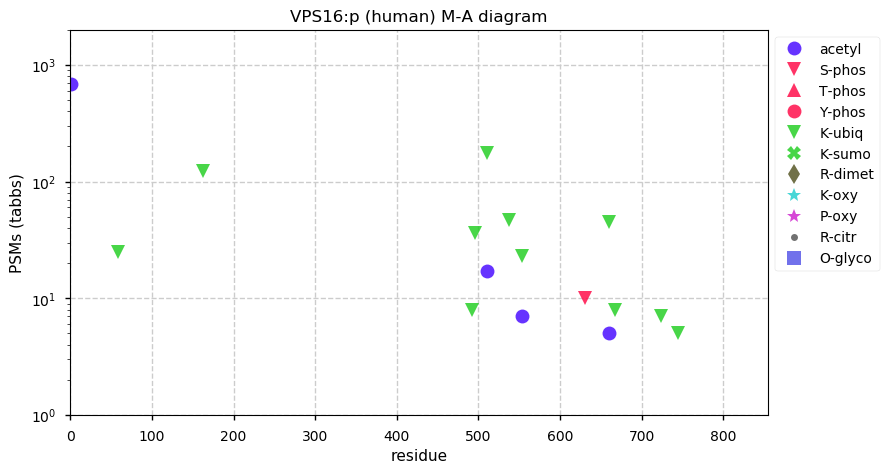
Fri Dec 04 14:15:13 +0000 2020VPS16:p θ(max) = 56. Found in HLA class I & class II peptide experiments. K+ubiquintyl grouped into two domains, (58-163) and (492-745). Abundant in cell lines and tissues, but not fluids.
Fri Dec 04 14:15:12 +0000 2020>VPS16:p, CORVET/HOPS core subunit (Homo sapiens) 🔗 Midsized subunit; CTMs: M1+acetyl; PTMs: 8× K+ubiqutinyl; aPTMs: K510,K544,K660+acetyl/ubiquitinyl; SAAVs: none; mature form: 1-839 [15,308×, 61 kTa]#ᗕᕱᗒ 🔗
Fri Dec 04 01:36:41 +0000 2020@olgavitek That is my concern about the study. I can't determine whether there are mix ups within the A or B groups, which are composed of multiple conditions that are compared with each other.
Fri Dec 04 01:33:38 +0000 2020@cenaptech The test is very definitive & easy to check with another, similarly definitive test.
Thu Dec 03 22:31:59 +0000 2020I'm looking at a fairly large data set (95 runs each from 2 different preparations, A & B). The files are clearly labelled. The results show that 5 of those labelled as part of A are really from B. Does this taint the whole study?
Thu Dec 03 22:03:03 +0000 2020@jimfinnis @cstross @jamesdnicoll Stone knives and bearskins
Thu Dec 03 16:03:22 +0000 2020DIA cognoscenti: does anyone have experience with how well spectrum prediction algorithms do when adding peptide-based modifications like K+GlyGly (ubiquitinyl) or K+QQTGG (SUMOyl)?
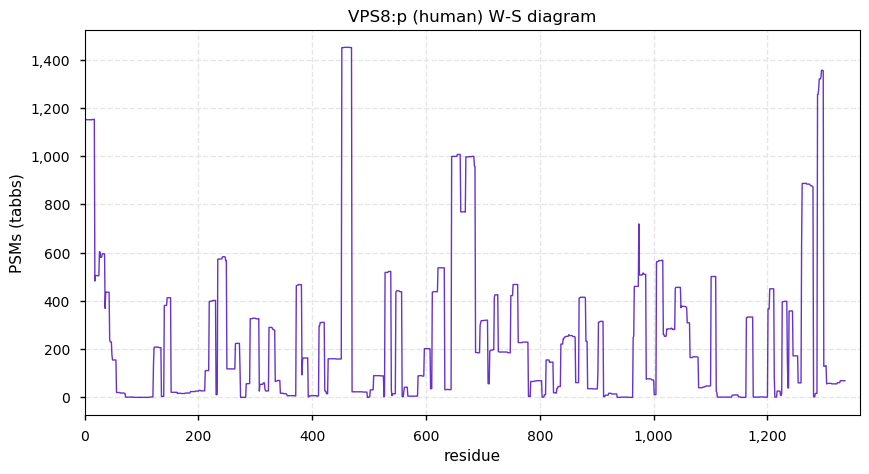
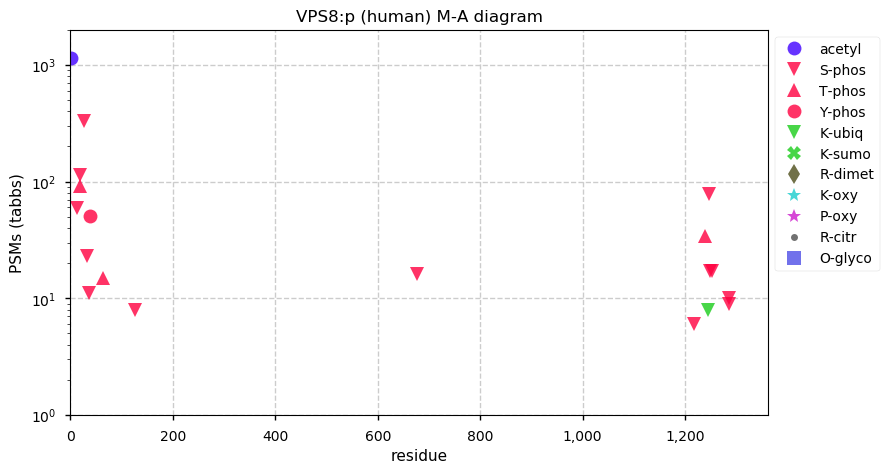
Thu Dec 03 15:52:06 +0000 2020VPS8:p θ(max) = 39. aka: FLJ32099, KIAA0804. Found in HLA class I (but not class II) peptide experiments. S,T+phosphoryl grouped into two phospho-domains, (13-127) & (1218-1236). Abundant in cell lines and tissues, but not fluids.
Thu Dec 03 15:52:06 +0000 2020>VPS8:p, vacuolar protein sorting 8 homolog (Homo sapiens) 🔗 Large subunit; CTMs: M1+acetyl; PTMs: 16× S,T+phosphoryl, Y38+phosphoryl; SAAVs: H1165Y (29%), S1319N (1%); mature form: 1-1428 [7,376 x, 26 kTa] #ᗕᕱᗒ 🔗
Thu Dec 03 14:14:16 +0000 2020@MHendr1cks Properly identifying email with students as a profit centre and pretty much every other use as a cost centre that requires charge-backs can go a long way. Requiring training into the legal issues associated with email can make people think a bit before pressing send.
Thu Dec 03 13:49:49 +0000 2020@MHendr1cks I am simultaneously horrified & not surprised. It can be fixed, but it requires both the faculty association and administration to put in some effort. Unfortunately neither will be willing to admit they have a problem.
Thu Dec 03 13:40:58 +0000 2020@MHendr1cks When I left academia a few years ago, email at my institution had become largely unusable because of its undisciplined use by both administrations and academics. I can only imagine how bad it must be now.
Thu Dec 03 13:33:53 +0000 2020@MHendr1cks I would assume that like most universities, yours does not have either email policies or training.
Wed Dec 02 17:29:18 +0000 2020@jwoodgett @realSMLewis As someone who has personally benefited from SBIR funding, having something like that in Canada would be great. None our funding agencies could run that type of program, but as a message to MPs it is an excellent case study.
Wed Dec 02 16:39:09 +0000 2020@jwoodgett @realSMLewis What specific investment-driven outcomes do you think would help? One of the reasons that the NIH is so successful politically is their conscious efforts to draw straight lines between their budget and economic activity all across the US.
Wed Dec 02 15:56:03 +0000 2020@jwoodgett @realSMLewis CIHR & Genome Canada's funding patterns have so reduced the number of potentially interested MPs that they have taken themselves out of the political conversation, leaving only arguments within the bureaucracy regarding funding.
Wed Dec 02 15:52:54 +0000 2020@jwoodgett @realSMLewis My point is until scientists are willing to contact MPs (repeatedly) in both their own riding & that of their institution with coherent demands wrt funding that will be positive for those MPs, national-level information campaigns will have little effect.
Wed Dec 02 15:15:14 +0000 2020@realSMLewis @jwoodgett At the moment, the majority of CIHR (& Genome Canada) funding really only affects 3 Liberal ridings, so there aren't a lot of voices in the caucus who care about the issue at a constituency level.
Wed Dec 02 15:03:04 +0000 2020@theoneamit The whole idea of doing an indexed search based on observed spectra (MS/MS peak intensities), along with all observed PTMs had no uptake in the proteomics community & continuing to work on it was a waste of time and money.
Wed Dec 02 14:18:49 +0000 2020@theoneamit I gave up on the entire spectrum library approach 3 or 4 years ago.
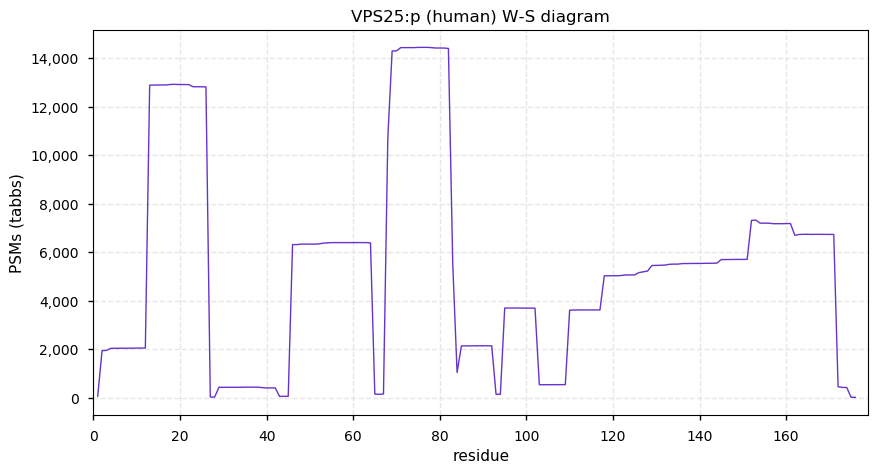
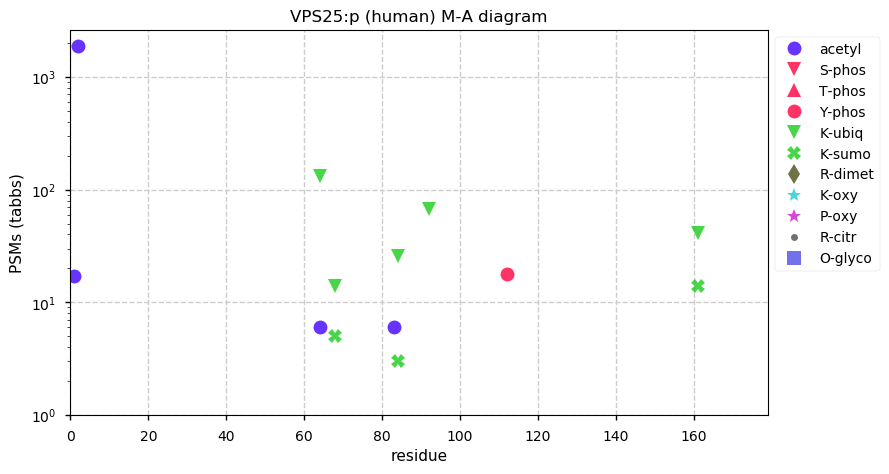
Wed Dec 02 13:57:49 +0000 2020VPS25:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
uacuacg[AUG]GCG[AUG]A
Wed Dec 02 13:57:49 +0000 2020VPS25:p θ(max) = 90. aka: MGC10540, EAP20, DERP9. Found in HLA class I & II peptide experiments, although rarely. The data shows that M1 & M3 can serve as translation initiation sites. Abundant in cell lines, common in urine but rare in blood plasma.
Wed Dec 02 13:57:48 +0000 2020>VPS25:p, vacuolar protein sorting 25 homolog (Homo sapiens) 🔗 Small subunit; CTMs: A2,S4+acetyl; aPTMs: 3x K+ubiquitinyl/SUMOyl, K64+acetyl/ubiquitinyl; SAAVs: none; mature form: 2,4-174 [14,262 x, 64 kTa] [20,693×, 93 kTa] #ᗕᕱᗒ 🔗
Tue Dec 01 19:05:19 +0000 2020I am genuinely excited to see the results of analyzing the data in PXD019258.
Tue Dec 01 17:54:39 +0000 2020@edemmott @eLife Nature journals seem determined to be the most innovative source for top hats and buggy whips.
Tue Dec 01 17:05:04 +0000 2020Revisiting PXD014845, it seems like an ideal data set to explore sensitivity vs. selectivity for PSM id algorithms.
Tue Dec 01 15:37:32 +0000 2020I know very little about the mechanics of protein structure prediction. Given the recent news, has anything changed that would allow structure predictions to easily determine changes associated with multiple PTM acceptor sites on a selected protein?
Tue Dec 01 13:57:57 +0000 2020Canadians (& especially our governments) can be so precious at times. I predict a 0.000 % chance of this happening. 🔗
Tue Dec 01 13:53:32 +0000 2020Well, that's the last time I go to the NYT web site. I don't like the level of cooperative surveillance between companies being quite so in-my-face.
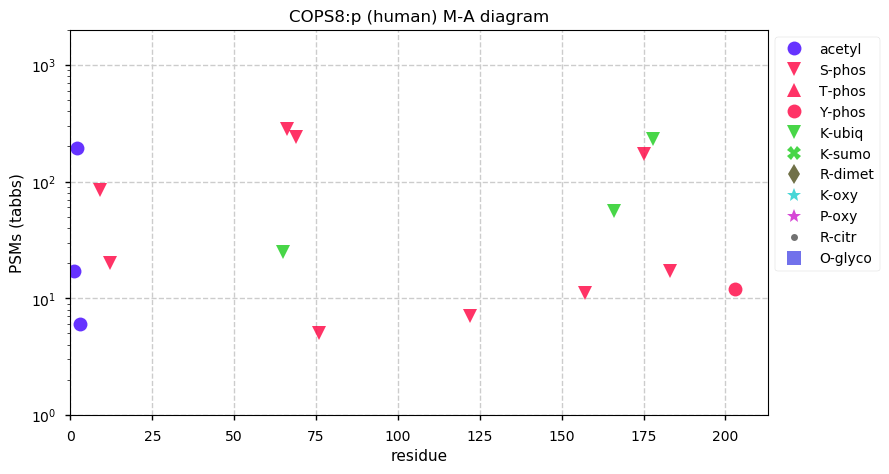
Tue Dec 01 13:27:28 +0000 2020COPS8:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
gcgaag[AUG]CCAGUGGCGGUG[AUG]G
Tue Dec 01 13:27:28 +0000 2020COPS8:p θ(max) = 90. aka: COP9, CSN8, MGC1297, SGN8. Found in HLA class I peptide experiments only. The data shows that M1 & M6 can serve as translation initiation sites. Abundant in most tissues and all cell lines.
Tue Dec 01 13:27:28 +0000 2020>COPS8:p, COP9 signalosome subunit 8 (Homo sapiens) 🔗 Small subunit; CTMs: P2,A7+acetyl; PTMs: 9× S+phorphoryl, 3× K+ubiquitinyl; SAAVs: none; mature form: 2,7-209 [20,693×, 93 kTa] #ᗕᕱᗒ 🔗
Mon Nov 30 18:39:34 +0000 2020IMHO, PXD020722 probably should have used single shot expts of individual urines to create the libraries instead of pooling them all and doing multidimensional chromatography. At least you would be able to sort out the effects of the various UTIs present in the pools.
Mon Nov 30 12:58:12 +0000 2020PKM:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
gcagcc[AUG]UCGAAGC … CGCAGCC[AUG]G
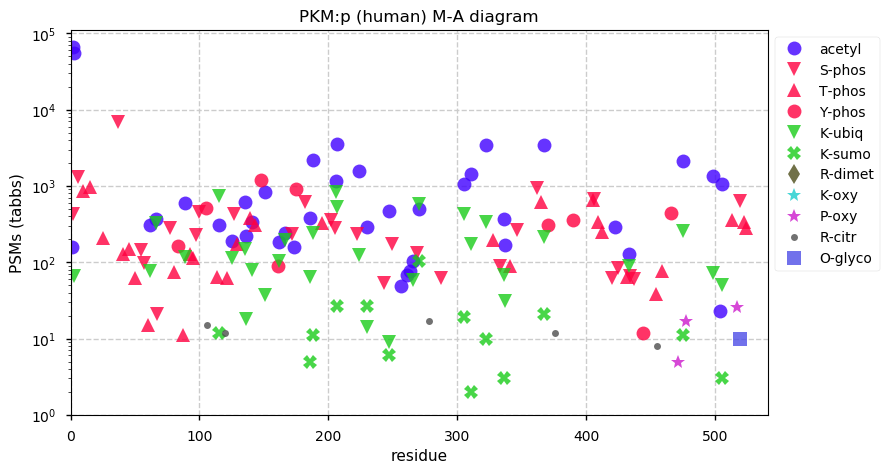
Mon Nov 30 12:58:11 +0000 2020PKM:p θ(max) = 90. aka: THBP1, OIP3, PK3, PKM2. Found in HLA class I & II peptide experiments. The data shows that M1 & M22 can serve as translation initiation sites. Abundant in most tissues and all cell lines.
Mon Nov 30 12:58:11 +0000 2020>PKM:p, pyruvate kinase M1/2 (H. sapiens) 🔗 Midsize enzyme; CTMs: S2,A23+acetyl; PTMs: 66×STY+phorphoryl; aPTMs: 15×K+acetyl/ubiquitinyl, 14×K+acetyl/ubiquitinyl/SUMOyl; SAAVs: V176E (8%); mature form: 2,23-531 [111,187×, 5650 kTa] #ᗕᕱᗒ 🔗
Sun Nov 29 19:26:43 +0000 2020@PastelBio Spooky!👻
Sun Nov 29 13:21:35 +0000 2020CRLF3:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
ggccag[AUG]AGGGGGGCG[AUG]G
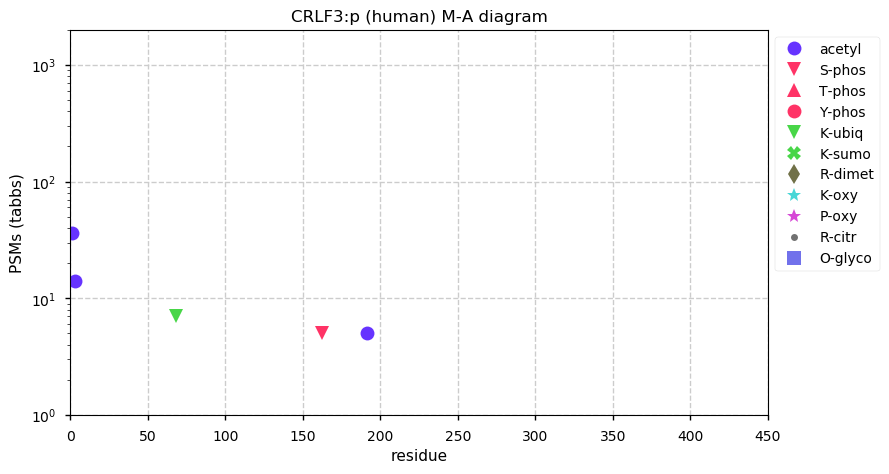
Sun Nov 29 13:21:34 +0000 2020CRLF3:p θ(max) = 71. Found in HLA class I & rarely in class II. The data shows that M1 & M5 can serve as translation initiation sites. The M5 site appears to be more commonly used than M1. Commonly found in cell lines and tissues: no known function.
Sun Nov 29 13:21:34 +0000 2020>CRLF3:p, cytokine receptor like factor 3 (Homo sapiens) 🔗 Small subunit; CTMs: M1,M5+acetyl; PTMs: no significant mods; SAAVs: L389P (13%); mature form: 1,5-442 [10,344×, 44 kTa] #ᗕᕱᗒ 🔗
Sat Nov 28 13:27:01 +0000 2020JUNB:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
gcccgg[AUG]UGCACUAAA[AUG]G
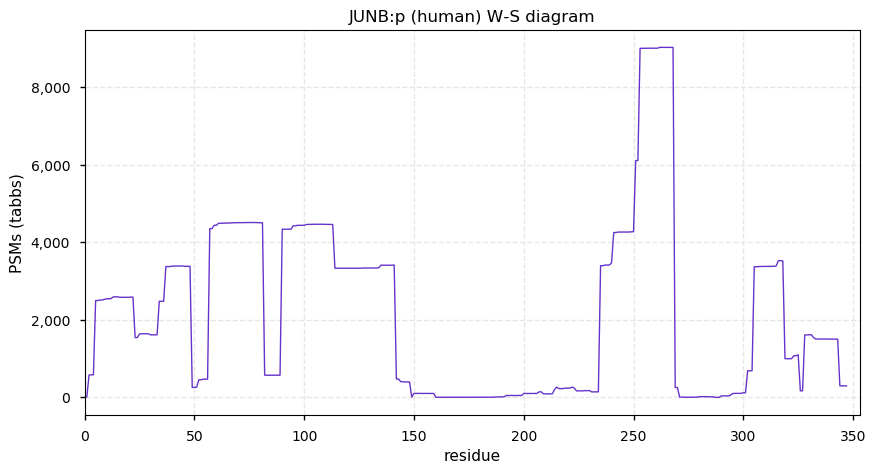
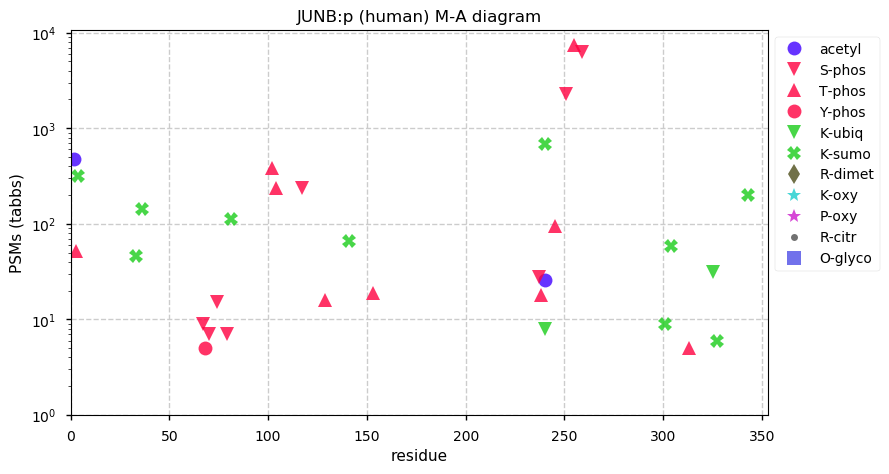
Sat Nov 28 13:27:00 +0000 2020JUNB:p θ(max) = 72. Only found in HLA class I. The data shows that M1 & M5 can serve as translation initiation sites. Too many GO annotations. Unusually large number of K+SUMOyl acceptor sites.
Sat Nov 28 13:27:00 +0000 2020>JUNB:p, JunB proto-oncogene, AP-1 transcription factor subunit (Homo sapiens) 🔗 Small subunit; CTMs: C2,M5+acetyl; PTMs: 9× SUMOYl, 17× S,T+phosphoryl; aPTMs: K240+acetyl/ubiquintyl/SUMOyl; SAAVs: none; mature form: 2,5-347 [8,949×, 40 kTa] #ᗕᕱᗒ 🔗
Fri Nov 27 21:09:41 +0000 2020GARS1:p is known to be present in both the mitochondrial matrix and cytoplasm. This alternate translation initiation mechanism would explain how this localization can occur.
Fri Nov 27 21:09:08 +0000 2020GARS1:p The mRNA context for this leaky-scanning problem is as follows:
caggcuc[AUG]CCCUCUCCGCGUCCAGUGCUGCUUAGAGGUGCUCGCGCCGCUCUGCUGCUGCUGCUGCCGCCCCGGCUCUUAGCCCGACCCUCGCUCCUGCUCCGCCGGUCCCUCAGCGCGGCCUCCUGCCCCCCGAUCUCCUUGCCCGCCGCCGCCUCCCGGAGCAGC[AUG]G
Fri Nov 27 18:38:51 +0000 2020@ucdmrt People who smell faintly of bananas.
Fri Nov 27 17:52:47 +0000 2020Added note: 5% only holds for cellular proteins. Most extracellular proteins do not have peptides from translation initiation events, at either M1 or alternate sites.
Fri Nov 27 16:57:33 +0000 2020@pwilmarth @MagnusPalmblad I think it would be better to use some type of deep learning (or other AI) to infer the author contributions. 🤔
Fri Nov 27 15:59:47 +0000 2020About 5% of peptides observed from translation initiation events should correspond to these "leaky-scanning" peptides. /fin
Fri Nov 27 15:59:47 +0000 2020m 33 STASVEIDDALYSR 46 q from UBA6:p, with S33+acetyl. 8/9
Fri Nov 27 15:59:46 +0000 2020m 9 ADKMDMSLDDIIK 21 l from ALYREF:p, with a 1 missed tryptic cleavage (K10) and several potential methionine oxidations; 7/9
Fri Nov 27 15:59:46 +0000 2020s 55 MDGAGAEEVLAPLR 68 l from GARS1:p, with M55+acetyl and M55+oxidation; 6/9
Fri Nov 27 15:59:46 +0000 2020If you want some data to test your new algorithm (or maybe an open search will find them?), try the "Keratinocyte" data from PXD019909. It has multiple observations of peptides generated by this mechanism, e.g.: 5/9
Fri Nov 27 15:59:46 +0000 2020Peptides beginning at the alternate M are tryptic peptides like any other. 4/9
Fri Nov 27 15:59:46 +0000 2020Alternate initiation M's are subject to the same co-translational modifications as M1 initiation: acetylation and/or removal of the M and acetylation of the following residue; and 3/9
Fri Nov 27 15:59:45 +0000 2020Almost all alternate initiation sites are the 2nd M in a protein sequence; 2/9
Fri Nov 27 15:59:45 +0000 2020If you are interested in adding an algorithm to an existing search engine to find alternate translation initiation sites, you should remember the following tips: 1/9
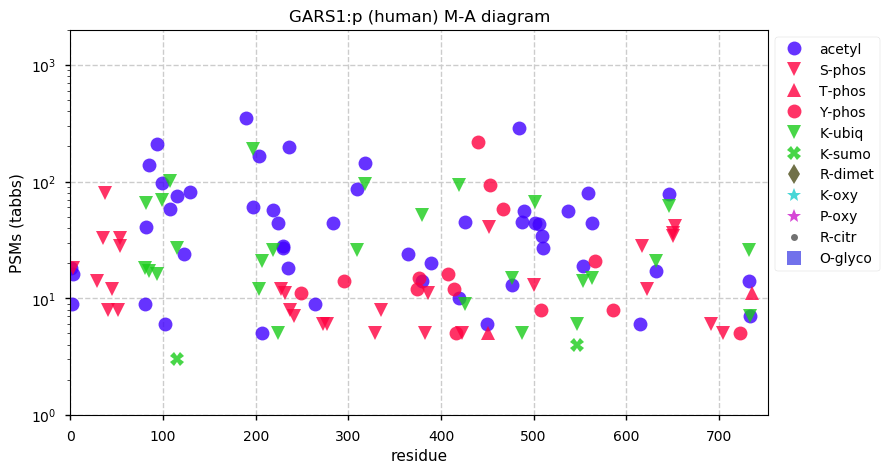
Fri Nov 27 13:49:40 +0000 2020GARS1:p θ(max) = 81. Found in HLA class I & II experiments. The data shows that M1 & M55 can serve as translation initiation sites. The M1 initiated sequence has a mitochondrial transit peptide (1-52), while the M55 initiated sequence does not.
Fri Nov 27 13:49:40 +0000 2020>GARS1:p, glycyl-tRNA synthetase 1 (Homo sapiens) 🔗 Midsized subunit; CTMs: M1,M55+acetyl; PTMs: 26× K+acetyl, 14× Y+phosphoryl; aPTMs: 26× K+acetyl/ubiquintyl; SAAVs: P4L (1%), P42A (35%); mature form: 53,55-739 [50,037×, 636 kTa] #ᗕᕱᗒ 🔗
Thu Nov 26 16:39:44 +0000 2020If true, it is kind of a nifty mechanism to use translational changes to determine the subcellular localization of enzyme isoforms.
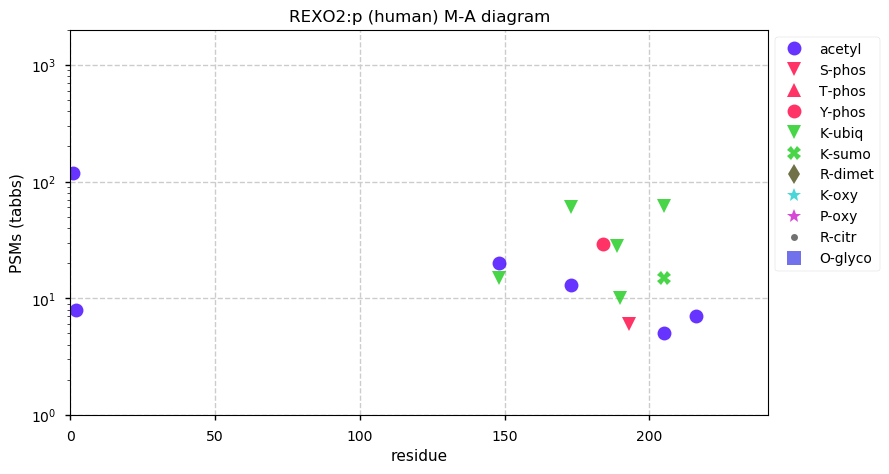
Thu Nov 26 13:06:16 +0000 2020REXO2:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
ccgggug[AUG]CUAGGCGGCUCCCUGGGCUCCAGGCUGUUGCGGGGUGUAGGUGGGAGUCACGGACGGUUCGGGGCCCGAGGUGUCCGCGAAGGUGGCGCAGCC[AUG]G
Thu Nov 26 13:06:16 +0000 2020REXO2:p The M1 initiated sequence has a mitochondrial targeting domain (1-25), which is removed when the protein enters the mitochondrial matrix. The M33 initiated sequence does not have that domain and is presumably the nuclear isoform of the protein.
Thu Nov 26 13:06:16 +0000 2020REXO2:p θ(max) = 70. aka DKFZP566E144, SFN, CGI-114. Commonly found in HLA class I & (less frequently) in class II peptide experiments. The data shows that M1 & M33 can serve as translation initiation sites.
Thu Nov 26 13:06:16 +0000 2020>REXO2:p, RNA exonuclease 2 (Homo sapiens) 🔗 Small protein; CTMs: A34+acetyl; aPTMs: K148,K173+acetyl/ubiquitinyl, K205+acetyl/ubiquintyl/SUMOyl; SAAVs: none; mature form: 26,34-189 [15,937×, 70 kTa] #ᗕᕱᗒ 🔗
Thu Nov 26 13:04:44 +0000 2020@InterProDB You should include "useful" on this list. It would be my only choice.
Wed Nov 25 19:28:14 +0000 2020@pwilmarth I would, if I could.
Wed Nov 25 17:32:48 +0000 2020@jwoodgett & they could divert money from SPOR to pay for them ...
Wed Nov 25 17:30:34 +0000 2020@jwoodgett It will be an issue. One solution is to move CIHR towards having the equivalent of program officers (or other type of grant management officers), who know and enforce current rules during a panel discussion.
Wed Nov 25 17:07:31 +0000 2020@astacus @slavov_n @c4pr_liv A lot of the data in PRIDE was submitted by the core personnel.
Wed Nov 25 15:38:36 +0000 2020@jwoodgett Canadian grants panels put more emphasis on this type of thing than those in other jurisdictions.
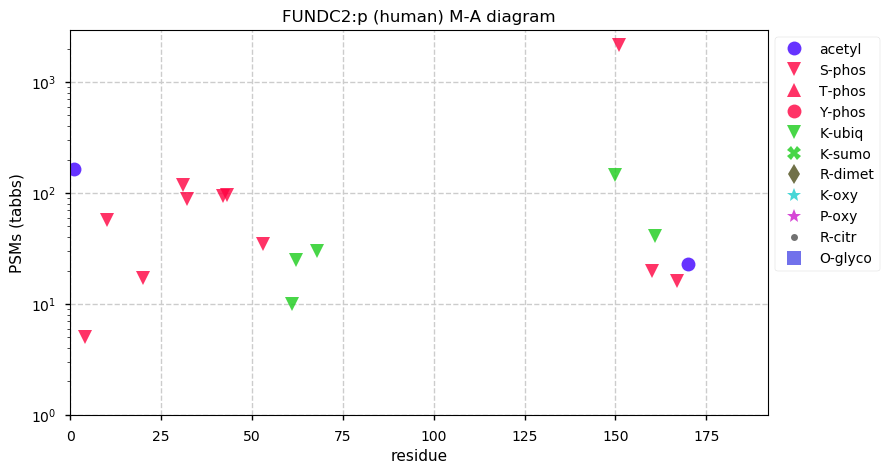
Wed Nov 25 13:56:39 +0000 2020FUNDC2:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
guggga[AUG]GAAACAUCUGCCCCACGUGCCGGAAGCCAAGUGGUGGCGACAACUGCGCGCCACUCCGCGGCCUACCGCGCAGAUCCUCUACGUGUGUCCUCGCGAGACAAGCUCACCGAA[AUG]G
Wed Nov 25 13:56:39 +0000 2020FUNDC2:p θ(max) = 65. aka HCBP6, DC44. Commonly found in HLA class I & (less frequently) in II peptide experiments. The data shows that M1 & M39 can serve as translation initiation sites. Too few GO annotations.
Wed Nov 25 13:56:39 +0000 2020>FUNDC2:p, FUN14 domain containing 2 (Homo sapiens) 🔗 Small protein; CTMs: M1,A40+acetyl; PTMs: 11x S+phosphoryl, 5x K+ubiquitinyl; SAAVs: none; mature form: 1,40-189 [8,735 x, 23 kTa] [14,819×, 114 kTa] #ᗕᕱᗒ 🔗
Tue Nov 24 21:06:22 +0000 2020My new candidate for least random looking protein sequence: trichohyalin 🔗 Almost fittingly, genetic variations to this protein cause "Uncombable hair syndrome" 🔗
Tue Nov 24 16:03:25 +0000 2020@AlexUsherHESA @UAlberta @UCalgary Their TRANSdisciplinary approach seems the equivalent of putting a cat & a sparrow in a confirmed space with no food in the hopes of developing a cat with wings.
Tue Nov 24 15:49:12 +0000 2020@PaoloDeLosRios @TrumanLab Nature Springer 🔗 puts the price at about the same dollar value as a 2016 Camry SE. Just the cost of trying to buy a tenure-track job I guess.
Tue Nov 24 15:22:52 +0000 2020@neely615 @MattWFoster I'd still vote for "saliva" as the worst. Urine is a solid runner up, though.
Tue Nov 24 13:53:19 +0000 2020Considering how variable the urine proteome can be, is library-based DIA the best way to follow disease-induced protein changes? I'd say "no" pretty emphatically.
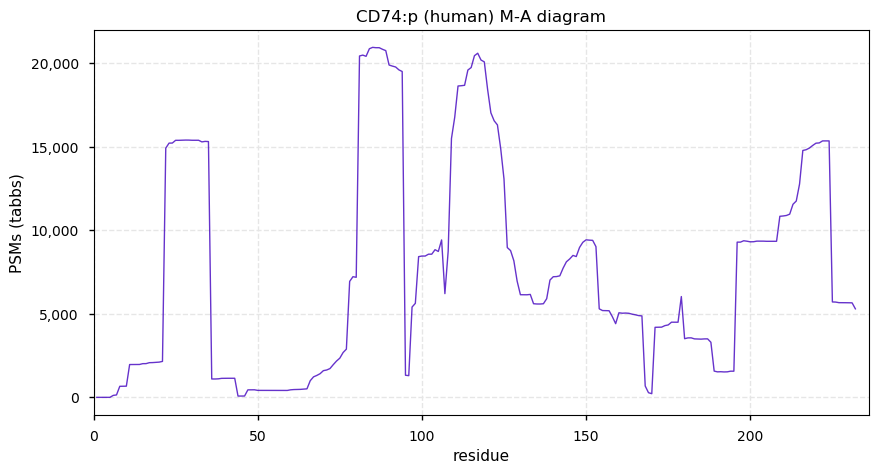
Tue Nov 24 12:51:57 +0000 2020CD74:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
ucccag[AUG]CACAGGAGGAGAAGCAGGAGCUGUCGGGAAGAUCAGAAGCCAGUC[AUG]G
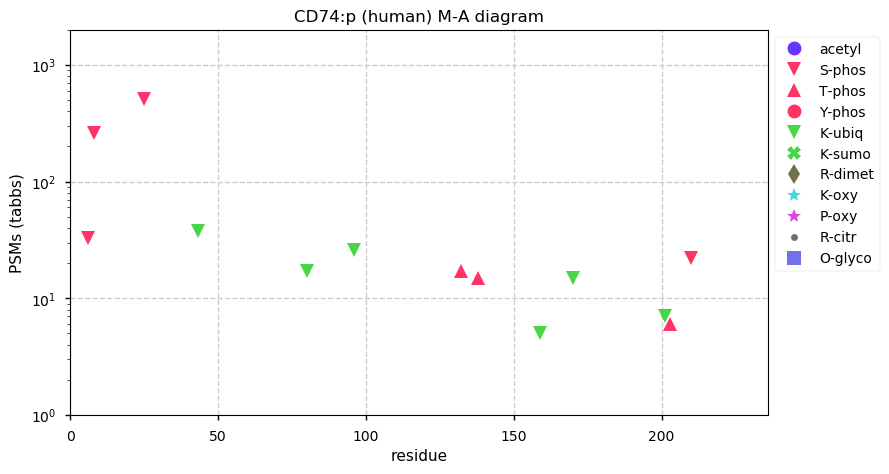
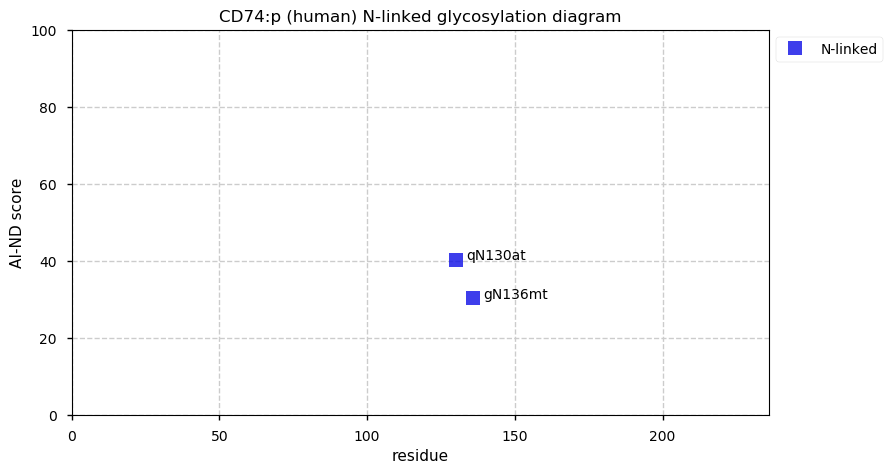
Tue Nov 24 12:51:56 +0000 2020CD74:p θ(max) = 66. aka DHLAG. Commonly found in both HLA class I & II peptide experiments. High occupancy phosphorylation acceptor sites (S8 & S25) on the cytoplasmic domain of the protein. The data shows that M1 & M18 can serve as translation initiation sites.
Tue Nov 24 12:51:56 +0000 2020>CD74:p, CD74 molecule (Homo sapiens) 🔗 Small type II membrane protein; CTMs: M17+acetyl; PTMs: N130,N136+glycosyl, 7× S,T+phosphoryl, 6× K+ubiquitinyl; SAAVs: none; mature form: 1,18,19-232 [14,819×, 114 kTa] #ᗕᕱᗒ 🔗
Mon Nov 23 19:11:37 +0000 2020Conversely, any acetylation or ubiquitination assigned to a peptide's C-terminal lysine is a false positive.
Mon Nov 23 17:46:32 +0000 2020Just a reminder: if you are analyzing lysine acetylation or ubiquitination data & set the number of missed cleavages to "1", you are actually setting it to "0" (most lysine mods result in abolishing tryptic cleavage at the modified lysine).
Mon Nov 23 16:25:31 +0000 2020@MattWFoster I'm not knocking the data though: this is probably the first thorough examination of the proteins present in the individual cell types that make up human skin. It is an excellent resource for both dermatology & cosmetics research.
Mon Nov 23 13:51:25 +0000 2020Leaving home to 'beat someone up' not a valid reason to break lockdown, Frenchman told 🔗 via @RFI_En
Mon Nov 23 13:15:55 +0000 2020EXOC6B:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
ccagucu[AUG]GAGCGGGGUAAG[AUG]G
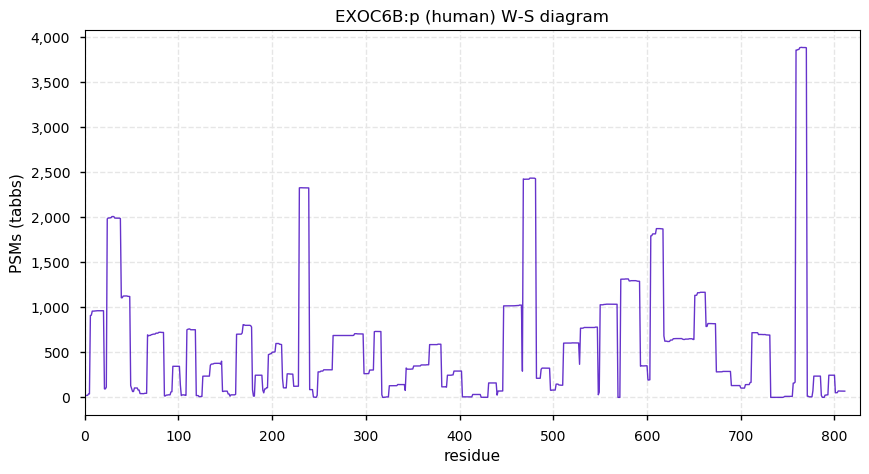
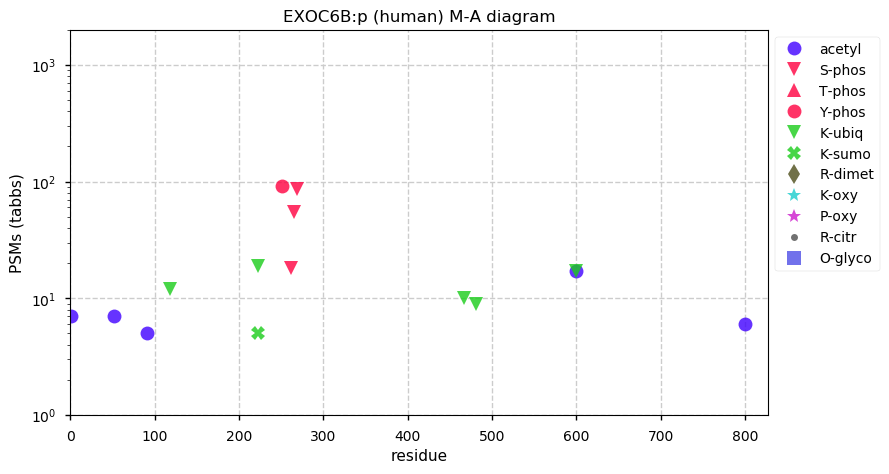
Mon Nov 23 13:15:55 +0000 2020EXOC6B:p θ(max) = 61. aka SEC15L2, SEC15B. Found in HLA class I & (very rarely) class II peptide experiments. The data shows that M1 & M6 can serve as translation initiation sites.
Mon Nov 23 13:15:54 +0000 2020>EXOC6B:p, exocyst complex component 6B (Homo sapiens) 🔗 Midsized subunit; CPTMs: M1,A7+acetyl; PTMs: Y251,S262,S265,S269+phosphoryl; aPMTs: K222+ubi/SUMOyl, K600+acetyl/ubi, SAAVs: none; mature form: 1,7-811 [9,506×, 35 kTa] #ᗕᕱᗒ 🔗
Mon Nov 23 01:23:36 +0000 2020@MattWFoster If there is lots of it, it probably isn't signalling.
Sun Nov 22 19:47:22 +0000 2020I've been using Windows via SSH for long enough it no longer seems unusual.
Sun Nov 22 19:08:32 +0000 2020Does anybody who hasn't screwed up royally ever use the word "hindsight"?
Sun Nov 22 16:32:27 +0000 2020@MattWFoster Such as cell-specific
1. alternate splicing;
2. translation initiation;
3. collagen modification;
4. citrulline formation (filaggrin and keratin);
5. structural (non-signaling) phosphorylation;
6. ubiquitination levels; etc.
Sun Nov 22 15:47:42 +0000 2020@MattWFoster The paper used a "traditional" style of analysis, that I refer to in my head as the "4-banger":
1. tryptic cleavage only;
2. protein N-terminal+acetylation;
3. M+oxidation; &
4. C+IAA.
coupled with the "Highlander" UniProt sequences, leaving lots of room for further analysis.
Sun Nov 22 14:06:46 +0000 2020ADPRS:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
gcgcgg[AUG]GCCGCAGCGGCG[AUG]G
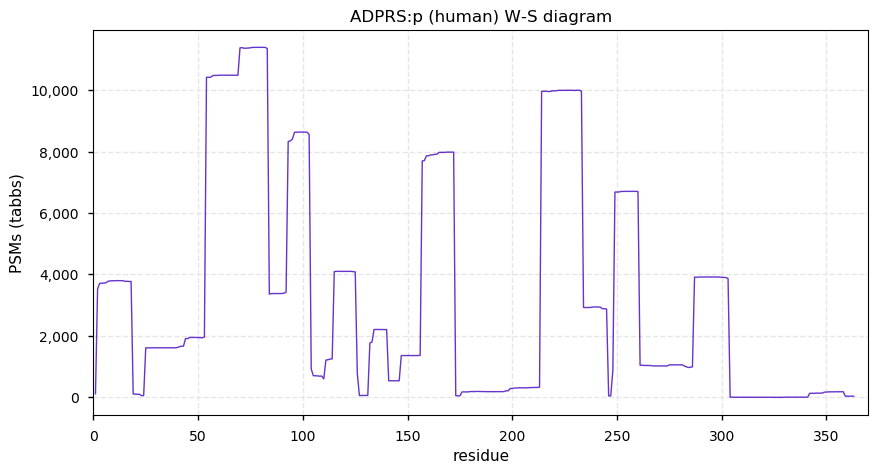
Sun Nov 22 14:02:56 +0000 2020ADPRS:p θ(max) = 71. aka ARH3, FLJ20446, ADPRHL2. Found in HLA class I & (rarely) class II peptide experiments. The data shows that M1 & M6 can serve as translation initiation sites.
Sun Nov 22 14:02:56 +0000 2020>ADPRS:p, ADP-ribosylserine hydrolase (Homo sapiens) 🔗 Small enzyme; CPTMs: A2,A7+acetyl; PTMs: T64,S67+phosphoryl; SAAVs: none; mature form: 2,7-363 [17,279×, 80 kTa] #ᗕᕱᗒ 🔗
Sun Nov 22 01:26:45 +0000 2020Not a lot of great trajectories amongst this group 🔗
Sat Nov 21 21:23:24 +0000 2020If you are feeling up for a challenge, the "dermis" data from PXD019909 (🔗) has one of the most naturally complex set of modifications I've run into recently.
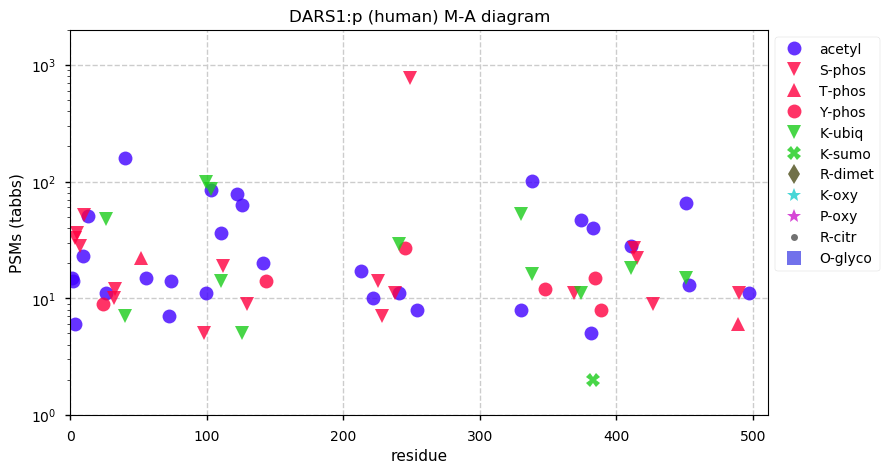
Sat Nov 21 12:54:12 +0000 2020DARS1:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
ugucccg[AUG]CCCAGCGCCAGCGCCAGCCGCAAGAGUCAGGAGAAGCCGCGGGAGAUC[AUG]G
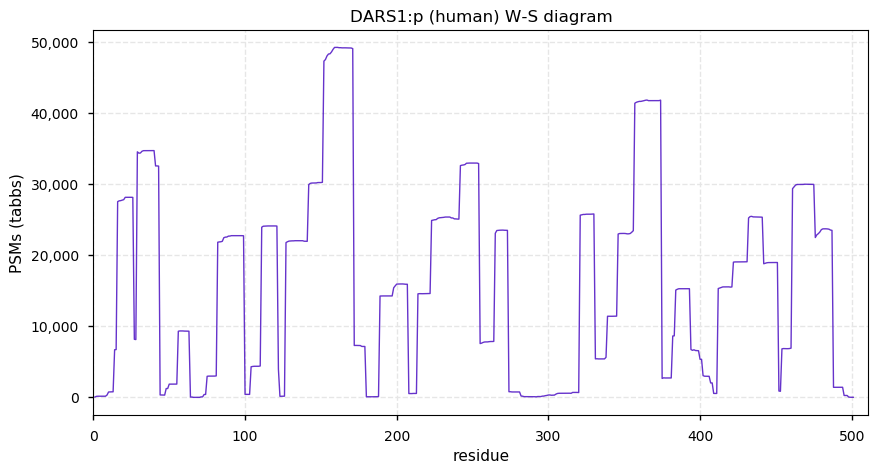
Sat Nov 21 12:54:12 +0000 2020DARS1:p θ(max) = 86. aka DARS. The abbreviation DARS1 is new: it was changed from DARS because of Excel-induced naming switching. Found in HLA class I & class II peptide experiments. The data shows that M1 & M18 can serve as translation initiation sites.
Sat Nov 21 12:54:11 +0000 2020>DARS1:p, aspartyl-tRNA synthetase 1 (Homo sapiens) 🔗 Midsized cenzyme; PTMs: 20x S,T+phosphoryl, 6x Y+phosphoryl; aPTMs: 12× K+ubiquitinyl/acetyl; SAAVs: none; mature form: 1,2,18,19-501 [56,674×, 628 kTa] [18,748, 97 kTa] #ᗕᕱᗒ 🔗
Fri Nov 20 18:41:49 +0000 2020@ypriverol @MiguelCos I created that list with ENSEMBL v. 100, but it should work with v. 101 😀
Fri Nov 20 18:34:11 +0000 2020@ypriverol @MiguelCos It is just the current version of ENSEMBL human proteins.
Fri Nov 20 17:14:54 +0000 2020@MiguelCos @ypriverol PPS: longer peptides are better for SAAV detection, so good quality Lys-C by itself is usually better than trypsin.
Fri Nov 20 17:04:45 +0000 2020@bkives How about newspapers?
Fri Nov 20 16:59:29 +0000 2020@MiguelCos @ypriverol PS: the other main lesson is that hemoglobin is really weird.
Fri Nov 20 16:53:45 +0000 2020@MiguelCos @ypriverol Maybe you can figure out what is going on with TMT studies!
Fri Nov 20 16:52:42 +0000 2020@MiguelCos @ypriverol I'm not discouraging looking for SAAVs: quite the opposite. But be careful & dig into the results if anything is even slightly hinkey. The lesson learned from looking at them for years is a bit of a tautology:
You are most likely to see the things you are most likely to see.
Fri Nov 20 16:43:15 +0000 2020@MiguelCos @ypriverol Either way. I'm not sure of the cause (I suspect side reactions), but the false positive rate for SAAVs is unacceptable in most of the TMT6+ data I have looked at.
Fri Nov 20 16:40:30 +0000 2020@MiguelCos If the SAAV is heterozygous, then a PSM with the reference sequence & the same PTMs is confirmatory evidence. However, you are most likely to observe SAAV with high frequency in the population (like TF:p.I448V) that are often homozygous.
Fri Nov 20 16:29:07 +0000 2020@ypriverol @MiguelCos It can be a problem. There are some types of data, for example data obtained from most TMT-dervatized samples, that simply can't be used for SAAV detection.
Fri Nov 20 16:26:39 +0000 2020@MiguelCos SAAVs are also rare: < 1% of PSMs correspond to SAAVs in a sample taken from an individual or a cell line. Unlike specific PTMs, they are all one-offs unless you have enough data to reproducibly detect the same SAAV repeatedly. 2/2
Fri Nov 20 16:21:32 +0000 2020@MiguelCos The problem with the brute force approach is that you must be careful of what I call "over-fitting". Because SAAVs correspond to the loss or addition of a range of small organic moieties, when you add it a PTM or 2 you can get solutions that add up to an SAAV that aren't. 1/2
Fri Nov 20 15:36:05 +0000 2020@MiguelCos That would be correct. I don't use that brute force approach very often any more: instead I use a list of SAAVs I made out of gnomAD nsSNV information, only checking for known SAAVs. The current list is available here: 🔗
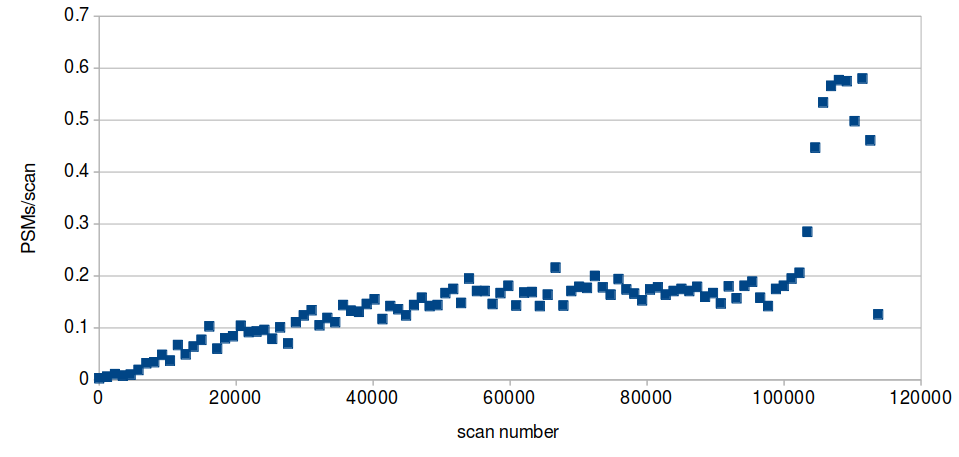
Fri Nov 20 15:26:13 +0000 2020Would you expect this type of chromatographic oddity to affect the utility of the library created from this data? 2/2
Fri Nov 20 15:25:14 +0000 2020Question for DIA tweeps: below is a histogram of the # of PSMs per scan (p), taken from an MS/MS data file meant to be used to create a library for DIA. The big step up in p is at the point where the gradient has switched to 98% B (a wash step). 1/2 🔗
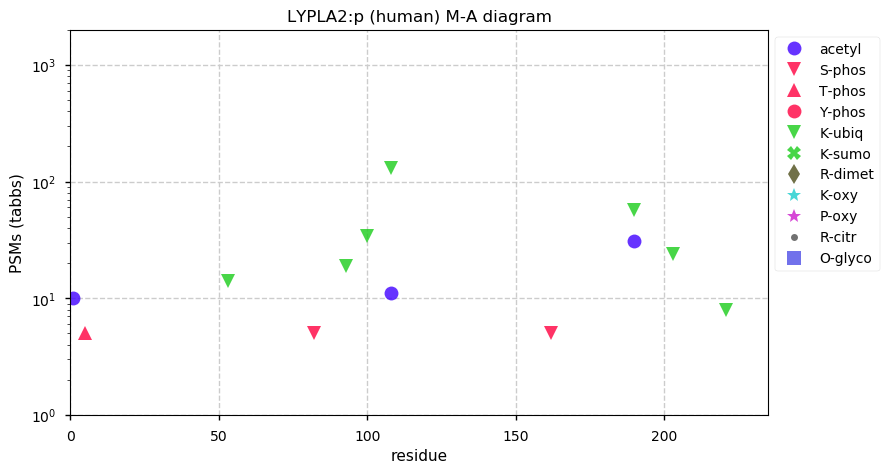
Fri Nov 20 13:04:46 +0000 2020LYPLA2:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
guggugu[AUG]UGUGGUAACACC[AUG]U
M6 appears to be in a significantly better context for initiation.
Fri Nov 20 13:04:46 +0000 2020LYPLA2:p θ(max) = 86. aka APT-2. Found in HLA class I & class II peptide experiments. The data shows that M1 & M6 can serve as translation initiation sites.
Fri Nov 20 13:04:45 +0000 2020>LYPLA2:p, lysophospholipase 2 (Homo sapiens) 🔗 Small dimeric enzyme; CPTMs: S7+acetyl; PTMs: 5× ubiquitinyl; aPTMs: K108,K190+ubiquitinyl/acetyl; SAAVs: none; mature form: 2,7-231 [18,748×, 97 kTa] #ᗕᕱᗒ 🔗
Thu Nov 19 15:03:40 +0000 2020It sounds like BJ was one of those kids who would make fun of my son for drawing a laser as an elongated rectangular box.
Thu Nov 19 15:02:02 +0000 2020As someone who spent years working with lasers, I really want to know what an "inexhaustible laser" might look like in practice. 🔗
Thu Nov 19 14:52:22 +0000 2020It involves over 300 separate experiments and uses A-539 cells (lung tumor origin) to study these interactions. If you want to try-before-you-buy, the identifications are all available at 🔗
Thu Nov 19 14:48:35 +0000 2020Anyone interested in SARS CoV and SARS COV2 protein-protein interactions in a cell should really take a close look at PXD020222 (🔗). While it has not received as much attention as some other studies, technically it is by far the most sophisticated.
Thu Nov 19 13:21:15 +0000 2020LYPLA1:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
gcggugu[AUG]UGCGGCAAUAAC[AUG]U
M6 appears to be in a significantly better context for initiation.
Thu Nov 19 13:21:15 +0000 2020LYPLA1:p θ(max) = 86. aka CRM1, CRM-1. Found in HLA class I & class II peptide experiments. The data shows that M1 & M6 can serve as translation initiation sites.
Thu Nov 19 13:21:15 +0000 2020>LYPLA1:p, lysophospholipase I (Homo sapiens) 🔗 Small dimeric enzyme; CPTMs: C2,S7+acetyl; PTMs: K89,K208+ubiquitinyl; aPTMs: K81,K174+ubiquitinyl/acetyl; SAAVs: I98M (1%); mature form: 2,7-214 [23,304 x, 110 kTa] #ᗕᕱᗒ 🔗
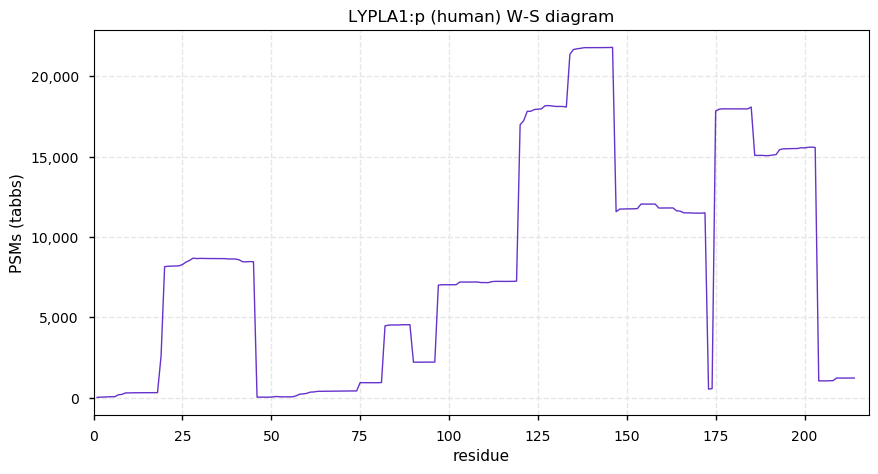
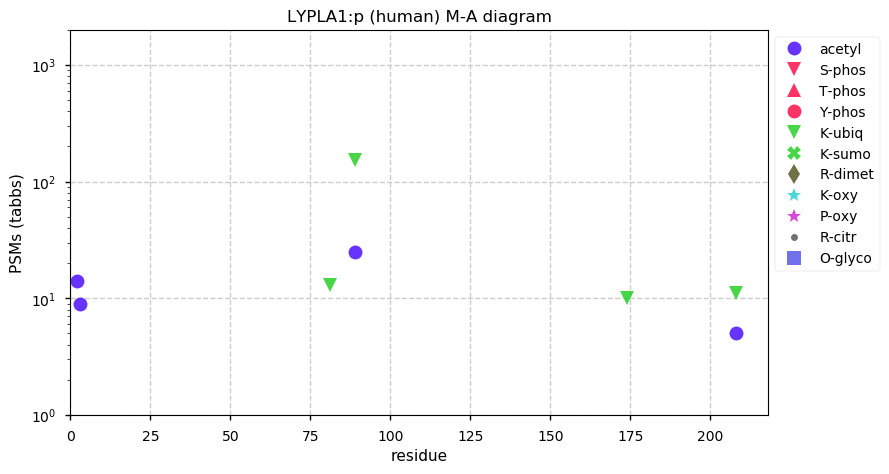
Thu Nov 19 13:18:59 +0000 2020@PastelBio While the picture may be an apt metaphor in some cases, illustrating scientific data as a midden that at best may contain a few tin cans is a fairly negative view of lab science.
Wed Nov 18 17:30:41 +0000 2020@neely615 @MattWFoster & the new "I am a super cool service" mantra on the ELIXER pages I find more than a little annoying. 🔗
Wed Nov 18 17:26:54 +0000 2020@neely615 @MattWFoster In the past, I had found that the PRIDE pages tended to lag the PX pages, but I guess their recent changes in back-end platforms may have shifted that around.
Wed Nov 18 17:20:23 +0000 2020@neely615 @MattWFoster I had only checked the PX page (🔗) which said it was locked. Thanks. I'm downloading it now ...
Wed Nov 18 17:02:08 +0000 2020For anyone who missed it, @gingraslab1 recommended the following reviews for anyone interested in this mRNA alternate translation mechanism:
🔗
🔗
Wed Nov 18 16:38:12 +0000 2020@MattWFoster Nope. I was unaware of that page. I have tried it now & we'll see if it works ...
Wed Nov 18 16:02:10 +0000 2020PS: I only ask because I rarely have any luck dealing directly with authors.
Wed Nov 18 15:56:59 +0000 2020Could somebody pester the authors of 🔗 to release their lock on PXD012615? The paper has been out since July but the data is still not publicly available.
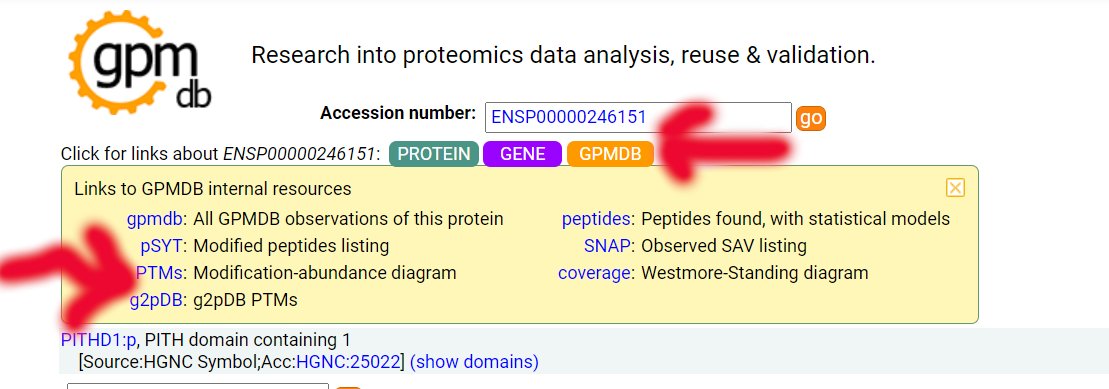
Wed Nov 18 13:59:38 +0000 2020@ypriverol If you go to the page for a specific accession, e.g. 🔗, then click on the orange "GPMDB" button just under the "Accession number" box to drop down a little menu. Select the "PTMs" link in that menu (Modification-abundance diagram). 🔗
Wed Nov 18 13:15:19 +0000 2020XPO1:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
uaaucu[ATG]CCAGCAAUU[AUG]ACA[AUG]U
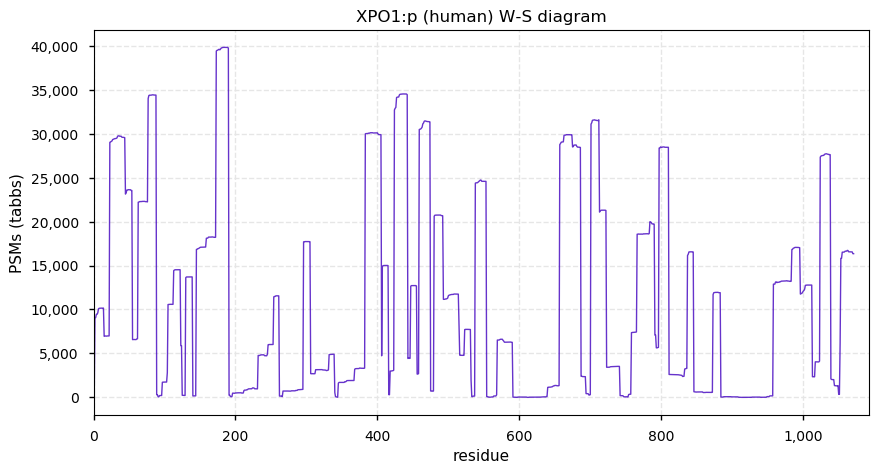
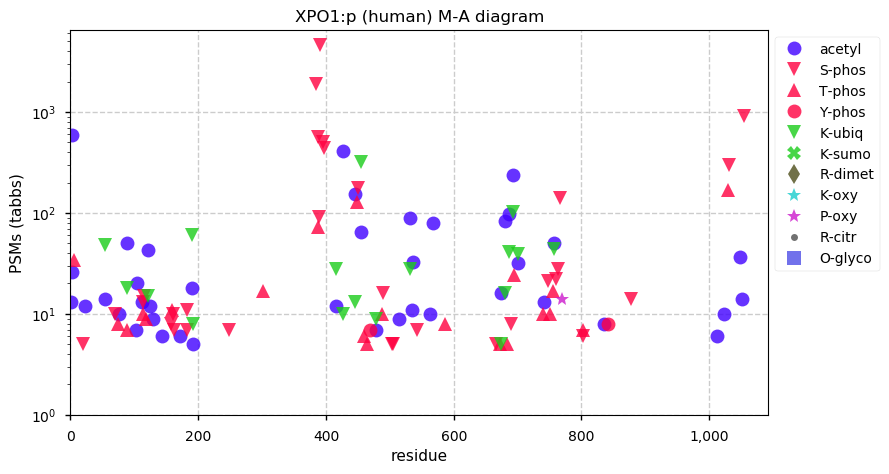
Wed Nov 18 13:15:19 +0000 2020XPO1:p θ(max) = 74. aka CRM1, CRM-1. Found in HLA class I peptide experiments, rarely in class II. The data shows that M1, M4 & M6 can serve as translation initiation sites.
Wed Nov 18 13:15:19 +0000 2020>XPO1:p, exportin 1 (Homo sapiens) 🔗 Large subunit; CPTMs: P2+acetyl; PTMs: 54x S,T,Y+phosphoryl; aPTMs: 17x ubiquitinyl/acetyl; SAAVs: none; mature form: 2,6,7-1071 [54,097 x, 802 kTa] [21,145×, 92 kTa] #ᗕᕱᗒ 🔗
Tue Nov 17 23:19:55 +0000 2020@deankoshelanyk @bkives Great!
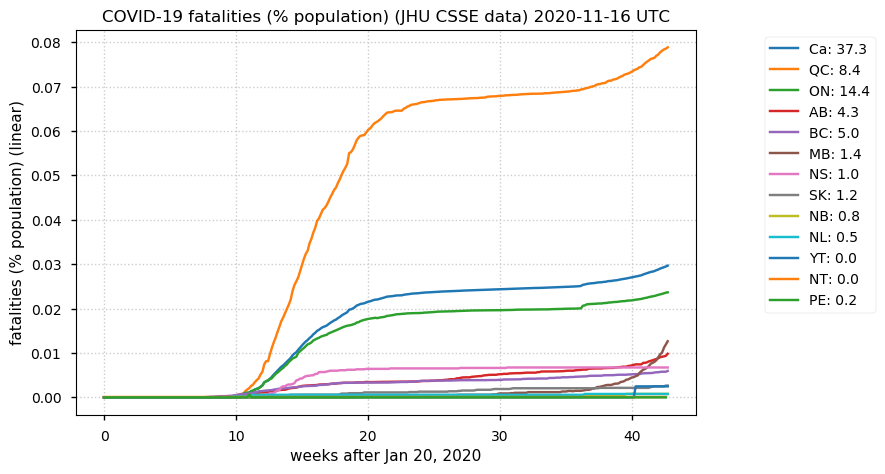
Tue Nov 17 18:52:41 +0000 2020QC (~0.08%) is closing in on the ND number (~0.1%), even though QC has 11× the population of ND.
🔗
Tue Nov 17 18:45:45 +0000 2020Le MB évolue dans la mauvaise direction, mais le QC est vraiment l'exception parmi les provinces canadiennes
Tue Nov 17 18:37:48 +0000 2020MB is trending in the wrong direction, but QC is really the exception among Canadian provinces 🔗
Tue Nov 17 18:14:40 +0000 2020@bkives Half of the way to Teulon is not the most convenient siting. I hear no one is using the Convention Centre at the moment: a big, well ventilated space downtown set up to manage large lines of people indoors might be a viable alternative.
Tue Nov 17 16:19:27 +0000 2020@MHendr1cks @jwoodgett @nictitate @grantsfacilitat @bcchr A simple test is to see if your name is eligible to show up on a "sunshine list". If it is, then you, my friend, are a provincial bureaucrat as far as the government is concerned.
Tue Nov 17 16:12:55 +0000 2020Very similar to my thinking wrt the direction for hardware in proteomics data analysis—so it must be right!
🔗
Tue Nov 17 15:59:17 +0000 2020@jwoodgett @MHendr1cks Drafting supplies.
Tue Nov 17 15:48:57 +0000 2020@slavov_n @ionicwoman Hopefully this type of investigation catches on.
Tue Nov 17 15:07:28 +0000 2020@jwoodgett & so ends the myth of meritocracy.
Tue Nov 17 14:15:40 +0000 2020@MHendr1cks @jwoodgett @nictitate @grantsfacilitat @bcchr Canadian universities are creatures of provincial governments & they are run like prov. gov. departments. There are concessions to the notion of academic freedom, but only to the extent that they don't create waves up the chain that can be felt by the premier's office.
Tue Nov 17 13:14:54 +0000 2020Any enterprising young investigator who is interested in mRNA & proteins could probably make a research specialty out of detecting these fairly common but generally ignored alternate translation products.
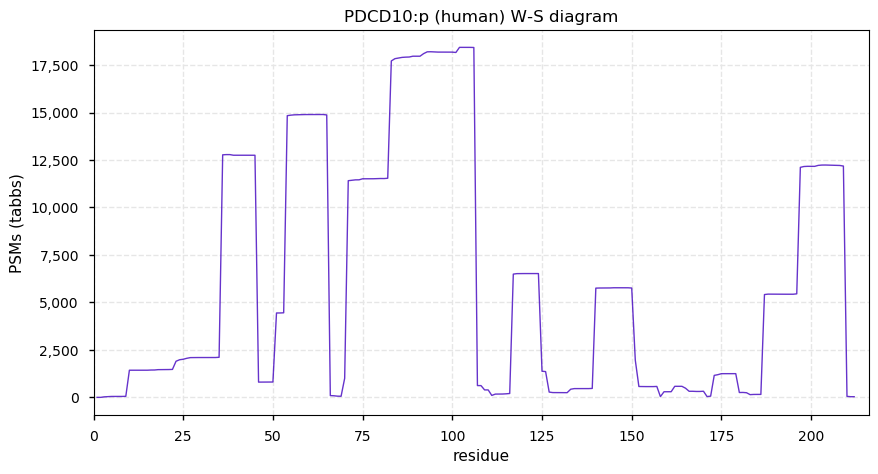
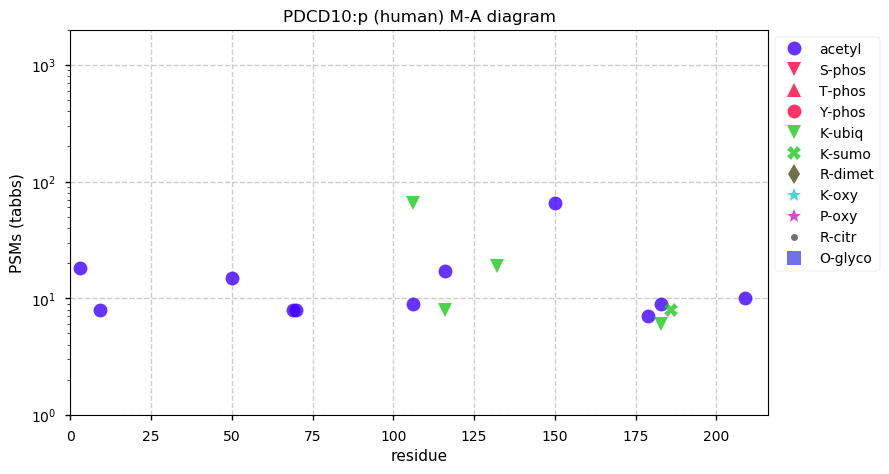
Tue Nov 17 12:55:29 +0000 2020PDCD10:p The mRNA context for this translation initiation leaky-scanning problem is as follows:
uuuuga[AUG]AGG[AUG]A
It is worth noting that the predicted M1 initiation codon [AUG] has a U at the -3 position, while the M3 [AUG] has an A at -3.
Tue Nov 17 12:53:07 +0000 2020PCDC10:p θ(max) = 74. aka TFAR15, CCM3. Found in HLA class I peptide experiments, not class II. Too many GO annotations. The data show that M3 in this sequence is a translation initiation site: there is no direct evidence of M1 being used for this purpose.
Tue Nov 17 12:53:07 +0000 2020>PDCD10:p, programmed cell death 10 (Homo sapiens) 🔗 Small protein; CPTMs: M3+acetyl; PTMs: 7× K+acetyl; aPTMs: 3× ubiquitinyl/acetyl; SAAVs: none; mature form: 1?,3,4-212 [21,145×, 92 kTa] #ᗕᕱᗒ 🔗
Mon Nov 16 23:34:32 +0000 2020Sometimes the names people chose for their data files look like some sort of SQL injection exploit.
Mon Nov 16 17:45:36 +0000 2020@byu_sam @JProteomeRes Calling software a "tool" is about the same as saying that my cat is a "mousetrap".
Mon Nov 16 15:19:58 +0000 2020@AlexUsherHESA Streamlining the "chain yanking" process is almost certainly the desired outcome. I'm surprised that the MB government hasn't put forward a similar plan, although it may be the Premier likes the current chain yanking regime.
Mon Nov 16 14:01:35 +0000 2020The biological origin/purpose of this raggedness is unknown.
Mon Nov 16 13:54:16 +0000 2020@pwilmarth @gingraslab1 Thanks, Phil. I had wanted the ref. because tomorrow's protein du jour is a good example of this fairly common phenom. & I wanted to use the correct language to describe the situation.
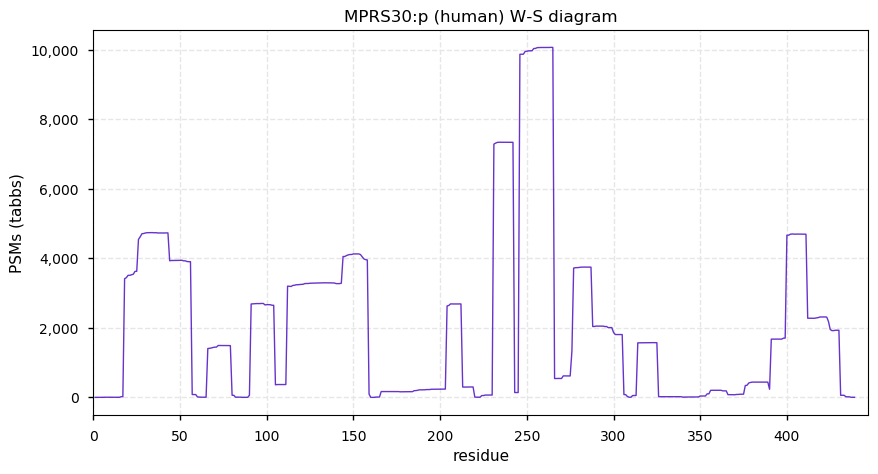
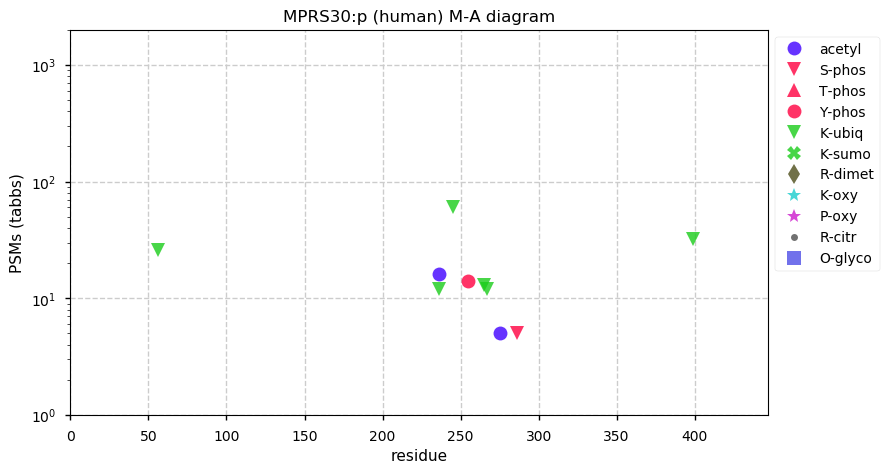
Mon Nov 16 13:17:13 +0000 2020MRPS30:p θ(max) = 60. aka PAP, PDCD9. Found in HLA class I peptide experiments, not class II. A mitochondrial targeting domain (1-17) is removed on entry to the mitochondrion. The protein's mature N-terminus is extraordinarily ragged, beginning anywhere from 18 to 34.
Mon Nov 16 13:17:13 +0000 2020>MRPS30:p, mitochondrial ribosomal protein S30 (Homo sapiens) 🔗 Midsized protein; PTMs: Y255+phosphoryl,6× K+ubiquitinyl; SAAVs: C33S (40%); mature form: 18-619 [12,542×, 58 kTa] #ᗕᕱᗒ 🔗
Sun Nov 15 14:11:50 +0000 2020@gingraslab1 I guess it is like alternate splicing: it doesn't square with proteomic's dominant "Highlander" approach to translated gene sequences so it is not publicly discussed.
Sun Nov 15 13:39:14 +0000 2020@gingraslab1 The articles were very helpful. I see the results of "leaky scanning" all the time in data, but it is something I don't think I've ever seen discussed in the proteomics literature.
Sun Nov 15 13:17:30 +0000 2020AIFM1:p is highly modified. The biological function of these PTMs and aPTMs is unknown. PTMs in the mitochondrial targeting domain suggest there may be some cytoplasmic role for this protein.
Sun Nov 15 13:17:30 +0000 2020AIFM1:p θ(max) = 70. aka AIF, CMTX4, DFNX5, PDCD8, NAMSD, AUNX1. Found in HLA class I & II peptide experiments. A mitochondrial targeting domain (1-54) is removed on entry to the mitochondrion. A proprotein domain (55-99,100) is removed to form the active protein.
Sun Nov 15 13:17:30 +0000 2020>AIFM1:p, apoptosis inducing factor mitochondria associated 1 (Homo sapiens) 🔗 Midsized protein; PTMs: 28× K+acetyl, 67× S,T+phosphoryl; aPTMs: 6× K+ubiquitinyl/acetyl; SAAVs: none; mature form: 100,101-619 [48,785×, 522 kTa] #ᗕᕱᗒ 🔗
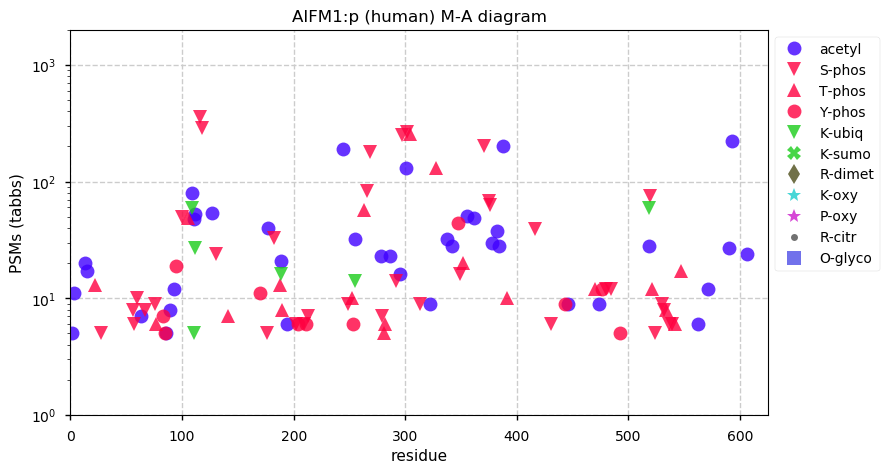
Sat Nov 14 23:53:14 +0000 2020@gingraslab1 Thanks.
Sat Nov 14 23:15:54 +0000 2020Does anyone know any good reviews on alterate translation initiation (translation starting at the second AUG in an mRNA ORF)?
Sat Nov 14 14:50:29 +0000 2020Looks like Theta is happy where it is, just west of Morocco, Iota is forming up nicely just north of Columbia and what is left of Eta is going to take a swipe at the Avalon Peninsula. 🔗
Sat Nov 14 13:37:55 +0000 2020PDCD7:p θ(max) = 38. aka HES18, ES18. Found in HLA class I peptide experiments, but not class II. The domain (2-129) has several proline-rich low complexity domains that are not observed in MS-based proteomics.
Sat Nov 14 13:37:55 +0000 2020>PDCD7:p, programmed cell death 7 (Homo sapiens) 🔗 Small nuclear protein; PTMs: 3× K+SUMOyl, 3× K+ubiquitinyl; SAAVs: none; mature form: 2-485 [2,103×, 4.6 kTa] #ᗕᕱᗒ 🔗
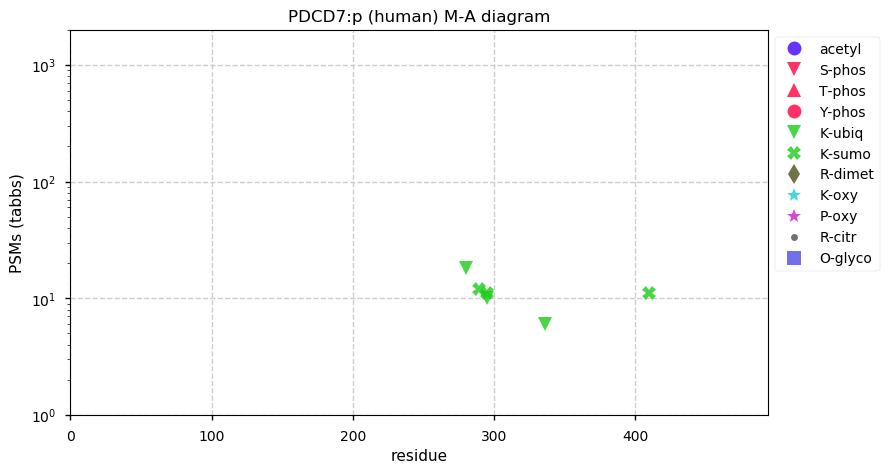
Sat Nov 14 13:23:49 +0000 2020@AJ_Brenes @JMarchingo People have mainly focussed on acetyl, phosphoryl and ubiquitinyl. There are a few big SUMOyl studies, but after that it gets pretty boutique.
Sat Nov 14 13:20:38 +0000 2020@AJ_Brenes @JMarchingo There are only a handful of methylation studies available in the public repositories.
Fri Nov 13 23:11:41 +0000 2020@AJ_Brenes 🔗
Fri Nov 13 15:49:01 +0000 2020@VATVSLPR It looks like that one is headed for the Miskito Coast, where Eta made landfall the first time. It will probably break up before it gets across to the Pacific.
Fri Nov 13 12:57:56 +0000 2020PDCD6:p θ(max) = 88. aka ALG-2, PEF1B. Observed in HLA class I & class II peptide experiments. Commonly observed in cell lines & tissues as well as urine extracellular vesicles. Associated with too many GO annotations.
Fri Nov 13 12:57:56 +0000 2020>PDCD6:p, programmed cell death 6 (Homo sapiens) 🔗 Small protein; CTMs: A2,A3+acetyl; PTMs: 4× Y+phosphoryl; aPTMs: 2× K+ubiquitinyl/acetyl; SAAVs: A3V (2%); mature form: 2,3-191 [32,575×, 209 kTa] #ᗕᕱᗒ 🔗
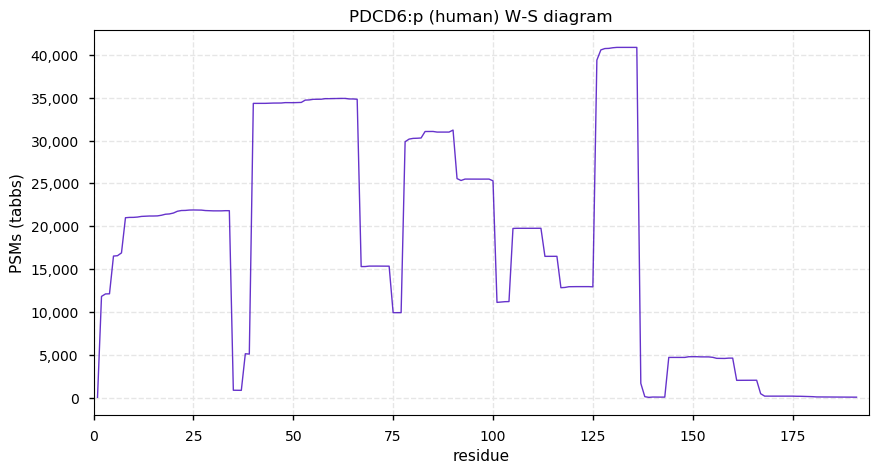
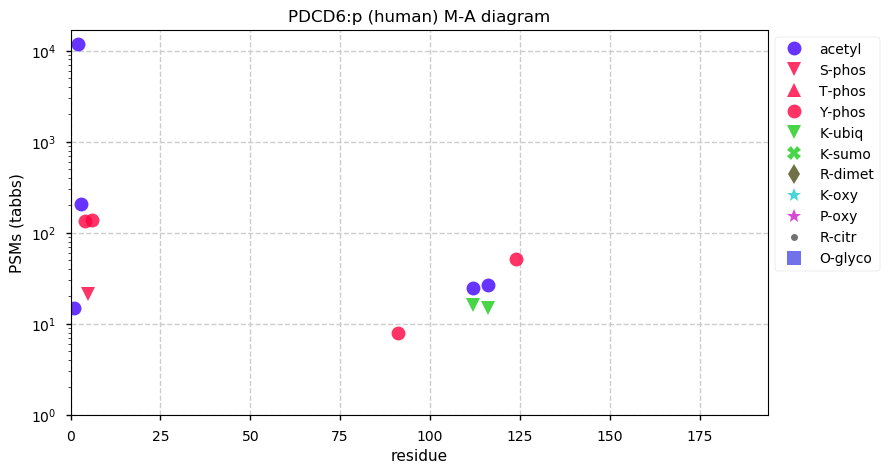
Thu Nov 12 15:33:44 +0000 2020@jwoodgett @epdevilla Reporters here (MB) are starting to take some pity on our equivalent provincial health officer: it is painfully clear that the elected people only show up for short-lived, dopey "good news" (#ReStartMB) & hide behind the PHO the rest of the time.
Thu Nov 12 15:17:41 +0000 2020@ypriverol If you don't mind mouse tissue, you have more options. For example, PXD018140 (Lui JJ Neuropharmacology, 181:108324, 🔗) has a lot of very nice high resolution spectra. It also has a phosphopeptide enrichment of > 95% with no labelling.
Thu Nov 12 13:40:52 +0000 2020PDCD5:p θ(max) = 93. aka TFAR19, MGC9294. Observed in HLA class I & class II peptide experiments. Commonly observed in cell lines & tissues. S119+phosphoryl is observed 60× more often than phosphorylated sites. Unusual density of K+ubiquitinyl/acetyl aPTM acceptors.
Thu Nov 12 13:40:52 +0000 2020>PDCD5:p, programmed cell death 5 (Homo sapiens) 🔗 Small nuclear subunit; CTMs: A2+acetyl; PTMs: 5× S+phosphoryl, Y80+phosphoryl; aPTMs: 7× K+ubiquitinyl/acetyl; SAAVs: none; mature form: 2-125 [29,221×, 159 kTa] #ᗕᕱᗒ 🔗
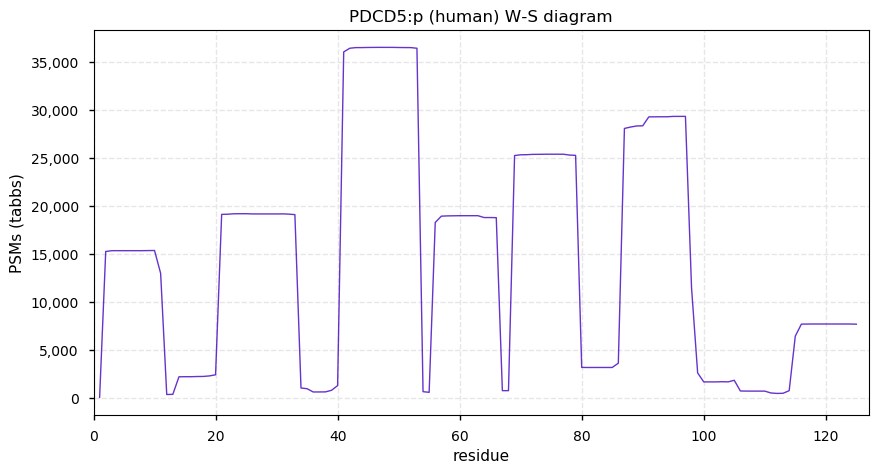
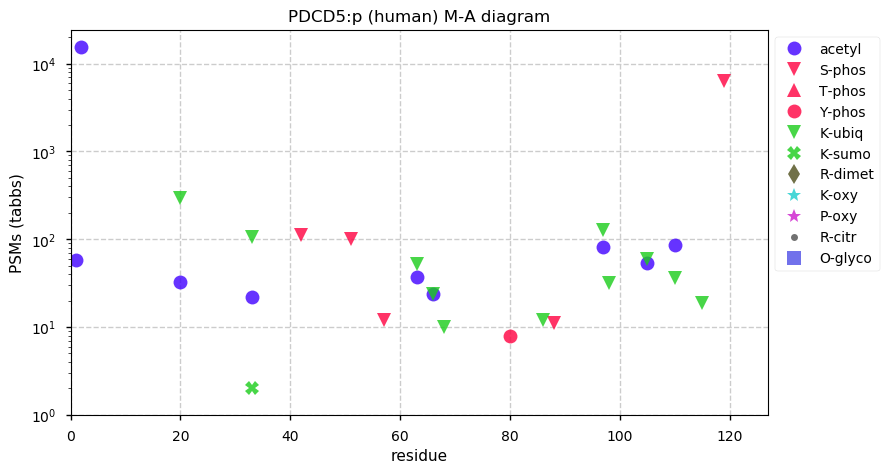
Thu Nov 12 13:39:07 +0000 2020@ypriverol These days healthy tissue phosphorylation data is almost always part of a TMT-type study that includes diseased tissue.
Thu Nov 12 13:36:48 +0000 2020@ypriverol I will look around for you. Most of the healthy tissue phosphorylation data sets come from a period around 2010, when constructing phosphorylation atlases was the rage. The results hold up, but a lot of that data lost in the TRANCHE-to-Massive shift.
Wed Nov 11 20:03:36 +0000 2020@VamsiMootha You guys are probably the right ones to ask: has there been any further insight into why there is no QARS2? I know that KARS1 & GARS1 serve both as cytoplasmic and mitochondrial tRNA synthetases, but I'm not sure what's up with Q.
Wed Nov 11 19:30:50 +0000 2020@UCDProteomics @theoneamit @KentsisResearch I actually do reanalyze DDA data for a living, so I do find it to be interesting, too.
Wed Nov 11 17:48:39 +0000 2020@mjmaccoss @UCDProteomics @theoneamit @KentsisResearch That sounds useful. I frequently harp on the idea that people have to be diligent about test their data for artifacts. Otherwise they can end up with data that is so messy it is hard to see anything through the unanticipated chemistry.
Wed Nov 11 16:34:37 +0000 2020@UCDProteomics @theoneamit @KentsisResearch Is there any way, at the moment, to determine levels of urea or IAA artifacts without doing a DDA experiment?
Wed Nov 11 16:24:47 +0000 2020@theoneamit @UCDProteomics @KentsisResearch Two main reasons in my mind:
1. DIA's main use at the moment is differential quant, so you need a "normal" & "treated/disease" measurement.
2. Any DIA measurement is "warts & all". The chances of someone else reproducing the details is low.
Wed Nov 11 15:54:54 +0000 2020@UCDProteomics @KentsisResearch Their many concern was always that the software would have trouble keeping up with hardware improvements (although history proved that their company's management would be the real problem).
Wed Nov 11 15:48:21 +0000 2020@UCDProteomics @KentsisResearch It was expectation of the guys at Sciex who came up with the idea (Tate & Bonner) that there was so much information in the data that it would take a long time to tease it all out.
Wed Nov 11 15:05:33 +0000 2020@AlexUsherHESA I don't think "win" is really an option. Staying competitive is about the best a country can achieve over time.
Wed Nov 11 14:26:37 +0000 2020PDCD4:p θ(max) = 81. aka H731. Observed in HLA class I peptide experiments (but not in class II). Many of the phosphorylation sites are high occupancy, forming several distinct domains. Commonly observed in cell lines and tissues.
Wed Nov 11 14:26:37 +0000 2020>PDCD4:p, programmed cell death 4 (Homo sapiens) 🔗 Small nuclear protein; CTMs: M1,D2+acetyl; PTMs: 7× K+ubiquitinyl, 38× S,T,Y+phosphoryl; aPTMs: K297+ubiquitinyl/SUMOyl; SAAVs: I36V (8%); mature form: 1,2-469 [28,948×, 191 kTa] #ᗕᕱᗒ 🔗
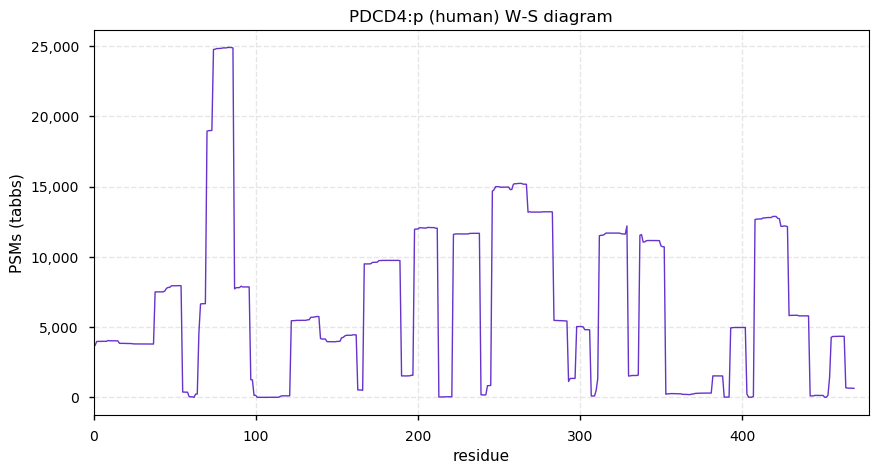
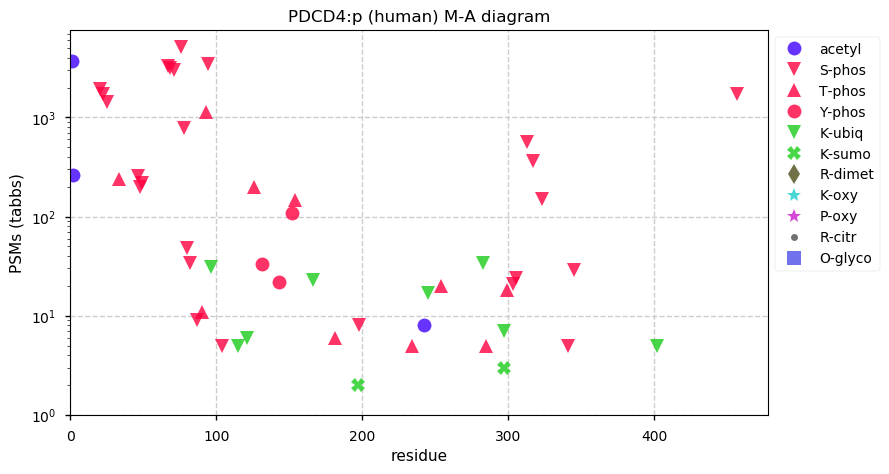
Tue Nov 10 21:38:38 +0000 2020@gangulyteena Also Remembrance Day in Canada & Australia and Veterans Day in the US.
Tue Nov 10 21:16:45 +0000 2020Doing some operating system upgrades: GPMDB will be down for about 30 minutes.
Tue Nov 10 20:37:29 +0000 2020I must admit to being someone who looks at marked up spectra quite a bit, but it rarely definitively answers the specific question that drives me to look in the first place.
Tue Nov 10 20:35:27 +0000 2020Thanks to everyone who participated in the poll. It appears the majority of respondents feel that visual inspection of marked up spectra is not a critical part of proteomics data interpretation.
Tue Nov 10 18:43:10 +0000 2020Eta and Theta 🔗
Tue Nov 10 15:40:39 +0000 2020@bkives The "lockdowns" recently used in Belgium and Holland were very effective at bringing down their case counts.
🔗
Tue Nov 10 15:35:44 +0000 2020I think the way this is shaping up is different than it would have been with the previous generation of proteomics researchers. 🔗
Tue Nov 10 14:19:10 +0000 2020Eta still marches on, at least until it makes landfall & sits still for a while on Wednesday evening. 🔗
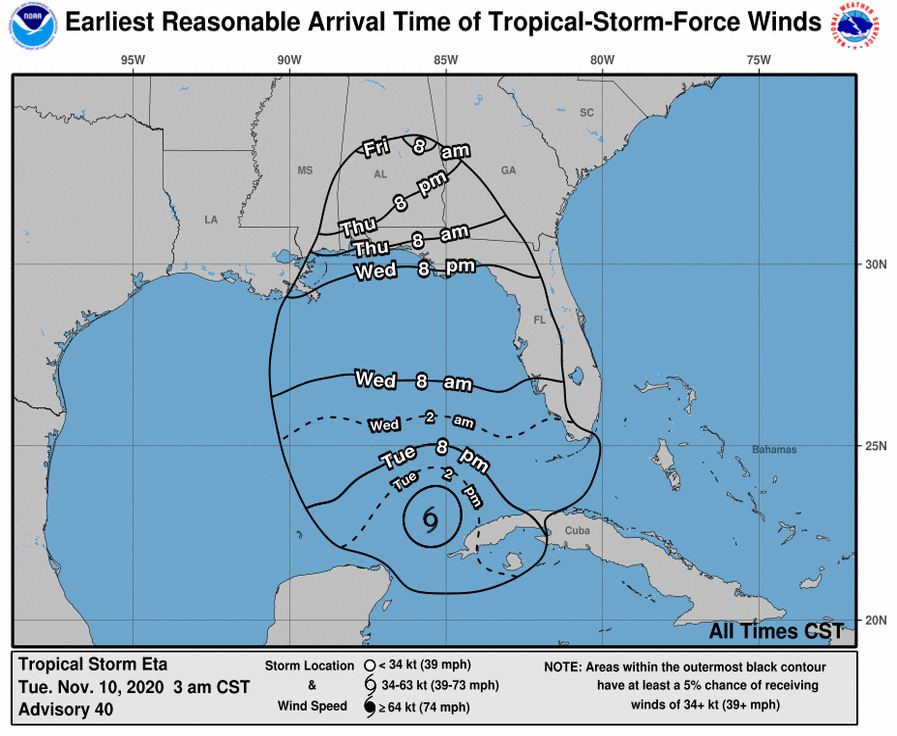
Tue Nov 10 13:03:38 +0000 2020PDCD2:p θ(max) = 53. aka ZMYND7 & RP8. Observed in HLA class I peptide experiments. Apart from its name, very little is known about the details of this protein's function.
Tue Nov 10 13:03:37 +0000 2020>PDCD2:p, programmed cell death 2 (Homo sapiens) 🔗 Small nuclear protein; PTMs: 5× K+ubiquitinyl; aPTMs: K157, K245+ubiquitinyl/SUMOyl; SAAVs: none; mature form: 2-344 [3,094×, 7.5 kTa] #ᗕᕱᗒ 🔗
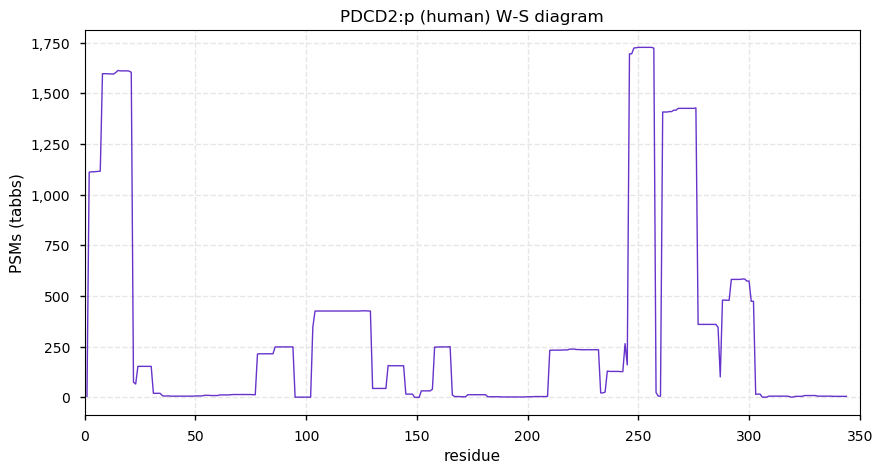
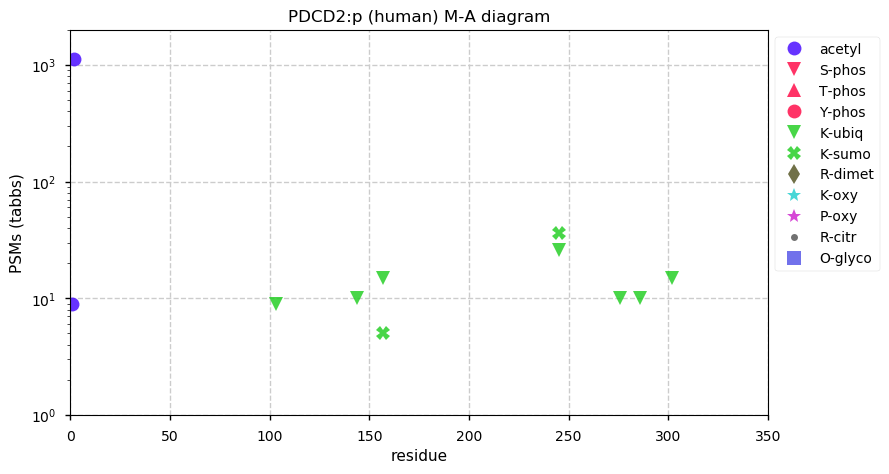
Mon Nov 09 18:54:15 +0000 2020Does looking at marked-up spectra of PSMs really help when interpreting large proteomics datasets or is it mainly a psychological crutch?
Mon Nov 09 16:56:12 +0000 2020Worth reading if you are currently relying on Zoom for confidentiality in meetings:
🔗
Mon Nov 09 14:56:09 +0000 2020Going down to 0% ids on both ends is good for reproducibility.
Mon Nov 09 14:02:55 +0000 2020From PXD021205: just the way you want data to work out in a good LC/MS/MS run. Nearly 40% id rate for most of the run, tapering down to 0% at both ends, with good retention time prediction correlations throughout. 🔗
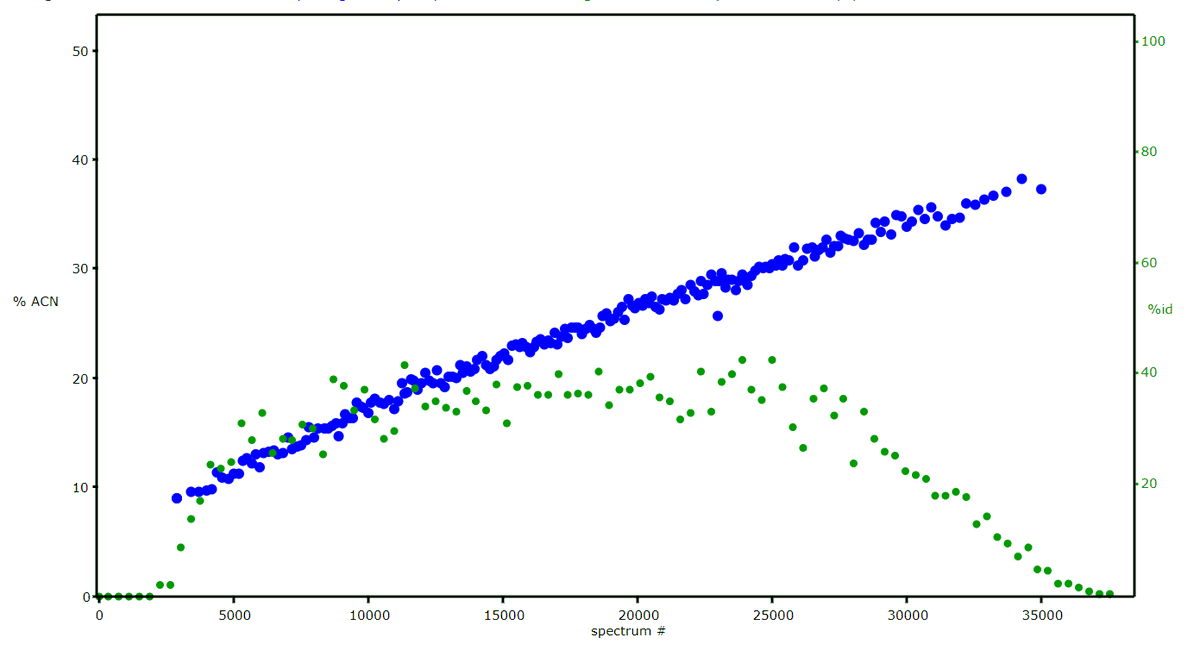
Mon Nov 09 12:55:58 +0000 2020PDCD1:p θ(max) = 13. The domain (79-112) has been observed in urine. The protein has been observed in lymphocytes, but rarely. This protein is a drug target, because of its immuno-regulatory role in T-cell apoptosis.
Mon Nov 09 12:55:58 +0000 2020>PDCD1:p, programmed cell death 1 (Homo sapiens) 🔗 Small plasma membrane protein; PTMs: T234, S261, S285+phosphoryl; SAAVs: none; mature form: 24?-288 [361×, 0.74 kTa] #ᗕᕱᗒ 🔗
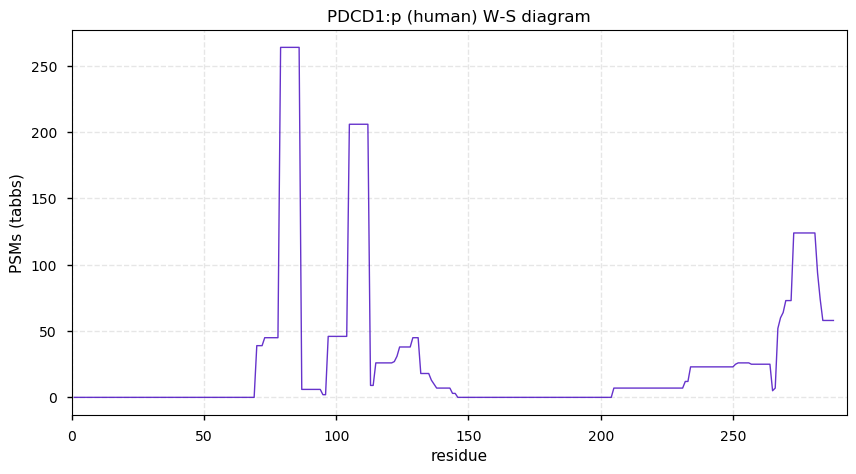
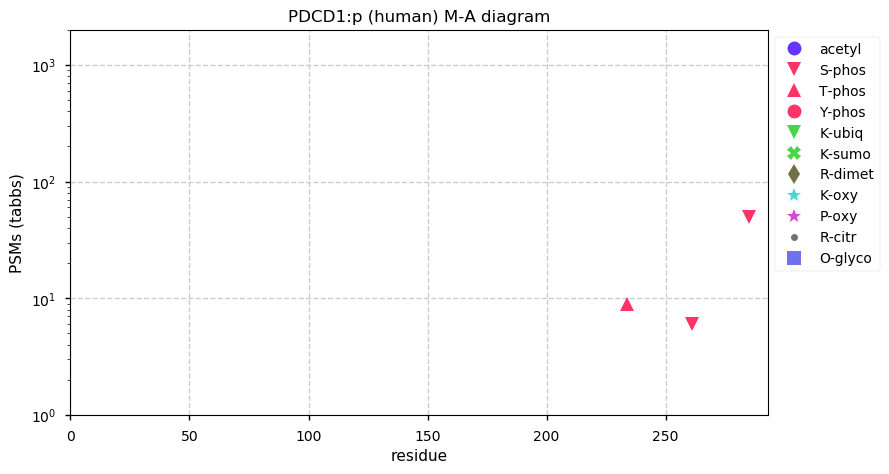
Mon Nov 09 01:39:57 +0000 2020Eta is the honey badger of storms: it is still running and causing storm conditions in Cuba and Florida 🔗
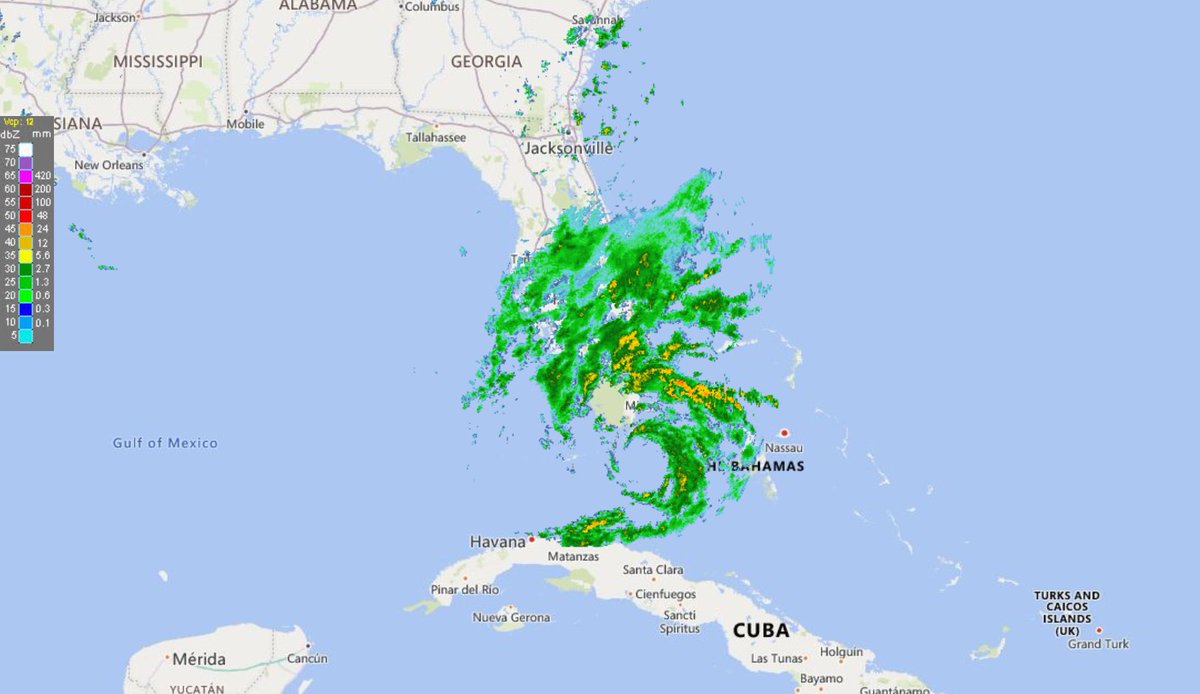
Sun Nov 08 14:18:27 +0000 2020@theoneamit Mine is only 11 months old.
Sun Nov 08 14:14:28 +0000 2020aceE:p Observations of domains starting at the protein N-terminus, e.g. (1-22), are interesting. Prokaryote initiator residues are normally N-formylmethionine, which are removed cotranslationally. aceE:p may be an exception or an artifact of strain variation.
Sun Nov 08 14:14:27 +0000 2020aceE:p θ(max) = 77. Necessary for nascient peptide elongation during ribosomal synthesis. Commonly detected in clinical unfractionated urine samples obtained from patients with P. aeruginosa UTIs.
Sun Nov 08 14:14:27 +0000 2020>aceE:p, pyruvate dehydrogenase subunit E1 (Pseudomonas aeruginosa PAO1) 🔗| Midsized intracellular subunit; PTMs: 8× K+acetyl; SAAVs: unknown; mature form: 1-822 [795×, 30 kTa] #ᗕᕱᗒ 🔗
Sat Nov 07 15:22:06 +0000 2020What would be your choice for the most self-important household appliance? I nominate the microwave oven: that thing will beep forever unless you come over and do what it wants you to do.
Sat Nov 07 14:23:13 +0000 2020As in so many things as a Canadian, I find myself torn between using the British spelling I grew up with or the American spelling my software screams at me to use.
Sat Nov 07 13:32:33 +0000 2020fusA1:p θ(max) = 96. Necessary for nascient peptide elongation during ribosomal synthesis. Commonly detected in clinical unfractionated urine samples obtained from patients with P. aeruginosa UTIs.
Sat Nov 07 13:32:33 +0000 2020>fusA1:p, elongation factor G (Pseudomonas aeruginosa) 🔗| Midsized intracellular subunit; PTMs: 7× K+acetyl; SAAVs: unknown; mature form: 2-706 [990×, 71 kTa] #ᗕᕱᗒ 🔗
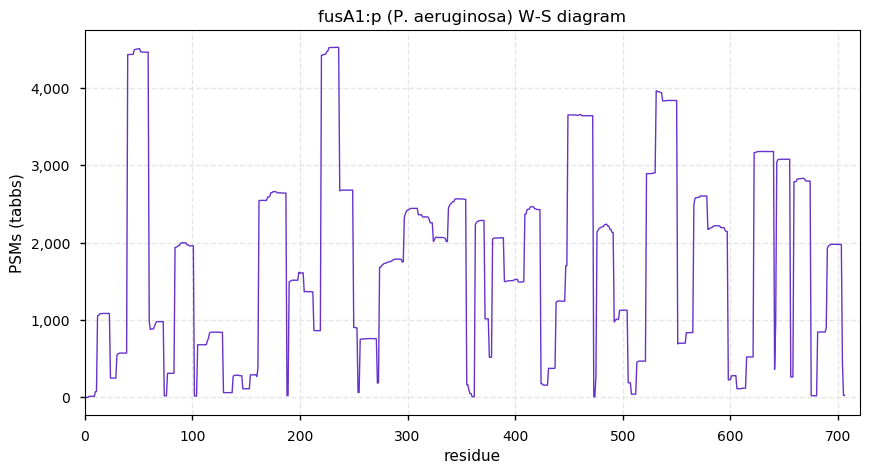
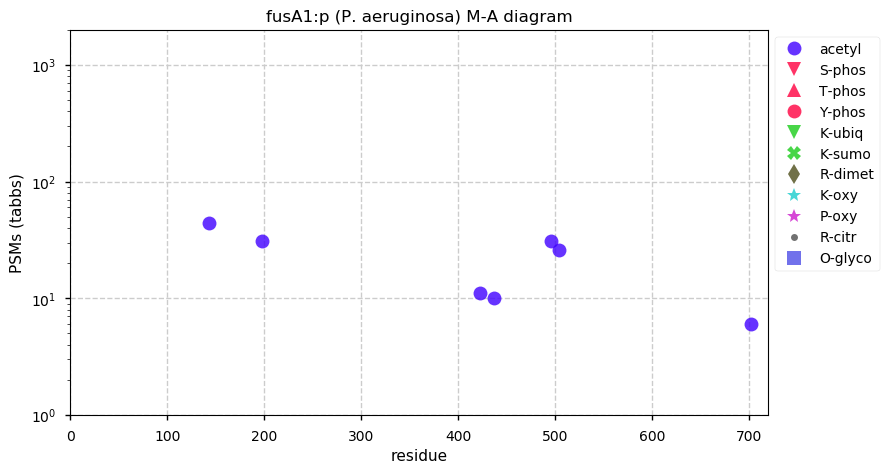
Fri Nov 06 20:55:46 +0000 2020@Smith_Chem_Wisc Bovine or porcine trypsin and/or endo Lys C?
Fri Nov 06 15:43:30 +0000 2020@AlexUsherHESA While still finding time to make a 1940's style reference to the women in his life.
Fri Nov 06 15:33:57 +0000 2020@pwilmarth @Sci_j_my By occupancy, I mean the number of modifications associated with a particular PSM. For example, does the algorithm treat a PSM with 5 modifications the same as a PSM with only 1.
Fri Nov 06 15:21:26 +0000 2020@Sci_j_my @pwilmarth Do any of the target-decoy algorithms segregate PSMs by peptide length, parent ion charge or PTM occupancy?
Fri Nov 06 14:52:16 +0000 2020I hope all Canadians remember this & chose to go to a different grocery store
🔗
Fri Nov 06 12:41:00 +0000 2020oprF:p θ(max) = 92. Homologous to pore forming proteins, but suspected of many other functions. Commonly detected in clinical unfractionated urine samples obtained from patients with P. aeruginosa UTIs.
Fri Nov 06 12:41:00 +0000 2020>oprF:p, porin (Pseudomonas aeruginosa) 🔗| Small extracellar/membrane subunit; PTMs: none; SAAVs: unknown; mature form: 25-350 [969×, 93 kTa] #ᗕᕱᗒ 🔗
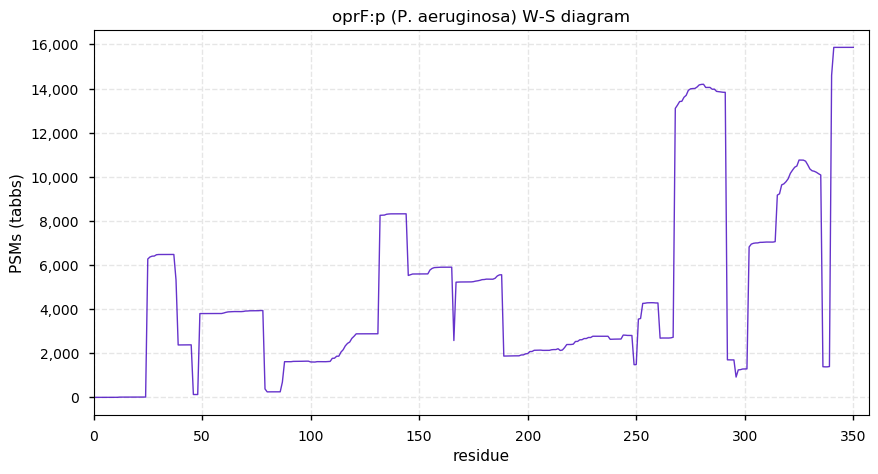
Thu Nov 05 14:11:31 +0000 2020Looking at a lot of data & papers has lead me to formulate a mangled version of a quote attributed to Mark Twain:
'Omics data doesn't exactly replicate itself, but at least it should rhyme.
Thu Nov 05 13:00:12 +0000 2020rpoB:p θ(max) = 87. A subunit in the protein complex that translates bacterial DNA into mRNA. All of the subunits of this complex are commonly detected in clinical unfractionated urine samples obtained from patients with P. aeruginosa UTIs.
Thu Nov 05 13:00:12 +0000 2020>rpoB:p, DNA-directed RNA polymerase subunit beta (Pseudomonas aeruginosa) 🔗| Large intracellular subunit; PTMs: 5 low occupancy K+acetyl; SAAVs: unknown; mature form: 2-1357 [946×, 84 kTa] #ᗕᕱᗒ 🔗
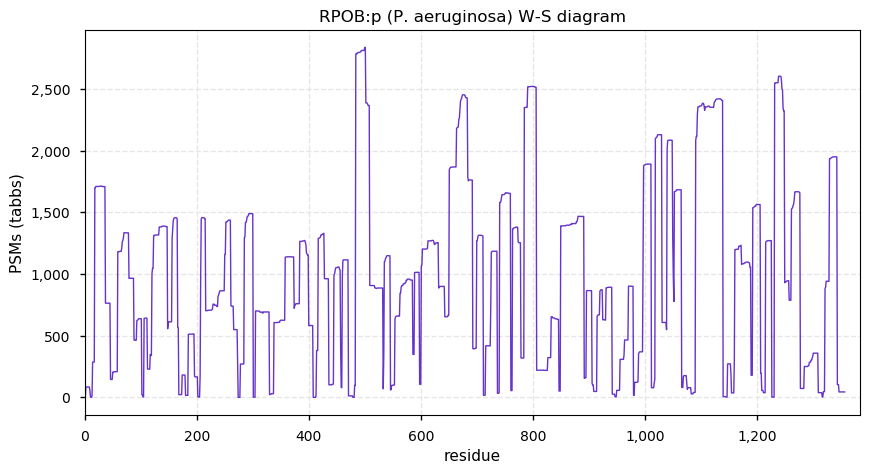
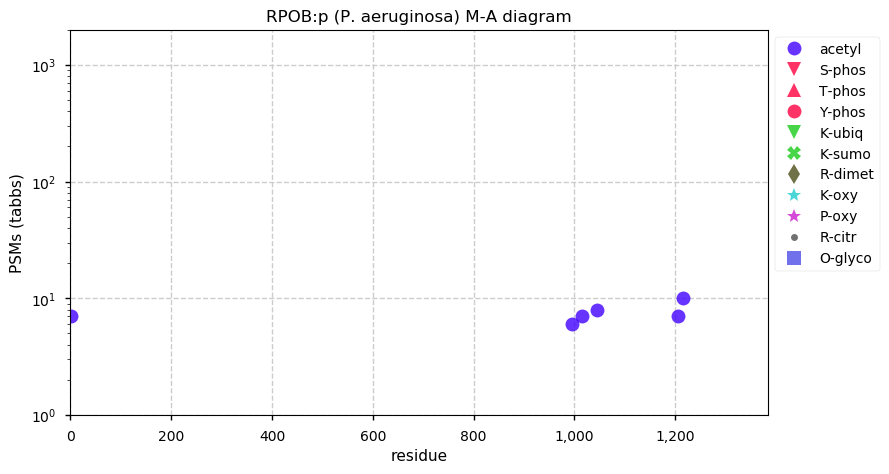
Wed Nov 04 22:04:10 +0000 2020@Sci_j_my @FrankSobott @JohnRYatesIII Glass is an insulator and quartz is conductive.
Wed Nov 04 16:34:01 +0000 2020But then, I wonder about strange things.
Wed Nov 04 16:33:30 +0000 2020I have wondered from time-to-time whether it is possible to prevent PA UTIs from happening in hospitalized patients in urinary biomarker studies, as it seems to be a very widespread problem.
Wed Nov 04 16:27:45 +0000 2020I've been following these stats for a while, but now that the population is generally engaged, the Canadian provincial numbers in terms of the fraction of the population seem out of line with the public discussion 🔗
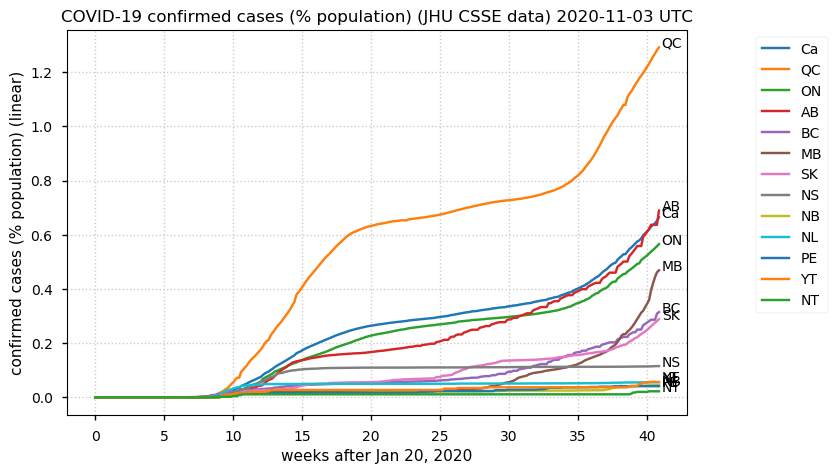
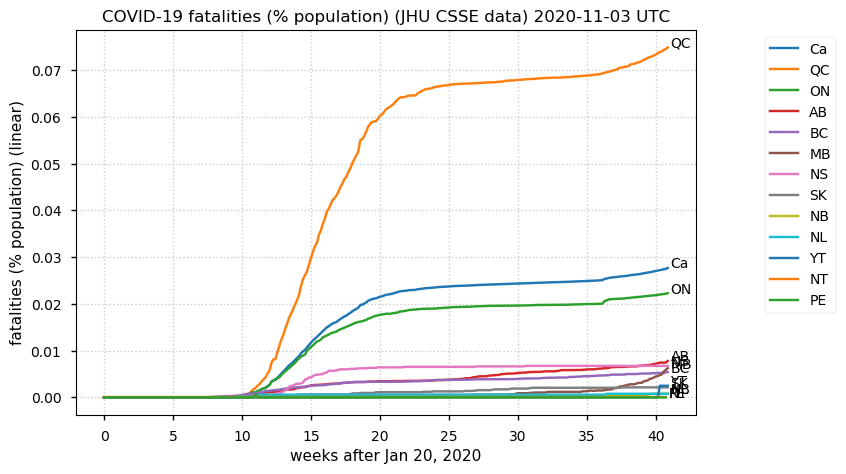
Wed Nov 04 15:46:53 +0000 2020Outside of hospitals, E. coli is the usual cause of UTIs & the source of detectable bacterial proteins in unfractionated urine. To a lesser extent, E. faecalis, K. pneumoniae, P. mirabilis, G. vaginalis & Lactobacillus sp. proteins may also be detectable.
Wed Nov 04 13:55:04 +0000 2020@dtabb73 Sorry, I made a mistake in the protein du jour tweet: I had to delete it and reload the corrected entry.
Wed Nov 04 13:53:32 +0000 2020Detection of P. aeruginosa DNAK:p in unfractionated urine is a good indicator of a hospital-induced urinary tract infection.
Wed Nov 04 13:53:32 +0000 2020DNAK:p θ(max) = 86. Similar function as HSP70 chaperones in eukaryotes. It is the answer to the question: "Which P. aeruginosa protein are you most likely to find in unfractionated human urine?"
Wed Nov 04 13:53:32 +0000 2020>DNAK:p, molecular chaperone DnaK (Pseudomonas aeruginosa) 🔗| Midsized intracellular subunit; PTMs: 13 K+acetyl; SAAVs: unknown; mature form: 2-637 [1,579×, 65 kTa] #ᗕᕱᗒ 🔗
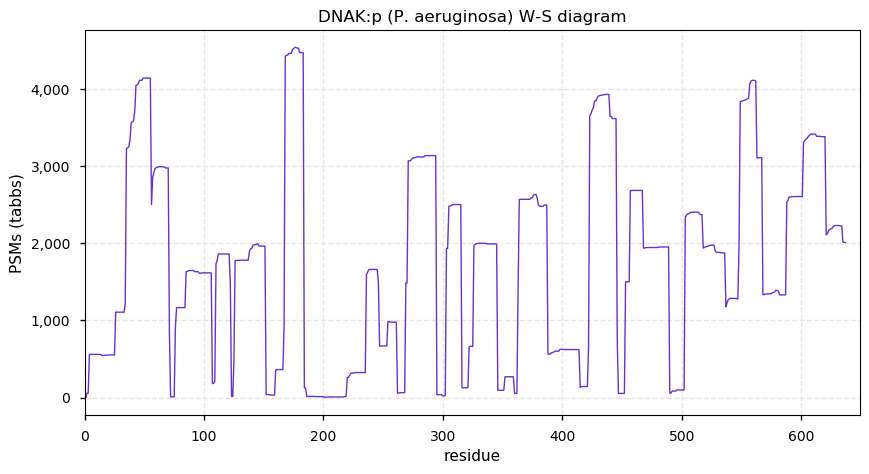
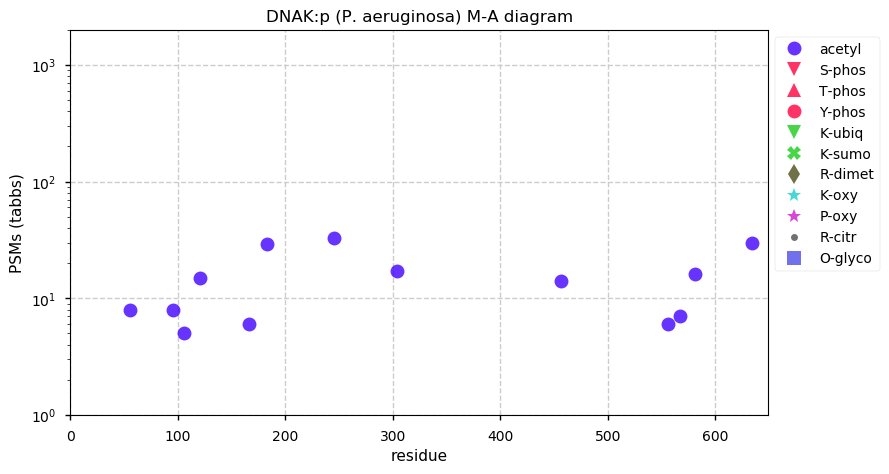
Tue Nov 03 20:57:36 +0000 2020@mikefeigin COVID-19 + SARS CoV2
Tue Nov 03 19:29:47 +0000 2020@Sci_j_my But I don't really recommend doing that experiment (until the cost per scan gets a lot lower). Really nice stuff like this study 🔗 doesn't require that type of brute force, just clear thinking, 6 LC/MS/MS runs & a few minutes of PSM id'ing.
Tue Nov 03 18:42:02 +0000 2020@marcoyannic 🔗
Great site if you at all interested in weather.
Tue Nov 03 18:17:09 +0000 2020🔗
Tue Nov 03 18:16:34 +0000 2020The wind field of the class 4 Hurricane Eta (220 km/h winds) making landfall in Nicaragua today ... 🔗
Tue Nov 03 18:04:30 +0000 2020@bkives Sounds like a clip that will feature prominently in opposition ads during the next election.
Tue Nov 03 17:12:27 +0000 2020@Sci_j_my Ask your department head's opinion. Assuming that you want to advance in some type of tenure track, your department head's take on things is pretty much all that matters.
Tue Nov 03 16:44:26 +0000 2020@Sci_j_my A truly thorough proteomics experiment on a cell lysate would require ~10 million PSMs (yet to be done).
Tue Nov 03 15:46:55 +0000 2020@Sci_j_my It is also collapsed down onto the "Highlander" version of the reference proteome (so ~20,000 protein sequences).
Tue Nov 03 15:43:30 +0000 2020@Sci_j_my This statement refers to what I know about H. sapiens samples.
Tue Nov 03 15:41:38 +0000 2020@Sci_j_my A state-of-the-art MS/MS experiment on a cell line lysate will end up with 0.5–0.6 million PMS, representing 0.2–0.3 million unique peptide sequences. The total number of available, detected unique peptides (incl. detected PTMs as variants) is about 5 million.
Tue Nov 03 14:35:04 +0000 2020@Sci_j_my Matthias can be a little over-the-top about things like that. You shouldn't take him too seriously.
Tue Nov 03 13:24:04 +0000 2020P. aeruginosa urinary infections are very common in catheterised patients and it can be a significant confounder in differential analysis unless urinary proteomics data is tested for the presence of bacterial proteins.
Tue Nov 03 13:24:03 +0000 2020PIV:p θ(max) = 64. Necessary protease for the species quorum sensing system. It is the answer to the question: "Which P. aeruginosa protein are you most likely to find in human urinary extracellular vesicles?"
Tue Nov 03 13:24:03 +0000 2020>PIV:p, protease IV (Pseudomonas aeruginosa) 🔗| Small extracellular protease; PTMs: none; SAAVs: unknown; mature form: 25-462 [379×, 26 kTa] #ᗕᕱᗒ 🔗
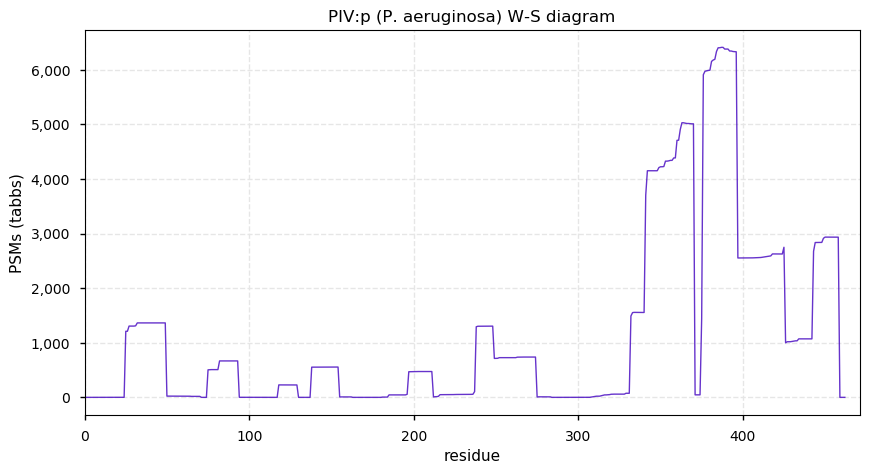
Mon Nov 02 18:54:16 +0000 2020@LindnerLab @idpemery But list them all. Some of the most successful academics I know list absolutely everything in their CV. Some things may seem trivial, but if you got up and spoke, list it.
Mon Nov 02 18:00:42 +0000 2020@zacmcd77 @lkpino @sophi_angie I really do sympathize: obtaining good clinical samples is difficult. But if you want to do any type of differential analysis between tissue samples, they really have to be samples of very similar things.
Mon Nov 02 15:31:50 +0000 2020Thanks to everyone who participated in the poll. It would seem that my fellow travelers share a low opinion of the current status of peer review, at least as applied to 'omics-type experiments.
Mon Nov 02 14:45:18 +0000 2020@sophi_angie Just analyzing the data. If the top 10 (in terms of assigned PSMs) are blood proteins (e.g. albumin, hemoglobin, transferrin, apolipoproteins, immunoglobulins), then it isn't a very good solid tissue sample.
Mon Nov 02 13:55:23 +0000 2020Still 1 hour to vote on this one ... 🔗
Mon Nov 02 13:54:45 +0000 2020This week's cold weather break-out is east of last week's: it has moved from the Great Plains to south of the Great Lakes. 🔗
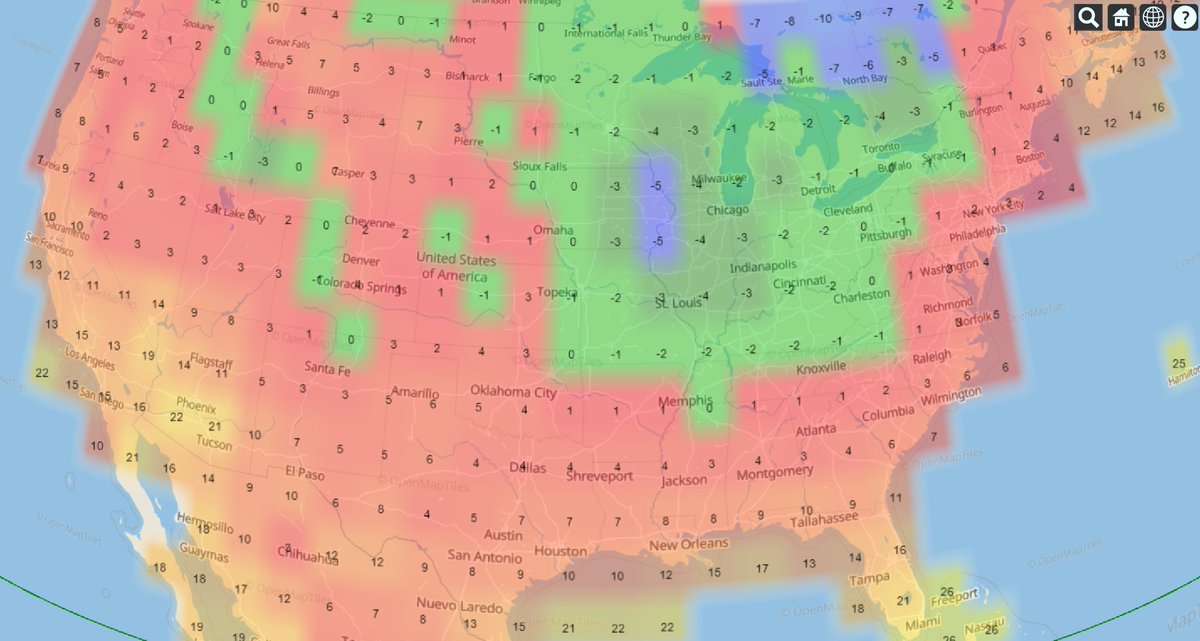
Mon Nov 02 12:47:41 +0000 2020PRPF6:p θ(max) = 69. aka TOM, bB152O15.1, ANT-1, U5-102K, Prp6, hPrp6, SNRNP102, RP60, C20orf14. Found in HLA class I experiments only. 13 Half A Tetratricopeptide repeat (HAT) domains. Very rarely observed in biological fluids.
Mon Nov 02 12:47:41 +0000 2020>PRPF6:p, pre-mRNA processing factor 6 (H. sapiens) 🔗 Midsized nuclear subunit; aPTMs: K178,K341,K618,K793+acetyl/ubiquitinyl/SUMOyl; PTMs: 37 ST+phosphoryl, R23+dimethyl; SAAVs: none; mature form: 1,2-941 [38,177×, 327 kTa] #ᗕᕱᗒ 🔗
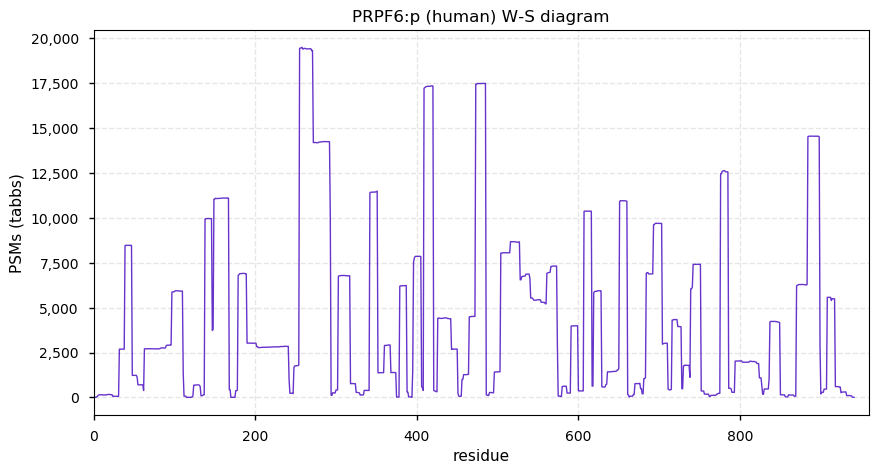
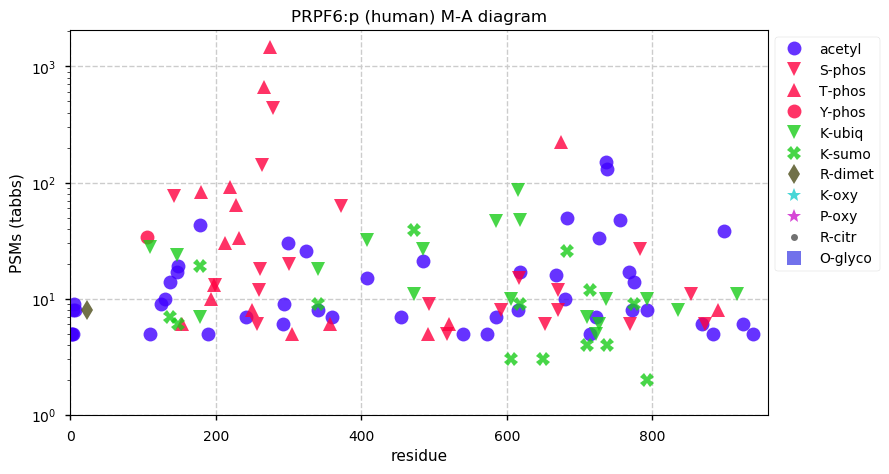
Sun Nov 01 15:25:08 +0000 2020I hate it when I go to the trouble of downloading a published data set involving clinical tissue samples only to discover that it is just blood with a dash of tissue. Is the current review process:
Sun Nov 01 14:22:00 +0000 2020DDX46:p θ(max) = 78. aka KIAA0801, FLJ25329, PRPF5, Prp5. Found in HLA class I experiments only. Low complexity domain (3-120) enriched in R & S; DEAD domain (529-532). Very rarely observed in biological fluids.
Sun Nov 01 14:21:59 +0000 2020>DDX46:p, DEAD-box helicase 46 (H. sapiens) 🔗 Large nuclear subunit; PTMs: 75 K+acetyl, 15 K+ubiquitinyl, 10 K+SUMOyl, 88 STY+phosphoryl; SAAVs: none; mature form: 2-1031 [41,612×, 956 kTa] #ᗕᕱᗒ 🔗
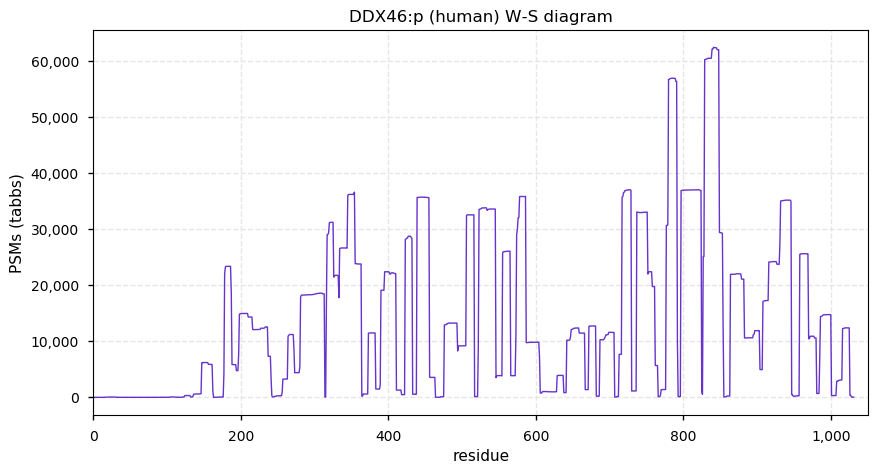
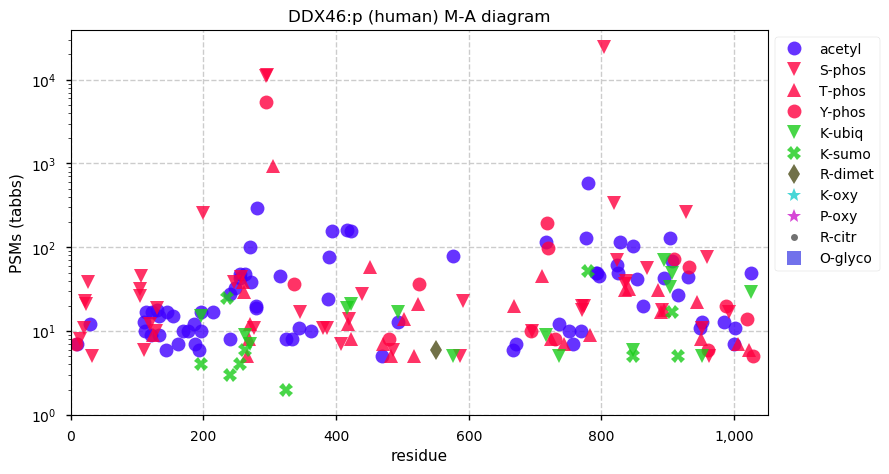
Sat Oct 31 16:29:12 +0000 2020@slavov_n @BiswapriyaMisra @JProteomeRes It is good, interesting data and merits careful examination. But their claims about "quantitation" & "10,438 proteins" are very enthusiastic academic hyperbole.
Sat Oct 31 15:34:19 +0000 2020@neely615 @ucdmrt As I have mentioned before, I have found the descriptions found in "Methods" sections to be of questionable value, so I have no problem with the "mistake" idea, per se. To the best of my knowledge, no one has adopted this method (incl. the authors).
Sat Oct 31 13:28:48 +0000 2020PRPF3:p θ(max) = 69. aka Prp3, hPrp3, SNRNP90, RP18. Found in HLA class I experiments only. Four high occupancy S+phosphoryl acceptors. Very rarely observed in biological fluids. Observed in samples with nucleated cells.
Sat Oct 31 13:12:51 +0000 2020>PRPF3:p, pre-mRNA processing factor 3 (H. sapiens) 🔗 Midsized nuclear subunit; PTMs: 37 K+acetyl, 35 S/T+phosphoryl, 4 K+ubiquitinyl, 6 K+SUMOyl; SAAVs: none; mature form: 2-683 [27,242×, 180 kTa] #ᗕᕱᗒ 🔗
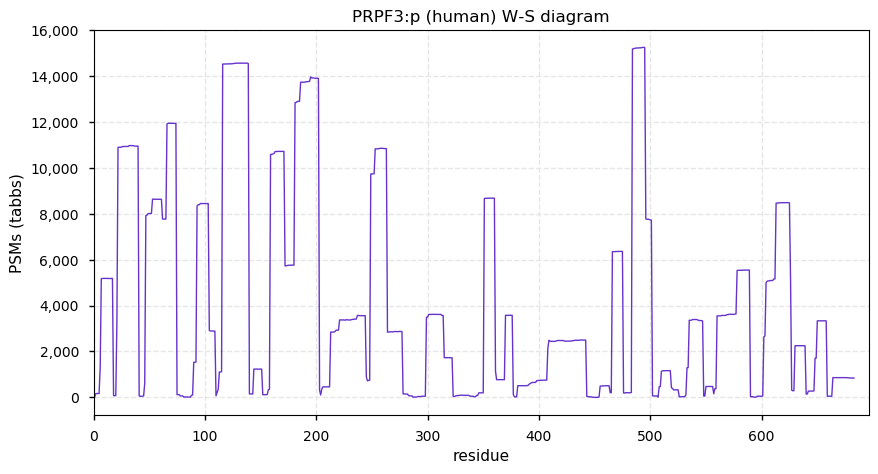
Fri Oct 30 17:44:38 +0000 2020I nominate PXD004452's 39 fraction trypsin digest experiment (🔗) to be the "best" HeLa cell proteomics data set yet.
Fri Oct 30 16:18:53 +0000 2020I'm looking through a large, multi-tissue data set & much to my surprise the tissue labeling in the data file names matches the sets of proteins found for all of the samples! 🥳👍⭐️
Fri Oct 30 12:40:40 +0000 2020NEXRAD ground clutter illustrating a cold front (& some New England coastal fog) 🔗
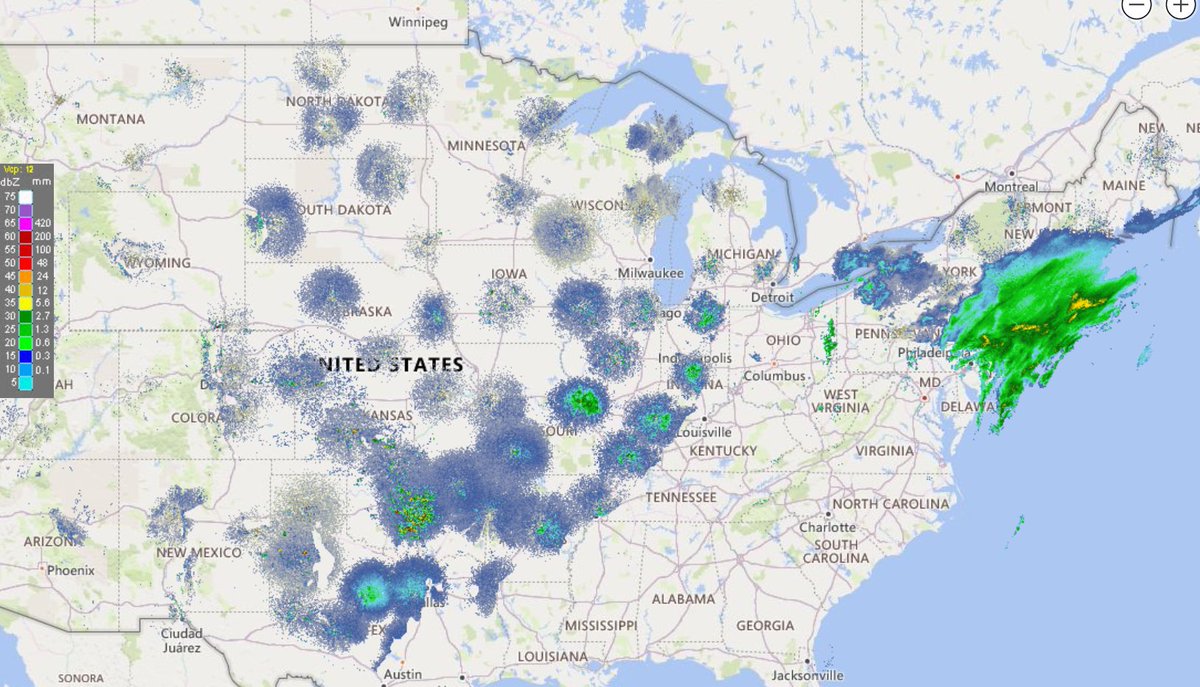
Fri Oct 30 12:30:10 +0000 2020DHX16:p θ(max) = 56. aka DBP2, Prp2, PRPF2, DDX16. Found in HLA class I experiments only. Two high occupancy phospho-domains near the N-terminus. Very rarely observed in biological fluids. DEAH domain located at (520-523). Present in samples with nucleated cells.
Fri Oct 30 12:30:09 +0000 2020>DHX16:p, DEAH-box helicase 16 (H. sapiens) 🔗 Large nuclear enzyme; PTMs: A2+acetyl, 40 S/T+phosphoryl, 9 K+ubiquitinyl, 4 K+SUMOyl; SAAVs: D566G (2%), V644A (1%); mature form: 2-688 [28,025×, 125 kTa] #ᗕᕱᗒ 🔗
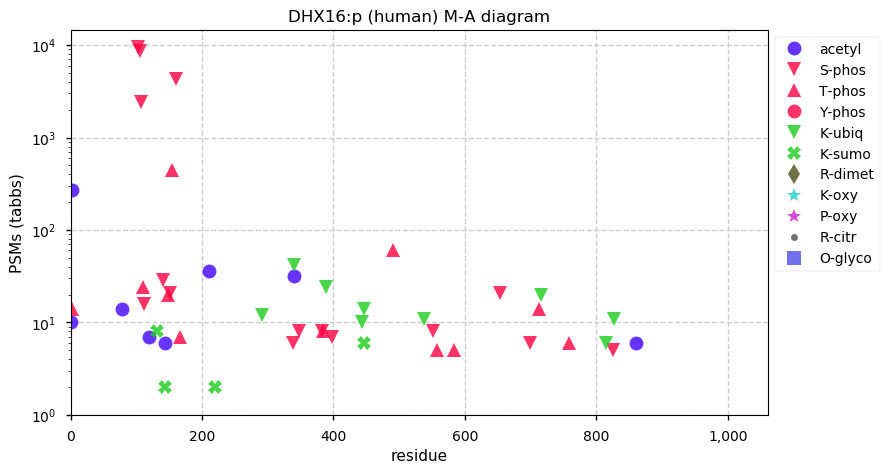
Thu Oct 29 19:38:29 +0000 2020@gangulyteena My recommendation would be KIF5B.
Thu Oct 29 15:26:36 +0000 2020The film trucks for a Hallmark MOW shoot are cluttering up the neighbourhood this morning. As though the friggin 'Burden of Truth' filming wasn't nuisance enough ...
Thu Oct 29 14:44:53 +0000 2020@kady He was only 5 at the time, so he wouldn't remember just how out-of-control the situation was following Laporte's kidnapping and murder.
Thu Oct 29 14:16:59 +0000 2020Once more I have had it proved to me that hopefulness is never as good as a backup copy.
Thu Oct 29 14:09:55 +0000 2020@pwilmarth @thermosci I guess things are better here in the land of winter, because it is only taking me Euler's number times as long here ... 🇨🇦
Thu Oct 29 12:32:16 +0000 2020SRPK2:p θ(max) = 76. aka SFRSK2. Found in HLA class I experiments & very rarely in class II. Long tryptic peptides (383-442), (445-489) & (552-594) make these domains difficult to observe.
Thu Oct 29 12:32:16 +0000 2020>SRPK2:p, SRSF protein kinase 2 (H. sapiens) 🔗 Midsized nuclear enzyme; PTMs: 5 K+acetyl/ubiquitinyl, 40 S/T+phosphoryl; SAAVs: none; mature form: (1,2,3)-688 [21,669×, 82 kTa] #ᗕᕱᗒ 🔗
Wed Oct 28 18:53:54 +0000 2020@doctorow It has many formal similarities with some of the grant applications I have reviewed for NCI, although the language in the ad has fewer caveats than the average grant.
Wed Oct 28 18:51:20 +0000 2020@bkives It would be useful for those of us outside of government to hear a clear statement from the Minister on this issue rather than one filtered through a spokesperson who has no direct responsibility.
Wed Oct 28 13:15:52 +0000 2020It may be 0 °C in Winnipeg, MB this morning (no surprise for this time of year), but it is only 3 °C in San Antonio & Austin, TX!
Wed Oct 28 12:57:40 +0000 2020SRPK1:p induced serine phosphorylation is believed to determine whether the modified proteins are localized, e.g., SRSF1 +phosphoryl is moved to the nucleus, while -phosphoryl it remains in the cytoplasm.
Wed Oct 28 12:57:39 +0000 2020SRPK1:p is one of three kinases believed to be primarily involved in multiple serine phosphorylations of serine/arginine rich splicing factors & other proteins with S/R enriched low complexity domains.
Wed Oct 28 12:57:39 +0000 2020SRPK1:p θ(max) = 74. aka SFRSK1. Found in HLA class I experiments & very rarely in class II. Long tryptic peptides (378-450) & (519-561) make these domains difficult to observe.
Wed Oct 28 12:57:39 +0000 2020SRPK1:p, SRSF protein kinase 1 (H. sapiens) 🔗 Small nuclear enzyme; PTMs: 2 high occupancy phosphodomains, 2 K+ubiquitinyl/SUMOyl, 6 K+ubiquitinyl; SAAVs: none; mature form: 1-655 [27,756×, 143 kTa] #ᗕᕱᗒ 🔗
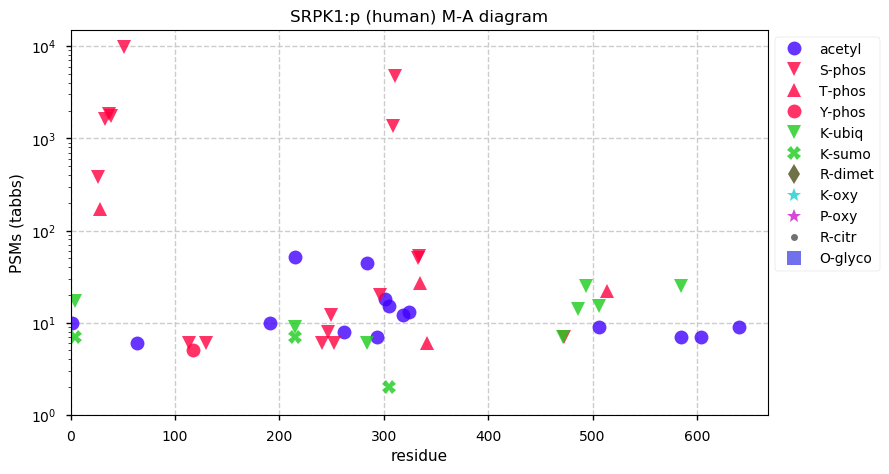
Wed Oct 28 12:24:47 +0000 2020A story that hasn't really gone away ... 🔗
Wed Oct 28 12:20:57 +0000 2020@PastelBio This is true.
Tue Oct 27 20:08:04 +0000 2020Dealing with large numbers of large files both requires & tests your patience.
Tue Oct 27 16:12:29 +0000 2020But I wish the HGVS documentation (🔗) didn't appear to stress using 3 letter symbols—even though it allows 1 letter—as it tends to annoy proteomics specialists.
Tue Oct 27 16:10:26 +0000 2020I really like the HGVS notation for SAAVs: anyone who is interested in the subject can figure out what it means without having to look it up more than once in a lifetime.
Tue Oct 27 15:42:15 +0000 2020@AraValentini Same for we OUS North Americans.
Tue Oct 27 15:09:02 +0000 2020@ypriverol I made the capability available for a while, but it takes quite a bit of effort to keep it up-to-date, only a few other people were interested & journals were completely against the notion. So, I really just do it as a hobby now (but NO pipelines involved).
Tue Oct 27 14:53:52 +0000 2020Big breakout of cold weather across the North American central plains this morning, from the Rocky Mountains in the west to a line running from Chicago, IL to Austin, TX in the east. 🔗
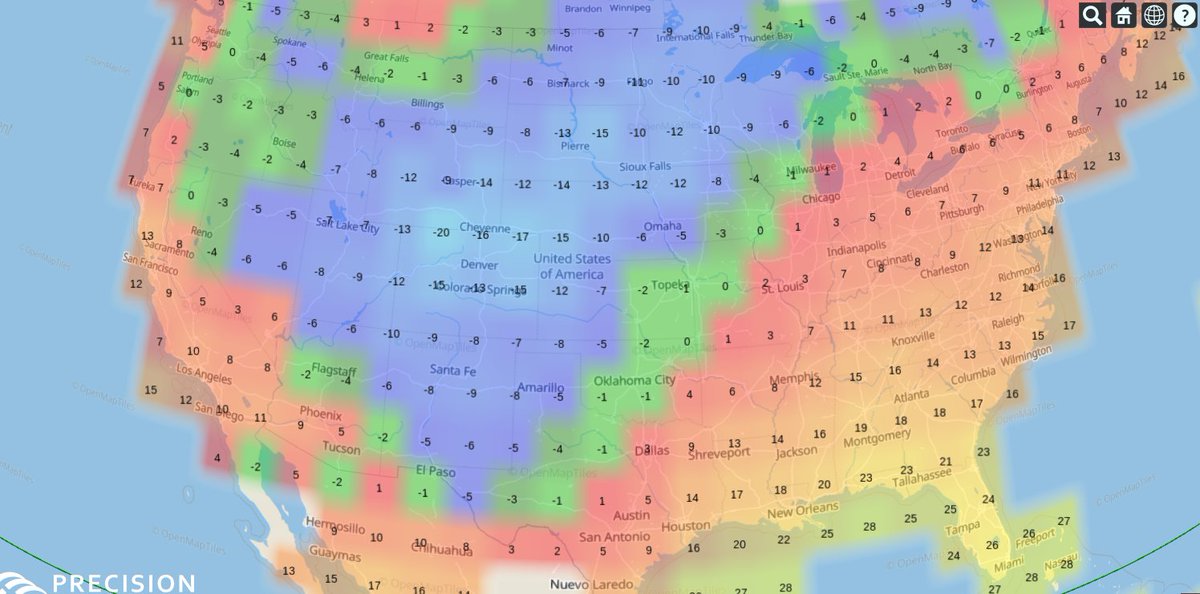
Tue Oct 27 14:46:05 +0000 2020The SAAVs are 0.5% of the PSMs and including such favorites as:
NID1:p.Q1113R (maf=0.3);
SEPTIN9:p.M576V (maf=0.9); and
DDX18:p.T94S (maf=0.4).
Tue Oct 27 14:37:03 +0000 2020The data is also very well suited for SAAV detection.
Tue Oct 27 14:27:12 +0000 2020Some of the clonal organiod proteome runs in PXD016582 (🔗) are excellent. Very clean sample preps & nice sharp chromatography with good recovery of R+dimethyl, S/T/Y+phosphoryl & H+diphthamide (which is rare!).
Tue Oct 27 13:29:06 +0000 2020It is also not up to your users to contact you if an error occurs (particularly if this site is throwing meaningless error messages): you should be logging this stuff & review it periodically.
Tue Oct 27 13:05:58 +0000 2020"Oops, something went wrong" is not an error message: it is just insulting your users.
Tue Oct 27 12:26:34 +0000 2020SON:p θ(max) = 40. aka DBP-5, NREBP, KIAA1019, BASS1, FLJ21099, FLJ33914, C21orf50. Found in HLA class I experiments only. The relatively low value for θ(max) is caused by low complexity domains that cannot be easily observed: (367-909), (1123-1485) & (1798-2048).
Tue Oct 27 12:26:34 +0000 2020SON:p, SON DNA binding protein (H. sapiens) 🔗 Large nuclear subunit; PTMs: highly modified with STY+phosphoryl, as well as K+acetyl/ubiquitinyl/SUMOyl; SAAVs: M1502I (15%), R1575C (72%); mature form: 2-2426 [40,954×, 605 kTa] #ᗕᕱᗒ 🔗
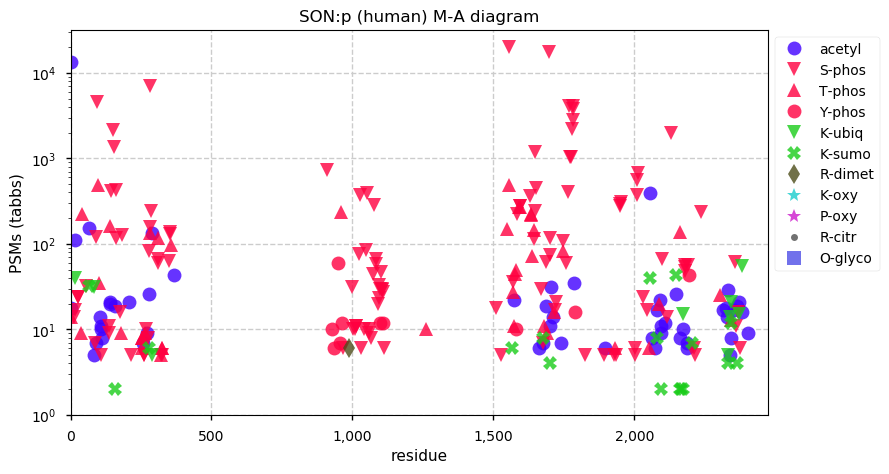
Mon Oct 26 20:14:24 +0000 2020@PastelBio Good article. Anyone interested in proteomics should read it.
Mon Oct 26 18:32:21 +0000 2020I see a lot of activity on Twitter regarding the invasive aspects of remote proctoring software: open book exams fix up most of the issues and require little, if any, surveillance.
Mon Oct 26 14:41:17 +0000 2020@MattWFoster @Smith_Chem_Wisc That is pretty much exactly what I did here, but it is still a moving target (although this is well-washed tissue rather than biofluids).
Mon Oct 26 13:00:57 +0000 2020🔗 interesting and on point for the understanding S/R proteins, splicing dynamics and the interpretation of well-established antibody-based identifications
Mon Oct 26 12:32:14 +0000 2020SRSF12:p θ(max) = 33. aka SRrp35, SFRS19, SFRS13B. Found in HLA class I & II experiments. (108-261) is enriched in R, K, H & S, but without the extended repetitive domains found in other SRSF proteins. Its PTM pattern is also different from other SRSF proteins.
Mon Oct 26 12:32:13 +0000 2020SRSF12:p, serine and arginine rich splicing factor 12 (H. sapiens) 🔗 Small nuclear subunit; PTMs: Y55+phosphoryl, (S105,S199,S219,S223)+phosphoryl; SAAVs: none; mature form: 2-261 [20,095×, 58 kTa] #ᗕᕱᗒ 🔗
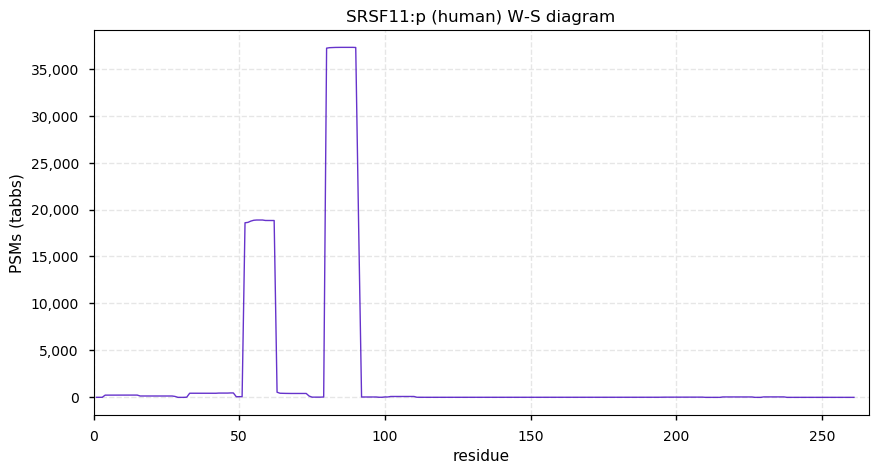
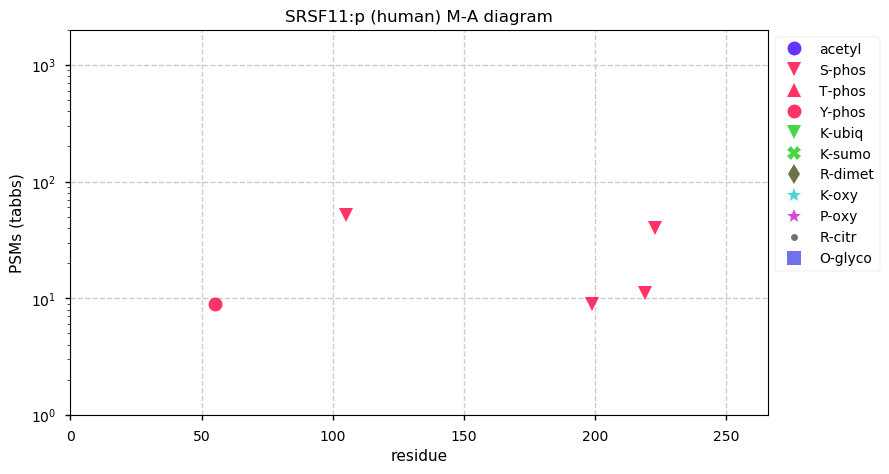
Sun Oct 25 16:55:36 +0000 2020This type of plot would probably would be more valuable if I knew what it should look like ...
Sun Oct 25 15:17:14 +0000 2020Frequency of mass shifts caused by SAAVs identified in PSMs from a human LC/MS/MS run. 🔗
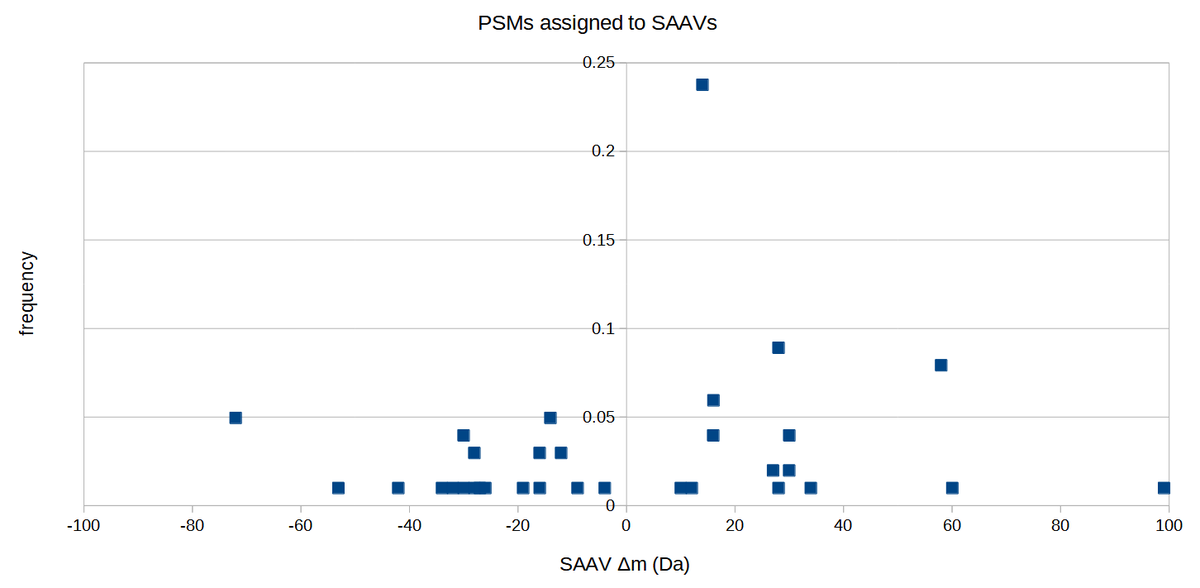
Sun Oct 25 13:05:38 +0000 2020SRSF11:p θ(max) = 59. aka p54, NET2, SFRS11 Found in HLA class I experiments only. Protein has multiple low complexity domains enriched in different residues: (12-31) G, (244-305) K/R/H and (345-424) K/R.
Sun Oct 25 13:05:37 +0000 2020SRSF11:p, serine and arginine rich splicing factor 11 (H. sapiens) 🔗 Small nuclear subunit; PTMs: 45 STY+phosphoryl in 3 high & 1 low occupancy domains, 5 K+ubiquitinyl, 4 K+SUMOyl; SAAVs: 0; mature form: 2-484 [32,446×, 295 kTa] #ᗕᕱᗒ 🔗
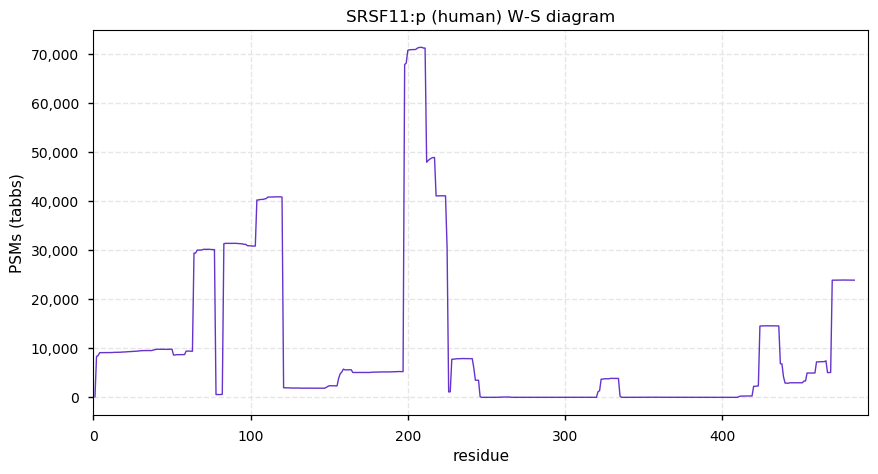
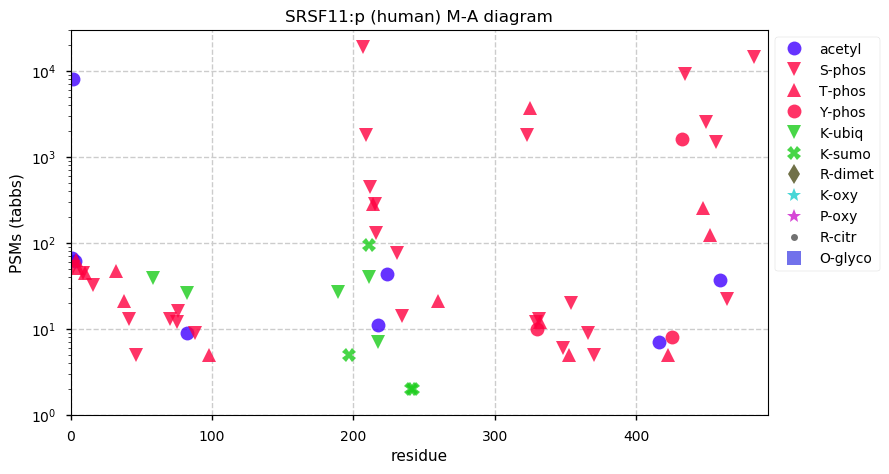
Sat Oct 24 18:09:23 +0000 2020Enough of my complaining. Back to our regularly scheduled programming ...
Sat Oct 24 15:28:26 +0000 2020@VATVSLPR I agree that poorly supervised lab work is often the underlying cause, but it doesn't explain how this stuff is still getting into the data associated with Cell and Nature branded articles.
Sat Oct 24 14:31:47 +0000 2020I frequently see papers describing statistical methods to improve reproducibility, but the problems I see most often in data are these meat-and-potatoes experimental goof-ups that can't be fixed by post hoc analysis.
Sat Oct 24 13:53:59 +0000 2020Why is it that some groups just can't seem to get IAA cysteine derivatization to work? It just bugs me to see it fail so often in published data after all of these years.
Sat Oct 24 13:09:16 +0000 2020SRSF10:p θ(max) = 72. aka TASR1, TASR2, SRp38, SRrp40, SFRS13, PPP1R149, FUSIP2, FUSIP1, SFRS13A. Found in HLA type I & II peptide experiments. Low complexity domain (102-159) with patterns of S, R & Y. C-terminal domain with high occupancy S+phosphoryl acceptors.
Sat Oct 24 12:51:48 +0000 2020SRSF10:p, serine and arginine rich splicing factor 10 (H. sapiens) 🔗 Small nuclear subunit; PTMs: 5 Y+phosphoryl, 16 S+phosphoryl, 4 T+phosphoryl, K9+SUMOyl; SAAVs: none; mature form: 2-182 [34,207×, 190 kTa] #ᗕᕱᗒ 🔗
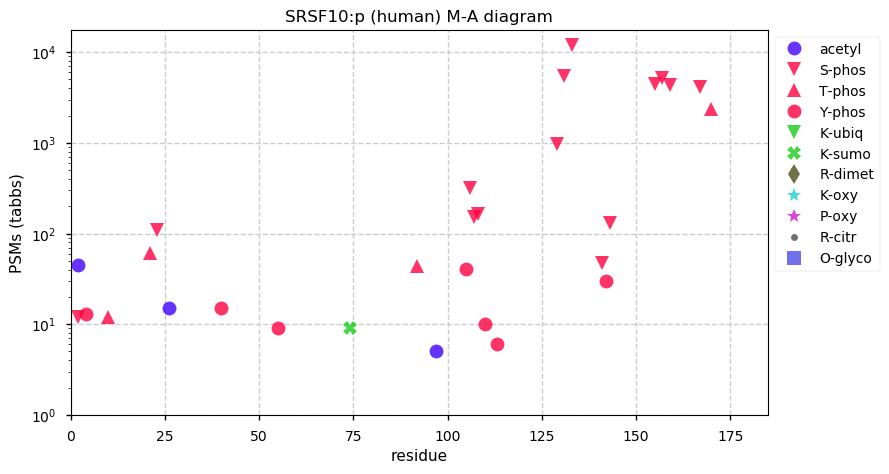
Fri Oct 23 21:39:58 +0000 2020@AlexUsherHESA I don't think it shows any causation, but for comparison: 🔗
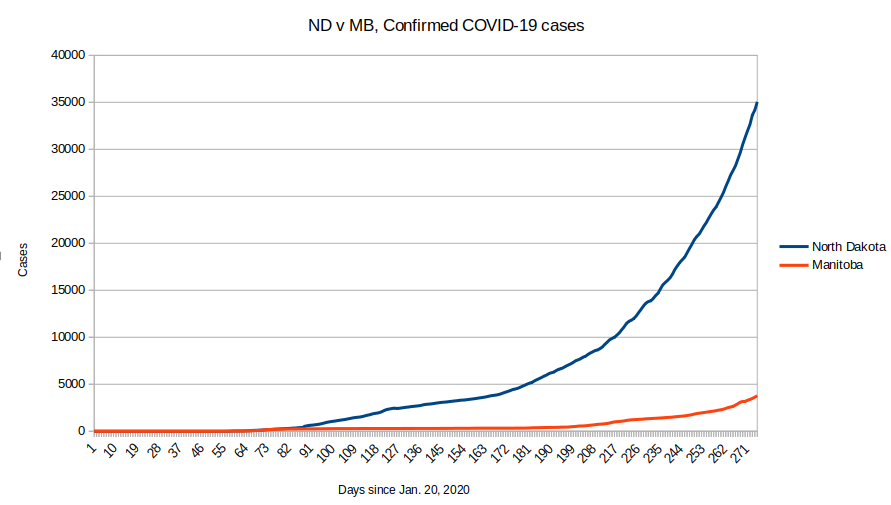
Fri Oct 23 18:02:37 +0000 2020@lstops But get your degree first.👨🏫
Fri Oct 23 17:43:55 +0000 2020@DigitalMapsAW Sea People
Fri Oct 23 17:27:51 +0000 2020@lstops To the extent that it is within my powers to grant such things - go off into the world and do good works.
Fri Oct 23 16:47:02 +0000 2020Does having a conference start over the weekend really help attendance at this point? I think people would be more likely to tune in if it was only on work days, preferably during work hours in their time zone. "Live" may be "dead". 🔗
Fri Oct 23 16:32:17 +0000 2020@a_makaju @lstops >= 6
Fri Oct 23 16:24:03 +0000 2020@lstops @a_makaju @lstops deduction "class I contamination of class II MHC Data" is the correct answer. ⭐️⭐️⭐️⭐️
Fri Oct 23 16:18:15 +0000 2020Winter is coming 🔗
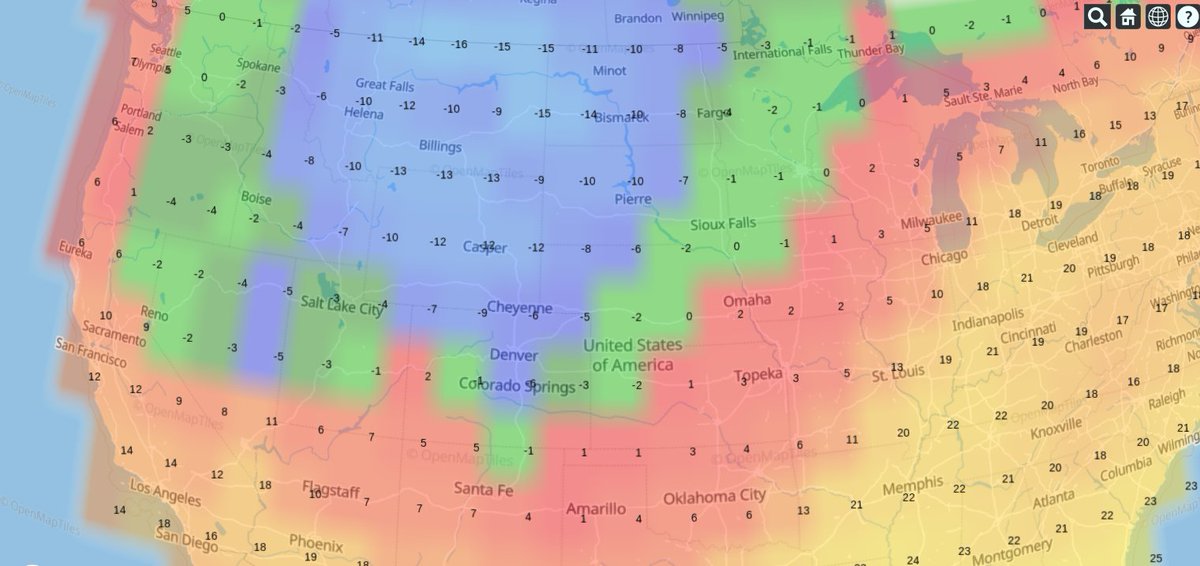
Fri Oct 23 15:40:42 +0000 2020@nesvilab I don't know if his current tactic will work—pointing out edge cases where the interpretation is very clearly wanting—but I do like his sticktoitiveness.
Fri Oct 23 15:09:10 +0000 2020@Sci_j_my A little warm on the 1st guess, stone cold on the 2nd.
Fri Oct 23 15:01:19 +0000 2020Advanced proteomics quiz: who can tell me what type of experiment produced this histogram of PSM peptide sequence lengths from a single LC/MS/MS run & what went wrong in the expt? (Note: this is real data from a Cell paper.) 🔗
Fri Oct 23 14:44:29 +0000 2020@nesvilab Bill's journey with the FDR interpretation of target-decoy simulations has been a twisty affair, but I do sort of admire him for following the math & trying to educate the community for such a long time.
Fri Oct 23 12:30:15 +0000 2020SRSF9:p θ(max) = 88. aka SRp30c, SFRS9. Found in HLA type I peptide experiments only. The sequence does not have the C-terminal, S/R repetitive low complexity domain found in most SRSF proteins & the highest density of Y+phosphoryl acceptors.
Fri Oct 23 12:30:15 +0000 2020SRSF9:p, serine and arginine rich splicing factor 9 (H. sapiens) 🔗 Small nuclear subunit; PTMs: 14 Y+phosphoryl, 16 S+phosphoryl, 2 T+phosphoryl, 8 K+ubiquitinyl; SAAVs: Y35F (1%); mature form: 2-221 [38,300×, 303 kTa] #ᗕᕱᗒ 🔗
Thu Oct 22 16:23:43 +0000 2020@astacus The Dutch data hasn't had much of that periodicity, from the start of the pandemic. In last few weeks, though, it is probably being masked (at least partially) by the rapid rate of increase.
Thu Oct 22 15:24:01 +0000 2020It would appear that things have gotten a little out of hand in Holland: 🔗
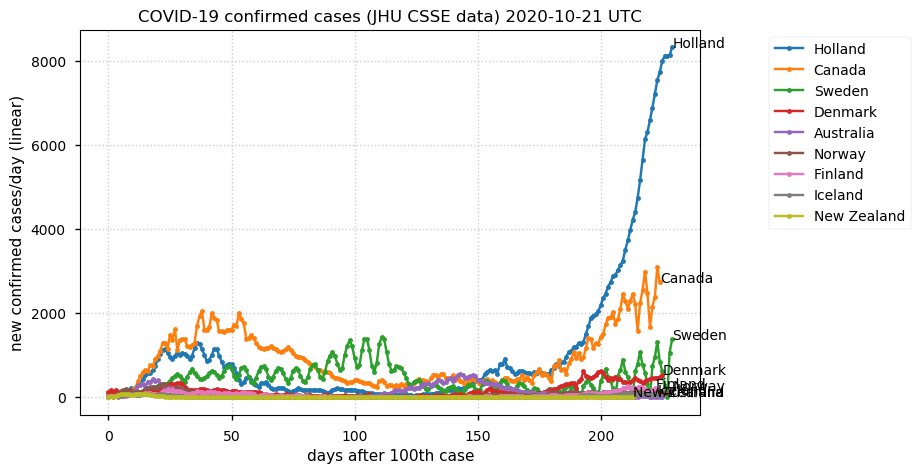
Thu Oct 22 15:12:13 +0000 2020@astacus @Sci_j_my I'm not sure what legacy email systems would do with an attachment with this type of file name, but I'm predicting some degree of data loss/system meltdown.
Thu Oct 22 15:07:29 +0000 2020@astacus @Sci_j_my √∛∜∪.☎☏☠
is also a perfectly valid file name (although rather tricky to type).
Thu Oct 22 14:37:21 +0000 2020@bkives Unless they are doing this because they want to spend more money in this sector, it will end up being a another example of Goodhart's (or Campbell's) law.
Thu Oct 22 14:21:11 +0000 2020@ucdmrt I'm pretty sure you would have to insert some type of photosynthetic pathway to get this to happen.
Thu Oct 22 14:02:36 +0000 2020@astacus @Sci_j_my Just wait until somebody in admin discovers that most Unicode characters can be used in file names, too.
"ΦΧΨΩ.αβγ" is a perfectly valid filename for any modern computer file system (I just created a file with this name on both Windows 10 and Ubuntu 18).
Thu Oct 22 11:55:20 +0000 2020SRSF8:p θ(max) = 31. aka SRP46, SFRS2B. Found in HLA type I peptide experiments only. It has several low complexity domains, with less enrichment of alternating R & S residues patterns than most SRSF proteins. The domain (150-200) has a Y every 5 residues.
Thu Oct 22 11:55:19 +0000 2020SRSF8:p, serine and arginine rich splicing factor 8 (H. sapiens) 🔗 Small nuclear subunit; PTMs: 1 Y+phosphoryl, 15 S+phosphoryl, 2 T+phosphoryl, K36+ubiquitinyl/acetyl; SAAVs: none; mature form: 2-282 [22,187×, 62 kTa] #ᗕᕱᗒ 🔗
Wed Oct 21 20:35:32 +0000 2020@btaplatt Good. 🇨🇦
Wed Oct 21 18:54:49 +0000 2020No election would be the best outcome.
🔗
Wed Oct 21 16:51:35 +0000 2020In my last note on the subject, using JSON description lines has the added benefit of automatically updating FASTA files to use UTF-8, rather than what ever ANSI code page happens to be on the originating computer.
Wed Oct 21 15:35:33 +0000 2020@VPrasadMDMPH "Iatrogenesis" is my new word for the day!
Wed Oct 21 15:22:03 +0000 2020@AlexUsherHESA @MHendr1cks Not really: it just shifts the political power around a bit.
Wed Oct 21 15:17:16 +0000 2020@MHendr1cks @AlexUsherHESA I prefer the German chair-based system for university organization. It is much more adaptable than the North American departmental system.
Wed Oct 21 14:43:42 +0000 2020@neely615 @dtabb73 In the parlance of the day, proteomics groups prefer to use "edge" computing resources.
Wed Oct 21 14:23:15 +0000 2020@neely615 @dtabb73 As well, proteomics groups rarely have access to HPC resources or the expertise to use them effectively. They really do prefer to use desktops (often with rather antique CPUs and memory).
Wed Oct 21 14:20:11 +0000 2020@neely615 @dtabb73 Proteomics groups mainly use Windows for all of their analytical software, which makes doing that type of thing challenging.
Wed Oct 21 13:15:32 +0000 2020SRSF7:p θ(max) = 56. aka 9G8, ZCRB2, HSSG1, AAG3, RBM37, ZCCHC20, SFRS7. Found in HLA type I peptide experiments only. It has a low complexity domain (121-238) enriched in patterns of alternating R & S residues. (192-233) is also a high occupancy serine phosphodomain.
Wed Oct 21 13:15:31 +0000 2020SRSF7:p, serine and arginine rich splicing factor 7 (H. sapiens) 🔗 Small nuclear subunit; PTMs: 5 Y+phosphoryl, 32 S+phosphoryl, 4 T+phosphoryl, 5 K+ubiquitinyl, 7 K+acetyl; SAAVs: none; mature form: 2-238 [50,634×, 329 kTa] #ᗕᕱᗒ 🔗
Tue Oct 20 23:36:23 +0000 2020@neely615 @astacus I got "software": as though I wasn't feeling old enough already.
Tue Oct 20 21:40:26 +0000 2020jFASTA specification: a valid FASTA file with:
1. 1st line (starts with ';') file info in JSON format,
2. descriptions (start with '>') in JSON format, &
3. last line (starts with ';') JSON format with hash value for preceding lines.
Tue Oct 20 21:21:01 +0000 2020And just to keep it from getting lonely, the mouse reference proteome sequence as a jFASTA file, again crafted with thanks from the FASTA file supplied by GenCode:
🔗
Tue Oct 20 20:18:36 +0000 2020The first (& probably only) jFASTA file ever produced. Reverently crafted from the current human reference proteome sequence FASTA supplied by GenCode.
🔗
Tue Oct 20 18:33:47 +0000 2020@UCDProteomics @lkpino @birgits61642917 Maybe it is because I check for new public datasets every day, but I see people using SILAC pretty routinely & getting the results published. They are usually using it to test a specific hypothesis, though, rather than trying to use quant to come up with one.
Tue Oct 20 16:01:02 +0000 2020While we North Americans tend to focus on ourselves, the trends in Europe don't look great either right now. 🔗
Tue Oct 20 14:54:41 +0000 2020@dtabb73 I told you to take the blue pill!
Tue Oct 20 14:50:07 +0000 2020@Smith_Chem_Wisc @pwilmarth I guess you could say that "persistently having no format" is "stable", but IMO FASTA has been a net negative for proteomics. I stopped using it for anything serious about 17 years ago, but every once in a while a client wants to use it, which always leads to hijinks.
Tue Oct 20 14:44:48 +0000 2020@Smith_Chem_Wisc @pwilmarth The description line is simply free ASCII text, with no type of formatting at all. The protein/DNA/RNA/cDNA lines (0-N) are also free ASCII text, although usually the single letter codes are upper case with no other characters, although "*" shows up pretty regularly. 2/2
Tue Oct 20 14:40:31 +0000 2020@Smith_Chem_Wisc @pwilmarth I wouldn't really use the word "stable" for FASTA. As currently understood, it has a comment line (>) that may or may not be followed by a multi-line sequence. There is no way to know the real length of either element, or whether they were cutoff somehow. 1/2
Tue Oct 20 12:14:39 +0000 2020SRSF6:p θ(max) = 52. aka SRP55, B52, SFRS6. Found in HLA type I peptide experiments only. It has a low complexity domain (184-344) enriched in patterns of alternating R & S residues. (291-316) is also a high occupancy serine phosphodomain.
Tue Oct 20 12:14:39 +0000 2020SRSF6:p, serine and arginine rich splicing factor 6 (H. sapiens) 🔗 Small nuclear subunit; PTMs: 5 Y+phosphoryl, 49 S+phosphoryl, 6 T+phosphoryl, 5 K+ubiquitinyl, 18 K+acetyl; SAAVs: none; mature form: 1-344 [48,854×, 376 kTa] #ᗕᕱᗒ 🔗
Mon Oct 19 21:59:55 +0000 2020While there is still some trouble with run-to-run holdover similar to the group's last effort, it is good data to take a look at if you are interested in virus-host interactions & what you can do with HEK 293T cells.
2/2
Mon Oct 19 21:58:54 +0000 2020PXD021588 is a new study, looking at protein-protein interactions between SARS CoV, SAR CoV2 and MERS viral protein baits and HEK 293T host proteins.
1/2
Mon Oct 19 20:44:26 +0000 2020Maybe by spring time ....
🔗
Mon Oct 19 19:08:11 +0000 2020@gingraslab1 There wasn't much other than glutathione. The usual suspects (e.g., trioxidation, propionamide, mercaptoethanol, cysteinylation) were all undetectable.
Mon Oct 19 18:34:47 +0000 2020@gingraslab1 does anybody there remember whether cysteine sidechain derivatization used for PXD018196? I couldn't find a mention of it in the bioRxiv PDF. The data looks like it is unmodified cysteine, with some adventitious glutathione modification.
Mon Oct 19 18:16:30 +0000 2020@VATVSLPR I'd put PEFF down as a "missed opportunity" because they chose to create their own custom RegEx-based system rather than utilize an existing, widely used text object structuring solution.
Mon Oct 19 14:08:25 +0000 2020If one could introduce a convention by which the file has a description line with no sequence at the top of the file that contains a JSON line with info about the file in general (incl. # of lines and a whole file hash), that would pretty much solve everything.
3/3
Mon Oct 19 14:06:27 +0000 2020e.g.,
>sp|P02769|ALBU_BOVIN Albumin OS=Bos taurus
becomes
>{"ids":["P02769","ALBU_BOVIN"], "desc":'"Albumin", "species":"Bos taurus", "mdh5": "432779d395a52bfc9f6574bc3e98afcd"}
2/3
Mon Oct 19 14:01:09 +0000 2020An extremely simple fix for many of the problems would be to convert the description line into a one line JSON object, making parsing and validation problems go away especially if the JSON included a hash value for the associated sequence. 1/2
Mon Oct 19 12:36:43 +0000 2020SRSF5:p θ(max) = 55. aka SRP40, HRS, SFRS5. Found in HLA type I peptide experiments only. It has a low complexity domain (180-272) enriched in patterns of alternating R & S residues. (225-259) is also a high occupancy serine phosphodomain.
Mon Oct 19 12:34:26 +0000 2020SRSF5:p, serine and arginine rich splicing factor 5 (H. sapiens) 🔗 Small nuclear subunit; PTMs: 3 Y+phosphoryl, 23 S+phosphoryl, 0 T+phosphoryl, 3 K+ubiquitinyl, 1 K+SUMOyl; SAAVs: none; mature form: 2-272 [35,099×, 143 kTa] #ᗕᕱᗒ 🔗
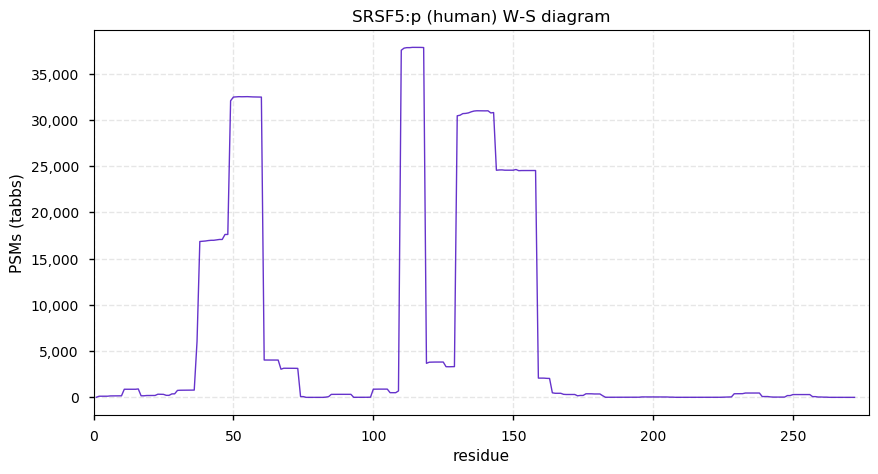
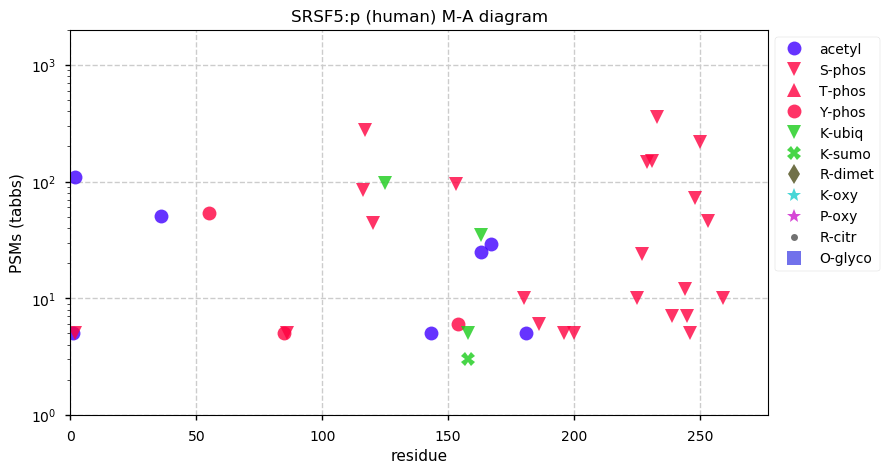
Sun Oct 18 16:51:43 +0000 2020I don't like to look at this any more ... 🔗
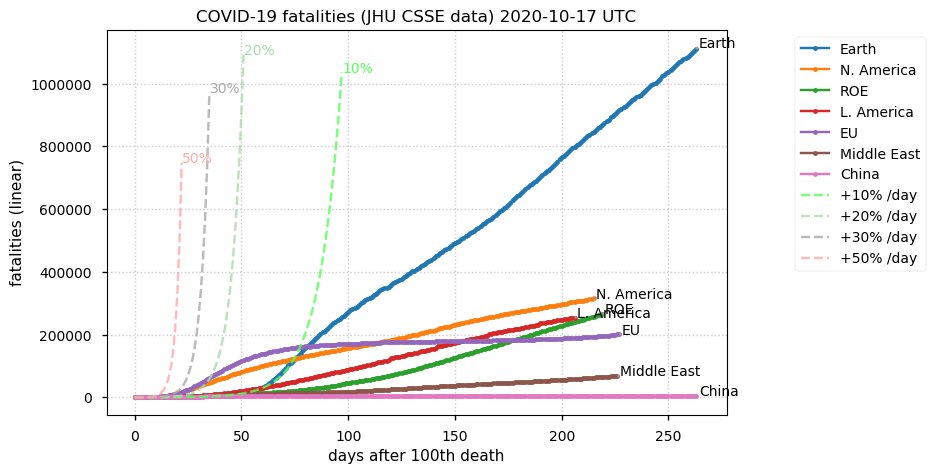
Sun Oct 18 14:47:24 +0000 2020Like high mobility group proteins, serine/arginine-rich splicing factors (🔗) are highly modified, commonly observed nucleic acid interacting proteins that get very little attention compared to their show-off cousins, the histones.
Sun Oct 18 12:33:40 +0000 2020SRSF4:p has 2 high MAF SAAVs, making it is unlikely that any individual will be homozygous for the reference protein sequence.
Sun Oct 18 12:33:39 +0000 2020SRSF4:p θ(max) = 40. aka SRP75, SFRS4. Found in HLA type I peptide experiments only. It has a low complexity domain (179-494) enriched in patterns of alternating R & S residues. (442-462) is also a high occupancy serine phosphodomain.
Sun Oct 18 12:33:39 +0000 2020SRSF4:p, serine & arginine rich splicing factor 4 (human) 🔗 Small nuclear subunit; PTMs: 56 STY+phosphoryl, (K58,K159)+acetyl/ubiquitinyl, (K256,K451)+SUMOyl; SAAVs: E253D (72%), G338A (50%); mature form: 2-494 [42,617×, 229 kTa] #ᗕᕱᗒ 🔗
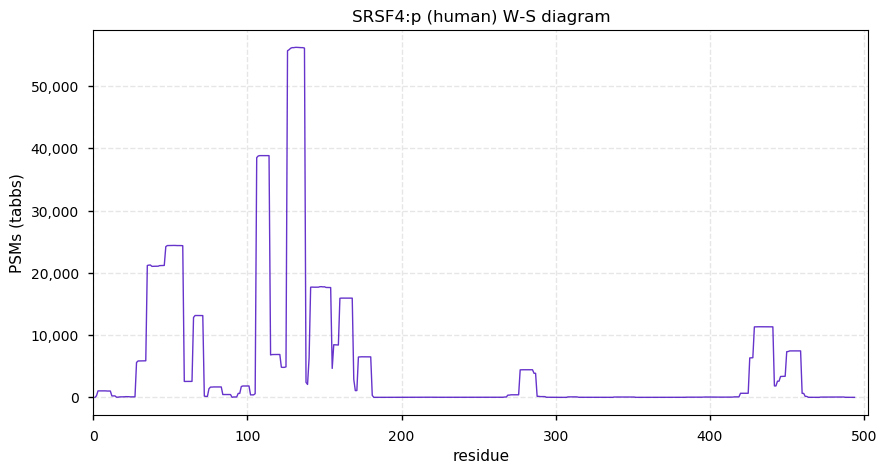
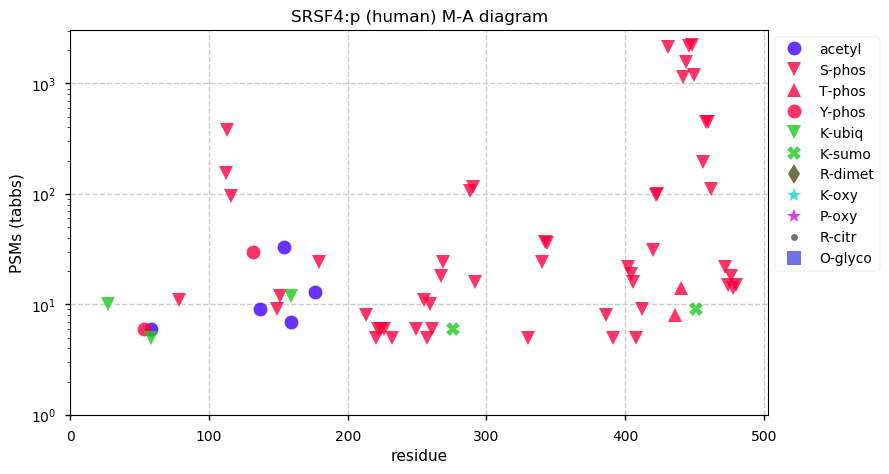
Sat Oct 17 15:04:43 +0000 2020Every once in a while I have to do something that reminds me just how awful FASTA files are for practical work.
Sat Oct 17 12:41:00 +0000 2020SRSF3:p θ(max) = 63. aka SRp20, SFRS3. Found in HLA type I peptide experiments only. It has a low complexity domain (86-164) highly enriched in patterns of R & S residues that are different from other SRSF proteins, e.g (105-136):
RRRSPPPRRRSPRRRSFSRSRSRSLSRDRRR
Sat Oct 17 12:41:00 +0000 2020SRSF3:p, serine and arginine rich splicing factor 3 (H. sapiens) 🔗 Small nuclear subunit; PTMs: M1+acetyl, 13 STY+phosphoryl, (K11,K23,K85)+acetyl/ubiquitinyl, K11+SUMOyl; SAAVs: none; mature form: 1-164 [57,141×, 496 kTa] #ᗕᕱᗒ 🔗
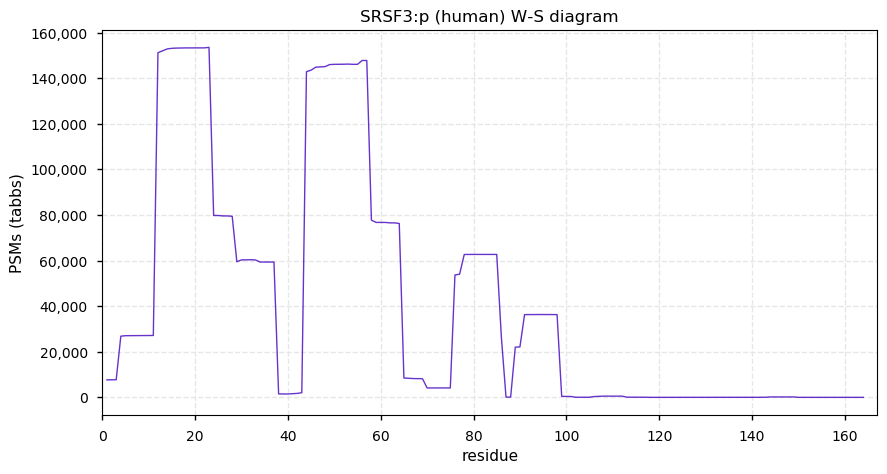
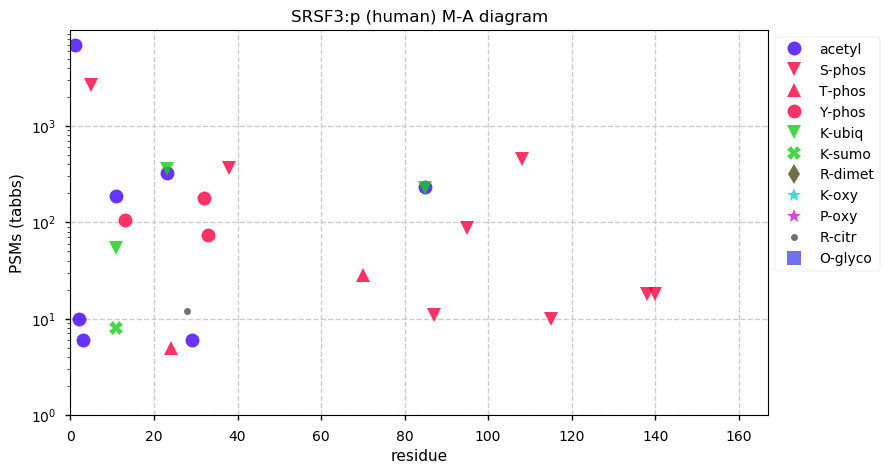
Fri Oct 16 18:47:12 +0000 2020@AlexUsherHESA Also great: "... as at all other research-intensive universities in this country, we are forced to struggle with the anomalous fact that we are neither resourced nor configured properly for half of the mission we have chosen to pursue ..."
Fri Oct 16 18:41:27 +0000 2020@AlexUsherHESA I particularly liked the "Research and Teaching" section. The phrase "... the deep aversion to risk everywhere discernible in the university ..." pretty much sums up my experience with Cdn. U15 shops.
Fri Oct 16 16:39:37 +0000 2020@astacus @cabin_pressure Google maps says the driving distance is only 3337 miles!
Fri Oct 16 15:30:18 +0000 2020The domain can be observed in phosphopeptide-enriched trypsin-based proteomics.
Fri Oct 16 15:30:18 +0000 2020SRSF2:p θ(max) = 51. aka SC-35, SC35, PR264, SFRS2A, SFRS2. Found in HLA type I peptide experiments only. It has a low complexity domain (117-208) highly enriched in alternating R & S residues. This domain is not observed in trypsin-based proteomics expts.
Fri Oct 16 15:30:17 +0000 2020SRSF2:p, serine and arginine rich splicing factor 2 (H. sapiens) 🔗 Small nuclear subunit; PTMs: S2+acetyl, 42 STY+phosphoryl, 3 K+ubiquitinyl, 1 K+SUMOyl; SAAVs: none; mature form: 2-221 [45,368×, 336 kTa] #ᗕᕱᗒ 🔗
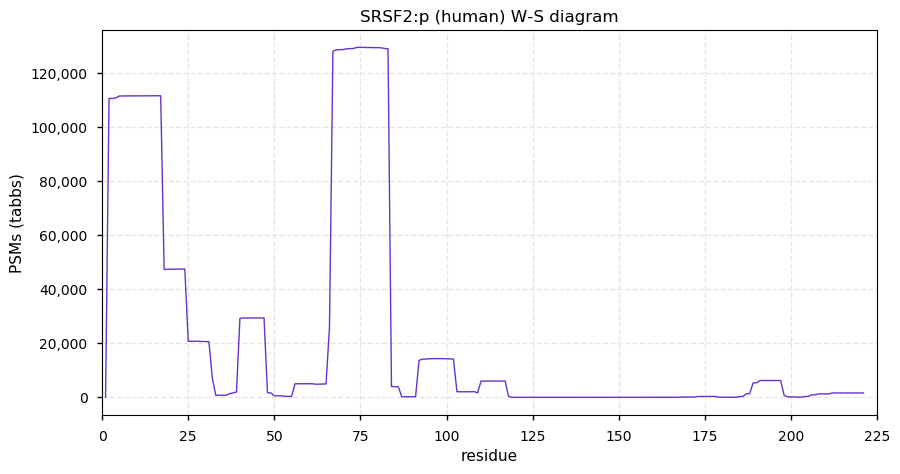
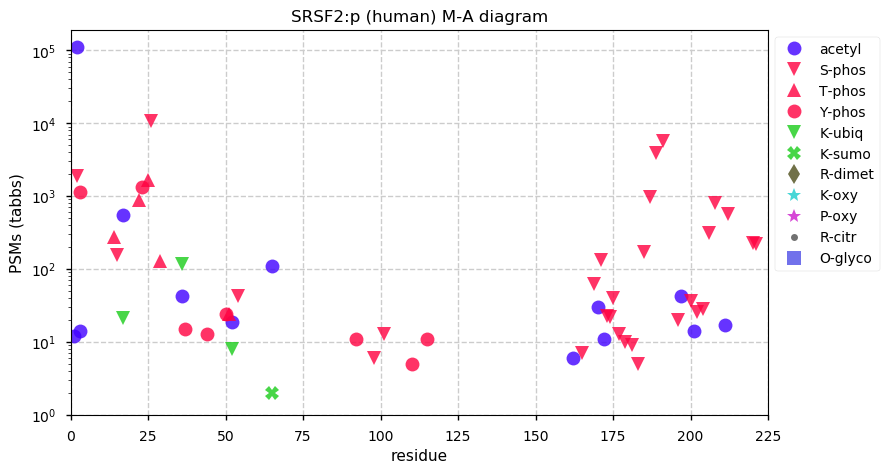
Fri Oct 16 15:20:56 +0000 2020@cdsouthan I take the 5th on that particular project.
Fri Oct 16 15:02:35 +0000 2020My stumbling block is even if the structure of the proteasome allowed this to happen, I don't see how can you get enough of any particular spliced peptide species to observe it. 2/2
Fri Oct 16 15:00:34 +0000 2020I personally can't get my head around proteasome splicing. I have no problem with the fact that proteases can be used to reverse hydrolysis, as I've actually used this in practice. 1/2
Fri Oct 16 14:34:43 +0000 2020Thanks to everyone who participate in the poll. There was significant support for proteasome splicing, small ORFs and the catchall "something else". The idea of using a non-canonical start codon only attracted 1 vote.
Thu Oct 15 19:22:15 +0000 2020@MattWFoster PXD012308 & PXD018998 are both pretty good, high resolution human class II datasets.
Thu Oct 15 17:56:01 +0000 2020Khanh Hoa banh mi for lunch. 😀👍
Thu Oct 15 17:31:39 +0000 2020@tomlau I would love to know if HUPO's HIPP project has an opinion.
Thu Oct 15 16:57:20 +0000 2020@tomlau People can get carried away by seeing what they want to see, but in this case I do think there is something to the observations. The mechanism generating the observations/peptides, though, is still up for debate.
Thu Oct 15 14:51:13 +0000 2020@dtabb73 Mine does that several times a day. My money is either on some sort of "eye-of-the-beholder" effect or elder-times pagan dark magic.
Thu Oct 15 14:24:01 +0000 2020Since I can't vote in my own poll, I should admit that I am fond of both the sORF and CUG start (🔗) ideas.
Thu Oct 15 13:27:59 +0000 2020It has been frequently reported that there are HLA class I peptides that cannot be explained using the reference human proteome. Which of the proposed mechanisms do you find the most likely source of these peptides:
Thu Oct 15 12:40:23 +0000 2020For anyone interested in such things, SRSF1:p is the only one of the 12 serine-arginine rich splicing factors that has observed arginine methylation.
Thu Oct 15 12:14:27 +0000 2020SRSF1:p has a C-terminal low complexity domain (204-248) highly enriched in alternating R and S residues:
RSRSRSRSRSRSRSRSNSRSRSYSPRRSRGSPRYSPRHSRSRSRT
Thu Oct 15 12:12:11 +0000 2020SRSF1:p θ(max) = 78. aka ZFM1, ZCCHC25, ZNF162. Found in HLA type I peptide experiments only. The protein has an unusually large number of observed R+dimethyl acceptor sites distributed across the sequence.
Thu Oct 15 12:12:11 +0000 2020SRSF1:p, serine and arginine rich splicing factor 1 (H. sapiens) 🔗 Small nuclear subunit; PTMs: S2+acetyl, 32 STY+phosphoryl, 7 K+ubiquitinyl, 2 K+SUMOyl, 27 R+dimethyl; SAAVs: none; mature form: 2-248 [55,406×, 718 kTa] #ᗕᕱᗒ 🔗
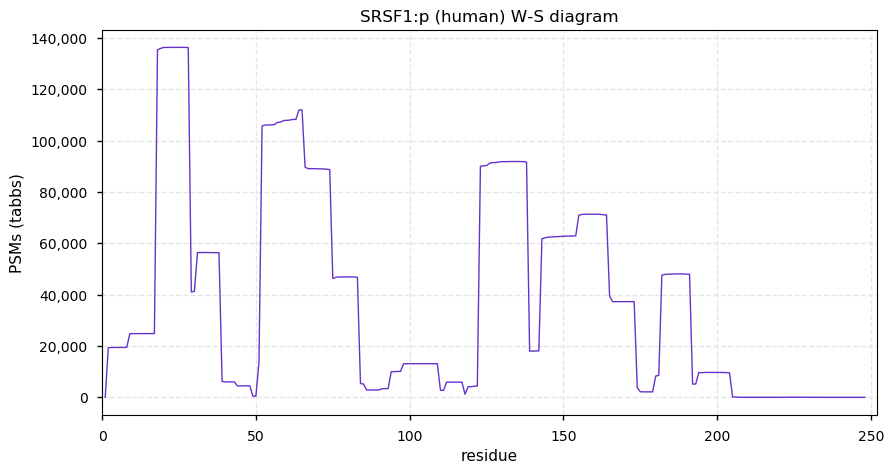
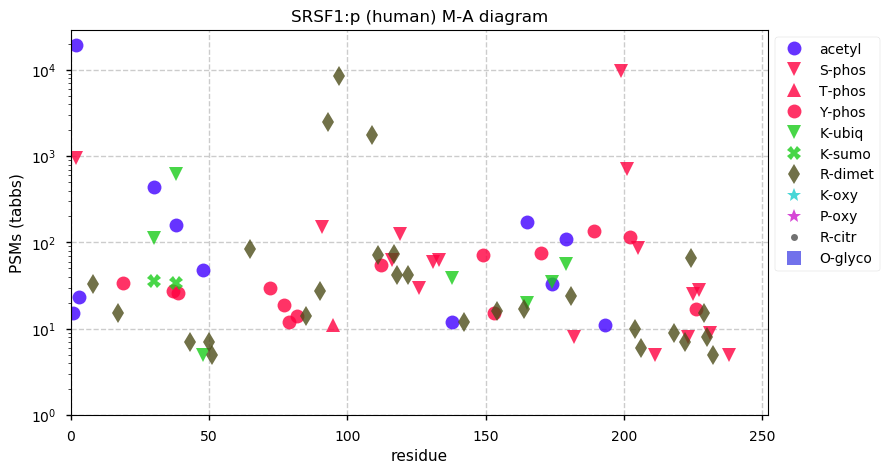
Wed Oct 14 12:15:56 +0000 2020SF1:p θ(max) = 47. aka ZFM1, ZCCHC25, ZNF162. Found in HLA type I peptide experiments only. The domain (307-639) has several proline-rich low complexity domains & it has few observable tryptic peptides. This domain can be observed using chymotrypsin cleavage.
Wed Oct 14 12:15:56 +0000 2020SF1:p, splicing factor 1 (H. sapiens) 🔗 Midsized nuclear subunit; PTMs: A2+acetyl, 34 STY+phosphoryl, 7 K+ubiquitinyl, 1 K+SUMOyl; SAAVs: none; 5 observed splice variants; mature form: 2-639 [44,234×, 389 kTa] #ᗕᕱᗒ 🔗
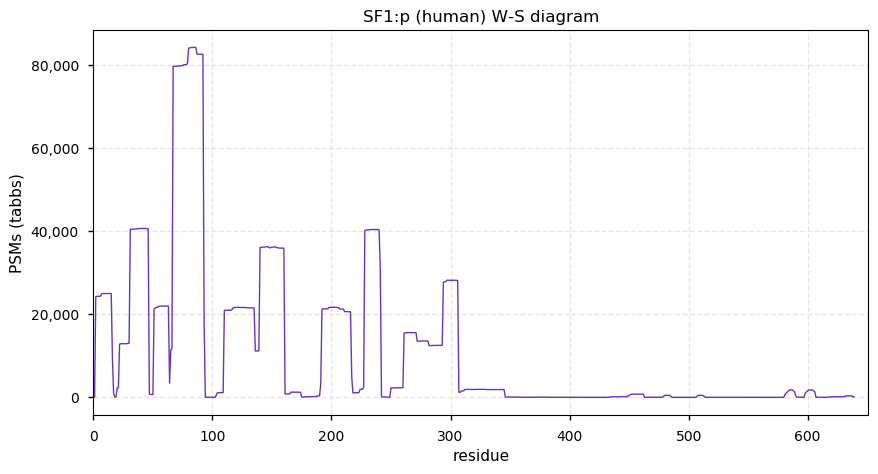
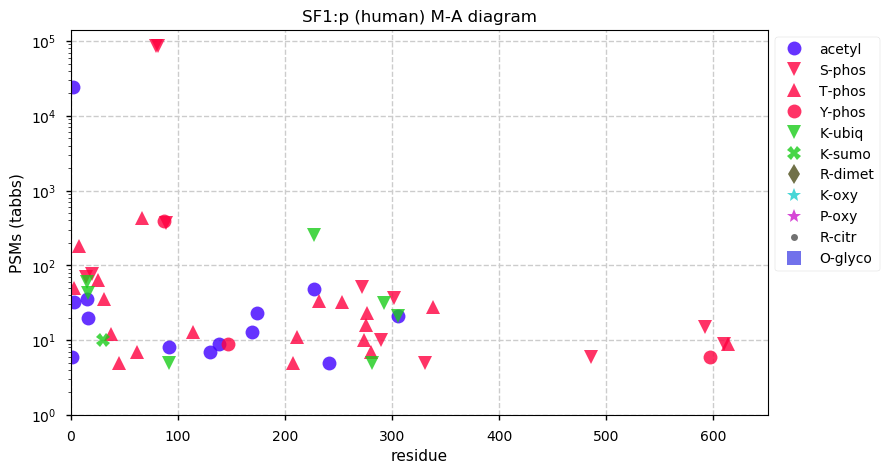
Tue Oct 13 19:35:44 +0000 2020@neely615 @byu_sam @ProteomicsNews @GrmyClair @PastelBio Sorry for my ignorance, but what does "ML" mean in this context? "Machine Learning" is all I can come up with ...
Tue Oct 13 15:27:32 +0000 2020@AlexUsherHESA About the only Canadian federal government agency that still values management with significant specialist knowledge is the uniformed military
Tue Oct 13 12:36:04 +0000 2020DDX42:p θ(max) = 68. aka RNAHP, RHELP, SF3b125, SF3B8. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells. DEAD domain (407-410) predicts RNA helicase activity.
Tue Oct 13 12:36:03 +0000 2020DDX42:p, DEAD-box helicase 42 (H. sapiens) 🔗 Midsized nuclear subunit; PTMs: A2+acetyl, 61 STY+phosphoryl, 1 K+ubiquitinyl, 8 K+SUMOyl; SAAVs: none; mature form: (1,2)-938 [32,504×, 78 kTa] [21,094, 78 kTa] #ᗕᕱᗒ 🔗
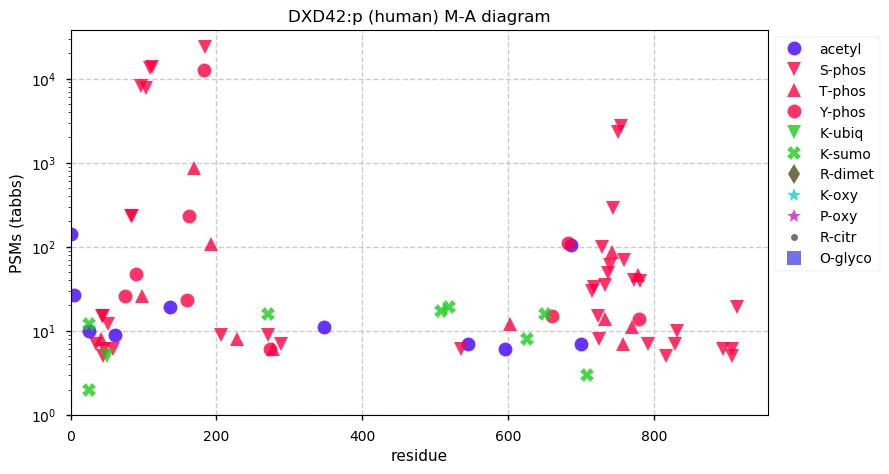
Mon Oct 12 13:12:31 +0000 2020PHF5A:p θ(max) = 80. aka MGC1346, SF3b14b, INI, bK223H9.2, Rds3, SAP14b, SF3B7. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells. It has 7 cysteine residues in the domain (23-49) that take part in a zinc finger structure.
Mon Oct 12 13:12:30 +0000 2020PHF5A:p, PHD finger protein 5A (H. sapiens) 🔗 Small nuclear subunit; PTMs: A2+acetyl, 5 Y+phosphoryl, 3 S+phosphoryl, 3 K+ubiquitinyl/acetyl, 2 K+ubiquitinyl/acetyl/SUMOyl; SAAVs: none; mature form: 2-110 [21,094×, 78 kTa] #ᗕᕱᗒ 🔗
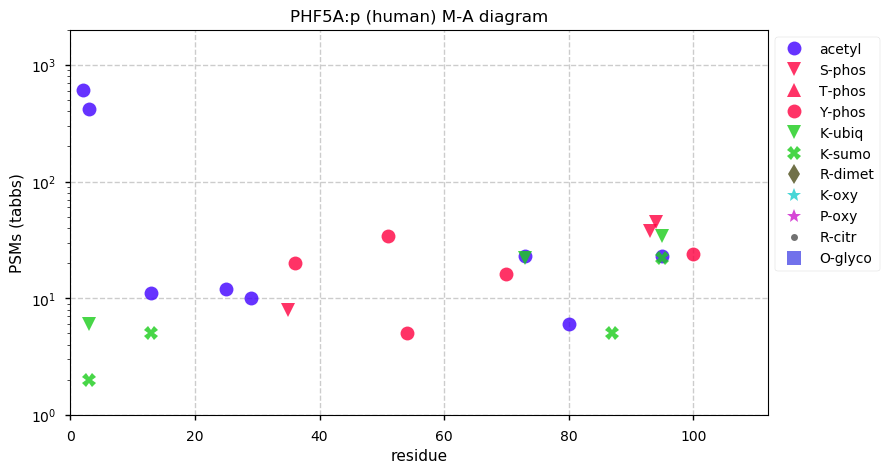
Sun Oct 11 12:24:00 +0000 2020SF3B6:p θ(max) = 74. aka P14, SF3B14a, Ht006, CGI-110, SAP14a. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells.
Sun Oct 11 12:24:00 +0000 2020SF3B6:p, splicing factor 3b subunit 6 (H. sapiens) 🔗 Small nuclear subunit; PTMs: A2+acetyl, 8 Y+phosphoryl, 3 K+ubiquitinyl/acetyl, 2 K+ubiquitinyl/acetyl/SUMOyl; SAAVs: none; mature form: 2-125 [26,011×, 102 kTa] #ᗕᕱᗒ 🔗
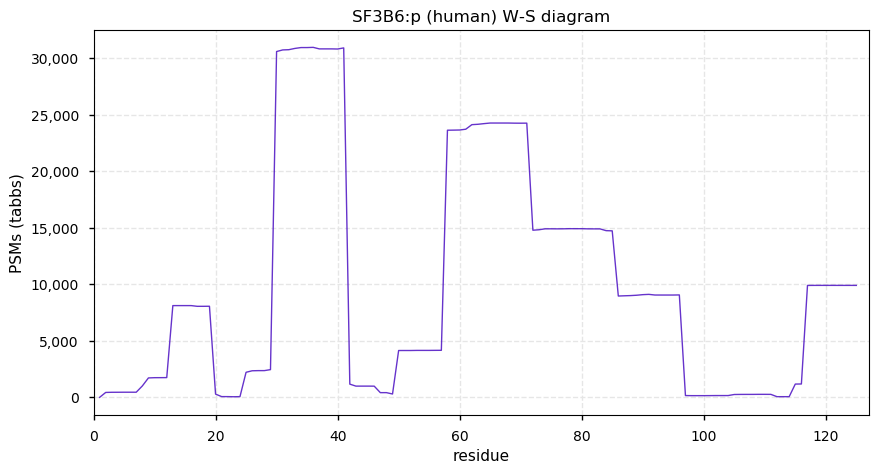
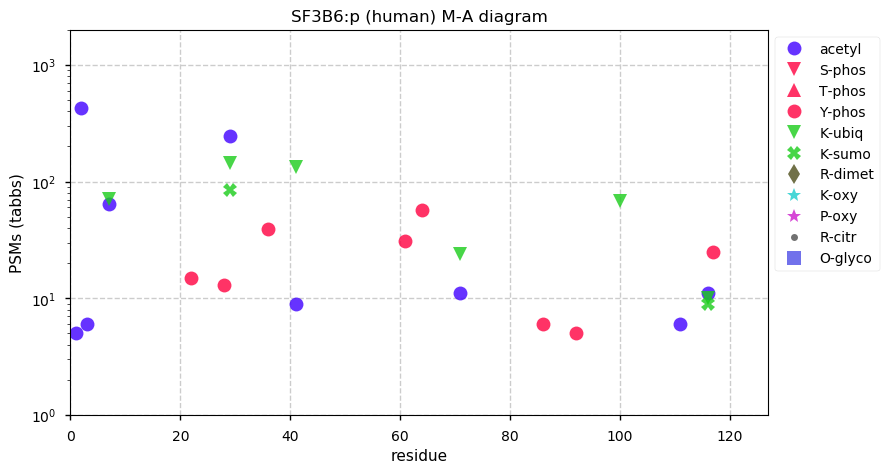
Sat Oct 10 13:21:53 +0000 2020SF3B5:p θ(max) = 94. aka SF3b10, MGC3133, Ysf3. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells.
Sat Oct 10 13:21:52 +0000 2020SF3B5:p, splicing factor 3b subunit 5 (H. sapiens) 🔗 Very small nuclear subunit; PTMs: S2+acetyl, (Y5,S9)+phosphoryl, (K17, K28)+ubiquitinyl/acetyl, K71+ubiquitinyl; SAAVs: none; mature form: 2-86 [22,829×, 89 kTa] #ᗕᕱᗒ 🔗
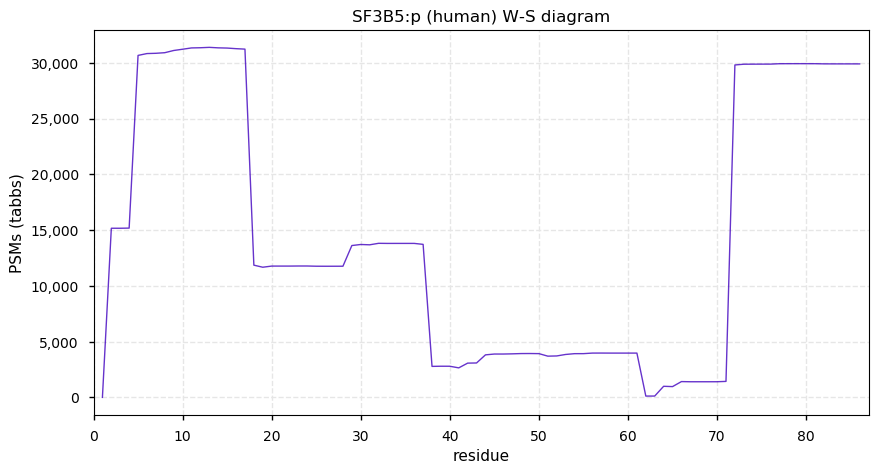
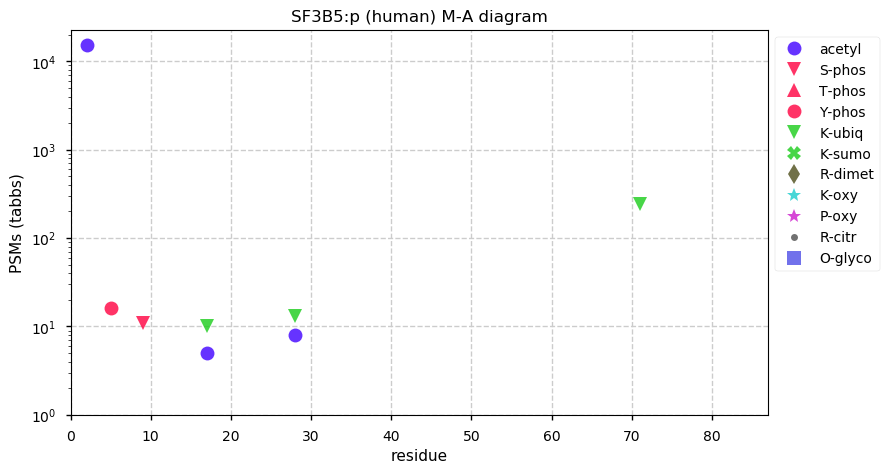
Fri Oct 09 15:50:51 +0000 2020Hurricane Delta's centre passed the Boomvang oil rig at about 7 AM local time this morning, ~300 km south of Houston in the Gulf of Mexico (27.3536°,-94.6253°) 🔗
Fri Oct 09 13:51:52 +0000 2020Looks like Hurricane Delta is just about to make landfall on the west Louisiana/east Texas coast (NEXRAD radar data). It doesn't appear that New Orleans will be affected by the storm, but Houston will be within its western edge. 🔗

Fri Oct 09 13:22:59 +0000 2020SF3B4:p has the fewest post-translational modifications of any SF3 subunit. It has a low complexity C-terminal domain (215-424) that is greatly enriched in proline, which has a subdomain (273-383) that is enriched in histidine as well.
Fri Oct 09 13:22:59 +0000 2020SF3B4:p θ(max) = 41. aka SAP49, SF3b49, Hsh49. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells.
Fri Oct 09 13:22:59 +0000 2020SF3B4:p, splicing factor 3b subunit 4 (H. sapiens) 🔗 Midsized nuclear subunit; PTMs: A2+acetyl, (Y16,Y69)+phosphoryl; SAAVs: none; mature form: 2-424 [31,697×, 136 kTa] #ᗕᕱᗒ 🔗
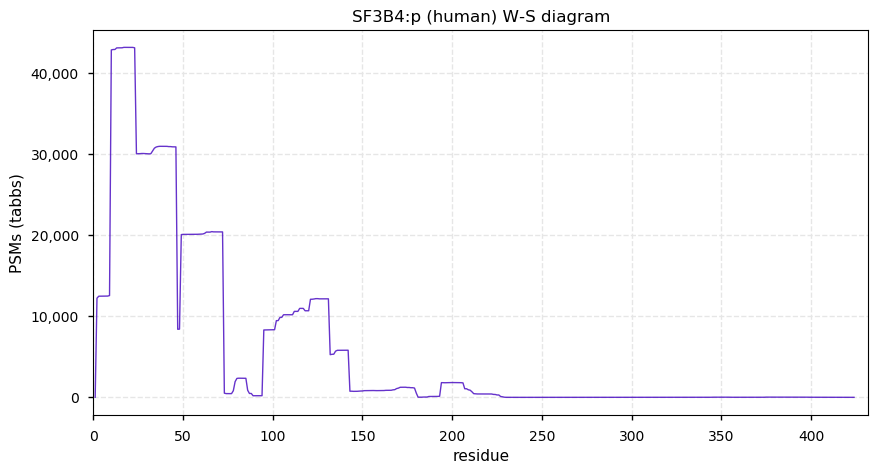
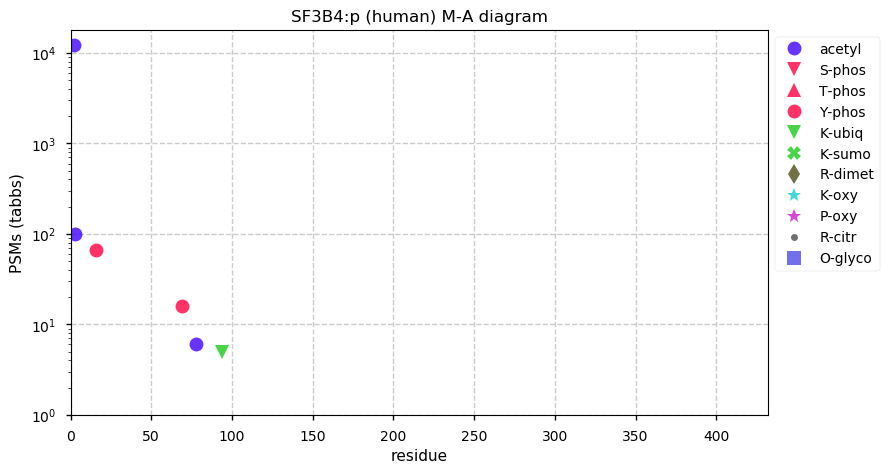
Thu Oct 08 21:06:50 +0000 2020But it is pretty hard to miss on the GOES visible satellite image 🔗

Thu Oct 08 21:04:14 +0000 2020The outer rain bands of Hurricane Delta are just starting to show up on US NEXRAD radar. You can't see the main body of the storm yet: it is still too far out in the Gulf of Mexico. 🔗

Thu Oct 08 19:45:44 +0000 2020PXD018140 (Liu JJ, et al., Neuropharmacology. 2020 Sep 22;181:108324, 🔗) has some excellent phosphopeptide-enriched data for 3 different mouse brain tissues.
Thu Oct 08 18:03:13 +0000 2020@CharlesDeschen1 @JR_Ottawa Getting America/New_York to go along seems like a long shot, though, as most of the Atlantic coast states use America/New_York rather than state-by-state timezones so it would involve quite a few changes.
Thu Oct 08 18:00:58 +0000 2020@CharlesDeschen1 @JR_Ottawa I used to work for a company that does remote telemetry and local time zones were a continuous nuisance.
Ontario unfortunately has 3 time zones:
America/Toronto (EST/EDT),
America/Atikokan (EST only) &
America/Winnipeg (CST/CDT).
Thu Oct 08 15:14:13 +0000 2020SF3B3:p has a significantly different PTM pattern than SF3B1:p or SF3B2:p. It has no SUMOYLation acceptor sites(SF3B1:p has 13 & SF3B2:p has 17). Its ratio of S:T phosphoryl acceptor sites is 1:23.
Thu Oct 08 15:14:13 +0000 2020SF3B3:p θ(max) = 70. aka SAP130, SF3b130, RSE1, KIAA0017. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells.
Thu Oct 08 15:14:12 +0000 2020SF3B3:p, splicing factor 3b subunit 3 (H. sapiens) 🔗 Large nuclear subunit; PTMs: 39 K+acetyl, 28 K+ubiquitinyl, 0 K+SUMOyl, 31 STY+phosphoryl; SAAVs: R128G (32%); mature form: 1-1217 [62,569×, 913 kTa] #ᗕᕱᗒ 🔗
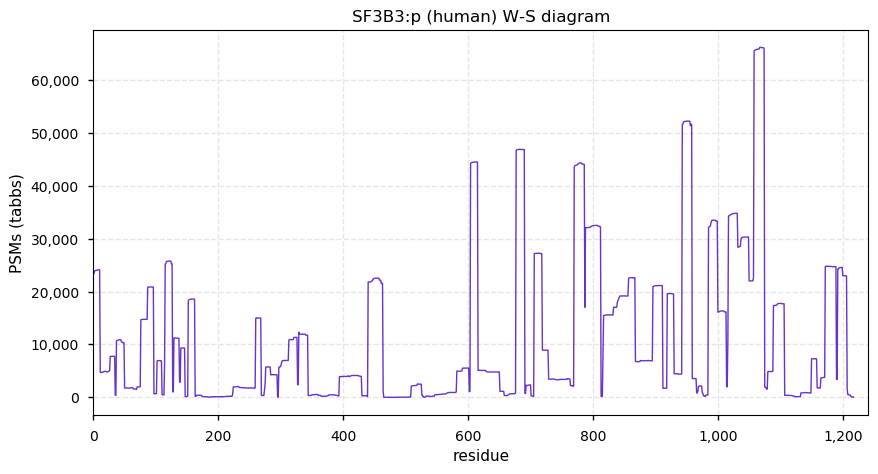
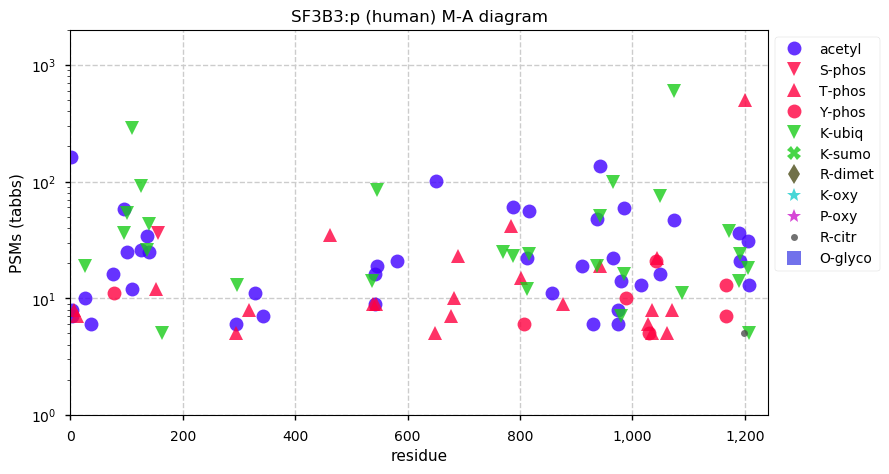
Wed Oct 07 20:44:33 +0000 2020@JR_Ottawa Does the proposed bill unify Ontario into a single timezone or does this just apply to America/Toronto?
Wed Oct 07 19:54:14 +0000 2020Hurricane Delta has a rather angry look to it, at least in the IR satellite imagery. 🔗

Wed Oct 07 12:16:38 +0000 2020SF3B2:p θ(max) = 68. aka SAP145, SF3b1, Cus1, SF3b145 . Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells.
Wed Oct 07 12:16:37 +0000 2020SF3B2:p, splicing factor 3b subunit 2 (H. sapiens) 🔗 Midsized nuclear subunit; PTMs: A2+acetyl, 65 K+acetyl, 24 K+ubiquitinyl, 17 K+SUMOyl, 62 STY+phosphoryl; SAAVs: none; mature form: 2-895 [55,003×, 901 kTa] #ᗕᕱᗒ 🔗
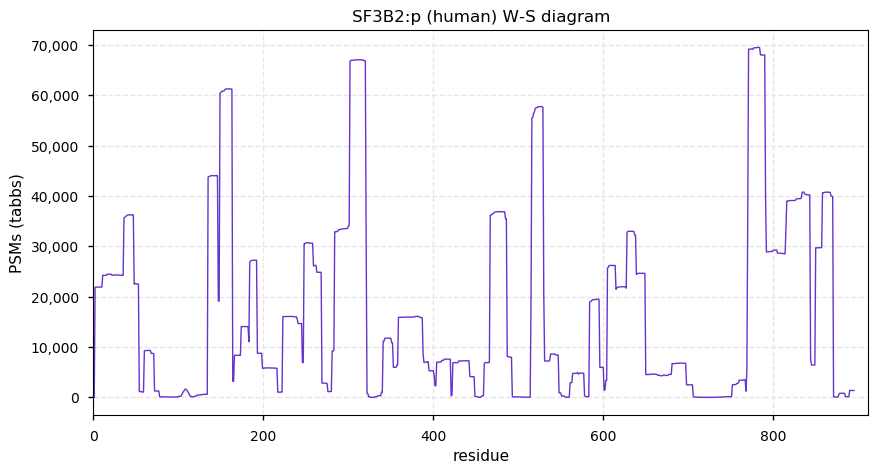
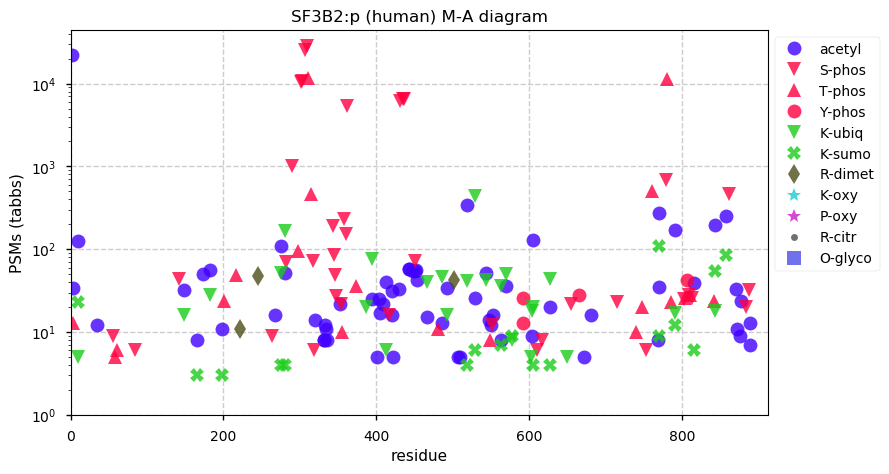
Tue Oct 06 18:44:36 +0000 2020Scientific papers reporting on experiments should always have a "things we f'd up" section.
Tue Oct 06 15:56:56 +0000 2020@nesvilab @goodlettlab1 But what we learned living in Germany was that the metric system was not as fully adopted there as we had thought. Using "pfund" for ordering meat at a butcher shop or specifying a pipe diameter in "zoll" was still pretty common.
Tue Oct 06 15:50:26 +0000 2020@nesvilab @goodlettlab1 When we lived in the US, for the first few years our son would ask what the real temperature was after hearing it on the radio in F.
Tue Oct 06 15:04:10 +0000 2020@AlexUsherHESA Makes the early election call a little less surprising.
Tue Oct 06 14:30:34 +0000 2020@goodlettlab1 -30 is cold. +10 is nice fall weather.
Tue Oct 06 12:11:05 +0000 2020SF3B1:p θ(max) = 72. aka SAP155, SF3b155, PRPF10, Prp10, Hsh155. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells.
Tue Oct 06 12:11:05 +0000 2020SF3B1:p, splicing factor 3b subunit 1 (H. sapiens) 🔗 Large nuclear subunit; PTMs: A2+acetyl, 69 K+acetyl, 22 K+ubiquitinyl, 13 K+SUMOyl, 127 STY+phosphoryl; SAAVs: none; mature form: 2-1304 [62,723×, 1227 kTa] #ᗕᕱᗒ 🔗
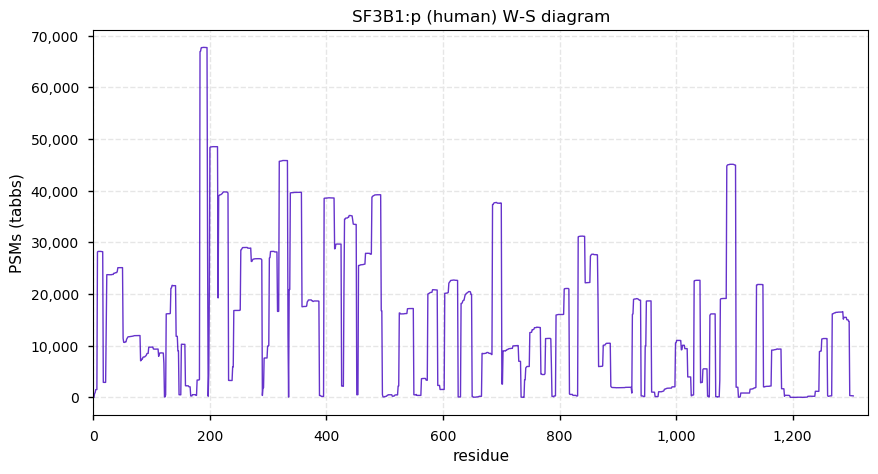
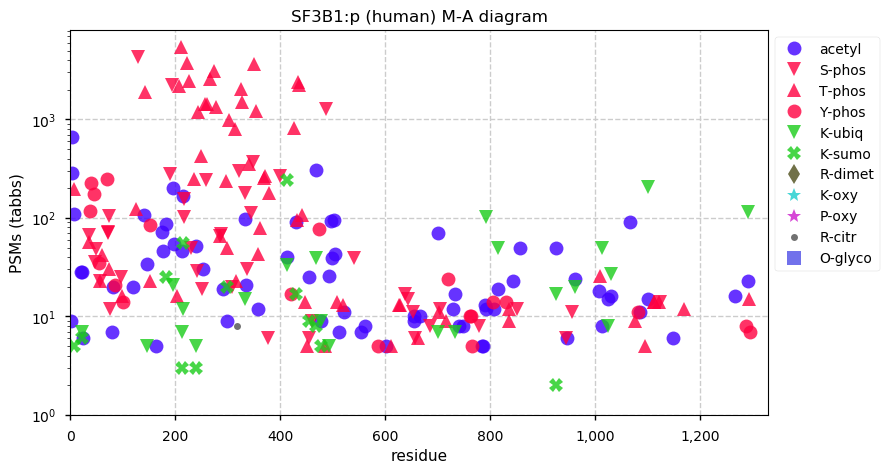
Mon Oct 05 21:06:34 +0000 2020Odd analytical finding of the day: a "global proteome" sample preparation method that has gotten rid of most of the ER, mitochondrial, lysosomal, golgi and chromatin proteins from tissue samples.
Mon Oct 05 12:07:59 +0000 2020SF3A3:p has a matrin-type zinc finger domain 406–437, an acid-rich low complexity domain 354-375 and a leucine-rich low complexity domain 255-275.
Mon Oct 05 12:07:59 +0000 2020SF3A3:p θ(max) = 77. aka SF3a60, SAP61, PRP9, PRPF9. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells.
Mon Oct 05 12:07:59 +0000 2020SF3A3:p, splicing factor 3a subunit 3 (H. sapiens) 🔗 Small nuclear protein; PTMs: M1+acetyl, 27 K+acetyl, 12 K+ubiquitinyl, 5 K+SUMOyl, 36 STY+phosphoryl; SAAVs: none; mature form: 1-601 [41,366×, 344 kTa] #ᗕᕱᗒ 🔗
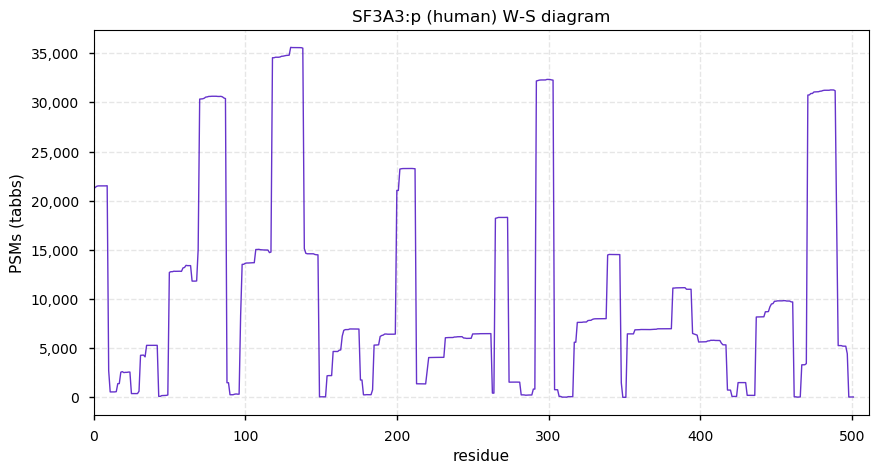
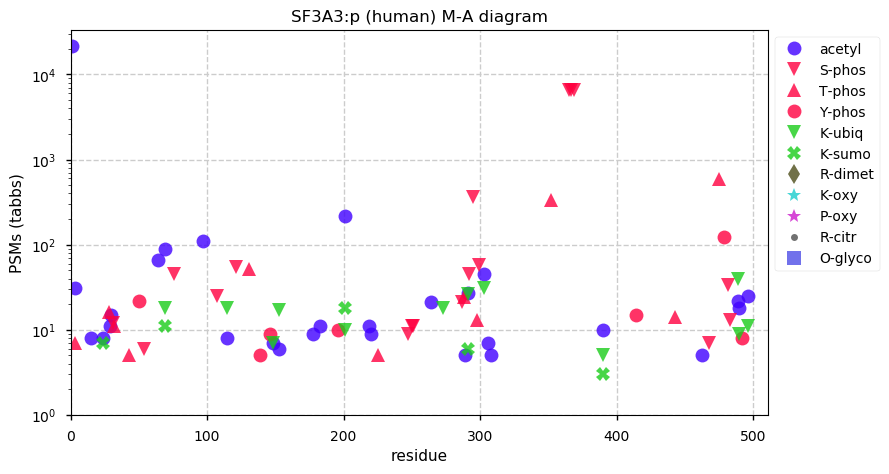
Sun Oct 04 19:28:06 +0000 2020SF3A2:p has at least 4 distinct domains: 1-216 (contains matrin-type zinc finger 54–84), 217-288 (proline rich with periodic leucines/valines), 289-443 (proline rich with 21 periodic histidines, spaced ~7 residues apart) & 445-464 (proline rich).
Sun Oct 04 17:57:20 +0000 2020Given what I know about internal mechanics of medical research, the outcome the RSA would out perform the USA in a public health response would never have occurred to me. 🔗
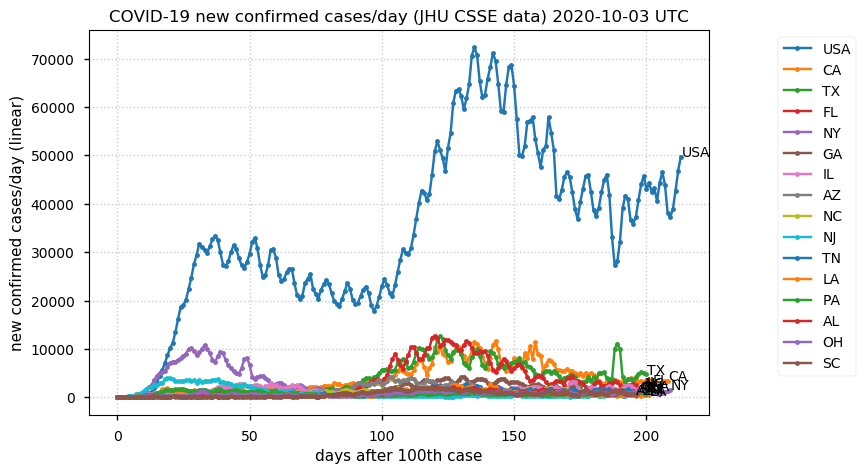
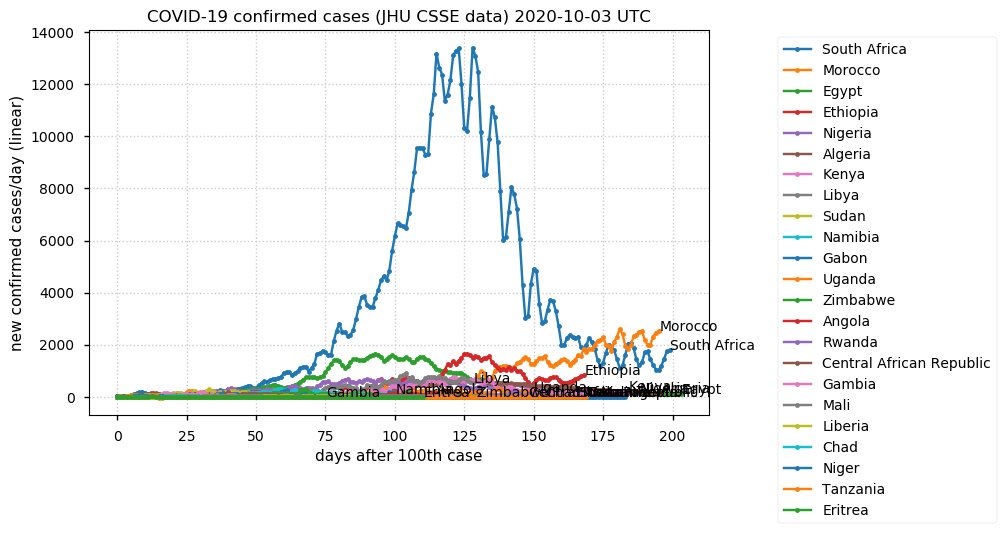
Sun Oct 04 17:50:52 +0000 2020@skathire @MHendr1cks I know someone who has to take dexamethasone or (even worse) prednisone from time to time: it does relieve symptoms quickly, but the mood swings and agitation can be very distressing.
Sun Oct 04 16:23:50 +0000 2020@astacus "googles" was better.
Sun Oct 04 15:15:58 +0000 2020I really should have a name for this, given how often I experience it. "Datentrauer" maybe?
Sun Oct 04 15:07:46 +0000 2020That feeling I get when, after reading an interesting paper, I start analyzing the data & realize that all of the interesting stuff was nonsense ...
Sun Oct 04 13:05:48 +0000 2020SF3A2:p θ(max) = 26. aka SF3a120, SAP114, PRPF21, Prp21. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells. The domain 217-464 does not produce tryptic peptides but it does contribute HLA type I peptides.
Sun Oct 04 13:05:47 +0000 2020SF3A2:p, splicing factor 3a subunit 2 (H. sapiens) 🔗 Small nuclear subunit; PTMs: M1+acetyl; SAAVs: none; mature form: 1-464 [27,915×, 168 kTa] #ᗕᕱᗒ 🔗
Sat Oct 03 12:32:36 +0000 2020SF3A1:p has multiple low complexity domains, with glutamic acid or proline being the most enriched residues.
Sat Oct 03 12:32:36 +0000 2020SF3A1:p θ(max) = 68. aka SF3a120, SAP114, PRPF21, Prp21. Its PTM pattern qualifies it as a hypermodified protein: it is over-represented in proteome phosphopeptide studies. Found in HLA type I peptide experiments only. Commonly in samples containing nucleated cells.
Sat Oct 03 12:32:36 +0000 2020SF3A1:p, splicing factor 3a subunit 1 (H. sapiens) 🔗 Midsized nuclear protein; PTMs: 53 STY+phosphoryl, 12 K+SUMOyl, 15 K+ubiquitinyl, 43 K+acetyl acceptors, R578+dimethyl; SAAVs: none; mature form: 2-793 [49,887×, 655 kTa] #ᗕᕱᗒ 🔗
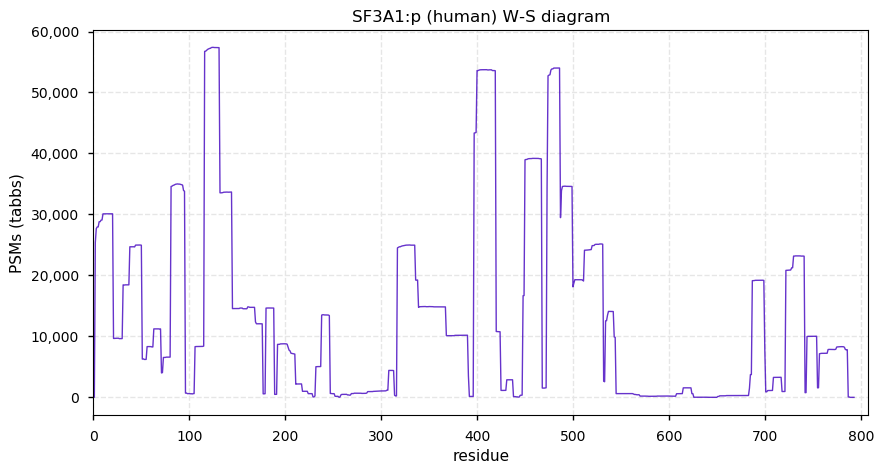
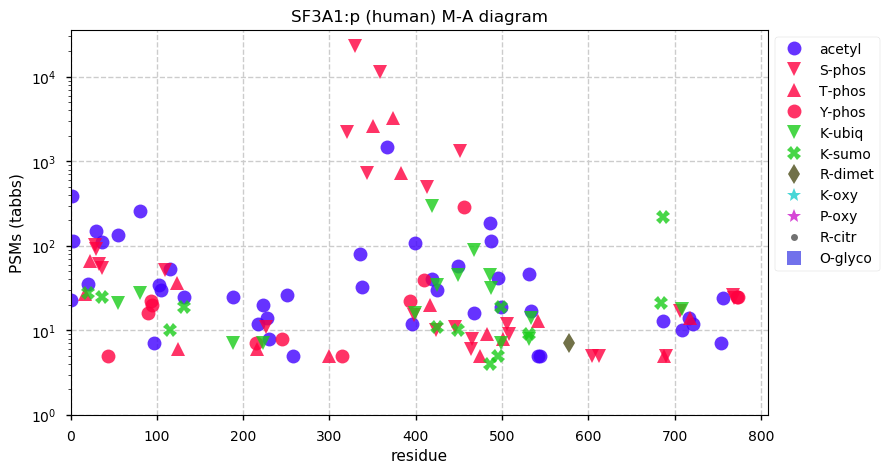
Fri Oct 02 16:00:46 +0000 2020It doesn't seem the community has a current consensus about the use of this term.
Fri Oct 02 16:00:10 +0000 2020Thanks to everyone who participated. The results show a dead-even tie between "hypermodified" implying ≥ 5% or ≥ 10% of a protein's residues serving as PTM acceptor sites, although a significant number of respondents were will to go as low as ≥ 1%.
Fri Oct 02 14:46:56 +0000 2020@AlexUsherHESA 🔗
Fri Oct 02 13:52:21 +0000 2020Interesting comment on the latest in phrenology, as per "Nature, that famous scientific tabloid". I am going to start using this description for that very tarnished old standard bearer. 🔗
Fri Oct 02 12:37:55 +0000 2020ACIN1:p has several low complexity domains, each one dominated by small numbers of specific residues, e.g.:
270 EEEEEEEEEEEEDDEEEEGDDE 291
573 SADSSSSRSSSSSSSSSRSRSRS 595
Fri Oct 02 12:35:31 +0000 2020ACIN1:p θ(max) = 53. aka KIAA0670, fSAP152, ACINUS. Its PTM pattern qualifies it as a hypermodified protein: it is over-represented in proteome phosphopeptide studies. Found in HLA type I peptide experiments only. Commonly observed in samples containing nucleated cells.
Fri Oct 02 12:32:07 +0000 2020ACIN1:p, apoptotic chromatin condensation inducer 1 (H. sapiens) 🔗 Large nuclear protein; PTMs: 163 STY+phosphoryl, 13 K+SUMOyl; SAAVs: R257K (2%), I311M (50%), S467P (43%), S478F (5%); mature form: 1-1341 [45,511×, 614 kTa] #ᗕᕱᗒ 🔗
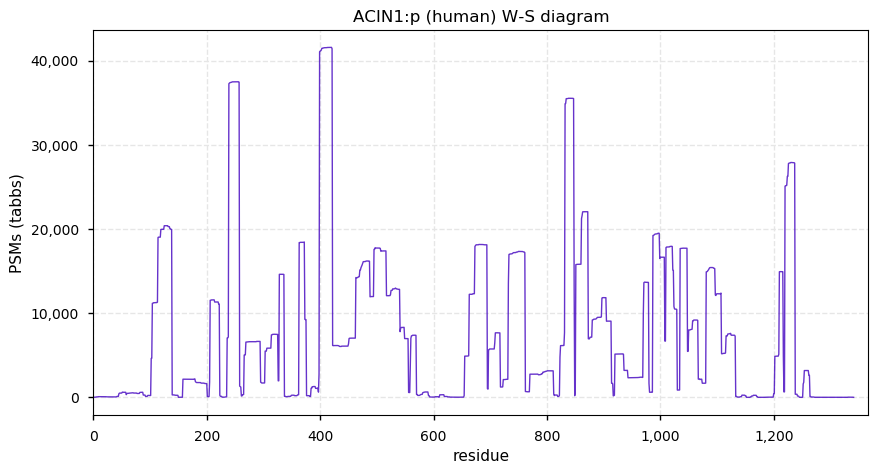
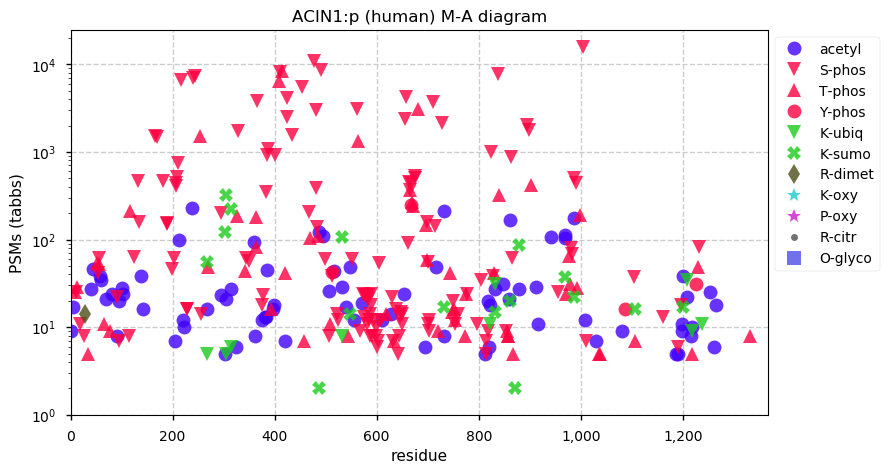
Fri Oct 02 12:14:31 +0000 2020No clear winner on this poll yet. If you have an opinion, there are only a few hours left! 🔗
Thu Oct 01 15:47:52 +0000 2020What fraction of residues need to be PTM acceptor sites for a protein to qualify as being "hypermodified"?
Thu Oct 01 13:59:54 +0000 2020If you are interested in doing a 3- channel SILAC study of phosphorylation (S,T & Y) in a cell line, PXD018566 is a good data set to examine. You will get a very good idea of the LOD/LOQ that can be obtained using this approach.
Thu Oct 01 12:10:50 +0000 2020LEF1:p is associated with too many biological functions and processes in ontologies. Has an N-terminal proline in the mature sequence (like HMGNs). Sequence has no PSM overlap with the other HMG proteins, except TCF7.
Thu Oct 01 12:10:50 +0000 2020LEF11:p θ(max) = 73. aka TCF1ALPHA, TCF10, TCF7L3. Its PTM pattern is unlike the other high mobility group proteins. Found in HLA type I peptide experiments only. Commonly observed in lymphocytes.
Thu Oct 01 12:10:50 +0000 2020LEF1:p, lymphoid enhancer-binding factor 1 (H. sapiens) 🔗 Small nuclear high mobility group protein; PTMs: STY+phosphoryl N-terminal domain, K+ubiquitinyl C-terminal domain; SAAVs: none; mature form: 2-399 [2,929×, 11 kTa] #ᗕᕱᗒ 🔗
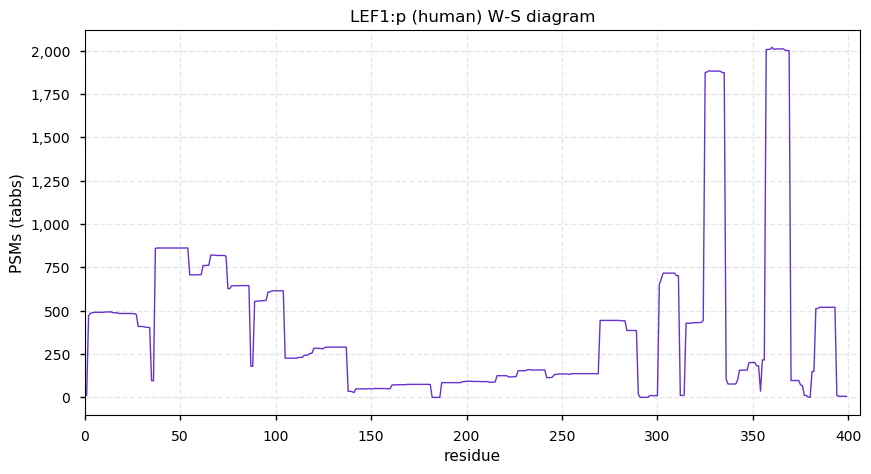
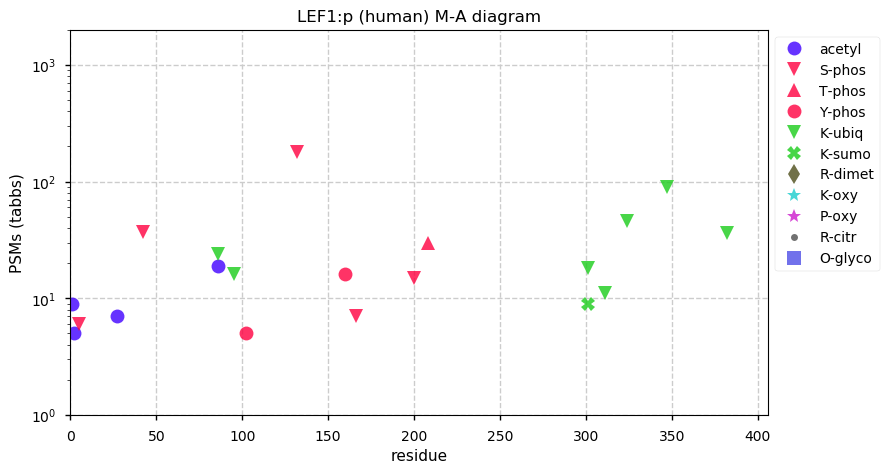
Wed Sep 30 17:47:12 +0000 2020@macro_momo I prefer Sorna. The wild life can be a little aggressive, but they are easier to deal with than bloody raccoons.
Wed Sep 30 15:16:06 +0000 2020@macro_momo That place is used in fiction but actually exists. My fake twitter locations are all purely fictional (UC Sunnydale, Paul Revere University, Miskatonic University, etc.). As is Isla Sorna, my new fake location.
Wed Sep 30 14:30:17 +0000 2020After last night, I'm moving my fake Twitter location out of the US, once and for all. Hello Costa Rica!
Wed Sep 30 12:59:19 +0000 2020HMGN1:p is associated with too many biological functions and processes in ontologies. All of the K+ubi acceptors are aPTM acceptors for acetyl. Unusual in having an N-terminal proline in the mature sequence. Sequence has no PSM overlap with the other 3 HMGN proteins.
Wed Sep 30 12:59:19 +0000 2020HMGN1:p θ(max) = 88. aka FLJ27265, FLJ31471, MGC104230, MGC117425, HMG14. Its PTM pattern is similar to HMGA1:p or HMGB1. Found in HLA type I and II peptide experiments. Common in all cell lines and tissue with nucleated cells.
Wed Sep 30 12:59:18 +0000 2020HMGN1:p, high mobility group nucleosome binding domain 1 (H. sapiens) 🔗 Small nuclear protein; PTMs: 17 K+acetyl, 7 K+ubiquitinyl, 13 ST+phosphoryl; SAAVs: A9T (6%); mature form: 2-100 [19,863 x, 156 kTa] #ᗕᕱᗒ 🔗
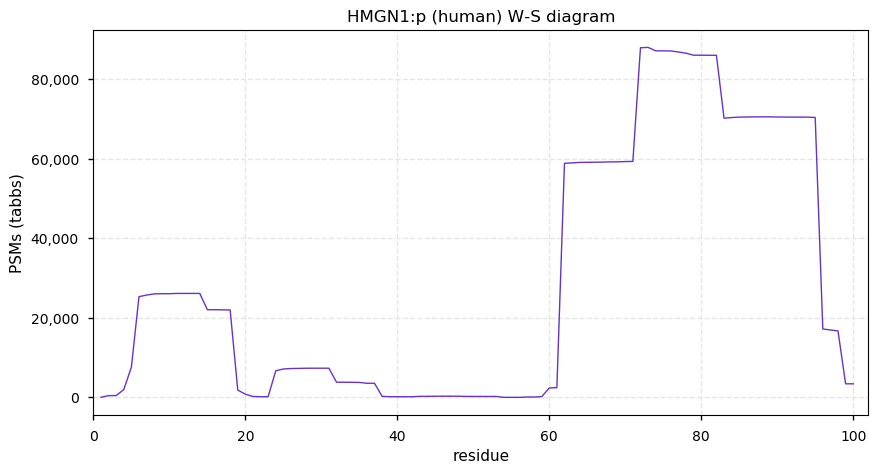
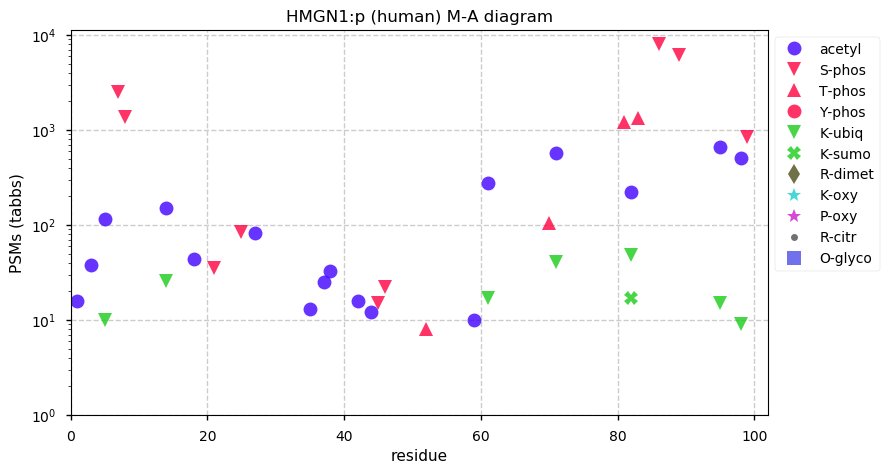
Wed Sep 30 12:49:08 +0000 2020Canadian (Ca) daily fatalities & broken out by province. Many Canadians feel their governments could have done a better job. 🔗
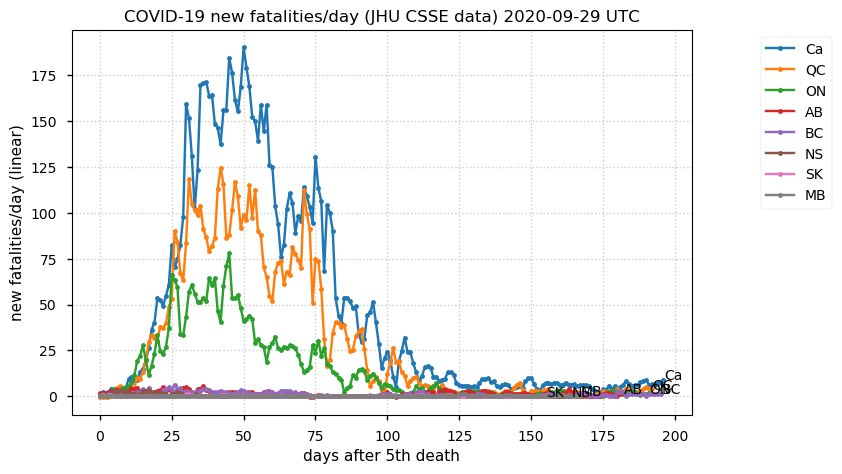
Tue Sep 29 19:38:04 +0000 2020@MiguelCos Many people do this sort of thing, but because I only deal with other people's data, I need to do it more comprehensively than most. It is more of a mind set than a routine set of steps. /…
Tue Sep 29 19:37:39 +0000 2020@MiguelCos These "best ways" tend to be indirect: using some property that can be derived from the results that was not directly measured or controlled for during the experiment. /3
Tue Sep 29 19:37:27 +0000 2020@MiguelCos Rather than accepting any part of a method (reagents, chromatography, MS/MS conditions, derivatizations, etc) as being true, I try to come up with the simplest way to test the data that will show the extent to which it was false. /2
Tue Sep 29 19:37:01 +0000 2020@MiguelCos What I call "adversarial analysis" is an attempt to deal with the fact that what actually happens in a lab is imperfectly recorded in written experimental methods (sometimes very imperfectly). /1
Tue Sep 29 14:29:42 +0000 2020Big thanks to Mikel Azkargorta, et al (PXD021140 & PXD021139) for including MGFs along with their timsTOF Pro raw data. Saves a lot of time & bandwidth for those of us primarily interested in the PSMs.
Tue Sep 29 14:22:58 +0000 2020Big thanks to MetaM for sharing the data so that I could indulge in my "adversarial analysis" methods! 🥳
Tue Sep 29 14:20:03 +0000 2020The "bad data" AAA shows the -1 residue has a similar balance between K & R, but there are other residues flanking the cleavage suggesting that trypsin is no longer dominating endogenous proteolysis. But the activity is not random, e.g. lots of L, but no I. 🔗
Tue Sep 29 14:14:10 +0000 2020The situation can be illustrated with two diagrams, representing the AAA of the -1 residue for the collection of PSMs in the 2 data sets. The "good data" AAA shows the -1 residue is mainly K| R, with 2% M for protein N-terminal peptides. Typical for a good trypsin digest. 🔗

Tue Sep 29 14:09:54 +0000 2020Just to close off the discussion, MetaM. kindly allowed me to take a look at the data. The problem—imho—was caused by slow tryptic cleavage in the "bad" sample, resulting in a big increase in PSMs with missed cleavage sites & non-tryptic cleavage. 🔗
Tue Sep 29 13:07:20 +0000 2020HMGA2:p does not share any tryptic PSMs with HMGA1:p. Unlike similarly numbered histones, A1 & A2 are not physically clustered on the same chromosome:
A1 - 6: 34,236,873-34,246,231
A2 - 12: 65,824,483-65,966,291
Tue Sep 29 11:55:21 +0000 2020HMGA2:p is associated with too many biological functions and processes in ontologies. It has a low complexity, acidic C-terminal domain (94-109)
94 QEETEETSSQESAEED 109
Tue Sep 29 11:55:21 +0000 2020HMGA2:p θ(max) = 78. aka BABL, LIPO, HMGIC. Its phosphorylation pattern has some similarities to HMGA1:p, but no corresponding acetylation or ubiquitinylation. Very rare in HLA peptide experiments. Common in some cell lines (HEK-293) but rare in others (HeLa).
Tue Sep 29 11:55:21 +0000 2020HMGA2:p, high mobility group AT-hook 2 (H. sapiens) 🔗 Small nuclear protein; PTMs: 11 ST+phosphoryl, many with high occupancy; SAAVs: none; mature form: 2-109 [8,792×, 51 kTa] #ᗕᕱᗒ 🔗
Mon Sep 28 19:32:54 +0000 2020@astacus An Argument for Sleeves (by Rob B.)
Mon Sep 28 17:43:19 +0000 2020@dtabb73 Luc Besson & his Cinéma du look could hit the right material out of the park.
Mon Sep 28 14:22:24 +0000 2020@Smith_Chem_Wisc I'd be happy to take a look, but I'd need access to the data.
Mon Sep 28 12:16:51 +0000 2020HMGA1:p binds to the minor groove of DNA with high AT content. It has a very low complexity C-terminal domain (91-101)
91 EKEEEEGISQESSEEEQ 101
Mon Sep 28 12:16:50 +0000 2020HMGA1:p θ(max) = 95. aka HMGIY. It is important for chromatin formation and maintenance. For a protein of its size, it has many high occupancy phosphorylation acceptors.
Mon Sep 28 12:16:50 +0000 2020HMGA1:p, high mobility group AT-hook 1 (H. sapiens) 🔗 Small nuclear protein; PTMs: 14 K+acetyl sites, 5 K+ubiquitinyl, 16 high occupancy ST+phosphoryl; SAAVs: none; mature form: 2-107 [25,805×, 274 kTa] #ᗕᕱᗒ 🔗
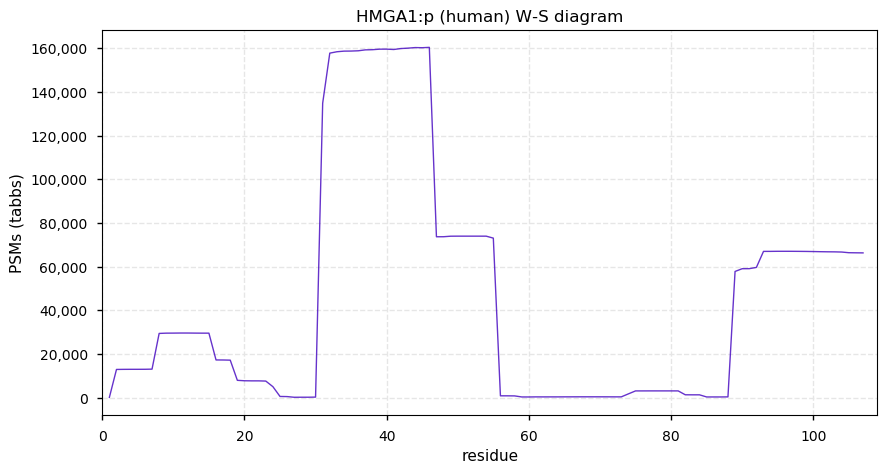
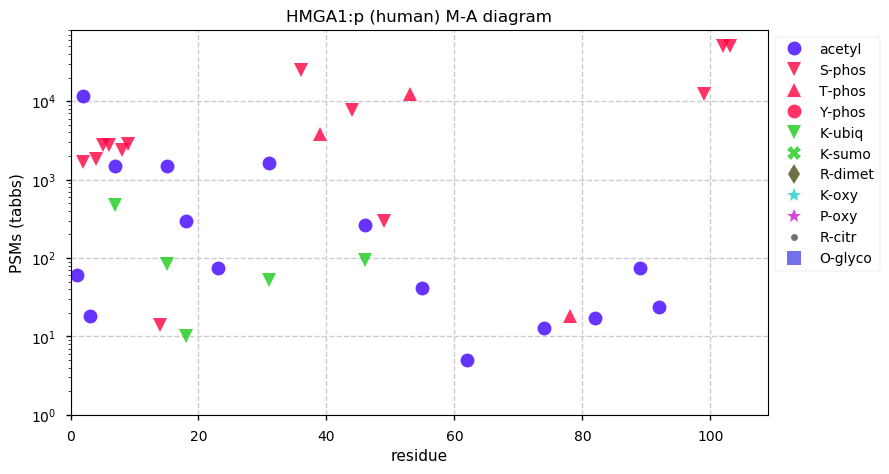
Sun Sep 27 13:04:10 +0000 2020High mobility group proteins are the red-headed stepchildren of chromatin proteomics: they are acknowledged as being necessary but dismissed as unimportant compared to histones.
Sun Sep 27 13:04:10 +0000 2020HMGB1:p has a very low complexity C-terminal domain (186-212)
186 EEEEDEEDEEDEEEEEDEEDEDEEEDDDDE 212
Sun Sep 27 13:04:10 +0000 2020HMGB1:p θ(max) = 90. aka HMG3, SBP-1, DKFZp686A04236. One of the most important proteins in chromatin formation and maintenance; very highly modified. The acetylation state determines whether the protein is in the cytosol or nucleus.
Sun Sep 27 13:04:10 +0000 2020HMGB1:p, high mobility group box 1 (H. sapiens) 🔗 Small nuclear/cytoplasmic protein; PTMs: 45 K+acetyl sites, 20 K+ubiquitinyl, 16 SY+phosphoryl; SAAVs: none; mature form: (1,2,3)-215 [54,050×, 1,042 kTa] #ᗕᕱᗒ 🔗
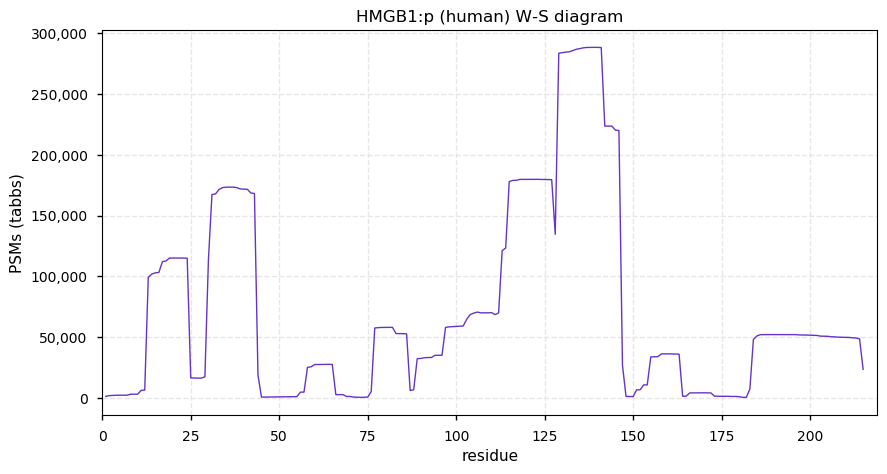
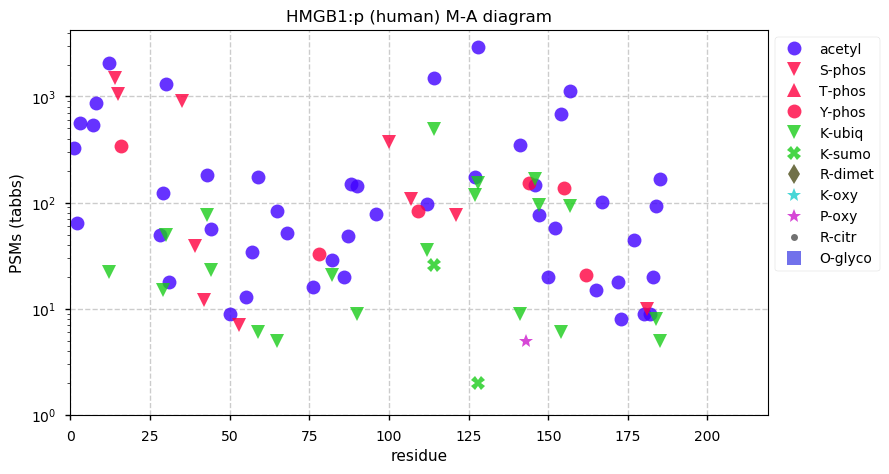
Sat Sep 26 12:14:30 +0000 2020EMC10:p has one transmembrane domain (222-242), leaving (24-221) in the ER lumen and (243-262) in the cytoplasm. The protein has been observed in a limited number of studies in urine, but not in studies of urinary microvesicles or exosomes.
Sat Sep 26 12:13:02 +0000 2020EMC10:p θ(max) = 55. aka INM02, HSS1, HSM1, C19orf63. May be present in HLA type 1 and type 2 peptide experiments. Observable in many cell lines, tissues and clinical fluids.
Sat Sep 26 12:13:02 +0000 2020EMC10:p, ER membrane protein complex subunit 10 (H. sapiens) 🔗 Small ER membrane protein; PTMs: N182+glycosyl; SAAVs: none; mature form: 24-262 [6,274×, 17 kTa] #ᗕᕱᗒ 🔗
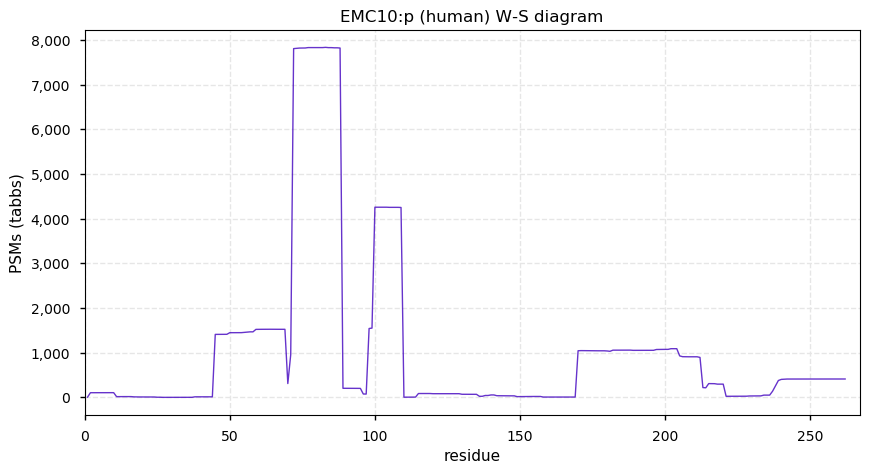
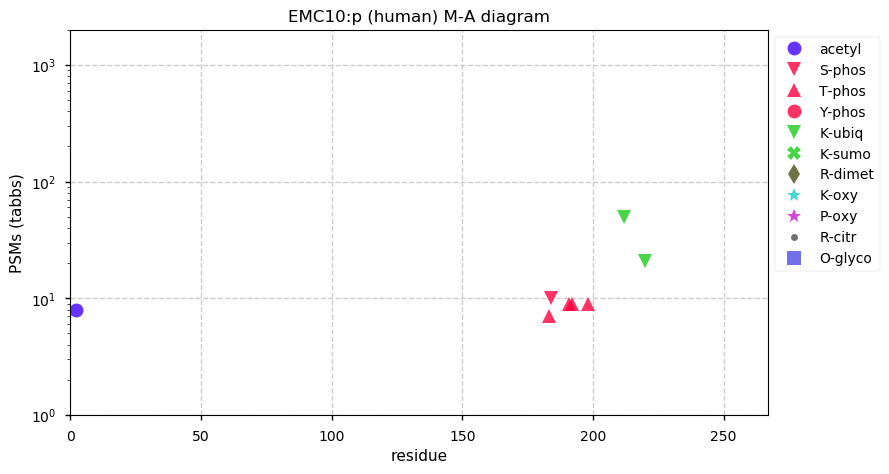
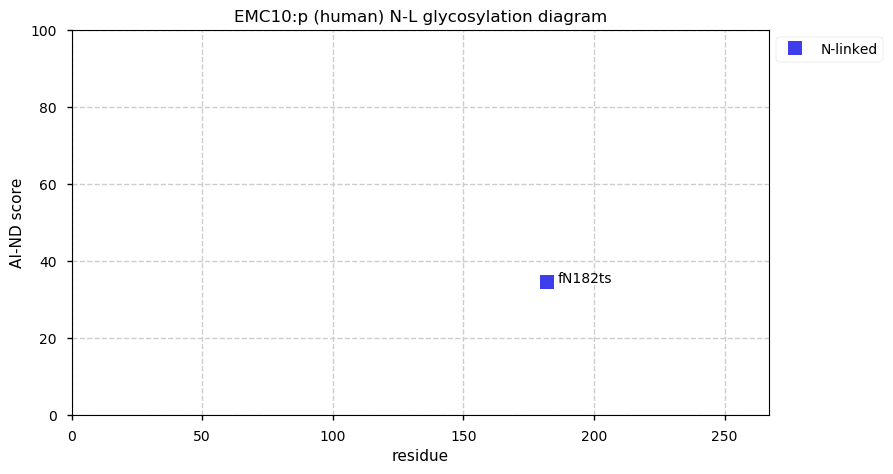
Fri Sep 25 18:11:54 +0000 2020@TrostLab @pride_ebi Maybe @ypriverol knows. There is certainly a lot of data in PRIDE without DOIs or PubMed IDs.
Fri Sep 25 18:00:06 +0000 2020The HLA type 1 & proteome data in PXD020011 are very good: nice job Fabio Marino, et al. Front. Immunol, 28 August 2020 🔗 The "proteome" data is excellent for testing SAAV-finding algos, once you allow for the 2% of PSMs with urea-induced carbamylation
Fri Sep 25 14:24:29 +0000 2020@dtabb73 Go Dave! Hopefully at least some of them are enjoyable. 🥳
Fri Sep 25 11:58:43 +0000 2020EMC9:p θ(max) = 42. aka CGI-112, C14orf122, FAM158A. May be present HLA type 1 peptide experiments. Observable in many cell lines & tissues but not in clinical fluids. It is the least frequently observed of the EMC proteins.
Fri Sep 25 11:58:43 +0000 2020EMC9:p, ER membrane protein complex subunit 9 (H. sapiens) 🔗 Small cytoplasmic protein; PTMs: G2+acetyl, no other modifications; SAAVs: none; mature form: 2-208 [1926×, 3.7 kTa] #ᗕᕱᗒ 🔗
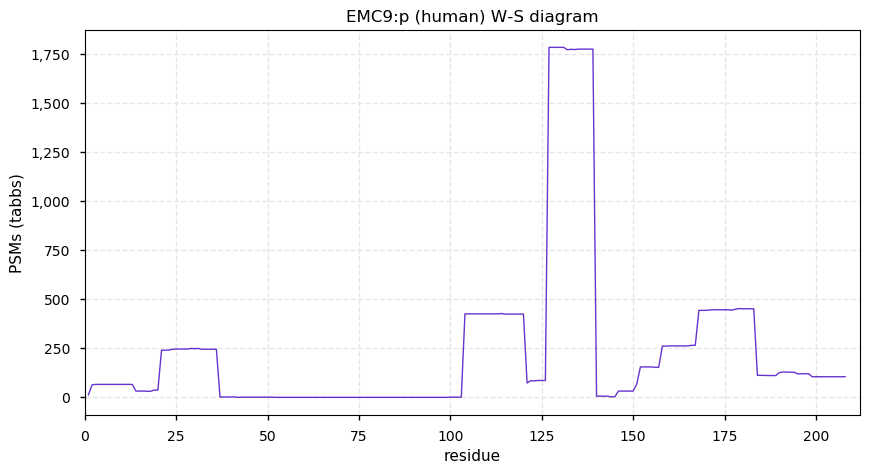
Thu Sep 24 23:50:10 +0000 2020@AlexUsherHESA Amen.
Thu Sep 24 12:07:03 +0000 2020EMC8:p θ(max) = 75. aka FAM158B, C16orf4, NOC4, C16orf2, COX4NB. Present HLA type 1 peptide experiments. Observable in most cell lines & tissues (rarely in urine, saliva & CSF). Probably associated with the ER membrane as part of the ER membrane complex.
Thu Sep 24 12:07:02 +0000 2020EMC8:p, ER membrane protein complex subunit 8 (H. sapiens) 🔗 Small cytoplasmic protein; PTMs: (S103, Y180)+phosphoryl, no glycosylation; SAAVs: none; mature form: 2-210 [10,253× , 38 kTa] #ᗕᕱᗒ 🔗
Wed Sep 23 17:37:18 +0000 2020Also, to no one in particular, if you are doing an HLA type II peptide experiment in human cells & the proteins with the most PSMs are Bos taurus C3, HBA, SERPINA1, ALB & C4A:
You are not doing it right.
Wed Sep 23 16:32:01 +0000 2020@Sci_j_my @edemmott @pwilmarth @ypriverol 34 is a little young to retire ...
Wed Sep 23 15:51:17 +0000 2020To anyone writing a proteomics paper, the terms "light" and "heavy" SILAC are meaningless unless you define the isotopically enriched residues you are using in the manuscript.
Wed Sep 23 14:27:35 +0000 2020@astacus Trypsin has 6 disulfide bonds that can hold it together, even with a few nips taken out of it.
Wed Sep 23 11:33:07 +0000 2020EMC7:p θ(max) = 63. aka C11orf3, C15orf24. Present HLA type 1 and type 2 peptide experiments. Observable in most cell lines & tissues (rarely in urine, saliva & CSF). It has 1 TM domains (160–180); (24-159) ER lumen & (181-242) cytoplasm.
Wed Sep 23 11:33:07 +0000 2020EMC7:p, ER membrane protein complex subunit 7 (H. sapiens) 🔗 Small ER membrane protein; PTMs: Y103+phosphoryl, 7 K+ubiquitinyl, no glycosylation; SAAVs: none; mature form: 24-242 [11,669×, 50 kTa] #ᗕᕱᗒ 🔗
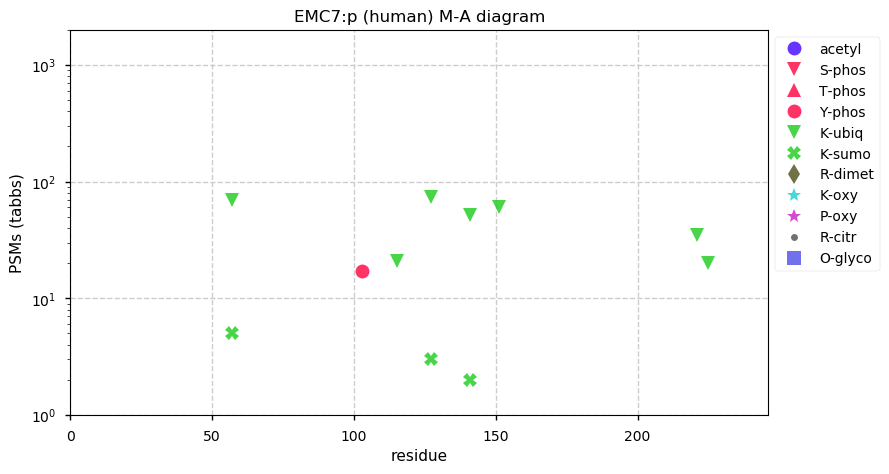
Wed Sep 23 01:48:56 +0000 2020@ProtifiLlc Thanks. 😀
Tue Sep 22 21:14:43 +0000 2020It could be that there is just a tonne of the stuff made, too.
Tue Sep 22 21:14:00 +0000 2020For anyone interested, this is full length human MT2A:
MDPNCSCAAG DSCTCAGSCK CKECKCTSCK KSCCSCCPVG CAKCAQGCIC KGASDKCSCC A
19 C's out of 61 total.
Tue Sep 22 21:11:50 +0000 2020Does anybody know why metallothionein, even though it is so small, ends up getting id'd in samples that are depleted of LMW proteins? I suspect its forms disulphide X-linked polymers after lysis that get reduced later on, but I don't know for sure.
Tue Sep 22 19:38:18 +0000 2020@byu_sam @Smith_Chem_Wisc "That's good. You've taken your first step into a larger world ..."
Tue Sep 22 19:25:23 +0000 2020@astacus But I doubt if any trace of that info made it on line. Monographs on specialist subjects (once the rage) have pretty much vanished.
Tue Sep 22 19:24:08 +0000 2020@astacus I remember reading a pre-internet paper back when I was interested in trypsin cleavage & it included a rating scheme (I, II, III, IV) for the amount of autolysis and the changes in specificity. This was quite a big deal when trying to sequence proteins using AAA.
Tue Sep 22 19:22:20 +0000 2020@astacus I always run at least semi-tryptic cleavage, so I tend to see these things. What seems to happen is that if the trypsin is a bit too old/stored incorrectly/inexpensive it undergoes autolysis, which "loosens up" its specificity to include H-X, R/K-P.
Tue Sep 22 18:45:53 +0000 2020@Smith_Chem_Wisc @byu_sam No problem. The code is pretty well documented.
Tue Sep 22 18:43:49 +0000 2020@Smith_Chem_Wisc @byu_sam Is he presenting 🔗 ?
Tue Sep 22 18:30:52 +0000 2020@astacus And I'm pretty sure it is these additional activities that make LysC effective for softening up proteins. Lysobacter enzymogenes isn't an ironic name: it secretes lots of enzymes with the aim of digesting the nearby environment into metabolically easy-to-use molecules.
Tue Sep 22 18:12:28 +0000 2020@astacus I've thought about it a lot, but it is pretty subtle. It depends on the exact conditions used (esp. time+temp) as well as the age, purity & storage of the trypsin. Adding endo-LysC knocks the whole thing into a cocked hat because of contaminating proteolytic activities.
Tue Sep 22 17:38:24 +0000 2020PXD018402, nice job! Good high-res data from C. elegans embryos, with minimal E. coli OMPs. Placentino M, et al. 🔗
Tue Sep 22 16:38:52 +0000 2020@astacus I sometimes need a trail of breadcrumbs myself.
Tue Sep 22 16:38:30 +0000 2020@astacus If you want to check for specific peptides, use 🔗 and enter the peptides either 1 at a time (top box) or a list of peptides 1 on a line (bottom box)
Tue Sep 22 16:37:02 +0000 2020@astacus if you want to see all of the peptides (non-tryptic and all) for a protein, the get the page for that protein (e.g. 🔗) and click the little green dot to get a listing of the peptides 🔗 /2
Tue Sep 22 16:34:46 +0000 2020@astacus The link had gotten mucked up: it is fixed now, but you may have to reload the page. That page is not what I suspect you want: the page gives you a TSV file of all of the peptides for a species, not a specific search. /1
Tue Sep 22 15:29:02 +0000 2020@astacus I'll take a look. I may have discontinued that and failed to remove it from that menu.
Tue Sep 22 14:58:17 +0000 2020@astacus I'm not quite sure what you are referring to by "PEPTIDES". Could you expand on this a bit?
Tue Sep 22 14:04:28 +0000 2020@Smith_Chem_Wisc & include the UTC time as well. Many people have a rather fuzzy notion of how time zones work.
Tue Sep 22 13:06:02 +0000 2020The EMC proteins have significant PPI data showing they may co-locate (PPIs for EMC1, from String, experimental data only). Note: MMGT1 was previously EMC5. 🔗
Tue Sep 22 12:21:11 +0000 2020EMC6:p θ(max) = 26. aka MGC2963, RAB5IFL, TMEM93. Present HLA type 1 peptide experiments. Observable in most cell lines & tissues (except urine, saliva & CSF). It has 2 TM domains (48–68) & (87–107); (2-47) cytoplasm, (69-86) ER lumen & (108-110) cytoplasm.
Tue Sep 22 12:21:11 +0000 2020EMC6:p, ER membrane protein complex subunit 6 (H. sapiens) 🔗 Small ER membrane protein; PTMs: A2+acetyl; aPTMs: K7+ubiquitinyl/acetyl; SAAVs: none; mature form: 2-110 [3,912×, 8.7 kTa] #ᗕᕱᗒ 🔗
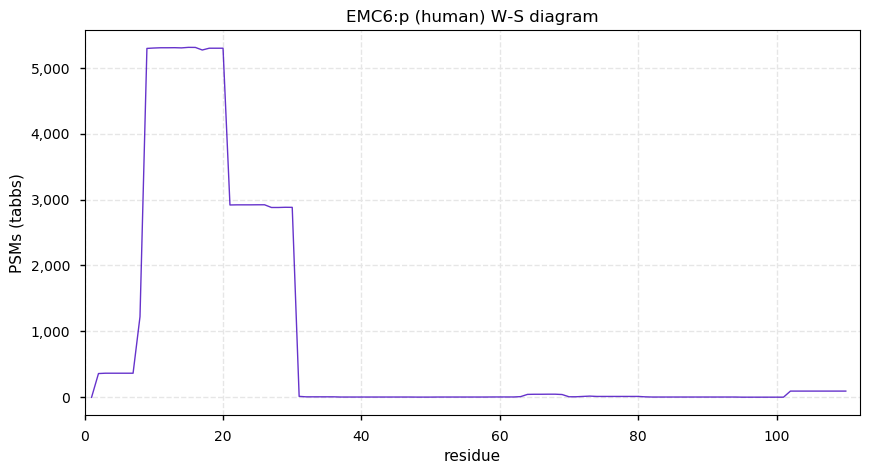
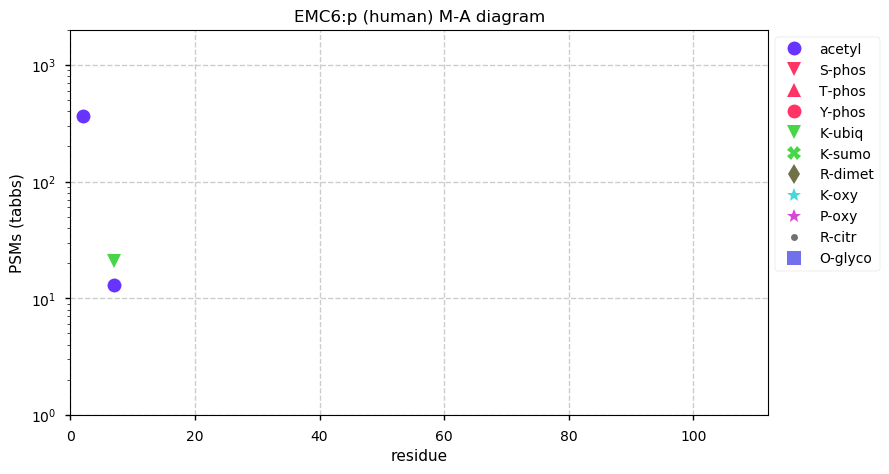
Mon Sep 21 15:25:38 +0000 2020@pwilmarth @KentsisResearch How does that affect the large number of PSMs associated with the calibration mixture channel only? Every sample has 1000's of PSMs associated with titin & thyroglobulin. when they are only in the cal. mix.
Mon Sep 21 12:18:06 +0000 2020MMGT1:p θ(max) = 64. aka EMC5, TMEM32. Found in HLA type 1 peptide experiments. Common in most cell lines, tissues; normally absent from urine and erythrocytes. It is predicted to have 2 atypical TM domains (5-25) & (45-65).
Mon Sep 21 12:16:05 +0000 2020MMGT1:p, membrane magnesium transporter 1 (H. sapiens) 🔗 Small ER membrane protein; PTMs: no N-terminal acetylation, C-terminal phosphodomain; SAAVs: none; mature form: (1,2)-131 [11,047×, 28 kTa] #ᗕᕱᗒ 🔗
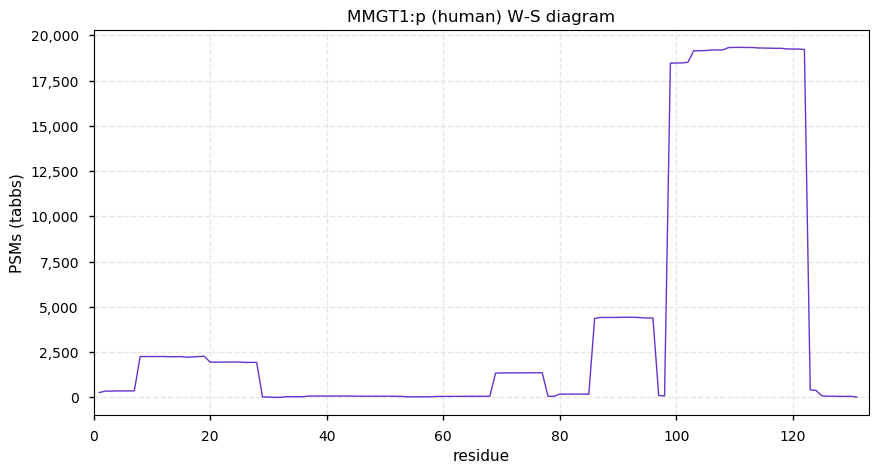
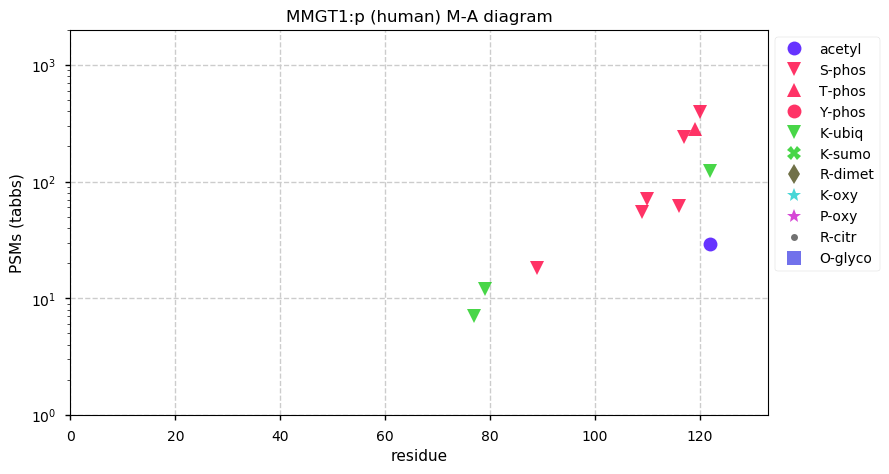
Sun Sep 20 18:26:58 +0000 2020@edward_marcotte @pwilmarth @ProteomicsNews 7-8 for big studies sounds pretty generous. The only studies I find have good metadata are small, focussed ones where there aren't many things that require tracking.
Sun Sep 20 16:49:30 +0000 2020Using certbot (from Let's Encrypt) once again makes dealing with HTTPS certificates easy (even on Windows)
Sun Sep 20 14:55:41 +0000 2020@KentsisResearch I have seen data sets from groups trying to use the mixed-cell-line-calibrant approach with tissues, but it doesn't work very well because so many of the abundant proteins in tissue are not present in cell lines.
Sun Sep 20 14:07:14 +0000 2020@KentsisResearch I guess that is where it started, but using tissue samples to make the mixture takes it to another level. Cell lines share a large number of proteins & exclude many others. They also have a much lower protein concentration range than tissue.
Sun Sep 20 12:57:41 +0000 2020Has anybody else looked at the data from 🔗 (PXD016999)? I'm particularly interested in what people think about the use of a calibration channel created from of a mixture of many samples. It seems to cause issues, but maybe it fixes more than it makes?
Sun Sep 20 12:33:02 +0000 2020EMC4:p θ(max) = 59. aka FLJ90746, MGC24415, PIG17, TMEM85. Found in HLA type 1 peptide experiments. Common in most cell lines, tissues; normally absent from urine and erythrocytes. It is predicted to have 2 atypical TM domains (84–104) & (130–150).
Sun Sep 20 12:33:02 +0000 2020EMC4:p, ER membrane protein complex subunit 4 (H. sapiens) 🔗 Small ER membrane protein; PTMs: A2+acetyl, (S22, S32, S36)+phosphoryl, (K16, K50, K67)+ubiquitinyl; SAAVs: none; mature form: 2-261 [10,091×, 30 kTa] #ᗕᕱᗒ 🔗
Sat Sep 19 14:25:01 +0000 2020If you want to do some automated data retrieval, please read these suggestions 🔗
Sat Sep 19 14:07:52 +0000 2020EMC3:p is predicted to have 2 atypical TM domains (14-34) & (118–138).
Sat Sep 19 14:07:52 +0000 2020EMC3:p θ(max) = 65. aka TMEM111. Found in HLA type 1 peptide experiments. Common in most cell lines, tissues; normally absent from urine and erythrocytes.
Sat Sep 19 14:07:51 +0000 2020EMC3:p, ER membrane protein complex subunit 3 (H. sapiens) 🔗 Small ER membrane associated subunit; PTMs: A2+acetyl, 9 K+ubiquitinyl, no glycosylation; SAAVs: none; mature form: 2-261 [11,952×, 44 kTa] #ᗕᕱᗒ 🔗
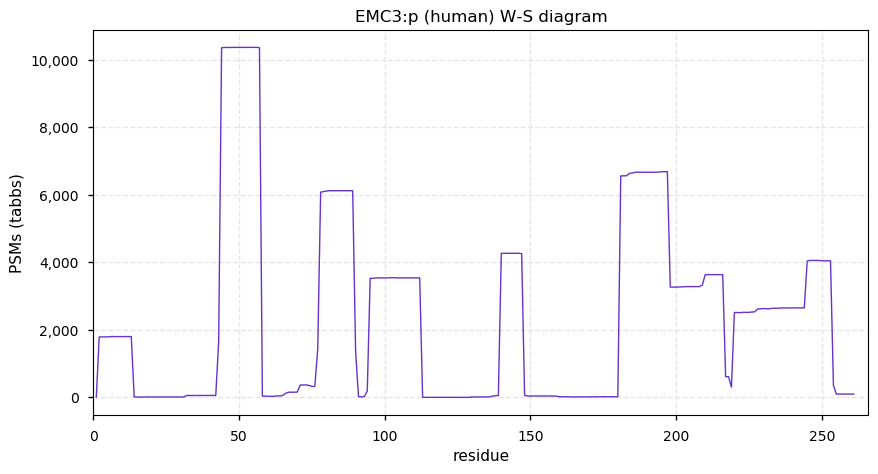
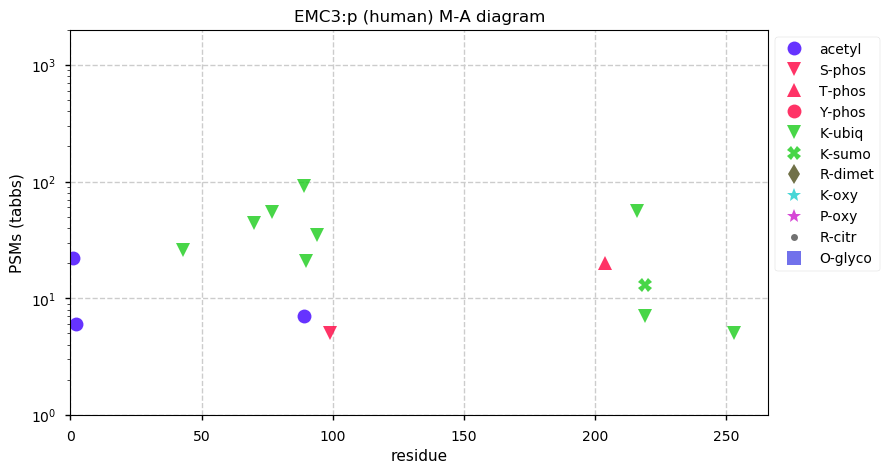
Sat Sep 19 14:04:58 +0000 2020And for last couple of years, almost all user-related problems are associated with some twit(s) using AWS to spider/scrape stuff, 1000's of threads at a time. 🤬
Sat Sep 19 12:58:35 +0000 2020I should say "other users". I tend to ignore problems I create for myself.
Sat Sep 19 12:28:04 +0000 2020Writing software and creating web resourses can be interesting and fun. Except for users: they are nothing but trouble.
Fri Sep 18 18:06:51 +0000 2020@pwilmarth "A darker grey is breaking through a lighter one"
Fri Sep 18 15:58:29 +0000 2020PXD021081, Jichang Huang, et al. 🔗 — nicely done. ⭐️
Fri Sep 18 14:52:54 +0000 2020@DanZiemianowicz It also makes load balancing a matter of adding more instances of slower modules. /4
Fri Sep 18 14:45:59 +0000 2020@DanZiemianowicz This approach makes debugging and asynchronous processing straightforward. It also simplifies the process of changing out a module that fills in a particular section. /3
Fri Sep 18 14:12:53 +0000 2020@DanZiemianowicz Once the document is complete, it gets kicked over to the database system for indexing and storage. /2
Fri Sep 18 14:11:50 +0000 2020@DanZiemianowicz I personally use a document based approach. There is a document format that can hold everything needed to run the analysis and report the results. Multiple modules can access the document and fill in their specific sections until the document is complete ... /1
Fri Sep 18 14:05:06 +0000 2020@DanZiemianowicz From an operational point of view, the layered approach leads to the capability of inserting a new hard drive, running a few diagnostics and being ready to go. The pipeline approach means that to try out a new module, you probably need a new pipeline.
Fri Sep 18 12:23:44 +0000 2020EMC2:p contains 3 tetratricopeptide repeats (TPR): (87-120), (155-188) & (192-225). These form helix-turn-helix structures usually associated with specific protein-protein interactions. No PTMs found within the TPR domains.
Fri Sep 18 12:23:44 +0000 2020EMC2:p θ(max) = 73. aka KIAA0103, TTC35. Found in HLA type 1 peptide experiments. Common in most cell lines, tissues; normally absent from urine and erythrocytes.
Fri Sep 18 12:23:44 +0000 2020EMC2:p, ER membrane protein complex subunit 2 (H. sapiens) 🔗 Small cytoplasmic protein; PTMs: A2+acetyl, no glycosylation; SAAVs: none; mature form: 2-297 [19,432×, 72 kTa] #ᗕᕱᗒ 🔗
Thu Sep 17 21:54:33 +0000 2020Human TTN has about 36,000 AA residues, so if a ribosome was running at an average rate of 5 AA per second, it would take
(36000/5) = 7200 secs = 120 minutes = 2 hours
to finish making one nascent TTN peptide.
Thu Sep 17 21:54:08 +0000 2020Thanks to everyone who participated in the poll. The literature indicates a prokaryote ribosome will add 17-20 amino acid (AA) residues per second to a nascent peptide. The slower eukaryote ribosome is has a range of between 2-8 AA added per second.
Thu Sep 17 16:22:55 +0000 2020The results are still pretty evenly spread, with 5 hours to go in the poll.
Thu Sep 17 15:08:07 +0000 2020PXD017766, nicely done (Navarro JF, et al. 🔗)
Thu Sep 17 14:15:54 +0000 2020Even though national news stories have moved on, the smoke & fires are still there (red = PM10 particles across North America today) 🔗
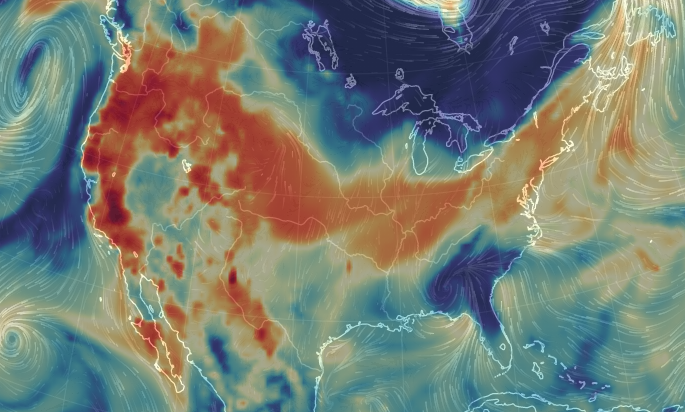
Thu Sep 17 14:05:18 +0000 2020@DanZiemianowicz I think some groups have effectively done this already, on a one-off basis. But they are still hampered by the community's insistence on inserting "pipeline" language when describing their architecture.
Thu Sep 17 14:02:08 +0000 2020Still offshore, but the winds are starting to kick up at the Kefalonia Airport (IATA: EFL, ICAO: LGKF) 🔗
Thu Sep 17 13:42:40 +0000 2020@DanZiemianowicz Probably the best way to refactor current "pipelines" into a more reliable architecture would be to adopt a layered approach, more like a protocol stack.
Thu Sep 17 12:02:05 +0000 2020Knowledge regarding the function of the proteins designated as being part of the Endoplasmic reticulum Membrane Complex (EMC) is still quite fragmentary, with contradictory models proposed in the literature.
Thu Sep 17 12:02:04 +0000 2020EMC1:p θ(max) = 74. aka KIAA0090. Found in HLA type 1 & type 2 peptide experiments. Transmembrane domain (959–979), (22-958) in ER lumen & (980-992) in cytoplasm. Common in most cell lines, tissues but rare in clinical fluids.
Thu Sep 17 12:02:04 +0000 2020EMC1:p, ER membrane protein complex subunit 1 (H. sapiens) 🔗 Midsized ER membrane protein; PTMs: (Y601, Y646, Y897)+phosphoryl, (N369, N912)+glycosyl; SAAVs: S344T (43%), R620C (1%); mature form: 22-992 [27,422×, 209 kTa] #ᗕᕱᗒ 🔗
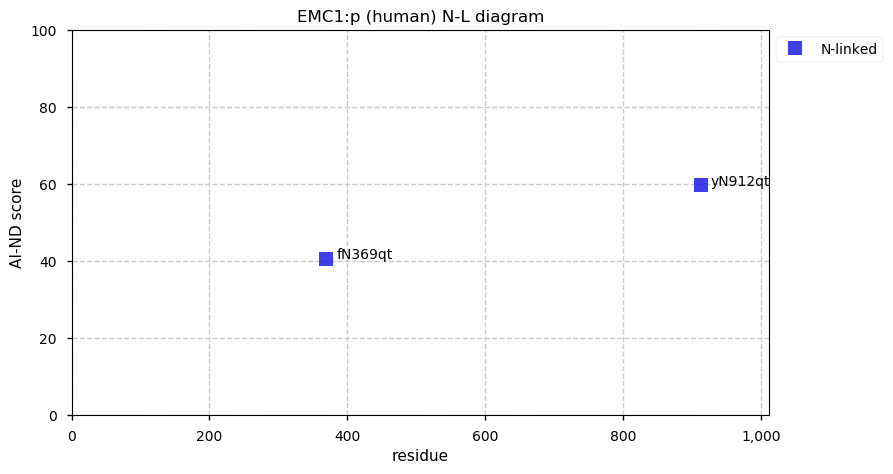
Wed Sep 16 21:50:32 +0000 2020From the time TTN mRNA translation by a ribosome begins, how long does it take to finish making a TTN nascent peptide?
(I had to discard the original poll: I left out an option 😟)
Wed Sep 16 19:37:52 +0000 2020Correction: it is will go over the Attic Peninsula, but it will make landfall on Peloponnesus.
Wed Sep 16 18:52:08 +0000 2020I've never even heard of a Medicane, but that doesn't mean that they don't exist. Seems to be aimed to make landfall on the Attic Peninsula. 🔗
Wed Sep 16 16:00:11 +0000 2020@cwvhogue I think it continues because it is a good model for academic developers: you can work on a module that is part of a system you don't understand. And because Celera promoted the idea so aggressively that it became uncool to think any other way.
Wed Sep 16 15:44:38 +0000 2020For anyone who may be using the NLM Genetics Home Reference site:
"As of October 1, 2020, the National Library of Medicine (NLM) will no longer offer Genetics Home Reference as a stand-alone website" 🔗
Wed Sep 16 15:04:32 +0000 20205. Pipelines encourage "faith-in-my-pipeline" rather than critical understanding of the results.
Wed Sep 16 15:04:31 +0000 20204. Pipelines cannot be effectively debugged. Because modules are daisy chained together with intermediate file formats, even the most obvious bugs in the pipeline can be hard to find. Unit testing modules cannot find pipeline problems.
Wed Sep 16 15:04:31 +0000 20203. Pipelines devalue broader understanding. A developer is encouraged to put all of their efforts into an individual task in minute detail and to complain about up-pipe modules while being dismissive down-pipe effects (it's not my problem).
Wed Sep 16 15:04:31 +0000 20202. Pipelines proliferate file formats. Each module in a pipeline has to communicate with the module up-pipe & the one down-pipe, usually using a custom file format that is never properly defined or tested.
Wed Sep 16 15:04:31 +0000 20201. Pipelines promote complexity, i.e., adding more modules to the pipeline. This is a property that is great for academic development groups but bad for anyone who wants to understand the results.
Wed Sep 16 14:43:50 +0000 20203. Pipelines devalue broader understanding. A developer is encouraged to put all of their efforts into an individual task in minute detail and to complain about up-pipe modules while being dismissive down-pipe effects (it's not my problem).
Wed Sep 16 14:43:50 +0000 20202. Pipelines proliferate file formats. Each module in a pipeline has to communicate with the module up-pipe & the one down-pipe, usually using a custom file format that is never properly defined or tested.
Wed Sep 16 13:03:31 +0000 2020Complicated image, but Holland, Canada and Denmark all seem to be heading into a 2nd wave of infections 🔗
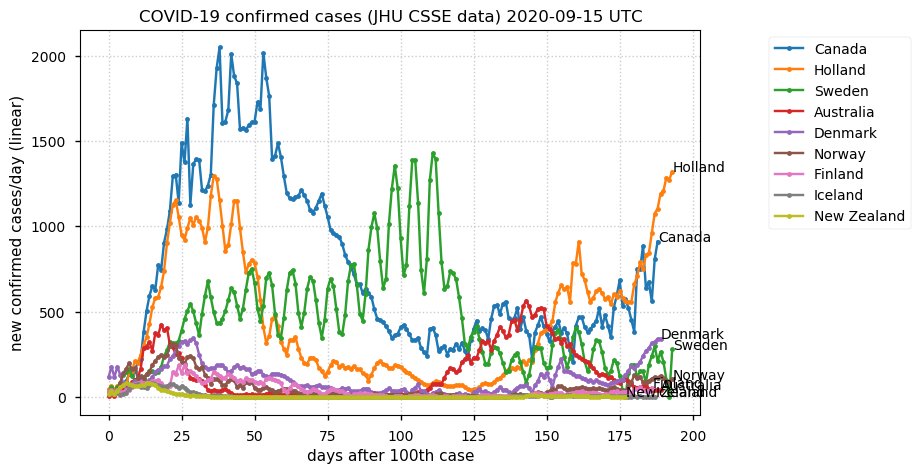
Wed Sep 16 12:42:03 +0000 2020Data analysis pipelines may be one of the worst ideas to come out of the 1990's.
Wed Sep 16 12:19:46 +0000 2020TFG:p θ(max) = 56. aka TF6, FLJ36137, SPG57. Found in HLA type 1 & type 2 peptide experiments. Low complexity domain (204-379): enriched in P's, Q's & Y's; depleted in basic and acidic residues. Common in most cell lines, tissues and clinical fluids.
Wed Sep 16 12:19:45 +0000 2020TFG:p, trafficking from ER to golgi regulator (H. sapiens) 🔗 Small ER associated cytoplasmic protein; PTMs: 26 S/T+phosphoryl sites, R381+dimethyl; SAAVs: T360P (1%); mature form: 1-396 [30,904×, 344 kTa] #ᗕᕱᗒ 🔗
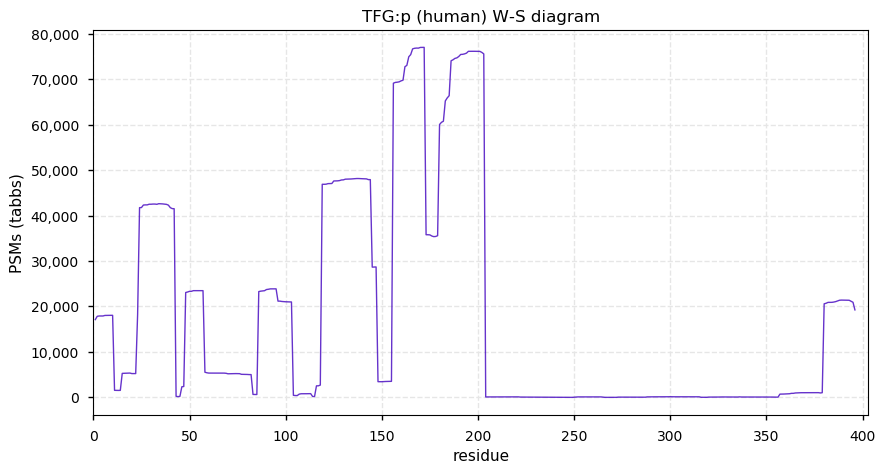
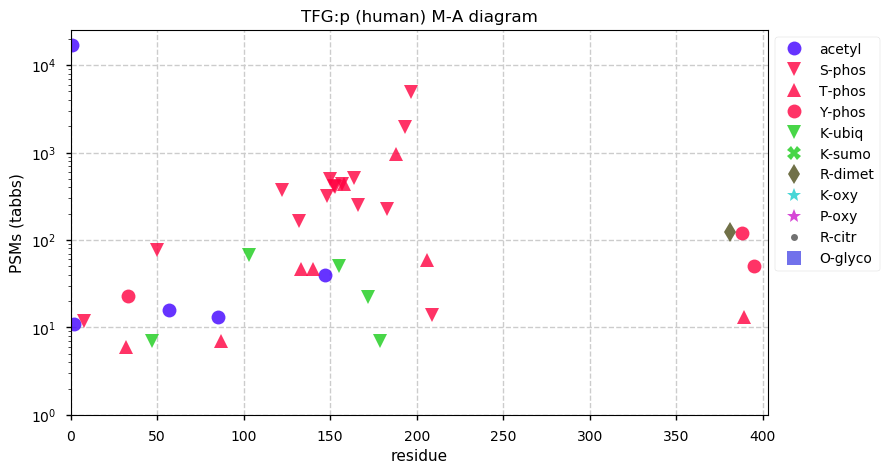
Tue Sep 15 23:17:43 +0000 2020The wind is now (as of 17:40) running at 146 kph gusting to 167 kph (91 mph/104 mph) & it is near the southeast edge of the eye wall: Viosca Knoll is the pink circled (A). The pressure has held at 983-985 mb since 12:30! 🔗

Tue Sep 15 20:01:59 +0000 2020The Viosca Knoll oil platform (29.2290°,-87.7810°) in the Gulf of Mexico near the center of Hurricane Sally is reporting sustained winds of 117 km/hr and gusts up to 144 km/hr (72 & 90 mph) as of 14:20 local time. 🔗

Tue Sep 15 18:38:38 +0000 2020@Sci_j_my Samsung and TSMC have been making them for a while now 🔗
Tue Sep 15 17:53:46 +0000 2020@VATVSLPR Could be. I don't even remember what year it was. I only really remember the long, slow, hot drive to and from the airport and the Dean (or some other academic official) telling me a kind of weird story about James Caan.
Tue Sep 15 16:06:07 +0000 2020Thank you to everyone who took part in this poll. The results were surprising to me: only 22% of respondents felt no money should be reserved while 52% thought that at least 20% should be set aside for this category of applicants.
Tue Sep 15 15:58:16 +0000 2020@VATVSLPR I hope it stays away. I almost took a job at City of Hope, many years ago, but I could not get the family to consider LA seriously.
Tue Sep 15 15:49:10 +0000 2020We are having hazy skies across Canada because of a complex of cyclonic storms off both the west & east coasts that are driving cross-continent winds at middle altitudes (illustrated here at ~ 5.5 km/18,000 ft high), pushing forest fire smoke all the way to Boston. 🔗
Tue Sep 15 14:14:03 +0000 2020@slashdot Sounds like one of those "life on Venus" type headlines.
Tue Sep 15 12:56:38 +0000 2020If you have an opinion on this idea, only 3 hours left to make it known. The results so far are a surprise to me ... 🔗
Tue Sep 15 12:05:31 +0000 2020GLG1:p θ(max) = 71. aka GP73, FLJ23608, bA379P1.3, GOLPH2, C9orf155. Found in HLA type 1 and type 2 peptide experiments. Transmembrane domain (13-36), (1-12) in cytoplasm (37-401) in the GOlgi lumen. Common in most cell lines, tissues and clinical fluids.
Tue Sep 15 12:05:31 +0000 2020GOLM1:p, golgi membrane protein 1 (H. sapiens) 🔗 Small Golgi membrane protein; PTMs: (N109, N144, S204, T218, T235)+glycosyl, 17 S/T+phosphoryl sites; SAAVs: H217R (23%), Q322H (5%); mature form: 1-401 [15,013×, 84.4 kTa] #ᗕᕱᗒ 🔗
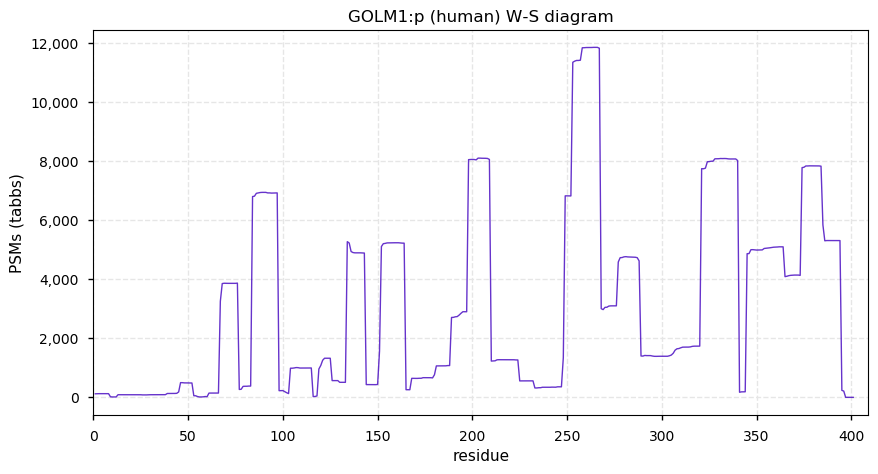
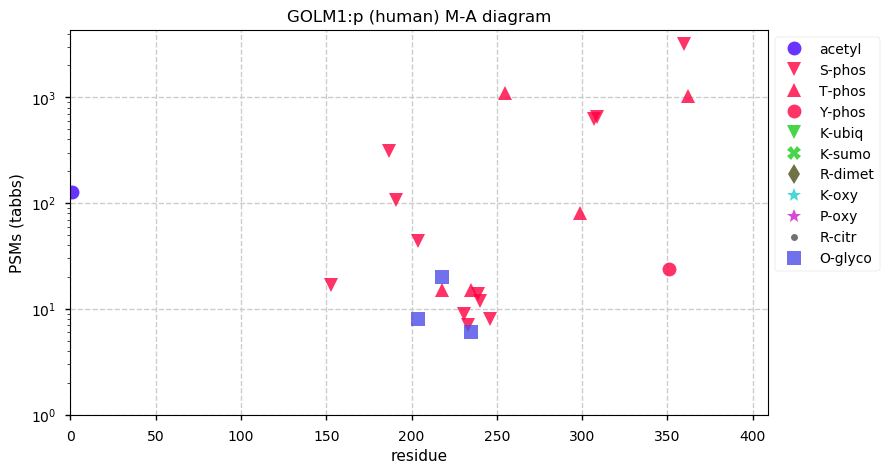
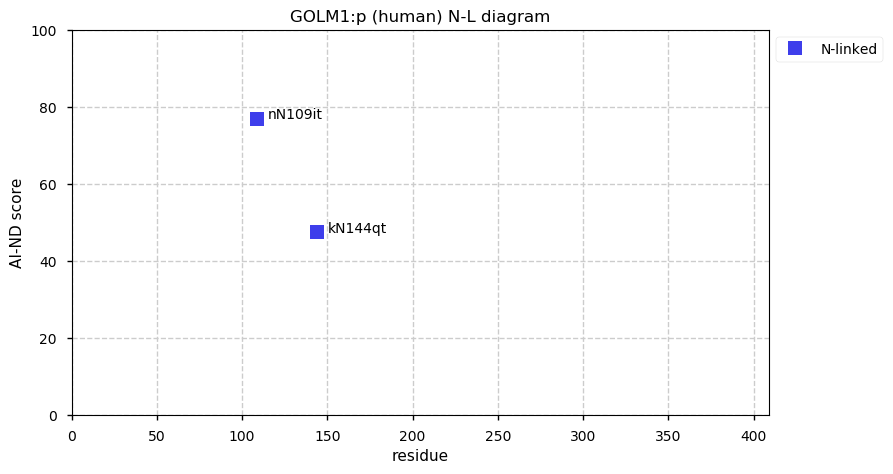
Mon Sep 14 20:18:54 +0000 2020This is starting to look biblical ... 🔗
Mon Sep 14 16:27:14 +0000 2020While only known to people who had the good fortune to work in Frank Field's lab, Field's Law states:
There is no such thing as an average protein.
Mon Sep 14 15:58:11 +0000 2020In any peer-reviewed system, how much grant money should be reserved for applicants that haven't been funded recently (e.g., no funding in the last 8 years):
Mon Sep 14 14:49:49 +0000 2020One of the odd things about "cartographic" proteome studies is that even though they mention previous studies in their manuscript, they don't seem to ever use those results to test a posteriori whether their new approach is going to work as well as hoped a priori.
Mon Sep 14 14:31:53 +0000 2020@MattWFoster @SnyderShot After analyzing a lot of the data, getting this information isn't really necessary (other than for the cat's casual interest).
Mon Sep 14 14:27:45 +0000 2020Seems to be back up and running.
Mon Sep 14 12:56:49 +0000 2020🔗 seems to be kind of stuck this morning
Mon Sep 14 12:08:39 +0000 2020Functions for GLG1:p has been the subject of some speculation in the literature, but no consensus has been reached. Note: GLG1:p in S. cervisiae is an unrelated protein.
Mon Sep 14 12:08:39 +0000 2020GLG1:p θ(max) = 61. aka MG-160, ESL-1, CFR-1. Found in HLA type 1 and type 2 peptide experiments. Transmembrane domain (1146–1166), (28-1145) in Golgi lumen, (1167-1179) in the cytoplasm. Common in most cell lines, tissues and clinical fluids.
Mon Sep 14 12:08:39 +0000 2020GLG1:p, golgi glycoprotein 1 (H. sapiens) 🔗 Large Golgi membrane protein; PTMs: (N210, N581, N677, T739, N786)+glycosyl; SAAVs: none; mature form: 28-1179 [30,051×, 272 kTa] [31,318, 272 kTa] #ᗕᕱᗒ 🔗
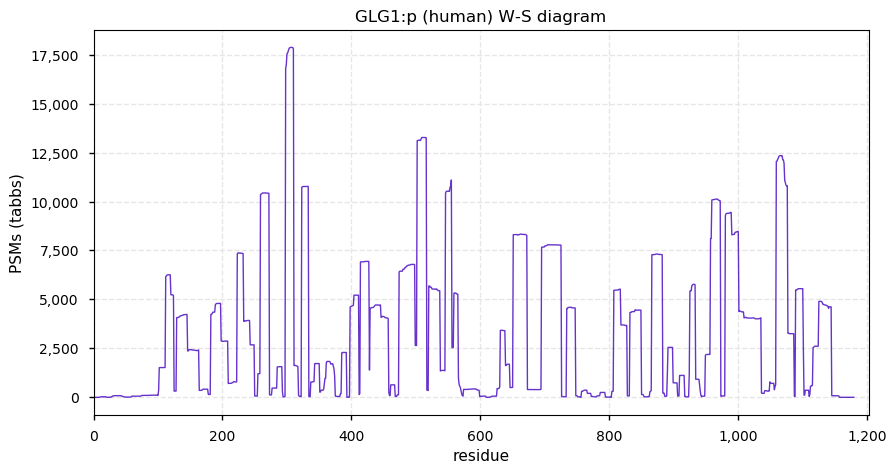
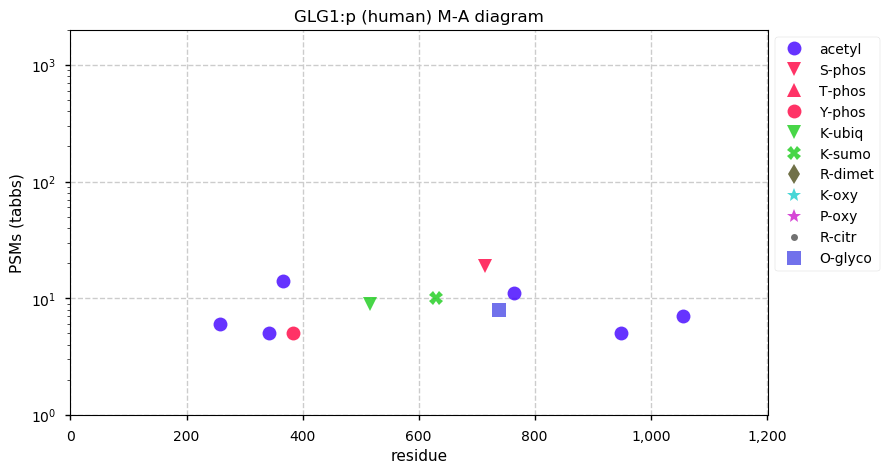
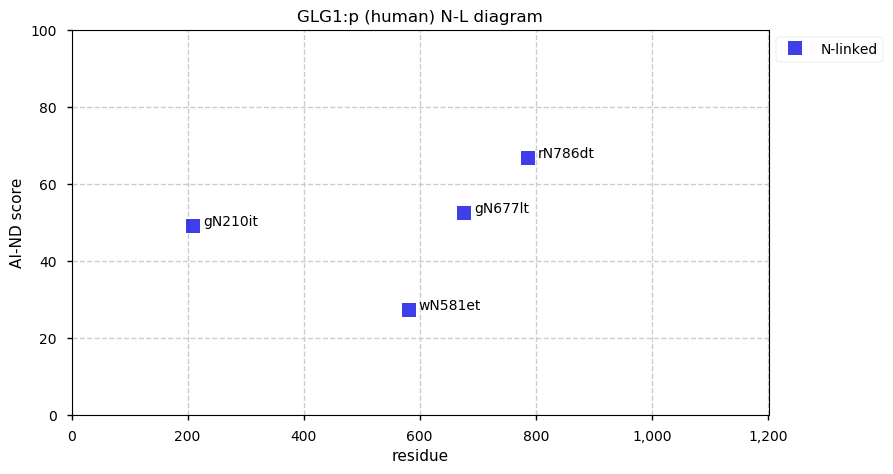
Sun Sep 13 19:14:35 +0000 2020@chrashwood I don't know if "best" is the right qualifier, but any of the ion exchange methods tend to strongly enrich N-terminal acetylation/carbamylation/carbamidomethyl, phosphorylation & deamidation in the most acidic fraction. With TMT, underivatized peptides end up there too.
Sun Sep 13 18:53:44 +0000 2020Sometimes I am still surprised by how well the 1st round of chromatography in a 2D experiment will segregate entire classes of PTMs into a single fraction.
Sun Sep 13 14:14:07 +0000 2020The experiments resulting in PXD020078 generated some nice data. Good work Gajanan Sathe, et al. 🔗
Sun Sep 13 12:40:26 +0000 2020GBF1:p θ(max) = 55. aka KIAA0248, ARF1GEF. Found in HLA type 1 and type2 peptide experiments. Present in most cell lines and tissues, but rare in fluids. Converts the GDP-bound form of ARF1:p to the active GTP-bound form.
Sun Sep 13 12:40:25 +0000 2020GBF1:p, golgi brefeldin A resistant guanine nucleotide exchange factor 1 (H. sapiens) 🔗 Large protein; PTMs: several high occupation phosphodomains; SAAVs: M375T (1%), G1693S (5%); mature form: 1,2-1009 [30,051×, 224 kTa] #ᗕᕱᗒ 🔗
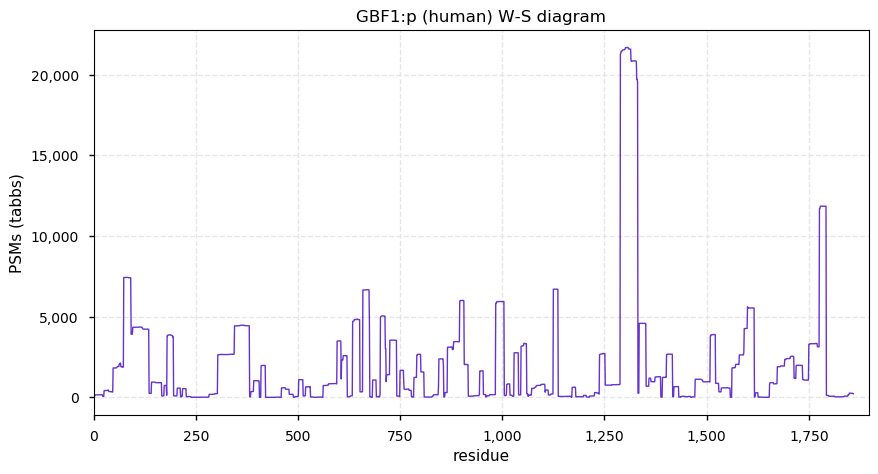
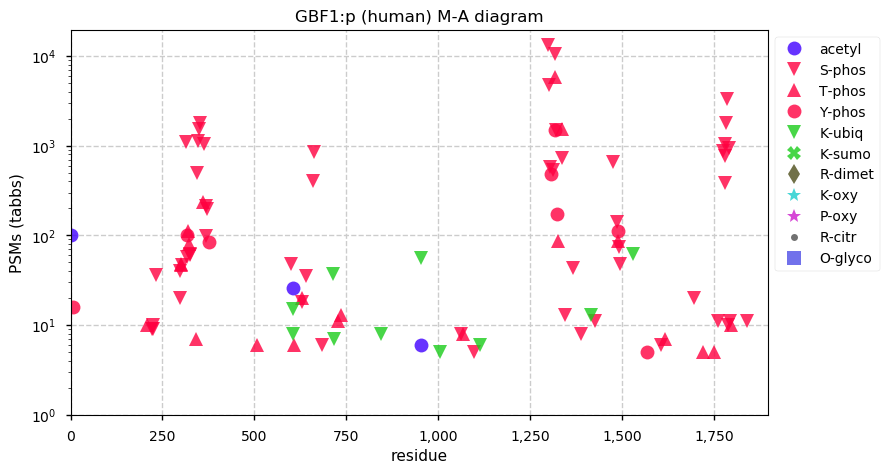
Sun Sep 13 00:26:48 +0000 2020@MattWFoster @SnyderShot There is no stopping the optimism of youth. We'll see what happens.
Sat Sep 12 23:09:07 +0000 2020@MattWFoster I have never (ever) had any luck getting info out of authors.

Sat Sep 12 15:08:45 +0000 2020The most Canadian story ever?
🔗
Sat Sep 12 14:41:52 +0000 2020Does anybody know if there is an index that associates the data files in PXD016999 (A Quantitative Proteome Map of the Human Body) with the specific tissues?
Sat Sep 12 12:33:36 +0000 2020MAN2B2:p has no PSM overlap with other human proteins. Like MAN2B1:p, it has a several unusually high MAF SAAVs, so most observations of the protein will not be homozygous for the reference sequence.
Sat Sep 12 12:33:36 +0000 2020MAN2B2:p θ(max) = 33. aka KIAA0935. Abundant in HLA type 1 and type2 peptide experiments. Observed in lymphocytes but not leukocytes. Present in most commonly used cell lines. Frequently found in urine and extracellular vesicles.
Sat Sep 12 12:33:35 +0000 2020MAN2B2:p, mannosidase alpha class 2B member 2 (H. sapiens) 🔗 Secreted enzyme; PTMs: (N226, N336, N516, N670, N675, N748)+glycosyl; SAAVs: R97L(1%), M446L/V(50%), N541S(46%), G624R(4%); mature form: 24,25-1009 [6,537×, 27.0 kTa] #ᗕᕱᗒ 🔗
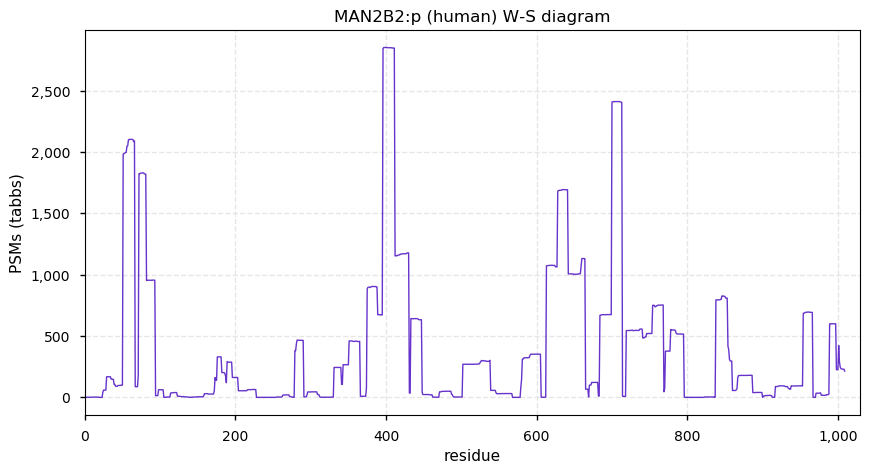
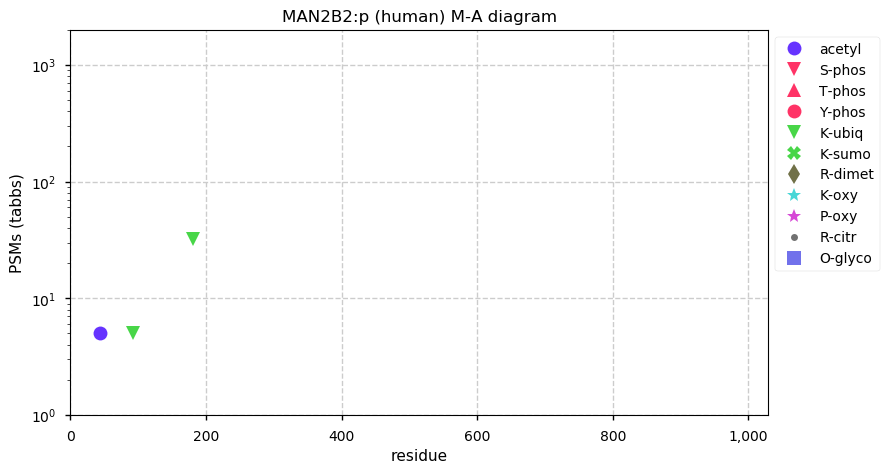
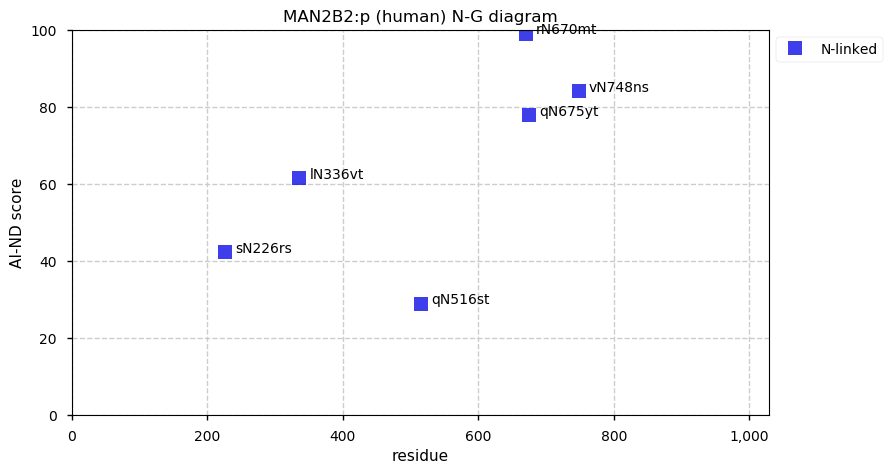
Sat Sep 12 12:28:43 +0000 2020@theoneamit @Smith_Chem_Wisc I wouldn't call it a fad. Immonium ions, like -NH3, -H20, a-y, or b-y, are there & assignable. But adding them in to a scoring algorithm results in a noticable reduction in performance. It is worth giving them a try, but they don't add enough new info to warrant inclusion.
Fri Sep 11 17:15:50 +0000 2020@AlexHgO @kusterlab I love the code docs. For example, on the top of 🔗:
/* Welcome to the second mess that is DataUpload.js */
// TODO:
// Make it work
They may want to do a code review prior to sending of to a journal 😀
Fri Sep 11 16:21:51 +0000 2020@Smith_Chem_Wisc I'm in the "no" category. I have tried with and without immonium several times over the years & I have been unable to find evidence of that including them improves the results: it has always tended to make things a little worse.
Fri Sep 11 15:52:09 +0000 2020Thanks to every one who participated in the poll. Among people with an opinion, 73% felt that immonium ions should be considered by any algorithm that assigns peptides to MS/MS spectra.
Fri Sep 11 15:29:11 +0000 2020@pwilmarth If you want to follow along in nearly real time, the data is in the "Luminosity" chart at the bottom of this page 🔗
Fri Sep 11 15:20:31 +0000 2020@pwilmarth Yesterday's solar radiation in Portland plotted on the same scale as Tuesday's measurements (from NOAA's DW6016 weather station). 🔗
Fri Sep 11 14:49:59 +0000 2020Only 1 hour left to vote on this one. 🔗
Fri Sep 11 14:40:03 +0000 2020But then again, I've never really understood the point of any of the proteomics "cartography" studies that have come out in the last 6 years.
Fri Sep 11 13:57:12 +0000 2020NDUFA11 is kind of a weird example ...
🔗
Fri Sep 11 12:09:30 +0000 2020MAN2C1:p is believed to be part of the cytosolic mechanism (which includes NGLY1:p) used to degrade N-linked protein glycosylation that is missed by the lysosomal mechanism.
Fri Sep 11 12:09:30 +0000 2020MAN2C1:p θ(max) = 65. aka MANA1, MANA. Abundant in HLA type 1 peptide experiments. Observed in most commonly used cell lines and many tissues. Compared to many other mannosidases, it is rarely found in urine.
Fri Sep 11 12:09:29 +0000 2020MAN2C1:p, mannosidase alpha class 2C member 1 (H. sapiens) 🔗 Large cytoplasmic protein; PTMs: (S160, S174, S480)+phosphoryl, 6 K+ubiquitinyl, no glycosylation; SAAVs: M336T (1%), R818H (1%); mature form: 2-1040 [6,792×, 27.5 kTa] #ᗕᕱᗒ 🔗
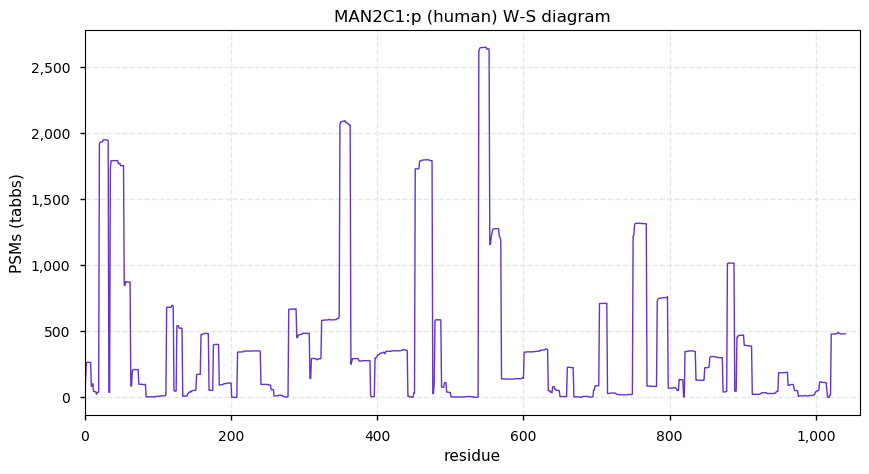
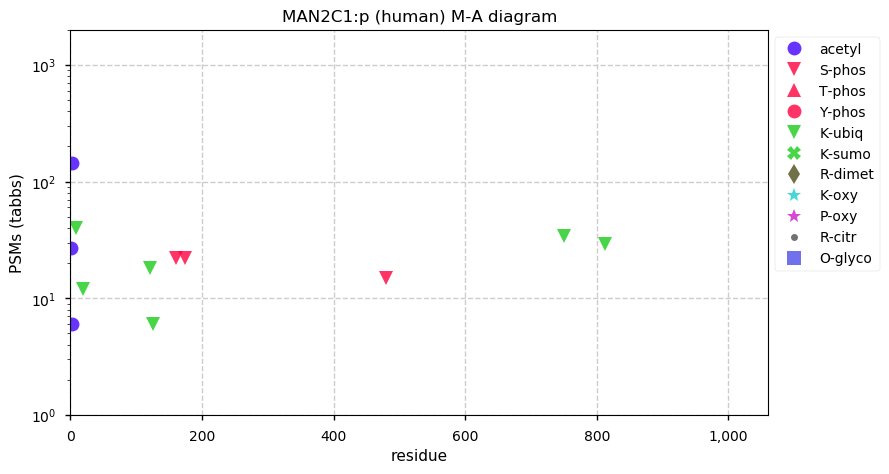
Fri Sep 11 11:47:22 +0000 2020Spain and France seem to be well into their 2nd wave of infection. 🔗
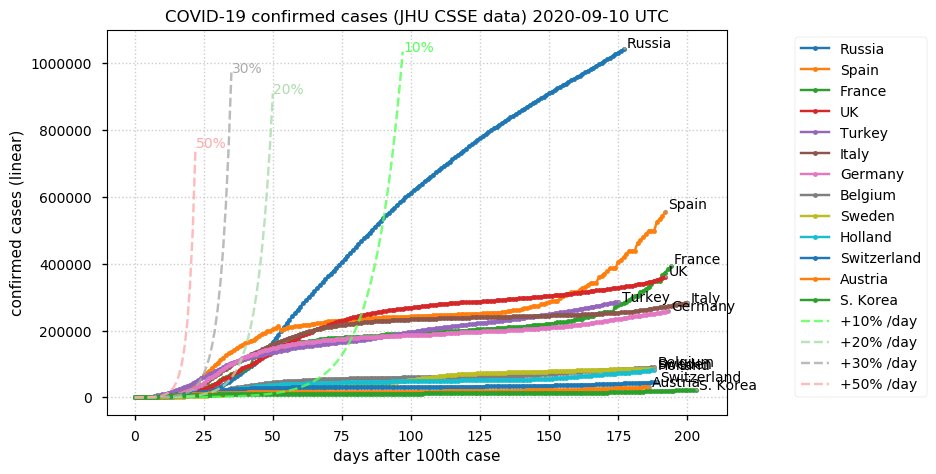
Thu Sep 10 19:03:08 +0000 2020@marcoyannic @Karl_Mechtler @hustniu It is human HSP90AB1:p 42-53 (it is also in HSP90B1:p 103-114)
Thu Sep 10 18:32:15 +0000 2020@ProteomicsNews In this context, what constitutes a "faker"?
Thu Sep 10 17:37:30 +0000 2020@birgits61642917 A little quantitation of how dark it was yesterday. Solar radiation for 2020/09/09 (in orange) compared to a 2020/09/07 (a sunny day, in blue) at the "NOVATO 1NE FIRE ROBINHOOD" weather station 🔗
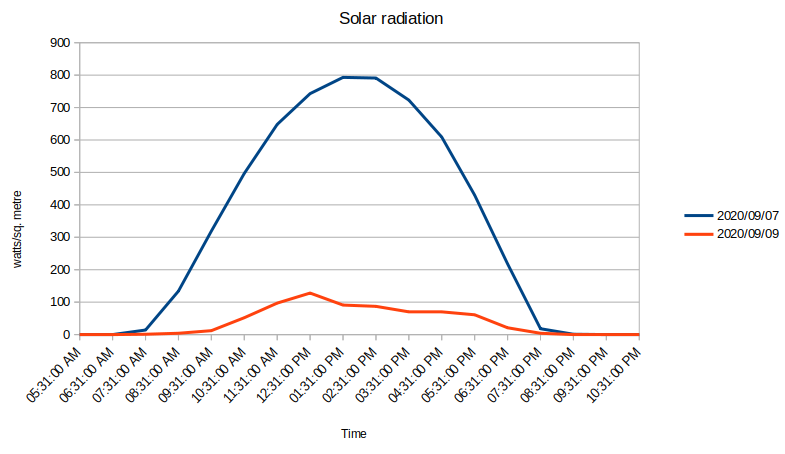
Thu Sep 10 16:00:35 +0000 2020@IonSource You may have been the first, but you seem to have started something.
Thu Sep 10 15:46:04 +0000 2020Should a search engine consider immonium ions when assigning a peptide sequence to an MS/MS spectrum?
Thu Sep 10 13:39:34 +0000 2020This display of surface winds and 10 um particle density pretty much shows why the sky will be red again today on the US West Coast 🔗
Thu Sep 10 12:20:49 +0000 2020MAN2B1:p has a rather high MAF for three of its SAAVs, meaning that it will be comparatively rare for an individual to be homozygous for the reference sequence at all three sites.
Thu Sep 10 12:20:49 +0000 2020MAN2B1:p θ(max) = 70. Large lysosome protein; aka LAMAN, MANB. Abundant in HLA type 2 peptide experiments. Observed in most commonly used cell lines, tissues and clinical fluids.
Thu Sep 10 12:20:49 +0000 2020MAN2B1:p, mannosidase alpha class 2B member 1 (H. sapiens) 🔗 PTMs: (N113, N310, N367, N497, N692, N766, N930)+glycosyl; SAAVs: L278V (24%), T312I (33%), R337Q (25%), A481S (6%), G741R (2%); mature form: 50-1011 [21,107×, 148 kTa] #ᗕᕱᗒ 🔗
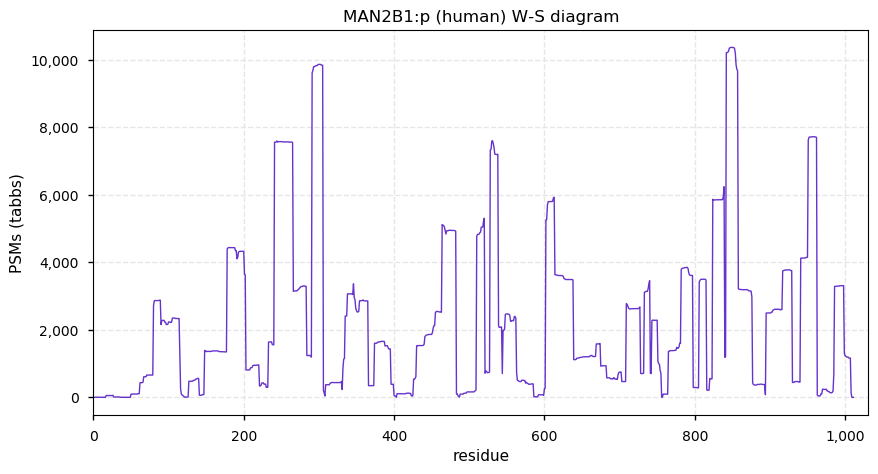
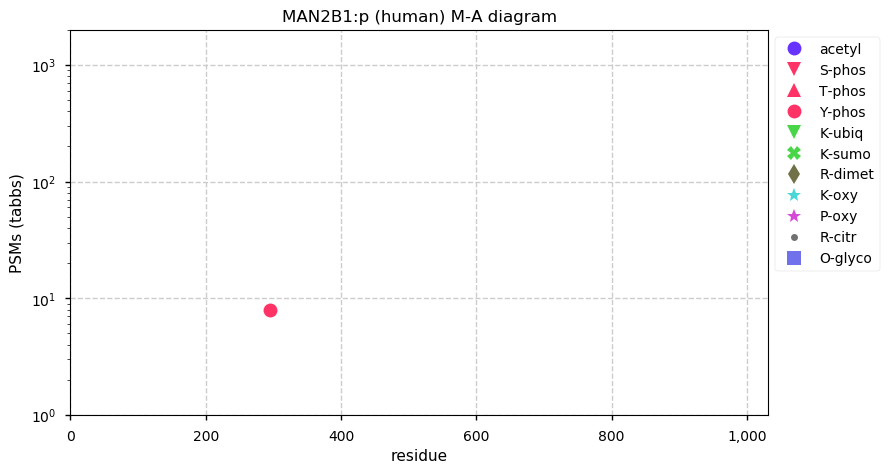
Wed Sep 09 21:25:49 +0000 2020@Sci_j_my @pwilmarth That is outlined in Revelation 4.
Wed Sep 09 20:22:55 +0000 2020@goodlettlab1 Most of my biology/biochemistry textbooks seem pretty quaint now, with the exception of the histology ones.
Wed Sep 09 17:17:56 +0000 2020I think I'm done now, but experience tells me there is bound to be yet-one-more-thing popping up ...
Wed Sep 09 17:15:53 +0000 2020GPMDB has been a little flakier than usual this morning, as I work through a bunch of OS + firmware changes and upgrades.
Wed Sep 09 16:57:24 +0000 2020@birgits61642917 According to a ground-based automated weather station nearby, at 8 AM local time the solar radiation was 7 W/m2. Normally, on a clear day that station reads 111 W/m2 at that time.
Wed Sep 09 16:49:35 +0000 2020Does anybody really use Flash anymore? I noticed Win10 was still updating it, but I can't imagine why other than some lingering contractual obligation.
Wed Sep 09 12:15:40 +0000 2020MAN2A1:p θ(max) = 59. aka GOLIM7, MANA2. 1 transmembrane domain (7-26), (1-6) cytoplasmic & (27-1144) lumenal. Present in both HLA type 1 and 2 peptide experiments. Observed in most commonly used cell lines and abundant in clinical fluids.
Wed Sep 09 12:02:29 +0000 2020MAN2A1:p, mannosidase alpha class 2A member 1 (H. sapiens) 🔗 Large Golgi membrane protein; PTMs: S80, S82, Y733+phosphoryl, N1125+glycosyl, 15 K+ubiqutinyl; SAAVs: F841Y (1%), P1027S (1%); mature form: 1-1144 [17,122×, 100 kTa] #ᗕᕱᗒ 🔗
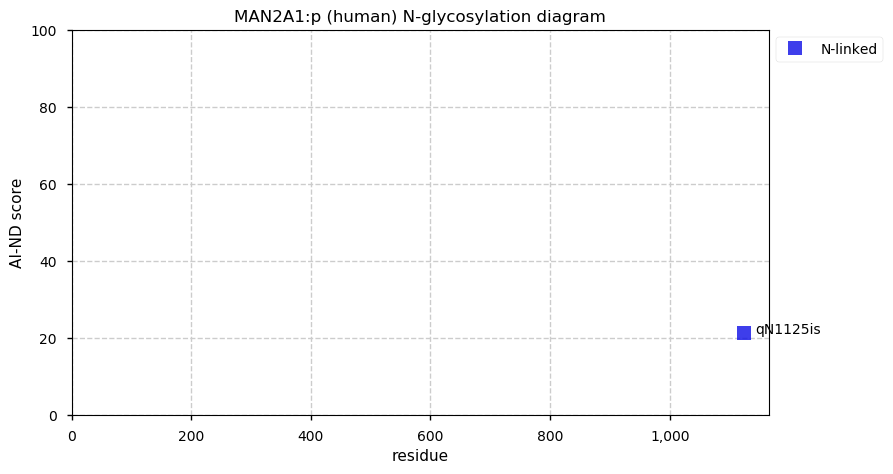
Tue Sep 08 16:28:08 +0000 2020An example of this type of simple graph 🔗
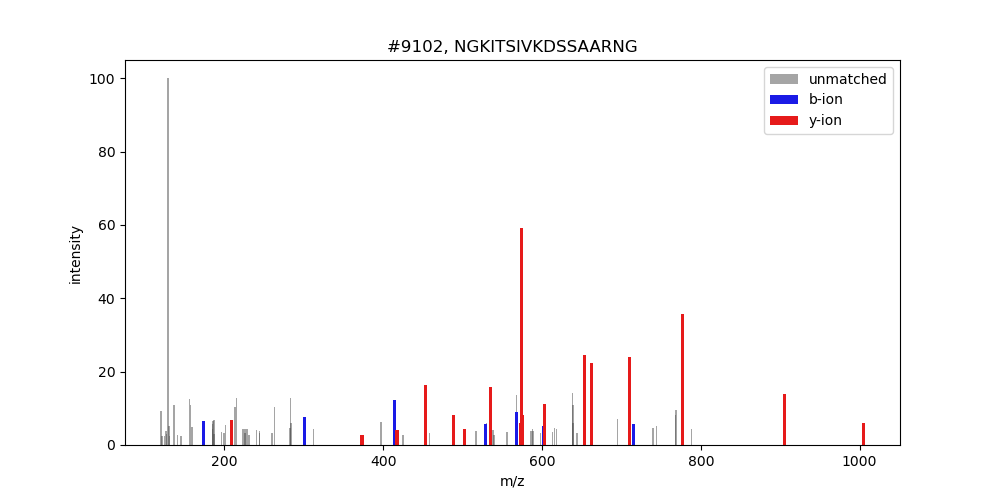
Tue Sep 08 16:23:10 +0000 2020The resulting graph is relatively simple, but anyone who knows (or is willing to learn) how matplotlib works can add bells and whistles.
Tue Sep 08 16:21:59 +0000 2020To anyone who was interested in this thread, I have uploaded a simple Python 3 solution to this problem (🔗). It uses pymsfilereader (🔗) to access the .raw file. 🔗
Tue Sep 08 12:13:31 +0000 2020MAN1C1:p θ(max) = 38. aka HMIC. 1 transmembrane domains (23-43), (1-22) cytoplasmic & (44-630) lumenal. Observed in both HLA type 1 and 2 peptide experiments. Absent from commonly used cell lines.
Tue Sep 08 12:13:31 +0000 2020MAN1C1:p, mannosidase alpha class 1C member 1 (H. sapiens) 🔗 Midsize Golgi membrane protein; PTMs: S164+phosphoryl; SAAVs: none; mature form: 1-630 [1,510×, 3.4 kTa] #ᗕᕱᗒ 🔗
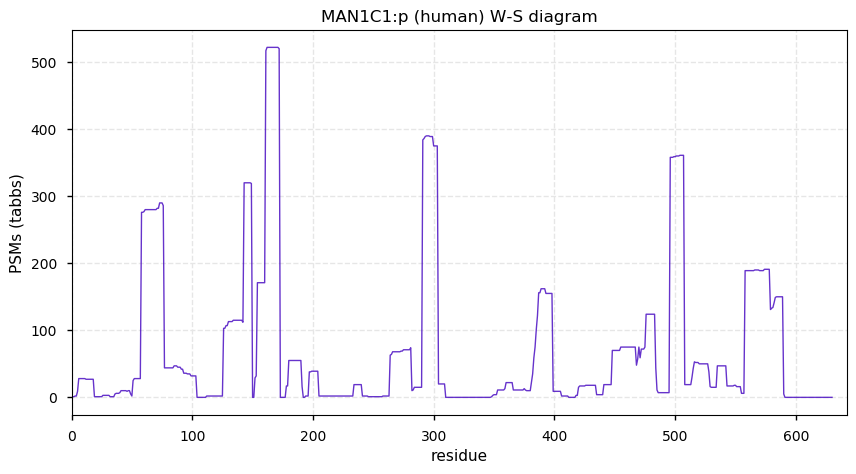
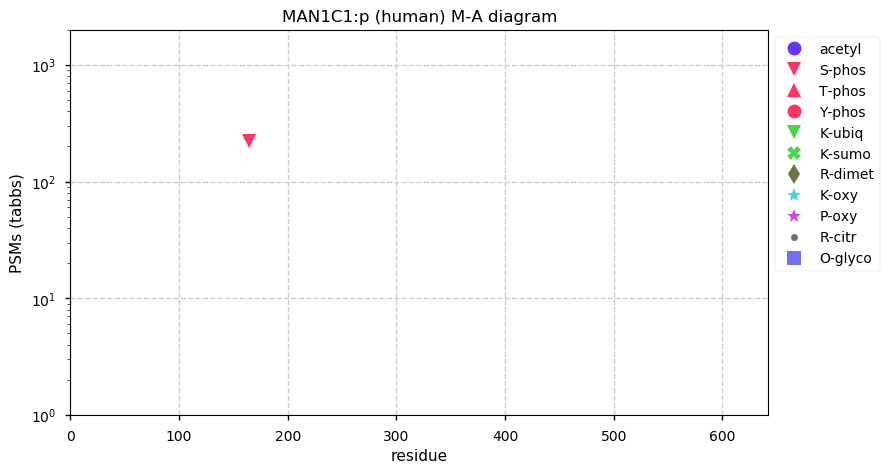
Mon Sep 07 20:45:24 +0000 2020While the title of the manuscript may be a little over-the-top, the data (PXD016477) associated with 🔗 is really pretty interesting
Mon Sep 07 12:23:28 +0000 2020MAN1B1:p is observed in most cells and tissues. Unlike MAN1A1:p, rarely observed in urine or blood plasma. There is no proteomics evidence for translation initiation at M37.
Mon Sep 07 12:07:22 +0000 2020MAN1B1:p θ(max) = 54. aka MANA-ER, MRT15, ERManI. 1 transmembrane domains (85-105), (2-84) cytoplasmic & (106-699) lumenal. May be involved in the proposed ER quality control compartment.
Mon Sep 07 12:07:21 +0000 2020MAN1B1:p, mannosidase alpha class 1B member 1 (H. sapiens) 🔗 Midsize ER membrane protein; PTMs: N-terminal phosphodoamin, T144, S154, T239+glycosyl; SAAVs: N59S (9%), G477R (1%); mature form: 2-699 [9,416×, 36 kTa] #ᗕᕱᗒ 🔗
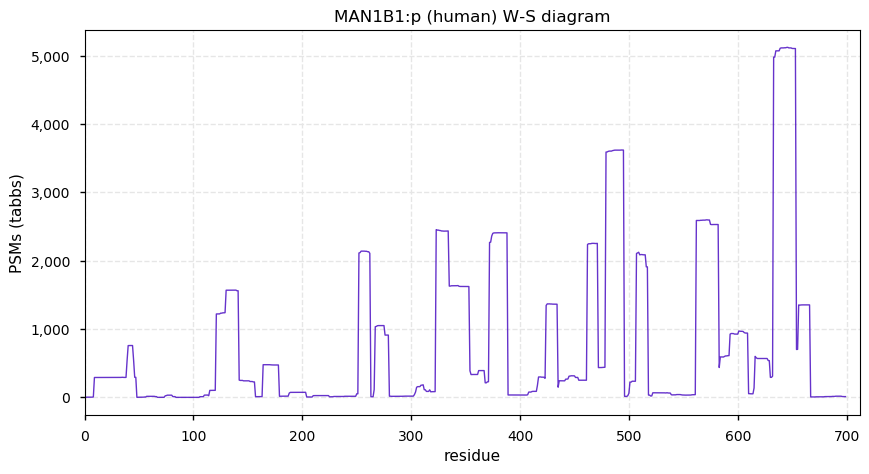
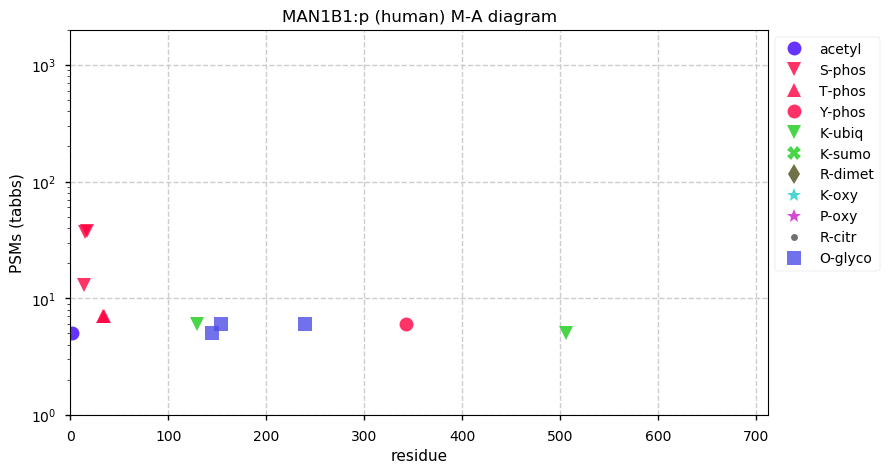
Sun Sep 06 22:34:32 +0000 2020MAN1A1:p is commonly observed in clinical fluid samples, e.g., urine, blood plasma and CSF.
Sun Sep 06 22:34:12 +0000 2020MAN1A1:p θ(max) = 57. 1 transmembrane domains (42–62), (2-41) cytoplasmic & (63-653) lumenal.
Sun Sep 06 15:06:43 +0000 2020@cdsouthan I would add low complexity sequences, esp. when it results in high or low pI proteins
Sun Sep 06 12:19:35 +0000 2020MAN1A1:p, mannosidase alpha class 1A member 1 (H. sapiens) 🔗 Midsize Golgi membrane protein; PTMs: S12+phosphoryl; SAAVs: none; mature form: 2-653 [11,208×, 72 kTa] #ᗕᕱᗒ 🔗
Sat Sep 05 18:07:10 +0000 2020@Popher After getting quite a few responses, I think you are right. So I had to write one myself, which I was hoping to avoid. Fortunately, the COM interface for the Thermo files isn't that hard to use (as compared to many COM interfaces), so it only took a few hours.
Sat Sep 05 18:02:43 +0000 2020@bffo @NCBI @emblebi @DDBJapan I was just wondering if they had any plans. Since I am interesting proteomics, downloading GenBank releases isn't useful (or practical) & most of NCBI (other than PubMed) isn't very relevant, so I haven't been following developments there for quite a while.
Sat Sep 05 16:54:10 +0000 2020Thanks to everyone who participated in the poll. It appears that there is still a significant minority of people who believe that the Mr should be considered by a bottom-up protein ID algorithm.
Sat Sep 05 16:22:45 +0000 2020@bffo @NCBI @emblebi @DDBJapan So, to your knowledge, no change in the near future.
Sat Sep 05 12:04:37 +0000 2020KRTCAP2:p θ(max) = 38. aka KCP2. 3 transmembrane domains (6–23), (35–55) & (76–108). It has a KKXX C-terminal endoplasmic reticulum retention motif (133-136), unlike the other OST subunits.
Sat Sep 05 12:04:37 +0000 2020KRTCAP2:p, keratinocyte associated protein 2 (M. musculus) 🔗 Small ER membrane protein; PTMs: T124+phosphoryl; SAAVs: none; mature form: 2-136 [2,083×, 3.7 kTa] #ᗕᕱᗒ 🔗
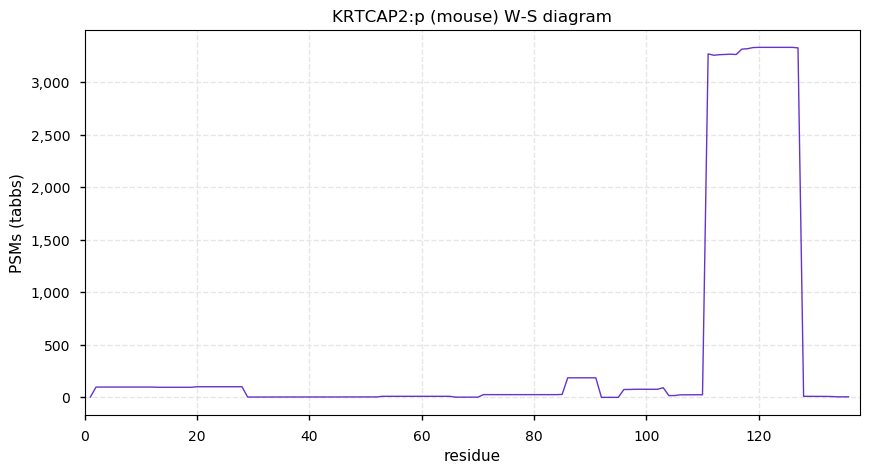
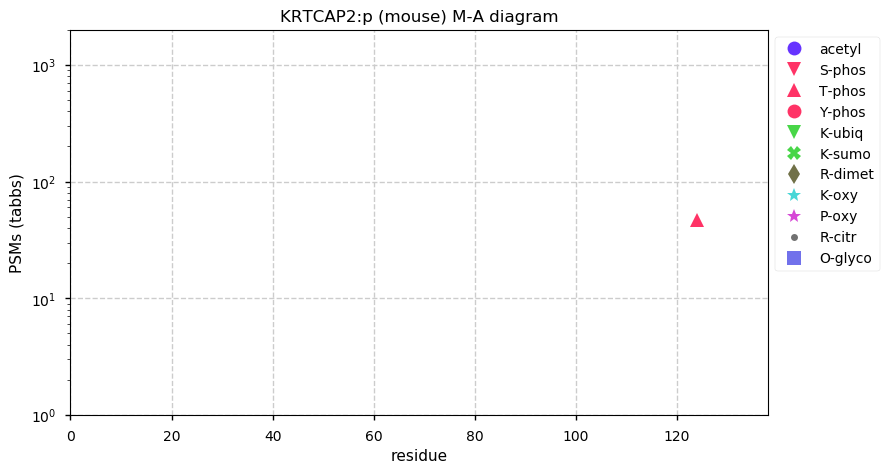
Fri Sep 04 21:11:00 +0000 2020@Popher Thermo actually makes the .dll available for free (with registration) on their software download site. There are also older versions kicking around on the net if backwards compatibility is an issue.
Fri Sep 04 17:50:15 +0000 2020@bffo @NCBI @emblebi @DDBJapan I have thought about it quite a bit. When do you think they will change from this model of information release to something more suited to its current scale & variety?
Fri Sep 04 16:36:14 +0000 2020When identifying proteins from SDS-PAGE gel bands using bottom-up MS/MS proteomics, is a protein's calculated intact mass a useful constraint for an identification algorithm? Assume that each band's Mr is also available.
Fri Sep 04 14:55:02 +0000 2020The HGNC names for the subunits of human oligosaccharyltransferase sound like the protein was assembled from the contents of a junk drawer.
Fri Sep 04 14:23:20 +0000 2020However, what I do like about looking at public data is that once in a while I'm wrong. Just additional proof that Intermittent reinforcement can make almost anything addictive.
Fri Sep 04 14:20:49 +0000 2020One thing I dislike about looking at public data is looking at the description of the method & saying to myself "that isn't going to work", then looking at the data and paper confirms my intuition.
Fri Sep 04 12:33:51 +0000 2020DAD1:p is an essential subunit of both OST-A and OST-B in humans. Its C-terminal transmembrane domain is unusual in that it has no extension into the ER lumen to anchor it to the membrane.
Fri Sep 04 12:33:50 +0000 2020DAD1:p θ(max) = 54%. 3 transmembrane domains (31–51), (53–73) & (93–113); domain (2-30) in the cytoplasm. Observed in HLA type I peptide studies, with several peptides from the domains (4-16), (73-85) & (99-107). All PTMs are on domains in the cytoplasm.
Fri Sep 04 12:33:50 +0000 2020DAD1:p, defender against cell death 1 (H. sapiens) 🔗 Small ER membrane protein; PTMs: S2+acetyl, Y16+phosphoryl; aPTMs: K82+acetyl/ubiquitinyl; SAAVs: none; mature form: 2-113 [21,035×, 76.6 kTa] #ᗕᕱᗒ 🔗
Thu Sep 03 17:18:46 +0000 2020So no avoiding it: I get to write one.
Thu Sep 03 17:17:28 +0000 2020After about a day, the answer seems to be "No". There may be solutions that can be assembled from components, but no existing, stand-alone OS software.
Thu Sep 03 16:21:13 +0000 2020@VATVSLPR So it is even more on point wrt to the COVID-19 discussion than I thought.
Thu Sep 03 16:11:50 +0000 2020@IonSource I find most people don't quite get "comorbidity", but it would be more accurate.
Thu Sep 03 16:01:16 +0000 2020@bittremieux @NatureBiotech @MarkusElsner1 @ProteomeXchange Maybe we have reached the point where "name & shame" has become a useful tactic when dealing with editors.
Thu Sep 03 15:47:20 +0000 2020They are both rather interesting (& well done) takes on SARS-CoV-2 proteomics.
Thu Sep 03 15:13:54 +0000 2020Globally, about 4,000 deaths per day attributed to TB & for the last 100 days, about 5,000 deaths per day attributed to COVID-19.
Thu Sep 03 15:12:12 +0000 2020Something to keep in mind when thinking about global health policy wrt COVID-19, given that while TB no vaccine, but there are effective therapies & public health measures against it.
🔗
Thu Sep 03 15:02:23 +0000 2020Does anybody know if these 2 data sets have an associated publication yet?
🔗
🔗
Thu Sep 03 12:37:44 +0000 2020Iran seems to be coming down from its 2nd phase of infection. Turkey seems to be heading in to a 2nd phase. 🔗
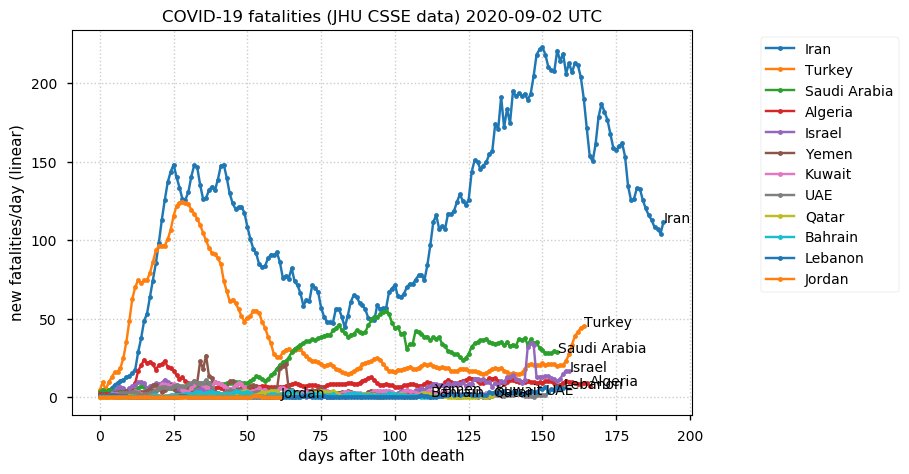
Thu Sep 03 12:06:17 +0000 2020TMEM258:p is an essential subunit of both OST-A and OST-B in humans. Under the name Kuduk, this protein has been associated with membrane stabilization in the nuclear membrane.
Thu Sep 03 12:06:16 +0000 2020TMEM258:p θ(max) = 61%. aka Kuduk, Kud, C11orf10. 2 transmembrane domains (16–38) & (54–77); (1-15) in the cytoplasm. Observed in HLA type I peptide studies, with several peptides from the domains (1-17) & (36-48).
Thu Sep 03 12:06:16 +0000 2020TMEM258:p, transmembrane protein 258 (H. sapiens) 🔗 Very small ER membrane protein; PTMs: M1+acetyl, no glycosylationl; SAAVs: none; mature form: 1-79 [5,903×, 13.6 kTa] #ᗕᕱᗒ 🔗
Wed Sep 02 17:49:46 +0000 2020Is there any stand-alone open source software that allows you to take:
1. a .raw file;
2. an MS/MS scan number; &
3. a peptide sequence + mods
and from that generate a nice looking spectrum with matched ions marked up.
Wed Sep 02 17:17:19 +0000 2020@ypriverol @byu_sam The example used in the image (human serum albumin) shows how tricky this type of "quantitative" interpretation can be in general.
Wed Sep 02 15:48:12 +0000 2020Changed 🔗 to use Raleway as the main font (from Open Sans), sticking with Roboto for tables/code/sequences. I particularly like the "l" character and the text figures.
Wed Sep 02 12:57:26 +0000 2020It looks like South Africa is nearly done turning around a bad situation. Keep it up, SA! 🔗
Wed Sep 02 12:13:50 +0000 2020OSTC:p is an essential subunit of both OST-A and OST-B in humans. All PTM sites occur on the N-terminal cytoplasmic domain.
Wed Sep 02 12:13:49 +0000 2020OSTC:p θ(max) = 40%. aka DC2. 3 transmembrane domains (33–53), (84–104), (118–138); (1-32) in the cytoplasms; (139-149) is ER lumenal. Observed in HLA type I peptide studies, with several peptides from the domain (62-73).
Wed Sep 02 12:13:49 +0000 2020OSTC:p, oligosaccharyltransferase complex non-catalytic subunit (H. sapiens) 🔗 Small ER membrane protein; PTMs: M1+acetyl, no glycosylation; aPTMs: K18+ubiqutinyl/acetyl; SAAVs: none; mature form: 1-149 [16,346×, 36.5 kTa] #ᗕᕱᗒ 🔗
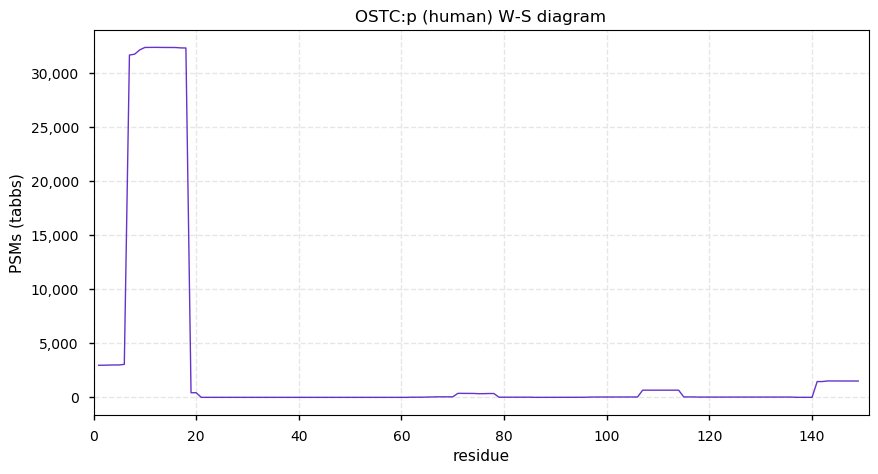
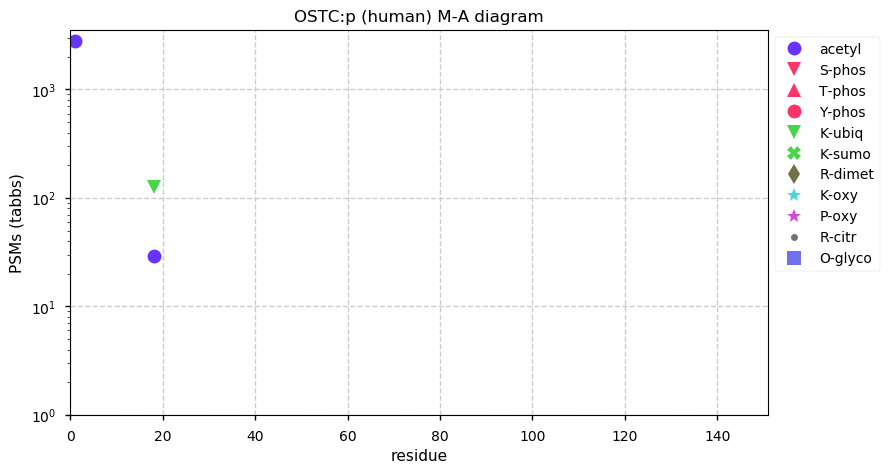
Tue Sep 01 20:46:20 +0000 2020@TheYCluster I agree: it was in the context of histones that I first heard the topic discussed, more than 20 years ago.
Tue Sep 01 17:47:04 +0000 2020I can't help but think this explanation of NES function 🔗 is missing something 🔗
Tue Sep 01 17:37:36 +0000 2020It is also good for accurately detecting N/Q deamidations (I realize this is my hill-to-die-on: I don't expect anyone else to be interested).
Tue Sep 01 15:55:16 +0000 2020The first draft of the blog post is up at 🔗 Feel free to discuss using #altptm.
Tue Sep 01 14:20:33 +0000 2020PXD015430 is my new favorite high resolution MS/MS phospho-peptide dataset. Does anybody know if it has been included in a publication yet?
Tue Sep 01 14:03:28 +0000 2020Thanks to everyone who added to this discussion. I'm going to summarize the thread in an blog post some time today. If you have any further comments, please include #altptm in your tweet. 🔗
Tue Sep 01 13:08:40 +0000 2020I've always found cigars excellent wrt promoting social distancing.
Tue Sep 01 12:14:25 +0000 2020DDOST:p is an essential subunit of both OST-A and OST-B in humans. All PTM sites occur on the N-terminal ER lumenal domain.
Tue Sep 01 12:14:25 +0000 2020DDOST:p θ(max) = 69%. aka OST, KIAA0115, OST48, WBP1, GATD6, AGER1, CDG1R, OKSWcl45. 1 transmembrane domain (428–447); (43–427) in the ER lumen; (428-456) is cytoplasmic. Commonly observed in HLA type I peptide studies & in a limited number HLA type II experiments.
Tue Sep 01 12:14:24 +0000 2020DDOST:p, dolichyl-diphosphooligosaccharide--protein glycosyltransferase non-catalytic subunit 🔗 Small ER protein; PTMs: Y289+phosphoryl, 4 K+ubiqutinyl, 3 shared with acetyl; SAAVs: R8G (3%); mature form: 43-456 [43,469×, 333 kTa] #ᗕᕱᗒ 🔗
Mon Aug 31 16:23:41 +0000 2020Is there a term (or terms) for the phenomenon of multiple PTMs possibly occupying the same protein residue?
For example, a lysine that may be either acetylated, SUMOylated or ubiquitinylated.
Mon Aug 31 15:23:15 +0000 2020When I'm researching proteins for my "protein-of-the-day" tweet, I've found ignoring anything attributed to a protein because of "CANCER!" to be a valuable strategy.
Mon Aug 31 14:03:05 +0000 2020RPN2:p is a subunit of both OST-A and OST-B. Its PTM pattern & topology is very different from RPN1:p, but both have extensive ubiquitin mods. All but 1 PTM site occur on the N-terminal ER lumenal domain. Its SUMOylation pattern is unique among OST subunits.
Mon Aug 31 14:02:44 +0000 2020RPN2:p θ(max) = 78%. aka SWP1, RPNII, RIBIIR & RPN-II. 3 transmembrane domains (541–561), (572–592) & (597–617); (23–540) is in the ER lumen. Not to be confused with PSMD1:p, which has been referred to as RPN2 in the past. Observed in both HLA type I & II peptide studies.
Mon Aug 31 12:50:44 +0000 2020RPN2:p, ribophorin II (H. sapiens) 🔗 Midsized ER membrane subunit; PTMs: Y267+phosphoryl, N106+glycosyl, 21 K+ubiquitinyl, 8 K-sites shared SUMOyl; SAAVs: G374D (1%), V501L (1%); mature form: 23-631 [52,446×, 695 kTa] #ᗕᕱᗒ 🔗
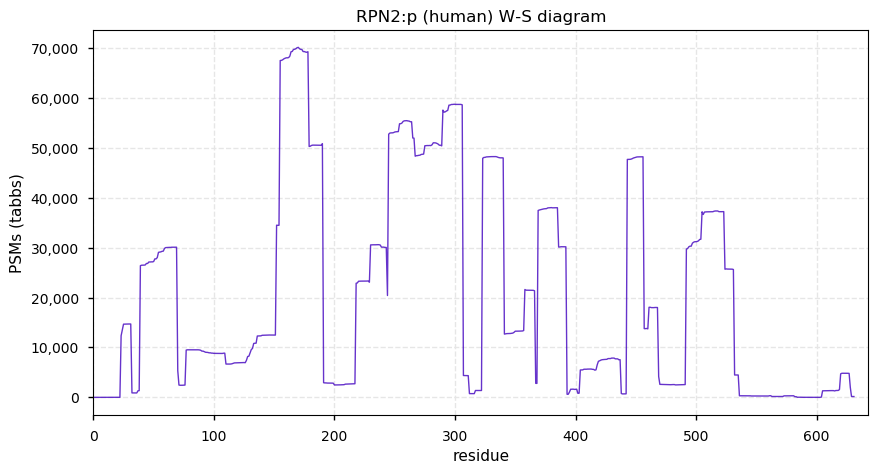
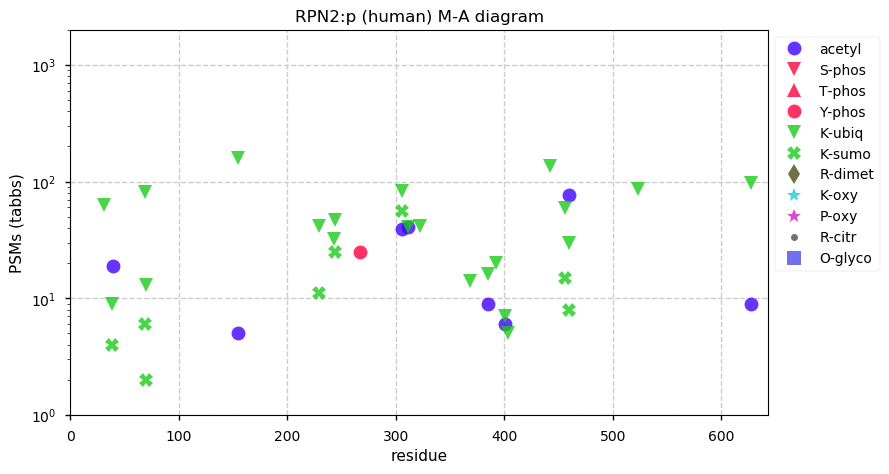
Sun Aug 30 22:13:32 +0000 2020@herrtschmidt You have to be careful about memory requirements. I don't use a "pipeline" in the usual sense, which make things easier, but so long as you are using C, C++, Python, Java, Perl or Rust there isn't any compilation or execution issues.
Sun Aug 30 17:39:24 +0000 2020I switched to mainly using ARM for proteomics data analysis a few years ago ... 🔗
Sun Aug 30 13:50:53 +0000 2020The functional significance of ten tyrosine phosphorylation sites on the ER lumenal domain of RPN1:p is a bit of a puzzle.
Sun Aug 30 13:41:16 +0000 2020RPN1:p θ(max) = 85%. Transmembrane domain (439–457), ER lumen (25-438) & cytoplasmic (458-607). Y+phosphoryl & N+glycosyl sites all on the lumenal domain. The protein is associated with binding the oligosaccharyltransferase-A complex to a ribosome in the rough ER.
Sun Aug 30 13:41:16 +0000 2020RPN1:p is a subunit of both OST-A and OST-B. The protein generates HLA type I and type II peptides, e.g, domain (1-9) in the ER signal sequence is commonly observed in type I experiments. The functional significance of the PTMs is unknown.
Sun Aug 30 13:41:15 +0000 2020RPN1:p, ribophorin I (H. sapiens) 🔗 Midsized ER membrane protein; PTMs: 10 Y+phosphoryl, 17 K+ubiqutinyl/acetyl, N299+glycosyl; SAAVs: T16S (1%), F203L (1%); mature form: 23,25-607 [62,618×, 1.1 MTa] #ᗕᕱᗒ 🔗
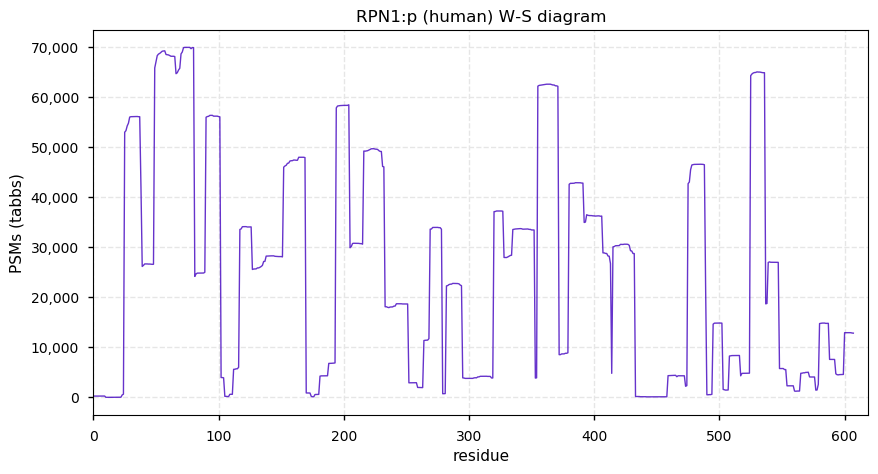
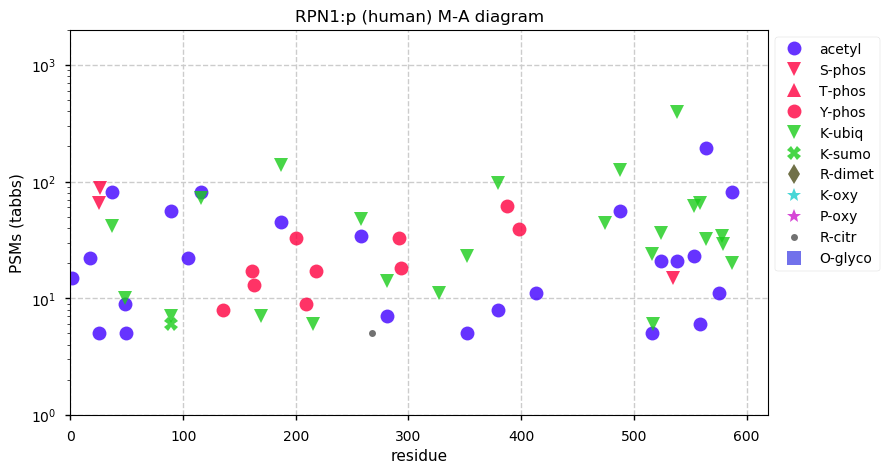
Sat Aug 29 13:21:47 +0000 2020Saw "Tenet" yesterday and I really think my Physics degree hampered my enjoyment of the film.
Sat Aug 29 12:46:21 +0000 2020OST4 is an odd little molecule that poses quite a few questions that have no answers. Its existence suggests there may be other small, non-catalytic ORFs that play important roles in larger complexes.
Sat Aug 29 12:26:06 +0000 2020OST4:p θ(max) = 100%. Transmembrane domain (5-25); cytoplasmic (1-4) & ER lumen (26-37). Known to be necessary for OST formation in many eukaryotes, from H. sapiens to S. cerevisiae. 1 of 5 non-catalytic subunits found in both forms of mammalian OST.
Sat Aug 29 12:26:06 +0000 2020OST4:p is a subunit of both OST-A and OST-B. Most proteomics observations are HLA type I and type II peptides from the domain (22-37). A limited number of observations are based on the intact subunit (1-37) or the only (nearly full length) tryptic peptide (1-34).
Sat Aug 29 12:26:05 +0000 2020OST4:p, oligosaccharyltransferase complex subunit 4, non-catalytic (H. sapiens) 🔗 Very small ER membrane protein; PTMs: M1+acetyl; SAAVs:none; gene has 3 short exons; mature form: 1-37 [496×, 1.4 kTa] #ᗕᕱᗒ 🔗
Fri Aug 28 20:58:53 +0000 2020@Smith_Chem_Wisc In Canadian it is pronounced "fastá".
Fri Aug 28 15:13:42 +0000 2020@MattWFoster @IonSource @JohnRYatesIII These guys 🔗 seemed to have some luck with fancy beads 🔗
Fri Aug 28 14:35:11 +0000 2020@dtabb73 It confuses me!🧐
Fri Aug 28 13:42:57 +0000 2020STT3B:p θ(max) = 32.1%. aka SIMP, FLJ90106, STT3-B . Has 13 transmembrane domains. The domain (2-41) is in the cytoplasm & the C-terminal domain (553–826) is in the ER lumen. The PTM pattern of STT3B:p is very different from the STT3A:p pattern.
Fri Aug 28 13:42:57 +0000 2020STT3B:p is a subunit in the enzyme complex that transfers the initial, large oligosaccharide from dolichol→protein in the protein glycosylation process. If STT3B:p is in the complex the transfer is post-translational (OST-B); if STT3A:p it is co-translational (OST-A)
Fri Aug 28 13:42:56 +0000 2020STT3B:p, STT3 oligosaccharyltransferase complex catalytic subunit B (H. sapiens) 🔗 Midsized ER membrane protein; PTMs: (N616, N627, N641)+glycosyl, 23 phospho & 14 ubiquitin sites; SAAVs:none; mature form: 2-826 [29,722×, 161 kTa] #ᗕᕱᗒ 🔗
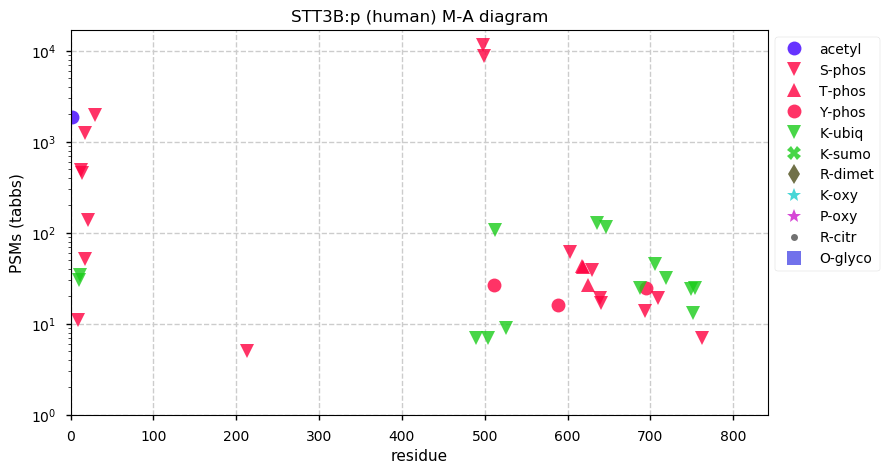
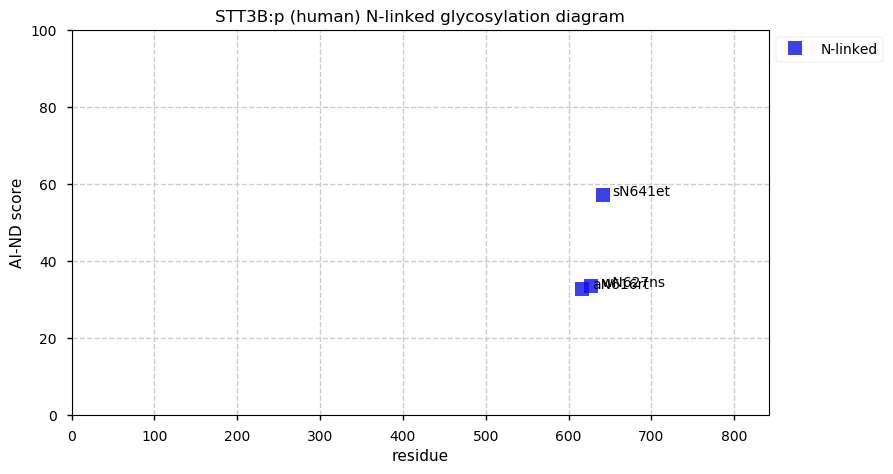
Fri Aug 28 11:56:32 +0000 2020@MattWFoster @IonSource @JohnRYatesIII Liquid nitrogen + mortar & pestle is the OG method.
Thu Aug 27 16:51:10 +0000 2020@JesseBrown Groundhogs can be a pain, but the tone of the article seems a bit arch considering the subject matter. Trying to compete with the TO media's raccoon obsession?
Thu Aug 27 16:23:23 +0000 2020Thanks to everyone who participated. It would appear that the use of "super-SILAC" for tissue sample protein quantitation is mildly controversial in the community-at-large.
Thu Aug 27 15:25:44 +0000 2020I don't really know who came up with the idea that membrane proteins were "hard to see", but it has persisted as received truth since at least the 1990's.
Thu Aug 27 15:04:03 +0000 2020@japantimes I had almost forgotten about "Space Force".
Thu Aug 27 12:16:28 +0000 2020STT3A:p θ(max) = 42%. aka TMC, MGC9042, STT3-A, ITM1. Has 13 transmembrane domains. The domain (1,2-17) is in the cytoplasm & the C-terminal domain (474-705) is in the ER lumen. A good counter example to the fallacy that membrane proteins are "difficult" to observe.
Thu Aug 27 12:16:28 +0000 2020STT3A:p is part of the enzyme complex that cotranslationally transfers the initial, large oligosaccharide from dolichol to the nascent protein chain in the rough ER at the beginning of the protein glycosylation process.
Thu Aug 27 12:16:28 +0000 2020STT3A:p, STT3 oligosaccharyltransferase complex subunit A (H. sapiens) 🔗 ER membrane subunit; PTMs: (N538, N548)+glycosyl, (Y66, Y331, Y338, Y617, Y618, Y651)+phosphoryl; SAAVs:none; mature form: (1,2)-705 [31,845×, 241 kTa] #ᗕᕱᗒ 🔗
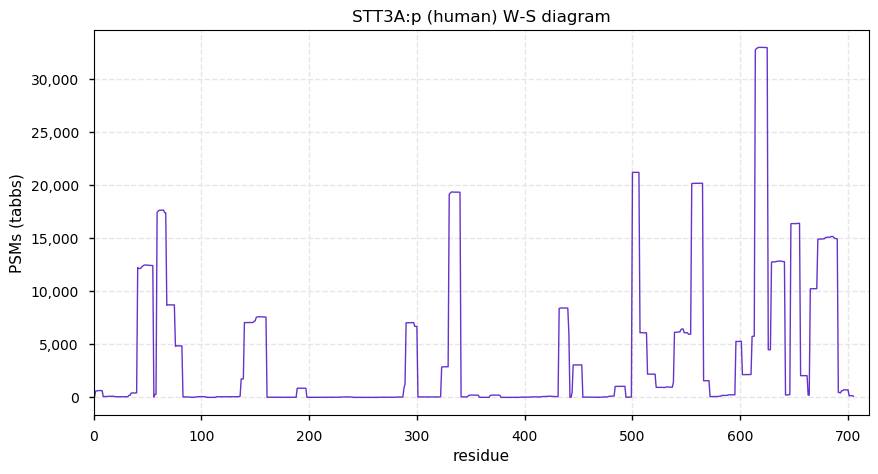
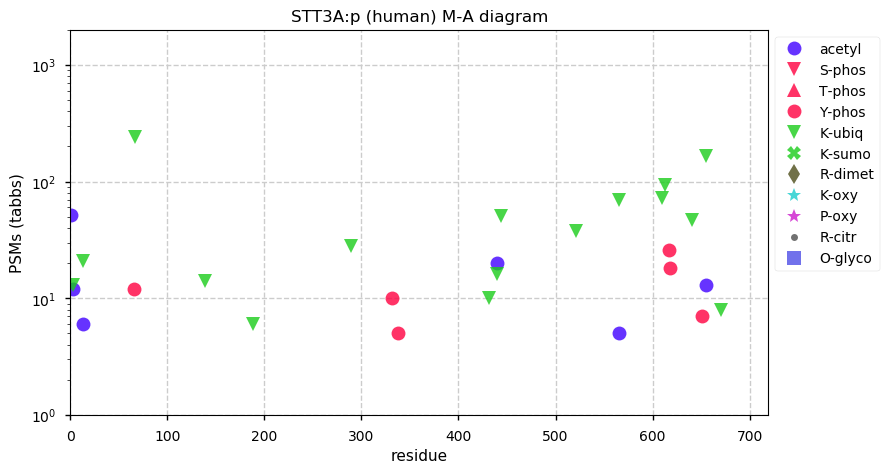
Wed Aug 26 16:18:34 +0000 2020Is "super-SILAC" a good approach to use for tissue sample protein quant?
Wed Aug 26 15:58:41 +0000 2020@SLIM_technology Back when RRKM theory was front and center in mass spec ...
Wed Aug 26 14:39:59 +0000 2020@neely615 The "ω %" column is the fraction of all PSM assignments for a particular protein that corresponds to a particular peptide. Using (20-35) as an example, that would be:
ω % = 100 ✕ (6,400/90,249) = 7.1
Wed Aug 26 14:36:21 +0000 2020@neely615 To generate a histogram for a new data set, if a PSM was assigned to the peptide (20–35), then the N would be 6400. base-10 log(6400) =3.8, so that PSM would add 1 to the histogram bin "3". That process is iterated for all of the PSMs in the data set to generate the histogram.
Wed Aug 26 14:31:22 +0000 2020@neely615 The histogram is based on the sort of numbers illustrated by this sort of chart for a protein (THY1:p in this case) 🔗
The "N" column is the number of times a peptide has been assigned as a PSM for this protein. /1
Wed Aug 26 12:34:18 +0000 2020CD84:p has 1 transmembrane domain (226-246), with a C-terminal intracellular domain. The PTM pattern strongly suggests a signalling/receptor role for the full length molecule.
Wed Aug 26 12:25:22 +0000 2020CD84:p θ(max) = 49%. aka SLAMF5, hCD84, mCD84. Short form found in urine. Prominent in HLA type II peptide studies of peripheral mononuclear cell samples. Found on platelets, lymphocytes & leukocytes; absent from common lines except JURKAT and THP-1 cells.
Wed Aug 26 12:25:22 +0000 2020CD84:p, CD84 molecule (H. sapiens) 🔗 Small protein; PTMs: (Y118, Y279, Y281, Y296, Y316)+phosphoryl; SAAVs:none; mature form: 22-345 and 22-131 (urine only) [4,696×, 17.4 kTa] #ᗕᕱᗒ 🔗
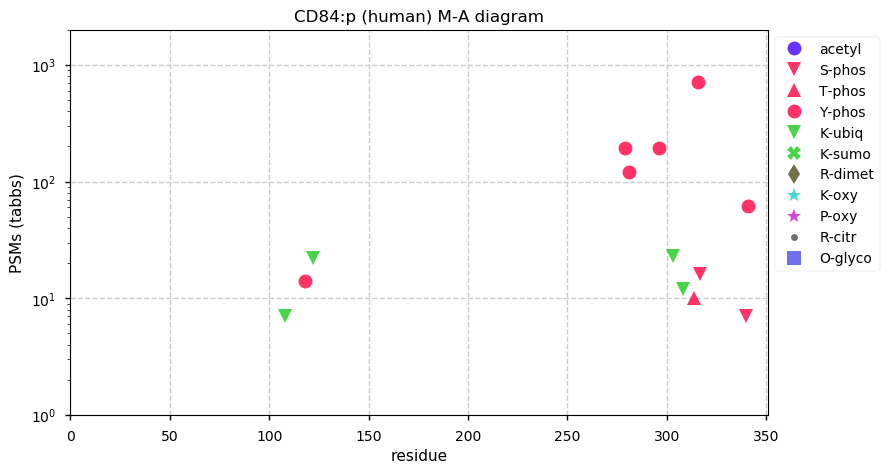
Tue Aug 25 19:05:01 +0000 2020@JoeLoo85614818 @ACSPublications @asmsnews Sitting: Ken Standing and John Fenn.
Tue Aug 25 16:23:50 +0000 2020Unless you are immune to embarrassment, avoid words like "gold standard" or "best practices" when referring to your own data.
Tue Aug 25 15:31:11 +0000 2020It isn't going to have any surprises, but it is a typical profile for running ~20,000 good quality spectra from a cell lysate. It quantifies the normally vague idea of how "deep" an experiment is sampling the proteins in a preparation.
Tue Aug 25 15:25:44 +0000 2020My favorite way to quickly assess a new LC/MS/MS run is to plot a histogram of the historical frequency of the PSMs. For this run, 1000 PSMs have been observed 10,000× previously, 3000 PSMs have been observed 100,000×, etc. 🔗
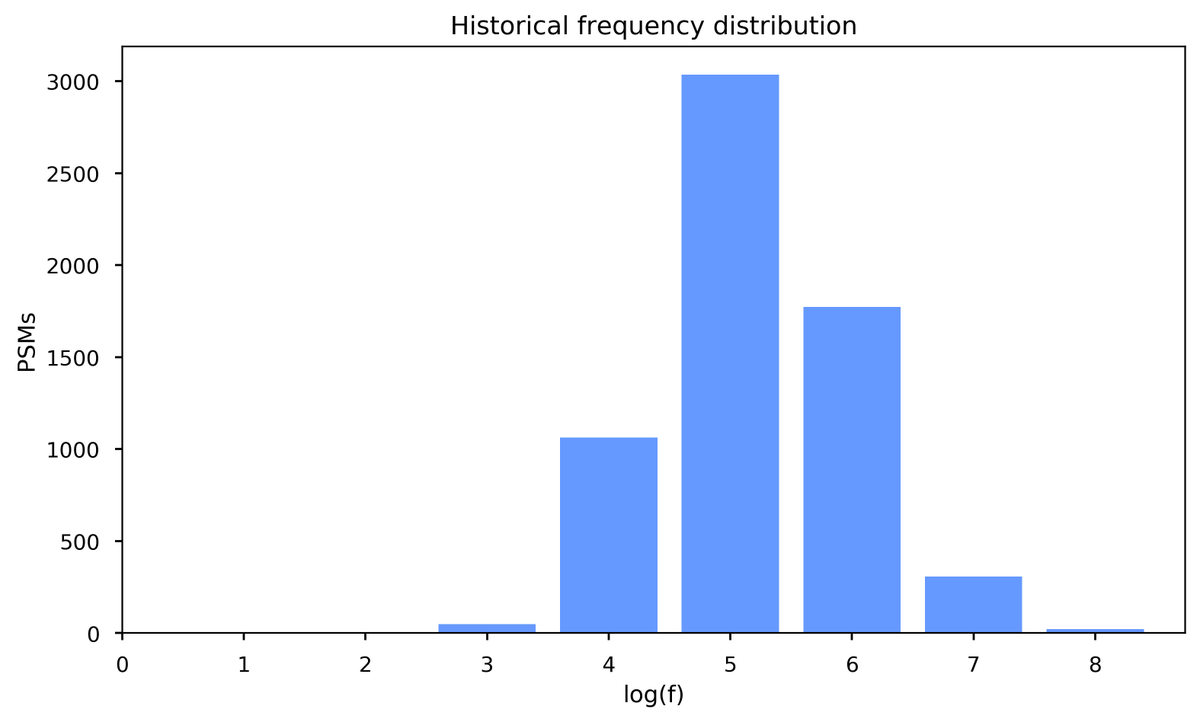
Tue Aug 25 14:59:53 +0000 2020Sometime I feel that Microsoft Edge was developed simply to annoy me personally ...
Tue Aug 25 14:55:48 +0000 2020@jvarga92 This latter method would imply an incomplete and somewhat stochastic process associated with the cotranslational glycosylation transfer, but it does fit in pretty well with cells' cavalier attitude towards making proteins and RNA.
Tue Aug 25 14:10:25 +0000 2020@jvarga92 There is always the possibility that proteins with inappropriate glycosylation end up being tossed out via ERAD, which wouldn't require additional site specificity, but that ends up having 2° & 3° structure determining the valid sites anyway.
Tue Aug 25 14:06:43 +0000 2020@jvarga92 No argument regarding the cotranslational aspect of the initial glycosylation transfer. But many eligible NXS/T don't end up glycosylated, so there must be some constraint beyond the sequence & 2° structure is about all that there is left.
Tue Aug 25 12:14:04 +0000 2020SELP:p θ(max) = 57%. aka CD62, PSEL, PADGEM, GMP140, CD62P, GRMP. Observed on platlets, megakaryocytes & cell lines derived from acute myeloid leukemia. One transmembrane domain (772-795) & 9 SUSHI domains.
Tue Aug 25 12:14:04 +0000 2020SELP:p, selectin P (H. sapiens) 🔗 Midsized protein; PTMs: (ST)617-652+glycosyl, N411+glycosyl, C-terminal phosphodomain; SAAVs: S331N (24%), E542K (1%), V640L (22%), N673S (1%), T756P (4%); mature form: 42-830 [2,762×, 26 kTa] #ᗕᕱᗒ 🔗
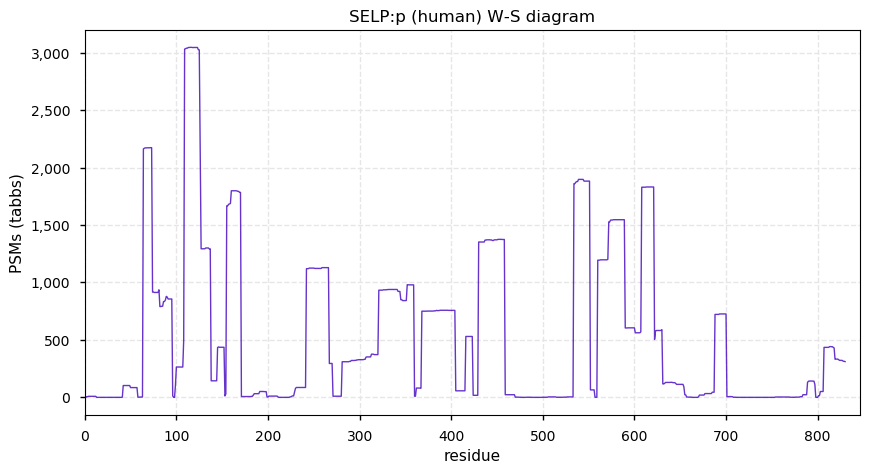
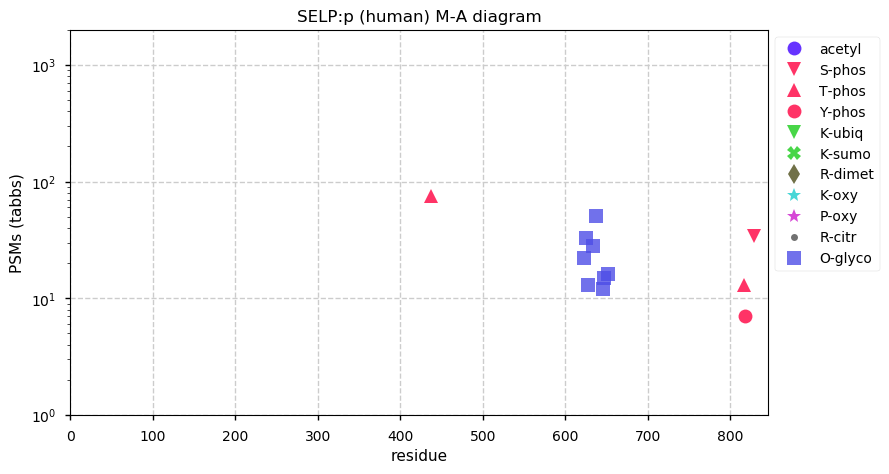
Mon Aug 24 21:22:48 +0000 2020@jvarga92 From my point of view, it would be more interesting to see the structure prior to glycosylation, as the carbohydrate may induce a change that could obscure the structure as seen by the oligosaccharyltransferase originally.
Mon Aug 24 20:51:20 +0000 2020@jvarga92 Thanks. Petrescu et al. is very much on point wrt to structural preferences. Figure 4B agrees with my own current prejudices, so it is my favorite ...
Mon Aug 24 14:38:32 +0000 2020It may be a good candidate for the awesome miraculous power of AI!
Mon Aug 24 14:35:25 +0000 2020I would have thought that by now someone would have systematically compared known N-linked sites to protein secondary structures & pulled out a useful correlation.
Mon Aug 24 13:19:06 +0000 2020I guess we are still in the "N-X-S/T sequon necessary but not sufficient" era.
Mon Aug 24 12:40:52 +0000 2020SELPLG:p θ(max) = 15%. aka PSGL-1, CD162. Transmembrane signalling receptor that binds P-selectin. Single transmembrane domain (321–343). Observed on lymphocytes; absent from most common cell lines, except JURKAT cells.
Mon Aug 24 12:40:52 +0000 2020SELPLG:p, selectin P ligand (H. sapiens) 🔗 Small membrane protein; PTMs: N302+glycosyl, (ST)287-292+glycosyl, (357-409) phosphodomain; SAAVs: none; (90-278) has no K/R residues; mature form: 18-412 [1,456×, 3.6 kTa] #ᗕᕱᗒ 🔗
Sun Aug 23 19:27:18 +0000 2020Is there any consensus protein 2ndary structure constraint for N-linked glycoslation sites?
Sun Aug 23 13:02:29 +0000 2020PROM2:p is rarely found in human solid tissue samples, but quite abundant in mouse cerebellum, kidney and skin. The low complexity phosphodomain 814-SSTSSEET-821 has been observed with 1, 2 or 3 simultaneously occupied phosphorylation sites.
Sun Aug 23 13:02:29 +0000 2020PROM2:p θ(max) = 38%. It has 5 transmembrane domains (107-127), (154-174), (427-447), (473-493) & (780-800) that are positioned very similarly to PROM1:p. The C-terminal phosphodomain is intracellular; the 2 N-glycosylation sites are in 1 extracellular domain.
Sun Aug 23 13:02:28 +0000 2020PROM2:p, prominin 2 (H. sapiens) 🔗 Midsized membrane protein; PTMs: (N707, N725)+glycosyl, (ST)814-821+phosphoryl; SAAVs: none; common in urinary EVs & several common cell lines; mature form: (18,20,21,22)-865 [4,152×, 28.5 kTa] #ᗕᕱᗒ 🔗
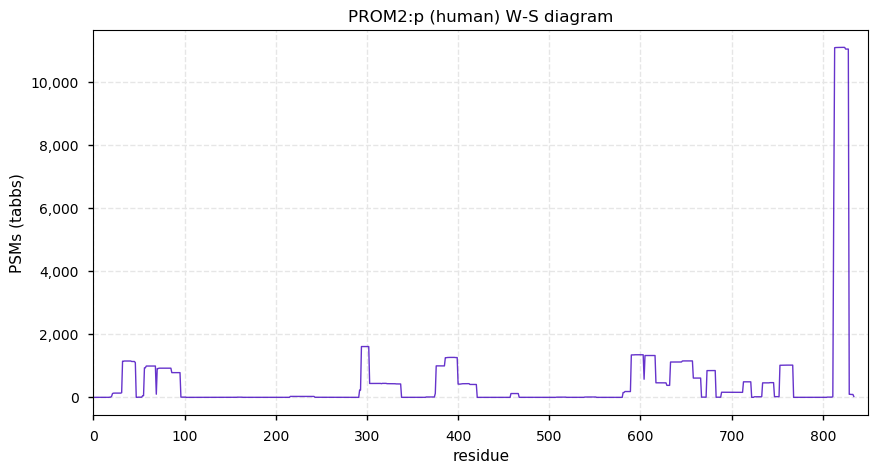
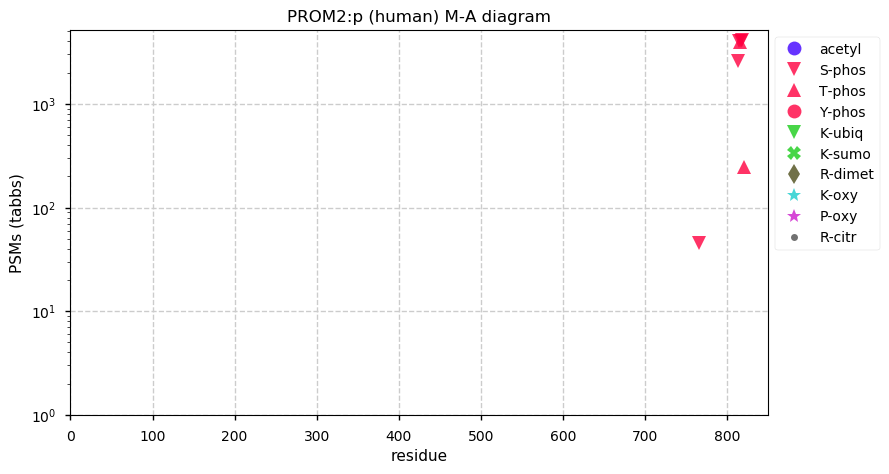
Sat Aug 22 16:29:43 +0000 2020And in more 2020 weather news:
🔗
Sat Aug 22 16:05:45 +0000 2020Thanks to everyone who participated.
30% of all respondents felt they were all equivalently difficult.
Among people willing to distinguish between the choices, MCP was rated as having the most difficult submission process by 84% of respondents.
Sat Aug 22 12:11:22 +0000 2020PROM1:p has no well defined function but it is used for sub-typing cancers. It is rarely found in human solid tissue samples, but quite abundant in mouse CNS tissue.
Sat Aug 22 12:02:33 +0000 2020PROM1:p θ(max) = 46%. aka AC133, CD133, RP41, CORD12, PROML1, MCDR2, STGD4. It has 5 transmembrane domains (109-129), (158-178), (434-454), (487-507) & (793-813). The C-terminal phosphodomain is intracellular; the 3 glycosites are in 1 extracellular domain.
Sat Aug 22 12:02:33 +0000 2020PROM1:p, prominin 1 (H. sapiens) 🔗 Midsized membrane protein; PTMs: (N220, N274, N395)+glycosyl, (Y828, Y852, S861, S863) + phosphoryl; SAAVs: none; common in stem cells & urinary EVs; mature form: 20?-865 [6,607×, 59 kTa] #ᗕᕱᗒ 🔗
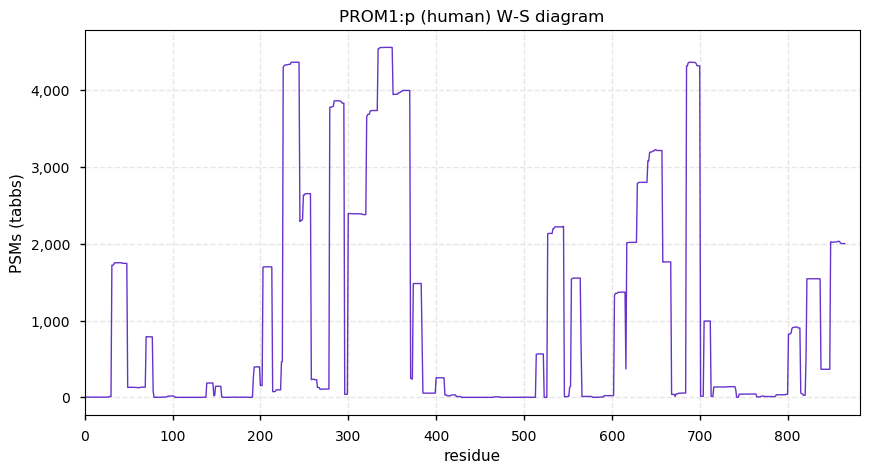
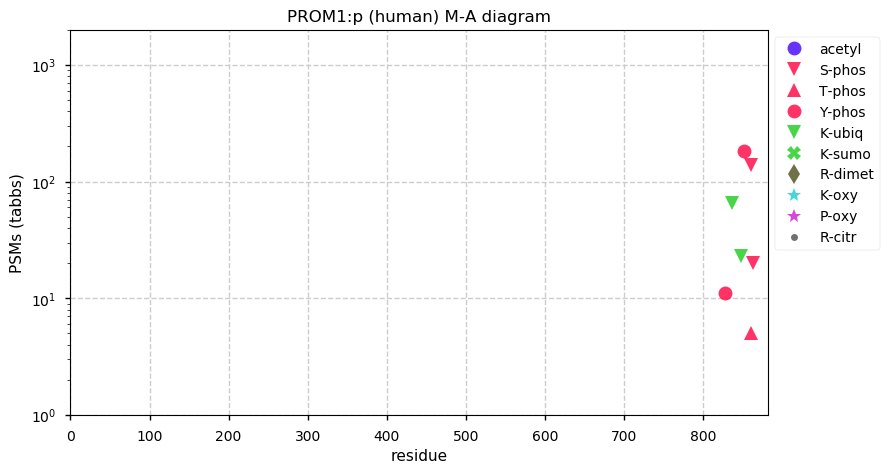
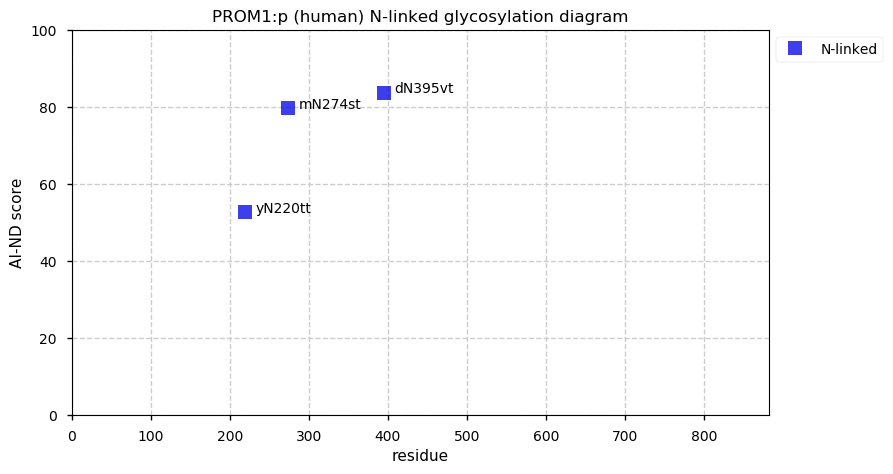
Fri Aug 21 19:02:23 +0000 20202020 seems obsessed with one-upping itself.
Fri Aug 21 19:01:33 +0000 2020Yup. This is really a thing ...
🔗
Fri Aug 21 16:49:36 +0000 2020Most of the classifications I can find in the literature are either based on physical properties (e.g., centrifugation sedimentation rate) or the tissue of origin/detection.
/fin
Fri Aug 21 16:46:34 +0000 2020There is quite a bit data associated with extracellular exosomes/vesicles in GPMDB (~4000 LC/MS/MS runs). Is there any effort to produce a systematic classification of these particles based on either the proteins they contain or have on their surfaces? /1
Fri Aug 21 16:41:20 +0000 2020@astacus I actually stopped reviewing when I realized I was pretty much the only one who did the download/reanalyze stuff (I also read code when available). It wasn't fair to authors who ended up getting stuck with me & I'm too stubborn to not do it.
Fri Aug 21 15:32:29 +0000 2020Which proteomics journal has the most difficult (or error prone) manuscript submission process? Feel free to nominate other magazines if yours isn't listed.
Fri Aug 21 12:54:16 +0000 2020THY1:p is also frequently observed in stem cells, particular iPSC. To make it a membrane protein, the C-terminal domain (131-161) is removed and a GPI anchor added.
Fri Aug 21 12:35:32 +0000 2020THY1:p θ(max) = 19%. aka CD90. THY1:p is quite rare in human T cell samples, but very common in mouse T cells. In both mouse and human it is frequently observed in CNS tissue.
Fri Aug 21 12:35:32 +0000 2020THY1:p, Thy-1 cell surface antigen (H. sapiens) 🔗 Small membrane protein; PTMs: (N42, N79, N119)+glycosyl; SAAVs: none; common in monocytes and brain tissue; mature form: 20-130 [14,028×, 90 kTa] #ᗕᕱᗒ 🔗
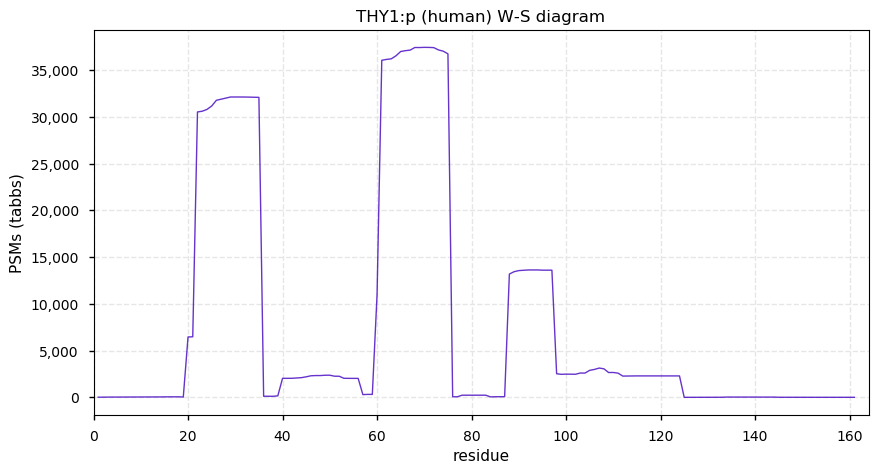
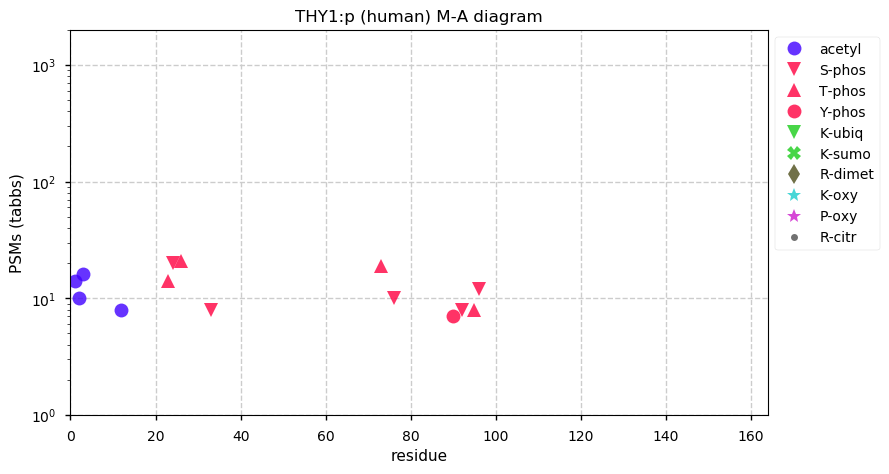
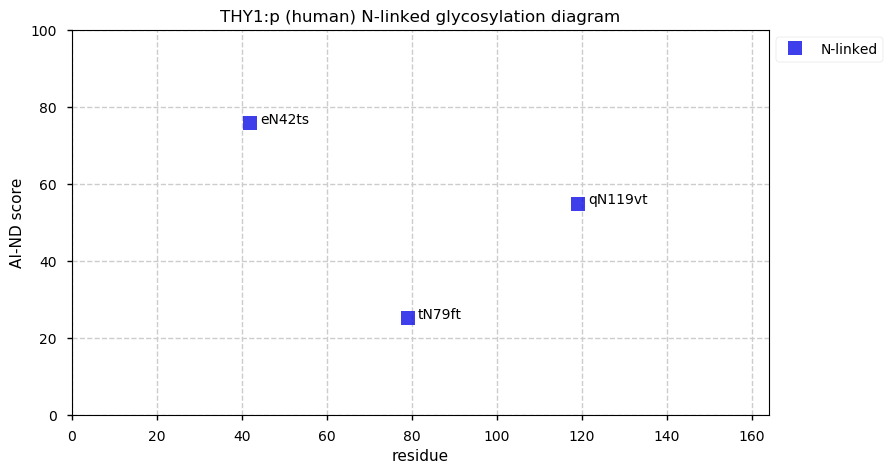
Thu Aug 20 19:11:18 +0000 2020@UCDProteomics While people are pretty creative about how they muddle up their samples, it almost always comes down to the dominance of a "blue team" attitude:
I wanted something to happen,
∴ it happened, &
I don't have to check that it did.
Thu Aug 20 14:50:55 +0000 2020Looking at data that could have been great with a little extra care but the publication ends up being nearly rubbish because of simple mistakes makes me a little sad. 😥
Thu Aug 20 12:04:55 +0000 2020CCR7:p θ(max) = 19%. aka BLR2, CDw197, CD197, CMKBR7 and EBI1. Mature protein has 7 TM domains; PTMs confirm that the C-terminal domain is intracellular. Functions as a G-protein coupled receptor. Most PSM observations based on HLA type II peptide experiments.
Thu Aug 20 12:04:55 +0000 2020CCR7:p, C-C motif chemokine receptor 7 (H. sapiens) 🔗 Small membrane protein; PTMs: (S365, S367)+phosphoryl, no glycosylation; SAAVs: none; observed on lymphocytes and leucocytes; mature form: 21?-378 [869×, 3.0 kTa] #ᗕᕱᗒ 🔗
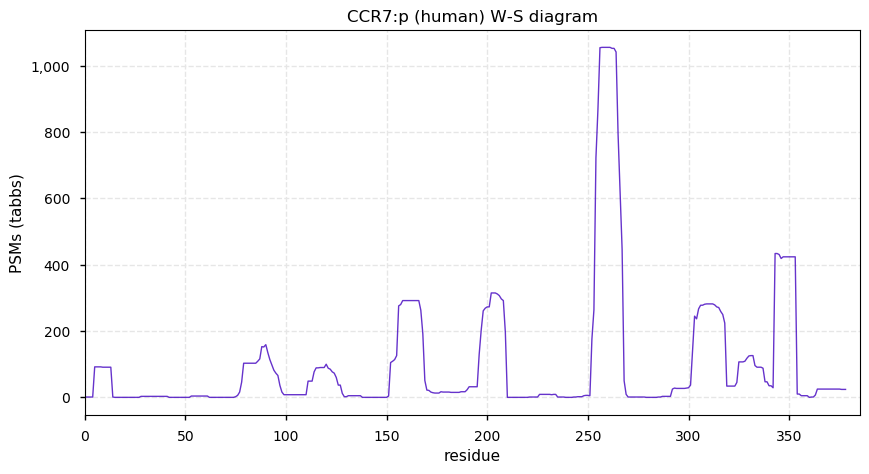

Wed Aug 19 15:21:59 +0000 2020@ypriverol @AJ_Brenes @pride_ebi Does "data set" in this context mean "raw file" or a collection of files with 1 PXD number?
Wed Aug 19 15:11:50 +0000 2020I've always considered Carl Linnaeus to be the 'inventor' of bioinformatics. Please let me know just how wrong I am ...
Wed Aug 19 14:38:48 +0000 2020@ypriverol @AJ_Brenes @pride_ebi So if I said > 5 million (PRIDE + MassIVE + jPOST + iPROX), I wouldn't be exaggerating.
Wed Aug 19 13:55:09 +0000 2020I was asked yesterday how many LC/MS/MS raw files there were kicking around in the public domain. I guessed > 1 million, but I didn't really have an authoritative answer. Any info from tweeters that run the repositories?
Wed Aug 19 11:51:40 +0000 2020CD48:p θ(max) = 59%. Observed on JURKAT cells, but absent from most commonly used cell lines. Most abundant on T cells. The C-terminal domain (221-243) is removed and a GPI membrane anchor attached to the new C-terminus.
Wed Aug 19 11:51:39 +0000 2020CD48:p, CD48 molecule (H. sapiens) 🔗 Small membrane protein; PTMs: (N40, N44, N104, N162, N189, N206)+glycosyl SAAVs: none; observed on lymphocytes and leukocytes; mature form: 35-220 [4,114×, 23 kTa] #ᗕᕱᗒ 🔗
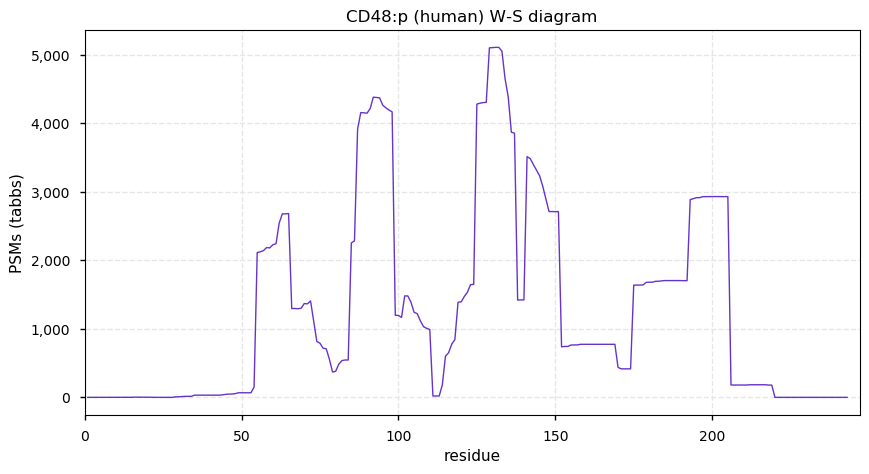
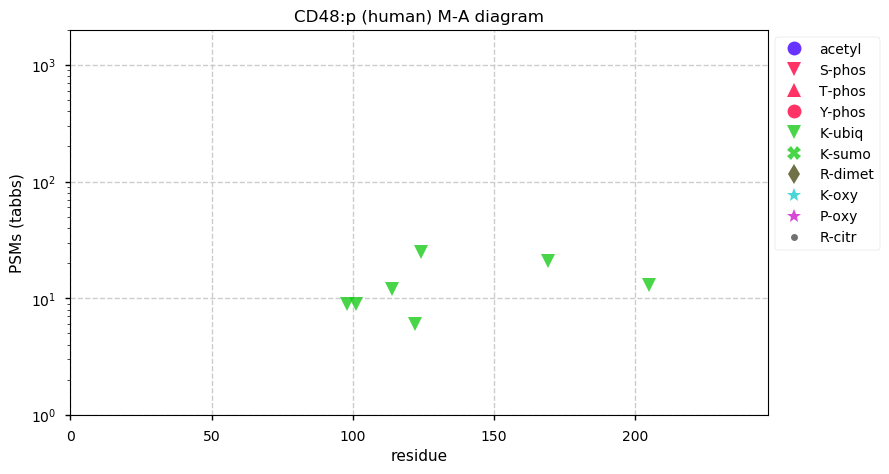
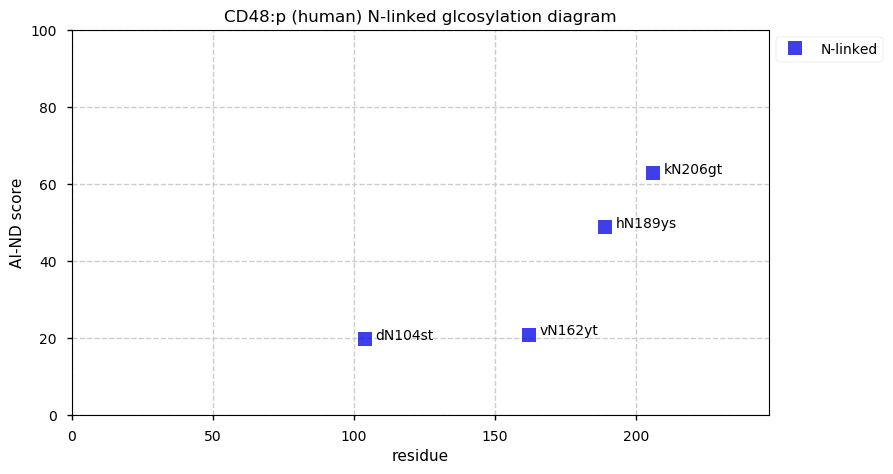
Tue Aug 18 18:00:07 +0000 2020@BobbyKlaus3 @UCDProteomics From what I can tell from public data, in infected cells it ends up being something like:
N > S > M >> any of the other products,
with PSM ratios of N:S:M being about 15:4:1.5
Tue Aug 18 15:31:56 +0000 2020@stephen_taylor decimals (or did you mean very, very quietly) 🧐
Tue Aug 18 15:17:02 +0000 2020When are US med school academic job postings going to start including the study section to which the lucky candidate must apply?
Tue Aug 18 14:24:55 +0000 2020This pattern of ubiquitin/SUMO modification on NEIL3:p is clearly trying to say something, but I don't know how to interpret the message (or even what language it is speaking) ...
🔗
Tue Aug 18 12:11:31 +0000 2020CD93:p observations in plasma & urine only confirm the N-terminal ½ of the sequence, suggesting a truncated, membrane free form of the protein. Not observable in common cell lines or T cells; present in B cells, monocytes, neutrophils & some endothelial cells.
Tue Aug 18 12:11:31 +0000 2020CD93:p θ(max) = 46%. Also known as C1qRP, C1qR(P), dJ737E23.1, CDw93, ECSM3, MXRA4, C1QR1. Transmembrane domain (581-606). Based on PTM pattern, (606-652) is intracellular & (22-580) is extracellular.
Tue Aug 18 12:11:31 +0000 2020CD93:p, CD93 molecule (H. sapiens) 🔗 Midsized membrane protein; PTMs: (Y628, Y644)+phosphoryl, T553+glycosyl, no N-glycosyl; SAAVs: P541S (48%); mature form: (22,24)-652 [3,565×, 17 kTa] #ᗕᕱᗒ 🔗
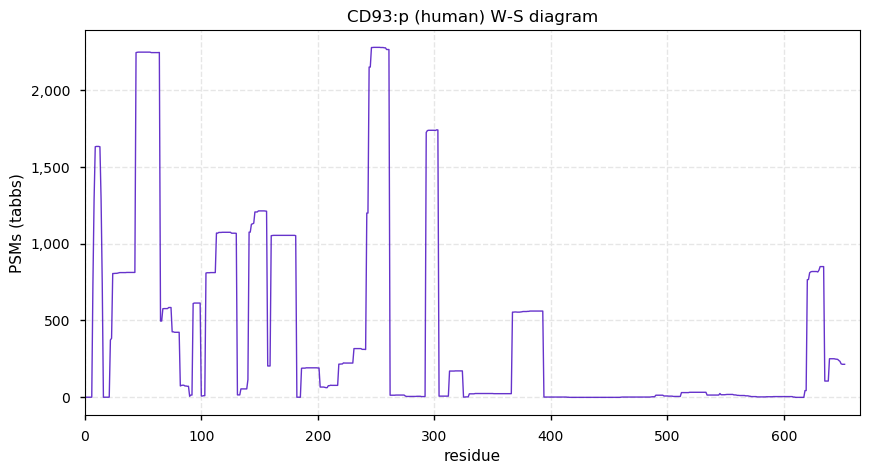
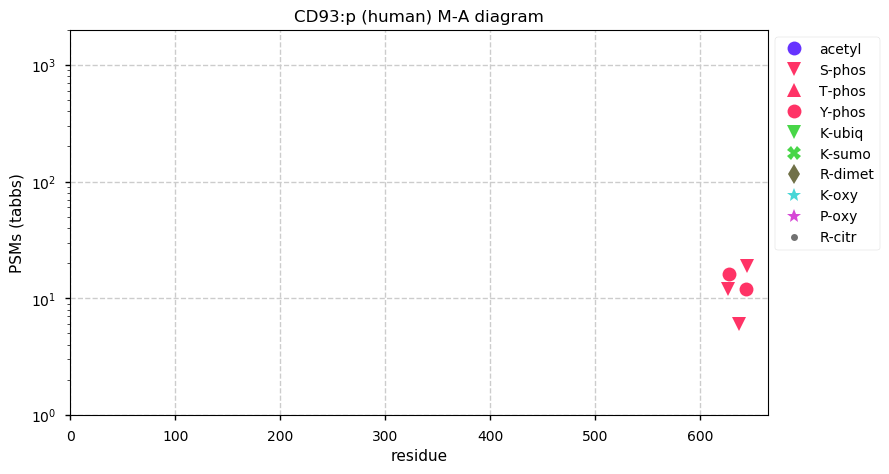
Mon Aug 17 12:21:16 +0000 2020CD63:p θ(max) = 50%. Also known as ME491, TSPAN30. 4 transmembrane domains: (12-32), (52-72), (82-102) & (204-224). The N+glycosyl PTMs are all found in the extracellular domain (103-203). Found on plasma, extracellular vesicle and lysosome membranes.
Mon Aug 17 12:21:16 +0000 2020CD63:p, CD63 molecule (H. sapiens) 🔗 Small membrane protein; PTMs: (N130, N150, N172)+glycosyl; SAAVs: none; observed in many common cell lines; mature form: (2-238) [16,532×, 75.4 kTa] #ᗕᕱᗒ 🔗
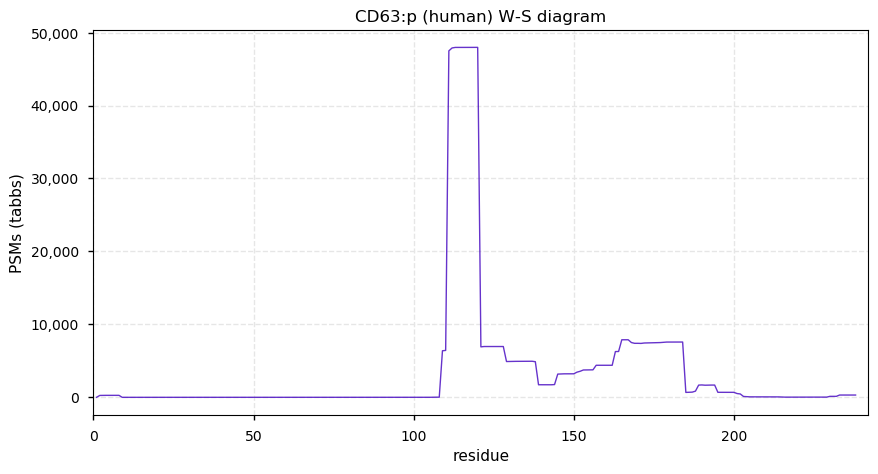
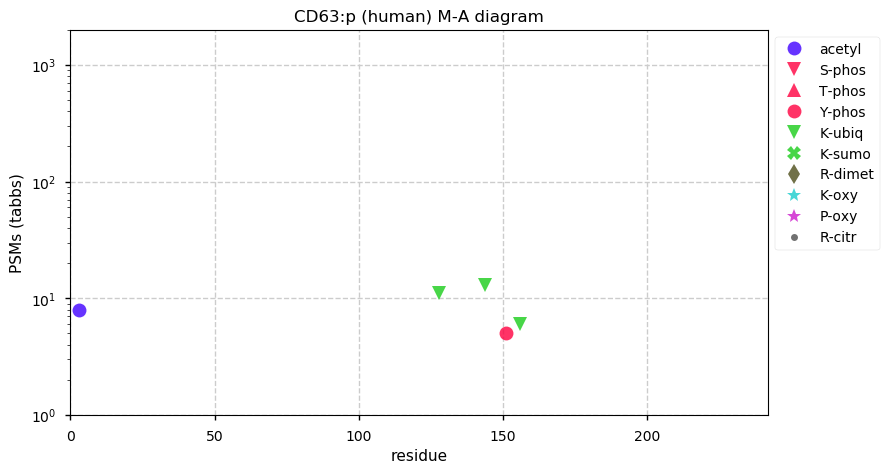
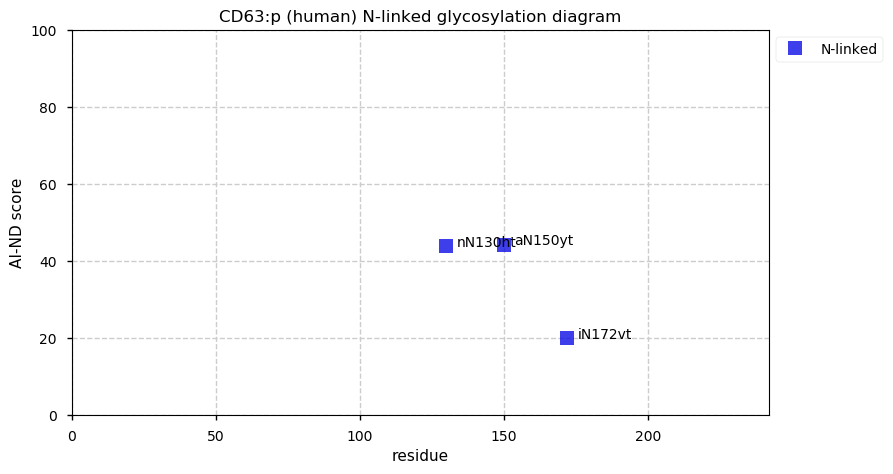
Sun Aug 16 17:26:13 +0000 2020Only ~200 lines left!
Sun Aug 16 12:34:43 +0000 2020ALCAM:p θ(max) = 75%. Also known as CD166 & MEMD. Commonly observed in most tissues and cell lines. Single transmembrane domain 528-551. Puzzling Y+phosphoryl in extracellular domain & K+ubiquintyl in intracellular domain. Abundant in HLA type II peptide experiments.
Sun Aug 16 12:34:43 +0000 2020ALCAM:p, activated leukocyte cell adhesion molecule (H. sapiens) 🔗 Midsize membrane protein; PTMs: (N95, N167, N265, N306, N361, N457, N480, N499)+glycosyl; SAAVs: N258S (8%), T301M (5%), M367I (2%); mature form: (28,33)-583 [22,897×, 207 kTa] #ᗕᕱᗒ 🔗
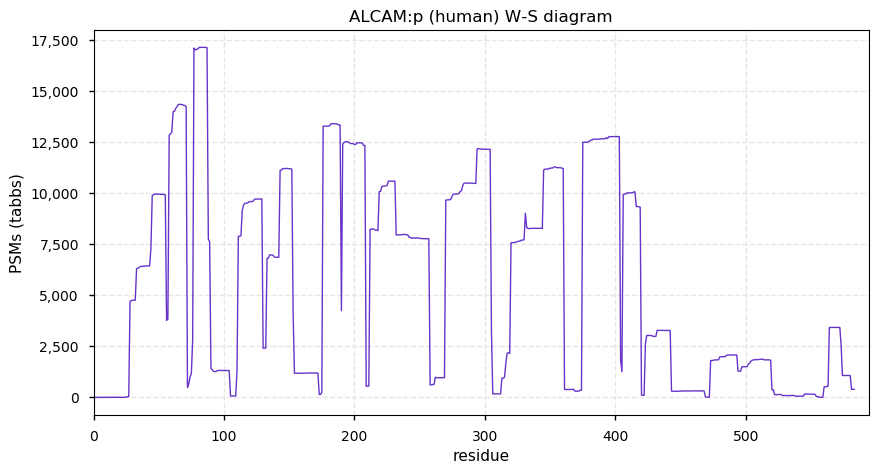
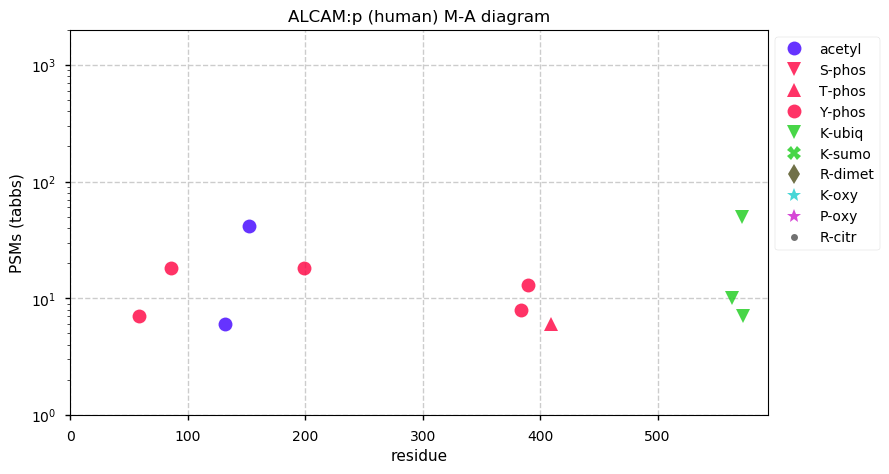
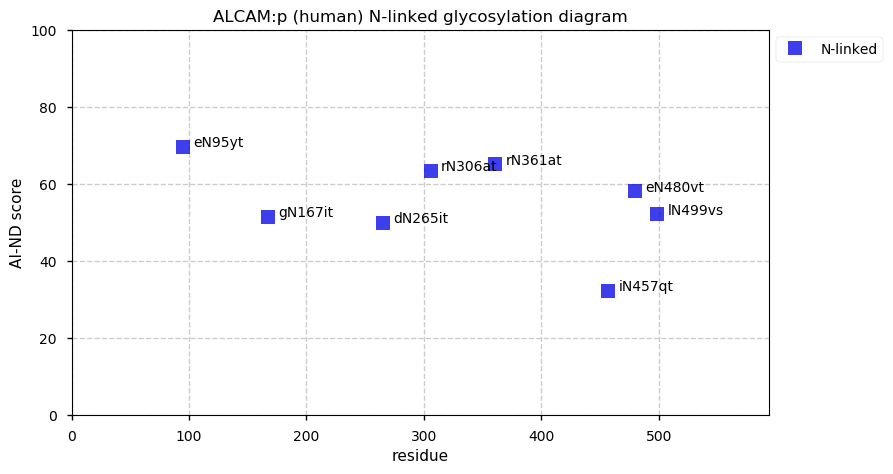
Sat Aug 15 17:39:48 +0000 2020@pwilmarth @theoneamit @Smith_Chem_Wisc Technically, it has worked pretty well, with steady improvements in data quality, quantity & the amount of information that can be extracted .
Conceptually, it is still stuck in the late 1990's.
Sat Aug 15 17:10:30 +0000 2020@theoneamit @Smith_Chem_Wisc When I began working on the first version (2006), I had already been working in MS-based protein analysis for 25 years and I was very interested in looking at the nuances in detail. I was also interested in whether or not the idea of proteomics had any merit.
Sat Aug 15 17:08:47 +0000 2020@theoneamit @Smith_Chem_Wisc No tutorials, I'm afraid. It is very much designed for a user base of 1 (me), based on my peculiar interests & the way I want the information to appear.
Sat Aug 15 12:11:25 +0000 2020CD109:p θ(max) = 45%. Observable in most tissue & clinical fluid samples, as well as most common cell lines such as HeLa and MCF10A. Prominent in platelet studies. Common in HLA type II studies, although some peptides—e.g., 1423-1431—are found in HLA type I data.
Sat Aug 15 12:11:25 +0000 2020CD109:p (H. sapiens) 🔗 Large protein; PTMs: (N41, N68, N118, N247, N279, N287, N291, N337, N365, N397, N419, N1086, N1355)+glycosyl; SAAVs: G377D (1%), Y703S (48%), V1009M (1%); mature form: 23-1422 [17,626×, 135 kTa] #ᗕᕱᗒ 🔗
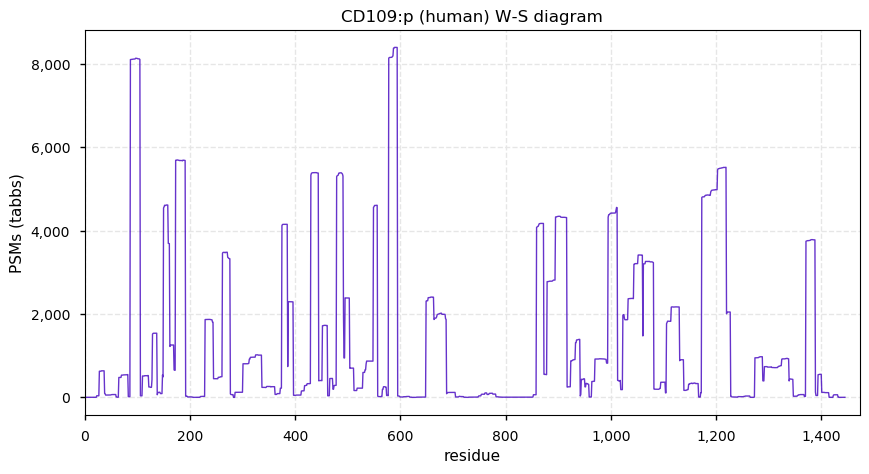
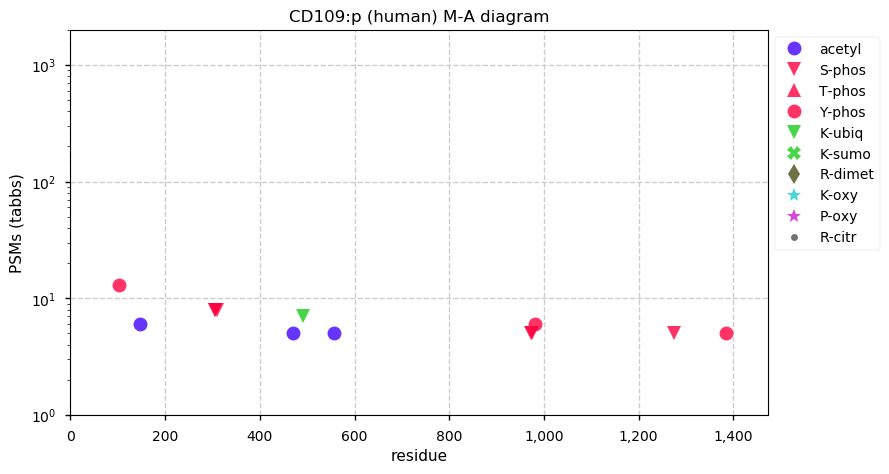
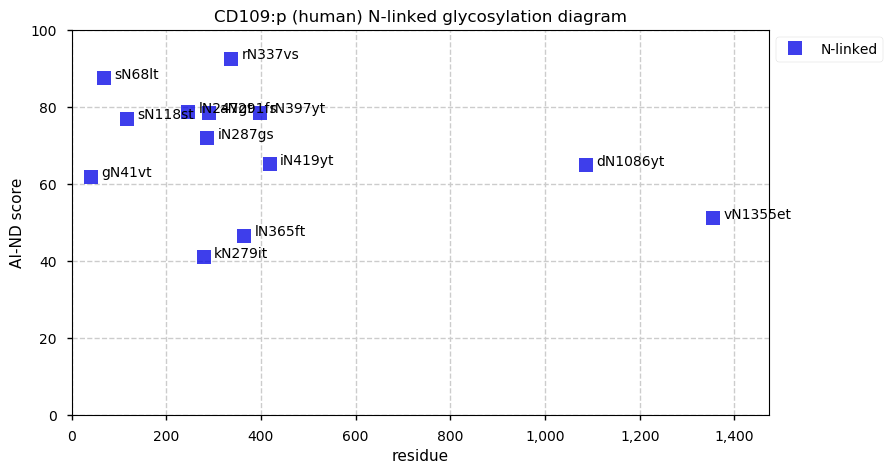
Fri Aug 14 18:09:13 +0000 2020@Smith_Chem_Wisc Actually, it is only the 1st protein if this was human data. If it was mouse, rat, zebrafish or chicken you may have to go further down the list.
Fri Aug 14 18:04:41 +0000 2020@Smith_Chem_Wisc 1. Go to 🔗
2. Enter your sequence in the top box and "Go"
3. Wait for the result
4. Go to the bottom of the page and click on the protein/splice of interest (in this case the 1st one) to get the list for that protein only.
Fri Aug 14 17:21:04 +0000 2020@chrashwood @CameronTFlower @pwilmarth @BiswapriyaMisra Is there an emoji for "bad memories"? 😱
Fri Aug 14 17:14:28 +0000 2020@Smith_Chem_Wisc And if I might be so bold, that peptide has been assigned 3900 times in public data, with 3822 of those PSMs being phosphorylated at that site, which IMHO adds to the confidence as prior knowledge.
Fri Aug 14 16:44:02 +0000 2020@CameronTFlower @pwilmarth @BiswapriyaMisra Without other evidence, probably not beyond what it can tell you about the molecular composition. It is like any other use of mass spec for id'ing a compound in organic chemistry.
Fri Aug 14 16:19:58 +0000 2020@pwilmarth @CameronTFlower @BiswapriyaMisra I'd personally be reluctant to accept an identification using a modern high res instrument with an unexplained parent ion mass difference without at least one hypothetical structure that gets you to within the demonstrated tolerance of the rest of the run.
Fri Aug 14 16:10:10 +0000 2020@pwilmarth @CameronTFlower @BiswapriyaMisra And as Phil indicates, it is hard to write up a general guide for the analysis, because it has to focus on the specific details of how the data was created & few labs follow anything resembling a general SOP.
Fri Aug 14 16:07:16 +0000 2020@pwilmarth @CameronTFlower @BiswapriyaMisra It is probably just the pedantic Canadian in me, but I usually refer to this type of interpretation as highly nuanced, rather than complicated. There are no really deep considerations, just a *lot* of simple ones that are commonly overlooked.
Fri Aug 14 15:43:22 +0000 2020@cdsouthan I ran two groups that had Edman sequencers as part of the lab's equipment, but I haven't seen one in person since '98. It would be great if that type of info was around on many of these proteins, but a lot of the seqs weren't known until Edman had largely died out.
Fri Aug 14 14:46:51 +0000 2020@SylviaC29659211 For high res parent & fragments, 20 ppm for both is a good place to start.
For high res parent & low res fragments, 20 ppm for the parent & the 0.4 Da for the fragments.
In both cases, examine the results and iterate to find the best setting for your data.
Fri Aug 14 14:14:14 +0000 2020@cdsouthan Regarding the experimental testing: almost all proteomics data sets have the information unless they have somehow excluded N-terminal peptides. While it is not routine for many groups to check for it, I always do when analyzing public data.
Fri Aug 14 14:11:31 +0000 2020@cdsouthan Any protein can be ragged and there are plenty of plasma proteins with several different N-terminal residues. Having 4 prominent ones is unusual (but not unprecedented).
Fri Aug 14 12:23:07 +0000 2020CD14:p is common in blood plasma, urine, CSF, milk, seminal plasma, vitreous humour and saliva, as well as monocytes, T-cells and dendritic cells. Rarely found in HeLa, JURKAT, Hek-293(T) or A-431 cell lines.
Fri Aug 14 12:23:07 +0000 2020CD14:p θ(max) = 79%. Unusually ragged N-terminus. The C-terminal cleavage events described in the literature and some database annotations are not supported by the proteomics data.
Fri Aug 14 12:23:07 +0000 2020CD14:p, CD14 molecule (H. sapiens) 🔗 Small extracellular protein; PTMs: (N37, N151, N282)+glycosyl; SAAVs: M234I (1%); mature form: (20,21,22,23)-375 [18,298×, 181 kTa] #ᗕᕱᗒ 🔗
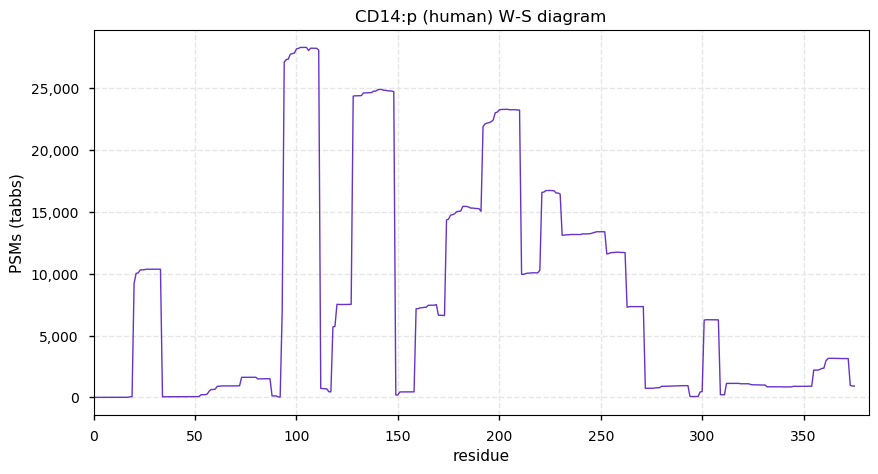
Thu Aug 13 18:50:11 +0000 2020@UCDProteomics I know I always got more traction for gel-like images of MS-based proteomics results from biologists than analytical chemists. The biologists would start picking out features within a few seconds of seeing it. 🔗
Thu Aug 13 18:41:22 +0000 2020@UCDProteomics What is the mass spec proteomics equivalent of Photoshop?
Thu Aug 13 16:37:32 +0000 2020Viruses are weird: data from "Proteome and phosphoproteome dynamics of CVB3 infected cells" (PXD011163)
🔗
Thu Aug 13 15:09:25 +0000 2020That aside, the phospho S:T ratio in the IMAC enrichment data was 6:1 and in the RXXpS/pT antibody enrichment data it was S:T ~ 4:1. The proteome average S:T is about 6½:5.
Thu Aug 13 14:11:03 +0000 2020The enrichment motif for the antibody was RXXS/T when the S/T was phosphorylated. The paper suggests that Lys-C was used as the proteolytic enzyme for this part of the study, but the results indicate trypsin was used instead.
Thu Aug 13 12:22:01 +0000 2020I should mention that this paper is the first public data (I know about) that used an S/T phosphorylation motif-specific antibody to enrich peptides & it worked pretty well.
Thu Aug 13 12:00:30 +0000 2020CD163:p is absent from most common cell lines (including JURKAT), but is observed in THP-1 cells. CD163:p has high affinity for hemoglobin bound to haptoglobin & lower affinity for free hemoglobin. Believed to be involved in innate immunity to bacteria.
Thu Aug 13 12:00:29 +0000 2020CD163:p θ(max) = 52%. AKA M130, MM130, SCARI1. Single transmembrane domain (1051-1071), separating an N-terminal extracellular domain from a C-terminal intracellular domain. Observations in plasma, CSF & urine do not have PSMs from the domain (999-1156).
Thu Aug 13 12:00:29 +0000 2020CD163:p, CD163 molecule (H. sapiens) 🔗 Large protein; PTMs: 2 phosphodomains, (N105, N123, N140, N445, N767, N1001, N1027)+glycosyl; SAAVs: I342V (24%), T901M (11%); mature form: 42?-1566 [9,918×, 62 kTa] #ᗕᕱᗒ 🔗
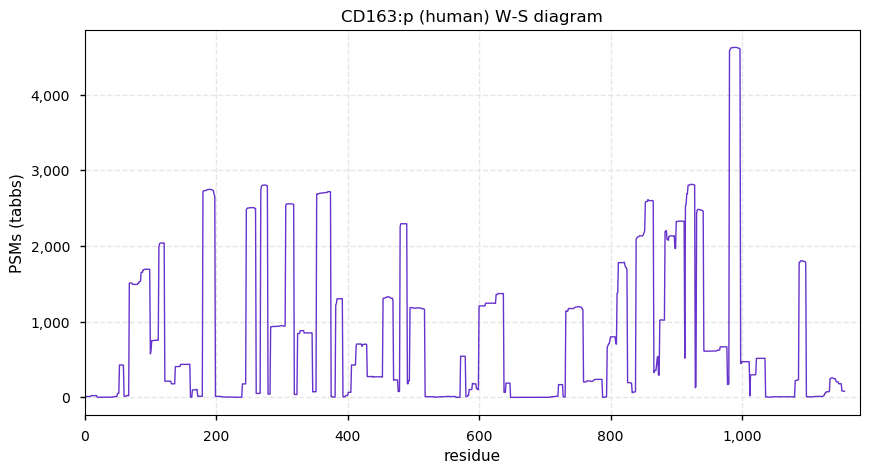
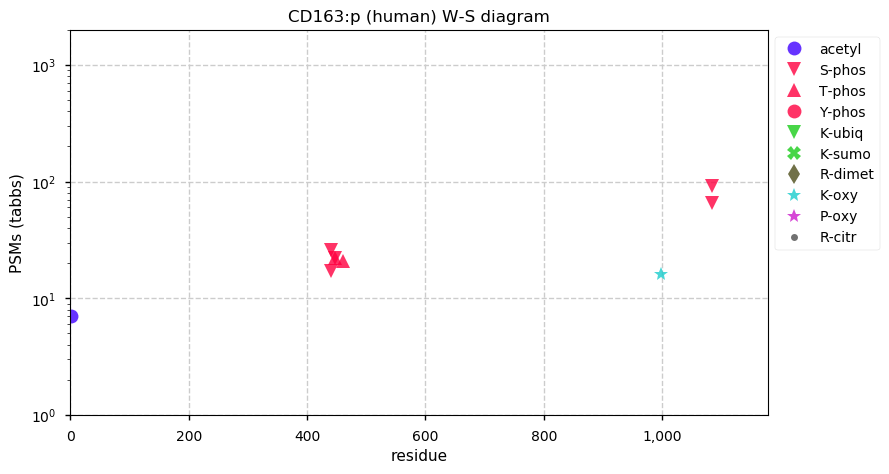
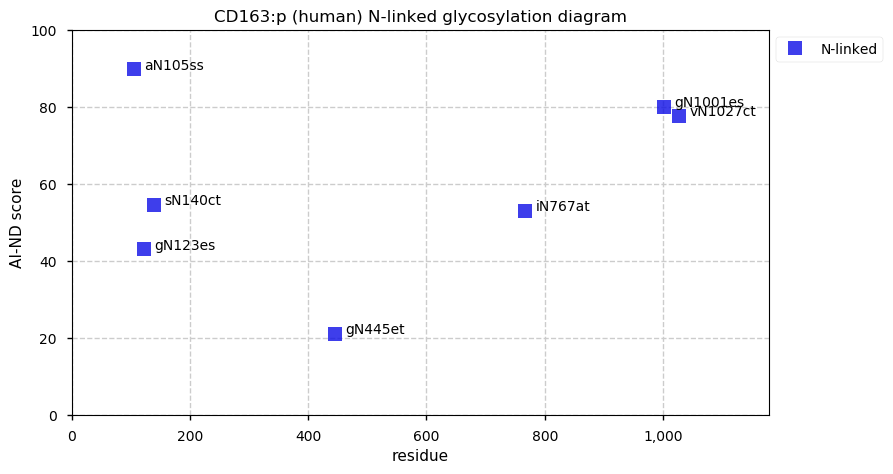
Thu Aug 13 01:40:11 +0000 2020The data they obtained using Akt substrate motif antibodies for phospho-AGC-motif enrichment is a more advanced topic (extra marks if you can figure it out).
Wed Aug 12 21:39:20 +0000 2020If you are interested in phospho-proteomics and would like to take some good high-res IMAC data out for a spin, PXD014832 from Bonucci M, et al. 🔗 is a great place to start ⭐️⭐️⭐️⭐️
Wed Aug 12 21:10:38 +0000 2020While this would be seasonal in Texas, it is pretty unusual in London UK:
🔗
Wed Aug 12 20:06:42 +0000 2020@MattWFoster It does have N-linked sugars in its central extracellular domain (N73, N292, N371), but none of the usual suspects that are commonly enriched & tested for in proteomics experiments.
Wed Aug 12 19:54:21 +0000 2020Maybe because with something like AFDN:p, it's hard to know where to start ...
🔗
Wed Aug 12 17:10:57 +0000 2020From a technical point of view, I find the lack of common PTMs observed for a protein like ENTPD1:p (which has been id'd in > 5,000 LC/MS/MS runs) more interesting than proteins covered with PTMs. 🔗
Wed Aug 12 12:26:54 +0000 2020ENTPD1:p catalyzes the conversion of extracellular ATP/ADP to cAMP, which is believed to be anti-inflamatory. It is not found in common cell lines. Absent from urine but observed on lymphocytes and leukocytes.
Wed Aug 12 12:26:54 +0000 2020ENTPD1:p θ(max) = 36%. It has two membrane spanning domains (17-37) & (477-499) leaving very short N- & C-terminal intracellular domains (1-16) & (500-510) and a longer, catalytic extracellular domain (38-476).
Wed Aug 12 12:26:54 +0000 2020ENTPD1:p, ectonucleoside triphosphate diphosphohydrolase 1 (H. sapiens) 🔗 Midsized membrane enzyme; aka CD39, NTPDase-1, ATPDase, SPG64; PTMs: (N73, N292, N371)+glycosyl; SAAVs: none; mature form: 1-510 [5,476×, 20 kTa] #ᗕᕱᗒ 🔗
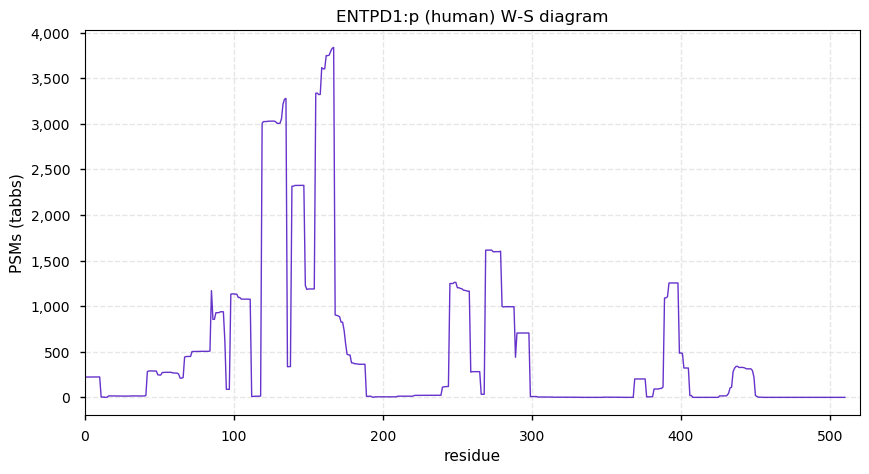
Tue Aug 11 15:25:16 +0000 2020CD38:p has no ER signal sequence, so the protein is made in the cytoplasm & inserted into a membrane following translation. Present on cells derived from hematopoetic stem cells, but rare on erythrocytes & platelets.
Tue Aug 11 12:41:38 +0000 2020CD38:p PTMs do not support any phosphorylation mediated signalling function. The protein is believed to be an enzyme catalyzes cyclic ADP-ribose synthesis & hydrolysis. The limited expression of the protein makes the purpose of the enzymatic function obscure.
Tue Aug 11 12:41:37 +0000 2020CD38:p θ(max) = 77%. Its topology is the reverse of most small CD's: a short unmodified intracellular N-terminal domain (2-23), a transmembrane domain (24-43) and a long N-glycoyslated extracellular C-terminal domain (44-300).
Tue Aug 11 12:41:37 +0000 2020CD38:p, CD38 molecule (H. sapiens) 🔗 Small membrane protein; PTMs: (N100, N164, N219)+glycosyl; SAAVs: none; absent from HeLa & HEK-293, but present in JURKAT, THP-1 and A-549 cells; mature form: 2-300 [5,528×, 30 kTa] #ᗕᕱᗒ 🔗
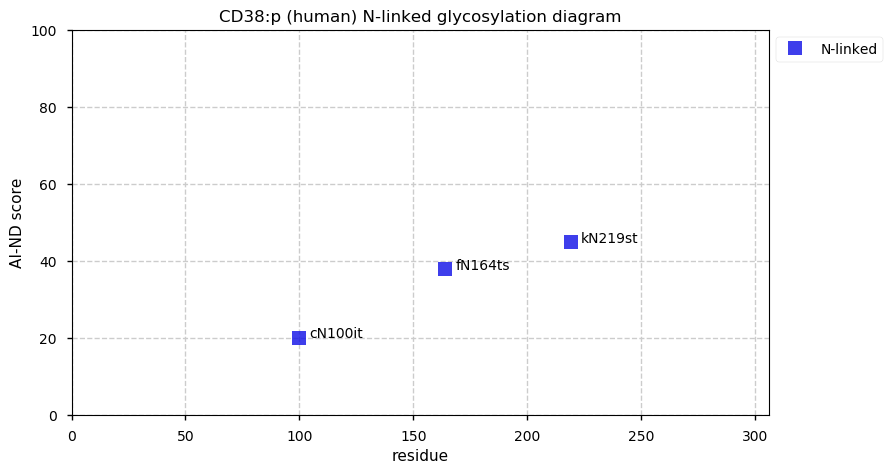
Mon Aug 10 16:14:41 +0000 2020@AlexUsherHESA It would have been great for the NDP & BQ if it had happened, though.
Mon Aug 10 15:41:10 +0000 2020Just got rid of my G-Suite accounts. Happy days!
Mon Aug 10 12:03:24 +0000 2020CD40:p θ(max) = 59%. Eyeballed transmembrane domain 195-215. Observed in U2-OS and THP-1 cells, but rare in most common cell lines. Also commonly found in urine, monocytes, platelets and hepatocytes.
Mon Aug 10 12:03:24 +0000 2020CD40:p, CD40 molecule (H. sapiens) 🔗 Small membrane protein; PTMs: (N153, N180)+glycosyl; SAAVs: S124L (5%); mature form: 21-277 [3,029×, 8.8 kTa] #ᗕᕱᗒ 🔗
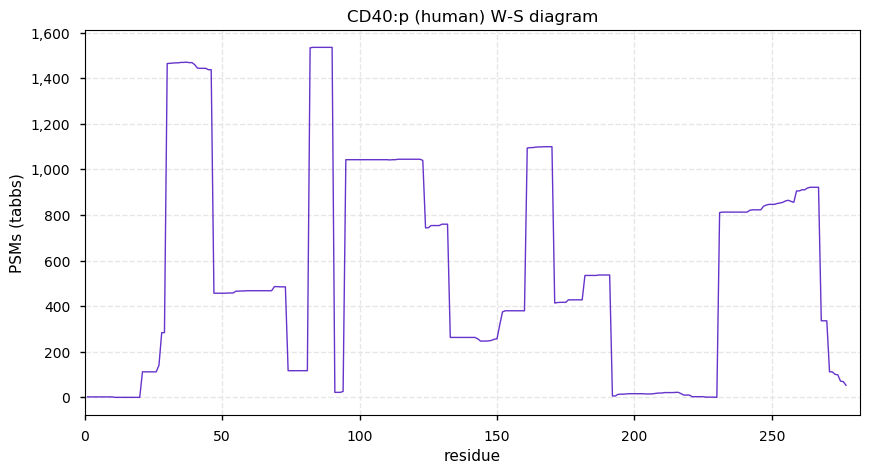
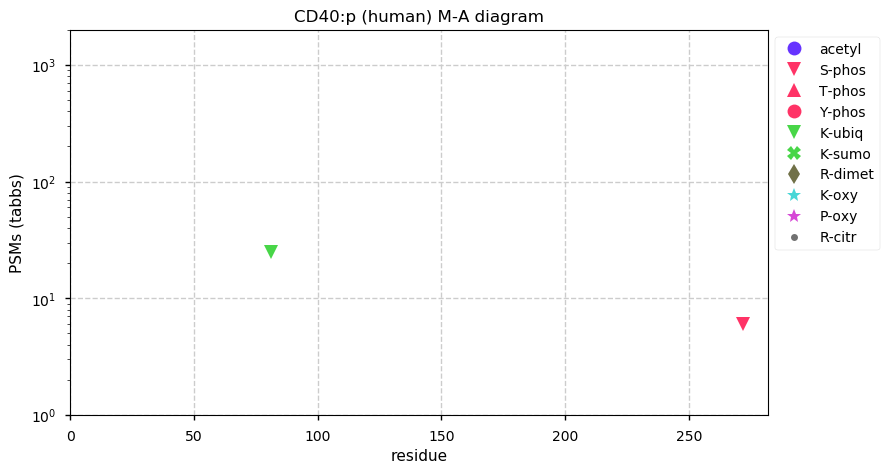
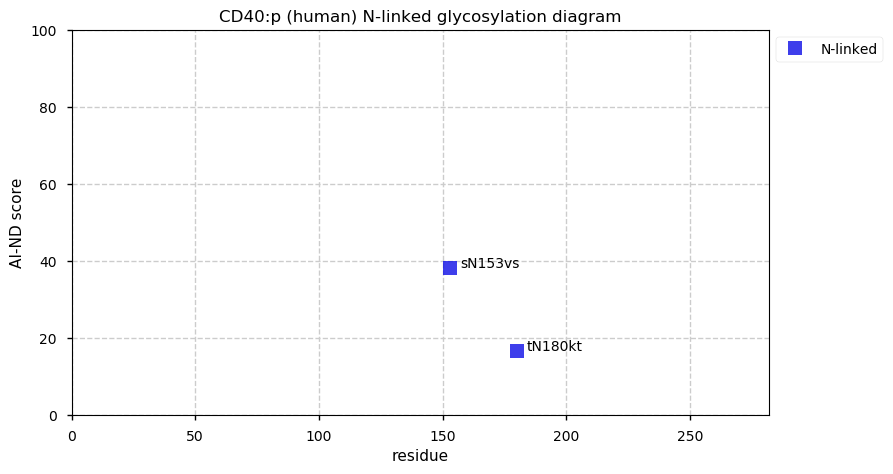
Sun Aug 09 13:36:32 +0000 2020CHD1:p θ(max) = 45%. Multiple long low complexity domains. May be abundant in cell lines and tissues. Sumoylation pattern is unusual in that it does not share K-sites with ubiquitination or acetylation.
Sun Aug 09 13:36:32 +0000 2020CHD1:p, chromodomain helicase DNA binding protein 1 (H. sapiens) 🔗 Large nuclear protein; PTMs: 5 major phosphodomains, 3 sumoylation domains; SAAVs: none; mature form: 1-1710 [17,372×, 94 kTa] #ᗕᕱᗒ 🔗
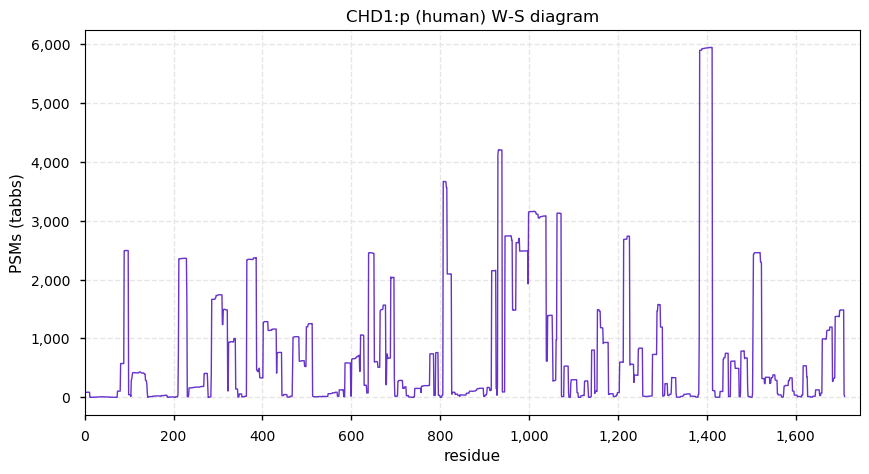
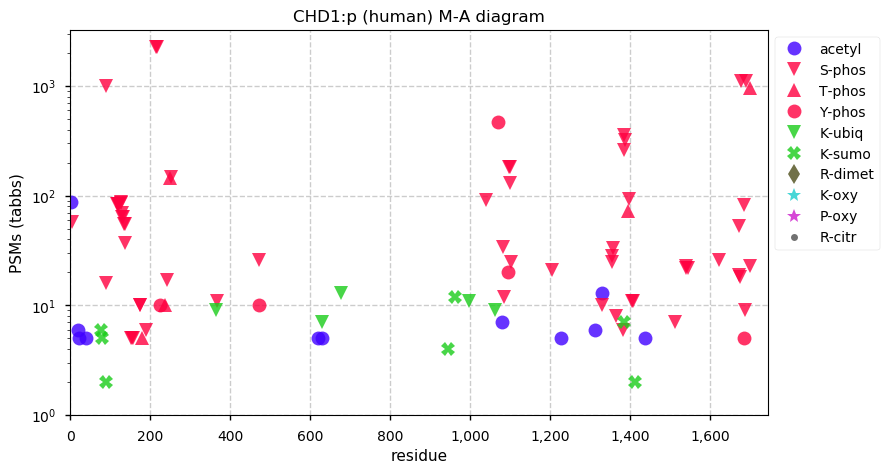
Sat Aug 08 18:54:15 +0000 2020@UCDProteomics @neely615 @h_i_g_s_c_h @olgavitek One can be both ... I know many exceptional a$$holes
Sat Aug 08 17:10:52 +0000 2020ENSEMBL's web system seems to be down, including the US mirrors. 😱
Sat Aug 08 16:39:16 +0000 2020If you ever need some really good ubiquitinylation site (K-GG) data, PXD018743 is a particularly elegant example (🔗).
Sat Aug 08 16:00:51 +0000 2020PXD018430 is a great data set if you are interested in examples of arginine citrullination (aka deimination).
Sat Aug 08 13:21:14 +0000 2020CD180:p's lack of phosphorylation & very short (651-661) cytoplasmic domain do not support a transmembrane signalling function for the protein.
Sat Aug 08 13:21:14 +0000 2020CD180:p θ(max) = 46%. Also known as: LY64, Ly78, RP105. Peptides from the mature protein are observed in HLA type II experiments. Very rare in most cell lines, except for the monocyte-derived line THP-1. Commonly observed in monocytes and B cells.
Sat Aug 08 13:21:14 +0000 2020CD180:p, CD180 molecule (H. sapiens) 🔗 Midsized membrane protein; PTMs: (N34, N70, N78, N201, N394, N402)+glycosyl; SAAVs: N53K (2%), S99R (9%); mature form: 24-661 [1,967×, 6.6 kTa] #ᗕᕱᗒ 🔗
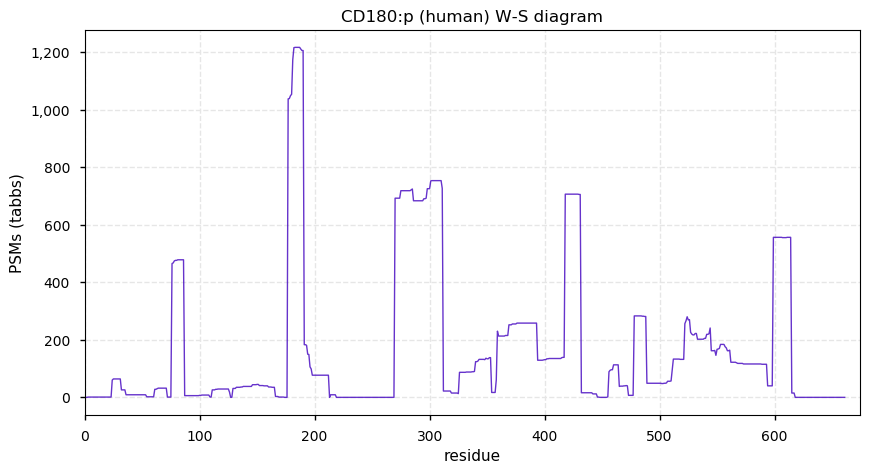
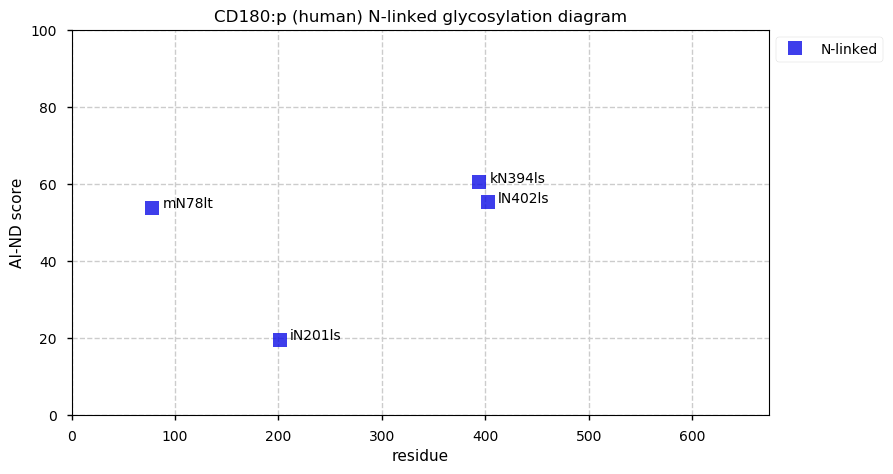
Fri Aug 07 12:10:18 +0000 2020ICAM3:p's phosphodomain structure combined with 1 transmembrane domain (486-510) strongly suggests a signalling/receptor-type function for the molecule, although none seems to described in the literature. Instead, most references seem to focus on its "stickiness".
Fri Aug 07 12:10:17 +0000 2020ICAM3:p θ(max) = 67%. Also known as: CDW50, ICAM-R, CD50. Peptides from the mature protein are observed in HLA type II experiments; peptides 2-12 & 4-12 (signal sequence) present in HLA type I data. Rare in most cell lines, other than JURKAT. Commonly observed in lymphocytes.
Fri Aug 07 12:10:17 +0000 2020ICAM3:p (H. sapiens) 🔗 Midsized protein; PTMs: (N52, N87, N110, N134, N206, N295, N320, N363, N389, N453)+glycosyl, 516-536 phosphodomain; SAAVs: I63V (1%), R115G (18%), D143G (18%), S525T (10%); mature form: 30-547 [6,600 ×, 35 kTa] #ᗕᕱᗒ 🔗
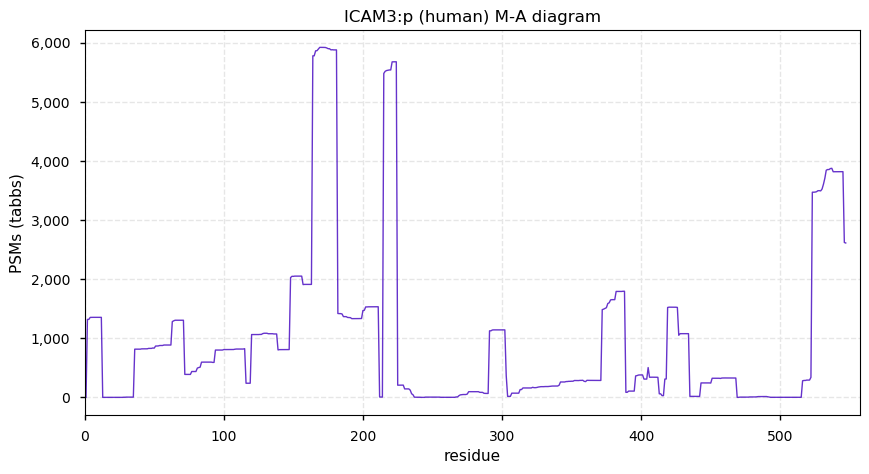
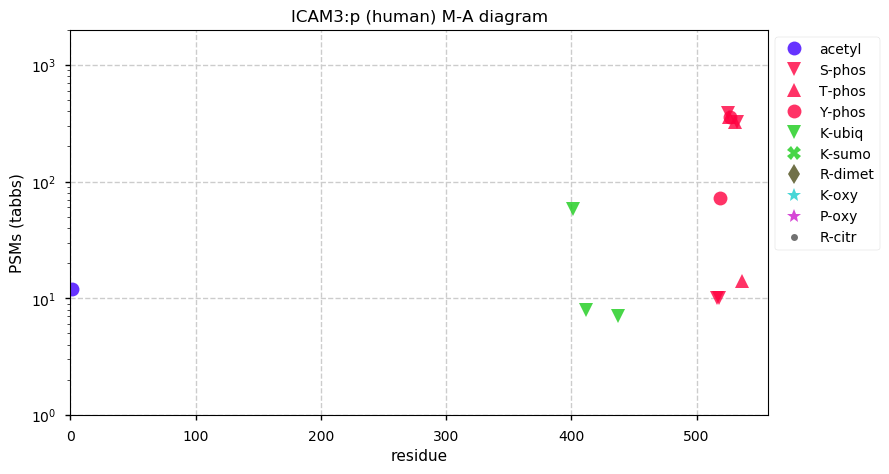
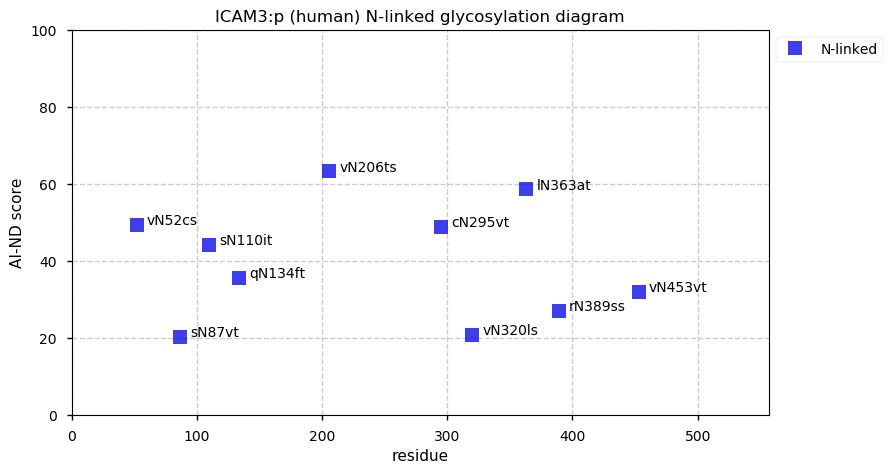
Thu Aug 06 17:33:23 +0000 20203. using the PSMs from that analysis, verify the cleavage sites described in the NCBI and UP entries that generate the mature proteins in Coxsackievirus B3 from the polyprotein. 4/4
Thu Aug 06 17:33:23 +0000 20202. reanalyze the data using human protein sequences and NCBI's CVB3 polyprotein sequence 🔗 (or for UP devotees, 🔗); and 3/4
Thu Aug 06 17:33:23 +0000 20201. download the data files OR11_20160122_PG_HeLa_CVB3_10h_A.raw, OR11_20160122_PG_HeLa_CVB3_10h_B.raw & OR11_20160122_PG_HeLa_CVB3_10h_C.raw from 🔗; 2/4
Thu Aug 06 17:33:22 +0000 2020Suggestion for a simple bioinformatics tutorial focussing on viral MS/MS-based proteomics, in 3 steps.
Have the students do the following: 1/4
Thu Aug 06 15:30:03 +0000 2020"But they say that not all scientists are aware of those nuances. There are plenty who have abandoned their main areas of expertise to explore Covid-19, including virologists who have never worked with coronaviruses before ..." 🔗
Thu Aug 06 14:29:41 +0000 2020Does anyone else find it annoying that the Gmail web interface is constantly trying to write your emails for you? Indicating spelling mistakes: good. Trying to finish my sentences: bad.
Thu Aug 06 12:22:28 +0000 2020ICAM2:p θ(max) = 54%. Also known as: CD102. Peptides from the mature protein are observed in HLA type II experiments; 1 peptide 6-13 from the signal domain is observed in HLA type I data.
Thu Aug 06 12:22:28 +0000 2020ICAM2:p, intercellular adhesion molecule 2 (H. sapiens) 🔗 Small membrane protein; PTMs: (N47, N82, N105, N153, N176)+glycosyl, Y260+phosphoryl; SAAVs: none; mature form: 25-275 [6,024 x, 24 kTa] #ᗕᕱᗒ 🔗
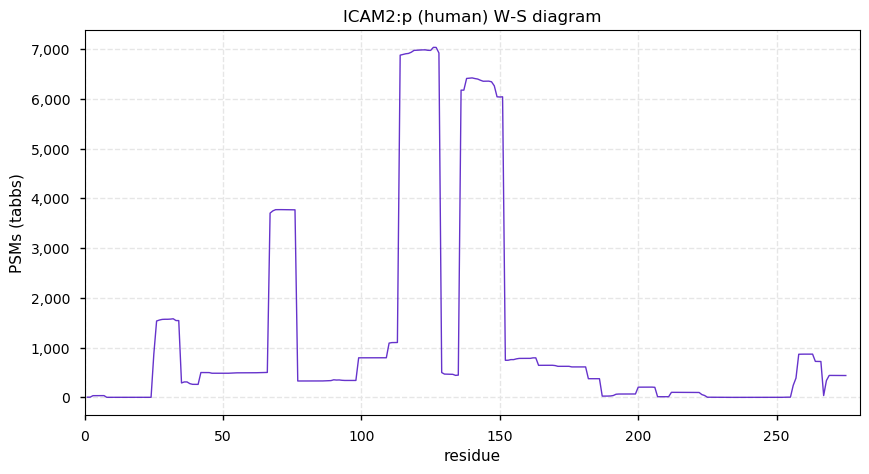
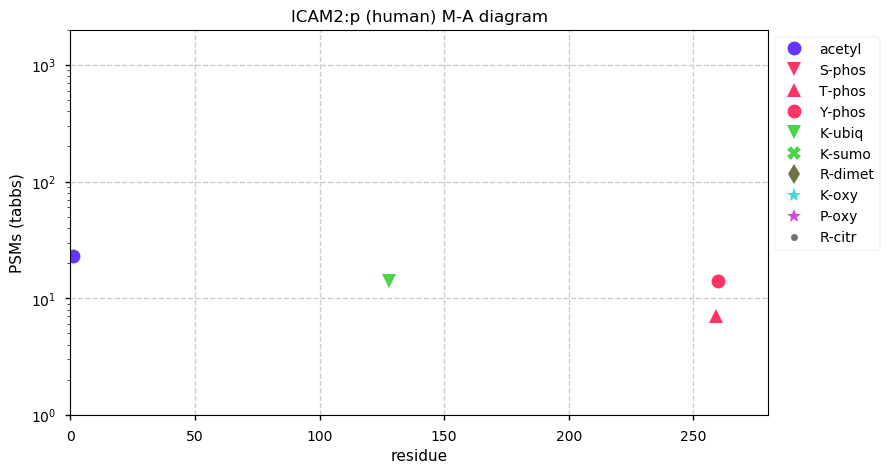
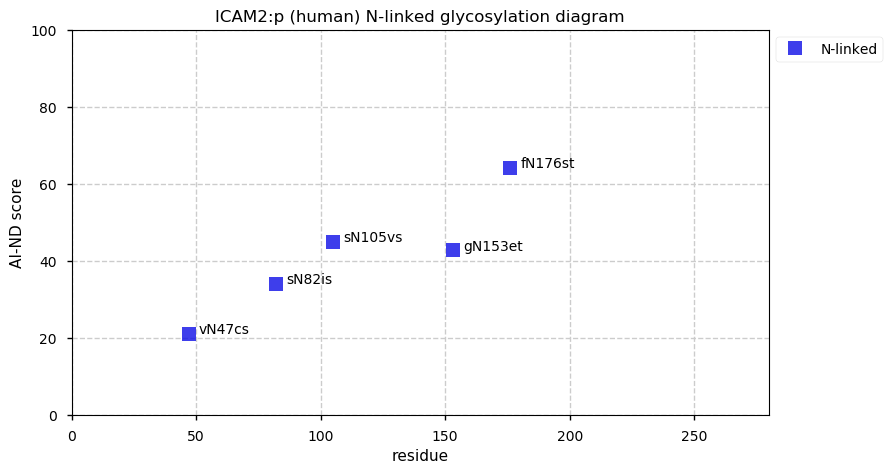
Wed Aug 05 19:56:45 +0000 2020@Smith_Chem_Wisc @wfondrie I used to have card boxes around, back in the 70's. The version info was written on the box in felt pen ...
Wed Aug 05 16:16:22 +0000 2020When you use a phrase like "Cells were lysed, reduced, and alkylated in lysis buffer" in your method, please include the name of the alkylation reagent used. 😡😡😡
Wed Aug 05 15:30:37 +0000 2020As someone who spent years learning & using quantum mechanics, I have no idea WTF this might entail (beyond the buzz words). 🔗
Wed Aug 05 15:13:03 +0000 2020ICAM1:p is the major protein used by human Rhinovirus A and B to enter cells for infection, resulting in the suite of symptoms often referred to as the "common cold".
Wed Aug 05 14:32:38 +0000 2020I can find quite a few systems that look like they were going to do this sort of thing, but they all have the feel of prototypes that either ran out of resources or interest before they were really populated enough to be useful.
Wed Aug 05 14:30:18 +0000 2020Is there a database query system somewhere that I can use to plug in a protein/gene name & get back the N-linked glycoforms that have been observed for that protein?
Wed Aug 05 12:45:32 +0000 2020ICAM1:p θ(max) = 67%. Also known as: BB2, CD54. Peptides from the mature protein are observed in HLA type II experiments, while 3 peptides (2-10, 2-11, 19-27) from the signal domain are observed in HLA type I data.
Wed Aug 05 12:45:32 +0000 2020ICAM1:p, intercellular adhesion molecule 1 (H. sapiens) 🔗 Midsized protein; PTMs: (N145, N183, N202, N267, N385)+glycosyl; SAAVs: K56M (8%), G241R (6%), P352L (1%), R397Q (1%); mature form: 28-532 [18,816 x, 119 kTa] #ᗕᕱᗒ 🔗
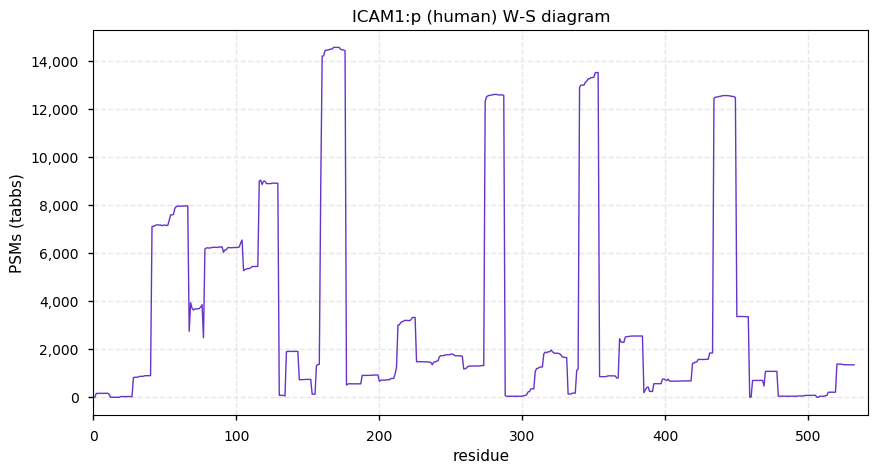
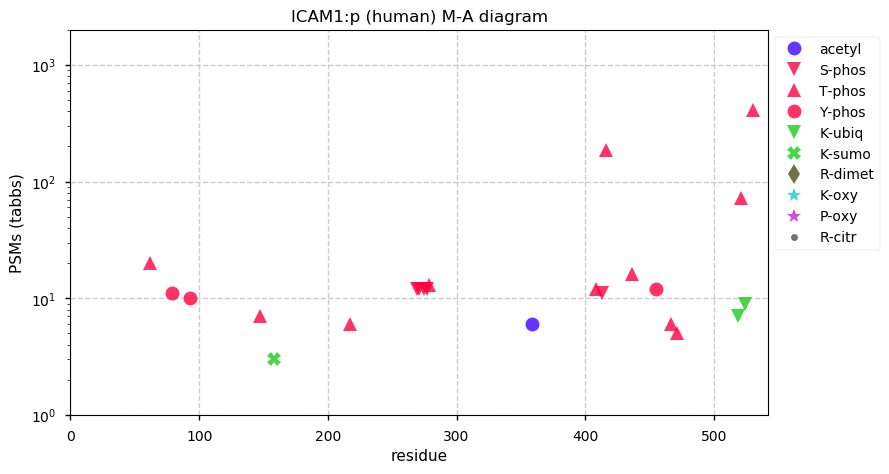
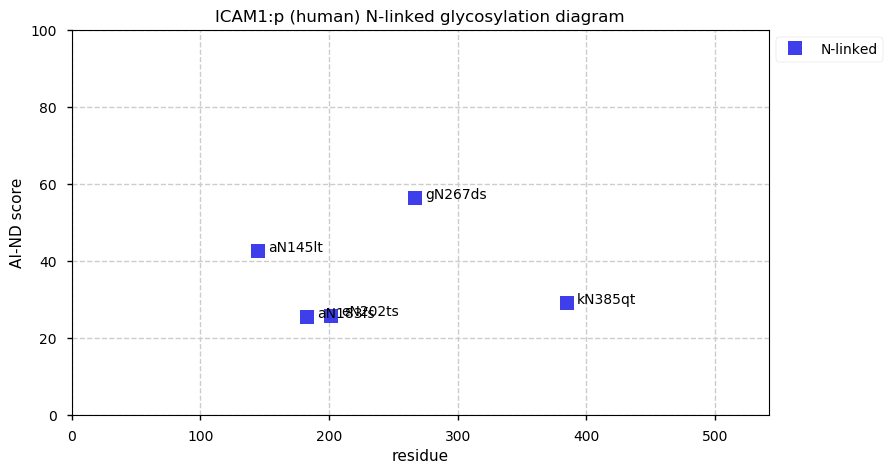
Tue Aug 04 17:06:47 +0000 2020I guess they don't have to go through the proteasome-TAP route as they are already in the ER, but they do have to be whittled down to 8-10 residue domains somehow.
Tue Aug 04 16:59:29 +0000 2020I had never noticed before how often peptides generated from protein ER signalling sequences end up in HLA/MHC type I data sets.
Tue Aug 04 12:03:18 +0000 2020CD58:p θ(max) = 44%. Also known as: LFA3. Eyeballed membrane spanning domain: 216-237. PTMs suggests no phosphorylation-mediated signalling function. Domain (61-184) is observed in urine. Found in HLA type II peptide experiments.
Tue Aug 04 12:03:18 +0000 2020CD58:p, CD58 molecule (H. sapiens) 🔗 Small membrane protein; PTMs: (N40, N169, N195)+glycosyl; SAAVs: none; common in urine, haematopoietic lineage cells & many cell lines; mature form: 31-250 [6,188 x, 14 kTa] #ᗕᕱᗒ 🔗
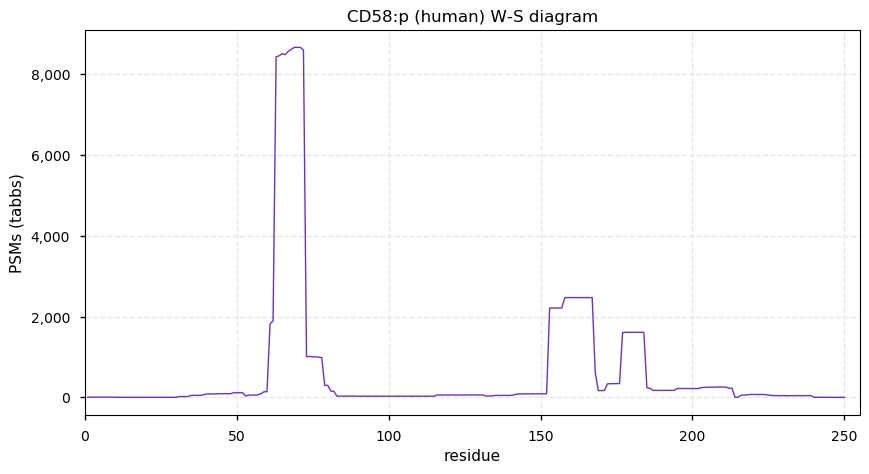
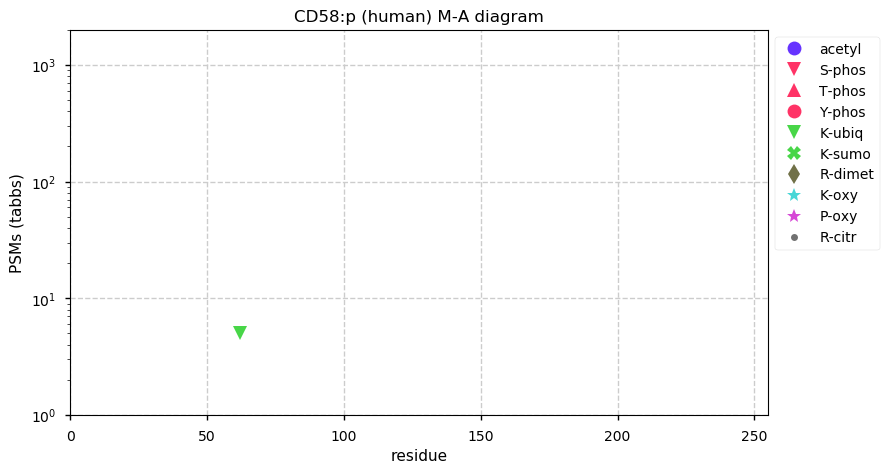
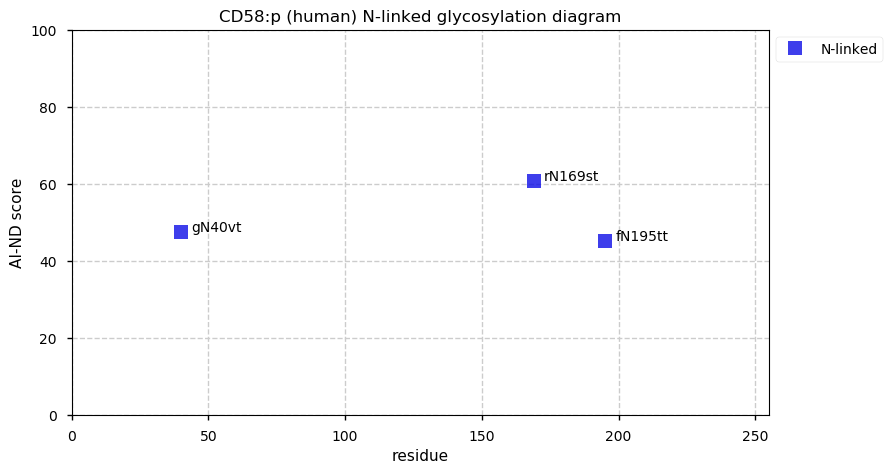
Mon Aug 03 17:51:42 +0000 2020@MiguelCos @Smith_Chem_Wisc IMO proteogenomics is the only straightforward way to id SAAVs. A maximum of 1% of the total number of good quality PSMs will have SAAVs: usually it is more like 0.5%. So for every 1000 PSMs assigned, you will pick up 5-10 PSMs with SAAVs.
Mon Aug 03 12:46:55 +0000 2020@HFazelinia @DocGatorDawg1 Thanks for all of the suggestions. 😎
Mon Aug 03 12:36:45 +0000 2020CD46:p θ(max) = 45%. Also known as: TRA2.10, MGC26544, TLX. Predicted membrane spanning domain: 329-349. Some viral & bacterial proteins bind to this molecule. PTM structure suggests a signalling function via Y-phosphorylation of the intracellular domain.
Mon Aug 03 12:36:45 +0000 2020CD46:p, CD46 molecule (H. sapiens) 🔗 Small membrane protein; PTMs: (Y362, Y372)+phosphoryl, (N83, N273)+glycosyl; (287-300) predicted O-glycosyl; SAAVs: S13F (1%); mature form: 35-384 [10,337 x, 39.7 kTa] #ᗕᕱᗒ 🔗
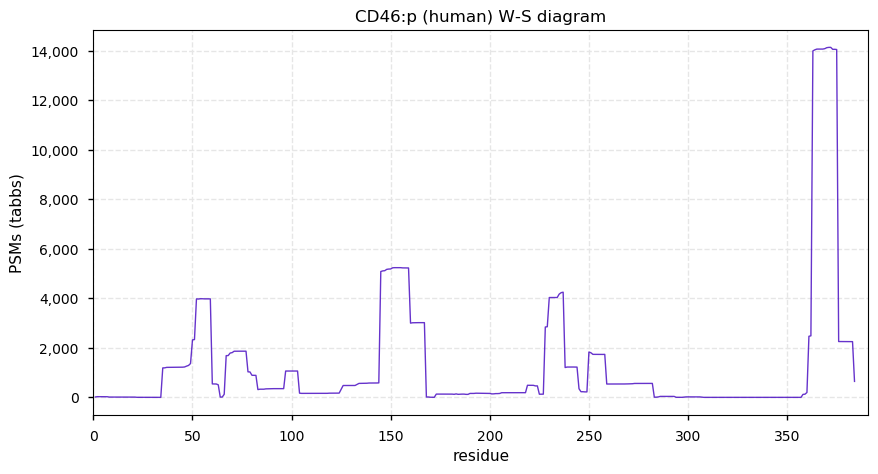
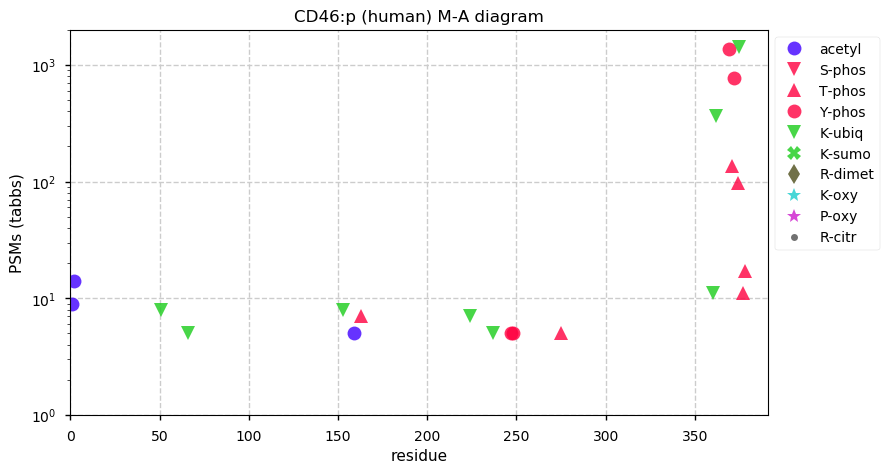
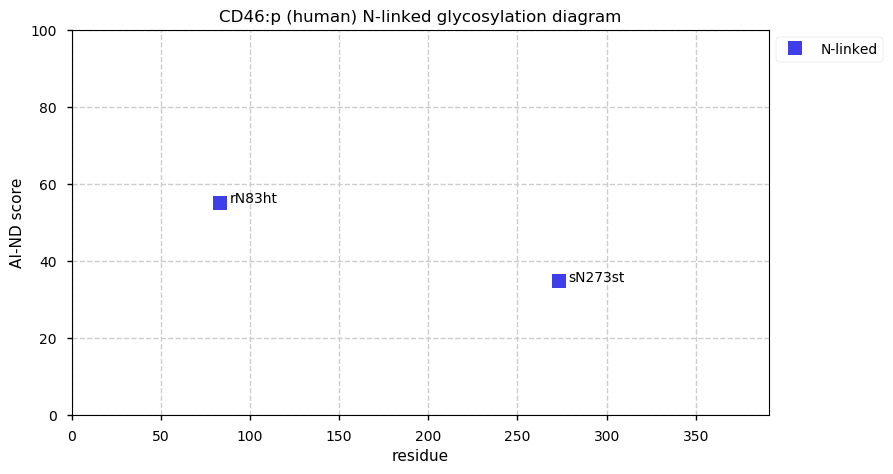
Sun Aug 02 17:09:19 +0000 2020What is the best small glycoform to use when searching data that contains lots of O-linked glycosylation? GalNAc, GalNAc+Gal, GalNAc+Gal+GlcNAc, ... ?
Sun Aug 02 13:54:30 +0000 2020As with many proteins, CD276:p may be involved in a lot of processes, but the details remain obscure.
Sun Aug 02 13:54:30 +0000 2020CD276:p θ(max) = 52%. Also known as: B7-H3, B7H3, B7RP-2. Predicted membrane spanning domain: 467-487. Frequently observed in many cell lines, CD8+ T cells and monocytes.
Sun Aug 02 13:54:30 +0000 2020CD276:p, CD276 molecule (H. sapiens) 🔗 Midsized protein; PTMs: C-term phosphodomain, (N91, N104, N215, N309, N322, N433)+glycosyl; SAAVs: P97L (20%), R267H (12%), A279T (20%), G508R(7%); mature form: 29-533 [11,685 x, 68.9 kTa] #ᗕᕱᗒ 🔗
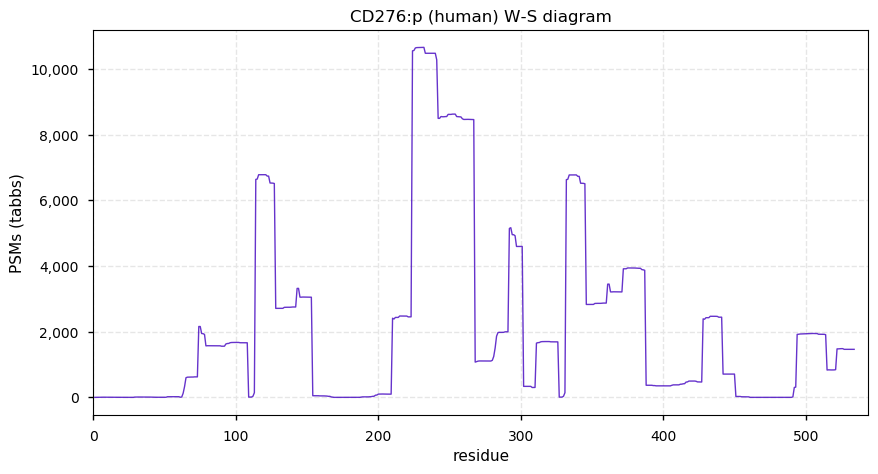
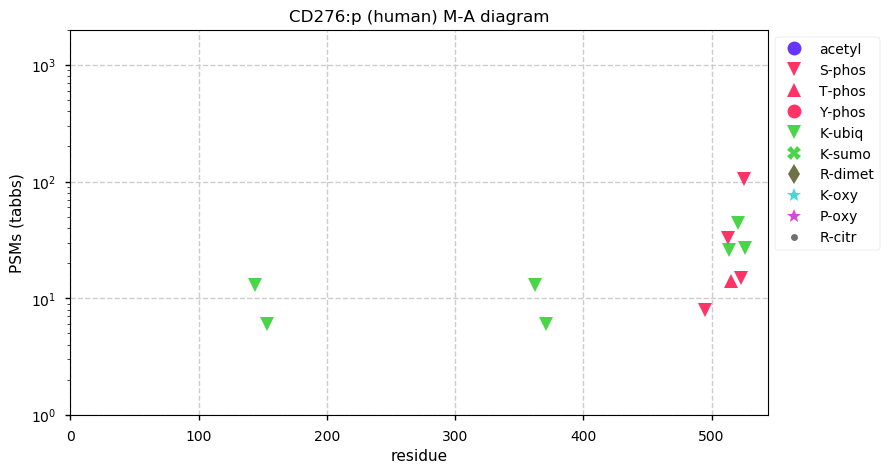
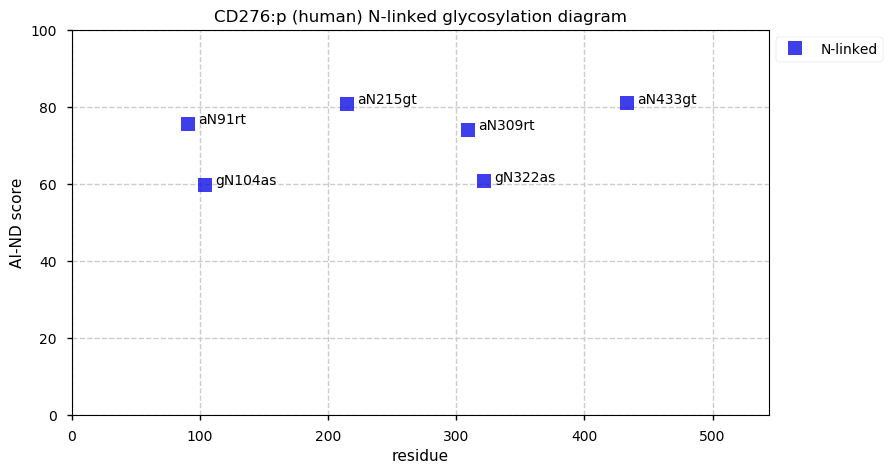
Sat Aug 01 17:47:06 +0000 2020@slashdot Sounds like the plot of a William Gibson short story from the late 80's.
Sat Aug 01 14:17:42 +0000 2020@OliverMBernhar1 @Smith_Chem_Wisc The strengths of commercial software are most important in non-academic/commercial labs, where things like ISO9000 compliance, reliability, support & QA/QC are required. At the moment the non-academic market is small, so freeware tends to dominate.
Sat Aug 01 12:58:49 +0000 2020CD99:p θ(max) = 58%. Also known as: "antigen identified by monoclonal antibodies 12E7, F21 and O13", MIC2. Predicted membrane spanning domains: 123-147. Observations in urine limited to the domain 21-135.
Sat Aug 01 12:58:49 +0000 2020CD99:p, CD99 molecule (H. sapiens) 🔗 Small plasma membrane protein; PTMs: (S167, S181)+phosphoryl, (S29, T41, S48)+glycosyl; SAAVs: M166V (7%), N173I (6%), N173S (6%); mature form: (21,22)-185 [14,240 x, 61 kTa] #ᗕᕱᗒ 🔗
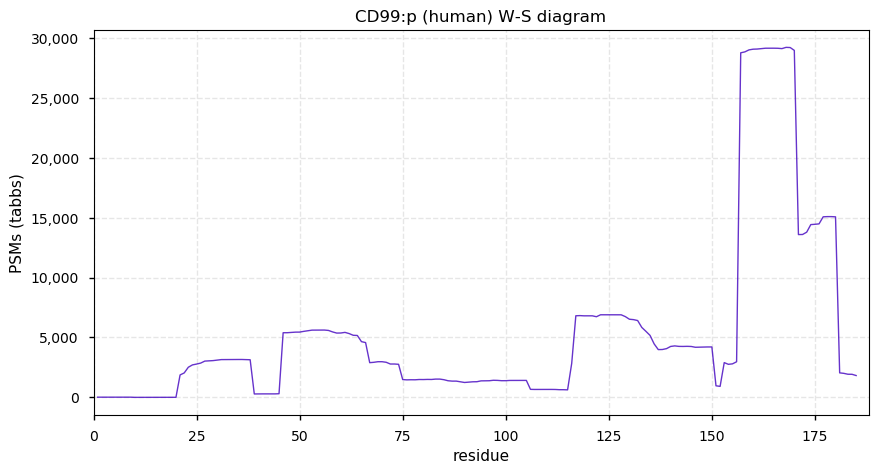
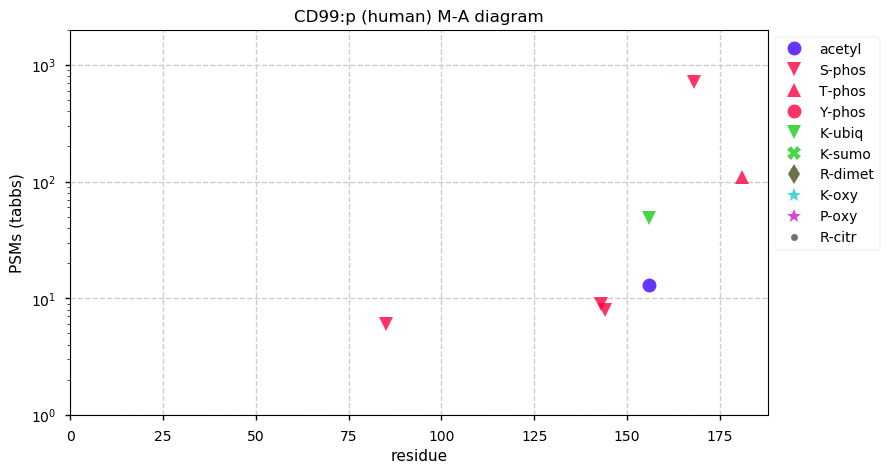
Fri Jul 31 23:00:44 +0000 2020@JoePlatelet @mjmaccoss It seems the host cell has an unfriendly attitude towards the type of folding present in the viral S protein.
Fri Jul 31 20:06:05 +0000 2020@JoePlatelet @mjmaccoss I can't really speak to the stoichiometry: this sort of summary gives you an idea of what you might see, not why. The same type of summary for the S protein has a different feel, taken from 122 LC/MS/MS runs out of the same set of experiments 🔗
Fri Jul 31 19:41:20 +0000 2020@mjmaccoss It is from 492 LS/MS/MS runs, but only 191 runs had observations of the N protein.
Fri Jul 31 19:39:38 +0000 2020@mjmaccoss It is a summary of the PTM results from all of the SARS_CoV2 studies to date (492 LC/MS/MS runs), not a single observation or experiment.
Fri Jul 31 18:53:46 +0000 2020@JesseBrown @telfordk At least there is nothing sketchy about its WHOIS record:
🔗
Fri Jul 31 17:01:11 +0000 2020Calling SARS CoV2 "N" a "phosphoprotein" doesn't really to it justice 🔗
Fri Jul 31 15:34:55 +0000 2020@slashdot Academic informatics groups take note!
Fri Jul 31 14:13:16 +0000 2020@MHendr1cks Forbin-McGill
Fri Jul 31 12:10:59 +0000 2020CD155:p θ(max) = 30%. Also known as SFA-1, PETA-3, TSPAN24, RAPH, MER2. Predicted membrane spanning domains: 19-39, 58-78, 92-112, 222-242. Observed HLA type II peptides chiefly from the domain 163-183. Observations in urine limited to the domain 179-221.
Fri Jul 31 12:10:59 +0000 2020CD151 molecule (Raph blood group) (H. sapiens) 🔗 Small plasma membrane protein; PTMs: N159+glycosyl & low occupancy K+ubiquitinyl; SAAVs: none; common in many tissues and cell lines; mature form: 2-253 [7,359 x, 24 kTa] #ᗕᕱᗒ 🔗
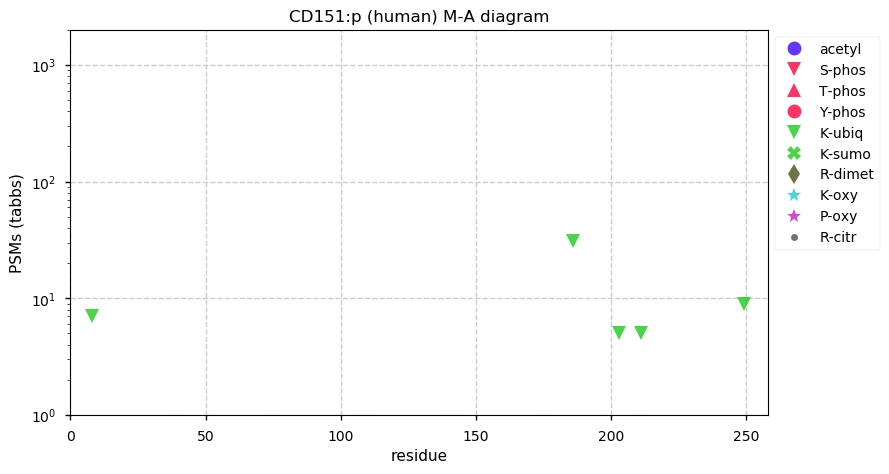
Thu Jul 30 15:37:53 +0000 2020CD109:p is found in many tissues and fluids by MS/MS-based proteomics, but only in the parathyroid gland and hair by antibody-based proteomics 🔗
Thu Jul 30 12:27:52 +0000 2020The two high MAF SAAVs (Y703S, T1241M) suggest that very few individuals will be homozygous for the reference sequence.
Thu Jul 30 12:27:52 +0000 2020CD109:p θ(max) = 44%. Membrane associated (GPI anchor), commonly observed in many but not all tissues & cell lines. The function of the protein is not well understood & its key feature—glycosylation—seems to be largely ignored in the literature.
Thu Jul 30 12:27:52 +0000 2020CD109:p (H. sapiens) 🔗 Large protein; PTMs: N41, N68, N118, N247, N279, N286, N291, N337, N365, N397, N419, N1086, N1355+glycosyl; SAAVs: G377D (1%), Y703S (48%), V1009M (1%), T1241M (41%); mature form: 23-1420 [17,199 x, 133 kTa] #ᗕᕱᗒ 🔗
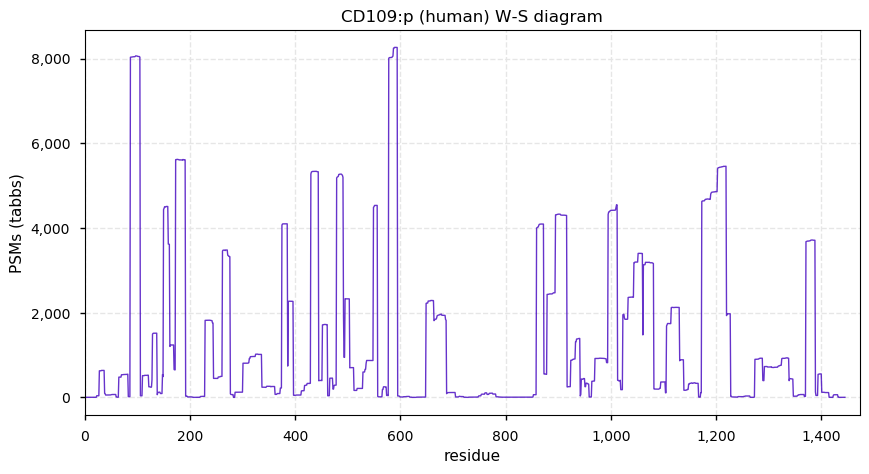
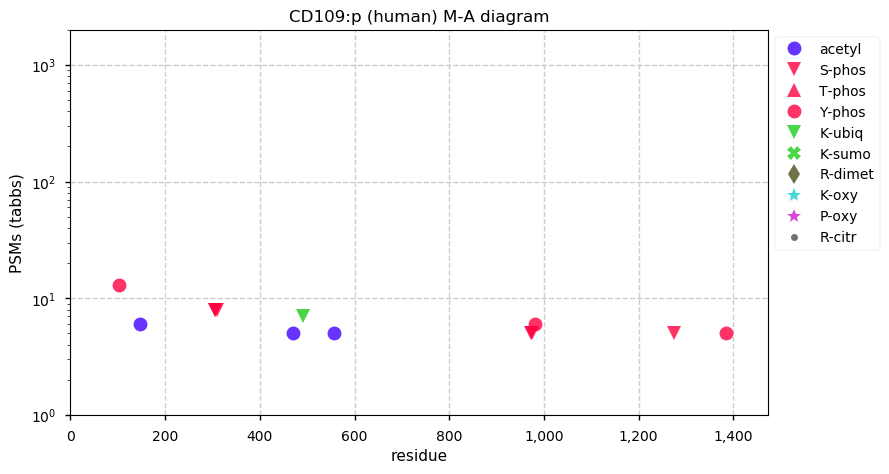
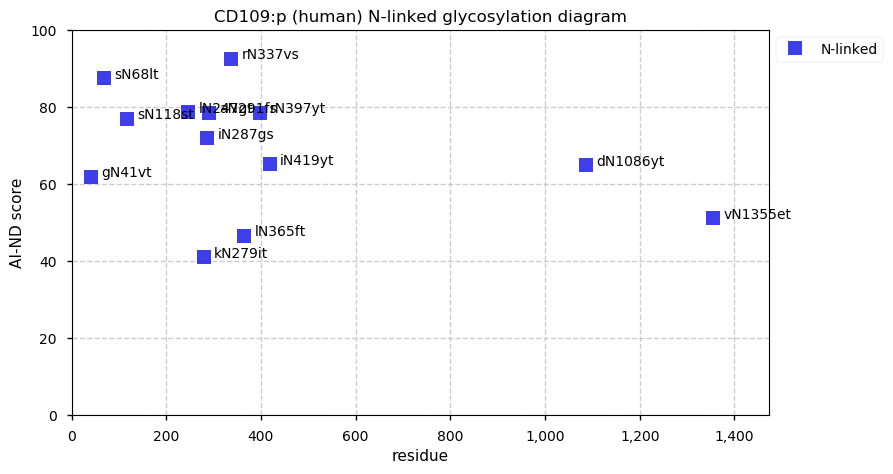
Wed Jul 29 14:51:59 +0000 2020@alistair604 IP only. For all three.
Wed Jul 29 12:45:04 +0000 2020CD9:p θ(max) = 37%. Enriched in extracellular vesicle studies, but common in most tissues and cell lines. Has 4 membrane spanning domains 13-33, 56-76, 88-111 & 196-221. PTM evidence strongly suggests the 112-195 is intracellular.
Wed Jul 29 12:45:03 +0000 2020CD9:p, CD9 molecule (H. sapiens) 🔗 Small plasma membrane protein; PTMs: K126, K169, K170, K179+ubiquitinyl/acetyl, no glycosylation; SAAVs: none; common in extracellular vesicles; mature form: 2-228 [20,624 x, 107 kTa] #ᗕᕱᗒ 🔗
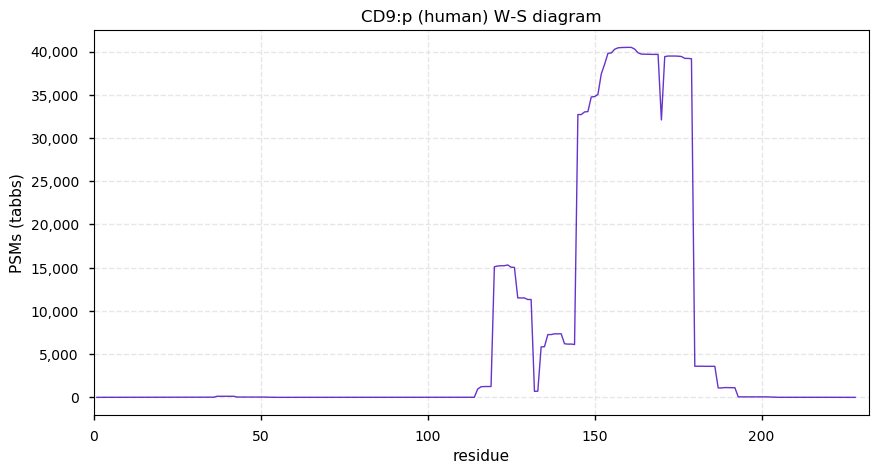
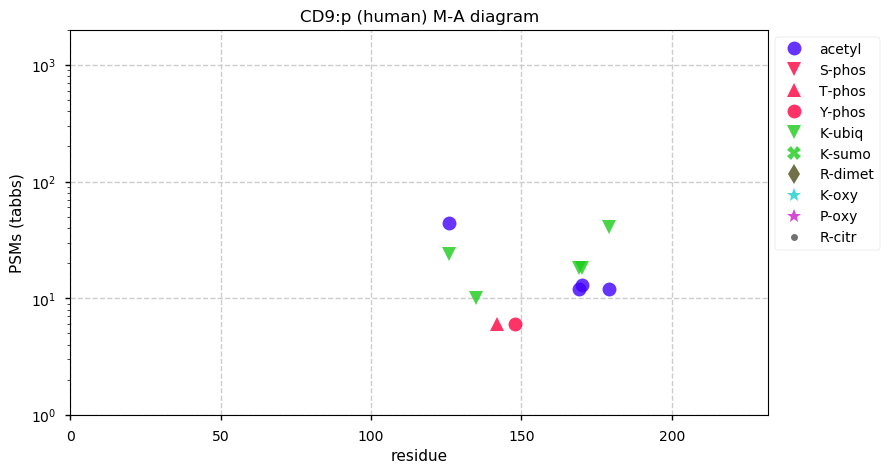
Tue Jul 28 19:59:02 +0000 2020@SciInstr1 It is a bit more complicated. For human (& mouse), all 3 use the same base sequence set supplied by GenCode 🔗 They then provide their own curation of that set, which may include sequences or variants not in the GenCode distribution.
Tue Jul 28 17:49:40 +0000 2020This histogram is an example where the purification did not work. The manuscript states that it was isolating HLA type I peptides, but the data shows it just isolated a set of cellular, non-HLA peptides. /fin 🔗
Tue Jul 28 17:49:40 +0000 2020This histogram demonstrates good purification of HLA type II peptides. The peptides form a skewed distribution (mode at ~15 aa). The distribution should drop off sharply for peptide lengths <11 aa or >20 aa./4 🔗
Tue Jul 28 17:49:39 +0000 2020This histogram demonstrates good purification of HLA type I peptides. The peptides form a tight distribution, with the mode at ~9 residues./3 🔗
Tue Jul 28 17:49:38 +0000 2020A simple way to see whether the experiments were properly done is to create a histogram of the number of PSMs versus their sequence length (in aa residues) for each LC/MS/MS run. The following 3 examples were created using public data from published manuscripts. /2
Tue Jul 28 17:49:38 +0000 2020For anyone who may be reviewing proteomics manuscripts involving HLA type I or type II peptides, it is worth keeping in mind that actually purifying these peptides is experimentally difficult and prone to mistakes./1
Tue Jul 28 17:11:22 +0000 2020@theoneamit @slavov_n @KentsisResearch @eLife I am not defending the practice. I am pointing out that tenure decisions are frequently based on social interactions. The technical aspects (publications, grants, talks) are often much less important than they are made out to be.
Tue Jul 28 16:09:46 +0000 2020@theoneamit @slavov_n @KentsisResearch @eLife It was a surrogate measure for answering the departmental tenure committee's main question: "Do I want to see this person every day for the next 25-30 years?"
Tue Jul 28 16:05:42 +0000 2020@theoneamit @slavov_n @KentsisResearch @eLife "Lunchability" was his term for whether or not he wanted to have lunch with somebody. If he liked have lunch with a junior faculty member, then they were a prime candidate for tenure. Otherwise, not so much.
Tue Jul 28 14:52:10 +0000 2020The combination of travel restrictions & visa uncertainty must make filling available academic positions unusually difficult right now in many countries.
Tue Jul 28 12:58:58 +0000 2020@slavov_n @KentsisResearch @eLife A very influencial department head I knew once spent quite a while discussing (over lunch) his only criterion for promotion: lunchability.
Tue Jul 28 12:00:14 +0000 2020CD81:p θ(max) = 38%. Enriched in extracellular vesicle studies, but common in most tissues and cell lines. Has 4 membrane spanning domains 13-33, 64-84, 90-112 & 202-224. PTM evidence strongly suggests the 113-201 is intracellular.
Tue Jul 28 11:50:53 +0000 2020CD81:p, CD81 molecule (H. sapiens) 🔗 Small plasma membrane protein; PTMs: Y127+phosphoryl, 8× [ST]149-168+phosphoryl , 6× [K]121-193+ubiquitinyl; SAAVs: none; mature form: (1,2)-236 [24,630 x, 158 kTa] #ᗕᕱᗒ 🔗
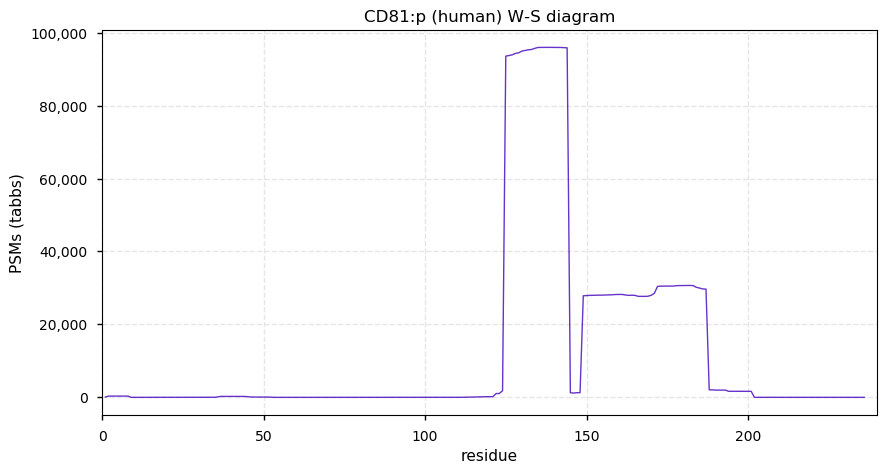
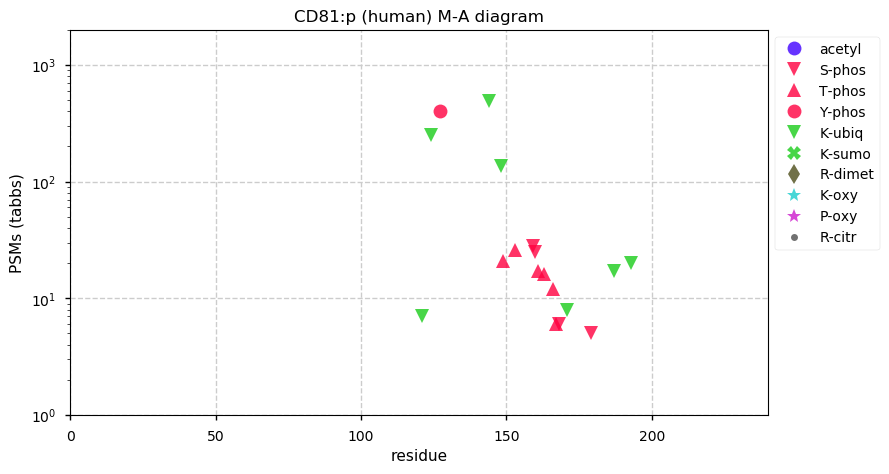
Mon Jul 27 17:43:59 +0000 2020@cdsouthan I'm afraid I don't go to conferences any more. We'll have to remain e-friends 😃
Mon Jul 27 14:56:12 +0000 2020@cdsouthan I am no fan of UniProt, but it is obviously very popular in the biomedical community.
Mon Jul 27 14:43:21 +0000 2020Thanks to every one who participated in this poll. UniProt is the favored source for human protein sequences & accessions by 63 out of 65 respondents, with 2 rebels using RefSeq instead.
Mon Jul 27 13:07:11 +0000 2020Seven different protein sequences are annotated as translated from GENCODE Basic TLS1 mRNA sequences.
Mon Jul 27 13:07:11 +0000 2020θ(max) is 45% for this splice variant. The literature describes a complex set of tissue/cell-specific splice patterns for CD44:g protein products, mainly involving the removal exons in the range exon 6-15 (out of 18).
Mon Jul 27 13:07:11 +0000 2020CD44:p, CD44 molecule (Indian blood group) (H. sapiens) 🔗 Midsized plasma membrane protein; PTMs: 33 phosphorylation sites, N57, N100+glycosyl; SAAVs: H63L (14%); mature form: 21-429 [42,313 x, 413 kTa] #ᗕᕱᗒ 🔗
Sun Jul 26 18:59:36 +0000 2020I really should have said "your source" rather than "primary source", as the named choices are all secondary sources for human sequences.
Sun Jul 26 14:36:38 +0000 2020If you were analyzing human proteomics data, which platform would you use as the primary source of protein sequences?
Sun Jul 26 12:51:20 +0000 2020CD8B:p θ(max) is 25%. The domain 148-210 is difficult to observe in tryptic peptide experiments. Peptides from the protein are found in HLA type I & type II experiments. CD8A and CD8B can form a heterodimer or separate homodimers.
Sun Jul 26 12:48:25 +0000 2020CD8B:p, CD8B molecule (H. sapiens) 🔗 Small integral plasma membrane subunit; PTMs: none; SAAVs: none; most abundant in T cells and mononuclear cells; mature form: 19-210 [267 x, 0.6 kTa] #ᗕᕱᗒ 🔗
Sat Jul 25 23:23:36 +0000 2020@AlexUsherHESA The same is true in Canadian academia.
Sat Jul 25 19:18:36 +0000 2020@macro_momo I often flip the switch when "Aristotelian Logic" gets trotted out, too. But Sod's Law is not disqualifying ...
Sat Jul 25 17:28:55 +0000 2020I stop reading a paper immediately when I discover:
1. it compares two or more methods against each other; or
2. it uses the concept "Occam's Razor" without mockery or derision.
Sat Jul 25 14:53:30 +0000 2020Jufran "Hot & Spicy" is truly my favorite banana ketchup 😃
Sat Jul 25 12:44:30 +0000 2020CD8A:p θ(max) is 41%. The domain 107-224 is difficult to observe in tryptic peptide experiments. Peptides from the protein are found in HLA type I & type II experiments.
Sat Jul 25 12:44:29 +0000 2020CD8A:p, CD8A molecule (H. sapiens) 🔗 Small membrane subunit; PTMs: S229+phosphoryl (low occupancy), no N-linked glycosylation; SAAVs: none; abundant in T cells & mononuclear cells; mature form: 22-235 [791 x, 3.3 kTa] #ᗕᕱᗒ 🔗
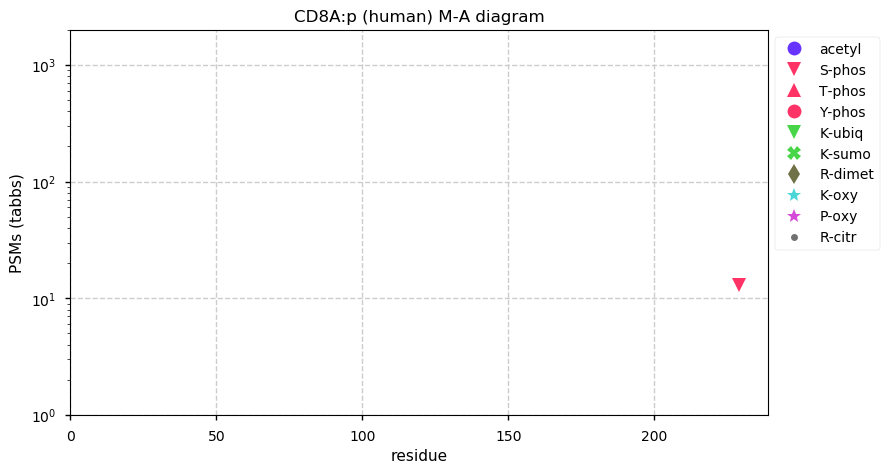
Fri Jul 24 19:45:22 +0000 2020@astacus @slavov_n 🔗
Fri Jul 24 14:18:48 +0000 2020Well, unless anyone has a good objection (speak now or forever hold your peace), θ it will be.
Fri Jul 24 11:59:09 +0000 2020The best sequence coverage observed for CD7:p is 25%. Its function is unknown, but the PTMs suggest signalling across the cell membrane. It is observed in HLA type I & type II peptide experiments.
Fri Jul 24 11:59:08 +0000 2020CD7:p, CD7 molecule (H. sapiens) 🔗 Small integral plasma membrane protein; PTMs: phosphodomain 216-239, N96+glycosyl; SAAVs: none; abundant in lymphocytes but commonly observed in urine; mature form: 26-240 [2,883 x, 12.6 kTa] #ᗕᕱᗒ 🔗
Thu Jul 23 18:01:19 +0000 2020But, if I was considering hiring the trainee/tech for a job or considering the PI for a grant that involved software development, I would not be impressed by the lack of organization, code documentation & supervision.
Thu Jul 23 17:58:47 +0000 2020That led me to look at the github repos for those projects. Looking at the code, it was pretty clear the PIs weren't providing much/any guidance at the code level either. There wasn't anything wrong with the code functionally.
Thu Jul 23 17:56:35 +0000 2020The reason I asked was I had done some grumbling about PIs that don't seem to take part in the design of web app interfaces, even when the app interface was the main outcome of a Nature-branded publication.
Thu Jul 23 17:53:47 +0000 2020Thank you to everyone who participated in the poll or who took a look. Most groups do not include code review as part of their normal lab meeting rotations.
Thu Jul 23 15:15:53 +0000 2020Is there any standard symbol for the observed sequence coverage of a protein in a proteomics measurement? I was thinking of using ψ or θ, but I'm open to anything (within reason).
Thu Jul 23 13:58:21 +0000 2020@AJ_Brenes @Sci_j_my I don't know of any, but I'd be surprised if it got LODs down into the concentration range of a lot of clinical interests. @mjmaccoss has been banging his head against the problems with plasma for quite a while & probably has the best perspective on the problem.
Thu Jul 23 13:33:10 +0000 2020@Sci_j_my The problem with plasma is the wide range of concentrations of biologically important proteins. Well known blood proteins, such as insulin, are often missed. Low level clinically important proteins, such as erythropoietin, are simply never seen.
Thu Jul 23 12:18:18 +0000 2020@Sci_j_my Most blood plasma studies are in the 200-600 protein range. I'd have to see what the proteins are to be sure, but if they really are seeing 2,000 that is better than standard practice in the field.
Thu Jul 23 11:56:17 +0000 2020The best sequence coverage observed for CD6:p is only 28%, due to long tryptic peptides & glycosylation. It is implicated in many processes, but its function is unknown. There is no proteomic evidence of secretion. It is observed in HLA type I peptide experiments.
Thu Jul 23 11:56:16 +0000 2020CD6:p, CD6 molecule (H. sapiens) 🔗 Midsized integral plasma membrane protein; PTMs: N49, N345+glycosyl; SAAVs: T217M (9%), A257V (54%), A271T (8%); abundant in T-cells; mature form: 18(?)-668 [1,208 x, 7.4 kTa] 🔗
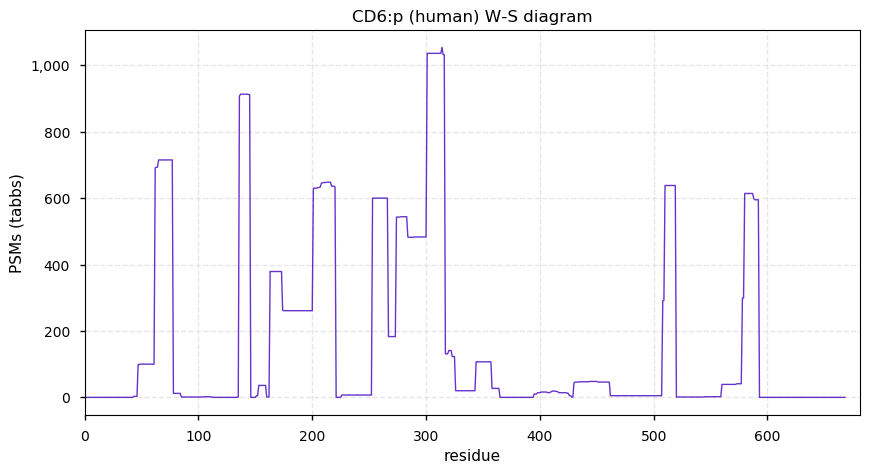
Wed Jul 22 17:29:14 +0000 2020For biomedical research labs that have bioinformatics trainees/techs/investigators: Are code reviews part of your normal lab meeting topic rotation?
Wed Jul 22 12:37:08 +0000 2020CD5:p is used as a immunohistochemical marker for T-cells. Its precise function is still a matter of speculation.
Wed Jul 22 12:37:08 +0000 2020CD5:p is abundant in T-cells and to a lesser extent in mononuclear cells. The phosphodomain 428-487 is part of the intracellular domain of the protein, which is C-terminal to the single membrane spanning domain.
Wed Jul 22 12:37:08 +0000 2020CD5:p, CD5 molecule (H. sapiens) 🔗 Midsized integral plasma membrane subunit; PTMs: 116N, 241N,+glycosyl, 18 phosphorylation sites in the domain 428-487; SAAVs: P224L (17%), H461R (3%); mature form: 25-495 [2,758 x, 19.7 kTa] 🔗
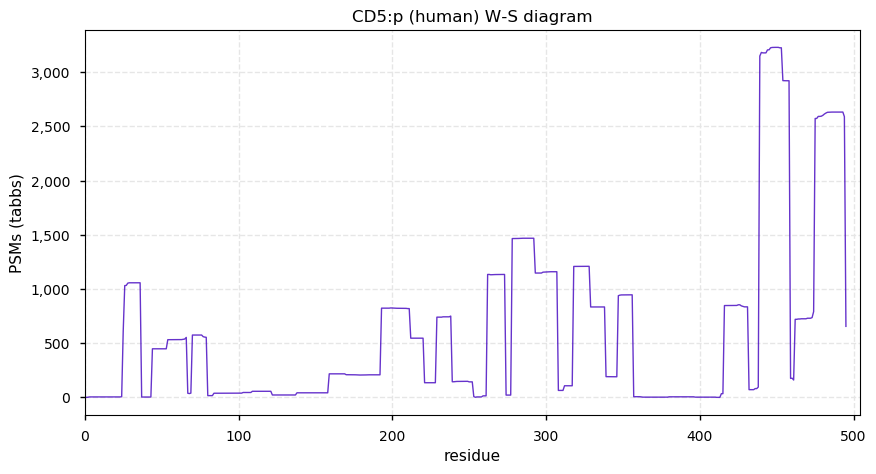
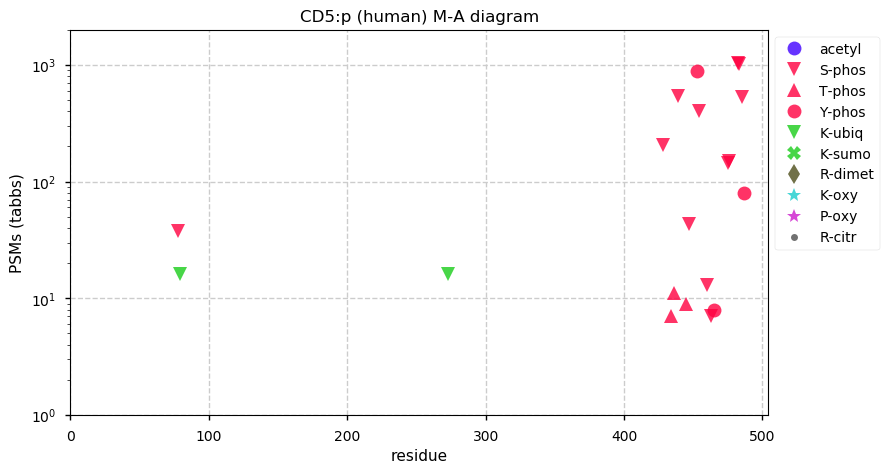
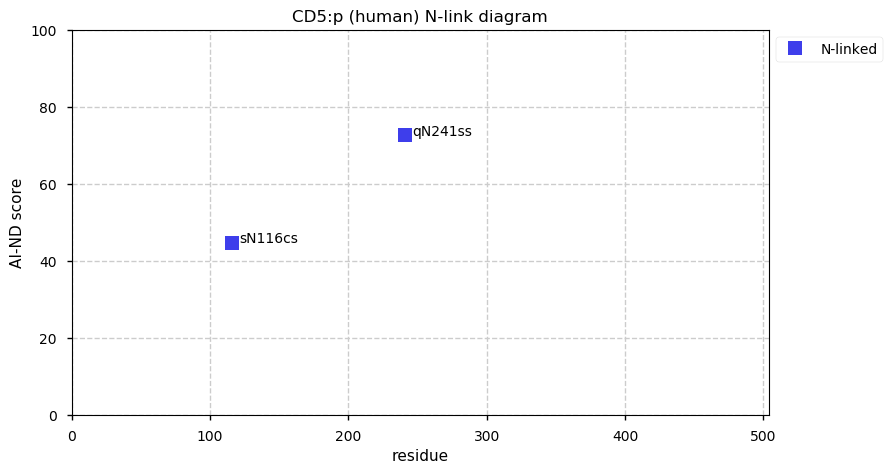
Tue Jul 21 20:37:15 +0000 2020@doctorow Überschwemmungfreude?
Tue Jul 21 19:37:49 +0000 2020@neely615 @pwilmarth @byu_sam At some point, though, Microsoft is most likely going to get more fussy about github projects that don't contain code.
Tue Jul 21 18:41:45 +0000 2020@DonMartinCTV @JustinTrudeau Neither of the main opposition parties have a "Tom Mulcair" equivalent available at the moment.
Tue Jul 21 17:34:08 +0000 2020@AJ_Brenes @font - add in a 'viewport' meta tag for all pages;
- left-align text by default;
- center-align numerical columns in tables;
- use italics & bold very sparingly and always for a purpose;
- include 'target' tags in links to other web sites so that they open a new window/tab.
Tue Jul 21 16:55:47 +0000 2020@AJ_Brenes Only some general advice:
- use @font-face in your CSS to import the font you want;
- use 'pt' to specify font sizes when practical, rather than measures like 'rem' or 'em' or 'px';
- have a few low resolution screens readily available to test layouts (older & cheaper = better)
Tue Jul 21 14:12:41 +0000 2020@AJ_Brenes @karthikskamath It is my contention that the PI, reviewers & editor should have suggested significant improvements to this interface, along the lines of my suggestions, prior to publication. To be clear, the problem here is supervision, NOT the junior investigator that created the interface.
Tue Jul 21 14:12:12 +0000 2020@AJ_Brenes @karthikskamath For example, the main point of 🔗 is a web interface. An example of the interface provided is 🔗, which is used to map phosphorylation sites across cell lines.
Tue Jul 21 14:11:58 +0000 2020@AJ_Brenes @karthikskamath Unfortunately, a considerable part of teaching is telling smart but inexperienced people the obvious and repeating it until they understand that it applies to them, too.
Tue Jul 21 12:08:36 +0000 2020CD36:p has an unusually ragged N-terminus. It is observed in HLA type II peptide experiments, with sequences from 5 loci: (38-52), (137-150), (83-213), (287-310) & (375-388).
Tue Jul 21 12:08:36 +0000 2020Also known as SCARB3, GPIV, FAT, GP4, GPIIIB, GP88 and GP3B. It has 214 annotations indicating involvement in GO functions and processes 🔗
Tue Jul 21 12:08:35 +0000 2020CD36:p, CD36 molecule (H. sapiens) 🔗 Midsized membrane subunit; PTMs: 79N, 205N, 220N, 235N, 321N, 417N+glycosyl, S/T+glycosyl; SAAVs: none; in platelets, fat cells, monocytes; mature form: (21,23,24,25)-472 [7,948 x, 58.0 kTa] 🔗
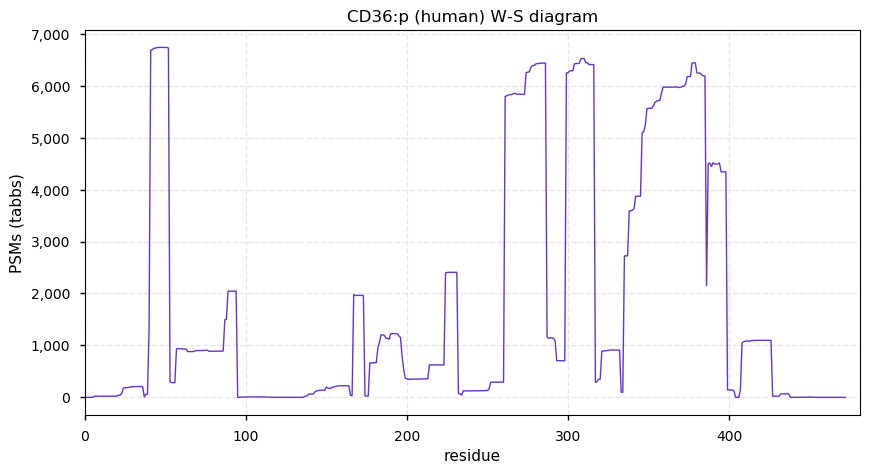
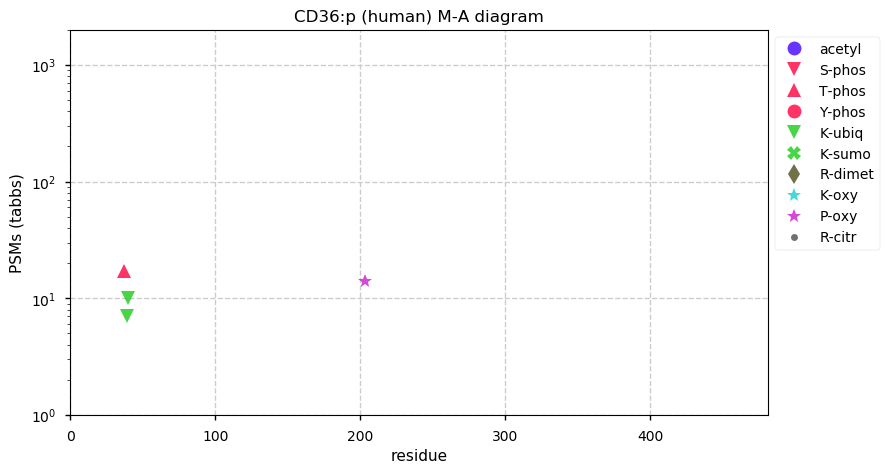
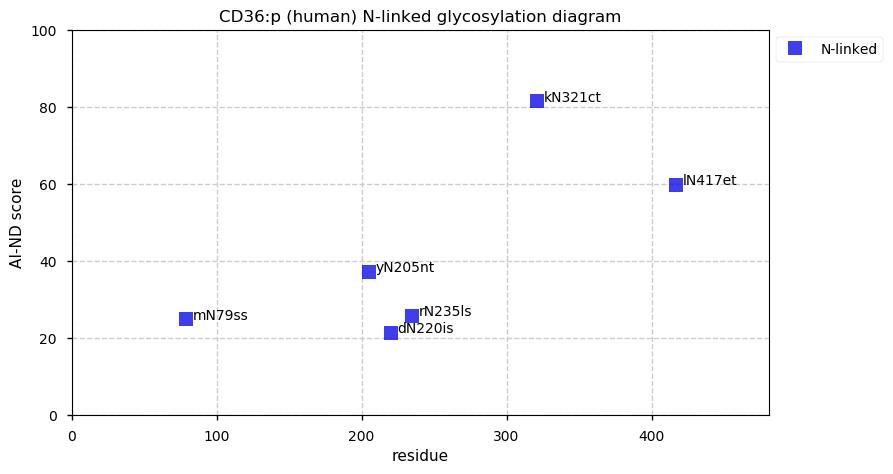
Mon Jul 20 17:32:39 +0000 2020Note to young people developing web-based bioinformatics apps:
use hints/help/examples for all text inputs;
place explanatory text on all input & output pages;
if something doesn't work or there is nothing to illustrate, don't just show a blank panel;
I could go on ...
Mon Jul 20 14:15:31 +0000 2020@KentsisResearch I'll do that one tomorrow, but I can't guarantee "interesting" ...
Mon Jul 20 11:57:55 +0000 2020CD4:p, CD4 molecule (H. sapiens) 🔗 Midsized integral plasma membrane subunit; PTMs: 453K+ubiquitinyl, 296N,325N+glycosyl; SAAVs: none; abundant in T-cells, monocytes and T-helper cells; mature form: 26-458 [1,964 x, 10.5 kTa] 🔗
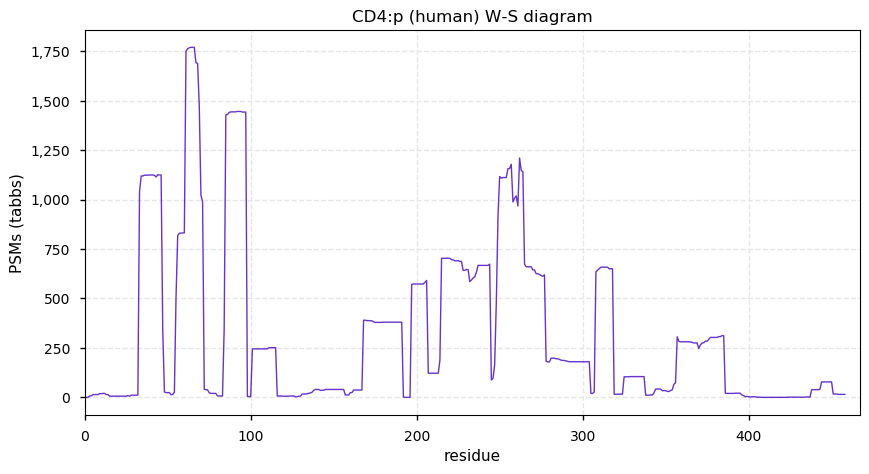
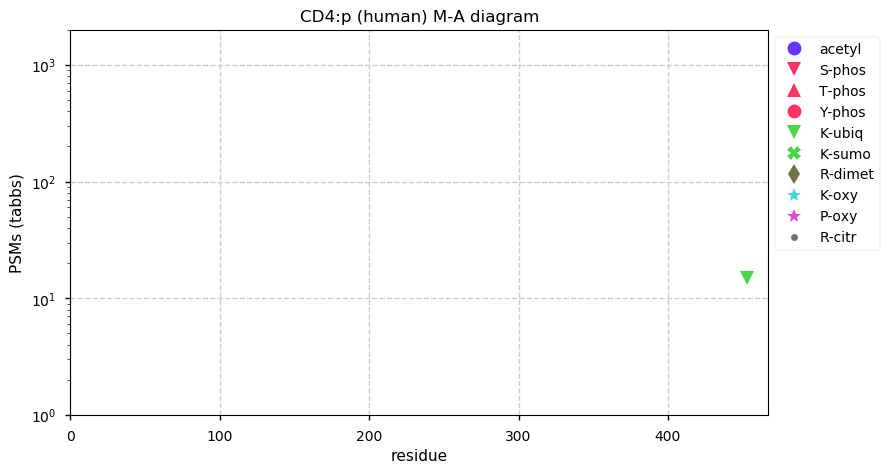
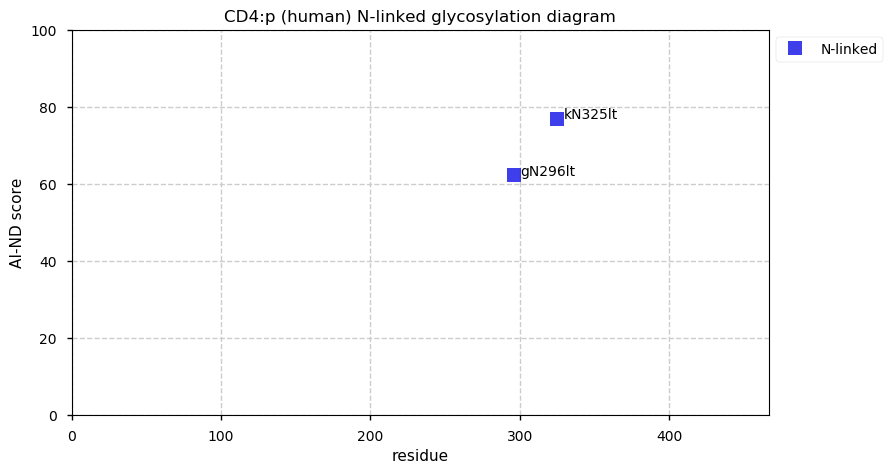
Sun Jul 19 13:43:19 +0000 2020The N-terminal extracellular domain is much shorter than CD3D/E/G, with phosphorylation on the C-terminal intracellular domain. Found in HLA type II data sets.
Sun Jul 19 13:43:19 +0000 2020May have the highest density of Y-phosphorylation of any protein. There are 3 pairs of Y+phosphoryl acceptors 11-12 residues apart: (72,83), (111,123), (142,153). 64Y+phosphoryl does not follow this pattern.
Sun Jul 19 13:43:19 +0000 2020CD247:p, CD247 molecule (H. sapiens) 🔗 Small integral plasma membrane subunit; aka CD3H, CD3Q, CD3Z; PTMs: 7 Y+phosphoryl, 8 K+ubiquitinyl sites, no glycosylation; SAAVs: none; abundant in T-cells; mature form: 22-164 [2080 x, 22.6 kTa] 🔗
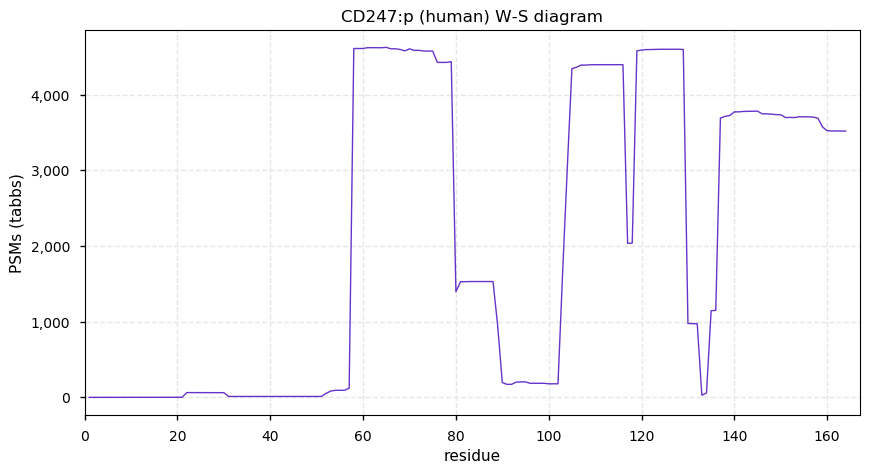
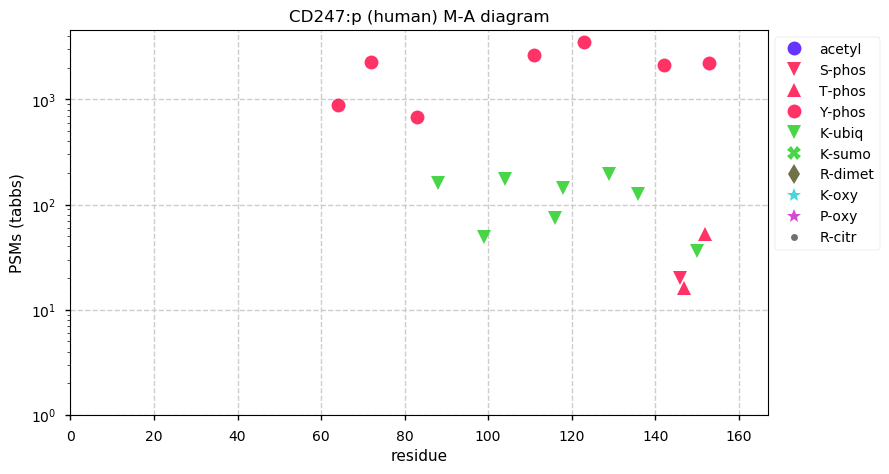
Sat Jul 18 14:05:51 +0000 2020I unfortunately have an obsessive need to make sense of the details of proteomics results (proteins & PTMs & SAAVs).
Sat Jul 18 13:46:41 +0000 2020I suspect the origin of K+ubiquintyl on the extracellular domains of CD3D/E/G subunits is misfolding in the ER. If the subunits do not fold properly a signficant fraction of the time, they would end up exported to the cytoplasm and tagged with ubiquitin via the ERAD mechanism.
Sat Jul 18 12:14:25 +0000 2020CD3G:p is commonly found in HLA type II peptide experiments, all peptides from the domain 165-182.
Sat Jul 18 12:14:25 +0000 2020CD3G:p tyrosine phosphorylation and N-linked glycosylation coincides with the intracellular domain. Ubiquitin ligation on the extracellular domain is a puzzle.
Sat Jul 18 12:14:25 +0000 2020CD3G:p, CD3G molecule (H. sapiens) 🔗 Small integral plasma membrane subunit; PTMs: 160Y,171Y+phosphoryl, 13 K+ubiquitinyl sites, 92N+glycosyl; SAAVs: none; abundant in T-cells; mature form: 23-182 [1,326 x, 6.4 kTa] 🔗
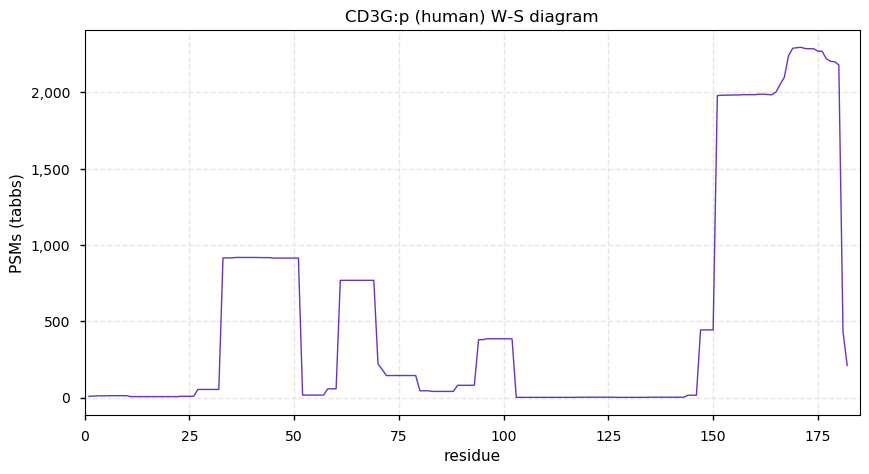
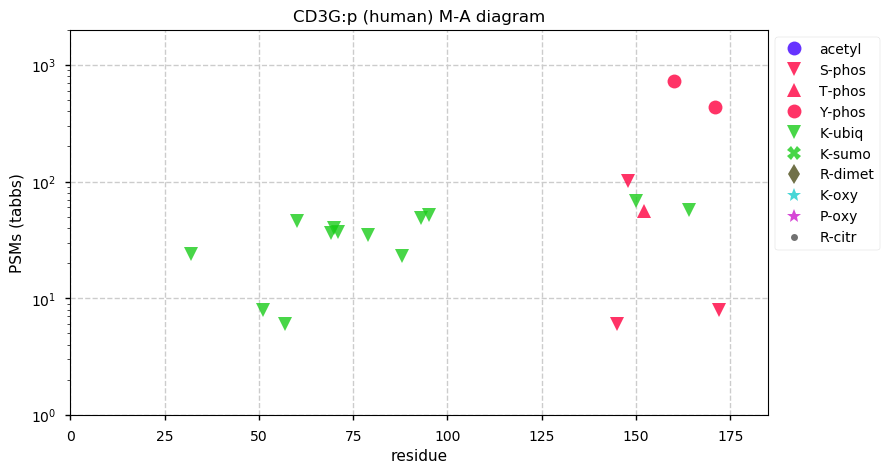
Fri Jul 17 21:21:11 +0000 2020@pwilmarth But the variation in the microbiome is really pretty nifty!
Fri Jul 17 21:20:30 +0000 2020@pwilmarth No doubt. A more thorough study of the human proteins wrt Sjögren is probably warranted. The salivary gland tissue has a significant amount of collagen in it, so that type of analysis would have missed quite a bit of even the high concentration stuff.
Fri Jul 17 20:59:14 +0000 2020@pwilmarth It depends. To start with for body fluids, fully non-specific cleavage. If the results show that to be a waste of processor time, then I switch to semi-tryptic. But never just-tryptic.
Fri Jul 17 20:51:02 +0000 2020@MattWFoster The companion data set from the same paper (PXD020222) used A-549 as annotated, with no traces of HEK-293T present in the data.
Fri Jul 17 20:33:41 +0000 2020@MattWFoster Combination of the proteins & SAAVs observed.
Fri Jul 17 19:27:22 +0000 2020The saliva data also contains many protein ids from oral bacteria (Prevotella, Veillonella, Aggregatibacter, etc.) not examined in the paper. If you are interested in oral microbiology, this data is a good example of what can be easily id'd from saliva, using clinical samples.
Fri Jul 17 19:25:23 +0000 2020PXD016231 (🔗) has well done proteomics data associated with plasma, salivary glands & saliva from patients with symptoms of Sjögren's syndrome. The authors have analyzed the human proteins detected to try to find biomarkers to aid in diagnosis.
Fri Jul 17 17:43:11 +0000 2020For anyone interested in analyzing the data, keep in mind:
1. there is carboxamidomethyl derivatization of amines as well as the desired reaction with cysteine sidechains; &
2. the cell line used was HEK-293T, not A-549 as per the PX annotation and the paper.
Fri Jul 17 17:40:56 +0000 2020PXD020019 (🔗) is the 1st publicly available data set to demonstrate the extent of K+ubiquitinyl of SARS COV-2 proteins in a cell line infected with the virus. It also has proteome & phosphoproteome data confirming the results of earlier studies.
Fri Jul 17 16:32:21 +0000 2020My best guess would be ~ 100 SAAVs in all, including special lists made for specific tissues. If heterozygous vs. homozygous SAAVs could be reliably distinguished, fewer may be necessary.
Fri Jul 17 16:27:54 +0000 2020Thanks to everyone who participated. Opinion seems fairly evenly split, except that no one likes "20". I agree with Prof. Tabb that if there were no tissue distributions, ~33 would be enough.
Fri Jul 17 12:38:16 +0000 2020CD3E:p is commonly found in HLA type II peptide experiments.
Fri Jul 17 12:38:16 +0000 2020CD3E:p tyrosine phosphorylation coincides with the intracellular domain. Ubiquitin ligation on the extracellular domain is a puzzle.
Fri Jul 17 12:38:16 +0000 2020CD3E:p, CD3E molecule (H. sapiens) 🔗 Small integral plasma membrane subunit; PTMs: 188Y,199Y+phosphoryl, 104K,177K+ubiquitinyl, no glycosylation; SAAVs: none; abundant in T-cells; mature form: 22-207 [2,640 x, 16.9 kTa] 🔗
Thu Jul 16 16:20:39 +0000 2020Postulate: it should be possible to select a small panel of human high MAF SAAVs observable using MS/MS-based proteomics that would be sufficient to identify any individual person. How many SAAVs would be required?
Thu Jul 16 13:42:43 +0000 2020@aarmey I think it speaks to the hands-off, blue-team-only analysis of 'omics data that this sort of easy-to-catch problem continues to exist, but it doesn't seem to be going away any time soon.
Thu Jul 16 13:15:01 +0000 2020CD3D:p is commonly found in HLA type II peptide experiments.
Thu Jul 16 13:15:01 +0000 2020CD3D:p phosphorylation coincides with the intracellular domain and glycosylation with the extracellular domain. Ubiquitin ligation on the extracellular domain is a puzzle.
Thu Jul 16 13:15:00 +0000 2020CD3D:p, CD3d molecule (H. sapiens) 🔗 Small integral plasma membrane subunit; PTMs: 149Y,160Y+phosphoryl, 23K,80K,82K+ubiquitinyl, 38N,75N+glycosyl; SAAVs: none; abundant in T-cells; mature form: 22-171 [2,030 x, 9.3 kTa] 🔗
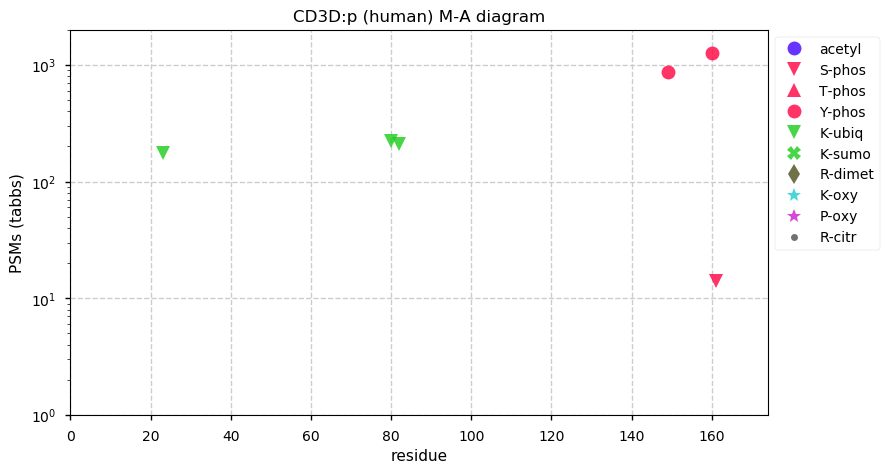
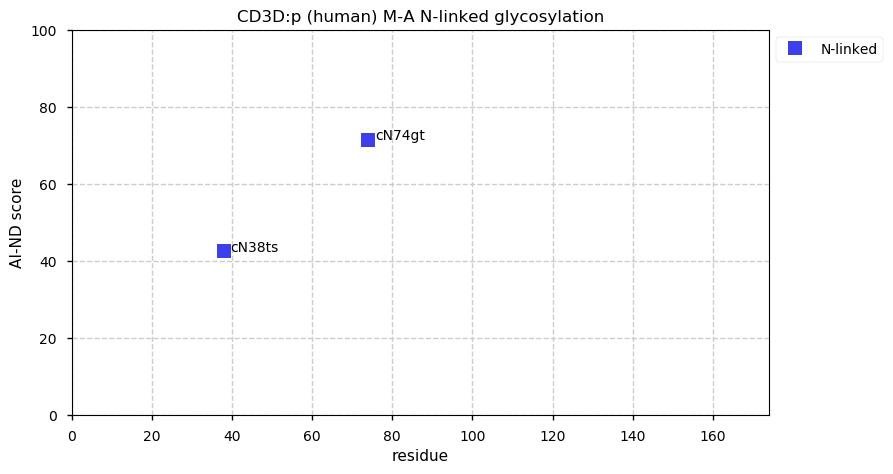
Thu Jul 16 13:00:17 +0000 2020Although it isn't covered a lot here, Mexico and Peru are still getting hit pretty hard, almost proportionally to the better known problem in Brazil. 🔗
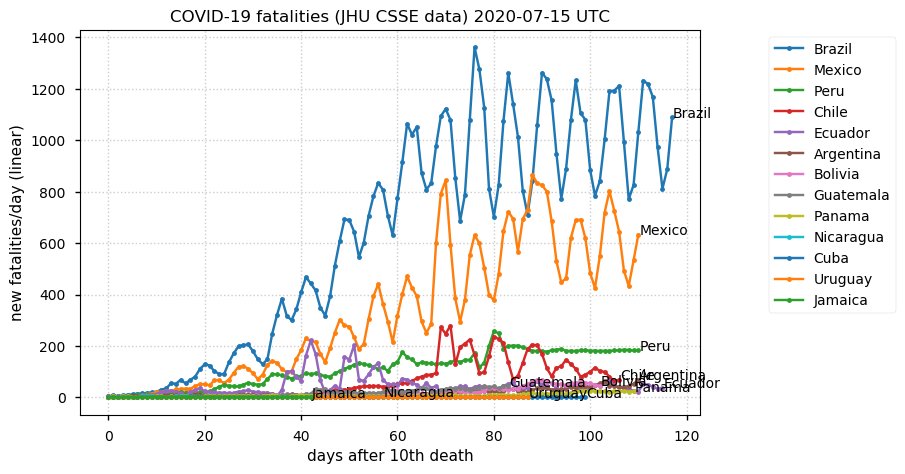
Wed Jul 15 22:20:52 +0000 2020Do A-549 cells look like HEK-293T cells in culture? People do seem to mix them up from time to time.
Wed Jul 15 16:32:09 +0000 2020@ucdmrt They are still doing the alkylation, they are just messing it up significant amounts of amine side reaction products.
Wed Jul 15 14:54:52 +0000 2020I don't understand why the 2 major proteomics mass spec labs in Munich seem to have both simultaneously lost the knack for doing clean carbamidomethyl cysteine side chain derivatizations. Reply with conspiracy theories only.
Wed Jul 15 12:06:29 +0000 2020This enzyme reduces specific double bonds in many steriod-derived bile acids and delta4-3-one structures in steroid hormones.
Wed Jul 15 12:06:28 +0000 2020Shares different subsets of observable tryptic peptides with Akr1c6:p, Akr1c18:p and Akr1cl:p.
Wed Jul 15 12:06:28 +0000 2020Akr1d1:p, aldo-keto reductase family 1, member D1 (M. musculus) 🔗 Small cytoplasmic enzyme; PTMs: significant K+acetylation; SAAVs: none; abundant in liver tissue; mature form: 1-325 [1,777 x, 46.8 kTa] 🔗
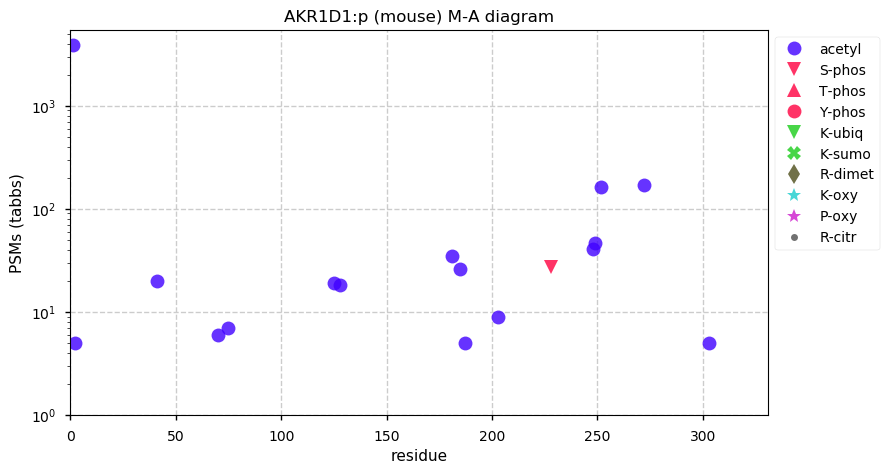
Tue Jul 14 21:21:13 +0000 2020I was able to figure out which ones corresponded to some experiments. The data quality was "non-archival" so I don't really need to know about the other files.
Tue Jul 14 18:13:00 +0000 2020@AlexUsherHESA Ireland would have become independent in 1801 (1802 at the latest).
Tue Jul 14 15:54:26 +0000 2020Does anybody know if there is an index describing the provenance of the raw files associated with PXD019645? Given the number of experiments involved in 🔗 & very generic files names, re-analysis of most of the data isn't possible.
Tue Jul 14 12:17:22 +0000 2020In humans, AKR1B1:p (aldose reductase) is the functional ortholog of mouse AKR1B3:p. While best known for its role in generating sorbitol from glucose, it can catalyze the reduction of many aldehydes and carbonyls.
Tue Jul 14 12:17:22 +0000 2020Shares different subsets of observable tryptic peptides with Akr1b7:p, Akr1b8:p, Akr1b10:p, Akr1c13:p, Akr1c12:p and Akr1c19:p.
Tue Jul 14 12:17:22 +0000 2020Akr1b3:p, aldo-keto reductase family 1, member B3 (M. musculus) 🔗 Small cytoplasmic enzyme; PTMs: significant K+acetyl; SAAVs: none; common in many tissues and cell lines; mature form: (2,3)-316 [14,298 x, 203 kTa] 🔗
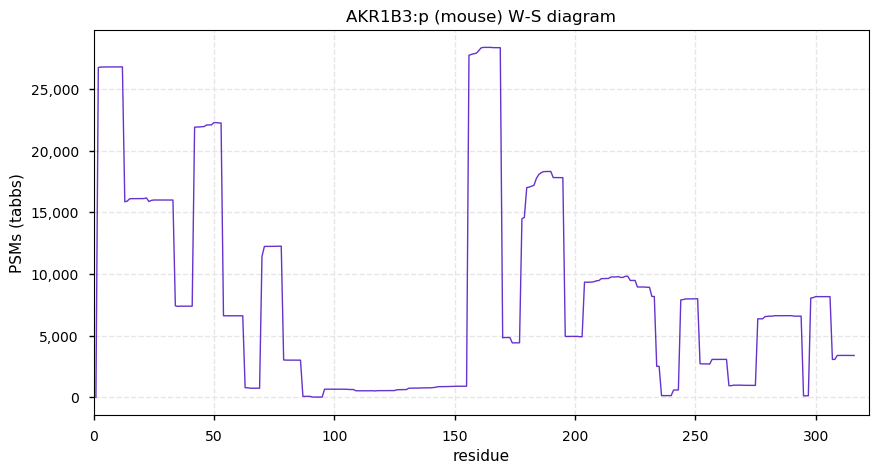
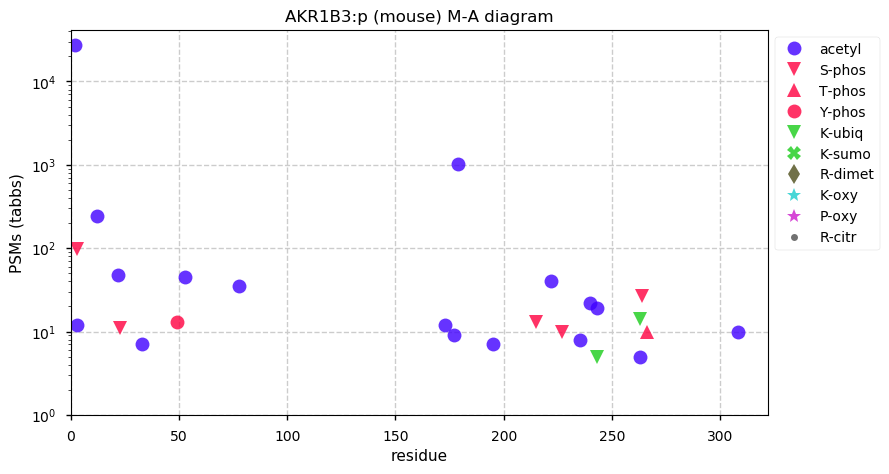
Mon Jul 13 19:32:23 +0000 2020🔗
Mon Jul 13 14:16:23 +0000 2020One of the small pleasures I take from science is its on-going validation of the principle that most plausible, well-reasoned, logical arguments are wrong.
Mon Jul 13 12:38:27 +0000 2020Reduces specific ketones in a variety of steroid hormones (e.g., 4-androstenedione & androsterone) and xenobiotics. In humans, AKR1C6 is a pseudogene with 11 exons at 10p15.1. Human AKR1C4 is the functional ortholog of mouse AKR1C6.
Mon Jul 13 12:38:27 +0000 2020Shares different subsets of observable tryptic peptides with Akr1cl:p, Akr1c14:p, Akr1c18:p, Akr1c19:p, Akr1c20:p, Akr1c21:p and Akr1d1:p.
Mon Jul 13 12:38:27 +0000 2020Akr1c6:p, aldo-keto reductase family 1, member C6 (M. musculus) 🔗 Small cytoplasmic enzyme; PTMs: significant K+acetylation and S+phosphoryl; SAAVs: none; common in liver tissue; mature form: 1-323 [2,933 x, 146 kTa] 🔗
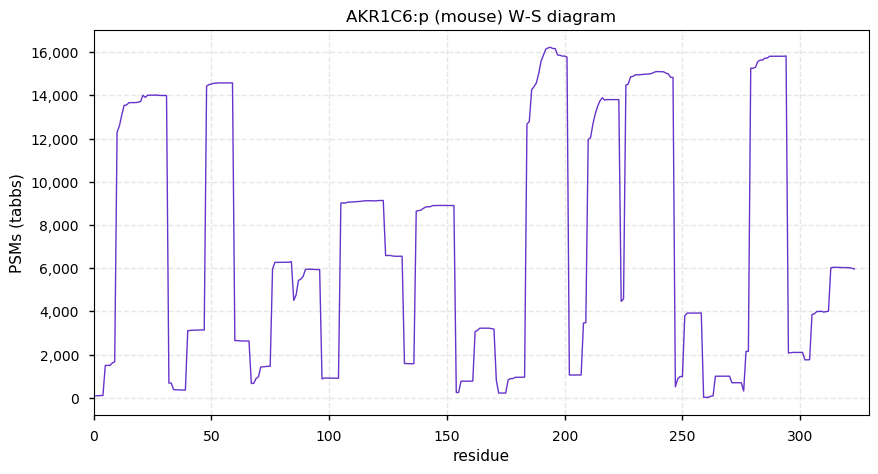
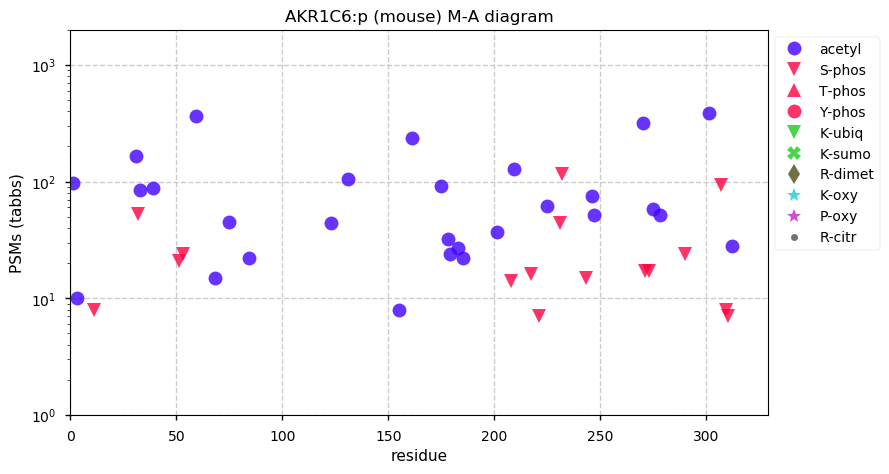
Sun Jul 12 18:34:55 +0000 2020long-winded-informatics-phrase-to-english dictionary entry:
"model based robust summarization strategy which models the log-transformed intensities directly through robust regression"
intransitive verb
:to make a best guess
Sun Jul 12 13:59:45 +0000 2020Catalyzes the reduction of an aldehyde to a primary alcohol for many substrates. Carries out an interim step in the synthesis of the ascorbic acid.
Sun Jul 12 13:22:03 +0000 2020A.K.A: aldehyde reductase, aldehyde reductase II, D-glucuronate dehydrogenase , D-glucuronate reductase, D-glucuronic reductase, hexonate dehydrogenase, L-glucuronate reductase, L-gulonate NAD-3-oxidoreductase, L-hexonate:NADP dehydrogenase & more ...
Sun Jul 12 13:22:03 +0000 2020Akr1a1:p, aldo-keto reductase family 1, member A1 (aldehyde reductase) (M. musculus) 🔗 Small cytoplasmic enzyme; PTMs: significant K+acetylation; SAAVs: none; common in many tissues and cell lines; mature form: (2,3)-325 [14,879 x, 228 kTa] 🔗
Sat Jul 11 14:36:18 +0000 2020@neely615 @pwilmarth Practically, no. The time for that was about 15 years ago. As things have developed, most published data sets are the results of one-off designs that don't suit themselves to comparison with "standard" experiments.
Sat Jul 11 13:47:36 +0000 2020The mechanism and result of losing this gene, leading to a dependence on dietary ascorbic acid (Vitamin C), has been the subject of considerable study and speculation (some of it pretty wacky).
Sat Jul 11 13:47:35 +0000 2020GULO:p is the last enzyme in the ascorbic acid synthesis pathway. Most vertebrates have a functional copy of this gene, but some primates (including humans), bats and guinea pigs are missing this enzyme. Humans have a non-functional GULO unitary pseudogene (chr 8p21).
Sat Jul 11 13:47:35 +0000 2020GULO:p, gulonolactone (L-) oxidase (M. musculus) 🔗 Small cytoplasmic enzyme; PTMs: 1 phosphodomain & significant K+acetyl; SAAVs: none; most abundant in liver, placenta & retina; mature form: (1,2)-440 [2,117 x, 66.1 kTa] 🔗
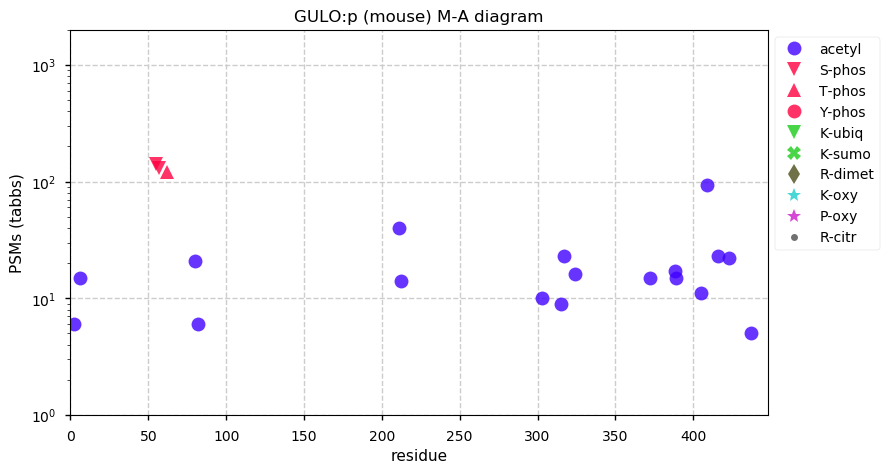
Fri Jul 10 17:35:58 +0000 2020I guess I should have asked "As a reviewer, do you ALWAYS question the enrichment-level ...".
Fri Jul 10 14:27:41 +0000 2020@HLAna_Marcu This is a promising start. Hopefully there are plans for this project to continue & broaden its coverage of MHC peptides. Good work.😃
Fri Jul 10 14:15:23 +0000 2020Also an argument for mandatory retirement.
Fri Jul 10 14:14:24 +0000 2020The NIH's own Statler and Waldorf (without the humor, stagecraft or timing). Pretty much a case study of why (very) senior bureaucrats should not have their own TV shows.
🔗
Fri Jul 10 11:42:03 +0000 2020ANAPC16:p, anaphase promoting complex subunit 16 (Homo sapiens) 🔗 Small intracellular subunit; PTMs: N- and C-terminal phosphodomains; SAAVs: none; low complexity N-terminal domain; mature form: 2-110 [3,432 x, 8.0 kTa] 🔗
Thu Jul 09 17:50:59 +0000 2020@wormmaps None of those things are bioinformatics.
Thu Jul 09 17:30:29 +0000 2020As a reviewer, do you ever question the enrichment-level obtained in affinity enrichment proteomics experiments?
Thu Jul 09 17:27:50 +0000 2020The published study doesn't mention anything about the level of enrichment, but does go to some lengths to analyze the pY levels detected in these cell culture samples, including some nifty looking, multi-panel graphics.
Thu Jul 09 17:26:08 +0000 2020I'm looking at a data set meant to monitor Y+phosphoryl PTM changes caused by a drug using an anti-pY antibody affinity enrichment. Normally, antibody-based pY studies end up with 80-90% of the PSMs having the modification. This study has 2% with the PTM.
Thu Jul 09 16:38:26 +0000 2020@neely615 @AOri_lab @MattWFoster @KentsisResearch @BiswapriyaMisra @Sci_j_my @AlexHgO I don't remember if you mentioned it, but what type of bat are you examining?
Thu Jul 09 15:28:58 +0000 2020@AJ_Brenes In humans, TCP11L2:p produces the most PSMs in the CPTAC LUAD study (human lung adenocarcinoma), triple negative breast cancer studies (but not in CPTAC breast cancer studies) & lymphocytes.
Thu Jul 09 15:02:16 +0000 2020@ypriverol As per our Privacy Statement:
"we do not store or track any information about your use of the material available on our sites, other than information required to ensure the overall security of the site."
Thu Jul 09 14:46:48 +0000 2020@ypriverol No. I don't keep track of use.
Thu Jul 09 13:45:43 +0000 2020ANAPC15:p, anaphase promoting complex subunit 15 (Homo sapiens) 🔗 Small intracellular subunit; PTMs: N-terminal acetylation; SAAVs: none; the C-terminal half of the sequence is an acidic, low complexity domain; mature form: 2-121 [696 x, 1.4 kTa] 🔗
Wed Jul 08 21:10:36 +0000 2020@scalzi We are at a peculiar pass in human relations when @axlrose has much more nuanced views on public health issues than the US VP.
Wed Jul 08 19:38:41 +0000 2020@dtabb73 @SuMBHG Nice. I really like the reversal of the classroom/conference style of having an enormous screen & a comparatively tiny human.
Wed Jul 08 17:48:16 +0000 2020@TrostLab @BrenesAlejandro @YKulathu It actually works just as well for programmed search algorithms. Looking for the assignment of impossible PTMs is my go to results-spreadsheet-quality-control metric.
Wed Jul 08 16:04:28 +0000 2020It will be interesting to see how many people are willing to pay to attend HUPO's 2020 online event.
Wed Jul 08 14:46:19 +0000 2020@slashdot Well put. Creating more difficult-to-distinguish ports with a bewildering range of options tends to be towards the end of a technology's natural development cycle.
Wed Jul 08 14:12:21 +0000 2020@TrostLab @BrenesAlejandro @YKulathu A good way to quickly assess papers describing new "open" search algorithms is the check how often they assign K or R post-translational modifications of this type to the C-terminal residue.
Wed Jul 08 14:09:08 +0000 2020@TrostLab @BrenesAlejandro @YKulathu A good study will usually end up with ~75% enrichment of K+GG. The main difference is that since the K+GG cannot be at the C-terminus of the peptide, there must be at least 1 "missed" cleavage. The same as K+acetyl (really K+anything) or R+methyl/dimethyl.
Wed Jul 08 12:51:20 +0000 2020ANAPC13:p, anaphase promoting complex subunit 13 (Homo sapiens) 🔗 Very small intracellular subunit; PTMs: Y26+phosphoryl; SAAVs: none; observed most often in cell lines; mature form: 1-74 [1,082 x, 2.6 kTa] 🔗
Tue Jul 07 18:19:00 +0000 2020I should note that because of the sample preparation used, there is a significant amount of cysteine SH acrylamide derivatization as well as IAA blocking.
Tue Jul 07 18:14:26 +0000 2020If you are interested in Mycobacterium smegmatis (a relatively easy to grow, non-pathogenic model species that can stand in for much nastier mycobacteria), PXD017602 has a particularly good sampling of the proteome. Published as part of 🔗
Tue Jul 07 17:07:30 +0000 2020Totally pointless statistic: GPMDB added 217 million PSMs in the last 30 days (0.217 gTa). Thanks to ProteomeXchange, JPOST, Massive and iPROX (& the folks that actually did the experiments)!
Tue Jul 07 15:05:43 +0000 2020@TrostLab @BrenesAlejandro @YKulathu Without enrichment, K+GG is almost never observed on other proteins.
Tue Jul 07 15:04:07 +0000 2020@TrostLab @BrenesAlejandro @YKulathu The number of K+GG modified ubiquitin tryptic peptides can be useful as a rough measure of protein turnover rate.
Tue Jul 07 14:54:33 +0000 2020@TrostLab @BrenesAlejandro @YKulathu An additional complication is that both UBB and UBC are polyproteins with multiple copies of ubiquitin that are cleaved out after translation, the number of copies depending on the species.
The most common PTM on ubiquitin (by far) is ubiquitination.
Tue Jul 07 12:44:01 +0000 2020CDC26:p, cell division cycle 26 (Homo sapiens) 🔗 Very small intracellular subunit; aka ANAPC12; PTMs: significant phosphorylation; SAAVs: none; commonly observed in cell lines and cancer tissue; mature form: 1-85 [4,444 x, 11.2 kTa] 🔗
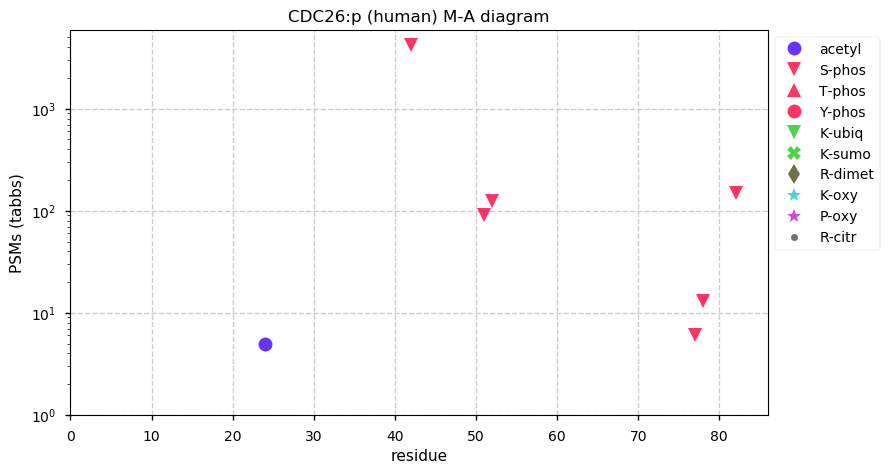
Mon Jul 06 15:34:03 +0000 2020@KislingerThomas IMHO amino acid variants caused by SNVs are the most under-appreciated aspect of proteomics measurements.
Mon Jul 06 15:27:36 +0000 2020@KislingerThomas The paper also has a good discussion of the requirements for the use of type of clinical marker. Many "clinical" proteomics papers tend to be quite naive about these requirements: clinical measurements have to be unambiguous, the less subtle the better.
Mon Jul 06 14:13:22 +0000 2020@KislingerThomas It is an easy to observe variant 🔗 Great to see it being used in a clinically relavent context.
Mon Jul 06 12:53:15 +0000 2020For anyone interested in the variability of ANAPC proteins across different taxa, 🔗 has a pretty good summary of knowledge on the subject.
Mon Jul 06 12:20:40 +0000 2020ANAPC11:p, anaphase promoting complex subunit 11 (Homo sapiens) 🔗 Very small intracellular subunit; PTMs: none; SAAVs: none; only observed in high sensitivity experiments; mature form: 1-84 [481 x, 1 kTa] 🔗
Sun Jul 05 17:17:12 +0000 2020@slashdot Better late than never.
Sun Jul 05 13:04:14 +0000 2020APC9 (ANAPC9) is only present in fungi. There is no homologous protein in other eukaryotes.
Sun Jul 05 13:04:14 +0000 2020ANAPC10:p, anaphase promoting complex subunit 10 (Homo sapiens) 🔗 Small intracellular subunit; PTMs: N-terminal phosphodomain and 4 K+ubiquitinyl sites; SAAVs: none; abundant in cell lines and cancer tissue; mature form: 2-185 [2,643 x, 6.2 kTa] 🔗
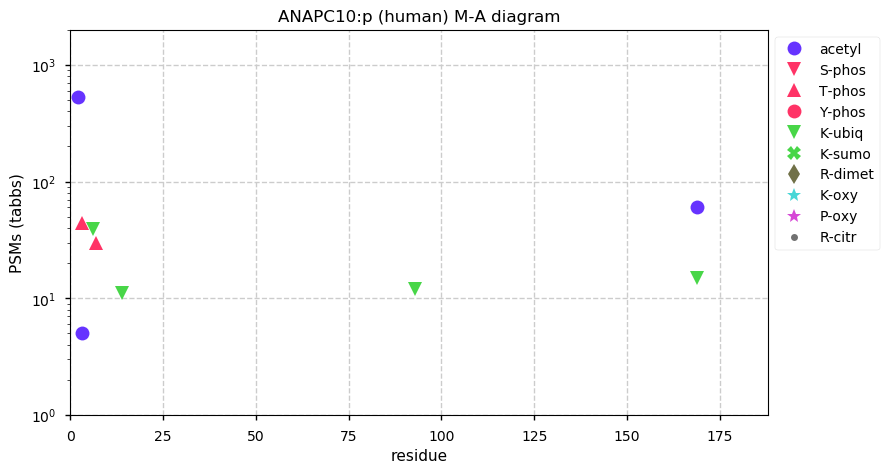
Sat Jul 04 20:54:54 +0000 2020@Freddyomics The PSMs are from samples prepared with Lys-C + trypsin.
Sat Jul 04 17:33:35 +0000 2020Actually, 2 replicates, A & B, no C.
Sat Jul 04 16:11:04 +0000 2020Calculated based on several large sets of PSMs from human cell line experiments.
Sat Jul 04 16:10:05 +0000 2020Fraction (in percent) of observable human tryptic peptides that contain at least 1 of the listed AA residues:
Res:%
A:63
C:18
D:63
E:66
F:37
G:60
H:35
I:49
K:56
L:72
M:28
N:44
P:52
Q:48
R:54
S:60
T:53
V:62
W:10
Y:31
Sat Jul 04 15:52:42 +0000 202012 patients, 3 replicates each, labelled with a patient number (1-12) and a replicate letter (A-C), but no where is there an indication of which are male or female.
Sat Jul 04 15:49:45 +0000 2020Perhaps I am being difficult, but was it impossible to label the data files associated with PXD015979 as being either "M" or "F", given that the title of the study is "Proteomics pinpoints alterations in grade I meningiomas of male versus female patients"?
Sat Jul 04 13:27:39 +0000 2020CDC23:p, cell division cycle 23 (Homo sapiens) 🔗 Midsized intracellular subunit; aka ANAPC8; PTMs: 2 phosphodomains and 22 K+ubiquitinyl sites; SAAVs: none; abundant in cell lines and cancer tissue; mature form: 2-599 [13,348 x, 53.3 kTa] 🔗
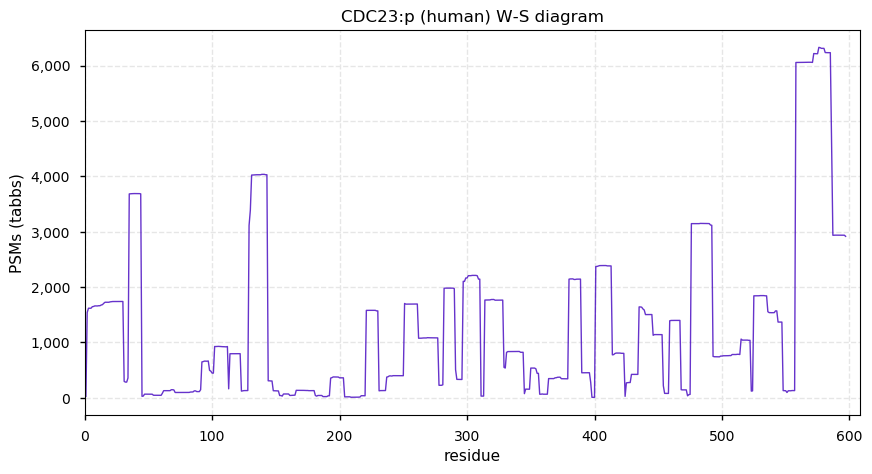
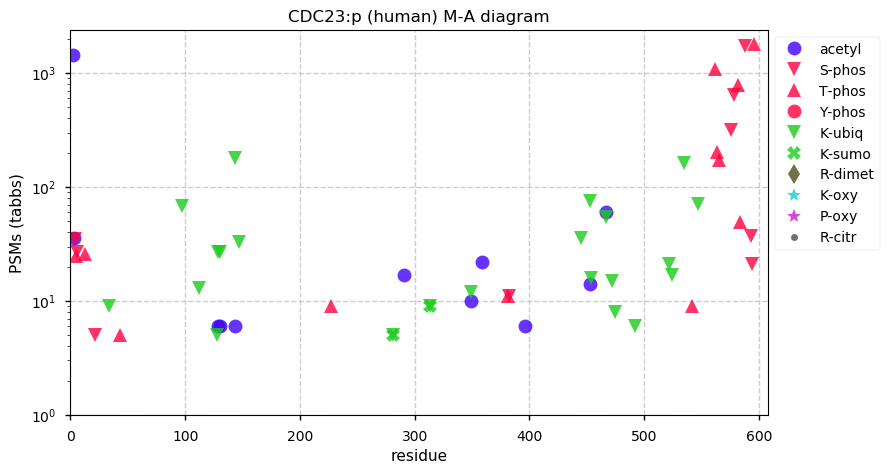
Sat Jul 04 13:26:35 +0000 2020@lucillabis @DuarteGouveia13 From a purely technical point of view, the best SARS-CoV-2 data set of infected calls, so far.
Sat Jul 04 02:44:28 +0000 2020@educhicano It is a GMP requirement (i.e., everywhere downstream of discovery in pharma). Not commonly used in academic labs, but if you want to do more serious QA, it is pretty much mandatory.
Fri Jul 03 16:42:57 +0000 2020@MattWFoster I'm not trying to suggest that screwing up your experiments is a great way to get into Nature - but it clearly doesn't hurt your chances ...
Fri Jul 03 16:09:11 +0000 2020@MattWFoster Another great example of both runaway amine carbamylation & carboxyamidomethylation is the data from Nature. 2014 May 29;509(7502):575-81 (🔗)
Fri Jul 03 15:56:53 +0000 2020@MattWFoster Although it would seem that the authors either failed to realize this was an issue with their experiments or neglected to mention it in their writeup.
Fri Jul 03 15:04:17 +0000 2020@pwilmarth @VATVSLPR Many groupsuse urea without detectable carbamylation, but as you say, they have to be careful about it and check their data to make sure that it isn't creeping back in. The same is true for off-target IAA derivatizations, non-tryptic cleavage, etc.
Fri Jul 03 14:46:16 +0000 2020@MattWFoster Even very well known groups have problems with that side reaction from time to time. The data from Nature. 2020 Jun;582(7813):592-596 (🔗) has ~15% of PSMs with that artifact.
Fri Jul 03 14:35:40 +0000 2020I guess many years of papers that only checked for 1 missed cleavage and methionine oxidation have educated students with the belief that peptides are stable compounds.
Fri Jul 03 14:33:06 +0000 2020My original intent behind the question was to elicit mods that had been initially identified through open search, but it sounds like many practitioners are unaware of common artifacts. 🔗
Fri Jul 03 14:14:52 +0000 2020@JesseBrown It will become the benchmark for "unforced errors" by a politician.
Fri Jul 03 13:35:55 +0000 2020Proteomics evidence suggests that translation initiates at M35 rather than M1.
Fri Jul 03 13:35:55 +0000 2020ANAPC7:p, anaphase promoting complex subunit 7 (Homo sapiens) 🔗 Midsized intracellular subunit, PTMs: 1 major phosphodomain and 23 K+ubiquitinyl sites; SAAVs: S33N (1%); abundant in cell lines and cancer tissue; mature form: (35,36)-599 [14,666 x, 64.5 kTa] 🔗
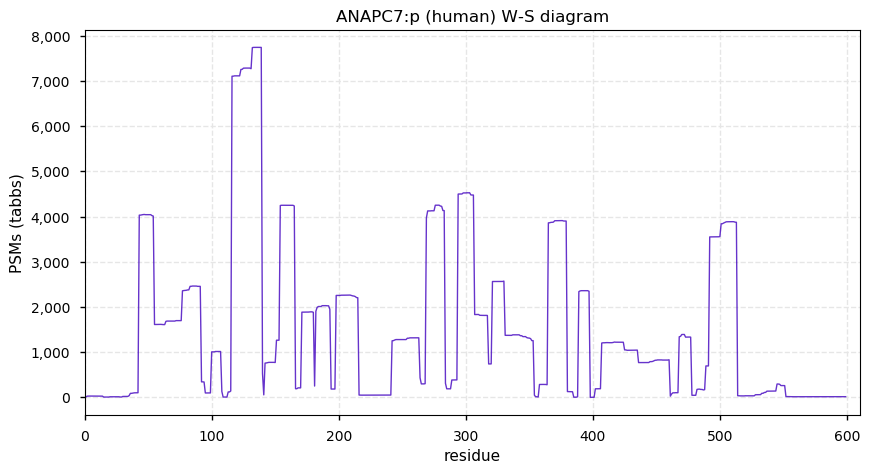
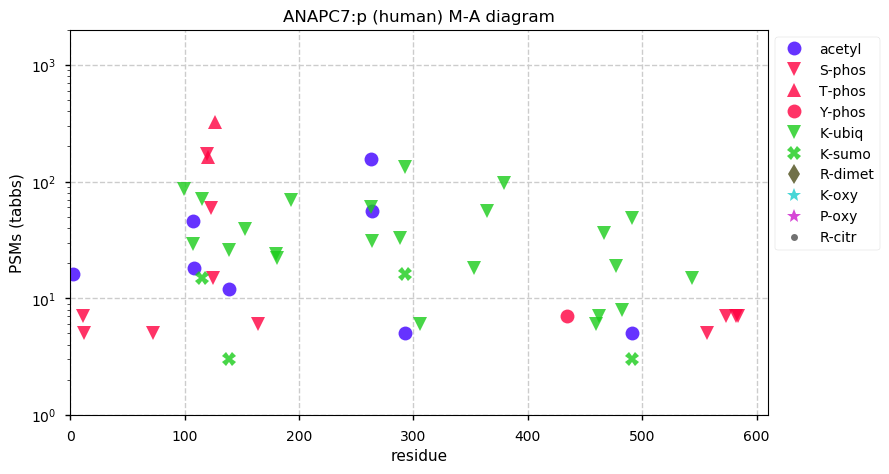
Thu Jul 02 17:15:25 +0000 2020@AJ_Brenes amen, brother, amen.
Thu Jul 02 14:49:38 +0000 2020Mine is the substitution of succinyl for TMT at amines on a peptide's N-terminus or lysine sidechain when using TMT derivatives in relative quantitation experiments.
Thu Jul 02 14:46:04 +0000 2020"Open" search strategies have been available to proteomics groups for more than a decade. What is your favorite PTM or experimental artifact that has been discovered by these methods?
Thu Jul 02 14:15:45 +0000 2020CDC16:p (ANAPC6:p), like CDC27:p (ANAPC3:p), retain their older "Cell Division Cycle" nomenclature rather than more function-based names because of long use in the literature. HGNC committees are rather inconsistent in their retention policies regarding trad. gene names.
Thu Jul 02 12:36:37 +0000 2020CDC16:p, cell division cycle 16 (Homo sapiens) 🔗 Midsized intracellular subunit; aka ANAPC6:p; PTMs: 1 N-terminal phosphodomain, 100-215 has multiple ubiquitin and SUMO transport tags; SAAVs: none; mature form: (1,2)-620 [10,822x, 45.4 kTa] 🔗
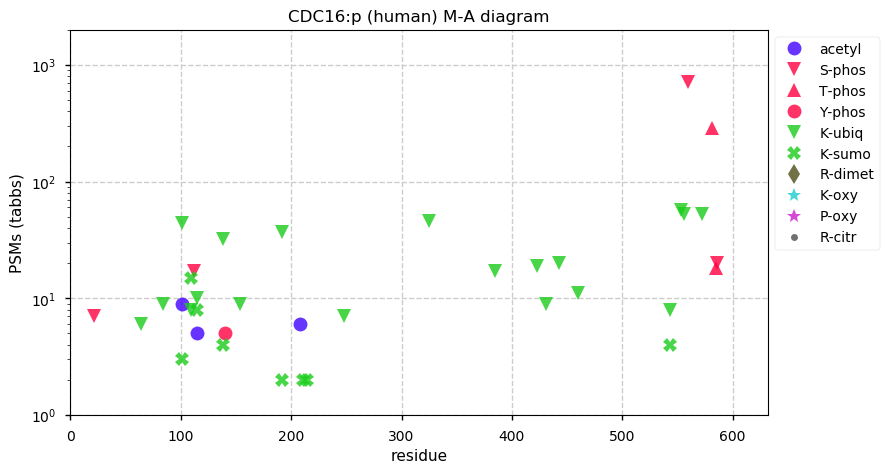
Wed Jul 01 17:50:52 +0000 2020And for anyone still in the "maybe it will vanish when it gets hot" camp, daytime temps are between 90-100 °F in Iran right now.
Wed Jul 01 16:51:56 +0000 2020@Smith_Chem_Wisc @dtabb73 Makes me happy I switched to using RPi4's.
Wed Jul 01 12:24:32 +0000 2020ANAPC5:p, anaphase promoting complex subunit 5 (Homo sapiens) 🔗 Midsized intracellular subunit, PTMs: 3 phosphodomain and 22 K+ubiquitinyl sites; SAAVs: none; abundant in cell lines and cancer tissue; mature form: 2-755 [9,776 x, 39.5 kTa] 🔗
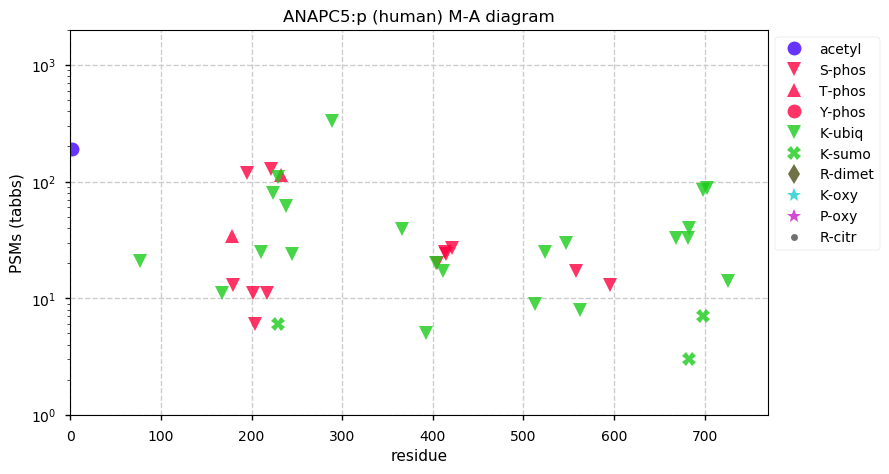
Wed Jul 01 12:22:33 +0000 2020Iran seems to be well in to its 2nd phase, that has lagged the initial phase by ~100 days. 🔗
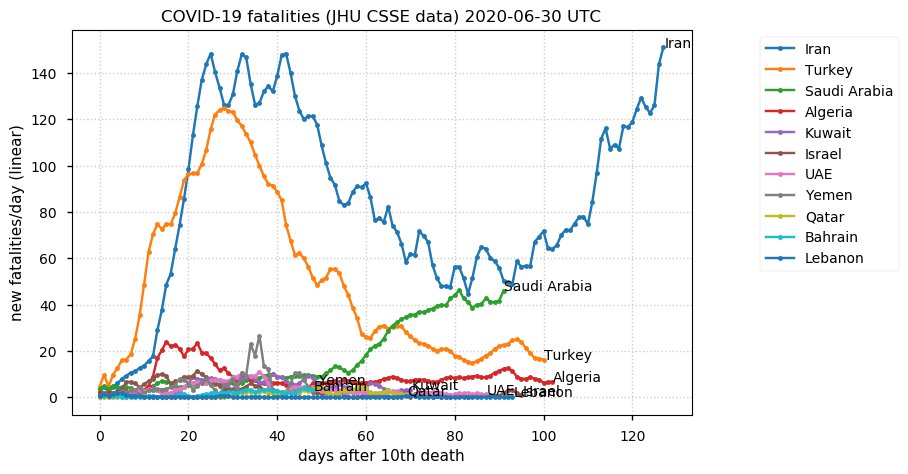
Tue Jun 30 14:40:16 +0000 2020For some reason, in 'omics studies "more" seems to be the relentless enemy of "better".
Tue Jun 30 14:07:08 +0000 2020What has changed so that it is now OK to publish a paper containing very little other than the fact that you have strung together a few very ordinary, one-off MDC runs of common cell lines?
Tue Jun 30 11:56:00 +0000 2020ANAPC4:p, anaphase promoting complex subunit 4 (Homo sapiens) 🔗 Midsized intracellular subunit, PTMs: 1 sig. phosphodomain and 7 K+ubiquitinyl sites; SAAVs: R465Q (7%); mature form: 1-808 [9,607 x, 39.3 kTa] 🔗
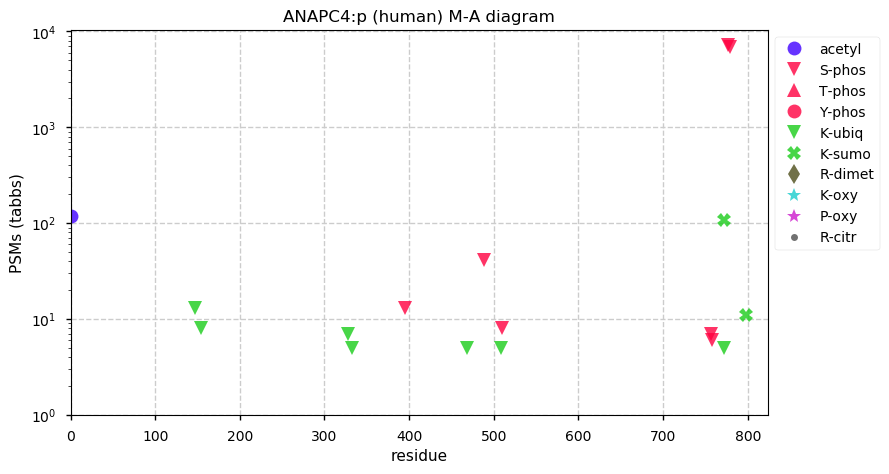
Mon Jun 29 17:46:25 +0000 2020After examining the data and re-reading the paper, it turns out this data isn't archival quality, so I don't need the file index after all.
Mon Jun 29 17:45:21 +0000 2020If you have any interest in the phosphorylation found in Toxoplasma gondii infected human cells, PXD019729 has the best public data available.
Mon Jun 29 15:11:28 +0000 2020Does anybody know if there is a master list for 🔗 (PXD019645) that describes the association of RAW files with their corresponding experiment?
Mon Jun 29 13:17:35 +0000 2020South Africa is about 1000/day less than Florida right now, but it is dominating African numbers 🔗
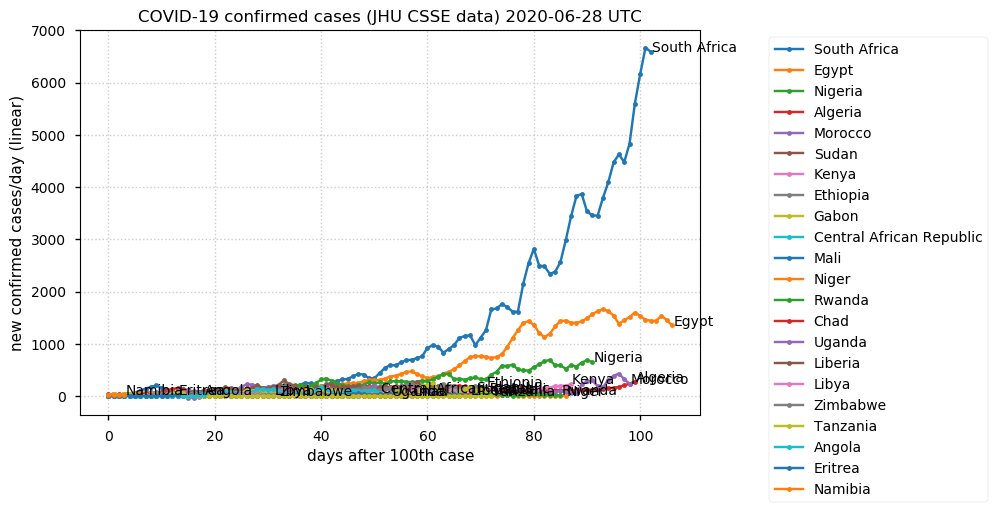
Mon Jun 29 12:57:41 +0000 2020If the story of regulating protein concentrations is told, it is a confusing narrative involving thousands of isolated processes with impossible-to-remember names, oddly stretched engineering analogies and "garbage collection" as the persistent theme.
Mon Jun 29 12:45:36 +0000 2020The story of building nascient polypeptides from nucleotides is always told in a simple, linear narrative with several heroic (but flawed) characters resolutely solving mysteries & cleverly breaking codes.
Mon Jun 29 12:16:53 +0000 2020CDC27:p, cell division cycle 27 (Homo sapiens) 🔗 Midsized intracellular subunit, aka ANAPC3; PTMs: 4 sig. phosphodomains and 20 K+ubiquitinyl sites; SAAVs: none; abundant in cell lines and cancer tissue; mature form: 2-824 [12,552 x, 53.2 kTa] 🔗
Sun Jun 28 12:44:27 +0000 2020I think one of the reasons that people believe protein concentration to be mainly dependent on RNA concentration is that everyone is taught about translation, but the lysosome & proteasome mechanisms are rarely-taught specialist topics.
Sun Jun 28 12:37:42 +0000 2020ANAPC2:p, anaphase promoting complex subunit 2 (Homo sapiens) 🔗 Midsized intracellular subunit; PTMs: significant S,T+phosphoryl and 7 K+ubiquitinyl sites; SAAVs: none; most abundant in cell lines and cancer tissue; mature form: 2-822 [6,732 x, 25.6 kTa] 🔗
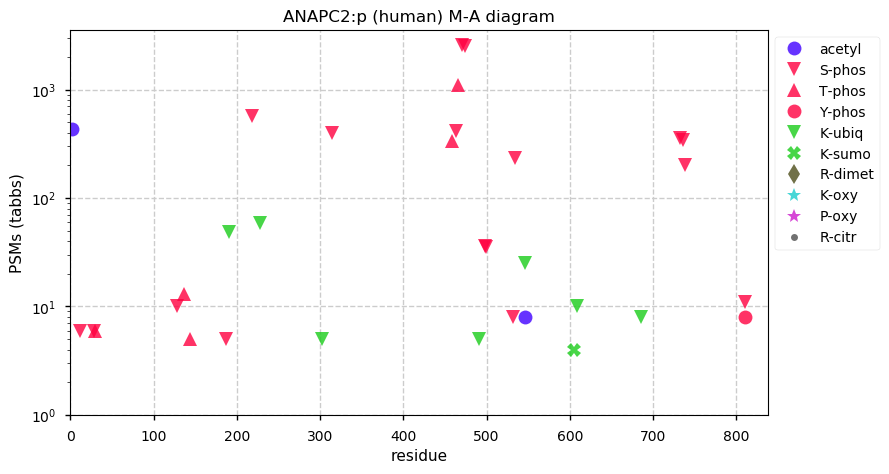
Sat Jun 27 15:19:43 +0000 2020@molcellprot @kusterlab Déjà vu, all over again ...
Sat Jun 27 12:22:14 +0000 2020The anaphase promoting complex (APC) is an E3 ligase protein with ~16 subunits in humans. It regulates the entry of a cell into anaphase by aggressively ubiquitinating a set of proteins simultaneously, leading to their rapid depletion via proteasome digestion.
Sat Jun 27 12:22:14 +0000 2020ANAPC1:p, anaphase promoting complex subunit 1 (Homo sapiens) 🔗 Large intracellular subunit; PTMs: 4 major phospho-domains & 28 K+ubiquitin sites; SAAVs: none; most abundant in cell lines; mature form: 2-1944 [15,853 x, 76.0 kTa] 🔗
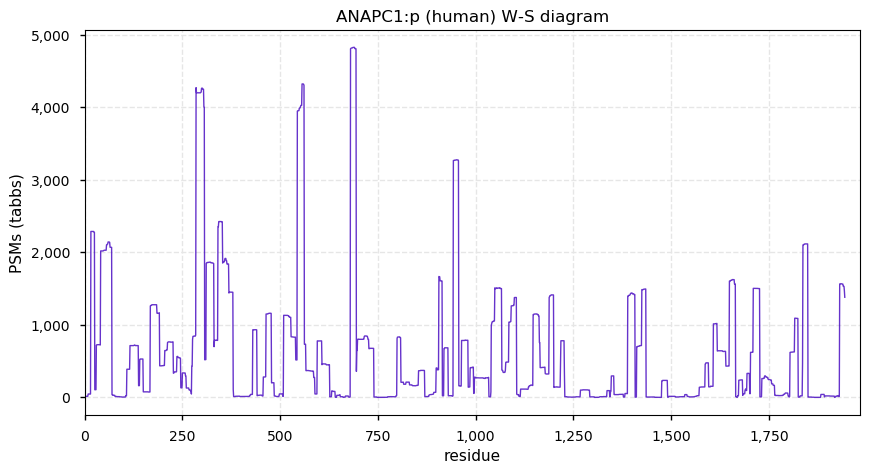
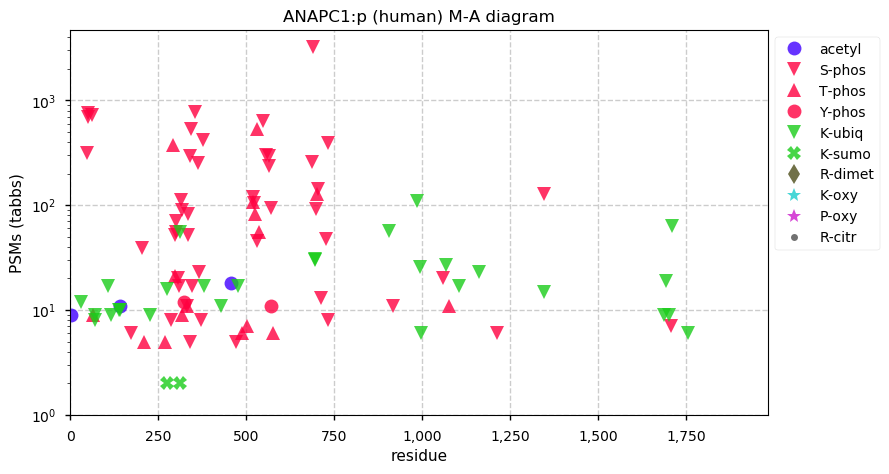
Fri Jun 26 21:32:48 +0000 2020@AlexUsherHESA Oui!
Fri Jun 26 21:21:51 +0000 2020Best version of "All Tomorrow's Parties"
Fri Jun 26 21:17:22 +0000 2020A lot of bending-over-backwards to defend a decision with terrible optics and very little chance of success
🔗
Fri Jun 26 20:53:12 +0000 2020@neely615 @MattWFoster @AOri_lab @KentsisResearch @BiswapriyaMisra @Sci_j_my @AlexHgO OK. Once it is on PX, I will, with permission, perspicaciously peruse the panoply of pinniped plasma proteasomal proteins produced.
Fri Jun 26 19:37:49 +0000 2020@neely615 @AOri_lab @MattWFoster @KentsisResearch @BiswapriyaMisra @Sci_j_my @AlexHgO P28062 (PSMB8, LMP7); P28065 (PSMB9, LMP2); Q06323(PSME1); and Q9UL46 (PSME2) are subunits of the immunoproteasome, which is an alternate assembly of the 19S proteasome regulatory particle. You don't seem to have any of the 20S core particle proteins (PSMCx & PSMDx).
Fri Jun 26 19:24:12 +0000 2020@slavovLab It may help, but the underlying problem is weak editors who routinely pass the buck on making decisions to reviewers.
Fri Jun 26 16:44:17 +0000 2020@slavovLab Peer review is (and always was) a crapshoot. Sometimes it is beneficial, usually it is just a cross to bear.
Fri Jun 26 14:50:39 +0000 2020@scottagerber If you like bubs, you are probably going to like the next few of these, focusing on the APC's
🔗
Fri Jun 26 12:32:15 +0000 2020Ten of twelve ubiquitinylation K-acceptors observed to be acetylation acceptors as well. Three of those sites also observed to be SUMOylation acceptors.
Fri Jun 26 12:32:15 +0000 2020BUB3:p, mitotic checkpoint serine/threonine kinase 3 (Homo sapiens) 🔗 Small intracellular protein; many PTMs; SAAVs: none; more frequently observed than BUB1 or BUB1B; mature form: 2-328 [37,657 x, 257 kTa] 🔗
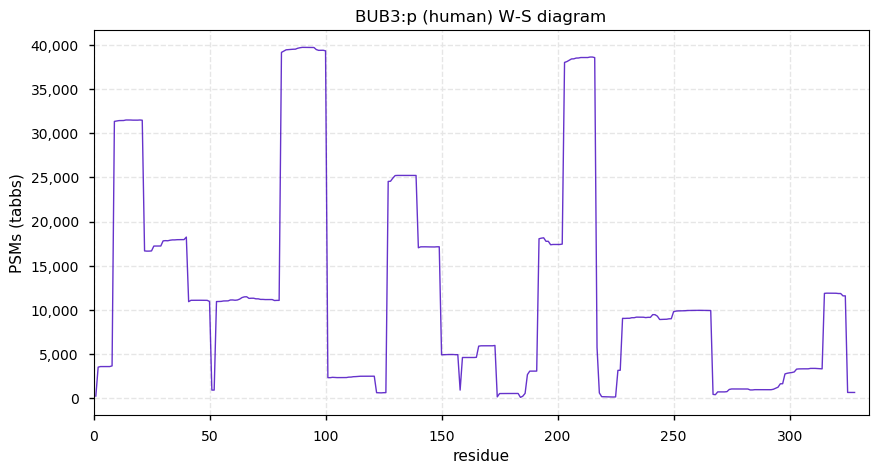
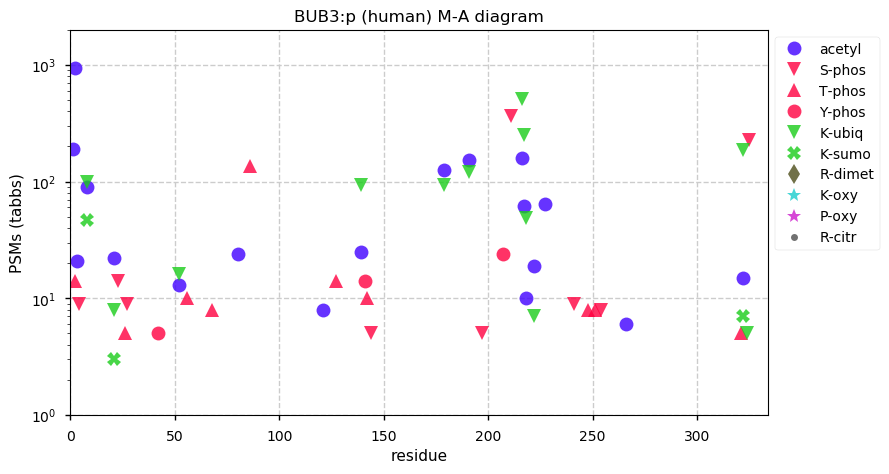
Thu Jun 25 15:59:02 +0000 2020It would appear that the proposed hypothesis of an anti-correlation between hot weather and new cases has not been confirmed experimentally. 🔗
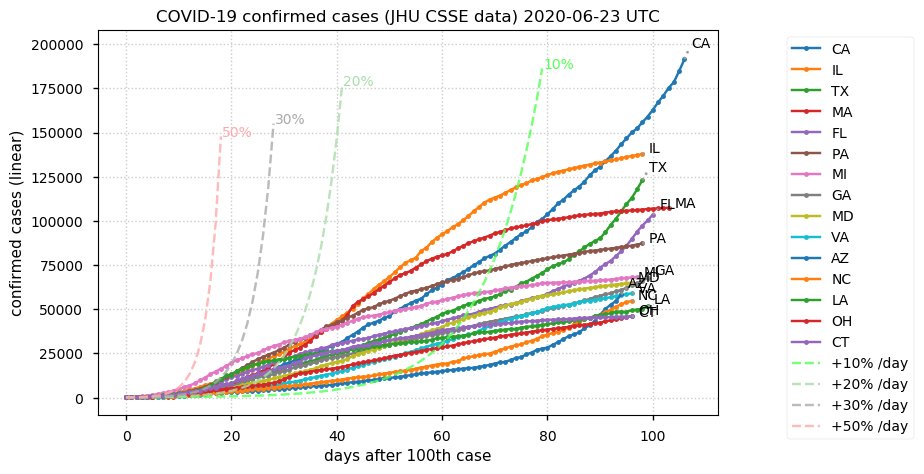
Thu Jun 25 13:04:19 +0000 2020No correlation between acetylation and ubiquitinylation K-acceptor site distributions.
Thu Jun 25 13:04:19 +0000 2020BUB1B:p, mitotic checkpoint serine/threonine kinase 1B (Homo sapiens) 🔗 Large intracellular protein; many PTMs; SAAVs: T40M (2%), R349Q (37%), E390D (4%), R550Q (1%), V618A (9%); mature form: 1-1050 [8,628 x, 37.6 kTa] 🔗
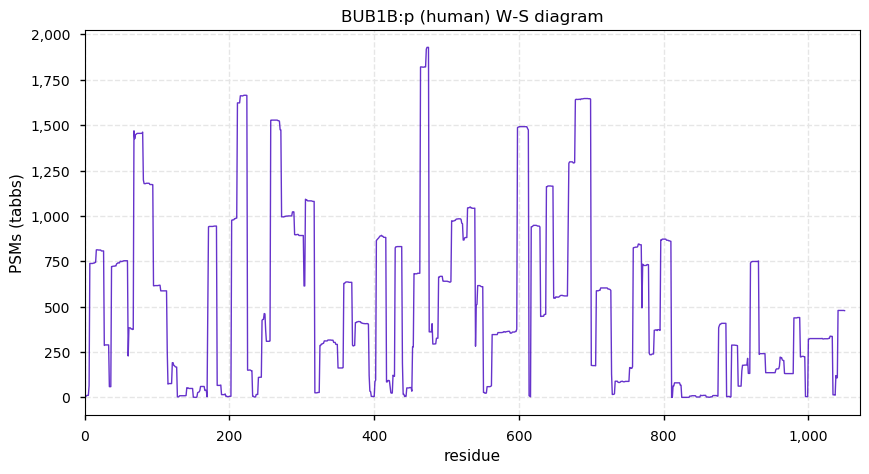
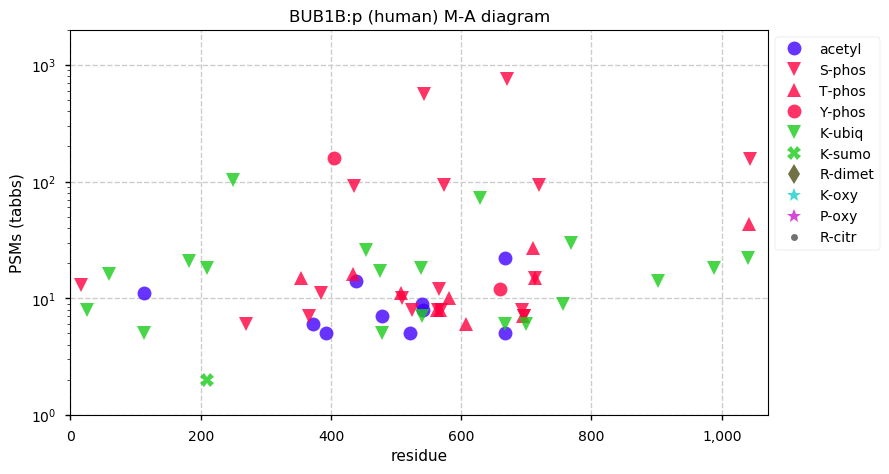
Wed Jun 24 22:34:38 +0000 2020@chrashwood Sounds like something Millenium would have done.
Wed Jun 24 17:50:39 +0000 2020@pitman_mark Everything I do requires SQL: the scripting language I wrap it with is largely a matter of task-specific convenience.
Wed Jun 24 17:03:22 +0000 2020@aprilfrommer @CodeWisdom I had unconsciously been using WTFs/min in examining information derived from data for as long as I can remember. It is particularly useful in the protein-protein interaction field, but any large scale protein/RNA/DNA study can be characterized effectively with this metric.
Wed Jun 24 15:28:25 +0000 2020@neely615 @MattWFoster @KentsisResearch @BiswapriyaMisra @Sci_j_my @AlexHgO The argument doesn't square with the data from plasma EVs (PXD001194). In that data, both 20 S hydrolytic subunits (🔗) and 19 S regulatory subunits (🔗) are well represented.
Wed Jun 24 13:23:54 +0000 2020@AlexUsherHESA In the other countries, are all universities so thoroughly integrated with provincial/state governments?
Wed Jun 24 12:33:22 +0000 2020More commonly observed in cell lines than in tissue samples, including cancer tissue.
Wed Jun 24 12:33:22 +0000 2020BUB1:p, mitotic checkpoint serine/threonine kinase (H. sapiens) 🔗 Large nuclear protein; many PTMs, no acetyl associated with ubiquitinyl sites; SAAVs: none; ; mature form: 1-1085 [6,609 x, 25.9 kTa] 🔗
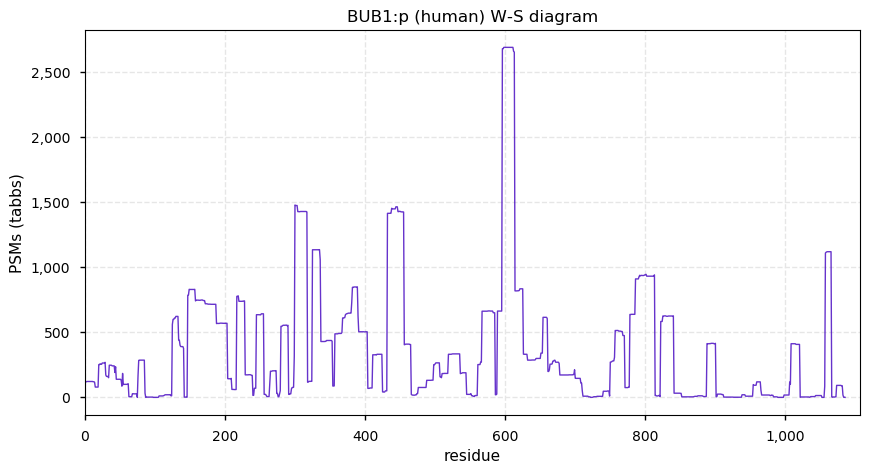
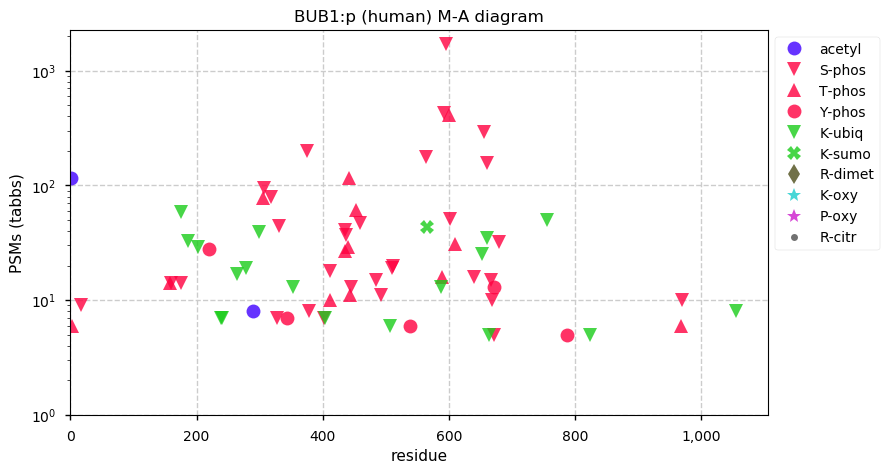
Tue Jun 23 18:26:59 +0000 2020@legalminimum @KLamrock @AlexUsherHESA Canadians are a deeply flawed people:
🔗
Tue Jun 23 17:52:05 +0000 2020@VATVSLPR @Sci_j_my Sorry: "Lora" not "Loro".
Tue Jun 23 17:48:27 +0000 2020@VATVSLPR @Sci_j_my It isn't a bad article, however they haven't seemed to convince the compositors of the Nature Index article: it uses the Google font "Loro" for some reason.
Tue Jun 23 16:37:59 +0000 2020@Sci_j_my At least they ended the article desperately spinning how wonderful a contribution their own font was to the scientific endeavor, with its "British formality and wit".
Tue Jun 23 15:05:00 +0000 2020Those UC Davis guys and their "analytical chemistry" are always spoiling things for others:
🔗
Tue Jun 23 14:42:05 +0000 2020Does anybody know of any good explanations as to why the K+ubiquitinyl sites in some proteins may also be acetylated, while they are not acetylated at all in other proteins?
Tue Jun 23 12:30:03 +0000 2020Commonly observed in urine extracellular vesicle/exosome studies as well as studies of ovarian and breast cancer tissue.
Tue Jun 23 12:30:03 +0000 2020SNCG:p, synuclein gamma (H. sapiens) 🔗 Small intracellular protein; PTMs: many phosphoryl acceptors (1×Y, 9×S, 2×T); SAAVs: E110V (20%); different tissue distribution than SNCA:p or SNCB:p; mature form: 1-127 [10,168 x, 76.2 kTa] 🔗
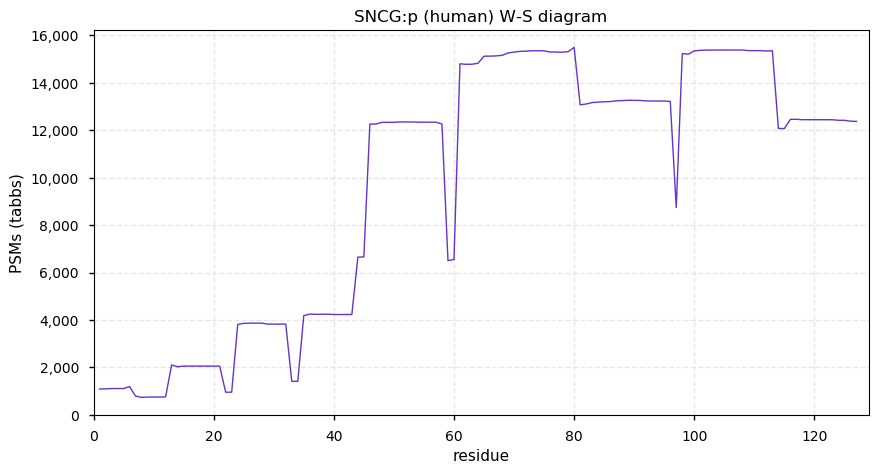
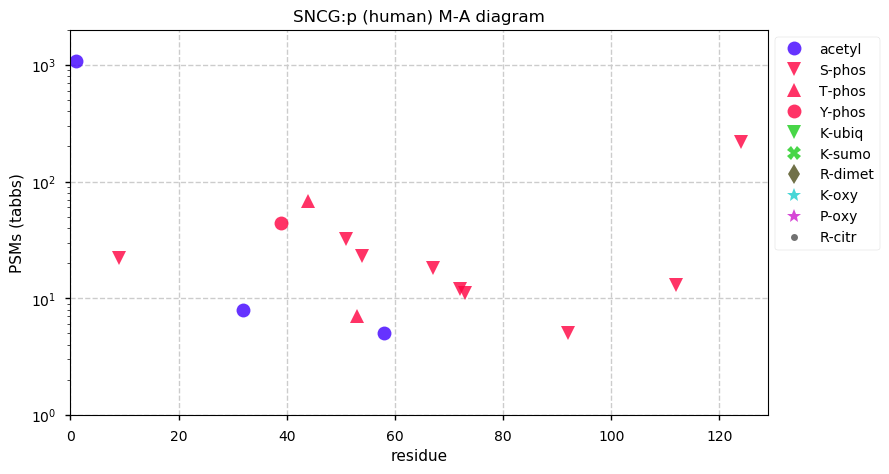
Tue Jun 23 00:08:08 +0000 2020@elspuddo @piefuchs I'm sticking with my answer. Positioning the plate under the laser was it's use. But it was for amusing Marvin.
Mon Jun 22 23:50:37 +0000 2020It was always a little weird when I was there, but it seems to be kind of metastable now
🔗
Mon Jun 22 21:33:39 +0000 2020@piefuchs Marvin's amusement?
Mon Jun 22 15:33:43 +0000 2020@astacus @edemmott ADH & Onions
Mon Jun 22 15:14:07 +0000 2020Nothobranchius furzeri (PXD016587): I'm not sure I buy the argument that the little guys are a good model for aging, but they are a great example of how adaptable even generic-looking vertebrates can be when there is an environment to exploit. 🔗

Mon Jun 22 13:20:45 +0000 2020SNCA:p and SNCB:p are commonly annotated as being nuclear proteins, however they are both frequently observed in platelets.
Mon Jun 22 13:10:23 +0000 2020SNCB:p, synuclein beta (Homo sapiens) 🔗 Small nuclear subunit; PTMs: 2 K-acetyl and 2 Y-phosphoryl acceptors; SAAVs: none; most abundant in brain tissue; mature form: 1-134 [5,510 x, 36.2 kTa] 🔗
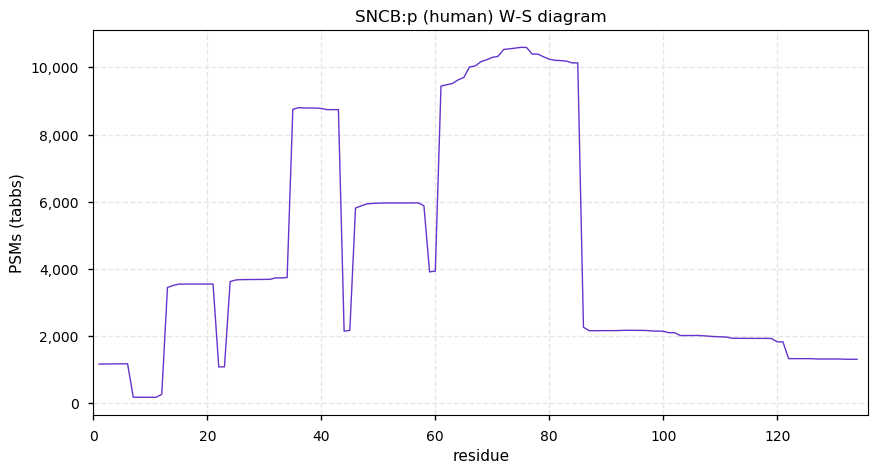
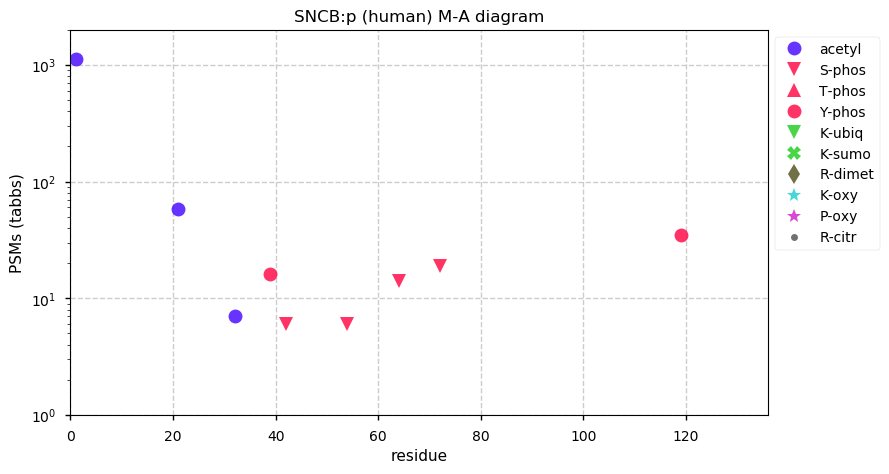
Sun Jun 21 13:29:25 +0000 2020@MattWFoster Although I admit that I occasionally get grumpy when a group that should know better publishes really flawed data and I mutter about it on Twitter.
Sun Jun 21 12:26:11 +0000 2020Mutations in SNCA:p have been associated with Parkinson's disease, however the function of the protein is unknown.
Sun Jun 21 12:21:20 +0000 2020SNCA:p, synuclein alpha (Homo sapiens) 🔗 Small nuclear subunit; aka PARK1, PARK4; PTMs: 9 K-acetyl and 3 Y-phosphoryl acceptors; SAAVs: none; mature form: 1-140 [11,392 x, 108 kTa] 🔗
Sat Jun 20 22:25:46 +0000 2020@MattWFoster I don't want to discourage people from making their data public.
Sat Jun 20 21:47:51 +0000 2020@MattWFoster The identification rate goes from 5% to (very rarely) 90%, depending on the data set. I always check for IAA & urea artifacts, but when they get to be more the 5% of PTMs, the entire data set gets rejected.
Sat Jun 20 17:20:21 +0000 2020Checking the overnight update, GPMDB just sailed through the 10,000,000,000 PSM (10 gTa) totally-made-up-milestone.
Sat Jun 20 13:11:27 +0000 2020SGSH:p is found in HLA type I peptide experiments and has K-ubiquitination sites, so it may be present outside of the lysosome.
Sat Jun 20 13:11:27 +0000 2020SGSH:p, N-sulfoglucosamine sulfohydrolase (Homo sapiens) 🔗 Midsize lysosomal enzyme; PTMs: N41, N264, N413+glycosyl; SAAVs: V361I (6%), R456H (36%); mature form: 21-502 [8,442 x, 39.6 kTa] 🔗
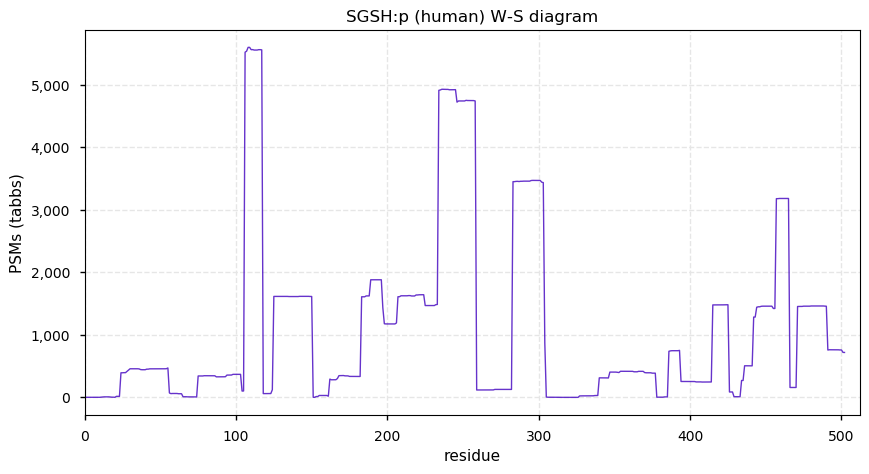
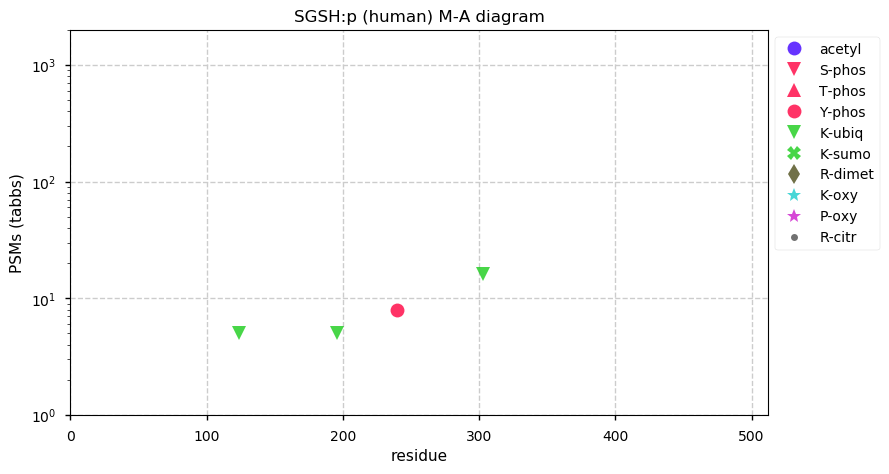
Fri Jun 19 14:03:07 +0000 2020Defects in this protein can lead to a number of autoimmune syndromes, e.g., juvenile idiopathic arthritis, rheumatoid arthritis, multiple sclerosis, Sjogren syndrome, systemic lupus erythematosus, type 1 diabetes, ulcerative colitis, or Crohn disease.
Fri Jun 19 14:03:06 +0000 2020SIAE:p, sialic acid acetylesterase (Homo sapiens) 🔗 Midsize lysosomal enzyme; PTMs: N138, N267, N290, N401, N422+glycosyl only; SAAVs: G64S (2%), M89V (2%); mature form: 24-523 [12,380 x, 61.1 kTa] 🔗
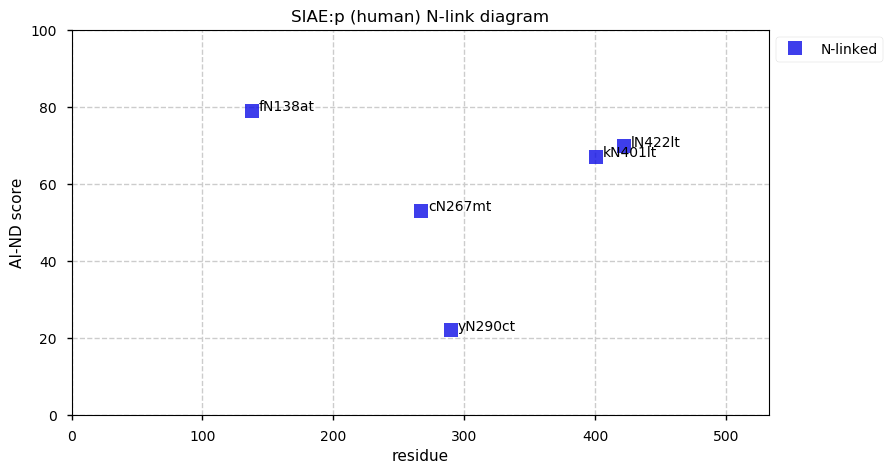
Fri Jun 19 01:00:16 +0000 2020@AJ_Brenes @astacus They are the "spherical human" sought after by theoretical physicists.
Thu Jun 18 21:46:11 +0000 2020Does anybody know of a proteomics data set that characterizes a hybrid mammal and both progenitors? For example, Equus caballus x asinus and the E. caballus and E. asinus parents.
Thu Jun 18 19:59:19 +0000 2020All done and no problems yet. My compliments, Apache.
Thu Jun 18 18:21:56 +0000 2020@UCDProteomics @chrashwood 🔗 (caution: also includes plantains)
Thu Jun 18 17:54:55 +0000 2020@astacus It is particularly difficult to justify when an 8 fraction MDC of HeLa cells is the "human" exemplar.
Thu Jun 18 17:26:14 +0000 2020Updating all my HTTPS web sites to only use TLS 1.2 and 1.3. I expect problems ...
Thu Jun 18 16:59:28 +0000 2020@UCDProteomics Yes it does: maybe emojis should be allowed in reviews 😉
Thu Jun 18 15:24:55 +0000 2020Why isn't "disgruntled former employer" a thing too?
Thu Jun 18 15:19:21 +0000 2020@theworldindex @gangulyteena One of the many reasons Canadians are not fond of their mobile phone service providers.
Thu Jun 18 14:55:54 +0000 2020My opinion is "No, it is not OK."
Thu Jun 18 14:48:45 +0000 2020Is it really OK to take a single 8 fraction MDC run labelled "dog" and say that you have characterized the Canis familiaris proteome well enough to compare it to other species?
Thu Jun 18 12:36:41 +0000 2020Involved in the further maturation of keratinocytes, particularly the disruption and removal of the nucleus. Mouse DNASE1L2:p does not have an ER-targeting signal sequence and is present in epidermal samples.
Thu Jun 18 12:36:41 +0000 2020DNASE1L2:p, deoxyribonuclease 1 like 2 (Homo sapiens) 🔗 Small intracellular enzyme; PTMs: none; SAAVs: none; present in hair in humans; mature form: 22-299 [294 x, 0.48 kTa] 🔗
Wed Jun 17 15:53:37 +0000 2020As someone who has written a lot of poorly documented, difficult to use web page interfaces that people complain about, these guys really seem to have taken the art of software obscurantism up a notch (or two) 🔗
Wed Jun 17 11:58:02 +0000 2020The literature has not come to any conclusions about the role of DNASE1L1:p. Probably prevents the buildup of adventitious DNA in the ER.
Wed Jun 17 11:58:02 +0000 2020DNASE1L1:p, deoxyribonuclease 1 like 1 (Homo sapiens) 🔗 Small endoplasmic reticulum enzyme; PTMs: none; SAAVs: V122I (1%); present at low levels in most cells and urine exosomes; mature form: 19-302 [2,452 x, 6.5 kTa] 🔗
Tue Jun 16 17:48:02 +0000 2020@neely615 @MattWFoster @KentsisResearch @BiswapriyaMisra @Sci_j_my This data set 🔗 that examines human plasma "microparticles" has the highest levels of proteasome proteins I've seen.
Tue Jun 16 12:56:04 +0000 2020@neely615 It is always worth keeping in mind that plasma & serum are artisanal blood products. Serum in particular can be surprisingly variable, depending on the recipe (& artisan).
Tue Jun 16 12:41:45 +0000 2020@neely615 If it is caused by platelets, then the typical platelet markers like CD41 or CD42a should be high. If it is nucleate cell contents, check for AARS or EPRS.
Tue Jun 16 12:32:18 +0000 2020An inhaled formulation of recombinant DNASE1:p (Dornase alfa) is used to treat DNA buildup in cystic fibrosis 🔗
Tue Jun 16 12:32:18 +0000 2020DNASE1:p (aka, serum endonuclease) is common in urine, seminal plasma & feces. It is not a common constituent of blood. There is no proteomics evidence of an intracellular version of this protein. It is commonly observed in rat saliva, but less often in human saliva.
Tue Jun 16 12:32:18 +0000 2020DNASE1:p, deoxyribonuclease 1 (Homo sapiens) 🔗 Small extracellular protein; PTMs: 1 low occupancy S-phosporyl; SAAVs: R2S (4%), R244Q (49%); mature form: 23-282 [3,000 x, 29.2 kTa] 🔗
Tue Jun 16 12:09:54 +0000 2020@neely615 Proteasomes are abundant in all blood cells, including platelets and erythrocytes.
Mon Jun 15 19:16:56 +0000 2020The only exception is DNASE1L3:p 286-291 (AFTNSK), which is shared with A2M:p.
Mon Jun 15 15:17:11 +0000 2020@chrashwood Rhetorical: maybe,
Exasperated: positively
Mon Jun 15 14:28:26 +0000 2020Is there any type of proteomics experiment that has not been referred to in the literature as "quantitative"?
Mon Jun 15 13:53:57 +0000 2020The 5 small DNA degradation enzymes from the genes
DNASE1, 1L1, 1L2, 1L3, 2 & 2B do not share any observed PSMs either between themselves or any other human protein.
Mon Jun 15 12:24:41 +0000 2020DNASE2B:p is annotated as being lysosomal, like DNASE2:p, however its signal sequence is marginal and it is not found in studies that are enriched in lysosomal proteins in human, mouse or rat. It has been observed in HLA type I peptide experiments.
Mon Jun 15 12:24:41 +0000 2020DNASE2B:p, deoxyribonuclease 2 beta (Homo sapiens) 🔗 Small cytoplasmic protein; no PTMs detected; very low abundance; mature form: 1-361 [68 x, 54 Ta] 🔗
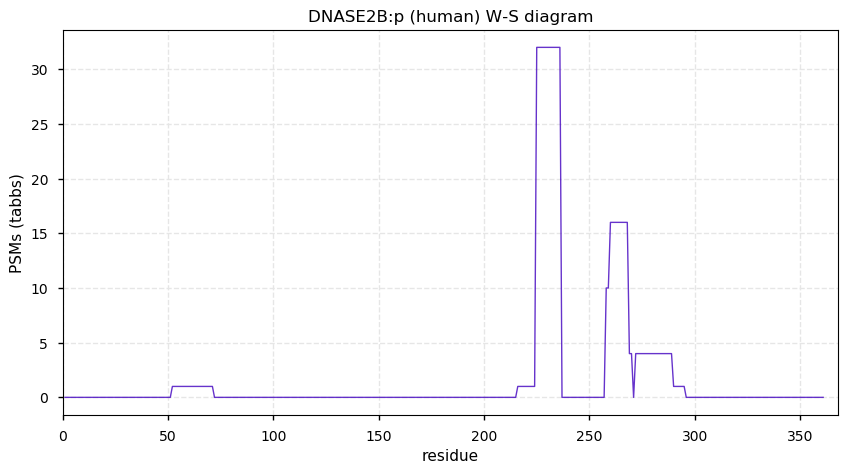
Sun Jun 14 12:30:22 +0000 2020DNASE2:p, deoxyribonuclease 2 (Homo sapiens) 🔗 Small lysosomal enzyme; PTMs: N86, N212, N266, N290+glycosyl; SAAVs: H204R (1%); mature form: 17-360 [10,398 x, 35.3 kTa] 🔗
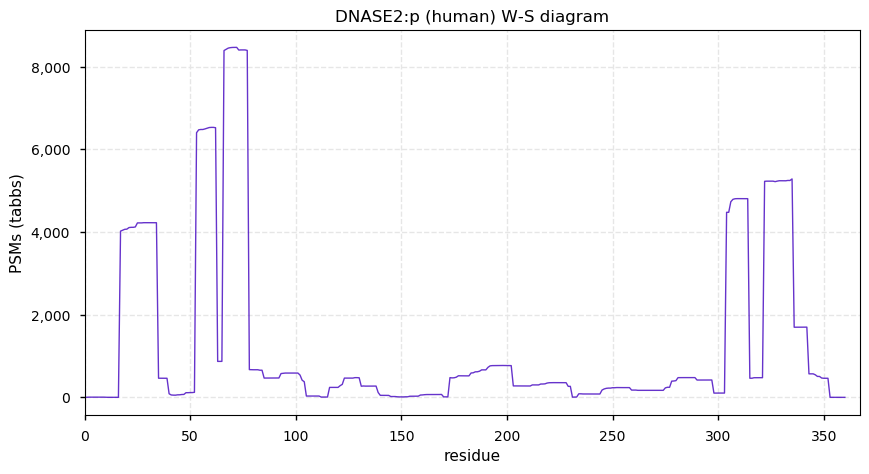
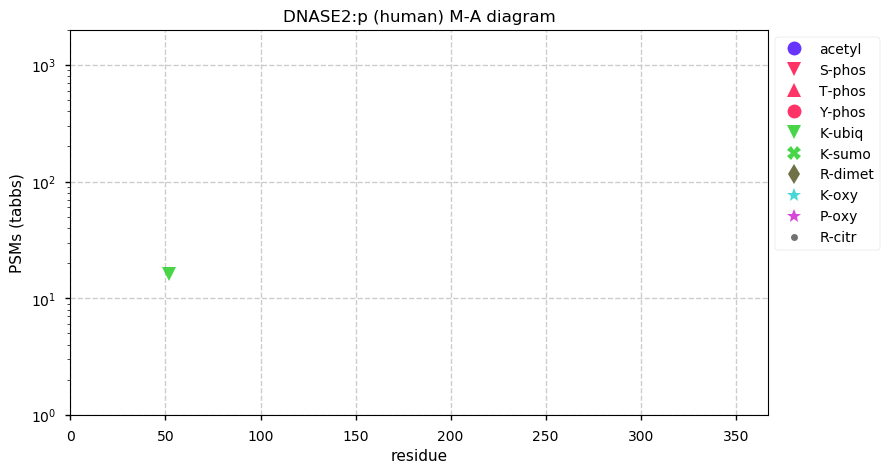
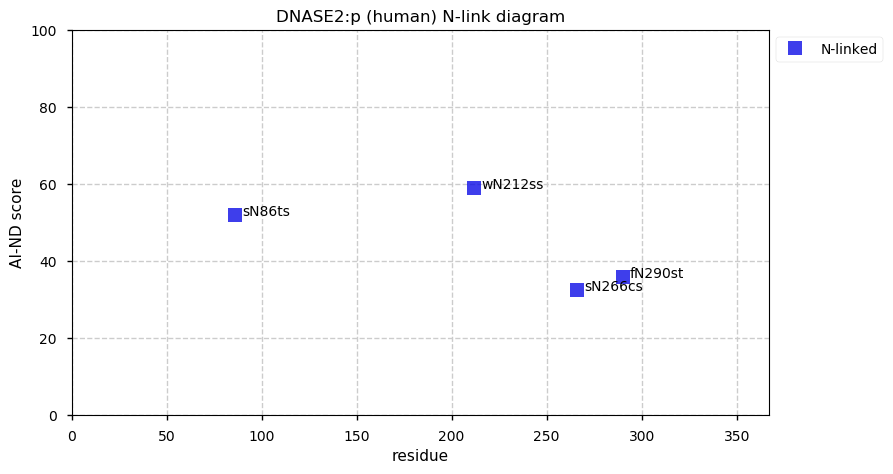
Sat Jun 13 13:04:31 +0000 2020@cdsouthan @uniprot @neXtProt_news Tried a few times and gave up.
Sat Jun 13 12:07:43 +0000 2020The mature protein is only present in mitochondria, but like most mitochondrial proteins it is translated in the cytosol (figure from 🔗) 🔗
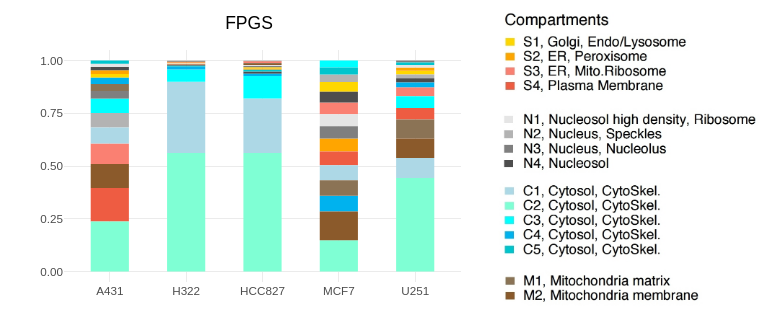
Sat Jun 13 12:07:42 +0000 2020This enzyme constructs the polyglutamate chains found attached to folate and folate analogues.
Sat Jun 13 12:07:42 +0000 2020FPGS:p, folylpolyglutamate synthase (Homo sapiens) 🔗 Midsized mitochondrial enzyme; PTMs: 9 K+ubiquityl in a domain 66-248, S539+phosphoryl; SAAVs: C6R (23%), I22V (22%); mature form: (32,33,34)-587 [4,080 x, 13.5 kTa] 🔗
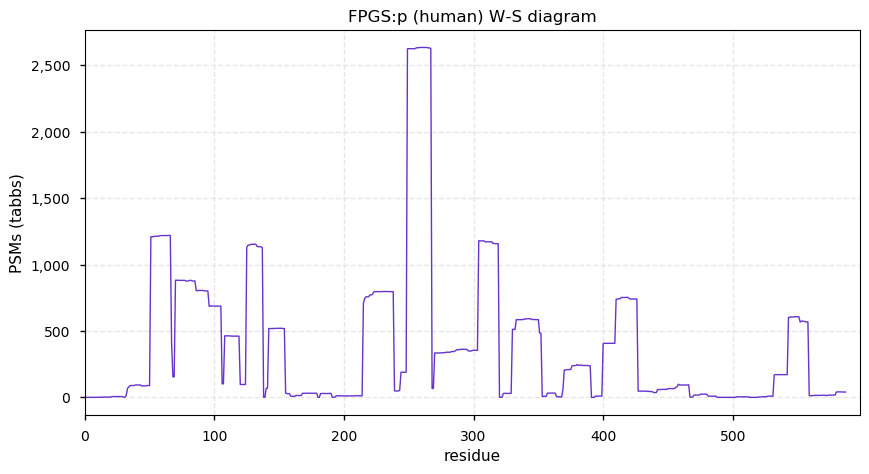
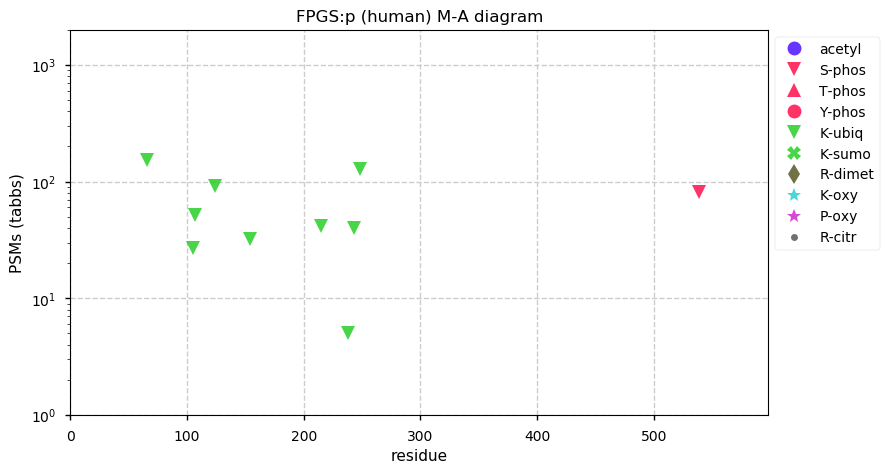
Fri Jun 12 17:51:22 +0000 2020PXD013724 has some very good chromatography and MS/MS, as well as describing a set of experiments that could be fairly easily reproduced in a lab tutorial/workshop
Fri Jun 12 12:04:27 +0000 2020This enzyme degrades the polyglutamate chains commonly found attached to folate and folate analogues. These chains are not peptides: the glutamate units are linked via their gamma-carboxylic acid groups.
Fri Jun 12 12:04:26 +0000 2020GGH:p, gamma-glutamyl hydrolase (H. sapiens) 🔗 Small extracellular/lysosomal enzyme; PTMs: N163, N203, N307+glycosyl; SAAVs: C6R(23%), A31T(20%); common in HLA II data sets, urine & plasma; mature form: (21,28,29)-318 [26,916x, 170 kTa] 🔗
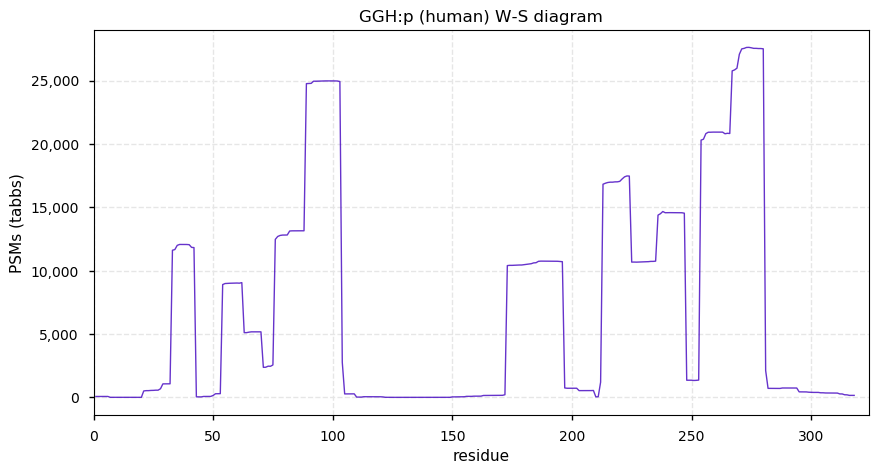
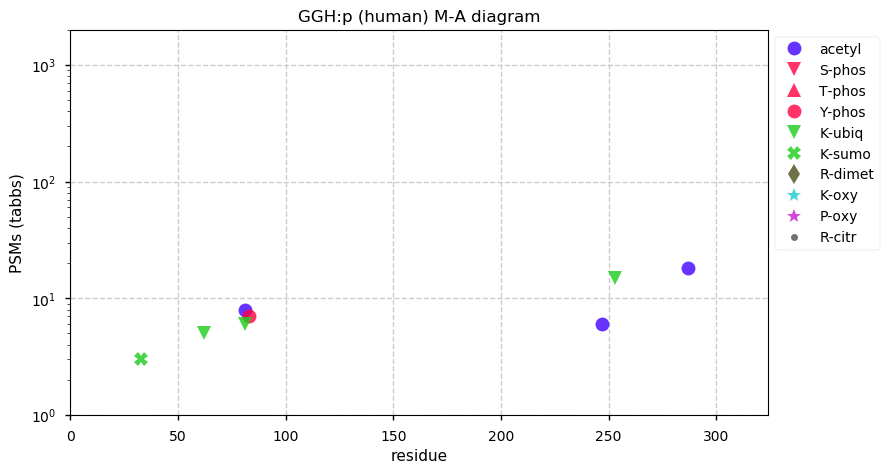
Thu Jun 11 18:18:20 +0000 2020@neely615 @JohnRYatesIII Does the Bruker or Thermo "real time" system give you feedback regarding experimental artifacts, e.g., urea-induced carbamylation or IAA modification of N-terminii?
Thu Jun 11 18:01:11 +0000 2020@neely615 @JohnRYatesIII It is interesting to see people get involved in "real-time" search again: hopefully it can improve data quality in the results.
Thu Jun 11 16:12:14 +0000 2020@bkives So Newfoundland does not require quarantine?
Thu Jun 11 14:49:33 +0000 2020@chrashwood It is just a consequence of working through the list of proteins assigned to the lysosome: almost all of them are N-glycosylated.
Thu Jun 11 12:32:09 +0000 2020The sulfate form of iduronic acid (a hexapyranose sugar) is a major component of sulfated mucopolysaccharides, e.g., heparan sulfate.
Thu Jun 11 12:32:09 +0000 2020IDS:p is annotated in several databases as an exclusively lysosomal protein. All evidence from large scale proteomics data shows that it is a secreted, extracellular protein. There is no evidence consistent with it being in the lysosome.
Thu Jun 11 12:32:09 +0000 2020IDS:p, iduronate 2-sulfatase (Homo sapiens) 🔗 Midsized extracellular enzyme; PTMs: N115, N246, N280+glycosyl only; SAAVs: none; found in urine, CSF and blood plasma; mature form: (33,34,36)-550 [2,680 x, 10 kTa] 🔗
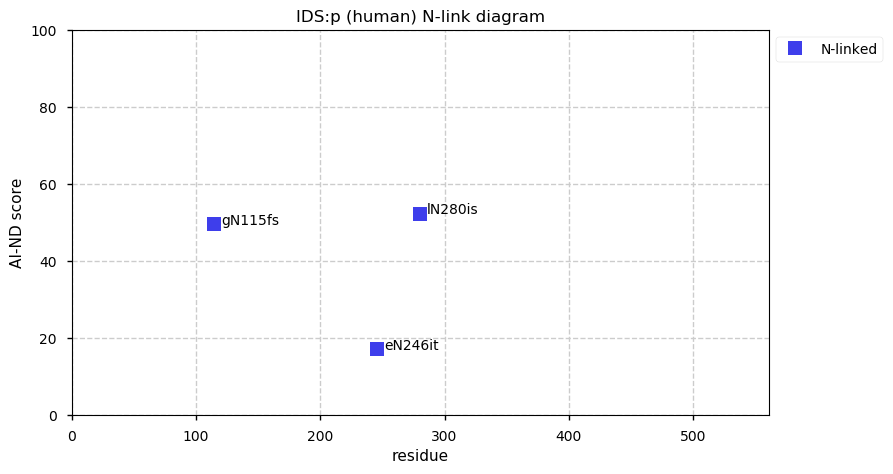
Wed Jun 10 14:06:23 +0000 2020@Sci_j_my I've tried to create bots to do it over the years, but the information is often either so noisy or with so little content that AI tends to get stuck in the ruts.
Wed Jun 10 14:01:33 +0000 2020@TrumanLab Often I will work through some type of theme, e.g. proteins annotated as being in the lysosome (current), mitochondrial matrix, Kreb's cycle, nuclear pores, etc. Sometimes it is just proteins with dopey names, in the news or that strike me as interesting in a data set.
Wed Jun 10 12:11:30 +0000 2020The most abundant SAAV for this protein, ACP2:p.R29Q (maf = 0.48), is part of the ER-targeting signal peptide so it is not present in the mature form of the enzyme.
Wed Jun 10 12:11:30 +0000 2020This enzyme was important in the original characterization of the lysosome by de Duve.
Wed Jun 10 12:11:30 +0000 2020ACP2:p, acid phosphatase 2, lysosomal (Homo sapiens) 🔗 Small lysosomal enzyme; PTMs: N92, N133, N167, N267 N331+glycosyl; SAAVs: R29Q (48%); common in HLA peptide data sets and urine; mature form: 31-423 [12,433 x, 62 kTa] 🔗
Tue Jun 09 18:25:06 +0000 2020@CarolynBertozzi @chrashwood Putting so much emphasis on NIH grantees makes it a lot less interesting for anyone OUS.
Tue Jun 09 15:54:47 +0000 2020@drsarahrhart It has varied. Sometimes it is by systematically going through a gene family, subcellular localization (I'm doing lysosomes at the moment), proteins in the news (like the SARS Cov-2 proteins) or just some that I noticed in a proteomics data set that didn't make sense to me.
Tue Jun 09 15:29:14 +0000 2020The main take-home message from doing this for 8 years: we know next-to-nothing about most proteins.
Tue Jun 09 15:29:14 +0000 2020Since 2012.01.04, every day I select a protein, do about an hour's worth of research on it (papers, database entries, etc) and make a little text summary about some aspect of the protein that is related to experimental proteomics.
Tue Jun 09 14:24:22 +0000 2020Does anyone know whether there has been a sociological study regarding why particular software platforms (& styles) become dominant in specific biomedical research disciplines?
Tue Jun 09 12:24:34 +0000 2020To anyone trying to access GPMDB at the moment, there is a DOS attack occurring that may take some time to clear up.
Tue Jun 09 12:08:11 +0000 2020The enzyme cleaves at GlcNAc-Asn amide bonds, leaving an Asp in the protein sequence. Also known as ASRG, GA and glycosylasparaginase
Tue Jun 09 12:06:32 +0000 2020AGA:p, aspartylglucosaminidase (Homo sapiens) 🔗 Small lysosomal hydrolase; PTMs: N38, N308+glycosyl, K228+acetyl; SAAVs: T149S (8%); common in HLA II data sets and urine; mature form: (24,25)-346 [9,117 x, 40.0 kTa] 🔗
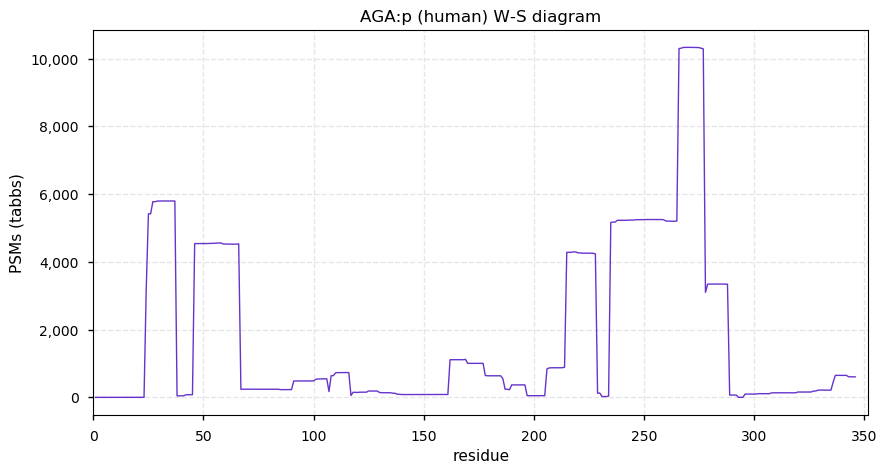
Mon Jun 08 16:11:13 +0000 2020@pitman_mark This description from PNNL uses "dark proteome" in the sense that I find an annoyance
🔗
Mon Jun 08 14:03:03 +0000 2020I am still irrationally triggered by terms like "dark proteome" when it refers to spectra with below threshold peptide sequence assignments.
Mon Jun 08 13:59:39 +0000 2020@bkives Sundown recorded 12.0 cm (via CoCoRaHS, station: CAN-MB-106)
Mon Jun 08 12:31:19 +0000 2020The extracellular vesicle fraction of urine is a good source of CTSA (and most other lysosomal proteins).
Mon Jun 08 12:31:19 +0000 2020CTSA:p, cathepsin A (Homo sapiens) 🔗 Small lysosomal enzyme; PTMs: N145, N333+glycosyl; SAAVs: none; common in HLA II data sets and urine; mature form: 29-480 [26,128 x, 137 kTa] 🔗
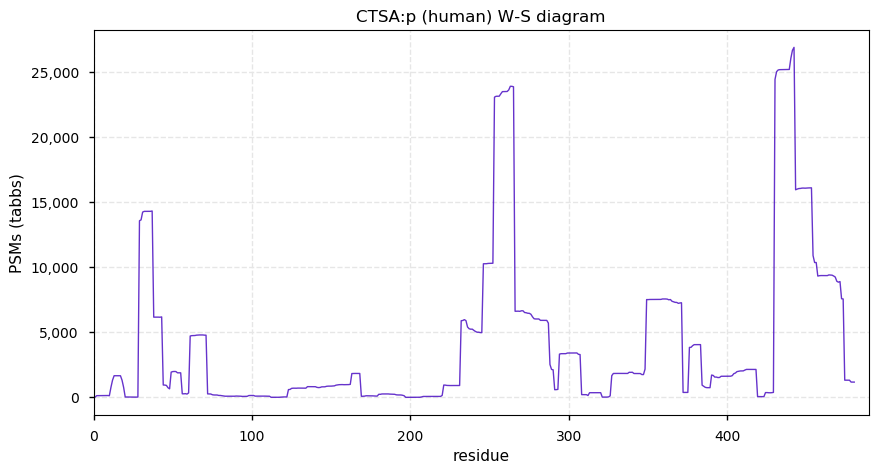
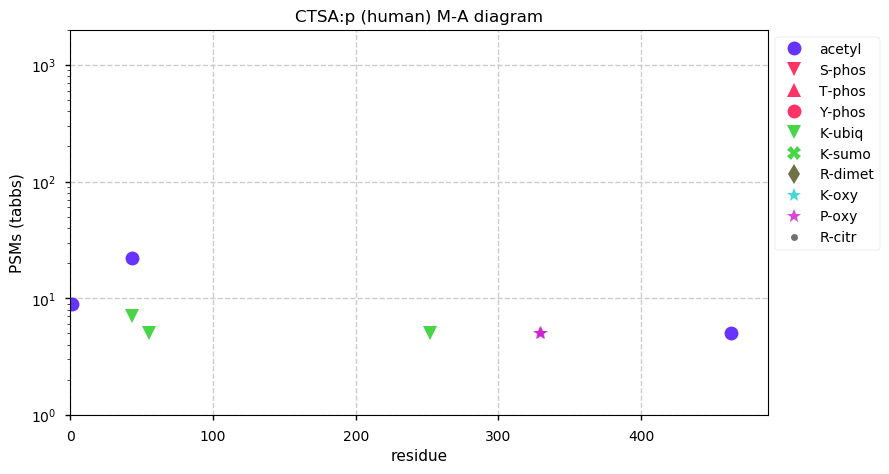
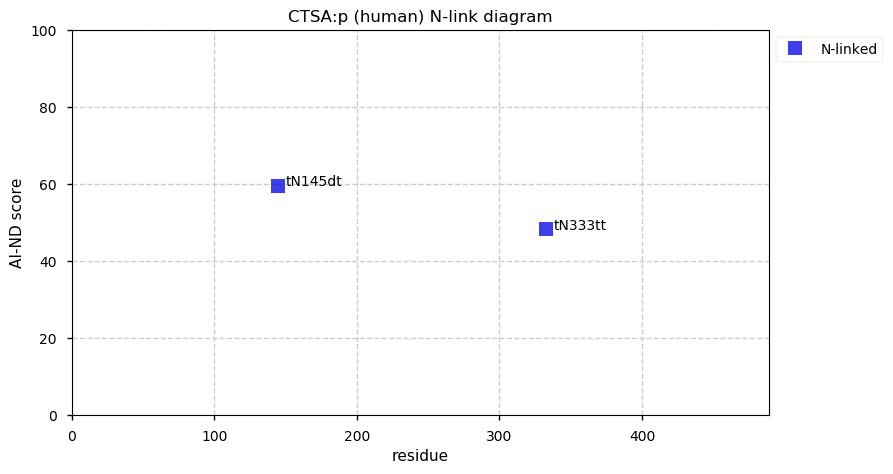
Sun Jun 07 12:41:04 +0000 2020The high MAF SAAV (p.P10L, maf=0.43) occurs in the signal sequence and is not part of the mature protein.
Sun Jun 07 12:41:03 +0000 2020GLB1:p, galactosidase beta 1 (Homo sapiens) 🔗 Midsized lysosomal enzyme; PTMs: N247, N464, N498+glycosyl; SAAVs: P10L (43%), C521R (7%); common in HLA II data sets and urine; mature form: 29-677 [24,887 x, 169 kTa] 🔗
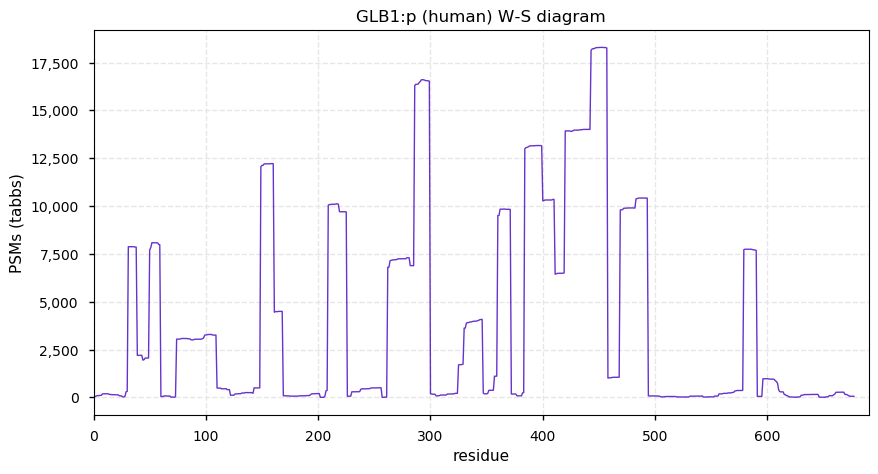
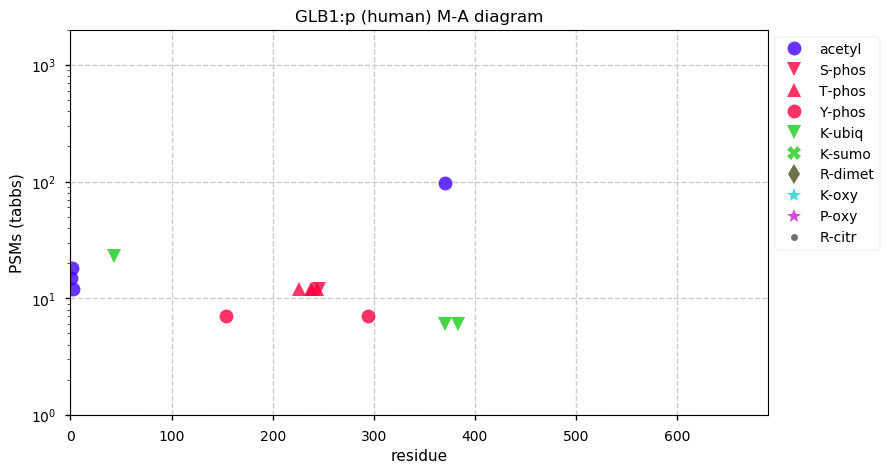
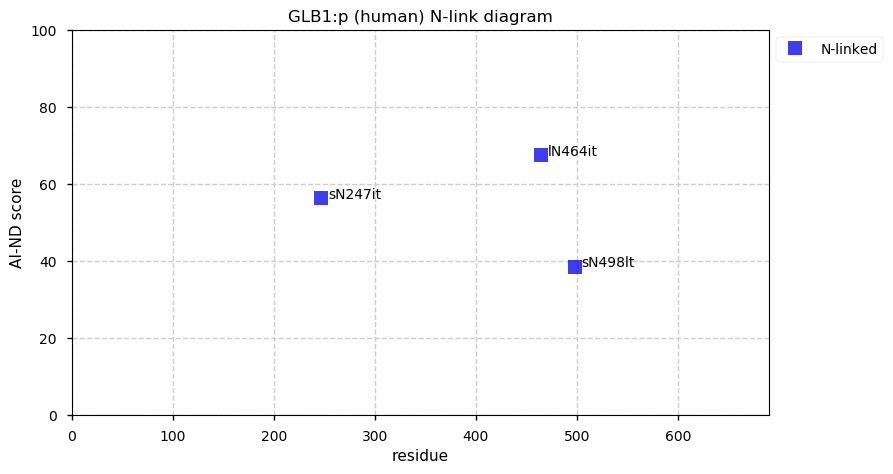
Sat Jun 06 15:10:40 +0000 2020@slavovLab @ASBMB @biorxivpreprint I have to agree that this was not good news 🔗
Sat Jun 06 11:57:58 +0000 2020NEU1:p forms a supramolecular structure (clump) with CTSA:p and GLB1:p that protects these enzymes from degradation in the lysosome.
Sat Jun 06 11:57:58 +0000 2020NEU1:p, neuraminidase 1 (Homo sapiens) 🔗 Small lysosomal enzyme; PTMs: N186, N352+glycosyl; SAAVs: G88A (2%); common in HLA II data sets and urine; mature form: 48-415 [7,806 x, 29 kTa] 🔗
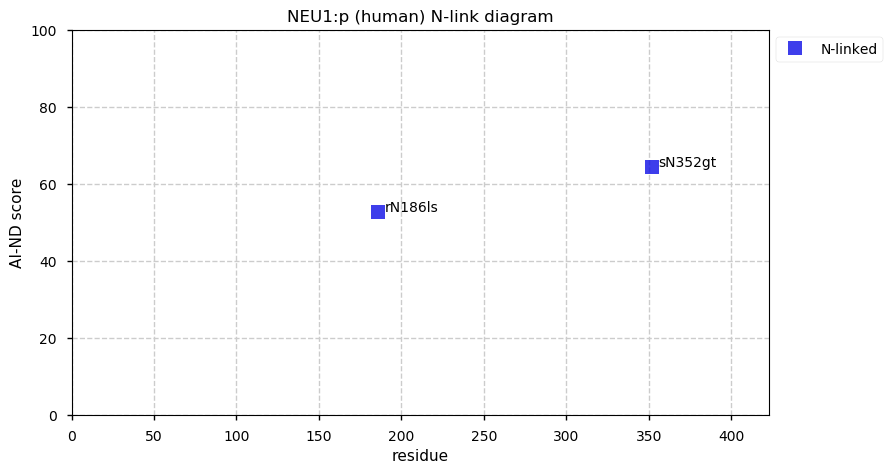
Fri Jun 05 16:27:06 +0000 2020PXD012921 is a nice addition to what is known about phosphorylation in human herpesvirus 5 (aka cytomegalovirus, HHV5 & HCMV) proteins. HHV5 has about 40 phosphoproteins, out of 190 protein coding genes.
Fri Jun 05 14:42:07 +0000 2020@byu_sam Rest-Of-Earth
Fri Jun 05 13:02:17 +0000 2020To hell in a handbasket 🔗
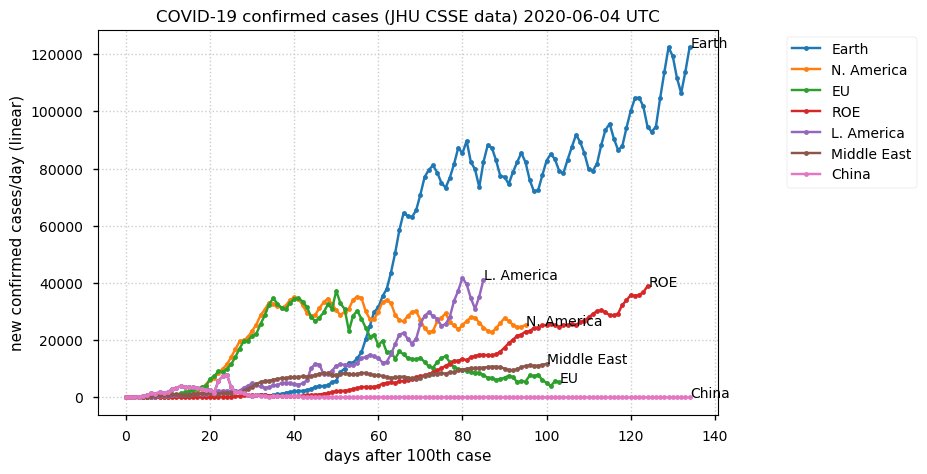
Fri Jun 05 11:53:38 +0000 2020These small, non-enzymatic glycoproteins enhance the processing of oligosaccharides and glycosphingolipids somehow.
Fri Jun 05 11:46:28 +0000 2020Prosaposin contains the sequences for Saposin A [60-142], Saposin B [195-273], Saposin C [311-390] and Saposin D [405–486]
Fri Jun 05 11:46:28 +0000 2020PSAP:p, prosaposin (Homo sapiens) 🔗 Midsized lysosomal/secreted polyprotein; PTMs: N80, N101, N215, N332, N426+glycosyl; SAAVs: Q190H (1%); common in HLA II data sets; signal sequence: 1-16 [38,799 x, 582 kTa] 🔗
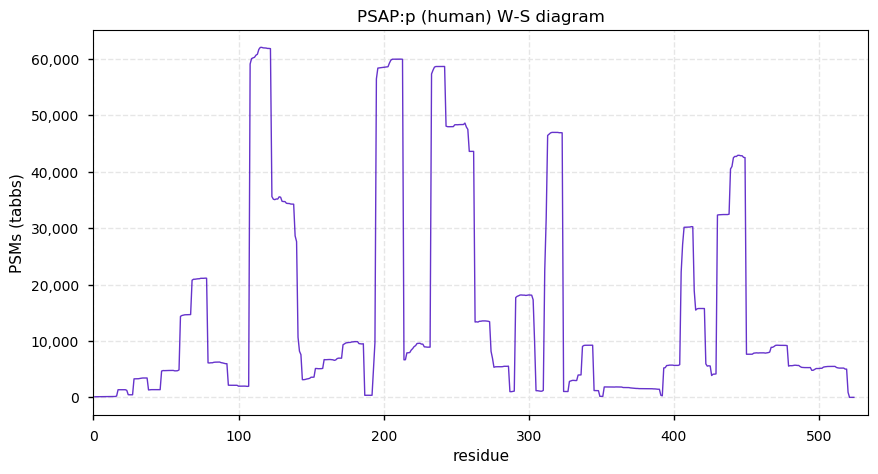
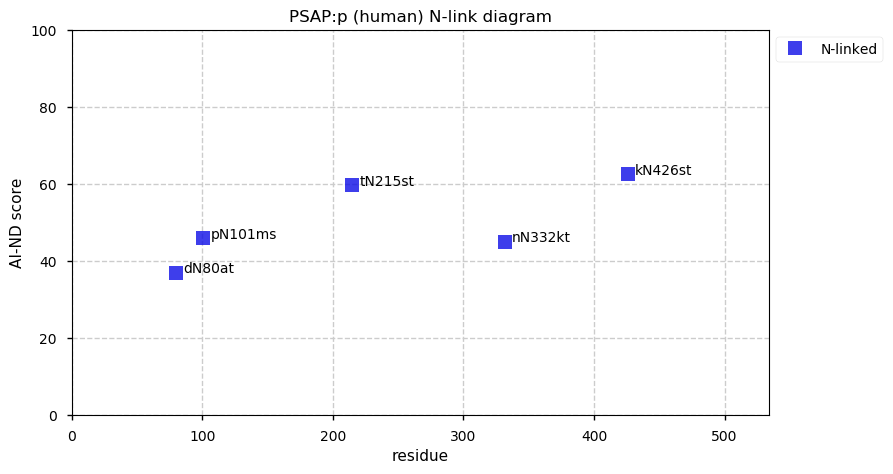
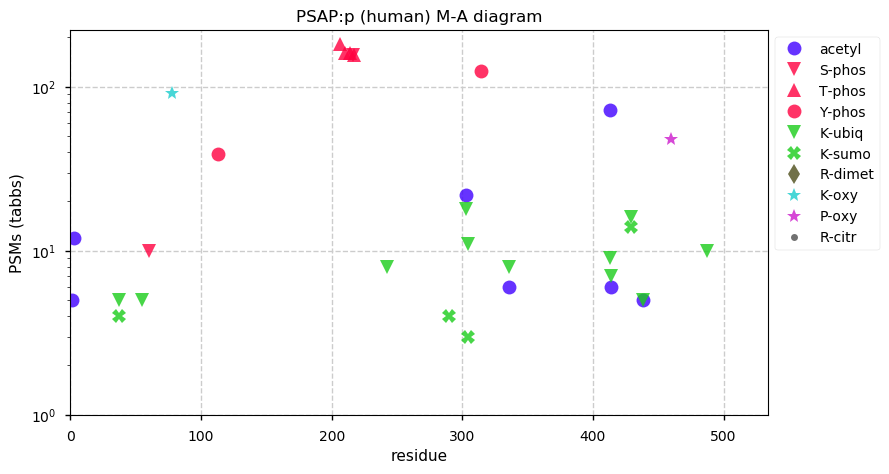
Thu Jun 04 16:59:43 +0000 2020Does anybody know if PXD015523 has an associated publication? The data represents a very interesting list of proteins, quite different than most (all?) of the results I've seen.
Thu Jun 04 14:49:26 +0000 2020@AlexUsherHESA PAIX, ORDRE ET BON GOUVERNEMENT!
Thu Jun 04 14:24:38 +0000 2020@kabalak @dtabb73 But even using ASCII symbols like "%" or "#" in a file name can generate uncaught errors in software pipelines. The filename will often return a "true" when it is tested for existence, but fails later on because of an untrapped error or exception.
Thu Jun 04 14:20:16 +0000 2020@kabalak @dtabb73 Windows versions have changed over time how they deal with non-ASCII characters in filenames, using ANSI, code-page approaches and now UTF-8: the result of using of those characters is unpredictable across versions.
Thu Jun 04 12:13:38 +0000 2020TRPV1:p is a temperature sensor that activates at >43 C, resulting in a "burning" sensation in humans. It can be activated by vanilloid compounds, such as capsaicin, making it the receptor targeted by pepper sprays. 🔗
Thu Jun 04 12:13:38 +0000 2020TRPV1:p, transient receptor potential cation channel subfamily V member 1 (Homo sapiens) 🔗 Midsized integral membrane protein; PTMs: none; SAAVs: M315I (27%); found in HLA I data sets; mature sequence: 1-839 [101 x, 0.5 kTa] 🔗
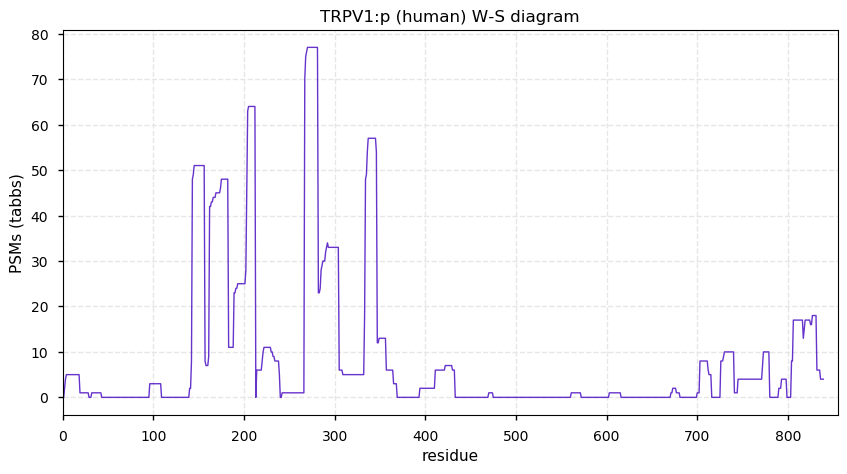
Wed Jun 03 14:27:19 +0000 2020To everyone who discovers you can use all sorts of characters (including most of UTF-8) to make fancy-looking file names: please don't.
Wed Jun 03 11:57:33 +0000 2020TRPA1:p is the primary receptor targeted by tear gas in humans.
🔗
Wed Jun 03 11:57:32 +0000 2020TRPA1:p, transient receptor potential cation channel subfamily A member 1 (Homo sapiens) 🔗 Large integral membrane protein; PTMs: Y22, S43, S1076+phosphoryl; SAAVs: R58T (10%); mature sequence: 1-1119 [335 x, 2.7 kTa] 🔗
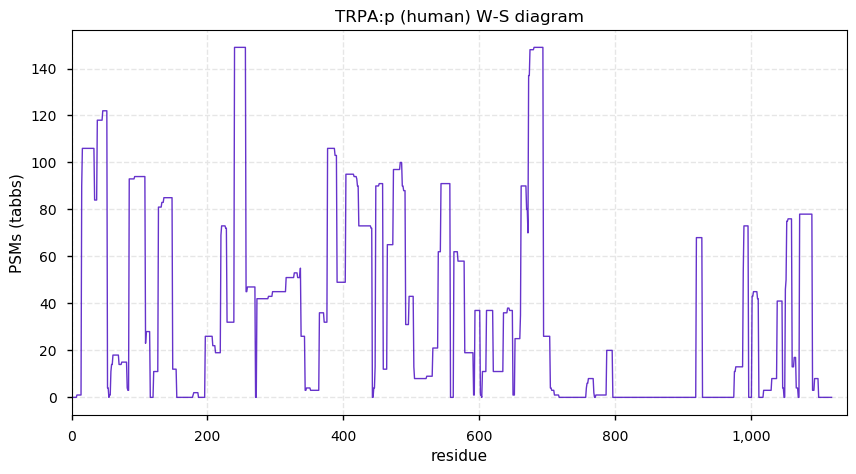
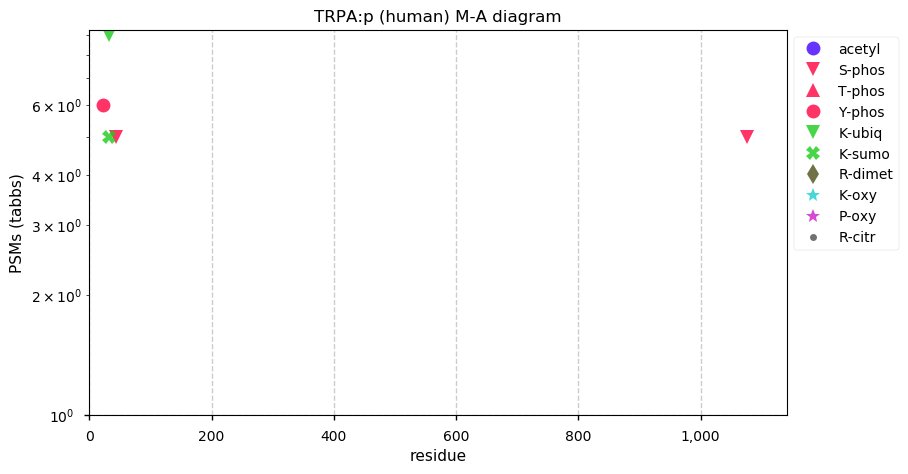
Tue Jun 02 14:52:27 +0000 2020FBN2:p is a secreted glycoprotein that is a constituent of elastic fibers in the extracellular matrix. However, when it ends up in the cytosol (presumably via ERAD), the cell seems to slap a ubiquitin or SUMO on it anywhere it can 🔗 🔗
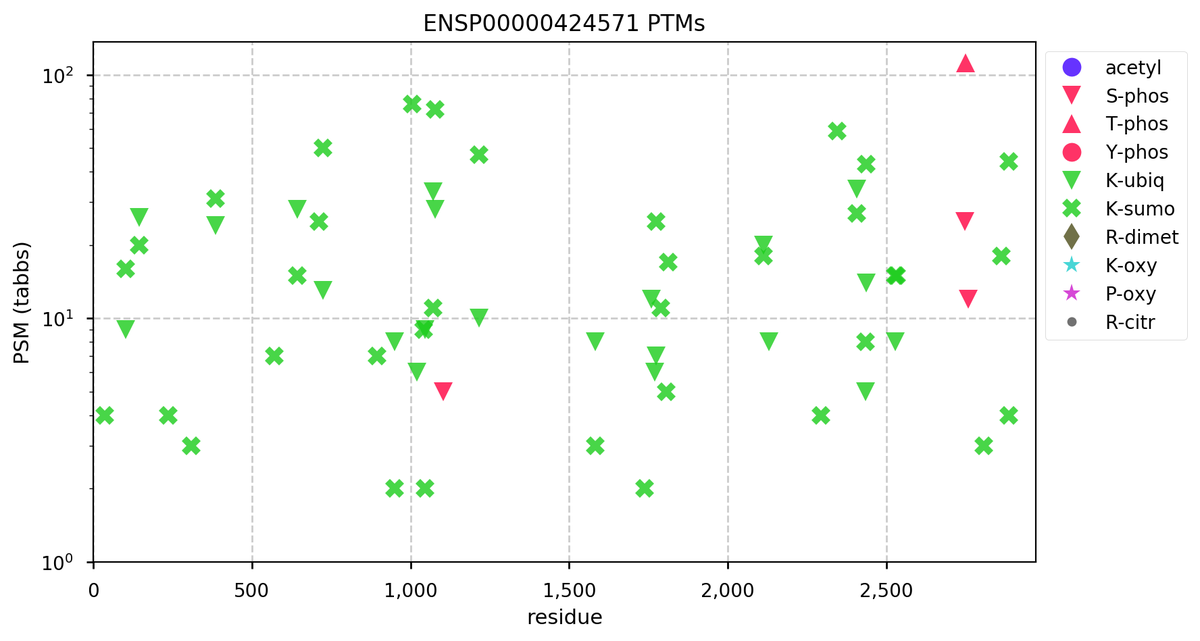
Tue Jun 02 12:11:33 +0000 2020GALC:p, galactosylceramidase (H. sapiens) 🔗 Midsized lysosomal protein; PTMs: N143, N379, N403+glycosyl only; SAAVs: T641A (6%); common in HLA type I and II data sets; mature form 43-685 [6,584 x, 26.4 kTa] 🔗
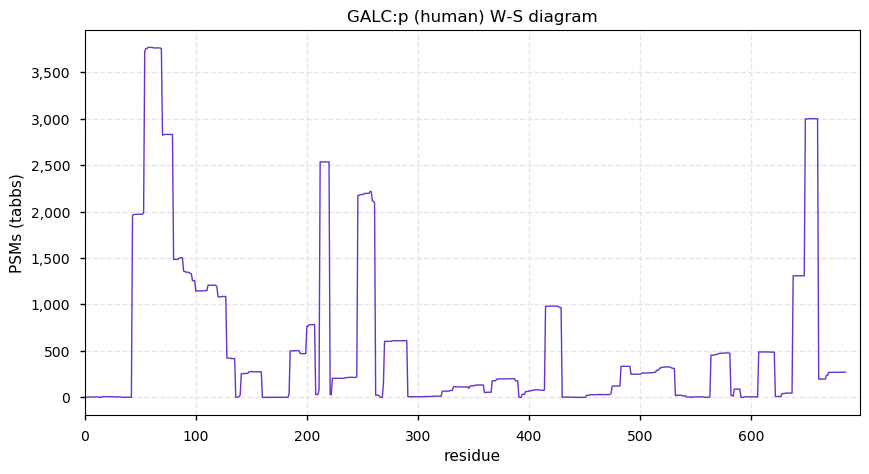
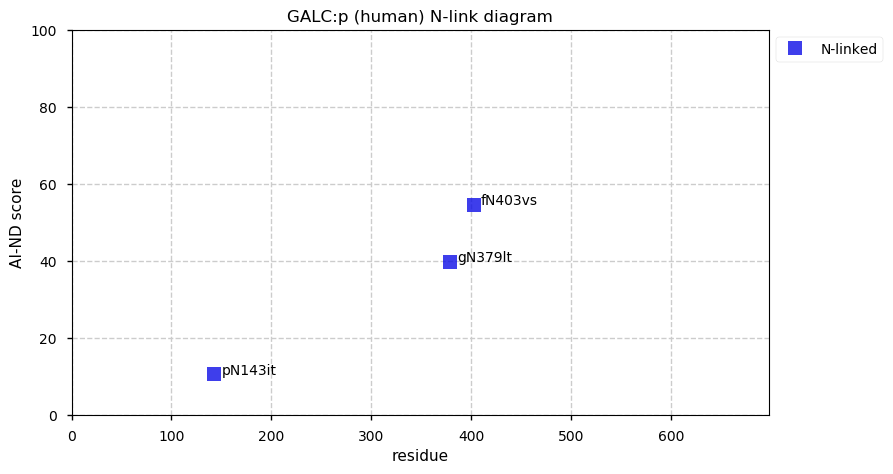
Mon Jun 01 21:36:53 +0000 2020If you ever want to see the extent to which collagens can dominate a human tumour tissue sample, PXD014980 is a great illustration of this phenomenon (S. Lee, et al., 🔗).
Mon Jun 01 16:35:23 +0000 2020I was looking through some PTM stats today when I realized how little public data there was about ubiquitin or SUMO ligation in any species other than H. sapiens.
Mon Jun 01 14:33:29 +0000 2020Exhaustive lists of the peptides from the most abundant proteins are here:
'N': 🔗
'S': 🔗
'M': 🔗
Mon Jun 01 14:33:29 +0000 2020The LC/MS/MS was very well done in these experiments. The cysteine derivatization reaction was not done well: <20% of Cys-containing peptides were recovered.
Mon Jun 01 14:33:29 +0000 2020PXD018594 has 2 reps of a time course, with virus originally loaded at 2 levels and sampled at days 1, 2, 3, 4 & 7 post-infection.
Mon Jun 01 14:33:29 +0000 2020These are very good examples of which viral proteins can be observed from virus grown in cell culture (Vero E6/C1008), using a simple 1D HPLC analysis.
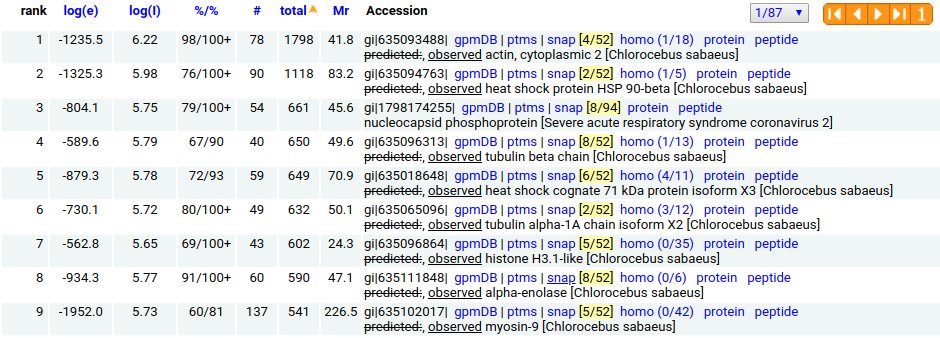
Mon Jun 01 12:25:48 +0000 2020DPP7:p, dipeptidyl peptidase 7 (H. sapiens) 🔗 Small lysosomal/secreted protein; PTMs: N50, N86, N315, N363, N428+glycosyl and 6 K+ubiquitin sites; SAAVs: A89G (18%); common in HLA type II data; mature form 26-492 [20,061 x, 120 kTa] 🔗
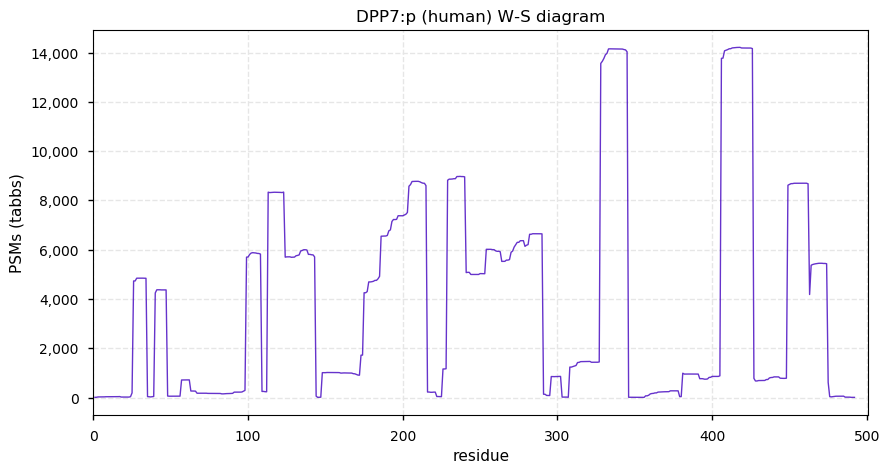
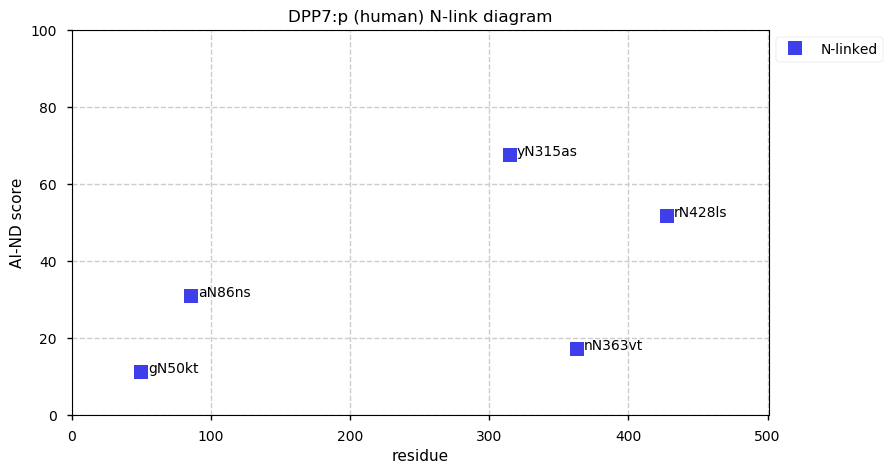
Sun May 31 18:19:12 +0000 2020In terms of new cases/day, India and Brazil are both on a bit of a tear 🔗
Sun May 31 15:45:55 +0000 2020I was able to easily find out about the plush dinosaur, though!
Sun May 31 15:45:05 +0000 2020Does anybody know if they managed to recover the reusable portion of the rocket after the launch yesterday? The US news is hard to sort through today.
Sun May 31 15:06:14 +0000 2020Until I started to profile individual lysosomal proteins, I didn't realize the extent of N-linked glycosylation associated with these proteins.
Sun May 31 12:27:53 +0000 2020CLN5:p, CLN5 intracellular trafficking protein (Homo sapiens) 🔗 Small lysosomal/secreted hydrolase; PTMs: N178, N203, N255, N271, N281+glycosyl; SAAVs: none; common in HLA type II data sets; mature form 51-358 [5,046 x, 9.6 kTa] 🔗
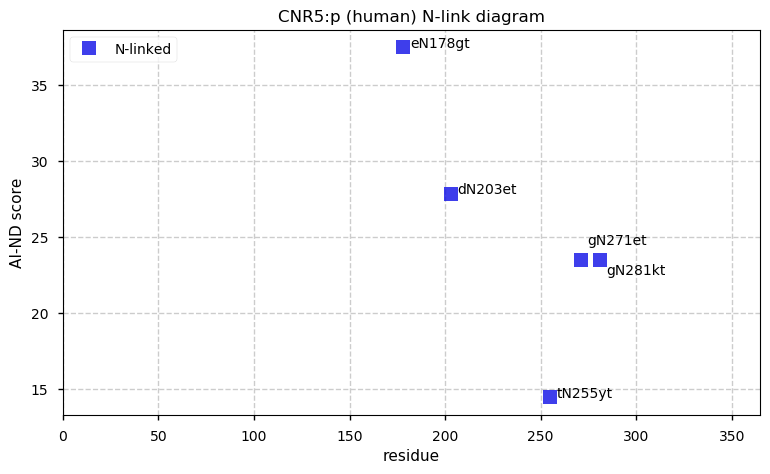
Sat May 30 11:42:46 +0000 2020CTSZ:p, cathepsin Z (Homo sapiens) 🔗 Small lysosomal hydrolase; PTMs: N184, N224+glycosyl; SAAVs: none; common in HLA type II data sets [26,734 x, 118 kTa] 🔗
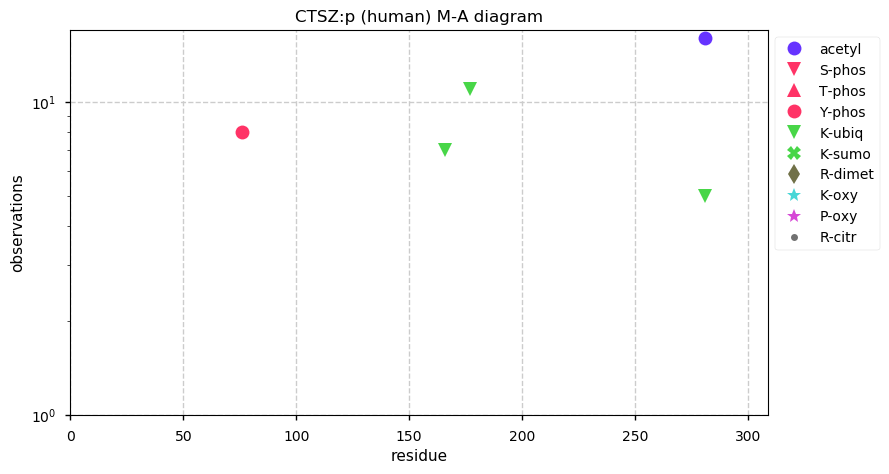
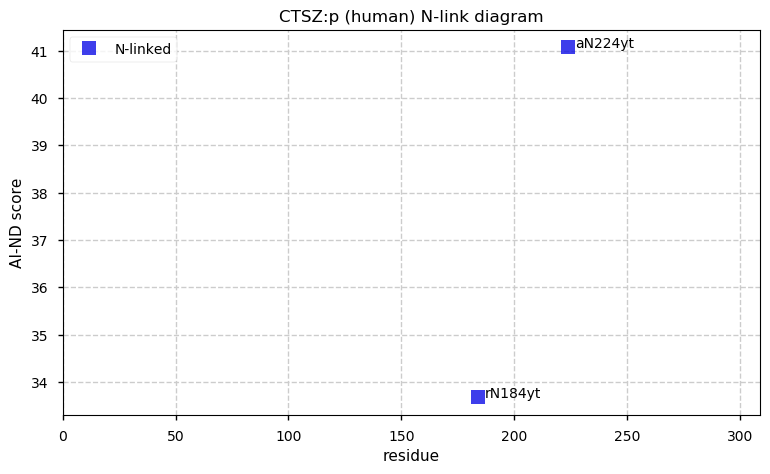
Fri May 29 15:07:21 +0000 2020@KentsisResearch @slavovLab @mbeisen @eLife I would suggest simply discounting the value of the paper. Trying to work through editors and authors only leads to frustration.
Fri May 29 15:00:41 +0000 2020I think filaggrin (FLG:p) is the king of citrullination, with 278 observed sites (most proteins have none):
🔗 🔗
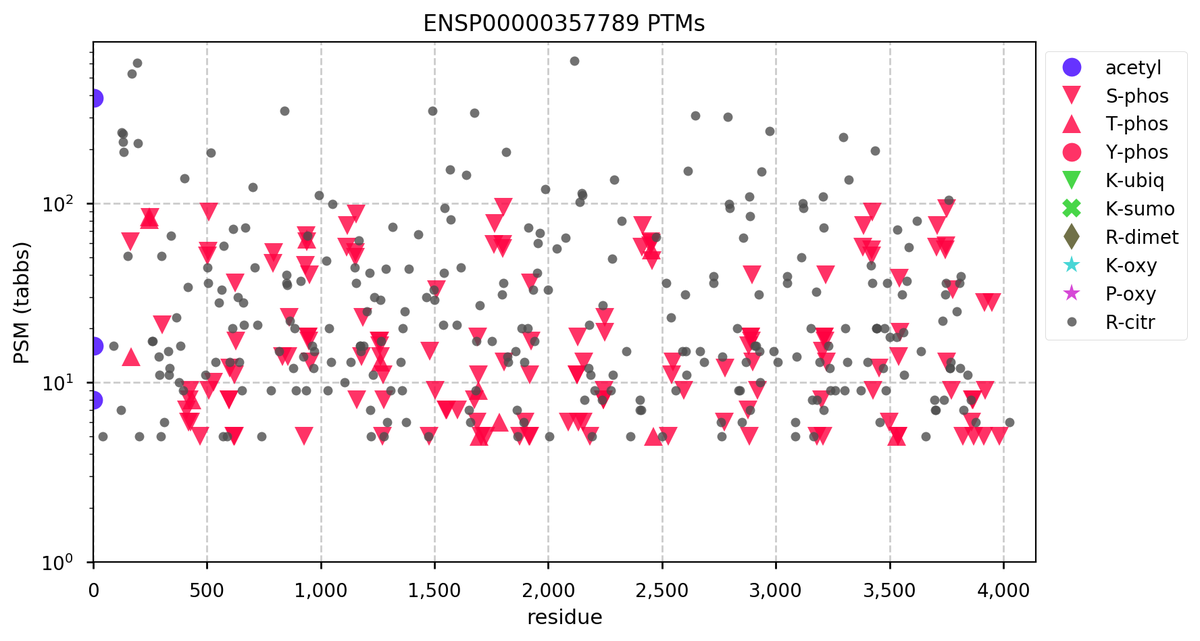
Fri May 29 13:32:31 +0000 2020@PastelBio @ProteomicsNews PXD018804 & PXD018594 from the Armengaud group's SARS-CoV-2 paper popped up on PRIDE this morning. It will be interesting to see what they found.
Fri May 29 11:44:08 +0000 2020CTSS:p, cathepsin S (Homo sapiens) 🔗 Small lysosomal hydrolase; PTMs: N104+glycosyl; SAAVs: R113G, R113W (38%); common in HLA type II data sets [11,668 x, 73.4 kTa] 🔗
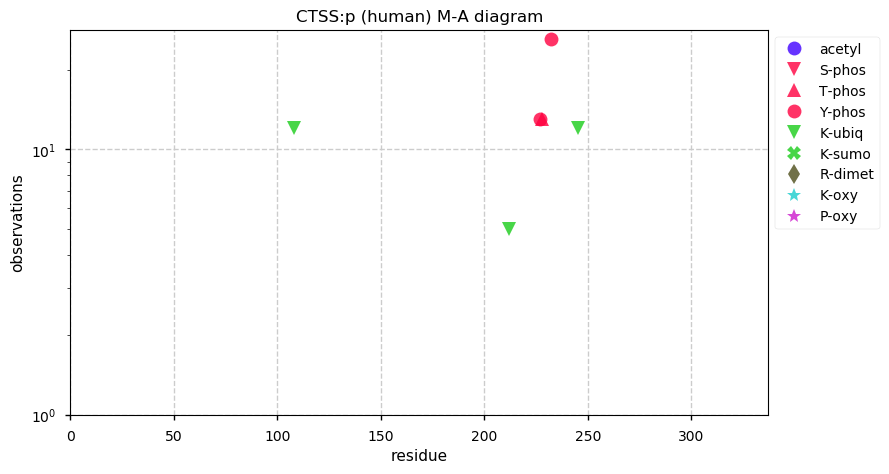
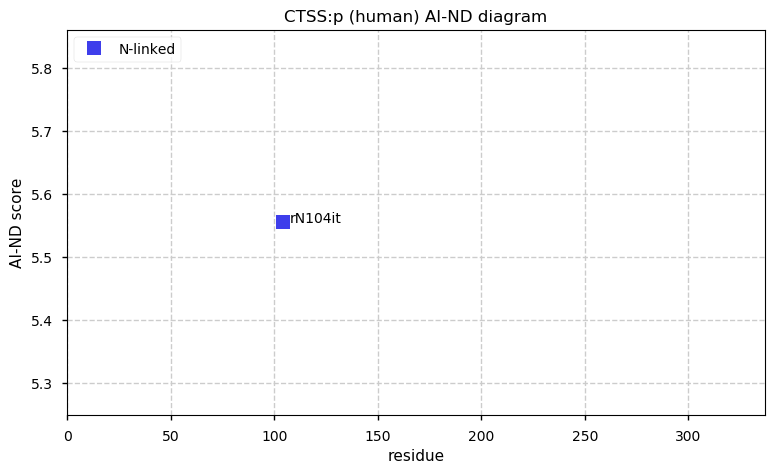
Thu May 28 16:14:15 +0000 2020@chrashwood They aren't simply deamidated: they are deamidated at least 10 σ more often than they should be (the observed deamidation rate for most N-sites is ~ 0.5%). And I must stress that you need at least 100 tabbs of data for a each N-site to apply this sort of analysis.
Thu May 28 15:44:35 +0000 2020@chrashwood Some are, most aren't.
Thu May 28 15:38:44 +0000 2020@nesvilab @chrashwood I agree. This is based on thousands of data sets, not individual observations.
I was surprised by it, too. But, every case I checked has come up agreeing with existing N-linked assignments.
Thu May 28 15:34:43 +0000 2020@chrashwood Maybe I should have used GLG1:p instead for the slide:
🔗 🔗
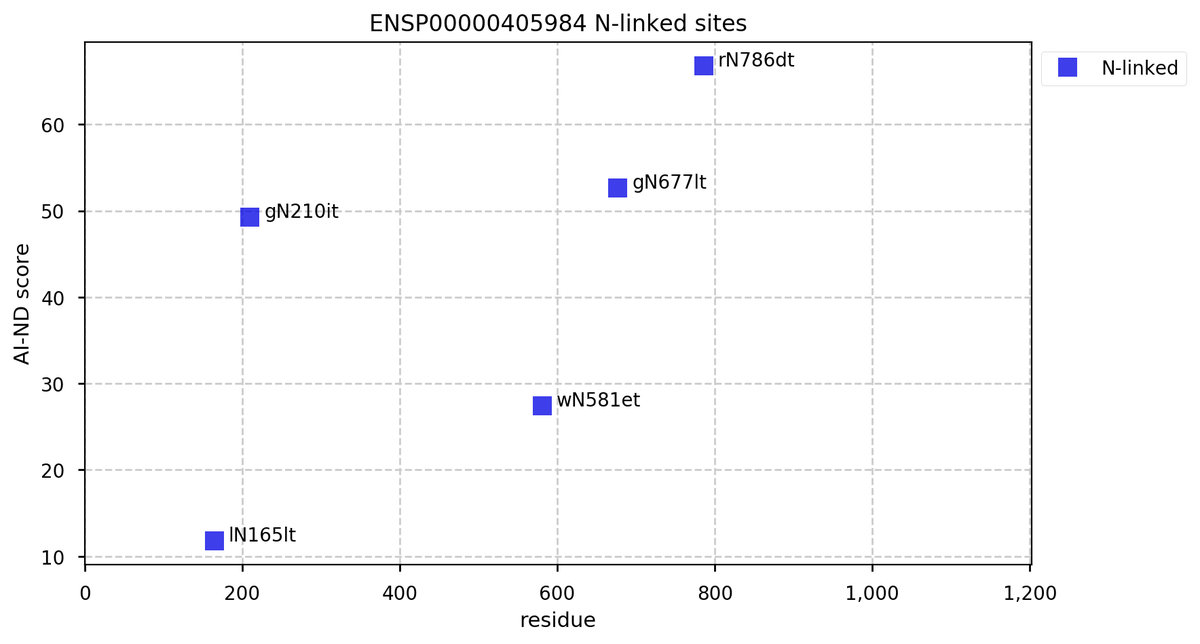
Thu May 28 15:32:14 +0000 2020@chrashwood No, they aren't assignment errors: I test for that. I also correct for the propensity of NG sites to deamidate (you'll notice that most of the sites in ITGB1 aren't NG's). This isn't something you could ever see from a single data set: it only appears if you have a lot of data.
Thu May 28 15:23:51 +0000 2020@chrashwood For some reason I can't seem to get NetNglyc to give me a result for ITGB1 using its Swissprot ID.
Thu May 28 15:22:39 +0000 2020@chrashwood It is based on N-deamidation anomalies in the protein's PSMs. I found that there were a few N-deamidation rates in the aggregated data that I couldn't explain, until I looked at the 2nd residue following the N: lo and behold it was almost always S/T. A bit of AI, et voilà.
Thu May 28 15:07:03 +0000 2020So what else needs to go on a page showing N-linked glycosylation site assignments?
🔗 🔗
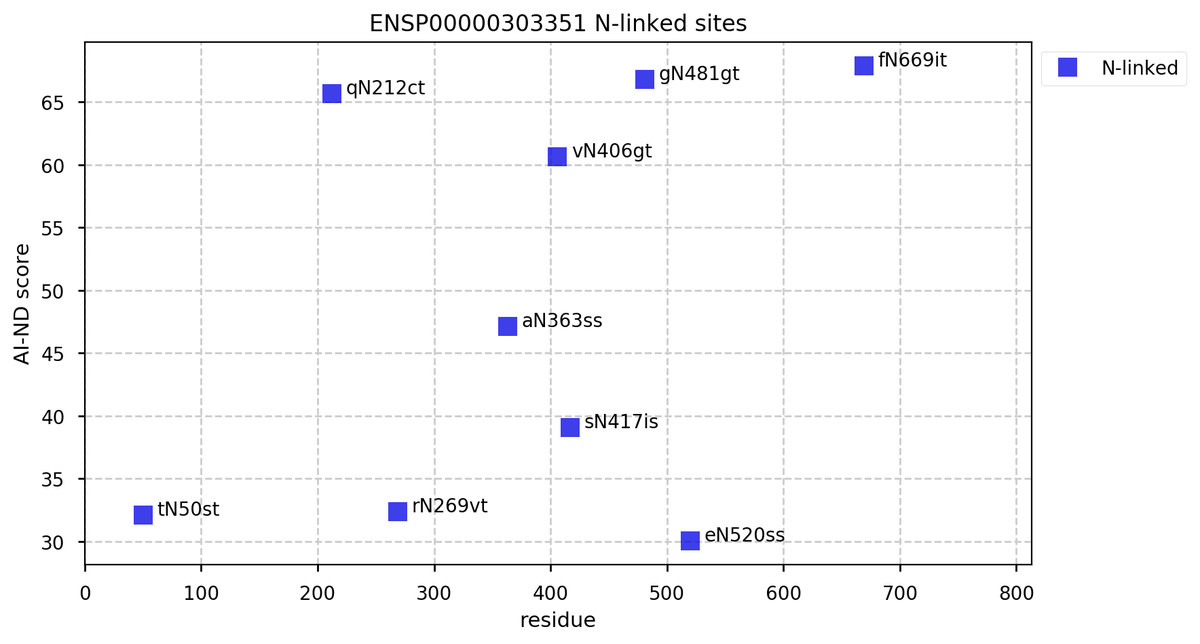
Thu May 28 13:02:00 +0000 2020@PastelBio @ProteomicsNews At least the data (PXD018804) should be released soon.
Thu May 28 12:39:32 +0000 2020@PastelBio @ProteomicsNews It is a pity, as Armengaud's lab is in the top 3 proteomics labs in Europe in terms of data quality (it may also be in the top 1). The article is available on sci-hub, though.
Thu May 28 11:52:17 +0000 2020CTSO:p, cathepsin O (Homo sapiens) 🔗 Small lysosomal hydrolase; PTMs: N105+glycosyl only; SAAVs: none; common in HLA type II data sets [746 x, 1.5 kTa] 🔗
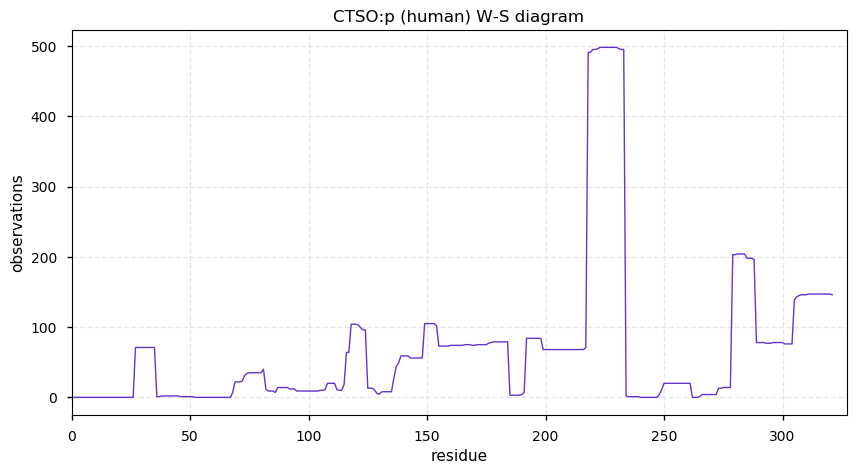
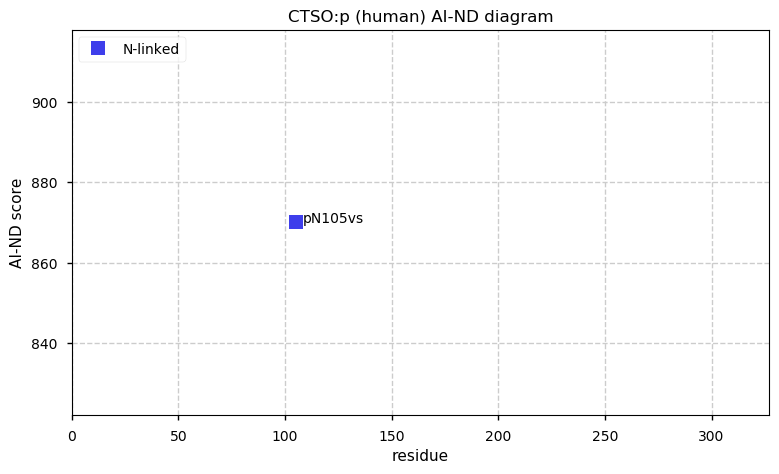
Wed May 27 23:48:30 +0000 2020@ypriverol The senior author is Peipei Ping from UCLA. She has been involved with HUPO since it started. Contact her directly and I'm sure she will put you in touch with the right team member.
Wed May 27 20:52:35 +0000 2020@TanentzapfLab And the reference for the tool paper is ...
Wed May 27 19:04:53 +0000 2020@chrashwood Thanks. I was sure there must be one.😀
Wed May 27 18:43:24 +0000 2020Is there an endogenous human enzyme, or pathway of enzymes, that removes N-linked glycosylation leaving an Asp residue (analogous to PNGase F)?
Wed May 27 15:09:32 +0000 2020@Sci_j_my It may be that this type of meta-definition is too broad (or subtle) for their algorithm to recognize it.
Wed May 27 15:07:48 +0000 2020@Sci_j_my I define a serial killer movie/tv show as being about 1 or more main characters who believe that they are on a mission & kill people in the name of completing that mission.
Wed May 27 14:29:27 +0000 2020@Sci_j_my Tried that for several months. It didn't make any difference.
Wed May 27 14:26:50 +0000 2020I would probably watch more Netflix if their algorithm for suggesting material that I might be interested in wasn't so horrible. Is there some way to tell them "I am NOT interested in serial killers (or their equivalents)"?
Wed May 27 11:53:49 +0000 2020The PTM pattern for CTSL:p (complementary ubiquitin/SUMO K-sites) suggests an intracellular role for the protein other than the usual lysosomal annotation.
Wed May 27 11:32:56 +0000 2020CTSL:p, cathepsin L (Homo sapiens) 🔗 Small lysosomal/exosomal hydrolase; PTMs: N221+glycosyl, (K103, K216)+ubiquitin and complementary SUMO; SAAVs: none; common in HLA type II data sets [13,165 x] 🔗
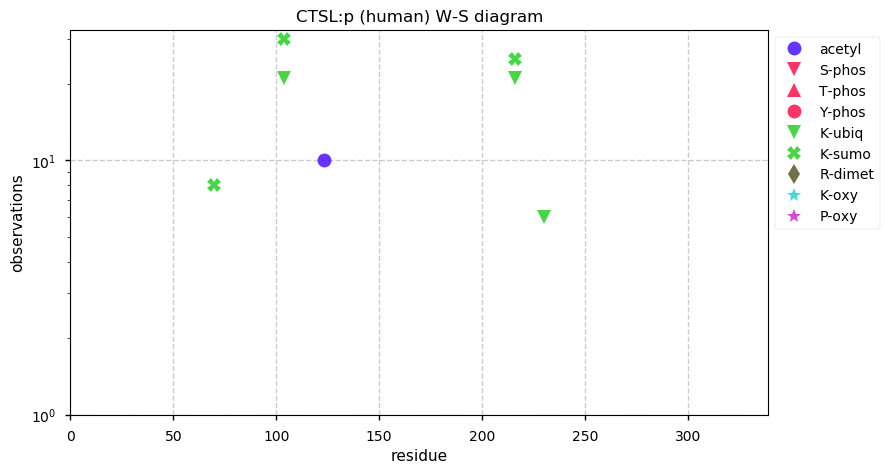
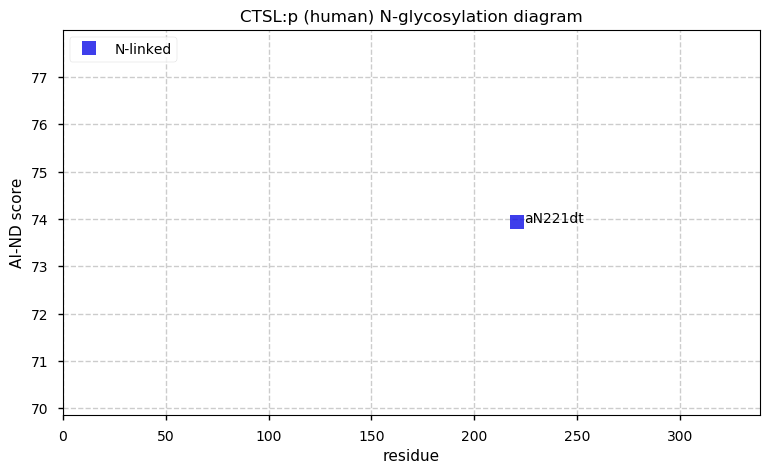
Tue May 26 23:35:08 +0000 2020@ypriverol It also uses the individual experiment XML files (I refer to them as "models") both as separate no-sql database-like objects for many purposes.
Tue May 26 23:20:00 +0000 2020@ypriverol MySql, with a lot of hardware, software and db optimization
Tue May 26 22:37:08 +0000 2020@Sci_j_my It will be a good place for your students to go to get hired. They aren't your competitors. At worst, they may be well-heeled collaborators
Tue May 26 22:22:32 +0000 2020@piefuchs We only tend to do that when it makes us look good, or at least makes Americans look bad.
Tue May 26 21:10:02 +0000 2020Once again, approaching a made-up milestone 🔗
Tue May 26 20:09:11 +0000 2020@JesseBrown It's an homage.
Tue May 26 19:08:59 +0000 2020@neely615 @ypriverol @pwilmarth @dtabb73 While there are many contradictory definitions of "big data" available on the web, there is a strong case to be made that "difficult/expensive to calculate" is a required, albeit ill-defined, part of many definitions.
Tue May 26 18:39:32 +0000 2020@VATVSLPR @Sci_j_my @OpenAcademics From my observations, that number seems to be "1" in almost all cases.
Tue May 26 17:44:49 +0000 2020@ypriverol @pwilmarth @dtabb73 @neely615 This suggests that solving a problem with a large data set is 'big data' only if it takes a lot of CPU-hours. If solving a 'big data' problem with 1 gTa of data takes 1000 CPU-hrs, is it still 'big data' if a new algorithm takes 0.1 CPU-hrs to solve it using the same 1 gTa?
Tue May 26 14:35:09 +0000 2020@pwilmarth @dtabb73 @neely615 Small letters with metric prefixes, e.g., my data resulted in 100 kilotabbs (kTa) of identifications (I know you were joking & maybe I am too, but maybe not ...) 🤔
Tue May 26 13:58:58 +0000 2020@neely615 I don't think any individual proteomics data set can be considered "big data": they are often too small even to detect the effects they are trying to measure. But, in aggregate, maybe?
Tue May 26 12:54:40 +0000 2020@neely615 I'm not clear on the temporal element, since proteomics data gathering is pretty low frequncy and non-parallel. Could you give an example?
Tue May 26 12:12:43 +0000 2020A consensus is forming about how big "big data" should be ... 🔗
Tue May 26 12:02:17 +0000 2020CTSK:p, cathepsin K (Homo sapiens) 🔗 Small lysosomal/extracellular hydrolase; PTMs: N103+glycosyl only; SAAVs: none; common in HLA type II data sets [2,332 x] 🔗
Mon May 25 15:23:49 +0000 2020@dtabb73 We'll see what the people think, but it looks like a done-deal right now.
Mon May 25 14:45:23 +0000 2020And if "PSM" in this context is a dimensionless unit representing the number of spectra assigned to a peptide sequence, should it have a name like other units, e.g., "dalton" or "mach"?
I could get behind calling it a "tabb".
Mon May 25 14:26:53 +0000 2020How big does a set of proteomics data have to be to qualify as "big data" (in giga PSMs, gPSM)?
Mon May 25 14:14:46 +0000 2020This thing is working so well I will probably have to do the hard part: coming up with some cutesy multi-word descriptive title that has a too-clever-by-half acronym 🔗
Mon May 25 12:15:44 +0000 2020CTSH:p, cathepsin H (Homo sapiens) 🔗 Small lysosomal hydrolase; PTMs: N72, N101, N230+glycosyl; SAAVs: G11R (8%), C26S (39%), K160R (1%); common in HLA type II data sets [2,998 x] 🔗
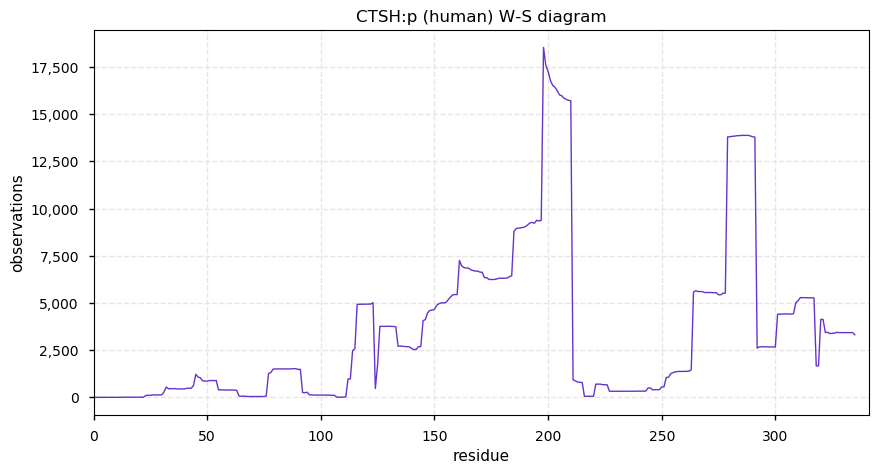
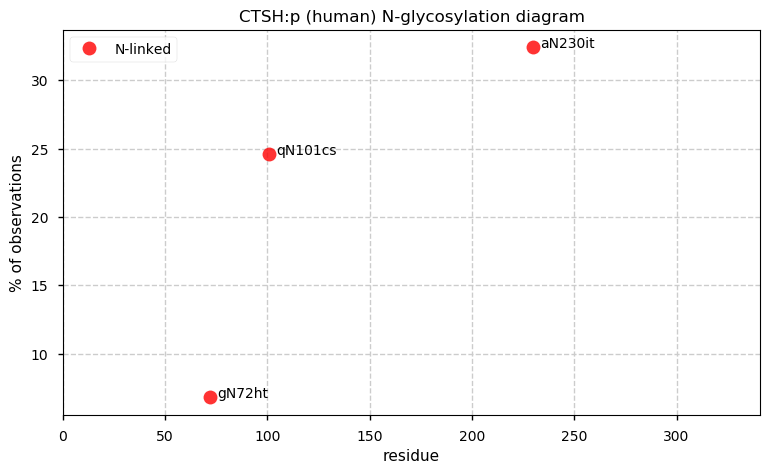
Sun May 24 17:45:51 +0000 2020@mhawkin2 @mekki Always has been, always will be.
Sun May 24 16:15:38 +0000 2020I think I've finally figured out a way to auto-curate observed N-linked sites out of GPMDB results (pats self on back) 🔗
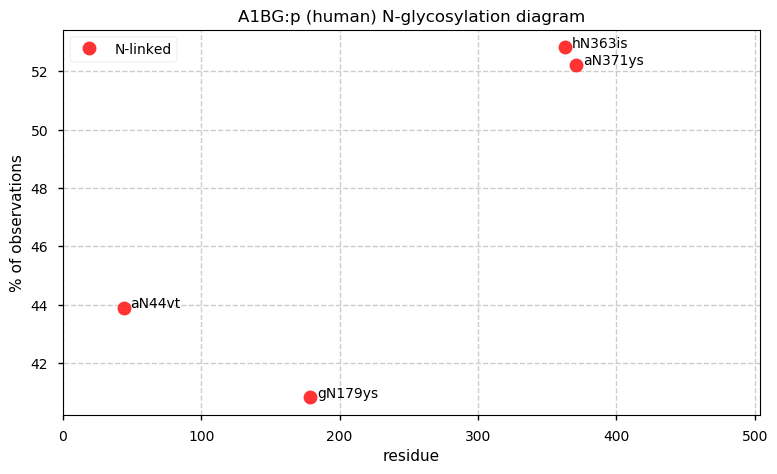
Sun May 24 15:56:36 +0000 2020Why am I always surprised that I can fix my own code?
Sun May 24 12:43:36 +0000 2020CTSF:p, cathepsin F (Homo sapiens) 🔗 Small lysosomal hydrolase; PTMs: N160, N195, N367, N378+glycosyl; SAAVs: R254S (41%); common in HLA type II data sets [2,998 x] 🔗
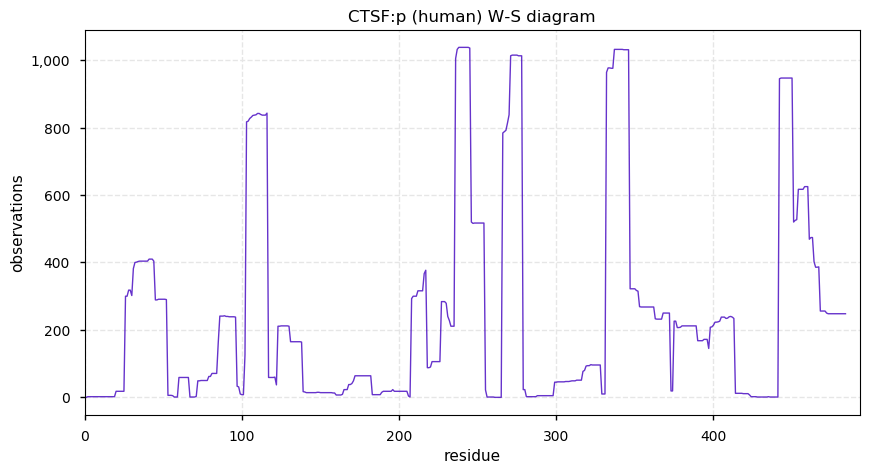
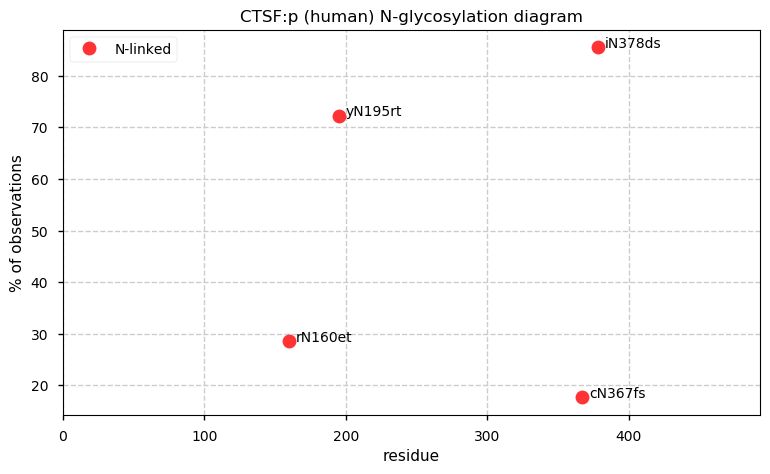
Sat May 23 14:20:21 +0000 2020@MattWFoster @veronicamars I alternate between Hearst and Barnett College NY, with occasional guest lectures at Miskatonic U. in Arkham MA. My vacation place is on the Costa Rican island, Isla Sorna.
Sat May 23 13:36:32 +0000 2020If you are interested in developing an MS-based SARS-CoV-2 clinical test, or just interested in what proteins are sampled by nasopharyngeal swabbing, this is the most interesting data I've seen so far 🔗
Sat May 23 12:45:02 +0000 2020The PTM pattern observed for CTSD strongly suggests an additional intracellular role for this protein outside of the lysosome.
Sat May 23 12:21:44 +0000 2020CTSD:p, cathepsin D (Homo sapiens) 🔗 Small lysosomal/extracellular protease; PTMs: N134, N263+glycosyl, 7 complementary K+acetyl/ubiquitinyl; SAAVs: G282R (1%); common in HLA type II data sets [55,365 x] 🔗
Fri May 22 19:46:16 +0000 2020A SARS-CoV-2 story, with an exciting subplot involving cysteine sidechain derivatization! 🔗 🔗

Fri May 22 19:40:34 +0000 2020Once again, cerbot saves the day (or at least makes the day much simpler).
Fri May 22 19:04:50 +0000 2020@Sci_j_my @VATVSLPR It does happen, but at much lower stoichiometry than eukaryotes & with species-dependent N-terminal residue specificity. To the best of my knowledge, protein N-terminal acetylation isn't a co-translational modification in prokaryotes: it is a PTM.
Fri May 22 16:38:43 +0000 20206. Unexpectedly, in 4/5 of the samples (pool-18,-34,-38,-51), the protocol led to good recovery of peptides with unmodified Cys (>10% of PSMs), similar to levels commonly found in protocols that produce Cys+carbamidomethyl. /eot
Fri May 22 16:38:43 +0000 20205. The IAA-Cys derivatization reaction did not work: there were effectively no PSMs with Cys+carbamidomethyl.
Fri May 22 16:38:43 +0000 20204. The protein digestion protocol used produced far less Met and Trp sidechain oxidation than is commonly generated by proteomics sample prep protocols, simplifying the results.
Fri May 22 16:38:43 +0000 20203. There is no significant problem caused by PSMs from the adventitious bacterial or viral microbiome in these swabbed samples.
Fri May 22 16:38:43 +0000 20203. The more intracellular protein PSMs in a sample, the more viral protein PSMs.
Fri May 22 16:38:43 +0000 20202. The SARS-CoV-2 "N" protein (nucleocapsid phosphoprotein) generates the most PSMs, >10× more than the next most abundant, the "S" (spike) protein. This agrees with other studies of infected cells and clinical samples.
Fri May 22 16:38:42 +0000 20201. The swabbing technique produces a variable mixture of plasma-derived extracellular fluid & intracellular proteins.
Fri May 22 16:38:42 +0000 2020The data set PXD019119 (Cardozo, Res. Sq. 2020 🔗) is full of interesting results.
Fri May 22 14:47:04 +0000 2020The human #sORF 12:96936664-96936723 (🔗), with the μ-protein sequence
MMEPLWLLDLPGGKYINMI* [obs. 52 x]
generates 1 PSM (7-15 [53 x]) when present in HLA type I peptide expts. 🔗

Fri May 22 13:46:31 +0000 2020@neely615 @Sci_j_my Anything that encourages venture investment in the field is a positive. Literally anything at all. A big player legitimizing the commercial prospects of proteomics of any type/technology is very, very good news.
Fri May 22 12:57:45 +0000 2020@Sci_j_my It also makes the government the funder, the approver and the consumer of the product.
Fri May 22 12:56:21 +0000 2020@Sci_j_my If there actually is something to buy: a government purchasing a huge number of doses of something that hasn't passed Phase III isn't normal practice. It distorts the economics of the development/approval process.
Fri May 22 12:50:11 +0000 2020@Sci_j_my @chenym Maybe there is another round of venture capital proteomics companies firing up: it has been almost 20 years since the first tranche.
Fri May 22 12:32:47 +0000 2020The whole vaccine business seems to be in chaos now 🔗
Fri May 22 12:25:03 +0000 2020I've been using Twython to do command line tweeting for a while now: probably won't go back to using the web/app interface
Fri May 22 12:01:14 +0000 2020CTSB:p, cathepsin B (Homo sapiens), Small lysosomal/extracellular protease; PTMs: N38, N192, N289+glycosyl; SAAVs: L26V (40%), S53G (6%), T75A (1%), S235N (4%), Q334P (1%); common in HLA type II data sets [35,501 x] 🔗
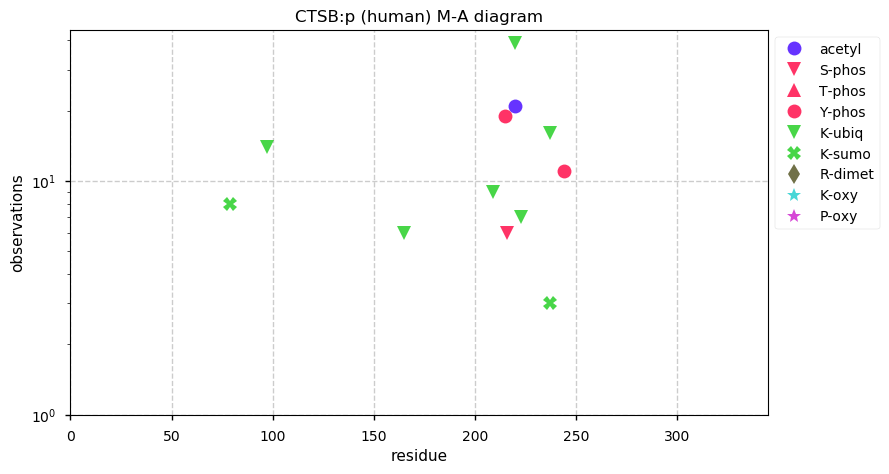
Thu May 21 19:19:22 +0000 2020Ground station rainfall amounts for 2020-05-17 and -18 in the vicinity of Midland, MI 🔗
Thu May 21 15:05:28 +0000 2020Seems like a bridge too far for me, but then again I can't stand Nat. Methods so maybe an improvement? 🔗
Thu May 21 14:48:52 +0000 2020@jwoodgett Virtual isn't going to work so well for fitting N95s or the other practical exercises that were part of the training as I remember it.
Thu May 21 14:39:20 +0000 2020@jwoodgett Does Mt. Sinai still enforce having new employees at the research institute take mandatory PPE/infection control training?
Thu May 21 14:35:06 +0000 2020The human #sORF 8:125083952-125091621 (🔗), with the μ-protein sequence
MTGSLMEEPISLHTWFHLIIYRHQL* [obs. 31 x]
generates 2 PSMs (6-15 [16 x], 7-15 [23 x]) when present in HLA type I peptide expts. 🔗

Thu May 21 12:16:36 +0000 2020DNASE2:p, deoxyribonuclease 2, lysosomal (H. sapiens) 🔗 Small lysosomal enzyme; PTMs: N86, N212, N266, N290+glycosyl; SAAVs: H204R (1%); mature sequence 17-360 [10,293 x] 🔗
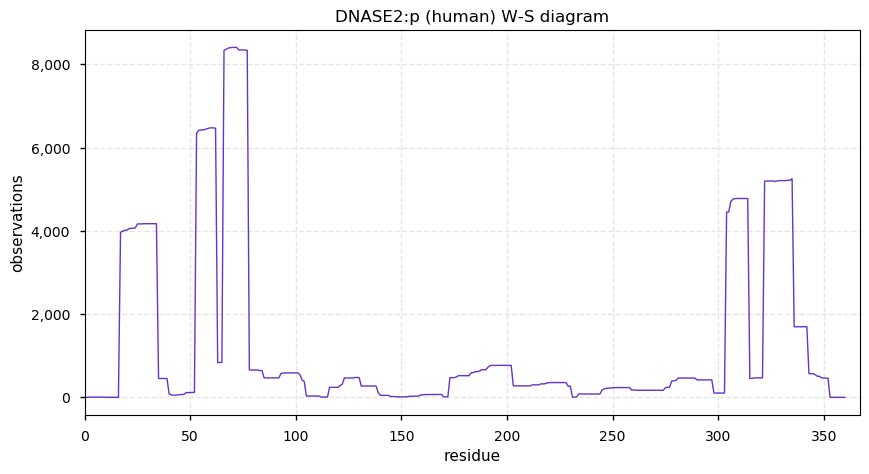
Wed May 20 20:13:44 +0000 2020BTW: anyone complaining that PubMed should "go back to the way it was yesterday" is now officially an "old-timer". Young people who work for you will enjoy your stories about how great things were back in the day.
Wed May 20 18:33:34 +0000 2020Sitting on buildings and honking at passersby has become a daily feature of goose life downtown.
Wed May 20 18:33:34 +0000 2020You have to look close, but there are 2 of them
Wed May 20 18:33:33 +0000 2020The goosegoyles of Winnipeg 🔗

Wed May 20 17:18:24 +0000 2020@pwilmarth The new version also has a tracker in it, probably not out of ill intent, but simple it-came-up-in-a-meeting-and-no-one-was-willing-to-say-no.
Wed May 20 15:37:20 +0000 2020@chrashwood @CarolynBertozzi While the N-glycans may be simply for lysosome targeting, it seems as though there are a lot of N-sites on the proteins when 1 should be enough to achieve the targeting function. It is also a rather elaborate mechanism compared to other compartment targeting methods.
Wed May 20 15:01:40 +0000 2020@AlexUsherHESA I rather liked
"You can reduce a seminar to a distortion-addled screen, sure, but that will never substitute for being there."
Wed May 20 14:38:11 +0000 2020Total made-up milestone:
GPMDB mean peptide redundancy = 500 x
Wed May 20 14:35:21 +0000 2020@MHendr1cks @AlexUsherHESA It is more like everyone is in the back row & not with the cool kids either.
Wed May 20 14:29:57 +0000 2020@MHendr1cks @AlexUsherHESA I had to endure that type of lecture in 1st year classes back in the 70's - although presented in a theatre rather than a small screen. They were awful then and I'm sure they have gotten worse since.
Wed May 20 14:18:43 +0000 2020@chrashwood It wouldn't surprise me if NIAID would be receptive to that sort of study. I was on the SAB of the U of Georgia NCRR glycomics center for about a decade & I don't remember anyone talking about the subject during the meetings.
Wed May 20 14:16:38 +0000 2020@chrashwood It seems to me that given the number of N-linked sites on some lysosomal matrix proteins & the current administration's fascination with lysosomes, really getting in to the detail of those glycoforms (& there potential effects on the HLA type II system) might be an astute move.
Wed May 20 12:59:10 +0000 2020Glyco-tweeps: are the N-linked glycoforms of lysosomal matrix proteins representative of the glycoforms of secreted (or cell surface) proteins or are they special in some way?
Wed May 20 12:38:23 +0000 2020The human #sORF 2:169572071-169572130 (🔗), with the μ-protein sequence
MKYMKEKLLFSLLIAPDD* [obs. 68 x]
generates 2 PSMs (2-10 [160 x], 3-10 [3 x]) when present in HLA type I peptide expts.
Wed May 20 12:29:52 +0000 2020I am finding lysosomal matrix proteins strangely interesting.
Wed May 20 11:56:00 +0000 2020GAA:p, glucosidase alpha, acid (H. sapiens) 🔗 Lysosomal enzyme; PTMs: N140,N233,N390,N470,N652,N882,N925+glycosyl; SAAVs: H199R (40%), R223H (40%), E689K (8%), V780I (29%), V816I (10%), T927I (5%); mature (70,123,204)-952 [26,862 x] 🔗
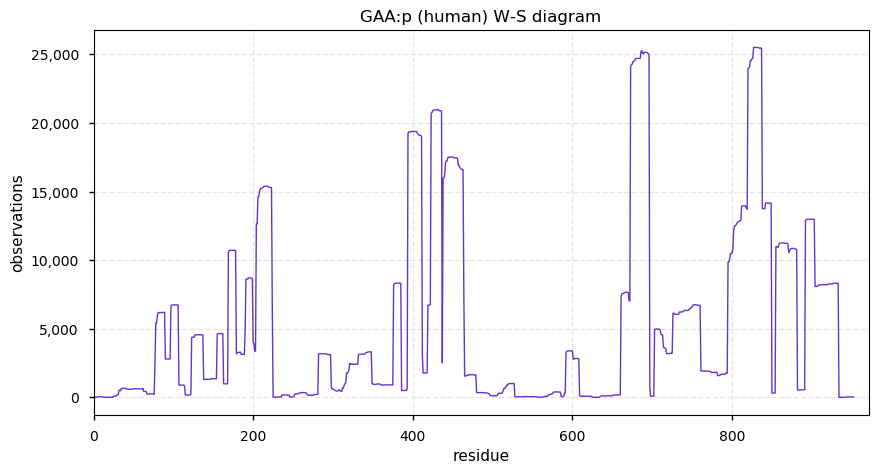
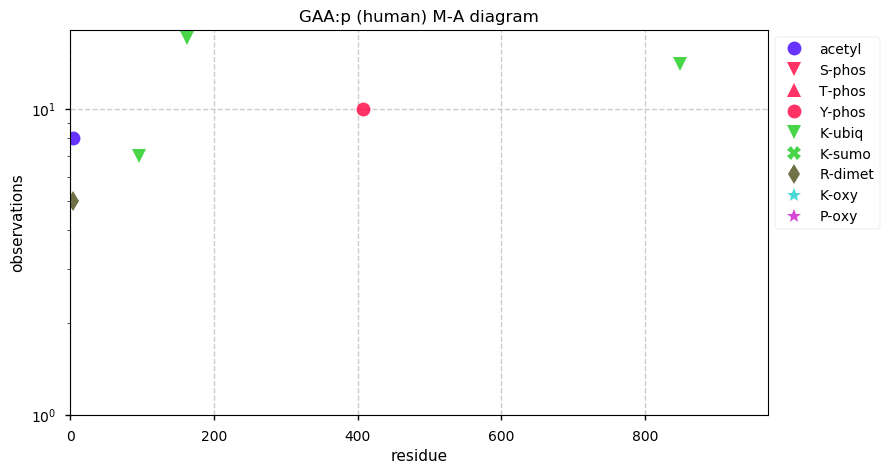
Tue May 19 17:42:43 +0000 2020@SpecInformatics Bad. No question.
Tue May 19 17:32:38 +0000 2020@SpecInformatics No, but it wouldn't surprise me if that became a thing.
Tue May 19 15:31:54 +0000 2020Has anybody else had trouble downloading data from iPROX lately? I keep on getting error messages, using either Aspera or HTTP.
Tue May 19 14:35:09 +0000 2020The UK daily fatality rate is starting to align with other, hard-hit European countries 🔗
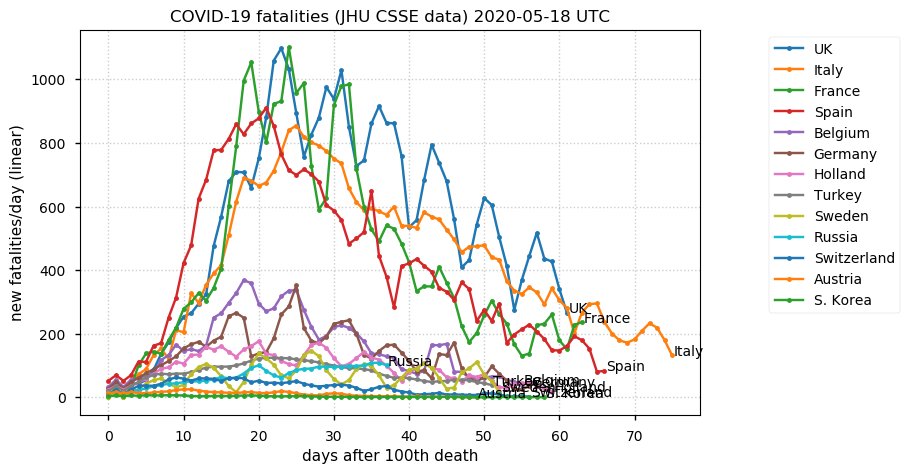
Tue May 19 12:34:02 +0000 2020The human #sORF 7:112790313-112790372 (🔗), with the μ-protein sequence
MAVAAGPVTEKVYADTGLY* [obs. 133 x]
generates 3 PSMs (8-17 [31 x],8-19 [220 x],11-19 [31 x]) when present in HLA type I peptide expts.
Tue May 19 12:26:51 +0000 2020ACPP:p, acid phosphatase prostate (Homo sapiens) 🔗 Small secreted/lysosomal enzyme; common in urine, prostate, ovary; no PSM overlap with other genes; PTMs: N94,N220,N333+glycosyl; SAAVs: D301E (1%); mature sequence 33-418 [7,056 x] 🔗
Mon May 18 15:01:29 +0000 2020@slavovLab My own experience with any type of "Resource" paper has been very hit and miss: they usually aren't archival quality. Quantity tends to win over quality. If the paper doesn't have a good QA/QC description, it probably isn't worth the bandwidth (or the postage).
Mon May 18 14:06:35 +0000 2020PXD016126 is an interesting study of the salivary proteome. The variability in the salivary microbiome is particularly nicely illustrated by the data. NOTE: the LC/MS/MS has much wider parent ion mass accuracy distributions than are common for this type of instrument.
Mon May 18 13:42:18 +0000 2020@behindthenet @AlexUsherHESA It will always be "Goon" for me.
Mon May 18 12:48:22 +0000 2020The human #sORF 1:179865721-179865783 (🔗), with the μ-protein sequence
MSSQDRLHWIHDQEDSARFF* [obs. 31 x]
generates 1 PSMs (10-20 [45 x]) when present in HLA type I peptide expts.
Mon May 18 12:42:43 +0000 2020The population distribution of ACP2:p.R29Q is interesting, given that R29 is in the signal sequence (ACP2:r is translated into the ER) & therefore not part of the mature enzyme 🔗
Mon May 18 12:36:12 +0000 2020ACP2:p, acid phosphatase 2 (Homo sapiens) 🔗 Small lysosomal enzyme; PTMs: N92,N133,N167,N177,N267,N331+glycosyl; SAAVs: R29Q (48%); mature sequence 31-423 [12,303 x] 🔗
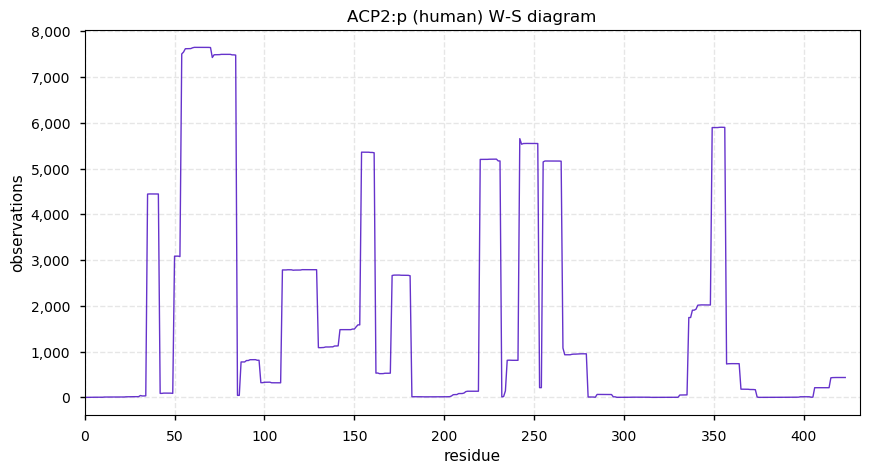
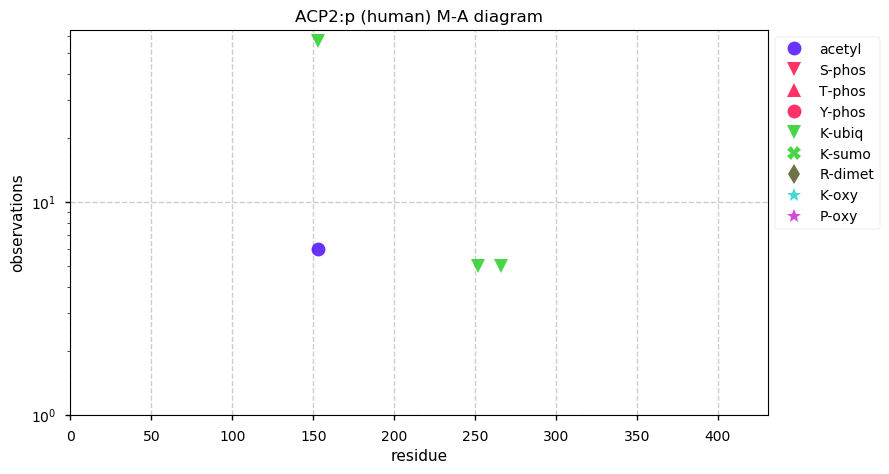
Sun May 17 13:24:40 +0000 2020@neely615 @Smith_Chem_Wisc What sort of barbarian pronounces SQL as a word?
Sun May 17 12:47:22 +0000 2020The human #sORF 10:43644655-43644771 (🔗), with the μ-protein sequence
MTLRLQDPQAGISKILSEERSWNKNPQIRRHYRKIHLE* [obs. 73 x]
generates 1 PSMs (4-14 [106 x]) when present in HLA type I peptide expts.
Sun May 17 12:33:03 +0000 2020LIPA:p, lipase A, lysosomal acid type (Homo sapiens) 🔗 Small lysosomal enzyme; PTMs: N72,N161,N273,N321+glycosyl; SAAVs: T16P (29%), G23R (12%); common in HLA type II peptide experiments [7,746 x] 🔗
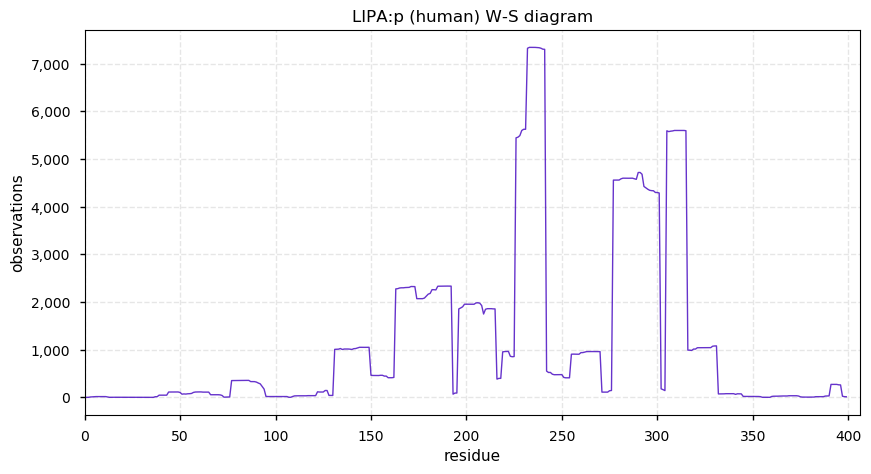
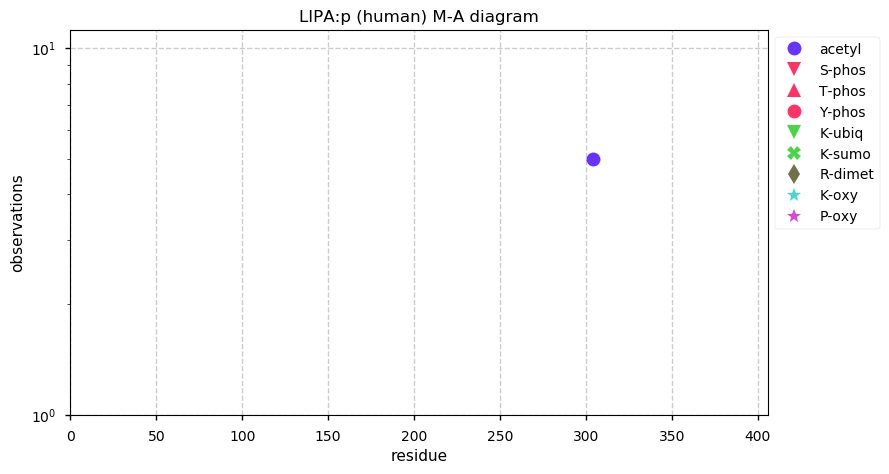
Sat May 16 18:09:07 +0000 2020@TanentzapfLab Not really a ringing endorsement of CIHR's current status in the Canadian academic biomedical research community.
Sat May 16 13:32:55 +0000 2020PS - I would vote for a 5% QA limit wrt non-tryptic PSMs for this type of experiment.
Sat May 16 13:05:32 +0000 2020The human #sORF 22:19068220-19089394 (🔗), with the μ-protein sequence
MRATKPTVQK* [obs. 116 x]
generates 2 PSMs (1-10 [10 x], 2-10 [282 x]) when present in HLA type I peptide expts.
Sat May 16 12:38:18 +0000 2020IFI30:p, lysosomal thiol reductase (Homo sapiens) 🔗 Small lysosomal enzyme; PTMs: T145,T174,T209,T219+phospho; SAAVs: R76Q (18%); common in HLA type II peptide experiments [10,613 x] 🔗
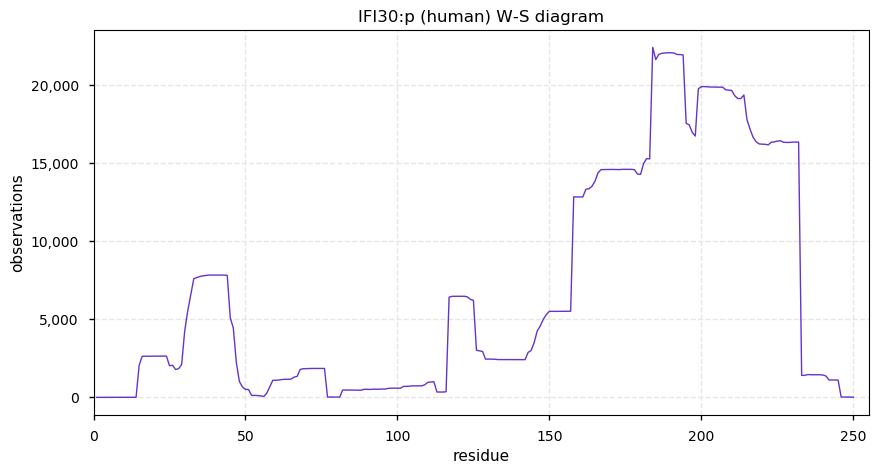
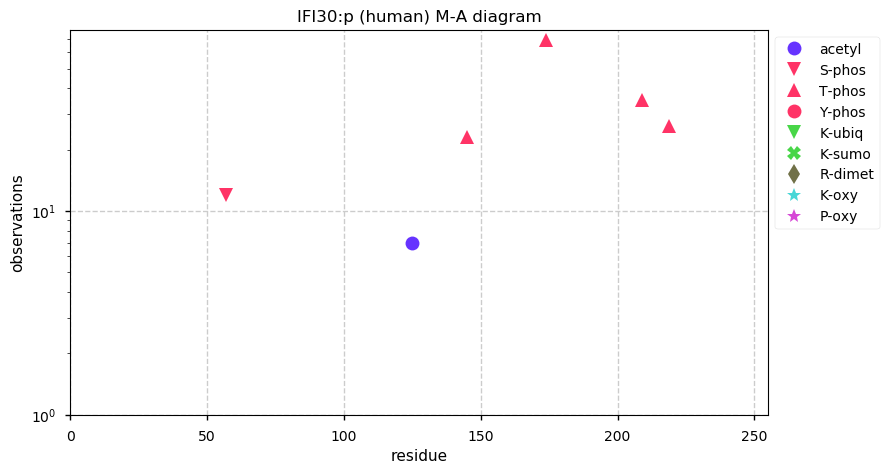
Sat May 16 02:28:54 +0000 2020Hmm, odd timing ... 🔗
Fri May 15 18:17:47 +0000 2020Thanks to everyone who provided an opinion for this poll. About 70% of respondents felt the need for QA requirement based on non-tryptic PSMs & 30% did not. The appropriate fraction of non-tryptic PSMs was pretty much evenly split between five and ten percent.
Fri May 15 16:21:46 +0000 2020@bkives And "Great Again"?
Fri May 15 15:22:25 +0000 20202 hours left and the results are very close to a 3-way tie!
Fri May 15 15:15:26 +0000 2020@jwoodgett In the future, when this little era of history is examined, the phrase "made a lot of sense" will be rarely used wrt to political leadership of any stripe.
Fri May 15 14:59:54 +0000 2020@cwvhogue @scottagerber Good. I never really know what to do with "no phenotype" mouse knockout results, as most studies don't involve a lot of real-world situations for mice.
Fri May 15 14:42:43 +0000 2020@cwvhogue @scottagerber I will take a look. Does it address the lack of a phenotype when MVP is knocked out?
Fri May 15 13:57:09 +0000 2020@lgatt0 @mvaudel @Bioconductor Code documentation usually provides a pretty good insight into the author(s) & their intentions wrt to the software's utility.
Fri May 15 13:21:23 +0000 2020Everything about vault particles is weird: usually a sign that our current understanding is incomplete/wrong.
Fri May 15 13:07:02 +0000 2020This type of high MAF SAAV accumulation would normally be associated with a pseudogene, rather than an abundantly translated gene.
Fri May 15 12:54:57 +0000 2020Like the other large vault protein TEF1:p, PARP4:p has an unusually large number of high MAF SAAVs, with 21 SAAVs with MAF ≥ 0.01 and 11 SAAVs with MAF ≥ 0.10.
Fri May 15 12:48:37 +0000 2020PARP4:p, poly(ADP-ribose) polymerase family member 4 (H. sapiens) 🔗 Large vault protein; many PTMs; SAAVs (>20%): P491L (38%), S873N (22%), A899T (35%), G1265A (39%), G1280C (39%), G1280R (39%), P1328T (39%), A1656P (42%); mature form (2,4)-1724 [18,285 x] 🔗
Fri May 15 12:43:27 +0000 2020The human #sORF 19:50799328-50802228 (🔗), with the μ-protein sequence
MPIQVLKGLTITH* (obs. 16x)
generates 1 PSM — 1-9 (21x) — when present in HLA type I peptide expts.
(1-9)|
Fri May 15 12:19:45 +0000 2020@Bioschema Yes, that one popped up overnight. I'll look at it today.
Thu May 14 23:10:26 +0000 2020@Bioschema You might also want to check for cysteine derivatized by acrylamide (+71 Da) in the gel band data, in addition to the IAA. It will give you about 10% more good PSMs.
Thu May 14 22:57:37 +0000 2020@Bioschema Sorry, I had meant to include the word "human" along with "cell line". The SARS-CoV-2 infected Vero cell data also has significant signals from virus proteins.
Thu May 14 18:20:22 +0000 2020@idpgrace You must love VCF files.
Thu May 14 18:07:43 +0000 2020In a TMT10 expt that used trypsin/lysC to generate peptides from a cell lysate, what is the maximum acceptable fraction of PSMs assigned to non-tryptic peptides:
Thu May 14 15:34:52 +0000 2020The Canadian fatality trends still show QC on an upwards trajectory, even after 2 months of travel, school & business restrictions. ON is not trending up, but it isn't trending down either. 🔗
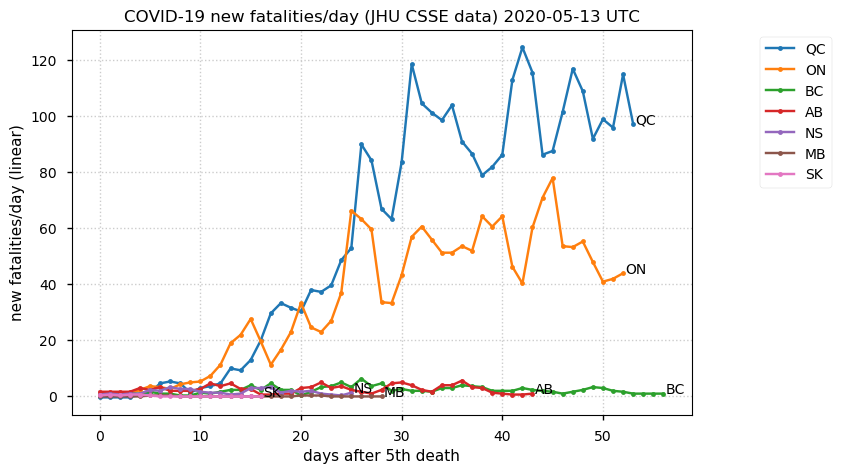
Thu May 14 14:09:31 +0000 2020Several months into the pandemic, PXD017710 remains the only publicly available data set with good signals from SARS-CoV-2 virus proteins that were obtained from an infected cell line (CACO-2, with easily observable ACE2:p).
Thu May 14 12:29:22 +0000 2020The human #sORF 3:131501588-131501683 (🔗), with the μ-protein sequence
MERVVHGGMSIFLKKMSHSLSSWSLMKIKPN* (obs. 78x)
generates 5 PSMs — 3-14 (24x), 3-15 (11x), 4-14 (150x), 7-15 (70x), 19-27 (13x) — when present in HLA type I peptide expts.
Thu May 14 12:04:27 +0000 2020TEP1:p has an unusually large number of high MAF SAAVs.
Thu May 14 11:56:39 +0000 2020TEP1:p, telomerase associated protein 1 (Homo sapiens) 🔗 Large vault protein; low occupancy phosphoryl and ubiquitin PTM sites, except S397+phospho; SAAVs (>30%): S116P (36%), R1055C (37%), V2214I (32%), I2486M (34%); mature form 1-2627 [5,343 x] 🔗
Wed May 13 16:57:08 +0000 2020Russia has overtaken other European countries in confirmed cases, with 11,000 new cases yesterday. 🔗
Wed May 13 16:00:35 +0000 2020@bkives I think BP probably holds some type of record as the angriest man in Canadian political history.
Wed May 13 14:19:53 +0000 2020Most of the listed protein-protein interactions for MVP are probably artifacts, except for PARP4 and TEP1 (picture from STRING v. 11) 🔗
Wed May 13 12:42:31 +0000 202078 MVP molecules form the exterior structure of a complex organelle commonly referred to as a "vault particle" 🔗 Abundant in eukaryotic cells, but of unknown function.
Wed May 13 12:36:07 +0000 2020MVP:p, major vault protein (Homo sapiens) 🔗 Midsized subunit; significant PTMs (23 phosphorylation sites); SAAVs: R27H (1%); abundant in cells; mature form 2-893 [38,084 x] 🔗
Wed May 13 11:59:23 +0000 2020The human #sORF 17:8379894-8382174 (🔗), with the μ-protein sequence
MCPSERMMKFRLYVDTIKVSKLAK*
generates 3 PSM (11-21, 14-21, 15-24) when present in HLA type I peptide expts (obs. 109 x).
Tue May 12 12:30:16 +0000 2020The human #sORF 1:179865721-179865783 (🔗), with the μ-protein sequence
MSSQDRLHWIHDQEDSARFF*
generates 1 PSM (10-20) when present in HLA type I peptide expts (obs. 31 x).
Tue May 12 12:21:16 +0000 2020I can put up with fraud in the scientific literature. There has been lots of it in the history of published papers & I don't think it has any lasting impact. For me, the modern threat to general confidence in this peculiar literary form is flagrant exaggeration.
Tue May 12 12:12:52 +0000 2020PEX26:p, peroxisomal biogenesis factor 26 (Homo sapiens) 🔗 Small peroxisomal membrane protein; PTMs: M1+acetyl, S(211,213)+phosphoryl; no SAAVs; mature form 1-305 [1,140 x] 🔗
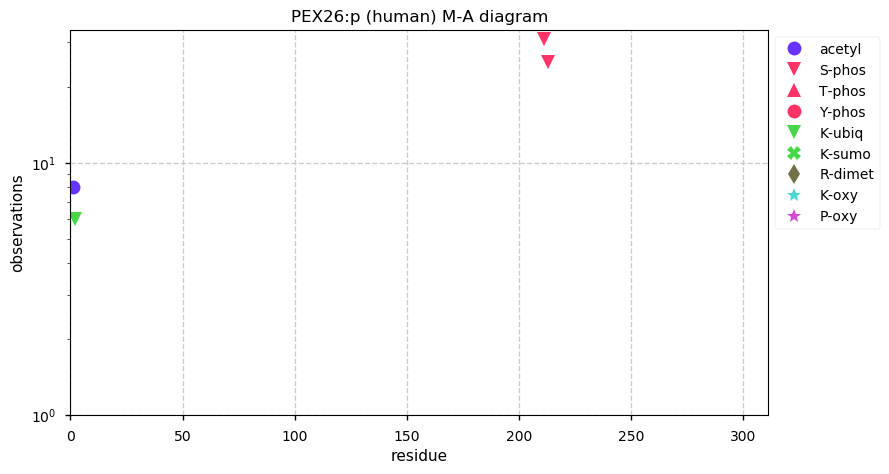
Mon May 11 13:32:21 +0000 2020PEX19:p, peroxisomal biogenesis factor 19 (Homo sapiens) 🔗 Small cytoplasmic protein; complex pattern of PTMs; no SAAVs; mature form (1,2,3)-283 [15,391 x] 🔗
Mon May 11 13:00:07 +0000 2020The human #sORF 12:118255649-118266698 (🔗), with the μ-protein sequence
MNLSNMEYFVPHTKRY*
generates 4 PSMs (6-16, 8-16, 9-16, 10-16 [18:151:7:3]) when present in HLA type I peptide expts (obs. 114 x).
Sun May 10 14:05:41 +0000 2020One of the many things I like about MHC/HLA peptide expts is that it is the only common style of proteomics expt that does not actively discriminate against short (< 10 kDa) protein sequences.
Sun May 10 13:34:18 +0000 2020The human #sORF 6:24720332-24720439 (🔗), with the μ-protein sequence
MGPRWGLSGGSSAGGPVTVRTAAGMEGLSWLVVFS*
generates 4 PSMs (11-20, 12-20, 15-23, 20-30) when present in HLA type I peptide expts (obs. 103 x).
Sun May 10 13:18:23 +0000 2020PEX16:p, peroxisomal biogenesis factor 16 (Homo sapiens) 🔗 Small peroxisomal membrane protein; PTMs: S158+phosphoryl (85 x), S183+phosphoryl (45 x); SAAVs: V103M (1%); mature form 1-336 [3,581 x] 🔗
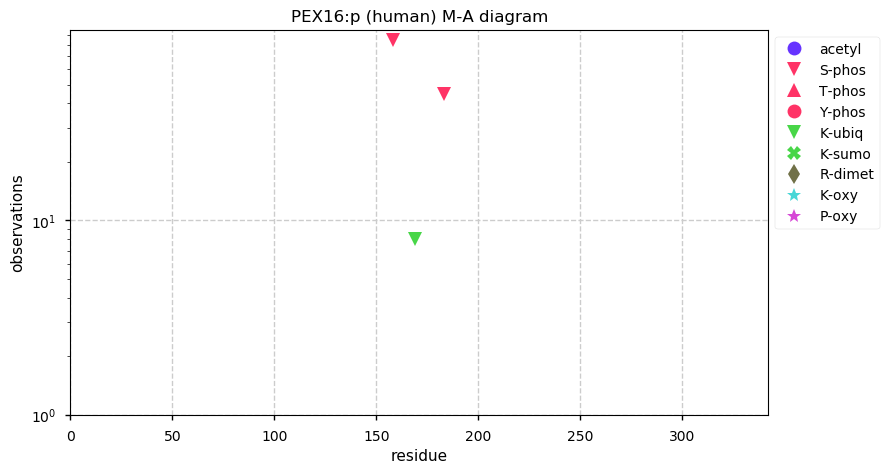
Sat May 09 22:45:12 +0000 2020For anyone who wants to know what a really good Trypanosoma brucei data set looks like, try PXD016370. Lots of proteins of unknown function, with very good signals and chromatography.
Sat May 09 15:30:56 +0000 2020@nesvilab Possibly, although I suspect most people intuitively under-estimate the fraction of PSMs that will contain C residues, assuming that it is close to cysteine's overall abundance in proteins (2%), rather than the actual number (20%).
Sat May 09 15:24:27 +0000 2020For everyone that participated in this poll, the correct answer is ~20% of PSMs should contain at least one cysteine residue, although it can be as high as ~30% for blood plasma studies.
Sat May 09 14:39:17 +0000 2020Thanks to everyone who participated in this poll. A large majority of the respondents (80%) do not check the efficacy of their cysteine sulfhydryl-blocking reaction prior to publishing the results.
Sat May 09 13:56:29 +0000 2020@leprevostfv Given Go's commercial popularity, it may be a good idea to train students to do more bioinformatics development using the language.
Sat May 09 13:21:37 +0000 2020The human #sORF 3:183717657-183717713 (🔗), with the microprotein sequence
MLLCRRLRLLSKNSLLLK*
generates 1 PSM (8-18) when present in HLA type I peptide experiments (obs. 198 x).
Sat May 09 13:09:05 +0000 2020Brazil is clearly having a serious problem, but so is much smaller Peru. 🔗
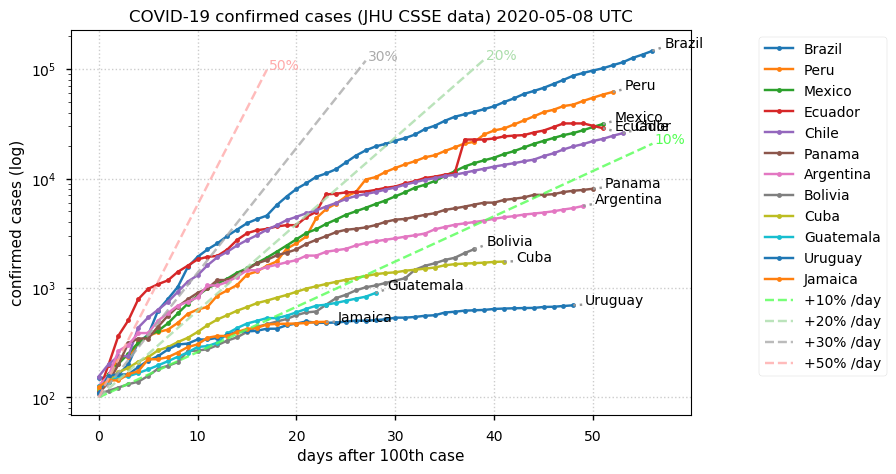
Sat May 09 12:46:16 +0000 2020Still one hour left, but the trend seems inevitable at this point.
Sat May 09 12:40:49 +0000 2020The number and pattern of PTMs observed on PEX14:p is very different than the other small PEX proteins with membrane spanning domains.
Sat May 09 12:33:18 +0000 2020PEX14:p, peroxisomal biogenesis factor 14 (Homo sapiens) 🔗
Small peroxisomal membrane protein; 3 phosphodomains; SAAVs: S70A (1%), V159M (3%), R320K (1%); mature form 2-377 [13,137 x] 🔗
Fri May 08 20:25:41 +0000 2020Wrote my first non-trival Go program. Not that hard a language to learn (if you already know half-a-dozen other languages with similar syntax).
Fri May 08 19:37:03 +0000 2020@jwoodgett There are other factors, aside from funding. There are very few virologists or epidemiologists at Canada's major biomedical research institutions. Without the PI's at influential places, things are going to go back to normal ASAP.
Fri May 08 18:26:26 +0000 2020@Sci_j_my As someone who has been married for a while, your significant other's opinion is correct & it doesn't matter what it is or how often it changes: it is correct by definition.
Fri May 08 18:10:21 +0000 2020This issue is similar to the toilet paper "hoarding" problem: commodity items meant for commercial markets are difficult to quickly re-purpose for household use.
🔗
Fri May 08 17:09:10 +0000 2020The human #sORF 17:59155144-59155179 (🔗), with the microprotein sequence
MSTIQHGGGGR*
generates 3 PSMs (2-11,3-11,4-11) when present in HLA type I peptide experiments.
Fri May 08 14:58:47 +0000 2020Follow-up: which choice is closest to the fraction of PSMs
that should contain at least one cysteine in a cell-contents type proteomics experiment, if the blocking reaction is complete?
Fri May 08 14:29:16 +0000 2020Using the software that you commonly use for proteomics data analysis, do you routinely check to determine the completeness of the cysteine side-chain-blocking reaction used in your experiments, prior to publication?
Fri May 08 13:59:15 +0000 2020@MHendr1cks It is a good chance to top-up funding for the SPOR Centres.
Fri May 08 13:00:11 +0000 2020PEX13:p, peroxisomal biogenesis factor 13 (Homo sapiens) 🔗
Small peroxisomal membrane protein; PTMs: A2+acetyl (564x), S205+phosphoryl (8x); SAAVs: L360V (1%); mature form 2-403 [2,802 x] 🔗
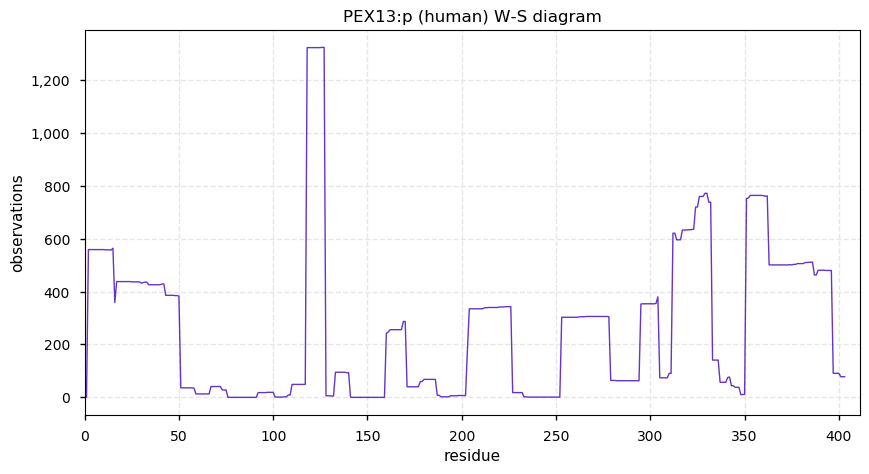
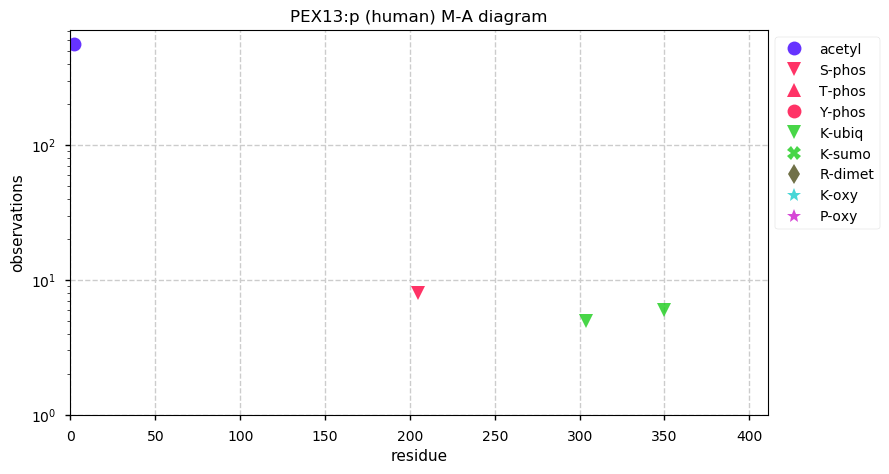
Thu May 07 18:05:08 +0000 2020The human #sORF 15:90875659-90875691 (🔗), with the microprotein sequence
MSRHLGAEAL*
generates 1 PSM (2-10) when present in HLA type I peptide experiments. The PSM N-terminal S2 is not acetylated.
Thu May 07 12:01:58 +0000 2020@juan_vizcaino @NatureMedicine It is a good example of faux open data. It appears to be available, but because of the way the system works it is practically inaccessible.
Thu May 07 11:43:50 +0000 2020PXD013649 is a pretty good set of data for anyone interested in understanding the detection of MHC I or II type peptides associated with different HLAs, from probably the best experimental group in this field.
Thu May 07 11:30:17 +0000 2020PEX12:p, peroxisomal biogenesis factor 12 (Homo sapiens) 🔗 Small peroxisomal membrane protein; PTMs: A2+acetyl; expt. PPIs with 6 PEX proteins (1,3,5,10,13,19); no SAAVs; mature form 2-359 [797 x] 🔗
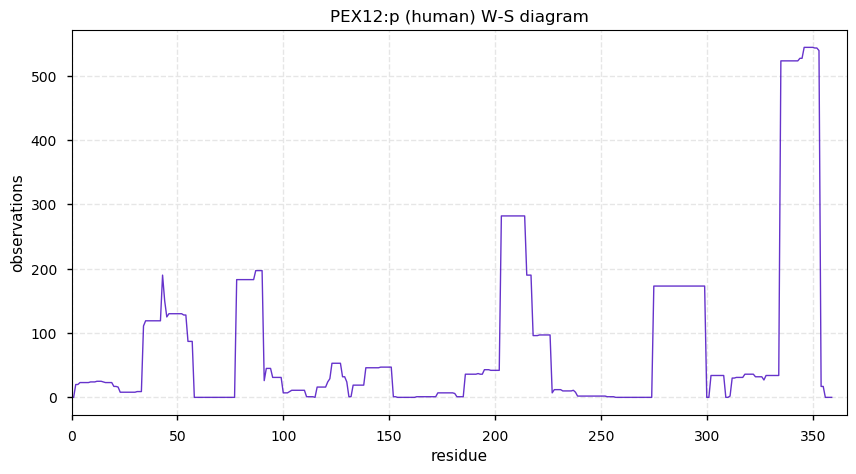
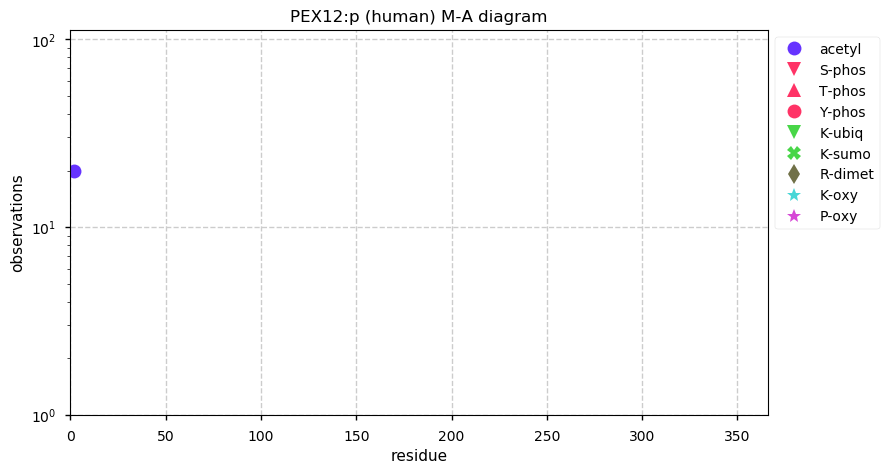
Thu May 07 00:11:08 +0000 2020@nbandeira @lkpino It should show up at 🔗 I had originally downloaded the data from Zendo, but it looks like it is the same files in the ProteomeXchange submission.
Wed May 06 23:59:17 +0000 2020@nbandeira @lkpino For that one, yes, about 5% ID rate for the phosphorylation data and significant phosphorylation for virus proteins.
Wed May 06 23:29:01 +0000 2020@nbandeira @lkpino My redo is at 🔗 The overall ID rate is ~20%, with 60-70% enrichment of phosphopeptide PSMs. I am using 20 ppm for the parent and 20 ppm for the fragments. No need for green monkey sequences with this data. It is just H1299 cells, but both SARS-COV & -COV2
Wed May 06 21:30:20 +0000 2020@nbandeira @lkpino What were you hoping to find? There are phosphopeptides, although none from SARS-COV2 proteins.
Wed May 06 17:11:30 +0000 2020An hour later, there are only 9h 37m left!
I really, really hate downloading large files via HTTP.😡
Wed May 06 16:04:20 +0000 2020Downloading the gnomad r3.0 VCF file: only 9 hours left to go!
Wed May 06 15:57:27 +0000 2020@NatureCustom @GrailBio Yup. This is what the "Nature" masthead has become, folks.
Wed May 06 15:08:49 +0000 2020When should the whole process of changing gene symbols every few years stop? Is there ever going to be an end to human genome releases & patches? There are plenty of species with genomes that need additional work, but Homo sapiens is not one of them.
Wed May 06 14:56:51 +0000 2020The human #sORF 6:34792125-34792172 (🔗), with the microprotein sequence
MAAAAAVSGAHAAAR*
generates up to 4 different PSMs when present in HLA type I peptide experiments. The N-terminal Met is not removed or acetylated.
Wed May 06 13:16:01 +0000 2020I noticed yesterday that HGCN had changed the names for the cytoplasmic tRNA synthetase genes. Rather than VARS or AARS, they are now VARS1 and AARS1. Really? Are they trolling us?
Wed May 06 12:54:26 +0000 2020The ratio of the number of observations in humans for the three PEX11 proteins is A:B:G, 597:10994:1192 (1:18:2). The three protein sequences do not share any PSMs.
Wed May 06 12:48:00 +0000 2020PEX11G:p, peroxisomal biogenesis factor 11 gamma (Homo sapiens) 🔗 Small peroxisomal membrane protein; PTMs: A2+acetyl, S164,T240+phosphoryl; ; SAAVs: C91W (46%); mature form 2-241 [1,192 x] 🔗
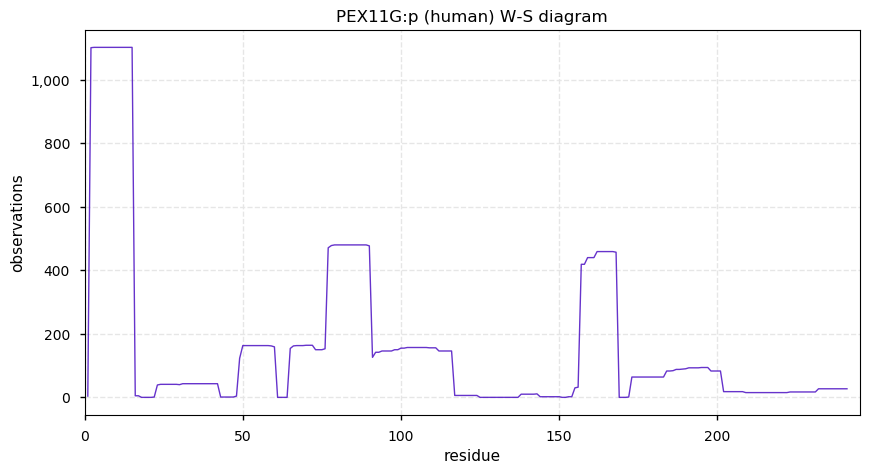
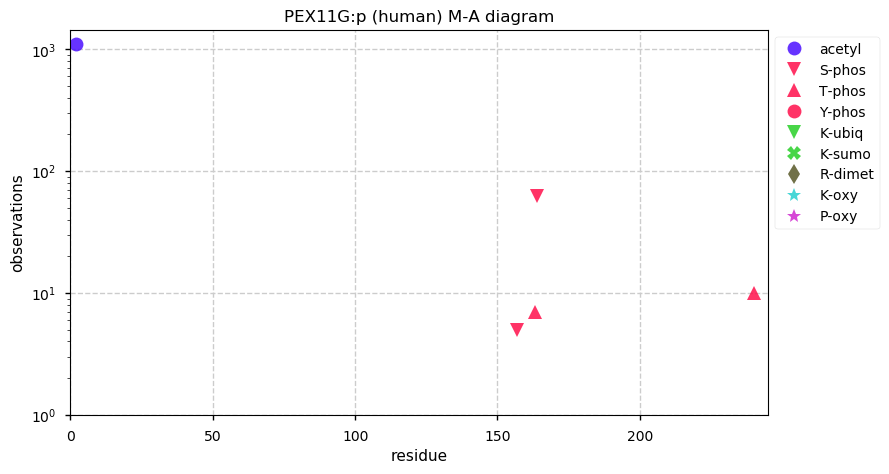
Wed May 06 12:31:12 +0000 2020The number of new cases/day is remaining high in North America & globally. 🔗
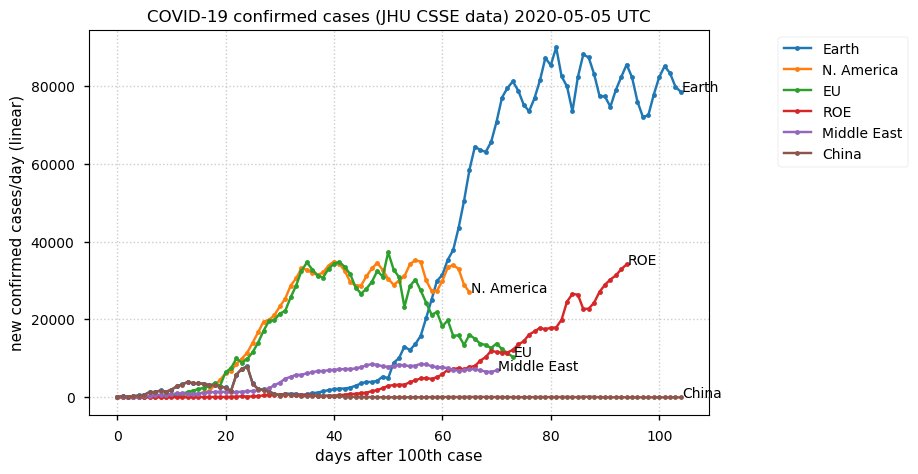
Tue May 05 21:25:40 +0000 2020The human sORF 8:11853325-11853372 (🔗), with the microprotein sequence
MSWSTMSTNGIPRGR*
produces the most PSMs of any sORF in HLA type I experiments (& I don't know why).
Tue May 05 14:44:51 +0000 2020@ypriverol The practical scientific use of ontologies (& controlled vocabularies) aren't as intuitive as most Comp. Sci. folks tend to think and some principles (e.g., rooted directed acyclic graphs) can be tough to adapt to how biological isolates are obtained.
Tue May 05 14:40:49 +0000 2020@ypriverol Thanks for the invitation, but I am not really the right guy for that type of project. I was only interested in finding out whether there was somewhere that scientists & biomed researchers could get some training wrt the practical use of ontologies to describe samples.
Tue May 05 12:39:19 +0000 2020PEX11B:p, peroxisomal biogenesis factor 11B (H. sapiens) 🔗 Small peroxisomal membrane protein; PTMs: M1+acetyl, S43,S160,S168+phosphoryl; commonly observed in tissues & cell lines; no SAAVs; mature form 1-259 [10,994 x] 🔗
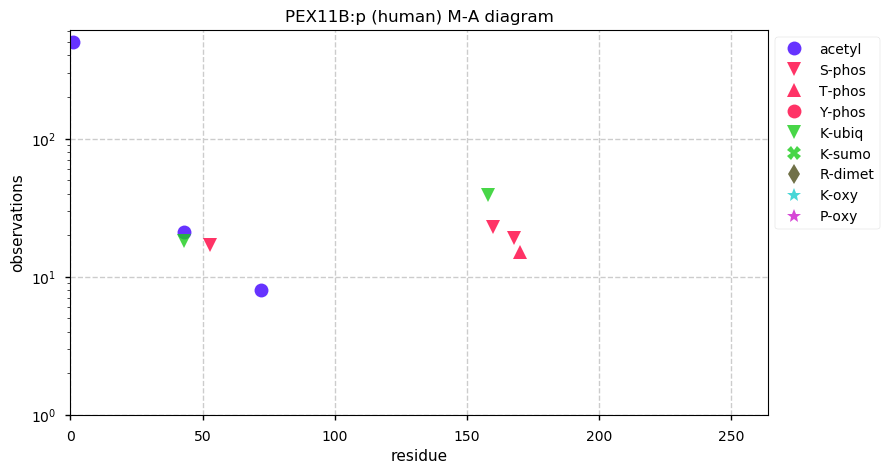
Mon May 04 15:26:00 +0000 2020Does any conference/meeting provide a short course on how to use ontologies (e.g., BRENDA) to describe samples? I see a lot of odd (and not very helpful) use of these terms.
Mon May 04 12:44:46 +0000 2020This data set 🔗 seems to have been announced a bit early. The associated iPROX entry is still restricted/not publicly available.
Mon May 04 12:31:41 +0000 2020@mzspectrum @Sci_j_my Thanks to everyone that contributed ideas. I will think about this a bit more.
Mon May 04 12:20:22 +0000 2020PEX11A:p, peroxisomal biogenesis factor 11A (Homo sapiens) 🔗 Small peroxisomal membrane protein; PTMs: M1+acetyl; expt. PPI with PEX19; no SAAVs; mature form 1-247 [597 x] 🔗
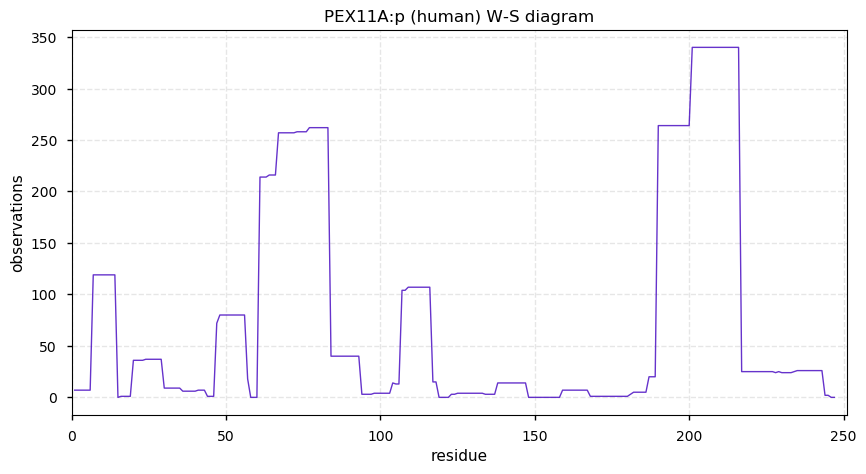
Sun May 03 22:55:53 +0000 2020@pwilmarth I can only speak for the ones I've written, but for those, the same as *, in the sense of an unknown gap in the sequence.
Sun May 03 17:45:16 +0000 2020@slashdot I don't think anyone who has been involved in a regulatory filing would answer "yes".
Sun May 03 14:06:21 +0000 2020Humans do not have PEX8 or PEX9, which are present in fungi.
Sun May 03 14:05:40 +0000 2020PEX10:p, peroxisomal biogenesis factor 10 (Homo sapiens) 🔗 Small peroxisomal membrane protein; no PTMs: N-terminal (A2) is not acetylated; expt. PPIs with 6 other PEX proteins; no SAAVs; mature form 1,2-346 [1,002 x] 🔗
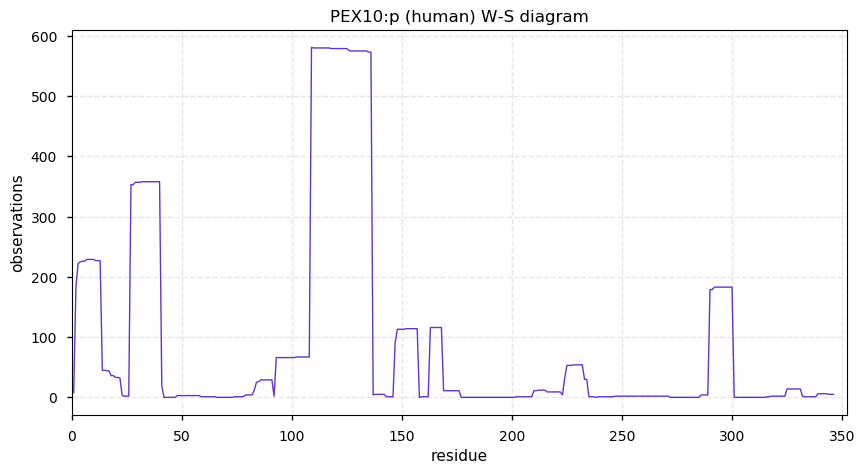
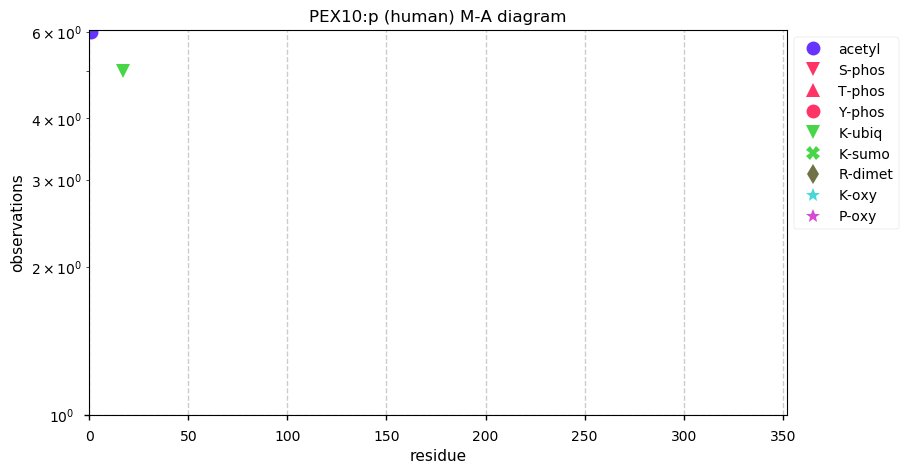
Sun May 03 13:43:04 +0000 2020@jjjotto The data is from a Nature Methods paper, so predictably, there is no information about the methods used.
Sun May 03 13:07:40 +0000 2020@Sci_j_my Oh, my HPLC work goes back further than that
🔗
Sun May 03 12:53:00 +0000 2020@Sci_j_my Yup. RT for all of the PSMs. The red dots indicate the RT for the peptide's PSMs
Sun May 03 12:34:37 +0000 2020@Sci_j_my The blue dots are binned averages for all of the PSMs, using an updated version of 🔗
Sun May 03 00:33:20 +0000 2020@mzspectrum @Sci_j_my It isn't my data. But it occurs throughout a couple hundred LC/MS files, with many different peptides.
Sun May 03 00:30:44 +0000 2020@ucdmrt @Sci_j_my Good Ids. No question about the assignments.
Sat May 02 16:20:46 +0000 2020It seems occur most often in "large-scale" studies. /fin
Sat May 02 16:18:50 +0000 2020The red dots are the occurrences of PSMs with the sequence "RFDEILEASDGIMVAR" (I just chose this peptide as an example: there are lots more with similar behavior in the data). Does this indicate some problem with the column, the pre-column, the gradient or something else?
Sat May 02 16:15:43 +0000 2020Chromatography tweeps: I see a phenomenon all of the time in data with iffy chromatography & I don't know what causes it. This graph is from an RP-LC/MS/MS run, where the blue dots are the predicted retention of a PSM & the green dots are the % of spectra leading to a PSM. /1 🔗
Sat May 02 15:02:29 +0000 2020So, to any editor complaining about a lack of positive responses to their automated email review requests, se regarder.
Sat May 02 13:26:01 +0000 2020If an "editor" is actually doing their job, there is simply no reason to have more than 2 reviews: in most cases, 1 or 2 reviews is sufficient.
Sat May 02 13:12:00 +0000 2020For me, it just got to the point that I just stopped wanting to participate.
Sat May 02 13:10:50 +0000 2020There always were bad editors, but the consolidation of scientific publishing seems to have had the effect of encouraging & rewarding bad behavior, leading a nearly uniform decline in the experience for reviewers & authors.
Sat May 02 12:57:04 +0000 2020The main reason I stopped reviewing for science magazines/journals was the uniformly bad behavior of "editors". Poor communications, lack of interest in anything but filling out forms, noblesse oblige, etc.
Sat May 02 12:50:52 +0000 2020@theoneamit Banana ketchup/sauce is a staple in the Philippines (my local grocery has about 3 meters of shelf space devoted to different brands). I like it on some things (great on fried rice), but I was wondering if anyone else has a preference.
Sat May 02 12:37:59 +0000 2020For anyone following along, PEX proteins are not related by genetics, structure or sequence: they are a group of proteins involved in the import of other proteins to the peroxisome
🔗
Sat May 02 12:12:48 +0000 2020PEX7:p, peroxisomal biogenesis factor 7 (Homo sapiens) 🔗 Small cytosolic protein; PTMs: A2+acetyl; no SAAVs; receptor for PTS2 motif ([RK][LVI].{5}[HQ][LA]) proteins; mature form 2-323 [960 x] 🔗
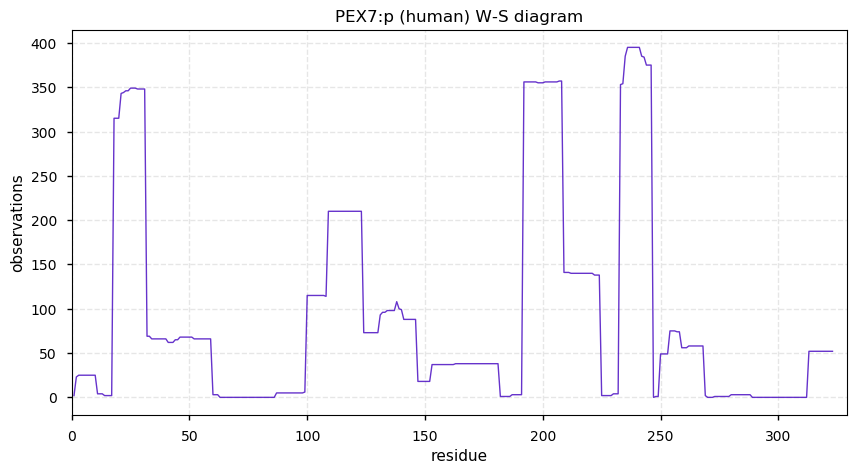
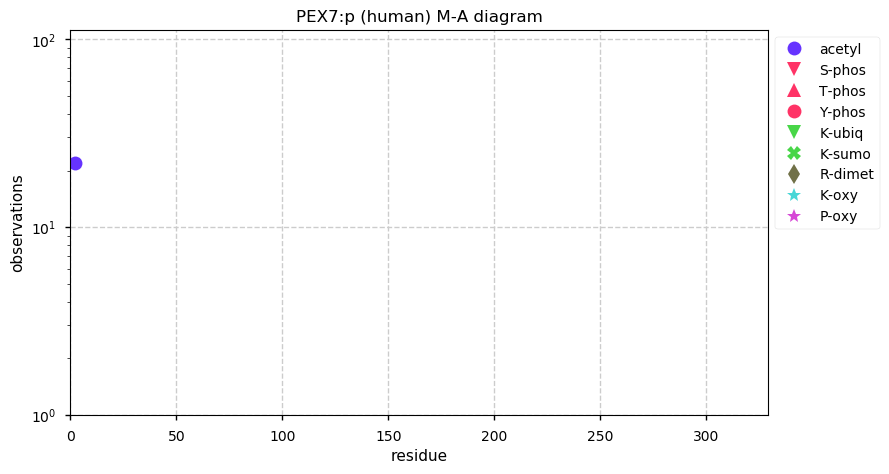
Fri May 01 19:57:06 +0000 2020@mvaudel @ypriverol @nesvilab @Smith_Chem_Wisc ENSEMBL has always included pseudogenes translations the ".all.fa" files (at least for as long as I can remember).
Fri May 01 19:35:01 +0000 2020@mvaudel @ypriverol @nesvilab @Smith_Chem_Wisc The * in the ENSEMBL ".all.fa" are stop codons, not splice junctions. They almost always indicate that the sequence is a pseudogene (nonsense mediated decay).
Fri May 01 18:59:07 +0000 2020@AnthonyCesnik @ypriverol @nesvilab @Smith_Chem_Wisc The initial X means that the 1st exon starts with an incomplete codon (1 or 2 bases rather than 3).
Fri May 01 15:45:38 +0000 2020Banana vs tomato ketchup. Opinions?
Fri May 01 15:37:21 +0000 2020A pretty good podcast about the out-sized impact of COVID-19 in long term care facilities in central Canada.
🔗
Fri May 01 14:56:51 +0000 2020Still working my way through AGC_BLOCK_TWO_SELF_CHECK.agc, from 🔗 — a good example of source documentation.
Fri May 01 14:47:30 +0000 2020@TrumanLab @Sci_j_my Some combination of kakistocracy, ennui & Dunning-Kruger would be my guess.
Fri May 01 14:40:06 +0000 2020I should know better by now, but my Charlie-Brown-like optimism leaves me convinced that this time there is going to be a football for me to kick.
Fri May 01 14:28:51 +0000 2020Nature Methods does it again!
Fri May 01 12:41:51 +0000 2020PEX6:p, peroxisomal biogenesis factor 6 (Homo sapiens) 🔗 Midsized peroxisomal membrane subunit; several low occupancy K+ubiquitin sites; SAAVs: A809V (8%), V882I (2%), P939Q (33%); mature form 2-980 [4,406 x] 🔗
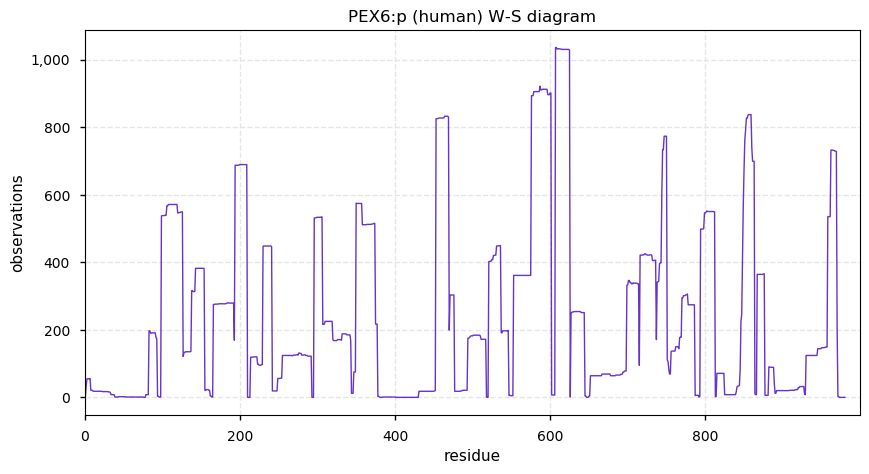
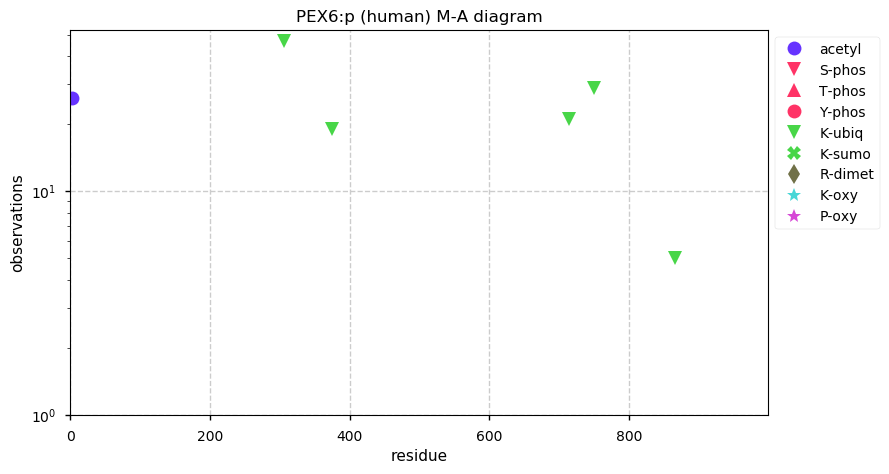
Fri May 01 01:43:53 +0000 2020@Smith_Chem_Wisc @ypriverol @nesvilab It may be a bit OG, but it was a standard interpretation back in the olden times 🔗
Fri May 01 00:45:08 +0000 2020@ypriverol @nesvilab @Smith_Chem_Wisc Yes, the same accession.
Thu Apr 30 23:46:07 +0000 2020@jwoodgett Yah. The trial is junk now.
Thu Apr 30 23:44:49 +0000 2020@ypriverol @nesvilab @Smith_Chem_Wisc Yes. It is treated as a stop (or gap), rather than a normal cleavage site, so even with no cleavage specificity, the sequence to the right of the * is treated like a new protein.
Thu Apr 30 23:24:36 +0000 2020@ypriverol @nesvilab @Smith_Chem_Wisc It would be interpreted as two peptides:
1. MGCCGCGSCGCSGG
2. GGGCGGGCGGGCGSCTTCR
The * symbol is considered to be the equivalent to a stop codon.
Thu Apr 30 21:36:30 +0000 2020Sometimes I have to remind myself how much better peptide chromatography is now, compared to 10-15 years ago. Much of it driven by the developments associated with DIA.
Thu Apr 30 18:04:37 +0000 2020@bkives As an alternative to "like", there should be some type of "grimly acknowledge" button on Twitter.
Thu Apr 30 17:58:00 +0000 2020@Sci_j_my It is one of the few books I own that cannot be effectively digitized & every time I read it I get a clearer understanding of how little I know about text.
Thu Apr 30 17:53:05 +0000 2020@pwilmarth I do a lot of AAA analysis on PSM sets to look for patterns in my own screw-ups.
Thu Apr 30 17:49:16 +0000 2020@pwilmarth The net effect is that the individual fractions in a batch go from acidic to basic peptides for this protocol. So anything that shifts a peptide towards being more acidic (deamidation, acetylation, phosphorylation, pyro-Glu) results in an enrichment in the early fractions.
Thu Apr 30 17:39:49 +0000 2020@Sci_j_my I do as I'm told by "The Elements of Typographic Style" (🔗).
Thu Apr 30 17:31:11 +0000 2020@pwilmarth Do you see the chromatographic effects? For example, the deamidated PSM fractions for the first batch of human samples range from:
UM_F_50cm_2019_0414.raw, 9.4% deamidated to
UM_F_50cm_2019_0421.raw, 1.8% deamidated.
And those are only assignments that are not 13C A1 artifacts.
Thu Apr 30 16:49:58 +0000 2020@pwilmarth I forgot to mention that PXD014414 also has 8-9% non-tryptic cleavage (no chromatographic enrichment). Other than N-terminal protein processing, most of the cleavage is due to a chymotrypsin-like activity ([FYWL]-X). Also very nice for algorithm development!
Thu Apr 30 16:25:12 +0000 2020@TanentzapfLab Definitely trolling via the virtue signalling route.
Thu Apr 30 16:13:08 +0000 2020@MattWFoster @pwilmarth The result, however, is that you end up with 2-3 x as many SAAV PSMs as you should find.
Thu Apr 30 16:12:44 +0000 2020@MattWFoster @pwilmarth No idea. I try to understand it from time to time, but it still eludes me (that is how I found the succinylation side reaction). My current suspects are other low level side reactions or impurities in the reagents.
Thu Apr 30 15:19:05 +0000 2020Section IX (page 24) of the Georgia state reopening plan seems kind of weird. It is the sort of thing that would give most Europeans (esp. Germans) cognitive dissonance when they first read it.
🔗
Thu Apr 30 14:59:49 +0000 2020@olgavitek @UCDProteomics @kusterlab You should talk to your institute/university legal department regarding how they view this type of IP. Lack of copyright ownership means it may be difficult to deal with recorded versions of the stream popping up online or being used as teaching material at other institutions.
Thu Apr 30 14:40:53 +0000 2020@pwilmarth But, as with most TMT-derivatized data, it is not suitable for SAAV detection.
Thu Apr 30 14:37:00 +0000 2020@pwilmarth The enrichment phenomenon is also very much in effect for peptide N-terminal cyclization, hydroxy-proline/lysine, protein N-terminal acetylation and TMT-related succinylation, also nice!
Thu Apr 30 14:34:35 +0000 2020@pwilmarth The deamidation rate isn't that unusual (~ 6% of PSMs), but because of the way the chromatography was done, they are enriched in specific fractions. The enrichment is also very nice from an algorithm development point of view.
Thu Apr 30 14:30:51 +0000 2020@pwilmarth PXD014414 is for some reason (prob. sample prep) depleted in S/T/Y phosphopeptides, but has a typical S:T site detection ratio (5:1), also making it good test data for accurately detecting rare (but there) PTMs. /2
Thu Apr 30 14:26:50 +0000 2020@pwilmarth The data set (PXD014414) has some additional nice properties for algorithm developers. It shows a nearly ideal ratio of N:Q deamidation (10:1), which make it useful for anyone who is interested in trying to reduce false positives for this rather tricky chemical modification /1
Thu Apr 30 13:01:33 +0000 2020Comparing trends for Holland, Canada & Sweden is puzzling: very different govt. policy responses and geographies but similar curves. 🔗
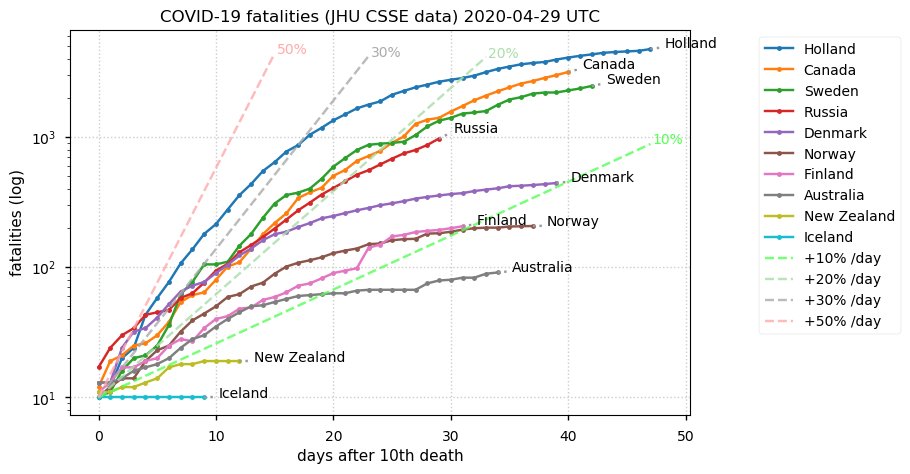
Thu Apr 30 12:34:40 +0000 2020PEX5:p recognizes the PTS1 C-terminal peroxisome targeting signal in cytoplasmic proteins & is necessary for the import of these proteins into the peroxisome. There is no PEX4 gene in animals (it is found in plants & fungi).
Thu Apr 30 12:30:16 +0000 2020PEX5:p, peroxisomal biogenesis factor 5 (Homo sapiens) 🔗
Midsized peroxisomal membrane subunit; several phosphodomains and ubiquitination sites; no SAAVs; mature form 2-639 [7,241 x] 🔗
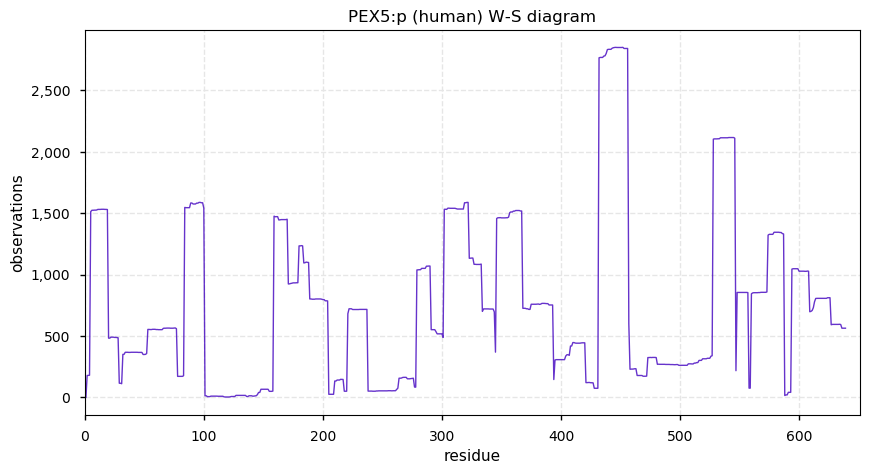
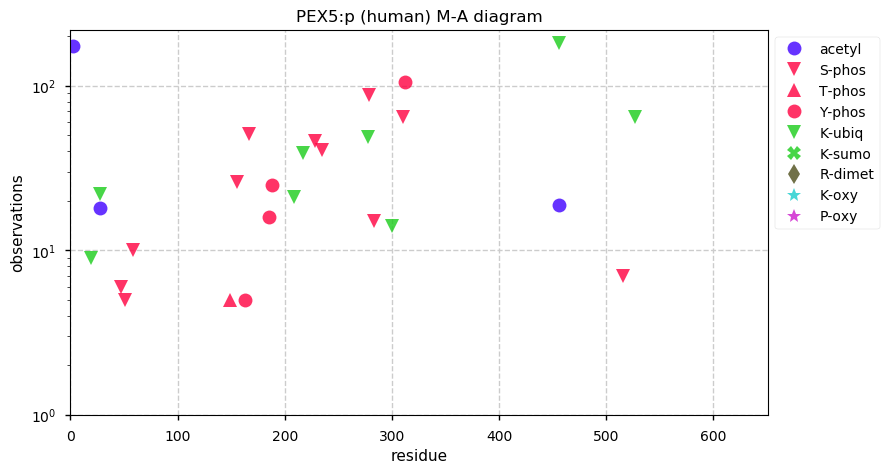
Thu Apr 30 00:10:46 +0000 2020@Sci_j_my @byu_sam There are specific programs that can get direct funding OUS, either as direct contracts or through Reseach Center-type programs: that is how UniProt gets NIH funding (or at least it did).
Wed Apr 29 18:52:33 +0000 2020@olgavitek @UCDProteomics @kusterlab One thing I was wondering about wrt to organizing an on-line event like yours: who owns the copyright to the video feed? Was it necessary to have the presenters sign waivers wrt to copyright ownership?
Wed Apr 29 18:32:20 +0000 2020PTM observation abundance diagrams for the 15 human PEX proteins (they traffic other proteins into the peroxisome) 🔗
Wed Apr 29 17:10:43 +0000 2020@UCDProteomics @olgavitek @kusterlab Will universities set up dedicated studio facilities with producers & techs to improve the quality of on-line talks. Will they offer video editing to enhance the performances?
Wed Apr 29 17:03:23 +0000 2020@UCDProteomics @olgavitek @kusterlab Will faculty and students be given time to "attend" a meeting for 3 – 7 days, even though they are still at their desks?
Wed Apr 29 16:45:20 +0000 2020@AlexUsherHESA Ukrainian settlers favored yurt-style tents & it just stuck.
Wed Apr 29 16:30:57 +0000 2020@UCDProteomics @olgavitek @kusterlab Can any particular field really sustain as many conferences when the cost of attending is low and many more people can attend?
Wed Apr 29 16:27:53 +0000 2020@UCDProteomics @olgavitek @kusterlab Will the PI's that talk at pretty much every conference (with very similar talks) end up like the vaudeville acts who couldn't sustain interest once movies came along?
Wed Apr 29 16:26:08 +0000 2020@UCDProteomics @olgavitek @kusterlab How a switch to on-line will change the way talks are delivered is going to be interesting to watch. Will tenure & promotion cmtes view an invited talk on-line the same way they would a conventional talk?
Wed Apr 29 13:24:41 +0000 2020PEX3:p, peroxisomal biogenesis factor 3 (Homo sapiens) 🔗 Small peroxisomal membrane subunit; central ubiquitination domain; no SAAVs; mature form 1-373 [5,388 x] 🔗
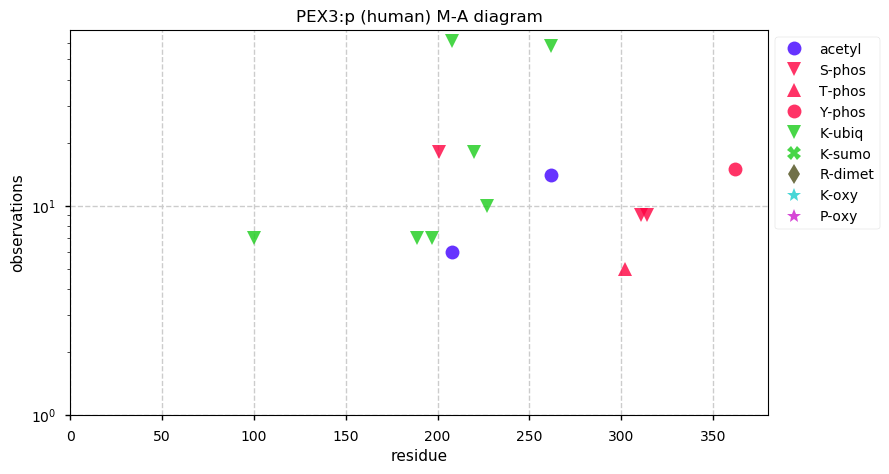
Tue Apr 28 16:59:03 +0000 2020@dtabb73 @MattWFoster Urine & CSF tend to have the most intact COL1 and COL3
Tue Apr 28 14:43:17 +0000 2020@dtabb73 It does kind of shoot holes in the warm-weather-and-sunshine theory 🔗
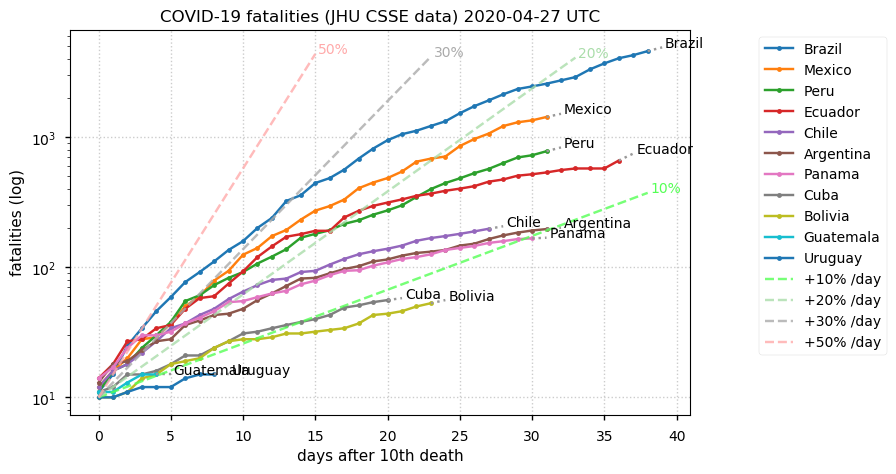
Tue Apr 28 14:23:38 +0000 2020@IonSource News organizations seem to be using a trailing-7-days average to mask the effect. I was just wondering if there has been any research/crazy-speculation-involving-aliens-and-the-Illuminati into why it is happening.
Tue Apr 28 13:33:23 +0000 2020Daily fatalities slowly trending down globally, but not smoothly. Has anyone explained the week-long period of the oscillations? 🔗
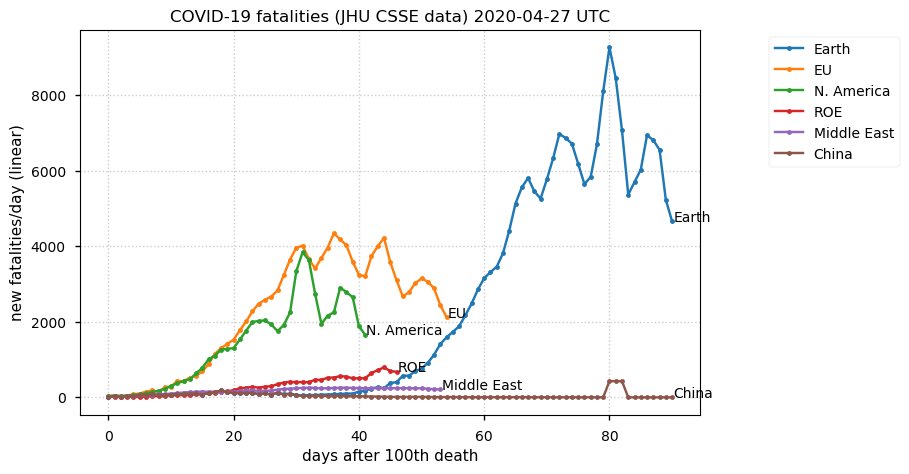
Tue Apr 28 12:42:28 +0000 2020PEX2:p, peroxisomal biogenesis factor 2 (Homo sapiens) 🔗 Small peroxisomal subunit; most abundant PTM: K84+acetyl (93 x); no SAAVs; mature form 2-305 [591 x] 🔗
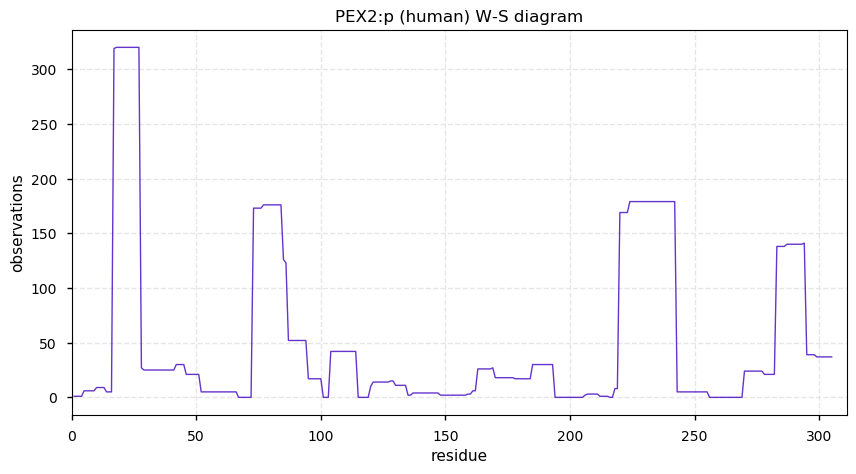
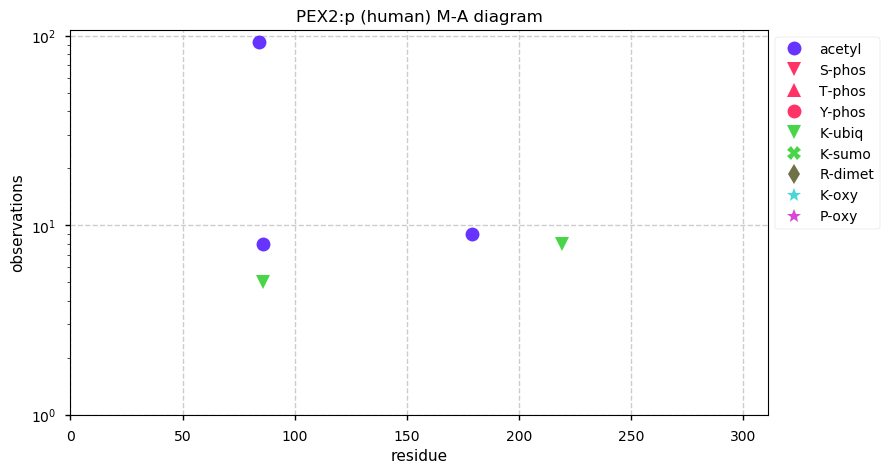
Mon Apr 27 18:15:23 +0000 2020PEX1:p ubiquitination is unusual in that there is no hint of complimentary acetylation.
Mon Apr 27 16:58:31 +0000 2020Unlike LysargiNase, TrypN is not a good exopeptidase, so peptides with C-terminal K or R are common with this enzyme.
Mon Apr 27 16:47:44 +0000 2020PXD017030 is an interesting demo of an enzyme TrypN (C. thermophilum) that cleaves N-terminal to R or K, similar to LysargiNase (M. acetivorans). Enzyme works great, but if you try the analysis yourself, the Cys-blocking didn't work & allow for 3 missed cleavages.
Mon Apr 27 15:03:32 +0000 2020But people always seem to leave Wolbachia pipientis wMelPop out of their search space for some reason.
Mon Apr 27 14:55:55 +0000 2020There seems to be a resurgence of interest in proteomics analysis of D. melanogaster.
Mon Apr 27 14:11:25 +0000 2020PEX1:p, peroxisomal biogenesis factor 1 (Homo sapiens) 🔗 Large peroxisomal membrane associated subunit; C-terminal phosphodomain, many K+ubiquitinyl sites; SAAVs: N271S (1%), W507C (1%), I696M (2%); mature form 1-1283 [7,146 x] 🔗
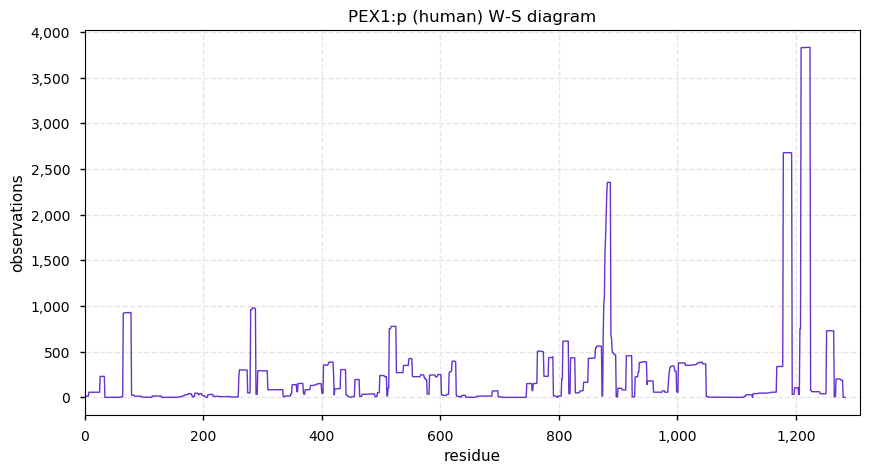
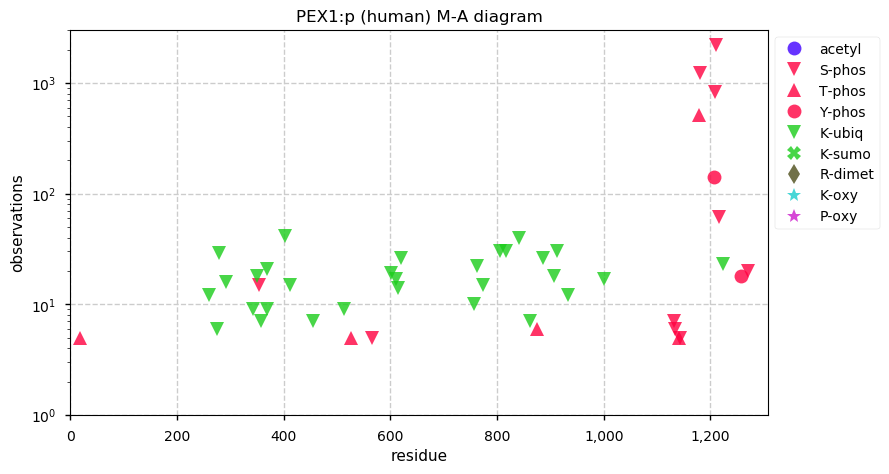
Mon Apr 27 13:03:48 +0000 2020@Sci_j_my 👍
Mon Apr 27 12:58:49 +0000 2020@Sci_j_my I have nothing but idle conjectures. It will take years to tease out why there are so many stark regional differences.
Mon Apr 27 12:47:15 +0000 2020@Sci_j_my The top line is QC = Quebec. BC is near the baseline.
Mon Apr 27 12:11:11 +0000 2020This chart of daily fatalities shows how differently COVID-19 is affecting Canada regionally 🔗
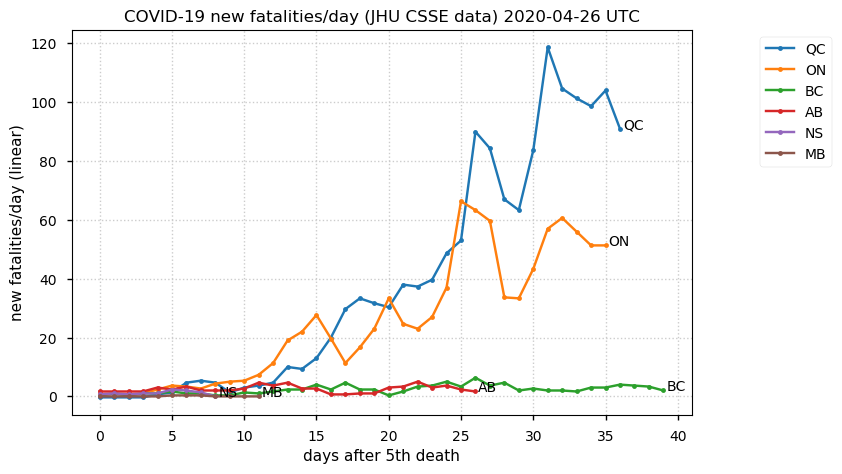
Sun Apr 26 15:25:32 +0000 2020@SinzAndrea @astacus @FrankSobott @Covid19Datalive @Covid19depot @Covid19DE @COVID19 @edemmott 🔗
Sun Apr 26 12:49:54 +0000 2020Decr2:p, 2-4-dienoyl-Coenzyme A reductase 2, peroxisomal (Mus musculus) 🔗 Small peroxisomal enzyme; abundant PTMs: A2+acetyl (2789 x), S287+phosphoryl (107 x); C-terminal targeting signal AKL; mature form 2-292 [3,272 x] 🔗
Sat Apr 25 14:59:40 +0000 2020The similarity in trajectory between France, Italy, Spain and the UK will make for interesting discussions in epidemiology seminars for a decade or two. 🔗
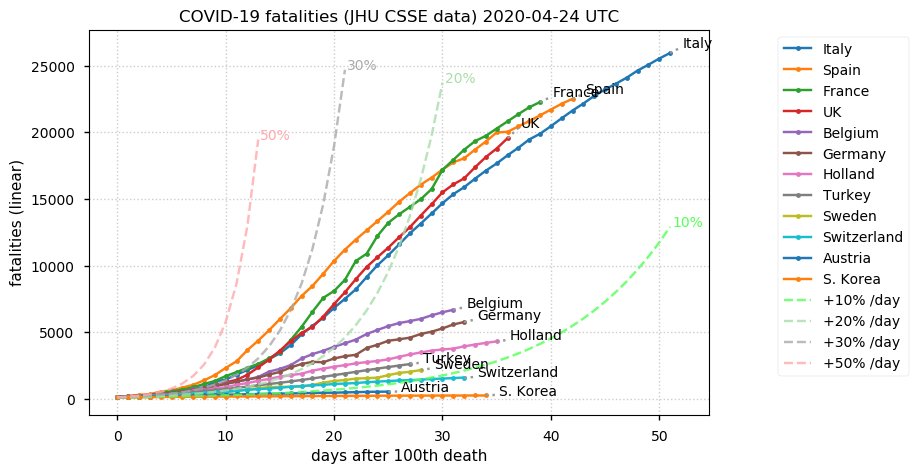
Sat Apr 25 14:14:03 +0000 2020You could probably turn how information about ECH1:p in protein-protein interaction, metabolomics & pathways networks is skewed by this annotation error into an interesting tutorial on how to view the interpretation of results using AI/DB-driven methods critically.
Sat Apr 25 14:00:44 +0000 2020@dtabb73 I have never done anything associated with mother-fracking memory without swearing.😠
Sat Apr 25 12:34:59 +0000 2020ECH1:p is a good example of how a simple bad annotation (mislocating this mitochondrial protein to the peroxisome) can propagate in strange ways through informatics systems that rely on text for finding relationships between proteins/genes.
Sat Apr 25 12:28:23 +0000 2020Ech1:p, enoyl coenzyme A hydratase 1, peroxisomal (Mus musculus) 🔗 Small mitochondrial enzyme; scattered K+acetyl sites; incorrectly annotated as being in peroxisome; mature form (34,35)-327 [8,477 x] 🔗
Fri Apr 24 21:59:08 +0000 2020@astacus @FrankSobott @SinzAndrea @Covid19Datalive @Covid19depot @Covid19DE @COVID19 @edemmott Just finished reanalyzing the data: found 7 unique sequences in 16 good PSMs assigned to the N protein in the 2 data files. Two of the sequences (210-226 & 210-233) are unique to SARS-CoV2, out of ~1000 Orthocoronavirinae species in GeneBank. Better than I would have thought.
Fri Apr 24 21:34:45 +0000 2020@sackloo @CellCellPress Try 🔗
Fri Apr 24 14:36:29 +0000 2020If these comments are too sparse, unintelligible or simply missing, insist that the authors do a better job. If the source code for the software is not available, ask for a written description of how the commenting was done. 3/3
Fri Apr 24 14:36:03 +0000 2020Take a look at the code (it is just text). About half of the text should be "comments", which are written descriptions of how the code works, meant to facilitate the use (and reuse) of the often inscrutable lines of software. 2/3
Fri Apr 24 14:35:40 +0000 2020Reviewer Pro-Tip℠:
If you are reviewing a manuscript with associated computer code you don't have to know anything about computer programming to provide a useful critique. 1/3
Fri Apr 24 14:05:39 +0000 2020Pecr:p, peroxisomal trans-2-enoyl-CoA reductase (Mus musculus) 🔗 Small peroxisomal enzyme; S2+acetyl & several K+acetyl domains; C-terminal targeting sequence AKL; mature form 2-303 [4,169 x] 🔗
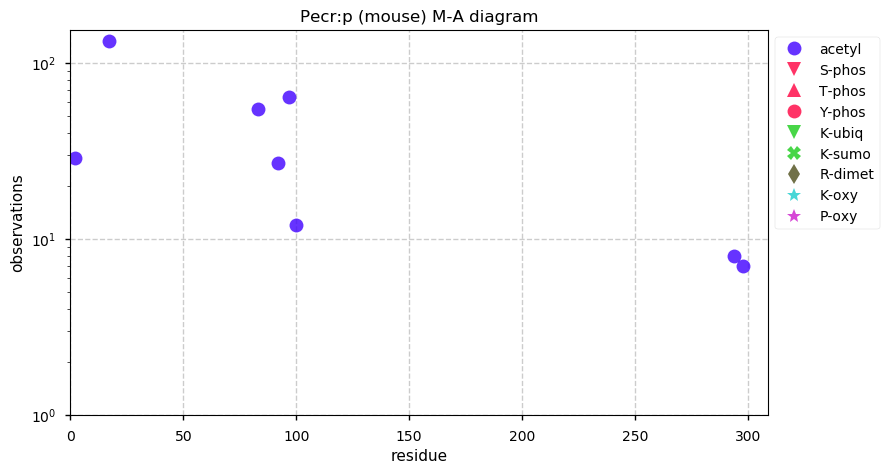
Thu Apr 23 19:35:17 +0000 2020@jwoodgett I'm sure they will take "lessons learned" from the Superclusters Initiative (& maybe CIHR's SPOR program) into account
Thu Apr 23 15:51:47 +0000 2020@olgavitek 🔗
Thu Apr 23 15:22:43 +0000 2020When did Northeastern University become the epicenter of crackpot speculation in the US?
Thu Apr 23 15:04:10 +0000 2020An easy (& useful) thing you can do is to insist that there is some straightforward way to determine which of the data files are associated with the specific experiments & figures described in the text. 2/2
Thu Apr 23 15:03:55 +0000 2020Reviewer Pro-Tip℠:
If you are reviewing a manuscript with publicly available data you don't have to download the data to provide a useful critique. 1/2
Thu Apr 23 12:47:50 +0000 2020Lonp2:p, lon peptidase 2, peroxisomal (Rattus norvegicus) 🔗 Midsized peroxisomal enzyme; frequently observed PTMS: S2+acetyl; C-terminal targeting sequence SKL; mature form 2-852 [1,175 x] 🔗
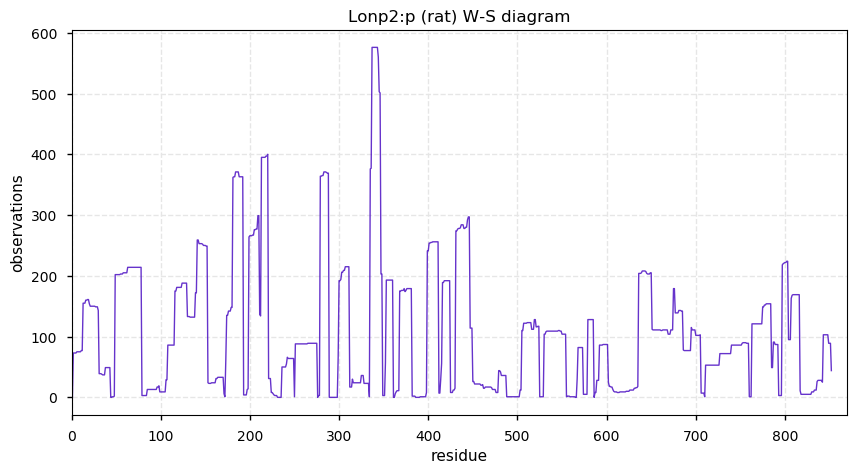
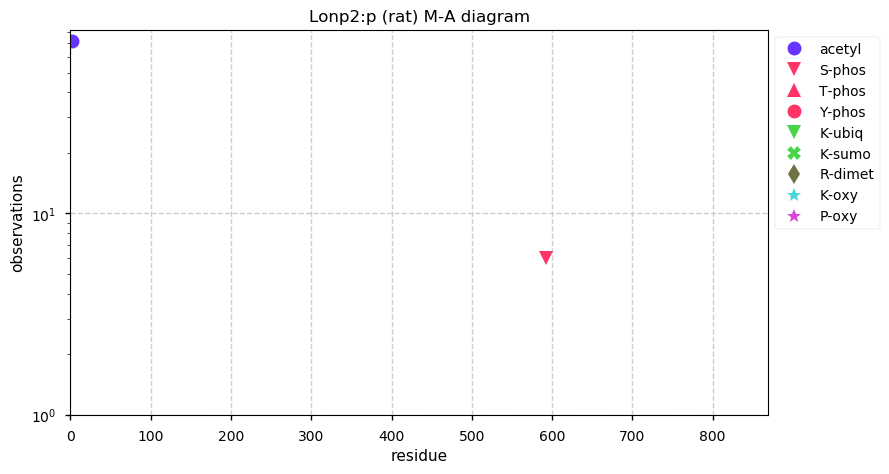
Wed Apr 22 20:34:13 +0000 2020@Smith_Chem_Wisc It was all stone-knives and bear-skins back then ...
Wed Apr 22 20:25:28 +0000 2020@Smith_Chem_Wisc You can just grab it at 🔗
Pre-genome proteome databases were not quite the same as they are now.
Wed Apr 22 16:01:37 +0000 2020@mollywood @kairyssdal The current Secretary of Labor has a history of arguing against regulating the provision of health care to workers.
Wed Apr 22 14:40:41 +0000 2020@MattWFoster @Eickelberg_MD Keep in mind that the soluble form, which is very commonly observed in urine, seems to be produced through a different mechanism and doesn't reflect cell surface concentrations.
Wed Apr 22 14:30:20 +0000 2020@MattWFoster @Eickelberg_MD It shows up in some of the "CPTAC Lung Adenocarcinoma (LUAD) Discovery Study" data sets. Otherwise, no. It is most frequently observed in testis, gut & kidney.
Wed Apr 22 13:16:51 +0000 2020Hao1:p, hydroxyacid oxidase 1 (Rattus norvegicus)
Small peroxisomal enzyme; frequently observed PTMS: M1+acetyl [86 ×], S194+phosphoryl [97 ×]; C-terminal targeting sequence VSKI; mature form 1-370 [1,157 ×] 🔗
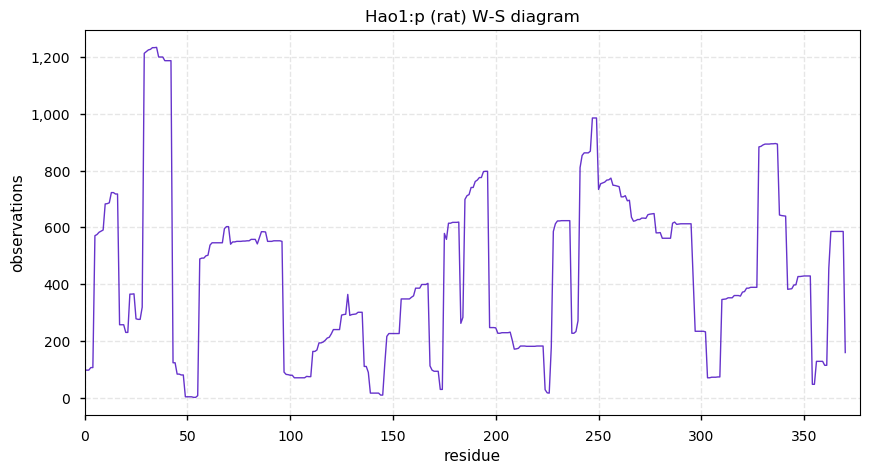
Tue Apr 21 13:07:25 +0000 2020Acot4:p, acyl-CoA thioesterase 4 (Mus musculus) 🔗 Small peroxisomal enzyme; frequently observed PTMS: A2+acetyl [442 x], S56+phosphoryl [1,219 x]; C-terminal targeting sequence CRL; present in humans; mature form (1,2)-421 [6,460 x] 🔗
Tue Apr 21 13:03:39 +0000 2020@ProteomicsNews These observations of this peptide show that it is very prone to oxidation and deamidation, which make it a problematic choice as a biomarker.
Tue Apr 21 13:01:43 +0000 2020@ProteomicsNews Making the data available certainly would help make the work more credible. However, the peptide they report "RPQGLPNNTASWFTALTQHGK" has been seen 483× in the 3 exisiting public datasets that did not use PNGase treatment 🔗
Tue Apr 21 12:25:29 +0000 2020Decreases in daily fatality numbers in the EU and North America have led to a significant drop in the global curve. 🔗
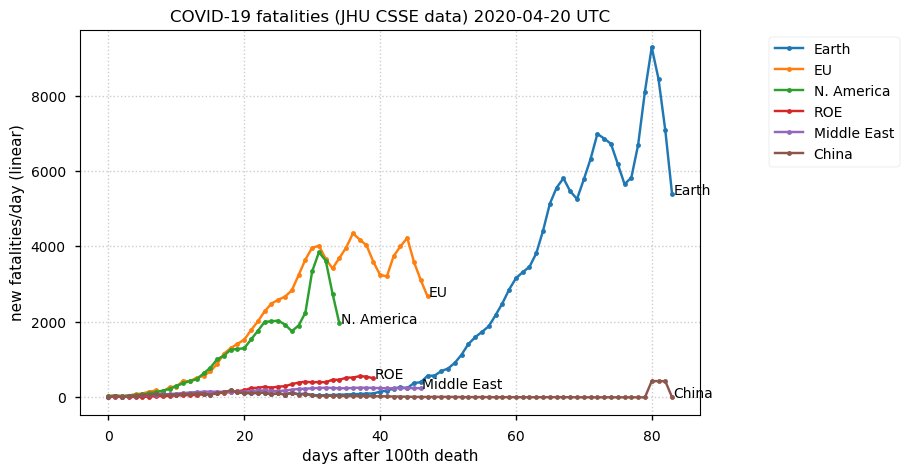
Tue Apr 21 12:17:27 +0000 2020Iran seems to on the downslope, while Singapore and India are ramping up. 🔗
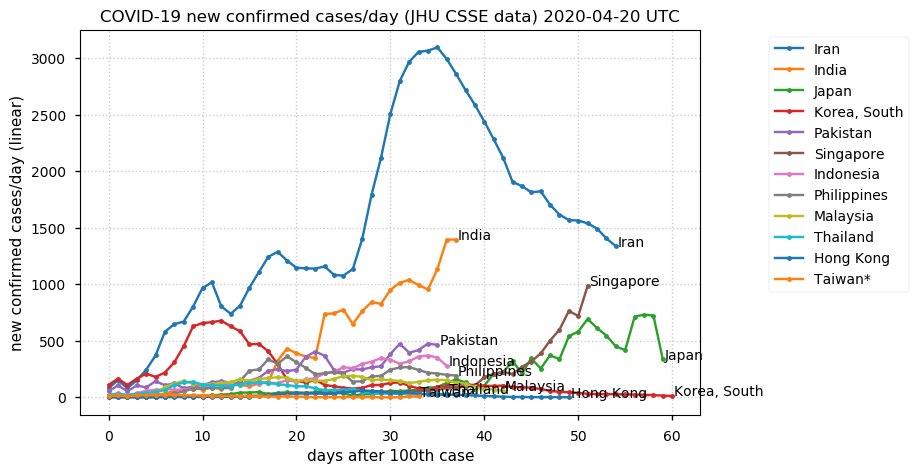
Mon Apr 20 22:23:06 +0000 2020Russia is currently bucking the trend of other northern countries 🔗
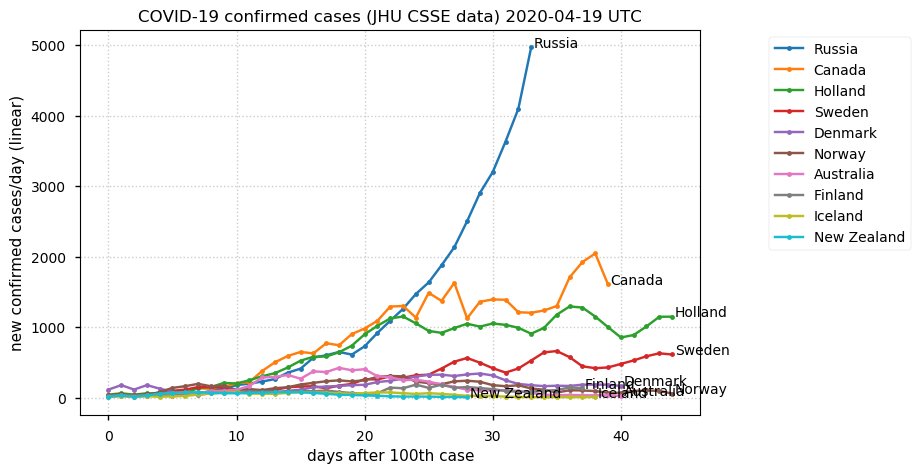
Mon Apr 20 16:33:24 +0000 2020@bkives Hopefully he can explain his thinking wrt MB university expenditure reductions.
Mon Apr 20 15:56:45 +0000 2020I thought it was just me
🔗
Mon Apr 20 14:51:41 +0000 2020Does HUPO have an award for "Least intuitive data file naming in a public dataset"?
There are many fine contenders.
Mon Apr 20 13:19:41 +0000 2020New cases going down in EU, with lots of day-to-day fluctuations. 🔗
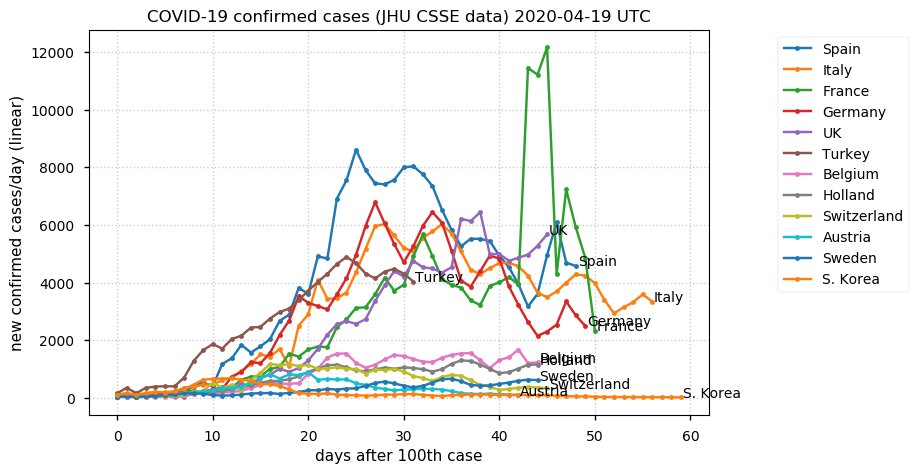
Mon Apr 20 12:30:47 +0000 2020Acot3:p, acyl-CoA thioesterase 3 (Mus musculus) 🔗 Small peroxisomal enzyme; acetyl: M1, K53, K301; phosphoryl: S67; C-terminal targeting sequence AKL; not present in humans; mature form 1-442 [5,816 x] 🔗
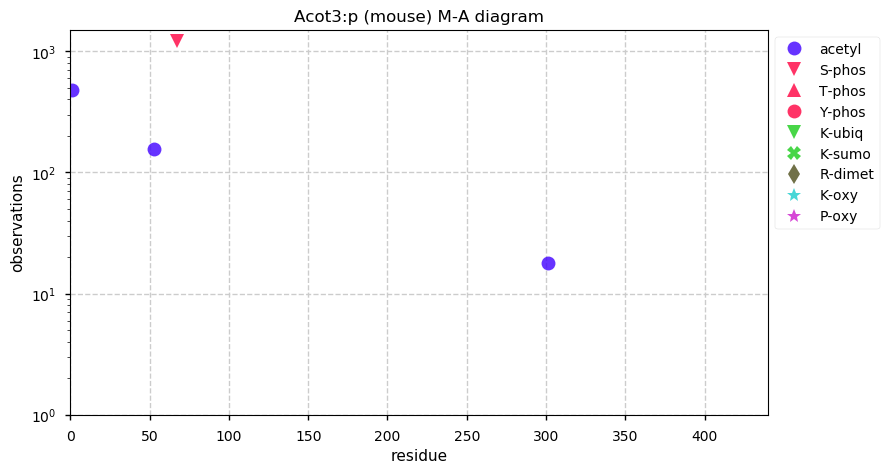
Mon Apr 20 12:02:57 +0000 2020Yah, sure, 30% should be no trouble 👨🎓
🔗
Mon Apr 20 00:14:18 +0000 2020@AlexUsherHESA It is standard GRC language. In custody does not necessarily mean alive, simply under the control of Crown.
Sun Apr 19 17:08:13 +0000 2020This data puts the Canadian government in a difficult position wrt short-term federal policy, as well as problems for provincial governments. 🔗
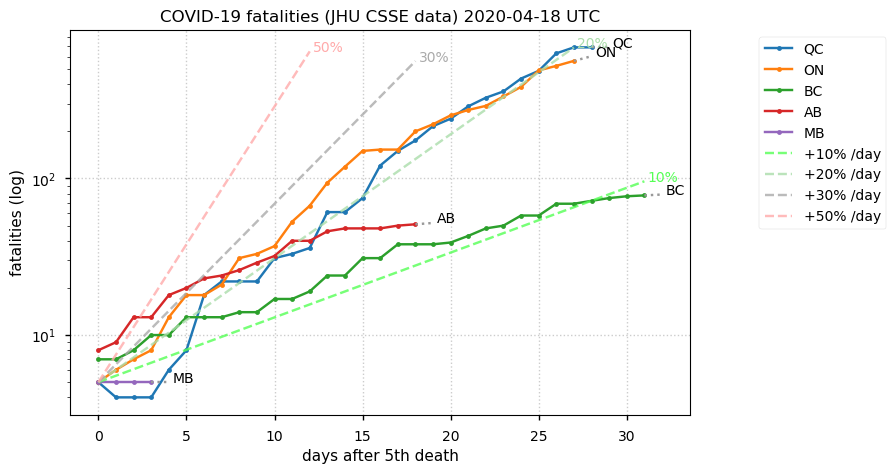
Sun Apr 19 15:03:49 +0000 2020For anyone interest in Alzheimer's Disease mouse model systems, PXD017916 provides a very interesting insight into all of the other proteins associated with insoluble Aβ deposits.
Sun Apr 19 12:40:11 +0000 2020In the human genome, ACNAT2:p is represented by a unitary pseudogene that is not translated because of a nonsense stop codon.
Sun Apr 19 12:37:08 +0000 2020Acnat2:p, acyl-coenzyme A amino acid N-acyltransferase 2 (Mus musculus) 🔗 Small peroxisomal enzyme; no significant PTMs; C-terminal targeting sequence SKL; aka glycine N-choloyltransferase; LOC313220; MGC108791; mature form 2-420 [1,104 x] 🔗
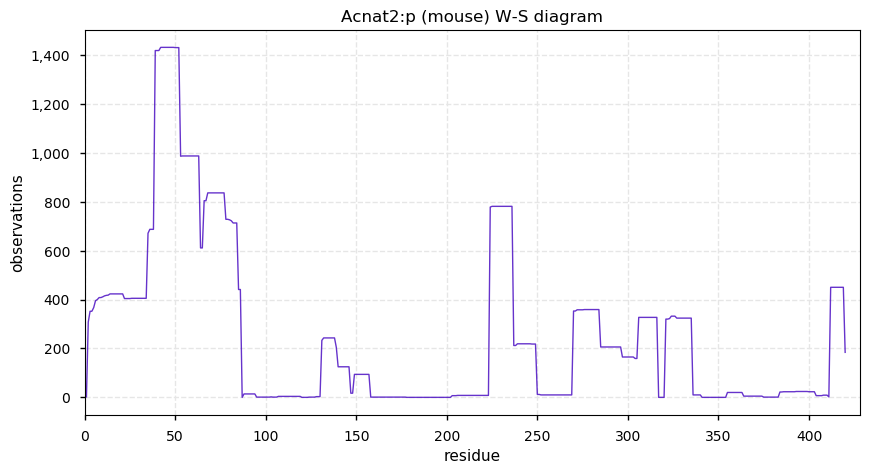
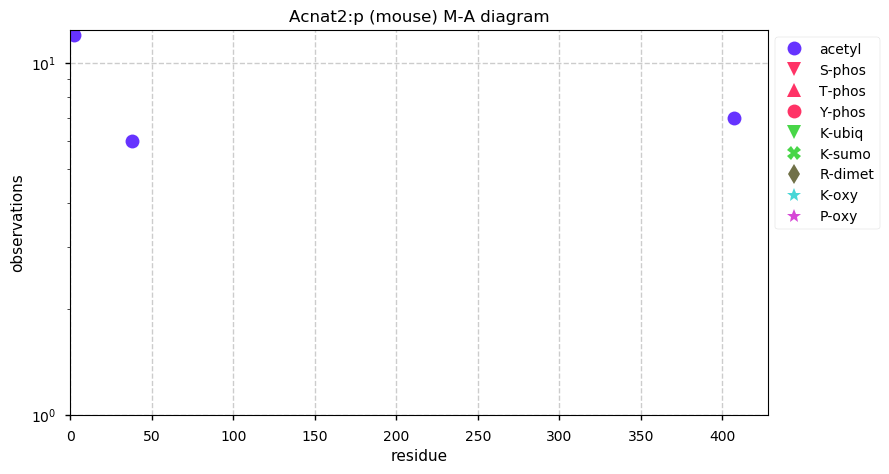
Sat Apr 18 18:47:23 +0000 2020@IonSource @gangulyteena I first heard it on "11-17-70" (UK title "17-11-70"), but "Tumbleweed Connection" was the first recording.
Sat Apr 18 16:52:56 +0000 2020@MattWFoster Thanks. I looked around but couldn't find anything on point. I knew someone would know.
Sat Apr 18 16:35:18 +0000 2020Does anybody know why endoC-βH1 cells (from 🔗) would be expressing large amounts of SV40 large T antigen? I am not familiar with the common uses and modifications associated with this particular cell line.
Sat Apr 18 15:47:43 +0000 2020@dtabb73 I can't help myself: unless you are using > 20 lines of CSS to do exactly the same thing, it is just not OK.
Sat Apr 18 14:12:54 +0000 2020@Sci_j_my A combination of automation, pattern matching and manual inspection when necessary.
Sat Apr 18 13:50:21 +0000 2020@Sci_j_my The methods used are generalizations of those described here 🔗
Sat Apr 18 12:40:35 +0000 2020Baat:p, bile acid-Coenzyme A:amino acid N-acyltransferase (Mus musculus) 🔗 Small peroxisomal enzyme; some acetylation; C-terminal targeting sequence SQL; aka BACAT, BAT, kan-1, choloyl-CoA hydrolase, glycine N-choloyltransferase; mature form 2-420 [1,732 x] 🔗
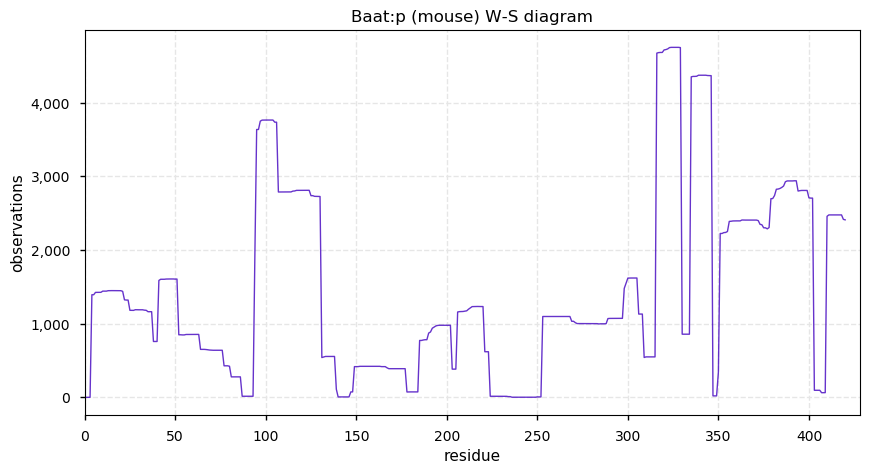
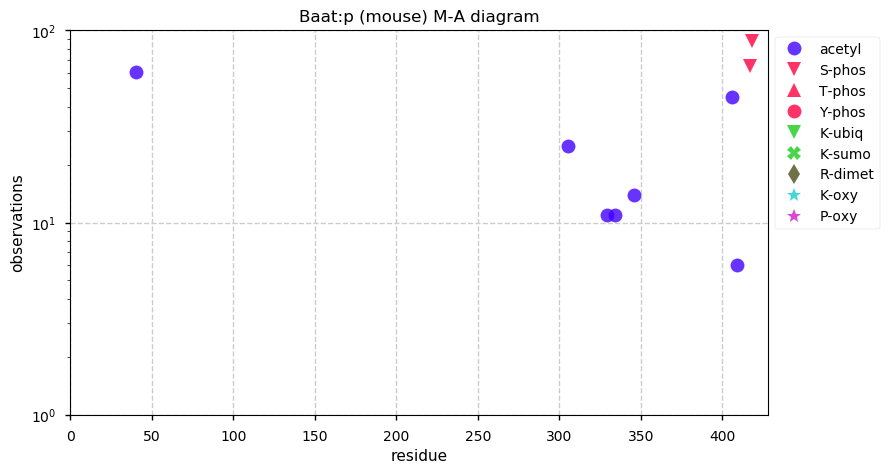
Sat Apr 18 12:28:37 +0000 2020@Sci_j_my My own research. It would be pretty tricky to compile a list a the number of times a site was observed, using the info in a typical paper. The best you may be able to do is count the number of papers that mention a site.
Fri Apr 17 17:14:27 +0000 2020@bkives I suspect the workers themselves may disagree.
Fri Apr 17 16:27:09 +0000 2020@astacus @Sci_j_my In a bit of after-the-fact pedantry, ER-targeted signal peptide removal and N-terminal processing (removal of the initiator methionine & acetylation) are co-translational modifications (not post-translational modifications).
Fri Apr 17 16:05:31 +0000 2020If this is true, on-line conferences will quickly turn in to vendor-sponsored webinars with presenters wearing NASCAR-driver-style jackets and hats covered with corporate advertising. So, no real change ...
Fri Apr 17 16:01:07 +0000 2020Based on these two polls, people who expressed an opinion would like conference fees for on-line conferences to be ≤ 20% of the fee for an in-person conference.
Fri Apr 17 14:57:03 +0000 2020While I'm not a fan of most qLC/MS data, PXD016166 does demonstrate some interesting effects, particularly the phosphorylation study. Rinfret Robert C, et al., J Proteome Res. 2020 Apr 8, 🔗
Fri Apr 17 12:56:46 +0000 2020And by "significant acetylation" I mean so far over-the-top that it looks like it must be fake (but it isn't).
Fri Apr 17 12:55:01 +0000 2020Scp2:p, sterol carrier protein 2, liver (Mus musculus) 🔗 Midsized peroxisomal protein; significant acetylation; C-terminal targeting sequence AKL; 2 common splice variants; mature form (1,2)-547 [10,901 x] 🔗
Thu Apr 16 17:40:26 +0000 2020@pwilmarth Most zoologists feel this way about birds, too.
Thu Apr 16 16:11:25 +0000 2020@jwoodgett They only ship to the US through Winnipeg 😡:
🔗
Thu Apr 16 15:50:20 +0000 2020It seems most people would like to see their grant/deparmental-fund budget for conferences reduced or abolished. For those willing to still pay something, what fraction of existing conference fees would be appropriate for on-line versions
(80% means a $100 fee ⇒ $80)?
Thu Apr 16 15:29:22 +0000 2020@NLKProteomics @dtabb73 @_Astro_Nerd_ Please don't feel you have to reply. I was just messing with you: solving those two little rascals would keep your lab busy for a generation or two.
Thu Apr 16 14:16:18 +0000 2020@astacus Saliva is horrible stuff to work with & testing for low-level proteins in saliva is dodgy, at best.
Thu Apr 16 13:59:57 +0000 2020If you want to vote, there is only an hour left. I find the result surprising, given that very few people actually pay for conferences themselves.
Thu Apr 16 13:54:53 +0000 2020@NLKProteomics @dtabb73 @_Astro_Nerd_ Have you mapped GYPA or GYPC yet?
Thu Apr 16 12:18:11 +0000 2020Note: Uox:p is an important protein in the purine degradation pathway in vertebrates, except hominoids (including H. sapiens). In humans, UOX is a unitary pseudogene with a nonsense stop code preventing its translation, truncating the pathway at the production of uric acid.
Thu Apr 16 12:15:49 +0000 2020Uox:p, urate oxidase (Mus musculus) 🔗 Small peroxisomal enyzme; significant acetylation; aka UOX-2, Uri, Uri2; C-terminal targeting sequence SRL; mature form 2-303 [2,645 x] 🔗
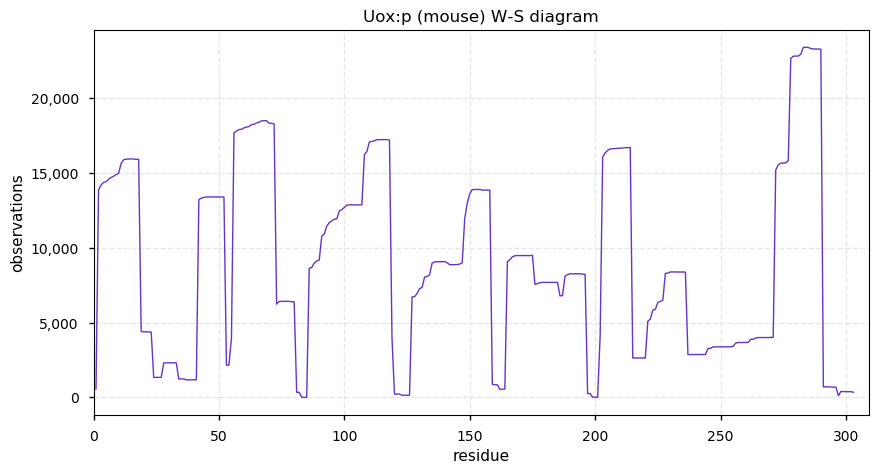
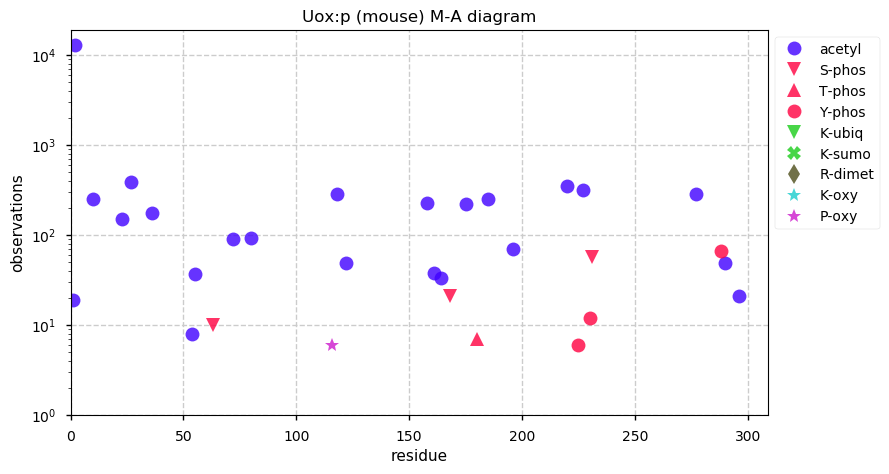
Wed Apr 15 17:47:26 +0000 2020@eliomen @doctorow I preferred to give "open book" exams. It did disadvantage the not-so-good students to a degree, but it took the idea of "cheating" out of the equation.
Wed Apr 15 17:43:09 +0000 2020Asian countries showing rather distinctive patterns in terms of total cases 🔗
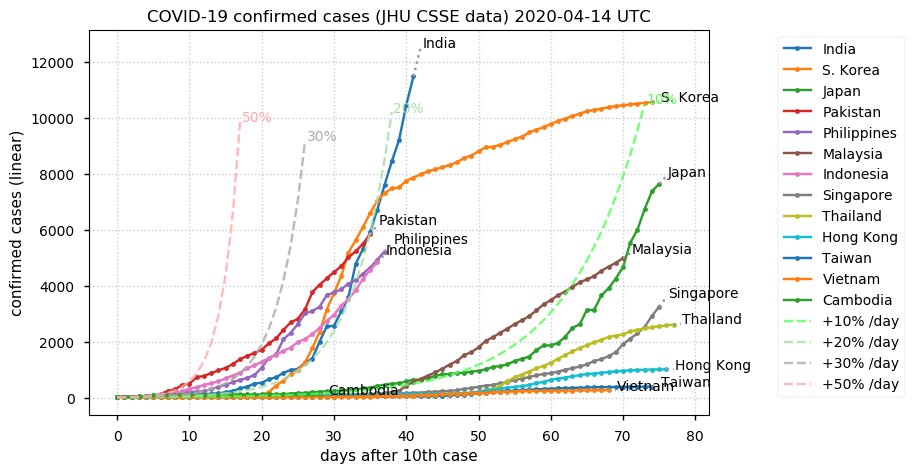
Wed Apr 15 15:37:34 +0000 2020How should on-line conferences be priced?
Wed Apr 15 14:55:40 +0000 2020This data was published as part of Rohlenova K, et al., Cell Metab. 2020 Apr 7;31(4):862-877 (🔗)
Wed Apr 15 14:53:48 +0000 2020PXD016678 has some very good examples of the effects of carbamylation on PSM assignment and protein quantitation. The MS/MS and chromatography are both very well done, making this a good example data set for studying this particular sample preparation artifact.
Wed Apr 15 14:15:02 +0000 2020@jdobbin @bkives The smaller Canadian provinces (except NS) are following a similar trend. 🔗
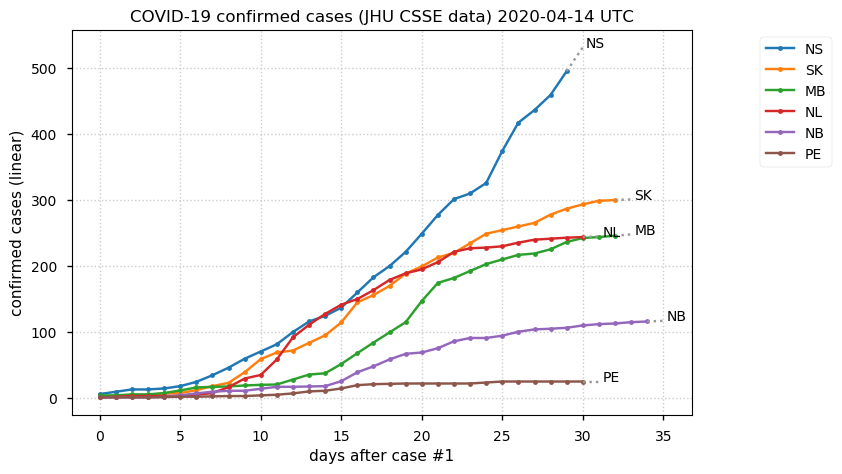
Wed Apr 15 12:10:52 +0000 2020Cat:p, catalase (Rattus norvegicus) 🔗 Midsized peroxisomal enyzme; N-terminal acetyl and scattered phosphorylation; aka Cas1, Cat01, Catl, Cs-1, CS1; C-terminal targeting sequence ANL; mature form 2-527 [3,272 x] 🔗
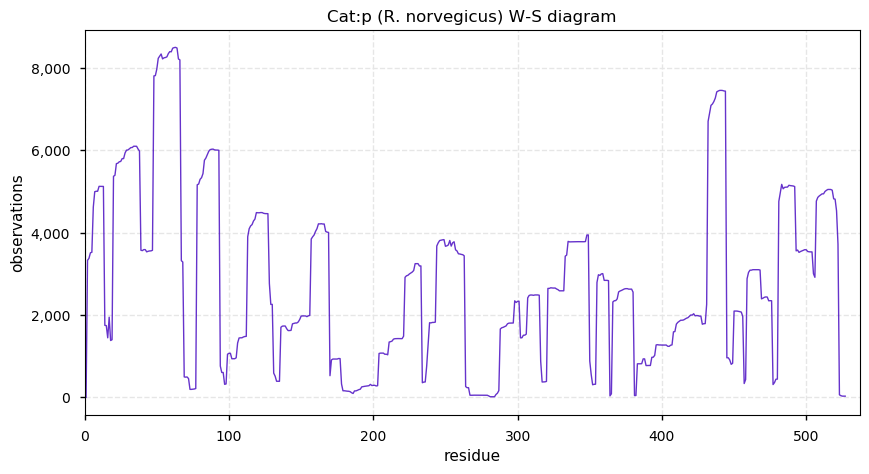
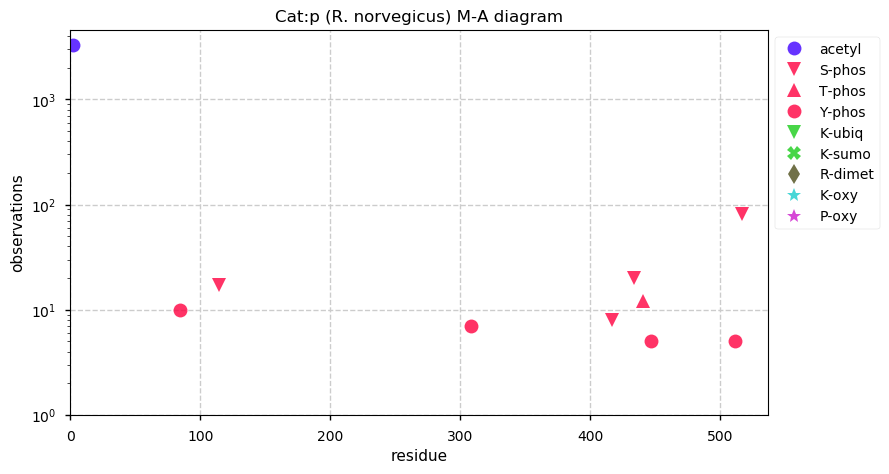
Wed Apr 15 12:00:13 +0000 2020And you can see the EU rate slowing on the total cases linear trend line. 🔗
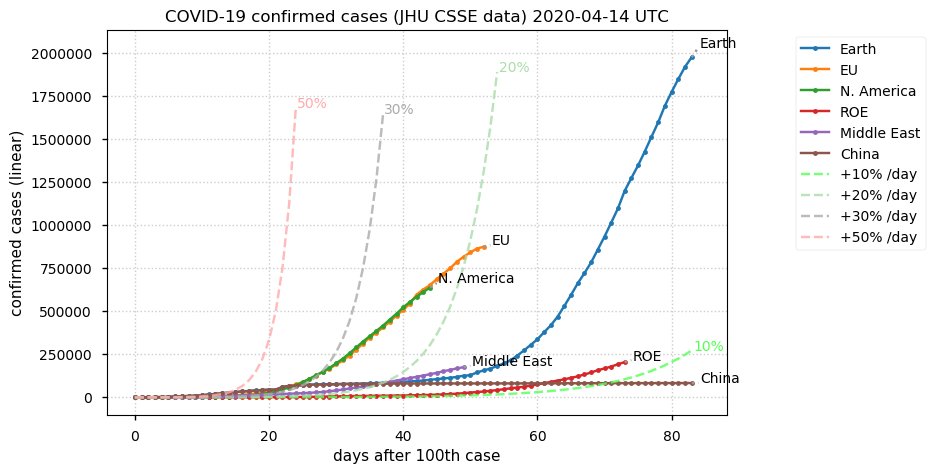
Wed Apr 15 11:54:49 +0000 2020The number of new cases in the EU yesterday dropped to about 20,000/day, 1/2 of its maximum 10 days ago of about 40,000/day. 🔗
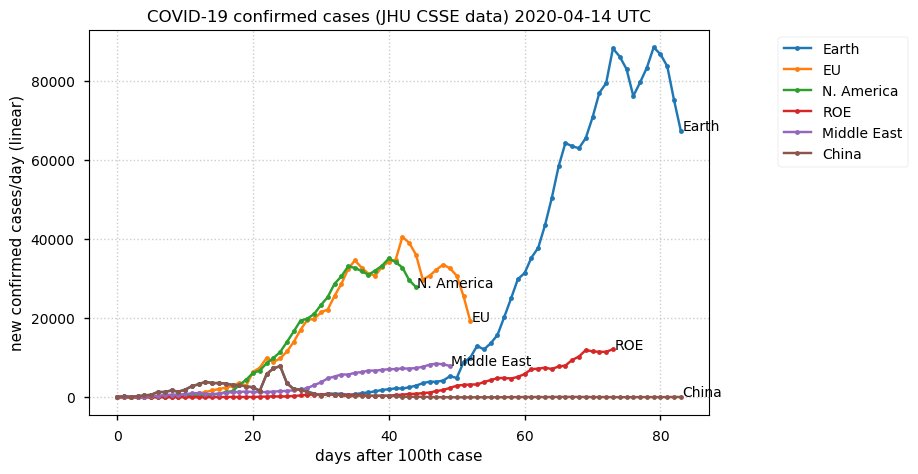
Wed Apr 15 00:21:05 +0000 2020@Keshava_Datta I had seen that, but I'm not sure which of the files on the site correspond to the study. The files require that I register, set up an account & then I might have access. I would then get to download the files one at a time. It is the same as hiding them in a closet.
Tue Apr 14 20:27:47 +0000 2020Pretty good news for Manitobans: so long as things don't go pear shaped for some other reason, we are now towards the good end of a logistic curve 🔗
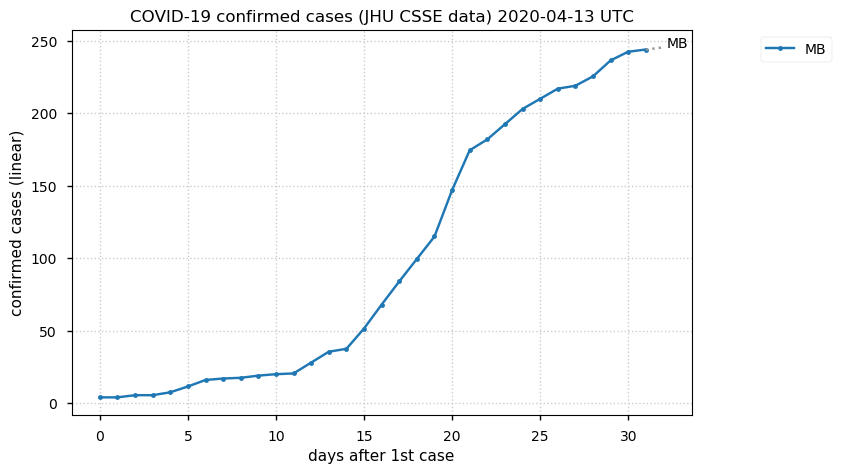
Tue Apr 14 18:24:54 +0000 2020At least my favorite pet food store is open: otherwise there were going to be some pointed looks directed my way later this evening.
Tue Apr 14 15:17:53 +0000 2020PXD017386 is an interesting study that you could probably use as a dry-lab data example (Mizukami H, et al., 2020, 🔗). Nothing high stakes about it, but lots of things that could use explaining.
Tue Apr 14 14:23:41 +0000 2020@Keshava_Datta The data is hidden, though. I had thought that the Nature masthead journals were against this sort of "publish-with-no-data" big projects.
Tue Apr 14 12:17:43 +0000 2020Ehhadh:p, enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase (Rattus norvegicus) 🔗 Midsized peroxisomal enyzme; N-terminal acetyl & scattered phosphorylation; aka LBP, MEF, Mfe, Mfe1, …; C-terminal targeting sequence SKL; mature form 2-722 [1,644 x] 🔗
Mon Apr 13 21:32:54 +0000 2020@MattWFoster @jke000 @pwilmarth @ProteomicsNews So you could use this API (with the JSON output specified) to filter out SNVs with the very low MAFs and studies that aren't really germane (like COSMIC). I guess it would take some coding, though.
Mon Apr 13 20:32:22 +0000 2020@jke000 @MattWFoster @pwilmarth @ProteomicsNews Just directed to the twitter-verse: thanks for getting back to me. My experience with this stuff is that you really have to get in to the source (& MAFs) of the original SNVs if you want to understand the sensitivity and specificity of the PSM assignment process.
Mon Apr 13 19:09:50 +0000 2020@KentsisResearch Thanks.
Mon Apr 13 19:01:59 +0000 2020@KentsisResearch Epstein-Barr. I was wondering about the possibility of class II being generated through the autophagocytosis of virons.
Mon Apr 13 18:25:19 +0000 2020Does anybody know whether intracellular viral proteins tend to be presented as peptides via the MHC class I or class II mechanism?
Mon Apr 13 17:33:37 +0000 2020If you take the NY and NJ lines out of the US graph, it makes a state-by-state linear plot of total cases easier for me to comprehend (NY is ~7.5 × the MA number) 🔗
Mon Apr 13 14:44:05 +0000 2020Pecr, peroxisomal trans-2-enoyl-CoA reductase (Rattus norvegicus) 🔗 Small peroxisomal enyzme; N-terminal acetylation; aka PX-2,4-DCR1, RLF98, TERP; C-terminal PEX5 targeting sequence ARL; mature form 2-305 [1,059 x] 🔗
Mon Apr 13 14:26:18 +0000 2020Probably not the best news for those of us in Canada:
🔗
Mon Apr 13 14:06:31 +0000 2020@jke000 @MattWFoster @pwilmarth @ProteomicsNews Does the variant annotation you are using include those derived from COSMIC SNVs?
Mon Apr 13 12:56:24 +0000 2020@RuneLinding Just noticed this paper of a group working on the technical aspects of qRT-PCR testing for SARS COV-2
🔗
Mon Apr 13 12:54:39 +0000 2020@RuneLinding Any of the numbers at this point are so hot-of-the-presses that using them as the basis for long-term predictions is simply speculation. They could take off like Turkey or roll over quickly like Thailand
Mon Apr 13 12:21:18 +0000 2020And N. America has diverged significantly from the EU wrt fatality trends 🔗
Mon Apr 13 12:18:17 +0000 2020Globally there has been linear growth in total cases for over a week 🔗
Sun Apr 12 15:00:44 +0000 2020Hsd17b4:p, hydroxysteroid (17-beta) dehydrogenase 4 (Rattus norvegicus) 🔗 Midsized peroxisomal enyzme; one phosphodomain; aka DBP, MFE-2, MPF-2; C-terminal targeting sequence YAKL; mature form 2-735 [2,939 x] 🔗
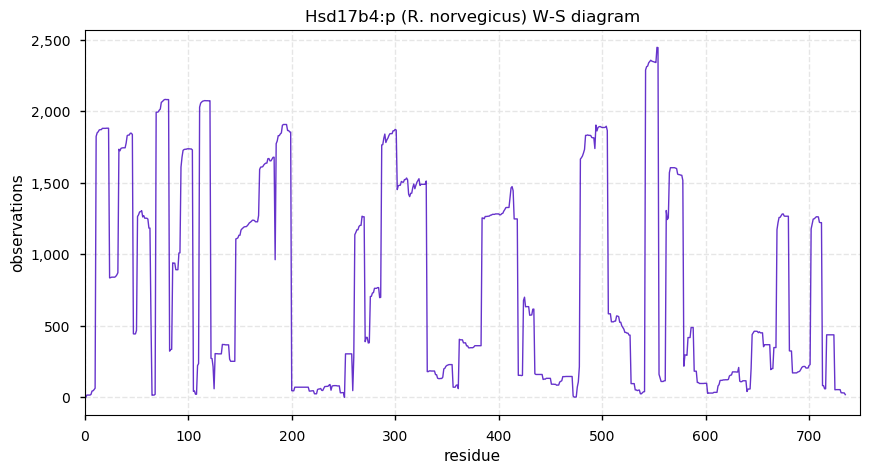
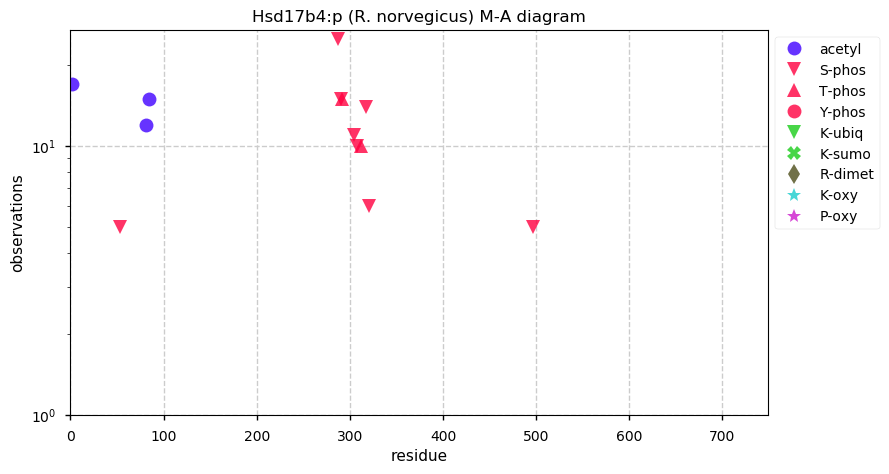
Sun Apr 12 12:34:21 +0000 2020Turkey is really getting socked compared to other Middle Eastern & African countries 🔗
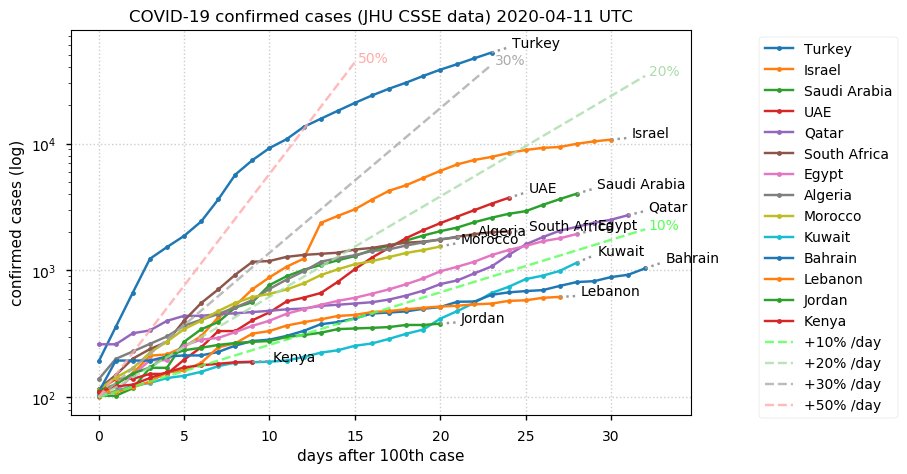
Sat Apr 11 19:36:56 +0000 2020@Sci_j_my The way I read the paper, they took the LC/MS/MS data and tried to find a spectrum that was the best fit to their desired modified sequence.
Sat Apr 11 19:30:57 +0000 2020@Sci_j_my The data in this case is different than a "normal" experiment. They added the amidated synthetic peptide and then tried to find the dopamine-derivatized form of the peptide after incubation. It isn't clear that they looked for any other solution to this particular spectrum.
Sat Apr 11 19:04:36 +0000 2020@Sci_j_my If it really occurs, it would be one of the very few glutamine side chain PTMs. I would have to see more than is presented in this paper to be convinced.
Sat Apr 11 18:56:26 +0000 2020A real problem for the Canadian U15 too:
🔗
Sat Apr 11 17:35:46 +0000 2020@pedrobeltrao @FPRoth @biorxivpreprint @intact_project Have they released the .raw files anywhere? The paper doesn't specifically mention depositing them.
Sat Apr 11 13:07:18 +0000 2020NSP16, 2'-O-ribose methyltransferase (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; no PTMs; methylates the 5' end of viral mRNA; generated by proteolysis of ORF1AB; mature form 1-298 [9 x] 🔗
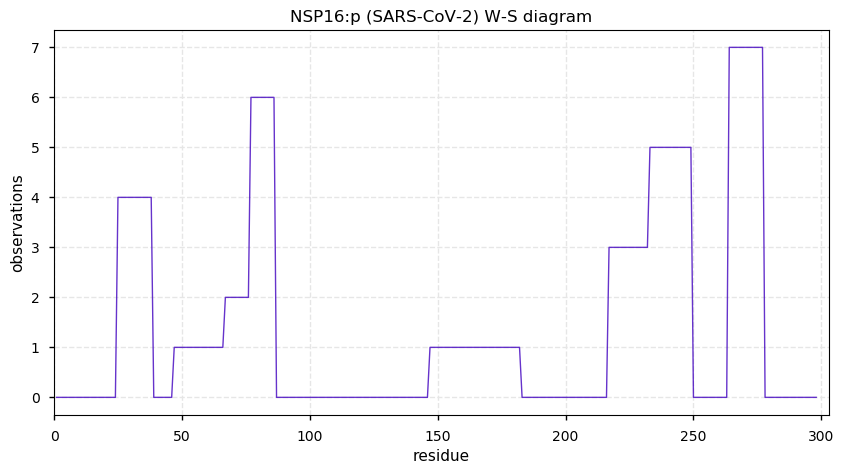
Sat Apr 11 02:49:36 +0000 2020N. America continues to diverge from the EU trend in daily fatalities 🔗
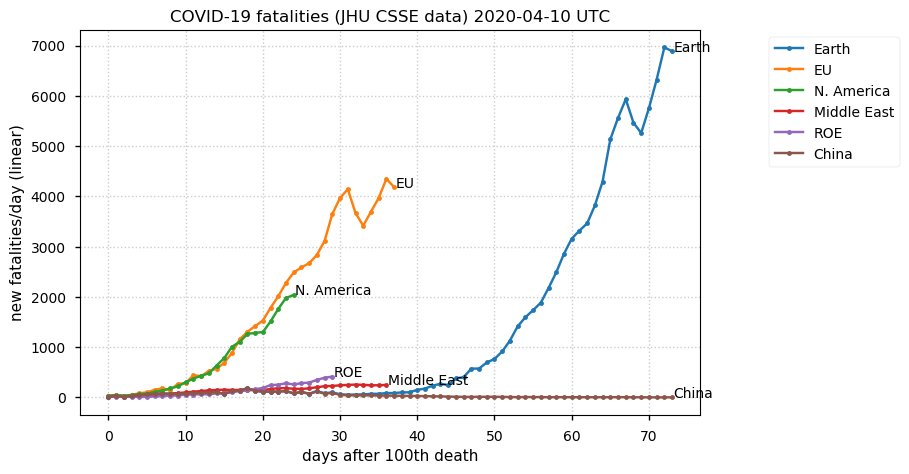
Fri Apr 10 19:34:59 +0000 2020For anyone getting started, remember that "C" in an HLA peptide stands for "cystine" (not "cysteine").
Fri Apr 10 19:09:53 +0000 2020If anyone is looking for a data set to use to get started with HLA peptide analysis (both I & II) from the view point of experiment design, data analysis or QA/QC, I'd recommend giving PXD017149 a good hard look. Lots of data, clinical samples and pretty uniform quality.
Fri Apr 10 18:40:53 +0000 2020It seems the EU is still driving the global trend in fatalities & the N. American trend is slowing 🔗
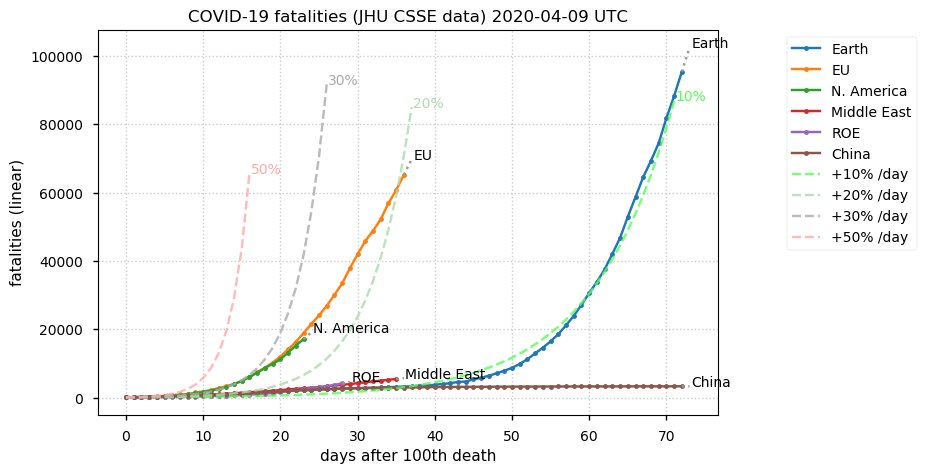
Fri Apr 10 17:33:30 +0000 2020@goodlettlab1 I guess I spoke too soon.
Fri Apr 10 15:31:19 +0000 2020Good for them
🔗
Fri Apr 10 15:29:07 +0000 2020I have to admit nitter is kind of nice
🔗
Fri Apr 10 15:14:56 +0000 2020If you are Canadian and have some spare time, 🔗 is a reasonable discussion of Canadian media's coverage the current situation, whether you agree with it or not.
Fri Apr 10 14:21:02 +0000 2020It isn't even one of the proteins commonly detected in whole urine from patients with P. aeruginosa infections, but it seems to be the only PA protein that ends up in uEVs.
Fri Apr 10 13:18:22 +0000 2020Why is piv:p the only P. aeruginosa protein selectively enriched in human urine extracellular vesicle preparations? Is it in the uEVs or some oddball EV sample prep artifact?
Fri Apr 10 12:27:40 +0000 2020@goodlettlab1 Although if you have Bluetooth turned on, you might as well keep location services on, too.
Fri Apr 10 11:51:59 +0000 2020NSP6 (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; no PTMs; several membrane spanning domains; generated by proteolysis of ORF1AB; mature form 1-290 [7 x] 🔗
Fri Apr 10 11:31:40 +0000 2020@goodlettlab1 Turn off location services! 😲
Fri Apr 10 01:05:50 +0000 2020@MattWFoster @jke000 @pwilmarth @ProteomicsNews It is a co-incidence that you mention a TF SAAV: it was TF:p.I448V (specifically ENSP00000385834:p.I448V) that got me interested in adding SNV information in to peptide id software.
Thu Apr 09 22:38:06 +0000 2020@MattWFoster Unfortunately no particular preference for one protein over another: it is generally modifying all proteins.
Thu Apr 09 22:24:22 +0000 2020@MattWFoster And cysteine persulfide in one data set from the Mann group.
Thu Apr 09 22:20:33 +0000 2020@MattWFoster There are actually lots of data sets that don't reduce or alkylate. Normally you see either trioxidation, reaction with some other reagent (acrylamide, beta-mercaptoethanol), cystine or unreacted cysteine, but I'd never seen glutathione before.
Thu Apr 09 21:45:40 +0000 2020I've always wondered why it didn't show up more often, but then again, I know I wonder about odd things ...
Thu Apr 09 20:43:28 +0000 2020PXD014963 is the first time I've seen significant derivatization of peptide cysteine sidechains with glutathione.
Thu Apr 09 15:22:12 +0000 2020@doctorow A French Press makes good cold coffee without any more mess than usual. You can brew it in a separate jar and decant into the FP to remove the grounds
Thu Apr 09 13:17:22 +0000 2020@alan_jarmusch Is there any way to link the existing SARS-CoV-2 analysis available in GPMDB into your knowledgebase?
Thu Apr 09 12:14:25 +0000 2020There are Xtal structures for NSP2 and it has been shown to be not necessary for viron generation in other coronaviruses, but it's precise function is still a blank spot in the literature.
Thu Apr 09 12:09:58 +0000 2020NSP2, protein of unknown function (Severe acute respiratory syndrome coronavirus 2) 🔗 Midsized accessory protein; no PTMs; generated by proteolysis of ORF1AB; mature form 1-638 [32 x] 🔗
Thu Apr 09 11:53:02 +0000 2020@MattWFoster @jke000 @pwilmarth @ProteomicsNews The entry for a specific TF splice looks like:
<protein id="ENSP00000385834">
<aa id="rs1049296" at="589" type="P" mut="S" />
+ 83 other <aa /> SAAVs
</protein>
Wed Apr 08 22:36:27 +0000 2020@MattWFoster @jke000 @pwilmarth @ProteomicsNews The number of PSMs with SAAVs should always be < 1% of the total number of PSMs (0.5% is typical).
Wed Apr 08 21:55:45 +0000 2020@MattWFoster @jke000 @pwilmarth @ProteomicsNews You really have to get rid of any variant that results in a mass shift of < 3 Da, as well as variants like M->F.
Wed Apr 08 19:09:57 +0000 2020I'm not going to pay any attention until they find super-huge phages
🔗
Wed Apr 08 16:52:28 +0000 2020@astacus A few specific domains of COL1A1 & A2 show up all of the time in HLA class II datasets, so I should not have been surprised, but it was just something I had never noticed.
Wed Apr 08 16:10:10 +0000 2020@KentsisResearch @neely615 I retrieve the standard sequences from ftp://ftp.ncbi.nlm.nih.gov/genomes/Viruses and add new viral sequences as necessary from RefSeq.
Wed Apr 08 15:55:22 +0000 2020@astacus There is just so many bits and pieces of it floating around in the ECM (because of remodeling) and it has to go somewhere ...
Wed Apr 08 15:04:09 +0000 2020Looking at a new dataset from the great Peter Lobel, I am surprised that I had never noticed how much collagen there is in lysosomal preparations. It make all sorts of sense, but still I was surprised.
Wed Apr 08 14:00:49 +0000 2020@KentsisResearch @neely615 I haven't used FASTA files for a long time. It isn't a good format, which becomes very clear if you try to do things in a distributed manner. I use a binary format that allows validation & allocate specified blocks of sequences for any particular PSM assignment session.
Wed Apr 08 13:50:38 +0000 2020@neely615 I'm not really sure what you are referring to here. Could you give me an example?
Wed Apr 08 13:46:12 +0000 2020Seems like someone has been watching "The Expanse" in their down time (note: it is not a documentary)
🔗
Wed Apr 08 12:53:13 +0000 2020@neely615 I also check the human endogenous retroviruses (but never see them, except for the rare observation of the gag protein of HERV K in some cancer tissues).
Wed Apr 08 12:49:57 +0000 2020@neely615 Less than human 50 viruses: HPVs, AdVs, the 7 herpesviruses, a few viruses commonly involved in cell line creation, CoVs & rhinoviruses, 5 parainfluenzaes, XMRV. It adds up to about less than 1000 protein sequences.
Wed Apr 08 12:25:48 +0000 2020I know another media-darling virus is hogging all the headlines, but I'm still curious about this odd HAdV C (human mastadenovirus C) observation
Wed Apr 08 12:03:38 +0000 2020NSP13, helicase (Severe acute respiratory syndrome coronavirus 2) 🔗 Midsized accessory protein; no PTMs; generated by proteolysis of ORF1AB; mature form 1-601 [15 x] 🔗
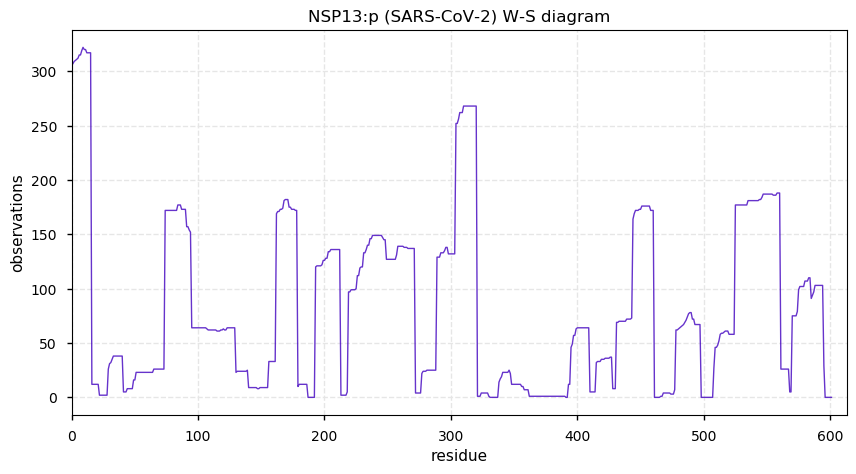
Wed Apr 08 00:32:02 +0000 2020For the 3rd day, new cases in the EU and globally are trending downwards (2 days for North America) 🔗
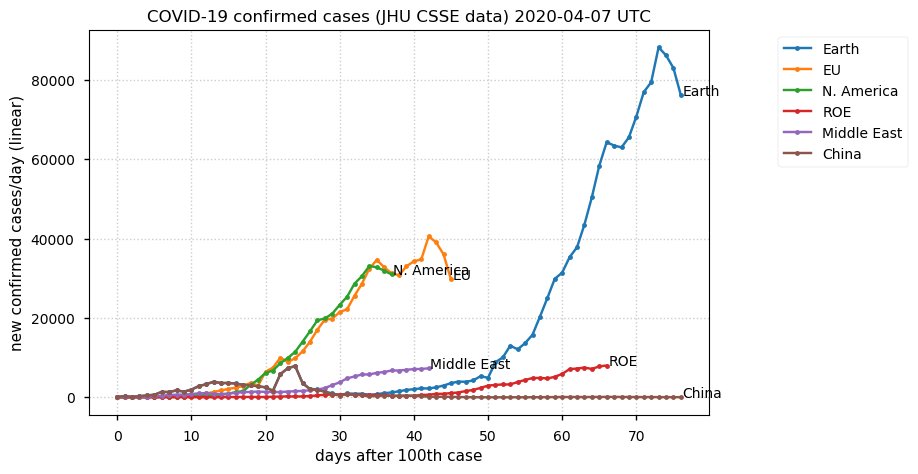
Wed Apr 08 00:29:55 +0000 2020New cases in NY and NJ are down for the 2nd day in a row 🔗
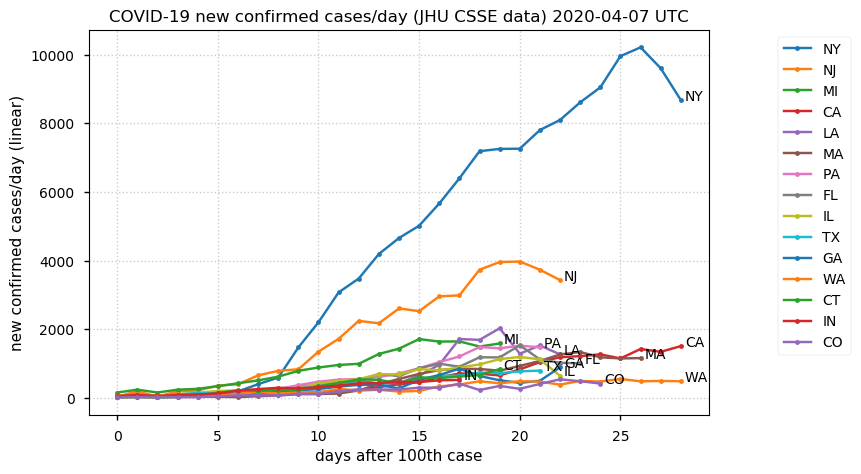
Tue Apr 07 17:21:32 +0000 2020@Sci_j_my @pwilmarth I know: the whole thing looks suspiciously like a setup ...
Tue Apr 07 17:20:27 +0000 2020@RuneLinding It might be fun to be a fly on the wall at the "who wants to volunteer to go to Wuhan" meeting.
Tue Apr 07 16:57:30 +0000 2020@Sci_j_my @pwilmarth So long as you are willing to go with ≥ 5 replicates.
Tue Apr 07 16:54:49 +0000 2020@Sci_j_my @pwilmarth While it does seems like a coincidence, 🔗 just popped up this morning. My preliminary QA shows it to be pretty good data that might fit your requirements.
Tue Apr 07 16:34:41 +0000 2020@pwilmarth There are a surprising number of poly E's: 41 genes code for 'EEEEEEEEEEEEE' and some have it twice.
Tue Apr 07 16:24:04 +0000 2020@VATVSLPR And does it help (or hurt) that these sequences only show up in 3 out of > 700 HLA preps (1 LCMS/MS per prep) that comprise the study in question?
Tue Apr 07 15:38:37 +0000 2020@VATVSLPR Does it help (or hurt) that this one also shows up, including a few adjacent residues from the same protein 🔗
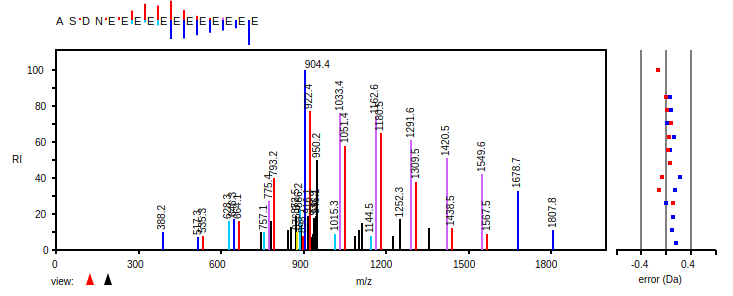
Tue Apr 07 15:26:26 +0000 2020@VATVSLPR It is an HLA class II peptide, so there are no peptide cleavage constraints.
Tue Apr 07 15:13:04 +0000 2020aka, real (but weird) or simply BS/BS.
Tue Apr 07 15:10:49 +0000 2020Alright, smart guys: is this PSM deterministic or stochastic? 🔗
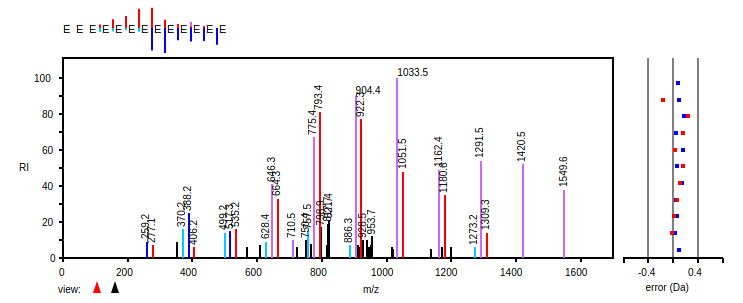
Tue Apr 07 14:52:40 +0000 2020🔗
Tue Apr 07 14:46:30 +0000 2020The geese have arrived and the cat wanted to go outside this morning (1st time since November for both). Spring comes to Winterfell!
Tue Apr 07 14:40:13 +0000 2020Does anybody know why there are so many HLA class I peptides from HAdV-C in clear cell renal cell carcinoma samples?
Tue Apr 07 13:31:21 +0000 2020NSP1, leader protein (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; N-terminal acetylation (contains the initiator M); generated by proteolysis of ORF1AB; mature form 1-180 [20 x] 🔗
Tue Apr 07 00:09:28 +0000 2020New global cases moving in a better direction, and European new fatalities in the hardest hit countries slowing down. 🔗
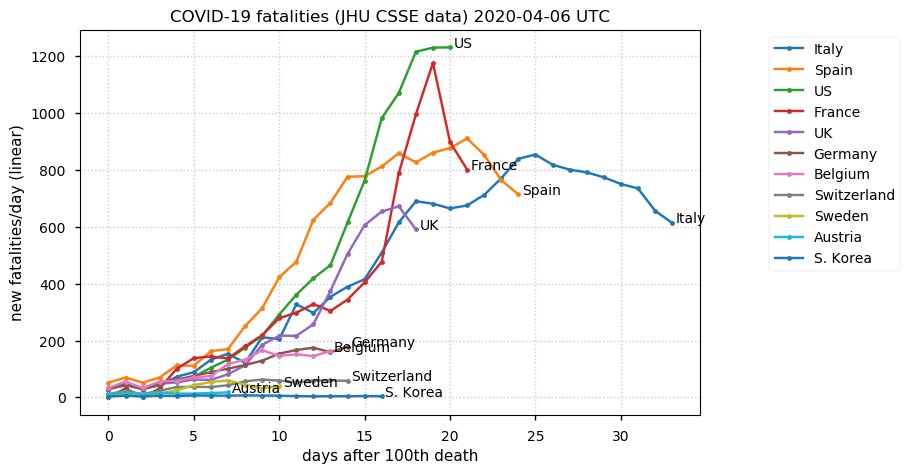
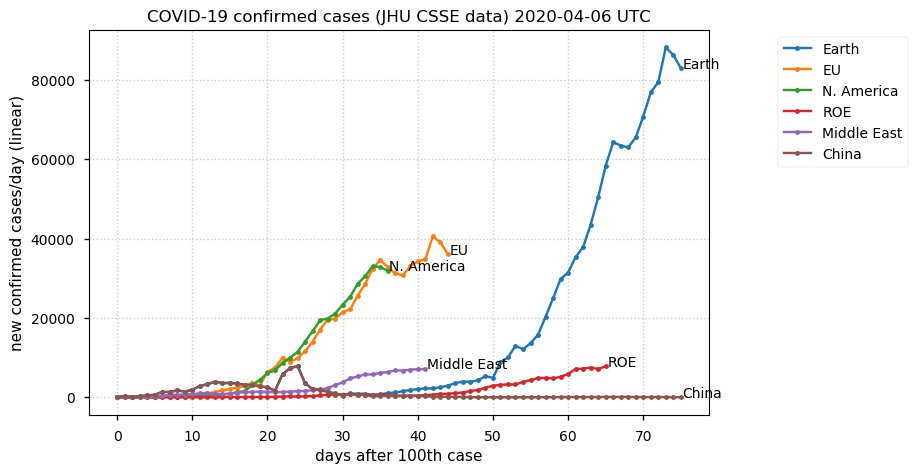
Mon Apr 06 21:32:21 +0000 2020@Sci_j_my @pwilmarth I can't think of one that meets all of the criteria, but I will have a look. Data sets with more than 3 replicates are pretty much always technical replicates of some sort.
Mon Apr 06 15:31:47 +0000 2020@slashdot If true, truly loopy.
Mon Apr 06 14:53:14 +0000 2020🔗
COBOL is a compiled language & for those too young to remember, here is a "Hello world example:
IDENTIFICATION DIVISION.
PROGRAM-ID. HELLO-WORLD.
* simple hello world program
PROCEDURE DIVISION.
DISPLAY 'Hello world!'.
STOP RUN.
Mon Apr 06 14:39:47 +0000 2020@Sci_j_my Patting a tiger will, however, remain ill advised ...
Mon Apr 06 14:24:18 +0000 2020For anyone who is still listening to press conferences for medical advice, please read 🔗
Mon Apr 06 14:05:45 +0000 2020Just a reminder, you should not use TMT for the quantitation of lysine succinylation.
Mon Apr 06 12:28:10 +0000 2020NSP15, endoribonuclease (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; scattered phosphorylation; generated by proteolysis of ORF1AB; mature form 1-346 [10 x] 🔗
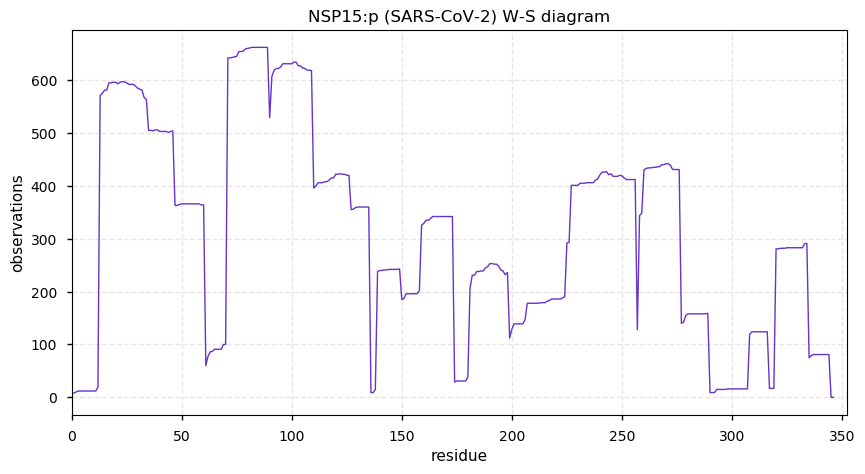
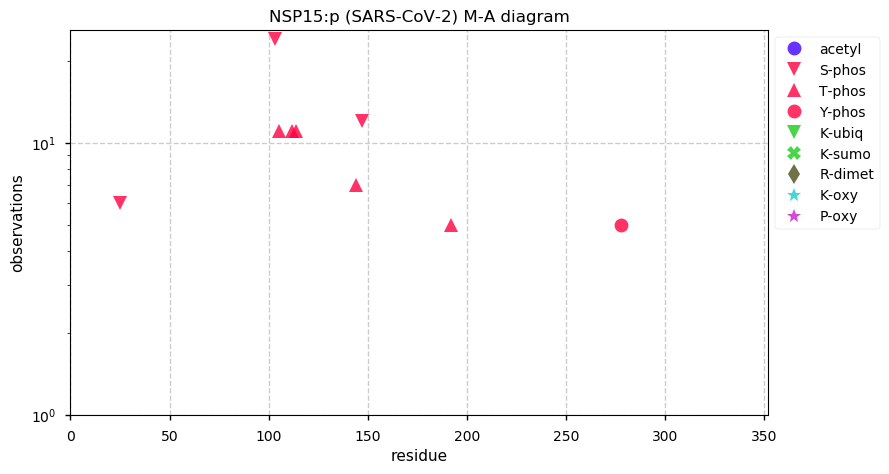
Mon Apr 06 00:12:03 +0000 2020Even the global growth in cases showed a better trend, with the US still following the EU with a lag, as has been the case for the last 2 weeks 🔗
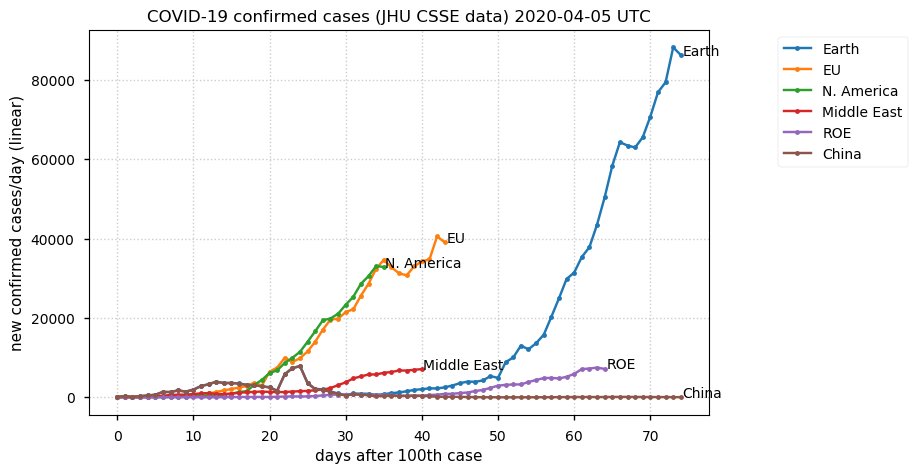
Mon Apr 06 00:09:02 +0000 2020Things slowed down today. 🔗
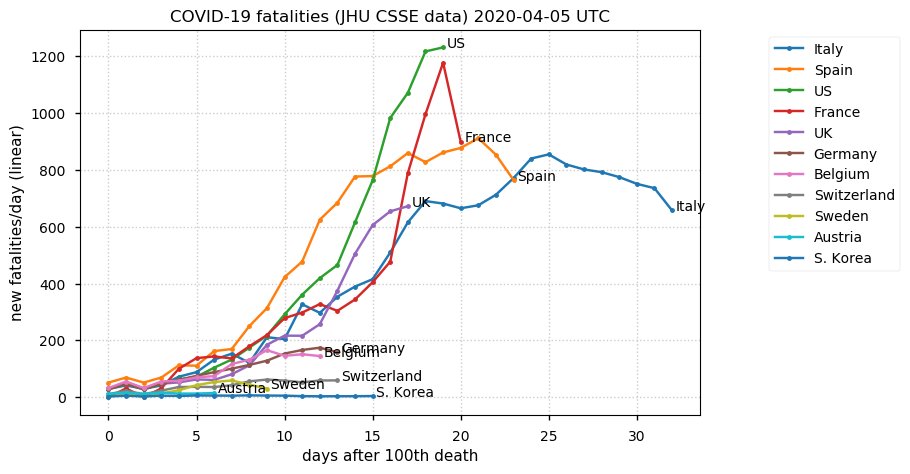
Sun Apr 05 17:26:05 +0000 2020@dtabb73 @friendofthesci @zacmcd77 I use a "red team/blue team" approach, which is nearly impossible in an academic lab, as PI will always be captain of the blue team.
Sun Apr 05 16:22:33 +0000 2020@friendofthesci @zacmcd77 I'm afraid it is too complicated for Twitter & too little general interest for a paper.
Sun Apr 05 14:16:47 +0000 2020@neely615 @byu_sam There are all sorts of weirdness in reporting: some politics but a lot simply people in an unanticipated stressful situation. I think that is why epidemiology is usually done years after the sturm-und-drang of the "event" is over.
Sun Apr 05 13:45:17 +0000 2020@doctorow Same problems here in Canada, but these guys are still at it 🔗
Sun Apr 05 13:04:22 +0000 2020@zacmcd77 Mainly automated QA metrics, and some manual inspection because I didn't believe how good the automated tests looked.
Sun Apr 05 12:36:38 +0000 2020NSP12, RNA-dependent RNA polymerase (Severe acute respiratory syndrome coronavirus 2) 🔗 Midsized accessory protein; scattered phosphorylation; generated by proteolysis of ORF1AB; mature form 1-932 [31 x] 🔗
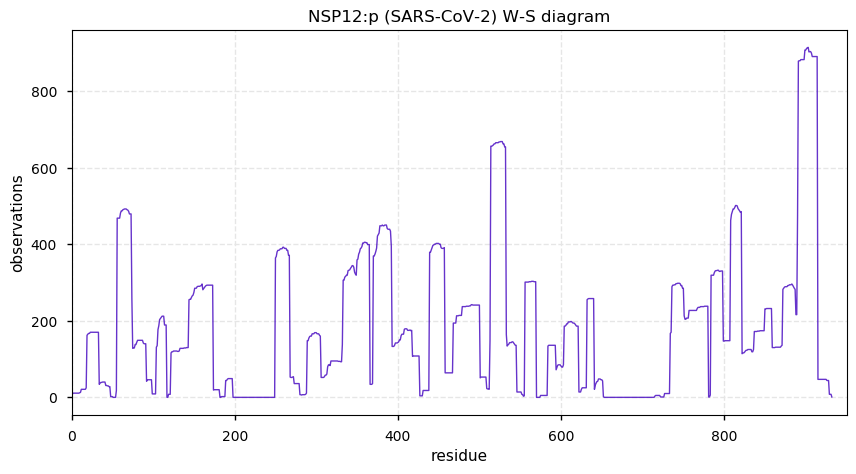
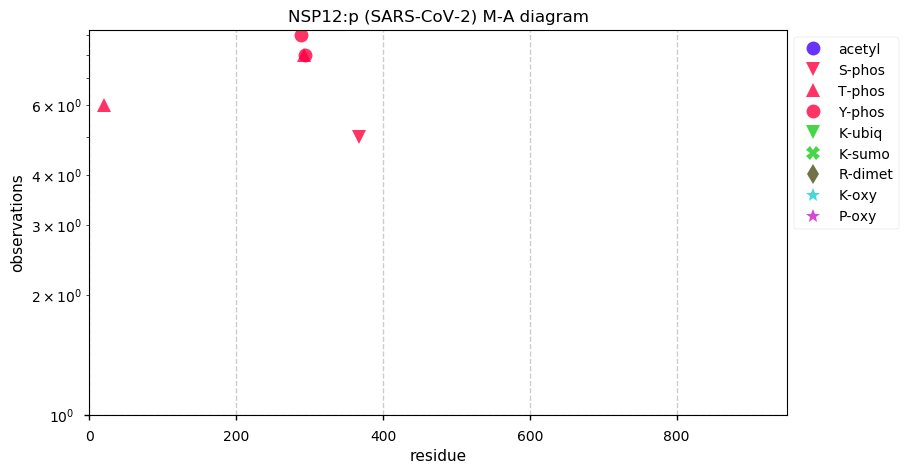
Sun Apr 05 01:40:03 +0000 2020@jwoodgett A lot of people who normally don't draw graphs are now drawing graph-like illustrations.
Sun Apr 05 00:12:24 +0000 2020Things are settling down in Spain and Italy, but they have gone pear shaped in France. Merde! 🔗
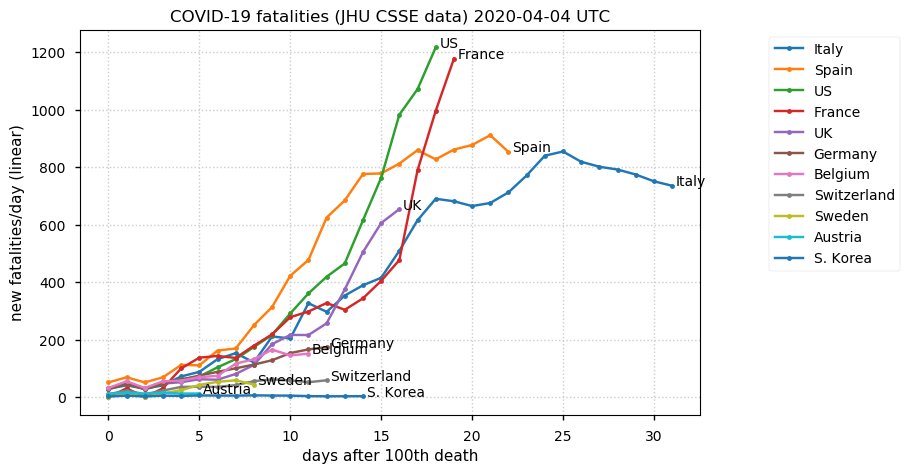
Sat Apr 04 19:08:13 +0000 2020Based on the manuscript, we know the versions of Ubuntu (16.04.5 LTS) and R (3.5.1) used to do the stats, but no idea who ran the instruments.
Sat Apr 04 19:01:03 +0000 2020It is a little mystery of modern proteomics /fin
Sat Apr 04 18:59:43 +0000 2020I'm pretty sure no one tests for its presence in cell lines or animals (tell me if I'm wrong), but it does produce easily detectable amounts of protein in infected cells. /3
Sat Apr 04 18:58:05 +0000 2020But it is still pretty much an enigma. Because of some early problems (described here 🔗), it is has fallen out of favor as a topic of study, but it regularly shows up in protein-protein pull down experiments. /2
Sat Apr 04 18:56:16 +0000 2020Xenotropic murine leukemia virus-related virus (XMRV) is a retrovirus that has been observed in > 6000 publicly available LC/MS/MS runs. It has been found in mouse cell line, mouse tissue & human cell line experiments, more often than any other virus. /1
Sat Apr 04 15:51:21 +0000 2020⭐️⭐️⭐️⭐️
Sat Apr 04 15:42:58 +0000 2020Whoever actually ran the instruments that generated the data for Atlasi, et al. 2020 (🔗 | 🔗) should be up for a HUPO award. By far the best MS/MS data made available in the last 12 months.
Sat Apr 04 14:04:38 +0000 2020@Sci_j_my You have clearly never met a statistician.
Sat Apr 04 13:25:18 +0000 2020Lots of interesting smORF peptides, too.🙂
Sat Apr 04 13:20:59 +0000 2020Textbook examples of observed HLA class I peptide length distributions. Congrats Reustle A 2020 (🔗 | 🔗). 🔗
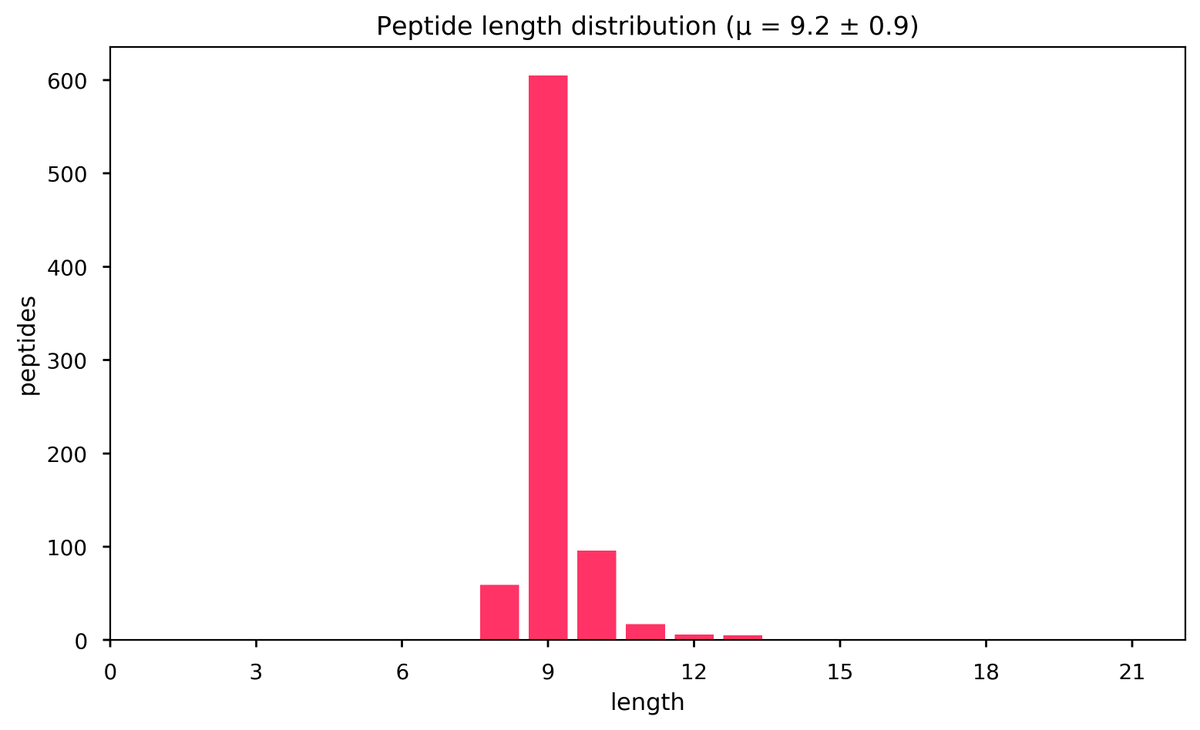
Sat Apr 04 12:35:03 +0000 2020NSP14, 3'-to-5' exonuclease (Severe acute respiratory syndrome coronavirus 2) 🔗 Midsized accessory protein; S53+phospho; generated by proteolysis of ORF1AB; mature form 1-527 [21 x] 🔗
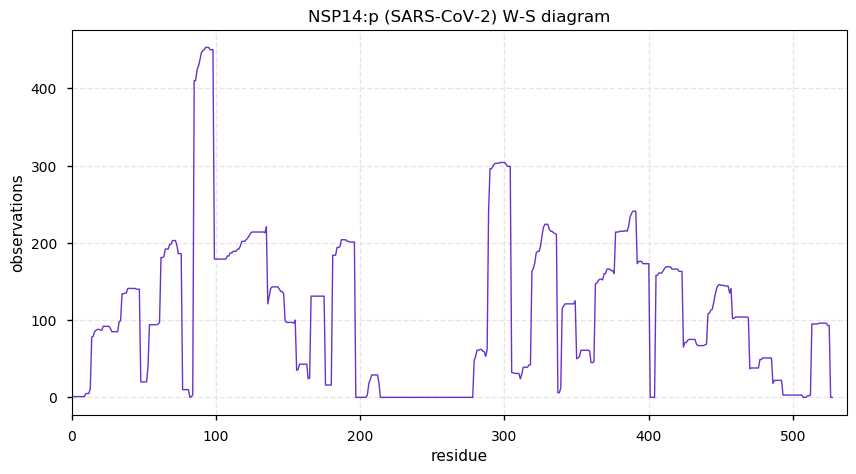
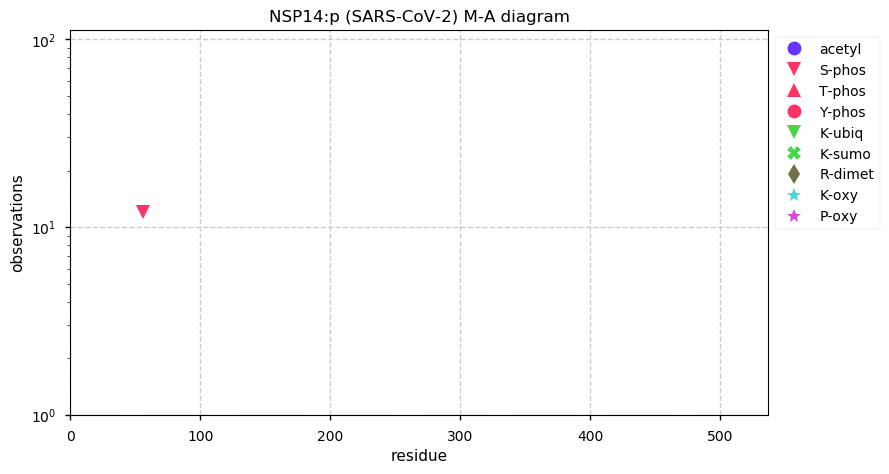
Sat Apr 04 11:53:27 +0000 2020@MikePrevost3 @NadelParis They are operating normally. The number of cases in Canada is still fairly low.
Sat Apr 04 01:16:44 +0000 2020Canada's reporting rates look screwy. In terms of cases, we are trending a little higher than Holland, but in fatalaties we are trending more like Denmark. Pourquoi? 🔗
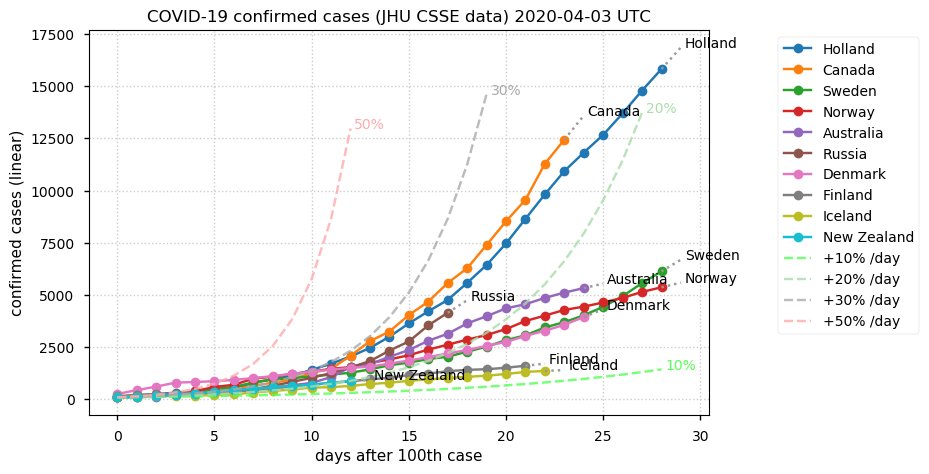
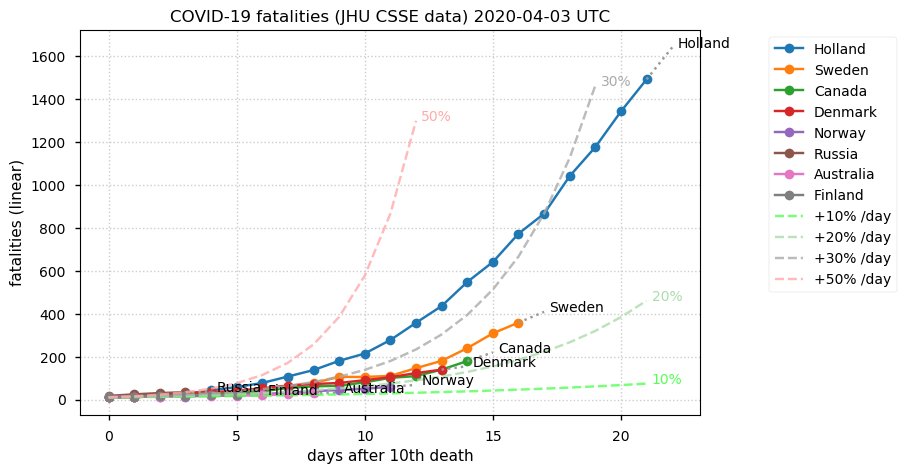
Sat Apr 04 00:48:10 +0000 2020@jwoodgett Soluble ACE2 can be pretty abundant in urine but largely absent from plasma ... apropos of nothing (fun fact)
Fri Apr 03 17:09:16 +0000 2020Hard times for the paparazzi business
🔗
Fri Apr 03 16:12:36 +0000 2020Also remember that if observable protein is still present in a sample that is about 1 million years old, it has been physically isolated from the environment for that time. Do not expect a lot of exotic modifications or extreme levels of oxidation.
Fri Apr 03 16:04:39 +0000 2020For anyone giving this a try, remember that the abundant enamel-specific protein amelogenin has an X-chromosome-coded protein AMELX:p with 3 observable splice variants & a Y-chromosome-coded protein AMELY:p that will only show up in male samples.
Fri Apr 03 15:19:17 +0000 2020If you want to test your peptide id skills, try re-analyzing PXD014342. Tooth enamel is difficult at the best of times, but from archeological samples it is double tough.
For extra points, try seeing how well they did in comparison to PXD009781.
Fri Apr 03 14:25:51 +0000 2020PXD016828 has some nice observations of thyroglobulin iodination in normal thyroid tissue as well as primary and metastatic tumors. Iodination is a complex, multi-step PTM that requires specialized intracellular vesicles, showing that the tumors retain this elaborate mechanism.
Fri Apr 03 14:14:24 +0000 2020@DougieGordie @AlexUsherHESA Why does anyone have their location services on while they are sitting around the house? It reduces battery life and does you no good at all.
Fri Apr 03 13:25:40 +0000 2020NSP4, non-structural protein 4 (Severe acute respiratory syndrome coronavirus 2) 🔗 Midsized accessory protein; no PTMs; formed by proteolysis of ORF1AB; potential N-T signal sequence + 4 adjacent membrane domains (250-398); mature form 34,35-500 [16 x] 🔗
Fri Apr 03 12:19:19 +0000 2020The trends in the total number of deaths and the number of deaths per day are still being driven by Europe, with N. America following the same trend, but delayed by about 12 days 🔗
Fri Apr 03 00:02:51 +0000 2020@Sci_j_my We do have several 4 kilo containers of antique mayonnaise you might be interested in.
Thu Apr 02 23:56:22 +0000 2020@Sci_j_my My wife would vote yes, I would vote no.
Thu Apr 02 21:52:57 +0000 2020@RuneLinding @EricTopol @FT But in a direct comparison, the US looks very similar to the EU, just about 12 days behind.
Thu Apr 02 21:52:00 +0000 2020@RuneLinding @EricTopol @FT It is always possible that the reason the FT doesn't use the EU numbers is that they don't have anyone on board who can parse that info out of the data, which is broken up by country and doesn't have an EU entry.
Thu Apr 02 21:30:06 +0000 2020@EricTopol @RuneLinding @FT Why compare the US to Spain? Wouldn't the EU be a better grouping for comparison?
Thu Apr 02 20:58:01 +0000 2020@byu_sam And I was amused by my interface complaining to me that their interface wasn't working. Sort of a bots-shading-bots situation.
Thu Apr 02 20:53:28 +0000 2020@byu_sam No, I'm using their numbers. They were about 2 hours late releasing yesterday's CSVs.
Thu Apr 02 20:31:58 +0000 2020@chrashwood All of the meeting systems are some variation of the same idea. I find them all even more tedious and less informative than there physical counterpart. Getting rid of all of the social cues & collective behavior associated with a physical meeting pretty much defeats the purpose.
Thu Apr 02 19:45:07 +0000 2020People really seem to have drunk the Kool-aid wrt Zoom meetings. The few I have been to are some combination of bad and worse.
Thu Apr 02 17:52:13 +0000 2020CIHR to the rescue
🔗
Thu Apr 02 17:23:56 +0000 2020@UCDProteomics It is where I get the numbers to make plots like those attached (& more at 🔗) 🔗
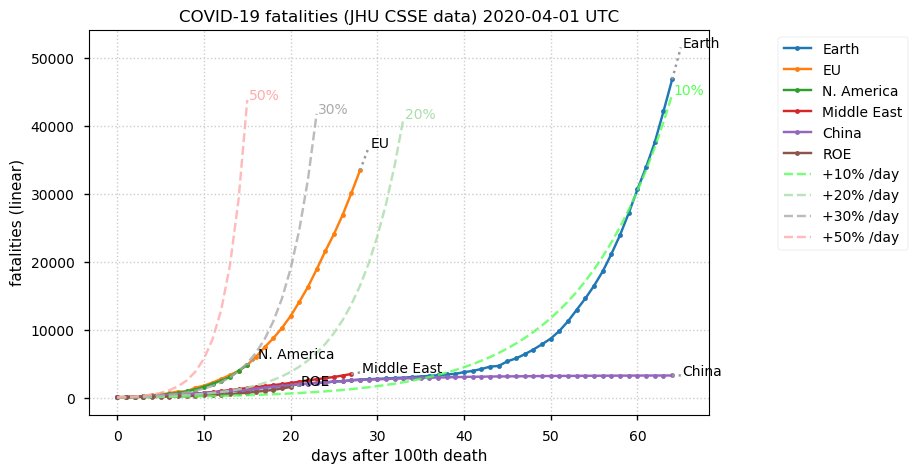
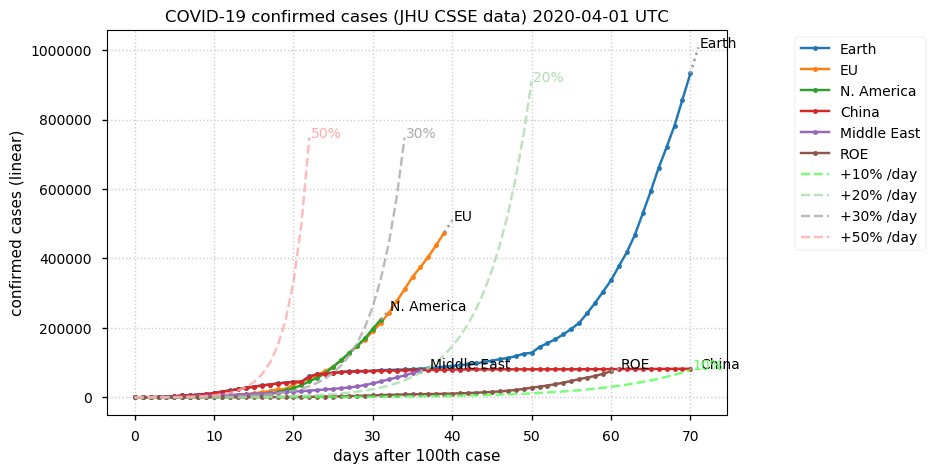
Thu Apr 02 17:14:05 +0000 2020@UCDProteomics But it has a vibrant "Issues" section!
Thu Apr 02 17:12:57 +0000 2020@UCDProteomics It is an odd github repository in that it contains some processed data (CSV files), but none of the code or database schemas.
Thu Apr 02 15:50:14 +0000 2020@cstross They are the guys you see on stage in what look like navy uniforms at US press conferences.
Thu Apr 02 15:47:30 +0000 2020@cstross This group 🔗 is an under-appreciated part of the US government that is very influential at the moment & comes out of the naval medical tradition. If you spend any time at the US NIH campus in Bethesda, you see these uniforms all over.
Thu Apr 02 15:35:14 +0000 2020NSP7, non-structural protein 7 (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; one phosphodomain; generated by proteolysis of ORF1AB; mature form 1-83 [71 x] 🔗
Thu Apr 02 14:04:35 +0000 2020@dtabb73 Try doing an amino acid analysis of the observed peptides and compare that to an amino acid analysis of the observed proteins. It is my go-to method for checking out new data.
Thu Apr 02 13:56:51 +0000 2020@dtabb73 And the lost of C-containing peptides is very common. At least 20% of public data has very-poor-to-no recovery of these peptides. It is rare (& remarkable) for a data set to show recovery of > 90% of observable cysteines.
Thu Apr 02 13:53:30 +0000 2020@dtabb73 One other thing: be sure to check for +57. Trust no one.
Thu Apr 02 13:20:42 +0000 2020You need to understand how SARS-CoV-2 generates ribosomal frame shifts to comprehend how orf1ab is used to generate accessory proteins
Thu Apr 02 13:14:35 +0000 2020Another article that deals directly with the subtleties of the virus de jour is Plant, et al. 2010
🔗
Thu Apr 02 13:06:55 +0000 2020If you have recently begun to wonder how a host cell ribosome translates protein from instructions provided by an RNA virus, this 2012 article by Firth & Brierley is a good place to start 🔗
Thu Apr 02 12:54:51 +0000 2020@dtabb73 Check for Cys+48 (3 O's, mono=47.984744). If there aren't any, then probably the reduction didn't work, either because of pH/stale reagent/pipetting error. If there are +48's, then it was the MMTS derivatization (pH/stale reagent/pipetting).
Thu Apr 02 01:07:55 +0000 2020Does it make me a bad person because I'm amused when someone else's automated API that I use has stalled out? I'm looking at you JHU CSSE 🙂
Wed Apr 01 22:01:14 +0000 2020I had not appreciated how bad an exopeptidase Lys-C really is until just now. It really leaves N-terminal K's dangling 🧐
Wed Apr 01 20:50:16 +0000 2020An exception to this is nsp4, which is processed by signal peptidase to create a new ragged N-terminus at residue 34 or 35. The signal peptide is removed after nsp4 is generated by nsp5's proteolysis of ORF1A:p, so it is not co-translational, but post-translational instead.
Wed Apr 01 18:28:14 +0000 2020PXD016519 used a particularly good batch of Lys-C (unfortunately no lot number in the paper).
Wed Apr 01 18:09:35 +0000 2020@bkives We might want to start thinking about appointing Gary Doer as pro tempore Premier until this is over.
Wed Apr 01 18:00:31 +0000 2020@IonSource @jwoodgett Some things never get old
🔗
Wed Apr 01 17:09:10 +0000 2020@JonAMichaels Mine's 40 and still causes the same effect.
Wed Apr 01 16:44:30 +0000 2020@FridoWelker @UCPH_health @CENIEH @NNFCPR @LFGeoGenetics @MPI_Biochem @NEaar_lab @IBE_Barcelona @uni_copenhagen @MuseumGNM When will the data in PRIDE be released?
Wed Apr 01 16:13:32 +0000 2020@slashdot May be the worst-kept-secret, ever.
Wed Apr 01 15:59:27 +0000 2020@jwoodgett Add it to the list
🔗
Wed Apr 01 15:15:04 +0000 2020@draganall The trend lines associated with COVID-19 deaths also show the same remarkable similarity between the EU and N. America, using the same simple alignment method. 🔗
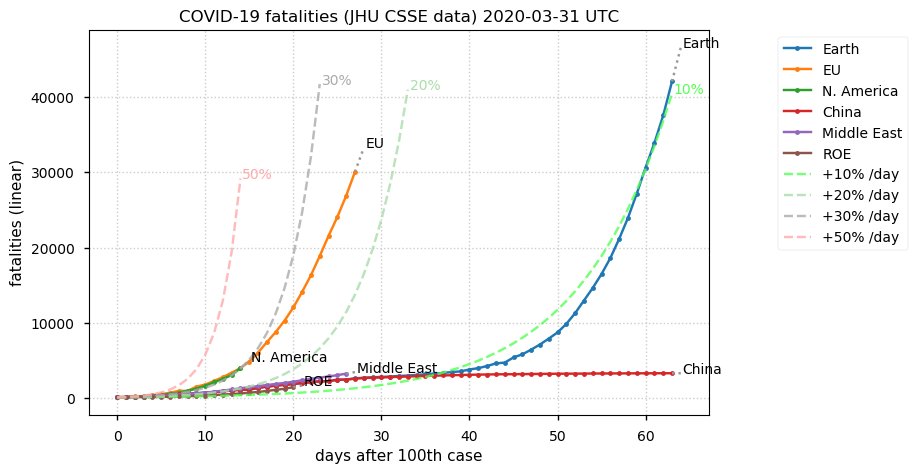
Wed Apr 01 15:06:44 +0000 2020The difference between the non-tryptic peptides in data files with large nsp5 signals and those in data files in which there is NO detectable nsp5 (this important) should give you some insight into how the viral protease affects host cell proteins. /fin
Wed Apr 01 14:30:26 +0000 2020Based on this graph, guess how many days ago Quebec (QC) changed the way they report cases to be different from the rest of Canada. 🔗
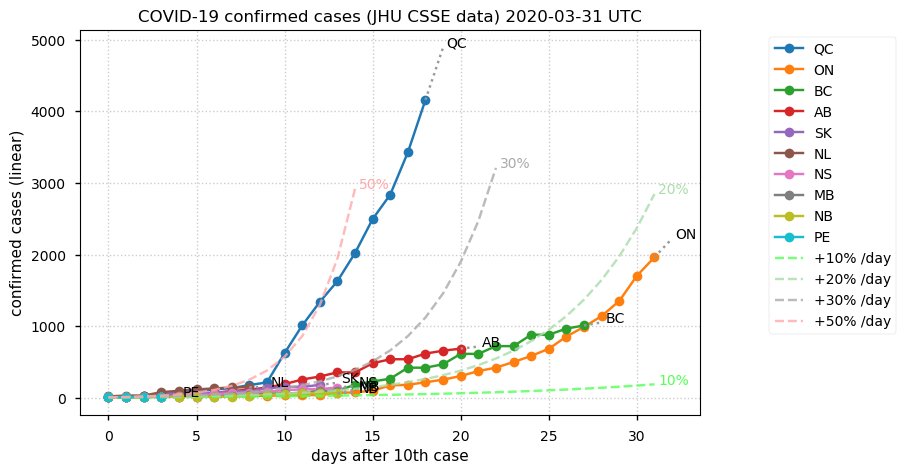
Wed Apr 01 14:11:20 +0000 2020Something that may be of interest to virology-proteomics-types, PXD018117 contains data where the viral protease (nsp5) is in high concentration, resulting in enhanced non-tryptic cleavage in the ID'd peptides (e.g., qx017090.raw, qx017091.raw, qx017092.raw). /1
Wed Apr 01 13:07:53 +0000 2020It looks like there is some additional "something" going on with the Strep tags: why do they even bother with Methods sections? 😾
Wed Apr 01 12:21:06 +0000 2020@draganall The ROE curve (Rest Of Earth) corresponds to everywhere outside of the other 4 geographies.
Wed Apr 01 12:09:18 +0000 2020The proteins generated by cleavage of a polyprotein do not have co-translational modifications, e.g., signal sequence removal or N-terminal acetylation
Wed Apr 01 12:06:56 +0000 2020NSP9, non-structural protein 9 (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; several phosphodomains; generated by proteolysis of ORF1AB; mature form 1-113 [50 x] 🔗
Wed Apr 01 11:51:07 +0000 2020@draganall The EU is a better comparator with China and N. America than individual countries like Italy or Spain.
Wed Apr 01 01:20:17 +0000 2020I'm surprised by how closely N. America is tracking the EU, with such a simple alignment (starting curves at the day with 100 cases) 🔗
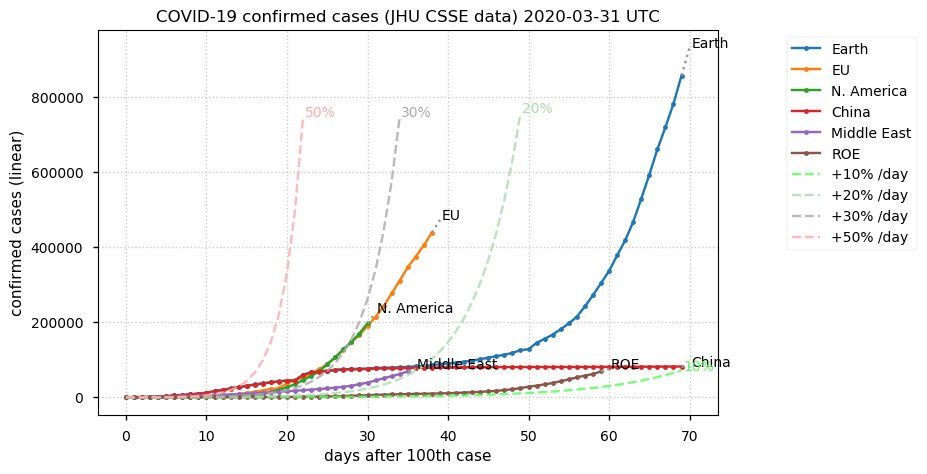
Wed Apr 01 01:05:39 +0000 2020Looks like the tag was on the C-terminus.
Tue Mar 31 21:49:55 +0000 2020Fire up the RPI4: there's check'n to do.
Tue Mar 31 21:49:02 +0000 2020Although I guess if the Strep II tags were attached to the C-terminus of the protein, that would explain it
Tue Mar 31 21:16:23 +0000 2020The data (PXD018117) indicates that most of the nsp proteins (from ORF1AB) are missing the last few residues of their C-termini. No idea why, since these are constructs.
Tue Mar 31 19:30:49 +0000 2020@bffo @NCBI How are they protecting the sequences from redistribution? Copyright or licenses?
Tue Mar 31 17:09:09 +0000 2020I guess it is too much of an "inside baseball" type question to expect a response.
Tue Mar 31 15:47:02 +0000 2020And here I thought we were using TP!
🔗
Tue Mar 31 15:17:35 +0000 2020@pwilmarth @ucdmrt Yup.
Tue Mar 31 14:47:32 +0000 2020@ucdmrt There has been a pretty big surge from the Cryo-folks.
Tue Mar 31 14:30:38 +0000 2020@jbiolchem This is coming down to wire. Time to stop lurking and vote.
Tue Mar 31 14:13:12 +0000 2020I realize they are free to do anything they want in terms of naming proteins, but anyone familiar with the more conventional nomenclature will most likely interpret this nomenclature incorrectly (thinking that NS8_SARS refers to nsp8). Why make this choice?/ fin
Tue Mar 31 14:12:31 +0000 2020What I find confusing is that UniProt is currently listing some of the ORF-translated protein, in a non-standard way. For instance, the ORF8 protein is listed as "NS8_SARS2", with the title "Non-structural protein 8". /3
Tue Mar 31 14:11:47 +0000 2020The protein cleaved out of the polyprotein translated from ORF1A that is 8th from the polyprotein's N-terminus is generally referred to as "nsp8" (non-structural protein 8). Both are non-structural proteins, i.e. they do not end up in the mature viron. /2
Tue Mar 31 14:11:33 +0000 2020Maybe someone can clear this up for me, as I am not a virologist. From my understanding of viral protein naming, the protein product of translating the RNA gene ORF8 (SARS-CoV-2) is generally referred to as "ORF8 protein". /1
Tue Mar 31 12:50:18 +0000 2020As I work through the accessory proteins for this virus, it is surprising to me how many phosphorylation sites have been observed.
Tue Mar 31 12:47:24 +0000 2020NSP10, non-structural protein 10 (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; several phosphodomains; generated by proteolysis of ORF1AB; mature form 1-139 [43 x] 🔗
Tue Mar 31 01:42:51 +0000 2020@THErealDVORAK Unless something changes, CA, MA & WA seem to be adding cases slowly compared to some other states 🔗
Tue Mar 31 01:11:58 +0000 2020In terms of total deaths, the EU remains the major contributor to the global trendline 🔗
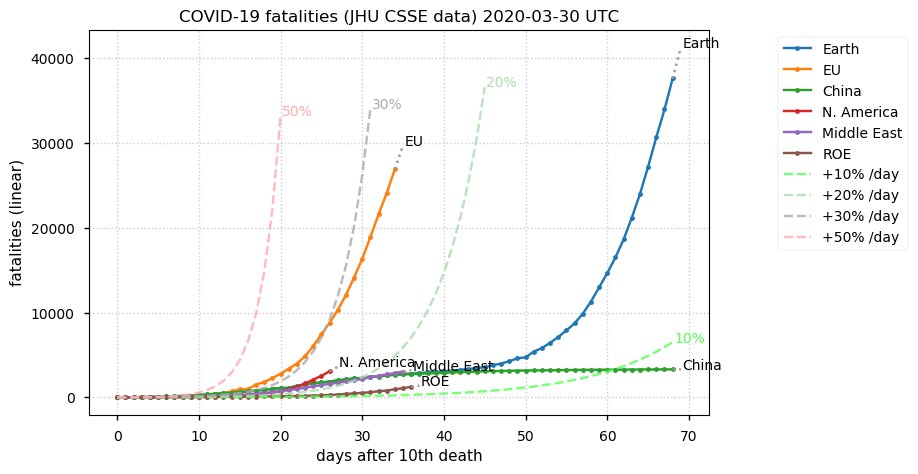
Tue Mar 31 00:23:06 +0000 2020And the decreased rate of increase shows up in the trends for the individual major European countries 🔗
Tue Mar 31 00:20:44 +0000 2020Well, the downward trend in the rate of increase of new cases in globally and in Europe continues for another day 🔗
Mon Mar 30 18:54:13 +0000 2020@Sci_j_my I'll be interested to see if the current trends continue today. The graphs update at 00:15 UTC.
Mon Mar 30 17:09:22 +0000 2020@AlexUsherHESA His main opponent's presence and leadership during the crisis may also keep him in longer, too.
Mon Mar 30 16:06:55 +0000 2020@FridoWelker Theorists should not touch things around the lab without mittens, a face shield and a minder.
Mon Mar 30 15:33:04 +0000 2020Never let an emergency go to waste
🔗
Mon Mar 30 14:43:11 +0000 2020NSP8, non-structural protein 8 (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; N-terminal phosphodomains; generated by proteolysis of ORF1AB: mature form 1-198 [58 x] 🔗
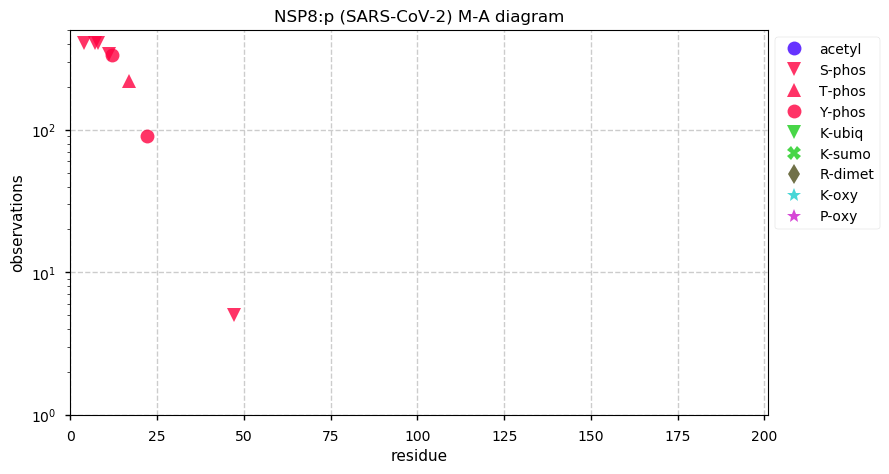
Mon Mar 30 14:20:36 +0000 2020@Sci_j_my Hopefully. The derivative of the individual EU countries case numbers (+ États-Unis), Spain, Germany, France & Italy are all showing decreases in the new cases/day. Italy shows a similar decrease last week, which bounced up again, so the decrease may not be monotonic. 🔗
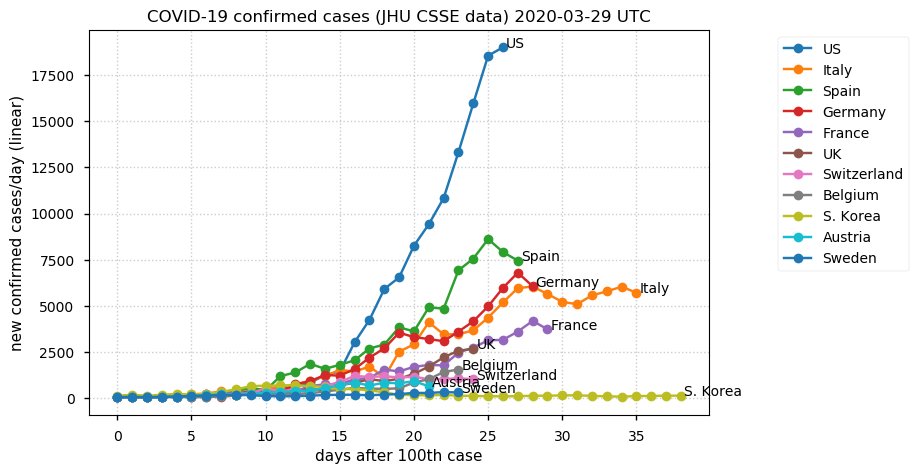
Mon Mar 30 11:47:43 +0000 2020And NY is still driving the numbers of US & North American cases and fatalities 🔗
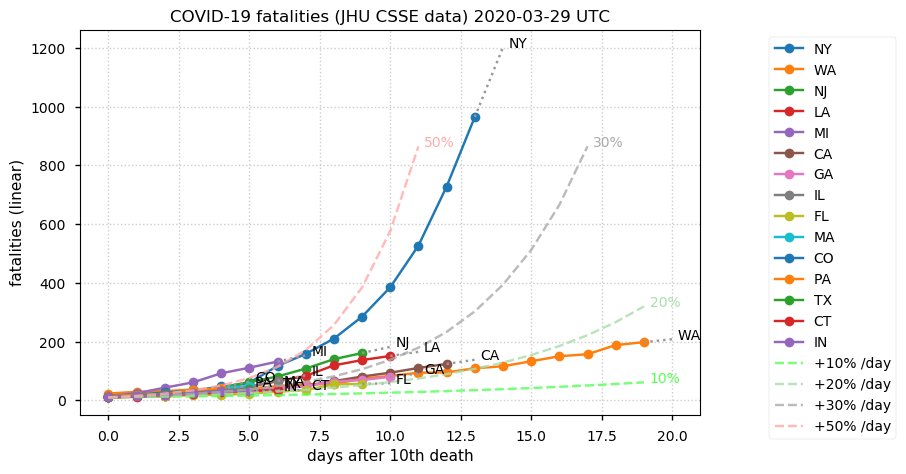
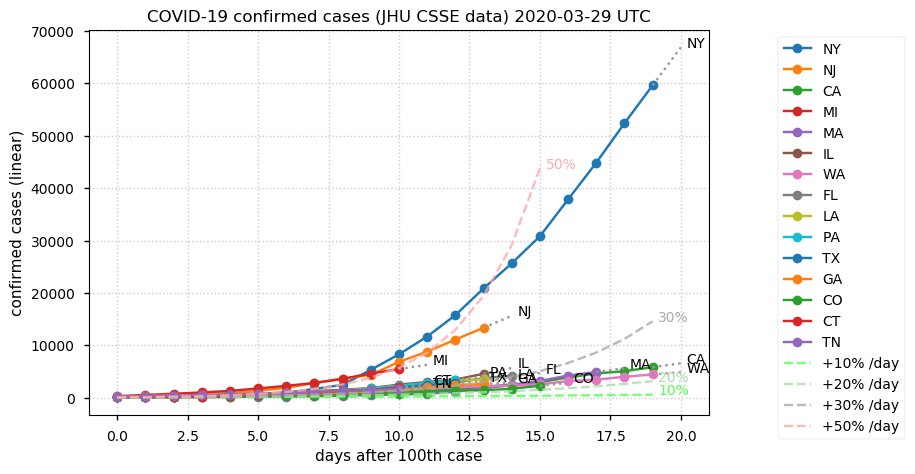
Mon Mar 30 11:28:00 +0000 2020Europe is still getting hit hard 🔗
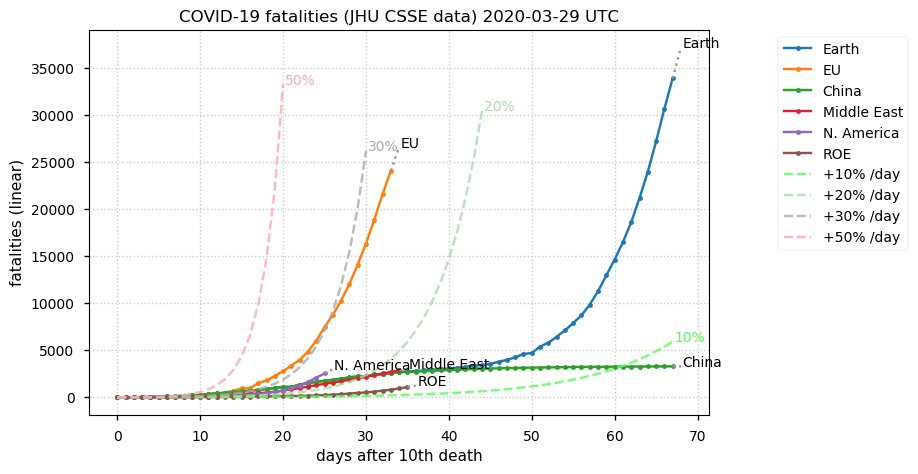
Mon Mar 30 00:27:48 +0000 2020Good news, although it may not last ... 🔗
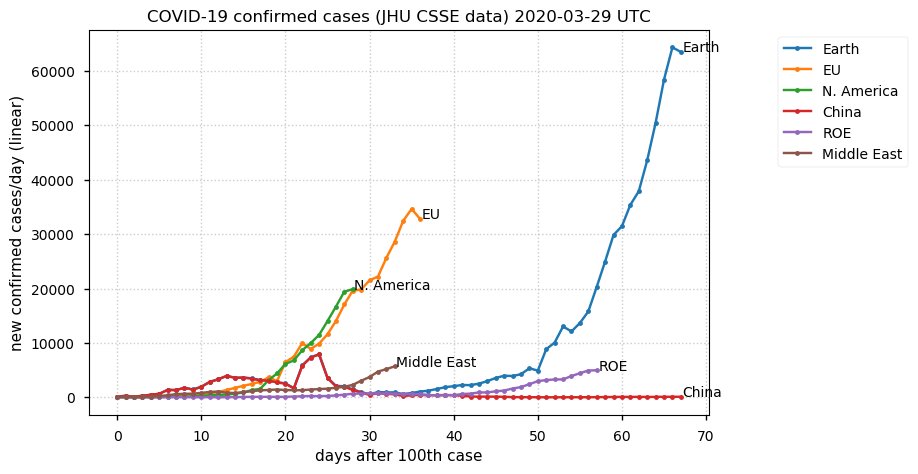
Sun Mar 29 15:31:42 +0000 2020Global fatalities per day (3 day moving average) 🔗
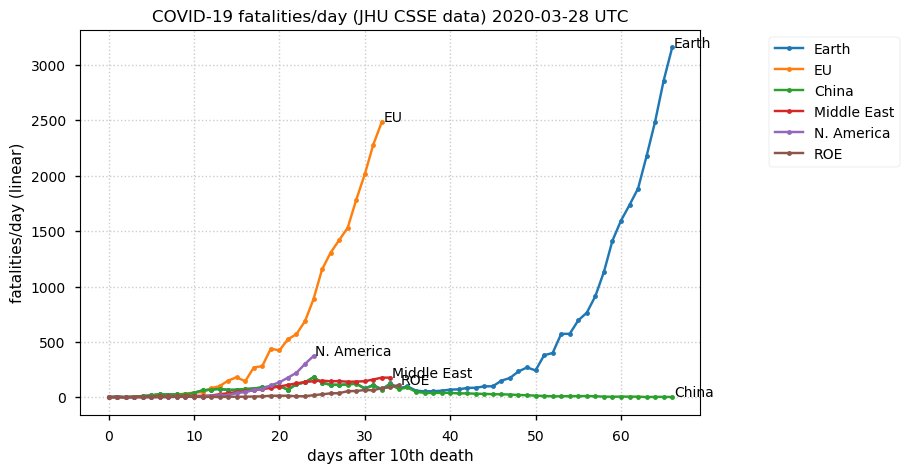
Sun Mar 29 13:05:57 +0000 2020Canada seems to be following along the same trend as Holland 🔗
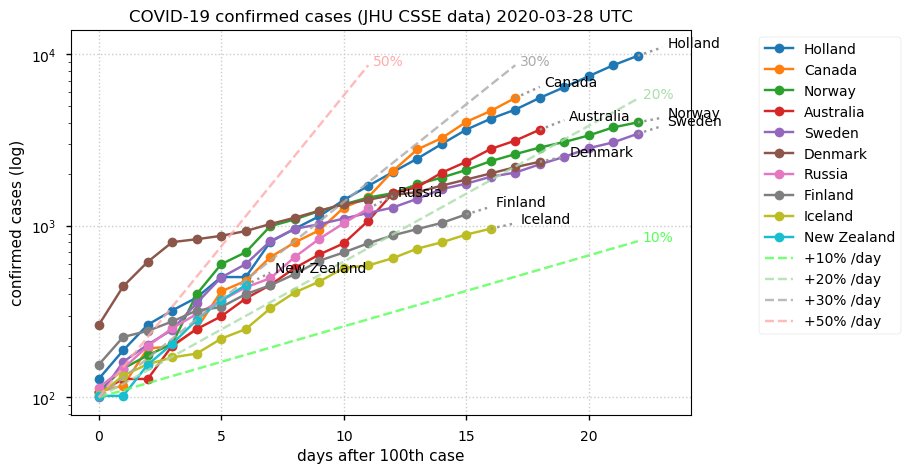
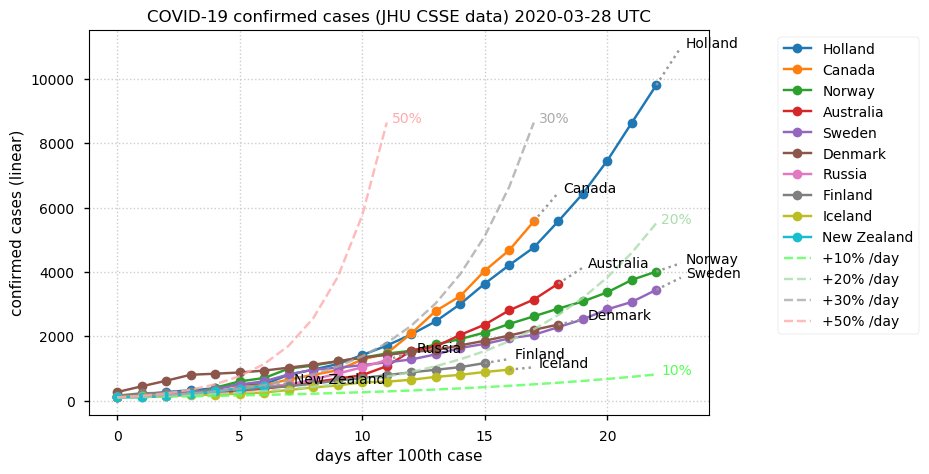
Sun Mar 29 12:37:26 +0000 2020Globally, the EU continues to be driving the curve wrt new cases & fatalities of COVID-19, although its rate of growth is slowing. 🔗
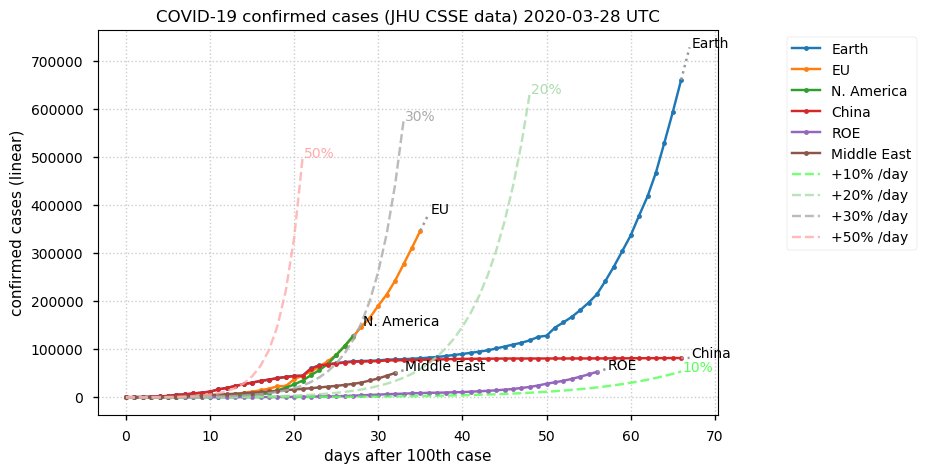
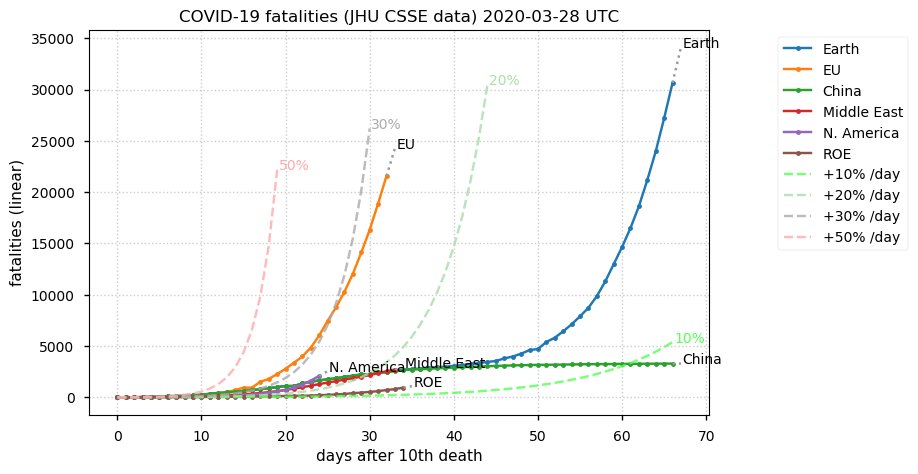
Sun Mar 29 12:20:21 +0000 20203C-like proteinase (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory protein; 3 phosphodomains; generated by proteolysis of ORFAB: domain 3264-3569; mature form 1-306 [62 x] 🔗
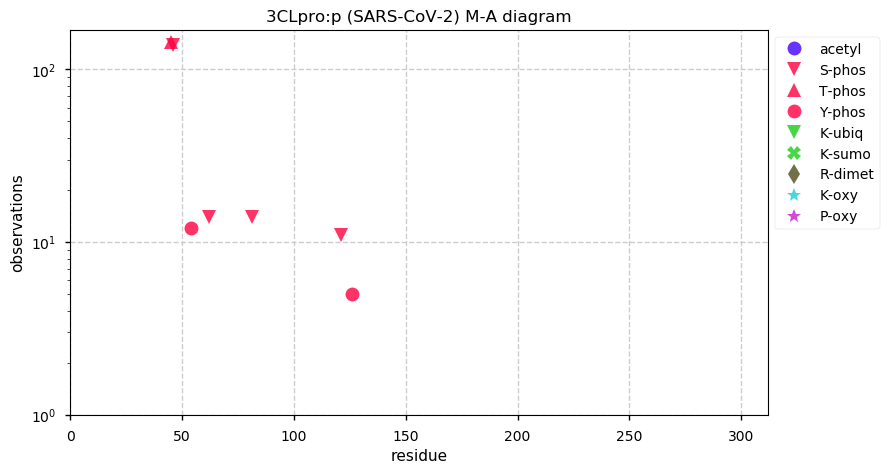
Sat Mar 28 17:24:23 +0000 2020My new pin on my favorite analog computer. 🔗
Sat Mar 28 16:23:11 +0000 2020The uneven distribution in the Westmore-Standing diagram is caused by the low number of observations & that most of the observations come from a study that expressed each component protein separately. Normally, all of the component proteins are expressed at identical levels.
Sat Mar 28 13:51:49 +0000 2020@VATVSLPR MI seems to be on the same track as NJ. However, the states that have 16 days data seem to be rolling over from their initial nearly exponential growth to be more like a normal logistic curve. 🔗
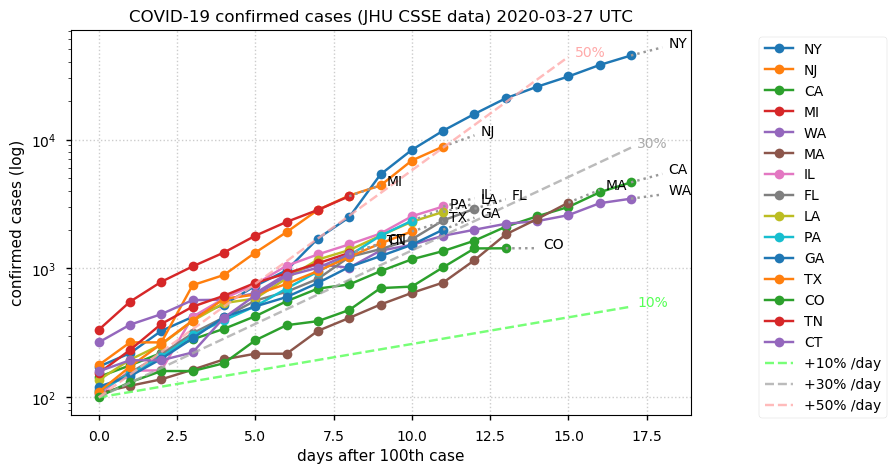
Sat Mar 28 13:41:52 +0000 2020GU280_gp01, orf1ab:p polyprotein (Severe acute respiratory syndrome coronavirus 2) 🔗 Very large viral polyprotein; contains 15 individual accessory proteins; translated form 1-7096 [124 x] 🔗
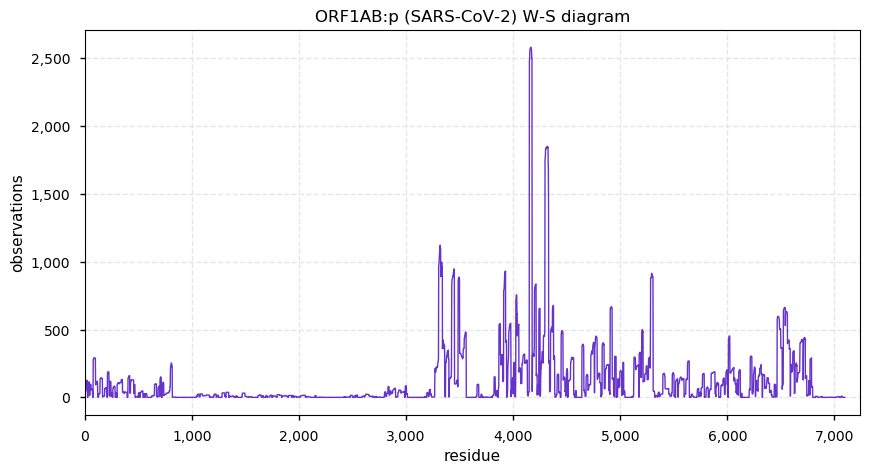
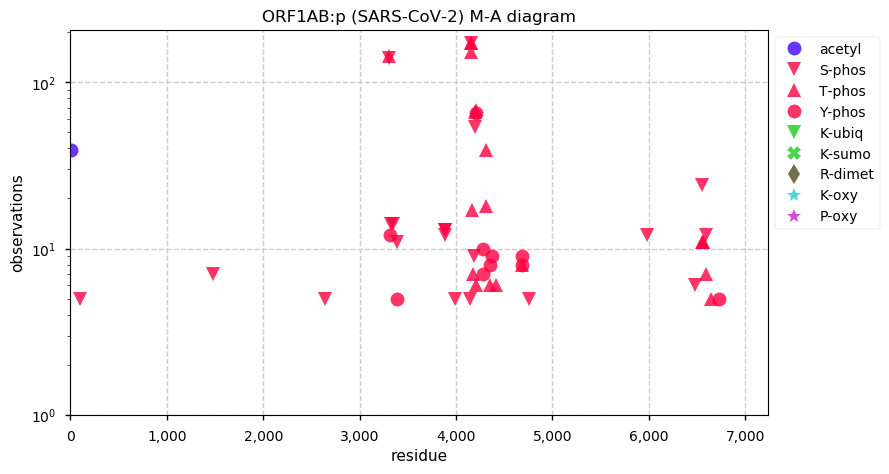
Sat Mar 28 13:12:53 +0000 2020And for the planet, North America is tracking Europe, where Europe has an 8 day lead. 🔗
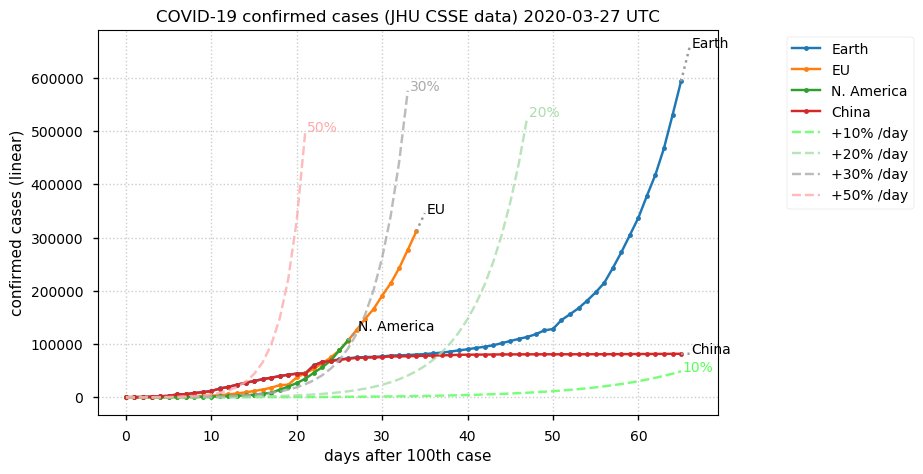
Sat Mar 28 12:45:32 +0000 2020NY is still the story in the US, but something bad is going on in Louisiana 🔗
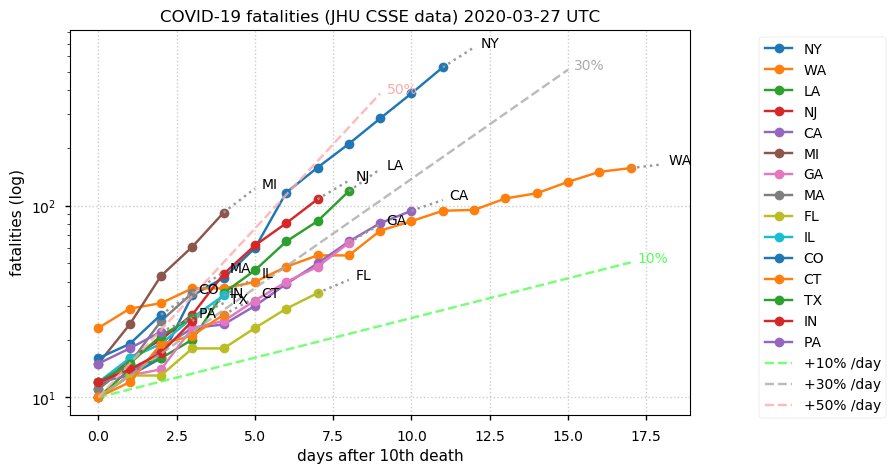
Sat Mar 28 12:33:30 +0000 2020Good news for me: looking past the sun just before dawn this morning I could clearly see Jupiter, Saturn and a dim Mars in a cluster low in the eastern sky.
Sat Mar 28 12:16:08 +0000 2020Quebec is the story here in Canada. 🔗
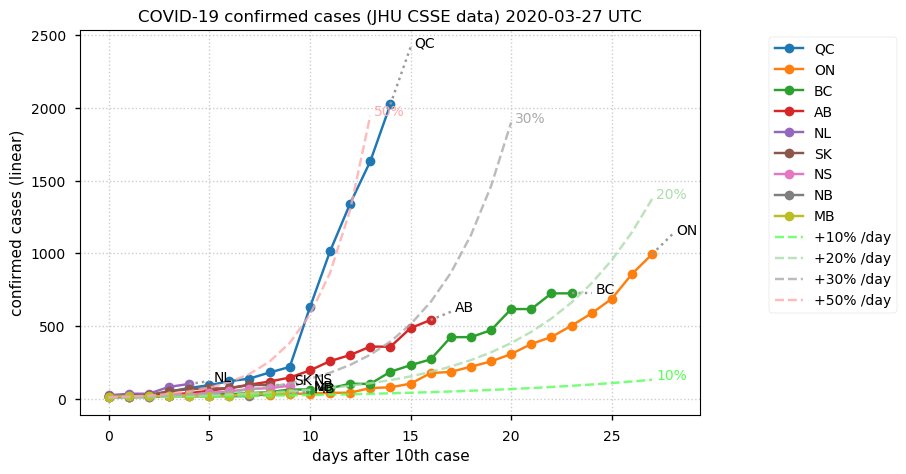
Fri Mar 27 23:46:36 +0000 2020@gingraslab1 @CianferaniS @KentsisResearch @JavR_Jabber @GeorgesBEDRAN3 Turns out I will be sticking with RefSeq. I get 5-10% more unique peptide sequences using RefSeq rather the ChlSab1.1 only collections (ENSEMBL & UniProt).
Fri Mar 27 20:12:20 +0000 2020@gingraslab1 @CianferaniS But I will probably switch over to the ENSEMBL proteome, ftp://ftp.ensembl.org/pub/release-99/fasta/chlorocebus_sabaeus/pep/
Fri Mar 27 20:02:53 +0000 2020@gingraslab1 @CianferaniS I just used RefSeq for C. sabaeus.
Fri Mar 27 19:10:25 +0000 2020I'd guess there are at least 4 kinases involved:
1. One that likes "SD" motifs;
2. One that likes "SP" motifs;
3. One that likes "T[NQ]"; &
4. At least one other Thr kinase.
Fri Mar 27 18:24:42 +0000 2020From a globalist perspective 🔗
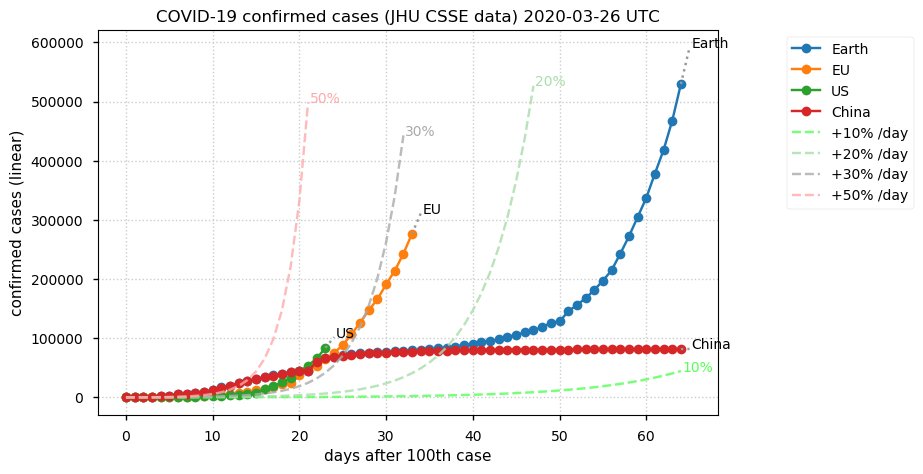
Fri Mar 27 16:36:56 +0000 2020For those who like sequences rather than tables: 🔗

Fri Mar 27 16:07:34 +0000 2020Anybody want to speculate on the cellular kinase(s) and/or phosphatase(s) involved?
Fri Mar 27 15:41:24 +0000 2020@jasonkeays @RosieBarton The Cuban Missile Crisis.
Fri Mar 27 14:27:34 +0000 2020Sorry, 2 papers on bioRxiv and 1 paper on researchsquare.
Fri Mar 27 14:15:43 +0000 2020Thanks to 3 papers released to bioRχiv that publicly released their data, we know a lot about the phosphorylation of SARS CoV-2 N protein (in terms of the # of PSMs supporting the modification): 🔗
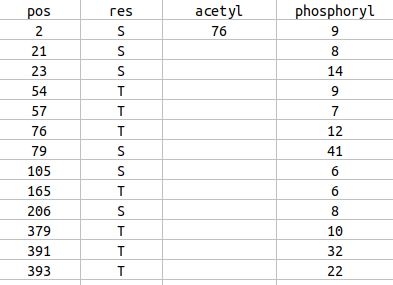
Fri Mar 27 13:51:53 +0000 2020🔗
Fri Mar 27 13:50:47 +0000 2020Although the main battle lines are still in the EU: 🔗
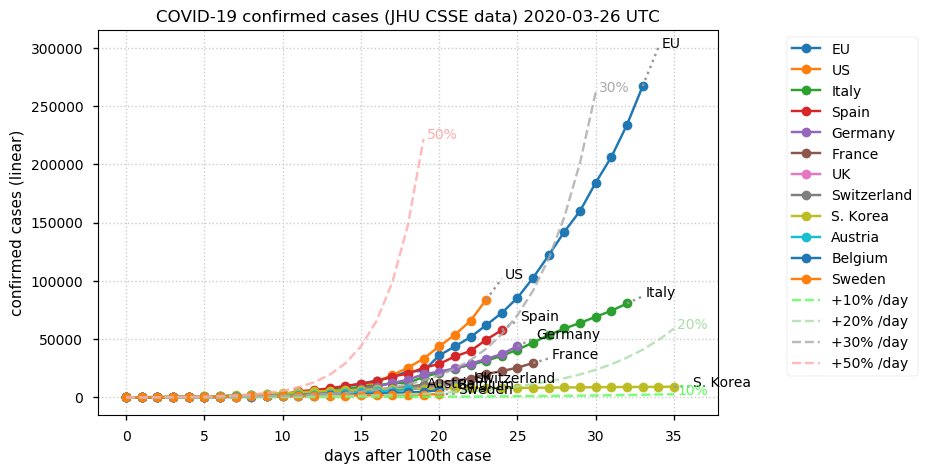
Fri Mar 27 12:24:21 +0000 2020🔗
Fri Mar 27 12:22:15 +0000 2020🔗
Fri Mar 27 12:18:34 +0000 2020G128_gp04, orf4a:p, NS4A protein (Human betacoronavirus 2c EMC/2012) 🔗 Small viral accessory protein; N-terminal acetylation; mature form 1-109 [113 x] 🔗
Thu Mar 26 22:48:02 +0000 2020@ypriverol No, I don't know them. I just saw someone reference the manuscript on Twitter, so I read it and followed the DOI's in the paper to the data, downloaded it and ran it through my RPI4 system. It is an English group, at the University of Bristol.
Thu Mar 26 22:15:24 +0000 2020@ypriverol It is in zenodo.
Thu Mar 26 20:44:29 +0000 2020@IonSource It was still a big deal to get a leucine enkephalin (YGGFL) spectrum.
Thu Mar 26 18:43:47 +0000 2020@UCDProteomics A friendly stranger suggests 🔗
Thu Mar 26 18:36:09 +0000 2020Be sure to include the propionamide (acrylamide) cysteine derivative in addition to the usual IAA cysteine modification for the gel band experiment.
Thu Mar 26 18:31:51 +0000 2020Setting aside my grousing about zenodo, the SARS-CoV-2 data from Davidson AD, et al. (🔗) is pretty good (incl. the phosphopeptide expts).
Thu Mar 26 17:01:55 +0000 2020@Gundrylab You would really go for a 1 year "grant" with a required 1:1 match and require "partner-led initiatives to advance a partner challenge"?
Thu Mar 26 15:27:58 +0000 2020@TheGonz79 @Sci_j_my @BFTJPRC Australia's and Canada's case numbers are growing at similar rates.
Thu Mar 26 14:40:45 +0000 2020@EdHuttlin @HMSBioPlex Probably, but there is no way to know for sure.
Thu Mar 26 14:24:48 +0000 2020@KentsisResearch @KroganLab Maybe, but I've found dealing directly with authors to be not-so-very useful in this type of situation. As I mentioned, normally I'd just QA the results without comment but given the subject matter the results will have some utility for a while, just not for PPI.
Thu Mar 26 14:09:28 +0000 2020@KentsisResearch @KroganLab I tried reanalyzing with different algorithms, but ended up with the same results. I've marked the results with a QC warning that they shouldn't be used for PPI determination. When more data for the virus accumulates, I will QA the results.
Thu Mar 26 13:21:34 +0000 2020@jwoodgett Trying to access the site from any internet connection protected by a properly configured DNS sinkhole server will end up being blocked.
Thu Mar 26 13:15:26 +0000 2020@jwoodgett The organization itself may be perfectly legit, but the web site has at least 17 trackers built in to the code. They seem to have used a "free" web-builder outfit to make their site, so the trackers probably weren't their idea: they are the price of "free".
Thu Mar 26 12:55:47 +0000 2020G128_gp08, M:p, membrane protein (Human betacoronavirus 2c EMC/2012) 🔗 Small viral protein; no peptides observed in the domain 1-104; part of viron; mature form 1?-219 [123 x] 🔗
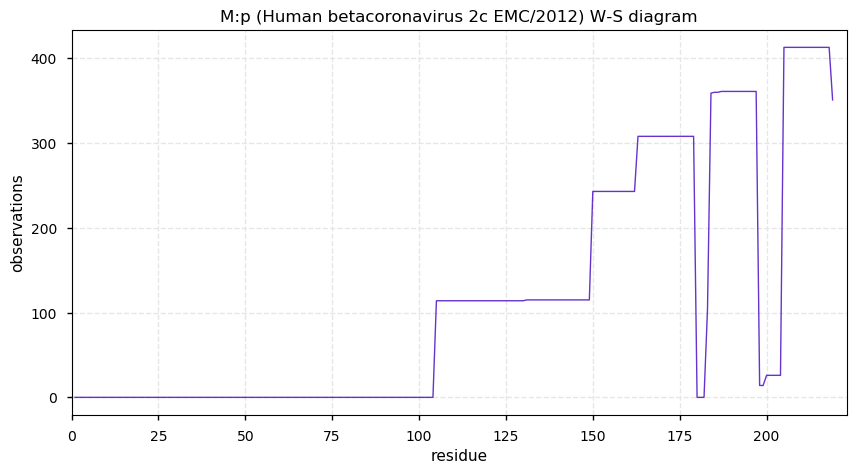
Thu Mar 26 12:52:37 +0000 2020Just finished downloading a small set of Thermo .RAW files that somebody had put on zenodo: nothing about it went well. Please use PRIDE or MASSIVE for mass spec raw data.
Thu Mar 26 01:25:39 +0000 2020@IonSource Minneapolis 1981. Everyone was very excited about FAB.
Wed Mar 25 19:55:38 +0000 2020@JesseBrown Bogosian is the only one that fits the situation.
Wed Mar 25 19:41:42 +0000 2020@jwoodgett All of my "bad site - stay away" systems raise alarms for the URL.
Wed Mar 25 18:27:01 +0000 2020@KentsisResearch I've re-read the paper a few more times and given the results another look. Unless something else presents itself, I'm going to mark the results "Not fit for purpose". I usually QA results for this type of thing, but given the moment, I'd like to keep them available.
Wed Mar 25 16:05:45 +0000 2020@KentsisResearch Hopefully it is something else, as carryover is NOT a good thing for affinity purification expts.
Wed Mar 25 15:59:26 +0000 2020@KentsisResearch That was my reading too, but many of the LC/MS/MS runs have strong signals from more than 1 viral proteins, including the EGFP experiments. I can't see how that can occur, given 1 viral protein construct per batch.
Wed Mar 25 15:22:33 +0000 2020Hopefully someone can answer this question, regarding #PXD018117. Are all of the viral protein constructs expressed simultaneously in one batch of cells or were the constructs expressed individually in separate batches? The manuscript isn't crystal clear about this point.
Wed Mar 25 12:33:44 +0000 2020Who knew NYC had its own flag? 🔗

Wed Mar 25 12:28:59 +0000 2020🔗
Wed Mar 25 12:22:04 +0000 2020G128_gp05, ORF4B:p, NS4B protein (Human betacoronavirus 2c EMC/2012) 🔗 Small viral acessory protein; N-terminal acetylation; interferes with host cell antiviral response; mature form 1-246 [127 x] 🔗
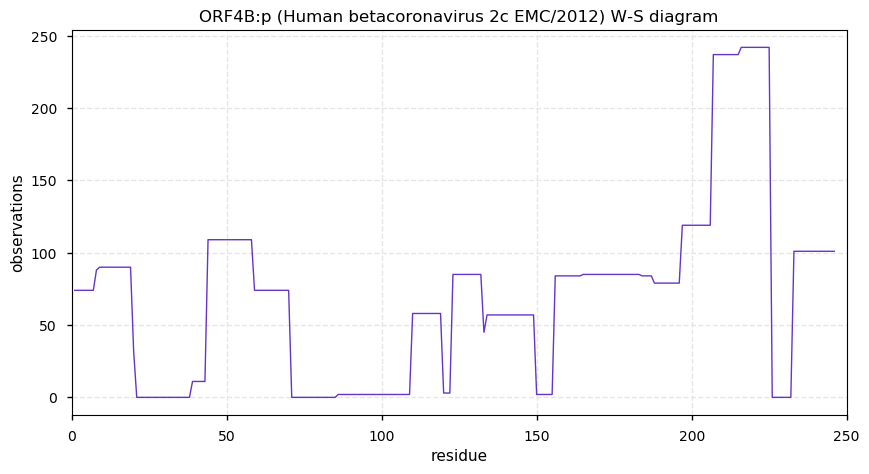
Wed Mar 25 11:38:42 +0000 2020This graph pretty clearly makes the argument that N.Y. has a serious public health problem right now. 🔗
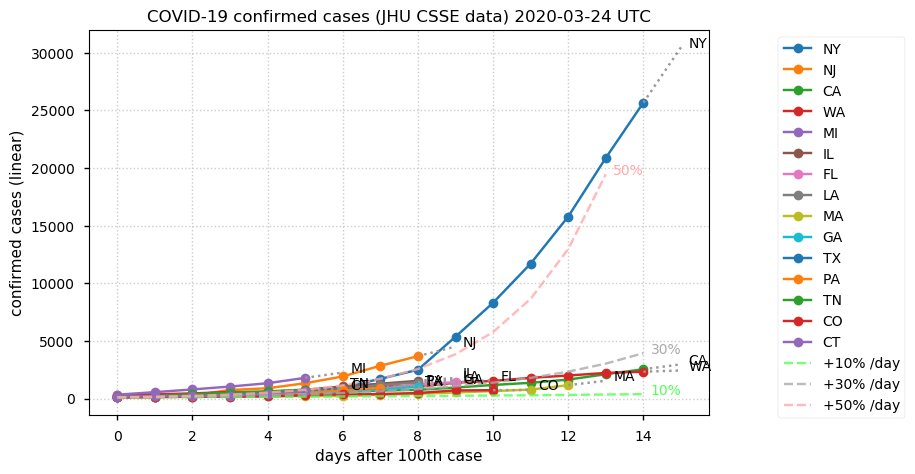
Tue Mar 24 19:36:20 +0000 2020Your choice: 🔗
Tue Mar 24 18:57:20 +0000 2020@neely615 And just because I got a deal on the zone name:
🔗
Tue Mar 24 17:50:03 +0000 2020@neely615 I didn't come up with it, but I can't remember exactly where I heard or read it either. There is some podcast/radio show/newpaper article/tweet out there that deserves the "credit".
Tue Mar 24 17:42:00 +0000 2020Israel and South Africa seem to be on trajectories similar to most EU countries, but some of the Gulf States are tracking closer to Singapore or Japan.
🔗
Tue Mar 24 16:02:28 +0000 2020After some hacking caused by the JHU data model changes, I was able to get the US states COVID-19 confirmed cases 🔗 and deaths 🔗 running again.
Tue Mar 24 15:56:16 +0000 2020@chrashwood @pwilmarth From the POV of societies, the problem is that there is nothing to replace the 1-on-1 sales time that companies pay for with sponsorships. Without some way to replace that interaction & on-site marketing, the whole business model of most conferences goes poof.
Tue Mar 24 14:49:08 +0000 2020Just got the analysis of PXD018117 from PRIDE 🔗 loaded into GPMDB 🔗 Uses sequences from: ENSEMBL (female human & FBS); RefSeq (SARS CoV2 & other common human viruses); and cRAP.
Tue Mar 24 12:49:36 +0000 2020Quebec took the lead overnight.
Tue Mar 24 12:36:01 +0000 2020G128_gp02, S:p, spike glycoprotein (Human betacoronavirus 2c EMC/2012) 🔗 Large viral protein; ER signal peptide removal; viron surface protein; mature form 18-1353 [127 x] 🔗
Tue Mar 24 12:33:12 +0000 2020F'ing Johns Hopkins CSSE. Changed their COVID-19 data structure overnight without advance notification and discontinued their previous data products. It is their stuff, so they can change it any way they want & I know they are under pressure, but😠
Mon Mar 23 21:09:53 +0000 2020Has anyone else reanalyzed PXD018117 yet?
Mon Mar 23 20:15:33 +0000 2020For the Canadians, it looks like Quebec or Alberta may overtake BC and Ontario in terms of confirmed COVID-19 cases within a few days
🔗
Mon Mar 23 15:48:49 +0000 2020@slavovLab @PhysRevLett @TokiwaGashi I usually keep pets to the Acknowledgements section, even though they often contribute more than many co-authors.
Mon Mar 23 14:51:01 +0000 2020The HUPO PSI Experiment begins in 10 minutes ...
Mon Mar 23 12:44:28 +0000 2020G128_gp09, N:p, nucleoprotein (Human betacoronavirus 2c EMC/2012) 🔗 Small viral protein; N-terminal acetylation; structural protein with several roles; mature form 2-413 [178 x] 🔗
Sun Mar 22 18:41:23 +0000 2020@doctorow Corrected: "This is particularly true right now, because the option of flying down to the US for concierge treatment has been removed: they really have no legal choice but to use their provincial health care systems."
Sun Mar 22 18:13:02 +0000 2020@doctorow This is particularly true right now, because the option of flying down to the US for concierge treatment has been removed: they really no legal choice but to use their province health care system.
Sun Mar 22 15:35:47 +0000 2020My favorite state (NY, mainly because it contains my favorite city) is on a bad trajectory at the moment
🔗
Sun Mar 22 15:03:37 +0000 2020The E1B gene was incorporated into the HEK 293 genome as part of its immortalization. E1B:p is the most abundantly detected viral protein in HEK 293 samples & it frequently observed in pull-down experiments that use this cell line.
Sun Mar 22 14:58:52 +0000 2020E1B, control protein 55K (Human mastadenovirus C) 🔗 Small viral accessory protein; observed phosphorylation and ubiquitylation; constitutively expressed in HEK 293 cells; mature form 1-495 [5,894 x] 🔗
Sat Mar 21 19:59:10 +0000 2020@goodlettlab1 @govsingapore @hongkong_news Done. Although I have to say that Hong Kong is looking a little wonky as of the last few days.
🔗
Sat Mar 21 19:20:30 +0000 2020@goodlettlab1 @govsingapore @hongkong_news Good idea. I just added US states, but adding good news helps.
Sat Mar 21 17:49:05 +0000 2020Any suggestions for country groupings or displays? 🔗
Sat Mar 21 14:00:01 +0000 2020GU280_gp07, ORF7a protein (Severe acute respiratory syndrome coronavirus 2) 🔗 Small accessory membrane protein; no observed PTMs; insufficient data to form a peptide observation pattern; mature form 1?-121 [2 x] 🔗
Sat Mar 21 13:48:10 +0000 2020Latest from World War C: the northern-most countries seem to be grouping up into a different track (way too early to be sure) 🔗
Sat Mar 21 00:20:30 +0000 2020@pwilmarth Unfortunately, a lot of people who don't like to be contradicted have insisted for a decade or two that the tautology
apple = orange
is correct & self-evident.
Fri Mar 20 21:46:25 +0000 2020We Canadians have missed the chance to be more like Singapore or Japan, but we do want to get on the South Korean off-ramp ASAP ... 🔗
Fri Mar 20 19:12:22 +0000 2020@DRAWheatcraft I couldn't attend the physical conference, either.
Fri Mar 20 16:09:29 +0000 2020I am looking forward to the results of HUPO PSI's first experiment: trying to pull off a multi-day, purely virtual conference with most of the conferees & presenters joining from home.
Fri Mar 20 15:50:05 +0000 2020Still entertaining myself with these 2 CSV data sets of fatalities and confirmed cases, broken down by date and geography. Fitting data to various logistic models is interesting ... 🤓🧑🔬👍
Fri Mar 20 13:46:44 +0000 2020Boy, CBC News Channel's new kaleidoscopic multi-panel flashing and scrolling screen layout is really off-putting. Should be used as a classroom example of how not to present information.
Fri Mar 20 12:53:33 +0000 2020NSP3, non-structural protein 3 (Severe acute respiratory syndrome coronavirus 2) 🔗 Large accessory protein; no observed PTMs; insufficient data to form a peptide observation pattern (ec = 2); mature form 1?-1945 [3 x] 🔗
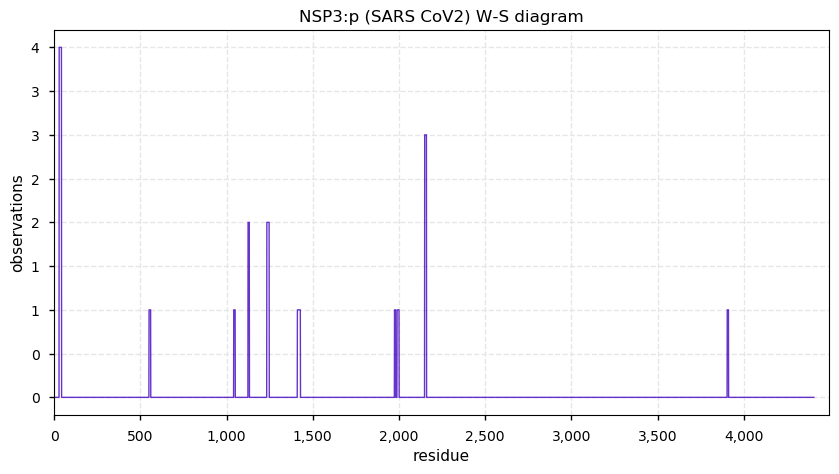
Thu Mar 19 21:07:05 +0000 2020It would also be a source of timely data (with geolocation) that would interest biomedical students learning R or Python to visualize data and model trends.
Thu Mar 19 20:59:25 +0000 2020I find the raw stats on this to be far more interesting than a whole bunch of bespoke new stories. Many thanks to Johns Hopkins CSSE & github
Thu Mar 19 17:56:14 +0000 2020With an appropriate peptide length distribution for this type of experiment. 🔗
Thu Mar 19 17:39:56 +0000 2020And they did a great job of getting peptides matching ^.[AP].+[VIL]$
Thu Mar 19 17:32:16 +0000 2020Compared to most MHC type I datasets, PXD013064 does an unusually good job at recovering cystine-containing peptides (Chen, et al. 🔗)👍
Thu Mar 19 16:10:03 +0000 2020If you want to do some further re-analysis of betacoronavirus proteomics data, PXD002358, PXD004716 & PXD004719 have a lots of data for MERS CoV infections
Thu Mar 19 15:30:49 +0000 2020@dtabb73 @ProteomicsNews @Smith_Chem_Wisc & 🔗

Thu Mar 19 15:24:06 +0000 2020@dtabb73 @ProteomicsNews @Smith_Chem_Wisc 🔗

Thu Mar 19 14:56:31 +0000 2020@dtabb73 @ProteomicsNews @Smith_Chem_Wisc It was all I could do to resist adding an appropriate GIF to emphasize this point.
Thu Mar 19 14:46:04 +0000 2020@dtabb73 @ProteomicsNews @Smith_Chem_Wisc Good point Dave. The proteomics-standard "Highlander" approach to sequences isn't the best way to deal with not very stable viral proteins.
Thu Mar 19 13:07:45 +0000 2020Like yesterday's ORF3a, ORF6 is an accessory (non-viron) protein that has been characterized in SARS CoV 🔗
Thu Mar 19 13:04:59 +0000 2020GU280_gp06, ORF6, protein (Severe acute respiratory syndrome coronavirus 2) 🔗 Very small membrane associated accessory protein; no observed PTMs; 1 splice variant; mature form 1?-63 [3 x] 🔗
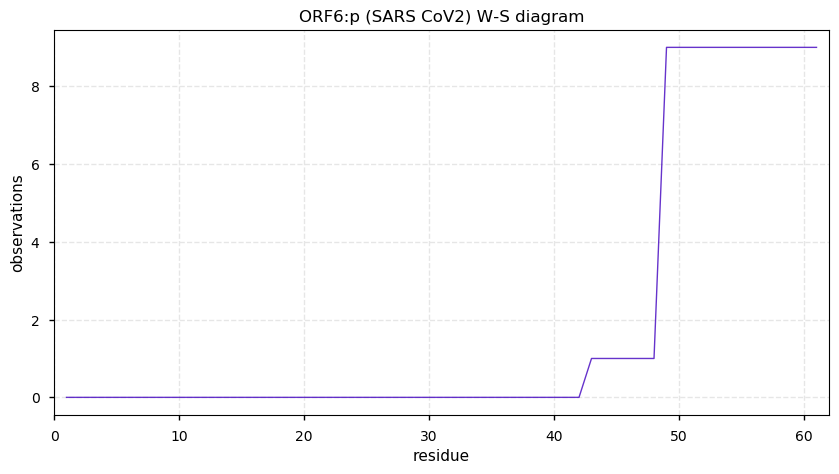
Wed Mar 18 17:36:35 +0000 2020@chrashwood And I would be remiss to neglect the CACO-2 data set that has ACE2:p with demonstrated positive viral receptor capability, all of PXD017710.
Wed Mar 18 16:42:01 +0000 2020@chrashwood If you are volunteering, PXD002121 would be a place to start, particularly the raw files prefixed:
"+WP vs -WP D9 to D22 TMT set7 B";
"+WP vs -WP D9 to D22 TMT set6 B";
"+WP vs -WP D9 to D22 TMT set7 A";
"+WP vs -WP D9 to D15 TMT set4 B"; &
"+WP vs -WP D9 to D15 TMT set4 A".
Wed Mar 18 16:10:50 +0000 2020ACE2:p also shows up in studies that pull down proteins with GPI anchors, although I don't think the existence of a GPI anchor on the molecule has been confirmed.
Wed Mar 18 15:30:32 +0000 2020Better characterization of its PTMs (particularly its N-linked glycoform & phosphorylation dynamics) would not be a bad thing.
Wed Mar 18 15:28:47 +0000 2020And for anyone who wants to contribute to SARS CoV2 work but doesn't want to work with viruses, its receptor ACE2:p is expressed abundantly in CACO-2 cell culture (to a much lesser extention in HeLa) and a truncated form is present in urine.
Wed Mar 18 14:20:33 +0000 2020An oldie, but additional relevance a month later. Note that the phosphorylation at Y781 is probably a good-old-fashioned signalling phosphosite (it is in the C-terminal cytoplasmic tail domain). 🔗
Wed Mar 18 11:52:33 +0000 2020All of the data used to generate the SARS CoV2 protein information featured this week was checked for phosphopeptides, but none of the SARS CoV2 viral proteins had phosphorylation signals.
Wed Mar 18 11:34:35 +0000 2020GU280_gp03, ORF3a, protein (Severe acute respiratory syndrome coronavirus 2) 🔗
Small viral protein; no observed PTMs; at least 3 membrane spanning domains; 1 splice variant; mature form 1?-275 [3 x] 🔗
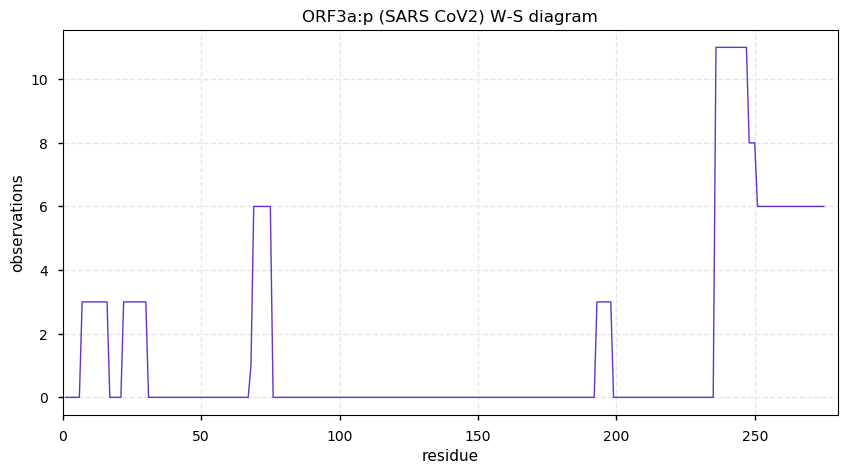
Tue Mar 17 19:14:08 +0000 2020@EdHuttlin @GygiLab If it is in process, I'll wait 'til it pops up.
Tue Mar 17 16:33:58 +0000 2020@EdHuttlin @GygiLab Will the data in PRIDE be released soon?
Tue Mar 17 12:43:25 +0000 2020GU280_gp05, M, membrane glycoprotein (Severe acute respiratory syndrome coronavirus 2) 🔗 Small viral protein; no observed PTMs: noteworthy lack of N-terminal acetylation; 1 splice variant; mature form 2-222 [3 x] 🔗
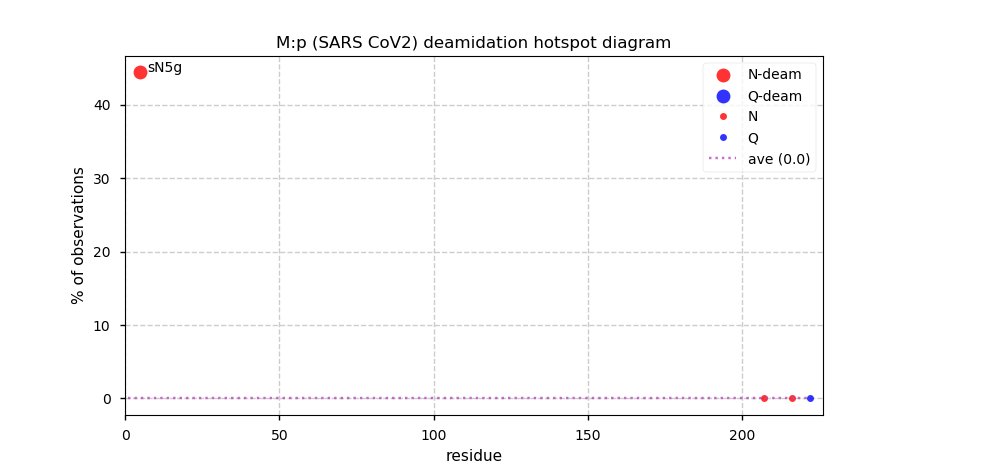
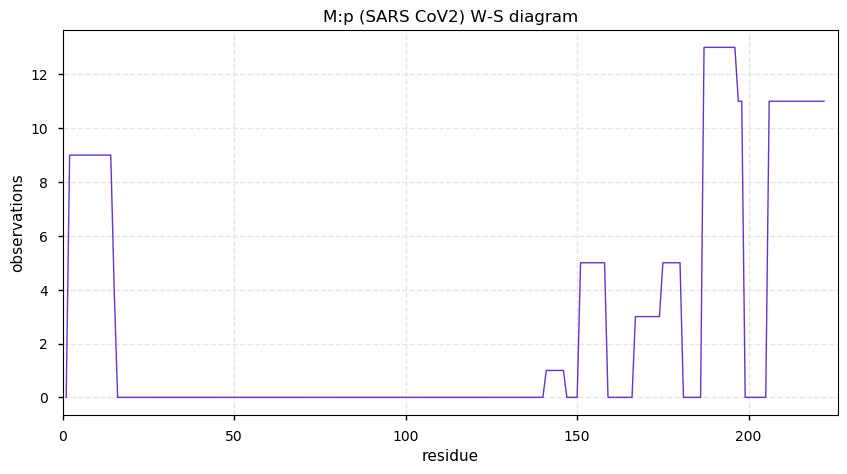
Mon Mar 16 15:54:07 +0000 2020NOTE: the SARS-CoV-22 proteins being featured this week were all observed in the data set PXD017710 using the RefSeq proteome sequence. The data was made available in conjunction with the preprint 🔗
Mon Mar 16 15:04:57 +0000 2020@AlexUsherHESA Canadian U15 outfits do not have the infrastructure or IT personnel to really have any significant number of simultaneous "on-line" (video) classes. Their IT is designed for administrative purposes only - simply preparing month end reports can clog their networks.
Mon Mar 16 12:20:43 +0000 2020GU280_gp02, S, surface glycoprotein (Severe acute respiratory syndrome coronavirus 2) 🔗 Large viral protein; no observed PTMs; 1 splice variant; mature form 1?-1273 [3 x] 🔗
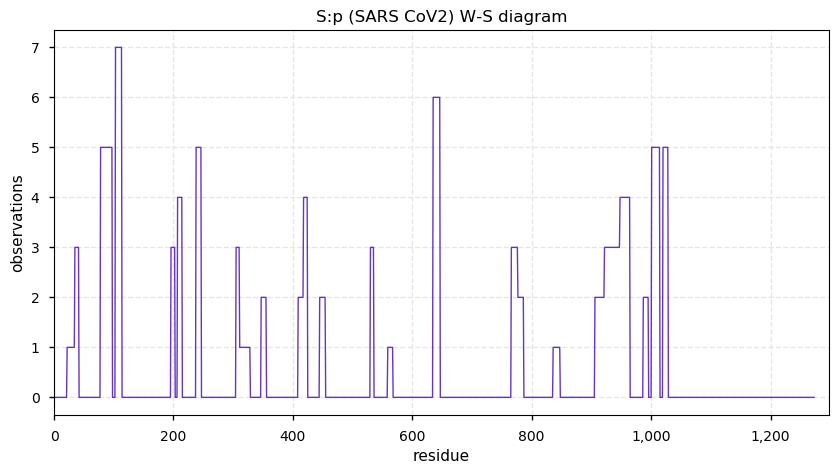
Sun Mar 15 18:14:02 +0000 2020GU280_gp10, ORF9, nucleocapsid phosphoprotein (Severe acute respiratory syndrome coronavirus 2) 🔗 Small viral protein; no observed PTMs; 1 splice variant; mature form 2?-414 [3 x] 🔗
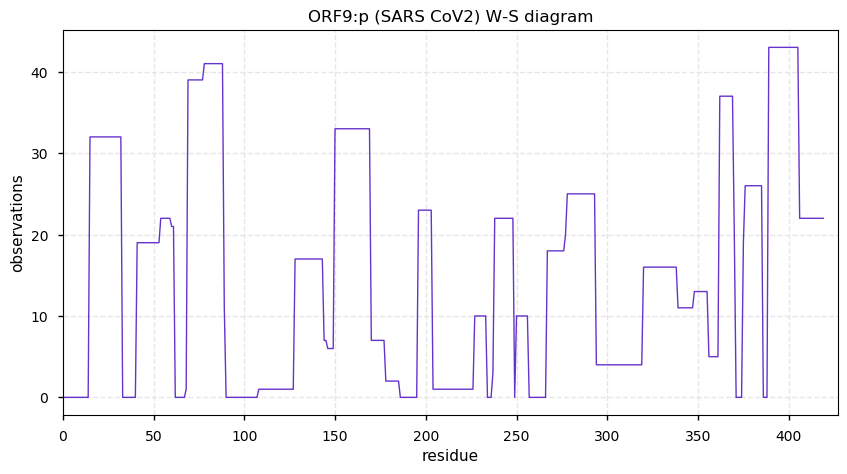
Sun Mar 15 13:11:58 +0000 2020My internet provider just sent me a marketing email, describing all the services I can buy from them that would "help" me during the CoVid19 outbreak. 😷👹
Sat Mar 14 17:27:58 +0000 2020@dtabb73 My triggers are "there will be a vaccine soon" & "scientists are hard at work on the problem"
Sat Mar 14 13:02:33 +0000 2020CCDC51:p, coiled-coil domain containing 51 (H. sapiens) 🔗 Midsized mitochondrial protein; few PTMs; 1 high maf SAAV: A232V (0.01); common in cell lines and tissues; 1 splice variant; mature form 37-411 [9,072 x] 🔗
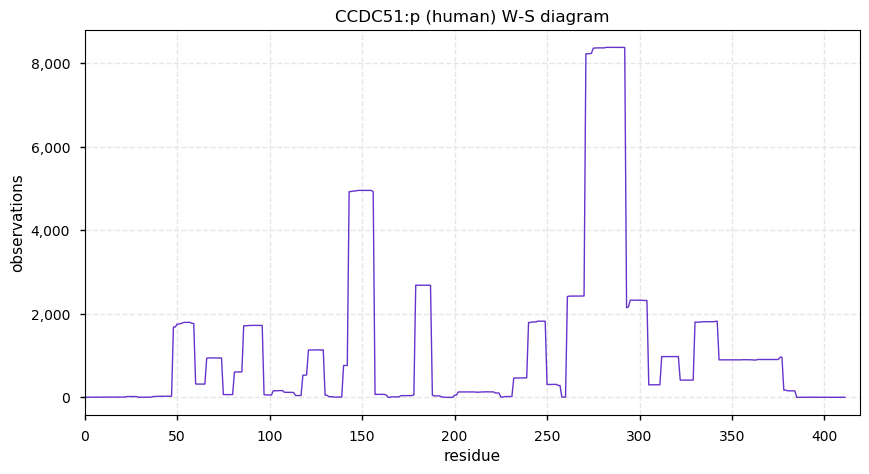
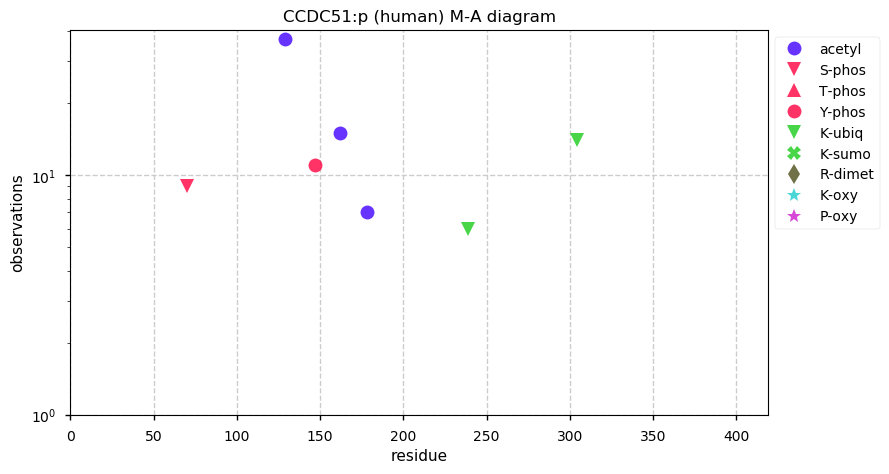
Fri Mar 13 19:11:57 +0000 2020Going to be watching Netflix buffer for the next few weeks ...
Fri Mar 13 16:58:57 +0000 2020Let's Encrypt has really transformed my use of HTTPS from dealing with overpriced-difficult-to-use-grifty-vendor-specific systems to running a simple, cross-platform open source utility.
Fri Mar 13 15:53:20 +0000 2020@stephen_taylor Possibly a conspiracy foisted on us by "Big Bidet"
Fri Mar 13 13:05:39 +0000 2020FAM136A:p, family with sequence similarity 136 member A (H. sapiens) 🔗 Small membrane protein; few PTMs other than ubiquitinylation; no high maf SAAVs; common in cell lines and tissues; 1 splice variant; mature form 2-138 [13,096 x] 🔗
Thu Mar 12 18:58:39 +0000 2020@slashdot Using a SCIF seems a little over the top. Although it fits in with a lot of the "this is the way it works in the movies" approaches being discussed as though they work in the real world.
Thu Mar 12 16:31:12 +0000 2020Event 201 is kind of "on the nose":
🔗
Thu Mar 12 14:58:18 +0000 2020@MuenchLab @BobbyKlaus3 @ProteomicsNews @pride_ebi The RP chromatography is also very good, with a nice long steady gradient and proper MS/MS start and stop times just clearing the edges of the peptide elution profiles.
Thu Mar 12 13:28:42 +0000 2020@MuenchLab @BobbyKlaus3 @ProteomicsNews @pride_ebi Thanks for making the data available through PRIDE. A quick re-analysis of your data shows that it is really good quality. The ion exchange chromatography worked very well, producing nice separation of peptide by charge.
Thu Mar 12 13:01:10 +0000 2020SMIM12:p, small integral membrane protein 12 (H. sapiens) 🔗 Very small membrane protein; no PTMs; no high maf SAAVs; common in cell lines and tissues; 1 splice variant; mature form 1-92 [1,745 x] 🔗
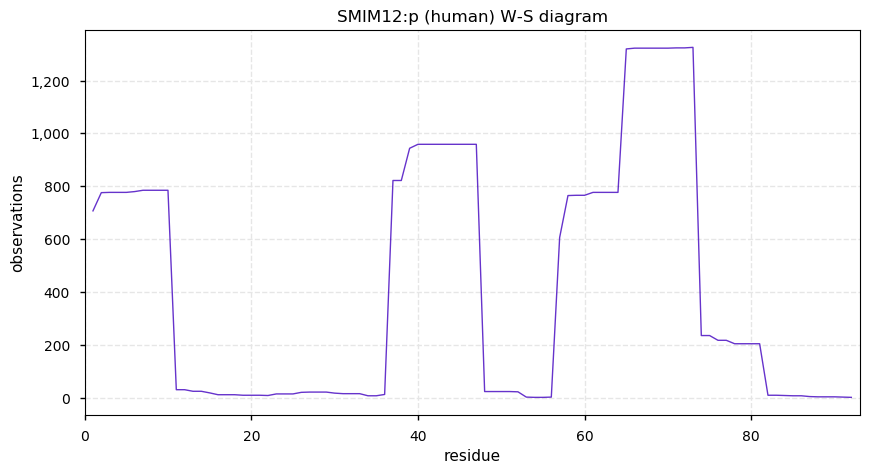
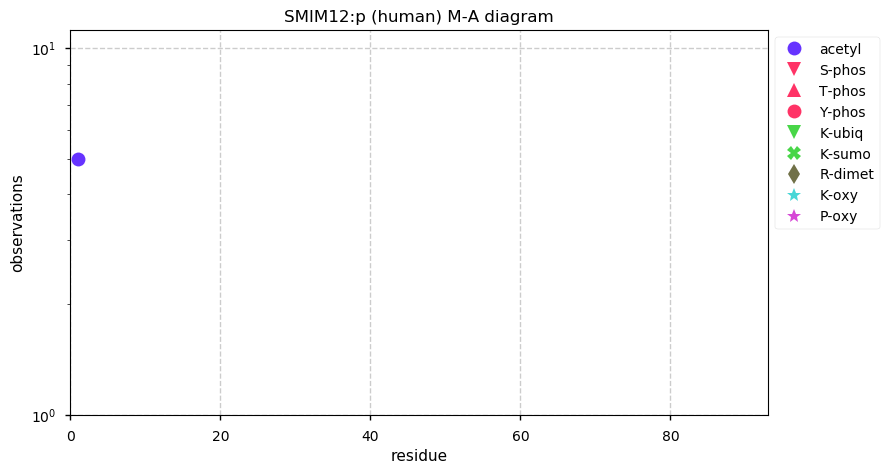
Wed Mar 11 19:44:06 +0000 2020@TrostLab @NHSuk Clinical testing has legal and procedural requirements that don't fit in with university research-oriented labs. It would be similar to having a university lab run lot release tests for a pharma company: it is just a non-starter.
Wed Mar 11 15:35:45 +0000 2020@pwilmarth @ypriverol @SpecInformatics @dtabb73 And it has simplified the mechanics of ensuring that automated processes recover gracefully from file errors.
Wed Mar 11 15:32:32 +0000 2020@pwilmarth @ypriverol @SpecInformatics @dtabb73 About a decade ago, I switched all files sent over a wire or archived within GPMDB to be gzip'd first. It has built-in file validation & it is platform independent. I realize it may not be practical for PRIDE, but it has made testing for file errors much easier for me.
Wed Mar 11 14:31:29 +0000 2020@jwoodgett And based on the press conference, it is unclear whether that is new money or simply rearranging the deck chairs at CIHR.
Wed Mar 11 14:21:44 +0000 2020@jwoodgett He just put $270 million more into the "disease-of-the-day" bucket a few minutes ago, just for political optics.
Wed Mar 11 14:13:58 +0000 2020@jwoodgett As the PM announces a bunch more money for politically-motivated research that Canada is ill-equipped to perform and cannot sustain past this tranche.
Wed Mar 11 12:54:55 +0000 2020ABHD11:p, abhydrolase domain containing 11 (H. sapiens) 🔗 Small cytoplasmic protein; no PTM domain structure; 1 high maf SAAV: V181M (0.05); common in cell lines and tissues; 1 splice variant; mature form 1-315 [12,528 x] 🔗
Tue Mar 10 15:43:28 +0000 2020@wenbostar As a final note, if you want to see these peptides you do not need a "special" data set of HLA type-I peptides. Grab any one of the HLA type-I data sets from ProteomeXchange — e.g., PXD015957 or PXD013831 — and you'll find lots (by which I mean 1-5% of PSMs)
Tue Mar 10 14:01:54 +0000 2020@wenbostar There is a little burst of papers on this subject that all make it sound like doing the analysis is much more intricate than it actually is in practice.
Tue Mar 10 11:43:47 +0000 2020THNSL1:p, threonine synthase like 1 (H. sapiens) 🔗 Midsized mitochondrial protein; no PTM domain structure; no high maf SAAVs; 1 splice variant; mature form 48-743 [9,327 x] 🔗
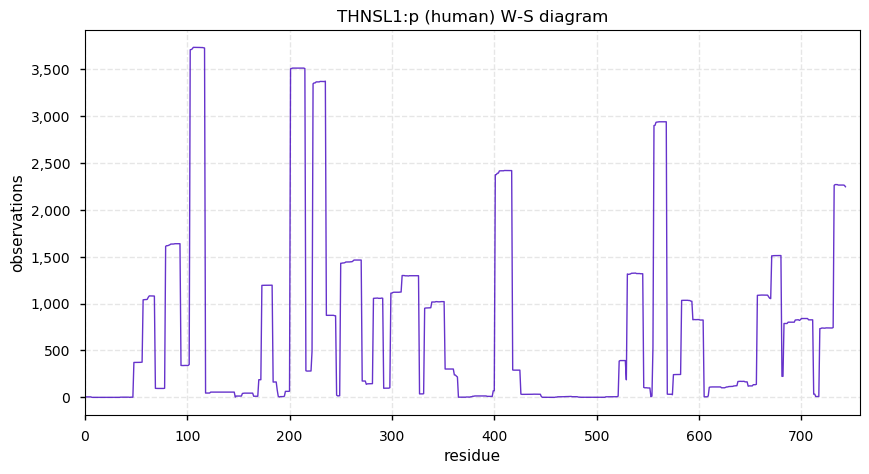
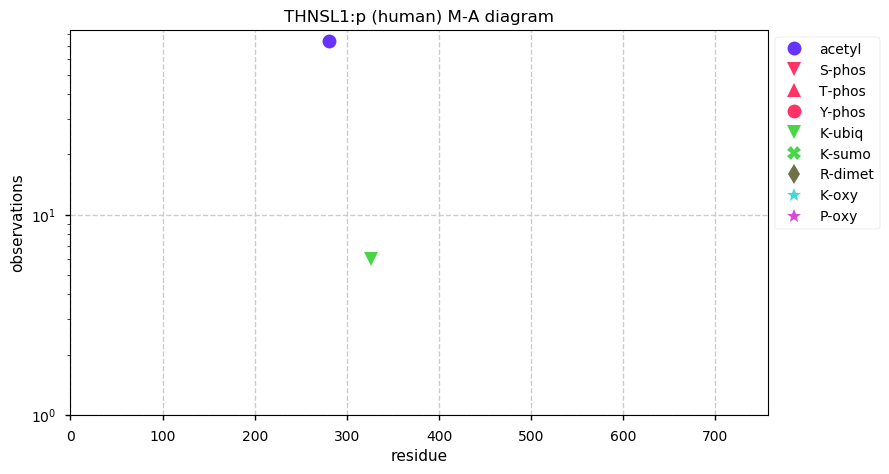
Tue Mar 10 00:41:55 +0000 2020@MattWFoster @ProteomicsNews I'm not arguing that the idea is a bad one, just that users don't seem to like it for some reason. I use it myself every day.
Tue Mar 10 00:34:14 +0000 2020@MattWFoster @ProteomicsNews Not in the FASTA header, but in a variety of other formats. The file format isn't very important to anybody but the implementer. Users just seem to have a problem with the idea.
Tue Mar 10 00:10:59 +0000 2020@MattWFoster @ProteomicsNews That sort of annotated-protein search has been available for quite a while. Most users don't seem to like it, or at least it makes them uneasy
Mon Mar 09 16:41:17 +0000 2020Previously unknown phrase soon to be part of the discussion amongst the chatterati: "bilateral interstitial pneumonia"
Mon Mar 09 13:00:23 +0000 2020@AlexUsherHESA Are there any examples of Canadian universities making pedagogical training available to professors? I've been a Prof at 3 Canadian unis and I've never been offered training of any type other how to use accounting/ordering software.
Mon Mar 09 12:41:44 +0000 2020OCIAD2:p, OCIA domain containing 2 (H. sapiens) 🔗 Small intracellular protein; several complementary acetyl/ubiquitinyl sites; no high maf SAAVs; obs. in many tissues and cell lines; 1 splice variant; mature form 2-154 [13,634 x] 🔗
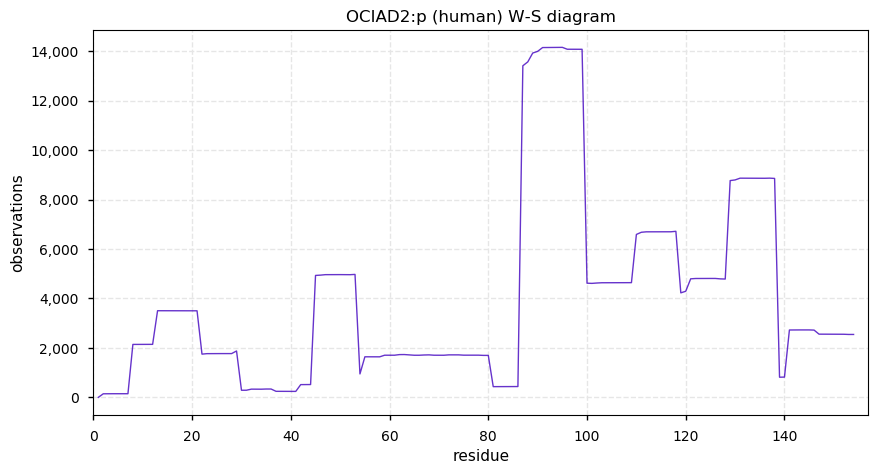
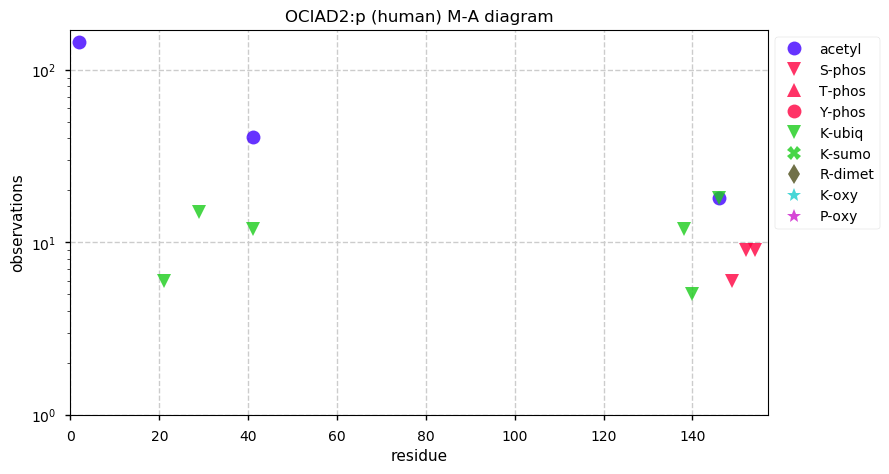
Sun Mar 08 17:06:14 +0000 2020@UCDProteomics Either that or they put the wrong data up on the repository (also a very real possibility)
Sun Mar 08 17:05:39 +0000 2020@UCDProteomics That may be, but in papers with many authors, giving one of the "authors" the job of making sure that the protocols, reagents and instrumentation were reported correctly isn't really asking too much (my 🙄 was over a paper that got all 3 wrong).
Sun Mar 08 16:40:45 +0000 2020I don't know why I even read the Methods section any more (sigh🙄) ...
Sun Mar 08 13:21:27 +0000 2020CCDC58:p, coiled-coil domain containing 58 (H. sapiens) 🔗 Small intracellular protein; several complementary acetyl/ubiquitinyl sites; 0 high maf SAAVs; obs. in many tissues and cell lines; 1 splice variant; mature form 2-144 [13,397 x] 🔗
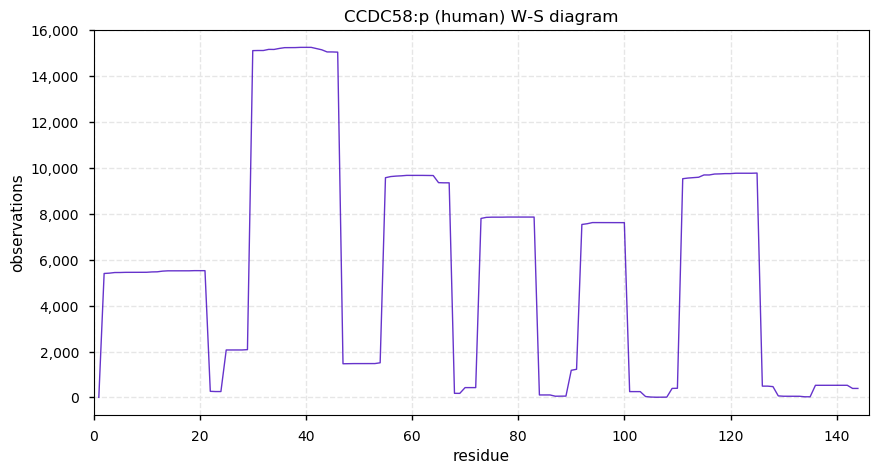
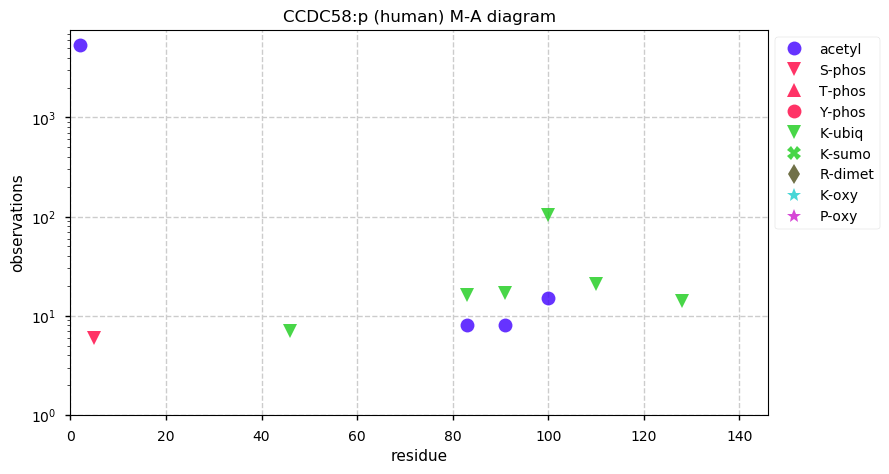
Sat Mar 07 19:00:29 +0000 2020@BiswapriyaMisra water
Sat Mar 07 16:46:12 +0000 2020PXD017614 does a good job of isolating and characterizing mitochondrial proteins. I can't locate an associated publication.
Sat Mar 07 12:49:45 +0000 2020HSDL1:p, hydroxysteroid dehydrogenase like 1 (H. sapiens) 🔗 Small intracellular protein; few PTMs; 1 high maf SAAV: S327C (0.25); obs. in many tissues and cell lines; 1 splice variant; mature form 2-330 [8,383 x] 🔗
Fri Mar 06 19:41:22 +0000 2020@macro_momo Which of the various "not-to-code" elements are you the most concerned with?
Fri Mar 06 19:08:54 +0000 2020PXD017025, nice data with unusually high id rates for human clinical samples. Reported in 🔗
Fri Mar 06 15:55:27 +0000 2020@goodlettlab1 @wfondrie @USHUPO You might want to try adding some video, since you already have a script, e.g. 🔗
Fri Mar 06 15:17:08 +0000 2020@TrumanLab @scottagerber @JohnRYatesIII @kevansf @asmsnews Most chaperonins are in the "deep-end of the pool" wrt to common PTMs 🔗
Fri Mar 06 15:06:39 +0000 2020@TrumanLab @JohnRYatesIII @kevansf @asmsnews The problem really needs at least 3 (S, T and Y) NCRR-type centers working on it.
Fri Mar 06 15:03:51 +0000 2020@TrumanLab @JohnRYatesIII @kevansf @asmsnews MS-based proteomics has been much better at detecting phosphorylation than anyone ever expected, with about 120k annotated. The methods for characterizing of these sites has not kept pace: they are still pretty much the same as when only a few hundred p-sites where known.
Fri Mar 06 13:23:22 +0000 2020DTNB:p, dystrobrevin beta (H. sapiens) 🔗 Midsized intracellular protein; several phosphodomains; no high maf SAAVs; obs. in many tissues and cell lines; 1 splice variant; mature form 1-627 [5,442 x] 🔗
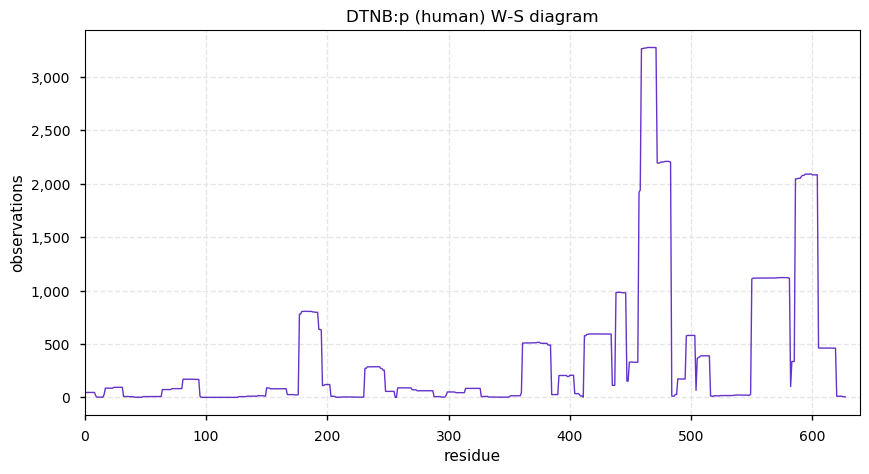
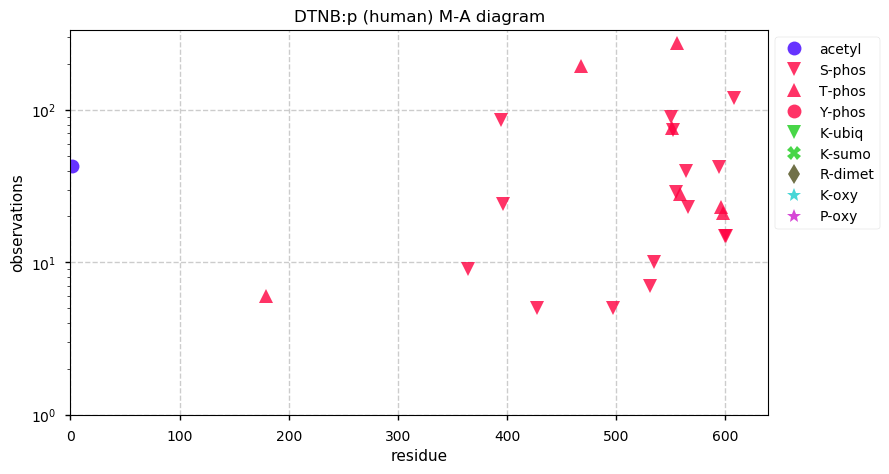
Thu Mar 05 18:27:31 +0000 2020I've read 5 recent papers over the last few days that self-identify as part of the "Human Proteome Project" and I'm stumped as to how these papers (& their related jargon) fit into any coherent project that has goals and endpoints.
Thu Mar 05 13:54:14 +0000 2020AKAP14:p, A-kinase anchoring protein 14 (H. sapiens)
Small intracellular protein; no PTMs; no high maf SAAVs; 1 of 3 spermatozoan-specific AKAPs; 1 splice variant; mature form 2-197 [70 x] 🔗
Wed Mar 04 20:10:06 +0000 2020@lgatt0 This site has a pretty good summary of the similarities & differences: 🔗 . For me, role-based permissions and CI/D features would be quite helpful. There are lots of similar comparisons, e.g. 🔗
Wed Mar 04 17:48:00 +0000 2020While I have only been using GitLab for a short while, I can see how it has some potential advantages over GitHub for longer term, multi-developer projects.
Wed Mar 04 16:26:06 +0000 2020@VATVSLPR A very reasonable hypothesis. But none of their software/wetware seems to have been capable of spotting this problem prior to publication ... or they simply decided to ignore it.
Wed Mar 04 16:17:33 +0000 2020Another QA indicator from the same multidimensional chromatography sample analysis, but a different fraction's LC/MS/MS run: 🔗
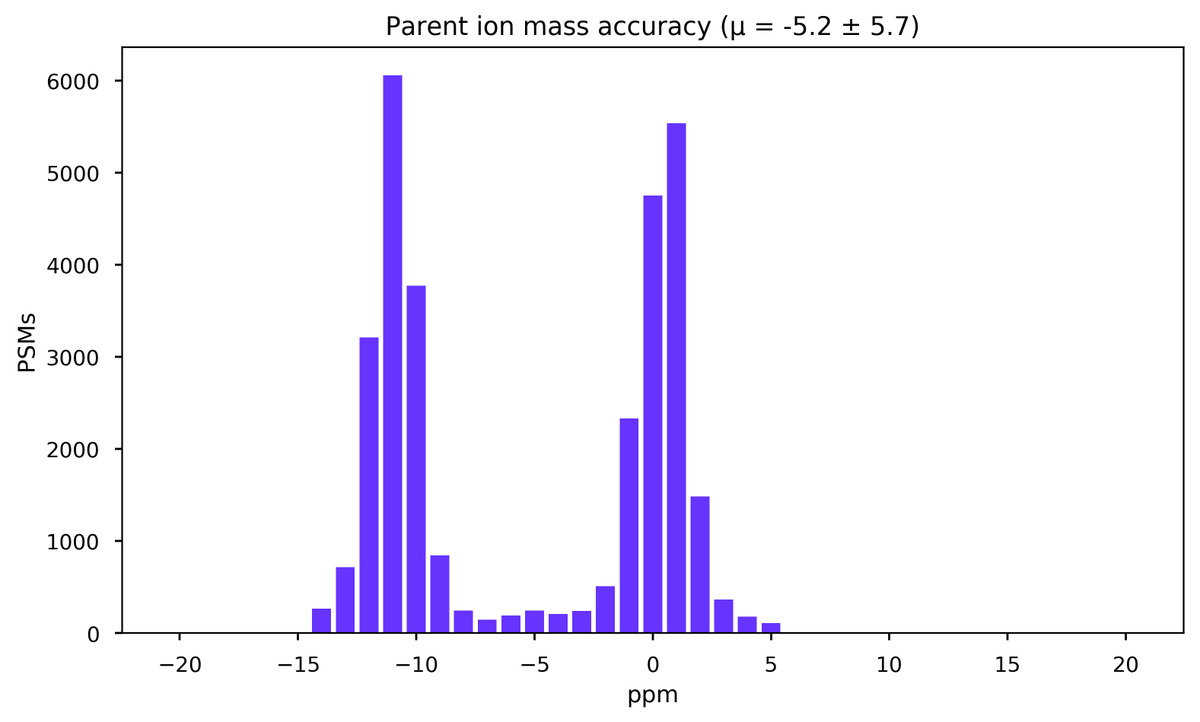
Wed Mar 04 16:07:53 +0000 2020@pwilmarth I saw a piece on military dog trainers a few weeks ago. They said the hardest thing to do was training the handler to pay attention to the dog, who has a lot more information about the environment than the handler. Also a reason dogs always have higher ranks than their handlers.
Wed Mar 04 13:41:37 +0000 2020AKAP13:p, A-kinase anchoring protein 13 (H. sapiens) 🔗
Large protein; intricate PTM structure; SAAVs: M452T (0.4), R574C (0.4), G624V (0.1), V845A/G (0.4), V897M (0.4), P1062A (0.4), E1106G (0.1), G2461S (0.1), N2793K (0.1); mature form 2-2813 [18,925 x] 🔗
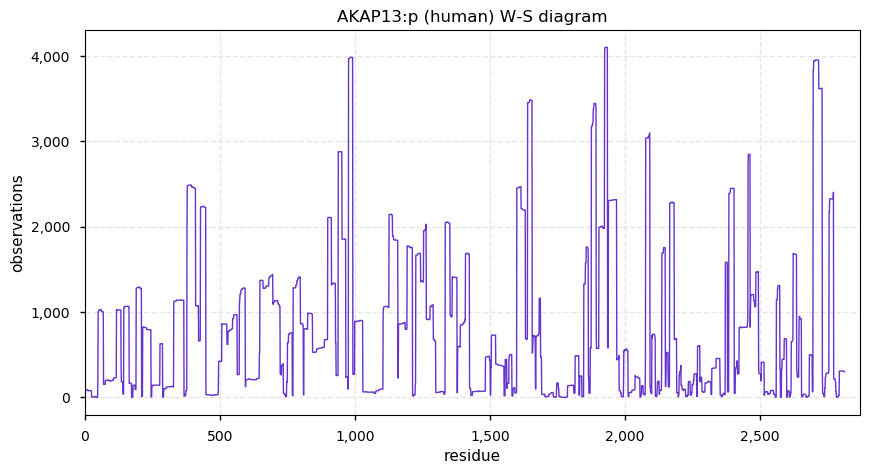
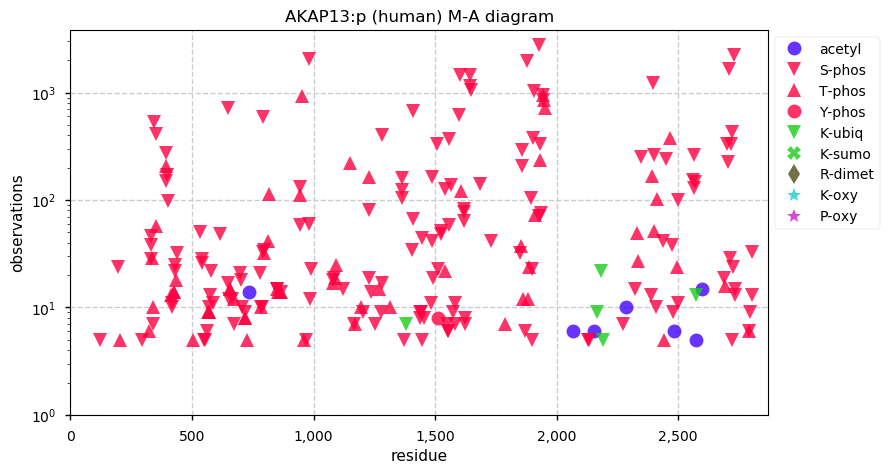
Wed Mar 04 13:37:09 +0000 2020Looks like there may be a QA problem here (plot of the id rate vs. scan #): 🔗
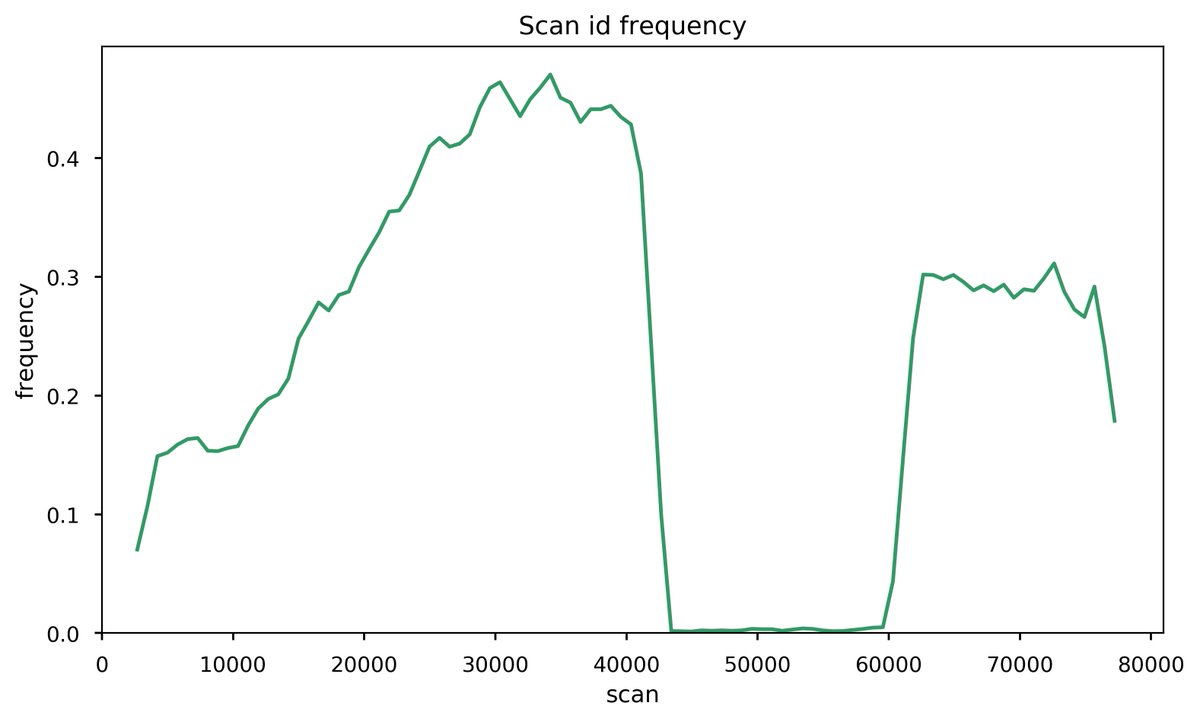
Tue Mar 03 18:39:49 +0000 2020@Freddyomics @Smith_Chem_Wisc A less excited explanation is available through the oracle-of-all-things:
🔗
The usage of the suffix "-ome" in scientific English is more nuanced than the 2002 paper would suggest — but every bit as annoying as a mechanism for generating nonce words.
Tue Mar 03 16:25:31 +0000 2020@lgatt0 @naturemethods @slavovLab Even if the binaries were sufficient (they aren't), the distribution contains many vendor libraries, with no indication that they have licenses to distribute them, e.g. Caliburn.Micro, SharpDX, .NET components, ThermoFisher.CommonCore, SciChart, QuickGraph, Ionic, GalaSoft
Tue Mar 03 15:47:22 +0000 2020Thanks to everyone who participated in this poll. I don't think there is a correct answer in general. I'm pretty sure that simply ignoring the issue can lead to problems in the differential analysis of quant data.
Tue Mar 03 15:11:59 +0000 2020@lgatt0 @slavovLab @naturemethods The title "Nature Methods" is meant to be taken ironically.
Tue Mar 03 13:02:50 +0000 2020AKAP12:p, A-kinase anchoring protein 12 (H. sapiens) 🔗 Large protein; intricate PTM structure; 2 very high maf SAAVs: K216Q (0.46), E1600D (0.44); 1 splice variant; mature form 2-1782 [26,418 x] 🔗
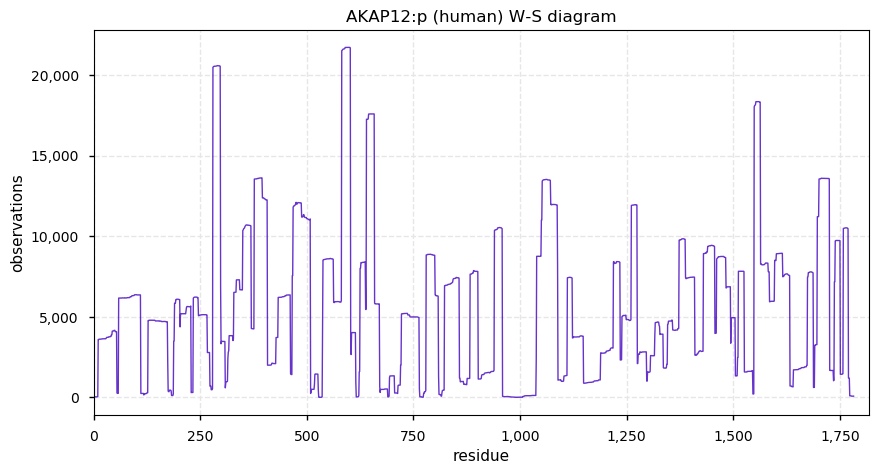
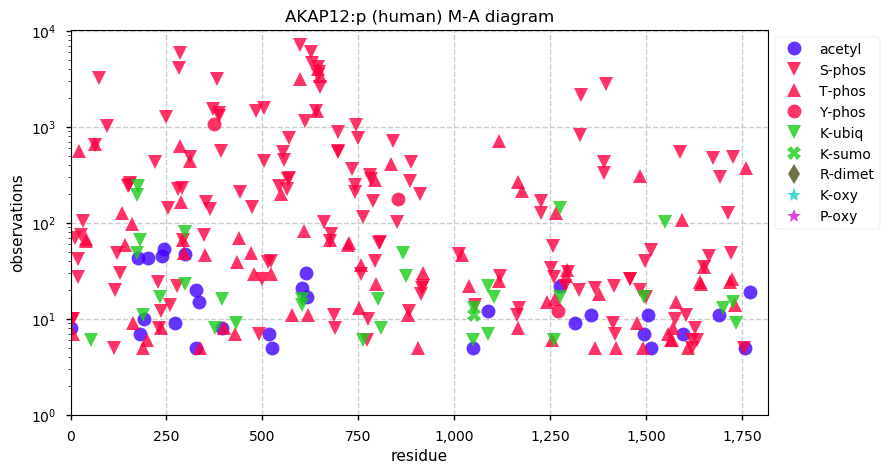
Mon Mar 02 19:12:24 +0000 2020@MHendr1cks And it is an attitude that university professors have been taking advantage of for generations.
Mon Mar 02 17:31:39 +0000 2020A pointless display of shameless skeuomorphism
Mon Mar 02 17:30:18 +0000 2020For example, the % fraction of Q's observed as the cyclic pyrrolidone form in NCOR1:p, nuclear receptor corepressor 1 — based on tryptic peptides from 17,953 data sets: 🔗
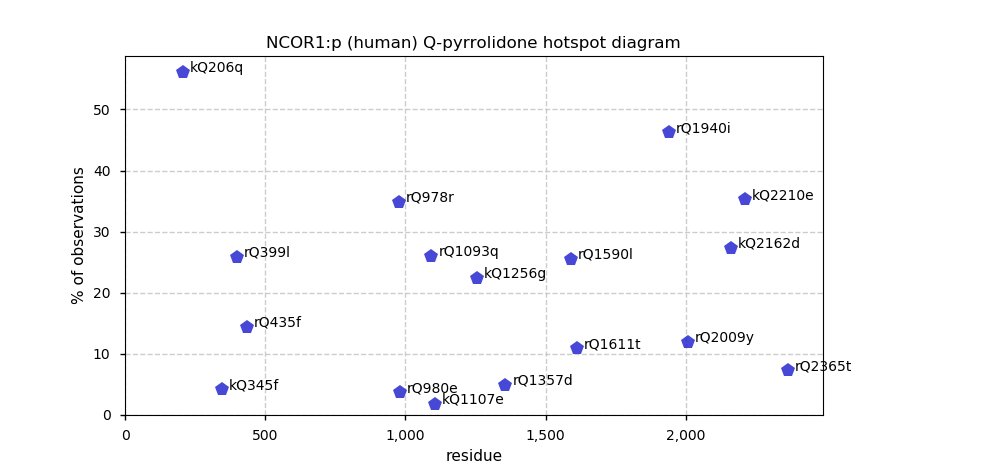
Mon Mar 02 15:40:01 +0000 2020What is the best way to deal with pyrrolidone-carboxylic acid formation at N-terminal Q residues in tryptic peptides when analyzing quantitative proteomics data?
Mon Mar 02 14:04:04 +0000 2020AKAP11:p, A-kinase anchoring protein 11 (H. sapiens) 🔗 Large protein; intricate phosphodomain structure; high maf SAAVs: R603H (0.01), R610C (0.02); 1 splice variant; mature form 2-1901 [9,207 x] 🔗
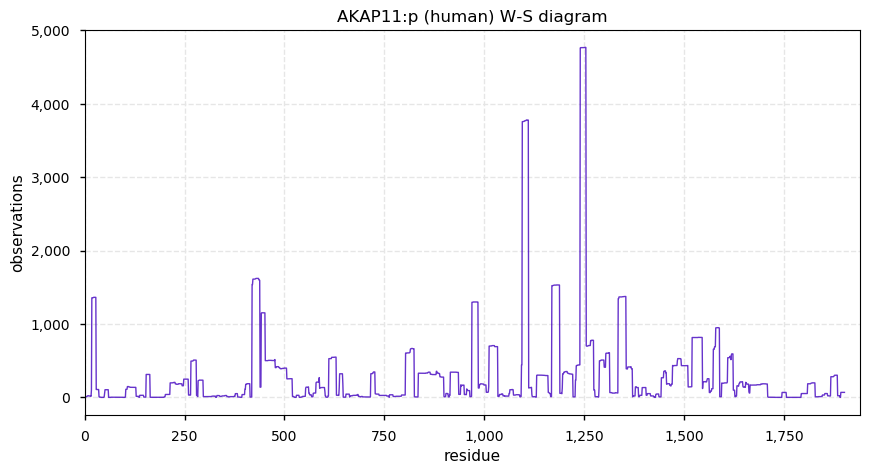
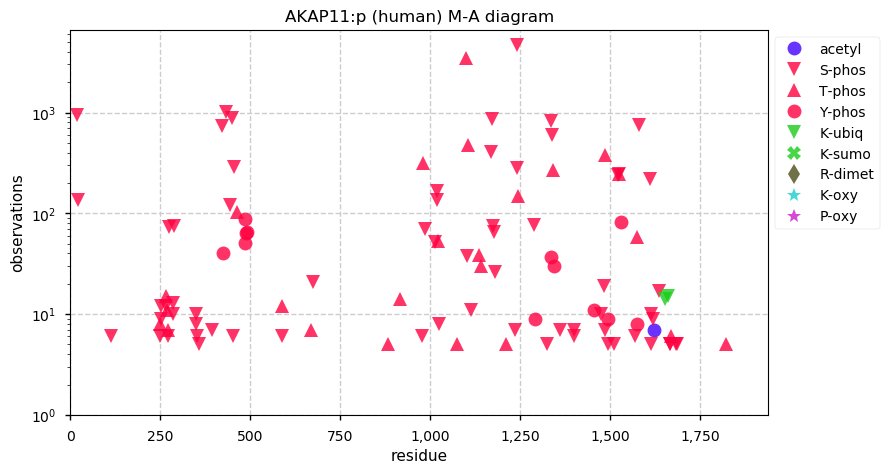
Sun Mar 01 15:42:07 +0000 2020@slashdot Kind of sums up why I gave up on trying to use Amazon to buy things.
Sun Mar 01 14:57:26 +0000 2020and quant folks should steer clear of N322 ... 🔗
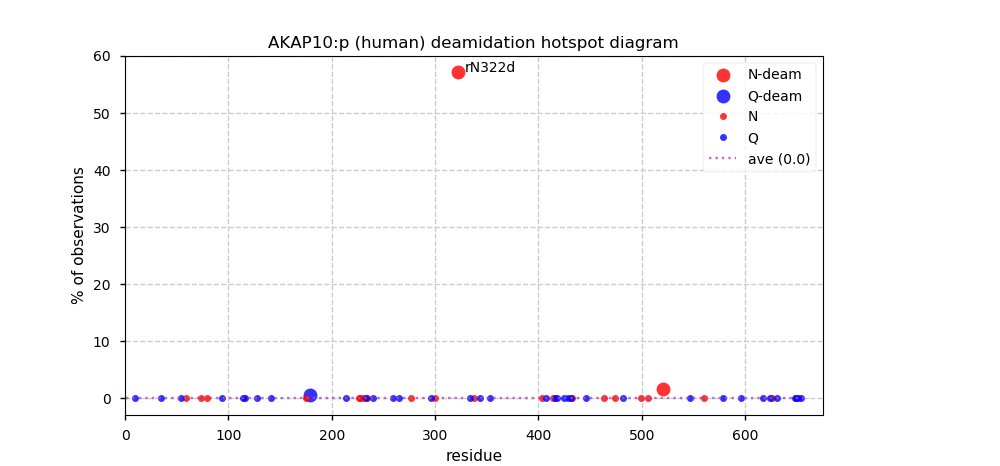
Sun Mar 01 14:54:34 +0000 2020AKAP10:p, A-kinase anchoring protein 10 (H. sapiens) 🔗 Midsized mitochondrial protein; several distinct phosphodomains; 2 very high maf SAAVs: R249H (0.6), I646V (0.6); 1 splice variant; mature form 14-662 [4,660 x] 🔗
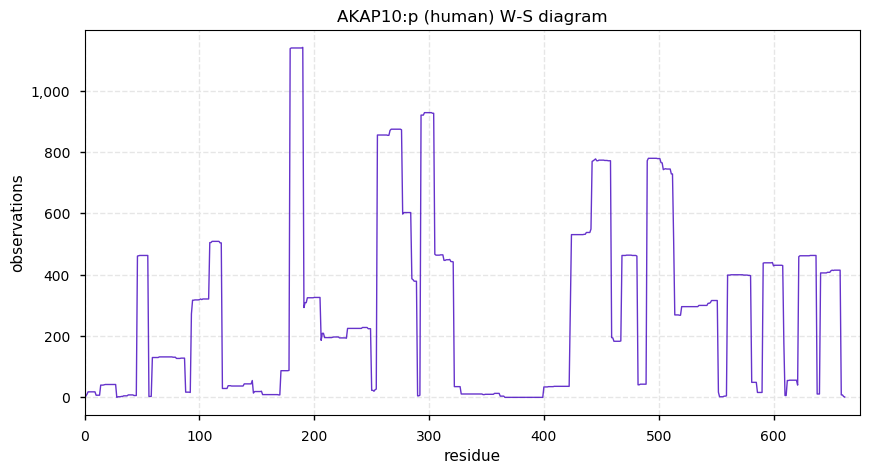
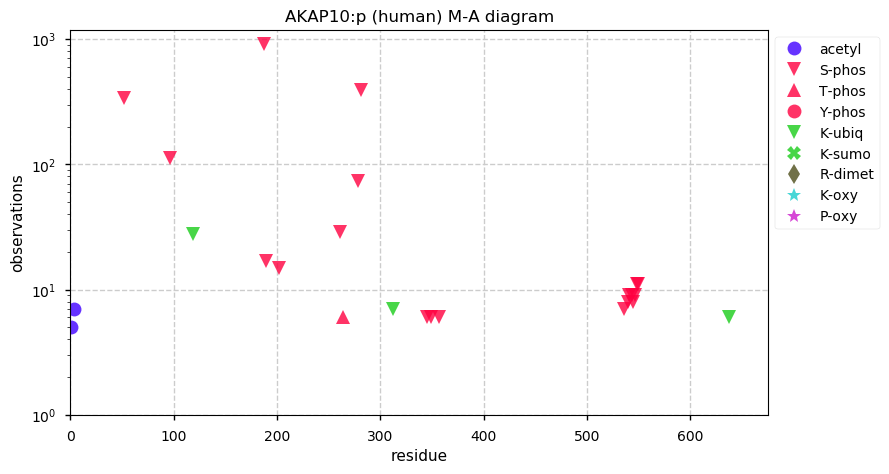
Sat Feb 29 19:02:18 +0000 2020That feeling when anticipation of a really interesting new data set turns into "would it really have been that difficult to have cleaned the damned ion source" 😡😢😭
Sat Feb 29 18:22:11 +0000 2020It is a good thing that "xit" isn't a Linux command.
Sat Feb 29 17:19:05 +0000 2020AKAP9 was previously known in the literature as KIAA0803, AKAP350, AKAP450, CG-NAP, YOTIAO, HYPERION, PRKA9, MU-RMS-40.16A, PPP1R45, and/or LQT11.
Sat Feb 29 16:12:30 +0000 2020@Smith_Chem_Wisc If I was younger, I'd be surprised by this sort of a apparent cluelessness, but still 😲
Sat Feb 29 15:55:10 +0000 2020@Smith_Chem_Wisc Boomer.
Sat Feb 29 15:15:18 +0000 2020AKAP9:p, A-kinase anchoring protein 9 (H. sapiens) 🔗 Large protein; extensive phosphorylation, with a PTM gap domain: 1802-2375; 2 high maf SAAVs: R1276Q (0.01), R1614P (0.01); 1 splice variant; mature form 1-3907 [15,546 x] 🔗
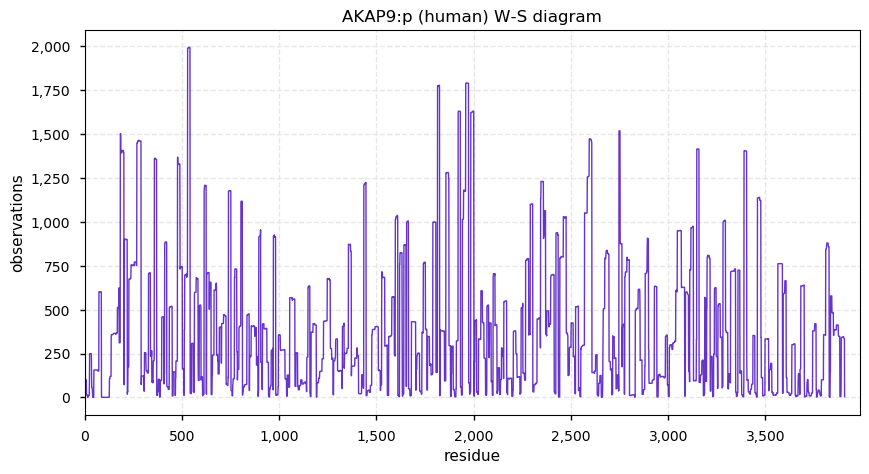
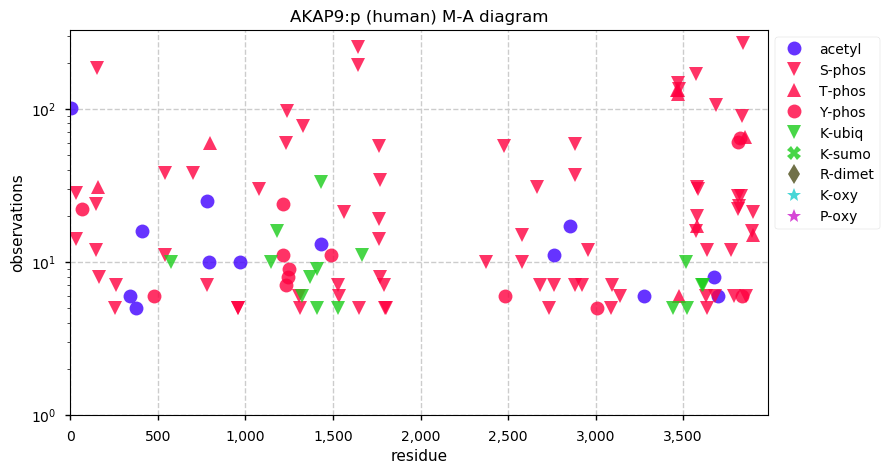
Fri Feb 28 22:50:04 +0000 2020The two prominent acetylation domains correspond to the zinc fingers ZF1 & ZF2 associated with chromatin binding and condensation
Fri Feb 28 17:15:05 +0000 2020@slashdot Come to think of it, I don't actually remember the last time I printed something.
Fri Feb 28 17:06:24 +0000 2020@slashdot I remember printing ... but it has been quite a while since I actually did it.
Fri Feb 28 15:20:23 +0000 2020Looking more carefully at the errors, it looks like my anti-snooping/anti-tracking system is just doing its job and preventing a lot of suspect stuff from loading, resulting in the warnings and errors.
Fri Feb 28 15:14:38 +0000 2020@ypriverol @ProteomicsNews While it is a significant issue facing many fields, the only real response to this problem by the publishing business has been at the publisher-level, with masthead extensions like "Scientific Data". Unfortunately, this approach hasn't really worked.
Fri Feb 28 15:11:03 +0000 2020@ypriverol @ProteomicsNews And I strongly encourage the completion and adoption of this & other proposals around the general problem of the standardization of reporting electronic data.
Fri Feb 28 15:05:46 +0000 2020The front page HTML code for 🔗 throws a ridiculous number of warnings and errors.
Fri Feb 28 13:22:33 +0000 2020And for fun ... 🔗
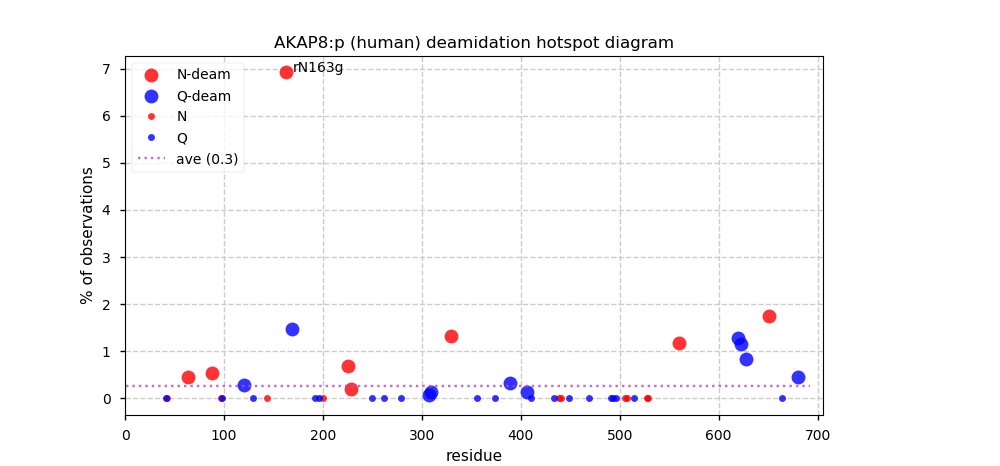
Fri Feb 28 13:21:53 +0000 2020AKAP8:p, A-kinase anchoring protein 8 (H. sapiens)
Midsized protein; prominent acetylation and phosphorylation domains; 1 high maf SAAVs: L350F (0.02); 1 splice variants; mature form 1?-692 [18,755 x] 🔗
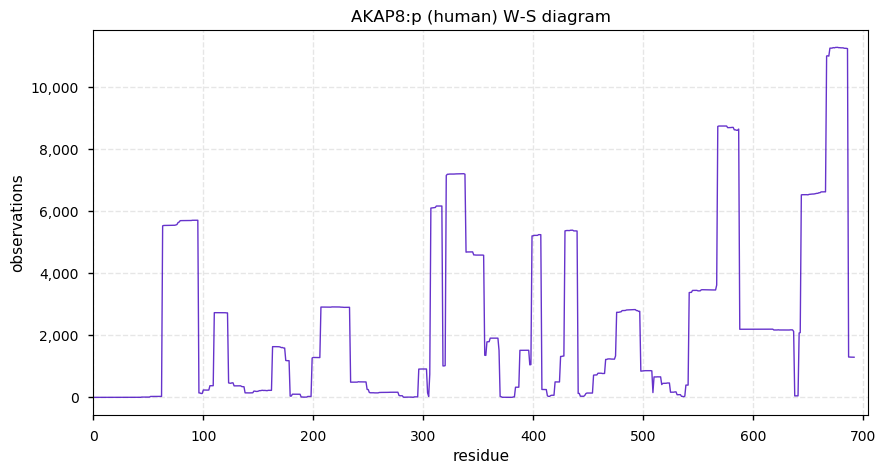
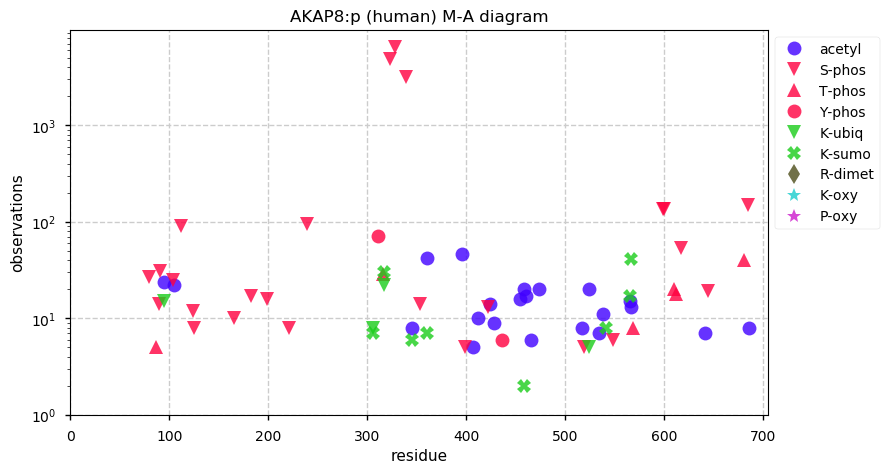
Thu Feb 27 19:58:50 +0000 2020@chrashwood There aren't any unassigned upper case English letters wrt to amino acid residues. If another symbol is needed, you really have to use lower case symbols, numbers or a non-English alphabet, e.g., Greek or Hebrew letters.
Thu Feb 27 19:44:59 +0000 2020@chrashwood The daffy reviewers should not have allowed that anyway. "J" is reserved as the ambiguity code for "I or L" and it should never be used for anything else in a publication.
Thu Feb 27 19:38:33 +0000 2020Why would California need more than 200 CoVid-19 test kits? Next they'll be asking for more than 64 KB of memory ...
🔗
Thu Feb 27 15:25:34 +0000 2020It is hard for me to understand why anyone thought the incoherent, babble-bot statements made by the US administration yesterday would lessen fears in financial markets. The only response team member that made any sense at all was Rear Admiral Schuchat.
Thu Feb 27 15:02:02 +0000 2020@ProteomicsNews as well as a naming convention that can be used to correlate these files with each specific experiment. This should be present in all papers, regardless of whether or not the data or results files have been made public. /fin
Thu Feb 27 14:58:40 +0000 2020@ProteomicsNews I don't disagree with this notion, but I would personally rather have a section in manuscripts entitled something like "Electronic data" that contains a list of the total number & format of all of the files necessary to generate the "Results" section /1
Thu Feb 27 13:42:52 +0000 2020AKAP7:p, A-kinase anchoring protein 7 (H. sapiens) 🔗 Small intracellular protein; no PTMs; no high maf SAAVs; found in many tissues and cell lines; at least 2 splice variants; mature form 1-384 [1,184 x] 🔗
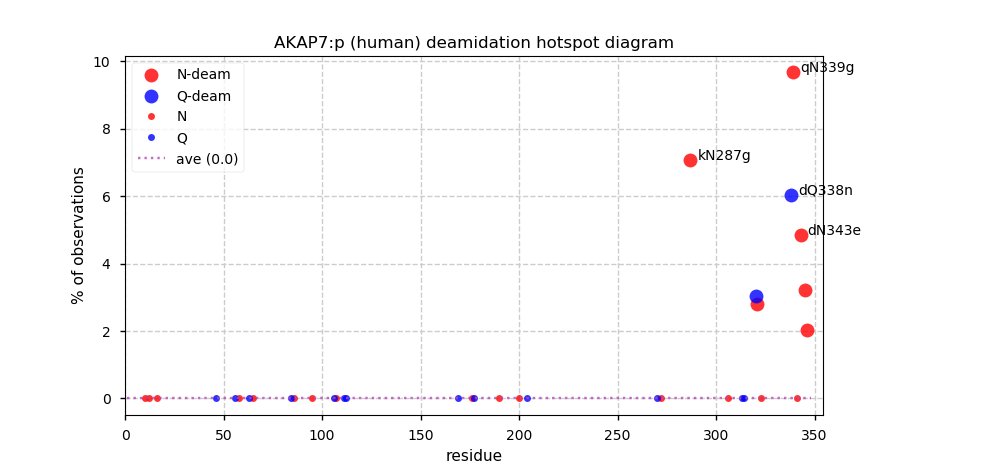
Wed Feb 26 21:53:05 +0000 2020Going slow at the superclusters: "$5.6 million in 2018-19 had been spent under the superclusters program out of nearly $244 million authorized for that fiscal year"
🔗
Wed Feb 26 18:33:42 +0000 2020@bkives MB is thoroughly post-culture and exploring the phenomenology of neo-culturalism. It takes others a long time to catch up.
Wed Feb 26 18:28:52 +0000 2020@hilaryagro @girlziplocked I'd want to know why he is using a Bulgarian (.bg) domain name???
Wed Feb 26 17:36:12 +0000 2020@AlexUsherHESA Does this story somehow involve Gary Filmon as a Locutus-like character? (very inside-curling MB-only reference)
Wed Feb 26 16:23:21 +0000 2020Are there any upcoming EU or US scientific conferences that are expected to be cancelled because of CoVid-19?
Tue Feb 25 17:43:50 +0000 2020@ypriverol @dtabb73 OK. Thanks for the response.
Tue Feb 25 17:36:37 +0000 2020@dtabb73 Those pages are my usual starting point, because 🔗 makes it easy for me to see which new data sets have popped up since yesterday.
Tue Feb 25 17:35:07 +0000 2020@dtabb73 There is a link to the FTP site on the ProteomeXchange page. In this case, ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2019/04/PXD012703. And the directory seems to be absent for some reason.
Tue Feb 25 17:10:13 +0000 2020Does anybody know why the FTP directory for 🔗 seems to be absent?
Tue Feb 25 15:22:26 +0000 2020@AlexUsherHESA As someone who has moved back to Manitoba and lived here long enough to re-establish bona fides, I would strongly second that response.
Tue Feb 25 13:57:12 +0000 2020AKAP5:p, A-kinase anchoring protein 5 (H. sapiens)
Small protein; some phosphorylation; 2 high maf SAAVs: P100L (0.07), R188Q (0.03); found in brain tissue and some cancers; 1 splice variant; mature form 1-427 [1,819 x] 🔗
Tue Feb 25 13:51:36 +0000 2020🔗 is an excellent TMT phosphopeptide dataset. Everything about it is well done. Please emulate.
Mon Feb 24 17:48:10 +0000 2020Thanks to everyone who participated in this poll. Trypsin an endopeptidase that may have some exopeptidase activity. If you view trypsin as only an endopeptidase, then the [mode,median,mean] tuple is [2,7,11]. If you include exopeptidase activity, the tuple is [1,6,10].
Mon Feb 24 13:16:33 +0000 2020AKAP4:p, A-kinase anchoring protein 4 (H. sapiens) 🔗 Midsized protein; no PTMs; 1 high maf SAAVs: A852T (0.01); present in spermatozoa and pericardial fluid; 1 splice variant; pro-protein 3-854; mature form 189-854 [884 x] 🔗
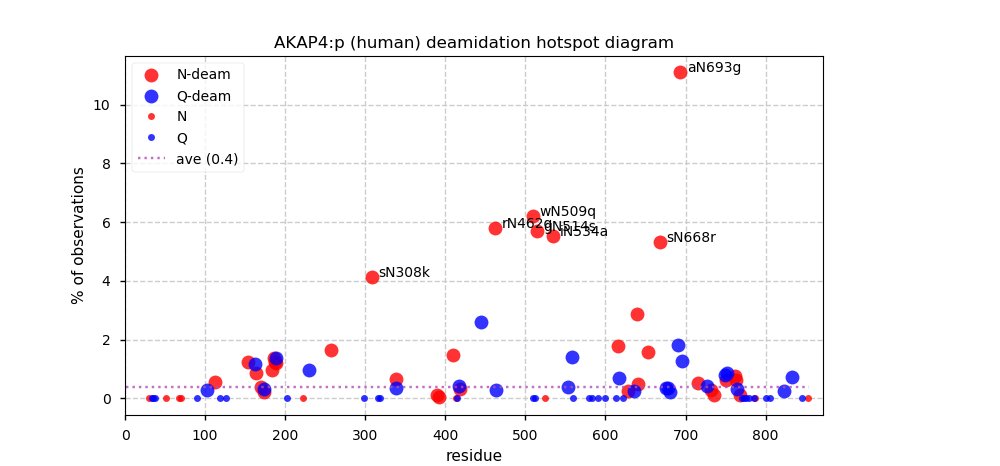
Sun Feb 23 17:43:28 +0000 2020Assume that you take the proteins in the human proteome and perform a theoretical tryptic digest, generating a histogram of the number of peptides vs length (# of residues). Which of the following are the closest values for that distribution's [mode, median,mean]:
Sun Feb 23 16:21:23 +0000 2020@pathogenomenick @BiswapriyaMisra Rule 1: do whatever it takes to get the support of your department head; and
Rule 2: nothing else really matters.
Sun Feb 23 14:14:40 +0000 2020AKAP3:p, A-kinase anchoring protein 3 (H. sapiens) 🔗
Midsized protein; protein N-terminal acetylation; 3 high maf SAAVs: G118E (0.29), I661T (0.18), S725L (0.15); only present in spermatozoa; 1 splice variants; mature form 2-853 [680 x] 🔗
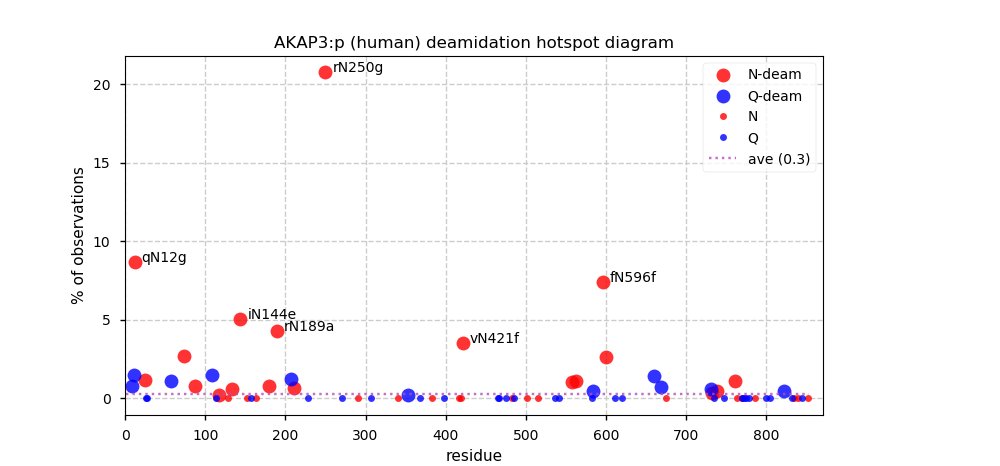
Sat Feb 22 21:44:02 +0000 2020PXD015361 provides an interesting wrinkle on the phosphopeptide isolation process that I'd never seen before.
Sat Feb 22 16:41:44 +0000 2020@TrumanLab It is usually awful. Most (all?) universities do not provide HR training for faculty & they tend to ask rather dopey, often borderline illegal, questions when they are 1-on-1. The questions are much better when there is an audience. 1-on-1 is more of a hazing than an interview.
Sat Feb 22 16:33:47 +0000 2020@TrumanLab You should do everything you can to avoid being questioned one-on-one during an academic interview. Nothing good comes of it.
Sat Feb 22 14:28:30 +0000 2020This is also a great data set if you still need to convince yourself that sORFs are really responsible for observable levels of MHC type I peptides
Sat Feb 22 14:14:47 +0000 2020🔗
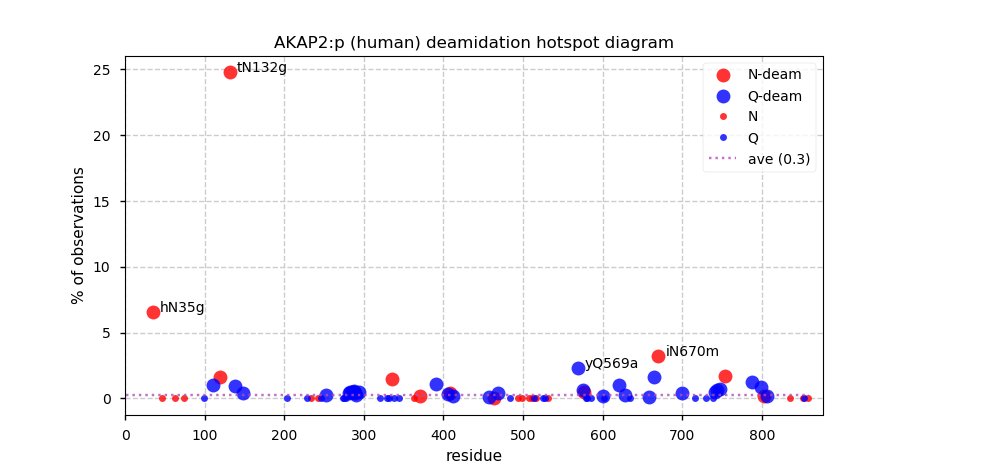
Sat Feb 22 14:14:15 +0000 2020AKAP2:p, A-kinase anchoring protein 2 (H. sapiens) 🔗
Midsized protein; phosphorylation (lots); 6 high maf SAAVs: P20L (0.01), C64Y (0.02), R109Q (0.02), R305C (0.01), L561S (0.54), A650V (0.02); 2 splice variants; mature form 1-859 [19,105 x] 🔗
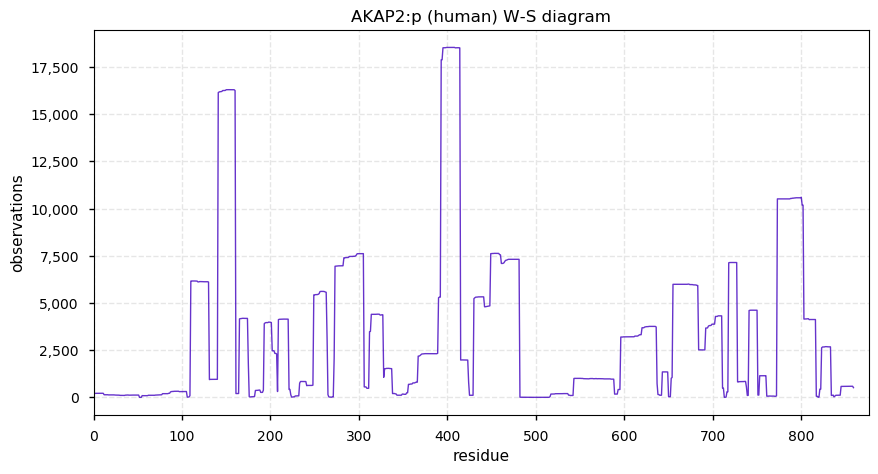
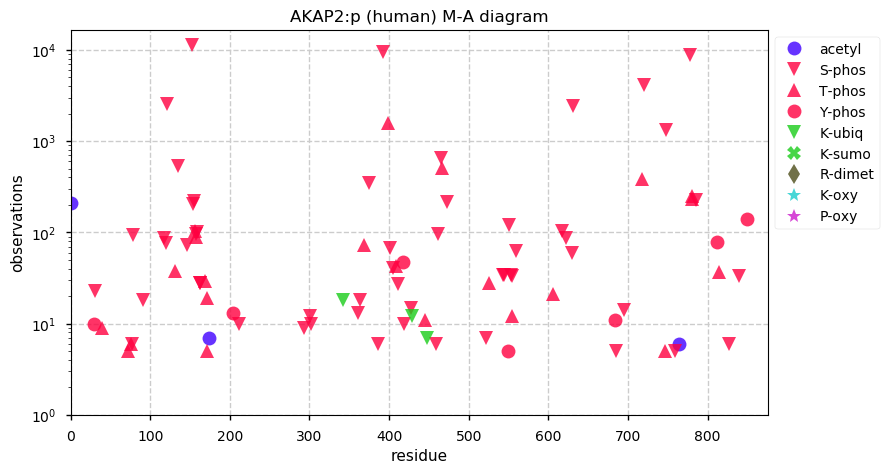
Fri Feb 21 23:17:20 +0000 2020@zhadu @JesseBrown @paulvieira @jamespmcleod I just watched a CN train with at least 100 cars loaded with intermodal containers go by on the main line thru downtown Wpg.
Fri Feb 21 22:59:47 +0000 2020The least-enticing email header line ever:
"Find out more about our updated Terms of Service"
Fri Feb 21 21:50:40 +0000 2020PXD015957 has some nearly ideal MHC type I peptide data sets. Good work 🔗 🔗
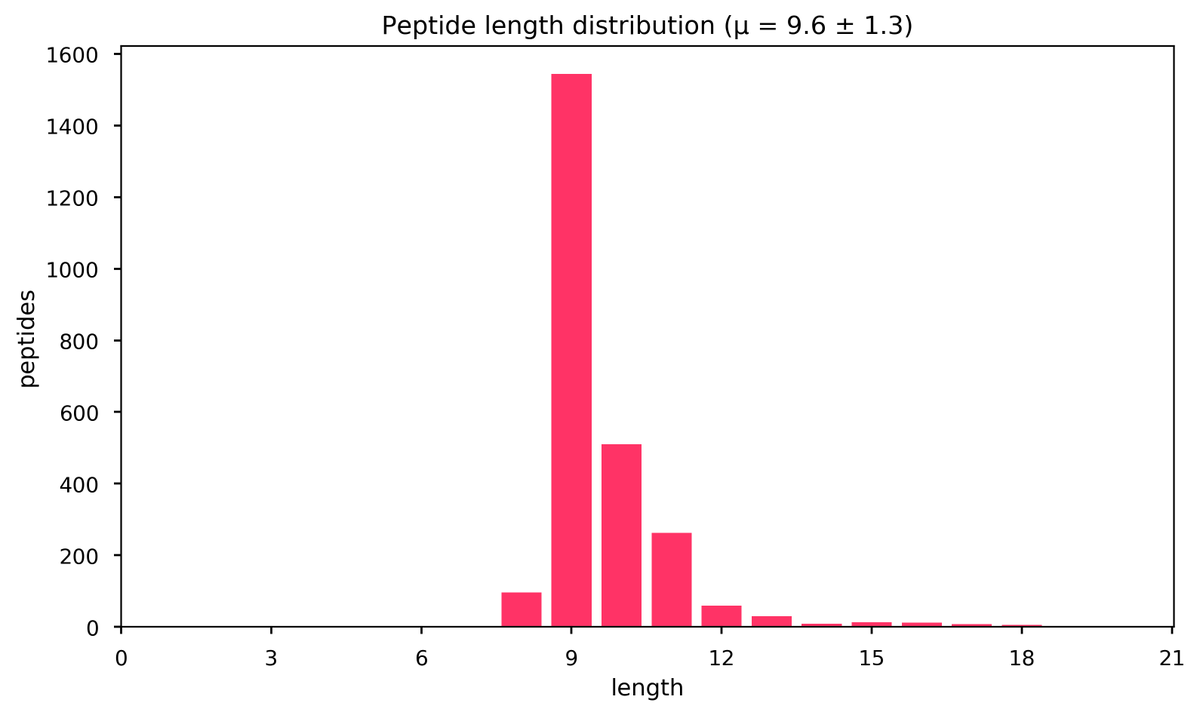
Fri Feb 21 17:01:35 +0000 2020@AlexUsherHESA @JJ_McCullough Reminiscent of something overlooked in the media discussion: 🔗
Fri Feb 21 15:43:13 +0000 2020@michaelhoffman @lenjf @CellsAndProts It is possibly the best method for BS'ing the boss ever invented.
Fri Feb 21 14:53:37 +0000 2020@cstross Anyone who knows anything about the subject knows it is changes in lunar gravity that cause the effect. The "light of the full moon" stuff is just a bunch of old-wives-tales.
Fri Feb 21 14:51:11 +0000 2020🔗
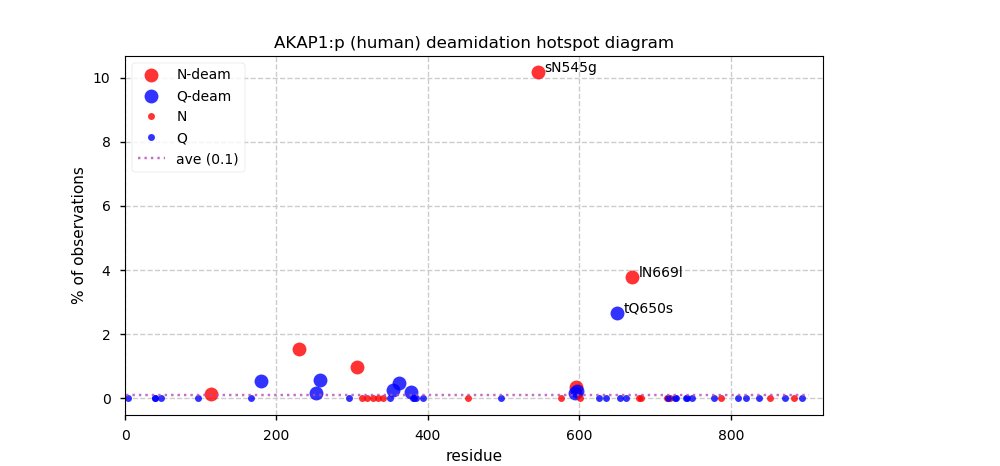
Fri Feb 21 14:50:35 +0000 2020AKAP1:p, A-kinase anchoring protein 1 (H. sapiens) 🔗
Midsized mito. protein; phosphorylation (lots); 6 high maf SAAVs: A18V (0.06), V60M (0.03), R124C (0.01), A218T (0.01), G323S (0.05), S330G (0.07); 1 splice variant; mature form 31-903 [14,588 x] 🔗
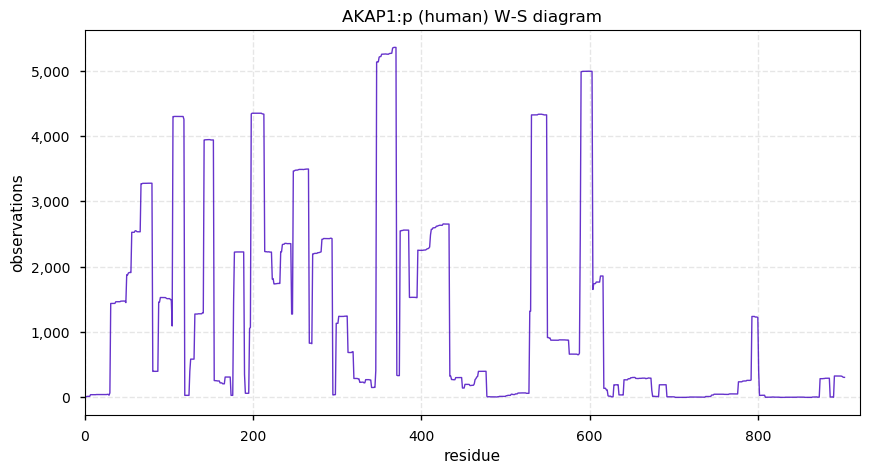
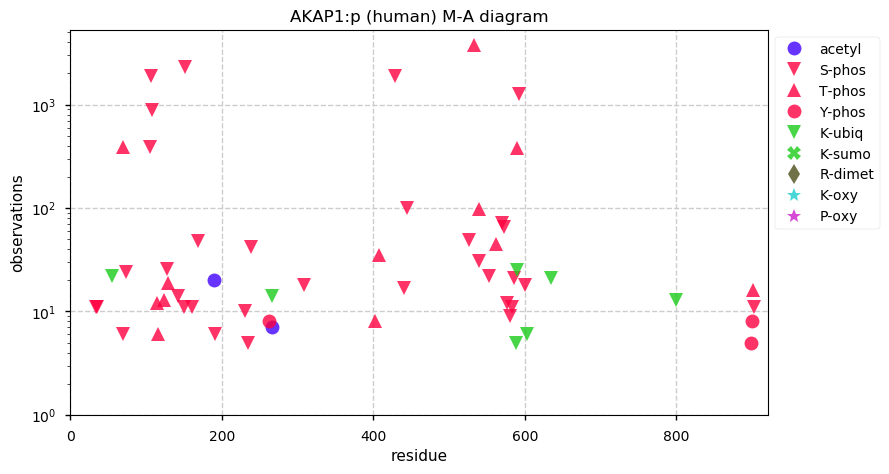
Fri Feb 21 13:49:17 +0000 2020@AlexUsherHESA Local triumphalism builds intensity as a function of distance west of the Manitoba border, building to a crescendo in the Lower Mainland
Thu Feb 20 19:38:41 +0000 2020@byu_sam @BiswapriyaMisra @OmicsPNNL But the variability inequality isn't bad either:
"extraction >> instrumental variance > instrumental stability > digestion"
Thu Feb 20 19:37:31 +0000 2020@byu_sam @BiswapriyaMisra @OmicsPNNL Nice quote: "for some peptides, nearly 100% of their variability is contributed by LC-MS analysis"
Thu Feb 20 19:25:51 +0000 2020@michaelhoffman @MHendr1cks @queensu @NSERC_CRSNG @QueensPrincipal Most universities expect faculty members to use the "Fight Club convention" whenever it comes to public statements.
Thu Feb 20 16:08:32 +0000 2020@neely615 @SciReports If you want to analyze it yourself, be sure to check for carbamylation and collagen hydroxyproline.
Thu Feb 20 15:59:29 +0000 2020@neely615 @SciReports Definitely the only publicly available grizzly bear data around. The results of our initial analysis of the raw data: 🔗
Thu Feb 20 13:48:41 +0000 2020WNT11:p, Wnt family member 11 (H. sapiens) 🔗 Small secreted protein; glycosylation: N40, N90, N160; no high maf SAAVs; 1 splice variant; most common in lung tissue; mature form 25?-354 [554 x] 🔗
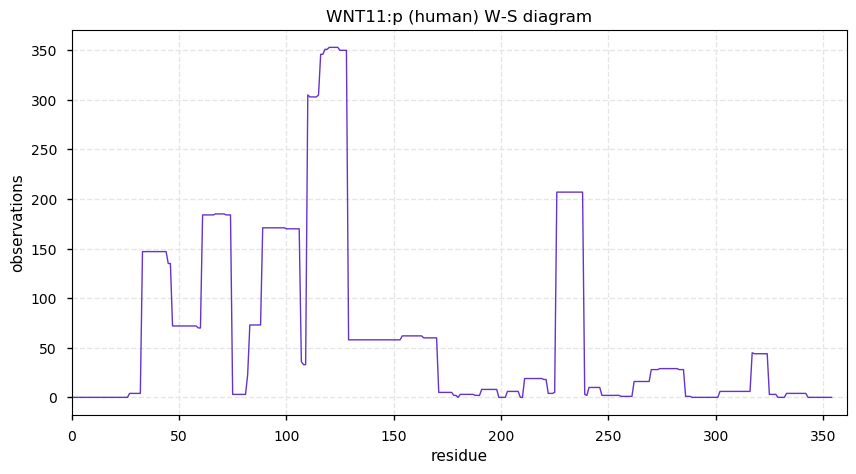
Wed Feb 19 22:33:50 +0000 2020@UCDProteomics Available on request?
Wed Feb 19 16:02:07 +0000 2020Please avert your eyes. A histogram showing the number of PSMs found in a human phosphopeptide LC/MS/MS run as a function of the log₁₀ of the # of times that the peptide sequence has been observed previously: 🔗
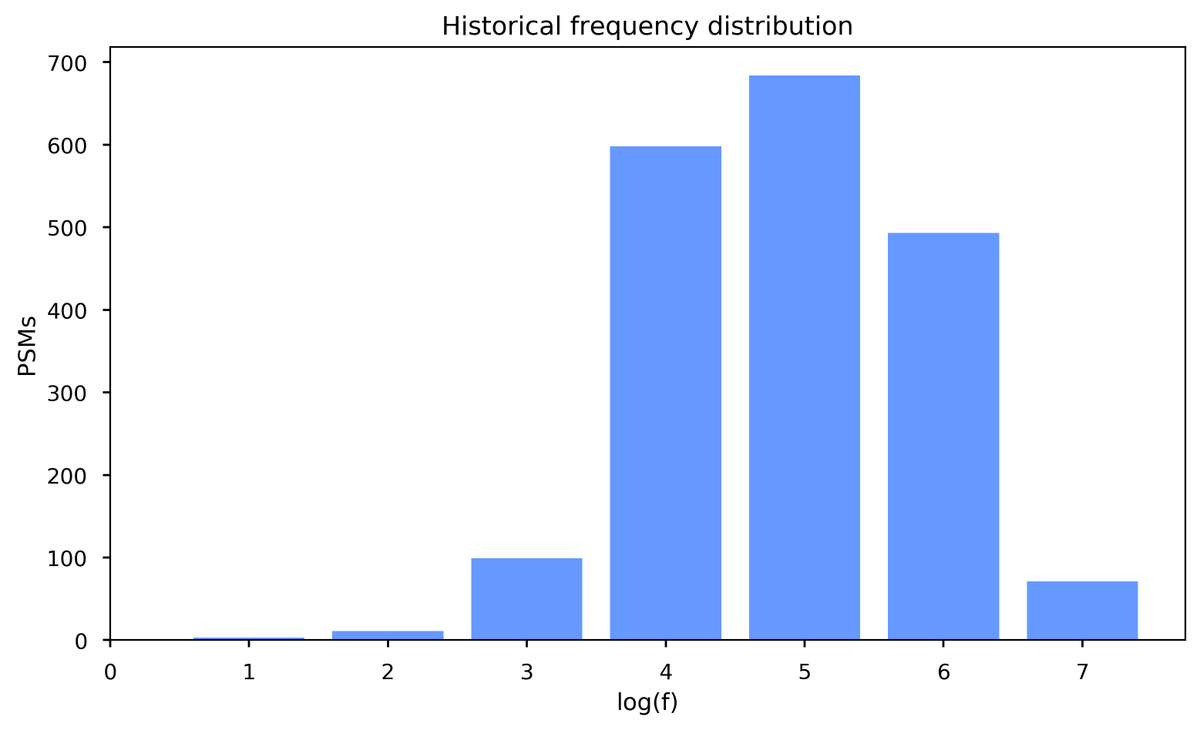
Wed Feb 19 14:15:06 +0000 2020WNT6:p, Wnt family member 6 (H. sapiens) 🔗 Small secreted protein; no PTMs; no high maf SAAVs; 1 splice variant; absent from common cell lines; mature form 25-365 [410 x] 🔗
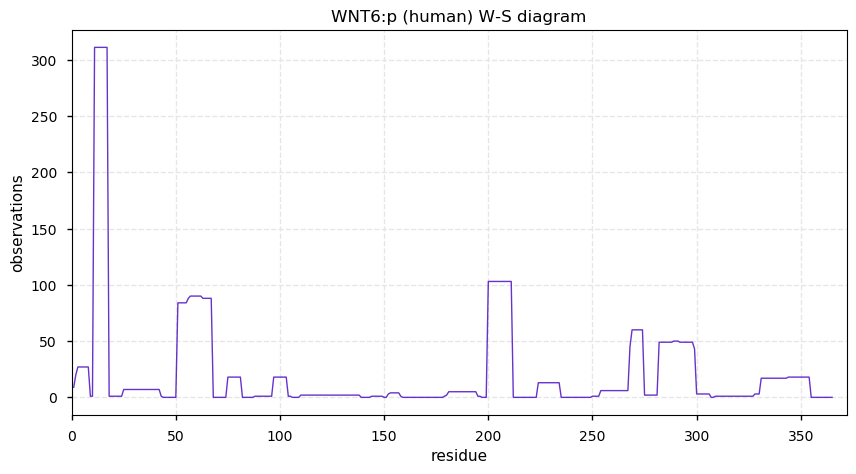
Wed Feb 19 01:49:31 +0000 2020@Sci_j_my Probably: it depends on the place.
Wed Feb 19 00:44:41 +0000 2020@Sci_j_my 5 spice.
Tue Feb 18 20:18:13 +0000 2020🔗
Tue Feb 18 15:36:05 +0000 2020And in case you were wondering, they do not elaborate on this point.
Tue Feb 18 15:27:44 +0000 2020Although there is a runner up just a few lines away:
"Quantification of differentially expressed proteins was analyzed further using standard procedure ..."
Tue Feb 18 15:27:02 +0000 2020Maybe the least helpful phrase ever in a proteomics Methods section:
"Tryptic peptides were labeled using the TMT kit/iTRAQ kit ..."
Tue Feb 18 13:46:18 +0000 2020WNT4:p, Wnt family member 4 (H. sapiens) 🔗 Small secreted protein; glycosylation: N88; no high maf SAAVs; 1 splice variant; absent from common cell lines; mature forms 23-351 [169 x] 🔗
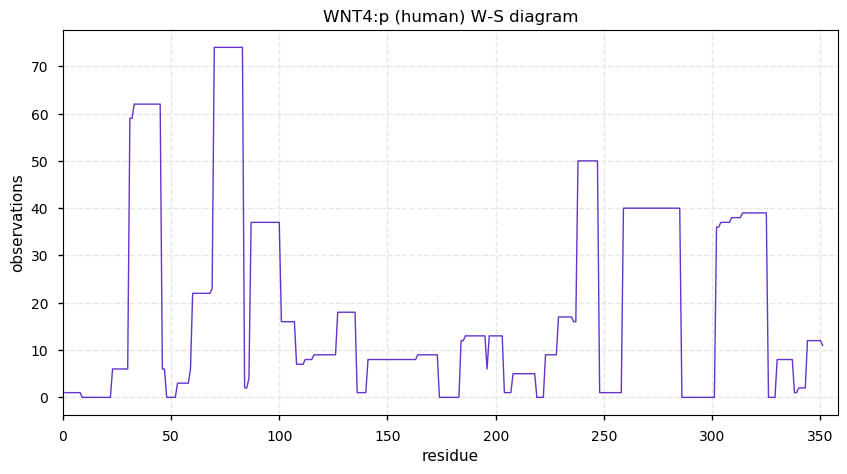
Mon Feb 17 17:12:40 +0000 2020@MetastasisRS @RuneLinding @NatureRevCancer It is hard to over-emphasize the value of understanding the proteins fibroblasts add to tumor tissue ECM as biomarkers. It is often such a large signal that proteomics studies tend to ignore it.
Mon Feb 17 15:49:26 +0000 2020@hecklab @chrashwood @UUBeta @epic_xs @asmsnews You might want to reword this one.
Mon Feb 17 14:20:36 +0000 2020@CVogelNYC Apophenia and pareidolia are common issues with AI classification systems. We seem to like to build in our own failings.
Mon Feb 17 14:08:01 +0000 2020WNT2B:p, Wnt family member 2B (H. sapiens) 🔗 Small secreted protein; glycosylation: N117, N283; no high maf SAAVs; 1 splice variant; absent from most cell lines; mature forms 19?-391 [724 x] 🔗
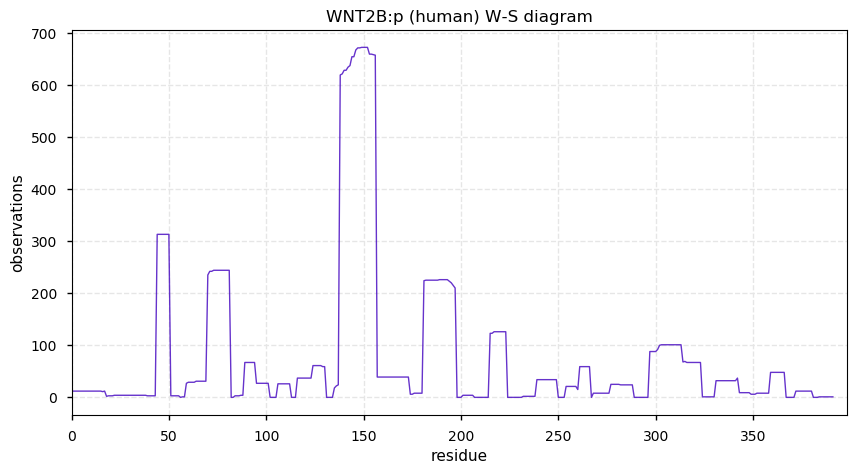
Sun Feb 16 19:34:16 +0000 2020@_Spectrumederp When the sample prep didn't work as imagined, leaving a high background of other peptides, generating non-stochastic false positives interpretted as genuine.
Sun Feb 16 14:37:37 +0000 2020WNT3:p, Wnt family member 3 (H. sapiens) 🔗 Small secreted protein; glycosylation: N301; no high maf SAAVs; 1 splice variant; most commonly observed in placenta; mature forms 22?-355 [190 x] 🔗
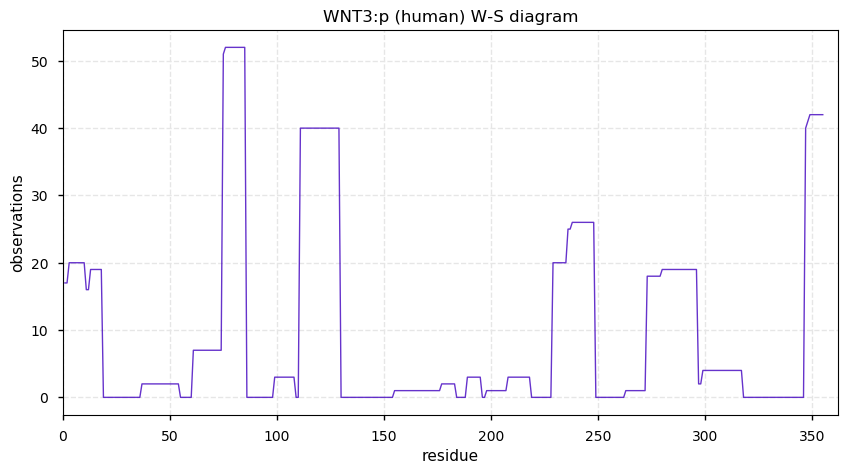
Sat Feb 15 16:21:10 +0000 2020And I'm currently in love with my new term "blinkered analysis".
Sat Feb 15 16:20:35 +0000 20203rd large data set this week where a combination of sample prep problems and blinkered analysis led to a difficult-to-interpret publication.
Sat Feb 15 14:13:29 +0000 2020WNT proteins are rarely observed in proteomics experiments.
Sat Feb 15 13:49:30 +0000 2020WNT2:p, Wnt family member 2 (H. sapiens) 🔗 Small secreted protein; no PTMs; no high maf SAAVs; 1 splice variant; most commonly observed in placenta and lung; mature forms 26?-360 [414 x] 🔗
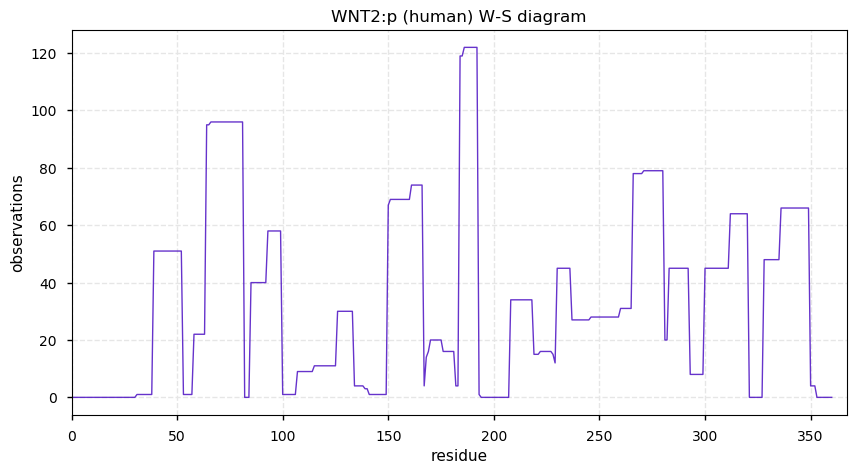
Fri Feb 14 16:32:05 +0000 2020@HFazelinia @PastelBio @ProteomicsNews When I made a non-redundant FASTA of smORF sequences found in MHC type I experiments that did not overlap with anything in ENSEMBL H. sapiens proteins, there were 11,131 smORFs with 501,965 total residues
Fri Feb 14 16:14:32 +0000 2020@HFazelinia @PastelBio @ProteomicsNews Most of the smORFs are very short (< 50 residues), making the total number of peptides is quite tractable. You end up with 1-3% of MHC type I peptide ids coming from these smORFs.
Fri Feb 14 14:54:32 +0000 2020@PastelBio @ProteomicsNews And that I probably never would have seen the article without @PastelBio 's good work.
Fri Feb 14 14:37:11 +0000 2020@PastelBio @ProteomicsNews I was actually trying to indicate my appreciation of the article: other than their renaming smORFs to be nuORFs they have observed the phenom that there are some ORFs whose only purpose seems to be producing peptides for MHC type I display.
Fri Feb 14 14:33:19 +0000 2020And, just for a bit of overkill: 🔗
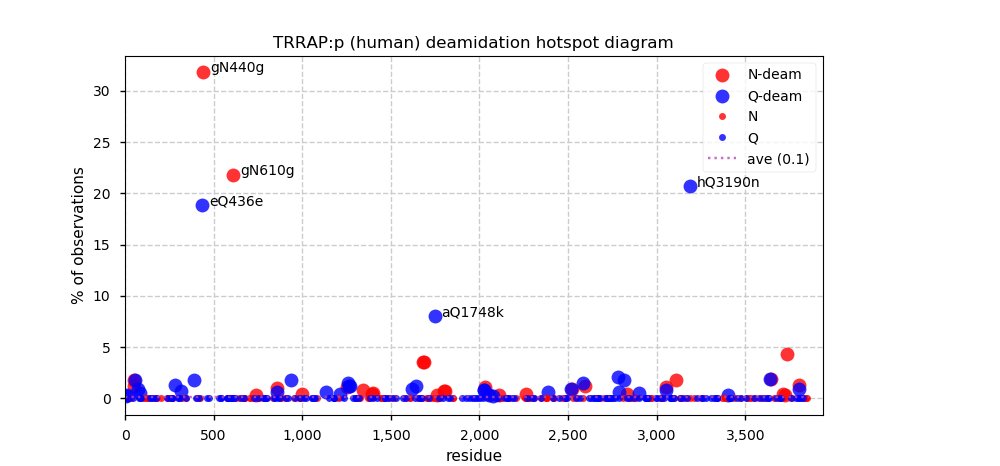
Fri Feb 14 14:32:09 +0000 2020TRRAP:p, transformation/transcription domain associated protein (H. sapiens) 🔗 Large protein; 3 distinctive phosphodomains, little association between acetyl & ubiquitinyl acceptors; no high maf SAAVs; 1 splice variant; mature forms 2-3859 [23,019 x] 🔗
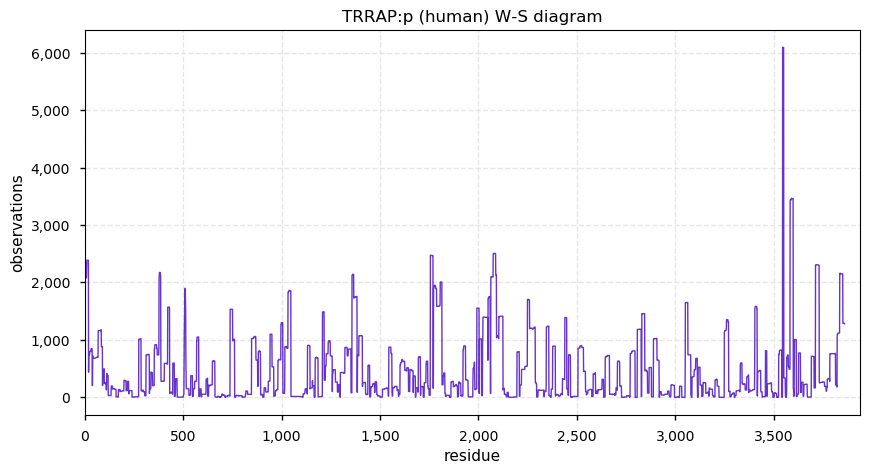
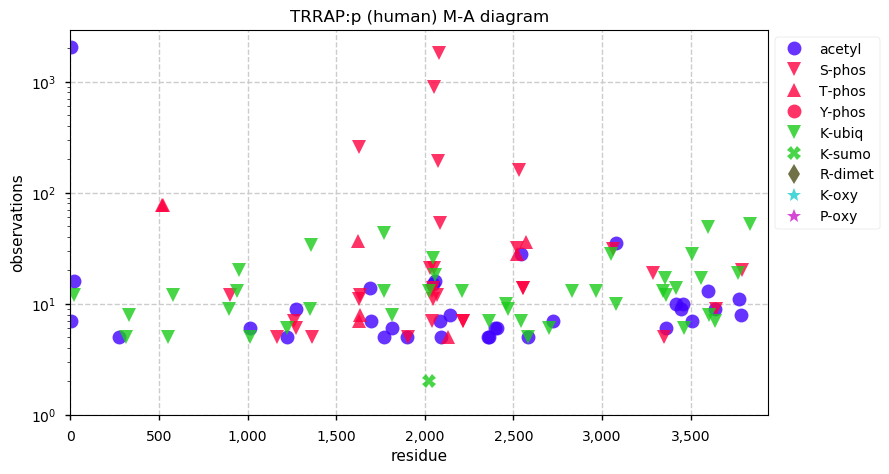
Thu Feb 13 22:58:41 +0000 2020@PastelBio 😁
Thu Feb 13 18:26:29 +0000 2020I guess Jürgen and Matthias are too busy to inspect the data themselves anymore.
Thu Feb 13 16:20:35 +0000 2020@AidanEstelle @idpgrace Bioinformaticians are H's.
Thu Feb 13 15:40:58 +0000 2020#PXD014644: swing-and-a-miss from a usually reliable group.
Thu Feb 13 15:20:53 +0000 2020Note: the blank spaces in the W-S diagram for SON:p are caused by hydrophilic low complexity domains, not hydrophobic membrane spanning domains.
Thu Feb 13 15:08:37 +0000 2020@WanunuLab @slavovLab The material used for seminar speaker introductions is almost always provided by the speaker themselves & simply read by the host representative.
Thu Feb 13 14:00:07 +0000 2020SON:p, SON DNA binding protein (H. sapiens) 🔗 Large protein; significant low complexity domains and extensive PTMs; high maf SAAVs: E1256G (0.02), M1502I (0.15), R1575C (0.72); 4 splice variants; mature forms 2-2426 [35,658 x] 🔗
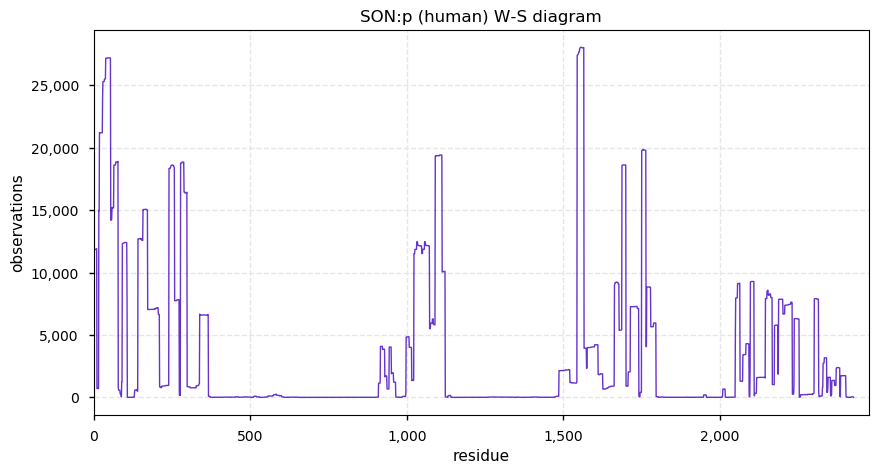
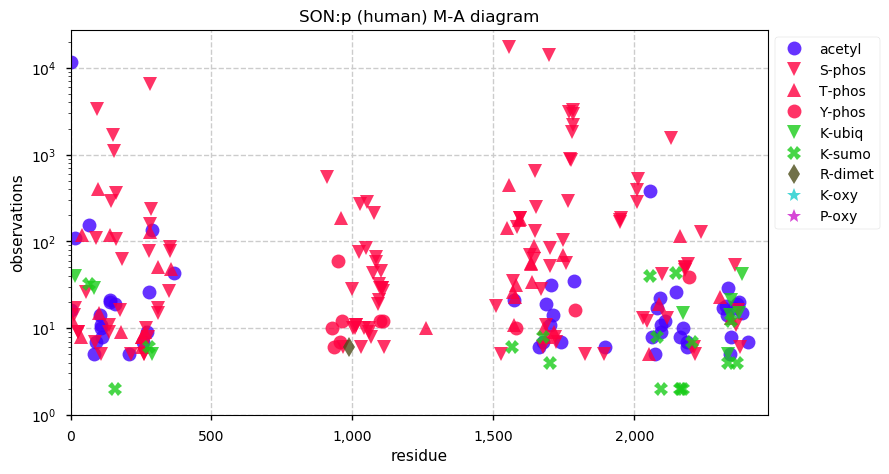
Wed Feb 12 22:56:44 +0000 2020@cstross There are a couple of pretty big North Atlantic cyclonic storms
🔗
heading your way
🔗
Wed Feb 12 18:40:18 +0000 2020@IvisonJ Do neither: simply go to Senegal and these problems will fade away.
Wed Feb 12 18:31:05 +0000 2020"Suchmaschinenoptimierung" may be my new favorite word.
Wed Feb 12 17:36:39 +0000 2020But it is how things work in the movies.
Wed Feb 12 17:36:07 +0000 2020🔗
This sentence makes no sense if you have ever been involved in biomedical research: "Speeding up research into drugs and vaccines for coronavirus is a priority for the World Health Organization's efforts to contain the outbreak centred in China."
Wed Feb 12 15:22:23 +0000 2020@Smith_Chem_Wisc No clue. I have never had any contact with the MPI group with respect to contaminant lists (or really anything at all). As far as I know, there has never been any sort of meeting or agreement as to what should be included on these lists. I just made mine up because I needed it.
Wed Feb 12 13:47:32 +0000 2020LRBA:p, LPS responsive beige-like anchor protein (H. sapiens) 🔗 Large protein; significant phosphodomains; many high maf SAAVs: e.g., A2692T (0.16), S2797L (0.14); abundant most tissues and cell lines; 1 splice variant; mature forms 2-2851 [28,499 x] 🔗
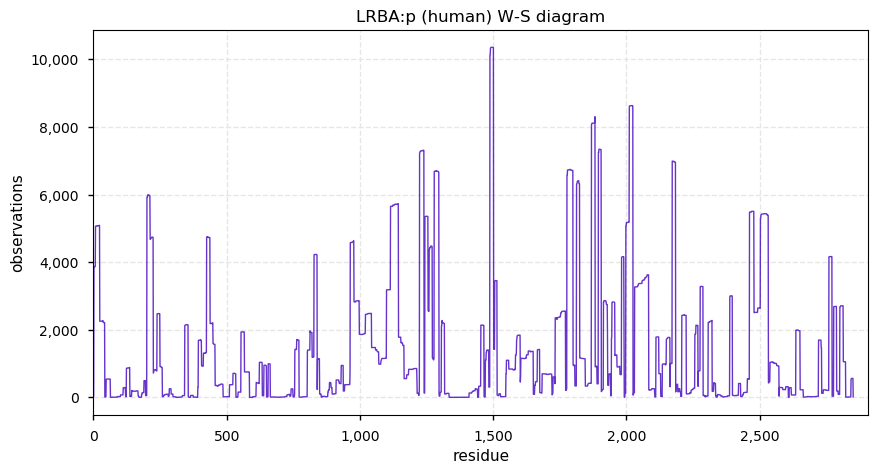
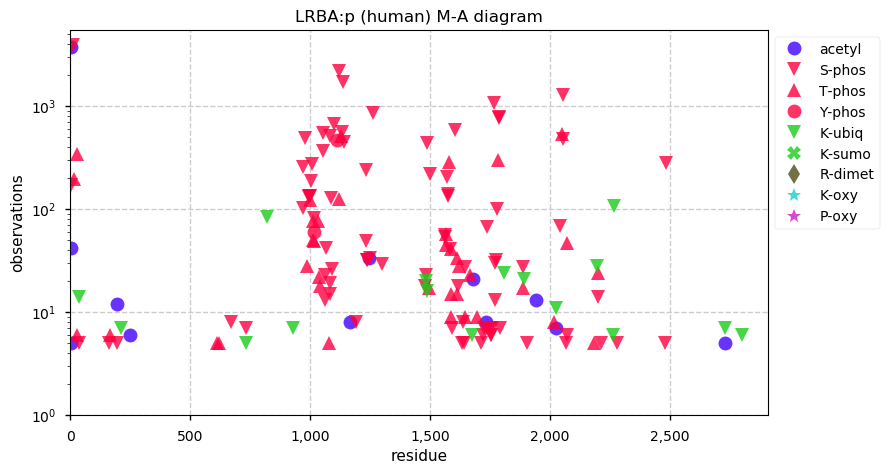
Tue Feb 11 21:20:41 +0000 2020@UCDProteomics But I should mention that many of the runs in PXD015087 with "Hela" in the file name contain consistent ids for large T antigen [Simian virus 40, gi:297591903], which is only usually found in HEK 293T cells.
Tue Feb 11 20:42:19 +0000 2020@UCDProteomics That is about what I found for the HeLa data: 🔗
Tue Feb 11 15:45:55 +0000 2020In an interesting recent article (Petyuk, et al., 🔗 ), the following statement is made:
"Most graduate programs now require statistics courses, where students learn tools like R and Python"
Is this really true? It runs contrary to my experience.
Tue Feb 11 13:46:16 +0000 2020DPEP2:p, dipeptidase 2 (H. sapiens) 🔗 Small intracellular protein; glycosylation: N176, N235, S438; no high maf SAAVs; abundant in blood plasma; 1 splice variant; mature form 33-463 [1,145 x] 🔗
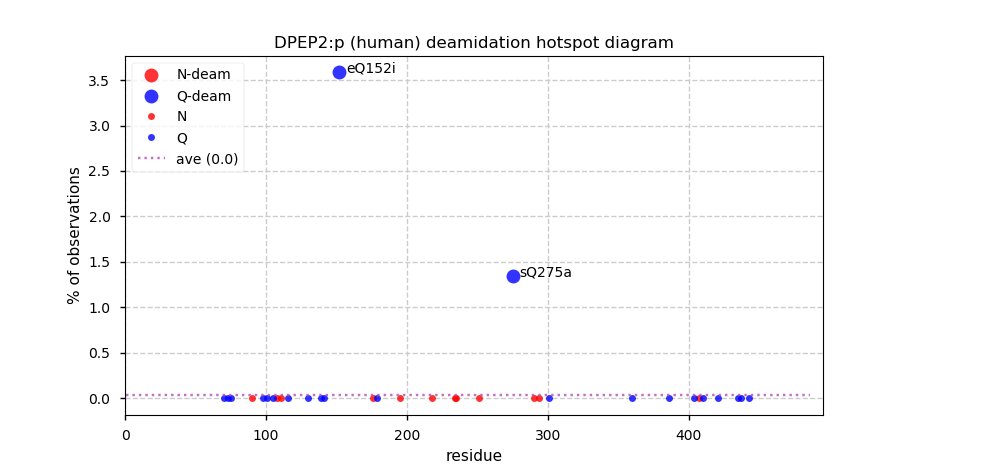
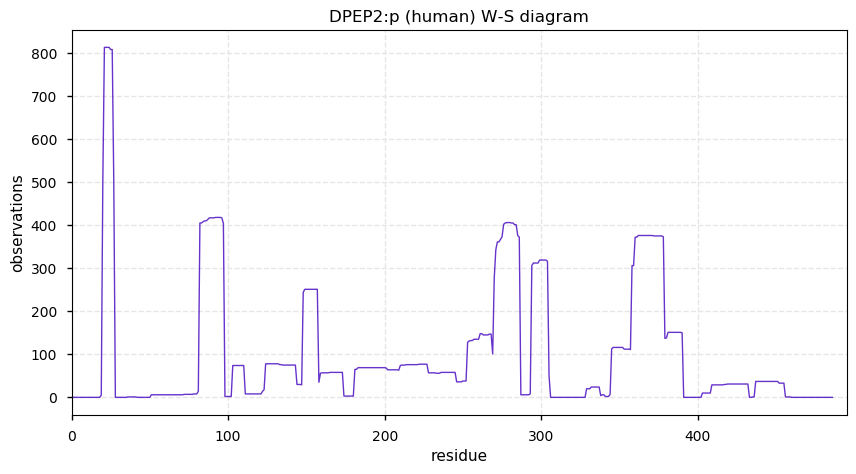
Mon Feb 10 17:49:09 +0000 2020@AJ_Brenes Less than unmodified|4-plex|8-plex|other?
Mon Feb 10 15:50:19 +0000 2020"Good programmers write good code, bad programmers write bad code, no matter the programming paradigm. However, the programming paradigm should constrain bad programmers from doing too much damage"
Mon Feb 10 15:47:37 +0000 2020🔗
Mon Feb 10 15:38:34 +0000 2020Thanks to everyone who participated in this poll. 85% of respondents (47 total) want to see human-readable source code made available when a scientific manuscript is primarily about new software. 11% were OK with supplying binary (executable) versions only.
Mon Feb 10 14:29:33 +0000 2020DPEP1:p, dipeptidase 1 (H. sapiens) 🔗 Small intracellular protein; N-glycosylation: N57, N279, N332, N358; no high maf SAAVs; abundant in urine as well as colorectal and lung cancer tissue; 1 splice variant; mature forms 17-385 [5,573 x] 🔗
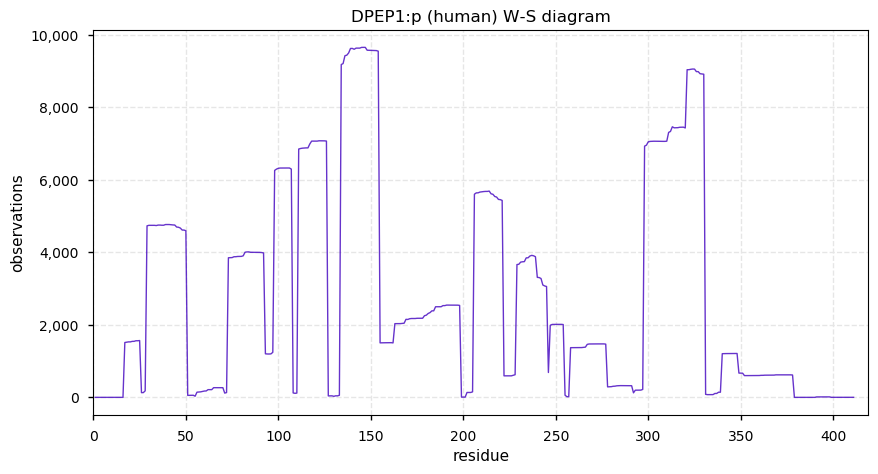
Sun Feb 09 18:53:16 +0000 2020There is still quite a bit of time left, but the results so far are not what I expected.
Sun Feb 09 13:27:53 +0000 2020CNDP2:p, carnosine dipeptidase 2 (H. sapiens) 🔗 Small intracellular protein; highly modified; SAAVs: R180Q (0.02); common in cell lines and tissues; at least 4 splice variants; mature forms 2-475 [39,299 x] 🔗
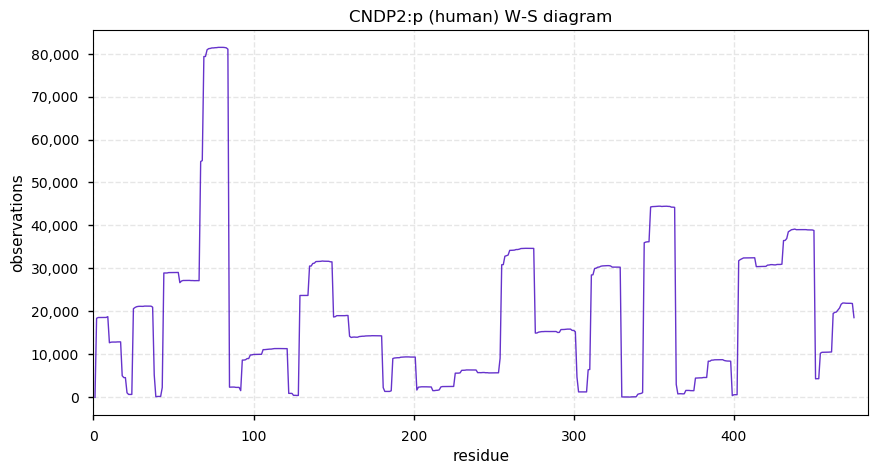
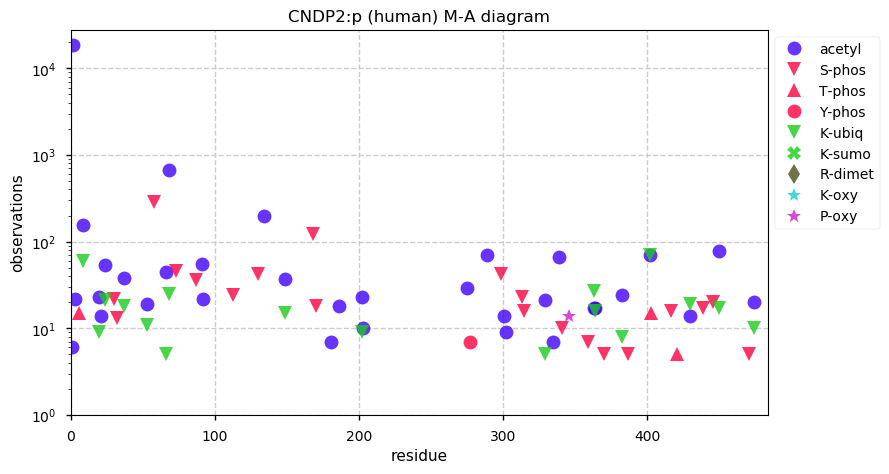
Sat Feb 08 21:26:07 +0000 2020@uwgenome Good for Jimmy. This seems to be one of the few society-type awards in that is not limited to people with academic appointments.
Sat Feb 08 21:02:33 +0000 2020@peaksdb Some one once showed me a Perl script that converted all of the variable and function names in C++ code into their MD5 hashes. It rendered the code unreadable (at least to me), even though it compiled just fine.
Sat Feb 08 21:00:20 +0000 2020@peaksdb I wouldn't be thrilled with something in assembler or reversed byte-code, as the "why" gets lost. I went with human-readable to emphasize that it should be something that is readily comprehensible by anyone familiar with the appropriate language.
Sat Feb 08 20:56:35 +0000 2020@peaksdb Depends on the language (& the reader). Would "source code" be better?
Sat Feb 08 18:43:46 +0000 2020@doctorow Anyone who has bought in to the "smart home" idea should also be paying attention to this issue. "Your" furnace, air conditioner, lighting, etc. may not be yours to sell.
Sat Feb 08 16:45:53 +0000 2020@pwilmarth But it is hard to avoid "OS-specific" because of things like path specification issues, platform specific threading standards and display widgets.
Sat Feb 08 16:41:14 +0000 2020@pwilmarth I was trying to think of a better way to put it, but many languages that don't require binaries actually have binary-like ways of distributing the executable, e.g. byte-code for Python or interpreter+code rollups for Perl. Maybe "executable version" would be more accurate.
Sat Feb 08 15:34:36 +0000 2020In a scientific manuscript primarily about a new software application, the authors should be required to provide:
Sat Feb 08 13:15:42 +0000 2020CNDP1:p, carnosine dipeptidase 1 (H. sapiens) 🔗 Midsized secreted protein; glycosylation: S27, N322, N382; not detectable in many cell lines, common in blood plasma; mature forms (27,29)-507 [5,988 x] 🔗
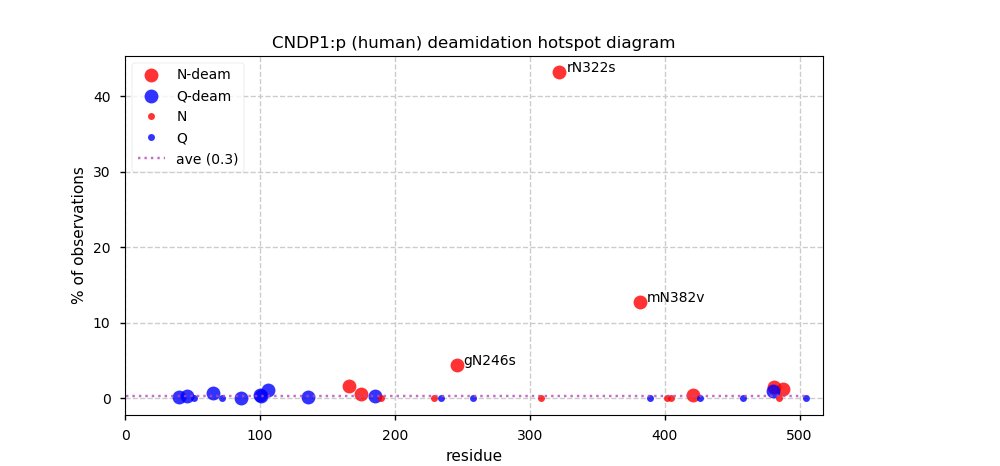
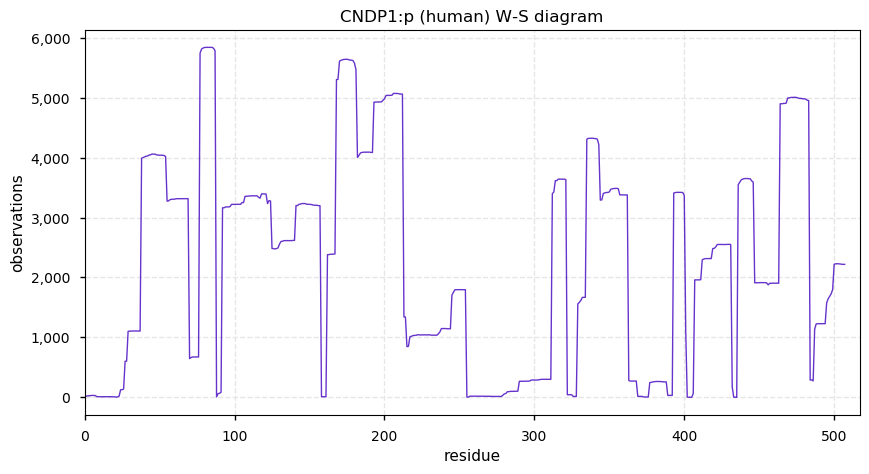
Sat Feb 08 02:17:11 +0000 2020@popgengoogling @BiswapriyaMisra This is the sentiment I normally associate with "available on request".
Fri Feb 07 20:47:19 +0000 2020@VATVSLPR @UCDProteomics I once consulted at a biotech company that had 4 identical high end instruments in a large lab area that had been purchased at the same time. Only 1 of the setups was actually used, because the other 3 weren't very reliable.
Fri Feb 07 16:23:25 +0000 2020@Smith_Chem_Wisc But as I said before: don't do this.
Fri Feb 07 16:22:44 +0000 2020@Smith_Chem_Wisc Do not try to use a complete list of proteins in a "microbiome". Proteomics is a biomass detector in this situation, so only the most abundant species will be detected. Including everything that might possibly be in there becomes a sequence homology nightmare.
Fri Feb 07 16:18:59 +0000 2020@Smith_Chem_Wisc E.g., GroEL, rpoB & rpoC. Get a list of these proteins for known pathogens and include it in the id run. For example, if you are analyzing urine, include proteins for E. coli, E. faecalis, K. pneumoniae, P. aeruginosa, P. mirabilis, G. vaginalis & a Lactobacillus
Fri Feb 07 16:13:34 +0000 2020@Smith_Chem_Wisc For prokaryotes, they are really only an issue in clinical samples (mycoplasma is almost always caught in cell culture). There are a few proteins that are very abundant in most species and are always id'd if the species is present.
Fri Feb 07 16:08:44 +0000 2020@Smith_Chem_Wisc Parainfluenza virus 5
Human adenovirus A - F
Human coronavirus 229E & HKU1
Human herpesvirus 1 - 7
Human immunodeficiency virus 1 & 2
Influenza A - C virus
Simian adenovirus 1
Simian virus 40
Xenotropic murine leukemia virus-related virus
Fri Feb 07 16:08:22 +0000 2020@Smith_Chem_Wisc If you are just passing through:
1. Viruses have small proteomes: adding 50 common viruses to an id run doesn't slow things down much. The following list is a good start:
Fri Feb 07 15:55:39 +0000 2020@Smith_Chem_Wisc I am truly not kidding about this. If you want to be in the field for any period of time, forget this problem exists.
Fri Feb 07 15:20:38 +0000 2020@Smith_Chem_Wisc Yes, but I would strongly suggest you drop the idea: it is information no one wants to know. This is particularly true of your medical collaborators.
Fri Feb 07 15:14:08 +0000 2020@VATVSLPR Only so much room in a tweet! There are at least 4 splices that can be differentiated by MS/MS and some peptide overlaps with the CEACAM5:p, CEACAM6:p & CEACAM8:p.
Fri Feb 07 14:08:07 +0000 2020Also a coronavirus receptor protein
Fri Feb 07 14:07:39 +0000 2020CEACAM1:p, carcinoembryonic antigen related cell adhesion molecule 1 (H. sapiens) 🔗 Midsized membrane protein; N-linked glycosylation: N152, N182, N197, N208, N224, N345, N351, N363, N378, N475; absent in common cell lines; mature form 35-526 [6,553 x] 🔗
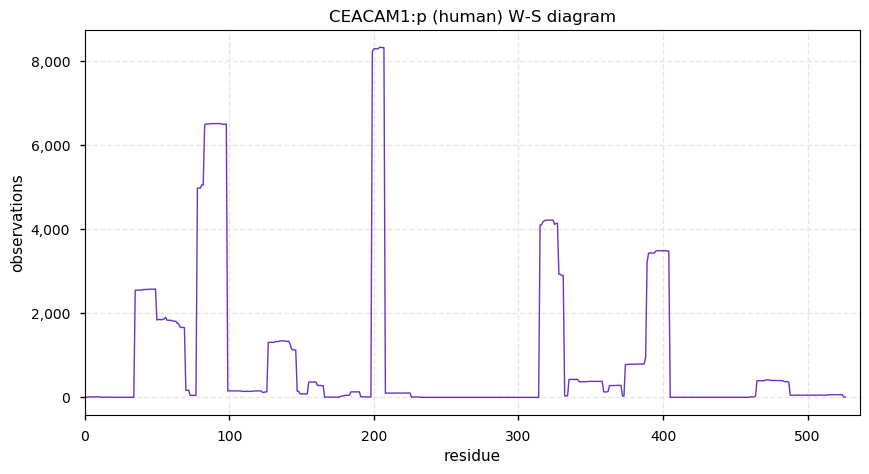
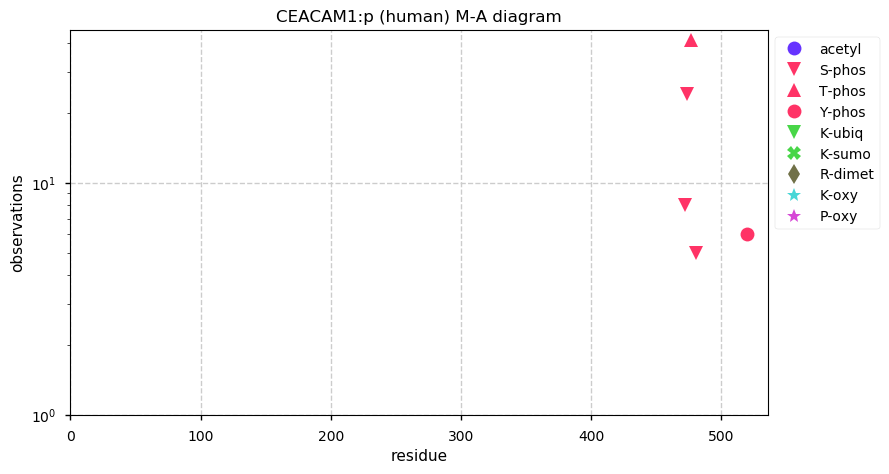
Thu Feb 06 18:40:56 +0000 2020The Iowa caucus debacle is a good example of what happens when you try to QC your way out of a QA failure.
Thu Feb 06 15:05:43 +0000 2020@PastelBio @olgavitek Great quote that UI developers everywhere should have on a prominently-placed Post-It note: "To our surprise, none of the test users were able to successfully complete task 1."
Thu Feb 06 14:05:48 +0000 2020Both ACE2 & ANPEP serve as receptors for human coronaviruses.
Thu Feb 06 13:59:23 +0000 2020ANPEP:p, alanyl aminopeptidase, membrane (H. sapiens) 🔗 Midsized membrane protein; N-linked glycosylation: N234, N265, N319, N527, N573, N681, N735, N818; 4 SAAVs: R86Q (0.6), A311V (0.1), I603M (0.2), S752N (0.7); mature forms 2-967 [7,762x] 🔗
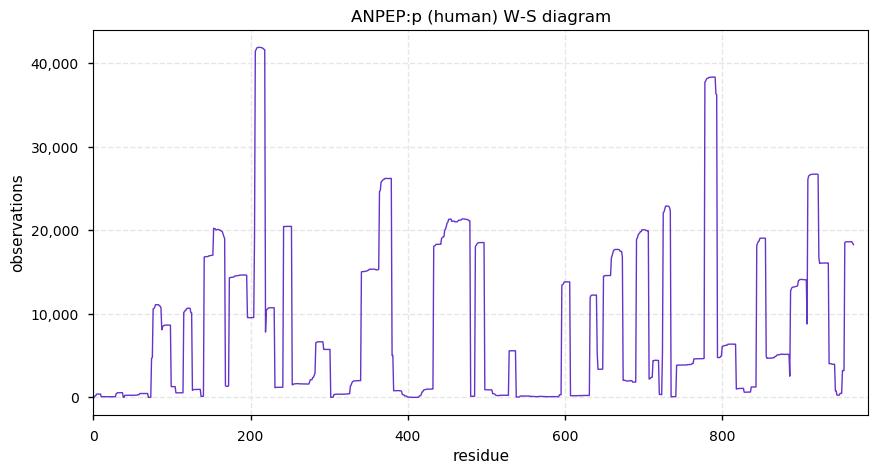
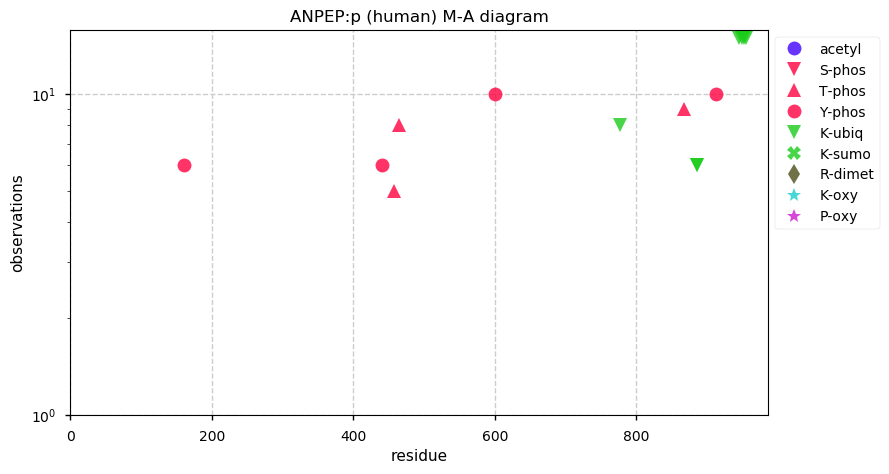
Wed Feb 05 16:45:04 +0000 2020@jwoodgett This just means they already have the grantees selected and they want to release the money ASAP.
Wed Feb 05 16:43:25 +0000 2020The most prominent N-linked glycosylation sites are N90, N103 and N546.
Wed Feb 05 15:01:18 +0000 2020@ypriverol @chrashwood Something like 🔗 has the idea: just think of "transaction" as representing any type of algorithmic data transformation.
Wed Feb 05 14:53:49 +0000 2020@ypriverol @chrashwood They are another thing I use all of the time to keep things straight.
Wed Feb 05 14:52:58 +0000 2020@ypriverol @chrashwood Merkle tree representations of sequential tasks can also be very useful. I know they have a bit of a stigma because of their use in "blockchain" applications, but their builtin verification makes them ideal for reproducibility applications.
Wed Feb 05 14:13:20 +0000 2020@ypriverol @chrashwood If you are thinking about reproducibility in this regard, you might want to include SHA256 (or equivalent) hashes for all of the files involved (spectra, sequence and output). I use them all the time & it simplifies many comparison tasks.
Wed Feb 05 13:19:20 +0000 2020ACE2:p, angiotensin I converting enzyme 2 (H. sapiens) 🔗 Midsized extracellular protein; significant N-linked glycosylation; no high maf SAAVs; 1 splice variant; mature forms 18-805, 24-805, 115-805 (urine) [5,129x] 🔗
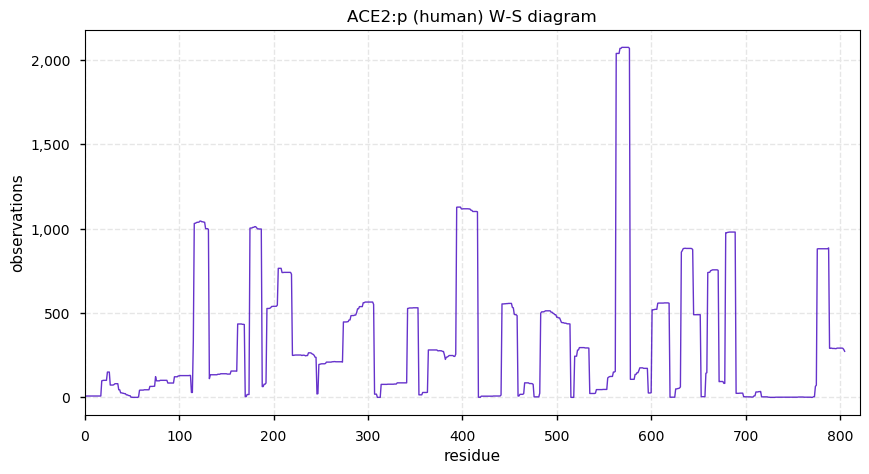
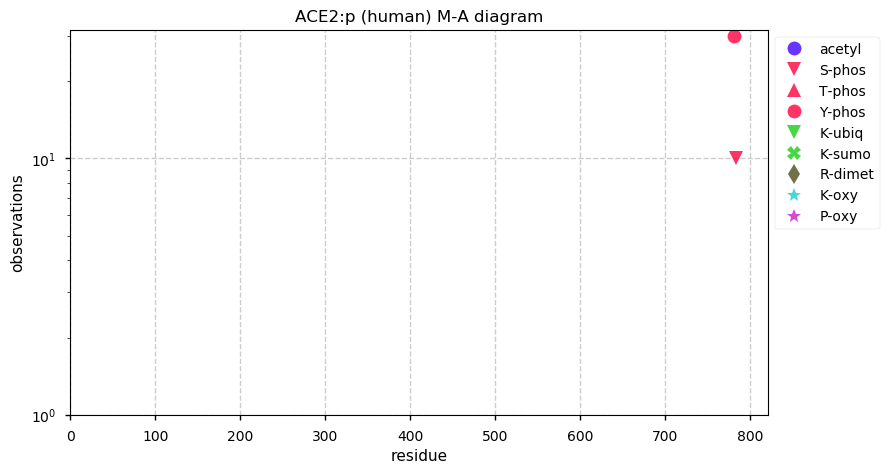
Tue Feb 04 21:17:23 +0000 2020@neely615 @Smith_Chem_Wisc It is almost as though they were encouraging you to use the paid version ...
Tue Feb 04 19:01:12 +0000 2020@chrashwood For an instrument that can do N expts simultaneously: yes. Although I wasn't thinking about models: I was thinking more of a functional spec, e.g. high res parent & fragment vs. high res parent & low res fragment. Those 2 options cover most of the data published.
Tue Feb 04 15:06:23 +0000 2020For modern instruments, only 4 parameters are required:
1. path to a spectrum file;
2. path to peptide sequence info file;
3. path to an output file; &
4. type of instrument used.
Tue Feb 04 15:01:21 +0000 2020@VATVSLPR Twitter doesn't allow you to vote in a poll you posted.
Tue Feb 04 14:13:07 +0000 2020PALM2:p, paralemmin 2 (H. sapiens) 🔗 Small plasma membrane protein; significant phosphorylation; no high maf SAAVs; 1 splice variant; mature form 1-411 [5,129x] 🔗
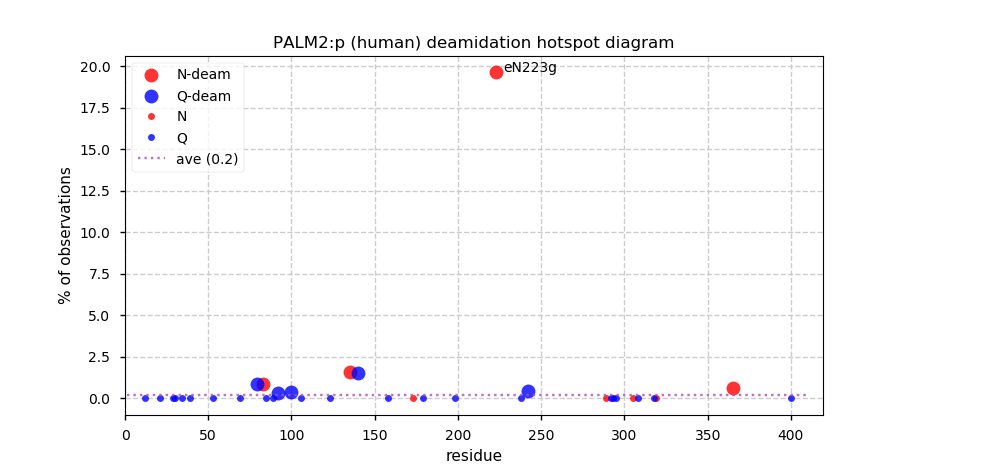
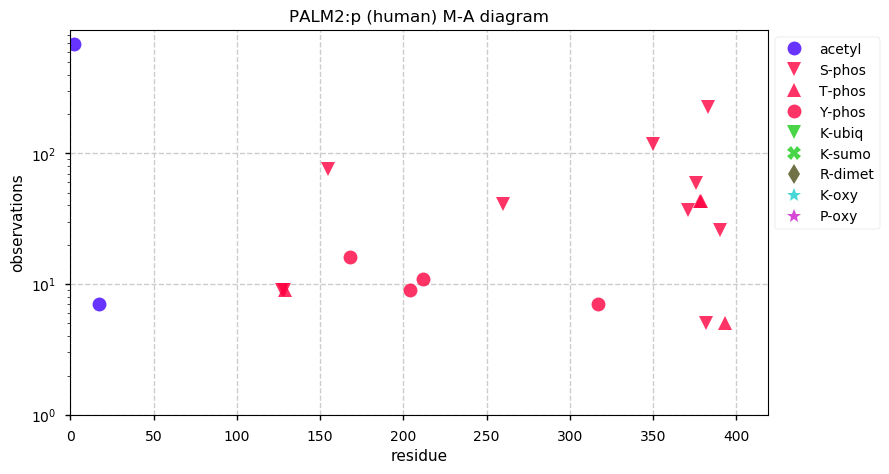
Mon Feb 03 23:28:59 +0000 2020Thanks to everyone who participated in this poll. It appears that the majority would prefer a simpler API than is commonly available on current PSM assignment software.
Mon Feb 03 20:37:32 +0000 2020@kadzuis @ChemRxiv I was two rooms over while Swapan and Vish spent years trying to get rid of ESI in-source fragmentation, so it should be easy to bring back ...
Mon Feb 03 19:14:51 +0000 2020Comparing a Threadripper vs RPIB4 for PSM assignment (I promise I will stop reporting this very esoteric stuff):
227 MGF files from PXD016632;
all times include loading spectra, PSM, FDR & p-value assignment + writing TSV final output and JSON metadata files. 🔗
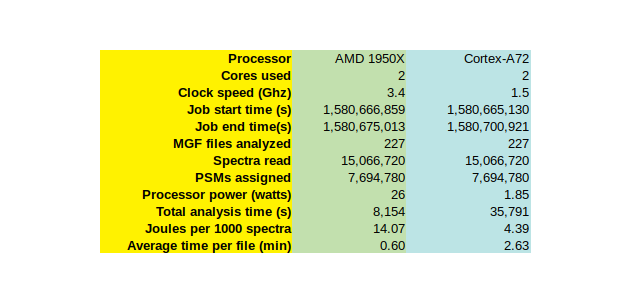
Mon Feb 03 15:15:30 +0000 2020If I got a vote, I would be firmly in the "< 5" camp.
Mon Feb 03 13:29:41 +0000 2020PALM:p, paralemmin (H. sapiens) 🔗 Small plasma membrane protein; significant phosphorylation; 1 high maf SAAV: T107A (0.45); 1 splice variant; mature form 1-387 [6,839 x] 🔗
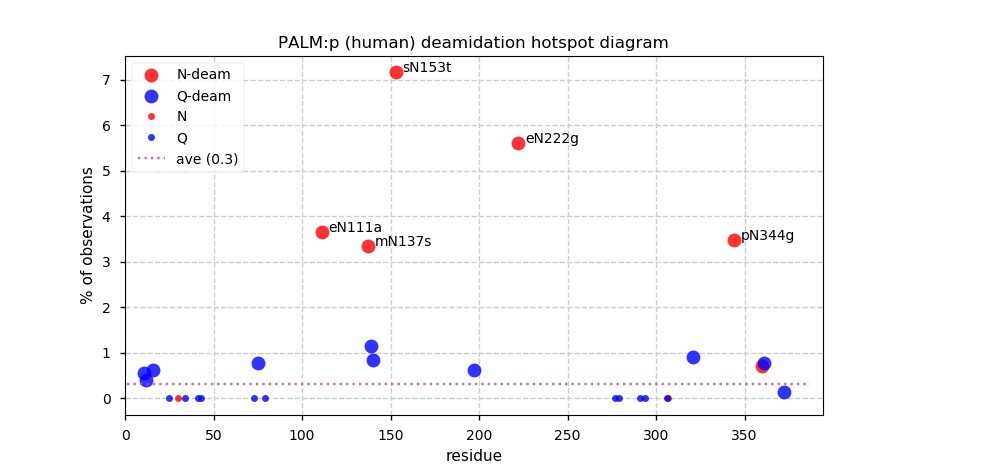
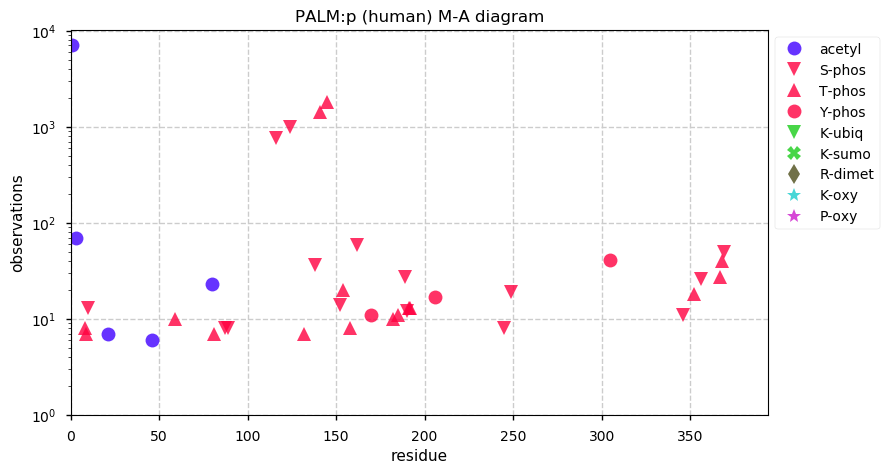
Sun Feb 02 23:07:53 +0000 2020How many user supplied parameters should a PSM assignment process require?
Sun Feb 02 14:37:58 +0000 2020CDC5L:p, cell division cycle 5 like (H. sapiens) 🔗 Midsized intracellular protein; highly modified in multiple domains; no high maf SAAV; 1 splice variant; mature form 1-802 [35,115 x] 🔗
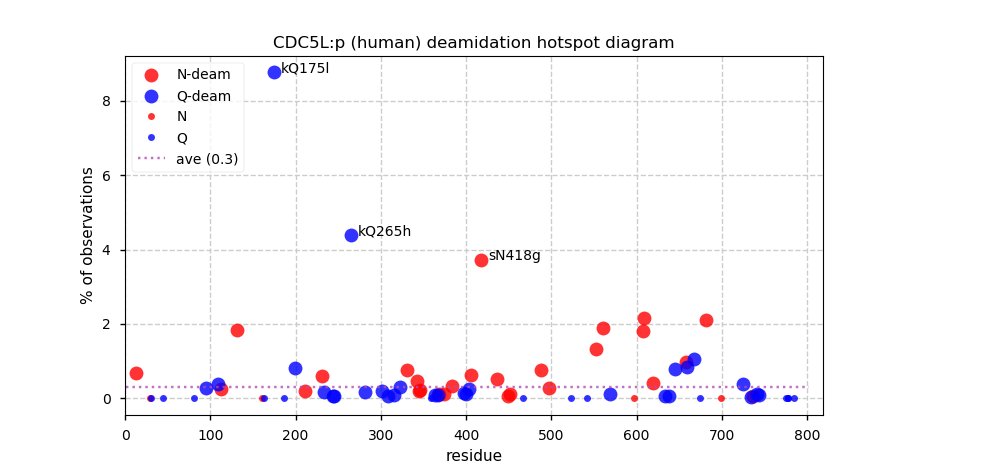
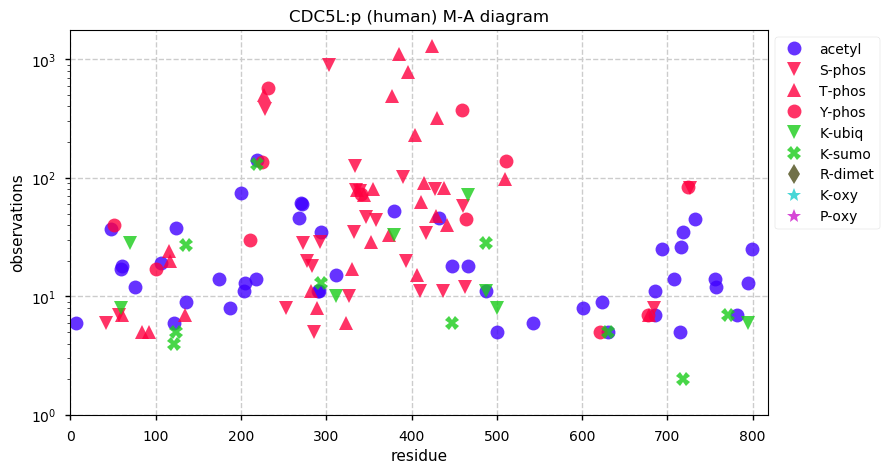
Sat Feb 01 16:49:05 +0000 2020@AlexUsherHESA Maybe signalling that -7 C is pretty warm for winterpeg in January?
Sat Feb 01 15:55:40 +0000 2020I have been pleasantly surprised by how smoothly an RPI4B functions when all 4 cores are showing nearly 100% use via htop.
Sat Feb 01 13:28:36 +0000 2020CCAR2:p, cell division cycle and apoptosis regulator 2 (H. sapiens) 🔗 Midsized intracellular protein; highly modified; no high maf SAAV; 7 splice variants; mature form 2-923 [42818 x] 🔗
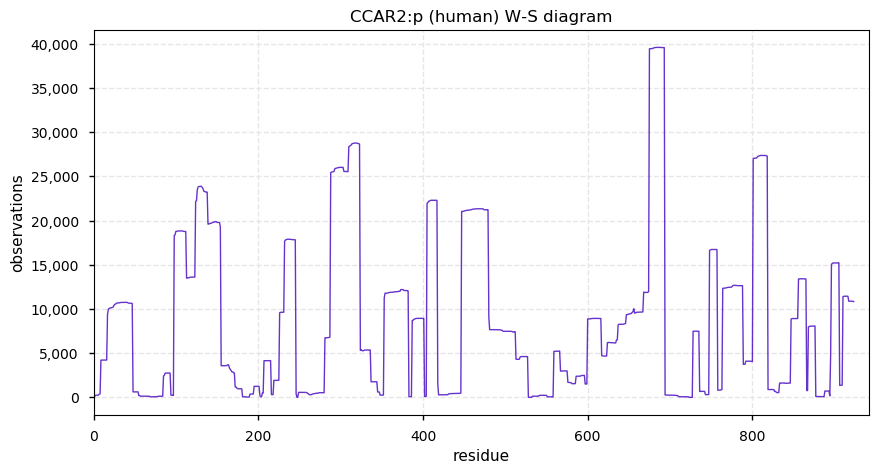
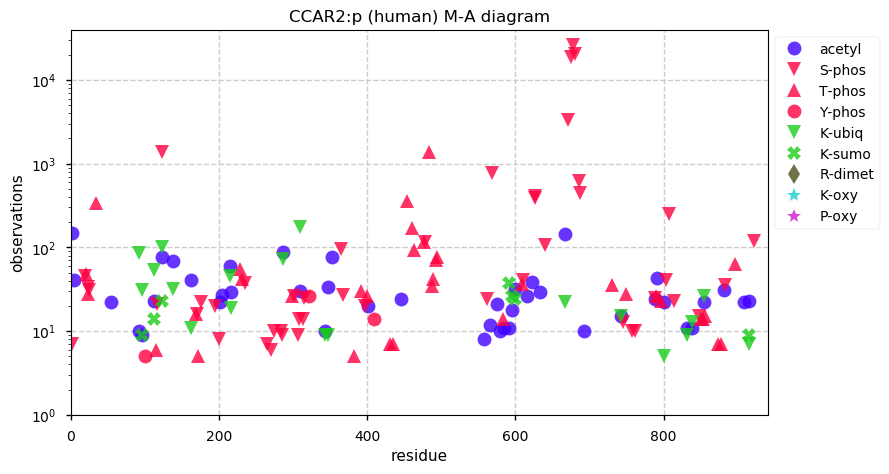
Fri Jan 31 16:10:18 +0000 2020Latest RPI4B run stats:
874 MGF files (PXD015943), phosphopeptide enriched
19,286,342 spectra
6,499,449 PSMs (>90% phosphopeptides)
66 sec/MGF
58,406 sec total time
11.2 Joules per 1000 spectra
incl. loading spectra, PSM, FDR & p-value assignment + writing TSV final output.
Fri Jan 31 15:54:23 +0000 2020@jwoodgett An editor adding themselves as a co-author was an innovation that I have never encountered: in my experience it has always been more of a Department Head/Chair move.
Fri Jan 31 15:48:38 +0000 2020@theoneamit More like garbage collectors.
Fri Jan 31 14:11:40 +0000 2020CCAR1:p, cell division cycle and apoptosis regulator 1 (H. sapiens) 🔗 Large intracellular protein; multiple periodic phosphodomains & C-terminal SUMOylated domain; no high maf SAAV; 1 splice variant; mature form 2-1150 [24,458 x] 🔗
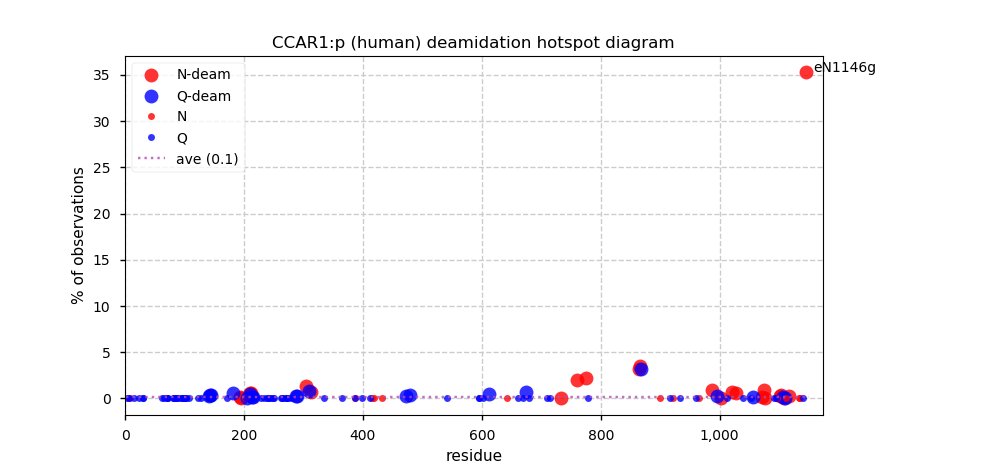
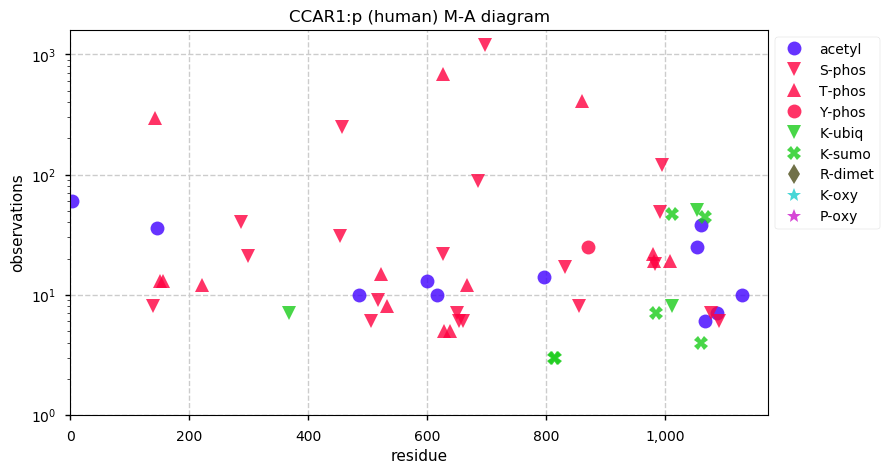
Fri Jan 31 13:47:21 +0000 2020@jwoodgett I do like "reviewer-coerced citation" as a term.
Thu Jan 30 19:11:32 +0000 2020@mjmaccoss @karthikskamath @byu_sam @Sci_j_my @dnusinow It also doesn't help that the relationship between the 2 is a differential equation, the simplest version of which is:
[rna] ∝ ∂[protein]/∂t
Thu Jan 30 17:48:23 +0000 2020@MHendr1cks @hwitteman A general misunderstanding of how CRCs work doesn't help and can create additional barriers (& resentments) to these lateral moves.
Thu Jan 30 17:01:31 +0000 2020@MHendr1cks @hwitteman Even lateral moves in Canada are unnecessarily difficult. Therefore, a program that tries to facilitate such moves without removing any of the barriers just doesn't seem to work very well.
Thu Jan 30 16:59:42 +0000 2020@MHendr1cks @hwitteman I think we have different reasons, but they result in the same conclusion. My problem with the general idea is that it is nearly impossible to make a lateral move from another system (e.g., German or US academia) into an equivalent position in Canada.
Thu Jan 30 16:11:49 +0000 2020So far, I am really liking #PXD015943 (🔗). It may be the best quality data set ever associated with a Nat. Biotech. publication.
Thu Jan 30 16:08:34 +0000 2020@hwitteman @MHendr1cks I'm afraid the entire CRC system/idea has never lived up to its initial goals and needs to be shutdown with extreme prejudice. It just doesn't fit in with the way Canadian academia works.
Thu Jan 30 15:36:25 +0000 2020@jwoodgett @upshur_ross It does follow the general pattern of reporting that neglects to mention HCoV-229E and HCoV-OC43 when discussing other Coronaviruses, focusing on SARS-CoV and MERS-CoV instead.
Thu Jan 30 14:27:26 +0000 2020FXYD7:p is only detected in CNS tissue and the SK-MEL24 cell line.
Thu Jan 30 14:25:00 +0000 2020FXYD7:p, FXYD domain containing ion transport regulator 7 (H. sapiens) 🔗 Very small intracellular protein; C-terminal phosphodomain; no high maf SAAV; 1 splice variant; mature form 2-80 [716 x] 🔗
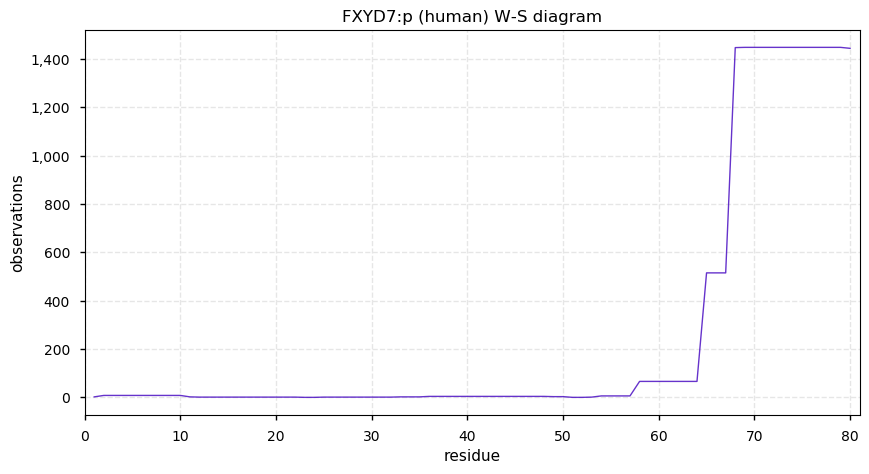
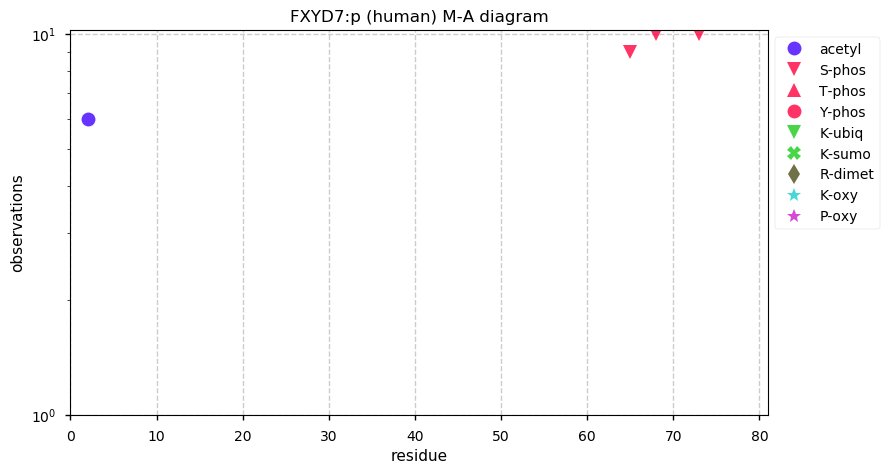
Wed Jan 29 17:13:13 +0000 2020Currently, QA is far more important in 'omics data production than QC.
Wed Jan 29 16:48:08 +0000 2020@juan_vizcaino @epic_xs Is the UK's departure from the EU going to affect EPIC-XS in any way?
Wed Jan 29 13:40:50 +0000 2020FXYD6:p is most frequently detected in CNS and reproductive tissue and several studies that used HEK-293T cells.
Wed Jan 29 13:30:34 +0000 2020FXYD6:p, FXYD domain containing ion transport regulator 6 (H. sapiens) 🔗 Very small secreted protein; no PTMs; no high maf SAAV; 1 splice variant; mature form 19-95 [1,465 x] 🔗
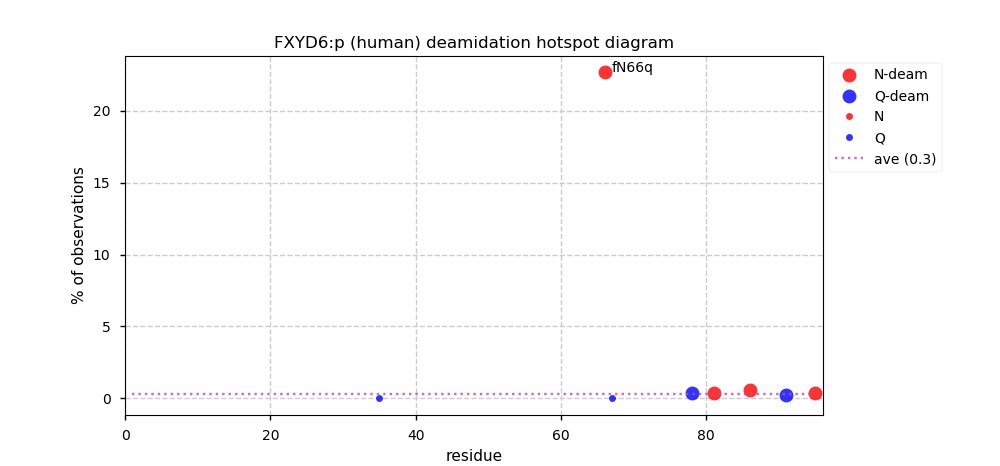
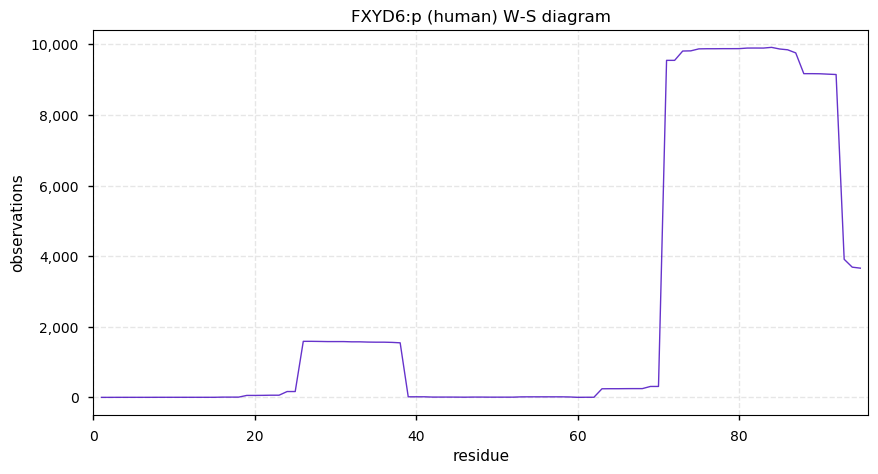
Tue Jan 28 22:07:18 +0000 2020@salemhorrorfest Guillermo del Toro remakes Steamboat Willie
Tue Jan 28 21:26:59 +0000 2020@slashdot !!!!
Tue Jan 28 18:14:29 +0000 2020@andy___jones Please note that I am not discounting the value of the paper that kicked off the discussion. If it causes people to give their heads a shake and realize that simply running a localization algorithm doesn't solve the problems created by insufficient data, I applaud their efforts.
Tue Jan 28 17:32:08 +0000 2020After thinking about this for a while and looking at more data, it seems possible that poly-proline peptides formed during normal protein turnover may not be properly disassembled by either proteasomes/lysosomes. Instead, they may be secreted into blood & end up in urine.
Tue Jan 28 15:48:12 +0000 2020@andy___jones And I have never met anyone on the biology side that really trusts these one-off site-specific localizations anyway: they are going to run an alanine scan if they are at all interested in a particular site. /fin
Tue Jan 28 15:46:31 +0000 2020@andy___jones and 10% where it is ambiguous, a "false localization rate" of 10% gives a misleading impression about the accuracy of most of the PTM assignments. /4
Tue Jan 28 15:46:11 +0000 2020@andy___jones Lumping these cases together and giving an overall "rate" is misleading, because each localization is a specific problem, unrelated to each other. For example, if you have 90% ideal PTM localizations (only 1 site on the peptide that could have the PTM) /3
Tue Jan 28 15:45:36 +0000 2020@andy___jones If you do have enough data, then it is settled. If you don't then no amount of stats or other calculations is going to help: the best you can do is check previous observations to see if they provide any clarity. /2
Tue Jan 28 15:45:27 +0000 2020@andy___jones Not at all. By providing a misleading overall stat, it simply confuses a bunch of specific issues. The "localization" of PTMs is a practical problem: either you have enough data to choose between alternatives or you don't. /1
Tue Jan 28 14:27:39 +0000 2020@andy___jones "False localization rate" is a new one for me. Sounds like a bit of a stretch/not-great-analogue ...
Tue Jan 28 14:05:12 +0000 2020Similar to FXYD1 in observed PTMs and peptide patterns, but they are observed in different tissues & cell lines.
Tue Jan 28 14:00:23 +0000 2020FXYD3:p, FXYD domain containing ion transport regulator 3 (H. sapiens) 🔗 Very small secreted protein; C-terminal phosphodomain; no high maf SAAV; 1 splice variant; mature form 21-113 [2,088 x] 🔗
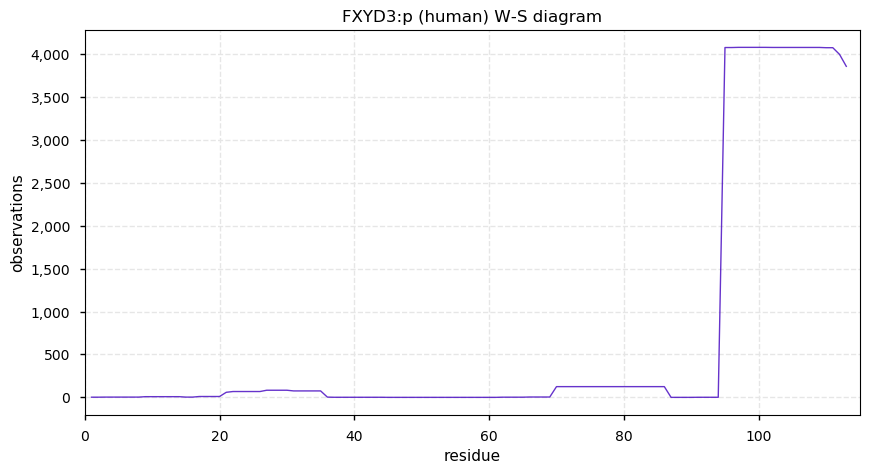
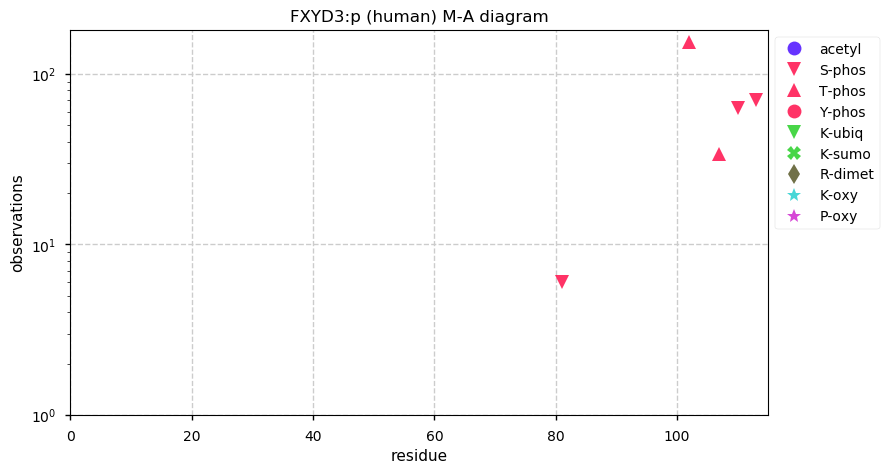
Mon Jan 27 21:07:59 +0000 2020Something new: a dataset where FFPE tissue is prep'd and the resulting tryptic peptide N-termini show significant formaldehyde (+COH2) reaction products, but nothing on the lysine side chains. Just when you think you've seen it all ...
Mon Jan 27 13:29:22 +0000 2020FXYD2:p, FXYD domain containing ion transport regulator 2 (H. sapiens) 🔗 Very small intracellular protein; no significant PTMs; no high maf SAAV; 1 splice variant; most commonly observed in urine; mature form (1,2,3)-66 [1,221 x] 🔗
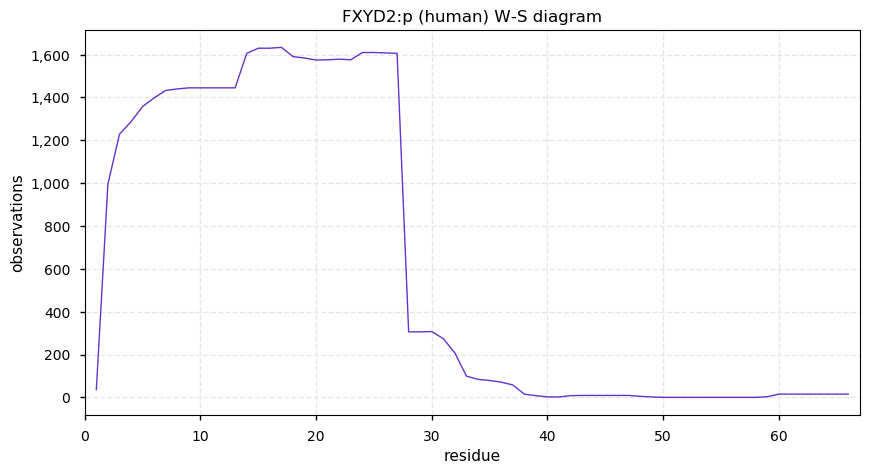
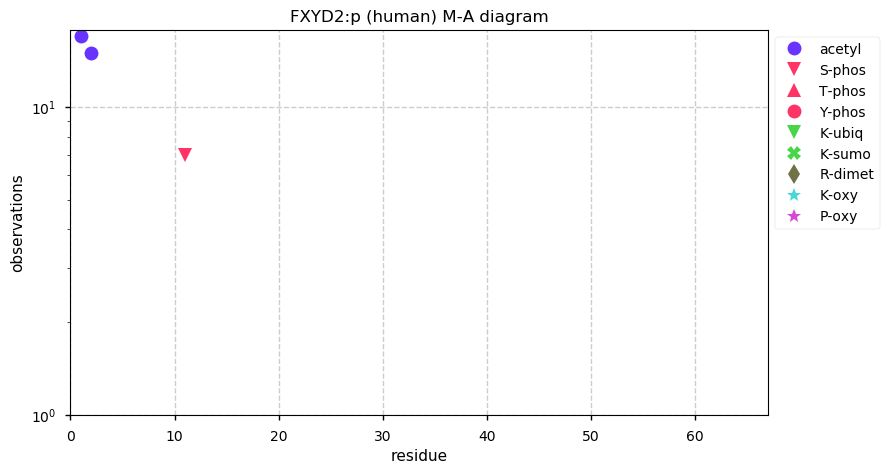
Sun Jan 26 17:46:40 +0000 2020Any epiphany on the subject would be welcome!
Sun Jan 26 17:30:10 +0000 2020"PPPPPPPPPPPPPPP" (& shorter versions of the same thing) also shows up a lot in urine peptidomics, but I doubt if any of the proteins that contain this sequence are really the source.
Sun Jan 26 16:17:47 +0000 2020@girlziplocked Contact @RobertPicardo directly. He seems like he would be aware of any good ideas on this subject.
Sun Jan 26 15:48:19 +0000 2020I guess it is the variability that causes surprise: in many samples it is undetectable, while in others it is #2 on the list, just behind COL1A1 peptides.
Sun Jan 26 15:45:03 +0000 2020I find I am still occasionally surprised by how prominent insulin C-peptide can be in urine samples that are subjected to peptidome-type analysis.
Sun Jan 26 15:20:55 +0000 2020FXYD1 is often only detected by a single tryptic peptide.
Sun Jan 26 15:19:44 +0000 2020FXYD1:p, FXYD domain containing ion transport regulator 1 (H. sapiens) 🔗 Very small extracellular protein; N-terminal phosphodomain; no high maf SAAV; 1 splice variant; mature form 21-92 [1,691 x] 🔗
Sun Jan 26 14:46:27 +0000 2020Will somebody give Toronto a lollipop and tell them to calm down?
🔗
Sat Jan 25 15:31:01 +0000 2020KPNA3:p, karyopherin subunit alpha 3 (H. sapiens) 🔗 Midsized nuclear membrane subunit; 56-82 high occupancy phosphodomain; 1 high maf SAAV: K121R (0.01); 1 splice variant; mature form 2-521 [27,774 x] 🔗
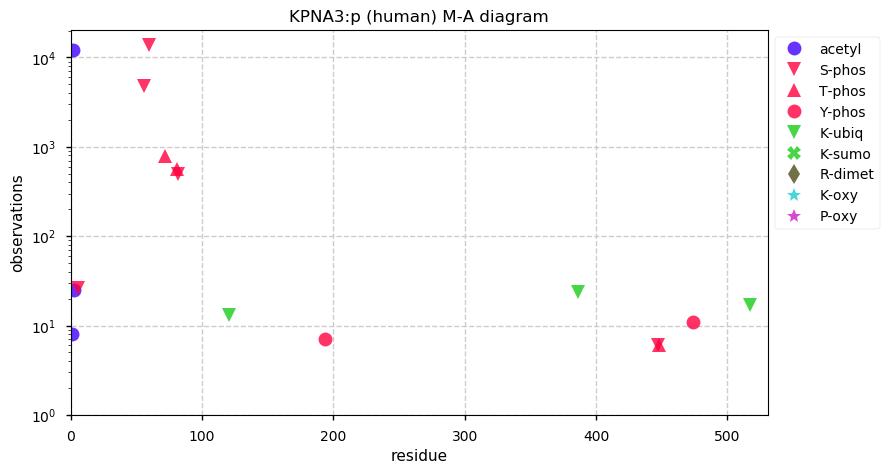
Fri Jan 24 21:25:36 +0000 2020@RuneLinding Of the several unintentionally funny quotes in this article, my favorite has to be:
"There is no software that is bugless"
Fri Jan 24 21:00:12 +0000 2020@ProteomicsNews @lstops @chrashwood I checked out the rat phosphorylation portion of #PXD014750 & the reagent clearly has worked. There is nearly complete coupling of the TMTpro to free peptide N-terminii and lysines & only a small amount of the succinylation side reaction in the data.
Fri Jan 24 20:16:59 +0000 2020This is a rather good article on how and why explosions cause traumatic brain injury 🔗
Fri Jan 24 15:44:17 +0000 2020FYI, reanalysis of #PXD014000 shows that the protocol for recovering unblocked cysteine-containing peptides in mammalian samples — J. R.Wiśniewski, et al., 🔗 — works as well as most protocols that use IAA/CAA blocking.
Fri Jan 24 13:34:37 +0000 2020KPNA2:p, karyopherin subunit alpha 2 (H. sapiens) 🔗 Midsized nuclear membrane subunit; extensive, low occupancy phosphorylation; 3 high maf SAAV: R117W (0.01), V506D (0.01), V506I (0.01); 1 splice variant; mature form 2-529 [40,637 x] 🔗
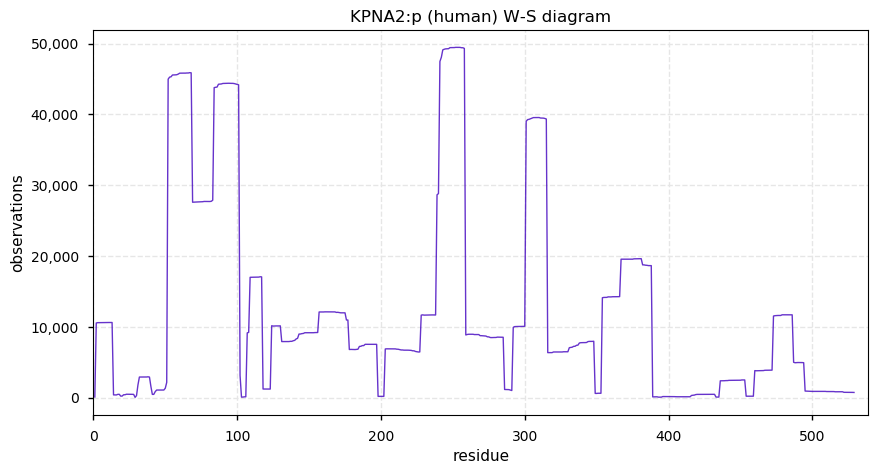
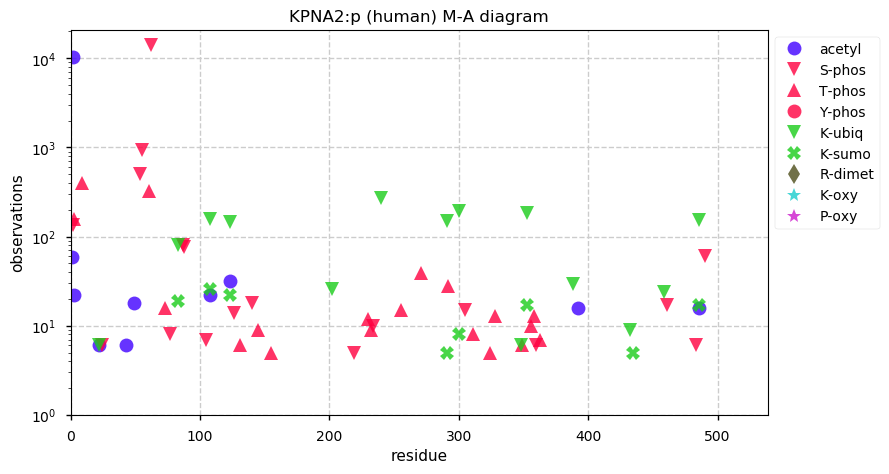
Fri Jan 24 00:58:30 +0000 2020@bkives Having lived in Chicago, New York and Paris, size and density are all that matters. Canada doesn't really have a big city.
Thu Jan 23 22:42:31 +0000 2020@ProteomicsNews @lstops @chrashwood Got it. Thanks. I will download the files and take a look. It may not have a direct bearing on the problem described, but it will be somewhere to start!
Thu Jan 23 21:34:31 +0000 2020How complete does a tryptic digest have to be so that the resulting data can be considered reproducible? And what metrics do you use to determine digestion completeness?
Thu Jan 23 19:16:03 +0000 2020@UCDProteomics Our friends on the nucleic acid side would rather be eaten by bears than admit that such a thing could be possible ...
Thu Jan 23 17:56:22 +0000 2020@nesvilab It uses port 33001 UDP by default, which many Uni IT departments don't allow on their networks.
Thu Jan 23 17:27:42 +0000 2020@jwoodgett Alternately, if viewed as a fraction of the country's total population, the equivalent of locking down Saskatoon.
Thu Jan 23 17:18:55 +0000 2020@jwoodgett Or Ontario & Quebec sealing their borders.
Thu Jan 23 17:12:57 +0000 2020@EricWDeutsch Client. I've been using it from time to time since CPTAC switched to using it exclusively. The most recent version (which it insisted that I install) is the first I've seen with this particular wrinkle.
Thu Jan 23 17:10:12 +0000 2020@lstops @chrashwood I haven't seen any in publicly available data yet. Is there any way I could get one of the problem .RAW files?
Thu Jan 23 16:46:46 +0000 2020Or, more importantly, does anybody really regularly use Aspera anymore?
Thu Jan 23 16:41:15 +0000 2020Has anybody else noticed that the latest version of Aspera seems to block all other IP access while it is running?
Thu Jan 23 15:20:29 +0000 2020KPNA1:p, karyopherin subunit alpha 1 (H. sapiens) 🔗 Midsized nuclear membrane subunit; several isolated phosphorylation sites; 1 high maf SAAV: S73N (0.99); 1 splice variant; mature form (1,2)-538 [26,830 x] 🔗
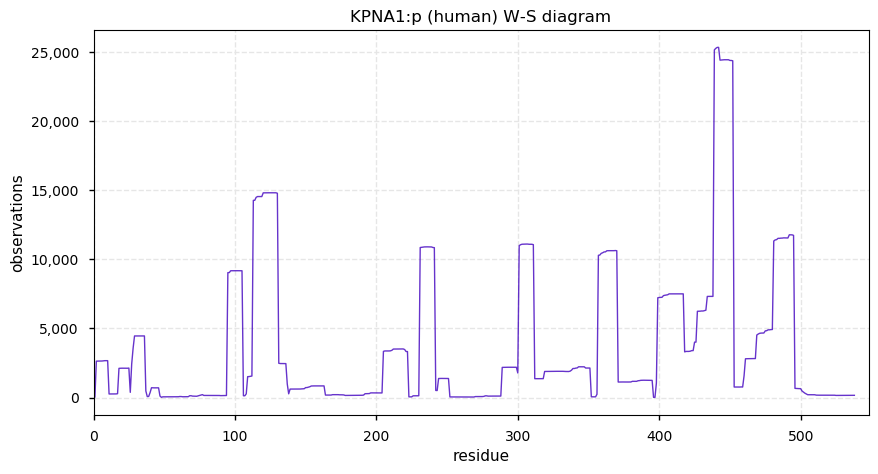
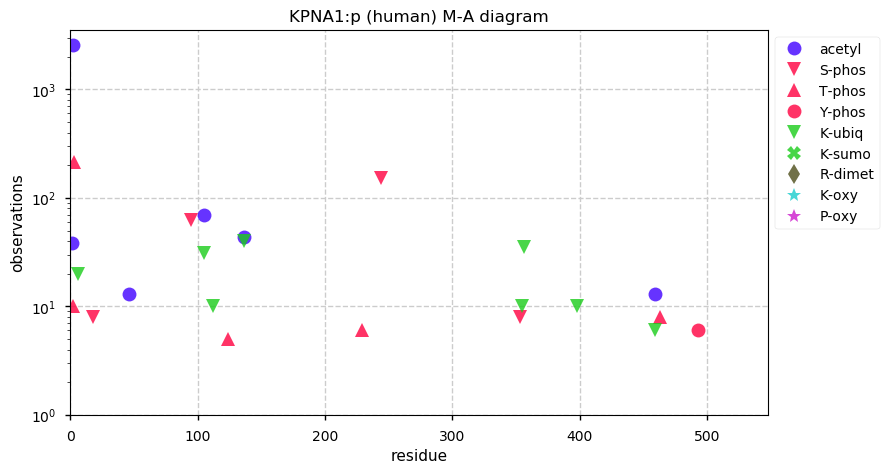
Wed Jan 22 15:59:02 +0000 2020The high dudgeon tone of the article is unintentionally (?) amusing. If only this poor, naive fellow could have access to some IT assistance to help him with his phone ...
🔗
Wed Jan 22 15:28:35 +0000 2020@DonMartinCTV It is actually common to have a few cold days around this time in Florida. The temperatures today are very similar to about this date in 2019 and 2018.
Wed Jan 22 15:12:49 +0000 2020Yikes! This guy had the office across the hall from me at the University of Manitoba Rady Faculty of Health Sciences.
🔗
Wed Jan 22 13:21:00 +0000 2020IPO13:p, importin 13 (Homo sapiens) 🔗 Midsized nuclear membrane subunit; few PTM sites; no high maf SAAVs; less commonly observed than other importins; 1 splice variant; mature form 1-963 [4,553 x] 🔗
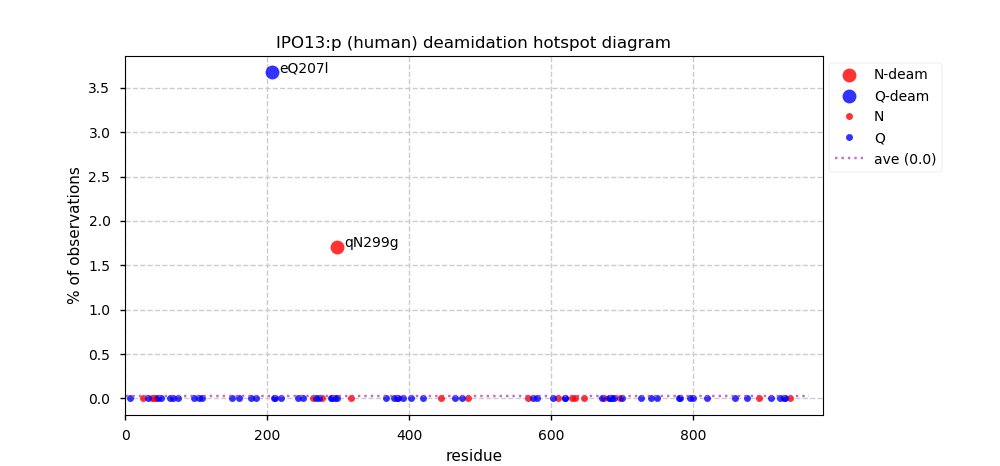
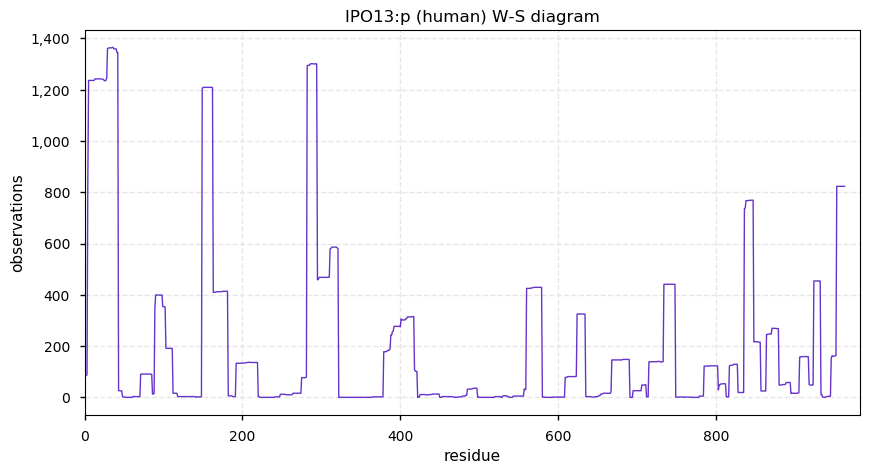
Tue Jan 21 16:04:44 +0000 2020@HMSBioPlex @EdHuttlin You should also take a look at the styling of the download page: the bait selection checkbox doesn't show up in Firefox and it clearly in the wrong place in MS Edge and Chrome.
Tue Jan 21 15:57:05 +0000 2020@EdHuttlin @HMSBioPlex The data download URL in the manuscript (🔗) is incorrect. It should be 🔗
Tue Jan 21 14:03:47 +0000 2020TNPO3:p, transportin 3 (H. sapiens) 🔗 MIdsized nuclear membrane subunit; 10 ubiquitinyl K-acceptor sites; no high maf SAAVs; aka IPO12; 1 splice variant; mature form 1-975 [14,318 x] 🔗
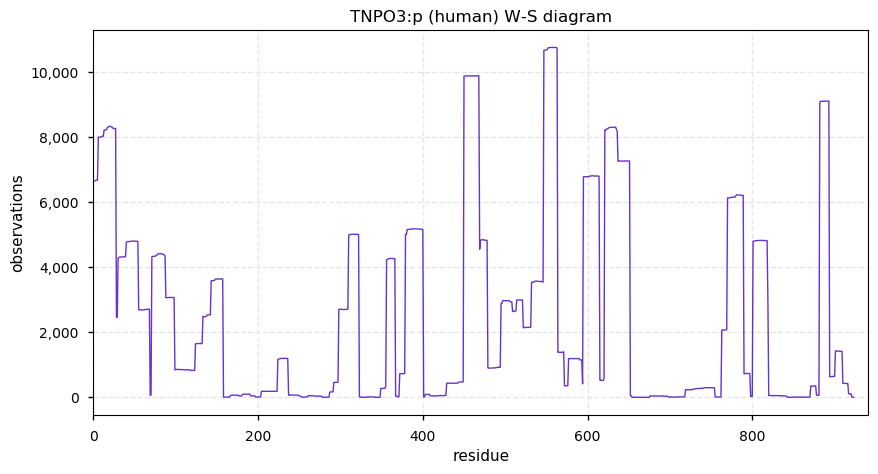
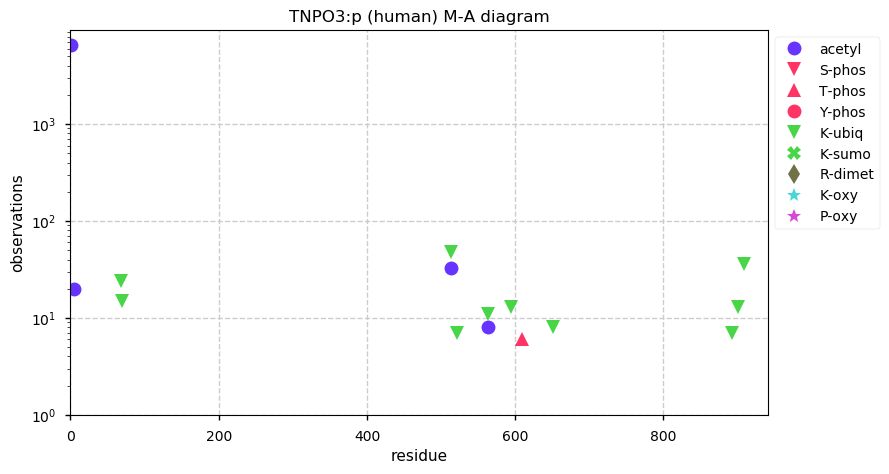
Tue Jan 21 13:26:05 +0000 2020@dtabb73 Been there, brother.
Mon Jan 20 18:42:20 +0000 2020@herrtschmidt A few times a year. I'm never really sure if it is truly a DOS attack, a spider that ignores robots.txt or simply an enthusiastic student who is trying to pull down a lot of information using too many simultaneous threads.
Mon Jan 20 18:20:09 +0000 2020The wall seems to be holding for the moment.
Mon Jan 20 17:51:24 +0000 2020Servers are back up, lets see what happens ...
Mon Jan 20 17:46:30 +0000 2020The attack started again, but this time we were able figure out the source. Once the servers can be restarted, hopefully the source of the attack will be effectively blocked.
Mon Jan 20 17:15:30 +0000 2020@gangulyteena Ten years ago, there was difference in technical quality between Chinese university-based research and similar US/EU efforts, but there is little, if any, difference now.
Mon Jan 20 16:58:10 +0000 2020The firewalls and servers have been restarted and the attack seems to be over, but continued monitoring will be necessary to be sure.
Mon Jan 20 16:30:18 +0000 2020GPMDB is currently being slowed down by a denial-of-service attack by an unknown party. We will try to get the attack blocked and the site back up to normal as soon as possible.
Mon Jan 20 13:49:40 +0000 2020IPO11:p, importin 11 (H. sapiens) 🔗 Large nuclear membrane subunit; scattered PTMs; no high maf SAAVs; 1 deamidation hotspot; 1 splice variant; mature form 1-975 [14,318 x] 🔗
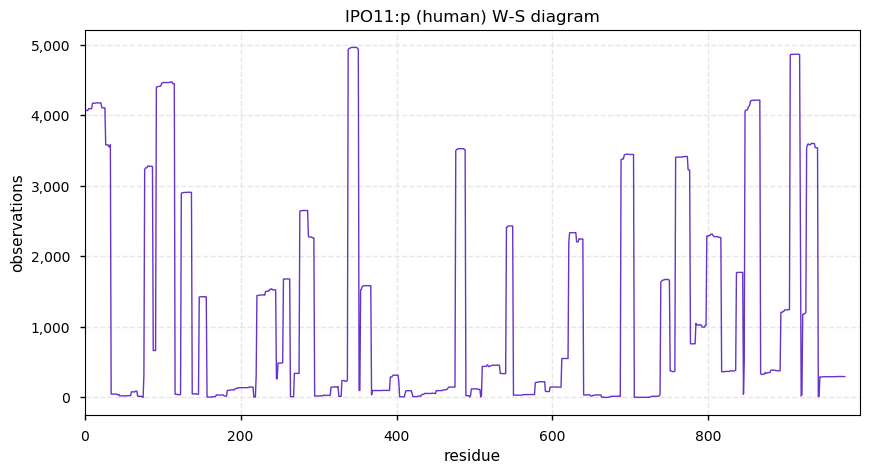
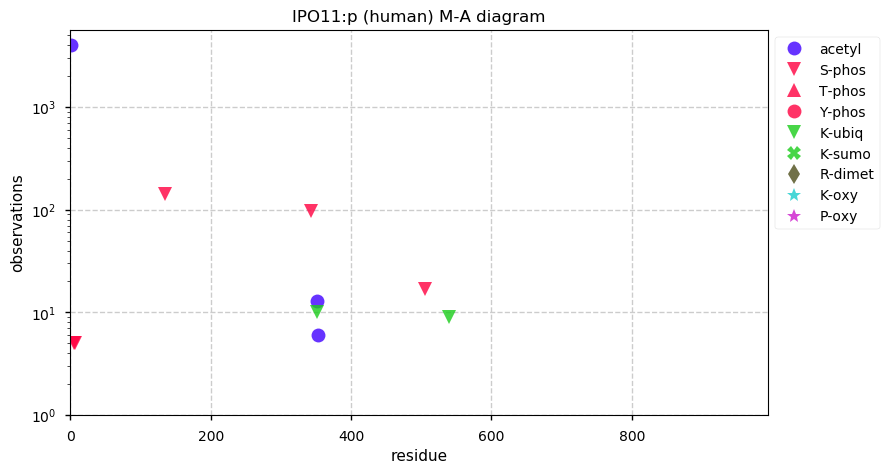
Sun Jan 19 14:39:32 +0000 2020IPO9:p, importin 9 (H. sapiens) 🔗 Large nuclear membrane subunit; phosphodomain 899-947; no high maf SAAVs; 1 deamidation hotspot; 1 splice variant; mature form (2,3)-1041 [29,701 x] 🔗
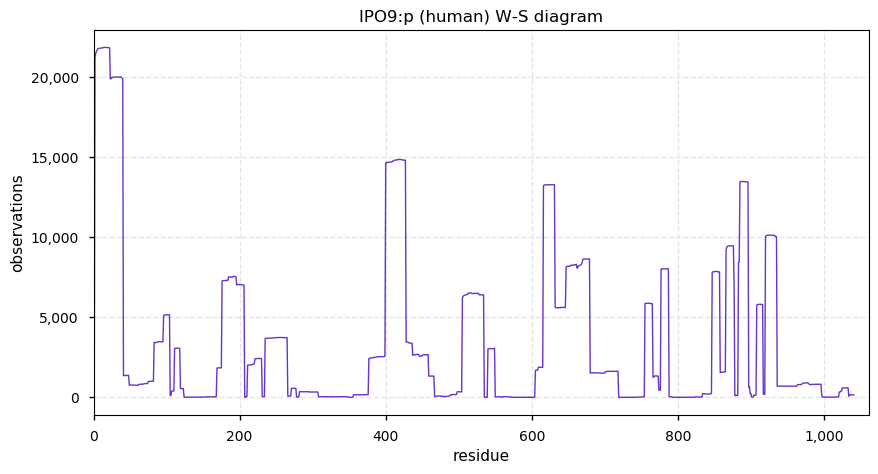
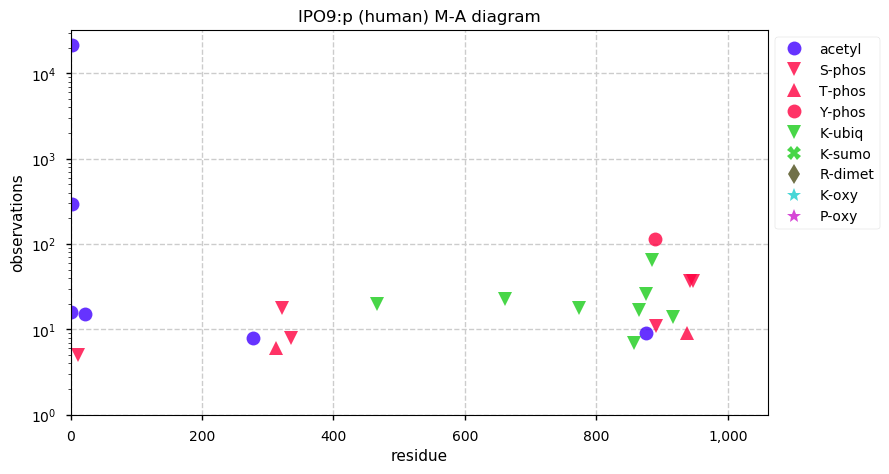
Sat Jan 18 16:31:02 +0000 2020IPO8:p, importin 8 (H. sapiens) 🔗 Large nuclear membrane subunit; high occupation phosphodomain 902-911; 1 high maf SAAV: I640V (0.01); mature form 1-1037 [17,886 x] 🔗
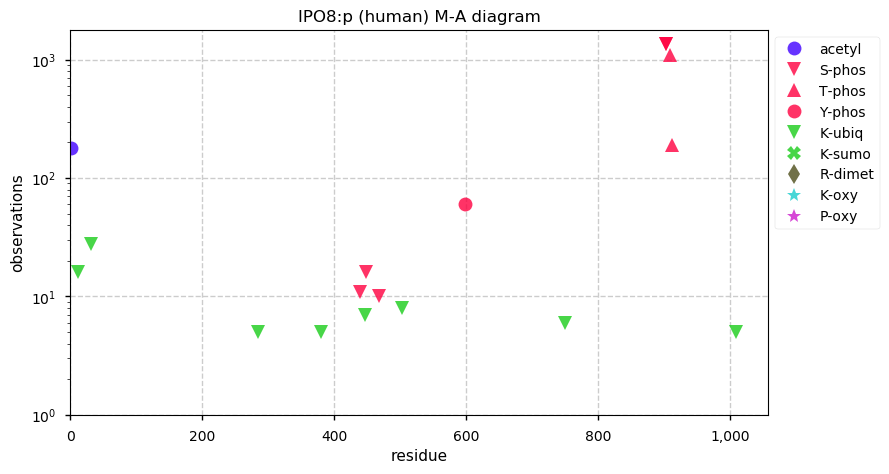
Sat Jan 18 14:09:00 +0000 2020@JuliaSkid11 @mudstonephoto Thanks for the suggestions. The data is already published. I was trying to understand what was wrong, to decide whether to archive it in GPMDB. The # of missed cleaves was OK, ~ 20% of PSM's. Non tryptic cleavage was high (15%), but within the range typical of tissue samples.
Fri Jan 17 21:27:12 +0000 2020@chrashwood Welcome to the weird world of trying to figure out what people actually did, based on their data rather than the Methods section, which often seem to be more like reminisces than the contents of an accurate diary.
Fri Jan 17 21:19:00 +0000 2020@goodlettlab1 You guys are brave 🧐
Fri Jan 17 21:18:14 +0000 2020@chrashwood Well, I guess it means the "not reproducible" pile then. Too bad, because based on the subject matter it would have been a good study to have available in the archive.
Fri Jan 17 20:58:07 +0000 2020@goodlettlab1 And once it gets going, it is the equivalent of whole-organism apoptosis.
Fri Jan 17 20:56:39 +0000 2020@goodlettlab1 When I worked at Eli Lilly they had a big program into dealing with septic shock. Of the many difficulties, "sepsis" isn't a specific thing: it is a collection of symptoms that can be triggered by many factors (including most bacteria that like to live in blood).
Fri Jan 17 20:43:28 +0000 2020@chrashwood If I can't figure it out, I'm going to have to throw it on the "not reproducible" pile & not import it into GPMDB. It is a shame to discard a large study like this one, but it wouldn't be the first time (or the hundredth) ...
Fri Jan 17 20:38:17 +0000 2020@chrashwood The method ended with 3 min wash at 100% B and 10 min wash at 0% B. "A" was 0.2% FA & "B" was 0.2% FA in 70% ACN. I guess the "gradient" they refer to may be oddly non-linear. There is no mention of a special gradient shape in the paper, but it can't be ruled out.
Fri Jan 17 20:32:57 +0000 2020@chrashwood The description in the paper sounds more linear:
"Samples were loaded onto the column for 12min at a flow rate of 0.35μL/min. Mobile phase B increased to 4% in the first min then increased on a gradient to 55% B at 75min. The method increased percent B to 100% by 76 min."
Fri Jan 17 19:59:01 +0000 2020@chrashwood Normal is a bit of an exaggeration, but more like the attached 🔗
Fri Jan 17 19:49:27 +0000 2020@chrashwood Could be a column issue, but it persists intermittently for the 240 HPLC runs in the study, so runs looking odd, others perfectly normal. Maybe the mixer?
Fri Jan 17 18:11:10 +0000 2020@mudstonephoto This is a typical run from the same lab, using the same type of instrument but from a different study 🔗
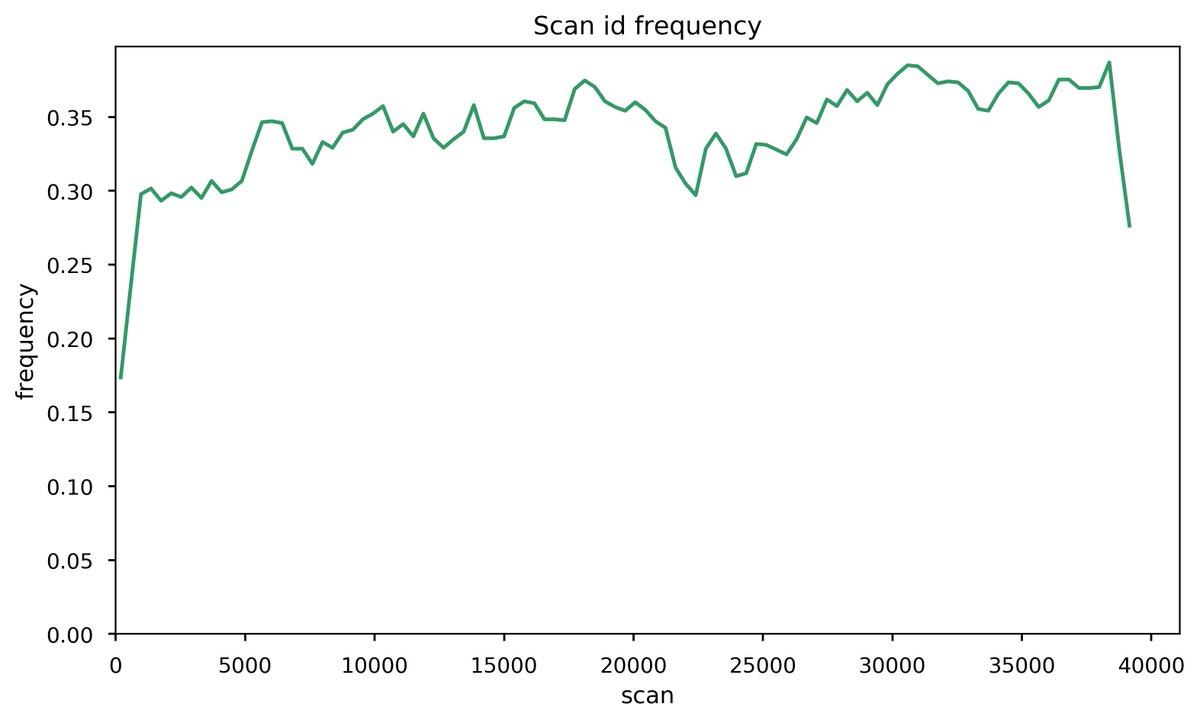
Fri Jan 17 17:25:48 +0000 2020Proteomics HPLC tweeps: I'm looking at a large dataset from human tissue LC data (run on a Lumos), where most of the RAW files have the odd behavior illustrated below. Any ideas? The graph is a running average of the PSM assignment frequency vs. scan #. 🔗
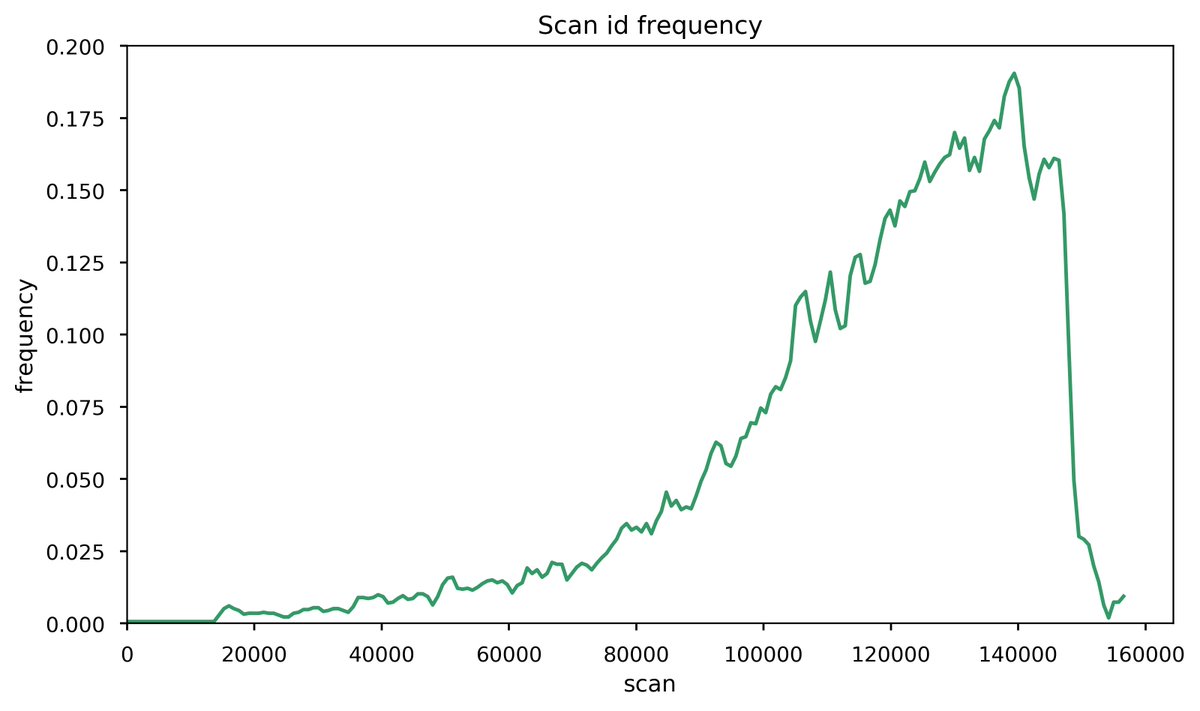
Fri Jan 17 15:58:52 +0000 2020@UCDProteomics @chrashwood @StoyanStoychev1 I was too focused on the dotp to notice, but it does seem similar. Keep in mind that 50% of useful tryptic peptides contain at least 1 proline (& 20% contain at least 2 prolines), so proline cis/trans-isomeration is not a rare effect.
Fri Jan 17 15:47:04 +0000 2020@UCDProteomics @chrashwood @StoyanStoychev1 It looks a lot like the illustration on p. 26 of 🔗 (with 3x the resolution)
Fri Jan 17 15:18:57 +0000 2020A bit more poking around resulted in 2 more deamidation motifs.
1. ...X[QN][KR] Q or N at the n-1 position of a tryptic peptide; &
2. ...[LIFW]Q[LIFW]... Q sandwiched between 2 large hydrophobic residues.
Fri Jan 17 13:39:01 +0000 2020IPO7:p, importin 7 (H. sapiens) 🔗 Large nuclear membrane subunit; high occupation phosphodomain 876-903; no high maf SAAVs; mature form 1-1038 [40,559 x] 🔗
Fri Jan 17 00:14:19 +0000 2020@byu_sam So I can't say that I would draw any general conclusions, especially about what happens in common edge cases, e.g., a bad injection.
Fri Jan 17 00:11:56 +0000 2020@byu_sam I read it again a few times and it seems they used a very limited set of RAW files for the simulations shown ( in most cases only 1). The PSM assignment parameters were also too simple for that data set: it would have resulted in about 30% false negatives.
Thu Jan 16 16:57:32 +0000 2020IPO5:p, importin 5 (H. sapiens) 🔗 Large nuclear membrane subunit; many PTMs with marked domain patterning; no high maf SAAVs; mature form 2-1097 [47,583 x] 🔗
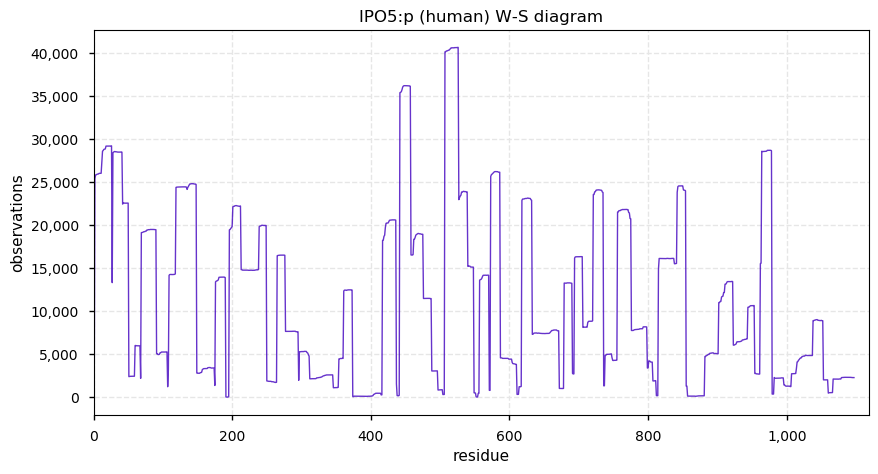
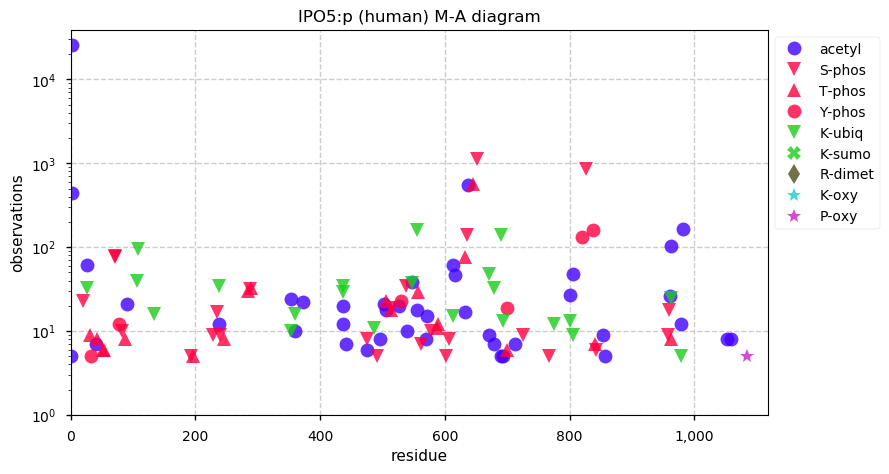
Thu Jan 16 15:23:13 +0000 2020@d_vivian Speed aside, the original motivation behind the development project was to create a PSM assignment system that was suitable for regulated tests, e.g., pharmaceutical R&D, drug batch QC tests or clinical tests. And that part has certainly worked.
Thu Jan 16 14:34:13 +0000 2020@d_vivian The Ryzen is clearly faster, but I'm not sure there is any significant value to getting to the answer 1 minute earlier. My takeaway from the research is that you can do fast, practical indexed PSM assignment on ARM processors with minimal memory requirements.
Thu Jan 16 13:08:33 +0000 2020@d_vivian If you mean Ryzen v RPI4, the Ryzen with 4 threads took 0.23 min per MGF and the RPI4 with 3 threads took 1.6 min per MGF. The Ryzen box cost about CA$5000 and the RPI4 cost CA$120.
Thu Jan 16 12:58:49 +0000 2020@d_vivian I'm not quite sure what you mean by comparison.
Thu Jan 16 12:24:11 +0000 2020@d_vivian It is doing the real work. I just gave the Ryzen a spin to see how well it would perform on a larger batch of MGFs.
Wed Jan 15 22:52:10 +0000 2020@leprevostfv If you interested, the C++ code for the PSM assignment app used is here: 🔗
Wed Jan 15 22:13:56 +0000 2020@leprevostfv For this data, about 1 GB per ID job, so running 4 simultaneous ID jobs used 4 GB.
Wed Jan 15 21:28:46 +0000 2020@byu_sam Can't say I understand the choices made in the "Data sets and Analysis" section, but I will think about it some more.
Wed Jan 15 21:14:29 +0000 2020@pwilmarth It isn't quite all penta-peptides. It is mostly peptides like:
Gly-Xxx-Asn/Gln-Yyy-Gly
where Xxx and Yyy are not Asn or Gln, and a few others. It must have cost them a lot to have that many peptides synthesized back then.
Wed Jan 15 20:25:08 +0000 2020Some more poking around shows that ...XNS... is also a likely peptide deamidation motif, as well as ...[KR][NQ]X... (i.e., N or Q at the N-terminus of a tryptic peptide).
Wed Jan 15 19:03:50 +0000 2020@byu_sam I was more wondering what people actually do in practice. Do you use Noble's approach?
Wed Jan 15 17:59:52 +0000 2020@chrashwood Just a normal 7200 RPM hard drive.
Wed Jan 15 17:25:34 +0000 2020New speed record (for me): 10.8 minutes to analyze 46 Orbitrap HF human proteome MGFs (14 s/MGF) with 22,400 spectra/MGF, using 4 cores of a Ryzen 1950X. I have officially geeked out.
Wed Jan 15 14:13:58 +0000 2020It turns out that in peptides, under the conditions commonly used in proteomics sample preparation, the N in ...XNA... is just about as likely to undergo deamidation as the better known ...XNG... motif.
Wed Jan 15 14:03:58 +0000 2020@friendofthesci @VATVSLPR It does seem like most other people are either shy or are not curious about the issue of target-decoy simulation confidence intervals.
Wed Jan 15 12:56:14 +0000 2020IPO4:p, importin 4 (H. sapiens) 🔗 Large nuclear membrane subunit; 12 ubiquitinyl K-sites, but only K788 paired with acetyl; 4 high maf SAAVs: R20C (0.01); A513V (0.3); P580A (0.3); A758T (0.01); mature form 1-1081 [26,439 x] 🔗
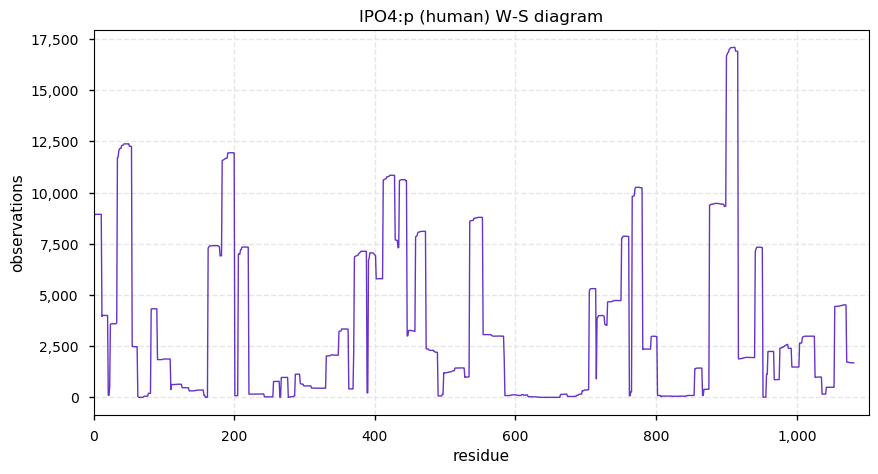
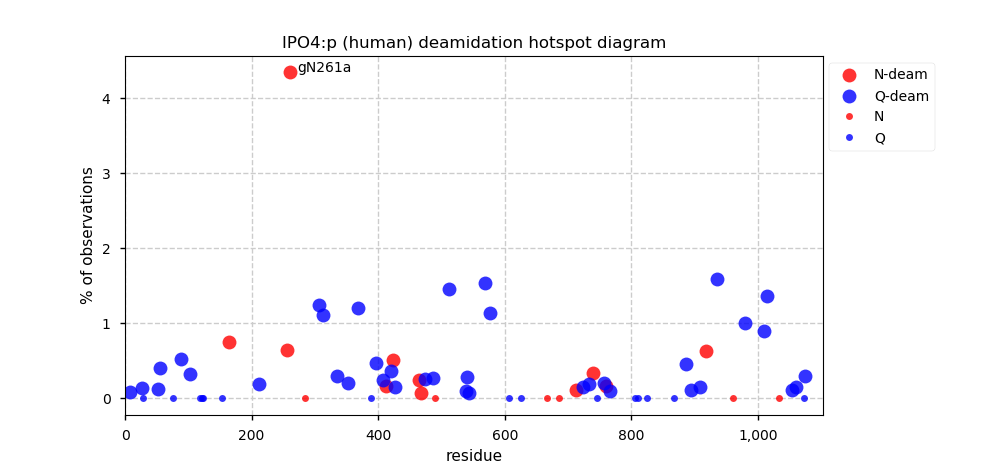
Wed Jan 15 02:50:43 +0000 2020@VATVSLPR @friendofthesci Thanks for the details.
Tue Jan 14 23:07:28 +0000 2020How do people calculate the error in a target-decoy simulation FDR, i.e., the "x" in
FDR = 0.01 ± x?
Tue Jan 14 21:52:36 +0000 2020I've really started to tire of learning new things all of the time, but there doesn't seem to be an alternative.
Tue Jan 14 20:17:20 +0000 2020@d_vivian Just to work out the kinks, I set up a folder that has the MGFs for those LC/MS/MS runs and the TSV + meta files generated by running idX using 1 thread on a Ryzen processor: ftp://ftp.thegpm.org/data/Kessel
Tue Jan 14 18:01:30 +0000 2020@IvisonJ I meant "It would seem ..."
Tue Jan 14 16:24:46 +0000 2020@IvisonJ I would seem that no one in the UK government has ever met a Scottish person.
Tue Jan 14 16:05:59 +0000 2020@d_vivian The experiments also used an instrument (Q-Exactive HF) that is not so bleeding edge as to be irrelevant to most current lab practice. /2
Tue Jan 14 16:04:01 +0000 2020@d_vivian Those particular experiments also have significant numbers of phosphopeptides, ubiquitinylated peptides, hypusine-containing peptides, dimethylated R peptides and selenocysteine-containing peptides, with very few experimental artifacts. /1
Tue Jan 14 15:29:52 +0000 2020@d_vivian That is a good suggestion (PXD004452). I particularly like the two 46 fraction MudPit runs with files labelled like "SH-SY5Y_REP1_46frac" & "SH-SY5Y_REP2_46frac". They are pretty consistent and were probably the best in terms of protein detection of the methods tried.
Tue Jan 14 13:53:40 +0000 2020I'm calling > 5% deamidation at an N/Q site a "hotspot" (for want of a better term)
Tue Jan 14 13:50:59 +0000 2020TNPO2:p, transportin 2 (H. sapiens) 🔗 Midsized nuclear membrane subunit; several phosphorylation sites and acetyl/ubiquitinyl pairs; 1 deamidation hotspot; no high maf SAAVs; mature form 1-887 [30,712 x] 🔗
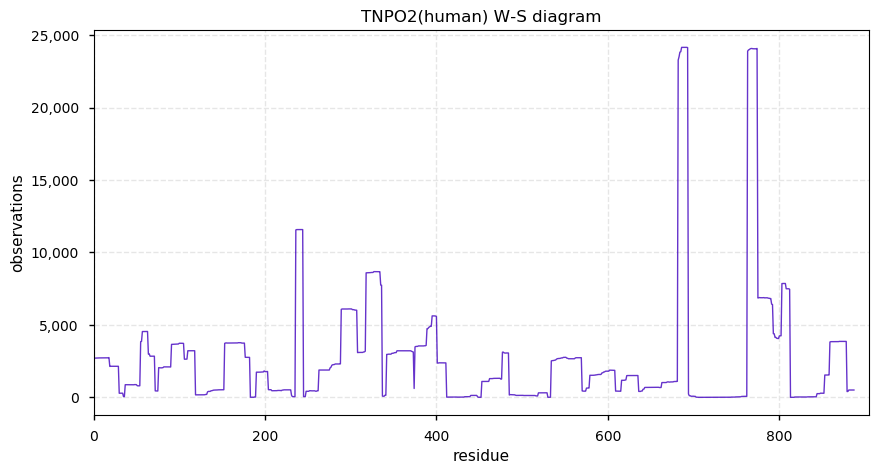
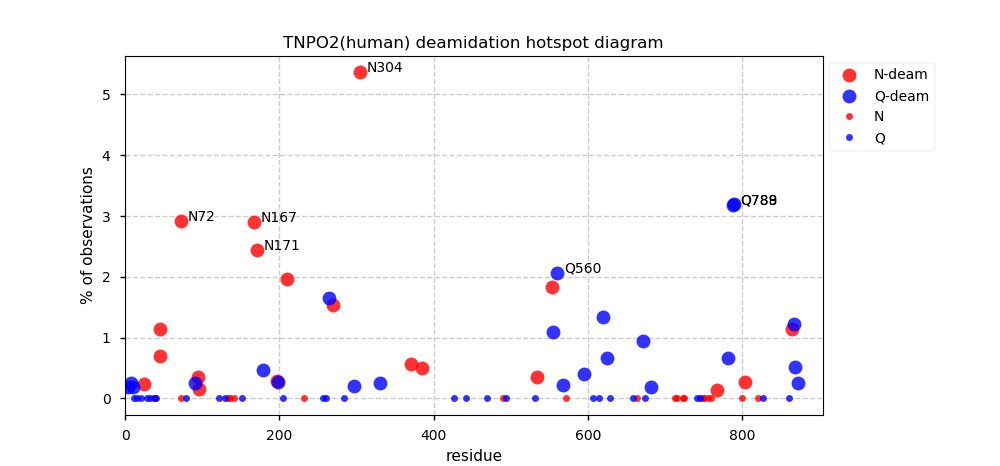
Tue Jan 14 00:38:43 +0000 2020Something that can serve as a Kessel Run for analysis (results may be in parsecs).
Mon Jan 13 22:52:06 +0000 2020@UCDProteomics @ProteomicsNews @nanopore I get the potential importance of the idea, but the paper doesn't make a great case for the authors' expertise in modern analytical methods.
Mon Jan 13 22:50:44 +0000 2020@UCDProteomics @ProteomicsNews @nanopore The Central Dogma was already considered old-fashioned when I was a Zoology undergrad (and that was several days ago). And the MS illustrated is a mass spectrograph (also a day-or-two out of date).
Mon Jan 13 22:39:23 +0000 2020Is there a publicly available proteomics MS/MS data set that the cool kids are using for analysis-speed benchmarking these days?
Mon Jan 13 21:13:38 +0000 2020@CSRpeerreview SRO's are one of the truly great things about the NIH system. Working for a while with a national system (CA) that has no equivalent heightens one's appreciation for just how much these folks improve the grants process.
Mon Jan 13 20:55:19 +0000 2020@rbharathkumar91 @KentsisResearch @Ciencia_2017 It is also kind of interesting from a technical point of view, as the experiment uses a super-SILAC approach to quantitation, but none of the E. coli proteins have SILAC-label incorporation (even though given the Methods they really should).
Mon Jan 13 20:51:24 +0000 2020@rbharathkumar91 @KentsisResearch @Ciencia_2017 A C. elegans dataset that maybe has a bit too much E. coli B for comfort is PXD004104 (🔗). One of the MudPit runs is given here, filtered to show the 1,641 E. coli protein ids 🔗
Mon Jan 13 19:09:00 +0000 2020@rbharathkumar91 @KentsisResearch @Ciencia_2017 I am sure there are groups that do this correctly, but many groups don't (at least based on publicly available data sets). I never analyze C. elegans data without an E. coli B proteome in the mix.
Mon Jan 13 19:02:23 +0000 2020@UCDProteomics @DNATech_Lutz Although about 5999 of them are going to be actin.
Mon Jan 13 18:40:34 +0000 2020@UCDProteomics @DNATech_Lutz Quick quiz: if you identify an average of 14 AA per PSM and you get an id on 30% of scans, how many AA's do you get per second?
Mon Jan 13 18:21:05 +0000 2020@GammaCounter Reviewers, editors and copywriters all go berserk if they see "I", "he" or "she" in a manuscript.
Mon Jan 13 17:30:21 +0000 2020@Smith_Chem_Wisc In any deep-ish C. elegans proteomics experiment, it is not unusual to id > 500 E. coli proteins corresponding to 15-20% of the PSMs.
Mon Jan 13 17:14:08 +0000 2020@Smith_Chem_Wisc Lab grown C. elegans are little bags full of E. coli in various states of digestion. I have never seen results from a C. elegans prep that did not have high levels of E. coli proteins easily detected in the data.
Mon Jan 13 16:48:49 +0000 2020For the life of me, I can't understand why researchers (& reviewers) think it is OK to identify C. elegans proteomics data using just C. elegans protein sequences.
Mon Jan 13 14:43:05 +0000 2020TNPO1:p, transportin 1 (H. sapiens) 🔗 Midsized nuclear membrane subunit; many low occupancy PTMs and 2 deamidation hotspots; no high maf SAAVs; mature form 1-890 [39,854 x] 🔗
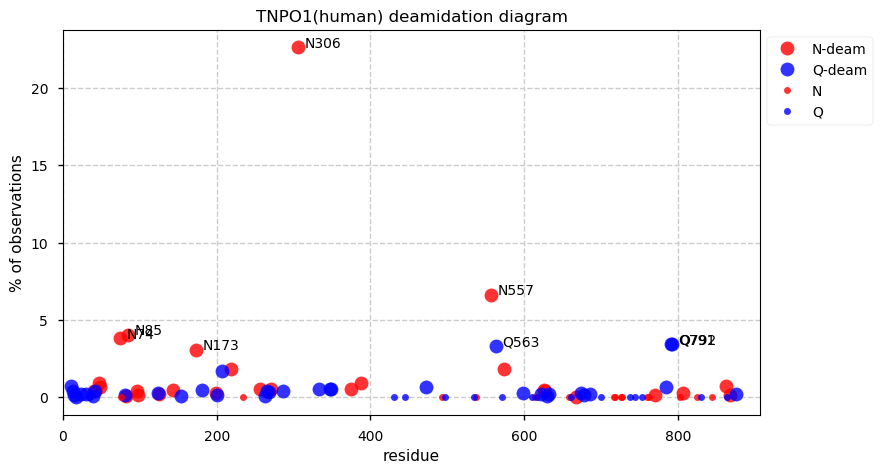
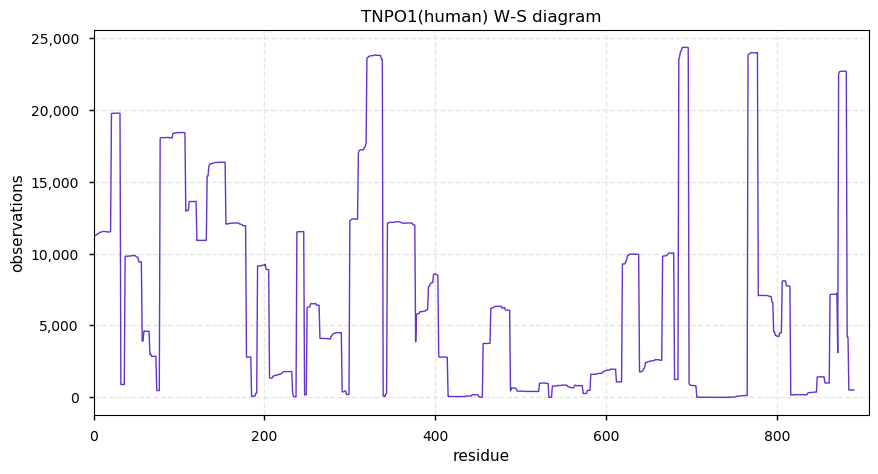
Sun Jan 12 14:22:13 +0000 2020KPNB1:p, karyopherin subunit beta 1 (H. sapiens) 🔗 Midsized nuclear membrane subunit; 19 complementary acetyl/ubiquitinyl K sites, but no SUMOyl; no high maf SAAVs; mature form 1-876 [63,977 x] 🔗
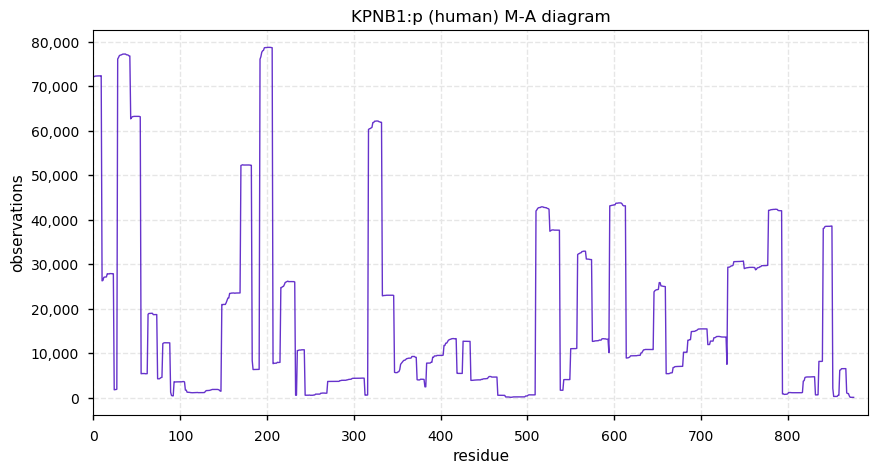
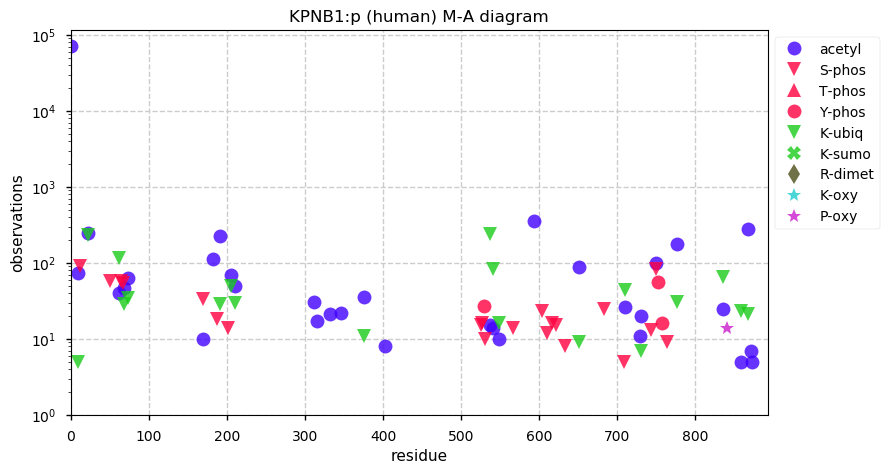
Sat Jan 11 23:25:04 +0000 2020@chrashwood How well ID algorithms do with deamidation is a good test of their accuracy. The ratio of Q:N deamidations is very useful for tuning up a search engine during development.
Sat Jan 11 21:41:28 +0000 2020Thanks to everyone who participated in this poll. No really clear winner here: quite a few respondents were skeptical about observed deamidations being real.
Sat Jan 11 17:27:19 +0000 2020@mudstonephoto Sure, if anyone was interested in investigating deamidation more deeply. The R&R paper deals with intact proteins: once you've generated tryptic peptides the deamidation rates will change because of relaxed steric constraints.
Sat Jan 11 17:03:22 +0000 2020A little food-for-thought on this one. The attached graphs show the fraction of observations in gpmDB where catalase (CAT) has a deamidated N or Q. 🔗
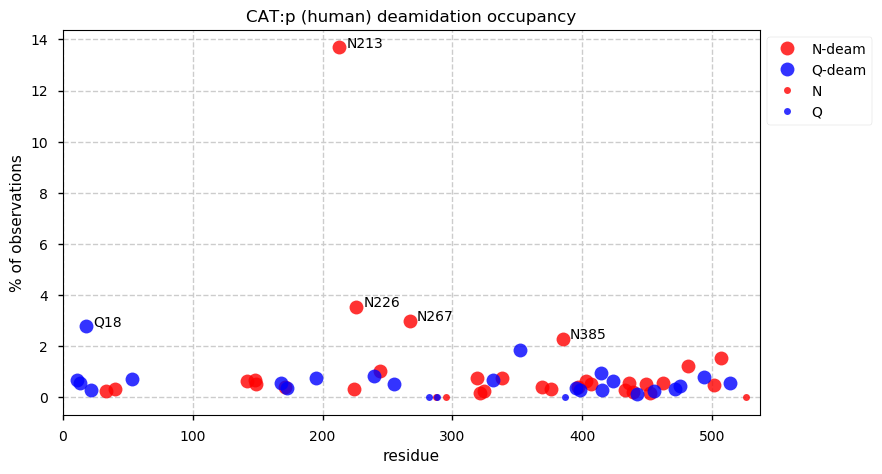
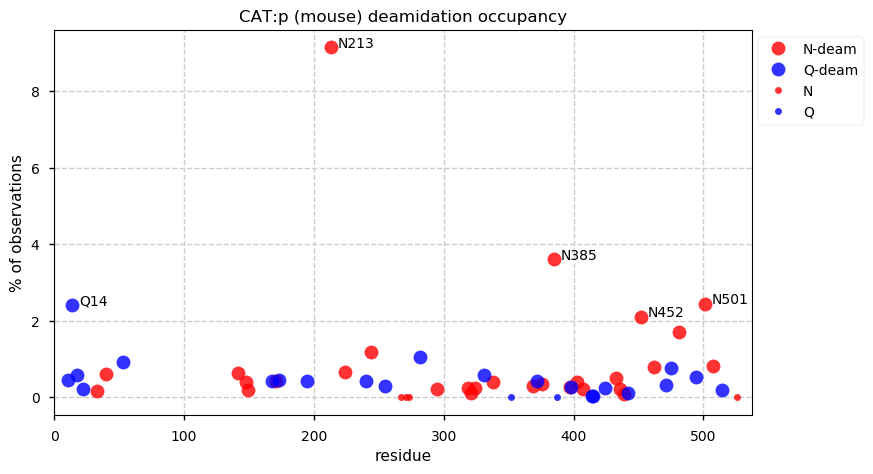
Sat Jan 11 13:13:19 +0000 2020COL1A2:p, collagen alpha-2(I) chain (Ursus arctos horribilis 🐻) 🔗| Large extracellular subunit; 107 hydroxyproline and hydroxylysine sites; significant endogenous proteolysis; mature form 86-1120 [16 x] 🔗
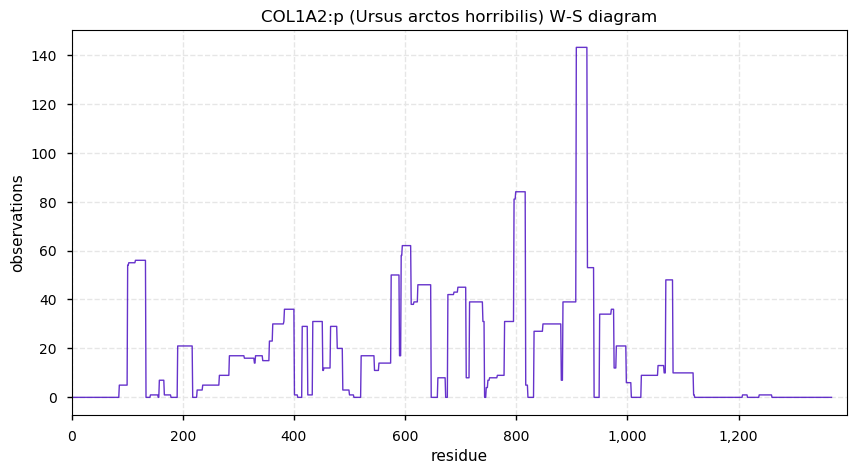
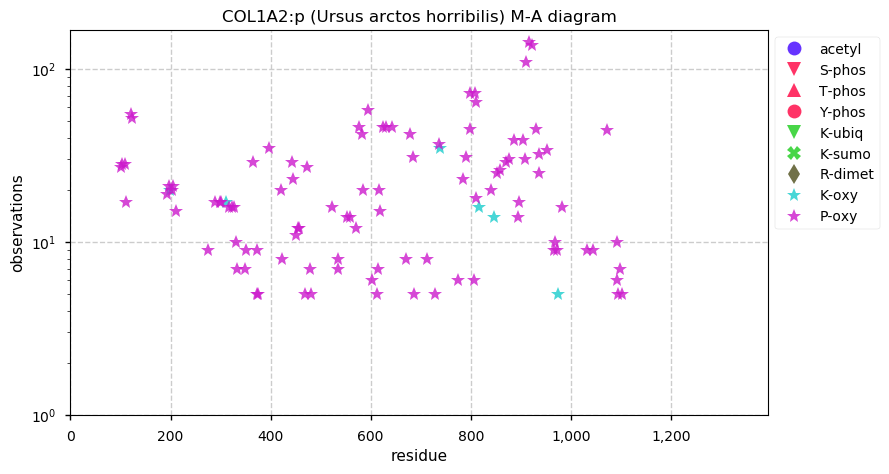
Fri Jan 10 20:48:54 +0000 2020Peptide deamidation at N and Q residues is frequently assigned in proteomics experiments. Is the site of the modification mainly:
Fri Jan 10 18:31:59 +0000 2020I'm still puzzled why CBC published this particular article and why it is still so prominent in its "Opinion" section
🔗
Fri Jan 10 18:10:09 +0000 2020@slashdot This has the potential to impact scientific computing rather significantly, especially if other countries adopt a similar position.
Fri Jan 10 17:16:04 +0000 2020I haven't written a protein inference script for a while, but I was pleasantly surprised at how easy it was using Python's built-in set methods.
Fri Jan 10 14:19:23 +0000 2020COL1A1:p, collagen alpha-1(I) chain isoform X1 (Ursus arctos horribilis) 🔗| Large extracellular subunit; 134 hydroxyproline and hydroxylysine sites; significant endogenous proteolysis; mature form 159-1254 [16 x] 🔗
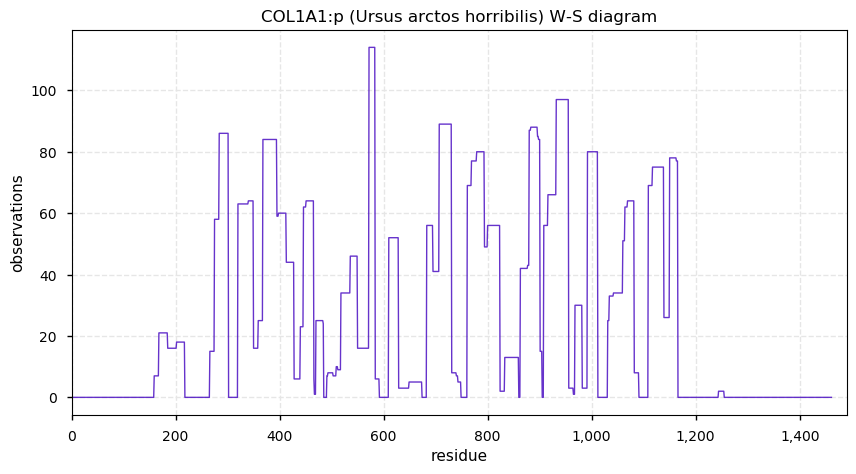
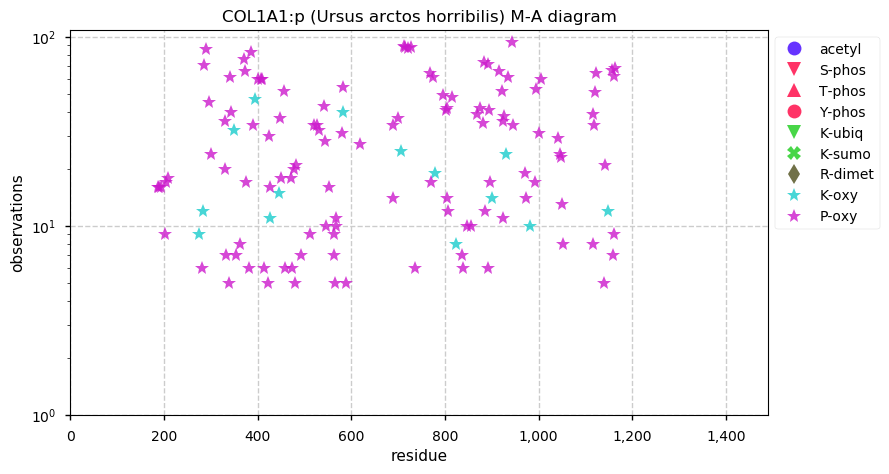
Fri Jan 10 13:21:20 +0000 2020@Picotti_Lab @PastelBio @kusterlab That aside, the methods used do a good job of detecting peptides that elute at low and high ACN. Failure to detect these peptides is a characteristic of many RP-HPLC methods used in proteomics.
Fri Jan 10 13:11:03 +0000 2020@Picotti_Lab @PastelBio @kusterlab The data has lots of good examples of the difficulties associated with running and reporting a lot of LC/MS/MS experiments.
Fri Jan 10 02:15:22 +0000 2020@dark_shark I had just finished listening to Heroes, not 2 minutes before seeing this reminder ...
Thu Jan 09 22:55:59 +0000 2020@UVicProteomics Problems don't seem to go away: Brian Chait's first project working in Ken Standing's lab was the analysis of wheat seed proteins.
Thu Jan 09 17:59:10 +0000 2020Oddly, PXD015442 seems to be the first rat blood plasma data set made publicly available.
Thu Jan 09 16:22:55 +0000 2020@Caroline_Bartma @dtabb73 ... often for the 3rd or 4th time.
Thu Jan 09 16:21:26 +0000 2020@AlexUsherHESA The incident has brought the Air India 182 crash to mind.
Thu Jan 09 13:26:31 +0000 2020@gtombline @PastelBio For practical reasons, few people in the field seem to care about this issue. I have always been fascinated by it, but the field is pretty happy ignoring the additional complexity.
Thu Jan 09 13:18:44 +0000 2020PRELP undergoes significant endogenous proteolysis. It is rare to observe the protein without multiple internal cleavages: there are 80 observed tryptic peptides and > 500 semi-tryptic peptides.
Thu Jan 09 13:12:23 +0000 2020PRELP:p, proline and arginine rich end leucine rich repeat protein (H. sapiens) 🔗 Small secreted protein; N-linked glycosylation at N124, N289, N320, N327; 2 high maf SAAVs: N334S (0.01), N348H (0.01); 1 splice variant; mature form 21-382 [13,639 x] 🔗
Wed Jan 08 15:58:51 +0000 2020NB: these results use the RefSeq Ursus arctos horribilis protein sequences rather than the Homo sapiens protein sequences used in the associated publication 🔗
Wed Jan 08 15:56:36 +0000 2020And now, thanks to 🔗, GPMDB has its first grizzly bear proteome entries 🔗
Wed Jan 08 14:07:49 +0000 2020@RaghunathAnand @pollyp1 @DrKYSR It is biomedical research jargon for a senior researcher (most often a professor) that serves as the "Principal Investigator" on a grant. It is often used in North American med schools as a generic term for a lab's senior professor.
Wed Jan 08 13:56:22 +0000 2020TGM5:p, transglutaminase 5 (H. sapiens) 🔗 Midsized epithelial enyzme; N-terminal acetylation; no high maf SAAVs; 1 splice variant; mature form 2-720 [2,787 x] 🔗
Wed Jan 08 13:36:38 +0000 2020@dtabb73 😎 I always had trouble with shredding when I used RAID 5. I swore off everything but RAID 0.
Wed Jan 08 13:29:08 +0000 2020@AlexUsherHESA I must admit to being in love with your phrase "nonsense boutique research ideas".
Wed Jan 08 02:43:16 +0000 2020@tfmPhD @HFazelinia I will leave counting smORFs to wiser people. My practical observation was that in a human MHC type I LC/MS run, depending on the HLA allele being studied, 1-5% of the PSMs correspond to peptides only present in smORF sequences.
Tue Jan 07 22:51:58 +0000 2020Frickn' big data
Tue Jan 07 22:51:33 +0000 2020Just changed some network topology in the unrealistic hope of speeding some things up: fingers crossed 👨🔧
Tue Jan 07 19:31:31 +0000 2020@d_vivian @UCDProteomics @mmkstarr MS/MS in proteomics tends to be better at hypothesis testing than it is at open ended investigations.
Tue Jan 07 19:29:30 +0000 2020@d_vivian @UCDProteomics @mmkstarr I have found it is easier to just take the genetic variant information from a genome sequence and add it into the PSM identification process. In my hands, MS/MS data hasn't a great way to discover variants de novo. It can be done, but the results are pretty noisy.
Tue Jan 07 18:00:35 +0000 2020@d_vivian @UCDProteomics @mmkstarr The point of the research is to come up with a very efficient system that can be used in a clinical lab, so the system and results have to be easily audited & contain validation information (from an informatics point of view).
Tue Jan 07 17:56:50 +0000 2020@d_vivian @UCDProteomics @mmkstarr Not planning to. The searches were for human, checking A0, A1 & A2 explicitly for phosphoryl (S|T|Y), acetyl (K), ubiquitinyl (K), deamidation (N), ammonia loss (nt Q|C), oxidation (M|W|P|K), dimethyl (R) & protein N-terminal processing (N-term cleave+acetyl|ER|SR|mito).
Tue Jan 07 17:10:18 +0000 2020@d_vivian @UCDProteomics @mmkstarr It is an option, but I don't think I'm going to need a cluster. My throughput test was running 40 mgf's from Orbitrap raw files and it finished in a little under 40 minutes. That is faster than the time to download the raw files and running msconvert to make the mgfs.
Tue Jan 07 13:41:42 +0000 2020@UCDProteomics @mmkstarr The RPI4 CPU (4 core, 1.5 GHz) and USB3 bridge are much more capable than the previous version. They cost US$55 each. NB: you have to be good with Debian-style Linux to use them for computations. But they have a standard Apache Web server, python3, SSH, g++, etc.
Tue Jan 07 13:27:59 +0000 2020The difference in the PTM profile between TGM2 & TGM4 is quite stark, even though the 2 proteins are the same length, have similar enzymatic activity, some tryptic peptide equivalence & have been observed a similar number of times.
Tue Jan 07 13:23:22 +0000 2020TGM4:p, transglutaminase 4 (H. sapiens) 🔗 Midsized enyzme; N-terminal acetylation; 5 high maf SAAV: E100D(0.4), Y244H (0.1), S249T (0.1), R372C (0.3), V409I (0.4); 1 splice variant; mature form (1,2)-684 [5,187 x] 🔗
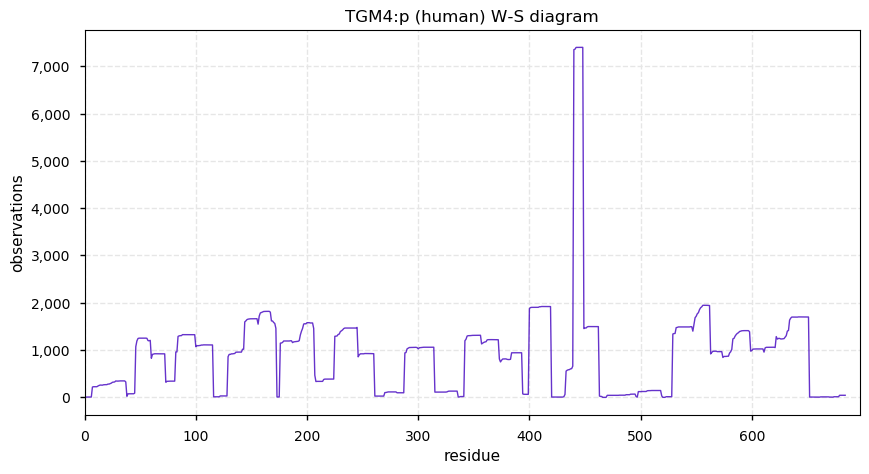
Mon Jan 06 18:34:41 +0000 2020@UCDProteomics @mmkstarr RPI = Raspberry PI (RPI 4 Model B in this case)
Mon Jan 06 14:39:52 +0000 2020@dtabb73 Sounds like when I lived in Vancouver: power optional.
Mon Jan 06 14:31:27 +0000 2020TGM2:p, transglutaminase 2 (H. sapiens) 🔗 Midsized enyzme; several Y-phosphodomains (17 sites); 1 high maf SAAV: V542F (0.01); 1 splice variant; mature form 2-687 [5,347 x] 🔗
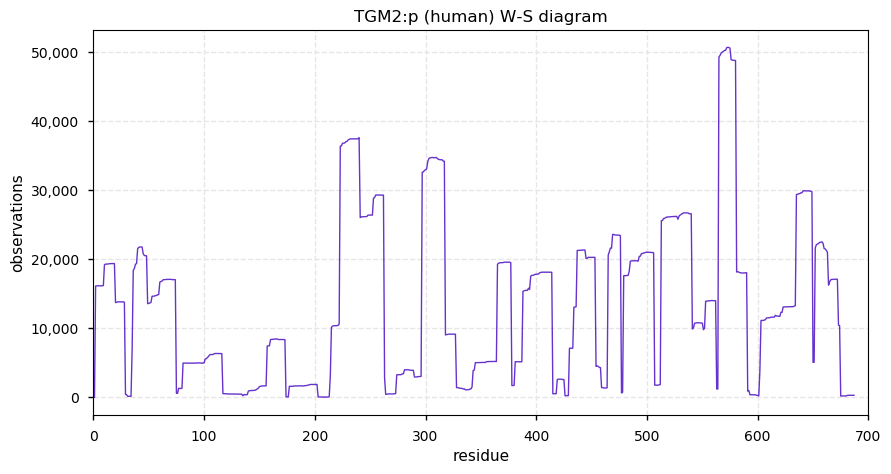
Mon Jan 06 14:11:07 +0000 2020@mmkstarr I've got some new algorithms (& software) that mean the new box runs PSM IDs faster than the heavyweight server using the previous generation of software. I'll use the Ryzen-based box for fancy simulations. Note: the Ryzen has an 850 W ps while the RPI has a 15 W ps.
Mon Jan 06 13:43:21 +0000 2020@dtabb73 I've personally never had much luck with RAID 5, but good luck to you!
Sun Jan 05 23:35:41 +0000 2020New proteomics ID server (white box) on top of the old 16 core Ryzen-based server it has replaced. 🔗

Sun Jan 05 14:45:21 +0000 2020TGM1:p, transglutaminase 1 (H. sapiens) 🔗 Midsized epithelium enyzme; N-terminal phosphodomain; 1 high maf SAAVs: S122L (0.01); 1 splice variant; mature form (1,2)-817 [5,347 x] 🔗
Sat Jan 04 17:43:11 +0000 2020@goodlettlab1 Frick'n raccoons 😠 🦝
Sat Jan 04 14:11:25 +0000 2020TGM3:p, transglutaminase 3 (H. sapiens) 🔗 Midsized epithelium enyzme; N-terminal acetylation; 3 high maf SAAVs: S249N (0.1), G650E (0.01), G654R (0.2); 1 splice variant; mature form 2-693 [7,814 x] 🔗
Sat Jan 04 13:21:20 +0000 2020It is currently warmer in Winnipeg CA than it is in Chihuahua MX. This is something that doesn't happen very often this time of the year.
Fri Jan 03 21:50:18 +0000 2020@KentsisResearch Raspian (the default Debian fork for the Pi's).
Fri Jan 03 21:02:36 +0000 2020Easier to configure than expected: headless install worked fine. It is up and doing ids.
Fri Jan 03 17:35:11 +0000 2020My Raspberry Pi 4b just arrived! I'm switching all of my QA/QC and routine PSM ID work to the RP next week, leaving my Ryzens free to do other work.
Fri Jan 03 14:01:46 +0000 2020PLA2G4E:p, phospholipase A2 group IVE (H. sapiens) 🔗 Midsized keratinocyte enyzme; no PTMs; no high maf SAAVs; 1 splice variant; mature form 1-868 [234 x] 🔗
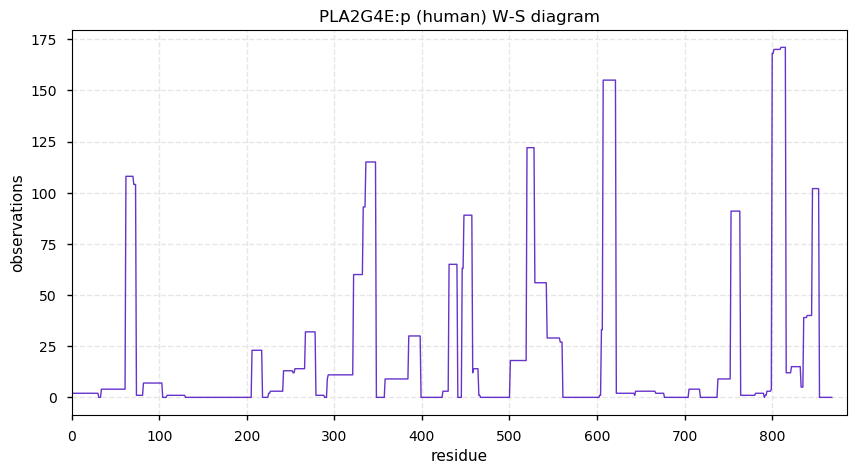
Thu Jan 02 17:08:49 +0000 2020Kind of disappointed in this one. A pretty routine application of dot-product-scoring spectrum library search that tries to connect itself to BLAST via a rather tortured metaphor while not mentioning a lot of relevant work 🔗
Thu Jan 02 17:04:46 +0000 2020@slavovLab And we know next-to-nothing about how or why this system works.
Thu Jan 02 13:54:30 +0000 2020ALOXE3:p, arachidonate lipoxygenase 3 (H. sapiens) 🔗
Midsized keratinocyte enyzme; no PTMs; no high maf SAAVs; 1 splice variant; mature form 1-711 [429 x] 🔗
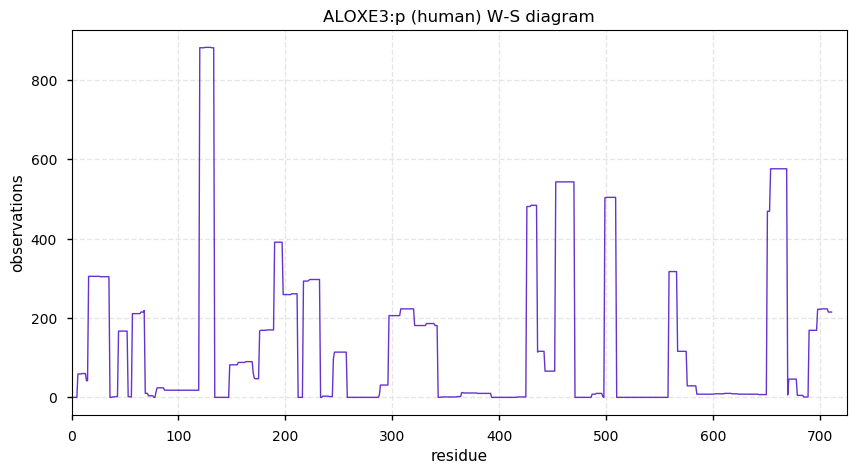
Wed Jan 01 20:07:21 +0000 2020@ProteomicsNews @PastelBio Post a few, with and without.
Wed Jan 01 15:04:46 +0000 2020@zacmcd77 @massspecpro The energy required for analyzing LCMS data will drop to 10 J per 1000 PSMs
Wed Jan 01 14:55:49 +0000 2020@zacmcd77 @massspecpro The LCMS cost for proteomics will drop to US$0.01 per 1000 PSMs.
Wed Jan 01 13:42:41 +0000 2020KPRP:p, keratinocyte proline rich protein (H. sapiens) 🔗
Midsized low complexity protein; N-terminal acetylation; 5 high maf SAAVs: Q14H (0.2), R168H (0.3), C413S (0.5), P532T (0.01), G538V (0.01); 1 splice variant; mature form 2-579 [6,848 x] 🔗
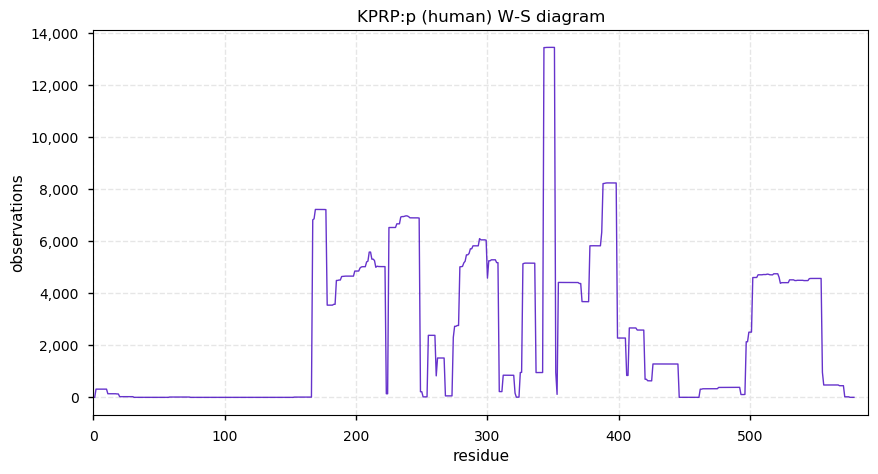
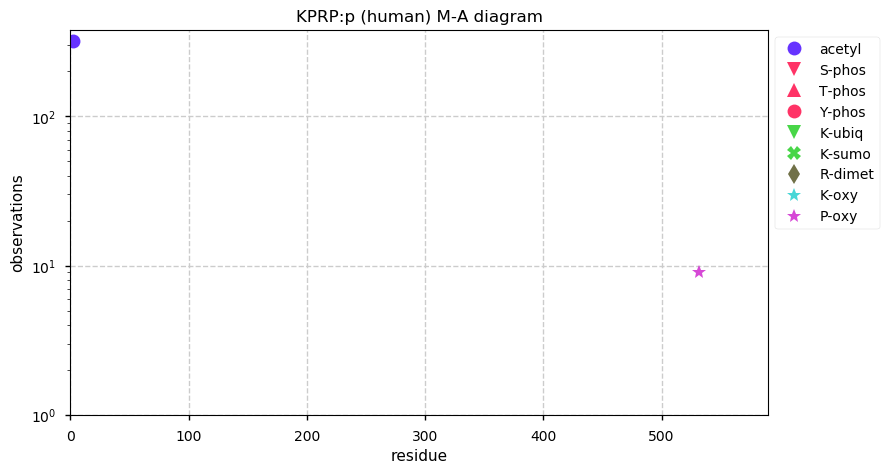
tweets = 4014