@norsivaeb Tweets 
| 2022
| 2021
| 2020
| 2019
| 2018
|
Tue Dec 31 19:49:34 +0000 2019@HFazelinia If you just want to check quickly, try running an MGF through idX 🔗 It will test for HLA peptides from these human microproteins, as well as the normal human proteome.
Tue Dec 31 19:35:12 +0000 2019& yes, there is a Fortran JSON API (there is a COBOL JSON API, too).
Tue Dec 31 19:24:50 +0000 2019To anyone who missed it the first time: this is a good exercise. Trying to do something with the data using Python or R (or Ruby or C++ or Fortran, etc) is also instructive.
Tue Dec 31 17:50:48 +0000 2019@slashdot A really interesting examination of the broad consequences of exploits aimed at the security models commonly used in cloud-based computing from a business perspective.
Tue Dec 31 15:42:11 +0000 2019@ljdursi I'm pretty sure there will never be a ribbon-cutting for any research that came up with a better approach to a "big" problem so that you could do the calculation on a Raspberry Pi.
Tue Dec 31 15:14:14 +0000 2019Commonly found in saliva and urine, due to the presence of shed keratinocytes.
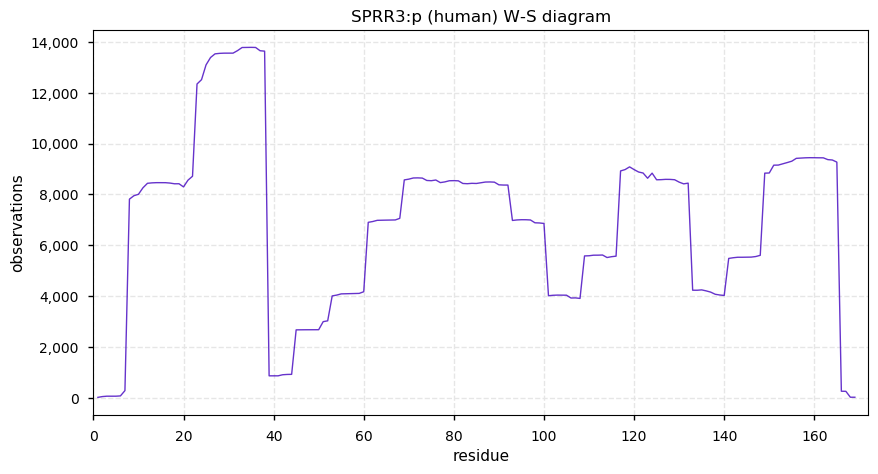
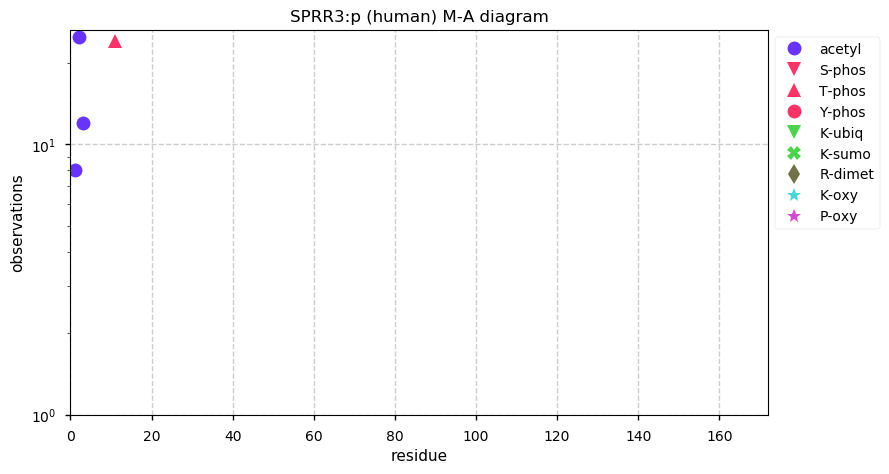
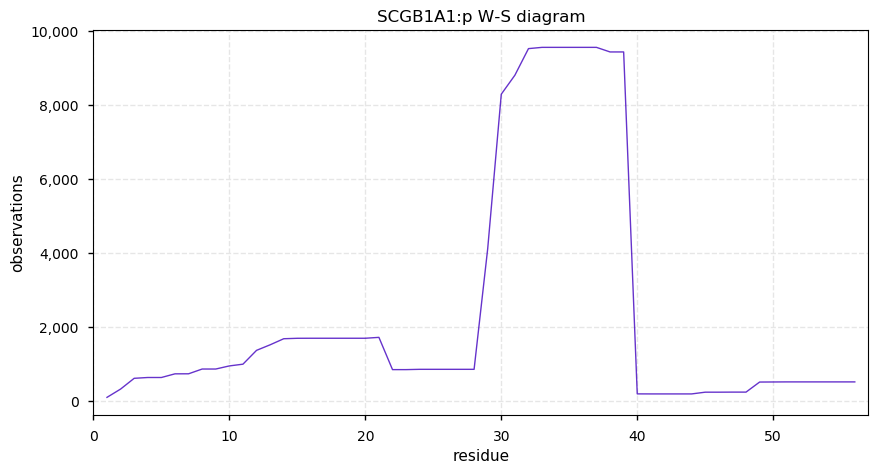
Tue Dec 31 15:11:34 +0000 2019SPRR3:p, small proline rich protein 3 (H. sapiens) 🔗 Small low-complexity cornified envelope protein; ragged N-terminus with acetylation; 3 high maf SAAVs: G105D (0.01), T147R (0.01), L149V (0.4); 1 splice variant; mature form (1,2,3)-169 [3,507 x] 🔗
Tue Dec 31 15:06:46 +0000 2019@neely615 @KentsisResearch @pwilmarth @ProteomicsNews Come on guys: complaining about shaky data analysis in proteomics papers is my shtick! And picking on a paper in SR-Nature is really shooting fish in a barrel.😀
Mon Dec 30 23:20:33 +0000 2019Just noticed a very bright Venus shining in through the office window. Time to go home.
Mon Dec 30 22:42:54 +0000 2019I am irrationally annoyed when I read a manuscript that uses the word "hypothesis" when they clearly should be using "postulate". I hope to get over this affliction in the New Year.
Mon Dec 30 16:32:45 +0000 2019@HFazelinia P.S. I added an entry to the microprotein FASTA that indicates whether the microprotein sequence PSMs overlap with a protein listed in human ENSEMBL
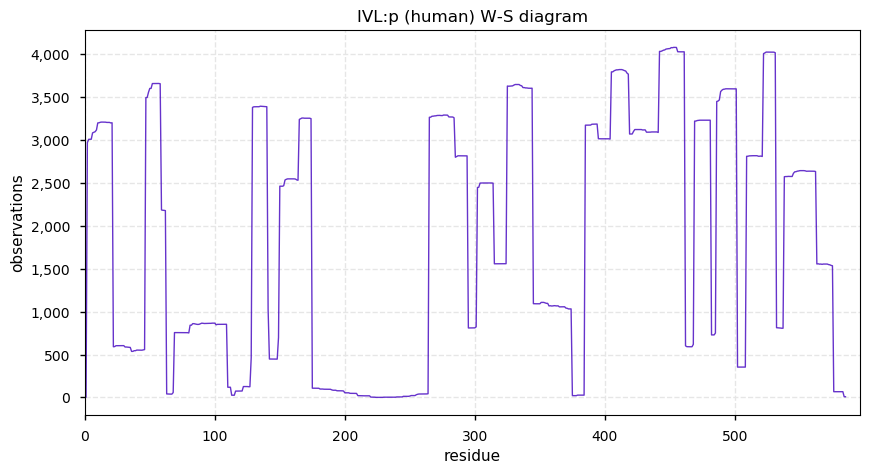
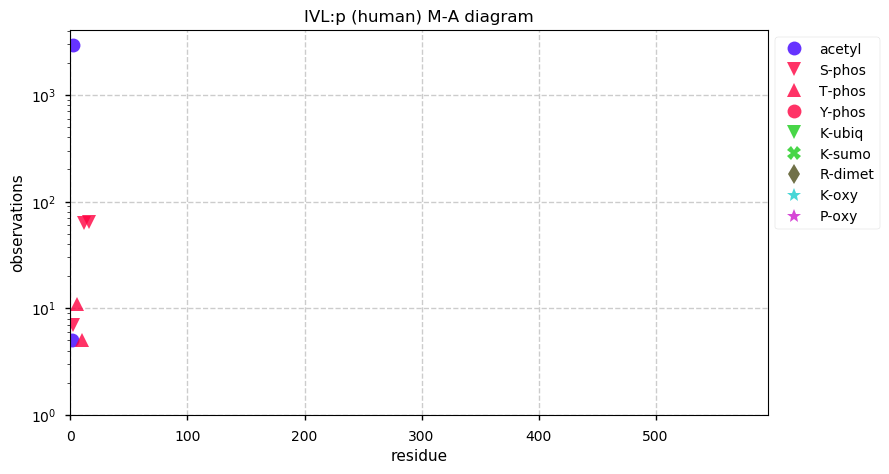
Mon Dec 30 13:40:08 +0000 2019Unlike LOR:p, IVL:p may be quite abundant in saliva and urine.
Mon Dec 30 13:38:26 +0000 2019IVL:p, involucrin (H. sapiens) 🔗 Midsized low-complexity cornified envelope protein; N-terminal acetylation & a compact phosphodomain; 2 high maf SAAVs: T113A (0.4), V480L (0.4); 1 splice variant; mature form 2-585 [4,502 x] 🔗
Mon Dec 30 02:33:10 +0000 2019@chrashwood @TheExpanseWR @uniprot Apolopoprotein? Otherwise the proteins all could be there, although the OBSC level looks high.
Mon Dec 30 01:18:38 +0000 2019Why is it that so few people seem to remember that decades begin in in years that end in "1", not "0"?
Sun Dec 29 23:05:31 +0000 2019@HFazelinia Now I have to figure out how to tell the Human Proteome Project guys they have 346,929 new protein sequences to check for ... 🥴
Sun Dec 29 20:58:29 +0000 2019@HFazelinia Let me know how it works out.
Sun Dec 29 18:37:11 +0000 2019@HFazelinia My first attempt at it is available at 🔗 You can either get the full list of all redundant microprotein sequences obtained from 🔗 or only the non-redundant microproteins that have been id'd in human MHC type I experiments.
Sun Dec 29 16:21:27 +0000 2019The cornified envelope (🔗) is made up of a set of proteins that can be low-complexity and resistant to trypsin digestion. Many also have a high density of disulphide bonds and other types of cross-links.
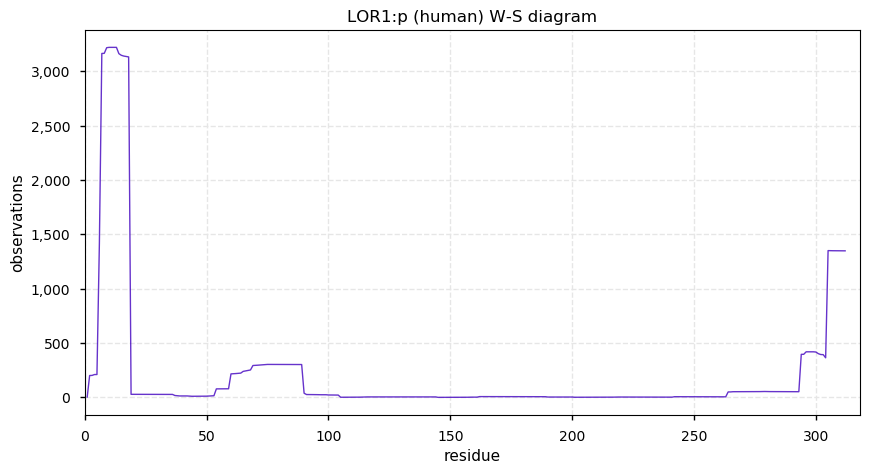
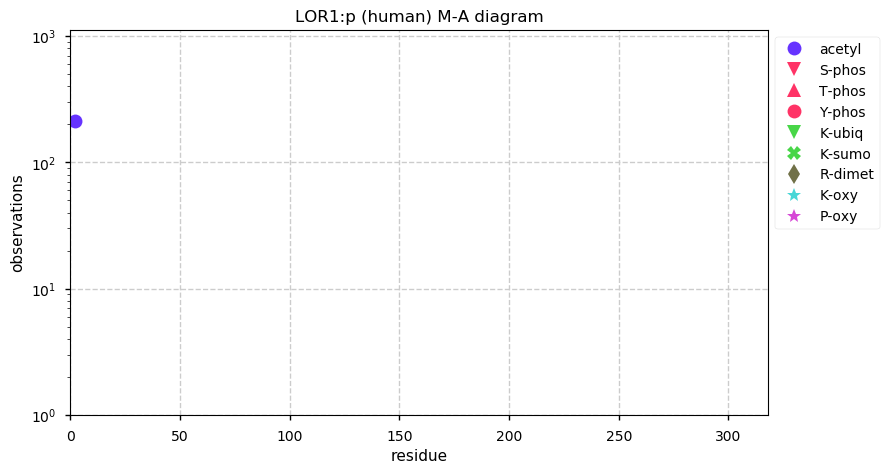
Sun Dec 29 16:10:35 +0000 2019LOR:p, loricrin (H. sapiens) 🔗 Small low-complexity cornified envelope protein; N-terminal acetylation; 1 high maf SAAVs: S29G (0.2); 1 splice variant; mature form 2-312 [695 x] 🔗
Sun Dec 29 14:03:23 +0000 2019@HFazelinia Would you like them in FASTA format or something else?
Sat Dec 28 18:51:44 +0000 2019If anyone is interested in checking an algorithm's ability to detect the normally rare arginine PTM that forms citrulline (deimidation), PXD012122 is a good data set to use.
Sat Dec 28 17:06:39 +0000 2019P.S., I have no axe to grind wrt the relevance of microproteins in MHC experiments. It would be much better for me if they did not exist; unfortunately they do.
Sat Dec 28 16:57:40 +0000 2019After reanalyzing a collection of ~ 200 good quality human MHC type I peptide MS/MS runs (includes various HLA alleles), there is good evidence for about 17,000 unique microprotein sequences contributing peptides that are presented.
Sat Dec 28 14:30:39 +0000 2019Is an email address valuable consideration? I'm leaning in the direction of "yes" at the moment.
Sat Dec 28 13:37:15 +0000 2019MTREX:p, Mtr4 exosome RNA helicase (H. sapiens) 🔗 Large nuclear protein; 53 K-acceptor sites for acetyl, ubiquitinyl and SUMOyl PTMs; no high maf SAAVs; 1 splice variant; mature form 2-1042 [33,858 x] 🔗
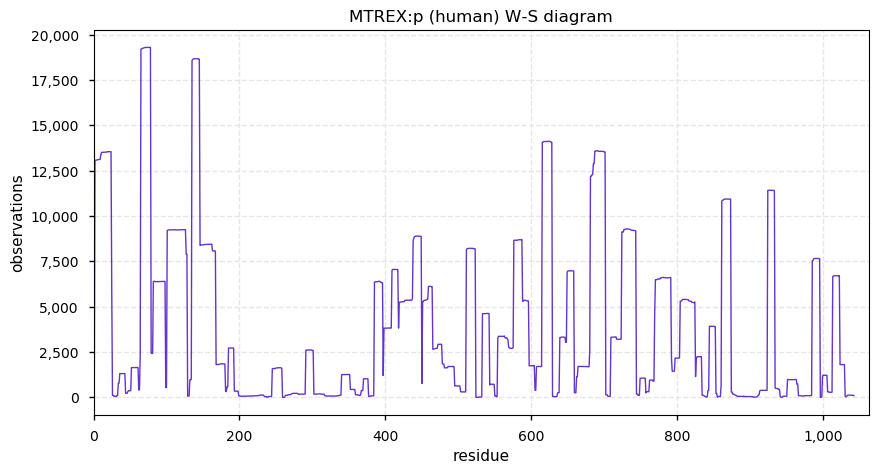
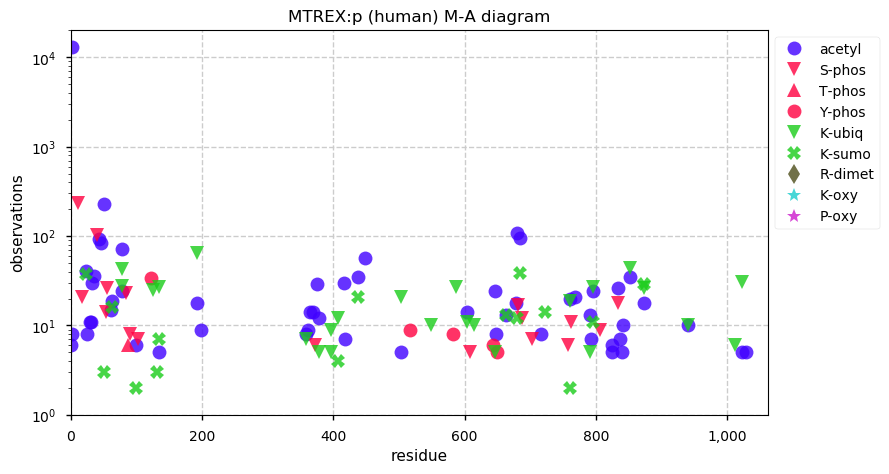
Fri Dec 27 18:26:35 +0000 2019@macro_momo The human & yeast MTREX proteins have rather different PTM site occupation distributions 🔗
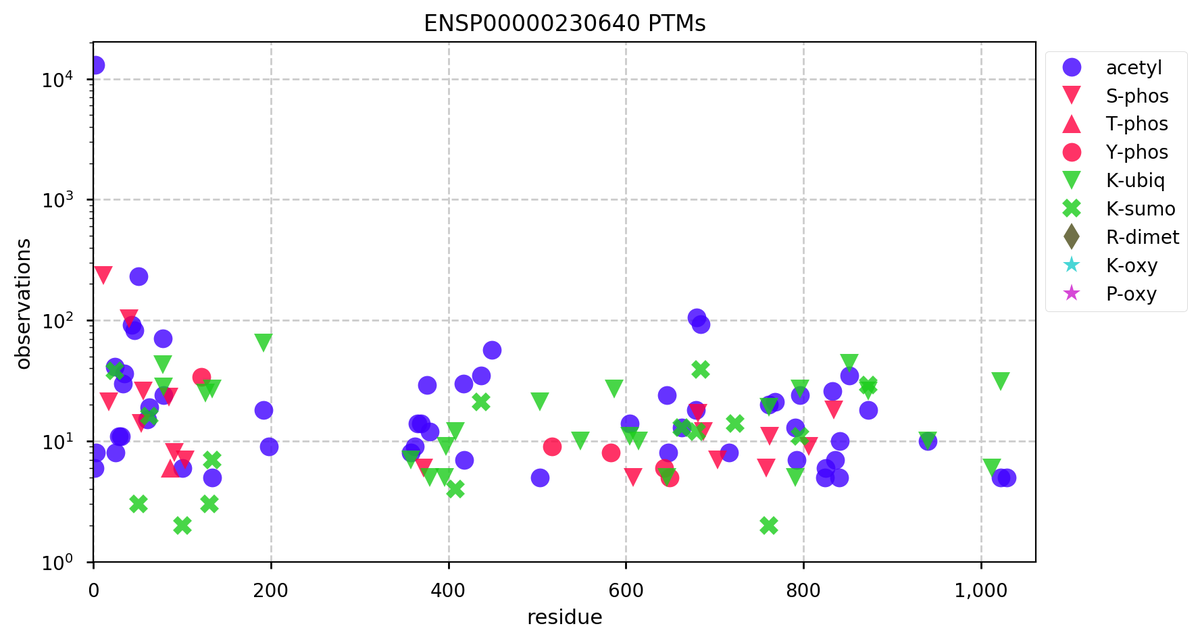
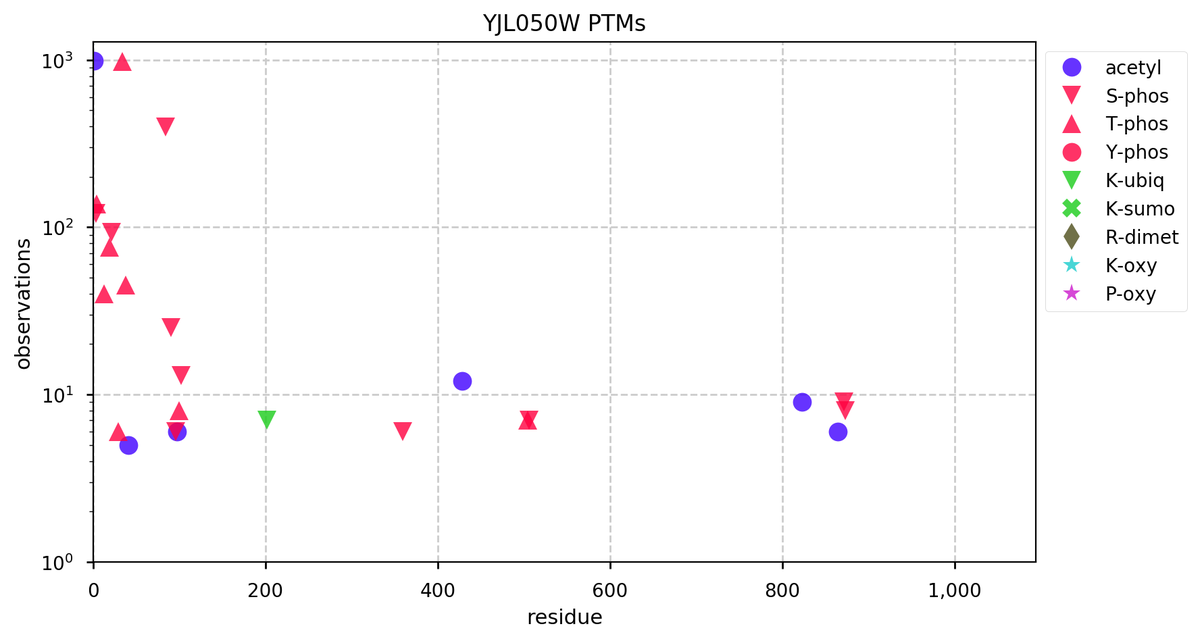
Fri Dec 27 13:59:59 +0000 2019DIS3:p, exosome endoribonuclease and 3'-5' exoribonuclease (Homo sapiens) 🔗 Midsized nuclear protein; many K-sites for acetyl, ubiquitinyl & SUMOyl PTMs; highest maf SAAVs: N269S (0.33), T326R (0.57); 1 splice variant; mature form 1-958 [22,168 x] 🔗
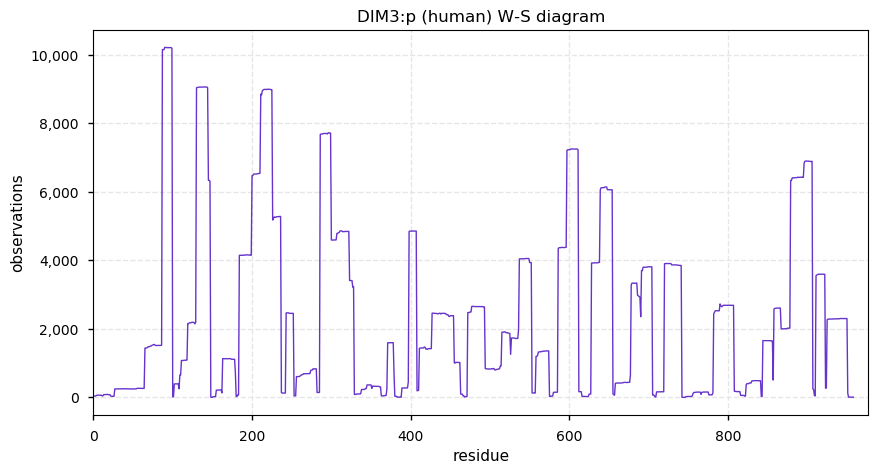
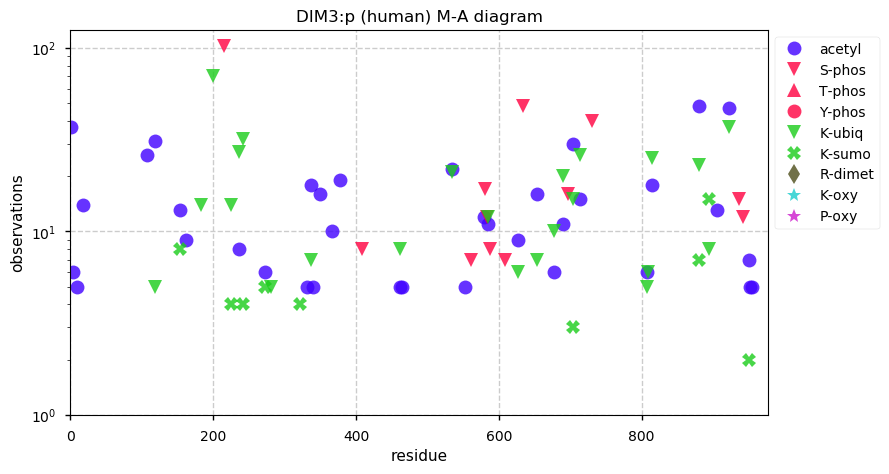
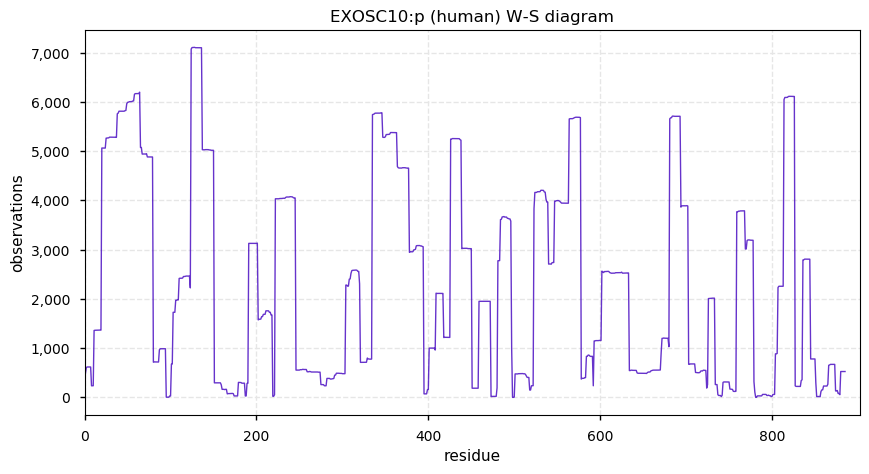
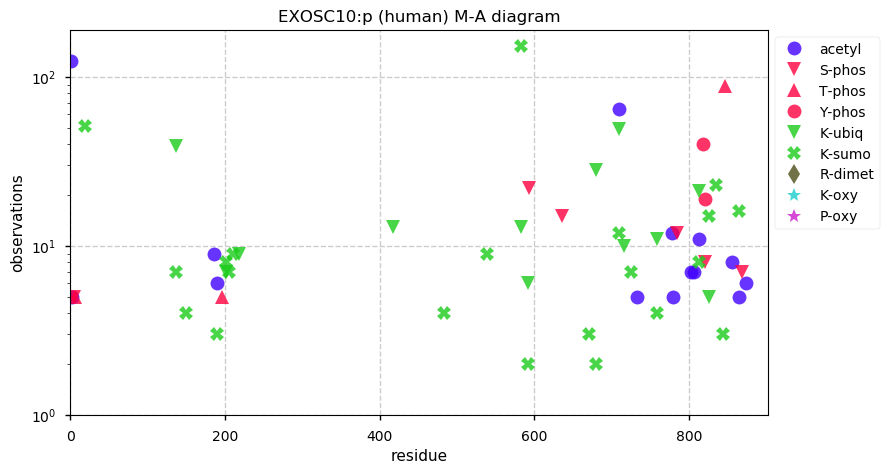
Thu Dec 26 15:01:21 +0000 2019EXOSC10:p, exosome component 10 (H. sapiens) 🔗 Midsized nuclear subunit; many ubiquitin and SUMO K-acceptor sites; no high maf SAAVs; 1 splice variant; mature form 1-20,449 [16,206 x] 🔗
Wed Dec 25 16:11:50 +0000 2019@jwoodgett @sxbegle Universities and governments value conventional thinking & behavior above all other things. Without exception.
Wed Dec 25 15:12:09 +0000 2019Interesting take on missing values: 🔗
Wed Dec 25 14:36:27 +0000 2019Is an email address valuable consideration?
Wed Dec 25 14:32:38 +0000 2019Dollars to doughnuts EXOSC9 is an exosome regulatory subunit.
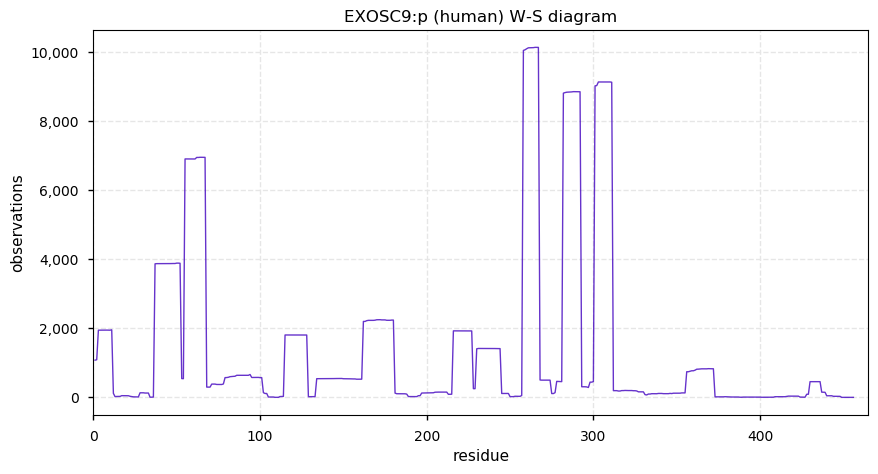
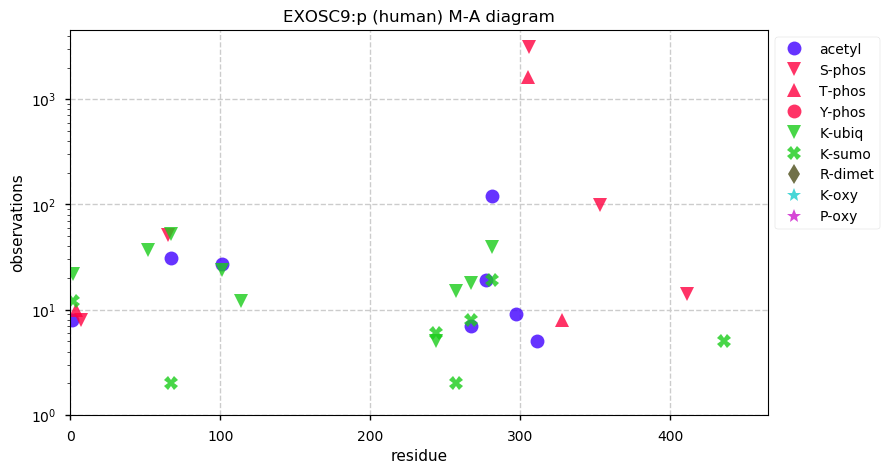
Wed Dec 25 14:27:43 +0000 2019EXOSC9:p, exosome component 9 (H. sapiens) 🔗 Small nuclear subunit; significant PTMs; 2 high maf SAAVs: I366V (0.1), S425T (0.1, found in HeLa); at least 2 splice variants; mature form 1-439 [16,206 x] 🔗
Tue Dec 24 21:18:02 +0000 2019It is the very model of a modern micro-protein 🔗
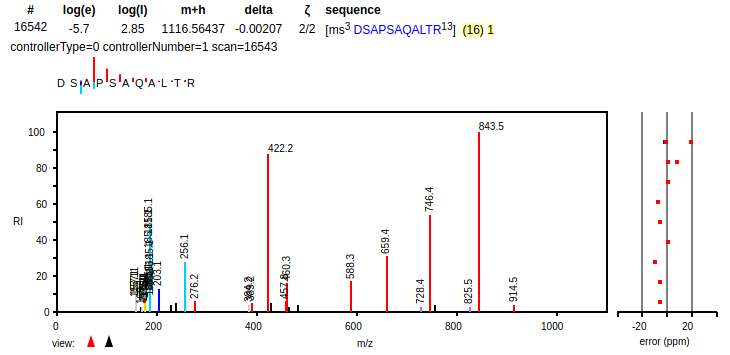
Tue Dec 24 17:38:16 +0000 2019Maybe suggesting Ulrich Laemmli or N. Leigh Anderson would have gotten some more traction.
Tue Dec 24 17:21:43 +0000 2019@astacus I don't know: I wouldn't be very offended if somebody told me I didn't deserve a Nobel prize. And all three did get some votes.
Tue Dec 24 16:09:29 +0000 2019A field-specific oddity of this position is that everyone I know who was involved in the early stages of genome sequencing is convinced they are owed at least 1 Nobel.
Tue Dec 24 16:01:23 +0000 2019Thanks to everyone who participated in this poll. The majority of respondents (58%) felt that nobody should be awarded a Nobel prize for proteomics.
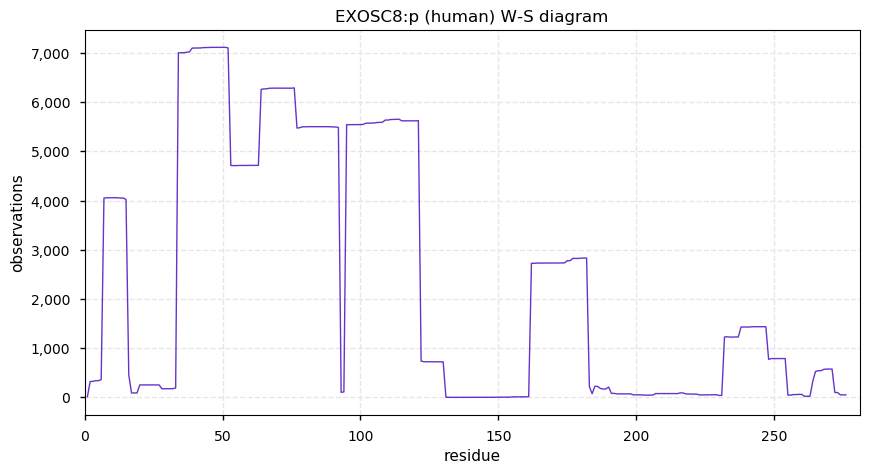
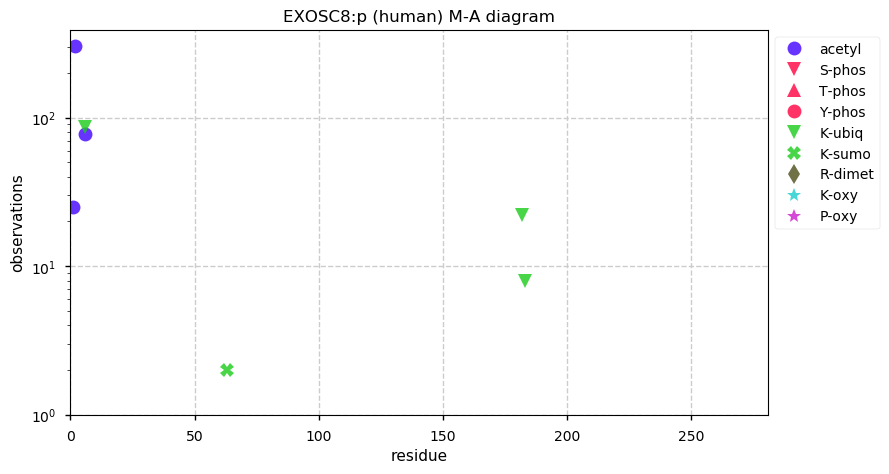
Tue Dec 24 14:57:16 +0000 2019EXOSC8:p, exosome component 8 (H. sapiens) 🔗 Small nuclear subunit; few PTMs; 1 high maf SAAV: P87H (0.01, homozygous in A-549 cells); 1 splice variant; mature form 2-276 [11,664 x] 🔗
Tue Dec 24 13:24:34 +0000 2019@dexivoje It was always just LD: no need for the I.
Mon Dec 23 15:37:31 +0000 2019@attilacsordas One of the people on the list probably isn't even eligible for a Nobel, because I know he has irritated an influential past Nobel laureate so much that any nomination (deserved or otherwise) would certainly be quashed.
Mon Dec 23 15:27:30 +0000 2019@attilacsordas I agree with you wrt to Nobels: they have always been political & do not represent any consensus among scientists. I don't like the idea of prizes for "science", at all. But the Nobels , with their faults, still resonate with the general public, political leaders & funders.
Mon Dec 23 14:47:33 +0000 2019@falcontigers Credit isn't really this issue: it is about the authors of a manuscript honestly acknowledging the original sources that influenced the work being described in the manuscript.
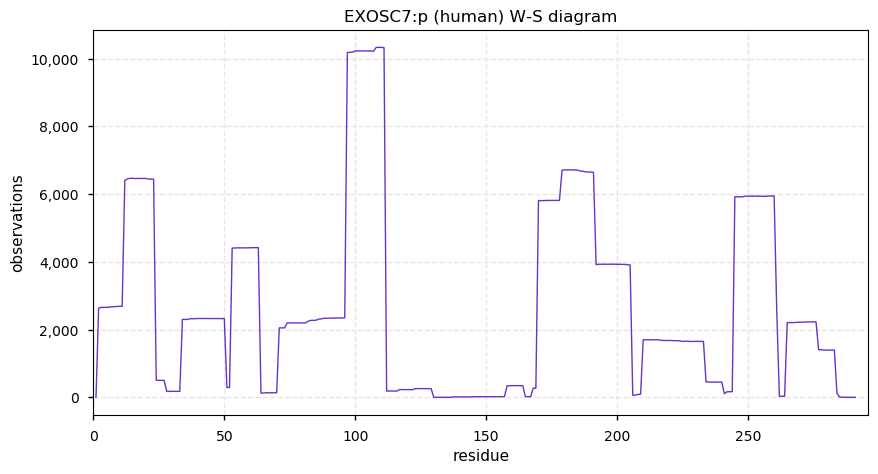
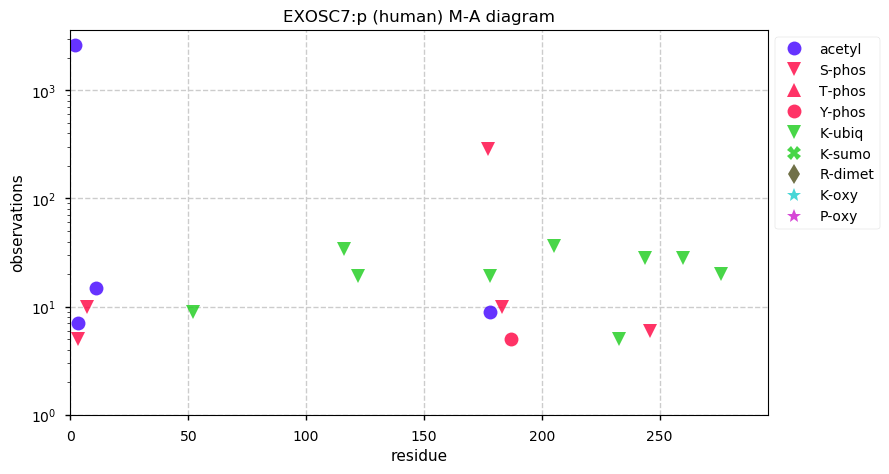
Mon Dec 23 14:38:36 +0000 2019EXOSC7:p, exosome component 7 (H. sapiens) 🔗 Small nuclear subunit; most significant PTMs: A2+acetyl and S177=phosphoryl; 2 high maf SAAV: T5A (0.05), V274L (0.3); 1 splice variant; mature form 2-291 [14,176 x] 🔗
Mon Dec 23 14:35:41 +0000 2019After the 2nd day of polling, "nobody" has become the majority candidate for a Nobel Prize in proteomics. Only 1 day left in the poll to voice your opinion.
Sun Dec 22 23:11:28 +0000 2019@astacus You should read Escoffier.
Sun Dec 22 16:32:30 +0000 2019"median ground truth-predicted correlation values between 0.2 and 0.5" 🔗
Sun Dec 22 15:34:17 +0000 2019Papers used to regularly credit people with ideas by citing a "private communication". Now, when many ideas become public through widely available platforms (like this one, blogs, GitHub, etc.), should people start citing the source of an idea as a "public communication"?
Sun Dec 22 13:41:51 +0000 2019After the 1st day of polling, "nobody" is currently the community's favorite choice for a Nobel Prize for proteomics. 2 days left to voice your opinion.
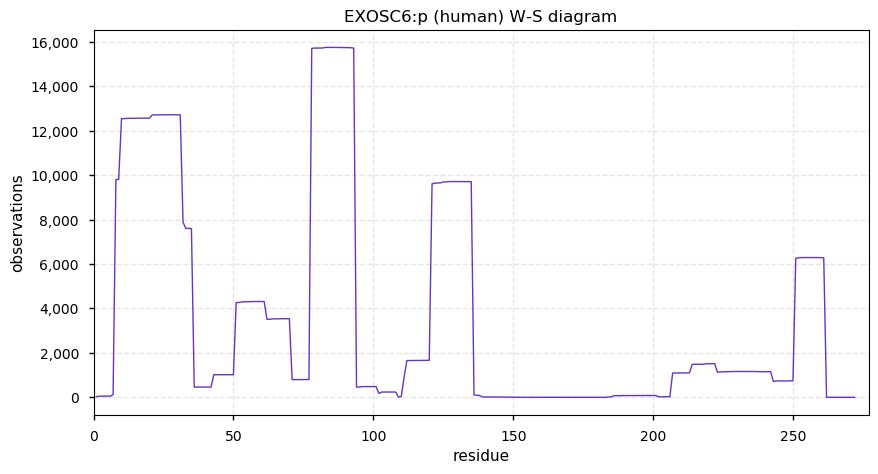
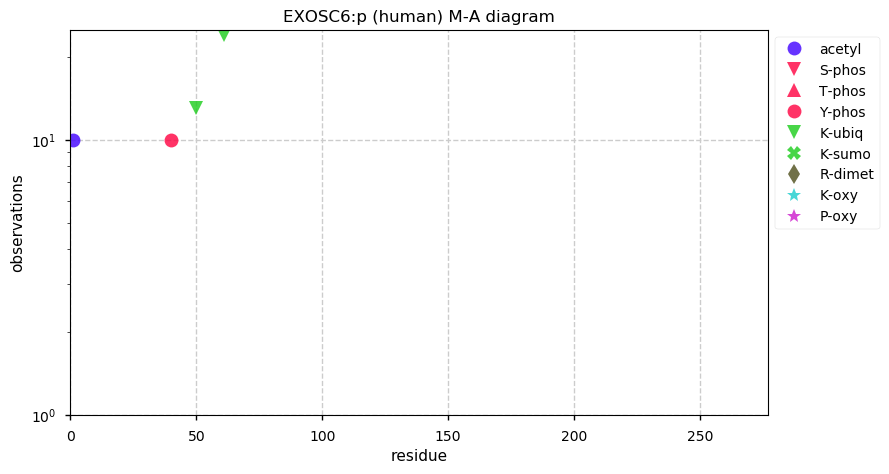
Sun Dec 22 13:34:53 +0000 2019EXOSC6:p, exosome component 6 (H. sapiens) 🔗 Small nuclear subunit; no significant PTMs; 1 high maf SAAV: R135P (0.01); 1 splice variant; mature form 1,2-272 [17,158 x] 🔗
Sat Dec 21 17:41:20 +0000 2019@UCDProteomics This is a pretty good essay about the current understanding of the curious world of how MHC type I peptides get chosen and presented
🔗
Sat Dec 21 17:03:45 +0000 2019@UCDProteomics They have only only showed up in MHC type I proteomics experiments, so far. Small, in this case, does means small though. Most protein prep protocols would lose them.
You can look through the FASTA files for human, mouse and rat at:
🔗
Sat Dec 21 15:52:30 +0000 2019Who deserves a Nobel Prize for proteomics (feel free to write in your own suggestions):
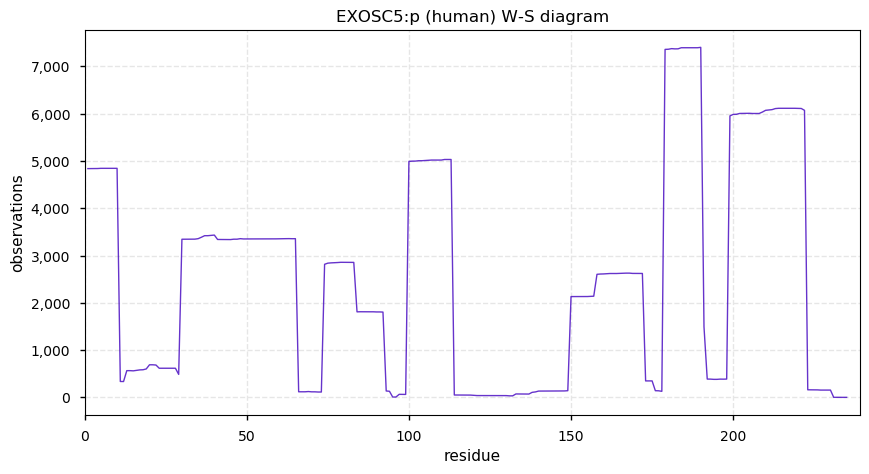
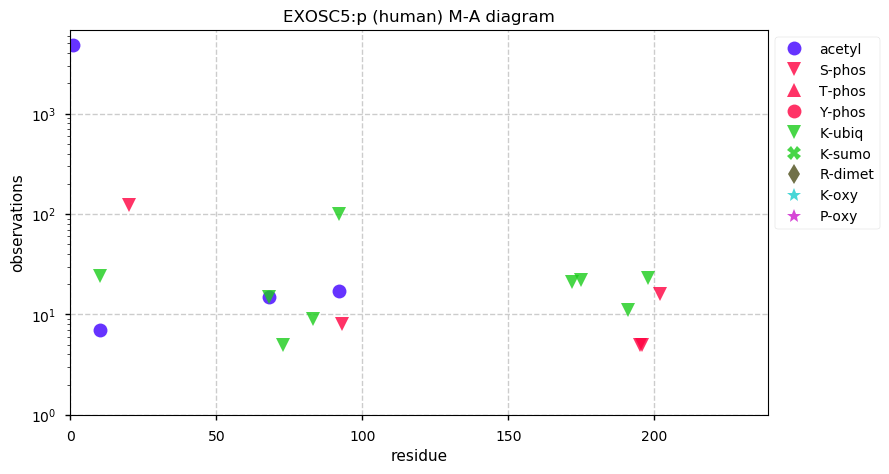
Sat Dec 21 14:16:09 +0000 2019EXOSC5:p, exosome component 5 (H. sapiens) 🔗 Small nuclear subunit; scattered ubiquitination; 1 high maf SAAV: T5M (0.4); 1 splice variant; mature form 1-235 [10,340 x] 🔗
Fri Dec 20 17:43:16 +0000 2019Another smORF gem. The full microprotein sequence:
>sorf|elkon_2015:851379|
MQRGPVTLVGR
the observed type I peptide:
MQRGPVTLVGR
Fri Dec 20 17:08:46 +0000 2019@ProteinMassSpec I press switches in a particular order that I dream up.
Fri Dec 20 16:05:27 +0000 2019Peptides from smORFs microproteins in MHC type I data are simply amazing (to me at least).
For example, for the full microprotein sequence:
>sorf|andreev_2015:501655|
MRATKPTVQK
and the observed type I peptide is:
RATKPTVQK
Yikes!🧐
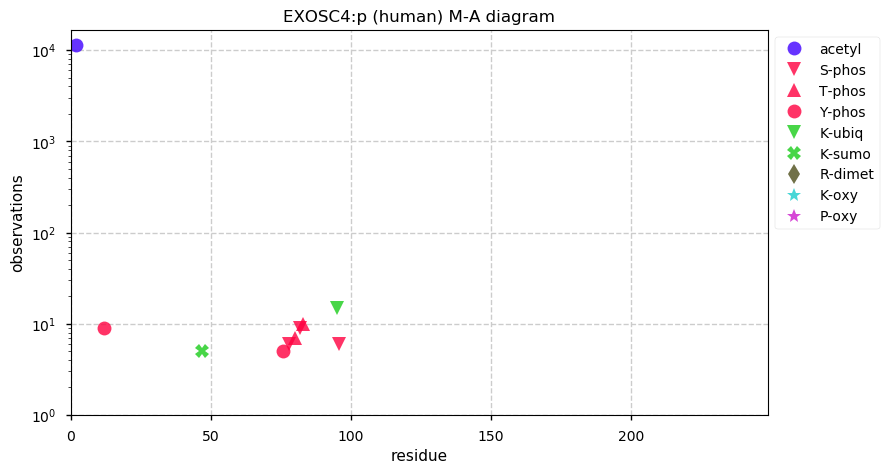
Fri Dec 20 13:24:56 +0000 2019EXOSC4:p, exosome component 4 (H. sapiens) 🔗 Small nuclear subunit; protein N-terminal acetylation; no observable SAAVs; 1 splice variant; mature form 2-245 [15,490 x] 🔗
Thu Dec 19 22:19:33 +0000 2019I'm going to go out on a limb and say that there may be some trends here ... 🔗
Thu Dec 19 14:32:21 +0000 2019@byu_sam @PastelBio That is true. Sometimes they even get decent recovery of cysteine containing peptides, although I've never been able to figure out why some groups do so much better at this than others.
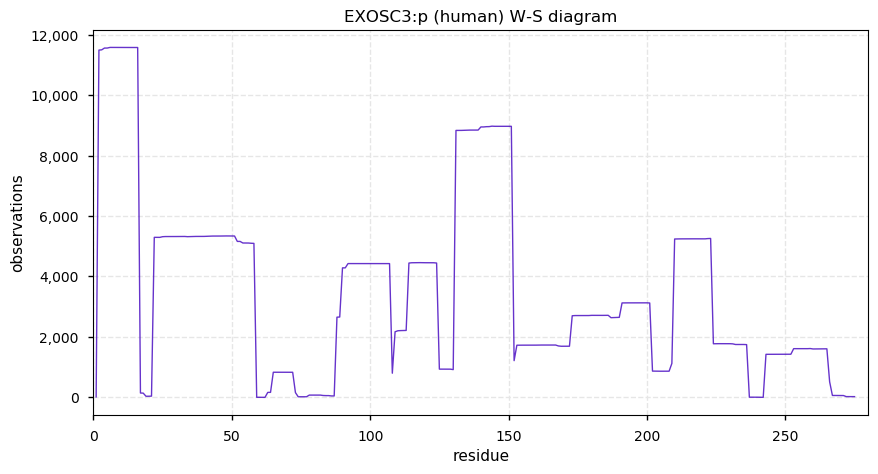
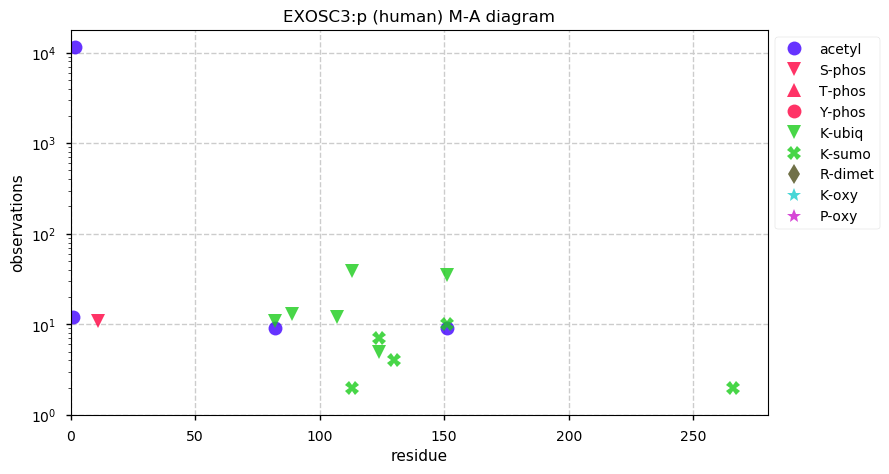
Thu Dec 19 14:08:21 +0000 2019EXOSC3:p, exosome component 3 (H. sapiens) 🔗 Small nuclear subunit; several complementary acetyl/ubiquitinyl/SUMOyl K-sites; 1 SAAV: Y225H (0.04); 1 splice variant; mature form 2-275 [15,023 x] 🔗
Wed Dec 18 17:04:44 +0000 2019@UCDProteomics Imputation is a screw-ball, Rube-Goldberg-style fix for a problem with many types of useful statistical tests. But, if you want to use the tests, you gotta use the fix.
Wed Dec 18 14:34:18 +0000 2019NOTE: exosome in this context refers to the 9 subunit RNA recycling complex (🔗), not the extracellular vesicles 🔗 of the same name.
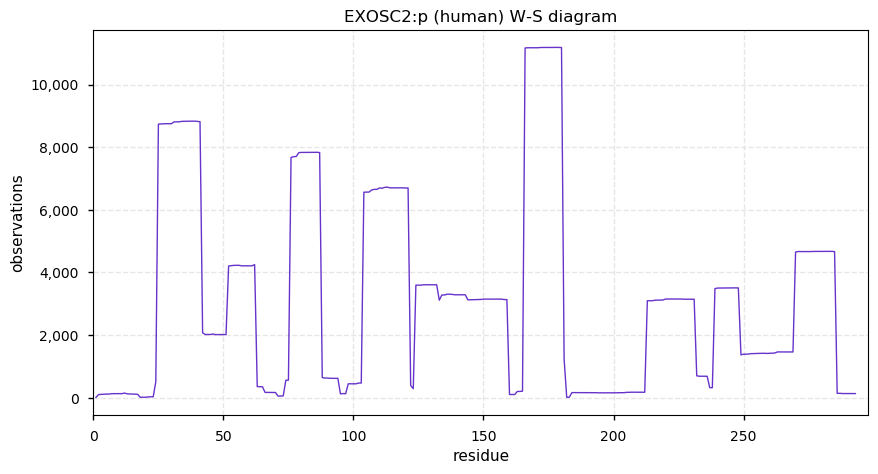
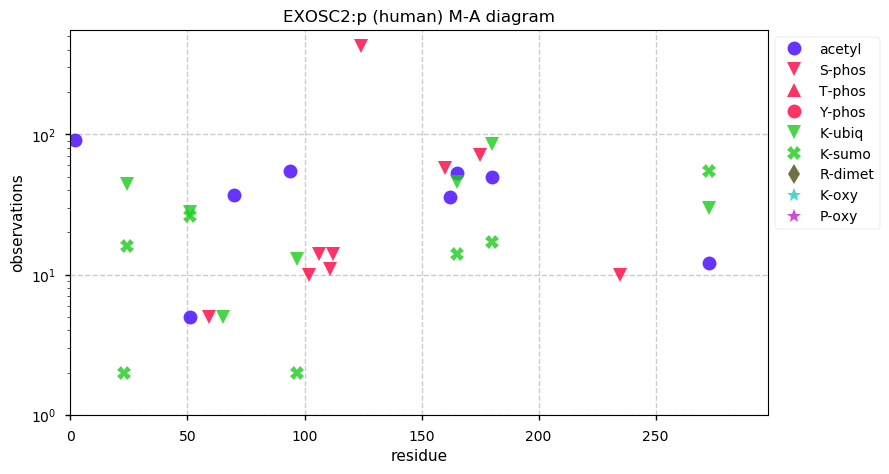
Wed Dec 18 14:29:18 +0000 2019EXOSC2:p, exosome component 2 (H. sapiens) 🔗 Small nuclear subunit; central S-phosphodomain & complementary acetyl/ubiquitinyl/SUMOyl K-sites; no high maf SAAVs; 1 splice variant; mature form 2-293 [15,724 x] 🔗
Tue Dec 17 15:20:57 +0000 2019@UCDProteomics @ProteomicsNews VERY SUBTLE .
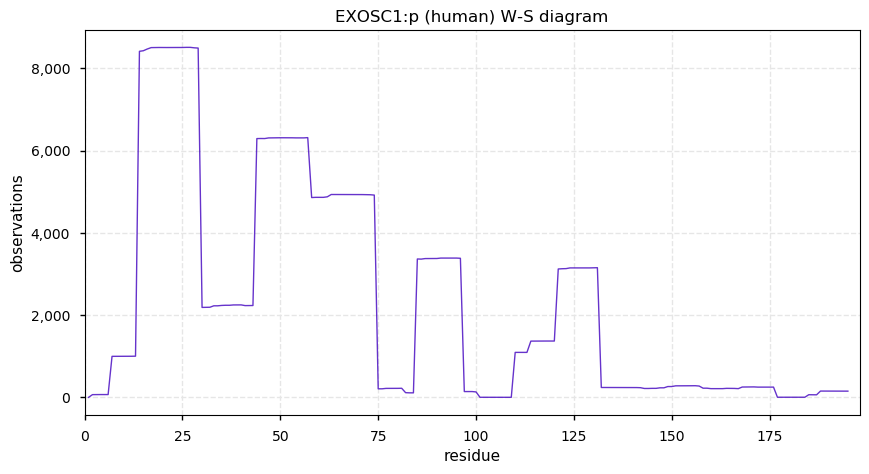
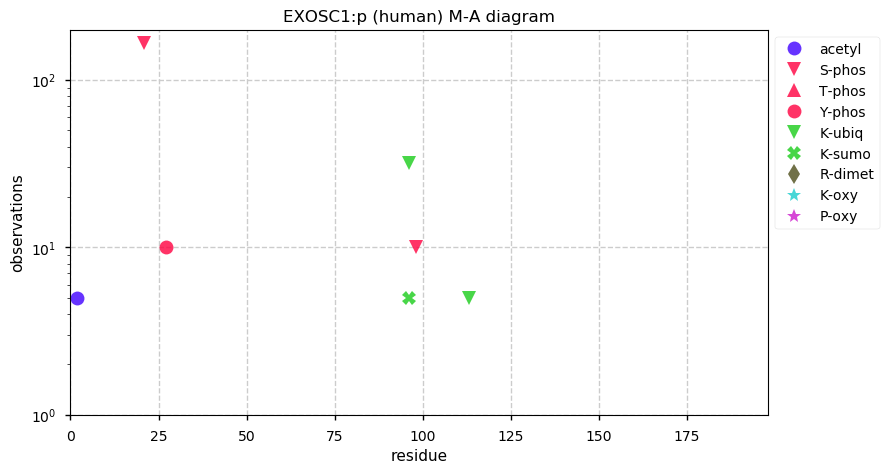
Tue Dec 17 14:27:25 +0000 2019EXOSC1:p, exosome component 1 (H. sapiens) 🔗 Small nuclear subunit; several PTMs; no high maf SAAVs; 1 splice variant; mature form 2-195 [9,790 x] 🔗
Tue Dec 17 14:09:06 +0000 2019Large population + GWAS = whatever was in the proposal
Mon Dec 16 15:52:13 +0000 2019And just to be a proteomics pedant: a list of protein sequences in a FASTA file is not a "database" any more than an HPLC is an ion source.
Mon Dec 16 15:42:54 +0000 2019After many years of working with FASTA formatted files, the only nice thing I can say about FASTA is that it must have worked great with punch cards.
Mon Dec 16 14:10:38 +0000 2019No one seems to be quite sure what happened with LIG2, or at the very least would rather not talk about it ...
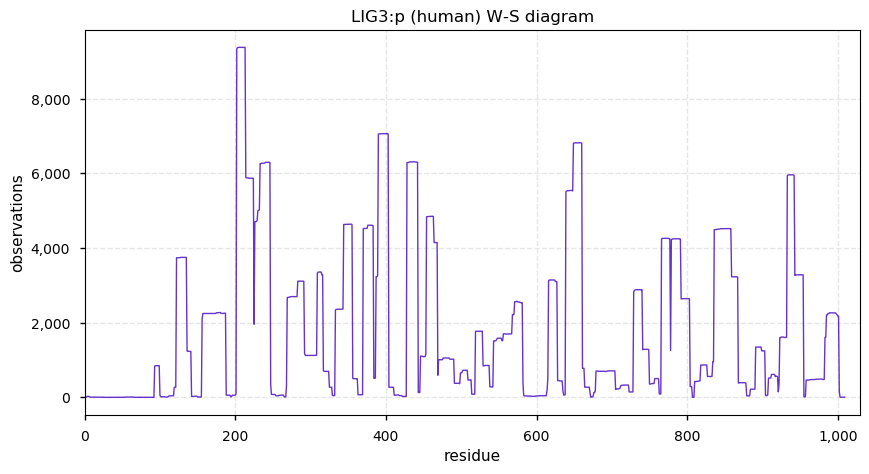
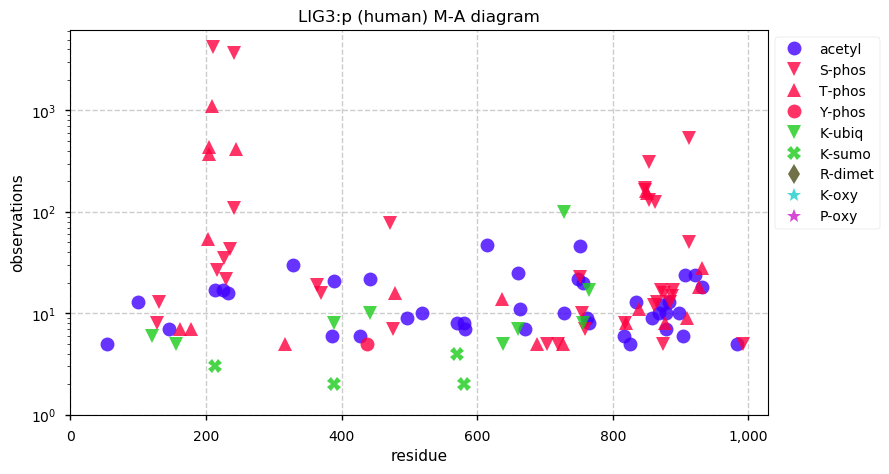
Mon Dec 16 14:09:04 +0000 2019LIG3:p, DNA ligase 3 (H. sapiens) 🔗 Midsized mitochondrial subunit; several high occupancy S/T-phosphodomains; 1 SAAV: R867H (maf=0.02); mature form 93-1009 [18,668 x] 🔗
Sun Dec 15 22:06:26 +0000 2019@nesvilab How would MSFragger or Sequest handle this situation?
Sun Dec 15 18:15:25 +0000 2019but not update the "missed sites counter" for either one, leaving it at 0 for EGPEKPNKK
3/3
Sun Dec 15 18:13:09 +0000 2019trypsin will cleave to about 1:1 EGPEKPNK:EGPEKPNKK, but it will not convert the longer form into the shorter one once it is formed. Because of this, I would suggest that an search engine should consider all three peptides to be potential solutions ...
/2
Sun Dec 15 18:09:14 +0000 2019Thanks to everyone who participated in this poll. The peptide has two sites that may result in peptides under experimental conditions:
1. EGPEK—P
2. EGPEKPNK—K
Both of these cleavages are encumbered though, as trypsin cleaves at a slow rate with a C-terminal P and ...
/1
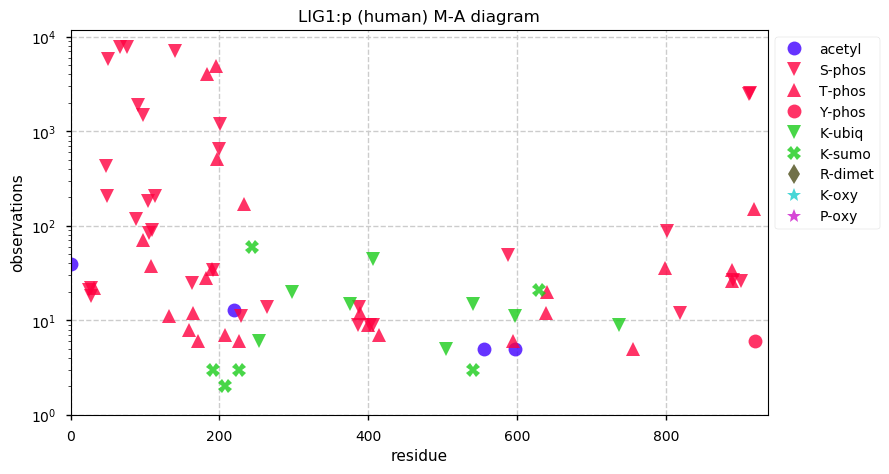
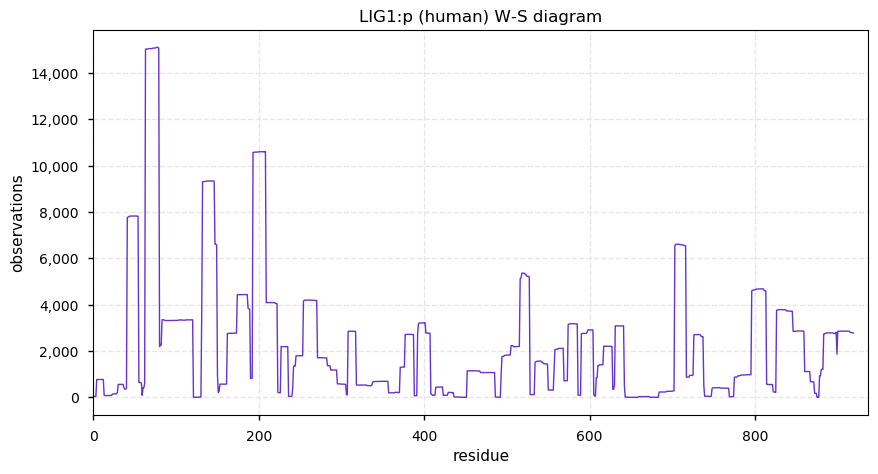
Sun Dec 15 14:48:15 +0000 2019LIG1:p, DNA ligase 1 (Homo sapiens) 🔗 Midsized nuclear subunit; many high occupancy S phosphosites; many low maf SAAVs; 1 splice variant; mature form 1-919 [18,557 x] 🔗
Sat Dec 14 17:32:56 +0000 2019Proteomics search engines normally consider the number of "missed" tryptic cleavage sites an important parameter for an identification run. How many "missed" sites should a search engine count for the following tryptic peptide:
EGPEKPNKK
Sat Dec 14 14:41:59 +0000 2019LIG4:p, ligase IV, DNA, ATP-dependent (H. sapiens) 🔗 Midsized nuclear subunit; few PTMs; 2 SAAVs: A3V (0.05), T9I (0.15); common in tissues and cell lines; mature form 2-911 [3,978 x] 🔗
Fri Dec 13 19:06:06 +0000 2019@aldosantin Budgeting requires open, public processes. Spending, not so much.🧐
Fri Dec 13 18:09:18 +0000 2019Does anyone know the exact moment at which graphs showing trends in data became "analytics"?
Fri Dec 13 17:23:15 +0000 2019@DonMartinCTV Apparently including a palette of allowed outer wear colours.
Fri Dec 13 15:20:18 +0000 2019I am growing to regret ever installing Mono: it takes forever to update
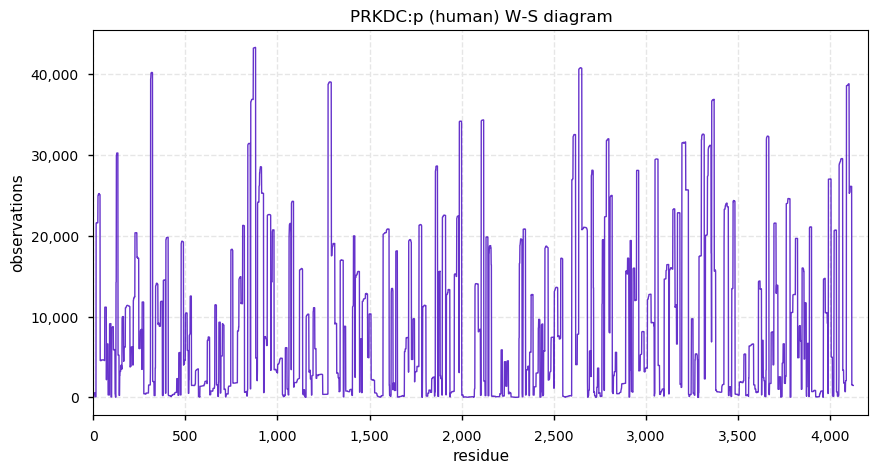
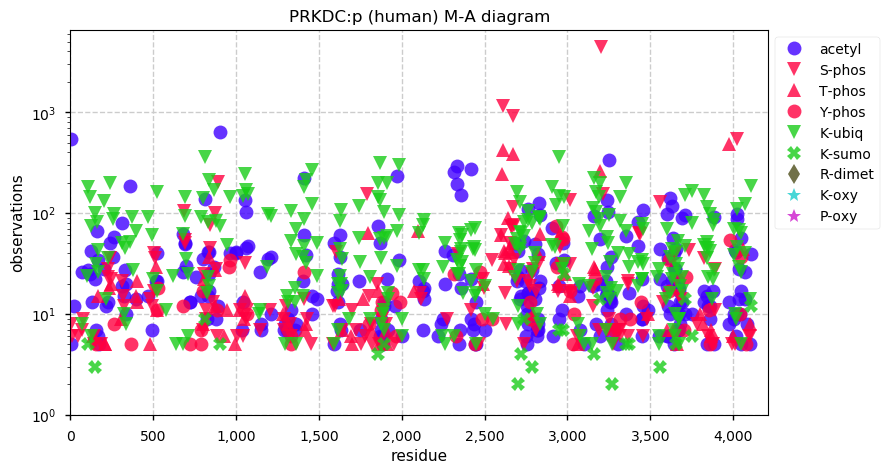
Fri Dec 13 14:14:44 +0000 2019PRKDC:p (XRCC7) is also part of the part of the non-homologous end-joining DNA repair pathway, like its other highly modified colleagues XRCC5 & 6. Why these proteins are have so many PTMs is a mystery, as is how to study PTMs in proteins with lots of them.
Fri Dec 13 14:10:06 +0000 2019PRKDC:p, protein kinase, DNA-activated, catalytic subunit (H. sapiens) 🔗 Very large nuclear subunit; aka XRCC7; heavily modified (513 PTM sites); many observable SAAVs; abundant in tissues and cell lines; mature form 2-4128 [62,406 x] 🔗
Thu Dec 12 22:34:35 +0000 2019PXD013453: good chance to test out my "# of phosphosites nearing an asymptote" hypothesis
Thu Dec 12 18:51:54 +0000 2019@Karl_Mechtler Faster.
Thu Dec 12 15:39:21 +0000 2019"Using good passwords and 2FA is good advice, but better advice is to never put networked cameras or microphones in your home, ever. " — Cory Doctorow 🔗
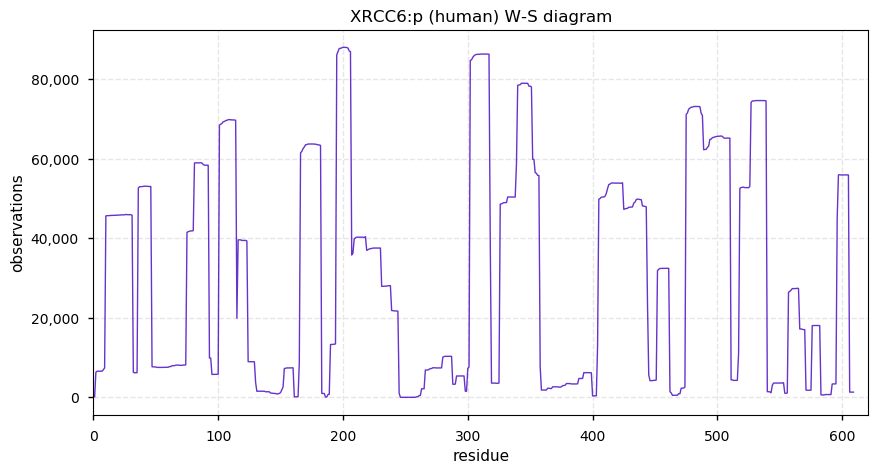
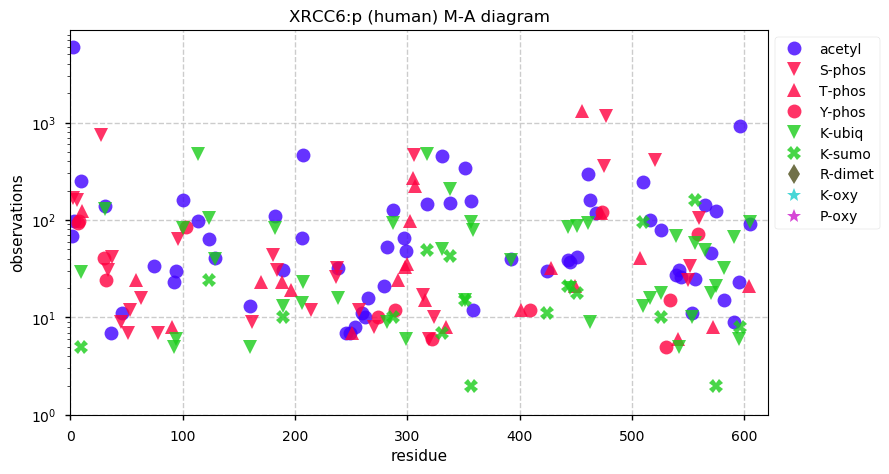
Thu Dec 12 15:05:47 +0000 2019It may be worth noting that XRCC5 & XRCC6 are subunits normally forming a heterodimeric protein –Ku– part of the non-homologous end-joining DNA repair pathway. Even with their garish PTM distributions, they have been observed about the same number of times (57,020:58,073)
Thu Dec 12 14:57:46 +0000 2019XRCC6:p, X-ray repair cross complementing 6 (H. sapiens) 🔗 Midsized nuclear subunit; heavily modified (125 PTM sites): 43 K's that may be either acetylated or ubiquitinylated; no SAAVs; abundant in tissues & cell lines; mature form 2-609 [58,022 x] 🔗
Wed Dec 11 17:24:48 +0000 2019@jwoodgett Are you implying that "Nature Pigeon Fancier" isn't a serious journal?
Wed Dec 11 15:05:25 +0000 2019@EricTopol @RuneLinding @salkinstitute @nchembio @alansaghatelian All of the methods I use to determine if subsets of PSMs are true checked out properly, so I have no hesitation about supporting the idea the smORF microproteins contribute to the peptides found in MHC type I experiments.
3/3
Wed Dec 11 15:02:20 +0000 2019@EricTopol @RuneLinding @salkinstitute @nchembio @alansaghatelian I also included (as usual) protein sequences from all ENSEMBL splices, common human viruses, human endogenous retroviruses, fetal bovine serum and cRAP. The results were that 1-3.5% of the PSMs correspond to smORF proteins.
/2
Wed Dec 11 15:00:07 +0000 2019@EricTopol @RuneLinding @salkinstitute @nchembio @alansaghatelian hyperbole update: I sampled LC/MS/MS runs from 4 good quality, public human MHC type I data sets and analyzed them including the translated human smORF sequence information from 🔗 in the analysis.
/1
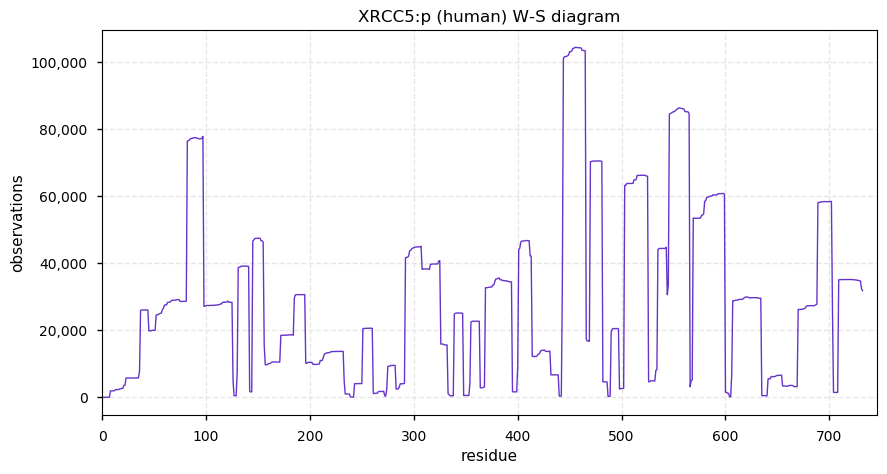
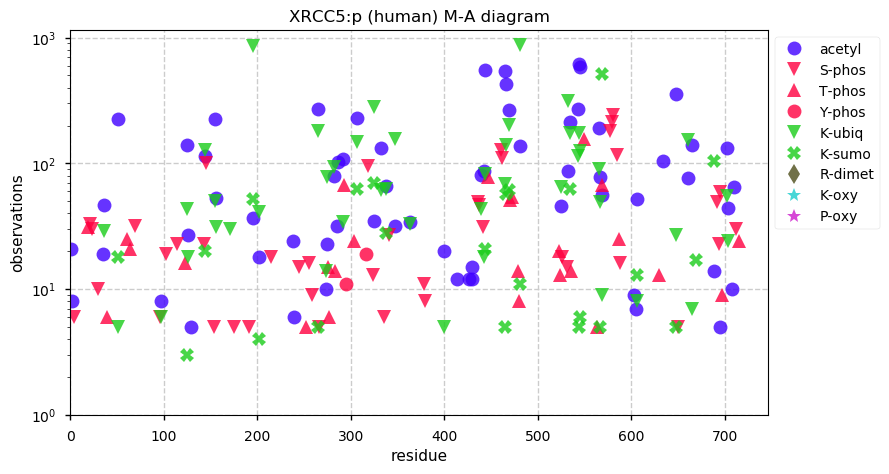
Wed Dec 11 13:34:28 +0000 2019XRCC5:p, X-ray repair cross complementing 5 (H. sapiens) 🔗 Midsized nuclear subunit; 130 PTMs sites: 45 K's may be acetylated or ubiquitinylated, 21 of which may be SUMOylated; no SAAVs; abundant in tissues & cell lines; mature form 1-732 [57,020 x] 🔗
Tue Dec 10 22:27:32 +0000 2019Thanks to everyone who participated in this poll. My personal answer would be "no": until sample preparation methods improve, the current set of human S/T/Y phosphorylation sites is very near an asymptote.
Tue Dec 10 16:36:20 +0000 2019@EricTopol @RuneLinding @salkinstitute @nchembio @alansaghatelian But there isn't enough evidence to justify the hyperbole surrounding (& included in) the article.
Tue Dec 10 16:35:32 +0000 2019@EricTopol @RuneLinding @salkinstitute @nchembio @alansaghatelian The hypothesis that there exist small translated ORFs that provide "self" MHC type I peptides is interesting & some preliminary evidence is provided. Enough so that a FASTA from 🔗 should be included when looking at MHC type I data to test the idea.
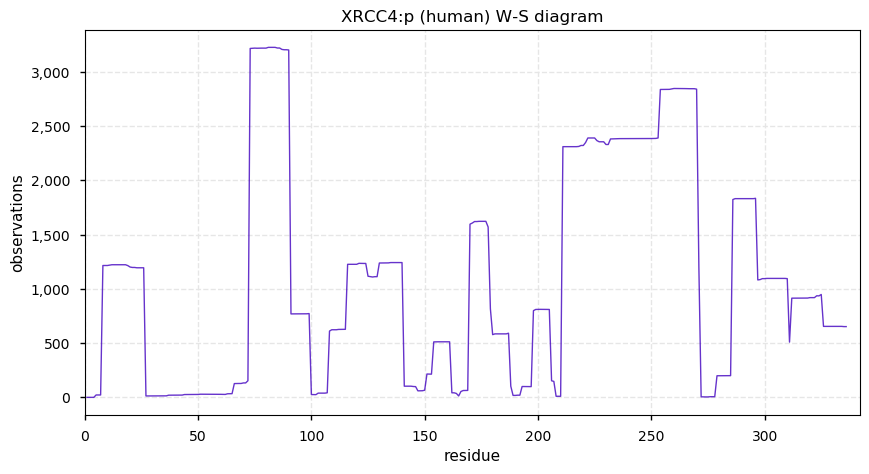
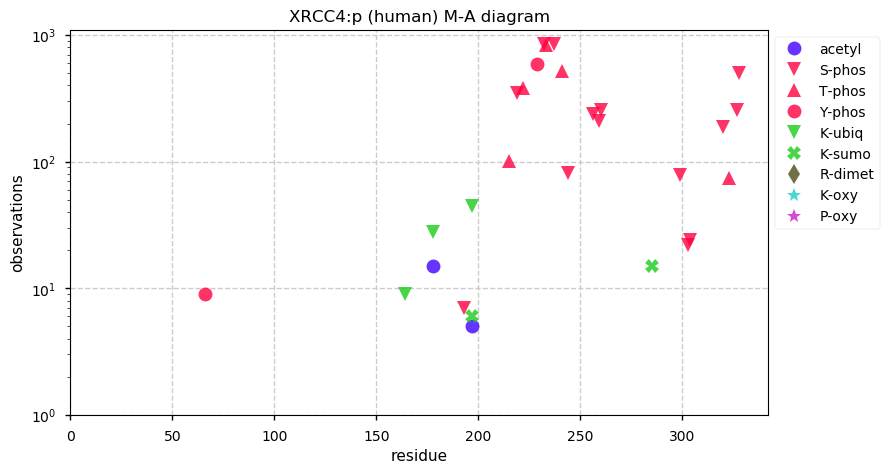
Tue Dec 10 13:34:55 +0000 2019In XRCC4:p, the PTMs are all localized to the C-terminal stalk domain, based on the crystal structure of unmodified protein: 🔗
Tue Dec 10 13:23:12 +0000 2019XRCC4:p, X-ray repair cross complementing 4 (H. sapiens) 🔗 Small nuclear subunit; significant C-terminal phosphodomains; SAAVs: I134T (0.01), A247S (0.03); mature form 1-336 [6,636 x] 🔗
Mon Dec 09 21:10:06 +0000 2019There have been several large data reanalysis projects on detectable, proteome-wide human S/T/Y phosphorylation acceptor sites. They have all come in at ~120 k reliable unique sites. Will taking more data generate significantly more unique sites?
Mon Dec 09 17:02:30 +0000 2019Junk science + bad reporting = HEADLINES!
🔗
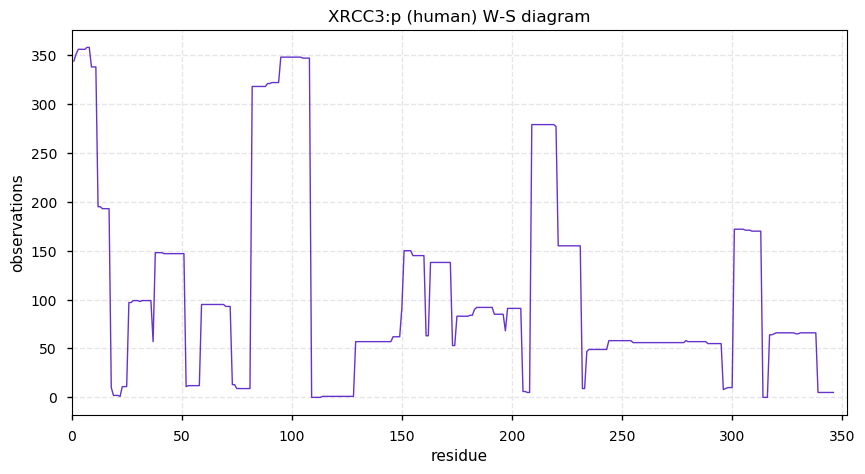
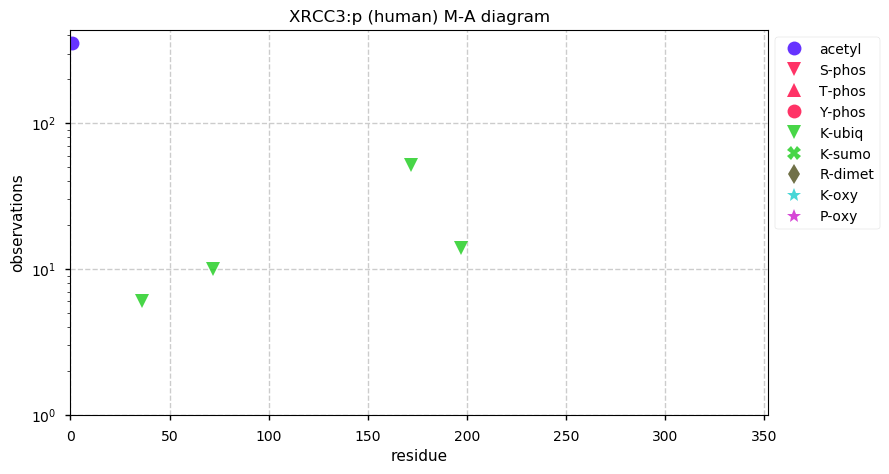
Mon Dec 09 13:39:41 +0000 2019XRCC3:p, X-ray repair cross complementing 3 (H. sapiens) 🔗 Small protein; only PTM: N-terminal acetylation; no SAAVs; found in mitochondrial studies; mature form 1-346 [1,285 x] 🔗
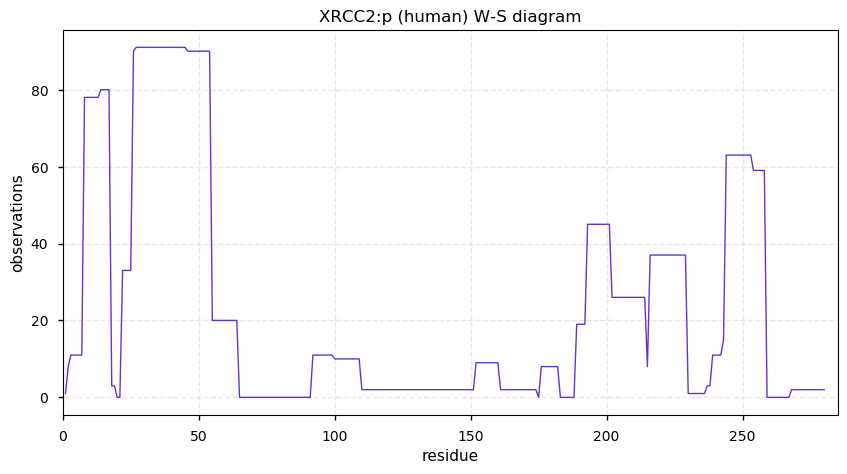
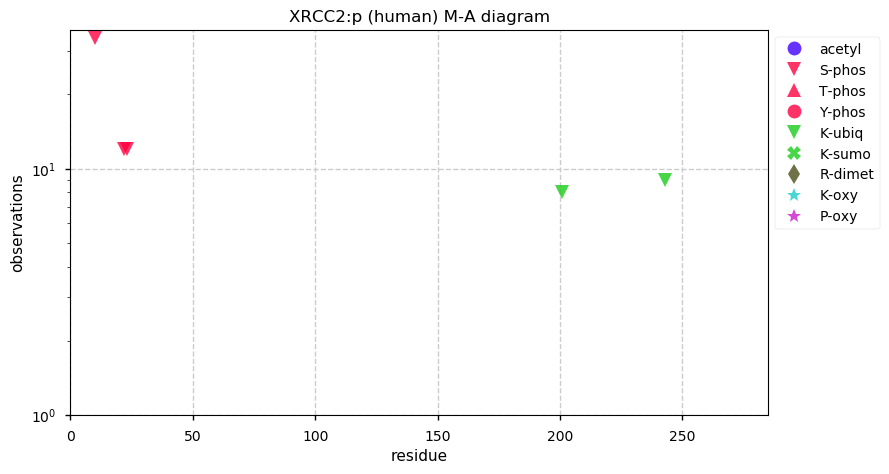
Sun Dec 08 14:35:56 +0000 2019XRCC2:p, X-ray repair cross complementing 2 (H. sapiens) 🔗 Small nuclear protein; limited PTMs; no SAAVs; very rarely observed in tissue; found in high sensitivity cell line studies; mature form 2-280 [248 x] 🔗
Sat Dec 07 18:31:21 +0000 2019@jwoodgett @CIHR_IRSC I suspect there is a well-deserved concern that a set of professional POs would interfere with political interference.
Sat Dec 07 17:36:42 +0000 2019I guess this rather specialized application isn't top-of-mind for developers.
Sat Dec 07 17:01:32 +0000 2019This is truly a failure of design. If you have ever spent time in an ICU or emergency room, the chorus of ignored, useless alarms can be overwhelming. 🔗
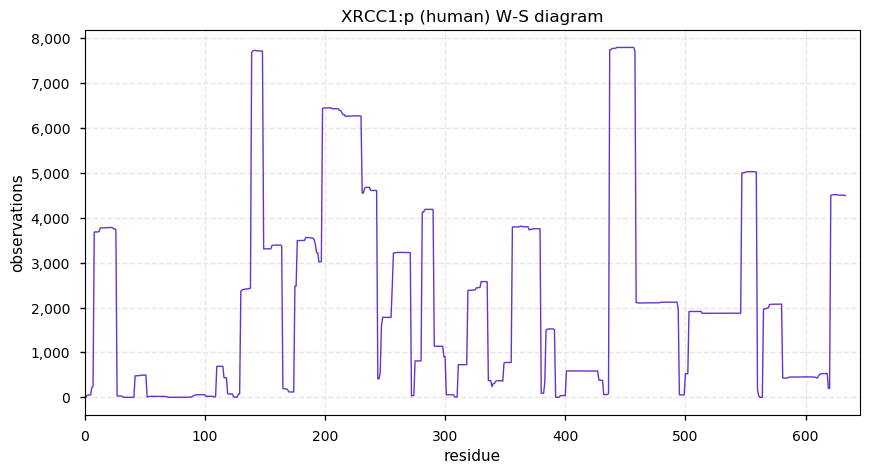
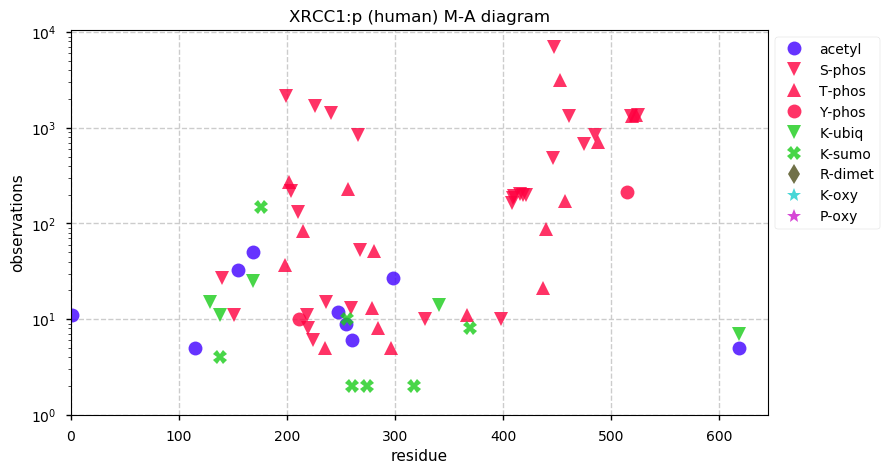
Sat Dec 07 13:59:56 +0000 2019XRCC1:p, X-ray repair cross complementing 1 (H. sapiens) 🔗 Midsized nuclear protein; 2 large phosphodomains; numerous SAAVs: R194W (0.12), R280H (0.07), T304A (0.03), Q399R (0.26) ; mature form 1-633 [17,274 x] 🔗
Fri Dec 06 18:42:45 +0000 2019@ProtifiLlc My own experience with indexing for these 2 types of samples has been better than I expected.
Fri Dec 06 18:41:38 +0000 2019@ProtifiLlc I was just wondering how well these strategies worked compared to other methods and whether there was any advantage to using them.
Fri Dec 06 18:16:59 +0000 2019Does anybody have any experience trying to use indexed and/or open search strategies on HLA type I or type II peptides?
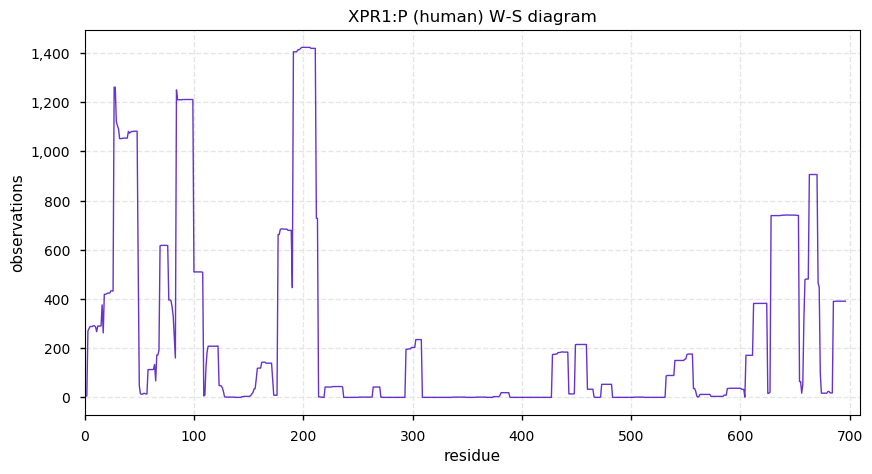
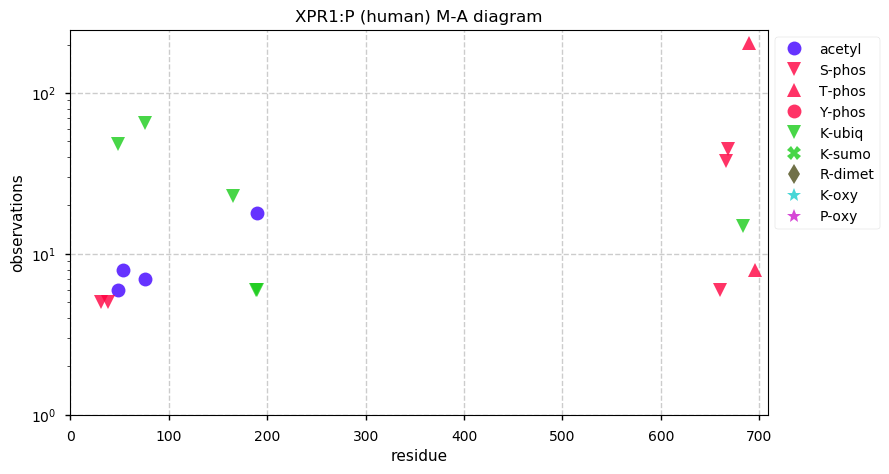
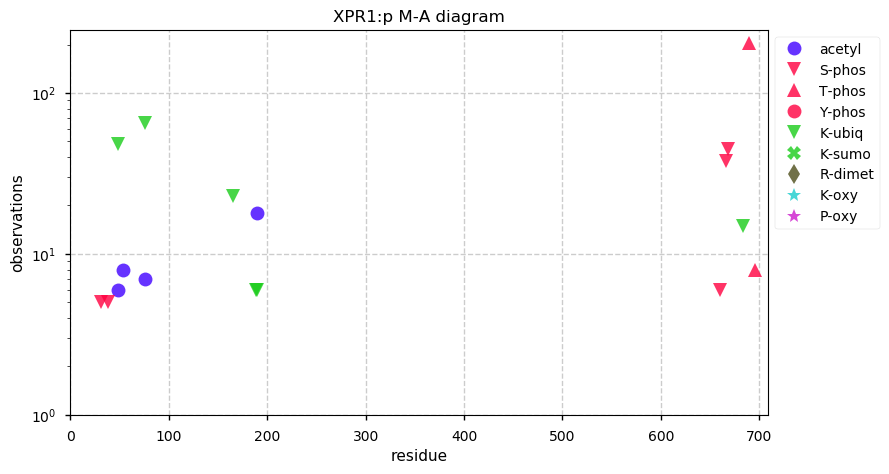
Fri Dec 06 13:14:28 +0000 2019XPR1:p, xenotropic and polytropic retrovirus receptor 1 (H. sapiens) 🔗 Midsized cytoplasmic protein; C-terminal phosphodomain; 8 transmembrane domains; no SAAVs; abundant in ovary and testes; mature form 1-696 [3,159 x] 🔗
Thu Dec 05 20:00:07 +0000 2019@Smith_Chem_Wisc I agree that things get more complicated when you want 5 sig figs for a specific peptide: different relative amounts of S and O change the composition enough to move the value and signal coalescence becomes in issue. How about a "practical" value of 1.003 u?
Thu Dec 05 17:49:58 +0000 2019oh boy! 8 TB of new SSD just showed up: now I get to spend hours copying files ...
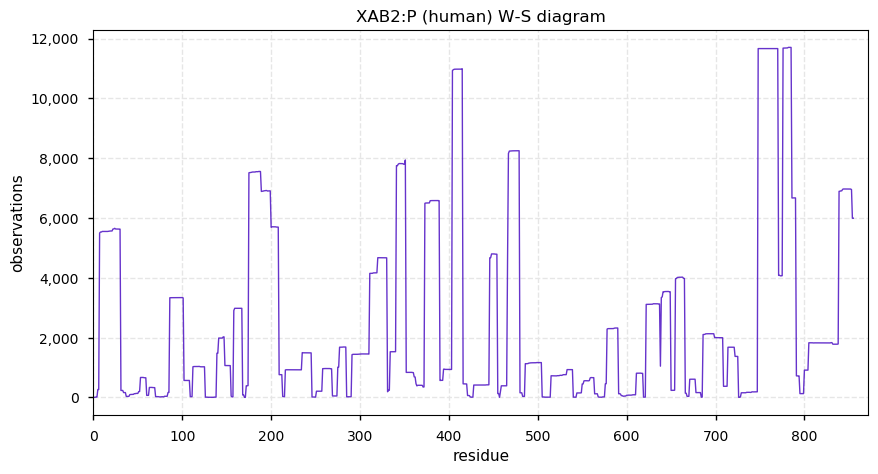
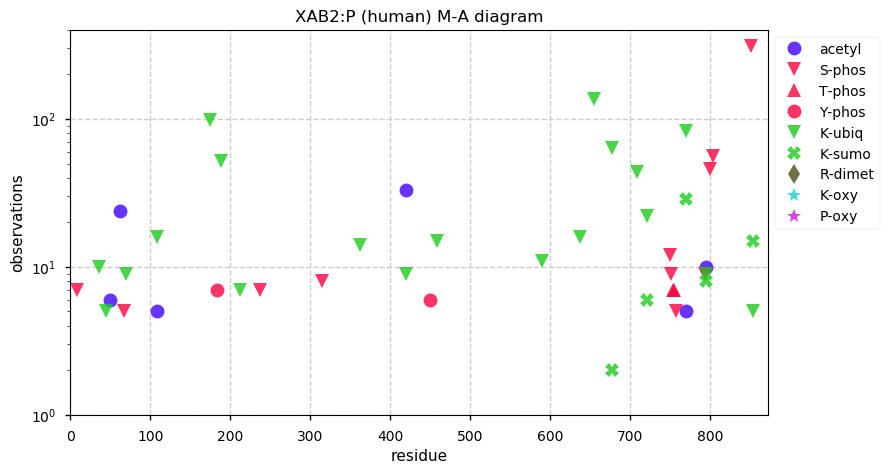
Thu Dec 05 14:16:33 +0000 2019XAB2:p, XPA binding protein 2 (H. sapiens) 🔗 Midsized cytoplasmic protein; low occupancy PTMs; 1 splice variant; no SAAVs; abundant in tissues and cell lines; mature form 5-855 [19,373 x] 🔗
Thu Dec 05 13:43:32 +0000 2019Thanks to everyone who participated in this poll. The best answer is 1.0034 u. ¹³C is ¹²C+1 neutron (1.0087 u), but 0.0053 u is converted into nuclear binding energy, reducing the mass of the resulting nucleus.
Wed Dec 04 15:30:49 +0000 2019@Smith_Chem_Wisc Me too!
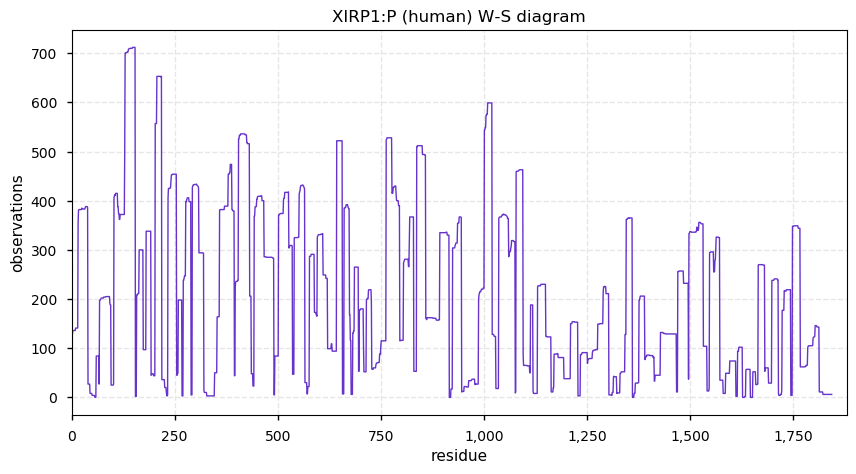
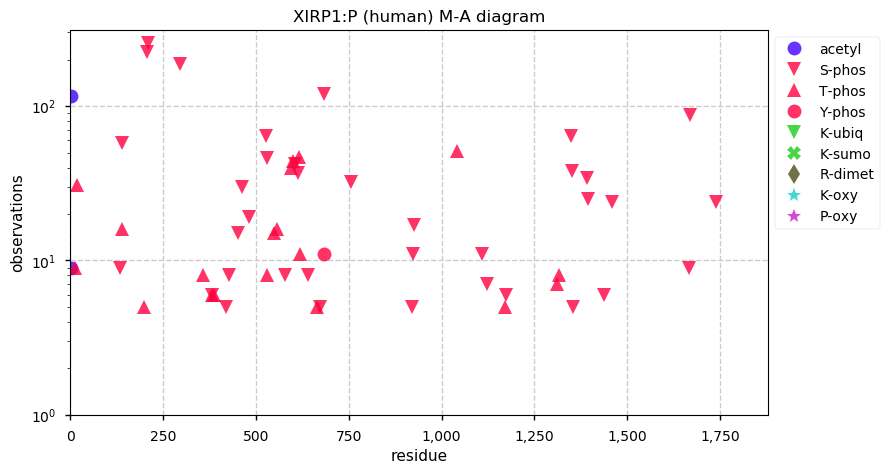
Wed Dec 04 13:14:12 +0000 2019XIRP1:p, xin actin binding repeat containing 1 (H. sapiens) 🔗 Very large cytoplasmic protein; highly phosphorylated; 1 splice variant; most abundant in heart muscle tissue; mature form 2-1843 [1,605 x] 🔗
Wed Dec 04 13:05:36 +0000 2019In MS-based proteomics, the ions observed produce a cluster of masses because about 1.1% of carbon is ¹³C. The lowest mass peak, A0, is all ¹²C, the next, A1, has 1 ¹³C, A2 has 2 ¹³C, etc. What is the correct mass difference to use for calculating mass(A1)-mass(A0)?
Tue Dec 03 18:10:21 +0000 2019Res ipsa loquitur 🔗
Tue Dec 03 18:06:14 +0000 2019@byu_sam I guess Science faculties should have an Office for the Prevention of Locally-Created Apocalypses and Supervillains (requires at least one additional vice-dean)
Tue Dec 03 18:01:36 +0000 2019@byu_sam Frickin' scientists
Tue Dec 03 17:54:00 +0000 2019@byu_sam Start of a dystopian sci-fi novel: "Scientists accidentally developed a bacteria that thrived on eating plastic, leading to the rapid destruction of most of the materials modern civilization was based on..."
Tue Dec 03 14:41:48 +0000 2019"As the saying goes, the problem with Bayes is the Bayesians ..." 🔗
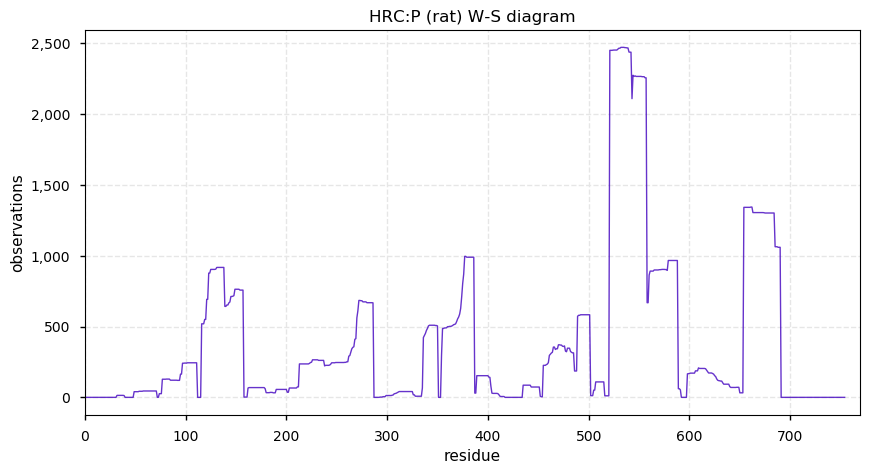
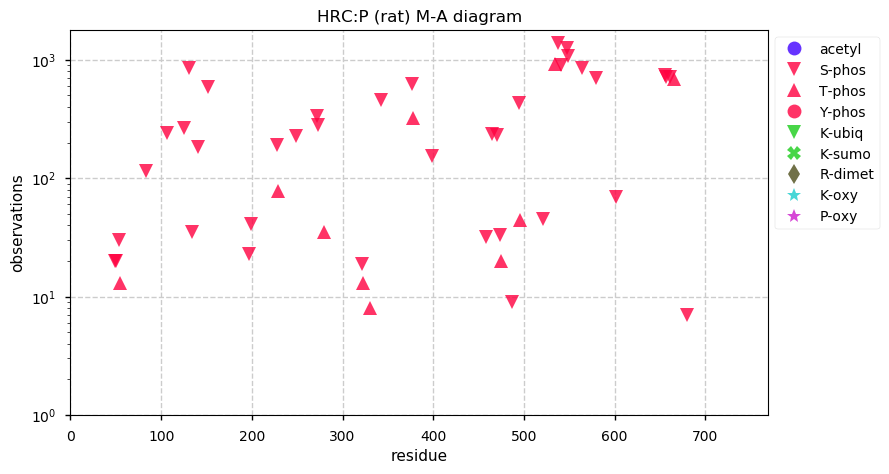
Tue Dec 03 13:39:59 +0000 2019Mature rat HRC:p has 11% H, 32% E/D and 7% phosphorylation acceptor sites.
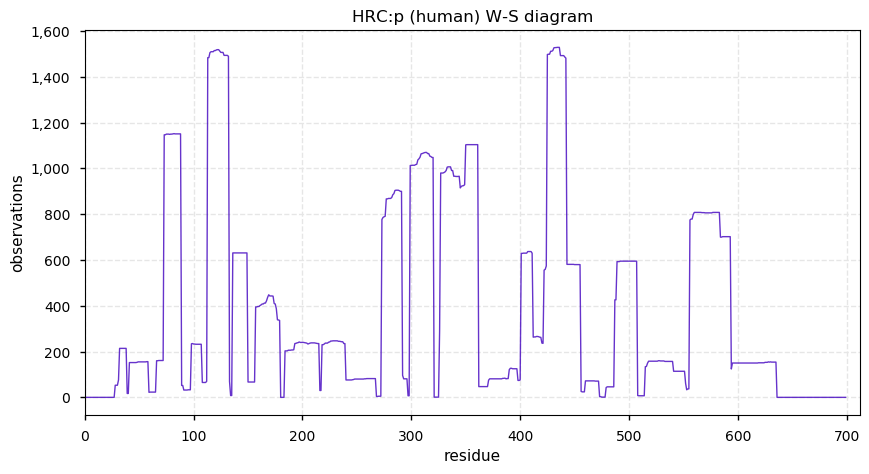
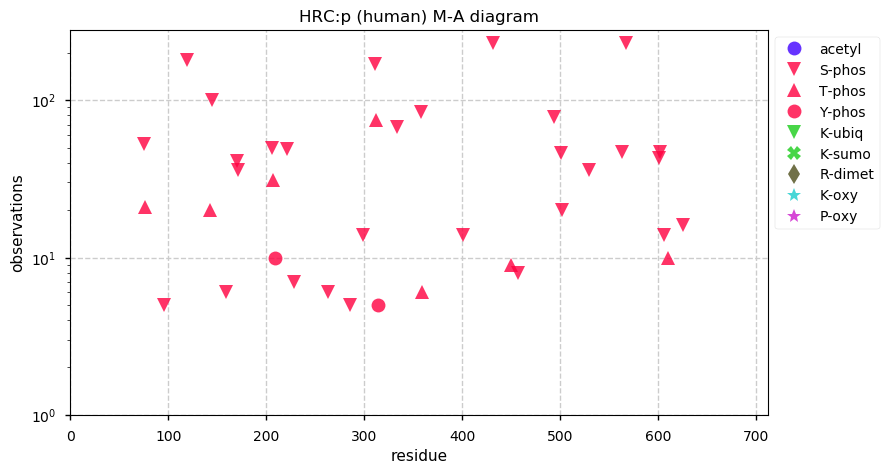
Tue Dec 03 13:34:11 +0000 2019HRC:p, histidine rich calcium binding protein (Rattus norvegicus) 🔗 Midsized sarcoplasmic reticulum protein; highly phosphorylated; multiple low complexity domains; very abundant in muscle tissue and H9C2 cells; no SAAVs; mature form 28-754 [276 x] 🔗
Tue Dec 03 13:28:07 +0000 2019@UCDProteomics @neely615 @Smith_Chem_Wisc While it is a lesser amount, hydroxylysine is still pretty common in collagens with hydroxproline. And unlike most lysine mods, it doesn't effect enzymatic cleavage at modified K residues.
Mon Dec 02 17:35:33 +0000 2019What could possibly go wrong with this idea?
🔗
Mon Dec 02 17:08:01 +0000 2019How many different ways are scan numbers and retention times recorded in MGF files?🤨
Mon Dec 02 15:52:46 +0000 2019I wouldn't trust these guys to build a toy truck that wouldn't fall apart, let alone widely disseminated, customer friendly nuclear reactors ... 🔗
Mon Dec 02 13:42:41 +0000 2019@pommie331 @TrostLab You should get familiar with sci-hub: solves the problem in most cases
Mon Dec 02 13:39:32 +0000 2019@pommie331 @TrostLab Why not?
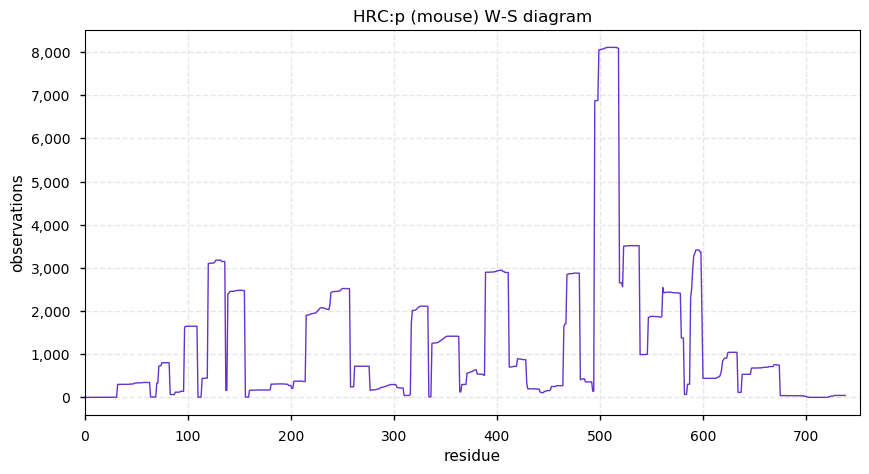
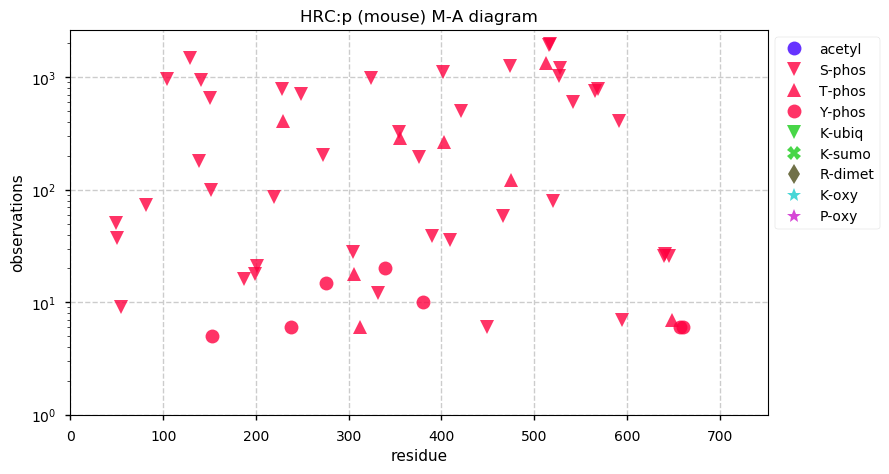
Mon Dec 02 12:57:54 +0000 2019Mature mouse HRC:p is 11% H, 32% E/D and 8% S/T phosphorylation acceptor sites.
Mon Dec 02 12:52:25 +0000 2019HRC:p, histidine rich calcium binding protein (Mus musculus) 🔗 Midsized sarcoplasmic reticulum protein; highly phosphorylated; multiple low complexity domains; very abundant in muscle tissue and C2C12 cells; no SAAVs; mature form 28-738 [1,692 x] 🔗
Mon Dec 02 12:48:36 +0000 2019@TrostLab I'm pretty sure the reason for this in proteomics is the conferences trying to put together a vendor-friendly set of speakers to attract & maintain sponsorships.
Mon Dec 02 12:37:36 +0000 2019@TrostLab I gave up on going to conferences 5 years ago for that very reason.
Sun Dec 01 23:22:21 +0000 2019@girlziplocked It was filmed in Canada, where such things can be true!
Sun Dec 01 19:18:42 +0000 2019@nesvilab @BrenesAlejandro So it is just a rule-of-thumb.
Sun Dec 01 18:52:54 +0000 2019@nesvilab @BrenesAlejandro Has anyone done any Monte Carlo type calculations to support this idea? I can't think of a reason that using larger libraries (e.g., 2x10⁶) would negatively affect the outcome.
Sun Dec 01 15:32:11 +0000 2019@nesvilab I don't really follow the DIA world very closely: is it common to use libraries with only 1 or 2x10⁵ peptides to characterize tissue proteomes in mammals?
Sun Dec 01 14:27:02 +0000 2019Mature human HRC:p is 13.3% H, 31.7% E/D and 5.7% S/T phosphorylation acceptor sites.
Sun Dec 01 14:18:18 +0000 2019HRC:p, histidine rich calcium binding protein (Homo sapiens) 🔗 Midsized sarcoplasmic reticulum protein; highly phosphorylated; multiple low complexity domains; very abundant in muscle tissue; no SAAVs; ; mature form 28-699 [1,533 x] 🔗
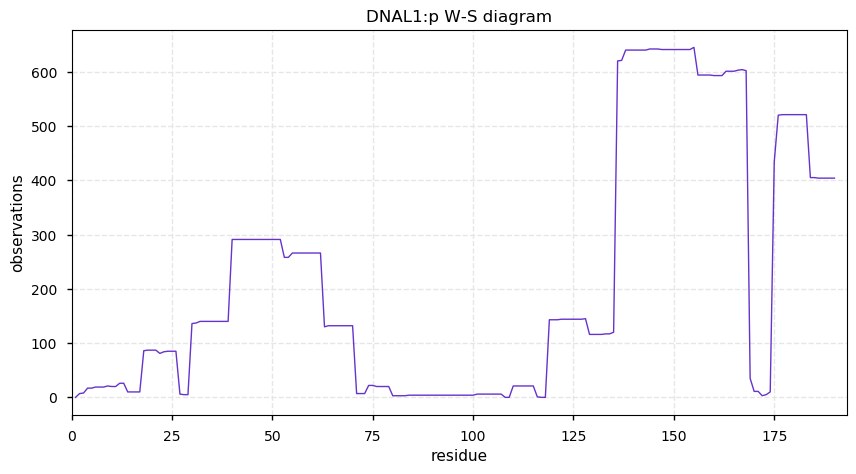
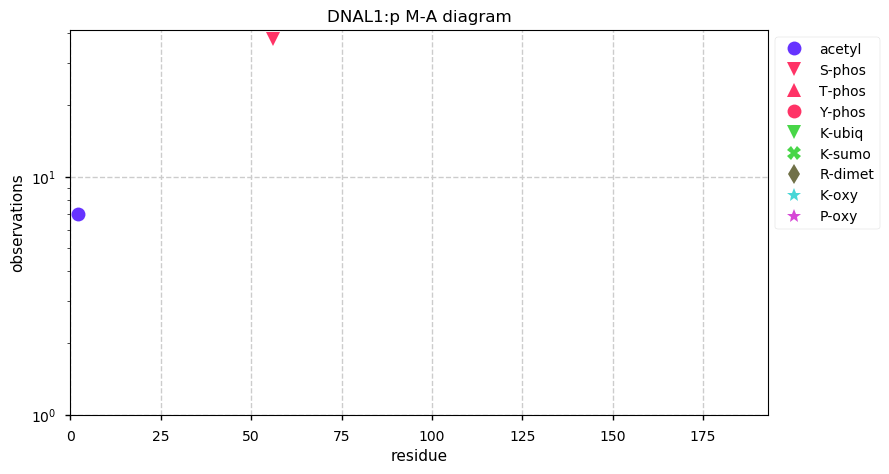
Sat Nov 30 13:15:19 +0000 2019DNAL1:p, dynein axonemal light chain 1 (H. sapiens) 🔗 Small cytoskeleton protein; PTMs: A2+acetyl, S56+phosphoryl; no SAAVs; abundant in cells with flagella/cilia; mature form 2-190 [995 x] 🔗
Sat Nov 30 12:51:27 +0000 2019But I did get it to work. I had always used preprocessor commands to #if/#elseif in Windows native threading, so just using pthreads for all platforms makes threading code easier to understand.
Fri Nov 29 22:31:59 +0000 2019Boy, getting pthreads to work on VC++ was more of a headache than I thought it was going to be - I should know better!
Fri Nov 29 17:09:11 +0000 2019It may also have pedagogical applications as an example of some good (& some bad) practices that can be turned into either a tutorial or straight lecture material.
Fri Nov 29 17:01:04 +0000 2019For anyone doing bioinformatics:
1. download your Twitter data (🔗); &
2. go through the files to see how they are structured.
It is a good example of how to organize text, metadata & images while retaining linkage information.
Fri Nov 29 15:35:31 +0000 2019@SamanthaLWilson @CJBellissimo However, the main problem with retaining mid-career researchers that are recruited into Canadian universities is that the universities (admin & faculty) have no clue what to do with someone who didn't enter as a tenure-track Assistant Professor.
Fri Nov 29 13:54:15 +0000 2019@SamanthaLWilson @CJBellissimo Both Runte and Bains are typical cocktail-party "innovation" cheerleaders. They believe in a "great researcher" model of R&D, rather than the more difficult to replicate (but more realistic) "hard work & good fortune" model.
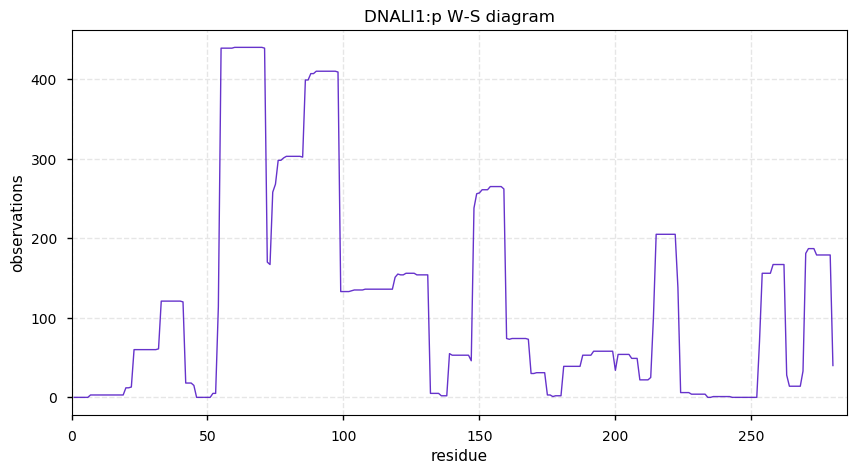
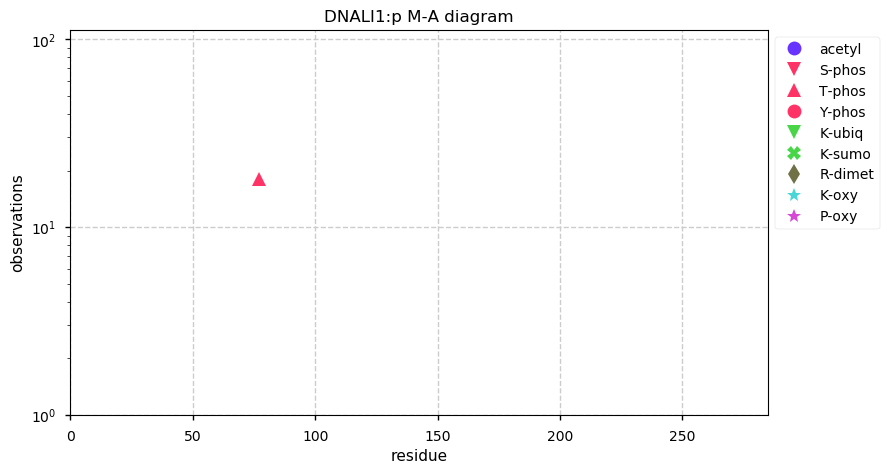
Fri Nov 29 13:37:14 +0000 2019DNALI1:p, dynein axonemal light intermediate chain 1 (H. sapiens) 🔗 Small cytoskeleton protein; S77+phosphoryl; no SAAVs; abundant in cells with flagella/cilia; mature form 1-280 [465 x] 🔗
Fri Nov 29 12:46:27 +0000 2019@MagnusPalmblad @nesvilab Doing it that way produced the proteins they wanted to see in the results.
Thu Nov 28 17:38:05 +0000 2019Anyone who wants an ideal QE MS/MS data set to use for testing algorithms should give PXD007940 a try.
Thu Nov 28 17:22:05 +0000 2019#PXD007940: wirklich nette Daten! 👍
Thu Nov 28 17:04:21 +0000 2019@zeitonline While Unabhängige Flugbegleiter Organisation is a perfectly good name for the union auf Deutsch, the hashtag #UFO doesn't really work out so well in English ...
Thu Nov 28 16:52:09 +0000 2019Thanks to everyone who participated in this poll. I think it is a decent snap-shot of people's experience with the current generation of PSM-assignment/statistical-significance-determination methods applied to high accuracy MS/MS fragment ion data.
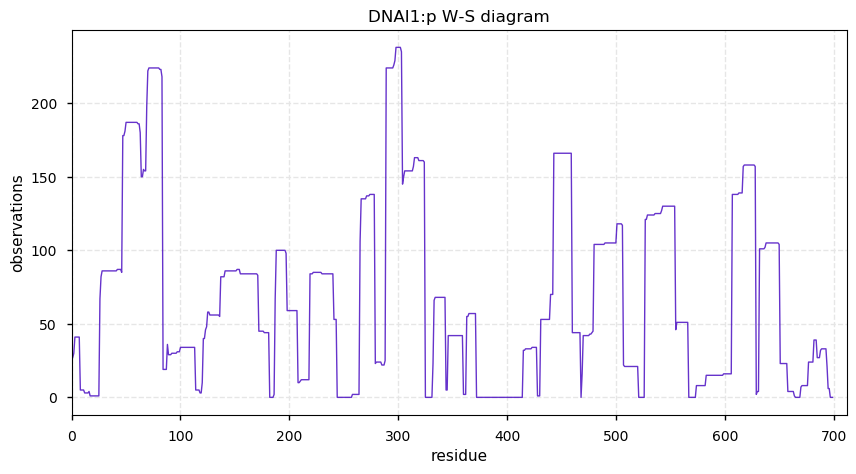
Thu Nov 28 13:13:59 +0000 2019DNAI1:p, dynein axonemal intermediate chain 1 (H. sapiens) 🔗 Midsize cytoskeleton protein; no PTMs; 2 SAAVs: V335I (maf=0.2), T535N (maf=0.02); abundant in cells with flagella/cilia; mature form 1-699 [370 x] 🔗
Wed Nov 27 19:35:06 +0000 2019🦃🦃🦃🦃🦃🇺🇸 🔗
Wed Nov 27 19:28:15 +0000 2019@nesvilab (puts on Boomer pants) I can remember a group that was absolutely certain that using a parent tolerance of ±5 ppm and a fragment tolerance of ±100 Da was the right way to analyze their data. They could not be talked out of it & they published results obtained that way.
Wed Nov 27 19:15:34 +0000 2019@nesvilab I realize the whole field is full of hacks around how to make various methods work best: I am trying to gauge how the community in general thinks about the consequences of making a particular choice for a parameter they are often called up to specify.
Wed Nov 27 17:41:00 +0000 2019@ProtifiLlc @nesvilab So your answer is more at 400 mDa?
Wed Nov 27 17:05:00 +0000 2019Assuming that the experiment used a DDA method and a maximum of 1 identifiable peptide sequence per isolation.
Wed Nov 27 16:42:51 +0000 2019The first version of this omitted "statistically significant", which was not what I was intending. I think of these algos as including the entire process (i.e., spectra → usable results) while others may consider the PSM assignment to be separate from assigning significance.
Wed Nov 27 16:36:46 +0000 2019If you have an instrument that measures fragment ion masses with high accuracy (e.g., ± 20 mDa), should a PSM assignment algorithm find more statistically significant PSMs when its fragment mass tolerance is set at ± 20 mDa or ± 400 mDa?
Wed Nov 27 16:33:23 +0000 2019@chrashwood All in. The whole calculation: spectra -> usable PSMs.
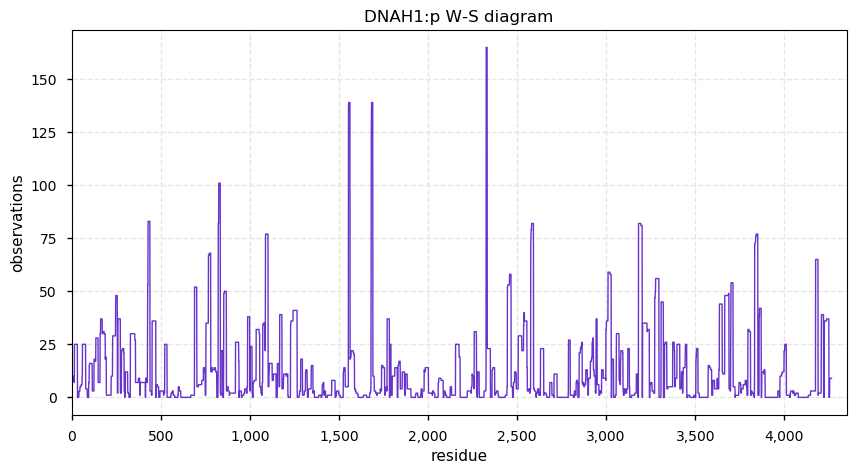
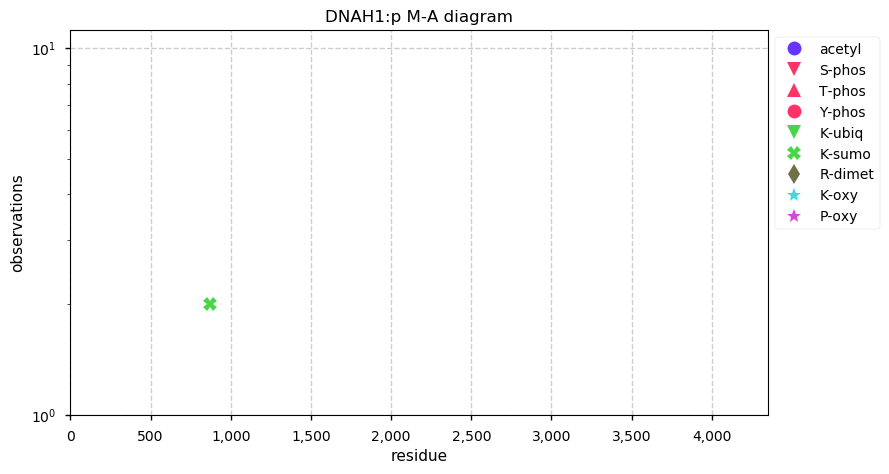
Wed Nov 27 16:17:13 +0000 2019DNAH1:p is a good example to bring up when someone speculates that MS-based PTM assignments are largely stochastic.
Wed Nov 27 13:55:14 +0000 2019DNAH1:p, dynein axonemal heavy chain 1 (H. sapiens) 🔗 Very large cytoskeleton protein; no PTMs; 2 SAAVs: R597C (maf=0.02), G3801D (maf=0.2); abundant in cells with flagella/cilia; mature form 1-4265 [822 x] 🔗
Tue Nov 26 18:52:24 +0000 2019@SpecInformatics @dtabb73 I agree. Once we've got something to propose, it would be helpful to get their buy in.
Tue Nov 26 18:51:36 +0000 2019@pwilmarth In my opinion, the field would be improved if there were some community-based standards (with examples) that you could point your new programmer/student at initially and that you could state was your standard when publishing or proposing a development process.
Tue Nov 26 18:48:40 +0000 2019@pwilmarth Thanks, Phil. The Google documentation guide is largely for text-based exposition, rather than code level documentation, but it does have many useful ideas. The practical problem is that without code-level docs, even the best text docs don't help you read the code.
Tue Nov 26 18:40:20 +0000 2019@SpecInformatics Then creating a set of files, marked up with what is generally considered to be best practices for use as exemplars. There will probably need to be 2 or 3 different styles of best practices, to accommodate different schools of thought.
Tue Nov 26 18:37:06 +0000 2019@SpecInformatics OK: lots of positive responses. My suggestion is to put several files of undocumented code on-line and let people mark them up. Then make the various styles of markup available and let the public cognoscenti decide which elements of documentation they like.
Tue Nov 26 17:40:24 +0000 2019@SpecInformatics Anyone else want to participate in coming up with some practical guidelines for code-level documentation in MS-based proteomics applications?
Tue Nov 26 17:27:27 +0000 2019@SpecInformatics A necessary evil. I like the idea about initially limiting the discussion to mass spec related code: it reduces the likelyhood of failure caused by too many conflicting interests. There are also probably some MS-only issues, such as the incorporation of vendor modules.
Tue Nov 26 16:29:39 +0000 2019@SpecInformatics Good suggestion. It would be good for bioinformatics if journals & granting agencies required at least some guidance for code documentation. This guidance would be particularly important for code that is not going to be made publicly available.
Tue Nov 26 16:22:02 +0000 2019Beta release of new PSM assignment software idX v. 2, including a demo web site 🔗
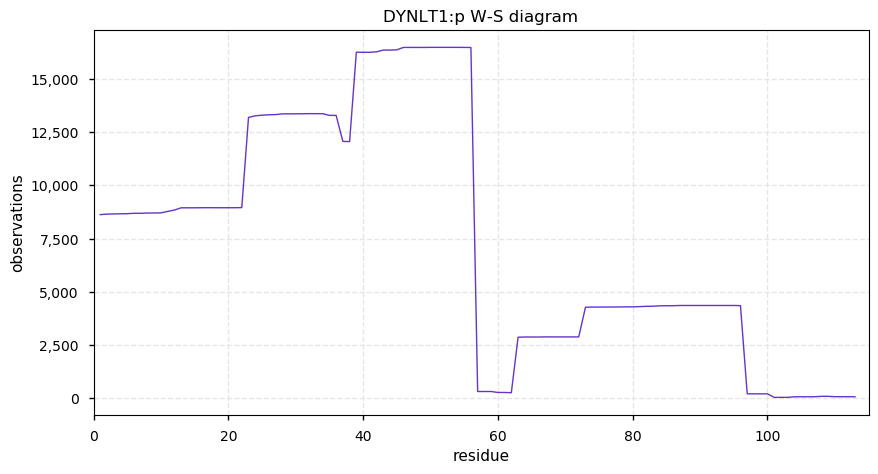
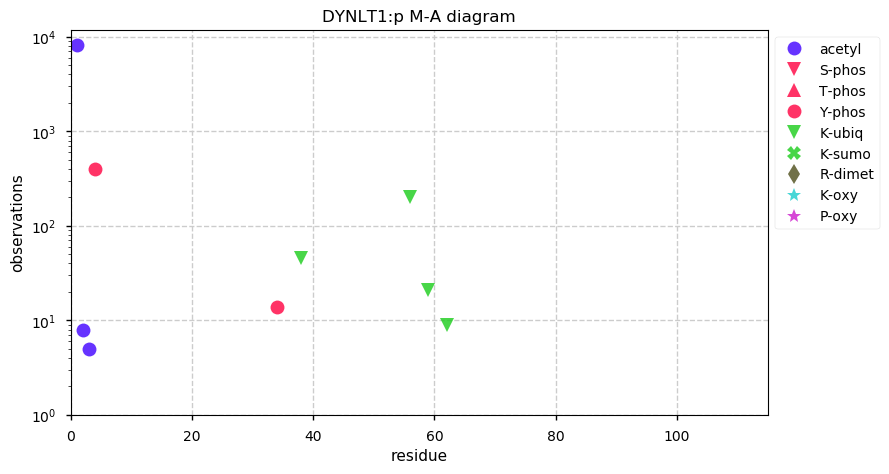
Tue Nov 26 14:39:19 +0000 2019DYNLT1:p, dynein light chain Tctex-type 1 (H. sapiens) 🔗 Very small cytosolic protein; few, low occupancy PTMs; no SAAVs; abundant in cell lines and tissues; mature form 1-113 [12,073 x] 🔗
Mon Nov 25 16:12:48 +0000 2019Are there any proteomics journals that have instructions to authors regarding code documentation?
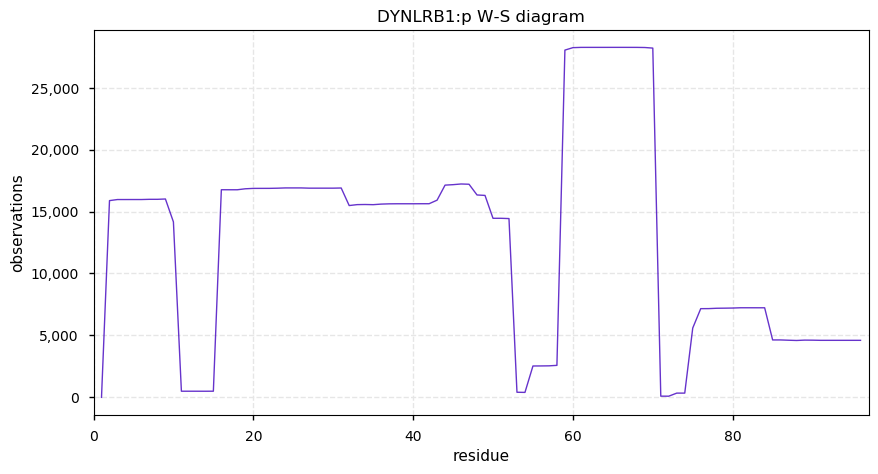
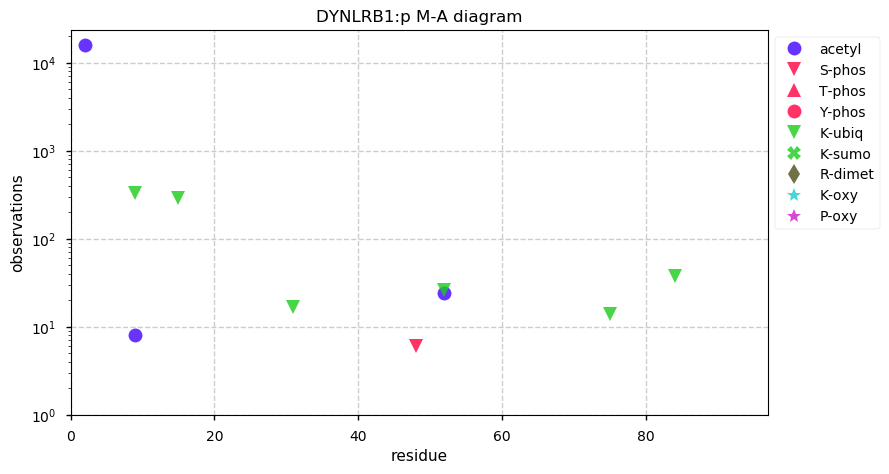
Mon Nov 25 13:15:17 +0000 2019DYNLRB1:p, dynein light chain roadblock-type 1 (Homo sapiens) 🔗 Very small cytosolic protein; few, low occupancy PTMs; no SAAVs; abundant in cell lines and tissues; mature form 2-94 [21,782 x] 🔗
Sun Nov 24 19:32:14 +0000 2019Given that many informatics tools in proteomics are practically limited to using Windows, this is a list we should all keep an eye on:
🔗
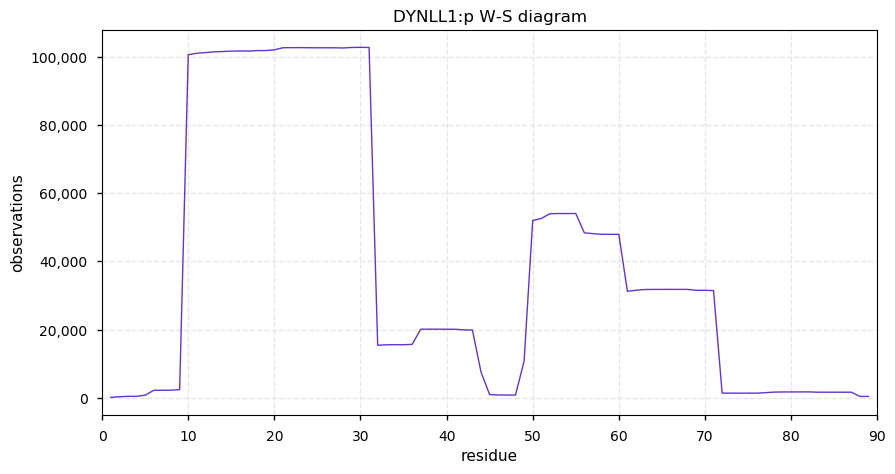
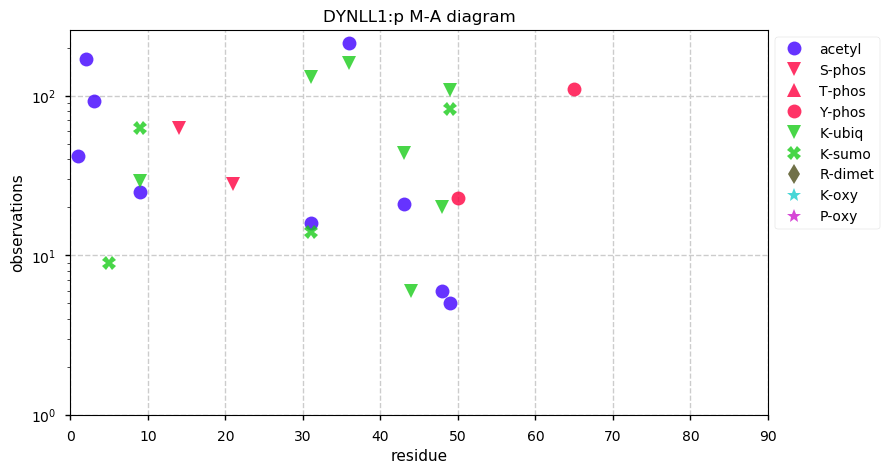
Sun Nov 24 14:13:17 +0000 2019DYNLL1:p, dynein light chain LC8-type 1 (H. sapiens) 🔗 Very small cytosolic protein; highly modified (more SUMO than might be expected); no SAAVs; abundant in cell lines and tissues; mature form 1-89 [30,446 x] 🔗
Sat Nov 23 13:55:50 +0000 2019Is there an ontology out there that allows the accurate description of an MS/MS MHC peptide experiment?
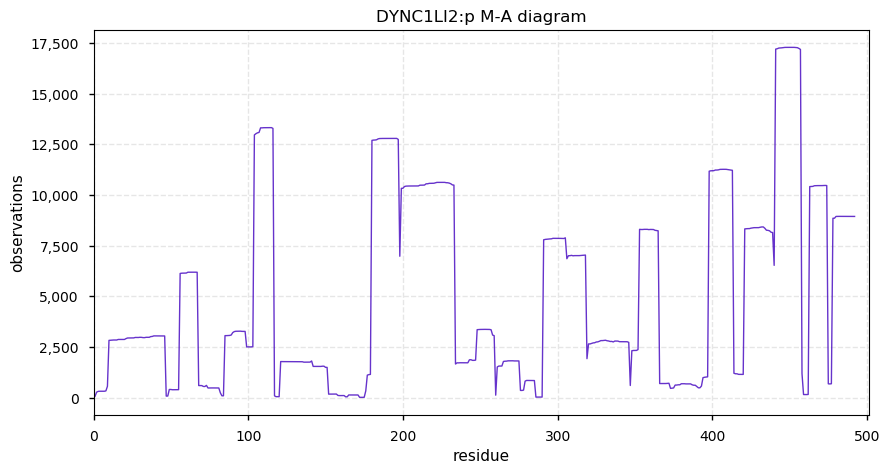
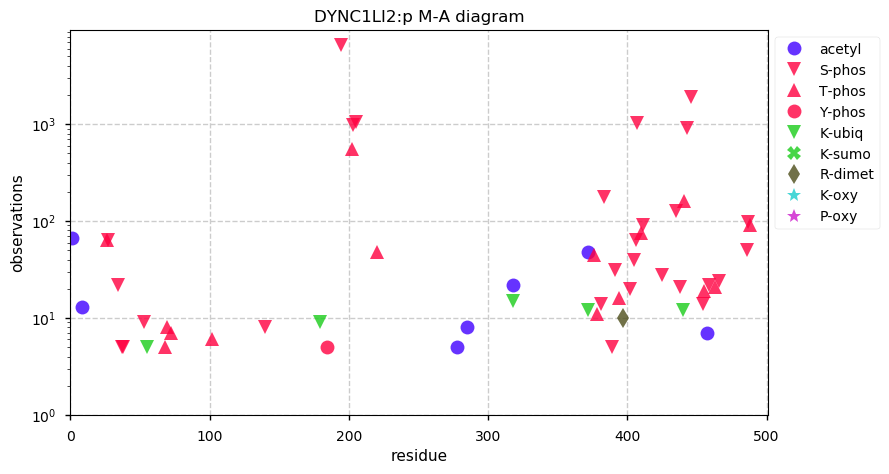
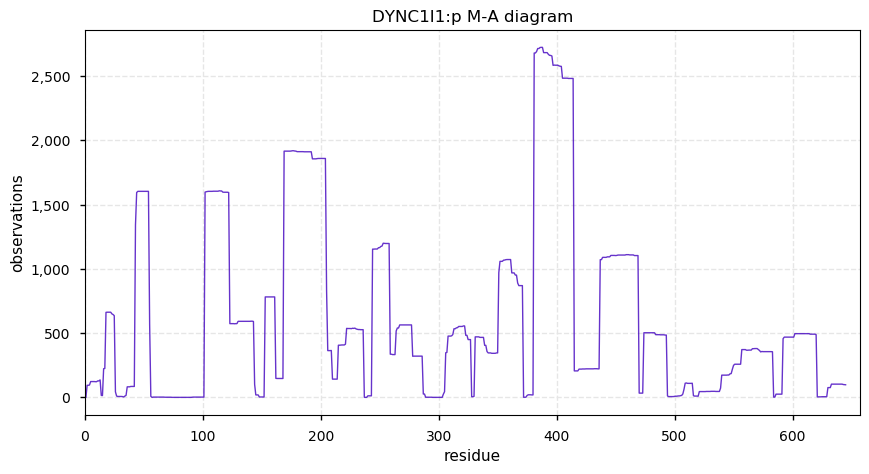
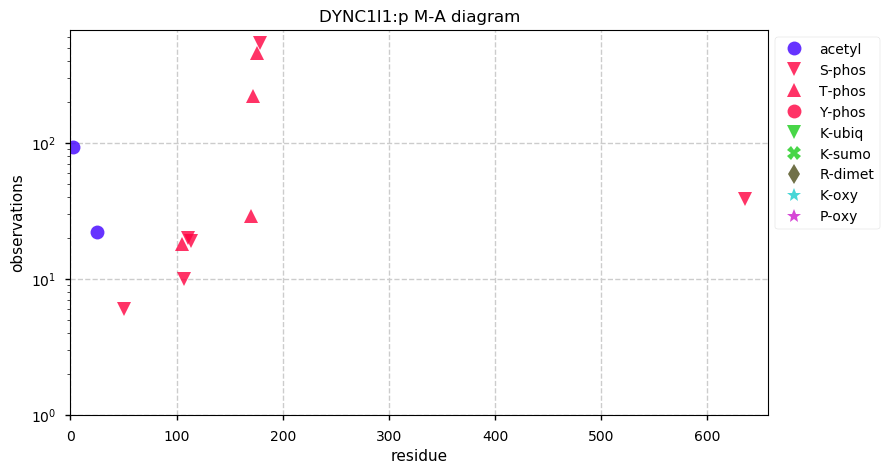
Sat Nov 23 13:22:57 +0000 2019DYNC1LI2:p, dynein cytoplasmic 1 light intermediate chain 2 (H. sapiens) 🔗 Small cytosolic protein; highly modified; no SAAVs; 1 splice variant; mature form 2-492 [23,827 x] 🔗
Fri Nov 22 16:45:08 +0000 2019Or even just the merits of "long" vs. "long long int" vs. "int64_t" ...
Fri Nov 22 16:44:21 +0000 2019Or, if anyone wants to express their opinions on the best warning level to use, please feel free ...
Fri Nov 22 16:43:21 +0000 2019Time to call it a day: clean compile with -Wall.
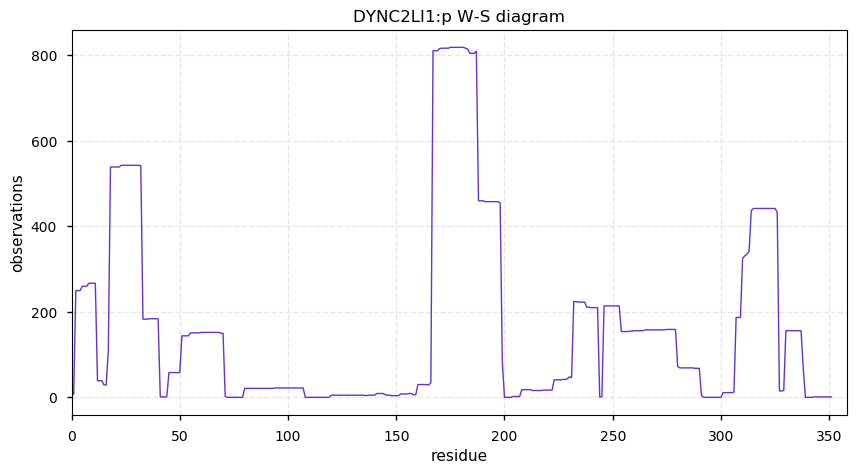
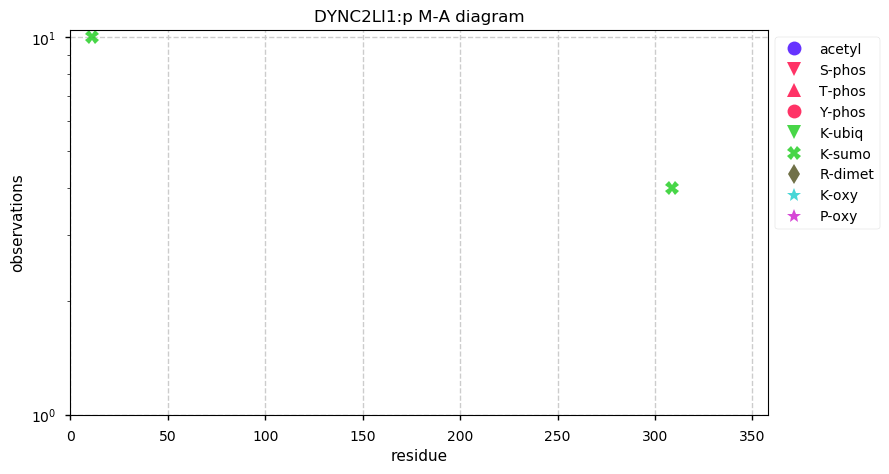
Fri Nov 22 13:10:00 +0000 2019DYNC2LI1:p, dynein cytoplasmic 2 light intermediate chain 1 (Homo sapiens) 🔗 Small cytosolic protein; no PTMs; 1 SAAV: F33S (maf=0.5); most abundant in testis; mature form 2-351 [1,310 x] 🔗
Thu Nov 21 22:25:43 +0000 2019No matter how many times I do it, I am always impressed by the degree to which testing software on different platforms forces you to think clearly about what you really wanted to do in the first place.
Thu Nov 21 14:27:53 +0000 2019DYNC1I1:p, dynein cytoplasmic 1 intermediate chain 1 (H. sapiens) 🔗 Midsized cytosolic protein; 2 short N-terminal phosphodomains; no SAAVs; most abundant in brain tissue; mature form 2-645 [3,491 x] 🔗
Wed Nov 20 21:25:25 +0000 2019I seem to have finally figured out an indexing-type PSM id algorithm that doesn't take much memory: ~ 300 MB of memory for 50,000 spectra matched to the human proteome. Small enough memory footprint to be practical on a Raspberry Pi.
Wed Nov 20 20:57:53 +0000 2019@AndrewWCraig @MHendr1cks Well at least we finally have a Minister of Middle Class Prosperity 😜
Wed Nov 20 13:44:05 +0000 2019DYNC1I2:p, dynein cytoplasmic 1 intermediate chain 2 (H. sapiens) 🔗 Very large cytosolic protein; high occupancy N-terminal phosphodomain; 1 SAAV: V289L (maf=0.07); mature form 2-612 [32,024 x] 🔗
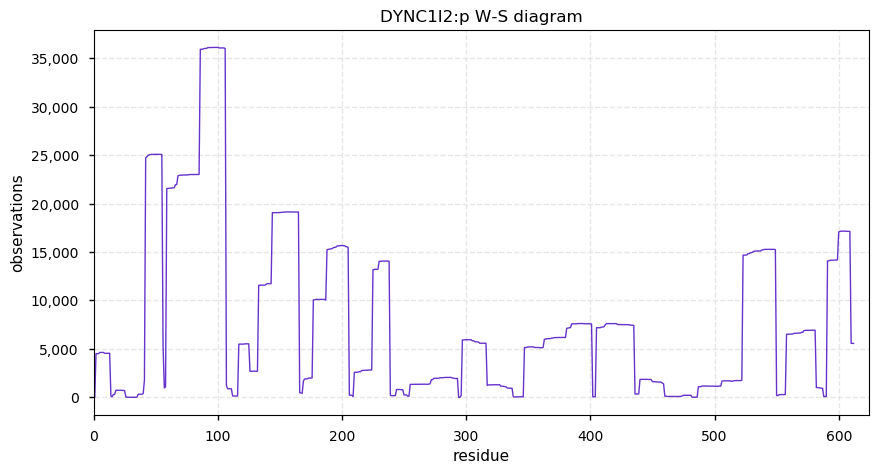
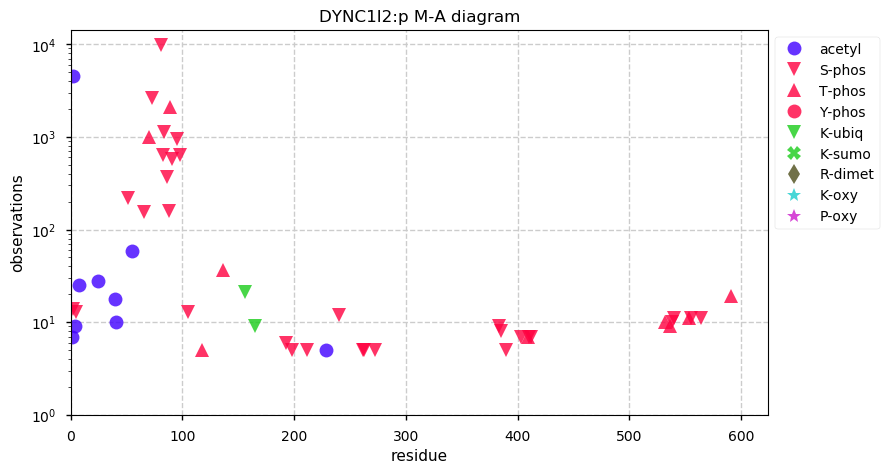
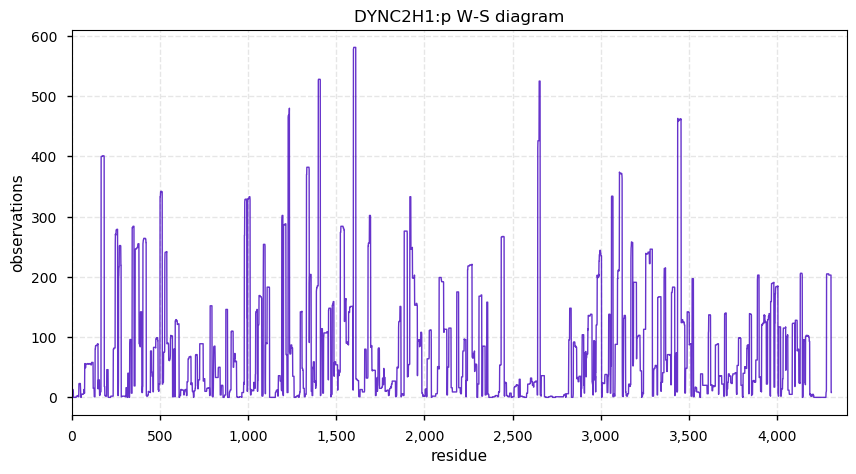
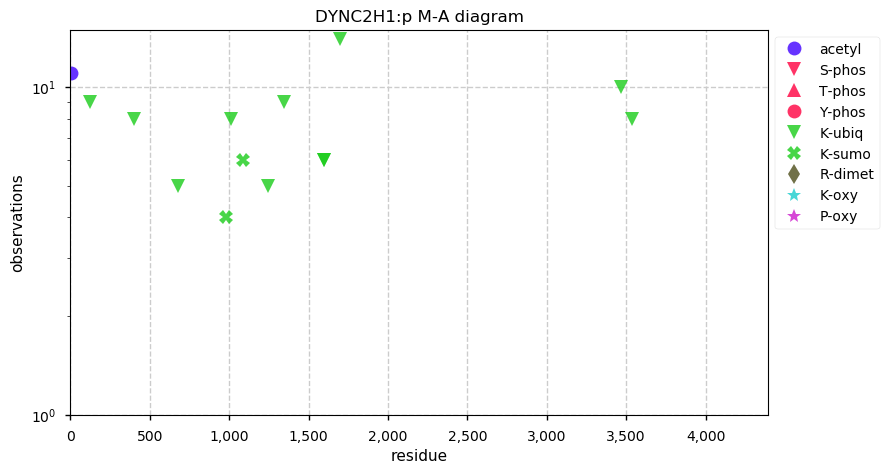
Tue Nov 19 14:07:02 +0000 2019DYNC2H1:p, dynein cytoplasmic 2 heavy chain 1 (Homo sapiens) 🔗 Very large cytosolic protein; no phosphorylation sites; highest maf SAAVs: K1413R (maf=0.3), R2871Q (maf=0.3), A3680V (maf=0.3); mature form 2-4307 [3,965 x] 🔗
Mon Nov 18 14:08:22 +0000 2019@kadzuis @an_chem I end up rejecting 20-30% of publicly available proteomics datasets because of quality issues (not counting data from blacklisted labs).
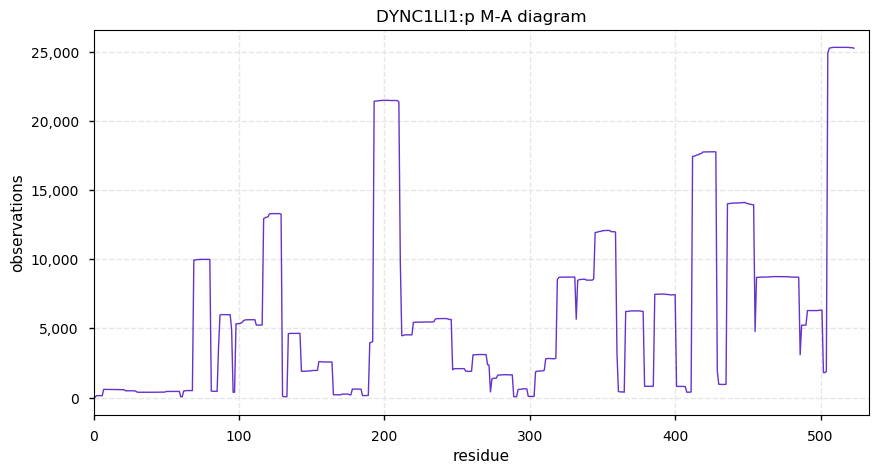
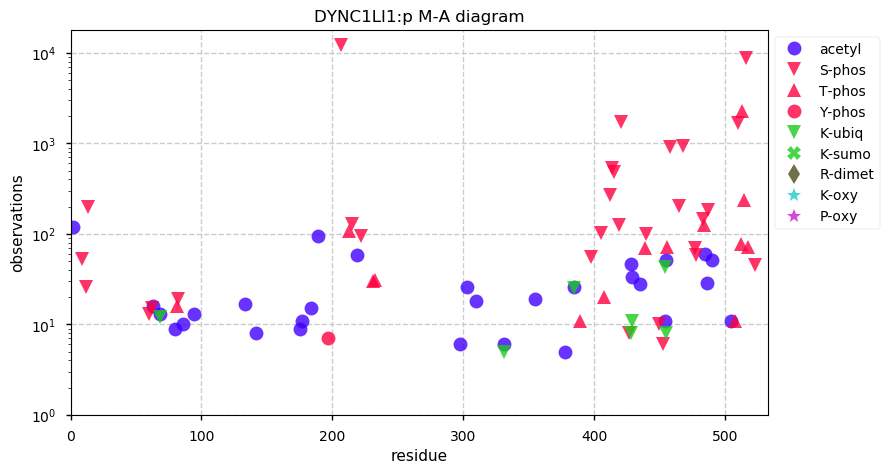
Mon Nov 18 13:43:40 +0000 2019DYNC1LI1:p, dynein cytoplasmic 1 light intermediate chain 1 (H. sapiens) 🔗 Midsized cytosolic protein; 2 phosphodomains; 1 SAAVs: M147T (maf=0.02); mature form 2-523 [30,585 x] 🔗
Sun Nov 17 20:02:37 +0000 2019Saved by Stirling's approximation (again)!
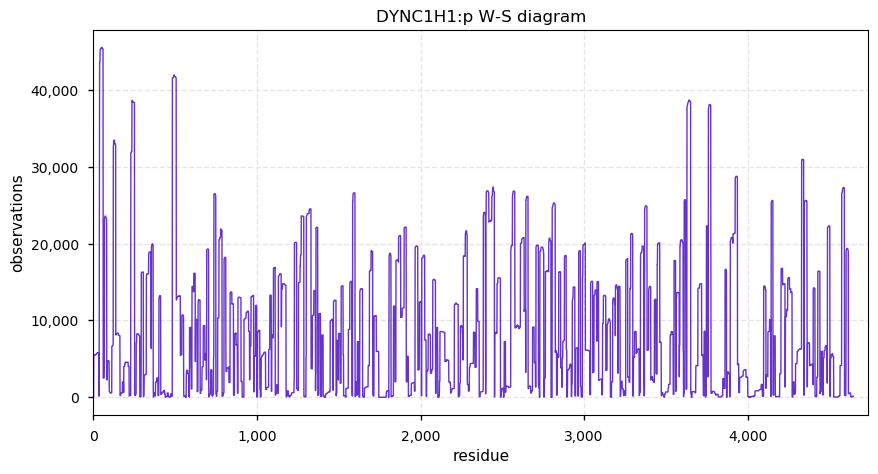
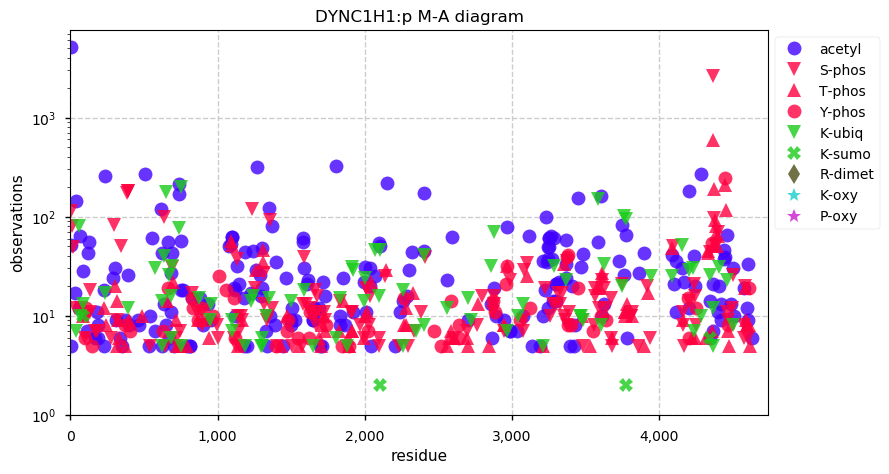
Sun Nov 17 14:26:18 +0000 2019DYNC1H1:p, dynein cytoplasmic 1 heavy chain 1 (H. sapiens) 🔗 Very large cytosolic protein; many low occupancy PTM sites; only 2 SAAVs: P3922L (maf=0.01), H4029Q (maf=0.1); mature form 2-4646 [59,814 x] 🔗
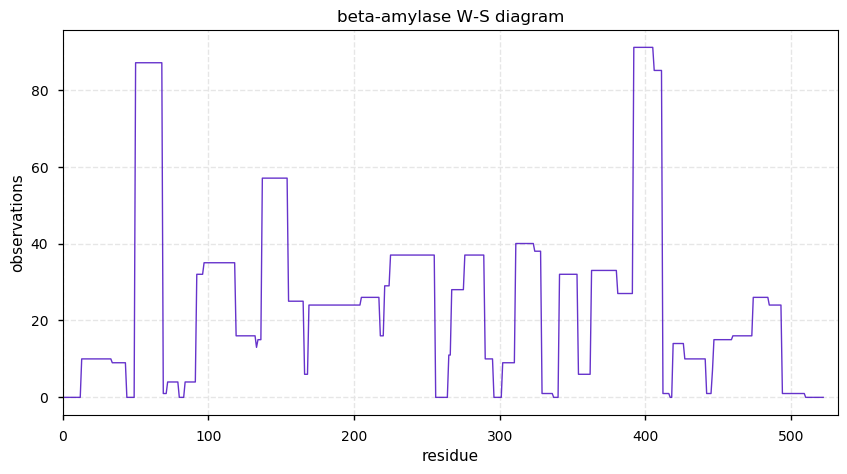
Sat Nov 16 14:22:43 +0000 2019beta-amylase (Cannibis sativa) 🔗| Midsized protein; abundant in seeds; catalyzes the removal of maltose from the non-reducing end of polysaccharides; mature form 1-522 [38 x] 🔗
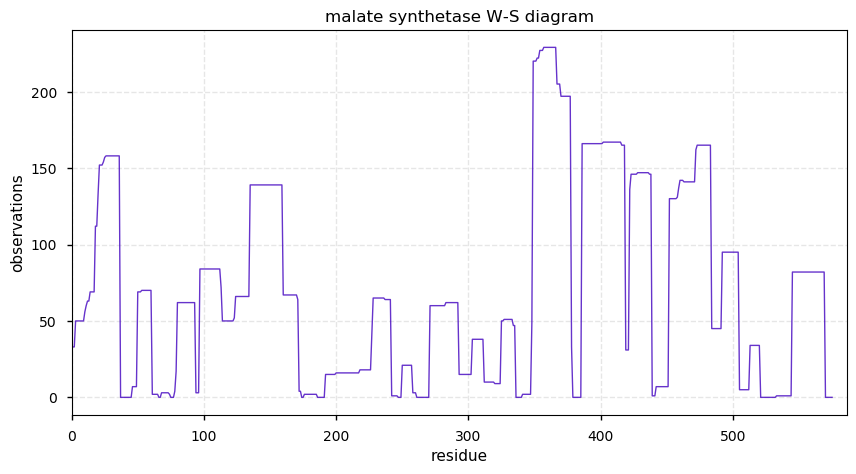
Fri Nov 15 14:16:23 +0000 2019malate synthase, glyoxysomal (Cannibis sativa) 🔗| Midsized protein; abundant in seeds; catalyzes acetyl-CoA + H2O + glyoxylate → (S)-malate + CoA; mature form 1-575 [49 x] 🔗
Thu Nov 14 14:00:06 +0000 201911 kDa late embryogenesis abundant protein (Cannibis sativa) 🔗| Small protein; abundant in seeds; function unknown (but lots of speculation); mature form 1-153 [43 x] 🔗
Thu Nov 14 02:00:14 +0000 2019@ProtifiLlc @JoelisSteele @tomlau voila
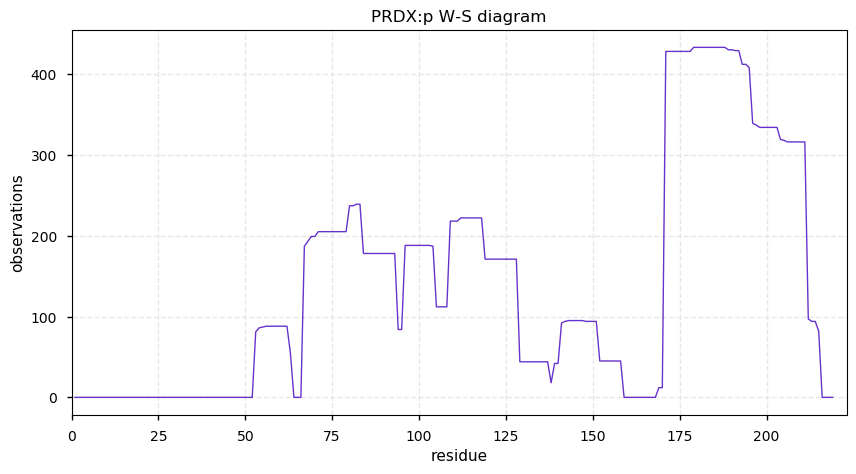
Wed Nov 13 14:03:44 +0000 2019PRDX:p, 1-Cys peroxiredoxin (Cannibis sativa) 🔗| Small protein; abundant in seeds; catalyzes the removal of peroxide; mature form 1-219 [49 x] 🔗
Wed Nov 13 00:59:59 +0000 2019@MHendr1cks Add ultra-super- & you may have something ...
Tue Nov 12 22:44:37 +0000 2019After working pretty much exclusively in Python for the last few years, doing a project in C++ 14 seems like a welcome homecoming.
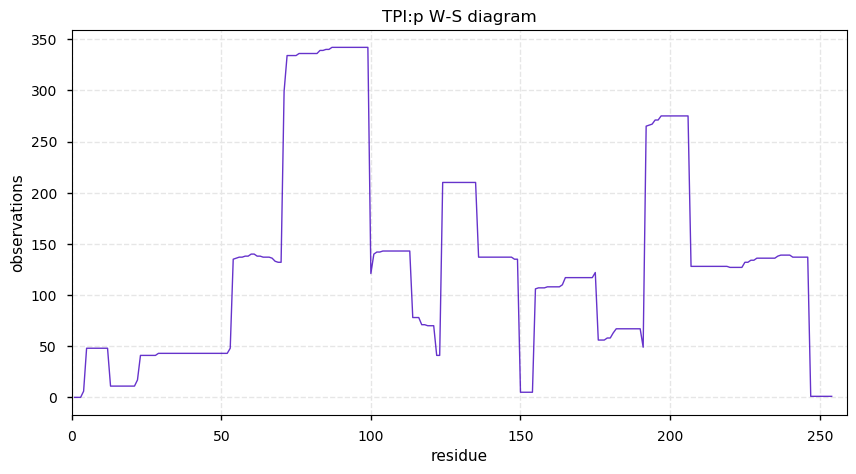
Tue Nov 12 14:32:02 +0000 2019TPI:p, triosephosphate isomerase, cytosolic-like (Cannibis sativa) 🔗| Small protein; abundant in seeds; catalyzes dihydroxyacetone phosphate → D-glyceraldehyde 3-phosphate; mature form 2-254 [68 x] 🔗
Tue Nov 12 14:13:22 +0000 2019@PoisonEcology @dtabb73 It is much easier to write software to use yourself than it is to write software for general use (which is nearly impossible).
Mon Nov 11 17:34:30 +0000 2019@jwoodgett @MHendr1cks Call out the army!
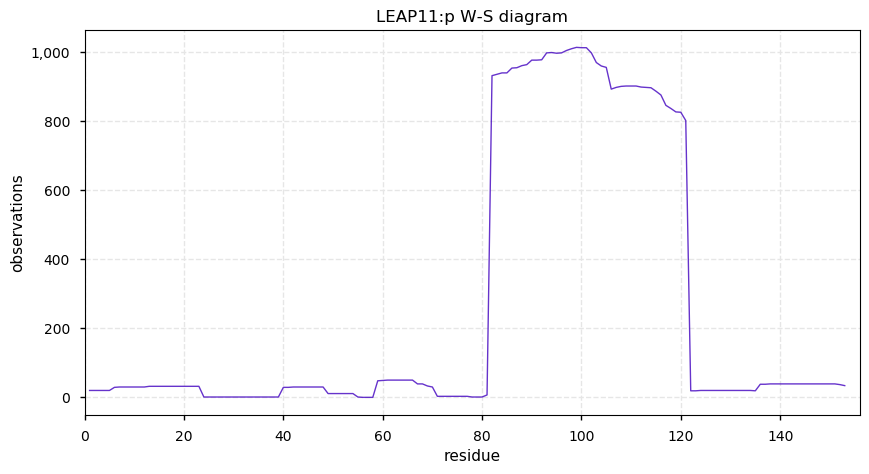
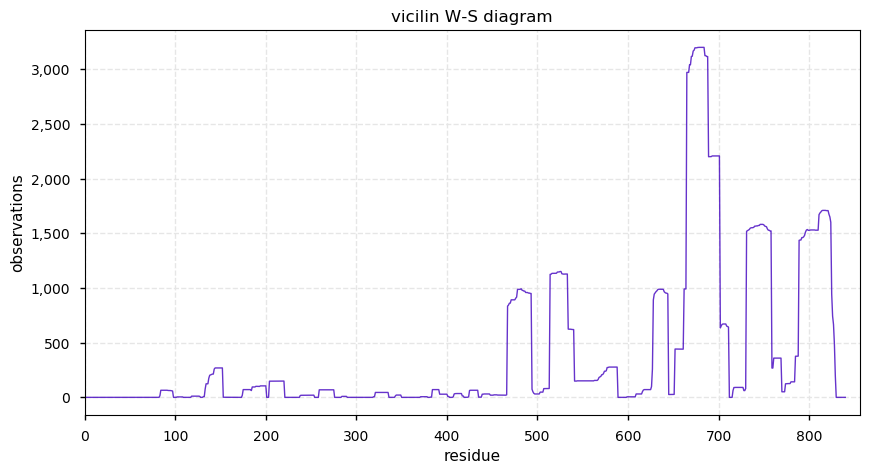
Mon Nov 11 14:06:06 +0000 2019CS7S:p, vicilin C72-like (Cannibis sativa) 🔗| Midsized protein; seed storage protein; multiple low complexity regions; mature form 83?-840 [49 x] 🔗
Sun Nov 10 22:07:25 +0000 2019Why is it that data sets created for manuscripts describing/testing/demo'ing search engines are always so awful?
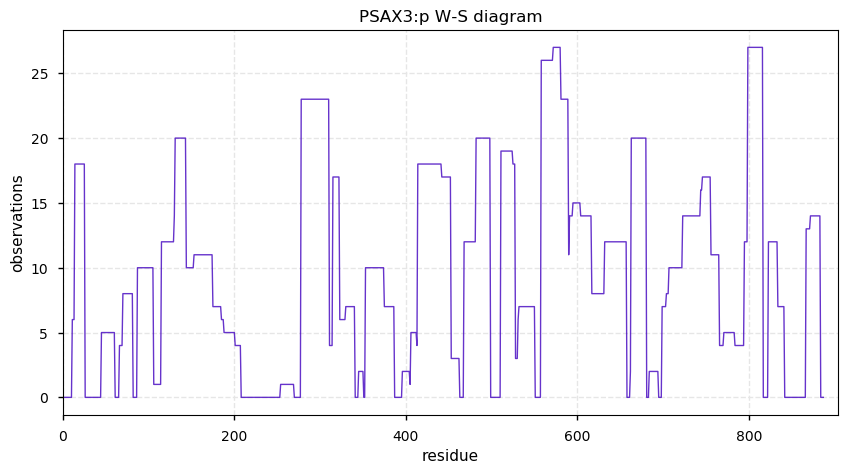
Sun Nov 10 14:17:10 +0000 2019PSAX3:p, puromycin-sensitive aminopeptidase isoform X3 (Cannibis sativa) 🔗| Midsized protein; abundant in seeds; cleaves other proteins during their maturation; mature form 2?-887 [22 x] 🔗
Sat Nov 09 19:57:23 +0000 2019@pwilmarth @UCDProteomics @PastelBio @duncoafc Possibly, but more likely working on surveillance data for some skevy giant advertising company
Sat Nov 09 18:29:00 +0000 2019@UCDProteomics @PastelBio @duncoafc Where are quantum computers when you need them?
Sat Nov 09 17:44:22 +0000 2019@UCDProteomics @PastelBio @duncoafc Thinking about the TIMS problem for a bit, it may not be easy to split into parallel processes unless someone can come up with a clever decomposition of the signals into independent problems.
Sat Nov 09 17:17:10 +0000 2019@UCDProteomics @PastelBio @duncoafc I was going to ask about whether the peak finding was using more than one core. I used to do quite a bit of TOF data handling & most people's first impulse is to do peak finding as a serial process.
Sat Nov 09 15:48:55 +0000 2019@MHendr1cks I saw a nice talk by a vice dean at UBC who had done a thorough network analysis of "collaboration" at the Med School. She found that the dominant form of "collaboration" was dept. heads being named on grants and papers authored by junior faculty.
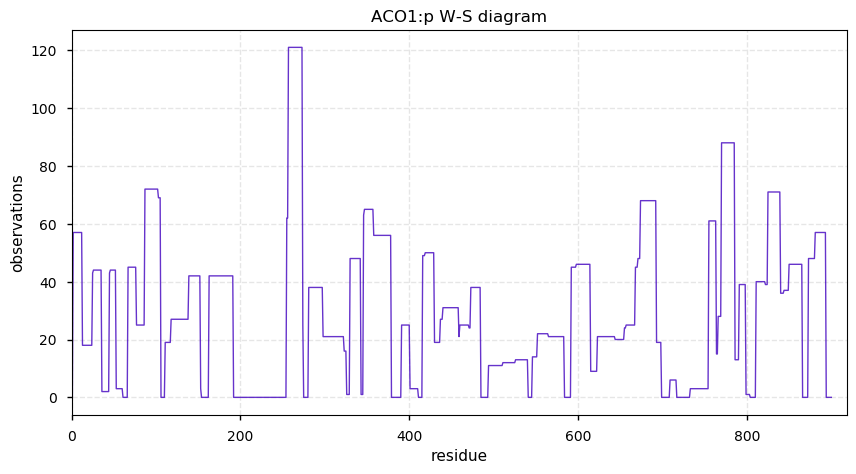
Sat Nov 09 15:33:07 +0000 2019ACO1:p, aconitate hydratase 1 (Cannibis sativa) 🔗| Very abundant in seeds; catalyzes citrate → isocitrate; mature form 2-900 [66 x] 🔗
Fri Nov 08 18:12:33 +0000 2019@UCDProteomics But at least the time estimate is to 10 sig figs!
Fri Nov 08 15:32:47 +0000 2019@mjmaccoss There does seem to be some sympathy for using a generic name like GVP, as opposed to specifying the mechanism of variation that has resulted in a detected peptide sequence change. Do you include splice variants under this category?
Fri Nov 08 15:06:02 +0000 2019@UCDProteomics You need at least a keg/sample. I saw guys in Germany doing GC/MS on wine who insisted they needed a case per analysis.
Fri Nov 08 14:21:23 +0000 2019Thanks to everyone who participated in this poll. The results are 58% for SAAV and 42% divided evenly between SNAP, SAP and SAV. SAAV is clearly the majority choice.
Fri Nov 08 14:07:35 +0000 2019@oceileachair To be more exact, any non-synonymous SNP that is detectable in a protein sequence using current MS/MS methods.
Fri Nov 08 13:56:06 +0000 2019@oceileachair Any non-synonymous SNP
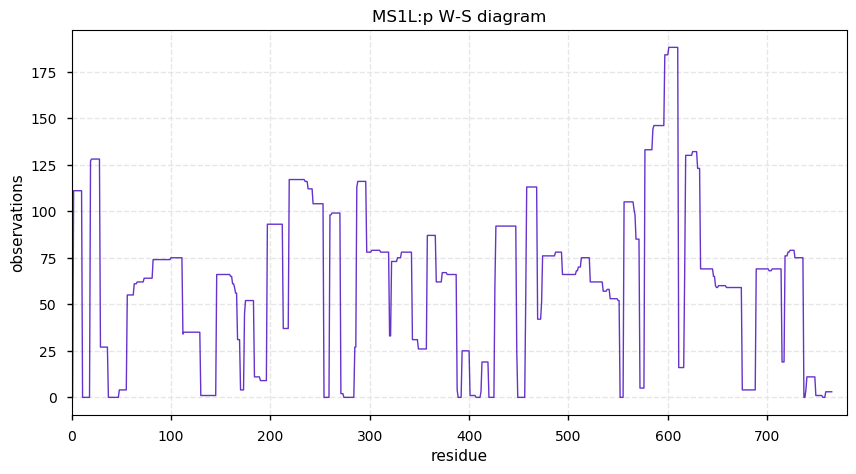
Fri Nov 08 13:22:45 +0000 2019MS1L:p, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase 1-like (Cannibis sativa) 🔗| Abundant in seeds; catalyzes 5-methyltetrahydropteroyltri-L-Glu + L-homoCys = L-Met + tetrahydropteroyltri-L-Glu; mature form 2-765 [68 x] 🔗
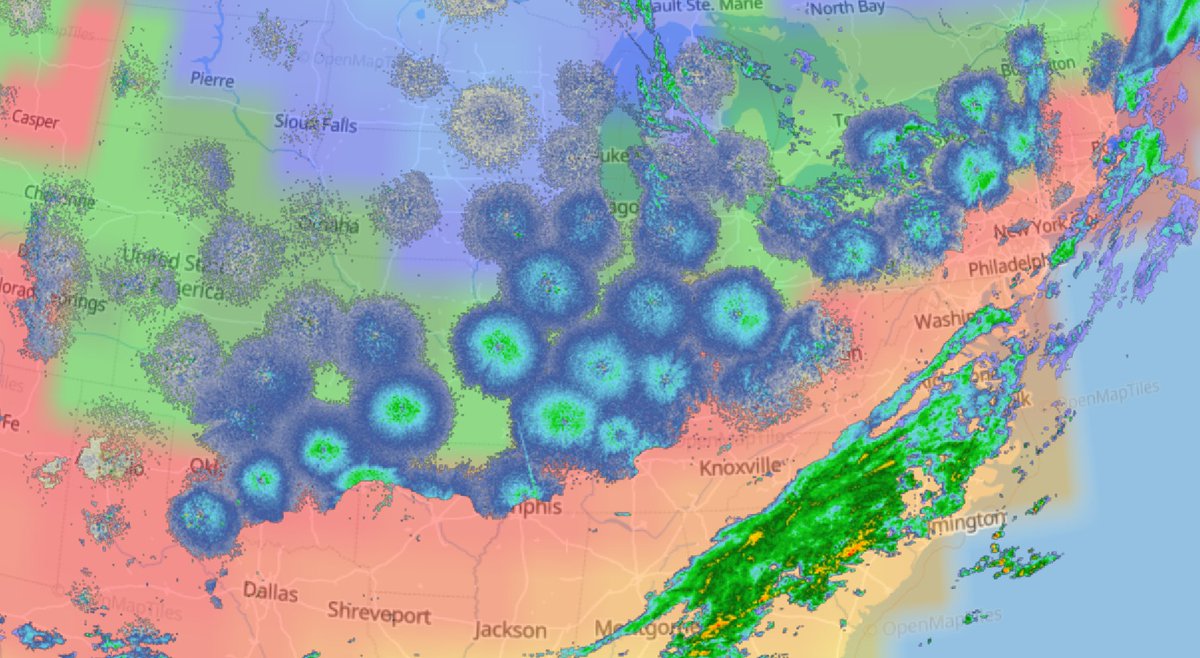
Fri Nov 08 03:53:23 +0000 2019Some over-the-top ground clutter signals from NEXRAD across the US Midwest at about 22:00 CST 🔗
Thu Nov 07 22:04:05 +0000 2019@AlexUsherHESA @univcan @KirstyDuncanMP I meant that it could be both the result of institutional blinkering and governmental policy mandates.
Thu Nov 07 21:54:07 +0000 2019@AlexUsherHESA @univcan @KirstyDuncanMP Can't it be both?
Thu Nov 07 21:02:37 +0000 2019So far (with 23 votes cast), it looks like the community favors calling YBX2:p.S63P (MAF =0.4) an SAAV, SAV, SNAP or SAP, in descending order of preference. Point or sequence variant as well as SAAP have also been suggested parenthetically.
Thu Nov 07 17:37:38 +0000 2019@chrashwood It is one word in many languages (German, Dutch, Spanish, Chinese, Russian). I've always thought the additional "A" was a bit too literal for an acronym, but I'm willing to go with the community's opinion.
Thu Nov 07 16:44:34 +0000 2019I now prefer using "variant" for this type of thing rather than polymorphism. I had the misfortune of taking several years of genetics classes (before sequenced genomes) & "polymorphism" has a lot of baggage from classical genetics that can confuse the issue.
Thu Nov 07 16:28:25 +0000 2019My wife had ongoing problems like this when I worked in the US, but there it is was refusing to believe that an OUS person (Canadian) could be a native English speaker ...
🔗
Thu Nov 07 14:14:27 +0000 2019Increasingly proteomics data is being used to confirm sequence changes caused by single nucleotide variants (aka SNV, SNP). What is the best acronym for these changes at the peptide sequence level?
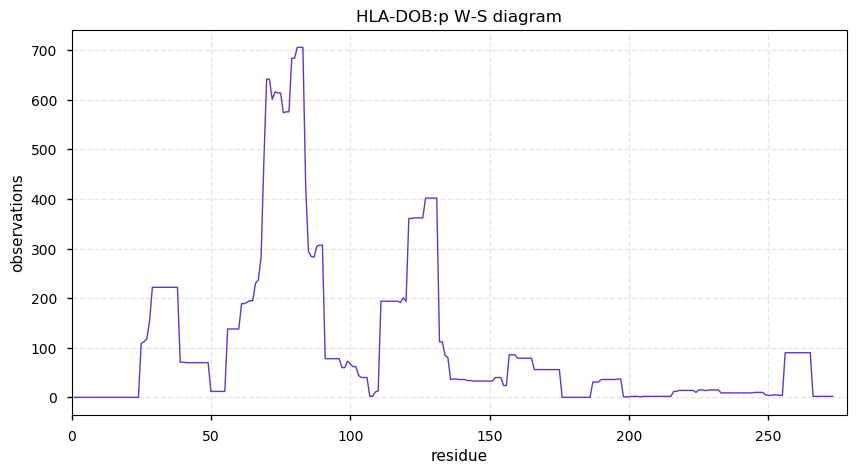
Thu Nov 07 14:03:38 +0000 2019HLA-DOB:p, major histocompatibility complex, class II, DO beta (H. sapiens) 🔗 Small membrane subunit; no PTMs; no SAVs; overrepresented in MHC class II peptide expts.; multiple haplotypes; mature form (2,28)-273 [654 x] 🔗
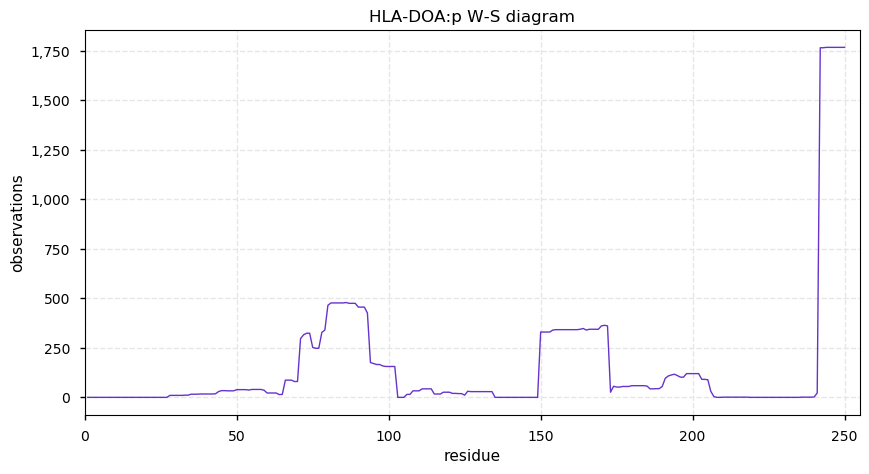
Wed Nov 06 13:49:19 +0000 2019HLA-DOA:p, major histocompatibility complex, class II, DO alpha (H. sapiens) 🔗 Small membrane subunit; no sig. PTMs; no SAVs; overrepresented in MHC class II peptide expts.; multiple haplotypes; mature form 26?-250 [1,109 x] 🔗
Wed Nov 06 13:09:54 +0000 2019@Sci_j_my Protons, neutrons & electrons (physics version)
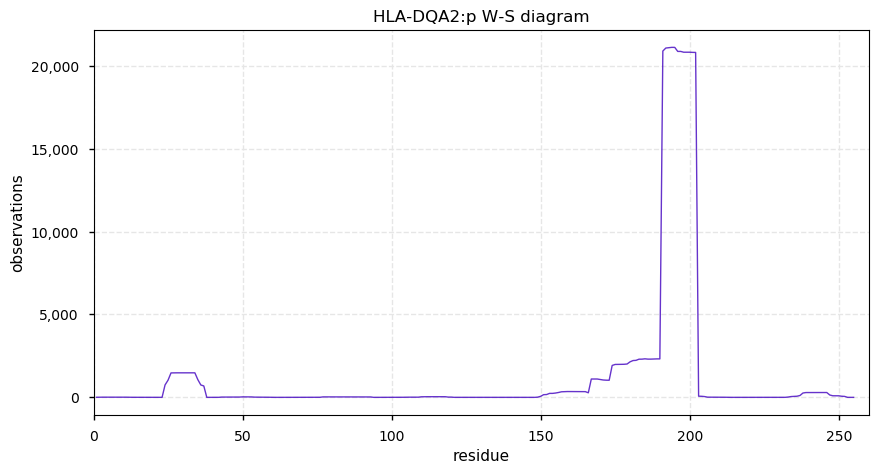
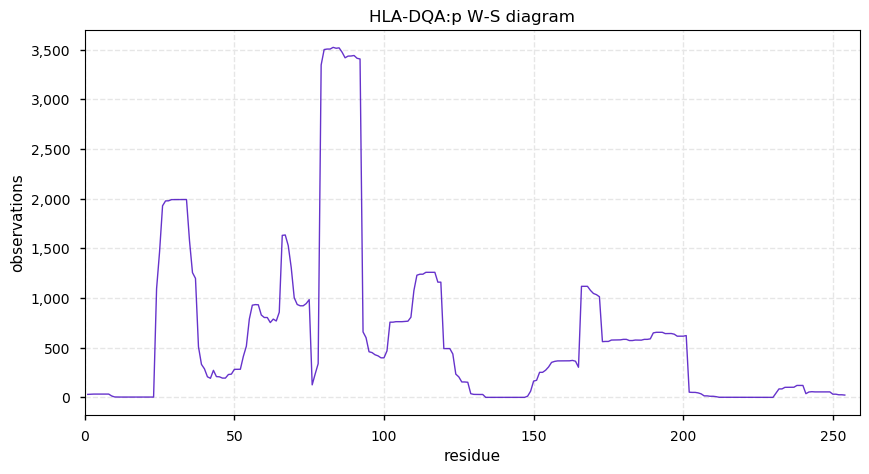
Tue Nov 05 14:02:13 +0000 2019HLA-DQA2:p, major histocompatibility complex, class II, DQ alpha 2 (H. sapiens) 🔗 Small membrane subunit; no PTMs; no SAVs; overrepresented in MHC class II peptide expts.; multiple haplotypes; mature form 24-255 [7,545 x] 🔗
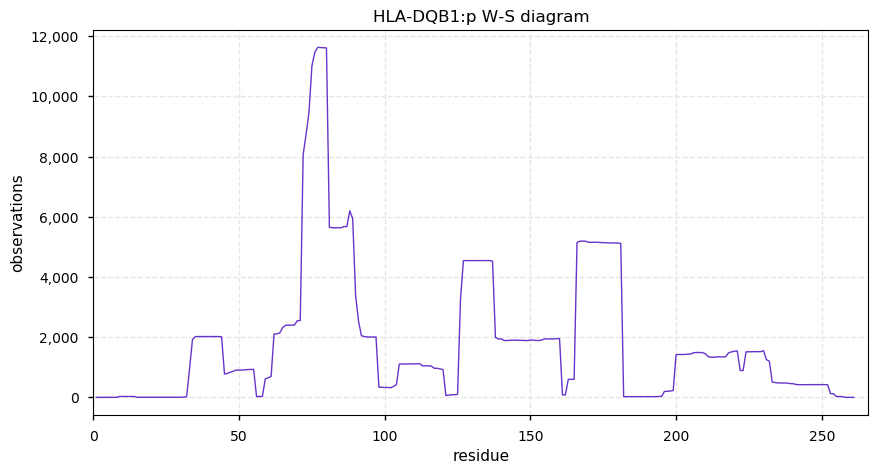
Mon Nov 04 13:37:40 +0000 2019HLA-DQB1:p, major histocompatibility complex, class II, DQ beta 1 (H. sapiens) 🔗 Small membrane subunit; no PTMs; many high MAF SAVs; overrepresented in MHC class II peptide expts.; multiple haplotypes; mature form 33-261 [5,762 x] 🔗
Sun Nov 03 15:22:20 +0000 2019@jwoodgett No, the 2008 economic collapse. It changed the way universities and government granting agencies were funded and accelerated the decline of departments as primary influences within University administrations
Sun Nov 03 15:05:22 +0000 2019@jwoodgett I'd put the date at pre-2008
Sun Nov 03 14:21:36 +0000 2019I still haven't seen any publicly available data from any of the new Thermo instruments announced at the ASMS this year.
Sun Nov 03 13:58:37 +0000 2019HLA-DQA1:p, major histocompatibility complex, class II, DQ alpha 1 (H. sapiens) 🔗 Small membrane subunit; no PTMs; 5 high MAF SAVs; overrepresented in MHC class II peptide expts.; multiple haplotypes; mature form 24-254 [2,941 x] 🔗
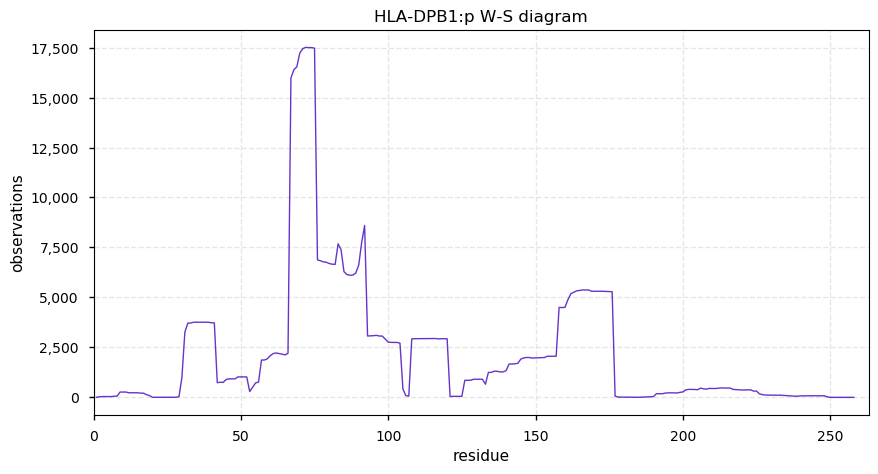
Sat Nov 02 15:44:02 +0000 2019HLA-DPB1:p, major histocompatibility complex, class II, DP beta 1 (H. sapiens) 🔗 Small membrane subunit; no PTMs; 21 high MAF SAVs; overrepresented in MHC class II peptide expts.; multiple haplotypes; mature form (30,31)-258 [8,175 x] 🔗
Fri Nov 01 19:14:22 +0000 2019@pwilmarth @UCDProteomics They will almost certainly be in the supplementary materials.
Fri Nov 01 19:09:09 +0000 2019@pwilmarth @UCDProteomics I would never think of trying to replace Nature Methods efforts to perform this great task. Although it could easily spawn Nature Jargon at any time.
Fri Nov 01 19:05:04 +0000 2019@kadzuis 6/6 People like to use simple keyword searches to find things, as Google has trained them to expect. Try to emulate (& facilitate) this approach as much as possible.
Fri Nov 01 19:04:21 +0000 2019@kadzuis 5/6 Ontologies can be helpful, but their singly-direct, non-cyclic graph interpretation doesn't fit well with a lot of experimental data.
Fri Nov 01 19:03:58 +0000 2019@kadzuis 4/6 Do not expect any chemist or biologist to understand that ontologies are anything but lists of keywords.
Fri Nov 01 19:03:36 +0000 2019@kadzuis 3/6 Breaking things up into a lot of columns looks like a good idea, but can become restrictive and it doesn't result in faster lookups.
Fri Nov 01 19:03:17 +0000 2019@kadzuis 2/6 For database storage: put as much metadata as possible in long strings of structured text (preferably JSON).
Fri Nov 01 19:02:40 +0000 2019@kadzuis Ron's metadata tips:
1/6 Use UTF-8. Always.
Fri Nov 01 18:51:10 +0000 2019@UCDProteomics It has been redefined repeatedly over the years (seems to be a bit of a hobby for Ruedi). I would say that it really doesn't have any well-established meaning: it is now mainly lab jargon.
Fri Nov 01 18:48:01 +0000 2019@chrashwood @kadzuis I pay for it with some money from software licenses and some additional top-up from myself. It has never had any funding that originated from government grants.
Fri Nov 01 16:10:45 +0000 2019@kadzuis Yup. It is my sole proprietorship.
Fri Nov 01 15:49:09 +0000 2019@Peptidome Do you know of any reference where the postulate that RNA transcript concentration should be an accurate surrogate for protein concentration was originally suggested? It seems to be a notion that many take for granted as true, but someone must have said it first.
Fri Nov 01 13:25:32 +0000 2019@darcianflux @SatchinPanda @NEJM But nature adores any number of false correlations.
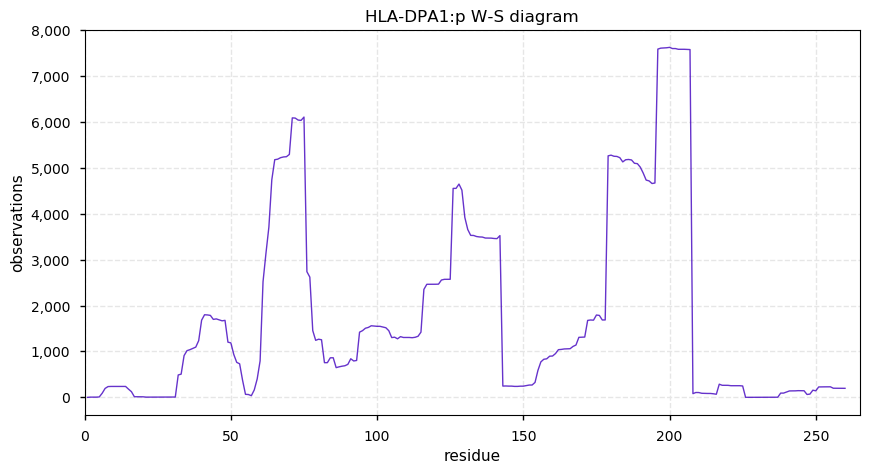
Fri Nov 01 13:07:57 +0000 2019HLA-DPA1:p, major histocompatibility complex, class II, DP alpha 1 (H. sapiens) 🔗 Small membrane subunit; no PTMs; major SAVs: A42T (maf=0.24), Q81R (maf=0.44), T114A/P/S (maf=0.44); multiple haplotypes; mature form 32-260 [5,573 x] 🔗
Thu Oct 31 20:20:12 +0000 2019@Smith_Chem_Wisc These are the hydroxyproline containing peptides from a typical prostate cancer tissue sample: 🔗
from CPCG_0346_1.raw at ftp://massive.ucsd.edu/MSV000081552/raw/RAW/
Thu Oct 31 19:58:03 +0000 2019@Smith_Chem_Wisc That range is normal for the collagens that have hydroxyproline & hydroxylysine. Many tissue samples will have that type of modified peptide, while almost all cell lines will not. It is particularly prevalent in tumour tissue samples.
Thu Oct 31 16:35:51 +0000 2019@Smith_Chem_Wisc New, improved, retains peptide redundancy!
Thu Oct 31 15:48:33 +0000 2019@Smith_Chem_Wisc And if you want a script that fully implements the idea:
🔗
Note: 🔗 outputs different FASTA file every time you run it, but preserves the length and AAA of tryptic peptides.
Thu Oct 31 15:08:53 +0000 2019@Smith_Chem_Wisc If you are will to use Python:
from random import shuffle
def s_p(_p):
kr = _p[len(_p)-1]
l = list(_p[:-1])
shuffle(l)
return ''.join(l)+kr
pep = 'DAMQYASESK'
npep = s_p(pep)
print(pep,npep)
Simple & it preserves AAA composition.
Thu Oct 31 14:47:02 +0000 2019It still surprises me that someone will go to great lengths designing & optimizing an experiment, uses the latest, most expensive gear & then uses the default settings on a search engine to analyze it — settings that haven't changed since 1997.
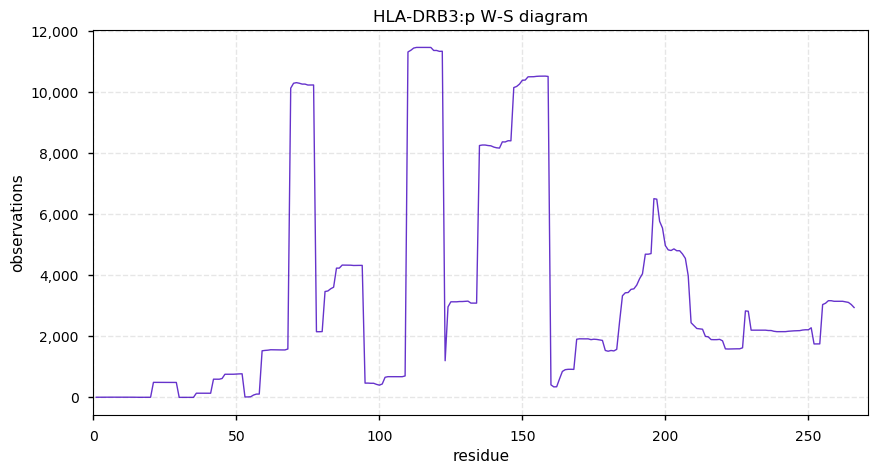
Thu Oct 31 13:29:59 +0000 2019HLA-DRB3:p, major histocompatibility complex, class II, DR beta 3 (H. sapiens) 🔗 Small membrane subunit; no significant PTMs; 1 SAV: Y76F (maf=0.14); over-represented in MHC class II peptide expts.; multiple haplotypes; mature form 30-266 [10,164 x] 🔗
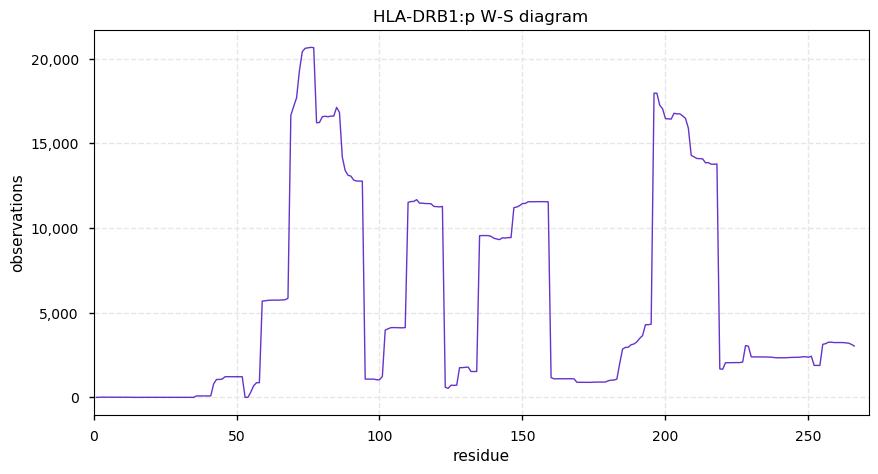
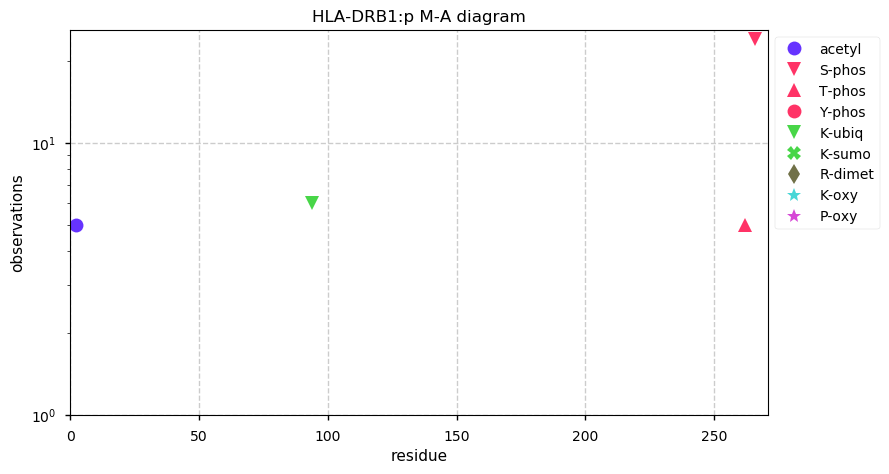
Wed Oct 30 13:08:34 +0000 2019HLA-DRB1:p, major histocompatibility complex, class II, DR beta 1 (H. sapiens) 🔗 Small membrane subunit; no significant PTMs; many SAVs; over-represented in MHC class II peptide expts.; multiple haplotypes; mature form 29-266 [11,985 x] 🔗
Wed Oct 30 12:42:37 +0000 2019A common mistake, but one too many A's in SAV ...
🔗
Tue Oct 29 17:08:54 +0000 2019Britain is at a point now where the Commonwealth should be thinking about intervening to re-establish order and good government 🔗
Tue Oct 29 16:11:09 +0000 2019🔗
🐕 or 🐴 ?
Tue Oct 29 15:13:37 +0000 2019@nesvilab @UCDProteomics @JustinWWalley I've used Aspera quite a bit (mainly because CPTAC requires it). While it can be made to work, it is not great for downloading large datasets.
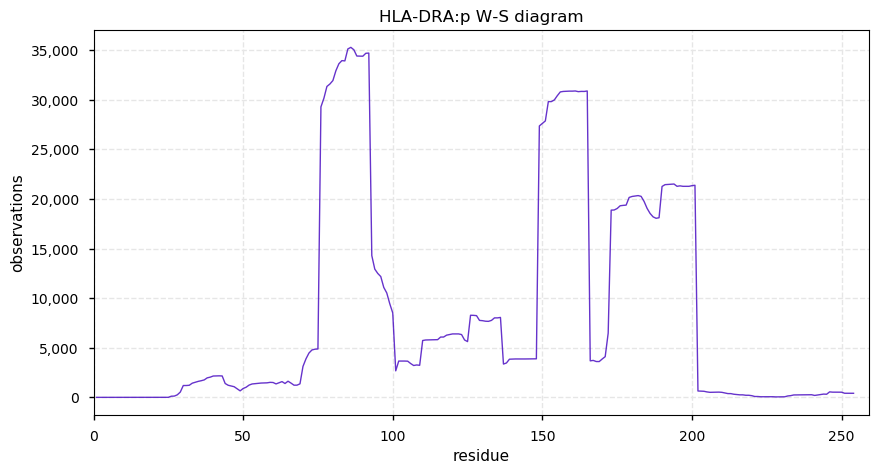
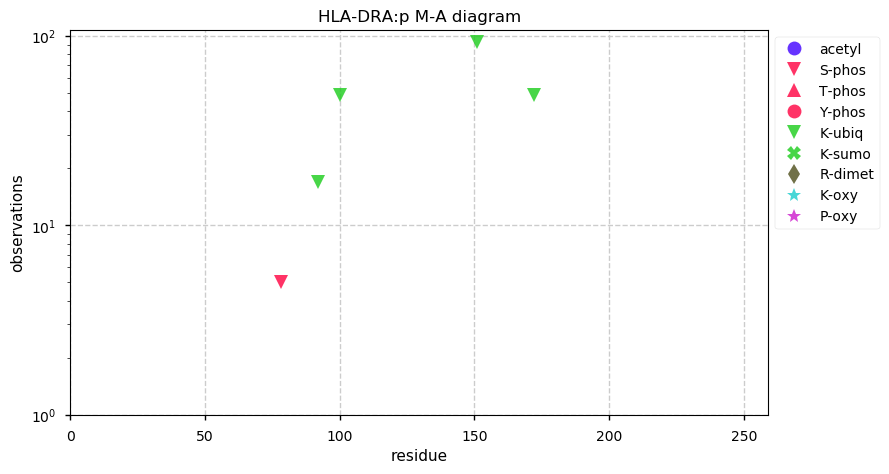
Tue Oct 29 13:17:10 +0000 2019HLA-DRA:p, major histocompatibility complex, class II, DR alpha (H sapiens) 🔗 Small membrane subunit; no significant PTMs; no SAVs; overrepresented in MHC class II peptide expts.; multiple haplotypes; mature form 26-254 [13,504 x] 🔗
Mon Oct 28 14:31:34 +0000 2019TAP1+TAP2 transport MHC I peptides from the cytoplasm into the ER, where the MHC I protein has been assembled on the inner side of the ER membrane. The membrane & protein+peptide are then exported intact via the Golgi so that the protein+peptide is on the outside of the cell.
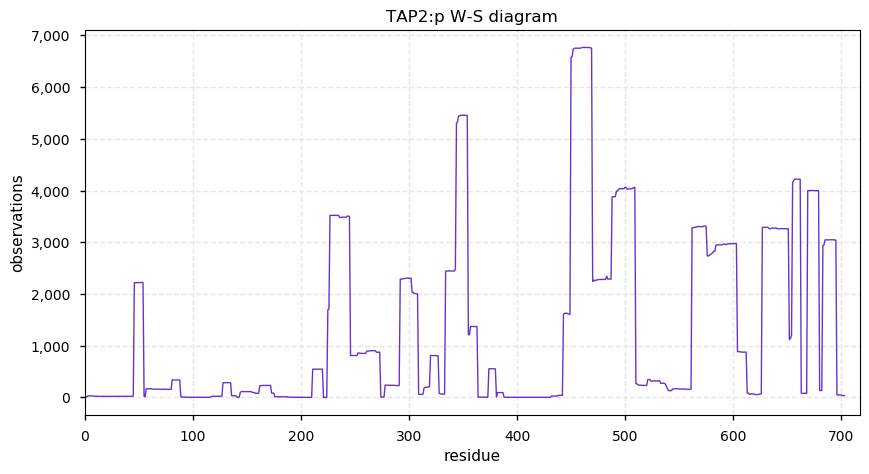
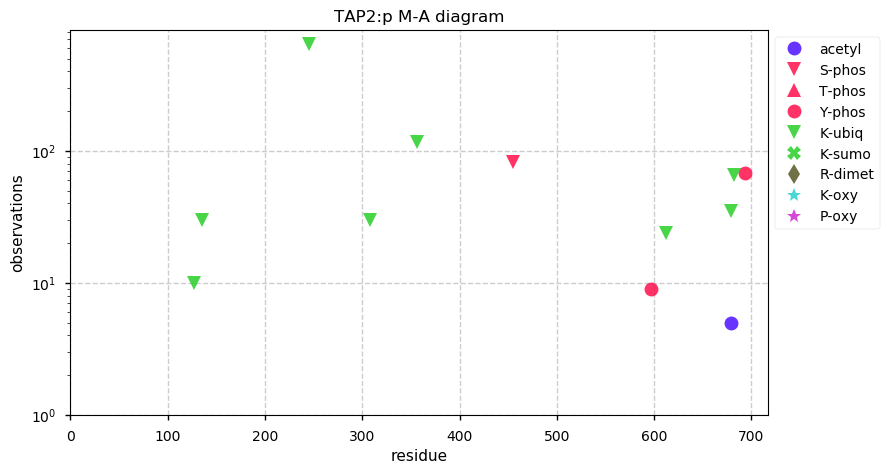
Mon Oct 28 13:24:54 +0000 2019TAP2:p, transporter 2, ATP binding cassette subfamily B member (Homo sapiens) 🔗 Midsized ER subunit; scattered PTMs; multiple SAVs: V379I (maf=0.2), A565T (maf=0.1), R651C (maf=0.1); 9 predicted TM domains; 2 splice variants; mature form 1-808 [13,115 x] 🔗
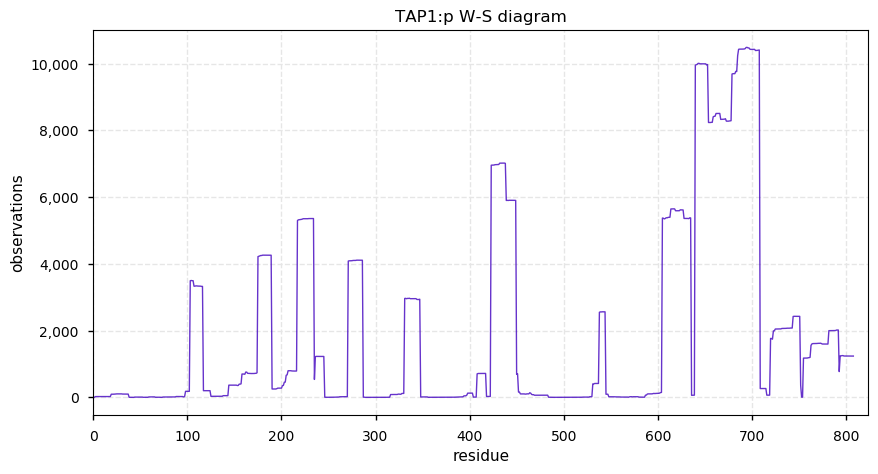
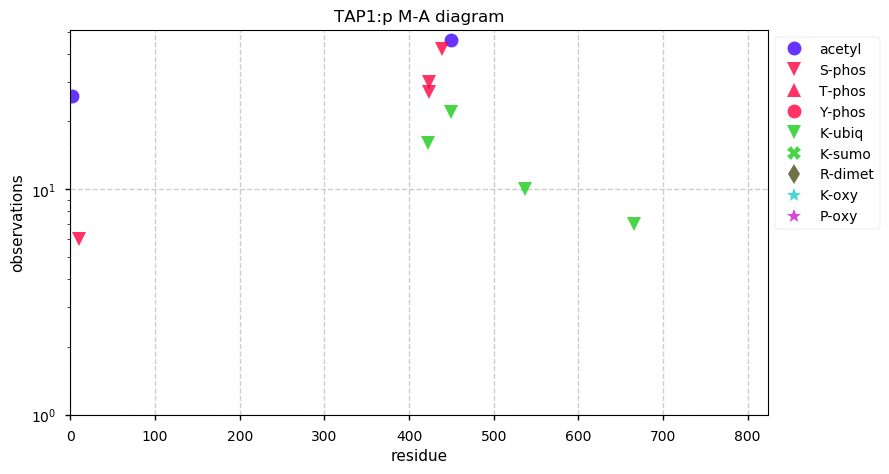
Sun Oct 27 13:20:42 +0000 2019TAP1:p, transporter 1, ATP binding cassette subfamily B member (H. sapiens) 🔗 Midsized ER membrane subunit; 1 phosphodomain; 3 SAVs: S346C (maf=0.01), S346F (maf=0.01), A430V (maf=0.03); 10 TM domains; 1 splice variant; mature form 1-808 [14,961 x] 🔗
Sun Oct 27 01:53:31 +0000 2019@JoelisSteele @Sci_j_my No. Do not tell them. They will not take it well.
Sat Oct 26 13:14:43 +0000 2019One B2M subunit and one subunit of either HLA-A, HLA-B, HLA-C, HLA-E, HLA-F or HLA-G forms the MHC class 1 antigen presentation protein in humans.
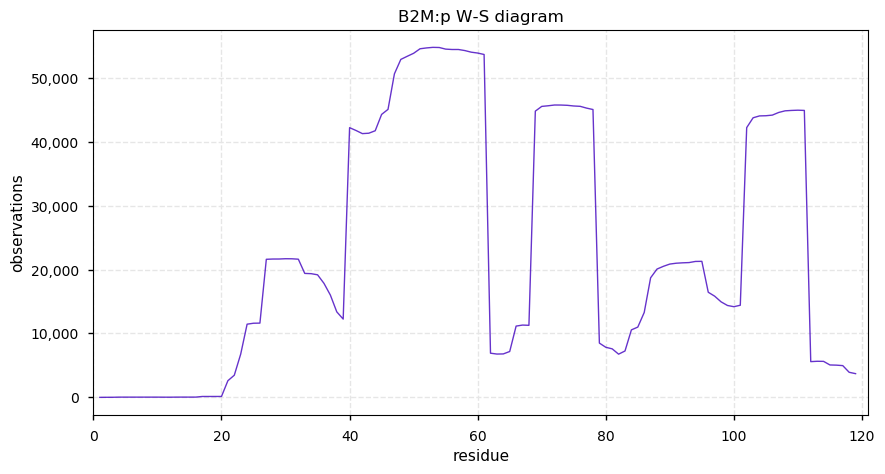
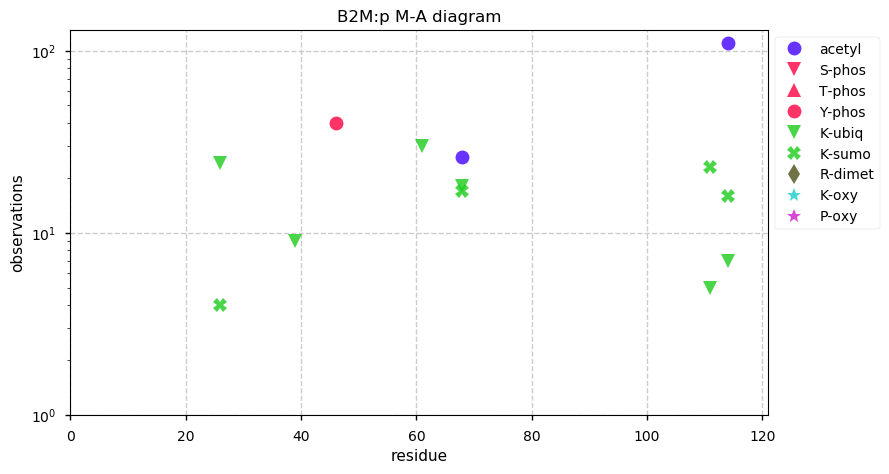
Sat Oct 26 13:08:37 +0000 2019B2M:p, beta-2-microglobulin (H. sapiens) 🔗 Very small membrane-associated subunit; sparse PTMs; no SAVs; over-represented in HLA II peptide expts; 1 splice variant; mature form 21-119 [24,721 x] 🔗
Fri Oct 25 22:28:01 +0000 2019I ran another HeLa digest: this time from home-brew cells (🔗) rather than from a bottle. The artisanal digest ended up with 2.8% of PSMs being bovine, compared with 5.1% of PSMs from the Pierce HeLa standard.
Fri Oct 25 21:55:05 +0000 2019@cstross I had a friend who had one and until he decommissioned it his office smelled like toasted plastic ...
Fri Oct 25 14:14:54 +0000 2019I just ran across this paper 🔗 that seems to be thinking in a similar way, although coming at it for a related-but-different experimental situation.
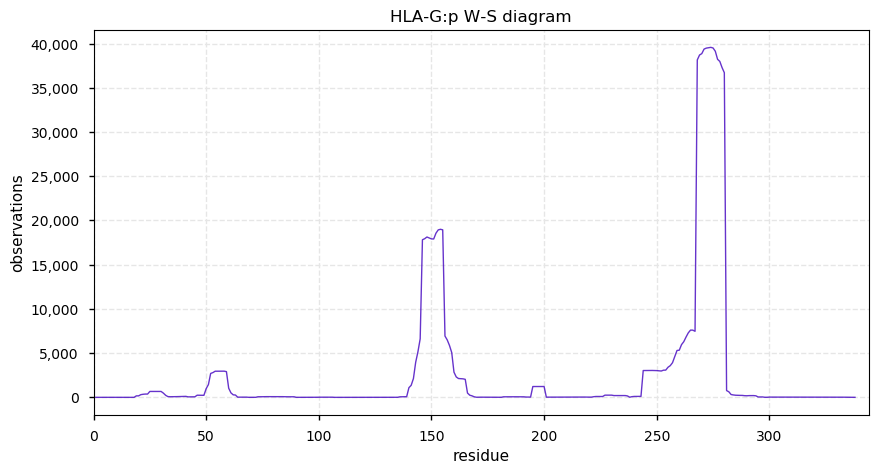
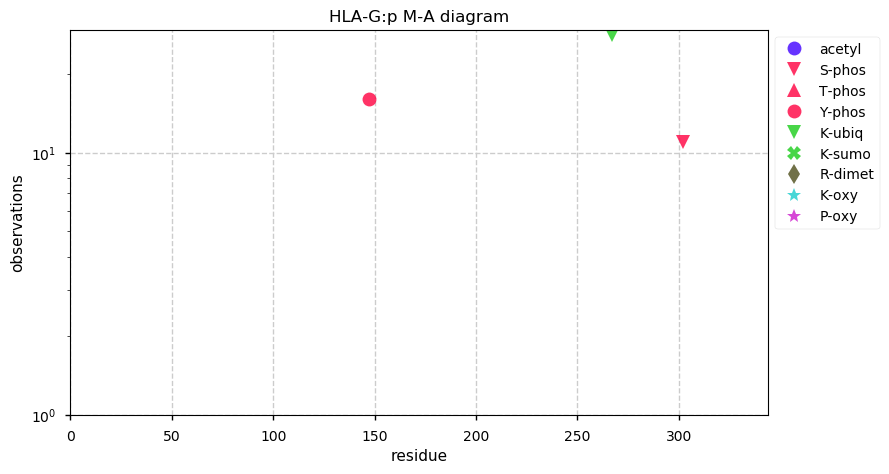
Fri Oct 25 13:00:16 +0000 2019HLA-G:p, major histocompatibility complex, class I, G (H. sapiens) 🔗 Small membrane-associated subunit; sparse PTMs; 1 SAV: T55S (maf=0.05); over-represented in HLA II peptide expts; multiple haplotypes; mature form (19,21)-338 [22,021 x] 🔗
Thu Oct 24 22:24:05 +0000 2019@cwvhogue coverage = 32% FOR C3, 33% for ALB, 68% for HBB, 49% for APOA1, 38% for SERPINA1, 38% for AHSG
Thu Oct 24 21:28:05 +0000 2019@cwvhogue I would agree, but I'm pretty sure there was way too much signal for it to be simply from endocytosis. Bovine C3 was the 15th most abundant protein (by spectrum counts).
Thu Oct 24 21:21:11 +0000 20193/3 I put together a short (38 entry) FASTA file that has the most abundant bovine proteins found in that extract, including almost all of the detected PSMs. If you want to check your sample for bovine contamination, try adding this to your search 🔗
Thu Oct 24 21:17:17 +0000 20192/3 So I analyzed a nice dataset that used the Pierce HeLa protein digest standard (cat #88328) & found prominent bovine proteins. The most abundant glycoprotein (by PSMs) was bovine C3. The bovine proteins were all from plasma and had no business being in a cell lysate.
Thu Oct 24 21:11:26 +0000 20191/3 I can't remember who was discussing this issue, but someone commented on a paper showing significant bovine glycosylation in a commercial HeLa extract. In response someone mentioned that they often find bovine proteins in HeLa-derived protein samples.
Thu Oct 24 18:36:00 +0000 2019And does anybody know how this is addressed in any of the secret-source apps (Mascot, Sequest, Andromeda, Byonic, Peaks, etc.)?
Thu Oct 24 16:50:05 +0000 2019Wow. What could possibly go wrong with this idea:
🔗
Thu Oct 24 16:09:10 +0000 2019Does anyone in the search engine design/implementation biz want to comment on this idea?
Thu Oct 24 16:07:56 +0000 2019Thanks to everyone who participated in this poll. 22 people expressed an opinion: 11 for "yes", 4 for "no" & 7 for "yes, depends on sequence". Put another way, 18 were in favor of using 2+ fragment ions to score PSMs in some way and 4 wanted only 1+ ions to be used.
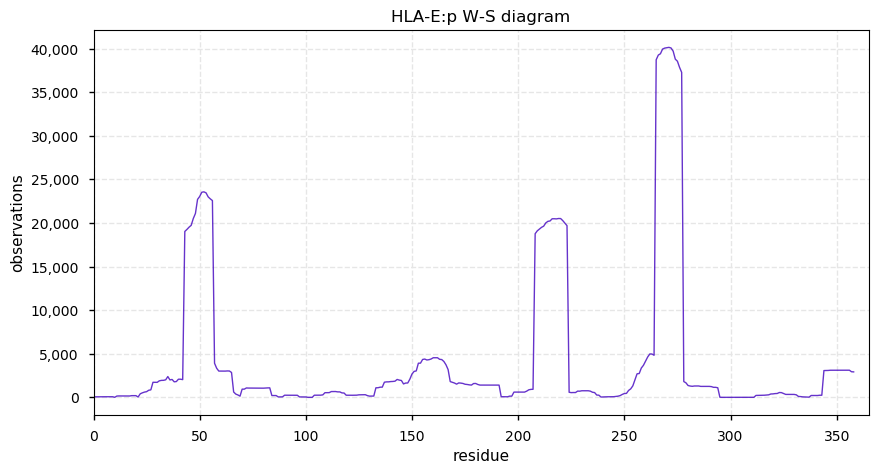
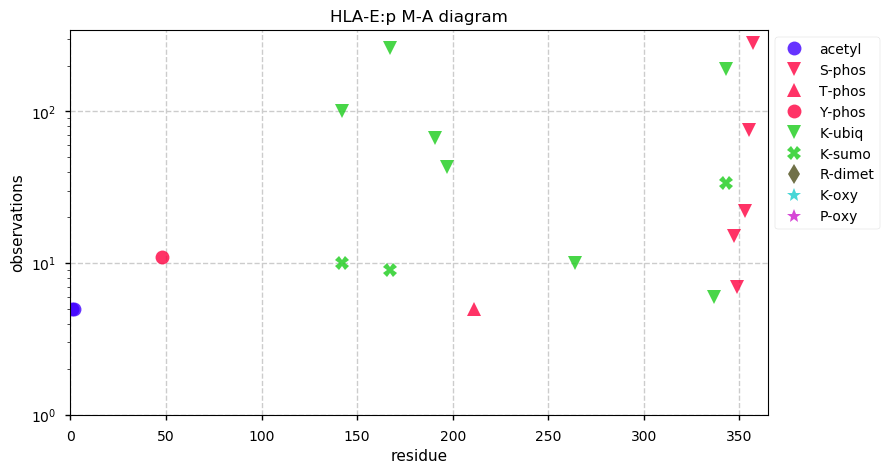
Thu Oct 24 14:11:08 +0000 2019HLA-E:p, major histocompatibility complex, class I, E (Homo sapiens) 🔗 Small membrane-associated subunit; C-terminal phosphodomain; no SAVs; over-represented in HLA II peptide expts; multiple haplotypes; mature form 22-358 [26,044 x] 🔗
Wed Oct 23 15:56:00 +0000 2019Most peptide identification algorithms check for singly-charged fragment ions when testing MS/MS spectra from parent ions with z=2. Should they also check for doubly-charged fragment ions from z=2 parents?
Wed Oct 23 15:29:05 +0000 2019Thanks to everyone who participated in this poll. Kolmogorov-Smirnov was the favorite. I don't know if the small number of respondents was because this sort of testing isn't commonly done or because people use platforms that don't describe the tests involved.
Wed Oct 23 14:21:01 +0000 2019@RuneLinding Can't happen soon enough for me: I hate optimizing code!
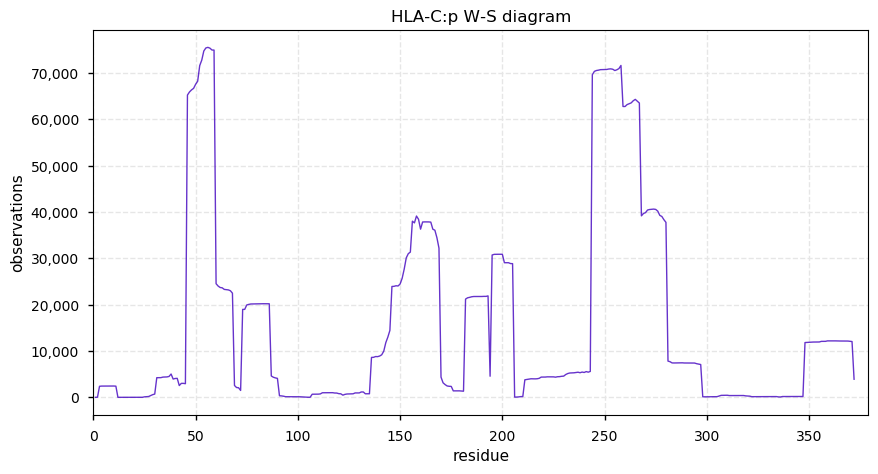
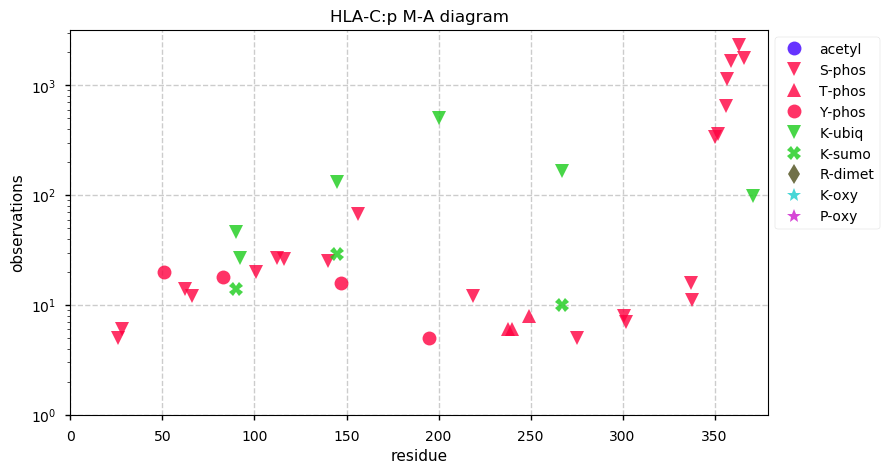
Wed Oct 23 14:00:26 +0000 2019HLA-C:p, major histocompatibility complex, class I, C (H. sapiens) 🔗 Small membrane-associated subunit; variably phosphorylated; many SAVs; over-represented in HLA II peptide expts; multiple haplotypes; mature form 19-372 [42,023 x] 🔗
Tue Oct 22 15:47:14 +0000 2019For instance, which of these tests do you use to determine if the PSMs corresponding to a rare PTM/SAV that you found in a data set were likely to be genuine or false positives?
Tue Oct 22 15:16:04 +0000 2019What mathematical test do you prefer for determining whether a subset of results conforms to the whole:
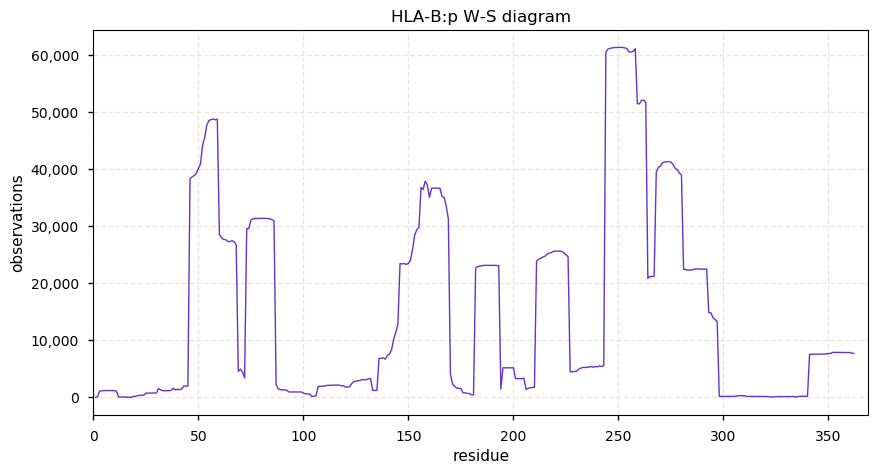
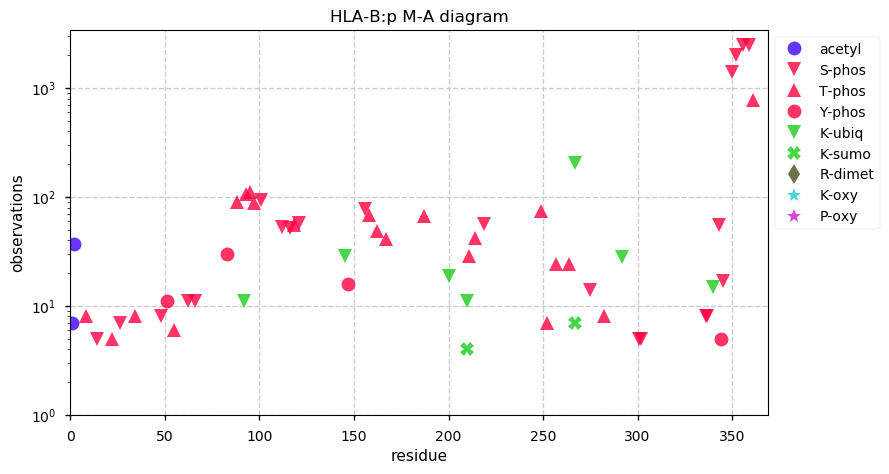
Tue Oct 22 12:26:19 +0000 2019HLA-B:p, major histocompatibility complex, class I, B (H.sapiens) 🔗 Small membrane-associated subunit; variably phosphorylated; many SAVs; over-represented in HLA II peptide expts; multiple haplotypes; mature form 19-362 [38,064 x] 🔗
Mon Oct 21 14:40:09 +0000 2019After suffering through the 40 days of campaigning associated with the Canadian federal election, I am confident in declaring it both the worst campaign ever & a triumph of stealth voter suppression by all parties.
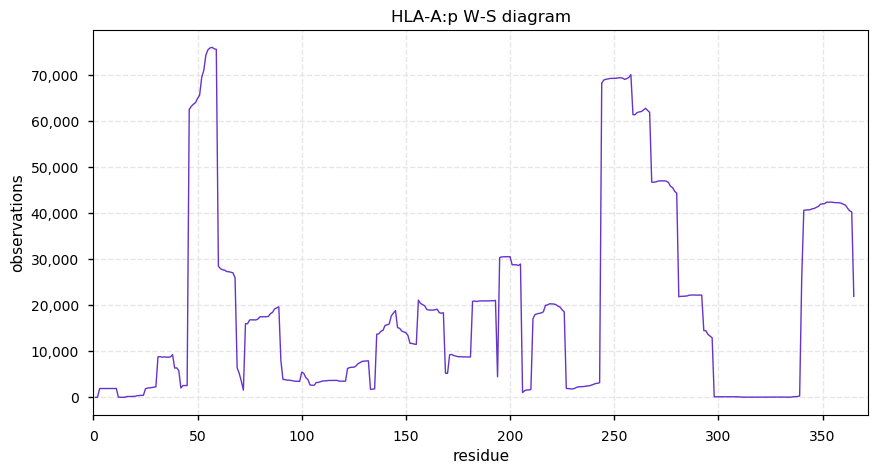
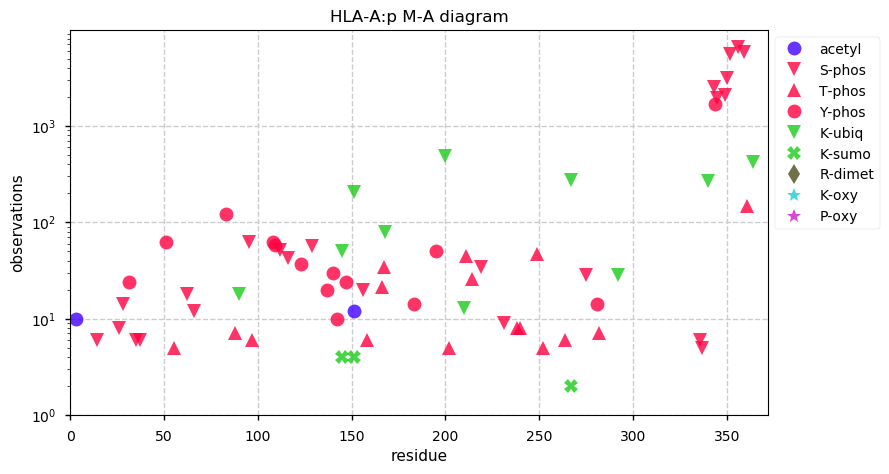
Mon Oct 21 13:02:45 +0000 2019HLA-A:p, major histocompatibility complex, class I, A (H. sapiens) 🔗 Small membrane-associated protein; variably phosphorylated; many SAVs; over-represented in HLA II peptide expts; multiple haplotypes; mature form 25-365 [40,661 x] 🔗
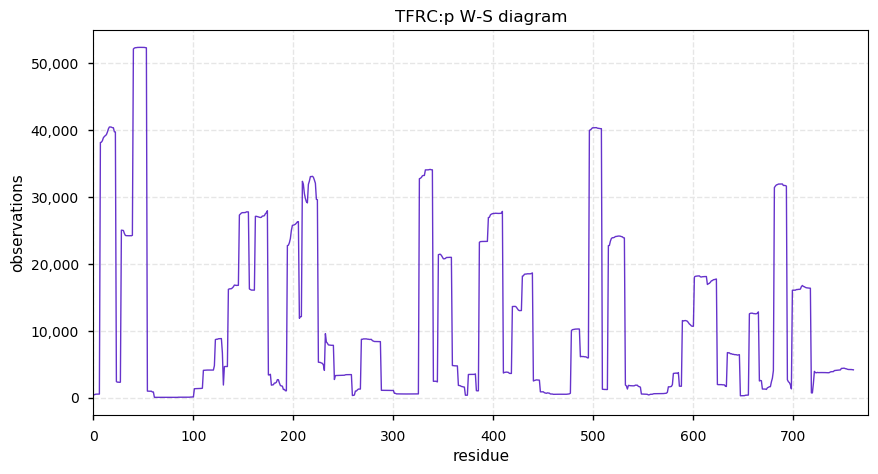
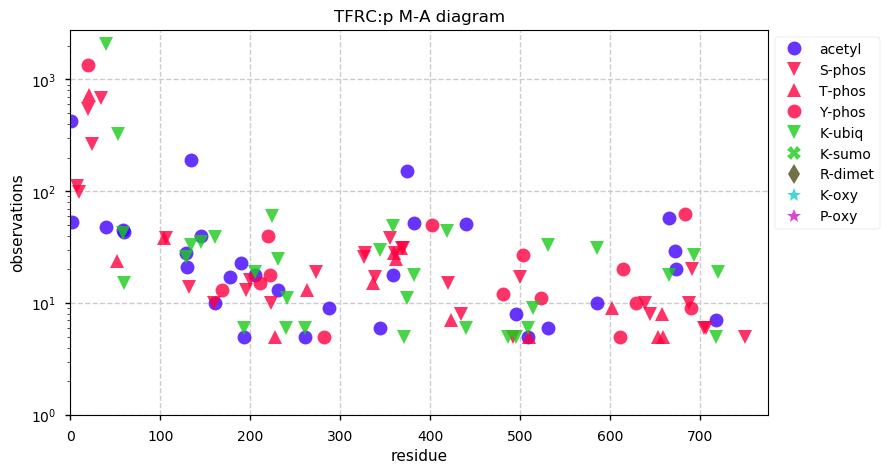
Sun Oct 20 11:31:44 +0000 2019TFRC:p, transferrin receptor (H. sapiens) 🔗 Midsized plasma membrane protein; highly modified; 2 SAVs: G142S (maf=0.3), G420S (maf=0.01); over-represented in HLA II peptide expts; 1 splice variants; mature form (1,2,101)-760 [45,015 x] 🔗
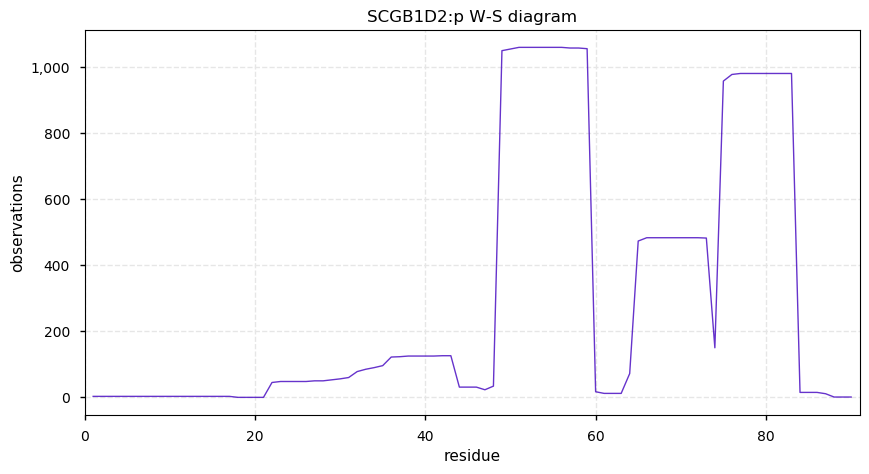
Sat Oct 19 12:15:32 +0000 2019SCGB1D2:p, secretoglobin family 1D member 2 (H. sapiens) 🔗 Very small protein; no PTMs; 1 SAV: P53L (maf=0.4); sHLA peptide 35-46; 1 splice variants; most abundant in sweat and uterine mucosa; mature form 22-90 [184 x] 🔗
Fri Oct 18 19:27:02 +0000 2019@neely615 @Smith_Chem_Wisc If you want to hunt for rare things (like SAVs), then it is worth it. Otherwise probably not.
Fri Oct 18 19:25:14 +0000 2019@neely615 @Smith_Chem_Wisc About 15% of peptides contain a W. As with M, all W oxidation is removed during the reduction step of C alkylation, so any W-ox you see is a sample prep artefact. Most sample prep ends up with about 5-10% of W-ox (i.e. ~1% of PSMs).
Fri Oct 18 16:12:05 +0000 2019A little bit of Javascript for obscuring text 🔗
Fri Oct 18 14:37:17 +0000 2019@theoneamit The method has changed over time, but the rule is that it chooses the X most intense peaks that pass all the tests. Tests include removing peaks from parent ions, parent ion small neutral losses, ions that are too small & A1's that are larger than the corresponding A0.
Thu Oct 17 19:15:11 +0000 2019@VATVSLPR Exactly. It is a widely used protocol here in Canada 😡🐦
Thu Oct 17 19:12:36 +0000 2019But it suggests that there may be more artefacts in the data that are created in situ because of the materials used in the cell preparation process
Thu Oct 17 19:00:07 +0000 2019My ISP seems to be relying on carrier pigeons today
Thu Oct 17 18:37:48 +0000 2019It still surprises me how often P02769 and O82803 peptides show up in HLA Class 2 studies. I guess they must become incorporated during cell isolation ...
Thu Oct 17 16:06:10 +0000 2019@doctorow I would hate to be a grad student in the current environment.
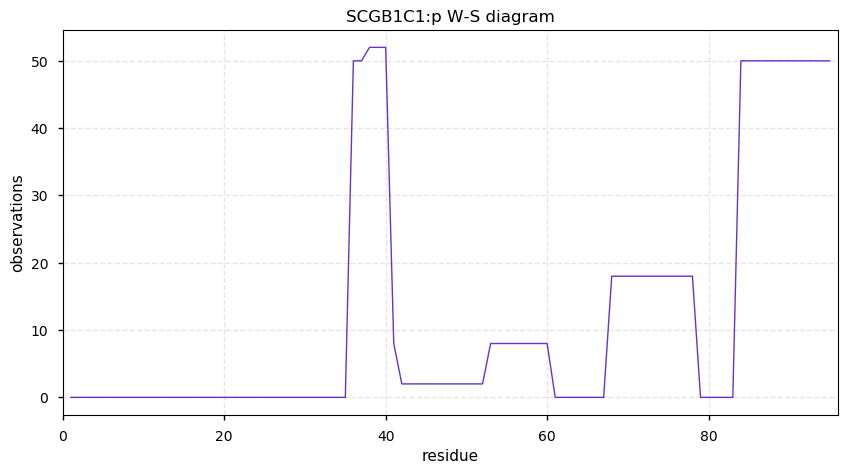
Thu Oct 17 12:10:23 +0000 2019SCGB1C1:p, secretoglobin family 1C member 1 (Homo sapiens) 🔗 Very small protein; no PTMs; no SAV; 1 splice variants; most abundant in nasal mucosa; mature form 24-95 [51 x] 🔗
Wed Oct 16 16:41:24 +0000 2019@neely615 You can use CAA, but you should change your alkylation protocols and check the recovery of Cys-containing peptides. As it turns out, even the group that promoted the change isn't immune to having CAA-alkylation fail without noticing it (e.g., PXD010697)
Wed Oct 16 15:14:31 +0000 2019@UCDProteomics When people do a lot of these experiments, they seem to lose interest in being quite so careful.
Wed Oct 16 14:41:59 +0000 2019@UCDProteomics The datasets I see strongly affected by carryover are affinity pull-down experiments trying to find protein-protein interaction partners.
Wed Oct 16 13:56:54 +0000 2019@KentsisResearch I mentioned it after seeing the 3rd public dataset in a week that was rendered irreproducible by a failed chloroacetamide alkylation (including 1 from the reagent's main proponent).
Wed Oct 16 13:12:46 +0000 2019@HHS_ORI @RuneLinding Regular (weekly at least) reading & review of lab notebooks and code.
Wed Oct 16 13:10:25 +0000 2019SCGB3A2:p, secretoglobin, family 3A, member 2 (Homo sapiens) 🔗 Very small protein; no PTMs; no SAV; 1 splice variants; most abundant in lung tissue; mature form 22-93 [176 x] 🔗
Tue Oct 15 15:57:42 +0000 2019Please stop using chloroacetamide when iodoacetamide would do. It has a much slower reaction rate and it doesn't effectively alkylate Cys residues in many commonly-used experimental protocols.
Tue Oct 15 14:55:52 +0000 2019@LewisGeer I had forgotten that they insisted on reminding you all the time that it was a registered trademark: caBIG®
Tue Oct 15 14:35:42 +0000 2019@jbrowaty @aldosantin @brent_bellamy I live downtown and I used to get vegetables and meat at the Forks several times a week. I simply don't go there any more because those things are no longer available and I don't want to eat at the beer hall (although I do like the cinnamon buns).
Tue Oct 15 14:28:57 +0000 2019Although I guess many people are too young to remember what an dragging-anchor on bioinformatics caBIG really was from 2004 to 2012 🔗
Tue Oct 15 14:25:34 +0000 2019@jwoodgett A laugh-out-loud tweet & article 🤣.
Tue Oct 15 14:07:45 +0000 2019From the guys who gave you caBIG, yet another project that will be done now+X years for as long as it is funded 🔗
Tue Oct 15 12:16:40 +0000 2019SCGB2A2:p, secretoglobin family 2A member 2 (Homo sapiens) 🔗 Very small protein; N53+glycosyl; no SAV; 1 splice variants; rare in cell lines; observed in nasal mucosa and breast tissue; mature form 19-93 [177 x] 🔗
Mon Oct 14 14:18:49 +0000 2019@christlet I have never met a Nobel laureate who did not tell some version of this story. Also, senior professors at research institutes favor recounting this type of anecdote about their struggles.
Mon Oct 14 12:55:28 +0000 2019SCGB2A1:p, secretoglobin family 2A member 1 (Homo sapiens) 🔗 Very small protein; no PTMs; no SAV; 1 splice variants; rare in cell lines, abundant in tears, uterine endometrium and nasal mucosa; mature form 19-95 [753 x] 🔗
Sun Oct 13 13:24:34 +0000 2019Secretoglobulins are a mammals-only family of very small proteins, largely secreted, with an odd set of trivial names (uteroglobin, mammaglobin, lacryglobin) that should be ignored 🔗
Sun Oct 13 13:17:54 +0000 2019@dtabb73 I've had nothing but good luck with Ryzens - I've made 6 computers based on them so far.
Sun Oct 13 13:02:25 +0000 2019SCGB1A1:p, secretoglobin family 1A member 1 (Homo sapiens) 🔗 Very small protein; no PTMs; no SAV; 2 splice variants (1 secreted, 1 intracellular); rare in cell lines; mature forms (19,20,21,22)-91 and (2,3)-56 [2,099 x] 🔗
Sat Oct 12 20:56:53 +0000 2019I regret ever purchasing a Microsoft product ...
🔗
Sat Oct 12 20:51:14 +0000 2019There is already a paper on the data (🔗), but it focuses on antibacterial peptides rather than the structure of the CMP. But if you love mucusoid structures, you could probably get 2 or 3 more papers out of the dataset.
Sat Oct 12 15:50:29 +0000 2019#PXD008600 is quite interesting. The composition of the cervical mucus plugs don't conform with my expectations of the constituents of a ball of mucus, but that may be just me. I would never have guessed that fibroblasts were involved in forming these little rascals.
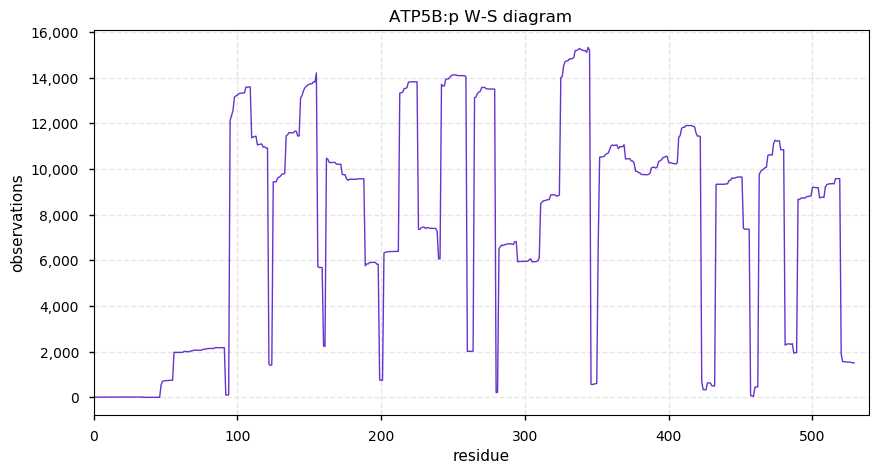
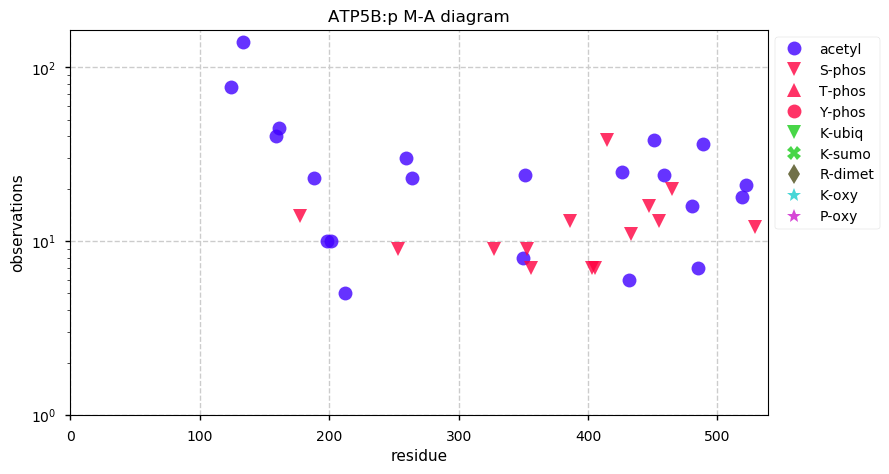
Sat Oct 12 13:50:57 +0000 2019Atp5b:p, ATP synthase F1 subunit beta (R. norvegicus) 🔗 Midsized mitochondrial subunit; significant acetylation; no SAV; 1 splice variant; mature form 47-529 [4,525 x] 🔗
Fri Oct 11 14:38:43 +0000 2019@jwoodgett Labeling things properly & keeping useful lab notebooks are the biggest adjustments new hires have to make when they get a job in industry.
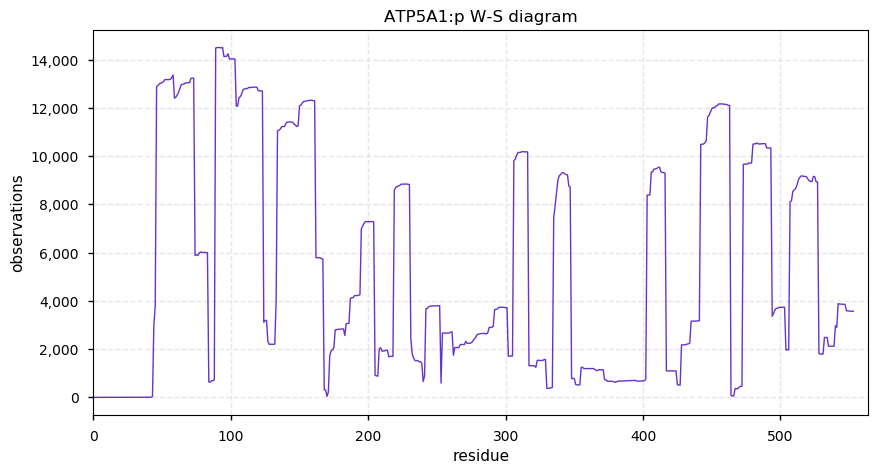
Fri Oct 11 12:52:05 +0000 2019Atp5a1:p, ATP synthase F1 subunit alpha (R. norvegicus) 🔗 Midsized mitochondrial subunit; significant acetylation; 1 SAV: R553P (maf=1.0); 1 splice variant; mature form 44-553 [4,613 x] 🔗
Thu Oct 10 14:44:24 +0000 2019@seandavis12 @HFazelinia OK. I promise not to talk about Fight Club any more.
Thu Oct 10 14:43:51 +0000 2019@slavovLab I love the line suggesting that people should "build bacteria that absorbs CO2" (apparently not knowing they have existed for quite a while). It really sums up why Silicon Valley simply burns money when it tries to be involved in any sort of scientific endeavor.
Thu Oct 10 13:32:21 +0000 2019@pwilmarth The phenomenon is called pareidolia 😉: 🔗
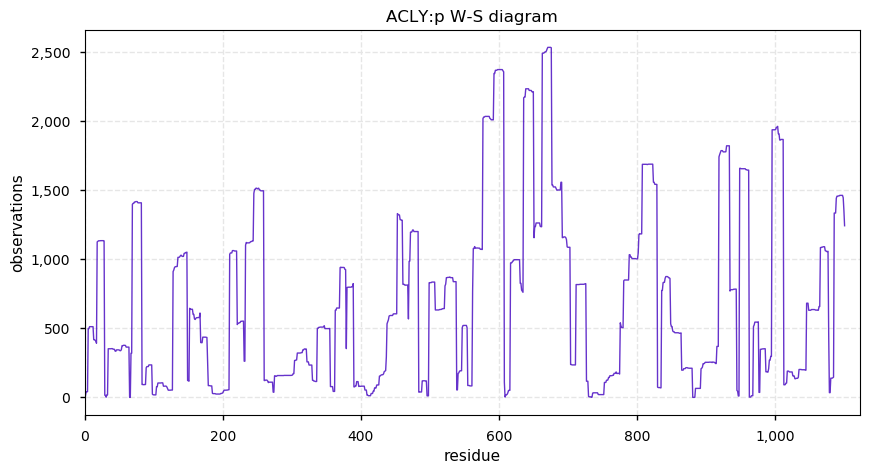
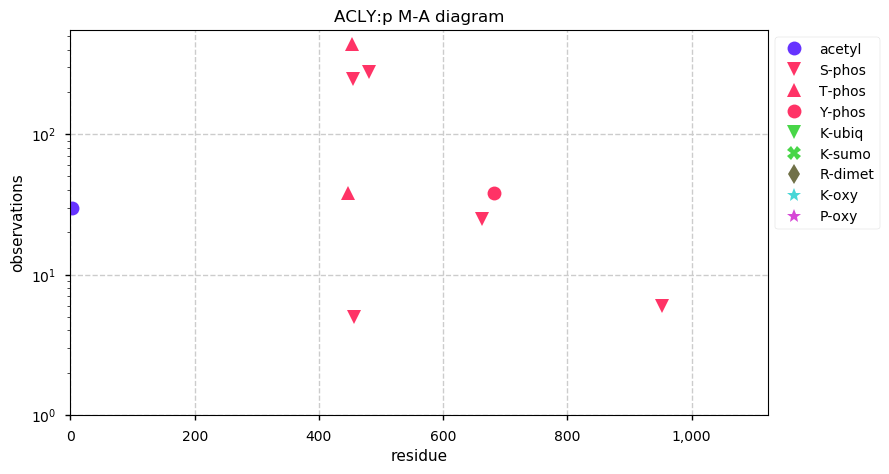
Thu Oct 10 12:10:23 +0000 2019Acly:p, ATP citrate lyase (Rattus norvegicus) 🔗 Large cytosolic enzyme; several phosphodomains; no SAVs; 1 splice variant; mature form 2-1101 [2,665 x] 🔗
Wed Oct 09 14:38:35 +0000 2019@seandavis12 @HFazelinia So, is this type of report:
Wed Oct 09 14:29:16 +0000 2019@seandavis12 @HFazelinia I'm not sure how to take this sort of report. On the one hand, they tried to do something at scale and put together a big team to do it. On the other, the data isn't available and I can't test what they say was found using a complex procedure, which is also secret.
Wed Oct 09 13:52:13 +0000 2019This is actually a significant problem in Canada 🔗
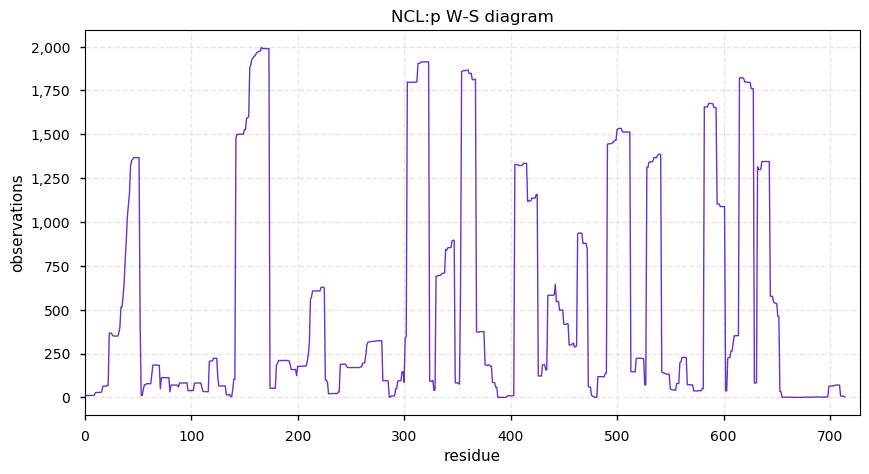
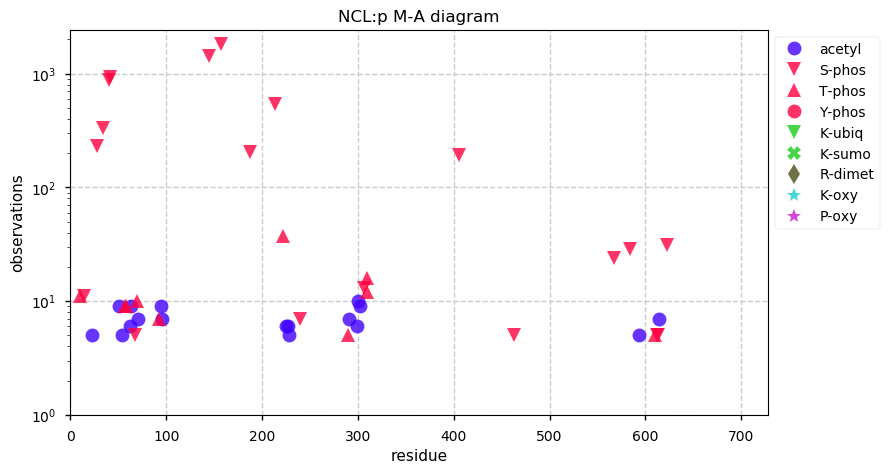
Wed Oct 09 11:22:58 +0000 2019Ncl:p, nucleolin (R. norvegicus) 🔗 Midsized nuclear protein; acetylation/phosphorylation clusters; no SAVs; 1 splice variant; mature form 1-714 [1,987 x] 🔗
Tue Oct 08 22:06:27 +0000 2019@Smith_Chem_Wisc @nesvilab @neely615 @fcyucn Try Mm2DLC_N_1_01.raw
scans: 42652, 43750 or 48869
they should solve to LAQFYGLPAFVAGTOSDAK, z=3
Tue Oct 08 15:04:08 +0000 2019@Smith_Chem_Wisc @nesvilab @neely615 @fcyucn Now I have to download it too ...
Tue Oct 08 14:19:13 +0000 2019@Smith_Chem_Wisc @nesvilab @neely615 @fcyucn As far as I know, no one has made proteomics data available from a bacteria that uses O in its proteome: it is in the active site of enzymes involved in generating methane in specific archaea and bacteria.
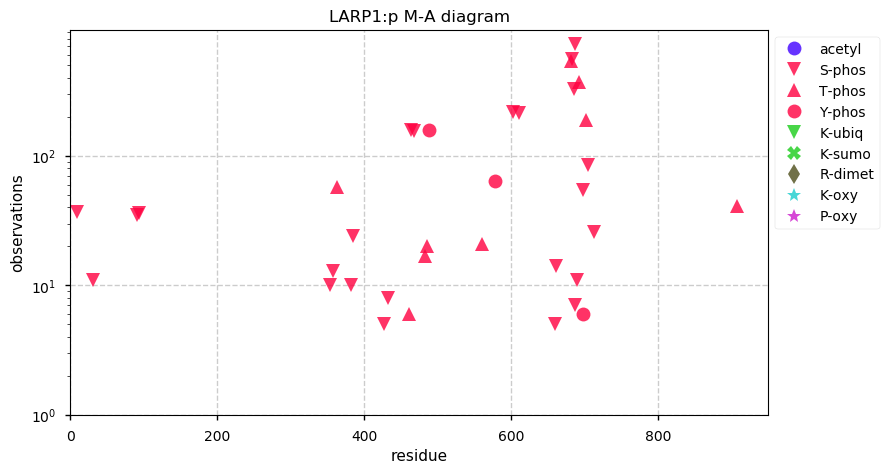
Tue Oct 08 12:31:05 +0000 2019LARP1:p, La ribonucleoprotein domain family, member 1 (R. norvegicus) 🔗 Midsized nuclear protein; significant phosphorylation; no SAVs; 1 splice variant; mature form 2-931 [1,171 x] 🔗
Tue Oct 08 01:10:10 +0000 2019@nesvilab @Smith_Chem_Wisc @neely615 @fcyucn If you are checking, you might as well check for O (pyrolysine) while you are at it.
Tue Oct 08 00:31:10 +0000 2019@Smith_Chem_Wisc @neely615 This exchange has been a pretty good example of quickly using public data to answer a question unrelated to the purpose for which the data was originally collected. Without the data, this sort of discussion could have dragged on (& on).
Mon Oct 07 21:02:55 +0000 2019I am not usually much for hash tags, but #InMyGreatAndUnmatchedWisdom really seems evocative of this moment in history
Mon Oct 07 17:37:22 +0000 2019Having the in-laws over for a few days, something that always makes me think of the rousing chorus of the 1996 release "Still".
Mon Oct 07 17:08:58 +0000 2019@Smith_Chem_Wisc @neely615 A good example is PXD010154, try "01087_C01_P010739_S00_N03_R1.raw", "01087_H01_P010739_S00_N08_R1.raw" and or "01087_F01_P010739_S00_N06_R1.raw".
Mon Oct 07 14:33:08 +0000 2019@Smith_Chem_Wisc @neely615 I don't mind harping on it: selenocysteine is not a PTM. It is a genome-encoded amino acid, the same as alanine or glycine. It does not require any special degree of confirmation: finding something other than U at the site would require investigation.
Mon Oct 07 14:11:56 +0000 2019@Smith_Chem_Wisc @neely615 But because of the general nature of the substitution, it is unlikely that any Se-Met would be detected using unspecialized methods.
Mon Oct 07 14:06:33 +0000 2019@Smith_Chem_Wisc @neely615 Getting back to the original post, the protein effects associated with Se poisoning do not involve selenocysteine. Se inhibits phosphorylation and causes the substitution of Se for S in methionine, so testing the data for Se-Met would be more informative.
Mon Oct 07 12:15:25 +0000 2019@neely615 🔗
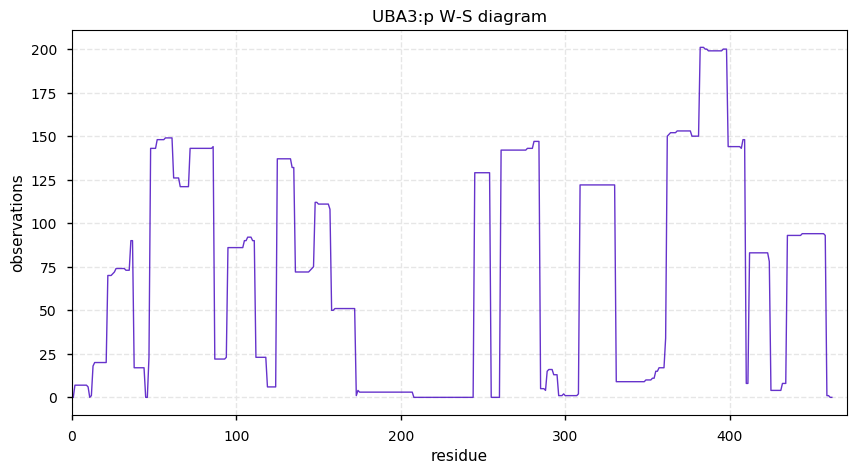
Mon Oct 07 11:51:29 +0000 2019Uba3, ubiquitin-like modifier activating enzyme 3 (R. norvegicus) 🔗 Small cytosolic protein; only PTM detected is N-terminal processing; no SAVs; 1 splice variant; mature form 2-462 [429 x] 🔗
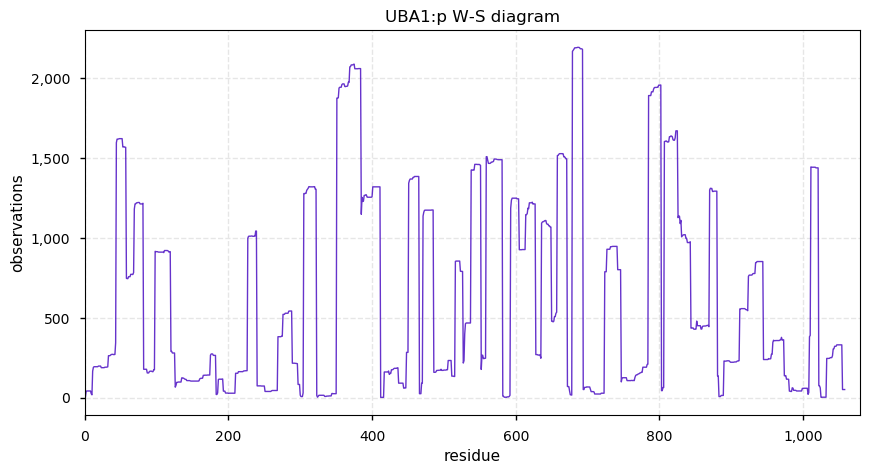
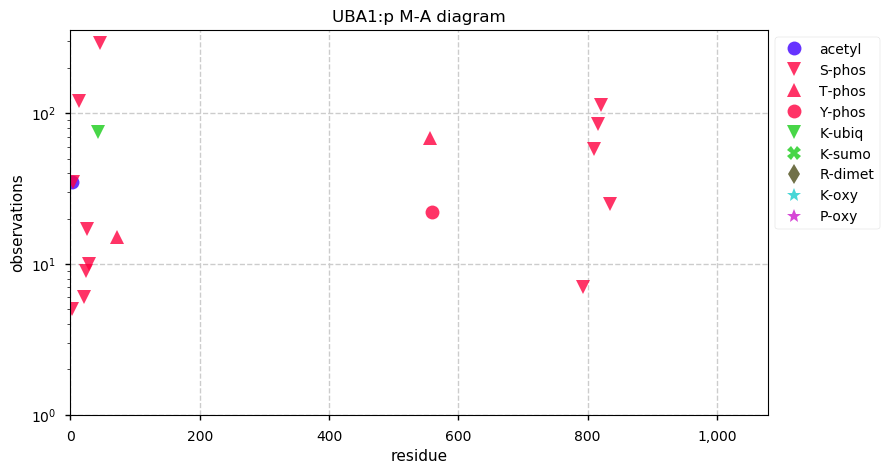
Sun Oct 06 13:14:18 +0000 2019Uba1, ubiquitin-like modifier activating enzyme 1 (R. norvegicus) 🔗 Large cytosolic protein; 3 phosphodomains; no SAVs; 1 splice variant; mature form 2-1058 [2,142 x] 🔗
Sat Oct 05 16:09:01 +0000 2019ProteomeXchange needs more rat phosphorylation data (S/T/Y). If you have some and you have been sitting on it for a while, please get it in there 🐀 !
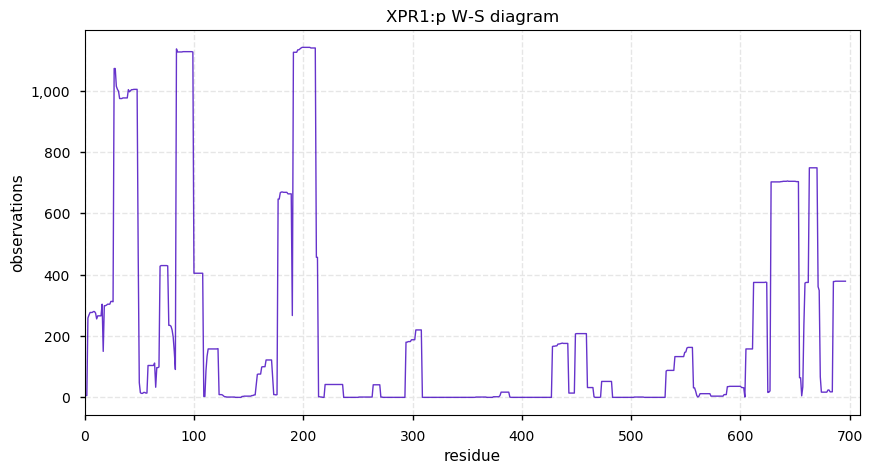
Sat Oct 05 12:00:04 +0000 2019XPR1:p, xenotropic and polytropic retrovirus receptor 1 (H. sapiens) 🔗 Midsized membrane protein, aka SLC53A1:p; 8 predicted TM domains; generates HLA class II peptides; no SAVs; 1 splice variant; mature form 1-696 [2,952 x] 🔗
Fri Oct 04 14:34:24 +0000 2019@lkpino A pretty good primer on how to think about experimental design in the context of large scale studies.
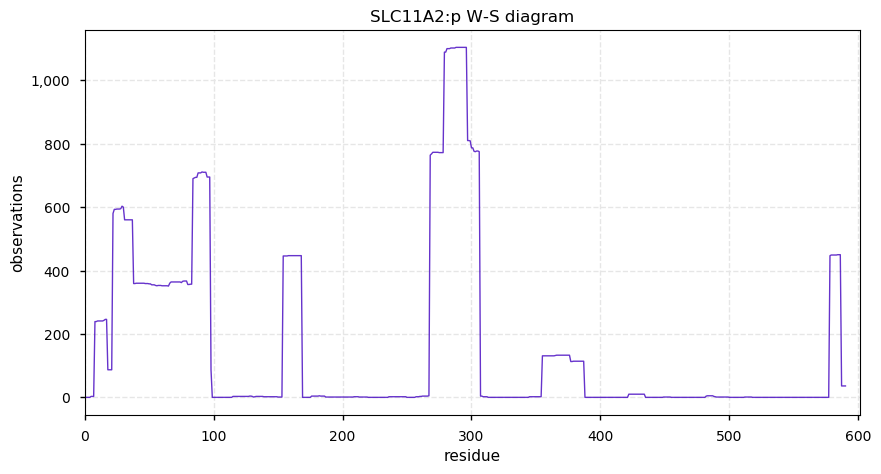
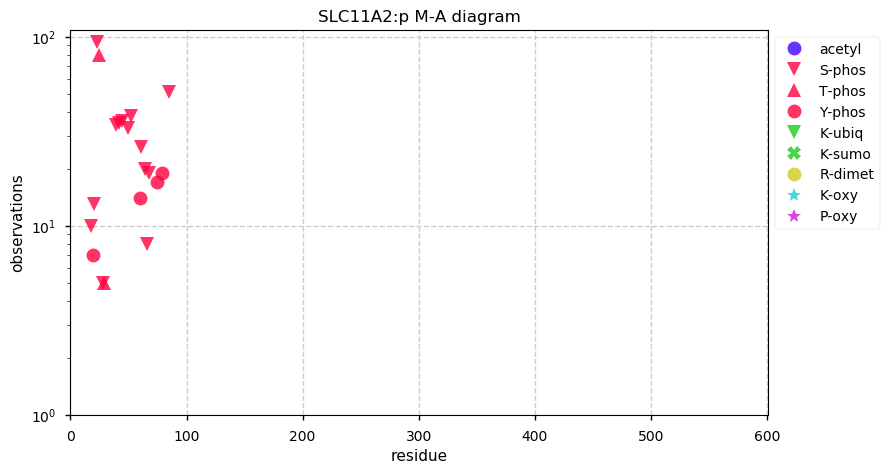
Fri Oct 04 13:12:02 +0000 2019SLC11A2:p, solute carrier family 11 member 2 (H. sapiens) 🔗 Midsized membrane protein; 12 predicted transmembrane domains; N-terminal phosphodomain; no SAVs; 1 splice variant; mature form 1-590 [1,840 x] 🔗
Thu Oct 03 18:48:03 +0000 2019When are academic grants going to be required to include a budget to purchase offsets for the carbon generated by research, travel and publications?
Thu Oct 03 16:49:48 +0000 2019@doctorow Might be a useful method for reviewing software when there is no source code provided.
Thu Oct 03 14:56:15 +0000 2019@Sci_j_my @NatureMedicine In my experience, its editorial standards are very similar to those of Nature Methods.
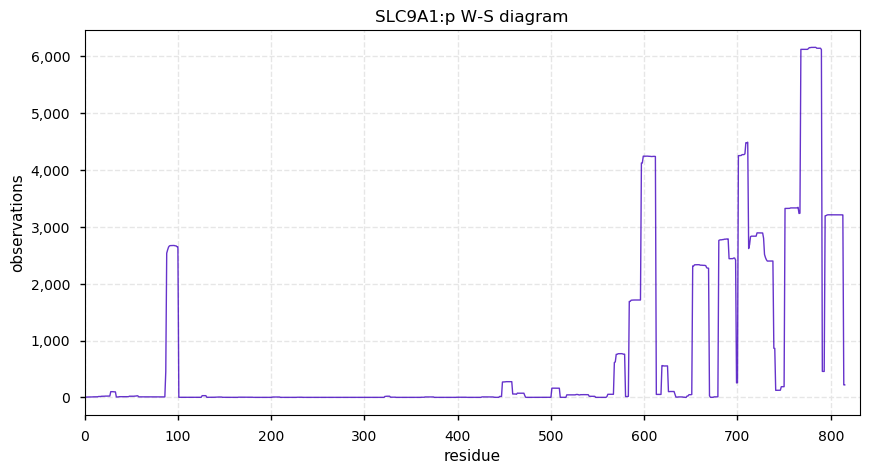
Thu Oct 03 12:25:48 +0000 2019SLC9A1:p, solute carrier family 9 member A1 (H. sapiens) 🔗 Midsized membrane protein; 12 predicted transmembrane domains; C-terminal phosphodomain; no SAVs; 1 splice variant; mature form 1-815 [7,942 x] 🔗
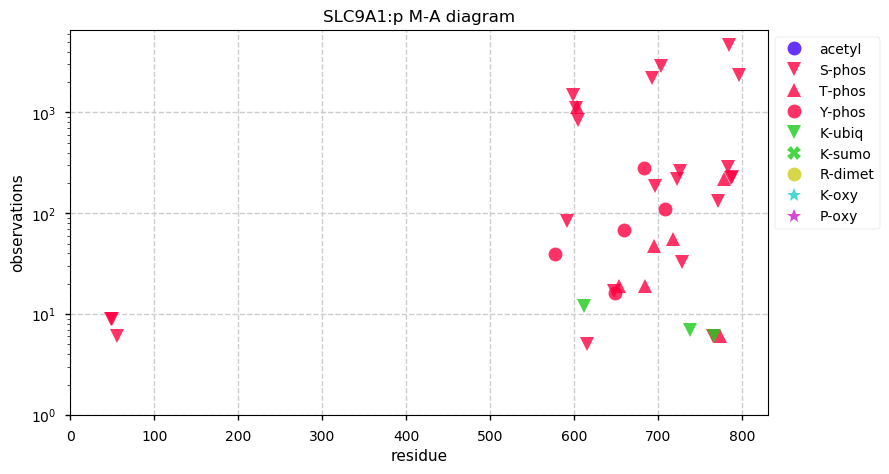
Thu Oct 03 12:14:26 +0000 2019@Sci_j_my @NatureMedicine You must admit, it is the simplest thing to do 🤡
Wed Oct 02 21:43:05 +0000 2019@VATVSLPR @ChasingMicrobes But if you are developing a new algorithm, an overly complicated standard output format that doesn't really support your "new thing" can be a bigger problem than the one you are actually trying to solve.
Wed Oct 02 21:42:17 +0000 2019@VATVSLPR @ChasingMicrobes I agree about standardizing when you are creating a larger system: I have stuck with a single file format (that I didn't invent) for input, output & temporary storage for years.
Wed Oct 02 19:53:52 +0000 2019@VATVSLPR @ChasingMicrobes FASTA is a little different, in that it is an input rather than an output format. The developer does gain quite a bit from using existing input formats, even if they are very limited in their capabilities.
Wed Oct 02 19:30:21 +0000 2019@VATVSLPR @ChasingMicrobes Also consider that there is little benefit for the developer associated with conforming to an existing output format and lots of downside (a lot of fiddly testing while wrestling with an under-specified format and dealing with the never-backwards-compatible v. 2.0)
Wed Oct 02 18:42:00 +0000 2019@KentsisResearch @Smith_Chem_Wisc @CompProteomics PXD010154 is also pretty deep and has a nice range of tissues, just be ready for lots of carbamylation.
Wed Oct 02 18:38:12 +0000 2019@KentsisResearch @Smith_Chem_Wisc @CompProteomics The raw files labelled "70frac_no_concatenation_rep1" or "293_REP1_46frac" in PXD004452 are pretty good for cell lines and "PROSTATE_Pt2_human_46frac" for tissue.
Wed Oct 02 15:02:47 +0000 2019@Smith_Chem_Wisc @neely615 @KentsisResearch @CompProteomics When it comes to speed, most modern algorithms are pretty non-linear: there is no generally applicable clocks-per-spectrum type parameter as there was with Sequest or Mascot.
Wed Oct 02 14:37:40 +0000 2019@Smith_Chem_Wisc @KentsisResearch @CompProteomics Comparative Proteomic Analysis of Eleven Common Cell Lines ... is good data and useful for testing how well you do with the non-tryptic cleavage associated with LysC. For me, it is rather shallow (at both the protein and peptide levels) for finding a lot of PTMs, though.
Wed Oct 02 14:31:22 +0000 2019@KentsisResearch @CompProteomics Chromatography is usually the least consistent aspect of a large study, but this study does it pretty well.
Wed Oct 02 14:27:54 +0000 2019@KentsisResearch @CompProteomics I suggest any one of the CPTAC LUAD data sets (🔗). It gives you a good idea of what to expect in a tissue sample using TMT quant. The MS & MS/MS resolution are pretty good (& consistent) and the chromatography is steady.
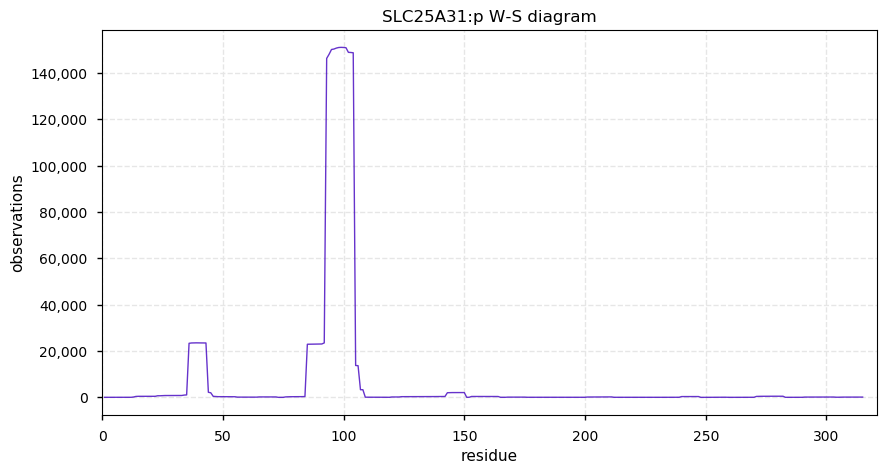
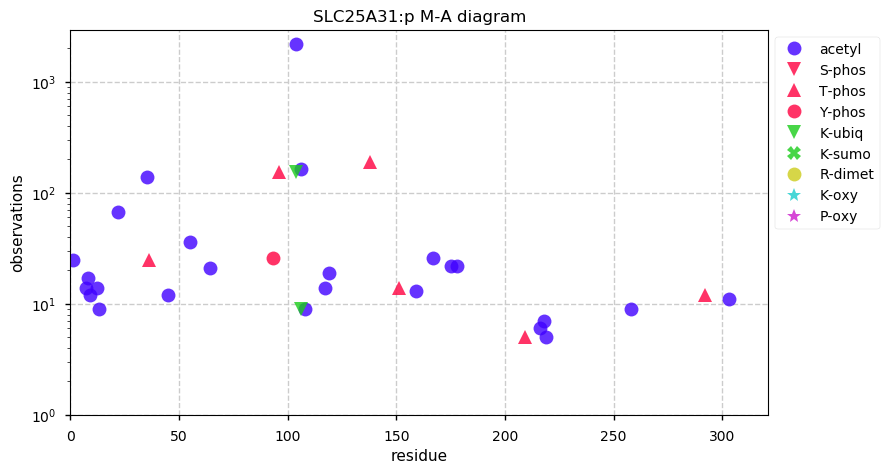
Wed Oct 02 12:07:50 +0000 2019SLC25A31:p, solute carrier family 25 member 31 (H. sapiens) 🔗 Small mitochondrial inner membrane protein; 6 predicted transmembrane domains; odd pattern of acetylation; 1 SAV: K9R (maf=0.03); 1 splice variant; mature form 1-315 [45,829 x] 🔗
Tue Oct 01 19:09:08 +0000 2019Just noticed GPMDB went over the 9 billion PSM mark over night. Doesn't really mean anything, other than that I think I should start planning to get some bigger disks.
Tue Oct 01 13:06:48 +0000 2019Two other proteins (SLC25A5 & SLC25A6) have similar sequences, PTM patterns and function. They can be distinguished at the peptide level in proteomics experiments.
Tue Oct 01 12:23:01 +0000 2019aka ANT1, responsible for moving ADP into the mitochondrial matrix and moving ATP out. Evidence suggests that K23+acetyl modulates translocation activity. Sequence does not have a mitochondrial targeting peptide.
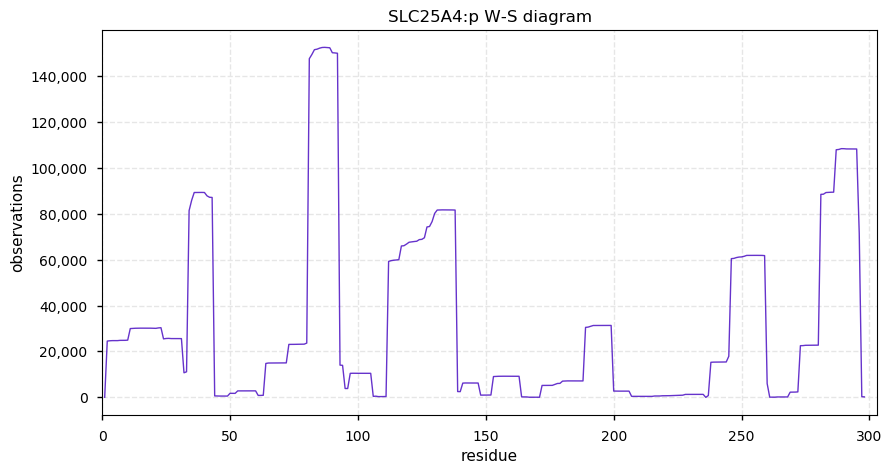
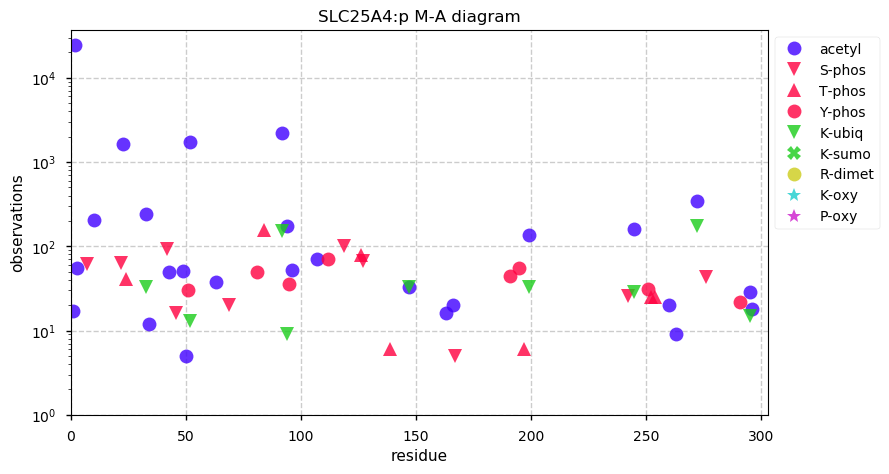
Tue Oct 01 12:12:53 +0000 2019SLC25A4:p, solute carrier family 25 member 4 (H. sapiens) 🔗
Small mitochondrial inner membrane protein; 6 predicted transmembrane domains; 9 sites shared between ubiquitinyl and acetyl; no SAVs; 1 splice variant; mature form 2-298 [58,403 x] 🔗
Mon Sep 30 14:37:29 +0000 2019Thank you everybody who took part in the poll and the associated discussion.
58% of participants felt 2 phosphorylation sites per peptide was the maximum they were comfortable reporting and 23% were OK with 3 sites.
Mon Sep 30 14:09:05 +0000 2019@bkives When the City redeveloped the back lane behind our building downtown, they carefully retained the existing trees and added feature lighting to display them at night. 4 months after that was finished Hydro came by and cut them all down.
Mon Sep 30 14:03:49 +0000 2019The criticism applies to a lot of cross-disciplinary research. 🔗
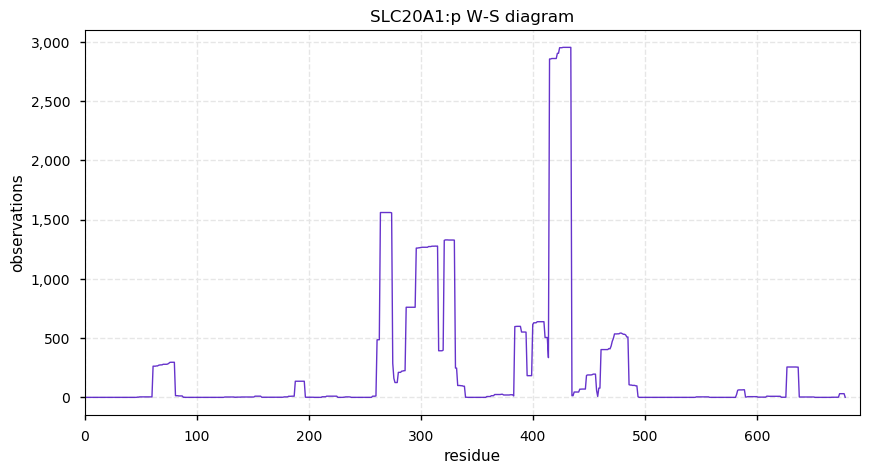
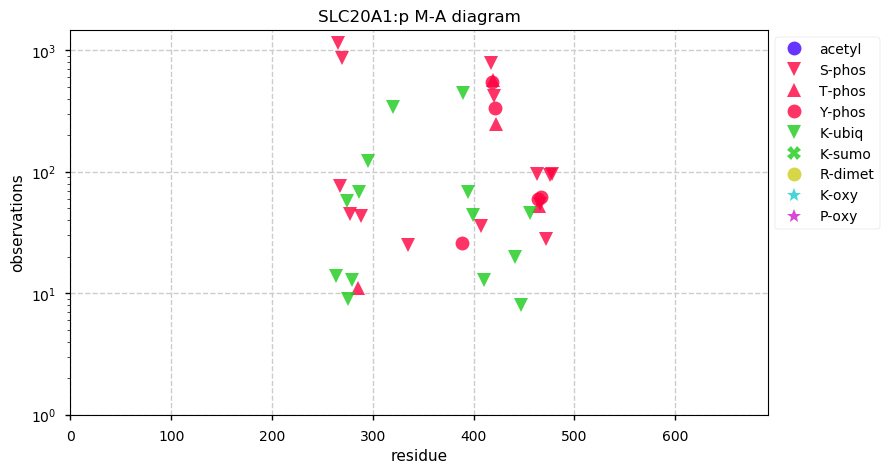
Mon Sep 30 12:12:32 +0000 2019SLC20A1:p, solute carrier family 20 (phosphate transporter), member 1 (Homo sapiens) 🔗 Midsized membrane protein; 10 predicted transmembrane domains; aka: gibbon ape leukemia virus receptor 1 (GLVR1); no SAVs; 1 splice variant; mature form 1(?)-679 [3,075 x] 🔗
Sun Sep 29 14:31:30 +0000 2019MS/MS search engines will sometimes return assignments with many too PTMs. What is the maximum number of phosphorylations on a single peptide that you feel comfortable reporting?
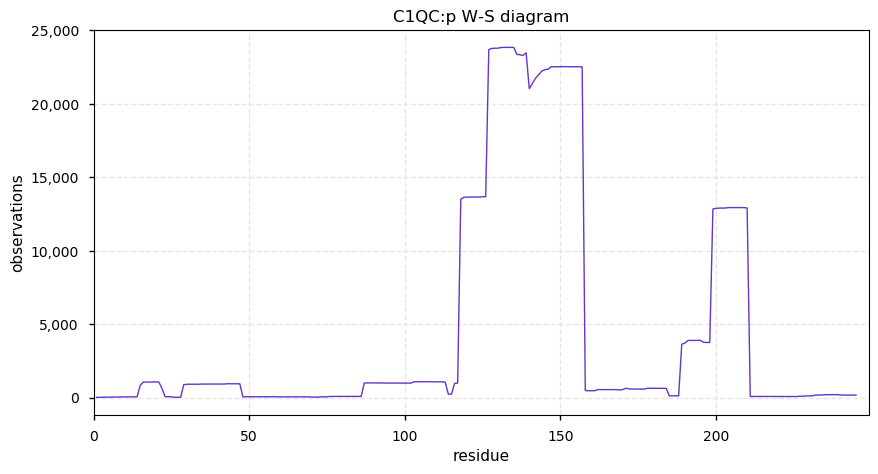
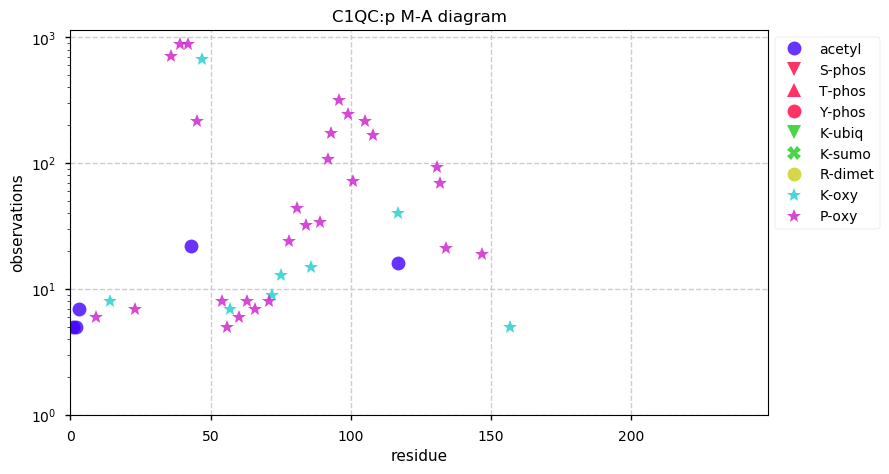
Sun Sep 29 13:14:44 +0000 2019C1QC:p, complement C1q C chain (H. sapiens) 🔗 Small blood plasma protein; unusual hydroxyproline-containing domain; rare in cell lines; 1 SAV: V187I (maf=0.01); 1 splice variant; mature form 29-245 [12,777 x] 🔗
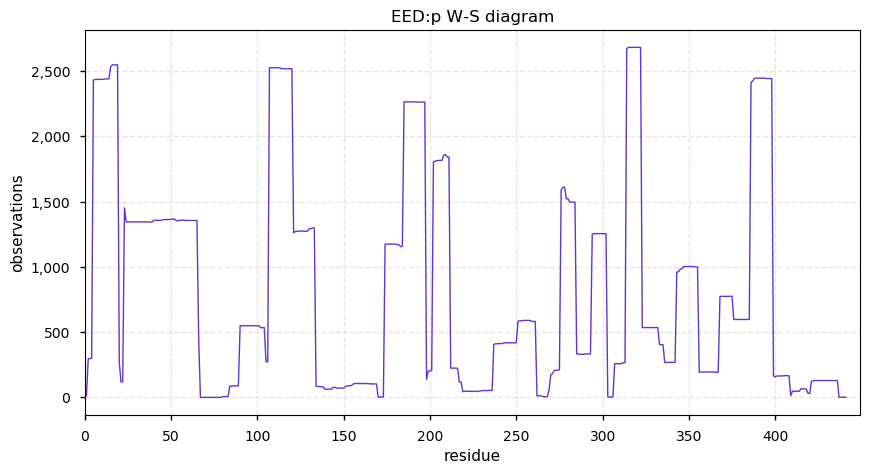
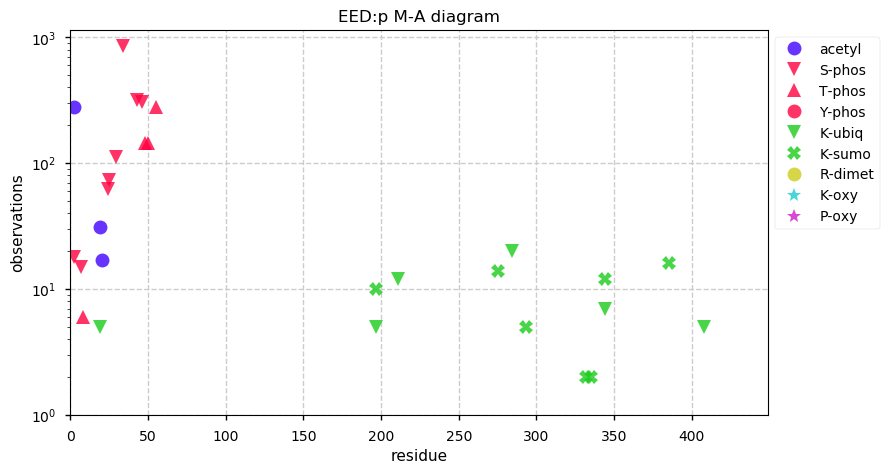
Sat Sep 28 12:43:24 +0000 2019EED:p has a classic mullet PTM pattern: all signalling in the front & transport in the back.
Sat Sep 28 12:41:43 +0000 2019EED:p, embryonic ectoderm development (H. sapiens) 🔗 Small nuclear protein; domain specific PTMs; one of the polycomb group; no SAVs; 2 splice variants; mature form 2-441 [7,609 x] 🔗
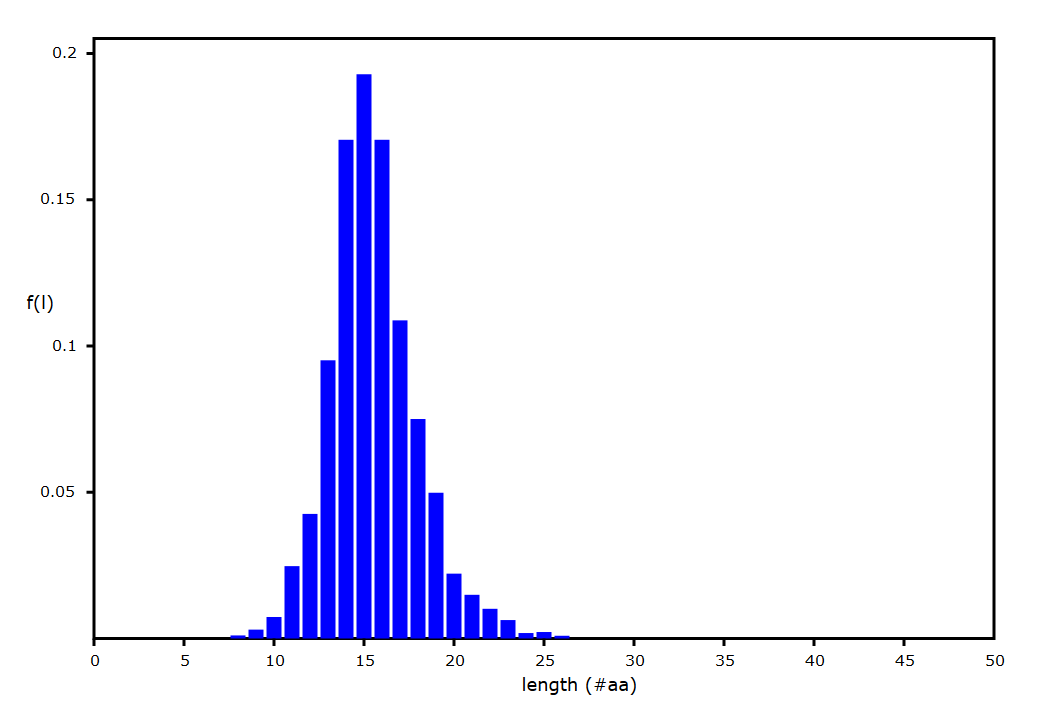
Sat Sep 28 02:12:05 +0000 2019A nearly ideal MHC II peptide length distribution, from an analysis of PXD014253 (q2133257c.raw), part of 🔗 🔗
Fri Sep 27 18:04:34 +0000 2019@Smith_Chem_Wisc @SpecInformatics @neely615 Are they used for the search or are they for display markup only?
Fri Sep 27 15:52:17 +0000 2019@PastelBio I was struck by this ad. I did not know that the NIH used Kelly to supply temp. contract bioinformatics researchers. No reason they shouldn't, but still I was surprised.
Fri Sep 27 12:25:06 +0000 2019The phosphorylation looks a bit more "Jackson Pollack" than "organized, coordinated signalling".
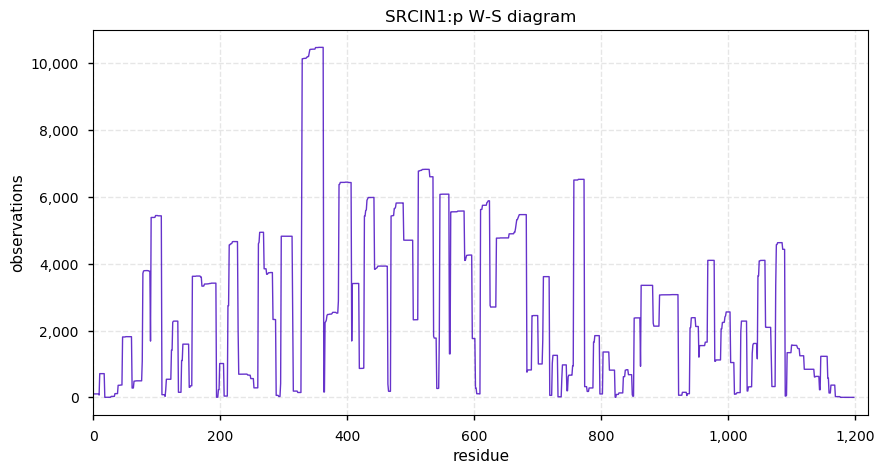
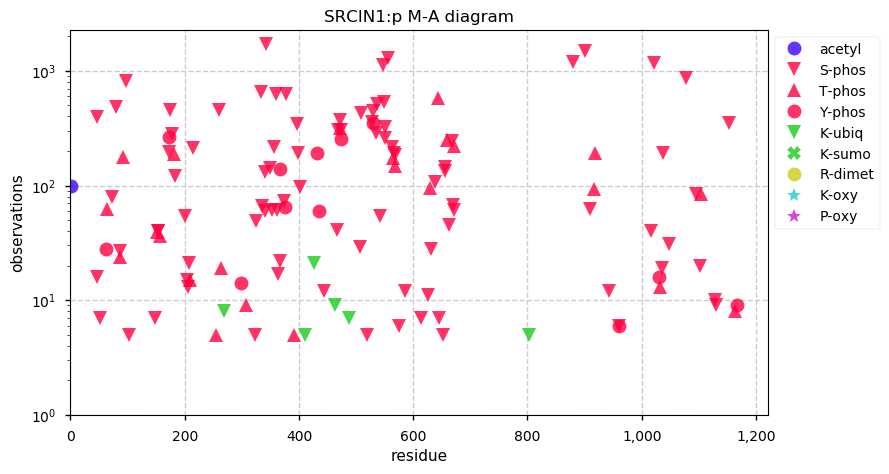
Fri Sep 27 12:08:12 +0000 2019Srcin1:p, SRC kinase signaling inhibitor 1 (M. musculus) 🔗 Large protein; extensive, high occupancy phosphorylation; found in CNS tissue; not found in common cell lines; no SAVs; 2 splice variants; mature form 1-1198 [5,765 x] 🔗
Thu Sep 26 17:24:12 +0000 2019@astacus Ever since polywater, studies involving the use of treated v untreated water are suspicious. Water is a tricky: it picks up (& loses) stuff easily. I have a hard time believing that D-depleted water has no other chemical differences when compared to "undepleted" water.
Thu Sep 26 15:16:17 +0000 2019@ClaireEEyers @astacus @chenym The main problem with WB is when people are following around the wrong protein because of a bad antibody. That is something that MS-based proteomics can correct by validating the WB initially and providing another line of evidence to confirm important findings.
Thu Sep 26 15:11:54 +0000 2019@ClaireEEyers @astacus @chenym While an SRM assay may tell you that parts of the protein are still present, it cannot give you information about whether or not the protein is still intact or if it has gone through some other biochemical transformation, e.g. glycosylation, ubiquitination
Thu Sep 26 13:20:16 +0000 2019Some times I see things that make it even more difficult than usual for me to take the "literature" & "peer review" seriously
🔗
Thu Sep 26 12:31:32 +0000 2019DNMT1:p, DNA methyltransferase 1 (H. sapiens) 🔗
Large nuclear protein; distinct PTM domains; several low complexity domains; 5 SAVs: H97R (maf=0.05), V120L (0.01), I327F (0.2), I327V (0.2), I544M (0.04) ; 2 splice variants; mature form 1-1632 [24,797 x] 🔗
Wed Sep 25 22:54:03 +0000 2019@astacus @chenym I have to agree with Rob. To get similar info to a WB using MS-based proteomics, you need to do an antibody pull-down, run it out on a gel, cut the right band, digest & run the peptides. Simply digesting the original sample without a pull-down wouldn't get the info.
Wed Sep 25 14:57:44 +0000 2019@BrenesAlejandro 4/ I find it is pretty rare to see much signal from the S27A portion of the sequence. It is very small so it is lost in many sample prep methods. It also is so low complexity that is does really have "good" tryptic peptides:
S27A = AKKRKKKSYTTPKKNKHKRKKVKLAVLKYY
Wed Sep 25 14:50:36 +0000 2019@BrenesAlejandro 3/ If the ubiquitin associated degradation pathway has been enhanced by a particular treatment, analyzing quant. results in terms of protein expression will probably not be very useful.
Wed Sep 25 14:48:29 +0000 2019@BrenesAlejandro 2/ Ubiquitin branching effectively amplifies this signal when the ubiquitin-related pathways are turned on, so the level of Gly-Gly tagged ubiquitin will tell you if one of these pathways has been activated.
Wed Sep 25 14:48:07 +0000 2019@BrenesAlejandro 1/ I have found that a sensitive test for protein ubiquitination levels is to check for Gly-Gly only on the genes that contain ubiquitin, at K's that are not at the C-terminus of the tryptic peptides.
Wed Sep 25 14:30:22 +0000 2019Some sort of Unicode character conversion issue seems to be affecting this particular ProteomeXchange entry's display: 🔗
Wed Sep 25 13:37:24 +0000 2019CBX4:p, chromobox 4 (H. sapiens) 🔗 Midsized nuclear protein; 26 sumoylation sites, 14 of which are shared with acetylation and/or ubiquitinylation; several low complexity domains; no SAVs; 1 splice variant; mature form 1-560 [5,204 x] 🔗
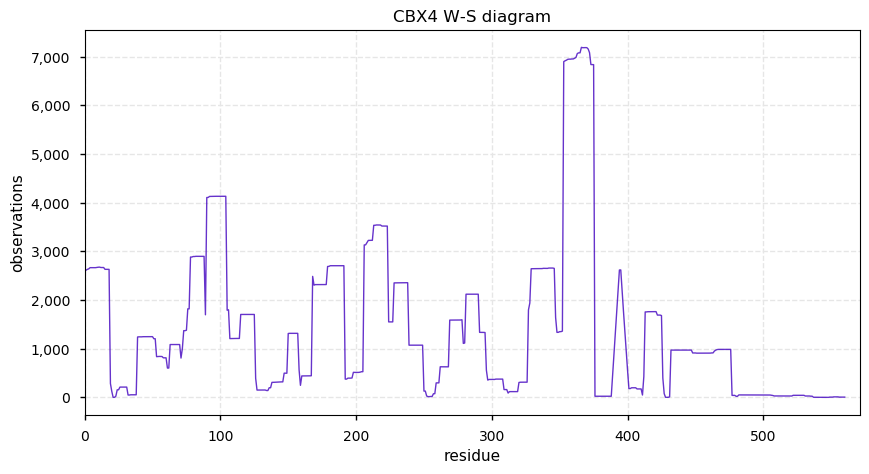
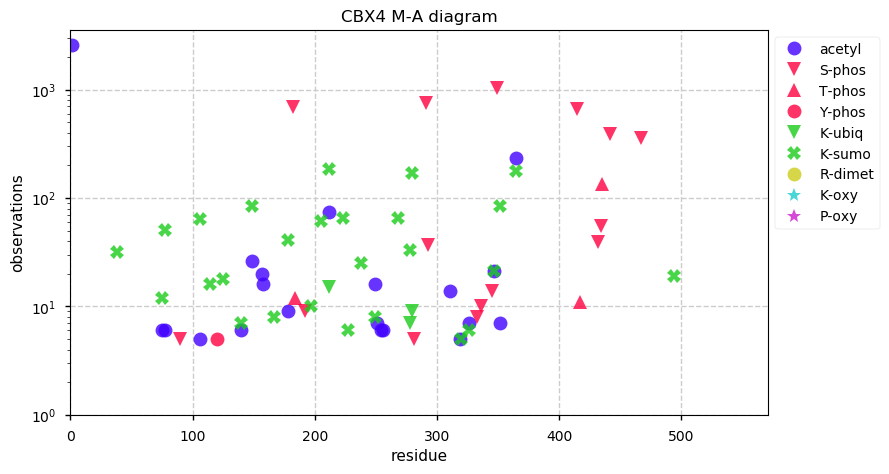
Tue Sep 24 21:36:02 +0000 2019@nesvilab There are some additional stats, so it is really the number of PSMs that pass all of the tests, not just the raw number.
Tue Sep 24 21:32:49 +0000 2019@nesvilab No, the abundance is the number of PSMs in GPMDB that have been assigned to a particular residue with a particular PTM.
Tue Sep 24 20:54:58 +0000 2019Just in case you were getting the impression that all proteins are covered with PTMs, here is a PTM cheat-sheet for the cyclins, an odd group of largely unrelated proteins named for when they are abundant during the cell cycle 🔗
Tue Sep 24 19:05:48 +0000 2019@PastelBio Cue the theremin music 😱
Tue Sep 24 17:34:57 +0000 2019@M_plus_H Given the group-to-group variability of the reaction, there should be a paper in there for someone who figures out a protocol that minimizes/eliminates/explains the formation of succinylated artifacts.
Tue Sep 24 17:25:40 +0000 2019@M_plus_H Succinylation is a common side reaction generated when using TMT reagents: you end up with 2-15% of the amines succinylated rather than TMTylated. This artifactual succinylation would be difficult-to-impossible to distinguish from biochemical succinylation.
Tue Sep 24 16:10:50 +0000 2019@hondanhon @andrewbusey You should at least try for an IRIS Indigo.
Tue Sep 24 13:51:20 +0000 2019Just be sure to remember that you can't use TMT reagents if you want to identify/quantify succinylation sites.
Tue Sep 24 12:06:22 +0000 2019ZNF281:p, zinc finger protein 281 (H. sapiens) 🔗 Midsized nuclear protein; more than 20 shared ubiquitinyl/sumoyl K-acceptor sites; commonly in nucleate cells; no SAVs; 2 splice variants; mature form 1-895 [7,202 x] 🔗
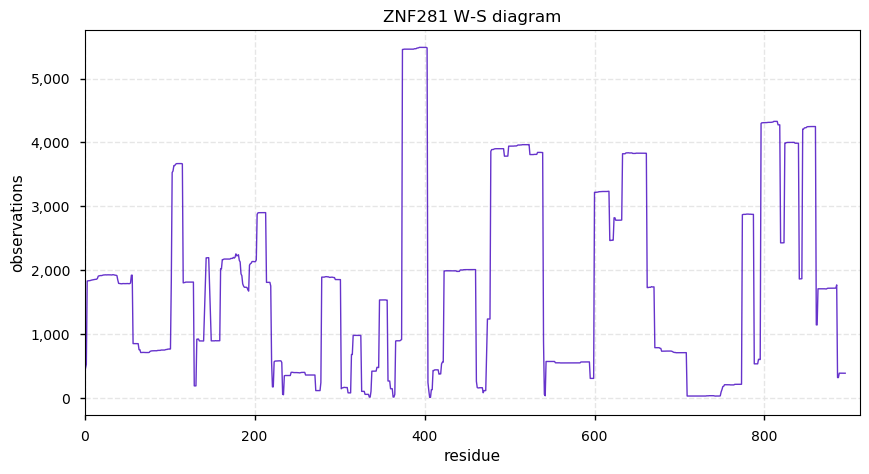
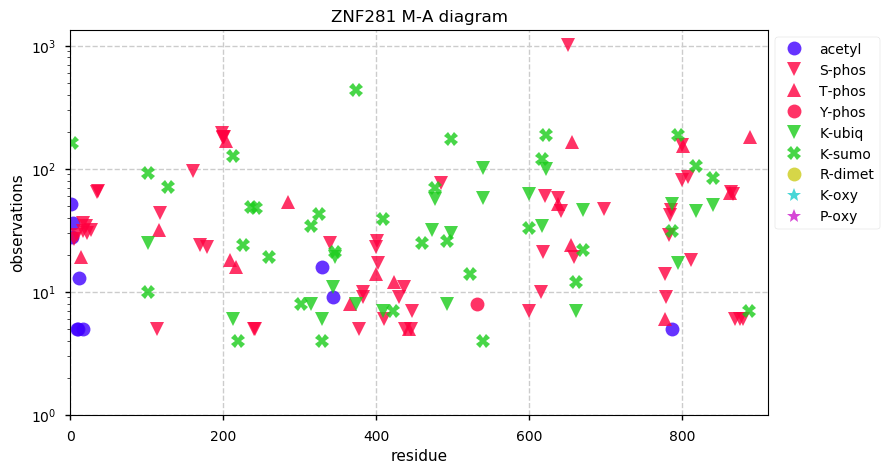
Mon Sep 23 18:47:45 +0000 2019@VATVSLPR @nesvilab @AnthonyCesnik I'd have never brought it up, except of Alexey's question. I consider it one of those historical oddities that hang around in scientific fields and become traditional, even though no one really remembers why it was done originally.
Mon Sep 23 18:22:13 +0000 2019If you are interested in succinylation and how to detect it in proteomics, #PXD013173 from 🔗 is a good data set to try out.
Mon Sep 23 17:38:57 +0000 2019Canadian bank cyber security would be hilarious, except for its horrible fallout for consumers:
🔗
🔗
Mon Sep 23 14:33:18 +0000 2019@Sci_j_my @mjmaccoss No.
Mon Sep 23 12:18:53 +0000 2019A good example of a protein we know a lot about, but have little idea of what it does.
Mon Sep 23 12:18:02 +0000 2019ZNF644:p, zinc finger protein 644 (H. sapiens) 🔗 Large nuclear protein; unusual pattern of K-sumoylation (51 sites) but no acetylation; commonly found in nucleate cells; no SAVs; 1 splice variant; mature form 1-1327 [4,675 x] 🔗
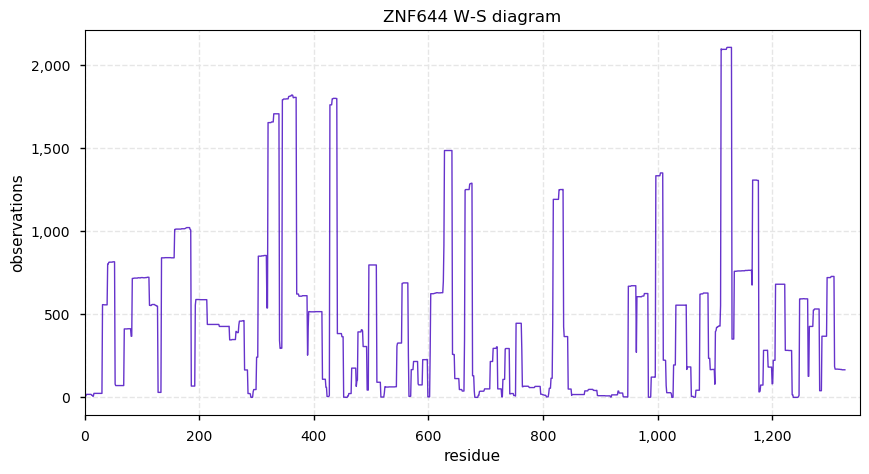
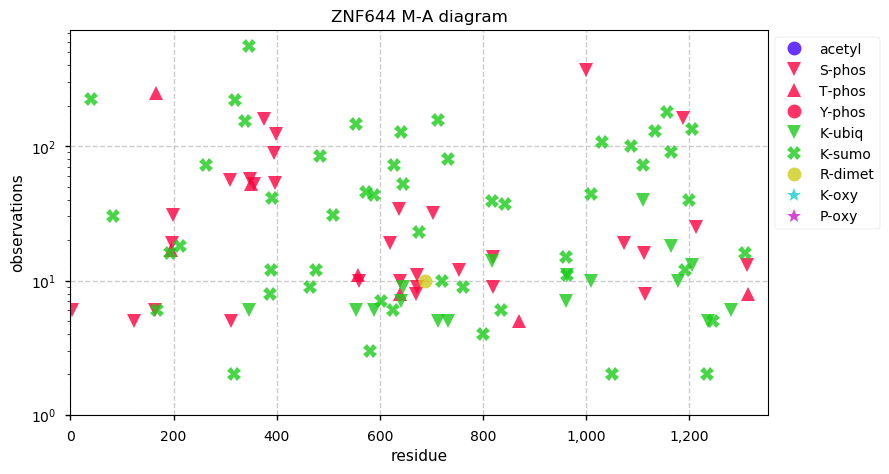
Sun Sep 22 19:26:07 +0000 2019@nesvilab @AnthonyCesnik It is also very easy to test its assumptions and results with Monte Carlo simulations and simulations using randomized sequences and spectra.
Sun Sep 22 19:22:34 +0000 2019@nesvilab @AnthonyCesnik I like the "apply_model" formulation particularly because it doesn't depend on the algorithm, it does pretty well with short (7-9 residue) peptides and it very naturally deals with the advantages of using high accuracy fragment ion masses.
Sun Sep 22 18:52:11 +0000 2019@nesvilab @AnthonyCesnik My current favorite PSM fitness test is in the "apply_model" method at 🔗 It uses Fisher's non-central Hypergeometric dist. and some boundary conditions. For large sets of PSMs matched to the same peptide & z, I use NBS 🔗
Sun Sep 22 16:52:54 +0000 2019@nesvilab @AnthonyCesnik 7/7 Since the "FDR" interpretation doesn't say anything about a specific PSM (or collection of selected PSMs) and it doesn't make any physical or mathematical predictions that can be tested, saying that a set of PSMs from an algorithm has an FDR=0.01 isn't helpful to me.
Sun Sep 22 16:52:20 +0000 2019@nesvilab @AnthonyCesnik 6/7 I normally deal with large numbers of PSMs that may have been created using different methods, so I have to use math and stats models that make testable estimates of the quality of any particular PSM or collection of PSMs, regardless of how the PSMs were generated.
Sun Sep 22 16:50:25 +0000 2019@nesvilab @AnthonyCesnik 5/7 Normally this sort of sensitivity to conditions isn't considered a positive for a statistical test, but people having been willing to look past these issues because it has become an accepted criterion for publication.
Sun Sep 22 16:50:11 +0000 2019@nesvilab @AnthonyCesnik 4/7 The "FDR" interpretation turned out to be very dependent on the number of peptide sequences being tested, the number of spectra being tested, the way that an algorithm actually generated a PSM, an algorithm's scoring system, etc.
Sun Sep 22 16:49:59 +0000 2019@nesvilab @AnthonyCesnik 3/7 No one was very happy with these values, so using a simple simulation to set "s" for you was considered to be better. There was a hand-waving argument about false positives made to justify the simulation, but it was never really proved.
Sun Sep 22 16:49:46 +0000 2019@nesvilab @AnthonyCesnik 2/7 At the time, this involved setting a lower acceptable score value (s) for each parent ion z, e.g. s>5.3 for z=3, s>3.7 for z=2 and s > 2.3 for z=1.
Sun Sep 22 16:49:37 +0000 2019@nesvilab @AnthonyCesnik 1/7 The "FDR" interpretation of a target-decoy simulation was originally motivated as a replacement for the arbitrary way that Sequest results where being interpretted at the time.
Sun Sep 22 14:55:53 +0000 2019@TrumanLab @NewPI_Slack You simply get used to it, as it will continue to happen. The only other alternative is to become paranoid and secretive.
Sun Sep 22 13:56:31 +0000 2019GTF2I:p, general transcription factor IIi (H. sapiens) 🔗 Midsized nuclear subunit; unusual pattern of K-sumoylation (43 sites); 1 SAV: T707I (maf=0.01); 1 splice variant; mature form 2-998 [33,786 x] 🔗
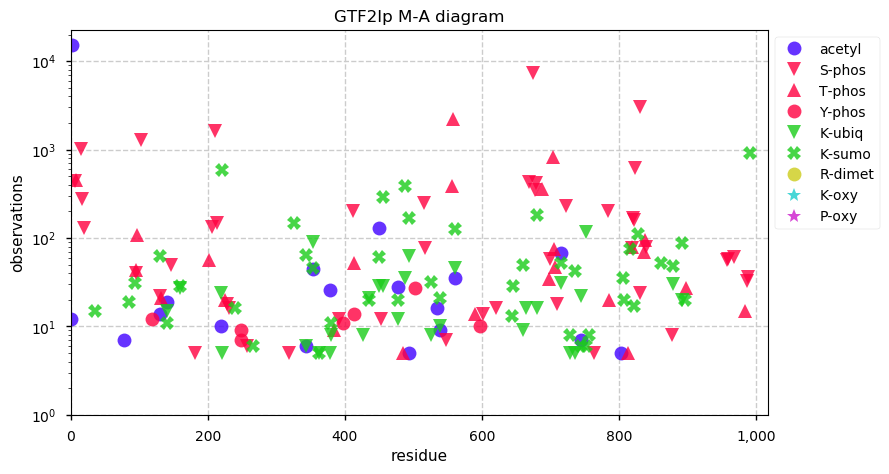
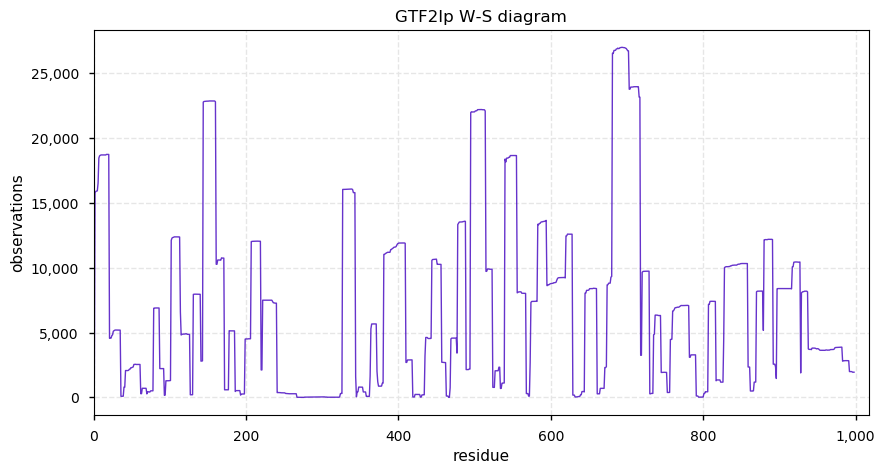
Sat Sep 21 23:09:00 +0000 2019@nesvilab @AnthonyCesnik Don't you mean shady FDR-based methods?
Sat Sep 21 15:16:31 +0000 2019@ProtifiLlc I will admit, though, that this suggestion has never been met with enthusiasm by Med. School colleagues or students.
Sat Sep 21 15:00:00 +0000 2019@ProtifiLlc Until some type of applied calculus course is required for biomedical researchers, this particular matter will reoccur. If the only relationship between two variables that you know about is "linear", that is the relationship you will try to apply to everything.
Sat Sep 21 11:56:08 +0000 2019RARA:p, retinoic acid receptor alpha (H. sapiens) 🔗 Small nuclear subunit; C-terminal phosphodomain; no SAVs; HLA I peptides: 46-53, 131-139, 208-217, 275-286; 2 splice variants; mature form 1-462 [839 x] 🔗
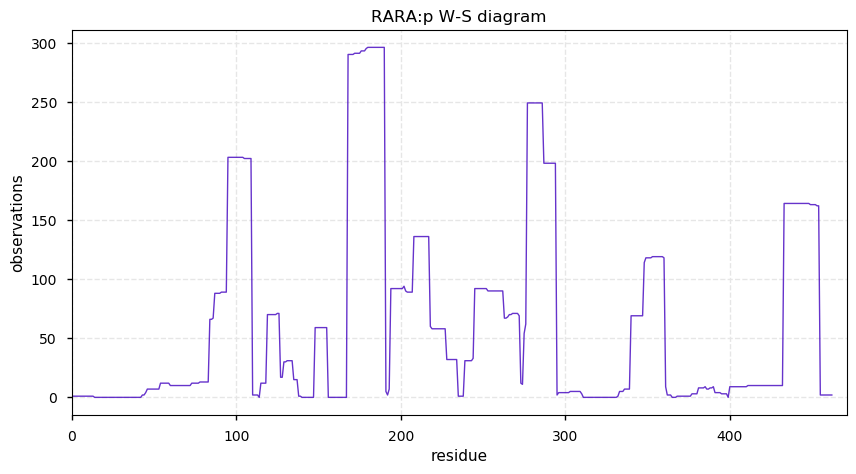
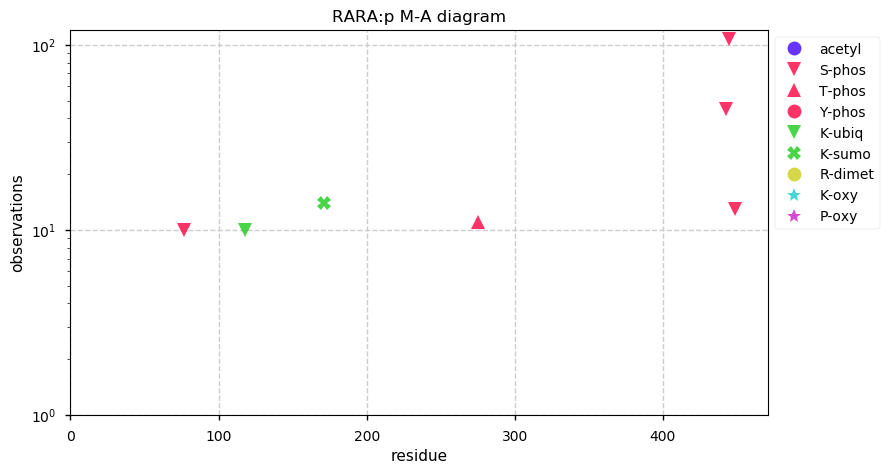
Fri Sep 20 16:46:29 +0000 2019@EvaChanda @MarcGarneau No doubt, but the Colt AR15 will no longer be available.
Fri Sep 20 16:33:12 +0000 2019@MarcGarneau It may be too late for the AR15 🔗
Fri Sep 20 12:06:28 +0000 2019PRAM1:p, PML-RARA regulated adaptor molecule 1 (H. sapiens) 🔗 Midsized nuclear subunit; odd dist. of phosphorylation sites; 2 SAVs: G135E (maf=0.16), A528V (maf=0.05); obs. in leukocytes, not in most cell lines; 1 splice; mature form 2-670 [1,978 x] 🔗
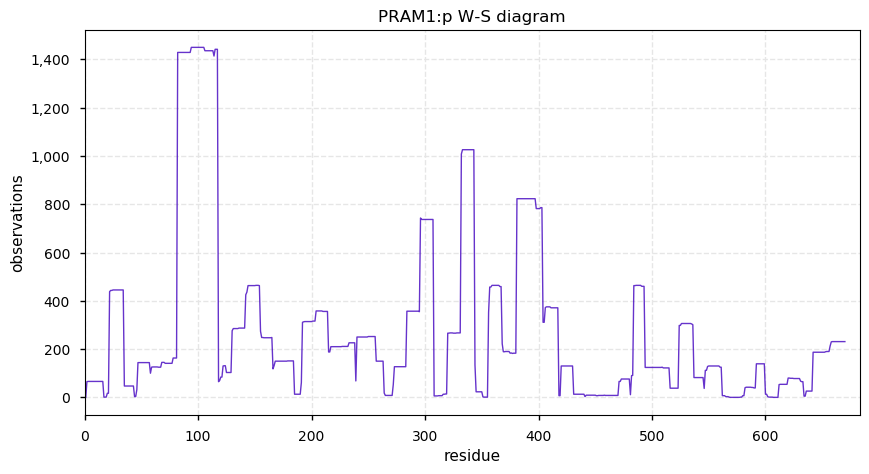
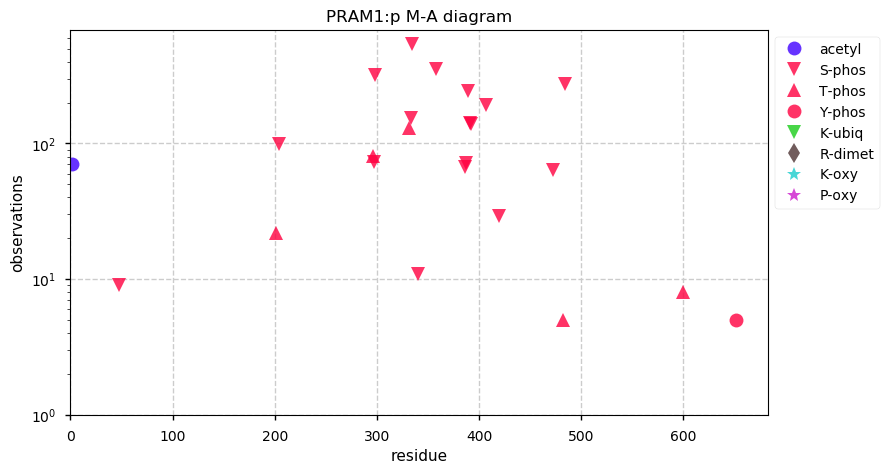
Thu Sep 19 16:11:46 +0000 2019@mjmaccoss @pwilmarth I think we have proved that Twitter isn't ideal for this type of thing. Unfortunately the structure of recent conferences has proven to be at least as bad.
Thu Sep 19 16:04:07 +0000 2019I should also mention that PML:p has 13 observed lysine acceptor sites for sumoylation, most of which are also observed ubiquitinylation sites. Many of these sites are known from the great data made available by Hendricks, et al. 🔗
Thu Sep 19 14:35:06 +0000 2019Our dear leader really seems to be taking it on the chin internationally about his often-callow demeanor:
🔗
Thu Sep 19 13:47:34 +0000 2019I love the audacity of naming this protein that polymerizes into still mysterious visible structures 🔗 after one of many negative phenotypes associated with a disruption of these structures.
Thu Sep 19 13:43:27 +0000 2019PML:p, promyelocytic leukemia (H. sapiens) 🔗 Midsized nuclear subunit; many PTMs: 4 phosphodomains; 1 SAV: F645L (maf=0.2); 5 obs. splice variants; mature form 1-882 [25,052 x] 🔗
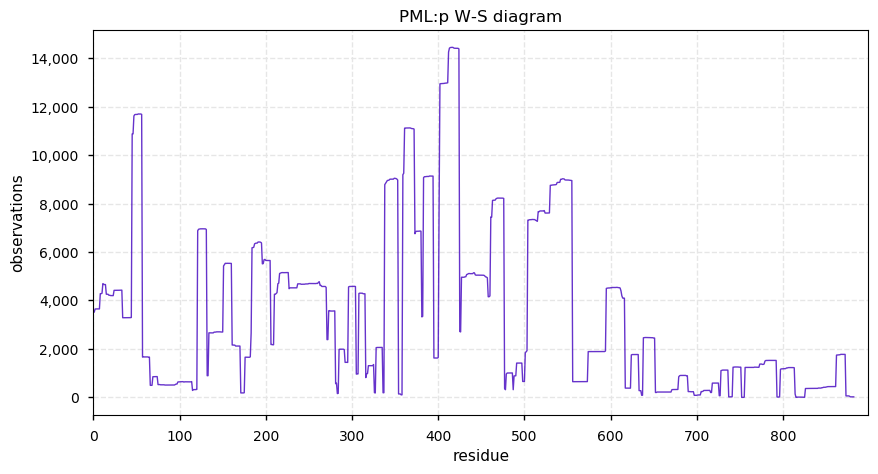
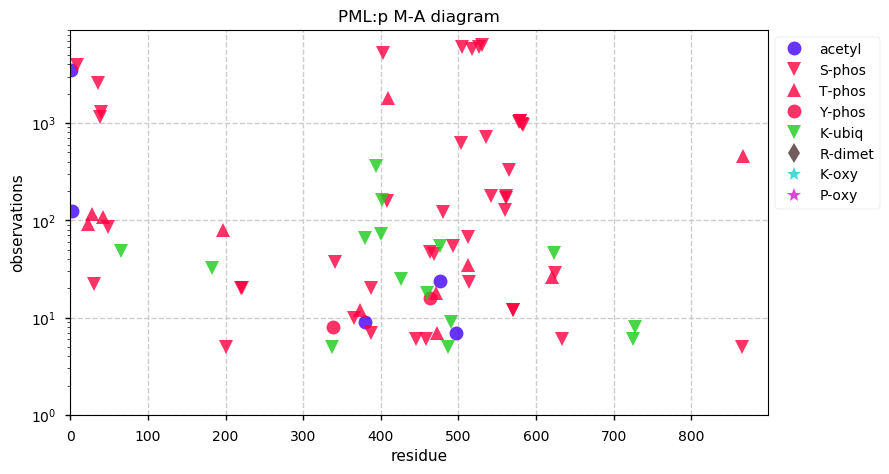
Thu Sep 19 12:23:24 +0000 2019@Sci_j_my @mjmaccoss @olgavitek @nesvilab @lukas_k @karthikskamath @chrashwood It has become common in emmulation of the perceived success/fundability of RNA-SEQ quant. expts where this type of subtlety is not applicable.
Wed Sep 18 23:33:18 +0000 2019@mjmaccoss @olgavitek @nesvilab @lukas_k @karthikskamath @chrashwood @Sci_j_my And simply averaging the signals for a protein together can lead to masking effects that make differential experiments less effective than they could be.
Wed Sep 18 23:30:11 +0000 2019@mjmaccoss @olgavitek @nesvilab @lukas_k @karthikskamath @chrashwood @Sci_j_my I also think we are pretty much saying the same thing (in short sentences). Proteins in vivo are significantly more complicated than they are generally thought to be, so expecting all peptide quant from the same protein to agree using current tech. is unrealistic.
Wed Sep 18 21:20:19 +0000 2019@nesvilab @mjmaccoss @lukas_k @karthikskamath @chrashwood @Sci_j_my I think that is a little strong, too. Mike has been doing quant. for a long time and knows only too well that peptide measurements aren't as consistent as they should be when you get down to specific cases.
Wed Sep 18 21:09:52 +0000 2019@mjmaccoss @lukas_k @karthikskamath @chrashwood @Sci_j_my Any examples of specific proteins where this idea has proven to be necessary?
Wed Sep 18 19:45:20 +0000 2019@mjmaccoss @lukas_k @karthikskamath @chrashwood @Sci_j_my But none of that will help with under-sampling.
Wed Sep 18 19:42:47 +0000 2019@mjmaccoss @lukas_k @karthikskamath @chrashwood @Sci_j_my Better ways of representing protein sequences as inputs for proteomics could fix this type of problem, but the most community is disinterested in even considering splice variants, let alone known backbone cleavages, PTM sites or high abundance SAVs.
Wed Sep 18 19:28:50 +0000 2019@mjmaccoss @lukas_k @karthikskamath @chrashwood @Sci_j_my I understand your point theoretically, but I don't see how we can get there from here, in a way that the biomedical community might profit from or understand.
Wed Sep 18 19:26:12 +0000 2019@mjmaccoss @lukas_k @karthikskamath @chrashwood @Sci_j_my Under-sampling in proteomics expts makes a lot of quantitative results difficult to interpret with accuracy, but suggesting that all peptide abundances should be uncoupled from their originating protein sequence removes what little structure an analyst has to rely on.
Wed Sep 18 19:00:56 +0000 2019@mjmaccoss @lukas_k @karthikskamath @chrashwood @Sci_j_my I assume you are taking this rather extreme position for some purpose. For many large, abundant proteins, e.g., COL1A1, there is nothing to be gained from trying to piece each possible proteoform back together (& top-down won't help)
Wed Sep 18 17:10:04 +0000 2019"Open Sans" versus "Merriweather Sans". Anyone have an opinion?
Wed Sep 18 12:08:23 +0000 2019MED18:p, mediator complex subunit 18 (H. sapiens) 🔗 Small nuclear protein; few PTMs; no SAVs; 1 splice variant; mature form 1-208 [2,391 x] 🔗
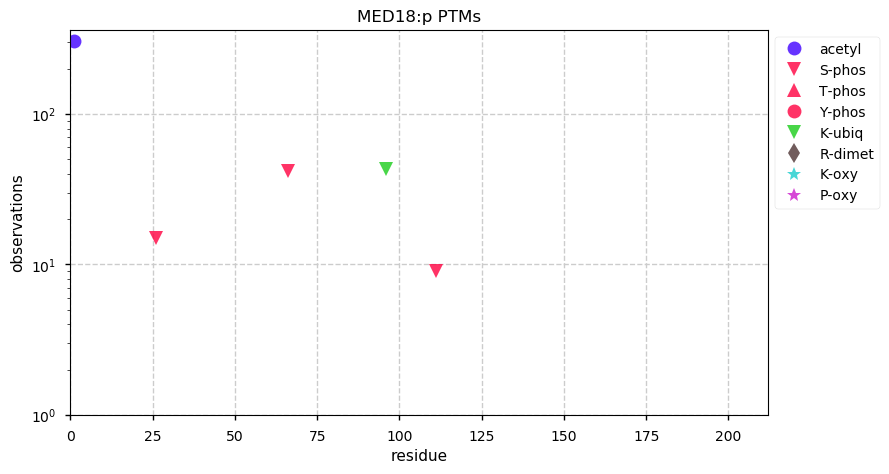
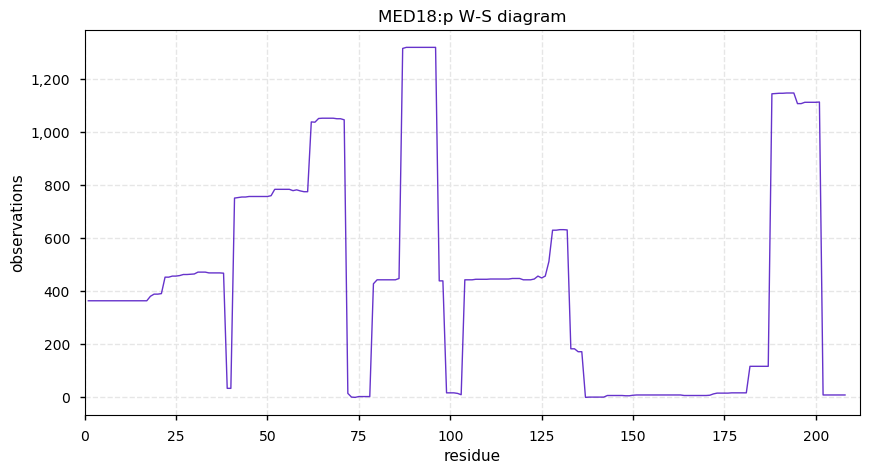
Tue Sep 17 19:26:10 +0000 2019I think in most cases site conservation is simply an attractive, convenient postulate that has gained currency through repetition.
Tue Sep 17 19:23:54 +0000 2019Is there any evidence that PTM acceptor sites are strongly conserved across species?
Tue Sep 17 16:02:44 +0000 2019A some-what improved format for a PTM cheat-sheet, featuring human signal recognition proteins 🔗
Tue Sep 17 12:08:20 +0000 2019MED17:p, mediator complex subunit 17 (H. sapiens) 🔗 Midsized nuclear protein; N-terminal phosphodomain; 1 SAV: E69D (maf=0.57); 1 splice variant; mature form 2-651 [5,955 x] 🔗
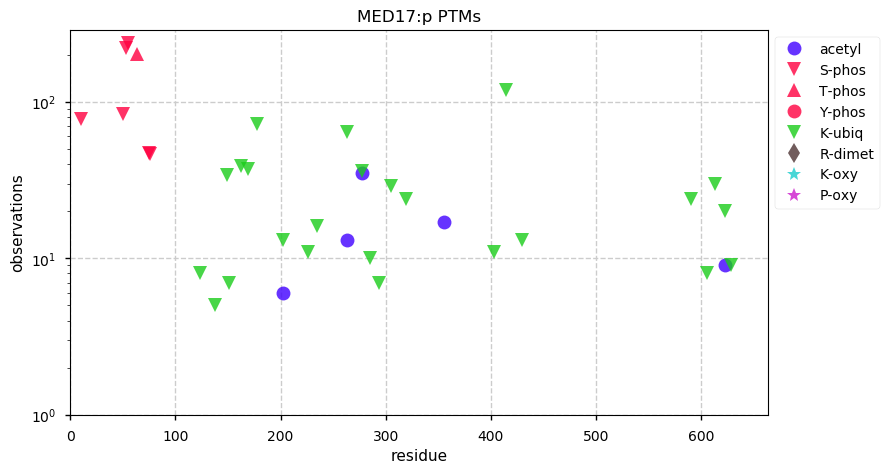
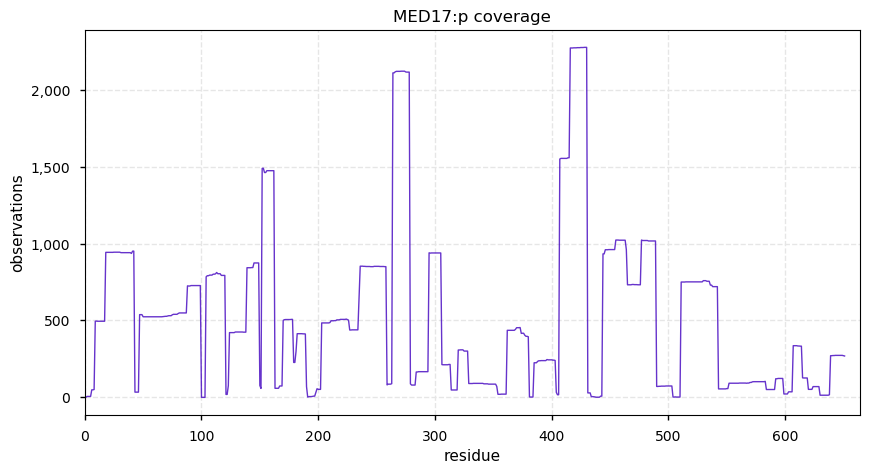
Mon Sep 16 20:47:16 +0000 2019It is f*ing peptides like this (from S. cerevisiae HHF1:p):
GKGGKGLGKGGAKR
that ends up being the exception that makes the rule.
Mon Sep 16 15:40:33 +0000 2019Is anyone keeping track of the number of tonnes of carbon being used by each party leader during the Canadian federal election?
Mon Sep 16 12:27:27 +0000 2019MED16:p, mediator complex subunit 16 (H. sapiens) 🔗 Midsized nuclear protein; many ubiquitination sites; no SAVs; 1 splice variant; mature form 2-860 [5,081 x] 🔗
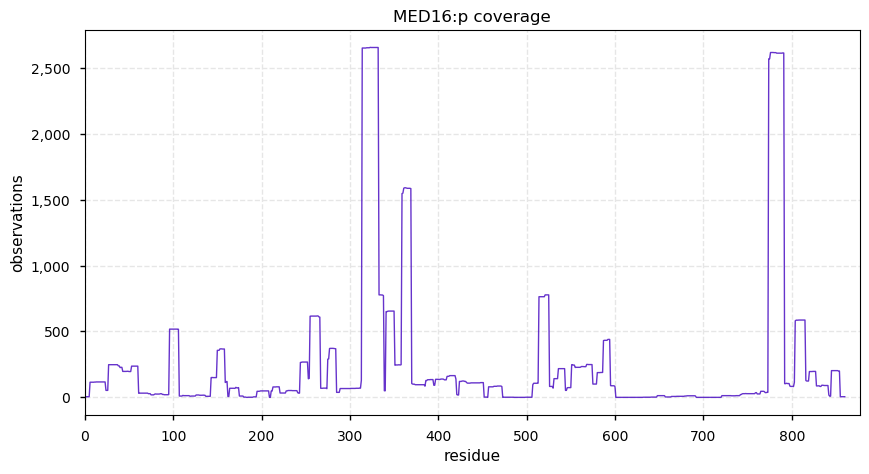
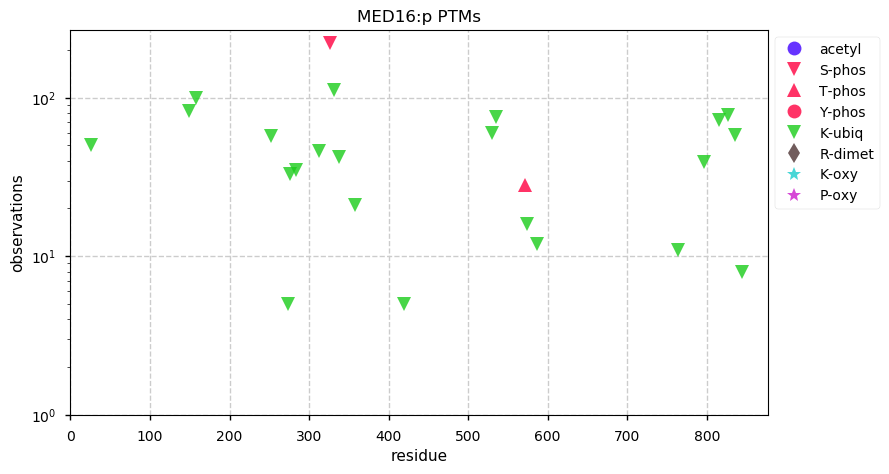
Sun Sep 15 14:39:01 +0000 2019@astacus I agree, but they are still in there pitching the idea.
Sun Sep 15 13:29:49 +0000 2019MED15:p, mediator complex subunit 15 (H. sapiens) 🔗 Midsized nuclear protein; several phosphodomains; low complexity domains between 105-496; no SAVs; 1 splice variant; mature form 1-788 [8,015 x] 🔗
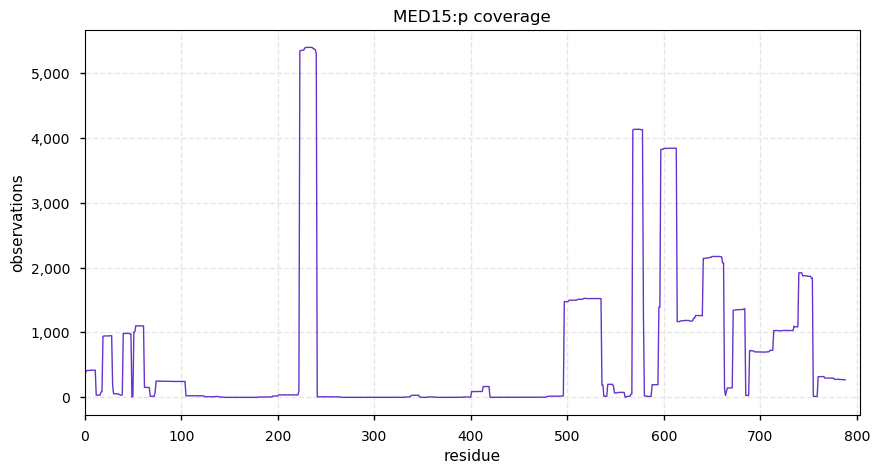
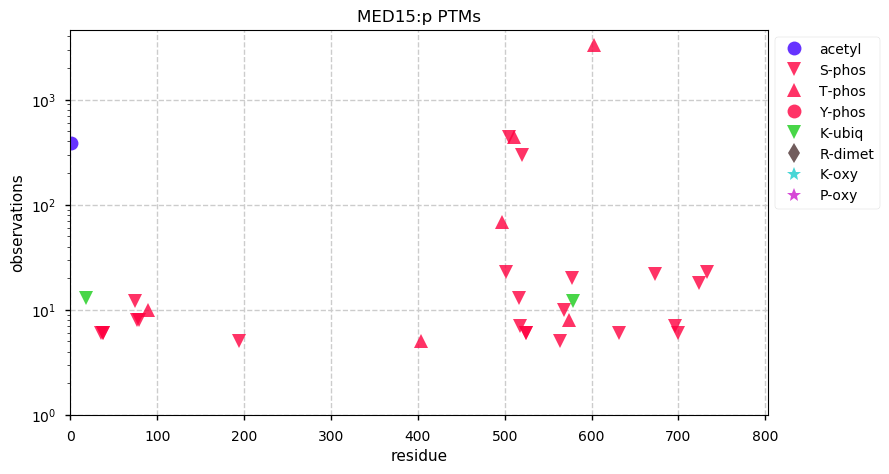
Sun Sep 15 13:11:57 +0000 2019It seems as though Nature Communications is trying to compete with JPR for HUPO papers. Nature has been trying to do this for years: those 2 dubious "draft human proteome" papers were an early salvo in the contest.
Sat Sep 14 23:00:30 +0000 2019A cheat-sheet for the PTMs observed on the 31 protein subunits of the human mediator complex (mediates interactions between transcription factors and RNA polymerase II): 🔗
Sat Sep 14 12:01:36 +0000 2019MED14:p, mediator complex subunit 14 (H. sapiens) 🔗 Large nuclear protein; 3 distinct phosphodomains; no SAVs; 1 splice variant; mature form 4-1454 [8,366 x] 🔗
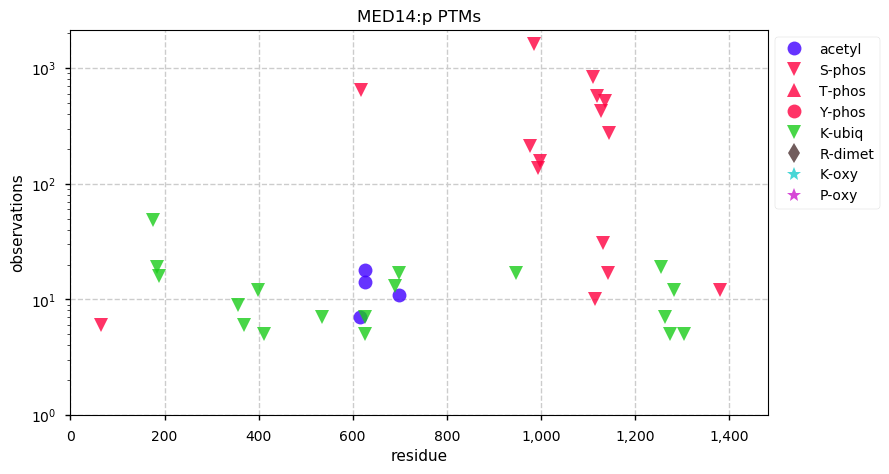
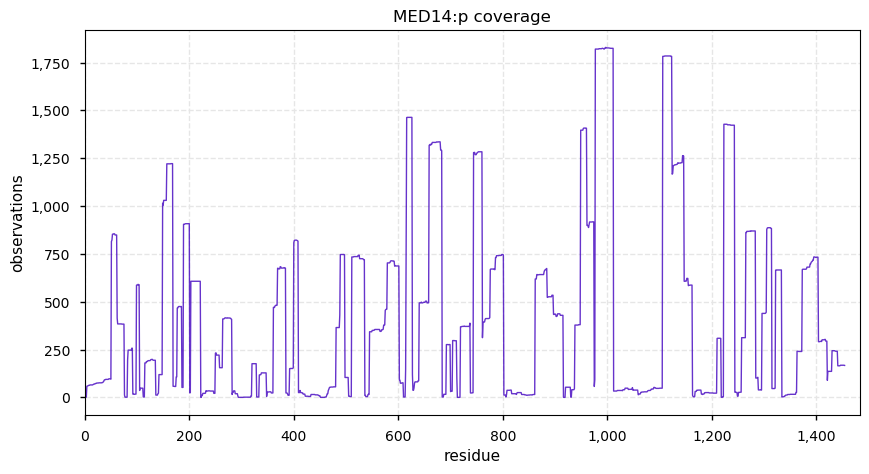
Fri Sep 13 12:44:41 +0000 2019MED13:p, mediator complex subunit 13 (H. sapiens) 🔗
Large nuclear protein; 4 distinct phosphodomains; few ubiquitinylation sites compared to MED12:p; no SAV; 1 splice variant; mature form 2-2174 [3,321 x] 🔗
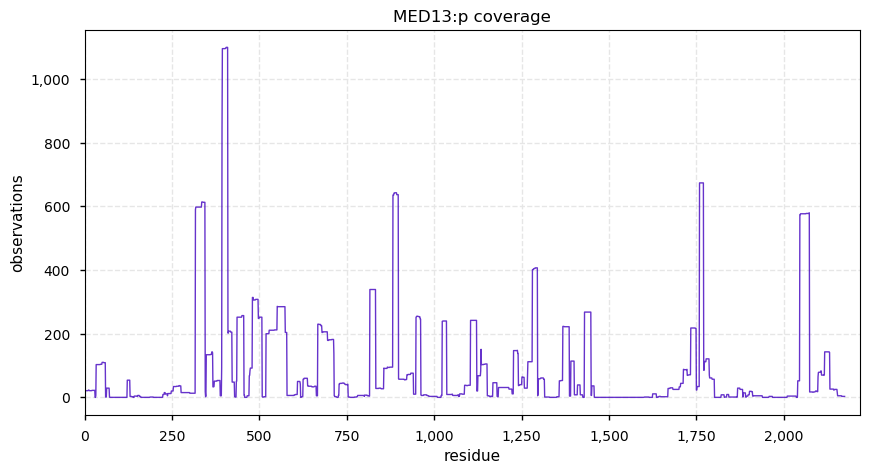
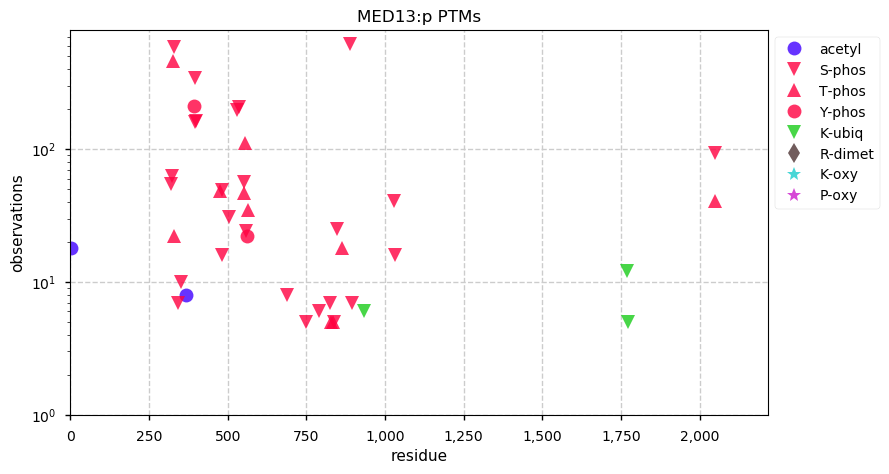
Thu Sep 12 19:05:39 +0000 2019Maybe "exploris-K" or "maxis-K" (I'm willing to sell naming rights).
Thu Sep 12 15:52:12 +0000 2019Is there any sort of catchy name for lysines that serve as acceptor sites for both acetylation and ubiquitinylation? Feel free to make one up.
I'm kind of partial to "death-acceptor" & if it is also a SUMOyl site "super-death-acceptor".
Thu Sep 12 12:52:21 +0000 2019MED12:p, mediator complex subunit 12 (H. sapiens) 🔗 Large nuclear protein; many PTMs, including R1899+dimethyl; 1 SAV: G1884S (maf=0.01); 1 splice variant; mature form 2-2177 [8,963 x] 🔗
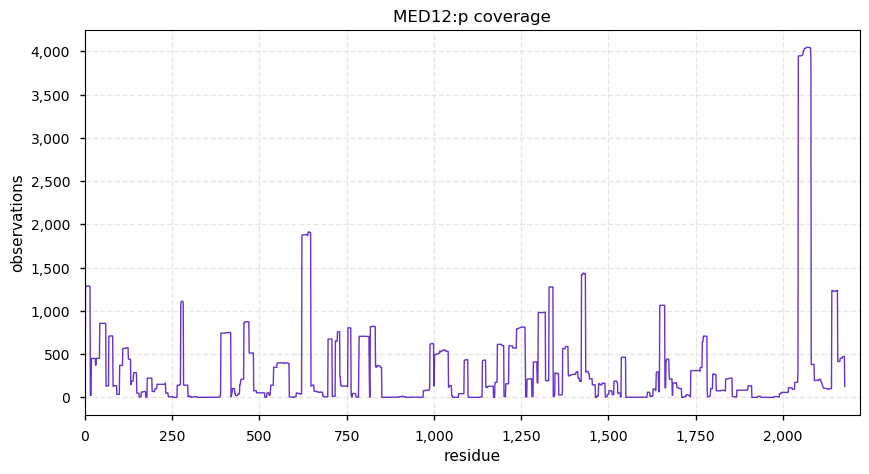
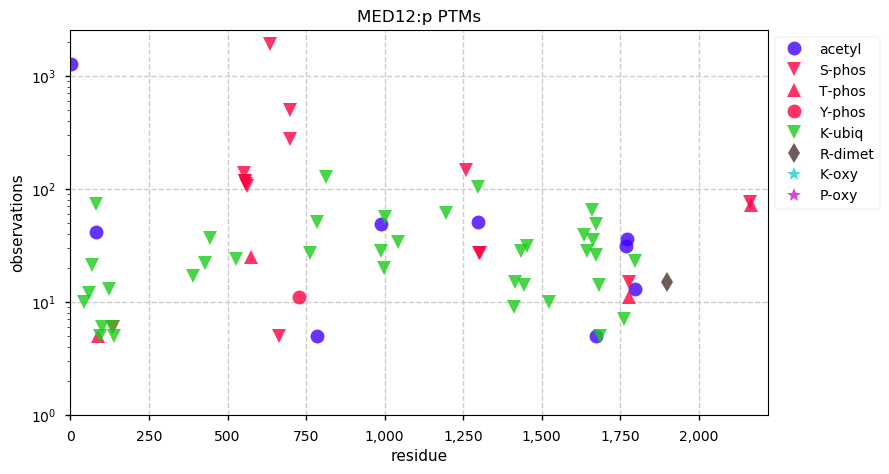
Wed Sep 11 15:31:02 +0000 2019@slavovLab And a 2nd hypothesis is that the mRNA ([R]) concentration should be proportional to the time-derivative to the protein ([P]) concentration: [R] ∝ ∂[P]/∂t, not [R] ∝ [P].
Wed Sep 11 12:53:26 +0000 2019MED11:p, mediator complex subunit 11 (H. sapiens) 🔗 Small intracellular protein; several low occupancy PTMs; no SAVs; 1 splice variant; mature form 2-117 [2,773 x] 🔗
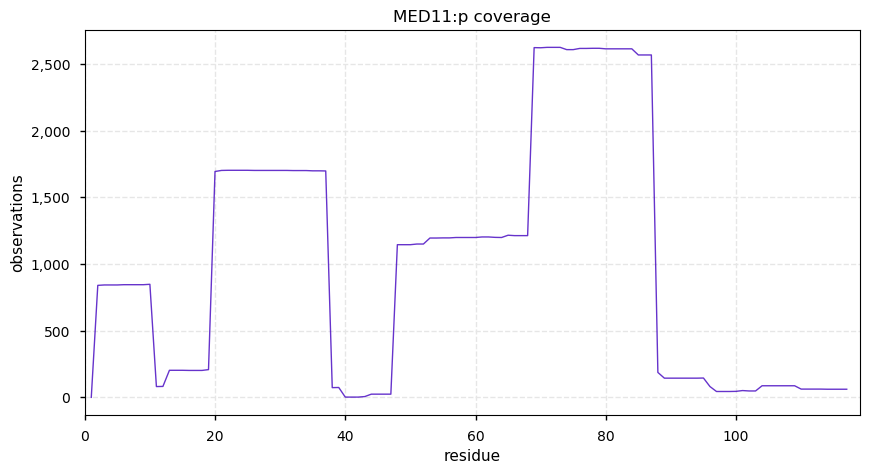
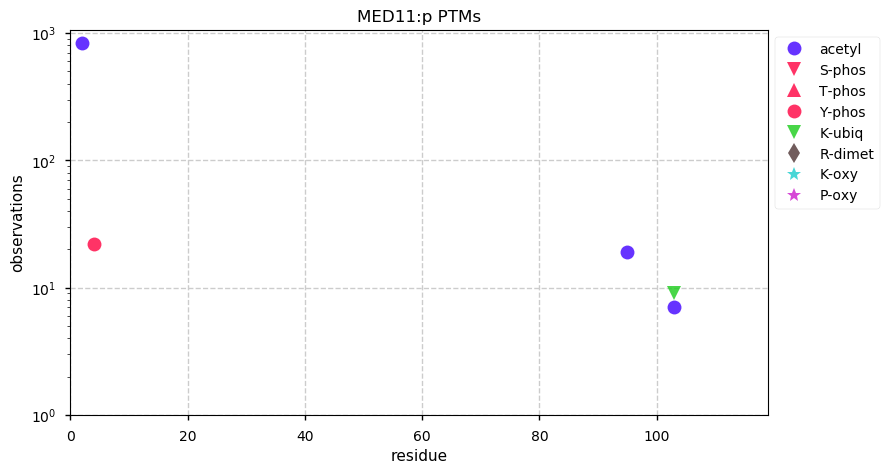
Wed Sep 11 12:45:59 +0000 2019@UCDProteomics I've analyzed some results using it. I haven't seen much off-target cleavage, but there isn't that much data available. 🔗
Wed Sep 11 02:25:38 +0000 2019@PastelBio One of the stories of my life. I'm sure it will be back.🤔
Tue Sep 10 23:32:15 +0000 2019@cwvhogue I would imagine it is an all-hands-on-deck situation!
Tue Sep 10 12:58:13 +0000 2019MED10:p, mediator complex subunit 10 (H. sapiens) 🔗 Small intracellular protein; no signficant PTMs; no SAVs; 1 splice variant; mature form 2-135 [2,837 x] 🔗
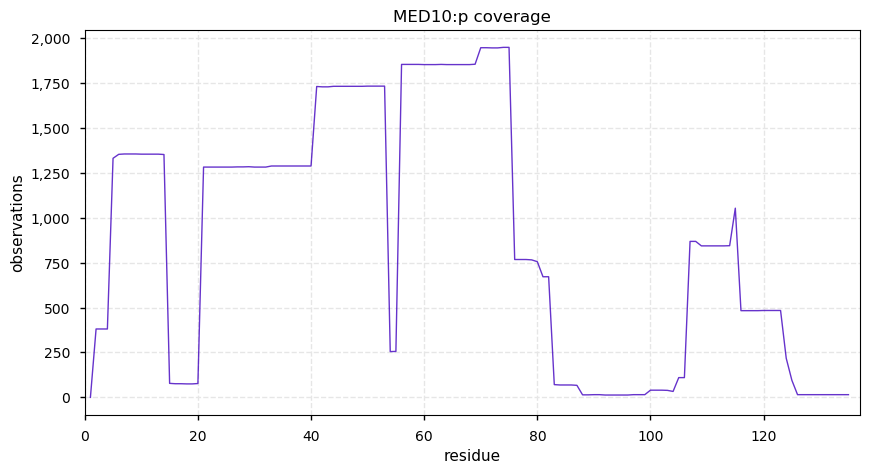
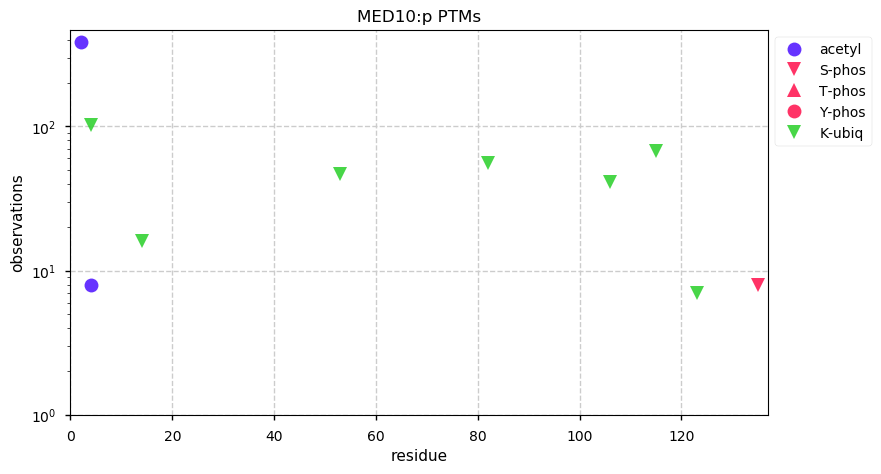
Mon Sep 09 13:12:12 +0000 2019MED9:p, mediator complex subunit 9 (H. sapiens) 🔗 Small intracellular protein; 2 major phosphorylation sites: S46 and S53; several low complexity domains; no SAVs; 1 splice variant; mature form 2-146 [2,508 x] 🔗
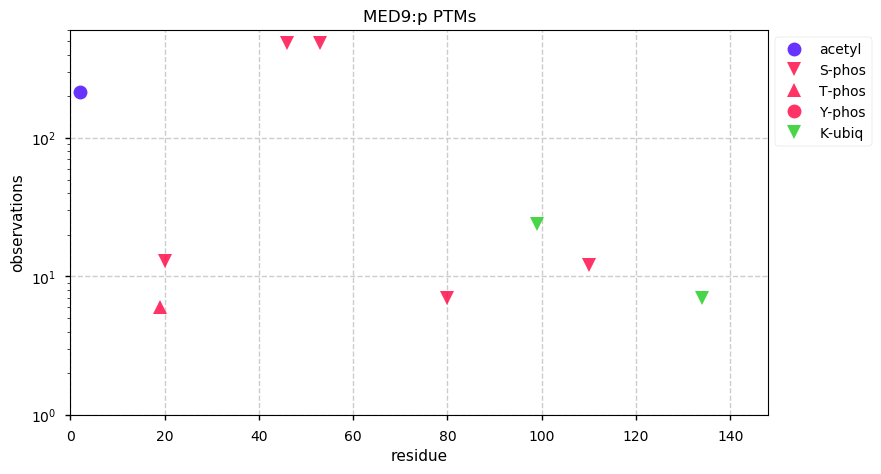
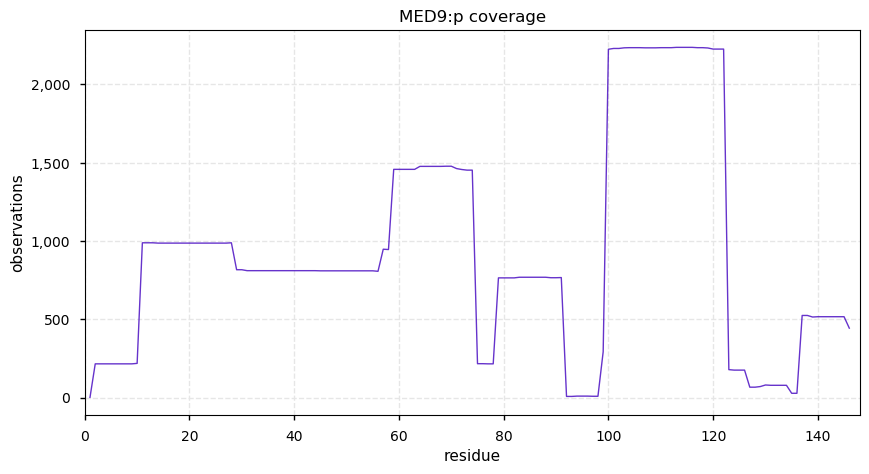
Mon Sep 09 13:06:48 +0000 2019@neXtProt_news @nesvilab Thanks. The instructions worked.
Sun Sep 08 12:58:35 +0000 2019MED8:p, mediator complex subunit 8 (H. sapiens) 🔗 Small intracellular protein; 2 phosphorylation sites: S11 and S82; no SAVs; 2 splice variants; mature form 1-268 [4,006 x] 🔗
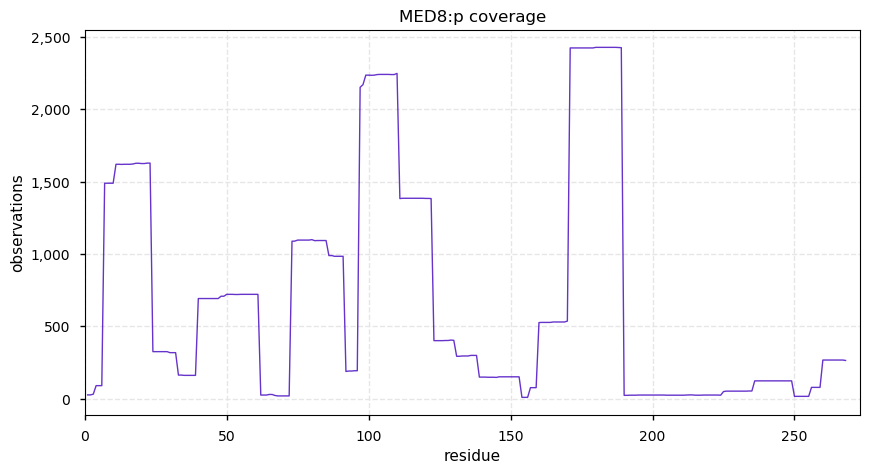
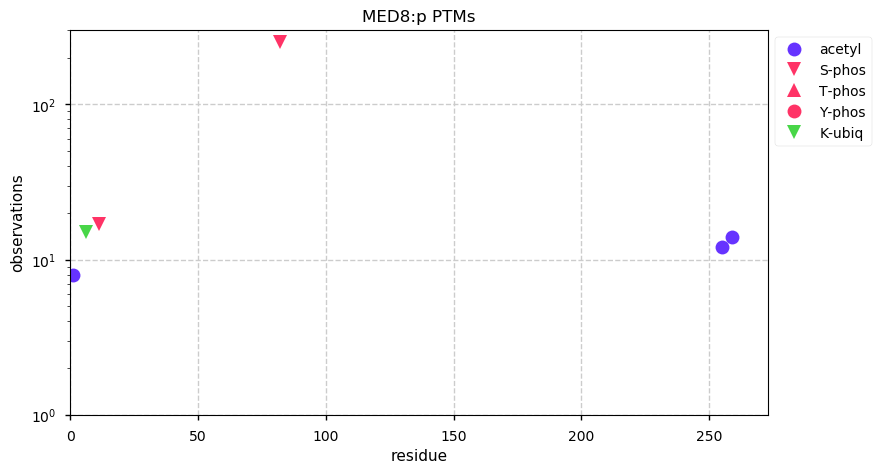
Sat Sep 07 23:12:17 +0000 2019I don't understand science reporting ... 🔗
Sat Sep 07 15:50:49 +0000 2019@nesvilab I was just trying to use nextprot for human PTM info for a protein (MED1) and couldn't figure out how to get it from their website. It used to be fairly easy, but I couldn't find it with the current interface. Any tips?
Sat Sep 07 14:39:35 +0000 2019Thanks to everyone who participated in the poll. I am surprised by the results (68% for UniProt): I had thought that the extensive campaign neXtProt has been conducting for years would have more effect on public opinion.
Sat Sep 07 13:47:40 +0000 2019MED7:p, mediator complex subunit 7 (H. sapiens) 🔗 Small intracellular protein; a few low occupancy PTMs; no SAVs; 1 splice variant; mature form 2-233 [2,136 x] 🔗
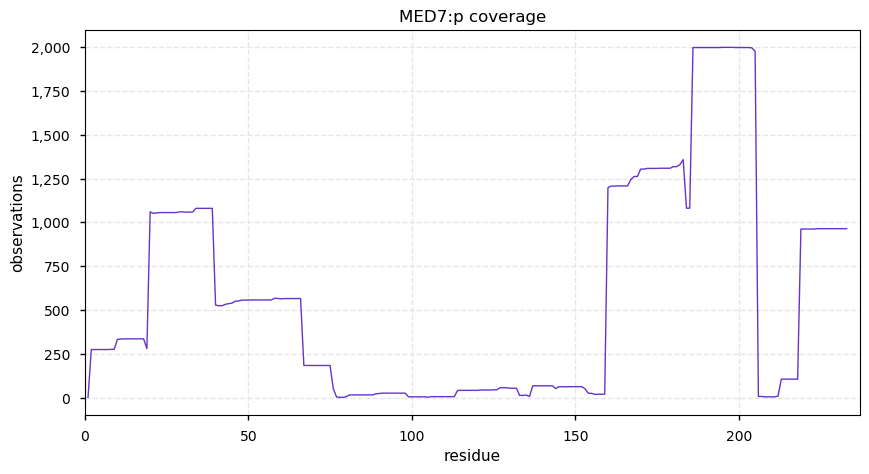
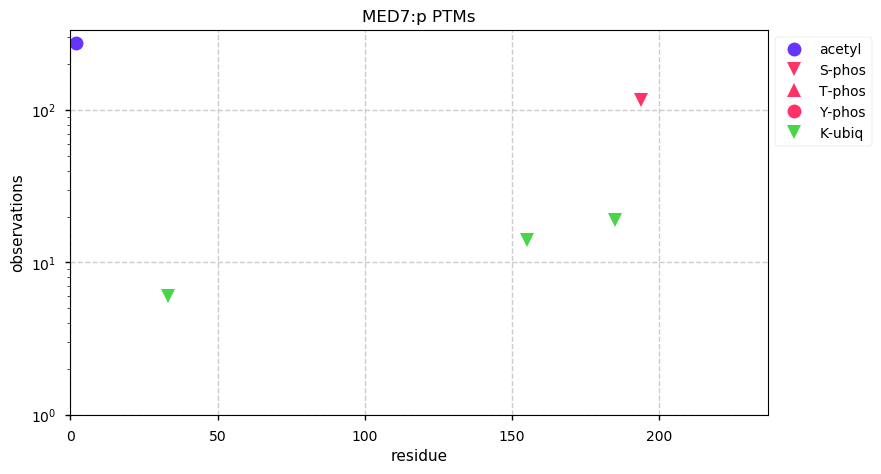
Fri Sep 06 14:30:03 +0000 2019Which resource for protein post-translational modification information do you consider to be the most reliable:
Fri Sep 06 12:28:28 +0000 2019MED6:p, mediator complex subunit 6 (H. sapiens) 🔗 Small intracellular protein; a few C-terminal acetylations; no SAVs; 1 splice variant; mature form 2-246 [2,659 x] 🔗
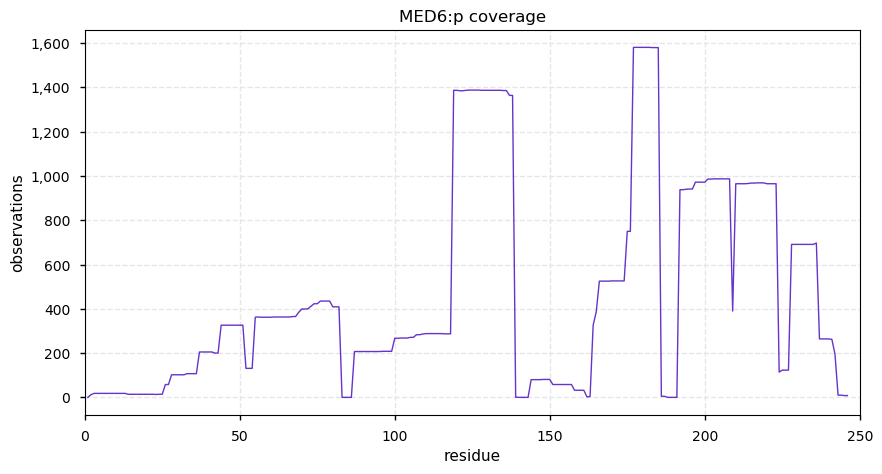
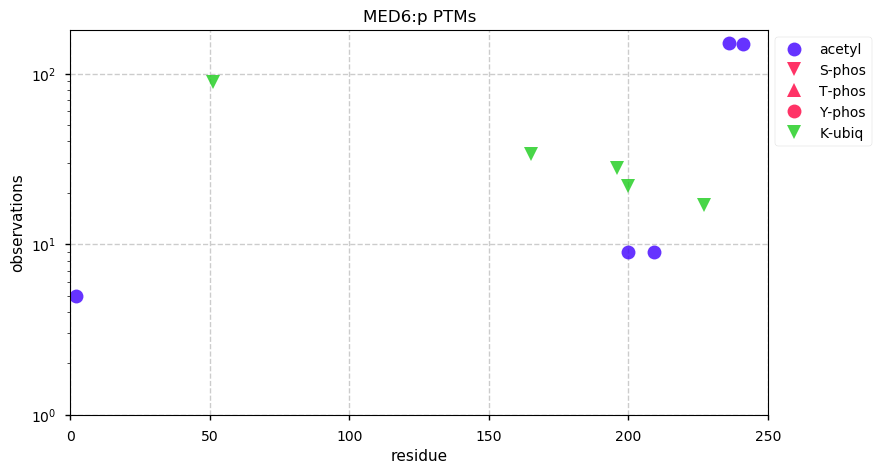
Thu Sep 05 15:51:43 +0000 2019@ypriverol To the best of my knowledge, there hasn't really been very much effort to deal with this issue on the PSM assignment level. To do it properly requires changes to both the way search engines work and how sequence collections for this purpose are curated.
Thu Sep 05 15:44:27 +0000 2019@_atanas_ Why refer to the SLC40A1gene as Fpn?
Thu Sep 05 12:20:47 +0000 2019MED4:p, mediator complex subunit 4 (H. sapiens) 🔗 Small intracellular protein; several low occupancy PTMs; no SAVs; 1 splice variant; mature form 2-270 [6,177 x] 🔗
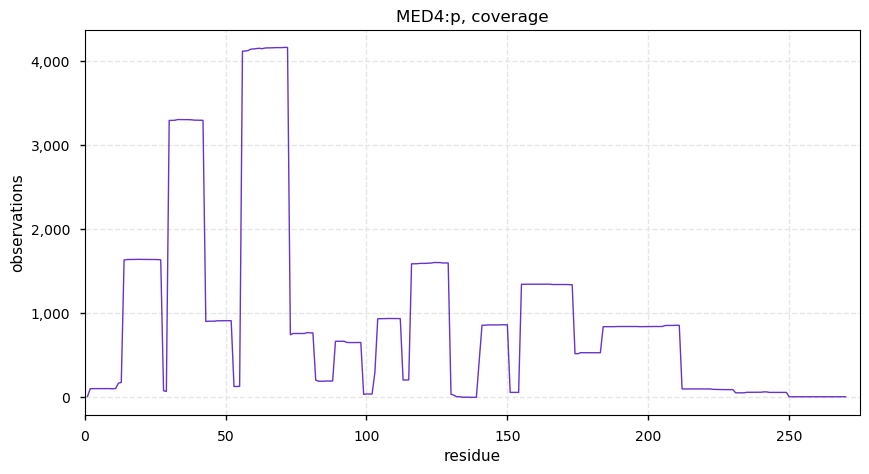
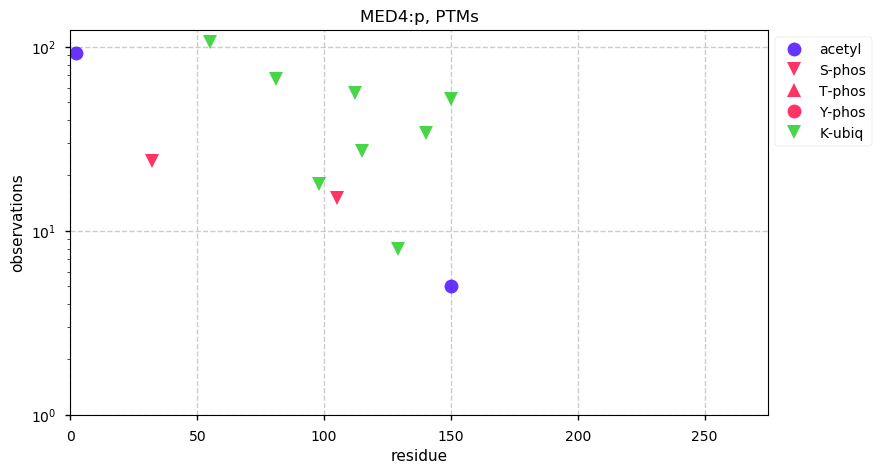
Wed Sep 04 17:15:18 +0000 2019@MHendr1cks @jwoodgett @robannan @graemedmoffat Is the annual budget expenditure & number of FTEs for CCV available somewhere? I tried to find it but probably just wasn't looking in the right places.
Wed Sep 04 14:23:50 +0000 2019@Smith_Chem_Wisc Most people in proteomics aren't very interested in how sequence collections get assembled or what is in them. But then, I still get a little confused when people refer to FASTA files as databases. It was a fortunate mistake made early on, but still ...
Wed Sep 04 12:19:43 +0000 2019MED1:p, mediator complex subunit 1 (H. sapiens) 🔗 Large intracellular protein; distinctive pattern of phosphodomains; 1 SAV: S1240G (maf=0.01); 1 splice variant; mature form 1-1581 [14,132 x] 🔗
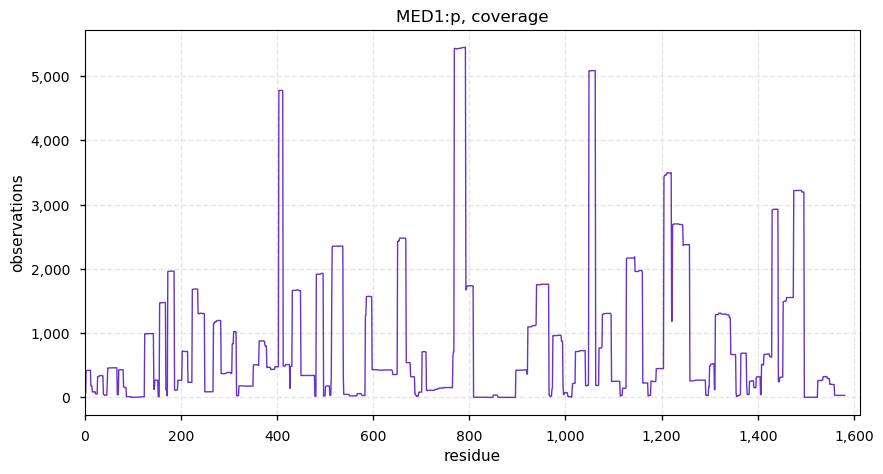
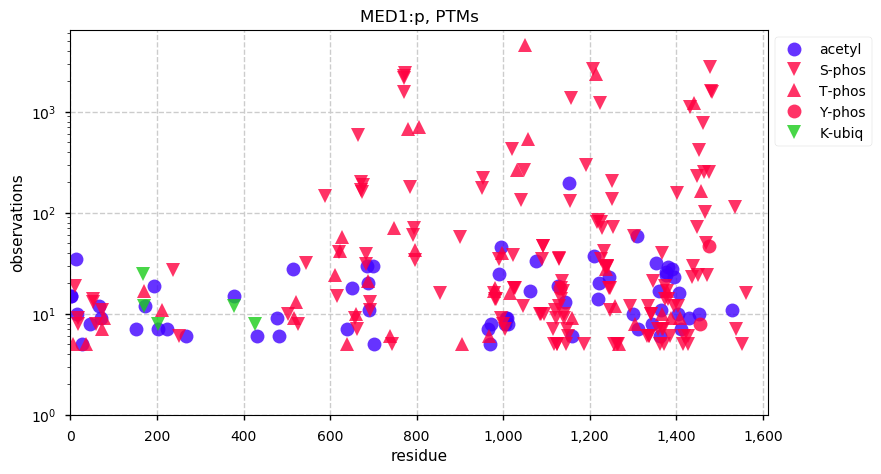
Tue Sep 03 14:40:09 +0000 2019@Smith_Chem_Wisc From what I can see in the literature, there isn't very much thought or consideration of proteins introduced to a sample as artifacts because of experimental design.
Tue Sep 03 12:35:36 +0000 2019STAB2:p, stabilin 2 (Homo sapiens) 🔗 Large membrane protein; most abundant in liver and spleen; no PTMs; no SAVs; 1 splice variant; mature form 20-2551 [888 x] 🔗
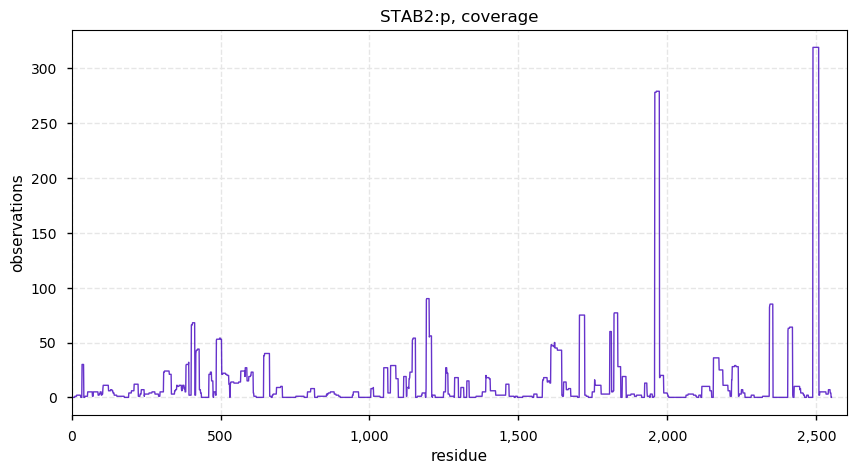
Mon Sep 02 13:14:06 +0000 2019Like so many proteins, its function seems to be in the eye of the beholder.
Mon Sep 02 13:04:58 +0000 2019Most commonly found in antigen-presenting cells, such as leukocytes, dendritic cells and sigmoid endothelium cells
Mon Sep 02 12:55:13 +0000 2019STAB1:p, stabilin 1 (Homo sapiens) 🔗 Large membrane protein; aka FEEL-1, CLEVER-1, FELE-1, FEX1, SCARH2; no sig. PTMs; 6 SAVs: e.g., M2506T (maf=0.55), I2282V (maf=0.34); not obs. in common cell lines; 1 splice variant; mature form 25-2570 [5,912 x] 🔗
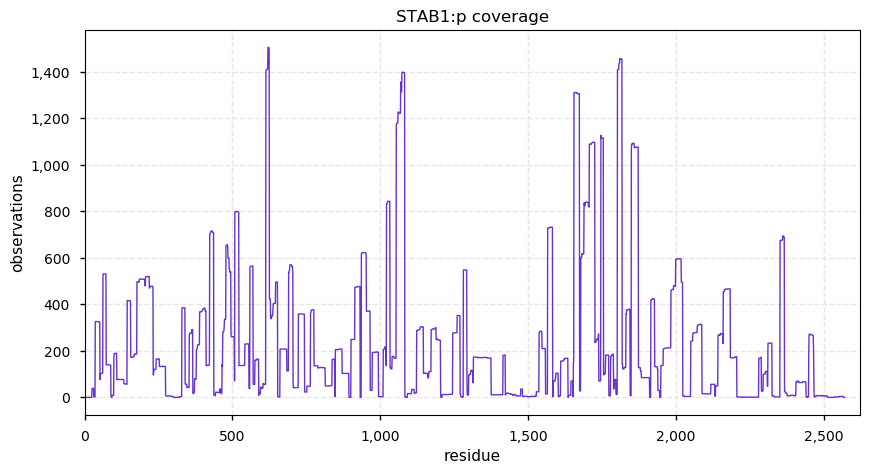
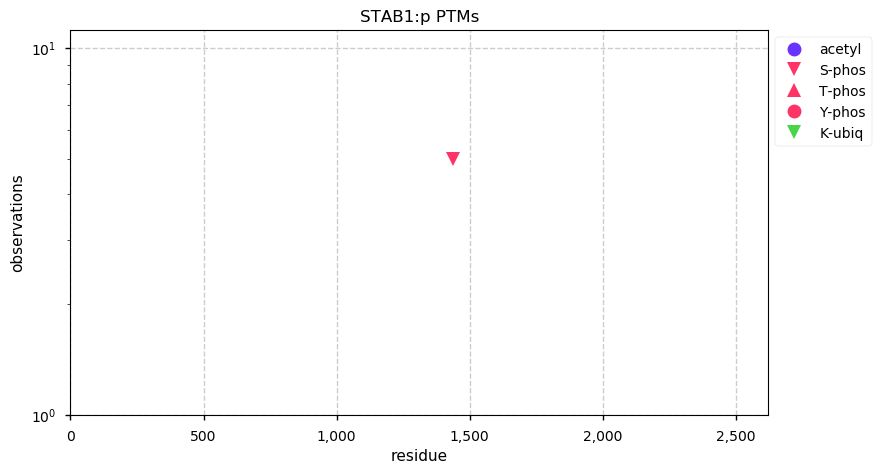
Sun Sep 01 13:25:19 +0000 2019@mailaved @ProteomicsNews What types of matrix molecule do people use for imaging work these days?
Sun Sep 01 13:12:42 +0000 2019An interesting little protein that attracts little interest.
Sun Sep 01 12:53:40 +0000 2019SURF6:p, surfeit 6 (Homo sapiens) 🔗 Small interacellular protein; S/T-phosphorylation; several low complexity (KRE) domains;1 splice variant; mature form 2-361 [9,085 x] 🔗
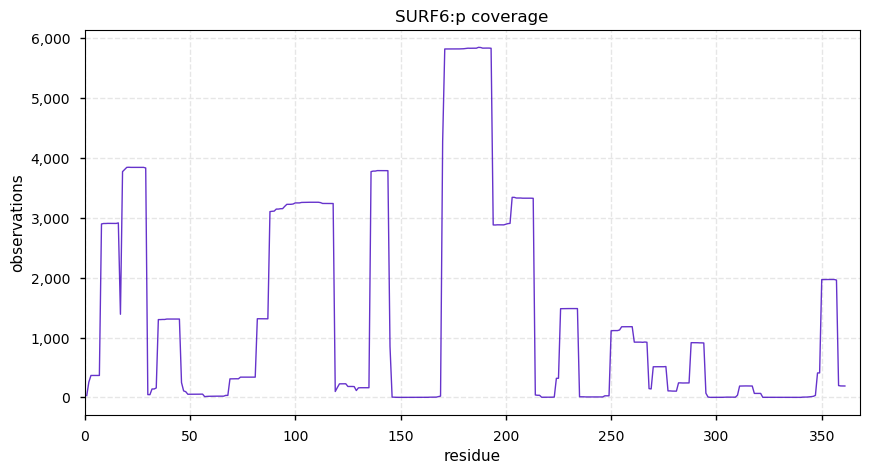
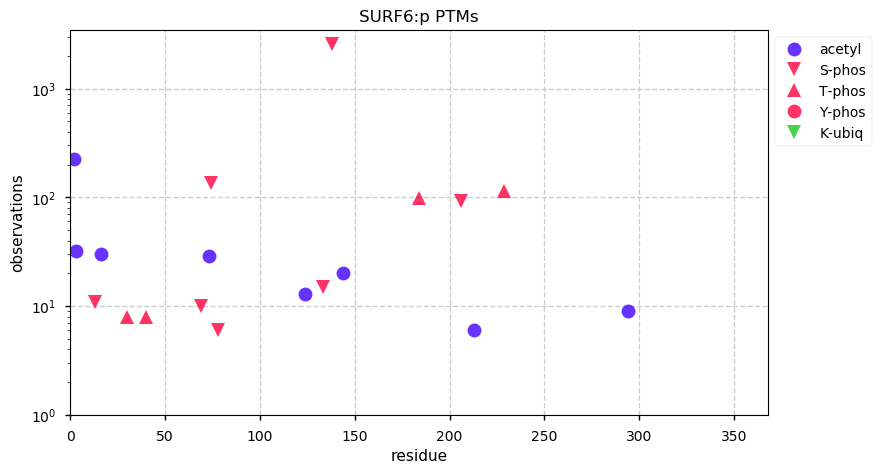
Sat Aug 31 20:35:08 +0000 2019@ProteomicsNews @chrashwood @educhicano I doubt if any two proteomics labs use the same combination of MS, chromatography and sample prep methods. It is often pretty easy to figure out where data comes from, even when people try to anonymize things.
Sat Aug 31 14:02:54 +0000 2019@SpecInformatics @neely615 Why do you want to do this type of analysis? It has fallen in and out of fashion since the mid 90's, but it hasn't been very successful at finding novel peptides.
Sat Aug 31 13:25:01 +0000 2019MED22:p, mediator complex subunit 22 (Homo sapiens) 🔗 Small nuclear protein; aka SURF5; no significant PTMs; gene has been difficult to map; no SAVs; mature form 2-200 [3,093 x] 🔗
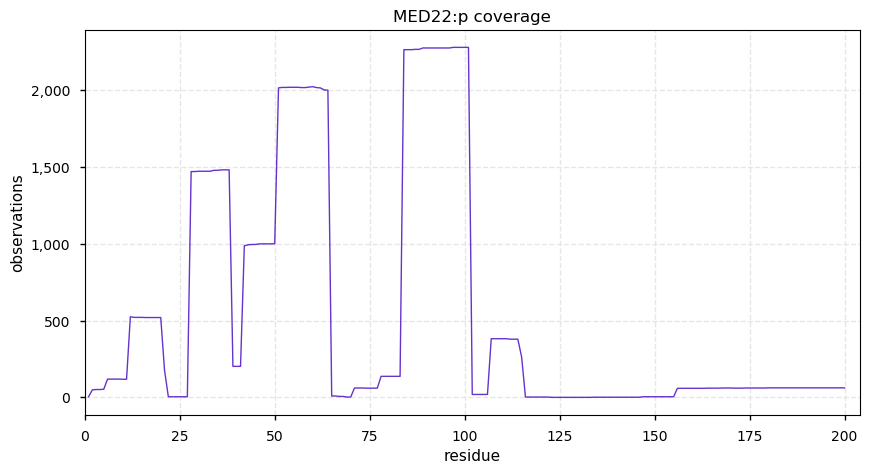
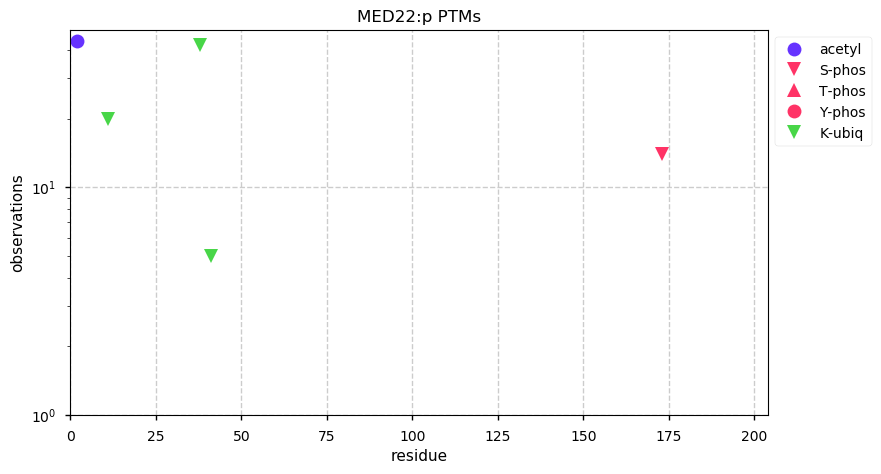
Fri Aug 30 14:12:47 +0000 2019@JoelisSteele That must be it ;-)
Fri Aug 30 12:51:06 +0000 2019@Sci_j_my @chrashwood And I'm experienced enough to know that there is no point bringing up this type of issue with journal editors once a paper has been published. Unless you enjoy frustration ...
Fri Aug 30 12:39:31 +0000 2019SURF4:p, surfeit 4 (Homo sapiens) 🔗 Small intracellular membrane protein; no significant PTMs; 5 membrane spanning domains; no SAVs; 1 splice variant; mature form 1,2,3,8-269 [29,783 x] 🔗
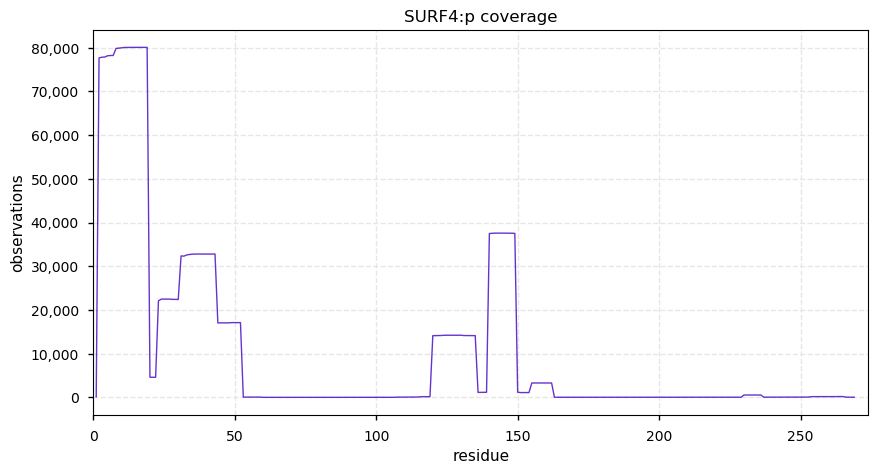
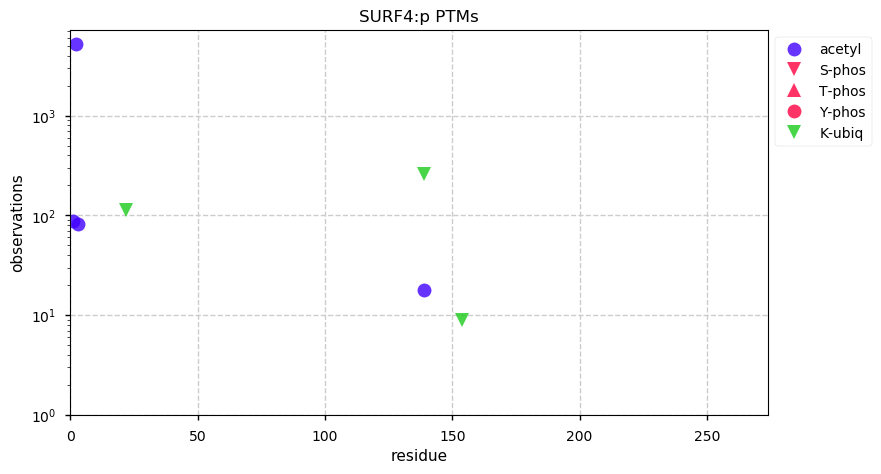
Fri Aug 30 12:27:40 +0000 2019@Sci_j_my @chrashwood One tried & true method is hacking the target-decoy simulation until the proteins of interest pass the test. Adding in custom filters after the initial standard analysis is pretty common in all types of hi throughput expts and they are prone to bias.
Fri Aug 30 12:07:06 +0000 2019@theoneamit True, but there are many proteins with much more puzzling PTM patterns.
Thu Aug 29 19:47:20 +0000 2019I will never understand why so many people are so enthralled by histone PTMs.
Thu Aug 29 16:11:01 +0000 2019Thanks to everyone who participated in this poll.
In the end, confident PTM assignments and speed of analysis were the most desirable traits in a novel proteomics search engine.
Thu Aug 29 14:46:40 +0000 2019@matthewhirschey @HHSGov Assuming WFT? means WTF?, I vote WFT?
Thu Aug 29 14:12:57 +0000 2019@IvisonJ The current UK PM reminds me of the foppish son of Edward I in Braveheart: full of strategies and ideas (& haircuts) but not really suited to the task of governing except by accident of birth.
Thu Aug 29 14:08:13 +0000 2019It's looking like speed and confidence may win: if you have an opinion, there is still 1 hr left to vote.
Thu Aug 29 13:09:17 +0000 2019One of the many proteins with a modification pattern you could spend a career trying to understand, but probably only scratch the surface.
Thu Aug 29 12:11:16 +0000 2019RPL7A:p, ribosomal protein L7a (Homo sapiens) 🔗 Small intracellular protein; aka SURF3; many acetyl/ubiquitinyl K acceptor sites; no SAVs; 1 splice variant; mature form 2-266 [54,923 x] 🔗
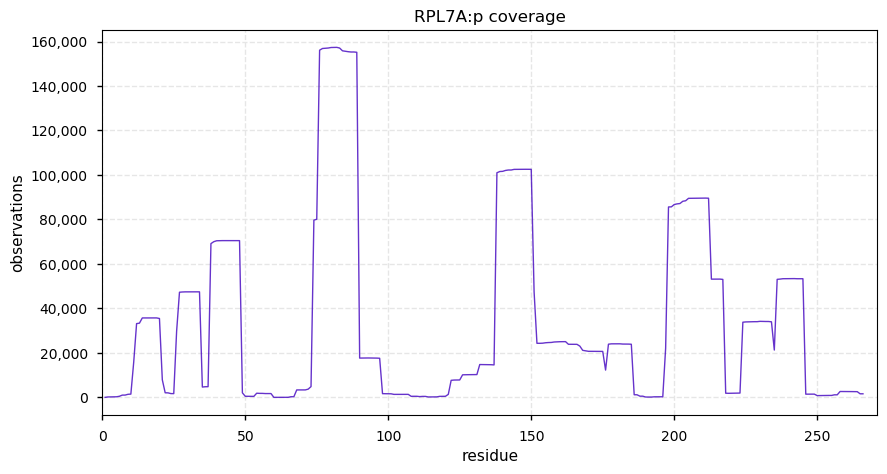
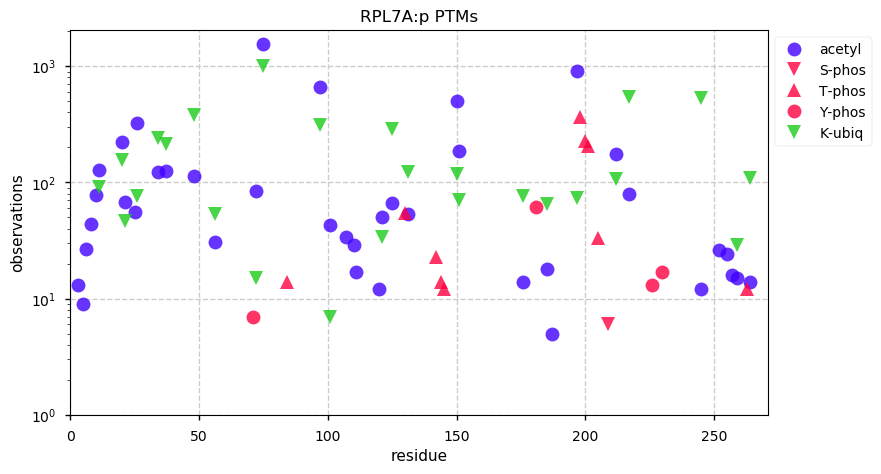
Wed Aug 28 16:45:55 +0000 2019@EconCalgary @jenditchburn @IRPP @aidanhollis Sell, every time.
Wed Aug 28 15:57:20 +0000 2019When you are looking at a new proteomics search engine, which of these is the most important parameter to you:
Wed Aug 28 13:39:52 +0000 2019@Smith_Chem_Wisc @NLKProteomics I later realized that if the morphism t:g→p explicitly includes "transcription + splicing + translation", then any further nit-picking about alternate splicing is dealt with as well.
Wed Aug 28 12:17:59 +0000 2019SURF2:p, surfeit 2 (Homo sapiens) 🔗 Small intracellular protein; phospho-domain 150-200; C-terminal low complexity domain 200-256; 2 SAVs: G213S (maf=0.32), R221Q (maf=0.02); 1 splice variant; mature form 2-256 [4,635 x] 🔗
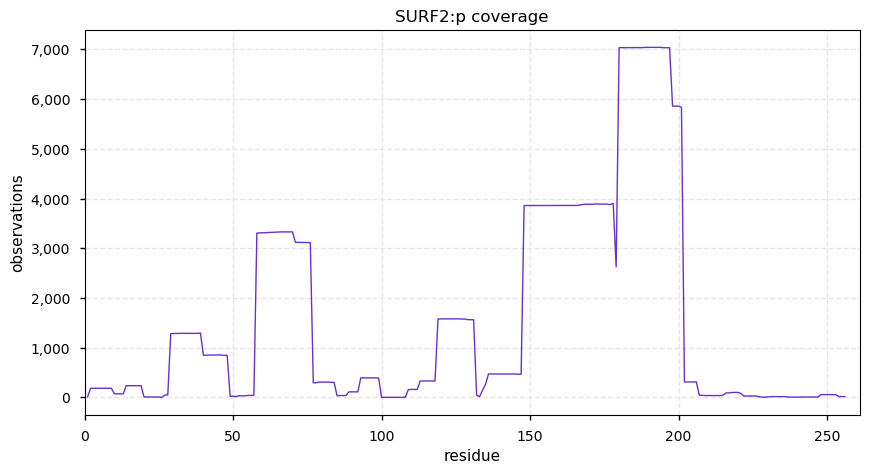
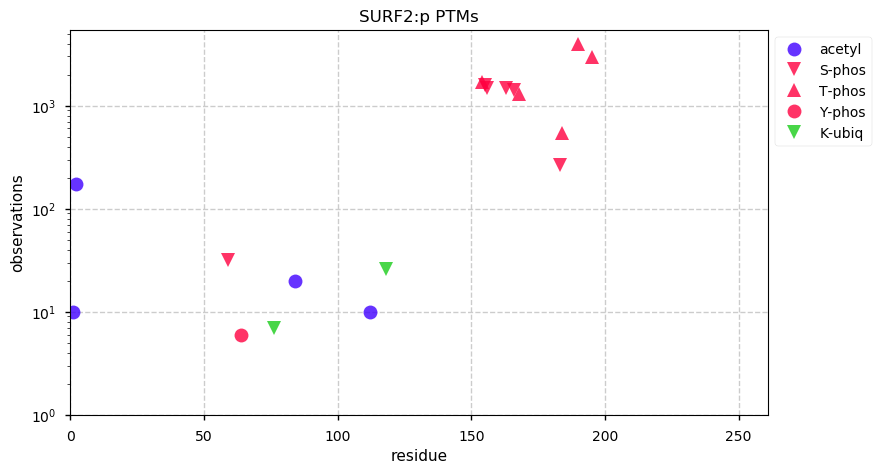
Wed Aug 28 00:22:38 +0000 2019@Smith_Chem_Wisc @NLKProteomics You could capture the HIST3 & similar cases if you broadened the idea of gene (g) to be a set of genes:
{g ∈ G | t:g → p = p0}
where G is the set of protein coding genes, p0 is the specific aa sequence and the morphism t:g→p is transcript+translation.
Tue Aug 27 22:47:29 +0000 2019@Smith_Chem_Wisc @NLKProteomics Actually, I wasn't thinking about PTMs when I mentioned histone 3 - histones have comparatively few PTMs. Histone 3's issue is there are many genes (🔗) with identical protein sequences, so you can't assign a specific gene to an obs protein.
Tue Aug 27 18:41:55 +0000 2019@Smith_Chem_Wisc @NLKProteomics OK, got it. Proteins like COL1A1:p or HIST2H3A:p are really outside of the scope of the discussion.
Tue Aug 27 16:30:58 +0000 2019Can anyone (e.g., @Smith_Chem_Wisc, @NLKProteomics) comment on how the new proteoform ranking scheme (Nat Meth) works for abundant proteins like collagen I A1 or histone 3? Based on the rules, it would seem like they are always going to be stuck at level ≥ 4.
Tue Aug 27 12:41:40 +0000 2019SURF1:p, cytochrome c oxidase assembly factor (Homo sapiens) 🔗 Small mitochondrial matrix protein; no PTMs; 1 SAV: D202H (maf=0.03); 1 splice variant; mature form 45-300 [5,124 x] 🔗
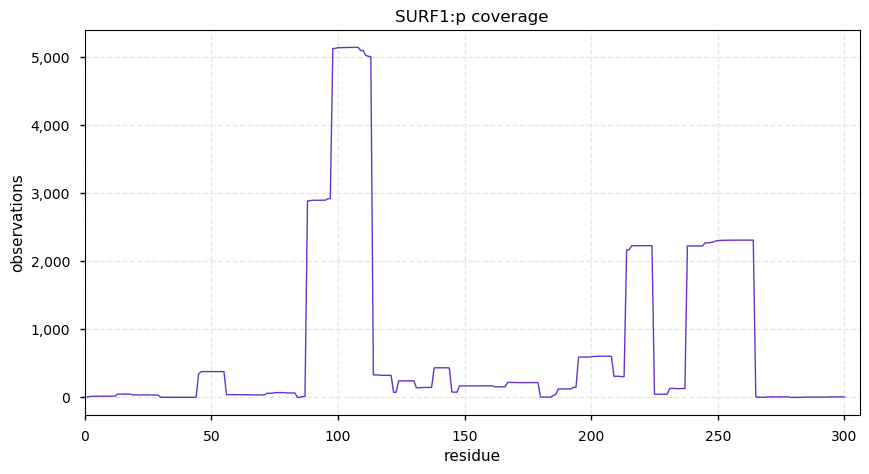
Mon Aug 26 18:58:36 +0000 2019Can anyone comment on how this scheme works for abundant proteins like collagen 1A1 or histone 3? Based on the rules, it would seem like they are going to be stuck in category 4, at best.
🔗
Mon Aug 26 12:42:08 +0000 2019SCO1:p, cytochrome c oxidase assembly protein (Homo sapiens) 🔗 Small mitochondrial protein; no PTMs; no SAVs; shares no tryptic peptides with SCO2; 256 obs. unique peptides; 1 splice variant; mature form 67,68,69-301 [6,788 x] 🔗
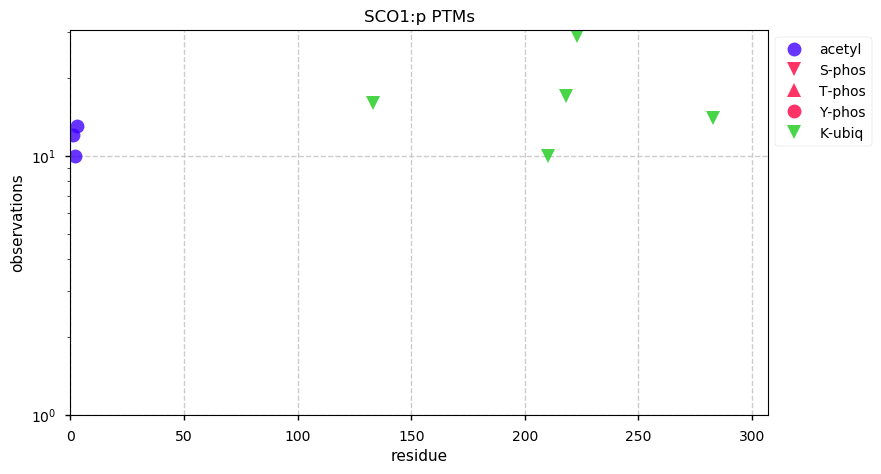
Sun Aug 25 16:41:21 +0000 2019Javascript + CSS programming to get browsers to do anything "fancy" using event handlers is the probably the most frustrating type of coding I engage in.
Sat Aug 24 12:59:44 +0000 2019SRPRB:p, SRP receptor subunit beta (H. sapiens) 🔗 Small ER-associated subunit; several distinct PTM domains; 3 SAVs: V9L (maf=0.17), F148S (maf=0.01), R252G (maf=0.01); 1 splice variant; mature form 2-271 [25,430 x] 🔗
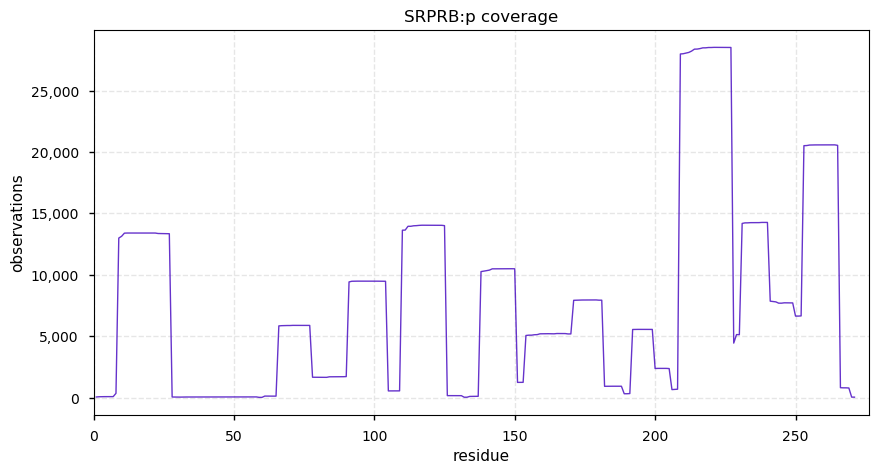
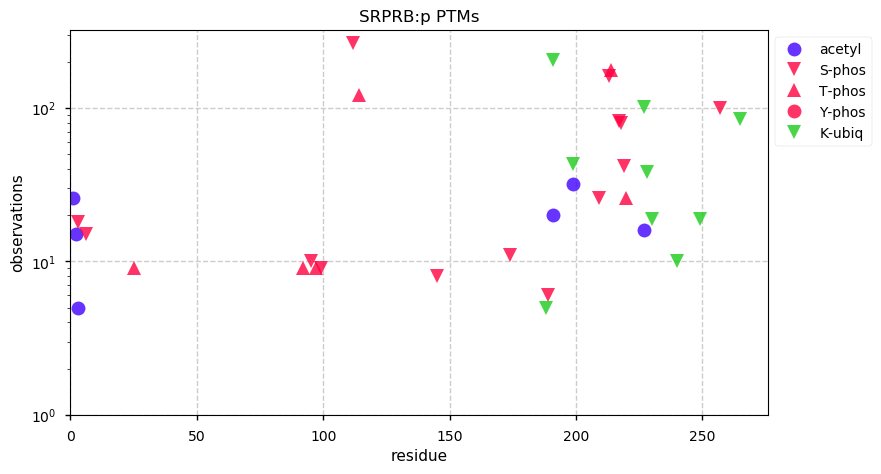
Fri Aug 23 21:36:24 +0000 2019If any proteomics organization is looking to start a new initiative, I would welcome one with the goal of "understanding and minimizing the formation of deleterious chemical artifacts during the sample preparation process".
Fri Aug 23 19:34:48 +0000 2019Global map of current atmospheric CO2 concentration (darker brown means more higher concentration)
🔗
Fri Aug 23 12:53:51 +0000 2019SRPRA:p, SRP receptor subunit alpha (H. sapiens) 🔗 Midsize ER-associated subunit; signficant phosphodomain (260-320); no SAVs ; 1 splice variant; mature form 1-638 [22,602 x] 🔗
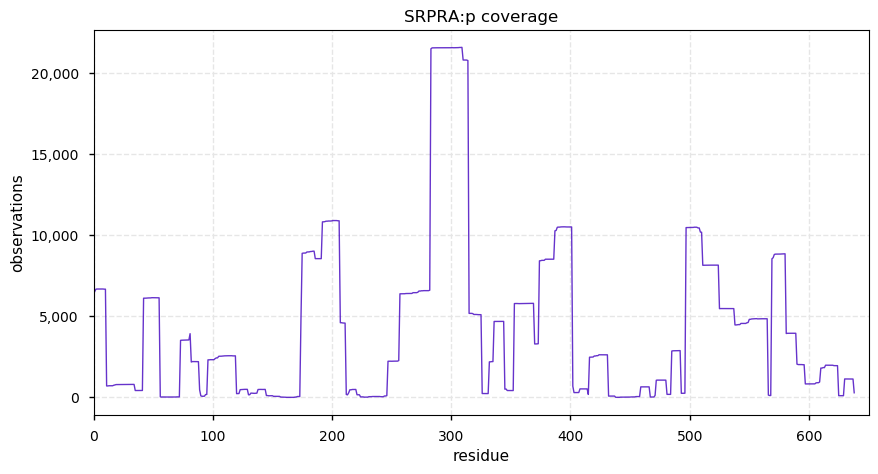
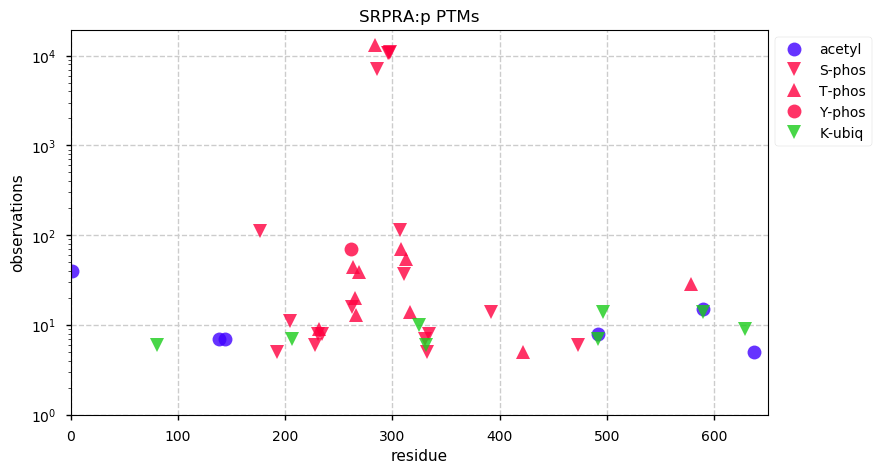
Thu Aug 22 21:16:06 +0000 2019@UCDProteomics @neely615 You, sir, are the King of New & Shiny Things.
In my role as "friendly stranger", here's another one:
🔗
Thu Aug 22 15:04:33 +0000 2019Not really breaking news, but it has bubbled up so that even the NY Times has noticed:
🔗
Thu Aug 22 13:24:59 +0000 2019@IlianAtanassov Given how far off the beaten path SRP72:p's PTM pattern places it, any idea is worth batting around. The SRP is an example of a common phenomenon in nature that is rare in academia: a collaboration between RNA and proteins.
Thu Aug 22 12:23:24 +0000 2019This one is pretty much daring us to figure out what on earth it is doing with so many PTMs, but sadly it is a non-histone "housekeeping" protein so its challenge will probably go unaddressed.
Thu Aug 22 12:20:09 +0000 2019SRP72:p, signal recognition particle 72 (H sapiens) 🔗 Midsize subunit; flamboyantly modified; multiple low complexity domains; 3 SAVs at residue 7: G7R (maf=0.01), G7W (maf=0.01), G7V (maf=0.01); 1 splice variant; mature form 2-671 [33,695 x] 🔗
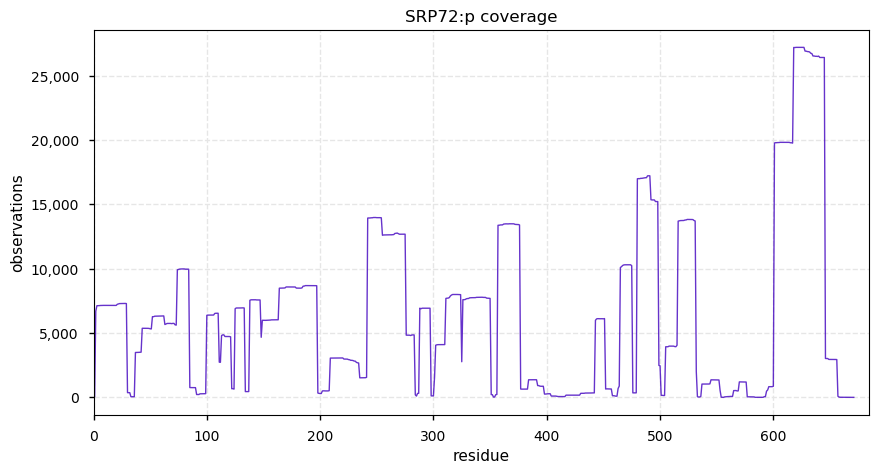
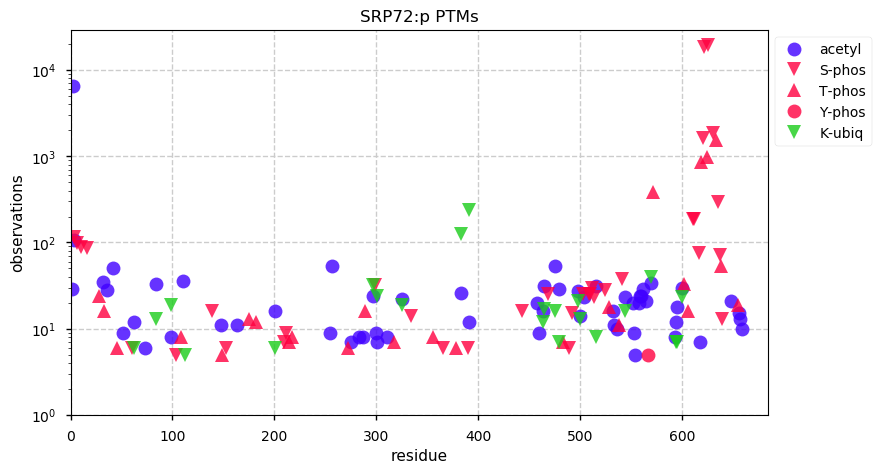
Thu Aug 22 12:12:57 +0000 2019@neely615 The searX system truly doesn't log anything about a query and masks your IP when it queries other systems for info.
Thu Aug 22 12:10:51 +0000 2019@neely615 I am understandably cautious of any faceb**k outfit saying they are going to 'simplify anything for me.
Thu Aug 22 11:44:35 +0000 2019@neXtProt_news @Mirshahvalady Thanks for the explanation.
Thu Aug 22 11:42:37 +0000 2019@neely615 I was unaware of the meta project you linked to in your post,
Wed Aug 21 19:00:17 +0000 2019@Mirshahvalady I'm afraid I don't quite understand your question regarding the use of SPARQL searches. Could you elaborate a bit?
Wed Aug 21 16:23:40 +0000 2019Thanks to everyone who participated in this poll. It turns out that there are no Y-linked proteins commonly observed in seminal plasma. The expression of Y-linked proteins is quite limited, while X-linked proteins are wide spread.
Wed Aug 21 12:57:48 +0000 2019SRP68:p, signal recognition particle 68 (H. sapiens) 🔗 Midsize intracellular subunit; highly acetylated; several low complexity domains; 1 SAV: A538T (maf=0.01); 1 splice variant; mature form 1,2-627 [33,695 x] 🔗
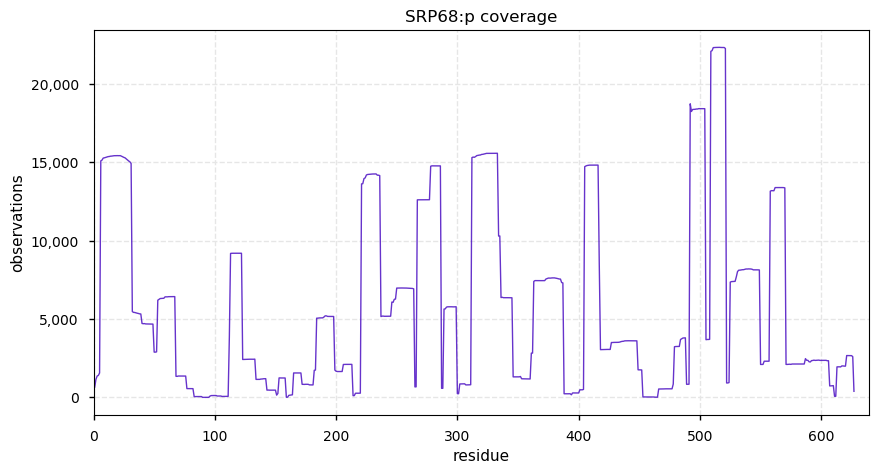
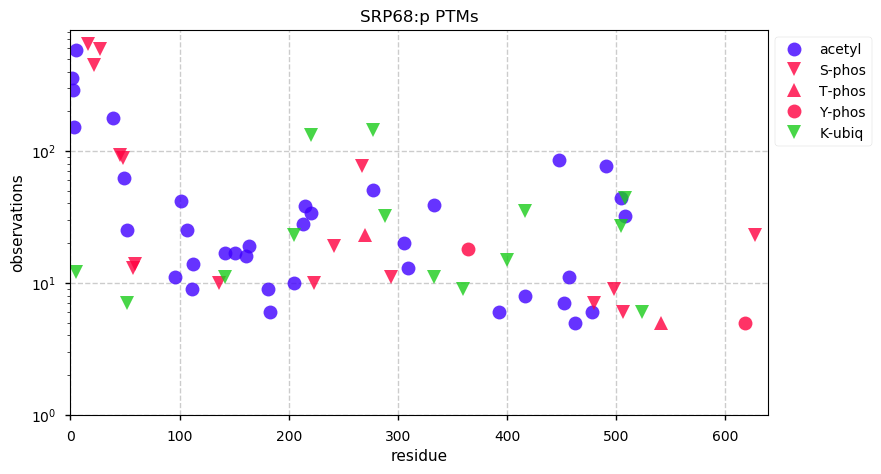
Tue Aug 20 19:14:39 +0000 2019@byu_sam I should have been clearer. I'm referring to the studies at
🔗
such as:
01CBTTC_PBT_Proteome_HMS_20180626
Tue Aug 20 17:25:32 +0000 2019@smolrobots @cstross I was wondering why so many guys have goatees ...
Tue Aug 20 16:16:53 +0000 2019@PastelBio Interesting study. I have occasionally seen data sets with significant amounts of persulfidation, but the associated manuscripts didn't recognize it was happening and I couldn't find any consistent reason for the phenomenon.
Tue Aug 20 15:00:17 +0000 2019If there are any CPTAC'ers listening, is there a reason that the 'Pediatric Brain Cancer Tumors Pilot Study' was done with such low sensitivity? This study has about 6,000 proteins per experiment, while other recent CPTAC studies have about 12,000.
Tue Aug 20 14:30:54 +0000 2019How many Y-linked proteins (i.e., proteins with genes on the Y-chromosome) are commonly observed in human seminal plasma?
Tue Aug 20 12:03:12 +0000 2019SRP54:p, signal recognition particle 54 (H. sapiens) 🔗 Midsize intracellular subunit; many PTMs; no SAVs; 2 splice variants; mature form 1,2-504 [20,910 x] 🔗
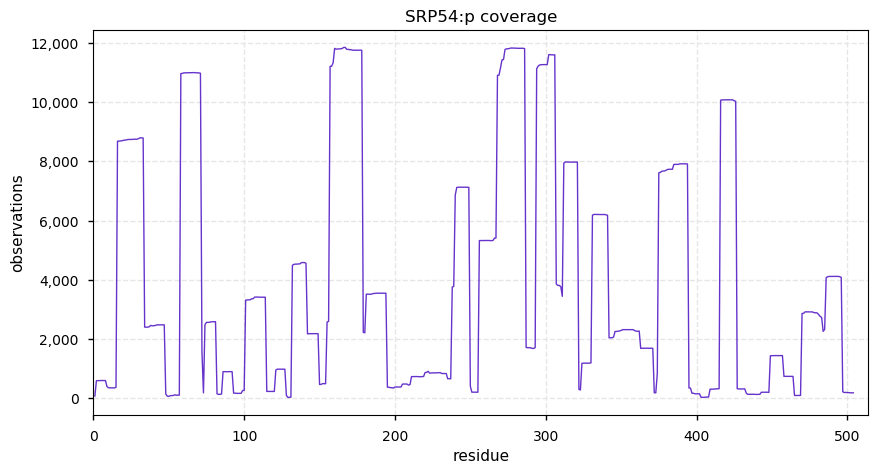
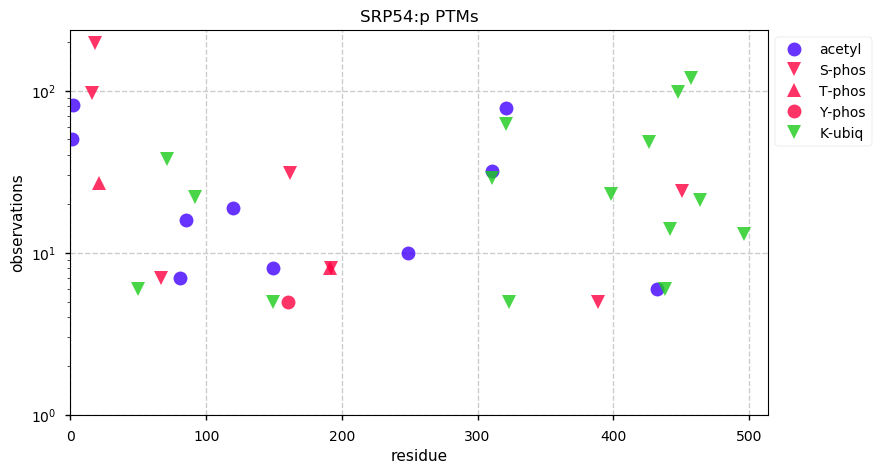
Mon Aug 19 12:29:41 +0000 2019SRP19:p, signal recognition particle 19 (H. sapiens) 🔗 Small intracellular subunit; several low occupancy PTM sites; no SAVs; 1 splice variant; mature form 2-144 [14,363 x] 🔗
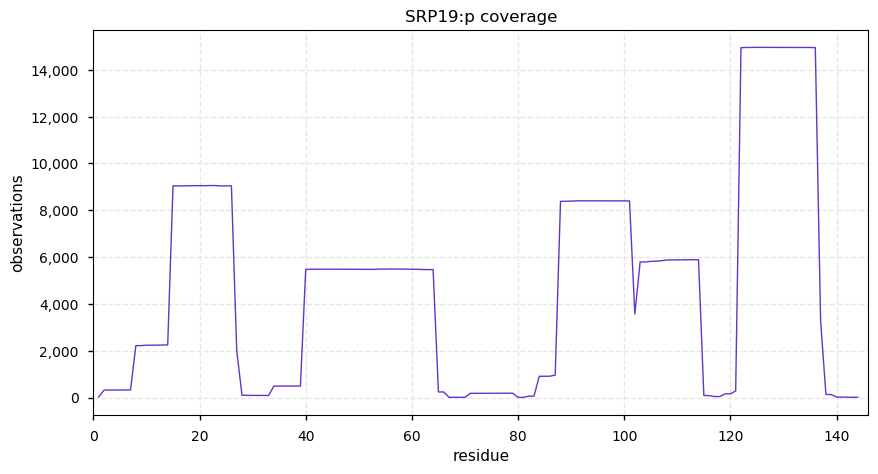
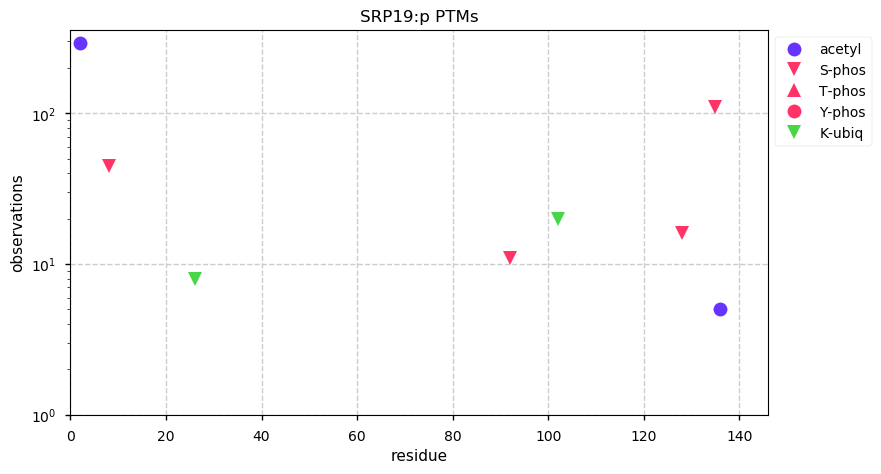
Sun Aug 18 13:58:40 +0000 2019@byu_sam @theoneamit @Smith_Chem_Wisc Good analogies. Someone fell in love with an idea and sought out evidence (regardless of how scant or improbable) to support it.
Sun Aug 18 13:17:24 +0000 2019You are not alone 🔗
Sun Aug 18 12:58:29 +0000 2019Normally, the ratio of S:T phosphorylation sites is > 3:1, so having so many T sites (& no S sites) is noteworthy. Also none of the sites is part of a TP motif.
Sun Aug 18 12:39:41 +0000 2019SRP14:p, signal recognition particle 14 (H. sapiens) 🔗 Small intracellular subunit; at least 7 T-phosphorylation sites; several low complexity domains; 1 SAVs: P124A (maf=0.12); 1 splice variant; mature form 1,2-136 [40,195 x] 🔗
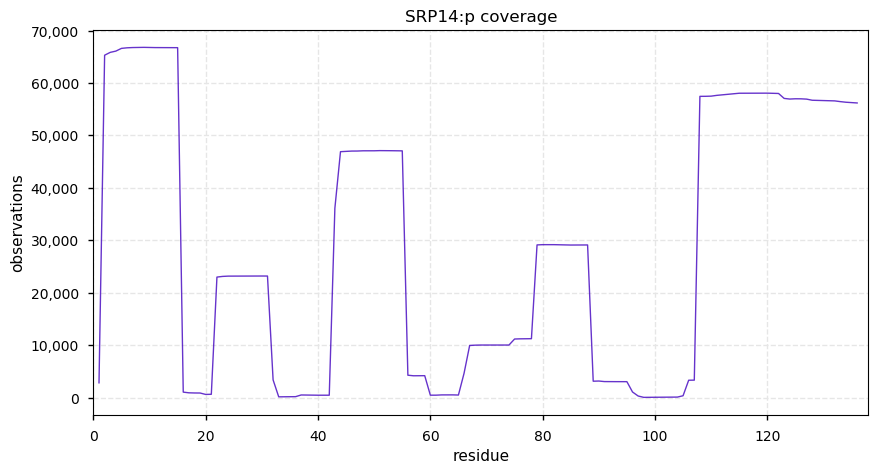
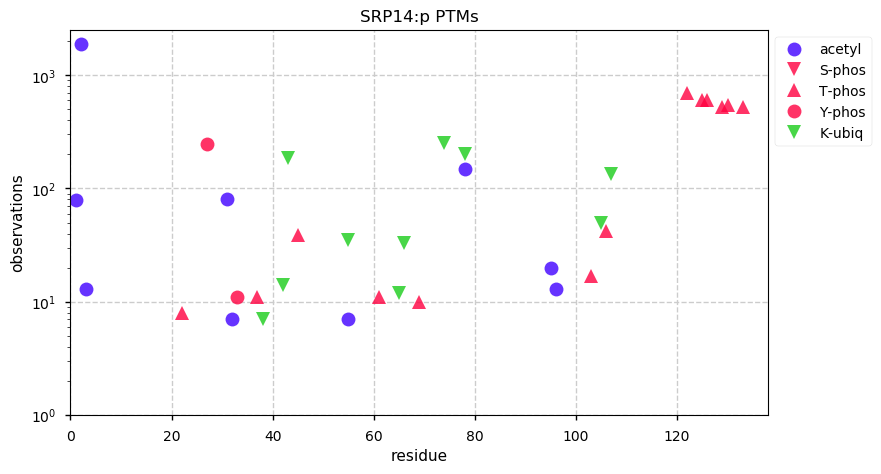
Sat Aug 17 16:55:01 +0000 2019It took a bit of doing to install my own searx instance but it does work 🤓 🔗
Sat Aug 17 14:30:04 +0000 2019It is still a little rough, but at least it avoids returning the highly repetitive database results
Sat Aug 17 14:16:15 +0000 2019Kind of falling in love with the searx meta-search platform. It does a nice job when you tell it to look for scientific results to a query, e.g. 🔗
Sat Aug 17 12:44:36 +0000 2019@slavovLab Platelets are a little smaller, but your point is well taken.
Sat Aug 17 12:41:47 +0000 2019SRP9:p, signal recognition particle 9 (H. sapiens) 🔗 Very small cytoplasmic subunit; highly acetylated; no SAVs; 1 splice variant; found in nucleated cells; mature form 2-86 [27,787 x] 🔗
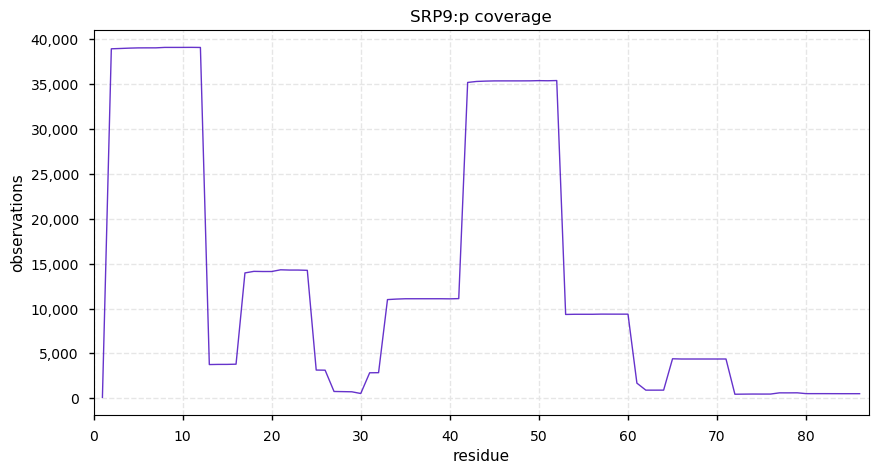
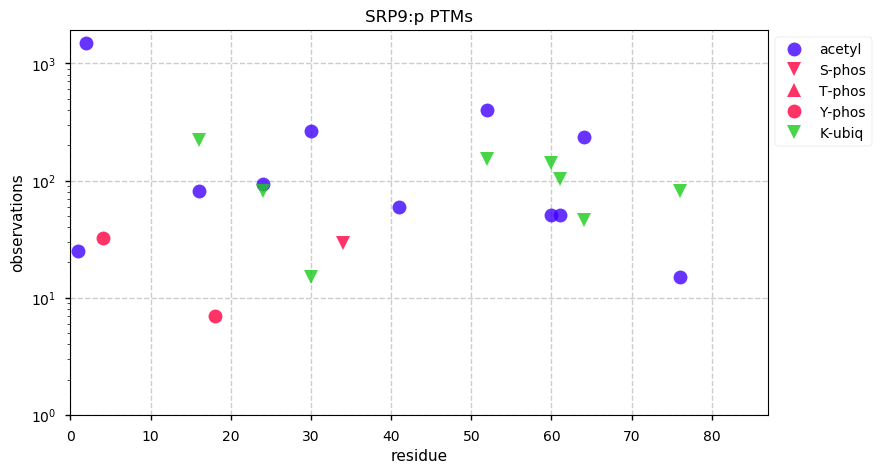
Fri Aug 16 21:32:38 +0000 2019@ZachRolfs You guys are right: there never really was any evidence for this idea, but it would have been kind of nifty if it was true.
Fri Aug 16 15:36:09 +0000 2019@ypriverol @Smith_Chem_Wisc @Sci_j_my @nesvilab @proteome_sw Do you mean that I should expand on the idea, or move the post to a different forum?
Fri Aug 16 15:32:09 +0000 2019@ypriverol @Smith_Chem_Wisc @Sci_j_my @nesvilab @proteome_sw My constructive comment is that some type of regular expression (motif) method of PTM site specification is necessary. Many PTMs benefit from motif specification, e.g., K+acetyl, K+(mono/di/tri)methyl, K+ubiquitin,hydroxy-Pro, hydroxy-Lys, R+(mono/di) methyl, etc.
Fri Aug 16 14:56:41 +0000 2019@nesvilab I think Liouville's Theorem is a better match to the situation (fixed volume for a solution in phase space). The uncertainty principle applies to any system with a wave-like representation, but I'm stumped as to how to represent DIA results as waves.
Fri Aug 16 14:30:49 +0000 2019@ypriverol @Smith_Chem_Wisc @Sci_j_my @nesvilab Try DM-ing @proteome_sw. They usually respond quite quickly.
Fri Aug 16 14:27:23 +0000 2019@ypriverol @Smith_Chem_Wisc @Sci_j_my @nesvilab And be sure get early buy-in from software vendors, e.g. Thermo, Bioinformatics Solutions, Proteome Software, Matrix Science, etc.
Fri Aug 16 14:17:46 +0000 2019@Smith_Chem_Wisc @ypriverol @Sci_j_my @nesvilab I am not discouraging you from having this discussion. However, part of the discussion should be how changing to a new, alternate way of representing these input parameters would genuinely benefit those who have to put in the non-trivial work of making this type of change.
Fri Aug 16 13:40:36 +0000 2019@Smith_Chem_Wisc @ypriverol @Sci_j_my @nesvilab There was a similar set of discussions back in the 2000's that came to a near consensus, but fell apart because nobody really had any reason to comply.
Fri Aug 16 13:36:21 +0000 2019@Smith_Chem_Wisc @ypriverol @Sci_j_my @nesvilab The discussion also illustrates why there hasn't been (and probably never will be) any agreement about how to represent this sort of thing. Too many equivalent representations & too many existing implementations that will resist making a change.
Fri Aug 16 12:17:45 +0000 2019SSR4:p, signal sequence receptor subunit 4 (H. sapiens) 🔗 Small intracellular protein; 3 Y-phosphorylation sites; no SAVs; 1 splice variant; 2 membrane spanning domains; mature form 24-173 [30,607 x] 🔗
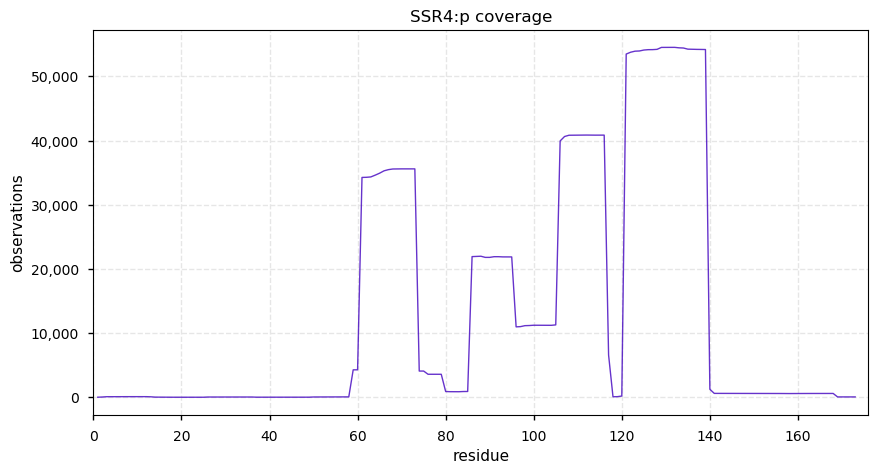
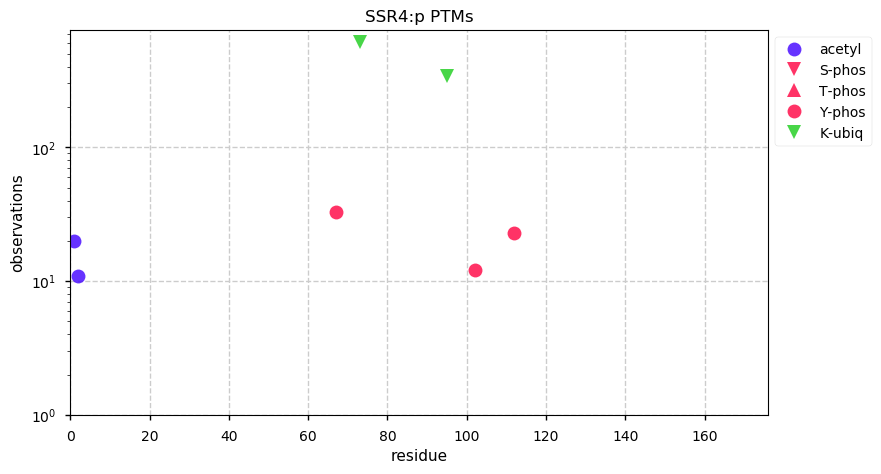
Thu Aug 15 12:23:22 +0000 2019SSR3:p, signal sequence receptor subunit 3 (H. sapiens) 🔗 Small intracellular protein; several PMTs; no SAVs; 1 splice variant; sparse coverage caused by low KR content; mature form 1,2,3-183 [20,261 x] 🔗
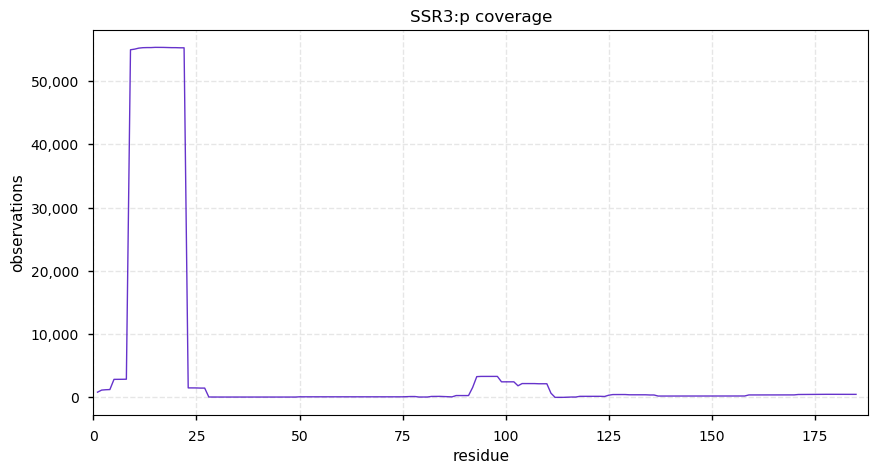
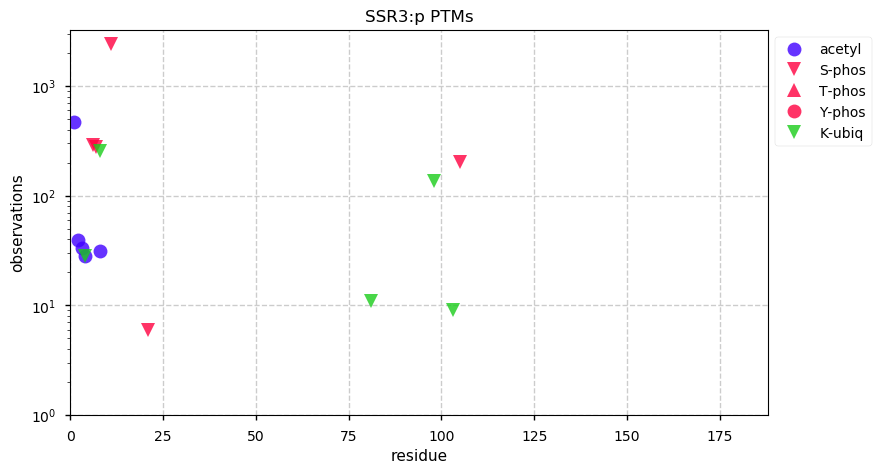
Wed Aug 14 13:05:28 +0000 2019SSR2:p, signal sequence receptor subunit 2 (H. sapiens) 🔗 Small intracellular glycoprotein; N88+glycosyl, N104+glycosyl; no SAVs; 1 splice variant; mature form 18-183 [1,636 x] 🔗
Tue Aug 13 12:48:23 +0000 2019SSR1:p, signal sequence receptor subunit 1 (H. sapiens) 🔗 Small intracellular protein; several PTM domains; 1 SAV: R6G (maf=0.02); commonly obs. in tissues, cell types and lines; evidence for 2 splice variants; mature form 33-286 [23,524 x] 🔗
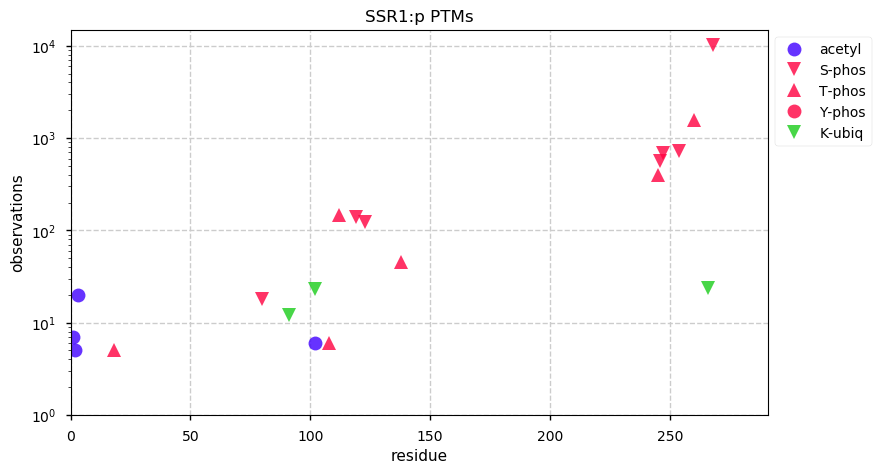
Mon Aug 12 14:57:01 +0000 2019The main 'insight' is reformulating the problem from using vector math to compare sequences to a spectrum, to using set math to generate intersections. The set approach benefits significantly from the efficient 'set' and 'dict' manipulation methods in Python.
Mon Aug 12 14:51:55 +0000 2019@theoneamit Risky claim on something 2 days old, but about 45 secs.
Mon Aug 12 14:06:50 +0000 2019@theoneamit For 60,000 spectra against Uniprot human, it took 4 GB. I can probably reduce the memory requirements a bit (at least manage it a bit better), as the code isn't very mature.
Mon Aug 12 12:51:07 +0000 2019PHB:p and PHB2:p are normally detected together, producing similar MS/MS signal amplitudes in cells and tissues. They do not share any tryptic peptides, though.
Mon Aug 12 12:45:10 +0000 2019PHB2:p, prohibitin 2 (H sapiens) 🔗 Small intracellular protein; extensive PTMs; no SAVs; commonly obs. in all tissues, cell types and lines; evidence for 2 splice variants; mature form 2-299 [47,140 x]
Sun Aug 11 18:49:05 +0000 20193/3 The prototype is at 🔗
🔗 - does the search;
🔗 - generates an output file;
🔗 - deals with spectrum file formats.
Sun Aug 11 18:47:59 +0000 20192/3 So I wrote a prototype in Python3, where the peptide sequence loading and PSM assignment takes about 170 code lines. Uses plenty of memory, but it cranks out ids in < 50 microseconds per spectrum.
Sun Aug 11 18:47:12 +0000 20191/3 It occurred to me yesterday that I could considerably simplify a PSM search engine by getting rid of almost all of the math and just use Python dicts and sets to do everything.
Sun Aug 11 13:36:18 +0000 2019PHB:p has a strong correlation between acetyl and ubiquitinyl acceptor sites
Sun Aug 11 12:45:44 +0000 2019PHB:p, prohibitin (H. sapiens) 🔗 Small intracellular protein; extensive PTMs; no SAVs; commonly obs. in all tissues, cell types and lines; 1 splice variant; mature form 1,2-272 [47,140 x] 🔗
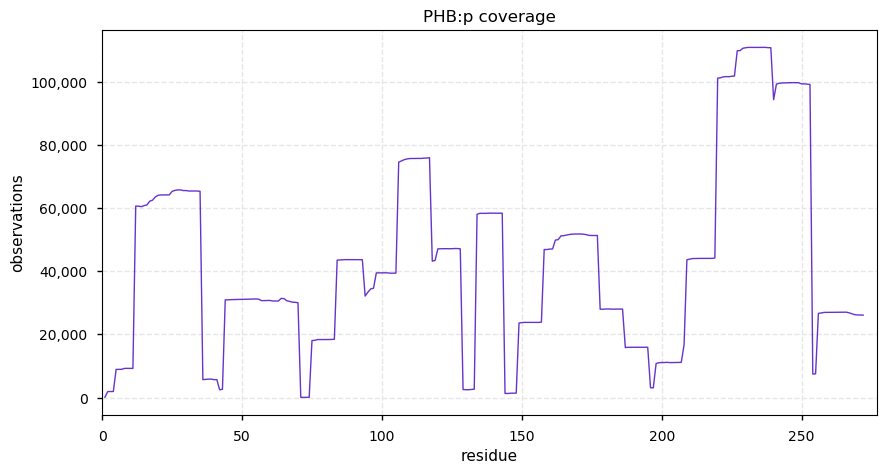
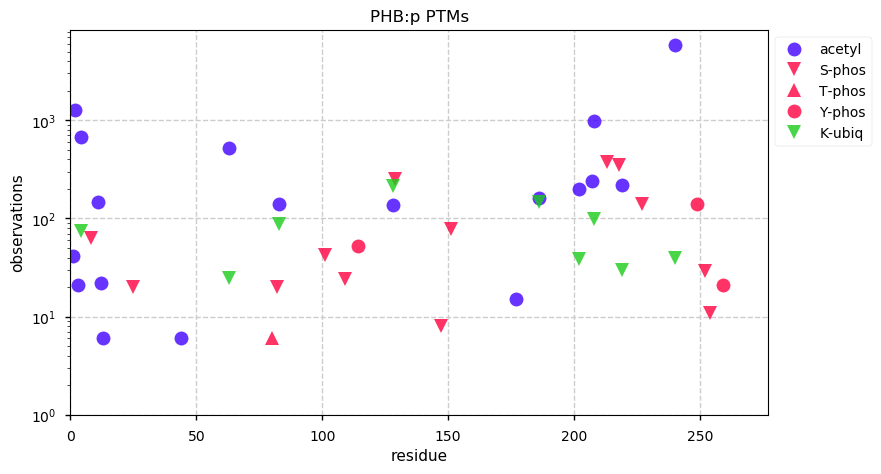
Sat Aug 10 13:02:45 +0000 2019CLEC14A:p, C-type lectin domain family 14 member A (H sapiens) 🔗 Small membrane glycoprotein; C-terminal S-phosphodomain; no SAVs; obs. in endothelium and endothelium-derived cell lines + urine; 1 splice variant; mature form 22-490 [1,635 x] 🔗
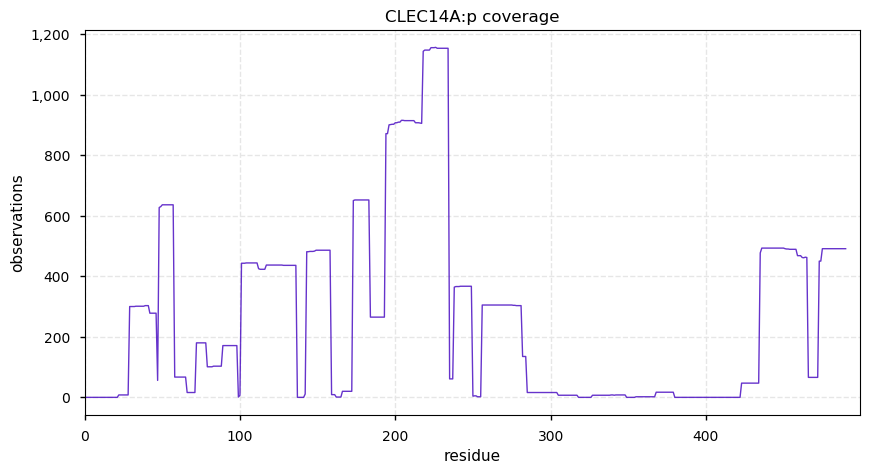
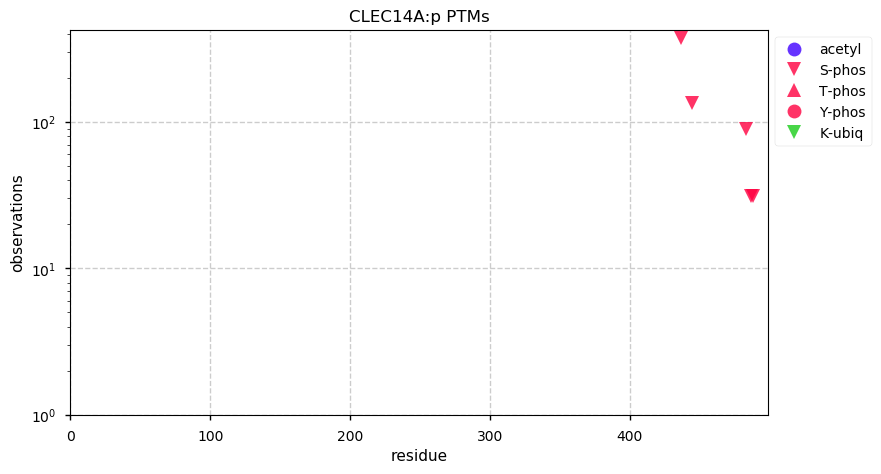
Fri Aug 09 17:46:36 +0000 2019It doesn't help much that 🔗 is hosted in Chicago, 🔗 is hosted in Dallas, 🔗 in Montreal and the 🔗 in Northern Virginia. #mbpoli
Fri Aug 09 17:41:18 +0000 2019Is there some sort of contest for "Worst Web Site for a Political Party" being waged in Manitoba that no one has mentioned? I just looked though the NDP, PC, Liberal and Green sites and they are just plain awful. #mbpoli
Fri Aug 09 14:41:09 +0000 2019The optical illusions generated by balls-on-strings demos (🔗) strike me as a strong argument against trying to use visual representations of complex data to search for meaningful interactions between components
Fri Aug 09 12:05:30 +0000 2019CLEC7A:p, C-type lectin domain family 7 member A (H sapiens) 🔗 Small membrane glycoprotein; S22+phosphoryl and S24+phosphoryl; no SAVs; obs. in monocytes & truncated form in urine; 1 splice variant; mature form 1-247 [227 x] 🔗
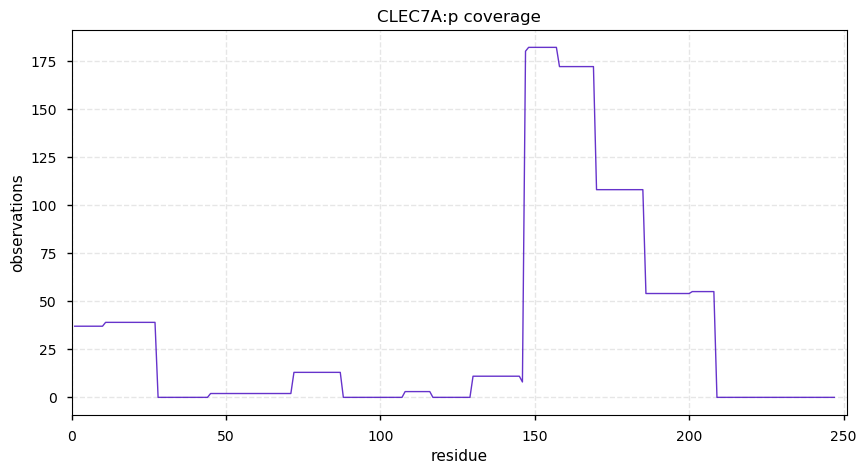
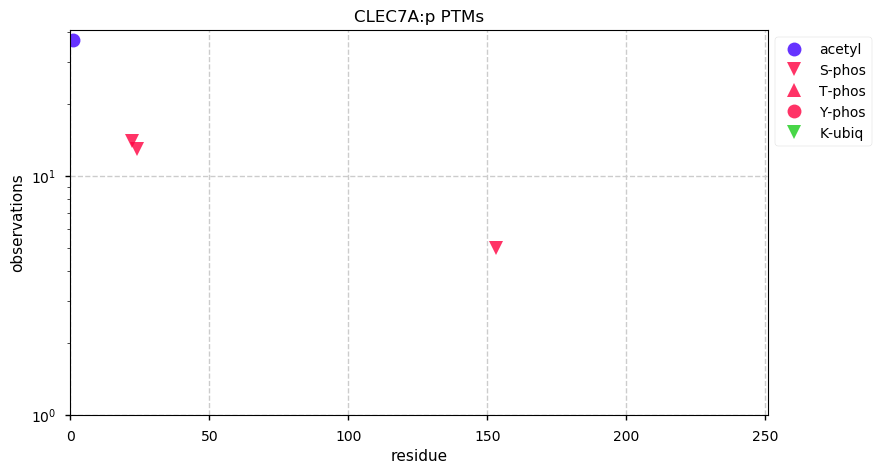
Thu Aug 08 17:17:01 +0000 2019@AlexUsherHESA Do you think this change in the proportion of funding will have any impact on the way Boards of Governors are appointed for the Unis where the province selects the majority of Governors (e.g., U of M, U of S, UBC)
Thu Aug 08 15:32:45 +0000 2019@KentsisResearch The study does have a bit of an issue, though, as many of the urine samples also show high levels of UTI bacterial proteins, particularly those from Pseudomonas aeruginosa.
Thu Aug 08 15:29:47 +0000 2019@KentsisResearch You may want to check out the data from Zhang C, et al (🔗, 🔗). They also found significant amounts of PHB in urine associated with tumor patients, although they did not draw any inferences from this particular information.
Thu Aug 08 13:03:07 +0000 2019ASGR2:p, asialoglycoprotein receptor 2 (H sapiens) 🔗 Small membrane glycoprotein (aka CLEC4H2:p); A2+acetyl, S12+phosphoryl, N102+glycosyl; no SAVs; mainly obs. in liver, blood, urine and hepatocyte cell lines; 1 splice variant; mature form 2-311 [1,517 x] 🔗
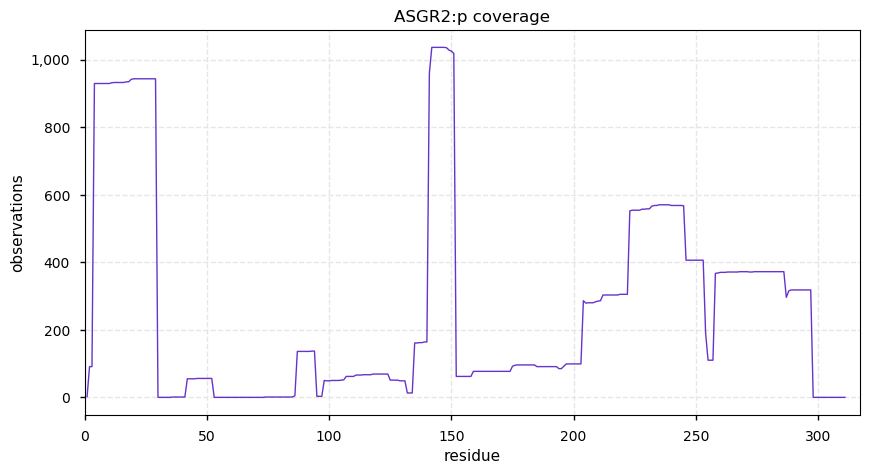
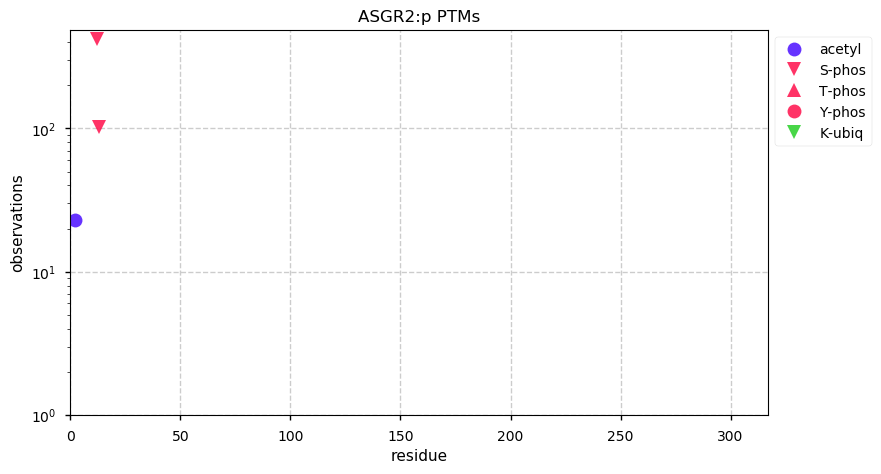
Wed Aug 07 17:55:20 +0000 2019Yikes! 🔗
Wed Aug 07 17:11:55 +0000 2019@UCDProteomics But this way way back in olden times (80's). The "N-terminus" and "C-terminus" notation isn't MS-based: it is based on even older-fashioned protein biochemistry notation.
Wed Aug 07 17:06:19 +0000 2019@UCDProteomics There were Y & B ions (🔗), but they included additional notation for the addition of hydrogens and protons to the backbone cleavage fragments. Lower case y & b were proposed by Klaus Biemann to simplify notation for the dominant forms of Y & B ions.
Wed Aug 07 16:23:58 +0000 2019@idpgrace I now will always bail immediately when a captcha appears.
Wed Aug 07 12:57:34 +0000 2019ASGR1:p, asialoglycoprotein receptor 1 (H sapiens) 🔗 Small membrane glycoprotein (aka CLEC4H1:p); T2+acetyl & T2+phosphoryl; no SAVs; mainly obs. in liver, urine and hepatocyte-derived cell lines; 1 splice variant; mature form 2-291 [1,228 x] 🔗
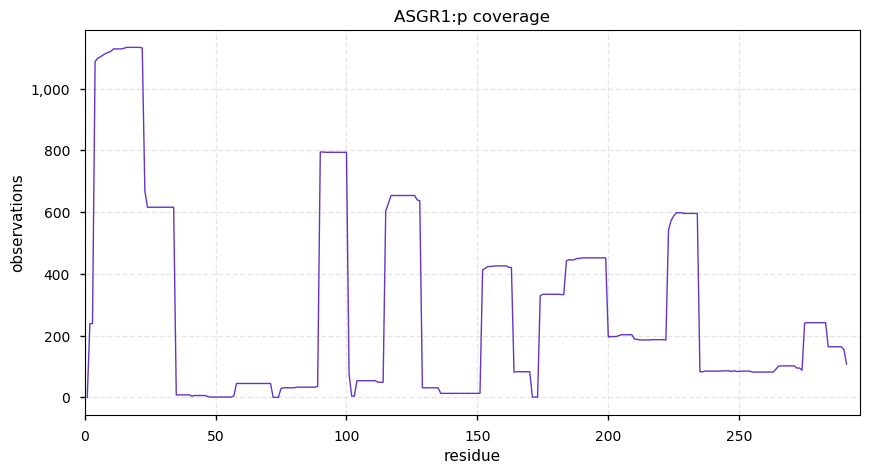
Wed Aug 07 12:31:38 +0000 2019@Sci_j_my They probably just went looking for something and they "found" it. If you aren't careful, looking at large scale data for odd things can lead even the best of us towards some very seductive mirages.
Wed Aug 07 03:08:53 +0000 2019@Sci_j_my It is derived from T-lymphocytes. None of the cell lines they used produce glucagon, but they say they found the same phosphopeptide in all of them.
Wed Aug 07 01:20:37 +0000 2019@slavovLab It is what the kids call "throwing shade".
Wed Aug 07 01:17:12 +0000 2019@slavovLab It is how people describe all proteins they are not personally interested in at the moment. It's use is analogous to how "junk" DNA (or PTMs) is used.
Tue Aug 06 19:22:11 +0000 2019PTM site observation frequency for human LINE-1 ORF1 (no special reason, just struck me as an interesting thing to check) 🔗
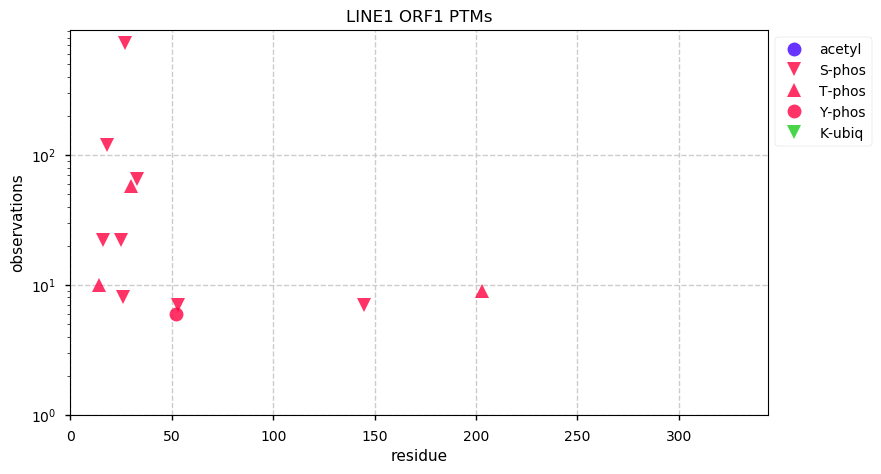
Tue Aug 06 18:13:06 +0000 2019@GNPS_UCSD @theoneamit Boy, I thought many of my tweets were obscure, inside-baseball geekery: @GNPS_UCSD takes it to a whole 'nother level. Hats off ...
Tue Aug 06 16:09:51 +0000 2019@Sci_j_my So I'm not sure what these folks found, but it isn't a GCG-derived peptide in a JURKAT cell sample.
Tue Aug 06 16:08:04 +0000 2019@Sci_j_my PG isn't the current gene name for glucagon, it is GCG. JURKAT cells don't have detectable levels of GCG. It is made in the pancreas & some other gut tissues. It is not detectable in common cell lines with a few exceptions, e.g. TOV-112D.
Tue Aug 06 15:20:14 +0000 2019@RL_Bearden ASMS is an unusually well run conference. If you use it as your conference-comparator, you will be often disappointed by other meetings.
Tue Aug 06 13:28:14 +0000 2019CLEC4G:p, C-type lectin domain family 4 member G (H sapiens) 🔗 Small membrane protein; 1 PTM: M1+acetyl; no SAVs; only obs. in liver, urine and dendritic cell EVs; 1 splice variant; mature forms 1-293 [815 x] 🔗
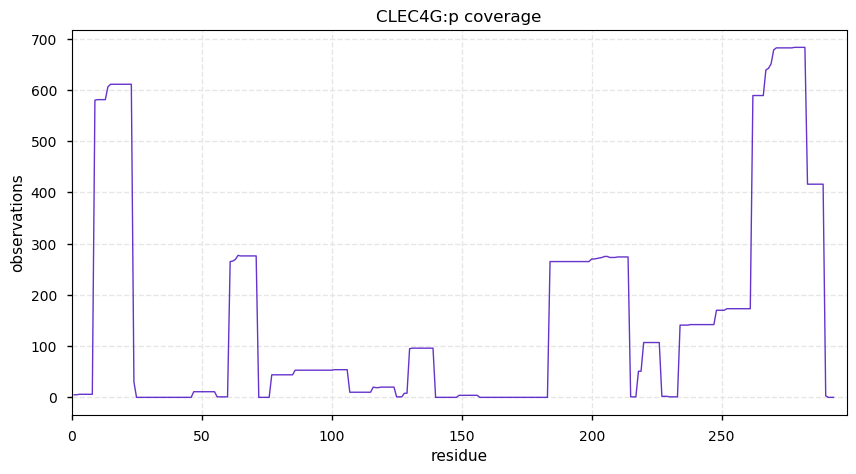
Mon Aug 05 16:28:07 +0000 2019It should be noted that this issue can be easily fixed by using technical replicates to discriminate against stochastic matching.
Mon Aug 05 13:11:50 +0000 2019CLEC4E:p, C-type lectin domain family 4 member E (H. sapiens) 🔗 Small membrane protein; N62+glycosylation; no SAVs; only detectable in monocytes, not obs. in common cell lines; 1 splice variant; mature form 1-219 [105 x] 🔗
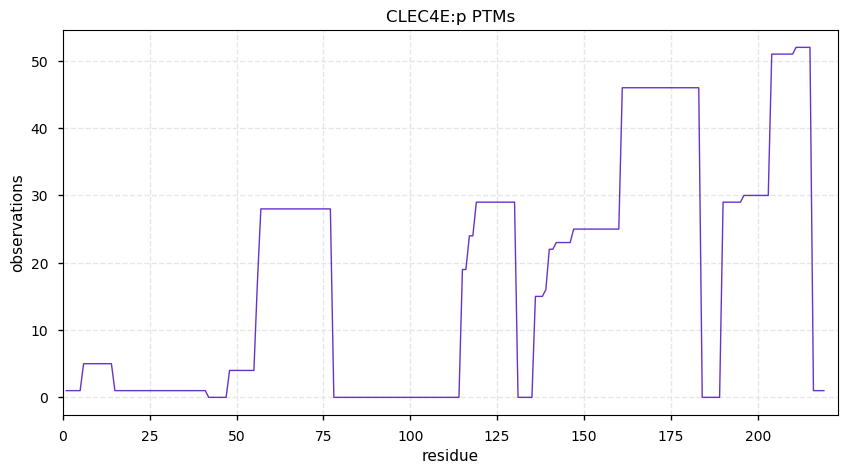
Sun Aug 04 22:12:31 +0000 2019@lukas_k Identification
Sun Aug 04 16:45:07 +0000 2019If the differential analysis found 50 proteins, then the minimum FPR (%) would be:
((10+10)/50)×100 = 40%
Sun Aug 04 16:43:07 +0000 2019You can use the individual FPRs to estimate a minimum FPR for the differential analysis. If there were 1000 proteins id'd in each run, with a 1% FPR the number of false positives in each run is ~10.
Sun Aug 04 16:26:06 +0000 2019Thank you to everyone who participated in this poll. The majority position was that it is not possible to know the FPR of this experiment's results without further investigation: simply knowing the individual runs' FPRs is insufficient.
Sun Aug 04 13:42:04 +0000 2019CLEC4A:p, C-type lectin domain family 4 member A (H. sapiens) 🔗 Small membrane protein; T2+acetyl & N185 glycosylation; no SAVs; only detectable in monocytes and dendritic cells; 1 splice variant; mature forms 2-237 [202 x] 🔗
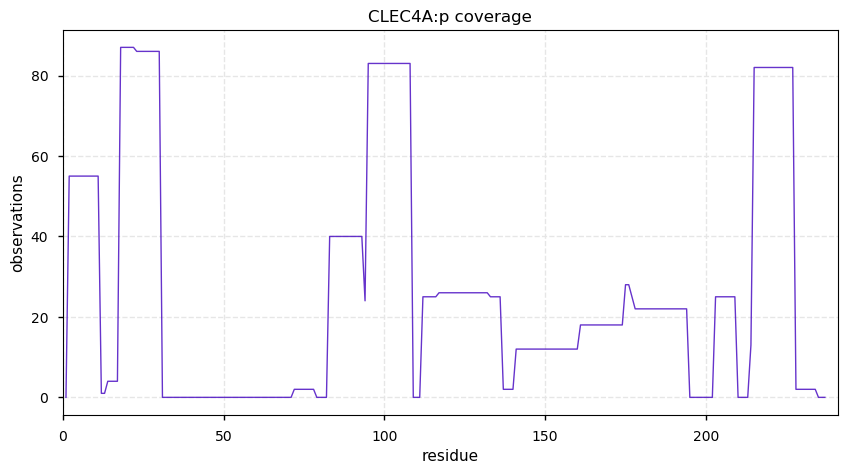
Sat Aug 03 15:52:24 +0000 2019You perform a proteomics experiment involving a differential analysis of 2 samples. The proteins in each sample are selected to have an FPR of 1%. The FPR of the protein subset found by taking the difference between the 2 samples is:
Sat Aug 03 12:38:18 +0000 2019CLEC3B:p, C-type lectin domain family 3 member B (H. sapiens) 🔗 Small glycoprotein; broadly distributed: present in many cell lines and tissues; 1 SAV: G106S (maf=0.44); 1 splice variant; mature forms 22-202 [14,242 x] 🔗
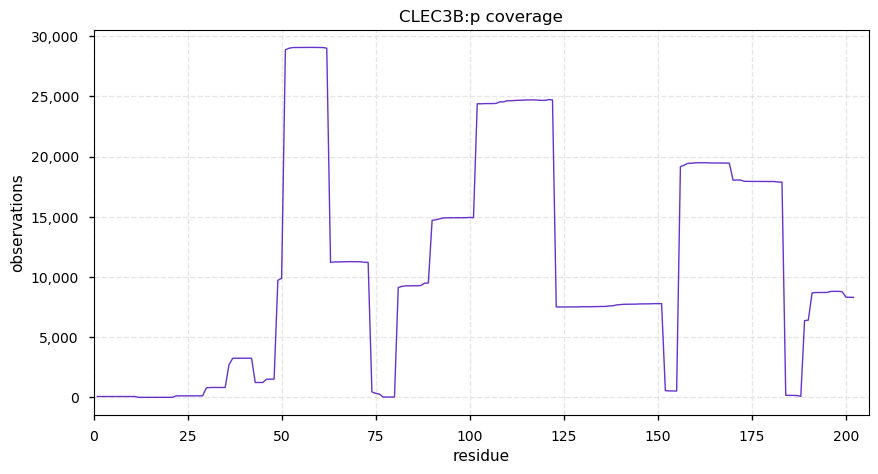
Fri Aug 02 16:50:41 +0000 2019@astacus Apparently. I'm not in love with the term myself, but since it seems to be part of the general vernacular for the field, its meaning can be in the eye of the beholder.
Fri Aug 02 14:19:01 +0000 2019Thanks to every who participated. The results are a pretty clear majority: 67% of the community find "proteoform" to be a useful term.
Fri Aug 02 12:39:40 +0000 2019@slavovLab @CanadaAir @AirCanada Most government employees must fly with them and because of limited competition, many Canadians have no choice but to use them.
Fri Aug 02 12:29:48 +0000 2019CLEC3A:p, C-type lectin domain family 3 member A (H sapiens) 🔗 Small extracellular protein; no PTMs; no SAVs; only obs. in breast tumor samples, not present in cell lines; 1 obs. splice variant; hypothetical mature form 22-206 [508 x] 🔗
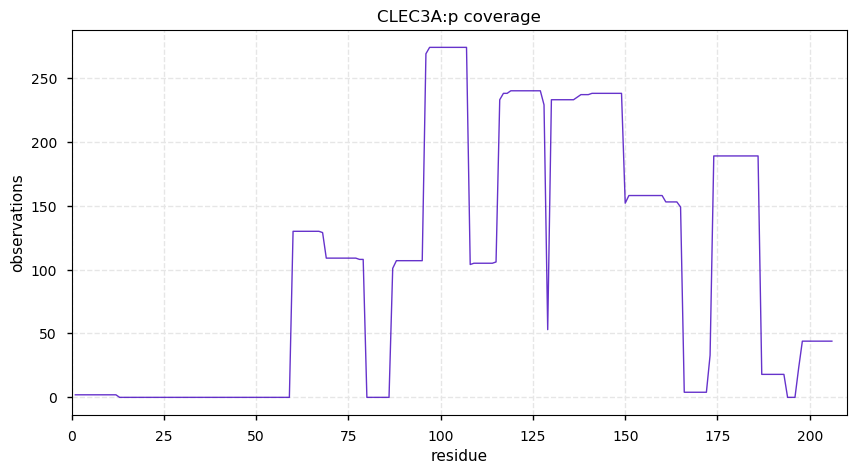
Fri Aug 02 02:20:36 +0000 2019@slavovLab @CanadaAir Maybe you mean @AirCanada? If so, I stopped using them 5 years ago.
Thu Aug 01 20:08:52 +0000 2019After 6 hours, it looks as though "proteoform" has value to the majority. Still 18 hours to go, though.
Thu Aug 01 14:57:14 +0000 2019@EmmMacfarlane The entry for the US is hilarious 🔗 Pretty much defines the term "smug": it recommends watching "Survivor" & "Bowling for Columbine" to understand the culture
Thu Aug 01 14:12:34 +0000 2019The term "proteoform" has become fairly common in the proteomics literature (see 🔗). Do you find it to be a useful term to use when describing your own research?
Thu Aug 01 12:21:12 +0000 2019CLEC1B:p, C-type lectin domain family 1 member B (H sapiens) 🔗 Small glycoprotein; N-terminal acetyl; 1 SAV: G64D (maf=0.98); best MHC I peptide: 113-121; only obs. in megakaryocytes, platelets & monocytes; 1 splice variants; mature form 1-229 [606 x] 🔗
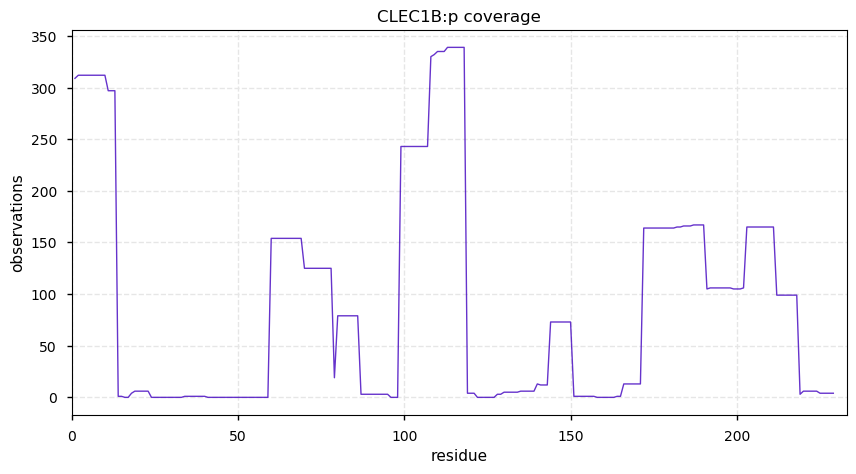
Wed Jul 31 18:06:51 +0000 2019@MHendr1cks It would be better to phase out CRCs rather than trying to fix the program's many issues.
Wed Jul 31 15:39:27 +0000 2019I'm going to spend the next few weeks highlighting some of the members in the 17 groups of C-type lectins (carbohydrate binding proteins)
Wed Jul 31 12:28:07 +0000 2019CHODL:p, chondrolectin (H. sapiens) 🔗 Small protein; no PTMs; no SAVs; mainly obs. in urine and testis, not present in cell lines; 2 obs. splice variants; mature form 22-273 [293 x] 🔗
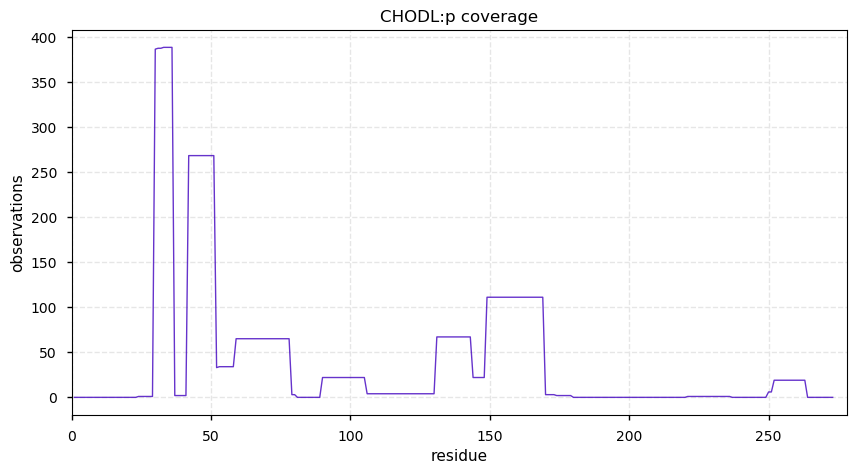
Tue Jul 30 12:40:24 +0000 2019SHROOM4:p, shroom family member 4 (H. sapiens) 🔗 Large intracellular phosphoprotein; many S phosphorylations; 1 high maf SAVs, A833T (0.02); not obs. in many cell lines, except those of endothelial origin; 1 splice variant; mature form 1-1493 [2,126 x] 🔗
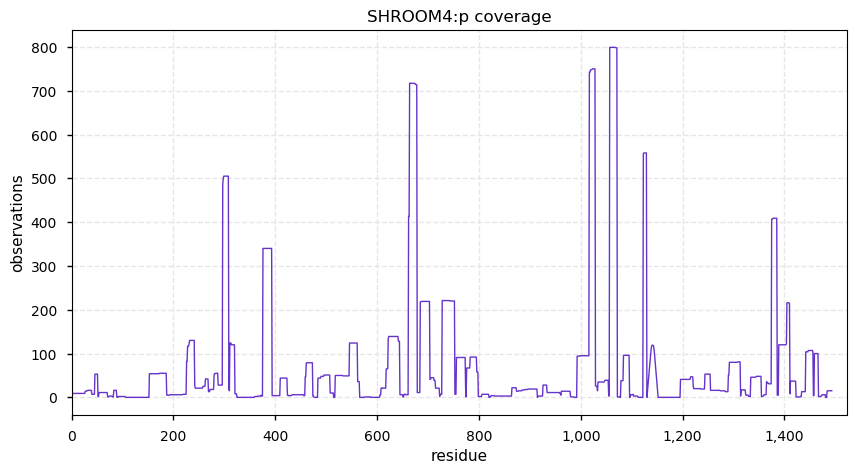
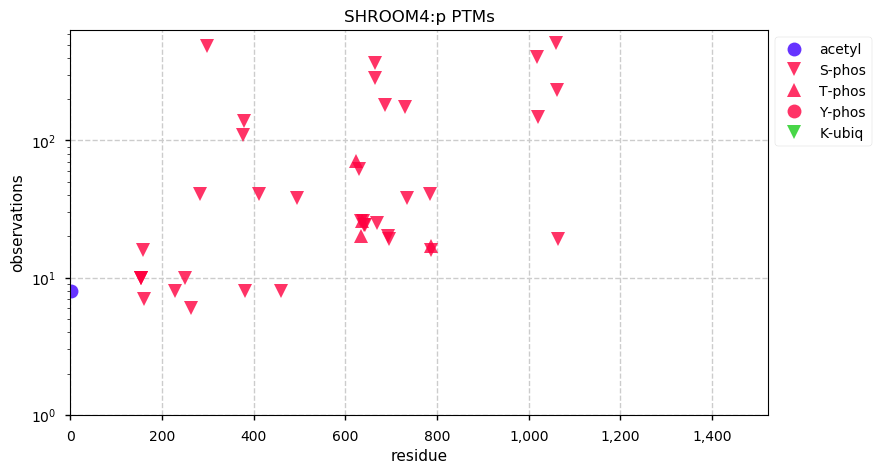
Mon Jul 29 21:23:11 +0000 2019@doctorow Looks very much like a mule rather than a donkey ...
Mon Jul 29 14:31:36 +0000 2019@idpgrace It seems to depend on the institution. At some places I've worked, it was either done by the student or the PI, in others it was unheard of.
Mon Jul 29 12:36:29 +0000 2019SHROOM3:p, shroom family member 3 (H. sapiens) 🔗 Large intracellular phosphoprotein; many S/T phosphorylations; 5 high maf SAVs, e.g., P469A (0.44) and P1290L (0.47); 1 splice variant; mature form 1-1996 [7,500 x] 🔗
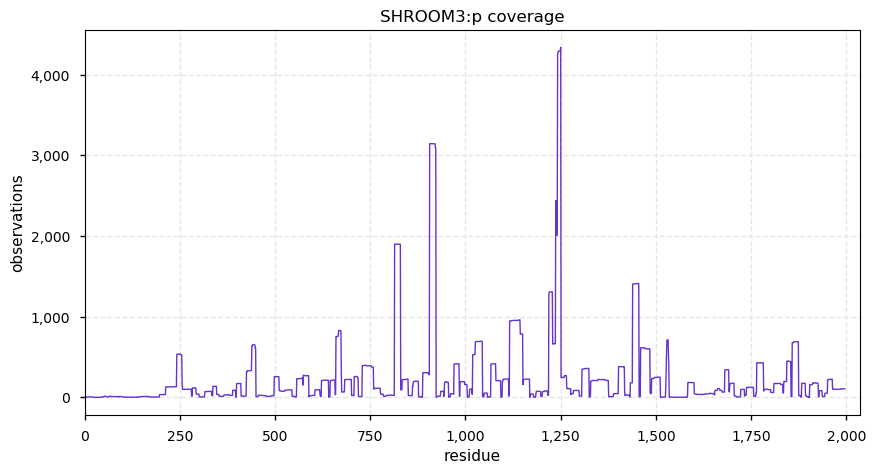
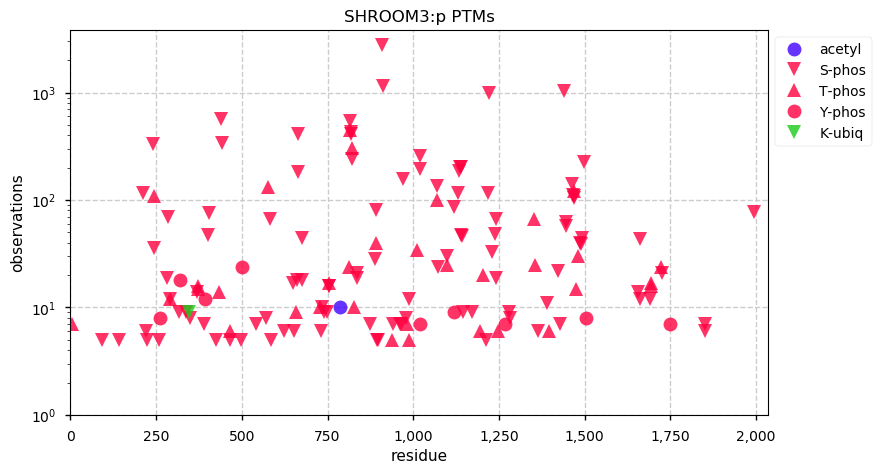
Sun Jul 28 13:57:46 +0000 2019SHROOM2:p, shroom family member 2 (H. sapiens) 🔗 Large intracellular phosphoprotein; many PTMs; 7 high maf SAVs, e.g., L1607F (0.46) and I1475V (0.15); 1 splice variant; mature form 1-1616 [4,353 x] 🔗
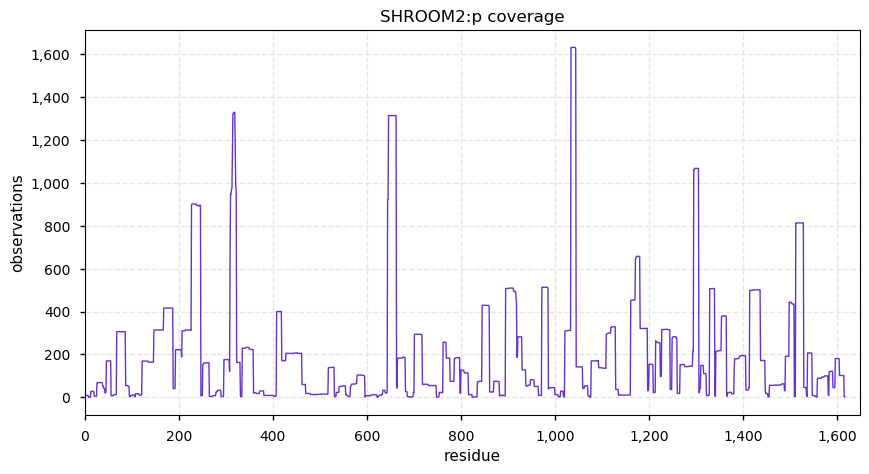
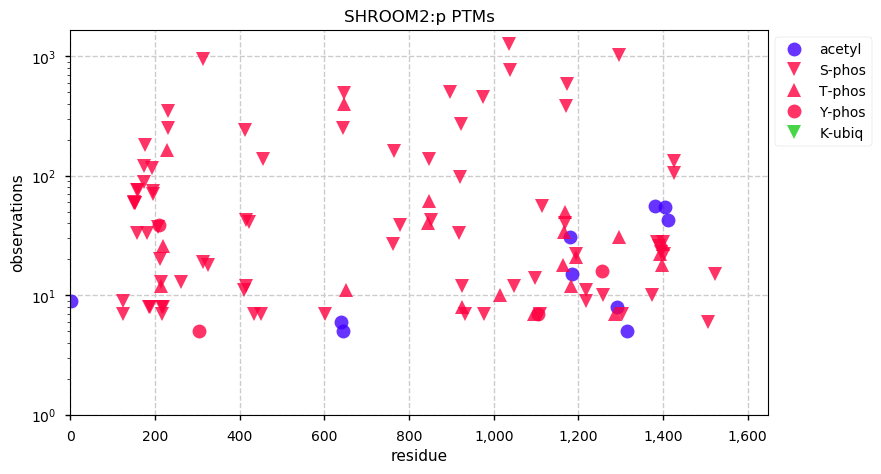
Sat Jul 27 13:54:47 +0000 2019Another protein where the observed tissue distribution does not make a lot of sense wrt the proposed function
Sat Jul 27 13:13:26 +0000 2019SHROOM1:p, shroom family member 1 (H sapiens) 🔗 Midsized cellular protein; 2 distinct phospho-domains; no high maf SAVs; 1 splice variant; 430-526 has no observable tryptic peptides; mature form 1-852 [3,304 x] 🔗
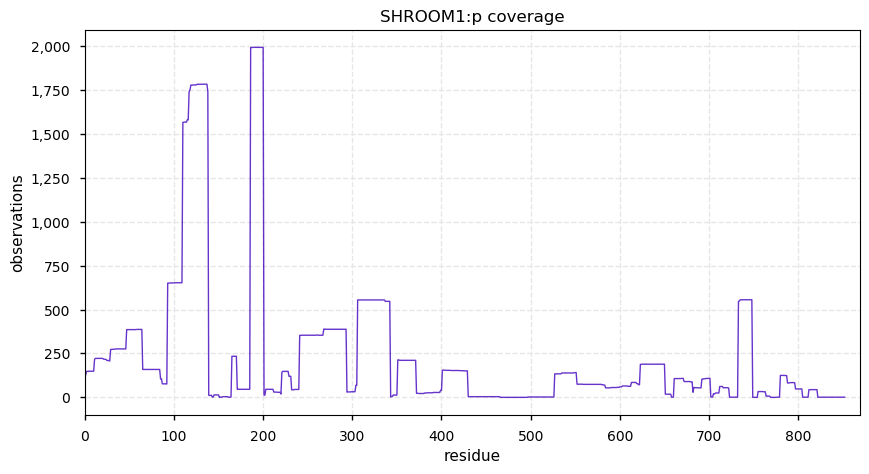
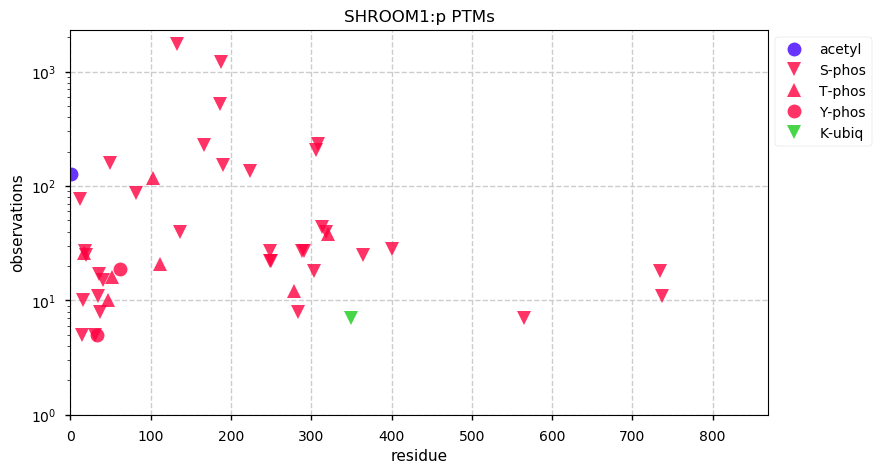
Fri Jul 26 12:21:16 +0000 2019QPCTL:p, glutaminyl-peptide cyclotransferase like (H. sapiens) 🔗 Small cellular protein; no PTMs; no high maf SAVs; 1 splice variant; no tryptic peptide overlap with QPCT; mature form 1-382 [4,765 x] 🔗
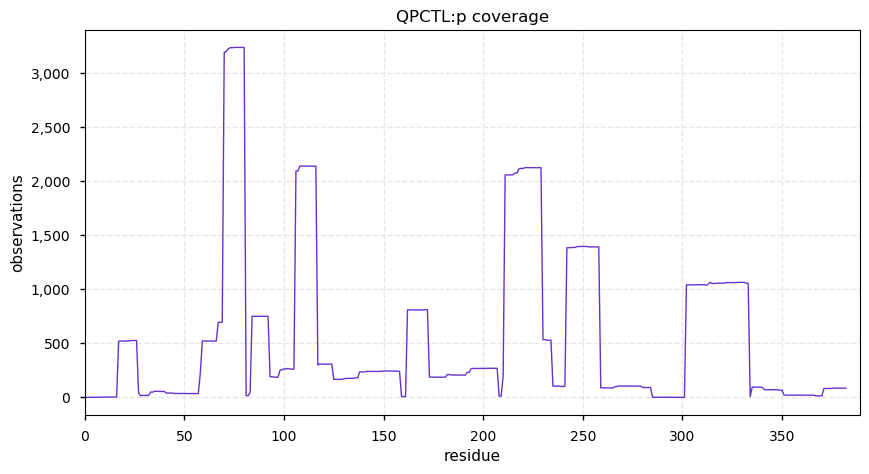
Thu Jul 25 15:14:59 +0000 2019@theoneamit @pwilmarth Computers may not be "accurate", but they are really good at being reproducible. I like to add in some randomness to calculations during testing to see if a particular implementation is stable to small unpredictable changes, but this sounds like something very different.
Thu Jul 25 14:09:53 +0000 2019The more you look at the information available about this protein, the less sense it seems to make.
Thu Jul 25 12:56:58 +0000 2019QPCT:p, glutaminyl-peptide cyclotransferase (H. sapiens) 🔗 Small extracellular protein; no PTMs; no high maf SAVs; 1 splice variant; mature form 29-361 [4,961 x] 🔗
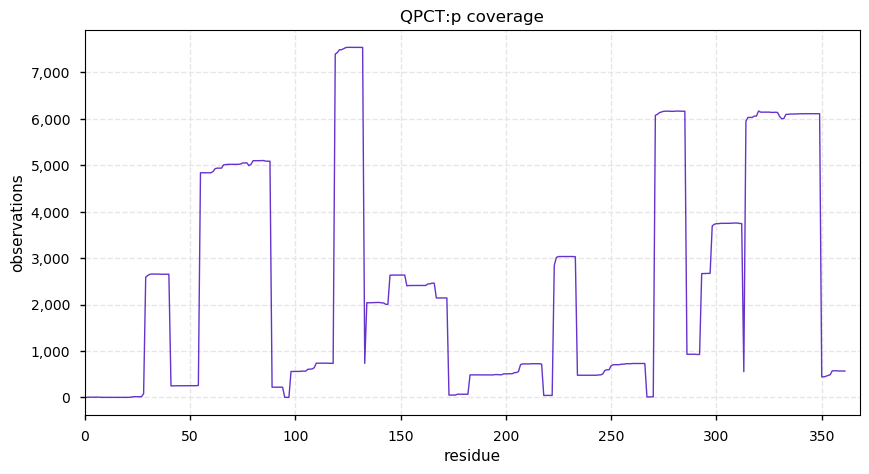
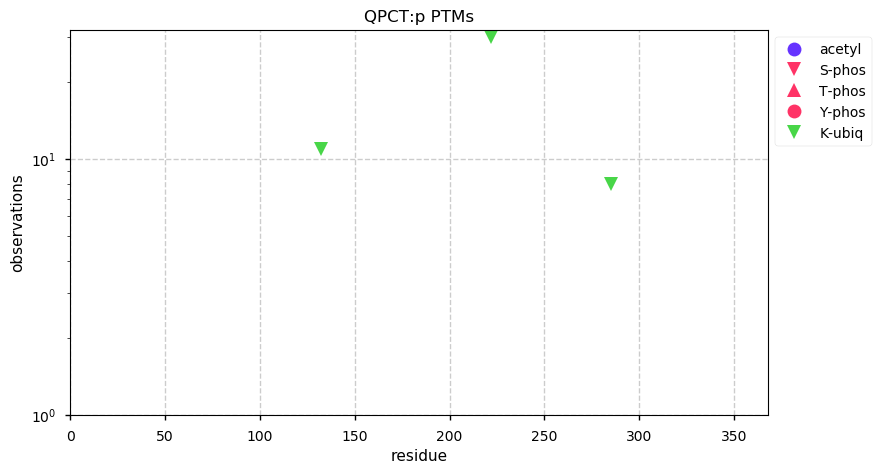
Wed Jul 24 12:55:31 +0000 2019STIL:p, SCL/TAL1 interrupting locus (H. sapiens) 🔗 Large cytoskeletal protein; multiple phosphodomains; no high maf SAVs; 1 splice variant; mature form 1-1287 [1,555 x] 🔗
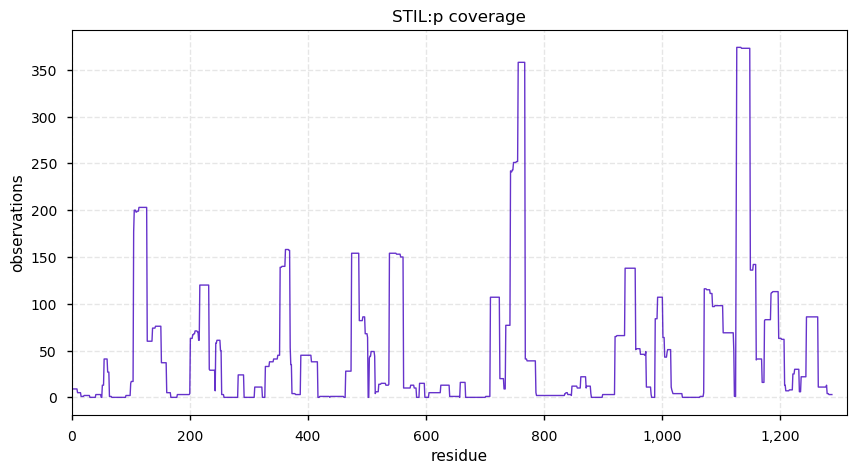
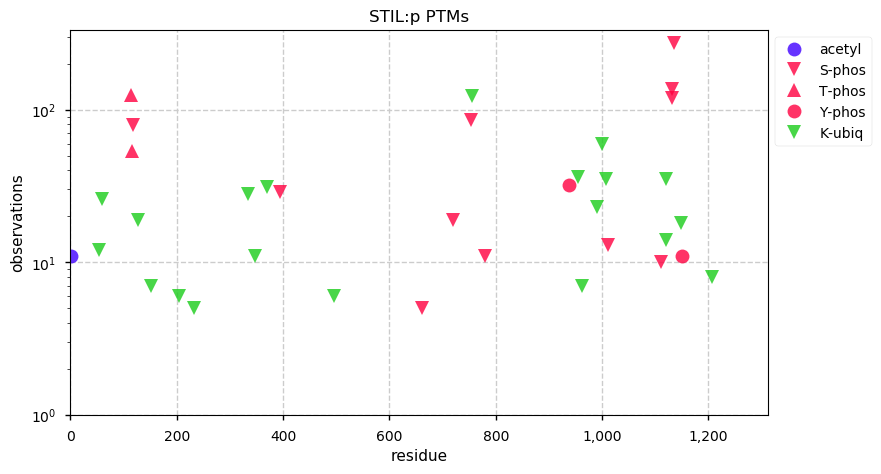
Tue Jul 23 21:40:44 +0000 2019When someone does a really good job on MHC class 2 peptides (e.g., PXD013599), the results are fascinating.
Tue Jul 23 18:20:56 +0000 2019@UCDProteomics @briansearle You could try using "tandem mass tag" for PubMed searching: it is much more specific for proteomics work.
Tue Jul 23 15:13:53 +0000 2019@chrashwood After reading the paper and looking at the supp. material, I'm sure that they are reporting this effect, but I am not sure that it is significant.
Tue Jul 23 12:26:25 +0000 2019SPICE1:p, spindle and centriole associated protein 1 (H. sapiens) 🔗 Midsized cytoskeletal protein; significant phosphorylation; no high maf SAVs; 1 splice variant; not obs in spermatozoa; mature form 2-855 [3,387 x] 🔗
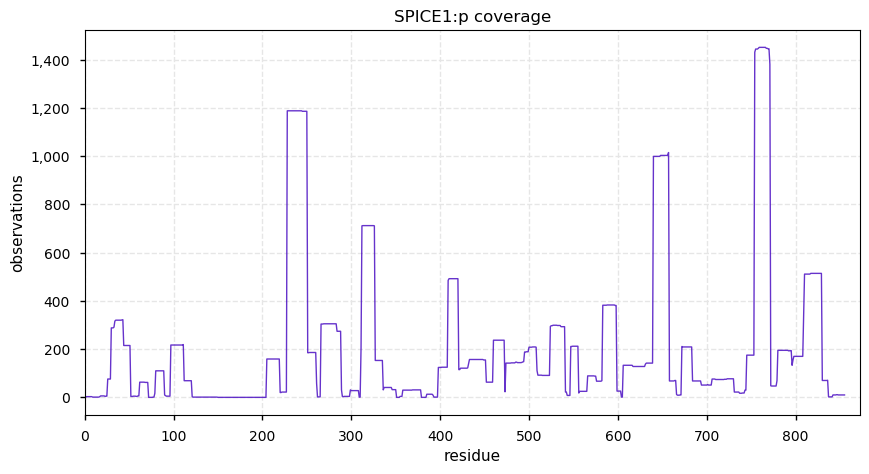
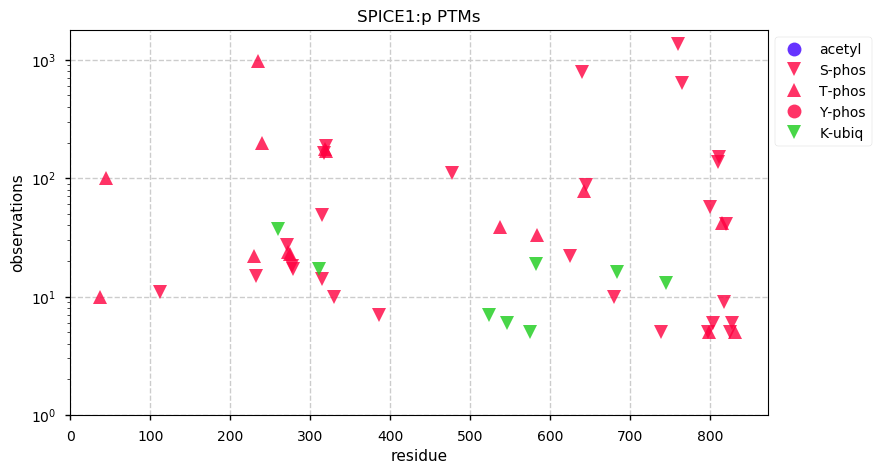
Mon Jul 22 17:45:45 +0000 2019🔗
Mon Jul 22 12:26:06 +0000 2019SPATC1:p, spermatogenesis and centriole associated 1 (Homo sapiens) 🔗 Midsized cytoskeletal protein; no obs. PTMs except N-terminal acetylation; no obs. SAVs; only obs. in sperm-containing tissue; 1 splice variant; mature form 2-597 [190 x] 🔗
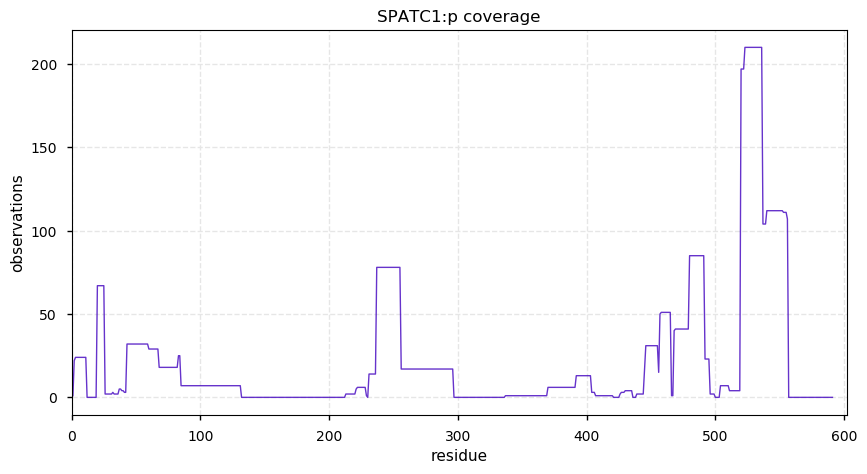
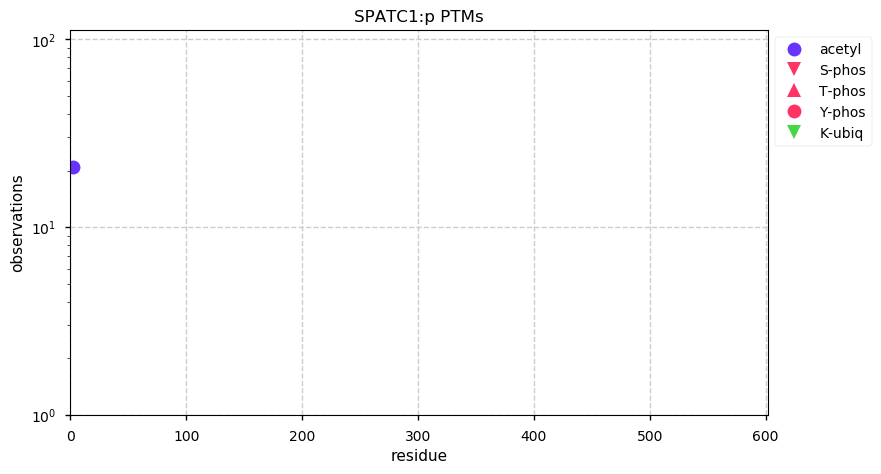
Sun Jul 21 17:38:22 +0000 2019At some point in a scientific career, one comes to the inescapable conclusion that unattributed reinvention is inevitable, so you might as well consider it a flattering demonstration of your own perspicacity (rather than grousing about it). 😀
Sun Jul 21 14:16:04 +0000 2019SASS6:p, SAS-6 centriolar assembly protein (H. sapiens) 🔗 Midsized cytoskeletal protein; several phosphodomains; no obs. SAVs; 1 splice variant; mature form 2-657 [2,718 x] 🔗
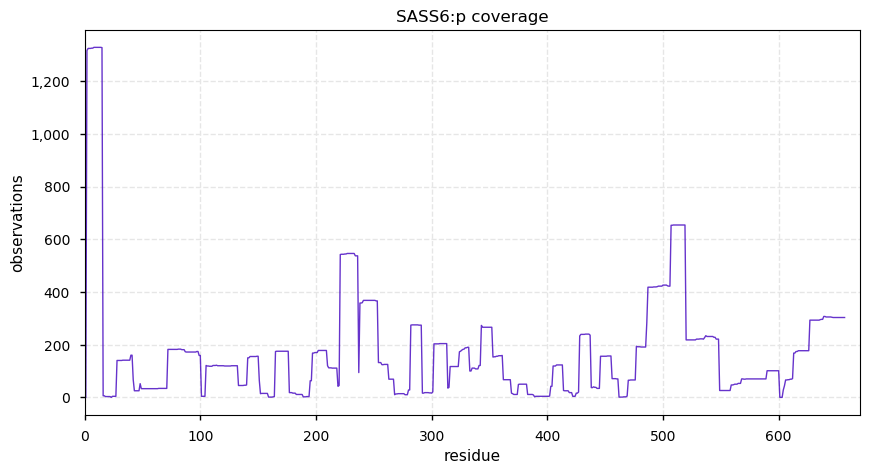
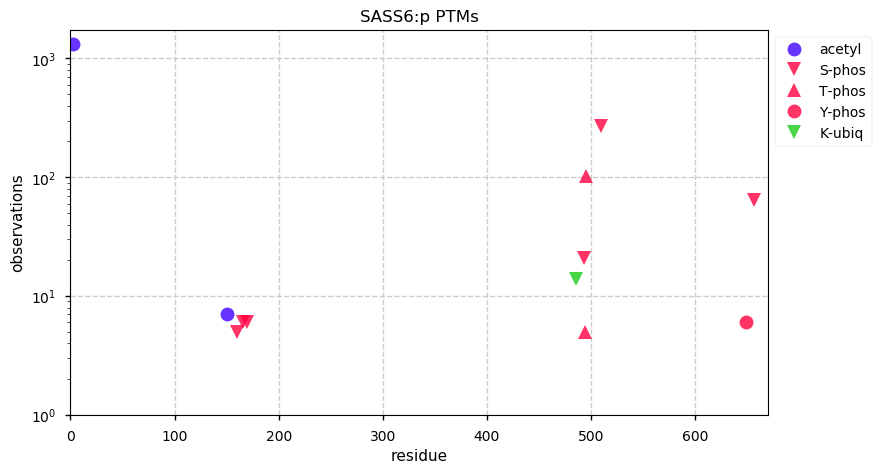
Sat Jul 20 17:48:52 +0000 2019@AlexUsherHESA Do you really want to hand the current president the opportunity to resurrect "54-40 or fight"?
Sat Jul 20 17:18:04 +0000 2019@astacus It is interesting, particular since it seems to be such a contentious issue in biomedical research. In physics and chemistry, author order often doesn't imply much of anything.
Sat Jul 20 12:35:57 +0000 2019POC5:p, POC5 centriolar protein (H. sapiens) 🔗 Midsized cytoskeletal protein; 2 distinct phosphodomains; 2 SAVs: H36R maf=0.4, I85T maf=0.2; 1 obs. splice variant; mature form 2-575 [1,439 x] 🔗
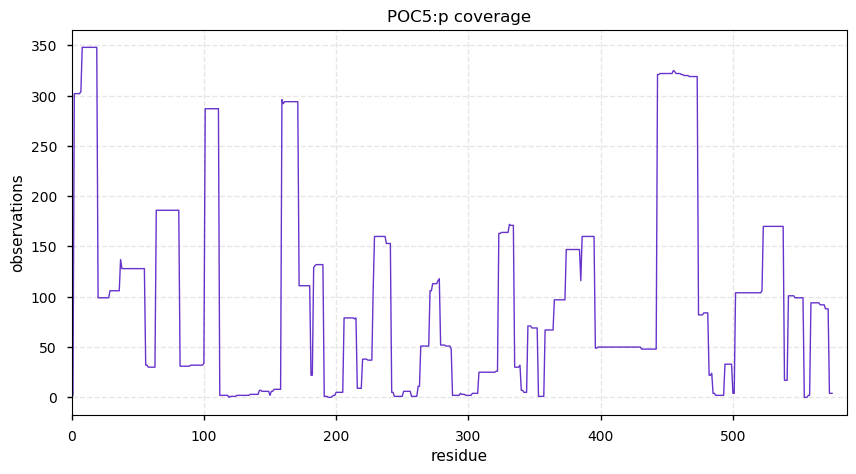
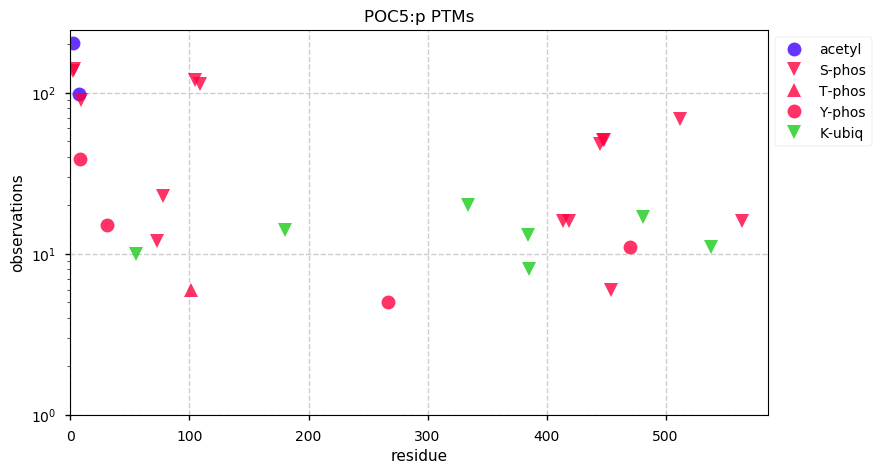
Fri Jul 19 21:29:10 +0000 2019@pwilmarth @dtabb73 After looking at the table, I have 3 suggestions:
1. Add a column for mods that are on a peptide's N-terminal residue (any AA type);
2. Replace the "K" and "R" columns with "C-term. K", "C-term. R", "other K", "other R"; &
3. Add a "U" column.
Fri Jul 19 19:57:32 +0000 2019@pwilmarth @dtabb73 It did. Thanks.
Fri Jul 19 19:51:14 +0000 2019@pwilmarth @dtabb73 It seems to need OHSU credentials ...
Fri Jul 19 19:33:56 +0000 2019@pwilmarth @dtabb73 Any chance of a screen shot?
Fri Jul 19 19:01:28 +0000 2019These guys just can't stop themselves from announcing bespoke R&D funding programs 🔗
Fri Jul 19 17:46:18 +0000 2019Open search peeps: what types of diagrams do you like to use to show the PSM results from a id run? I've seen some correlation diagrams (parent ion mass - assigned peptide sequence mass) in the literature, but not much else. Any favs?
Fri Jul 19 15:51:46 +0000 2019@AlexUsherHESA Sorry, I should have waited for the rest of the thread.
Fri Jul 19 15:50:37 +0000 2019@AlexUsherHESA I'm pretty sure you know it is because of the Morrill Land-Grant Acts that gave federal land to the states if they used some of the land (& revenue derived from the rest) to establish a university, leading to the "land-grant college" system.
Fri Jul 19 12:42:24 +0000 2019POC1B:p, POC1 centriolar protein B (H. sapiens) 🔗 Small cytoskeletal protein; several S-phosphorylation sites; no SAVs; 1 obs. splice variant; most abundant in spermatozoa; mature form 2-478 [770 x] 🔗
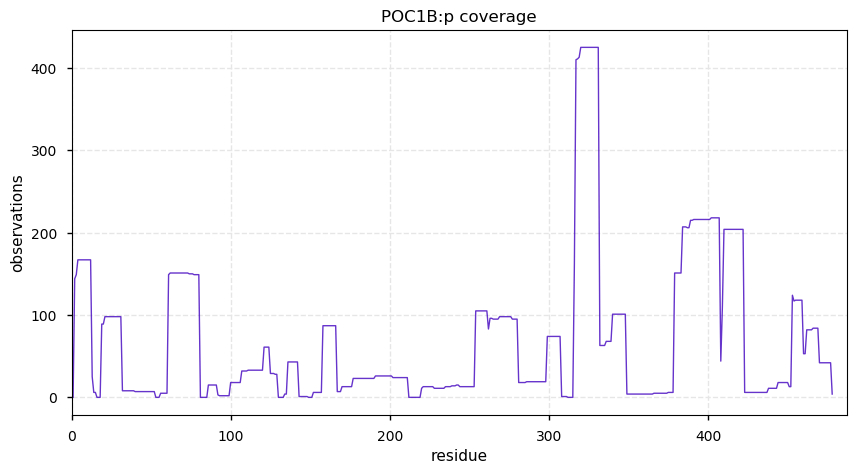
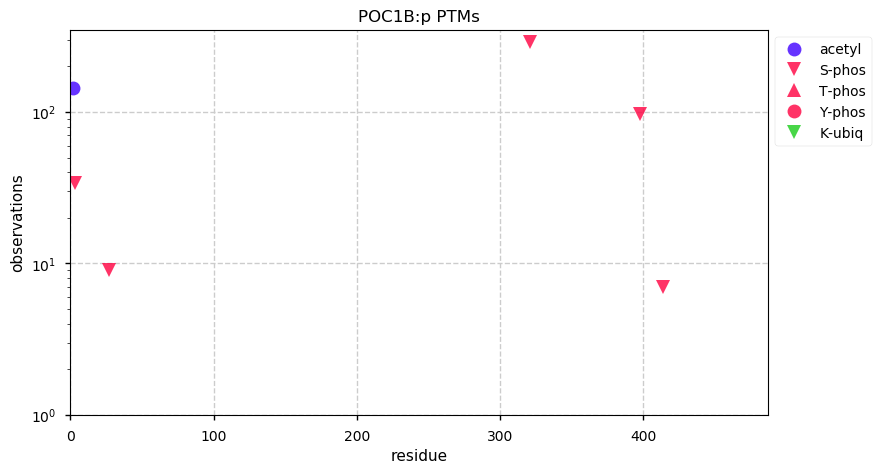
Thu Jul 18 21:08:05 +0000 2019@chrashwood @nesvilab As far as I know, MaxQuant is free to download, but it isn't open source. If I remember correctly, the license expressly forbids any attempt to discover anything about the source.
Thu Jul 18 20:53:53 +0000 2019@chrashwood @nesvilab MQ was originally developed to have Mascot as the search engine & the Max Planck lab was very much a Mascot-only operation. Then, the MP lab developed a closed source search engine that had very (very) similar characteristics to Mascot & used it in MQ instead.
Thu Jul 18 20:21:44 +0000 2019🔗
Thu Jul 18 17:39:20 +0000 2019@olgavitek @nesvilab And commercial software comes with support.
Thu Jul 18 15:44:17 +0000 2019@nesvilab @AnthonyCesnik They are not from an open/extended search: they are just from a nothing-special X! Tandem ID run.
Thu Jul 18 15:25:38 +0000 2019@nesvilab Everyone in the business knows what happened in the Mascot -> Andromeda development cycle, so I can't see any company being enthusiastic about having the same thing happen to them.
Thu Jul 18 15:17:26 +0000 2019@nesvilab This comparison is just between two closed-source apps, where you have the option of improving your code during the testing process. I just can't see how any company would be motivated to go along with it, unless you were a customer.
Thu Jul 18 14:32:54 +0000 2019@AnthonyCesnik The PSM's used for this second correlation diagram are here: 🔗
Thu Jul 18 14:29:10 +0000 2019@AnthonyCesnik Looking back through some data, this one from a gall bladder sample generates an even more complex pattern, mainly due to collagen hydroxy-P/K and some experimental artifacts. 🔗
Thu Jul 18 14:11:53 +0000 2019@nesvilab If I was them, I wouldn't cooperate unless you were a customer. Comparisons are hard to do properly, even with neutral 3rd parties. In a case like this, you are clearly motivated to show your app is better than theirs.
Thu Jul 18 14:02:15 +0000 2019@AnthonyCesnik The search run was just one I was doing as part of the GPMDB data reanalysis project. The PSMs that generated the correlation diagram are displayed here: 🔗
Thu Jul 18 12:12:33 +0000 2019POC1A:p, POC1 centriolar protein A (H. sapiens) 🔗 Small cytoskeletal protein; few, low occupancy PTMs; no SAVs; 1 obs. splice variant; mature form 2-407 [799 x] 🔗
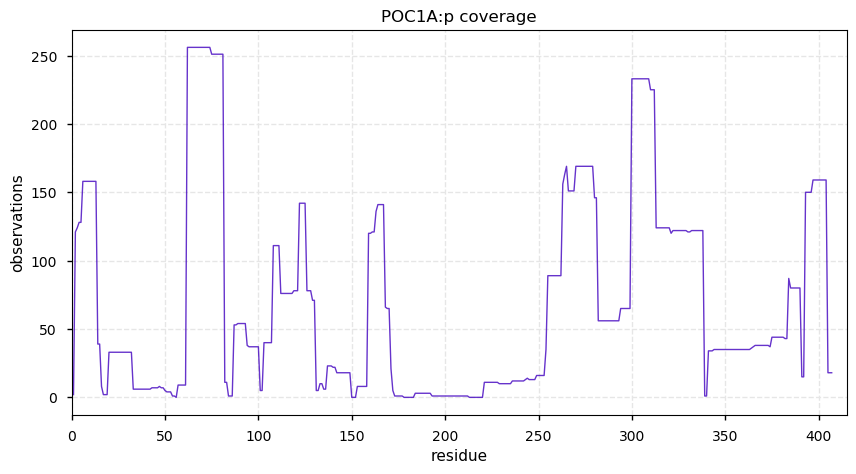
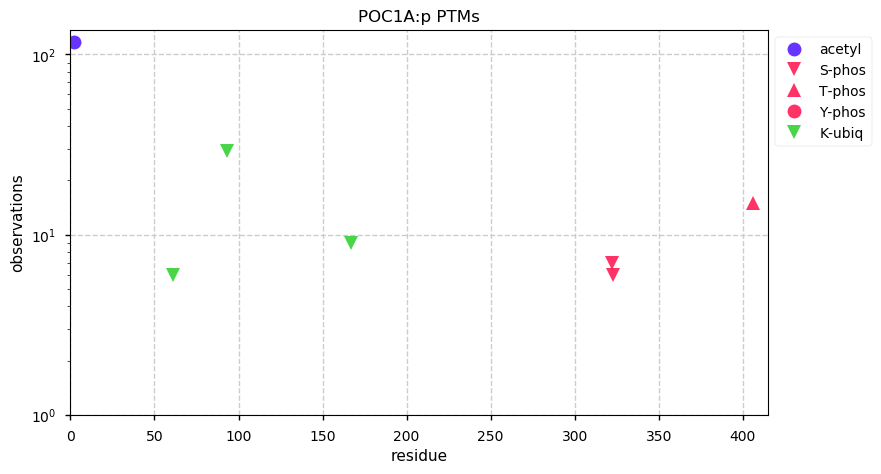
Wed Jul 17 22:53:32 +0000 2019@AnthonyCesnik You got what was plotted correct. It is not an open search, though. It is a conventional search of a CPTAC phosphopeptide run using TMT10.
Wed Jul 17 21:35:24 +0000 2019It is surprising (at least to me) how complex the conventionally-assigned PSMs for a single HPLC run look when plotted this way: 🔗
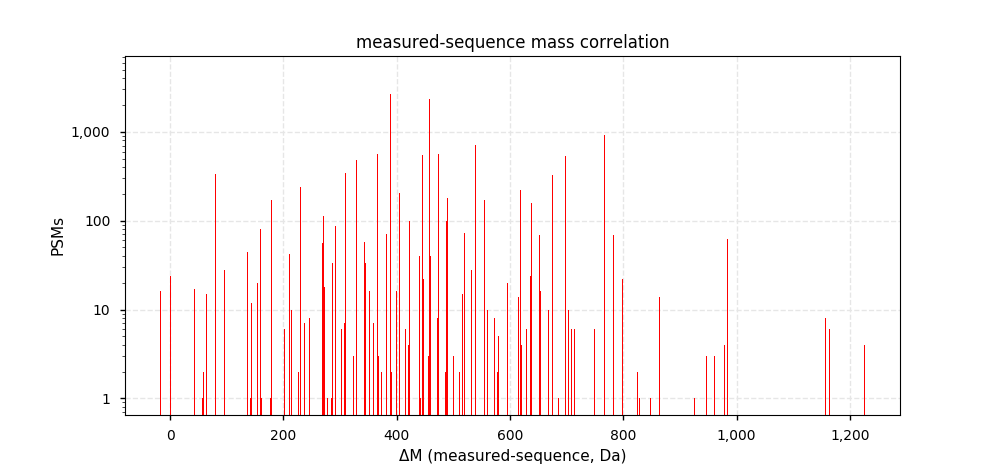
Wed Jul 17 17:07:55 +0000 2019@Sci_j_my Good luck with your job search. My only advise is that when you go to an interview, be sure to really quiz your potential department head. If anything about the head seems even a little off, think hard before accepting an offer.
Wed Jul 17 15:25:34 +0000 2019🔗 A case study in how not to design and execute a warning system that involves sensors, software and people.
Wed Jul 17 14:11:57 +0000 2019@UCDProteomics @pwilmarth Those little m.2 babies are the closest thing I've seen to magic in quite a while. I started using them 2 years ago and wouldn't think about setting up a new computer without them.
Wed Jul 17 14:08:58 +0000 2019PCM1:p, pericentriolar material 1 (H. sapiens) 🔗
Large cytoskeletal protein; distinctive PTM pattern; obs. SAVs: N159S maf=0.3, M597V maf=0.3, A691S maf=0.14, R1251H maf=0.01; 5 obs. splice variants; mature form 2-2024 [19,870 x] 🔗
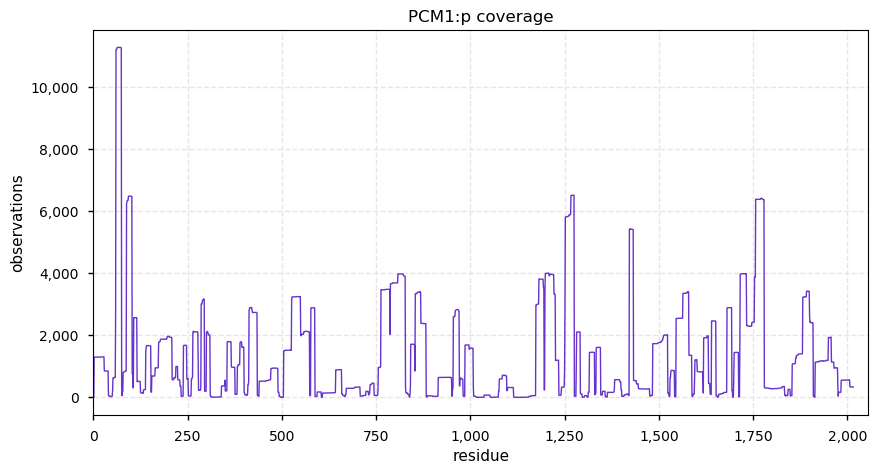
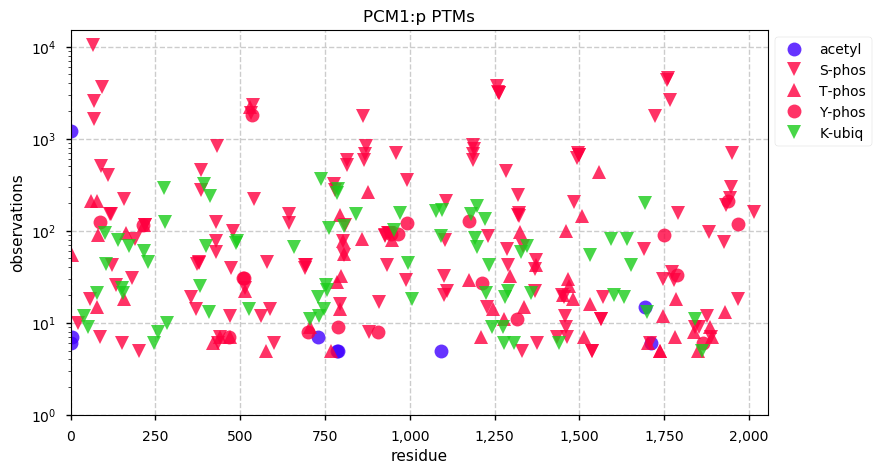
Tue Jul 16 23:14:18 +0000 2019@SpecInformatics I was a little unclear. I didn't mean that RAID would help in some circumstances: it won't. It is the high speed SSD that helps for IO bound jobs.
Tue Jul 16 16:32:35 +0000 2019@SpecInformatics They really only help for disk I/O bound issues, such as database queries or raw file conversions. They will not impact on CPU-bound things: peptide id searches & peak finding. Large SQLite files tend to perform rather badly, but that isn't really a hardware issue.
Tue Jul 16 16:25:01 +0000 2019@SpecInformatics No, RAID wouldn't help at all for NVMe-type devices.
Tue Jul 16 15:48:00 +0000 2019@SpecInformatics There is no such thing as too much NVMe M.2 PCI-E storage. It improves disk I/O bound processes by at least an order of magnitude. Memory speed doesn't make that much difference: I'd suggest buying more, lower speed memory.
Tue Jul 16 14:44:17 +0000 2019@nesvilab @astacus It means that if JHU was in Canada, to retain the same pace it would require all of the funding available from the Canadian "tricouncil" as well as all of CFI and Genome Canada funding & still need to find some more money.
Tue Jul 16 14:38:16 +0000 2019@nesvilab @astacus It doesn't actually change the math a lot. PI salaries are not the largest component of funded grants (at least not in 2 rotations I did on an NIH study section). And JHUs total yearly R&D spending is US$2,562,000,000 🔗
Tue Jul 16 14:20:50 +0000 2019PS, calling the PTM pattern "distinctive" is classic Canadian understatement: either the PTM observations are wrong (note the log scale) or the current understanding of ubiquitinyl and S/T/Y phosphoryl mods needs an overhaul.
Tue Jul 16 14:19:49 +0000 2019OFD1:p, centriole and centriolar satellite protein (H. sapiens) 🔗 Large cytoskeletal protein; distinctive PTM pattern; no SAVs; 1 obs. splice variant; mature form 3-1012 [4,445 x] 🔗
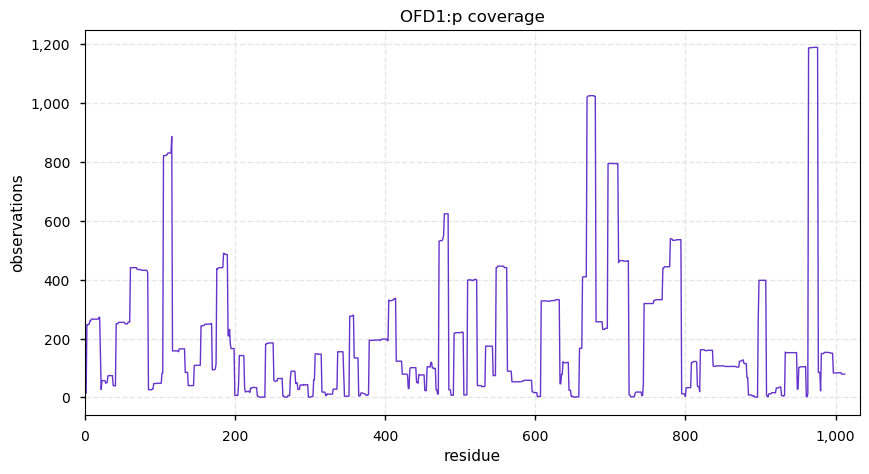
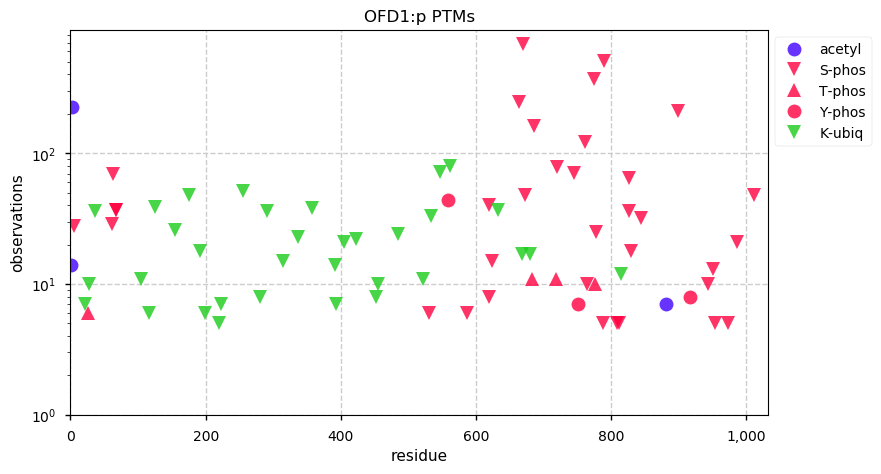
Mon Jul 15 22:46:33 +0000 2019@nesvilab Every once in a while I like to reminder my Canadian colleagues that 1 US university gets more funding than the entire amount of "discretionary" funding available for the entire country through CIHR.
Mon Jul 15 21:51:46 +0000 2019Did you know that Johns Hopkins University received US$674,583,550 in NIH funding in 2018? 🔗
Mon Jul 15 21:45:08 +0000 2019@doctorow Or Canada's.
Mon Jul 15 19:45:04 +0000 2019@astacus @MuscleScience @DannyJamesWard @TheMuscleTussle I'd be surprised if there is one, because it isn't true in general. People often confuse the standard deviation of a distribution with the standard error of its mean and really get into trouble when they discover that both of those values have errors, too.
Mon Jul 15 16:36:09 +0000 2019@TrostLab But there are lots of studies into mouse and rat brains that could be used to provide methods & compare results.
Mon Jul 15 16:34:00 +0000 2019@TrostLab There are genomes (& predicted proteomes) for a couple of ground squirrel-like species (Ictidomys tridecemlineatus, Urocitellus parryii, Marmota flaviventris and Marmota marmota marmota), but none specifically for grey tree squirrels.
Mon Jul 15 16:08:30 +0000 2019@TrostLab I was going to refer to the culinary possibilities (which would have never occurred to me except I spent some time in Indiana), but I can see that has been mentioned. Squirrels can have a spongiform encephalopathy, so I would be cautious about trying any recipes.
Mon Jul 15 16:05:34 +0000 2019@TrostLab Do you know the species ? Lots of very different rodents are referred to as "squirrels".
Mon Jul 15 13:11:56 +0000 2019HYLS1:p, centriolar and ciliogenesis associated (H. sapiens) 🔗 Small cytoskeletal protein; several phosphorylation sites; 1 SAV: C31R maf=0.3; present at low levels in many cell lines; mature form 1-299 [411 x] 🔗
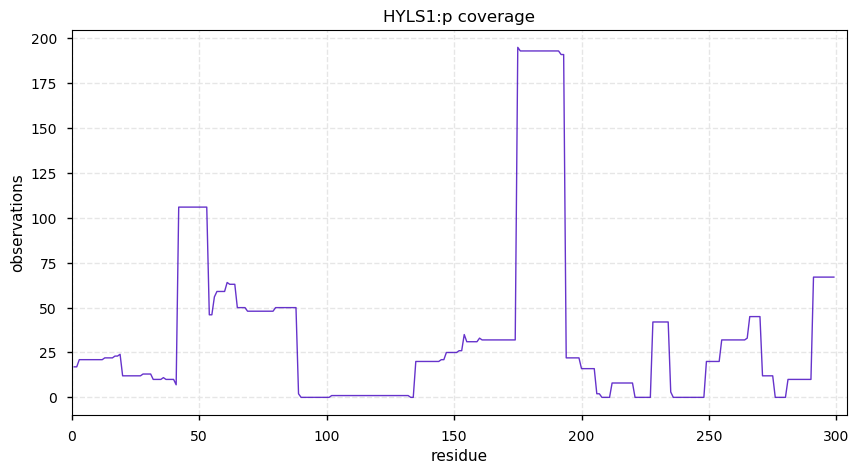
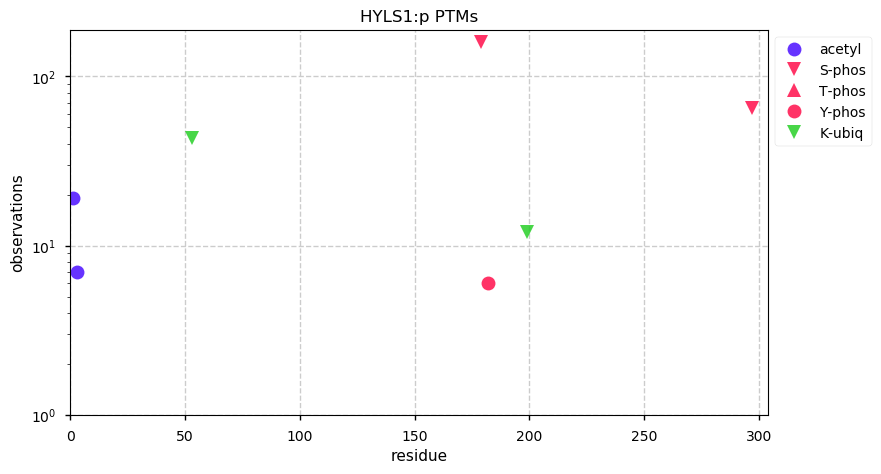
Sun Jul 14 13:43:23 +0000 2019@pwilmarth Amen, brother, amen. It seems as though most tissue samples that are primarily acellular have similar characteristics.
Sun Jul 14 13:02:37 +0000 2019CNTROB:p, centrobin, centrosomal BRCA2 interacting protein (H. sapiens) 🔗 Midsized cytoskeletal protein; 3 S-phosphodomains; no SAVs; 1 splice variant observed; mature form 2-925 [1,597 x] 🔗
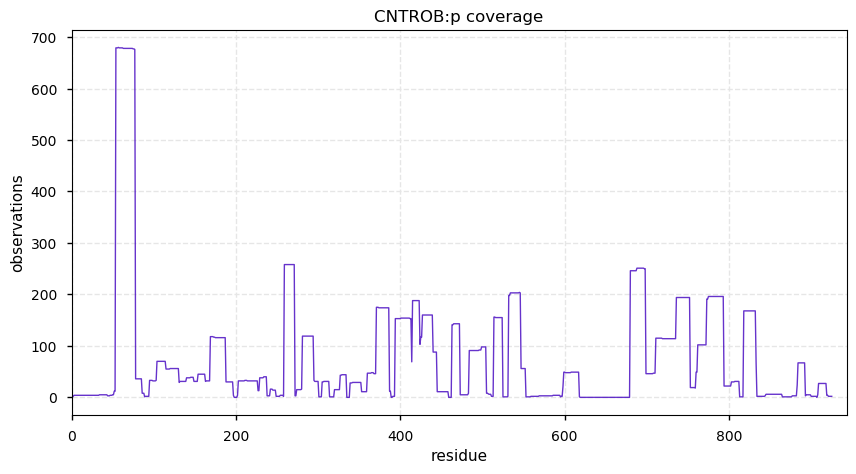
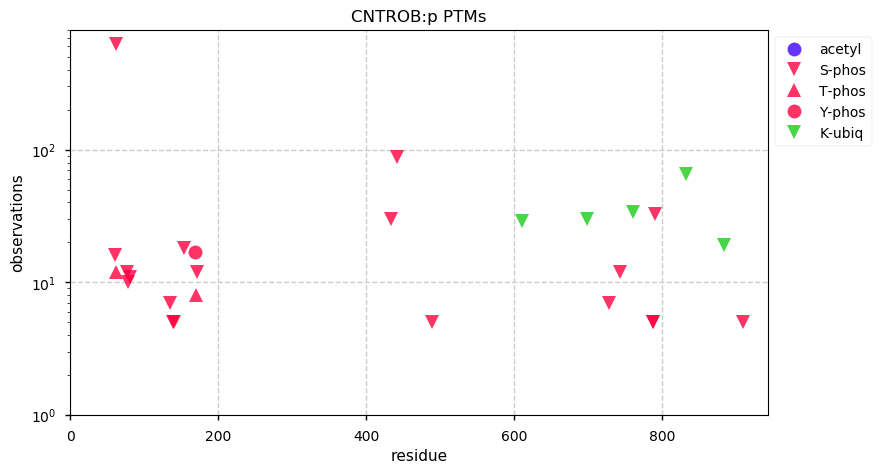
Sat Jul 13 16:07:31 +0000 2019@farmgirlphd @MHendr1cks @jwoodgett Even back in the 2000's, when I was on the CIHR Genomics panel for a couple of cycles, there was a very significant bias against any project that involved non-mammalian model species.
Sat Jul 13 15:38:23 +0000 2019@manohar_dange 5. Why so much Y-phosphorylation, when it is rare compared to S/T-phosphorylation in non-mitochondrial proteins?
Sat Jul 13 15:33:12 +0000 2019@manohar_dange 4. If you were to ask most biochemists about the role of PTMs in the TCA cycle, they would say there is none. Is there something wrong with the experiments that show PTM involvement or is the prevailing understanding of this biochemistry incomplete?
Sat Jul 13 15:29:36 +0000 2019@manohar_dange 3. It appears as though significant amounts of acetylation is occurring at non-random acceptor sites on most of the enzymes: do these have a regulatory effect or are they related to the ubiquitin modifications?
Sat Jul 13 15:25:38 +0000 2019@manohar_dange 2. There is significant phosphorylation on a few of the important enzymes in the citric acid cycle: do these have a regulatory role in mitochondrial metabolism?
Sat Jul 13 15:24:05 +0000 2019@manohar_dange 1. Mitochondrial matrix proteins can be modified by ubiquitin, even though they are mechanically isolated from the proteosome (so why is this happening).
Sat Jul 13 13:12:52 +0000 2019CNTRL:p, centriolin (H. sapiens) 🔗 Large cytoskeletal protein; S/Y-phosphorylation; no SAVs; 1 splice variant observed; significant tryptic peptide overlaps with other proteins; mature form 1-2,325 [2,699 x] 🔗
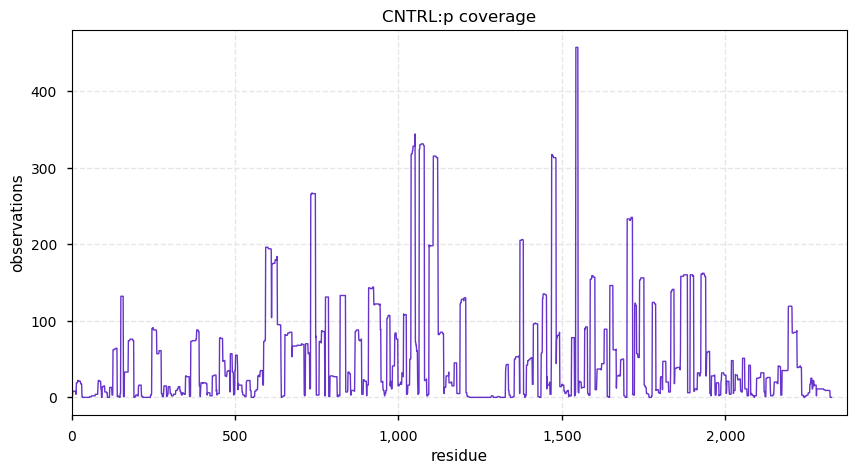
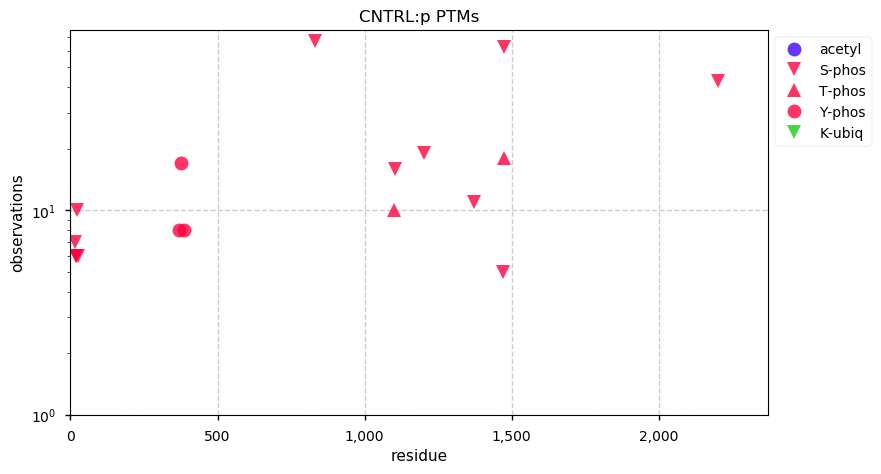
Fri Jul 12 21:04:09 +0000 2019and the credits:
🔗
Fri Jul 12 21:02:06 +0000 2019the director's cut:
🔗
Fri Jul 12 20:14:56 +0000 2019The observation frequency of 5 PTMs on the gene products involved in Kreb's cycle: a short film. 🔗

Fri Jul 12 12:29:39 +0000 2019CCSAP:p, centriole, cilia and spindle associated protein (H. sapiens) 🔗 Small cytoskeletal protein; S/T-phosphorylation sites; no SAVs; 1 splice variant observed; abundant in brain tissue; mature form 1-270 [719 x] 🔗
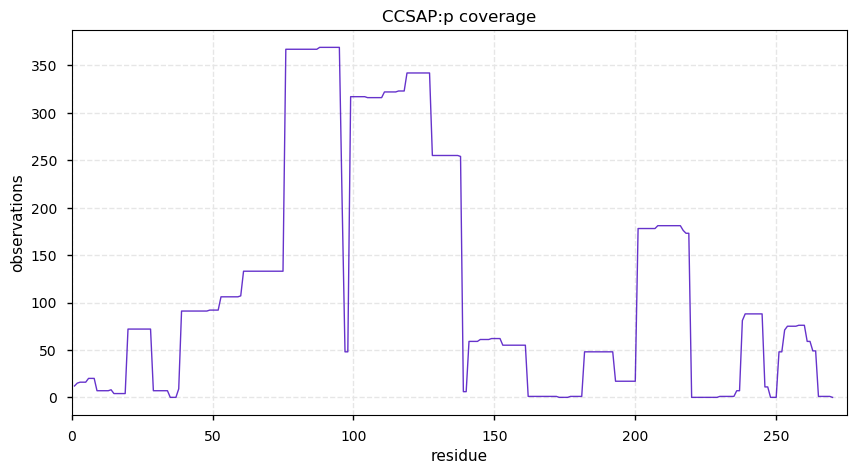
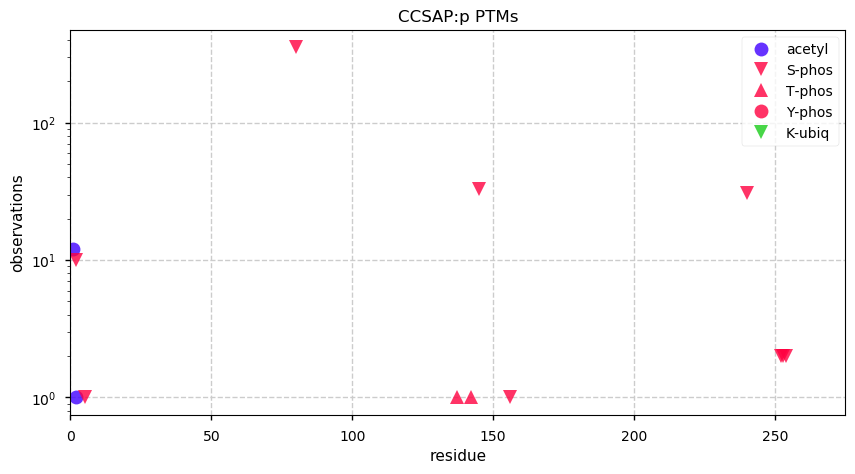
Thu Jul 11 22:01:44 +0000 2019For the life of me, I've never understood the Canadian fascination with French Gothic Revival architecture. It always looks out of place to me when I see it in a Canadian city.
Thu Jul 11 21:47:25 +0000 2019@thomdunn @doctorow Sounds more like Mira Grant to me.
Thu Jul 11 18:04:16 +0000 2019🔗
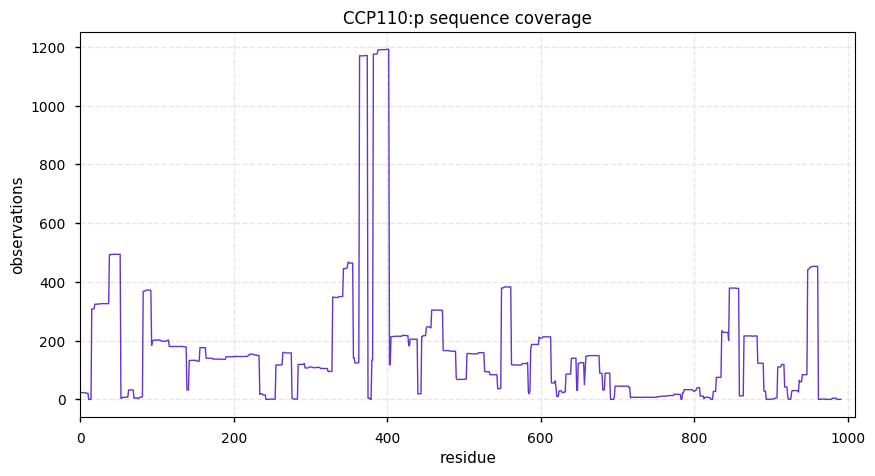
Thu Jul 11 16:28:29 +0000 2019CCP110:p, centriolar coiled-coil protein 110 (H. sapiens) 🔗 Large cytoskeletal protein; many PTMs; 1 SAVs: P171L maf=0.24; 1 splice variant observed; mature form 1-991 [3,121 x] 🔗
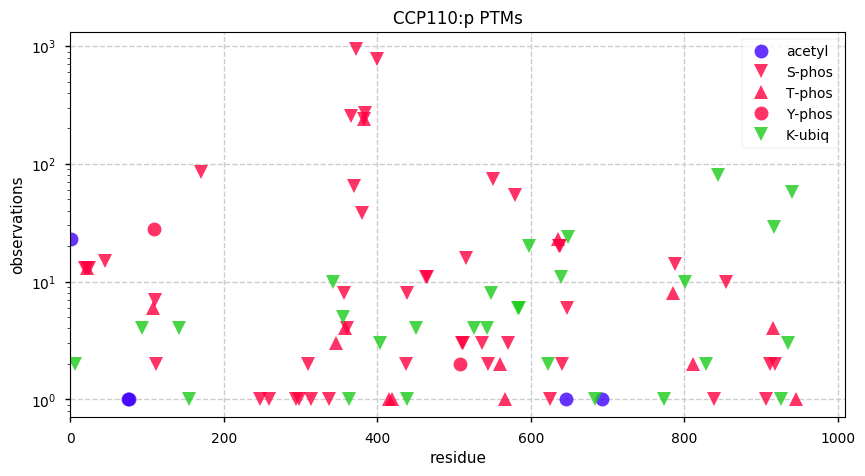
Thu Jul 11 00:01:36 +0000 2019@pwilmarth I didn't know that there was any format other than spreadsheets that was being used to report information to biomedical researchers. What other formats are being pushed as alternatives?
Wed Jul 10 23:25:58 +0000 2019@UCDProteomics Serum can be quite a bit more variable, especially lab-to-lab. The process of making plasma into serum can pull out some proteins that may affect differential biomarker investigations.
Wed Jul 10 21:00:34 +0000 2019My own pet peeve is that it encourages single solutions to spectra or groups of spectra (i.e., solutions that only require 1 line), often resulting in rather contorted logic to exclude or hide other solutions that may be just as good.
Wed Jul 10 19:04:47 +0000 2019What is the biggest drawback to using Excel-type spreadsheets as the final reporting format for all types of proteomics results and information.
Wed Jul 10 12:28:15 +0000 2019CEP83:p, centrosomal protein 83 (H. sapiens) 🔗 Midsized cytoskeletal protein; 1 phosphodomain; no SAVs; 1 splice variant observed; putative mature form 1-701 [610 x] (image from SubCell BarCode) 🔗
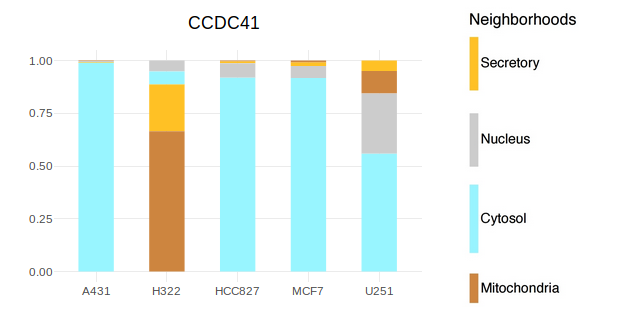
Tue Jul 09 16:15:10 +0000 2019It is as though evolution typed it out on a mechanical keyboard with a couple of sticky keys
Tue Jul 09 15:55:00 +0000 2019The next time some one asks me for an "example", "typical", "average", "normal" or "representative" protein sequence, I am going to point them to this fruit fly masterpiece 🔗
Tue Jul 09 13:23:05 +0000 2019CEP295NL:p, CEP295 N-terminal like (H. sapiens) 🔗 Midsized putative cytoskeletal protein; most abundant in spermatozoa; no PTMs; no SAVs; mature form 1-621 [114 x] 🔗
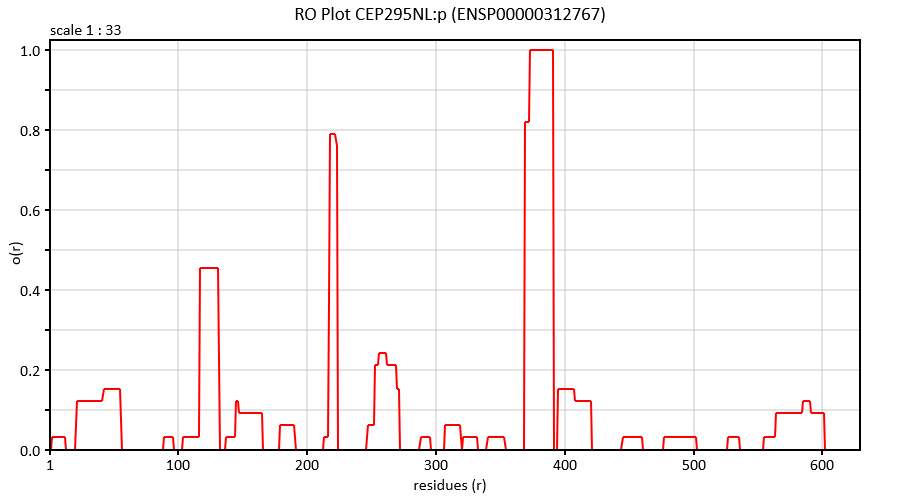
Mon Jul 08 21:08:28 +0000 2019@macro_momo I thought, rather naïvely, that the research literature was to tell us things we don't already know.
Mon Jul 08 16:49:23 +0000 2019And by this I mean K-malonylation of non-histone proteins.
Mon Jul 08 16:00:32 +0000 2019PXD006503 is pretty interesting data. It suggests that it would be valuable to have a lot more K-malonylation data to compare with what is known about K-ubiquitinylation.
Mon Jul 08 13:09:25 +0000 2019CEP19:p, centrosomal protein 19 (H. sapiens) 🔗 Small cytoskeletal protein; rarely observed; no PTMs; no SAVs; mature form 1-167; some evidence that M3 may be a translation initiator [53 x] (image from SubCell Barcode) 🔗
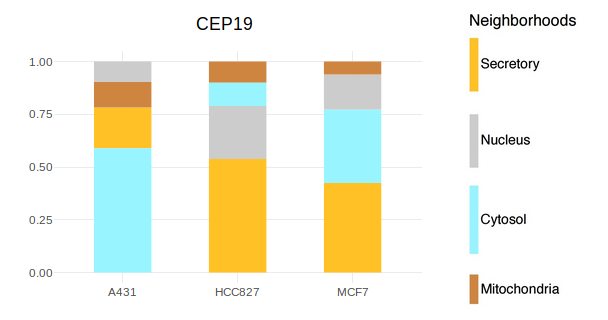
Sun Jul 07 12:50:07 +0000 2019CEP164:p, centrosomal protein 164 (H. sapiens) 🔗 Large cytoskeletal protein; many S/T phosphorylation sites; 1 splice variant observed; 1 SAV: T988S (maf=0.3); mature form 1-1460 [1,256 x] 🔗
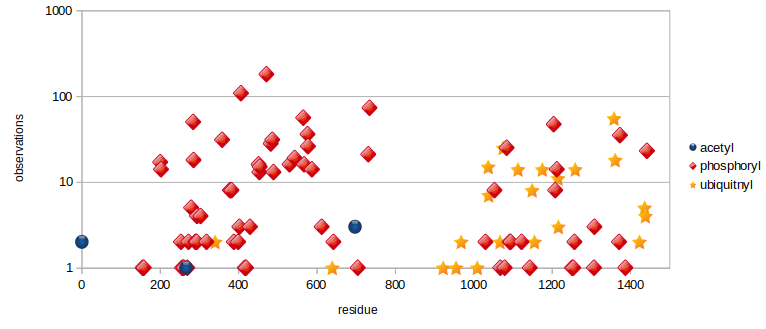
Sat Jul 06 14:22:23 +0000 2019Although AI is also running neck-and-neck in the misplaced confidence sweepstakes.
Sat Jul 06 13:56:05 +0000 2019About the only human activity where AI has exceeded its creators is "dithering".
Sat Jul 06 12:27:52 +0000 2019CEP126:p, centrosomal protein 126 (H. sapiens)
Large putative cytoskeletal protein; no splice variants confidently observed (NBS EC=3); proposed mature form 1-1117 (image from String) 🔗

Fri Jul 05 22:39:36 +0000 2019@ypriverol Point 2 should read
2. NCBI endogenous retrovirus + LINE-1 proteins;
rather than simply "retrovirus"
Fri Jul 05 18:28:52 +0000 2019@macro_momo @ypriverol I use this one for ORF1 🔗 and gi|74705367| for ORF2. I don't check for additional heterogeneity, as I'm not really an aficionado of the subject (more of a cognoscenti)
Fri Jul 05 15:14:47 +0000 2019@ypriverol I don't think any of the existing sequence collections are ideal for proteomics: depending on your application & species mix you really have to kludge things together to do a good job.
Fri Jul 05 14:05:58 +0000 2019@ypriverol I use ENSEMBL protein sequences for reanalysis, but that isn't the whole enchilada. For human, I normally use:
1. ENSEMBL;
2. NCBI retrovirus + LINE-1 proteins;
3. NCBI common viruses;
4. SwissProt-based cRAP;
5. dbSNP SAVs; &
6. GPMDB PTMs.
Fri Jul 05 12:28:24 +0000 2019CEP112:p, centrosomal protein 112 (H. sapiens) 🔗 Midsized cytoskeletal protein; 1 phosphodomain, no ubiquitin acceptor sites; 1 splice variant observed; no high maf SAVs; mature form 1-955 [744 x] 🔗
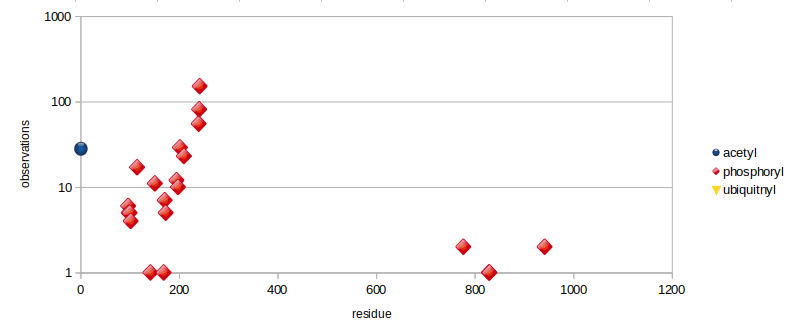
Thu Jul 04 18:35:23 +0000 2019@macro_momo @dtabb73 I would also say "yes", but I admit to having no qualifications that would allow me to have an opinion.
Thu Jul 04 18:05:05 +0000 2019@scottmccain5 @pwilmarth @ProteomicsNews @olgavitek And probably quite a few false negatives, too.
Thu Jul 04 17:59:27 +0000 2019@scottmccain5 @pwilmarth @ProteomicsNews @olgavitek You might be able to use sequence tag calling software as some type of 1st pass peptide-sequence-detector, but it would produce many false positives.
Thu Jul 04 17:51:17 +0000 2019@scottmccain5 @pwilmarth @ProteomicsNews @olgavitek I'm not trying to be either mean or sarcastic: when I read it I laughed spontaneously. It would be great if such a thing existed.
Thu Jul 04 17:48:12 +0000 2019@scottmccain5 @pwilmarth @ProteomicsNews @olgavitek The answer is "no" (but the question did make me laugh).
Thu Jul 04 17:08:25 +0000 2019For the dog owners, 🔗
Thu Jul 04 15:30:34 +0000 2019@jervenbolleman @BrenesAlejandro @UCDProteomics That makes sense: UniProt (and its predecessor) has always had an inexplicably awkward relationship with nucleic-acid-derived sequences, particularly wrt the human genome.
Thu Jul 04 14:48:20 +0000 2019I love it when a news site complains about my network security; it confirms that the network is doing its job and that I should never use that site again.
Thu Jul 04 12:37:11 +0000 2019CEP104:p is the only centrosomal protein prominently observed in urine
Thu Jul 04 12:32:22 +0000 2019CEP104:p, centrosomal protein 104 (H. sapiens) 🔗 Midsized cytoskeletal protein; some low occupancy PTMs; 1 splice variant observed; no high maf SAVs; mature form 1-925 [1,586 x] 🔗
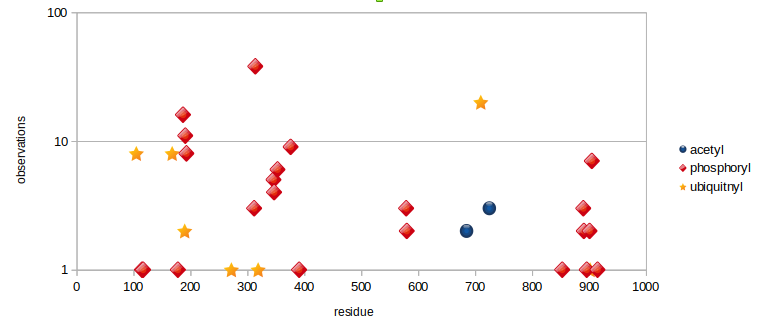
Wed Jul 03 14:46:32 +0000 2019I was just re-reading Schrödinger's paper criticizing the Copenhagen interpretation of quantum mechanics (🔗). Really (really) insightful, but only remembered because of a silly joke that people with no sense of humor take seriously.
Wed Jul 03 13:11:47 +0000 2019Simply calling this one a "phosphoprotein" really doesn't give you an accurate picture of how "phospho" this particular "protein" is in vitro ..
Wed Jul 03 13:07:44 +0000 2019CEP170B:p, centrosomal protein 170B (H. sapiens) 🔗 Large cytoskeletal protein; highly phosphorylated; some tryptic peptide overlap with CEP170; no high maf SAVs; mature form 2-1554 [7,362 x] 🔗
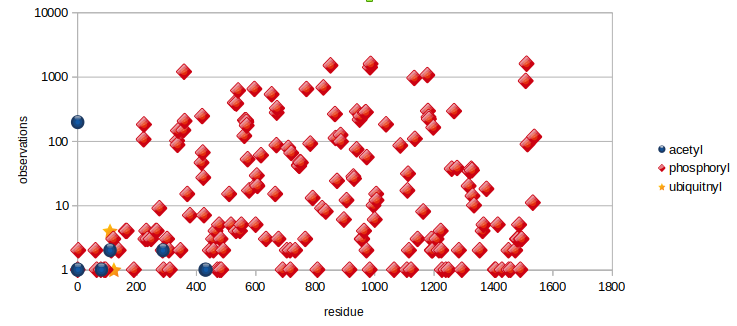
Tue Jul 02 16:34:56 +0000 2019@pwilmarth It isn't OK, but it is customary.
Tue Jul 02 15:11:38 +0000 2019What methods do people use to keep track of how well parent ion declustering is working in their instruments?
Tue Jul 02 13:19:50 +0000 2019CEP95:p, centrosomal protein 95 (H. sapiens) 🔗 Midsize cytoskeletal protein; 1 splice variant observed; no high maf SAVs; mature form 2-821 [912 x] 🔗
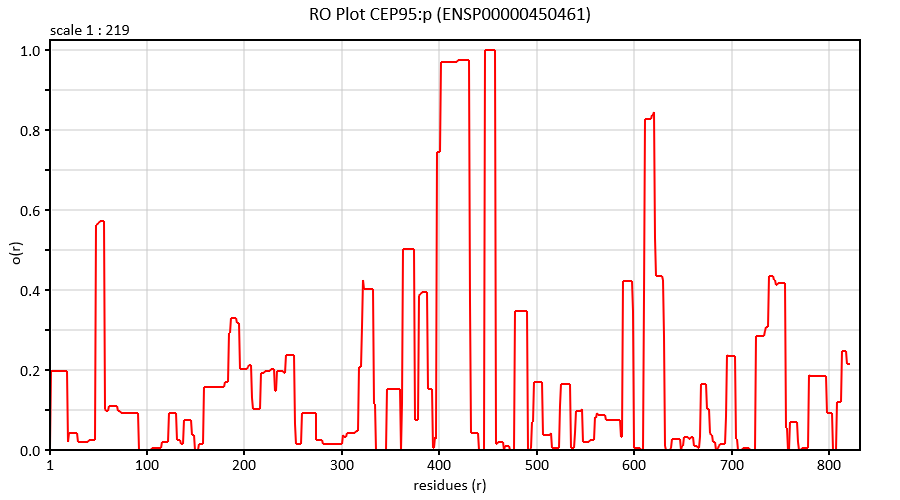
Mon Jul 01 12:53:31 +0000 2019CEP85L:p is a rarity: a protein where there genuinely is a proteotypic peptide.
Mon Jul 01 12:51:30 +0000 2019CEP85L:p, centrosomal protein 85 like (H. sapiens) 🔗 Midsize putative cytoskeletal protein; 1 splice variant observed; 1 high maf SAVs: S137G maf=0.50; mature form 1-805 [742 x] 🔗
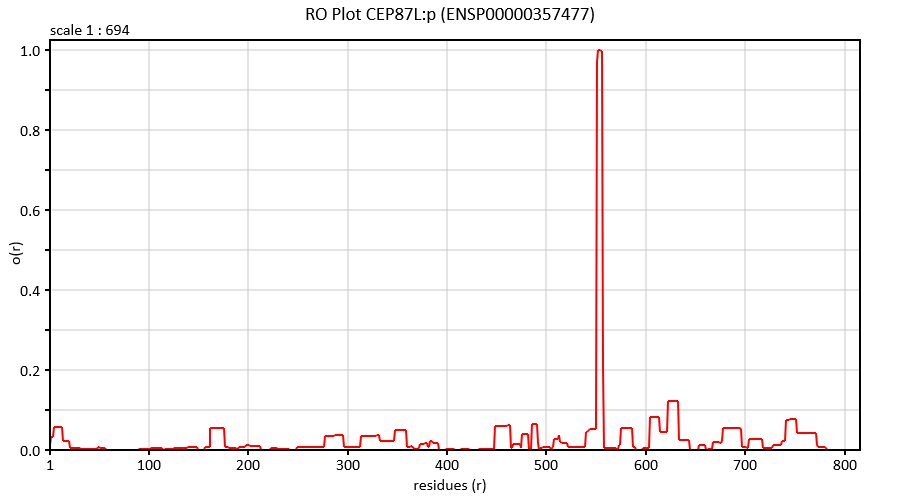
Sun Jun 30 20:32:25 +0000 2019@cwvhogue Don't forget the version for 8-bit Atari.
Sun Jun 30 20:29:37 +0000 2019@mekki @doctorow @Microsoft The old "art-as-a-service" business model.
Sun Jun 30 20:28:01 +0000 2019@AlexUsherHESA Extra points for working in Glen Murray!
Sun Jun 30 16:53:51 +0000 2019@chrashwood @theoneamit @byu_sam @Ciencia_2017 Or simply relocating bands because they aren't where they should be.
Sun Jun 30 16:50:15 +0000 2019@chrashwood @theoneamit @byu_sam @Ciencia_2017 It is the same thing as using Photoshop to clean up a gel image by "correcting" wavy bands so that they look better for a publication.
Sun Jun 30 16:09:22 +0000 2019@chrashwood @theoneamit @byu_sam @Ciencia_2017 You should not use guesses to alter data so as to conform with themselves. If you have calibrants (knowns with acceptance criteria) then no problem. But talking yourself into using best-guesses as though they were known is not OK.
Sun Jun 30 15:28:12 +0000 2019@theoneamit @byu_sam @Ciencia_2017 @chrashwood And by field, I mean proteomics software development.
Sun Jun 30 15:27:24 +0000 2019@theoneamit @byu_sam @Ciencia_2017 @chrashwood I guess I'm one of the few people in the field that has ever run a GMP-regulated analytical lab, so I have a rather different perspective on the rigor required for accepting the results of a measurement.
Sun Jun 30 15:15:48 +0000 2019@byu_sam @Ciencia_2017 @chrashwood It has become a fixture in the literature, but it is still very bad practice. Viewed dispassionately, the method is as follows:
1. Take data;
2. Make best-guesses as to what was measured; and
3. Alter the data to conform to those guesses.
Sun Jun 30 12:19:27 +0000 2019CEP78:p, centrosomal protein 78 (H. sapiens) 🔗 Midsize cytoskeletal phosphoprotein; 1 splice variant observed; 0 high maf SAVs; mature form 1-722 [2,441 x] 🔗
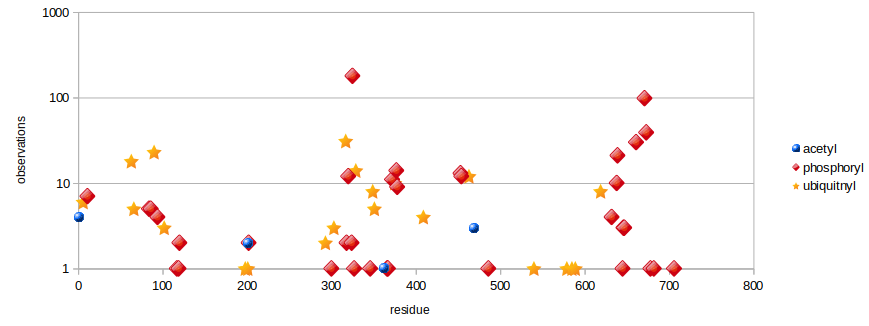
Sat Jun 29 20:02:05 +0000 2019@nesvilab @chrashwood Clearly, I disagree.
Sat Jun 29 19:23:29 +0000 2019@nesvilab @chrashwood Running a search with a wide parent ion mass tolerance window, using those results to recalibrate the parent ion masses and then re-running the search with a narrow parent ion mass tolerance window.
Sat Jun 29 15:46:18 +0000 2019@chrashwood The letter has merit: internal standards are no replacement for real QA/QC calibration curves. But at least the use of standards is better than the awful practice in proteomics of using search engine identification results to recalibrate spectra.
Sat Jun 29 12:38:24 +0000 2019CEP76:p, centrosomal protein 76 (H. sapiens) 🔗 Midsize cytoskeletal phosphoprotein; 1 splice variant observed; 0 high maf SAVs; mature form 1,2,3-659 [1,907 x] 🔗
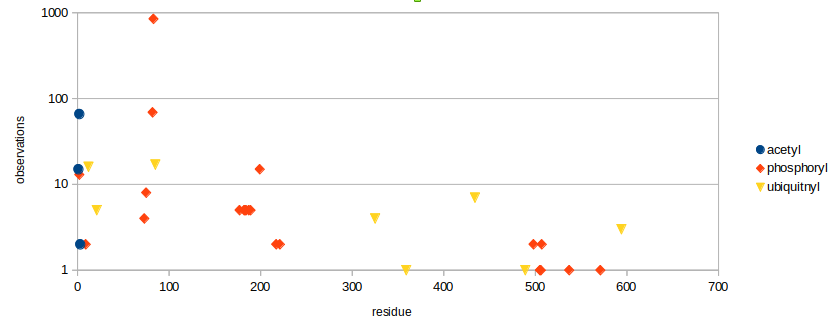
Sat Jun 29 12:29:24 +0000 2019@KentsisResearch @ProtifiLlc @UCDProteomics @paolocifani Peptide are pretty stable in neat TFA. It is used in peptide synthesis to cleave the peptide from resin and it doesn't cause any significant amount of backbone cleavage. Once the data becomes public, we'll know for sure.
Fri Jun 28 18:21:33 +0000 2019It turns out that "open" searches can be used to qualify data sets for SAV analysis pretty easily 😀. Haven't found any new PTMs though 😟.
Fri Jun 28 17:08:05 +0000 2019@chrashwood @lukas_k In that case (part of a distribution) may be able to protect the library as a necessary part of the software installation or as something generated on the fly.
Fri Jun 28 17:01:06 +0000 2019@chrashwood @lukas_k Although I should emphasize "MAY BE" strongly, as courts have been back and forth on similar types of protection.
Fri Jun 28 16:59:21 +0000 2019@chrashwood @lukas_k If you then distribute that as a file ("🔗") and state that you can only access the data via:
>python3 🔗
then your copyright protection may be OK under DCMA rules
Fri Jun 28 16:57:13 +0000 2019@chrashwood @lukas_k # Copyright © 2019 Ronald C. Beavis
def dn():
d = '''190,70
172,145
173,147'''
return d
def main():
ls = dn().split('\n')
for l in ls:
print(l)
if __name__== "__main__":
main()
Fri Jun 28 16:56:03 +0000 2019@chrashwood @lukas_k All is not lost, though, if you want to be a bit creative. Let's say you have some data:
190,70
172,145
173,147
You can embed that data into software code and then a copyright will be applicable.
Fri Jun 28 15:23:29 +0000 2019@chrashwood @lukas_k If you have some time, listen to the discussion of property rights and copyright here 🔗 (and/or read John Locke's Second Treatise on Government) to get some idea of how legal thought works wrt the ownership of things & ideas.
Fri Jun 28 14:46:20 +0000 2019@chrashwood @lukas_k But I'm not sure that the files created from annotation databases like Pfam or Uniprot have defendable copyrights, even though they claim them as part of their licenses.
Fri Jun 28 14:43:25 +0000 2019@chrashwood @lukas_k Software is protected by copyright, pretty much in all instances - generating software is an inherently creative act involving the writing of some type of text.
Fri Jun 28 13:57:30 +0000 2019@lukas_k @chrashwood "but the not numbers themselves" should have been "but not the numbers themselves"
Fri Jun 28 13:45:22 +0000 2019@lukas_k @chrashwood Courts have held repeatedly that any representation of numbers generated by instruments cannot be protected by copyright. If you make a table of numbers and put it in a paper, then the image of the table can be protected, but the not numbers themselves.
Fri Jun 28 12:06:16 +0000 2019CEP72:p, centrosomal protein 72 (H. sapiens) 🔗 Midsize cytoskeletal phosphoprotein; 1 splice variant observed; 2 high maf SAVs: P412T maf=0.09, T509A maf=0.27; mature form 2-646 [1,936 x] 🔗
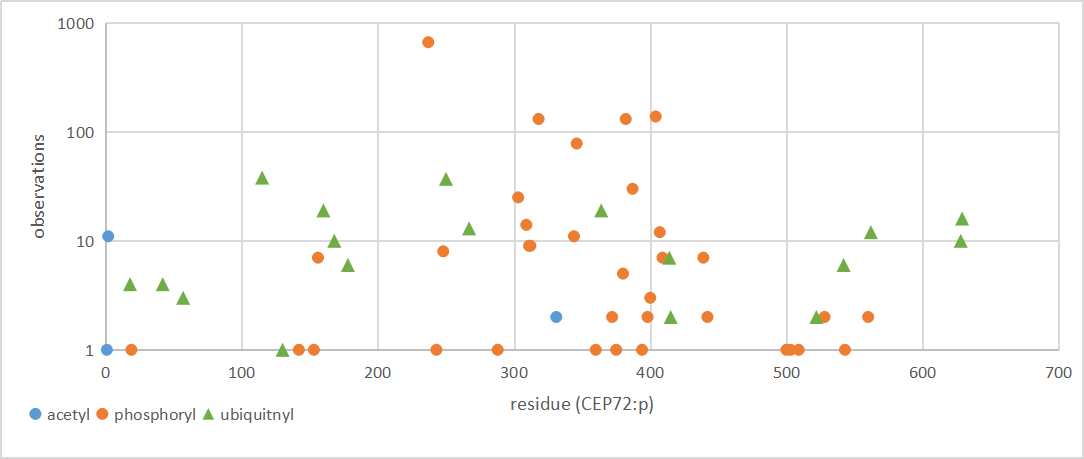
Thu Jun 27 21:25:14 +0000 2019@paulozoom The definition of AI is particularly apt.
Thu Jun 27 20:25:30 +0000 2019@chrashwood Licensing doesn't usually apply to something that is a set of numbers generated by a instrument, as it can't be protected by copyright. CC-type licensing only applies to the use of material protected by a copyright 🔗
Thu Jun 27 16:21:00 +0000 2019I guess I'm alone in being interested in this type of thing.
Thu Jun 27 12:18:44 +0000 2019CEP70:p, centrosomal protein 70 (H. sapiens) 🔗 Midsize cytoskeletal phosphoprotein; 1 splice variant observed; 1 high maf SAV: S135N maf=0.31; mature form 1-597 [469 x] 🔗
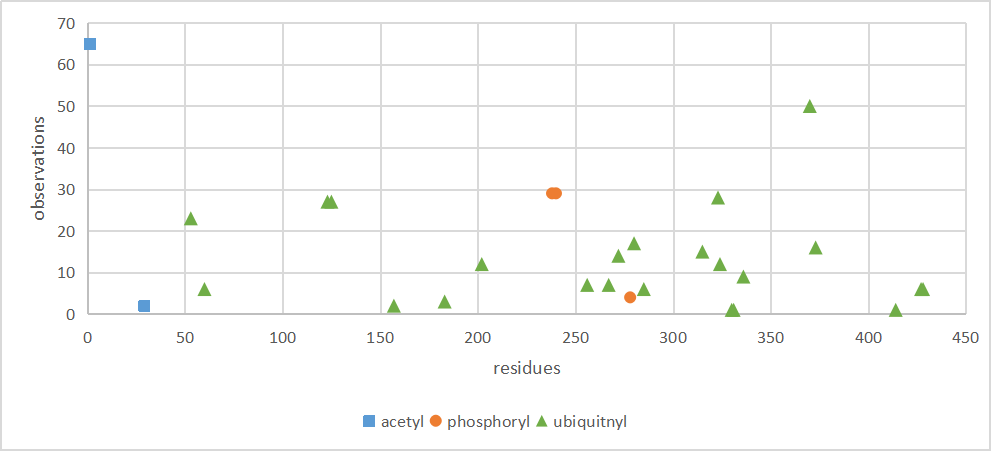
Wed Jun 26 18:03:37 +0000 2019@UCDProteomics I'd suggest making up a small MAPT-only FASTA & adding it to the search, using the sequences for:
ENSP00000413056
ENSP00000460965
ENSP00000458742
ENSP00000389250
ENSP00000485913
ENSP00000408975
ENSP00000334886
They should cover all identifications.
Wed Jun 26 14:46:54 +0000 2019I worked for Lilly in protein product development (including insulin) before these price increases began 🔗
Wed Jun 26 13:06:39 +0000 2019CEP68:p, centrosomal protein 68 (Homo sapiens) 🔗 Midsize cytoskeletal phosphoprotein; 1 splice variant observed; 1 high maf SAV: G74S maf=0.12; mature form 2-757 [1,514 x] 🔗
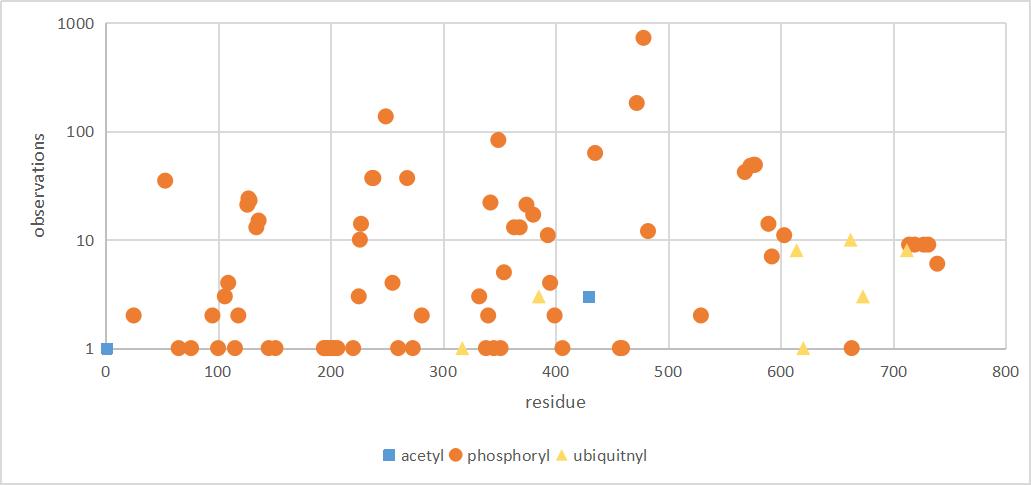
Tue Jun 25 23:23:33 +0000 2019@Peptidome I don't know much about it.
Tue Jun 25 19:33:51 +0000 2019@dtabb73 @ProteomicsNews @mjhelf @MetabOseek @xkcd I feel I must say "yes", although I have no peer-reviewed evidence upon which to base my assertion.
Tue Jun 25 19:09:06 +0000 2019@mjmaccoss You'll get no argument from me on both counts. Batch, experiment ordering & instrument maintenance effects are underappreciated, but easily observable in many data sets.
Tue Jun 25 18:22:35 +0000 2019PS: I really don't know how to make this type of resource multiple-license-compliant, or even if "license compliance" is a meaningful concept in this case. Maybe someone who has been involved in FAIR discussions would have some insights?
Tue Jun 25 17:09:20 +0000 2019@Smith_Chem_Wisc There are 2 different relative scales.
For one's own lab:
from 1 (very good) to 5 (excellent)
For all other labs:
from 1 (mainly noise) to 5 (barely adequate)
Tue Jun 25 15:45:33 +0000 20194/4 Given this situation, is there a straightforward method to make a JSON exchange doc (e.g., 🔗) truly compliant with the licenses of original information sources?
Tue Jun 25 15:44:47 +0000 20193/4 Each of the sources has a license that allows for the public use of its information, but each of the licenses is different & most require some type of linking to their license text in order for a use to be compliant.
Tue Jun 25 15:44:29 +0000 20192/4 The information is drawn from a number of information sources: IntAct, OMIM, UniProt, NextProt, Pfam, etc. For me, it raises an interesting practical problem in CC-type licensing.
Tue Jun 25 15:44:13 +0000 20191/4 Recently, a browser extension called "Gene Info" (🔗) became available that allows a user to quickly bring up information about a protein by double-clicking on the name of its gene on any web page.
Tue Jun 25 14:40:22 +0000 2019An interesting take on one of the potential consequences of current attempts to reduce GHG emissions 🔗
Tue Jun 25 14:24:21 +0000 2019Note: there is no proteomics evidence that the CEP63:p splice variant annotated in UniProt and NextProt (ENSP00000336524, 703 aa, TSL:5) is translated.
Tue Jun 25 14:12:19 +0000 2019CEP63:p, centrosomal protein 63 (H. sapiens) 🔗 Small cytoskeletal phosphoprotein; 2 splice variants observed; no high maf SAVs; mature form 1-495 [903 x] 🔗
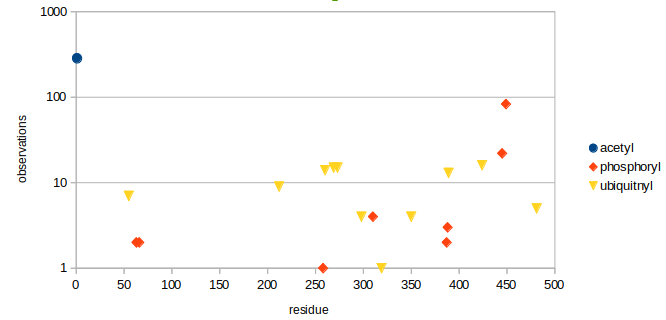
Mon Jun 24 13:29:16 +0000 2019CEP57L1:p, centrosomal protein 57 like 1 (H. sapiens) 🔗 Small cytoskeletal phosphoprotein; rarely identified; no PSM overlap with CEP57; 1 splice variant observed; no high maf SAV; mature form 1-460 [297 x] 🔗

Sun Jun 23 15:15:28 +0000 2019@ucdmrt While they only mention one journal, most high profile journals have very similar editorial practices.
Sun Jun 23 13:44:22 +0000 2019CEP57:p, centrosomal protein 57 (H. sapiens) 🔗 Midsized cytoskeletal protein; many PTMs; 1 splice variant observed; 1 high maf SAV: R448G (maf=0.80); mature form 2-500 [1,734 x] 🔗

Sat Jun 22 17:18:55 +0000 2019My little warning (& last word) regarding succinylation side reactions found when using NHS-ester-based peptide quantitation reagents 🔗
Sat Jun 22 13:37:27 +0000 2019CEP44:p, centrosomal protein 44 (H. sapiens) 🔗 Small cytoskeletal protein; C-terminal phosphodomain; 1 splice variant observed; no high maf SAVs; mature form 2-390 [3,079 x] 🔗
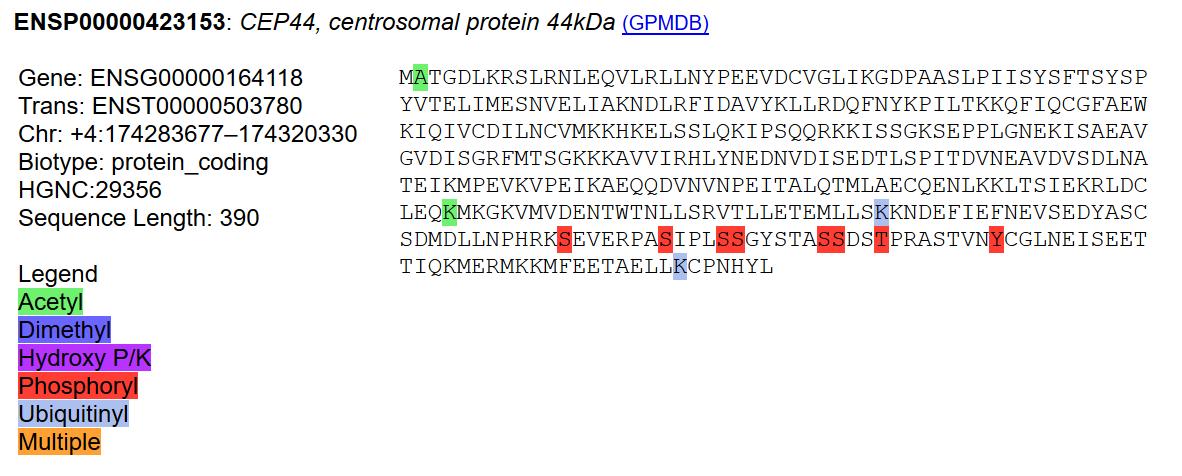
Fri Jun 21 19:24:35 +0000 2019PRIDE should add some sort of question that gently inquires as to how often the submitter cleans their ion sources. Also: Was there an ion source cleaning during the course of acquiring a particular data set?
Fri Jun 21 16:02:45 +0000 2019@PastelBio The list wasn't very interesting. Yours is better 🙂
Fri Jun 21 15:19:07 +0000 2019I have been experimenting with using gab & dissenter as a way of annotating on-line information. Looks very promising.
Fri Jun 21 14:27:41 +0000 2019@UCDProteomics @AlexHgO Most motherboards for those processors allow 128 GB of memory. You may have to be a bit careful about tuning things when you get up to that much memory, but it does work.
Fri Jun 21 12:41:48 +0000 2019CEP41:p, centrosomal protein 41 (H. sapiens) 🔗 Small cytoskeletal phosphoprotein; 11 S-phosphorylation sites; 1 splice variant observed; no high maf SAVs; mature form 2-373 [6,116 x] 🔗

Thu Jun 20 23:32:16 +0000 2019@UCDProteomics I do love my Threadrippers
Thu Jun 20 18:54:44 +0000 2019@UCDProteomics People who use yeast in biomedical research don't like to talk about this subject: they get kind of a far-away look in their eyes and wander off when it is brought up.
Thu Jun 20 18:52:25 +0000 2019@UCDProteomics Almost all yeast strains have significant infections of L-A and L-BC viruses. I've only run across 2 or 3 yeast datasets without strong signals from L-A, L-BC or both. I made them default members of the yeast proteome years ago.
Thu Jun 20 14:58:49 +0000 2019Some simple, solid advice to avoid deserialization problems: 🔗
Thu Jun 20 12:40:23 +0000 2019CEP295:p, centrosomal protein 295 (H. sapiens) 🔗 Large cytoskeletal phosphoprotein; more than 40 K-ubiquitinylation sites; 1 splice variant observed; no high maf SAVs; mature form 1-2601 [1,300 x] 🔗
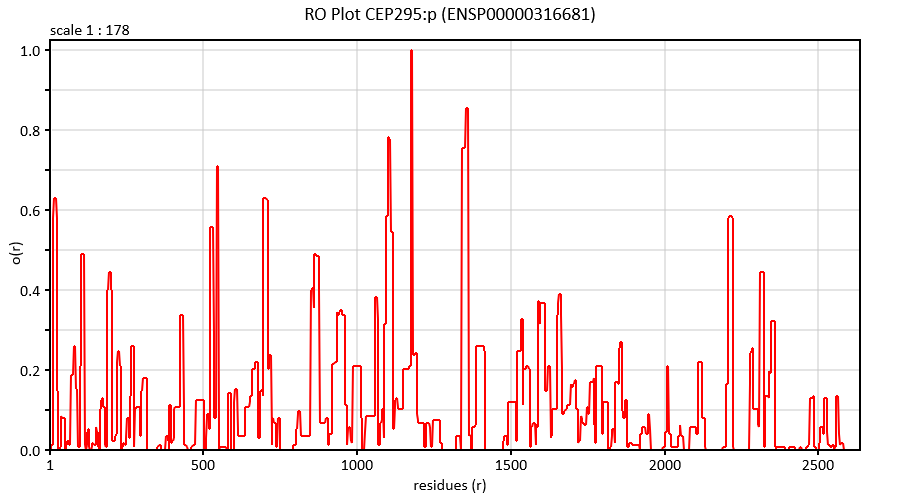
Wed Jun 19 17:29:03 +0000 2019@pwilmarth The caption is "Artist's impression of scientist doing DNA science."
Wed Jun 19 16:54:51 +0000 2019This article lost me at the first image 🔗
Wed Jun 19 12:00:36 +0000 2019CEP162:p, centrosomal protein 162 (H. sapiens) 🔗 Large cytoskeletal protein; 6 S-phosphorylation sites; 1 splice variant observed; no high maf SAVs; mature form 1-1327 [712 x] 🔗
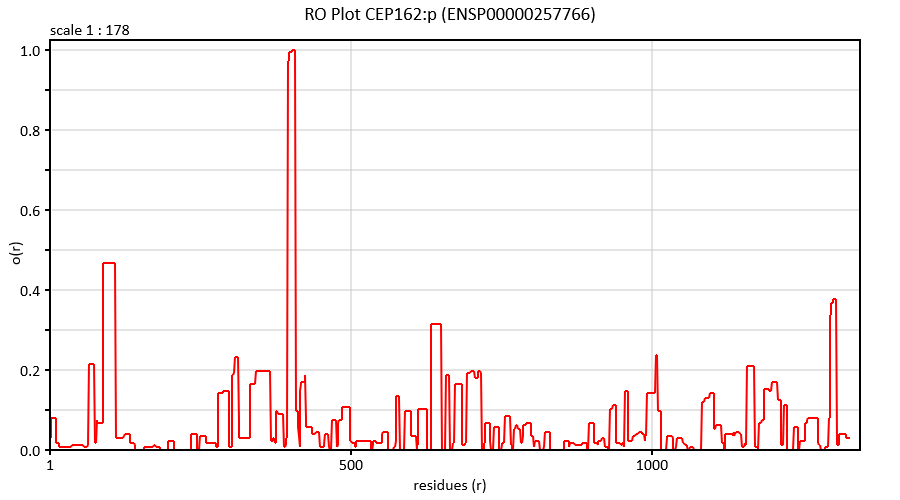
Tue Jun 18 19:38:21 +0000 2019And there it is, loud and proud, in a TMT6 data set.
Tue Jun 18 15:07:27 +0000 2019@astacus @GEEEJAYELL @julieherniman As per 🔗
Tue Jun 18 14:39:15 +0000 2019@astacus @GEEEJAYELL @julieherniman It has been available in the US for several years. Thankfully it has stayed out of the Canadian market so far.
Tue Jun 18 14:19:29 +0000 2019@byu_sam I'll just correct for it in TMT studies in the future. Hopefully it clears up some of the problems using these studies for more detailed PTM and SAV analysis.
Tue Jun 18 14:18:09 +0000 2019@byu_sam No I haven't. It turns out a succinylation artifact is not unusual in TMT10 datasets: it is present at similar levels with similar peptide retention time shifts in all the ones I've checked so far.
Tue Jun 18 12:45:21 +0000 2019CEP128:p, centrosomal protein 128 (H. sapiens) 🔗
Large cytoskeletal protein; significant PTMs; 1 splice variant observed; p.R16Q maf=0.03, p.A451V = 0.01; mature form 2-1094 [1,796 x] (image from String) 🔗
Mon Jun 17 16:58:23 +0000 2019I originally thought it was an in-source decay reaction of the TMT10 derivative, but subsequent investigation of the data showed clearly that it was a reaction that occurred during sample preparation
Mon Jun 17 16:03:08 +0000 2019@DanielJDrucker @jwoodgett @DiabetesCanada @CIHR_IRSC 💔🖤💘
Mon Jun 17 15:24:05 +0000 2019or at least something isobaric with succinylation (+100.016 Da)
Mon Jun 17 15:18:59 +0000 2019Does anybody know what is causing peptide N-terminal succinylation to displace the TMT10 reagent in the latest batch of CPTAC data (LUAD)? The reaction affects 2-5% of the total PSMs.
Mon Jun 17 13:55:57 +0000 2019#required - Error propagation calculations and anything-close-to-correct use of significant figures.
Mon Jun 17 13:54:19 +0000 2019#banned - Unreadable, very detailed hierarchical dendrograms that aren't the basis for a significant conclusion in the paper.
Mon Jun 17 12:48:29 +0000 2019CEP120:p, centrosomal protein 120 (H. sapiens) 🔗 Midsized cytoskeletal protein; significant PTMs; 1 splice variant observed; p.L602V maf=0.41, p.Q879H = 0.21; mature form 1,2-986 [1,442 x] 🔗
Sun Jun 16 17:41:15 +0000 2019What type of informatics analysis would you like to see either #banned or #required in proteomics papers?
Sun Jun 16 13:28:15 +0000 2019CEP55:p, centrosomal protein 55 (Homo sapiens) 🔗 Small cytoskeletal protein; significant PTMs; 1 splice variant observed; p.H378L maf=0.62; mature form 2-464 [6,466 x] 🔗
Sat Jun 15 15:16:25 +0000 2019@pwilmarth In-source chemistry went out of fashion when ion source development money dried up (early 2000's) so no surprise if it was never studied. There were also some nasty legal issues as to who owned the iTRAQ/TMT idea, which wouldn't make it an attractive thing to work on.
Sat Jun 15 15:12:39 +0000 2019@pwilmarth Thanks, Phil. I did a pretty thorough literature search and came up with nothing. I was hoping that somebody looked into it but used some new set of terms to describe what they were doing.
Sat Jun 15 15:08:38 +0000 2019CEP89:p, centrosomal protein 89 (H. sapiens) 🔗 Midsized cytoskeletal protein; N-terminal phosphodomain; 1 splice variant observed; p.V398A maf=0.30; mature form 1,2-783 [1,100 x] 🔗
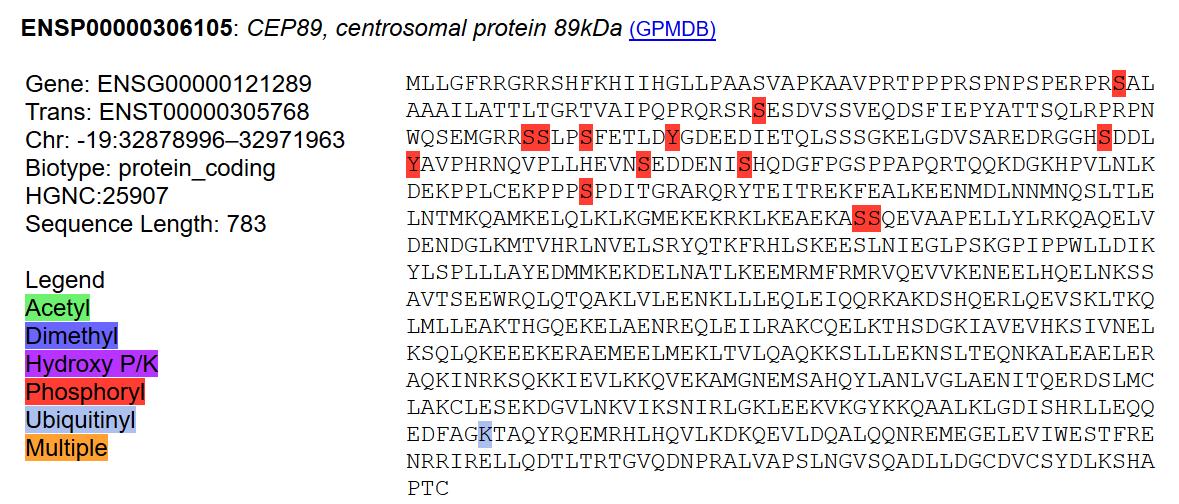
Fri Jun 14 23:14:04 +0000 2019Does anybody know about any studies into in-source decay products generated by TMT10-derivatized peptides?
Fri Jun 14 14:52:53 +0000 2019@MHendr1cks 🔗 Canadian Brain Research Strategy. It looks like an academic lobbying group.
Fri Jun 14 12:56:44 +0000 2019CEP152:p, centrosomal protein 152 (H. sapiens) 🔗 Large cytoskeletal phosphoprotein; 1 splice variant observed; no high maf SAVs; mature form 2-1710 [2,112 x] 🔗
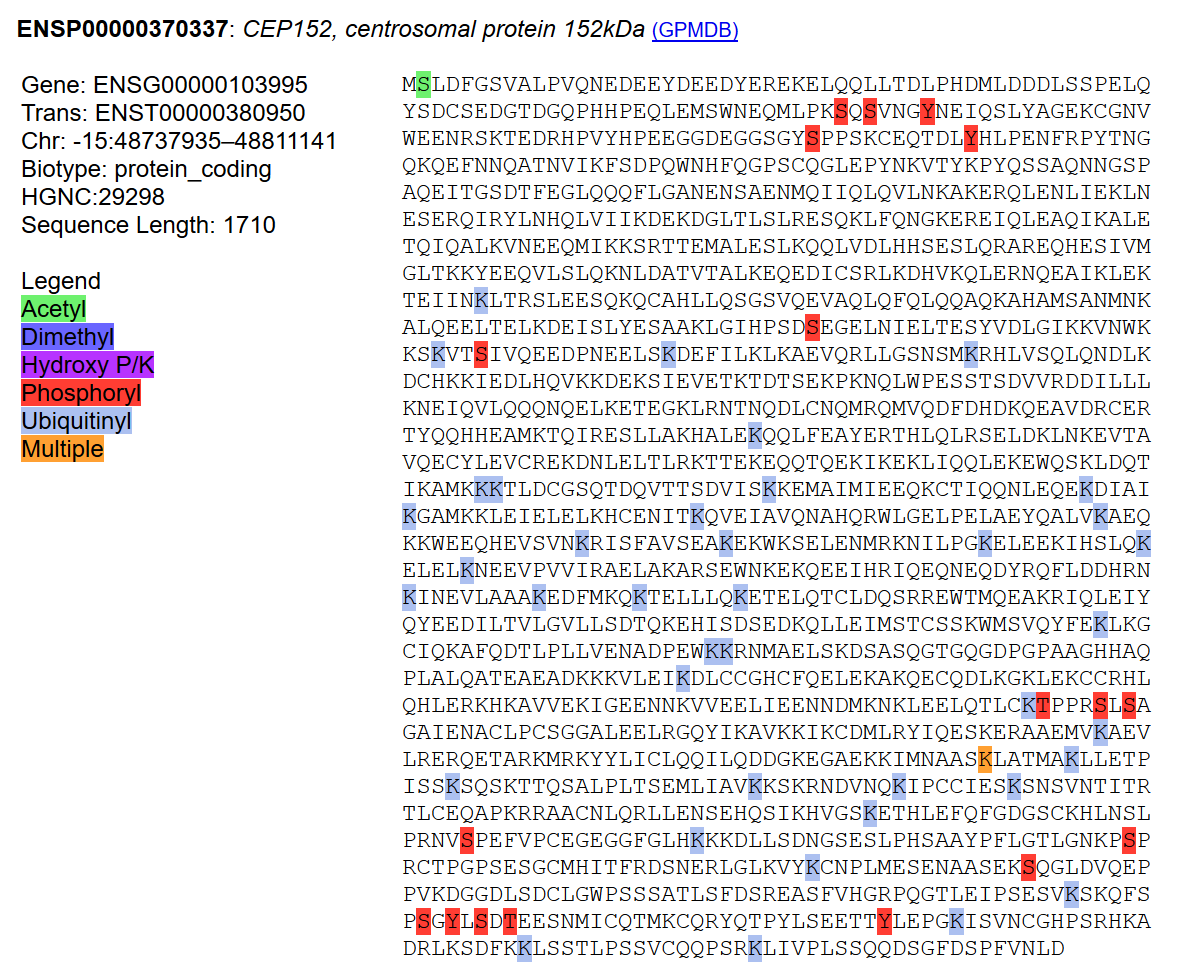
Thu Jun 13 13:32:40 +0000 2019CEP135:p, centrosomal protein 135 (Homo sapiens) 🔗 Large cytoskeletal phosphoprotein; only 1 splice variant observed; 1 high maf SAV, p.I769V (0.14); mature form 2-1140 [2,743 x] 🔗

Wed Jun 12 13:26:29 +0000 2019@nesvilab How does the Elixir program differ from an STTR-type funding mechanism?
Wed Jun 12 13:09:16 +0000 2019CEP192:p, centrosomal protein 192 (Homo sapiens) 🔗 Large cytoskeletal phosphoprotein; many PTMs; only 1 splice variant observed; 8 SAVs with maf > 0.05, e.g. p.R2115Q (0.81), p.R2449L (0.78), p.L2121P (0.77); mature form 1-2537 [4,059 x] 🔗
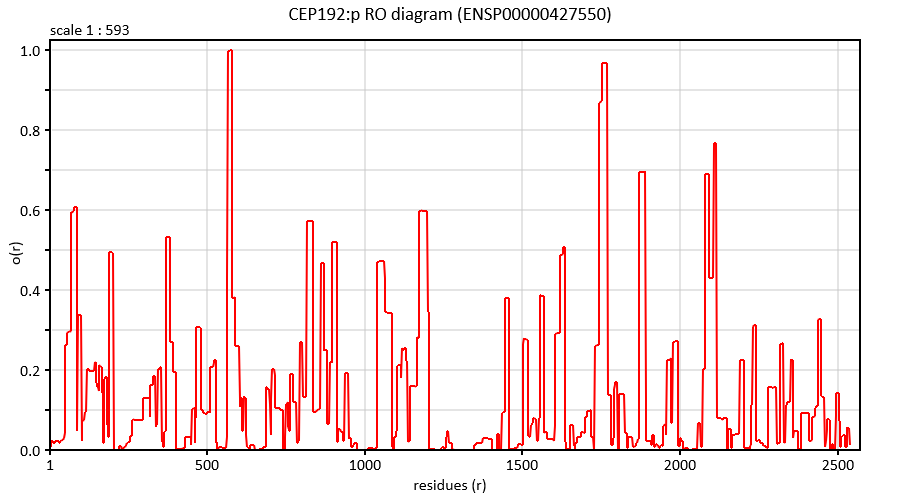
Wed Jun 12 12:32:32 +0000 2019@AlexUsherHESA Whenever this has happened to me, it has always been the result of a wrong turn at Albuquerque
Tue Jun 11 14:00:29 +0000 2019CEP131:p, centrosomal protein 131 (H. sapiens) 🔗 Large cytoskeletal protein; many PTMs; 3 observed splice variants; p.T397A maf = 0.95, p.P446S maf = 0.97; mature form 1-1080 [8,947 x] 🔗
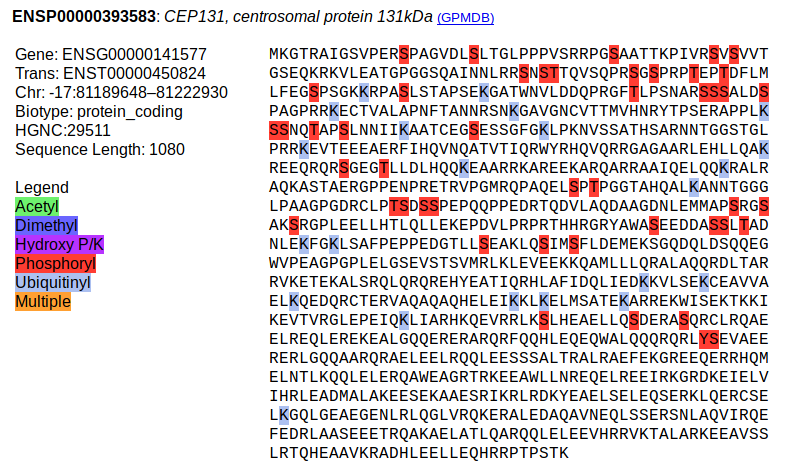
Mon Jun 10 13:27:14 +0000 2019CEP290:p, centrosomal protein 290 (H. sapiens) 🔗 Large cytoskeletal protein; several low occupancy PTMs; aka KIAA0373, FLJ13615, 3H11Ag, rd16, NPHP6, JBTS5, SLSN6, LCA10, MKS4, BBS14, CT87, POC3; mature form 2-2481 [2,238 x] 🔗
Sun Jun 09 14:04:38 +0000 2019CEP85:p, centrosomal protein 85 (H. sapiens) 🔗 Midsized cytoskeletal protein; at least 15 S/T phosphorylation and 7 ubiquitinylation sites; mature form 2-762 [3,903 x] 🔗
Sat Jun 08 16:23:42 +0000 2019CEP97:p, centrosomal protein 97 (H. sapiens) 🔗 Large cytoskeletal protein; at least 35 S/T phosphorylation sites; most common SAV = p.V319L, maf = 0.02; mature form 2-865 [5,131 x] 🔗
Fri Jun 07 21:02:52 +0000 2019@dtabb73 I think you left out the word "evil".
Fri Jun 07 20:02:12 +0000 2019Some days a few overly enthusiastic REST users seem to take up all of my time.
Fri Jun 07 14:27:05 +0000 2019@slavovLab The whole article is simply nonsense.
Fri Jun 07 12:44:47 +0000 2019CEP350:p, centrosomal protein 350 (H. sapiens) 🔗 Very large cytoskeletal phosphoprotein; more than 30 ubiquitinylation sites; no commonly observed SAVs; mature form 1-3117 [5,840 x] 🔗
Thu Jun 06 12:41:14 +0000 2019CEP170:p, centrosomal protein 170 (H. sapiens) 🔗 Large cytoskeletal protein; highly phosphorylated; most abundant SAV: p.M1275V maf = 0.01; mature form 2-1584 [19,743 x] 🔗

Wed Jun 05 14:29:27 +0000 2019TO: any one who makes E. coli proteomics data available on PRIDE
PLEASE: include strain/biovar information.
E. coli (+ Shigella) is a generic name for many different genomes and proteomes 🔗
Wed Jun 05 12:53:21 +0000 2019CEP250:p, centrosomal protein 250kDa (H. sapiens) 🔗 Large cytoskeletal protein; only significant PTMs in C-terminal phosphorylated domain; no high maf SAVs; mature form 1-2442 [5,264 x] (image from String) 🔗

Tue Jun 04 15:09:03 +0000 2019@jwoodgett It should be a part of grad student training. Any time a job candidate tried to show me a PowerPoint when visiting me in my office, I lost interest in them as a potential colleague/employee.
Tue Jun 04 15:06:20 +0000 2019Oh those Silicon Valley guys ... 🔗
Tue Jun 04 13:32:07 +0000 2019PCM1:p, pericentriolar material 1 (H. sapiens) 🔗 Large cytoskeletal protein; extensive PTMs: more than 60 ubiquitination sites; many SAVs: e.g. M597V maf = 0.8; mature form 2-2024 [19,670 x]; image: HLA peptides from glioblastoma (red) 🔗
Mon Jun 03 14:50:40 +0000 2019GLIPR2:p, GLI pathogenesis-related 2 (H. sapiens) 🔗
Small membrane protein; significant PTMs; no high maf SAVs; common in most tissues and cell lines; mature form 2-154 [12,409 x] 🔗

Sun Jun 02 16:38:45 +0000 2019@pauldauenhauer I'm pretty sure that any character that has to be escaped for regular expression matching will have the same effect, e.g. (, ), + or \
Sun Jun 02 13:43:31 +0000 2019Image shows HLA class I peptides (red) found for GLIPR1:p in melanoma tissue: data from 🔗 🔗
Sun Jun 02 13:40:20 +0000 2019GLIPR1:p, GLI pathogenesis-related 1 (H. sapiens) 🔗 Small protein with a signal peptide sequence; no significant PTMs; no high maf SAVs: found in hematopoietic cells, stem cells, some cancers and cell lines; mature form 22-266 [1,598 x]
Sat Jun 01 12:39:09 +0000 2019MAP10:p, microtubule associated protein 10 (H. sapiens) 🔗 Large cytosolic protein; no observed PTMs; least frequently observed of the MAPs: not found in most common cell lines; mature form 1-1047 [179 x] (image from String) 🔗
Fri May 31 13:31:53 +0000 2019@bkives Great story. Any chance you could identify at least 1 person who confided in the premier about how upset they were the prospect of a provincial election in the same year as Manitoba's 150th anniversary? I suspect their initials are all BWP ...
Fri May 31 12:47:55 +0000 2019MAP9:p, microtubule associated protein 9 (H. sapiens) 🔗 Large protein; 7 S-phosphorylation acceptor sites; no high maf SAVs; mature form 2-647 [979 x]; rarely observed compared to other MAPs (image from SubCellBarCode) 🔗
Thu May 30 14:31:49 +0000 2019MAP1S:p, microtubule associated protein 1S (H. sapiens) 🔗 Large proprotein; extensive PTMs; several high maf SAVS, p.S411C maf=0.11, p.C440Y maf=0.14; mature form 2-829 (HC), 830-1059 (LC) [20,047 x]; aka MAP8:p 🔗
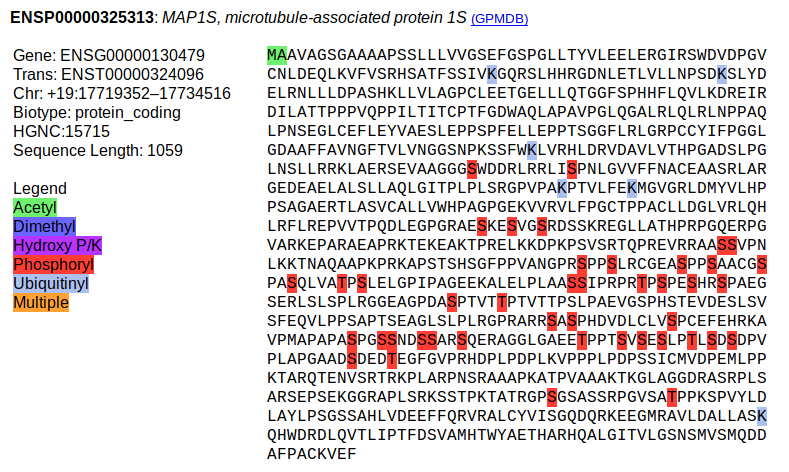
Wed May 29 16:21:31 +0000 2019@jwoodgett @CIHR_IRSC The language and rights granted to 'Bang the Table Pty Ltd' (particularly in section 19) are very different than CIHR's own terms of use 🔗
Wed May 29 15:52:43 +0000 2019@jwoodgett @CIHR_IRSC You should carefully read the terms of use 🔗 for that site before entering your information into it. You may not realize exactly who is getting the information and how it is being stored and distributed.
Wed May 29 12:42:22 +0000 2019MAP7D3:p, MAP7 domain containing 3 (H. sapiens) 🔗 Midsize protein; extensive PTMs; p.E502A maf=0.55; several low complexity domains; mature form 3-876 [6,492 x]; observed in most tissues and cell lines (image from String) 🔗

Tue May 28 13:19:30 +0000 2019MAP7D2:p, MAP7 domain containing 2 (H. sapiens) 🔗 Midsize protein; few PTMs; few SAVs; several low complexity domains; mature form 1-773 [2,096 x]; observed in testis and CNS tissue & many cancers/cell lines (image from String) 🔗
Tue May 28 12:24:00 +0000 2019@dtabb73 It is a pretty common motif in plants
Mon May 27 17:16:23 +0000 2019@nesvilab I thought Nature Methods had jumped the shark years ago, but they truly are setting new standards: publishing two trendy sounding methods for gaming FDR-based calculations.
Mon May 27 15:04:13 +0000 2019@IRPP @ISED_CA @FraunhoferISI A nice discussion of Canada's current situation with respect to the federal government's under-performing interventions in technology development.
Mon May 27 13:24:12 +0000 2019MAP7D1:p, MAP7 domain containing 1 (H. sapiens) 🔗 Midsize putative nuclear protein; highly phosphorylated; many low maf SAVs; significant low complexity domains; mature form 1-841 [15,356 x] 🔗
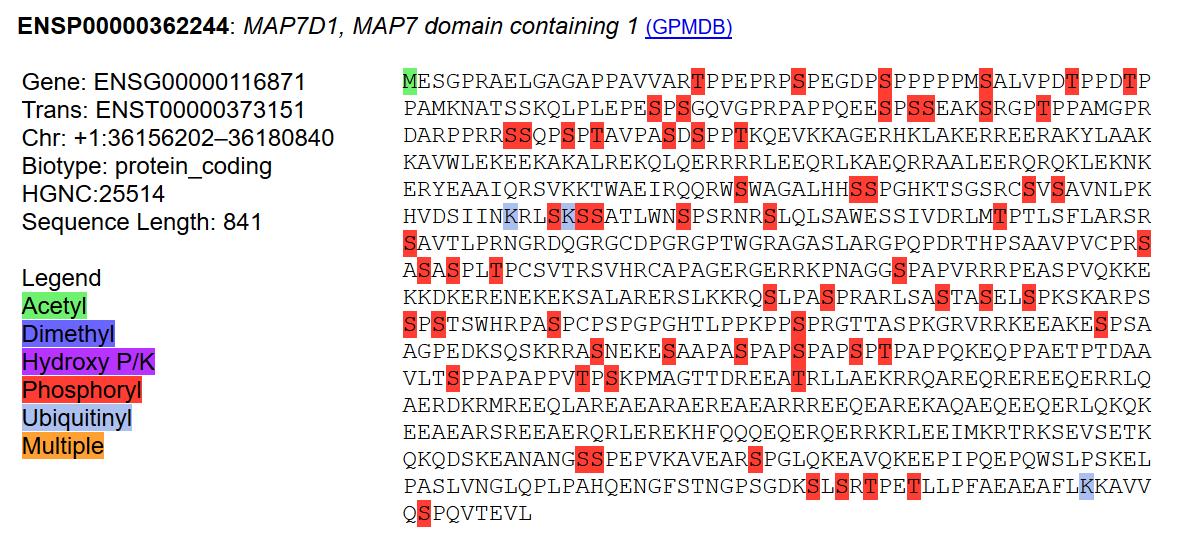
Sun May 26 14:22:14 +0000 2019MAP7:p, microtubule associated protein (H. sapiens) 🔗 Midsize cytoskeleton protein; many phosphorylations in 3 domains; many low maf SAVs; found in many tissues and cell lines; mature form 2-749 [15,488 x] 🔗
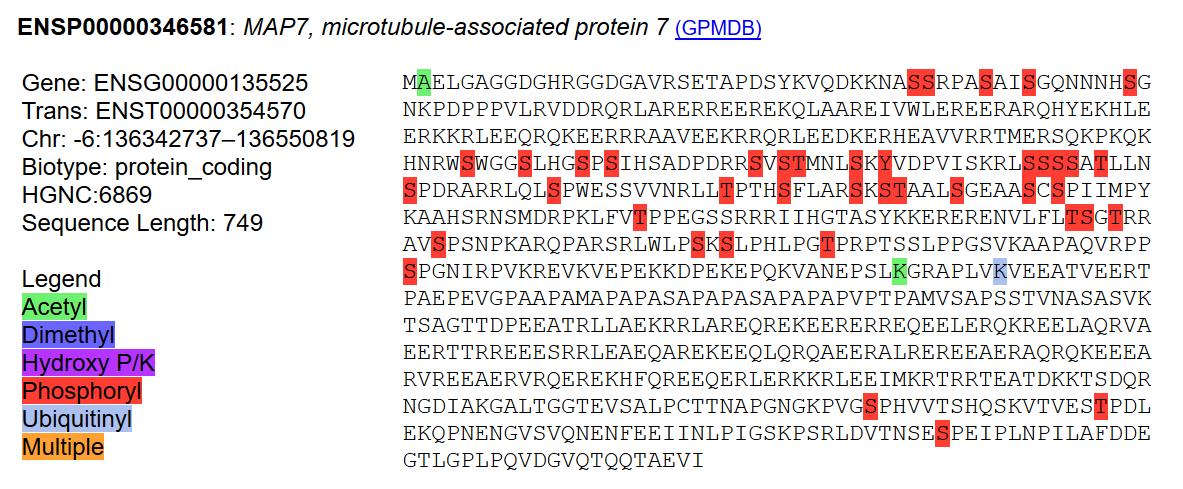
Sat May 25 13:37:51 +0000 2019MAP6D1:p, MAP6 domain containing 1 (H. sapiens) 🔗 Small putative cytoskeleton protein; few PTMs; no high maf SAVs; only observed in CNS tissue and some tumors; mature form 2-199 [363 x] 🔗
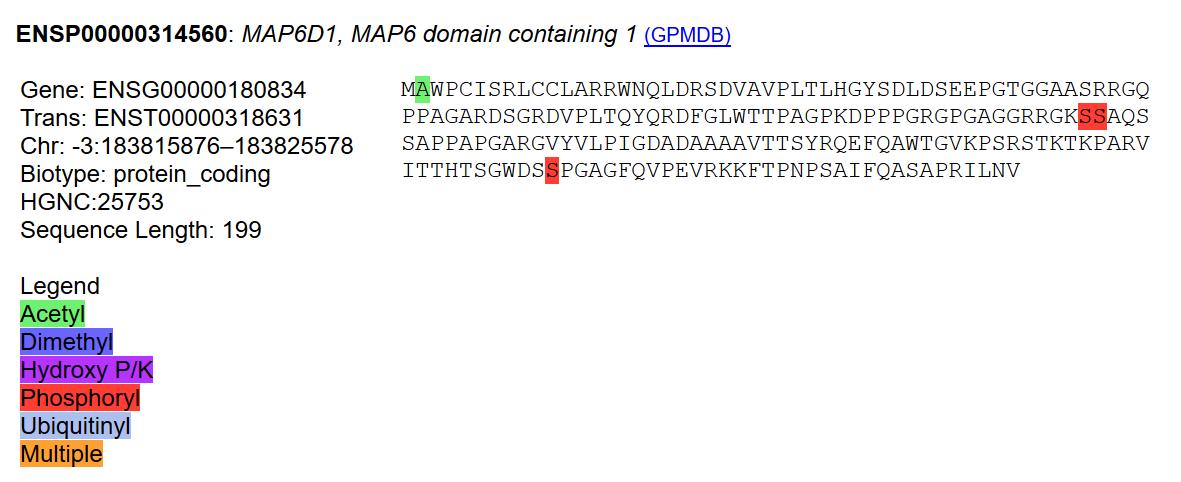
Fri May 24 15:02:10 +0000 2019The process sounds a lot like what happened to create the current "Supercluster" train wreck 🔗
Fri May 24 14:56:57 +0000 2019Only found in CNS tissue and some tumors, particularly in melanoma and colon cancer samples. A shorter (484 aa) splice variant is also commonly observed along with the 813 aa form.
Fri May 24 14:50:38 +0000 2019MAP6:p, microtubule associated protein 6 (H. sapiens) 🔗 Midsized cytoskeleton protein; few PTMs; p.I247M maf = 0.45; mature form 2-813 [1,293 x] 🔗
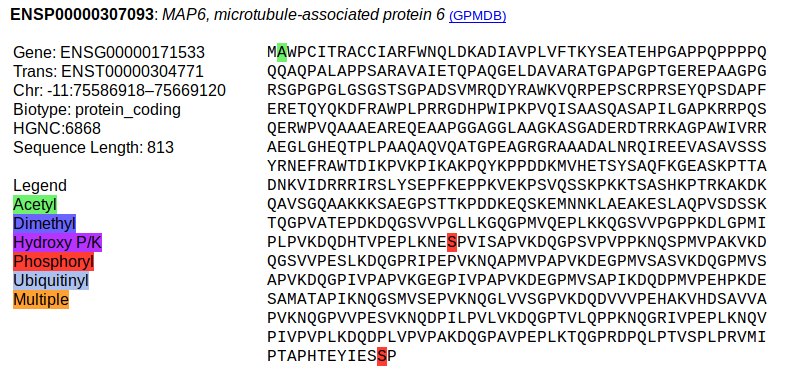
Thu May 23 15:30:01 +0000 2019The dataset PXD010982 (🔗, 🔗, human hair) is a good example of how complicated it can be to distinguish between closely related proteins even with lots of sample & relatively few distinct gene products present.
Thu May 23 13:12:28 +0000 2019MAP4:p, microtubule associated protein 4 (H. sapiens) 🔗 Large cytoskeleton protein; many PTMs, many low maf SAVs, highest maf = 0.4 p.V628I; mature form 2-1152; more than 1 splice variant frequently observed [47,141 x] 🔗

Wed May 22 14:32:52 +0000 2019MAP2:p, microtubule associated protein 2 (H. sapiens) 🔗 Large cytoskeleton protein; about 70 S and 30 T phosphorylation sites and only N-terminal acetylation; many low maf SAVs, highest maf = 0.04 p.M1099V; mature form 2-1827 [7,013 x] 🔗
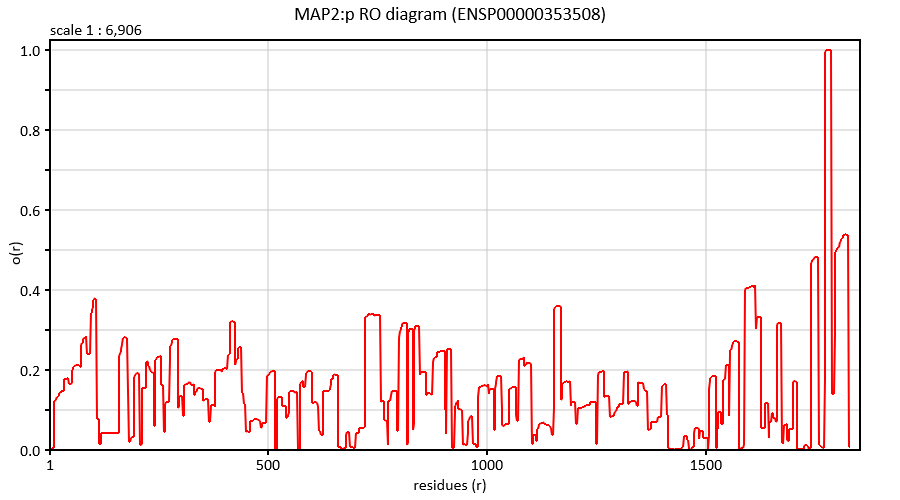
Tue May 21 21:00:08 +0000 2019Nothing stays the same (not even the kilogram, second, meter, ampere, kelvin or mole) 🔗
Tue May 21 14:20:17 +0000 2019Fool me once ... 🔗
Tue May 21 13:23:23 +0000 2019MAP1B:p, microtubule associated protein 1B (H. sapiens) 🔗 Large cytoskeleton proprotein; >250 phosphoryl and >90 acetyl acceptor sites; many low maf SAVs, except for p.I594V maf=0.90; mature forms 2-2206 (HC) 2207-2468 [16,650 x]
Mon May 20 13:29:49 +0000 2019MAP1A:p, microtubule associated protein 1A (H. sapiens) 🔗 Large cytoskeleton protein; >100 phosphorylation acceptor sites; many low maf SAVs, e.g., p.T1277A maf=0.01; mature forms 2-2803 (HC), 2567-2803 (LC) [16,650 x] image from String 🔗
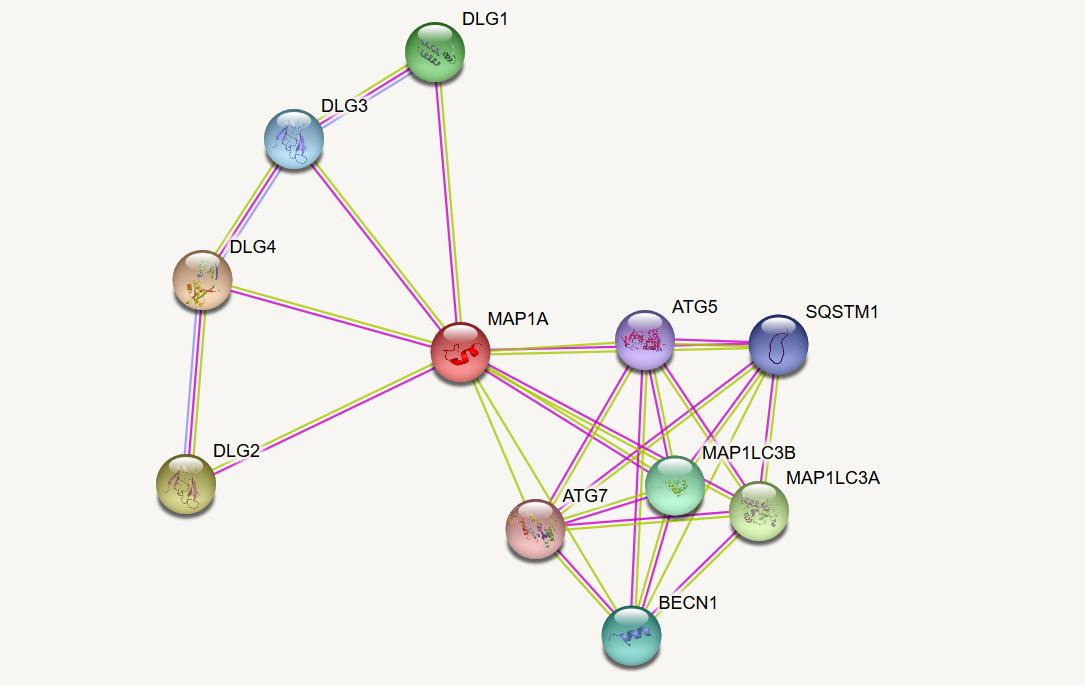
Sun May 19 21:02:47 +0000 2019@pwilmarth It was a requirement I never really understood, as I have found that different people "experience" the same annotated images differently.
Sun May 19 19:36:44 +0000 2019@pwilmarth How many journals still require manuscript packages that contain images of annotated spectra?
Sun May 19 16:28:42 +0000 2019Thanks to everyone who participated in the poll and the associated discussion. The results are interesting: apparently the descriptive text in a FASTA file is, at best, of secondary interest. What is desired is an identifier for use in performing further analysis.
Sun May 19 13:55:30 +0000 2019MTMR4:p, myotubularin related protein 4 (H. sapiens) 🔗 Large enzyme; some S/T phosphorylation; 2 high frequency maf SAVs, p.L170V and p.S280G; mature form 2-1195 [2,175 x] 🔗

Sat May 18 19:14:06 +0000 2019I've probably complained about this before, by why is it so hard for people to understand/remember the difference between quality assurance and quality control?
Sat May 18 17:14:11 +0000 2019@nesvilab Many people still privately hope for the return of the halcyon days the "Central Dogma". Replacing "protein" with "gene" quietly moves things in that direction. And practically, many downstream resources (GO, pathways, etc) are already there: they only use gene names.
Sat May 18 15:01:37 +0000 2019What information obtained from a protein's description line in a FASTA-formatted sequence listing file is most important to you?
Sat May 18 13:57:04 +0000 2019MTMR3's size and PTM pattern suggest it is significantly different from MTM1, MTMr1 and MTMR2.
Sat May 18 13:53:09 +0000 2019MTMR3:p, myotubularin related protein 3 (H. sapiens) 🔗 Large enzyme; extensive S/T phosphorylation; highest maf SAV: p.N960S maf=0.04; mature form 1-1198 [6,055 x] 🔗
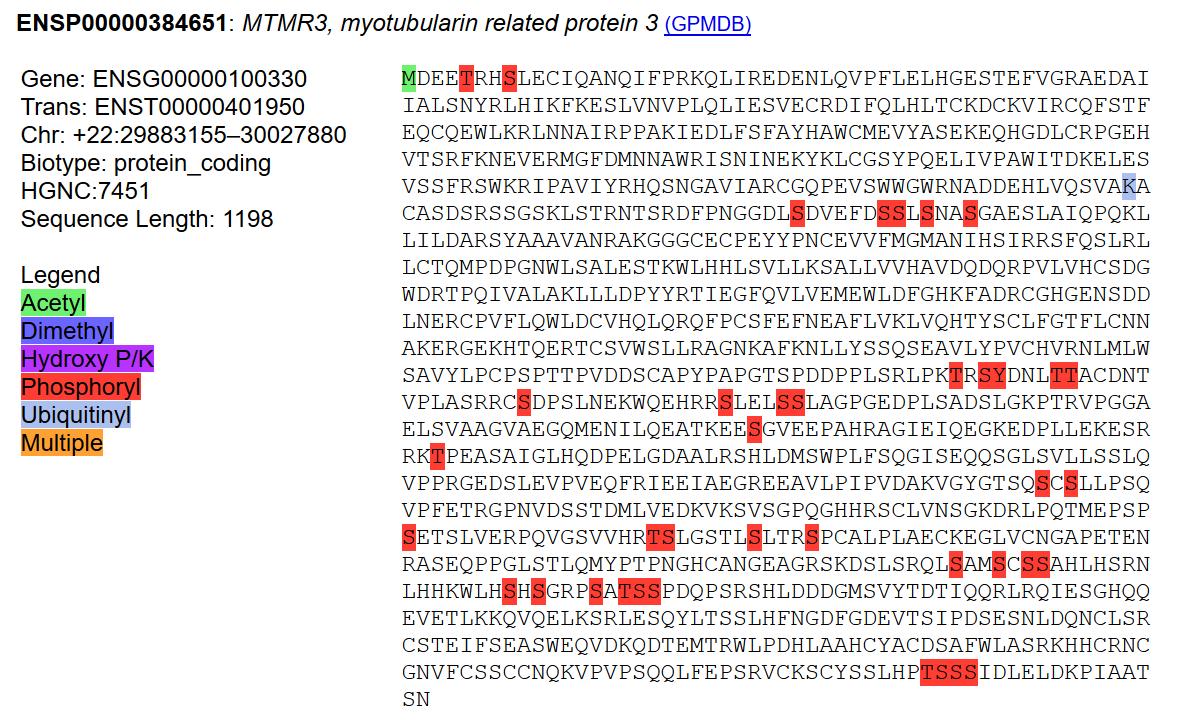
Fri May 17 12:25:08 +0000 2019MTMR2:p, myotubularin related protein 2 (H. sapiens) 🔗 Midsized phosphatase; N-terminal and C-terminal phosphodomains; SAV p.K3T maf=0.2; mature form 1-643 [5,213 x] 🔗
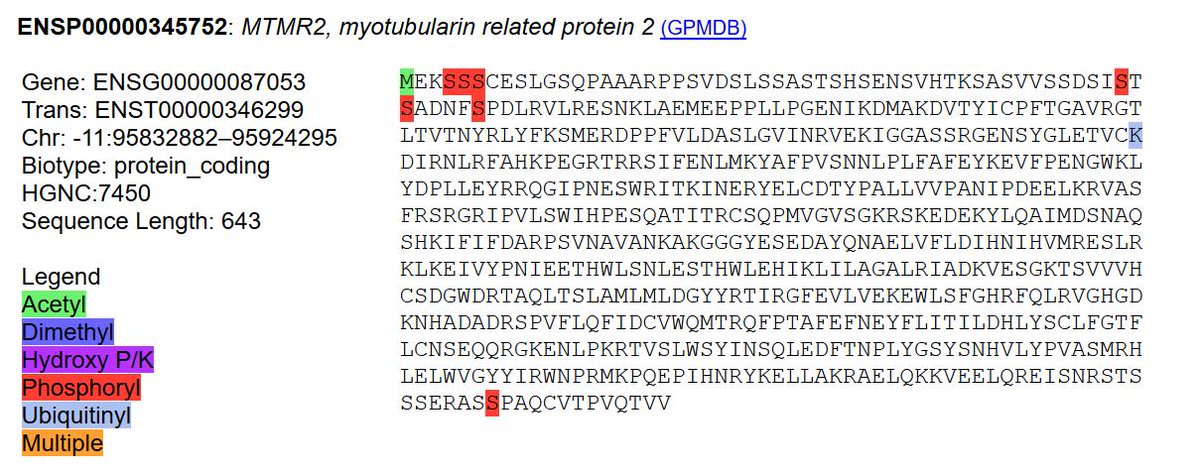
Thu May 16 20:22:18 +0000 2019@AlexUsherHESA The lack of research funding has lead to an increase in activities that coincidentally are more likely to generate citations than new research, such as writing review articles.
Thu May 16 18:03:26 +0000 2019For non-Canadians, the original contract was for $5.7 million to fully replace an existing payroll management system: 🔗
Thu May 16 13:01:12 +0000 2019MTMR1:p, myotubularin related protein 1 (H. sapiens) 🔗 Midsized phosphatase; N-terminal and C-terminal phosphodomains; no high maf SAVs; mature form 1-665 [7,621 x] 🔗
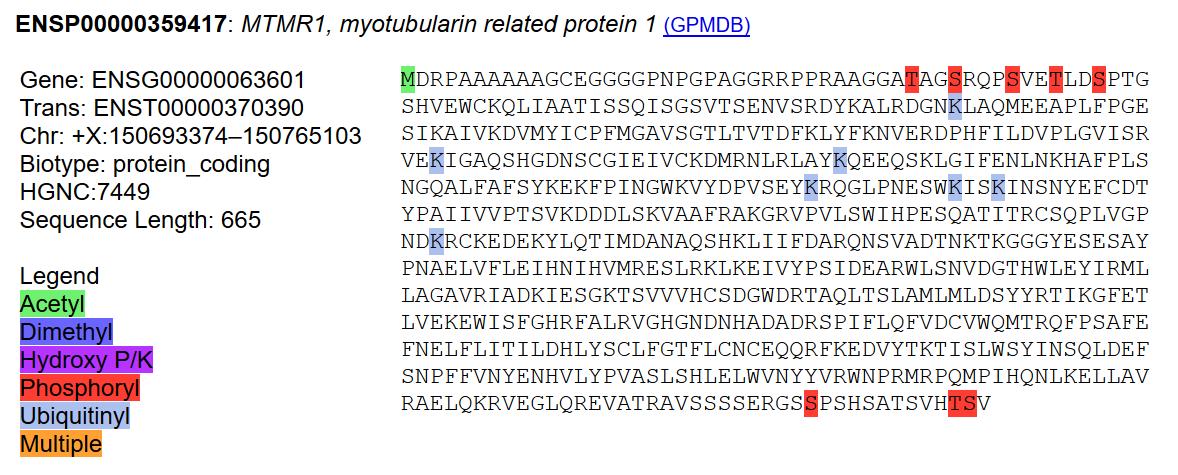
Wed May 15 12:32:48 +0000 2019MTM1:p, myotubularin 1 (H. sapiens) 🔗 Endosomal phosphatase; N-terminal and C-terminal phosphodomains; no high maf SAVs; mature form 2-603 [6,606 x] 🔗
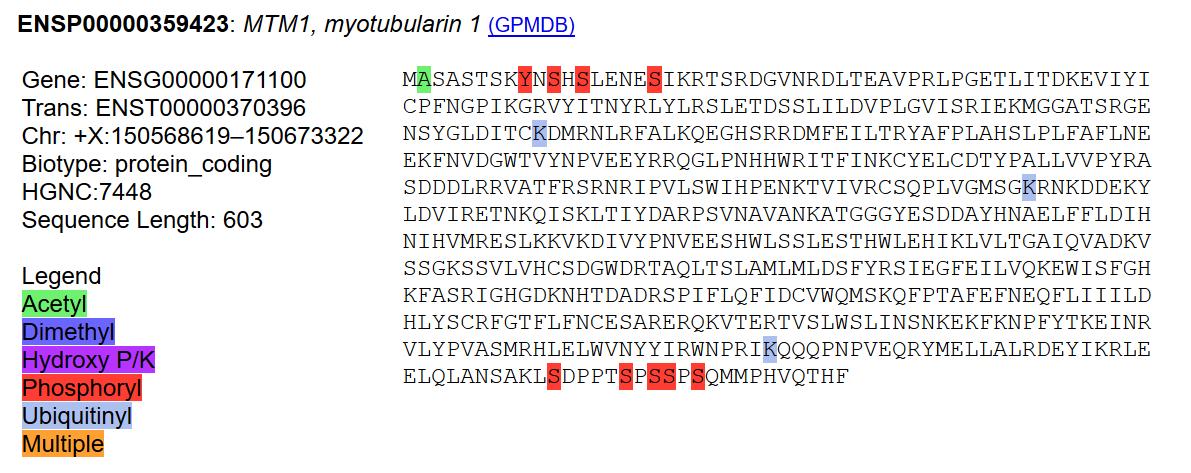
Tue May 14 13:02:10 +0000 2019PTPMT1:p, protein tyrosine phosphatase, mitochondrial 1 (H. sapiens) 🔗 Putative mitochrondrial matrix enzyme; only PTM is A2+acetyl; no high maf SAVs; proteomics information does not support a MT targeting sequence; mature form 2-201 [6,290 x] 🔗
Mon May 13 17:46:37 +0000 2019@Gundrylab @jvaneyk1 The discoverer of Omenn Syndrome?
Mon May 13 15:59:46 +0000 2019@slavovLab @emblebi @CellPressNews There were a lot of sore feelings when IPI was discontinued. I guess some people still haven't gotten over it.
Mon May 13 14:19:39 +0000 2019COX7C:p, cytochrome c oxidase subunit 7C (H. sapiens) 🔗 Very small mitochondrial complex IV matrix subunit; no PTMs; no high maf SAVs; mature form 17-63 [5,837 x]; one of the smallest protein subunits commonly observed. 🔗
Sun May 12 15:36:36 +0000 2019The alpha release of my most recent PSM assignment software (SE) is now available at 🔗 It is a relatively straightforward proteomics MS/MS search engine, written in Python 3.
Sun May 12 12:56:33 +0000 2019COX8A:p, cytochrome c oxidase subunit 8A (H. sapiens) 🔗 Very small mitochondrial complex IV matrix subunit; no PTMs; no high maf SAVs; mature form 26-69 [1,855 x] 🔗
Sat May 11 17:00:57 +0000 2019@chrashwood @JohnRYatesIII JPR seems to be doing a pretty good job of it already!
Sat May 11 12:19:28 +0000 2019COX7B:p, cytochrome c oxidase subunit 7B (H. sapiens) 🔗 Very small mitochondrial complex IV matrix subunit; no PTMs; no high maf SAVs; mature form 25-80 [1,221 x] 🔗
Fri May 10 16:53:34 +0000 2019@TrostLab @KentsisResearch To be sure, I grabbed a run from PXD010621 & ranked the proteins by # of phospho-PSMs detected. Of the top 20
SRRM2
NCL
TRIM28
HSP90AA1
SRRM1
TCOF1
MKI67
AKAP12
BCLAF1
ATRX
MSH6
PCM1
RANBP2
ZC3H13
MAP1B
TRP53BP1
NPM1
TJP2
NUCKS1
RIF1
all but 2 are nuclear.
Fri May 10 15:45:32 +0000 2019@TrostLab @KentsisResearch SRRM1, SRRM2, NUCKS1, BRACA1, BRACA2, TP53, all of the heterogeneous ribonuclear proteins, all of the HISTs, all of the SNRNPs, NPM1, TRIM28, AHNAK, NUMA1, all of the PRPFs, most DEAH-box and DEAD-box proteins, all CHDs, all MCMs, etc.
Fri May 10 14:01:57 +0000 2019@TrostLab @KentsisResearch The most abundant (& most extensively modified) phospho-proteins tend to be localized to the nucleus, not the cytosol.
Fri May 10 12:43:15 +0000 2019COX7A2:p, cytochrome c oxidase subunit 7A2 (H. sapiens) 🔗 Very small mitochondrial complex IV matrix subunit; some Y-phosphorylation; commonly observed; mature form 56-115 [16,237 x] 🔗

Thu May 09 19:52:26 +0000 2019Working on one of my least-favorite-things-to-do: writing release documentation.
Thu May 09 15:41:35 +0000 2019But for me the most interesting thing is that there are effectively no new phosphorylation sites discovered in all of this data. It may be that the phase of discovering new phosphorylation sites is now over.
Thu May 09 15:38:51 +0000 2019The phosphorylation data in 🔗 is really good. About 98% of the PSMs are phosphopeptides, the MS/MS and chromatography are excellent and there is lots of it: each mESC sample has 12 fractions and each MS/MS run has > 27,000 PSMs.
Thu May 09 13:01:48 +0000 2019COX7A1:p, cytochrome c oxidase subunit 7A1 (H. sapiens) 🔗 Very small mitochondrial complex IV matrix subunit; no PTMs; rarely observed in cell lines; mature form 22-79 [1,086 x] 🔗
Wed May 08 18:05:44 +0000 2019@UCDProteomics @chrashwood @olgavitek @KhouryCollege @Northeastern You should never underestimate the paranoia of biomedical researchers.
Wed May 08 17:33:22 +0000 2019@cstross Google, for me, gave a somewhat more on-the-nose answer in first place, "Fulminant acute colitis following a self-administered hydrofluoric acid enema"
🔗
Wed May 08 14:42:53 +0000 2019COX6B2:p, cytochrome c oxidase subunit 6B2 (H. sapiens) 🔗 Small mitochondrial complex IV intermembrane subunit; no PTMs; no high maf SAVs; very rarely observed [32 x] 🔗
Tue May 07 12:53:23 +0000 2019COX6B1:p, cytochrome c oxidase subunit 6B1 (H. sapiens) 🔗 Small mitochondrial complex IV intermembrane subunit; surprising number of PTMs; no high maf SAVs; mature form 2-86 [18,208 x] 🔗

Mon May 06 22:36:10 +0000 2019@Ciencia_2017 @Smith_Chem_Wisc @ProtifiLlc No idea at all. But there was zero interest in using it by anyone, except one guy at Sciex (back in 2005 when it was still MDS-Sciex).
Mon May 06 20:55:05 +0000 2019@Smith_Chem_Wisc @ProtifiLlc I still use library searches myself (I don't think I could run GPMDB without them), but I gave up on trying to promote the idea.
Mon May 06 20:18:40 +0000 2019@Smith_Chem_Wisc @ProtifiLlc After about a decade of trying as hard as possible to promote the method, I finally realized no one wants to use library searches and showing them how well it works would never change their minds.
Mon May 06 18:53:27 +0000 2019@ProtifiLlc It is unfortunate that the proteomics community is so dead set against using spectrum library ID methods.
Mon May 06 16:51:05 +0000 2019Thanks to everyone who voted. The rankings in the human proteome for these four proteins is as follows (#1 would have the highest PSM count):
TP53: #997
NUP214: #1715
SOD1: #1780
INS: #6592
Mon May 06 15:12:00 +0000 2019For anyone who is in Winnipeg and would like to attend, Ken Standing's memorial will be held at the Pavilion Atrium in Assiniboine Park, 55 Pavilion Crescent, this evening starting at 7:00 PM
Mon May 06 14:33:12 +0000 2019There are still two hours left in this poll and insulin seems to really need a friend ...
Mon May 06 14:31:56 +0000 2019COX6A1 and COX6A2 do not share any observable tryptic peptides. In tissues where COX6A2 is expressed, it does not replace COX6A1: both are present in the tissue. COX6A2 is not observable in any commonly used human cell line.
Mon May 06 12:31:32 +0000 2019COX6A2:p, cytochrome c oxidase subunit 6A2 (H. sapiens) 🔗 Small mitochondrial complex IV matrix subunit; no PTMs; no high maf SAVs; only observed in striated muscle [182 x] 🔗
Sun May 05 19:31:35 +0000 2019@chrashwood @nesvilab Bioinformatics is just like any other field: if you have one group doing something exclusively (no matter how well they are doing it), funding agencies can get a little squirrelly about support because of a perceived lack of interest in the community.
Sun May 05 18:45:27 +0000 2019@chrashwood @nesvilab A bit of competition isn't going to hurt anything. There are lots of search engines, so why not a few raw file convertors? The file conversion process is often a bit of a bottle-neck.
Sun May 05 18:08:52 +0000 2019@nesvilab The new parser has worked for me on Linux. It still has an issue or two, but nothing that should be a show-stopper.
Sun May 05 16:43:45 +0000 2019If you look across all public proteomics data, which human protein in the following list has the most identified PSMs:
Sun May 05 12:49:24 +0000 2019COX6A1:p, cytochrome c oxidase subunit 6A1 (H. sapiens) 🔗 Small mitochondrial complex IV matrix subunit; no PTMs; no high maf SAVs; mature form 24,25-109 [3,486 x] (image from String) 🔗
Sat May 04 13:17:37 +0000 2019COX5B:p, cytochrome c oxidase subunit 5B (H. sapiens) 🔗 Small mitochondrial complex IV matrix subunit; several PTMs; no high maf SAVs; mature form 31,32,33-129 [22,168 x] 🔗

Fri May 03 18:29:36 +0000 2019@Peptidome Thanks for letting me know. Best of luck with your experimental design efforts.
Fri May 03 18:21:44 +0000 2019@Peptidome If you'd let me know when you've got a copy of the file so I can take it off-line, I would appreciate it.
Fri May 03 16:54:39 +0000 2019@ptrainnv You would think this was from April 1st, but unfortunately it is not.
Fri May 03 16:09:32 +0000 2019@Peptidome A selection of semi-tryptic peptides (mainly caused by protein N-terminal processing) is also in the list.
Fri May 03 16:08:29 +0000 2019@Peptidome The column heads:
Protein: UP accession
Mass (z=0): neutral mass (mDa)
Start: position of the 1st residue
End: position of the last residue
Pre: residue preceding
Peptide: sequence
Post: residue following
n(z=+1): # observations with z = +1
& n(z=+2),n(z=+3),n(z=+4)
Fri May 03 16:04:39 +0000 2019@Peptidome The list found at 🔗 gives all of the tryptic peptides observed from human proteins aligned with the latest human UP accessions, along with the number of times they have been observed with z= +1,+2,+3 or +4.
Fri May 03 15:23:32 +0000 2019@nesvilab @pwilmarth Simply providing a short list of alternate parent ion tuples (m/z,z,I) for an MS/MS spectrum would cover most cases.
Fri May 03 14:47:56 +0000 2019@pwilmarth It certainly wouldn't hurt to have that additional information.
Fri May 03 14:24:09 +0000 2019COX5A:p, cytochrome c oxidase subunit 5A (H. sapiens) 🔗 Small mitochondrial complex IV matrix subunit; several PTMs; no high maf SAVs; mature form 41,42,44-150 [24,305 x] 🔗

Thu May 02 15:19:01 +0000 2019@nesvilab In the words of Patrick Swayze's character (Dalton) in the movie Roadhouse: opinions vary.
Thu May 02 14:55:17 +0000 2019Something else I don't understand about COX4I2 is its String interaction diagram. Even though I2 is very rare, it seems to have a very similar interaction diagram compared to I1.
Thu May 02 14:33:26 +0000 2019@Smith_Chem_Wisc And thanks to everyone who participated, both in the poll and the discussion.
Thu May 02 14:32:53 +0000 2019@Smith_Chem_Wisc I think this poll got buried a bit, but a majority stated that they don't use the "two peptide rule" when assigning PSMs to proteins. It has strong defenders, however, so I would have to conclude that opinion on the heuristic is both mixed and polarized.
Thu May 02 14:07:52 +0000 2019COX4I2:p is not found in cell lines: it is only found in brain and some female-specific tissues. It was not found by the Human Protein Atlas project. Unlike I1, I2 lacks a mitochondrial targeting peptide, so it's proposed localization to mitochondria is hard to explain.
Thu May 02 13:58:53 +0000 2019COX4I2:p is a bit of an oddball. It is named as isoform 2 (I2) of COX4, but it is very rarely found in humans; the ratio of observations is COX4I1:COX4I2 = 27,880:71. I2 doesn't share any observable tryptic peptides with I1, so there is no chance of confusion.
Thu May 02 12:57:02 +0000 2019COX4I2:p, cytochrome c oxidase subunit 4I2 (H. sapiens) 🔗 Small putative mitochondrial complex IV matrix subunit; no PTMs; no high maf SAVs; mature form 1-171 [71 x] (image from String) 🔗
Thu May 02 00:20:18 +0000 2019@Smith_Chem_Wisc Still no one voting for the two peptide rule: I guess it may have died out.
Wed May 01 18:44:27 +0000 2019@TheFirstNuomics @ypriverol It installed and ran without any problems on Ubuntu 18.04. Thanks, guys.
Wed May 01 18:06:07 +0000 2019@BrenesAlejandro It may be useful, but PRIDE may then be liable to somehow verify that a user will agree to the software license that you must agree to when downloading MaxQuant from its distribution site.
Wed May 01 17:47:17 +0000 2019Is it OK that the FTP directory corresponding to this entry seems to have a distribution copy of MaxQuant in it? 🔗
Wed May 01 17:37:56 +0000 2019@UCDProteomics @TrostLab How about 3 decimal places & state that masses are to the nearest mDa?
Wed May 01 16:35:56 +0000 2019@Smith_Chem_Wisc So far there isn't a lot of love for the "two peptide rule" ...
Wed May 01 15:29:15 +0000 2019@pwilmarth @Smith_Chem_Wisc As it is a rule-of-thumb, I think it's use is more a matter of belief (the dreaded "common sense") than a product of thoughtful study. I do know people who have been forced to use it by reviewers, though.
Wed May 01 14:13:59 +0000 2019@Smith_Chem_Wisc Does your group use the "two peptide rule" as an input when deciding the validity of the protein assignments generated from a set of PSMs?
Wed May 01 12:46:38 +0000 2019COX4I1:p, cytochrome c oxidase subunit 4I1 (H. sapiens) 🔗 Small mitochondrial complex IV matrix subunit; many PTMs; no high maf SAVs; mature form 23-169 [27,880 x] 🔗

Wed May 01 12:12:32 +0000 2019@Smith_Chem_Wisc I've never used it either. I agree that it would be interesting to know how widely used it is in practice.
Tue Apr 30 18:58:13 +0000 2019@HFazelinia @BrenesAlejandro And I am not mocking "value-priced" Lys-C. The use of Lys-C with trace enzymes that cleave at other sites has considerably widened the list of observable peptides and consequently the sequence coverage for proteins with membrane-spanning domains.
Tue Apr 30 18:41:03 +0000 2019@HFazelinia @BrenesAlejandro Doll S, et al. Nat Commun. 2017 (PXD006675, 🔗) generated the best MT-ND6 ids in heart that I've seen. There is only 1 good tryptic peptide, but there are several semi-tryptic peptides that show up if you use "value-priced" Lys-C.
Tue Apr 30 17:47:21 +0000 2019@Smith_Chem_Wisc I think the CV reference citation would be well worth the shade. Most people (at a minimum all of the authors) don't care about the consequences of heavy-tailed distributions in protein biochemistry.
Tue Apr 30 17:19:29 +0000 2019Bad things always happen when chemists try to count things other than atoms or electrons 🔗
Tue Apr 30 15:29:11 +0000 2019@BrenesAlejandro Then you should really go back and try to figure out why you are missing MT-CO3 and MT-ND4 (human MT-ND4L has no observable tryptic peptides), because they are both present in similar amounts to their other colleagues.
Tue Apr 30 14:55:36 +0000 2019@BrenesAlejandro Check to see how many of the other MT-genome encoded proteins are present in the results (MT-ND1,MT-ND2,MT-ND3,MT-ND4,MT-ND4L,MT-ND5,MT-ND-6,MT-CO1,MT-CO2,MT-CYB,MT-ATP6 & MT-ATP8). Most data sets will have MT-CO2, but get dodgy fast with the others.
Tue Apr 30 14:11:42 +0000 2019@lkpino Yes, it is a mess - particularly the protein-protein interaction correlations. But, to be fair, none of the authors are really experts in biological data interpretation.
Tue Apr 30 13:04:25 +0000 2019This peculiarity (shared with a set of other proteins) also means it has a tendency to "come-and-go" from data sets, giving the impression of differential expression when naïvely interpreting experiments with low numbers of replicates.
Tue Apr 30 12:25:51 +0000 2019Note: this protein will often be excluded from results because of the "2 peptide" heuristic. The protein is present at in every cell that has mitochondria, but it has few observable peptides.
Tue Apr 30 12:16:07 +0000 2019MT-CO3:p, mitochondrially encoded cytochrome c oxidase III (H. sapiens) 🔗 Small mitochondrial complex IV inner membrane subunit; Y67+phosphoryl; no SAVs; 7 membrane spanning domains; mature form 1,2-221 [3,140 x] 🔗
Mon Apr 29 19:01:52 +0000 2019@georgecolombo @doctorow I have a soft spot for TOS on the Atari ST line: it was about a decade ahead of its time for a personal computer.
Mon Apr 29 14:46:42 +0000 2019MT-CO2:p, mitochondrially encoded cytochrome c oxidase II (Homo sapiens) 🔗 Small mitochondrial complex IV inner membrane subunit; a very-hard-to-explain PTM, K171+ubiquitinyl; no high maf SAVs; 3 membrane spanning domains; mature form 1-227 [28,725 x] 🔗
Mon Apr 29 14:19:35 +0000 2019@MikeTheBiochem I try to move things into protein (& genome) coordinates as soon as possible. It may seem like an unnecessary complication, but it lessens confusion later on, particularly when dealing with protein sequence variants due to alternate splicing.
Mon Apr 29 14:07:53 +0000 2019This notational heterogeneity must make it challenging for anyone trying to compare results between publications or data obtained from different facilities.
Mon Apr 29 14:05:17 +0000 2019Thanks to everyone who sent in answers to my question yesterday about PTM notation in spreadsheets. The consensus seemed to be using notation inline with the PSM peptide sequence, with individual groups using different approaches to represent the mods.
Sun Apr 28 18:13:16 +0000 2019@ProtifiLlc @pwilmarth How do you deal with multiple mods on a single residue using this style of annotation?
Sun Apr 28 17:36:38 +0000 2019@MikeTheBiochem So something like:
"M625+Oxidation;M628+Oxidation"
in a cell to indicate 2 oxidized Met residues at protein coordinates 625 and 628 in the peptide
621 DNSTMGYMAAK 631.
Sun Apr 28 13:31:32 +0000 20192/2 Does anyone have a favorite method of jotting down all of the modifications in a clear format to achieve this end? I am currently using 🔗, but I'm open to any other suggestions.
Sun Apr 28 13:28:35 +0000 20191/2 One of the most popular output formats for proteomics PSM results is a rectangular table. This format is a challenge when trying to report the modifications associated with a particular PSM, because the mods must be all reported in 1 cell.
Sun Apr 28 12:39:20 +0000 2019One of the 13 mitochondrial chromosome encoded proteins; few observable tryptic peptides because of numerous hydrophobic domains.
Sun Apr 28 12:31:35 +0000 2019MT-CO1:p, mitochondrially encoded cytochrome c oxidase I (Homo sapiens) 🔗 Midsized mitochondrial complex IV inner membrane subunit; no PTMs; no SAVs; 12 membrane spanning domains; mature form 1-513 [3,688 x] 🔗
Sat Apr 27 13:52:35 +0000 2019Top of a yellow letter on a white background, photo from 10x magnification, Samsung Galaxy Tab S2 screen. 🔗
Sat Apr 27 13:17:13 +0000 2019Why are biomedical researchers so fond of using Roman numerals when naming things? Was it a fad by some group of people who were forced to take Latin when they were at school? Or maybe a cynical ploy to evade text mining?
Sat Apr 27 13:14:14 +0000 2019UQCR11:p, ubiquinol-cytochrome c reductase, complex III subunit XI (H. sapiens) 🔗 Very small mitochondrial complex III intermembrane subunit; no PTMs; no SAVs; aka QCR10; mature form 1,2-56 [2,293 x] 🔗
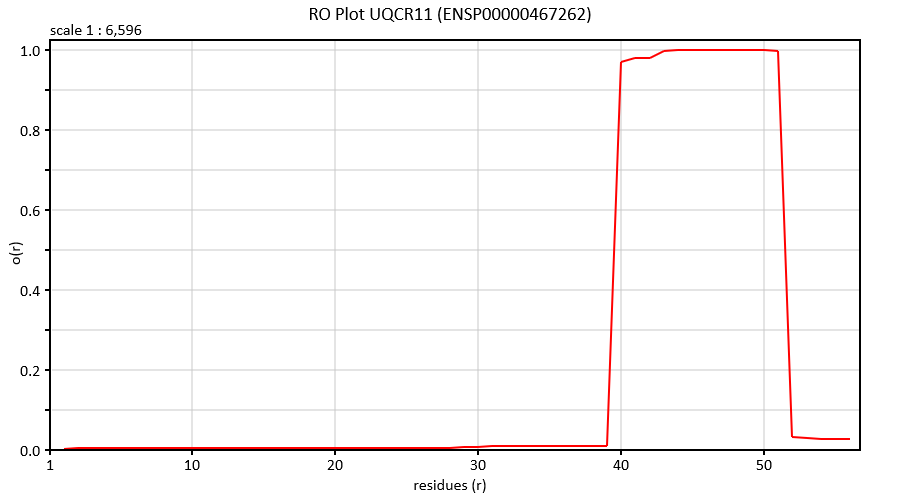
Fri Apr 26 12:17:45 +0000 2019UQCR10:p, ubiquinol-cytochrome c reductase, complex III subunit X (H. sapiens) 🔗 Very small mitochondrial complex III intermembrane subunit; 1 low occupation PTM, Y44+phosphoryl; p.I47V maf=0.07; aka UQCR9; mature form 2-63 [8,239 x] 🔗
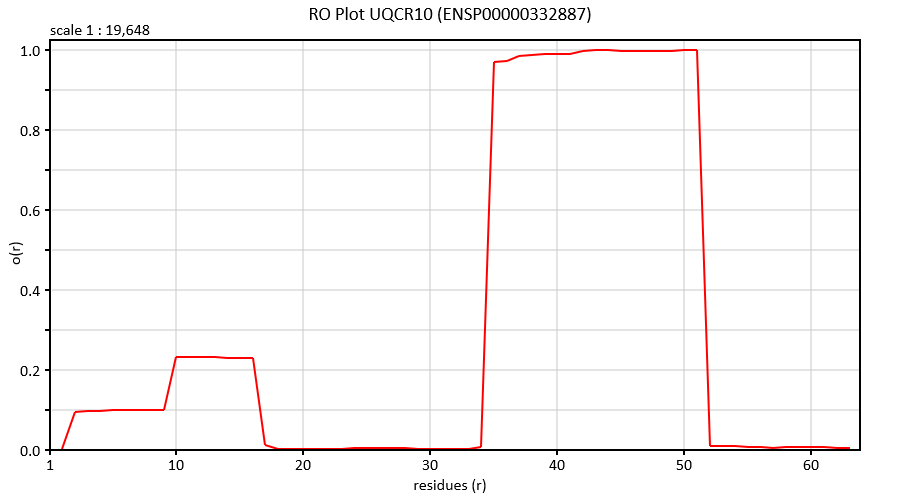
Thu Apr 25 23:35:00 +0000 2019UQCRH:p, ubiquinol-cytochrome c reductase hinge protein (H. sapiens) 🔗 Very small mitochondrial complex III intermembrane subunit; many PTMs; no high maf SAVs; mature form 1,2-91 [12,917 x] 🔗
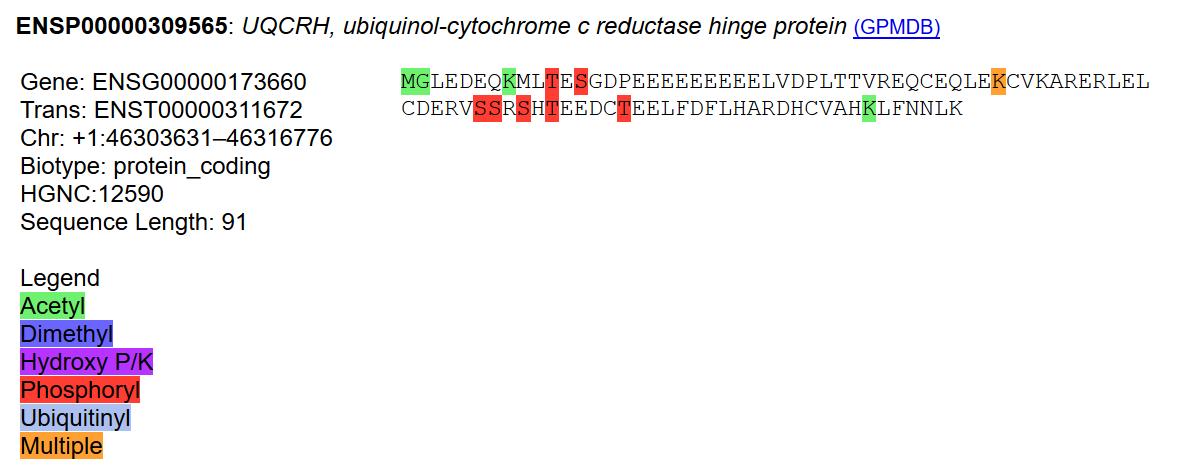
Thu Apr 25 22:48:10 +0000 2019@UCDProteomics @nesvilab @byu_sam @pwilmarth Disease-causing missense alleles are pretty rare in the population. And finding a specific SAV in proteomics data is unlikely, but you can find a set of higher MAF ones in any good data set. You only need ~10 SAVs with MAFs < 0.1 to ID a person.
Thu Apr 25 22:16:34 +0000 2019@UCDProteomics @nesvilab @byu_sam @pwilmarth The only ones relevant for identifying individuals are the neutral missense variants, which is a more manageable 66,191.
Thu Apr 25 16:42:11 +0000 2019@UCDProteomics @pwilmarth I would say we are able to identify people with proteomics data right now so long as the data is good enough, based on detected SAVs. You don't have to find very many SAVs with significant MAFs in the population to ID an individual.
Wed Apr 24 16:10:17 +0000 2019Thanks to everyone who participated in the poll as well as those who added comments.
Wed Apr 24 12:54:33 +0000 2019UQCRB:p, ubiquinol-cytochrome c reductase binding protein (Homo sapiens) 🔗 Small mitochondrial complex III intermembrane subunit; highly modified (4 Y+phosphoryl!); no SAVs with significant mafs; mature form 2-111 [16,830 x] 🔗

Tue Apr 23 18:58:49 +0000 2019@nesvilab Unfortunately Twitter polls only allow 4 choices.
Tue Apr 23 17:54:48 +0000 2019@neely615 What makes collagen an issue is that there is so much of it in many types of tissue. It's abundance in solid tissue samples is akin to albumin's abundance in plasma. 🔗
Tue Apr 23 17:11:41 +0000 2019@neely615 I think any protein with a collagen-like domain and some GXPG sites that goes through the right cellular export machinery can end up with hydroxyproline and hydroxylysine PTMs
Tue Apr 23 16:18:51 +0000 2019@PastelBio @ProteomicsNews And for another view of the same issue: 🔗
Tue Apr 23 15:59:52 +0000 2019Collagen peptides with the PTM hydroxyproline and can be > 10% of IDs in many cancer tissue samples.
Tue Apr 23 15:55:47 +0000 2019How does your proteomics PSM-assignment system cope with the fact that collagens (e.g., COL1A1) are often the most abundant proteins in tissue and they have many high-occupation sites for rare PTMs?
Tue Apr 23 12:57:37 +0000 2019Note: UQCRFS1 is listed in some sources as having a mitochondrial transit peptide removed in the mature form. This is not the case: the protein does not enter the mitochondrial matrix (it stays in the intermembrane region), so the transit peptide is not cleaved.
Tue Apr 23 12:52:50 +0000 2019UQCRFS1:p, ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 (H. sapiens) 🔗 Small mitochondrial complex III intermembrane subunit; 3 phosphorylation sites; p.S6A maf=0.9; mature form 1-274 [22,956 x] 🔗

Mon Apr 22 17:09:16 +0000 2019UQCRC2:p, ubiquinol-cytochrome c reductase core protein 2 (H. sapiens) 🔗 Small mitochondrial complex III subunit; 8 T and 4 Y phosphorylation sites; R254H maf=0.07-0.26; mature form 15-453 [36,876 x] 🔗

Sun Apr 21 13:54:56 +0000 2019@AlexUsherHESA The whole debate reminds me of watching Réal Caouette on TV explaining Social Credit with visual aids: lots of talk, numbers and arrows pointing every-which-way, but no one being convinced on either side.
Sun Apr 21 13:49:02 +0000 2019UQCRC1:p, ubiquinol-cytochrome c reductase core protein 1 (H. sapiens) 🔗 Small mitochondrial complex III subunit; 4 Y+phosphoryl sites; no high maf SAVs; mature form 35-480 [33,915 x] 🔗
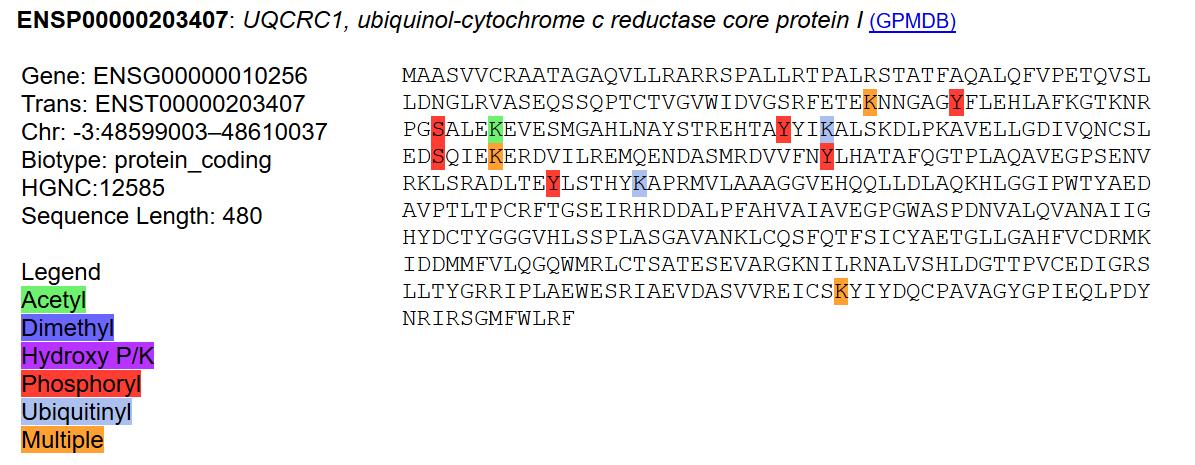
Sat Apr 20 13:50:04 +0000 2019@manojkushwaha63 Are you asking whether or not a search engine should select which ion series to consider, based on whether there is a pattern of matches?
Sat Apr 20 12:09:47 +0000 2019UQCRQ:p, ubiquinol-cytochrome c reductase, complex III subunit VII (H. sapiens) 🔗 Very small mitochondrial complex III subunit; N-terminal acetyl & low occupancy phosphorylations; no high maf SAVs; mature form 2-82 [12,427 x] 🔗
Fri Apr 19 22:04:43 +0000 2019SDHA:p, succinate dehydrogenase complex flavoprotein subunit A (H. sapiens) 🔗 Midsized mitochondrial matrix subunit; multiple acetylations and phosphorylations; V657I, maf = 0.13; mature form 33-664 [38,850 x] 🔗

Fri Apr 19 14:58:24 +0000 2019It seems there is no consensus in the community about interpreting the significance of fragment ion counts, although many had an opinion. The same number of people (within error) voted for ">4" as voted for ">7".
Thu Apr 18 18:11:31 +0000 2019The early results are looking pretty stochastic ...
Thu Apr 18 17:07:17 +0000 2019@pwilmarth @UCDProteomics @byu_sam Hopefully most student (& post docs) will have to ask what those extensions mean ...
Thu Apr 18 16:08:26 +0000 2019Based on people's comments on their feelings about an ion count of 4, I guess I should have had ">3" on the list.
Thu Apr 18 15:42:32 +0000 2019@UCDProteomics While the information and conclusions generated from the data should stand the test of time, the data itself starts to look "quaint" after about 5 years.
Thu Apr 18 15:40:51 +0000 2019@UCDProteomics When anyone asks my advice about this topic (& hopefully that is the case here), I suggest having a stated policy on data retention. For example: The Facility will retain data for 5 years after the analysis was performed.
Thu Apr 18 14:20:52 +0000 2019PS: I know "it depends on a lot of things". But as a general heuristic, what choice would you be comfortable with telling a trainee or collaborator.
Thu Apr 18 14:18:53 +0000 2019When interpreting PSM's generated by a search engine, how many "ion counts" (matches between observed MS/MS fragment ions and the y/b ions predicted from a peptide sequence) are necessary to have confidence in the result?
Thu Apr 18 12:43:53 +0000 2019Highly unusual in that of the 13 confidently assigned phosphorylation sites, all 13 are on threonine residues. Also note that the SAV H445R has a maf = 0.99.
Thu Apr 18 12:40:20 +0000 2019GC:p, GC, vitamin D binding protein (H. sapiens) 🔗 Found in blood and in plasma-derived fluids; multiple acetylations and phosphorylations; 3 high maf SAVs: H445R, D432E, T436K; mature form 17-474 [24,453 x] 🔗
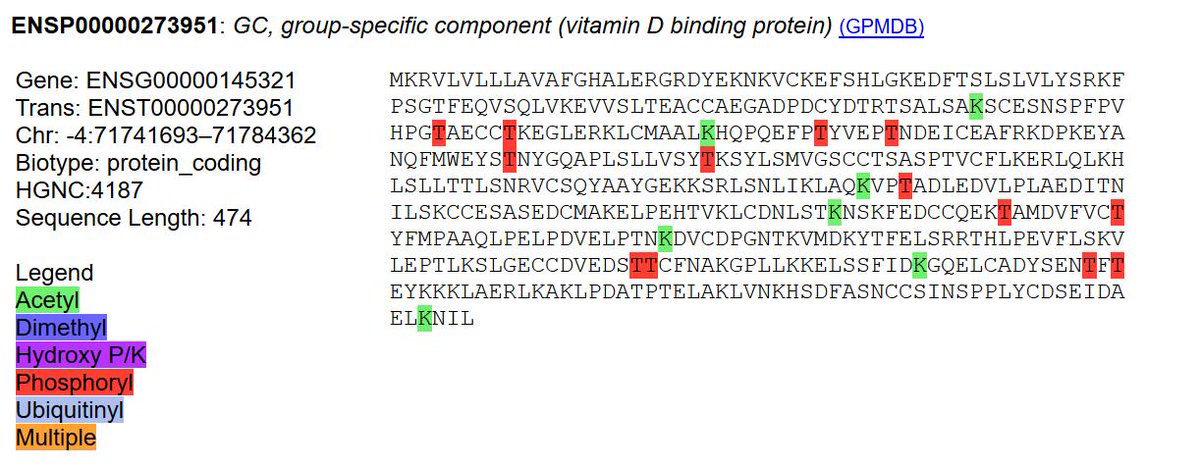
Wed Apr 17 15:37:23 +0000 2019@slavovLab @francesarnold After having had numerous conversations with Nobel laureates during my career, my best advice to any student would be "DO NOT TALK TO A NOBEL LAUREATE UNDER ANY CIRCUMSTANCES"
Wed Apr 17 13:39:12 +0000 2019NDUFB2:p, NADH:ubiquinone oxidoreductase subunit B2 (H. sapiens) 🔗 Very small mitochondrial inner membrane subunit; no PTMs; no high maf SAVs; observed mature form 33,34-105 [1,058 x] (image from String) 🔗
Wed Apr 17 13:35:41 +0000 2019Thanks everybody who participated in the poll as well as everybody that contributed to the discussion. I found it informative.👩🔬👨🔬
Wed Apr 17 01:55:02 +0000 2019It has tightened up a bit, but b & y +2 ions are still out in front ...
Tue Apr 16 16:30:26 +0000 2019It seems as though using b & y +2 ions has taken the lead, although it was tied with neutral losses earlier on ...
Tue Apr 16 14:11:58 +0000 2019@Smith_Chem_Wisc The practice of annotating these ions in displays (I do this too) also muddy the water: users tend to believe these annotations are from the associated search engine, rather than post hoc assignments made by the spectrum drawing software.
Tue Apr 16 14:07:05 +0000 2019@Smith_Chem_Wisc I have found that people that might be considered as "power users" (as opposed to "developers") often have adamant beliefs on this topic. Both you and Dave seem to be voting "None", but we'll see how the poll works out.
Tue Apr 16 13:14:08 +0000 2019What ion types should be considered by a peptide MS/MS search engine when assigning a PSM in addition to b/y (or c/z) +1 ions:
Tue Apr 16 12:45:43 +0000 2019NDUFB1:p, NADH:ubiquinone oxidoreductase subunit B1 (H. sapiens) 🔗 Very small mitochondrial inner membrane subunit; N-terminal acetylation; no high maf SAVs; observed mature form 1,2-58 [5,285 x]
Tue Apr 16 00:51:54 +0000 2019Or if you distrust/hate ftp, try 🔗
Mon Apr 15 23:45:14 +0000 2019Anyone who is interested in making constructive suggestions about columns to add to a search engine result TSV file, please take a look at ftp://ftp.thegpm.org/projects/se/results/se_example.tsv and let me know.
Mon Apr 15 21:14:41 +0000 2019I was pleasantly surprised by how easy it was to use Cython to convert my Python modules into compiled C binaries (both on Linux and Windows).
Mon Apr 15 16:24:20 +0000 2019@dtabb73 When I have taught intro R workshops, the first thing I told the class is that students having previous experience with other programming languages are probably going to be at a disadvantage.
Mon Apr 15 12:31:57 +0000 2019METAP1D:p, methionyl aminopeptidase type 1D, mitochondrial (Homo sapiens) 🔗 Small mitochondrial matrix enzyme; no PTMs; no high maf SAVs; observed mature form 43,45-335 [1,125 x] 🔗
Sun Apr 14 13:55:01 +0000 2019PDF:p, peptide deformylase, mitochondrial (H. sapiens) 🔗 Small mitochondrial matrix enzyme; no PTMs; no high maf SAVs; observed mature form 29,30,31,32,33-243 [3,668 x] 🔗
Sat Apr 13 18:06:31 +0000 2019@MiguelCos I particularly like the attempted copyright declaration.
Sat Apr 13 17:36:47 +0000 2019@theoneamit @ankitYYZ The current scoring function is quite compact: I'm sure we can add a few more terms for a k-score function (_s = spectrum, _p = peptide).
def score_id(_s,_p):
c = 0
for p in _p:
if p in _s:
c += 1
return c
Sat Apr 13 15:33:50 +0000 2019@UCDProteomics PS - eukaryotic mitochondria contain the 3 enzymes necessary to formylate initiator M's, remove the formylation and cleave off the initiator M's during the in-mitochondrial translation of the 13 mt-encoded proteins
Sat Apr 13 14:31:32 +0000 2019@PastelBio To the extent that "synergy " means "ruthless-cut-throat-competition-for-money-and-resources"
Sat Apr 13 13:36:18 +0000 2019@UCDProteomics If you just want to see some examples of what happens if you are a little to aggressive with formic acid use, this data set (🔗) has peptide NT, K and R formylation.
Sat Apr 13 13:00:39 +0000 2019MTFMT:p, mitochondrial methionyl-tRNA formyltransferase (Homo sapiens) 🔗 Small mitochondrial matrix enzyme; no PTMs; no high maf SAVs; observed mature form 32,50-389 [1,480 x] 🔗
Sat Apr 13 12:43:09 +0000 2019@UCDProteomics In prokaryotes, the formyl-methionine is removed so efficiently I've never been able to find it there either.
Sat Apr 13 12:35:11 +0000 2019@UCDProteomics I've never seen any study that enriched for protein NT formylation. Whenever I've looked for it, all I've seen are false positives. Are you looking for it in eukaryotes?
Fri Apr 12 12:59:52 +0000 2019NDUFA13:p, NADH:ubiquinone oxidoreductase subunit A13 (H. sapiens) 🔗 Small mitochondrial intermembrane subunit; N-terminal A2+acetyl; no high maf SAVs; observed mature form 2-144 [14,394 x] (image from String) 🔗
Thu Apr 11 13:29:14 +0000 2019@theoneamit @ankitYYZ Not yet. I am testing how well it works for the range of expts, instruments and file format subtypes commonly used.
Thu Apr 11 12:50:49 +0000 2019NDUFA12:p, NADH:ubiquinone oxidoreductase subunit A12 (H. sapiens) 🔗 Small mitochondrial intermembrane subunit; no significant PTMs; no high maf SAVs; observed mature form 1,3-145 [12,512 x] 🔗
Wed Apr 10 15:42:25 +0000 2019@ankitYYZ WRT to the output information, since it is written in Python, it is fairly straightforward to have the output format altered to conform to any style you wish.
Wed Apr 10 15:39:52 +0000 2019NDUFA11:p, NADH:ubiquinone oxidoreductase subunit A11 (H. sapiens) 🔗 Small mitochondrial intermembrane subunit; no significant PTMs; no high maf SAVs; observed mature form 2-141 [7,918 x] 🔗

Wed Apr 10 15:33:32 +0000 2019So it appears to be a statistical "dead heat" between the PSI formats and CSV as the favoured output format. I think I've got a coin around here somewhere so that I can make a decision ...
Wed Apr 10 15:30:22 +0000 2019@ankitYYZ The new one (SE) is written in pure Python, rather than C++. It is about 20x faster than X! Tandem and it is written in a tutorial style so it can be used to show anyone interested how it works.
Wed Apr 10 15:24:12 +0000 2019@KislingerThomas The last sentence should be THE mandatory last sentence in all journal paper abstracts (unless the article demonstrates otherwise).
Tue Apr 09 14:41:23 +0000 2019I was expecting a bit more diversity of response wrt output formats. So far it seems like:
Tue Apr 09 12:48:29 +0000 2019NDUFA10:p, NADH:ubiquinone oxidoreductase subunit A10 (H. sapiens) 🔗 Small mitochondrial matrix subunit; several low occupancy PTMs; no high maf SAVs; observed mature form 36-355 [18,632 x] 🔗

Tue Apr 09 01:48:56 +0000 2019@UCDProteomics @nesvilab It seems to be a Nordic thing. There are several groups that use that method, with mixed results.
Mon Apr 08 23:19:57 +0000 2019I've just finished a new peptide ms/ms search engine. What output format(s) would be the best for people who want to plug the results into their data handling systems?
Mon Apr 08 12:40:56 +0000 2019NDUFA9:p, NADH:ubiquinone oxidoreductase subunit A9 (H. sapiens) 🔗 Small mitochondrial matrix subunit; several low occupancy PTMs; no high maf SAVs; observed mature form 36-377 [19,774 x] 🔗

Sun Apr 07 17:30:25 +0000 2019NDUFA8:p, NADH:ubiquinone oxidoreductase subunit A8 (H. sapiens) 🔗 Small mitochondrial intermembrane subunit; 2 PTMs, P2+acetyl and T83+phosphoryl; no high maf SAVs; observed mature form 2-172 [15,268 x] 🔗

Sat Apr 06 17:17:05 +0000 2019NDUFA7:p, NADH:ubiquinone oxidoreductase subunit A7 (H sapiens) 🔗 Very small mitochondrial subunit; one PTM, K80+acetyl; no high maf SAVs; observed mature form 11-113 [8,995 x] 🔗
Fri Apr 05 20:31:24 +0000 2019The experiments had very good recovery of mitochondrial proteins, some nice ubiquitinated ubiquitin PSMs and > 5000 phosphorylated PSMs per experiment. The data is virus-free, with LINE1 ORF1 just detectable in 2 fo the 3 runs.
Fri Apr 05 20:23:34 +0000 2019#PXD010175 (Yang M, et al. 🔗) is really solid, thorough proteomics data. If you want to know what proteins are observable using TMT6 in human B cells, this one is about as good as it gets 🔗
Fri Apr 05 14:49:42 +0000 2019NDUFA6:p, NADH:ubiquinone oxidoreductase subunit A6 (H. sapiens) 🔗 Small mitochondrial subunit; 2 acetylation sites K111+acetyl and K147+acetyl; p.A35V maf = 0.14; observed mature form 32,34-154 [10,988 x] 🔗
Thu Apr 04 19:40:00 +0000 2019@nesvilab Note, I mean 0.5 to 2 megabytes (MB) per second, not megabits (Mb) per second.
Thu Apr 04 19:26:23 +0000 2019@nesvilab I regularly get somewhere between 0.5 and 2 MB/sec from PRIDE's FTP site. MASSIVE's FTP site is usually between 5 and 8 MB/sec. I am OUS, but still in NA.
Thu Apr 04 15:00:30 +0000 2019@nesvilab I went to a conference hosted by 🔗, a govt funded ($200 million) protein technology research supercluster. In 2 days with 8 hours of lectures/day, no specific protein was named and no mention of what proteins were, other than food.
Thu Apr 04 13:23:02 +0000 2019NDUFA5:p, NADH:ubiquinone oxidoreductase subunit A5 (H. sapiens) 🔗 Very small mitochondrial subunit; multiple acetylation sites; no mitochondrial transit peptide; observed mature form 1,2-116 [18,442 x] 🔗

Wed Apr 03 23:48:03 +0000 2019@karthikskamath @nesvilab People do like to see the numbers: undefined superlatives are too frequent in this business (imho)
Wed Apr 03 19:21:08 +0000 2019@nesvilab But are they ultrafast?
Wed Apr 03 18:36:13 +0000 2019@PastelBio How fast is "Ultrafast"?
Wed Apr 03 13:32:46 +0000 2019NDUFA4:p, NADH:ubiquinone oxidoreductase subunit A4 (H. sapiens)
Very small mitochondrial subunit; some low occupancy PTMs ; no high maf SAVs; aka MLRQ, CI-9k, COXFA4; no mitochondrial transit peptide; observed mature form 1-88 [16,108 x] 🔗

Tue Apr 02 17:01:38 +0000 2019The same holds true for lysine acetylation, methylation and ubiquitination at the C-terminus of tryptic peptides
Tue Apr 02 16:58:24 +0000 2019I'm pretty sure that trypsin does not cleave at methyl- or dimethyl-arginine. While it seems to be OK to report PSMs with methylation on the C-terminal R of a tryptic peptide, to my way of thinking those PSMs should be considered false positives.
Tue Apr 02 14:31:11 +0000 2019NDUFA3:p, NADH:ubiquinone oxidoreductase subunit A3 (H. sapiens) 🔗 Very small mitochondrial subunit; N-terminal acetylation is the only PTM; no high maf SAVs; mature form ambiguous [4,123 x] 🔗
Mon Apr 01 14:51:52 +0000 2019NDUFA2:p, NADH:ubiquinone oxidoreductase subunit A2 (H sapiens) 🔗 Very small mitochondrial subunit; observed 10x more often than NDUFA1; no high maf SAVs or mitochondrial targeting signal; observed mature form 2-99 [10,758 x] (image from String) 🔗
Sun Mar 31 14:07:18 +0000 2019NDUFA1:p, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa (H. sapiens) 🔗 Very small mitochondrial subunit; no PTMs; no high maf SAVs; no mitochondrial targeting signal; observed mature forms 1-70 [1,039 x] (image from String) 🔗
Sat Mar 30 15:32:31 +0000 2019Thanks for the input, everybody 👨🔬👩🔬
Sat Mar 30 14:27:01 +0000 2019NDUFS8:p, NADH:ubiquinone oxidoreductase core subunit S8 (H. sapiens) 🔗 Small mitochondrial matrix subunit; one low occupancy PTM: Y74+phosphoryl; no high maf SAVs; observed mature forms 35-210 [14,784 x] 🔗
Sat Mar 30 13:46:57 +0000 2019@dtabb73 And how much does the motherboard allow?
Sat Mar 30 13:44:43 +0000 2019@dtabb73 How much memory do you currently have?
Fri Mar 29 21:41:26 +0000 2019@pwilmarth So a value of about 5% unlabeled is normal in your experience. At what value would you pull the plug on a sample & insist that it be done again?
Fri Mar 29 19:51:45 +0000 2019A WinRAR vulnerability to keep in mind when downloading large datasets that contain .rar compressed files:
🔗
Fri Mar 29 19:45:33 +0000 2019I really don't know: I'd like to hear some opinions.
Fri Mar 29 19:45:01 +0000 2019To the quant-types: a fraction of the peptides in an iTRAQ/TMT experiment are always underivatized. How big does that fraction have to be before it affects the interpretation of the data?
Fri Mar 29 13:15:35 +0000 2019NDUFS7:p, NADH:ubiquinone oxidoreductase core subunit S7 (Homo sapiens) 🔗 Small mitochondrial matrix subunit; no PTMs; no high maf SAVs; observed mature forms 30,39-213 [11,844 x] 🔗
Thu Mar 28 21:23:19 +0000 2019This should never have been allowed to happen ... 🔗
Thu Mar 28 14:56:28 +0000 2019#PXD009040 is a good quality, very interesting data set (🔗). Has the high ZNF-domain protein concentration (~10% of ids) characteristic of sumoylation affinity capture studies.
Thu Mar 28 13:23:20 +0000 2019NDUFS6:p, NADH:ubiquinone oxidoreductase core subunit S6 (H. sapiens) 🔗 Very small mitochondrial matrix subunit; several rarely occupied PTMs; no commonly observed SAVs; observed mature form 29-124 [11,224 x] 🔗

Wed Mar 27 15:14:00 +0000 2019I don't really have an opinion regarding TCM, but this seems to be an odd choice for JPR to be taking right now 🔗
Wed Mar 27 14:23:08 +0000 2019NDUFS5:p, NADH:ubiquinone oxidoreductase core subunit S5 (H. sapiens) 🔗 Small mitochondrial matrix subunit; no PTMs, high maf SAVs, or mitochondrial membrane transit domain; observed mature form 2-106 [9,898 x] 🔗
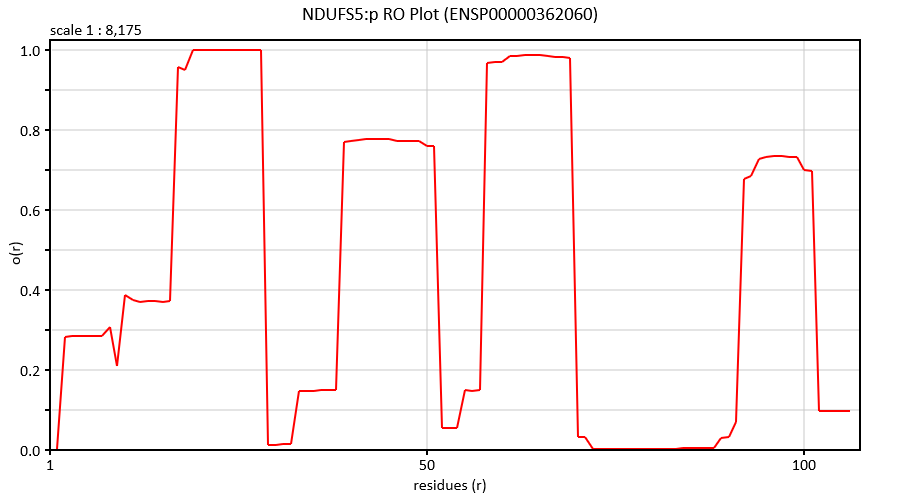
Tue Mar 26 14:20:13 +0000 2019NDUFS4:p, NADH:ubiquinone oxidoreductase core subunit S4 (Homo sapiens) 🔗 Small mitochondrial matrix subunit; no PTMs; no commonly observed SAVs; observed mature form 43-175 [9,544 x] 🔗
Mon Mar 25 19:03:51 +0000 2019I often feel like adding "human corneocytes" to the list of tissues and/or cell types present in a sample.
Mon Mar 25 13:08:21 +0000 2019NDUFS3:p, NADH:ubiquinone oxidoreductase core subunit S3 (Homo sapiens) 🔗 Small mitochondrial matrix subunit; distinctive pattern of 4 Y+phosphoryl PTMs; no commonly observed SAVs; observed mature form 37-263 [25,930 x] 🔗

Mon Mar 25 02:18:46 +0000 2019@MSBioworks I reanalyzed the data. The preliminary analysis in the paper didn't check for the modification.
Sun Mar 24 12:52:10 +0000 2019env:p, viral envelope protein (Human endogenous retrovirus K) 🔗| Midsized glycoprotein; very rarely observed; found in some cancer cell lines; many copies of gene present in the human genome [23 x] 🔗

Sat Mar 23 14:01:55 +0000 2019gag:p, group specific antigen protein (Human endogenous retrovirus K) 🔗 Midsized polyprotein; very rarely observed; found in some cancer tissues and induced pluripotent stem cells; many copies of gene present in the human genome [37 x] 🔗

Fri Mar 22 19:08:00 +0000 2019@pwilmarth Nature articles are for entertainment purposes only.
Fri Mar 22 17:41:07 +0000 2019NDUFS2:p, NADH:ubiquinone oxidoreductase core subunit S2 (Homo sapiens) 🔗 Small mitochondrial membrane subunit; no significant PTMs; most common SAV p.P352A maf=0.1; observed mature form 34-463 [19,964 x] 🔗
Thu Mar 21 16:26:27 +0000 2019@UCDProteomics For example, the following shows the bacterial proteins found in a single clinical urine sample used in a cancer biomarker study (🔗)
🔗
Thu Mar 21 15:53:27 +0000 2019@UCDProteomics Bacteria are often really abundant in saliva. Urine can be less so: normally only low levels of E. coli are detectable. But if the person has a UTI, the bacterial proteins are easily detected, particularly if they have been catheterized in hospital.
Thu Mar 21 15:48:18 +0000 2019@HFazelinia The microbiome resource is somewhere to start, but a practical FASTA resource would need to:
1. indicate expected abundance (by species);
2. include pathogens (e.g., P. aeruginosa in urine); &
3. have standard protein-level annotation.
Thu Mar 21 14:24:41 +0000 2019NDUFS1:p, NADH:ubiquinone oxidoreductase core subunit S1 (H. sapiens) 🔗 Midsized mitochondrial membrane subunit; at least 6 Y phosphorylation sites but no S/T sites; observed mature form 24-727 [30,419 x] 🔗

Wed Mar 20 14:46:42 +0000 2019MTHFD2:p, methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase (H. sapiens) 🔗 Small mitochondrial matrix enzyme; some low occupancy PTM; observed mature form 36-350 [13,550 x] 🔗

Wed Mar 20 14:39:28 +0000 2019Why do people still analyze clinical proteomics data from urine & saliva samples as though there are no significant bacterial populations in those fluids?
Tue Mar 19 16:38:14 +0000 2019This really looks like it must mean something, but I have yet to figure out just what that might be ... 🔗
Tue Mar 19 14:18:50 +0000 2019MAP2K1:p, mitogen-activated protein kinase kinase 1 (H. sapiens) 🔗 Small cytoplasmic phosphoprotein; extensive S/T phosphorylation; observed mature form 2-393 [24,258 x] 🔗
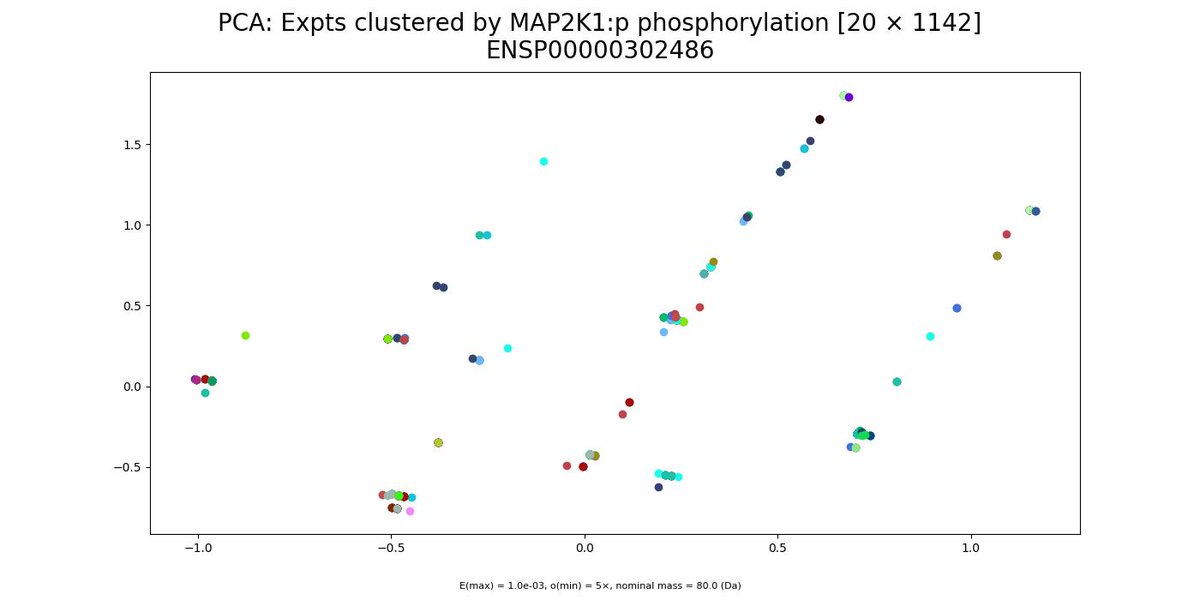
Mon Mar 18 14:54:19 +0000 2019MAVS:p, mitochondrial antiviral signaling protein (H. sapiens) 🔗 Midsized mitochondrial outer membrane protein; extensive S/T phosphorylation; observed mature form 2-540 [17,172 x] 🔗
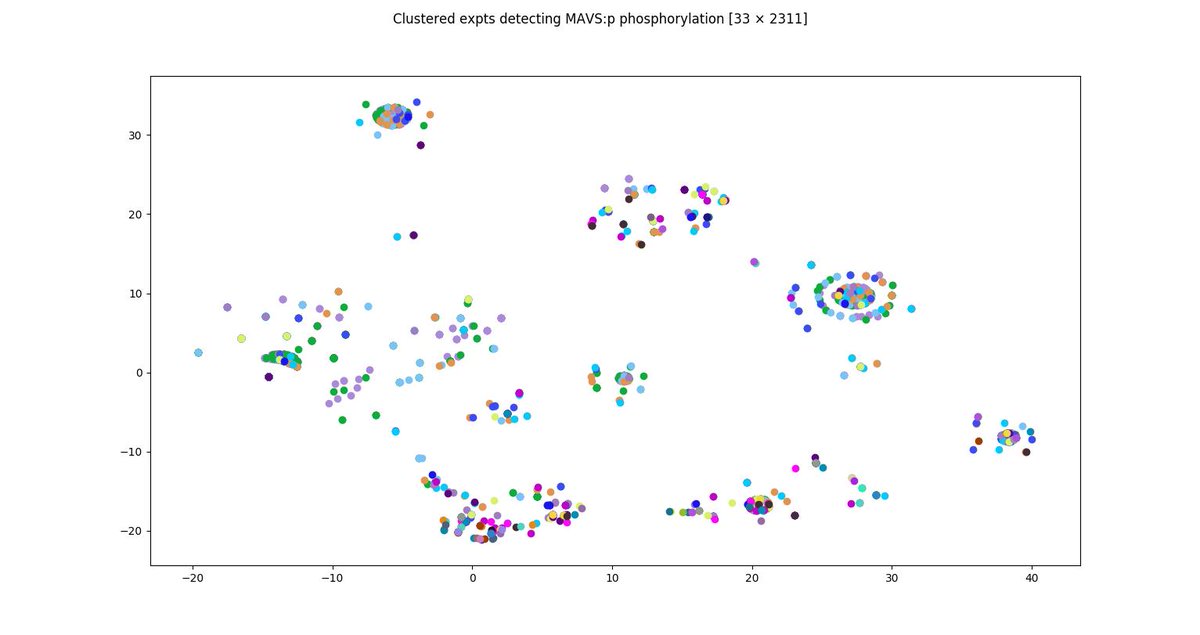
Sun Mar 17 13:51:52 +0000 2019GCSH:p, glycine cleavage system protein H (H. sapiens) 🔗 Small mitochondrial matrix enzyme; H-protein in the glycine cleavage system; limited PTM; observed mature form 43-173 [9,978 x] 🔗

Sat Mar 16 15:55:40 +0000 2019@MSBioworks The files "GluC27.mzml" and "GluC28.mzml" in 🔗 have some pretty good ids of the peptide EIF5A2 (ENSP00000295822) 43–70 that have K50+hypusine and/or K47+acetyl modifications
Sat Mar 16 13:29:52 +0000 2019GLDC:p, glycine decarboxylase (H. sapiens) 🔗 Large mitochondrial matrix enzyme; P-protein in the glycine cleavage system; one PTM, S445+phospho; observed mature form 37-1020 [5,727 x] 🔗
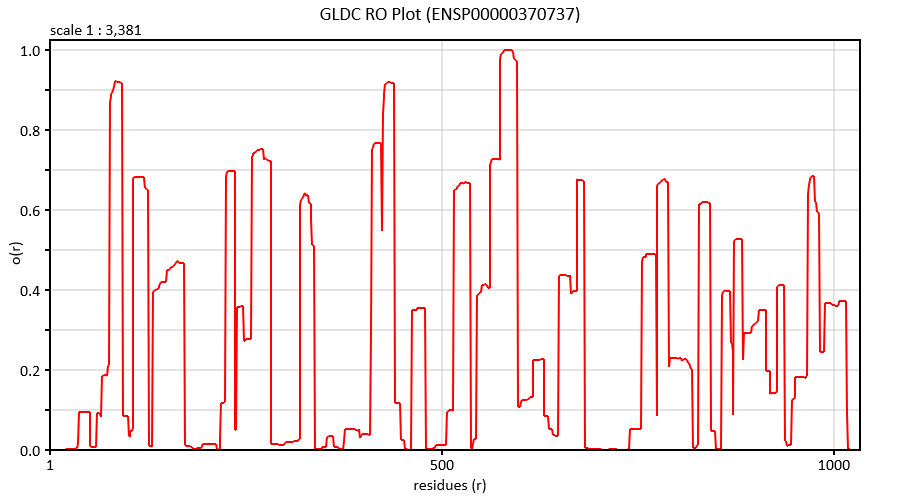
Fri Mar 15 14:40:04 +0000 2019@AnthonyCesnik @Smith_Chem_Wisc +12.000 is described here 🔗 & a practical list of formalin-protein reactions here 🔗
Fri Mar 15 14:21:01 +0000 2019DLD:p, dihydrolipoamide dehydrogenase (H. sapiens) 🔗 Midsized mitochondrial matrix subunit; L-protein in glycine cleavage system; vital in many metabolic pathways; extensive PTM; no high maf SAVs; observed mature form 36-504 [38,499 x] 🔗
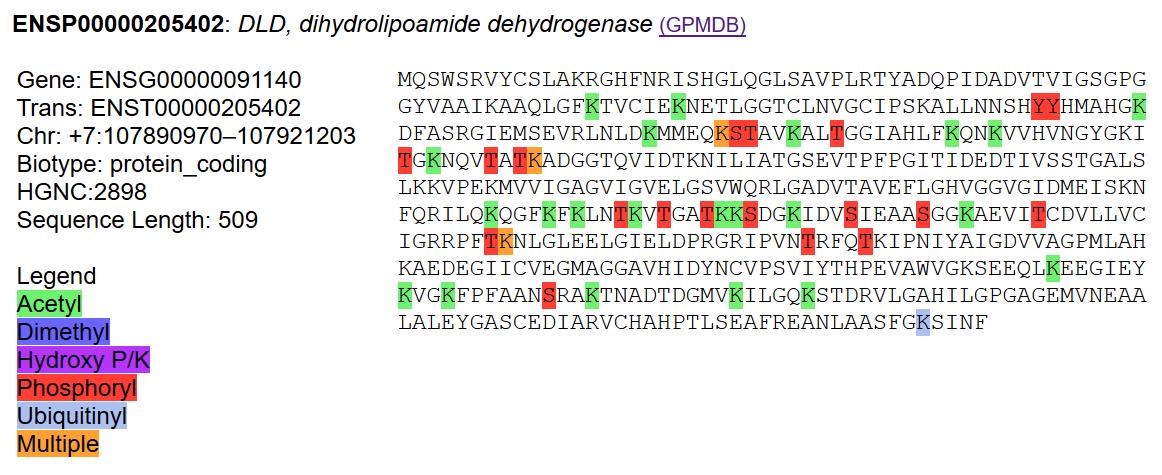
Fri Mar 15 13:13:42 +0000 2019@dtabb73 @ProteomicsNews @Eskom_SA @CityPowerJhb @City_Ekurhuleni @CityofCT @CityofJoburgZA @eThekwiniM @CityTshwane @TrafficSA @ewnreporter @SABCNewsOnline @IOL Reminds me of living in Vancouver: there the problem was wind storms that would knock out power for days in the fall and winter
Fri Mar 15 12:51:31 +0000 2019Thanks, everybody. Good discussion.
Fri Mar 15 12:50:14 +0000 2019@ypriverol GPMDB records all PTM's as mass shifts: they are interpreted into names on the fly for display purposes
Fri Mar 15 12:43:40 +0000 2019@AnthonyCesnik @Smith_Chem_Wisc It is a single carbon. I once came across the chemical reaction - involves a resonant rearrangement following formaldehyde adduction - but I don't remember where I found it (it was 20 years ago).
Thu Mar 14 21:38:27 +0000 2019@Smith_Chem_Wisc @ProteomicsNews 😀🥕
Thu Mar 14 21:37:19 +0000 2019From the responses, I'm not quite sure whether no one is aware of a list of commonly observed but unexplained modification masses or whether the community is backing away from the idea of prevalent unexplained modifications.
Thu Mar 14 21:26:38 +0000 2019@Smith_Chem_Wisc @ProteomicsNews I get the two "M"s, but what does the carrot signify?
Thu Mar 14 21:09:22 +0000 2019@cstross 🔗
Thu Mar 14 20:49:05 +0000 2019@dtabb73 I would guess that 12.000 and 30.011 are somewhere on the list. I spent a not-very-enjoyable 6 months tracking down every significant formaldehyde-related protein modification back when I was at Eli Lilly in the 90's.
Thu Mar 14 17:55:47 +0000 2019(2/2) but that was based on the results from one study. I've been trying the masses on the Unimod list, but I've had limited success finding anything. Any help would be appreciated.
Thu Mar 14 17:52:09 +0000 2019(1/2) I've got a question for the open search crowd. Is there a list somewhere that has the most commonly found unknown mod masses obtained from an open search strategy? I've tried using the list on Unimod (based on 🔗) ...
Thu Mar 14 12:48:25 +0000 2019AMT:p, aminomethyltransferase (H. sapiens) 🔗 Small mitochondrial matrix subunit; T-protein in the glycine cleavage system; no PTMs; no high maf SAVs; observed mature form 29-403 [1,663 x] 🔗

Wed Mar 13 19:48:32 +0000 2019@Sci_j_my However, K122+acetyl has been id'd in the slightly longer peptide 115-123 (GELLEAIKR) by these guys 🔗
Wed Mar 13 19:31:39 +0000 2019@Sci_j_my Yah, the K+acetyl that they show in 2F is a false positive: trypsin or Lys-C will not cut with that mod in place
Wed Mar 13 19:22:49 +0000 2019@UCDProteomics Frick'n plants and their stupid chlorophyll 🤢
Wed Mar 13 15:53:01 +0000 2019@byu_sam Good one!
Wed Mar 13 15:40:41 +0000 2019@theoneamit I like it, too. It makes some types of ubiquitination experiments ambiguous, but that's just the way biochemistry rolls ...
Wed Mar 13 15:08:42 +0000 2019Thyroglobulin is present in several other tissues, but TG in non-thyroid tissues are not iodinated. TG is present at surprisingly high levels in some breast tumours (but with no Y+iodine).
Wed Mar 13 15:06:02 +0000 2019The protein is produced in the thyroid and through a fairly complex process the iodine is added, iodinated Y residues are covalently linked together and cleaved out of the protein to form thyroxine and triiodothyronine, which is then secreted into blood.
Wed Mar 13 15:01:52 +0000 2019Another favorite rare PTM is iodination. It only happens on one protein (thyroglobulin), although there are ~25 Y mono- or di-iodination sites in this protein (mature form 20-2768).
Wed Mar 13 14:17:41 +0000 2019You have to be nice to your animals: 🔗
Wed Mar 13 12:08:08 +0000 2019MTHFD2:p, methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase (H. sapiens) 🔗 Small mitochondrial matrix enzyme; few, low occupancy PTMs; no high maf SAVs; observed mature form 36-350 [13,495 x] 🔗
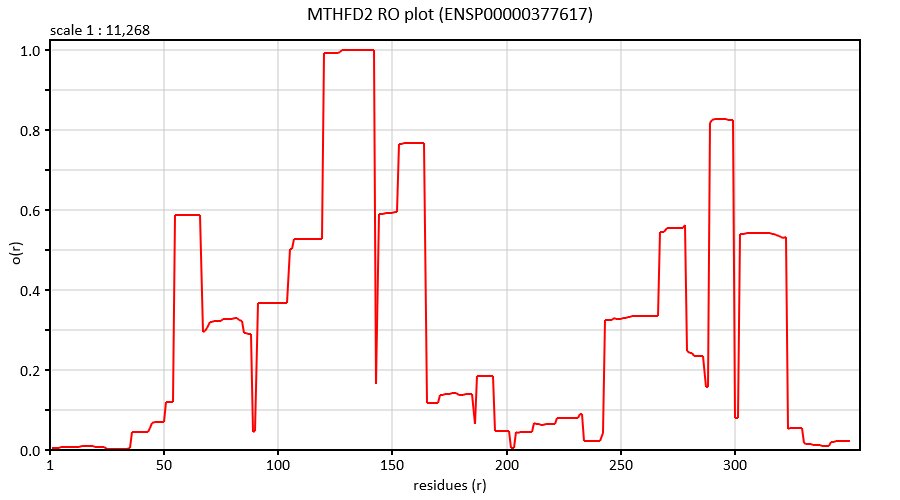
Tue Mar 12 22:01:49 +0000 2019Even though hypusine is so rare, it requires 2 dedicated enzymes, DHS and DOHH, that appear to do nothing other than synthesize hypusine in place at K50 🔗
Tue Mar 12 21:58:01 +0000 2019You can only observed it in cases where K47 is acetylated, so it can't be cleaved, resulting in a longer peptide that is in a better parent ion mass range and the 2 modified K's are not charged.
Tue Mar 12 21:49:23 +0000 2019It is practically difficult to observe, even though EIF5A is a very commonly observed protein. K50 is in very +vely charged domain, resulting in tryptic peptides that are small and highly charged, which are difficult to see in a standard trypsin-cleavage experiment.
Tue Mar 12 21:42:39 +0000 2019@UCDProteomics @ypriverol @JProteomeRes @Karl_Mechtler It is one of the commonly observed porcine trypsin autolysis products, 🔗
Tue Mar 12 19:36:56 +0000 2019@debcha @GreatDismal Technically, it is actually a pretty accurate way to describe it 🔗
Tue Mar 12 18:16:35 +0000 2019I will freely admit that I may be the only person on earth that actually has "favorite PTMs".
Tue Mar 12 18:11:57 +0000 2019What is your favorite mammalian PTM that only occurs at a very small number of sites (< 10)? Mine is hypusine (the name of the modified residue, not the modification): in humans it only occurs at EIF5A:pm.K50=hypusine or EIF5A2:p:pmK50=hypusine.
Tue Mar 12 12:49:11 +0000 2019SHMT2:p, serine hydroxymethyltransferase 2 (H. sapiens) 🔗 Midsized mitochondrial matrix enzyme; significantly modified; no high maf SAVs; observed mature form 27-504 [39,327 x] 🔗

Mon Mar 11 18:48:30 +0000 2019@theoneamit That particular spelling is trademarked 🔗 There is a "Roar Shack Design" company, but their logo, marks & business are probably different enough there shouldn't be an issue.
Mon Mar 11 17:45:12 +0000 2019@nesvilab Yes, it does. I don't think this sort of thing has much value unless some type of confidence interval can be assigned to an identification.
Mon Mar 11 16:31:06 +0000 2019The black dots are the MS/MS spectra to be id'd and the other points are from the training data set: 0.5 million MS/MS spectra obtained from samples of the same organism using the same type of mass spec.
Mon Mar 11 16:20:21 +0000 2019The first identifications of unknowns using my experimental t-SNE-based search engine, tentatively named "Roar Shack" 🔗
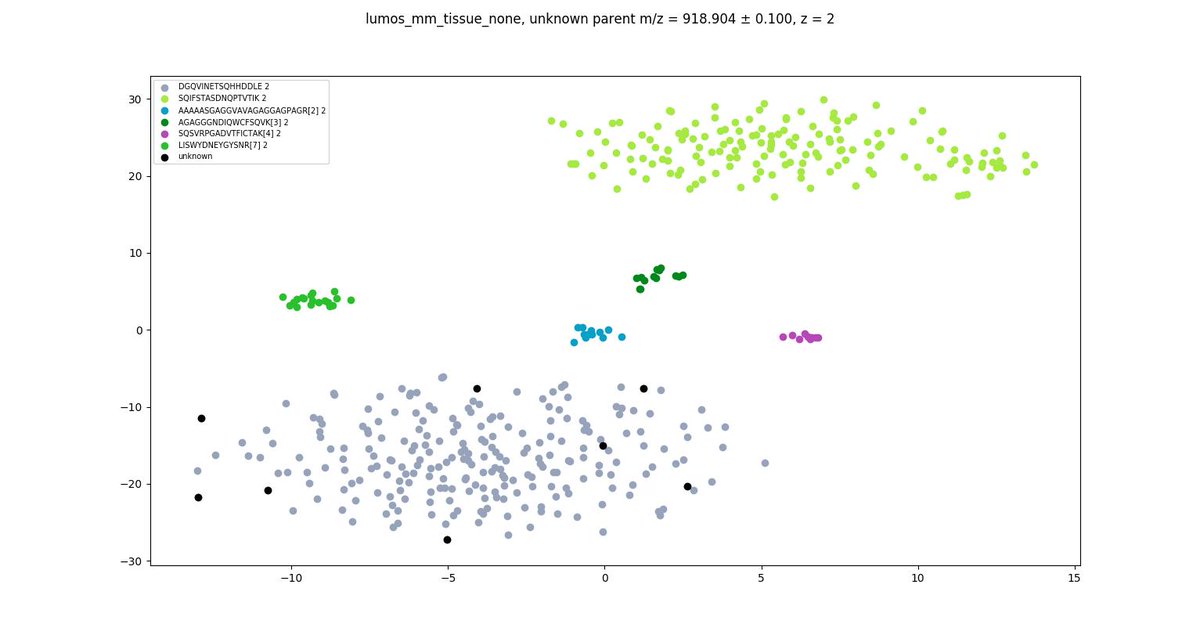
Mon Mar 11 13:46:38 +0000 2019AGXT2:p, alanine-glyoxylate aminotransferase 2 (H. sapiens) 🔗 Midsized mitochondrial matrix enzyme; no PTMs; p.V140I maf = .4;most abundant in liver and kidney tissue; absent in common cell lines; observed mature form 48-514 [859 x] 🔗
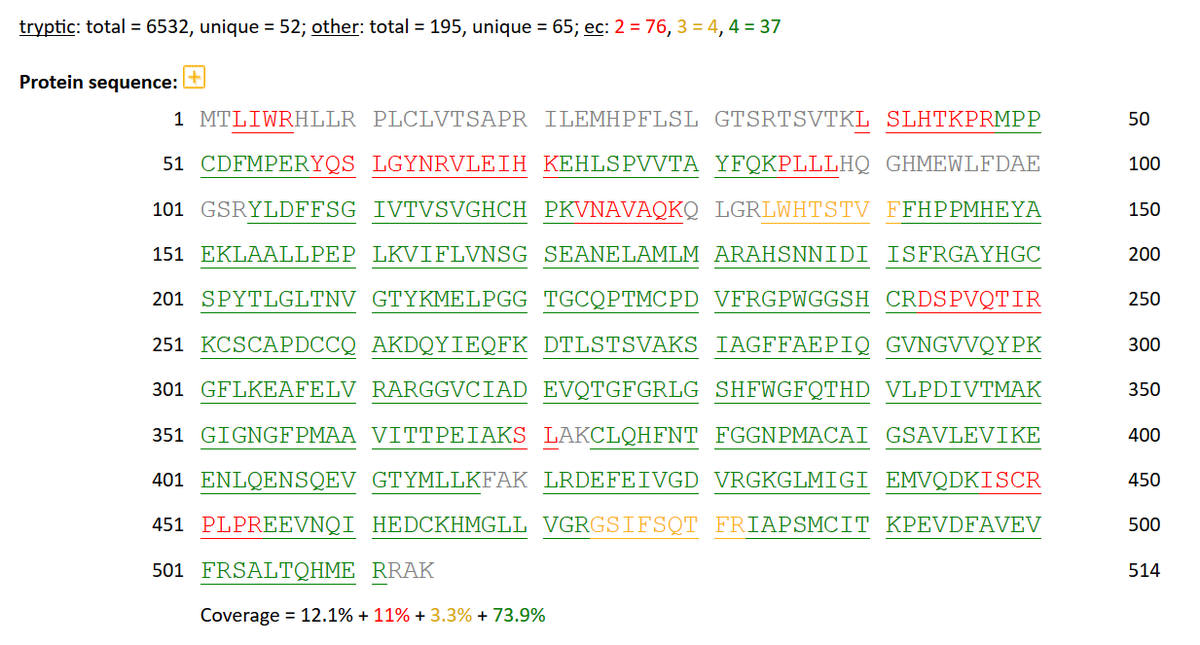
Sun Mar 10 13:50:56 +0000 2019The colors indicate the status of each residue, based on the NBS v. 2 identification classification system (ratings run from 1 to 4)
Sun Mar 10 13:47:52 +0000 2019GATM:p, glycine amidinotransferase (H. sapiens) 🔗 Small mitochondrial matrix enzyme; no PTMs; p.Q110H (rs1288775) maf = 0.23; observed mature form (37,38)-423 [6,527 x] 🔗
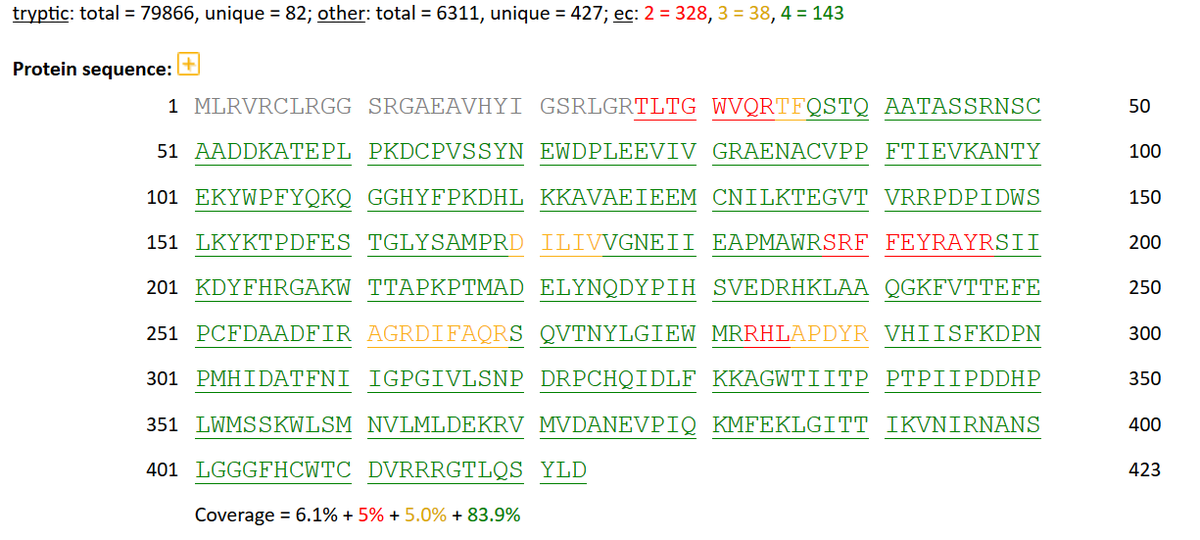
Sat Mar 09 23:40:40 +0000 2019@slavovLab No. Too many government services & licenses depend on age.
Sat Mar 09 16:23:35 +0000 2019@AlexUsherHESA You may want to consider including "both" as a choice in your poll.
Sat Mar 09 13:33:52 +0000 2019ALAS1:p, 5'-aminolevulinate synthase 1 (H. sapiens)
Midsized mitochondrial matrix enzyme; catalyzes glycine + succinyl-CoA -> delta-aminolevulinic acid [2,256 x] 🔗

Fri Mar 08 14:00:35 +0000 2019@nesvilab The plot shows the clustering of ~20,000 spectra that had been assigned to 100 distinct modified peptides in many QE LC/MS/MS runs from different studies. The tSNE takes about 500 passes to converge (~8 min). It was run on a single thread (Python)
Fri Mar 08 13:21:56 +0000 2019CPOX:p, coproporphyrinogen oxidase (H. sapiens) 🔗 Small mitochondrial matrix enzyme; p.N272H and p.V294I are readily observable SAVs; observed mature form (110,111,112)-454 [15,199 x] 🔗
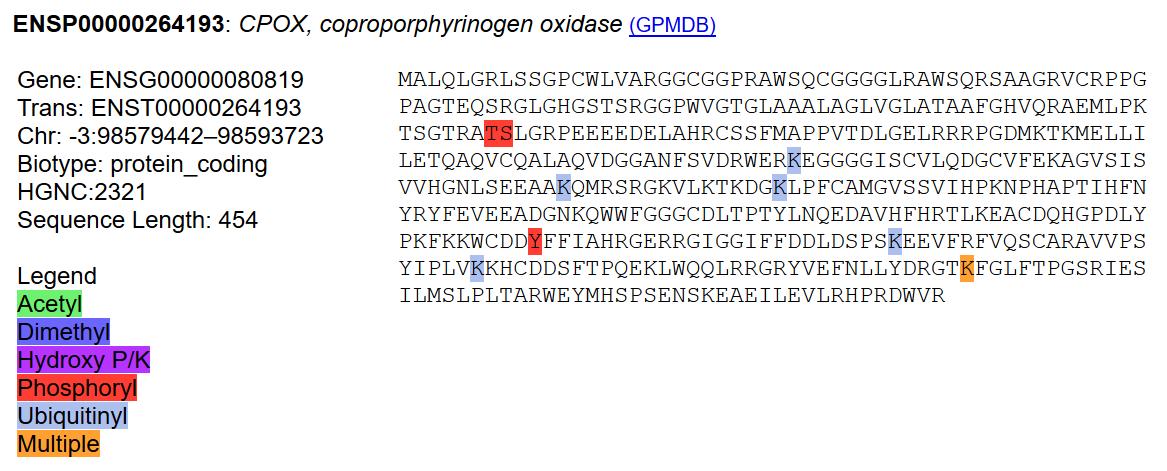
Fri Mar 08 02:28:54 +0000 2019@nesvilab It remains to be seen if it can scale: I doubt that it can. It does seem to be pretty good for visualizing the effects of changing parameters.
Thu Mar 07 22:12:52 +0000 2019And now it appears to be useful/interesting (probably) 🔗
Thu Mar 07 22:08:13 +0000 2019@nationalpost I guess now we know why China decided to go after canola.
Thu Mar 07 18:30:23 +0000 2019This is more like it. Just a bit of totally-reworking-everything-from-scratch sharpens up the t-SNE plot quite a bit. 🔗
Thu Mar 07 15:20:32 +0000 2019@lkpino @damon_may Thanks for the reference.
Thu Mar 07 13:35:33 +0000 2019FECH:p, ferrochelatase (H. sapiens) 🔗 Small mitochondrial matrix enzyme; no PTMs; aka protoporphyrin ferrochelatase, observed mature form (37,38)-423 [15,199 x] 🔗
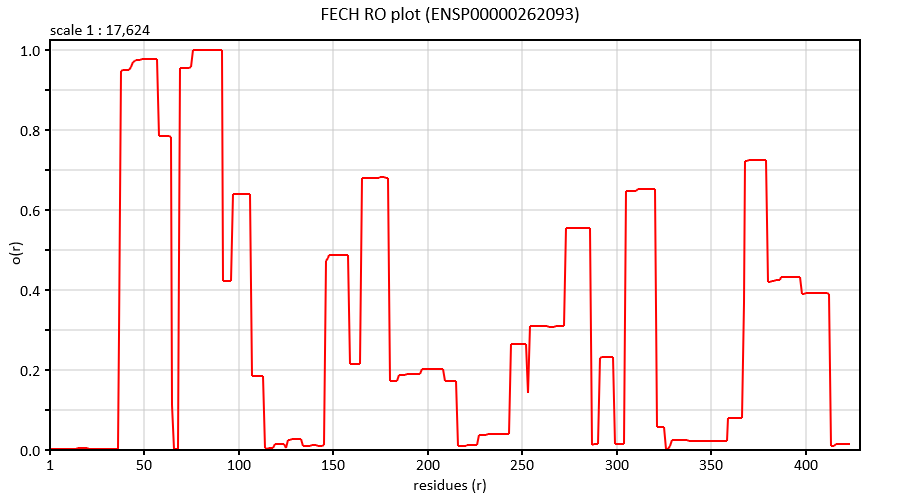
Thu Mar 07 00:21:50 +0000 2019@chrashwood Yes. The legend is the peptide sequence, the parent ion z and the number of spectra associated with the sequence/z pair
Wed Mar 06 23:52:48 +0000 2019I'm experimenting with the use of T-distributed stochastic neighbor embedding plots to improve spectrum clustering algorithms: it seems to be a promising approach – so long as you keep your head about you. 🔗
Wed Mar 06 17:26:48 +0000 2019@DonMartinCTV @ElizabethMay @gmbutts @Puglaas Have there been any job losses or job transfers out of Canada in the 6 month period following the decision against using a DPA for SNC-Lavalin?
Wed Mar 06 17:22:20 +0000 2019@TondaMacC His whole testimony does have a Captain Renault quality to it: possibly he watched Casablanca at some point recently.
Wed Mar 06 17:00:07 +0000 2019I can understand using zip archives to group files from individual experiments, but lumping everything together is unhelpful.
Wed Mar 06 16:58:29 +0000 2019Is there any way to discourage people from making PRIDE depositions like 🔗 (PXD011839)? By placing all of the raw files in one very large zip file, it makes downloading the data unnecessarily difficult and error prone.
Wed Mar 06 14:34:07 +0000 2019@kadzuis It is seductive
Wed Mar 06 14:27:58 +0000 2019The enzyme's function is now established to be the same in yeast as it is in other eukaryotes: the nitrogen fixation function has been abandoned (except for its persistence in the gene's name).
Wed Mar 06 14:22:51 +0000 2019NFS1:p, cysteine desulfurase (H. sapiens) 🔗 Midsized mitochondrial matrix protein; nitrogen fixation 1 (S. cerevisiae, homolog); observed mature form 38-457 [11,134 x] 🔗
Tue Mar 05 22:52:45 +0000 2019@TheGonz79 As a true pedant, I feel honour bound to point out that the most frequently id'd peptide that contains 'TWEET' is 'ALSTWEETLTSFK', found in the yeast protein LHS1:p (orf YKL073W).
Tue Mar 05 20:36:41 +0000 2019rs1047891 is a nice little SAV for testing: maf of ~ 0.30; present in CPS1, one of the most abundant proteins in hepatocytes; easy to detect AA change ENSP00000402608:p.T1412N 🔗
Tue Mar 05 20:12:43 +0000 2019The Vancouver supercluster seems to be the first one out of the gate: 🔗
Tue Mar 05 19:03:36 +0000 2019@TrostLab @nesvilab @ProtifiLlc And so are David Fenyö and Eric Deutsch.
Tue Mar 05 18:54:16 +0000 2019@TrostLab @nesvilab @ProtifiLlc And yes, I am a physicist, too.
Tue Mar 05 18:52:36 +0000 2019@TrostLab @nesvilab @ProtifiLlc The real issue is when did Alexey become the field's premier photobombing specialist?
Tue Mar 05 13:43:28 +0000 2019FDXR:p, ferredoxin reductase (H sapiens) 🔗 Midsized mitochondrial matrix protein; a single, low occupation phosphorylation site: S317+phosphoryl; observed mature form 33-491 [14,985 x] 🔗
Mon Mar 04 19:35:40 +0000 2019I am at a loss: 🔗
Mon Mar 04 18:46:15 +0000 2019A nice, focused critique of how poorly the current patent system can perform with respect to biomedical devices and discoveries 🔗
Mon Mar 04 14:05:12 +0000 2019FDX1:p, ferredoxin 1 (H. sapiens) 🔗 Small mitochondrial matrix protein; 5 potential S-phosphoryl sites; found in most tissues and cell lines; observed mature form (61,62)-184; aka adrenodoxin; adrenal ferredoxin [5,223 x] 🔗
Sun Mar 03 14:33:40 +0000 2019The longer versions of FXN, (42,56,78)-210, mentioned in the literature are not supported by the proteomics data as mature forms. There is some evidence that 42-210 may be transiently present as an intermediate form.
Sun Mar 03 14:24:51 +0000 2019FXN:p, frataxin (H. sapiens) 🔗 Small mitochondrial matrix protein; has a low-occupation N-terminal S/T phosphorylation domain; observed mature form 81-210 [6,002 x]
Sun Mar 03 14:22:50 +0000 2019@michaelhoffman @CIHR_IRSC @NSERC_CRSNG @SSHRC_CRSH @jwoodgett CCV is a good example of how "feature creep" causes a good system to rapidly become unusable — CCV was actually pretty good when it was originally rolled out as a editable PDF.
Sat Mar 02 22:26:02 +0000 2019@nesvilab @pwilmarth I spent > 1 hr yesterday trying to get a RAID array on a new version of SELinux to work the way I wanted it to (I've done this many times in the past). Saying that someone couldn't install something doesn't mean there was anything wrong with it when it was created.
Sat Mar 02 22:19:52 +0000 2019@nesvilab @pwilmarth It would also be a miracle if a package designed to install perfectly on Solaris 1 could be installed by anyone today. Or if you could get the best designed installer for Mac OS 8 to work now. Let alone trying to run a Python 1.6 script or a Perl 4 script.
Sat Mar 02 20:23:55 +0000 2019@indranilp @Smith_Chem_Wisc I would read the code. It is far more informative than simply running the compiled version.
Sat Mar 02 17:28:22 +0000 2019@Smith_Chem_Wisc I know from experience that maintaining SW availability at any URL for more than 5 years is difficult and may be quite impractical in an academic setting.
Sat Mar 02 17:24:42 +0000 2019@Smith_Chem_Wisc The paper has a narrow view of why software (SW) is published and how it is maintained. There is a big difference between SW published to demonstrate the utility/feasibility of a new idea versus SW meant to be an app for non-technical users.
Sat Mar 02 14:07:46 +0000 2019PMPCB:p, peptidase, mitochondrial processing beta subunit (H. sapiens) 🔗 Midsized mitochondrial matrix subunit; with an S/T phosphorylation domain at the N-terminus; observed mature form 44-489 [19,868 x] 🔗
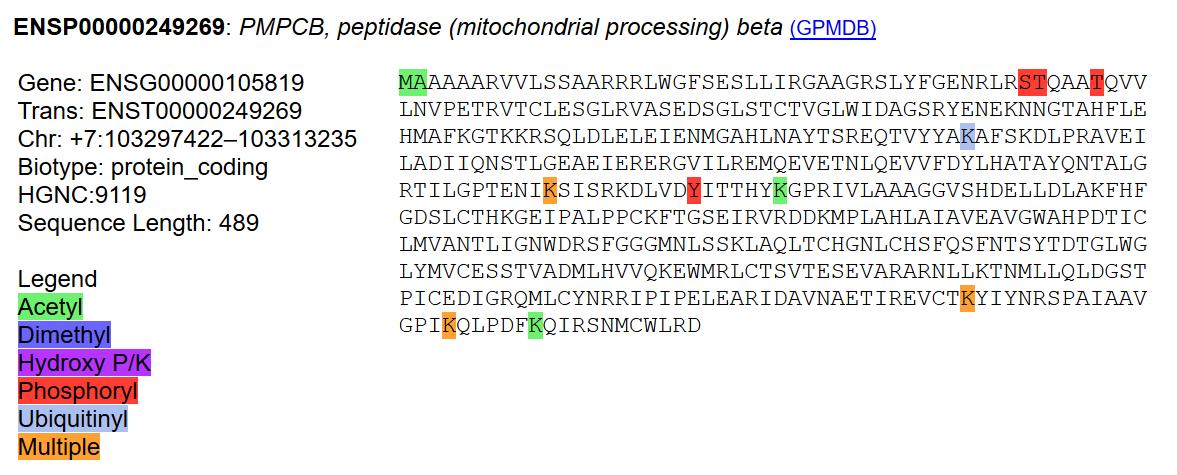
Fri Mar 01 15:54:43 +0000 2019@AlexUsherHESA But it may mean that high-prestige programs are irrationally biased against people that appreciate a large, bold font.
Fri Mar 01 15:35:08 +0000 2019@PastelBio I was a little surprised after reading the article that the word "mouse" never came up.
Fri Mar 01 15:15:18 +0000 2019PMPCA:p, peptidase, mitochondrial processing alpha subunit (H. sapiens) 🔗 Midsized mitochondrial matrix subunit; at least 4 K-aceytl and 12 K-ubiquitinyl acceptors; observed mature form (33,34)-525 [20,602 x] 🔗

Fri Mar 01 13:52:14 +0000 2019@UCDProteomics Totally toast.
Thu Feb 28 18:43:24 +0000 2019@SusanDelacourt It is the equivalent of loudly saying "Squirrel!" to distract the masses.
Thu Feb 28 18:41:40 +0000 2019@davidreevely Didn't Israel just launch a mission to the Moon? Even if it fails, that is more "throwing a punch" than this proposed Canadian contribution to the US effort.
Thu Feb 28 17:19:55 +0000 2019@EricWDeutsch Is the code for the PEFF validator available? It took 8 minutes to validate the nextprot_chromosome_1.peff file (from neXtProt's site), which only has 4,486 sequences. The file was declared valid, but it threw 5,473 warnings.
Thu Feb 28 15:41:09 +0000 2019@EricWDeutsch Sorry. I found the validator at 🔗
Thu Feb 28 14:30:54 +0000 2019@EricWDeutsch My apologies if it is mentioned on the web site, but is there validation software available that will allow a developer to test a PEFF file for compliance with the standard?
Thu Feb 28 13:58:38 +0000 2019LCE1A:p, late cornified envelope 1A (H. sapiens) 🔗 Small low-complexity protein; very rarely identified; part of the epidermal differentiation complex (EDC) on chromosome 1 [89 x]
Wed Feb 27 16:51:26 +0000 2019import json
import sys
def jsms_parse(_in):
sp = []
f = open(_in,'r')
for l in f:
o = json.loads(l)
if 'lv' in o:
sp.append(o)
f.close()
return sp
ss = jsms_parse(sys.argv[1])
for s in ss:
print(s['pm'])
Wed Feb 27 16:49:34 +0000 2019I'm probably a bit too proud of the minimal parser, which takes a JSMS file name on the command line, parses all of the data into objects and lists the parent ion m/z values from those objects in 15 lines.
Wed Feb 27 16:47:29 +0000 2019After fiddling around with it for a few days, JSMS (a JSON Lines MGF alternative) is nearly complete (🔗). The GitHub repo 🔗 has MGF & mzML to JSMS convertors, a validator and a minimal JSMS file parser.
Wed Feb 27 16:00:06 +0000 2019@LM_Orre That is true for most proteomics studies that are trying to find all of the observable proteins/peptides in a tissue or cell line. Protein ids approach an asymptote at about 10⁶ PSMs and peptide ids at about 10⁷ PSMs.
Wed Feb 27 14:32:47 +0000 2019@LM_Orre Yes, that is the case. Based on the Monte Carlo simulations, you would need about 10^7 ids per sample to fully sample the peptides available to the method.
Wed Feb 27 14:26:39 +0000 2019The observations of this protein (in the same family as yesterday's) also does not agree with the mRNA data, which suggest that it should be much more abundant.
Wed Feb 27 13:52:15 +0000 2019PGLYRP4:p, peptidoglycan recognition protein 4 (H. sapiens) 🔗 Small extracellular protein; extremely rarely identified; may be present in ovarian ascites and saliva (weak evidence) [23 x]
Wed Feb 27 13:50:21 +0000 2019@LM_Orre Good old fashioned mislabeling. The unique peptide MCS plot should have been labeled blue (lower curve) PXD010154 and the orange (upper curve) PXD006895.
Wed Feb 27 13:47:49 +0000 2019This plot is mislabeled. The blue (lower curve) should be PXD010154 and the orange (upper curve) should be PXD006895.
Tue Feb 26 15:19:27 +0000 2019This is an example of a protein where the mRNA evidence contradicts the proteomics evidence: based on the mRNA there should be lots of this protein in some tissues 🔗
Tue Feb 26 15:16:48 +0000 2019PGLYRP3:p, peptidoglycan recognition protein 3 (H. sapiens) 🔗 Small extracellular protein; extremely rarely identified; may be present in ovarian ascites and skin (weak evidence) [13 x]
Mon Feb 25 22:23:17 +0000 2019@ypriverol OK. If you can give me the credentials for the repo, I can take a look. Is the JSON going to make use of ontologies in a similar manner as other PSI formats?
Mon Feb 25 22:04:25 +0000 2019@PastelBio This sort of cell line/tissue mix up happens all the time: this group is one of the few I've seen acknowledge and try to fix the problem.
Mon Feb 25 21:38:54 +0000 2019@pwilmarth Phil, you have a real talent for understatement. Are you sure you aren't Canadian?
Mon Feb 25 21:00:34 +0000 2019@ypriverol Where can I go to get a look at your proposal?
Mon Feb 25 20:20:52 +0000 2019I am thinking about how (& whether) to create an easy-to-parse, easy-to-extend, UTF-8 compatible MGF replacement using JSON Lines for use in behind-the-scenes APIs 🔗
Mon Feb 25 13:43:07 +0000 2019PGLYRP2:p, peptidoglycan recognition protein 2 (H. sapiens) 🔗 Midsized extracellular phosphoprotein; only S-acceptor phosphorylation; rare most cell lines, common in urine, plasma and some cancer tissues; observed mature form (21,22,23)-576 [13,526] 🔗
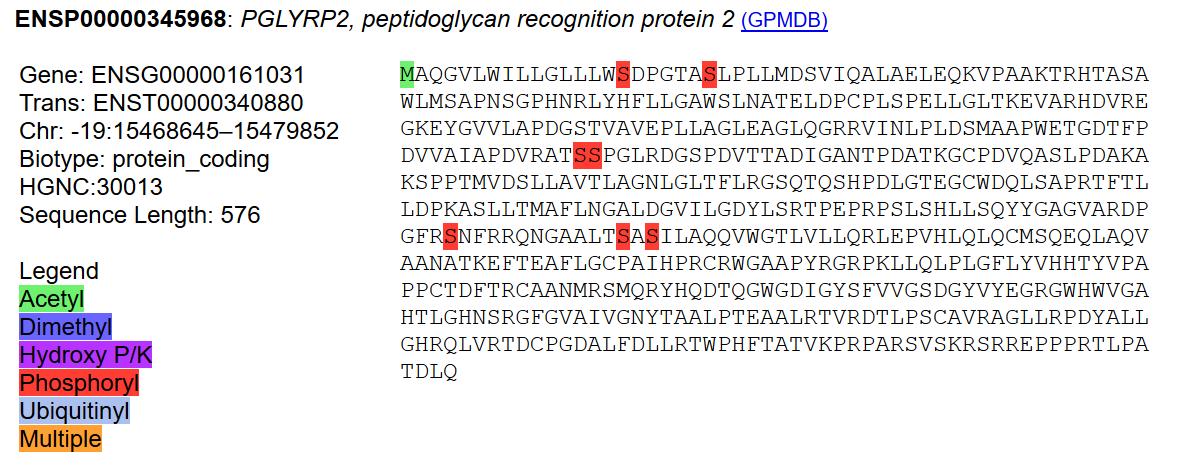
Sun Feb 24 14:13:54 +0000 2019PGLYRP1:p, peptidoglycan recognition protein 1 (H. sapiens) 🔗 Small extracellular protein; no PTMs; absent from most cell lines, common in urine, neutrophils and some cancer tissues; observed mature form 22-196 [3,893] 🔗
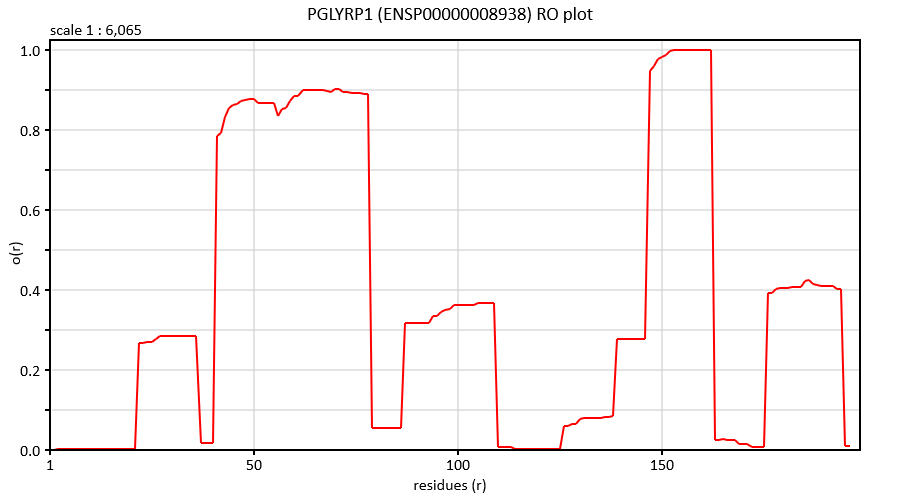
Sat Feb 23 14:50:34 +0000 2019SPG7:p, paraplegin matrix AAA peptidase subunit (H. sapiens) 🔗 Midsized mitochondrial matrix enzyme subunit; no PTMs; aka spastic paraplegia 7; mature form is a homohexameric metallopeptidase 🔗
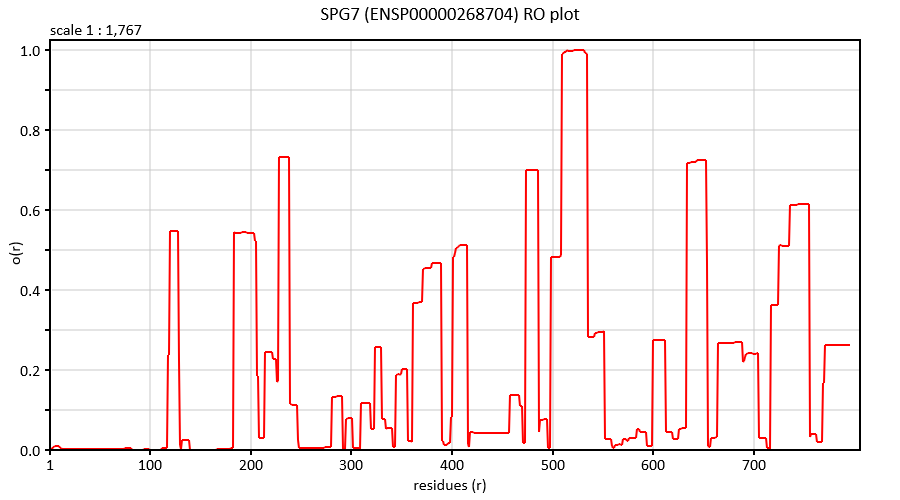
Fri Feb 22 21:34:23 +0000 2019@ypriverol Any idea how many of these are being used on Win64, Linux and/or OsX?
Fri Feb 22 17:16:39 +0000 2019@ypriverol That graph surprises me.
Fri Feb 22 15:58:02 +0000 2019@ProteomicsNews This can't be emphasized enough.
Fri Feb 22 13:58:10 +0000 2019MAIP1:p, matrix AAA peptidase interacting protein 1 (H. sapiens) 🔗 Small mitochondrial matrix enzyme subunit; one rarely occupied PTM site: K173+acetyl; aka C2orf47; observed mature form 97-291 [7,963 x] 🔗
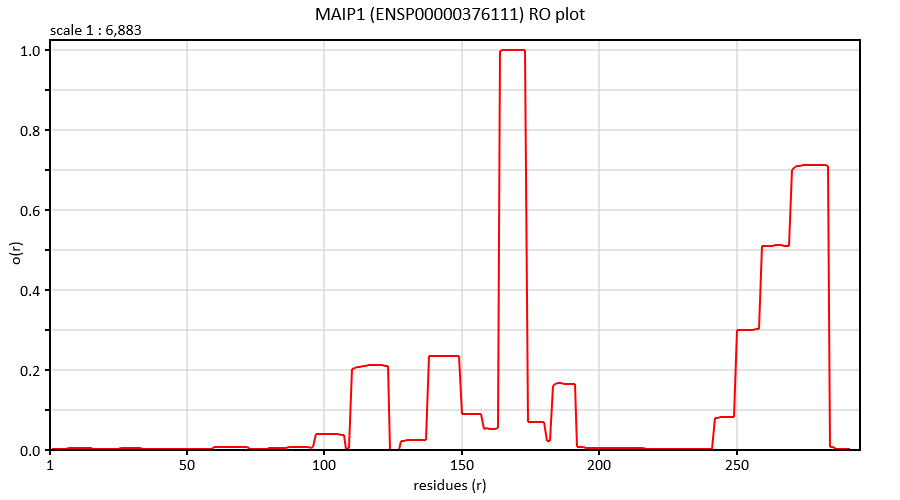
Thu Feb 21 17:45:17 +0000 2019@nesvilab @ypriverol @leprevostfv @BrenesAlejandro I am not an absolutist for open or closed source: I have written considerable more closed source code than open source. However I have found that for a complex, difficult-to-test-adequately project – like a search engine – open source has some benefits.
Thu Feb 21 17:29:11 +0000 2019@nesvilab @ypriverol @leprevostfv @BrenesAlejandro Students have also used the project as a starting point for their work in developing new search engines and/or for porting to different computational platforms such as various types of computer clusters or GPU-based systems.
Thu Feb 21 17:23:33 +0000 2019@nesvilab @ypriverol @leprevostfv @BrenesAlejandro X! Tandem was mainly written by me, but other people contributed code to do things that they wanted to be included, particularly handling specific file formats and generalizing the scoring mechanism. Others did thorough code reviews and pointed out errors.
Thu Feb 21 17:19:42 +0000 2019@nesvilab @ypriverol @leprevostfv @BrenesAlejandro That has always been the case for open-source utilities. A small number of people (often only one) write the majority of the code and act as the editor for contributions by others.
Thu Feb 21 15:52:33 +0000 2019@btaplatt About to do his best John Banner impersonation, I'd wager.
Thu Feb 21 13:53:23 +0000 2019AFG3L2:p, AFG3 like matrix AAA peptidase subunit 2 (H. sapiens) 🔗 Midsized mitochondrial matrix enzyme subunit; many PTM sites; observed mature form 52-797 [21,553 x] 🔗

Wed Feb 20 17:00:50 +0000 2019@Smith_Chem_Wisc That is, the biological replicates are trying to understand whether or not the specific growth conditions for a particular flask are biasing the results.
Wed Feb 20 16:58:50 +0000 2019@Smith_Chem_Wisc IMHO: Technical replicates are from the same flask. Biological replicates are from different flasks, preferably cultured at different times and/or locations.
Wed Feb 20 15:36:27 +0000 2019Neither study is near obtaining the maximum number of unique peptides available to their respective protocols.
Wed Feb 20 15:34:24 +0000 2019Both studies are near the maximum number of unique proteins that can be discovered using their experimental protocols. The experimental procedure used in PXD006895 generated almost 60% more unique protein ids.
Wed Feb 20 15:29:25 +0000 2019This plots show the unique peptide detection performance for the same two studies. 🔗
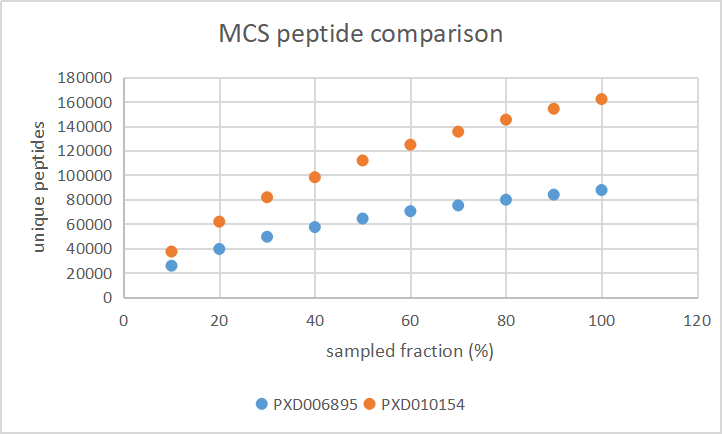
Wed Feb 20 15:28:36 +0000 2019This plot shows the unique protein detection performance of 2 recent large-scale proteomics experiments (PXD010154 🔗 & PXD006895 🔗). Both studies used about 1.3 million MS/MS spectra per sample. 🔗
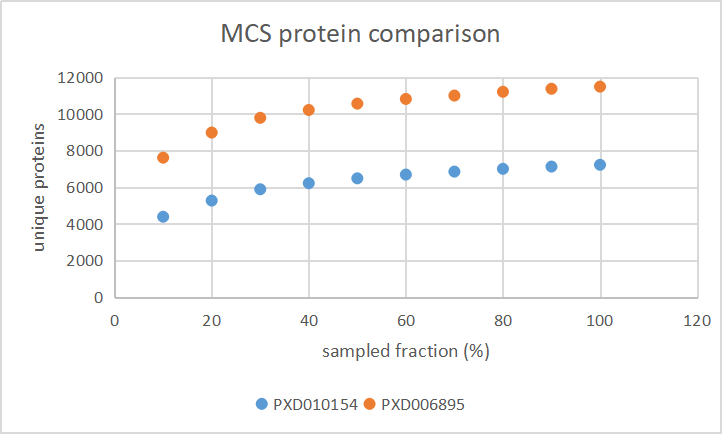
Wed Feb 20 13:28:25 +0000 2019LONP1:p, lon peptidase 1, mitochondrial (H. sapiens) 🔗 Midsized mitochondrial matrix subunit; at least 7 occasionally occupied acetylation sites; observed mature form 68-959 [33,327 x] 🔗
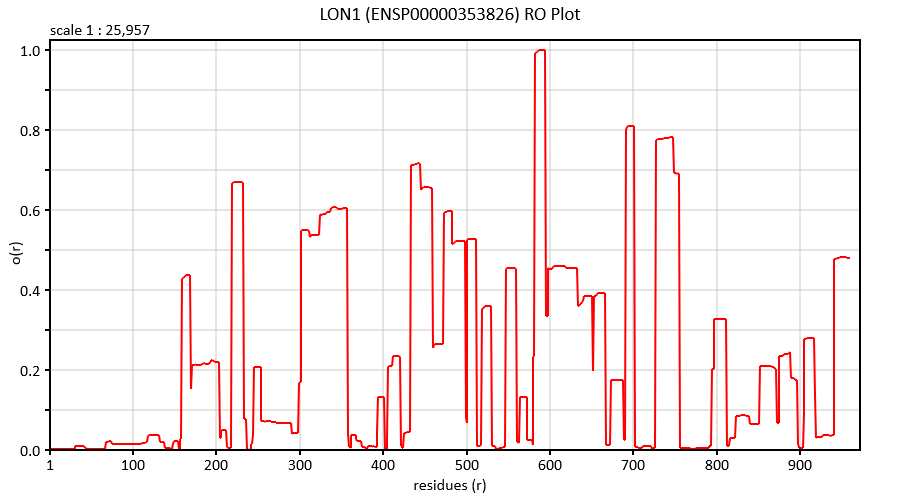
Tue Feb 19 22:44:18 +0000 2019@nesvilab Taking more spectra mostly fills-in-the-blanks for the proteins with concentrations above their LOD: increased sequence coverage and detection of PTMs and SAVs.
Tue Feb 19 22:29:28 +0000 2019@pwilmarth I've seen quite a few data sets out of that lab. I would describe their experimental design paradigm to be 'seat-of-the-pants' (in the best bayerische Tradition). They tend to be a pretty jazzy when it comes to following an SOP.
Tue Feb 19 21:52:20 +0000 2019@nesvilab My experience is that the experimental design and chromatographic methods determine the protein LOD for a particular study. Large numbers of MS/MS spectra have a marginal effect on the # of unique protein ids.
Tue Feb 19 20:57:55 +0000 2019The votes are in. With 50 people/robots participating, "hype" is the winner with 66% of the votes and "9000" is in second place with 20%.
Tue Feb 19 15:29:32 +0000 2019GOT2:p, glutamic-oxaloacetic transaminase 2 (H. sapiens) 🔗 Small mitochondrial matrix enzyme; 15 K-acetylation and 7 (!!!!) Y-phosphorylation sites; p.V346G maf=0.79; observed mature form 29-430 [35,760 x] 🔗

Tue Feb 19 14:15:22 +0000 2019Hype is still leading the pack, with 9,000 a distant second ...
Mon Feb 18 21:37:05 +0000 2019We are off to a good start, with "hype" in the early lead!
Mon Feb 18 20:50:54 +0000 2019What is the minimum number of proteins from a tissue sample that have to be observed with good confidence for a result to be considered "deep proteomics"?
Sun Feb 17 20:46:43 +0000 2019After another 4 days of use, I've ordered the parts to build 2 more of these things.
Sun Feb 17 18:17:53 +0000 2019Does anybody know if the samples in this study (🔗) were kept at elevated temperature and/or low pH following the tryptic digest for an extended period of time?
Sun Feb 17 17:24:42 +0000 2019D2HGDH:p, D-2-hydroxyglutarate dehydrogenase (H. sapiens) 🔗 Midsized mitochondrial matrix enzyme; no observed PTMs; observed mature form 38-521 [2,834 x] 🔗
Sat Feb 16 15:14:36 +0000 2019ETFDH:p, electron transfer flavoprotein dehydrogenase (H. sapiens) 🔗 Midsized mitochondrial subunit; no observed PTMs; observed mature form 36-617 [10,690 x] 🔗
Fri Feb 15 16:47:09 +0000 2019@AlexUsherHESA It is "unwillingness" not "inability"
Fri Feb 15 13:57:29 +0000 2019There may be something wrong with this sequence: it is located in mitochondria but it does not have a mitochondrial targeting peptide (MTP). There is a longer splice, but the unobserved portion of the N-terminus doesn't really look like an MTP.
Fri Feb 15 13:50:49 +0000 2019ETFB:p, electron transfer flavoprotein beta subunit (H. sapiens) 🔗 Small mitochondrial subunit; many observed PTMs; observed mature form 2-255 [28,047 x] 🔗
Thu Feb 14 15:58:19 +0000 2019@InklessPW Lest we forget 🔗
Thu Feb 14 15:36:54 +0000 2019ETFA:p, electron transfer flavoprotein subunit alpha (H. sapiens) 🔗 Small mitochondrial subunit; many observed PTMs; observed mature form 20-333 [32,126 x] 🔗

Wed Feb 13 19:27:25 +0000 2019@kady I would be more interested if they hauled the relevant lobbyists from SNV-L before the committee and asked them what they wanted from the Government and what they obtained from the Government.
Wed Feb 13 17:59:10 +0000 2019The Chicago Tribune endorses Bill Daley for mayor, although I don't know why an endorsement is really necessary in this particular case 🔗
Wed Feb 13 17:14:29 +0000 2019@chrashwood Maybe it is just a load issue.
Wed Feb 13 17:10:43 +0000 2019@chrashwood It has started working again, although it is responding very slowly.
Wed Feb 13 16:42:05 +0000 2019Does anyone know why 🔗 is down at the moment?
Wed Feb 13 13:23:27 +0000 2019FASTKD3:p, FAST kinase domains 3 (H. sapiens) 🔗 Midsized mitochondrial matrix kinase; no observed PTMs; observed mature form 69-662 [1,430 x] 🔗
Tue Feb 12 22:21:58 +0000 2019I've only been using a Raspberry Pi implementation of pi-hole for 24 hours, but based on what I've seen so far, I probably will end up building several more for my other networks.
Tue Feb 12 15:28:55 +0000 2019@UCDProteomics @BenoitKunath @ypriverol @mvaudel @byu_sam @lukas_k @ProtifiLlc @dtabb73 @ProteomicsNews @pwilmarth @Bioschema You may be doing this already, but if you are transferring a directory of files, or want to update a remote directory with new files, try the Windows command line utility robocopy:
>robocopy "local_dir" "remote_dir" /MIR
It is much better than a drag-and-drop
Tue Feb 12 15:14:46 +0000 2019@AlexUsherHESA You might also reflect on the 1 year anniversary of the Superclusters funding announcement (on the 14th), which so far has resulted in 5 generic-looking web sites & little else.
Tue Feb 12 13:49:23 +0000 2019@BenoitKunath @ypriverol @mvaudel @byu_sam @lukas_k @ProtifiLlc @dtabb73 @ProteomicsNews @pwilmarth @UCDProteomics @Bioschema The simplest way I know of to check the file integrity is to gzip the files prior to transfer. gzip generates and stores a CRC value for the data that can be checked for consistency following transfer.
Tue Feb 12 13:44:36 +0000 2019MTO1:p, mitochondrial tRNA translation optimization 1 (H. sapiens) 🔗 Midsized mitochondrial matrix subunit; lot of ubiquitination; observed mature form (25,34)-732 [3,383 x] 🔗
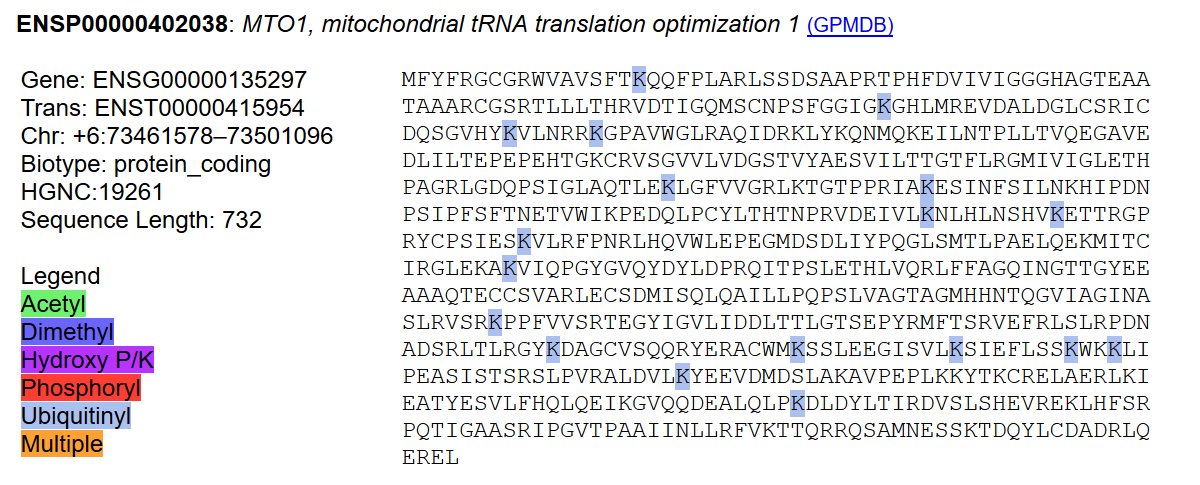
Tue Feb 12 13:32:59 +0000 2019@BenoitKunath @ypriverol @mvaudel @byu_sam @lukas_k @ProtifiLlc @dtabb73 @ProteomicsNews @pwilmarth @UCDProteomics @Bioschema What file formats are involved?
Mon Feb 11 16:05:23 +0000 2019@AlexUsherHESA Twitter is the new Hansard.
Mon Feb 11 13:15:56 +0000 2019TSFM:p, Ts translation elongation factor, mitochondrial (H. sapiens) 🔗 Small mitochondrial matrix subunit; 4 phosphorylation sites; observed mature form 42-325 [16,168 x] 🔗
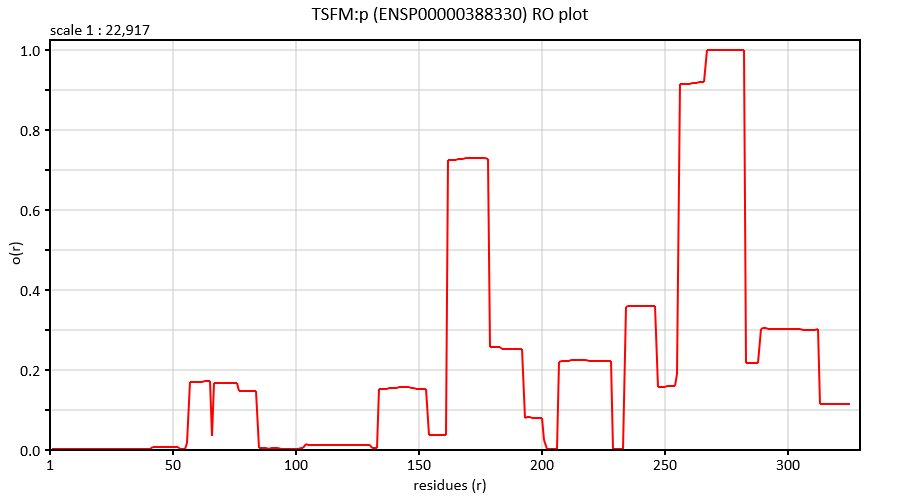
Sun Feb 10 19:41:04 +0000 2019@ForrestMichelle Hope you like it 😀
Sun Feb 10 18:38:24 +0000 2019I don't think anyone in the province of Manitoba is surprised either that the Premier is in CR, or that he doesn't do his own media posts. Most would be surprised if he regularly used anything more advanced than a bakelite single-line POTS phone.
🔗
Sun Feb 10 18:20:03 +0000 2019@AlexUsherHESA And superclusters! In Toronto!
Sun Feb 10 14:50:15 +0000 2019@AaronWherry At least the PMO is getting its money's worth out of the new AG.
Sun Feb 10 14:35:37 +0000 2019TUFM:p, Tu translation elongation factor, mitochondrial (H. sapiens) 🔗 Small mitochondrial matrix subunit; heterogeneously modified; one of the most commonly ID'd mitochondrial proteins; observed mature form 47-455 [52,576 x] 🔗
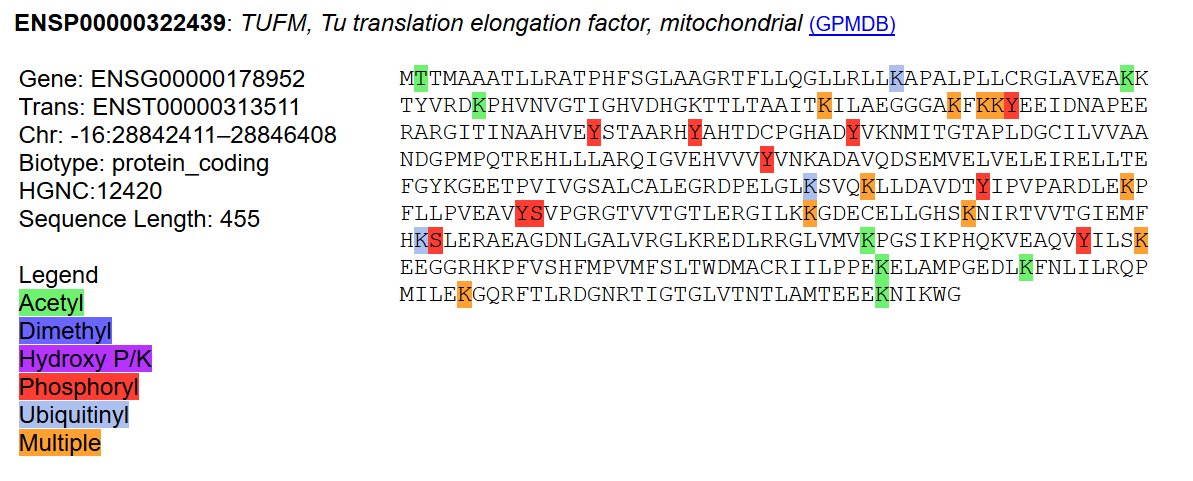
Sat Feb 09 18:27:13 +0000 2019My suggestions for making use of a biomedical database RESTful API 🔗
Sat Feb 09 16:37:30 +0000 2019U.S. steel tariff 'boondoggle' offers more exclusions to China than Canada 🔗
Sat Feb 09 14:32:36 +0000 2019Sometimes I wish that people using a REST API could make their requests in some manner that does not so closely simulate a denial-of-service attack.
Sat Feb 09 14:10:03 +0000 2019GFM1:p, G elongation factor mitochondrial 1 (H sapiens) 🔗 Midsized mitochondrial matrix protein; most frequently occupied PMT S91+phosphoryl; observed mature form 36-751 [18,729 x] 🔗
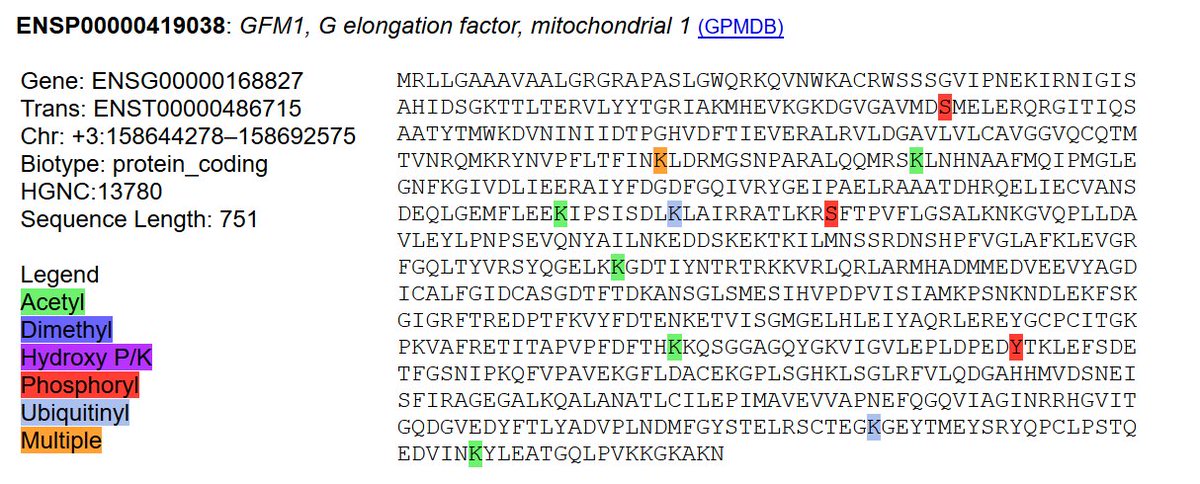
Sat Feb 09 00:45:08 +0000 2019@UCDProteomics @pwilmarth @neely615 @ProtifiLlc @ypriverol @mvaudel @byu_sam @lukas_k @dtabb73 @ProteomicsNews @Bioschema @MagnusPalmblad The discussion seems to have taken a turn to the dark side.
Fri Feb 08 21:12:50 +0000 2019@PastelBio @ProteomicsNews And a more typical Class II result, that seems to be a mixture of at least 2 different length distributions 🔗
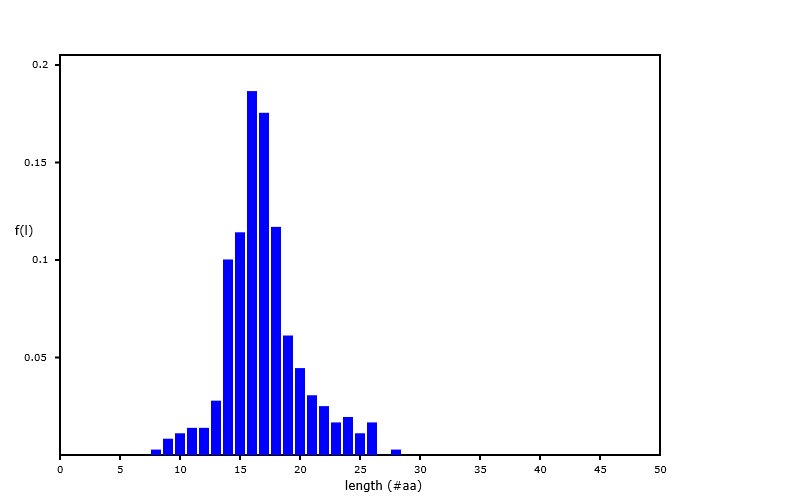
Fri Feb 08 21:11:17 +0000 2019@PastelBio @ProteomicsNews And a really good Class II result 🔗
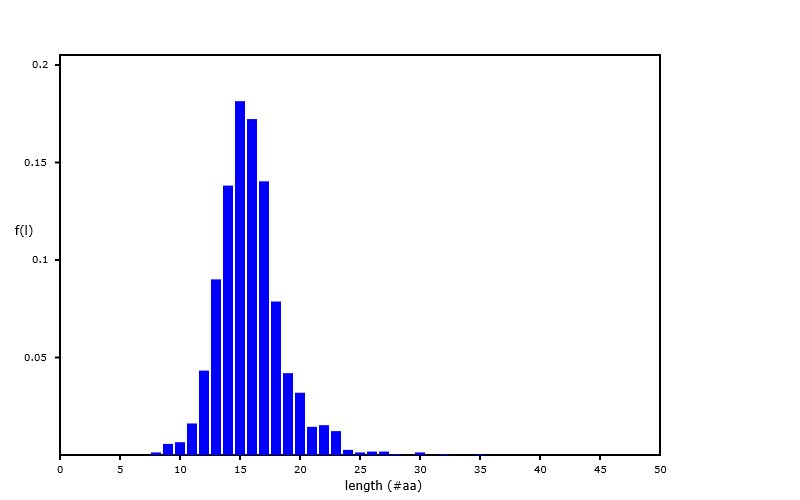
Fri Feb 08 21:10:13 +0000 2019@PastelBio @ProteomicsNews And another typical Class I result, with a sharp distribution sitting on a broad background 🔗
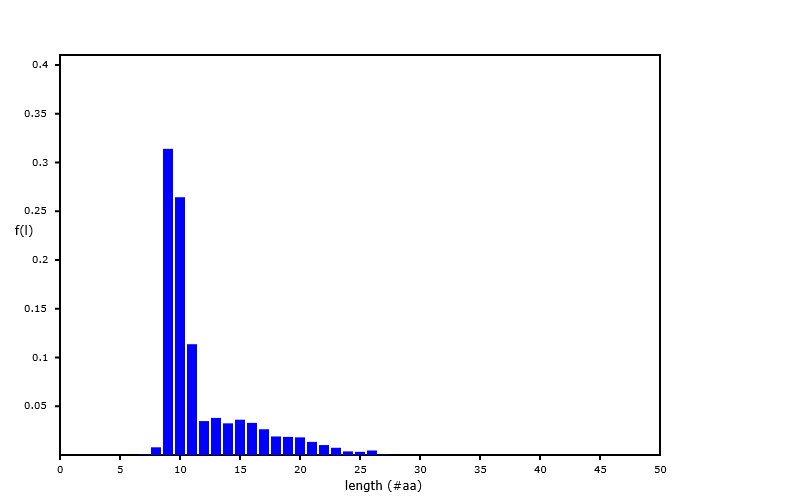
Fri Feb 08 21:09:03 +0000 2019@PastelBio @ProteomicsNews A more typical MHC Class I result, with a shoulder towards higher length 🔗
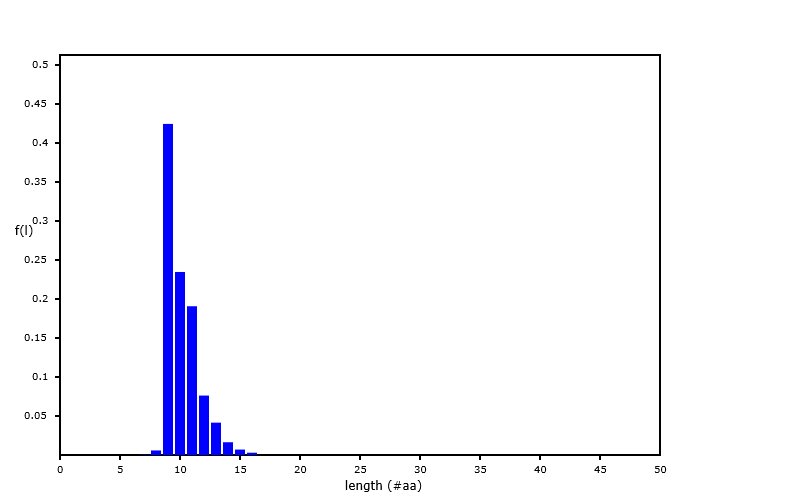
Fri Feb 08 21:08:01 +0000 2019@PastelBio @ProteomicsNews A really good MHC Class I result, showing the observed peptide lengths 🔗
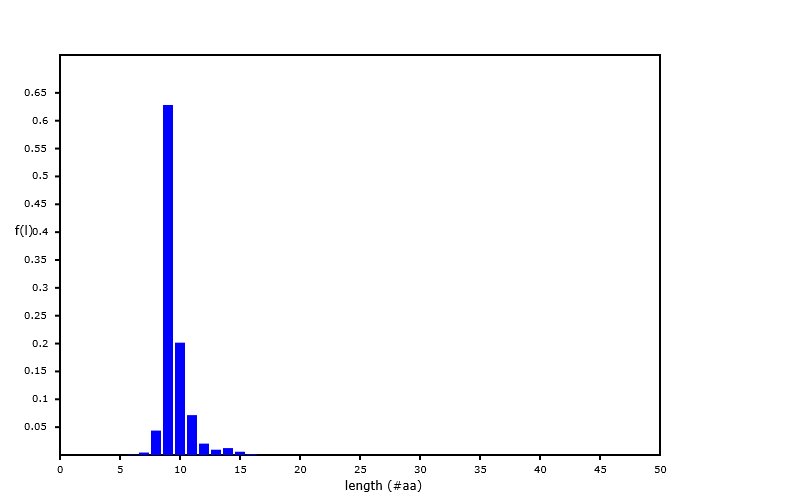
Fri Feb 08 17:44:05 +0000 2019@PastelBio @ProteomicsNews It also isn't unusual that a particular MHC peptide prep doesn't work, resulting in a set of peptides that don't conform to length constraints for genuine MHC peptide
Fri Feb 08 17:42:25 +0000 2019@PastelBio @ProteomicsNews I'd suggest +1: 700-2500; +2: 350-1250; etc. Also note that MHC Class 2 peptides can be highly charged.
Fri Feb 08 15:50:36 +0000 2019@UCDProteomics @ypriverol @mvaudel @byu_sam @lukas_k @ProtifiLlc @dtabb73 @ProteomicsNews @pwilmarth @Bioschema I don't know how to write generic quant software for proteomics data. Every time I do it I have to customize things to take into account the peculiarities of the experimental protocol, data and protein sequences involved in the measurement.
Fri Feb 08 14:18:18 +0000 2019LETM1:p, leucine zipper and EF-hand containing transmembrane protein 1 (H. sapiens) 🔗 Midsized mitochondrial interior membrane protein; many PTM acceptor sites; observed mature form 116-739 [24,386 x] 🔗
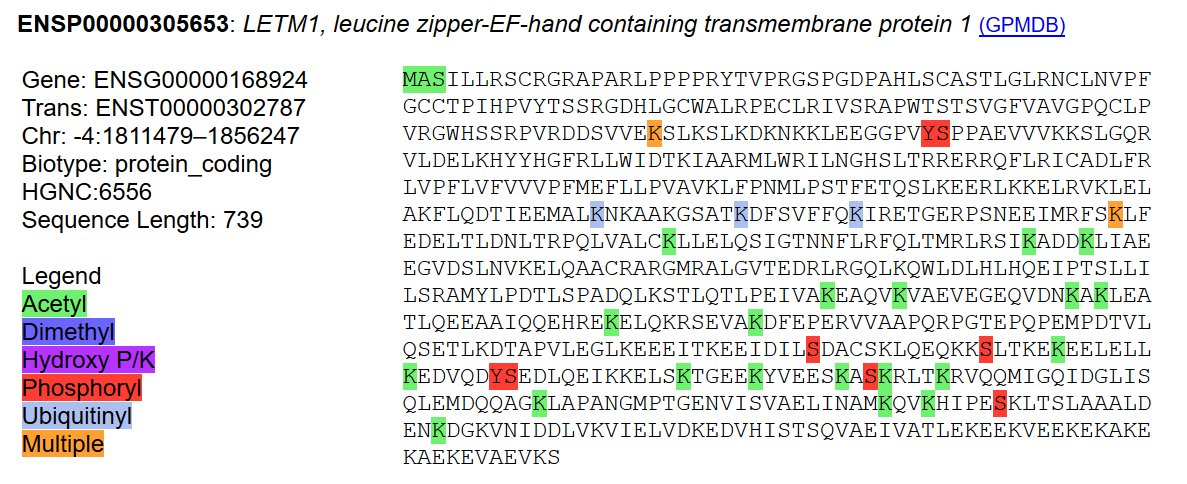
Fri Feb 08 14:05:51 +0000 2019@ypriverol @mvaudel @byu_sam @lukas_k @ProtifiLlc @dtabb73 @ProteomicsNews @pwilmarth @UCDProteomics @Bioschema I use my own software, which is not publicly available.
Fri Feb 08 02:19:16 +0000 2019Always something unexpected: PXD011762 is the first data set I've seen where the tryptic peptides have their cysteines cysteinylated (the method says they should be IAA derivatized). It is common in MHC peptides, but not in tryptic digests.
Thu Feb 07 16:38:38 +0000 2019@PastelBio Thanks, 😀
Thu Feb 07 16:25:24 +0000 2019I wonder what elliptical terms the PMO used in emails and notes to refer to SNC-****** 🔗
Thu Feb 07 13:20:18 +0000 2019MICU2:p, mitochondrial calcium uptake 2 (H sapiens) 🔗 Small mitochondrial interior membrane subunit; forms a heterodimer with MICU1; observed mature form 31-434 [7,157 x]; aka EF hand domain family A1 🔗
Thu Feb 07 13:12:28 +0000 2019@neely615 The system is broken.
Wed Feb 06 16:13:24 +0000 2019@slavovLab Bergseng E, et al, "Different binding motifs of the celiac disease-associated HLA molecules DQ2.5, DQ2.2, and DQ7.5 revealed by relative quantitative proteomics of endogenous peptide repertoires " 🔗
Wed Feb 06 16:01:38 +0000 2019@girlziplocked The Strategic Defense Initiative
Wed Feb 06 15:33:53 +0000 2019good data ✓; interesting sample ✓; charismatic species ✓
Wed Feb 06 15:30:55 +0000 2019"13_honeys_proteomics" (🔗) is one of those data sets where the title is really all you need to know about the experiments.
Wed Feb 06 14:16:52 +0000 2019MICU1:p, mitochondrial calcium uptake 1 (H. sapiens) 🔗 Small mitochondrial interior membrane subunit; 5 K-ubiquitinylation acceptor sites; observed mature form 58-476 [6,386 x]; aka calcium binding atopy-related autoantigen 1 🔗
Tue Feb 05 18:26:33 +0000 2019@drtongue96 @bkives Electric bikes are great. Electric scooters are great. Electric scooter "rental" companies are evil.
Tue Feb 05 18:20:14 +0000 2019@bkives Simply make it illegal for them to operate in Winnipeg.
Tue Feb 05 16:41:39 +0000 2019@PastelBio My position is that such data cannot be truly anonymized, as it is always possible at a later date to find it unambiguously, using a similar sample from the same individual.
Tue Feb 05 16:16:11 +0000 2019@PastelBio It does beg the question, however, as to whether proteomics data that contains readily identifiable SAVs can ever be truly anonymized.
Tue Feb 05 16:14:39 +0000 2019@PastelBio This is a pretty good article on the problems associated with GDPR and clinical data for proteomics & related fields. Anyone designing data storage, analysis or interpretation algorithms, software or databases should read this article.
Tue Feb 05 14:26:05 +0000 2019MCU:p, mitochondrial calcium uniporter (H. sapiens) 🔗 Small mitochondrial interior membrane subunit; 3 low occupancy K-acetylation and 1 Y-phosphorylation site; observed mature form 67-351 [11,303 x] 🔗
Mon Feb 04 20:58:11 +0000 2019@UCDProteomics @zacmcd77 But of course I've never tried to get the reaction to run myself. I leave that to wiser men ...
Mon Feb 04 20:57:05 +0000 2019@UCDProteomics @zacmcd77 The enzyme works pretty well. The spectra take a little getting used to: 2+ are almost always just a ladder of b-ions & maybe a few small y's. Not uncommon to see an a-ion ladder too.
Mon Feb 04 18:57:07 +0000 2019@Smith_Chem_Wisc @byu_sam The N:Q deamidation ratio is also useful when tuning algorithms: any change that results in a significant reduction of the ratio is going in the wrong direction.
Mon Feb 04 18:32:55 +0000 2019@bkives For after Brexit breakfast?
Mon Feb 04 17:35:49 +0000 2019Thanks to Sarah at the NYU Institute for Systems Genetics for fixing a problem with the g2pDB REST API!
Mon Feb 04 15:23:42 +0000 2019@Smith_Chem_Wisc @byu_sam For QC purposes with SAVs, the peptide artifact modification distribution for SAV PSMs (M/W mono and di-oxidation, N-term/K carbamylation, N-term pyro-Glu, etc.) should be within error of the distribution of the non-SAV PSMs.
Mon Feb 04 15:20:01 +0000 2019@Smith_Chem_Wisc @byu_sam For QA purposes, yes the N:Q deamidation ratio for the whole data set. It depends on just about everything done to the sample: storage time + temp during and after cleavage, MS resolution and peak shape, deisotoping/parent ion assignment & PSM assignment.
Mon Feb 04 13:52:28 +0000 2019ME2:p, malic enzyme 2 (Homo sapiens) 🔗 Midsized mitochondrial enzyme; highest occupancy PTMs are K156+acetyl and K240+acetyl; shares some tryptic peptides with ME1 and ME3 [22,185 x] 🔗
Sun Feb 03 13:54:57 +0000 2019ME1:p, malic enzyme 1 (H. sapiens) 🔗 Midsized cytosolic enzyme; several low-occupancy PTM sites; shares some tryptic peptides with ME2 and ME3 [15,162 x] 🔗
Sun Feb 03 01:42:43 +0000 2019@Smith_Chem_Wisc @byu_sam The best QA measure I've been able to come up with for SAV-worthiness is the N:Q deamidation ratio. If it is N:Q > 4, then everything is going to be OK. If N:Q < 2 and the data doesn't come from an archeological sample, don't get your hopes up.
Sat Feb 02 17:39:21 +0000 2019@Smith_Chem_Wisc @byu_sam My experience with the subject has made me much more skeptical. I too have convinced myself that TMT-derived SAVs were correct, only to find out later that I was wrong.
Sat Feb 02 14:34:19 +0000 2019@byu_sam More generally, if you are using any type of quantitation reagent (TMT, iTRAQ, dimethyl), you will have a very hard time finding any legitimate SAVs in the resulting data
Sat Feb 02 12:50:11 +0000 2019GRPEL1:p, GrpE like 1, mitochondrial (H sapiens) 🔗 Small mitochondrial matrix subunit; involved in protein recruitment to the HSP70 chaperonin; many K-acetylation acceptor sites [19,720 x] 🔗
Sat Feb 02 12:38:15 +0000 2019@byu_sam My experience is that it is very difficult to see SAVs when you are using TMT6 or 10.
Fri Feb 01 17:11:25 +0000 2019@ypriverol File format adoption is almost as difficult to understand as file format development.
Fri Feb 01 16:55:01 +0000 2019@pwilmarth I like #3, with its muted call back to the '90s AAA analysis boom. You can still find proteins that to this day have been named and assigned to particular tissues based on a AAA result from that period.
Fri Feb 01 16:20:23 +0000 2019@pwilmarth The current rationale for the "canonical" selection process is described here 🔗
Fri Feb 01 15:52:25 +0000 2019@pwilmarth I kind of like the whimsical, Dark Ages sound of "canonical". "Anointed" (or "blessed") would certainly carry the same connotations.
Fri Feb 01 13:26:40 +0000 2019Note: this is not the "canonical" sequence (Q9H078-1, ENSP00000294053, 1-707). There is no evidence that the canonical sequence has been observed in proteomics experiments.
Fri Feb 01 13:20:10 +0000 2019CLPB:p, ClpB homolog, mitochondrial AAA ATPase chaperonin (H. sapiens) 🔗 Midsized mitochondrial matrix enzyme subunit; involved in protein unfolding and disaggregation; observed mature form 127-677 [12,536 x] 🔗
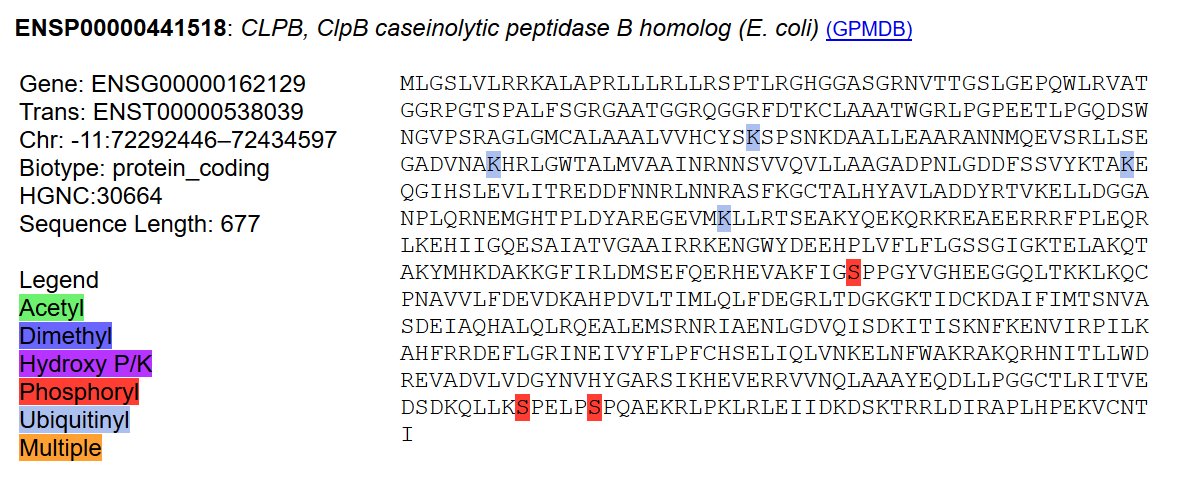
Thu Jan 31 13:54:44 +0000 2019@slavovLab It is a lot, but I am not in any way surprised. I have very little respect for the integrity of the peer review process, even though I have benefited from it.
Thu Jan 31 13:22:13 +0000 2019CLPX:p, caseinolytic mitochondrial matrix peptidase chaperone subunit (H. sapiens) 🔗 Midsized mitochondrial matrix enzyme subunit; significant acetylation and phosphorylation; observed mature form 108-633 [15,550 x] 🔗
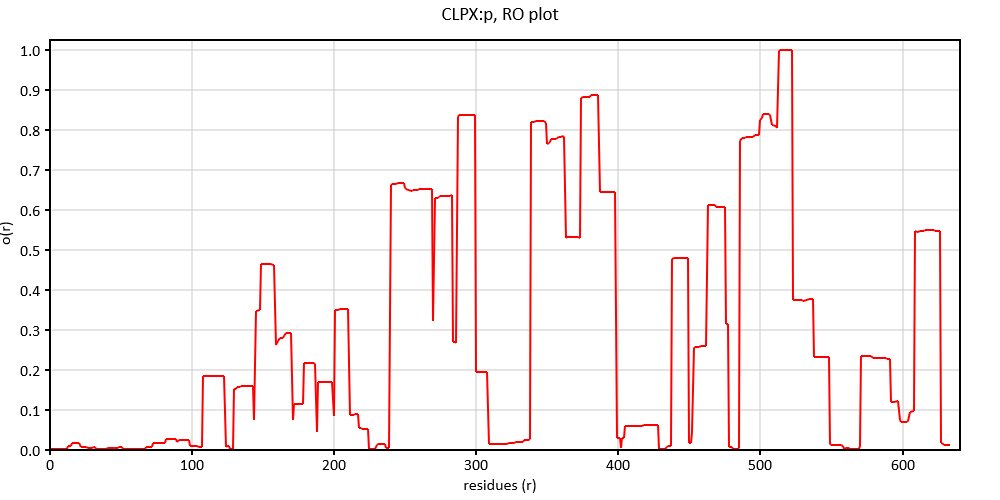
Thu Jan 31 13:16:55 +0000 2019@slavovLab I've seen it myself quite a few times. Even editors get in on the game.
Wed Jan 30 16:01:52 +0000 2019The measured temperature across Europe: 2019-01-30 15:00 UTC - this is why winter-time cycling is more popular in Europe than it is in most of North America. 🔗
Wed Jan 30 16:00:43 +0000 2019The measured temperatures across North America: 2019-01-30 15:00 UTC 🔗
Wed Jan 30 15:38:08 +0000 2019CLPP:p forms a 14mer toroid that is the proteolytic part of the mitochondrial equivalent of the proteasome, i.e., it recycles misfolded or damaged proteins.
Wed Jan 30 14:19:30 +0000 2019@AlexUsherHESA I guess now we know where the SCALE AI supercluster money is going.
Wed Jan 30 13:35:33 +0000 2019CLPP:p, caseinolytic mitochondrial matrix peptidase proteolytic subunit (H. sapiens) 🔗 Small mitochondrial matrix enzyme subunit; small number of relatively rare PTMs; observed mature form 57-277 [14,865 x] 🔗
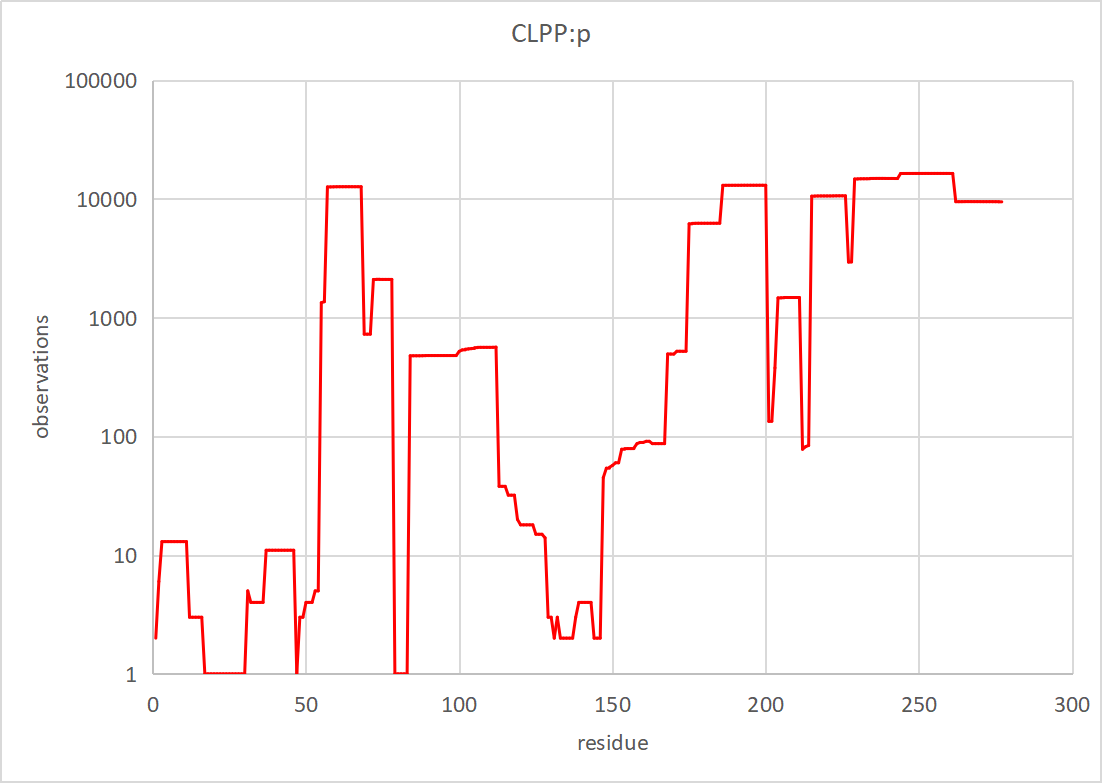
Tue Jan 29 22:35:29 +0000 2019On further reading and a few calculations, this paper has a lot of issues that need to be addressed and it probably didn't discover any new PTMs.
Tue Jan 29 16:51:51 +0000 2019"in-source chemistry" would probably be a better description than "in-source decay" as a source for unexpected ion species.
Tue Jan 29 16:11:23 +0000 2019Most of them are pretty easy to explain, particularly the Cys mods, but there may still be some interesting things in there, once you take into account non-tryptic cleavage, alternate splicing and in-source decay.
Tue Jan 29 16:06:35 +0000 2019Good for these guys 🔗 They have reported all of the variants they have not been able to explain.
Tue Jan 29 15:30:14 +0000 2019AUH:p residue observation diagram - the number of times a specific residue has been part of an identified PSM 🔗
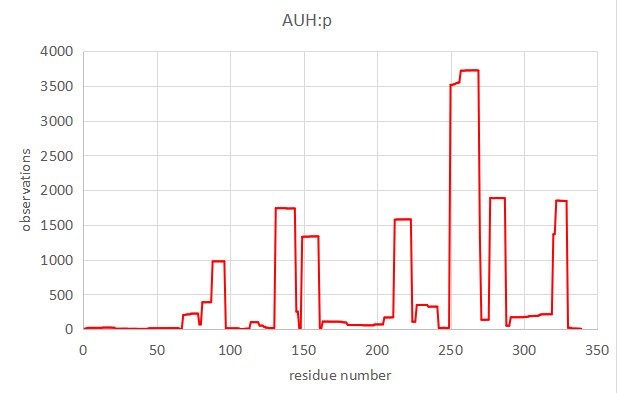
Tue Jan 29 13:50:27 +0000 2019AUH:p, AU RNA binding methylglutaconyl-CoA hydratase (H. sapiens) 🔗 Small mitochondrial matrix subunit; only 1 PMT, pm.K204+acetyl; observed mature form 68-399 [4,536 x]
Mon Jan 28 18:50:54 +0000 2019@byu_sam Although it is also possible that I am just an old guy yelling at the TV.
Mon Jan 28 18:50:16 +0000 2019@byu_sam If there are new PTMs, great! But often these claims are made without any conclusions regarding new biochemistry that has been discovered. Statistical distributions and heuristics aren't very good at proving that something is really deterministic.
Mon Jan 28 18:42:26 +0000 2019@byu_sam I often read papers that suggest that a new method/algorithm has found many new IDs in a data set, but there is very little discussion as to whether these new IDs are consistent with "old" IDs or whether they uncover any new information about proteins in the sample.
Mon Jan 28 18:39:27 +0000 2019@byu_sam My concern is that the field has become so consumed with producing "more" IDs that little attention is paid to whether the additional IDs improve the accuracy of determining the details of the proteins present in a particular sample.
Mon Jan 28 16:32:32 +0000 2019SUOX:p, sulfite oxidase (H. sapiens) 🔗 Midsized mitochondrial matrix subunit; no observed PTMs; observed mature form 78-545 [4,805 x]
Mon Jan 28 16:16:35 +0000 2019I would suggest that the "finding more PSMs" (based on a target-decoy simulation) approach to evaluating proteomics data analysis methods is the major reason the field has gotten stuck in a rut. 🔗
Sun Jan 27 15:08:19 +0000 2019DIABLO:p, diablo IAP-binding mitochondrial protein (H. sapiens) 🔗 Small mitochondrial matrix protein; involved in the initiation of apoptosis; observed mature form 56-239 [16,929 x] 🔗
Sat Jan 26 16:28:26 +0000 2019@kadzuis Stick with it, Gary. You've got to do this sort of thing for yourself: if others benefit, great, but primarily it has to satisfy your needs and curiosity.
Sat Jan 26 14:08:34 +0000 2019MSRA:p, methionine sulfoxide reductase A (H. sapiens) 🔗 Small mitochondrial matrix metalloenzyme; has an unusually ragged N-terminus; observed mature form 25,26,27,28-235 [5,328 x] 🔗

Fri Jan 25 13:49:12 +0000 2019MSRB2:p, methionine sulfoxide reductase B2 (H. sapiens) 🔗 Small mitochondrial matrix metalloenzyme; no observed PTMs; observed mature form 21-182 [3,464 x] 🔗
Thu Jan 24 22:31:30 +0000 2019@ypriverol @pwilmarth @leprevostfv @byu_sam @mvaudel @EricWDeutsch @OpenMSTeam Just for completeness, you can also state the list of potential phosphorylation sites as a range rather than an enumerated list:
🔗
Thu Jan 24 22:24:14 +0000 2019Even after living in the US for many years, sometimes the citizens of that great republic do give me pause ... 🔗
Thu Jan 24 20:41:58 +0000 2019@byu_sam Good. Thanks for the info.
Thu Jan 24 19:59:09 +0000 2019@byu_sam I haven't been in CPTAC for some time, but NIST was central to a lot of data processing/storage/dissemination at one point. Sounds like the endless phone calls haven't changed, though.
Thu Jan 24 19:13:09 +0000 2019Does anybody know if CPTAC has been affected by the NIST shutdown?
Thu Jan 24 19:06:52 +0000 2019@ypriverol @pwilmarth @leprevostfv @byu_sam @mvaudel @EricWDeutsch @OpenMSTeam OK. Is there anything available online regarding this effort?
Thu Jan 24 17:42:06 +0000 2019PXD009283: really good data ✓, interesting problem ✓, charismatic species ✓, 🔗
Thu Jan 24 16:59:56 +0000 2019@Smith_Chem_Wisc I suspect in most cases they can't find the data, even if they wanted to make it available. Phil Andrews (Tranche) used to get really upset about this stuff. I have a record of 0 successes in the last 15 years of making such requests.
Thu Jan 24 16:42:46 +0000 2019@Smith_Chem_Wisc My experience is that "data available on request" means that you can make a request for the data, but you will only get back some vague excuse as to why you will never get it.
Thu Jan 24 15:26:17 +0000 2019@ypriverol @pwilmarth @leprevostfv @byu_sam @mvaudel @EricWDeutsch @OpenMSTeam If you want the numbers rather than the frequencies, add "&w=n" to the URL.
Thu Jan 24 15:24:58 +0000 2019@ypriverol @pwilmarth @leprevostfv @byu_sam @mvaudel @EricWDeutsch @OpenMSTeam If there is no information about the sites, the frequency values are 'null'. This also happens if the accession is misspelled.
Thu Jan 24 15:20:55 +0000 2019@ypriverol @pwilmarth @leprevostfv @byu_sam @mvaudel @EricWDeutsch @OpenMSTeam You get back a JSON result:
{"2314":0.032,"2316":0.963,"2319":0.006}
which lists the set of sites being tested and the normalized frequency of observation for that set of sites. If using UniProt accs, use the splice variant identifier (-1,-2, etc) whenever possible.
Thu Jan 24 15:17:29 +0000 2019@ypriverol @pwilmarth @leprevostfv @byu_sam @mvaudel @EricWDeutsch @OpenMSTeam With no attempt at humour, an alternate resource you can use is available by using REST:
🔗
or if you want to use Uniprot:
🔗
Thu Jan 24 13:38:20 +0000 2019NAXE:p, NAD(P)HX epimerase (H. sapiens) 🔗 Small mitochondrial matrix protein; conflicting information about protein function; observed mature form 47-288 [19,862 x] 🔗
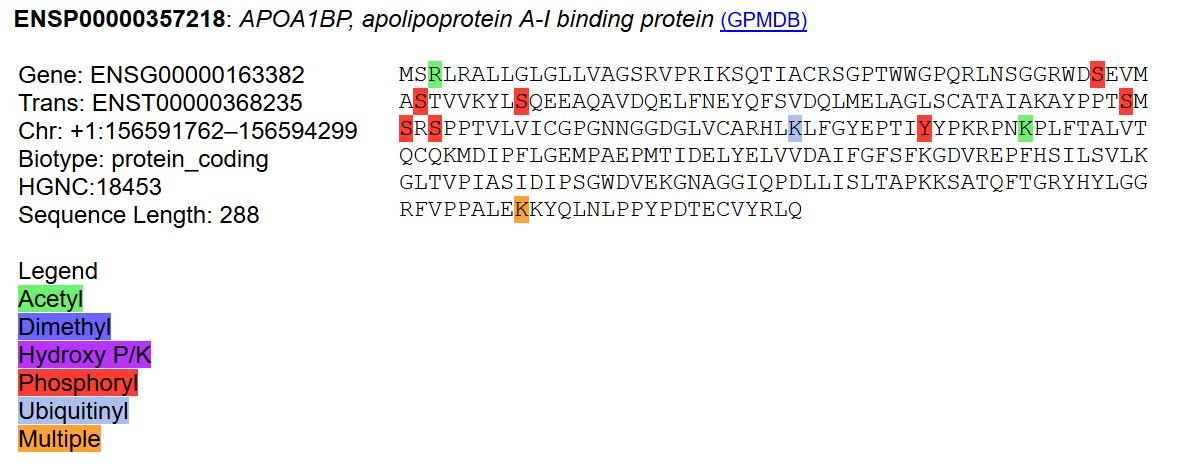
Wed Jan 23 14:35:35 +0000 2019MRRF:p, mitochondrial ribosome recycling factor (H. sapiens) 🔗 Small mitochondrial matrix protein; only 1 PTM, pm.K100+acetyl; involved in mitoribosome disassembly [8,242 x] 🔗

Tue Jan 22 16:54:29 +0000 2019@ypriverol @mvaudel @byu_sam import random
p_sites = '36,37,39'
sites = p_sites.split(',')
true_site = sites[random.randint(0,len(sites)-1)]
Tue Jan 22 14:58:32 +0000 2019TSFM:p, Ts translation elongation factor, mitochondrial (H. sapiens) 🔗 Small mitochondrial matrix protein; the most prominent SAV p.V252I (maf=0.01); observed mature form 42-325 [16,049 x] 🔗

Tue Jan 22 13:41:55 +0000 2019@AlexUsherHESA It is the very essence of Canadian political thought.
Mon Jan 21 16:03:31 +0000 2019@22_Minutes @DonMartinCTV It is much nicer in St. John's today. Actually, it is always nicer in St. John's (but it really is warmer there today).
Mon Jan 21 14:15:35 +0000 2019GFM1:p, G elongation factor mitochondrial 1 (H. sapiens) 🔗 Midsized mitochondrial matrix protein; highest occupancy PTM is pm.S91+phosphoryl; observed mature form 36-751 [18,610 x] 🔗
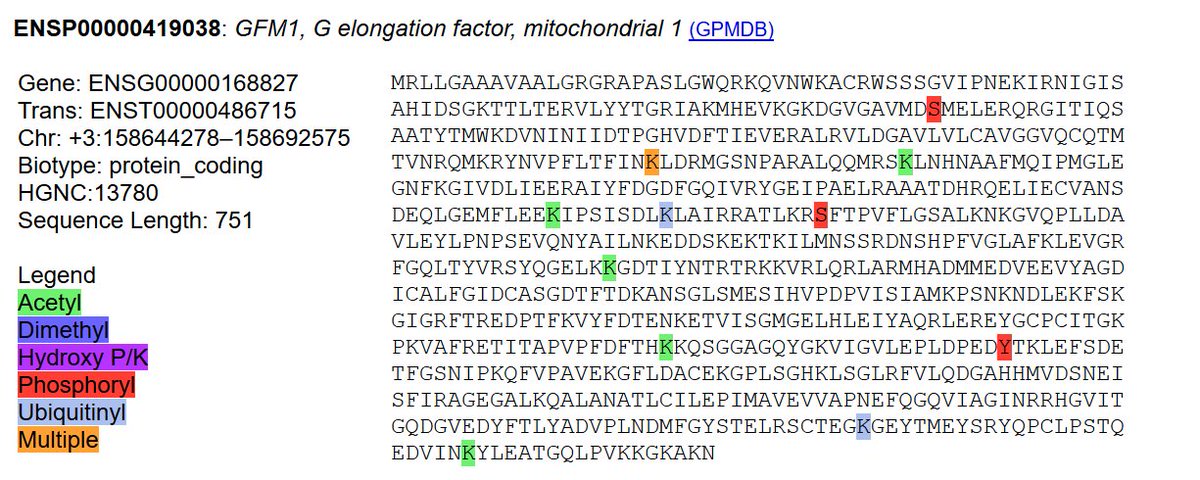
Sun Jan 20 15:06:24 +0000 2019DGLUC:p, D-glutamate cyclase (H. sapiens) 🔗 Midsized mitochondrial matrix protein; no PTMs; function very recently hypothesized (formerly C14orf159); observed mature form 29,30,32-616 [7,612 x]
Sat Jan 19 14:37:40 +0000 2019ECI1:p, enoyl-CoA delta isomerase 1 (H. sapiens) 🔗 Small mitochondrial matrix subunit; pm.K283+acetyl is the highest occupancy PTM site; observed mature form 42-302 [19,127 x] 🔗

Fri Jan 18 18:56:23 +0000 2019@BrenesAlejandro @vapetyuk @ProteomicsNews The article does suffer from mainly quoting either instrument company reps or people who take data, with very little input from anyone who thinks about how to deal with proteomics data->information->knowledge on a more global scale.
Fri Jan 18 15:31:51 +0000 2019ECI2:p, enoyl-CoA delta isomerase 2 (H. sapiens) 🔗 Small mitochondrial matrix subunit; no ID peptides shared with ECI1; observed mature form 38-394 [15,715 x] 🔗

Thu Jan 17 14:12:32 +0000 2019HSPD1:p, heat shock protein family D (Hsp60) member 1 (H. sapiens) 🔗 Midsized mitochondrial matrix subunit; extensively modified; observed mature form 26,27-573 [78,225 x] 🔗
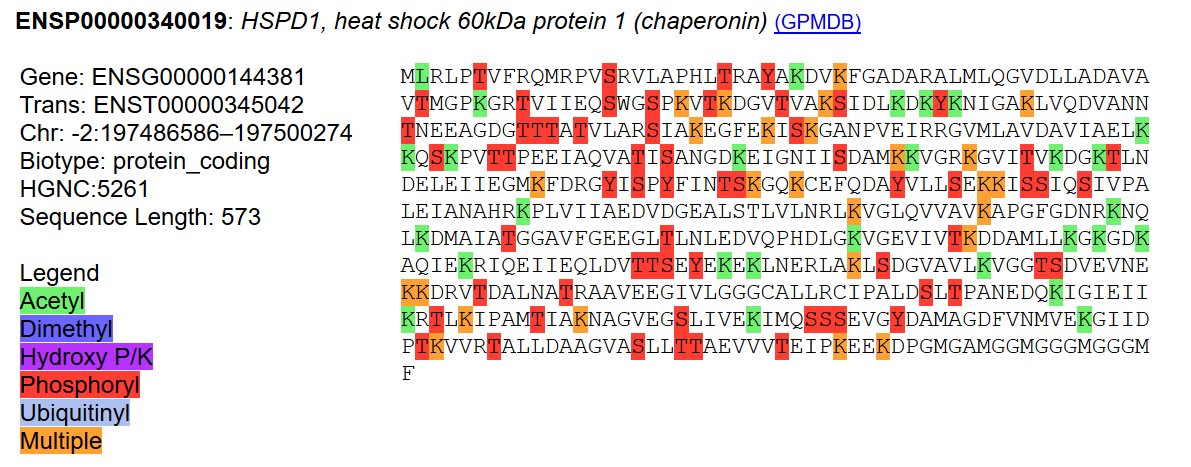
Wed Jan 16 19:19:33 +0000 2019While it isn't surprising that an app company is actually selling data it said that it would never sell, good for California for suing these guys 🔗
Wed Jan 16 13:50:07 +0000 2019ATP5F1D:p, ATP synthase F1 subunit delta (H. sapiens) 🔗 Small mitochondrial membrane associated subunit; only one PTM, pm.K138+acetyl; observed mature form 23-168 [19,672 x] 🔗

Tue Jan 15 21:48:38 +0000 2019#PXD003912 is one of the very (very) few data sets I have seen in which the experimental protocol results in complete recovery of cysteine-containing tryptic peptides. Good work 🔗
Tue Jan 15 14:00:44 +0000 2019ATP5F1C:p, ATP synthase F1 subunit gamma (H. sapiens) 🔗 Small mitochondrial membrane associated subunit; extensively modified; commonly observed; observed mature form 24-298 [38,282 x] 🔗
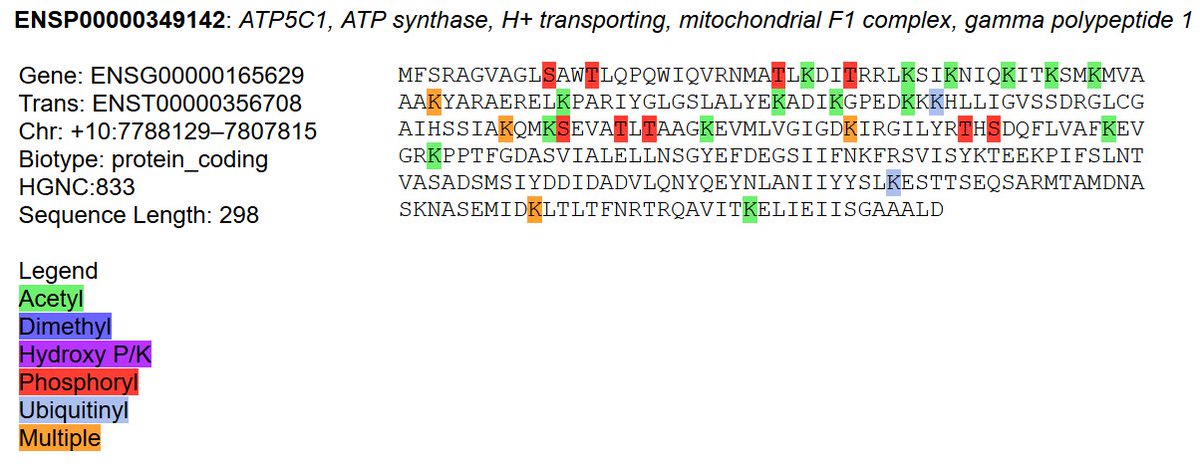
Mon Jan 14 14:08:16 +0000 2019ATP5F1B1:p, ATP synthase F1 subunit beta (H. sapiens) 🔗 Midsized mitochondrial membrane associated subunit; extensively modified; very commonly observed; observed mature form 47-529 [68,041 x] 🔗
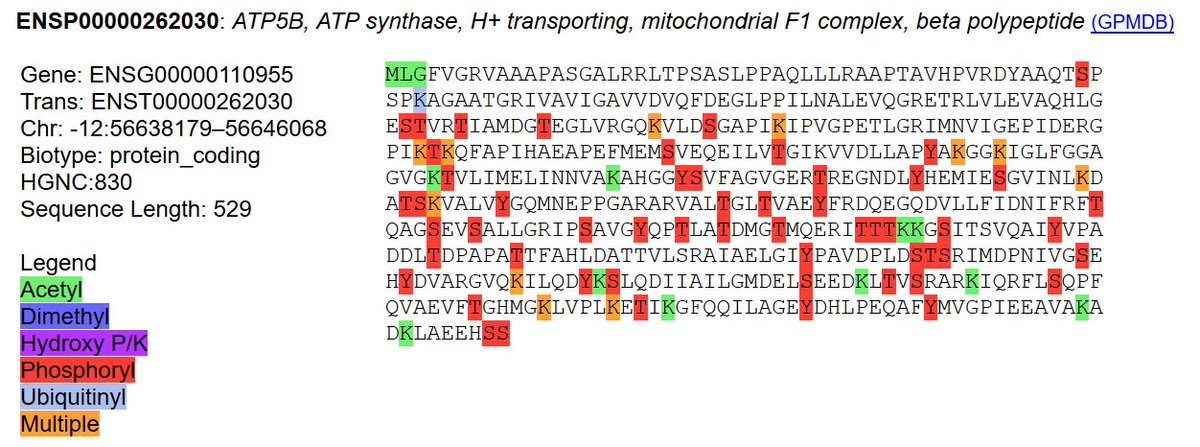
Sun Jan 13 16:25:45 +0000 2019There are no functions (even speculative ones) associated with the PTMs on ATP5F1A, even though these modifications are present in pretty much every study of acetylation, phosphorylation and/or ubiquitination.
Sun Jan 13 16:23:09 +0000 2019ATP5F1A:p, ATP synthase F1 subunit alpha (H. sapiens) 🔗 Midsized mitochondrial membrane associated subunit; extensively modified; very commonly observed; observed mature form 44-553 [69,324 x] 🔗
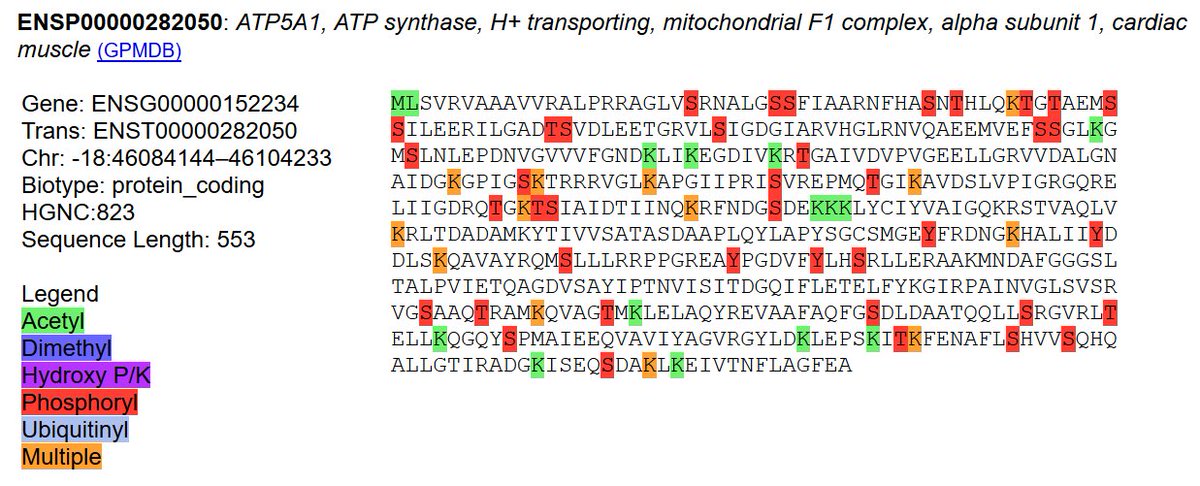
Sat Jan 12 19:05:06 +0000 2019CYCS:p, cytochrome c, somatic (H. sapiens) 🔗 Small mitochondrial protein; unusual PTM pm.C15,C18+heme; very commonly observed; observed mature form 2-105 [29,692 x] 🔗
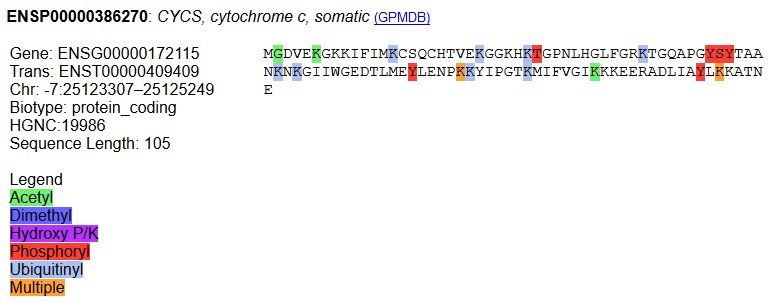
Fri Jan 11 14:37:24 +0000 2019FTMT:p, ferritin mitochondrial (H. sapiens) 🔗 Small mitochondrial subunit; no observed PTMs; rare except in testis/spermtozoon; observed mature form 52,53,54-242; shares no observable tryptic peptides with FTL or FTH1 [160 x]
Thu Jan 10 13:40:25 +0000 2019DHODH:p, dihydroorotate dehydrogenase (quinone) (H. sapiens) 🔗 Small mitochondrial inner membrane protein; pm.Y194+phosphoryl and p.A341V may be present; observed mature form 36-395 [7,628 x] 🔗

Wed Jan 09 18:45:35 +0000 2019Truly one of my favorite papers, "Unskilled and unaware of it: how difficulties in recognizing one's own incompetence lead to inflated self-assessments" 🔗
Wed Jan 09 16:35:18 +0000 2019@bkives When I was living in Chicago, they had a similar program. Seemed to increase the frequency of parts theft (wheels, handlebars, seats).
Wed Jan 09 16:23:34 +0000 2019Are there ontology terms for MHC Class I or Class II peptides?
Wed Jan 09 13:56:54 +0000 2019FECH:p, ferrochelatase (H. sapiens) 🔗 Midsized mitochondrial enzyme; no observed PTMs; observed mature form 37,38-423; ENSP00000262093:p.R96Q is the most commonly observed SAV [15,021 x]
Tue Jan 08 14:19:32 +0000 2019CRAT:p, carnitine O-acetyltransferase (H. sapiens) 🔗 Midsized mitochondrial enzyme; one low occupancy acetylation site; observed mature form 28,29-626 [9,589 x] 🔗
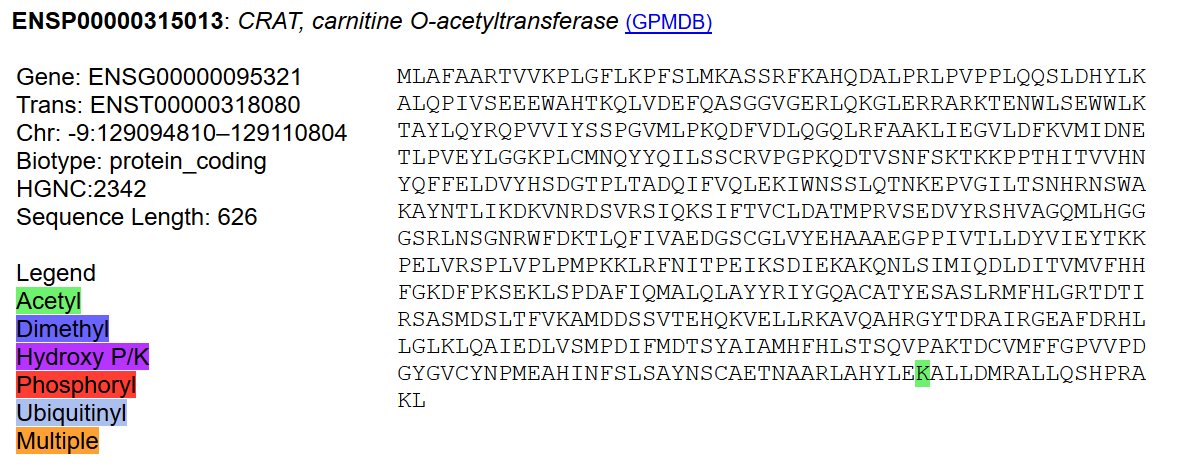
Mon Jan 07 20:14:13 +0000 2019@UCDProteomics It will take at least a generation to go away, probably two or three.
Mon Jan 07 13:46:06 +0000 2019FDXR:p, ferredoxin reductase (H. sapiens) 🔗 Midsized mitochondrial inner membrane associated enzyme; a small number of low occupancy PTM sites; observed mature form 33-491 [13,120 x] 🔗

Sun Jan 06 14:46:53 +0000 2019CPT2:p, carnitine palmitoyltransferase 2 (H. sapiens) 🔗 Midsized mitochondrial intrinsic membrane enzyme; involved in long-chain fatty acid metabolism; observed mature form 25,26-658 [17,664 x] 🔗
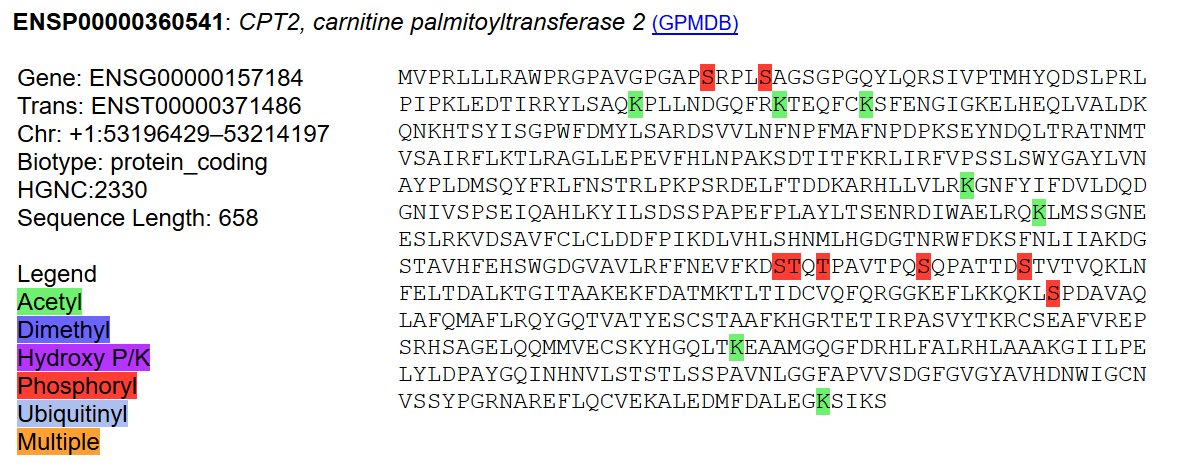
Sat Jan 05 15:03:37 +0000 2019In what is a real tour de force of misnaming, apolipoprotein O (APOO) and apolipoprotein O-like (APOOL) are not apolipoproteins at all: they are components of the superstructure that maintains mitochondrial cristea
Sat Jan 05 14:56:47 +0000 2019APOO:p, apolipoprotein O (H. sapiens) 🔗 Small mitochondrial complex subunit; several low occupancy phosphorylation acceptor sites; observed mature form 24-198 [9,351 x] 🔗

Fri Jan 04 13:58:15 +0000 2019APOOL:p, apolipoprotein O like (H. sapiens) 🔗 Small mitochondrial complex subunit; several low-occupancy phosphorylation acceptor sites; observed mature form 20-268 [9,818 x] 🔗

Thu Jan 03 20:01:56 +0000 2019@UCDProteomics Such language!
Thu Jan 03 15:20:22 +0000 2019@Sci_j_my No one has ever demonstrated what aspects of a protein's life cycle can be adequately described by a directed acyclic graph.
Thu Jan 03 14:18:44 +0000 2019IMMT:p, inner membrane mitochondrial protein (H. sapiens) 🔗 Midsized mitochondrial protein subunit; many PTM acceptor sites; observed mature form 34-747 [37,547 x] 🔗
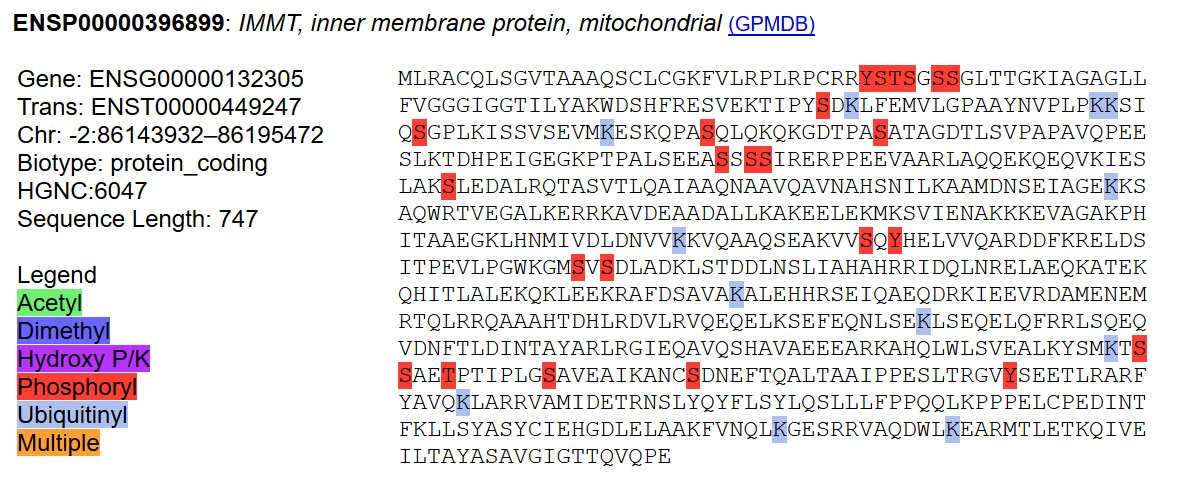
Wed Jan 02 19:50:19 +0000 20193 prominent unproven postulates in proteomics data analysis:
1. the FDR interpretation of the target-decoy simulation;
2. the idea that the PSM score alone is enough for an id; &
3. the idea that proteins can be described using ontologies.
Wed Jan 02 15:22:56 +0000 2019And the PTM diagram, featuring 12 K+acetyl, 3 Y+phosphoryl and 1 K+ubiquitinyl 🔗
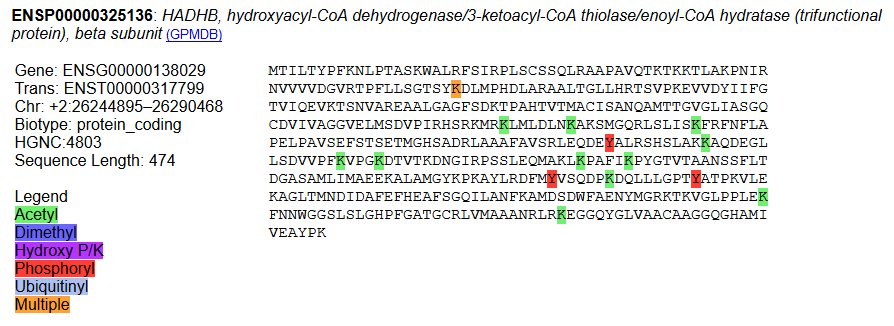
Wed Jan 02 14:25:57 +0000 2019Follows the general pattern of highly modified mitochondrial metabolic enzymes. I'm unaware of any explanation for this type of PTM patterning.
Wed Jan 02 14:24:18 +0000 2019HADHB:p, hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta (H. sapiens) 🔗 Small mitochondrial protein subunit; distinctive pattern of Tyr-phosphorylation; observed mature form 34-474 [38,312 x]
Tue Jan 01 14:38:41 +0000 2019HADHA:p, hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha (H. sapiens) 🔗 Midsized mitochondrial protein subunit; many PTM acceptor sites; observed mature form 37-763 [46,040 x] 🔗
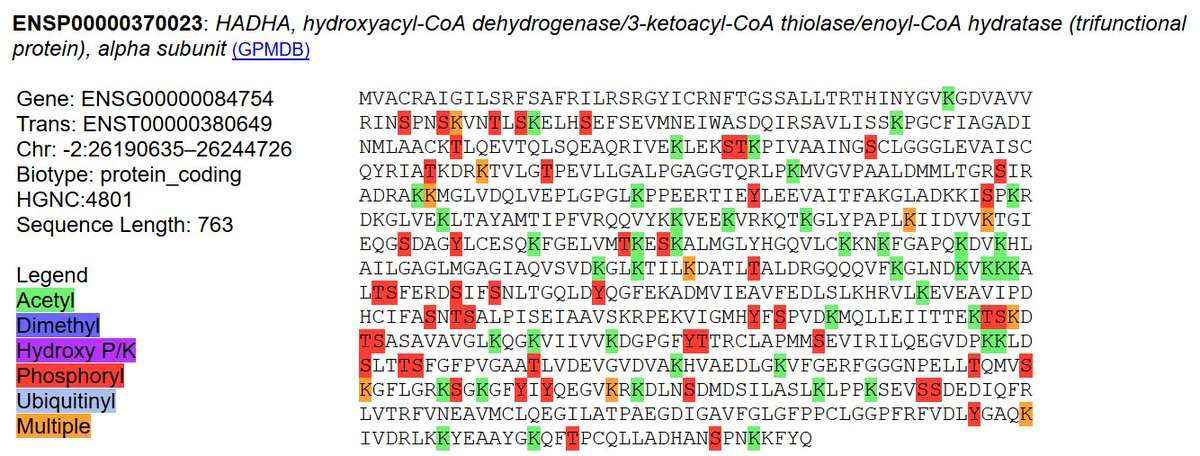
tweets = 2202